
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 31 March 2023
Sec. Plant Nutrition
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1172275
This article is part of the Research TopicNovel Insights into the Regulatory Role of Sugar and Amino Acids Signaling in Plant-Microbe InteractionsView all 6 articles
Chloroplasts have important roles in photosynthesis, stress sensing and retrograde signaling. However, the relationship between chloroplast peptide chain release factor and ROS-mediated plant growth is still unclear. In the present study, we obtained a loss-of-function mutant dig8 by EMS mutation. The dig8 mutant has few lateral roots and a pale green leaf phenotype. By map-based cloning, the DIG8 gene was located on AT3G62910, with a point mutation leading to amino acid substitution in functional release factor domain. Using yeast-two-hybrid and BiFC, we confirmed DIG8 protein was characterized locating in chloroplast by co-localization with plastid marker and interacting with ribosome-related proteins. Through observing by transmission electron microscopy, quantifying ROS content and measuring the transport efficiency of plasmodesmata in dig8 mutant, we found that abnormal thylakoid stack formation and chloroplast dysfunction in the dig8 mutant caused increased ROS activity leading to callose deposition and lower PD permeability. A local sugar supplement partially alleviated the growth retardation phenotype of the mutant. These findings shed light on chloroplast peptide chain release factor-affected plant growth by ROS stress.
Photosynthesis in plant chloroplasts, the most important organelles in higher plants, converts light energy to chemical energy and fixes carbon, which are stored into sugar. Chloroplasts are differentiated plastids evolved from a cyanobacterial ancestor by endosymbiosis in a eukaryotic host cell (Raven and Allen, 2003; Dyall et al., 2004). The chloroplast genome in higher plants encodes about 100 genes, but the overwhelming majority of chloroplast-located proteins are encoded by nuclear genes (Abdallah et al., 2000).
A chloroplast genome contains about 40 photosynthesis-related and 60 housekeeping genes that are essential for protein synthesis (Sato et al., 1999). To date, protein synthesis events in chloroplast have been intensively explored, while the mechanism of translation termination remaining largely unknown (Zoschke and Bock, 2018). Three types of peptide chain release factors have been characterized in Escherichia coli (Scolnick et al., 1968; Craigen and Caskey, 1986; Gao et al., 2007; Zaher and Green, 2011) and two types, prfA and prfB, have been characterized in Arabidopsis plastids (Meurer et al., 1998; Meurer et al., 2002; Stoppel et al., 2011). prfA and prfB are responsible for UAA/UAG and UAA/UGA, respectively, and are recognized by the first and third amino acids in the highly conserved motifs in P-A(V/Q)-T of prfA and S-P-F of prfB (Scolnick et al., 1968; Nakamura and Ito, 1998; Ito et al., 2000; Nakamura et al., 2000; Nakamura and Ito, 2002). The GGQ motif in both prfA and prfB is essential for peptidyl-tRNA hydrolysis (Frolova et al., 1999; Dinçbas-Renqvist et al., 2000; Song et al., 2000; Vestergaard et al., 2001; Mora et al., 2003). Peptide chain release factors have two functions in translation termination, viz. recognition of the stop codon and catalysis of peptidyl-tRNA hydrolysis. Mutations in peptide chain release factor genes cause abnormal phenotypes exampled by dysfunction of chloroplasts in Arabidopsis (Meurer et al., 2002; Stoppel et al., 2011; Nick et al., 2013).
The chloroplast is an important site for reactive oxygen species (ROS) production (Foyer and Noctor, 2005; Asada, 2006). ROS are reactive forms of molecular oxygen, including singlet oxygen (1O2), superoxide radicals (), hydrogen peroxide (H2O2) and hydroxyl radicals (OH.). The photosynthetic electron transport chain (PET) in chloroplasts is a constant source of ROS, and these ROS cause oxidative stress when at excessive levels (Woodson, 2016). ROS plays important roles in transduction of intracellular signals and in control of gene expression and activity of antioxidant systems (Desikan et al., 2001; Apel and Hirt, 2004; Mullineaux et al., 2006; Foyer and Shigeoka, 2011). Exposure to stress in plants can lead to a sudden drastic increase in ROS production which was called an “oxidative burst” (Desikan et al., 2001). Generation and accumulation of ROS caused by imbalanced metabolic activities that are very sensitive to environmental stimuli lead to changes in ion fluxes and gene expression, oxidation of cellular components, disrupted organelle integrity and consequent effects on plant developmental processes such as embryogenesis, leaf growth, root branching, stoma opening and closing, and flowering. Therefore, ROS production must be fine-tuned to maintain redox homeostasis (Suzuki et al., 2012).
Here we report on the characterization of the dig8 (drought inhibited growth of lateral roots) mutant originally isolated by its abnormal lateral root development (Xiong et al., 2006). In addition to fewer lateral roots, the mutant has pale green leaves, dwarf stature and other abnormalities. The DIG8 gene was identified by map-based mapping and cloning and found to encode a chloroplast-localized peptide chain release factor with vital roles in chloroplast development and function. Dysfunction molecular transportation in the dig8 mutant caused abnormal chloroplast development causing higher levels of ROS activity leading to plant growth retardation. We provide evidence linking the chloroplast development and ROS-mediated callose deposition, and propose that the normal chloroplasts are essential for transportation through plasmodesmata in Arabidopsis.
Osmotic stress is an influential environmental stimulus that can inhibit lateral root growth in Arabidopsis. This effect was used to establish a screening system for mutants affecting osmotic stress response. Mutant dig8 was isolated from an M2 generation of an EMS-mutagenized population in the Columbia (Col) background by selecting for defective lateral root development. As shown in Figure 1A, the dig8 mutant had a shorter primary root and fewer lateral roots compared to the wildtype (WT). The lateral root density of the dig8 mutant was one-seventh that of Col (Figure 1B). Seedlings of the dig8 mutant had smaller leaves and shorter petioles with the same rosette number (Figures 1C, E). Reduced chlorophyll contents in the dig8 mutant (Figure 1D) caused pale green cotyledons and leaves (Figure S1).
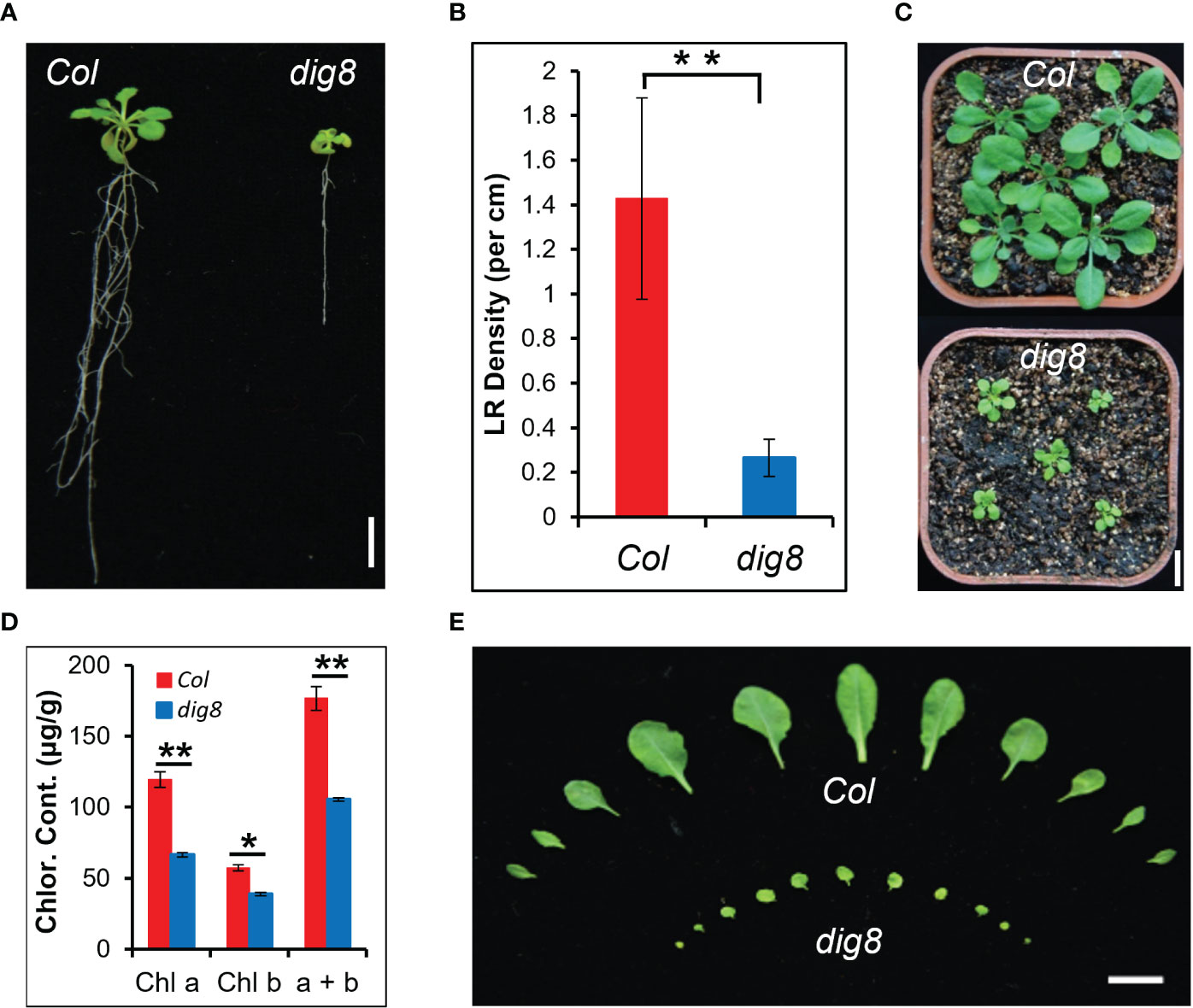
Figure 1 Identification of the dig8 mutant. (A), The dig8 mutant has fewer lateral roots. Scale bar, 1 cm. (B), Lateral root (LR) density in the dig8 mutant and Col. Error bars represent SD (n=30). **P < 0.01 (Student’s t test). (C), Morphology of Col and dig8 seedlings in soil. Scale bar, 1 cm. (D), Chlorophyll contents in Col and dig8 mutant. Shown are means of chlorophyll a, chlorophyll b and their total contents. Error bars represent SD from three replicates. *P < 0.05 and **P < 0.01 (Student’s t test). (E), Comparison of the rosette leaves at transition from the vegetative to the reproductive stage. Scale Bar, 1 cm.
