- 1Institute of Crop Science, National Agriculture and Food Research Organization, Tsukuba, Japan
- 2Institute of Agrobiological Sciences, National Agriculture and Food Research Organization, Tsukuba, Japan
Root system architecture affects the efficient uptake of water and nutrients in plants. The root growth angle, which is a critical component in determining root system architecture, is affected by root gravitropism; however, the mechanism of root gravitropism in rice remains largely unknown. In this study, we conducted a time-course transcriptome analysis of rice roots under conditions of simulated microgravity using a three-dimensional clinostat and following gravistimulation to detect candidate genes associated with the gravitropic response. We found that HEAT SHOCK PROTEIN (HSP) genes, which are involved in the regulation of auxin transport, were preferentially up-regulated during simulated microgravity conditions and rapidly down-regulated by gravistimulation. We also found that the transcription factor HEAT STRESS TRANSCRIPTION FACTOR A2s (HSFA2s) and HSFB2s, showed the similar expression patterns with the HSPs. A co-expression network analysis and an in silico motif search within the upstream regions of the co-expressed genes revealed possible transcriptional control of HSPs by HSFs. Because HSFA2s are transcriptional activators, whereas HSFB2s are transcriptional repressors, the results suggest that the gene regulatory networks governed by HSFs modulate the gravitropic response through transcriptional control of HSPs in rice roots.
1 Introduction
The root is an essential organ for absorbing water and nutrients from the soil in terrestrial plants. Root system architecture is vital for the efficient acquisition of water and nutrients that are unevenly distributed throughout the ground. The root growth angle, which affects the root system architecture, is controlled by root gravitropism. DEEPER ROOTING 1 (DRO1) and its homologs, positively regulate the root gravitropism and control the root growth angle in monocots (Uga et al., 2013; Ashraf et al., 2019; Kitomi et al., 2020; Feng et al., 2022; Nakano et al., 2022). A functional allele of DRO1 causes deep-rooting and enhances the ability of drought avoidance (Uga et al., 2013). A non-functional allele of quantitative trait locus for SOIL SURFACE ROOTING 1 (qSOR1) causes shallow-rooting and these roots are more effective for surface layer phosphorus absorption (Kitomi et al., 2020; Oo et al., 2021). Thus, genetic improvement of the root growth angle by manipulating root gravitropism contributes to the enhanced absorption of water and nutrients in the soil.
Gravistimulation by rotating plants 90° is a standard approach to dissecting the gravitropic response (Luschnig et al., 1998; Sedbrook et al., 1999; Boonsirichai et al., 2003; Guan et al., 2003). A three-dimensional (3D) clinostat is a device that minimizes the effects of gravity by rotating in all directions. The mounted sample is rotated three-dimensionally in two orthogonal axes. By continuously changing the law of gravity before the mounted sample is subjected to gravitational stimulation, the gravity vector is dispersed, and the effect of gravity is reduced. This condition is called “simulated microgravity” because it is imperfect. Thus, the 3D clinostat allows for simulated microgravity experiments in the laboratory on plant samples that are slow to respond to gravity (Herranz et al., 2013; Movie S1). Simulated microgravity treatment (SMT) using a 3D clinostat has also been used to examine the gravitropic response (Hoson et al., 1997; Herranz et al., 2013). Because all plants on the earth experience gravity, the former method captures the response to changes in the direction of gravity (dGS: directional gravistimulation). In contrast, the latter method can mimic space flight experiments and also produce the transition from simulated microgravity to forced gravity (fGS: forced gravistimulation).
The directional gravitropic mechanism of roots may be classified into four processes based on studies in the dicotyledonous model plant Arabidopsis: 1) graviperception in gravity-sensing columella cells (Su et al., 2017; Nakamura et al., 2019b), 2) gravity signaling following dGS mediated by LAZY1 (LZY)-like proteins (Nakamura et al., 2019a; Furutani and Morita, 2021), 3) redistribution of auxin resulting from differential polar auxin transport by PIN-FORMED (PIN) transporters (Adamowski and Friml, 2015; Han et al., 2021), and 4) differential growth in the elongation zone resulting from auxin signaling-dependent apoplast alkalinization (Barbez et al., 2017; Li et al., 2022). These processes are likely conserved in monocotyledonous plants, including rice (Uga et al., 2013; Ashraf et al., 2019; Zhang et al., 2019; Kitomi et al., 2020; Feng et al., 2022; Nakano et al., 2022). Mutations in Defective In Outer Cell Layer Specification 1 (DOCS1) in rice, a leucine-rich repeat receptor-like kinase, inhibits the formation of the gravity-sensing root cap, which decreases the response to gravity (Bettembourg et al., 2017). Actin binding protein RICE MORPHOLOGY DETERMINANT (RMD) suppresses the gravitropic response of the crown root by linking actin filaments and gravitropic perception organelle amyloplasts, and rmd mutants show faster gravitropism (Huang et al., 2018). DRO1 and qSOR1 are homologs of the Arabidopsis LZY gene family and are involved in gravity signaling in rice (Uga et al., 2013; Kitomi et al., 2020). OsPIN2/LARGE ROOT ANGLE1 (LRA1) plays an essential role in polar auxin transport in the rice root tip (Inahashi et al., 2018; Wang et al., 2018). The E3 ubiquitin ligase SOIL-SURFACE ROOTING 1 (SOR1) targets a noncanonical Aux/IAA protein OsIAA26 to the 26S proteasome pathway and is involved in auxin signaling in rice (Chen et al., 2018). Mutations in these genes affect the gravitropic response and alter the root growth angle; however, the gene regulatory network involved in the root gravitropic response is not well understood in rice.
In this study, we conducted a time-series RNA-seq of rice roots under simulated microgravity conditions (SMC) using a 3D clinostat and fGS to identify gravity state-responsive genes in rice roots. Heat shock transcription factors (HSFs) and HEAT SHOCK PROTEINs (HSPs) were up-regulated by SMT and rapidly down-regulated upon release from SMT. Genes in the co-expression module, including the HSFs and HSPs share HSF-binding cis-elements. These results suggest that HSFs govern the gene regulatory network during the transition from simulated microgravity to forced gravity in rice roots.
2 Materials and methods
2.1 Simulated microgravity treatment using a 3D clinostat
Sasanishiki (lowland japonica rice) with a functional allele of qSOR1 and a near-isogenic line with a non-functional allele of qSOR1 (qsor1-NIL; Kitomi et al., 2020) with a Sasanishiki background was used. Hulled seeds were washed three times with sterile water. The seeds were then soaked in 1.0% (v/v) PLANT PRESERVATIVE MIXTURE (PPM™; Plant Cell Technology, Inc., USA) and incubated at 30°C to germinate for 24 hours. The germinated seeds were sown in a 0.4% (w/v) agarose gel (Sigma-Aldrich, USA) in a microplate-type Petri dish (Stem, Japan) (Figure 1A; Seeding). Roots protruding from the medium were susceptible to desiccation, rather than gravitropic stimulation, so the Petri dish was tilted 60 degrees and placed in the dark at 28°C for 14 h to allow the roots to elongate into the agar medium (Figure 1A; Preculture). They were then rotated with a 3D clinostat PMS-VI (AES, Japan) in the dark for 6.0 h at 28°C (Figures 1A; S1; Movie S1; Clinorotation). SMC was mimicked by adjusting the rotational speed of the 3D clinostat (X-axis 11.0 RPM/Y-axis 13.0 RPM). However, the influence of other physical stimuli that occurred by this device, such as vibrations, cannot be excluded as a possibility in this clinostat treatment. After simulated microgravity treatment (SMT), the Petri dish was placed vertically and gravistimulation was applied at 90 degrees (Figure 1A; Gravistimulation). Eighteen seeds were sown on each plate, and 10-16 seminal root tips with 2-3 mm were collected and pooled for RNA-seq. Seminal roots that were too short were excluded from the sampling (Figure S2). The root tips were collected at 0.0, 0.5, 1.5, and 3.0 h after fGS. For the control plot without SMT, the Petri dish was tilted 60 degrees for 14 h, then placed vertically, and the root tips were sampled after 6.0 h in the dark (Figure 1B). Therefore, the control samples are the same age as the clinorotated sample at 0h after fGS.
