
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 June 2023
Sec. Plant Systematics and Evolution
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1186598
This article is part of the Research Topic Innovations in Natural Dye Production: Bridging Tradition and Modern Technology View all 4 articles
Introduction: Indigofera L. is the third largest genus in Fabaceae and includes economically important species that are used for indigo dye-producing, medicinal, ornamental, and soil and water conservation. The genus is taxonomically difficult due to the high level of overlap in morphological characters of interspecies, fewer reliability states for classification, and extensive adaptive evolution. Previous characteristic-based taxonomy and nuclear ITS-based phylogenies have contributed to our understanding of Indigofera taxonomy and evolution. However, the lack of chloroplast genomic resources limits our comprehensive understanding of the phylogenetic relationships and evolutionary processes of Indigofera.
Methods: Here, we newly assembled 18 chloroplast genomes of Indigofera. We performed a series of analyses of genome structure, nucleotide diversity, phylogenetic analysis, species pairwise Ka/Ks ratios, and positive selection analysis by combining with allied species in Papilionoideae.
Results and discussion: The chloroplast genomes of Indigofera exhibited highly conserved structures and ranged in size from 157,918 to 160,040 bp, containing 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Thirteen highly variable regions were identified, of which trnK-rbcL, ndhF-trnL, and ycf1 were considered as candidate DNA barcodes for species identification of Indigofera. Phylogenetic analysis using maximum likelihood (ML) and Bayesian inference (BI) methods based on complete chloroplast genome and protein-coding genes (PCGs) generated a well-resolved phylogeny of Indigofera and allied species. Indigofera monophyly was strongly supported, and four monophyletic lineages (i.e., the Pantropical, East Asian, Tethyan, and Palaeotropical clades) were resolved within the genus. The species pairwise Ka/Ks ratios showed values lower than 1, and 13 genes with significant posterior probabilities for codon sites were identified in the positive selection analysis using the branch-site model, eight of which were associated with photosynthesis. Positive selection of accD suggested that Indigofera species have experienced adaptive evolution to selection pressures imposed by their herbivores and pathogens. Our study provided insight into the structural variation of chloroplast genomes, phylogenetic relationships, and adaptive evolution in Indigofera. These results will facilitate future studies on species identification, interspecific and intraspecific delimitation, adaptive evolution, and the phylogenetic relationships of the genus Indigofera.
Fabaceae is considered the third largest family of angiosperms, which is traditionally divided into three subfamilies including Caesalpinioideae, Mimosoideae, and Papilionoideae (Lewis et al., 2005; LPWG, 2013). However, six subfamilies (i.e., Caesalpinioideae, Cercidoideae, Detarioideae, Dialioideae, Duparquetioideae, and Papilionoideae) were proposed based on a phylogenetic framework reconstructed from the chloroplast matK gene (LPWG, 2017). Indigofera L. is the third largest genus in Fabaceae, comprising approximately 750 species with a pantropical distribution (Schrire, 2005). The genus has four centers of diversity in Africa and Madagascar (c. 550 species), Asia, especially the temperate Sino-Himalayan region (c. 105 species), Australia (c. 50 species), and the New World (c. 45 species) (Schrire et al., 2009). The genus Indigofera includes economically important species (Gerometta et al., 2020), used for a variety of purposes. Indigofera tinctoria L. and I. suffruticosa Mill. are the main sources for the production of natural indigo (Marquiafável et al., 2009; Baran et al., 2010; Calvo et al., 2011; Schrire, 2013; Zhang et al., 2019; Gerometta et al., 2020), which has the advantages of low toxicity and abundant availability compared to chemically synthesized dyes (Shahid-ul-Islam et al., 2013; Pattanaik et al., 2019). Moreover, indigo production is one of indispensable economic industries in India (Gulrajani, 2001; Bechtold et al., 2002). Indigofera also includes antiarthritic, anthelmintic, anticancer, antibacterial, and antidiabetic medicinal plants (Rajkapoor et al., 2009; Kumar et al., 2011; Dkhil et al., 2020), and a host of forage crops, ornamental, and soil conservation plants (Hassen, 2007; Marquiafável et al., 2009; Schrire, 2013). Due to the large number of species, often homogeneous morphology, overlap in the morphology of related species, and wide distribution, Indigofera is a taxonomically difficult genus at both the morphological and molecular levels (Schrire, 1995; Zhao, 2016). To evaluate generic relationships in the tribe Indigofereae, Barker et al. (2000) conducted the first molecular phylogenetic analyses of Indigofereae based on nuclear ITS and two chloroplast DNA (cpDNA) regions (trnL and trnK introns). Generic relationships were not well resolved because of the inconsistent phylogenetic topologies from cpDNA and ITS sequences. These early efforts suggested that these two cpDNA regions were inadequate for phylogenetic reconstruction within Indigofereae due to their low resolution. Subsequently, phylogenetic analyses of Indigofera have been performed using nuclear ITS sequences (Schrire et al., 2003, 2009; Zhao, 2016). Schrire et al. (2003, 2009). classified Indigofera into four large monophyletic clades based on a combined molecular (nuclear ITS) and morphological data set, i.e., the Pantropical, Palaeotropical, Tethyan/Boreotropical, and Cape clades. All of these studies have laid an important foundation for the taxonomy and identification of Indigofera species. However, the lack of cpDNA data limits our understanding of the phylogenetic relationships, biogeography, and evolutionary history of Indigofera. Based on these studies, Zhao (2016) conducted a large-scale phylogeny of 295 Indigofera species based on ITS sequences, with a focus on Sino-Himalayan endemics, and a rapid radiation in the Sino-Himalayan region was detected.
Chloroplasts play critical roles in the survival, adaptation, and evolution of plants (Wicke et al., 2011; Zhao et al., 2019; Dopp et al., 2021). Chloroplast DNA has an independent transcription and transport system and encodes ribosomal proteins related to photosynthesis (Jansen and Ruhlman, 2012). The chloroplast genome generally is a circular quadripartite structure with a length of 107 kb (Cathaya argyrophylla Chun & Kuang)—218 kb (Pelargonium hortorum L. H. Bailey) (Sato et al., 1999; Lin et al., 2010; Daniell et al., 2016), and composed of a pair of inverted repeat regions (IRs) separated by a large single-copy (LSC) region and a small single-copy (SSC) region (Kolodner and Tewari, 1979; Sugiura, 1992; Daniell et al., 2016). Given its uniparental inheritance, moderate mutation rate, and relative ease of sequencing, the chloroplast genome is widely used as an effective resource for exploring the origin and evolution of plants, understanding the phylogenetic relationships of different taxonomic categories, and identifying species (Provan et al., 2001; Bock and Knoop, 2012; Schwarz et al., 2015; Lei et al., 2016; Asaf et al., 2017a; Liang et al., 2020; Chen et al., 2022). Recent advances in next-generation sequencing (NGS) and computational resources have enabled unparalleled phylogenomic analyses (Asaf et al., 2017b; Choi et al., 2022). Complete chloroplast genome data have been sporadically applied in legume phylogenomic analyses at a range of different taxonomic levels, such as in Campylotropis Bunge (Feng et al., 2022b), Sophora L. (Liao et al., 2021), Dalbergia L. f. (Song et al., 2019), Millettioid/Phaseoloid clade (Oyebanji et al., 2020), Papilionoideae (Choi et al., 2022), and Fabaceae (Zhang et al., 2020b). The chloroplast genome also provides insights into other aspects of evolutionary processes, such as adaptive evolution driven by natural selection. Recent studies have detected a number of positively selected chloroplast genes associated with adaptive evolution (Xie et al., 2019; Zhao et al., 2021; Yang et al., 2022). As a large and widespread genus, Indigofera occupies diverse pantropical habitats, where ecology and geography uniquely shaped the Indigofera phylogeny (Schrire et al., 2005a, 2005b). The environment, herbivores, and pathogens have imposed strong pressures on Indigofera species (Schrire, 2013). However, few studies have investigated the adaptive evolution of Indigofera, and to date, only six complete chloroplast genomes of Indigofera species have been reported (Zhang et al., 2020b, 2022; Zhao et al., 2023).
In this study, we sequenced, assembled, and annotated plastomes of 17 Indigofera species based on a taxonomically representative sampling. By leveraging another 19 previously published cp genomes from Papilionoideae, we aimed to: (1) examine the diversity of chloroplast genomes of Indigofera, (2) identify promising molecular markers for future study, (3) preliminarily explore the systematic position and phylogenetic relationships of the genus Indigofera based on cp genomes, and (4) investigate selective or adaptive evolution in the cp genomes of Indigofera species.
Seventeen Indigofera species were selected for sampling based on their morphological characteristics and prior phylogenetic studies of Indigofera, which covered four ITS-based monophyletic clades (i.e., the Pantropical, Palaeotropical, Tethyan, and East Asian clades) of Indigofera (Schrire et al., 2003, 2009; Zhao, 2016), as well as nine morphology-based subsections (Fang and Zheng, 1994) (Figure 1). Fresh and healthy leaves were collected from natural populations, and then directly dried with silica gel. The voucher specimens were identified by Dr. Xue-Li Zhao and Prof. Xin-Fen Gao according to Flora of China (Gao and Schrire, 2010) and the speciemens of CVH (Chinese Virtual Herbarium; https://www.cvh.ac.cn/) and JSTOR (https://www.jstor.org/), and deposited at the Herbarium of Chengdu Institute of Biology, Chinese Academy of Sciences (CDBI). Voucher information and GenBank accession numbers of newly generated cp genomes are listed in Table 1.
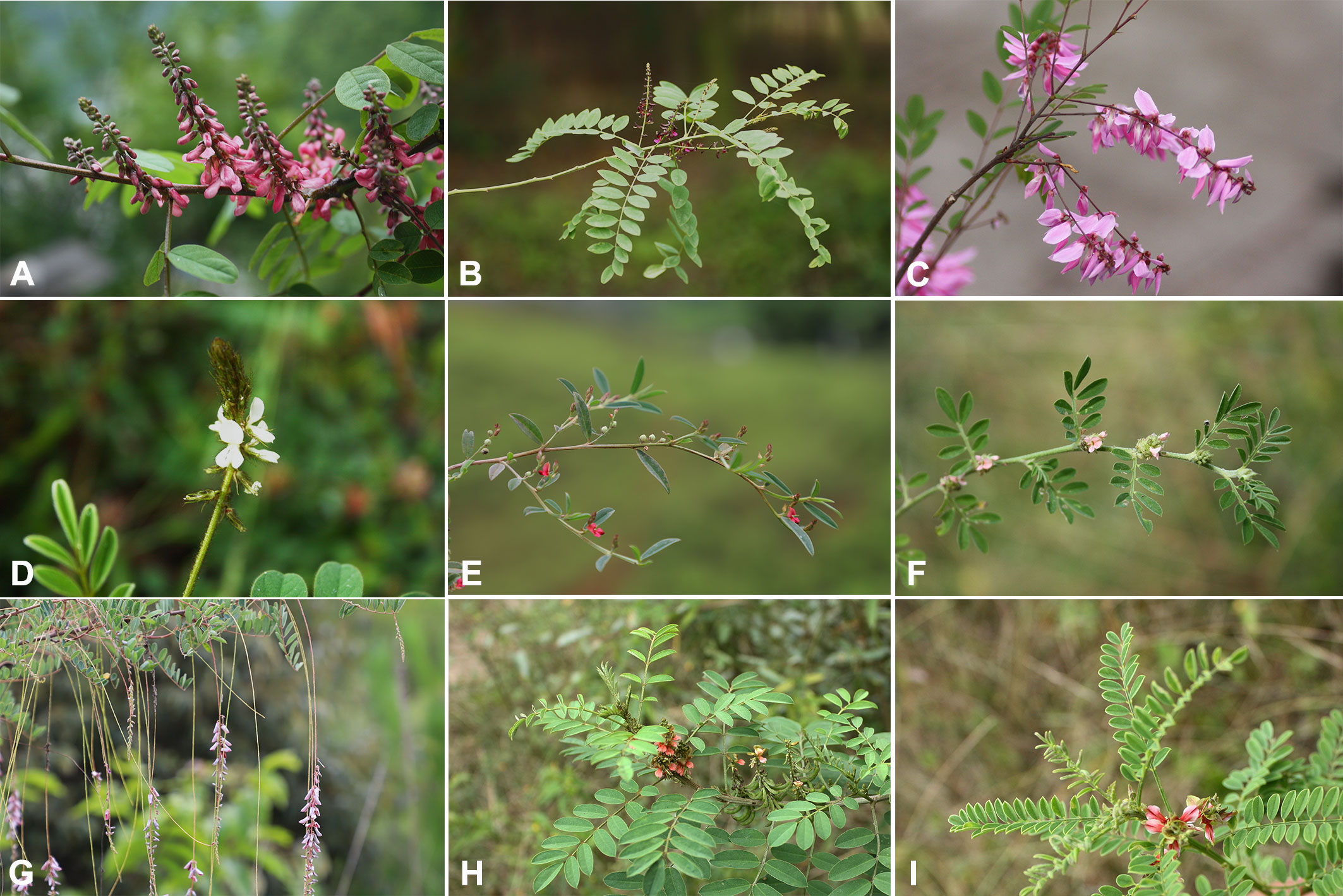
Figure 1 Morphological characteristics of Indigofera species. (A) I. amblyantha, (B) I. atropurpurea, (C) I. decora, (D) I. hirsuta, (E) I. linifolia, (F) I. linnaei, (G) I. pendula, (H) I. suffruticosa, (I) I. wightii.