To compare growth traits, we transferred 10-day-old WT and mutant seedlings to the vertical plate and made daily measurements of root growth. The root growth rate of the dig8 mutant was much slower than that of Col (Figure 2A). Following transplanting to soil in pots the growth rate of the mutant was also much slower than that of the WT (Figure 2B). Overall the mutant showed extreme dwarfness, but the flowering timing was similar to that of Col (Figure 2C); the mutant had a sparse inflorescence with few small flower buds (Figures 2D, E), tiny petals, short styles, and short stamens (Figures 2F, G). The siliques of dig8 mutant were shorter than those of Col but the seeds were significantly larger (Figures 2H, S2).
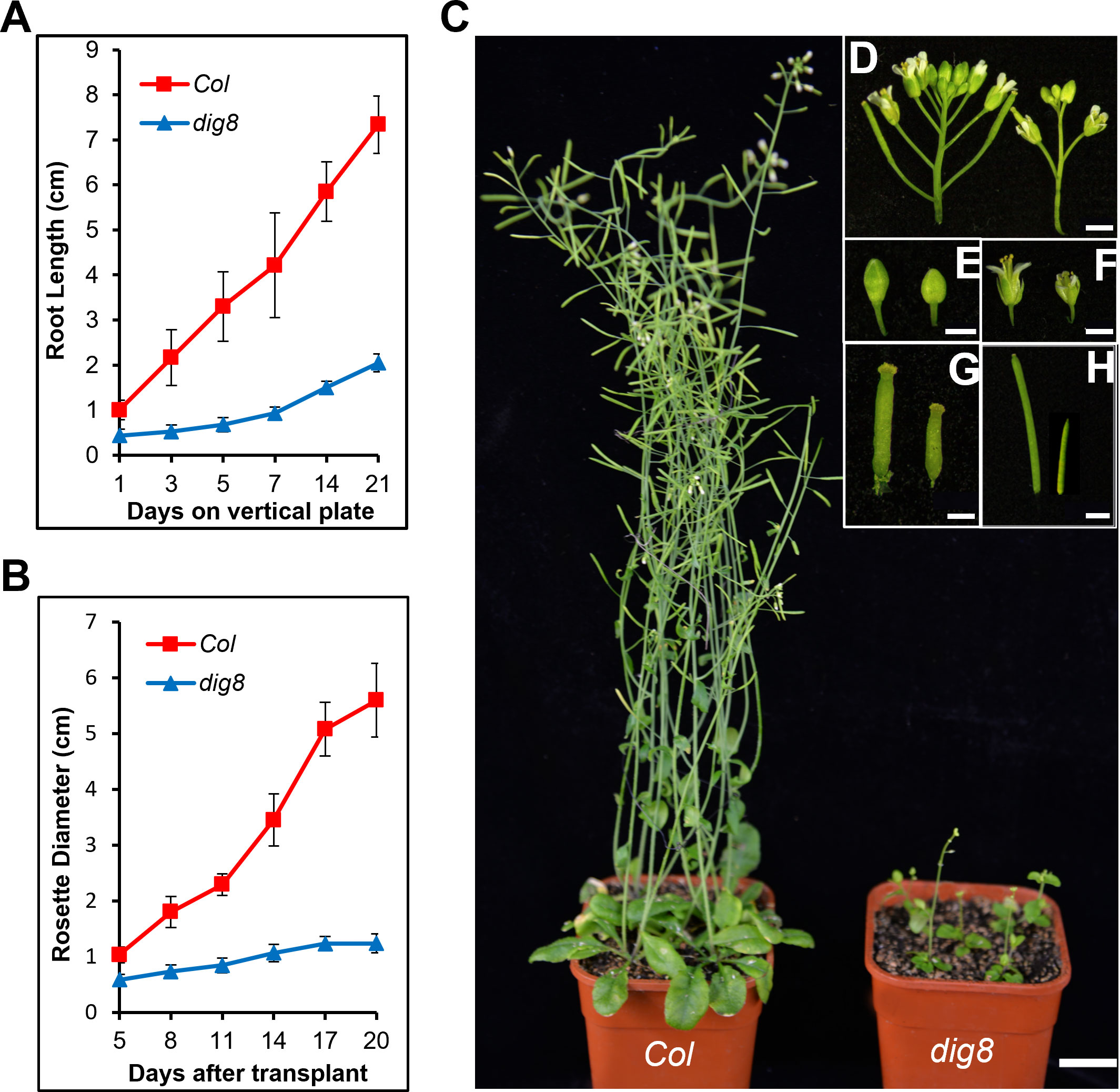
Figure 2 Retarded growth of the dig8 mutant. (A), Root growth rates of Col and dig8 mutant on a vertical plate. 10-day-old seedlings were transferred to a vertically placed plate and root length was recorded daily for 3 weeks. Error bars represent SD (n=20). (B), Growth rate measured daily by rosette diameters. Error bars represent SD (n=20). (C), Phenotypes of Col and dig8 mutant adult plants. Scale bar, 2 cm. (D–H), Enlarged view of flowers and siliques of Col (on the left) and dig8 mutant (on the right). (D), inflorescences; (E), flower buds; (F), pollinated flowers; (G), developing siliques; (H), siliques. Scale bars, 0.2 cm.
To map the DIG8 locus, a mapping population was generated by crossing the dig8 mutant with the ecotype Landsberg erecta (Ler). The mutation was first mapped to the end of Chromosome 3 between BAC clones T16L24 and MAA21. Fine mapping located DIG8 gene in clone F26K9. Sequencing identified a single G-to-A nucleotide change in the gene AT3G62910. This mutation was predicted to cause a glycine to arginine conversion at amino acid residue 147 in the translated peptide (Figures 3A, S3). This gene encodes a peptide chain release factor and a mutant in this gene was earlier named APG3 (albino or pale green mutant 3) based the pale green phenotype (Motohashi et al., 2007). The three-dimensional structure of the protein predicted by Phyre2 showed that the basic structure was made up of alpha-helices (Figure S4A). As seen in Figure S4B the conserved PCRF (peptide chain release factor) domain of the DIG8 protein is located at 119-234 a.a. and the mutant 147 site is highly conserved relative to other surrounding residues. This indicated that the mutant protein could have weakened or partial loss-of-function in translation termination. Alignment with the peptide chain release factors in both nuclei and chloroplasts revealed that DIG8 was a chloroplast-located protein (Figure S5A). With a conserved GGQ motif and P-A(V/Q)-T motif, DIG8 was considered to be a prfA protein (Figure S5B).
