- 1College of Horticulture, Hebei Agricultural University, Baoding, China
- 2Research Center of Chinese Jujube, College of Horticulture, Hebei Agricultural University, Baoding, China
The Ziziphus mauritiana Lam. and Z. jujuba Mill. are the two most economically important members of the genus Ziziphus. The fruit color of Z. mauritiana remains green throughout fruit development in the majority of commercial cultivars, whereas its close relative, Z. jujuba Mill. turns from green to red in all cultivars. However, the lack of transcriptomic and genomic information confines our understanding of the molecular mechanisms underlying fruit coloration in Z. mauritiana (Ber). In the present study, we performed the transcriptome-wide analysis of MYB transcription factors (TFs) genes in Z. mauritiana and Z. jujuba, and identified 56 ZmMYB and 60 ZjMYB TFs in Z. mauritiana and Z. jujuba, respectively. Through transcriptomic expression analysis, four similar MYB genes (ZmMYB/ZjMYB13, ZmMYB/ZjMYB44, ZmMYB/ZjMYB50, and ZmMYB/ZjMYB56) from Z. mauritiana and Z. jujuba were selected as candidate key genes regulating flavonoid biosynthesis. Among these genes, the ZjMYB44 gene was transiently highly expressed in fruit, and flavonoid content accumulation also increased, indicating that this gene can influence flavonoid content during the period of fruit coloration in Z. jujuba. The current study adds to our understanding of the classification of genes, motif structure, and predicted functions of the MYB TFs, as well as identifying MYBs that regulate flavonoid biosynthesis in Ziziphus (Z. mauritiana and Z. jujuba). Based on this information, we concluded that MYB44 is involved in the flavonoids biosynthesis pathway during the fruit coloring of Ziziphus. Our research results provide an important understanding of the molecular mechanism of flavonoid biosynthesis resulting in fruit coloration and laying a foundation for further genetic improvement of fruit color in Ziziphus.
1 Introduction
Transcription factors (TFs) are important regulators of gene expression in plants, directing a variety of physiological mechanisms such as growth, development, cell division, stress response, and fruit color development (Muthamilarasan et al., 2014; Zhang et al., 2018; Qing et al., 2019; Fan et al., 2020; Muhammad et al., 2022; Muhammad et al., 2022a; Muhammad et al., 2023). As a result, interpreting the molecular and physiological functions of these TFs is now a major research focus. Furthermore, TF research gained traction, and a slew of new studies had been published. In plants, MYB (myeloblastosis) transcription factors are ubiquitous in eukaryotic systems, and their function in plant flavonoids like anthocyanin biosynthesis was first discovered in maize (Paz-Ares et al., 1987).
After the initial identification of MYB in Zea mays, numerous MYB TFs genes were gradually discovered in a variety of different plants, including the Arabidopsis thaliana (Stracke et al., 2001). These MYBs are recognized by the family-specific function of a highly conserved MYB domain at the N-terminus (Yanhui et al., 2006; Stracke et al., 2007). Plant MYB proteins have an MYB DNA-binding domain that is extremely conserved, the MYB domain is a 50-53 amino acid segment or region that interacts with DNA in a sequence-specific fashion (Zhang et al., 2018; Qing et al., 2019). The MYB TFs are divided into four subfamilies depending on the number of highly conserved imperfect repeats in the DNA-binding domain: R3 MYB (MYB1R) has one repeat, R2R3 MYB has 2 repeats, R1R2R3 MYB (MYB3R) has 3 repeats, and 4R MYB has 4 repeats (Lin-Wang et al., 2010; Muhammad et al., 2022). Similarly, the R2R3-MYB is the biggest TF gene family in plants, with 126 genes discovered in A. thaliana (Stracke et al., 2001; Yanhui et al., 2006). In strawberries, the R2R3 MYB gene FaMYB1 regulates anthocyanin and flavonol production (Aharoni et al., 2001; Lin-Wang et al., 2010).
According to numerous reports, MYB TFs are involved in physiological and metabolic pathways in plant species, as well as responding to various biotic and abiotic stresses (Qing et al., 2019). The role of these MYB TFs has already been studied in some plant species like Physcomitrella patens (Pu et al., 2020), potato (Li et al., 2020), Casuarina equisetifolia (Wang et al., 2021), Dendrobium catenatum (Zhang et al., 2021), Hedychium coronarium (Abbas et al., 2021), Woodland strawberry (Li et al., 2021), Brassica napus L (Li et al., 2021a), Sunflower (Li et al., 2020a), Pitaya (Xie et al., 2021), Pineapple (Liu et al., 2017), Vaccinium corymbosum (Wang et al., 2021a), Chinese jujube (Qing et al., 2019), and so on. Similarly, the role of MYB TFs has also been reported in color or pigment production in the plant species like Liriodendron (Yang et al., 2021), kiwi (Li et al., 2017), peach (Zhang et al., 2018), apple (Jiang et al., 2019), Chinese bayberry (Cao et al., 2021), Pistacia chinensis (Song et al., 2021), Fragaria ananassa (Liu et al., 2021a), Populus deltoids (Zhuang et al., 2021), and pepper (Liu et al., 2021). However, none of the comparative studies have been conducted in Z. mauritiana and Z. jujuba that describe the role of MYBs in flavonoid biosynthesis during fruit coloration.
The Ziziphus Mill. is a monetarily important genus of the Rhamnaceae family with roughly one hundred and seventy species found all over this planet (Liu et al., 2020). Z. jujuba and Z. mauritiana are the most economically important members of this genus. Z. mauritiana is native to the Indian and Pakistan Subcontinent (Muhammad et al., 2020; Muhammad et al., 2022b). Out of the 14 native Ziziphus species in China, the jujube is thought to have evolved straightforwardly from wild sour jujube, making it an excellent source for the introduction of a variety of useful attributes for the increase in jujube fruit quality (Liu et al., 2011; Zhao et al., 2014; Liu et al., 2020; Muhammad et al., 2022c). Jujube is also one of the oldest domesticated fruit trees, with findings of cultivation stretching back over 7,000 years (Uddin et al., 2021; Uddin et al., 2022a). Aside from being a major dry fruit crop, it is also a good resource of herbal remedies and a source of income for some 20 million Asian people (Liu and Wang, 2009; Liu et al., 2020; Uddin et al., 2021; Uddin et al., 2022a). Because of their high nutritive quality and assorted phytochemical composition, both species (Z. jujuba and Z. mauritiana) have become important cash crops (Wang et al., 2018; Muhammad et al., 2022c; Uddin et al., 2022). The primary bioactive elements of jujube fruits, which have been widely used as food, food additives, and flavoring, are vitamin C, triterpenoids, amino acids, carbohydrates, phenolics, flavonoids, or anthocyanin (Wu et al., 2012; Wang et al., 2018).
Our completion of the whole-genome sequence of Z. jujuba Mill. in 2014 (Liu et al., 2014) for the first time provided evidence for genome-wide profiling of the MYB superfamily. However, the lack of whole-genome sequencing of Z. mauritiana limits our insights in this context. MYB transcription factors play a role in many aspects of plant growth, development, metabolism, and fruit coloration. But the comparative study of MYB TFs in Z. jujuba, and Z. mauritiana have not yet been completely known and characterized, and the expression of MYB genes in different developmental stages of the fresh fruit of these two species is undefined. The motifs analysis and phylogenetic trees are all reported in this article. Furthermore, the transient overexpression of ZjMYB44 significantly increased flavonoid accumulation in the Z. jujuba fruit. Similarly, the ZjMYB44 gene was transiently highly expressed in fruit, and flavonoid content accumulation increased, indicating that this gene can influence flavonoid content leading to color regulation in Ziziphus fruit. Our findings can help characterize ZjMYB and ZmMYB genes, and this research will aid future further functional studies of ZjMYB and ZmMYB superfamily genes involved in the color development of Z. jujuba and Z. mauritina fruits.
2 Materials and methods
2.1 Identification of ZmMYB and ZjMYB protein sequences and database searches
The transcriptome data from Z. mauritiana and Z. jujuba was used to identify all of the presumed MYBs proteins. The Pfam database (http://pfam.xfam.org/) was searched to obtain the hidden Markov Model (HMM) profile for the MYB binding domain (Finn et al., 2016; Zhang et al., 2018). The presence of an MYB domain in the selected MYB proteins was further verified using the online program HMMER (https://www.ebi.ac.uk/Tools/hmmer/). The Arabidopsis Information Resource (TAIR) database was used to obtain Arabidopsis MYBs protein sequences and was then blasted using Blastp. The ZmMYB and ZjMYB proteins were identified using the Arabidopsis MYBs domain sequences as a reference. To evade repetition, multiple alignments were accomplished among the identified ZmMYB and ZjMYB protein sequences.
2.2 Sequence analysis and construction of the phylogenetic tree
The MEGA version 7.0 was used to create a phylogenetic tree employing the neighbor-joining approach (Tamura et al., 2013; Cui et al., 2018). Furthermore, the MYB protein sequences from Arabidopsis were obtained, and ClustalW was used to align Arabidopsis MYB protein sequences and the deduced amino-acid sequences of ZmMYBs and ZjMYBs (Chenna et al., 2003; Larkin et al., 2007; Cui et al., 2018). The phylogenetic tree between ZmMYBs and ZjMYBs and the known MYBs from Arabidopsis was then constructed using the neighbor-joining method and the MEGA7.0 program, and bootstrap analysis was performed with 1,000 replicates (Yang et al., 2015). The Compute pI/Mw online tool (http://web.expasy.org/compute pi/) was used to calculate the theoretical isoelectric point (pI) and molecular weight (Mw) of ZmMYB and ZjMYB proteins, and Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) was used to predict their subcellular localization.
2.3 Conserved domain and motif analysis in MYB protein sequences
The MEME Suit 5.5.0 program (http://meme-suite.org/tools/meme) was used to recognize the motifs manifest in the MYBs proteins, and the conserved motifs of ZmMYB and ZjMYB proteins were identified (Bailey et al., 2009). The distribution of motifs was set to zero or one per sequence, the maximum number of motifs to be found was 10, and the default parameters are being used. Pfam (http://pfam.xfam.org) was used to define the conserved domains of ZmMYB and ZjMYB proteins. Thereafter, we used the TBTools software (https://github.com/CJ-Chen/TBtools) to visualize the Pfam, and MEME, results (Chen et al., 2020).