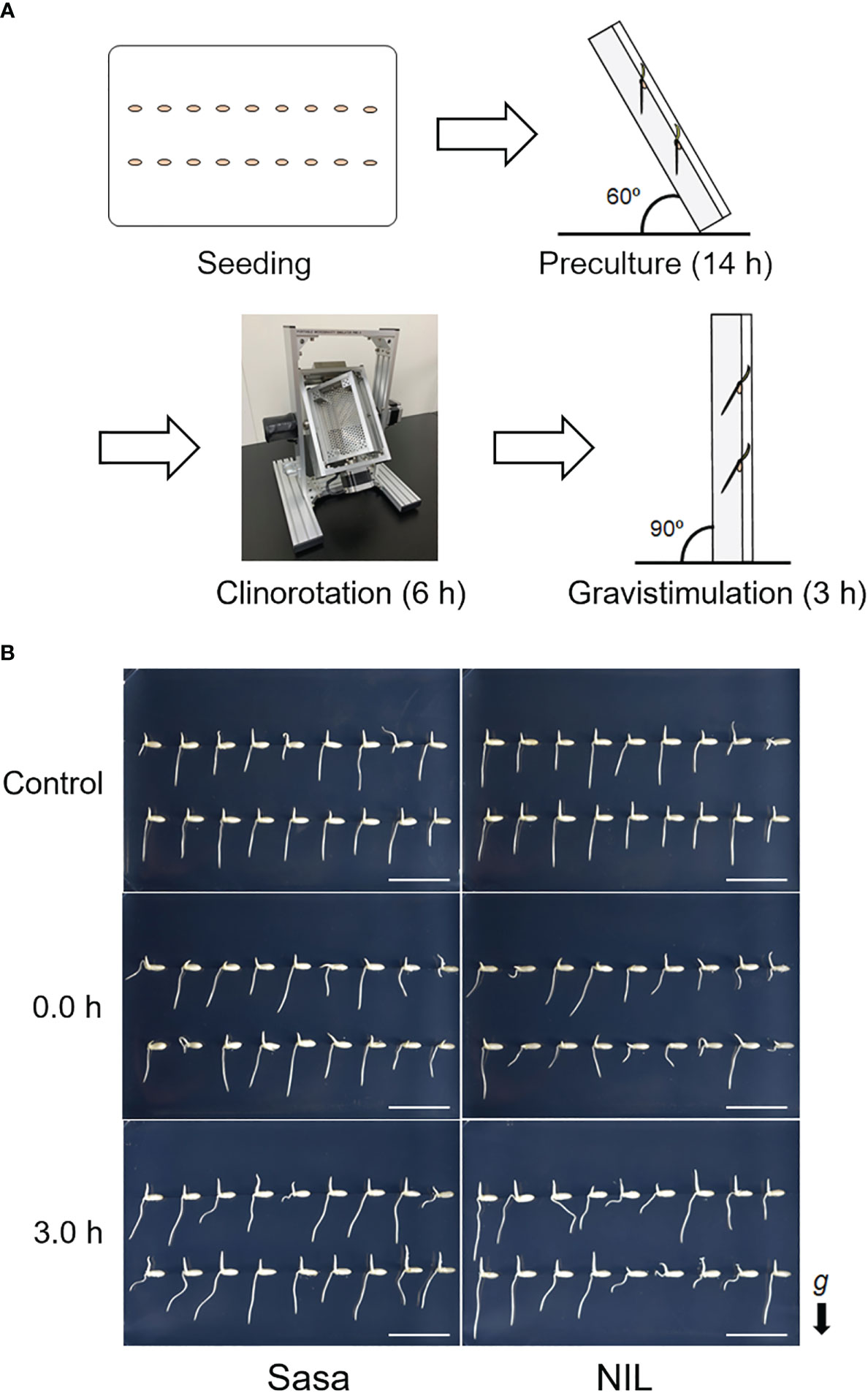
Figure 1 Simulated-microgravity treatment for rice roots. (A) Overview of simulated-microgravity treatment (SMT) and gravity treatment. Seeding: Germinated seeds were sown onto 0.4% (w/v) agarose gels in a microplate-type Petri dish. Preculture: Petri dishes were tilted 60 degrees and placed at 28°C in the dark for 14 h to allow the roots to grow down to the center of the medium. Clinorotation: The seedlings were rotated on a 3D clinostat for 6.0 h at 28°C in the dark. Gravistimulation: After SMC, gravistimulation was applied by placing the Petri dish vertically. (B) Representative photos after each treatment. 0.0 h and 3.0 h are different individuals. g: gravity vector, Sasa: Sasanishiki, NIL: qsor1-NIL. Bar = 2 cm.
2.2 RNA extraction
Root tips were immediately frozen in liquid nitrogen and ground to a fine powder with a ShakeMaster BMS-A20TP (BMS, Japan). RNA was extracted with the RNeasy Plant Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions.
2.3 RNA-seq analysis
RNA-seq libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (E7760, New England Biolabs, USA) according to the manufacturer’s instructions. Sequencing of the libraries was performed with 150 bp paired-end reads on the S4 flow cells of the Illumina NovaSeq6000 platform at Macrogen Japan. The reads were mapped to the IRGSP-1.0 genome assembly with MSU7 gene model annotation using the STAR aligner program (ver. 2.7.3a) with options “–outFilterMultimapNmax 1 –quantMode GeneCounts” (Dobin et al., 2013). Differentially expressed genes (DEGs; |log2[fold-change]| >1, the false discovery rate <0.05) were called using the glmLRT of R package edgeR (ver. 3.26.8; Robinson et al., 2010) and transcripts per kilobase million (TPM) values were computed. Genes with TPM values >2 were considered expressed. Pearson’s correlation coefficients between samples were calculated using expression levels (log2[FPKM + 1]) of all expressed genes. Relative expression in Figures 2A; S3A is shown as log2[FC to average FPKM of each gene in all samples]. Principal component analysis (PCA) was performed using TPM values of all expressed genes. All heatmaps were plotted using ComplexHeatmap (ver. 2.0.0; Gu et al., 2016). Gene ontology (GO) enrichment analysis was performed using clusterProfiler (ver 3.12.0; Yu et al., 2012) and visualized using corrplot (ver 0.84; Wei and Simko, 2017). An initial quality check using PCA and heatmap detected the wired behavior in one replicate of Sasanishiki, Sasa3 (Figures S3A–C). Therefore, Sasa3 was excluded from subsequent analyses, although we could not determine this reason.
