- 1College of Agronomy, Xinjiang Agricultural University, Urumqi, China
- 2Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Agronomy, Shanxi Agricultural University, Jinzhong, China
Nitrate transporters (NRTs) are crucial for the uptake, use, and storage of nitrogen by plants. In this study, 42 members of the GhNRT2 (Nitrate Transporter 2 family) were found in the four different cotton species. The conserved domains, phylogenetic relationships, physicochemical properties, subcellular localization, conserved motifs, gene structure, cis-acting elements, and promoter region expression patterns of these 42 members were analyzed. The findings confirmed that members of the NRT2 family behaved typically, and subcellular localization tests confirmed that they were hydrophobic proteins that were mostly located on the cytoplasmic membrane. The NRT2 family of genes with A.thaliana and rice underwent phylogenetic analysis, and the results revealed that GhNRT2 could be divided into three groups. The same taxa also shared similar gene structure and motif distribution. The composition of cis-acting elements suggests that most of the expression of GhNRT2 may be related to plant hormones, abiotic stress, and photoreactions. The GhNRT2 gene was highly expressed, mainly in roots. Drought, salt, and extreme temperature stress showed that GhNRT2 gene expression was significantly up-regulated or down-regulated, indicating that it may be involved in the stress response of cotton. In general, the genes of the NRT2 family of cotton were comprehensively analyzed, and their potential nitrogen uptake and utilization functions in cotton were preliminarily predicted. Additionally, we provide an experimental basis for the adverse stress conditions in which they may function.
1 Introduction
Nitrogen (N), mainly in the form of nitrate, is a key factor in plant development, affecting various biological processes in plants, such as photosynthesis and protein synthesis (Frink et al., 1999; Evans and Clarke, 2019). Nitrogen fertilizer use has made a significant contribution to agricultural output and global food security, but this use is also accompanied by substantial environmental consequences such as soils acidification and water eutrophication (Guo et al., 2010; Zhang et al., 2015). With the expansion of planting area and the nutritional needs of crops, nitrogen fertilizer use has increased. However, the absorption efficiency of nitrogen is often less than 40%, and the residual nitrogen left in the field can result in serious environmental pollution (Liu et al., 2022). Therefore, it is particularly important to improve the efficiency of nitrogen fertilizer. To gain maximum yields, an enhancement of the nitrogen use efficiency (NUE) of the crop with low nitrogen inputs is necessary (Liu et al., 2021). Plants mainly absorb nitrogen from soil, with nitrate and ammonium nitrogen being the most effective forms for plant nitrogen; these forms of nitrogen are known as available nitrogen and can be absorbed and used directly by plant roots. Although these two forms of nitrogen have similar nutritional effects on higher plants, there are a few differences in absorption and utilization, and the degree of variation differs with plant species and environmental conditions. Insufficient nitrogen supplies can cause changes in the external morphology and internal metabolism of the plant (Wen et al., 2020).
A study found that there were four gene families of transporters involved in absorption, transportation, and storage: NPF/NRT1 (Nitrate transporter 1(NRT1)/Peptide Transporter (PTR) family (NPF), Transporters protein 1/Peptide family), NRT2 (Nitrate transporter 2 family, Transporters protein 2), CLC (Chloride Channel), and SLAC/SLAH (Slowly Activating Anion Channel) (Taochy et al., 2015). The nitrate uptake process in plants is carried out by NRT2 related proteins, which are typical high-affinity nitrate transporters in the plant (Du et al., 2022). The larger rice genome only possesses four NRT2 genes and two NAR2 genes, compared to seven NRT2 genes and two NAR2 genes in the Arabidopsis genome (Wang et al., 2018). Even though all of the NRT2 family genes are involved in the transport of , their roles and transcriptional patterns vary. For example, AtNRT2.1 was found to be mainly expressed in the cortex and epidermis of the roots (Guan et al., 2021). AtNRT2.3 and AtNRT2.6 were mainly expressed in roots and young leaves (Dechorgnat et al., 2012), while AtNRT2.4 was mainly expressed in roots, and plays an important role in nutrient absorption and transportation of through phloem (Kiba et al., 2012). The homologous protein OsNPF2.4 in rice was found to be involved in the acquisition of low-affinity nitrates. OsNPF2.4 mediates not only nitrate acquisition, but also root-to-stem nitrate transport and nitrogen reactivation from source to sink organs (Xia et al., 2015). After prolonged starvation, the expression of AtNRT2.5 was highly induced, was the main transporter of high affinity nitrate uptake, and played a major role in nitrate acquisition and it’s reuse in nitrogen-deficient plants (Lezhneva et al., 2014). Additionally, AtNRT2.7 regulates nitrate content and dormancy in seeds. AtNRT2.7 was shown to be specifically expressed in seeds and was the only NRT2 transporter located in the vacuole membrane, where it was involved in the storage of (Chopin et al., 2007). In addition to its role in various processes related to , overexpression of NRT2.1 and NRT 2.2 in wheat resulted in increased yield. Similarly, overexpression of NRT2.1, NRT2.2, and NRT2.3a in rice increased crop yields by 10-20% (He et al., 2015; Chen et al., 2017). Furthermore, NRT2 family proteins were also shown to be involved in responding to drought and salinity stresses (Akbudak et al., 2022).
In the case of insufficient nitrogen concentration, the chlorophyll concentration of the plant decreases, turning the leaves yellow and weakening its photosynthetic capacity, which ultimately leads to a significant loss of crop yield (Wen et al., 2020). The distribution of nitrogen in plants is concentrated in the most active part of life activities, so adequate nitrogen supply greatly affects the growth and development of plants (Kant, 2018; Vidal et al., 2020). Cotton is the most significant fiber and cash crop, and nitrogen deficiency can lead to dwarfing of cotton plants, loss of branches and bolls, yellowing of leaves, and severe premature aging and early maturation. Excessive non-protein nitrogen in the plant body results in overly luxuriant growth, large leaves, long petioles, light green leaves, continuous birth of axillary buds, more bud shedding, late maturation, susceptibility to diseases and insect attack, an increase in cotton rotten bolls, and poorer fiber quality (Shah et al., 2022). Nitrogen has an effect on the growth and development of cotton in characteristics such as plant growth morphology, leaf morphology and physiological characteristics, and effects on dry matter accumulation and yields (Iqbal et al., 2020). Nitrogen fertilizers showed different degrees of effects on cotton fiber strength, length, uniformity, and macron value. The increase of nitrogen use efficiency helps increase the fiber length and uniformity of cotton, but excessive nitrogen fertilizer dosage can lead to significant decreases in fiber length and uniformity. Fiber strength and macron value tended to decrease with the increase of nitrogen application rate (Khan et al., 2017).
Modern breeding programs often aim to achieve high yields with high nitrogen supply, which can lead to the prevalence of low nitrogen use efficiency varieties (Liu et al., 2021). Although nitrogen use efficiency can be improved through field management, genetic improvement to produce varieties with high nitrogen use efficiency represent a more basic strategy. As a family of high-affinity nitrogen transporters, the NRT2 family is essential for the nitrogen utilization in cotton. In this study, we performed bioinformatics analysis of two major cultivars, G.hirsutum and G.barbadense, as well as their possible parental materials G.arboreum and G.raimondii. By applying nitrate, we were able to determine the expression pattern and response of the NRT2 family, as well as the nitrogen supply level of G.hirsutum. We also used the expression of the NRT2 family under stress conditions to characterize their coping mechanisms. The results presented here represent a foundation for enhancing the ability to use nitrogen by providing evidence and recommendations for further describing the function of the NRT2 family in cotton.
2 Materials and methods
2.1 Identification of NRT2 genes in cotton
The complete genome sequence data of G.hirsutum (HAU), G.barbadense (HAU), G.arboreum (CRI), and G.raimondii (JGI) were downloaded from the CottonFGD (https://cottonfgd.net/(accessed on 1 September 2022)). The protein-conserved domain PF07690 (MFS_1) of NRT2s was downloaded from the PFAM database (http://pfam.xfam.org), and the identified A.thaliana NRT2 gene in the TAIR (https://www.arabidopsis.org/) was also downloaded. The A.thaliana NRT2s (AT1G08090, AT1G08100, AT5G60780, AT5G60770, AT1G12940, AT3G45060, AT5G14570) sequences were then used to perform a BUPP search in CottonFGD with an e-value set to 1e-10, in order to obtain the protein database of cotton NRT2. The proteins were aligned using HMM and Blastp, and the domain was reconfirmed using the CCD database (http://www.NCBI.nlm.nih.gov/CDD/) on the NCBI website. Proteins that did not meet the conditions were eliminated, and the remaining non-redundant proteins were listed as cotton NRT2 proteins. The physical and chemical parameters of the four cotton NRT2 protein sequences, including molecular weight, isoelectric point (pI), and protein length, were calculated using the ExPAsy online tool (https://web.expasy.org/protparam/) (Wilkins et al., 1999). Subcellular localization prediction was carried out using the 1) UniPort (https://www.uniprot.org/) 2) WoLF PSORT (https://wolfpsort.hgc.jp/) 3) Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) for Subcellar localization prediction. online functional tool.
