- 1College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an, China
- 2State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, China
- 3Shandong Collaborative Innovation Center for Fruit and Vegetable Production with High Quality and Efficiency, Shandong Agricultural University, Tai’an, China
Drought stress is an adverse stimulus that affects agricultural production worldwide. NAC transcription factors are involved in plant development and growth but also play different roles in the abiotic stress response. Here, we isolated the apple MdNAC29 gene and investigated its role in regulating drought tolerance. Subcellular localization experiments showed that MdNAC29 was localized to the nucleus and transcription was induced by the PEG treatment. Over-expression of MdNAC29 reduced drought tolerance in apple plants, calli, and tobacco, and exhibited higher relative conductivity, malondialdehyde (MDA) content, and lower chlorophyll content under drought stress. The transcriptomic analyses revealed that MdNAC29 reduced drought resistance by modulating the expression of photosynthesis and leaf senescence-related genes. The qRT-PCR results showed that overexpression of MdNAC29 repressed the expression of drought-resistance genes. Yeast one-hybrid and dual-luciferase assays demonstrated that MdNAC29 directly repressed MdDREB2A expression. Moreover, the yeast two-hybrid and bimolecular fluorescence complementation assays demonstrated that MdNAC29 interacted with the MdPP2-B10 (F-box protein), which responded to drought stress, and MdPP2-B10 enhanced the repressive effect of MdNAC29 on the transcriptional activity of the MdDREB2A. Taken together, our results indicate that MdNAC29 is a negative regulator of drought resistance, and provide a theoretical basis for further molecular mechanism research.
1 Introduction
Plants are exposed to a variety of adverse environmental stressors in their natural habitats, including biotic stressors (fungal, bacterial, viral, and insect pest) and abiotic stressors (high salinity, drought, flooding, heat, cold, and injury) (Zhang et al., 2012). Abiotic stressors are significant during plant growth and development, particularly in the face of global warming, limited water supplies, and soil desertification. Among the adverse external stimuli, drought stress has become a major environmental factor limiting crop productivity and management (Zhu and Ni, 2016; Basia and Arie, 2005). When plants are subjected to drought stress, they regulate gene expression and produce new proteins through the perception and transmission of drought signals, resulting in several morphological, physiological, and biochemical changes to ensure normal development and growth (Kavi et al., 2005).
NAC (NAM-ATAF1/2-CUC2) is one of the largest transcription factor (TF) families in plants and is widely distributed in terrestrial plants (Puranik et al., 2012; An et al., 2018). The first NAC TF was cloned from Petunia (Souer et al., 1996) and subsequently found in Arabidopsis and rice. NAC TFs share a highly conserved NAC domain of about 150 amino acids (aa) at the N-terminal end and a highly variable transcriptional regulatory region at the C-terminal end (Olsen et al., 2005). Studies have shown that NAC TFs are involved in the regulation of plant growth and development (Sun et al., 2012; Wen et al., 2022). In apples, MdNAC52 regulates the biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11 (Sun et al., 2019). In Arabidopsis, AtNAC029 is involved in leaf senescence, and overexpression of AtNAC029 causes premature senescence (Ma et al., 2020). NAC TFs play an important regulatory role in the abiotic stress response (Shao et al., 2015). Drought, salt stress, or abscisic acid (ABA) induce the expression of OsNAC5 and OsNAC10 in rice and AtAF1, ANAC19, ANAC55, and ANAC72 in Arabidopsis, demonstrating the role of these NAC TFs in drought and salt resistance (Tran et al., 2004; Lu et al., 2007; Jeong et al., 2010; Takasaki et al., 2010). The expression level of wheat TaNAC2 increases significantly under drought stress and enhances drought tolerance by activating drought stress response genes (Mao et al., 2014). Overexpression of OsNAC106 reduces salt stress tolerance in rice and the deletion mutation results in increased salt tolerance (Sakuraba et al., 2015). In summary, NAC TFs have different functions in response to abiotic stressors.
Other TF families, including AP2/ERF, MYB, TCP, AREB, and WRKY play important roles in abiotic stress (Li et al., 2020; Li et al., 2021b). DREBs are a subfamily of AP2/EREBP TFs that have a conserved AP2 domain structure. The DREB gene has been extensively studied during abiotic stress in plants (Agarwal et al., 2006; Khan, 2014; Eckardt, 2019). DREB2A members are mainly involved in drought and salt stress. Over-expressing the wheat TaDREB2 gene increases the survival of wheat under severe drought conditions (Morran et al., 2011). Over-expressing PgDREB2A results in better tolerance to high salt and drought stress in tobacco (Agarwal et al., 2010). Overexpressing MdDREB2A in apple calli enhances drought tolerance (Lian et al., 2021). In addition, MdAREB1A, MdRD22, MdRD29A, and MdRD29B are responsive to drought or osmotic stress in apples (Ma et al., 2017; An et al., 2018; Shao et al., 2019).
F-box proteins contain an F-box motif and have substrate recognition properties during ubiquitin-mediated protein hydrolysis. These proteins are called F-box because the motif was first identified in CyclinF (Risseeuw et al., 2003). Several studies have shown that F-box proteins play important roles in various signaling pathways, such as stress resistance, photomorphogenesis, development of flower organs, regulation of circadian rhythms, and growth hormone signaling (Sonneveld et al., 2005; Zhang et al., 2008; Sheard et al., 2010; Koops et al., 2011). Apple F-box protein MdAMR1L1 regulates the degradation of the ascorbic acid (Asc) synthesis catalase MdGMP1 protein through the ubiquitination pathway, affecting Asc synthesis and thus fruit quality and stress tolerance (Ma et al., 2022). KUF1 (KARRIKIN-upregulated F-box protein) negatively regulates drought tolerance in Arabidopsis by modulating various physiological traits, morphological adjustments, and ABA responses (Dubois, 2022; Tian et al., 2022). Overexpression of the F-box protein AtPP2-B11 reduces drought tolerance in Arabidopsis (Li et al., 2014). The soybean F-box-Like protein GmFBL144 interacts with a small heat shock protein to negatively regulate plant drought stress tolerance (Xu et al., 2022). Loss-of-function of ARABIDOPSIS F-BOX PROTEIN HYPERSENSITIVE TO ABA 1 enhances drought tolerance and delays germination (Kim et al., 2021).
Apples are important cultivated fruit trees in China. Apples often suffer from drought stress during growth, which seriously limits fruit quality and yield. NAC TFs play an important role in the apple response to abiotic stress. MdNAC29 plays a role in cold tolerance (An et al., 2018); however, its role in drought tolerance in apples has not been investigated. In this study, we cloned the NAC TF MdNAC29 (Phytozome Gene accession number: MDP0000481448) from apples. Over-expressing MdNAC29 reduced drought tolerance. The quantitative real-time polymerase chain reaction (qRT-PCR) results showed that overexpressing MdNAC29 reduced drought resistance by repressing the expression of drought tolerance genes. The yeast-one-hybrid (Y1H) and luciferase (Luc) assays demonstrated that MdNAC29 negatively regulates drought tolerance by directly binding to the MdDREB2A promoter and repressing its expression. Furthermore, MdNAC29 interacted with the F-box protein MdPP2-B10. Further studies showed that MdPP2-B10 responded to drought stress and enhanced the repressive effect of MdNAC29 on the transcriptional activity of the MdDREB2A. Our study provides new insight into the role of MdNAC29 in regulating drought tolerance in apples.
2 Materials and methods
2.1 Plant materials and growth conditions
The plant materials used in this study included ‘GL-3’ (Malus domestica) apple tissue culture seedlings (Wen et al., 2022), ‘Orin’ apple calli, and tobacco (Nicotiana benthamiana) seeds. The ‘GL-3’ apple seedlings were cultured in an incubator (16 h-light/8 h-dark) at 24°C. The apple calli were cultured on MS medium supplemented with 0.5 mg/L 6-BA and 1.5 mg/L 2,4-D at 24°C in the dark and subcultured at 12–16 day intervals. The sterilized tobacco seeds were planted on MS medium and incubated in the dark at 4°C for 72–96 h, then transferred to an incubator (incubation conditions: 14 h-light/10 h-dark; temperature: 24/22°C). The apple calli and tissue-cultured tobacco were used for genetic transformation. The substrate-cultivated transgenic tobacco and apple seedlings were used for functional identification.
2.2 RNA extraction and real-time quantitative PCR analysis
Total RNA was extracted using the FastPure® Plant Total RNA Isolation Kit-RC401 (Vazyme, Nanijing, China). RNA integrity was examined by agarose gel electrophoresis. Using RNase-Free ddH2O as the control, 1 µL of RNA solution was pipetted onto a NanoPhotometer P-360 ultra-microspectrophotometer (Munich, Germany) for concentration and purity, A260/A280 was in the range of 1.8-2.0, indicating that a high-quality RNA solution was obtained. Reverse transcription was performed using the HiScript III RT SuperMix kit R323-01 (Vazyme) to obtain cDNA. qRT-PCR was performed according to the fluorescent dye provided in the Ultra SYBR Mixture RT-qPCR CW0957 Kit (CW Biotech, Taizhou, China). The comparative Ct (2–ΔΔCt) method was used for analysis. MdACTIN was the internal apple reference gene (An et al., 2018), and qRT-PCR primers are listed in Supplementary Table 1.
2.3 Subcellular localization analysis
MdNAC29 was constructed on the PRI101-GFP vector and the recombinant plasmid was transferred into GV3101 Agrobacterium. Onion (Allium cepa) bulbs were selected and the inner epidermis was cut into multiple rectangular square pieces with a sterile scalpel blade. The pre-cultured onion inner epidermis was added to the prepared infection solution and infiltrated at 150 rpm for 20 min at 28°C. The excess bacterial solution was blotted off the inner epidermis with sterile blotting paper and re-laid on new MS solid medium on an ultra-clean bench. The inner onion epidermis was placed on a slide, and fluorescence of the target fusion protein was observed under a scanning confocal microscope LSM-880 (Zeiss AG, Zena, Germany) and photographed.
