
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 July 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1188149
This article is part of the Research Topic Crop Improvement in the Era of Next-Generation Sequencing View all 14 articles
Cytoplasmic male sterility (CMS) is predominantly used for F1 hybrid breeding and seed production in Sorghum. DNA markers to distinguish between normal fertile (CMS-N) and sterile (CMS-S) male cytoplasm can facilitate F1 hybrid cultivar development in Sorghum breeding programs. In this study, the complete chloroplast (cp) genome sequences of CMS-S and Korean Sorghum cultivars were obtained using next-generation sequencing. The de novo assembled genome size of ATx623, the CMS-S line of the chloroplast, was 140,644bp. When compared to the CMS–S and CMS-N cp genomes, 19 single nucleotide polymorphisms (SNPs) and 142 insertions and deletions (InDels) were identified, which can be used for marker development for breeding, population genetics, and evolution studies. Two InDel markers with sizes greater than 20 bp were developed to distinguish cytotypes based on the copy number variation of lengths as 28 and 22 bp tandem repeats, respectively. Using the newly developed InDel markers with five pairs of CMS-S and their near isogenic maintainer line, we were able to easily identify their respective cytotypes. The InDel markers were further examined and applied to 1,104 plants from six Korean Sorghum cultivars to identify variant cytotypes. Additionally, the phylogenetic analysis of seven Sorghum species with complete cp genome sequences, including wild species, indicated that CMS-S and CMS-N contained Milo and Kafir cytotypes that might be hybridized from S. propinquum and S. sudanese, respectively. This study can facilitate F1 hybrid cultivar development by providing breeders with reliable tools for marker-assisted selection to breed desirable Sorghum varieties.
Sorghum (Sorghum bicolor [L.] Moench) is the fifth most important major cereal cultivated worldwide and it is used not only for human nourishment, but also for animal fodder and feed, construction material, fencing, and brooms (Doggett, 1988; Mundia et al., 2019). Sorghum is a diploid C4 plant with outstanding tolerance to most types of abiotic stress (Tari et al., 2013). Sorghum’s genome is significantly smaller genome than maize (around 800 vs. 2,500 Mb), and it has recently undergone high-quality diploid genome sequencing, making it an emerging model for highly productive C4 crops (Paterson et al., 2009; Mccormick et al., 2018). Heterosis, or hybrid vigor, is the ability of hybrids to outperform elite inbred line parents and is probably the most important strategy to increase grain yield in various crops, including Sorghum (Kim and Zhang, 2018). In several vegetable and cereal crops, commercial seed production is based on F1 hybrids produced using the cytoplasmic genetic male sterility (CMS) system. The CMS system relies on a set of male sterility-inducing cytoplasm that are complemented by alleles at genetic loci in the nuclear genome that either restore fertility or maintain sterility. In 1952, a milo CMS cytoplasm was identified in the offspring of a hybrid between two cultivars, milo and kafir, wherein milo as the female and kafir as the male. The A1 CMS line sourced from milo is the predominant CMS line used to produce hybrids in Sorghum. The presence of restorer genes enabling the production of fertile F1 hybrids using the CMS approach is essential for the cost-effective production of hybrid Sorghum seeds. To date, a total of nine distinct resources of CMS, namely, A1 (milo), A2, A3, A4, Indian A4 (A4M, A4VZM, and A4G), A5, A6, 9E, and KS cytoplasm, have been identified in Sorghum (Schertz, 1977; Li and Li, 1998). Although most grain Sorghum types currently utilized in agricultural production are hybrids in the world, hybrid cultivars are still not widely used in Korea, which explains the low average yields.
The chloroplast is the primary site of photosynthesis and carbon fixation in plants, is an essential organelle for land plants (Daniell et al., 2016), and is inherited maternally (Asaf et al., 2017). Some cp genome sequences have been used to distinguish between species and conduct phylogenetic studies, as the chloroplast (cp) genome is more conserved and shorter in length than the nuclear and mitochondrial (mt) genomes. The cp and mt genomes are often used to study plant evolution (Cho et al., 2015; Lu et al., 2016; Hong et al., 2017; Hong et al., 2019). Although the cp genome’s specific structure and sequences remain conserved, the larger mt genome (200−2400 kb) has a significantly different structure and various isoforms, even within a single plant cell (Sugiyama et al., 2005; Cho et al., 2022). Therefore, polymorphic DNA originating from the cp genome is favorable for developing CMS markers, even though several CMS genes are mitochondrial as they are maternally inherited (Cho et al., 2006).
The mt and cp genomes of the normal fertile (CMS-N) of S. bicolor, BTx623, have previously been sequenced and are under the GenBank accession numbers NC_008360 and NC_008602, respectively. Recently, ergonomically important genes, such as those of the flowering time (Casto et al., 2019), dwarf (Hilley et al., 2017) and brown midrib (Bout and Vermerris, 2003) have been isolated using genetic mapping and comparative genomic studies between Sorghum and other crops. The mt genome rearrangement between CMS-S and CMS-N in Sorghum was analyzed, and it was discovered that the coding region of the coxI gene in CMS9E was found to be extended at the 3’- end by 303 nucleotides, resulting in an extension of 101 amino acids at the C-terminal of the protein. A novel chloroplast DNA deletion has been reported in most CMS lines of Sorghum and this deletion occurred in the middle of the gene rpoC2, coding for the ß”-subunit of RNA polymerase (Bailey-Serres et al., 1986; Chen et al., 1993). Chen et al. (1995) also reported that rpoB, rbcL, and rpoC2 transcripts are low in inflorescence tissues and pollen of CMS. Molecular characterization of the cytoplasm using mitochondrial DNA probes revealed sufficient diversity to broaden the cytoplasmic base of Sorghum hybrids (Xu et al., 1995; Sivaramakrishnan et al., 1997). A previous study demonstrated the strict maternal inheritance of mt and cp DNA in the Sorghum cytoplasm (Pring et al., 1982). All genes encode proteins with a mitochondrial transit peptide and numerous penta-tatricopeptide repeats (Kante et al., 2018; Praveen et al., 2018). The cytoplasmic male sterile line (S rfrf) and its near-isogenic maintainer line (S or N RfRf) are essential for breeding F1 hybrids using CMS systems. The test cross is the most popular traditional method to identify the cytoplasmic type in Sorghum. DNA markers have been used for the indirect selection of major cultivation traits that distinguish the fertile and sterile individuals in several crops, such as onion, maize, wheat, cotton, and others (Bosacchi et al., 2015; Melonek et al., 2021).
This study aimed to: (i) Obtain complete chloroplast (cp) genome sequences of CMS-S and Korean Sorghum cultivars using next-generation sequencing, (ii) Identify single nucleotide polymorphisms (SNPs) and insertions and deletions (InDels) in the cp genomes that can serve as DNA markers for breeding, and phylogenetic studies, (iii) Develop InDel markers, including tandem repeats, to accurately distinguish between cytotypes (CMS-S and CMS-N) based on copy number variation, and validate their effectiveness in identifying cytotypes in Korean Sorghum cultivars.
One male sterile line (ATx623) and four Korean cultivars of S. bicolor were used for the complete cp genome sequencing (Table 1). Five pairs of S. bicolor near-isogenic lines (male sterile and maintainer lines) and 1,104 individual plants from six Korean cultivars were used to identify cytoplasmic types with insertion and deletion (InDel) markers (Tables 2, 3). To conduct comparative genome analysis, the cp genome sequence information in Sorghum species was retrieved from the National Center for Biotechnology Information (Table 1). All plants were grown at the Department of Southern Area Crop Institute in Miryang, Korea.
