
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 June 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1184058
This article is part of the Research Topic Climate Change and Stress Mitigation Strategy in Plants View all 9 articles
 Zhou Zhang1†
Zhou Zhang1† Sunil S. Gangurde2†
Sunil S. Gangurde2† Songbin Chen1
Songbin Chen1 Rushil Ramesh Mandlik3
Rushil Ramesh Mandlik3 Haiyan Liu4
Haiyan Liu4 Rupesh Deshmukh3
Rupesh Deshmukh3 Jialing Xu1
Jialing Xu1 Zhongkang Wu1
Zhongkang Wu1 Yanbin Hong4*
Yanbin Hong4* Yin Li1*
Yin Li1*The 14-3-3 protein is a kind of evolutionary ubiquitous protein family highly conserved in eukaryotes. Initially, 14-3-3 proteins were reported in mammalian nervous tissues, but in the last decade, their role in various metabolic pathways in plants established the importance of 14-3-3 proteins. In the present study, a total of 22 14-3-3 genes, also called general regulatory factors (GRF), were identified in the peanut (Arachis hypogaea) genome, out of which 12 belonged to the ε group, whereas 10 of them belonged to the non- ε-group. Tissue-specific expression of identified 14-3-3 genes were studied using transcriptome analysis. The peanut AhGRFi gene was cloned and transformed into Arabidopsis thaliana. The investigation of subcellular localization indicated that AhGRFi is localized in the cytoplasm. Overexpression of the AhGRFi gene in transgenic Arabidopsis showed that under exogenous 1-naphthaleneacetic acid (NAA) treatment, root growth inhibition in transgenic plants was enhanced. Further analysis indicated that the expression of auxin-responsive genes IAA3, IAA7, IAA17, and SAUR-AC1 was upregulated and GH3.2 and GH3.3 were downregulated in transgenic plants, but the expression of GH3.2, GH3.3, and SAUR-AC1 showed opposite trends of change under NAA treatment. These results suggest that AhGRFi may be involved in auxin signaling during seedling root development. An in-depth study of the molecular mechanism of this process remains to be further explored.
In multicellular organisms, the exchange of information via signaling within the cell and between cells can stimulate the recognition of biotic and abiotic factors so that the plant system can respond to stresses and regulate growth and development. The growth and genetic potential of a plant are affected by a variety of environmental factors such as light, temperature, humidity, moisture, lodging, wind velocity, and the internal plant atmosphere, which includes plant hormones, signaling molecules, chlorophyll content, and stomatal movement (Fulgosi et al., 2002). There are small molecular proteins called 14-3-3 proteins in plant cells that play an important role in cell signal transduction in response to biotic and abiotic stresses and regulation of plant growth and development (Grienenberger and Fletcher, 2015; Hajibarat et al., 2022; Huang et al., 2022). In addition, it also participates in material transport and metabolism during various metabolic processes and plant hormone signaling. These signaling proteins interact with plant defense protein factors, and kinase activation also plays an important role in the inhibition of cell division and differentiation and oil transport proteins. The 14-3-3 proteins, which are phosphorylation-dependent or phosphorylation-independent binding protein, also stimulate gibberellin transcription protein (GASA), etc. (Herzog et al., 1995; Hughes and Dunn, 1996; Boutrot et al., 2008; Marshall et al., 2011). 14-3-3 proteins act as hubs for a cellular web, transducing and integrating diverse hormone signals (Camoni et al., 2018). Moreover, plant 14-3-3 proteins are also considered to be indirectly involved in auxin signaling through the protein kinase pathway (Takahashi et al., 2012).
The 14-3-3 proteins were initially cataloged by Moore and Perez (1967) during an extensive study of bovine brain proteins. The name 14-3-3 proteins originated from phase chromatography using diethylaminoethyl cellulose (DEAE) fraction elution in the position of fragments and starch migration rate in gel electrophoresis. Further, studies on 14-3-3 proteins demonstrated that these proteins are highly conserved in eukaryotes and form heterologous or homologous dimeric molecules with monomeric protein molecules. The 14-3-3 proteins were extensively studied in Arabidopsis and are also known as GF14s or general regulatory factors (GRFs) (Rooney and Ferl, 1995; Wu et al., 1997). A varying number of 14-3-3 genes are identified in the genomes of different plant species, for example, 10 in Arabidopsis and 18 in soybean, which is a polyploid plant species (Wu et al., 1997; Li and Dhaubhadel, 2011).
The 14-3-3 interacts with target proteins through a specific phosphorylation site, resulting in protein–protein interactions and regulating signal transduction (Denison et al., 2011; Paul et al., 2012). In plants, more than 300 target proteins were identified to interact with the 14-3-3 protein (Oecking and Jaspert, 2009). These target proteins are involved in a variety of important physiological processes during abiotic and biotic stress tolerance, activity and stability confirmation of proteins, protein localization, and transport and regulation of cell division (Yang et al., 2014). There are several reports on the expression and functional diversity of H+-ATPase membrane proteins. The 14-3-3 protein is involved in the regulation of the activity of H+-ATPase (Duby et al., 2008). Nitrate reductase is a key nitrate metabolic enzyme. The activity of nitrate reductase is dependent on the process of phosphorylation. In the dark, 14-3-3 protein inhibits the activity of nitrate reductase and reduces the production and metabolism of toxic nitrate (Bachmann et al., 1996). Sucrose phosphate synthase is a key enzyme in carbon metabolism in plants; the 14-3-3 can inhibit the activity of sucrose phosphate synthase. On the other hand, with the inhibition of 14-3-3 activity, the corresponding starch and sucrose content increases (Moorhead et al., 1999; Toroser et al., 2000). In barley, 14-3-3 protein interacts with transcription factors such as AREB, ABF/ABI5, which are involved in abscisic acid (ABA) metabolism. The VP1 gene mutations in ABI5 cannot be combined without 14-3-3 proteins; studies reported that binding VP1 and ABI5 needs 14-3-3 as a bridge to form a complex binding to the ABA-responsive element (Himmelbach et al., 2003). 14-3-3 negatively regulates BRZ1 and BRZ2/BES1 transcription in Arabidopsis thaliana and rice (Bai et al., 2007; Gampala et al., 2007). The 14-3-3 protein is also involved in the signaling of brassinosteroids (Wang et al., 2011) and gibberellins (Ishida et al., 2004), using a photographic approach (Taoka et al., 2011). Plant 14-3-3 proteins and target protein binding during the stress response stimulate signaling mechanisms in transgenic plants, which include the non-biological responses to the ROS reaction, salt stress tolerance, osmotic stress tolerance, and high and low-temperature stress responses (Kidou et al., 1993; Chen et al., 1994; Roberts et al., 2002; Elmayan et al., 2007). In addition, the biological response of plant resistance genes (R-genes) in response to pathogen attack (Yang et al., 2009). Peanuts are an important oilseed and legume crop. Most of the research efforts in peanut are carried out to improve the resistance against devastating biotic stresses such as stem rot (Dodia et al., 2019), bacterial wilt (Luo et al., 2019) and late leaf spot (Gangurde et al., 2021), bacterial wilt (Luo et al., 2019). Around 90% of peanuts are cultivated in semi-arid zones of the world where drought is an important yield-limiting factor (Gangurde et al., 2019; Pandey et al., 2021). As discussed above, several reports have revealed that the 14-3-3 gene involves the regulation of growth and development and the stimulation of responses to biotic and abiotic stresses in plants. Therefore, it is necessary to perform structural and functional analysis of the peanut 14-3-3 gene so that it can be used as a molecular tool to improve peanut varieties resistant to biotic and abiotic stresses. In this context, in the present study, 22 14-3-3 genes, named AhGRFs, were identified through a genome-wide identification approach. Among the identified genes, AhGRFi was cloned and transformed into A. thaliana to analyze its function. The sequence similarity of the peanut 14-3-3 protein was investigated, and the expression pattern of the 14-3-3 genes in different plant tissues of peanut during different developmental stages was studied. The expression of the 14-3-3 gene was validated using quantitative real-time quantitative PCR.
