- 1Key Laboratory of Quality and Safety Control for Subtropical Fruit and Vegetable, Ministry of Agriculture and Rural Affairs, Collaborative Innovation Center for Efficient and Green Production of Agriculture in Mountainous Areas of Zhejiang, College of Horticulture Science, Zhejiang Agriculture and Forestry University, Hangzhou, Zhejiang, China
- 2School of Life Science and Shanxi Key Laboratory for Research and Development of Regional Plants, Shanxi University, Taiyuan, Shanxi, China
- 3Zhejiang Provincial Key Laboratory of Bioremediation of Soil Contamination, College of Environment and Resources, College of Carbon Neutrality, Zhejiang Agriculture and Forestry University, Hangzhou, Zhejiang, China
Numerous studies have revealed the gasotransmitter functions of hydrogen sulfide (H2S) in various biological processes. However, the involvement of H2S in sulfur metabolism and/or Cys synthesis makes its role as a signaling molecule ambiguous. The generation of endogenous H2S in plants is closely related to the metabolism of Cys, which play roles in a variety of signaling pathway occurring in various cellular processes. Here, we found that exogenous H2S fumigation and Cys treatment modulated the production rate and content of endogenous H2S and Cys to various degrees. Furthermore, we provided comprehensive transcriptomic analysis to support the gasotransmitter role of H2S besides as a substrate for Cys synthesis. Comparison of the differentially expressed genes (DEGs) between H2S and Cys treated seedlings indicated that H2S fumigation and Cys treatment caused different influences on gene profiles during seedlings development. A total of 261 genes were identified to respond to H2S fumigation, among which 72 genes were co-regulated by Cys treatment. GO and KEGG enrichment analysis of the 189 genes, H2S but not Cys regulated DEGs, indicated that these genes mainly involved in plant hormone signal transduction, plant-pathogen interaction, phenylpropanoid biosynthesis, and MAPK signaling pathway. Most of these genes encoded proteins having DNA binding and transcription factor activities that play roles in a variety of plant developmental and environmental responses. Many stress-responsive genes and some Ca2+ signal associated genes were also included. Consequently, H2S regulated gene expression through its role as a gasotransmitter, rather than just as a substrate for Cys biogenesis, and these 189 genes were far more likely to function in H2S signal transduction independently of Cys. Our data will provide insights for revealing and enriching H2S signaling networks.
1 Introduction
Following the original paper postulating the novel concept of “gasotransmitter” nearly 20 years ago, the importance of these gaseous signaling molecules in biological pathways has been widely reported (Wang, 2002; Wang, 2014; Feng et al., 2019; Corpas and Palma, 2020; Aroca et al., 2021; Liu et al., 2021; Yang et al., 2022). Hydrogen sulfide (H2S) is qualified as the third member of gasotransmitter family following the identification of nitric oxide and carbon monoxide (Wang, 2002; Yang et al., 2008). For hundreds of years since its discovery, H2S has been well-known as a gas with an unpleasant odor and high toxicity (Fu et al., 2018; Lefer, 2019). Even the occasional discovery that H2S can increase plant resistance to pests and pathogens has been attributed to the cytotoxic properties of H2S (Wang, 2012). Until recent decades, the endogenously generation of H2S in animals and plants reignited scientists’ thinking on the significance of endogenous H2S in organism.
The generation of endogenous H2S in plants is closely related to Cys metabolism, and it can be roughly summarized into two pathways, one is that Cys desulfhydrases (CDes) catalyze the degradation of Cys to produce H2S, and the other is that H2S is generated as a side reaction product during O-acetylserine(thiol)lyase (OAS-TL) mediated Cys biosynthesis (Papenbrock et al., 2007; Heeg et al., 2008; Alvarez et al., 2010). For the former pathway, L/D-CDes (L-CDes; EC 4.4.1.28 and D-CDes; EC 4.4.1.15), requiring pyridoxal 5’-phosphate (PLP) as a cofactor, primarily responsible for the generation of endogenous H2S in plant, catalyze the desulfuration of Cys to H2S plus ammonia and pyruvate in a stoichiometric ratio of 1:1:1 (Jin and Pei, 2015; Liu et al., 2021). There were some CDes genes have been reported in Arabidopsis, including L-Cys desulfhydrase (LCD, AT3G62130), D-Cys desulfhydrase 1 (DCD1, AT1G48420) and D-Cys desulfhydrase 2 (DCD2, AT3G26115) (Papenbrock et al., 2007). Specially, the L-Cys desulfhydrase 1 (DES1, AT5G28030), identified from a previously reported minor cytosolic OAS-TL protein CS-LIKE, is demonstrated as a novel L-Cys desulfhydrase (Alvarez et al., 2010). DES1 and LCD using L-Cys as the substrate are the most widespread in plants. The substrate of DCD1 is D-Cys, while DCD2 can degrade the two isomers of Cys. In addition, another Cys desulfuration reaction occurs in Fe-S cluster biosynthesis and involves the formation of L-Ala and elemental sulfur or H2S from Cys through an enzyme-bound persulfide (-SSH) intermediate, which now has been known catalyzed by NifS-like proteins (Zheng and Dean, 1994; Rydz et al., 2021). In Arabidopsis, AtNFS1 (AT5G65720) encodes a cysteine desulfurase and AtNFS2 (AT1G08490) is a chloroplastic NifS-like protein, and both AtNFS1 and AtNFS2 require PLP as cofactor for proper folding. H2S can be produced with the availability of an appropriate amount of reducing agent to provide electrons during AtNFS1 and AtNFS2 mediating the Fe-S clusters formation (Leon et al., 2002; Pilon-Smits et al., 2002). For the latter pathway, H2S can be generated as a side product of Cys biosynthesis, accomplished by two sequential reactions catalyzed by Ser acetyltransferase (SAT; EC 2.3.1.30) and OAS-TL (EC 2.5.1.47). OAS-TL catalyzes the incorporation of sulfide (i.e., H2S) to O-acetyl-serine (OAS) to induce the last step of Cys synthesis (Romero et al., 2014), which is a reversible reaction where Cys could be decomposed to H2S and OAS (Gotor et al., 2019; Sehar et al., 2022). In Arabidopsis, the OAS-A1 (AT4G14880), OAS-B (AT2G43750), and OAS-C (AT3G59760) encode authentic OAS-TL proteins located in the cytosol, plastids, and mitochondria of Arabidopsis cells, respectively (Wirtz et al., 2004; Wirtz and Hell, 2006). The CYS-C1 (AT3G61440) encodes another OAS-TL isoform, which actually functions as the β-cyanoalanine synthase (β-CAS; EC 4.4.1.9) and catalyzes the reaction between L-Cys and HCN to synthesize β-cyanoalanine and H2S, a process that linking cyanide detoxification and H2S generation (Alvarez et al., 2012; Birke et al., 2012; Fang et al., 2022). The remaining OAS-TL-like proteins encoding genes include CYS-C1 (AT3G61440), CYS-D1 (AT3G04940), CYS-D2 (AT5G28020), DES1 (AT5G28030), and CS26 (AT3G03630) (Yamaguchi et al., 2000). It’s worth noting that DES1, mentioned above, is an OAS-TL homology but has nonconservative amino acid changes in its β8A-β9A loop, an important structure for interaction with SAT, so DES1 can not interact with SAT to mediate Cys biosynthesis and has more than 10 times lower affinity for OAS as a substrate than that for L-Cys, therefore, DES1 has been confirmed as a L-Cys desulfhydrase to produce endogenous H2S in plants (Alvarez et al., 2010). Coherently, H2S generation occurs during the regulation of Cys homeostasis, a process that Cys synthesis and degradation in the cell are coordinated mainly through the activities of OAS-TL and CDes.
H2S fulfills all criteria for being a gasotransmitter, such as being a gaseous molecule, generated endogenously, small and generally reactive, exerting important signaling roles. Evidence has been accumulating to elucidate the physiological functions of H2S in numerous physiology processes from the perspective of gaseous signaling molecule (Lu et al., 2013; Wang, 2014; Aroca et al., 2021; Li et al., 2022; Yang et al., 2022), including crosstalk with phytohormone signals (Jia et al., 2018; Shen et al., 2020; Zhou et al., 2021), interaction with Ca2+ signaling (Fang et al., 2017) and NO signals (Lu et al., 2013; Liu et al., 2021), regulation of gene transcription, and mediation of protein modification (Paul and Snyder, 2015; Aroca et al., 2018; Ma et al., 2021), etc. Mediating protein S-persulfidation, converting the mercapto groups (-Cys-SH) into hydropersulfide groups (-Cys-SSH), has been confirmed to be a key route for H2S bioactivity and signaling transduction, and involves in a myriad of cellular processes in plants linked to growth, development, stress responses, and phytohormone signals (Aroca et al., 2018; Shen et al., 2020; Ma et al., 2021; Sun et al., 2021). Nonetheless, there are still a few objections that the positive functions of H2S just due to its role as a substrate for S metabolism, especially for the synthesis of Cys, which is the first organic compound containing reduced S synthesized by the plants (Takahashi et al., 2011; Romero et al., 2014). Cys occupies a central position in plant metabolism because it per se and its derivative molecules play roles in a variety of signaling pathway occurring in various cellular processes (Paulsen and Carroll, 2013; Romero et al., 2014). Therefore, the controversy centers on the possibility that H2S acts only as a substrate to promote the synthesis of Cys or endogenous H2S production disturbs the Cys homeostasis, and the H2S-induced Cys, not H2S itself, is the core molecule that plays the role of signal transduction.
In this study, we provide some evidence through comprehensive transcriptome analysis to support the conclusion that H2S functions as a gasotransmitter, besides as the substrate for Cys biogenesis, providing insights for revealing and enriching H2S signal transduction networks.
2 Materials and methods
2.1 Plants and treatments
The Columbia wild type Arabidopsis thaliana (Col-0) was used in this study, and the seeds were surface sterilized by soaking in 70% ethanol for 5 min and rinsed well with sterile distilled water (3 times). These surface sterilized seeds were planted on 1/2 Murashige and Skoog (½MS) solid medium with 0.7% (w/v) agar and 1.5% sucrose. After 2 days stratification in darkness at 4°C, seeds were transferred to plant incubators with a 10 h-light (130 μmol m-2 s-1)/14 h-dark photoperiod at 22°C.
For H2S fumigation treatment, the 7-day-old seedlings were transferred aseptically to ½MS medium and successively fumigated with H2S released by NaHS. The NaHS solution-containing tube was placed in the Petri dish and the H2S fumigation concentration was 50 μmol L-1 (the volume was calculated by subtracting the volume of culture medium from the total volume of Petri dish). For Cys treatment, the 7-day-old seedlings were transferred aseptically to ½MS containing 1 mmol L-1 Cys, and the Cys containing ½MS was prepared by directly adding corresponding volume of Cys mother liquor, sterilized by filtration, to the ½MS medium. In order to enhance the scientific credibility of the one single variable principle, the 7-day-old seedlings were also transferred aseptically to ½MS medium for the control.
