
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 10 July 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1182685
Panax ginseng Meyer is one of the most valuable plants and is widely used in China, while ginseng anthracnose is one of the most destructive diseases. Colletotrichum panacicola could infect ginseng leaves and stems and causes serious anthracnose disease, but its mechanism is still unknown. Here, transcriptome and metabolism analyses of the host leaves were conducted to investigate the ginseng defense response affected by C. panacicola. Upon C. panacicola infection, ginseng transcripts altered from 14 to 24 h, and the expression of many defense-related genes switched from induction to repression. Consequently, ginseng metabolites in the flavonoid pathway were changed. Particularly, C. panacicola repressed plant biosynthesis of the epicatechin and naringin while inducing plant biosynthesis of glycitin, vitexin/isovitexin, and luteolin-7-O-glucoside. This work indicates C. panacicola successful infection of P. ginseng by intervening in the transcripts of defense-related genes and manipulating the biosynthesis of secondary metabolites, which might have antifungal activities.
Ginseng, Panax ginseng, is one of the most important medicinal herbs in Northeast China with high values in nutrition and medicine (Kim et al., 2010; Liu et al., 2020). Ginseng is a perennial plant with a long cultivation period and is widely planted in China, Korea, and Japan (Sun et al., 2018; Wang et al., 2018). Because of its long life, P. ginseng now suffers from various biotic stresses with fungal diseases as one of the main sources (Kim et al., 2019). There are many fungal diseases in ginseng fields including anthracnose, gray mold, Alternaria blight, damping-off, and root rot (Kim et al., 2010; Choi et al., 2011; Gao et al., 2014; Guan et al., 2014; Liu et al., 2020; Chen et al., 2022; Lu et al., 2019). Among these, anthracnose is one of the most destructive diseases, causing ginseng seedling blight in nurseries and promoting ginseng leaf spots in permanent beds (Choi et al., 2011; Chung and Bae, 1979).
Colletotrichum panacicola was reported as a major fungal pathogen causing ginseng anthracnose disease in Korea (Choi et al., 2011). Recently, C. panacicola was confirmed in all provinces in Northeast China such as Jilin, Liaoning, and Heilongjiang (Liu et al., 2020). The C. panacicola was able to infect both ginseng leaves and stems more aggressively than other species (Liu et al., 2020). Usually, chemical pesticides were used to control ginseng disease. However, deleterious pesticide residues accumulate in both ginseng roots and the surrounding soil, which has become a serious environmental problem (Ryu et al., 2014). The outcome of the interaction between ginseng and C. panacicola has not been well characterized so far. Detailed molecular and comparative transcriptome analyses during ginseng–C. panacicola interactions are becoming feasible, as the genome of P. ginseng has been reported (Jayakodi et al., 2018).
During plant–microbe interaction, plants have developed an innate immune system to recognize potentially infectious pathogens (Jones and Dangl, 2006; Jones et al., 2016; Ngou et al., 2021; Yuan et al., 2021). Downstream of innate recognition, the complex defense signaling/responses are activated including, calcium (Ca2+) burst, reactive oxygen species (ROS) burst, mitogen‐activated protein kinase (MAPK) activation, plant hormone signaling, transcription factors (TFs), and secondary metabolism (Tsuda et al., 2009; Birkenbihl et al., 2017; Wang et al., 2020). Generally, plant hormones such as salicylic acid (SA), jasmonate (JA), ethylene (ET), and abscisic acid (ABA) are repeatedly observed upon pathogen infection (Bari and Jones, 2009; Chen and Yu, 2014; Verma et al., 2016; Chen et al., 2020). Depending on the pathogen and plant styles, the role of hormones is different from time to time (Glazebrook, 2005; Liu et al., 2015; Liu et al., 2017). SA signaling has been traditionally associated with defense against hemibiotrophic or biotrophic pathogens, whereas JA/ET signaling appears to be more important to necrotrophic pathogens and insects (Glazebrook, 2005). The JA signaling was also involved in plant–biotrophic pathogen interaction, while SA/JA signaling collaborated during effectors that triggered immunity response (Guerreiro et al., 2016; Liu et al., 2016). ABA signaling has emerged as an important modulator of plant stress networks (Chen and Yu, 2014; Chen et al., 2020). In plants, ABA positively or negatively regulates plant defense depending on the lifestyle of the pathogen (Denancé et al., 2013).
Global expression analyses proposed that plant cells’ transcriptional re-programming is a key step to mounting an efficient defense response (Birkenbihl et al., 2017; Chen et al., 2022; Liu et al., 2015). Several transcription factor families are repeatedly reported to be involved in the regulation of gene expression during plant–pathogen interaction (Birkenbihl et al., 2017). Next to the plant hormones and TFs, plant secondary metabolites play crucial roles in defense. Low-molecular-mass secondary metabolites with antimicrobial activity that are induced by various microbes are called phytoalexins (Pedras et al., 2011), which are usually considered molecular markers of disease resistance (Ahuja et al., 2012). Compared with many plant–microbe interaction systems, however, so far, there has been no such information reported during ginseng–C. panacicola interaction. Here, the C. panacicola was assumed to derive a strategy to overcome ginseng defense response and promote disease development.
This work performed transcriptome analysis in ginseng leaves during C. panacicola infection. A further study compared the data derived from the same treatments and revealed the mechanism of disease development in C. panacicola by suppressing defense-related gene expression and modulating certain antifungal flavonoid biosynthesis. These results will not only increase understanding of the complexity of P. ginseng–C. panacicola interaction but also enhance efforts to identify functional genes in plant defense response and disease development.
In nature, P. ginseng grows slowly and faces multiple biotic and abiotic stresses including anthracnose. In the field, the anthracnose disease in ginseng leaves usually contains several tiny spots with a small yellow halo at the beginning, the lesion develops, the yellow halo extends from time to time, and finally, necrotic spots with an empty hole in the middle area are present in ginseng leaves (Figure 1A). The spots with empty holes seriously influence the growth of ginseng leaves. C. panacicola strain FSL was isolated from ginseng leaves in Fusong, Jilin Province. The spores of C. panacicola FSL were then incubated on garden ginseng leaves in the lab. At the early stage, there was no obvious symptom observed on ginseng leaves (Supplementary Figure 1). The typical necrotic spots were observed after a 1-week infection (Figure 1A, arrows). It is indicated that ginseng is susceptible to C. panacicola FSL.
