
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 28 June 2023
Sec. Plant Proteomics and Protein Structural Biology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1187372
This article is part of the Research Topic Regulation of Plant Immunity by Immune Receptors View all 7 articles
 Liwei Guo1,2†
Liwei Guo1,2† Yuanyu Mu1,2†
Yuanyu Mu1,2† Dongli Wang3
Dongli Wang3 Chen Ye1,2
Chen Ye1,2 Shusheng Zhu1,2
Shusheng Zhu1,2 Hong Cai1,2
Hong Cai1,2 Youyong Zhu1,2
Youyong Zhu1,2 Youliang Peng3
Youliang Peng3 Junfeng Liu3*
Junfeng Liu3* Xiahong He1,2,4*
Xiahong He1,2,4*Plant nucleotide-binding and leucine-rich repeat (NLR) proteins are immune sensors that detect pathogen effectors and initiate a strong immune response. In many cases, single NLR proteins are sufficient for both effector recognition and signaling activation. These proteins possess a conserved architecture, including a C-terminal leucine-rich repeat (LRR) domain, a central nucleotide-binding (NB) domain, and a variable N-terminal domain. Nevertheless, many paired NLRs linked in a head-to-head configuration have now been identified. The ones carrying integrated domains (IDs) can recognize pathogen effector proteins by various modes; these are known as sensor NLR (sNLR) proteins. Structural and biochemical studies have provided insights into the molecular basis of heavy metal-associated IDs (HMA IDs) from paired NLRs in rice and revealed the co-evolution between pathogens and hosts by combining naturally occurring favorable interactions across diverse interfaces. Focusing on structural and molecular models, here we highlight advances in structure-guided engineering to expand and enhance the response profile of paired NLR-HMA IDs in rice to variants of the rice blast pathogen MAX-effectors (Magnaporthe oryzae AVRs and ToxB-like). These results demonstrate that the HMA IDs-based design of rice materials with broad and enhanced resistance profiles possesses great application potential but also face considerable challenges.
Rice (Oryza sativa) is one of the three main cash crops worldwide and a staple food of more than half the world’s population. However, rice production is constantly threatened by diseases. Magnaporthe oryzae that causes rice blast, also known as “the cancer of rice,” was ranked first in the top 10 list of fungal plant pathogens, annually destroying enough rice to feed more than 200 million people for a year (Dean et al., 2012; Fisher et al., 2012; Savary et al., 2019). Therefore, identifying effective approaches to reduce rice yield loss is a clear ongoing need to secure rice supplies and meet the high global demand for food.
Plants differ from animals because they lack circulating immune cells for the interception of microbial signals. Therefore, during their interactions with microbes, plants depend on the cell-autonomous innate immune system of each individual cell to sense and respond to the signaling molecules of microbes. The activation of plant defense responses is triggered by the recognition of invading organisms by immune receptors. However, when pathogens evolve strateges to interfere with host defenses, the immune system is rendered nullified. The ensuing failure of the plant immune system allows further ingress of invading pathogens, resulting in disease susceptibility in plants (Lapin and Van den Ackerveken, 2013).
Molecular mechanisms of plant–pathogen interactions have demonstrated that most nucleotide-binding and leucine-rich repeat (NLR) domains are crucial downstream components of host resistance (Ausubel, 2005; Jones and Dangl, 2006; Jacob et al., 2013; Böhm et al., 2014; Zhang et al., 2022). Plants possess several R genes, most of which encode NLR receptors, and serve important functions, such as molecular exchange and signal transduction in the regulation of immune responses, biological stress, and apoptosis (Collier and Moffett, 2009; Jacob et al., 2013; Bernoux et al., 2016). However, the continuous evolution of virulent strains of pathogens hinders the management of gene-for-gene diseases such as rice blast caused by M. oryzae. Several strategies have been developed to consider resistance erosion, including genetic engineering of R genes. In recent years, many researchers have confirmed that several NLR proteins contain non-canonical integrated domains (IDs), which recognize avirulence (AVR) effectors and trigger immune responses. In particular, the heavy metal-associated (HMA) IDs in rice have been targeted as candidate-engineering elements for anti-rice blast. Therefore, a deep understanding of the molecular mechanisms of plant–pathogen interactions and further exploration and use of plant immunity for disease control are crucial to promote the sustainable development of green agriculture. In this review, we discuss the immune systems of plants and distinctive structures of NLR pairs. We also highlight the progress made in exploiting the structural basis of interactions between paired NLR-HMA IDs of rice and MAX-effectors (M. oryzae AVRs and ToxB-like) for R gene engineering. Finally, we discuss the prospects that need to be addressed to pave the way for the development of broad-race, spectrum-resistant cultivars.
With the co-evolution of hosts and host-adapted microbes, plants have developed diverse immune strategies to combat microbial pathogens (Eitas and Dangl, 2010; Yang et al., 2013). In the prepenetration phase, the cell wall is the first natural physical and defensive barrier of plants and is known to exert basal immune responses. Once pathogens are recognized, cell wall-associated defense appears to operate via the inhibition of fungal cell wall-degrading enzymes, secretion of fungitoxic peptides and phytoalexins, and cell wall strengthening (Hückelhoven, 2007). When pathogens enter host cells, a crosstalk between classical pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) is coordinated to initiate immune responses (Böhm et al., 2014; Zipfel, 2014). Signaling initiated by pattern recognition receptors (PRRs) and NLR proteins leads to overlapping downstream cellular responses, including defense gene expression, production of reactive oxygen species (ROS), and callose deposition, which may cause local cellular suicide and prevent further growth of biotrophic pathogens (Jones and Dangl, 2006; Tsuda and Katagiri, 2010).
On the cell surface, receptor-like kinases (RLKs) and receptor-like proteins (RLPs) function as PRRs that act in the first tier of the plant immune system to respond to PAMPs (Boller and Felix, 2009). The extracellular domains (ECDs) of PRRs are highly diverse, and PRRs can be subdivided according to the nature of their ligand-binding ectodomains (Boller and Felix, 2009; Couto and Zipfel, 2016; Sun et al., 2022). One large class of PRRs includes leucine-rich repeats (LRR) (Boller and Felix, 2009; Couto and Zipfel, 2016; Sun et al., 2022). LRRs are ECDs of RLKs and RLPs that bind chitin and peptidoglycan (Kaku et al., 2006; Williams et al., 2011; Willmann et al., 2011; Williams et al., 2014). The main difference between LRR-RLKs and LRR-RLPs is that the former comprises ECDs, transmembrane domain, and intracellular kinase domain (IKD), whereas the latter lacks an IKD (Ade et al., 2007; Monaghan and Zipfel, 2012). Other existing ECDs include lectin motifs and epidermal growth factor-like domains (Böhm et al., 2014). Generally, LRR-containing RLKs and RLPs act as sensors of peptide PAMPs (Monaghan and Zipfel, 2012). The initiated PPR signaling results in PTI responses, like stomatal closure, ion flow, ROS burst, gene expression changes, and aggregation of antimicrobial active substances (Göhre and Robatzek, 2008; Wirthmueller et al., 2013) that ward off microorganisms.
