
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 June 2023
Sec. Plant Systematics and Evolution
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1183653
 Ri-Hong Jiang1†
Ri-Hong Jiang1† Si-Qi Liang2,3,4†
Si-Qi Liang2,3,4† Fei Wu3,5,6
Fei Wu3,5,6 Li-Ming Tang7
Li-Ming Tang7 Bo Qin1
Bo Qin1 Ying-Ying Chen1
Ying-Ying Chen1 Yao-Heng Huang1
Yao-Heng Huang1 Kai-Xiang Li1*
Kai-Xiang Li1* Xian-Chun Zhang2,3*
Xian-Chun Zhang2,3*Germplasm resources are the source of herbal medicine production. The cultivation of superior germplasm resources helps to resolve the conflict between long-term population persistence and growing market demand by consistently producing materials with high quality. The fern species Cibotium barometz is the original plant of cibotii rhizoma (“Gouji”), a traditional Chinese medicine used in the therapy of pain, weakness, and numbness in the lower extremities. Long-history medicinal use has caused serious wild population decline in China. Without sufficient understanding of the species and lineage diversity of Cibotium, it is difficult to propose a targeted conservation scheme at present, let alone select high-quality germplasm resources. In order to fill such a knowledge gap, this study sampled C. barometz and relative species throughout their distribution in China, performed genome skimming to obtain plastome data, and conducted phylogenomic analyses. We constructed a well-supported plastome phylogeny of Chinese Cibotium, which showed that three species with significant genetic differences are distributed in China, namely C. barometz, C. cumingii, and C. sino-burmaense sp. nov., a cryptic species endemic to NW Yunnan and adjacent regions of NE Myanmar. Moreover, our results revealed two differentiated lineages of C. barometz distributed on the east and west sides of a classic phylogeographic boundary that was probably shaped by monsoons and landforms. We also evaluated the resolution of nine traditional barcode loci and designed five new DNA barcodes based on the plastome sequence that can distinguish all these species and lineages of Chinese Cibotium accurately. These novel findings on a genetic basis will guide conservation planners and medicinal plant breeders to build systematic conservation plans and exploit the germplasm resources of Cibotium in China.
Traditional Chinese herbal medicine plays an indispensable role in the treatment of multiple diseases in China and other developing countries (Newman et al., 2008). Apart from the traditional usage, many medicinal plants, such as Artemisia annua L. (artemisinin, Tu, 2016), Huperzia javanica (Sw.) C. Y. Yang (Huperzine A, Zangara, 2003; it was mistakenly named as Huperzia serrata (Thunb.) Trevis in many studies, Chen et al., 2021), and Panax notoginseng (Burk.) F. H. Chen (Notoginseng triterpenes, Huang et al., 2021), are also the source of modern pharmaceuticals generating increasing attention. Although China harbors abundant medicinal plant diversity, the original species of many commonly used herbal medicines are facing the risk of population decline and even extinction under growing demand (Chen et al., 2016). Germplasm resources are a major determinate of medicine production (Ma and Xiao, 1998; Huang et al., 2008; Zhang and Jiang, 2021; Meng et al., 2023). Cultivation of specific high-quality germplasm resources will not only resolve the present conflict between conservation and exploitation but also ensure a steady production of high-quality medicines (Ma and Xiao, 1998; Chen et al., 2016). Therefore, clarifying genetic background and diversity is the basic and crucial step in achieving sustainable utilization of medicinal plants and provides implications for the collection, identification, evaluation, and conservation of germplasm resources (Schoen and Brown, 1993; Ma and Xiao, 1998; Yu et al., 2013; Khoury et al., 2022).
Cibotium barometz (L.) J. Sm. is the original species (Figures 1A–I) of traditional medicine, cibotii rhizoma (“Gouji”, Figure 1A), the processed rhizome of which can be used in the therapy of pain, weakness, and numbness of the lower extremities (Chinese Pharmacopoeia Commission, 2020). Phytochemical research has shown that the extract of its rhizomes is rich in active compounds such as pterosins, terpenes, steroids, flavonoids, glucosides, phenolic acids, and pyrones (Xu et al., 2012). Bioactivity experiments supported the efficacy, including the treatment of osteoporosis and osteoarthritis, antioxidant and antimicrobial activities, as well as abirritation (Ju et al., 2005; Cuong et al., 2009; Zhao et al., 2011; Li et al., 2014; Fu et al., 2017; Heng et al., 2020; Sun, 2021). Pot cultures and crafts of this species are also popular on the market because of their large, elegant evergreen fronds and stump-like rhizomes covered with long, soft, golden hairs resembling gold-hair dogs (Figures 1B–D). Medicinal and ornamental values have resulted in the destructive plunder of abundant natural resources by C. barometz in China. Investigation has shown that uncontrolled collection and habitat deconstruction are major threats to its population survival (Zhang et al., 2002).

Figure 1 Morphology of Cibotium plants from China. (A) Dry sliced rhizomes of C. barometz, “Gouji.” (B) Rhizome, stipes, and young fronds of C. barometz covered with golden filiform hairs. (C) Rhizomes of C. barometz sold as medicinal herbs at a village fair. (D) Fronds of C. barometz. (E) Veins, hairs, and unopened sori on the abaxial surface of the pinnae of C. barometz. (F) Opened sori of C. barometz. (G, H) Cross section and basal part of the stipe of C. barometz. (I, J) Basal part of pinna in C. barometz and C. cumingii showing the difference of basal pinnules on the basiscopic side. Photographs by R.-H. Jiang (A, B, D, F), X.-C. Zhang (C, I, J), and Q.-K. Ding (E, G, H).
C. barometz is listed in Appendix II of CITES (Zhang et al., 2002; https://cites.org/eng/app/appendices.php), and the genus Cibotium is listed in the Grade II Category of the List of National Key Protected Wild Plants of China (State Forestry and Grassland Administration and the Ministry of Agriculture and Rural Affairs, P. R. China, 2021). Although the Chinese government has attached great importance to this genus, researchers are unable to specify which species or populations are key units awaiting conservation grounded in present knowledge. Such a phenomenon could lead to the waste of protective efforts and affect the maximization of medical value. Previous studies have shown that the genus Cibotium (Cibotiaceae, a member of the tree fern clade) comprises ca. 9–12 species distributed in tropical and subtropical regions of Asia, Central America, and the Hawaiian Islands (Holttum, 1963; Palmer, 1994; Korall et al., 2006; Smith et al., 2006; Geiger et al., 2013), three Asian members of which form a monophyletic clade (Geiger et al., 2013). Two species, C. barometz and C. cumingii Kunze, were recognized from China (Zhang and Nishida, 2013). The former is widespread in southern China and extends to northeastern India and Malaysia, while the latter is only known from Taiwan Island in China, as well as the Ryukyu and Philippine Islands (Holttum, 1963; Zhang and Nishida, 2013). However, the geographical pattern of genetic diversity and differentiation of C. barometz has not been explored throughout its wide distribution, let alone talking about specific populations awaiting conservation and the variation of medicinal values among different regions accurately. In addition, though individuals can grow up to three meters tall, merely a handful of morphological traits can be applied to the discrimination of C. barometz and C. cumingii (Holttum, 1963; Zhang and Nishida, 2013). Therefore, whether recognized phenotypes could adequately reflect the diversity of this genus also remains to be verified.
In previous studies, several chloroplast DNA (cpDNA) fragments have been applied to the phylogenetic construction of the tree fern clade, including Cibotium (Korall et al., 2006; Geiger et al., 2013). However, the informative variation sites provided by these loci are insufficient to illuminate the relationship within Chinese Cibotium. With advantages including low requirements for material quality, low costs, and rich variable sites, the chloroplast genome (plastome) has been widely utilized for phylogenetic reconstruction at different levels as well as species delimitation of closely related species in recent years (e.g., Hammer et al., 2019; Wei and Zhang, 2020; Ji et al., 2021; Du et al., 2022; Xi et al., 2022; Zhang et al., 2022; Yang et al., 2023). Therefore, analyzing genetic variations of plastomes obtained from multiple samples representing the major distribution will help us clarify species diversity and the phylogeographical pattern of Chinese Cibotium, which are crucial to suggesting reasonable conservation units and germplasm resources suitable for sustainable medicine production. Furthermore, plastome data can not only be applied to develop traditional DNA barcodes but also used as a single genetic marker, namely ultra-barcodes (Nock et al., 2011; Kress et al., 2015; Hollingsworth et al., 2016), which largely benefits the identification and evaluation of medical plant products, especially partial organs and tissues lacking diagnostic phenotypic features (Park et al., 2021; Qin et al., 2022; Wang et al., 2022; Wei et al., 2022).
In this study, we performed genome skimming and assembled complete plastomes of representative samples of C. barometz and relatives throughout the distribution range in China and adjacent areas. We aimed to 1) compare structure and composition variations of plastomes among Chinese Cibotium species; 2) propose a phylogeny-based species delimitation; 3) investigate the geographical pattern of variation of C. barometz based on plastome data; and 4) suggest candidate barcodes for specific species and lineage identification of Chinese Cibotium. We believed that our findings would benefit the conservation and breeding of this important medicinal plant taxon and provide insights into the systematics and evolution of ferns.