2.4 Vector construction and plant transformation
The MdNAC29 coding sequence (CDS) was ligated into the PRI101 vector (35S promoter, GFP) to obtain the MdNAC29 overexpressing vector and then transformed into Agrobacterium LBA-4404. Good-growing ‘GL-3’ apple tissue cultures seedlings were selected, the leaf tips and petioles were removed, the leaves were scratched vertically on the veins, laid flat on pre-culture medium, and incubated for 2-3 d protected from light. The leaves were incubated overnight at 220 rpm and 28°C, then 1 mL of activated bacteria was added to 30 mL of YEP+Rif+Kan liquid medium and shaken until the OD600 nm = 0.6-0.8. The mixture was centrifuged and the supernatant was discarded. A 30 mL portion of the MS suspension was added to resuspend the bacteria, followed by centrifugation; the supernatant was discarded. This was repeated three times. A 30 μL aliquot of acetosyringone was added. The ‘GL-3’apple leaves ‘were pre-cultured for 2 d and then transferred to the above Agrobacterium (28°C, 150 rpm for 30 min). After 2 d of dark culture, the apple leaves were transferred to apple leaf differentiation medium (MS + 0.5 mg/L NAA + 2.0 mg/L TDZ). The seedlings were excised from the leaf wound and inserted into the apple subculture medium to obtain transgenic apple seedlings. Transgenic plants were screened with kanamycin (kan). Three independent highly expressing lines were selected for the experiments. Transgenic apple calli were obtained by Agrobacterium-mediated transformation. Wild-type calli were incubated with Agrobacterium for 0.5 h, then transferred to selective medium for 30 d (An et al., 2020). The genetic transformation steps of tobacco leaves were the same as apple leaves. The gene CDSs are listed in Supplementary Table 2.
2.5 Measurements of relative electrical conductivity and malondialdehyde and chlorophyll contents
The appropriate amount of plant leaves were weighed, cut into small pieces, put in a centrifuge tube with ddH2O, soaked for 2 h at 24°C, and R1 conductivity was measured with the DDS-307A conductivity meter. Then, the leaf mixture was heated in a boiling water bath for 25-30 min, cooled to room temperature, and R2 final conductivity was determined. Relative electrical conductivity (%) = (R1/R2) × 100 (%). The thiobarbituric acid-based method was used to determine MDA content. Absorbance was determined at 450, 532, and 600 nm using a UV-vis spectrophotometer-2450 (Ma et al., 2017). Chlorophyll was extracted in 95% ethanol; the details are provided in An et al. (2021).
2.6 Yeast two-hybrid assays
The MdNAC29-pGBKT7 vector was constructed and transferred into receptor cells and cultured on -Trp/X-α-gal medium at 30°C to verify autonomous activity. Screening of MdNAC29-PGBKT7 vector yeast two-hybrid cDNA libraries: Denature 20 µL of Carrier DNA at 98°C for 5 min,4°C for 5 min, and repeat the process. Add 600 µL of Y2H Gold yeast receptor cells, 20 µL of pre-cooled MdNAC29-PGBKT7 vector plasmid and 20 µL of apple cDNA library, 2.5 mL of PEG/LiAc to 15 mL centrifuge tubes under ice bath conditions. Mix lightly with a water bath at 30°C for 45 min (turn and mix every 15 min). Add 160 µL of DMSO to the centrifuge tube, turn and mix, then mix in a water bath at 42°C for 20 min (turn and mix every 5 min). Centrifuge at 5400 rpm for 5 min and discard the supernatant. Add 3 mL of YPDA liquid medium to the centrifuge tube, 30°C, 220 rpm for 90 min. Centrifuge at 5400 rpm for 5 min and discard supernatant. Add 1 mL of sterile 0.9% NaCl solution to the centrifuge tube and apply 100 µL of the suspension to SD/-T-L medium and incubate for 3-5d at 30°C. Select the monoclonal clones that turn blue and re-line them to SD/-Ade/-His/-Leu/-Trp/X-α-gal medium. Screened MdPP2-B10 (NCBI: XM_008361688.3) was inserted into the pGADT7 vector following the Y2H assay steps as described previously (Li et al., 2021a).
2.7 Bimolecular fluorescence complementation assays
The MdNAC29 CDS was cloned into the pSPYCE-35S/pUC-SPYCE vector to produce MdNAC29-YFPC. The full-length MdPP2-B10 CDS was cloned into the pSPYNE-35S/pUC-SPYNE vector to produce MdPP2-B10-YFPN. The recombinant plasmids were transferred into Agrobacterium GV3101 and infected with onion epidermal cells using the same method as described in the Subcellular localization section.
2.8 Yeast one-hybrid assays
Full-length MdNAC29 was inserted into the pGADT7 vector, and the MdDREB2A promoter fragment (1264-1463bp upstream of ATG) was ligated into the pAbAi vector. The linearized plasmids were transferred into Y1H Gold yeast receptor cells and verified using Matchmaker one (Takara Bio, Shiga, Japan) to screen the bait strains for the AbA minimum inhibitory concentration. Different combinations of recombinant were co-transformed into Y1H yeast strains and cultured at -L-U+AbAmin, observed, and photographed. (Li et al., 2017).
2.9 Dual-luc assays
The MdDREB2A promoter fragment was inserted into the Luc vector, and MdNAC29 was inserted into the pGreenII0029-62SK vector. The different combinations of plasmids were transformed into Agrobacterium GV3101 strain, which were transiently injected into tobacco leaves, placed in the dark for 12 h, and incubated in a light incubator. The injected tobacco leaves were sprayed with D-luciferin sodium salt after 48-72 h and observed for fluorescence using a live imager (Xenogen, Alameda, CA, USA).
2.10 RNA sequencing
The RNA used for RNA-seq was extracted from Wild type (WT) and MdNAC29 transgenic ‘Orin’ apple calli. Transcriptome sequencing included RNA extraction, detection, library construction, and sequencing. First, Pearson’s correlation coefficient (R2) analysis was used as an indicator to assess the correlations in the samples, with an R2 value closer to 1 indicating that the two replicate samples were more correlated (Supplementary Figure S1). DEGseq2/edgeR was used to screen differentially expressed genes (DEGs) with q value < 0.05 and | log2 FoldChange | ≥ 1, and the total number of DEGs, the number of upregulated genes and the number of downregulated genes were counted. The sequencing data were filtered to obtain the clean data, which was compared with the apple genome to obtain the mapped data for structural analysis, such as variable splicing analysis, novel gene mining and gene structure optimization, differential expression analysis, KEGG functional annotation of differentially expressed genes, and functional enrichment based on the expression of genes in the samples or groups (Wang et al., 2010; Pertea et al., 2015; Yi et al., 2016; Chen et al., 2018).
2.11 Statistical analyses
Three biological repetitions were performed for each treatment, and each biological repetition had three technical replicates. Different letters represent significant differences (one-way ANOVA was performed using SPSS; SPSS Inc., Chicago, IL, USA, Tukey-Kramer test, p < 0.05). The data analysis was carried out using Student’s t-test, and asterisks denote significant differences. GraphPad Prism 6 software (GraphPad Software Inc., La Jolla, CA, USA) was used for drawing.
3 Results
3.1 MdNAC29 is induced by drought stress and localized in the nucleus
NAC29 gene expression is induced by multiple abiotic stressors in plants (Huang et al., 2015; Wang et al., 2015; Seok et al., 2017). To further investigate the response of MdNAC29 to drought stress, we subjected ‘GL-3’ apple seedlings to 18% PEG6000 treatment, as shown in Figure 1A. The MdNAC29 transcript level increased under the PEG treatment to reach its highest level at 3h followed by a decrease, indicating that MdNAC29 may play an important role in the drought stress response. The subcellular localization experiments revealed that MdNAC29 was localized in the nucleus (Figure 1B).
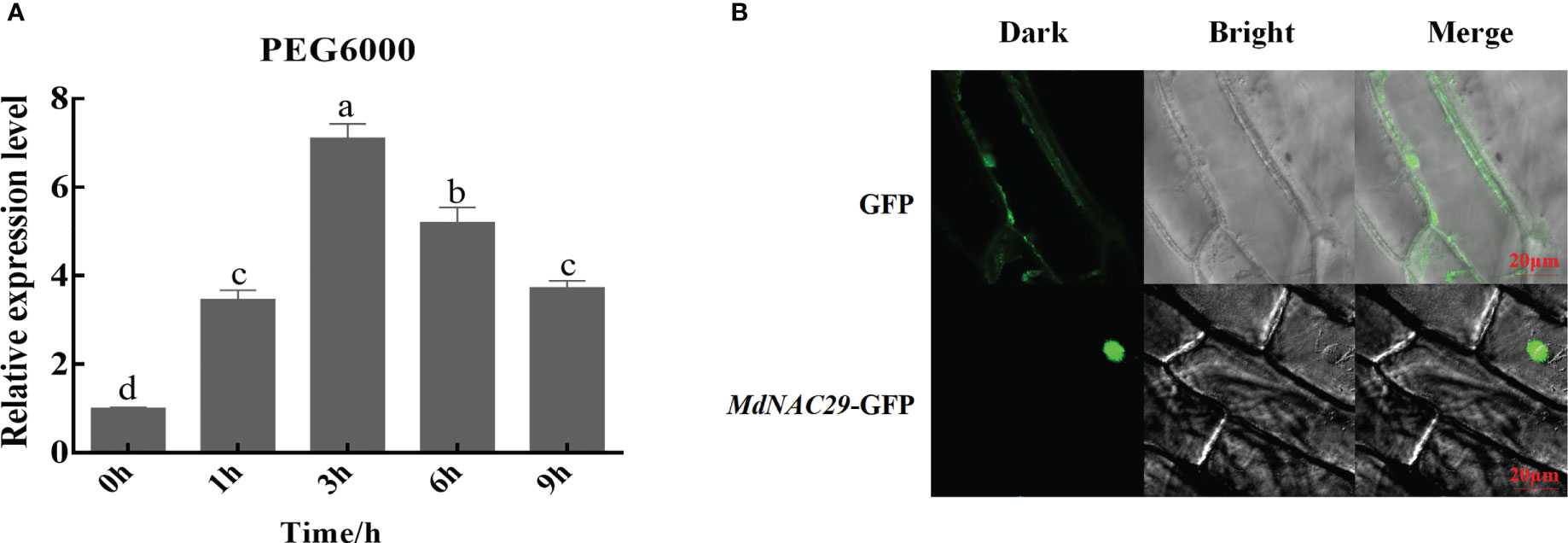
Figure 1 Effects of drought stress on the MdNAC29 transcript level and subcellular localization analysis. ‘GL-3’ apple seedlings grown at room temperature were treated with 18% PEG6000. (A) The transcript level of MdNAC29 was determined using qRT-PCR. (B) Subcellular localization of MdNAC29 in onion epidermal cells. Different letters represent significant differences (one-way ANOVA was performed using SPSS; SPSS Inc., Chicago, IL, USA, Tukey-Kramer test, p < 0.05).