The genome information for peanut (Arachis hypogaea v1.0) was obtained from the phytozome database (Goodstein et al., 2012) at https://phytozome-next.jgi.doe.gov/. A local protein database was created in BioEdit (Hall et al., 2011) using protein sequences obtained from the phytozome database (Ahypogaea_530_v1.0). Previously identified 14-3-3 proteins from rice (Yashvardhini et al., 2018), soybean (Li and Dhaubhadel, 2011), and Arabidopsis (Wu et al., 1997) were used as a query for BLASTp against the local protein database for peanut. To identify significant candidates, the minimum cut-off value was kept to E-value 10−5 and a bit score >100. The top-hit candidates with the highest bit score were selected, and redundant ones were filtered. The non-redundant candidates were used for further analysis. A genetic map was generated by the online tool MG2C (http://mg2c.iask.in/mg2c_v2.0). The identified AhGRF proteins were further confirmed through HMMER (https://www.ebi.ac.uk/Tools/hmmer/) and the Pfam database (https://pfam.xfam.org/).
The molecular weight and isoelectric focusing point for the identified proteins were estimated by using Expasy tools (https://web.expasy.org/compute_pi/). The TBtools (https://bio.tools/tbtools) were used to predict the gene structure. The subcellular localization of the identified protein was predicted using CELLO (http://cello.life.nctu.edu.tw/) and Wolfpsort (https://wolfpsort.hgc.jp/) servers. The transmembrane domains were predicted using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and SOSUI Server (http://harrier.nagahama-i-bio.ac.jp/sosui/). The motifs were identified by the MEME tool (http://meme-suite.org/tools/meme).
Multiple sequence alignments for peanut AhGRF proteins were performed in the CLUSTAL W function in MEGA X. The phylogenetic tree was generated based on 14-3-3 protein sequence alignment via the MEGA X program using the maximum likelihood method. The robustness of the tree was confirmed by using a 1,000-bootstrap value.
The RNA-seq data for expression analysis of 22 AhGRF genes were downloaded from the NCBI Sequence Read Archive (Bio Project IDs: PRJNA243319, PRJNA773958, PRJNA687108, PRJNA638812, PRJNA555172, PRJNA525247, PRJNA511663, PRJNA517600, PRJNA503795, PRJNA498570, and PRJNA291488). The CLC workbench (https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/) was used to process the raw reads. The reads were mapped onto the peanut reference genome sequence. The reads per kilobase of the transcript per million (RPKM) mapped read values were calculated, and a heatmap for the same was generated using the R package ‘pheatmap’ (Kolde and Kolde, 2018). Hierarchical clustering was utilized to cluster genes.
The cultivated peanut (A. hypogaea L.) variety of Shanyou 523 seeds was provided by the Guangdong Academy of Agricultural Sciences, Guangzhou, China. A. thaliana (Columbia-0 Ecotype) was used for peanut AhGRFi gene transformation and functional characterization. The seeds of transgenic A. thaliana were sterilized with 70% (v/v) ethyl alcohol for 2 min and then transferred to a 10% (v/v) sodium hypochlorite solution for 10 min. Further, the seeds were rinsed with sterile water five times on a clean bench. A. thaliana seeds were placed on one-half MS solid culture medium (pH 5.8, 1 mM KOH) after surface sterilization (Murashige and Skoog, 1962) and incubated at 4°C temperature for 3 days. They were further grown at 22°C with a 16 h/8 h photoperiod and 6,000 lx light intensity (lux). After two weeks, seedlings were transplanted to the soil for growth and maturation under the same growth conditions (peat soil:vermiculite ratio 3:1).
The full-length cDNA sequence of the AhGRFi gene, which is identical to that of LOC112798274 14-3-3-like protein A in A. hypogaea with Gene ID: 112798274 in the gene database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/gene/112798274), was amplified using gene-specific primers (Table S1) from the peanut cotyledons cDNA library (Li et al., 2005). The AhGRFi amplicon was extracted and purified from agarose gel using a quick gel extraction kit (QIAGEN) as per the manufacturer’s instructions. The purified AhGRFi gene cDNA sequence was cloned into the pMD18-T vector for sequencing.
The yeast two-hybrid assay was performed according to the manufacturer’s instructions for the Matchmaker GAL4-based two-hybrid system 3 (Clontech) (Zhou et al., 2013). The full-length AhGRFi cDNA was subcloned into the pGADT7 (AD) and pGBKT7 (BD) vectors. All primer sequences used for construction are listed in Table S1. Constructs were transformed into yeast strain AH109 by the lithium acetate method, and yeast cells were grown on a minimal medium/-Leu-Trp at 30°C for 3 days. Transformed colonies were plated onto a minimal medium/-Leu/-Trp/-His/-Ade containing 20 µg/ml 5-bromo-4-chloro-3-indolyl-a-D-galactopyranoside to test for possible interactions.
Total RNA isolation was carried out from the roots, stems, and leaves of the 12-day-old seedling and the 20, 30-, 40-, 50-, and 70-day-old embryos after pegging into soil (DAP) using the TRIZOl reagent (Invitrogen). The cDNA was synthesized from isolated RNA using the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa).
Expression patterns of the AhGRFi gene in peanut tissues were investigated using qRT-PCR, according to Li et al. (2009). The qRT-PCR was performed on the LightCycler480 (RoChe) using the SYBR Green real-time PCR Master Mix (Toyobo) with gene-specific primers and 18S rRNA (Guo et al., 2012) as the internal control gene. Primer sequences used for this analysis are listed in Table S1.
The coding region without a stop codon for the AhGRFi gene from peanut was cloned into the intermediate vector pA7-YFP, and the AhGRFi : YFP cassette was further subcloned into the pBI121 vector, replacing the GUS gene. Primer sequences used for the construction are listed in Table S1. The recombinant vector was transformed into Agrobacterium tumefaciens strain EHA105. Tobacco (Nicotiana tabacum var. Xanthi) leaf epidermal cells were infiltrated with Agrobacterium carrying a recombinant vector with 35Spro : AhGRFi-YFP. Simultaneously, in the control experiment, the tobacco epidermal cells were infiltrated with Agrobacterium (35Spro : YFP). The infiltrated plants were maintained at under 22°C with 16 h of photoperiod, and the light intensity was 6,000 lx. The YFP signal was subsequently detected by the LSM 710 duo laser scanning confocal microscope (ZEISS, Germany) with a filter set of 514 nm for excitation and 527 nm for emission after 24 h of agroinfiltration.
For the heterologous expression, the coding sequence of AhGRFi was cloned into the pBI121 vector under the control of the CaMV 35S promoter. The recombinant plasmid was transformed into the Agrobacterium tumefaciens EHA105 strain. The recombinant plasmid was transformed into A. thaliana by the floral dip method (Clough and Bent, 1998). Transgenic seeds were selected on a one-half MS medium containing 50 mg/L kanamycin. According to the segregation ratio of the kanamycin selection marker, homozygous transgenic lines for Arabidopsis were identified and further confirmed by leaf PCR (Kasajima et al., 2004) using gene-specific primers of AhGRFi.
Seeds of transgenic Arabidopsis carrying the AhGRFi gene and wild-type Arabidopsis were surface sterilized and placed on one-half MS medium containing 1 uM Naphthalene acetic acid (NAA) and incubated at 4°C for 48 h under normal environmental conditions at 22°C at 16 h light/8 h dark, vertical 6,000 lx for vertical training for 2 weeks. Later, the root length was recorded for both control and transgenic Arabidopsis to study the effect of the AhGRFi gene on root growth. Three biological repeats were carried out for each treatment; 15 plants were used each time. Statistical analyses were performed using the Student’s t-test. All data were expressed as means ± standard error (SE). p <0.05 and p <0.01 are considered statistically significant.
Total RNA was isolated from Arabidopsis seedlings cultured on one-half MS medium for 2 weeks using the RNAprep Pure Plant Kit (TIANGEN). The cDNA was synthesized from isolated RNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). Real-time PCR analysis of overexpression in transgenic lines was performed with specific primers for AhGRFi and with Actin2 and UBQ10 as internal control genes. Auxin-responsive gene expression in transgenic Arabidopsis was analyzed with gene-specific primers and the Actin2 gene as an internal control (Yang et al., 2014). Primer sequences are shown in Table S1.