After 10 days of treatment, the seedling phenotypes were observed, and the whole plants were taken for subsequent anthocyanin level analysis, the determination of endogenous H2S and Cys contents as well as the enzymatic activities. The aerial tissue of Arabidopsis seedlings was obtained for RNA preparation and transcriptome sequencing.
2.2 Determination of the enzymatic activities of CDes and OAS-TL
Aliquots of 500 mg of seedlings were powdered in liquid nitrogen for small-scale extraction of proteins, and the power was dissolved in 0.5 mL of extraction buffer (50 mmol L-1 HEPES-KOH, pH 7.4, 10 mmol L-1 KCl, 1 mmol L-1 EDTA, 1 mmol L-1 EGTA, 10% [v/v] glycerin, 10 mmol L-1 DTT, and 0.5 mmol L-1 phenylmethylsulfonyl fluoride [PMSF]). The proteins were extracted at 4°C for 1 hour with frequent shaking, and the centrifugation at 4°C for 10 min and 12,000 g was performed to get the supernatant containing total proteins, which were then used to detect the enzymatic activity (Heeg et al., 2008).
The OAS-TL proteins catalyze the last step of Cys biosynthesis, so the enzymatic activity of OAS-TL was detected to represent the intensity of Cys production in plants. The enzymatic activity of OAS-TL was determined according to method reported previously (Liu et al., 2019). Briefly, the protein extraction was added to a reaction mixture of 50 mmol L-1 Tris-HCl (pH 7.5), 5 mmol L-1 DTT, 5 mmol L-1 OAS and 5 µmol L-1 PLP. The reaction was initiated by adding 10 mmol L-1 Na2S. After incubating for 10 min at 25°C, the reaction was terminated by adding 100 µL absolute acetic acid. Then, 200 µL ninhydrin reagent (25 mg/ml in acetic acid:HCl, 60:40, v/v) was added to detect the generated Cys. The mixture was boiled for 10 min before being cooled rapidly and 200 µL absolute ethanol was added to stop the reaction. Finally, the absorbance of the reaction was measured at 560 nm to determine the cysteine concentration.
The CDes are the most important enzyme that catalyzes the decomposition of Cys to pyruvate, ammonia, and H2S, so the activity of CDes was ascertained by measuring the production rate of H2S from Cys (Fang et al., 2017). The enzyme activity was determined in a reaction mixture (1 mL) containing 100 mmol L-1 Tris-HCl (pH 9.0), 0.8 mmol L-1 L-Cys or D-cysteine, 2.5 mmol L-1 DTT and 100 µL protein extraction. The reaction was incubated at 37°C for 15 min, and the generated H2S was absorbed by 0.5 mL Zn(AC)2 when placed together in a closed vial with the aforementioned reaction mixture. Then, 100 µL of 20 mmol L-1 N, N-dimethyl-p-phenylenediamine and 100 µL of 30 mmol L-1 FeCl3 were added to the Zn(AC)2 solution. After incubation in dark for 15 min, the absorbance was measured at 670 nm to determine the level of produced H2S.
2.3 Measurement of the contents of endogenous Cys and H2S
The endogenous H2S content was measured according to previously described method based on the principle that H2S reacts with N, N-dimethyl-p-phenylenediamine in the presence of the FeCl3 to produce blue methylene blue, which has the maximum absorption at 670 nm (Qiao et al., 2015; Fang et al., 2016). Cys can react specifically with acid ninhydrin to form a pink product, which has a maximum absorbance at 560 nm. The reaction is highly sensitive for Cys determination, so the Cys content was determined according to the method based on this reaction (Gaitonde, 1967; Fang et al., 2016).
2.4 Analysis of anthocyanin level
Anthocyanin content was measured as described previously (Rabino and Mancinelli, 1986). Briefly, 100 mg Arabidopsis seedlings were incubated in 1 mL extraction buffer (methanol containing 1% HCl, v/v) for 24 h in darkness at 4°C with occasional shaking. After extraction, the mixture was then centrifuged for 15 min at 5000 rpm, and then the supernatants were collected to detect the absorbance at 530 and 657 nm. The concentration of the anthocyanin was presented as mg g-1 dry weight of the differently treated plants using the following equation: [OD530-0.25*OD657] * volume of the extract (mL)/fresh weight (g).
2.5 RNA extraction and library preparation for transcriptome sequencing
The RNA extraction, library preparation and transcriptome sequencing were performed by Beijing Biomarker Biotechnology Co., Ltd., Beijing, China. Briefly, total RNA was isolated from seedlings using RNAiso-plus (TaKaRa), and the total RNA was analyzed by NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE) for concentration and purity, and by the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 (Agilent Technologies, CA, USA) for RNA integrity. Then 1 μg total RNA was used as input to prepare the non-strand-specific RNA-seq library by using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) and index codes were added to attribute sequences to each sample following manufacturer’s recommendations. Briefly, mRNA was purified from total RNA by using poly-T oligo-attached magnetic beads, and then fragmented using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). Subsequently, first strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase, and second strand cDNA synthesis was then performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ends of DNA fragments, NEBNext adaptor with hairpin loop structure were ligated to prepare for hybridization. The library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA) to select cDNA fragments of preferentially 240 bp in length. With these operations, we generated the size-selected and adaptor-ligated cDNA, which was then incubated with 3 μL of USER Enzyme (NEB, USA) at 37°C for 15 min followed by 5 min at 95°C before PCR. Then, library amplification was performed with PCR using Phusion High-Fidelity DNA polymerase, universal PCR primers and index (X) primer. Finally, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
After cluster generation of the index-coded samples, which was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumia), the library sequencing was carried out on an Illumina platform.
2.6 Data processing and analysis
After sequencing, the paired-end raw reads were generated, and the raw reads were firstly processed through in-house perl scripts to remove low quality reads, adapter containing reads, ploy-N containing reads. Then, Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality. Only clean reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. Hisat2 tools soft were used to map with reference genome. Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); GO (Gene Ontology).
2.7 Quantification of gene expression levels and differential expression analysis
Gene expression levels were estimated by FPKM. Differential expression analysis of two conditions/groups was performed using the edgeR. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 found by edgeR and Fold Change >1.5 were assigned as differentially expressed.
2.8 GO enrichment analysis
GO enrichment analysis of the DEGs was implemented using the GOseq R packages based Wallenius non-central hyper-geometric distribution, which can adjust for gene length bias in DEGs (Young et al., 2010).
2.9 KEGG enrichment analysis
KEGG (Kanehisa et al., 2008) is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). The statistical enrichment of differential expression genes in KEGG pathways was analyzed by using KOBAS software (Mao et al., 2005).
2.10 Protein-protein interaction analysis
PPI is the basis on which many biological pathways are built, so PPI prediction is essential for understanding the function mechanism of proteins. DEGs were blast (blastx) to the genome of a related species (the protein-protein interaction of which exists in the STRING database: http://string-db.org/) to get the predicted PPI of these DEGs. Then the PPI networks of these DEGs were visualized in Cytoscape (Shannon et al., 2003).
2.11 Validation of transcriptomic data by qRT-PCR
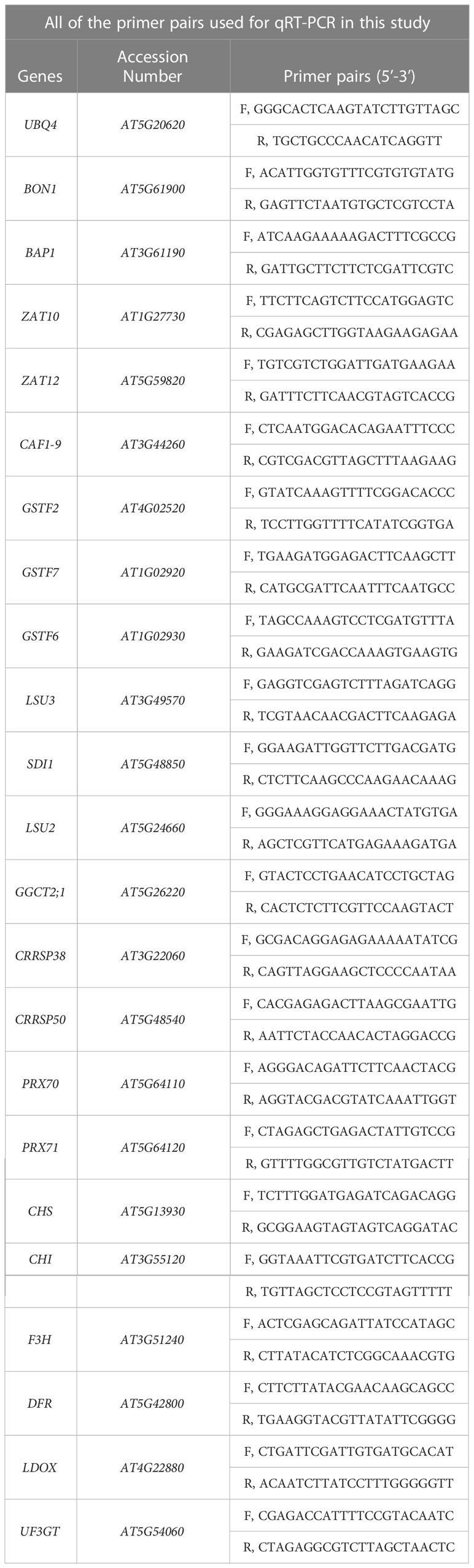
Total RNA was isolated from seedlings by using RNAiso-plus (TaKaRa, Shiga, Japan, Cat9109) according to the manufacturer’s instructions. The cDNA was synthesized using a reverse transcription system kit (PrimeScript RT Reagent Kit, TaKaRa, RR037B) and oligo (dT) primers, and then the qRT-PCR was performed to validate the transcriptomic data of target genes according to the instructions of the Bio-Rad Real-Time System (CFX96TM C1000 Thermal Cycler). In our study, the UBQ4 (AT5G20620), a housekeeping reference gene, was used as the normalizer. All of the primer pairs used for qRT-PCR were checked for amplification specificity and were listed in Table 1.
3 Results
3.1 Effects of H2S and Cys treatment on seedling development in Arabidopsis
To analyze the correlation and distinction of exogenous H2S and Cys treatment on seedling development, we observed and compared the Arabidopsis seedlings phenotypes after 10 days of 50 μmol L-1 of H2S fumigation and 1 mmol L-1 of Cys treatment. It was found that no obvious stress phenotypes were produced in H2S and Cys treated seedlings (Figure 1A). Exogenous H2S fumigation had little effect on the relative speed of plant growth, while Cys treatment seemed to cause a slower leaf growth rate of seedlings with smaller leaves at the same days after planting (Figure 1A). Anthocyanins are natural water-soluble pigments extensively exist in plants, and the anthocyanin level in leaves provides valuable information about the physiological status of plants. In our study, we found that Cys treatment, but not H2S fumigation, could lead to darker red and purple colors in leaves, especially in the petioles and center veins of leaves (Figure 1B). Moreover, analysis of anthocyanin content indicated that the anthocyanin level of the leaves was increased by Cys treatment but not affected by H2S fumigation (Figure 2B), which was consistent with the phenotype that darker anthocyanin color in leaves of Cys-treated seedlings. All these results mentioned above indicated the specificity of H2S and Cys in regulating seedling development.
