- 1Department of Biotechnology, ICAR-Indian Institute of Rice Research, Hyderabad, India
- 2Crop Improvement, Council of Scientific & Industrial Research (CSIR)-Centre for Cellular and Molecular Biology, Hyderabad, India
- 3Seeds/Biotech R&D Division, Rallis India Limited, Bangalore, India
- 4Institute of Molecular Physiology, Heinrich Heine University, Düsseldorf, Germany
- 5Crop Improvement, Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India
- 6Department of Plant Sciences, School of Life Sciences, University of Hyderabad, Hyderabad, India
- 7Rice Breeding Innovation Platform, International Rice Research Institute (IRRI), Los Banos, Philippines
- 8School of Biotechnology, Kalinga Institute of Industrial Technology (KIIT) University, Bhubaneswar, Odisha, India
- 9Department of Genetics, Osmania University, Hyderabad, India
- 10Safe and Sustainable Value Chain, World Vegetable Center, Tainan, Taiwan
Bacterial blight disease of rice caused by Xanthomonas oryzae pv. oryzae (Xoo) is one of the most serious constraints in rice production. The most sustainable strategy to combat the disease is the deployment of host plant resistance. Earlier, we identified an introgression line, IR 75084-15-3-B-B, derived from Oryza officinalis possessing broad-spectrum resistance against Xoo. In order to understand the inheritance of resistance in the O. officinalis accession and identify genomic region(s) associated with resistance, a recombinant inbred line (RIL) mapping population was developed from the cross Samba Mahsuri (susceptible to bacterial blight) × IR 75084-15-3-B-B (resistant to bacterial blight). The F2 population derived from the cross segregated in a phenotypic ratio of 3: 1 (resistant susceptible) implying that resistance in IR 75084-15-3-B-B is controlled by a single dominant gene/quantitative trait locus (QTL). In the F7 generation, a set of 47 homozygous resistant lines and 47 homozygous susceptible lines was used to study the association between phenotypic data obtained through screening with Xoo and genotypic data obtained through analysis of 7K rice single-nucleotide polymorphism (SNP) chip. Through composite interval mapping, a major locus was detected in the midst of two flanking SNP markers, viz., Chr11.27817978 and Chr11.27994133, on chromosome 11L with a logarithm of the odds (LOD) score of 10.21 and 35.93% of phenotypic variation, and the locus has been named Xa48t. In silico search in the genomic region between the two markers flanking Xa48t identified 10 putatively expressed genes located in the region of interest. The quantitative expression and DNA sequence analysis of these genes from contrasting parents identified the Os11g0687900 encoding an NB-ARC domain-containing protein as the most promising gene associated with resistance. Interestingly, a 16-bp insertion was noticed in the untranslated region (UTR) of the gene in the resistant parent, IR 75084-15-3-B-B, which was absent in Samba Mahsuri. The association of Os11g0687900 with resistance phenotype was further established by sequence-based DNA marker analysis in the RIL population. A co-segregating PCR-based INDEL marker, Marker_Xa48, has been developed for use in the marker-assisted breeding of Xa48t.
1 Introduction
Rice is one of the world’s main staple food crops. Sustainable rice production, which aims to reduce production losses due to pests and diseases, is an important element in ensuring global food security (FAO, 2004). Rice production is affected significantly by the number of diseases caused by bacteria, viruses, fungi, nematodes, etc.; bacterial blight (BB) caused by Xanthomonas oryzae pv. oryzae (Xoo) is one of the major constraints on higher productivity. The reduction in yield due to BB has recorded up to 81% in a severe case of infection (Zhang and Wang, 2013).
Host plant resistance through the breeding of one or more resistance genes is the most potent way of managing the yield losses due to the disease (Kumar et al., 2020). As the pathogen is highly dynamic in nature, it is equally important to extend the genetic repertoire from diverse novel sources including wild relatives of rice in response to the BB (Brar et al., 1997). Identification of genetic resources through the analysis of wild relatives and introgression of two or more major resistance genes with divergent functions into elite rice varieties are two promising approaches that can lead to durable resistance. Till now, 47 resistance genes have been mapped on various chromosomes for the BB, and many of the resistance genes have been identified and characterized from wild rice lines like Xa21 (Oryza longistaminata), Xa23 and Xa30 (Oryza rufipogon), Xa27 (Oryza minuta), Xa29 (Oryza officinalis), Xa33 and Xa38 (Oryza nivara), and others (Song et al., 1995; Zhang et al., 2001; Gu et al., 2004; Tan et al., 2004; Natrajkumar et al., 2012). O. officinalis, one of the wild rice species, is the key resource for identifying numerous potential genes against many diseases due to their abundant genetic diversity. According to Zhang et al. (1994), O. officinalis makes up 50% of the highly resistant materials among the Chinese wild rice species. A recently published article by Matt Shenton et al. (2020) suggests that the genome size of O. officinalis is 1.6 times larger than the genome size of cultivated Oryza sativa, and genome analysis also revealed the prevalence of their stress tolerance to environmental biotic and abiotic changes. Expression pattern analysis of known BB resistance genes in O. officinalis indicated the possibility of the presence of new genes or quantitative trait loci (QTLs) in this wild species. There has been limited investigation on bacterial blight resistance in O. officinalis, and the molecular mechanism underlying the resistance has not been studied so far (Jiang et al., 2019). The wild rice introgression line derived from O. officinalis provides the solid foundation to identify the novel gene(s) or QTL against BB resistance.
In our study, keeping the above-mentioned information, we have conceptualized the experiment to identify the potential donor with broad-spectrum resistance and to study the pattern of inheritance of resistance. This study aims to map a novel resistance gene/QTL conferring resistance against BB disease in O. officinalis and identify putative candidate genes and associated markers through high-resolution genetic linkage mapping, in silico identification of putative candidate gene(s), and their validation through sequencing and expression analysis. The study holds immense significance, as it provides valuable insights into the genetic basis and molecular mechanisms associated with resistance against BB disease, as it can be leveraged for developing resistant cultivars through marker-assisted breeding.
2 Material and methods
2.1 Plant material and development of mapping population
The wild rice introgression line, IR 75084-15-3-B-B, containing introgressions from O. officinalis, displayed a high level of resistance against many virulent isolates of Xoo, viz., IX-020, IX-007, IX-212, and IX-206, collected from various parts of India. It was crossed with Samba Mahsuri, a high-yielding, fine grain-type rice variety, which is highly susceptible to BB disease. The true F1s derived from the cross were advanced by repeated selfing to develop a recombinant inbred line (RIL) population consisting of 400 individuals. In the F7 generation, a set of 94 lines consisting of 47 homozygous resistant and 47 homozygous susceptible was used for molecular mapping and identification of the genomic regions associated with the resistance to BB disease.
2.2 Screening of mapping population against Xoo strains
A virulent strain of Xoo isolate, i.e., IX-020 (collected from ICAR-IIRR, Hyderabad, Telangana, GPS coordinates 17.3201° N, 78.3939° E) [Yugander et al., 2017], was used for phenotypic evaluation of the mapping population developed from the above-mentioned cross. The isolate was cultured on modified Wakimoto’s agar (MWA) plates and incubated at 28°C for 72–96 hours (Laha et al., 2009). The bacterial cells were collected after the incubation period and diluted with sterile distilled water to a final concentration of approximately 108 cfu/ml, which served as the inoculum. Under field conditions, the plants were inoculated at the maximum tillering stage through the leaf clipping method (Kauffman, 1973), and a minimum of five leaves per plant were clip inoculated.