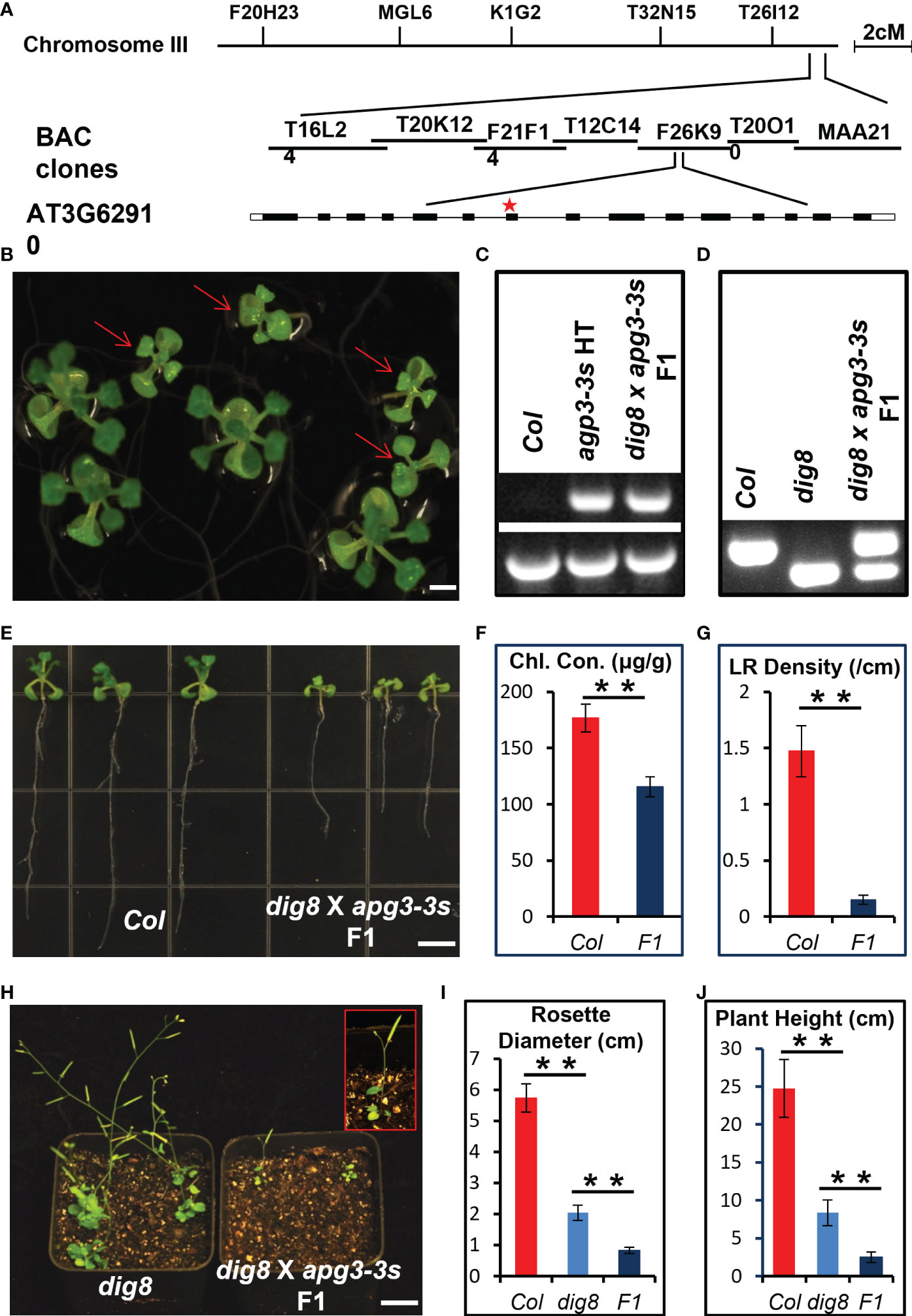
Figure 3 Mapping and allelic verification of DIG8. (A), Map-based cloning of the DIG8 locus. The dig8 mutant was crossed with the Ler ecotype and the F2 population was used for mapping. DIG8 was localized on Chromosome 3. Schematic structure of the DIG8 gene (AT3G62910); black boxes, exons; white boxes, UTRs; lines, introns; star, dig8 mutation site. (B), Allelic verification of DIG8 gene. Then dig8 mutant was crossed with an apg3-3s heterozygote. F1 seedlings on MS medium showed segregation ratio of 1 normal: 1 defective (arrows) seedling as expected for allelic mutations. Scale bars, 0.2 cm. (C), PCR verification of T-DNA insertion in the apg3-3s mutant allele. Lower bands were generated by PCR using primers LP and RP; upper bands by PCR using primers LB and RP. (D), dCAPS verification of dig8 mutant site. The BglII site was designed in reverse primer for dig8 mutant site. PCR products were digested with BglII to verify the point mutation in dig8 mutant. (E), Lateral root and pale green phenotypes of dig8 x apg3-3s F1 seedlings compared to Col. (F), Total chlorophyll contents in Col and dig8 x apg3-3s F1 seedlings. Error bars represent SD from three independent replicates. (G), Lateral root (LR) densities of Col and F1 mutant seedlings. Error bars represent SD (n=20). Scale bars, 0.5 cm. (H), Comparison of dig8 x apg3-3s F1 with dig8 mutant. Enlarged view of a dig8 x apg3-3s F1 plant in red rectangle showing 3 rosette leaves and 1 silique at the reproductive stage. Scale bars, 1 cm. I and J, Comparisons of rosette width (I) and plant height (J) of Col, dig8 and dig8 x apg3-3s F1 seedlings. Error bars represent SD (n=20). Asterisks indicate a significant difference between the indicated samples (t-test, **P < 0.01).
Genetic allelism test was performed to determine whether DIG8 and APG3 were alleles. We crossed the dig8 mutant with an apg3-3s (T-DNA insertion mutant SAIL_1251_H05), heterozygote since the apg3-3s homozygous was lethal (Figure S6). The phenotypes of dig8 mutant x apg3-3s heterozygote F1 seedlings segregated 1 normal: 1 mutant (Figure 3B). We confirmed that the dig8 x apg3-3s F1 plants with mutant phenotype were heterozygous for the T-DNA insertion and heterozygous for dig8 point mutation, revealing that DIG8 and APG3 were alleles (Figures 3C, D). As shown in Figures 3E–G, the mutant F1 seedlings had pale green cotyledons and leaves, and a shorter primary root and hardly any lateral roots compared with Col. Interestingly, these plants had only 3 rosette leaves, 1 silique and were approximately 2 cm in height. These phenotypes were much more severe than those found in the dig8 mutant (Figures 3H–J).
Since DIG8 was predicted to be a peptide release factor in chloroplasts, we generated a DIG8-GFP fusion construct and co-transferred it with a chloroplast localization marker into chloroplasts. As shown in Figure 4A, DIG8 co-localized with the marker in chloroplasts. To reveal the temporal-spatial expression pattern of DIG8, we performed qRT-PCR and promoter-GUS analysis. DIG8 was dominantly expressed in shoot tissue at the seedling stage, in leaves and flowers at the reproductive stage (Figures 4B, C).
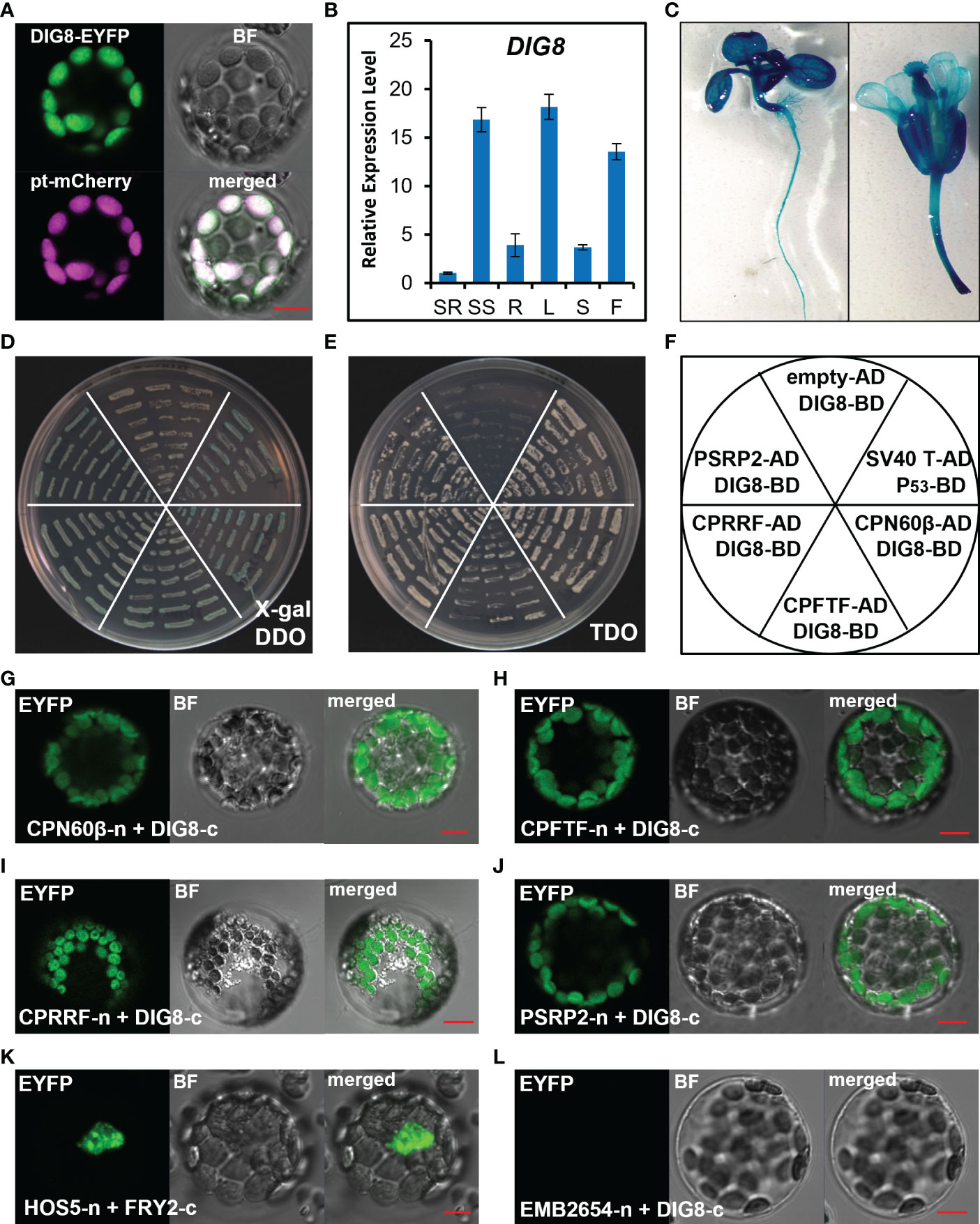
Figure 4 Characterization of the DIG8 protein. (A), DIG8 co-localized with a plastid marker. A DIG8-EYFP construct was co-transformed with plastid-localized marker pt-mCherry in protoplasts. The fluorescence signals were detected by confocal microscopy. BF, bright field. (B), qRT-PCR analysis of DIG8 expression. SR, seedling root; SS, seedling shoot; R, root; L, leaf; S, stem; F, flower. ACT2 was used as the internal control in qRT-PCR assays. Error bars represent the standard deviations (n=3). (C), DIG8 promoter-driven GUS expression. Transgenic plants expressing GUS under control of the DIG8 promoter were stained with X-Gluc and imaged under a microscope. (D–F), Confirmation of interactions between DIG8 and predicted DIG8- interacting proteins in the yeast 2-hybrid system. DIG8 was subcloned into the BD vector. CPN60β, CPFTF, CPRRF, PSRP2 were subcloned into the AD vector. DIG8-BD was co-transformed into yeast with each AD construct. The interactions were expressed on DDO medium with X-gal (D) and TDO medium (E). (F), Arrangement of yeast strains shown in (D) and (E). The empty-AD and DIG8-BD pair was used as a negative control, and the SV40T-AD and P53-BD pair was used as a positive control. (G–L), Confirmation of interactions between DIG8 and predicted DIG8-interacting proteins in protoplasts. DIG8 was subcloned into the c-EYFP vector; the others were subcloned into the n-EYFP vector. DIG8-c was co-transformed into protoplasts with each n-YFP construct. The EYFP signals of DIG8-c with CPN60β-n (G), CPFTF-n (H), CPRRF-n (I), PSRP2-n (J) were observed by confocal microscopy. (K), Interaction of HOS5-n and FRY2-c was used as the positive control. (L), Interaction of EMB2654-n and DIG8-c was used the negative control. Scale bars=10 µm.