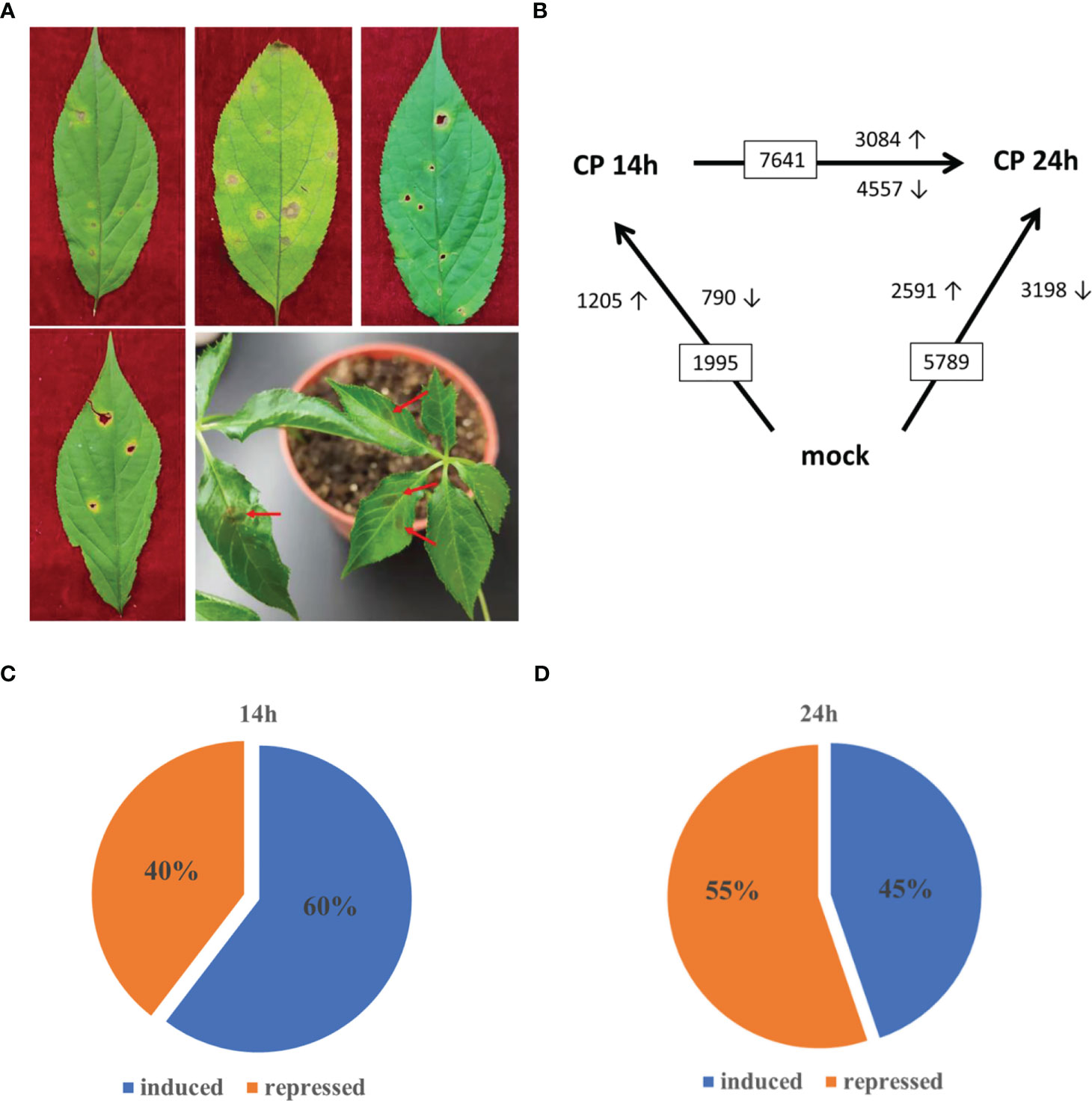
Figure 1 Transcription analysis of Colletotrichum panacicola-infected Panax ginseng plants. (A) Ginseng anthracnose disease phenotype in the forest and C. panacicola FSL infection phenotype on 2-year-old ginseng leaves (7 days post-infection). (B) Numbers of differentially expressed genes (≥2-fold; p ≤ 0.05) in ginseng at 14 and 24 h after spray inoculation with spores of C. panacicola FSL (CP) or mock treatment (CK) identified by RNA-seq. (C) Percentage of C. panacicola FSL induced and repressed significantly differentially expressed genes at 14 h compared with control (total number of SSTF gene is 1,995). (D) Percentage of C. panacicola FSL induced and repressed significantly differentially expressed genes at 24 h compared with control (total number of SSTF gene is 5,789).
To understand the molecular mechanism of P. ginseng–C. panacicola interaction, RNA sequencing was performed. Ginseng leaves incubated with C. panacicola were harvested at 14 h (Cp14h) and 24 h (Cp24h). For the control (CK), ginseng leaves without any treatment were harvested. RNA-seq data were analyzed as previously indicated (Chen et al., 2022). A total of 365 million validated high-quality reads were obtained from all nine libraries (Supplementary File 1) and then aligned to the P. ginseng genome (Jayakodi et al., 2018).
To identify genes involved in ginseng response to C. panacicola at the genome-wide level under different times, the differentially changed genes (statistically significantly, p ≤ 0.05, altered at least twofold (SSTF)) were compared between C. panacicola-treated (CP) and un-treated (CK) ginseng. At 14 h, a total of 1,995 SSTF genes were identified in CP-treated plants compared with CK: 1,205 (approximately 60%) were increased, while 790 (40%) were decreased (Figures 1B, C). Not surprisingly, a total of 5,789 SSTF genes were identified at 24 h compared with CK, which were about threefold more than at 14 h (Figure 1B). This indicates that more genes were affected in response to C. panacicola treatment. To our surprise, approximately 55% of genes (3198) were repressed while 45% of genes (2591) were induced (Figure 1C), suggesting the balance between induction and repression of host genes changed from 14 to 24 h (Figures 1C, D). The comparison of transcripts between 24 and 14 h was also performed. As indicated in Figure 1B, 7,641 SSTF genes were observed, with 3,084 (40.4%) increased and 4,557 (59.6%) decreased, further confirming the repression of host genes by C. panacicola at 24 h.
Since pathogen incubation often up- or downregulates host gene expressions during the interaction, the transcription diversity was further analyzed in ginseng upon C. panacicola infection at 14 and 24 h. The differentially expressed genes (DEGs) were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) methods.
For the SSTF genes at 14 h, GO terms about defense response, protein serine/threonine kinase activity, cell wall, unsaturated fatty acid biosynthetic process, killing of cells of other organisms, and plasma membrane were enriched (Supplementary Figures 2A). KEGG analysis indicated the genes associated with the plant–pathogen interaction, metabolism, and biosynthesis were enriched (Supplementary Figure 2B). For the SSTF genes at 24 h, GO and KEGG terms about similar pathways were enriched as those at 14 h (Supplementary Figures 3A, B).
Next, when comparing the SSTF genes at both 14 and 24 h, 904 genes were observed. It was indicated that 904 genes were consistently significantly affected upon C. panacicola treatment, not only at 14 h but also at 24 h (Figure 2A). A heatmap analysis was performed, and different expression patterns among these 904 genes are shown in Figure 2B. The red-framed genes (I and III) indicated the differentially expressed genes in CK and CP treatment. SSTF genes in group I was induced upon CP infection compared with CK, while genes in group III were decreased. Interestingly, the highest expression level of genes in group II was observed in Cp14h, while the lowest expression level of genes in group IV was also observed in Cp14h. KEGG analysis of SSTF genes observed at both 14 and 24 h indicated that defense- and metabolism biosynthesis-related genes are enriched after C. panacicola infection. These pathways included plant–pathogen interaction, biosynthesis of unsaturated fatty acids, and metabolisms such as fatty acid, linoleic acid, alpha-linolenic acid, phenylalanine, anthocyanin, tryptophan, and flavonoid (Figure 2C). The enrichment of these pathways at both 14 and 24 h suggests their role in ginseng–C. panacicola interaction.
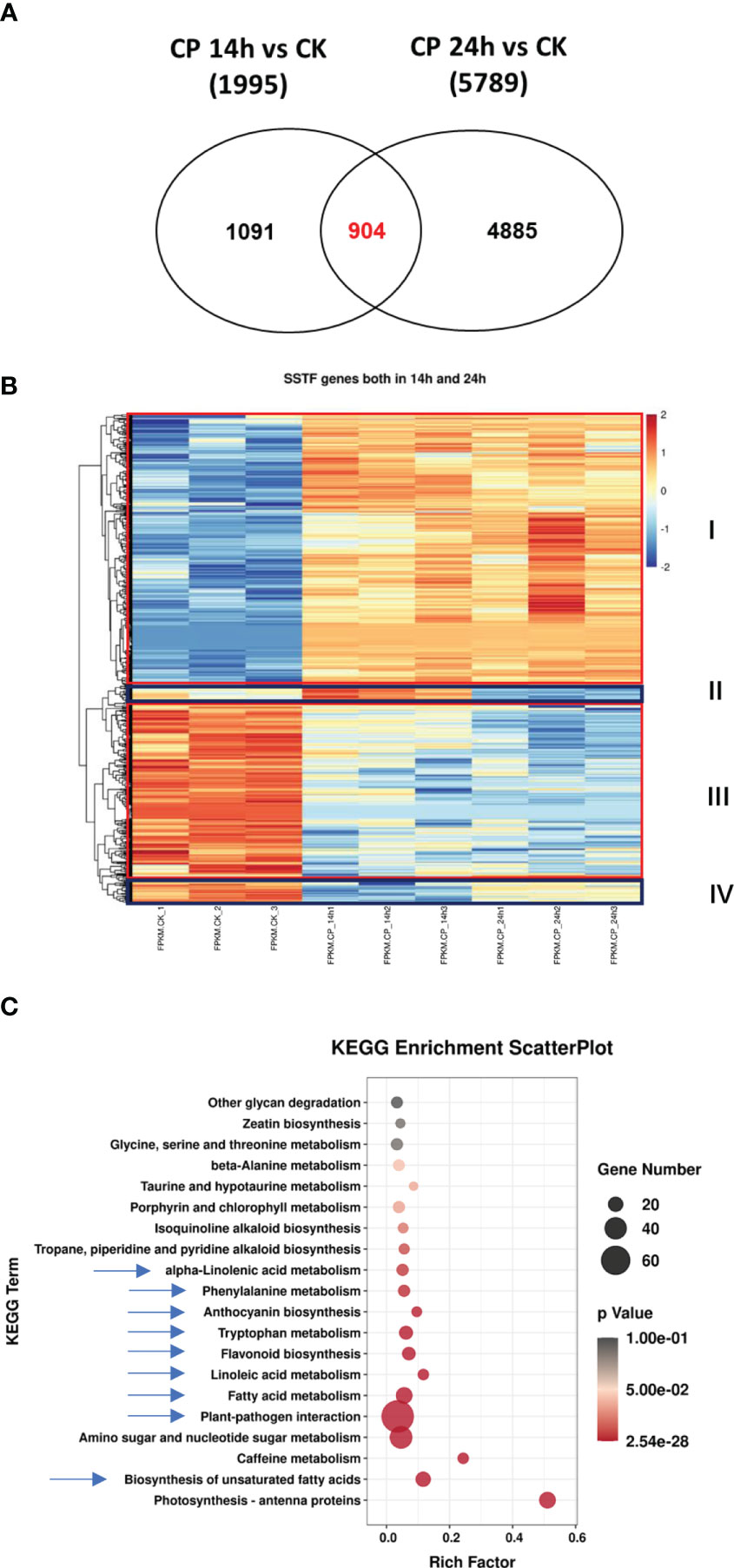
Figure 2 Characterization of the overlap genes upon Colletotrichum panacicola FSL infection at different times. (A) Venn diagram illustrating the total number and the number of common genes affected in ginseng 14 and 24 h post-C. panacicola FSL inoculation. (B) A heatmap analysis of common genes affected in ginseng 14 and 24 h post-C. panacicola FSL inoculation. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of common genes observed in ginseng at both 14 and 24 h post-C. panacicola FSL inoculation.