2.2 Multi-sequence alignment and phylogenetic analysis
To explore the evolutionary relationship between NRT2 proteins in different species, the identified NRT2 family sequences of rice (O.sativa) and tomato (Solanum lycopersicum) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) (Cai et al., 2008; Akbudak et al., 2022). The Arabidopsis NRT2 sequences and the cotton NRT2 protein sequences were also obtained. Using these sequences, a phylogenetic tree was constructed using the neighbor-joining method in MEGA 7.0, with a bootstrap value of 1000 and other parameters set to the default value. The resulting tree was then decorated using EVOLVIEW (https://www.omicsclass.com/article/671). Multiple sequence alignments of the protein sequences were generated using MUSCLE with default parameters and subsequently trimmed using trimAl (Kumar et al., 2016).
2.3 Conservation motif and gene structure analysis of NRT2 gene in cotton
The protein sequences of cotton were downloaded from cottonFGD and calibrated using MUSCLE with default parameters. A phylogenetic tree was then constructed using the adjacency ligation method with 1000 guided replications in MEGA7.0. To search for conservative motifs, the online tool MEME was used with optimization parameters set to a maximum of 10 conserved domains in each gene, a minimum and maximum width of each motif of 6-50 amino acid sequences, and default settings for other parameters. The gene structure of NRT2 in cotton was predicted using the online tool GSDS (http://GSDS.cbi.pku.edu.cn/) (Hu et al., 2015). Finally, TBtools were used to visualize the results of conserved motif analysis, genome annotation, motif information, and the constructed evolutionary tree of cotton.
2.4 Repeat and chromosomal localization analysis of NRT2 gene in cotton
The gene annotation file for four cotton types (G.arboretum, G.raimondii, G.hirsutum, G.barbadense) was downloaded from cottonFGD. The chromosomal distribution information of the NRT2 gene was obtained from the reference cotton database, and the distribution of NRT2 genes on the four cotton chromosomes was visualized using TBtools and plotted on chromosome location images using MapChart (Chen et al., 2020). To detect gene duplication, the CDS sequences of the NRT2 gene were compared, and gene pairs that met the criteria for tandem repeat genes (i.e., two or more genes within 200 kb and not less than 75% similarity) were identified. The synonymous (Ks) and non-synonymous (Ka) substitution ratios were calculated using TBtools for the gene pairs and CDS sequences of G.hirsutum, G.arboreum, and G.raimondii. The Ka/Ks value was used to assess the selection pressure for each gene pair during evolution, with Ka/Ks ratios of >1, = 1, and<1 indicating positive selection pressure, neutral evolution, and purifying selection pressure, respectively. To analyze the homology relationship of the orthologous NRT2 gene obtained from cotton and other selected species, a homology analysis plot was constructed using the Multicollinearity Scanning Toolkit MCScanX (Wang et al., 2012).
2.5 Analysis of cis-acting elements in the promoter region of the NRT2 gene in cotton
Using cottonFGD, the genomes of four different cotton species were obtained. The promoter sequence of the cotton NRT2 family was extracted from the 1500bp upstream region of each gene with the start codon (ATG) based on the location of gene on the chromosome. PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was then used to identify the cis-acting regulatory elements in the promoter sequences. The identified elements were visualized using TBtools.
2.6 Expression profiles of NRT2 gene in different tissues of cotton
RNA-seq data for the TM-1 cultivar (PRJNA248163) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) to observe the expression patterns of the GhNRT2 family genes. The RNA-Seq data for TM-1 includes various growth stages such as root, stem, leaf, torus, bract, sepal, petal, filament, and anther at different stages of development.
2.7 NRT2 gene subcellular localization analysis
The open reading frames (ORF) of GhNRT2 family were inserted into the pCAMBIA1302 vector. This construct was introduced into A.tumefaciens EHA105 (pSoup), which was subsequently transformed into cotton protoplast (Wang et al., 2022). The protoplast was analyzed by confocal microscope (Laser confocal super-resolution microscope LSM980) with bright field and fluorescence imaging. All the primers used for this experiment are shown in Supplementary Table S1.
2.8 Spatial-temporal expression analysis of plant material and stress treatment
XLZ-36 (a common G.hirsutum) plants were grown in a mixture of vermiculite and nutrient rich soil (1:2, w/w) at 28°C/25°C (light/dark) under a 16 hour photoperiod. Plants grown for three weeks were used in the experiments. For stress treatment, seedlings were treated with 300mM NaCl solution as salt, 4% PEG6000 solution as drought, 42°C as heat, and 10°C as cold for five days, and each treatment consisted of three replications. After treatment, leaves were excised, immediately frozen with liquid nitrogen, and stored at -80°C until analysis.
2.9 RNA isolation, cDNA synthesis, and qRT−PCR
Total RNA was extracted from the samples using the FastPure Plant Total RNA Isolation Kit (Polysaccharides and Polyphenolics-rich, RC401-01, Vazyme Biotech, Nanjing, China). The RNA was treated with DNase I to remove any DNA contamination. Reverse transcription was performed on 2 μg of total RNA using the HiScript III All-in-one RT SuperMix Perfect kit for qPCR, R333-01 (Vazyme). Quantitative real-time PCR was performed on three biological replicates using the ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme) with the ABI 7500 Real-Time PCR system. The expression levels of the target genes were normalized to the expression levels of cotton histone H3, which is known for its stable expression across different tissues, developmental stages, and environmental conditions (Zhu et al., 2017). The 2−ΔΔCT method (Livak and Schmittgen, 2001) was used to analyze the relative expression levels of the target genes. The primer sequences are provided in Supplementary Table S2.
3 Results
3.1 Identification of NRT2 genes in G.raimondii, G.arboreum, G.barbadense, and G.hirsutum
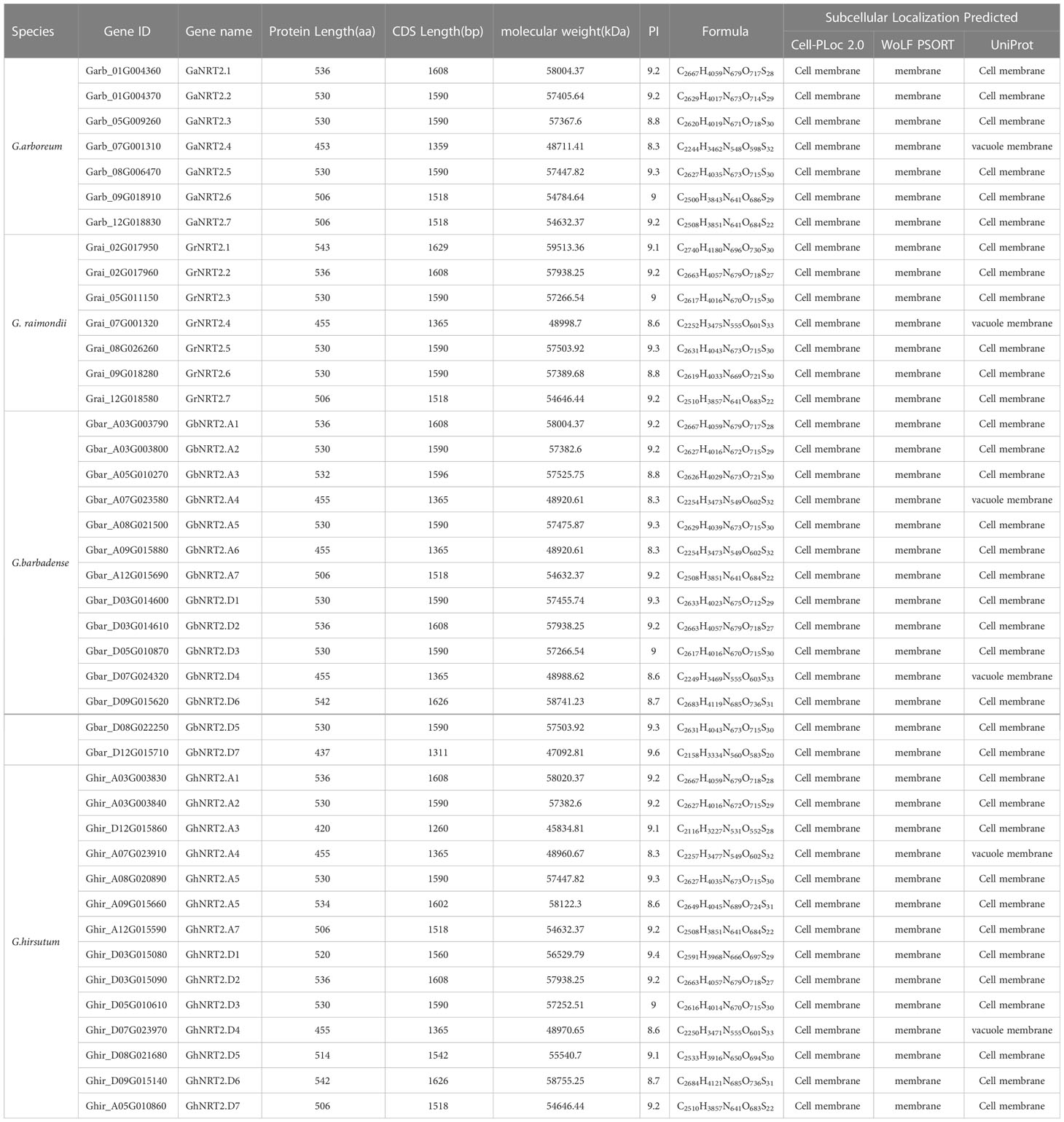
To identify NRT2 genes in four cotton species (G.arboreum, G.raimondii, G.barbadense, and G.hirsutum), the protein sequences of NRT2 genes in A.thaliana were used as a query in BLASTp. A total of 42 NRT2 domain-containing genes were identified, with 7, 7, 14, and 14 genes found in G.arboreum, G.raimondii, G.hirsutum, and G.barbadense, respectively. In G.hirsutum and G.barbadense, seven genes were identified in the A genome and seven in the D genome. The genes were named according to their chromosome locations in cotton, with names such as GaNRT2.1-2.7 (G.arboreum), GrNRT2.1-2.7 (G.raimondii), GhNRT2.A1-7, GhNRT2.D1-7 (G.hirsutum), GbNRT2.A1-7, and GbNRT2.D1-7 (G.barbadense). The NRT2 gene in G.arboreum (AA) and G.raimondii (DD) represented half the number of NRT2 genes in G.hirsutum and G.barbadense (AADD) in two tetraploid cottons, indicating that the NRT2 family remained conserved during evolution. Table 1 provides details of the gene ID number, coding region (CDS) length, protein length, molecular weight (MW), isoelectric point (pI), and other properties of the identified cotton NRT2 family proteins. The proteins encoded by the 42 genes showed different physicochemical properties, with protein length varying between 420-540 amino acids, molecular size between 45,834.81-59,513.36, and theoretical isoelectric points (pl) between 8.27-9.61. All of the discovered members of the cotton NRT2 family were found to be basic proteins, and the prediction results of subcellular localization indicate that all 42 cotton NRT2 proteins were localized to the membrane.