The plants were scored for their reaction against Xoo following the Standard Evaluation System scale developed by International Rice Research Institute (IRRI-SES; http://www.knowledgebank.irri.org/images/docs/rice-standard-evaluation-system.pdf). Disease reaction was recorded 15 days post inoculation on the basis of average lesion length as well as disease score for each plant. The plants with a lesion length of less than 3 cm were classified as resistant (R; score 1), those with a lesion length of within 3–5 cm were classified as moderately resistant (MR; score 3), and those with a lesion length of more than 5 cm were classified as susceptible (S; score 5 or 7). The resistant parent IR 75084-15-3-B-B and susceptible parent Samba Mahsuri, their F1 progeny, and F2 individuals were screened and evaluated for the genetic inheritance of resistance. The disease scores for each F2 individual (approximately 900) were recorded to compute the segregation ratio using the chi-square test and for determining the number of genes involved in resistance at F3 generation; approximately 20 plants for each F3 line were raised, and all were screened for their disease reaction (minimum of five leaves per plant) using the isolate IX-020. Based on this observation, entries were considered as homozygous resistant if all the 20 plants showed resistance reaction, homozygous susceptible if susceptible reaction was recorded for all the 20 plants, and segregating if both resistant and susceptible reactions were observed within the plants. The homozygous lines were advanced through selfing to F7 generation. In the F7 generation, 94 homozygotes RILs were selected including 47 homozygous resistant and 47 homozygous susceptible lines for molecular mapping using single-nucleotide polymorphism (SNP) markers. In order to confirm the pattern of resistance observed in the resistant parent, a set of 40 lines consisting of 20 homozygous resistant lines and 20 homozygous susceptible lines along with the parents was screened against isolates, including IX-007, IX-212, and IX-206. The objective of this screening was to determine if the resistance pattern observed in the resistant parent was replicated in the selected lines.
2.3 SNP genotyping and construction of linkage map
Based on phenotypic screening for consecutive generations, 94 lines from the F7 generation consisting of 47 each of homozygous resistant and homozygous susceptible lines along with the parents Samba Mahsuri and IR 75084-15-3-B-B, were selected for molecular mapping. The selected lines were genotyped using a 7k rice SNP chip [platform Illumina iScan and SNP Chip details: Cornell-IR_RiceLD (Ref:15069608 X 339319)], which was composed of 6,658 SNPs (Morales et al., 2020). The polymorphic SNPs between the parents were identified from the software QTL IciMapping to construct the genetic linkage map through the inclusive composite interval mapping method (ICIM) using the Kosambi function in the software (https://www.isbreeding.net).
2.4 QTL mapping
For mapping the QTL associated with the BB resistance, genotypic data obtained from the 7k SNP chip were compared with the phenotypic data, i.e., BB resistance score as well as lesion length of the selected 94 entries. The analysis of QTL was executed using inclusive composite interval mapping for additive QTL (ICIM-ADD) using the software QTL IciMapping. As per the ICIM software, a logarithm of the odds (LOD) score of 3 was set as a minimum threshold based on the permutation test for the identification of QTLs with a significant effect on the phenotype.
2.5 In silico analysis for annotated putative candidate genes
The identified QTL region related to the BB resistance was analyzed using the Batch retrieval function from RAP-DB (https://rapdb.dna.affrc.go.jp/) as well as the Rice genome annotation project (http://rice.plantbiology.msu.edu/) for in silico analysis. The putative candidate genes were identified from the Rice annotation project Database website (https://rapdb.dna.affrc.go.jp) from the genomic region specific for the resistance QTL/gene.
2.6 RNA extraction and quantitative real-time PCR
To analyze the expression of the gene(s) present within the QTL interval as well as to identify the most likely putative candidate gene(s) for Xa48t, real-time PCR primers (Supplementary Table 1) were designed from the exonic regions of the genes using qPCR primer design tool QuantPrime (Arvidsson et al., 2008). Resistant parent IR 75084-15-3-B-B, susceptible parent Samba Mahsuri, and two homozygotes (one homozygous resistant and one homozygous susceptible) were grown on hydroponics media under controlled conditions with precise temperature, lighting, humidity, nutrient solution, water quality, etc., favoring Xoo spread on rice plant. Twenty-five-day- old seedlings were inoculated with Xoo isolate IX-020, and samples at 0 hpi (mock-inoculated plant) (hour post inoculation), 24 hpi, and 48 hpi were harvested for the analysis. Total RNA was extracted from each genotype in three replicates using the NucleoSpin® RNA Plant kit and converted into cDNA using PrimeScript™ 1st strand cDNA Synthesis Kit according to the protocol for the real-time PCR. TB Green® Premix Ex Taq™ II along with diluted cDNA was analyzed under the qPCR system for gene expression analysis.
2.7 Marker development
To develop PCR-based diagnostic markers for Xa48t, we utilized the chromosome walking method (Chinault and Carbon, 1979). This method involves sequence analysis of the genes located within the region of interest, which were shortlisted based on the results of expression analysis through amplification using overlapping primers. To identify sequence differences between the resistant parent IR 75084-15-3-B-B and the susceptible parent Samba Mahsuri, overlapping primer pairs were designed for PCR amplification and sequencing. It was ensured that the expressed regions of the shortlisted genes, along with approximately 1-kb upstream and 500-bp downstream sequences of the putative candidate genes, were also sequenced. We also utilized the available simple sequence repeat (SSR) and INDEL markers within the region of interest on Chr. 11L. These markers were identified from the Rice Annotation Project Database (RAP-DB, http://rapdb.dna.affrc.go.jp/), and the synthesized SSR and INDEL markers were used for amplification of the parents to identify polymorphic markers. Detailed information regarding the markers is presented in Supplementary Table 2.
3 Results
3.1 BB resistance evaluation of mapping population
The resistant parent was observed to show a high level of resistance against the Xoo isolate with a lesion length of less than 1 cm, while the susceptible parent Samba Mahsuri recorded a susceptibility reaction with a lesion length of more than 9 cm. The F1s derived from the cross IR 75084-15-3-B-B/Samba Mahsuri were resistant with a lesion length of below 1 cm, indicating the dominant nature of BB resistance in IR 75084-15-3-B-B.
The F2 population was segregated in the ratio of 3:1, including 598 resistant and 229 susceptible lines. The observed frequency of the population segregation fitted the expected phenotypic ratio of 3:1, which was confirmed statistically through the chi- square test (Table 1).
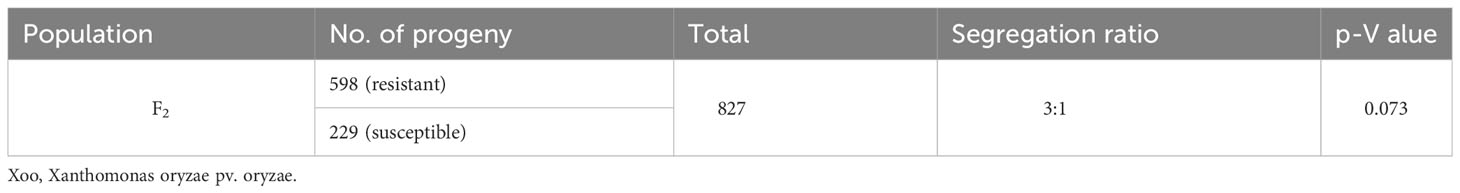
Table 1 Frequency of resistance reaction against the Xoo isolate of F2 from the cross between IR 75084-15-3-B-B and Samba Mahsuri.
3.2 Identification of homozygous resistant and homozygous susceptible lines for QTL mapping
From F2s, a population consisting of 400F3 lines (300 resistant and 100 susceptible) was selected and screened for BB resistance under field conditions along with the parents. Among them, 160 F3 lines derived displayed resistance, while the 100 F3s were susceptible to the BB disease. The selected lines were advanced further through selfing to F7 generation, and finally, a set of 94 lines (47 homozygous resistant lines and 47 homozygous susceptible lines) were identified and used for QTL mapping. The selected 94 F7 lines (i.e., recombinant inbred lines) along with the parents were also screened under controlled glasshouse conditions in order to confirm their homozygosity and were then used for mapping (Figure 1). Again, the screening of 40 homozygous sets displayed a resistance pattern of 20 homozygous resistant lines similar to that of the resistant parent when exposed to Xoo isolates IX-007, IX-212, and IX-206.

Figure 1 Bacterial blight disease reaction on identified homozygous resistant and identified homozygous susceptible lines along with the parents. RP-IR 75084-15-3-B-B, SP- BPT5204, homozygous resistant (HR), and homozygous susceptible (HS).
However, the homozygous susceptible lines showed a consistent susceptibility pattern across the isolates tested (Supplementary Table 4).