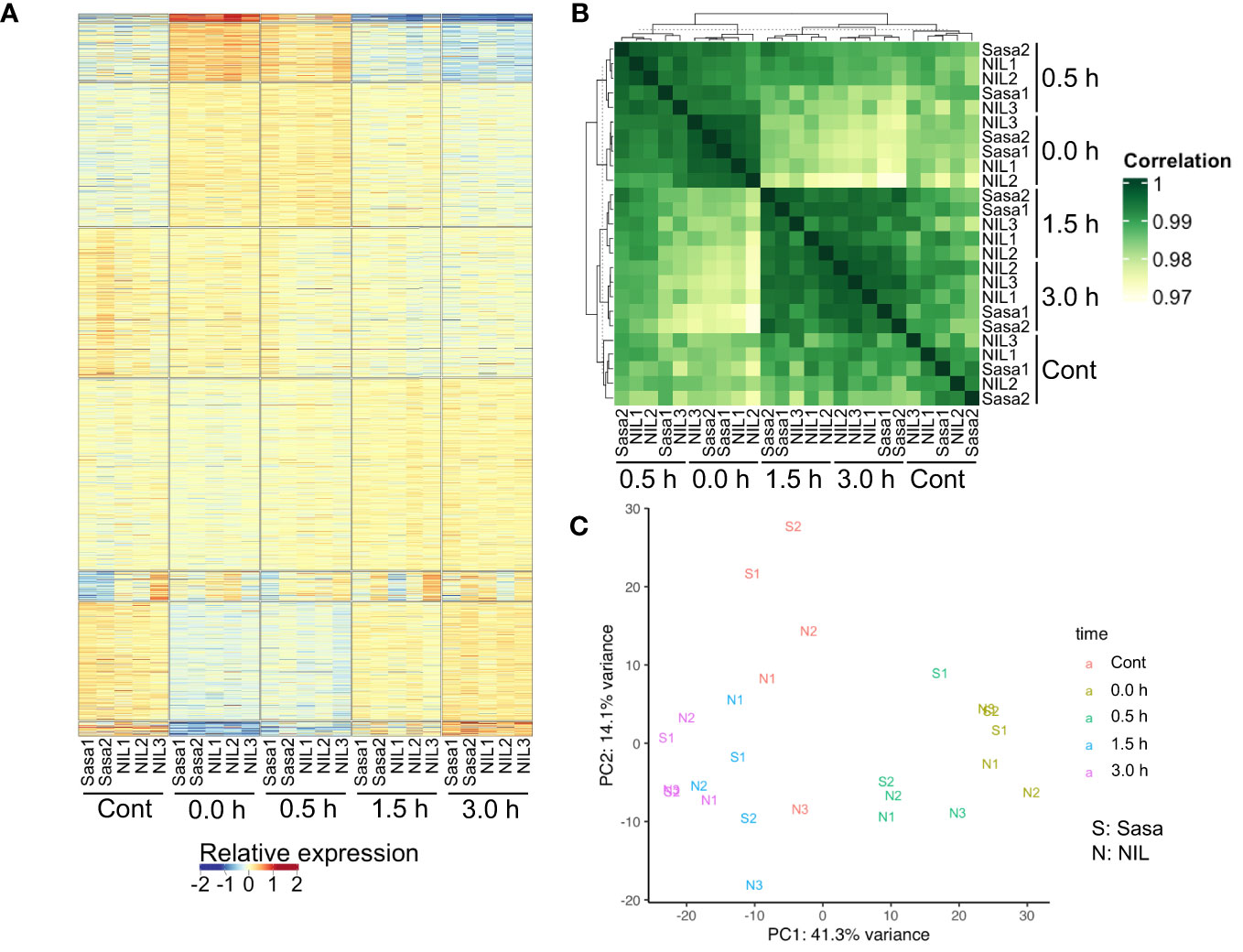
Figure 2 Summary of RNA-seq results. (A) Heatmap of gene expression levels of two and three replicates for Sasanishiki (Sasa) and qsor1-NIL (NIL), respectively. Samples were ordered by k-means clustering. (B) Heatmap of the correlation matrix of Sasanishiki and qsor1-NIL. (C) PCA plot based on TPM values of all expressed genes. We excluded one replicate (Sasa3) from the analysis because this replicate showed abnormal behavior (Figure S1).
2.4 Alignment of amino acid sequences
Clustal Omega (Sievers et al., 2011) was used to calculate the homology between HSF proteins.
2.5 Co-expression network analysis
Pairwise Pearson’s correlation coefficients between the genes were calculated and gene pairs with correlation coefficients >0.9 were extracted for further analysis. Co-expression modules were analyzed using the “fastgreedy.community” function of igraph (ver. 1.3.4; Csardi and Nepusz, 2006). Modules with less than five genes were discarded. Cis-motifs enriched in the upstream regions of genes within each co-expression module were searched using MEME (ver. 5.4.1; Bailey et al., 2015) using the option “-mod zoops -nmotifs 3 -minw 6 -maxw 13 -revcomp -markov_order 0” and the detected motifs were compared with the Arabidopsis DAP-seq motif dataset (Bailey et al., 2015) using TOMTOM (ver. 5.4.1; Kitomi et al., 2020) and the options “-no-ssc -verbosity 1 -min-overlap 5 -dist pearson -thresh 1e-4.”
3 Results
3.1 Root gravitropic response in Sasanishiki and qsor1-NIL
An overview of the SMT using a 3D clinostat and fGS treatment is presented in Figure 1A. Images of the roots at control, 0.0 h, and 3.0 h after fGS are shown in Figure 1B. The qsor1-NIL showed weaker gravitropism in the seminal roots compared with Sasanishiki (Kitomi et al., 2020). To identify genes downstream of qSOR1, we compared Sasanishiki and qsor1-NIL. Root elongation tended to be suppressed under SMC (Figure S4A). In the control without SMT, the root tips of both Sasanishiki and qsor1-NIL elongated toward the direction of gravity (Figures 1B; S4B). In contrast, the roots of Sasanishiki and qsor1-NIL at 0.0 h after fGS showed various directions of elongation (Figures 1B; S4). After 3.0 h of fGS, the root tips of both Sasanishiki and qsor1-NIL elongated in the direction of gravity (Figures 1B; S4B).
3.2 Gene expression patterns under SMC and following fGS in Sasanishiki and qsor1-NIL
To identify genes associated with the gravitropic response in rice roots, we performed a time-series RNA-seq analysis along with fGS after SMC. We considered 0.0 h after fGS as under SMC. A heatmap of 16,648 expressed genes showed that transcriptional changes mainly occurred at 0.0 h and 0.5 h after fGS in qsor1-NIL and Sasanishiki (Figure 2A). The correlation matrix of expression levels revealed that replicates for each condition exhibited similar patterns (r > 0.99; Mann-Whitney U test P = 3.8e-9) and the time-series gene expression profile for qsor1-NIL broadly resembled that of Sasanishiki (r > 0.99; Mann-Whitney U test P = 2.0e-10; Figure 2B). The controls without SMT were clustered with later-stage samples (1.5 h and 3.0 h) rather than early-stage samples (0.0 h and 0.5 h). PCA based on TPM values of the top 5% genes with the highest variability revealed that the difference in time points, rather than genotype, had more of an effect on gene expression (Figure 2C). This indicates that SMT-induced transcriptional changes were similar in both genotypes.
The expression of qSOR1 tended to be slightly lower in qsor1-NIL compared with that in Sasanishiki. Although the expression levels of qSOR1 were constant throughout the time-series in qsor1-NIL, qSOR1 expression in Sasanishiki was down-regulated at 1.5 h after fGS (Figure S5). In contrast, no difference in DRO1 expression between Sasanishiki and qsor1-NIL was observed (Figure S5). This suggests that the expression of qSOR1 and DRO1 in the root tips under SMC and subsequent fGS are similar between functional and non-functional qSOR1 genetic backgrounds. We identified only a few DEGs between Sasanishiki and qsor1-NIL after fGS (Figure S1D; Tables S1, S2). The maximum number of DEGs that were up-regulated in Sasanishiki compared with qsor1-NIL was seven at 0.0 h, whereas the maximum number of DEGs that were down-regulated was 11 at 0.5 h and 1.5 h. Among these, no rice genes known to be involved in gravitropism was found. We also examined auxin-related genes associated with gravitropism; however, the expression patterns of these genes were almost identical between Sasanishiki and qsor1-NI (Figure S6). These results indicate that qSOR1 is not involved in the regulation of gene expression patterns in the root tip during SMC and subsequent fGS.
3.3 Differentially expressed genes during fGS compared to under SMC
We identified 152 and 117 DEGs in which expression was up-regulated during fGS in Sasanishiki and qsor1-NIL, respectively (Table S3). In addition, we identified 286 and 333 DEGs in which expression was down-regulated during fGS in Sasanishiki and qsor1-NIL, respectively (Table S4). GO enrichment analysis revealed that the down-regulated genes that were common to both genotypes were associated with response to heat, response to temperature stimulus, response to abiotic stimulus, cellular response to heat, response to hydrogen peroxide, protein folding, chaperone-mediated protein folding, and response to unfolded protein (Figure 3). We found that the three GOs (response to heat, response to temperature stimulus, and response to abiotic stimulus) were detected in qsor1-NIL, but not in Sasanishiki, at 0.5 h after fGS. Because the expression levels of the six genes in these GOs were almost the same between the two genotypes, we concluded that this discrimination was dependent upon less statistical power in Sasanishiki at 0.5 h after fGS because of the number of replicates (Figure S7). Based on these results, we focused our analysis on the DEGs that were common to both genotypes.