3.2 MdNAC29 negatively regulates drought tolerance
We further validated the biological functions of MdNAC29 by constructing the overexpression vector (35S::MdNAC29) and obtaining the transgenic materials. First, control and MdNAC29- over-expressing apple plants were exposed to drought conditions for 25 d. More leaf wilt symptoms were observed in response to drought stress in MdNAC29 transgenic apple plants than in the control (Figure 2A). The qRT-PCR was used to detect the expression level of MdNAC29, and MdNAC29 expression was significantly upregulated in the overexpressing MdNAC29 transgenic apple plants (Figure 2B). To assess the drought tolerance of MdNAC29 transgenic apple plants, we measured relative electrical conductivity as well as malondialdehyde (MDA) and chlorophyll contents and found that the overexpressing MdNAC29 transgenic apple plants had higher MDA content (Figure 2C), higher relative electrical conductivity (Figure 2D), lower chlorophyll content (Figure 2E) than the control. These results suggest that overexpressing MdNAC29 reduced drought tolerance in apples. Next, 7-day-old WT and transgenic (35::MdNAC29) apple calli were transferred to MS medium supplemented with 6% PEG and cultured for 15 d. As shown in Figure 2F, overexpressing MdNAC29 apple calli resulted in more pronounced drought-sensitive phenotype, with lower fresh weight and higher MDA content compared to the WT (Figures 2G–I). In addition, consistent with the results of the transgenic apple plants, overexpressing MdNAC29 in tobacco (Figures 2J, K) by heterologous transformation resulted in lower chlorophyll content (Figure 2L) and higher relative electrical conductivity (Figure 2M) compared to the WT. Taken together, these results suggest that MdNAC29 acts as a negative regulator in drought tolerance.
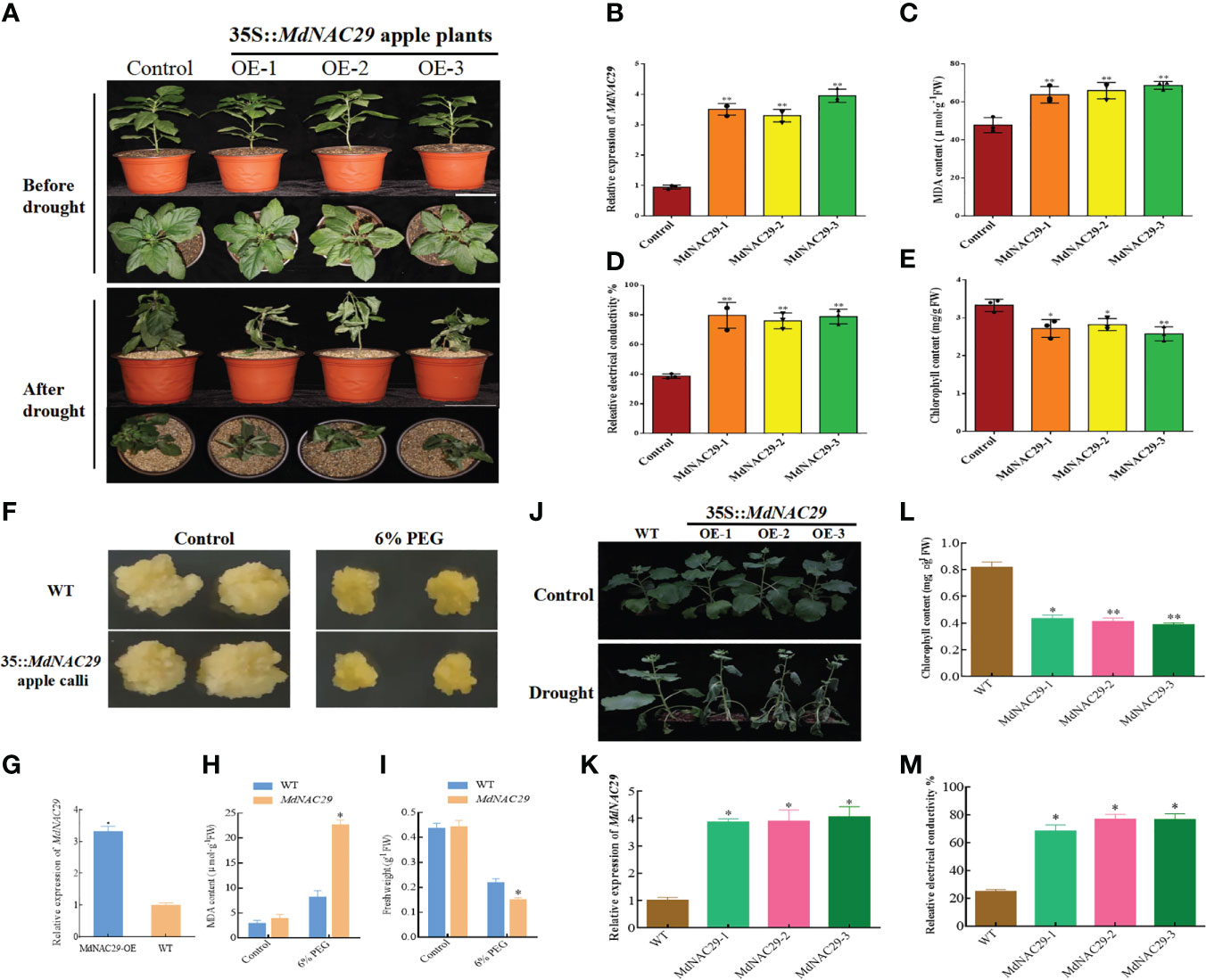
Figure 2 MdNAC29 reduces drought resistance in apple plants, calli and tobacco. (A) Phenotypes of control and MdNAC29 transgenic apple plants after 25d of drought treatment. Scale bar: 10 cm. (B) The transcript level of MdNAC29 was determined using qRT-PCR in transgenic apple. (C–E) MDA, relative electrical conductivity and chlorophyll content of control and MdNAC29 transgenic apple after drought treatment. (F) Phenotypes of WT and MdNAC29 transgenic apple calli after 15d of 6% PEG. (G) The transcript level of MdNAC29 was determined using qRT-PCR in transgenic apple calli. (H, I) MDA, fresh weight of WT and MdNAC29 transgenic apple calli after 15d of 6% PEG. (J) Phenotypes of WT and MdNAC29 transgenic tobacco after 10d of drought treatment. (K) The transcript level of MdNAC29 was determined using qRT-PCR in transgenic tobacco. (L, M) Chlorophyll content and relative electrical conductivity of WT and MdNAC29 transgenic tobacco after drought treatment. “*” denote significant differences p < 0.05, “**”denote significant differences p < 0.01.
3.3 Transcriptome analysis of overexpressing MdNAC29 in apple calli
3.3.1 Differential gene analysis
To further investigate the regulatory pathways associated with MdNAC29 drought tolerance, we performed RNA-seq analysis. Compared with the control (WT), 5,498 DEGs were detected, of which 2,689 genes were upregulated and 2,809 genes were downregulated (Figure 3A). The DEGs in the groups (MdNAC29 and WT) were taken and set together as differential gene sets for hierarchical cluster analysis, and the data were normalized using the Z-score. A cluster heat map was plotted for each grouping (Figure 3B). The FPKMs of all differential gene concatenations were normalized using R language to plot the Kmeans cluster analysis. The same class of genes had similar trends under the different treatments and may have similar functions (Figure 3C).
