
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 May 2023
Sec. Plant Bioinformatics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1189038
This article is part of the Research TopicCrop Improvement by Omics and BioinformaticsView all 17 articles
 Wenchen Tang1,4
Wenchen Tang1,4 Yupeng Hao1,4
Yupeng Hao1,4 Xinyu Ma1,4
Xinyu Ma1,4 Yiqi Shi1,4
Yiqi Shi1,4 Yongmeng Dang1,4
Yongmeng Dang1,4 Zeyu Dong1,4
Zeyu Dong1,4 Yongyan Zhao1,4
Yongyan Zhao1,4 Tianlun Zhao1,4
Tianlun Zhao1,4 Shuijin Zhu1,4
Shuijin Zhu1,4 Zhiyuan Zhang1,4
Zhiyuan Zhang1,4 Fenglin Gu2*
Fenglin Gu2* Ziji Liu3*
Ziji Liu3* Jinhong Chen1,4*
Jinhong Chen1,4*The CCCH zinc finger gene family encodes a class of proteins that can bind to both DNA and RNA, and an increasing number of studies have demonstrated that the CCCH gene family plays a key role in growth and development and responses to environmental stress. Here, we identified 57 CCCH genes in the pepper (Capsicum annuum L.) genome and explored the evolution and function of the CCCH gene family in C. annuum. Substantial variation was observed in the structure of these CCCH genes, and the number of exons ranged from one to fourteen. Analysis of gene duplication events revealed that segmental duplication was the main driver of gene expansion in the CCCH gene family in pepper. We found that the expression of CCCH genes was significantly up-regulated during the response to biotic and abiotic stress, especially cold and heat stress, indicating that CCCH genes play key roles in stress responses. Our results provide new information on CCCH genes in pepper and will aid future studies of the evolution, inheritance, and function of CCCH zinc finger genes in pepper.
Zinc finger proteins (ZFPs), which are named for their ability to bind zinc to form a stable finger-like structure, are sequence-specific transcription factors that usually contain varying numbers of cysteine (Cys) and histidine (His) residues. Cys and His are used to chelate zinc ions to form a zinc finger structure, which can recognize and bind to DNA (Hall, 2005). Zinc finger proteins are also associated with the metabolism of different types of RNAs in organisms (Hall, 2005) and can specifically bind to DNA, RNA, and DNA–RNA complexes to regulate gene expression. Several gene families have been identified in plants based on their function and structure, including the RING finger (Freemont, 1993; Kosarev et al., 2002), CCCH (Li et al., 2001), DOF (Lijavetzky et al., 2003), WRKY (Zhang and Wang, 2005), ERF (Nakano et al., 2006), and LIM (Arnaud et al., 2007) families. Zinc finger protein motifs can be divided into different types according to the number of conserved Cys and His residues and the spacing between these residues, such as C2H2, C8, C6, C3HC4, C2HC5, C4, C4HC3, and CCCH (Berg and Shi, 1996; Takatsuji, 1998; Moore and Ullman, 2003; Schumann et al., 2007). CCCH zinc finger proteins generally contain at least one zinc finger motif. Three Cys and one His residue are the most important components of this motif. The common sequence of the CCCH motif can be defined as C-X4-15-C-X4-6C-X3-4-H (where X stands for any amino acid, numbers indicate the number of amino acids, C is Cys, and H is His), and C-X7-8-C-X5-C-X3-H is the largest sequence among CCCH proteins (Wang et al., 2008).
CCCH zinc finger proteins are involved in plant development, adaptation, hormonal regulation, and the regulation of processes related to physiological adversity, especially responses to biotic and abiotic stress. In Arabidopsis, AtTZF1, which consists of two zinc finger motifs separated by 18 amino acids, is a CCCH-type zinc finger protein (Iuchi and Kuldell, 2005). Overexpression of AtTZF1 enhances the tolerance of Arabidopsis thaliana to cold and drought stress and affects the growth and stress responses mediated by abscisic acid (ABA) and gibberellic acid (GA) (Lin et al., 2011). The expression patterns of AtTZF1, AtTZF2, and AtTZF3 are similar (Lee et al., 2012). AtC3H49/AtTZF3 and AtC3H20/AtTZF2 can regulate growth rate, plant size, leaf and flower morphology, as well as aging and lifespan. Overexpression of these two genes can attenuate transpiration, enhance drought tolerance, alter growth patterns, and delay senescence (Lee et al., 2012). In addition, the CCCH zinc finger proteins HUA1 and HUA2 play a role in AGAMOUS pre-mRNA processing and in floral reproductive organ identity (Li et al., 2001; Cheng et al., 2003). In rice, OsTZF1 improves stress tolerance by regulating the RNA metabolism of stress-responsive genes (Jan et al., 2013). GhZFP1 in cotton contains two typical zinc finger motifs (C-X8-C-X5-C-X3-H and C-X5-C-X4-C-X3-H) that improve drought and disease resistance in transgenic tobacco (Guo et al., 2009). The overexpression of GmZF351 in transgenic soybeans activates lipid biosynthesis genes, accelerates the accumulation of seed oil, and thus increases the seed oil content (Li et al., 2017). In cucumber, CsSEF1 encodes protein containing three conserved zinc finger motifs, two of which are CCCH motifs. The expression of CsSEF1 is up-regulated in leaves and flowers; it plays a role in later developmental stages after embryogenesis and the signal transduction pathway of fruits from photoassimilate limitation to the sink organs (Grabowska et al., 2009; Tazuke and Asayama, 2013). In pepper, the CCCH zinc finger protein CaC3H14 regulates antagonistic interactions between salicylic acid (SA) and jasmonic acid (JA)/ethylene (ET) signaling, which enhances the resistance of plants to Ralstonia solanacearum infection (Qiu et al., 2018).
A total of 68, 67, 68, 91, 34, 62, 80, 89, 103, 116, 31, and 86 CCCH zinc finger family genes have been identified in Arabidopsis (Wang et al., 2008), rice (Wang et al., 2008), maize (Peng et al., 2012), poplar (Chai et al., 2012), Medicago truncatula (Zhang et al., 2013), citrus (Liu et al., 2014), tomato (Xu, 2014), banana (Mazumdar et al., 2017), cabbage (Brassica rapa) (Pi et al., 2018), soybean (Hu and Zuo, 2021), rose (Li et al., 2021), and tobacco (Tang C. et al., 2022), respectively. Although CCCH zinc finger proteins play an important role in many aspects of plant growth and development, no systematic studies have been conducted to analyze and identify members of the CCCH gene family in pepper to date.
Pepper has the highest vitamin C content among all vegetables, which can promote appetite and improve digestion. Whole-genome sequencing and bioinformatics analysis can be used to identify and analyze CCCH zinc finger genes involved in the growth and development, metabolism, and adaptation to stress in pepper plants (Kim et al., 2014; Qin et al., 2014). Here, we identified 57 CCCH zinc finger genes in the pepper genome. We also systematically analyzed the phylogenetic structure, domains, conserved motifs, chromosome localization, duplication events, collinearity, and tissue-specific expression patterns of these CCCH zinc finger genes, and this provided insights into the roles of CCCH gene family members in the growth and development of pepper plants. Finally, the published RNA sequencing (RNA-seq) data were used to investigate the expression of CCCH genes in different tissues, such as the roots, stems, and leaves, and the expression patterns of the genes were validated using quantitative real-time polymerase chain reaction (qRT-PCR). These results provide new insights that will aid future studies of the functions of candidate genes involved in the growth, development, adaptation, hormone regulation, and stress physiology of pepper plants.
In this study, we used genomic data from Capsicum annuum cv. CM334. First, we downloaded amino acid sequences for all Capsicum proteins from the Phytozome database1 (Tuskan et al., 2006; Goodstein et al., 2012) and amino acid sequences for CCCH (PF00642, Zinc finger C-X8-C-X5-C-X3-H type, and similar sequences) from the Pfam database2 (El-Gebali et al., 2019). The CCCH motif was used to retrieve the amino acid sequence of peppers in hmmsearch3 with a threshold of E-value < 1 × 10-5. All the obtained protein sequences were submitted to the Pfam database and SMART domain search database3 to confirm the structural integrity of the zf_CCCH domain. Furthermore, we made use of the Pfam2 and SMART4 databases to clarify the structural integrity of the ZF_CCCH domain (Schultz et al., 2000). We extracted sequences of the conserved domains from the identified pepper CCCH proteins. We used the ExPASy tool5 (Gasteiger et al., 2005) to calculate the number of amino acids, isoelectric point (pI), molecular weight (Mw), and other physical and chemical properties of the zinc finger CCCH protein sequences.
We downloaded amino acid sequences for pepper, tomato, and rice from the Phytozome database1. Arabidopsis CCCH zinc finger genes were identified from the Arabidopsis information resource website6. Sequences were aligned using the neighbor-joining method, and the evolutionary tree was constructed in MEGA 11 software (Kumar et al., 2018). Branch support was tested by performing 1,000 bootstrap replications. The phylogenetic tree was uploaded in Newick format to the EvolView web server7 to visualize the tree. The subfamily classification of the Capsicum CCCH gene family was based on a previously published classification for Arabidopsis thaliana (Wang et al., 2008). MCScanX8 was used to characterize syntenic relationships among CCCH genes in Arabidopsis, tomato, and pepper.
We downloaded genome sequences and coding sequences from the Phytozome database1 to analyze the structure of CCCH gene family members. The structure of the CCCH genes was plotted using TBtools (Chen et al., 2020). MEME Suite Version 5.4.19 was used to identify the conserved motifs of CCCH gene family members in pepper, with the maximum motif search number set to 10, and other parameters set to their default values. Any repetitions were considered a motif position that was distributed throughout the sequence (Bailey et al., 2009).