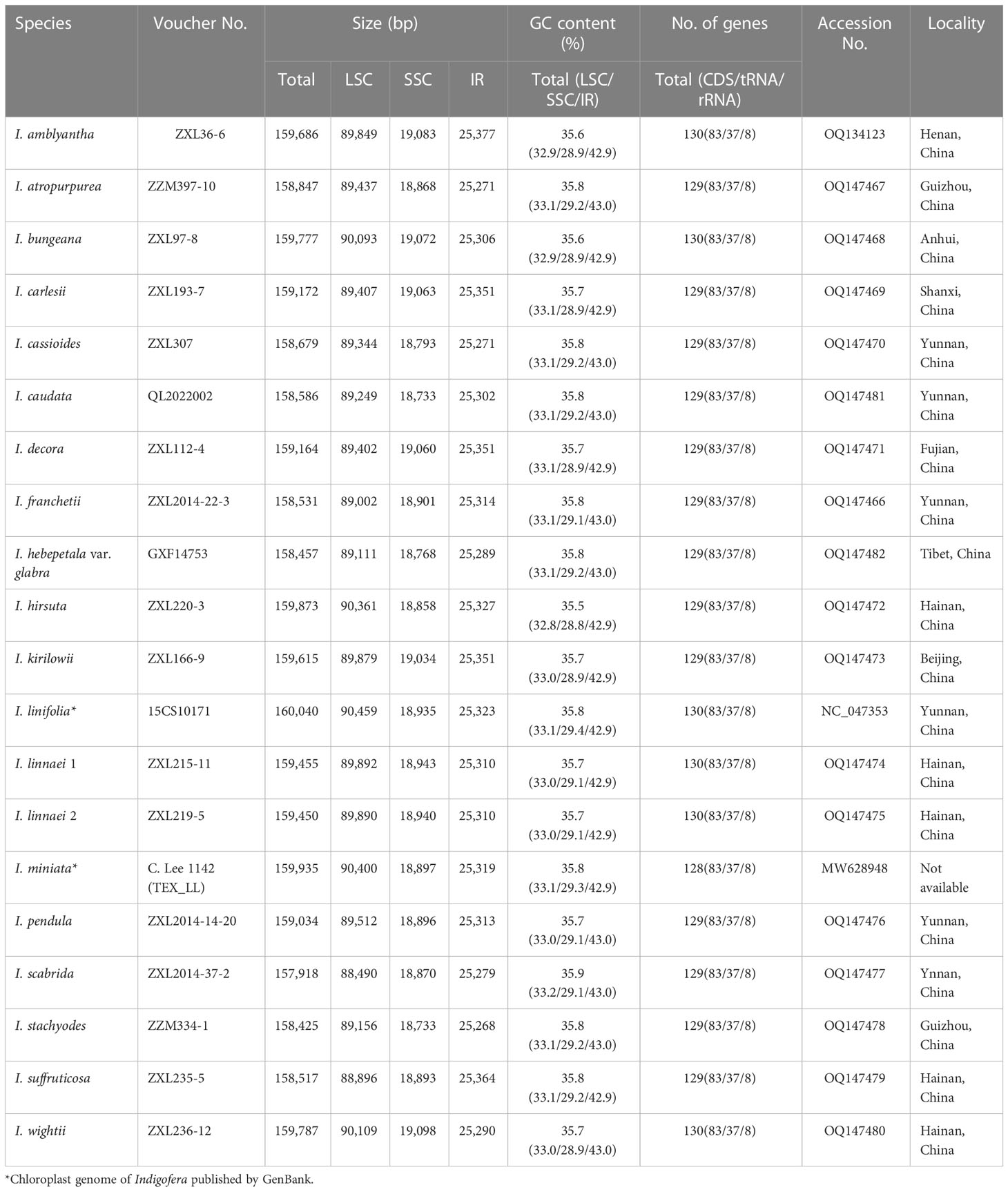
Table 1 Characteristics of the 20 complete chloroplast genomes of Indigofera, including 18 newly generated plastomes and two published plastomes.
Total genomic DNA was extracted from leaf tissues using a Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China). The DNA concentration, purity, and integrity were evaluated using the Nanodrop 2000 spectrophotometry (Thermo Scientific, US) and 1% agarose gel electrophoresis. High-quality DNA samples were used to construct paired-end libraries. The paired-end libraries were subsequently sequenced on the Illumina NovaSeq 6000 platform (PE150) at Annoroad Gene Technology Co., Ltd (Beijing, China).
The raw data were trimmed to remove adapters and low-quality reads using Trimmomatic v.0.39 (Bolger et al., 2014). The clean reads were then assembled using GetOrganelle v1.7.6.1 (Jin et al., 2020) with the k-mers of 21, 45, 65, 85, 105, and 121. De novo assembly graphs were visualized using Bandage v0.8.1 (Wick et al., 2015). Geneious Prime 2022.1.1 (Biomatters, Auckland, New Zealand) was used to identify the location of IRs. Gene annotation was conducted using Plastid Genome Annotator (Qu et al., 2019) with a published chloroplast genome of Indigofera linifolia (L. f.) Retz. (NC_047353) as a reference, and the start/stop codons and pseudogenes were further manually checked. The physical chloroplast genome maps were drawn using OGDRAW v1.3.1 (Greiner et al., 2019).
A total of 18 newly assembled and two published Indigofera cp genomes (I. linifolia and I. miniata Ort.) (Table 1) were analyzed. Sequences were aligned using the program MAFFT v7 (Katoh and Standley, 2013) with default parameters. The boundaries of LSC, SSC, and IRs in the cp genomes were identified and visualized using IRscope (Amiryousefi et al., 2018). The GC content, gene components, and length of the whole cp genome, LSC, SSC, and IRs were analyzed using Geneious Prime 2022.1.1 (Biomatters Ltd. Auckland, New Zealand). Variable and parsimony-informative sites were detected by MEGA v11.0.11 (Tamura et al., 2021). Nucleotide diversity (Pi) was calculated using DnaSP v6.12.03 (Rozas et al., 2017) with a window length of 600 bp and step size of 200 bp.
Simple sequence repeats (SSRs) were detected using MISA v2.1 (Beier et al., 2017). Thresholds of 11, 6, 5, 4, 3, and 3 repeat units were set for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively. Long repeat sequences (forward, reverse, palindromic, and complementary) were identified using REPuter (Kurtz et al., 2001), with a minimum repeat size of 30 bp and a Hamming distance of 3.
To investigate the systematic placement and phylogenetic relationships of the Indigofera species, 19 publicly available cp genome sequences in Papilionoideae were downloaded from NCBI (Table 1 and Supplementary Table S1). A total of 37 cp genomes were used to construct phylogenetic relationships. Phylogenetic analyses were conducted using maximum likelihood (ML) and Bayesian inference (BI) methods based on two data sets: once using the complete plastome, and once constraining analysis to the concatenated protein-coding genes (PCGs). The PCGs were extracted from each cp genome, then concatenated and aligned with PhyloSuite v1.2.2 (Zhang et al., 2020a). Multiple sequence alignment was aligned by MAFFT v7 (Katoh and Standley, 2013), and then trimmed using trimAL v1.2 (Capella-Gutiérrez et al., 2009) with automatic mode to reduce potentially poorly aligned regions. The trimmed alignment was visually examined and manually adjusted in Geneious Prime 2022.1.1 (Biomatters Ltd. Auckland, New Zealand).
The best-fit models of nucleotide substitution were estimated by jModelTest v2.1.10 (Guindon and Gascuel, 2003; Darriba et al., 2012) using a corrected Akaike Information Criterion (AIC) score (Akaike, 1974). ML analyses were implemented using IQ-TREE v2.2.0 (Minh et al., 2020) under the GTR+I+J model with 1,000 bootstrap replicates. Bayes inference was performed using MrBayes v3.2.7 (Ronquist et al., 2012) under the GTR+I+G model. Markov Chain Monte Carlo (MCMC) algorithms have two parallel runs with 10,000,000 generations independently, and sampling every 1,000 generations. The initial 25% of trees were discarded as burn-in, and the remaining trees were used to construct a 50% majority-rule consensus tree and calculate the posterior probability (PP) values using Tracer v1.7.1 (Rambaut et al., 2018). The phylogenetic trees were visualized and processed using Interactive Tree Of Life (iTOL) v5 (Letunic and Bork, 2021) and Figtree v1.4.4 (https://github.com/rambaut/figtree/releases/tag/v1.4.4).
Each shared PCG sequence of all species was extracted and aligned with MAFFT v7 (Katoh and Standley, 2013). Pairwise Ka/Ks of all species were calculated based on the concatenated PCGs alignment using DnaSP v6.12.03 (Rozas et al., 2017).
Positive selected analyses of shared genes were performed with the branch-site model (Yang and Reis, 2011) and Bayesian Empirical Bayes (BEB) methods (Yang et al., 2005). A total of 70 shared PCGs of all species were extracted and aligned with MACSE v2 (Ranwez et al., 2018). Before calculation, stop codons and gaps were removed. The branch-site model was implemented to assess potential positive selection in the CODEML algorithm from the PAML v4.10.6 package (Yang, 1997, 2007). A null hypothesis (model = 2, NSsites = 2, omega = 1, fix_omega = 1) and an alternative hypothesis (model = 2, NSsites = 2, omega = 0, fix_omega = 1.5) models were applied separately. The ratio (ω) of the non-synonymous substitution rate to the synonymous substitution rate was used to measure the selective pressure. Whether ω > 1, ω = 1, or ω < 1 indicates positive selection, neutral evolution, and purifying selection, respectively (Yang and Nielsen, 2002). The likelihood-ratio tests (LRT) were performed according to Lan et al. (2017). The posterior probabilities of amino acid sites were calculated with the BEB method to identify specific positions experiencing positive selection (Yang et al., 2005). A gene with a test p-value < 0.05 at a site associated with positive selection was thus considered a positively selected gene (Xie et al., 2019).
Complete chloroplast genomes of 17 Indigofera species were newly sequenced and annotated in the present study, of which four species (I. pendula Franch., I. franchetii X. F. Gao & Schrire, I. amblyantha Craib and I. carlesii Craib) are endemic to China. We studied the basic information of plastomes of 17 species of Indigofera using the published sequences from I. linifolia and I. miniata in GenBank. Illumina sequencing generated about 3 Gb of paired-end raw sequence data for each newly sampled Indigofera species. The complete cp genome ranged from 157,918 bp (I. scabrida Dunn) to 160,040 bp (I. linifolia). Every species exhibited a typical quadripartite genome structure, consisting of an LSC region (88,490—90,459 bp) and an SSC region (18,733—19,098 bp) separated by a pair of inverted repeats (25,268—25,377 bp) (Table 1 and Figure 2). The GC content of these complete cp genomes ranged from 35.5% (I. hirsuta L.) to 35.9% (I. scabrida), in which the IR regions possessed the highest GC content (42.9%—43%), followed by the LSC (32.8%—33.2%), and SSC regions (28.8%—29.4%). The annotated cp genomes of Indigofera contained a total of 128—130 genes, including 83 protein-coding genes (10 in the IR regions), 37 tRNA genes (14 in the IR regions), and eight rRNA genes (all in the IR region) (Table 1 and Figure 2). Of these genes, four tRNA genes (trnG-UCC, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron, and two tRNA genes (trnA-UGC and trnI-GAU) contained two introns (Table 2). Nineteen genes contained two copies, including eight protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1, and ycf2), four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5), and seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC). In these cp genomes, rps12 was identified as a trans-spliced gene, with the 5’-end in the LSC region, and the 3’-end in the IR regions.