3.2 Conserved motif and gene structure analysis of NRT2 in cotton
To investigate the evolutionary relationship between NRT2 members in G.arboreum, G.raimondii, G.barbadense, and G.hirsutum, a phylogenetic tree was created using 42 cotton NRT2 genes, which were categorized into three subfamilies. The resulting tree showed that NRT2 genes from the four cotton species were closely grouped in each subfamily. In order to study the gene structure of the cotton NRT2 family genes, coding sequences were mapped onto the genome sequences of exon/intron sequences. The analysis revealed that the exon/intron structure of the cotton NRT2 gene was generally well-conserved across different varieties (Figure 1).
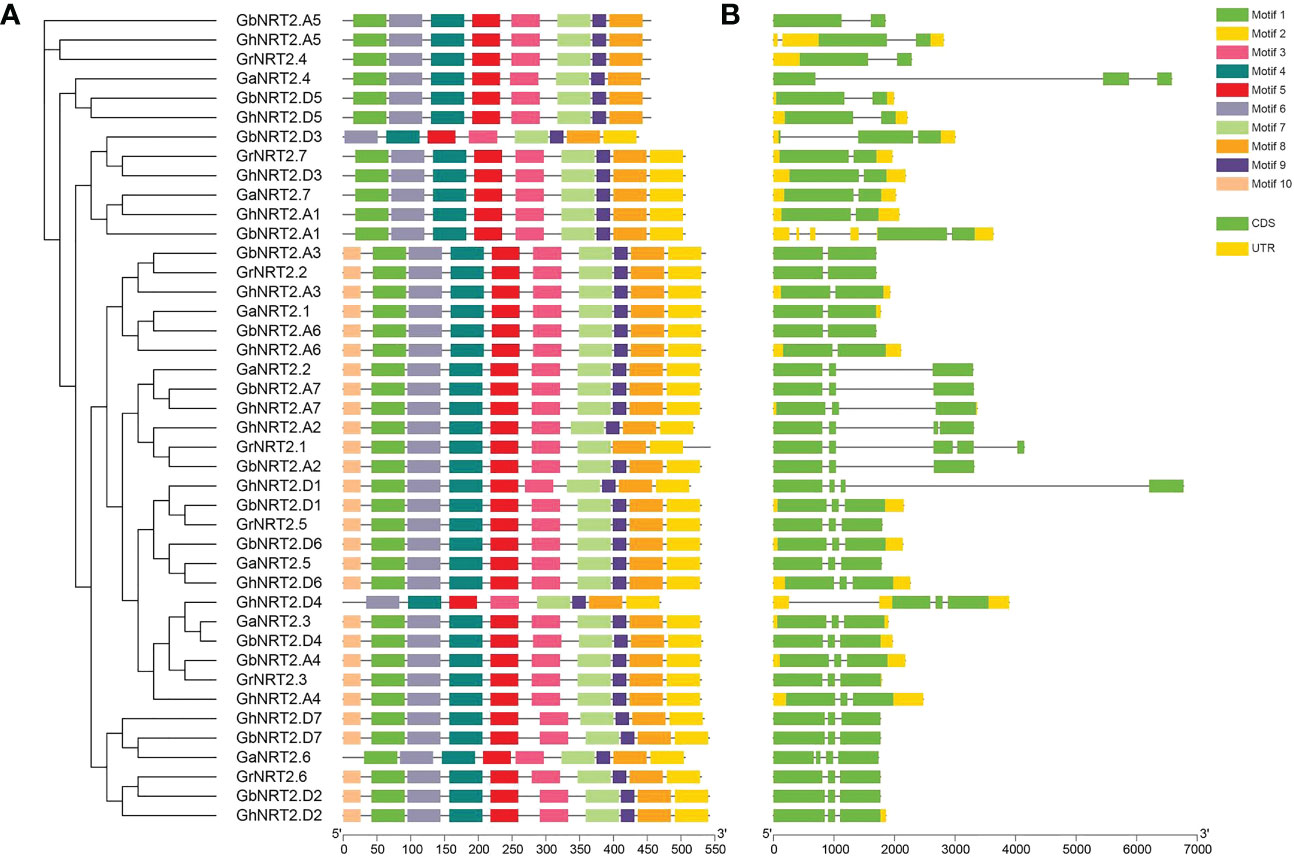
Figure 1 Phylogenetic tree and sequence structures of the candidate NRT2s in four cotton species (G.arboreum, G.raimondii, G.barbadense, and G.hirsutum). (A) Neighbor-joining (NJ) phylogenetic tree of NRT2s in four cotton species. Different colors represent different motifs. (B) Gene structures of NRT2s in cotton. CDS are shown as green boxes, UTR are shown as yellow boxes, and the lines between the colored boxes correspond to the introns. Numbers below represent the position.
To gain further insight into the structural characteristics of the NRT2 genes in cotton, we employed the MEME server to identify ten conserved motifs. We found that motifs 2, 3, 4, 5, 6, 7, 8, and 9 were present in all NRT2 protein sequences (Figure 1). Groups I and II contained the same motifs (1, 3, 4, 5, 6, 7, 8, 9), whereas GbNRT2.D3 and GhNRT2.D4 in Group III lacked motifs 1 and 10. This result suggests that members of the same family have similar functions.
To further examine the phylogenetic relationships, the arrangement of exons and introns of the cotton NRT2 genes were studied. The findings showed that the number of exons and introns in the cotton NRT2 gene ranged from 2 to 5. Group I and II genes shared a similar structure, with one intron and two exons, while most members of Group III had a comparable exon-intron structure and gene length (Figure 1).
3.3 Chromosomal location, gene duplication and evolutionary relationships of Gossypium NRT2 family genes
Cotton NRT2 genes are not distributed on all 13 chromosomes. Based on the annotation of four cotton genomes, 14 GhNRT2s and 14 GbNRT2s genes were assigned to 12 chromosomes, of which 7 were located on the A-genome, 2 genes on chromosome A03 (GbNRT2A1, GbNRT2A2 and GhNRT2.A1, GhNRT2.A2), and one gene on chromosome A05, 7, 8, 9, 12 (GbNRT2.A3, GbNRT2.A4, GbNRT2.A5, GbNRT2.A6, GbNRT2.A7 and GhNRT2.A3, GhNRT2.A4, GhNRT2.A5, GhNRT2.A6, GhNRT2.A7). The remaining 7 were located on the D-genome, 2 genes on chromosome D03 (GbNRT2.D1, GbNRT2.D2 and GhNRT2.D1, GhNRT2.D2), D05, 7, 8, 9, and a gene on chromosome 12 (GbNRT2.D3, GbNRT2.D4, GbNRT2.D5, GbNRT2.D6, GbNRT2.D7 and GhNRT2.D3, GhNRT2.D4, GhNRT2.D5, GhNRT2.D6, GhNRT2.D7). The NRT2 gene of G.arboreum was assigned to chromosomes 1 (GaNRT2.1 and GaNRT2.2), 5 (GaNRT2.3), 7 (GaNRT2.4), 8 (GaNRT2.5), 9 (GaNRT2.6), and 12 (GaNRT2.7). A total of 7 genes of G.raimondii were assigned to two genes on chromosome 02 (GrNRT2.1 and GrNRT2.2) and one gene on 05 (GrNRT2.3). There was one gene each on chromosomes 7 (GrNRT2.4), 8 (GrNRT2.5), 9 (GrNRT2.6), and 12 (GrNRT2.7) (Figure 2). Previous studies have shown that G.arboreum and G.raimondii were donor species for the A and D genomes.