Detailed chromosomal mapping was obtained from GFF genomic files downloaded from the Phytozome database1 to visualize the chromosomal distribution of the CCCH genes in pepper in TBtools (Chen et al., 2020). We also identified tandem duplication events in CCCH family genes using MCScanX in TBtools. MCScanX in TBtools and BLASTP searches were used to identify the segmental duplication events of CCCH genes in pepper and clarify collinearity relationships between genes in different species (Wang et al., 2012; Chen et al., 2020). The non-synonymous (Ka) and synonymous (Ks) substitutions between gene pairs was calculated by using TBtools.
We analyzed the expression profiles of pepper CCCH zinc finger genes in different tissues, under different types of biotic stress and abiotic stress, and in the presence of different phytohormones by downloading the following RNA-seq datasets from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus10: flower, root, stem, placenta, and pericarp (stage 1, 2, and 3) of pepper plants during the mature green (MG) stage, breaker (B) stage, and 5 and 10 days after the breaker stage (BioProject ID: PRJNA223222); 30 min, 4 h, 1 day, 2 days, and 3 days after infection with PepMoV and TMV (BioProject ID: PRJNA223222); 1, 3, 6, 12, and 24 h under cold, heat, drought, and salt stress (BioProject ID: PRJNA525913); and 1, 3, 6, 12, and 24 h after MeJA, SA, ET, and ABA treatment (BioProject ID. PRJNA634831) (Kim et al., 2014; Kang et al., 2020; Lee et al., 2020). The fragments per kilobase of exon model per million mapped reads (FPKM) values were calculated using Hisat2 (v2.0.5) and Sringtie (v2.1.7) software with the following formula: log(FPKM+1). These data were then visualized using the ‘pheatmap’ package in R software.
In this experiment, gene expression levels of CCCH genes were detected using the pepper cultivar CM334. All pepper plants were sown and grown under greenhouse conditions (16 h light/8 h dark, 25-28°C). When peppers had six true leaves, the experimental groups were subjected to cold treatment (16 h light/8 h dark, 10°C) and heat treatment (16 h light/8 h dark, 40°C) in the incubator, and the leaves were collected at 0, 3, 6, 12, 24, and 72 h after the treatment. Three replicates were collected from three different plants, immediately frozen in liquid nitrogen, and then stored in a -80°C refrigerator.
The RNA sample was extracted using an RNAprep Pure Plant Plus Kit (Tiangen) according to the manufacturer’s instructions. The DNAase-treated RNA was reverse-transcribed with M-MLV (RNase H-) reverse transcriptase. qRT-PCR was performed using a CFX96TM Real-Time system (Applied Biosystems). Primers (20-24 bp) were designed using the Primer-BLAST tool in NCBI, and the amplicon lengths were 80-220 bp (Supplementary Table 1). All settings were set to their default values. Three technical replicates were performed for each gene, and UBI3 was used as the internal reference gene. The total volume of each reaction was 20 µL, which consisted of 2 µL of cDNA, 1 µL of gene-specific primers, 7 µL of ddH2O, and 10 µL of 2× ChamQ Universal SYBR qPCR Master Mix reagent. The thermal cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. At the end of the cycle, a solubility-free curve was generated to analyze the expression of each gene tested.
In this study, 57 CCCH genes were identified from the C. annuum cv. CM334 genome using the Hidden Markov Model of LEA against the genome database of C. annuum. These CCCH genes were renamed from PEPTY1 to PEPTY57 according to their order on chromosome 1-12 (Supplementary Table 2). All identified CCCH genes encoded proteins ranging from 295 to 1015 amino acids, and their predicted isoelectric points (pI) ranged from 4.7 to 9.39. To investigate the sequence characteristics of the most common CCCH motifs in the pepper CCCH zinc finger proteins, we extracted amino acid sequences from CCCH conserved regions (Thompson et al., 1997). The CCCH domain mainly consisted of a triple cysteine and a histidine, and the following motif was commonly observed (C-X7-8-C-X5-C-X3-H) (Supplementary Figure 1).
We constructed a phylogenetic tree using the entire amino acid sequence of each member of pepper, Arabidopsis, tomato, and rice to explore the evolutionary relationships among CCCH zinc finger genes. As shown in Figure 1, the pepper CCCH zinc finger genes were divided into 12 groups based on previous studies of Arabidopsis. The number of CCCH zinc finger genes in each group was uneven. Group XII was the largest (13 CCCH zinc finger genes), followed by Group I (8 CCCH zinc finger genes) and Group II, VII, and VIII (each with 2 CCCH zinc finger genes). Group III, IV, V, VI, IX, X, and XI have 3, 4, 5, 3, 3, 6, and 6 CCCH zinc finger genes, respectively.
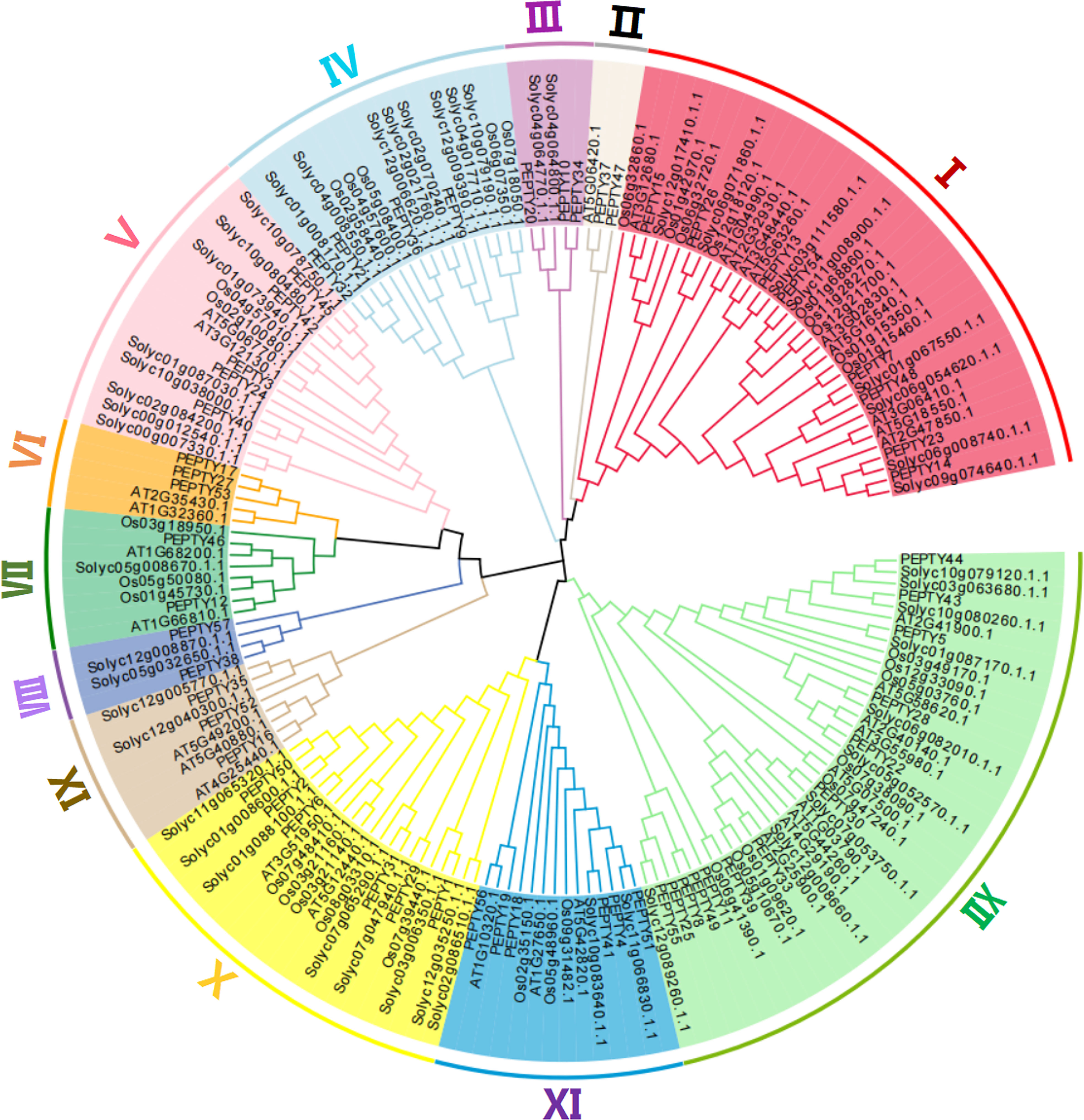
Figure 1 Evolutionary tree of CCCH genes in Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, and Capsicum annuum. The different shades of color correspond to different subgroups.
We performed a structure analysis of the 57 CCCH zinc finger genes in pepper. All the CCCH genes had introns and exons, but they varied greatly in size and number. The number of exons ranged from 1 to 14 (Supplementary Table 3). Most of the genes had less than 10 exons. The average number of exons per gene was 5.4. Genes in Group VI and VII both had two exons, and genes in Group XII contained only one exon. However, genes in Group VIII had 10 exons. Subsequently, the conserved motifs of the CCCH genes in pepper were identified using the online MEME suite program. Ten conserved motifs were detected, ranging from 6 to 50 amino acids in length (Figure 2; Supplementary Table 3). Unsurprisingly, the structure of the genes in the same subclade was similar. The five conserved motifs 1, 5, 6, 7, and 8, were all found in Group I. Motif 5, 7, and 8 had the C-X8-C-X5-C-X3-H structure. Motif 4 was only present in Group X, motif 5 was widely present in Group V and VI, motif 9 was only present in Group XII, and motif 10 (C-X7-C-X5-C-X3-H) was only present in Group XI. Most genes in the same branch had similar conserved motif compositions and structures, which suggests that they were functionally similar.
