- 1Institute of Food Crops, Hubei Academy of Agricultural Sciences, Hubei, Wuhan, China
- 2Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement, Hubei, Wuhan, China
- 3Key Laboratory of Crop Molecular Breeding, Ministry of Agriculture and Rural Affairs, Hubei, Wuhan, China
- 4School of Life Science and Technology, Wuhan Polytechnic University, Hubei, Wuhan, China
- 5Ministry of Agriculture and Rural Affairs (MARA) Key Laboratory of Sustainable Crop Production in the Middle Reaches of the Yangtze River (Co-construction by Ministry and Province), College of Agriculture, Yangtze University, Hubei, Jingzhou, China
- 6Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 7Western Crop Genetics Alliance, College of Science, Health, Engineering and Education, Murdoch University, Perth, WA, Australia
Introduction: Multiple nodes and dwarf mutants in barley are a valuable resource for identifying genes that control shoot branching, vegetative growth and development.
Methods: In this study, physiological, microscopic and genetic analysis were conducted to characterize and fine-map the underling gene of a barley mutant with Multiple Stem Nodes and Spikes and Dwarf (msnsd), which was selected from EMS- and 60Co-treated barley cv. Edamai 934.
Results and discussion: The msnsd mutant had more stem nodes, lower plant height and a shorter plastochron than Edamai 934. Moreover, the mutant had two or more spikes on each tiller. Microscopic analysis showed that the dwarf phenotype of msnsd resulted from reduced cell lengths and cell numbers in the stem. Further physiological analysis showed that msnsd was GA3-deficient, with its plant height increasing after external GA3 application. Genetic analysis revealed that a single recessive nuclear gene, namely, HvMSNSD, controlled the msnsd phenotype. Using a segregating population derived from Harrington and the msnsd mutant, HvMSNSD was fine-mapped on chromosome 5H in a 200 kb interval using bulked segregant analysis (BSA) coupled with RNA-sequencing (BSR-seq), with a C-T substitution in the exon of HvTCP25 co-segregating with the msnsd phenotype. RNA-seq analysis showed that a gene encoding gibberellin 2-oxidase 8, a negative regulator of GA biosynthesis, was upregulated in the msnsd mutant. Several known genes related to inflorescence development that were also upregulated and enriched in the msnsd mutant. Collectively, we propose that HvMSNSD regulates the plastochron and morphology of reproductive organs, likely by coordinating GA homeostasis and changed expression of floral development related genes in barley. This study offers valuable insights into the molecular regulation of barley plant architecture and inflorescence development.
1 Introduction
Globally, barley (Hordeum vulgare L.) is the second most important temperate cereal crop after wheat. Shoot architecture is a major determinant of the function, diversification and adaptation of barley. It is largely defined by plant height, leaf arrangement, branching patterns and inflorescence morphology (Wang et al., 2018). These features are determined by the activity and fate of the shoot apical meristem, axillary meristems (AXMs), leaf meristems and intercalary meristems (McKim, 2019). Typically, a single AXM initiates and forms in each leaf axil, which together with the leaf, node and subtending internode, generates a phytomer unit (Mcmaster, 2005). Plastochron refers to the time interval between the formation of two successive leaves, representing the temporal pattern of leaf initiation. Plastochron and phyllotaxy are two major contributors to plant architecture (Hibara et al., 2021). In rice (Oryza sativa L.), three genes responsible for regulating rapid leaf development—PLASTOCHRON 1 (PLA1) (Miyoshi et al., 2004), PLA2 (Kawakatsu et al., 2006) and PLA3 (Kawakatsu et al., 2009)—have been identified, encoding a plant-specific cytochrome P450 protein family, the MEI2-like RNA-binding protein and glutamate carboxypeptidase, respectively.
Barley multiple nodes and dwarf (mnd) mutants have more stem nodes but significantly lower plant heights than wild-type plants. Several mnd mutants, such as 93–597 derived from barley line ‘6121’ (Zhang and Zhai, 1995) and 76–2104 derived from ‘Zaoshu 3’ (Kawakatsu et al., 2006), have been reported since 1922 (Harlan and Pope, 1922). Apart from abnormal stem structure, the mnd mutations always exert pleiotropic effects on other agronomic traits, producing shorter leaf blades and aberrant inflorescences. Consistent with the increased stem node and leaf numbers, a shorter plastochron is usually observed in these mutants (Mascher et al., 2014). For these mutants, three underlying genes have been identified in barley—MND1, MND4 and MND8. MND4 is a member of the CYP78A family of cytochrome P450 enzymes, an ortholog of the rice PLA1 gene. Variations in MND4 were associated with the “multiple nodes but dwarf” phenotypes in barley, such as SNPs, which change the encoded amino acids, introducing premature stop codons, disrupting the splice site and partially or completely deleting this gene (Mascher et al., 2014). MND1 and MND8 were identified recently. MND1 located on chromosome 7H and encoded an Acyl-CoA N-acetyltransferase-like protein (Walla et al., 2020), while MND8 located on chromosome 5H and encoded a MATE transporter-family protein (Hibara et al., 2021).
Spike morphology is another determining factor for grain yield in the grass family. Barley inflorescence architecture differs from other Poaceae species, such as rice, wheat and maize. Barley specifically develops a branchless, spike-shaped inflorescence, where grain-producing spikelets are attached directly to the main axis (Gauley and Boden, 2019; Koppolu and Schnurbusch, 2019). Two-row and six-row differentiation is specific to barley spikes, with Vrs1 (Komatsuda et al., 2007), Vrs2 (Youssef et al., 2017), Vrs3 (Bull et al., 2017), Vrs4 (Koppolu et al., 2013) and Vrs5 (Ramsay et al., 2011) being the known row-type genes in barley. Besides row-type, branched spike is another variation observed in barley spike morphology. The grass-specific Teosinte branched 1/Cycloidea/Proliferating cell factor (TCP) family genes that encode transcription factors, such as the CYC/TB1-type TCP (Shang et al., 2020) and COMPOSITUM1 (Poursarebani et al., 2020), play a crucial role in branched spike formation in barley. Moreover, loss-of-function of the barley SEPALLATA MADS-box protein HvMADS1 is responsible for maintaining the branched inflorescence-like structure at high ambient temperatures (Li et al., 2021). In addition to these genes, a novel branched-spike gene in barley has been mapped on chromosome 2H in a 5 cM interval (Wang et al., 2020), with major and minor QTLs also identified for lateral spikelet indeterminacy and supernumerary spikelet phenotype in the central spikelets based on the genetic analysis of the multiflorus2.b mutant (Koppolu et al., 2022). However, for most mnd mutants, more than one spike per tiller, especially on the main tiller, can be produced, significantly different from the typical branched spike and potentially beneficial for improving barley yield.
A barley mutant with multiple stem nodes, spikes and dwarf (msnsd) was identified from chemically and physically treated cv. Edamai 934 (E934 hereafter) (Qin et al., 2021), an elite feed barley variety developed by Hubei Academy of Agricultural Sciences. Here, we undertook phenotypic and genetic characterization of msnsd and its causal gene HvMSNSD using BSR-seq. This study will not only enrich our understanding of barley plant architecture and inflorescence development, but also provide valuable resources for barley dwarf and yield breeding.
2 Materials and methods
2.1 Comparison of agronomic traits of msnsd and its parent E934
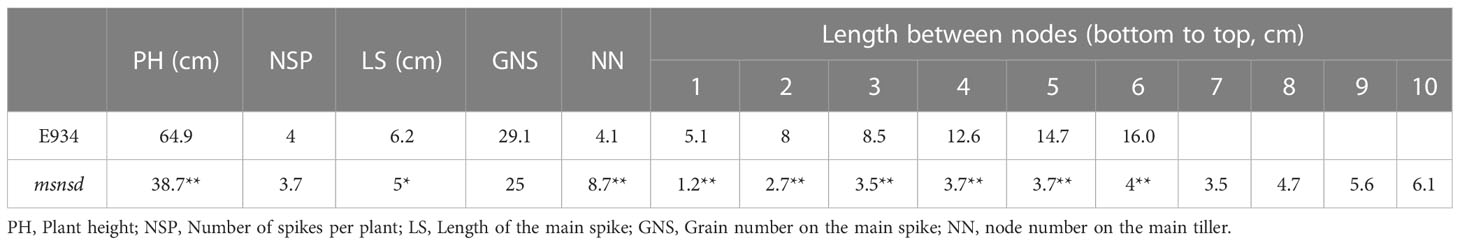
The msnsd mutant, characterized with “Multiple Stem Nodes, Spikes and Dwarf”, was generated from ethyl methane sulfonate (EMS)- and 60Co-treated feed barley variety E934 (Qin et al., 2021). The msnsd mutant and E934 were grown in the field at the experimental station of Hubei Academy of Agricultural Sciences, Wuhan, China, with management practices following the local standard practices. At maturity, plant height (PH), node number on the main tiller (NN), number of spikes per plant (NSP), length of the main spike (LS) and grain number on the main spike (GNS) were measured on ten plants. The average values for each parameter were used to determine differences between msnsd and E934. All tillers were removed at the five-leaf stage to determine the plastochrons of msnsd and E934. The fifth leaf on the main tiller was labeled, with leaf number recorded weekly.
2.2 Microscopic analysis of stem cells and immature spikes of msnsd and E934
Stem sections in the center of msnsd and E934 plants at the heading stage were sliced lengthwise into 10 mm sections and fixed in 10% formalin solution for 48 h at room temperature. Next, the tissue was dehydrated in a series of graded ethanol solutions to displace the water and then infiltrated with wax. The infiltrated tissues were then embedded into wax blocks and placed in the microtome for sectioning. Thereafter, sections were deparaffinized and rehydrated with distilled water. The slides were immersed in Safranin O solution for 1–2 h, rinsed in tap water to remove excess dye and dehydrated successively with a graded series of alcohol (50%, 70% and 80%), each for 3–8 s. The slides were then immersed in the fast green solution for 30–60 s and dehydrated in absolute alcohol three times. Finally, they were cleared in xylene for 5 min and mounted with the resin mounting medium. Cells of msnsd and E934 were observed in a bright field using an Olympus BX51 photomicroscope. All these procedures were carried out at the Wuhan Servicebio Technology Co., Ltd (Wuhan, China). Transverse sections of the stem were checked following the same procedure.