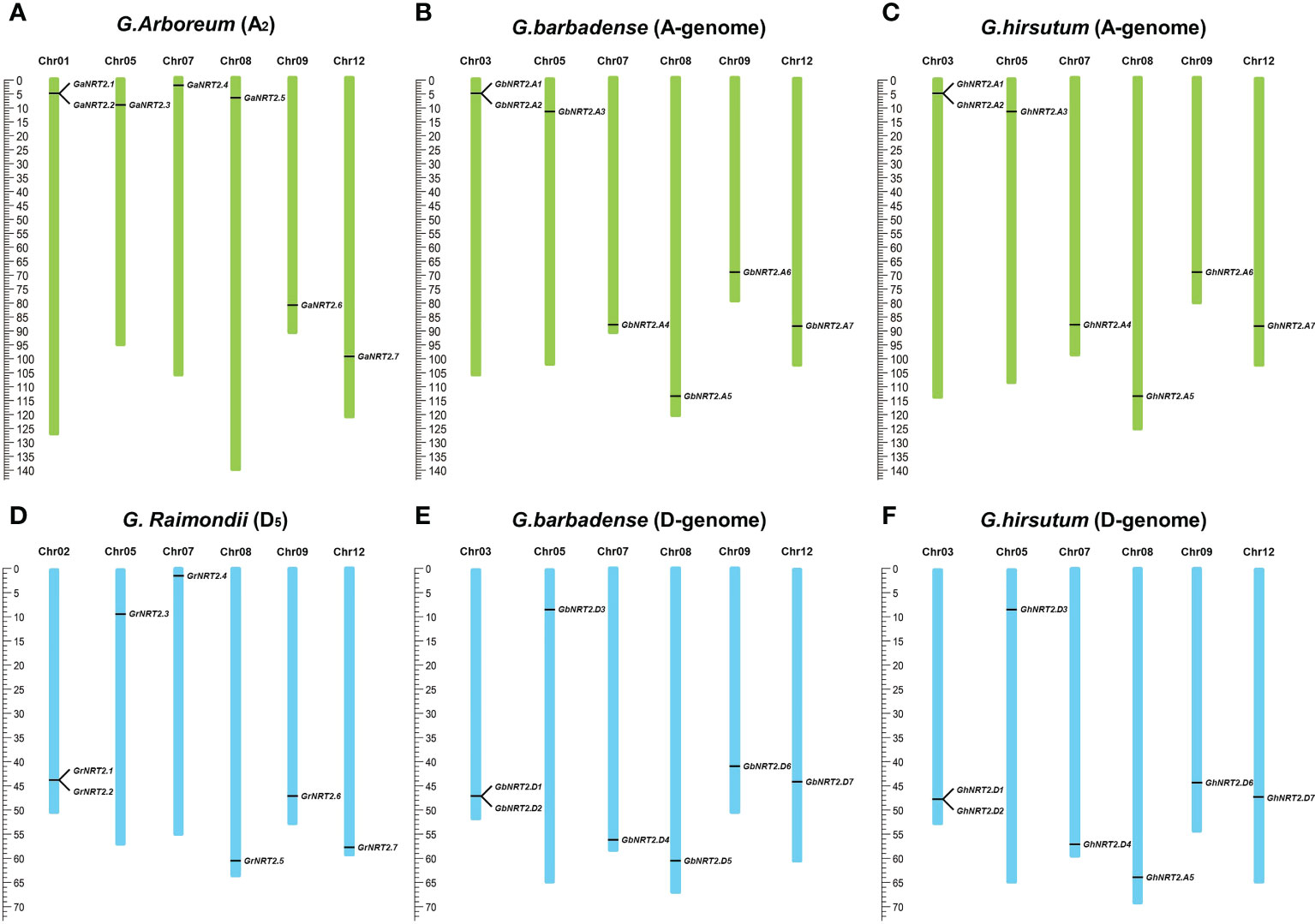
Figure 2 Chromosome distribution and duplication of GhNRT2s. (A) 7 GaNRT2s were mapped on 6 chromosomes in G.arboreum; (B) 7 GbNRT2s were mapped on 6 chromosomes in G.barbadense (A-genome); (C) 7 GhNRT2s were mapped on 6 chromosomes in G.hirsutum (A-genome); (D) 7 GrNRT2s were mapped on 6 chromosomes in G.raimondii; (E) 7 GbNRT2s were mapped on 6 chromosomes in G.barbadense (D-genome); (F) 7 GhNRT2s were mapped on 6 chromosomes in G.hirsutum (D-genome).
The number of GhNRT2 and GbNRT2 genes present in the A and D genome of cotton was consistent with the number of GaNRT2 and GrNRT2 genes. Tandem repeats of NRT2.1 and 2.2 were observed in all four types of cotton, indicating possible gene duplication. To gain insight into the relationship between NRT2 genes and potential gene duplication within the cotton genome, we analyzed the occurrence of tandem duplication and segmental duplication during the evolution of this gene family. We calculated the non-synonymous differentiation level (Ka) and the synonymous differentiation degree of 35 homology pairs to infer possible selection pressure. The Ka/Ks ratio was used to assess the selection pressure on repeating genes. A Ka/Ks ratio of 1 suggests pseudo genes with natural selection; a Ka/Ks ratio<1 indicates that duplicate genes had a tendency to purify, and a Ka/Ks ratio >1 indicates accelerated evolution with positive selection. Our analysis revealed that the Ka/Ks ratio of all 25 gene pairs was less than 0.5, leading us to hypothesize that the NRT2 gene family in cotton experienced strong pressure for purification selection.
To further investigate the genetic origins and evolutionary relationships of NRT2s in cotton, we conducted collinear analyses of cotton in comparison with A.thaliana, rice, and tomato. The collinear analysis results of four cotton varieties were mainly divided into two forms within species. In G. arboreum and G. raimondii, NRT2.1 and NRT2.2 were collinear with NRT2.3, NRT2.5, and NRT2.6, while NRT2.3 was collinear with NRT2.5 and NRT2.6. In G. barbadense and G. hirsutum, NRT2.A1/A2 was collinear with NRT2.A5, NRT2.D1/D2, NRT2.D3, NRT2.D4, NRT2.D5, and NRT2.D6 (Figure 3). These results suggest that the internal gene sequencing of the homologous fragment was conserved and may also be functionally conserved. Additionally, we compared different species and found that AtNRT2.1, AtNRT2.4, and AtNRT2.6 were collinear with GhNRT2.1, GhNRT2.3, and GhNRT2.5 in G. hirsutum, while AtNRT2.5 was collinear with GhNRT2.6 and GhNRT2.7, and AtNRT2.7 was collinear with GhNRT2.4 (Figure 3). These results indicate that these genes possess high similarities in structure and function across species.
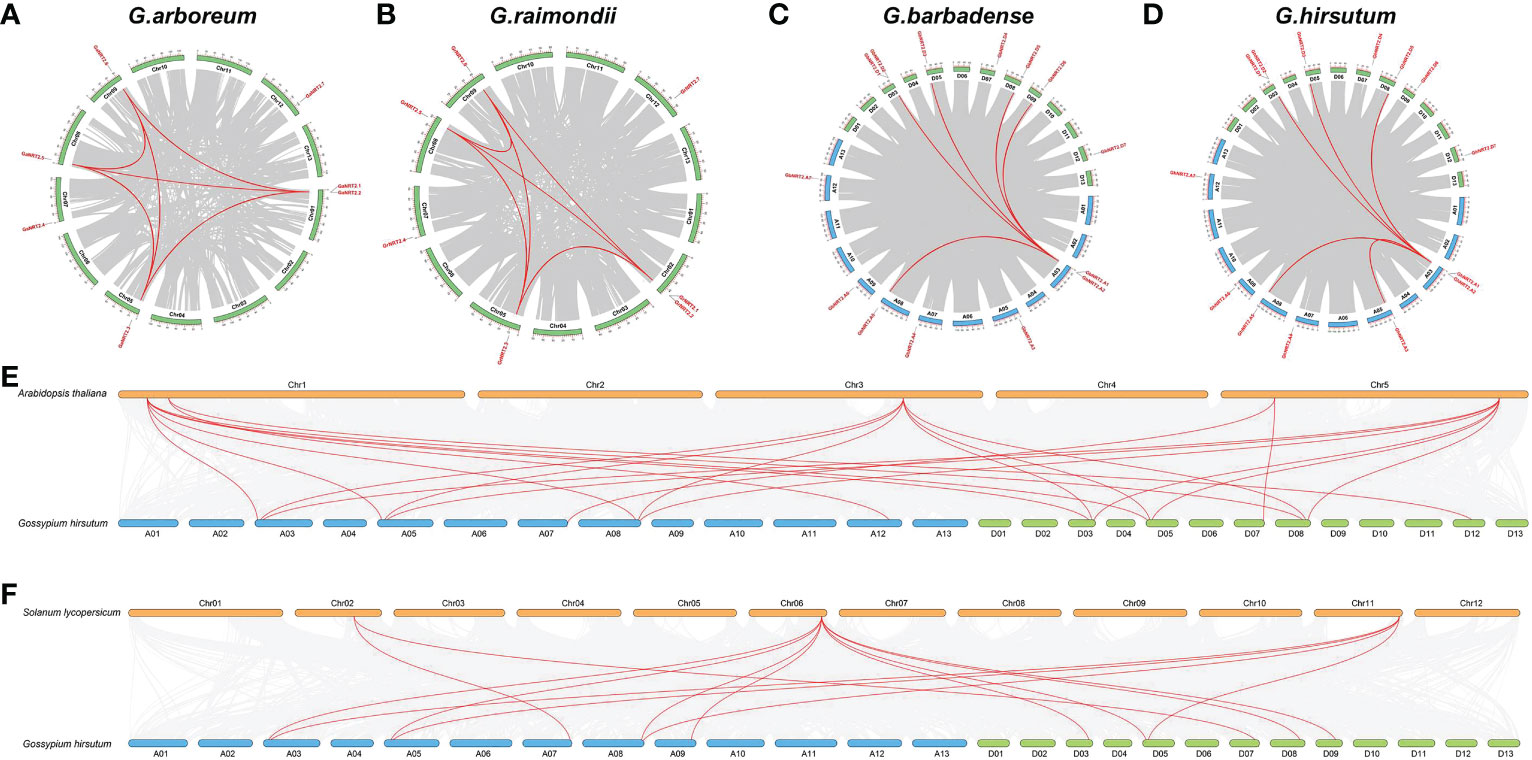
Figure 3 Synteny analysis of the GhNRT2s genes. (A) G.arboreum; (B) G.raimondii; (C) G.barbadense; (D) G.hirsutum; (E) Arabidopsis thaliana and G.hirsutum; (F) Solanum lycopersicum L. and G.hirsutum.