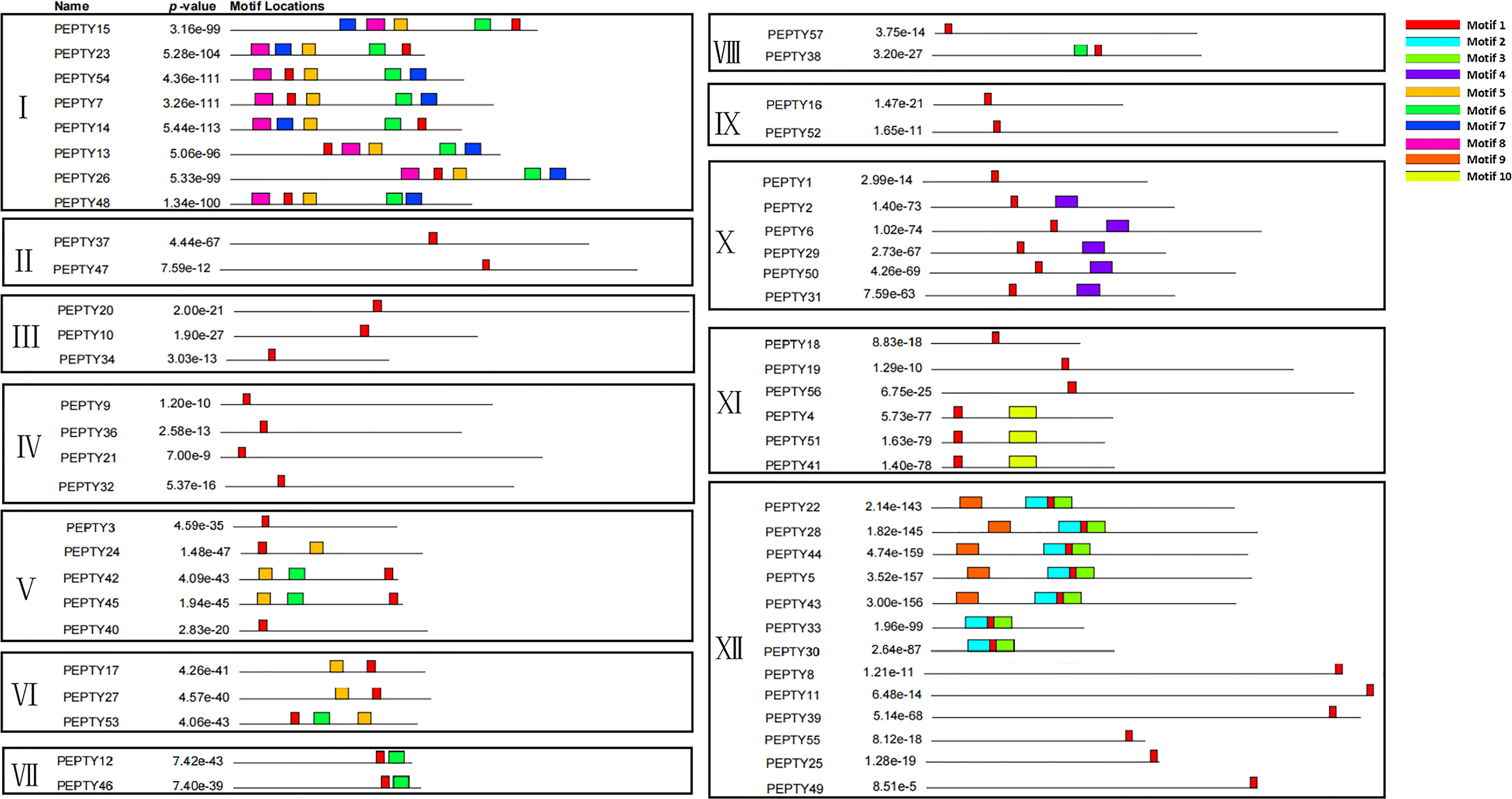
Figure 2 Protein motifs of the CCCH gene family in pepper. The colorful boxes delineate different motifs. The clustering was performed according to the results of the phylogenetic analysis.
Using the pepper genome annotation information and TBtools (Tuskan et al., 2006; Chen et al., 2020), we characterized the chromosomal distribution of CCCH zinc finger genes. A total of 55 of the 57 CCCH zinc finger genes identified could be mapped on chromosomes; PEPTY56 and PEPTY57 were the two genes that could not be mapped. As shown in Figure 3, these 55 CCCH genes were unevenly distributed across the 12 chromosomes, and the number of genes on each chromosome was not related to chromosome size. For example, the largest chromosome (Chr 01) contained seven CCCH genes; however, the chromosome containing the most genes was Chr 11, which had eight CCCH genes. Chr 05 and 12 had only two CCCH genes, which was the same number of CCCH genes contained on the shortest chromosome (Chr 08).
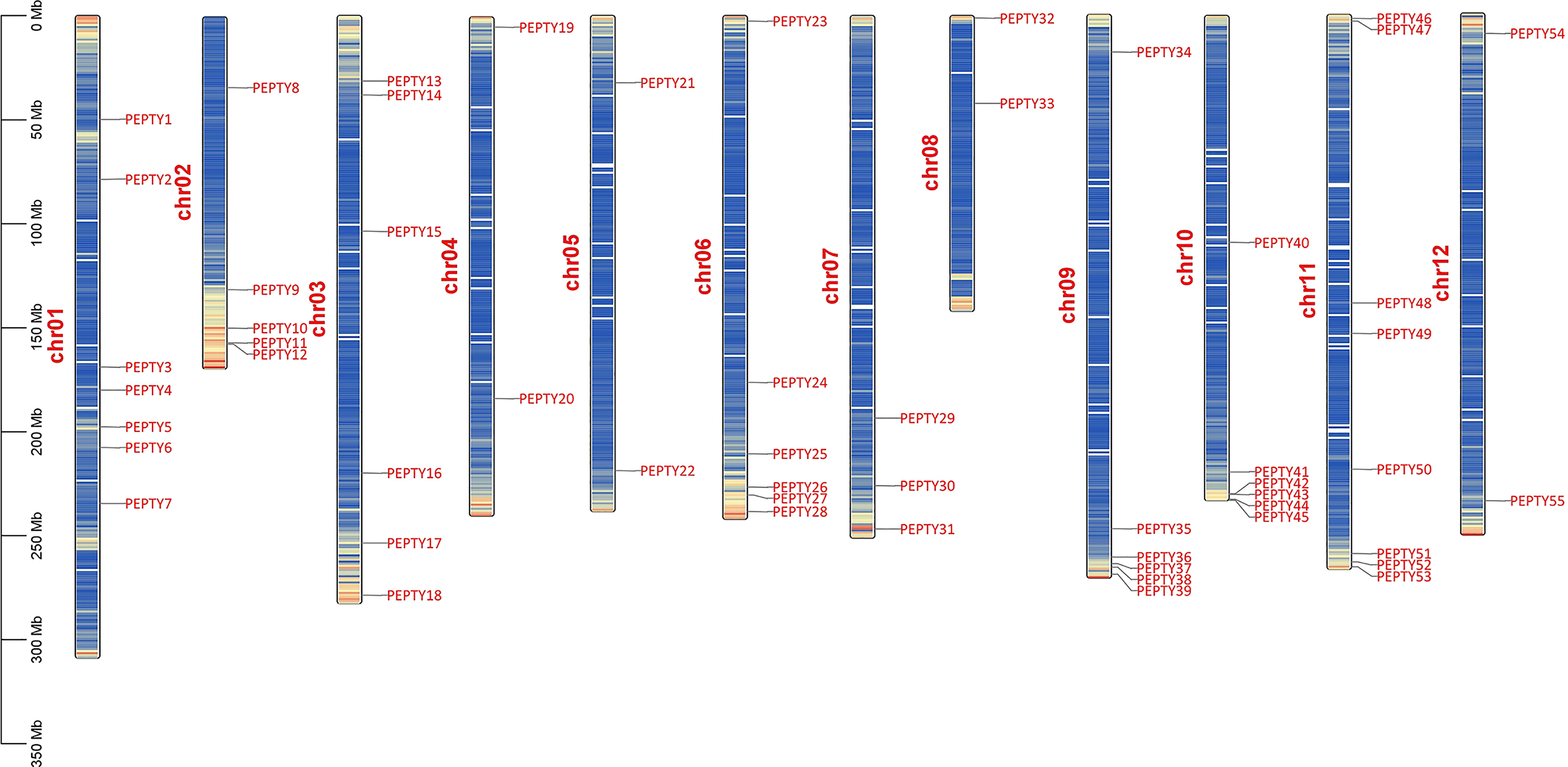
Figure 3 Chromosomal distribution of CCCH genes in pepper. Chr01–12 indicate chromosomes 01–12. Bands on the chromosomes indicate gene density.
Next, we identified tandem duplication events using the Multiple Collinearity Scan toolkit (MCScanX) in TBtools. No tandem duplication events were identified. Thus, we identified segmental duplication events using MCScanX in TBtools and BLASTP searches (Wang et al., 2012; Chen et al., 2020). A total of 5 segmentally duplicated gene pairs were detected, and these were detected across nine chromosomes (Figure 4). On chromosomes 10, 2 pairs of genes (PEPTY42/PEPTY45 and PEPTY43/PEPTY44) on the same chromosomes appear to be products of segmental duplication events. Segmental duplication events were not detected on Chr 01, 04, 07, 09, and 12. These findings indicate that segmental duplication events appear to have played a key role in shaping the diversity of CCCH genes in pepper.