Based on sequence homology of previously characterized 14-3-3 protein-encoding genes in rice, soybean, and Arabidopsis a total of 22 14-3-3 genes were identified in the peanut genome (Table 1). A genetic map was generated by MG2C, showing the overall distribution of AhGRF gene members and their corresponding positions on chromosomes (Figure S1). HMMER and Pfam confirmed the presence of 14-3-3 domains in the identified gene-encoded proteins. The identified genes encode proteins ranging from 196 to 300 amino acids. Their molecular weight ranged from 22,103 to 34,357 Da, while their isoelectric point ranged from 4.4 to 4.8. The TMHMM and SOSUI servers confirmed the absence of transmembrane domains in peanut 14-3-3 proteins. The genes AhGRFi and AhGRFj, AhGRFo, and AhGRFp encoded similar proteins with identical amino acid sequences.
The phylogenetic tree generated using the maximum likelihood method in the MEGA X program gives insight into the evolution pattern of peanut AhGRF genes. The phylogenetic tree was constructed using the AhGRF protein sequences from peanut, rice, soybean, and Arabidopsis (Figure 1). As per previous studies, the 14-3-3 proteins were divided into ε and non-ε groups. The AhGRFu, AhGRFv, AhGRFq, AhGRFo, AhGRFp, AhGRFr, AhGRFs, AhGRFk, AhGRFl, AhGRFm, AhGRFn, and AhGRFt constituted the ε group, whereas AhGRFd, AhGRFi, AhGRFj, AhGRFc, AhGRFb, AhGRFa, AhGRFf, AhGRFe, AhGRFh, and AhGRFg belonged to the non-ε group. Both the ε and non-ε groups were divided into four branches, which were subsequently divided into further subbranches. The gene pairs AhGRFi and AhGRFj and AhGRFo and AhGRFp were closely related and encoded for similar proteins with identical amino acid sequences (Figure 2, Table 2). The similarity between the respective gene sequence pairs was found to be 100%.
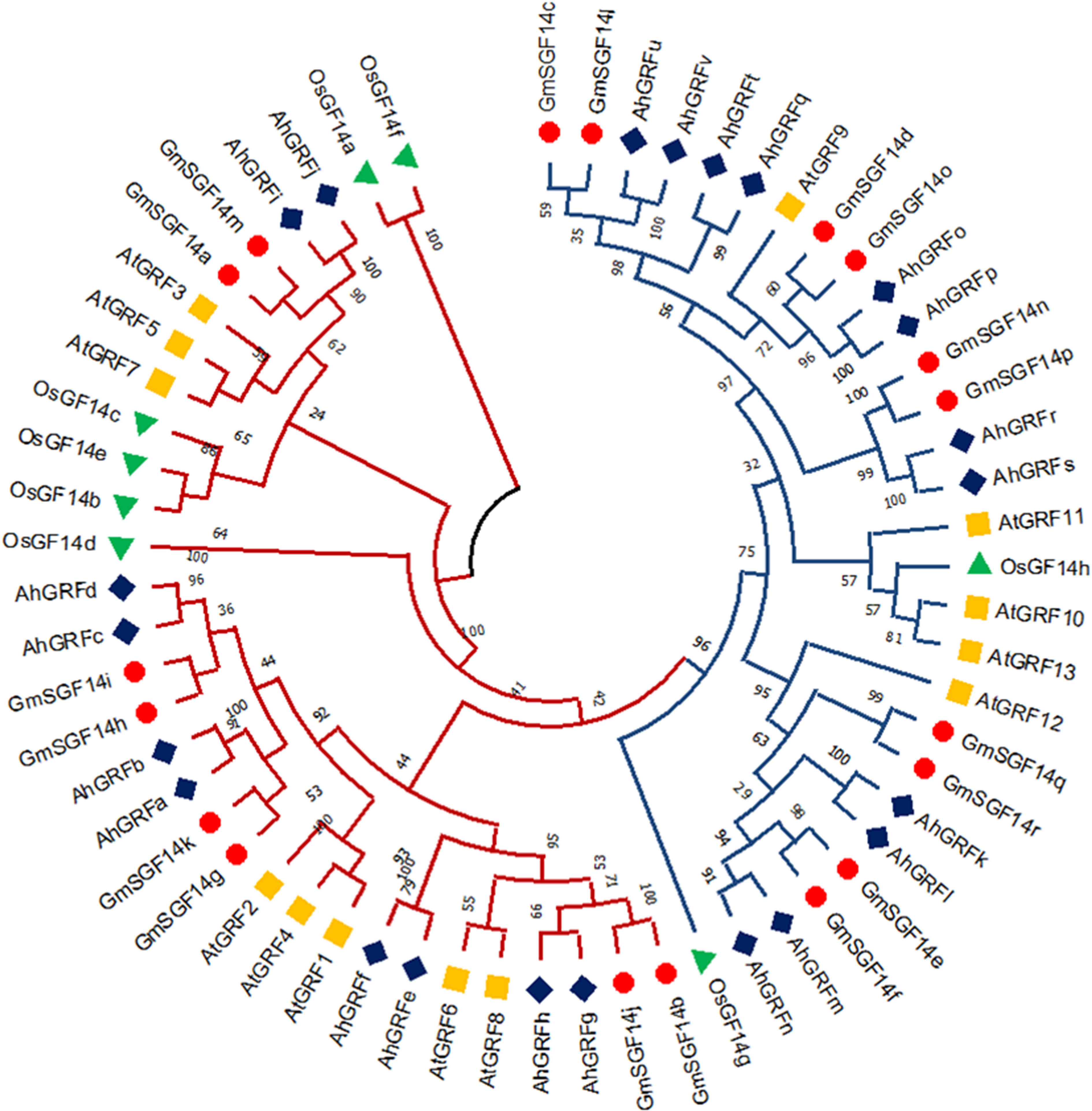
Figure 1 Phylogenetic tree showing the evolutionary relationship among AhGRF proteins identified in peanut, soybean, Arabidopsis, and rice.
The number of introns varied from two to 10 in AhGRF genes identified in peanuts. Some of the gene pairs displayed a similar number of introns and exons, which were also similar in length. AhGRFi and AhGRFj, AhGRFo, and AhGRFp genes encoded for identical proteins also have similar gene structures with the same number of introns and exons (Figure 3).
The expression of the identified AhGRF genes was determined using transcriptomic data from various developmental stages and stress conditions. The genes AhGRFe and AhGRFf showed expression in almost all tissues except reproductive tissues. The expressions of AhGRFk, AhGRFb, AhGRFm, and AhGRFl were found to be specific to the pistil and flower, respectively. The AhGRFh was found to be mainly expressed in seed and pod tissues, while the AhGRFr was primarily expressed in peg tissues. In comparison, AhGRFp expression was restricted to the testa, roots, and some seed tissues, whereas AhGRFi expression was restricted to leaf, root, and some pod tissues. The AhGRFj is primarily expressed in the testa, roots, pod, and lateral tissues. The AhGRFs, AhGRFu, and AhGRFt were primarily expressed in the root and some of the pod tissues only. Aluminum stress did not show any significant change in root gene expression. Iron–cadmium treatment showed expression of AhGRFg, AhGRFc, AhGRFd, and AhGRFs in the roots. A water deficit increased the expression of AhGRFi and AhGRFk in leaf tissue. While drought stress at the seedling stage increased the expression of AhGRFe, AhGRFf, and AhGRFt genes while reducing the expression of AhGRFr, AhGRFc, and AhGRFd (Figure 4).
In a previous analysis of a peanut cotyledon cDNA library (Li et al., 2005), a 14-3-3-like protein gene sequence was identified. Our further cloning and analysis found that it was an AhGRFi gene identical to that of LOC112798274 14-3-3-like protein A with Gene ID: 112798274 in the NCBI gene database. In this study, a 944-bp sequence containing the 783-bp full CDS of the AhGRFi gene was amplified from the corresponding peanut cDNA library using gene-specific primers by PCR (Figure S2). The resulting AhGRFi sequence was cloned by the T-A cloning methodology into the pMD18-T vector and sequenced for confirmation of the presence of the target gene.