To establish successful infections, pathogens have evolved a diverse repertoire of effectors that are delivered to plant cells to interfere with PTI (Jones and Dangl, 2006). These effectors are characterized by low molecular weight (≤300 amino acids), presence of a signal peptide at the N-terminus, high cysteine content, and low sequence similarity (Donofrio and Raman, 2012). To counter effector infection, plants use specific NLR receptor proteins to detect pathogen-secreted effector proteins either through direct or indirect binding and activate high-level defense responses of the host (Hogenhout et al., 2009; Tsuda and Katagiri, 2010; Cesari et al., 2014a; Cesari et al., 2014b; Selin et al., 2016). Recognition of pathogen effectors results in effector-triggered immunity (ETI).
Although the two classes of immune receptors involve different activation mechanisms and appear to require different early signaling components, a crosstalk between PTI and ETI appears to be mediated by an integrated signaling network (Tsuda and Katagiri, 2010; Yuan et al., 2021a; Yuan et al., 2021b). ETI enhances, but does not initiate, PTI-induced defense responses by upregulating PTI signaling components. In turn, PTI potentiates ETI-induced cell death. Thus, immune pathways activated by the cell surface and intracellular receptors in plants mutually potentiate the activation of strong defenses against pathogens (Ngou et al., 2021; Yuan et al., 2021b).
In most cases, the recognition and interactions between plants and pathogens are believed to be consistent with the canonical gene-for-gene hypothesis (Flor, 1971). However, several recent studies have demonstrated that single R genes are inadequate for defense against pathogen invasion and necessitate complementary paired R genes encoding for defense responses (Bomblies et al., 2007; Ashikawa et al., 2008; Cesari et al., 2013; Cesari et al., 2014a; Cesari et al., 2014b). NLR protein pairs comprise two structurally and functionally different proteins encoded by paired R genes closely linked in a head-to-head configuration that can form homologous or heterologous complexes within the host (Eitas and Dangl, 2010; Kanzaki et al., 2012; Adachi et al., 2019). NLR proteins are multidomain proteins that possess a conserved architecture, including a C-terminal LRR domain, a central nucleotide-binding and oligomerization domain (NOD), and a variable N-terminal domain (Takken and Goverse, 2012). The N-terminal domains mainly include the Toll-interleukin 1 receptor (TIR)-like or coiled-coil (CC) types; thus, NLR proteins can be largely classified into TIR-NLR (TNL) and CC-NLR (CNL) based on differences in N-terminal structure (Pan et al., 2000; DeYoung and Innes, 2006). Many studies have established the CC and TIR domains as signaling modules (Lapin et al., 2019; Lapin et al., 2022; Zhang et al., 2022).
In the absence of pathogens, NLR paired proteins are maintained in an inactive state. Structure-function analyses have shown that the particular motifs of NLR play critical roles in their auto-inhibition mechanisms. For a number of NLR proteins, the “inactive” state has been shown to be associated with ADP binding (Wang et al., 2019a). The NOD of NLRs contains ADP/ATP-binding motif (P-loop) and is believed to function as a nucleotide switch (Lukasik and Takken, 2009; Wang and Chai, 2020). An ADP molecule binds to pocket formed by tight packing of the three subdomains, nucleotide binding domain (NBD), helical domain 1 (HD1), and winged helical domain (WHD) from the NOD module. The intramolecular interaction indicates that CNL ZAR1 in Arabidopsis adopts an inactive conformation (Wang et al., 2019a). In addition, ZAR1LRR further inhibits ZAR1 by sequestering the NLR in a monomeric state, although the LRR-mediated inhibition is not essential for autoinhibition of some NLRs (Steele et al., 2019; Wang and Chai, 2020). A pair of Arabidopsis TNL proteins, RPS4 and RRS1, was shown to be required for recognition of AvrRps4 from the bacterial pathogen Pseudomonas syringae and PopP2 from Ralstonia solanacearum. RPS4 TIR domain forms homo-dimer and signals an effector-independent cell death. Co-expression of the RRS1 TIR domain, a RRS1/RPS4 TIR hetero-dimer competes with the formation of the RPS4 TIR homo-dimer to maintain the paired NLRs in an inactive state (Narusaka et al., 2009; Cesari et al., 2014a). Striking similarities are apparent between the RPS4/RRS1 and the RGA4 and RGA5 from rice functional models (Cesari et al., 2013; Cesari et al., 2014a; Cesari et al., 2014b).
After AVR recognition, the co-regulation of both NLR proteins exerts synergistic induction of immune responses for disease resistance (Eitas and Dangl, 2010; Okuyama et al., 2011; Zhai et al., 2014; Nishimura et al., 2015). The effector-triggered assembly of the CNL ZAR1 into pentameric resistosomes results in the formation of a calcium-permeable ion channel that integrates into the plasma membrane and initiates immune responses (Wang et al., 2019a; Wang et al., 2019b; Bi et al., 2021). Activated TNR and TNL use nicotinamide adenine dinucleotide (NAD+) or NAD+ with ATP as substrates to produce ADP-ribosylated adenosine triphosphate (ADPr-ATP) and ADPr-ADPR (di-ADPR) through ADP-ribosylation reactions, which are likely to be further hydrolyzed to pRib-ADP and pRib-AMP (2′-(5′′-phosphoribosyl)-5′-adenosine diphosphate and monophosphate). However, TNLs converge on the enhanced disease susceptibility 1 (EDS1) family of lipase-like proteins, which plays a key link between TNL activation and resistance pathway induction (Lapin et al., 2019). EDS1 forms heterodimers with phytoalexin deficient 4 (AtPAD4) and uses the same surface to interact with PAD4-related senescence associated gene 101 (SAG101). Together with the N requirement gene 1 (NRG1), a coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors (Lapin et al., 2019). The cryo-electron microscopy (cryo-EM) structure of TNL-activated EDS1-SAG101 revealed that TIR-catalyzed bioactive compounds, ADPr-ATP/ADPR and pRib-ADP/AMP, bind specifically between EDS1-PAD4 and EDS1-SAG101 dimers. The triggered conformational changes in PAD4 and SAG101 EP domains allosterically induce interactions with helper NLR NRG1A for plant resistance and cell death, respectively (Jia et al., 2022).