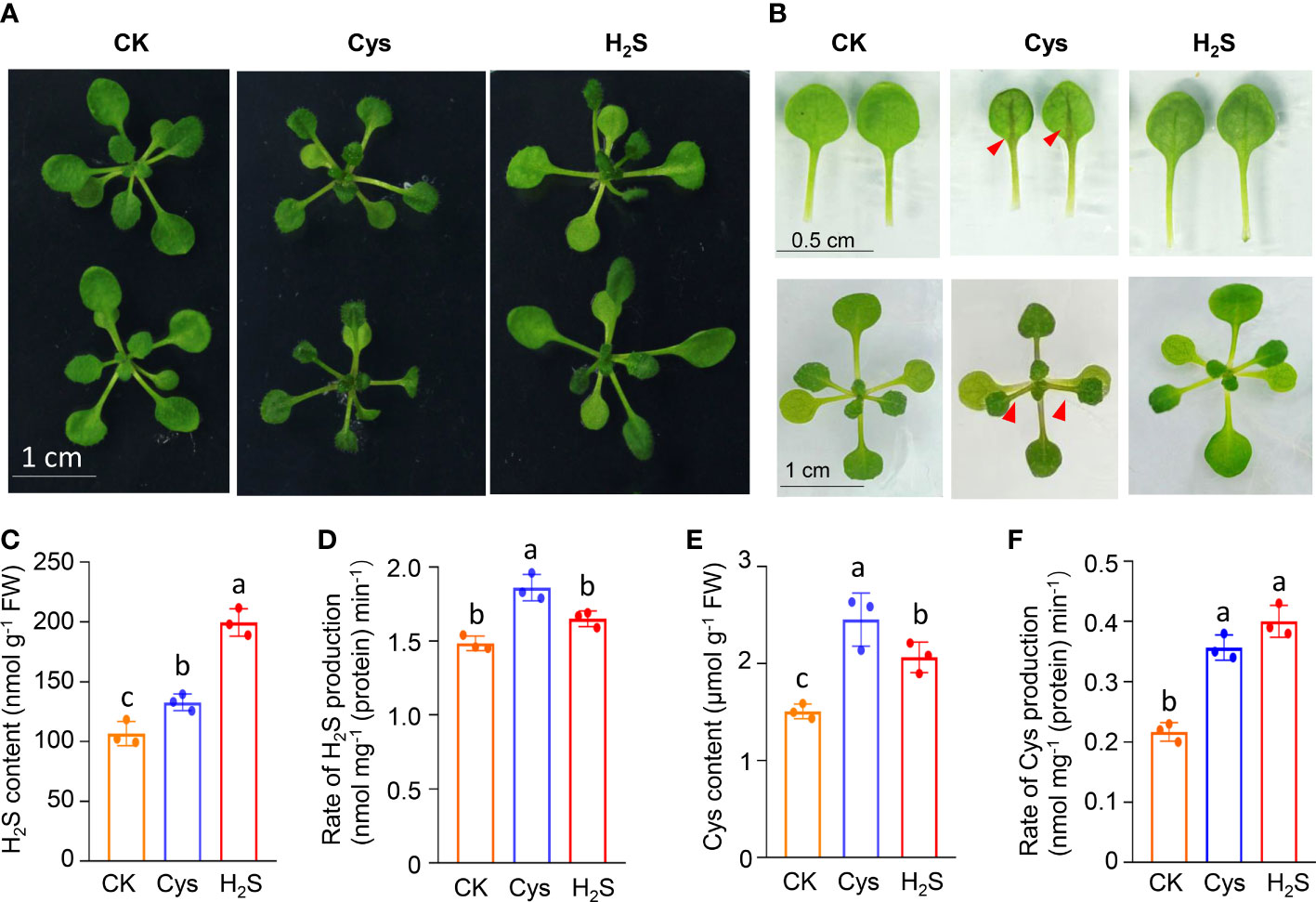
Figure 1 Responses of seedling development as well as endogenous H2S and Cys generation to exogenous H2S and Cys treatment in Arabidopsis. (A) Phenotypes of Arabidopsis seedlings after H2S and Cys treatment; (B) Leaf size and anthocyanidin color in leaves of H2S fumigated and Cys treated seedlings. (C, E) Endogenous H2S and Cys content in H2S fumigated and Cys treated seedlings. (D, F) Production rate of endogenous H2S and Cys in H2S fumigated and Cys treated seedlings. Red triangle indicates the anthocyanin accumulation in the center vein of leaves in Cys treated seedlings. Different letters represent statistically significant differences (p < 0.05).
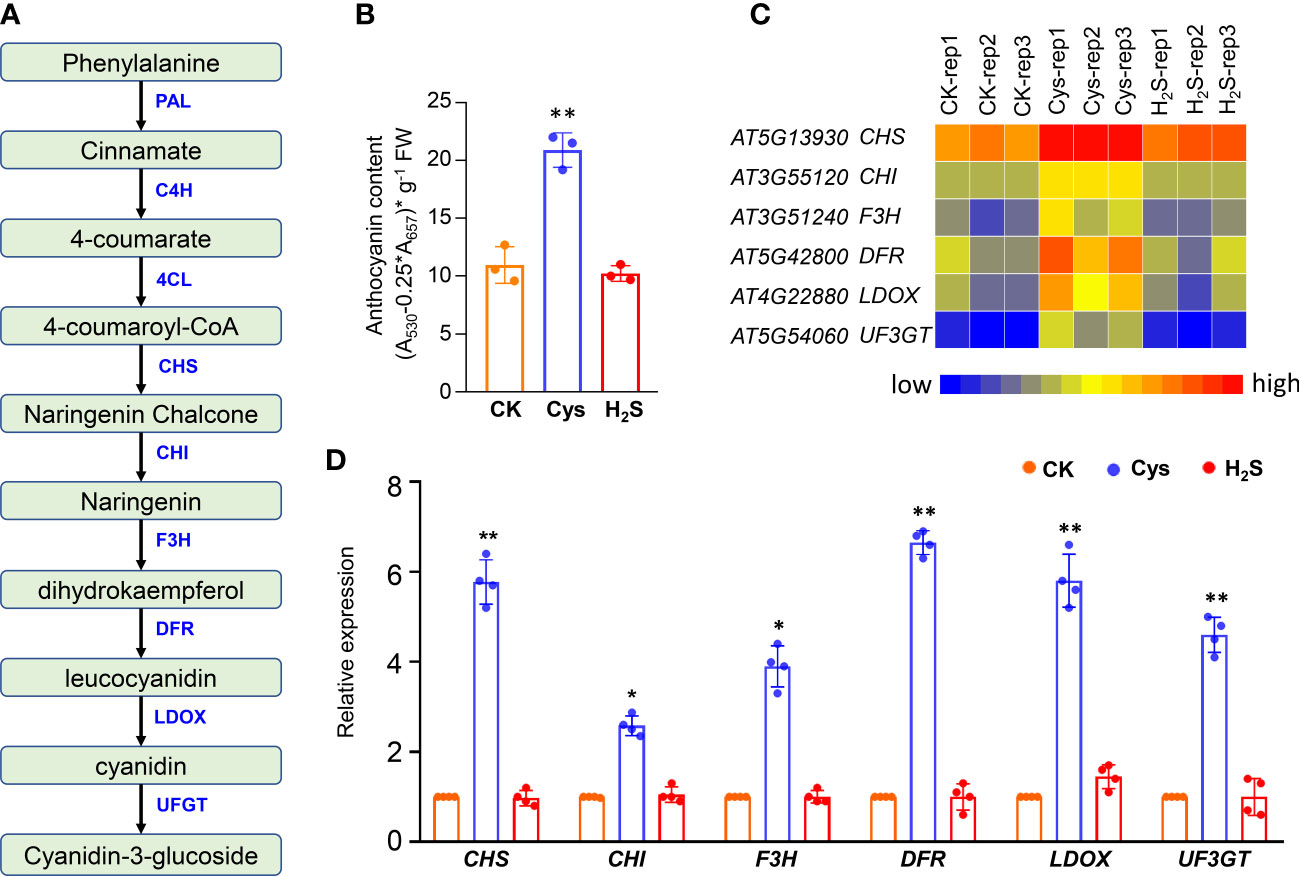
Figure 2 Analysis of anthocyanin level and anthocyanin biosynthesis genes in H2S and Cys treated seedlings. (A) Schematic representation of anthocyanin biosynthesis in plants. (B) Anthocyanin content of H2S and Cys treated seedlings. (C, D) Heatmap representation (C) and qRT-PCR analysis (D) of the anthocyanin biosynthesis genes (CHS, CHI, F3H, DFR, LDOX, UF3GT) in H2S and Cys treated seedlings. The symbol * indicates significant difference at the 0.05 level (p < 0.05) and ** indicates significant difference at the 0.01 level (p < 0.01).
3.2 Responses of endogenous H2S and Cys generation to exogenous H2S and Cys treatment
Exogenous application of signaling molecule could induce endogenous signal, so we detected the content and production rate of endogenous H2S and Cys in exogenous H2S and Cys treated seedlings. The data showed that both H2S fumigation and Cys treatment could simultaneously increase the content of endogenous H2S and Cys to a certain degree (Figures 1C, E), suggesting that the two kinds of exogenous treatments in our study activated both the endogenous H2S and Cys signals to various degrees. Our data also demonstrated that exogenous H2S and Cys treatment regulated the endogenous Cys-H2S cycle by modulating the production intensity of endogenous H2S and Cys. Both the production rate of H2S (Figure 1D), mediated by Cys degradation, and the production rate of Cys (Figure 1F) could be enhanced by Cys treatment, indicating that Cys treatment promoted the cyclic metabolism of Cys-H2S. However, H2S fumigation didn’t affect the production rate of H2S (Figure 1D), suggesting that the CDes activities and endogenous H2S generation were not activated in H2S fumigated seedlings. Because of the property of small molecular gas, H2S can be directly uptaken by leaves, so our data suggested that the increase of endogenous H2S content in seedlings fumigated by H2S probably due to the direct uptake of H2S by the leaves, rather than stimulating CDes mediated endogenous H2S production. The increased production rate of Cys in H2S fumigated seedlings (Figure 1F) might because of the higher level of endogenous H2S content.
3.3 Overview of the transcriptome profiles in Cys and H2S treated seedlings
To reveal the correlation and distinction between H2S as a gasotransmitter and as a substrate for Cys synthesis, we performed comprehensive transcriptomic analysis based on RNA sequencing data of 9 libraries generated from RNA samples of Control (CK), H2S fumigated (H2S), and Cys treated (Cys) Arabidopsis seedlings with three replicates. The library construction and RNA sequencing data analysis flow chart was briefly summarized in the Figure 3A. We obtained a total of 57.76 Gb clean data, and the clean data of each library reached at least 5.81 Gb with average base quality (Q30) more than 92.36% (Table S1) and sample correlation coefficients more than 98% (Figure 3B), which indicated that the quality and accuracy of sequencing data were sufficient and reliable for subsequent analysis although the sampling method probably caused slight variation in replications of the same treatment. The reads of each sample were mapped to the designated reference genome, and the alignment efficiency ranged from 96.18% to 97.61% (Table S1), so the selected reference genome can meet our needs for information analysis.
