- 1Laboratoire des Grandes Cultures, Institut National de la Recherche Agronomique de Tunisie, Université de Carthage, Ariana, Tunisia
- 2INRA, Unité Mixte de Recherche (UMR) 1349 Institut de Génétique, environnement et Protection des Plantes (IGEPP), Le Rheu, France
- 3Institut Agro-Rennes-Angers, UP ESP, Rennes, France
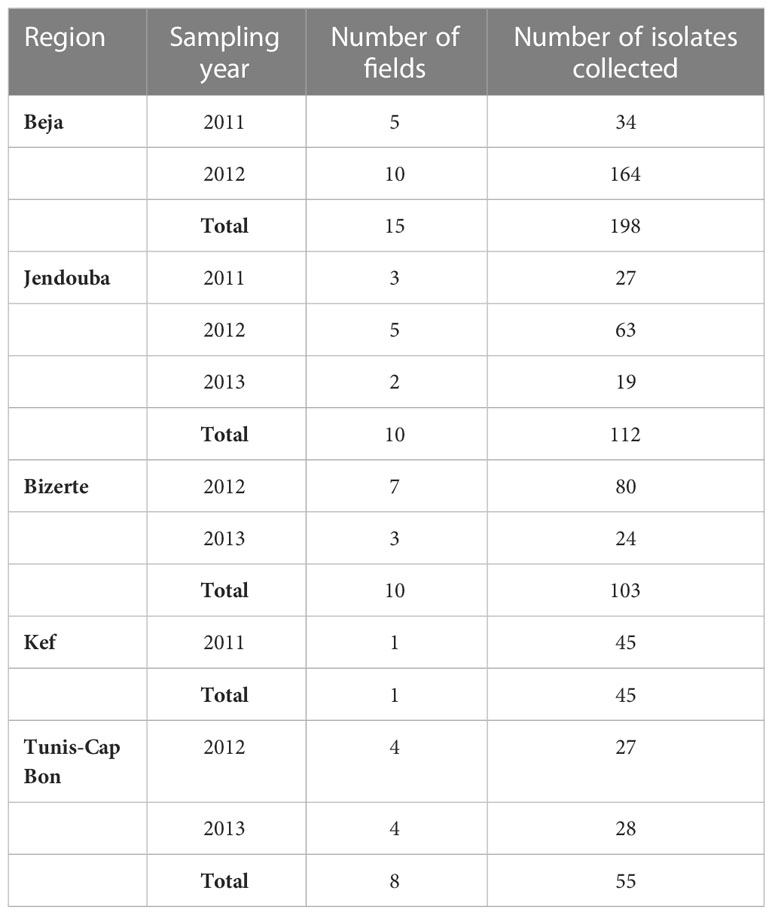
Faba bean ascochyta blight, caused by Ascochyta fabae Speg. (teleomorph: Didymella fabae Punith.), is one of the most devastating diseases of the crop. It can cause yield losses that reach 95% in conducive weather conditions. Surveys were carried out in five regions of Tunisia: Beja, Bizerte, Jendouba, Kef and Tunis-Cap Bon. A total of 513 fungal isolates were collected from 2011 to 2013. A molecular characterization was conducted to identify the mating type of each individual using a mating type specific PCR. Results revealed that the two mating types MAT1-2 and MAT1-1 coexisted in all surveyed regions. An imbalance in favor of MAT1-2 was observed particularly in Bizerte and Jendouba regions (sex ratio was 18:85 and 32:80, respectively). Moreover, morphological and pathogenic characterization of 122 isolates among the collection revealed a significant variability in conidia type (one celled or two celled conidia) frequency, in conidia mean size and in aggressiveness toward Badii faba bean cultivar (incubation period, IP; percentage necrotic leaf area, S; and area under disease progression curve, AUDPC). A principal component analysis (PCA) performed on morphologically studied parameters (frequency of conidia cell number and conidia mean size) identified three groups of isolates based on morphological traits: one celled (1C) and two celled (2C) conidia rates, one celled and two celled conidia length and width (1L, 1W, 2L and 2W, respectively). A second PCA using aggressiveness parameters (IP: Incubation period, S1, S4 and S9: percentage of necrotic leaf area respectively 5, 20 and 45 days after inoculation) identified three distinct pathogenic groups: poorly pathogenic AG1, moderately pathogenic AG2 and highly pathogenic AG3. Morphological and pathogenic groups and mating type data were used to conduct a multiple factorial correspondence analysis (MFCA) which revealed a correlation between the variables studied. Five groups were identified, each associated with a morphological and pathogenic trait and mating type. The most pathogenic group belonged to MAT1-2 suggesting that in locations where MAT1-2 is prevalent the epidemic risk is more important.
Introduction
Faba bean is one of the most commonly grown legumes in Tunisia. Ascochyta blight caused by Didymella fabae Jellis and Punith (anamorph Ascochyta fabae Speg.) is a widely spread disease throughout the world particularly in temperate regions (Maurin et al., 1990). It is also one of the most destructive foliar diseases of faba bean fields in Tunisia (Kharrat et al., 2006). The pathogen infects all aerial part of the plant causing necrotic lesions on leaves and pods and enlarged lesions on the stem and the petiole. It leads to yield losses estimated at 10 - 30% which may reach 95% in favorable conditions (Hanounik, 1980; Kharrat et al., 2006).
D. fabae (anamoph: Ascochyta fabae Speg.) was first described in England in 1991 by Jellis and Punithalingam. The genus Didymella is part of Didymellaceae Family, Pleosporales Order, Dothideomycetes Class, Pezizomycotina Sub-phylum and Ascomycota Phylum (Nasraoui, 2015). The anamorph, A. fabae, Speg. first described in Argentina by Spegazzini in 1899 (Maurin et al., 1990), belongs to the group of Deuteromycetes (imperfect fungi). It is a phialospore species that is part of the subgroup of anamorphic pycnidia fungi (Nasraoui, 2015). A. fabae grows on oat agar (OA) medium, in a yellowish colony (Jellis and Punithalingam, 1991). Pycnidia represent fruiting bodies of asexual reproduction that produce hyaline, elliptical, straight or slightly curved conidia with a truncated or rounded base and a rounded apex (Jellis and Punithalingam, 1991). They are predominantly two celled (97%) with a nucleus in each cell (Peever, 2007). Their dimensions are 16-19 x 3.5-4.5 µm (Jellis and Punithalingam, 1991; Omri Ben Youssef et al., 2012). Several studies have reported significant variability of morphological characters within populations of A. fabae (Kharbanda and Bernier, 1980; Maurin and Tivoli, 1992; Kohpina et al., 1999; Kharrat et al., 2000). Various parameters relating to A. fabae development were considered, including mycelium aspect (aerial or diffuse…), pycnidia pigmentation (brown, black, orange) and colony margin (regular or irregular). The characterization of 30 isolates of A. fabae from Tunisia, France and Algeria, carried out by Kharrat et al. (2000), confirmed this morphological variability. However, the diversity of these criteria appeared to be not linked to the geographic origin of the strains especially since Maurin (1989) observed significant variability even among conidia originating from the same pycnidia.
The pathogenicity variation of A. fabae has been reported in numerous studies (Hanounik and Robertson, 1989; Maurin, 1989; Rashid et al., 1991; Onfroy et al., 1999; Kharrat et al., 2000; Omri Ben Youssef et al., 2016; Blake et al., 2022). All these studies showed significant different interactions of faba bean lines when inoculated with different isolates suggesting significant pathogenic variability of the fungus and the presence of physiological specialization (Tivoli et al., 2006). However, physiological races in A. fabae remain a subject of controversy as some authors support the hypothesis of race presence (Hanounik and Robertson, 1989; Rashid et al., 1991), while others agree on the presence of a certain specialization without being able to identify physiological races (Maurin, 1989; Beed et al., 1994; Kharrat et al., 2000). In fact, Vicia faba resistance to A. fabae is polygenic and involves simultaneously major genes and other minor genes, making the distinction between the host resistance reaction and sensitivity difficult (Kharrat, 1999).