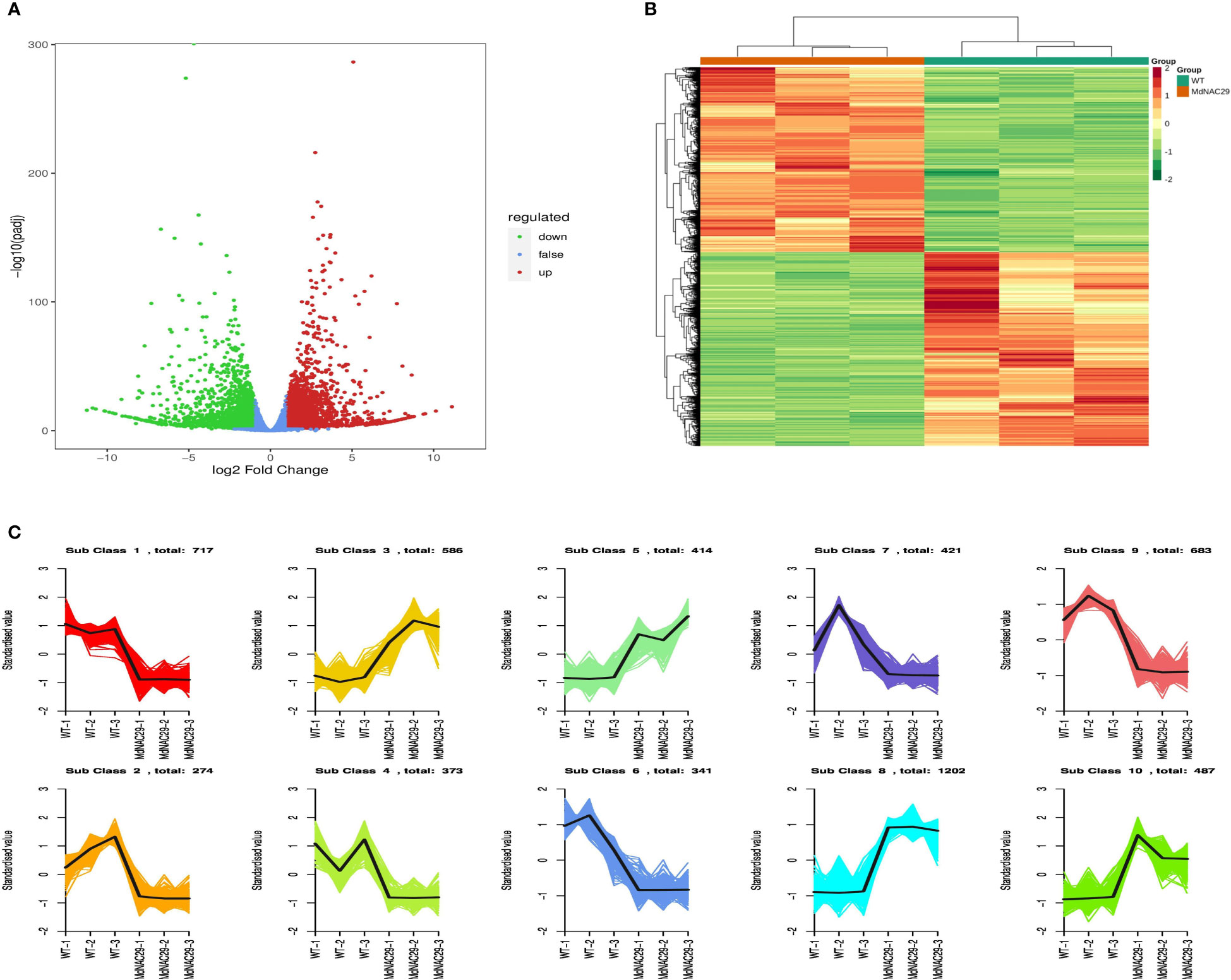
Figure 3 Screening and classification of differentially expressed genes between WT and MdNAC29 transgenic apple calli. (A) Scatterplot of differentially expressed RNAs. (B) Heat map of clustering of differentially expressed genes. (C) Kmeans analysis plot of differentially expressed genes.
3.3.2 GO classification and KEGG annotation of the DEGs
The GO classification analysis revealed that overexpressing MdNAC29 significantly enriched the DEGs in the biological processes of cellular composition and molecular functions (Figure 4A). We performed KEGG annotation and GO classification analyses of the DEGs generated by overexpressing MdNAC29 to further identify the functions of the genes. The KEGG enrichment analysis showed that the DEGs produced by overexpressing MdNAC29 were mainly enriched in the pathways of photosynthesis, plant hormone signal transduction, plant-pathogen interaction, and porphyrin and chlorophyll metabolism compared to the WT (Figure 4B). We selected the 50 GO-Terms with the lowest q-values in the GO enrichment analysis results and found that 11 genes regulated leaf senescence and 17 genes were related to chlorophyll binding (Supplementary Figure S2), and these biological processes and molecular functions were closely related to drought tolerance.
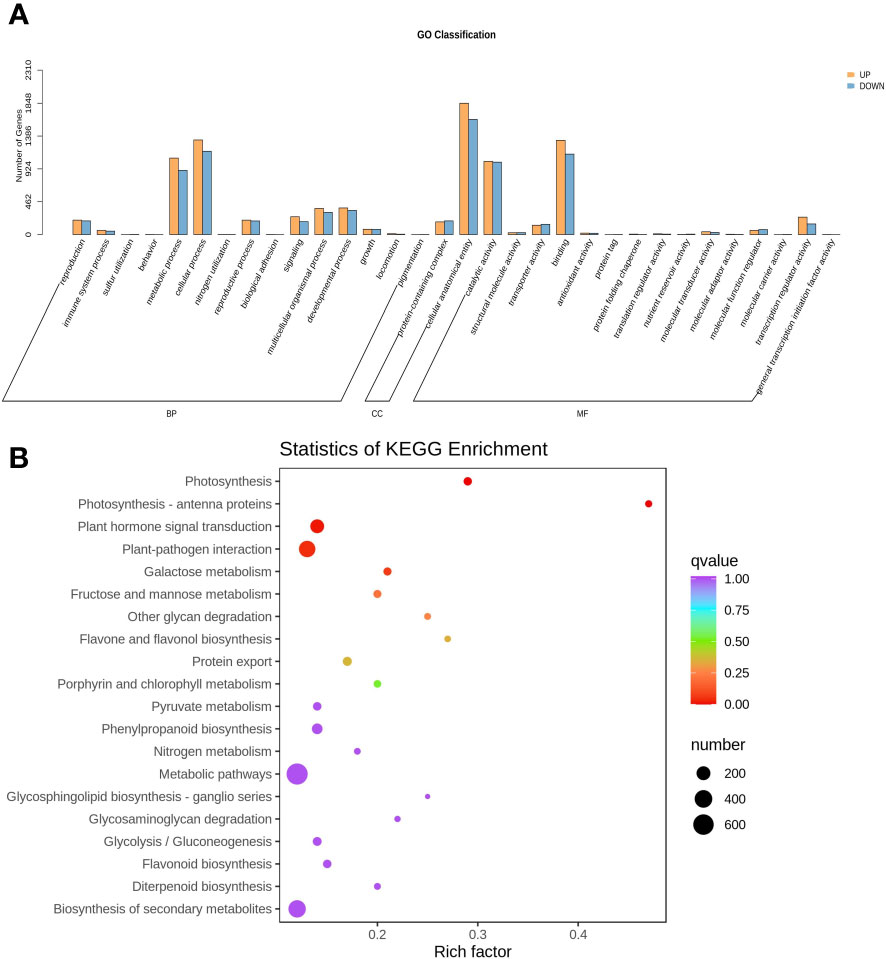
Figure 4 GO classification and KEGG annotation analyses of differentially expressed genes between the WT and MdNAC29 transgenic apple calli. (A) GO category histogram. (B) KEGG enrichment scatterplot.
3.4 Overexpression of MdNAC29 represses the expression of drought tolerance genes
To further analyze the effect of overexpressing MdNAC29 on genes associated with drought stress, we selected the key genes associated with drought stress and analyzed their expression levels in MdNAC29 transgenic apple plants by qRT-PCR (Supplementary Table 3). As shown in Figure 5, overexpressing MdNAC29 in apple plants resulted in a significant decrease in the expression of genes (MdERD5, MdRD22, MdRD29A, MdAREB1, MdDREB2A, MdMYB46) that positively regulate drought tolerance (Chen et al., 2019; Lian et al., 2021; Li et al., 2022; Liu et al., 2022), further indicating that MdNAC29 negatively regulates drought tolerance.
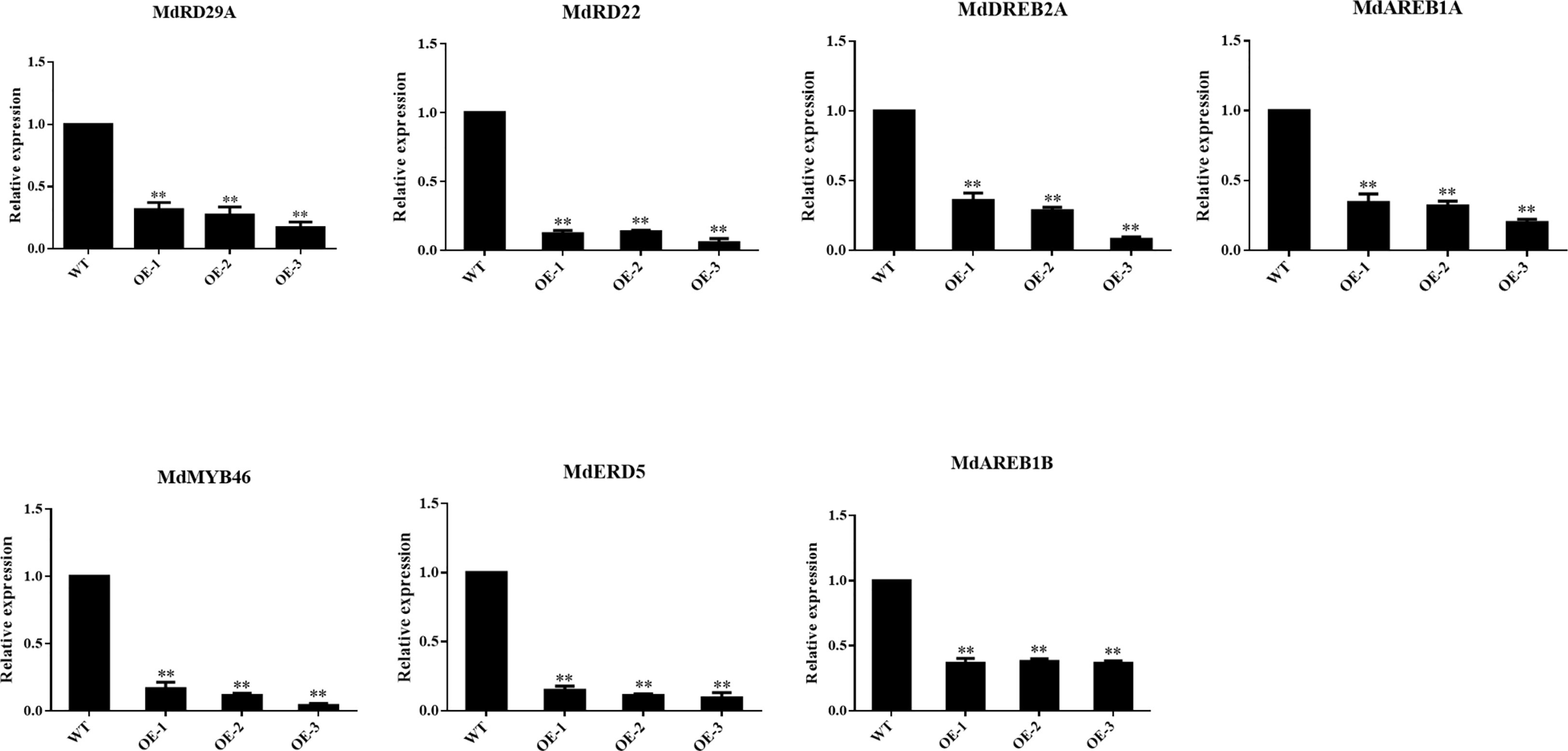
Figure 5 Expression analysis of key genes associated with drought tolerance in MdNAC29 transgenic apple plants. MdERD5: MD09G1080200, MdRD22: MD15G1098800, MdRD29A: MD01G1201000, MdAREB1B: MD05G1082000, MdAREB1A: MD15G1081800, MdDREB2A: MD01G1158600, MdMYB46: MD03G1176000. “*” denote significant differences p < 0.05, “**”denote significant differences p < 0.01.
