
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 April 2023
Sec. Plant Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1170825
This article is part of the Research Topic Physiological and Molecular Mechanisms of Woody Plants Under Abiotic and Biotic Stresses View all 8 articles
 Manman Zhang1,2
Manman Zhang1,2 Fusheng Wang1,2
Fusheng Wang1,2 Zhou Hu1,2
Zhou Hu1,2 Xiaoli Wang1,2
Xiaoli Wang1,2 Qian Yi1,2
Qian Yi1,2 Jipeng Feng1,2
Jipeng Feng1,2 Xiaochun Zhao1,2*
Xiaochun Zhao1,2* Shiping Zhu1,2*
Shiping Zhu1,2*Response regulator (RR) is an important component of the cytokinin (CK) signal transduction system associated with root development and stress resistance in model plants. However, the function of RR gene and the molecular mechanism on regulating the root development in woody plants such as citrus remain unclear. Here, we demonstrate that CcRR5, a member of the type A RR, regulates the morphogenesis of root through interacting with CcRR14 and CcSnRK2s in citrus. CcRR5 is mainly expressed in root tips and young leaves. The activity of CcRR5 promoter enhanced by CcRR14 was proved with transient expression assay. Seven SnRK2 family members with highly conserved domains were identified in citrus. Among them, CcSnRK2.3, CcSnRK2.6, CcSnRK2.7, and CcSnRK2.8 can interact with CcRR5 and CcRR14. Phenotypic analysis of CcRR5 overexpressed transgenic citrus plants indicated that the transcription level of CcRR5 was associated with root length and lateral root numbers. This was also correlated to the expression of root-related genes and thus confirmed that CcRR5 is involved in the root development. Taken together, the results of this study indicate that CcRR5 is a positive regulator of root growth and CcRR14 directly regulates the expression of CcRR5. Both CcRR5 and CcRR14 can interact with CcSnRK2s.
Root system is an important organ for plant anchorage, support, absorption, and stress resistance (Zhuang et al., 2021). Plants initially perceive various stresses through their roots, modulating root system architecture through cell division, elongation, and differentiation to fight for chances to survive (Gupta et al., 2020). Plant hormones are known to affect root growth and development (Cai et al., 2022; Jia et al., 2022; Jiu et al., 2022). Phytohormones such as auxin and brassinosteroids (BRs) promote the elongation of primary root and initiation of lateral root (Yu et al., 2015; Wei and Li, 2016; Li et al., 2020b; Liu et al., 2021), whereas abscisic acid (ABA), cytokinin (CK), and jasmonic acid (JA) inhibit the root growth (Yang et al., 2017a; Deng et al., 2022; Liu et al., 2022). In general, CK is considered as a vital factor of root plasticity, as it not only influences root growth and development but also plays important roles in regulating the response to abiotic stress of root (Ha et al., 2012; Li et al., 2022).
In plants, CK signaling is mediated by a typical two-component system (TCS), which consists of histidine kinases (HKs), histidine phosphotransfer proteins (HPs), and response regulators (RRs) (Liang et al., 2012). RRs play a bridging role in receiving upstream phosphorylation signals and regulating the expression of downstream target genes (Yan et al., 2021). In Arabidopsis thaliana, ARR1 and ARR12 are involved in root plasticity development under low temperature by reducing the accumulation of auxin in roots (Zhu et al., 2015). ARR7 and ARR15 are essential to maintain the normal development of root stem-cell system by negatively regulating CK signaling (Muller and Sheen, 2008). ARR16 and ARR17 were proved to be key components in CK-mediated root hydrotropic response (Chang et al., 2019). Overexpression of OsRR3 and OsRR5 produced longer roots and more lateral roots compared with wild-type plants when treated with exogenous CK (Cheng et al., 2010). OsRR2 is repressed by WUSCHEL-RELATED HOMEOBOX11 (WOX11), and they were involved in the processes of crown root elongation, lateral root primodium initiation, and root hair formation (Zhao et al., 2015; Cheng et al., 2016). In woody plants, overexpression of the type A RR RcRR1 resulted in optimized root system with longer primary root and more lateral roots (Gao et al., 2013). In Populus, PtRR13 negatively regulates the development of adventitious root (Ramirez-Carvajal et al., 2009).
Abiotic stress-responsive hormone ABA also plays an important role in mediating root growth and development. ABA enables its receptors to bind and sequester the type 2C protein phosphatases (PP2Cs), which induces auto-activation of class III SNF1-RELATED PROTEIN KINASE 2 (SnRK2s) to phosphorylate downstream transcription factors and regulates the expression of ABA-responsive gene (Sheard and Zheng, 2009; Wang et al., 2017). In A. thaliana, SnRK2.1, SnRK2.5, and SnRK2.9 were reported to control root growth under non-stress conditions, while SnRK2.4 and SnRK2.10 function mostly in root development under salt stress (Kawa et al., 2019). SnRK2.2 played an important role in root hydrotropism (Dietrich et al., 2017). In addition, NtSnRK2.2 positively regulated lateral root development to enhance salt tolerance (Liu et al., 2020). Overexpression of TaSnRK2.4 or TaSnRK2.7, which are highly expressed in roots, resulted in better root system structure in Arabidopsis (Mao et al., 2010; Zhang et al., 2011).
The cross-talk between CK and ABA plays a vital role in root plasticity, and CK signaling probably act at downstream of ABA pathway (Basu et al., 2016; Rowe et al., 2016). In Arabidopsis, SnRK2.2, SnRK2.3, and SnRK2.6 directly interacted with phosphorylated ARR5 to mediate drought tolerance (Huang et al., 2018). However, there are few reports on the RRs in citrus and little knowledge on the interactions between RRs and SnRK2s.
CcRR5, a type A RR family member in citrus, was previously revealed to be involved in root morphogenesis, and its expression was upregulated at the early stages of PEG treatment (Zhang et al., 2022). In this study, the role of CcRR5 was further explored to clarify the regulatory mechanism in the process of root development in citrus. Through stable genetic transformation mediated by Agrobacterium tumefaciens, CcRR5 overexpressing and suppressed transgenic citrus plants were generated for further investigation of the function of CcRR5 on growth and development of the roots. The relationships among CcRR5, CcRR14, and CcSnRK2s were analyzed in order to understand the molecular mechanism of root development in citrus.
Volkamer (Citrus limonia) was used for cloning the gene for vector constructions. Trifoliate orange (Poncirus trifoliata) was used for genetic transformation. Nicotiana benthamiana was used for transient transformation and BiFC (bimolecular fluorescence complementation) assay. Experimental plants were grown in a greenhouse under a 16/8h light/dark photoperiod at 28°C.
For subcellular localization, a coding sequence (CDS) fragment without termination codon of CcRR5 was fused to the BamHI/SacI-digested Cam35S-GFP vector. The primers are shown in Supplementary Table S1. The fusion vector 35S::CcRR5-GFP and the green fluorescent protein (GFP) control were transformed into tobacco leaves (4–5 weeks old). GFP fluorescence was observed under a FV3000 confocal microscope (Olympus) as described by Du et al. (2023).
The promoter of CcRR5 was ligated to the BamHI and HindIII sites of p1300GNGM-GUS vector to drive the GUS reporter gene. The CDS of CcRR14 was cloned into the pFGC5941MDB3F-GN plasmid to obtain the 35S:CcRR14 recombinant plasmid. The combination was injected into tobacco leaves to measure the GUS expression as described by Li et al. (2020a).
The promoter of CcRR5 was ligated to the BamHI and HindIII sites of p1300GNGM-GUS vector and introduced into P. trifoliata plants. Roots, stems, and leaves were detached from transgenic plants and submerged in GUS dye solution at 37°C for 12h. The stained tissues were decolorized with 70% alcohol at 37°C for no less than 48h.
The CDS of CcRR5 without termination codon was fused into the BamHI/SwaI-digested pFGC5941MDB3F-GN vector to generate the 35S:CcRR5. For construction of RNAi vector, a 200–300 bp cDNA fragment of CcRR5 was amplified and integrated into the pFGC5941MDB3F-GN vector. Subsequently, the constructs were introduced into Agrobacterium tumefaciens EHA105 by electroporation. Epicotyl explants of 1-month-old P. trifoliata were used to conduct A. tumefaciens–mediated transformation. The G418-containing MS medium was used to screen the transgenic plants. The positive transgenic citrus plants were identified by GFP fluorescence and genomic polymerase chain reaction (PCR). The expression of CcRR5 in transformed plants was further confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis.
The shoots in the same length were cut from both transgenic and control plants of P. trifoliata. Cuttings were placed into a substance of mixed expanded perlite and vermiculite for rooting under 28°C. Rooted plantlets were grown in a greenhouse under a 16/8h light/dark photoperiod. Root indexes such as the length of root and number of root tips were determined.