To test the possible function of DIG8 in plastid protein synthesis regulation, we carried out yeast-two-hybrid assays to verify for interaction of DIG8 with ribosome components and a new peptide folding chaperonin. The β subunit of the chloroplast chaperonin 60 (CPN60β, AT1G55490), chloroplast protein folding trigger factor (CPFTF, AT2G30695), plastid-specific ribosomal protein 2 (PSRP2, AT3G52150), and chloroplast ribosome recycling factor (CPRRF, AT3G63190) were selected to confirm interaction with DIG8. Co-transformed yeast clones were transferred to the DDO medium with X-gal and TDO medium along with positive and negative controls. All selected yeast clones turned blue in the presence of X-gal and grew on the TDO medium, indicating that DIG8 interacted with these chloroplast ribosome-related proteins (Figures 4D–F). To confirm the locations of interactions in chloroplasts, we generated BiFC constructs in which DIG8 was fused with C-YFP and other proteins were fused with N-YFP. Signals for interaction in chloroplasts were detected by transient expression in protoplasts. Consistent with earlier results in yeast, DIG8 showed strong interaction with CPN60β, CPFTF, PSRP2 and CPRRF in plant cells (Figures 4G–L).
Since DIG8 was localized in chloroplasts and had an essential role in chloroplast protein synthesis, mutation in the DIG8 allele would likely cause changes in the chloroplast structure. Transmission electron microscopy of leaf cross-sections of Col and dig8 mutant showed intact and evenly distributed thylakoid layers in Col, whereas aggregations of thylakoid that formed 4-5 giant stacks were evident in the mutant (Figures 5A, B). This abnormal thylakoid stacking alone would lead to chloroplast dysfunction. Although DIG8 was a component of the peptide translation process, mutation of DIG8 would obviously affect expression of chloroplast encoded genes. Indeed, we found some of chloroplast genes were down-regulated, whereas others were up-regulated (Figure S7).

Figure 5 The dig8 mutation causes aberrant grana structure in chloroplasts. A and B, images of transmission electron microscopy of grana structures in leaf chloroplasts of Col (A) and dig8 mutant (B). Scale bar, 2 μm. (C), Fluorescence of Col and dig8 mutant measured by a Joliot-type spectrophotometer. Leaves of Col and dig8 mutant were dark-adapted for 15 min, illuminated with either low (80 μmol photons m-2 s-1) or medium (320 μmol photons m-2 s-1) orange actinic light (630 nm), and allowed to recover in darkness. Saturating pulses (7,900 μmol photons m-2 s-1, 200 ms duration) were applied to measure maximum fluorescence Fm and Fm’. (D), maximum PSII quantum yield of Col and dig8 mutant in darkness and light. Darkness adapted Arabidopsis leaves were exposed to different intensities of actinic light followed by recovery in darkness. Maximum quantum yield of PSII was calculated. LL: 80 μmol photons m-2 s-1, ML: 320 μmol photons m-2 s-1. **P < 0.01 (Student’s t test). (E), Chlorophyll a fluorescence quenching of Col and dig8 mutant. Leaves of Col and dig8 mutant were darkness-adapted for 15 min. NPQ was induced by 500 s of orange actinic light (630 nm, LL: 80 μmol photons m-2 s-1, ML: 320 μmol photons m-2 s-1), followed by recovery in darkness. Four independent biological replicates were analyzed. (F), P700 oxidation and reduction in Col and dig8 mutant. Darkness adapted (15 min) leaves of Col and dig8 mutant were illuminated with orange (630 nm, 320 μmol photons m-2 s-1) or far-red (725 nm, 1400 μmol photons m-2 s-1) actinic light. P700 oxidation and reduction were monitored by absorbance changes at 705 nm. Four independent biological replicates were analyzed.
Since the grana in the dig8 mutant were significantly changed, the photosynthetic properties of Col and dig8 mutant plants were analyzed. PSII performance was monitored by chlorophyll a fluorescence. The dig8 mutant showed a much lower chlorophyll fluorescence level than the WT (Figure 5C). The maximum PSII quantum yield of dig8 mutant was about 75% of Col. The steady-state PSII quantum yield (Fv’/Fm’) of dig8 under low light (80 μmol photons m-2 s-1) was reduced to 30% of the Col. With light intensity increased to 320 μmol photons m-2 s-1, Fv’/Fm’ of the dig8 mutant dropped further to 13% of Col (Figure 5D). Under 80 μmol photons m-2 s-1 illumination, Col did not induce obvious NPQ, whereas the dig8 had much higher NPQ. Under moderate light illumination (320 μmol photons m-2 s-1), NPQ was significantly induced in Col compared to the low light conditions, whereas the dig8 mutant showed no change in NPQ value compared to its low light value and was significantly lower than that of the WT. After illumination NPQ was allowed to relax in darkness. The recovery of the dig8 mutant was slower than that of Col (Figure 5E).
P700 oxidation in light followed by reduction in dark was analyzed to examine the function of PSI. When leaves were illuminated with saturating far-red light, only PSI was excited. The amplitude of absorbance change reflects the amount of excited PSI reaction centers. As shown in Figure 5F, the amount of active PSI reaction centers in the dig8 mutant was about 45% of that in the WT, levels that agreed with the amounts of chlorophyll in the respective plants (Figure 1D). When Col samples were illuminated with orange light, both PSI and PSII were excited. Considerable amounts of electrons fluxed from PSII to the PQ pool, which reduced to P700, caused a decrease in the final amount of oxidized PSI reaction centers. However, the dig8 mutant showed almost the same amount of when excited by orange light and far-red light, suggesting there was much less electron input from PSII in the dig8 mutant.
Excessive light can damage relevant organelles involved in photosynthesis. Therefore, plants need NPQ for protection. Under the action of NPQ, excess light energy is lost in the form of heat. Since dig8 mutant induced high NPQ even under low light conditions (Figure 5E), we speculated that low light intensity for Col was equivalent to strong light stress for the dig8 mutant. Thus, the dig8 mutant would exhibit stress symptoms of high light intensities even under conditions of normal light intensity.
To test this hypothesis, ROS levels were detected by DAB and NBT staining. As shown in Figures 6A, B, tissues of the dig8 mutant stained much more darkly than Col, indicating that ROS generation in dig8 mutant cells was much greater than in Col. The stained signal intensity was quantified by Image J (Figure S8). The ROS-related gene expression levels were mostly increased in the dig8 mutant, consistent with higher ROS levels (Figure 6C).