Paired NLR proteins exist widely in crops, such as Oryza sativa, Zea mays, Triticum aestivum, Hordeum vulgare, Aegilops tauschii, Triticum urartu, Brachypodium distachyon, and Setaria italica. In addition to having an intact CC/TIR-NB-ARC-LRR domain, they possess certain non-canonical IDs (Nishimura et al., 2015; Bailey et al., 2018; Stein et al., 2018). Most IDs are fused to the N- or C-terminus of NLR proteins (Cesari et al., 2013; Nishimura et al., 2015; Sarris et al., 2015). However, IDs are also present at both termini in certain NLR proteins (Ellis, 2016), or are located between the CC and NB domains in rare cases, such as the Pik-1 R protein of rice (Maqbool et al., 2015). NLR proteins carrying IDs can recognize pathogen effector proteins, known as sensor NLR (sNLR) proteins, and the protein partner that participates in the activation of downstream immune signaling is known as the helper NLR (hNLR) protein (Cesari et al., 2013; Maqbool et al., 2015; Sarris et al., 2015). Paired NLR proteins recognize pathogen effectors through direct physical contact with IDs or indirect interactions with the hNLR protein. Direct and indirect interactions between NLR proteins and pathogen effectors have been explained by various models, such as the receptor-ligand model (Keen, 1990), guard model (van der Biezen and Jones, 1998; Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008), decoy model (Collier and Moffett, 2009; Dodds and Rathjen, 2010), and bait model (Collier and Moffett, 2009).
Rice and M. oryzae constitute one of the model systems for studying plant–pathogen interactions. Currently, more than 100 R genes conferring resistance to blast disease have been mapped to the rice genome, of which more than 20 have been cloned. Except for the R genes Pi-d2 and pi21, all the other genes encode NLR proteins (Liu and Wang, 2016). Interactions between the rice R genes and avirulence genes of M. oryzae, such as Pib/AvrPib, Pi-ta/AvrPi-ta, Pi9/AvrPi9, and Piz-t/AvrPiz-t, are consistent with the canonical gene-for-gene hypothesis (Flor, 1971). Researchers have also found that R gene pairs in rice, such as Pik-1/Pik-2, RGA4/RGA5, Pi5-1/Pi5-2, and Pii-1/Pii-2, jointly participate in defense responses (Lee et al., 2009; Yuan et al., 2011; Zhai et al., 2011; Cesari et al., 2013; Cesari et al., 2014a; Cesari et al., 2014b; Zhai et al., 2014; Maqbool et al., 2015; Takagi et al., 2017). Particularly, RGA5 and Pik-1, the respective sNLR proteins of RGA4/RGA5 and Pik-1/Pik-2, depend on the conserved HMA IDs for the specific direct binding of effector proteins secreted by M. oryzae, thereby regulating hNLR-protein-triggered immune defense responses (Cesari et al., 2013; Zhai et al., 2014). HMA IDs are homologous to heavy metal-associated plant proteins and isoprenylated plant proteins (HIPPs). The conserved α/β sandwich structure of these proteins is comprised of two α-helices and four antiparallel β-sheets (topology: β1-αA-β2-β3-αB-β4), and heavy metal binding occurs at the conserved Cys-XX-Cys motif (Robinson and Winge, 2010; de Abreu-Neto et al., 2013). HMA IDs exist not only in the RGA4/RGA5, Pik-1/Pik-2, and pi21 R proteins of rice and participate in immune responses, but also in the NLR receptor proteins of flowering crops belonging to the Brassicaceae, Fabaceae, and Rosaceae families (Kroj et al., 2016; Sarris et al., 2016). They have also been identified in other plant proteins that are unrelated to NLR proteins (de Abreu-Neto et al., 2013).
Accumulating evidence demonstrated that the relative location and sequence diversity of HMA IDs in the RGA4/RGA5 and Pik-1/Pik-2 proteins of rice exert important effects on their biological functions (Table 1) (Cesari et al., 2013; Zhai et al., 2014; Maqbool et al., 2015). HMA IDs are located separately downstream of the RGA5 LRR domain (RGA5-HMA), but between the CC and NB-ARC domains at the N-terminus of Pik proteins (Pik-HMA), they showed sequence similarities of up to 54% (Cesari et al., 2013; Maqbool et al., 2015; Guo et al., 2018). The hNLR RGA4 acts as an auto-active inducer of immune signaling (Cesari et al., 2014b). RGA4 and RGA5 form homo- and hetero-complexes and interact through their CC domains. While the RGA5 CC domain is necessary but not sufficient for repression of RGA4-mediated cell death, its HMA IDs is dispensable (Cesari et al., 2013; Cesari et al., 2014a; Cesari et al., 2014b). RGA5-HMA IDs can simultaneously recognize the effectors AVR1-CO39 and AVR-Pia through directly binding, which is repressed by RGA5 in the absence of effectors (Cesari et al., 2014b). Upon M. oryzae invasion, the binding of AVR1-CO39 and AVR-Pia to the self-interacting αα/β2 interfaces of RGA5-HMA IDs competes with RGA5-HMA self-interaction, which forms a heterocomplex with 1:1 stoichiometry and relieves RGA4 repression (Guo et al., 2018). The β2 of RGA5-HMA IDs is aligned in an antiparallel manner with the β2 of AVR1-CO39, forming a continuous antiparallel β-sheet comprising the four-stranded β-sheet of RGA5-HMA IDs and β1, 2, and 6 of AVR1-CO39 (Guo et al., 2018). However, in AVR-Pia, the HMA-binding interface is more extended. In addition to the β2 of RGA5-HMA IDs, both β3 and the loop formed between β2 and β3 also participate in interactions during binding to AVR-Pia (Guo et al., 2018).
Co-evolutionary dynamics have driven the emergence of five different Pik alleles, Pikh, Pikp, Pikm, Piks, and Pik*, to display the extended recognition of AVR-Pik (A to F) variants (Yoshida et al., 2009; Kanzaki et al., 2012; Zhai et al., 2014; De la Concepcion et al., 2021; Xiao et al., 2022). AVR1-CO39, AVR-Pia, and AVR-Pik variants have no apparent sequence identity but are highly similar to the MAX (Magnaporthe AVRs and ToxB-like) effector family, which adopts a six β-sandwich fold stabilized by buried hydrophobic residues and commonly, but not always, disulfide bonds (de Guillen et al., 2015; Zhang et al., 2018). Although RGA5 and Pik-1 both display canonical HMA IDs formed in dimer states, structure-function analyses involving nuclear magnetic resonance and x-ray crystallography reveal that the intramolecular interaction mechanisms and binding interfaces are completely different in the two HMA IDs/MAX-effector systems (Maqbool et al., 2015; Cesari, 2018; De la Concepcion et al., 2018; Guo et al., 2018).