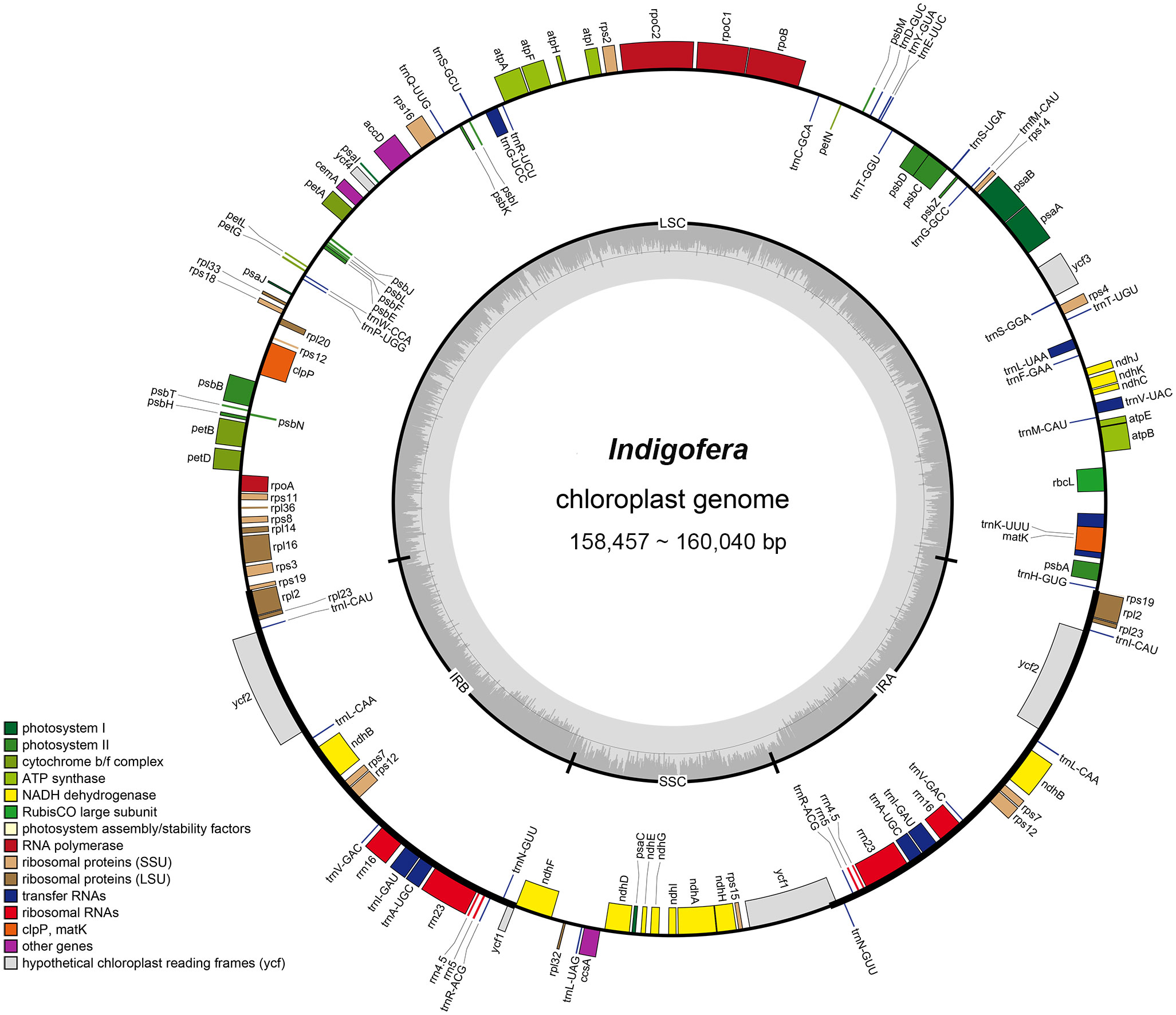
Figure 2 The chloroplast genome map of Indigofera species. Genes inside and outside of the circle are transcribed clockwise and counterclockwise, respectively. Genes belonging to different functional groups are color-coded. The darker and lighter gray color in the inner circle corresponds to the GC content and AT content, respectively.
We observed both contraction and expansion of IRs at the SC-IR boundary. Among these 20 cp genomes, the IRs regions varied from 25,268 to 25,377 bp (Table 1). The rps19 and rpl2 were located at the JLB (LSC/IRb) boundary, and the ycf1 pseudogene (except for I. miniata) and ndhF at the JSB (SSC/IRb) boundary (Supplementary Figure S1). At the JLB boundary, the rps19 region in I. amblyantha, I. bungeana Walp., I. linifolia, I. linnaei Ali, I. miniata, and I. wightii Graham expanded 2—6 bp toward the IRb region and led to the production of rps19 pseudogene in the IRa (except in the case of I. miniata). The ycf1 crossed the JSA (SSC/IRa) boundary with 4,881—4,920 bp in the SSC region and expanded from 456 to 478 bp into the IRa region, which led to the ycf1 pseudogene (except for I. miniata). At the JSB boundary, the ndhF region of I. amblyantha, I. atropurpurea Bench.-Ham. ex Hornem., I. bungeana, I. cassioides Rottler ex Candolle, I. hebepetala var. glabra Ali, I. hirsuta, I. linifolia, I. stachyodes Lindl., I. suffruticosa, and I. wightii expanded 3—12 bp toward IRb.
To explore variable regions with high resolution for species identification in Indigofera, we conducted a sliding windows analysis of the aligned sequences of 20 cp genomes. The sliding windows analysis observed 13 highly variable regions, comprising the intergenic spacer regions and introns: trnK-rbcL (1,542 bp), ndhJ-trnF (761 bp), trnL-rps4 (1,846 bp), ycf3-psaA (1,075 bp), rpoC2 (621 bp), atpI-atpH (1,163 bp), atpH-atpF (983 bp), psbK-rps16 (734 bp), petA-psbJ (748 bp), trnW-psaJ (1,449 bp), ndhF-trnL (1,699 bp), ndhG-ndhI (1,153 bp), and ycf1 (627 bp) (Table 3 and Figure 3). The 13 highly variable regions had remarkably higher Pi values (0.02329 to 0.03725), and the largest Pi value was for trnK-rbcL (0.03725). All these highly variable regions were located in the LSC and SSC regions (Figure 3 and Supplementary Table S2).
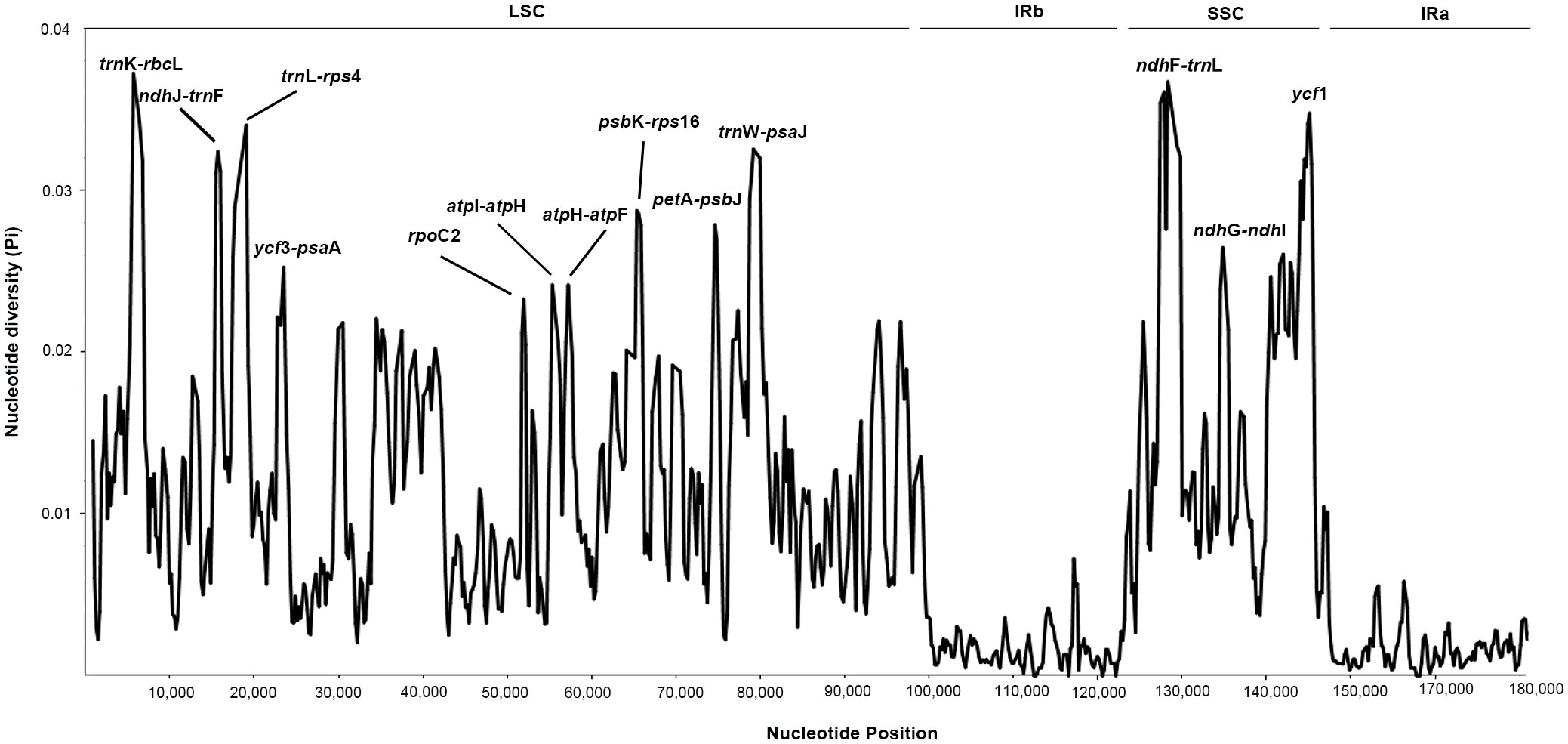
Figure 3 The nucleotide diversity (Pi) value (Y-axis) with their positions (X-axis) in each window of the Indigofera species chloroplast genomes based on sliding window analysis. The 13 regions with high diversity are indicated above the peaks.
The number of SSRs in 20 Indigofera plastomes varied from 55 in I. linifolia to 97 in I. hirsuta. The most abundant SSRs were mononucleotide repeats, where the number varied from 43 to 73, followed by dinucleotide repeats (6—24) (Figure 4A and Supplementary Table S3). Moreover, 94.03%—100% of SSRs were distributed in the LSC and SSC regions (Supplementary Table S4). Mononucleotides and dinucleotides were found in all species of Indigofera and trinucleotides and pentanucleotides were present in all species except I. wightii or I. caudata. Tetranucleotides, hexanucleotides, and complex nucleotides were detected in some species (Figure 4A and Supplementary Table S3). We found that SSRs in the plastomes of Indigofera contained a large number of A/T pairs and AT/AT repeats (Figure 4B and Supplementary Table S5).
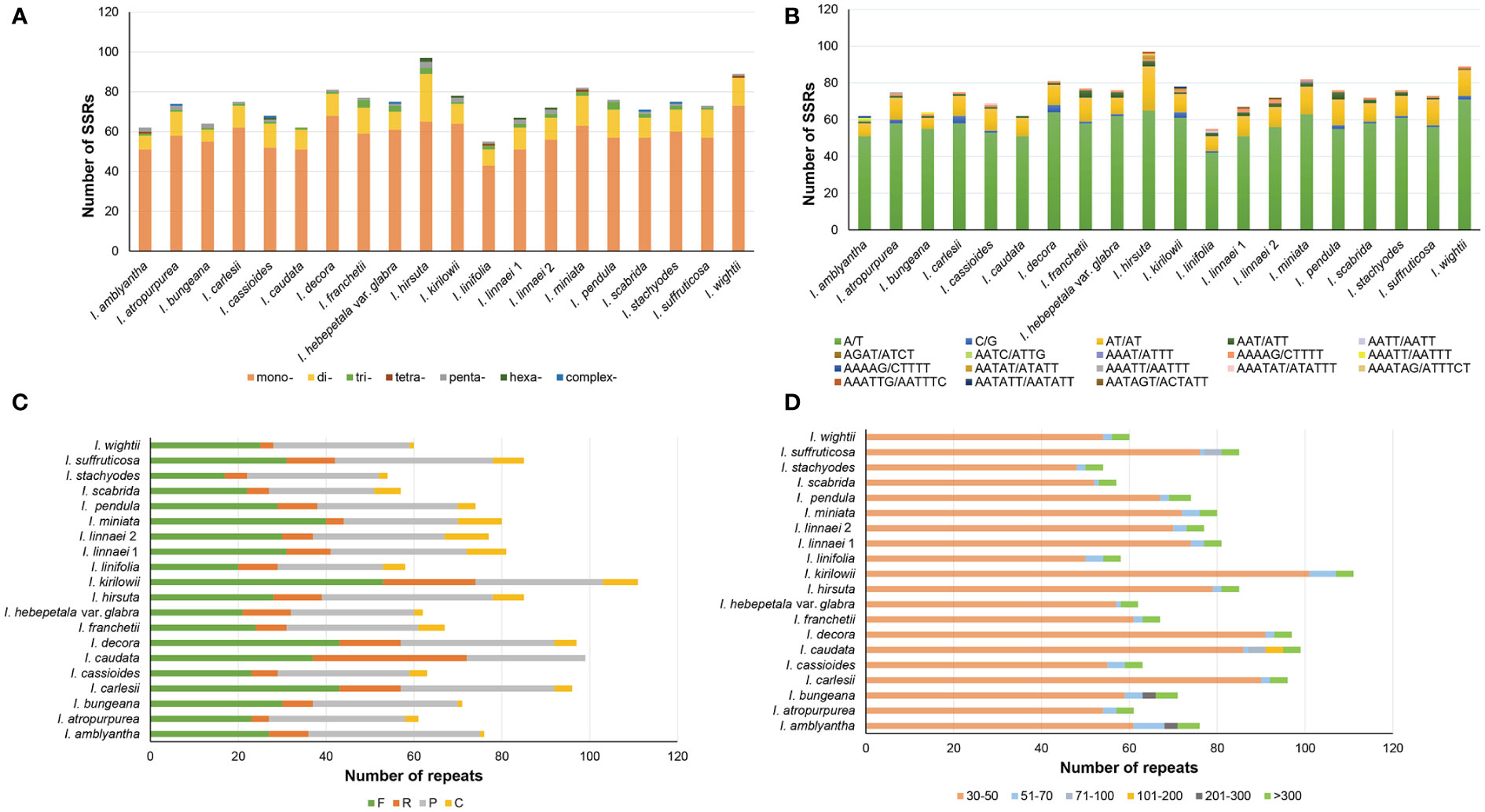
Figure 4 Analysis of SSRs and long sequence repeat in each species. (A) The number of different SSR types. (B) The number of the identified SSRs in different repeat class types. (C) The type and number of long repeat sequences. (D) Length distribution of long repeat sequences.