Frond tissues of 25 Cibotium individuals were collected for genome skimming sequencing throughout the distribution range of China and adjacent regions (Table 1; Figure 2). Most accessions were fresh fronds dried using silica gel and preserved at 4 °C, except for five samples obtained from specimens deposited in the herbarium PE (Table 1). Based on the presence or lack of basal pinnules on the basiscopic side of pinnae on voucher specimens (Figures 1I, J, Zhang and Nishida, 2013), 25 samples were sorted into 23 individuals of C. barometz and two individuals of C. cumingii preliminarily.
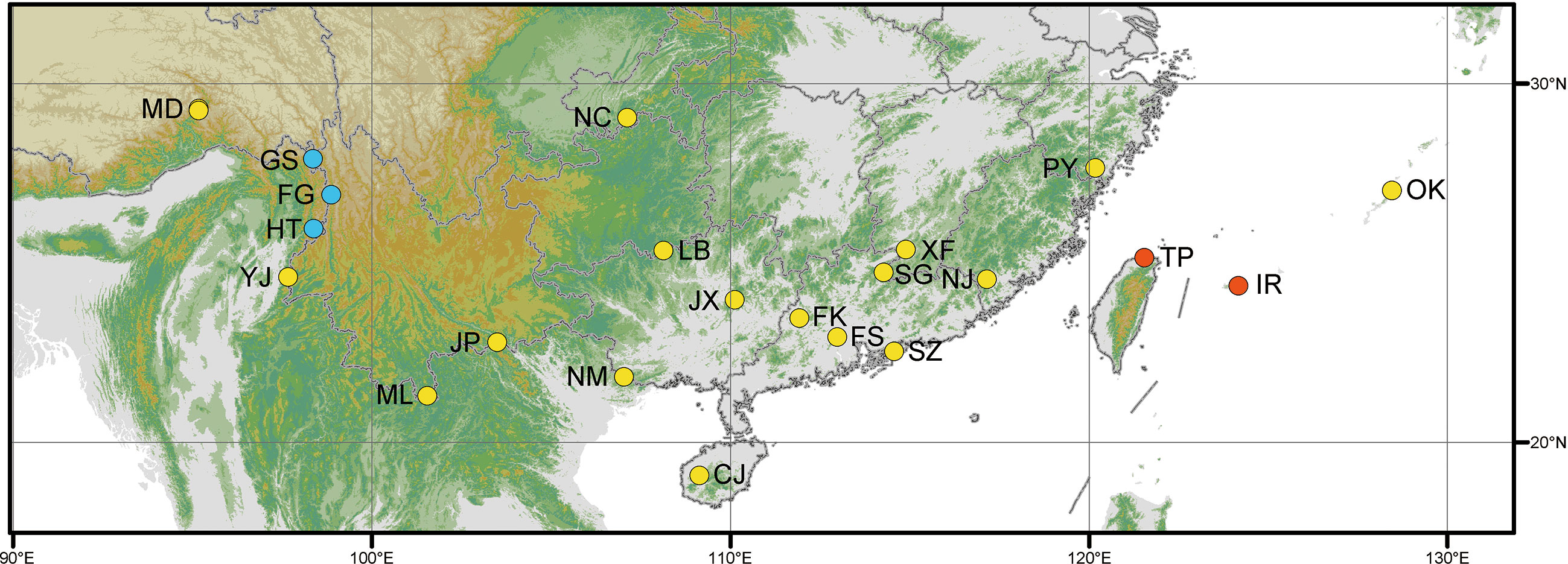
Figure 2 Map showing the distribution of Cibotium samples in this study. Yellow, blue, and red dots represent localities of C. barometz, C. sino-burmaense, and C. cumingii, respectively. The code of each sampling locality means the same as in Table 1.
All the tissue samples were sequenced at the Novogene Corporation (Beijing, China). Total genomic DNA was extracted with a modified CTAB procedure (Doyle and Doyle, 1987). Libraries with an insert size of 350 bp were constructed using a TruSeq Nano DNA HT Sample Preparation Kit (Illumina, San Diego, California, USA) following the manufacturer’s recommendations. Paired-end reads (PE150) were then sequenced on an Illumina NovaSeq 6000 platform. After quality control of raw reads using ng_QC v.2.0 developed by Novogene Corporation with the default settings, we obtained ca. 2 to 4 Gb of clean reads for each sample.
We de novo assembled plastomes of all our samples with clean reads using the GetOrganelle toolkit (Jin et al., 2020) with recommended parameters. The complete plastome of C. barometz (NC_037893, Liu et al., 2018) downloaded from the GenBank database was used as a reference during assembly and annotation. Assembly errors were identified in the initial assembly contigs and manually corrected by mapping raw reads to assembled sequences with Geneious v.11.1.4 (Kearse et al., 2012). Boundaries of large single-copy (LSC), small single-copy (SSC), and two inverse repeat regions (IRs) were detected using RepeatFinder v.1.0.1 (Volfovsky et al., 2001). Genome annotation was performed with GeSeq (Tillich et al., 2017) and Geneious v.11.1.4 (Kearse et al., 2012). Protein-coding sequences were checked against the National Center for Biotechnology Information (NCBI) database and manually corrected. tRNAs were confirmed with tRNAscan-SE v2.0.3 (Lowe and Chan, 2016). The final circular map of the plastome was visualized using OGDraw v.1.3.1 (Greiner et al., 2019). We also used the program LAGAN (Brudno et al., 2003) in mVISTA to compare the gene order and structure among different species with the plastome sequence alignment generated by MAFFT v.7.313 (Katoh and Standley, 2013).
The full-length plastome sequences of all Cibotium samples and the reference (NC_037893), as well as three outgroup species from the tree fern clade, i.e., Alsophila spinulosa (NC_012818), Sphaeropteris brunoniana (NC_051561), and Plagiogyris euphlebia (NC_046784), were aligned with MAFFT v.7.313 (Katoh and Standley, 2013) after the removal of one IR region. According to the phylogeny of Korall et al. (2006), Alsophila, Sphaeropteris, and Cibotium all belong to the “core” tree fern clade, while Plagiogyris is more distantly related to the three genera. The alignment was then filtered using GBLOCKS v.0.91b (Castresana, 2000) to remove ambiguously aligned regions. We also extracted the protein-coding genes of each plastome with a python script (https://github.com/Kinggerm/PersonalUtilities/blob/master/get_annotated_regions_from_gb.py) and concatenated all these single gene alignments to build a protein-coding gene dataset for phylogenetic analyses. The best-fitting nucleotide substitution models of the full-length and protein-coding gene alignments were determined as TVM + F + G4 and GTR + F + G4, respectively, based on the Bayesian information criterion (BIC) by ModelFinder (Kalyaanamoorthy et al., 2017). Maximum likelihood (ML) analysis was performed with both datasets using IQ-TREE v.1.6.8 (Nguyen et al., 2015) with 10,000 ultrafast bootstrap replicates (Minh et al., 2013). Bayesian inference (BI) analysis was performed with the protein-coding gene dataset using MrBayes v.3.2.6 (Ronquist et al., 2012). One cold and three hot chains were run for 2,000,000 generations, with sampling taken every 1,000 generations and a burn-in of 25%. The convergence of Markov chain Monte Carlo runs was checked with Tracer v.1.7.1 (Rambaut et al., 2018) to ensure that the effective sampling size (ESS) of all parameters was above 200. Phylogenetic trees were all visualized, rooted with P. euphlebia, and edited in FigTree v.1.4.2 (Rambaut, 2014).
To identify candidate regions for species and even lineage discrimination in Chinese Cibotium plants, we first used DnaSP v.6.12.03 (Rozas et al., 2017) to evaluate π of the plastome sequence alignment of C. barometz with a window length of 800 bp and a step size of 200 bp. Nucleotide polymorphism sites fixed to specific species and lineages were also identified by checking the alignment, including all Cibotium samples. Additionally, the feasibility and convenience of PCR amplification in practice were also taken into consideration; therefore, the chosen barcode regions are all shorter than 800 bp in length and have conservative flanks suitable for primers to combine with. Candidate loci meeting all these requirements were finally selected, PCR primers of which were designed using Primer3 v.2.3.7 (Koressaar and Remm, 2007; Untergasser et al., 2012).
We extracted sequences of newly selected loci and nine cpDNA markers (atpA, atpB, rbcL, rps4, rbcL-accD, rbcL-atpB, trnG-trnR, trnL-trnF, and rps4-trnS) applied in previous studies (Korall et al., 2006; Geiger et al., 2013) from all our samples, including other accessible data of C. barometz and C. cumingii on GenBank and aligned them. We counted the number of variable sites with MEGA v.10.1.6 (Kumar et al., 2018) and performed ML analysis on each alignment of the 14 barcode loci and some of their combinations, including outgroups, following the same procedure as mentioned above. We compared the topologies of the resulting phylogenetic trees to the ones built with the plastome dataset to evaluate the effectiveness of these loci in species and lineage discrimination. Multiple individuals of a specific taxon resolved as monophyletic with bootstrap support over 50% and were treated as successfully discriminated.