Similarly, when comparing healthy ginseng and red skin ginseng, GO enrichment analysis showed that DEGs were significantly more clustered into “response to chitin”, “transcription, DNA-templated”, and “defense response” terms, while the KEGG analysis indicated DEGs were greatly enriched in pathways such as “biosynthesis of unsaturated fatty acids” and “plant–pathogen interaction” (Luo et al., 2022). It was also reported that there was a significant difference in the soil microorganisms between healthy P. ginseng and heavily ginseng red skin root syndrome-diseased groups; the GO and KEGG term enrichment was very likely due to the different soil microorganisms (Dong et al., 2022). These studies together with our previous studies on ginseng–Botrytis cinerea interaction (Chen et al., 2022) revealed that the transcriptome associated with plant defense changed with diversity in P. ginseng upon C. panacicola infection.
Since C. panacicola infection downregulated approximately 59.6% of host genes 24 h after infection, we next wanted to know which pathways are involved. Supplementary Figure 4 reveals the GO terms in biological process, cellular component, and molecular function at 14 and 24 h. The upregulated and downregulated genes in each GO term were also included. As mentioned previously, C. panacicola infection altered the balance between induction and repression of many genes in ginseng. Supplementary Figure 4B clearly indicates the repression of genes in many GO terms upon pathogen infection at 24 h. These included the nucleus, DNA-templated regulation of transcription, DNA-templated transcription, DNA-binding transcription factor activity, and DNA binding.
We next analyzed the downregulated SSTF genes at 24 h. Approximately 3,200 genes were used in the heatmap analysis. As shown in Figure 3A, most genes were significantly decreased at 24 h when compared with CK. However, the expression level of many genes at 14 h was not seriously decreased and even remained at a higher level. It suggests that the downregulation of these genes happened from 14 to 24 h.
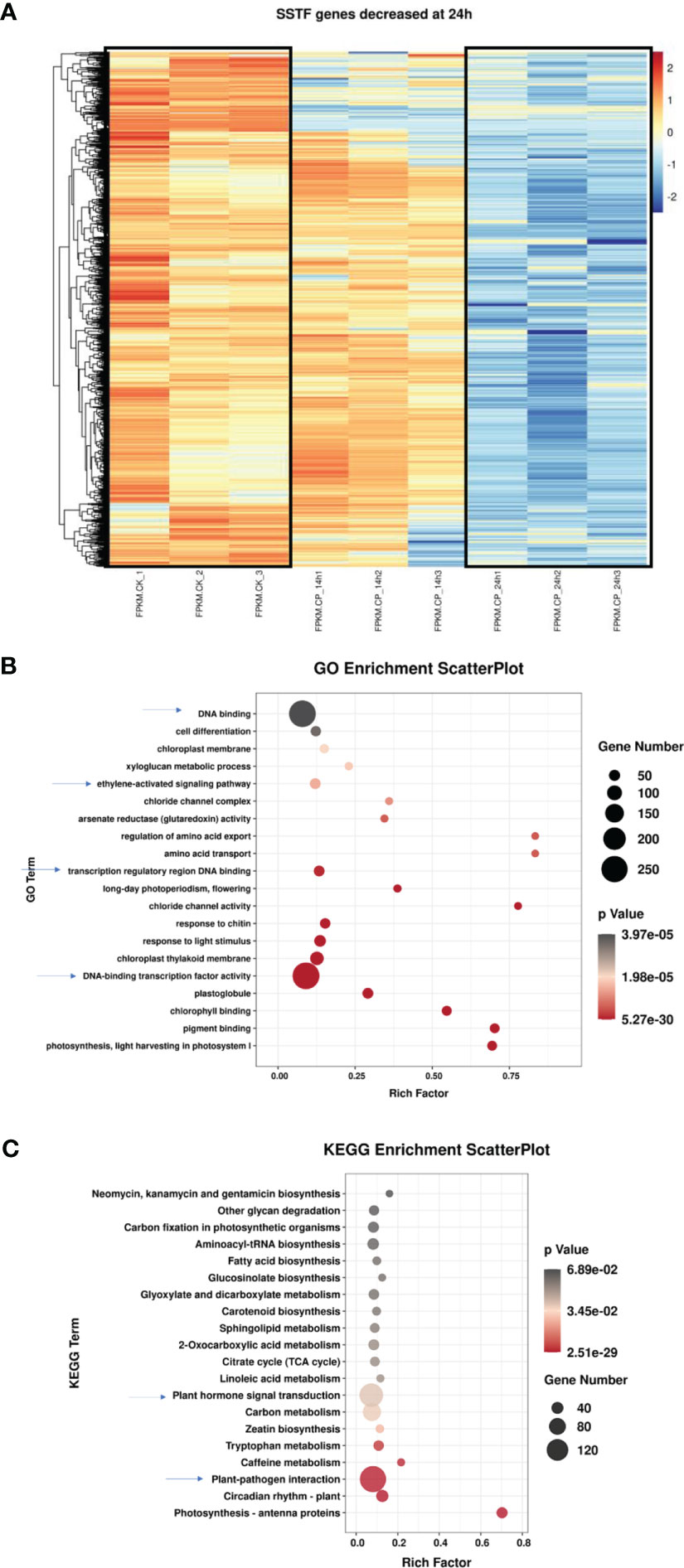
Figure 3 Characterization of the differentially expressed genes in ginseng upon Colletotrichum panacicola FSL infection at 24 h. (A) A heatmap analysis of differentially expressed genes in ginseng upon C. panacicola FSL infection. The indicated genes in ginseng were decreased at 24 h upon C. panacicola FSL infection. (B) Gene Ontology (GO) analysis of C. panacicola FSL repressed genes in ginseng at 24 h after infection. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of C. panacicola FSL repressed genes at 24 h after infection.
To obtain detailed pathway information of the downregulated SSTF genes at 24 h, GO and KEGG analyses were further performed based on 3,200 genes (Figures 3B, C). Plant–pathogen interaction, plant hormone signal transduction, transcription factors, and ethylene-activated signaling pathways were observed, suggesting their potential role in disease development. Similarly, the downregulated genes associated with defense responses, defense response to fungus, certain TFs, secondary metabolite biosynthetic process, and plant–pathogen interaction are enriched during the compatible interaction between ginseng and B. cinerea (Chen et al., 2022). Since TFs and hormones play an important role in plant defense against pathogens, here, the downregulation of TFs and hormones pathway genes in ginseng by C. panacicola might contribute to disease development.
Since secondary metabolites are involved in plant–pathogen interaction, the differentially expressed genes in the flavonoid biosynthesis pathway were checked. Genes involved in the flavonoid pathway were enriched at both 14 and 24 h. Approximately 52 genes were used for the heatmap analysis (Figure 4). At least three groups were observed. In group I, genes were decreased at 24 h when compared with CK and 14 h, suggesting their expression was repressed at a late stage. In group II, genes at 24 h were the highest compared with CK and 14 h, suggesting the induction of these genes at a late stage. In group III, the highest gene expression level was observed at 14 h, indicating an earlier response toward C. panacicola. The differential expression of genes in the flavonoid pathway suggests a differential role in ginseng–C. panacicola interaction. Similarly, when ginseng was attacked by B. cinerea, the GO term “regulation of flavonoid biosynthetic process” was enriched in downregulated genes (Chen et al., 2022). With this, many metabolites associated with flavonoids were decreased including kaempferol, quercetin, and luteolin. The decrease in flavonoids suggested that these compounds played a role in plant defense. Indeed, B. cinerea BcQdo is involved in the degradation of flavonoids such as kaempferol and quercetin, causing diseases (Chen et al., 2022).