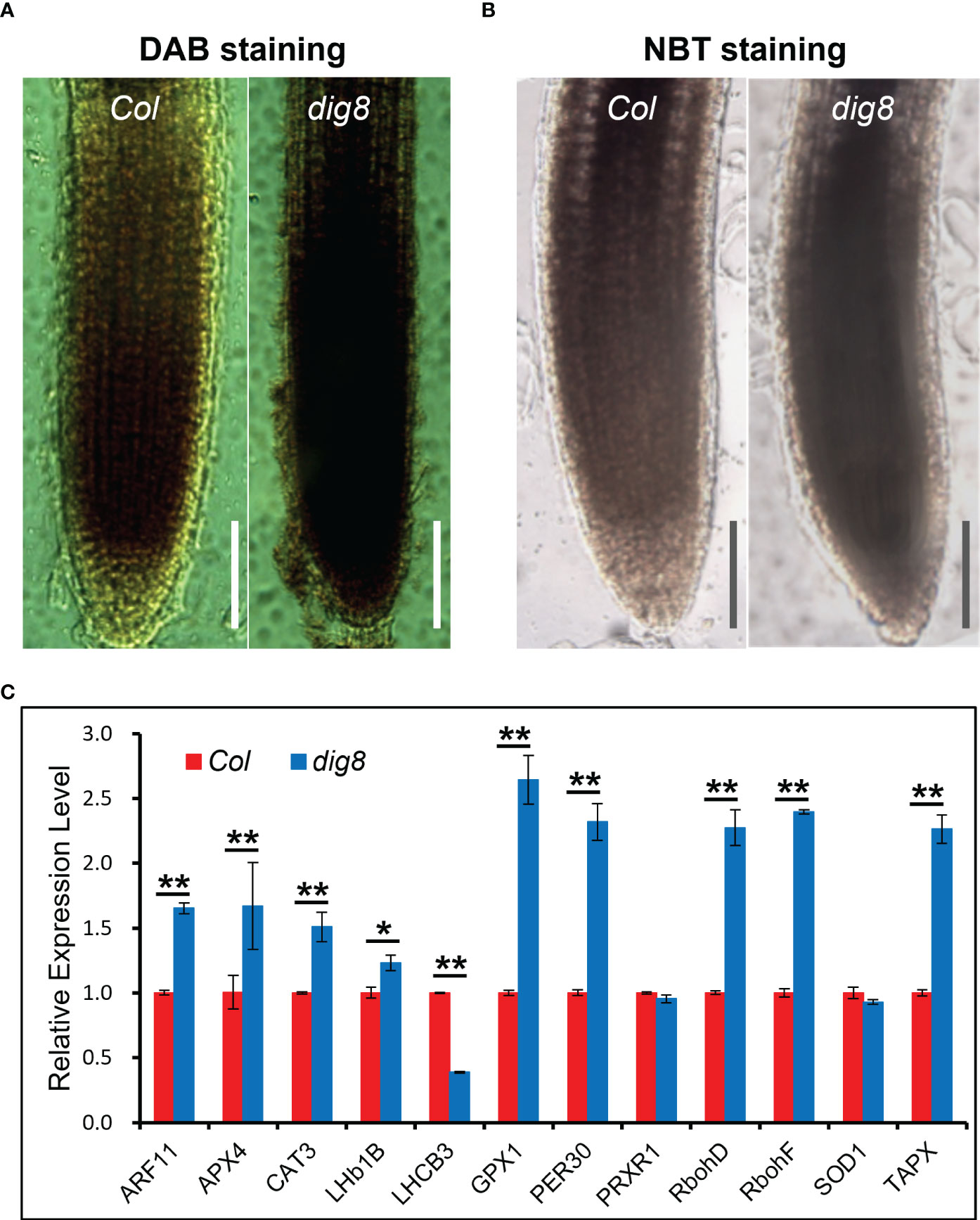
Figure 6 Higher ROS levels in the dig8 mutant. A and B, ROS was detected by DAB (A) and NBT (B) staining. Seedlings of Col and dig8 mutant were immersed in DAB and NBT solution at room temperature, and decolorization in ethanol. The images were captured with a Leica M80 stereo microscope. Scale Bar, 1 mm. (C), qRT-PCR analysis of ROS-related genes in Col and dig8 mutant. Error bars represent SDs among three independent replicates. Asterisks indicate a significant difference between the indicated samples (t-test, *P < 0.05, **P < 0.01).
Aniline blue staining of roots of Col and dig8 mutant was undertaken to test if excessive ROS levels in the dig8 mutant caused increased callose deposition. The mutant showed much more callose deposition than Col (Figures 7A, B). Among the 12 GSL genes in Arabidopsis, GSL8 was the major one responsible for callose synthesis and mutations of GSL8 resulted in severe growth retardation. Expression levels of GSL genes in Col and dig8 mutant seedlings were determined under normal growth conditions. As shown in Figure S9, 6 of these GSL genes were upregulated in the dig8 mutant and among them, GSL8 was expressed fourfold more that in Col. Thus excessive accumulation of callose in surrounding walls could restrict phloem unloading and cause constriction of intercellular symplastic channels. To test the PD permeability among plant cells, CFDA (5, 6-carboxyfluorescein diacetate), a fluorescent dye capable of penetrating cell membrane and plasmodesmata, was used as a probe for cell labeling. 10-d-old seedlings were wounded on cotyledon by scissors and CFDA solution was applied to the wound. The fluorescent signals were detected after incubation for 10 min allowing molecular transportation through plasmodesmata. As shown in Figures 7C, D, the CFDA signal could be detected at root tip of Col, but hardly any signal was observed at the whole root of dig8. These observations suggest that the molecular transportation through plasmodesmata from shoot to root was largely blocked in dig8 mutant.
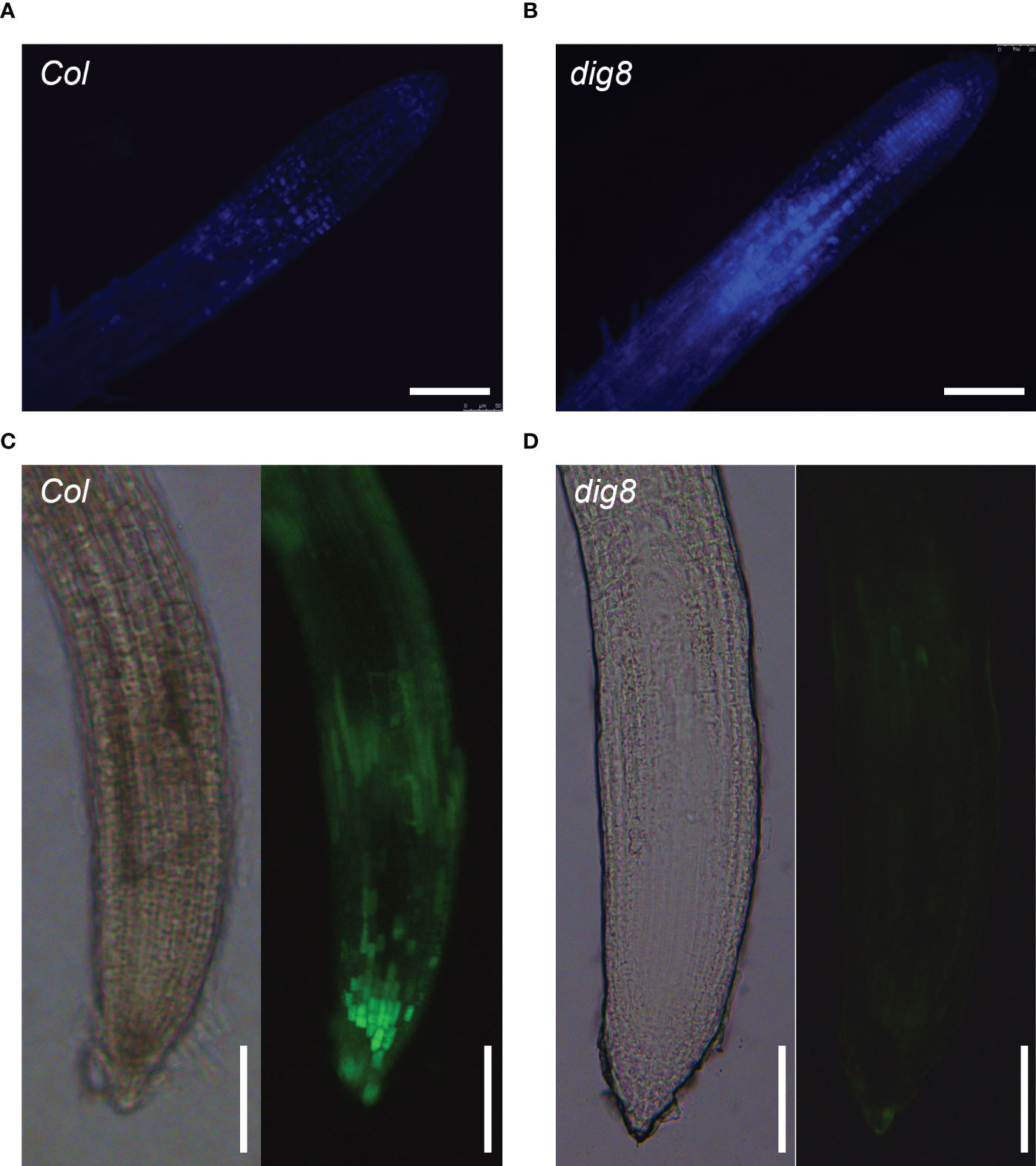
Figure 7 ROS caused lower PD permeability in the dig8 mutant. (A, B), Callose deposition in Col and dig8 mutant. Seedlings were cleared and stained with aniline blue. Roots images were taken with fluorescence microscopy. (C, D), PD permeability of Col and dig8 mutant. Cotyledons of 10-day-old seedlings were wounded and CFDA dye solution was applied to the wound. Roots images of Col and dig8 mutant (D) were taken with a microscope. Scale bar, 1 mm.
Since lower PD permeability was found in dig8 mutant caused by increased ROS level and callose deposition, we tested whether an increased local carbon source in the growth medium could rescue the mutant phenotype. Sucrose at 0.5%, 1%, 2% and 3% (W/V) was added into the growth medium and the growth status were recorded 14d after the seedlings were transferred to the plates. As shown in Figures 8A–D, comparing with Col, the dig8 mutant exhibited significant increases in primary root length, lateral root number, and biomass, particularly at higher sucrose levels. With biomass of Col values reach peak at 1% sucrose, dig8 biomass increased 2 times without sucrose concentration limitation. The similar tendencies were found in root weight, LR number and PR length (Figures 8E–H). These results demonstrated that a locally nutrient supplement largely rescued the cell energy deficit.