Citrus genomic DNA was extracted with a CTAB method (Zhang et al., 2020). The methods of RNA extraction, reverse transcription into cDNA, and qRT-PCR analysis were the same as described by Zhang et al. (2022).
The protein sequences of citrus (C. clementina) were downloaded from Phytozome database (http://phytozome.jgi.doe.gov/pz/portal.html). Ten SNF1-related protein kinase 2s’ protein sequences of Arabidopsis thaliana (http://www.arabidopsis.org/) (AtSnRK2s) were used as reference sequences to identify the members of SnRK2 family in C. clementina (CcSnRK2s). The confirmation of SnRK2 family members, the gene structure and motifs analysis, the physicochemical properties, and the subcellular localization prediction of CcSnRK2s were performed as described by Zhang et al. (2022). Based on the Maximum Likelihood Method (MJ), the phylogenetic tree with protein sequences of SnRK2s from A. thaliana and citrus was constructed by using MEGA7.0 software (Kumar et al., 2016).
To test the autoactivation of CcRR5, CcRR14, and CcSnRK2s, the vectors of pGBDT7-CcRR5, pGBDT7-CcRR14, and pGBDT7-CcSnRK2s were transferred into the cells of Y2HGold yeast strain, respectively. The yeast cells were cultured on synthetic defined (SD)/-Trp and SD/-Trp/-His/-Ade solid medium at 28°C for 2–3 days to observe the growth of the cells.
The CDS of CcRR5 and CcRR14 were inserted into the prey vector pGADT7, respectively, while the CDS of CcSnRK2s was cloned into the bait vector pGBKT7. Each pair of constructs was transformed using the lithium acetate method and then cultured on SD/-Trp/-Leu and SD/-Leu/-Trp/-His/-Ade with X-a-gal solid medium at 28°C.
The CDS of CcRR14 and CcSnRK2s were cloned into pCV-cYFP and pCV-nYFP vectors, respectively. Recombinant plasmids were transformed into A. tumefaciens EHA105 cells, and each pair of combinations was co-transformed into tobacco leaves. YFP fluorescence was observed under a FV3000 confocal microscope (Olympus).
Statistical analysis was performed by using the SPSS Statistics 17.0 based on Duncan’s test. Significant differences between different transgenic lines and wild-type plants were also subjected to Student’s t-test. The correlation analysis was conducted through bioinformatics (http://www.bioinformatics.com.cn/) with Pearson’s calculation method.
Transient expression assay was performed to validate subcellular localization of CcRR5 (Figure 1A). The 35S::GFP protein fluorescence signal was observed throughout the cell of tobacco leaves, while the 35S::CcRR5-GFP fusion protein fluorescence signal was observed only in the nuclear and cytomembrane, demonstrating that the protein of CcRR5 is localized in the nuclear and cytomembrane.
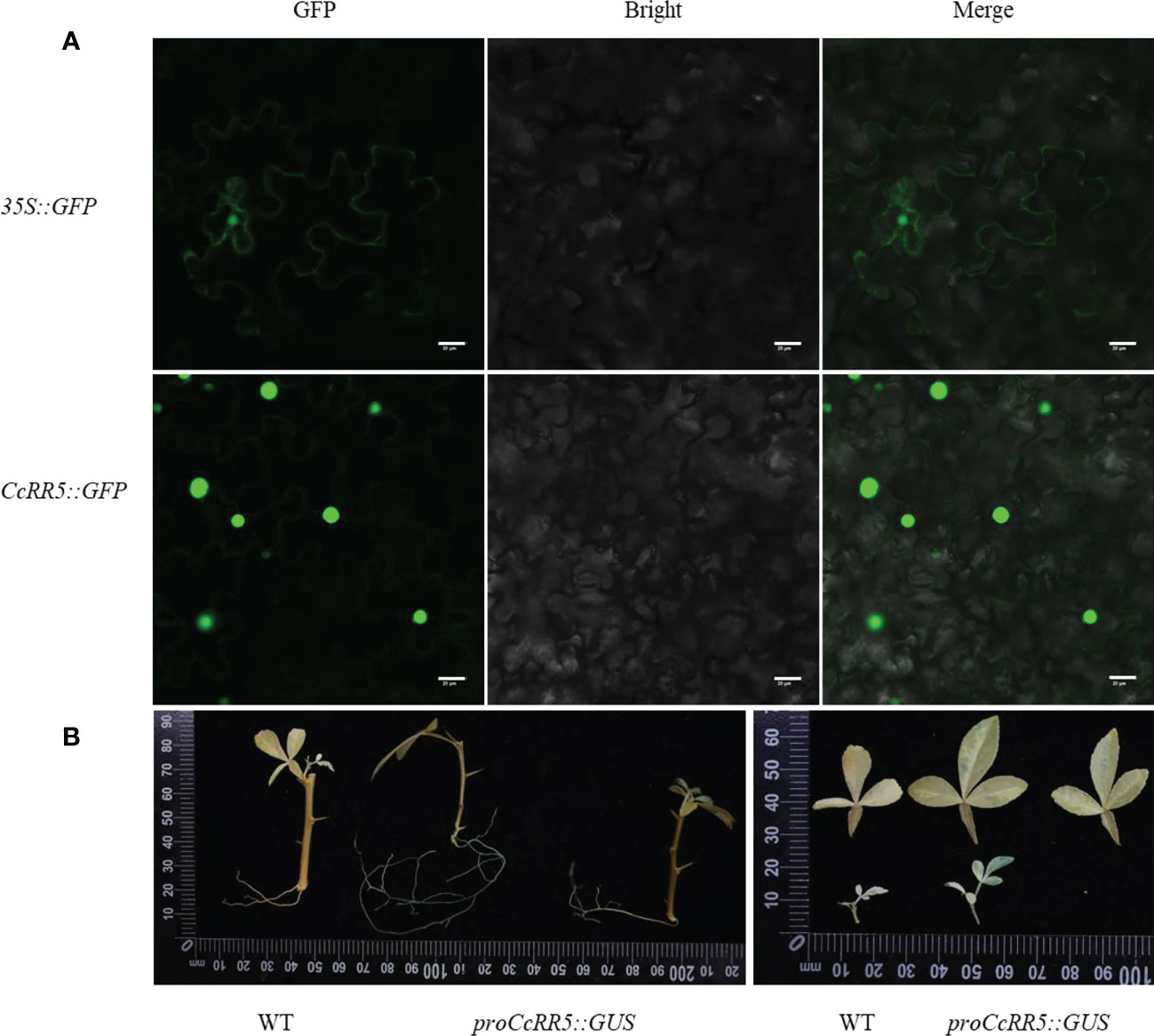
Figure 1 Subcellular localization and GUS assay of CcRR5. (A) Subcellular localization. (B) GUS assay. GFP, green fluorescent protein fluorescence; merge, merge of GFP fluorescence; bright field image. Bars = 20 µm. WT, wild type; proCcRR5::GUS, proCcRR5::GUS transgenic plants.
Previous studies have shown that the expression of CcRR5 is not typical tissue specific, with higher expression in leaves and roots (Zhang et al., 2022). To further analyze the expression pattern of CcRR5, the CcRR5pro::GUS transgenic plants were generated in P. trifoliata background (Figure 1B). The GUS staining was strongly observed in both primary and lateral root tips of four-week-old plantlets,while weakly detected in old leaves.
Previous study showed that the expressions of type A CcRR5 and type B CcRR14 in different citrus rootstocks were both correlated with the length of primary root (Zhang et al., 2022). In view of the relationship between type A and type B RRs in Arabidopsis, it could be speculated that CcRR14 may directly regulate the expression of CcRR5. The promoter of CcRR5 was sequenced in this study (Supplementary Figure S1). The binding site of CcRR14 was found in the promoter of CcRR5, which indicated that CcRR5 could be the direct target of CcRR14. To further examine whether CcRR14 is able to enhance the activity of the CcRR5 promoter, we performed the transient expression assay with tobacco leaves. As shown in Figure 2, the expression of GUS was significantly higher in proCcRR5::GUS plus CcRR14 combination (with strong staining) than that of the control (proCcRR5::GUS, with weak staining). The results of promoter sequence analysis and GUS transient expression assays proved that CcRR14 can directly regulate the expression of CcRR5 in citrus.

Figure 2 Protein–DNA interaction between CcRR14 and the CcRR5 promoter. (A) GUS staining. (B) The expression level of GUS. Each error bar indicated the mean ± SD (n = 3). Different lowercase letters (a–c) mean significant difference, p < 0.05.