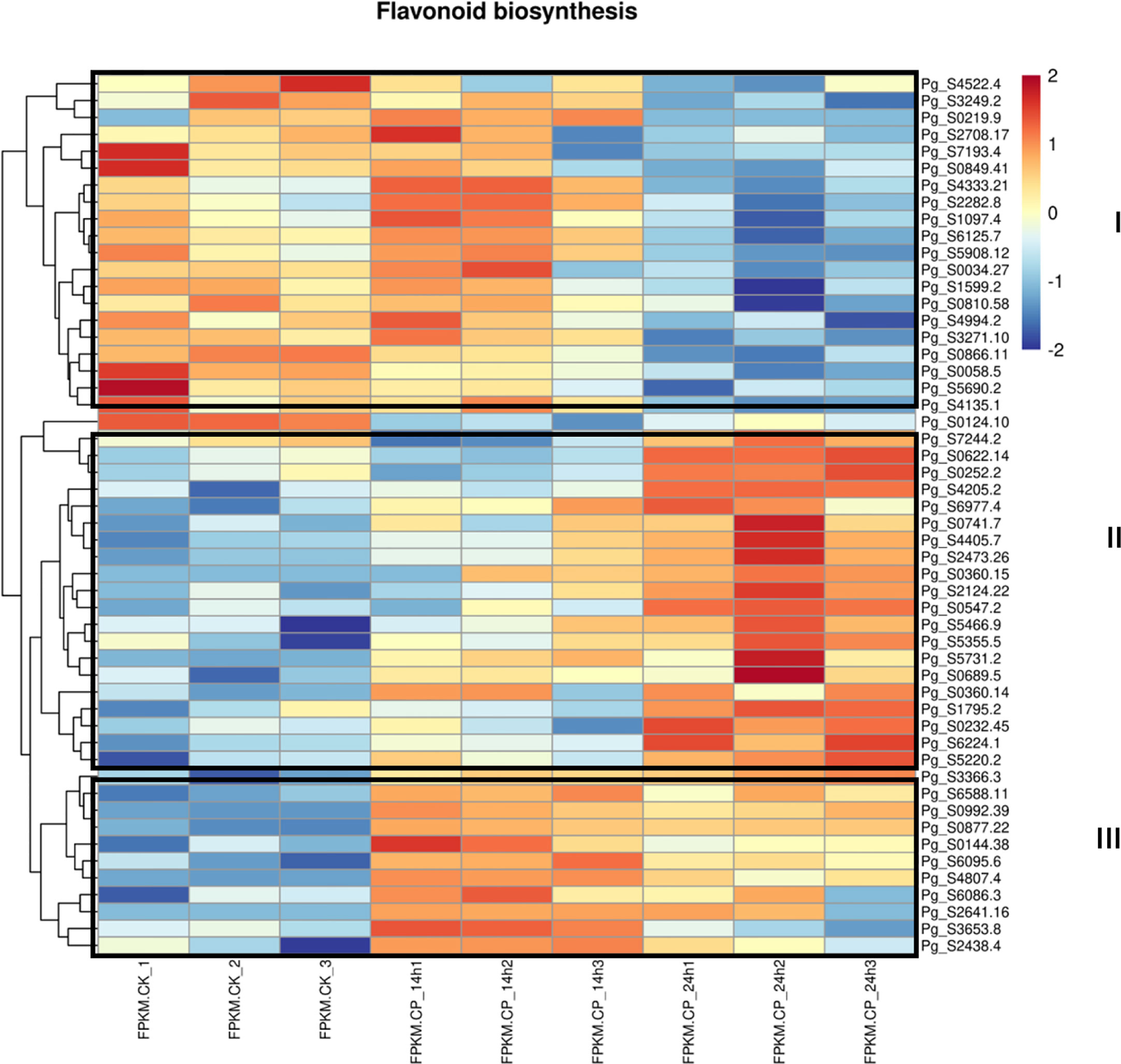
Figure 4 Heatmap analysis of differentially expressed genes in flavonoid pathway. Differentially expressed genes (DEGs) were observed in CK and Colletotrichum panacicola FSL-treated ginseng plants at 14 and 24 h.
Since genes in the flavonoid pathway are differentially expressed in ginseng upon C. panacicola infection, the study next analyzed if ginseng-related metabolites are affected. Two-year-old ginseng was sprayed with C. panacicola spores for 14 h (CpFSL) and harvested for ultrahigh-performance liquid chromatography–electrospray ionization–tandem mass spectrometry (UPLC-ESI-MS/MS) analysis (Supplementary Table 1; Supplementary Figure 5; Supplementary File 2). The plants without fungi treatment were used as control (CK). Four replicates were performed for each treatment. Seventeen metabolites in the flavonoid pathway were detected in ginseng leaves under this condition (Supplementary Table 2; Supplementary File 2). The heatmap of differentially accumulated compounds in the flavonoid pathway is shown in Figure 5A. At least two groups of differentially accumulated compounds were observed (framed). Group I indicated the increased chemicals in ginseng after C. panacicola infection, while group II indicated the decreased chemicals in ginseng. In particular, epicatechin, naringin, etc., were significantly decreased, suggesting that these compounds were suppressed by fungi (Figures 5B, C, Supplementary Figures 6, 7, Supplementary Table 2). In contrast, glycitin, vitexin/isovitexin, luteolin-7-O-glucoside, etc., were significantly induced by fungi (Figures 5D–F, Supplementary Figures 6, 7, Supplementary Table 2). Interestingly, ginseng compounds associated with flavonoids were also observed upon B. cinerea B05.10 infection (Chen et al., 2022). The accumulation of kaempferol, quercetin, and luteolin was reduced, while the concentration of hesperetin increased in ginseng by B. cinerea (Chen et al., 2022). Further studies indicated that kaempferol, quercetin, and luteolin had antifungal activity toward B. cinerea, suggesting that the reduction of these compounds could be a strategy for fungal virulence (Chen et al., 2022). Here, the changes of ginseng compounds in the flavonoid pathway upon C. panacicola treatment may play a similar role.