Immature spikes from msnsd and E934 plants at the booting stage were pre-fixed with 2.5% glutaraldehyde in a phosphate buffer (pH 7.0) at 4°C overnight. Subsequent treatments of the samples were conducted at the Wuhan Servicebio Technology Co., Ltd, which were then observed by scanning electron microscope (Hitachi TM-100) according to the manufacturer’s instructions (Qin et al., 2015).
2.3 Hormone response test
The msnsd and E934 plants at the five-leaf stage on Jan 13, 2021, were tested for their responses to five phytohormones, including 50 mg/L gibberellic acid 3 (GA3), 200 µM 3-indoleacetic acid (IAA), 50 µM 6-benzyl aminopurine (6-BA), 50 µM 1-aminocyclopropane-1-carboxylic acid (ACC) and 100 µM ABA. Around 15 mL of each phytohormone was poured directly into the surrounding soil, with water-treated seedlings serving as the control. The msnsd and E934 plants were sprayed with 50 mg/L GA3 and Tween 20 after the appearance of spikes. All treatments were applied to five plants of similar seedling length. After 20 days of treatment, plant height of the treated and control plants was recorded weekly until maturity. Agronomic traits, including tiller number, stem node number, internode length on the main tiller, spike number on the main tiller, leaf number and spikelet number per spike were determined after harvest.
2.4 Quantification of endogenous GAs
E934 and msnsd were grown in the field under normal conditions in autumn 2020. At the booting stage, GA contents (GA1, GA3, GA4, GA7 and GA20) in the young apical meristem (excluding inflorescence) of msnsd and E934 were determined using LC-MS/MS (Mubarak et al., 2012) at the Wuhan Greensword Creation Technology Co. Ltd. Three independent biological replicates and three technical replicates were measured for each sample.
2.5 Genetic analysis of the gene controlling the msnsd phenotype in barley
Genetic characterization of the gene (s) controlling the msnsd phenotype used the phenotypes of the F1 hybrid, F2 population and F2:3 family lines derived from Harrington and msnsd plants. Harrington is a two-row, malting barley variety from Canada. Chi-squared test was used to determine the suitability of observed data with the expected segregation ratios.
2.6 BSR-sequencing analysis based on BSA and RNA-sequencing
To map the causal gene for msnsd, bulked segregant RNA-seq (BSR-seq) based on bulked segregant analysis (BSA) and RNA-sequencing (RNA-seq) was conducted in two pools with distinct phenotypes from the F2 population. Five seeds from 40 wild-type and 40 mutants with multiple stem nodes and multiple spikes from F2 individuals were sown and grown at 25°C for two weeks to extract high-quality RNA. A half centimeter’s leaves from all the 200 wild-type or mutant seedlings were pooled and collected to construct wild-type or mutant bulks for RNA isolation. RNA was extracted following the TRIzol protocol (Rio et al., 2010). RNA-seq was undertaken using the HiSeq 2500 platform (Illumina) following the manufacturer’s protocol. RNA extraction and RNA-seq were conducted at the Beijing Novogene Bioinformatics Technology Co. Ltd. Implementation of single nucleotide polymorphism (SNP) calling and the filtration and identification of SNPs and Indels associated with the msnsd phenotype were performed as described elsewhere (Wang et al., 2017; Zhang et al., 2020b). High-quality reads were aligned to the barley Morex-V1 assembly. Thereafter, the mapping results were filtered using in-house Perl Scripts. Only uniquely mapped reads with a Phred quality value > 40 were kept.
2.7 Validation of the BSR-seq results and preliminary mapping of HvMSNSD
Seedling DNA from the F3 progeny of 46 wild-type (24 individuals were homozygous and 22 were heterozygous according to performance of F2:3) and 37 mutants of F2 individuals was extracted using CTAB methods to validate the BSR-seq analysis. Indel markers in the target region were developed according to the BSR-seq analysis and published results (Zhou et al., 2015). These markers were then applied to the two parents, separated on 8% non-denaturing polyacrylamide gels and visualized after silver staining. Subsequently, polymorphic markers were used to genotype the resulting sub-pools of F3 (each derived from one F2 individual) to validate the BSR-seq data and preliminarily map the candidate gene.
2.8 Fine-mapping of HvMSNSD
Indel markers linked to HvMSNSD were applied to the F2 population comprising of 460 progenies to fine-map the gene. The phenotype of F2 individuals was confirmed by evaluating their own and F2:3 performance under field conditions. Several SNP markers in the target region were further developed based on the BSR-seq analysis and applied to the recombinant lines using the Penta-primer amplification refractory mutation system (PARMS) at Gentides Biotech Co., Ltd, Wuhan, China. Furthermore, 1,869 F2:3 and F3:4 homozygous mutant individuals derived from the heterozygous F2 and F3 lines were used to confirm and fine-map the candidate gene.
2.9 Cloning and analysis of candidate genes
High-confidence genes in the target region were identified according to the annotated barley reference genome (Morex V3 assembly) (Mascher et al., 2021). Gene-specific overlapping primers (Supplementary File 1) were designed using DNAMAN5.0 software. Specific PCR products from genomic DNA and cDNA of E934, msnsd and Harrington were sequenced and analyzed using DNAMAN5.0.
2.10 RNA preparation and transcriptome sequencing
Immature spikes of msnsd and E934 from the glume primordium to the lemma primordium stages, approximately 0.5–1 cm in length, were collected and frozen in liquid nitrogen, with three biological replicates for each sample. Total RNA extraction, library construction and RNA-seq were performed at Beijing Novogene Bioinformatics Technology Co. Ltd, Beijing, China. Clean reads of each sample were mapped to the reference Morex V3 assembly (Mascher et al., 2021). Transcript quantification from RNA-seq data was performed using the Salmon software package (1.9.0). Expression levels of mapped reads were quantified based on transcripts per kilobase of exon per million mapped reads (TPM) (Patro et al., 2017). The Bioconductor package DESeq2 (Love et al., 2014) was used to perform differential expression analysis. Differentially expressed transcripts (DETs) in msnsd and E934 were defined as those with |log2 fold change (LFC)| ≥ 1.5 and adjusted P values ≤ 0.05. The GO enrichment analysis of DETs was conducted online (http://wheat.cau.edu.cn/TGT/). GO terms with a corrected FDR < 0.05 were considered significantly enriched. The percentage of DETs for each GO term was calculated based on all transcripts in the Morex V3 assembly.
3 Results
3.1 Phenotype of msnsd compared with wild-type E934
Compared with E934, the msnsd phenotype was characterized as a mutant with multiple stem nodes and spikes per tiller but dwarf (Figure 1; Table 1), when grown under field and glasshouse conditions. In particular, the msnsd mutant had at least four times more stem nodes and leaves than the wild-type (Figure 1A) due to a higher rate of leaf initiation (Figure 1F). Consequently, the distance between two nodes in msnsd decreased significantly relative to E934, such that msnsd plants were nearly 40% shorter than wild-type plants (Figure 1B; Table 1). Apart from the shortened plastochron, msnsd had more tillers (Figure 1B), but smaller diameter and spike length than E934 (Table 1). Moreover, msnsd had more than one fertile spike per tiller, located at the base of the main spike or the lower stem nodes, somewhat different from typical branched spikes (Figure 1D). These extra mini spikes at the base of the main spike appeared at the beginning of the reproductive stage (Figures 1C, D), but all msnsd spikes had lower fertility rates than E934. Further analysis showed that msnsd had smaller stamens and pistils than E934 (Figure 1E). Moreover, all trait alterations in msnsd were stable and repeatable, despite being sown one month later or earlier than the normal sowing time in Wuhan, China.
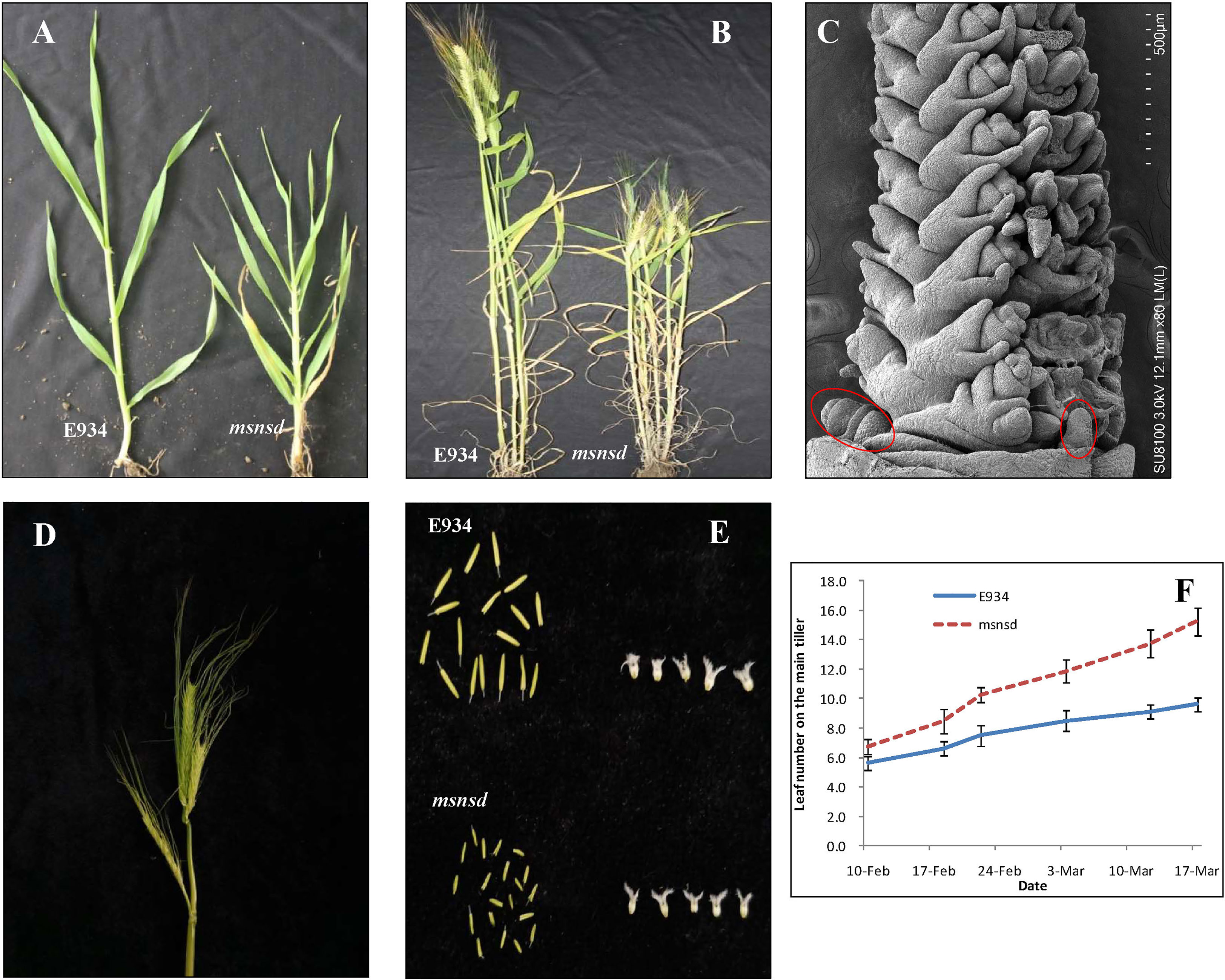
Figure 1 Phenotypes of msnsd and E934; (A): E934 and msnsd at the jointing stage; (B): Edamai 934 and msnsd before maturity; (C): Young inflorescences of msnsd (circles indicate an extra inflorescence primordium); (D): Mature spikes of msnsd; (E): Stamens and pistils of E934 and msnsd. (F): Leaf number on the main tiller of E934 and msnsd on different dates.