To understand the role of the AhGRFi gene in peanut growth and development, RNA was extracted from different peanut developmental stages and tissues, and the expression pattern of the AhGRFi gene was studied using real-time fluorescent quantitative PCR (Figure 5). The AhGRFi showed the highest expression level in peanut seeds and seedling roots. In different developmental stages of peanut seeds, the expression of AhGRFi was relatively stable, and the expression level at 10 d and 20 d after the pegging stage was slightly lower as compared to the later stages.
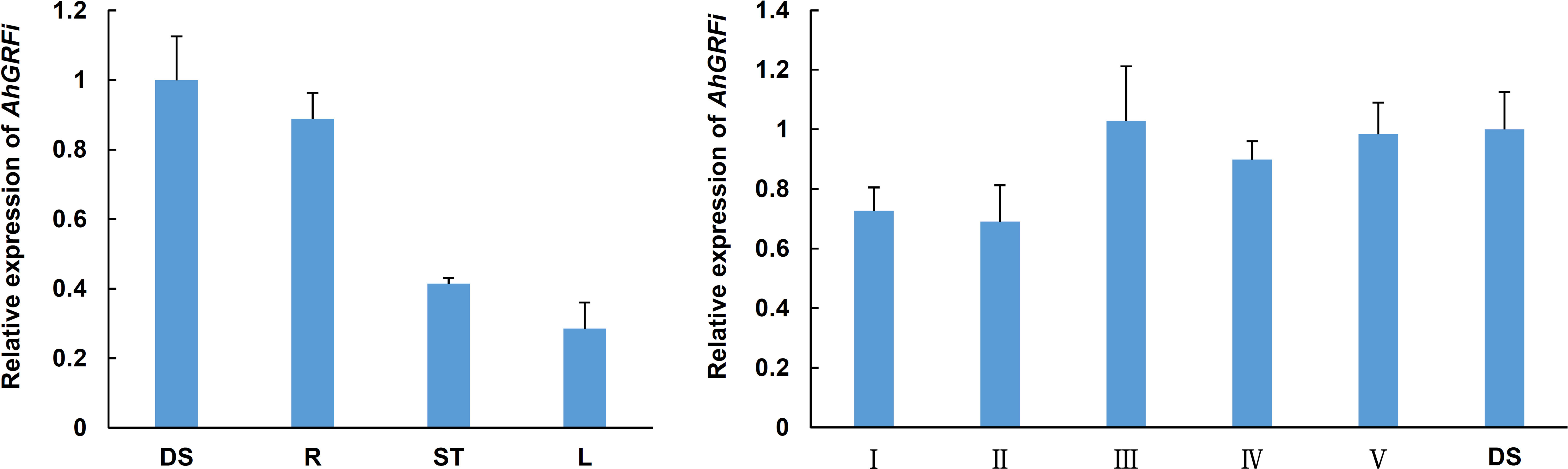
Figure 5 Analysis of the expression pattern of the AhGRFi gene. Error bars represent the means ± SE of three replications. Different organizations of peanut RNA: DS, dry seeds; R, stem; L, leaf; Different development periods of peanut RNA: I, 20 DAP; II, 30 DAP; III, 40 DAP; IV, 50 DAP; V, 70 DAP; Ah18S RNA reference gene. Error bars represent the means ± SE of three replications.
In eukaryotic cells, 14-3-3 proteins are usually bound to two target proteins to function as homologous or heterologous dimers. To understand the interaction mechanism of the peanut AhGRFi protein, the AhGRFi gene was cloned into pGADT7 and pGBKT7 vectors and transformed into yeast-competent cells (AH109). The yeast could grow in the SD/-Trp-Leu medium, but the colony in the SD/-Trp-Leu medium did not grow in the SD/-Trp-Leu-His-Ade medium (Figure 6). In vitro experimental results showed that the peanut protein AhGRFi did not interact with itself, suggesting that the AhGRFi protein does not function as a homodimer.
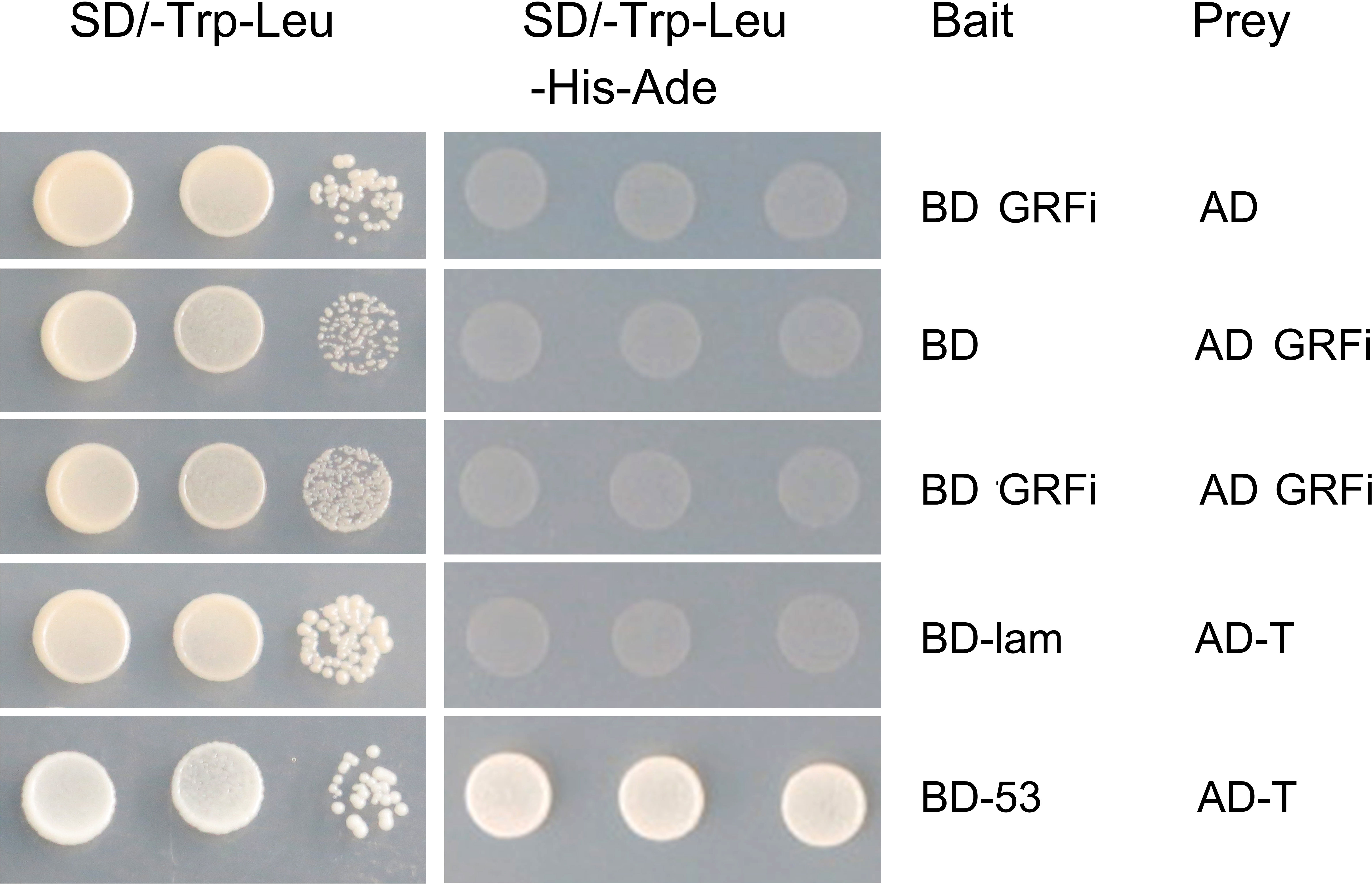
Figure 6 AhGRFi protein interaction analyses in vitro. Interaction was determined by growth assays on medium lacking adenine. Dilutions (1.10−1, and 10−2 for control and without dilution for lacking adenine) of saturated cultures were spotted onto the plates.