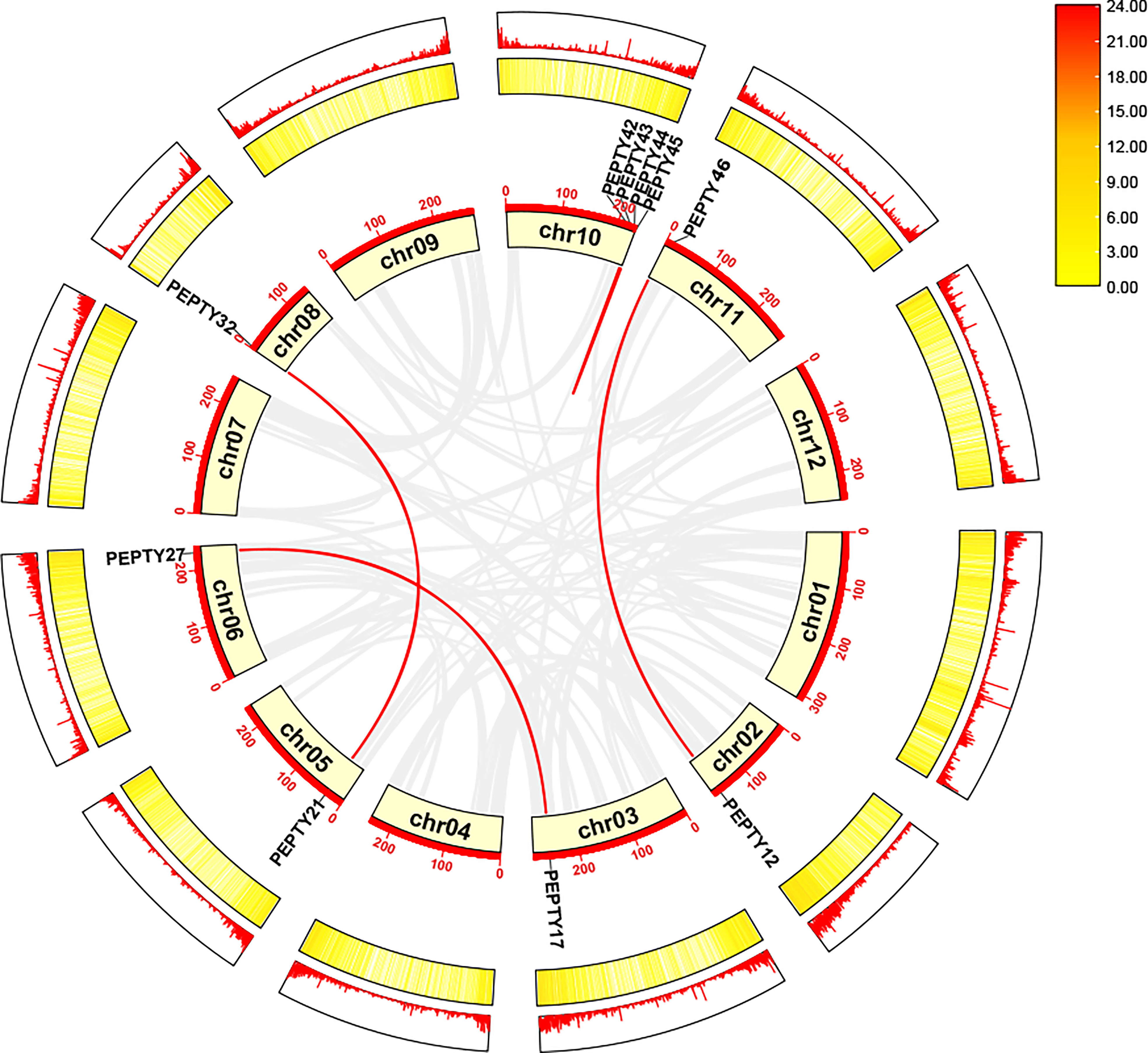
Figure 4 Collinearity analysis of the CCCH gene family in pepper. Chromosomes 01–12 are represented by yellow rectangles. The gray lines indicate syntenic blocks in the pepper genome, and the red lines between chromosomes delineate segmentally duplicated gene pairs.The outermost heatmap and lines represent gene density on the chromosomes.
We also investigated collinearity relationships between pepper CCCH genes and associated genes from Arabidopsis and Solanum lycopersicum to identify homologous genes. Collinearity relationships were observed between 14 pepper genes and 20 Arabidopsis genes and between 40 pepper genes and 42 tomato genes. A total of 21 pairs of homologous genes were identified between pepper and Arabidopsis, and 47 pairs of homologous genes were identified between pepper and tomato (Supplementary Figure 3). The logarithm of homologous genes with tomato was twice that of homologous genes with Arabidopsis; and this is likely because the closer phylogenetic relationship between pepper and tomato (both in the family Solanaceae) than between pepper and Arabidopsis. To assess the selective constraint pressure of gene pairs, Ka/Ks calculations were performed in TBtools (Supplementary Table 4). Most gene pairs have Ka/Ks ratios below 1, indicating that purification selection may have been undertaken during evolution.
We characterized the expression of pepper CCCH genes in five tissues: flower, root, stem, placenta, and pericarp tissue (Figure 5; Supplementary Table 5). PEPTY24 was expressed at high levels in flowers and at low levels in the roots and stems; PEPTY12 and PEPTY46 were expressed at high levels in stems, but their expression gradually decreased in the roots and flowers as development advanced. PEPTY29 was most highly expressed in the flowers, followed by the roots and stems. In placenta period, the expression of PEPTY10 gradually increased with developmental stage. The expression of PEPTY30 was the highest in the initial breaker stage. The expression of PEPTY35 was up-regulated at the early developmental stage in the placenta and was down-regulated at the breaker stage. In pericarp period, the expression of PEPTY10 was significantly up-regulated at day 10 of the breaker stage. The expression of PEPTY2 was high at stage 1 in both the placenta and pericarp period (PL1 and PR1) and decreased thereafter. The expression of CCCH might vary among organs and at different growth and developmental stages. Some of these genes such as PEPTY24 and PEPTY30 are likely involved in the growth and development of pepper.
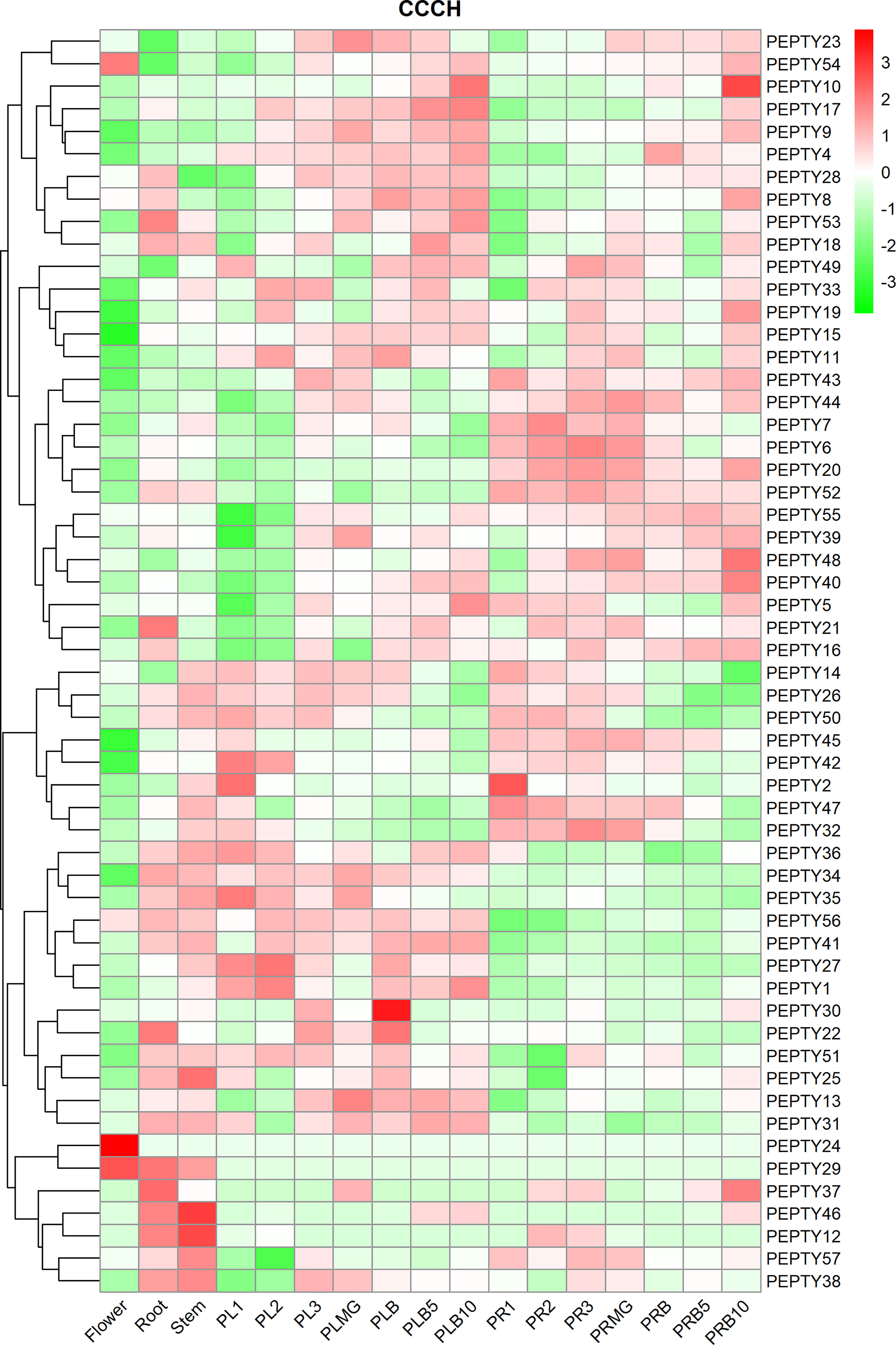
Figure 5 Hierarchical clustering of expression profiles of pepper CCCH genes in different organs. The heatmap was constructed using the ‘pheatmap’ package in R software, and the fragments per kilobase of exon model per million mapped reads (FPKM) values of the CCCH genes were converted to log(FPKM+1) values. The different tissues included flower, root, stem, placenta (PL), and pericarp (PR). MG denotes mature green, and B denotes breaker. 1, 2, and 3 indicate stage. 5 and 10 indicate days. Red indicates a high relative abundance of transcripts. Green indicates a low relative abundance of transcripts.