3.5 MdNAC29 binds directly to the MdDREB2A promoter to repress its expression
The NAC TFs that bind to the DREB2A promoter to regulate drought tolerance in plants have been widely studied (Jin et al., 2017; Lian et al., 2021). We speculate that MdNAC29 in apple may also bind to the MdDREB2A promoter to regulate drought tolerance. We analyzed the MdDREB2A promoter sequence (Supplementary Table 4) and found that it contained the NAC TF binding site (ACACGT) (Figure 6A). Y1H assays were performed to test whether MdNAC29 directly binds to the MdDREB2A promoter. Only the co-transformed with PAbAi-MdDREB2A and PGADT7-MdNAC29 grew normally on -L-U+200AbA deficient medium (Figure 6B), indicating that MdNAC29 directly binds to the MdDREB2A promoter. To further verify how MdNAC29 regulates the expression of MdDREB2A, we performed the dual luciferase reporter assay. The LUC/REN relative activity of the co-expressed MdNAC29-62SK+MdDREB2A-Luc combination decreased significantly compared to the combination of 62SK+MdDREB2A-Luc (Figure 6C), the mutation of the NAC binding site (ACACGT) in the MdDREB2A promoter (ACCCGG), MdNAC29 did not affect MdDREB2A expression, indicating that MdNAC29 inhibited MdDREB2A expression. In addition, qRT-PCR analysis revealed that MdDREB2A expression reached its highest level after 1 h of PEG treatment and then gradually decreased (Figure 6D).
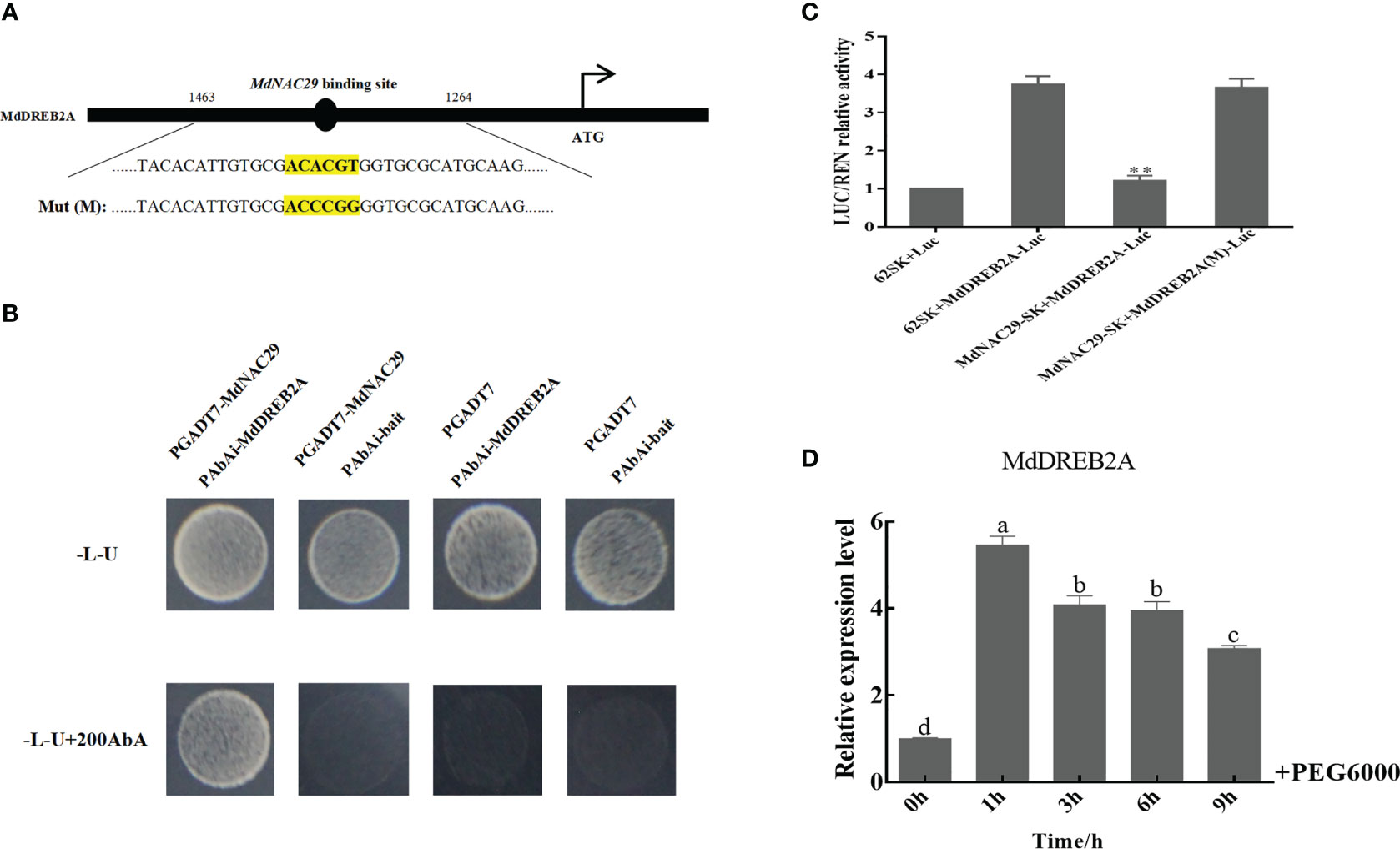
Figure 6 MdNAC29 repressed expression of the drought tolerance gene MdDREB2A. (A) Structural analysis of MdDREB2A. (B) YIH assays of MdNAC29 bound to the MdDREB2A promoter in vitro. (C) LUC/REN relative activity was detected from the effects of MdNAC29-62SK on the expression of MdDREB2A-Luc. (D) Expression pattern of MdDREB2A in response to PEG treatment. Different letters represent significant differences (one-way ANOVA was performed using SPSS; SPSS Inc., Chicago, IL, USA, Tukey-Kramer test, p < 0.05). **”denote significant differences p < 0.01.
3.6 MdNAC29 interacts with MdPP2-B10 in response to drought stress
To further analyze whether MdNAC29 interacts with MdPP2-B10 to co-regulate drought tolerance, we analyzed the MdNAC29 structure and determined that the NAC domain was located at 10-134 aa (Figure 7A). MdNAC29 was tested for autonomous activity and had strong autonomous activation activity (Figure 7B). We verified by segmentation (1-150aa and 150-280 aa) and found no autonomous activation at 1-150 aa, which could be used in subsequent Y2H assays. After preliminary screening by the Y2H assay, only the combination of MdNAC291-150aa-pGADT7 + MdPP2-B10-pGBKT7 grew normally on QDO/X-α-Gal medium, indicating that MdNAC29 interacts with MdPP2-B10 (Figure 7C), and the interaction between MdNAC29 with MdPP2-B10 was further confirmed by BiFC assay (Figure 7D). The Y2H and BiFC primers are listed in Supplementary Table 1. Furthermore, the expression of MdPP2-B10 was significantly higher in overexpressing MdNAC29 apple plants compared to WT (Figure 7E). and we analyzed the MdPP2-B10 expression pattern after the PEG6000 treatment and found that MdPP2-B10 responded to drought stress (Figure 7F). To verify whether MdPP2-B10 had an effect on MdNAC29-mediated transcription of MdDREB2A, it was further analyzed by LUC/REN relative activity test, which showed that MdPP2-B10 enhanced the repressive effect of MdNAC29 on the transcriptional activity of the downstream target gene MdDREB2A (Figure 7G).