A total of 1,514 repeat sequences were identified in 20 Indigofera plastomes using REPuter, including 597 forward (39.43%), 202 reverse (13.34%), 620 palindromic (40.95%), and 95 complementary (6.27%) repeats (Figure 4C and Supplementary Table S6). The proportion of long repeats detected in the LSC, SSC, IRa, and IRb regions was 25.25%—77.59%, 3.33%—52.53%, 1.72%—7.06%, and 12.07%—25%, respectively (Supplementary Table S7). Repeat lengths less than 100 bp were the most common, accounting for 88.73%—96.40% of total repeats. Among these, long repeat lengths of 30—50bp were the most common. Repeat lengths of 101—200 bp were only observed in I. caudata, and 201—300 bp repeats only in I. amblyantha and I. bungeana. However, repeat lengths greater than 300 bp were detected in all species (Figure 4D and Supplementary Table S8).
The data matrix of the complete genome used in phylogenetic analyses consisted of 129,037 nucleotide sites, of those, 34,002 (26.35%) were parsimony informative. The PCG data matrix consisted of 69,282 nucleotide sites, of these, 8,906 (12.85%) were parsimony informative. The ML and BI analyses generated identical tree topologies of Indigofera, whereas the topological discrepancies were observed in the Robinioid and Genistoid clades among phylogenies from two datasets (Figure 5 and Supplementary Figures S2, S3).
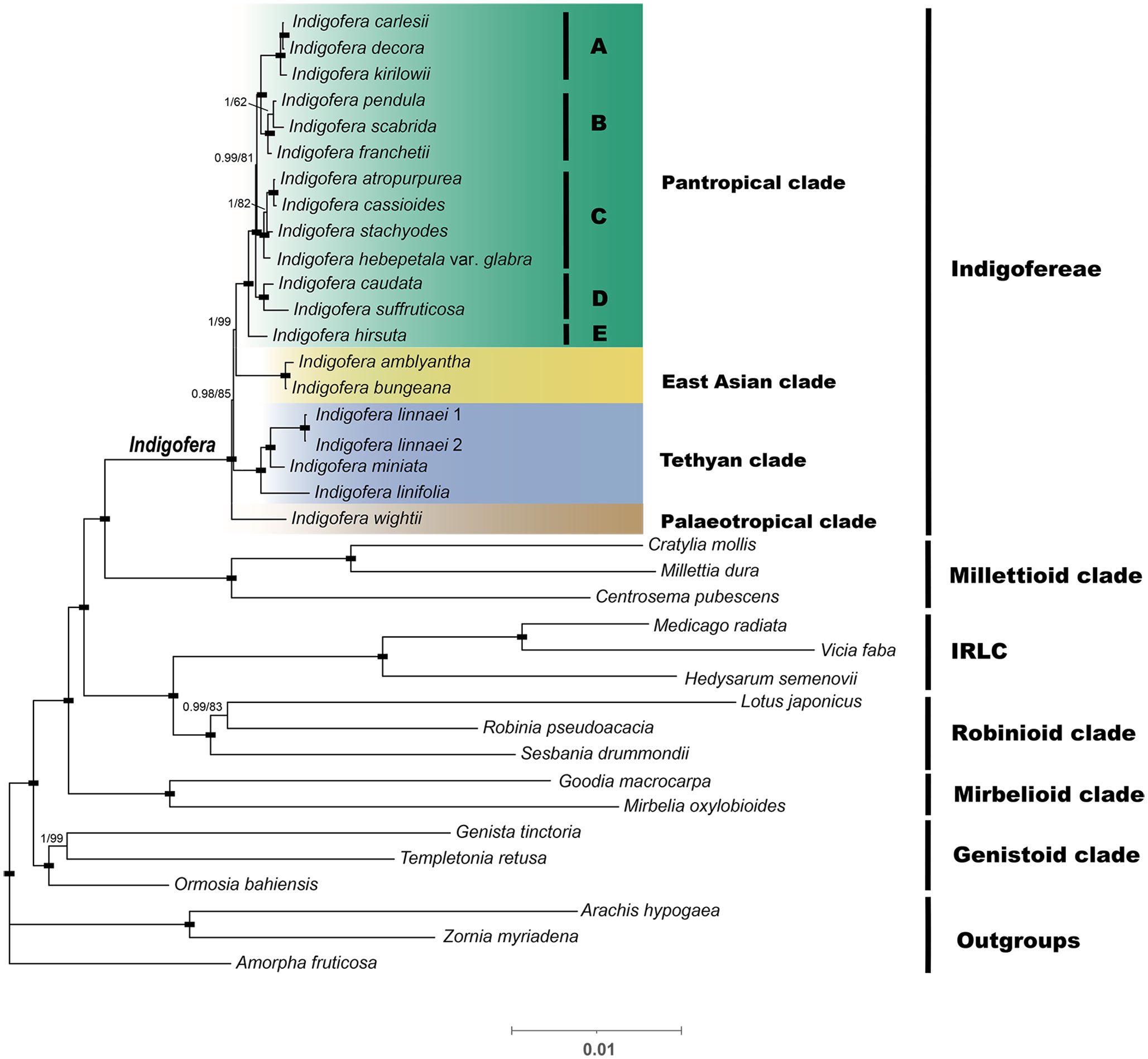
Figure 5 A Bayesian inference (BI) tree based on a concatenated dataset of protein-coding genes (PCGs). The numbers above the branches are Bayesian posterior probabilities (PP; before the slash) and ML bootstrap supports values (BS; after the slash). The black rectangle on the node indicates support for BS=100 and PP=1. The paleotropical clade includes five lineages, i.e., A-E.
The plastid phylogenomic analyses generated highly supported phylogeny with six monophyletic clades, i.e., Millettioid, IRLC, Robinioid, Mirbelioid, Genistoid, and Indigofera clades, with the Millettioid clade being the most closely related to Indigofera. All phylogenetic topologies fully supported a monophyletic Indigofera [100% bootstrap support (BS) and 1 posterior probability (PP); Figure 5 and Supplementary Figures S2, S3]. Nineteen Indigofera species were resolved into four monophyletic clades, namely Pantropical, East Asian, Tethyan, and Palaeotropical clades according to Schrire et al. (2009) and Zhao (2016). Within the Pantropical clade, 13 species formed five monophyletic lineages (Figure 5A-E). The East Asian clade was composed of I. amblyantha and I. bungeana. Within the Tethyan clade, I. linnaei was sister to I. miniata and related to I. linifolia. The Palaeotropical clade included only I. wightii. In contrast to the PCGs phylogenetic trees, the ML and BI trees based on the complete cp genome suggested I. pendula is a highly supported sister to I. franchetii (100% BS, 1 PP) in the Pantropical clade. Moreover, our analyses suggested the Tethyan clade was moderately (85%/66% BS, 0.98/0.99 PP) supported as a sister clade of the Pantropical and East Asian clades (Figure 5 and Supplementary Figures S2, S3).
The pairwise Ka/Ks ratios of each species pair were calculated, and the ratios ranged from 0 to 0.6 (Figure 6 and Supplementary Table S9). Higher pairwise Ka/Ks ratios were observed in Indigofera species pairs than non-Indigofera species pairs. In addition, high Ka/Ks ratios were detected in the species pairs associated with the Pantropical clade, such as I. franchetii vs. I. pendula, I. decora Lindl. vs. I. carlesii, I. kirilowii Maxim. ex Palibin vs. I. decora and I. kirilowii vs. I. carlesii.
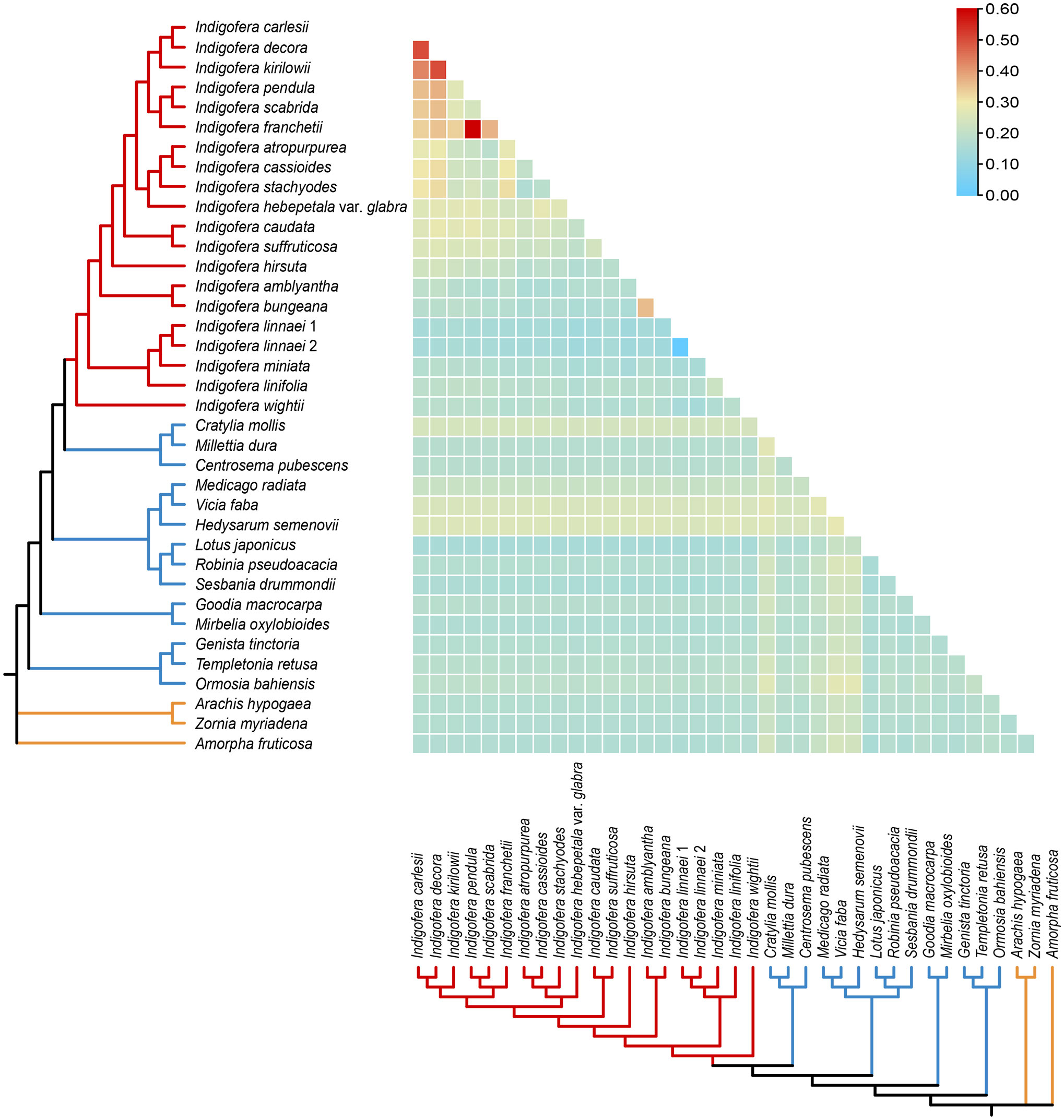
Figure 6 Pairwise Ka/Ks ratios in Indigofera and allied species. This heatmap shows pairwise Ka/Ks ratios between every concatenated shared PCGs sequence in the multigene nucleotide alignment. The scale factors associated with each value are shown on the top right side of the figure.
A total of 70 PCGs were used for positive selection analysis with the branch-site model (Table 4). No p-values were significant in each gene range by the likelihood ratio test, however, 13 PCGs (accD, ccsA, matK, ndhA, ndhE, ndhF, ndhI, petA, petN, psaA, psaB, rpoB, and rpoC1) showed significant posterior probabilities, indicating sites with positive selection based on the BEB test. Notably, the posterior probability of the amino acid residues of petN was as high as 0.968. Among these genes, most genes only had one positive selective site, whereas accD, ccsA, matK, and psaA have two positive selective sites.
Comparative analysis of the 20 Indigofera cp genomes showed highly conserved genes and structures. The genome size, gene content, and gene organization of Indigofera varied little and GC content was consistent with observations in other legume taxa (Table 1) (Keller et al., 2017; Liao et al., 2021; Feng et al., 2022b). Like most angiosperms, the cp genomes of Indigofera exhibited the typical quadripartite structure, consisting of the LSC, SSC, IRa, and IRb regions (Figure 2). IR regions were more conserved compared with single-copy regions, consistent with most higher plants, something largely caused by repeated corrections caused by gene transformations between the two IR regions (Zhang et al., 2016).