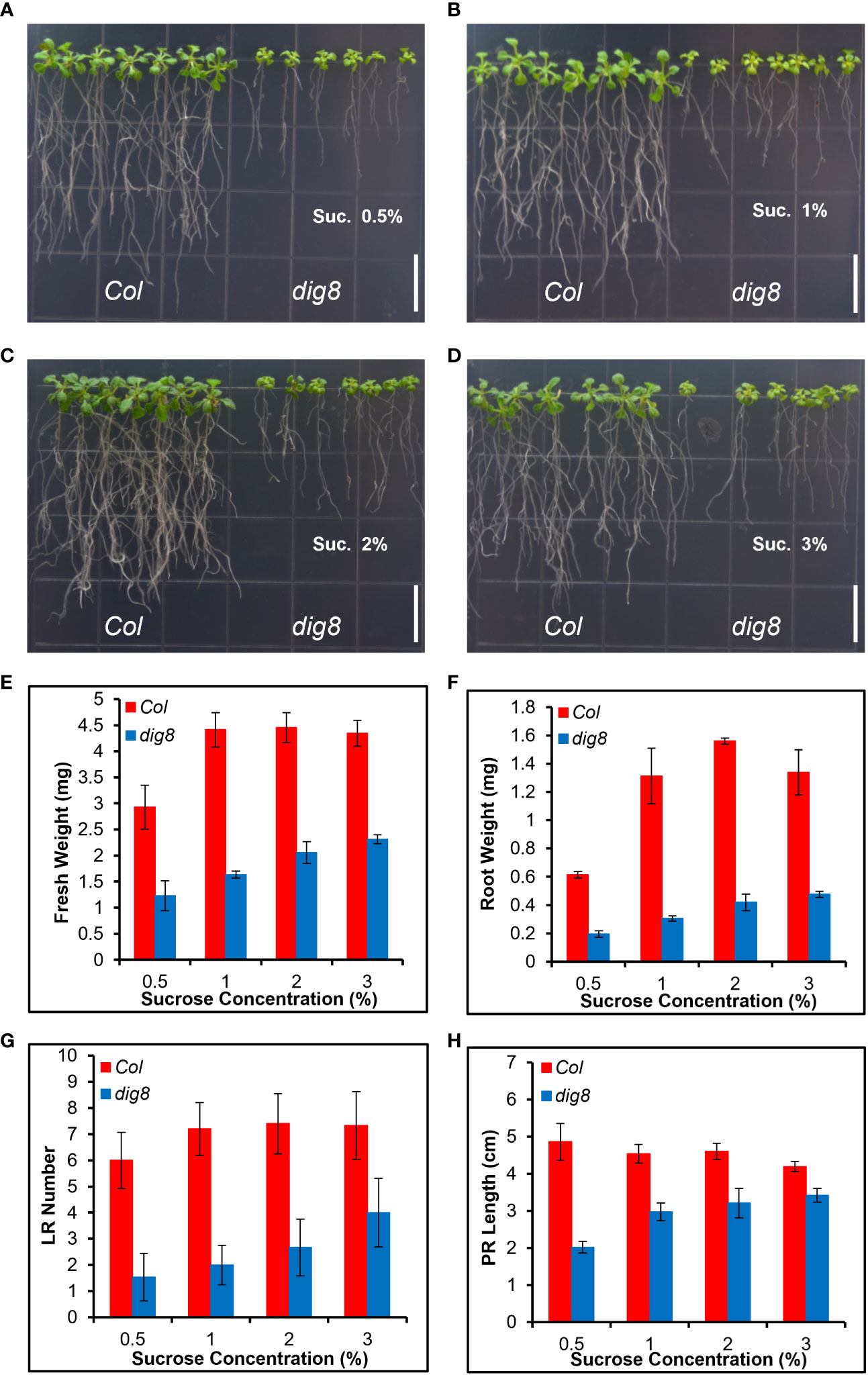
Figure 8 The dig8 mutant phenotype can be partially rescued by sugar supplementation. (A–D), Col and dig8 mutant grown on MS media supplemented with sucrose at the rate (w/v) of 0.5% (A), 1% (B), 2% (C), or 3% (D), respectively. Scale bar, 1 cm.10-d-old seedlings were transferred to the shown vertical plates and pictures were taken 2 weeks after the transfer. (E–H), fresh weight (E), root weight (F), lateral root number (G) and primary root length (H) of Col and dig8 mutant seedlings at different sucrose concentrations. Error bars represent SD (n=20).
In this study we identified and characterized a mutant dig8 that is defective in lateral root development. Map-based cloning showed that DIG8 encodes a peptide chain release factor in chloroplasts that is essential for plant growth and development. The DIG8 gene is identical to the APG3 gene whose mutation caused an albinotic or pale green phenotype. While homozygous T-DNA insertion mutant apg3-3s (Motohashi et al., 2007; Satou et al., 2014) is not viable, the dig8 mutation is viable albeit grew poorly. The dig8 mutation caused a single base change in the functional PCRF domain of the protein (Figure S4) and had a less drastic phenotype. The more severe phenotypes seen with the F1 plants derived from the dig8 and apg3-3s cross were likely due to the dosage effect of the single copy of the dig8 weak allele whereas the other apg3-3s copy (a T-DNA insertion) was null.
In addition to ribosome release factors many plastid-located proteins involved in protein translation affect chloroplast development and plant growth with somewhat similar effects. These include chloroplast chaperonin 60 (Apuya et al., 2001; Suzuki et al., 2009; Ke et al., 2017), chloroplast translation elongation factor G (Albrecht et al., 2006; Ruppel and Hangarter, 2007), chloroplast ribosome recycling factor (Wang et al., 2010), and plastid ribosomal protein (Ma et al., 2015). Accuracy and efficiency of the chloroplast translation system is essential for chloroplast function, and termination of protein translation must occur for protein release and ribosome recycling. It seems likely that peptide chain release factor DIG8 in the chloroplast translation system has a similar function to bacterial release factors, which promote hydrolysis of peptidyl-tRNA during termination of translation (Adio et al., 2018).
Chloroplast-to-nucleus retrograde signaling is essential for plant cell function. The chloroplast is a major site of reactive oxygen species (ROS) production (Koussevitzky et al., 2007). As a chloroplastic protein with important function, DIG8 affects chloroplast development and functions. Mutations of DIG8 may result in slower dissociation of nascent peptides from ribosomes in chloroplasts, and in turn reduce the translation efficiency of chloroplast-coded genes, most of which are core components of photosynthesis. Indeed, the dig8 mutation leads to abnormal thylakoid stack (Figures 5A, B) and extremely poor PSI and PSII performance (Figures 5C–F).
The link between ROS levels and callose deposition has been demonstrated in many studies. Callose biosynthesis and deposition is induced by ROS which can be triggered by dysmorphism of chloroplasts, exposure to stress conditions and pathogen attack (Dat et al., 2000; Bolwell et al., 2002; Bussotti et al., 2005; Kissoudis et al., 2016; D'Ambrosio et al., 2017). We found that, with most of callose biosynthesis gene GSLs highly expressed, callose is highly deposited in dig8 mutant (Figure S9).
As the most important transportation form of sugars, sucrose transportation is very important for plant growth and biomass accumulation. By adding sucrose in growth medium, the dig8 mutant can be largely rescued, demonstrating that chloroplast has a strong correlation with PD-mediated sugar transportation (Figure 8). Similarly, a sucrose transportation reporter system pSUC2-GFP was used to screen trafficking mutants in maize. Maize gat1 mutant was isolated and mapped to a gene encoding a plastid thioredoxin (TRX-m3) that has antioxidant function in post-translational protein redox status, indicating chloroplast affects sucrose transportation by redox-mediated PD dimension(Benitez-Alfonso et al., 2009).
In addition, DIG8 also regulates plant growth by affecting photosynthesis efficiency. As well as photosynthesis, light can damage photosynthetic apparatus. Irreversible photosynthetic apparatus inactivation produces an inhibitory effect on photosynthesis, which is called photoinhibition. To reduce photoinhibition, plants have the rapid quality control of PSII against photooxidative damage. In this repair cycle, D1 protein synthesis is the key step. Mutation of DIG8 will decrease the D1 protein dissociation efficiency from ribosomes, and in turn can repress PSII system reestablishment for photosynthesis. Actually, researchers had allotropic expressed of the psbA gene in Arabidopsis, tobacco and rice, and found that more D1 protein significantly stimulates transgenic plant growth by enhancing net CO2 assimilation rates with increases in biomass and grain yield (Chen et al., 2020). Overexpression of DIG8 may promote plant growth and could be exploited in crop breeding.
In this study, we found that the functional loss of the peptide chain releasing factor DIG8 in Arabidopsis thaliana led to abnormal development and function of chloroplasts and over accumulation of ROS and callose, resulting in obstruction of redox-mediated PD transportation between plant cells which ultimately led to the abnormal development of plants. Our research shows that DIG8 is essential for plant growth.
The Arabidopsis dig8 mutant in Col background was isolated from an EMS mutant library. The EMS mutant library was generated in our lab by ethyl methanesulphonate (EMS) mutagenesis of Col ecotype. An apg3-3s (SAIL_1251_H05) heterozygous mutant was ordered from ABRC (Arabidopsis Biological Resource Center, Ohio State University, Columbus OH, U.S.A.). Transgenic PDIG8-GUS lines were generated by floral dipping. Seeds were sterilized in 50% bleach and rinsed with water before sowing on half MS medium and incubated at 4°C for stratification. Seedlings were grown in growth rooms at 21°C with a 14 h-light/10h-darkness photoperiod.
Leaves were harvested, weighted and placed in 1.5 mL tubes; 80% acetone was added and the samples were incubated in darkness for 2 h. The samples were centrifuged and supernatants were transferred to new tubes. Light absorptions at 665 nm and 649 nm were measured with a microplate reader and were used to determine chlorophyll concentrations.