3.2 Microscopic analysis of stem cells in msnsd and E934
To determine whether the shorter msnsd stems were caused by cell elongation or proliferation changes, we examined the cell morphology of mature stem sections in the center of msnsd and E934 plants longitudinally and horizontally under a microscope. As expected, the msnsd stems comprised shorter cells than E934, with the mean length of msnsd stem cells about three-fourths of those in E934 (Figure 2A). While msnsd had more tillers than E934, it had much thinner stems than E934 (Figure 2B). Microscopic analysis of the transverse section showed that cell size decreased in msnsd, especially on the inner side of the stem (Figure 2C), as did cell layers (Figure 2C).
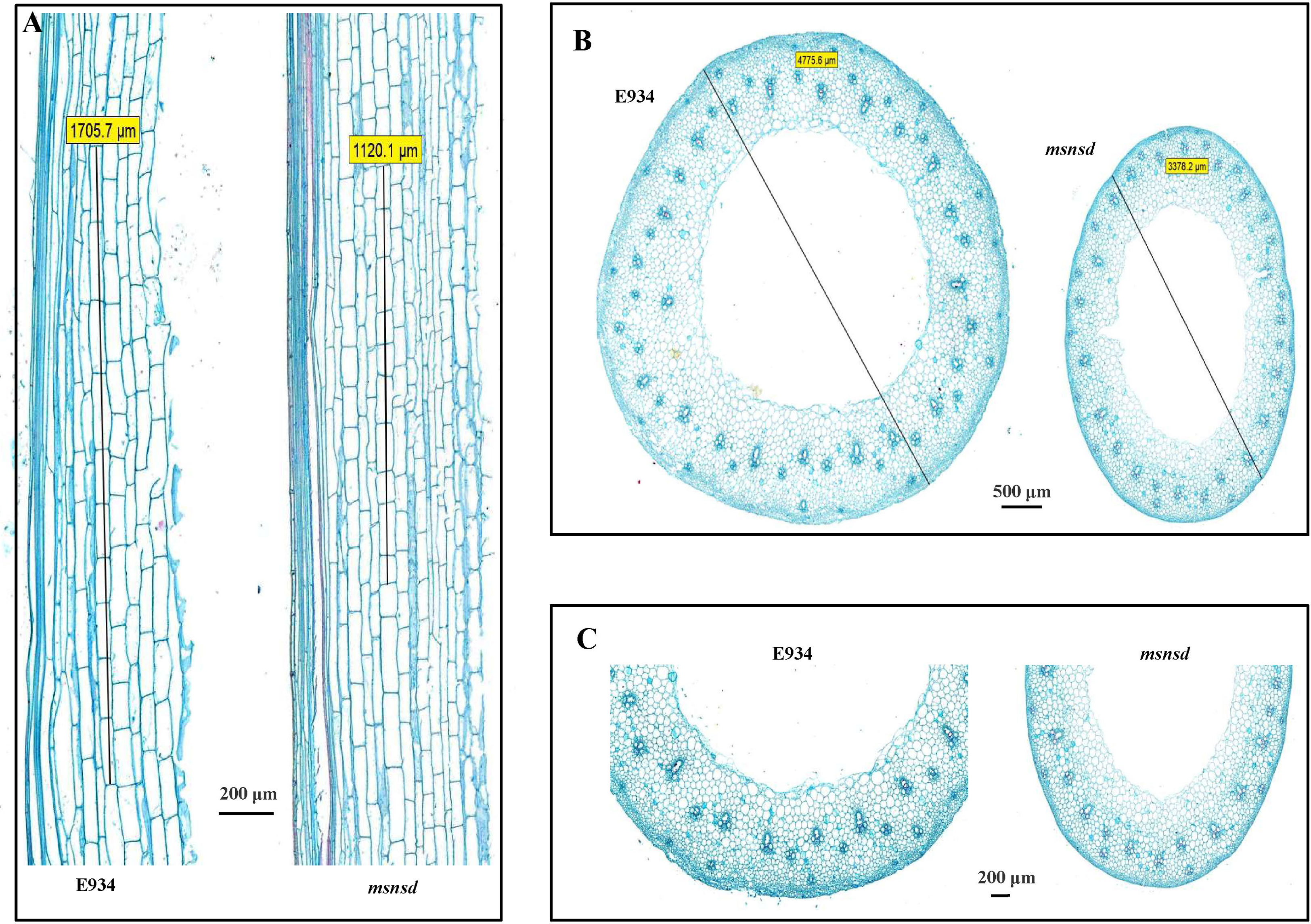
Figure 2 Microscopic analysis of E934 and msnsd. (A): the longitudinal culm; (B) Transverse sections; (C): Enlarged transverse section. Lines in (A) indicate the total length of ten cells. Lines in (B) indicate stem diameter.
3.3 Analysis of hormone response and quantification of endogenous GA levels
We investigated the response of E934 and msnsd to five different hormones to determine whether the dwarf msnsd phenotype was caused by a deficiency in GAs or other phytohormones. At the seedling stage, seven days of GA3 application significantly increased E934 and msnsd seedling lengths relative to the water treatment (Figures 3A, B), with the msnsd seedling length almost the same as E934 (Figures 3A, B①). However, msnsd plants grew slower than E934 plants after about two weeks of GA3 application (Figure 3B②). Furthermore, the GA and control treatments produced similar internode numbers at maturity for E934 and msnsd (Figure 3C), contributing to the similar plant heights in both treatments at the same stage (Figures 3B, C③). GA3 sprayed on E934 and msnsd at the heading stage increased internode length and plant height in both of them (Figure 3C). However, msnsd plants remained shorter than E934 plants in all treatments (Figure 3C). Except for plant height, all other investigated traits did not differ between the water and hormone treatments and neither msnsd nor E934 plants responded to treatment with other hormones in terms of plant height.
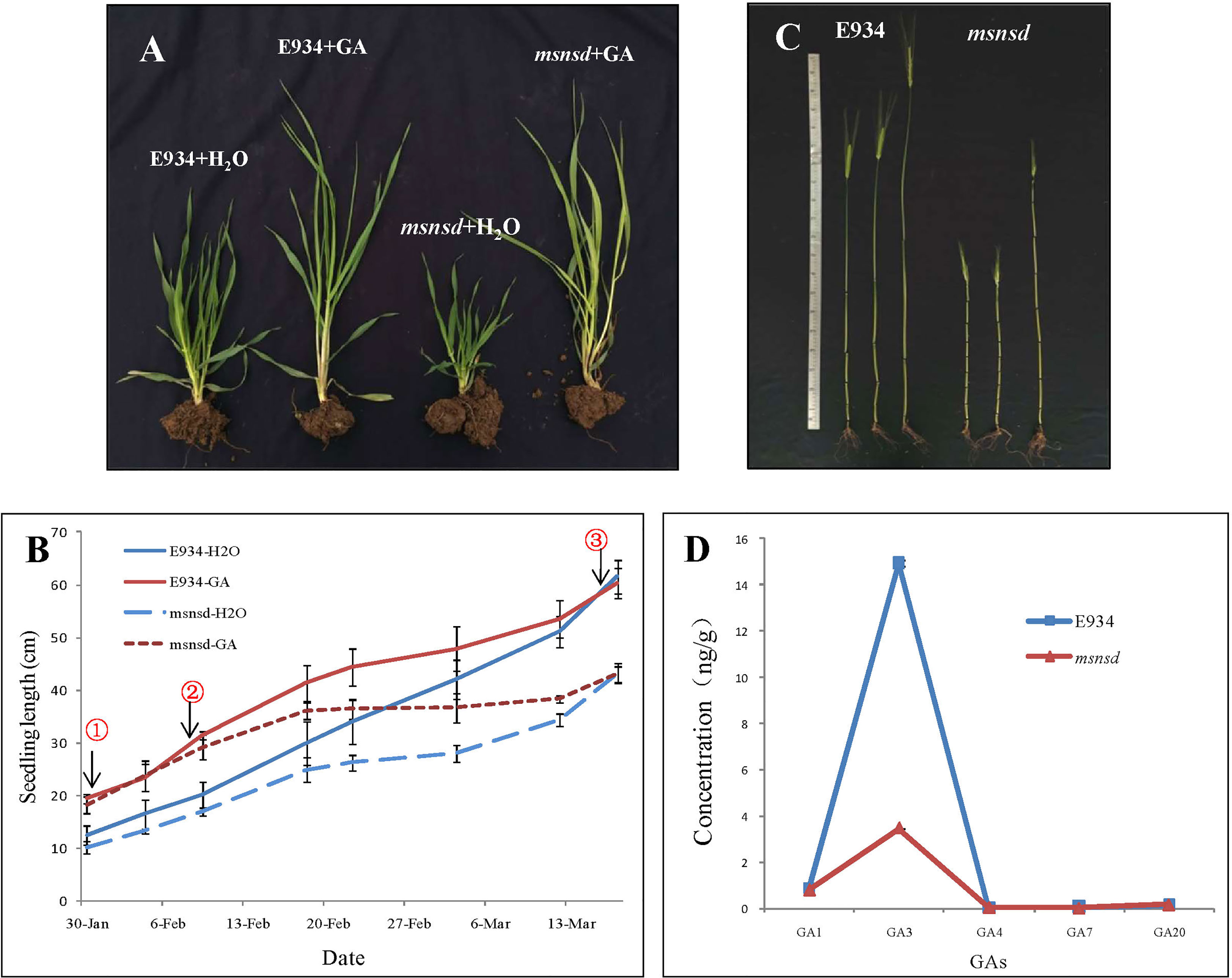
Figure 3 Application and quantification of GA in E934 and msnsd. (A): Seedlings of msnsd and E934 in the 50 mg/L GA3 and control (H2O) treatments; (B): Seedling length of msnsd and E934 in the 50 mg/L GA3 and control (H2O) treatments (① Seedling length of msnsd and E934 nearly the same with GA3 application, ② Growth of msnsd plants in the GA3 treatment slows, ③ Plant height of msnsd and E934 plants nearly the same in the GA3 and water control treatments); (C): Internodes of E934 and msnsd in the 50 mg/L GA3 and control (H2O) treatments (Left to right: control, GA3 application at the seeding stage, GA3 application at the heading stage); (D): GA concentration in the young apical meristem of msnsd and E934 plants.