2.4 Treatment for plant materials (fruit)
The fruit samples of green-colored cultivars of Z. mauritiana (Gaolong) at different developmental stages were collected from Yunan Province, China. Fifteen cDNA libraries were generated using total RNA extracted from the Z. mauritiana green cultivars fruit peel and Z. jujuba red fruit cultivar (Jinsixiaozao) (three replicates per tissue; Junior fruit (JF: JF1, JF2, JF3), Before white riping fruit (BWRF: BWRF1, BWRF2, BWRF3); Half ripening fruit (HRF: HRF1, HRF2, HRF3); White-ripening fruit (WRF: WRF1, WRF2, WRF3), and Ripen fruit (RF: RF1, RF2, RF3). At the age of 30 (junior fruit, JF), 50 (before ripening fruit, BRF), 80 (white ripening fruit, WRF), 90 (half ripening fruit, HRF), 100 (full ripening fruit, RF) days after pollination (DAP), fruit specimens were harvested, and the samples were collected. The fruit skin samples (at least 10 fruits per sample) were promptly frozen in liquid nitrogen and preserved at -80°C for further investigation. Each tissue or fruit skin sample consisted of three biological replicates.
2.5 Transient expression of ZjMYB13 ZjMYB44, ZjMYB50, and ZjMYB56 in Z. jujuba fruit
The red fruit cultivar was used at the white ripening stage in the transient expression assay. The ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56 were inserted into the pCambia1302 vector, and the recombinant plasmids (pCambia1302 vector) of ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56 were constructed and transferred to Escherichia coli, the recombinant plasmids ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56 were transformed into Agrobacterium tumefaciens GV3101. Fruits of ‘Dongzao’ jujube collected at the white ripe stage were injected with transformed A. tumefaciens containing ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56 respectively. Similarly, a pCambia1302 control was also set, and 20 ‘Dongzao’ fruits of the same (size, color value, hardness, etc.) and 1 mL of resuspended bacteria were injected along the shoulder, and one fruit was injected on two sides. The injection ended the dark treatment for 24h and was sampled after every 24h intervals, then after, fruits were ground and stored at -80˚C for subsequent flavonoids test and real-time quantitative assay.
2.6 Measurement of flavonoids
The total flavonoid content was determined by using the procedure as follows.
2.6.1 Standard preparation of rutin solution
0.2 mg/ml of rutin solution was prepared. 0 mL, 1 mL, 2 mL, 3 mL, 4 mL, and 5 mL of rutin solution were taken and added to 0.6 mL of 5% sodium nitrite (NaNO2) solution. After allowing the solution to stand for 6 minutes mixed well, 0.4 mL 10% aluminum nitrate solution [10% Al(NO3)3] was added, and mixed properly, and after 6 min, 2 mL of 4% sodium hydroxide (NaOH) was added. The absolute ethanol was adjusted to 10 mL, then mixed, and after 30 min, the absorbance was measured using a UV-vis spectrophotometer at 511 nm.
2.6.2 Sample determination
The sample material was mixed in 50 mL centrifuge tubes at 1:30 (g sample: mmL ethanol) and sonicated at 50°C for 40 min after 15 min centrifugation 1mL supernatant was removed. Poured 1 mL of sample (supernatant) into the tube, 0.6 mL of 5% sodium nitrite solution was added and was allowed to stand for 6 min. the 0.4 mL of 10% aluminum nitrate solution was added and mixed well. After 6 min 2 mL of 4% sodium hydroxide was added. The absolute ethanol was adjusted to 10 mL, then mixed, and after 30 min, the absorbance was measured using a UV-vis spectrophotometer at 511 nm.
2.7 Real-time quantitative detection
RNA was extracted from fruit peel at five developmental stages of fruit. Real-time quantitative primers were designed with the NCBI website. Three pairs were designed to screen the best primers and were used for subsequent real-time quantitative detection as shown in Table S1. qRT-PCR analysis was performed on a Bio-Rad real-time quantitative instrument. A real-time quantification kit (Tiangen Biotechnology, China) was used. The 20 µL reaction system contained 10 µL of 2 SYBR Premix, 10 µM of primers 0.4 µL each, 1 µL of diluted cDNA, and 8.2 µL ddH2O. They were incubated at 94°C for 15 min, followed by 15 s at 94°C, 55 to 63°C, and 15 s at 72°C. The 2−ΔΔCT method (Livak and Schmittgen, 2001) was used to analyze gene expression levels, and ZjActin was used as an internal control (Zhang et al., 2015; Shi et al., 2020; Zhang et al., 2020). All qRT-PCR primers are presented the Table S1.
3 Results
3.1 Phylogenetic analyses and function predictions of ZmMYBs and ZjMYBs
In the present study, we performed the transcriptome-wide analysis of MYB TFs genes in Ziziphus and identified 56 ZmMYB (ZmMYB1 through ZmMYB56) and 60 ZjMYB (ZjMYB1 through ZjMYB60) transcription factors (TFs) in Z. mauritiana and Z. jujuba, respectively. To assess the evolutionary pattern among Ziziphus species (Z. mauritiana and Z. jujuba) and Arabidopsis MYB proteins, the deduced amino acid sequences of the MYB proteins identified from Arabidopsis and Ziziphus transcriptomes were completely aligned. After that, a combined phylogenetic tree was constructed using the neighbor-joining method and bootstrap analysis (1,000 reiterations) (Figure S1).
By aligning the entire set of predicted MYB protein sequences from A. thaliana (171), Chinese jujube (60), and Z. mauritiana (56), a phylogenetic tree of AtMYB, ZjMYB, ZmMYBs protein was constructed (Figure S1). Furthermore, in a combined phylogenetic tree of Z. jujuba and Z. mauritiana, 13 pairs of putative orthologous proteins (ZmMYB12 and ZjMYB13, ZmMYB11 and ZjMYB39, ZmMYB42 and ZjMYB48, ZmMYB9 and ZjMYB9, ZmMYB7 and ZjMYB6, ZmMYB22 and ZjMYB32, ZmMYB2 and ZjMYB2, ZmMYB21 and ZjMYB26, ZmMYB23 and ZjMYB24, ZmMYB8 and ZjMYB41, ZmMYB44 and ZjMYB44, ZmMYB26 and ZjMYB21, ZmMYB50 and ZjMYB50) were identified. In contrast, 2 pairs of paralogous MYB family proteins were identified in Z. mauritiana: ZmMYB53 and ZmMYB51, ZmMYB25 and ZmMYB20. Similarly, 10 pairs of paralogous MYB family proteins were identified in Z. jujuba, i.e., ZjMYB51 and ZjMYB36, ZjMYB57 and ZjMYB40, ZjMYB52 and ZjMYB37, ZjMYB55 and ZjMYB38, ZjMYB46 and ZjMYB31, ZjMYB47 and ZjMYB35, ZjMYB20 and ZjMYB19, ZjMYB43 and ZjMYB28, ZjMYB45 and ZjMYB30, ZjMYB35 and ZjMYB49 as shown in Figure 1.
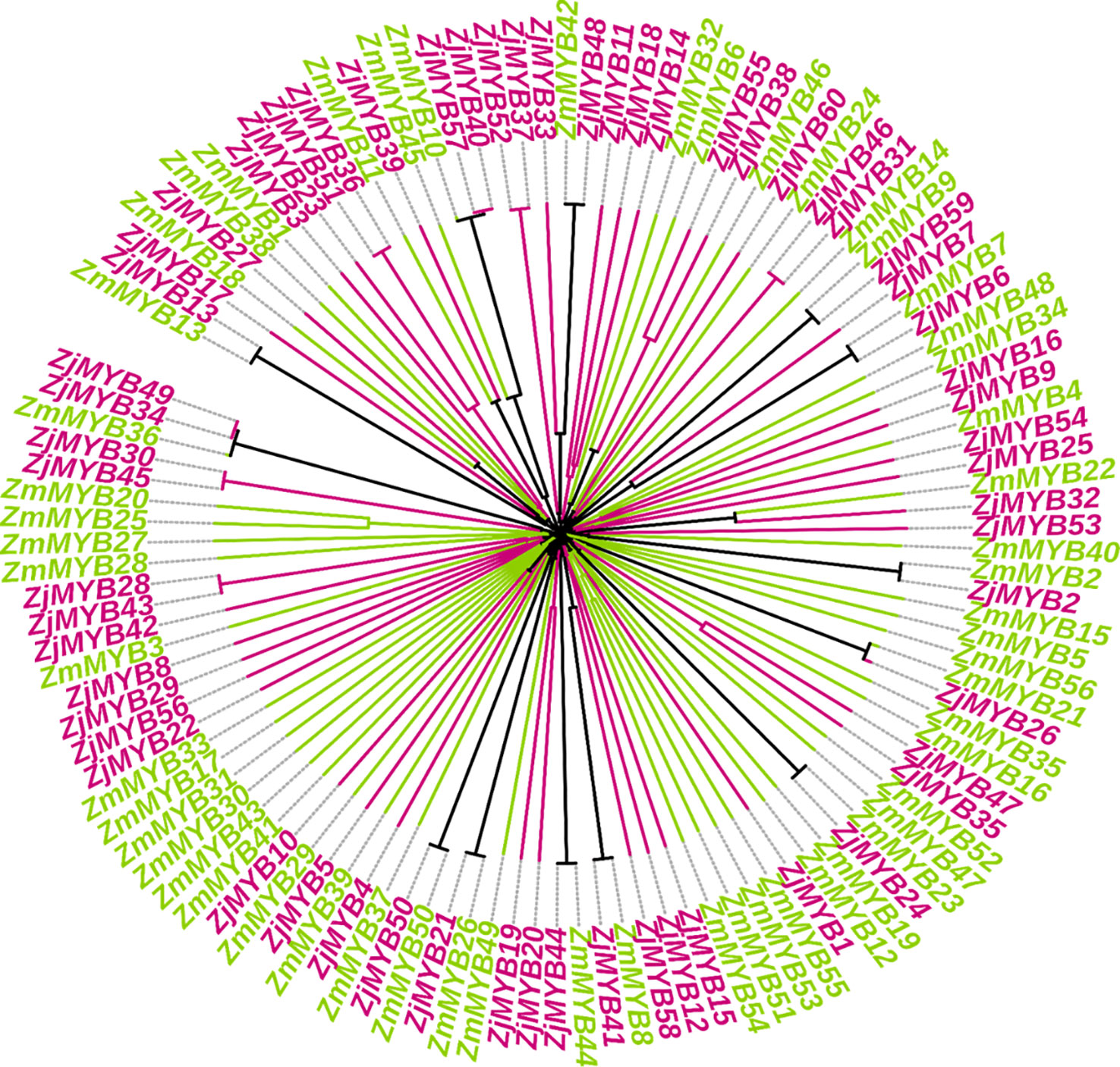
Figure 1 Comparative phylogenetic analysis of Z. mauritiana and Z. jujuba MYB proteins and are labeled by two different colors. The ZmMYB and ZjMYB represent Z.mauritiana and Z. jujuba MYB proteins respectively. ZmMYBs are in yellow-green and ZjMYBs are represented in red-violet color.