In contrast to RGA4, expression of Pik-2 dose not constitutively activate cell death. HMA IDs in sNLR Pik-1 requires the hNLR Pik-2 to initiate signaling, suggesting that the assembly of an active signaling complex requires all three proteins (Kanzaki et al., 2012; Maqbool et al., 2015). Structure-informed similarity searches showed that the co-evolution between pathogens and hosts has driven the emergence of two different binding modes among the Pik-HMA/AVR-Pik systems through multiple interfaces (Figure 1) (Maqbool et al., 2015; De la Concepcion et al., 2018; De la Concepcion et al., 2019; Xi et al., 2022). (1) Like the RGA5-HMA/effector binding systems, AVR-PikD/E/A competes with Pikm-HMA self-interaction in a 1:1 binding mode (De la Concepcion et al., 2019). (2) Pikp-HMA/AVR-PikD/E and AVR-Pikh/AVR-PikC form a 2:1 complex, but do not compete with Pikp/h-HMA self-interaction (Figure 1). The figure is optimized basing the functional models of RGA4/RGA5 NLR pair (Cesari et al., 2014a). Further analysis showed that the overall orientations of each component in the Pikm-HMA/effector complexes are similar to each other, and to determine Pikp-HMA/AVR-PikD/E and Pikh-HMA/AVR-PikC. However, the core residues contributing to the interaction are significantly different. Within the Pikp-HMA/AVR-PikD complex, Pikp-HMASer218,Gly230 binds to AVR-PikDHis46 via hydrogen bonds, forming the core region for the regulation of their interactions. In the Pikm-HMA/AVR-PikD complex, G230 is replaced by Val231 at a structurally equivalent position in the Pikp-HMA structure. Therefore, Pikm-HMAVal231 cannot directly form salt bridges with AVR-PikDHis46. In the Pikm-HMA/AVR-PikE complex, the amino acid at position 46 of the effector protein is asparagine (Asn). Consequently, Asn is rotated out of the binding pocket and is located well away from PikmVal231, and hydrogen bonds are formed between AVR-PikE(Asn46:N<σπ>δ</σπ>2) and both Pikm-HMA(Ser219:OH) and the new water molecule. Such a configuration affects the position of AVR-PikmPhe44-Gly48 and pushes them away from Pikm-HMA IDs, further altering the interactions across the core interface. Such structural changes are related to the decreased binding affinity of AVR-PikE to Pikm-HMA compared with that of AVR-PikD (De la Concepcion et al., 2018). In the Pikm-HMA/AVR-PikA coordination complex, Asn46 is rotated further out of the HMA pocket. Although a hydrogen bond is still formed with Pikm-HMA(Ser219:OH), a considerable difference occurs in orientation. These changes cause the residues of AVR-PikAAsn46 - Pro50 to be located a distance from Pikm-HMA, and such structural observations are again related to decreased effector binding affinity (De la Concepcion et al., 2018). The research results described above indicate that sequence diversity is a key factor affecting the interactions between MAX-effectors of M. oryzae and paired NLR-HMA IDs in rice and the recognition of MAX-effectors, thereby affecting biological functions of paired NLR-HMA IDs.
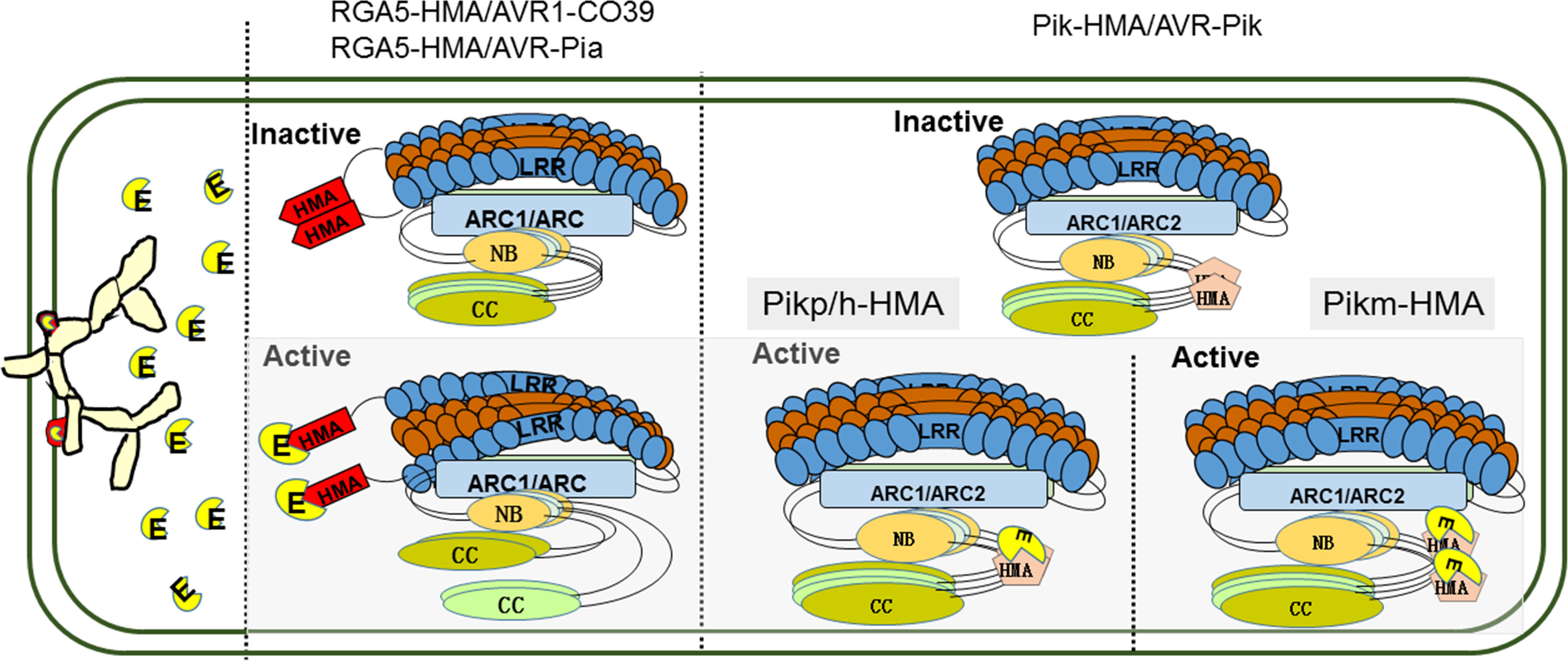
Figure 1 Binding modes for the diverse recognition of paired NLR-HMA IDs/MAX-effectors. In the absence of pathogens, NLR paired proteins are kept in an inactive state by forming a hetero-complex, and the HMA IDs forms a homodimers. After pathogen infected, the secreted effector proteins directely bind to sNLR like molecular traps and activate the immune. For RGA4/RGA5 (blue and brown), the effectors of AVR1-CO39 and AVR-Pia (yellow) binding to RGA5-HMA IDs (red) competes with RGA5-HMA self-interaction, which forms a heterocomplex with a 1:1 stoichiometry and relieves RGA4 repression. Like the RGA5-HMA/effector binding systems, AVR-PikD/E/A compete with Pikm-HMA self-interaction in a 1:1 binding mode; but directly bind to Pikp/h-HMA forming a 2:1 complex. E; effectors secreted by Magnaporthe oryzae. HMA IDs; heavy metal-associated (HMA) integrated domains (IDs). NLR; nucleotide-binding and leucine-rich repeat. MAX-effectors; Magnaporthe oryzae AVRs and ToxB-like.