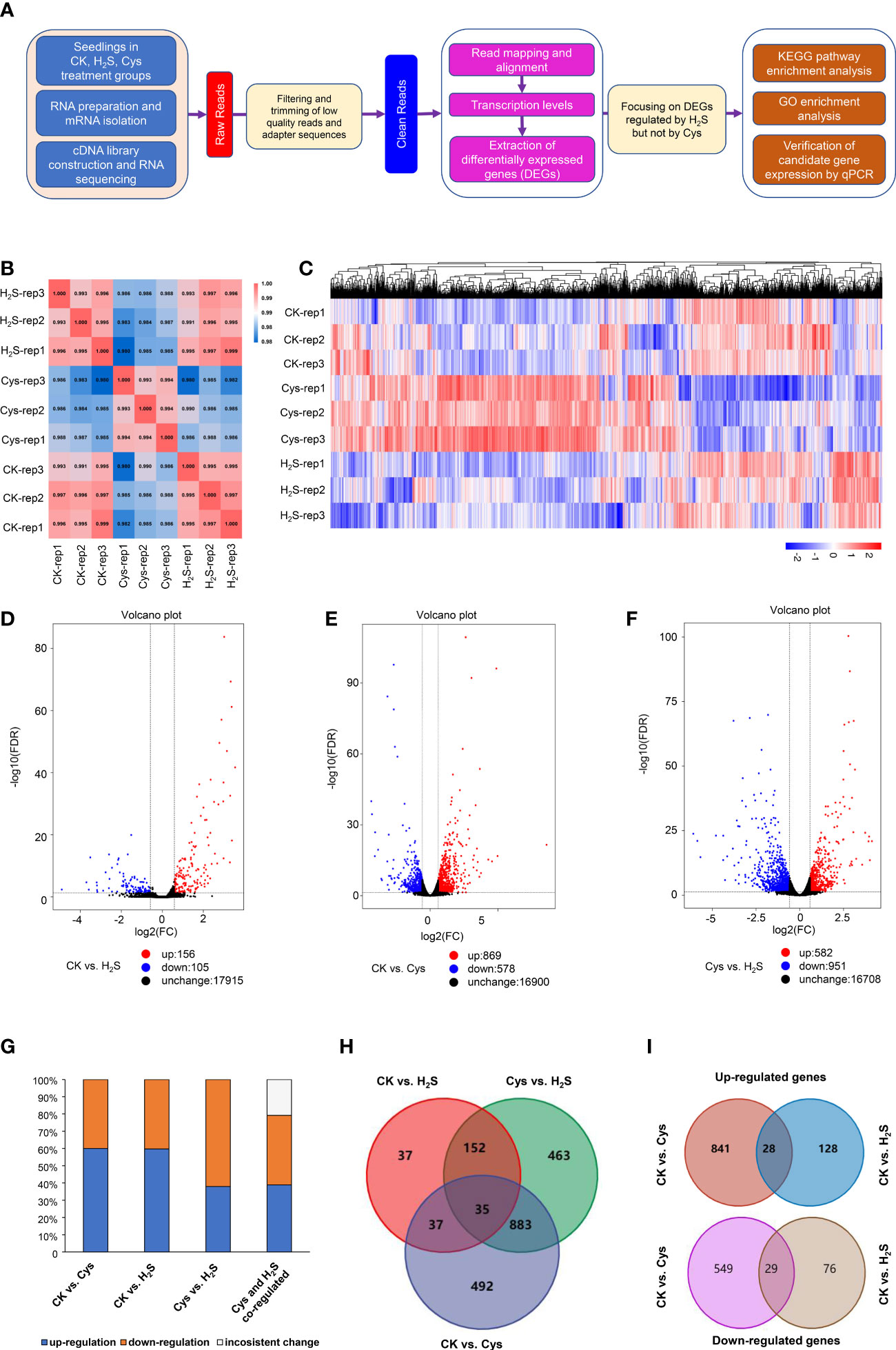
Figure 3 Characterization of transcriptome profiles in seedlings responding to H2S and Cys treatment. (A) Flow chart of analyzing transcriptome sequencing data. (B) Heat diagram of correlation coefficient between the 9 mRNA samples. (C) Heatmap representation of a one-dimensional hierarchical clustering of DEGs as determined by mRNA sequencing. (D–F) Volcano Plot showing the number of differentially expressed mRNAs (FDR < 0.05 and Fold Change >1.5) in different compared groups, (D) CK vs. H2S, (E) CK vs. Cys, (F) Cys vs. H2S. (G) Percentage of up-regulated and down-regulated genes in Cys-regulated, H2S-regulated, as well as H2S and Cys co-regulated DEGs. (H) Venn diagram showing the overlapping of DEGs in different compared groups, CK vs. Cys, CK vs. H2S, and Cys vs. H2S. (I) Venn diagram showing the overlapping of up-regulated DEGs and down-regulated DEGs in the comparison groups of CK vs. Cys and CK vs. H2S.
3.4 Analysis of differentially expressed genes between H2S and Cys treated seedlings
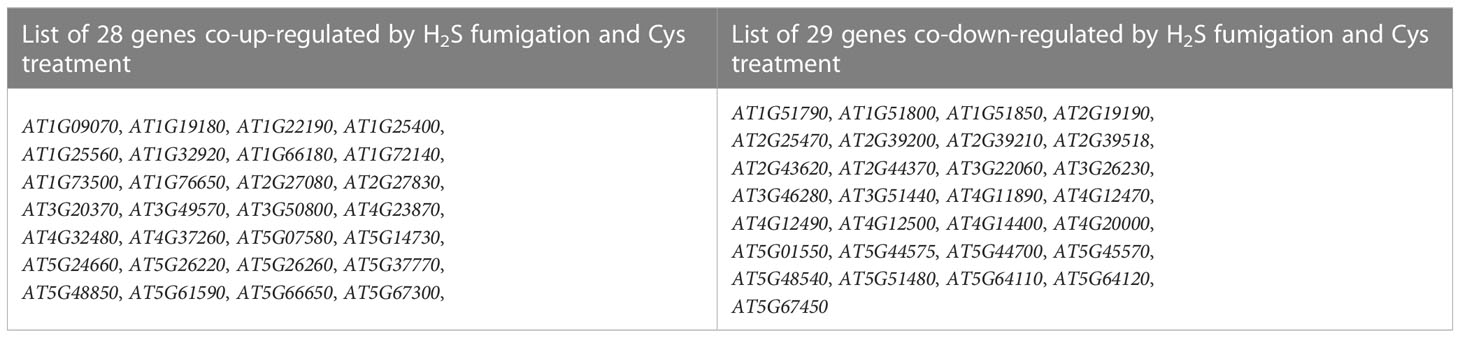
Gene expression profiles calculated based on the fragments per kilobase of transcript per million fragments mapped (FPKM) value showed that there was no significant difference in the overall FPKM expression levels of transcripts among CK, H2S, Cys treated seedlings (Figures S1A, B). The principal component analysis (PCA) plot was used to provide a systematic view of the consistency and variance of our RNA sequencing dataset, and the result showed that the CK, Cys and H2S groups could be distinguished (Figure S1C). Furthermore, we identified the DEGs according to the gene expression profiles in H2S fumigated and Cys treated seedlings. EdgeR was used to normalize the data and extract DEGs with FDR < 0.05 and Fold Change >1.5. The Heatmap represented the one-dimensional hierarchical clustering of differential gene expression among these three groups (Figure 3C). The volcanic maps indicated that a total of 261 DEGs were identified in H2S fumigated seedlings, of which 156 genes (59.77%) were upregulated and 105 genes (40.23%) were downregulated (Figures 3D, G). Moreover, among these H2S-regulated genes, 72 genes were co-regulated by Cys treatment (Figure 3H), hinting that H2S might regulate the transcription of the other 189 genes through its role of gasotransmitter rather than as the substrate for Cys synthesis. There were 1447 DEGs caused by Cys treatment, including 869 (60.06%) upregulated genes and 578 (39.94%) downregulated genes (Figures 3E, G). Among these Cys-regulated genes, only 4.9% (72) of these DEGs could also respond to H2S fumigation and up to 95.1% (1375) of the genes could be regulated only by Cys treatment (Figure 3H), suggesting that Cys might play its role in regulating the expression of these genes through itself or its derivative molecules, bypass degrading to generate endogenous H2S. In the Cys and H2S co-regulated genes, 28 genes were co-up-regulated and 29 genes were co-down-regulated in Cys-treated and H2S-fumigated seedlings (Table 2, Sheet 2 and 3 of Supp Excel S5, and Figure 3I), however, 9 genes up-regulated by Cys treatment were down-regulated by H2S-fumigation, and 6 genes down-regulated by Cys treatment were up-regulated by H2S-fumigation (Sheet 4 of Supp Excel S5 and Figure 3G). We also identified 1533 DEGs between H2S-fumigated and Cys-treated seedlings, including 582 (37.96%) up-regulated genes and 951 (62.04%) down-regulated genes (Figures 3F, G). Consequently, H2S and Cys could induce different DEGs in Arabidopsis seedlings, indicating that H2S performed its functions not only by serving as a substrate of Cys but also by participating in other physiological pathways, possibly involving its gasotransmitter functions.
3.5 Function enrichment analysis of H2S but not Cys regulated DEGs
In order to further explore the functions of H2S as a gasotransmitter, we then focused on these 189 genes regulated only by H2S fumigation but not by Cys treatment, including 122 up-regulated genes and 67 down-regulated genes. KOG analysis of these genes showed that the most mapped functional categories were “general function prediction only”, “posttranslational modification, protein turnover and chaperones”, “transcription” and “signal transduction mechanisms” (Figure 4A). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment showed that these genes mainly focused on plant hormone signal transduction, plant-pathogen interaction, phenylpropanoid biosynthesis, and MAPK signaling pathway (Figure 4B). This indicated that the mechanisms for H2S functioning as a gasotransmitter might involve cross-talk with hormone signals and MAPK signals, and regulating the interaction of plant and pathogen as well as the biosynthesis of phenylpropanoid, which has important functions in plant development and plant-environment interaction (Dong and Lin, 2021). Protein-protein interaction (PPI) plays essential roles in many biological processes, and we predicted 24 PPIs in these H2S but not Cys regulated DEGs (Figure 4C), such as BON1 and BON association protein BAP1, CAF1-9 and CAF1-11, which may provide important insights for revealing H2S signaling pathway.