To investigate the subcellular localization of AhGRFi, YFP (yellow fluorescent protein)-tagged AhGRFi, driven by the CaMV 35S promoter, was agroinfiltrated into tobacco epidermal cells to express the fusion protein. The AhGRFi : YFP fusion proteins were examined with a Zeiss confocal laser scanning microscope (at 514 nm for the excitation spectra and at 527 nm for the emission spectra). We observed that the fluorescence signal was mainly distributed in the cytoplasm, as shown in Figure 7. The coincidence of yellow fluorescent protein signals in the nucleus was confirmed by DAPI staining. Therefore, we concluded that the peanut AhGRFi protein is mainly distributed in the cytoplasm and not in the nucleus.
To study the biological function of peanut AhGRFi, it was heterologously expressed in A. thaliana via Agrobacterium-mediated transformation. Transgenic lines were confirmed by PCR (Figure S3). qRT-PCR was performed to detect the overexpression levels in transgenic lines, and three high-expression lines were used for further study (Figure S4). Under normal culture conditions, when transgenic A. thaliana with overexpression of the AhGRFi gene was compared with a wild-type control, there was no difference in Arabidopsis growth and seed development in both overexpression and wild-type Arabidopsis lines (Figure 8A). In the presence of exogenous NAA treatment, two-week-old overexpression lines showed significant enhancement of root growth inhibition as compared to wild-type (Figures 8B–D). The root growth of overexpression and wild-type Arabidopsis lines was estimated by root length and fresh root weight. The statistical analysis showed that the root length and fresh weight of overexpression Arabidopsis lines were significantly inhibited as compared to wild-type Arabidopsis, as shown in Figures 8E, F. Therefore, we conclude that the root growth of transgenic Arabidopsis carrying the peanut AhGRFi gene is sensitive to NAA.
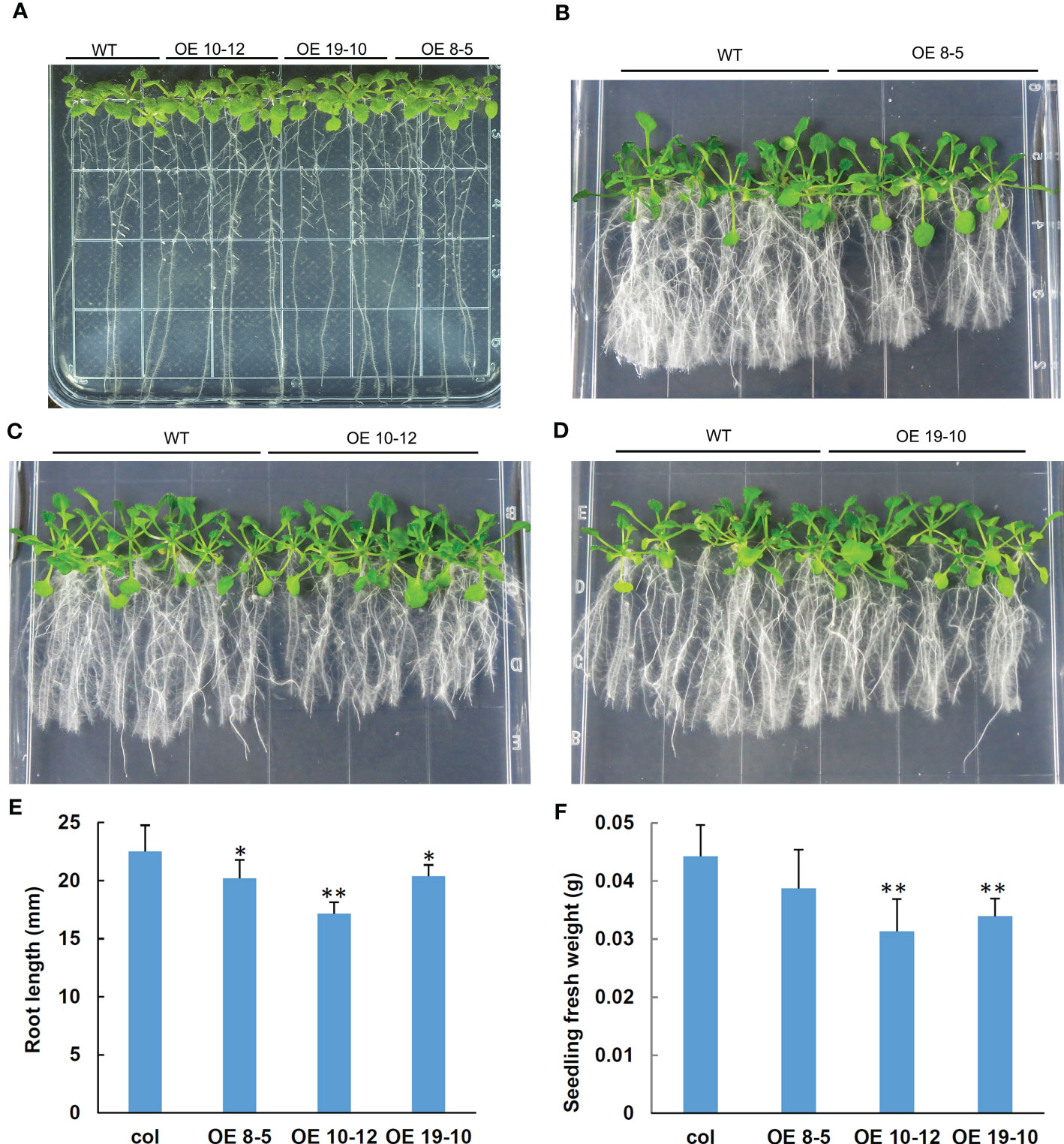
Figure 8 Effect of NAA on root lengths of transgenic Arabidopsis overexpressing AhGRFi. (A) Phenotypes of transgenic Arabidopsis in one-half MS medium under NAA treatment (A) control group; (B–D) treatment group). Measurement of the Arabidopsis root length (E) and fresh weight of the Arabidopsis seedlings (F) Error bars represent the means (n = 15) ± SE of three replications. * and ** above the error bars indicate statistical significance at P <0.05 and P <0.01, respectively.
The effect of AhGRFi overexpression on auxin-responsive genes such as IAA3, IAA7, IAA17, GH2.2, GH2.3, and SAUR-AC1 was further investigated by qRT-PCR. As shown in Figure 9, the expression of auxin-responsive genes such as IAA3, IAA7, IAA17, and SAUR-AC1 was upregulated in AhGRFi-overexpressing Arabidopsis, while GH3.2 and GH3.3 were downregulated. Under NAA treatment, the expression levels of IAA3, IAA7, and IAA17 were still relatively high, but the expression of GH3.2 and GH3.3 showed opposite trends of change, whereas the upregulation of SAUR-AC1 expression is slightly reduced compared to the wild-type plant.