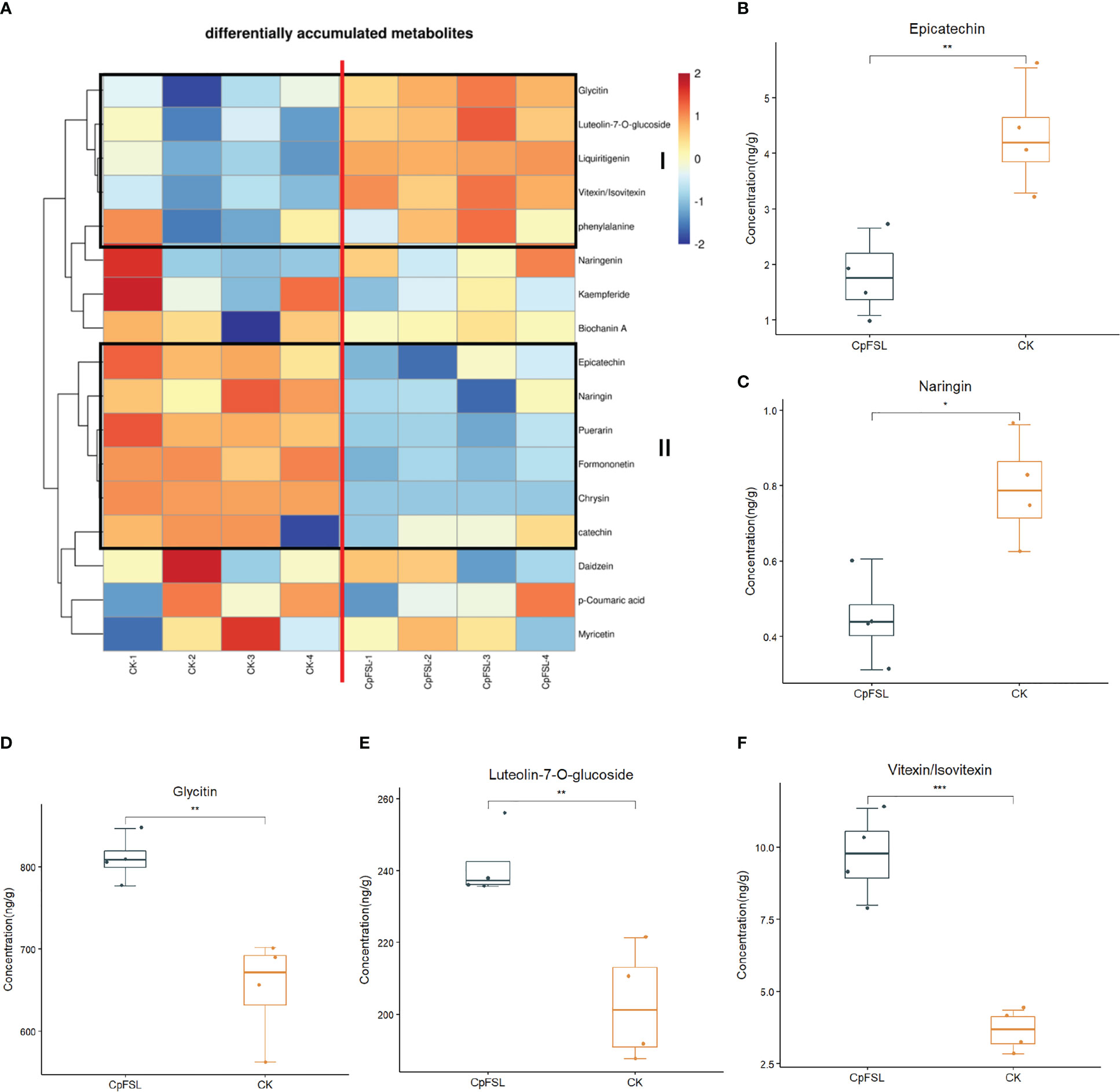
Figure 5 Differentially accumulated chemicals in ginseng plant associated with flavonoid pathway upon Colletotrichum panacicola FSL infection. (A) Heatmap analysis of differentially accumulated chemicals in flavonoid pathway upon C. panacicola FSL infection. (B–F) Differentially accumulated compounds in CK and C. panacicola FSL-treated ginseng including epicatechin (B), naringin (C), glycitin (D), luteolin-7-O-glucoside (E), and vitexin/isovitexin (F). Four replicates were performed, and the significance between different treatments was indicated as * p < 0.05, ** p < 0.01 and *** p < 0.001.
To know if the altered compounds have antifungal activity toward C. panacicola, the fungi were incubated on potato dextrose agar (PDA) plates with several chemicals under the concentration of 1.0 mg/ml or 10.0 μmol/ml. These compounds include p-coumaric acid, puerarin, naringin, myricetin, and chrysin. The fungus without any chemicals was used as a control. The diameters and the inhibition rates were calculated at 48, 72, 96, or 120 h. As indicated in Figure 6A, under PDA concentration of 1.0 mg/ml, many compounds could suppress C. panacicola growth in the early stage. p-Coumaric acid could significantly highly suppress C. panacicola growth since the colony sizes were the smallest at 72, 96, and 120 h post-incubation (Figures 6A, B). The inhibition rates for p-coumaric acid were approximately 100% at 72 h, 97.2% at 96 h, and 96.5% at 120 h after incubation (Figure 6D). Puerarin, naringin, and myricetin could also significantly suppress the fungal growth at different times (72, 96, and 120 h) (Figure 6), but the inhibition rates were lower than those of p-coumaric acid. These results showed that p-coumaric acid, puerarin, naringin, and myricetin could inhibit C. panacicola growth, which further indicated that certain components in flavonoids have antifungal activity. However, chrysin could only inhibit fungal growth at the early stage (Figures 6A, C, D). Similarly, under PDA concentration of 10.0 μmol/ml, p-coumaric acid could completely inhibit the fungal growth at different times (Supplementary Figures 9A, F), while puerarin exhibited the second higher antifungal activity toward C. panacicola (Supplementary Figures 9B, F). The other three compounds could also inhibit fungal growth to a certain degree at the early stage (Supplementary Figures 9C–F). Considering the reduction of naringin (Figure 5C, Supplementary Figure 6, Supplementary Table 2), puerarin (Supplementary Table 2), and p-coumaric acid (Supplementary Figure 8) during ginseng–C. panacicola interaction at the early stage, C. panacicola very likely developed a strategy to reduce the biosynthesis of these antifungal metabolites.
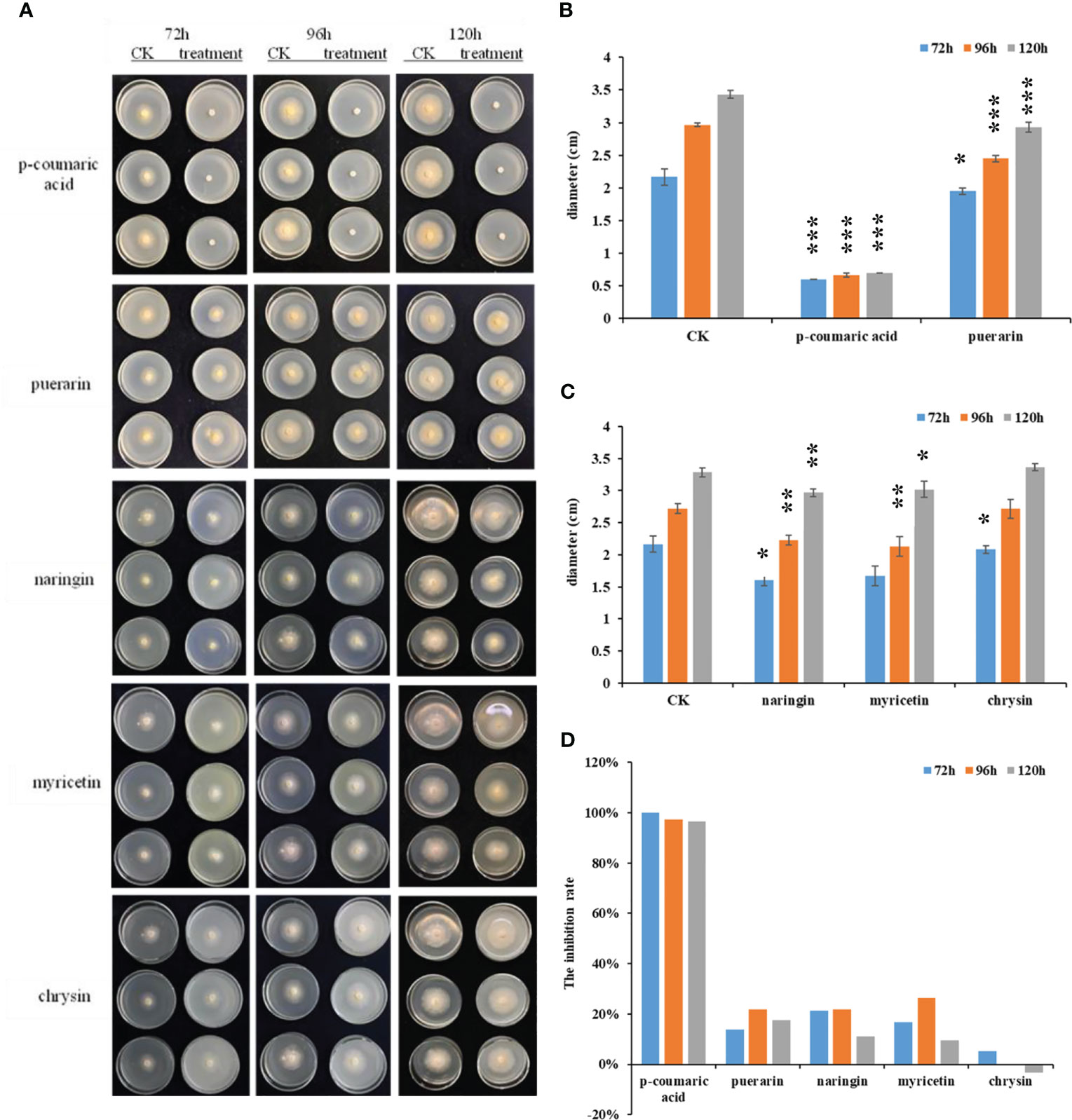
Figure 6 Antifungal activity of indicated chemicals toward Colletotrichum panacicola FSL. (A) Mycelial plugs of the C. panacicola FSL were inoculated on a potato dextrose agar (PDA) medium with different compounds including p-coumaric acid, puerarin, naringin, myricetin, and chrysin under the concentration of 1.0 mg/ml. The growth of the fungi was observed and photographed at 72, 96, and 120 h. (B) The colony diameters of p-coumaric acid and puerarin incubated fungus were determined at 72, 96, and 120 h. Asterisks indicate significant differences between untreated (CK) and chemicals treated (* p < 0.05, *** p < 0.001). (C) The colony diameters of naringin, myricetin, and chrysin incubated fungus were determined at 72 96, and 120 h. Asterisks indicate significant differences between untreated (CK) and chemicals treated (* p < 0.05; ** p < 0.01). (D) The relative inhibition rates of different compounds toward C. panacicola FSL were determined at 72, 96, and 120 h. All data represent means ± SD from at least three replicates.
