
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Plant Sci., 19 April 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1188832
This article is part of the Research TopicFunctional Genomics in Fruit Trees: from ‘Omics to Sustainable Biotechnologies, Volume IIView all 11 articles
Editorial on the Research Topic
Functional genomics in fruit trees: from ‘omics to sustainable biotechnologies, volume II
The Research Topic collects 10 manuscripts focused on cutting-edge topics in functional genomics of fruit trees, that can be grouped into three main themes: advancements in genome editing technologies, availability of tools and approaches for understanding gene family structure and function, and study of gene regulation through long non-coding RNA (lncRNA), transgenesis, and transcriptomic analysis (Figure 1). The first set of manuscripts showcases perspectives and limitations of transgene-free genome editing in fruit trees, the use of CRISPR/Cas9 constructs for the reduction of stomatal density in grapevine, and the generation of edited citrus varieties enriched in antioxidant compounds. A second group describes the use of comparative gene family analysis tools, of novel workflows for the Rosaceae, the creation of a comprehensive platform for germplasm innovation and functional genomics in Macadamia, and the construction of a high-density genetic linkage map to identify genetic loci responsible for seedlessness in mandarin. The third set of articles includes studies on citrus focused on the role of lncRNA in response to Huanglongbing (HLB), the fruit-specific expression of Ruby to improve anthocyanin accumulation, and the exploitation of the transcriptome relating to growth and palatability.
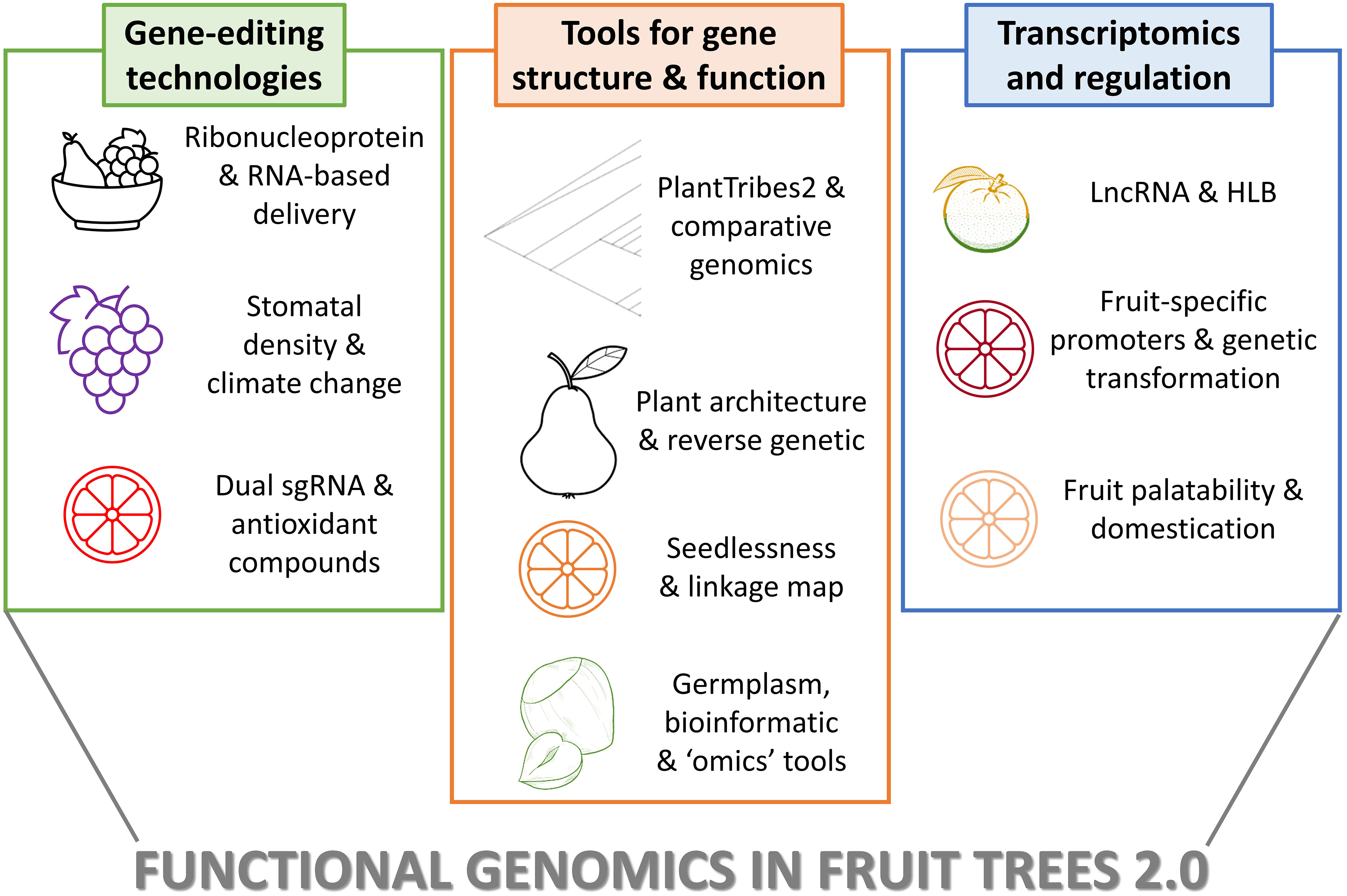
Figure 1 Methods and applications showed in the Editorial Functional Genomics in fruits trees 2.0. The figure summarizes the main approaches reported in the 10 papers collected in Research Topic, organized in the three main groups as summerized in the Editorial. The plant species and the applications are briefly reported.