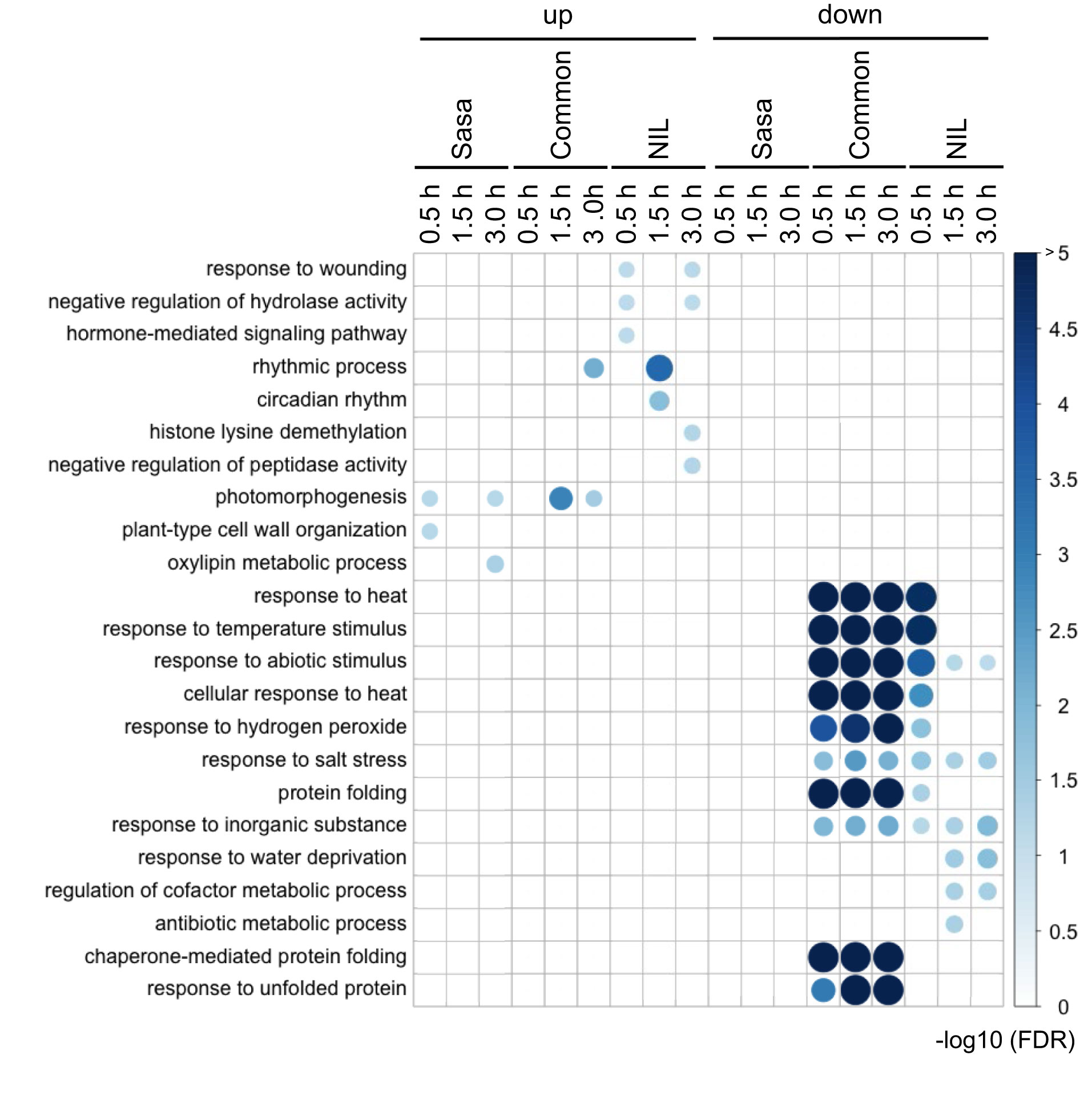
Figure 3 Gene ontologies enriched in each DEG cluster. The numbers on the right side of the figure indicate an adjusted p-value of -log10 at false discovery rate (FDR). The size and the intensity of the color for the circle represent the degree of the FDR value. Sasa: Sasanishiki, NIL: qsor1-NIL, common: common DEGs between Sasanishiki and qsor1-NIL.
Common DEGs between Sasanishiki and qsor1-NIL in representative GOs were investigated (Figure 4). One of the most prominent GO-enriched DEGs was protein folding. In Arabidopsis, HEAT SHOCK PROTEINs (HSPs) are involved in gravitropism through the regulation of auxin transport. HSP genes were enriched in the down-regulated DEGs (Figure 4B), which included five HSP20s, five HSP40s, eight HSP70s, and six HSP90s. The expression of most HSPs was higher at 0.0 h after fGS, but lower at 1.5 h and 3.0 h after fGS compared with those of the control without SMT (Figure 4B). These results indicate that the expression of HSP genes was induced by SMT, but repressed by prolonged fGS, likely reflecting an adaptive gravitropic response.
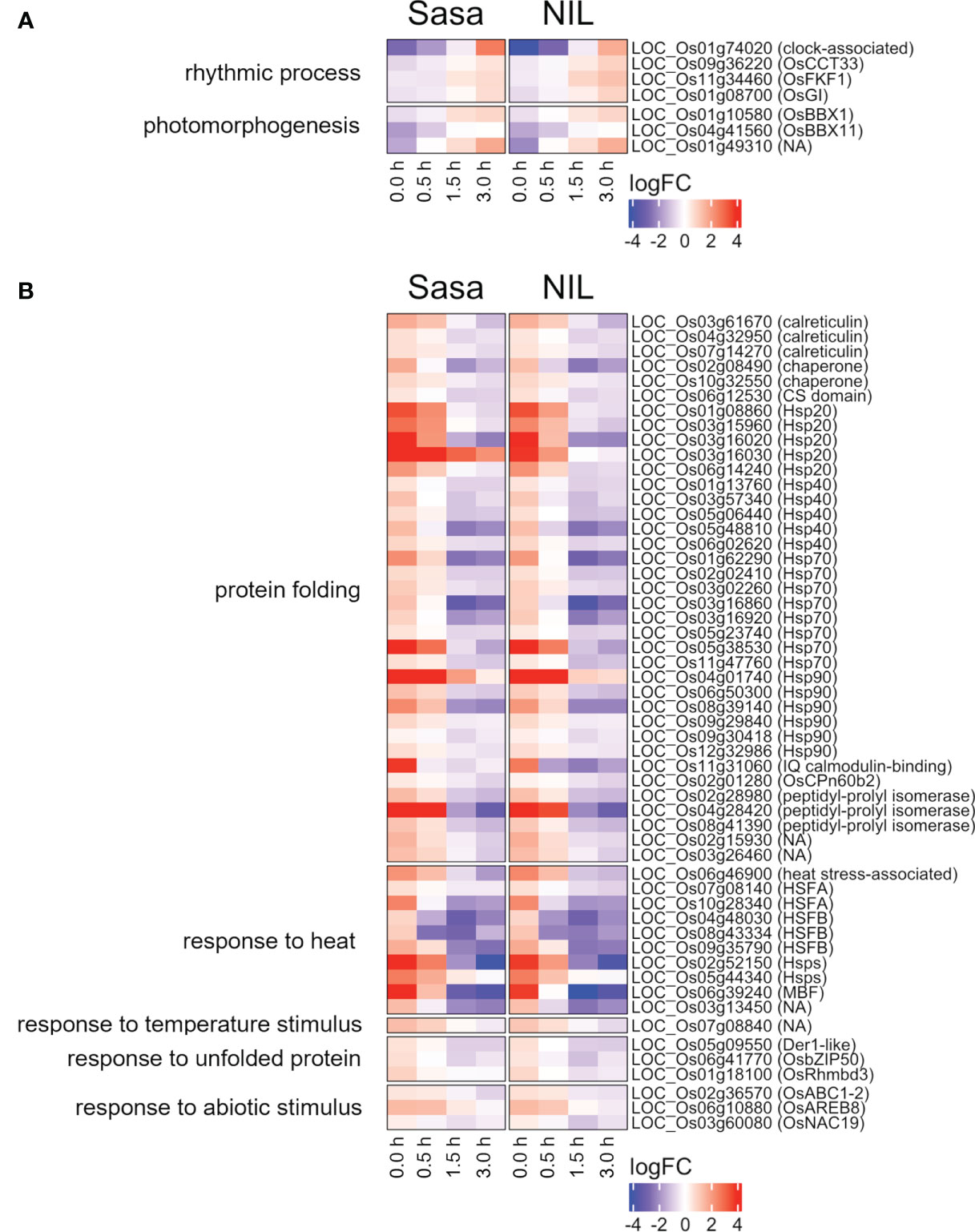
Figure 4 Common DEGs between Sasanishiki and qsor1-NIL in representative GOs. (A) Genes up-regulated during fGS. (B) Genes down-regulated during fGS. Gene expression is shown as logFC relative to each control. Sasa: Sasanishiki, NIL: qsor1-NIL.