Ascochyta fabae is a haploid heterothallic pathogen that can maintain itself during the off-season on different sources such as seeds and plant debris (Kaiser, 1997; Tivoli and Banniza, 2007). It is maintained on plant debris under both anamorph and teleomorph forms for at least one season (Gossen and Morrall, 1986; Pedersen et al., 1993; Rubiales and Trapero-Casas, 2002; Omri Ben Youssef et al., 2012). In heterothallic species, the mating-type locus contains one of two dissimilar forms of sequences called idiomorphs since the alternative alleles of mating-type locus are dissimilar, but are located at the same chromosomal location within the genome (Metzenberg and Glass, 1990). Conventionally, mating type idiomorphs of complementary isolates are called MAT1-1 and MAT1-2 (Turgeon and Yoder, 2000). In Dothideomycetes and particularly in Didymella genus, the idiomorph MAT1-1 corresponds to a unique regulatory gene MAT1-1-1 characterized by the presence of an ORF (Open Reading Frame) region encoding a protein with a motif called the α domain, initially discovered in the MATα1 transcription factor of Saccharomyces cerevisiae (Barve et al., 2003; Chérif et al., 2006). The idiomorph MAT-1-2 corresponds to the MAT-1-2-1 gene, characterized by the presence of an ORF zone encoding a regulatory protein with a high mobility domain (HMG) (Turgeon et al., 1993; Coppin et al., 1997; Turgeon, 1998; Turgeon and Yoder, 2000; Barve et al., 2003; Chérif et al., 2006; Debuchy and Turgeon, 2006). Idiomorphs are flanked by DNA sequences, common to both sex types (Kronstad and Staben, 1997), with unknown molecular function but not essential for reproduction (Debuchy and Turgeon, 2006).The development of specific primers to each idiomorphic sequence at the MAT locus of A. fabae (Chérif et al., 2006) made it possible to address questions such as the spatial and temporal distribution of these mating types, their respective prevalence within populations and finally the impact of this distribution on the diversity and genetic structure of pathogen populations (Milgroom, 1996; Barve et al., 2003; Chérif et al., 2006). The first studies dealing with A. fabae mating type distribution and involvement in genetic structure were those of Ozkilinc et al. (2015) and Omri Ben Youssef et al. (2019) who studied the role of mating types in the population diversity of Syrian and Tunisian collections of A. fabae. Omri Ben Youssef et al. (2019) showed that among their 240 Tunisian isolates MAT1-2 was more common in Tunisia than MAT1-1, and Tunisian populations had a skewed distribution (1:2 ratio) for MAT1-1:MAT1-2. Of the four locations studied, two (Beja and Tunis) gave a 1:1 distribution while for the other two locations, MAT1-2 was more common than MAT1-1. However, Ozkilinc et al. (2015) showed that 1:1 ratio could not be rejected for two populations of A. fabae from Syria although a random mating hypothesis was rejected in these populations. In contrast, a study conducted on 311 isolates of A. fabae collected between 1991 and 2018 in South Australia by Blake et al. (2022) showed an equal ratio of MAT1–1 and MAT1–2. Moreover, this study came to a conclusion that there is no relationship between isolate aggressiveness and mating type.
The presence of two mating types can induce teleomorph development of D. fabae and by genetic recombination, new virulent strains can emerge. However, the presence of two mating types in the same region or field does not lead automatically to sexual recombination as revealed by Ozkilinc et al. (2015) and Omri Ben Youssef et al. (2019). The trend toward clonal reproduction in the same region or field could be due to a temporal and/or spatial lag in the development of the two mating types. The time lag could originate from different optima of the climatic parameters for the development of the two sexual types, particularly temperature and humidity, two key parameters for reproduction (Maurin et al., 1990). Variability in optimal temperature was reported for A. fabae (Kharrat, 2003) and other Ascochyta species (Le May et al., 2012). However, this variability has so far not been linked to mating type. A spatial lag would work on a lower scale than the field (plant and/or its different organs). Indeed, mating types may develop in the same field but on different plants or different organs (stem or leaf) making their recombination difficult as suggested by Omri Ben Youssef et al. (2019).
The present study aimed to: (i) assess the frequency and the distribution of both A. fabae mating types among different locations in a larger Tunisian population to confirm the findings of Omri Ben Youssef et al. (2019); (ii) define the aggressiveness level and the morphological features of A. fabae isolates belonging to each mating type, and (iii) define if the membership to a mating type is associated with particular phenotypic traits or aggressiveness level.
Material and methods
Fungal isolates
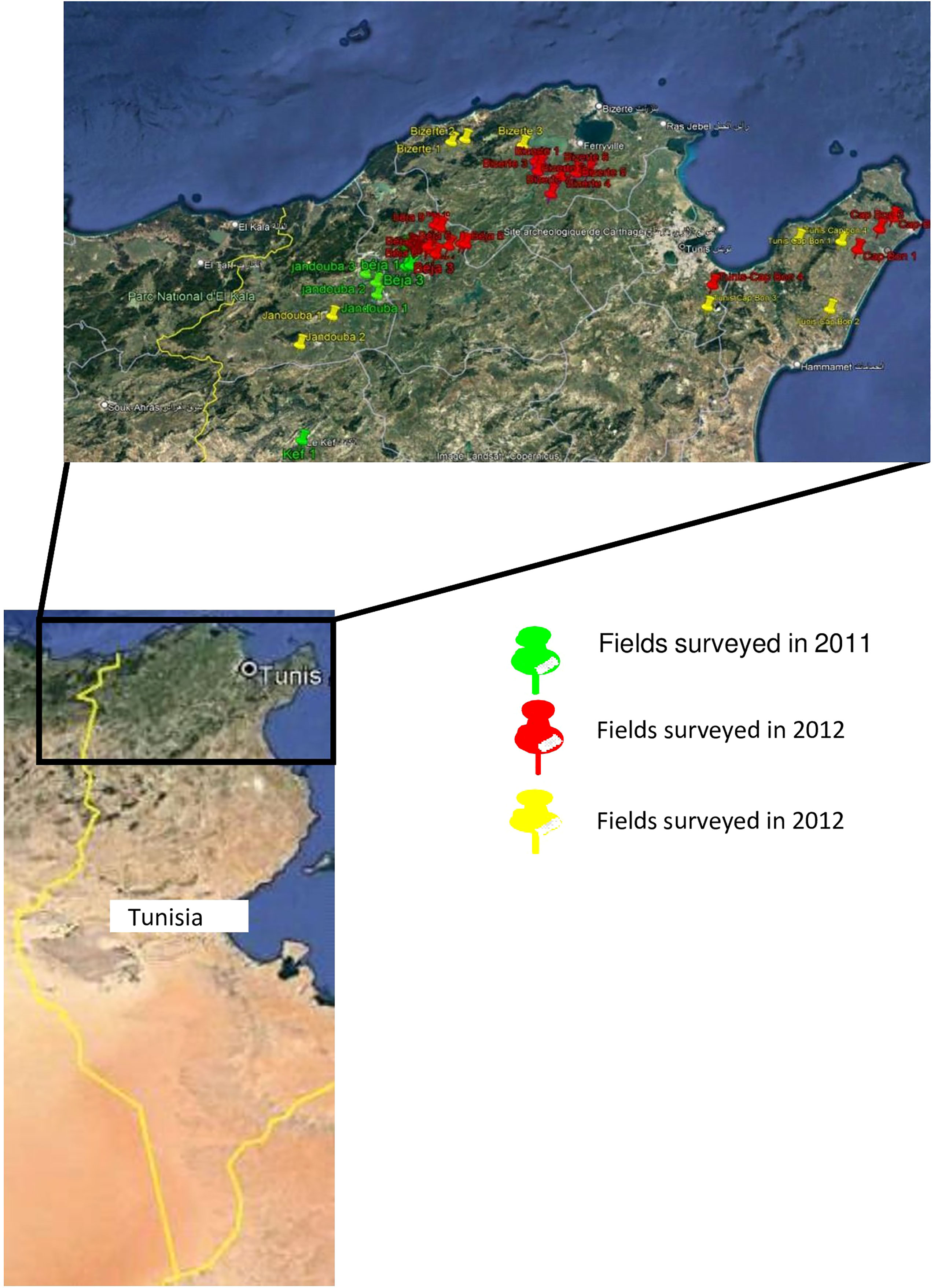
A total of 513 A. fabae isolates were used in this study. All the isolates were collected between 2011 and 2013 in several fields from five different geographical region locations in Tunisia, namely Beja (198 isolates: B1-B198), Bizerte (103 isolates: Bz1-Bz103), Jendouba (112 isolates: J1-J112), Kef (45 isolates: K1-K45) and Tunis-Cap Bon (55 isolates: TC1-TC55) (Table 1, Figure 1). All isolates were sampled from infected leaves. Faba bean tissue with lesions were collected randomly at intervals of 3 to 5 km in each region without considering the cultivar. Lesions were cultured on V8 medium in Petri dishes. Resulting conidia were spread onto malt agar and single germinating conidia were transferred to potato dextrose agar (PDA) and incubated at 20°C with a 12h photoperiod under cool white fluorescent lamps as described in Omri Ben Youssef et al. (2019).
Each single spore isolate was then grown in 75ml of peptone liquid (LP) medium supplemented with streptomycin (1.5g) and penicillin (0.75g). Each culture was raised from four pieces (approximately 1cm² each) cut from the margin of an actively growing culture on PDA. Inoculated vials were incubated, under agitation, for 14 days at 20°C with a 12h photoperiod under cool white fluorescent lamps. Mycelia were harvested by vacuum filtration through two layers of sterilized Miracloth (Calbiochem CN Biosciences, Inc., La Jolla, CA), rinsed twice in sterile water and stored at -80°C until lyophilized. DNA was extracted from 1g fungal mycelium and isolated using Nucleospin Plant II Kit (Macherey-Nagel, France) according to manufacturer’s instructions.