Expressions of RR genes in citrus were strongly induced by ABA, suggesting a close relationship between CcRRs and ABA signaling pathway (Zhang et al., 2022). SnRK2s, the key components of ABA signaling pathway, play important roles in the formation of root system. Referring to the interaction between ARR and AtSnRK2s in Arabidopsis, CcRRs might also interact with some members of SnRK2s in citrus. To comprehensively explore the potential interactions, analysis of the gene family is an essential method (Wang et al., 2015). Therefore, we performed genome-wide identification of SnRK2 family members in citrus. Seven SnRK2 members were identified from clementine genome after removing redundancy and alternative splicing sequences. The predicted physical and chemical properties of the seven members are shown in Supplementary Table S2. SnRK2s encode the proteins of 341–365 amino acids with molecular weights of 38.3–41.2 kD. They are hydrophilic proteins and rich in acidic amino acids, most of them are localized in the nucleus. In phylogenetic analysis (Figure 3), SnRK2s from Arabidopsis and citrus were divided into three groups. The members in citrus were termed based on their relationship with SnRK2s from Arabidopsis. Considering that Ciclev10020980m had no homolog in Arabidopsis, it was termed as CcSnRK2a alone. All the seven CcSnRK2s members possess the motif 1–7 (Supplementary Figure S2). Among them, CcSnRK2.3 and CcSnRK2.6 possessing the motif 8 were clustered in group 3, while CcSnRK2.9 and CcSnRK2.10 were placed into group 1 with motif 10. CcSnRK2a, CcSnRK2.7, and CcSnRK2.8 were classified in group 2. However, the motif 9 is only present in CcSnRK2.7 and CcSnRK2.8 but absent from CcSnRK2a. Sequence analysis showed that the N-terminal kinase domain of seven CcSnRK2s is relatively conserved, while the C-terminal domain, which plays a vital role in response to abiotic stress and ABA induction, is highly divergent (Figure 4).
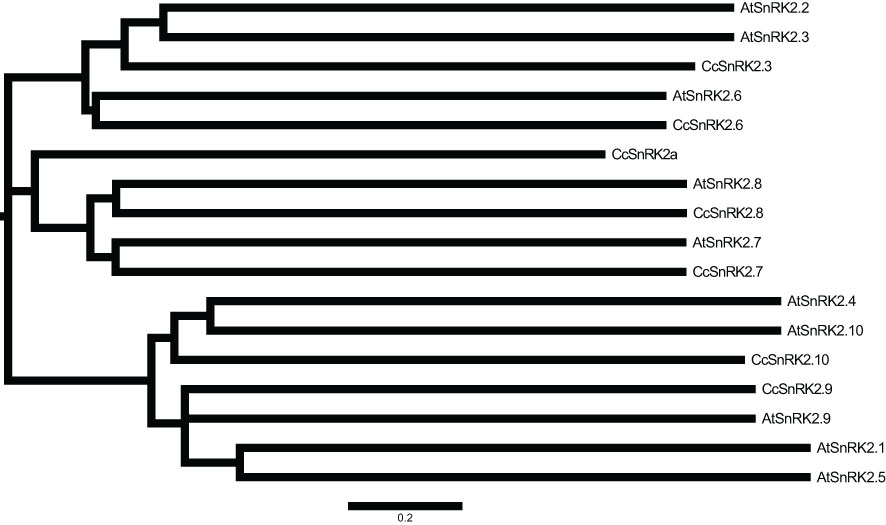
Figure 3 Phylogenetic tree of SnRK2 proteins from Citrus clementina and Arabidopsis thaliana CcSnRK2s: C. clementina; AtSnRK2s: A. thaliana. The scale bar was the distance scale.
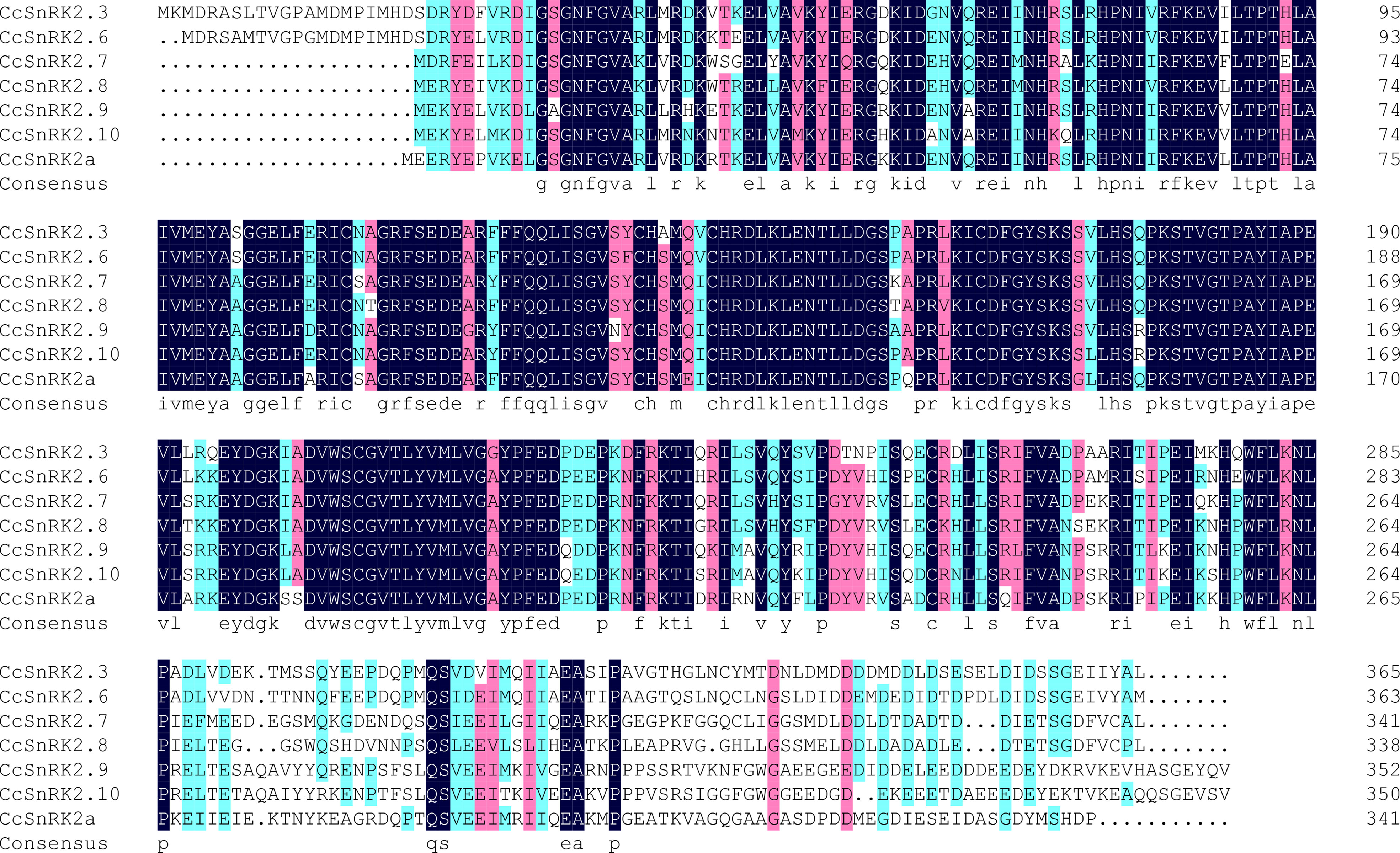
Figure 4 Amino acid sequence comparison of CcSnRK2s. Blueblack background: 100% similarity; blue: > 80% similarity; pink: > 50% similarity.
The RR interaction network in Arabidopsis was well studied previously, such as ARR involved in cytokinin and auxin signaling via AHP-ARR and ARR-PIN pathways (Hwang et al., 2012), and cytokinin and ABA signaling via ARR-AtSnRK2.2/2.3/2.6 pathway (Huang et al., 2018). However, little information on RR interaction network is available in woody plants. Therefore, it is necessary to explore whether the CcRR-CcSnRK2 network in citrus also works in the same way as that in Arabidopsis and how many members of the CcSnRK2 family can interact with CcRRs.