Analysis of the relative transcript abundance of PEPTY genes under different types of abiotic stress revealed that the expression of many of these genes was significantly up-regulated under cold, heat, drought (D-mannitol) and salt (sodium chloride, NaCl) stress (Figure 6; Supplementary Table 5). The expression of PEPTY2, PEPTY5, PEPTY7, PEPTY8, PEPTY11, PEPTY16, PEPTY36, PEPTY45, and PEPTY57 was up-regulated under cold stress. The expression of PEPTY4, PEPTY9, PEPTY26, PEPTY32, PEPTY34, PEPTY42, PEPTY51, and PEPTY52 was significantly up-regulated at all time points under heat stress. The expression of PEPTY6, PEPTY31, PEPTY32, and PEPTY48 was highest at 12, 6, 24, and 12 h, respectively. By contrast, the expression of PEPTY14, PEPTY30, PEPTY40, and PEPTY46 was up-regulated at 24, 72, 24, and 72 h, respectively, under salt stress. Under drought stress, the expression of PEPTY5, PEPTY10, PEPTY14, PEPTY23, PEPTY39, and PEPTY40 was up-regulated.
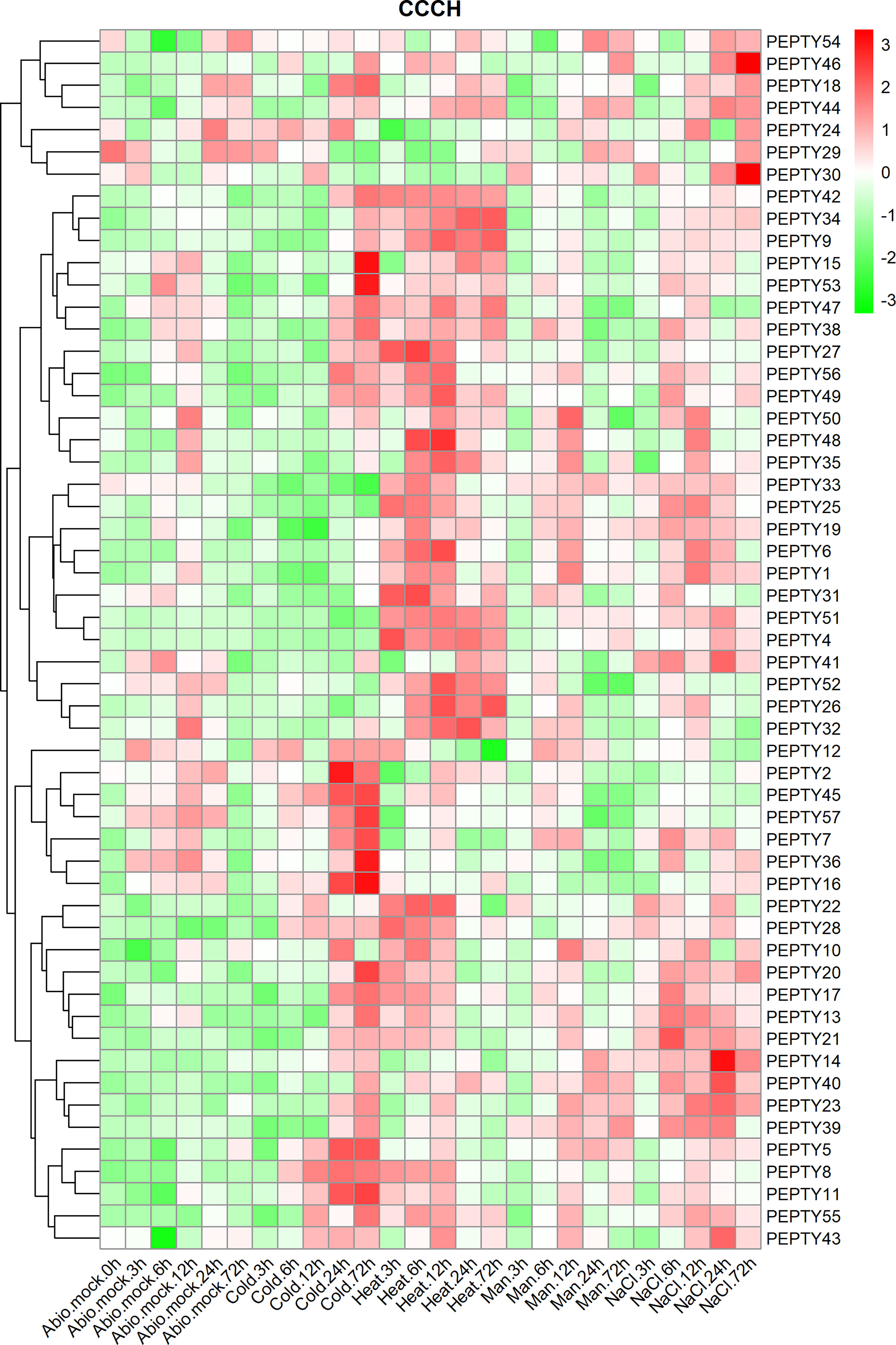
Figure 6 Expression profiles of pepper CCCH genes under different types of abiotic stress. Abiotic stresses included cold, heat, drought (D-mannitol), and salt (NaCl). Time points include 1, 3, 6, 12, and 24 h. The control group is indicated by Abio.mock labels. Red indicates a high relative abundance of transcripts. Green indicates a low relative abundance of transcripts.
The expression of CCCH genes after treatment with two viruses was performed to clarify their responses to biotic stress (Figure 7; Supplementary Table 5). The expression of PEPTY22 following pepper mottle virus (PepMoV) treatment was highest 30 min post-treatment and decreased thereafter. The expression of most genes, such as PEPTY8, PEPTY11, and PEPTY54, was up-regulated 4 h post-treatment. By contrast, the expression of PEPTY22 was significantly up-regulated 30 min after treatment with tobacco mosaic virus (TMV), which was consistent with its response to PepMoV treatment. The expression of PEPTY4 and PEPTY46 was high 4 h after TMV treatment. In addition, the expression of PEPTY20, PEPTY28, PEPTY30, PEPTY40, and PEPTY53 was high 2 days after TMV treatment. The expression of PEPTY25 and PEPTY33 was high 3 days after TMV treatment. The responses of most CCCH genes were more pronounced to TMV treatment than to PepMoV treatment.
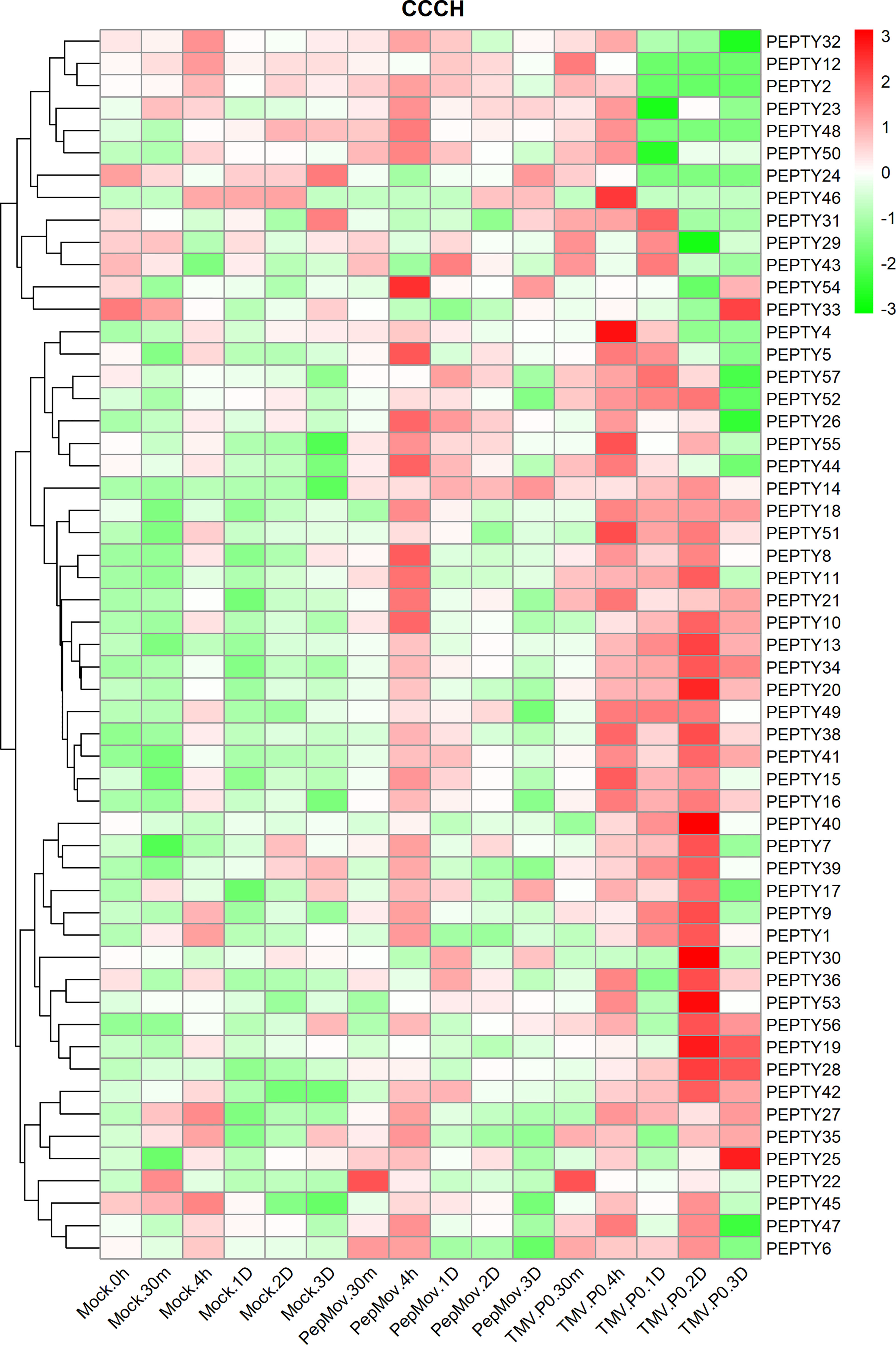
Figure 7 Expression profiles of pepper CCCH genes under different types of biotic stress. Biotic stresses included pepper mottle virus (PepMoV) and tobacco mosaic virus (TMV). Time points include 30 min, 4 h, 1 d, 2 d, and 3 d. The control group is indicated by mock labels. Red indicates high relative abundance of transcripts. Green indicates low relative abundance of transcripts.