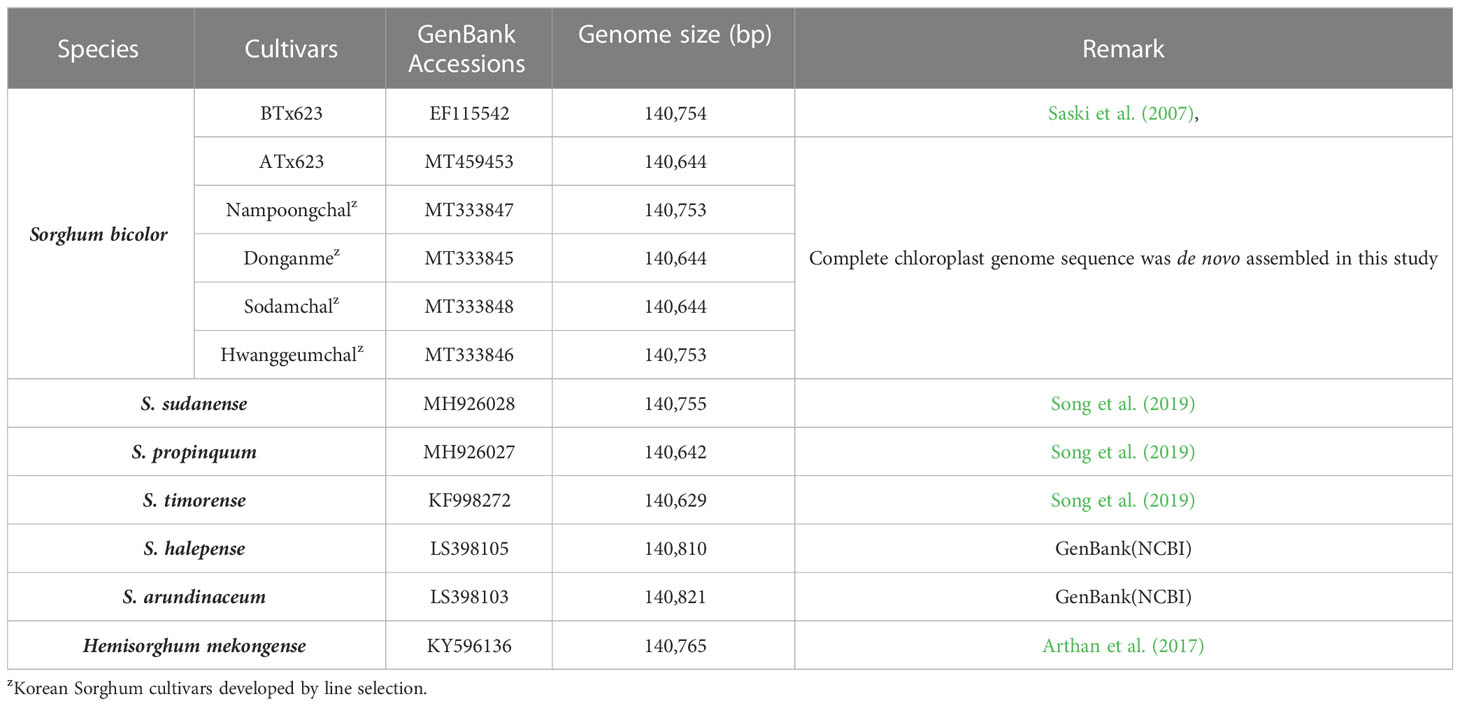
Table 1 List of Sorghum species and GenBank accession numbers of complete chloroplast genome sequences.
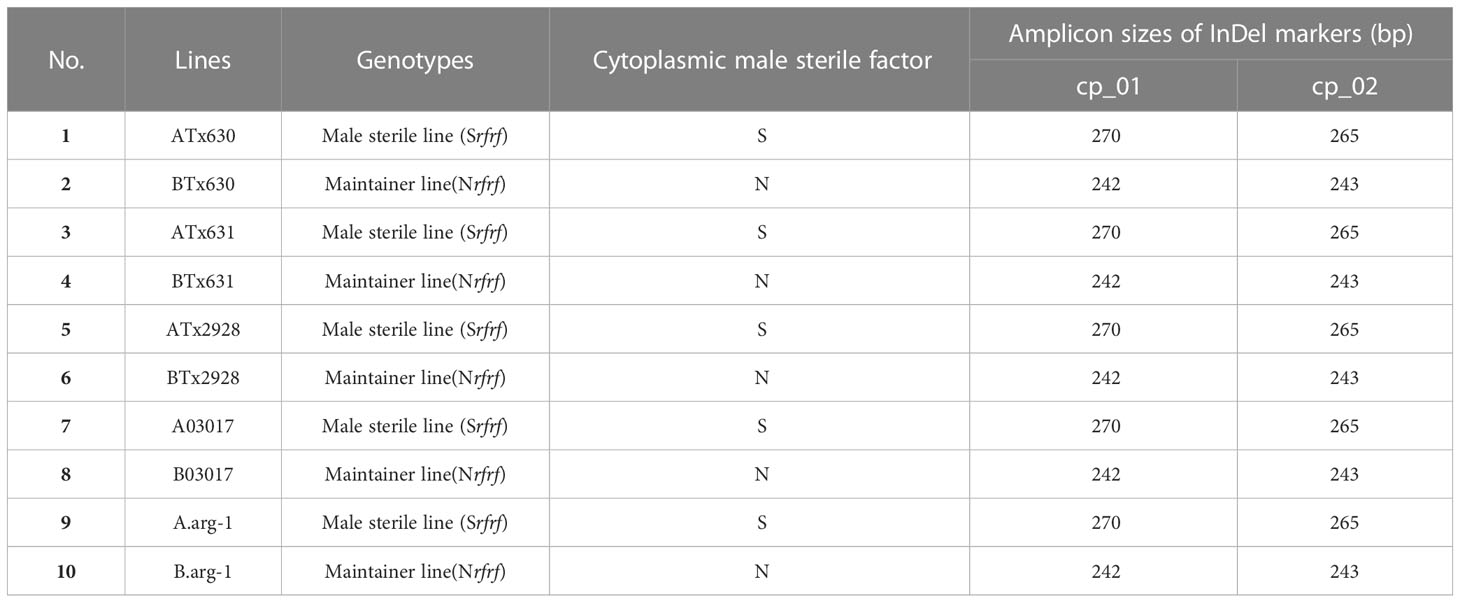
Table 2 Identification of cytoplasmic male sterile factors in Sorghum bicolor using chloroplast specific insertion and deletion (InDel) markers.
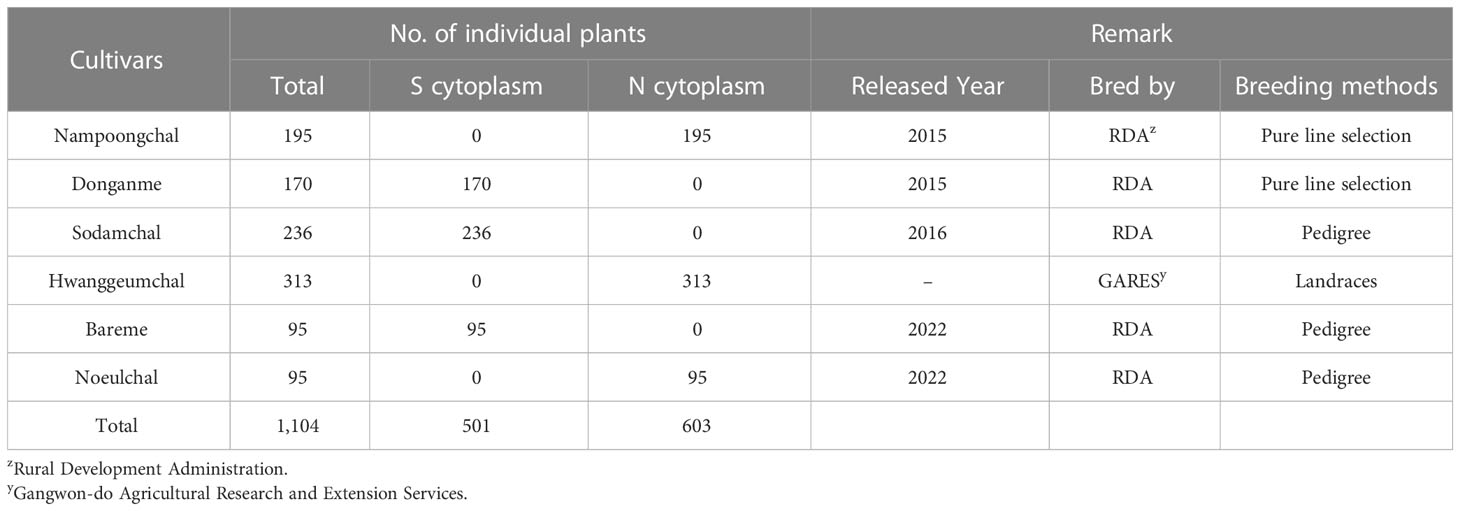
Table 3 Application of cytoplasmic male sterile factors identification using the chloroplast genome specific markers (InDel cp_01) in the Korean Sorghum bicolor cultivars.