Mating type determination and distribution
Mating type of all the 513 A. fabae isolates was determined using the multiplex MAT-specific
PCR assay (Chérif et al., 2006). Primer combinations AL2p2SeqF4 (5’GCAACATCCTAGCATGATG3’) specific to MAT1-1, AL1p1SeqF5 (5’CTGTCTCACCCAAGGCAAAC3’) specific to MAT1-2 and ACom1A1AvAfAp
(5’CACATCACCCCACAAGTCAG3’), specific to an aligning flanking 3’ region of A. lentis, A. viciae-villosae, A. fabae and A. pisi were used. Single PCR was carried out in 25μl containing 10 ng of genomic DNA, 1X PCR buffer (containing 1.5 mM MgCl2), 0.2 mMdNTPs, 1unit Taq DNA polymerase (Promega, USA) and 0.2 µM each of the primers. Amplification was performed in Bio-Rad thermal cycler (Bio-Rad Laboratories, USA) and cycling conditions consisted of an initial denaturation at 95°C for 3 min followed by 35 cycles of 94°C for 20 sec, 58°C for 20 sec, 72°C for 40 sec, and a final extension at 72°C for 10 min. DNA amplicons were separated in 1.5% ethidium bromide-stained agarose gels and visualized under UV light on a ChemiDOCTM XRS documentation system (Bio-Rad, USA). Amplicon size was estimated using a DNA ladder (Hyperladder II, USA). Collected data on mating type were then submitted to Chi-Square test in PROC FREQ of SAS 9.2.
Isolate phenotyping
122 isolates were sampled randomly among the 513 isolates for phenotyping considering regions and mating types (11 to 13 isolates were selected for each region and each mating type). Shapiro Wilk test, Fischer test and Student t-test were respectively used to assess sample normality to compare variances and independence of subsamples (from each region) for each scored parameter for phenotyping. All these tests gave no significant difference between regions (P>0.05) allowing regional data to be combined in subsequent analyses.
Morphological characterization
Morphological phenotyping consisted of evaluating size, shape and conidial septation of the isolates under compound microscope (objective x40). Two slides were prepared from each isolate. Septum type (one celled, 1C and two celled, 2C) was evaluated on two microscope fields and conidial dimensions (one celled and two celled conidia length and width, 1L, 1W, 2L and 2W, respectively) were evaluated on 15 conidia of each preparation, and data transformed to percentage in the two preparation fields. To test individual variation the data of each parameter were subjected to an analysis according to a completely random design using PROCGLM procedure of SAS 9.2 software.
To determine if conidia-associated traits can separate and classify isolates into different groups, a Principal Component Analysis (PCA) was performed considering the mean septum type (1C and 2C) and mean dimensions of each type of conidia (1L, 1W, 2L and 2W). The analysis was performed using the PROC PRINCOMP procedure of the SAS 9.2 software.
Aggressiveness characterization
In a total of 366 pots containing potting soil, 2 seeds of Badii faba bean cultivar (released by Field Crop Laboratory of National Agronomic Research Institute of Tunisia in 2006 and known to be sensitive to ascochyta blight) were sown per pot and grown with irrigation under controlled conditions until three leaf stage. To evaluate the aggressiveness level of the 122 selected A. fabae isolates, at the three-leaf stage, each of three pots (one pot is considered an experimental unit repeated 3 times) was inoculated with 6 ml of a spore suspension of one of the 122 isolates adjusted to a concentration of 105 spores per ml. The pots were then maintained under 20°C and 16h of photoperiod with misting thrice a day to maintain favorable humidity. For each isolate, the incubation period (IP) was evaluated by measuring the number of days required for the onset of the first symptoms. The disease was also evaluated by visually estimating the percentage of necrotic leaf area (S) on the three first inoculated leaves at a regular interval of five days up to 45 days after inoculation. S1 to S9 were recorded on 5, 10, 15, 20, 25, 30, 35, 40 and 45 days after inoculation, respectively. Each set of data was then submitted to an analysis according to a randomized complete block design to test individual variation.
In order to check if aggressiveness parameters can separate the isolates into distinct groups, a PCA was conducted considering the mean incubation period (IP) and the percentage of necrotic leaf area for S1, S4, and S9. The analysis was performed, using the PROC PRINCOMP procedure of the SAS 9.2 software.
Statistical analysis of aggressiveness and morphological traits with mating type
In order to identify a link between mating type and phenotypic traits (morphology and aggressiveness) of A. fabae individuals, an MFCA (multiple factorial correspondence analysis) was conducted in SAS 9.2, through a multidimensional contingency table (Burt matrix) of all two-way cross-tabulations across all variables. MFCA decomposed the Burt matrix to find the pair wise associations which account for the greatest inertia proportion and displayed them on a reduced number of dimensions. It was carried out considering (i) three morphological groups (MG) obtained from the PCA performed on mean septum type and mean dimensions of each type of conidia, (ii) three aggressiveness groups (AG) obtained from the PCA conducted on the mean incubation period and the percentage of necrotic leaf area, and (iii) two mating types.
Results
Mating type occurrence and distribution within different Tunisian locations
Mating type of isolates was determined in PCR analysis using specific mating type primers which amplified one band of 700bp for Mat1-1or 450bp for Mat1-2 (Figure 2). Results showed that among the 513 Tunisian isolates, MAT1-2 was more common in the combined Tunisian collection than MAT1-1 (Table 2) with the Chi square analysis rejecting random mating (χ2 = 39.86, P-value <0.0001). However, after combining the populations of all years for regions where we sampled more than one year (having tested the two/three years independently and identifying no differences) results were different from one region to another.
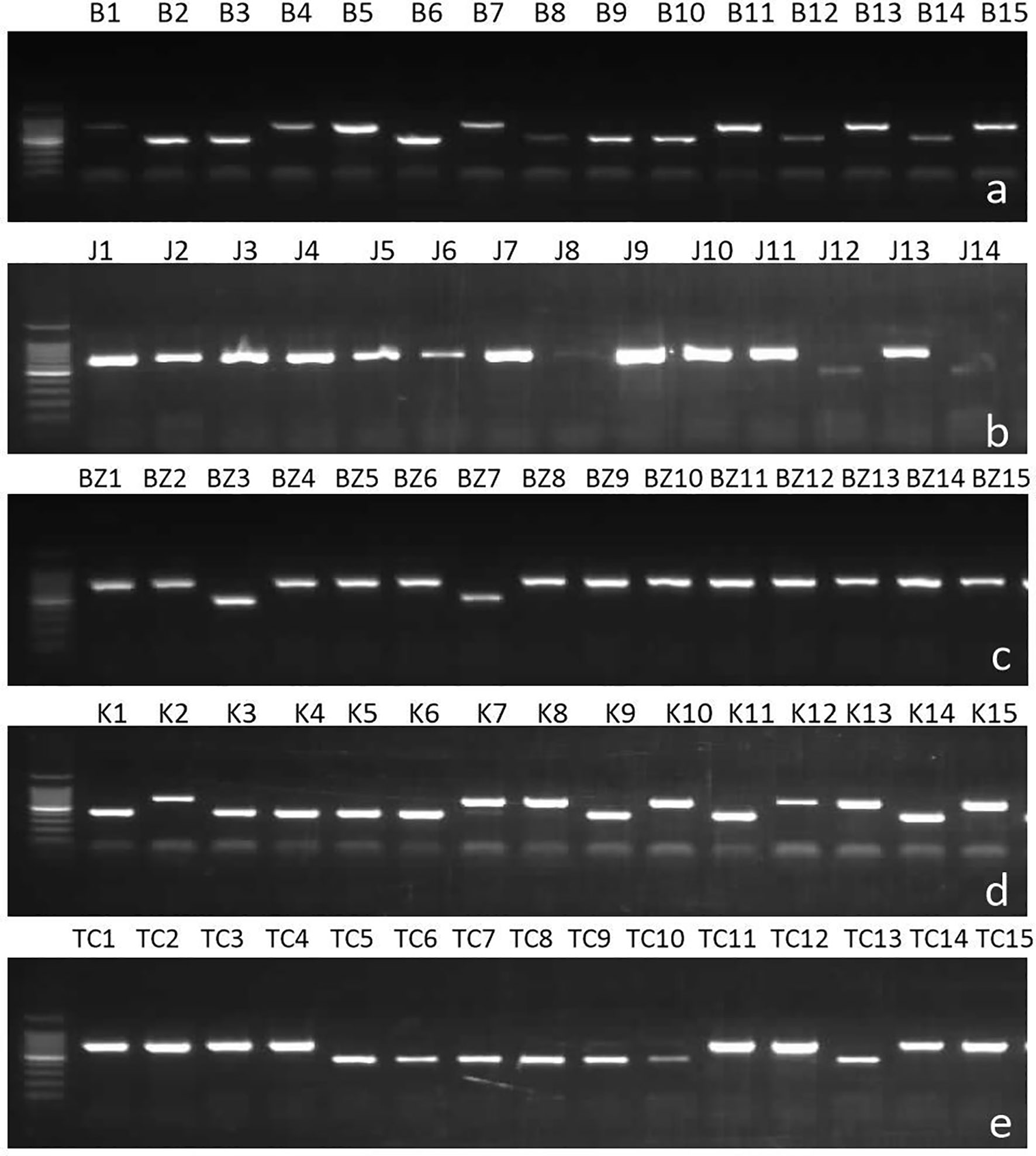
Figure 2 Samples of gels showing result of multiplex PCR analysis of Ascochyta fabae mating types in Tunisia with primers AL2p2SeqF4 specific to MAT1-1, AL1p1SeqF5 specific to MAT1-2 and ACom1A1AvAfAp specific to the flanking region. Bands of size 700 bp correspond to MAT 1-2 and bands of 450 bp correspond to MAT 1-1. (A) Beja, (B) Jendouba, (C) Bizerte, (D) Kef and (E) Tunis-Cap Bon.