3.2 Conserved sequences in Ziziphus MYB proteins
The MEME and TBtool were used to find the conserved motifs. The findings demonstrated that the majority of R2R3-MYB proteins belonging to the same subfamily shared similar motifs, further demonstrating how closely related these proteins’ evolutionary relationships are to those of the phylogenetic tree (Figure 2). MYB TFs contain the MYB DNA-binding domain, which is usually found near the protein’s N-terminus. Based on MEME analysis, and hidden Markov Model (HMM) profile for the MYB binding domain, the MYB domains were found in all of the deduced MYB protein sequences in Ziziphus (Z. mauritiana and Z. jujuba), and each domain sequence contained 50-53 amino acid residues (Figure 3). Further, the sequence logos showed the presence of conserved sequences at specific positions. A to J depicted the repeats of Z. mauritiana MYB protein sequences, and M to V presented Z. jujuba MYB protein sequences. These findings indicated that the majority of amino acids are conservatively represented in each repeat of two Ziziphus species (Figure S2).
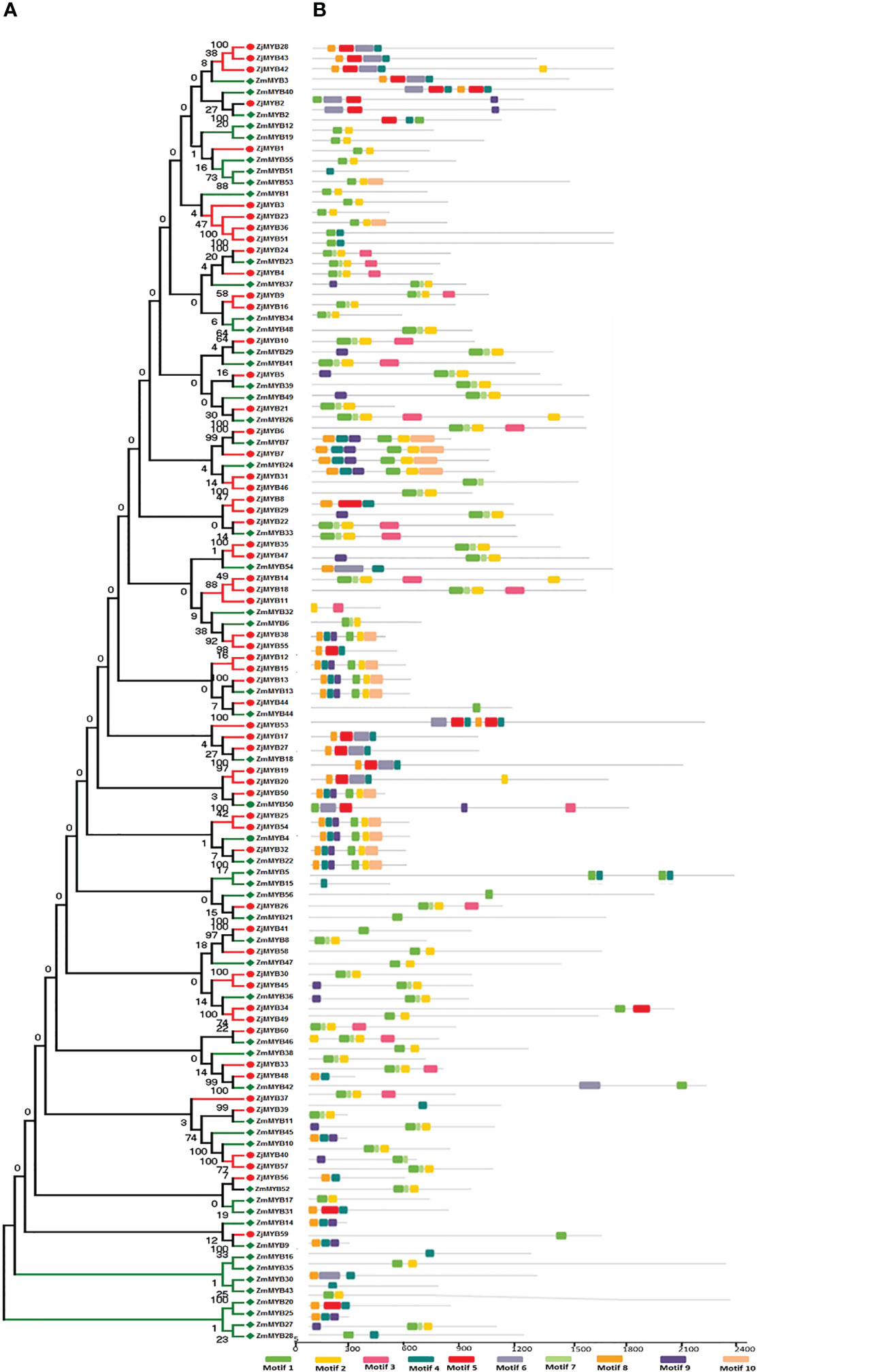
Figure 2 Phylogenetic relationships, the architecture of conserved protein motifs in R2R3-MYB proteins from Z. mauritiana and Z. jujuba. (A) The phylogenetic tree was constructed with 56 and 60 R2R3-MYB proteins from Z. mauritiana and Z. jujuba, respectively. The ZjMYBs are represented by red color while ZmMYBs are represented by green color. (B) Architecture of conserved protein motifs in different subfamilies. The colored boxes designated the different motifs as listed at the bottom of the figure.
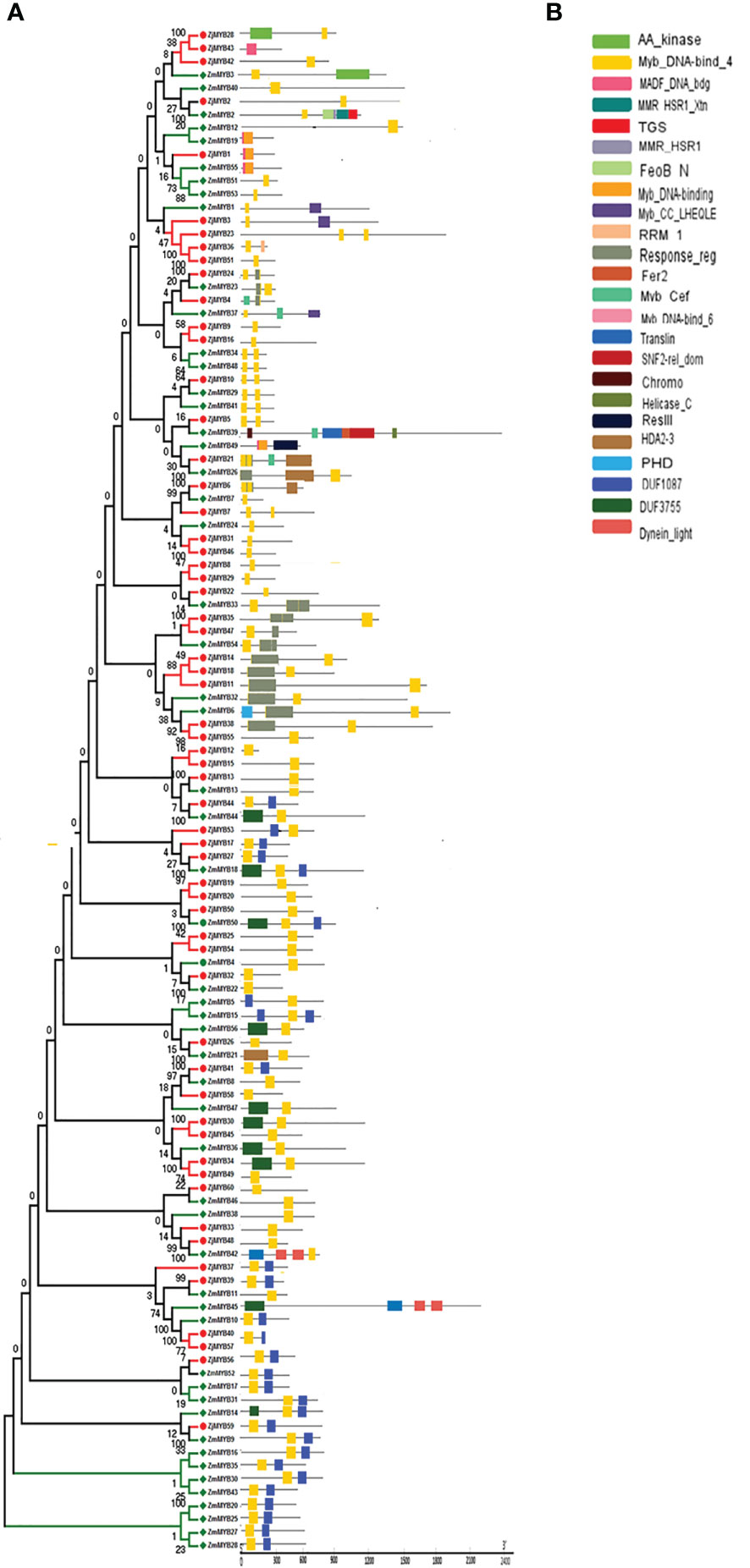
Figure 3 Phylogenetic relationships, the architecture of conserved MYBs domains in R2R3-MYB proteins from Z. mauritiana and Z. jujuba. (A) The phylogenetic tree was constructed with 56 and 60 R2R3-MYB proteins from Z. mauritiana and Z. jujuba respectively. (B) The architecture of conserved MYBs domains in different subfamilies. The colored boxes designated the different MYBs domains as shown in the figure.