The contraction and expansion of the IR regions has been proposed as an important source of size variation in cp genomes (Aii et al., 1997; Huang et al., 2014). However, in our present study, all taxa exhibited a highly conserved pattern of IR boundaries with only slight structural variations. Consistent with other research, we suggested that contraction and expansion of the IR regions can lead to the creation of pseudogenes (Raman et al., 2017; Gao et al., 2018; Ruang-Areerate et al., 2021; Han et al., 2022; Lian et al., 2022; Li et al., 2022c). A comparison of 20 Indigofera plastomes revealed that rps19, ycf1, and ndhF demonstrated pronounced expansion or contraction, as reported in Salvia L., Amygdalus L., Iris L., Prunus L. subg. Cerasus Mill., and Asteraceae (Du et al., 2021; Pascual-Diaz et al., 2021; Du et al., 2022; Feng et al., 2022a; Li et al., 2022c). These subtle differences in boundaries could potentially be used for species identification.
The chloroplast genome has a relatively conserved structure and moderate evolutionary rate. It is also inherited uniparentally and thus less affected by paralogous genes when constructing phylogenetic relationships (Wolfe et al., 1987; Jansen and Ruhlman, 2012; Gitzendanner et al., 2018). Because of these reasons, the cp genome has often been used to construct phylogenies (Aecyo et al., 2021; Yang et al., 2021; Chen et al., 2022; Tong et al., 2022). DNA barcodes based on cp genome screening have been widely applied for species identification, wide-range phylogenetic analyses, and population genetics, such as rbcL, matK, rpoC1, atpF-atpH, psbK-psbI, rpoB, and trnH-psbA (Kress et al., 2005; Newmaster et al., 2006; Chase et al., 2007; Hollingsworth et al., 2011; Dong et al., 2012; Li et al., 2021; Chen et al., 2022).
Previous studies of Indigofera and its allied genera mainly include five plastid regions, comprising three intergenic regions of ndhJ-trnF, atpH-atpF, trnD-trnT, and two introns of trnL and matK (Barker et al., 2000; Peng et al., 2015; Zhao and Gao, 2015; Zhao et al., 2017). Among these hypervariable regions identified in the present study, only two intergenic regions (ndhJ-trnF, atpH-atpF) have ever been used as genetic markers in previous studies, i.e., phylogenetic analyses of Indigofera (Zhao and Gao, 2015), and population genetic analyses of both the I. bungeana (Zhao et al., 2017) and I. decora complexes (Peng et al., 2015). Although most genes in the cp genomes of Indigofera were observed to be conserved in the present study, the intergenic spacers of trnK-rbcL, ndhJ-trnF, trnL-rps4, ycf3-psaA, atpI-atpH, atpH-atpF, psbK-rps16, petA-psbJ, trnW-psaJ, ndhF-trnL, and ndhG-ndhI, and introns of ycf1, rpoC2 exhibited divergence and were identified as potential biomarkers to indicate species (Table 3 and Figure 3). The intergenic regions of trnK-rbcL and ndhF-trnL, which were predicted to have the highest nucleotide diversity, could be used as candidate DNA barcodes for fast species identification in Indigofera. In addition, the ycf1 gene, a large open reading frame, is commonly detected in land plant plastomes and is generally considered to play crucial roles in plant development (Drescher et al., 2000; Martinez-Alberola et al., 2013). The high variation rate of the ycf1 gene is valuable for intrageneric phylogenetic reconstruction, and it is comparable to the matK in terms of consistency (Neubig et al., 2009; Dong et al., 2015). Therefore, ycf1 has been used as a potentially promising genetic marker in Astragalus L. (Dastpak et al., 2018; Zaveska et al., 2019), Orchidaceae (Whitten et al., 2014), and Pinus L. (Olsson et al., 2018). Additionally, ycf1 has also proposed to be a plastid candidate barcode in Papilionoideae, such as in Sophora (Liao et al., 2021), Dalbergia (Li et al., 2022b), Onobrychis Mill. (Moghaddam et al., 2022) and Quercus L. (Yang et al., 2017), etc. Consistent with these observations, we found that ycf1 exhibited high nucleotide diversity and could be used as for potential chloroplast marker for phylogenetic and evolutionary studies of Indigofera species.
In addition to highly variable regions, SSRs that are widely distributed in the genome are regarded as powerful tools for many aspects of evolutionary population biology, given their polymorphism (Tautz and Renz, 1984; Li et al., 2002). SSRs developed from transcriptome or genome have been used in Indigofera (Guo et al., 2016; Otao et al., 2016; Chikmawati et al., 2019; Royani et al., 2022). Chloroplast SSRs are typically non-recombinant, uni-parentally inherited, and effectively haploid, and as such have been widely used to fully understand the population genetic diversity and evolutionary history with biparental inherited molecular markers (Echt et al., 1998; Provan et al., 2001; Hatmaker et al., 2018; López-Villalobos and Eckert, 2018; Salmona et al., 2020; Yao et al., 2021). The abundant cp SSRs identified in the present study laid the foundation for the identification of assays detecting polymorphisms at the population level of Indigofera.
The monophyly and phylogenetic placement of the genus Indigofera was evaluated. The plastid phylogenomic analyses generated a highly resolved phylogeny in the present study. Species of the Genistoid clade formed the most basal clade, followed by the Mirbelioid clade. The IRLC and Robinioid clade formed a monophyletic clade (Figure 5). Within the sampled species, Indigofera species showed a close relationship with the Millettioid clade, consistent with the previous studies (Cardoso et al., 2013; de Queiroz et al., 2015; Oyebanji et al., 2020; Zhang et al., 2020b; Choi et al., 2022). However, phylogenetic relationships of the Robinioid clade and Genistoid clade in the complete plastome-based and PCGs-based phylogenetic trees exhibited different topologies (Figure 5 and Supplementary Figures S2, S3). In addition, both the ML and BI trees constructed based on complete plastomes suggested that species of the Genistoid and Robinioid clades clustered together, except for Templetonia retusa (Vent.) R. Br. and Lotus japonicus (Regel) K. Larsen, which was inconsistent with the previous results (Zhang et al., 2020b; Choi et al., 2022). Rapidly evolution sites that tend to accumulate non-phylogenetic signals and potentially misaligned loci in the plastome dataset are often suggested to explain discordance (Burleigh and Mathews, 2004; Rodriguez-Ezpeleta et al., 2007; Philippe et al., 2011; Zhang et al., 2017). Copy and loss of gene duplication and lineage sorting can cause inconsistencies between gene trees, which in turn leads to conflicts between gene trees and species trees (Swenson and El-Mabrouk, 2012; Goncalves et al., 2019; Walker et al., 2019; Wei et al., 2021). It has been reported that a fewer misplaced leaves in the gene tree can lead to a completely different history, with significantly more duplications and losses (Hahn, 2007; Fang et al., 2010; Doroftei and El-Mabrouk, 2011).
The monophyly of Indigofera and its four clades was all strongly supported based on datasets of the complete cp genome and PCGs. However, the phylogenetic relationships of the Tethyan, East Asian, and Palaeotropical clades were inconsistent with nuclear ITS-based phylogenies (Schrire et al., 2003, 2009; Zhao, 2016). Within the Pantropical clade, sampled species exhibited a pantropical, Sino-Japanese, or Sino-Himalayan distribution consistent with previous studies (Schrire et al., 2009; Zhao, 2016). Species in this clade formed five monophyletic lineages, where I. hirsuta, a species with a widespread pantropical distribution, was the earliest diverging taxon. Three species (I. carlesii, I. decora, and I. kirilowii) of the I. decora complex, which were previously assigned to subsect. Decorae, formed a robustly supported clede, which is consistent with previous classifications (Fang and Zheng, 1994; Peng et al., 2015). These three species have shared numerical characteristics, with leaves and flowers significantly larger than other species of Indigofera, e.g. leaflets 1.5—6.5 (—7.5) × 1—3.5 cm, flowers 1.2—1.5 (—1.8) × 0.7—0.9 cm, wings and keels ca. 1.2—1.4 cm, and specifically distributed in eastern, central, and northern China, and extending to Korea and Japan (Fang and Zheng, 1994; Peng et al., 2015). Interestingly, four species (i.e., I. atropurpurea, I. cassioides, I. stachyodes, and I. hebepetala var. glabra) grouped closely in our phylogeny, which is inconsistent with the morphology-based classification (Fang and Zheng, 1994). Fang and Zheng (1994) classified I. atropurpurea and I. hebepetala var. glabra into subsect. Bracteatae based on the erect recemes and conspicuous bracts at anthesis, whereas I. cassioides and I. stachyodes were classified into subsect. Pendulae because their racemes are both pendulous and much longer than leaves. In addition, three Sino-Himalayan endemics (i.e., I. franchetii, I. pendula, and I. scabrida) showed close relationships. As a global biodiversity hotspot, the temperate Sino-Himalayan region harbors high levels of species richness (ca. 105 species) and morphological diversity of Indigofera (Fang and Zheng, 1994; Schrire et al., 2009; Gao and Schrire, 2010), and comprehensive phylogenetic relationships of Indigofera species in this region need a comprehensive sampling in the further study. Within the Palaeotropical clade, our samples involved only one species (I. wightii) distributed in tropical Asia. Within the East Asian clade, two species of subsect. Pseudotinctoriae (i.e., I. bungeana and I. amblyantha) showed close affinities, as reported in previous studies (Zhao, 2016; Zhao et al., 2017). Within the Tethyan clade, I. linifolia, I. linnaei, and I. miniata, which are mainly distributed in the tropics of Asia, Africa, Australia, and America, are all herbs with simple or 1-foliate leaves, or compound leaves with alternate leaflets, and keel beard loss, and their close affinity was also supported the previous phylogenetic studies based on ITS sequences (Schrire et al., 2009; Zhao, 2016). Overall, most of the Indigofera lineages were resolved with high support values in this study, which indicated that the chloroplast genome may be a suitable component in the construction of a robust phylogeny for Indigofera, and therefore represent an effective tool for resolving taxonomic controversy in this genus.
Positive selection is thought to play a key part in the adaptation of organisms to diverse environments (Moseley et al., 2018; Feng et al., 2022b), however, purifying selection is more common than positive selection due to the constant elimination of deleterious mutations (Ogawa et al., 1999; Wu et al., 2020; Huang et al., 2021). The low Ka/Ks ratios within Indigofera species suggested that most genes are undergoing purifying selection to retain conserved functions in Indigofera species.
This analysis of adaptive evolution contributes to a deep understanding of genetic variation and changes in protein structure and function (Wicke et al., 2014; Li et al., 2022d). To investigate the differences in selective pressures between two different evolutionary branches of Indigofera and non-Indigofera, the PAML v4.10.6 package (Yang, 1997, 2007) was used to analyze selective pressures in a branch-site model for 70 shared PCGs. Thirteen genes with significant posterior probabilities for codon sites were identified in the BEB test. Codon sites with higher posterior probabilities can be considered as sites undergoing positive selection (Yang et al., 2005; Tyagi et al., 2020). These genes included two photosystem subunit genes (psaA and psaB), four NADH-dehydrogenase subunit genes (ndhA, ndhE, ndhF, and ndhI), two subunits of cytochrome b/f complex genes (petA and petN), two DNA-dependent RNA polymerase genes (rpoB and rpoC1), and the accD, ccsA, and matK genes.
Four genes (ndhA, ndhE, ndhF, and ndhI) encode a subunit of the NADH dehydrogenase complex and were responsible for the electron transport chain necessary to generate ATP during photosynthesis (Weiss et al., 1991; Kofer et al., 1998; Green, 2011; Peng et al., 2011; Zhang et al., 2020c; Xie et al., 2021). Previous studies have suggested that the ndh gene family may be involved in the protection of chloroplasts against photooxidative stress (Martin et al., 1996) and that the antioxidant stress capacity of plants is also closely related to their resistance to other environmental conditions (Shaaltiel et al., 1988). Species of Indigofera, which are mainly distributed in tropical and subtropical regions (Schrire et al., 2009; Peng et al., 2015; Zhao and Gao, 2015; Zhao, 2016), generally have strong heat tolerance and adaptability, and adapt to dry and hot environments by producing smaller leaves and fewer leaflets (Schrire et al., 2009; Niu et al., 2020; Zhao et al., 2020). In drought habitats, plants will reduce water transpiration by reducing leaf area, and this reduction of leaf area has a necessary trade-off of photosynthetic capacity (Peng et al., 2007; Niu et al., 2020). Environmental differences had a greater effect on the leaf blade phenotype of some species of I. bungeana complex (Niu et al., 2020; Zhao et al., 2020). We speculate that chloroplast functional genes involved in plant photosynthesis may play a key role in the ecological adaptation of Indigofera species to drought stress. In addition, petA and petN are membrane components necessary for the transport of respiratory and photosynthetic electrons (Gray, 1992; Yamori and Shikanai, 2016). Notably, the posterior probability value for the amino acid sites of petN was as high as 0.968 in this study, indicating that petN was strongly positively selected. Moreover, accD is known to code the β-carboxylase subunit of acetyl-CoA carboxylase, which is essential for leaf growth and development (Madoka et al., 2002; Kode et al., 2005; Rousseau-Gueutin et al., 2013; Tyagi et al., 2020; Chen et al., 2021; Li et al., 2022a). Positive selection pressure on accD might be the adaptive evolution of Indigofera species to selection pressures imposed by their herbivores and pathogens (Schrire, 2013). We also identified positively selected sites in the rpoB, rpoC1, and matK, which might have played key roles in the adaptive evolution of Indigofera species (Hao et al., 2010; Hertel et al., 2013; Shi et al., 2019). In summary, 13 PCGs showed significant positive selection markers, but the adaptive evolution of Indigofera in specific ecological environments needs to be further explored through molecular, physiological, and ecological studies.