DNA was extracted from approximately 100 mg of fresh leaf samples using the NucleoSpin Plant II Mini Kit (Macherey-Nagel, Germany), following the manufacturer’s instructions. The quality and quantity of the genomic DNAs were examined using agarose gel electrophoresis and a NanoDrop 8000 spectrophotometer (Thermo Fisher, USA). Total DNA was sequenced using an Illumina HiSeq 2000 (Illumina, San Diego, USA), and raw reads ranged from 1.9 to 2.7 Gb (Supplementary Table 1). The cp genome sequences were determined from the de novo assembly of low-coverage whole-genome sequences according to previous reports (Hong et al., 2017; Hong et al., 2019). In particular, trimmed paired-end reads (Phred score > 20) were assembled using CLC Assembly Cell Packages (ver. 4.2.1, https://www.qiagenbioinformatics.com/products/clc-assembly-cell/) using default parameters. The cp genome sequence contigs were selected from the initial assembly through the Basic Local Alignment Search Tool using the S. bicolor cp genome sequence as a reference (GenBank accession number: EF115542). Gaps and ambiguous sequences were manually adjusted using Sanger sequencing. PCR amplification and Sanger sequencing were performed to verify the four junction regions between the inverted repeats (IRs) and large single copy (LSC)/small single copy (SSC). The cp genome annotation was conducted using GeSeq (Tillich et al., 2017) with the reference sequences of S. bicolor from GenBank. The cp genome map was illustrated using the OGDraw software (Lohse et al., 2007).
Single nucleotide polymorphisms (SNPs) and InDels between the male sterile and maintainer lines were precisely identified using a variant calling process through MAFFT (Katoh and Standley, 2013). To amplify the InDel regions, 20 ng of genomic DNA was used in 20 µL PCR mixture comprising 2× TOP simple preMix-nTaq master mix (Enzynomics, Seoul, Korea) consisting of 0.2 U/µL of n-taq DNA polymerase, 3 mM of Mg2+, and 0.4 mM of each deoxynucleotide triphosphate mixture with 10 pmol of each primer. The primer sequences used are listed in Table 4. PCR was conducted in a thermocycler (Veriti, Applied Biosystems, CA, USA) using the following cycling parameters: 95°C (5 min); 35 cycles at 95°C (20 s), 55°C (20 s), and 72°C (1 min); and the final extension was conducted at 72°C (5 min). The PCR products were analyzed by capillary electrophoresis (QIAxcel Advanced System, Qiagen, Germany) following the manufacturer’s protocol. PCR products were purified with the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, USA) and sequenced by direct sequencing in Bioneer Co. (Bioneer, Daejeon, South Korea). Sequences were aligned using ClustalW in MEGA 11.
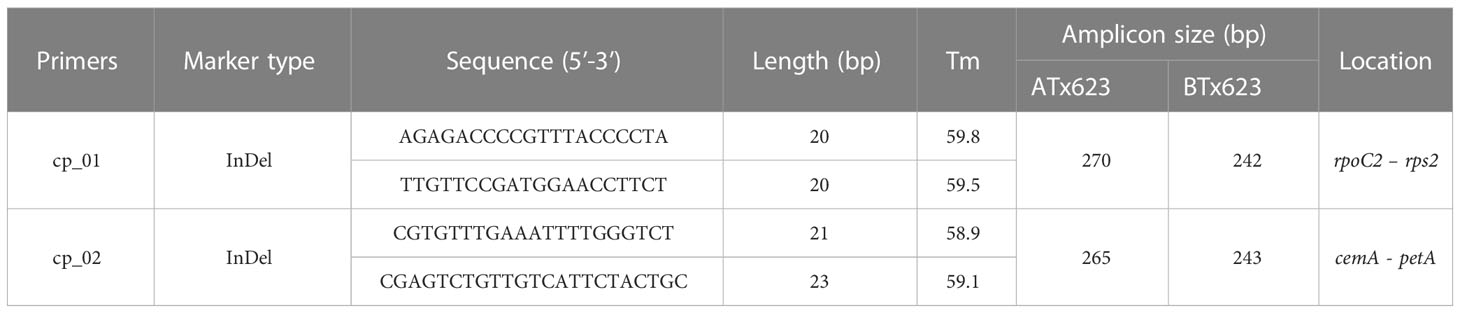
Table 4 The information of primers used in this study for the identification of cytoplasmic male sterile factors in Sorghum bicolor using the chloroplast genome sequences between the male sterile (ATx623) and maintainer (BTx623) lines.
To investigate the phylogenetic position of Sorghum depending on the cytotype, we used eight complete cp genome sequences. Six complete cp genome sequences of the Sorghum species were retrieved from GenBank (Table 1). Phylogenetic analysis was conducted using the maximum composite likelihood model with 1,000 bootstrap replicates in MEGA 11 (Tamura et al., 2021). A phylogenetic tree was constructed using the neighbor-joining method (Tamura et al., 2004) with MEGA 11.
We sequenced and assembled the complete cp genomes of one isogenic line (CMS-S, ATx623) and four Korean Sorghum varieties using the Illumina HiSeq 2000 system. The complete sequences of the five cp genomes were generated using de novo and reference-based assemblies. Sequencing with approximately 1,657–20,373X coverage generated 130.28 Gbp of paired-end reads (Supplemental Table 1). The complete size of CMS-N is 140,754 bp, as reported by Saski (Saski et al., 2007). We found that the complete cp genome sizes of S. bicolor ATx23 and BTx623 were 140,644 and 140,754 bp, respectively and included a pair of IRs of 22,259 bp (CMS-N) and 22,782 bp (CMS-S) separated by SSC regions of 12,503 bp (CMS-N) and 12,506 bp (CMS-S) and LSC regions of 82,685 bp (CMS-N) and 82,574 bp (CMS-S) (Figure 1), respectively. The comparison of cp genomes of two inbred lines (ATx623 and BTx623) and four South Korean Sorghum cultivars (Nampoongchal, Donganme, Sodamchal, and Hwanggeumchal) showed no significant differences in their gene and gene order (Figure 1 and Supplementary Table 2). All six cp genome structures had a typical quadratic structure.