Ultimately, the expression profiles of CCCH genes were further analyzed under treatment with four phytohormones. The results are shown in Figure 8. The expression of PEPTY8, PEPTY14, PEPTY22, PEPTY35, PEPTY44, PEPTY55, and PEPTY56 was increased after methyl jasmonate (MeJA) treatment. The expression of 13 genes (PEPTY4, PEPTY13, PEPTY15, PEPTY26, PEPTY27, PEPTY28, PEPTY34, PEPTY35, PEPTY41, PEPTY42, PEPTY43, PEPTY53, and PEPTY56) increased after SA treatment. The expression of PEPTY35, PEPTY41, PEPTY42, PEPTY43, PEPTY53, and PEPTY56 was up-regulated after SA treatment. The expression of PEPTY37 significantly increased 3 h after ET treatment. This gene was not expressed in the other treatments or the control. However, the expression of PEPTY9, PEPTY11, PEPTY20, PEPTY21, and PEPTY49 was down-regulated. The expression of PEPTY21 and PEPTY43 was up-regulated after ABA treatment, especially at 12 h, and the expression of PEPTY46 was more significantly up-regulated at 24 h. These results suggest that CCCH genes play a role in the response to phytohormones.
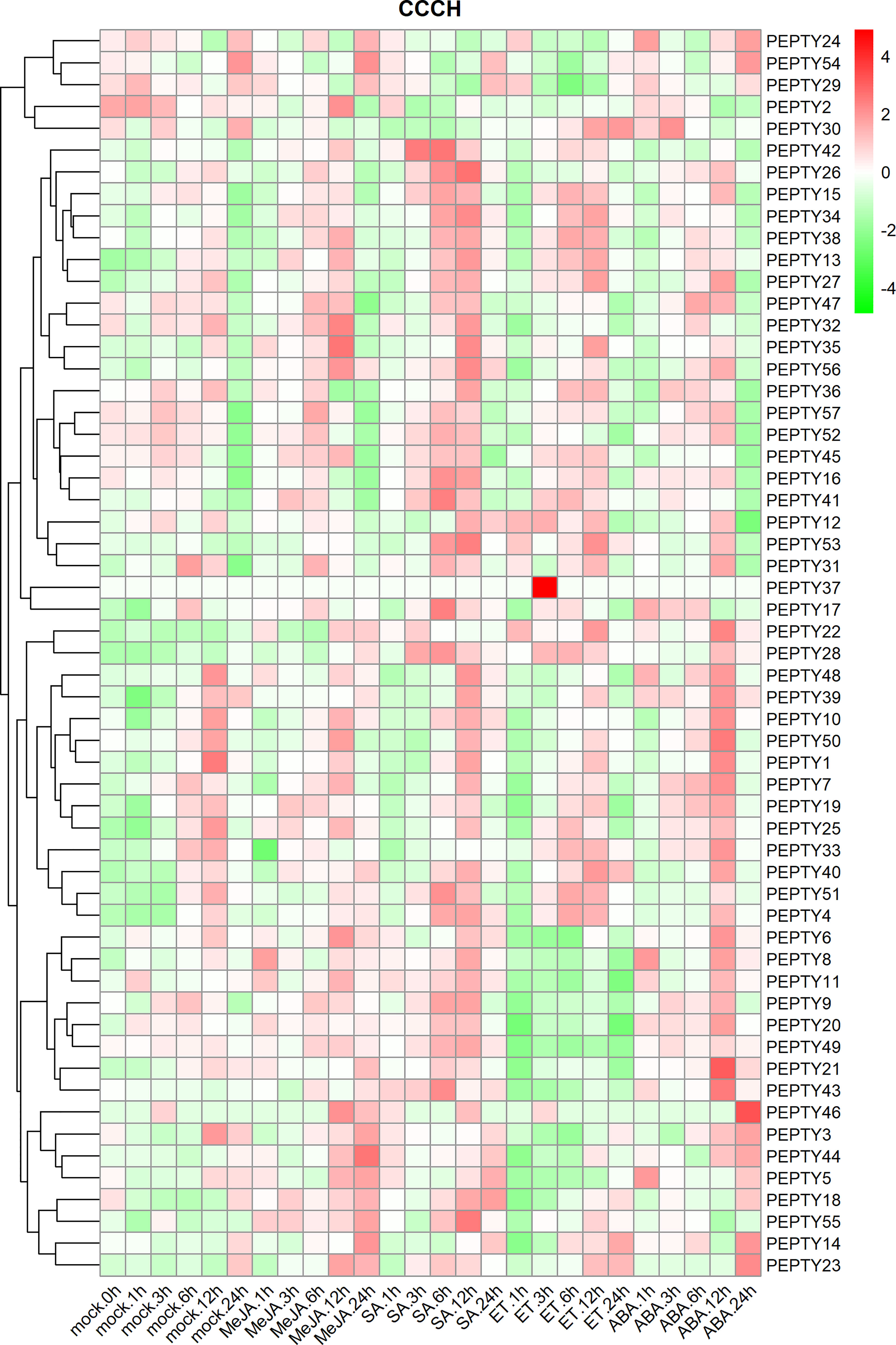
Figure 8 Expression profiles of pepper CCCH genes under phytohormone treatments. The phytohormone treatments included methyl jasmonate (MeJA), salicylic acid (SA), ethylene (ET), and abscisic acid (ABA). Time points include 1, 3, 6, 12, and 24 h. The control group is indicated by a mock label. Red indicates a high relative abundance of transcripts. Green indicates a low relative abundance of transcripts.
We conducted qRT-PCR analysis on 5 genes that were significantly up-regulated under cold treatment and 7 genes with expression patterns that varied under heat treatment in the heat map (Figure 9). Under cold stress, the expression of four genes (PEPTY12, PEPTY16, PEPTY36, and PEPTY57) peaked at 72 h, whereas the expression of PEPTY45 peaked at 24 h. The expression of all these genes did not significantly differ from that of the control under cold treatment in the early stage; however, at 72 h, the expression of genes under cold treatment was at least two-fold higher than that of genes in the control group. A similar pattern was observed for PEPTY4, PEPTY9, PEPTY26, PEPTY27, PEPTY34, PEPTY51, and PEPTY52 under heat treatment, and the significance of differences was even more pronounced. The expression of EPTY4, PEPTY9, PEPTY27, PEPTY34, PEPTY51, and PEPTY52 peaked at 72 h, whereas the expression of PEPTY26 peaked at 24 h. Differences in the expression of PEPTY4, PEPTY9, and PEPTY51 between the heat treatment and control group gradually increased over time.
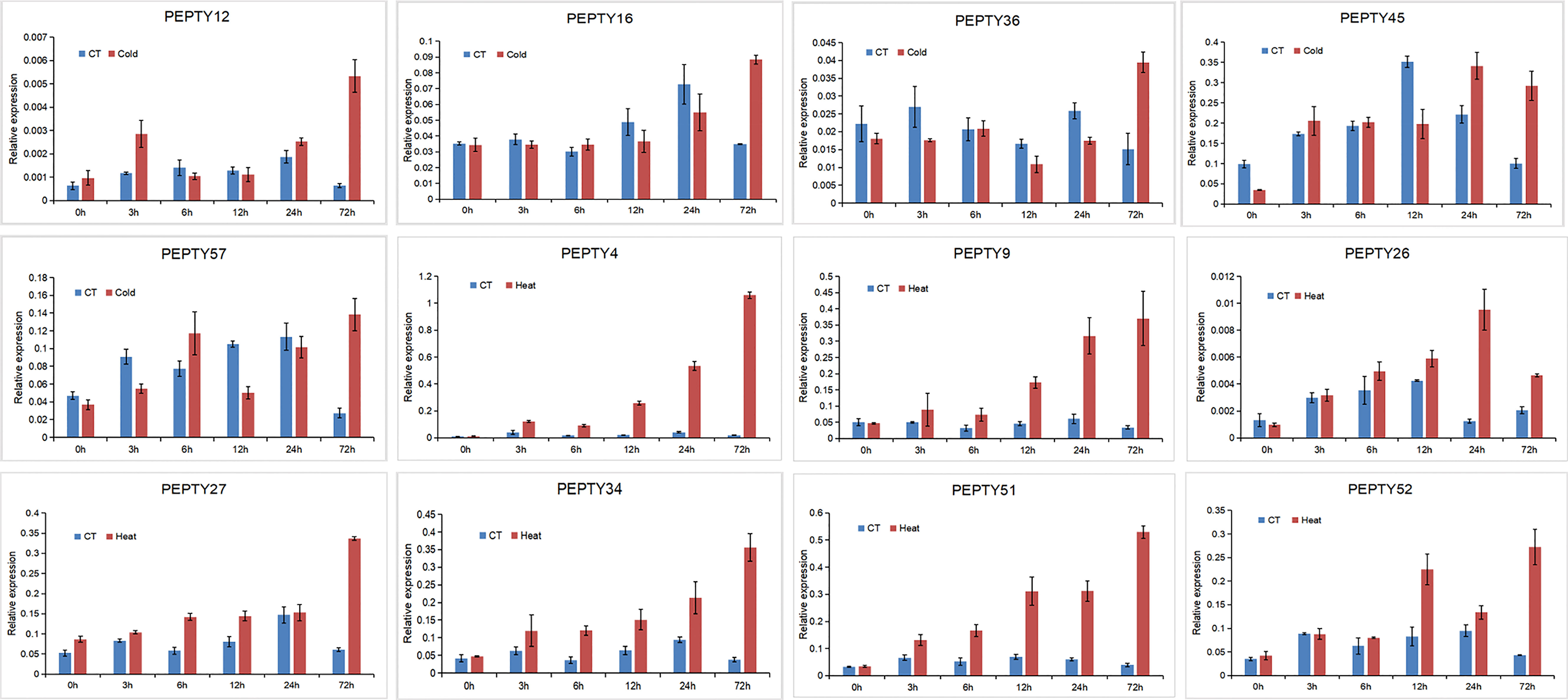
Figure 9 qRT-PCR analysis of 12 pepper CCCH genes under different stress treatments. The x-axis shows the time points after stress treatments. The y-axis shows the relative expression levels normalized to the reference gene UBI3. Data are mean ± SD of three technical replicates.