We provided insights into the structural variation of the chloroplast genomes as well as the phylogenetic relationships in the genus Indigofera in the present study. To better perform phylogenetic construction, population genetics, and species identification for Indigofera, we screened promising molecular markers both from intergenic and coding regions. The monophyly of Indigofera and its four monophyletic clades was supported using a robust plastid phylogenomic framework. We found that cp genome data was effective in improving the resolution of phylogenies in Indigofera. Conflicting topologies were detected by complete cp genome and PCGs sequences. These topological incongruences deserve further exploration of the underlying biologically relevant evolutionary history, using nuclear and plastome datasets. We will also expand the genomic sampling to analyze the phylogenetic relationships, and biogeography of Indigofera in future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OQ134123, OQ147467, OQ147468, OQ147469, OQ147470, OQ147481, OQ147471, OQ147466, OQ147482, OQ147472, OQ147473, OQ147474, OQ147475, OQ147476, OQ147477, OQ147478, OQ147479, OQ147480.
S-MZ and X-LZ conceived and designed the study. S-MZ, FW, X-LZ and X-FG collected the samples. S-MZ, FW and S-YY performed the experiments. S-MZ analyzed the sequence data and drafted the manuscript. X-FG, Z-MZ and X-LZ participated in data analysis and revised the manuscript. All the authors contributed to and approved the submitted version.
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31800170), and the Scientific Research Fund Project of Yunnan Provincial Department of Education (Grant No. 2021Y241 and 2022Y585).
We acknowledge Chun-Qiao Peng, Lu Qu, Bo Xu, Meng Li, and Li-Na Guo for their help in the materials collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1186598/full#supplementary-material
Aecyo, P., Marques, A., Huettel, B., Silva, A., Esposito, T., Ribeiro, E., et al. (2021). Plastome evolution in the Caesalpinia group (Leguminosae) and its application in phylogenomics and populations genetics. Planta 254, 1–19. doi: 10.1007/s00425-021-03655-8
Aii, J., Kishima, Y., Mikami, T., Adachi, T. (1997). Expansion of the IR in the chloroplast genomes of buckwheat species is due to incorporation of an SSC sequence that could be mediated by an inversion. Curr. Genet. 31, 276–279. doi: 10.1007/s002940050206
Akaike, H. (1974). A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723. doi: 10.1109/TAC.1974.1100705
Amiryousefi, A., Hyvönen, J., Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Asaf, S., Khan, A. L., Aaqil Khan, M., Muhammad Imran, Q., Kang, S. M., Al-Hosni, K., et al. (2017a). Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PloS One 12, e0182281. doi: 10.1371/journal.pone.0182281
Asaf, S., Khan, A. L., Khan, M. A., Waqas, M., Kang, S. M., Yun, B. W., et al. (2017b). Chloroplast genomes of arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: structures and comparative analysis. Sci. Rep. 7, 1–15. doi: 10.1038/s41598-017-07891-5
Baran, A., Fiedler, A., Schulz, H., Baranska, M. (2010). In situ raman and IR spectroscopic analysis of indigo dye. Anal. Methods 2, 1372–1376. doi: 10.1039/C0AY00311E
Barker, N. P., Schrire, B. D., Kim, J. H. (2000). “Generic relationships in the tribe indigofereae (Leguminosae: papilionoideae) based on sequence data and morphology,” in Advances in legume systematics, 9. Eds. Herendeen, P. S., Bruneau, A. (Richmond, UK: Royal Botanic Gardens, Kew), 311–337.
Bechtold, T., Turcanu, A., Geissler, S., Ganglberger, E. (2002). Process balance and product quality in the production of natural indigo from Polygonum tinctorium ait. applying low-technology methods. Bioresour. Technol. 81, 171–177. doi: 10.1016/S0960-8524(01)00146-8
Beier, S., Thiel, T., Münch, T., Scholz, U., Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Bock, R., Knoop, V. (2012). Genomics of chloroplasts and mitochondria (Berlin, Germany: Springer Science and Business Media).
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burleigh, J. G., Mathews, S. (2004). Phylogenetic signal in nucleotide data from seed plants: implications for resolving the seed plant tree of life. Am. J. Bot. 91, 1599–1613. doi: 10.3732/ajb.91.10.1599
Calvo, T. R., Cardoso, C. R. P., da Silva Moura, A. C., Dos Santos, L. C., Colus, I. M. S., Vilegas, W., et al. (2011). Mutagenic activity of Indigofera truxillensis and I. suffruticosa aerial parts. J. Evidence-Based Complementary Altern. Med. 12, e0182281. doi: 10.1093/ecam/nep123
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cardoso, D., Pennington, R. T., De Queiroz, L., Boatwright, J. S., Van Wyk, B. E., Wojciechowski, M., et al. (2013). Reconstructing the deep-branching relationships of the papilionoid legumes. S. Afr. J. Bot. 89, 58–75. doi: 10.1016/j.sajb.2013.05.001
Chase, M. W., Cowan, R. S., Hollingsworth, P. M., Berg, C., Madriñán, S., Petersen, G., et al. (2007). A proposal for a standardised protocol to barcode all land plants. Taxon 56, 295–299. doi: 10.1002/tax.562004
Chen, Q., Hu, H., Zhang, D. (2022). DNA Barcoding and phylogenomic analysis of the genus Fritillaria in China based on complete chloroplast genomes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.764255
Chen, J., Zang, Y., Shang, S., Liang, S., Zhu, M., Wang, Y., et al. (2021). Comparative chloroplast genomes of zosteraceae species provide adaptive evolution insights into seagrass. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.741152
Chikmawati, T., Rifai, M., Ariyanti, N. (2019). Genetic diversity of the dye-producing Javanese tom Indigofera tinctoria l. from three Indonesian islands based on simple sequence repeat markers. Pak. J. Bot. 51, 903–908. doi: 10.30848/PJB2019-3(33
Choi, I. S., Cardoso, D., de Queiroz, L. P., de Lima, H. C., Lee, C., Ruhlman, T. A., et al. (2022). Highly resolved papilionoid legume phylogeny based on plastid phylogenomics. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.823190
Daniell, H., Lin, C. S., Yu, M., Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 1–29. doi: 10.1186/s13059-016-1004-2
Darriba, D., Taboada, G. L., Doallo, R., Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Dastpak, A., Osaloo, S. K., Maassoumi, A. A., Safar, K. N. (2018). Molecular phylogeny of Astragalus sect. Ammodendron (Fabaceae) inferred from chloroplast ycf1 gene. Ann. Bot. Fenn. 55, 75–82. doi: 10.5735/085.055.0108
de Queiroz, L. P., Pastore, J. F., Cardoso, D., Snak, C., de, C., Lima, A. L., et al. (2015). A multilocus phylogenetic analysis reveals the monophyly of a recircumscribed papilionoid legume tribe diocleae with well-supported generic relationships. Mol. Phylogenet. Evol. 90, 1–19. doi: 10.1016/j.ympev.2015.04.016
Dkhil, M. A., Zreiq, R., Hafiz, T. A., Mubaraki, M. A., Sulaiman, S., Algahtani, F., et al. (2020). Anthelmintic and antimicrobial activity of Indigofera oblongifolia leaf extracts. Saudi J. Biol. Sci. 27, 594–598. doi: 10.1016/j.sjbs.2019.11.033
Dong, W., Liu, J., Yu, J., Wang, L., Zhou, S. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PloS One 7, e35071. doi: 10.1371/journal.pone.0035071
Dong, W., Xu, C., Li, C., Sun, J., Zuo, Y., Shi, S., et al. (2015). ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 5, 1–5. doi: 10.1038/srep08348
Dopp, I. J., Yang, X., Mackenzie, S. A. (2021). A new take on organelle-mediated stress sensing in plants. New Phytol. 230, 2148–2153. doi: 10.1111/nph.17333
Doroftei, A., El-Mabrouk, N. (2011). “Removing noise from gene trees,” in Lecture notes in computer science, vol. 6833. Eds. Przytycka, T. M., Sagot, M. F. (Heidelberg, Germany: Springer), 76–91.
Drescher, A., Ruf, S., Calsa, T., Jr., Carrer, H., Bock, R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22, 97–104. doi: 10.1046/j.1365-313x.2000.00722.x
Du, Z., Lu, K., Zhang, K., He, Y., Wang, H., Chai, G., et al. (2021). The chloroplast genome of Amygdalus l. (Rosaceae) reveals the phylogenetic relationship and divergence time. BMC Genomics 22, 1–16. doi: 10.1186/s12864-021-07968-6
Du, Q., Yang, H., Zeng, J., Chen, Z., Zhou, J., Sun, S., et al. (2022). Comparative genomics and phylogenetic analysis of the chloroplast genomes in three medicinal Salvia species for bioexploration. Int. J. Mol. Sci. 23, 12080. doi: 10.3390/ijms232012080
Echt, C. S., DeVerno, L. L., Anzidei, M., Vendramin, G. G. (1998). Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa. ait. Mol. Ecol. 7, 307–316. doi: 10.1046/j.1365-294X.1998.00350.x
Fang, G., Bhardwaj, N., Robilotto, R., Gerstein, M. B. (2010). Getting started in gene orthology and functional analysis. PloS Comput. Biol. 6, e1000703. doi: 10.1371/journal.pcbi.1000703
Fang, Y. Y., Zheng, C. Z. (1994). “Indigofera linnaeus,” in Flora of China, vol. 40 . Ed. Wei, Z. (Beijing, China: Science Press), 239–326.
Feng, Y., Gao, X. F., Zhang, J. Y., Jiang, L. S., Li, X., Deng, H. N., et al. (2022b). Complete chloroplast genomes provide insights into evolution and phylogeny of Campylotropis (Fabaceae). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.895543
Feng, J. L., Wu, L. W., Wang, Q., Pan, Y. J., Li, B. L., Lin, Y. L., et al. (2022a). Comparison analysis based on complete chloroplast genomes and insights into plastid phylogenomic of four Iris species. Biomed. Res. Int. 2022, 2194021. doi: 10.1155/2022/2194021
Gao, X. F., Schrire, B. D. (2010). “Indigofera Linnaeus,” in Flora of China, vol. 10. Eds. Wu, Z. Y., Raven, P. H. (Beijing, China and St. Louis, Missouri: Beijing and Missouri Botanical Garden Press), 137–164.
Gao, X., Zhang, X., Meng, H., Li, J., Zhang, D., Liu, C. (2018). Comparative chloroplast genomes of Paris sect. Marmorata: insights into repeat regions and evolutionary implications. BMC Genomics 19, 878. doi: 10.1186/s12864-018-5281-x
Gerometta, E., Grondin, I., Smadja, J., Frederich, M., Gauvin-Bialecki, A. (2020). A review of traditional uses, phytochemistry and pharmacology of the genus Indigofera. J. Ethnopharmacol. 253, 112608. doi: 10.1016/j.jep.2020.112608
Gitzendanner, M. A., Soltis, P. S., Yi, T. S., Li, D. Z., Soltis, D. E. (2018). Plastome phylogenetics: 30 years of inferences into plant evolution. Adv. Bot. Res. 85, 293–313. doi: 10.1016/bs.abr.2017.11.016
Goncalves, D. J., Simpson, B. B., Ortiz, E. M., Shimizu, G. H., Jansen, R. K. (2019). Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Mol. Phylogenet. Evol. 138, 219–232. doi: 10.1016/j.ympev.2019.05.022
Gray, J. C. (1992). Cytochrome f: structure, function and biosynthesis. Photosynth. Res. 34, 359–374. doi: 10.1007/BF00029811
Green, B. R. (2011). Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66, 34–44. doi: 10.1111/j.1365-313X.2011.04541.x
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guindon, S., Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Guo, L. N., Zhao, X. L., Gao, X. F. (2016). De novo assembly and characterization of leaf transcriptome for the development of EST-SSR markers of the non-model species Indigofera szechuensis. Biochem. Syst. Ecol. 68, 36–43. doi: 10.1016/j.bse.2016.06.010
Hahn, M. W. (2007). Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biol. 8, R141. doi: 10.1186/gb-2007-8-7-r141
Han, C., Ding, R., Zong, X., Zhang, L., Chen, X., Qu, B. (2022). Structural characterization of Platanthera ussuriensis chloroplast genome and comparative analyses with other species of orchidaceae. BMC Genomics 23, 1–13. doi: 10.1186/s12864-022-08319-9
Hao, D. C., Chen, S. L., Xiao, P. G. (2010). Molecular evolution and positive Darwinian selection of the chloroplast maturase matK. J. Plant Res. 123, 241–247. doi: 10.1007/s10265-009-0261-5
Hassen, A. (2007). Characterization and evaluation of indigofera species as potential forage and cover crops for semi-arid and arid ecosystems (Pretoria, South Africa: University of Pretoria).