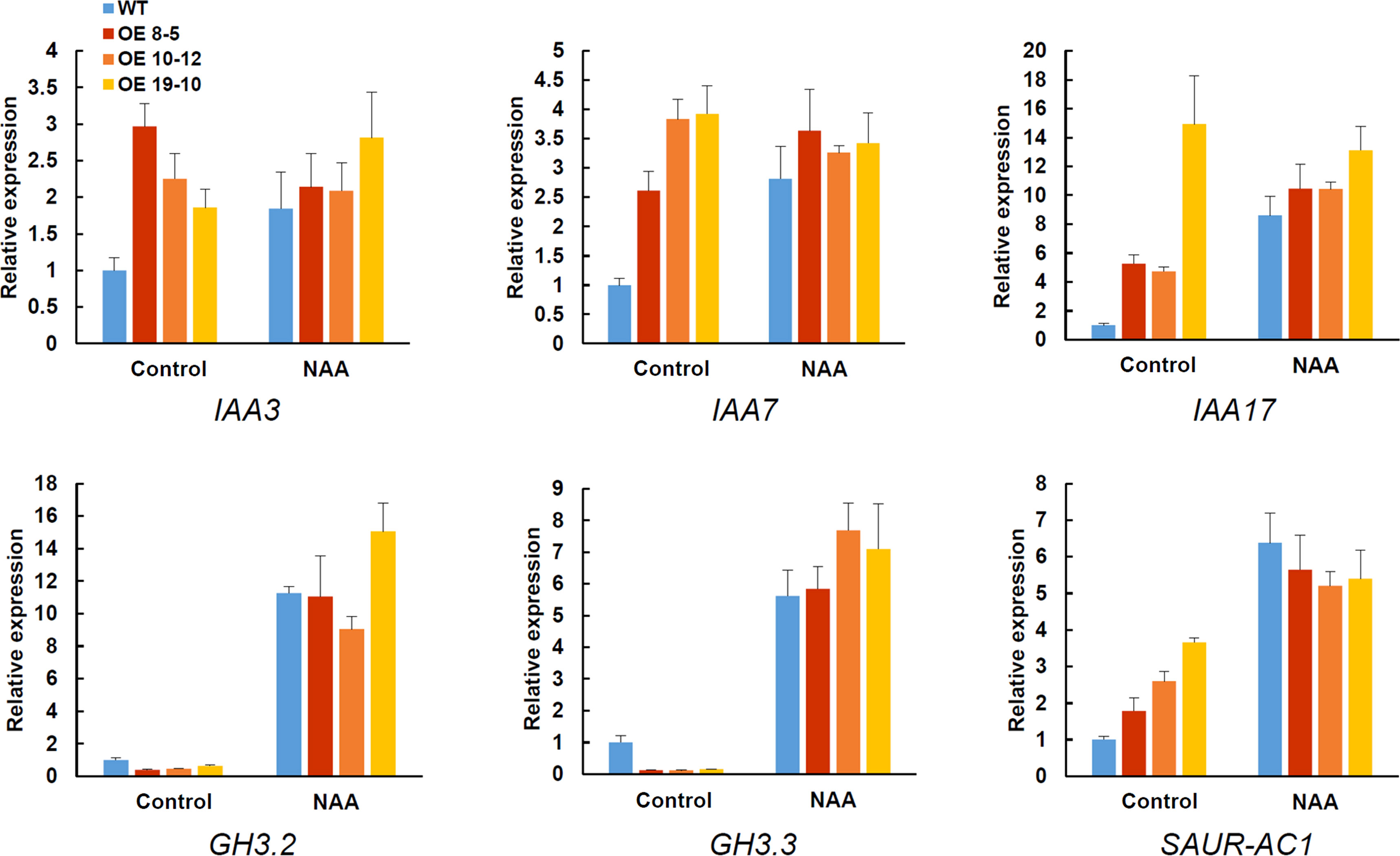
Figure 9 Real-time PCR analysis of auxin-responsive genes in transgenic Arabidopsis overexpressing AhGRFi. The relative expression values were normalized to the Arabidopsis Actin2 gene. Error bars represent the means ± SE of three replications.
The AhGRFi–YFP fusion protein transiently expressed in tobacco epidermal cells showed a yellow fluorescent signal distribution in the cytoplasm but not in the nucleus. 14-3-3 protein is a kind of widespread protein in eukaryotic cells called “scaffold protein,” because the protein monomer state usually has no biological function. Through the formation of homologous or heterologous dimers, 14-3-3 protein is involved in the physiological regulation of cellular processes, including phosphorylation and signal transduction processes, by binding to phosphorylated proteins (Boer et al., 2013). There are two cases about the localization of 14-3-3 protein in cells: one is that the 14-3-3 protein is in the intracellular protein itself, and another is that the 14-3-3 protein binds to other proteins with the aid of the target protein to change the location of the information itself, which is involved in the regulation of complex biological processes (Paul et al., 2005). Bioinformatics structure prediction of peanut 14-3-3 proteins revealed that the protein has no signal peptide or transmembrane domain; thus, protein translation without modifications may be directly involved in the regulation of cell activities. The fusion protein of Arabidopsis 14-3-3 and fluorescent protein, expression, and localization of 14-3-3 proteins in cells were identified in tobacco protoplasts and onion cells, and it was found that different members of the 14-3-3 family of A. thaliana and their locations are different, which are located in the cytoplasm and nucleus and presumably play a different role in cell signal transduction processes (Sehnke et al., 2002). The rice 14-3-3 proteins were localized in the nucleus (Chen et al., 2006). Therefore, different members of the family of 14-3-3 proteins in the same species have different functions; the 14-3-3 proteins of different species have high homology, but their functions are also likely to differ very far. The positioning mechanism of 14-3-3 protein, in the aspect of plant hormone signal transduction and adversity stress response, is not only able to regulate the activity of the target protein and be functional but is also indirectly involved in gene transcriptional regulation (Himmelbach et al., 2003; Ishida et al., 2004; Wang et al., 2011).
Previous studies have found that 14-3-3 protein binds to target proteins via phosphorylation recognition sequence specificity, to participate in a complex regulatory network of cell biology. In plants, 14-3-3 genes are involved in plant growth and development processes including biotic and abiotic stress responses, plant hormone signal transduction, metabolism process regulation, cell growth and division, etc. (Yaffe and Elia, 2001). For instance, in this study, expression analysis showed very high expressions of AhGRFc and AhGRFd under aluminum stress. The expression pattern of the cotton 14-3-3 gene found that the cotton 14-3-3 proteins are involved in cotton root growth and drought stress response. In the case of corn, the 14-3-3 gene expression of corn ZmGF14-6 as well as tomato and rape 14-3-3 gene expression is affected by stress (Sun et al., 2014). Gene expression pattern showed that AhGRFs had the highest expression level in peanut seeds and seedling root; in different developmental stages of peanut seeds, the expression of AhGRFs was relatively stable, and the expression level at 20 d and 30 d after pegging stage was slightly lower than in the later stages (Figure 5). The reason behind this is that 14-3-3 protein is involved in the process of plant hormone signal transduction, such as the ABA and GA signaling pathways, which can affect the growth and development of plants (Sehnke et al., 2002). It is reported that overexpression of the Ta14-3-3 gene in Arabidopsis can inhibit root and seedling growth (Li et al., 2013). In a study of wild soybean GsGF14o, overexpression causes changes in Arabidopsis morphology, including small pores and short hair, under drought stress in transgenic A. thaliana, in addition to slower root growth (Sun et al., 2014).
Plant 14-3-3 proteins have been found to be associated with plant stress response (Hajibarat et al., 2022; Huang et al., 2022) and involved in a network of interactions that finely regulate signaling and homeostasis of multiple hormones such as abscisic acid (ABA) (Schoonheim et al., 2009), gibberellins (GAs) (Ito et al., 2014), ethylene (Catalá et al., 2014), and auxin (Takahashi et al., 2012; Camoni et al., 2018). In our previous studies, stress treatments such as drought, salt, high and low temperatures, and osmotic testing were applied to AhGRFi-overexpressing Arabidopsis lines, but we did not find significant phenotypic differences (data not shown). ABA, 1-aminocyclopropane-1-carboxylic acid (ACC), NAA, GA3, and paclobutrazol (PAC) treatments had been applied to transgenic plants, but except for NAA, there were no significant changes in other treatments (data not shown). The experimental results demonstrated that under NAA treatment, AhGRFi overexpression significantly inhibited root growth in Arabidopsis seedlings. Further analysis indicated that the expression of auxin-responsive genes in transgenic Arabidopsis behaved differently under NAA treatment. The early auxin-responsive genes GH3.2 and GH3.3 were downregulated under normal conditions but upregulated under treatment. GH3 genes inactivate auxin to maintain the dynamic balance of auxin in plants (Lin et al., 2022), and upregulation of GH3 genes enhances plant response to low concentrations of auxin (Chapman and Estelle, 2009). The combined effect of multiple genes may lead to the inhibition of root growth under exogenous NAA treatment, but it did not affect the growth of roots under normal culture conditions. The result suggests that this 14-3-3 protein in peanuts may be involved in the regulation of root growth by auxin signaling.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject accession numbers: PRJNA243319, PRJNA773958, PRJNA687108, PRJNA638812, PRJNA555172, PRJNA525247, PRJNA511663, PRJNA517600, PRJNA503795, PRJNA498570, and PRJNA291488.