The concentrations of GA1, GA3, GA4, GA7 and GA20 in the young apical meristem (excluding inflorescence) in msnsd and E934 were determined at the booting stage. The GA3 concentration significantly decreased in msnsd to about one-fifth of that in E934 (Figure 3D). However, the concentration of other GAs did not significantly differ between E934 and msnsd.
3.4 Genetic analysis of the gene controlling multiple stem nodes and spikes but dwarf phenotype of barley
The msnsd mutant was crossed with Harrington to characterize the gene underlying the msnsd phenotype. All F1 plants showed the wild-type phenotype as Harrington, but individuals of the F2 population showed segregation between the wild-type and mutant (multiple stem nodes, dwarf and multiple spikes per tiller) phenotypes. The segregation ratio of wild-type individuals to mutants was 340/109, or 3: 1 ratio using a Chi-square test. Moreover, among the 283 F2:3 lines derived from wild-type F2 individuals, 99 had the homozygous wild-type phenotype and 184 showed segregation of the phenotype within the family. All 89 F2:3 families derived from the msnsd-type F2 individuals showed a homozygous mutant phenotype, suggesting that the multiple nodes and spikes phenotype in msnsd followed a simple Mendelian inheritance pattern, with traits behaving in a single and recessive pattern.
3.5 Initial mapping of HvMSNSD by BSR-seq analysis
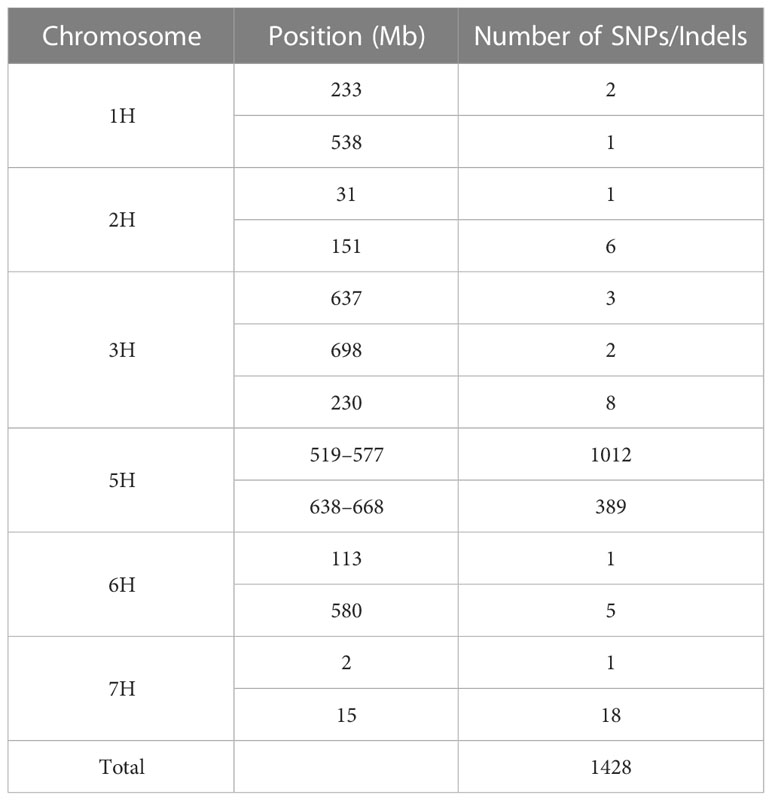
Using pooled RNA samples from 40 wild-type and 40 mutant individuals derived from F2:3 families and two parents, 1,445 SNPs or Indels were identified as likely associated with the HvMSNSD gene, with most (1,409) located on chromosome 5H (Table 2; Supplementary File 2). Further analysis showed that among these markers, 1,012 were between the 519 Mb and 577 Mb positions based on the Morex V1 assembly, whereas 389 were between the 638 Mb and 668 Mb positions, suggesting that the HvMSNSD gene was on chromosome 5H.
3.6 Sequence of MND4 and MND8 in msnsd and E934
Studies have shown that MND4 (HORVU5Hr1G081060) and MND8 (HORVU5Hr1G118820) are on chromosome 5H. Further analysis revealed that MND4 and MND8 are in the intervals between 562934428–562936259 and 653835747–653839974 bp, respectively, coincident with the two intervals of the candidate gene HvMSNSD identified by BSR-seq. Therefore, MND4 and MND8 were cloned from E934 and msnsd to verify whether they caused the msnsd phenotype. The results showed that msnsd and E934 had the same sequence for the coding region of MND4 and MND8, suggesting that they might not be the causal genes for msnsd in the present study.
3.7 Validation of candidate gene markers and preliminary mapping of HvMSNSD
Several markers on chromosome 5H were designed based on the Indels identified in the BSR-seq analysis, with their flanking sequences and other Indel markers around the candidate region obtained from published results. The markers were firstly used to screen Harrington and the msnsd mutant, with 14 (Supplementary File 1; Supplementary Sheet 1) showing polymorphisms between the two parents. The markers were then used to genotype 83 F2:3 families derived from 46 wild-type and 37 mutant F2 individuals. The results showed that 10 of the 83 families were recombinant families, representing four types of genotypes and phenotypes (Figure 4A). According to the phenotypic and genotypic data of the 10 recombinant families, the HvMSNSD gene was then mapped to the region between Indel5170 (557, 861, 942 bp) and Indel5181 (578, 321, 468 bp) on chromosome 5H (Figure 4A), which excluded MND8 from further analysis.
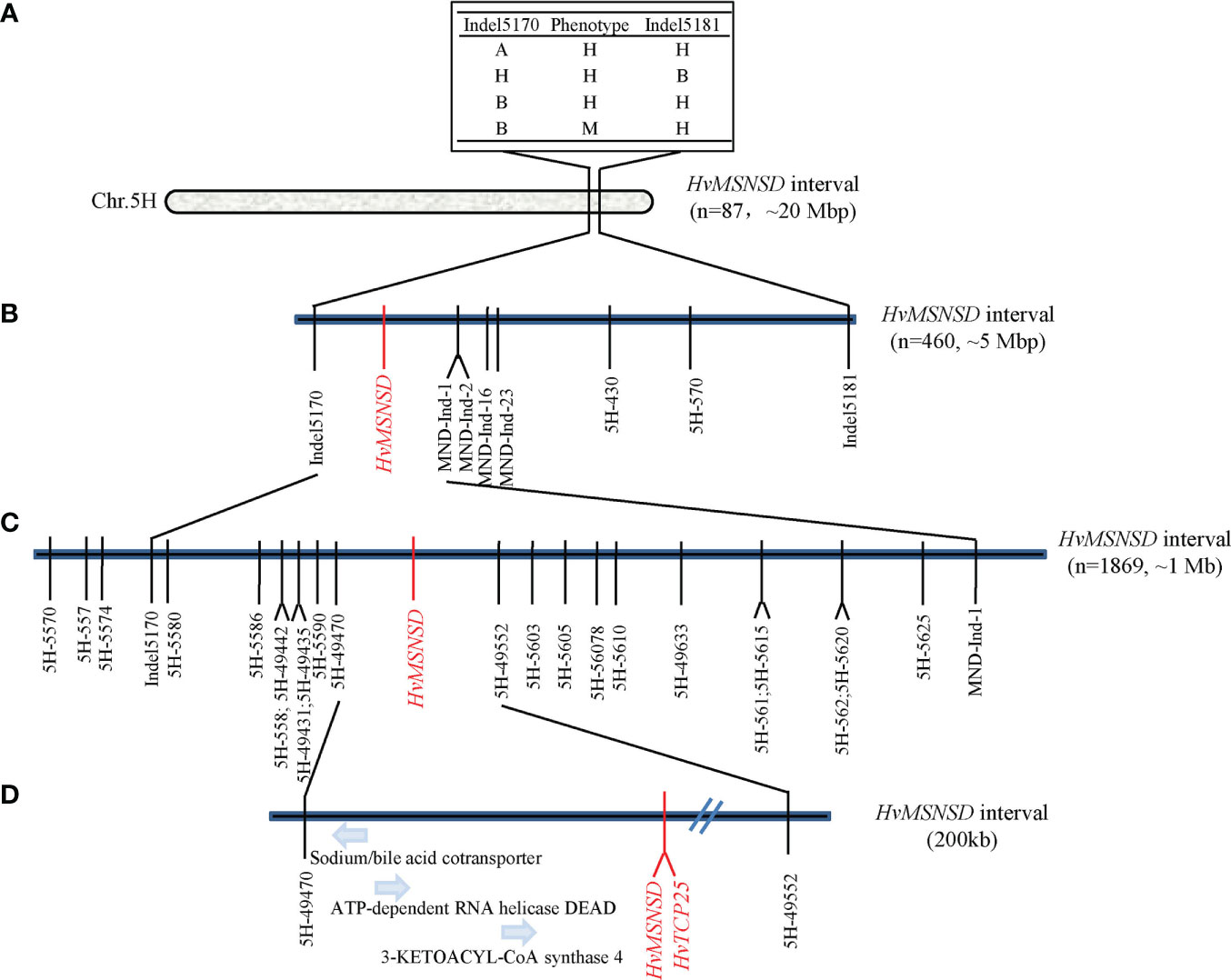
Figure 4 Map-based cloning of HvMSNSD; (A): Preliminary mapping of HvMSNSD; (B, C): Fine-mapping of HvMSNSD; (D): Candidate genes in the target region.