To clarify these speculates, we studied the interactions between CcRR5 and CcSnRK2 subfamily proteins using yeast two-hybrid assay. First, the transcriptional activation of CcRR5, CcRR14, and CcSnRK2s was tested. The pGBKT7-CcRR5, pGBKT7-CcRR14, pGBKT7-CcSnRK2.3, pGBKT7-CcSnRK2.6, pGBKT7-CcSnRK2.7, pGBKT7-CcSnRK2.8, pGBKT7-CcSnRK2.9, and pGBKT7-CcSnRK2.10 fusion vectors (CcSnRK2a was not cloned from root sample of Volkamer) were constructed and transformed into Y2HGold yeast cells, respectively. As shown in Supplementary Figure S3, auto-activation was only observed in the yeast transformed with pGBKT7-CcRR14 in transactivation activity assay. In Figure 5A, CcRR5 showed significant interactions with CcSnRK2.3, CcSnRK2.6, CcSnRK2.7, and CcSnRK2.8 in Y2H assay. The results of yeast two-hybrid assay confirmed that CcRR14 also interacts with these four members of SnRK2. Those results were further confirmed by BiFC assay (Figure 5B, Supplementary Figure S4).
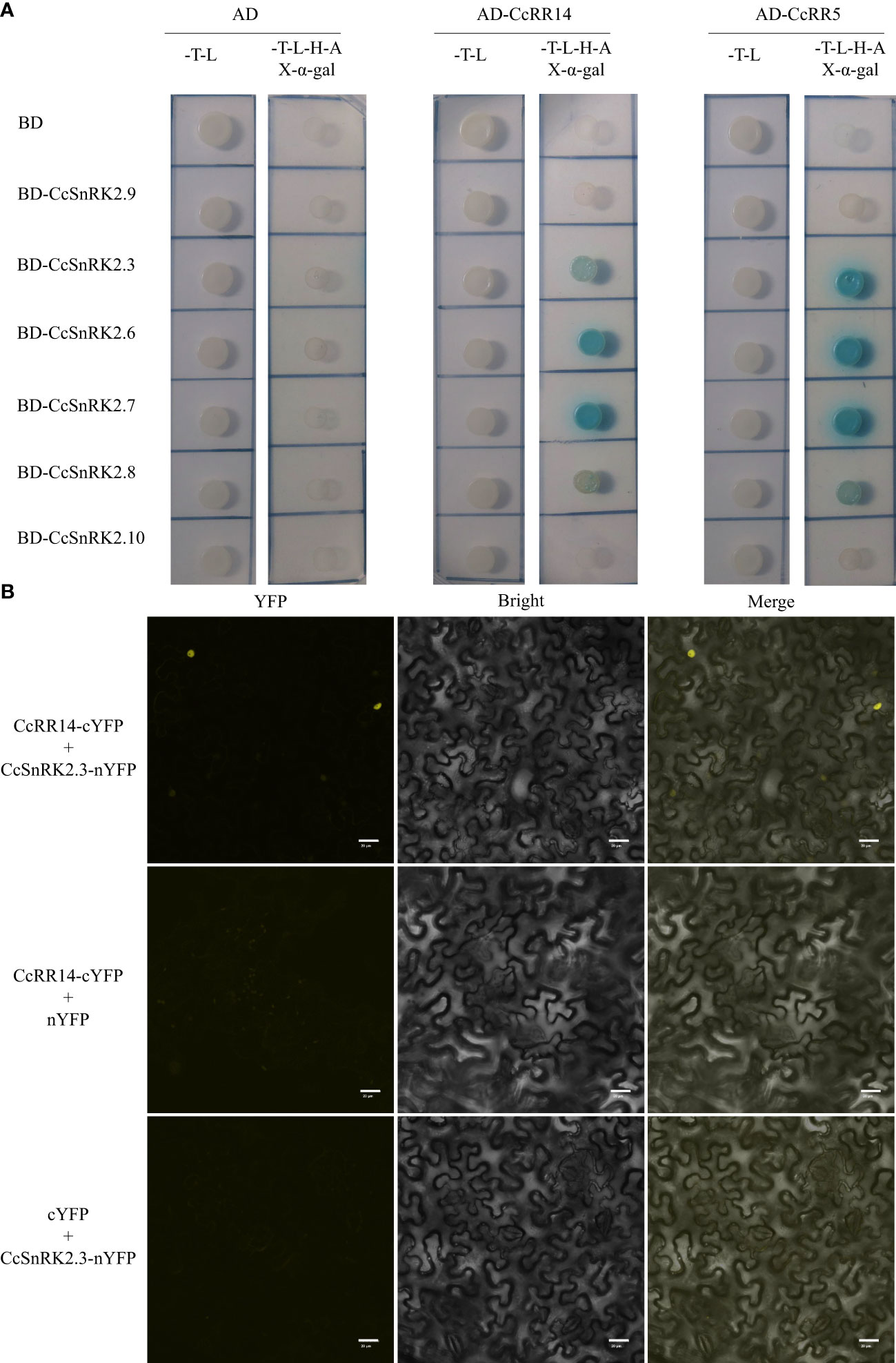
Figure 5 Interactions between CcRR5, CcRR14, and CcSnRK2s proteins. (A) Yeast two-hybrid assays between CcRR5, CcRR14, and CcSnRK2s. SD/-T-L, synthetic dropout medium lacking Trp and Leu. SD/-T-L-H-A, synthetic dropout medium lacking Trp, Leu, His, and Ade. (B) BiFC assays between CcRR14 and CcSnRK2.3. YFP, YFP fluorescence; merge, merge of YFP fluorescence; bright field image. Bars = 20 µm. .
The 35S::CcRR5 and CcRR5-RNAi vectors were transformed into citrus rootstock P. trifoliata. Three overexpression lines (35S::CcRR5 OE-3, OE-5, and OE-7) and four RNAi-mediated gene knockdown lines (CcRR5-RNAi-2, RNAi-4, RNAi-9, and RNAi-15) were selected for further study (Figure 6). Compared with the control, the expression of CcRR5 in overexpression lines increased more than three times, while that of in RNAi lines decreased to 0.13, 0.16, 0.12, and 0.44 times. The expressions of CcRR14 and CcSnRK2s in transgenic plants were also analyzed. The expression of CcRR14 was significantly upregulated in three overexpression plants and suppressed in three RNAi plants (RNAi-2, -4, and -9). The correlation between the expressions of these two genes was extremely significant (Table 1). The expressions of CcSnRK2s in transgenic plants were not consistent. The expressions of CcSnRK2.3 and CcSnRK2.6 in three RNAi plants (RNAi-2, -9, and -15) were significantly upregulated. The correlation between the expression of CcSnRK2.3 and CcRR5 was extremely significant, while the correlation between the expression of CcSnRK2.6 and CcRR5 was significant. Even though the expression of CcSnRK2.7 was upregulated in overexpression plants and corresponded with the expression of CcRR5, the correlation was not significant.
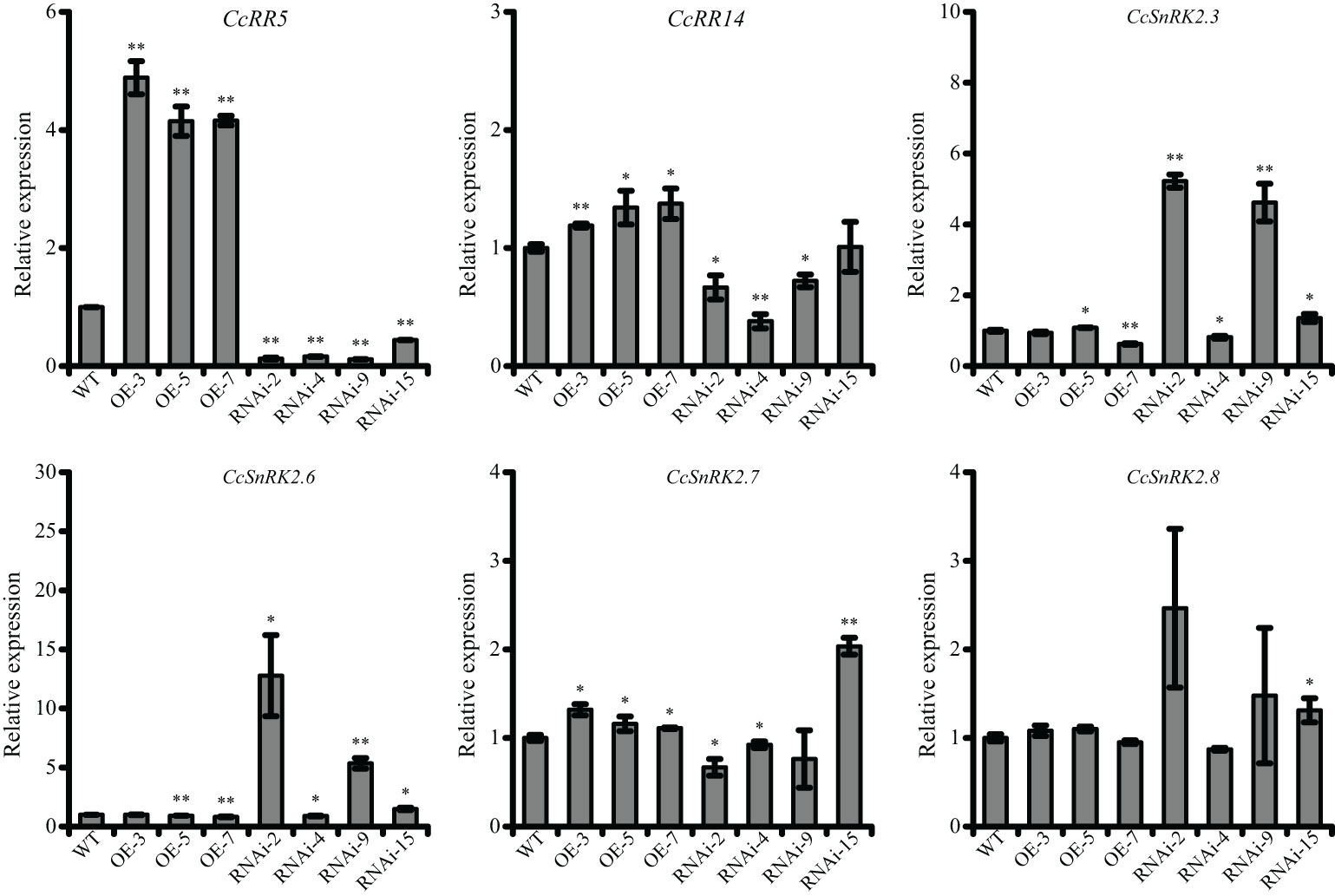
Figure 6 The expression level of CcRR5, CcRR14, and CcSnRK2s in transgenic plants. WT, wild type; OE, CcRR5 overexpression plants; RNAi, CcRR5 silence plants. Each error bar indicated the mean ± SD (n = 3). The asterisk indicated that the expression of CcRRs or CcSnRK2s in overexpression and silence plant was significantly different with that in the WT plants, *p < 0.05; **p < 0.01.