Complete chloroplast genomes of 25 sampled individuals of Cibotium were obtained and assembled into circular molecules comprising one LSC, one SSC, and two IRs (Figure 3; Table 1), which are all typical quadripartite structures. Complete plastomes of C. cumingii and the majority of C. barometz ranged from 165,077 to 166,443 bp in length with very similar GC contents of ca. 41.7%, except for five “C. barometz” samples probably of an unknown species collected from NW Yunnan and NE Myanmar with significantly shorter length (162,108–162,206 bp) and lower GC content (41.4%). The length of LSC (85,634–85,781 bp) and SSC (22,017–22,067 bp) is rather stable among all accessions, whereas IR size varies among samples of C. cumingii (28,681–28,759 bp), most C. barometz (28,944–29,349 bp), and those Yunnan–Myanmar samples (27,206–27,248 bp) with clearly no difference. The boundaries of IRs are the same among all samples, without any expansion or contraction. In comparison, the intergeneric region between rrn16 and rps12 varies greatly among species (Figure 4), which mainly results in IR size variation.
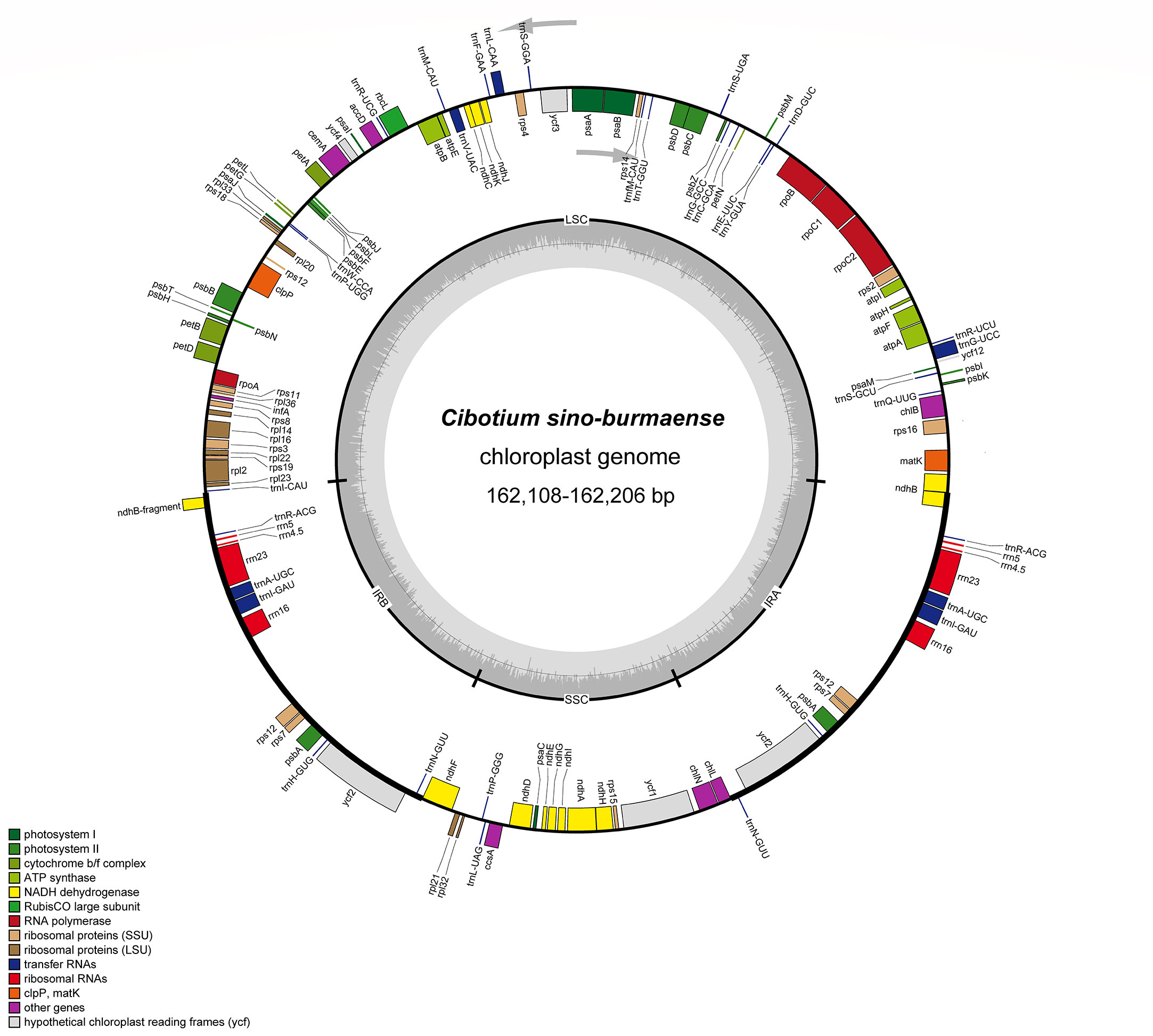
Figure 3 Plastome map of Cibotium sino-burmaense. Arrows indicate the direction of gene transcription. The dark gray area of the inner circle shows GC content variation among different regions of the plastome.
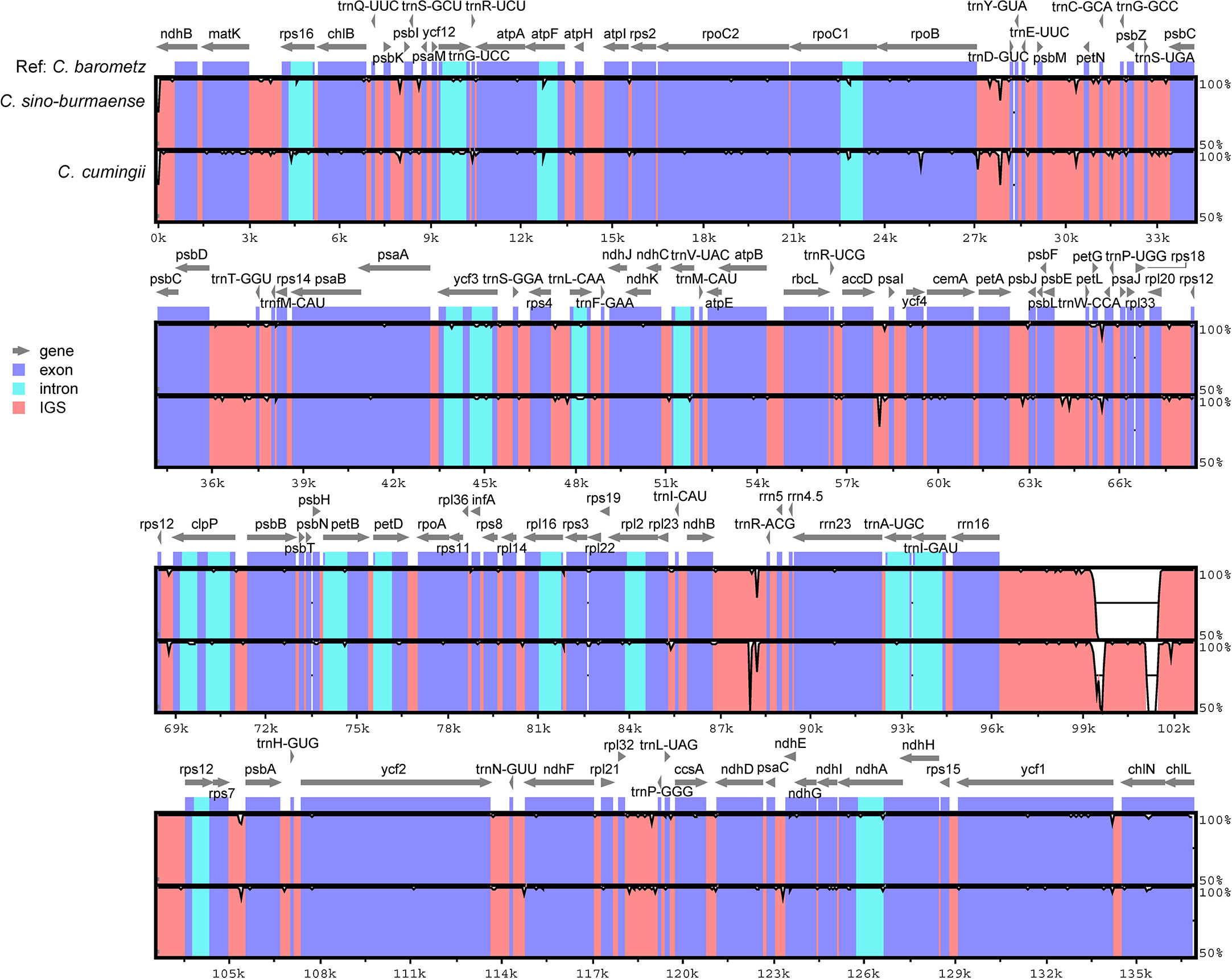
Figure 4 Sequence identity plot comparing the plastome sequence and constitution of three Chinese Cibotium species with C. barometz as a reference. Each sequence starts with the beginning of LSC and ends at the end of SSC. Gray arrows indicate genes by their orientation. A cut-off of 50% identity was used for the plot, and the Y-axis represents the percent identity ranging from 50% to 100%.