3.8 Fine-mapping of HvMSNSD
Indel5170 and Indel5181 were applied to the F2 population comprising 460 individuals to fine-map HvMSNSD. Five polymorphic markers—three Indel markers and two SNP markers in the target region—were further developed according to the BSR-seq analysis results, delimiting HvMSNSD to the region between Indel5170 and MND-Ind-1, spanning about 5 Mb (Figure 4B; Supplementary File 1).
Furthermore, 24 polymorphic SNP markers around the target region were developed and applied to 1,869 F2:3 and F3:4 homozygous mutant progenies derived from the heterozygous lines. Finally, the gene was narrowed to the region between 5H-49470 (559, 135, 754) and 5H-49552 (560, 194, 831), spanning about 1 Mb on chromosome 5H (Figure 4C), further indicating that the reported MND4 gene was not the causal gene for msnsd in this study.
The barley reference genome sequence identified 13 genes within the 1 Mb target region of HvMSNSD, with only TCP family members reportedly associated with the spikelet morphology of barley. Therefore, this TCP gene was considered the most likely candidate causal gene for msnsd and subjected to further cloning and sequence analysis.
3.9 Cloning and analysis of HvTCP25
Based on the barley reference genome sequence, the TCP gene in the target region is the previously identified HvTCP25 gene, which was 999 bps without intron with a TCP conserved domain at the N-terminal of an amino acid coding sequence (interval of 65th-126th amino acid). The coding regions and the 800 bps upstream regions of HvTCP25 from msnsd, E934 and Harrington were cloned and sequenced, revealing that the coding region was 1005 bps without intron and msnsd had a C-T substitution at +663 in the nearly 2000 bps compared to E934 and Harrington, with no change in amino acids (TTC to TTT).
A PARMS marker (Supplementary File 1) was developed based on the C-T variation and used for genotyping the F2, F3 and F4 populations comprising 2,329 individuals. Interestingly, the C-T SNP co-segregated with the msnsd phenotype in the large population. Consequently, the MSNSD interval was delimited to the 200 kb interval between 5H-49470 and 5H-TCP (Figure 4D), which also confirmed that the known MND4 was not the underling gene for msnsd. In addition to HvTCP25, three other known genes were identified in the 200 kb region, including sodium/bile acid cotransporter, ATP-dependent RNA helicase DEAD and 3-ketoacyl-CoA synthase 4 (Figure 4D). However, none of these genes had sequence variations likely associated with the msnsd phenotype after their sequencing in E934, Harrington and msnsd.
3.10 Transcriptional profiling by RNA sequencing
Transcriptome analysis was conducted on 0.5–1 cm young panicles of msnsd and E934 to determine the potential target genes of HvMSNSD that regulated barley development. A set of 118 DETs were discovered between msnsd and E934 (Supplementary Files 3, 4), with 83 and 35 DETs upregulated and downregulated, respectively, in msnsd compared with E934. According to the RNA-seq data, HvTCP25 had a slightly but not significant lower expression level in msnsd than E934, but interestingly, the HvMSNSD mutation upregulated the recently cloned MND1 gene (HORVU.MOREX.r3.7HG0742750) more than four-fold, while the other two MNDs (MND4 and MND8) were not responsive. Moreover, expression of the other three genes in the target region, including sodium/bile acid cotransporter, ATP-dependent RNA helicase DEAD and 3-ketoacyl-CoA synthase 4, was not changed by the mutation of HvMSNSD.
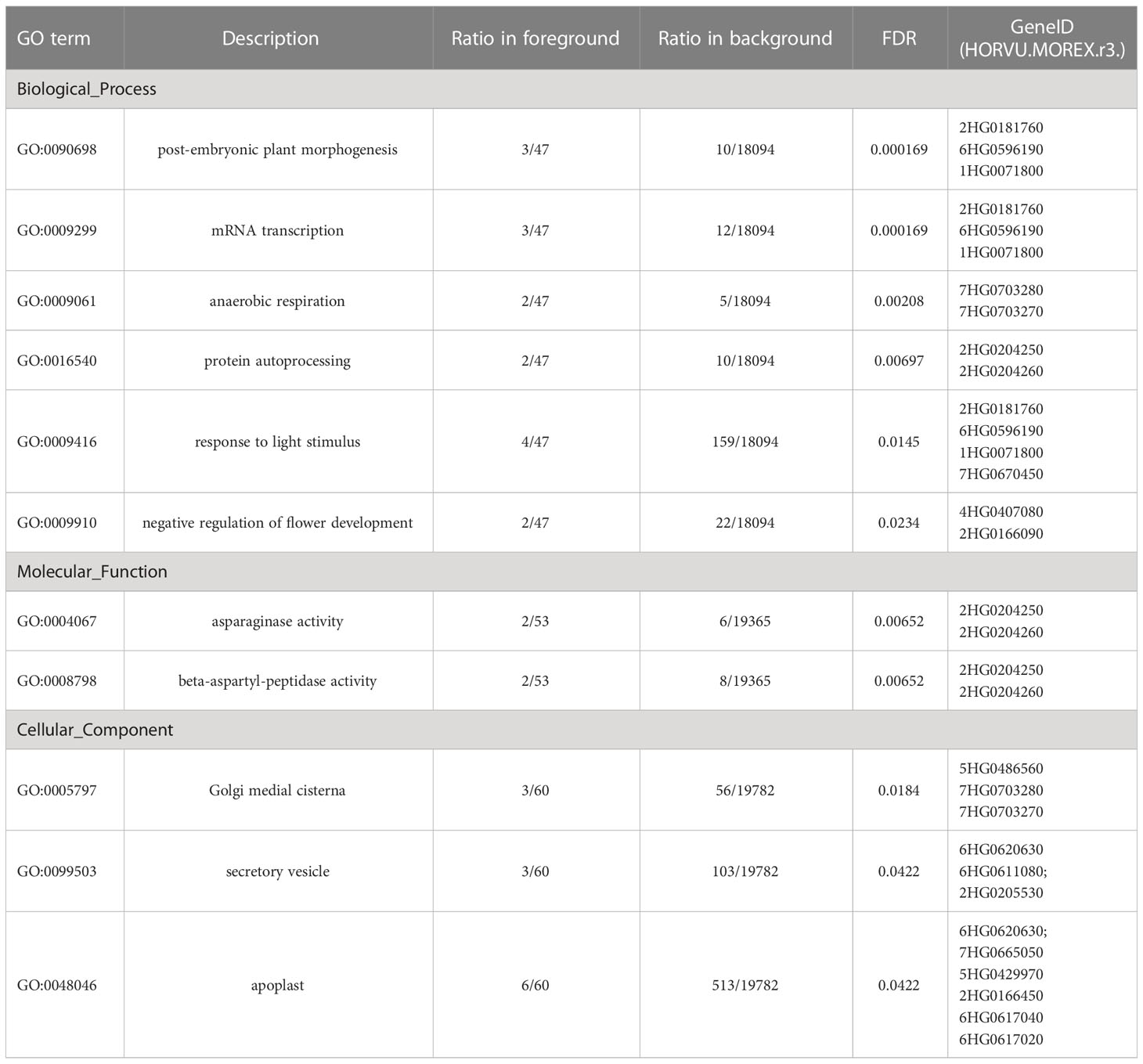
Though msnsd was GA-deficient and responded to GA application (GA-responsive), no GA biosynthesis and GA metabolisome related genes was identified to be DETs. Next, the 118 DETs were subjected to GO enrichment analysis to identify pathways likely regulated by HvMSNSD. Eleven GO terms were significantly enriched (Table 3). Strikingly, three of the six enriched GO terms in Biological Process—”post-embryonic plant morphogenesis”, “mRNA transcription” “ and “response to light stimulus” —were related to three light-dependent short hypocotyl (LSH)-like proteins/genes, which were upregulated in msnsd compared with E934. In addition, two TERMINAL FLOWER1 (TFL1) genes belonging to the GO term “negative regulation of flower development” were upregulated in msnsd (Supplementary File 4). Moreover, some known genes controlling inflorescence development in barley showed differential expression in msnsd and E934. For example, the expression of a member of MADS-box gene family (HORVU.MOREX.r3.7HG0664320) increased in msnsd, while the expression of BTB/POZ and TAZ domain-containing protein (HORVU.MOREX.r3.2HG0176250) decreased (Supplementary File 4).