3.4 Differentially expressed transcription factor genes during SMT and fGS
Transcription factors (TFs) regulate the expression of downstream genes and often play an important role in morphogenesis. Nine TFs were commonly up-regulated and 19 TFs were down-regulated during fGS in Sasanishiki and qsor1-NIL (Figure 5). Of the 19 TFs, seven were HSFs, including HEAT SHOCK TRANSCRIPTION FACTOR A2D (HSFA2D), which acts upstream of LAZY1 and is involved in shoot gravitropism in rice (Zhang et al., 2018) (Figure 5B). The expression of these HSFs was higher at 0.0 h after fGS compared with those in the control without SMT, indicating that the expression of these HSFs was induced by SMT (Figure 6). These HSFs belong to the HSFA2 and HSFB2 sub-groups. HSFs in other sub-groups were not induced by SMT, suggesting that HSFs in sub-groups HSFA2 and HSFB2 are specifically involved in the forced gravitropic response in rice (Figure S8; Table S5).
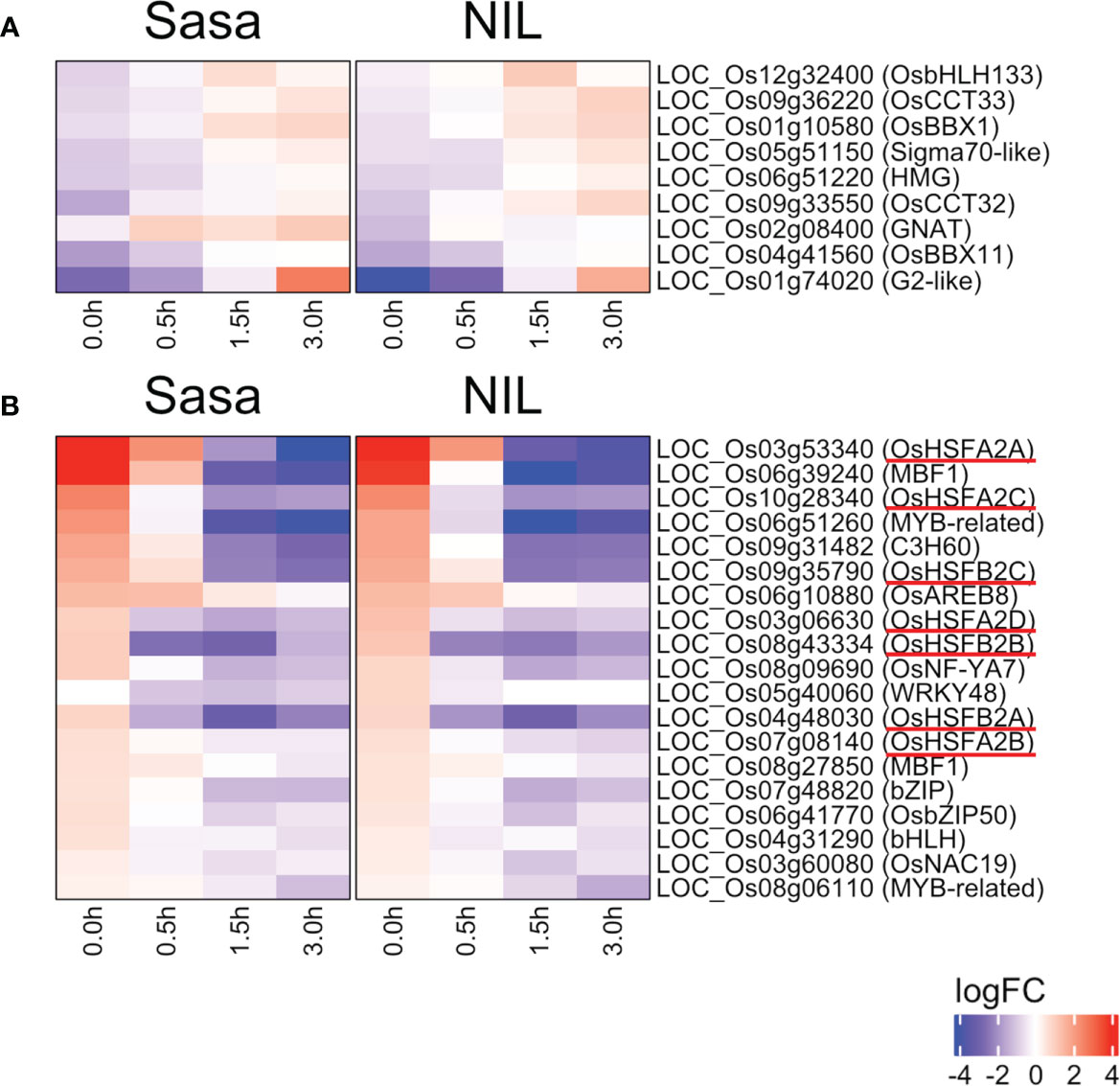
Figure 5 Changes in transcription factor expression during fGS. (A) Transcription factors in the DEGs whose expression was up-regulated during fGS. (B) Transcription factors in the DEGs whose expression was down-regulated during fGS. Gene expression is shown as logFC relative to each control. Sasa: Sasanishiki, NIL: qsor1-NIL. HSFs were underlined in red.