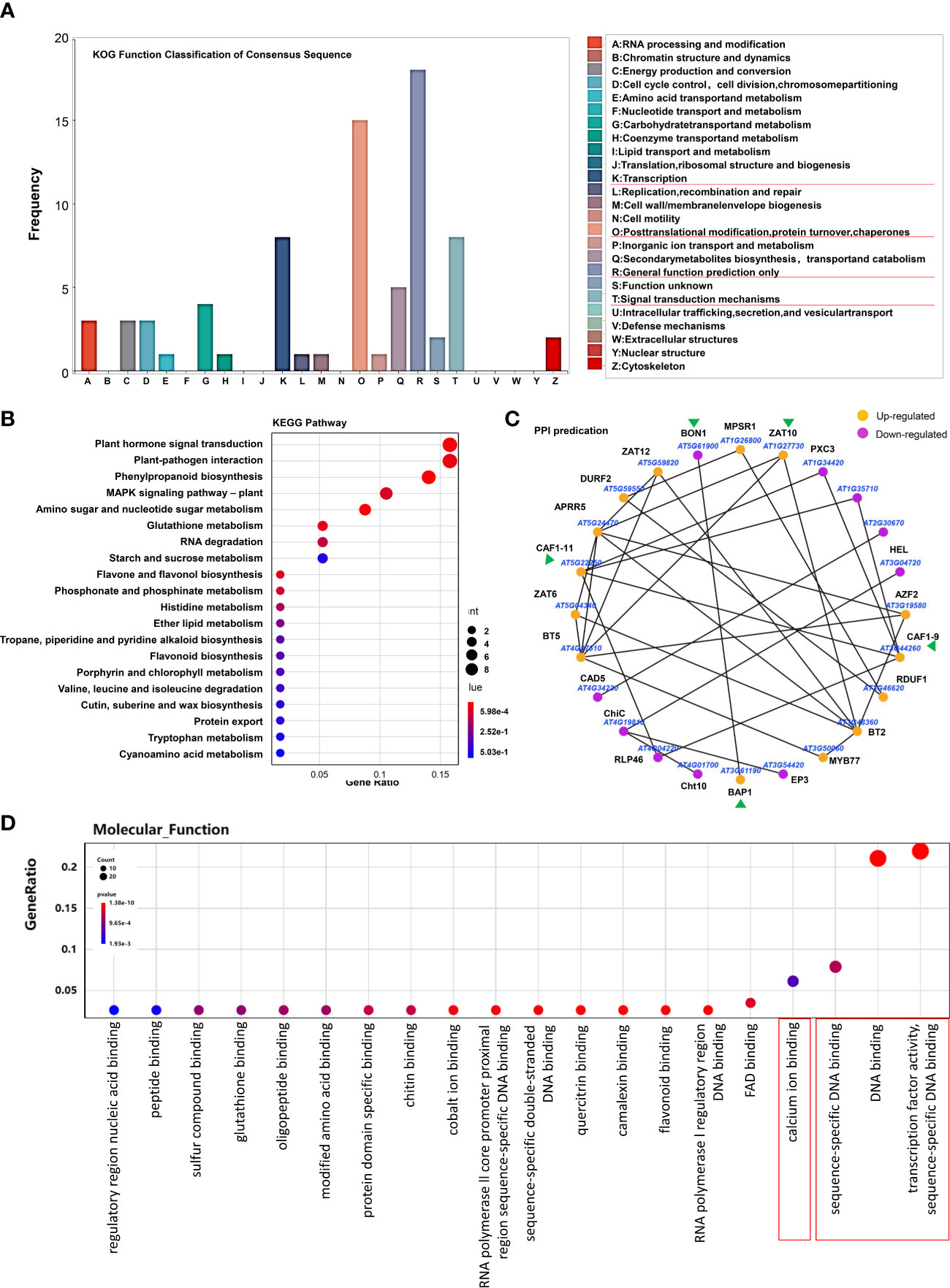
Figure 4 Overall analysis of the DEGs regulated by H2S fumigation but not by Cys treatment. (A, B) KOG function classification (A) and KEGG pathway (B) of the 189 DEGs that could be regulated by H2S fumigation but not by Cys treatment. (C) PPI network of proteins encoded by these DEGs. (D) GO enrichment analysis of these 189 DEGs.
In addition, GO enrichment analysis showed that more than half of these H2S but not Cys regulated genes encode DNA binding proteins (Figure 4D), including AP2 domain ethylene-responsive transcription factors (TFs), AP2/ERF and B3 domain-containing TFs, RING-H2 finger proteins, Zinc finger proteins, Zinc finger CCCH domain-containing proteins, MYB TFs, etc. (Figure 5A). Some Ca2+ and CaM binding proteins could also be regulated by exogenous H2S fumigation, including Ca2+-binding proteins, KRP1 (AT4G27280) and PBP1 (AT5G54490), Calmodulin like proteins, CML13 (AT1G66400) and CML16 (AT3G25600), and CaM-binding protein CAMBP25 (AT3G56880) (Figure 5B). Furthermore, some stress-responsive genes were also included in these H2S but not Cys regulated DEGs, and were represented by the Heatmap (Figure 5C). It was indicated that these genes involved in a variety of stress-responsive pathways, including hypoxia, temperature stress, wounding, drought, dehydration, salt and osmotic stress, oxidative stress, etc., also involved in plants defense reactions to pathogen attack as well as response to endogenous signals, such as hormone, carbohydrate, and organic substance (Figure 5C).
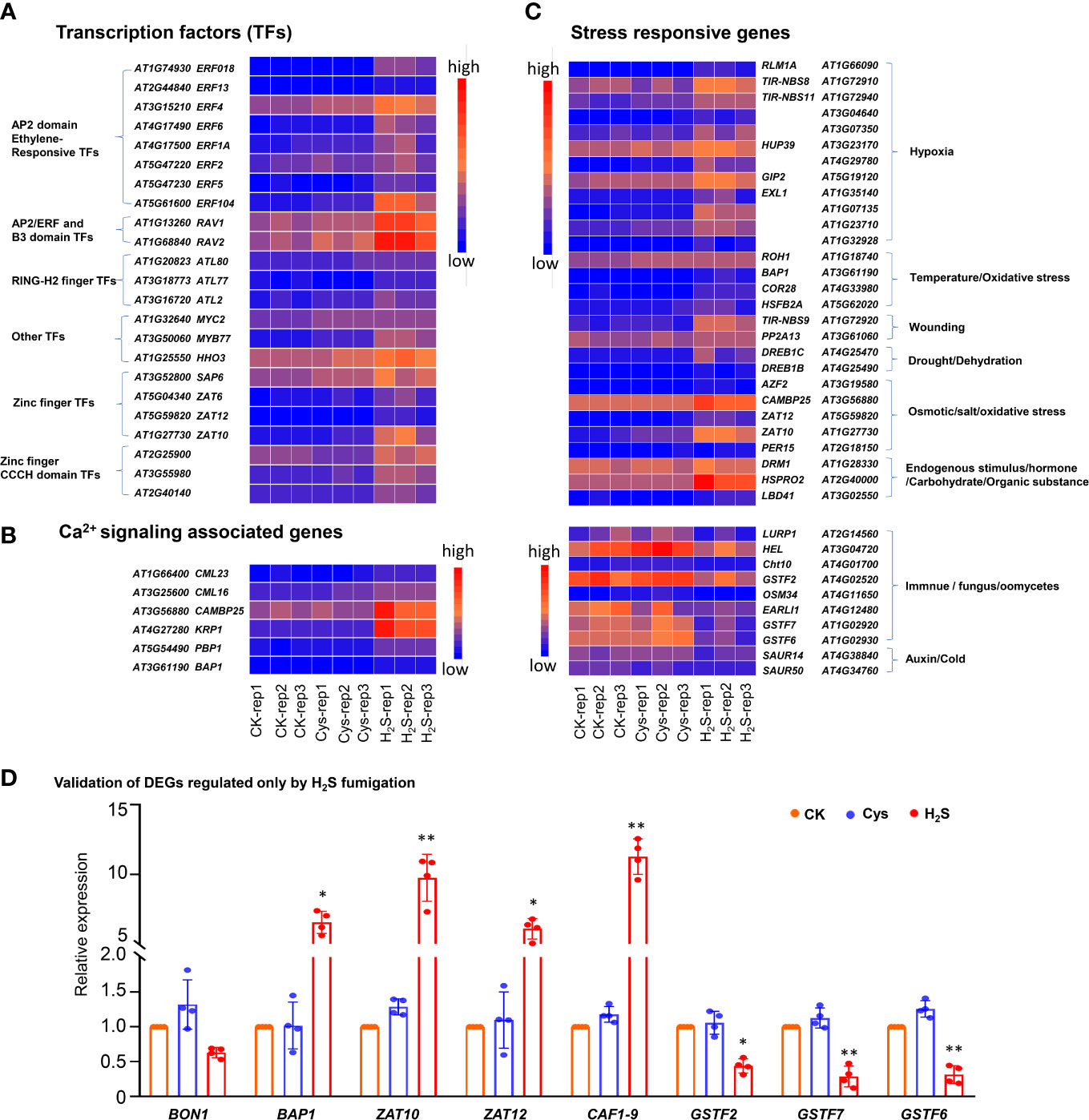
Figure 5 Heatmap representation and qRT-PCR analysis of some DEGs regulated by H2S but not by Cys treatment. (A) Heatmap representation of the up-regulated DEGs encoding transcription factors. (B) Heatmap representation of the DEGs encoding stress responsive genes. (C) Heatmap representation of the up-regulated DEGs encoding Ca2+signaling associated proteins. (D) qRT-PCR analysis of the transcript abundance of BON1, BAP1, ZAT10, ZAT12, CAF1-9, GSTF2, GSTF7, and GSTF6 in H2S and Cys treated seedlings. The symbol * indicates significant difference at the 0.05 level (p < 0.05) and ** indicates significant difference at the 0.01 level (p < 0.01).
Furthermore, the expression levels of some DEGs were analyzed by qRT-PCR, and the expression patterns for all chosen mRNAs were consistent with the results based on RNA sequencing (Figure 5D). The BON1 (AT5G61900) could be slightly down-regulated by H2S fumigation with no statistical difference, but a BON1-associated gene BAP1 (AT3G61190) could be significantly up-regulated by H2S fumigation (Figure 5D). As chloroplast ROS marker genes, both BAP1 and ZAT10 (AT1G27730) were up-regulated by H2S fumigation (Figure 5D), indicating that the chloroplasts ROS signaling might be activated by H2S signaling. The expression of CAF1-9 (AT3G44260), probable CCR4-associated factor 1 (CAF1) homolog 9, was enhanced nearly by 12 times in H2S fumigated seedlings (Figure 5D). It has been reported that CCR4-CAF1 complex is the major enzyme complex that catalyzes mRNA degradation through initiating mRNA deadenylation (Liang et al., 2009), so the regulation of H2S on CAF1-9 might enrich the signal transduction pathway of H2S from the perspective of deadenylation induced mRNA degradation. Additionally, three glutathione S-transferase coding genes (GSTF2, GSTF6, GSTF7) were obviously down-regulated by H2S fumigation, but not affected by Cys treatment (Figures 5C, D).