Engineering made-to-order plant immune receptors has been confirmed as a potential method to confer plant disease resistance (Kourelis et al., 2023). Targeted point mutations or random mutational screens have succeeded in extending NLR recognition specificity or increasing their activation properties to create sensitized NLR (Harris et al., 2013; Segretin et al., 2014). Therefore, the use of molecular engineering to design R genes for the creation of new specificities or expansion of NLR protein recognition profiles has great potential in crop genetics and breeding (Kim et al., 2016; Cesari, 2018; Grund et al., 2019; Kourelis et al., 2021; Cesari et al., 2022). A classic example is the alternative strategy for modifying the protein kinase PBS1 in Arabidopsis thaliana (Kim et al., 2016). It is believed that PBS1 acts as a bait in the system, similar to the mousetrap model. The NLR protein RPS5 (resistance to Pseudomonas syringae) forms a pre-activation complex with PBS1. The cleavage of PBS1 by AvrPphB (Gly-Asp-Lys-Ser-His-Val-Ser) secreted by the pathogenic oomycete Phytophthora infestans leads to conformational changes in PBS1, which serve as a key factor in triggering RPS5-induced HR (DeYoung et al., 2012). Thus, replacement of the cleavage site of PBS1 with that of protease effector proteins of other bacteria and viruses (PBSRCS2 and PBSTuMV) can also trigger RPS5 activity, thereby expanding the recognition specificity of the host towards pathogens (Kim et al., 2016; Pottinger et al., 2020).
The use of structural biology to elucidate the molecular basis of the interactions between plant NLR proteins and pathogen effector proteins at the atomic level provides new approaches to achieve precise NLR protein design through targeted mutations. The highlights of the potential of HMA-IDs engineering in rice are as follows: (1) in the RGA4/RGA5 and Pik-1/Pik2 systems, HMA IDs can act as sNLR, binding MAX-effectors like molecular traps and activating the immune response of the system; (2) RGA5-HMA IDs and Pik-HMA IDs show 54% sequence similarity; (3) structure-function analysis demonstrated high plasticity in HMA ID and MAX-effectors interactions. Because their sequence is unrelated but structurally similar, MAX-effectors bind to different surfaces of structurally conserved HMA IDs. Therefore, the design of HMA IDs with expanded recognition profiles based on HMA IDs/MAX-effector complex structures has immense potential and has achieved promising progress (Table 2).
De la Concepcion et al. (2019) analyzed and compared the key residues that participate in AVR-Pik interactions in the crystal structures of the complexes. By constructing key mutations in the interface 2 and 3 regions of Pik-HMA, or swapping residues found in Pikm into Pikp, the researchers obtained the mutant PikpAsn261Lys,Lys262Glu, with an expanded AVR-Pik recognition profile for the first time. Besides having enhanced AVR-PikD recognition ability, the mutation could also recognize AVR-PikE and AVR-PikA. Maidment et al. (2021) demonstrated that the OsHIPP19-HMA protein in rice, which is related to cold/drought tolerance and possesses conserved domains, can bind to multiple effector proteins (AVR-PikD, C, F) in a similar HMA IDs/MAX-effector mode. This study provides an important molecular basis for designing Pik-1 receptor mutants similar to OsHIPP19-HMA IDs, with an expanded Pik resistance profile. RGA5-HMA-ID modifications have also received considerable research attention, with researchers successfully creating the RGA5-HMASel1027Val,Gly1009Asp (RGA5-HMA2) mutant with new AVR-Pib recognition sites (Liu et al., 2021) and obtaining RGA5-HMA(E1029A,I1030L,T1031V,E1033D,D1034L,K1035R,R1037K,L1038I,V1039E) (RGA5-HMAm1) and RGA5-HMA(E1029A,I1030L,T1031V,E1033D,D1034L,K1035R,R1037K,L1038I,V1039E,M1065Q,E1067S,L1068Q) (RGA5-HMAm1m2) with expanded MAX-effector recognition profiles (Cesari et al., 2022). Moreover, the HMA-IDs of Pikm-1 have been successfully swapped with nanobody (single-domain antibody fragment) fusions that bind either green fluorescent protein (GFP) or mCherry (Kourelis et al., 2023). Pikobody is shortened for Pikm-1-nanobody fusions with GFP or mCherry. The bioengineered Pikobody confers new-to-nature resistance activities against Potato virus X (PVX) expressing GFP or mCherry (Kourelis et al., 2023). These results demonstrate that the HMA-IDs-based design of rice materials with broad resistance profiles possesses great application potential but also faces considerable challenges.
For the structure-guided engineering of immune receptors, R gene stacking deserves concern. Functional stacking of resistance genes has been used in plant breeding strategy to increase durability and confer broad-spectrum resistance (Ghislain et al., 2019; Luo et al., 2021; Kourelis et al., 2023). Stacking of three late blight R genes in potato varieties confers extreme resistance to late blight (Ghislain et al., 2019). A five-transgene cassette has been successfully introduced into bread wheat and shows very high level of resistance to a fungal rust pathogen in wheat (Luo et al., 2021). However, overaccumulation of NLRs often leads to autoimmune responses, suggesting that the levels of these immune receptors must be tightly controlled (Cheng et al., 2011). In addition, gain-of-function mutants of NLRs, constitutive expression of defense marker Pathogenesis Related (PR) genes, enhanced pathogen resistance, and altered plant development, such as dwarfism, are general features of plant autoimmunity (Cheng et al., 2011). Therefore, NLRs should be maintained at proper levels to prevent autoimmunity (Cheng et al., 2011). Besides, in some hybrid plants, independently evolved immune system components are mismatched and trigger inappropriate immune reactions in the absence of pathogens (Tran et al., 2017). NLR misactivation has been linked to autoimmunity and induces a typical hybrid necrosis (Chakraborty et al., 2018).
Coordinated development of structural biology and genetics has provided an important molecular basis for elucidating the interactions between plant NLR proteins and pathogens. In this review article, we summarize the importance of the plant immune system and the progress in genetic engineering strategies for the targeted design of HMA IDs with expanded, enhanced, or new MAX-effector recognition abilities based on the structural information of paired NLR-HMA IDs in rice and MAX-effectors of M. oryzae. Therefore, adequate use of the immune systems of crops and in-depth exploration of the interaction modes of NLR proteins and effectors provide an important molecular basis for the mining and designing of R genes and resistant crop varieties. However, the following issues require further consideration and investigation:
● Transgenic rice varieties conferring increased or extended anti-rice blast specificity by modified HMA IDs are yet to be developed.