3.4 Multiple sequence alignment and phylogenetic analysis
In order to study the evolutionary relationship between cotton NRT2 genes (G.arboreum, G.raimondii, G.barbadense and G.hirsutum) and other species, we identified 57 NRT2 genes in 7 genomes (cotton, Arabidopsis, rice, tomato), including 14 each for G.barbadense and G.hirsutum, 7 each for G.arboreum and G.raimondii, 4 for rice, 4 for tomatoes, and 7 for Arabidopsis. We then performed phylogenetic analysis of these 57 NRT2 protein sequences, and constructed an evolutionary tree using the Neighbor-Joining (NJ) method. Phylogenetic analysis revealed the relatively deep evolutionary origins of these genes as well as recent duplications.
According to the topology and boot values of the tree, the candidates were divided into three groups: group I–III and group I had 7 members, including Arabidopsis AtNRT2.5, 2 GhNRT2s, 2 GbNRT2s, GrNRT2.7 and GaNRT2.7; Group II had 9 members (OsNRT2.4, AtNRT2.7, SINNRT2.1, 2 GbNRT2s, 2 GhNRT2s and GaNRT2.4); Group III was the largest and included 3 SINNRT2s, 5 AtNRT2s, 3 OsNRT2s, 10 GbNRT2s, 10 GhNRT2s, 5 GrNRT2s, and 5 GaNRT2s (Figure 4). The results showed that the genes located in group I were functionally similar to AtNRT2.5 of A.thaliana, and played a role in nitrate acquisition and reuse in nitrogen-deficient plants (Chopin et al., 2007). The related genes located in group II were consistent with AtNRT2.5 of A.thaliana and OsNRT2.4 of rice. Acquisition and root-to-stem nitrate transport had the same function (Lezhneva et al., 2014; Xia et al., 2015).
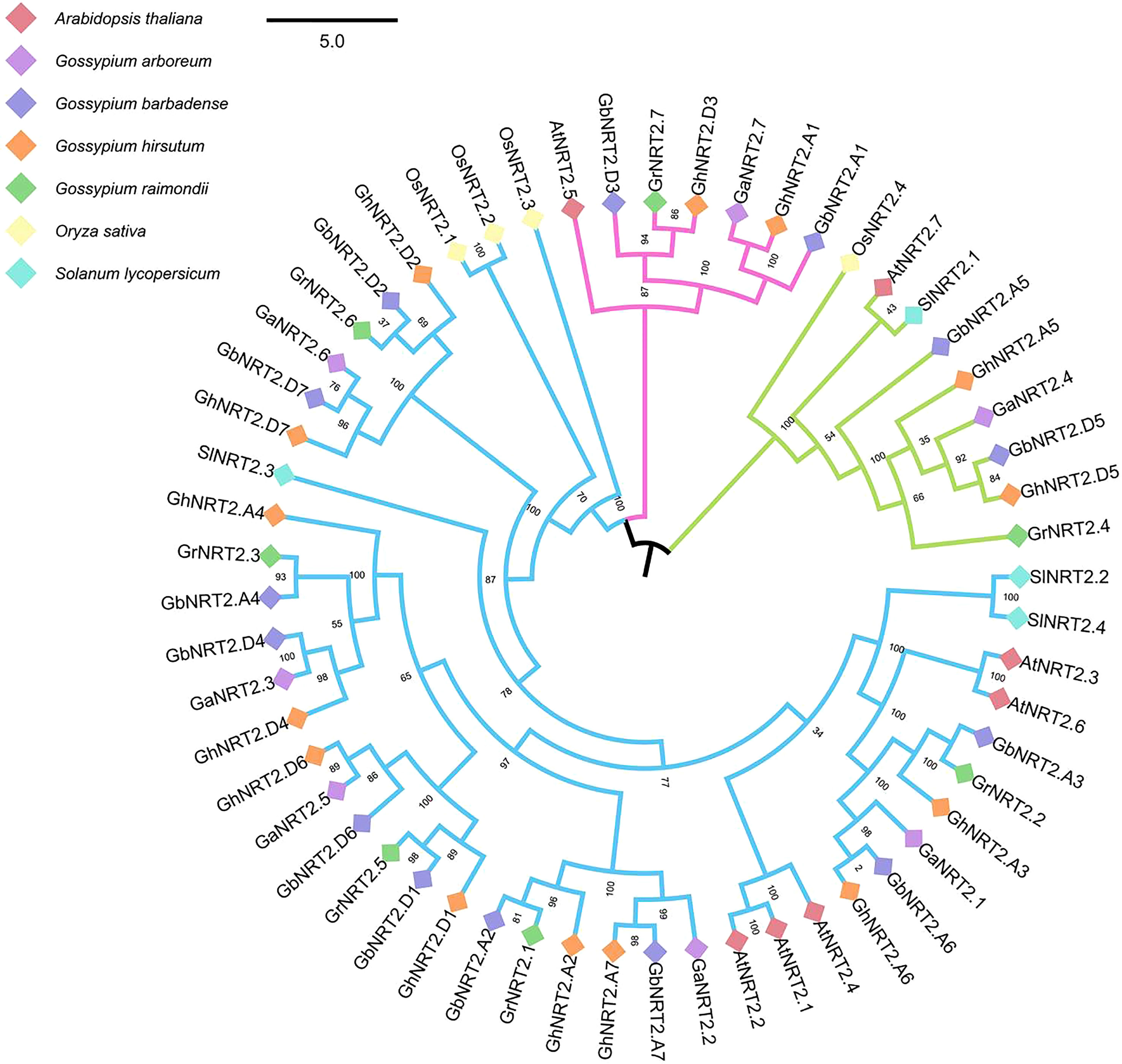
Figure 4 Phylogenetic analysis of NRT2 gene families Phylogenetic analysis of the NRT2 gene families in cotton, Arabidopsis, rice, and tomato. Different groups are colored by a special line. Different plants use small squares of different colors. The number beside the branch indicates the bootstrap value of each group from 1,000 replicates.
3.5 Cis-element analysis in the promoter regions of cotton NRT2 genes
To understand the underlying regulatory mechanism, we predicted cis-acting elements and analyzed the 1,500bp upstream promoter region of 42 cotton NRT2s via PlantCARE. In addition to some common basic core elements (e.g., A-box, TATA-box) and photo-responsive elements (e.g., ACE), the remaining elements were divided into five broad categories, including hormone-responsive elements, protein-binding sites, special response elements, tissue-specific expression elements, and biotic and abiotic response elements. Hormonal response elements included abscisic acid (ABRE), salicylic acid (TCA-element), gibberellin (P-box, TATC-box), jasmonic acid (TGACG motif, CGTCA motif), and auxin (AuxRR-core). In the group of protein binding sites, 13 cis-acting elements were found to interact specifically with the C-terminal domain of 60K, and in the group of special reaction elements, 4 cotton NRT2s may be involved in the regulation of zein metabolism (O2 site). In the tissue-specific expression element group, 21 cotton NRT2s may be associated with meristem activation and expression (GCN4_motif, HD-Zip 1, and CAT-box). In the biotic and abiotic cis-responsive cis-component group, 3, 12, and 17 cotton NRT2s may be involved in hypoxia (GC-motif), low temperature (LTR), and defense and stress (TC-rich repeats) response processes, and 23 cotton NRT2s were associated with sites in drought (MBS) response.