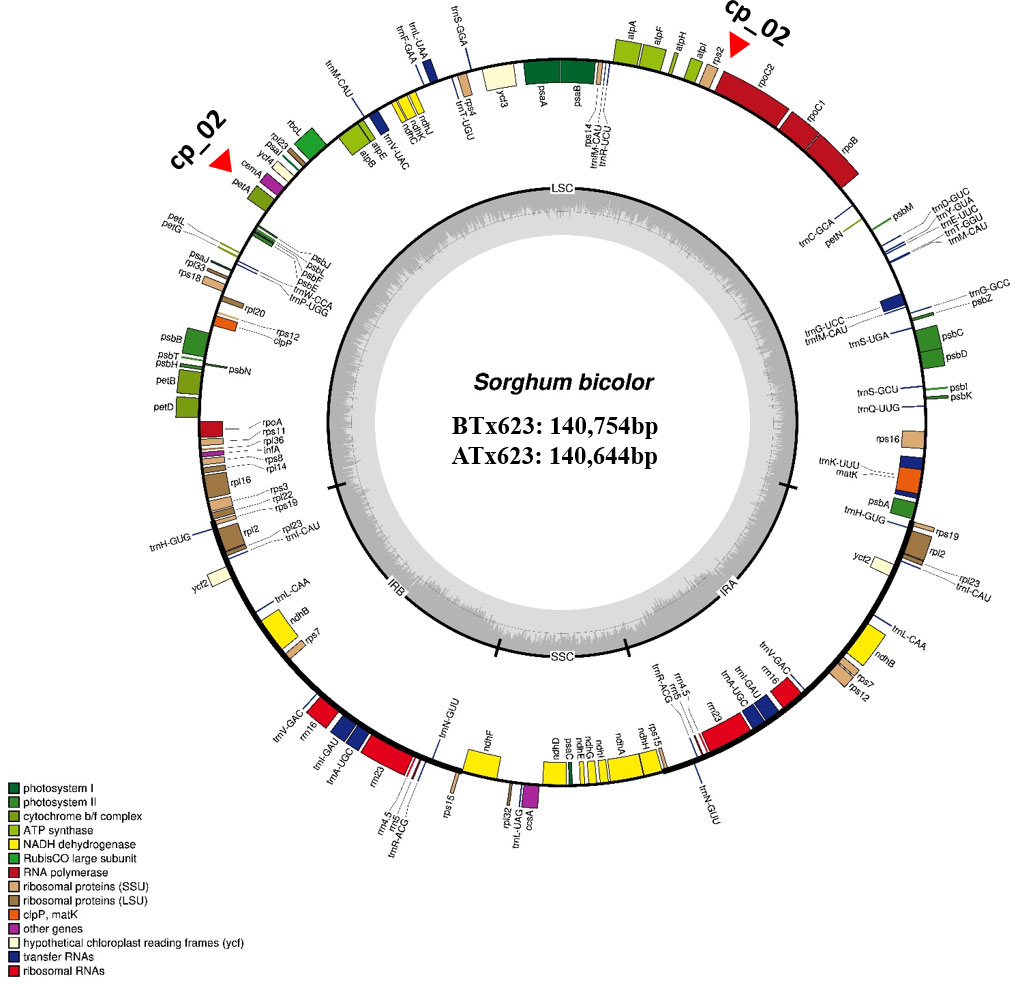
Figure 1 The complete chloroplast genome map of Sorghum bicolor in the male sterile (ATx623) and maintainer line (BTx623). The genes inside and outside the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional groups are represented using different colors. Thick lines indicate the extent of the inverted repeats (IRa and IRb) that separate the genomes into small single-copy (SSC) and large single-copy (LSC) regions. The red-filled red triangles indicate the region of insertion and deletion (InDel) markers.
A total of 103 genes were identified in the Sorghum cp genome, including 40 photosynthesis-related genes, 29 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. Sixteen genes contained one, two, or three introns, and six of these were tRNAs (Supplemental Table 2). Notably, six protein-coding genes (rps12, rps15, rps19, rps7, rpl2, and rpl23), eight tRNA genes (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and all rRNA genes were duplicated in the IR regions, which is common in most Poaceae genomes. The Sorghum cp genome contained 16 intron-containing genes. Among them, ten protein-coding genes (petB, petD, atpF, ndhB, ndhA, rpoC1, rps12, rps16, rpl16, and rpl2) and six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) had a single intron, and two genes (rps12, and ycf3) contained two introns (Supplemental Table 2).
Comparing the cp genome sequences of four Sorghum cultivars revealed that Nampungchal and Hwanggeumchal were near, whereas Donganme and Sodamchal were identical. This might be attributed to cultivar development from the same cytoplasmic background genetic resources. Phenotypically, these cultivars showed diverse traits, such as seed or grain color, lodging tolerance, culm length, plant height, and waxy endosperm. Thus, these cultivars could be important materials for further genetic research and new cultivar development.
Although the content and genes order in the cp genomes of the two Sorghum inbred lines and four South Korean Sorghum cultivars were very similar, numerous polymorphic sites were found among them. In the cytoplasm of cp genomes, 19 single nucleotide polymorphisms (SNPs) and 142 InDels in the genic region that can be used for marker development for breeding, population genetics, and evolution studies (Supplementary Table 3). When complete cp genomes were aligned to identify polymorphisms that may distinguish between S- and N-cytoplasms, wherein the difference is more than 20bp, we found that differences of 28 and 22 bp in the intergenic regions of rpoC2-rps2 and cemA-petA, respectively, were due to those in copy number variation with two major tandem repeats (Figure 2). The alignment showed that InDels with lengths of 28 and 22 bp were present at the same position in the CMS-S cytotype (Figure 2). Thus, two pairs of primers were designed based on the InDels with length of 28 and 22 bp to amplify the regions (Table 4). To evaluate the accuracy of this marker, we tested five pairs of near isogenic lines (NILs) of Sorghum and compared the results with those obtained with the CMS specific InDel markers. PCR amplification showed that Sorghum cytotypes could be clearly distinguished using gel electrophoresis (Figure 3). All male sterile lines known to contain S-cytoplasm had upper bands of 270 and 265 bp with the cp_01 and cp_02 InDel markers, respectively. In contrast, all maintainer lines with N-cytoplasm had lower bands of 242 and 243 bp (Table 2). We also identified the cytoplasmic male sterile factors, namely, S or N, by InDel markers with 1,104 plants from six South Korean Sorghum cultivars (Figure 3 and Table 3). The results of marker analysis revealed that individuals of each cultivar, Donganme, Sodamchalm, and Baremae, had 100% S-cytoplasm. In contrast, Hwanggeumchal, Nampungchal, and Noeulchal only contained N-cytoplasm types (Table 3). All cultivars containing S-cytoplasm were found to have a 28 bp insertion.
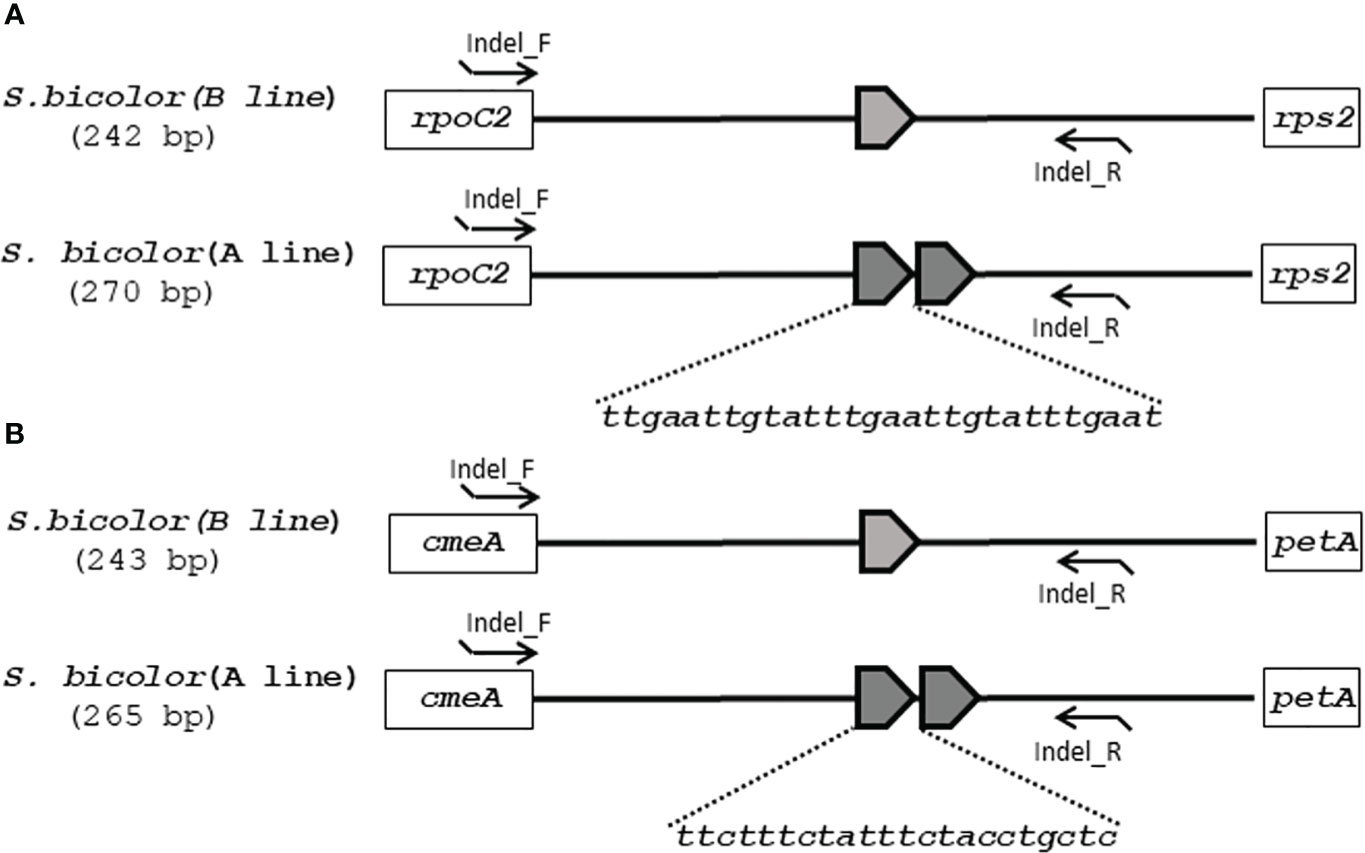
Figure 2 Schematic diagrams of (A) InDel cp_01 and (B) InDel cp_02 with cytoplasmic male sterile and maintainer lines of Sorghum bicolor in chloroplast genomes. (A) 28 and (B) 22 base pairs of tandem repeats (TRs) are represented as pentagons. The InDel cp_01 and InDel cp_02 marker primers are indicated as arrowed lines.