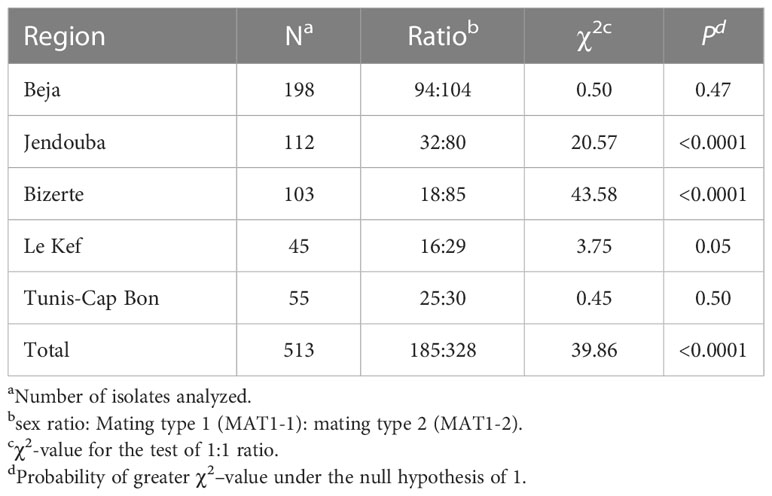
Table 2 Mating type ratios and χ2test for random mating within Ascochyta fabae populations from Tunisia.
Random mating was rejected using the χ2ratio test in Bizerte and Jendouba (χ2= 43.58, P- value <0.0001 and χ2= 20.57, P-value <0.0001 respectively) but was not rejected in the remaining three regions, Beja (χ2= 0.50, P-value = 0.47), Kef (χ2= 3.75, P-value = 0.0526) and Tunis-Cap Bon (χ2= 0.45, P-value = 0.50) (Table 2). For the 103 isolates from Bizerte and the 112 isolates from Jendouba, the two mating type ratios were 18:85 and 32:80, respectively, with greater numbers of MAT1-2 in both these regions.
Isolate phenotyping
Morphological characterization
Analysis of variance showed a significant effect (P-value < 0.0001) of the isolate on the different parameters (1C and 2C conidia rates, and conidia dimension: 1L, 1W, 2L and 2W). A PCA was performed on these data to illustrate correlation between these variables and to classify A. fabae isolates into morphological groups (MG). Results showed that the first two dimensions explained more than 71% of variation. Component 1 accounted for 44% of data variance and component 2 accounted for 27.60% of data variance (Figure 3). The first component was positively correlated with 2C, 2L, 2W, and 1W. It was negatively correlated with 1C and 1L. The second component was positively correlated with 1C, 1W and 2W. It was negatively correlated with 2C, 1L, and 2L (Table 3). All morphological parameters were correlated with each other (Table 4).
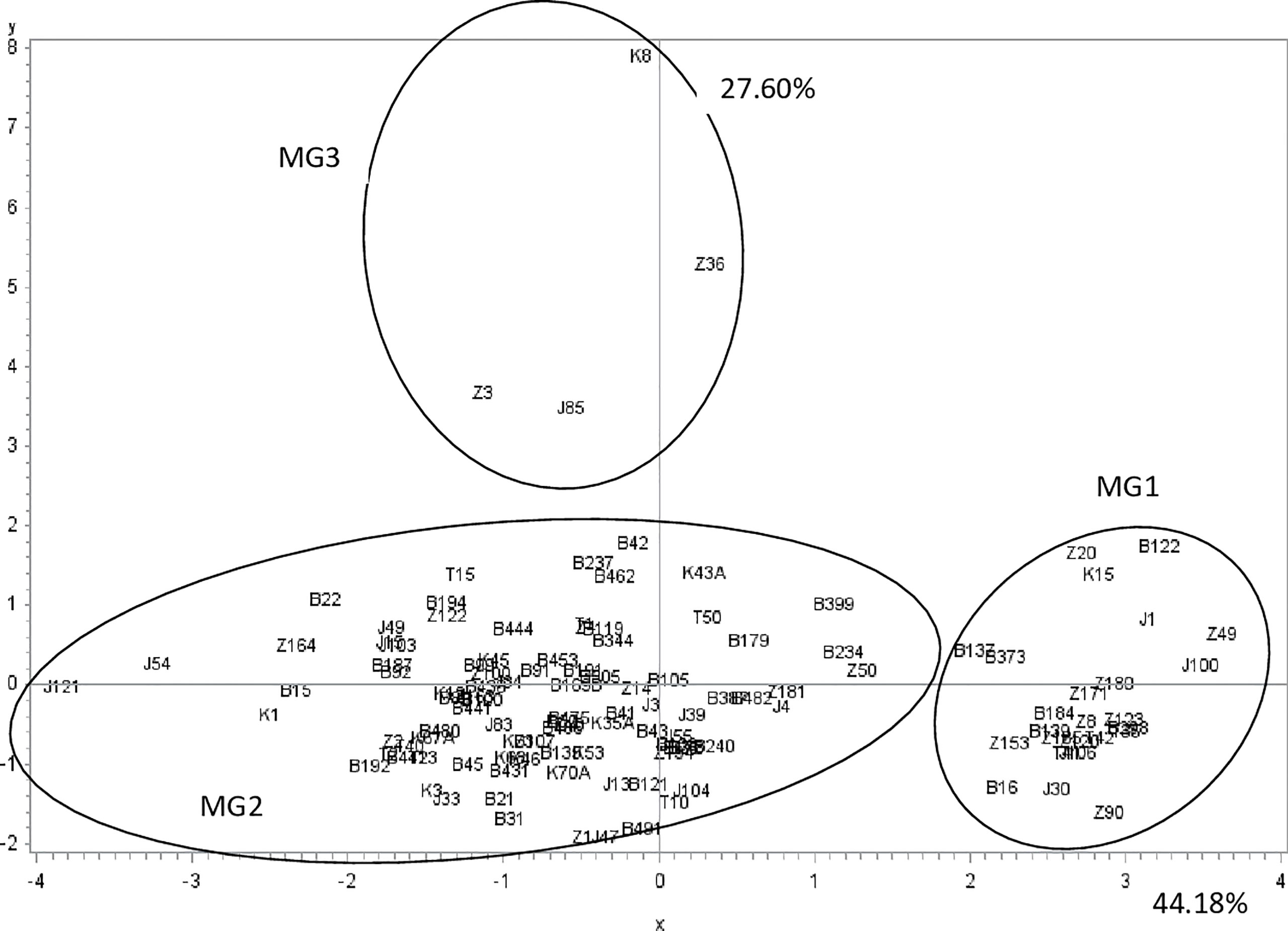
Figure 3 Ascochyta fabae isolate position on the two first most representative components (x and y) representing respectively 44.2 and 27.6 percent of variability in morphological parameters (one celled, two celled rate conidia, length and width of one celled and two celled conidia). Z, Bizerte; B, Beja; J, Jendouba; K, Kef and T, Tunis-Cap Bon.
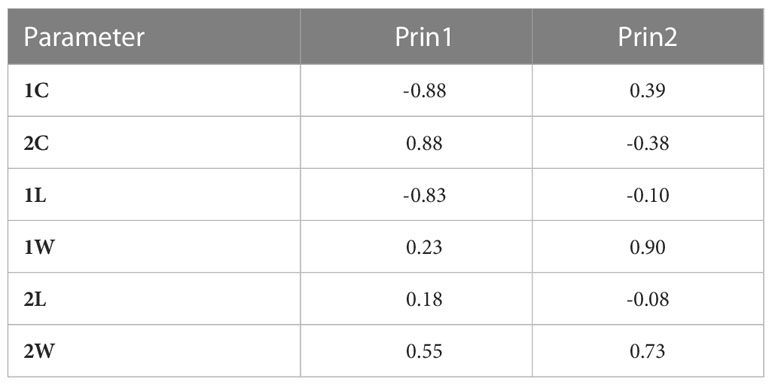
Table 3 Pearson Correlation Coefficients of morphological parameters of Ascochyta fabae isolates with the first two components (Number of isolates=122, one-celled conidia rate, 1C; two-celled conidia rate, 2C; one-celled conidia length, 1L, one-celled conidia width, 1W; two-celled conidia length, 2L and two celled conidia width, 2W).
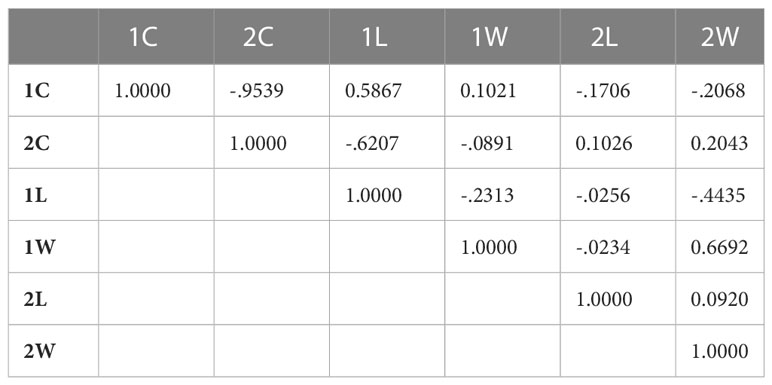
Table 4 PCA Correlation Matrix of morphological parameters of Ascochyta fabae isolates (Number of isolates=122, one-celled conidia rate, 1C; two-celled conidia rate, 2C; one-celled conidia length, 1L, one-celled conidia width, 1W; two-celled conidia length, 2L and two celled conidia width, 2W).