Indeed, the expression of many genes in the metabolite biosynthesis pathway was differentially regulated upon C. panacicola interaction (Supplementary Figures 6, 7). For example, the expressions of Pg_S3366.3, Pg_S6977.4, Pg_S0360.15, Pg_S5731.2, and Pg_S6588.11 were increased upon C. panacicola infection, while the expression of Pg_S4522.4 was decreased (Supplementary Figures 6A, B). Here, Pg_S3366.3 encoded type III polyketide synthase B, Pg_S6977.4 and Pg_S6588.11 encoded enzymes belonging to EC:1.14.11.9 (flavanone 3-dioxygenase), Pg_S0360.15 encoded isoflavone reductase, Pg_S5731.2 encoded caffeoyl-CoA O-methyltransferase, while Pg_S4522.4 encoded chalcone isomerase. These enzymes are involved in flavonoid biosynthesis. The induction and suppression of these genes suggested their role in the changes of flavonoids during ginseng–C. panacicola interaction. However, two genes in the isoflavonoid biosynthesis pathway, Pg_S2855.12 and Pg_S0785.34, which encoded 2-hydroxyisoflavone dehydratase (EC:4.2.1.105), were differentially regulated upon C. panacicola infection (Supplementary Figures 7A, B). While the expression of Pg_S2855.12 was decreased at 14 h, Pg_S0785.34 was induced by C. panacicola at the same time. The increase and the decrease of the genes suggested their different roles in regulating isoflavonoid biosynthesis. Indeed, we observed that the accumulated metabolites in the isoflavonoid pathway were different upon C. panacicola infection (Supplementary Figure 7C).
The comparative transcriptome and metabolite analyses in this work provide molecular and chemical clues to understanding the complex interaction between garden ginseng and C. panacicola FSL. Here, C. panacicola strain FSL was originally isolated from ginseng leaves under forest conditions, which had higher virulence toward garden ginseng. Similarly, C. panacicola had also been reported to cause anthracnose disease in P. ginseng in Korea based on morphological, pathogenicity, cultural, and molecular studies (Choi et al., 2011). Recently, Liu et al. obtained 232 Colletotrichum isolates from 12 main ginseng-producing areas in Northeast China (Liu et al., 2020). Based on morphological characteristics and sequence analyses of specific domains in the fungal genome, 135 out of 232 isolates belonged to C. panacicola, while the other isolates were grouped with Colletotrichum lineola (Liu et al., 2020). Further work revealed the isolates of C. panacicola presented in all three provinces (Jilin, Liaoning, and Heilongjiang) in Northeast China, suggesting that C. panacicola was the widely distributed Colletotrichum species in China (Liu et al., 2020). Inoculations of ginseng showed that the isolates of C. panacicola could infect both leaves and stems with higher aggressiveness (Liu et al., 2020). In addition, Cui also isolated a virulent strain of C. panacicola from the wild ginseng in Jilin Province (Cui et al., 2020). All reports indicated that C. panacicola caused garden ginseng anthracnose disease.
Illumina-based RNA-seq of ginseng leaves revealed that plant transcripts changed upon C. panacicola FSL infection at different times. However, more than half of the transcripts in ginseng showed decreased expression at 24 h, indicating that ginseng defense responses were suppressed by C. panacicola from 14 to 24 h. This is consistent with our previous report that B. cinerea B05.10 infection downregulated approximately 60% of SSTFs in ginseng at 14 h, suggesting that these genes play a special role in plant–fungus interaction (Chen et al., 2022). Therefore, B. cinerea B05.10 promotes disease development in P. ginseng by suppressing many defense-related genes’ expression at the early stage. Here, C. panacicola FSL may use a similar strategy. Other reports also indicated the transcript changes in red skin root-affected ginseng fibrous roots (Gong et al., 2020). The fibrous roots of red skin root-affected ginseng showed 1,856 upregulated genes and 1,581 downregulated genes when compared with the healthy ginseng roots (Gong et al., 2020). Thus, P. ginseng changed its transcripts upon different stresses. Here, the differentially expressed genes in garden ginseng upon C. panacicola infection suggest their potential roles in defense. Indeed, we observed that the transcripts and the accumulation of antifungal compounds in the flavonoid pathway changed upon C. panacicola treatment.
Previous works indicate that plants had various mixtures and high amounts of antifungal compounds (Whitehead and Bowers, 2013), some of which provide the plants with a certain degree of basic resistance against pests (Schijlen et al., 2004; Treutter, 2005). For example, camalexin is a major phytoalexin in Arabidopsis thaliana, which is induced by a great number of biotrophic and necrotrophic plant pathogens (Glazebrook, 2005). Camalexin biosynthetic capacity is observed in both leaf and root upon infection with the Pythium sylvaticum (Bednarek et al., 2005). High camalexin concentrations were observed at the infection site by Alternaria alternata or B. cinerea (Kliebenstein et al., 2005; Ahuja et al., 2012). Camalexin can also be induced by PAMPs. For example, in oomycete-derived necrosis and ethylene-induced peptide 1 (Nep1)-like proteins, bacteria-derived peptidoglycans were involved in camalexin accumulation (Qutob et al., 2006; Gust et al., 2007). In addition, the changes in flavonoids have also been identified from various plant species to date (Li et al., 2018; Sudheeran et al., 2019; Yu et al., 2018). These compounds are not only involved in the innate defense response but also induced in the host’s response to pathogen attack (Heale, 1992; Jeandet et al., 1995). Metabolite analysis of P. ginseng indicates that in many of the secondary metabolites in the flavone and flavonol biosynthesis pathway, isoflavonoid biosynthesis is influenced by B. cinerea B05.10 (Chen et al., 2022). It also indicated the red mango fruit had more anthocyanin and flavonols than the green mango fruit, which had antifungal activity against the fungus Colletotrichum gloeosporioides (Sudheeran et al., 2020). Previous reports have also shown that quercetin and cyanidin aglycones could inhibit C. gloeosporioides hyphal growth and conidial germination (Puupponen-Pimiä et al., 2001; Ramos et al., 2006; Mikulic-Petkovsek et al., 2013). Since certain flavonoids could inhibit C. panacicola growth; here, the alteration of ginseng flavonoids at the early stage would be a candidate strategy for C. panacicola to cause disease development.
The comparative transcriptome analysis in this work provides molecular clues to understanding the mechanism of ginseng susceptibility toward C. panacicola. Illumina-based RNA-seq of ginseng leaves revealed that plant transcripts changed upon C. panacicola infection at different times. More than half of the transcripts in ginseng showed decreased expression at 24 h, indicating that ginseng defense responses were suppressed by C. panacicola from 14 to 24 h. In particular, the transcripts and the accumulation of certain antifungal compounds in the flavonoid pathway changed upon C. panacicola treatment. We hope this finding can help in understanding the pathogenicity of C. panacicola in promoting P. ginseng anthracnose disease development.
Two-year-old garden ginseng was grown in a pathogen-free chamber. C. panacicola strain FSL was isolated from ginseng leaves in the Fusong forest, Jilin Province, and was grown on a PDA plate. Infection of C. panacicola on ginseng leaves was performed (Chen et al., 2022). For ginseng mRNA sequencing and gene qPCR analysis, the spores were spray incubated on healthy, undetached ginseng at different times. For mRNA sequencing, ginseng leaves were harvested at 14 h (Cp14h) and 24 h (Cp24h). For control, ginseng leaves were only incubated with ddH2O. For qPCR analysis, the leaves were harvested at 14, 24, and 48 h. Three independent biological replicates were performed. All samples were frozen at −80°C until use. The primers for qPCR are shown in Supplementary Table 3.
Total RNA extraction, purification, monitoring, cDNA library construction and sequencing, raw data cleaning, and analyzing were performed as previously indicated (Chen et al., 2022). Reference genome information was observed from the ginseng genome database website (http://ginsengdb.snu.ac.kr/). An index of the reference genome was built, and paired-end clean reads were aligned by using the HISAT package, while the FPKM of each gene was finally calculated (Chen et al., 2022).