Furthermore, the MYB domain is the core motif of MYB transcription factors, and it is intimately associated with binding to the promoters of their target genes. Multiple sequence alignment analyses of 116 MYBs from Ziziphus (56 from Z. mauritiana and 60 from Z. jujuba) were used to generate sequence logos to investigate conservation at specific positions in the MYB domain. The R2 and R3 repeats contain many conserved amino acids, including the distinctive Trp (W) residues that are regarded landmarks of the MYB domain, as shown in Figure S2. The R2 repeat contains three conserved Trp residues as sown in Figure S2.
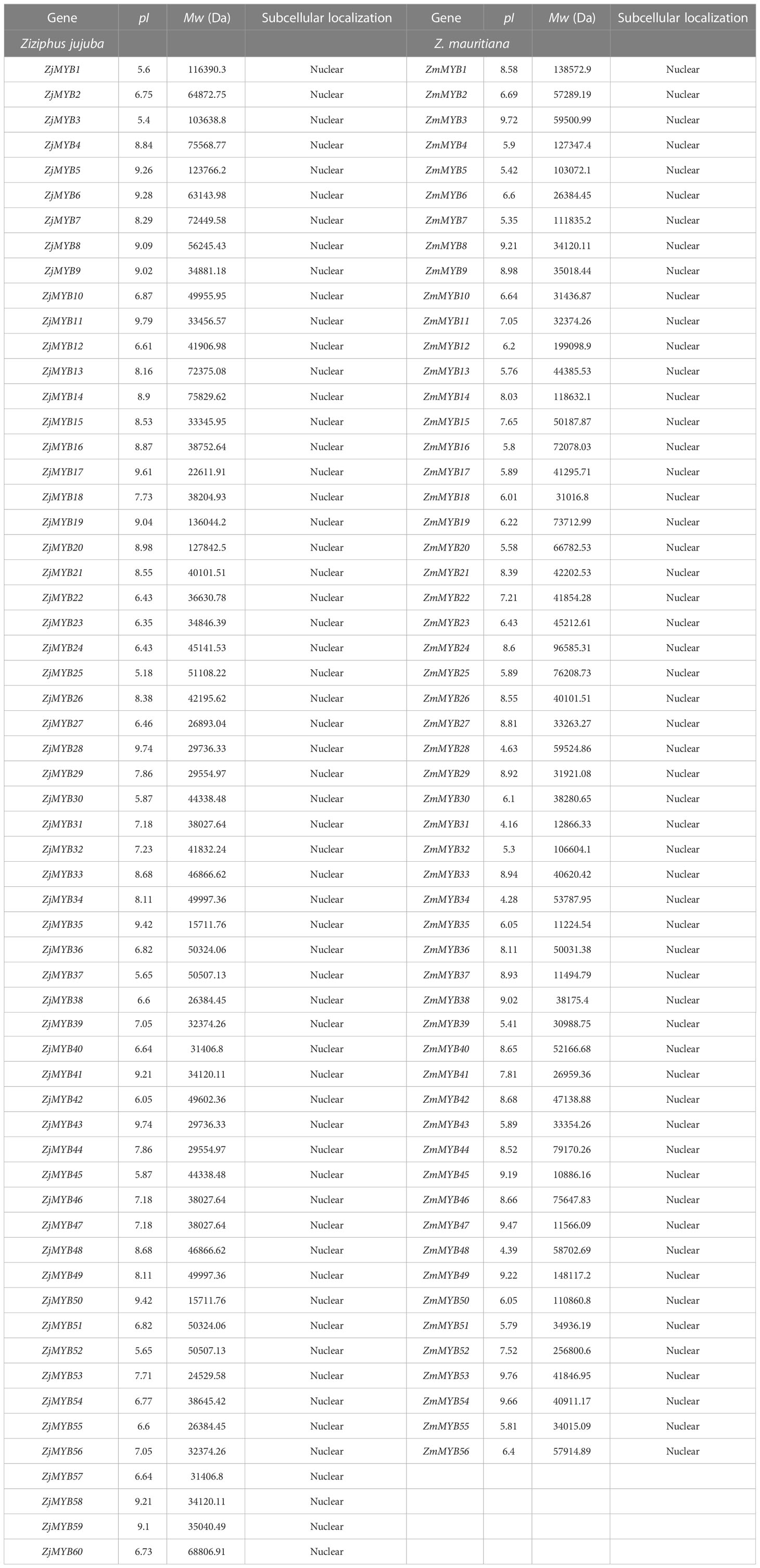
3.3 Subcellular localization prediction analysis
The 60 Z. jujuba and 56 Z. mauritiana isolated MYB protein sequences from the transcriptome were designated as ZjMYB1 through ZjMYB60 and ZmMYB1 through ZmMYB56, respectively. The 60 ZjMYB and 56 ZmMYB proteins were all predicted to be localized in the nucleus as shown in Table 1.
3.4 Expression patterns of MYB genes from transcriptome of Z. mauritiana and Z. jujuba involved in flavonoid biosynthesis/fruit coloration
The concentration levels of plant flavonoid pigments play a significant role in fruit color change. Our present transcriptomic study of MYB unveiled the important potential ZmMYB/ZjMYB genes, i.e., ZmMYB/ZjMYB13, ZmMYB/ZjMYB44, ZmMYB/ZjMYB50, and ZmMYB/ZjMYB56, involved in the flavonoid biosynthesis during the fruit developmental stages or during the period of fruit color change in Z. mauritiana (green color fruit) and Z. jujuba (red color fruit). The four important potential MYB genes (ZmMYB/ZjMYB13, ZmMYB/ZjMYB44, ZmMYB/ZjMYB50, and ZmMYB/ZjMYB56) were dramatically down-regulated or with low expression in Z. mauritiana, which is probably concomitant with the lack of flavonoid color pigments content in green fruit cultivars of Z. mauritiana. The ZmMYB44 was significantly downregulated in fruit developmental stages of Z. mauritiana while this gene (ZjMYB44) was upregulated in fruit developmental stages of Z. jujuba as shown in Figure 4. Moreover, in Z. mauritiana, the expression level of this gene was highest in the junior fruit (JF) developmental stage (Figure 4), whereas in Z. jujuba, the high expression level was observed in the full ripening fruit (RF) stage (Figure 4). Similarly, the high expression of ZmMYB56 was found in JF (Junior fruit) and BWRF (Before white ripening fruit) stages and then decreased the expression in WRF (White ripening fruit), HRF (Half ripening fruit), and RF (Ripen fruit) stages of fruit development in Z. mauritiana. Further, the expression of ZmMYB/ZjMYB13 and ZmMYB/ZjMYB50 genes were downregulated in Z. mauritiana fruit developmental stages and was upregulated in Z. jujuba fruit as shown in Figure 4. The rest of the genes showed no significant expression in the fruit developmental stages of both Z. mauritiana and Z. jujuba. Thus these four ZmMYB/ZjMYB genes of Z. mauritiana and Z. jujuba may be the possible key genes for the fruit color change in Ziziphus Mill.
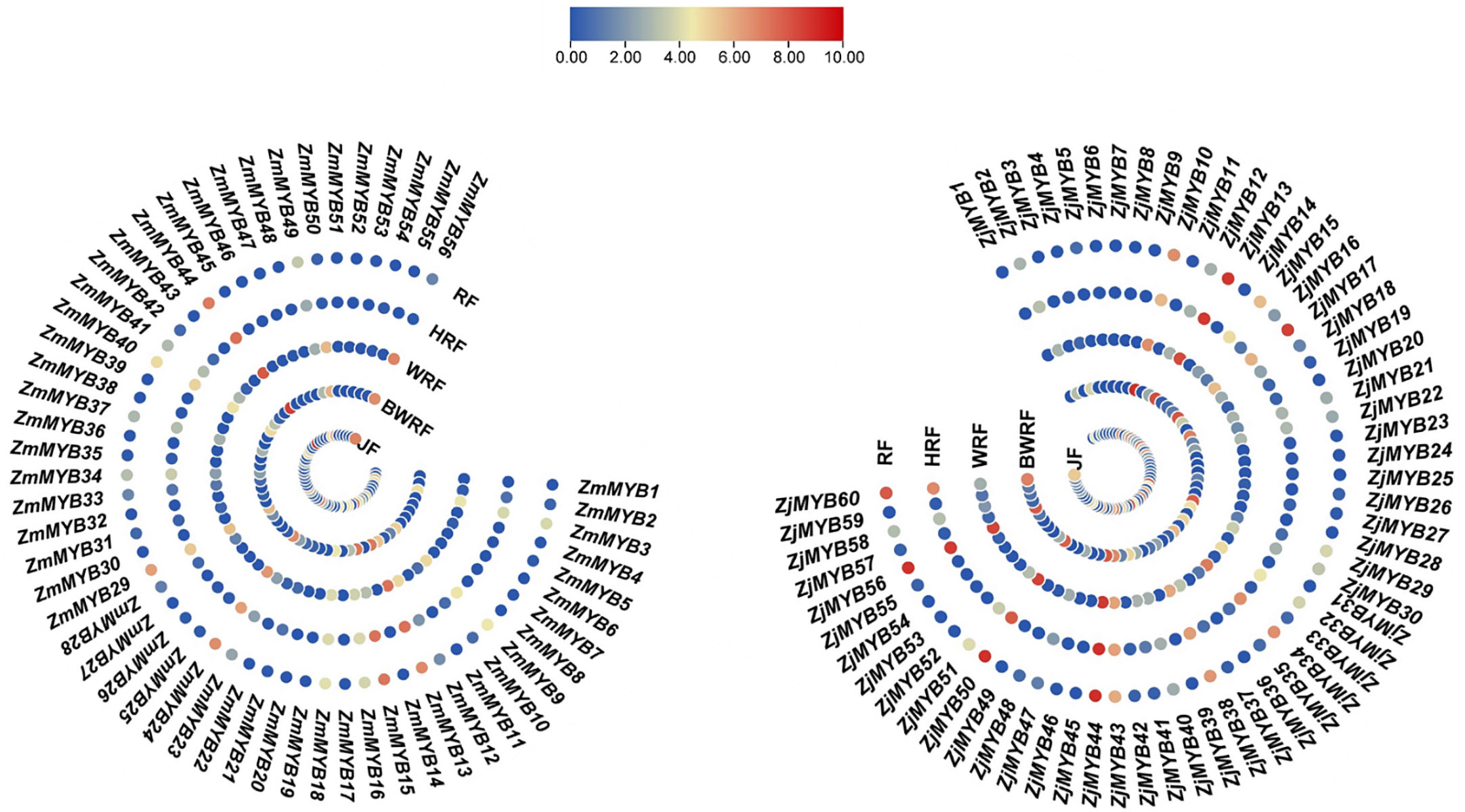
Figure 4 Differential expression of ZmMYB and ZjMYB genes in different developmental stages of Ziziphus mauritiana and Z. jujuba fruit. The Non-significance is blue and the Significant is shown as red in color. The ZmMYB gene that has been up-regulated is red, while the down-regulated is shown as blue in this figure. JF, BWRF, WRF, HRF, and RF represent junior fruit, before white ripening, white ripening, half ripening, and ripen fruit developmental stages, respectively.