These two components distinguished three groups of isolates (Figures 3). The first group (MG1) was composed of isolates with a high frequency of two-celled, large and long conidia (Table 3, Figure 3). The second group (MG2) was composed of isolates with one and two celled isolates with medium dimensions (Table 3, Figure 3). The third group (MG3) was composed of isolates with a high frequency of large one celled conidia (Table 3, Figure 3).
Aggressiveness characterization
Aggressiveness was significantly different between the tested isolates (P-value <0.0001) for all the parameters assessed (IP, S1, S4, and S9). PCA analysis showed that the two first components explained more than 92% of variation. Component 1 accounted for 81.12% of data variance and component 2 accounted for 11.37% of data variance (Figure 4). The first component was positively correlated with S1, S4 and S9, and was negatively correlated with IP. The second component was positively correlated with IP, S1, and S4. It was negatively correlated with S9 (Table 5). All scored aggressiveness parameters were correlated with each other (Table 6).
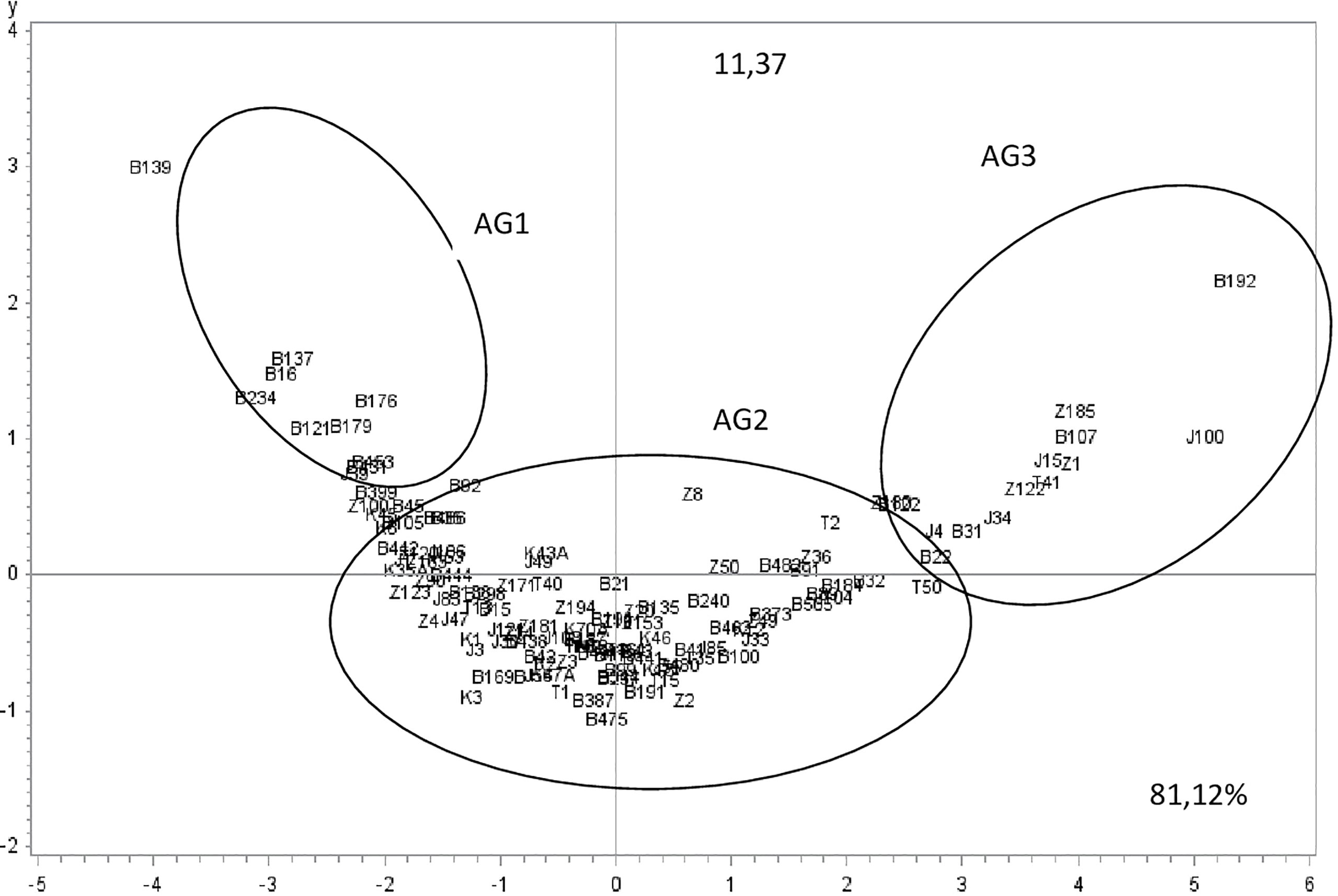
Figure 4 Ascochyta fabae isolate position on the two first most representative components (x and y) representing respectively 81.1 and 11.3% of variability in aggressiveness parameters (Incubation period: IP, the percentage of necrotic leaf area: 5,20 and 45 days after inoculation respectively S1, S4, and S9). Z, Bizerte; B, Beja; J, Jendouba; K, Kef and T, Tunis-Cap Bon.
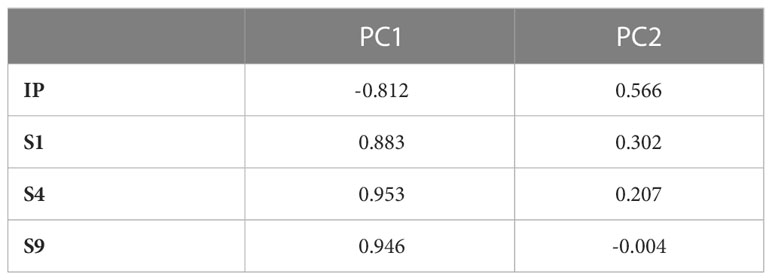
Table 5 Pearson correlation coefficients of aggressiveness parameters of Ascochyta fabae isolates with the first two components (PC1 and PC2) revealed by PCA conducted on 122 isolates and considering incubation length period in days, IP: and scoring of the percentage of necrotic leaf area: 5, 20 and 45 days after inoculation of faba bean cultivar Badii, respectively S1, S4 and S9.
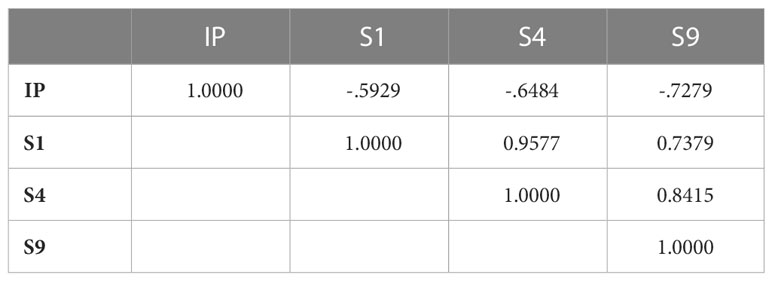
Table 6 PCA Correlation Matrix of aggressiveness parameters of Ascochyta fabae isolatesrevealed by PCA conducted on122 isolates and considering incubation length period in days, IP: and scoring of the percentage of necrotic leaf area: 5, 20 and 45 days after inoculation of faba bean cultivar Badii, respectively S1, S4 and S9.
The two components distinguished three isolate groups (Figures 4, 5). The first group: aggressiveness group1 (AG1), was composed of 12 isolates with long incubation period and low disease scores (S1, S4 and S9) and mostly originated from Beja region. The second group: aggressiveness group2 (AG2), was composed of 95 isolates with medium values of all parameters (IP and S1, S4 and S9) and categorized as moderately pathogenic. Isolates belonging to this group originated from all regions. The third group: aggressiveness group3, (AG3) was composed of 15 isolates having high disease scores (S1, S4 and S9) and low IP. This group was categorized as the most pathogenic. Isolates belonging to this group mostly originated from Beja, Bizerte and Jendouba regions.

Figure 5 Reaction of faba bean Badii cultivar to non-aggressive (A, B), moderately aggressive (C, D) and highly aggressive (E, F) Ascochyta fabae isolates.
Statistical analysis of aggressiveness and morphological traits with mating type
The purpose in using MFCA was to establish whether there was a correlation between morphological traits, mating type and aggressiveness groups. Based on the values of inertia and Chi square, the first dimensions were considered the most representative and accounted for almost 49% of variation, with dimension 1 and 2 representing respectively more than 25% and more than 23% of the variation. Dimension 1 is positively correlated with MAT1-1, MG1 and AG1, and negatively correlated with MAT1-2, MG2, MG3, AG2 and AG3. Dimension 2 is positively correlated with MAT1-2, MG1, AG1 and AG3. It is negatively correlated with MAT1-1, MG2, MG3 and AG2 (Table 7, Figure 6). The two components distinguished five groups of isolates (G1 to G5) according to their pathogenicity, morphological traits and mating type (Figure 6, Table 8). G1 was composed of isolates belonging to MAT1-1, to the moderately to poorly pathogenic group1 (AG2 and AG1) and mostly MG1 morphological traits of two-celled, large and long conidia. G2 was composed of isolates belonging to MAT1-1, to the moderately pathogenic group (AG2) and mostly MG2 morphological traits of one and two-celled isolates with medium dimensions and isolates with long one-celled conidia. G3 was composed of isolates belonging to MAT1-2, to the moderately pathogenic group (AG2) and mostly MG2 and MG3 morphological traits. G4 was composed of isolates belonging to MAT1-2, to the moderately pathogenic group (AG2) and MG1 morphological traits. G5 was composed exclusively of the most pathogenic isolates toward cv. Badii, belong to MAT1-2 group and mostly MG2 morphological traits (isolates with one and two-celled isolates with medium dimensions).