3.6 Analysis of H2S and Cys co-regulated DEGs
Involvement of H2S as a substrate in the synthesis of Cys is an important way for H2S participating in S metabolism. The H2S and Cys co-regulated DEGs might be genes regulated by both H2S and Cys signals, or H2S-induced Cys signals, or Cys-regulated H2S signals, so we analyzed these DEGs to provide evidences for crosstalk between H2S and Cys signals pathway. KEGG analysis indicated that the biological functions of these genes mainly involved in environmental information processing (MAPK signaling pathway, Phosphatidylinositol signaling system, Plant hormone signal transduction), organismal system (plant-pathogen interaction), and metabolism pathway (phenylpropanoid biosynthesis, amino sugar and nucleotide sugar metabolism, pentose and glucuronate interconversions, etc.) (Figure 6A). GO function classification indicated that these genes involved in 34 subcategories of biological processes, cellular components, and molecular functions. For the biological process, “cellular process”, “metabolic process”, and “response to stimulus” were the most representative groups. In the subcategory of cellular components, “membrane”, “cell part”, “membrane part” were the predominant groups. For the molecular function, “binding” and “catalytic activity” were the most common (Figure 6B).
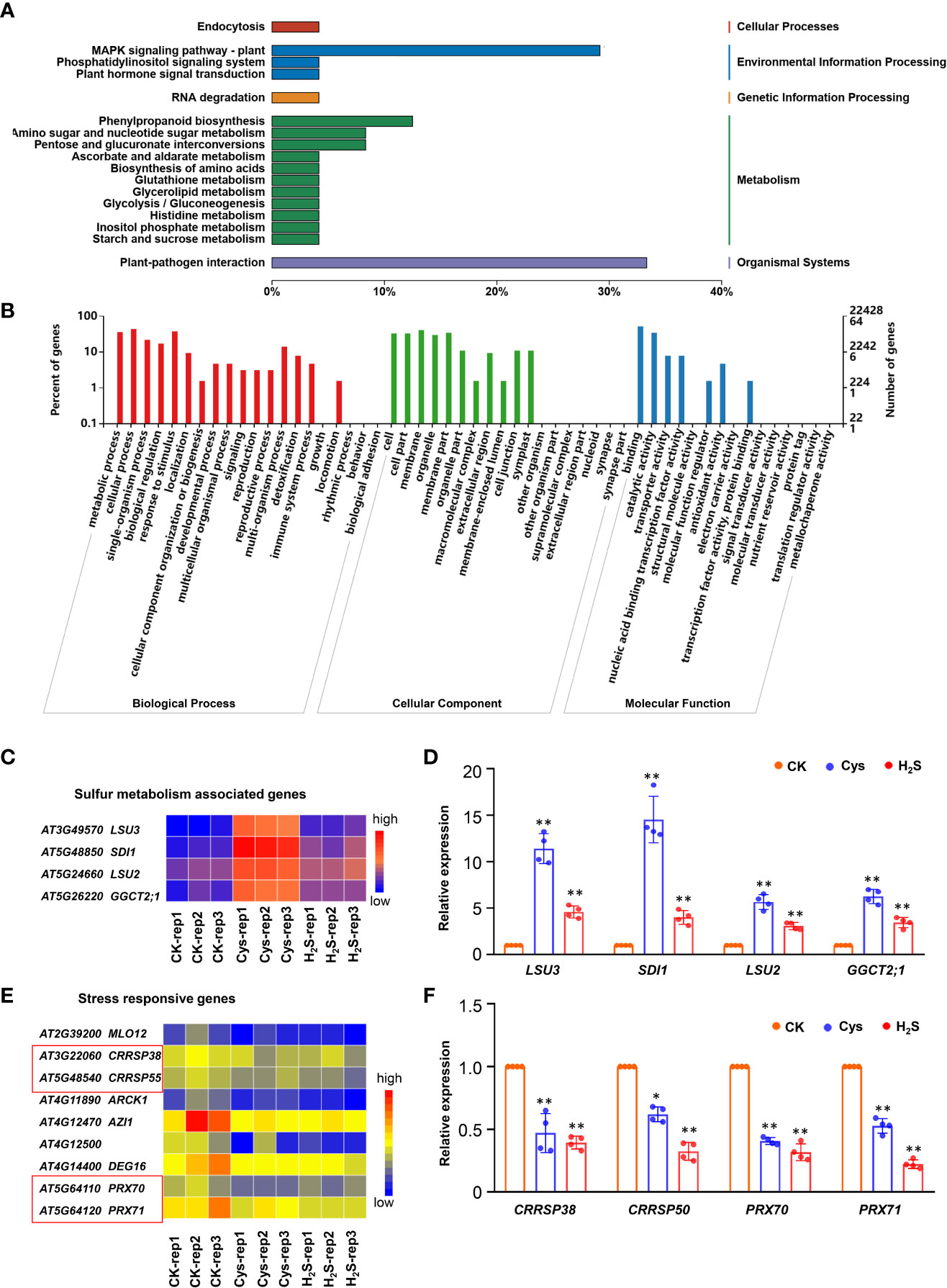
Figure 6 Overall analysis of the DEGs co-regulated by both H2S and Cys treatment. (A, B) KEGG analysis (A) and GO function classification (B) of the 72 DEGs that could be co-regulated by both H2S and Cys treatment. (C, D) Heatmap representation (C) and qRT-PCR analysis (D) of the sulfur metabolism associated genes (LSU3, SDI1, LSU2, GGCT2;1). (E, F) Heatmap representation (E) and qRT-PCR analysis (F) of the stress responsive genes (CRRSP38, CRRSP50, PRX70, PRX71). The symbol * indicates significant difference at the 0.05 level (p < 0.05) and ** indicates significant difference at the 0.01 level (p < 0.01).
Sulfur is essential for all living organisms on earth as a key component of amino acids (i.e., Cys and methionine), polypeptide glutathione, several group transfer coenzymes and vitamins (Romero et al., 2014). An in-depth study of these genes showed that the transcription of some S metabolism associated genes (LSU2, LSU3, SDI1, and GGCT2;1), which involve in response to low sulfur and sulfur deficiency, were significantly increased in both H2S and Cys treated seedlings (Figures 6C, D), suggesting that the treatment of exogenous S compounds (H2S and Cys) might regulate S absorption and metabolism (Sirko et al., 2014; Garcia-Molina et al., 2017), which might involve the feedback regulation of S absorption and metabolism. H2S-Cys cycle may participate in the feedback regulation of S-containing compound on S response. Some stress responsive genes could be down-regulated by both H2S and Cys treatments, including MILDEW RESISTANCE LOCUS O (MLO12), cysteine-rich receptor-like secreted protein encoding genes (CRRSP38 and CRRSP55), and the peroxidase superfamily genes (PRX70 and PRX71), this might be attribute to the enhancement of endogenous Cys and Cys-related signaling pathways (Figures 6E, F). The expression changes of some DEGs in H2S and Cys treated seedlings detected by qRT-PCR were consistent with the results from RNA sequencing data (Figures 6D, F), which further validated the reliability of the high-throughput sequencing results in this study.
3.7 Activation of anthocyanin synthesis induced by Cys but not H2S treatment
In plants, anthocyanins play a role not only in reproduction, by attracting pollinators and seed disseminators, but also in protecting plants from various abiotic and biotic stresses. More and more evidences show that anthocyanins have health-promoting properties, which makes anthocyanin metabolism an interesting target for breeders and researchers (Pojer et al., 2013; Liu et al., 2018). Anthocyanins are synthesized from 4-coumaroyl-CoA, which is generated from phenylalanine via the general phenylpropanoid pathway, involving the phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H) and 4-coumaryol CoA ligase (4CL). One molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA are condensed by chalcone synthase (CHS), which is the starting reaction of flavonoid biosynthesis. A coloured anthocyanidin (pelargonidin) are synthesized through sequential enzyme reactions involving the chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS, also known as LDOX), and UDP-glucose: flavonoid glucosyltransferase (UFGT) (Figure 2A).
Our data indicated that the anthocyanin level of the leaves was increased by Cys treatment but not affected by H2S fumigation (Figure 2B), which consistent with the phenotype that darker anthocyanin color in leaves of Cys-treated seedlings, especially in the petioles and center veins of leaves (Figure 1B). Both the RNA sequencing and the qRT-PCR data indicated that these anthocyanin-synthesis-associated genes (CHS, CHI, F3H, DFR, LDOX, UF3GT) were significantly up-regulated by Cys treatment, but had no obvious changes in H2S fumigated seedlings (Figures 2C, D), indicating that Cys enhanced anthocyanin level through up-regulating the expression of genes responsible for anthocyanin synthesis. The expression of DFR, encoding the key enzyme in the anthocyanin biosynthetic pathway, was even increased nearly sevenfold in Cys treated seedlings (Figure 2D). H2S fumigation enhanced endogenous Cys content but didn’t promote anthocyanin accumulation, which suggested the distinction of H2S and Cys signals, indicating that H2S could regulates biological processes as a gasotransmitter in a Cys-independent manner.
4 Discussion
H2S is well-known because of its unpleasant odor and high toxicity, and the toxicological properties of H2S were focused in most of the early researches (Wang, 2002; Fu et al., 2018). Until recent decades, increasing evidence revealed that H2S functions as a signaling molecule in plant growth and development as well as in biotic and abiotic stress responses (Wang, 2014; Paul and Snyder, 2015; Aroca et al., 2018; Liu et al., 2021; Zhang et al., 2021; Li et al., 2022; Yang et al., 2022). However, some scientists remain skeptical about its direct role as gasotransmitter and maintain that the positive function of H2S may largely depend on S-containing organic molecules, especially Cys, which has been demonstrated to play roles in various cellular processes (Paulsen and Carroll, 2013; Romero et al., 2014). During S metabolism in plants, H2S, uptaken directly from the atmosphere or generated form sulfate, could act as the substrate to generate Cys, while Cys can be degraded to produce H2S, so the changes of H2S level will undoubtedly influence Cys content and metabolism. Therefore, revealing the difference and relationship between H2S signaling and Cys-related signaling will be of great significance for clarifying the gasotransmitter functions of H2S.
Based on our study, H2S fumigation obviously increased both the endogenous H2S and Cys content, meanwhile Cys treatment enhanced endogenous Cys and H2S content to various degrees (Figures 1C, E), indicating that the endogenous H2S and Cys signals could be activated by exogenous H2S and Cys treatments. In addition, based on our transcriptome data, we analyzed and compared the FPKM value of genes associated Cys synthesis and degradation, including AtOAS-A1, AtOAS-B, AtOAS-C, AtCYS-C1, AtCYS-D1, AtCYS-D2, AtCS26, AtDES1, AtLCD, AtDCD1, AtDCD2, AtNFS1, and AtNFS2, in Control, Cys-treated, and H2S-fumigated seedlings, and data indicated that both Cys and H2S treatment can not affect the expression of these genes obviously (Table S2). Therefore, it might be speculated that Cys or H2S might regulate Cys and H2S metabolism by regulating the activities of corresponding enzymes rather than the expressions of corresponding genes.
Comprehensive transcriptome analysis indicated that H2S and Cys treatment can induced different DEGs in seedlings, and there were 189 genes could be regulated only by H2S fumigation but not response to Cys treatment (Figure 3H), hinting that H2S involves in regulating gene transcription through its gasotransmitter function, rather than only as a substrate of Cys, and there is a high probability that these 189 genes function importantly in H2S signal transduction pathways.