3.3 Construction of linkage map
A high-density genetic linkage map was constructed using IciMapping software (https://www.isbreeding.net) with the Kosambi function from the phenotyping data pertaining to BB scores of the mapping population and the genotype data obtained from 7k data analysis. An aggregate of 6,658 SNP markers were called for genotyping of 94 selected RILs and the parents. The SNP markers with insertion/deletion or either heterozygous condition in either of the parents were filtered out from the polymorphic survey. Based on shortlisting of the polymorphic survey of the SNP markers, which were in the homozygous state, we identified 2,562 SNPs for further linkage analysis. Out of these, 1,514 redundant SNPs were found, which overlapped the same position on each chromosome with 0 cM through linkage map information supported by the software. The linkage map was finally constructed with a total of 1,048 SNPs, with approximately 88 SNPs anchored in each chromosome on average (Table 2; Figure 2).
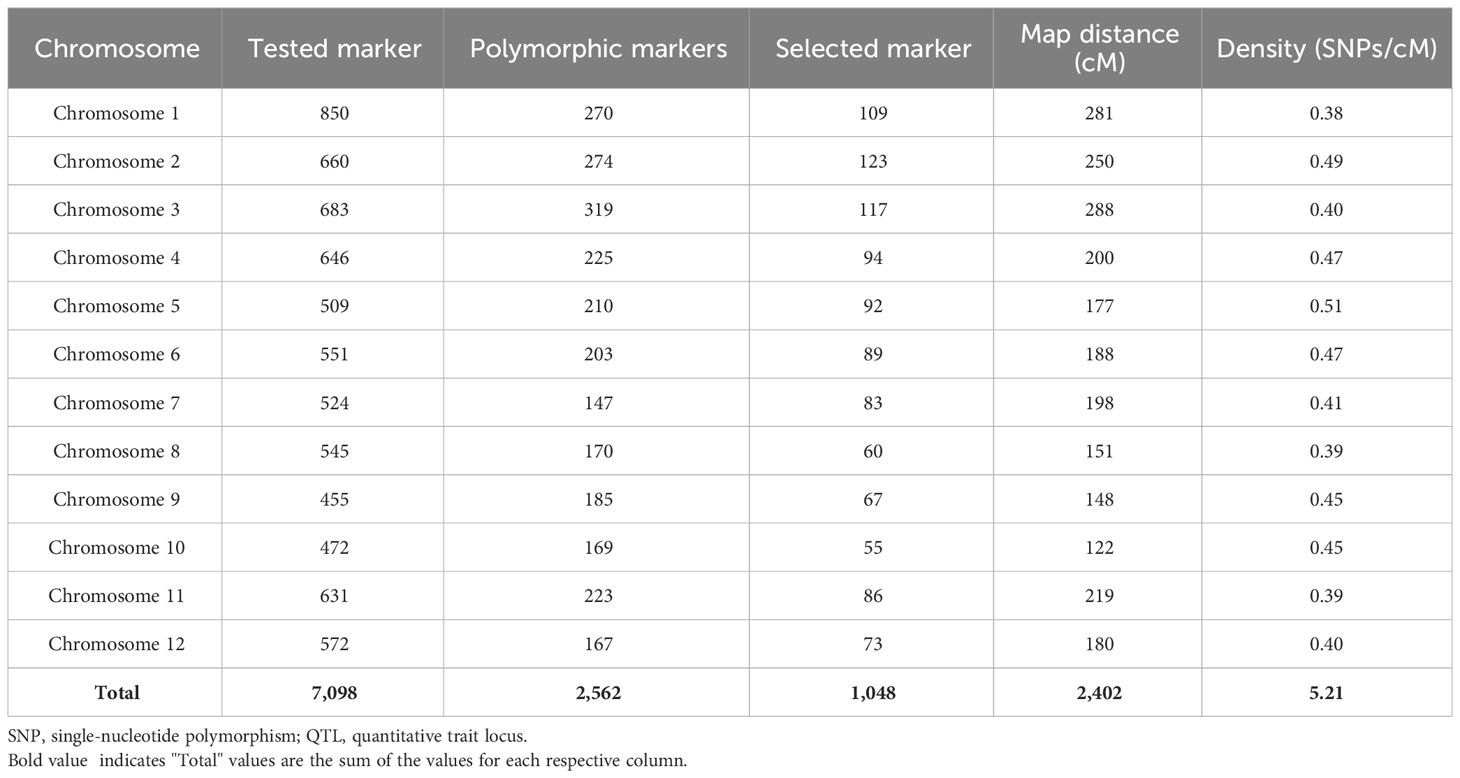
Table 2 Distribution of SNP markers on all the chromosomes used for construction of linkage map and QTL analysis.
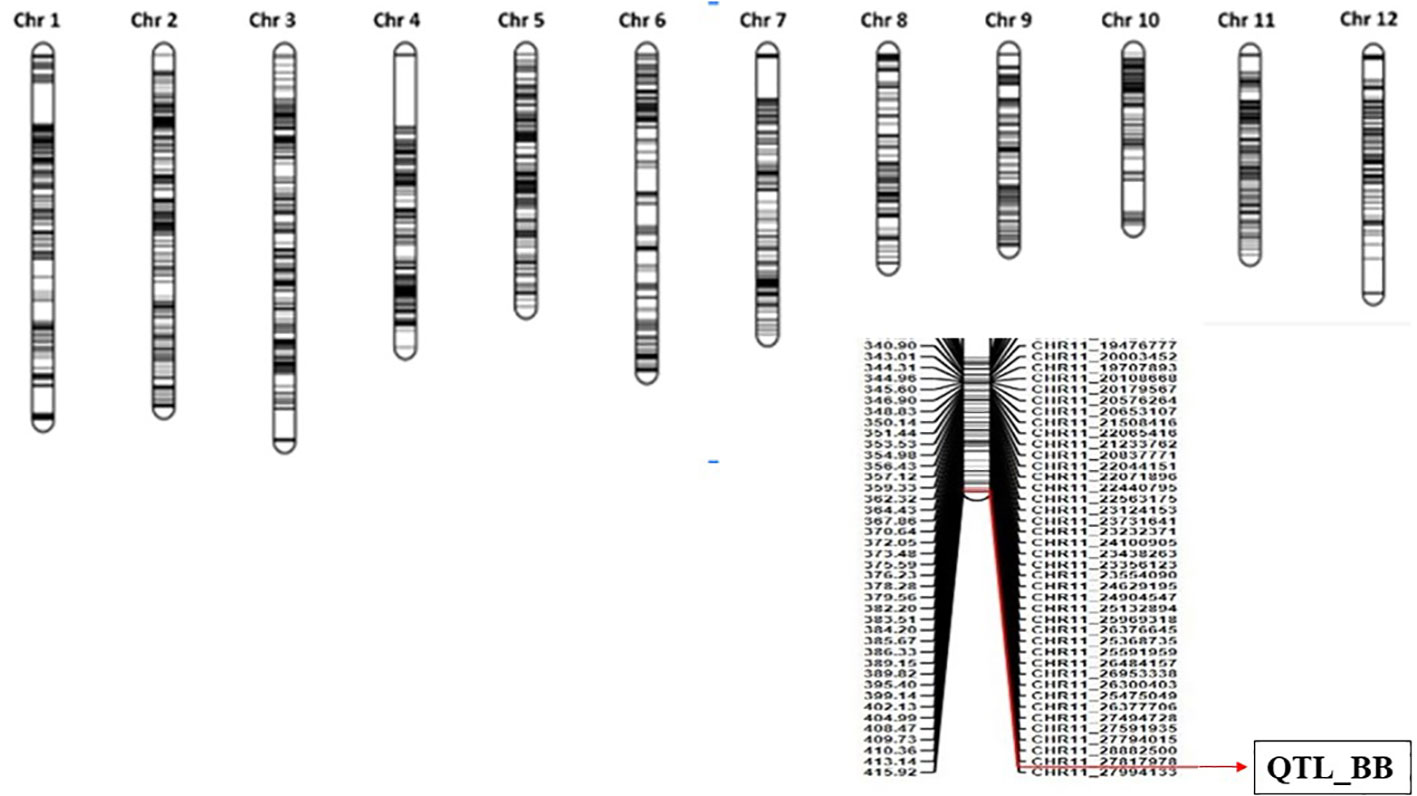
Figure 2 Genetic linkage map of the rice genome constructed based on the recombination frequencies of 1,014 SNP markers. The major QTL associated with bacterial blight resistance was located on chromosome 11. SNP, single-nucleotide polymorphism; QTL, quantitative trait locus.