● Exploration of the feasibility of designing HMA IDs through modification or exchange to confer recognition profiles for MAX-effectors of other pathogens besides M. oryzae. It is unclear whether continuous resistance can be provided by transplanting engineered HMA IDs into crops belonging to related or unrelated genera of rice.
● Elucidation of the molecular mechanisms of synergistic interactions in the paired NLR proteins of rice to determine how paired NLR-HMA IDs activate sNLR proteins to induce molecular transduction of hNLR immune signals.
● Exploration of the molecular mechanisms and modification strategies for HMA IDs/MAX-effectors as models for further application to other crops for in-depth investigation of the diversity of NLR proteins in recognizing effectors and activating immune mechanisms.
XH and JL conceived and designed the review. LG and YM wrote the manuscript. Other authors listed have made a substantial direct and intellectual contribution to the work and approved it for publication.
This work was supported by the Natural Science Foundation of China (31901870), the Major Science and Technology Project of Yunnan Province (202202AE090036), High-level Talents Introduction Plan of Yunnan Province-Young Talents Special Project, and Yunnan Chen xuewei Expert Workstation(202305AF150124).
We thank the Key Laboratory of Agro-Biodiversity and Pest Management of Education Ministry of China and Yunnan Agricultural University for providing a working place.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adachi, H., Derevnina, L., Kamoun, S. (2019). NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50, 121–131. doi: 10.1016/j.pbi.2019.04.007
Ade, J., DeYoung, B. J., Golstein, C., Innes, R. W. (2007). Indirect activation of a plant nucleotide binding site–leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. U S A 104, 253–2536. doi: 10.1073/pnas.0608779104
Ashikawa, I., Hayashi, N., Yamane, H., Kanamori, H., Wu, J., Matsumoto, T., et al. (2008). Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer pikm-specific rice blast resistance. Genetics 180, 2267–2276. doi: 10.1534/genetics.108.095034
Ausubel, F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? nat. Immunol 6 (10), 973–979. doi: 10.1038/ni1253
Bailey, P. C., Schudoma, C., Jackson, W., Baggs, E., Dagdas, G., Haerty, W., et al. (2018). Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 1–18. doi: 10.1186/s13059-018-1392-6
Bernoux, M., Burdett, H., Williams, S. J., Zhang, X., Chen, C., Newell, K., et al. (2016). Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28, 146–159. doi: 10.1105/tpc.15.00303
Bi, G., Su, M., Li, N., Liang, Y., Dang, S., Xu, J., et al. (2021). The ZAR1 resistosome is a calcium- permeable channel triggering plant immune signaling. Cell 184, 3528—3541.e12. doi: 10.1016/j.cell.2021.05.003
Böhm, H., Albert, I., Fan, L., Reinhard, A., Nürnberger, T. (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 20, 47–54. doi: 10.1016/j.pbi.2014.04.007
Boller, T., Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J. L., et al. (2007). Autoimmune response as a mechanism for a dobzhansky-muller-type incompatibility syndrome in plants. PloS Biol. 5, e236. doi: 10.1371/journal.pbio.0050236
Cesari, S. (2018). Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 219, 17–24. doi: 10.1111/nph.14877
Cesari, S., Bernoux, M., Moncuquet, P., Kroj, T., Dodds, P. N. (2014a). A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy hypothesis”. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00606
Cesari, S., Kanzaki, H., Fujiwara, T., Bernoux, M., Chalvon, V., Kawano, Y., et al. (2014b). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959. doi: 10.15252/embj.201487923
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Cesari, S., Xi, Y., Declerck, N., Chalvon, V., Mammri, L., Pugnière, M., et al. (2022). New recognition specificity in a plant immune receptor by molecular engineering of its integrated domain. Nat. Commun. 13, 1524. doi: 10.1038/s41467-022-29196-6
Chakraborty, J., Ghosh, P., Das, S. (2018). Autoimmunity in plants. Springer Link 248, 751–767. doi: 10.1007/s00425-018-2956-0
Cheng, Y. T., Li, Y., Huang, S., Huang, Y., Dong, X., Zhang, Y., et al. (2011). Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. 108, 14694–14699. doi: 10.1073/pnas.1105685108
Collier, S. M., Moffett, P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529. doi: 10.1016/j.tplants.2009.08.001
Couto, D., Zipfel, C. (2016). Regulation of patter recognition receptor signaling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Dangl, J. L., Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
de Guillen, K., Ortiz-Vallejo, D., Gracy, J., Fournier, E., Kroj, T., Padilla, A. (2015). Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PloS Pathog. 11, e1005228. doi: 10.1371/journal.ppat.1005228
de Abreu-Neto, J. B., Turchetto-Zolet, A. C., de Oliveira, L. F., Zanettini, M. H., Margis-Pinheiro, M. (2013). Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 280, 1604–1616. doi: 10.1111/febs.12159
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
De la Concepcion, J. C., Franceschetti, M., MacLean, D., Terauchi, R., Kamoun, S., Banfield, M. J. (2019). Protein engineering expands the effector recognition profile of a rice NLR immune receptor. eLife 8, e47713. doi: 10.7554/eLife.47713
De la Concepcion, J. C., Franceschetti, M., Maqbool, A., Saitoh, H., Terauchi, R., Kamoun, S., et al. (2018). Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat. Plants 4, 576–585. doi: 10.1038/s41477-018-0194-x
De la Concepcion, J. C., Maidment, J. H. R., Longya, A., Xiao, G., Franceschetti, M., Banfield, M. J. (2021). The allelic rice immune receptor pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface. PloS Pathog. 17, e1009368. doi: 10.1371/journal.ppat.1009368
DeYoung, B. J., Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
DeYoung, B. J., Qi, D., Kim, S. H., Burke, T. P., Innes, R. W. (2012). Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Proc. Natl. Acad. Sci. U S A 14, 1071–1084. doi: 10.1111/j.1462-5822.2012.01779.x
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Donofrio, N. M., Raman, V. (2012). Roles and delivery mechanisms of fungal effectors during infection development: common threads and new directions. Curr. Opin. Microbiol. 15, 692–698. doi: 10.1016/j.mib.2012.10.004