It has been reported that H2S interacts widely with phytohormones in diverse processes during the developmental and environmental responses of plants. H2S treatment down-regulated the ethylene biosynthesis genes, while up-regulated the ethylene receptor genes in banana fruit (Ge et al., 2017). H2S induced persulfidation on ACOs to inhibit ethylene synthesis involving osmotic stress response (Jia et al., 2018). The endogenous biosynthesis and/or signaling of ethylene and auxin could be modulated by H2S to inhibit the process of petiole abscission in tomato (Liu et al., 2020). Moreover, H2S might act as a downstream component of auxin signaling to trigger lateral root formation in tomato (Fang T, et al., 2014). The interaction between H2S and ABA signaling has been well studied in regulating stomatal movement. As early as 2012, Jin et al. reported that H2S interacting with ABA in the stomatal regulation responds to drought stress in Arabidopsis (Jin et al., 2013). Recently, it was found that during stomatal response to ABA induction, H2S mediated DES1 persulfidation to amplify H2S signal, which then drive the persulfidation of the NADPH oxidase RBOHD to produce ROS and induce stomatal closure (Shen et al., 2020). H2S induces the persulfidation of SnRK2.6/OST1 to positively regulate ABA signaling mediated stomatal movement (Chen et al., 2020). H2S mediated the persulfidation of ABI4, a key positive regulator of ABA signaling, to improve its ability for activating MAPKKK18 transcription, which is important for ABA response in Arabidopsis (Zhou et al., 2021). Above all, it seems that H2S functions not only as an integral molecule in the hormone signaling network of plants but also as a “referee” to harmonize the interaction between hormones. Therefore, revealing the detailed mechanism of the interactions between H2S and these phytohormones during the respective physiological processes will be valuable. Focusing on these 189 genes, regulated only by H2S fumigation but not Cys treatment, we found that many of these genes involve in plant hormone signal transduction (Figure 4B), including ethylene responsive TFs, auxin-responsive genes, salicylic acid responsive genes, ABA signaling associated genes, etc. (Figure 5A). Moreover, more than half of these genes encoded protein providing DNA binding and transcription factor activities (Figures 4D, 5A), participating or having crosstalk with many phytohormones signals. Our transcriptome data indicated that these 189 genes might function as the alternative downstream components in H2S signal transduction and crosstalk with hormone signal pathways, providing some insights for determining the mechanism of H2S interaction with phytohormones.
H2S also interacts with Ca2+ and calcium signaling, especially in the process of plant response to metal stress. Our previous study indicated that Ca2+/CaM2 physically interacts with bZIP transcription factor TGA3 to enhance its ability for activating LCD transcription, then improve the production efficiency of endogenous H2S in Arabidopsis response to Cr6+ stress (Fang et al., 2017). Similarly, H2S dependent pathway is also a component of Ca2+ mediated activation of the antioxidant system and upregulation of the expression of genes associating with heavy metal chelation in Setaria italica coping with Cr6+ stress (Fang H, et al., 2014). Additionally, H2S can strongly enhanced Ca2+ induced upregulation of CaM and CBL expressions in Setaria italica under Cr6+ stress (Fang H, et al., 2014). NaHS treatment increased the CDPK transcripts in seedling leaves of zucchini under Ni stress (Valivand et al., 2019), while CDPK3 enhanced LCD activity and intensified H2S signal to enhance the tolerance of Arabidopsis to Cd stress (Qiao et al., 2016). These researches suggest that H2S and Ca2+ signal do not have a simple upstream and downstream relationship, but a complex cross interaction in plants response to metal stress. In this study, some Ca2+ signal transduction associated genes (CML23, CML16, CAMBP25, KRP1, PBP1 and BAP1) could be upregulated by H2S fumigation (Figure 5B). BAP1, encoding a membrane-associated protein containing a Ca2+-dependent phospholipid-binding C2 domain, involves in programmed cell death (PCD) and defense reaction across the kingdoms (Yang et al., 2007), so the upregulation of H2S fumigation on this gene strongly hinted the function of H2S in regulating PCD and defense, and the underlying mechanism is probably related to the interaction with Ca2+ signaling.
H2S has been widely reported to participate and/or regulate the ROS signal transduction, such as activating some antioxidant enzymes through inducing persulfidation, increasing GSH content, interacting with H2O2 signal, etc. (Choudhary and Chaudhary, 2021; Liu et al., 2021; Zhang et al., 2021). Chloroplast ROS signaling is a major driving force in chloroplast to nucleus retrograde signaling and plays important roles in plant stress responses (Foyer and Hanke, 2022; Li and Kim, 2022). Based on our data in this study, the chloroplast ROS marker genes ZAT10 and BAP1 were up-regulated by H2S fumigation, but not affected by Cys treatment, indicating that H2S might induce the chloroplast ROS signaling independently of Cys.
Comprehensively, our results provide evidence for H2S as a gasotransmitter besides as the substrate for Cys synthesis in regulating plant response to both developmental and environmental cues, and further enrich and deepen the H2S signal transduction networks. However, the underlying mechanism of H2S mediating the transcriptional regulation of these DEGs is still worth of further exploration. H2S-induced persulfidation has been proved to be an important post-translational modification (PTM) in plants (Aroca et al., 2018; Gotor et al., 2019; Chen et al., 2020; Ma et al., 2021; Zhou et al., 2021), so H2S might regulating genes expression through inducing persulfidation on proteins that function as transcription factors or involve in transcriptional regulatory complexes of target genes. Determining the connection between H2S mediated regulation of protein persulfidation and gene transcription is very significant for revealing the detailed mechanism of H2S signal transduction.
Data availability statement
The transcriptomic data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, and the BioProject accession number PRJNA971675.
Author contributions
HF conceived the present idea and wrote the manuscript. ZY, KX, and LZhou performed the experiment most. HF, ZY, and KX performed the data analysis, HF, YP and LZhang supervised the project, provided critical feedback and helped shape the final manuscript. All authors helped perform the analysis with constructive discussions, and proofread the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a fund from the Natural Science Foundation of Zhejiang Province (Grant No. Q21C060002) and the National Natural Science Foundation of China (32102367) to HF.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1184991/full#supplementary-material
Supplementary Figure 1 | Analysis of transcriptome profiles in seedlings responding to H2S and Cys treatment. (A, B) Boxplot and density distribution diagram showing the gene expression features in 9 transcriptome samples of CK, H2S fumigated and Cys treated seedlings. FPKM, fragments per kilobase million. (C) Principal component analysis (PCA) of gene expression in 9 transcriptome samples of CK, H2S fumigated and Cys treated seedlings.
References
Alvarez, C., Calo, L., Romero, L. C., Garcia, I., Gotor, C. (2010). An O-acetylserine(thiol)lyase homolog with l-cysteine desulfhydrase activity regulates cysteine homeostasis in arabidopsis. Plant Physiol. 152, 656–669. doi: 10.1104/pp.109.147975
Alvarez, C., Garcia, I., Romero, L. C., Gotor, C. (2012). Mitochondrial sulfide detoxification requires a functional isoform O-acetylserine(thiol)lyase c in arabidopsis thaliana. Mol. Plant 5, 1217–1226. doi: 10.1093/mp/sss043
Aroca, A., Gotor, C., Romero, L. C. (2018). Hydrogen sulfide signaling in plants: emerging roles of protein persulfidation. Front. Plant Sci. 9, 1369. doi: 10.3389/fpls.2018.01369
Aroca, A., Zhang, J., Xie, Y., Romero, L. C., Gotor, C. (2021). Hydrogen sulfide signaling in plant adaptations to adverse conditions: molecular mechanisms. J. Exp. Bot. 72, 5893–5904. doi: 10.1093/jxb/erab239
Birke, H., Haas, F. H., De Kok, L. J., Balk, J., Wirtz, M., Hell, R. (2012). Cysteine biosynthesis, in concert with a novel mechanism, contributes to sulfide detoxification in mitochondria of arabidopsis thaliana. Biochem J. 445, 275–283. doi: 10.1042/BJ20120038
Chen, S., Jia, H., Wang, X., Shi, C., Wang, X., Ma, P., et al. (2020). Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Mol. Plant 13, 732–744. doi: 10.1016/j.molp.2020.01.004
Choudhary, K. K., Chaudhary, N. (2021). Hydrogen sulfide and reactive oxygen species crosstalk and acquisition of abiotic stress tolerance. Hydrogen Sulfide Plant Biol. 201–212. doi: 10.1016/B978-0-323-85862-5.00016-6
Corpas, F. J., Palma, J. M. (2020). H2S signaling in plants and applications in agriculture. J. Adv. Res. 24, 131–137. doi: 10.1016/j.jare.2020.03.011
Dong, N. Q., Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Fang, T., Cao, Z., Li, J., Shen, W., Huang, L. (2014). Auxin-induced hydrogen sulfide generation is involved in lateral root formation in tomato. Plant Physiol. Biochem. 76, 44–51. doi: 10.1016/j.plaphy.2013.12.024
Fang, H., Jing, T., Liu, Z., Zhang, L., Jin, Z., Pei, Y. (2014). Hydrogen sulfide interacts with calcium signaling to enhance the chromium tolerance in Setaria italica. Cell Calcium 56, 472–481. doi: 10.1016/j.ceca.2014.10.004
Fang, H., Liu, Z., Jin, Z., Zhang, L., Liu, D., Pei, Y. (2016). An emphasis of hydrogen sulfide-cysteine cycle on enhancing the tolerance to chromium stress in arabidopsis. Environ. Pollut. 213, 870–877. doi: 10.1016/j.envpol.2016.03.035
Fang, H., Liu, Z., Long, Y., Liang, Y., Jin, Z., Zhang, L., et al. (2017). The Ca2+/calmodulin2-binding transcription factor TGA3 elevates LCD expression and H2S production to bolster Cr6+ tolerance in arabidopsis. Plant J. 91, 1038–1050. doi: 10.1111/tpj.13627
Fang, H., Liu, R., Yu, Z., Shao, Y., Wu, G., Pei, Y. (2022). Gasotransmitter H2S accelerates seed germination via activating AOX mediated cyanide-resistant respiration pathway. Plant Physiol. Biochem. 190, 193–202. doi: 10.1016/j.plaphy.2022.09.003