YL and YH conceived this study and designed the experiment. ZZ, SG, SC, HL, JX, ZW, and YL performed the experiments and data analysis. SG, RM, and RD performed bioinformatics analysis. ZZ and SG wrote the manuscript. YL, SG, and YH completed the final revision. All authors contributed to the article and approved the submitted version.
This research was supported by grants from the National Natural Science Foundation of China (No. 31171625), the Science and Technology Planning Project of Guangdong Province, China (Nos. 2020A1414040005 and 2021A0505030047), the Research and Development projects in key areas of Guangdong Province (No. 2022B0202060004), and the Guangdong Modern Agro-industry Technology Research System (2022KJ136-02).
We thank Professor Shangzhi Huang of School of Life Sciences at Sun Yat-sen University for providing us with the peanut cotyledon cDNA library. We are also grateful to all the laboratory members for their technical advice and helpful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1184058/full#supplementary-material
Supplementary Figure 1 | Genetic map of AhGRF gene family.
Supplementary Figure 2 | Cloning of AhGRFi. M:DL2000 Marker, 1:cDNA of AhGRFi
Supplementary Figure 3 | Detection of AhGRFi gene by leaf PCR of Aradopsis plants. M: DL2000 Marker, lane 1: vector control, lane 2: wild-type control, lanes 3-24: AhGRFi transgenic samples.
Supplementary Figure 4 | qRT-PCR analysis of AhGRFi gene expression in Arabidopsis transgenic lines. The relative expression values were normalized to the Arabidopsis Actin2 and UBQ10 gene. Error bars represent the means ± SE of three replication.
Bachmann, M., Huber, J. L., Liao, P. C., Gage, D. A., Huber, S. C. (1996). The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett. 387, 127–131. doi: 10.1016/0014-5793(96)00478-4
Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong., K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. 104, 13839–13844. doi: 10.1073/pnas.0706386104
Boer, A., van Kleeff, P. J., Gao, J. (2013). Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma 250 (2), 425–440. doi: 10.1007/s00709-012-0437-z
Boutrot, F., Chantret, N., Gautier, M. F. (2008). Genome-wide analysis of the rice and arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9, 86. doi: 10.1186/1471-2164-9-86
Camoni, L., Visconti, S., Aducci, P., Marra, M. (2018). 14-3-3 proteins in plant hormone signaling: doing several things at once. Front. Plant Sci. 13 9. doi: 10.3389/fpls.2018.00297
Catalá, R., López-Cobollo, R., Mar Castellano, M., Angosto, T., Alonso, J. M., Ecker, J. R., et al. (2014). The arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342. doi: 10.1105/tpc.114.127605
Chapman, E. J., Estelle, M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43, 265–285. doi: 10.1146/annurev-genet-102108-134148
Chen, Z., Fu, H., Liu, D., Chang, P. F., Narasimhan, M., Ferl, R., et al. (1994). A NaCl-regulated plant gene encoding a brain protein homology that activates ADP ribosyltransferase and inhibits protein kinase c. Plant J. 6 (5), 729–740. doi: 10.1046/j.1365-313x.1994.6050729.x
Chen, F., Li, Q., Sun, L., He, Z. (2006). The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 13 (2), 53–63. doi: 10.1093/dnares/dsl001
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Denison, F. C., Paul, A.-L., Zupanska, A. K., Ferl, R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727. doi: 10.1016/j.semcdb.2011.08.006
Dodia, S. M., Joshi, B., Gangurde, S. S., Thirumalaisamy, P. P., Mishra, G. P., Dayama Narandrakumar, D. (2019). Genotyping-by-sequencing based genetic mapping reveals large number of epistatic interactions for stem rot resistance in groundnut. Theor. Appl. Genet. 132, 4, 1001–1016. doi: 10.1007/s00122-018-3255-7
Duby, G., Poreba, W., Piotrowiak, D., Bobik, K., Derua, R., Waelken, S. E., et al. (2008). Activation of plant plasma membrane h+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the h+-ATPase c-terminal region. J. Biol. Chem. 284, 4213–4221. doi: 10.1074/jbc.M807311200
Elmayan, T., Fromentin, J., Riondet, C., Alcaraz, G., Blein, J. P., Simon-Plas, F. (2007). Regulation of reactive oxygen species production by a 14-3-3 protein in elicited tobacco cells. Plant Cell Environ. 30 (6), 722–732. doi: 10.1111/j.1365-3040.2007.01660.x
Fulgosi, H., Soll, J., de Faria Maraschin, S., Korthout, H. A., Wang, M., Testerink, C. (2002). 14-3-3 proteins and plant development. Plant Mol. Biol. 50 (6), 1019–1029. doi: 10.1023/a:1021295604109
Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in arabidopsis. Dev. Cell. 13 (2), 177–189. doi: 10.1016/j.devcel.2007.06.009
Gangurde, S. S., Kumar, R., Pandey, A. K., Burow, M., Laza, H. E., Nayak, S. N., et al. (2019). Climate-smart groundnuts for achieving high productivity and improved quality: current status, challenges, and opportunities. In Genomic Designing of Climate-Smart Oilseed Crops. Springer International Publishing. pp 133–172. doi: 10.1007/978-3-319-93536-2_3
Gangurde, S. S., Nayak, S. N., Joshi, P., Purohit, S., Sudini, H. K., Chitikineni, A., et al. (2021). Comparative transcriptome analysis identified candidate genes for late leaf spot resistance and cause of defoliation in groundnut. Int. J. Mol. Sci. 22 (9), 4491. doi: 10.3390/ijms22094491
Guo, D., Liang, J., Qiao, Y., Yan, Y., Li, L., Dai, Y., et al. (2012). Involvement of G1-to-S transition and AhAUX-dependent auxin transport in abscisic acid-induced inhibition of lateral root primodia initiation in Arachis hypogaea L. J Plant Physiol. 169 (11), 1102–1111. doi: 10.1016/j.jplph.2012.03.014
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40 (D1), D1178–D1186. doi: 10.1093/nar/gkr944
Grienenberger, E., Fletcher, J. C. (2015). Polypeptide signaling molecules in plant development. Curr. Opin. Plant Biol. 23, 8–14. doi: 10.1016/j.pbi.2014.09.013
Hajibarat, Z., Saidi, A., Zohreh Hajibarat, Z. (2022). Genome-wide identification of 14-3-3 gene family and characterization of their expression in developmental stages of solanum tuberosum under multiple biotic and abiotic stress conditions. Funct. Integr. Genomics 22 (6), 1377–1390. doi: 10.1007/s10142-022-00895-z
Hall, T., Biosciences, I., Carlsbad, C. (2011). BioEdit: an important software for molecular biology. GERF Bull. Biosci. 2 (1), 60–61.