Figure 3 Capillary gel electrophoresis of Korean Sorghum bicolor cultivars with cytoplasmic male sterile factor-specific InDel cp_01 markers. (A) Nampoongchal, (B) Donganme, (C) Sodamchal, (D) Hwanggeumchal, (E) Bareme, and (F) Noeulchal. Ten individual plants (1 to 10) of each cultivar and cytoplasmic male sterile (ATx623) and maintainer (BTx623) lines are depicted.
As Sorghum has a wide range of mating rates of more than 7-30% (Djè et al., 2000; Barnaud et al., 2008), we expected genetic variation in the cytoplasmic genome, but they were all identical. In Sorghum breeding, paper bags are used for repeated self-pollination and generation advancement to avoid outcrossing. Consequently, the developed varieties appear to have the same cytoplasmic type, implying that the breeding cultivars are genetically fixed, and the pure line is well maintained. Hwannggeumchal and Nampungchal contain the S-cytoplasm type, whereas Donganme and Sodmchal contain the N-cytoplasm type (Table 1). In Sorghum, the first report described a 165 bp deletion in the middle of rpoC2 in CMS lines containing A1, A2, A5, and A6 cytoplasm (Chen et al., 1995). Consistent with previous results, all male sterile lines used in this study were included in the A1 cytoplasm.
In Korea, Sorghum is usually bred by landrace selection and utilization of landraces in a breeding program. The cytoplasm types in Korean Sorghum varieties have not yet been identified. Through cp genome sequencing, the cytotypes of Korean varieties, Hwanggeumchal and Nampungchal contained 140,753 bp, whereas those of Donganme and Sodamchal cytotypes contained 140,644 bp (Table 1).
Typically, IR regions have identical lengths; however, they can extend or contract inside the chloroplast. Therefore, we compared the cp genomes of LSC, SSC, and IR (IRa and IRb) among ATx623, S. sudanense, S. propinquum, and BTx623 (Table 5 and Figure 4). The total lengths of the cp genome of ATx623 and S.propinquum were nearly identical (140,644 and 140,642 bp, respectively), whereas those of S.sudanens and BTx623 were also identical (140,755 and 140,754 bp, respectively) (Table 5). However, the total cp genome lengths of S. sudanense and BTx623 were slightly longer (111-112 bp) than those of ATx623 and S. propinquum respectively. The guanine-cytosine content in the cp genome of S. bicolor (ATx623 and BTx623) and congeneric species (S. sudanense and S. propinquum) was 38.5%, and all the species contained 114 unique genes (Table 5). Additionally, we discovered that the size of intergenic spacers (IGSs) between the rpl22-rps19 genes of ATx623 in the junction between the LSC and IRb regions (LSC-IRb) were similar to those of S. sudanense, S.propinquum and BTx623 (Figure 4). Similarly, the IGSs between the rps19- psbA genes, located in the IRa-LSC junction, of ATx623, S sudanense, S propinquum and BTx623 were similar (Figure 4). The boundaries between the IRa regions were similar in size (1,182 bp) in all the compared species (Figure 4). Similarly, the ndhH gene spanned the IRb-SSC region, and the fragment located in the IRb region was equal in size (2,188 bp) among the compared species.
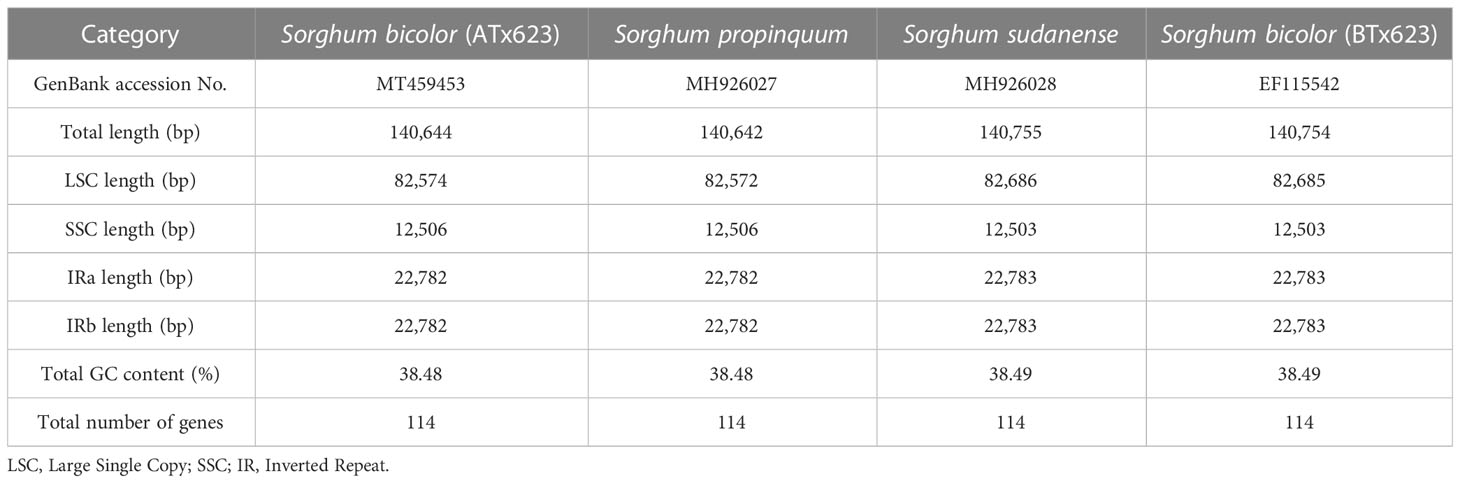
Table 5 Summary of chloroplast genome characteristics for four Sorghum genera containing the male sterile (ATx623) and maintainer line (BTx623) of Sorghum bicolor.