Phenotypes of different transgenic plants were shown in the Figure 7A. The qRT-PCR analysis of self-rooted plants showed that compared with the control, the expression of CcRR5 was significantly upregulated in the roots of the overexpressed plants but had no difference in the roots of the silenced plants (Figure 7B). In CcRR5 overexpression lines, the primary root lengths were significantly longer than those of the control plants (Figure 7C). Total numbers of root tips of CcRR5-OE lines were also significantly greater than those of the control plants (Figure 7D). However, silence of CcRR5 did not affect the growth of root in RNAi plants.
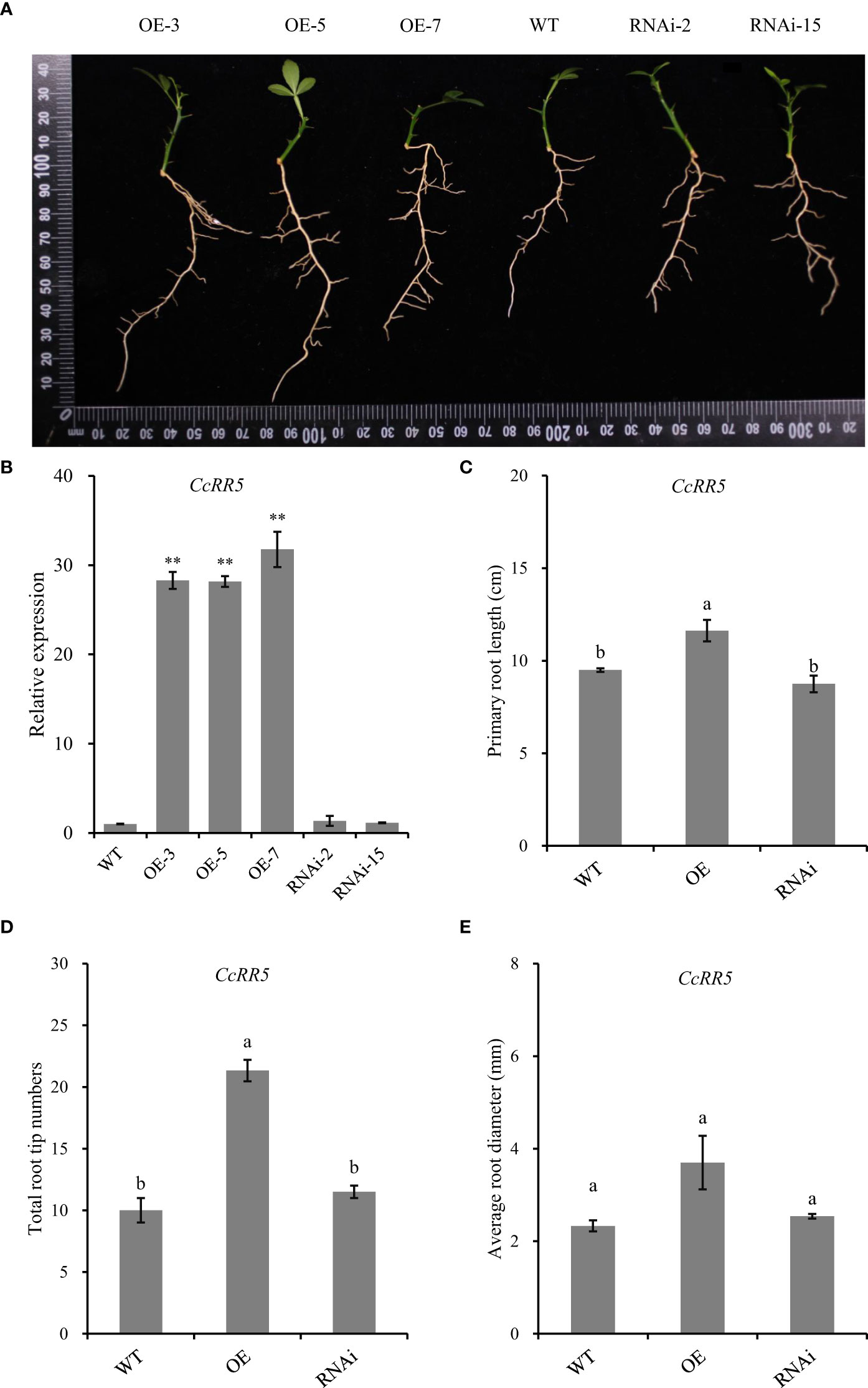
Figure 7 Functional analysis of CcRR5 in citrus. (A) Morphological observations of roots in CcRR5 overexpression (CcRR5-OE) and CcRR5 RNA interference (CcRR5-RNAi) transgenic plants. (B) Relative expression of CcRR5 in transgenic plants, (C) primary root length of plantlet, (D) total root tip numbers of plantlet, and (E) average root diameter of plantlet. The error bars in (B) indicated the mean ± SD (n = 3). The asterisk indicated that the expression of CcRR5 in overexpression and silence plants was significantly different with that in the WT plants. **p < 0.01. The error bars in (C–E) indicated the mean ± SE. Means followed by different letters (a, b) indicate significantly different at p < 0.05.
To further investigate the involvement of CcRR5 in root development, the expression levels of some genes might relate to root growth and development were determined by qRT-PCR (Figure 8). Results showed that the expression of CcMYB77 in the roots and leaves of CcRR5-OE lines was significantly suppressed. The correlation between the expression of CcMYB77 and CcRR5 was extremely significant in both roots and leaves (Figure 9). The expression of CcIAA17 showed the opposite trend with that of CcMYB77 in leaves, but the same trend in roots, which was extremely negatively corresponded to the expression of CcRR5. In addition, the expressions of CcPIN3 and CcSHY2 were also extremely significantly correlated to the expression of CcRR5 in leaves, while the expressions of CcPIN3 and CcPIN7 were significantly correlated to the expression of CcRR5 in roots. These results suggested that CcRR5 involved in the regulation of root growth and played a positive role in root morphogenesis.