3.5 qRT-PCR expression analysis
To validate the possible role of the 4 key MYB genes obtained from transcriptome data of Z. mauritiana and Z. jujuba, we conducted the qRT-PCR expression analysis. The expression of ZmMYB13 was high at the junior fruit developmental stage of Z. mauritania but gradually decreased its expression at BWRF (before white ripening), WRF (white ripening), HRF (half ripening), RF (ripe fruit) stages of fruit development. While the expression of this gene (ZjMYB13) was high and upregulated in the fruit developmental stages of Z. jujuba except for WRF (white ripening) stage. Similarly, the expression of ZmMYB44 was significantly downregulated in the fruit developmental stages of Z. mauritiana while the same gene ZjMYB44 was significantly upregulated in the fruit developmental stages of Z. jujuba, confirming its expression level presented by the transcriptomic data. Similarly, ZmMYB/ZjMYB56 presented downregulated trend in Z. mauritiana and upregulated trend in Z. jujuba. Moreover, the expression pattern of ZmMYB/ZjMYB50 was not significant at all as shown in Figure 5.
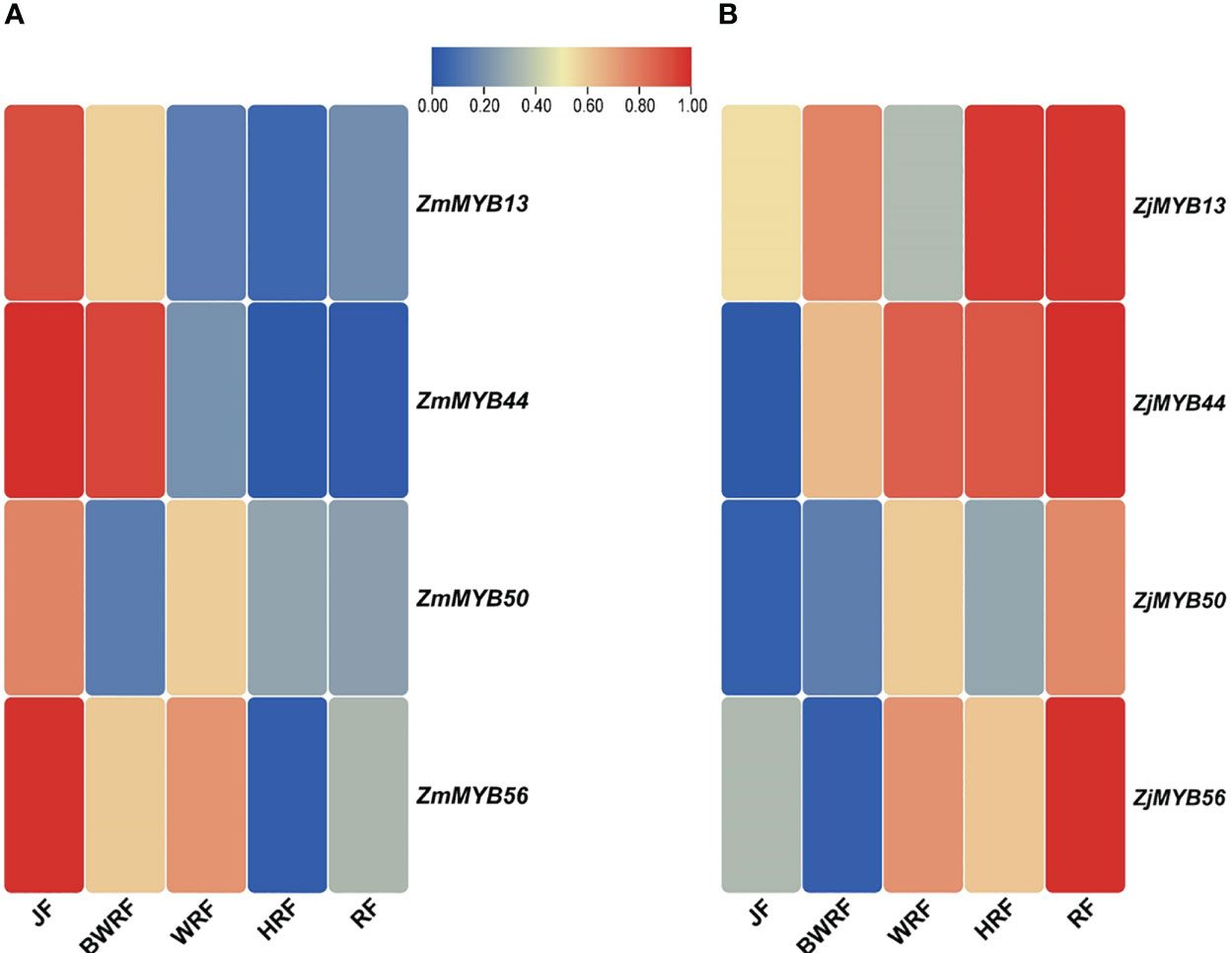
Figure 5 Validation of differential expression of the MYB genes (ZmMYB13, ZmMYB44, ZmMYB50, and ZmMYB56 and ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56) among five stages of fruit development of Z. mauritiana and Z. jujuba using qRT-PCR. The red color represents the high expression of the MYB genes and the blue color represents the low-level expression in corresponding fruit developmental stages. The (A) represents the expression level of MYB (ZmMYB13, ZmMYB44, ZmMYB50, and ZmMYB56) genes in Z. mauritiana while (B) represents the expression level of the same MYB (ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56) genes in Z. jujuba. The JF stands for (Junior fruit), BWRF stands for (Before white ripening stage), WRF represents (White ripening stage), HRF stands for (Half ripening stage), and RF represents (ripen stage) of fruit development.
3.6 Transient expression assay and flavonoid determination
To further validate the transcriptome and qRT-PCR-based expression analysis results of the selected MYB genes (ZmMYB13, ZmMYB44, ZmMYB50, and ZmMYB56 and ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56), we performed in vivo transient Agrobacterium infiltration assays on jujube fruit. We injected the plasmids ZjMYB44-pCambia1302, ZjMYB50-pCambia1302, ZjMYB56-pCambia1302, and ZjMYB13- pCambia1302 into the pericarps of jujube fruits. Compare to the control, the high transient expression of ZjMYB44 significantly increased flavonoid accumulation in the fruit around the injection site (Figure 6A). Flavonoid biosynthetic ZjMYB44 gene was consistently up-regulated in the fruit near the injection site. Thus, high transient expression of the ZjMYB44 gene in fruit increased flavonoid accumulation, indicating that this gene is a key influencer of flavonoid content during the period of fruit color change in Z. jujuba as shown in Figure 6A. On the other hand, ZjMYB50 gene was transiently expressed in fruit only at 96h after the injection with a constant decrease of flavonoid content, indicating that this gene cannot improve the flavonoid content accumulation (Figure 6B). Furthermore, ZjMYB56 was transiently expressed in fruit, but the gene was not highly expressed in fruit and the flavonoid content was decreased, thus indicating that this gene cannot affect the flavonoid accumulation in the fruit peel (Figure 6C). Moreover, the ZjMYB13 gene was transiently expressed in the fruit, the expression of ZjMYB13 gene was high in the fruit, and the flavonoid content first decreased and then rose after 72h, demonstrating that this gene might indirectly induce the flavonoid content in the fruit peel of jujube (Figure 6D).