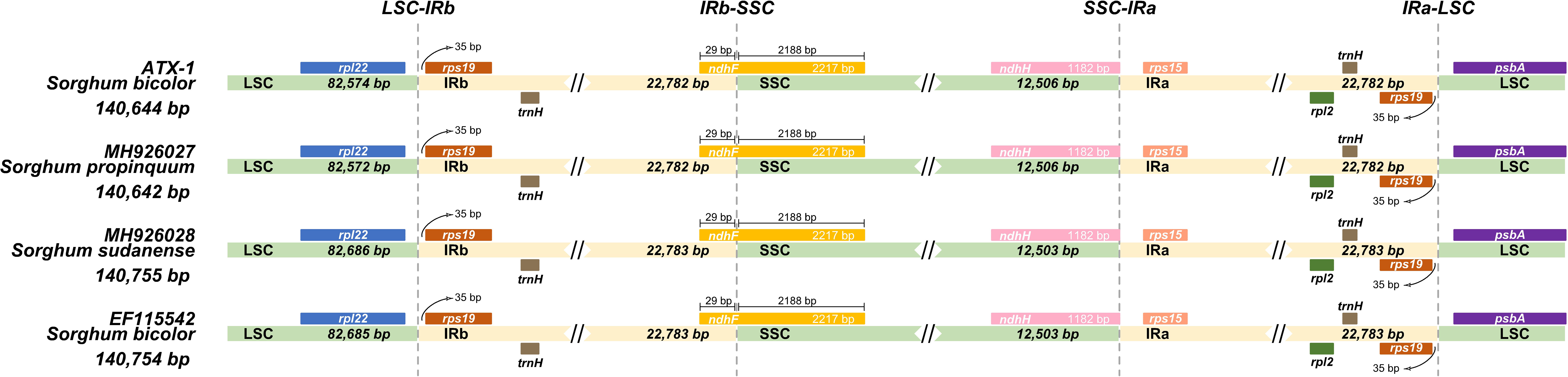
Figure 4 Comparison of the border position and size of LSC, SSC, and IR regions in the chloroplast genome of four Sorghum species. Gene names of each border are designated in boxes. LSC, Large Singe Copy; SSC, Small Single Copy; IRs, Inverted Repeats.
Molecular phylogenetic analysis offers new perspectives on the evolutionary linkages between species. Thus, phylogenetic analysis was conducted using the complete cp genome sequences of the eight Sorghum species. The results of the maximum composite likelihood analysis are shown in the phylogenetic tree (Figure 5). The phylogenetic tree was monophyletic and formed two clades within these eight Sorghum species, wherein S. timorense was the outgroup. A strong bootstrap value (100%) was observed for three of the five nodes. Eusorghum species, such as S. bicolor (ATx623), S. propinquum, S. halepense, S. sudanense, S. bicolor (BTx623), and S. arundinaceum were grouped into one clade, whereas hemisorghum mekongense formed another group clade. In the Eusorghum species clade, S. biclor (ATx623) was the sister to S.propinquum in the same branch, whereas S. sudanense was the sister to S. bicolor (BTx623), with a short branch length, indicating a dispersed evolutionary history or that they are a closer ancestor. Genetic distance analysis revealed the considerably less genetic distance between the analyzed Sorghum species. The lowest genetic distance value of 0.0000 was observed between S. propinquum and S. color (ATx 623) followed by the second lowest value of 0.00002 between S. bicolor (BTx623) and S. sudanese (Supplementary Table 4).
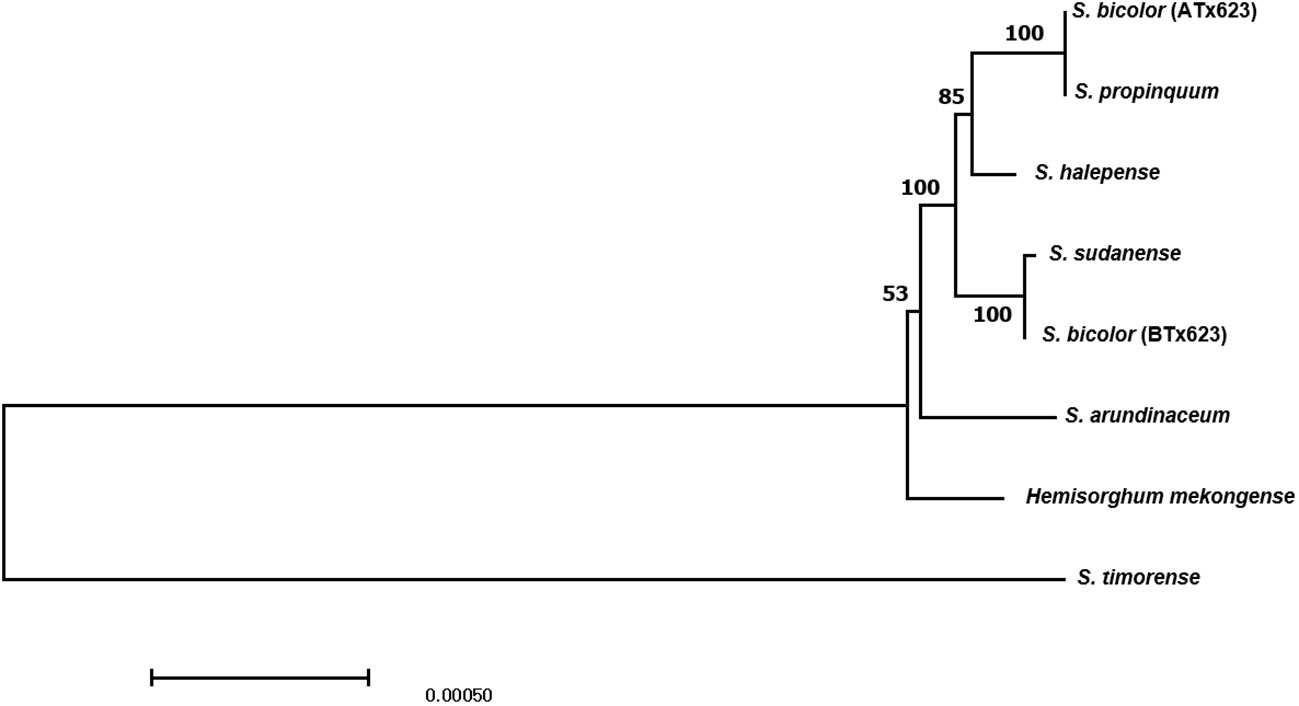
Figure 5 The phylogenic tree of six congeneric Sorghum species, including ATx623 (male sterile line) and BTx623 (maintainer line) of Sorghum bicolor, constructed using the maximum composite likelihood model. Bootstrap values are shown below each clade.
The cp genome is a useful tool for analyzing the evolutionary relationships among species. This is due to the fact that photosynthesis-related organelles such as chloroplasts, contain a circular genome that is comparatively stable and is passed along from the mother to offspring. Moreover, recent research has focused on the cp genome, as it provides essential genetic information to investigate the evolutionary links between related species.
The overall structural organization and introns, genes, and gene order of the analyzed cp genome of a CMS-S line and South Korean cultivars of Sorghum were conserved and showed no significant difference in the cp genome size. Similarly, the SSC, LSC, IR regions, and GC content of cp genomes (38.5%) (Table 5) were also found to be similar among the Eusorghum species. These results are consistent with previous studies on the cp genomes of Sorghum and other species from the Poaceae family (Lu et al., 2016; Song et al., 2017; Song et al., 2019). Recently, numerous taxonomists have focused on the cp genome to investigate the phylogenetic relationships of related species. For example, cp genomes can provide sufficient genetic information for species identification. In this study, we developed InDel markers based on sequence variation in the cp genome for the accurate cytoplasm identification of species and developed InDel markers for further cytoplasm evaluation of species.
In the chloroplasts, the ndhD gene is a component of the NADH dehydrogenase complexes. The specific mutations in the ndhD gene hinder the NADH dehydrogenase complex’s ability to operate normally, which reduces the anther’s ability to produce energy. In wheat, a mutation in the ndhD gene causes male sterility in wheat. The mutation is a single-nucleotide substitution that changes a cytosine to a thymine results in a frameshift that leads to the production of a truncated ndhD protein (Han et al., 2022). PsaA and psaB gene are the subunit of photosystem I (PSI) and PSI complex of proteins that uses light energy to drive the transfer of electrons from water to NADPH. These complex has been shown to play a role in the regulation of gene expression in plants (Azarin et al., 2020). Therefore, we analyzed the ndhD, psaA, psaB gene sequences (nucleotide and amino acids) with Clutal W and we found that there is no genetic variation such as SNP or InDel between male sterile line and maintainer line (data not shown).