3.4 QTL analysis
Correlating the genotypic data analyzed from 1,048 polymorphic SNP markers with the phenotypic data on the basis of lesion length as well as BB score for the selected 94 RILs, we have identified the major locus on the long arm of chromosome 11 to be associated with BB resistance reaction. Inclusive composite interval mapping for additive QTL through the IciMapping software marks the presence of QTL between the two SNP markers, Chr11.27817978 and Chr11.27994133, with a LOD score of 10.21 explaining 35.93% of phenotypic variations (Table 3). The markers were located at 415 cM on the linkage map and span a physical distance of 176 kb referred on the Nipponbare reference genome IRGSP v1 (Figure 2) (Supplementary Figure 1).
3.5 Identification of candidate genes and their expression analysis
We analyzed the QTL region of interest for the presence of genes based on the Rice Genome Annotation Project Database and resource. The region indicated the presence of 10 genes encoding various biological functions, which includes two NBS-LRR class disease resistance proteins, two NB- ARC-containing protein, one von Willebrand factor-containing protein, one MLA10 protein, and four hypothetical conserved proteins. Among them, two of the genes are associated with blast disease resistance. The details of the putative candidate genes are described in Supplementary Table 1; Figure 3A.
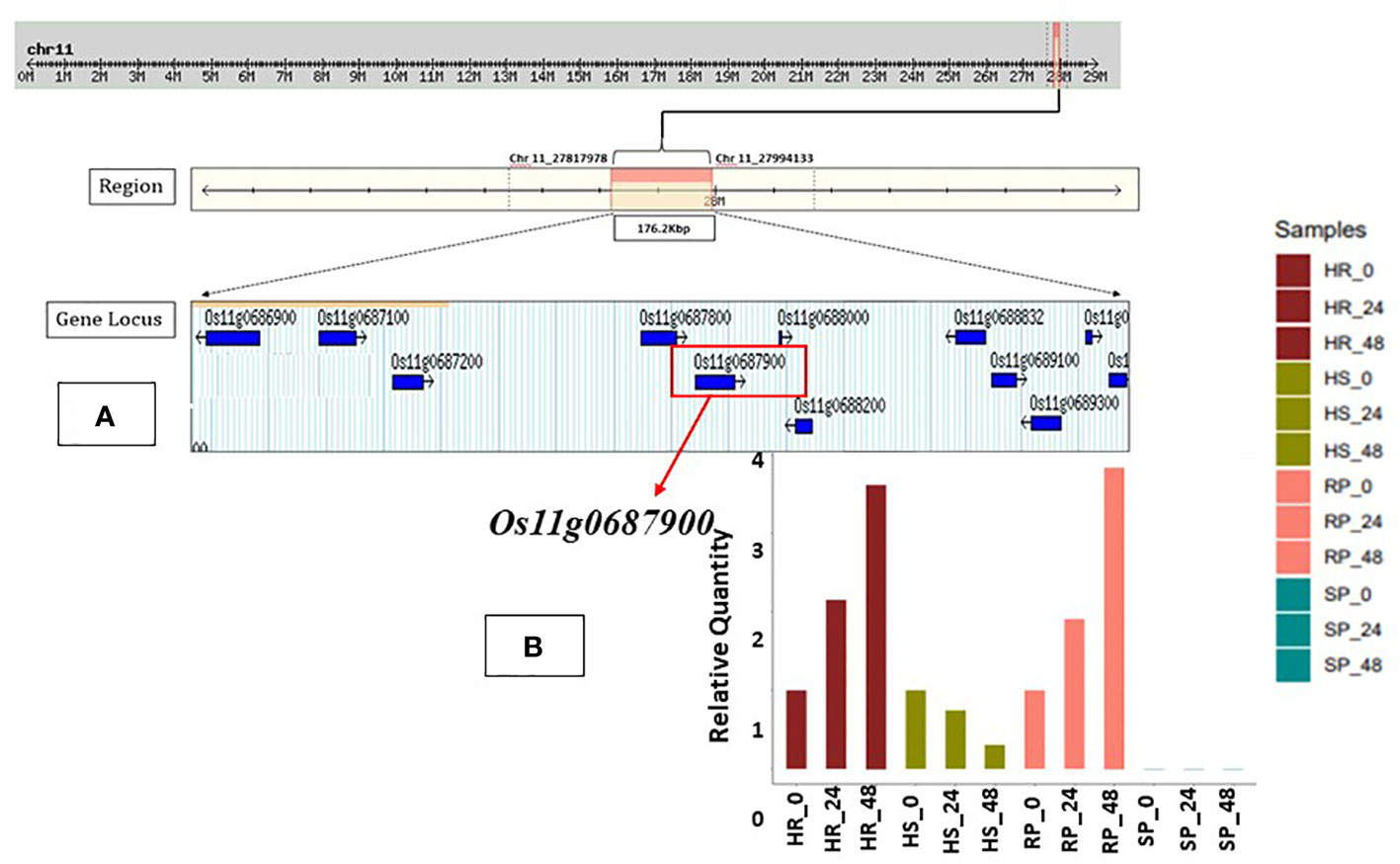
Figure 3 (A) In silico analysis for the identified QTL associated with resistance against bacterial blight through Rice Genome Annotation Project Database (RAP-DB). Ten putative candidate genes encoding various biological functions were present in the region of major QTL. Source RAP-DB (IRGSP-1.0). (B) Fold change and expression level of putative candidate gene Osllg0687900. Gene Osllg0687900 encoding for NB-ARC domain-containing protein; expression level shows upregulation at 24 hpi and 48 hpi for both resistant parent (RP) and homozygous resistant (HR) lines and possibly associated with resistance against bacterial blight. SNP, single-nucleotide polymorphism; QTL, quantitative trait locus.
The expression analysis of 10 genes from the QTL region associated with BB resistance was investigated through the relative quantification method (also known as the 2−ΔΔCt method) in the parents as well as a highly resistant RIL (RIL-1) and a highly susceptible RIL (RIL-94) line. Notably, the putative candidate gene Os11g0687900 showed a substantially upregulated expression level (two-fold) in RP and HR in an exponential pattern after inoculation with the Xoo isolate IX-020 at 24 hpi and 48 hpi (Figure 3B). The expression of other potential genes was either low or absent at other times, depending on the circumstances. The expression details of other candidate genes are also provided in Supplementary Table 3. The expression level of gene Os11g0687900 as well as functional annotation correlated with the identified resistance QTL and was hence considered to be the most probable candidate gene.
3.6 Analysis of sequences of the candidate gene Os11g0687900
The putative candidate gene Os11g0687900 was sequenced in the resistant parent, IR 75084-15-3-B-B, and susceptible parent Samba Mahsuri using a set of seven designed overlapping primers. The sequence analysis revealed that there was 16-bp insertion in the 5′UTR of resistant parent IR 75084-15-3-B-B with respect to the Nipponbare sequence. The insertion is located 23 bp upstream of the start codon of the gene, within the 5′UTR. Notably, PCR amplification using the overlapping primers did not yield any product with Samba Mahsuri. Further, a comparative analysis of the genome of Samba Mahsuri (unpublished and Reddy and Ulaganathan, 2015), Nipponbare, and the resistant parent revealed that the locus of the gene and its upstream region is absent in Samba Mahsuri. The sequence alignment with the O. officinalis genome available in the National Center for Biotechnology Information (NCBI) database (Oryza_officinalis_v1.0 GCA_008326285.1) showed the presence of the 16-bp insertion in the O. officinalis allele of Os11g0687900. The alignment result is depicted in Figure 4.