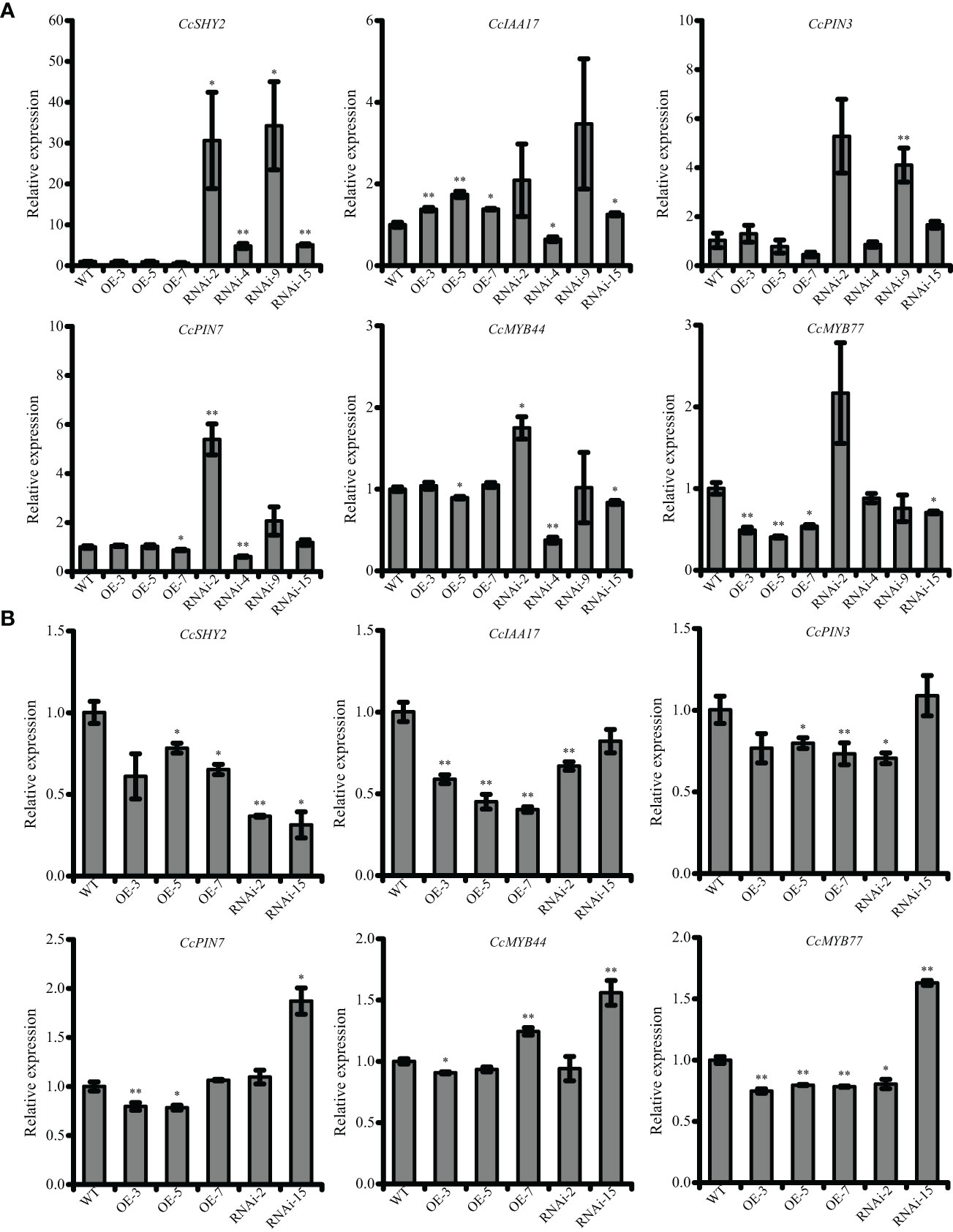
Figure 8 The expression levels of root-related genes in the leaves (A) and roots (B) of CcRR5 transgenic plants. WT, wild type; OE, CcRR5 overexpression plants; RNAi, CcRR5 silence plants. Each error bar indicated the mean ± SD (n = 3). The asterisk indicated that the expression of genes in overexpression and silence plant was significantly different with that in the WT plants. *p < 0.05; **p < 0.01.
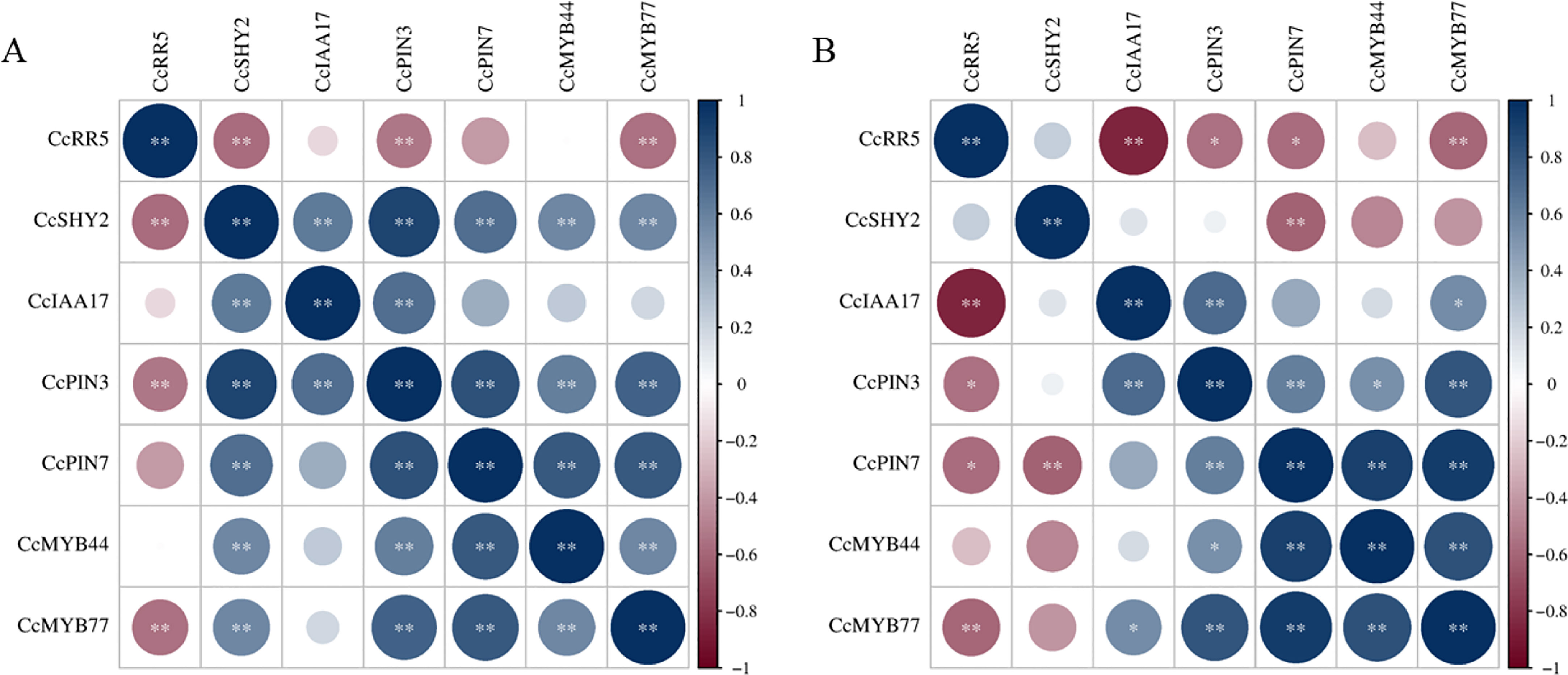
Figure 9 The correlation between expressions of CcRR5 and root-related genes in the leaves (A) and roots (B) of CcRR5 transgenic plants. The color bar represented the range of correlation coefficient (r). *p < 0.05; **p < 0.01.
It was reported that type B ARR family members serve as transcriptional regulators of type A ARRs. They usually bind to the cytokinin response motif or extended motif (Tajima et al., 2004; Ramireddy et al., 2013). Both type A ARR5 and ARR15 are direct targets of the transcriptional type B RR, ARR2 (Che et al., 2008). Interestingly, the expression of type B CcRR14, in the same group of a phylogenic tree with ARR1 and ARR2, was significantly correlated with the expression of type A CcRR5 in our previously study (Zhang et al., 2022), suggesting that CcRR14 might regulate the expression of CcRR5. In this study, we identified that promoter of CcRR5 contains several binding elements of type B RRs. In CcRR5 transgenic plants, the expression of CcRR14 was significantly correlated to the expression of CcRR5. In GUS assay, the activity of CcRR5 promoter was enhanced by CcRR14. All those findings confirmed the hypothesis that CcRR14 directly regulates the expression of CcRR5.