Herzog, M., Dorne, A. M., Grellet, F. (1995). GASA, a gibberellin-regulated gene family from arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol. Biol. 27 (4), 743–752. doi: 10.1007/BF00020227
Himmelbach, A., Yang, Y., Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 (5), 470–479. doi: 10.1016/s1369-5266(03)00090-6
Huang, Y., Wang, W., Yu, H., Peng, J., Hu, Z., Chen, L. (2022). The role of 14-3-3 proteins in plant growth and response to abiotic stress. Plant Cell Rep. 41 (4), 833–852. doi: 10.1007/s00299-021-02803-4
Hughes, M. A., Dunn, M. A. (1996). The molecular biology of plant acclimation to low temperature. J. Exp. Bot. 47 (296), 291–305. doi: 10.1093/JXB%2F47.3.291
Ishida, S., Fukazawa, J., Yuasa, T., Takahashi, Y. (2004). Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell. 16, 2641–2651. doi: 10.1105/tpc.104.024604
Ito, T., Nakata, M., Fukazawa, J., Ishida, S., Takahashi, Y. (2014). Scaffold function of Ca2+-dependent protein kinase: tobacco Ca2+-dependent protein kinase1 transfers 14-3-3 to the substrate repression of shoot growth after phosphorylation. Plant Physiol. 165, 1737–1750. doi: 10.1104/pp.114.236448
Kasajima, I., Ide, Y., Ohkama-Ohtsu, N., Yoneyama, T., Fujiwara, T. (2004). A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol. Biol. Rep. 22, 49–52. doi: 10.1007/BF02773348
Kidou, S., Umeda, M., Kato, A., Uchimiya, H. (1993). Isolation and characterization of a rice cDNA similar to the bovine brain-specific 14-3-3 protein gene. Plant Mol. Biol. 21 (1), 191–194. doi: 10.1007/BF00039631
Li, X., Dhaubhadel, S. (2011). Soybean 14-3-3 gene family: identification and molecular characterization. Planta 233 (3), 569–582. doi: 10.1007/s00425-010-1315-6
Li, C., Wu, K., Fu, G., Li, Y., Zhong, Y., Lin, X., et al (2009). Regulation of oleosin expression in developing peanut (Arachis hypogaea L.) embryos through nucleosome loss and histone modifications. J. Exp. Bot. 60 (15), 4371–4382. doi: 10.1093/jxb/erp275
Li, J., Song, S. S., Zhao, Y. S., Guo, W. W., Guo, G. H., Peng, H. R., et al. (2013). Wheat 14-3-3 protein conferring growth retardation in arabidopsis. J. Integr. Agric. 12 (2), 209–217. doi: 10.1016/S2095-3119(13)60220-8
Li, H.-G., Wang, L., Zhang, Y.-S., Lin, X.-D., Liao, B., Yan, Y.-S., et al. (2005). Cloning and sequencing of the gene ahy-b encoding a subunit of peanut conarachin. Plant Sci. 168, 1387–1392. doi: 10.1016/j.plantsci.2004.09.028
Lin, Q., Jiaxin Gong, J., Zhang, Z., Meng, Z., Wang, J., Wang, S., et al. (2022). The arabidopsis thaliana trehalose-6-phosphate phosphatase gene AtTPPI regulates primary root growth and lateral root elongation. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1088278
Luo, H., Pandey, M. K., Khan, A. W., Wu, B., Guo, J., Ren, X. (2019). Next-generation sequencing identified genomic region and diagnostic markers for resistance to bacterial wilt on chromosome B02 in peanut (Arachis hypogaea l.). Plant Biotechnol. J. 17 (12), 2356–2369. doi: 10.1111/pbi.13153
Marshall, E., Costa, L. M., Gutierrez-Marcos, J. (2011). Cysteine-rich peptides (CRPs) mediate diverse aspects of cell–cell communication in plant reproduction and development. J. Exp. Bot. 62, 1677–1686. doi: 10.1016/j.pbi.2014.09.013
Moore, B. W., Perez, V. J. (1967). “Specific acidic proteins of the nervous system,” in Physiological and biochemical aspects of nervous integration. Ed. Carlson, F. D. (Woods Hole, MA: Prentice-Hall, Inc, The Marine Biological Laboratory), 343–359.
Moorhead, G., Douglas, P., Cotelle, V., Harthill, J., Morrice, N., Meek, S., et al. (1999). Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J. 18, 1–12. doi: 10.1046/j.1365-313x.1999.00417.x
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Oecking, C., Jaspert, N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12, 760–765. doi: 10.1016/j.pbi.2009.08.003
Pandey, M. K., Gangurde, S. S., Sharma, V., Pattanashetti, S. K., Naidu, G. K., Faye, I. (2021). Improved genetic map identified major QTLs for drought tolerance-and iron deficiency tolerance-related traits in groundnut. Genes 12, 37. doi: 10.3390/genes12010037
Paul, A. L., Denison, F. C., Schultz, E. R., Zupanska, A. K., Ferl, R. J. (2012). 14-3-3 phosphoprotein interaction networks-does isoform diversity present functional interaction specification? Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00190
Paul, A. L., Sehnke, P. C., Ferl, R. J. (2005). Isoform-specific subcellular localization among 14-3-3 proteins in arabidopsis seems to be driven by client interactions. Mol. Biol. Cell. 16 (4), 1735–1743. doi: 10.1091/mbc.e04-09-0839
Roberts, M. R., Salinas, J., Collinge, D. B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50 (6), 1031–1039. doi: 10.1023/a:1021261614491
Rooney, M. F., Ferl, R. J. (1995). Sequences of three arabidopsis general regulatory factor genes encoding GF14 (14-3-3) proteins. Plant Physiol. 107 (1), 283–284. doi: 10.1104/pp.107.1.283
Schoonheim, P. J., Costa Pereira, D. D., de Boer, A. H. (2009). Dual role for 14-3-3 proteins and ABF transcription factors in gibberellic acid and abscisic acid signalling in barley (Hordeum vulgare) aleurone cells. Plant Cell Environ. 32, 439–447. doi: 10.1111/j.1365-3040.2009.01932.x
Sehnke, P. C., DeLille, J. M., Ferl, R. J. (2002). Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell. 14 (Suppl), S339–S354. doi: 10.1105/tpc.010430
Sun, X., Luo, X., Sun, M., Chen, C., Ding, X., Wang, X., et al. (2014). A glycine soja 14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 55 (1), 99–118. doi: 10.1093/pcp/pct161
Takahashi, K., Hayashi, K., Kinoshita, T. (2012). Auxin activates the plasma membrane h+-ATPase by phosphorylation during hypocotyl elongation in arabidopsis. Plant Physiol. 159, 632–641. doi: 10.1104/pp.112.196428
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476 (7360), 332–335. doi: 10.1038/nature10272
Toroser, D., Athwal, G. S., Huber, S. C. (2000). Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 435, 110–114. doi: 10.1016/s0014-5793(98)01048-5
Wang, H., Yang, C., Zhang, C., Wang, N., Lu, D., Wang, J., et al. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell. 21, 825–834. doi: 10.1016/j.devcel.2011.08.018
Wu, K., Rooney, M. F., Ferl, R. J. (1997). The arabidopsis 14-3-3 multigene family. Plant Physiol. 114 (4), 1421–1431. doi: 10.1104/pp.114.4.1421
Yaffe, M. B., Elia, A. E. (2001). Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13 (2), 131–138. doi: 10.1016/s0955-0674(00)00189-7
Yang, F., Song, Y., Yang, H., Liu, Z., Zhu, G., Yang, Y. (2014). An auxin-responsive endogenous peptide regulates root development in arabidopsis. J. Integr. Plant Biol. 56 (7), 635–647. doi: 10.1111/jipb.12178
Yang, X., Wang, W., Coleman, M., Orgil, U., Feng, J., Ma, X., et al. (2009). Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 60 (3), 539–550. doi: 10.1111/j.1365-313X.2009.03978.x
Yashvardhini, N., Bhattacharya, S., Chaudhuri, S., Sengupta, D. N. (2018). Molecular characterization of the 14-3-3 gene family in rice and its expression studies under abiotic stress. Planta 247 (1), 229–253. doi: 10.1007/s00425-017-2779-4
Keywords: Arachis hypogaea, 14-3-3, GRF, transgenic Arabidopsis thaliana, root, auxin signaling
Citation: Zhang Z, Gangurde SS, Chen S, Mandlik RR, Liu H, Deshmukh R, Xu J, Wu Z, Hong Y and Li Y (2023) Overexpression of peanut (Arachis hypogaea L.) AhGRFi gene enhanced root growth inhibition under exogenous NAA treatment in Arabidopsis thaliana. Front. Plant Sci. 14:1184058. doi: 10.3389/fpls.2023.1184058
Received: 10 March 2023; Accepted: 24 May 2023;
Published: 21 June 2023.
Edited by:
Anirudh Kumar, Indira Gandhi National Tribal University, IndiaReviewed by:
Prasad Gandham, Louisiana State University, United StatesCopyright © 2023 Zhang, Gangurde, Chen, Mandlik, Liu, Deshmukh, Xu, Wu, Hong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanbin Hong, aG9uZ3lhbmJpbkBnZGFhcy5jbg==; Yin Li, bGl5aW5AbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.