All the plastomes encode a total of 117 unique genes in identical order, including 85 protein-coding genes, 28 tRNA genes, and four rRNA genes (Table 2; Figure 3), which are generally consistent with the reference (Liu et al., 2018). In most samples of C. barometz, the annotated matK gene region could not be successfully translated into protein (pseudogenization) because of an early termination resulting from the missing one or two nucleotides (Table 1). The gene ycf2, which was predicted as a pseudogene (6,250 bp) due to a codon shift mutation in the reference plastome of C. barometz (NC_037893, Liu et al., 2018), is normal (6,249 bp) in all the samples of this study. All four rRNA genes, five tRNA genes (trnA-UGC, trnH-GUG, trnI-GAU, trnN-GUU, and trnR-ACG), and three protein-coding genes (rps7, psbA, and ycf2) are totally duplicated, whereas ndhB and rps12 have only one incomplete duplication.
Phylogenetic trees (Figures 5; S1) built with ML and BI analyses based on both full-length (136,298 bp) and protein-coding genes (73,080 bp) datasets showed generally similar topologies and strongly supported the monophyly of C. cumingii, most C. barometz, as well as the five “C. barometz” samples from NW Yunnan and NE Myanmar. The clade formed by the five Yunnan–Myanmar samples (Clade A, Figure 5) was sister to the clade including all the other accessions of C. barometz. The remaining samples of C. barometz except the one collected on Hainan Island could be further divided into two lineages, i.e., Subclade E, including samples from SE China (Zhejiang, Jiangxi, Fujian, Guangdong, and Guizhou) and the Ryukyu Islands, and Subclade W, including samples from SW China (Chongqing, Guangxi, Yunnan, and Xizang). The Hainan sample clustered within Subclade E based on the protein-coding gene dataset with low support value (Figure 5) but became a sister to the combination of Subclade E and Subclade W (MLBS = 26) based on the full-length dataset (Figure S1). Therefore, although the relationship was not consistently resolved, both results reflected the divergence among the southeastern and southwestern subclades as well as the Hainan sample within C. barometz.
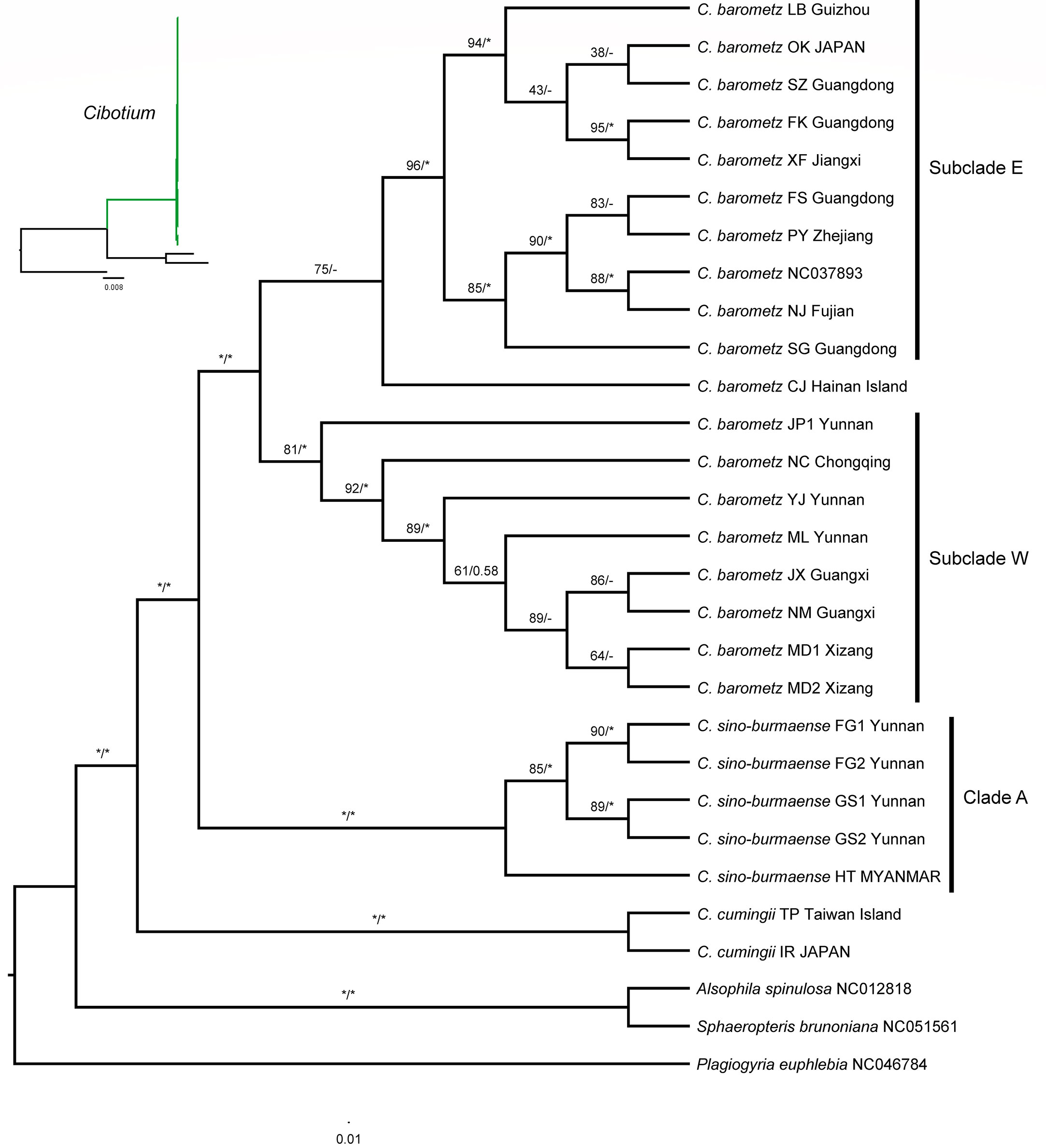
Figure 5 Maximum likelihood cladogram of the Cibotium plant from China and adjacent regions inferred from 85 concatenated protein-coding genes. The numbers above branches are bootstrap values (MLBS) and posterior probabilities (BIPP). An asterisk (*) indicates MLBS = 100% or BIPP = 1.0. En-dash (–) indicates the lack of support value. The corresponding phylogram showing branch length is placed in the upper left corner.
Based on phylogenetic results (Figures S2A–I), only four (trnL-trnF, trnG-trnR, rps4-trnS, and rbcL-accD) of the nine traditional cpDNA loci are effective in the identification of C. cumingii, while only the first two of the four could further discriminate Yunnan–Myanmar Cibotium correctly. None of them accurately depicted the intraspecies divergence within C. barometz. We also tested the concatenated dataset of trnL-trnF, trnG-trnR, rps4-trnS, and rbcL-accD and found poor resolution among all C. barometz, including those Yunnan–Myanmar samples (Figure S2J).
The nucleotide variability of C. barometz plastomes is shown in Figure 6. Variable regions were distributed evenly along the plastome with π value less than 0.002 except for a highly variable region within IR between rrn16 and rps12. Five fragments (Figure 6) with moderate variation for species and lineage discrimination as well as suitable length and flanks for PCR amplification were chosen as candidate DNA barcode loci. Comparing with the nine old cpDNA loci, these new barcodes showed higher variability among the Chinese Cibotium species (Table 3) and were all capable of correctly assigning individuals of C. cumingii, Yunnan–Myanmar Cibotium, and two different lineages within C. barometz into respective clades (Figures S2K–O). The performances of rps3-rps19 and psaC-ndhG are the best among the five because each taxon was resolved as monophyletic with bootstrap support over 50%. The Hainan sample with an uncertain phylogenetic position was clustered with samples of the western lineage by rps3-rps19 and ndhA but clustered with the samples of the eastern lineage by chlB-trnQ, petD-rpoA, and psaC-ndhG. The concatenated dataset of the five new loci also supported a closer relationship between the Hainan sample and the western lineage and revealed a higher support value compared with each single locus phylogeny (Figure S2P).