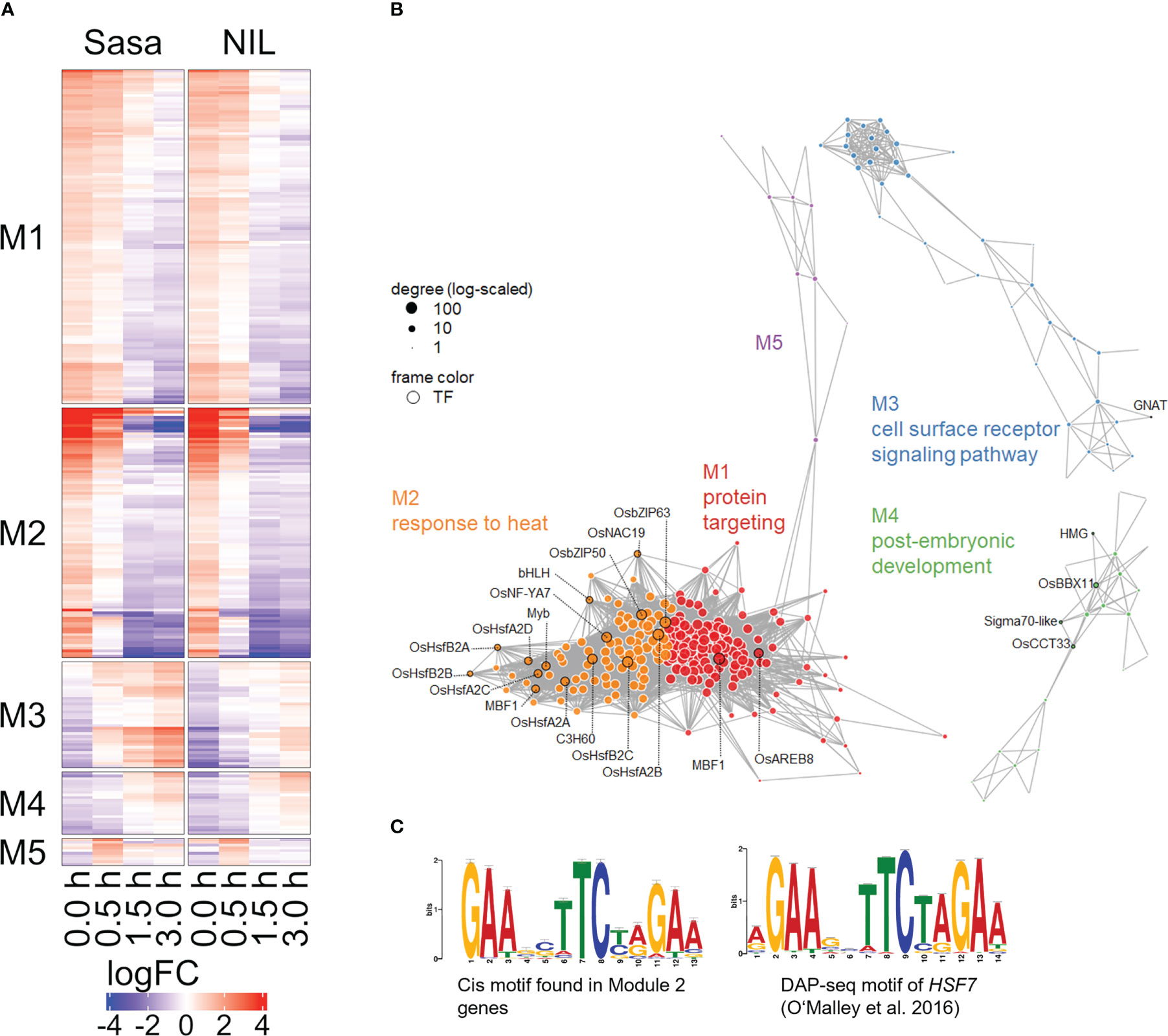
Figure 6 Co-expression network model of simulated-microgravity induced DEGs. (A) Heatmap of relative expression levels of DEGs commonly detected in Sasanishiki and qsor1-NIL. Genes were clustered by fast greedy, based on logFC relative to each control. (B) Five major modules (M1 to M5) presented in different colors together with the representative GO term. Transcription factors are framed in black and labeled with their gene symbols. (C) A representative motif found in the 200-bp upstream sequences of M2 genes (left; MEME program) and the corresponding DAP-seq result (right; TOMTOM program; O’Malley et al., 2016).
3.5 Similarity of detected DEGs between 3D clinostat treatment and space flight
The 3D clinostat is designed to minimize the effect of gravity by randomly and continuously changing the direction of gravity; however, the extent by which SMT reproduces real microgravity in space is not guaranteed. Transcriptome analysis of plants under microgravity in space have been conducted (Zupanska et al., 2013; Vandenbrink et al., 2019; Kruse et al., 2020; Barker et al., 2023). We compared genes detected as DEGs between our SMT experiment in rice and the previous space experiment in Arabidopsis (Zupanska et al., 2013). Many HSFs and HSPs were commonly detected DEGs in the two studies (Table S6) as well as other studies (Correll et al., 2013; Choi et al., 2019; Barker et al., 2023; Paul et al., 2005; Paul et al., 2012; Kwon et al., 2015; Li et al., 2017). These results support that our SMT treatment properly mimics real microgravity in space.
3.6 Co-expression analysis of genes detected as DEGs after fGS
To infer the gene regulatory networks under SMCs and following fGS, we performed a co-expression analysis using DEGs commonly expressed in Sasanishiki and qsor1-NIL. The DEGs were divided into five co-expression modules (Figures 6A, B; Table S7). The seven HSFs that were altered under the SMC were included in module 2 (M2), which was enriched with genes associated with “response to heat.” HSP40 genes (LOC_Os01g13760, LOC_Os03g57340, LOC_Os05g48810, LOC_Os06g02620), HSP70 genes (LOC_Os01g62290, LOC_Os03g16860, LOC_Os03g16920, LOC_Os05g38530), and HSP90 genes (LOC_Os04g01740, LOC_Os08g39140) were also included in M2, suggesting a possible interaction between HSFs and HSPs. We identified the enriched motif within a 200 bp upstream region of the genes belonging to M2 (55 out of 92 genes, 59.8%), which closely resembles the AtHSFA7 binding motif in the Arabidopsis cistrome dataset (q-value = 4.29e-9; Figure 6C) (O’Malley et al., 2016), whereas TCP motifs (M1, q = 8.21e-6) and MYB-related motifs (M4, q = 2.45e-4) were identified in the other modules (Figure S9, q < 1e-4). Because Arabidopsis HSF-binding motifs are nearly identical to one another irrespective of class or sub-group, we anticipate that rice HSFs also recognize similar sequences (Figure S10). These results suggest that HSFA2s and HSFB2 directly regulate the expression of genes in the M2 module and shape the M2 module.
4 Discussion
Gravitropism is a key determinant of root system architecture. We performed a time-series RNA-seq study under SMCs and following fGS, which revealed dynamic transcriptional changes and identified genes involved in the gravitropic response in rice roots. A co-expression network analysis revealed that the HSF-HSP pathway may contribute to the gravitropic response in rice roots. HSFA2D is a positive regulator of gravitropism and acts upstream of asymmetric auxin distribution in rice shoots (Zhang et al., 2018). The dGS induces the expression of HSFA2D and HSFA2D activates the rice LZY1 homolog LAZY1 (LA1), which is involved in gravity signaling and asymmetric distribution of auxin in shoots (Zhang et al., 2018). HSP40s and HSP90s are involved in auxin perception and transport (Harrison and Masson, 2008; Wang et al., 2016). Therefore, the HSF-HSP pathway may be responsible for the asymmetric auxin distribution followed by differential growth during the gravitropic response in roots as in shoots.
Sasanishiki was more responsive to fGS compared with qsor1-NIL with respect to root growth; however, we did not observe clear differences in the expression of genes associated with root growth during SMT and following fGS between Sasanishiki and qsor1-NIL. This indicates that qSOR1 acts independently of transcriptional control. AtLZYs, Arabidopsis homologs of qSOR1 and DRO1, are expressed in columella cells and AtLZYs recruit RLD proteins to the plasma membrane in the direction of gravity, which results in polar localization of the auxin efflux carrier PIN3. Thus, qSOR1 may be involved in the regulation of auxin transport or gravity signaling, in a similar manner. Interestingly, AtLZYs also interact with HSP70s and its rice homologs were up-regulated by SMT and repressed following fGS (Furutani et al., 2020). Whether HSP70 is involved in auxin transport, however, is unknown, although HSP40s and HSP90s are involved in auxin transport. HSP40s regulate the localization of PIN3 (Harrison and Masson, 2008). HSP90s stabilize PIN1 and auxin receptor TIR1 with the co-chaperones, HOPs and SGT1b (Figure S11; Samakovli et al., 2021; Muñoz et al., 2022). All differentially expressed HSP40s, HSP90s, HOPs, and SGT1b were assigned to the closely related modules M1 and M2 (Figure 6B), which indicates that these genes are tightly co-expressed under SMC and following fGS. Because HSP70s interact with co-chaperone HSP40s and cooperate with protein folding with HSP90s, HSP70s may also be involved in auxin transport. These suggest a possible crosstalk between LZYs, including qSOR1 and the HSF-HSP pathway in regulating the localization and stabilization of PINs and auxin transport. qSOR1 is negatively regulated by auxin signaling and qSOR1 was transiently repressed by fGS (Figure S5; Kitomi et al., 2020). This suggests the existence of a negative feedback loop between qSOR1-mediated auxin transport and auxin signaling-dependent repression of qSOR1.