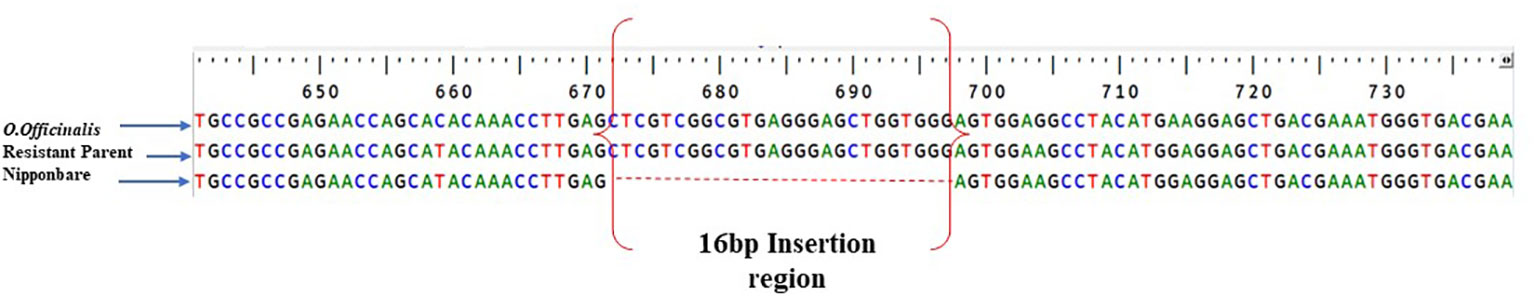
Figure 4 The alignment result depicts the presence of 16-bp insertion region in Oryza officinalis genome as well as resistant parent, while Samba Mahsuri revealed the absence of 16-bp insertion and also sequence variation in the region flanking the gene.
Population genomics analysis using the tool RiceVarMap (Zhao et al., 2021) indicated that approximately 42.74% of the 4,726 sequenced rice varieties do not possess this locus, and approximately 47.20% of the varieties do not possess the 16-bp insertion. Additionally, the population analysis indicated that insertion at this locus is sparsely distributed (~8.6%) in the rice germplasm.
Considering the following observations in the present study (viz., i) gene expression of Os11g0687900 was exponentially upregulated upon Xoo infection and ii) a 16-bp insertion is present in the 5′UTR of the gene only in the BB-resistant donor parent), it is indeed compelling to speculate that this insertion may have a regulatory role with respect to the induction of the gene observed only in resistant plants. The 16-bp insertion sequence was predicted to harbor AP2/ERF transcription factor binding site using the PLACE database (New PLACE —A Database of Plant Cis-acting Regulatory DNA Elements) (Higo et al., 1999), which is frequently used for the identification of plant cis-acting regulatory DNA elements.
3.7 Development of functional marker for Xa48t
The functional markers for the mapped gene were developed and validated in the population. In the first set, primers were designed from the 16-bp insertion region that clearly showed 150-bp amplification in the resistant parent and homozygous resistant RILs, while the PCR amplicon was absent in the susceptible parent and homozygous susceptible RILs. These results further indicated that the 16-bp insertion-associated region was present only in the RP and HR and absent in the SP and HS (Figure 5A).
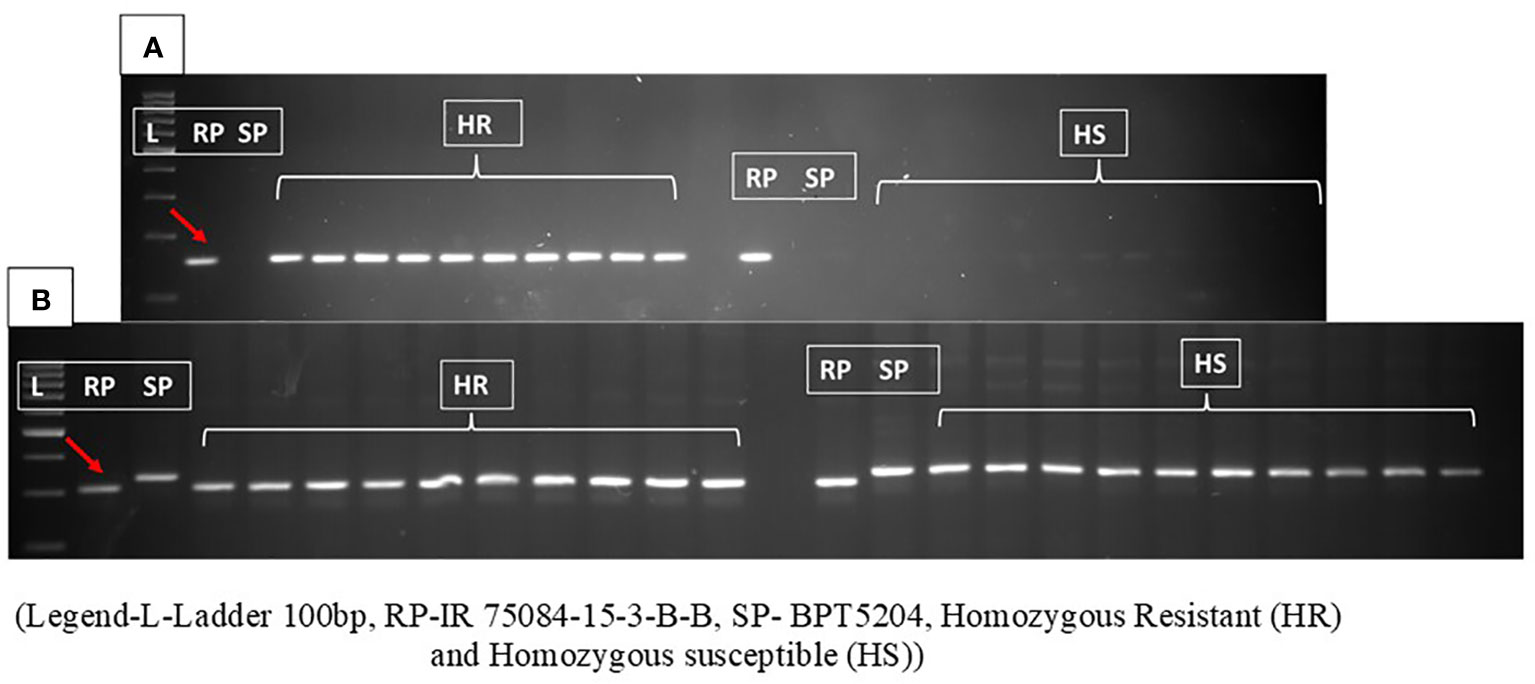
Figure 5 (A) The amplification pattern of primer designed 16-bp insertion shows band size of approximately 150 bp representing the resistance in parent and homozygous resistant RILs, while PCR was absent in susceptible parent and homozygous susceptible RILs. (B) Amplification pattern of functional Marker_Xa48 in resistant parent, susceptible parent, homozygous resistant (HR), and homozygous susceptible (HS) RILs. The polymorphic INDEL primers amplify a fragment of size of 316 bp in the susceptible parent and all the susceptible RILs and a 300-bp fragment in the resistant parent and the resistant RILs with complete cosegregation in the mapping population. RILs, recombinant inbred lines. Red arrow represents the band size of resistant parent.
In order to develop co-dominant markers for marker-assisted breeding, five SSR markers and eight INDEL markers in the vicinity of the candidate gene for Xa48t, viz., Os11g0687900, were used. One of the INDEL markers was found to be polymorphic between the resistant parent and the susceptible parent. The polymorphic INDEL marker was located at a distance of approximately 78 kb from locus Os11g0687900. The polymorphic INDEL primers amplify the band size of 310 bp in the susceptible parent and 300 bp in the resistant parent and also showed complete co-segregation with the phenotype in the RIL mapping population (Figure 5B). The INDEL marker is named Marker_Xa48 and can be used as a functional marker for gene Xa48t.