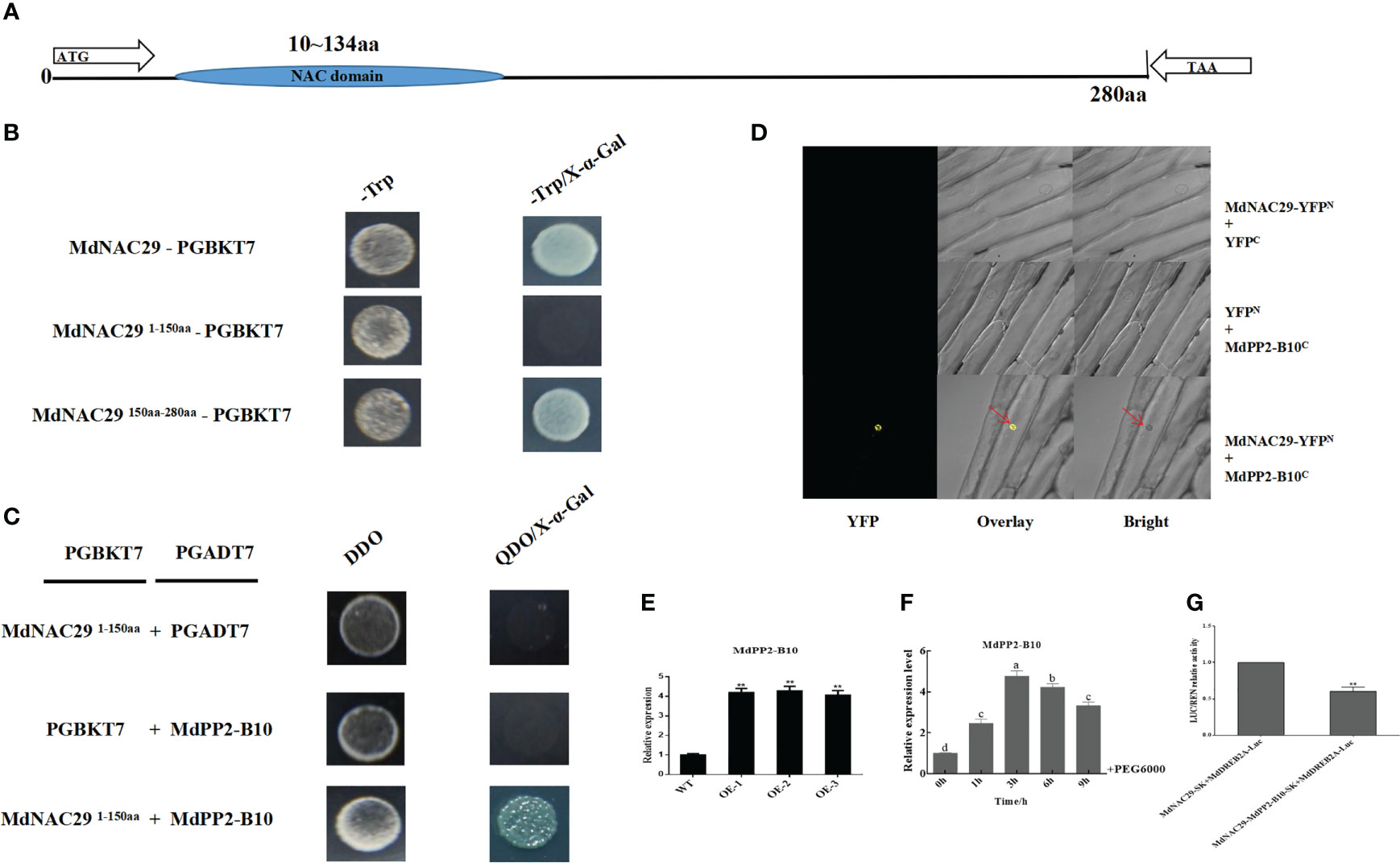
Figure 7 MdNAC29 directly interacts with MdPP2-B10 and MdPP2-B10 responds to PEG treatment. (A) Structural analysis of MdNAC29. (B) The MdNAC29 protein sequence (1-150 aa) fragment had no autonomous activation activity. (C) Y2H assays verified the interaction between MdNAC29 and MdPP2-B10. The PGADT7 and PGBKT7 empty vectors were used as negative controls, DDO:-T-L medium, QDO:-T-L-A-H medium. (D) BiFC assays to verify the interaction between MdNAC29 and MdPP2-B10. (E) Expression analysis of MdPP2-B10 in MdNAC29 transgenic apple plants. (F) Expression pattern of MdPP2-B10 in response to PEG treatment. (G) LUC/REN assay to analyze the effect of MdPP2-B10 on MdNAC29-mediated transcriptional activation of MdDREB2A. Different letters represent significant differences (one-way ANOVA was performed using SPSS; SPSS Inc., Chicago, IL, USA, Tukey-Kramer test, p < 0.05). **”denote significant differences p < 0.01.
4 Discussion
Plants have evolved morphological, physiological, and molecular resistance mechanisms in response to abiotic stress, and among these molecular mechanisms are the regulation of gene expression by TFs and the roles of some functional genes. NAC genes are TFs specific to plants that affect plant growth and development while also playing a role in transcriptional regulation of various abiotic stressors, such as drought, salt, and cold stress as well as pathogen infection (Sun et al., 2012; Tak et al., 2017). Members of the NAC gene family are differentially expressed in response to abiotic stress. Here, we isolated the NAC TF MdNAC29 in apples. Our results show that MdNAC29 was localized to the nucleus and responded to drought stress (Figure 1). Overexpressing MdNAC29 negatively regulated drought tolerance in transgenic apple plants, calli, and tobacco (Figure 2). MdERD5, MdRD22, MdRD29A, MdAREB1, MdDREB2A and MdMYB46 have been widely reported to enhance drought tolerance in plants (An et al., 2018; Chen et al., 2019; Li et al., 2022; Liu et al., 2022). We speculate that MdNAC29 regulates the expression of these drought-responsive genes. The qRT-PCR results showed that overexpressing MdNAC29 in apple plants significantly downregulated the expression of these genes (Figure 5).
The DREB TF family is essential in improving stress-resistant plant varieties (Zhao et al., 2012; Yoshida et al., 2014). The DREB transcription factor (TaDTG6-B) ameliorates drought tolerance in wheat (Mei et al., 2022). MdDREB2A is a key drought-tolerance gene in apples (Chen et al., 2020; Li et al., 2022). In our study, we analyzed the MdDREB2A promoter sequence and detected a NAC binding site (ACACGT), which further confirmed that MdNAC29 could bind the MdDREB2A promoter and inhibit its expression (Figure 6). The RNA-seq results showed that overexpressing MdNAC29 positively regulated leaf senescence (Supplementary Figure S2). NAC29 also positively regulates leaf senescence in Arabidopsis and tobacco (Liang et al., 2014; Ma et al., 2020), suggesting that the NAC29 gene is highly conserved across species. Hence, our study provides a new basis for future analysis of MdNAC29 regulation of leaf senescence. Furthermore, DREB2A may bind to the DRE element (A/GCCGAC) of target gene promoters (e.g., COR, RD, LTI, and DHN) to regulate their expression during tolerance to stress (Liu et al., 2014). Interestingly, the DRE motifs were present on the MdAREB1A and MdRD22 promoters (Supplementary Table 5). We hypothesized that MdDREB2A regulates the expression of MdAREB1A and MdRD22 to affect drought tolerance in apples.
F-box proteins are widely found in eukaryotes and are important components of the SCF-E3 ubiquitin ligase complex. These proteins are involved in important physiological processes, such as cell cycle regulation, gene transcriptional regulation, apoptosis, and signal transduction, through the ubiquitin-proteasome pathway (Nakayama and Nakayama, 2006). In Arabidopsis, BAF1 (F-box E3 ubiquitin ligase BES1-ASSOCIATED F-BOX1) mediates the degradation of the BES1 (Brassinosteroid-activated transcription factor) through selective autophagy (Wang et al., 2021). F-box proteins respond to a wide range of abiotic stressors. In Arabidopsis, the F-box protein DOR specifically interacts with ASK14 and CUL1 to negatively regulate drought tolerance. DOR inhibits ABA-induced stomatal closure under drought stress (Zhang et al., 2008). Overexpression of the grapevine F-box protein VvF-box5 significantly increases the survival rate of transgenic Arabidopsis under drought treatment (Yu et al., 2018). Our study revealed that MdNAC29 interacted with MdPP2-B10, the expression of MdPP2-B10 was significantly higher in overexpressing MdNAC29 apple plants compared to WT (Figure 5), and MdPP2-B10 responds to drought stress (Figure 7E). Moreover, LUC/REN assays showed that MdPP2-B10 enhanced the repressive effect of MdNAC29 on the transcriptional activity of the MdDREB2A (Figure 7F). Most of the reported F-box proteins had been shown to function via SCF-dependent protein degradation, some F-box proteins that do not depend on the formation of the SCF complex to perform their functions, but rather alter the expression of downstream genes by regulating transcription factor activity, thereby affecting the plant’s resistance response. (Jia et al., 2018). We speculate that MdPP2-B10 may modify the MdNAC29 protein and affecting its expression without causing its degradation. Therefore, investigating whether MdPP2-B10 forms the SCF complex and the molecular mechanism of its interaction will be the focus of our future research.
In conclusion, based on the results of this study, we propose a working model for the regulation of drought tolerance by MdNAC29 in response to drought stress (Figure 8). Drought stress induced the expression of MdNAC29, which repressed its expression by binding to the MdDREB2A promoter motif (ACACGT), which, in turn, affected the expression of its downstream target genes to negatively regulate drought tolerance. Drought stress induced the expression of MdPP2-B10, which interacted with MdNAC29 to co-regulate drought tolerance. Our findings provide new insight into the response mechanism of MdNAC29 to drought stress.