Rice LA1, a homolog of qSOR1 and DRO1, is required for gravitropism in shoots, but not in roots. HSFA2D acts upstream of LA1 and regulates gravitropism in shoots. Seven HSFs, including HSFA2D, were up-regulated by SMT and down-regulated by fGS (Figure 5B). Five of these HSFs are responsive to gravity changes in shoots (Zhang et al., 2018, Figure S12), suggesting that transcriptional regulation during the gravitropic response of rice shoots and roots is similar. The expression of LA1 is reduced in hsfa2d, suggesting that HSFA2D is a positive regulator of LA1. Whether qSOR1 is positively regulated by HSFs, including HSFA2D, in roots is unknown; however, the expression of qSOR1 was not increased by SMT, although the expression of HSFs was increased. This is the same situation that was observed in shoots, in which LA1 was not induced by dGS when HSFs were up-regulated (Zhang et al., 2018; Figure S13). It is possible that the transcriptional regulation of LZYs by HSFs is indirect or requires co-factors. HSF TFs include both transcriptional activators and repressors. Members of the HSFA subfamily are activators, whereas members of the HSFB subfamily are either activators or repressors. Of the seven HSFs induced by SMT and repressed by fGS, three OsHSFB2s are predicted to function as repressors based on the presence of a repression domain (Lavania et al., 2018). In Arabidopsis, HsfB1/B2b represses the expression of HSPs, HsfA2, and HsfB1/B2b themselves under normal conditions (Ikeda et al., 2011). Under heat stress conditions, HsfA1s are rapidly activated by translocation from the cytoplasm to the nucleus and induce the expression of HsfA2 and HsfB1/B2b. Activated HsfA2 induces HSPs to acquire thermotolerance; however, HsfB1/B2b represses the expression of HsfA2 and HsfB1/B2b, attenuating the heat stress response to normal conditions (Ikeda et al., 2011). We hypothesize that a similar scenario may control the adaptive gravitropic response in rice roots. OsHSFs other than OsHSFA2s and OsHSFB2s, whose expression is constant, may be activated by translocation during perturbed gravity and induce the expression of OsHSFA2s and OsHSFB2s. Activated OsHSFA2s induce the expression of HSPs and OsHSFBs for auxin redistribution and its subsequent attenuation. At this point, OsHSFBs may be inactivated by unknown mechanisms. During fGS, OsHSFBs may be promptly activated and repress the expression of OsHSFA2s and HSPs, resulting in the attenuation of the adaptive gravitropic response. However, further studies are needed to clarify the mechanism of gravitropism in rice root and utilize the knowledges to improve the root traits of monocotyledonous plants.
Data availability statement
We have released our RNA-seq data of accession number DRA013338 on the DDBJ repository (https://ddbj.nig.ac.jp/search).
Author contributions
YU conceived and supervised the research. YU and YK designed the research, NK and YK performed the experiments. NK, RN and TK analyzed the transcriptome data. NK, RN, TK, and YU wrote the manuscript with contributions from all authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Japan Science and Technology Agency CREST [grant number JPMJCR17O1] and the Japan Society for the Promotion of Science KAKENHI [grant numbers JP19H02936].
Acknowledgments
We thank the Advanced Analysis Center of the National Agriculture and Food Research Organization (NARO) for the use of facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1193042/full#supplementary-material
References
Adamowski, M., Friml, J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell. 27, 20–32. doi: 10.1105/tpc.114.134874
Ashraf, A., Rehman, O. U., Muzammil, S., Léon, J., Naz, A. A., Rasool, F., et al. (2019). Evolution of deeper rooting 1-like homoeologs in wheat entails the c-terminus mutations as well as gain and loss of auxin response elements. PloS One 14, e0214145. doi: 10.1371/journal.pone.0214145
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43(W1), W39–W49. doi: 10.1093/nar/gkv416
Barbez, E., Dünser, K., Gaidora, A., Lendl, T., Busch, W. (2017). Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, E4884–E4893. doi: 10.1073/pnas.1613499114
Barker, R., Kruse, C. P. S., Johnson, C., Saravia-Butler, A., Fogle, H., Chang, H. S., et al. (2023). Meta-analysis of the space flight and microgravity response of the arabidopsis plant transcriptome. NPJ Microgravity 9, 21. doi: 10.1038/s41526-023-00247-6
Bettembourg, M., Dal-Soglio, M., Bureau, C., Vernet, A., Dardoux, A., Portefaix, M., et al. (2017). Root cone angle is enlarged in docs1 LRR-RLK mutants in rice. Rice (NY) 10, 50. doi: 10.1186/s12284-017-0190-1
Boonsirichai, K., Sedbrook, J. C., Chen, R., Gilroy, S., Masson, P. H. (2003). ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell. 15, 2612–2625. doi: 10.1105/tpc.015560
Chen, H., Ma, B., Zhou, Y., He, S. J., Tang, S. Y., Lu, X., et al. (2018). E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc. Natl. Acad. Sci. U.S.A. 115, 4513–4518. doi: 10.1073/pnas.1719387115
Choi, W. G., Barker, R. J., Kim, S. H., Swanson, S. J., Gilroy, S. (2019). Variation in the transcriptome of different ecotypes of arabidopsis thaliana reveals signatures of oxidative stress in plant responses to spaceflight. Am. J. Bot. 106, 123–136. doi: 10.1002/ajb2.1223
Correll, M. J., Pyle, T. P., Millar, K. D. L., Sun, Y., Yao, J., Edelmann, R. E., et al. (2013). Transcriptome analyses of arabidopsis thaliana seedlings grown in space: implications for gravity-responsive genes. Planta 238, 519–533. doi: 10.1007/s00425-013-1909-x
Csardi, G., Nepusz, T. (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9. Available at: https://igraph.org.