Recently, cp genome sequence analysis has been successfully used to reconstruct phylogenetic relationships among plant lineages. Previous phylogenetic studies based on entire cp genomes have been used to resolve the difficult phylogenetic relationships among closely related species. In this study, the whole cp genomes of a Sorghum CMS-S line and South Korean cultivars were sequenced and assembled using next-generation sequencing. In a previous study, four Sorghum species were grouped into two groups. S. sudanense, S. bicolor, and S. propinquum formed groups. S. sudanense, S. bicolor, and S. propinquum belong to the subgenus Sorghum which contains 10 species (Song et al., 2019). Phylogenetic analysis using the complete cp genome of seven Sorghum species, including wild species, revealed that CMS-S and CMS-N of the S. bicolor cytoplasm were highly similar to S. propinquum and S. sudanense, respectively. These results were consistent with those of a previous study (Song et al., 2019; Ananda et al., 2021). S. propinquum is a wild perennial diploid rhizomatous species distributed across Southeast Asia and the Indian subcontinent. Various traits are potentially useful for the introgression of S. propinquum into S. bicolor. The primary gene pool of modern Sorghum cultivars contains the wild species, S. propinquum (De Wet and Harlan, 1971; De Wet, 1978). Previous results found that S. propinquum showed increased height, early maturity, and high yield (Ananda et al., 2020). This might explain the close relationship between S. bicolor (ATx623) and S. propinquum.
In this study, the S cytoplasm of S. bicolor Milo was found to have genetic exchange with S. propinquum. In contrast, S. sudanense is believed to be segregated from a natural hybrid of S. bicolor and S.arundinaceum. Our findings revealed that S. sudanense is closely related to S. bicolor, which represents CMS-N, including the maintenance and restoration lines; hence, these results are consistent with those of previous studies. In S. bicolor, the milo cytoplasm (A1) has been widely used in hybrid production, and kafir has been used as a maintainer line as it produces fully fertile hybrids when crossed with the milo parent. These results phylogenetically support the fact that the milo and kafir cytotypes originated from S. propinquum and S. Sudanese, respectively. Bayesian inference analysis indicated that the Sorghum genus diverged from Miscanthus about 19.5 million years ago (mya). Smaller spikelets are a distinctive feature of S. propinquum. This is consistent with the morphology of the small anther, and pollen depleted exone caused by a 165 bp deletion of the rpoC2 region, such as in the A1, A2, A5, and A6 cytoplasms. Doggett (1988) proposed that durra (milo) originated in Ethiopian because it contains the entire set of wild-type bicolor-durra crosses.
In the CMS system, the breeding programs were divided into two groups. One group was devoted to the development of the female inbred line (A/B-line) and the second was devoted to the development of the male inbred line (R-line). Prior to the hybrid development program, testcrossing, or sterilization, a new line should be identified as maintainers or restorers by a testcross. If the lines with the S cytoplasm have a dominant allele present in the nuclear genome, the plant will be an R-line to restore male fertility. Unless the line lacks the dominant allele for fertility restoration, the plant will be male-sterile (Senthil et al., 1994). Maintainer lines have the N cytoplasm and lack a dominant Rf allele. It is easy to discover a new B-line or predict the male infertility gene type by simply identifying the cytoplasmic type using a marker prior to crossbreeding. When developing B-lines, resources with the N-cytoplasm are first selected as markers, and new lines can then be cultivated through crossbreeding between B-lines. N (Rfrf) can also be used if the combinatorial ability test is conducted on a lineage with the N-cytoplasm. In conclusion, the newly developed InDel markers based on the cp genome variation can facilitate a new F1 hybrid breeding system in Korea.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
M-EC conceived the design of the study, analyzed the data, and drafted the manuscript. J-YK and S-IH collected and grew Sorghum cultivars and lines in Miryang, Korea. RS conducted the bioinformatics work and was engaged in drafting the manuscript. K-SC was responsible for data analysis and writing of the manuscript. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
This work was conducted with the support of the “Co-operative Research Program for Agriculture Science & Technology Development (Project No. PJ01505601),” Rural Development Administration, Republic of Korea. This project funded by Korean government not commercial affiliation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1188149/full#supplementary-material
Supplementary Figure 1 | The comparison of the chloroplast genome structure of Sorghum genus using the mVISTA program.
Supplementary Figure 2 | Multiple sequence alignment of InDel region in the chloroplast genome for the identification of cytoplasmic male sterile factors in Sorghum bicolor. (A) InDel cp_01 and (B) InDel cp_02. Each tandem repeat is shown as a black dotted line.
Ananda, G. K. S., Myrans, H., Norton, S. L., Gleadow, R., Furtado, A., Henry, R. J. (2020). Wild sorghum as a promising resource for crop improvement. Front. Plant Sci. 11, 1108. doi: 10.3389/fpls.2020.01108
Ananda, G., Norton, S., Blomstedt, C., Furtado, A., Møller, B., Gleadow, R., et al. (2021). Phylogenetic relationships in the sorghum genus based on sequencing of the chloroplast and nuclear genes. Plant Genome 14 (3), e20123. doi: 10.1002/tpg2.20123
Arthan, W., Mckain, M. R., Traiperm, P., Welker, C. a. D., Teisher, J. K., Kellogg, E. A. (2017). Phylogenomics of Andropogoneae (Panicoideae: Poaceae) of Mainland Southeast Asia. Syst. Bot. 42, 418–431.
Asaf, S., Khan, A. L., Khan, M. A., Waqas, M., Kang, S. M., Yun, B. W., et al. (2017). Chloroplast genomes of arabidopsis halleri ssp. gemmifera and arabidopsis lyrata ssp. petraea: structures and comparative analysis. Sci. Rep. 7, 7556. doi: 10.1038/s41598-017-07891-5
Azarin, K., Usatov, A., Makarenko, M., Kozel, N., Kovalevich, A., Dremuk, I., et al. (2020). A point mutation in the photosystem I P700 chlorophyll a apoprotein A1 gene confers variegation in helianthus annuus l. Plant Mol. Biol. 103, 373–389. doi: 10.1007/s11103-020-00997-x
Bailey-Serres, J., Hanson, D. K., Fox, T. D., Leaver, C. J. (1986). Mitochondrial genome rearrangement leads to extension and relocation of the cytochrome c oxidase subunit I gene in sorghum. Cell 47, 567–576. doi: 10.1016/0092-8674(86)90621-5
Barnaud, A., Trigueros, G., Mckey, D., Joly, H. (2008). High outcrossing rates in fields with mixed sorghum landraces: how are landraces maintained? Heredity 101, 445–452. doi: 10.1038/hdy.2008.77
Bosacchi, M., Gurdon, C., Maliga, P. (2015). Plastid genotyping reveals the uniformity of cytoplasmic male sterile-T maize cytoplasms. Plant Physiol. 169, 2129–2137. doi: 10.1104/pp.15.01147
Bout, S., Vermerris, W. (2003). A candidate-gene approach to clone the sorghum brown midrib gene encoding caffeic acid O-methyltransferase. Mol. Genet. Genomics 269, 205–214. doi: 10.1007/s00438-003-0824-4
Casto, A. L., Mattison, A. J., Olson, S. N., Thakran, M., Rooney, W. L., Mullet, J. E. (2019). Maturity2, a novel regulator of flowering time in sorghum bicolor, increases expression of SbPRR37 and SbCO in long days delaying flowering. PloS One 14, e0212154. doi: 10.1371/journal.pone.0212154
Chen, Z., Muthukrishnan, S., Liang, G. H., Schertz, K. F., Hart, G. E. (1993). A chloroplast DNA deletion located in RNA polymerase gene rpoC2 in CMS lines of sorghum. Mol. Gen. Genet. 236, 251–259. doi: 10.1007/BF00277120
Chen, Z., Schertz, K. F., Mullet, J. E., Dubell, A., Hart, G. E. (1995). Characterization and expression of rpoC2 in CMS and fertile lines of sorghum. Plant Mol. Biol. 28, 799–809. doi: 10.1007/BF00042066
Cho, K.-S., Lee, H.-O., Lee, S.-C., Park, H.-J., Seo, J.-H., Cho, J.-H., et al. (2022). Mitochondrial genome recombination in somatic hybrids of solanum commersonii and s. tuberosum. Sci. Rep. 12 (1), 8659. doi: 10.1038/s41598-022-12661-z
Cho, K., Yang, T., Hong, S., Kwon, Y., Woo, J., Park, H. (2006). Determination of cytoplasmic male sterile factors in onion plants (Allium cepa l.) using PCR-RFLP and SNP markers. Molecules Cells 21, 411.