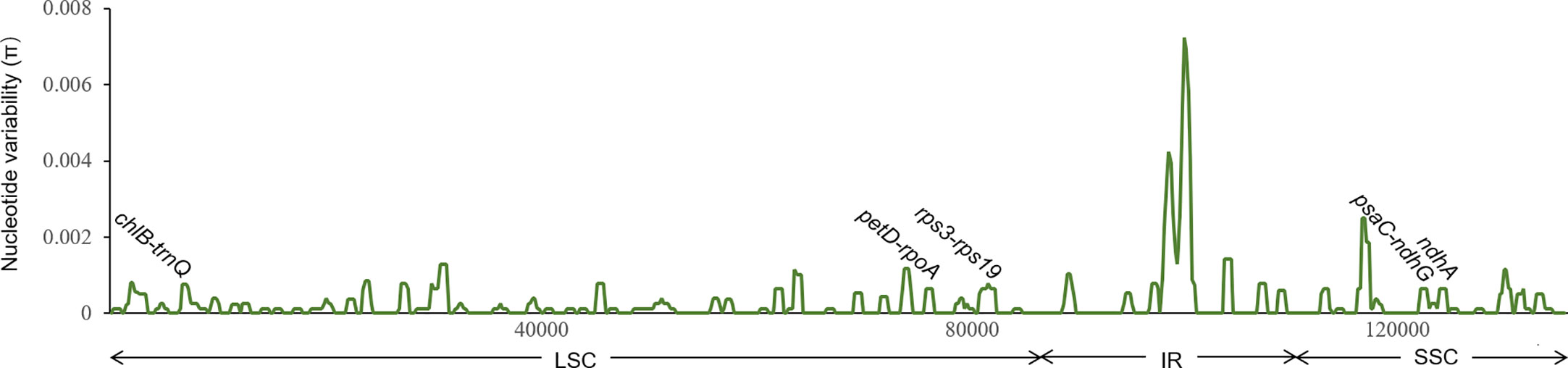
Figure 6 Sliding window analysis of 19 plastomes of Cibotium barometz (window length: 800 bp, step size: 200 bp). X and Y axes indicate the position of the midpoint of a window and nucleotide variability (π) of each window, respectively. Those marked fragments show the position of five newly designed DNA barcodes for inter- and intra-species discrimination in Chinese Cibotium.
Our study newly assembled plastomes of 25 Cibotium samples from China and adjacent regions and presented the conserved structure and composition of which among and within species. A well-supported phylogeny was constructed based on the plastome dataset and revealed three monophyletic clades corresponding to three species, i.e., C. barometz, C. cumingii, and C. sino-burmaense, a new species from NW Yunnan and NE Myanmar. The phylogeny also showed two diverging lineages within C. barometz with distinct geographical distribution regions. Additionally, we suggested several DNA barcodes that could accurately identify all these Chinese species and lineages and may benefit conservation and medicinal quality evaluation in practice.
Based on our plastome-based phylogenetic relationship, C. cumingii is the one that diverged first among the Chinese Cibotium species. The result further distinguished two well-supported sister clades from the remaining samples, one comprising samples distributed in S. China and the Ryukyu Islands corresponding to traditionally recognized C. barometz, and the other comprising five samples from NW Yunnan and NE Myanmar (Clade A, Figure 5). Plastome characteristics, including IR size and GC content, also support the genetic difference between these Yunnan–Myanmar Cibotium and the widespread C. barometz. Therefore, we named these Yunnan–Myanmar samples a new species, Cibotium sino-burmaense, hereafter.
We compared specimens of C. sino-burmaense with C. barometz and found obvious differences in pinnules and sori characters (see details in the taxonomic treatment part). We checked the spores of C. sino-burmaense and found they shared similar perine features with the two known Chinese species photographed by Gastony (1982), with strongly developed equatorial and distal ridges. However, the equatorial diameter of exospores is significantly larger (41–55 μm) than that of C. barometz from S. China (30–45 μm). Because larger spore size is an indicator of higher ploidy levels in some fern taxa (Barrington et al., 1986; Henry et al., 2014), we also compared the genome size of the new species with that of C. barometz. The nuclear DNA content of C. sino-burmaense was estimated at 4.79 pg/C by flow cytometry with Capsicum annuum var. annuum (3.38 pg/C, Moscone et al., 2003) as the internal standard (Figure S3). This result is very close to the record of C. barometz (4.58 pg/C, diploid, Clark et al., 2016), showing no significant ploidy variation signal between the two species.
Three known populations of C. sino-burmaense are all endemic to the border of SW China and NE Myanmar. Narrow distribution might be a signal of low genetic diversity and population size reduction (bottleneck), which may lead to inbreeding, affect adaptive potential, facilitate the accumulation of deleterious mutations, and finally hinder the long-term survival of species (Ellegren and Galtier, 2016). Considering the limited sample size of this study, more comprehensive field exploration of adjacent regions is needed to illuminate the accurate habitat range, population size, and anthropogenic influences of this new species. Our newly designed DNA barcode, together with phenotypic differences, could be applied to the research by accurately distinguishing C. sino-burmaense and C. barometz (Figures S2K–P). In addition, genetic information from the nuclear genome should also be utilized to further evaluate the genetic diversity, effective population size, demographic history, and probable conservation necessity of this endemic Cibotium species.
In recent decades, integrating the principles and methodologies of disciplines such as taxonomy, phylogeny, and evolutionary ecology has become a powerful approach to aiding medicinal discovery, identification, and conservation (Sun et al., 2021; Xu et al., 2021; Zaman et al., 2021). Here, by means of phylogenomic analyses, we clarified the species boundary of the highly demanded medicinal plant C. barometz, as our findings supported the phylogenetic difference between it and C. cumingii and revealed a new species, C. sino-burmaense, distributed at the west edge of its geographical range. Moreover, the plastome-based phylogeny also presented two subclades within C. barometz occupying different areas in China (Subclade E and Subclade W, Figure 5). Due to the lack of other significant divergence in plastome constitution and morphological features between the two subclades, we considered this finding to be an intraspecific lineage divergence with a geographical pattern. Comparing with previous studies constrained by limited sampling areas (e.g., Wu et al., 2007; You and Deng, 2012), the results of this study revealed the east–west divergence throughout the whole distribution region in S. China. The geographical boundary is close to two general phylogeographic breaks in the Sino-Japanese floristic region, i.e., ca. 105°E and the boundary between the Second and Third ladders of landform in China, as reviewed by Ye et al. (2017). The east and west sides of 105°E are dominated by Pacific and Indian monsoons, respectively (Qiu et al., 2011), while altitude is significantly varied between and within different ladders (Li et al., 2013), which also shapes diverse ecological conditions (Fang et al., 2004). Heterogeneous climate and landform as well as refugia isolation resulted from intensity changes of monsoons may have contributed to the east–west genetic split of C. barometz, as demonstrated in other plant lineages (e.g., Bai et al., 2014; Sun et al., 2014; Kou et al., 2016). Additionally, the genetic difference also suggested that the east and west lineages should be considered as at least two management units with respective genetic characters and geographical distributions for conservation (Palsbøll et al., 2007). Due to the limited sampling size and uniparentally inherited feature of the plastome, our study could neither investigate further within-population diversity nor reveal probable hybridization or introgression among linages. In the future, studies with a larger population sampling size of C. barometz and biparentally inherited nuclear genome data would evaluate population diversity on a finer scale, detect genetic communications among lineages, trace demographic history backwards, and predict the vulnerability of different lineages under the influence of habitat fragmentation and changing climate.
Germplasm resources play a fundamental role in high-quality genuine medicine production (Ma and Xiao, 1998; Huang et al., 2008; Yao et al., 2020; Cheng et al., 2021; Zhang and Jiang, 2021; Meng et al., 2023; Xu et al., 2023). At present, all the cibotii rhizome slices sold on the market come from natural sources without domestication, which seriously affects medicinal quality (Ju et al., 2012; Yang et al., 2015). Environmental conditions affect the synthesis and accumulation of secondary metabolites, which are usually medicinal components in plants (Li et al., 2020). Therefore, it is expected that C. barometz populations growing in habitats with diverse climates and ecology may also exhibit pharmacodynamic differences. The genetic divergence observed in this study will be beneficial for selecting specific high-quality germplasm resources from natural populations for cultivation and for elucidating the influence of multiple external factors on the synthesis pathways of metabolites. Combined with the efficacy information of plants from different locations, the newly designed barcode regions could be applied to the identification of geographical origin, which will probably become a method of medicinal quality evaluation in the future.
1a. Pinnules on basiscopic side of lower pinnae present or only one absent, rarely with two absent; sori 1–10 per pinnule segments........................................................................................2
2a. Pinnules on acroscopic and basisicopic sides of a pinna nearly equal in length; apex of pinnule segments apiculate; sori oblong, usually 1–5 pairs per pinnule segment; average exospore equatorial diameter less than 43 μm...1. C. barometz
2b. Pinnules on basiscopic side of a pinna much shorter (c. 1/2) than those on the acroscopic side; apex of pinnule segments acute; sori oblong to spherical, usually 4–8 and sometimes over 10 pairs per pinnule segment; average exospore equatorial diameter more than 45 μm...............................................................2. C. sino-burmaense
1b. Pinnules on basiscopic side of lower pinnae usually three lacking; sori usually one or two per pinnule segment...................................................................3. C. cumingii
(1) Cibotium barometz (L.) J. Sm., London J. Bot. 1 (1842) 437.
≡ Polypodium barometz L., Sp. Pl. 2 (1753) 1092.
≡ Aspidium barometz (L.) Willd., Sp. Pl., ed. 4 [Willdenow] 5 (1810) 268.
≡ Nephrodium barometz (L.) Sweet, Hort. Brit. [Sweet], ed. 2. (1830) 580.
≡ Dicksonia barometz (L.) Link, Fil. Spec. (1841) 166.
Neotype (designated by Mazumdar in Nordic J. Bot. 34(4): 465. 2016): —China. Guangxi, SE of Shang-sze (Shangsi) District, Shap Man Taai Shan (Shiwandashan), near Hoh Lung village, 16 Jun 1933, W.T. Tsang 22473 (S No. S14-33459 [image!])