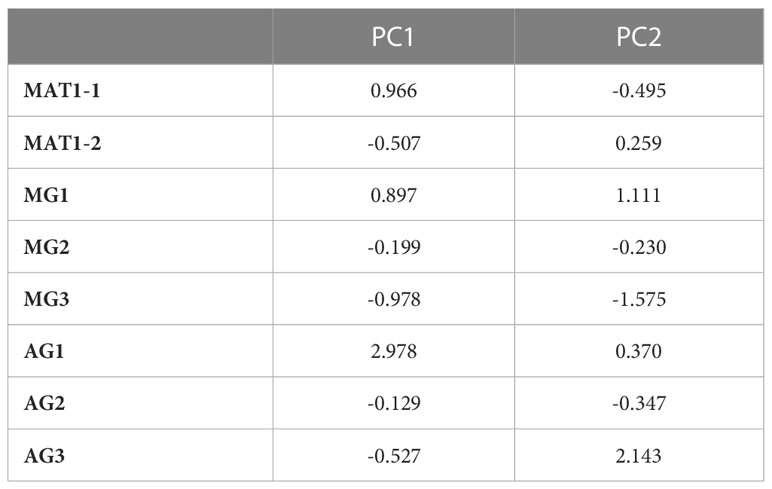
Table 7 Coordinates of the different groups of Ascochyta fabae isolates with the first two components revealed by FMCA conducted on 122 isolates and considering mating type 1, MAT1-1; mating type 2, MAT1-2; Morphological group 1,2 and 3 respectively, MG1, MG2, MG3 and, Aggressiveness group 1, 2 and 3 respectively, AG1, AG2, AG3.
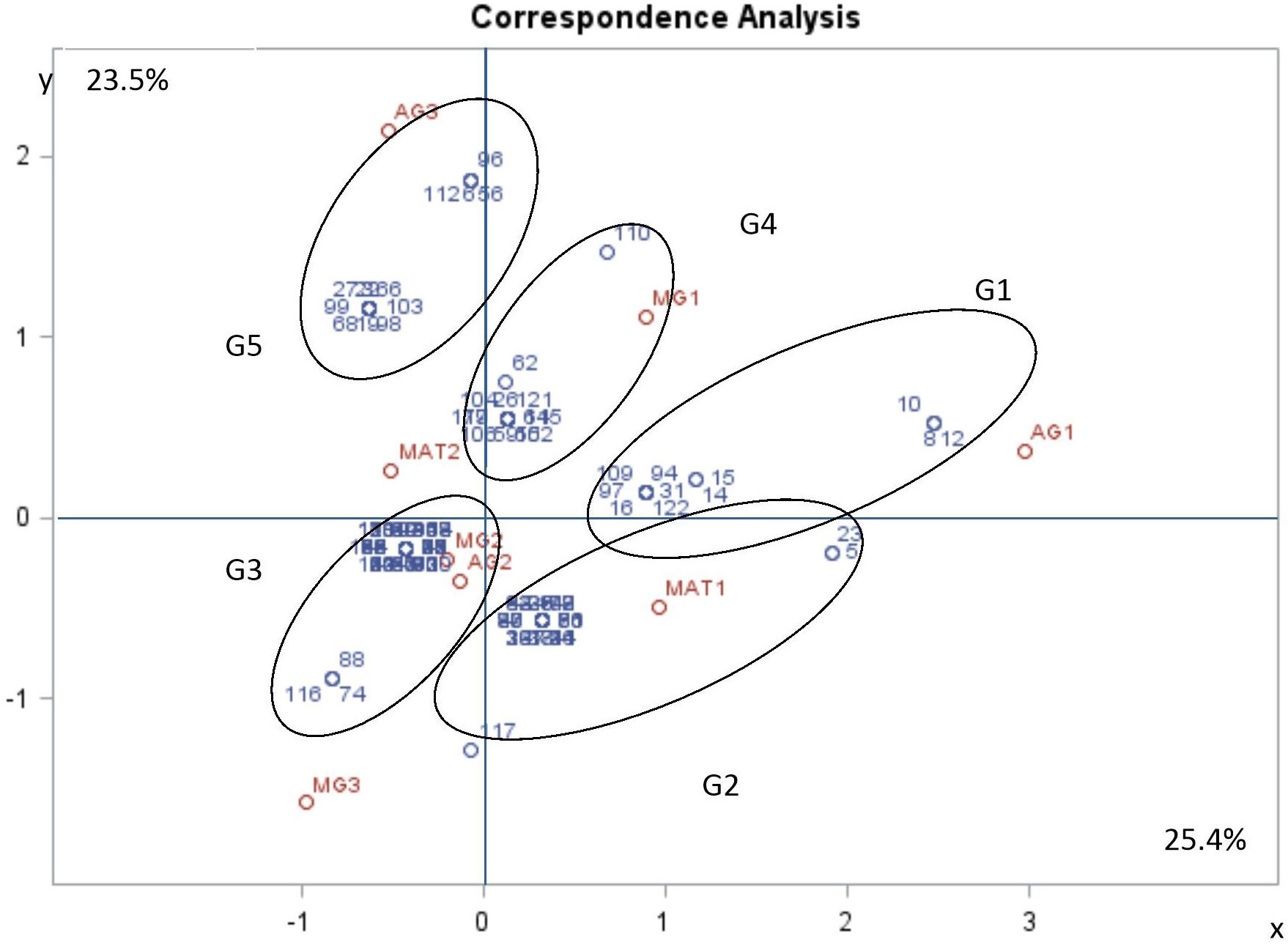
Figure 6 Multiple factorial correspondence analysis (MFCA) performed on Ascochyta fabae isolates regarding their mating type (MAT1-1 and MAT1-2), their aggressiveness group (AG1, AG2 and AG3) and their morphological group (MG1, MG2 and MG3). Aggressiveness group (AG1: poorly pathogenic, AG2: moderately pathogenic and AG3: highly pathogenic), Mating type (MAT1: mating type 1 and MAT2: mating type 2) and morphological group (MG1, MG2 and MG3). The two first most representative components (x and y) represent respectively 25.4 and 23.5 percent of variability. Variable correlation with two components (in red) and Isolate position on two components (in blue).
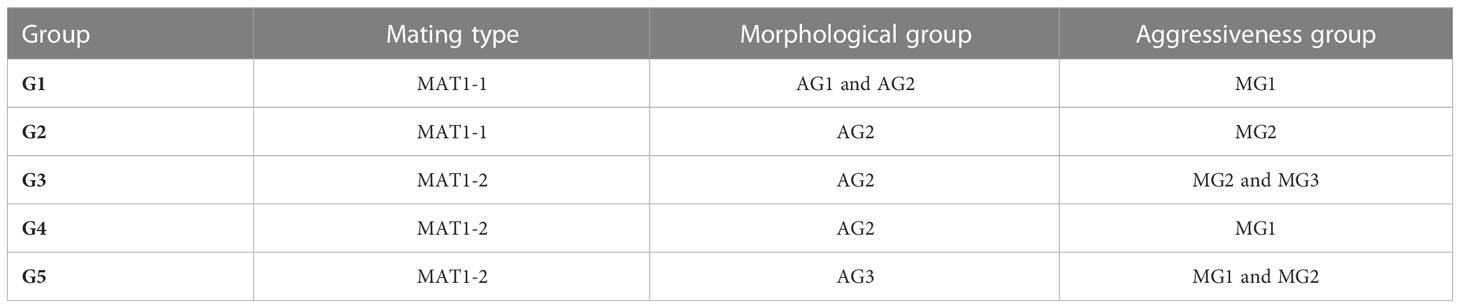
Table 8 Morphological traits, aggressiveness group and mating type of isolate groups defined by projection on the first two components (MAT1-1: mating type1, MAT1-2: mating type 2, MG1, MG2, MG3: Morphological group 1,2 and 3 respectively, AG1, AG2, AG3: Aggressiveness group 1, 2 and 3, respectively.