4 Discussion
Deployment of a diverse set of resistance genes represents the most feasible strategy to ensure durable management of BB disease of rice. In previous studies, at least 47 resistance genes have been mapped from diverse sources. Few major resistance genes have been mapped from wild species of Oryza and also characterized. These include O. longistaminata; Xa21 (Song et al., 1995), O. minuta; Xa27, Xa35(t) (Gu et al., 2004; Guo et al., 2010), O. rufipogon; Xa30 (Tan et al., 2004), O. officinalis; Xa29(t) (Tan et al., 2004), O. nivara; Xa33 (Natrajkumar et al., 2012), O. nivara; Xa38, Oryza australiensis; and Xa40(t) (Kim et al., 2015). However, because of the breakdown of resistance due to the existence/emergence of virulent races of Xoo, the breeding of rice cultivars that have durable resistance is a challenge (Busungu et al., 2016; Neelam et al., 2019; Chen et al., 2020). In this regard, expanding the repertoire of resistance genes through tapping the wild rice gene resources and pyramiding two or more resistance genes to develop BB resistance cultivars represent the ideal strategy for the management of the disease (Sundaram et al., 2009). In this study, we have identified and mapped the novel gene locus conferring resistance against bacterial blight disease on the long arm of chromosome 11 from the rice breeding line having introgression segments from O. officinalis.
Previously, we screened a set of wild rice introgression using different hypervirulent isolates of Xoo to identify the potential donor for the development of BB-resistant cultivars. The rice introgression line, IR-75084-15-3-B-B, possessing introgressions from O. officinalis, displayed broad-spectrum resistance against the four hypervirulent isolates of the pathogen, namely, IX-020, IX-007, IX-212, and IX-206, under controlled (glasshouse) conditions, indicating broad-spectrum resistance against the disease (Sinha et al., 2021). In order to understand the inheritance of resistance in the rice line and also to undertake molecular mapping of the resistant locus/loci, we developed a mapping population by crossing the resistant rice line IR-75084-15-3-B-B and Samba Mahsuri, which is susceptible to bacterial blight. Based on the segregation pattern of resistance trait in the F2 generation, it was inferred that the population is segregating in the ratio of 3: 1, and the resistance is governed by a single, dominant gene.
The RIL mapping population derived from the cross was subsequently phenotypically evaluated against Xoo. A subset of 94 RILs in F7 generation displaying extreme phenotype reaction (i.e., 47 RILs that are homozygous resistant and 47 RILs that are homozygous susceptible) was selected for high-throughput SNP-based genotyping using 7k rice SNP chip (Morales et al., 2020). The screening of 20 homozygous resistant RILs with four hypervirulent Xoo strains showed a similar resistance pattern as that of the resistant parent, suggesting that all the resistant RILs carried the genetic loci for broad-spectrum resistance. Recombinant inbred lines are highly suited for molecular mapping of genes/QTLs, as they do not show any segregation of individuals within RILs (Rockman and Kruglyak, 2008). The high-density linkage mapping and the gene/QTL analysis based on the high-throughput SNP genotyping of 94 RILs laid a strong foundation in our study for the identification of genomic regions associated with bacterial blight resistance trait. This approach has been adopted in earlier studies aimed at the molecular mapping of genes/QTLs associated with agronomically important genes/QTLs in rice (Kim, 2018; Kim and Reinke, 2019; Neelam et al., 2019).
From the correlation analysis of phenotypic and genotypic data of 94 RILs, we identified and mapped a novel genetic locus that governs resistance against bacterial blight on the long arm of chromosome 11 (https://www.isbreeding.net). The resistance locus, which is named Xa48, was flanked by SNP markers 11955796 and 11963686 spanning a distance of 176 kb (i.e., from 27.81 Mbp to 27.99 Mbp) on chromosome 11. So far, about 17 of the previously identified bacterial blight resistance genes including Xa3/Xa26, Xa4, Xa10, Xa21, Xa22, Xa23, xa26, Xa30, Xa32, Xa35, Xa36, Xa39, Xa40, Xa43(t), xa44(t), and Xa47(t) were also localized on chromosome 11 (Yoshimura et al., 1983; Song et al., 1995; Kaku and Ogawa, 2001; Wang et al., 2001; Zhang et al., 2001; Chen et al., 2002; Wang et al., 2003; Sun et al., 2004; Tan et al., 2004; Guo et al., 2010; Miao et al., 2010; Zhang et al., 2014; Kim et al., 2015; Kim, 2018; Kim and Reinke, 2019 and Xing et al., 2021). Among these genes, the gene locus identified in this study was found to be located in the vicinity of five genes, namely, Xa3/Xa26, Xa4, Xa40, Xa43(t), and Xa47(t) (Wang et al., 2001; Sun et al., 2004; Kim et al., 2015; Kim and Reinke, 2019). Among them, the earlier identified genes, Xa3/Xa26, Xa4, and Xa40, have been mapped and characterized in a region not very close to the gene identified in this study, i.e., Xa48, while Xa43(t) is located in the proximity of Xa48 in the upstream region, and Xa47(t) is located in the downstream region. However, the PCR-based diagnostic markers developed for Xa43(t), i.e., IBb27os11_14 and S_BB11.ssr_9, did not show polymorphism among the parents used in this present study (Kim and Reinke, 2019). Also, while Xa43(t) was identified from the rice line, P8 using a Japonica MAGIC population (Bandillo et al., 2013), the gene identified in this study, i.e., Xa48, has been identified from a wild species of rice, O. officinalis. Another newly identified gene locus Xa47(t) was identified from introgression line G252 from wild rice O. rufipogon against virulent strains of China, Japan, and the Philippines. A lso, the identified candidate gene for Xa47(t) gene is Os11g0688832, which is different from Xa48 putative candidate gene. This indicates that Xa48 is indeed a novel BB resistance gene.
The information obtained through the Rice gene annotation project database (Sakai et al., 2013) is useful for identifying putative candidate genes located in the genomic region spanning genes/QTLs (Kale et al., 2021). Through in silico analysis, a total of 10 putative candidate genes encoding different biological functions were found to be located in the region of interest. The functional annotation indicates the presence of a distinct class of genes encoding proteins with various biological functions in the 176 kb region. The NBS-LRR domain plays an important role in signal transduction and mediates effector-triggered immunity against a diverse array of pathogens (Hammond Kosack and Jones 1996). Another gene encoding for the NB-ARC domain is also involved in activating defense response by acting as signaling motifs (van Ooijen et al., 2008). The gene encoding for MLA10 functions in regulating cell compartment-specific activity in cell death and disease resistance in barley (Bai et al., 2012). Other genes were involved in the molecular and structural activity via protein binding. These genes are worth investigating further through expression analysis to establish their putative role in bacterial blight resistance.
Among the genes underlying mapped QTL, expression analysis was carried out to identify the promising candidate gene(s) associated with the resistance phenotype. The results based on the relative quantification of genes in different samples revealed that the expression of Os11g0687900 was significantly higher in the resistant parent IR-75084-15-3-B-B and homozygous resistant lines as compared to the bacterial blight- susceptible parent Samba Mahsuri and homozygous susceptible line. The functional annotation of this gene suggests an NB-ARC domain-containing protein, which is known to be associated with disease resistance (van Ooijen et al., 2008). Further investigation through sequencing of the resistance and susceptible alleles of Os11g0687900 among the resistant and susceptible parents revealed the insertion of 16 bp in the 5′UTR of the resistant parent. The 16-bp insertion is unique to the resistant parent and also identified in the O. officinalis genome, suggesting that this cis-acting motif of 16 bp might play a critical role in bacterial blight resistance. Notably, this motif is predicted to harbor transcription factor binding sites/cis-elements, which might be associated with the induction of expression of the gene upon Xoo infection (Jiang et al., 2020). Further, analysis of the 16-bp motif in the publically available genome sequence of O. officinalis, along with Nipponbare and several Indica rice varieties, indicated the absence of 16-bp insertion region in the majority of Indica rice cultivars including Samba Mahsuri. It would be interesting to further validate the precise function of Os11g0687900 in BB resistance by over-expression or knockout through genome editing. In particular, the cis-acting motif of 16 bp is an interesting subject for further investigation. Knock-in (using homology-directed repair (HDR) of genome editing) of the 16-bp motif in popular and susceptible rice cultivars can help develop resistant alleles in susceptible cultivars. A PCR-based INDEL marker, named Marker_Xa48, which is located at a distance of 78 kb from the gene locus, has been developed and shown to be co-segregating with the resistant phenotype. This marker can be used for introgression of resistant alleles of the Xa48 gene through MAB.