4 Discussion
4.1 msnsd may be a valuable resource for barley breeding and study
The multiple nodes and dwarf (mnd) mutants of barley are a valuable resource for detecting genes that control shoot branching and vegetative versus reproductive growth (Walla et al., 2020), with several mnd mutants identified in the last century. msnsd is a “multiple stem nodes and spikes and dwarf” mutant of barley identified from the 60Co- and EMS-treated E934 variety. The pleiotropic alteration is independent of external conditions and sowing date in msnsd, which differs from mnd, whose dwarf phenotype could only be observed under field conditions but not in glasshouse (Mascher et al., 2014). In addition, msnsd provides a valuable genetic resource for lodging resistance according to its field performance. Spikes per plant and grains per spike are two of the three most important yield components for the grass family, therefore, the increased tillers and multiple spikes per tiller in msnsd could be favorable traits for barley breeding. However, the higher rate of infertility of these external spikes due to decrescent reproductive organs decreases the grain number in msnsd. While several mnd mutants have been identified, only three genes associated with mnd have been cloned (Mascher et al., 2014; Walla et al., 2020; Hibara et al., 2021). Therefore, characterizing msnsd and its causal gene will be helpful for illustrating barley morphogenesis and breeding barley varieties with lodging resistance and higher yield potential by optimizing the balance between spike number and fertility rate.
4.2 Dwarf phenotype of msnsd caused by inhibited cell elongation and shortened phyllochron
It is well known that dwarf phenotypes of plants are induced by shortened cells, inhibited cell division, or both. In wheat, peduncle tissues of DD399 comprised shorter cells than the wild-type ND399, which may have reduced its plant height (Wu et al., 2021). Likewise, the length of parenchyma cells significantly decreased in the maize dwarf mutant dnl2 (Han et al., 2022). In barley, the significantly reduced length of the uppermost internode in the mutant Sheathed Spike 1 (SS1) was attributed mainly to suppressed cell elongation (Pu et al., 2021). However, the reduced plant height of two maize dwarf mutants, short internode length1 (sil1) and short internode length2 (sil2), was caused by decreased cell numbers and longitudinal cell size (Li et al., 2020). In the present study, the cell length of mature stems decreased in msnsd, suggesting inhibited cell elongation. Moreover, successive leaves on the main culm of msnsd appeared faster than E934, with a shorter time interval from node initiation to maturity. Thus, it was inferred that an inhibited cell elongation and shorter phyllochron of msnsd caused shorter internodes and the dwarf phenotype. In addition, the decreased cell size and cell number may contribute to the reduced stem diameter and thickness in msnsd.
4.3 msnsd was gibberellin-deficient but GA-sensitive
GA is a well-known phytohormone that plays a key role in plant growth and development, especially plant height. If exogenous GA application promoted plant growth, the plant was considered GA-sensitive; otherwise, it was called GA-insensitive (Ellis et al., 2004). Many studies have been conducted to study the effect of GA on plant height. The known dwarfing genes Rht-B1b and Rht-D1b in wheat were considered GA-insensitive dwarfing genes due to their reduced response to GA (Peng et al., 1999), whereas the plant height of wheat Rht12 dwarf lines increased by more than 50% in the GA3 treatment (Chen et al., 2014) and the wheat dwarf mutant DD399 also had significantly longer seedling and coleoptile lengths in the GA treatment than the non-treated control (Wu et al., 2021). Some dwarf mutants in plants, like the dwarf and narrow-leaf mutant dnl2 in maize, are GA biosynthesis-deficient mutants, with significantly lower GA concentrations than the wild-type mutant (Han et al., 2022). Inhibition of UI parenchyma cell extension in barley SS1 was possibly related to insufficient amounts of endogenous bioactive gibberellins (Pu et al., 2021). In the present study, GA3 application promoted stem elongation in E934 and msnsd plants at the vegetative and reproductive stages, suggesting that these plants are GA-sensitive, with msnsd mutants deficient in GA biosynthesis rather than GA signal transduction. Further quantification of GAs in apical meristems showed that the GA biosynthesis, particularly GA3, was significantly inhibited in msnsd. Interestingly, the increase in plant height in msnsd slowed after 14 days of GA application at the seedling stage and almost stopped after 28 days, indicating that the GA sprayed on msnsd seedlings had been used up after one month. GA application at the heading stage also increased plant height, confirming that the GA amount was insufficient, but external GA application could trigger GA signaling in msnsd. The GA3 treatment did not change spike performance in msnsd; however, the crosstalk between hormones such as auxin, cytokinin, gibberellins and abscisic acid along the spike sections played a decisive role in spike and spikelet development of barley (Youssef and Hansson, 2019), suggesting that the morphology of stem and spike of msnsd formed through independent pathways. In barley, GA3-treated vrs2 also exhibited typical symptoms of gibberellin application, such as elongation of stem and spike internodes, but still showed gradients in lateral spikelet fertility along the spike, as observed in the non-treated control plants (Youssef et al., 2017). Genetic studies also confirmed that the rice PLASTOCHRON genes PLA1 and PLA2 acted downstream of the GA signal transduction pathway to regulate leaf growth by affecting cellular proliferation (Mimura et al., 2012; Mimura and Itoh, 2014).
4.4 HvTCP25 might be the causal gene for msnsd, with pleiotropic effects on stem and spike development
Among the identified barley multiple stem nodes and dwarf mutants in the current study, some mutants had similar spike performance to the msnsd mutant, developing more than one spike on each tiller. A genetic analysis showed that two independent but closely linked recessive genes controlled some of the multiple nodes and dwarf mutant phenotypes (Xu and Huang, 1999). However, some previous studies (Li and Li, 1994) and our study showed that a single recessive gene controlled ‘spike variation and multiple nodes and dwarf’ phenotypes, which exerted pleiotropic effects on stem and spike morphology in barley.
To date, three genes responsible for this phenotype in barley (MND1, MND4 and MND8) encoded Acyl-CoA N-acetyltransferase-like protein, cytochrome P450 and MATE family transporter protein, respectively. While MND4 and MND8 were on chromosome 5H, the mapping and sequencing analysis showed they were not the causal gene for the msnsd mutant in this study. Screened by BSR-seq and map-based cloning, the MSNSD gene was mapped on chromosome 5H in a 1 Mb interval. Among the 13 high-confidence genes in the 1 Mb target region, only plant-specific TCP transcription factor family genes have been reported responsible for branched spike formation in barley. According to published public datasets, 20 TCP genes encoding mature proteins have been identified in barley (Gao et al., 2021). Mutation in the CYC/TB1-type TCP on chromosome 5H produced a branched inflorescence in the bdi1 mutant (Shang et al., 2020), while another member COMPOSITUM 1 specified branch-inhibition in barley (Triticeae) versus branch-formation in non-Triticeae grasses (Poursarebani et al., 2020). Apart from the identity of the spikelet meristem, TCP members also participated in barley seed germination (Seven and Akdemir, 2020) and selection from wild barley (Hordeum vulgare ssp. spontaneum) to cultivated barley (Hordeum vulgare subsp. vulgare) (Gao et al., 2021). In the present study, the TCP gene in the target region is the known HvTCP25 gene (Gao et al., 2021), belonging to subgroup class I, as distinguished by the deletion of four amino acids at the N-terminal. Based on the publicly released RNA-seq datasets, HvTCP25 had a very low expression level across the 21 investigated tissues and developmental stages (Gao et al., 2021), but its function remains unclear. However, the TCP domain of the members of this gene family was quite conserved between barley and its relatives, suggesting that they play similar roles in plant development. In this study, while the multiple spikes per tiller in msnsd differed from the branched spike mentioned earlier, re-sequencing of HvTCP25 in msnsd, E934 and Harrington identified a C-T SNP on it. While the C-T variation was a nonsense mutation, it co-segregated with the msnsd phenotype in the large mapping population comprising 2,329 progenies, indicating that HvTCP25 is most likely the causal gene for the msnsd phenotype in the current study. However, HvTCP25’s precise function and mechanisms in the msnsd phenotype remain under investigation.