3.6 NRT2 gene subcellular localization analysis
Three G.hirsutum genes were arbitrarily chosen for subcellular localization in order to assess the accuracy of the software’s predictions. The pCAMBIA1302-GhNRT2.A1, GhNRT2.A3, and GhNRT2.A5 vectors were constructed and transformed into the Agrobacterium tumefaciens strain EHA105 (pSoup) to determine the localization of the GhNRT2.A1, GhNRT2.A3, GhNRT2.A5 proteins by transient expression in cotton protoplast. The GhNRT2.A1, GhNRT2.A3, and GhNRT2.A5 proteins appeared to be localized to the cell membrane, according to the GFP fluorescence signal generated by laser confocal microscopy (Figure 5).
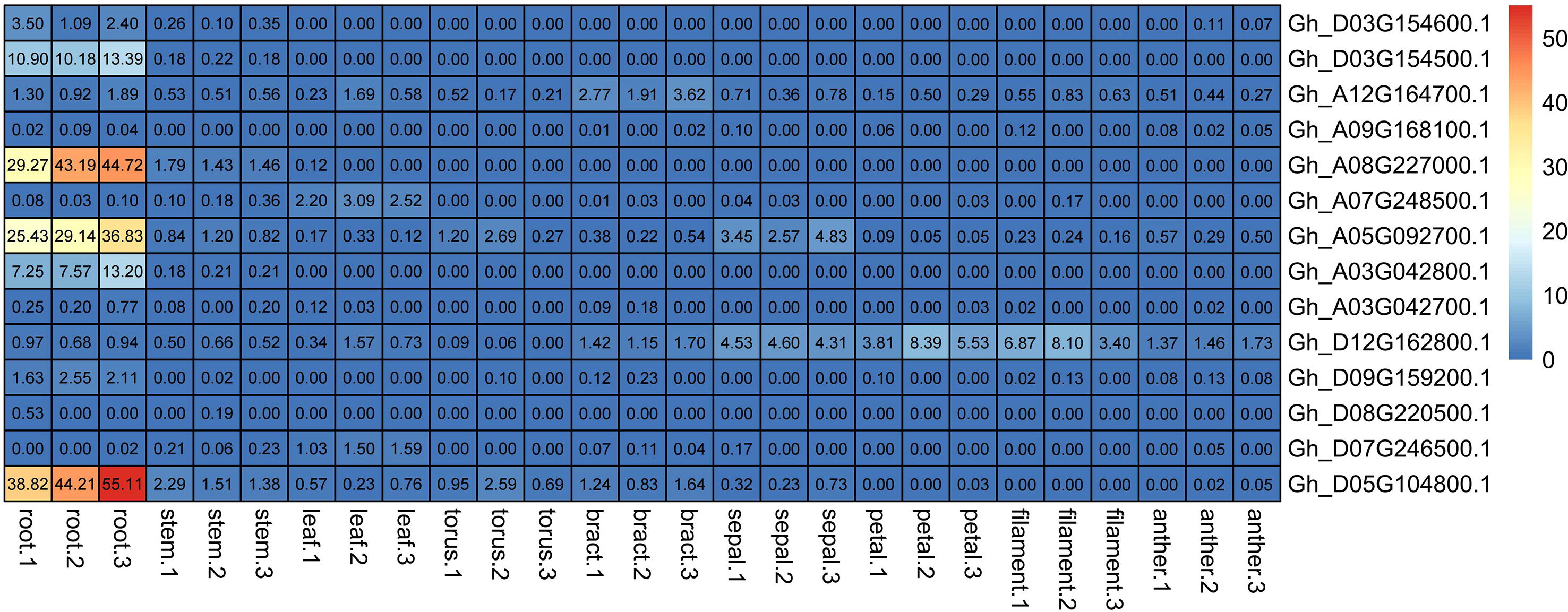
Figure 5 GhNRT2s genes expression in different organ. Expression heat map of GhNRT2s family genes in different organ transcriptomes. All data were downloaded in cottonFGD (https://cottonfgd.net/, on October 18, 2022).
3.7 Analysis of NRT2 gene expression in different tissues of cotton
RNA-seq expression data in different tissues of the cotton NRT2 gene family were obtained from the cotton database CottonFGD. Due to the lack of data, there was only relevant data for G.hirsutum in the database, so we chose G.hirsutum for characterization. From the RNA-seq data, the NRT gene family showed obvious root specificity. Almost all of the NRT genes had high expression specifically in the roots. In addition to this, the NRT genes were also expressed in the leaves and stems. Among GhNRT2 genes, GhNRT2.A4 showed specific high expression in leaves, while GhNRT2.A6 and GhNRT2.D7 genes were expressed as a specificity of flower organs (Figure 6). To verify the accuracy of these data, we verified this result by qRT-PCR. Considering the homologous relationship, we chose GhNRT2.A1-A7 to determine their expression level (Figure 7). The results were almost identical to the RNA-seq data, which showed expression at specific high levels in the root and in both leaves and stems (Figure S1). Among GhNRT2 genes, GhNRT2.A4 showed high expression in leaves, and these results were consistent with the correlation between the expression pattern of these genes and their driving function.
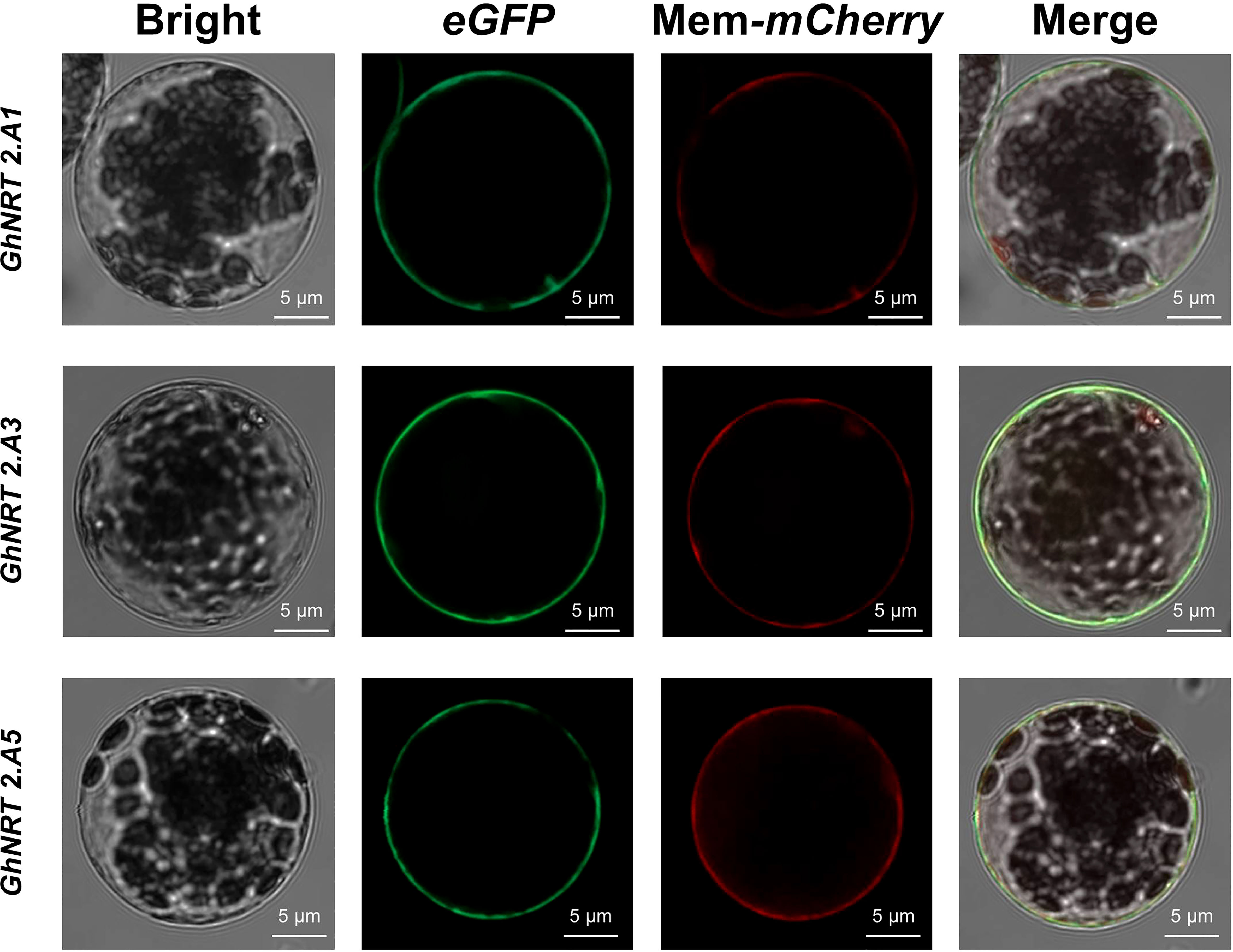
Figure 6 GhNRT2s genes Subcellular localization. Subcellular localization assay with cotton protoplast using the pCaMV35S::GhNRT2.A1/GhNRT2.A3/GhNRT2.A5-eGFP construct. Red fluorescence is mCherry of membrane localization. Scale bars = 5 µm.