5 Conclusion
Based on the findings in our study, the major locus on the long arm of chromosome 11 associated with BB resistance reaction has been identified from O. officinalis introgression lines through 7k SNP chip-based ICIM mapping and was named Xa48t. The resistance locus was observed to be located between two SNP markers, Chr11.27817978 and Chr11. 27994133, explaining 35.93% of phenotypic variation and with a LOD score of 10.21. The most promising candidate gene associated with the resistant trait is Os11g0687900, which codes for the NB-ARC domain-containing protein, and its candidacy has been validated through expression analysis. The PCR-based INDEL marker tightly linked to gene Xa48t has been developed for the deployment of the gene into elite rice cultivars through marker-assisted breeding.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov; OQ559125 (BankIt2678609 Oryza; OQ559125).
Author contributions
PS carried out disease screening, mapping, expression analysis, and functional marker development and drafted the manuscript. DKT carried out disease screening and crossing program and drafted the manuscript. MS interpreted the expression analysis data. CG assisted with analysis and functional marker development. HS, AD, AM, RG, PE, KM, MS, CK, and YA helped with disease screening in the field. SKM guided the mapping and expression analysis and provided critical input in the manuscript. LG and CN guided the mapping and expression analysis and reviewed the manuscript. HP, IG, S-RK, KJ, SRH, and RO revised the manuscript. SRM conceptualized the experiment; guided disease screening, mapping, expression analysis, and functional marker development; and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by DST (Department of Science and Technology) through DST-INSPIRE Scheme as a doctoral study fellowship from a period of April 2017 to March 2022 and ICAR-Indian Institute of Rice Research (ICAR-IIRR).
Acknowledgments
PS thanks the DST (Department of Science and Technology) Government of India for providing funding support through the DST INSPIRE scheme. The authors gratefully acknowledge the support extended by the ICAR-Indian Institute of Rice Research (ICAR-IIRR) by providing facilities.
Conflict of interest
Author RG was employed by the company Rallis India Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1173063/full#supplementary-material
References
Arvidsson, S., Kwasniewski, M., Riaño-Pachón, D. M., Mueller-Roeber, B. (2008). QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinf. 9 (1), 1–15. doi: 10.1186/1471-2105-9-465
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., et al. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoSPathog 8 (6), e1002752. doi: 10.1371/journal.ppat.1002752
Bandillo, N., Raghavan, C., Muyco, P. A., Sevilla, M. A., Lobina, I. T., Dilla-Ermita, C. J., et al. (2013). Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6, 11. doi: 10.1186/1939-8433-6-11
Brar, D. S., Elloran, R. M., Talag, J. D., Abbasi, F., Khush, G. S. (1997). Cytogenetic and molecular characterization of an intergeneric hybrid between Oryza sativa L. and Porteresiacoarctata (Roxb.) Tateoka. Rice Genet. Newsl 14, 43–44.
Busungu, C., Taura, S., Sakagami, J. I., Ichitani, K. (2016). Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed. Sci. 66 (4), 636–645. doi: 10.1270/jsbbs.16062
Chen, S., Wang, C., Yang, J., Chen, B., Wang, W., Su, J., et al. (2020). Identification of the novel bacterial blight resistance gene Xa46 (t) by mapping and expression analysis of the rice mutant H120. Sci. Rep. 10 (1), 1–11. doi: 10.1038/s41598-020-69639-y
Chen, H., Wang, S., Zhang, Q. (2002). New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 92 (7), 750–754. doi: 10.1094/PHYTO.2002.92.7.750
Chinault, A. C., Carbon, J. (1979). Overlap hybridization screening: isolation and characterization of overlapping DNA fragments surrounding the leu2 gene on yeast chromosome III. Gene 5, 111–126. doi: 10.1016/0378-1119(79)90097-0
FAO (2004) Species description Oryza sativa L. Available at: http://www.fao.org/ag/AGP/AGPC/doc/GBASE/data/pf000274.htm.
Gu, K., Tian, D., Yang, F. W., Wu, L., Sreekala, C., Wang, D., et al. (2004). High-resolution genetic mapping of Xa27 (t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor. Appl. Genet. 108, 800–807. doi: 10.1007/s00122-003-1491-x
Guo, S., Zhang, D., Lin, X. (2010). Identification and mapping of a novel bacterial blight resistance gene Xa35 (t) originated from Oryza minuta. Scientia Agricultura Sin. 43 (13), 2611–2618.
Hammond-Kosack, K. E., Jones, J. D. (1996). Resistance gene-dependent plant defense responses. The Plant cell, 8 (10), 1773–1791. doi: 10.1105/tpc.8.10.1773
Higo, K., Ugawa, Y., Iwamoto, M., Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 (1), 297–300. doi: 10.1093/nar/27.1.297
International Rice Research Institute Standard Evaluation System Scale Developed. Available at: http://www.knowledgebank.irri.org/images/docs/rice-standard-evaluation-system.pdf.
Jiang, C., Xiao, S., Li, D., Chen, L., Zhong, Q., Yin, F., et al. (2019). Identification and Expression Pattern Analysis of Bacterial Blight Resistance Genes in Oryza officinalis Wall ex Watt Under Xanthomonas oryzaePv. oryzae Stress. Plant Mol. Biol. Rep. 37, 436–449. doi: 10.1007/s11105-019-01164-3
Jiang, G., Yin, D., Shi, Y., Zhou, Z., Li, C., Liu, P., et al. (2020). OsNPR3.3-dependent salicylic acid signaling is involved in recessive gene xa5-mediated immunity to rice bacterial blight. Sci. Rep. 10 (1), 6313. doi: 10.1038/s41598-020-63059-8
Kaku, H., Ogawa, T. (2001). Genetic Analysis of the relationship between the browning reaction and bacterial blight resistance gene Xa3 in rice. J. Gen. Plant Pathol. 67, 228–230. doi: 10.1007/PL00013017
Kale, R. R., Durga Rani, C. V., Anila, M., Mahadeva Swamy, H. K., Bhadana, V. P., Senguttuvel, P., et al. (2021). Novel major QTLs associated with low soil phosphorus tolerance identified from the Indian rice landrace, Wazuhophek. PLoS One 16 (7), e0254526. doi: 10.1371/journal.pone.0254526
Kauffman, H. E., Reddy, A.P.K., Hsieh, S.P.Y., Merca, S.D. (1973). An improved technique for evaluat-ing resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Kim, S. (2018). Identification of novel recessive gene xa44(t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip. Theor. Appl. Genet. 131, 2733–2743. doi: 10.1007/s00122-018-3187-2
Kim, S. M., Reinke, R. F. (2018). Identification of QTLs for tolerance to hypoxia during germination in rice. Euphytica 214 (9), 1–10. doi: 10.1007/s10681-018-2238-8
Kim, S.-M., Reinke, R. F. (2019). A novel resistance gene for bacterial blight in rice Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PloS One 14 (2), e0211775. doi: 10.1371/journal.pone.0211775
Kim, S. M., Suh, J. P., Qin, Y., Noh, T. H., Reinke, R. F., Jena, K. K. (2015). Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.). Theor. Appl. Genet. 128, 1933–1943. doi: 10.1007/s00122-015-2557-2
Kumar, A., Kumar, R., Sengupta, D., Das, S. N., Pandey, M. K., Bohra, A., et al. (2020). Deployment of Genetic and Genomic Tools Toward Gaining a Better Understanding of Rice-Xanthomonas oryzaepv. oryzae Interactions for Development of Durable Bacterial Blight Resistant Rice. Front. Plant Sci. 11, 1152. doi: 10.3389/fpls.2020.01152
Laha, G. S., Reddy, C. S., Krishnaveni, D., Sundaram, R. M., Srinivas, P. M., Ram, T., et al. (2009). Bacterial blight of rice and its management. DRR Tech. Bull. 41, 37.
Miao, L., Wang, C., Zheng, C., Che, J., Gao, Y., Wen, Y., et al. (2010). Molecular mapping of a new gene for resistance to rice bacterial blight. Sci. Agric. Sin. 43 (15), 3051–3058.