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Feng, X., Jia, L., Cai, Y., Guan, H., Zheng, D., Zhang, W., et al. (2022). ABA-inducible DEEPER ROOTING 1 improves adaptation of maize to water deficiency. Plant Biotechnol. J. 20, 2077–2088. doi: 10.1111/pbi.13889
Furutani, M., Hirano, Y., Nishimura, T., Nakamura, M., Taniguchi, M., Suzuki, K., et al. (2020). Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat. Commun. 11, 76. doi: 10.1038/s41467-019-13729-7
Furutani, M., Morita, M. T. (2021). LAZY1-LIKE-mediated gravity signaling pathway in root gravitropic set-point angle control. Plant Physiol. 187, 1087–1095. doi: 10.1093/plphys/kiab219
Gu, Z., Eils, R., Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Guan, C., Rosen, E. S., Boonsirichai, K., Poff, K. L., Masson, P. H. (2003). The ARG1-LIKE2 gene of arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol. 133, 100–112. doi: 10.1104/pp.103.023358
Han, H., Adamowski, M., Qi, L., Alotaibi, S. S., Friml, J. (2021). PIN-mediated polar auxin transport regulations in plant tropic responses. New Phytol. 232, 510–522. doi: 10.1111/nph.17617
Harrison, B. R., Masson, P. H. (2008). ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 53, 380–392. doi: 10.1111/j.1365-313X.2007.03351.x
Herranz, R., Anken, R., Boonstra, J., Braun, M., Christianen, P. C., de Geest, M., et al. (2013). Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiol. 13, 1–17. doi: 10.1089/ast.2012.0876
Hoson, T., Kamisaka, S., Masuda, Y., Yamashita, M., Buchen, B. (1997). Evaluation of the three-dimensional clinostat as a simulator of weightlessness. Planta 203, S187–S197. doi: 10.1007/PL00008108
Huang, G., Liang, W., Sturrock, C. J., Pandey, B. K., Giri, J., Mairhofer, S., et al. (2018). Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 9, 2346. doi: 10.1038/s41467-018-04710-x
Ikeda, M., Mitsuda, N., Ohme-Takagi, M. (2011). Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254. doi: 10.1104/pp.111.179036
Inahashi, H., Shelley, I. J., Yamauchi, T., Nishiuchi, S., Takahashi-Nosaka, M., Matsunami, M., et al. (2018). OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant 164, 216–225. doi: 10.1111/ppl.12707
Kitomi, Y., Hanzawa, E., Kuya, N., Inoue, H., Hara, N., Kawai, S., et al. (2020). Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields. Proc. Natl. Acad. Sci. U.S.A. 117, 21242–21250. doi: 10.1073/pnas.2005911117
Kruse, C. P. S., Meyers, A. D., Basu, P., Hutchinson, S., Luesse, D. R., Wyatt, S. E. (2020). Spaceflight induces novel regulatory responses in arabidopsis seedling as revealed by combined proteomic and transcriptomic analyses. BMC Plant Biol. 20, 1–16. doi: 10.1186/s12870-020-02392-6
Kwon, T., Sparks, J. A., Nakashima, J., Allen, S. N., Tang, Y., Blancaflor, E. B. (2015). Transcriptional response of arabidopsis seedlings during spaceflight reveals peroxidase and cell wall remodeling genes associated with root hair development. Ame. J. Bot. 102, 21–35. doi: 10.3732/ajb.1400458
Lavania, D., Dhingra, A., Grover, A. (2018). Analysis of transactivation potential of rice (Oryza sativa l.) heat shock factors. Planta. 247, 1267–1276. doi: 10.1007/s00425-018-2865-2
Li, L., Chen, H., Alotaibi, S. S., Pěnčík, A., Adamowski, M., Novák, O., et al. (2022). RALF1 peptide triggers biphasic root growth inhibition upstream of auxin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 119, e2121058119. doi: 10.1073/pnas.2121058119
Li, H. S., Lu, J. Y., Zhao, H., Sun, Q., Yu, F., Pan, Y., et al. (2017). The impact of space environment on gene expression in arabidopsis thaliana seedlings. Sci. China Tech. Sci. 60, 902–910. doi: 10.1007/s11431-016-0232-7
Luschnig, C., Gaxiola, R. A., Grisafi, P., Fink, G. R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in arabidopsis thaliana. Genes Dev. 12, 2175–2187. doi: 10.1101/gad.12.14.2175
Muñoz, A., Mangano, S., Toribio, R., Fernández-Calvino, L., Del Pozo, J. C., Castellano, M. M. (2022). The co-chaperone HOP participates in TIR1 stabilisation and in auxin response in plants. Plant Cell Environ. 45, 2508–2519. doi: 10.1111/pce.14366
Nakamura, M., Nishimura, T., Morita, M. T. (2019a). Bridging the gap between amyloplasts and directional auxin transport in plant gravitropism. Curr. Opin. Plant Biol. 52, 54–60. doi: 10.1016/j.pbi.2019.07.005
Nakamura, M., Nishimura, T., Morita, M. T. (2019b). Gravity sensing and signal conversion in plant gravitropism. J. Exp. Bot. 70, 3495–3506. doi: 10.1093/jxb/erz158
Nakano, Y., Konishi, J., Ito, H., Tanaka, T., Seki, M., Aoki, H., et al. (2022). Polymorphism of HvDRO1 and HvqSOR1 associated with root growth angle in barley accessions. Plant Root. 16, 1–10. doi: 10.3117/plantroot.16.1
O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166, 1598. doi: 10.1016/j.cell.2016.04.038
Oo, A. Z., Tsujimoto, Y., Mukai, M., Nishigaki, T., Takai, T., Uga, Y. (2021). Synergy between a shallow root system with a DRO1 homologue and localized p application improves p uptake of lowland rice. Sci. Rep. 11, 9484. doi: 10.1038/s41598-021-89129-z
Paul, A. L., Popp, M. P., Gurley, W. B., Guy, C., Norwood, K. L., Ferl, R. J. (2005). Arabidopsis gene expression patterns are altered during spaceflight. Space Life Sci. 36, 1175–1181. doi: 10.1016/j.asr.2005.03.066
Paul, A. L., Zupanska, A. K., Ostrow, D. T., Zhang, Y., Sun, Y., Li, J. L., et al. (2012). Spaceflight transcriptomes: unique responses to a novel environment. Astrobiology 12, 40–46. doi: 10.1089/ast.2011.0696
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Samakovli, D., Roka, L., Dimopoulou, A., Plitsi, P. K., Žukauskait, A., Georgopoulou, P., et al. (2021). HSP90 affects root growth in arabidopsis by regulating the polar distribution of PIN1. New Phytol. 231, 1814–1831. doi: 10.1111/nph.17528
Sedbrook, J. C., Chen, R., Masson, P. H. (1999). ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 96, 1140–1145. doi: 10.1073/pnas.96.3.1140
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Su, S. H., Gibbs, N. M., Jancewicz, A. L., Masson, P. H. (2017). Molecular mechanisms of root gravitropism. Curr. Biol. 27, R964–R972. doi: 10.1016/j.cub.2017.07.015
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Vandenbrink, J. P., Herranz, R., Poehlman, W. L., Feltus, F. A., Villacampa, A., Ciska, M., et al. (2019). RNA-Seq analyses of arabidopsis thaliana seedlings after exposure to blue-light phototropic stimuli in microgravity. Am. J. Bot. 106, 1466–1476. doi: 10.1002/ajb2.1384
Wang, L., Guo, M., Li, Y., Ruan, W., Mo, X., Wu, Z., et al. (2018). LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J. Exp. Bot. 69, 385–397. doi: 10.1093/jxb/erx427
Wang, R., Zhang, Y., Kieffer, M., Yu, H., Kepinski, S., Estelle, M. (2016). HSP90 regulates temperature-dependent seedling growth in arabidopsis by stabilizing the auxin co-receptor f-box protein TIR1. Nat. Commun. 7, 1. doi: 10.1038/ncomms10269
Wei, T., Simko, V. (2017) R package “corrplot”: visualization of a correlation matrix (Version 0.84). Available at: https://github.com/taiyun/corrplot.
Yu, G., Wang, L. G., Han, Y., He, Q. Y. (2012). clusterProfiler: an r package for comparing biological themes among gene clusters. Omics. 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, Y., Xiao, G., Wang, X., Zhang, X., Friml, J. (2019). Evolution of fast root gravitropism in seed plants. Nat. Commun. 10, 3480. doi: 10.1038/s41467-019-11471-8
Zhang, N., Yu, H., Yu, H., Cai, Y., Huang, L., Xu, C., et al. (2018). A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell. 30, 1461–1475. doi: 10.1105/tpc.18.00063
Keywords: clinostat, gravitropism, heat shock transcription factor, rice (Oryza sativa. L.), RNA-Seq, simulated microgravity treatment
Citation: Kuya N, Nishijima R, Kitomi Y, Kawakatsu T and Uga Y (2023) Transcriptome profiles of rice roots under simulated microgravity conditions and following gravistimulation. Front. Plant Sci. 14:1193042. doi: 10.3389/fpls.2023.1193042
Received: 24 March 2023; Accepted: 24 May 2023;
Published: 09 June 2023.
Edited by:
Karl H. Hasenstein, University of Louisiana at Lafayette, United StatesReviewed by:
Baris Uzilday, Ege University, TürkiyeJohn Z. Kiss, University of North Carolina at Greensboro, United States
Joshua Vandenbrink, Louisiana Tech University, United States
Copyright © 2023 Kuya, Nishijima, Kitomi, Kawakatsu and Uga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yusaku Uga, eXVnYUBhZmZyYy5nby5qcA==; Taiji Kawakatsu, cml2ZXJ3aW5AYWZmcmMuZ28uanA=
†These authors have contributed equally to this work and share first authorship
‡Present address: Ryo Nishijima, Department of Bioscience and Biotechnology, Fukui Prefectural University, Fukui, Japan
 Noriyuki Kuya
Noriyuki Kuya Ryo Nishijima2†‡
Ryo Nishijima2†‡ Taiji Kawakatsu
Taiji Kawakatsu Yusaku Uga
Yusaku Uga