ABA signaling cascade serves as crucial factor in aspects of plant growth and development (Wang et al., 2017). Currently, stress response mediated by SnRK2s pathway has been widely studied in model plants (Miao et al., 2021). Here, seven SnRK2 members were identified from C. clementina, which was consistent with the number of SnRK2s in P. trifoliate, but less than that of in Arabidopsis, maize, cotton, and apple, due to the retraction or diversification of gene and the duplication of whole-genome (Huai et al., 2008; Liu et al., 2017; Shao et al., 2014; Song et al., 2022). The Group III members of SnRK2 were reported to strongly interact with ARR1 and ARR5 in Arabidopsis (Huang et al., 2018). In this research, we verified the interaction between CcRRs and SnRK2 family members and had some new discoveries. In addition to CcSnRK2.3 and CcSnRK2.6, group II members CcSnRK2.7 and CcSnRK2.8, homologous to AtSnRK2.7 and AtSnRK2.8, can also interact with CcRR5 and CcRR14. It can be assumed that CcRR5, CcRR14, and CcSnRK2s in citrus should work in the similar way as that in Arabidopsis (Huang et al., 2018), that the type B CcRR14 positively regulates the cytokinin signaling and inhibits the activity of CcSnRK2s, while the type A CcRR5, upregulated by CcRR14 in a negative feedback loop, negatively regulates the cytokinin signaling and be phosphorylated by CcSnRK2s to regulate the expression of ABA-responsive genes.
CcRR5, a type A cytokinin RR in citrus, is homologous to ARR5/6/7/15, and its expression was significantly correlated with primary root length (Zhang et al., 2022). In this research, overexpression of CcRR5 in citrus resulted longer primary root and greater number of lateral roots compared to the control, which is in agree with the findings of Kiba et al. (2003) and To et al. (2007) in Arabidopsis. Recent studies have reported that ARR1 is a negative regulator of primary root growth under aluminum and salt stresses (Yang et al., 2017b; Yan et al., 2021). This suggested that CcRR14 might directly bind to the promoter of CcRR5 and regulate its expression to affect the elongation of citrus roots in response to abiotic stresses. Overexpression of AtSnRK2.8, a root-specific protein kinase, increased the tolerance of drought (Umezawa et al., 2004). AtSnRK2.4 and AtSnRK2.10 can maintain root growth under salt stress (McLoughlin et al., 2012). Better root architecture with enhanced salt-stress tolerance was also achieved in tobacco by overexpression of NtSnRK2.2, a group II member in Nicotiana tabacum L. (Liu et al., 2020). The interactions between CcSnRK2s and CcRRs were further confirmed that CcRR5 contributed to the root growth. More evidences have been observed that expressions of auxin-related genes such as PINFORMEDs (PINs), auxin signal inhibitor SHY2, and auxin/indole-3-acetic acid IAA17 were significantly correlated to the expression of CcRR5. Those genes are the key factors in regulating root meristem size and adventitious root formation in Arabidopsis (Hwang et al., 2012; Liu et al., 2015; Lakehal et al., 2019). The expression of MYB77 was significantly decreased in CcRR5-overexpression citrus plants, which was reported to be a negative factor of primary root growth strongly expressed in lateral and primary roots in Arabidopsis (Shin et al., 2007). All those findings indicated that CcRR5 is involved in formation of root system architecture, which might be regulated by some genes involved in other hormone signal pathways.
The type A RR CcRR5 is mainly localized in the nucleus and expressed in root tips and young leaves in citrus. Phenotypic comparison and expression analysis of transgenic citrus plants indicated that CcRR5 could enhance the growth of roots and formation of lateral roots. The promoter activity of CcRR5 can be enhanced by CcRR14. They both can interact with CcSnRK2s.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
SZ and XZ conceived the project. MZ, SZ and XZ designed the experiments. MZ, FW, ZH, XW, QY and JF performed the experiments. MZ, SZ and XZ wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2018YFD1000101), China Agriculture Research System (CARS-Citrus), the “Double World-classes” Development Plan of Southwest University, and 111 Project (B18044).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1170825/full#supplementary-material
Basu, S., Ramegowda, V., Kumar, A., Pereira, A. (2016). Plant adaptation to drought stress [version 1; referees: 3 approved]. F1000Research 5, 1554. doi: 10.12688/f1000research.7678.1
Cai, B., Wang, T., Sun, H., Liu, C., Chu, J., Ren, Z., et al. (2022). Gibberellins regulate lateral root development that is associated with auxin and cell wall metabolisms in cucumber. Plant Sci. 317, 110995. doi: 10.1016/j.plantsci.2021.110995
Chang, J., Li, X., Fu, W., Wang, J., Yong, Y., Shi, H., et al. (2019). Asymmetric distribution of cytokinins determines root hydrotropism in Arabidopsis thaliana. Cell Res. 0, 1–10. doi: 10.1038/s41422-019-0239-3
Che, P., Lall, S., Howell, S. H. (2008). Acquiring competence for shoot development in Arabidopsis. Plant Signal. Behav. 3 (2), 99–101. doi: 10.4161/psb.3.2.4958
Cheng, X., Jiang, H., Zhang, J., Qian, Y., Zhu, S., Cheng, B. (2010). Overexpression of type-a rice response regulators, OsRR3 and OsRR5, results in lower sensitivity to cytokinins. Genet. Mol. Res. 9 (1), 348–359. doi: 10.4238/vol9-1gmr739
Cheng, S., Zhou, D. X., Zhao, Y. (2016). WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 11 (2), e1130198. doi: 10.1080/15592324
Deng, J., Kong, L., Zhu, Y., Pei, D., Chen, X., Wang, Y., et al. (2022). BAK1 plays contrasting roles in regulating abscisic acid-induced stomatal closure and abscisic acid-inhibited primary root growth in Arabidopsis. J. Integr. Plant Biol. 64 (6), 1264–1280. doi: 10.1111/jipb.13257
Dietrich, D., Pang, L., Kobayashi, A., Fozard, J. A., Boudolf, V., Bhosale, R., et al. (2017). Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat. Plants 3 (6), 17057. doi: 10.1038/nplants.2017.57
Du, J., Wang, Q., Shi, H., Zhou, C., He, J., Wang, X. (2023). A prophage-encoded effector from “Candidatus liberibacter asiaticus” targets ASCORBATE PEROXIDASE6 in citrus to facilitate bacterial infection. Mol. Plant Pathol. 00, 1–15. doi: 10.1111/mpp.13296
Gao, B., Fan, L., Li, X., Yang, H., Liu, F., Wang, L., et al. (2013). RcRR1, a Rosa canina type-a response regulator gene, is involved in cytokinin-modulated rhizoid organogenesis. PloS One 8 (8), e72914. doi: 10.1371/journal.pone.0072914
Gupta, A., Rico-Medina, A., Cao-Delgado, A. I. (2020). The physiology of plant responses to drought. Science 368 (6488), 266–269. doi: 10.1126/science.aaz7614
Ha, S., Vankova, R., Yamaguchi-Shinozaki, K., Shinozaki, K., Phan Tran, L. S. (2012). Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 17 (3), 172–179. doi: 10.1016/j.tplants.2011.12.005
Huai, J., Wang, M., He, J., Zheng, J., Dong, Z., Lv, H., et al. (2008). Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 27, 1861–1868. doi: 10.1007/s00299-008-0608-8
Huang, X., Hou, L., Meng, J., You, H., Li, Z., Gong, Z., et al. (2018). The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 11 (7), 970–982. doi: 10.1016/j.molp.2018.05.001
Hwang, I., Sheen, J., Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63 (1), 353–380. doi: 10.1146/annurev-arplant-042811-105503
Jia, Z., Giehl, R. F. H., von Wiren, N. (2022). Nutrient-hormone relations: Driving root plasticity in plants. Mol. Plant 15 (1), 86–103. doi: 10.1016/j.molp.2021.12.004
Jiu, S., Xu, Y., Xie, X., Wang, J., Xu, J., Liu, X., et al. (2022). Strigolactones affect the root system architecture of cherry rootstock by mediating hormone signaling pathways. Environ. Exp. Bot. 193, 104667–104667. doi: 10.1016/j.envexpbot.2021.104667
Kawa, D., Meyer, A. J., Dekker, H. L., Abd-El-Haliem, A., Gevaert, K., Van De Slijke, E., et al. (2019). SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 182 (1), 361–377. doi: 10.1104/pp.19.00818
Kiba, T., Yamada, H., Sato, S., Kato, T., Tabata, S., Yamashino, T., et al. (2003). The type-a response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44 (8), 868–874.