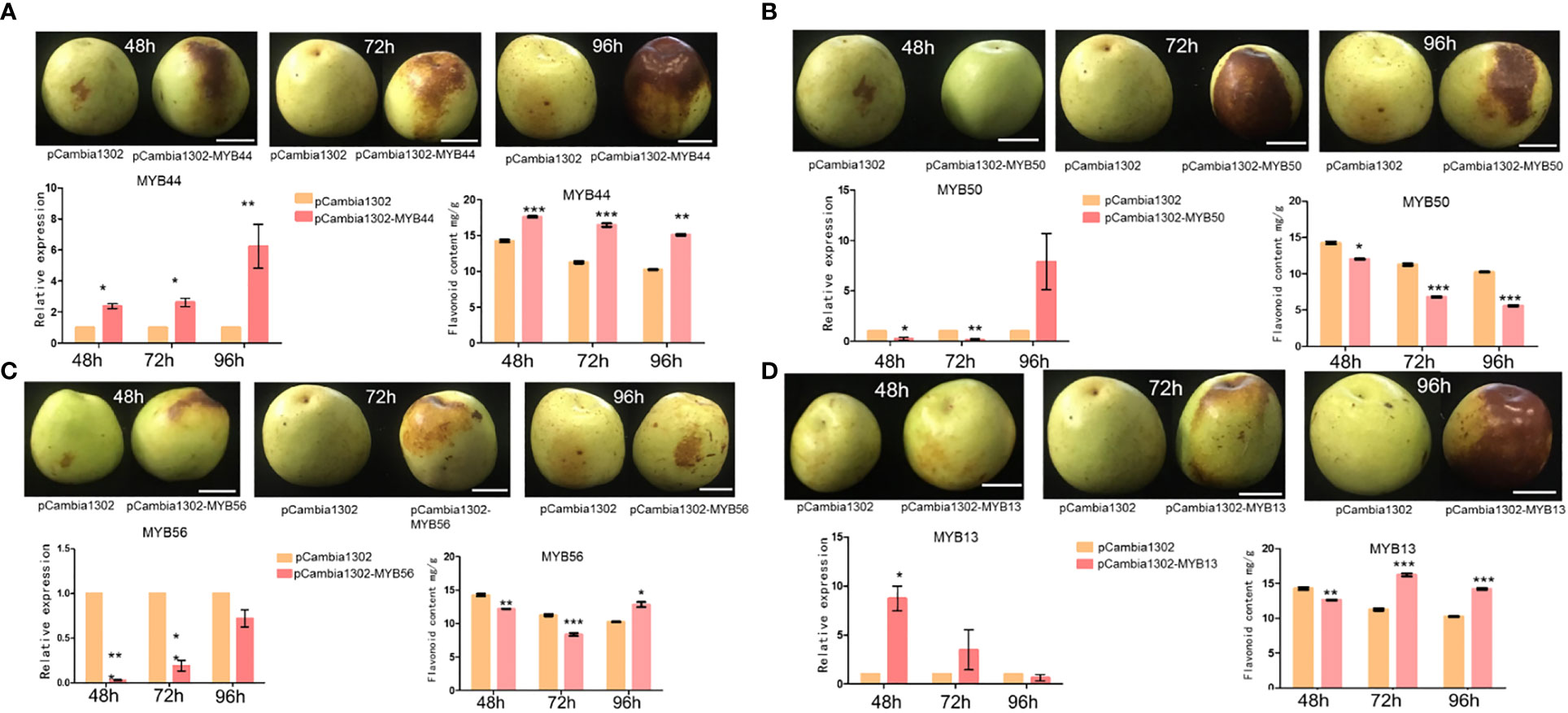
Figure 6 The transient expression of 4 MYB genes (ZjMYB13, ZjMYB44, ZjMYB50, and ZjMYB56) in jujube fruit. pCambia1302 was used as an empty vector. (A) The phenotype of pCambia1302, pCambia1302-MYB44, expression and flavonoid content of pCambia1302, pCambia1302-MYB44 of Z. jujuba fruits. (B) The phenotype of pCambia1302, pCambia1302-MYB50, expression and flavonoid content of pCambia1302, pCambia1302 MYB50 of Z. jujuba fruits. (C) The phenotype of pCambia-1302, pCambia1302-MYB56, expression and flavonoid content of pCambia-1302, pCambia1302-MYB56 of Z. jujuba fruits. (D) The phenotype of pCambia-1302, pCambia1302-MYB13, relative expression level and flavonoid content of pCambia1302, pCambia1302-MYB13 of Z. jujuba fruits.
4 Discussion
The MYB TFs are engaged in physiological and biochemical mechanisms in plants, as well as responses to abiotic and biotic stresses (Raffaele et al., 2008; Qing et al., 2019), and the function of MYB has been extensively studied. MYB TFs are also engaged in flavonoid biosynthesis. For example, overexpression of the Arabidopsis AtMYB12 gene increased flavonoid content accumulation under low temperatures in a light-dependent fashion (Bhatia et al., 2018). The MYB10 gene in Fragaria ananassa plays a general regulatory role in the flavonoid/phenylpropanoid pathway during fruit ripening (Medina-Puche et al., 2014; Qing et al., 2019).
The MYB is the largest TF gene family in plants that is divided into four subfamilies based on the number of MYB imperfect tandem repeats (Rs) in the proteins: R3 MYB (MYB1R) has one repeat, R2R3 MYB has 2 repeats, R1R2R3 MYB (MYB3R) has 3 repeats, and 4R MYB has 4 repeats (Lin-Wang et al., 2010). Only R2R3-MYB plays a role in flavonoid or anthocyanin biosynthesis, with 126 genes discovered in A. thaliana (Stracke et al., 2001; Yanhui et al., 2006). In a genome-wide characterization and expression analysis of the MYB superfamily genes in jujube Qing et al. (2019) discovered 99 R2R3-MYB genes and 171 MYB genes in total. Due to the lack of genomic information on Z. mauritiana, we used transcriptome-wide analysis of the fruit peel and identified 60 MYBs in jujube (Z. jujuba) and 56 MYBs in Ber (Z. msuritiana). The number of MYBs decreased in jujube, this may be due to the fact that the genome contains all of the genes in a cell’s DNA (Hamilton and Robin Buell, 2012; Chen et al., 2019), whereas the transcriptome only contains those that are expressed within a specific tissue of the plant species (Ward et al., 2012).
The evolution of common ancestors for plant species and subsequent biological evolution research showed that orthologous genes could have contributed to the evolutionary developments of inert, and novel active genes (Fan et al., 2020). Orthologs are genes in different genomes that originated from the splitting of taxonomic lineages, whereas paralogs are genes in the same genome that originated from gene duplication (Thornton and DeSalle, 2000; Song et al., 2016). Paralogs typically have different functions, whilst orthologs may have the same function (Tatusov et al., 1997; Song et al., 2016). Our current comparative and phylogenetic analysis revealed 13 pairs of putative orthologous proteins between Z. jujuba (ZjMYBs) and Z. mauritiana (ZmMYBs). In contrast, 10 pairs of paralogous MYB family proteins were identified in Z. jujuba. Similarly, 2 pairs of paralogous MYB family proteins were also identified in Z. mauritiana (Figure 1). MEME was used to identify conserved motifs in both species’ MYB proteins.
A number of flavonoid-related MYB TFs have already been discovered in plants that regulate secondary metabolism in the plant species (Chezem et al., 2016; Wang et al., 2017). For example, the TT2, the first PA-related MYB TF discovered, activates DFR, ANS, and ANR, causing proanthocyanidin biosynthesis in the seed coats of A. thaliana (Nesi et al., 2001). The unique regulators (VvMYBA1 and VvMYBA2), activate the UFGT genes of the anthocyanin pathway in grapevines (Kobayashi et al., 2002). Moreover, in grapevines, VvMYBPA2 was discovered to be a direct regulator of numerous structural flavonoid pathway genes (Terrier et al., 2009). The MdMYB10 is an important regulator during apple fruit coloring (Espley et al., 2007). In apple fruits, MdMYB3 triggers flavonoid biosynthesis-related genes such as CHI, CHS, FLS, and UFGT (Wang et al., 2017). Similarly, the FaMYB10 controls anthocyanin pathway-related genes in strawberries, such as the majority of EBGs and LBGs in matured fruit receptacles during ripening (Medina-Puche et al., 2014). The MYB genes found in H. brasiliensis, HbMYB1 overexpression in tobacco reduces stress-induced cell death (Peng et al., 2011; Wang et al., 2017). Furthermore, PeMYB2, PeMYB11, and PeMYB12 have all been found to be involved in the development of red color in Phalaenopsis spp. flowers (Hsu et al., 2015).
The flavonoids like anthocyanin structural genes control plant pigmentation both temporally and spatially. The transcriptional level is primarily responsible for the regulation of structural genes (Li et al., 2017; Li et al., 2017a). R2R3-MYBs regulate anthocyanin transcription by influencing the transcripts of anthocyanin structural genes. Numerous anthocyanin R2R3-MYB TFs in various fruit plants have been isolated and categorized (Feng et al., 2015). A homolog of PyMYB10, PyMYB10.1, was cloned from ‘Aoguan’ (Feng et al., 2015). The transcriptome analysis on ‘Hongyang’ kiwifruit, revealed 9 R2R3 MYBs to be involved in anthocyanin synthesis during fruit development (Li et al., 2015). The MYB5 and MYBA have been shown to be positive potent inducers in the early stages of development of ‘Hongyang’ fruit (7 days after anthesis, DAA), where kiwifruit undergo a temporary anthocyanin formation, as well as later during fruit ripening (Li et al., 2015; Li et al., 2017; Li et al., 2017a). Some findings suggested that VvMYB5a and VvMYB5b in grapes, as well as MdMYBA in apples, could be stimulated by low temperatures, which then stimulated the expression of ANS, resulting in flavonoids like anthocyanin formation in fruit (Li et al., 2017).
The recognition of key MYB TFs associated with flavonoid accumulation during the period of fruit coloration in various species has greatly improved our understanding of the control mechanism of flavonoid biosynthesis in fruits. Based on our present transcriptome expression analysis four MYB genes (ZmMYB/ZjMYB13, ZmMYB/ZjMYB44, ZmMYB/ZjMYB50, and ZmMYB/ZjMYB56), were dramatically down-regulated or with low expression in Z. mauritiana green color fruit cultivar, which is probably concomitant with the lack of flavonoid color pigments content in green fruit cultivars of Z. mauritiana. For example, the ZmMYB44 was significantly downregulated in Z. mauritiana fruit developmental stages while this gene (ZjMYB44) was upregulated in fruit developmental stages of Z. jujuba as shown in Figure 4. Furthermore, the expression of this gene was high in the junior fruit (JF) developmental stage in Z. mauritiana (Figure 4) while the highest expression was found in the full ripening fruit (RF) stage in Z. jujuba (Figure 4). Similarly, the high expression of ZmMYB56 was found in JF (Junior fruit) and BWRF (Before white ripening fruit) stages and then decreased the expression in WRF (White ripening fruit), HRF (Half ripening fruit), and RF (Ripe fruit) stages of fruit development in Z. mauritiana. Further, the expression of ZmMYB/ZjMYB13 and ZmMYB/ZjMYB50 genes were downregulated in Z. mauritiana fruit developmental stages and was upregulated in Z. jujuba fruit as presented earlier in Figure 4. Moreover, the transient overexpression of AcMYB5-1/5-2/A1-1 in N. benthamiana leaves increased expression patterns of NtANS and NtDFR, suggesting that those MYB TFs may be involved in the flavonoid synthesis pathway (Li et al., 2017). Furthermore, the total flavonoid content of jujube decreased as the fruit ripened (Wu et al., 2012), indicating the low expression of corresponding genes. Since the transient expression assay in Z. jujuba fruit, ZjMYB44 significantly increased flavonoid accumulation indicating that this gene can stimulate the flavonoid content accumulation in the jujube fruit as described in Figure 6A.