Hatmaker, E. A., Staton, M. E., Dattilo, A. J., Hadziabdic, Ð., Rinehart, T. A., Schilling, E. E., et al. (2018). Population structure and genetic diversity within the endangered species Pityopsis ruthii (Asteraceae). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00943
Hertel, S., Zoschke, R., Neumann, L., Qu, Y., Axmann, I. M., Schmitz-Linneweber, C. (2013). Multiple checkpoints for the expression of the chloroplast-encoded splicing factor matK. Plant Physiol. 163, 1686–1698. doi: 10.1104/pp.113.227579
Hollingsworth, P. M., Graham, S. W., Little, D. P. (2011). Choosing and using a plant DNA barcode. PloS One 6, e19254. doi: 10.1371/journal.pone.0019254
Huang, H., Shi, C., Liu, Y., Mao, S. Y., Gao, L. Z. (2014). Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol. Biol. 14, 1–17. doi: 10.1186/1471-2148-14-151
Huang, R., Xie, X., Chen, A., Li, F., Tian, E., Chao, Z. (2021). The chloroplast genomes of four Bupleurum (Apiaceae) species endemic to southwestern China, a diversity center of the genus, as well as their evolutionary implications and phylogenetic inferences. BMC Genomics 22, 714. doi: 10.1186/s12864-021-08008-z
Jansen, R. K., Ruhlman, T. A. (2012). “Plastid genomes of seed plants,” in Genomics of chloroplasts and mitochondria, vol. 35 . Eds. Bock, R., Respiration, V. (Dordrecht, Netherlands: Springer), 103–126.
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 1–31. doi: 10.1186/s13059-020-02154-5
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Keller, J., Rousseau-Gueutin, M., Martin, G. E., Morice, J., Boutte, J., Coissac, E., et al. (2017). The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses of four novel legume chloroplast genomes from Lupinus. DNA Res. 24, 343–358. doi: 10.1093/dnares/dsx006
Kode, V., Mudd, E. A., Iamtham, S., Day, A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44, 237–244. doi: 10.1111/j.1365-313X.2005.02533.x
Kofer, W., Koop, H. U., Wanner, G., Steinmüller, K. (1998). Mutagenesis of the genes encoding subunits a, c, h, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol. Genet. Genomics 258, 166–173. doi: 10.1007/s004380050719
Kolodner, R., Tewari, K. (1979). Inverted repeats in chloroplast DNA from higher plants. Proc. Natl. Acad. Sci. U. S. A. 76, 41–45. doi: 10.1073/pnas.76.1.41
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., Janzen, D. H. (2005). Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U. S. A. 102, 8369–8374. doi: 10.1073/pnas.0503123102
Kumar, R. S., Rajkapoor, B., Perumal, P. (2011). In vitro and in vivo anticancer activity of Indigofera cassioides rottl. ex. DC. Asian Pac. J. Trop. Med. 4, 379–385. doi: 10.1016/S1995-7645(11)60108-9
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Lan, Y., Sun, J., Tian, R., Bartlett, D. H., Li, R., Wong, Y. H., et al. (2017). Molecular adaptation in the world’s deepest-living animal: insights from transcriptome sequencing of the hadal amphipod Hirondellea gigas. Mol. Ecol. 26, 3732–3743. doi: 10.1111/mec.14149
Lei, W., Ni, D., Wang, Y., Shao, J., Wang, X., Yang, D., et al. (2016). Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci. Rep. 6, 1–13. doi: 10.1038/srep21669
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lewis, G. P., Schrire, B., Mackinder, B., Lock, M. (2005). Legumes of the world (Richmond, UK: Royal Botanic Gardens, Kew).
Li, Y. C., Korol, A. B., Fahima, T., Beiles, A., Nevo, E. (2002). Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 11, 2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x
Li, B., Liu, T., Ali, A., Xiao, Y., Shan, N., Sun, J., et al. (2022a). Complete chloroplast genome sequences of three aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics 23, 218. doi: 10.1186/s12864-022-08400-3
Li, C., Liu, Y., Lin, F., Zheng, Y., Huang, P. (2022b). Characterization of the complete chloroplast genome sequences of six Dalbergia species and its comparative analysis in the subfamily of papilionoideae (Fabaceae). PeerJ 10, e13570. doi: 10.7717/peerj.13570
Li, M., Song, Y. F., Sylvester, S. P., Sylvester, S. P., Wang, X. R. (2022c). Comparative analysis of the complete plastid genomes in Prunus subgenus Cerasus (Rosaceae): molecular structures and phylogenetic relationships. PloS One 17, e0266535. doi: 10.1371/journal.pone.0266535
Li, Y., Wang, T. R., Kozlowski, G., Liu, M. H., Yi, L. T., Song, Y. G. (2022d). Complete chloroplast genome of an endangered species Quercus litseoides, and its comparative, evolutionary, and phylogenetic study with other Quercus section Cyclobalanopsis species. Genes (Basel) 13, 1184. doi: 10.3390/genes13071184
Li, H., Xiao, W., Tong, T., Li, Y., Zhang, M., Lin, X., et al. (2021). The specific DNA barcodes based on chloroplast genes for species identification of orchidaceae plants. Sci. Rep. 11, 1–15. doi: 10.1038/s41598-021-81087-w
Lian, C., Yang, H., Lan, J., Zhang, X., Zhang, F., Yang, J., et al. (2022). Comparative analysis of chloroplast genomes reveals phylogenetic relationships and intraspecific variation in the medicinal plant Isodon rubescens. PloS One 17, e0266546. doi: 10.1371/journal.pone.0266546
Liang, H., Zhang, Y., Deng, J., Gao, G., Ding, C., Zhang, L., et al. (2020). The complete chloroplast genome sequences of 14 Curcuma species: insights into genome evolution and phylogenetic relationships within zingiberales. Front. Genet. 11. doi: 10.3389/fgene.2020.00802
Liao, M., Gao, X. F., Zhang, J. Y., Deng, H. N., Xu, B. (2021). Comparative chloroplast genomics of Sophora species: evolution and phylogenetic relationships in the early–diverging legume subfamily papilionoideae (Fabaceae). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.778933
Lin, C. P., Huang, J. P., Wu, C. S., Hsu, C. Y., Chaw, S. M. (2010). Comparative chloroplast genomics reveals the evolution of pinaceae genera and subfamilies. Genome Biol. Evol. 2, 504–517. doi: 10.1093/gbe/evq036
López-Villalobos, A., Eckert, C. (2018). Consequences of multiple mating–system shifts for population and range–wide genetic structure in a coastal dune plant. Mol. Ecol. 27, 675–693. doi: 10.1111/mec.14484
LPWG (2013). Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species–rich clades. Taxon 62, 217–248. doi: 10.12705/622.8
LPWG (2017). A new subfamily classification of the leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66, 44–77. doi: 10.12705/661.3
Madoka, Y., Tomizawa, K. I., Mizoi, J., Nishida, I., Nagano, Y., Sasaki, Y. (2002). Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol. 43, 1518–1525. doi: 10.1093/pcp/pcf172
Marquiafável, F. S., Ferreira, M. D. S., de Pádua Teixeira, S. (2009). Novel reports of glands in Neotropical species of Indigofera l. (Leguminosae, papilionoideae). Flora 204, 189–197. doi: 10.1016/j.flora.2008.01.012
Martin, M., Casano, L. M., Sabater, B. (1996). Identification of the product of ndhA gene as a thylakoid protein synthesized in response to photooxidative treatment. Plant Cell Physiol. 37, 293–298. doi: 10.1093/oxfordjournals.pcp.a028945
Martinez-Alberola, F., Del Campo, E. M., Lazaro-Gimeno, D., Mezquita-Claramonte, S., Molins, A., Mateu-Andres, I., et al. (2013). Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts. PloS One 8, e79685. doi: 10.1371/journal.pone.0079685
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Moghaddam, M., Ohta, A., Shimizu, M., Terauchi, R., Kazempour-Osaloo, S. (2022). The complete chloroplast genome of Onobrychis gaubae (Fabaceae–papilionoideae): comparative analysis with related IR-lacking clade species. BMC Plant Biol. 22, 75. doi: 10.1186/s12870-022-03465-4
Moseley, R. C., Mewalal, R., Motta, F., Tuskan, G. A., Haase, S., Yang, X. (2018). Conservation and diversification of circadian rhythmicity between a model crassulacean acid metabolism plant Kalanchoë fedtschenkoi and a model C3 photosynthesis plant Arabidopsis thaliana. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01757
Neubig, K. M., Whitten, W. M., Carlsward, B. S., Blanco, M. A., Endara, L., Williams, N. H., et al. (2009). Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst. Evol. 277, 75–84. doi: 10.1007/s00606-008-0105-0
Newmaster, S., Fazekas, A., Ragupathy, S. (2006). DNA Barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Botany 84, 335–341. doi: 10.1139/b06-047
Niu, X. J., Nie, J., Yang, Z. Y., Zhao, X. L. (2020). Leaf morphological responses of Indigofera bungeana to drought stress. Acta Bot. Boreal. Occident. Sin. 40, 613–623. doi: 10.7606/j.issn.1000-4025.2020.04.0613
Ogawa, T., Ishii, C., Kagawa, D., Muramoto, K., Kamiya, H. (1999). Accelerated evolution in the protein-coding region of galectin cDNAs, congerin I and congerin II, from skin mucus of conger eel (Conger myriaster). Biosci. Biotechnol. Biochem. 63, 1203–1208. doi: 10.1271/bbb.63.1203
Olsson, S., Grivet, D., Cid-Vian, J. (2018). Species-diagnostic markers in the genus Pinus: evaluation of the chloroplast regions matK and ycf1. For. Syst. 27, e016. doi: 10.5424/fs/2018273-13688
Otao, T., Kobayashi, T., Uehara, K. (2016). Development and characterization of 14 microsatellite markers for Indigofera pseudotinctoria (Fabaceae). Appl. Plant Sci. 4, 1500110. doi: 10.3732/apps.1500110
Oyebanji, O., Zhang, R., Chen, S. Y., Yi, T. S. (2020). New insights into the plastome evolution of the Millettioid/Phaseoloid clade (Papilionoideae, leguminosae). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00151
Pascual-Diaz, J. P., Garcia, S., Vitales, D. (2021). Plastome diversity and phylogenomic relationships in asteraceae. Plants (Basel) 10, 2699. doi: 10.3390/plants10122699
Pattanaik, L., Naik, S. N., Hariprasad, P. (2019). Valorization of waste Indigofera tinctoria l. biomass generated from indigo dye extraction process–potential towards biofuels and compost. Biomass Conv. Bioref. 9, 445–457. doi: 10.1007/s13399-018-0354-2
Peng, Y., Jiang, G., Liu, X., Niu, S., Liu, M., Biswas, D. (2007). Photosynthesis, transpiration and water use efficiency of four plant species with grazing intensities in hunshandak sandland, China. J. Arid Environ. 70, 304–315. doi: 10.1016/j.jaridenv.2007.01.002
Peng, L., Yamamoto, H., Shikanai, T. (2011). Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta 1807, 945–953. doi: 10.1016/j.bbabio.2010.10.015
Peng, C. Q., Zhao, X. L., Gao, X. F. (2015). Study on the population genetics of Indigofera decora complex based on cpDNA and nrDNA ITS sequences. Plant Sci. J. 33, 425–437. doi: 10.11913/PSJ.2095-0837.2015.40425
Philippe, H., Brinkmann, H., Lavrov, D. V., Littlewood, D. T., Manuel, M., Worheide, G., et al. (2011). Resolving difficult phylogenetic questions: why more sequences are not enough. PloS Biol. 9, e1000602. doi: 10.1371/journal.pbio.1000602
Provan, J., Powell, W., Hollingsworth, P. M. (2001). Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol. Evol. 16, 142–147. doi: 10.1016/s0169-5347(00)02097-8
Qu, X. J., Moore, M. J., Li, D. Z., Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 1–12. doi: 10.1186/s13007-019-0435-7
Rajkapoor, B., Kavimani, S., Ravichandiran, V., Sekhar, K., Kumar, R. S., Kumar, M. R., et al. (2009). Effect of Indigofera aspalathoides on complete freund’s adjuvant-induced arthritis in rats. Pharm. Biol. 47, 553–557. doi: 10.1080/13880200902902489
Raman, G., Park, V., Kwak, M., Lee, B., Park, S. (2017). Characterization of the complete chloroplast genome of Arabis stellari and comparisons with related species. PloS One 12, e0183197. doi: 10.1371/journal.pone.0183197
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ranwez, V., Douzery, E. J., Cambon, C., Chantret, N., Delsuc, F. (2018). MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 35, 2582–2584. doi: 10.1093/molbev/msy159
Rodriguez-Ezpeleta, N., Brinkmann, H., Roure, B., Lartillot, N., Lang, B. F., Philippe, H. (2007). Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 56, 389–399. doi: 10.1080/10635150701397643
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rousseau-Gueutin, M., Huang, X., Higginson, E., Ayliffe, M., Day, A., Timmis, J. N. (2013). Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 161, 1918–1929. doi: 10.1104/pp.113.214528
Royani, J. I., Sinaga, O. F. B., Aliyah, K. N., Hardianto, D., Agustina, T., Rofiq, M. N., et al. (2022). Screening of simple sequence repeats (SSR) primers from mutated Indigofera zolligeriana miq plants. IOP Conf. Series: Earth Environ. Sci. 1114, 12106. doi: 10.1088/1755-1315/1114/1/012106
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Ruang-Areerate, P., Kongkachana, W., Naktang, C., Sonthirod, C., Narong, N., Jomchai, N., et al. (2021). Complete chloroplast genome sequences of five Bruguiera species (Rhizophoraceae): comparative analysis and phylogenetic relationships. PeerJ 9, e12268. doi: 10.7717/peerj.12268
Salmona, J., Olofsson, J. K., Hong-Wa, C., Razanatsoa, J., Rakotonasolo, F., Ralimanana, H., et al. (2020). Late Miocene origin and recent population collapse of the Malagasy savanna olive tree (Noronhia lowryi). Biol. J. Linn. Soc 129, 227–243. doi: 10.1093/biolinnean/blz164
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Tabata, S. (1999). Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 6, 283–290. doi: 10.1093/dnares/6.5.283
Schrire, B. D. (1995). “Cladistic analysis of the tribe indigofereae (Leguminosae),” in Advances in legume systematics, vol. 7 . Eds. Crisp, M. D., Doyle, J. J. (Richmond, UK: Royal Botanic Gardens, Kew), 161–244.