Feng, J., Chen, L., Zuo, J. (2019). Protein s-nitrosylation in plants: current progresses and challenges. J. Integr. Plant Biol. 61, 1206–1223. doi: 10.1111/jipb.12780
Foyer, C. H., Hanke, G. (2022). ROS production and signalling in chloroplasts: cornerstones and evolving concepts. Plant J. 111, 642–661. doi: 10.1111/tpj.15856
Fu, Y., Tang, J., Yao, G. F., Huang, Z. Q., Li, Y. H., Han, Z., et al. (2018). Central role of adenosine 5'-phosphosulfate reductase in the control of plant hydrogen sulfide metabolism. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01404
Gaitonde, M. K. (1967). A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104, 627–633. doi: 10.1042/bj1040627
Garcia-Molina, A., Altmann, M., Alkofer, A., Epple, P. M., Dangl, J. L., Falter-Braun, P. (2017). LSU network hubs integrate abiotic and biotic stress responses via interaction with the superoxide dismutase FSD2. J. Exp. Bot. 68, 1185–1197. doi: 10.1093/jxb/erw498
Ge, Y., Hu, K. D., Wang, S. S., Hu, L. Y., Chen, X. Y., Li, Y. H., et al. (2017). Hydrogen sulfide alleviates postharvest ripening and senescence of banana by antagonizing the effect of ethylene. PloS One 12, , e180113. doi: 10.1371/journal.pone.0180113
Gotor, C., Garcia, I., Aroca, A., Laureano-Marin, A. M., Arenas-Alfonseca, L., Jurado-Flores, A., et al. (2019). Signaling by hydrogen sulfide and cyanide through post-translational modification. J. Exp. Bot. 70, 4251–4265. doi: 10.1093/jxb/erz225
Heeg, C., Kruse, C., Jost, R., Gutensohn, M., Ruppert, T., Wirtz, M., et al. (2008). Analysis of the arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20, 168–185. doi: 10.1105/tpc.107.056747
Jia, H., Chen, S., Liu, D., Liesche, J., Shi, C., Wang, J., et al. (2018). Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01517
Jin, Z., Pei, Y. (2015). Physiological implications of hydrogen sulfide in plants: pleasant exploration behind its unpleasant odour. Oxid. Med. Cell. Longev. 2015, 397502. doi: 10.1155/2015/397502
Jin, Z., Xue, S., Luo, Y., Tian, B., Fang, H., Li, H., et al. (2013). Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in arabidopsis. Plant Physiol. Biochem. 62, 41–46. doi: 10.1016/j.plaphy.2012.10.017
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Lefer, D. (2019). Redox pioneer: professor hideo kimura. Antioxid. Redox Signal. 30, 1699–1708. doi: 10.1089/ars.2018.7618
Leon, S., Touraine, B., Briat, J. F., Lobreaux, S. (2002). The AtNFS2 gene from arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem. J. 366, 557–564. doi: 10.1042/BJ20020322
Li, H., Chen, H., Chen, L., Wang, C. (2022). The role of hydrogen sulfide in plant roots during development and in response to abiotic stress. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23031024
Li, M., Kim, C. (2022). Chloroplast ROS and stress signaling. Plant Commun. 3, 100264. doi: 10.1016/j.xplc.2021.100264
Liang, W., Li, C., Liu, F., Jiang, H., Li, S., Sun, J., et al. (2009). The arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 19, 307–316. doi: 10.1038/cr.2008.317
Liu, D., Li, J., Li, Z., Pei, Y. (2020). Hydrogen sulfide inhibits ethylene-induced petiole abscission in tomato (Solanum lycopersicum l.). Hortic. Res. 7, 14. doi: 10.1038/s41438-019-0237-0
Liu, D., Lu, J., Li, H., Wang, J., Pei, Y. (2019). Characterization of the O-acetylserine(thiol)lyase gene family in Solanum lycopersicum l. Plant Mol. Biol. 99, 123–134. doi: 10.1007/s11103-018-0807-9
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L., Visser, R., Bovy, A. (2018). Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: a review. Front. Chem. 6. doi: 10.3389/fchem.2018.00052
Liu, H., Wang, J., Liu, J., Liu, T., Xue, S. (2021). Hydrogen sulfide (H2S) signaling in plant development and stress responses. aBIOTECH. 2, 32–63. doi: 10.1007/s42994-021-00035-4
Lu, C., Kavalier, A., Lukyanov, E., Gross, S. S. (2013). S-sulfhydration/desulfhydration and s-nitrosylation/denitrosylation: a common paradigm for gasotransmitter signaling by H2S and NO. Methods 62, 177–181. doi: 10.1016/j.ymeth.2013.05.020
Ma, X., Zhang, L., Pei, Z., Zhang, L., Liu, Z., Liu, D., et al. (2021). Hydrogen sulfide promotes flowering in heading Chinese cabbage by s-sulfhydration of BraFLCs. Hortic. Res. 8, 19. doi: 10.1038/s41438-020-00453-3
Mao, X., Cai, T., Olyarchuk, J. G., Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Papenbrock, J., Riemenschneider, A., Kamp, A., Schulz-Vogt, H. N., Schmidt, A. (2007). Characterization of cysteine-degrading and H2S-releasing enzymes of higher plants - from the field to the test tube and back. Plant Biol. (Stuttg) 9, 582–588. doi: 10.1055/s-2007-965424
Paul, B. D., Snyder, S. H. (2015). H2S: a novel gasotransmitter that signals by sulfhydration. Trends Biochem. Sci. 40, 687–700. doi: 10.1016/j.tibs.2015.08.007
Paulsen, C. E., Carroll, K. S. (2013). Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679. doi: 10.1021/cr300163e
Pilon-Smits, E. A., Garifullina, G. F., Abdel-Ghany, S., Kato, S., Mihara, H., Hale, K. L., et al. (2002). Characterization of a NifS-like chloroplast protein from arabidopsis. implications for its role in sulfur and selenium metabolism. Plant Physiol. 130, 1309–1318. doi: 10.1104/pp.102.010280
Pojer, E., Mattivi, F., Johnson, D., Stockley, C. S. (2013). The case for anthocyanin consumption to promote human health: a review. Compr. Rev. Food Sci. Food Saf. 12, 483–508. doi: 10.1111/1541-4337.12024
Qiao, Z., Jing, T., Jin, Z., Liang, Y., Zhang, L., Liu, Z., et al. (2016). CDPKs enhance cd tolerance through intensifying H2S signal in arabidopsis thaliana. Plant Soil 398, 99–110. doi: 10.1007/s11104-015-2643-x
Qiao, Z., Jing, T., Liu, Z., Zhang, L., Jin, Z., Liu, D., et al. (2015). H2S acting as a downstream signaling molecule of SA regulates cd tolerance in arabidopsis. Plant Soil 393, 137–146. doi: 10.1007/s11104-015-2475-8
Rabino, I., Mancinelli, A. L. (1986). Light, temperature, and anthocyanin production. Plant Physiol. 81, 922–924. doi: 10.1104/pp.81.3.922
Romero, L. C., Aroca, M. A., Laureano-Marin, A. M., Moreno, I., Garcia, I., Gotor, C. (2014). Cysteine and cysteine-related signaling pathways in arabidopsis thaliana. Mol. Plant 7, 264–276. doi: 10.1093/mp/sst168
Rydz, L., Wrobel, M., Jurkowska, H. (2021). Sulfur administration in fe-s cluster homeostasis. Antioxidants (Basel) 10. doi: 10.3390/antiox10111738
Sehar, Z., Gautam, H., Iqbal, N., Alvi, A. F., Jahan, B., Fatma, M., et al. (2022). The functional interplay between ethylene, hydrogen sulfide, and sulfur in plant heat stress tolerance. Biomolecules 12. doi: 10.3390/biom12050678
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome. Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shen, J., Zhang, J., Zhou, M., Zhou, H., Cui, B., Gotor, C., et al. (2020). Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell 32, 1000–1017. doi: 10.1105/tpc.19.00826
Sirko, A., Wawrzynska, A., Rodriguez, M. C., Sektas, P. (2014). The family of LSU-like proteins. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00774
Sun, H. J., Wu, Z. Y., Nie, X. W., Bian, J. S. (2021). Role of hydrogen sulfide and polysulfides in neurological diseases: focus on protein s-persulfidation. Curr. Neuropharmacol. 19, 868–884. doi: 10.2174/1570159X18666200905143550
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., Hell, R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Valivand, M., Amooaghaie, R., Ahadi, A. (2019). Seed priming with H2S and Ca2+ trigger signal memory that induces cross-adaptation against nickel stress in zucchini seedlings. Plant Physiol. Biochem. 143, 286–298. doi: 10.1016/j.plaphy.2019.09.016
Wang, R. (2002). Two's company, three's a crowd: can H2S be the third endogenous gaseous transmitter? FASEB J. 16, 1792–1798. doi: 10.1096/fj.02-0211hyp
Wang, R. (2012). Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol. Rev. 92, 791–896. doi: 10.1152/physrev.00017.2011
Wang, R. (2014). Gasotransmitters: growing pains and joys. Trends Biochem. Sci. 39, 227–232. doi: 10.1016/j.tibs.2014.03.003
Wirtz, M., Droux, M., Hell, R. (2004). O-Acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in arabidopsis thaliana. J. Exp. Bot. 55, 1785–1798. doi: 10.1093/jxb/erh201
Wirtz, M., Hell, R. (2006). Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J. Plant Physiol. 163, 273–286. doi: 10.1016/j.jplph.2005.11.013
Yamaguchi, Y., Nakamura, T., Kusano, T., Sano, H. (2000). Three arabidopsis genes encoding proteins with differential activities for cysteine synthase and beta-cyanoalanine synthase. Plant Cell Physiol. 41, 465–476. doi: 10.1093/pcp/41.4.465
Yang, Z., Wang, X., Feng, J., Zhu, S. (2022). Biological functions of hydrogen sulfide in plants. Int. J. Mol. Sci. 23. doi: 10.3390/ijms232315107
Yang, G., Wu, L., Jiang, B., Yang, W., Qi, J., Cao, K., et al. (2008). H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science 322, 587–590. doi: 10.1126/science.1162667
Yang, H., Yang, S., Li, Y., Hua, J. (2007). The arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol. 145, 135–146. doi: 10.1104/pp.107.100800
Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, J., Zhou, M., Zhou, H., Zhao, D., Gotor, C., Romero, L. C., et al. (2021). Hydrogen sulfide, a signaling molecule in plant stress responses. J. Integr. Plant Biol. 63, 146–160. doi: 10.1111/jipb.13022
Zheng, L., Dean, D. R. (1994). Catalytic formation of a nitrogenase iron-sulfur cluster. J. Biol. Chem. 269, 18723–18726. doi: 10.1016/S0021-9258(17)32225-1
Keywords: hydrogen sulfide, gasotransmitter, cysteine, transcriptome sequencing, Arabidopsis
Citation: Fang H, Yu Z, Xing K, Zhou L, Shao Y, Zhang X, Pei Y and Zhang L (2023) Transcriptomic analysis reveals the functions of H2S as a gasotransmitter independently of Cys in Arabidopsis. Front. Plant Sci. 14:1184991. doi: 10.3389/fpls.2023.1184991
Received: 13 March 2023; Accepted: 04 May 2023;
Published: 02 June 2023.
Edited by:
Judy Brusslan, California State University, Long Beach, United StatesReviewed by:
Jérémy Couturier, Université De Lorraine, FranceCecilia Gotor, Spanish National Research Council (CSIC), Spain
Copyright © 2023 Fang, Yu, Xing, Zhou, Shao, Zhang, Pei and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huihui Fang, ZmFuZ2hoQHphZnUuZWR1LmNu; Lu Zhang, MjAxNDAwNDZAemFmdS5lZHUuY24=
†These authors have contributed equally to this work
‡ORCID: Huihui Fang, orcid.org/0000-0002-1774-2436
 Huihui Fang
Huihui Fang Zhenyuan Yu1†
Zhenyuan Yu1† Lingyi Zhou
Lingyi Zhou Yuke Shao
Yuke Shao Yanxi Pei
Yanxi Pei