Cho, K.-S., Yun, B.-K., Yoon, Y.-H., Hong, S.-Y., Mekapogu, M., Kim, K.-H., et al. (2015). Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PloS One 10, e0125332. doi: 10.1371/journal.pone.0125332
Daniell, H., Lin, C. S., Yu, M., Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
De Wet, J. M. J. (1978). Systematics and evolution of sorghum sect. sorghum (Gramineae). Am. J. Bot. 65, 477–484. doi: 10.1002/j.1537-2197.1978.tb06096.x
De Wet, J. M. J., Harlan, J. R. (1971). The origin and domestication of sorghum bicolor. Economic Bot. 25, 128–135. doi: 10.1007/BF02860074
Djè, Y., Heuertz, M., Lefebvre, C., Vekemans, X. (2000). Assessment of genetic diversity within and among germplasm accessions in cultivated sorghum using microsatellite markers. Theor. Appl. Genet. 100, 918–925. doi: 10.1007/s001220051371
Doggett, H. (1988). Sorghum 2nd edition tropical agriculture. Ser. Longman Sci. Technical Essex England 231 (4), 243–254.
Han, Y., Gao, Y., Li, Y., Zhai, X., Zhou, H., Ding, Q., et al. (2022). Chloroplast genes are involved in the Male-sterility of K-type CMS in wheat. Genes (Basel) 13 (2), 310. doi: 10.3390/genes13020310
Hilley, J. L., Weers, B. D., Truong, S. K., Mccormick, R. F., Mattison, A. J., Mckinley, B. A., et al. (2017). Sorghum Dw2 encodes a protein kinase regulator of stem internode length. Sci. Rep. 7, 4616. doi: 10.1038/s41598-017-04609-5
Hong, S. Y., Cheon, K. S., Yoo, K. O., Lee, H. O., Cho, K. S., Suh, J. T., et al. (2017). Complete chloroplast genome sequences and comparative analysis of chenopodium quinoa and c. album. Front. Plant Sci. 8, 1696. doi: 10.3389/fpls.2017.01696
Hong, S.-Y., Cheon, K.-S., Yoo, K.-O., Lee, H.-O., Mekapogu, M., Cho, K.-S. (2019). Comparative analysis of the complete chloroplast genome sequences of three amaranthus species. Plant Genet. Resour. 17, 245–254. doi: 10.1017/S1479262118000485
Kante, M., Rattunde, H. F. W., Nébié, B., Weltzien, E., Haussmann, B. I. G., Leiser, W. L. (2018). QTL mapping and validation of fertility restoration in West African sorghum A1 cytoplasm and identification of a potential causative mutation for Rf2. Theor. Appl. Genet. 131, 2397–2412. doi: 10.1007/s00122-018-3161-z
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, Y. J., Zhang, D. (2018). Molecular control of Male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. doi: 10.1016/j.tplants.2017.10.001
Li, Y., Li, C. (1998). Genetic contribution of Chinese landraces to the development of sorghum hybrids. Euphytica 102, 47–57. doi: 10.1023/A:1018374203792
Lohse, M., Drechsel, O., Bock, R. (2007). OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274. doi: 10.1007/s00294-007-0161-y
Lu, D., Zhao, Y., Han, R., Wang, L., Qin, P. (2016). The complete chloroplast genome sequence of the purple feathergrass stipa purpurea (Poales: poaceae). Conserv. Genet. Resour. 8, 101–104. doi: 10.1007/s12686-016-0519-x
Mccormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., et al. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354. doi: 10.1111/tpj.13781
Melonek, J., Duarte, J., Martin, J., Beuf, L., Murigneux, A., Varenne, P., et al. (2021). The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat. Commun. 12, 1036. doi: 10.1038/s41467-021-21225-0
Mundia, C. W., Secchi, S., Akamani, K., Wang, G. (2019). A regional comparison of factors affecting global sorghum production: the case of north America, Asia and africa’s sahel. Sustainability 11, 2135. doi: 10.3390/su11072135
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. doi: 10.1038/nature07723
Praveen, M., Uttam, A. G., Tonapi, V. A., Madhusudhana, R. (2018). Fine mapping of Rf2, a major locus controlling pollen fertility restoration in sorghum A1 cytoplasm, encodes a PPR gene and its validation through expression analysis. Plant Breed. 137, 148–161. doi: 10.1111/pbr.12569
Pring, D. R., Conde, M. F., Schertz, K. F., Levings, C. S. (1982). Plasmid-like DNAs associated with mitochondria of cytoplasmic male-sterile Sorghum. Mol. Gen. Genet. MGG 186, 180–184. doi: 10.1007/BF00331848
Saski, C., Lee, S. B., Fjellheim, S., Guda, C., Jansen, R. K., Luo, H., et al. (2007). Complete chloroplast genome sequences of Hordeum vulgare, sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 115, 571–590. doi: 10.1007/s00122-007-0567-4
Schertz, K. (1977). Registration of A2 Tx2753 and B Tx2753 sorgum germplasm 1(Reg.No. GP 30 and 31). Crop Sci. 17 (6), 983–983. doi: 10.2135/cropsci1977.0011183X001700060056x
Senthil, N., Rangasamy, S. S., Palanisamy, S. (1994). Male Sterility inducing cytoplasm in sorghum classification, genetics of sterility and fertility restoration studies. Cereal Res. Commun. 22 (3), 179–184.
Sivaramakrishnan, S., Seetha, K., Reddy, B. V. (1997). Characterization of the a 4 cytoplasmic male-sterile lines of sorghum using RFLP of mtDNA. Euphytica 93, 301–305. doi: 10.1023/A:1002906606333
Song, Y., Chen, Y., Lv, J., Xu, J., Zhu, S., Li, M. (2019). Comparative chloroplast genomes of Sorghum species: sequence divergence and phylogenetic relationships. BioMed. Res. Int. 2019, 5046958. doi: 10.1155/2019/5046958
Song, Y., Chen, Y., Lv, J., Xu, J., Zhu, S., Li, M., et al. (2017). Development of chloroplast genomic resources for Oryza species discrimination. Front. Plant Sci. 8, 1854. doi: 10.3389/fpls.2017.01854
Sugiyama, Y., Watase, Y., Nagase, M., Makita, N., Yagura, S., Hirai, A., et al. (2005). The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genomics 272, 603–615. doi: 10.1007/s00438-004-1075-8
Tamura, K., Nei, M., Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tari, I., Laskay, G., Takács, Z., Poór, P. (2013). Response of sorghum to abiotic stresses: a review. J. Agron. Crop Sci. 199, 264–274. doi: 10.1111/jac.12017
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Keywords: Sorghum bicolor, chloroplast genome, CMS, phylogenetic tree, InDel
Citation: Choe M-E, Kim J-Y, Syed Nabi RB, Han S-I and Cho K-S (2023) Development of InDels markers for the identification of cytoplasmic male sterility in Sorghum by complete chloroplast genome sequences analysis. Front. Plant Sci. 14:1188149. doi: 10.3389/fpls.2023.1188149
Received: 17 March 2023; Accepted: 26 June 2023;
Published: 17 July 2023.
Edited by:
Umesh K. Reddy, West Virginia State University, United StatesReviewed by:
Mehboob-ur Rahman, National Institute for Biotechnology and Genetic Engineering, PakistanCopyright © 2023 Choe, Kim, Syed Nabi, Han and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwang-Soo Cho, a3NjaG9sdm9lQGtvcmVhLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.