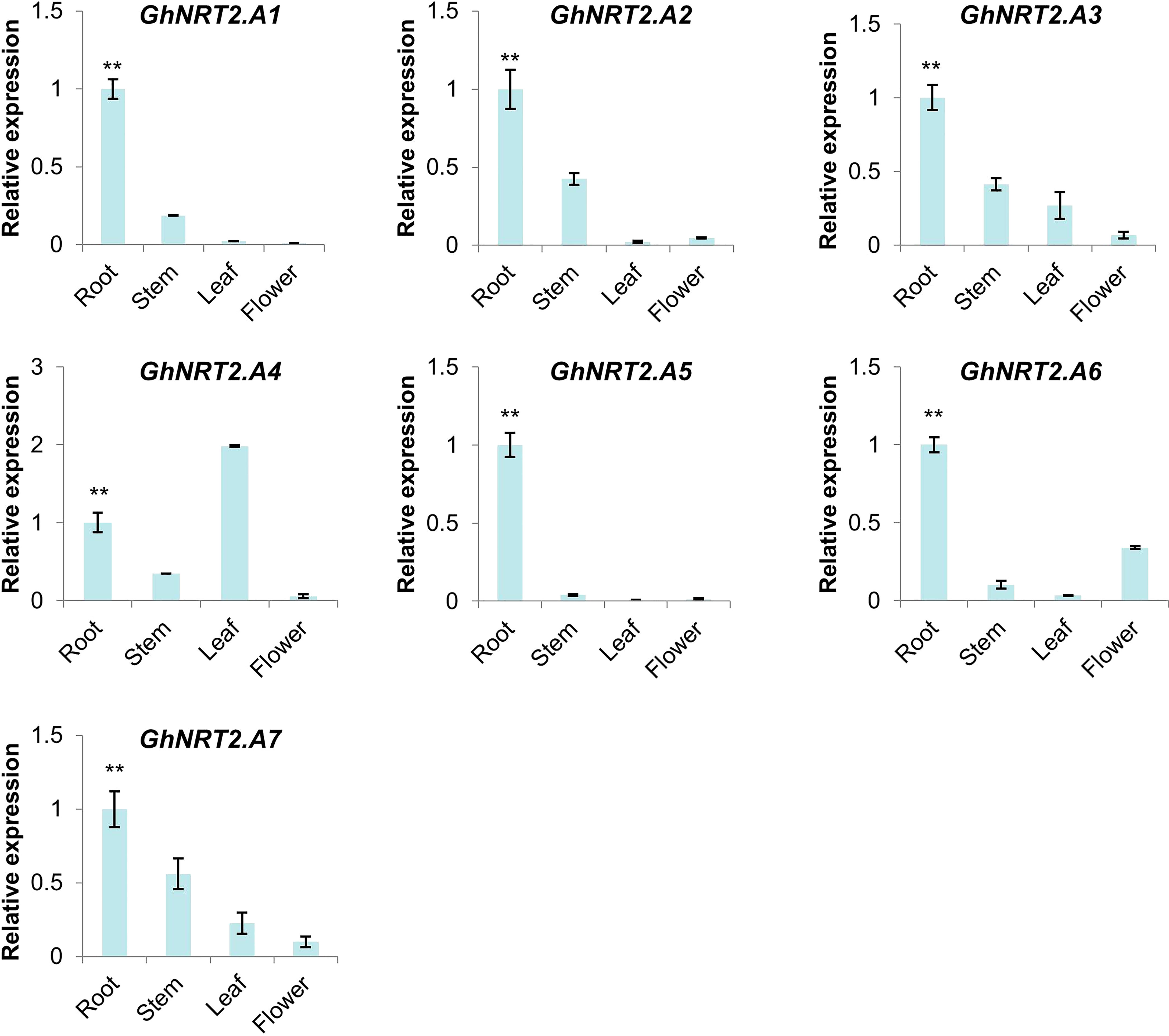
Figure 7 The expression of GhNRT2.A1-A7 in different organ. The qRT-PCR result of GhNRT2.A1-A7 in root, stem, leaf, and flower of G.hirsutum. Values represent the mean ± S.D (n = 3 replicates). **p< 0.01, Dunn’s test.
3.8 Analysis of NRT2 gene expression in different stress of cotton
To ascertain the response of the NRT2 family genes in cotton to salt, drought, and temperature stress, salt and drought processes by NaCl were simulated. Then, using PEG6000 to imitate drought and salt processes at a high temperature of 42°C and a low temperature of 10°C, expression analysis was carried out. The results showed that the expression levels of GhNRT2.A1-A7 responded to stress, among which GhNRT2.A1, GhNRT2.A3, and GhNRT2.A6 were up-regulated under stress condition, and the expression of GhNRT2.A4 and GhNRT2.A7 was down-regulated. Expression levels of GhNRT2.A2 and GhNRT2.A5 did not change significantly (Figure 8). The results were almost identical to the RNA-seq data (Figure S2).
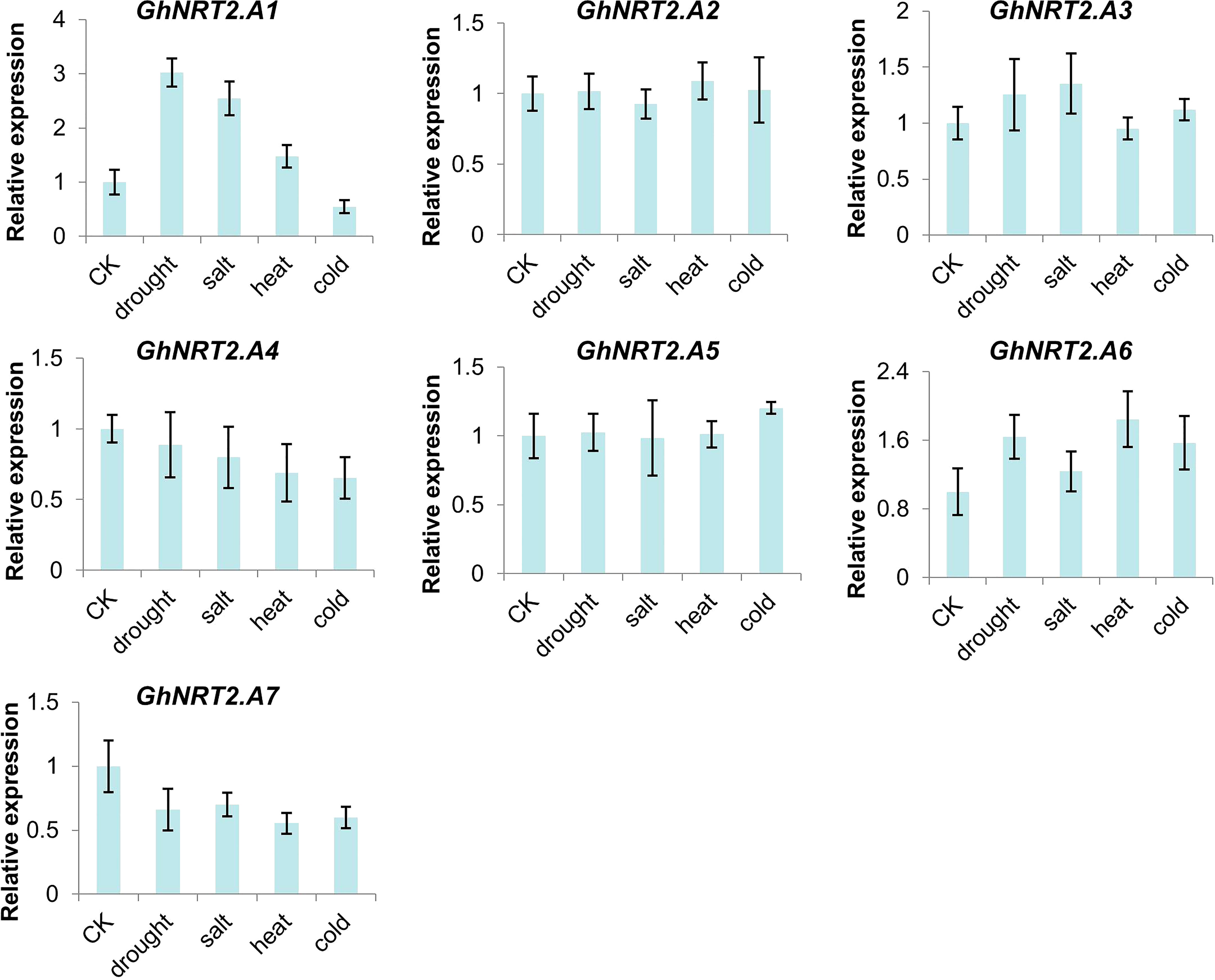
Figure 8 The expression of GhNRT2.A1-A7 in different treatments. The qRT-PCR result of GhNRT2.A1-A7 in root of G.hirsutum treated with drought (4% PEG6000), salt (200 mM NaCl), heat (42°C), and cold (10°C). Values represent the mean ± S.D (n = 3 replicates).