The recent advancements of new gene-editing tools opens new opportunities for functional studies in fruit trees. Gouthu et al. report the use of a transgene-free gene editing via Ribonucleoprotein (RNP) delivery and the ectopic application of RNA-based products; these approaches are mainly addressed to a sustainable and an eco-friendlier environment for a crop production system that could potentially replace the use of chemicals. Both technologies are strictly dependent on the foundational knowledge of gene-to-trait relationships, and the potential and limitations are carefully reviewed.
Through a genome editing approach based on CRISPR/Cas9 technology, Clemens et al. showed the potential of manipulating stomatal density for optimizing grapevine adaptation under changing climate conditions. By inactivation of the VvEPFL9-1, a positive regulator of stomata formation, different edited lines of the table grape variety ‘Sugraone’ with a significant reduction in stomatal density and a significant increase in pore length were produced. Interestingly, epfl9-1 mutants showed an improved intrinsic water-use efficiency, a desirable trait to improve plant water conservation and to delay early sugar accumulation.
Salonia et al. used a dual sgRNA approach to knockout the fruit-specific β-LCY2 to introduce lycopene in five different Tarocco and Sanguigno sweet orange varietal groups. The approach revealed to be highly efficient in introducing point or short mutations, large deletions and the inversion of the region between the cutting site of both sgRNAs. No altered phenotype in vegetative tissues of edited plants has been observed. This work represents the first example of the use of a genome editing approach to potentially improve qualitative traits of citrus fruit.
In an effort to address accessibility and computational challenges in genome-scale research and to rely on comparative genomic approaches that integrate across plant community resources and data types, Wafula et al. provided a valuable tool for the research community working on plant genomics. PlantTribes2 is a scalable, easily accessible, highly customizable, and broadly applicable bioinformatic framework useful for comparative and evolutionary analyses of gene families from any type of organism, including fungi, microbes, animals, and plants. Examples of application are the evaluation of targeted gene family assembly and genome quality. Such as example, Zhang et al. showed an application of PlantTribes2 making simpler the acquisition and the analysis of genome-scale data, through an iterative processes of reverse genetics aimed to understand pear architecture genes. To individuate putative architecture genes in pear, it could be possible to start with genes of interest and the workflow proposed provides a comparative genome approach to efficiently identify, investigate, and then improve and/or validate genes of interest across genomes and genome resources.
Macadamia is an important nut crop, but it’s becoming difficult for researchers to process and use the vast amount of genomic data available. As a central portal, Wang et al. have developed MacadamiaGGD, a database integrating data from germplasm, genomes, transcriptomes, genetic linkage maps, and SSR markers. The database is freely available online and includes bioinformatic tools to conveniently analyze data of interest. The database is expected to broaden the understanding of the germplasm, genetics, and genomics of macadamia species and facilitate molecular breeding efforts.
In citrus, Kumar et al. identified two closely associated SNPs, AX-160417325 and AX-160536283, in Fs-locus on LG5 of ‘Mukaku Kishu’ mandarin. These SNPs reduced the population size and positively predicted seedlessness in 25.0-91.9% of the progenies in studied populations. These markers should be strategic in reducing the effective population size at seedling stage in crosses involving ‘MK’ paternity. Further work will be done, but the availability of these SNPs opens the way in the production of seedless citrus fruits, highly appreciated by consumers.
LncRNAs serve as crucial regulators in plant response to various diseases. Zhuo et al. identified and characterized 8,742 lncRNAs among HLB-tolerant rough lemon and HLB-sensitive sweet orange. LNC_28805 was identified as one of the most important candidate lncRNAs; on the other hands, WRKY33 and SYP121 are two candidate genes targeted by miRNA5021 developing a key role in the bacteria pathogen responses based on the prediction of protein-protein interaction network. This study will be useful in understanding the role of lncRNAs involved in citrus HLB regulation and opens the road for further investigation of their regulatory functions.
Tissue specific promoters are important tools for the precise genetic engineering of crops. Thilmony et al., in the framework of four fruit-preferential promoters, found that CitWax exhibited high fruit-preferential expression of Ruby in Mexican lime. In some of the transgenic trees with high levels of flower and fruit anthocyanin accumulation, leaves deeply coloured at juvenile phase, lost the coloration at maturity. CitWax promoter could control the expression of Ruby increasing the nutritional value and health benefits of citrus fruit.
Pérez-Roman et al. analyzed the transcriptomes of developing fruitlets of wild and domesticated citrus to identify key traits brought about by domestication. Domestication promoted growth processes at the expense of chemical defenses, also impacting in nitrogen and carbon allocation, presumably leading to major differences in organoleptic properties. The production of unpleasant secondary metabolites and acidity, for instance, decreased considerably improving palatability. The results also appear to suggest that domesticated mandarins evolved through progressive refining of other relevant palatability properties.
All authors drafted the manuscript, revised and approved the final version.
We greatly appreciate the active contribution of all authors and reviewers for their precious work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: gene-editing, seedlessness, palatability, stomatal density, plant architecture, database, huanglongbing, fruit trees
Citation: Licciardello C, Perrone I, Gambino G, Velasco R and Talón M (2023) Editorial: Functional genomics in fruit trees: from ‘omics to sustainable biotechnologies, volume II. Front. Plant Sci. 14:1188832. doi: 10.3389/fpls.2023.1188832
Received: 17 March 2023; Accepted: 03 April 2023;
Published: 19 April 2023.
Edited and Reviewed by:
Adnane Boualem, Université Paris-Saclay, CNRS, INRAE, Université Evry, Institute of Plant Sciences Paris-Saclay (IPS2) Gif sur Yvette, FranceCopyright © 2023 Licciardello, Perrone, Gambino, Velasco and Talón. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Concetta Licciardello, Y29uY2V0dGEubGljY2lhcmRlbGxvQGNyZWEuZ292Lml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.