C. annuum is one of the most widely grown solanaceous vegetables worldwide and capsaicin produced from seed of C. annuum is an economically important spice, medicine, vegetable, and biopesticide. However, previous studies have shown that pepper plants are highly sensitive to biotic and abiotic stresses, such as pathogens, drought, cold, and heat (Kim et al., 2014; Kang et al., 2020; Lee et al., 2020). CCCH proteins have been identified in plants. These proteins are rather unusual in that they can regulate the expression of genes by binding to mRNA in addition to DNA (Kim et al., 2014; Qin et al., 2014). Functional analyses of CCCH genes in Arabidopsis, rice, maize, poplar, alfalfa (Medicago truncatula), citrus, tomato, banana, cabbage, soybean, rose, tobacco, and other plants have been conducted (Wang et al., 2008; Chai et al., 2012; Peng et al., 2012; Zhang et al., 2013; Liu et al., 2014; Xu, 2014; Mazumdar et al., 2017; Pi et al., 2018; Hu and Zuo, 2021; Li et al., 2021; Tang C. et al., 2022).
We identified 57 CCCH zinc finger genes in the genome of C. annuum cv. CM334. A total of 80 CCCH genes have been identified in tomato belonging to (Xu, 2014), which is also a member of the family Solanaceae. We searched for CCCH genes in the C. annuum L. Zunla-1 genome. However, this species only had 69 CCCH genes (Supplementary Table 6), which was lower than in tomato. The CCCH genes in CM334 could be divided into 12 subfamilies, and Group III and VIII genes were only present in pepper and tomato, but not in Arabidopsis thaliana and rice (Figure 1). These subfamilies are likely unique to the Solanaceae family.
Structural analysis of the CCCH genes revealed that the CCCH motifs are highly conserved, motif type and motif position were highly similar within each subfamily, but motif type and motif position varied among most subfamilies. The similarity and specificity within and between subfamilies, respectively, indicated that genes in the same subfamily may have similar functions, and genes in different subclades may perform different functions. No motifs in PEPTY35 were in Group IX, and 56.1% of pepper CCCH genes had at least two motifs. The main structures present were C-X5-C-X4-C-X3-H and C-X7-8-C-X5-C-X3-H.
Gene duplication is one of the primary drivers of the evolution of genomic and genetic systems. Duplicated genes have the potential to develop new functions. Gene family expansion in the genome generally stems from tandem and segmental duplication events (Moore and Purugganan, 2003; Cannon et al., 2004; Levasseur and Pontarotti, 2011). In Group V, there are five pepper CCCH genes (PEPTY3, PEPTY24, PEPTY40, PEPTY42, and PEPTY45), but only two Arabidopsis CCCH genes (AtC3H36 and AtC3H52) and two rice CCCH genes (OsC3H14 and OsC3H31). Two homologs of Arabidopsis or rice were likely generated by segmental duplication, and the pepper CCCH genes likely underwent one round of whole-genome duplication and one tandem duplication.
The expression levels of CCCH genes in pepper varied significantly among tissues and developmental stages (Chai et al., 2012; Li et al., 2021). Only the expression of PEPTY24, PEPTY29, and PEPTY54 was up-regulated in flowers. The expression of PEPTY24 was specific to flowers, which may be involved in the regulation of flowering in pepper. PEPTY29 was expressed in flower, root, and stem, but not in placenta and pericarp; this gene might thus be involved in regulating flower, root, and stem development. Twenty-five genes were expressed in the roots, and 27 genes were expressed in the stems. The expression patterns of CCCH genes in pepper differ from those of CCCH genes in Arabidopsis and rice, where most CCCH genes are expressed in the roots, inflorescences, leaves, and seeds (Wang et al., 2008).
The expression profiles of CCCH genes under biotic stress, abiotic stress, and phytohormone treatments showed that most PEPTY genes were highly expressed under these conditions. Comparison with other studies confirmed that the activity of most CCCH zinc finger proteins can be induced by hormones such as ABA and GA; they may play a role in hormone-mediated signaling pathways (Verma et al., 2016; Han et al., 2021). This pattern of activity is similar to that observed under biotic and abiotic stress; it is even likely that a particular gene could respond to multiple different treatments. For example, in rice, the OsTZF1 gene responds to GA, MeJA, and salicylate (Jan et al., 2013). In Arabidopsis, the expression of AtOZF1 was highly induced by ABA and salinity treatment (Huang et al., 2011). High expression of AtTZF2 and AtTZF3 enhances tolerance to high salt stress, and the silencing of these two genes reduces the tolerance of plants to salt and drought stress (Huang et al., 2011; Huang et al., 2012; Lee et al., 2012). In addition, AtTZF4, 5, and 6 are positive regulators of ABA (Bogamuwa and Jang, 2013). These results enhance our understanding of the growth of pepper plants, as well as the response of pepper to various types of stress and hormone treatments.
After identifying CCCH genes in pepper that play significant roles in responses to cold and heat stress, the expression patterns of five candidate genes that were highly induced by cold stress and seven candidate genes that were highly induced by heat stress were validated by qRT-PCR. PEPTY4 and PEPTY51, which were both in Group XI, were not expressed under cold stress and in the control environment, but they were highly expressed under heat stress. However, both PEPTY16 and PEPTY52 belonged to Group XI; the former was highly expressed under cold stress, and the latter was highly expressed under heat stress. PEPTY36 in Group IV was highly expressed under cold treatment at 72 h. PEPTY9, which also belongs to the same subfamily as PEPTY36, was not significantly expressed under cold stress, but its expression was gradually up-regulated under heat stress. Thus, the expression patterns were not always the same among each subfamily member of each CCCH gene in pepper. One plausible explanation for this observation is that pepper is more sensitive to low-temperature and high-temperature stress. In addition, the responses of different genes to cold and heat might vary (Wang et al., 2019; Wang et al., 2021; Yang et al., 2021; Gao et al., 2022; Tang B. et al., 2022; Zhang et al., 2022). Therefore, further functional studies of these CCCH genes are needed to clarify the pathways underlying their responses to cold stress and heat stress.
In this study, the phylogenetic relationships, structure, conserved motifs, chromosomal localization, duplication events, and expression profiles of CCCH genes were analyzed and 57 CCCH zinc finger genes were identified in pepper. A phylogenetic tree was constructed using CCCH sequences from Arabidopsis, tomato, and rice. Based on studies of Arabidopsis, we divided the pepper CCCH genes into 12 subfamilies. The exon/intron structure and motif composition were conserved in most subfamily. These genes were unevenly distributed on 12 chromosomes, and segmental duplication events appear to have been the major driver of gene expansion in the CCCH family. We characterized the expression profiles of CCCH genes in different tissues of pepper and under various types of stress and validated these expression patterns using qRT-PCR analysis. We found that CCCH zinc finger genes play important roles in biological processes such as growth and development and adaptation to stress. Overall, our findings will aid future studies aimed at examining the evolution, inheritance, and function of CCCH zinc finger genes in pepper and other plants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JC, ZZ, SZ, TZ, ZL, FG and WT designed the research. WT, YZ, XM, YS and YD performed the research. WT, YH, and ZD analyzed the data. WT, ZZ, ZL, FG, and JC wrote the manuscript. All authors contributed to the article and approved the submitted version.