Discussion
The main objectives of this study were to assess frequency and distribution of both A. fabae mating types among different locations in Tunisia, to define aggressiveness level and morphological features of A. fabae isolates belonging to each mating type, and to check if the membership to a mating type would be associated to particular phenotypic traits or aggressiveness level. The D. fabae teleomorph has been reported in many countries: Canada (Kharbanda and Bernier, 1980), Poland (Filipowicz, 1983), United Kingdom (Jellis and Punithalingam, 1991), Australia (Kaiser, 1997), Syria (Bayaa and Kabbabeh, 2000), Spain (Rubiales and Trapero-Casas, 2002) and more recently in Tunisia (Omri Ben Youssef et al., 2012). Since the discovery of the teleomorph form in Tunisia, several questions arose regarding the spatial distribution of these mating types and their respective prevalence within populations (Ozkilinc et al., 2015; Omri Ben Youssef et al., 2019; Blake et al., 2022). Our study showed that MAT1-2 was more common than MAT1-1 in three Tunisian locations (Bizerte, Jendouba and Kef), and for two locations (Beja and Tunis-Cap Bon) sexual reproduction was regularly observed. Indeed, Beja and Tunis-Cap Bon were known to be hot spots of A. fabae and are the main faba bean growing regions. The minor type of faba bean (Vicia faba minor) are most commonly grown in Beja and Tunis-Cap Bon and possibly the majority of disease samples in those regions came from the minor type crop. In contrast, both minor and major type (Vicia faba major) are grown in Jendouba and Bizerte so here the disease samplings were likely to be collected equally from both types. One hypothesis is that mating type may be linked to host type (minor or major) since pathogens can be subjected to host selection as mentioned by McDonald and Linde (2002) and De Meeûs et al. (2007). Host cultivar or faba bean type was not considered in the data analyses in this study but it could be an area of further study. These results confirmed an earlier study conducted by Omri Ben Youssef et al. (2019) on a smaller population which concluded that sexual recombination does not play an important role in fungus diversity through these regions. The same conclusion was reported by Ozkilinc et al. (2015) in their study on Syrian populations. In contrast, Blake et al. (2022) reported an equal ratio of MAT1–1 and MAT1–2 in a collection of 311 isolates of A. fabae from South Australia. This result may be due to the sample size and the origin of the isolates that was mainly from field trials rather than commercial crops.
Morphological characterization of 122 isolates among the whole population revealed a high level of diversity. These results support those of Kharrat et al. (2000) who reported morpho-biological diversity after evaluating the aggressiveness and some morphological traits of an international collection of A. fabae isolates. This diversity was also reported by Maurin (1989) on conidia originating from the same pycnidia. Kharrat et al. (2000) and Maurin (1989) did not establish any link between the studied parameters. Contrarily, our study was able to establish correlations between the different parameters through a PCA. The correlation established between the different phenotypic parameters through PCA concluded with three groups (MG1, MG2 and MG3) according to their morphological parameters: the frequency of one-celled and two celled conidia and the dimensions of the different conidia types (length and width). Similarly, pathogenic variability was revealed between 122 isolates on the faba bean cv. Badii. This variability was observed for incubation period (IP), and for the disease scores (S1, S4, and S9). Our results confirmed those obtained in previous studies (Hanounik and Robertson, 1989; Maurin, 1989; Rashid et al., 1991; Kharrat et al., 2000; Bessaidi et al., 2017). Maurin (1989) assessed pathogenic variability as well as sculptural criteria (morphology, growth, fruiting in vitro). However, he was not able to define pathotypes given the diversity of characters and the instability of the strains since the colonies presented variant sectors, possibly due to the progeny of single spore isolates being heterogeneous. Hanounik and Robertson (1989) showed consistently significant differential interactions among faba bean lines tested against eight isolates of A. fabae from Syria. A similar result was obtained by Rashid et al. (1991) when they tested 19 inbred lines against 5 isolates of A. fabae. Kharrat et al. (2000) scored six lines for their resistance to three isolates of A. fabae on both stems and leaves and they concluded that there was a differential interaction between lines and isolates and also a differential interaction between organ (stem or leaves) and isolate. None of these studies used the PCA tool to group the isolates into aggressiveness groups. This study started initially with 144 isolates but only 122 isolates gave usable results. However according to Pallant (2011) ideally, there should be at least 150 cases for PCA and there should be a ratio of at least five cases for each variable which is not the case for morphological groups in this study. Hence the morphological traits of MG3 may not be a good indicator of aggressiveness or mating type and a study on a larger population may provide a clearer response. MFCA analysis distinguished five different groups based on morphological traits, mating type and aggressiveness level. The most interesting result is that all the highly aggressive isolates were classified in one group: G5 which is composed of MAT1-2 isolates. Some of these have the same frequency of one or two-celled conidia, medium dimensions and relatively long one-celled conidia (MG2) while others in G5 have a high frequency of two-celled conidia, large and long conidia (MG1). Most isolates with a high frequency of one-celled conidia were grouped in G3 and linked to MAT1-2 mating type and were moderately pathogenic (AG2). Isolates from G2 and G4 have the same aggressiveness level (moderately aggressive: AG2) but have different morphological traits. Such results suggest that there is no specific traits linked to aggressiveness since the most aggressive isolates had diverse and the most frequent morphological traits (MG1 and MG2). But it seems that aggressiveness is linked to MAT1-2. Where MAT1-2 is predominant, the spatial and temporal dynamics of the epidemic is rapid and the faba bean crop is exposed to a high disease risk because of the high aggressiveness of this mating type. Rigorous monitoring of the fields is needed in such cases to overcome the epidemic. As supposed by Omri Ben Youssef et al. (2019) our results are an additional argument to support the hypothesis that MAT1-2 fitness seems to be higher than that of MAT1-1, particularly in Bizerte, Jendouba and Kef regions. This hypothesis can be confirmed by further studies to compare the biology of MAT1-1 and MAT1-2 that can identify how climatic conditions and host affect both in vitro mating types, mycelium growth, sporulation and germination, and in vivo necrosis development, pycnidium genesis, host infection and spatial dispersion. The high aggressiveness linked to MAT1-2 revealed in this study suggests a higher risk for faba bean if contaminated with MAT1-2 than with MAT1-1. As seeds constitute one of the main inoculum sources (Tivoli and Banniza, 2007), seed movement could be a risk of long-distance dispersion by transferring highly pathogenic MAT1-2 isolates from one region to another. Limiting seed transfer and seed health management could avoid such risk. Furthermore, faba bean breeding program should orientate disease resistance selection mainly toward MAT1-2 to be effective at the on-farm level.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZB, IH, MK, and AM generated the experimental data. NO designed supervised the experiments, and performed statistical analysis and drafted the manuscript. KM and CLM: coordinated and supervised the manuscript redaction. All authors contributed to the article and approved the submitted version.
Funding
This work was co-funded by the Ministry of Agriculture, Hydraulic Resources and Fisheries, the Ministry of Higher Education and Scientific Research of Tunisia and MEDILEGARIMNET project (2012-2015; Proposal ID 396: Breeding, agronomic and biotechnological approaches for reintegration and revalorization of legumes in Mediterranean agriculture). This work was supported by the UMR 1349 IGEPP, INRA, France.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1176517/full#supplementary-material
References
Barve, M. P., Arie, T., Salimath, S. S., Muehlbauer, F. J., Peever, T. L. (2003). Cloning and characterization of the mating type (MAT) locus from Ascochyta rabiei (teleomorph: Didymella rabiei) and a MAT phylogeny of legume-associated Ascochyta spp. Fungal Gen. Biol. 39 (2), 151–167. doi: 10.1016/S1087-1845(03)00015-X
Bayaa, B., Kabbabeh, S. (2000). First record in Syria of Didymella fabae, the teleomorph of Ascochyta fabae and causal organism of faba bean blight. Plant Dis. 84, 1140. doi: 10.1094/PDIS.2000.84.9.1044C
Beed, F. D., Sue, R. E., Strange, R. N. (1994). Variation in the production of ascochitine by Ascochyta fabae. Mycol. Res. 98(9), 1069–1076. doi: 10.1016/S0953-7562(09)80435-8
Bessaidi, Z., Omri Ben Youssef, N., Touati, R., Timoumi, S., Jammezi, N., Halila, I., et al. (2017). Ascochyta fabae Speg. Mating type phenotyping. Communication. Scientific Séminaire, Laboratory of Field Crops, INRAT (Tunis Tunisia: des grandes cultures et changements climatiques), 18 -20 April 2017.
Blake, S. N., Lee, R. C., Russ, M. H., Farquharson, E. A., Rose, J. A., Herdina, et al. (2022). Phenotypic and genotypic diversity of Ascochyta fabae populations in Southern Australia. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.918211
Chérif, M., Chilvers, M. I., Akamatsu, H., Peever, T. L., Kaiser, W. J. (2006). Cloning of the mating type locus from Ascochyta lentis (teleomorph: Didymella lentis) and development of a multiplex PCR mating assay for Ascochyta species. Curr. Genet. 50, 203–215. doi: 10.1007/s00294-006-0085-y
Coppin, E., Debuchy, R., Arnaise, S., Picard, M. (1997). Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol.Rev. 61(4), 411–428. doi: 10.1128/mmbr.61.4.411-428.1997
Debuchy, R., Turgeon, B. (2006). “Mating-type structure, evolution, and function in Euascomycetes,” in Growth, Differentiation and Sexuality, the mycota (1). Eds. Debuchy, R., Turgeon, B. (Berlin: Springer Berlin Heidelberg), 293–323.
De Meeûs, T., McCoy, K. D., Prugnolle, F., Chevillon, C., Durand, P., Hurtrez-Boussès, S., et al. (2007). Population genetics and molecular epidemiology or how to “débusquer la bête”. Infect. Genet. Evol. 7 (2), 308–332. doi: 10.1016/j.meegid.2006.07.003
Filipowicz, A. (1983). The pathogenicity of isolates of Ascochyta fabae Speg. on horse bean (Vicia faba L. var. minor Harz) (abstr.) Faba Bean Abstr. Commonwealth. Agric. Bureaux. 4, 47.