= Balantium glaucescens Link, Fil. Spec. (1841) 40.
Type: —Not designated.
= Cibotium glaucescens Kunze, Farnkräuter 1 (1841) 63, t.31.
Type: —Not designated.
= Cibotium assamicum Hook., Sp. Fil. [W. J. Hooker] 1 (1844) 83, t.29B.
Holotype: —India. Assam, Mrs. Mack s.n. (not traced).
= Dicksonia assamicum Griff., Notul. 2 (1849) 607.
Lectotype (designated here): —India. Assam, Griffith s.n. (K barcode K001090393 [image!]).
= Cibotium djambianum Hassk., Fil. Jav. 1 (1856) 61.
Type: —Not designated.
Distribution: —China (Chongqing, Fujian, Guangdong, Guangxi, Guizhou, Hainan, Hunan, Jiangxi, Sichuan, Taiwan, Xizang, Yunnan, Zhejiang), Japan (Ryukyu Islands), Indonesia (Java to Sumatra), Malaysia, Myanmar, Thailand, Vietnam.
(2) Cibotium sino-burmaense X.C. Zhang & S.Q. Liang, sp. nov. (Figure 7)

Figure 7 Habitat and morphology of Cibotium sino-burmaense sp. nov. from Dulongjiang, Gongshan, Yunnan, China. (A, B) Habitat. (C) Habit. (D) Pinnae on the abaxial side. (E) Pinnules on the adaxial surface. (F) Pinnules with opened sori. (G, H) Spores under light microscopy. (I) Golden filiform hairs on the stipe base. Photographs by X.-C. Zhang (A–F, I), and S.-Q. Liang (G, H).
Diagnosis: —This new species resembles C. barometz and C. cumingii, differing from the former in the significantly shortened pinnule length on basiscopic side, as well as acute apex and more sori of pinnule segments, and from the latter in the denser sori per pinnule segment and presence of the second and third pinnules on the basiscopic side of lower pinnae.
Holotype: —China. Yunnan: Gongshan county, Dulongjiang Township, 2 May 2022, X.C. Zhang 12880 (PE!).
Note: —The holotype consists of a single large frond mounted on fifteen herbarium sheets, labeled “sheet 1” to “sheet 15”.
Description: —Rhizome prostrate, stout, densely covered with shiny yellowish brown long hairs. Stipes thick, up to 80 cm or more, dark brown to purplish black at base and becoming green upwards, covered with long hairs similar to those on rhizome at base, upper part covered with small, appressed flaccid hairs. Lamina ovate, 2-pinnate-pinnatifid, up to 3 m, subleathery, adaxial surface deep green, abaxial surface glaucous, with small flaccid hairs on midrib; pinna 8–10 pairs, alternate, stalked, medial pinnae 60–80 × 20–30 cm, basal pinna pairs reduced slightly; pinnules more than 30 pairs per lower pinna, shortly stalked, up to 20 cm on the acroscopic side, 10-14 cm on the basiscopic side; pinnule segments, alternate, slightly falcate, with acute apex, margins crenulate to serrulate-serrate. Sori oblong to spherical, usually 4–8 and sometime over 10 pairs at base of lower pairs of pinnule segments; indusia bivalvate, outer indusia larger, orbicular, inner significantly smaller, oblong. Spores pale yellowish, with strongly developed equatorial and distal ridges.
Etymology: —Sino-burmaense is derived from the known distribution of this species along China–Myanmar border.
Additional Specimens Examined: —China. Yunnan: Gongshan county, Dulongjiang Township, 23 Jan 2017, X.C. Zhang & al. 8134; Fugong County, 26 April 2022, X.C. Zhang 12831. Myanmar. Kachin: Htawgaw, April 1925, G. Forrest 26496 (PE barcode 01654827, 01654828, 00388348).
Distribution and habitat: —China (NW Yunnan), Myanmar (Kachin). On cliff with open canopy.
(3) Cibotium cumingii Kunze, Farrnkräuter 1 (1841) 64, 65.
≡ Cibotium barometz var. cumingii (Kunze) C. Chr., Index Filic. 3 (1905) 183.
Lectotype (designated here): —Philippines. Luzon, H. Cuming 123 (K barcode K000376224 [image!]; isolectotypes: K barcode K000376225 [image!], K000376228, K000376229, K000376231, K000376232; BM barcode BM001048122 [image!]; E barcode E00822366 [image!], E00822367 [image!], E00822369 [image!], E00822373 [image!]; P barcode P00633260 [image!], P00633261 [image!], P00633262 [image!]; US barcode 00134826 [image!]; Z barcode Z-000002072 [image!]).
= Cibotium crassinerve Rosenst., Meded. Rijks-Herb. 31 (1917) 4.
Lectotype (designated here): —Philippines. Luzon, Benguet, Dec 1908, H. M. Curran & M. L. Merritt 15800 (L barcode L 0051165 [image!]; isolectotype: MICH No. 1190172 [image!]).
= Cibotium taiwanense C. M. Kuo, Taiwania 30 (1985) 56, 57.
Lectotype (designated here): —China. Taiwan, Hsinchu, Chu-tong, August 1972, C. M. Kuo 1703 (TAI No. 149443 [image!]; isolectotypes: TAI No. 148725 [image!], 150173 [image!]).
Distribution: —China (Taiwan), Japan (Ryukyu Islands), Philippines.
This study presents the conserved structure and gene composition of the chloroplast genome within Cibotium from China. Based on phylogenomic analyses, we constructed a well-supported phylogeny of Chinese Cibotium and indicated that there are three species distributed in China, namely C. barometz, C. cumingii, and C. sino-burmaense, an overlooked cryptic species from NW Yunnan and NE Myanmar. Moreover, our results uncovered the east-west lineage divergence in C. barometz. We also evaluated the species resolution of nine old cpDNA loci and suggested five new cpDNA barcodes that are capable of identifying all the above-mentioned species and lineages of Chinese Cibotium accurately. In conclusion, our findings will improve people’s understanding of the germplasm resource diversity of this endangered medicinal plant group and play a guiding role in its wild population conservation and medical value exploitation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OQ721080-OQ721104.
X-CZ, K-XL and R-HJ designed this study. FW, L-MT, BQ, Y-YC and Y-HH collected and cultivated plant materials of this study. S-QL performed experiments, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by “Evaluation of the Germplasm Resources of the Protected Plant Cibotium barometz” project (GuiLinYan[RC]2302) of Guangxi Forestry Research Institute, the National Plant Specimen Resource Center Project (NPSRC) (E0117G1001), the “Field Survey and Conservation Studies of some State Key Protected Fern Species” project in National Forestry and Grassland Administration, the “2022 Central Financial Forestry Reform and Development Funds: Collection, Conservation and Use of Germplasm Resources of Cibotium barometz” of Department of Forestry of Guangxi Zhuang Autonomous Region, as well as “Survey and Collection of Germplasm Resources of Woody and Herbaceous Plants in Guangxi, China” (GXFS-2021-34).
We appreciate Dr. Xiang-Yun Zhu’s valuable discussion about taxonomic treatment. We thank Dr. Jie Yang, Mr. Jun-Yong Tang, and Mr. Ji-Gao Yu for their help in plastome data analyses. We also thank Jin‐Dan Zhang from the Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences, for the technical assistance on flow cytometry.
Author L-MT was employed by the company Guangxi Forestry Industry Group Stock Corporation, Nanning, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1183653/full#supplementary-material
Bai, W. N., Wang, W. T., Zhang, D. Y. (2014). Contrasts between the phylogeographic patterns of chloroplast and nuclear DNA highlight a role for pollen-mediated gene flow in preventing population divergence in an East Asian temperate tree. Mol. Phylogenet. Evol. 81, 37–48. doi: 10.1016/j.ympev.2014.08.024
Barrington, D. S., Paris, C. A., Ranker, T. A. (1986). Systematic inferences from spore and stomate size in the ferns. Am. Fern J. 76, 149–159. doi: 10.2307/1547723
Brudno, M., Do, C. B., Cooper, G. M., Kim, M. F., Davydov, E., NISC Comparative Sequencing Program, et al. (2003). LAGAN and multi-LAGAN: efficient tools for Large-scale multiple alignment of genomic DNA. Genome Res. 13, 721–731. doi: 10.1101/gr.926603
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chen, S. L., Yu, H., Luo, H. M., Wu, Q., Li, C. F., Steinmetz, A. (2016). Conservation and sustainable use of medicinal plants: problems, progress, and prospects. BMC Chin. Med. 11, 37. doi: 10.1186/s13020-016-0108-7
Chen, S. S., Zhang, M. H., Wang, J. X., Zhang, X. C. (2021). Original plant and research progress of the medicinal plant. Huperzia javanica. Guihaia 4111, 1794–1809. doi: 10.11931/guihaia.gxzw202103069
Cheng, R. Y., He, W. R., Shen, X. F., Xiang, L., Liang, Y., Meng, Y., et al. (2021). Analysis on content differences of artemisinin and arteannuin b in different provenances of Artemisia annua under indoor hydroponic conditions. Chin. J. Exp. Tradit. Med. Formulae 27, 145–151. doi: 10.13422/j.cnki.syfjx.20211547
Chinese Pharmacopoeia Commission [CPC] (2020). Chinese Pharmacopoeia. 1st Edn (Beijing: Chinese Medical Science and Technology Press).