Col expressing the DIG8 promoter (2000 bp) driven GUS (PDIG8-GUS) was used to determined expression pattern of the DIG8 gene. For GUS-staining, plant tissues were immersed in the GUS staining buffer and incubated overnight at 37°C. After removal of the staining buffer the samples were washed with 70% ethanol and GUS signals were captured with a Leica M80 stereo microscope.
Protoplast isolation and transient expression assays were performed followed standard protocols (Yoo et al., 2007). Briefly, 3-week-old young leaves were cut to 1 mm strips and the cell walls were digested by the fungal Cellulose R10® and Macerozyme R10®. Centrifuge the digested mixture at 100g to pellet the protoplasts in a 30-ml round-bottomed tube for 1-2 min. Re-suspend protoplasts in W5 solution. Rest the protoplasts by keeping on ice for 30 min. Remove the W5 solution as much as possible without touching the protoplast pellet. Re-suspend protoplasts in MMG solution and keep at room temperature. Transient expression was performed in a 2-mL microfuge tube, 10 µL DNA were added to tube and then gently mixed with 100 µL protoplasts. After adding 110 µL of PEG, the tube was gently tapped to allow completely mixing. Incubate the transfection mixture at room temperature for up to 15 min. Dilute the transfection mixture with 400-440 µL W5 solution and mix well to stop the transfection process. Centrifuge at 100g for 2 min using a bench-top centrifuge and remove the supernatant. Re-suspend protoplasts gently with 1 ml WI in each well of a 6-well tissue culture plate. Incubate protoplasts at room temperature (20-25°C) for 16 h. For subcellular localization, the DIG8-EYFP construct and chloroplast-localized pt-mCherry marker were co-transformed into protoplasts. EYFP and mCherry signals were detected by using a Zesis LSM710 confocal microscope. For protein-protein interaction (BiFC) analysis, the DIG8-c-EYFP construct was co-transformed in combinations with n-EYFP constructs. EYFP signals were detected by confocal microscopy.
For gene quantification analysis, 14-d-old seedlings of Col and dig8 lines were harvested and total RNA was extracted. After reverse transcription, the expression levels of each gene were checked by qRT-PCR. ACT2 was used as control.
Yeast transformation was conducted as described previously (Cheng et al., 2018). DIG8 was subcloned into pGBK-T7 and CPN60β, CPFTF, CPRRF and PSRP2 were subcloned into pGAD-T7. All construct combinations, including positive and negative controls, were co-transformed into yeast strain AH109 and selected on the DDO (SD-TL) medium. Protein interactions were displayed on X-gal DDO and TDO (SD-TLH) media. Briefly, pick one colony of Y2Hgold yeast cells in 2 ml YPD, incubate at 30°C overnight with shaking at 200 rpm. Centrifuge at 10,000 rpm for 1 min, discard supernatant. The pellet was resuspended in LiAc and brief centrifugated, and resuspended in 450 μl LiAc. After centrifugation at 10,000 rpm for 1 min and discarding supernatant, yeast cells were mixed with DNA. After adding 40 μl DMSO, the mixture was vortexed briefly and incubated at 42°C in a water bath for 20 min. The mixture was chilled on ice for 3 min and briefly centrifuged. After suspending in 100-150 µl H2O, cells were spread on the SD drop-out medium.
Leaves of Col and dig8 mutant were collected and cut into small pieces before fixation in glutaraldehyde solution followed by OsO4 treatment. After washing with PBS buffer, the samples were dehydrated in ethanol and embedded into Epon 812 resin. Chloroplast structures of Col (n=12) and dig8 (n=15) in ultrathin sections were observed with a transmission electron microscope, at least 3 ultrathin sections of each were examined.
PSII fluorescence quenching and oxidation-reduction were monitored using a Joliot spectrometer JTS-10 (Bio-Logic Scientific Instruments). Arabidopsis leaves were dark-adapted for 15 min prior to initiating measurements. Chlorophyll a fluorescence traces of leaves were recorded in fluorescence mode. Non-photochemical Quenching (NPQ) was induced by 500 s of orange actinic light (630 nm, low intensity: 80 μmol photons m-2 s-1 or moderate intensity: 320 μmol photons m-2 s-1) and plants were allowed to recover in darkness. Saturating pulses (7,900 μmol photons m-2 s-1, 200 ms duration) were applied to measure maximum fluorescence Fm and Fm’. The maximum PSII quantum yield (Fv/Fm) and steady-state PSII quantum yield in light (Fv’/Fm’) were calculated. NPQ was calculated by (Fm – Fm’)/Fm’. For P700 measurements, dark-adapted leaves were illuminated with orange (630 nm, 320 μmol photons m-2 s-1) or far-red (725 nm, 1400 μmol photons m-2 s-1) actinic light. The amount of was recorded by absorbance change at 705 nm during the actinic pulse and following darkness. All experiments were repeated with 10 leaves of each Col and dig8 mutant.
ROS levels were detected by DAB and NBT staining. For DAB staining, seedlings of Col and dig8 mutant were immersed in DAB solution for 24 h at room temperature. The seedlings were decolorized in 95% ethanol for 0.5 h and the remaining brown polymerization product produced by DAB interaction with ROS was observed using a Leica M80 stereomicroscope. For NBT staining, seedlings of Col and dig8 mutant were immersed in NBT solution at room temperature for 24 h without light. The following procedures were the same as for DAB staining. For ROS signal quantification, the DAB and NBT staining images of the seedlings to be analyzed were imported to ImageJ, and converted into 8-bit grayscale images. Then adjust the threshold to select the stained part of the seedling and calculate the stained area to quantify ROS signal intensity of Col (n=5) and dig8 (n=5) mutant.
Callose structures were revealed by aniline blue staining. Tissue samples destained overnight in 95% ethanol were washed in PBS buffer before immersion in callose staining buffer with 0.01% aniline blue for 2 hours and imaged in Citifluor solution under fluorescence microscope with UV filters. For PD permeability test, cotyledons of 10-day-old Col and dig8 seedlings were wounded with scissors, and CFDA dye solution was dropped on the wound. After 10 min incubation, the samples were observed under fluorescence microscope.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
TC and LX conceived and designed the research. XZ, YH, and XH performed the experiments. TC wrote the manuscript. SZ provided technical assistant. XZ, YH, and XH contributed equally to this work. All authors contributed to the article and approved the submitted version.
This work was supported by the Fundamental Research Funds for central public welfare scientific research institutes (Y2019XK15-04) and the Starting Research Fund from the Hangzhou Normal University (2019QDL015).
The authors thank Dr. Guangchao Wang for assistance with TEM, and ABRC for providing the apg3-3s seeds.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1172275/full#supplementary-material
Abdallah, F., Salamini, F., Leister, D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of arabidopsis. Trends Plant Sci. 5, 141–142. doi: 10.1016/s1360-1385(00)01574-0
Adio, S., Sharma, H., Senyushkina, T., Karki, P., Maracci, C., Wohlgemuth, I., et al. (2018). Dynamics of ribosomes and release factors during translation termination in e. coli. Elife 7:e34252. doi: 10.7554/eLife.34252
Albrecht, V., Ingenfeld, A., Apel, K. (2006). Characterization of the snowy cotyledon 1 mutant of arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60, 507–518. doi: 10.1007/s11103-005-4921-0
Apel, K., Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/Annurev.Arplant.55.031903.141701
Apuya, N. R., Yadegari, R., Fischer, R. L., Harada, J. J., Zimmerman, J. L., Goldberg, R. B. (2001). The arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60alpha gene. Plant Physiol. 126, 717–730. doi: 10.1104/Pp.126.2.717
Asada, K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396. doi: 10.1104/Pp.106.082040
Benitez-Alfonso, Y., Cilia, M., San Roman, A., Thomas, C., Maule, A., Hearn, S., et al. (2009). Control of arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. U.S.A. 106, 3615–3620. doi: 10.1073/Pnas.0808717106
Bolwell, G. P., Bindschedler, L. V., Blee, K. A., Butt, V. S., Davies, D. R., Gardner, S. L., et al. (2002). The apoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 53, 1367–1376. doi: 10.1093/jxb/53.372.1367
Bussotti, F., Agati, G., Desotgiu, R., Matteini, P., Tani, C. (2005). Ozone foliar symptoms in woody plant species assessed with ultrastructural and fluorescence analysis. New Phytol. 166, 941–955. doi: 10.1111/J.1469-8137.2005.01385.X
Chen, J. H., Chen, S. T., He, N. Y., Wang, Q. L., Zhao, Y., Gao, W., et al. (2020). Nuclear-encoded synthesis of the D1 subunit of photosystem ii increases photosynthetic efficiency and crop yield. Nat. Plants 6, 570–580. doi: 10.1038/S41477-020-0629-Z