Gossen, B. D., Morrall, R. A. A. (1986). Transmission of Ascochyta lentis from infected lentil seed and plant residue. Can. J. Plant Pathol. 8, 28–32. doi: 10.1080/07060668609501837
Hanounik, S. B. (1980). Effect of chemical treatments and host genotypes on disease severity/yield relationships of Ascochyta blight in faba beans. FABIS. Newslett. 2, 50.
Hanounik, S. B., Robertson, L. D. (1989). Resistance in Vicia faba germplasm to blight caused by Ascochyta fabae. Plant Dis. 73, 202–205. doi: 10.1094/PD-73-0202
Jellis, G. J., Punithalingam, E. (1991). Discovery of Didymella fabae sp. nov., teleomorph of Ascochyta fabae, on faba bean straw. Plant Pathol. 40, 150–157. doi: 10.1111/j.1365-3059.1991.tb02305.x
Kaiser, W. J. (1997). Inter- and intranational spread of ascochyta pathogens of chickpea, faba bean, and lentil. Can. J. Plant Pathol. 19, 215–224. doi: 10.1080/07060669709500556
Kharbanda, P. D., Bernier, C. C. (1980). Cultural and pathogenic variability among isolates of Ascochyta fabae. Can. J. Plant Pathol. 2 (3), 139–142. doi: 10.1080/07060668009501429
Kharrat, M. (1999). Analyse génétique de la résistance de Vicia faba L. à différents pathotypes d’Ascochyta fabae Speg. Conséquences pour la mise en œuvre de stratégies de sélection. Thèse de doctorat (France: ENSAR, Université de Renne), 149 p.
Kharrat, M. (2003). Sélection de lignées de féverole résistantes l’anthracnose causée par Ascochyta fabae. Proceedings du 2ème séminaire du réseau REMAFEVE /REMALA.
Kharrat, M., Le Guen, J., Tivoli, B. (2006). Genetics of resistance to 3 isolates of Ascochyta fabae on Faba bean (Vicia faba L.) in controlled conditions. Euphytica 151(1), 49. doi: 10.1007/s10681-006-9127-2
Kharrat, M., Tivoli, B., Le Guen, J. (2000). Characterisation of Tunisian Ascochyta fabae isolates, causal agent of blight of Faba bean. Annales. l’INRAT. 73, 91–104.
Kohpina, S., Knight, R., Stoddard, F. L. (1999). Variability of Ascochyta fabae in South Australia. Aust. J. Agric. Res. 50, 1475–1481. doi: 10.1071/AR98204
Kronstad, J. W., Staben, C. (1997). Mating type in filamentous fungi. Annu. Rev. Genet. 31, 245–276. doi: 10.1146/annurev.genet.31.1.245
Le May, C., Guibert, M., Leclerc, A., Andrivon, D., Tivoli, B. (2012). A Single, Plastic Population of Mycosphaerella pinodes Causes Ascochyta Blight on Winter and Spring Peas (Pisum sativum) in France. App. Environ. Microbiol. 78, 8431–8440. doi: 10.1128/AEM.01543-12
Maurin, N. (1989). Biologie d’Ascochyta fabae Speg. et étude des relations hôte-parasite en vue de l’appréciation de la résistance de la féverole à l’anthracnose (France: Université de Rennes I).
Maurin, N., Tivoli, B. (1992). Variation in the resistance of Vicia faba to Ascochyta fabae in relation to disease development in field trials. Plant Pathol. 41 (6), 737–744. doi: 10.1111/j.1365-3059.1992.tb02557.x
Maurin, N., Tivoli, B., Onfroy, C. (1990). Mieux connaître les maladies et leurs agents pathogènes pour mieux les combattre: exemple de l’anthracnose de la féverole. Perspect. Agricoles. 146, 36.
McDonald, B. A., Linde, C. A. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40 (1), 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
Metzenberg, R. L., Glass, N. L. (1990). Mating-type and mating strategies in Neurospora. Bio Essays. 12, 53–59. doi: 10.1002/bies.950120202
Milgroom, M. G. (1996). Recombination and the multilocus structure of fungal populations. Ann. Rev. Phytopathol 34, 457–477. doi: 10.1146/annurev.phyto.34.1.457
Nasraoui, B. (2015). Les champignons et pseudo-champignons pathogènes des plantes cultivées: Biologie, Nouvelle systématique, Interaction pathologique (Tunisie: Publication de l’Institut National Agronomique de Tunisie).
Omri Ben Youssef, N., Bessaidi, Z., Halila, I., Mbazia, A., Jammezi, N., Kharrat, M., et al. (2016). Genetic and pathogenic diversity of Ascochyta fabae Speg. Population. Presentation in the 4thInternational Ascochyta Workshop held in Troia, Portugal from 10th to 14th October 2016. doi: 10.13128/Phytopathol_Mediterr-23562
Omri Ben Youssef, N., Kerdraon, L., Mieuzet, L., Halila, I., Jammezi, N., Mbazia, A., et al. (2019). Population structure of the faba bean blight pathogen Ascochyta fabae (teleomorph, Didymella fabae) in Tunisia. Phytopathol. Mediterranea. 58 (1), 81–94. doi: 10.13128/Phytopathol_Mediterr-23562
Omri Ben Youssef, N., Le May, C., Mlayeh, O., Kharrat, M. (2012). First report of Didymella fabae, teleomorph of Ascochyta fabae, on faba bean crop debris in Tunisia. Phytopathol. Mediterranea. 51 (2), 369–373. doi: 10.14601/Phytopathol_Mediterr-9496
Onfroy, C., Tivoli, B., Corbie, R., Bouznad, Z. (1999). Cultural, molecular and pathogenic variability of Mycosphaerella pinodes and Phoma medicaginis var. pinodella isolates from dried pea (Pisum sativum) in France. Plant Pathol. 48 (2), 218–229.
Ozkilinc, H., Thomas, K., Abang, M., Peever, T. L. (2015). Population structure and reproductive mode of Didymella fabae in Syria. Plant Pathol. 64, 1005–1257. doi: 10.1111/ppa.12359
Pallant, J. (2011). Multivariate analysis of variance. SPSS survival manual. Crows. Nest.: Allen. Unwin. 20 (11), 283–296.
Pedersen, E. A., Bedi, S., Morrall, R. A. A. (1993). Gradient of ascochyta blight in Saskatchewan lentil crops. Plant Dis. 77, 143–149. doi: 10.1094/PD-77-0143
Peever, T. L. (2007). Role of host specificity in the speciation of Ascochyta pathogens of cool season food legumes. Eur. J. Plant Pathol. 119, 119–126. doi: 10.1007/s10658-007-9148-2
Rashid, K. Y., Bernier, C. C., Conner, R. L. (1991). Evaluation of faba bean for resistance to Ascochyta fabae and development of host differentials for race identification. Plant Dis. 75, 852–855. doi: 10.1094/PD-75-0852
Rubiales, D., Trapero-Casas, A. (2002). Occurrence of Didymella fabae, the Teleomorph of Ascochyta fabae on Faba Bean straw in Spain. J. Phytopathol. 150, 146–148. doi: 10.1046/j.1439-0434.2002.00727.x
Tivoli, B., Banniza, S. (2007). Comparison of the epidemiology of ascochyta blights on grain legumes. Eur. J. Plant Pathol. 119 (1), 59–76. doi: 10.1007/s10658-007-9117-9
Tivoli, B., Baranger, A., Avila, C. M., Banniza, S., Barbetti, M., Chen, W. D., et al. (2006). Screening techniques and sources of resistance to foliar diseases caused by major necrotrophic fungi in grain legumes. Euphytica 147 (1-2), 223–253. doi: 10.1007/s10681-006-3131-4
Turgeon, B. G. (1998). Applications of mating-type technology to problems in fungal biology. Ann. Rev.Phytopathol. 36, 115–137. doi: 10.1146/annurev.phyto.36.1.115
Turgeon, B. G., Bohlmann, H., Ciuffetti, L. M., Christiansen, S. K., Yang, G., Schäfer, W., et al. (1993). Cloning and analysis of the mating type genes from Cochliobolus heterostrophus. Mol. Genom. Gen. 238, 270. doi: 10.1007/BF00279556
Keywords: faba bean, ascochyta blight, morphological diversity, conidia, Didymella fabae, mating type
Citation: Omri Ben Youssef N, Halila I, Mbazia A, Bessaidi Z, Missaoui K, Kharrat M and Le May C (2023) Didymella fabae Punith.: mating type occurrence, distribution and phenotyping of the anamorph Ascochyta fabae Speg. in Tunisia. Front. Plant Sci. 14:1176517. doi: 10.3389/fpls.2023.1176517
Received: 28 February 2023; Accepted: 18 July 2023;
Published: 04 September 2023.
Edited by:
Jennifer Davidson, South Australian Research and Development Institute, AustraliaReviewed by:
Robert Campbell Lee, Curtin University, AustraliaSara N. Blake, South Australian Research and Development Institute, Australia
Copyright © 2023 Omri Ben Youssef, Halila, Mbazia, Bessaidi, Missaoui, Kharrat and Le May. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Noura Omri Ben Youssef, b21yaS5ub3VyYUBpbnJhdC51Y2FyLnRu
†These authors have contributed equally to this work and share senior authorship
 Noura Omri Ben Youssef
Noura Omri Ben Youssef Imen Halila1
Imen Halila1 Khawla Missaoui
Khawla Missaoui