4.5 HvMSNSD may regulate barley infloresence development by known genes associated with floral development
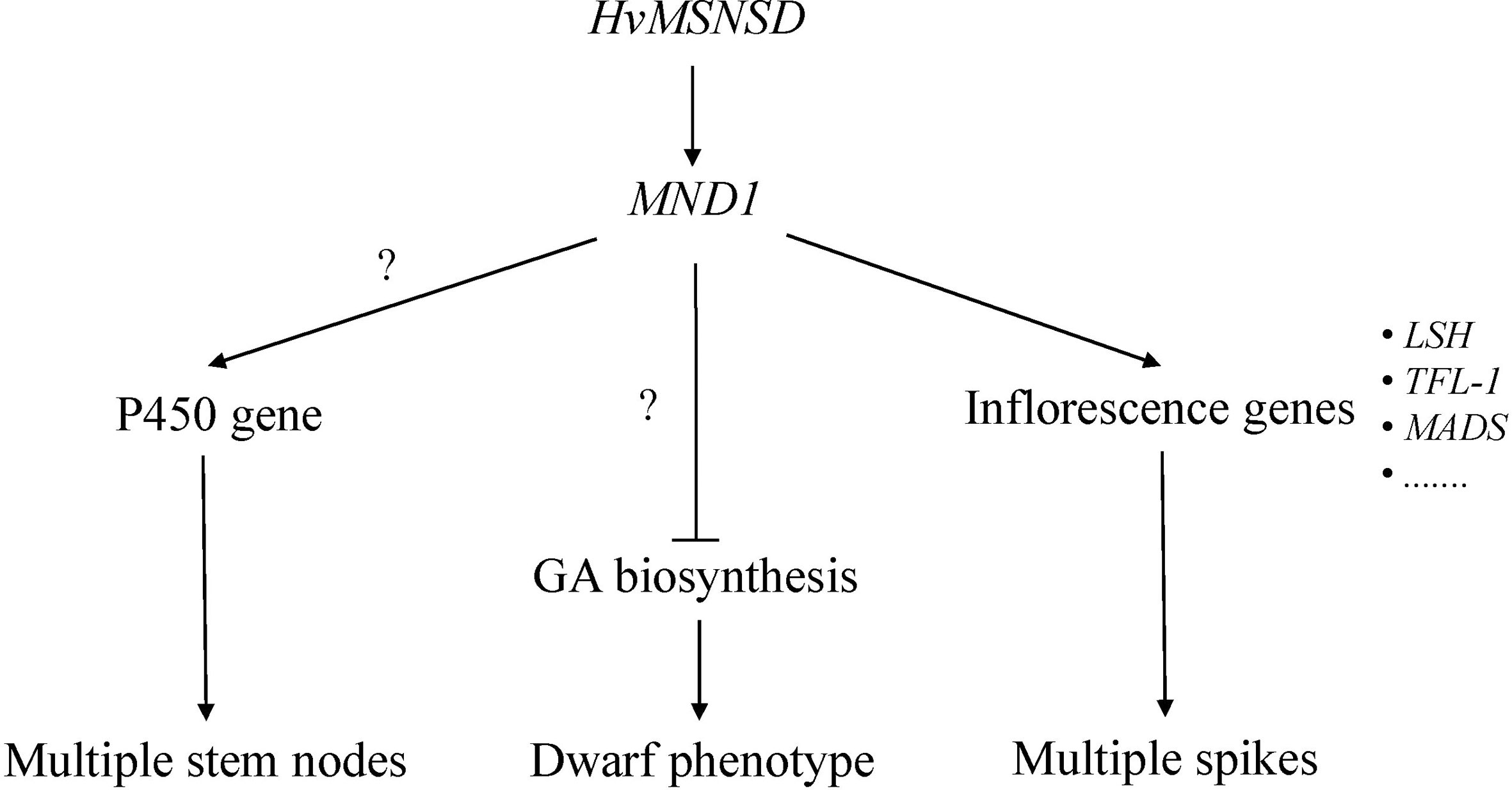
Studies on the rice plastochron and barley multiple nodes and dwarf mutants confirmed that the responsible genes independently regulated these traits (Kawakatsu et al., 2006; Hibara et al., 2021). Among the three investigated MNDs based on our RNA-seq results, the HvMSNSD mutation induced MND1 expression. Further analysis demonstrated that MND8 was not expressed in young panicles (0.5–1 cm) of E934 or msnsd, while MND4 was expressed in E934 and msnsd with no difference between them. These results suggest that HvMSNSD regulates barley development through MND1. However, both MND4 and rice PLA1 encoded plant-specific cytochrome P450 protein family genes, expression of three P450 genes (Supplementary File 3) were up-regulated in msnsd, implying that they might cause the shorter plastochron in msnsd.
While all mnd mutants showed a dwarf phenotype, the transcriptomic analysis of mnd1.a detected no differential expression of genes related to GA biosynthesis or signaling in GA-signaling pathways (Walla et al., 2020). To be consistent with this, we didn’t detect any differential expression of GA biosynthesis and metabolism related genes either. It probably because that it was young infloresence but not stems was used in transcriptome analysis, which also suggested that HvMSNSD participated in development of barley plant height and inflorescences via different pathways.
For the mnd1.a mutant, a core set of DETs comprising 43 transcripts was discovered at all three developmental stages, including developing inflorescences, compared with the wild-type Bowman (Walla et al., 2020). The msnsd mutant had more DETs than the wild-type E934. Further analysis identified several overlapped DETs between msnsd and mnd1.a. For example, msnsd and mnd1.a upregulated three and four LIGHT-DEPENDENT SHORT HYPOCOTYLS-like (LSH) proteins, respectively, compared with their corresponding wild-type mutants. In addition, both mutants strongly upregulated two TERMINAL FLOWER1-like (TFL-1) genes. However, mnd1.a downregulated seven MADS-box genes in inflorescences, with only two upregulated in msnsd. All these common genes are known to participate in inflorescence development. Studies on Arabidopsis revealed that plants with increased TCP2 expression displayed the light-dependent short-hypocotyl phenotype (He et al., 2016). The TFL1 gene from perennial ryegrass involved in floral transition and axillary meristem identity (Jensen et al., 2001) is an essential molecular switch for regulating endosperm cellularization and seed size in Arabidopsis (Zhang et al., 2020a), while the rice TFL1 gene acted downstream of ethylene in regulating flowering time (Yin et al., 2017). Additionally, studies on Asteraceae proposed that the C class MADS-box TF GAGA1 contributed to stamen development upstream of GhCYC3, belonging Mascher et al., 2021 to the TCP protein family (Zhao et al., 2020). Recently, loss-of-function in HvMADS1 was responsible for developing the branched inflorescence-like structure in barley at high ambient temperatures (Li et al., 2021). Based on the common DETs identified in msnsd and mnd1.a, HvMSNSD and HvMND1 act as upstream regulators to control inflorescence development in barley through some shared molecular pathways. We proposed a possible pathway for HvMSNSD in barley stem and spike morphology development in this study (Figure 5).
5 Conclusions
A “multiple stem nodes and spikes but dwarf” mutant, msnsd, was identified from EMS- and 60Co-treated barley variety E934. Using the F2 population derived from Harrington and msnsd, the causal gene was fine-mapped on chromosome 5H in a 200 kb interval, based on BSR-seq analysis results, with the C-T substitution on exon of HvTCP25 co-segregating with the msnsd phenotype in the population. Transcriptomic analysis showed altered expressions of MND1 and several genes associated with inflorescence development and GA biosynthesis in msnsd. We propose that HvMSNSD, probably the known HvTCP25, regulates the plastochron and morphology of reproductive organs, likely by coordinating the expression of genes involved in GA biosynthesis and floral development in barley.
Data availability statement
The data presented in the study are deposited in GEO (Gene Expression Omnibus) on NCBI, accession number is GSE235622.
Author contributions
DQ conducted all experiments and drafted the manuscript. RL performed gene cloning and expression analysis. FX performed the fieldwork. GD analyzed the genomic sequence. QX, YP, LX, and HC took part in DNA extraction. GG, JD, and CL conceived the experiment and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Key Research and Development Program of China (2018YFD1000700, 2018YFD1000706), the National Natural Science Foundation of China (31771774), the Young Top-notch Talent Cultivation Program of Hubei Province, Hubei Hongshan Laboratory and the China Agriculture Research System of MOF and MARA (CARS-05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1189743/full#supplementary-material
Supplementary File 1 | List and sequence of primer pairs used.
Supplementary File 2 | Distribution and p value of putative HvMSNSD associated variations.
Supplementary File 3 | Distribution of Differential expressed transcripts.
Supplementary File 4 | Differentially expressed transcripts in msnsd and E934 plants.
References
Bull, H., Casao, M. C., Zwirek, M., Flavell, A. J., Thomas, W. T. B., Guo, W., et al. (2017). Barley SIX-ROWED SPIKE3 encodes a putative jumonji c-type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nat. Commun. 8, 936. doi: 10.1038/s41467-017-00940-7
Chen, L., Hao, L., Condon, A. G., Hu, Y. G. (2014). Exogenous GA3 application can compensate the morphogenetic effects of the GA-responsive dwarfing gene Rht12 in bread wheat. PloS One 9, e86431. doi: 10.1371/journal.pone.0086431
Ellis, M. H., Rebetzke, G. J., Chandler, P., Bonnett, D., Spielmeyer, W., Richards, R. A. (2004). The effect of different height reducing genes on the early growth of wheat. Funct. Plant Biol. 31, 583–589. doi: 10.1071/fp03207
Gao, G., Kan, J., Jiang, C., Ahmar, S., Zhang, J., Yang, P. (2021). Genome-wide diversity analysis of TCP transcription factors revealed cases of selection from wild to cultivated barley. Funct. Integr. Genomics 21, 31–42. doi: 10.1007/s10142-020-00759-4
Gauley, A., Boden, S. A. (2019). Genetic pathways controlling inflorescence architecture and development in wheat and barley. J. Integr. Plant Biol. 61, 296–309. doi: 10.1111/jipb.12732
Han, L., Jiang, C., Zhang, W., Wang, H., Li, K., Liu, X., et al. (2022). Morphological characterization and transcriptome analysis of new dwarf and narrow-leaf (dnl2) mutant in maize. Int. J. Mol. Sci. 23, 795. doi: 10.3390/ijms23020795
Harlan, H. V., Pope, M. N. (1922). The use and value of back-crosses in small-grain breeding. J. Heredity 7, 319–322. doi: 10.1093/oxfordjournals.jhered.a102237
He, Z., Zhao, X., Kong, F., Zuo, Z., Liu, X. (2016). TCP2 positively regulates HY5/HYH and photomorphogenesis in arabidopsis. J. Exp. Bot. 67, 775–785. doi: 10.1093/jxb/erv495
Hibara, K. I., Miya, M., Benvenuto, S. A., Hibara-Matsuo, N., Mimura, M., Yoshikawa, T., et al. (2021). Regulation of the plastochron by three many-noded dwarf genes in barley. PloS Genet. 17, e1009292. doi: 10.1371/journal.pgen.1009292
Jensen, C. S., Salchert, K., Nielsen, K. K. (2001). A TERMINAL FLOWER1-like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol. 125, 1517–1528. doi: 10.1104/pp.125.3.1517
Kawakatsu, T., Itoh, J., Miyoshi, K., Kurata, N., Alvarez, N., Veit, B., et al. (2006). PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell 18, 612–625. doi: 10.1105/tpc.105.037622
Kawakatsu, T., Taramino, G., Itoh, J., Allen, J., Sato, Y., Hong, S. K., et al. (2009). PLASTOCHRON3/GOLIATH encodes a glutamate carboxypeptidase required for proper development in rice. Plant J. 58, 1028–1040. doi: 10.1111/j.1365-313X.2009.03841.x
Komatsuda, T., Pourkheirandish, M., He, C., Azhaguvel, P., Kanamori, H., Perovic, D., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104, 1424–1429. doi: 10.1073/pnas.0608580104
Koppolu, R., Anwar, N., Sakuma, S., Tagiri, A., Lundqvist, U., Pourkheirandish, M., et al. (2013). Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. USA 110, 13198–13203. doi: 10.1073/pnas.1221950110