Cheng, X., Zhang, S., Tao, W., Zhang, X., Liu, J., Sun, J., et al. (2018). Indeterminate Spikelet1 recruits histone deacetylase and a transcriptional repression complex to regulate rice salt tolerance. Plant Physiol. 178, 824–837. doi: 10.1104/Pp.18.00324
Craigen, W. J., Caskey, C. T. (1986). Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322, 273–275. doi: 10.1038/322273a0
D'Ambrosio, J. M., Couto, D., Fabro, G., Scuffi, D., Lamattina, L., Munnik, T., et al. (2017). Phospholipase C2 affects mamp-triggered immunity by modulating ros production. Plant Physiol. 175, 970–981. doi: 10.1104/Pp.17.00173
Dat, J., Vandenabeele, S., Vranová, E., Van Montagu, M., Inzé, D., Van Breusegem, F. (2000). Dual action of the active oxygen species during plant stress responses. Cell Mol. Life Sci. 57, 779–795. doi: 10.1007/S000180050041
Desikan, R., A-H.- Mackerness., S., Hancock, J. T., Neill, S. J. (2001). Regulation of the arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172. doi: 10.1104/Pp.127.1.159
Dinçbas-Renqvist, V., Engström, A., Mora, L., Heurgué-Hamard, V., Buckingham, R., Ehrenberg, M. (2000). A post-translational modification in the ggq motif of Rf2 from escherichia coli stimulates termination of translation. EMBO J. 19, 6900–6907. doi: 10.1093/Emboj/19.24.6900
Dyall, S. D., Brown, M. T., Johnson, P. J. (2004). Ancient invasions: From endosymbionts to organelles. Science 304, 253–257. doi: 10.1126/Science.1094884
Foyer, C. H., Noctor, G. (2005). Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/Tpc.105.033589
Foyer, C. H., Shigeoka, S. (2011). Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 155, 93–100. doi: 10.1104/Pp.110.166181
Frolova, L. Y., Tsivkovskii, R. Y., Sivolobova, G. F., Oparina, N. Y., Serpinsky, O. I., Blinov, V. M., et al. (1999). Mutations in the highly conserved ggq motif of class 1 polypeptide release factors abolish ability of human Erf1 to trigger peptidyl-trna hydrolysis. Rna 5, 1014–1020. doi: 10.1017/S135583829999043x
Gao, H., Zhou, Z., Rawat, U., Huang, C., Bouakaz, L., Wang, C., et al. (2007). Rf3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129, 929–941. doi: 10.1016/J.Cell.2007.03.050
Ito, K., Uno, M., Nakamura, Y. (2000). A tripeptide 'Anticodon' deciphers stop codons in messenger rna. Nature 403, 680–684. doi: 10.1038/35001115
Ke, X., Zou, W., Ren, Y., Wang, Z., Li, J., Wu, X., et al. (2017). Functional divergence of chloroplast Cpn60α subunits during arabidopsis embryo development. PloS Genet. 13, E1007036. doi: 10.1371/Journal.Pgen.1007036
Kissoudis, C., Seifi, A., Yan, Z., Islam, A. T., Van Der Schoot, H., Van De Wiel, C. C., et al. (2016). Ethylene and abscisic acid signaling pathways differentially influence tomato resistance to combined powdery mildew and salt stress. Front. Plant Sci. 7. doi: 10.3389/Fpls.2016.02009
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. doi: 10.1126/science.1140516
Ma, Z., Wu, W., Huang, W., Huang, J. (2015). Down-regulation of specific plastid ribosomal proteins suppresses Thf1 leaf variegation, implying a role of Thf1 in plastid gene expression. Photosynth Res. 126, 301–310. doi: 10.1007/S11120-015-0101-5
Meurer, J., Lezhneva, L., Amann, K., Gödel, M., Bezhani, S., Sherameti, I., et al. (2002). A peptide chain release factor 2 affects the stability of uga-containing transcripts in arabidopsis chloroplasts. Plant Cell 14, 3255–3269. doi: 10.1105/Tpc.006809
Meurer, J., Plücken, H., Kowallik, K. V., Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem ii in arabidopsis thaliana. EMBO J. 17, 5286–5297. doi: 10.1093/Emboj/17.18.5286
Mora, L., Heurgué-Hamard, V., Champ, S., Ehrenberg, M., Kisselev, L. L., Buckingham, R. H. (2003). The essential role of the invariant ggq motif in the function and stability In vivo of bacterial release factors Rf1 and Rf2. Mol. Microbiol. 47, 267–275. doi: 10.1046/J.1365-2958.2003.03301.X
Motohashi, R., Yamazaki, T., Myouga, F., Ito, T., Ito, K., Satou, M., et al. (2007). Chloroplast ribosome release factor 1 (Atcprf1) is essential for chloroplast development. Plant Mol. Biol. 64, 481–497. doi: 10.1007/S11103-007-9166-7
Mullineaux, P. M., Karpinski, S., Baker, N. R. (2006). Spatial dependence for hydrogen peroxide-directed signaling in light-stressed plants. Plant Physiol. 141, 346–350. doi: 10.1104/Pp.106.078162
Nakamura, Y., Ito, K. (1998). How protein reads the stop codon and terminates translation. Genes Cells 3, 265–278. doi: 10.1046/J.1365-2443.1998.00191.X
Nakamura, Y., Ito, K. (2002). A tripeptide discriminator for stop codon recognition. FEBS Lett. 514, 30–33. doi: 10.1016/S0014-5793(02)02330-X
Nakamura, Y., Ito, K., Ehrenberg, M. (2000). Mimicry grasps reality in translation termination. Cell 101, 349–352. doi: 10.1016/S0092-8674(00)80845-4
Nick, S., Meurer, J., Soll, J., Ankele, E. (2013). Nucleus-encoded light-harvesting chlorophyll A/B proteins are imported normally into chlorophyll b-free chloroplasts of arabidopsis. Mol. Plant 6, 860–871. doi: 10.1093/Mp/Sss113
Raven, J. A., Allen, J. F. (2003). Genomics and chloroplast evolution: What did cyanobacteria do for plants? Genome Biol. 4, 209. doi: 10.1186/Gb-2003-4-3-209
Ruppel, N. J., Hangarter, R. P. (2007). Mutations in a plastid-localized elongation factor G alter early stages of plastid development in arabidopsis thaliana. BMC Plant Biol. 7, 37. doi: 10.1186/1471-2229-7-37
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Tabata, S. (1999). Complete structure of the chloroplast genome of arabidopsis thaliana. DNA Res. 6, 283–290. doi: 10.1093/Dnares/6.5.283
Satou, M., Enoki, H., Oikawa, A., Ohta, D., Saito, K., Hachiya, T., et al. (2014). Integrated analysis of transcriptome and metabolome of arabidopsis albino or pale green mutants with disrupted nuclear-encoded chloroplast proteins. Plant Mol. Biol. 85, 411–428. doi: 10.1007/S11103-014-0194-9
Scolnick, E., Tompkins, R., Caskey, T., Nirenberg, M. (1968). Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. U.S.A. 61, 768–774. doi: 10.1073/Pnas.61.2.768
Song, H., Mugnier, P., Das, A. K., Webb, H. M., Evans, D. R., Tuite, M. F., et al. (2000). The crystal structure of human eukaryotic release factor Erf1–mechanism of stop codon recognition and peptidyl-trna hydrolysis. Cell 100, 311–321. doi: 10.1016/S0092-8674(00)80667-4
Stoppel, R., Lezhneva, L., Schwenkert, S., Torabi, S., Felder, S., Meierhoff, K., et al. (2011). Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petb transcript stability in arabidopsis chloroplasts. Plant Cell 23, 2680–2695. doi: 10.1105/Tpc.111.085324
Suzuki, N., Koussevitzky, S., Mittler, R., Miller, G. (2012). Ros and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/J.1365-3040.2011.02336.X
Suzuki, K., Nakanishi, H., Bower, J., Yoder, D. W., Osteryoung, K. W., Miyagishima, S. Y. (2009). Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in arabidopsis thaliana. BMC Plant Biol. 9, 38. doi: 10.1186/1471-2229-9-38
Vestergaard, B., Van, L. B., Andersen, G. R., Nyborg, J., Buckingham, R. H., Kjeldgaard, M. (2001). Bacterial polypeptide release factor Rf2 is structurally distinct from eukaryotic Erf1. Mol. Cell 8, 1375–1382. doi: 10.1016/S1097-2765(01)00415-4
Wang, L., Ouyang, M., Li, Q., Zou, M., Guo, J., Ma, J., et al. (2010). The arabidopsis chloroplast ribosome recycling factor is essential for embryogenesis and chloroplast biogenesis. Plant Mol. Biol. 74, 47–59. doi: 10.1007/S11103-010-9653-0
Woodson, J. D. (2016). Chloroplast quality control - balancing energy production and stress. New Phytol. 212, 36–41. doi: 10.1111/Nph.14134
Xiong, L., Wang, R. G., Mao, G., Koczan, J. M. (2006). Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol. 142, 1065–1074. doi: 10.1104/Pp.106.084632
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/Nprot.2007.199
Zaher, H. S., Green, R. (2011). A primary role for release factor 3 in quality control during translation elongation in escherichia coli. Cell 147, 396–408. doi: 10.1016/J.Cell.2011.08.045
Keywords: chloroplast, peptide chain release factor, ROS, callose, sugar transportation
Citation: Zhang X, Han Y, Han X, Zhang S, Xiong L and Chen T (2023) Peptide chain release factor DIG8 regulates plant growth by affecting ROS-mediated sugar transportation in Arabidopsis thaliana. Front. Plant Sci. 14:1172275. doi: 10.3389/fpls.2023.1172275
Received: 23 February 2023; Accepted: 14 March 2023;
Published: 31 March 2023.
Edited by:
Yuanhu Xuan, Shenyang Agricultural University, ChinaReviewed by:
Shou-qiang Ouyang, Yangzhou University, ChinaCopyright © 2023 Zhang, Han, Han, Zhang, Xiong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Chen, Y2hlbnRhb0Boem51LmVkdS5jbg==; Liming Xiong, bHhpb25nQGhrYnUuZWR1Lmhr
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.