Morales, K. Y., Singh, N., Perez, F. A., Ignacio, J. C., Thapa, R., Arbelaez, J. D., et al. (2020). An improved 7K SNP array, the C7AIR, provides a wealth of validated SNP markers for rice breeding and genetics studies. PloS One 15 (5), e0232479. doi: 10.1371/journal.pone.0232479
Natrajkumar, P., Sujatha, K., Laha, G. S., Srinivasa Rao, K., Mishra, B., Viraktamath, B. C., et al. (2012). Identification and fine-mapping of Xa33, a novel gene for resistance to Xanthomonas oryzaepv. oryzae. Phytopathology 102, 222. doi: 10.1094/PHYTO-03-11-0075
Neelam, K., Mahajan, R., Gupta, V., Bhatia, D., Gill, B. K., Komal, R., et al. (2019). High-resolution genetic mapping of a novelbacterial blight resistance gene xa-45 (t) identified from Oryza glaberrima and transferred to Oryzasativa. Theor. Appl. Genet. 6, 1–7. doi: 10.1007/s00122-019-03501-2
Reddy, M. M., Ulaganathan, K.. (2015). Draft genome sequence of Oryza sativa elite indica cultivar RP Bio-226. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00896
Rockman, M. V., Kruglyak, L. (2008). Breeding designs for recombinant inbred advanced intercross lines. Genetics 179 (2), 1069–1078. doi: 10.1534/genetics.107.083873
Sakai, H., Lee, S. S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., et al. (2013). Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54 (2), e6. doi: 10.1093/pcp/pcs183
Shenton, M., Kobayashi, M., Terashima, S., Ohyanagi, H., Copetti, D., Hernández-Hernández, T., et al. (2020). Evolution and diversity of the wild rice Oryza officinalis Complex, across continents, genome types, and ploidy levels Genome Biol. Evol. 12 (4), 413–428. doi: 10.1093/gbe/evaa037
Sinha, P., Kumar, T. D., Hajira, S. K., Ayyappa, D., Aleena, D., Masthani, S. K., et al. (2021). Identification of novel sources of resistance against bacterial blight disease through intensive screening of wild rice introgression lines. Indian J. Plant Prot. 49 (2), 131–137. Retrieved from https://epubs.icar.org.in/index.php/IJPP/article/view/115221.
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. science 270 (5243), 1804–1806. doi: 10.1126/science.270.5243.1804
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527. doi: 10.1046/j.1365-313X.2003
Sundaram, R. M., Vishnupriya, M. R., Laha, G. S., Rani, N. S., Rao, P. S., Balachandran, S. M., et al. (2009). Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnol. Journal: Healthcare Nutr. Technol. 4 (3), 400–407. doi: 10.1002/biot.200800310
Tan, G. X., Ren, X., Weng, Q. M., Shi, Z. Y., Zhu, L. L., He, G. C. (2004). Mapping of a new resistance gene to bacterial blight in rice line introgressed from Oryza officinalis. Acta GeneticaSinica 31 (7), 724–729.
van Ooijen, G., Mayr, G., Kasiem, M. M., Albrecht, M., Cornelissen, B. J., Takken, F. L. (2008). Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 59 (6), 1383–1397. doi: 10.1093/jxb/ern045
Wang, C., Tan, M., Xu, X., Wen, G., Zhang, D., Lin, X. (2003). Localizing the bacterial blight resistance gene, Xa22(t), to a 100-Kilobase bacterial artificial chromosome. Phytopathol 93, 1258–1262. doi: 10.1094/PHYTO.2003.93.10.1258
Wang, W., Zhai, W., Luo, M., Jiang, G., Chen, X., Li, X., et al. (2001). Chromosome landing at the bacterial blight resistance gene Xa4 locus using a deep coverage rice BAC library. Mol. Gen. Genomics 265, 118–125. doi: 10.1007/s004380000382
Xing, J., Zhang, D., Yin, F., Zhong, Q., Wang, Bo, Xiao, S., et al. (2021). Identification and fine-mapping of a new bacterial blight resistance gene, Xa47(t), in G252, an introgression line of Yuanjiang common wild rice (Oryza rufipogon). Plant Dis. 105 (12), 4106–4112. doi: 10.1094/PDIS-05-21-0939-RE
Yoshimura, A., Mew, T. W., Khush, G. S., Omura, T. (1983). Inheritance of resistance to bacterial blight in rice cultivar Cas 209. Phytopathology 73, 1409–1412. doi: 10.1094/Phyto-73-1409
Yugander, A., Sundaram, R. M., Ladhalakshmi, D., Hajira, S. K., Prakasam, V., Prasad, M. S., et al. (2017). Virulence profiling of Xanthomonas oryzae pv. oryzae isolates, causing bacterial blight of rice in India. Eur. J. Plant Pathol. 149, 171–191. doi: 10.1007/s10658-017-1176-y
Zhang, H., Wang, S. (2013). Rice versus Xanthomonas oryzae pv oryzae: A unique pathosystem. Curr. Opin. Plant Biol. 16 (2), 188–195. doi: 10.1016/j.pbi.2013.02.008
Zhang, Q., Wang, C., Shi, A., Bai, J., Ling, S.C., Li, D.Y., et al. (1994). Evaluation of wild rice species resistance to rice bacterial blight (Xanthomonas oryza epv. oryzae) in wild rice species. Sci. Agric. Sin. 27 (5), 1–9.
Zhang, Q., Wang, C. L., Zhao, K. J., Zhao, Y. L., Caslana, V. C., Zhu, X. D., et al. (2001). The effectiveness of advanced rice lines with new resistance gene Xa23 to rice bacterial blight. Rice Genet. Newslett. 18, 71–72.
Zhang, F., Zhuo, D. L., Zhang, F., Huang, L. Y., Wang, W. S., Xu, J. L., et al. (2014). Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzaepv. oryzae rice. Plant Pathol. 64, 568–575. doi: 10.1111/ppa.12283
Keywords: bacterial blight (BB), Xanthomonas oryzae pv. oryzae(Xoo), Oryza officinalis, recombinant inbred line (RIL), mapping, single nucleotide polymorphism (SNP), nucleotide-binding adaptor (NB-ARC)
Citation: Sinha P, Kumar T. D, Sk H, Solanki M, Gokulan CG, Das A, Miriyala A, Gonuguntala R, Elumalai P, M. B. V. N K, S. K. M, Kumboju C, Arra Y, G. S. L, Chirravuri NN, Patel HK, Ghazi IA, Kim S-R, Jena KK, Hanumanth SR, Oliva R, Mangrauthia SK and Sundaram RM (2023) Fine mapping and sequence analysis reveal a promising candidate gene encoding a novel NB-ARC domain derived from wild rice (Oryza officinalis) that confers bacterial blight resistance. Front. Plant Sci. 14:1173063. doi: 10.3389/fpls.2023.1173063
Received: 24 February 2023; Accepted: 04 August 2023;
Published: 25 August 2023.
Edited by:
Harmeet Kaur, Indian Council of Agricultural Research, IndiaReviewed by:
Kanti Kiran, Gujarat Biotechnology University, IndiaNannan Yang, Wagga Wagga Agricultural Institute (WWAI), Australia
Gerbert Sylvestre Dossa, Food and Agriculture Organization of the United Nations, Italy
Andres Mäe, Estonian Crop Research Institute, Estonia
Meng Yuan, Huazhong Agricultural University, China
Copyright © 2023 Sinha, Kumar T., Sk, Solanki, Gokulan, Das, Miriyala, Gonuguntala, Elumalai, M. B. V. N, S. K., Kumboju, Arra, G. S., Chirravuri, Patel, Ghazi, Kim, Jena, Hanumanth, Oliva, Mangrauthia and Sundaram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raman Menakshi Sundaram, cm1zXzI4QHJlZGlmZm1haWwuY29t; Satendra K. Mangrauthia, c2ttZHJyQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Pragya Sinha
Pragya Sinha Dilip Kumar T.1†
Dilip Kumar T.1† Hajira Sk
Hajira Sk Yugander Arra
Yugander Arra N. Neerja Chirravuri
N. Neerja Chirravuri Hitendra Kumar Patel
Hitendra Kumar Patel Irfan Ahmad Ghazi
Irfan Ahmad Ghazi Sung-Ryul Kim
Sung-Ryul Kim Surekha Rani Hanumanth
Surekha Rani Hanumanth Satendra K. Mangrauthia
Satendra K. Mangrauthia Raman Menakshi Sundaram
Raman Menakshi Sundaram