Koppolu, R., Jiang, G., Milner, S. G., Muqaddasi, Q. H., Rutten, T., Himmelbach, A., et al. (2022). The barley mutant multiflorus2.b reveals quantitative genetic variation for new spikelet architecture. Theor. Appl. Genet. 135, 571–590. doi: 10.1007/s00122-021-03986-w
Koppolu, R., Schnurbusch, T. (2019). Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J. Integr. Plant Biol. 61, 278–295. doi: 10.1111/jipb.12771
Li, G., Kuijer, H. N. J., Yang, X., Liu, H., Shen, C., Shi, J., et al. (2021). MADS1 maintains barley spike morphology at high ambient temperatures. Nat. Plants. 7, 1093–1107. doi: 10.1038/s41477-021-00957-3
Li, P., Li, W. (1994). Morphological and genetic analysis of natural mutants in multinode branches of barley. Scientia Agricultura Sin. 27, 10. doi: 10.3864/j.issn.0578-1752.1994-27-04-1-10
Li, C., Tang, J., Hu, Z., Wang, J., Yu, T., Yi, H., et al. (2020). A novel maize dwarf mutant generated by Ty1-copia LTR-retrotransposon insertion in Brachytic2 after spaceflight. Plant Cell Rep. 39, 393–408. doi: 10.1007/s00299-019-02498-8
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mascher, M., Jost, M., Kuon, J. E., Himmelbach, A., Aßfalg, A., Beier, S., et al. (2014). Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol. 15, R78. doi: 10.1186/gb-2014-15-6-r78
Mascher, M., Wicker, T., Jenkins, J., Plott, C., Lux, T., Koh, C. S., et al. (2021). Long-read sequence assembly: a technical evaluation in barley. Plant Cell 33, 1888–1906. doi: 10.1093/plcell/koab077
McKim, S. M. (2019). How plants grow up. J. Integr. Plant Biol. 61, 257–277. doi: 10.1111/jipb.12786
Mcmaster, G. S. (2005). Phytomers, phyllochrons, phenology and temperate cereal development. J. Agric. Sci. 143, 137–150. doi: 10.1017/S0021859605005083
Mimura, M., Itoh, J. (2014). Genetic interaction between rice PLASTOCHRON genes and the gibberellin pathway in leaf development. Rice 7, 25. doi: 10.1186/s12284-014-0025-2
Mimura, M., Nagato, Y., Itoh, J. (2012). Rice PLASTOCHRON genes regulate leaf maturation downstream of the gibberellin signal transduction pathway. Planta 235, 1081–1089. doi: 10.1007/s00425-012-1639-5
Miyoshi, K., Ahn, B. O., Kawakatsu, T., Ito, Y., Itoh, J., Nagato, Y., et al. (2004). PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl. Acad. Sci. USA 101, 875–880. doi: 10.1073/pnas.2636936100
Mubarak, A., Bondonno, C. P., Liu, A. H., Considine, M. J., Rich, L., Mas, E., et al. (2012). Acute effects of chlorogenic acid on nitric oxide status, endothelial function, and blood pressure in healthy volunteers: a randomized trial. J. Agric. Food Chem. 60, 9130–9136. doi: 10.1021/jf303440j
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Poursarebani, N., Trautewig, C., Melzer, M., Nussbaumer, T., Lundqvist, U., Rutten, T., et al. (2020). COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat. Commun. 11, 5138. doi: 10.1038/s41467-020-18890-y
Pu, X., Tang, Y., Zhang, M., Li, T., Qiu, X., Zhang, J., et al. (2021). Identification and candidate gene mining of HvSS1, a novel qualitative locus on chromosome 6H, regulating the uppermost internode elongation in barley (Hordeum vulgare l.). Theor. Appl. Genet. 134, 2481–2494. doi: 10.1007/s00122-021-03837-8
Qin, D., Dong, J., Xu, F., Guo, G., Ge, S., Xu, Q., et al. (2015). Characterization and fine mapping of a novel barley stage green-revertible albino gene (HvSGRA) by bulked segregant analysis based on SSR assay and specific length amplified fragment sequencing. BMC Genomics 16, 838. doi: 10.1186/s12864-015-2015-1
Qin, D., Xu, F., Xu, Q., Peng, Y., Ge, S., Dong, J., et al. (2021). Construction of a mutant library in barley and preliminary cytological analysis of leaf width mutants. J. Nucl. Agric. Sci. 35, 9. doi: 10.11869/j.issn.11100-18551.12021.11802.10262
Ramsay, L., Comadran, J., Druka, A., Marshall, D. F., Thomas, W. T., Macaulay, M., et al. (2011). INTERMEDIUM-c, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43, 169–172. doi: 10.1038/ng.745
Rio, D. C., Ares, M., Jr., Hannon, G. J., Nilsen, T. W. (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. pdb.prot5439. doi: 10.1101/pdb.prot5439
Seven, M., Akdemir, H. (2020). DOF, MYB and TCP transcription factors: their possible roles on barley germination and seedling establishment. Gene Expr. Patterns. 37, 119116. doi: 10.1016/j.gep.2020.119116
Shang, Y., Yuan, L., Di, Z., Jia, Y., Zhang, Z., Li, S., et al. (2020). A CYC/TB1-type TCP transcription factor controls spikelet meristem identity in barley. J. Exp. Bot. 71, 7118–7131. doi: 10.1093/jxb/eraa416
Walla, A., Wilma van Esse, G., Kirschner, G. K., Guo, G., Brünje, A., Finkemeier, I., et al. (2020). An acyl-CoA n-acyltransferase regulates meristem phase change and plant architecture in barley. Plant Physiol. 183, 1088–1109. doi: 10.1104/pp.20.00087
Wang, W., He, J., Chen, S., Peng, P., Zhong, W., Wang, X., et al. (2020). Construction of a high-density genetic map and fine mapping of a candidate gene locus for a novel branched-spike mutant in barley. PloS One 15, e0227617. doi: 10.1371/journal.pone.0227617
Wang, B., Smith, S. M., Li, J. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
Wang, Y., Xie, J., Zhang, H., Guo, B., Ning, S., Chen, Y., et al. (2017). Mapping stripe rust resistance gene YrZH22 in Chinese wheat cultivar zhoumai 22 by bulked segregant RNA-seq (BSR-seq) and comparative genomics analyses. Theor. Appl. Genet. 130, 2191–2201. doi: 10.1007/s00122-017-2950-0
Wu, Q., Chen, Y., Xie, J., Dong, L., Wang, Z., Lu, P., et al. (2021). A 36 Mb terminal deletion of chromosome 2BL is responsible for a wheat semi-dwarf mutation. Crop J. 9, 9. doi: 10.1016/j.cj.2020.1006.1015
Xu, D., Huang, Z. (1999). Studies on multinode dwarfing traits in barley-chromosome mapping of multinode dwarfing traits. Crop Sci. 25, 150–156. doi: 10.3321/j.issn:0496-3490.1999.02.003
Yin, C. C., Zhao, H., Ma, B., Chen, S. Y., Zhang, J. S. (2017). Diverse roles of ethylene in regulating agronomic traits in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01676
Youssef, H. M., Eggert, K., Koppolu, R., Alqudah, A. M., Poursarebani, N., Fazeli, A., et al. (2017). VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 49, 157–161. doi: 10.1038/ng.3717
Youssef, H. M., Hansson, M. (2019). Crosstalk among hormones in barley spike contributes to the yield. Plant Cell Rep. 38, 1013–1016. doi: 10.1007/s00299-019-02430-0
Zhang, P., Guo, G., Wu, Q., Chen, Y., Xie, J., Lu, P., et al. (2020b). Identification and fine mapping of spot blotch (Bipolaris sorokiniana) resistance gene Sb4 in wheat. Theor. Appl. Genet. 133, 2451–2459. doi: 10.1007/s00122-020-03610-3
Zhang, B., Li, C., Li, Y., Yu, H. (2020a). Mobile TERMINAL FLOWER1 determines seed size in arabidopsis. Nat. Plants 6, 1146–1157. doi: 10.1038/s41477-020-0749-5
Zhang, J., Zhai, D. (1995). Polyknobby dwarf branched polyspike mutant in barley "93-597". Barley Sci. 03, 46. doi: 10.14069/j.cnki.32-1769/s.1995.03.020
Zhao, Y., Broholm, S. K., Wang, F., Rijpkema, A. S., Lan, T., Albert, V. A., et al. (2020). TCP And MADS-box transcription factor networks regulate heteromorphic flower type identity in gerbera hybrida. Plant Physiol. 184, 1455–1468. doi: 10.1104/pp.20.00702
Keywords: barley, BSR-seq, TCP, multiple stem nodes and spikes and dwarf, fine-mapping
Citation: Qin DD, Liu R, Xu F, Dong G, Xu Q, Peng Y, Xu L, Cheng H, Guo G, Dong J and Li C (2023) Characterization of a barley (Hordeum vulgare L.) mutant with multiple stem nodes and spikes and dwarf (msnsd) and fine-mapping of its causal gene. Front. Plant Sci. 14:1189743. doi: 10.3389/fpls.2023.1189743
Received: 20 March 2023; Accepted: 13 June 2023;
Published: 06 July 2023.
Edited by:
Thomas Hartwig, Heinrich Heine University of Düsseldorf, GermanyReviewed by:
Qin Yang, Northwest A&F University, ChinaYawen Zeng, Yunnan Academy of Agricultural Sciences, China
Filiz Gürel, University of Maryland, College Park, United States
Copyright © 2023 Qin, Liu, Xu, Dong, Xu, Peng, Xu, Cheng, Guo, Dong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengdao Li, Qy5MaUBtdXJkb2NoLmVkdS5hdQ==; Jing Dong, ZG9uZ2ppbmdzaXIwNjFAMTYzLmNvbQ==; Ganggang Guo, Z3VvZ2FuZ2dhbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work and share first authorship
 Dandan D. Qin
Dandan D. Qin Rui Liu1,4†
Rui Liu1,4† Guoqing Dong
Guoqing Dong Qing Xu
Qing Xu Yanchun Peng
Yanchun Peng Le Xu
Le Xu Ganggang Guo
Ganggang Guo Jing Dong
Jing Dong Chengdao Li
Chengdao Li