Clark, J., Hidalgo, O., Pellicer, J., Liu, H., Marquardt, J., Robert, Y., et al. (2016). Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 210, 1072–1082. doi: 10.1111/nph.13833
Cuong, N. X., Minh, C. V., Kiem, P. V., Huong, H. T., Ban, N. K., Nhiem, N. X., et al. (2009). Inhibitors of osteoclast formation from rhizomes of cibotium barometz. J. Nat. Prod. 72, 1673–1677. doi: 10.1021/np9004097
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. Bot. Soc Amer. 19, 11–15. doi: 10.1016/0031-9422(80)85004-7
Du, X. Y., Kuo, L. Y., Zuo, Z. Y., Li, D. Z., Lu, J. M. (2022). Structural variation of plastomes provides key insight into the deep phylogeny of ferns. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.862772
Ellegren, H., Galtier, N. (2016). Determinants of genetic diversity. Nat. Rev. Genet. 17, 422–433. doi: 10.1038/nrg.2016.58
Fang, J. Y., Shen, Z. H., Cui, H. T. (2004). Ecological characteristics of mountains and research issues of mountain ecology. Biodiversity Sci. 12, 10–19. doi: 10.17520/biods.2004003
Fu, C. L., Zheng, C. S., Lin, J., Ye, J. X., Mei, Y. Y., Pan, C. B., et al. (2017). Cibotium barometz polysaccharides stimulate chondrocyte proliferation in vitro by promoting G1/S cell cycle transition. Mol. Med. Rep. 15, 3027–3034. doi: 10.3892/mmr.2017.6412
Gastony, J. (1982). Spore morphology in the dicksoniaceae. II. the genus Cibotium. Can. J. Bot. 60, 955–972. doi: 10.1139/b82-121
Geiger, J. M. O., Korall, P., Ranker, T. A., Kleist, A. C., Nelson, C. L. (2013). Molecular phylogenetic relationships of Cibotium and origin of the Hawaiian endemics. Am. Fern J. 103, 141–152. doi: 10.1640/0002-8444-103.3.141
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Hammer, T. A., Zhong, X., des Francs-Small, C. C., Nevill, P. G., Small, I. D., Thiele, K. R. (2019). Resolving intergeneric relationships in the aervoid clade and the backbone of Ptilotus (Amaranthaceae): evidence from whole plastid genomes and morphology. Taxon 68, 297–314. doi: 10.1002/tax.12054
Heng, Y. W., Ban, J. J., Khoo, K. S., Sit, N. W. (2020). Biological activities and phytochemical content of the rhizome hairs of Cibotium barometz (Cibotiaceae). Ind. Crops Prod. 153, 112612. doi: 10.1016/j.indcrop.2020.112612
Henry, T. A., Bainard, J. D., Newmaster, S. G. (2014). Genome size evolution in Ontario ferns (Polypodiidae): evolutionary correlations with cell size, spore size, and habitat type and an absence of genome downsizing. Genome 57, 555–566. doi: 10.1139/gen-2014-0090
Hollingsworth, P. M., Li, D. Z., van der Bank, M., Twyford, A. D. (2016). Telling plant species apart with DNA: from barcodes to genomes. Philos. Trans. R. Soc B 371, 20150338. doi: 10.1098/rstb.2015.0338
Holttum, R. E. (1963). “Cibotium,” in Flora malesiana. Eds. Van Steenis, C. G. G. J., Holttum, R. E. (Netherlands: Martinus Nijhoff. Dr W. Junk Publishers), 164–166.
Huang, L. Q., Guo, L. P., Hu, J., Shao, A. J. (2008). Molecular mechanism and genetic basis of geoherbs. China J. Chin. Mater. Med. 33, 2303–2308.
Huang, Z., Luo, X., Zhang, Y., Ying, Y., Cai, X., Lu, W., et al. (2021). Notoginseng triterpenes inhibited autophagy in random flaps via the beclin-1/VPS34/LC3 signaling pathway to improve tissue survival. Front. Bioeng. Biotechnol. 9. doi: 10.3389/fbioe.2021.771066
Ji, Y., Yang, J., Landis, J. B., Wang, S., Yang, Z., Zhang, Y. (2021). Deciphering the taxonomic delimitation of Ottelia acuminate (Hydrocharitaceae) using complete plastomes as super-barcodes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.681270
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., de Pamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Ju, C. G., Cao, C. X., Shi, L., Li, J., Jia, T. Z. (2005). Study on the analgesic and haemostatic effects of cibotii rhizoma, its concoction products and hairs of cibotii rhizome. Chin. Tradit. Pat. Med. 27, 1279–1281.
Ju, C. G., Xu, G., Song, Y. J., Zhao, W. L., Jia, T. Z. (2012). Comparison of the content of total phenolic acid in Cibotium barometz and its processed products from different areas. Chin. J. Exp. Tradit. Med. Formulae 18, 24–26.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Khoury, C. K., Brush, S., Costich, D. E., Curry, H. A., de Haan, S., Engels, J. M. M., et al. (2022). Crop genetic erosion: understanding and responding to loss of crop diversity. New Phytol. 233, 84–118. doi: 10.1111/nph.17733
Korall, P., Pryer, K. M., Metzgar, J. S., Schneider, H., Conant, D. S. (2006). Tree ferns: monophyletic groups and their relationships as revealed by four protein-coding plastid loci. Mol. Phylogenet. Evol. 39, 830–845. doi: 10.1016/j.ympev.2006.01.001
Koressaar, T., Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kou, Y. X., Cheng, S. M., Tian, S., Li, B., Fan, D. M., Chen, Y. J., et al. (2016). The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late early Miocene. J. Biogeogr. 43, 351–360. doi: 10.1111/jbi.12635
Kress, W. J., García-Robledo, C., Uriarte, M., Erickson, D. L. (2015). DNA Barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 30, 25–35. doi: 10.1016/j.tree.2014.10.008
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, Y. Q., Kong, D. X., Fu, Y., Sussman, M. R., Wu, H. (2020). The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 148, 80–89. doi: 10.1016/j.plaphy.2020.01.006
Li, T. Q., Lei, W., Ma, Z. S., Wang, L., Wu, Z. X., Zhao, X. (2014). Experimental study of the effect of Cibotium barometz extract on ovariectomy-induced osteoporosis in rats. Chin. J. Osteoporosis 20, 736–740. doi: 10.3969/j.issn.1006-7108.2014.07.004
Li, B. Y., Pan, B. T., Cheng, W. M., Han, J. F., Qi, D. L., Zhu, C. (2013). Research on geomorphological regionalization of China. Acta Geogr. Sin. 68, 291–306.
Liu, S., Wang, Z., Wang, T., Su, Y. (2018). The complete chloroplast genome of Cibotium barometz (Cibotiaceae), an endangered CITES medicinal fern. Mitochondrial DNA Part B: Resour. 3, 464–465. doi: 10.1080/23802359.2018.1462128
Lowe, T. M., Chan, P. P. (2016). tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Ma, X. J., Xiao, P. G. (1998). Genetic diversity of germplasm resources in the importance of genetic diversity in the development of medicinal plants. China J. Chin. Mater. Med. 23, 579–581.
Meng, X. C., Deng, D. Q., Du, H. W., Guan, Y. (2023). Scientific connotation of high-quality genuine medicinal materials. Chin. Tradit. Herb. Drugs 54, 939–947. doi: 10.7501/j.issn.0253-2670.2023.03.028
Minh, B. Q., Nguyen, M. A. T., von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Moscone, E. A., Baranyi, M., Ebert, I., Greilhuber, J., Ehrendorfer, F., Hunziker, A. T. (2003). Analysis of nuclear DNA content in Capsicum (Solanaceae) by flow cytometry and feulgen densitometry. Ann. Bot. 92, 21–29. doi: 10.1093/aob/mcg105
Newman, D. J., Kilama, J., Bernstein, A., Chivian, E. (2008). “Medicines from nature,” in Sustaining life: how human health depends on biodiversity. Eds. Chivian, E., Bernstein, A. (New York: Oxford University Press), 117–161.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nock, C. J., Waters, D. L., Edwards, M. A., Bowen, S. G., Rice, N., Cordeiro, G. M., et al. (2011). Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 9, 328–333. doi: 10.1111/j.1467-7652.2010.00558.x
Palsbøll, P. J., Bérubé, M., Allendorf, F. W. (2007). Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16. doi: 10.1016/j.tree.2006.09.003
Park, I., Song, J. H., Yang, S., Chae, S., Moon, B. C. (2021). Plastid phylogenomic data offers novel insights into the taxonomic status of the Trichosanthes kirilowii complex (Cucurbitaceae) in south Korea. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.559511
Qin, M., Zhu, C. J., Yang, J. B., Vatanparast, M., Schley, R., Lai, Q., et al. (2022). Comparative analysis of complete plastid genome reveals powerful barcode regions for identifying wood of Dalbergia odorifera and D. tonkinensis (Leguminosae). J. Syst. Evol. 60, 73–84. doi: 10.1111/jse.12598
Qiu, Y. X., Fu, C. X., Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Rambaut, A. (2014). FigTree v 1.4. 2 molecular evolution, phylogenetics and epidemiology. Edinburgh: univ. Edinburgh. Inst. Evol. Biol.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarisation in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a Large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sánchez-Delbarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Schoen, D. J., Brown, H. D. (1993). Conservation of allelic richness in wild crop relatives is aided by assessment of genetic markers. Proc. Natl. Acad. Sci. U.S.A. 910, 10623–10627. doi: 10.1073/pnas.90.22.10623
Smith, A. R., Pryer, K. M., Schuettpelz, E., Korall, P., Schneider, H., Wolf, P. G. (2006). A classification for extant ferns. Taxon 55, 705–731. doi: 10.2307/25065646
State Forestry and Grassland Administration and the Ministry of Agriculture and Rural Affairs, P. R. China (2021) List of wild plants under state protection. Available at: http://www.forestry.gov.cn/main/5461/20210908/162515850572900.html.