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Lakehal, A., Chaabouni, S., Cavel, E., Le Hir, R., Ranjan, A., Raneshan, Z., et al. (2019). A molecular framework for the control of adventitious rooting by TIR1/AFB2-Aux/IAA-Dependent auxin signaling in Arabidopsis. Mol. Plant 12 (11), 1499–1514. doi: 10.1016/j.molp.2019.09.00
Li, T., Lei, W., He, R., Tang, X., Han, J., Zou, L., et al. (2020b). Brassinosteroids regulate root meristem development by mediating BIN2-UPB1 module in Arabidopsis. PloS Genet. 16 (7), e1008883. doi: 10.1371/journal.pgen.1008883
Li, J., Luan, Q., Han, J., Zhang, C., Liu, M., Ren, Z. (2020a). CsMYB60 directly and indirectly activates structural genes to promote the biosynthesis of flavonols and proanthocyanidins in cucumber. Hortic. Res. 7, 103. doi: 10.1038/s41438-020-0327-z
Li, L., Zheng, Q., Jiang, W., Xiao, N., Zeng, F., Chen, G., et al. (2022). Molecular regulation and evolution of cytokinin signaling in plant abiotic stresses. Plant Cell Physiol. 63 (12), 1787–1805. doi: 10.1093/pcp/pcac071
Liang, Y., Wang, X., Hong, S., Li, Y., Zuo, J. (2012). Deletion of the initial 45 residues of ARR18 induces cytokinin response in Arabidopsis. J. Genet. Genomics 39 (1), 37–46. doi: 10.1016/j.jgg.2011.12.004
Liu, Z., Ge, X., Yang, Z. (2017). Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum l.). BMC Genet. 18, 54. doi: 10.1186/s12863-017-0517-3
Liu, W., Li, R. J., Han, T. T., Cai, W., Fu, Z. W., Lu, Y. T. (2015). Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 168 (1), 343–356. doi: 10.1104/pp.15.00030
Liu, S., Strauss, S., Adibi, M., Mosca, G., Yoshida, S., Ioio, R. D., et al. (2022). Cytokinin promotes growth cessation in the arabidopsis root. Curr. Biol. 32 (9), 1974–1985.e3. doi: 10.1016/j.cub.2022.03.019
Liu, M., Wang, J., Gou, J., Wang, X., Li, Z., Yang, X., et al. (2020). Overexpression of NtSnRK2.2 enhances salt tolerance in Nicotiana tabacum by regulating carbohydrate metabolism and lateral root development. Funct. Plant Biol. 47 (6), 537. doi: 10.1071/FP19299
Liu, L., Yu, Z., Xu, Y., Guo, C., Zhang, L., Wu, C., et al. (2021). Function identification of MdTIR1 in apple root growth benefited from the predicted MdPPI network. J. Integr. Plant Biol. 63 (04), 723–736. doi: 10.1111/jipb.12996
Mao, X., Zhang, H., Tian, S., Chang, X., Jing, R. (2010). TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum l.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 61 (3), 683–696. doi: 10.1093/jxb/erp331
McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., van der Does, D., Laurière, C., et al. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72 (3), 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
Miao, R., Yuan, W., Wang, Y., Garcia-Maquilon, I., Xu, W. (2021). Low ABA concentration promotes root growth and hydrotropism through relief of ABA INSENSITIVE 1-mediated inhibition of plasma membrane h+-ATPase 2. Sci. Adv. 7, eabd4113. doi: 10.1126/sciadv.abd4113
Muller, B., Sheen, J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453(7198), 1094–1097. doi: 10.1038/nature06943
Ramireddy, E., Brenner, W. G., Pfeifer, A., Heyl, A., Schmülling, T. (2013). In planta analysis of a cis-regulatory cytokinin response motif in arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol. 54 (7), 1079–1092. doi: 10.1093/pcp/pct060
Ramirez-Carvajal, G. A., Morse, A. M., Dervinis, C., Davis, J. M. (2009). The cytokinin type-b response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 150 (2), 759–771. doi: 10.1104/pp.109.137505
Rowe, J. H., Topping, J. F., Liu, J., Lindsey, K. (2016). Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 211, 225–239. doi: 10.1111/nph.13882
Shao, Y., Qin, Y., Zou, Y. J., Ma, F. W. (2014). Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene 552 (1), 87–97. doi: 10.1016/j.gene.2014.09.017
Sheard, L. B., Zheng, N. (2009). Signal advance for abscisic acid. Nature 462 (7273), 575–576. doi: 10.1038/462575a
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., et al. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19 (8), 2440–2453. doi: 10.1105/tpc.107.050963
Song, J., Sun, P., Kong, W., Xie, Z., Li, C., Liu, J. H. (2022). SnRK2.4-mediated phosphorylation of ABF2 regulates ARGININE DECARBOXYLASE expression and putrescine accumulation under drought stress. New Phytol. 238 (1), 216–236. doi: 10.1111/nph.18526
Tajima, Y., Imamura, A., Kiba, T., Amano, Y., Yamashino, T., Mizuno, T. (2004). Comparative studies on the type-b response regulators revealing their distinctive properties in the his-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 45 (1), 28–39. doi: 10.1093/pcp/pcg154
To, J., Deruere, J., Maxwell, B., Morris, V., Hutchison, C., Ferreira, F., et al. (2007). Cytokinin regulates type-a Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell. 19 (12), 3901–3914. doi: 10.1105/tpc.107.052662
Umezawa, T., Yoshida, R., Maruyama, K., Yamaguchi-Shinozaki, K., Shinozaki, K. (2004). SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. p. Natl. Acad. Sci. U.S.A. 101 (49), 17306–17311. doi: 10.1073/pnas.0407758101
Wang, L., Hu, W., Sun, J., Liang, X., Yang, X., Wei, S., et al. (2015). Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 237, 33–45. doi: 10.1016/j.plantsci.2015.05.008
Wang, T., Li, C., Wu, Z., Jia, Y., Wang, H., Sun, S., et al. (2017). Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01121
Wei, Z., Li, J. (2016). Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 9 (001), 86–100. doi: 10.1016/j.molp.2015.12.003
Yan, Z., Wang, J., Wang, F., Xie, C., Lv, B., Yu, Z., et al. (2021). MPK3/6-induced degradation of ARR1/10/12 promotes salt tolerance in Arabidopsis. EMBO Rep. 22 (10), e52457. doi: 10.15252/embr.202152457
Yang, Z. B., He, C., Ma, Y., Herde, M., Ding, Z. (2017a). Jasmonic acid enhances Al-induced root growth inhibition. Plant Physiol. 173 (2), 1420–1433. doi: 10.1104/pp.16.01756
Yang, Z. B., Liu, G., Liu, J., Zhang, B., Meng, W., Müller, B., et al. (2017b). Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep. 18 (7), 1213–1230. doi: 10.15252/embr.201643806
Yu, C. L., Sun, C. D., Shen, C., Wang, S., Liu, F., Liu, Y., et al. (2015). The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in cd stress responses in rice (Oryza sativa l.). Plant J. 83 (5), 818–830. doi: 10.1111/tpj.12929
Zhang, P., Liu, X., Yu, X., Wang, F., Long, J., Shen, W., et al. (2020). The MYB transcription factor CiMYB42 regulates limonoids biosynthesis in citrus. BMC Plant Bio. 20 (1), 254. doi: 10.1186/s12870-020-02475-4
Zhang, H., Mao, X., Jing, R., Chang, X., Xie, H. (2011). Characterization of a common wheat (Triticum aestivum l.) TaSnRK2.7 gene involved in abiotic stress responses. J. Exp. Bot. 62 (3), 975–988. doi: 10.1093/jxb/erq328
Zhang, M., Wang, F., Wang, X., Feng, J., Yi, Q., Zhu, S., et al. (2022). Mining key genes related to root morphogenesis through genome-wide identification and expression analysis of RR gene family in citrus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1068961
Zhao, Y., Cheng, S., Song, Y., Huang, Y., Zhou, S., Liu, X., et al. (2015). The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27 (9), 2469–2483. doi: 10.1105/tpc.15.00227
Zhu, J., Zhang, K. X., Wang, W. S., Gong, W., Liu, W. C., Chen, H. G., et al. (2015). Low temperature inhibits root growth by reducing auxin accumulation via ARR1/12. Plant Cell Physiol. 56 (4), 727–736. doi: 10.1093/pcp/pcu217
Keywords: citrus, response regulators, SnRK2s, root development, abiotic stresses
Citation: Zhang M, Wang F, Hu Z, Wang X, Yi Q, Feng J, Zhao X and Zhu S (2023) CcRR5 interacts with CcRR14 and CcSnRK2s to regulate the root development in citrus. Front. Plant Sci. 14:1170825. doi: 10.3389/fpls.2023.1170825
Received: 21 February 2023; Accepted: 27 March 2023;
Published: 17 April 2023.
Edited by:
Qinglong Dong, Hebei Agricultural University, ChinaReviewed by:
Xu Yuantao, Huazhong Agricultural University, ChinaCopyright © 2023 Zhang, Wang, Hu, Wang, Yi, Feng, Zhao and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaochun Zhao, emhhb3hpYW9jaHVuQGNyaWMuY24=; Shiping Zhu, emh1c2hpcGluZ0BjcmljLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.