Eitas, T. K., Dangl, J. L. (2010). NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13, 472–477. doi: 10.1016/j.pbi.2010.04.007
Ellis, J. G. (2016). Integrated decoys and effector traps: how to catch a plant pathogen. BMC Biol 14, 13. doi: 10.1186/s12915-016-0235-8
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Ghislain, M., Byarugaba, A. A., Magembe, E., Njoroge, A., Rivera, C., Roman, M. L., et al. (2019). Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol. J. 17, 1119–1129. doi: 10.1111/pbi.13042
Göhre, V., Robatzek, S. (2008). Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. doi: 10.1146/annurev.phyto.46.120407.110050
Grund, E., Tremousaygue, D., Deslandes, L. (2019). Plant NLRs with integrated domains: unity makes strength. Plant Physiol. 179, 1227–1235. doi: 10.1104/pp.18.01134
Guo, L., Cesari, S., de Guillen, K., Chalvon, V., Mammri, L., Ma, M., et al. (2018). Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces. Proc. Natl. Acad. Sci. U S A 115, 11637–11642. doi: 10.1073/pnas.1810705115
Harris, C. J., Slootweg, E. J., Goverse, A., Baulcombe, D. C. (2013). Stepwise artificial evolution of a plant disease resistance gene. Proc. Natl. Acad. Sci. U S A 110, 21189–21194. doi: 10.1073/pnas.131113411
Hogenhout, S. A., van der Hoorn, R. A., Terauchi, R., Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22, 115–122. doi: 10.1094/MPMI-22-2-0115
Hückelhoven, R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. doi: 10.1146/annurev.phyto.45.062806.094325
Jacob, F., Vernaldi, S., Maekawa, T. (2013). Evolution and conservation of plant NLR functions. Front. Immunol. 4. doi: 10.3389/fimmu.2013.00297
Jia, A., Huang, S., Song, W., Wang, J., Meng, Y., Sun, Y., et al. (2022). TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity. Science 377, eabp8180. doi: 10.1126/science.abq8180
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U S A 103, 11086–11091. doi: 10.1073/pnas.0508882103
Kanzaki, H., Yoshida, K., Saitoh, H., Fujisaki, K., Hirabuchi, A., Alaux, L., et al. (2012). Arms race co-evolution of Magnaporthe oryzae AVR-pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907. doi: 10.1111/j.1365-313X.2012.05110.x
Keen, N. T. (1990). Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24, 447–463. doi: 10.1146/annurev.ge.24.120190.002311
Kim, S. H., Qi, D., Ashfield, T., Helm, M., Innes, R. W. (2016). Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351, 684–687. doi: 10.1126/science.aad3436
Kourelis, K., Marchal, C., Kamoun, S. (2021). NLR immune receptor-nanobody fusions confer plant disease resistance. bioRxiv 379, 934–939. doi: 10.1101/2021.10.24.465418
Kourelis, J., Marchal, C., Posbeyikian, A., Harant, A., Komoun, S. (2023). NLR immune receptor–nanobody fusions confer plant disease resistance. Science 379, 934–939. doi: 10.1126/science.abn4116
Kourelis, J., van der Hoorn, R. A. L., Sueldo, D. J. (2016). Decoy engineering: the next step in resistance breeding. Trends Plant Sci. 21, 371–373. doi: 10.1016/j.tplants.2016.04.001
Kroj, T., Chanclud, E., Michel-Romiti, C., Grand, X., Morel, J.-B. (2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626. doi: 10.1111/nph.13869
Lapin, D., Johanndrees, O., Wu, Z., Li, X., Parker, J. E. (2022). Molecular innovations in plant TIR-based immunity signaling. Plant Cell 34, 1479–1496. doi: 10.1093/plcell/koac035
Lapin, D., Kovacova, V., Sun, X., Dongus, J. A., Bhandari, D., von Born, P., et al. (2019). A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31, 2430–2455. doi: 10.1105/tpc.19.00118
Lapin, D., Van den Ackerveken, G. (2013). Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci. 18, 546–554. doi: 10.1016/j.tplants.2013.05.005
Lee, S. K., Song, M. Y., Seo, Y. S., Kim, H. K., Ko, S., Cao, P. J., et al. (2009). Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181, 1627–1638. doi: 10.1534/genetics.108.099226
Liu, W., Wang, G. L. (2016). Plant innate immunity in rice: a defense against pathogen infection. Natl. Sci. Rev. 3, 295–308. doi: 10.1093/nsr/nww015
Liu, Y., Zhang, X., Yuan, G., Wang, D., Zheng, Y., Ma, M., et al. (2021). A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors. Proc. Natl. Acad. Sci. U.S.A. 118, e2110751118. doi: 10.1073/pnas.2110751118
Lukasik, E., Takken, F. L. W. (2009). STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12, 427–436. doi: 10.1016/j.pbi.2009.03.001
Luo, M., Xie, L., Chakraborty, S., Wang, A., Matny, O., Jugovich, M., et al. (2021). A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. 39, 561–566. doi: 10.1038/s41587-020-00770-x
Maidment, J. H. R., Franceschetti, M., Maqbool, A., Saitoh, H., Jantasuriyarat, C., Kamoun, S., et al. (2021). Multiple variants of the fungal effector AVR-pik bind the HMA domain of the rice protein OsHIPP19, providing a foundation to engineer plant defense. J. Biol. Chem. 296, 100371. doi: 10.1016/j.jbc.2021.100371
Maqbool, A., Saitoh, H., Franceschetti, M., Stevenson, C. E., Uemura, A., Kanzaki, H., et al. (2015). Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 4, e08709. doi: 10.7554/eLife.08709
Monaghan, J., Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Narusaka, M., Shirasu, K., Noutoshi, Y., Kubo, Y., Shiraishi, T., Iwabuchi, M., et al. (2009). RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. doi: 10.1111/j.1365-313X.2009.03949.x
Ngou, B. P. M., Ahn, H.-K., Ding, P., Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Nishimura, M. T., Monteiro, F., Dangl, J. L. (2015). Treasure your exceptions: unusual domains in immune receptors reveal host virulence targets. Cell 161, 957–960. doi: 10.1016/j.cell.2015.05.017
Okuyama, Y., Kanzaki, H., Abe, A., Yoshida, K., Tamiru, M., Saitoh, H., et al. (2011). A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66, 467–479. doi: 10.1111/j.1365-313X.2011.04502.x
Pan, Q., Wendel, J., Fluhr, R. (2000). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50, 203–213. doi: 10.1007/s002399910023
Pottinger, S. E., Bak, A., Margets, A., Helm, M., Tang, L., Casteel, C., et al. (2020). Optimizing the PBS1 decoy system to confer resistance to potyvirus infection in Arabidopsis and soybean. Mol. Plant Microbe Interact. 33, 932–944. doi: 10.1094/MPMI-07-19-0190-R
Robinson, N. J., Winge, D. R. (2010). Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562. doi: 10.1146/annurev-biochem-030409-143539
Sarris, P. F., Cevik, V., Dagdas, G., Jones, J. D. G., Krasileva, K. V. (2016). Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14, 8. doi: 10.1186/s12915-016-0228-7