Sun, Q. Z. (2021). Cibotium barometz polysaccharide regulates IL-1β-mediated proliferation and apoptosis in osteoarthritis chondrocyte via miR-181c. Chin. J. Gerontol. 41, 2398–2402. doi: 10.3969/j.issn.1005-9202.2021.11.046
Sun, Y., Hu, H., Huang, H., Vargas-Mendoza, C. F. (2014). Chloroplast diversity and population differentiation of Castanopsis fargesii (Fagaceae): a dominant tree species in evergreen broad-leaved forest of subtropical China. Tree Genet. Genomes 10, 1531–1539. doi: 10.1007/s11295-014-0776-3
Sun, J. H., Liu, B., Guo, L. P., Huang, L. Q. (2021). Significance of plant taxonomy in Chinese material medica resources: the changes of family and genus category and standardization of scientific names in Chinese pharmacopoeia. Sci. Sin. Vitae 51, 579–593. doi: 10.1360/SSV-2020-0345
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq – versatile and accurate annotation of organelle genomes. Nucl. Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Tu, Y. (2016). Artemisinin–a gift from traditional Chinese medicine to the world (Nobel lecture). Angew. Chem. Int. Ed. Engl. 55, 10210–10226. doi: 10.1002/anie.201601967
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Res. 40, e115. doi: 10.1093/nar/gks596
Volfovsky, N., Haas, B. J., Salzberg, S. L. (2001). A clustering method for repeat analysis in DNA sequences. Genome Biol. 2, 1–11. doi: 10.1186/gb-2001-2-8-research0027
Wang, J., Fu, C. N., Mo, Z. Q., Möller, M., Yang, J. B., Zhang, Z. R., et al. (2022). Testing the complete plastome for species discrimination, cryptic species discovery and phylogenetic pesolution in Cephalotaxus (Cephalotaxaceae). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.768810
Wei, R., Zhang, X. C. (2020). Phylogeny of Diplazium (Athyriaceae) revisited: resolving the backbone relationships based on plastid genomes and phylogenetic tree space analysis. Mol. Phylogenet. Evol. 143, 106699. doi: 10.1016/j.ympev.2019.106699
Wei, X. P., Zhang, X. Y., Dong, Y. Q., Cheng, J. L., Bai, Y. J., Liu, J. S., et al. (2022). Molecular structure and phylogenetic analyses of the complete chloroplast genomes of three medicinal plants Conioselinum vaginatum, Ligusticum sinense, and Ligusticum jeholense. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.878263
Wu, L., Deng, H. P., Xu, J., Song, Q. Z., Liu, G. H., Zhang, L. R., et al. (2007). AFLP analysis of genetic diversity in Cibotium barometz populations. China J. Chin. Mater. Med. 32, 1468–1469.
Xi, J., Lv, S., Zhang, W., Zhang, J., Wang, K., Guo, H., et al. (2022). Comparative plastomes of Carya species provide new insights into the plastomes evolution and maternal phylogeny of the genus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.990064
Xu, Y. Y., Li, K., Jiao, R., Wang, Q., Wang, Z., Shi, B., et al. (2023). Evaluation and screening of superior germplasms in Paris polyphylla var. yunnanensis. J. Southwest For. Univ. 43, 36–41. doi: 10.11929/j.swfu.202112068
Xu, J. X., Wang, Y. L., Wang, J. J., Tang, W., Chen, L. J., Long, C. L. (2012). Chemical constituents of Cibotium barometz and their bioactivities. Nat. Prod. Res. Dev. 24, 134–140.
Xu, Y. L., Wang, Z. Y., Yang, S. D., Lu, L. (2021). Application and prospect of evolutionary ecology in evaluation of germplasm resources of medicinal plants. Chin. Tradit. Herb. Drugs 52, 1221–1233. doi: 10.7501/j.issn.0253-2670.2021.05.001
Yang, C. Z., Liu, X. F., Cai, D. L., Fan, S. M. (2015). Investigation on resource and quality assessment of cibotii rhizoma. China J. Chin. Mater. Med. 40, 1919–1924. doi: 10.4268/cjcmm20151014
Yang, J., Xiang, Q. P., Zhang, X. C. (2023). Uncovering the hidden diversity of the rosette-forming Selaginella tamariscina group based on morphological and molecular data. Taxon 72, 8–19. doi: 10.1002/tax.12817
Yao, J., Wang, W. R., Guo, S. L., Meng, X. C. (2020). Different germplasm of paeoniae radix alba and paeoniae radix rubra. Mod. Chin. Med. 22, 1933–1937.
Ye, J. W., Zhang, Y., Wang, X. J. (2017). Phylogeographic breaks and the mechanisms of their formation in the sino-Japanese floristic region. Chin. J. Plant Ecol. 41, 1003–1019. doi: 10.17521/cjpe.2016.0388
You, Y. F., Deng, H. P. (2012). Analysis of genetic diversity of the rare and endangered species Cibotium barometz by SRAP markers. Acta Bot. Boreal. –Occident. Sin. 32, 688–692.
Yu, Y. B., Wang, Q. L., Kell, S., Maxted, N., Ford-Lloyd, B. V., Wei, W., et al. (2013). Crop wild relatives and their conservation strategies in China. Biodiversity Sci. 21, 750–757. doi: 10.3724/SP.J.1003.2013.08138
Zaman, W., Ye, J., Saqib, S., Liu, Y., Shan, Z., Hao, D., et al. (2021). Predicting potential medicinal plants with phylogenetic topology: inspiration from the research of traditional Chinese medicine. J. Ethnopharmacol. 281, 114515. doi: 10.1016/j.jep.2021.114515
Zangara, A. (2003). The psychopharmacology of huperzine a: an alkaloid with cognitive enhancing and neuroprotective properties of interest in the treatment of alzheimer's disease. Pharmacol. Biochem. Behav. 75, 675–686. doi: 10.1016/s0091-3057(03)00111-4
Zhang, X. C., Jia, J. S., Zhang, G. M. (2002). Survey and evaluation of the natural resources of Cibotium barometz (L.) j. smith in China, with reference to the implementation of the CITES convention. Fern Gaz. 16, 383–387.
Zhang, Y. J., Jiang, Y. Y. (2021). Research approach and progress in term of the quality of Chinese medicinal. Acta Chin. Med. Pharmacol. 49, 106–112.
Zhang, X. C., Nishida, H. (2013). “Cibotiaceae,” in Flora of China, vol. 2–3 (Pteridophytes). Eds. Wu, Z. Y., Raven, P. H., Hong, D. Y. (Beijing: Science Press; St. Louis.: Missouri Botanical Garden Press), 132–133.
Zhang, Z. R., Yang, X., Li, W. Y., Peng, Y. Q., Gao, J. (2022). Comparative chloroplast genome analysis of Ficus (Moraceae): insight into adaptive evolution and mutational hotspot regions. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.965335
Keywords: chloroplast genome, Cibotium barometz, Cibotium sino-burmaense, conservation, DNA barcoding, endangered species, germplasm resource, species diversity
Citation: Jiang R-H, Liang S-Q, Wu F, Tang L-M, Qin B, Chen Y-Y, Huang Y-H, Li K-X and Zhang X-C (2023) Phylogenomic analysis, cryptic species discovery, and DNA barcoding of the genus Cibotium in China based on plastome data. Front. Plant Sci. 14:1183653. doi: 10.3389/fpls.2023.1183653
Received: 10 March 2023; Accepted: 02 May 2023;
Published: 06 June 2023.
Edited by:
Qiu-Yun(Jenny) Xiang, North Carolina State University, United StatesReviewed by:
Zhechen Qi, Zhejiang Sci-Tech University, ChinaCopyright © 2023 Jiang, Liang, Wu, Tang, Qin, Chen, Huang, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Chun Zhang, emhhbmd4Y0BpYmNhcy5hYy5jbg==; Kai-Xiang Li, bGt4MjAyQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.