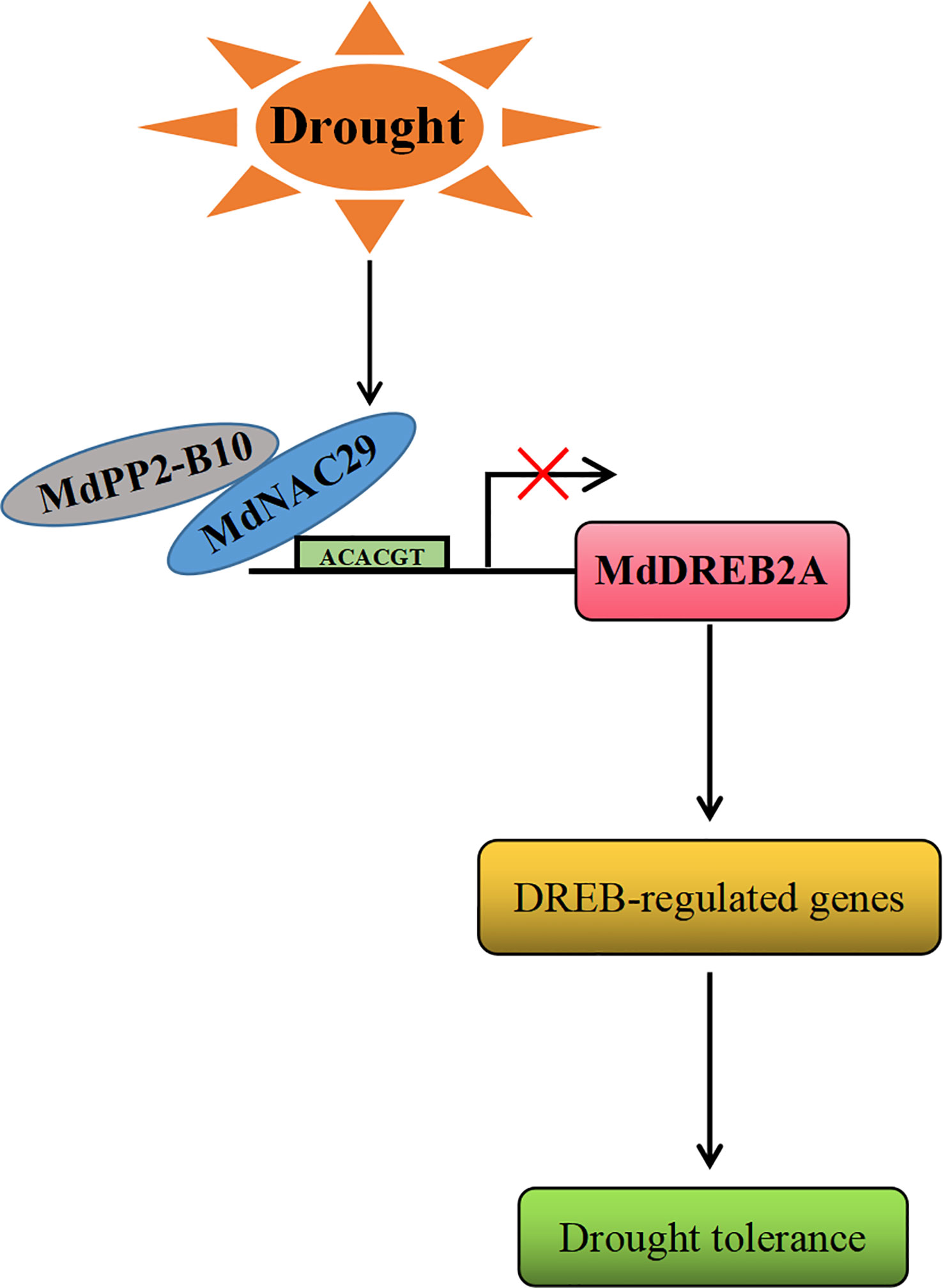
Figure 8 A working model for the regulation of drought tolerance by MdNAC29. Drought stress induced the expression of MdNAC29, which was repressed to negatively regulate drought tolerance by binding to the MdDREB2A promoter element (ACACGT). Drought stress induced MdPP2-B10 expression, which interacted with MdNAC29 to jointly regulate drought tolerance.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) repository, accession number PRJNA946317.
Author contributions
LL, WX, SL, and XJ designed the study. SL and XJ performed the experiments. SL, XJ, QT, and BW analyzed the data. SL and XJ wrote the paper. XF, DL, XC, WX, and LL participated in the manuscript amending. SL and XJ are the co-first author and contributed equally to this work. LL and WX are the co-corresponding author. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by Natural Science Foundation of Shandong Province (ZR2020ZD18), Shandong Provincial Fruit Industry Technology System -Cultivation and Soil Fertilization Post (SDAIT-06-04), Shandong Province Major Science and Technology Innovation Project (2021LZGC021).
Acknowledgments
We are grateful to Wuhan Metware Biotechnology Co., Ltd for assisting in sequencing and orbioinformatics analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1173107/full#supplementary-material
References
Agarwal, P., Agarwal, P. K., Joshi, A. J., Sopory, S. K., Reddy, M. K. (2010). Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Mol. Biol. Rep. 37, 1125–1135. doi: 10.1007/s11033-009-9885-8
Agarwal, P. K., Agarwal, P., Reddy, M. K., Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25, 1263–1274. doi: 10.1007/s00299-006-0204-8
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2018). An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 221, 74–80. doi: 10.1016/j.jplph.2017.12.009
An, J. P., Wang, X. F., Espley, R. V., Lin, W. K., Bi, S. Q., You, C. X., et al. (2020). An apple b-box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergisticallywith MdMYBs and MdHY5. Plant Cell Physiol. 61, 130–143. doi: 10.1093/pcp/pcz185
An, J. P., Zhang, X. W., Liu, Y. J., Zhang, J. C., Wang, X. F., You, C. X., et al. (2021). MdABI5 works with its interaction partners to regulate abscisic acid-mediated leaf senescence in apple. Plant J. 105 (6), 1566–1581. doi: 10.1111/tpj.15132
Basia, V., Arie, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Chen, K. Q., Guo, Y. N., Song, M. R., Liu, L. F., Xue, H., Dai, H. Y., et al. (2020). Dual role of MdSND1 in the biosynthesis of lignin and in signal transduction in response to salt and osmotic stress in apple. Horticulture Res. 7 (1), 283–295. doi: 10.1038/s41438-020-00433-7
Chen, K. Q., Song, M. R., Guo, Y. N., Liu, L. F., Xue, H., Dai, H. Y., et al. (2019). MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 17 (12), 2341–2355. doi: 10.1111/pbi.13151
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Dubois, M. (2022). KUFfed by drought: a KARRIKIN-upregulated f-box protein compromises plant growth and survival under drought. Plant Physiol. 190 (4), 2087–2089. doi: 10.1093/plphys/kiac413
Eckardt, N. A. (2019). DREB duo defines distinct drought and cold response pathways. Plant Cell. 31, 1196–1197. doi: 10.1105/tpc.19.00331
Huang, Q. J., Wang, Y., Li, B., Chang, J. L., Chen, M. J., Li, K. X., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic arabidopsis. BMC Plant Biol. 15 (1), 268. doi: 10.1186/s12870-015-0644-9
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S. H., Choi, D. Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153 (1), 185–197. doi: 10.1104/pp.110.154773
Jia, Q., Sun, S., Sun, T. H., Lin, W. X. (2018). Mechanism of f-box protein family in plant resistance response to environmental stress. Chin. J. Eco-Agriculture 26 (8), 1125–1136. doi: 10.13930/j.cnki.cjea.171170
Jin, C., Li, K. Q., Xu, X. Y., Zhang, H. P., Chen, H. X., Chen, Y. H., et al. (2017). A novel nac transcription factor, pbenac1, of pyrus betulifolia confers cold and drought tolerance via interacting with pbedrebs and activating the expression of stress-responsive genes. Front. Plant Sci. 8, 01049. doi: 10.3389/fpls.2017.01049
Kavi, K. P. B., Sangam, S., Amrutha, R. N., Sri, L. P., Naidu, K. R., Rao, K. R. S., et al. (2005). Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr. Science. 88, 424–438.
Khan, M. S. (2014). The role of dreb transcription factors in abiotic stress tolerance of plants. Biotechnol. Biotechnol. Equip. 25, 2433–2442. doi: 10.5504/BBEQ.2011.0072
Kim, H., Song, E., Kim, Y., Choi, E., Hwang, J., Lee, J. H. (2021). Loss-of-function of ARABIDOPSIS f-BOX PROTEIN HYPERSENSITIVE TO ABA 1 enhances drought tolerance and delays germination. Physiologia Plantarum 173 (4), 2376–2389. doi: 10.1111/ppl.13588
Koops, P., Pelser, S., Ignatz, M., Klose, C., Marrocco, S. K., Kretsch, T. (2011). EDL3 is an f-box protein involved in the regulation of abscisic acid signalling in arabidopsis thaliana. J. Exp. Botany. 62 (15), 5547–5560. doi: 10.1093/jxb/err236
Li, Y. Z., Jia, F. J., Yu, Y. L., Luo, L., Huang, J. G., Yang, G. D., et al. (2014). The SCF E3 ligase AtPP2-B11 plays a negative role in response to drought stress in arabidopsis. PMB Reporter. 32, 943–956. doi: 10.1007/s11105-014-0705-5
Li, S., Wang, Q. J., Chen, X. D., Zhang, R., Li, L., Du, G. Y., et al. (2021b). Cloning and expression analysis of transcription factor PpWRKY18 in peach. J. Nucl. Agric. Sci. 35 (9), 1987–1993. doi: 10.11869/j.issn.100-8551.2021.09.1987
Li, S., Wang, Q. J., Wen, B. B., Zhang, R., Jing, X. L., Xiao, W., et al. (2021a). Endodormancy release can be modulated by the GA4-GID1c-DELLA2 module in peach leaf Buds.Front. Plant Sci. 12, 713514. doi: 10.3389/fpls.2021.713514
Li, T., Xu, Y., Zhang, L., Ji, Y., Tan, D., Yuan, H., et al. (2017). The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29, 1316–1334. doi: 10.1105/tpc.17.00349
Li, F., Zhang, L., Ji, H., Xu, Z., Zhou, Y., Yang, S. (2020). The specific W-boxes of GAPC5 promoter bound by TaWRKY are involved in drought stress response in wheat. Plant Sci. 296, 110460. doi: 10.1016/j.plantsci.2020.110460
Li, X. W., Zhou, S. X., Liu, Z. Y., Lu, L. Y., Dang, H., Li, H. M., et al. (2022). Fine-tuning of SUMOylation modulates drought toleranceof apple. Plant Biotechnol. J. 20, 903–919. doi: 10.1111/pbi.13772
Lian, X. Y., Zhao, X. Y., Zhao, Q., Wang, G. L., Li, Y. Y., Hao, Y. J. (2021). MdDREB2A in apple is involved in the regulation of multiple abiotic stress responses. Hortic. Plant J. 7 (3), 197–208. doi: 10.1016/j.hpj.2021.03.006
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U. S. A. 111, 10013–10018.