The research was supported by the Project of Central Public-interest Scientific Institution Basal Research Fund, Grant No: 1630032022009, Project of Sanya Yazhou Bay Science and Technology City, Grant No: SCKJ-JYRC-2022-05 and SCKJ-JYRC-2022-25, and Hainan Provincial Natural Science Foundation of China (No. 322MS132).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZZ declared a shared affiliation with the authors FG, ZL at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1189038/full#supplementary-material
Arnaud, D., Déjardin, A., Leplé, J. C., Lesage-Descauses, M. C., Pilate, G. (2007). Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 14, 103–116. doi: 10.1093/dnares/dsm013
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Berg, J. M., Shi, Y. (1996). The galvanization of biology: a growing appreciation for the roles of zinc. Science 271, 1081–1085. doi: 10.1126/science.271.5252.1081
Bogamuwa, S., Jang, J. C. (2013). The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ. 36, 1507–1519. doi: 10.1111/pce.12084
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chai, G., Hu, R., Zhang, D., Qi, G., Zuo, R., Cao, Y., et al. (2012). Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics 13, 253. doi: 10.1186/1471-2164-13-253
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: was plotted using TBtools analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, Y., Kato, N., Wang, W., Li, J., Chen, X. (2003). Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev. Cell 4, 53–66. doi: 10.1016/s1534-5807(02)00399-4
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Freemont, P. S. (1993). The RING finger. a novel protein sequence motif related to the zinc finger. Ann. N Y Acad. Sci. 684, 174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x
Gao, C., Mumtaz, M. A., Zhou, Y., Yang, Z., Shu, H., Zhu, J., et al. (2022). Integrated transcriptomic and metabolomic analyses of cold-tolerant and cold-sensitive pepper species reveal key genes and essential metabolic pathways involved in response to cold stress. Int. J. Mol. Sci. 23 (12), 6683. doi: 10.3390/ijms23126683
Gasteiger, E., Hoogland, C., Alexandre, G., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” In: Walker, J.M. (ed) The proteomics protocols handbook. Humana press: Springer protocols Handbooks. 571–607. doi: 10.1385/1-59259-890-0:571
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Grabowska, A., Wisniewska, A., Tagashira, N., Malepszy, S., Filipecki, M. (2009). Characterization of CsSEF1 gene encoding putative CCCH-type zinc finger protein expressed during cucumber somatic embryogenesis. J. Plant Physiol. 166, 310–323. doi: 10.1016/j.jplph.2008.06.005
Guo, Y. H., Yu, Y. P., Wang, D., Wu, C. A., Yang, G. D., Huang, J. G., et al. (2009). GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 183, 62–75. doi: 10.1111/j.1469-8137.2009.02838.x
Hall, T. M. (2005). Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 15, 367–373. doi: 10.1016/j.sbi.2005.04.004
Han, G., Qiao, Z., Li, Y., Wang, C., Wang, B. (2021). The roles of CCCH zinc-finger proteins in plant abiotic stress tolerance. Int. J. Mol. Sci. 22 (15), 8327. doi: 10.3390/ijms22158327
Hu, X., Zuo, J. (2021). The CCCH zinc finger family of soybean (Glycine max l.): genome-wide identification, expression, domestication, GWAS and haplotype analysis. BMC Genomics 22, 511. doi: 10.1186/s12864-021-07787-9
Huang, P., Chung, M. S., Ju, H. W., Na, H. S., Lee, D. J., Cheong, H. S., et al. (2011). Physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 1, a plasma membrane protein involved in oxidative stress. J. Plant Res. 124, 699–705. doi: 10.1007/s10265-010-0397-3
Huang, P., Ju, H. W., Min, J. H., Zhang, X., Chung, J. S., Cheong, H. S., et al. (2012). Molecular and physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2-mediated signaling pathway. Plant Cell Physiol. 53, 193–203. doi: 10.1093/pcp/pcr162
Iuchi, S., Kuldell, N. (2005). “Zinc finger proteins: from atomic contact to cellular function,”, vol. 276. (New York: Kluwer Academic/Plenum Publishers). doi: 10.1007/b139055
Jan, A., Maruyama, K., Todaka, D., Kidokoro, S., Abo, M., Yoshimura, E., et al. (2013). OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 161, 1202–1216. doi: 10.1104/pp.112.205385
Kang, W. H., Sim, Y. M., Koo, N., Nam, J. Y., Lee, J., Kim, N., et al. (2020). Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum l. Sci. Data 7, 17. doi: 10.1038/s41597-020-0352-7
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kosarev, P., Mayer, K. F., Hardtke, C. S. (2002). Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3, RESEARCH0016. doi: 10.1186/gb-2002-3-4-research0016
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, S. J., Jung, H. J., Kang, H., Kim, S. Y. (2012). Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 53, 673–686. doi: 10.1093/pcp/pcs023
Lee, J., Nam, J. Y., Jang, H., Kim, N., Kim, Y. M., Kang, W. H., et al. (2020). Comprehensive transcriptome resource for response to phytohormone-induced signaling in capsicum annuum l. BMC Res. Notes 13, 440. doi: 10.1186/s13104-020-05281-1
Levasseur, A., Pontarotti, P. (2011). The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics. Biol. Direct 6, 11. doi: 10.1186/1745-6150-6-11
Li, C.-H., Fang, Q.-X., Zhang, W.-J., Li, Y.-H., Zhang, J.-Z., Chen, S., et al. (2021). Genome-wide identification of the CCCH gene family in rose (Rosa chinensis jacq.) reveals its potential functions. Biotechnol. Biotechnol. Equip. 35, 517–526. doi: 10.1080/13102818.2021.1901609
Li, J., Jia, D., Chen, X. (2001). HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13, 2269–2281. doi: 10.1105/tpc.010201
Li, Q. T., Lu, X., Song, Q. X., Chen, H. W., Wei, W., Tao, J. J., et al. (2017). Selection for a zinc-finger protein contributes to seed oil increase during soybean domestication. Plant Physiol. 173, 2208–2224. doi: 10.1104/pp.16.01610
Lijavetzky, D., Carbonero, P., Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis dof gene families. BMC Evol. Biol. 3, 17. doi: 10.1186/1471-2148-3-17
Lin, P. C., Pomeranz, M. C., Jikumaru, Y., Kang, S. G., Hah, C., Fujioka, S., et al. (2011). The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 65, 253–268. doi: 10.1111/j.1365-313X.2010.04419.x
Liu, S., Khan, M. R., Li, Y., Zhang, J., Hu, C. (2014). Comprehensive analysis of CCCH-type zinc finger gene family in citrus (Clementine mandarin) by genome-wide characterization. Mol. Genet. Genomics 289, 855–872. doi: 10.1007/s00438-014-0858-9
Mazumdar, P., Lau, S. E., Wee, W. Y., Singh, P., Harikrishna, J. A. (2017). Genome-wide analysis of the CCCH zinc-finger gene family in banana (Musa acuminata): an insight into motif and gene structure arrangement, evolution and salt stress responses. Trop. Plant Biol. 10, 177–193. doi: 10.1007/s12042-017-9196-5
Moore, R. C., Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Moore, M., Ullman, C. (2003). Recent developments in the engineering of zinc finger proteins. Brief Funct. Genomic Proteomic 1, 342–355. doi: 10.1093/bfgp/1.4.342
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Peng, X., Zhao, Y., Cao, J., Zhang, W., Jiang, H., Li, X., et al. (2012). CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PloS One 7, e40120. doi: 10.1371/journal.pone.0040120
Pi, B., He, X., Ruan, Y., Jang, J. C., Huang, Y. (2018). Genome-wide analysis and stress-responsive expression of CCCH zinc finger family genes in Brassica rapa. BMC Plant Biol. 18, 373. doi: 10.1186/s12870-018-1608-7
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Qiu, A., Lei, Y., Yang, S., Wu, J., Li, J., Bao, B., et al. (2018). CaC3H14 encoding a tandem CCCH zinc finger protein is directly targeted by CaWRKY40 and positively regulates the response of pepper to inoculation by Ralstonia solanacearum. Mol. Plant Pathol. 19, 2221–2235. doi: 10.1111/mpp.12694
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P., Bork, P. (2000). SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. doi: 10.1093/nar/28.1.231
Schumann, U., Prestele, J., O’geen, H., Brueggeman, R., Wanner, G., Gietl, C. (2007). Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 104, 1069–1074. doi: 10.1073/pnas.0610402104
Takatsuji, H. (1998). Zinc-finger transcription factors in plants. Cell Mol. Life Sci. 54, 582–596. doi: 10.1007/s000180050186
Tang, C., Deng, Z., Liu, Q., Jin, W., Xiang, S., Xie, P., et al. (2022). Genome-wide identification and expression analysis of CCCH type zinc-finger gene family in Nicotiana tabacum. J. Henan Agric. Sci. 51, 48–58. doi: 10.15933/j.cnki.1004-3268.2022.04.006
Tang, B., Li, X., Zhang, X., Yin, Q., Xie, L., Zou, X., et al. (2022). Transcriptome data reveal gene clusters and key genes in pepper response to heat shock. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.946475
Tazuke, A., Asayama, M. (2013). Expression of CsSEF1 gene encoding putative CCCH zinc finger protein is induced by defoliation and prolonged darkness in cucumber fruit. Planta 237, 681–691. doi: 10.1007/s00425-012-1787-7
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Wang, D., Guo, Y., Wu, C., Yang, G., Li, Y., Zheng, C. (2008). Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9, 44. doi: 10.1186/1471-2164-9-44
Wang, J., Liang, C., Yang, S., Song, J., Li, X., Dai, X., et al. (2021). iTRAQ-based quantitative proteomic analysis of heat stress-induced mechanisms in pepper seedlings. PeerJ 9, e11509. doi: 10.7717/peerj.11509
Wang, J., Lv, J., Liu, Z., Liu, Y., Song, J., Ma, Y., et al. (2019). Integration of transcriptomics and metabolomics for pepper (Capsicum annuum l.) in response to heat stress. Int. J. Mol. Sci. 20 (20), 5042. doi: 10.3390/ijms20205042
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Xu, R. (2014). Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in Solanum lycopersicum. Mol. Genet. Genomics 289, 965–979. doi: 10.1007/s00438-014-0861-1
Yang, Y., Guang, Y., Wang, F., Chen, Y., Yang, W., Xiao, X., et al. (2021). Characterization of phytochrome-interacting factor genes in pepper and functional analysis of CaPIF8 in cold and salt stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.746517
Zhang, J., Liang, L., Xie, Y., Zhao, Z., Su, L., Tang, Y., et al. (2022). Transcriptome and metabolome analyses reveal molecular responses of two pepper (Capsicum annuum l.) cultivars to cold stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.819630
Zhang, Y., Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5, 1. doi: 10.1186/1471-2148-5-1
Keywords: gene family, CCCH, phylogenetic analysis, Capsicum annuum, stress
Citation: Tang W, Hao Y, Ma X, Shi Y, Dang Y, Dong Z, Zhao Y, Zhao T, Zhu S, Zhang Z, Gu F, Liu Z and Chen J (2023) Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in Capsicum annuum L.. Front. Plant Sci. 14:1189038. doi: 10.3389/fpls.2023.1189038
Received: 18 March 2023; Accepted: 26 April 2023;
Published: 30 May 2023.
Edited by:
Yan Zhao, Shandong Agricultural University, ChinaReviewed by:
Fang Wang, Shandong Agricultural University, ChinaCopyright © 2023 Tang, Hao, Ma, Shi, Dang, Dong, Zhao, Zhao, Zhu, Zhang, Gu, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhong Chen, amluaG9uZ2NoZW5Aemp1LmVkdS5jbg==; Ziji Liu, TGl1emlqaTE5ODJAMTYzLmNvbQ==; Fenglin Gu, WGlhb2d1NDExN0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.