Differential expression analysis of genes in all samples (CK1-3, Cp14h1-3, and Cp24h1-3) was performed as previously described (Chen et al., 2022). The differentially expressed genes were identified with log2 (fold change) >1 or log2 (fold change) <−1 and with statistical significance (p-value < 0.05) using the R package.
GO enrichment and KEGG analyses of differentially expressed genes were then performed (Chen et al., 2022).
The flavonoids were extracted and measured according to the methods described previously, with minor modifications (Mi et al., 2022). Approximately 100 mg of ginseng leaves was extracted two times with 300 µl of methanol:acetonitrile:water (2:2:1, v/v) in an ultrasonic sonicator for 0.5 h at 4°C. The sample was incubated at −20°C for 1 h for protein deposition. After centrifugation (14,000 rpm, 20 min, 4°C), the supernatants were collected, freeze-dried, and stored at −80°C. Then, the supernatants were filtered, collected, and separated on a Waters C18 column (2.1 mm × 150 mm, 1.7 µm) with a flow rate of 400 µl/min. For the separation, solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid) were used under the gradient, as follows: 0–3.0 min, 5%–20% B; 3.0–4.3 min, 20% B; 4.3–9.0 min, 20%–45% B; 9.0–11.0 min, 45%–98% B; 11.0–13.0 min, 98% B; 13.0–13.1 min, 98%–5% B; and 13.1–15.0 min, 5% B. The auto-sampler temperature was set to 4°C, and 2 µl of the extraction was injected for analysis.
Mass spectrometric detection was carried out using an ABSCIEX QTRAP 6500 mass spectrometer coupled with an electrospray ionization (ESI) source under both negative (−) and positive (+) ion modes. The multiple reaction monitoring (MRM) scan mode was used to quantify the individual flavonoid compounds. All samples were equally mixed for quality control (QC) to indicate stability and repeatability during the experiments. Relative standard deviation (RSD) in QC samples was calculated, and the RSD% below 15% was considered and further analyzed. Metabolites ferulic acid, p-coumaric acid, glycetein, glycitin, daidzein, puerarin, formononetin, daidzin, biochanin A, genistin, genistein, apigenin, myricetin, quercetin, isorhamnetin, luteolin, kaempferol, kaempferide, sakuranetin, quercetin 3-glucoside, liquiritigenin, catechin, dihydrokaempferol, quercitrin, eriodictyol, gallocatechin, epicatechin, naringenin, rutin, epigallocatechin, isoliquiritigenin, chrysin, luteolin-7-O-glucoside, naringin, taxifolin, isovitexin/vitexin, and butin were used as standards for sample quantification (Sigma Chemical Company, St. Louis, MO, USA; Supplementary Table 1).
The antifungal activity of flavonoids toward C. panacicola was assessed by measuring the mycelial plug growth (Chen et al., 2022). The indicated flavonoids (98%) were first dissolved in methanol or ethanol and then pre-mixed with a sterile melting PDA medium to obtain the final concentration (1.0 mg/ml or 10.0 μmol/ml). The incubation of fungi and the measurement of mycelial growth were performed by Chen et al. (2022). Each experiment was performed in three replicates.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE226837.
SL designed the research plan and wrote the article, JX, NL, JH, JS, and SL performed the research. All authors contributed to the article and approved the submitted version.
This work was partly supported by the National Natural Science Foundation of China (No. 32171801 to SL), the Jilin province science and technology development project of China (No. 20210202095NC to NL), and the Cross-Disciplinary Innovation Founding of Jilin University (No. JLUXKJC2020313 to SL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1182685/full#supplementary-material
Supplementary Figure 1 | C. panacicola FSL infection phenotype on 2-years old ginseng leaves at 14 hours, 24 hours and 48 hours, respectively.
Supplementary Figure 2 | GO/KEGG analysis of significantly differentially expressed genes in ginseng upon C. panacicola FSL treatment at 14 hours.
Supplementary Figure 3 | GO/KEGG analysis of significantly differentially expressed genes in ginseng upon C. panacicola FSL treatment at 24 hours.
Supplementary Figure 4 | GO enrichment analysis of up- and down- regulated SSTF genes in ginseng upon C. panacicola FSL treatment at 14 hours and 24 hours, respectively.
Supplementary Figure 5 | XIC map of standard mixtures and QC RSD percentage of ginseng samples in this work. QC RSD% < 15% was considered.
Supplementary Figure 6 | Differentially accumulated chemicals and genes in flavonoid biosynthesis pathway upon C. panacicola FSL treatment.
Supplementary Figure 7 | Differentially accumulated chemicals and genes in isoflavonoid biosynthesis pathway upon C. panacicola FSL treatment.
Supplementary Figure 8 | Differentially accumulated chemicals in phenylpropanoid biosynthesis pathway upon C. panacicola FSL treatment.
Supplementary Figure 9 | Anti-fungi activity of indicated chemicals towards C. panacicola FSL under the concentration of 10.0 umol/mL.
Supplementary File 1 | Total validated high-quality reads were obtained from all libraries.
Supplementary File 2 | XIC maps of selected compounds observed in untreated control (CK) or C. panacicola FSL treated ginseng leaves (CpFSL).
Ahuja, I., Kissen, R., Bones, A. M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90. doi: 10.1016/j.tplants.2011.11.002
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Bednarek, P., Schneider, B., Svatos, A., Oldham, N. J., Hahlbrock, K. (2005). Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in arabidopsis roots. Plant Physiol. 138, 1058–1070. doi: 10.1104/pp.104.057794
Birkenbihl, R. P., Liu, S., Somssich, I. E. (2017). Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9. doi: 10.1016/j.pbi.2017.04.004
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., Zhao, Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Chen, L., Yu, D. (2014). “ABA regulation of plant response to biotic stresses,” in Abscisic acid: metabolism, transport and signaling. Ed. Zhang, D.-P. (Dordrecht: Springer Netherlands), 409–429. doi: 10.1007/978-94-017-9424-4_20
Chen, H., Zhang, S., He, S., Wang, M., Liu, S. (2022). The necrotroph Botrytis cinerea promotes disease development in Panax ginseng by manipulating plant defense signals and antifungal metabolites degradation. J. Ginseng Res. 46, 790–800. doi: 10.1016/j.jgr.2022.03.005
Choi, K. J., Kim, W. G., Kim, H. G., Choi, H. W., Lee, Y. K., Lee, B. D., et al. (2011). Morphology, molecular phytogeny and pathogenicity of Colletotrichum panacicola causing anthracnose of Korean ginseng. Plant Pathol. J. 27, 1–7. doi: 10.5423/ppj.2011.27.1.001
Chung, H.S., Bae, H.W. (1979). Ginseng anthracnose in Korea: factors affecting primary inoculum, growth of the pathogen, disease development and control. Kor. J. Plant Protect. 18, 35–41.
Cui, L., Feng, Z., Song, M., Ding, X. (2020). Isolation and identification of anthracnose pathogen in wild ginseng. Special Wild Economic Anim. Plant Res. 042, 25–29. doi: 10.16720/j.cnki.tcyj.2020.03.006
Denancé, N., Sánchez-Vallet, A., Goffner, D., Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00155
Dong, L., Bian, X., Zhao, Y., Yang, H., Xu, Y., Han, Y., et al. (2022). Rhizosphere analysis of field-grown Panax ginseng with different degrees of red skin provides the basis for preventing red skin syndrome. BMC Microbiol. 22, 12. doi: 10.1186/s12866-021-02430-9
Gao, J., Wang, Y., Guan, Y. M., Chen, C. Q. (2014). Fusarium cerealis, a new pathogen causing ginseng (Panax ginseng) root rot in China. Plant Dis. 98, 1433. doi: 10.1094/pdis-03-14-0328-pdn
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Gong, L., Gao, J., Xu, T., Qu, J., Wang, Z., Yang, Z., et al. (2020). Transcriptome analysis of field-grown Asian ginseng provides clues to environmental conditions and developmental mechanisms related to red skin root syndrome. Ind. Crops Prod. 153, 112486. doi: 10.1016/j.indcrop.2020.112486
Guan, Y. M., Lu, B. H., Wang, Y., Gao, J., Wu, L. J. (2014). First report of root rot caused by Fusarium redolens on ginseng (Panax ginseng) in jilin province of China. Plant Dis. 98, 844. doi: 10.1094/pdis-09-13-0922-pdn
Guerreiro, A., Figueiredo, J., Sousa Silva, M., Figueiredo, A. (2016). Linking jasmonic acid to grapevine resistance against the biotrophic oomycete Plasmopara viticola. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00565
Gust, A. A., Biswas, R., Lenz, H. D., Rauhut, T., Ranf, S., Kemmerling, B., et al. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in arabidopsis. J. Biol. Chem. 282, 32338–32348. doi: 10.1074/jbc.M704886200
Heale, J. B. (1992). “Activation of host defence mechanisms in response to botrytis cinerea,” in Recent advances in botrytis research. Eds. K Verhoeff, N. M., Williamson, B. (Wageningen, the Netherlands: Pudoc Scientific), 48–58.