Liu, Y. J., An, J. P., Gao, N., Wang, X., Chen, X. X., Wang, X. F., et al. (2022). MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple. Plant Cell Environ. 45 (11), 3233–3248. doi: 10.1111/pce.14429
Liu, H., Li, D. J., Deng, Z. (2014). Advances in research of transcriptional regulatory network in response to cold stress in plants. Scientia Agricultura Sinica. 47 (18), 3523–3533. doi: 10.3864/j.issn.0578-1752.2014.18.001
Lu, P. L., Chen, N. Z., An, R., Su, Z., Qi, B. S., Ren, F., et al. (2007). A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in arabidopsis. Plant Mol. Biol. 63 (2), 289–305. doi: 10.1007/s11103-006-9089-8
Ma, L., Li, R. P., Ma, L. Y., Song, N., Xu, Z., Wu, J. S. (2020). Involvement of NAC transcription factor NaNAC29 in alternaria alternata resistance and leaf senescence in nicotiana attenuata. Plant Diversity. 43 (6), 502–509. doi: 10.1016/j.pld.2020.11.003
Ma, S. Y., Li, H. X., Wang, L., Li, B. Y., Wang, Z. Y., Ma, B. Q., et al. (2022). F-box protein MdAMR1L1 regulates ascorbate biosynthesis in apple by modulating GDP-mannose pyrophosphorylase. Plant Physiol. 188, 653–669. doi: 10.1093/plphys/kiab427
Ma, Q. J., Sun, M. H., Lu, J., Liu, Y. J., Hao, Y. J. (2017). An apple CIPK protein kinase targets a novel residue of AREB transcription factor for ABA-dependent phosphorylation. Plant Cell Environ. 40 (10), 2207–2219. doi: 10.1111/pce.13013
Mao, X., Chen, S., Li, A., Zhai, C., Jing, R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in arabidopsis. PloS One 9 (1), 84359. doi: 10.1371/journal.pone.0084359
Mei, F. M., Chen, B., Du, L. Y., Li, S. M., De, H., Chen, N., et al. (2022). A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. Plant Cell 34 (11), 4472–4494.
Morran, S., Eini, O., Pyvovarenko, T., Parent, B., Singh, R., Ismagul, A., et al. (2011). Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. doi: 10.1111/j.1467-7652.2010.00547.x
Nakayama, K. I., Nakayama, K. (2006). Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 6, 369–381. doi: 10.1038/nrc1881
Olsen, A. N., Ernst, H. A., Leggio, L. L., Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87.
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from rna-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Puranik, S., Sahu, P. P., Srivastava, P. S., Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Risseeuw, E. P., Daskalchuk, T. E., Banks, T. W., Liu, E., Cotelesage, J., Hellmann, H., et al. (2003). Protein interaction analysis of SCF ubiquitin E3 ligase subunits from arabidopsis. Plant J. 6 (34), 753–767. doi: 10.1046/j.1365-313X.2003.01768.x
Sakuraba, Y., Kim, Y. S., Han, S. H., Lee, B. D., Paek, N. C. (2015). The arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27, 1771–1787.
Seok, H. Y., Woo, D. H., Nguyen, L. V., Tran, H. T., Tarte, V. N., Mehdi, S. M., et al. (2017). Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 245, 329–341.
Shao, H. B., Wang, H. Y., Tang, X. L. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6, 00902. doi: 10.3389/fpls.2015.00902
Shao, Y., Zhang, X., Nocker, S., Gong, X., Ma, F. (2019). Overexpression of a protein kinase gene MpSnRK2.10 from malus prunifolia confers tolerance to drought stress in transgenic arabidopsis thaliana and apple. Gene 692, 26–34. doi: 10.1016/j.gene.2018.12.070
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468 (7322), 400–405. doi: 10.1038/nature09430
Sonneveld, T., Tobutt, K. R., Vaughan, S. P., Robbins, T. P. (2005). Loss of pollen-s function in two self-compatible selections of prunus avium is associated with deletion/mutation of an s haplotype-specific f-box gene. Plant Cell. 17 (1), 37–51. doi: 10.1105/tpc.104.026963
Souer, E., Houwelingen, V. A., Kloos, D., Mol, J., Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordial boundaries. Cell 85, 159–170. doi: 10.1016/S0092-8674(00)81093-4
Sun, Q. G., Jiang, S. H., Zhang, T. L., Xu, H. F., Fang, H. C., Zhang, J., et al. (2019). Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 289, 110286. doi: 10.1016/j.plantsci.2019.110286
Sun, L. J., Li, D. Y., Zhang, H. J., Song, F. M. (2012). Functions of NAC transcription factors in biotic and abiotic stress responses in plants. HEREDITAS 34 (8), 993–1002. doi: 10.3724/SP.J.1005.2012.00993
Tak, H., Negi, S., Ganapathi, T. R. (2017). Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 254, 803–816. doi: 10.1007/s00709-016-0991-x
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284 (3), 173–183. doi: 10.1007/s00438-010-0557-0
Tian, H. T., Watanabe, Y., Nguyen, K. H., Tran, C. D., Abdelrahman, M., Liang, X. H., et al. (2022). KARRIKIN UPREGULATED f-BOX 1 negatively regulates drought tolerance in arabidopsis. Plant Physio. 190, 4, 2671–2687. doi: 10.1093/plphys/kiac336
Tran, L. S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of arabidopsis stress inducible NAC transcription factors that bind to a drought responsive cis-element in the early responsive to dehydration stress1promoter. Plant Cell. 16 (9), 2481–2498. doi: 10.1105/tpc.104.022699
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant journal: Cell Mol. Biol. 83 (2), 224–236. doi: 10.1111/tpj.12879
Wang, L. K., Feng, Z. X., Wang, X., Wang, X. W., Zhang, X. G. (2010). DEGseq: an r package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26 (1), 136–138. doi: 10.1093/bioinformatics/btp612
Wang, P., Nolan, T. M., Clark, N. M., Jiang, H., MontesSerey, C., Guo, H. Q., et al. (2021). The f-box E3 ubiquitin ligase BAF1 mediates the degradation of the brassinosteroid-activated transcription factor BES1 through selective autophagy in arabidopsis. Plant Cell. 33 (11), 3532–3554. doi: 10.1093/plcell/koab210
Wen, B. B., Gong, X. Y., Tan, Q. P., Zhao, W. Z., Chen, X. D., Li, D. M., et al. (2022). MdNAC4 interacts with MdAPRR2 to regulate nitrogen DeficiencyInduced leaf senescence in apple (Malus domestica). Front. Plant Science. 13, 925035. doi: 10.3389/fpls.2022.925035
Xu, K. H., Zhao, Y., Zhao, Y., Feng, C., Zhang, Y. H., Wang, F. W., et al. (2022). Soybean f-Box-Like protein GmFBL144 interacts with small heat shock protein and negatively regulates plant drought stress tolerance. Front. Plant Science. 13, 823529. doi: 10.3389/fpls.2022.823529
Yi, Z., Chen, J., Sun, H., Rosli, H. G., Pombo, M. A., Zhang, P. (2016). iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670. doi: 10.1016/j.molp.2016.09.014
Yoshida, T., Mogami, J., Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yu, L. F., Zhou, S., Zhang, S. X., Ji, Y. J., Liu, Y. W., et al. (2018). Gene structure and expression analysis of f-box gene VvF-box5 in grapevine (Vitis vinifera l.). 19(2), 361–369.
Zhang, J. Y., Wang, Q. J., Guo, Z. R. (2012). Progresses on plant AP2/ERF transcription factors. HEREDITAS 34 (7), 835–847. doi: 10.3724/SP.J.1005.2012.00835
Zhang, Y., Xu, W., Li, Z., Deng, X. W., Wu, W., Xue, Y. (2008). F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in arabidopsis. Plant Physiol. 148 (4), 2121–2133. doi: 10.1104/pp.108.126912
Zhao, T., Liang, D., Wang, P., Liu, J., Ma, F. (2012). Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol. Genet. Genomics 287 (5), 423–436. doi: 10.1007/s00438-012-0687-7
Keywords: apple, MdNAC29, MdDREB2A, drought tolerance, F-box protein, RNA-seq
Citation: Li S, Jing X, Tan Q, Wen B, Fu X, Li D, Chen X, Xiao W and Li L (2023) The NAC transcription factor MdNAC29 negatively regulates drought tolerance in apple. Front. Plant Sci. 14:1173107. doi: 10.3389/fpls.2023.1173107
Received: 24 February 2023; Accepted: 05 April 2023;
Published: 06 July 2023.
Edited by:
Ping Wang, Iowa State University, United StatesCopyright © 2023 Li, Jing, Tan, Wen, Fu, Li, Chen, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Xiao, Z3VsaWdodDk4NjkxOEAxNjMuY29t; Ling Li, bGlsaW5nc2RhdUAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Sen Li
Sen Li Xiuli Jing
Xiuli Jing Qiuping Tan
Qiuping Tan Binbin Wen
Binbin Wen Xiling Fu1,2,3
Xiling Fu1,2,3 Xiude Chen
Xiude Chen Ling Li
Ling Li