Sarris, P. F., Duxbury, Z., Huh, S. U., Ma, Y., Segonzac, C., Sklenar, J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100. doi: 10.1016/j.cell.2015.04.024
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Segretin, M. E., Pais, M., Franceschetti, M., Chaparro-Garcia, A., Bos, J. I., Banfield, M. J., et al. (2014). Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol. Plant Microbe Interact. 27, 624–637. doi: 10.1094/MPMI-02-14-0040-R
Selin, C., de Kievit, T. R., Belmonte, M. F., Fernando, W. G. (2016). Elucidating the role of effectors in plant-fungal interactions: progress and challenges. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00600
Steele, J. F. C., Hughes, R. K., Banfield, M. J. (2019). Structural and biochemical studies of an NB-ARC domain from a plant NLR immune receptor. PloS One 14, e0221226. doi: 10.1371/journal.pone.0221226
Stein, J. C., Yu, Y., Copetti, D., Zwickl, D. J., Zhang, L., Zhang, C., et al. (2018). Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus oryza. Nat. Genet. 50, 285–296. doi: 10.1038/s41588-018-0040-0
Sun, Y., Wang, Y., Zhang, X., Chen, Z., Xia, Y., Wang, L., et al. (2022). Plant receptor-like protein activation by a microbial glycoside hydrolase. Nature 610, 335–342. doi: 10.1038/s41586-022-05214-x
Takagi, H., Abe, A., Uemura, A., Oikawa, K., Utsushi, H., Yaegashi, H., et al. (2017). Rice blast resistance gene Pii is controlled by a pair of NBS-LRR genes Pii-1 and Pii-2. bioRxiv, 1–32. doi: 10.1101/227132
Takken, F. L., Goverse, A. (2012). How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15, 375–384. doi: 10.1016/j.pbi.2012.05.001
Tran, D. T. V., Chung, E.-H., Habring-Muller, A., Demar, M., Schwab, R., Dangl, J. L., et al. (2017). Activation of a plant NLR complex through heteromeric association with an autoimmune risk variant of another NLR. Curr. Bio 27, 1148–1160. doi: 10.1016/j.cub.2017.03.018
Tsuda, K., Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
van der Biezen, E. A., Jones, J. D. (1998). The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R228. doi: 10.1016/s0960-9822(98)70145-9
van der Hoorn, R. A., Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
Wang, J., Chai, J. (2020). Molecular actions of NLR immune receptors in plants and animals. Sci. China Life Sci. 63, 1303–1316. doi: 10.1007/s11427-019-1687-6
Wang, J., Hu, M., Wang, J., Qi, J., Han, Z., Wang, G., et al. (2019b). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, aav5870. doi: 10.1126/Science
Wang, J., Wang, J., Hu, M., Wu, S., Qi, J., Wang, G., et al. (2019a). Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, 5868–5870. doi: 10.1126/science.aav5868
Williams, S. J., Sohn, K. H., Wan, L., Bernoux, M., Sarris, P. F., Segonzac, C., et al. (2014). Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303. doi: 10.1126/science.1247357
Williams, S. J., Sornaraj, P., deCourcy-Ireland, E., Menz, R. I., Kobe, B., Ellis, J. G., et al. (2011). An autoactive mutant of the m flax rust resistance protein has a preference for binding ATP, whereas wild-type m protein binds ADP. Mol. Plant Microbe Interact. 24, 897–906. doi: 10.1094/MPMI-03-11-0052
Willmann, R., Lajunen, H. M., Erbs, G., Newman, M. A., Kolb, D., Tsuda, K., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U S A 108, 19824–19829. doi: 10.1073/pnas.1112862108
Wirthmueller, L., Maqbool, A., Banfield, M. J. (2013). On the front line: structural insights into plant-pathogen interactions. Nat. Rev. Microbiol. 11, 761–776. doi: 10.1038/nrmicro3118
Xi, Y., Cesaria, S., Kroj, T. (2022). Insight into the structure and molecular model of action of plant paired NLR immune receptors. Essays Biochem. 66, 513–526. doi: 10.1042/EBC20210079
Xiao, G., Wang, W., Liu, M., Li, Y., Liu, J., Franceschetti, M., et al. (2022). The Piks allele of the NLR immune receptor Pik breaks the recognition of AvrPik effectors of rice blast fungus. J. Integr. Plant Biol. 00, 1–16. doi: 10.1111/jipb.13375
Yang, S., Li, J., Zhang, X., Zhang, Q., Huang, J., Chen, J. Q., et al. (2013). Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 110, 18572–18577. doi: 10.1073/pnas.1318211110
Yoshida, K., Saitoh, H., Fujisawa, S., Kanzaki, H., Matsumura, H., Yoshida, K., et al. (2009). Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591. doi: 10.1105/tpc.109.066324
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021a). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Yuan, M., Ngo, B. P. M., Ding, P., Xin, X.-P. (2021b). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030. doi: 10.1016/j.pbi.2021.102030
Yuan, B., Zhai, C., Wang, W. J., Zeng, X., Xu, X., Hu, H., et al. (2011). The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genet. 122, 1017–1028. doi: 10.1007/s00122-010-1506-3
Zhai, C., Lin, F., Dong, Z., He, X., Yuan, B., Zeng, X., et al. (2011). The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 189, 321–334. doi: 10.1111/j.1469-8137.2010.03462.x
Zhai, C., Zhang, Y., Yao, N., Lin, F., Liu, Z., Dong, Z., et al. (2014). Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PloS One 9, e98067. doi: 10.1371/journal.pone.0098067
Zhang, X., He, D., Zhao, Y., Cheng, X., Zhao, W., Taylor, I. A., et al. (2018). A positive-charged patch and stabilized hydrophobic core are essential for a virulence function of AvrPib in the rice blast fungus. Plant J. 96, 133–146. doi: 10.1111/tpj.14023
Zhang, B., Liu, M., Wang, Y., Yuan, W., Zhang, H. (2022). Plant NLRs: evolving with pathogen effectors and engineerable to improve resistance. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1018504
Keywords: protein engineering, molecular structure, paired NLRs, HMA IDs, rice
Citation: Guo L, Mu Y, Wang D, Ye C, Zhu S, Cai H, Zhu Y, Peng Y, Liu J and He X (2023) Structural mechanism of heavy metal-associated integrated domain engineering of paired nucleotide-binding and leucine-rich repeat proteins in rice. Front. Plant Sci. 14:1187372. doi: 10.3389/fpls.2023.1187372
Received: 16 March 2023; Accepted: 09 June 2023;
Published: 28 June 2023.
Edited by:
Wei Wang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Liang Xiangxiu, South China Agricultural University, ChinaCopyright © 2023 Guo, Mu, Wang, Ye, Zhu, Cai, Zhu, Peng, Liu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junfeng Liu, amxpdUBjYXUuZWR1LmNu; Xiahong He, aGV4aWFob25nQGhvdG1haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.