Jayakodi, M., Choi, B. S., Lee, S. C., Kim, N. H., Park, J. Y., Jang, W., et al. (2018). Ginseng genome database: an open-access platform for genomics of Panax ginseng. BMC Plant Biol. 18, 62. doi: 10.1186/s12870-018-1282-9
Jeandet, P., Bessis, R., Sbaghi, M., Meunier, P. (1995). Production of the phytoalexin resveratrol by grapes as a response to Botrytis attack under natural conditions. J. Phytopathol. 143, 135–139. doi: 10.1111/j.1439-0434.1995.tb00246.x
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, J. D., Vance, R. E., Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395. doi: 10.1126/science.aaf6395
Kim, Y. C., Lee, J. H., Bae, Y.-S., Sohn, B.-K., Park, S. K. (2010). Development of effective environmentally-friendly approaches to control Alternaria blight and anthracnose diseases of Korean ginseng. Eur. J. Plant Pathol. 127, 443–450. doi: 10.1007/s10658-010-9610-4
Kim, H., Mohanta, T. K., Park, Y.-H., Park, S.-C., Shanmugam, G., Park, J.-S., et al. (2019). Complete genome sequence of the mountain-cultivated ginseng endophyte Burkholderia stabilis and its antimicrobial compounds against ginseng root rot disease. Biol. Control 140, 104126. doi: 10.1016/j.biocontrol.2019.104126
Kliebenstein, D. J., Rowe, H. C., Denby, K. J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J. 44, 25–36. doi: 10.1111/j.1365-313X.2005.02508.x
Li, Y., Fang, J., Qi, X., Lin, M., Zhong, Y., Sun, L., et al. (2018). Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 19:1471. doi: 10.3390/ijms19051471
Liu, L., Sonbol, F. M., Huot, B., Gu, Y., Withers, J., Mwimba, M., et al. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7, 13099. doi: 10.1038/ncomms13099
Liu, L., Zhang, L., Wang, Y., Zhan, H., Yang, J., Peng, L., et al. (2020). Distribution, identification and characterization of Colletotrichum lineola and C. panacicola causing anthracnose on ginseng in northeast China. Crop Protection 137, 105265. doi: 10.1016/j.cropro.2020.105265
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P., Somssich, I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4, e07295. doi: 10.7554/eLife.07295
Liu, S., Ziegler, J., Zeier, J., Birkenbihl, R. P., Somssich, I. E. (2017). Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ 40, 2189–2206. doi: 10.1111/pce.13022
Lu, B. H., Wang, X. H., Wang, R., Wang, X., Yang, L. N., Liu, L. P., et al. (2019). First report of Botrytis pelargonii causing postharvest gray mold on fresh ginseng roots in China. Plant Dis. 103, 149. doi: 10.1094/PDIS-01-17-0031-PDN
Luo, M., Li, A.X., Wang, F.Q., You, J.F. (2022). Integrative analysis of multiple metabolomes and transcriptome revealed color expression mechanism in red skin root syndrome of panax ginseng. Ind Crop Prod 177, 114491. doi: 10.1016/j.indcrop.2021.114491
Mi, S., Zhang, X., Wang, Y., Zheng, M., Zhao, J., Gong, H., et al. (2022). Effect of different genotypes on the fruit volatile profiles, flavonoid composition and antioxidant activities of chilli peppers. Food Chem. 374, 131751. doi: 10.1016/j.foodchem.2021.131751
Mikulic-Petkovsek, M., Slatnar, A., Schmitzer, V., Stampar, F., Veberic, R., Koron, D. (2013). Chemical profile of black currant fruit modified by different degree of infection with black currant leaf spot. Sci. Horticult. 150, 399–409. doi: 10.1016/j.scienta.2012.11.038
Ngou, B. P. M., Ahn, H. K., Ding, P., Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Pedras, M. S., Yaya, E. E., Glawischnig, E. (2011). The phytoalexins from cultivated and wild crucifers: chemistry and biology. Nat. Prod. Rep. 28, 1381–1405. doi: 10.1039/c1np00020a
Puupponen-Pimiä, R., Nohynek, L., Meier, C., Kähkönen, M., Heinonen, M., Hopia, A., et al. (2001). Antimicrobial properties of phenolic compounds from berries. J. Appl. Microbiol. 90, 494–507. doi: 10.1046/j.1365-2672.2001.01271.x
Qutob, D., Kemmerling, B., Brunner, F., Küfner, I., Engelhardt, S., Gust, A. A., et al. (2006). Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell. 18, 3721–3744. doi: 10.1105/tpc.106.044180
Ramos, F. A., Takaishi, Y., Shirotori, M., Kawaguchi, Y., Tsuchiya, K., Shibata, H., et al. (2006). Antibacterial and antioxidant activities of quercetin oxidation products from yellow onion (Allium cepa) skin. J. Agric. Food Chem. 54, 3551–3557. doi: 10.1021/jf060251c
Ryu, H., Park, H., Suh, D. S., Jung, G. H., Park, K., Lee, B. D. (2014). Biological control of Colletotrichum panacicola on Panax ginseng by Bacillus subtilis HK-CSM-1. J. Ginseng Res. 38, 215–219. doi: 10.1016/j.jgr.2014.05.001
Schijlen, E. G., Ric de Vos, C. H., van Tunen, A. J., Bovy, A. G. (2004). Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65, 2631–2648. doi: 10.1016/j.phytochem.2004.07.028
Sudheeran, P. K., Ovadia, R., Galsarker, O., Maoz, I., Sela, N., Maurer, D., et al. (2020). Glycosylated flavonoids: fruit's concealed antifungal arsenal. New Phytol. 225, 1788–1798. doi: 10.1111/nph.16251
Sun, Z., Yang, L., Zhang, L., Han, M. (2018). An investigation of Panax ginseng Meyer growth promotion and the biocontrol potential of antagonistic bacteria against ginseng black spot. J. Ginseng Res. 42, 304–311. doi: 10.1016/j.jgr.2017.03.012
Treutter, D. (2005). Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. (Stuttg). 7, 581–591. doi: 10.1055/s-2005-873009
Tsuda, K., Sato, M., Stoddard, T., Glazebrook, J., Katagiri, F. (2009). Network properties of robust immunity in plants. PloS Genet. 5, e1000772. doi: 10.1371/journal.pgen.1000772
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Wang, W., Feng, B., Zhou, J. M., Tang, D. (2020). Plant immune signaling: advancing on two frontiers. J. Integr. Plant Biol. 62, 2–24. doi: 10.1111/jipb.12898
Wang, D., Wang, S. R., Yang, M. J., Lu, B. H., Liu, L. P., Gao, J. (2018). Detection of the resistance of Alternaria panax and cross-resistance on ginseng in jilin province. Agrochemicals 057, 603–605. doi: 10.16820/j.cnki.1006-0413.2018.08.016
Whitehead, S. R., Bowers, M. D. (2013). Evidence for the adaptive significance of secondary compounds in vertebrate-dispersed fruits. Am. Nat. 182, 563–577. doi: 10.1086/673258
Yu, C., Luo, X., Zhan, X., Hao, J., Zhang, L., YB, L. S., et al. (2018). Comparative metabolomics reveals the metabolic variations between two endangered Taxus species (T. fuana and T. yunnanensis) in the Himalayas. BMC Plant Biol. 18, 197. doi: 10.1186/s12870-018-1412-4
Keywords: Panax ginseng, Colletotrichum panacicola, transcriptome, anthracnose disease, flavonoids
Citation: Xia J, Liu N, Han J, Sun J, Xu T and Liu S (2023) Transcriptome and metabolite analyses indicated the underlying molecular responses of Asian ginseng (Panax ginseng) toward Colletotrichum panacicola infection. Front. Plant Sci. 14:1182685. doi: 10.3389/fpls.2023.1182685
Received: 09 March 2023; Accepted: 19 June 2023;
Published: 10 July 2023.
Edited by:
Simone Ferrari, Sapienza University of Rome, ItalyCopyright © 2023 Xia, Liu, Han, Sun, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouan Liu, c2hvdWFuLmNhbWVsbGlhQG91dGxvb2suY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.