5 Conclusion
In this study, we conducted the first transcriptome-wide analysis of Z. mauritiana and Z. jujuba MYB superfamily genes, which included 56 ZmMYB and 60 ZjMYB transcription factors (TFs) genes, in Z. mauritiana and Z. jujuba, respectively. The phylogenetic analysis revealed 13 pairs of putative orthologous proteins between Z. jujuba (ZjMYBs) and Z. mauritiana (ZmMYBs). In contrast, 10 pairs of paralogous MYB family proteins were identified in Z. jujuba. Similarly, 2 pairs of paralogous MYB family proteins were identified in Z. mauritiana. Four similar MYB genes (ZmMYB/ZjMYB13, ZmMYB/ZjMYB44, ZmMYB/ZjMYB50, and ZmMYB/ZjMYB56) from Z. mauritiana and Z. jujuba were identified as candidate key regulating flavonoid biosynthesis genes through transcriptomic expression analysis. Moreover, in vivo transient expression of ZjMYB44 significantly increased flavonoid accumulation around the injection site, and the ZjMYB44 gene was also transiently highly expressed in fruit, indicating that this gene can influence flavonoid content and lead to coloration in Ziziphus fruit. Our research results provide an important understanding of the molecular mechanism of flavonoid biosynthesis resulting in fruit coloration in Ziziphus, laying the groundwork for further functional characterization and genetic improvement of Ziziphus (Z. mauritiana and Z. jujuba) fruit coloration.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
NM wrote this article, NM and ZLuo did the laboratory research work. XZ helped in the laboratory research work. MY, ZLiu, and ML critically revised this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Ten Thousand Talents Plan Leadership Project and Beijing Municipal Education Commission (CEFF-PXM2019_014207_000099), National Key R&D Program of China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1171288/full#supplementary-material
References
Abbas, F., Ke, Y., Zhou, Y., Yu, Y., Waseem, M., Ashraf, U., et al. (2021). Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 12, 623742. doi: 10.3389/fpls.2021.623742
Aharoni, A., De Vos, C. R., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28 (3), 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Bailey, T. L., Bodén, M., Buske, F. A., Frith, M. C., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bhatia, C., Pandey, A., Gaddam, S. R., Hoecker, U., Trivedi, P. K. (2018). Low temperature-enhanced flavonol synthesis requires light-associated regulatory components in arabidopsis thaliana. Plant Cell Physiol. 59 (10), 2099–2112. doi: 10.1093/pcp/pcy132
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Sci. 12, 691384. doi: 10.3389/fpls.2021.691384
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Song, Y., Li, X., Chen, J., Mo, L., Zhang, X., et al. (2019). Genome sequences of horticultural plants: past, present, and future. Hort Res. 6, 1–23. doi: 10.1038/s41438-019-0195-6
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31 (13), 3497–3500. doi: 10.1093/nar/gkg500
Chezem, W. R., Nicole, K., Clay, N. K. (2016). Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochem. 131, 26–43. doi: 10.1016/j.phytochem.2016.08.006
Cui, X., Wang, Y. X., Liu, Z. W., Wang, W. L., Li, H., Zhuang, J. (2018). Transcriptome-wide identification and expression profile analysis of the bHLH family genes in Camellia sinensis. Funct. Integr. Genomics 18, 489–503. doi: 10.1007/s10142-018-0608-x
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49 (3), 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Fan, H., Cui, M., Li, N., Li, X., Liang, Y., Liu, L., et al. (2020). Genome-wide identification and expression analyses of R2R3-MYB transcription factor genes from two orchid species. PeerJ 8, e9781. doi: 10.7717/peerj.9781
Feng, S., Sun, S., Chen, X., Wu, S., Wang, D., Chen, X. (2015). PyMYB10 and PyMYB10. 1 interact with bHLH to enhance anthocyanin accumulation in pears. PloS One 10 (11), e0142112. doi: 10.1371/journal.pone.0142112
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 (D1), D279–D285. doi: 10.1093/nar/gkv1344
Hamilton, J. P., Robin Buell, C. (2012). Advances in plant genome sequencing. Plant J. 70 (1), 177–190. doi: 10.1111/j.1365-313X.2012.04894.x
Hsu, C. C., Chen, Y. Y., Tsai, W. C., Chen, W. H., Chen, H. H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in phalaenopsis orchids. Plant Physiol. 114-254599, 175–191. doi: 10.1104/pp.114.254599
Jiang, S., Chen, M., He, N., Chen, X., Wang, N., Sun, Q., et al. (2019). MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hort Res. 6, 1–14. doi: 10.1038/s41438-019-0118-6
Kobayashi, S., Ishimaru, M., Hiraoka, K., Honda, C. (2002). Myb-related genes of the kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta. 215, 924–933. doi: 10.1007/s00425-002-0830-5
Larkin, M. A., Blackshields, G., Brown, N., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal wand clustal X version 2.0. Bioinformatics. 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, W., Ding, Z., Ruan, M., Yu, X., Peng, M., Liu, Y. (2017). Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 7 (1), 16861. doi: 10.1038/s41598-017-16905-1
Li, J., Lin, K., Zhang, S., Wu, J., Fang, Y., Wang, Y. (2021a). Genome-wide analysis of myeloblastosis-related genes in Brassica napus l. and positive modulation of osmotic tolerance by BnMRD107. Front. Plant Sci. 12, 678202. doi: 10.3389/fpls.2021.678202
Li, Y., Lin-Wang, K., Liu, Z., Allan, A. C., Qin, S., Zhang, J., et al. (2020). Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum l.). Int. J. Biol. Macromol 148, 817–832. doi: 10.1016/j.ijbiomac.2020.01.167
Li, J., Liu, H., Yang, C., Wang, J., Yan, G., Si, P., et al. (2020a). Genome-wide identification of MYB genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus l. Ind. Crops Prod. 143, 111924. doi: 10.1016/j.indcrop.2019.111924
Li, W., Liu, Y., Zeng, S., Xiao, G., Wang, G., Wang, Y., et al. (2015). Gene expression profiling of development and anthocyanin accumulation in kiwifruit (Actinidia chinensis) based on transcriptome sequencing. PloS One 10 (8), e0136439. doi: 10.1371/journal.pone.0136439
Li, B., Xia, Y., Wang, Y., Qin, G., Tian, S. (2017a). Characterization of genes encoding key enzymes involved in anthocyanin metabolism of kiwifruit during storage period. Front. Plant Sci. 8, 341. doi: 10.3389/fpls.2017.00341
Li, H., Zhou, Y., Ma, Z., Lu, X., Li, Y., Chen, H. (2021). Genome-wide identification and expression analysis of myb transcription factors and their responses to abiotic stresses in woodland strawberry (Fragaria vesca). Horticulturae. 7 (5), 97. doi: 10.3390/horticulturae7050097
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., McGhie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in rosaceae. BMC Plant Biol. 10 (1), 1–17. doi: 10.1186/1471-2229-10-50
Liu, M. J., Liu, P., Liu, G. N. (2011). Advances of research on germplasm resources of Chinese jujube. Acta Hortic. 993, 15–20. doi: 10.17660/ActaHortic.2013.993.1
Liu, M. J., Wang, M. (2009). Chinese Jujube germplasm resources (Beijing: China Forestry Publishing House), 46–47.