Schrire, B. D. (2005). “Tribe indigofereae,” in Legumes of the world. Eds. Lewis, G., Schrire, B., Mackinder, B., Lock, M. (Richmond, UK: Royal Botanic Gardens, Kew), 361–365.
Schrire, B. D. (2013). A review of tribe indigofereae (Leguminosae–papilionoideae) in southern Africa (including south Africa, Lesotho, Swaziland & namibia; excluding Botswana). S. Afr. J. Bot. 89, 281–283. doi: 10.1016/j.sajb.2013.06.014
Schrire, B. D., Lavin, M., Barker, N., Cortes-Burns, H., Von Senger, I., Kim, J. (2003). “Towards a phylogeny of indigofera (Leguminosae–papilionoideae): identification of major clades and relative ages,” in Advances in legume systematics, vol. 10 . Eds. Klitgaard, B. B., Bruneau, A. (Richmond, UK: Royal Botanic Gardens, Kew), 269–302.
Schrire, B. D., Lavin, M., Barker, N. P., Forest, F. (2009). Phylogeny of the tribe indigofereae (Leguminosae–papilionoideae): geographically structured more in succulent-rich and temperate settings than in grass-rich environments. Am. J. Bot. 96, 816–852. doi: 10.3732/ajb.0800185
Schrire, B. D., Lavin, M., Lewis, G. P. (2005a). “Global distribution patterns of the leguminosae: insights from recent phylogenies,” in Plant diversity and complexity patterns-local, regional and global dimensions, vol. 55 . Eds. Friis, I., Balslev, H. (Biol. Skr), 375–422.
Schrire, B. D., Lewis, G. P., Lavin, M. (2005b). “Biogeography of the leguminosae,” in Legumes of the world. Eds. Lewis, G., Schrire, B., Mackinder, B., Lock, M. (Richmond, UK: Royal Botanic Gardens, Kew), 21–54.
Schwarz, E. N., Ruhlman, T. A., Sabir, J. S., Hajrah, N. H., Alharbi, N. S., Al-Malki, A. L., et al. (2015). Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 53, 458–468. doi: 10.1111/jse.12179
Shaaltiel, Y., Glazer, A., Bocion, P., Gressel, J. (1988). Cross tolerance to herbicidal and environmental oxidants of plant biotypes tolerant to paraquat, sulfur dioxide, and ozone. Pestic. Biochem. Physiol. 31, 13–23. doi: 10.1016/0048-3575(88)90024-7
Shahid-ul-Islam, Shahid, M., Mohammad, F. (2013). Perspectives for natural product based agents derived from industrial plants in textile applications-a review. J. Clean. Prod. 57, 2–18. doi: 10.1016/j.jclepro.2013.06.004
Shi, H., Yang, M., Mo, C., Xie, W., Liu, C., Wu, B., et al. (2019). Complete chloroplast genomes of two Siraitia Merrill species: comparative analysis, positive selection and novel molecular marker development. PloS One 14, e0226865. doi: 10.1371/journal.pone.0226865
Song, Y., Zhang, Y., Xu, J., Li, W., Li, M. (2019). Characterization of the complete chloroplast genome sequence of Dalbergia species and its phylogenetic implications. Sci. Rep. 9, 20401. doi: 10.1038/s41598-019-56727-x
Sugiura, M. (1992). The chloroplast genome. Plant Mol. Biol. 19, 149–168. doi: 10.1007/978-94-011-2656-4_10
Swenson, K. M., El-Mabrouk, N. (2012). Gene trees and species trees: irreconcilable differences. BMC Bioinform. 13, 15. doi: 10.1186/1471-2105-13-S19-S15
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tautz, D., Renz, M. (1984). Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 12, 4127–4138. doi: 10.1093/nar/12.10.4127
Tong, R., Gui, C., Zhang, Y., Su, N., Hou, X., Liu, M., et al. (2022). Phylogenomics, plastome structure and species identification in Mahonia (Berberidaceae). BMC Genomics 23, 1–21. doi: 10.1186/s12864-022-08964-0
Tyagi, S., Jung, J. A., Kim, J. S., Won, S. Y. (2020). Comparative analysis of the complete chloroplast genome of mainland Aster spathulifolius and other Aster species. Plants (Basel) 9, 568. doi: 10.3390/plants9050568
Walker, J. F., Walker-Hale, N., Vargas, O. M., Larson, D. A., Stull, G. W. (2019). Characterizing gene tree conflict in plastome-inferred phylogenies. PeerJ 7, e7747. doi: 10.7717/peerj.7747
Wei, N., Pérez-Escobar, O. A., Musili, P. M., Huang, W. C., Yang, J. B., Hu, A. Q., et al. (2021). Plastome evolution in the hyperdiverse genus Euphorbia (Euphorbiaceae) using phylogenomic and comparative analyses: large-scale expansion and contraction of the inverted repeat region. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.712064
Weiss, H., Friedrich, T., Hofhaus, G., Preis, D. (1991). The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Eur. J. Biochem. 197, 563–576. doi: 10.1111/j.1432-1033.1991.tb15945.x
Whitten, W. M., Neubig, K. M., Williams, N. H. (2014). Generic and subtribal relationships in Neotropical cymbidieae (Orchidaceae) based on matK/ycf1 plastid data. Lankesteriana 13, 375–392. doi: 10.15517/lank.v13i3.14425
Wick, R. R., Schultz, M. B., Zobel, J., Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wicke, S., Schäferhoff, B., Depamphilis, C. W., Müller, K. F. (2014). Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous lentibulariaceae. Mol. Biol. Evol. 31, 529–545. doi: 10.1093/molbev/mst261
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Müller, K. F., Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wolfe, K. H., Li, W. H., Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U. S. A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Wu, Z., Liao, R., Yang, T., Dong, X., Lan, D., Qin, R., et al. (2020). Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae). BMC Genomics 21, 621. doi: 10.1186/s12864-020-07045-4
Xie, X., Huang, R., Li, F., Tian, E., Li, C., Chao, Z. (2021). Phylogenetic position of Bupleurum sikangense inferred from the complete chloroplast genome sequence. Gene 798, 145801. doi: 10.1016/j.gene.2021.145801
Xie, D. F., Yu, H. X., Price, M., Xie, C., Deng, Y. Q., Chen, J. P., et al. (2019). Phylogeny of Chinese Allium species in section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, allioideae) species revealed by the chloroplast complete genome. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00460
Yamori, W., Shikanai, T. (2016). Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu. Rev. Plant Biol. 67, 81–106. doi: 10.1146/annurev-arplant-043015-112002
Yang, Z. (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics 13, 555–556. doi: 10.1093/bioinformatics/13.5.555
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Q., Fu, G. F., Wu, Z. Q., Li, L., Zhao, J. L., Li, Q. J. (2022). Chloroplast genome evolution in four montane zingiberaceae taxa in China. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.774482
Yang, Z., Nielsen, R. (2002). Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917. doi: 10.1093/oxfordjournals.molbev.a004148
Yang, Z., Reis, M. D. (2011). Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 28, 1217–1228. doi: 10.1093/molbev/msq303
Yang, J., Vazquez, L., Chen, X., Li, H., Zhang, H., Liu, Z., et al. (2017). Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus subgenus Quercus) and assessment of their utility as DNA barcodes. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00816
Yang, H., Wang, L., Chen, H., Jiang, M., Wu, W., Liu, S., et al. (2021). Phylogenetic analysis and development of molecular markers for five medicinal Alpinia species based on complete plastome sequences. BMC Plant Biol. 21, 1–16. doi: 10.1186/s12870-021-03204-1
Yang, Z., Wong, W. S., Nielsen, R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yao, Z., Wang, X., Wang, K., Yu, W., Deng, P., Dong, J., et al. (2021). Chloroplast and nuclear genetic diversity explain the limited distribution of endangered and endemic Thuja sutchuenensis in China. Front. Genet. 12. doi: 10.3389/fgene.2021.801229
Zaveska, E., Maylandt, C., Paun, O., Bertel, C., Frajman, B., Schönswetter, P., et al. (2019). Multiple auto- and allopolyploidisations marked the pleistocene history of the widespread Eurasian steppe plant Astragalus onobrychis (Fabaceae). Mol. Phylogenet. Evol. 139, 106572. doi: 10.1016/j.ympev.2019.106572
Zhang, Y., Du, L., Liu, A., Chen, J., Wu, L., Hu, W., et al. (2016). The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00306
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020a). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, S. D., Jin, J. J., Chen, S. Y., Chase, M. W., Soltis, D. E., Li, H. T., et al. (2017). Diversification of rosaceae since the late Cretaceous based on plastid phylogenomics. New Phytol. 214, 1355–1367. doi: 10.1111/nph.14461
Zhang, N., Long, J. L., Wu, Y., Zhang, Y. P., Wu, Z. K. (2022). The complete chloroplast genome of Indigofera stachyodes (Fabaceae), a traditional Chinese medicinal plant. Mitochondrial DNA Part B. 7, 474–475. doi: 10.1080/23802359.2022.2050472
Zhang, L., Wang, L., Cunningham, A. B., Shi, Y., Wang, Y. (2019). Island blues: indigenous knowledge of indigo-yielding plant species used by hainan miao and Li dyers on hainan island, China. J. Ethnobiol. Ethnomed. 15, 1–9. doi: 10.1186/s13002-019-0314-3
Zhang, R., Wang, Y. H., Jin, J. J., Stull, G. W., Bruneau, A., Cardoso, D., et al. (2020b). Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of leguminosae. Syst. Biol. 69, 613–622. doi: 10.1093/sysbio/syaa013
Zhang, R., Xu, B., Li, J., Zhao, Z., Han, J., Lei, Y., et al. (2020c). Transit from autotrophism to heterotrophism: sequence variation and evolution of chloroplast genomes in orobanchaceae species. Front. Genet. 11. doi: 10.3389/fgene.2020.542017
Zhao, X. L. (2016). A systematic study of Chinese indigofera (Fabaceae) (Chengdu, China: Institute of Biology, Chinese Academy of Sciences).
Zhao, J., Chen, J., Xiong, Y., He, W., Xiong, Y., Xu, Y., et al. (2023). Organelle genomes of Indigofera amblyantha and Indigofera pseudotinctoria: comparative genome analysis, and intracellular gene transfer. Ind. Crops Prod. 198, 116674. doi: 10.1016/j.indcrop.2023.116674
Zhao, X. L., Gao, X. F. (2015). Indigofera pseudonigrescens (Fabaceae: papilionoideae): a new species from sichuan, China. Phytotaxa 222, 251–258. doi: 10.11646/phytotaxa.222.4.2
Zhao, X. L., Gao, X. F., Zhu, Z. M., Gao, Y. D., Xu, B. (2017). The demographic response of a deciduous shrub (the Indigofera bungeana complex, fabaceae) to the pleistocene climate changes in East Asia. Sci. Rep. 7, 1–13. doi: 10.1038/s41598-017-00613-x
Zhao, K., Li, L., Quan, H., Yang, J., Lan, X. (2021). Comparative analyses of chloroplast genomes from 14 zanthoxylum species: identification of variable DNA markers and phylogenetic relationships within the genus. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.605793
Zhao, C., Wang, Y., Chan, K. X., Marchant, D. B., Franks, P. J., Randall, D., et al. (2019). Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land. Proc. Natl. Acad. Sci. U. S. A. 116, 5015–5020. doi: 10.1073/pnas.1812092116
Keywords: Indigofera, plastid genome, phylogeny, positive selection, Papilionoideae, Leguminosae
Citation: Zhou S-M, Wang F, Yan S-Y, Zhu Z-M, Gao X-F and Zhao X-L (2023) Phylogenomics and plastome evolution of Indigofera (Fabaceae). Front. Plant Sci. 14:1186598. doi: 10.3389/fpls.2023.1186598
Received: 15 March 2023; Accepted: 10 May 2023;
Published: 06 June 2023.
Edited by:
Roman A. Volkov, Chernivtsi University, UkraineReviewed by:
Ruth Clark, Royal Botanic Gardens, Kew, United KingdomCopyright © 2023 Zhou, Wang, Yan, Zhu, Gao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Fen Gao, eGZnYW9AY2liLmFjLmNu; Xue-Li Zhao, emhhb3h1ZWxpQHN3ZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.