Liu, M., Wang, J., Wang, L., Liu, P., Zhao, J., Zhao, Z., et al. (2020). The historical and current research progress on jujube–a superfruit for the future. Hortic. Res. 7, 1–17. doi: 10.1038/s41438-020-00346-5
Liu, J., Wang, J., Wang, M., Zhao, J., Zheng, Y., Zhang, T., et al. (2021a). Genome-wide analysis of the R2R3-MYB gene family in Fragaria× ananassa and its function identification during anthocyanins biosynthesis in pink-flowered strawberry. Front. Plant Sci. 12, 702160. doi: 10.3389/fpls.2021.702160
Liu, Y., Wu, Y., Wang, Z., Zhang, S., Liu, X., Tikunov, Y., et al. (2021). Genome-wide characterization of the R2R3-MYB transcription factors in pepper (Capsicum spp.) unveils the role of CaMYB101 as repressor in anthocyanin biosynthesis. bioRxiv, 2021–2008. doi: 10.1101/2021.08.24.457473
Liu, C., Xie, T., Chen, C., Luan, A., Long, J., Li, C., et al. (2017). Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 18 (1), 1–16. doi: 10.1186/s12864-017-3896-y
Liu, M. J., Zhao, J., Cai, Q. L., Liu, G. C., Wang, J. R., Zhao, Z. H., et al. (2014). The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 5 (1), 5315. doi: 10.1038/ncomms6315
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodríguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of fragaria× ananassa fruits. J. Exp. Bot. 65 (2), 401–417. doi: 10.1093/jxb/ert377
Muhammad, N., Luo, Z., Yang, M., Li, X., Liu, Z., Liu, M. (2022). The joint role of the late anthocyanin biosynthetic UFGT-encoding genes in the flowers and fruits coloration of horticultural plants. Sci. Hortic. 301, 111110. doi: 10.1016/j.scienta.2022.111110
Muhammad, N., Luo, Z., Yang, M., Liu, Z., Liu, M. (2022b). The nutritional, medicinal, and drought-resistance properties of ziziphus mill. make it an important candidate for alleviating food insecurity in arid regions–a case of Pakistan. Horticulturae. 8 (10), 867. doi: 10.3390/horticulturae8100867
Muhammad, N., Luo, Z., Yang, M., Liu, Z., Liu, M. (2023). The underlying molecular mechanisms of external factors influencing fruit coloration in fruit trees. Scientia Hortic. 309, 111615. doi: 10.1016/j.scienta.2022.111615
Muhammad, N., Uddin, N., Khan, M. K. U., Ali, N., Ali, K., Jones, D. A. (2022a). Diverse role of basic helix-Loop-Helix (bHLH) transcription factor superfamily genes in the fleshy fruit-bearing plant species. Czech J. Genet. Plant Breed. 59 (1), 1–13. doi: 10.17221/2/2022-CJGPB
Muhammad, N., Uddin, N., Khan, M. K. U., Mengjun, L., Xuan, Z., Ali, N., et al. (2020). Ethnomedicinal and cultural uses of ziziphus species in flora of malakand division KP, Pakistan. Singapore J. Sci. Res. 10, 1–7. doi: 10.3923/sjsres.2020.1.7
Muhammad, N., Zhao, Z. H., Yang, M., Liu, M. J. (2022c). Current status of ziziphus species: their distribution, cultivation, and production in pakistan. SHS acta horticulturae 1350 V international jujube symposium. Acta Hortic. 1350, 23–34. doi: 10.17660/ActaHortic.2022.1350.4
Muthamilarasan, M., Khandelwal, R., Yadav, C. B., Bonthala, V. S., Khan, Y., Prasad, M. (2014). Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica l.). PloS One 9 (10), e109920. doi: 10.1371/journal.pone.0109920
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., Lepiniec, L. (2001). The arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 13, 2099–2114. doi: 10.1105/TPC.010098
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., Saedler, H. (1987). The regulatory c1 locus of zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6 (12), 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Peng, S. Q., Wu, K. X., Huang, G. X., Chen, S. C. (2011). HbMyb1, a myb transcription factor from hevea brasiliensis, suppresses stress induced cell death in transgenic tobacco. Plant Physiol. Biochem. 49 (12), 1429–1435. doi: 10.1016/j.plaphy.2011.09.007
Pu, X., Yang, L., Liu, L., Dong, X., Chen, S., Chen, Z., et al. (2020). Genome-wide analysis of the MYB transcription factor superfamily in physcomitrella patens. Int. J. Mol. Sci. 21 (3), 975. doi: 10.3390/ijms21030975
Qing, J., Dawei, W., Jun, Z., Yulan, X., Bingqi, S., Fan, Z. (2019). Genome-wide characterization and expression analyses of the MYB superfamily genes during developmental stages in Chinese jujube. PeerJ. 7, e6353. doi: 10.7717/peerj.6353
Raffaele, S., Vailleau, F., Leger, A., Joubès, J., Miersch, O., Huard, C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in arabidopsis. Plant Cell. 20 (3), 752–767. doi: 10.1105/tpc.107.054858
Shi, Q., Du, J., Zhu, D., Li, X., Li, X. (2020). Metabolomic and transcriptomic analyses of anthocyanin biosynthesis mechanisms in the color mutant ziziphus jujuba cv. tailihong. J. Agric. Food Chem. 68, 15186–15198. doi: 10.1021/acs.jafc.0c05334
Song, A., Gao, T., Li, P., Chen, S., Guan, Z., Wu, D., et al. (2016). Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 7, 199. doi: 10.3389/fpls.2016.00199
Song, X., Yang, Q., Liu, Y., Li, J., Chang, X., Xian, L., et al. (2021). Genome-wide identification of pistacia R2R3-MYB gene family and function characterization of PcMYB113 during autumn leaf coloration in pistacia chinensis. Int. J. Biol. Macromol 192, 16–27. doi: 10.1016/j.ijbiomac.2021.09.092
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the arabidopsis thaliana seedling. Plant J. 50 (4), 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 (5), 447–456. doi: 10.1016/S1369-5266(00)00199-0
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 (12), 2725–2729. doi: 10.1093/molbev/mst197
Tatusov, R. L., Koonin, E. V., Lipman, D. J. (1997). Agenomic perspective on protein families. Science 278, 631–637. doi: 10.1126/science.278.5338.631
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verries, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Thornton, J. W., DeSalle, R. (2000). Gene family evolution and homology: genomics meets phylogenetics. Annu. Rev. Genomics Hum.Genet 1, 41–73. doi: 10.1146/annurev.genom.1.1.41
Uddin, N., Ali, N., Nisar, M., Liu, M., Liu, Z., Muhammad, N., et al. (2021). SSR-based population structure, molecular diversity and identity cards of ziziphus species from Pakistan and China. Genet. Resour. Crop Evol. 68, 2391–2409. doi: 10.1007/s10722-021-01138-z
Uddin, N., Muhammad, N., Ali, N., Nisar, M., Liu, M. (2022). Genomic constitution and relationships of ziziphus species collected from malakand division, Pakistan. Physiol. Mol. Biol. Plants 28, 1939–1953. doi: 10.1007/s12298-022-01249-6
Uddin, N., Muhammad, N., Nisar, M., Ali, N., Ullah, R., Ali, E. A., et al. (2022a). Distribution of polyphenolic compounds, antioxidant potential, and free amino acids in ziziphus fruits extract; a study for determining the influence of wider geography. Food Sci. Nutr. 10 (5), 1414–1430. doi: 10.1002/fsn3.2726
Wang, A., Liang, K., Yang, S., Cao, Y., Wang, L., Zhang, M., et al. (2021a). Genome-wide analysis of MYB transcription factors of Vaccinium corymbosum and their positive responses to drought stress. BMC Genom. 22 (1), 1–17. doi: 10.1186/s12864-021-07850-5
Wang, Y., Xu, Y. E., Ma, X., Liu, X., Yang, M., Fan, W., et al. (2018). Extraction, purification, characterization and antioxidant activities of polysaccharides from zizyphus jujuba cv. linzexiaozao. Int. J. Biol. Macromol 118, 2138–2148. doi: 10.1016/j.ijbiomac.2018.07.059
Wang, Y., Zhan, D. F., Li, H. L., Guo, D., Zhu, J. H., Peng, S. Q. (2017). Transcriptome-wide identification and characterization of MYB transcription factor genes in the laticifer cells of Hevea brasiliensis. Front. Plant Sci. 8, 1974. doi: 10.3389/fpls.2017.01974
Wang, Y., Zhang, Y., Fan, C., Wei, Y., Meng, J., Li, Z., et al. (2021). Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 21 (1), 1–17. doi: 10.1186/s12870-021-03083-6
Ward, J. A., Ponnala, L., Weber, C. A. (2012). Strategies for transcriptome analysis in nonmodel plants. Am. J. Bot. 99 (2), 267–276. doi: 10.3732/ajb.1100334
Wu, C. S., Gao, Q. H., Guo, X. D., Yu, J. G., Wang, M. (2012). Effect of ripening stage on physicochemical properties and antioxidant profiles of a promising table fruit ‘pear-jujube’(Zizyphus jujuba mill.). Sci. Hortic. 148, 177–184. doi: 10.1016/j.scienta.2012.09.026
Xie, F., Hua, Q., Chen, C., Zhang, Z., Zhang, R., Zhao, J., et al. (2021). Genome-wide characterization of R2R3-MYB transcription factors in pitaya reveals a R2R3-MYB repressor HuMYB1 involved in fruit ripening through regulation of betalain biosynthesis by repressing betalain biosynthesis-related genes. Cells. 10 (8), 1949. doi: 10.3390/cells10081949
Yang, Z. P., Li, H. L., Guo, D., Peng, S. Q. (2015). Identification and characterization of the EIN3/EIL gene family in hevea brasiliensis. Tree Genet. Genomes. 11, 1–11. doi: 10.1007/s11295-015-0877-7
Yang, L., Liu, H., Hao, Z., Zong, Y., Xia, H., Shen, Y., et al. (2021). Genome-wide identification and expression analysis of r2r3-myb family genes associated with petal pigment synthesis in liriodendron. Int. J. Mol. Sci. 22 (20), 11291. doi: 10.3390/ijms222011291
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB transcription factor superfamily of arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Zhang, T., Cui, Z., Li, Y., Kang, Y., Song, X., Wang, J., et al. (2021). Genome-wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front. Genet. 12, 714696. doi: 10.3389/fgene.2021.714696
Zhang, C., Huang, J., Li, X. (2015). Identification of appropriate reference genes for RT-qPCR analysis in Ziziphus jujuba mill. Sci. Hortic. 197, 166–169. doi: 10.1016/j.scienta.2015.09.026
Zhang, C., Ma, R., Xu, J., Yan, J., Guo, L., Song, J., et al. (2018). Genome-wide identification and classification of MYB superfamily genes in peach. PloS One 13 (6), e0199192. doi: 10.1371/journal.pone.0199192
Zhang, Q., Wang, L., Liu, Z., Zhao, Z., Zhao, J., Wang, Z., et al. (2020). Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba mill. peel coloration. Food Chem. 312, 125903. doi: 10.1016/j.foodchem.2019.125903
Zhao, J., Jian, J., Liu, G., Wang, J., Lin, M., Ming, Y., et al. (2014). Rapid SNP discovery and a RAD-based high-density linkage map in jujube (Ziziphus mill.). PloS One 9 (10), e109850. doi: 10.1371/journal.pone.0109850
Keywords: Ziziphus, MYB TFs, transcriptional regulation, flavonoid biosynthesis, transient expression, fruit coloration
Citation: Muhammad N, Luo Z, Zhao X, Yang M, Liu Z and Liu M (2023) Transcriptome-wide expression analysis of MYB gene family leads to functional characterization of flavonoid biosynthesis in fruit coloration of Ziziphus Mill. Front. Plant Sci. 14:1171288. doi: 10.3389/fpls.2023.1171288
Received: 22 February 2023; Accepted: 21 April 2023;
Published: 12 May 2023.
Edited by:
Xitong Fei, Northwest A and F University, ChinaReviewed by:
Lin Zhang, Hubei University of Chinese Medicine, ChinaHaiyan Ren, Shanxi Agricultural University, China
Copyright © 2023 Muhammad, Luo, Zhao, Yang, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiguo Liu, bGl1emhpZ3VvQGhlYmF1LmVkdS5jbg==; Mengjun Liu, bG1qMTIzNDU2N0BhbGl5dW4uY29t
†These authors have contributed equally to this work
 Noor Muhammad
Noor Muhammad Zhi Luo
Zhi Luo Xin Zhao1,2
Xin Zhao1,2 Mengjun Liu
Mengjun Liu