4 Discussion
Nitrate is an essential component for plant growth since it is both a significant nutrient in the nitrogen absorption route and an important signal for plant development. Crop yield is directly related to the availability of nitrogen; in the event of its deficiency, a plant’s chlorophyll concentration level declines, which results in poor photosynthesis activity (Zhao et al., 2022).
In cotton, we found that the NRT2 gene in G.arboreum (AA) and G.raimondii (DD) were essentially half of the total G.hirsutum and G.barbadense (AADD) NRT2 genes in two tetraploid cottons. We showed that there was no copy number variation in the NRT gene during doubling from diploid to tetraploid. Additionally, based on where these genes were located on the genome, they remained almost identical to their diploid parents, which indicates that the NRT2 family remained conserved during the process of evolution. The NRT2 gene family in cotton has experienced strong purification selection pressure.
Cis-acting regulatory element analysis of the cotton NRT2 promoter regions revealed that it has an abundant number of hormone-responsive elements, protein-binding sites, special response elements, tissue-specific expression elements, and biotic and abiotic response elements. Different hormonal response elements like abscisic acid, salicylic acid, gibberellin (P-box, TATC-box), jasmonic acid, and auxin were also part of the cis-acting regulatory elements. Interestingly, although almost all genes in the GhNRT family are specifically expressed in the roots, most GhNRTs still have photoresponsive factors in the promoters of the genes, indicating that the expression of most GhNRTs is also regulated by light. The latest research proves that ET (ethylene)/JA (jasmonic acid)-NRT signaling module participates in − distribution between plant roots and aerial parts in a nitrate reductase-dependent manner, regulating stress tolerance and plant growth (Zhang et al., 2014)
Under stress conditions, the cotton NRT2 gene family responded differently i.e., GhNRT2.A1, GhNRT2.A3 and GhNRT2.A6 were up-regulated under stress conditions, while the expression of GhNRT2.A4 and GhNRT2.A7 were down-regulated. Although we have not performed expression or knockout experiments to demonstrate that genes from the GhNRT family increase stress tolerance in cotton, changes in the expression of these genes in response to stress suggest that they may be involved in the stress response. Zhao and colleagues have demonstrated that the plant may have a Cl/NO3 cooperative transport system through the control of NRTs, which is crucial for salt resistance, especially for Cl sensitive plants (Zhao et al., 2022). In a previous report, the expression of OsNRT2.3 in rice was shown to increase during high temperature stress, increasing high temperature tolerance and nitrogen uptake levels (Zhang et al., 2022), In tomato (Solanum lycopersicum L.), the SlNRT2 family proteins were also involved in the response to drought and salinity (Akbudak et al., 2022). The responses of the SlNRT2 genes to drought and salinity stresses were diverse, and they were neither stress- nor tissue-specific (Akbudak et al., 2022). In wheat (Triticum aestivum L.), the TaNRT2 gene family improved high salt tolerance (Shi et al., 2022).
The expression levels of the NRT2 family of genes changed by various degrees as a result of our four adversity treatments. GhNRT2.A1 and GhNRT2.A7 in particular demonstrated consistent up- or down-regulation of the two genes under various stresses. This implies that dealing with various adversities may involve the same biological pathways. Additionally, because there was only a single sample time in this investigation, it would be possible to more accurately characterize the pattern of gene expression in the NRT2 family in response to stress. These mechanisms still need to be investigated further. The genetic resources available for enhancing cotton resistance to environmental stresses could be greatly increased by focusing on specific NRT2 members that coordinate nitrate absorption and transport in response to signals from various stress response pathways.
Based on our findings, we suggest that NRT2 transporters could be considered as potential targets for plant adaption to varied forms of nitrogen supply and for improving NUE. Since previous studies have shown that the NRT2 family has a role in responding to drought and waterlogging stresses, this same functionality should also be investigated for cotton. Thorough study of the gray leaf spot and bacterial wilt RNA-seq data confirmed that the expression of the NRT1/PTR subfamily members was down-regulated after pathogen attack (Xia et al., 2022). Reports are also available which imply that NRT2 family proteins play a role in plant response to light, which ultimately increases photosynthesis. Previous studies on A.thaliana revealed that AtNRT2.1 was up-regulated during the day and down-regulated at night, which might be hindered by sucrose (Lejay et al., 1999). Our next step is to investigate introgression or expressing altered forms of NRT2 with higher uptake or sensing activity in local cultivars, which may optimize NUE and other genetic traits, and hence can increase the crop yield with minimized fertilizer application.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Conceptualization, YP and PW. Methodology, YP. Software, YP. Validation, PW. Formal analysis, JX. Investigation, TZ and YY. Resources, KZ and GS. Data curation, MK. Writing original draft preparation, YP and PW. Writing review and editing, PW and MA. Visualization, QC. Supervision, GS. Project administration, GS. Funding acquisition, GS. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Hainan Yazhou Bay Seed Lab, grant number B21HJ8201, Basic scientific research business expenses of the Chinese Academy of Agricultural Sciences, grant number YBXM13 and was funded by The National Natural Science Foundation of China, grant number 31771851.
Acknowledgments
We thank Mr. Ye Yulu for his help in revising the manuscript.We thank the core Facility Platform, Institute of Crop Sciences, CAAS for their assistance with Confocal analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor ZG declared a shared affiliation with the authors PW, YP, MA, MK, JX, YY, TZ, GS at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1170048/full#supplementary-material
Supplementary Figure 1 | The expression of GhNRT2.A1-A7 in different organs compared by RNA-seq data and RT-PCR data (For easy comparison, all root data were set to 1).
Supplementary Figure 2 | The expression of GhNRT2.A1-A7 in different treatments compared by RNA-seq data and RT-PCR data (For easy comparison, all CD data were set to 1).
References
Akbudak, M. A., Filiz, E., Çetin, D. (2022). Genome-wide identification and characterization of high-affinity nitrate transporter 2 (NRT2) gene family in tomato (Solanum lycopersicum) and their transcriptional responses to drought and salinity stresses. J. Plant Physiol. 272, 153684. doi: 10.1016/j.jplph.2022.153684
Cai, C., Wang, J. Y., Zhu, Y. G., Shen, Q. R., Li, B., Tong, Y. P., et al. (2008). Gene structure and expression of the high-affinity nitrate transport system in rice roots. J. Integr. Plant Biol. 50 (4), 443–451. doi: 10.1111/j.1744-7909.2008.00642.x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J., Fan, X., Qian, K., Zhang, Y., Song, M., Liu, Y., et al. (2017). pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 15, 1273–1283. doi: 10.1111/pbi.12714
Chopin, F., Orsel, M., Dorbe, M. F., Chardon, F., Truong, H. N., Miller, A. J., et al. (2007). The arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell. 19, 1590–1602. doi: 10.1105/tpc.107.050542
Dechorgnat, J., Patrit, O., Krapp, A., Fagard, M., Daniel-Vedele, F. (2012). Characterization of the Nrt2.6 gene in arabidopsis thaliana: A link with plant response to biotic and abiotic stress. PloS One 7 (8), e42491. doi: 10.1371/journal.pone.0042491
Du, R. J., Wu, Z. X., Yu, Z. X., Li, P. F., Mu, J. Y., Zhou, J., et al. (2022). Genome-wide characterization of high-affinity nitrate transporter 2 (NRT2) gene family in brassica napus. Int. J. Mol. Sci. 23 (9), 4965. doi: 10.3390/ijms23094965
Evans, J. R., Clarke, V. (2019). The nitrogen cost of photosynthesis. J. Exp. Bot. 70 (1), 7–15. doi: 10.1093/jxb/ery366
Frink, C. R., Waggoner, P., Ausubel, J. H. (1999). Nitrogen fertilizer: Retrospect and prospect. Proc. Natl. Acad. Sci. U S A. 96, 1175–1180. doi: 10.1073/pnas.96.4.1175
Guan, M., Chen, M., Cao, Z. (2021). NRT2.1, a major contributor to cadmium uptake controlled by high-affinity nitrate transporters. Ecotoxicol. Environ. Saf. 218, 112269. doi: 10.1016/j.ecoenv.2021.112269
Guo, J. H., Liu, X. J., Zhang, Y., Shen, J. L., Han, W. X., Zhang, W. F., et al. (2010). Significant acidification in major Chinese croplands. Science 327 (5968), 1008–1010. doi: 10.1126/science.1182570
He, X., Qu, B., Li, W., Zhao, X., Teng, W., Ma, W., et al. (2015). The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 169 (3), 1991–2005. doi: 10.1104/pp.15.00568
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Iqbal, A., Qiang, D., Zhun, W., Xiangru, W., Huiping, G., Hengheng, Z., et al. (2020). Growth and nitrogen metabolism are associated with nitrogen-use efficiency in cotton genotypes. Plant Physiol. Biochem. 149, 61–74. doi: 10.1016/j.plaphy.2020.02.002
Kant, S. (2018). Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Semin. Cell Dev. Biol. 74, 89–96. doi: 10.1016/j.semcdb.2017.08.034
Khan, A., Tan, D., Munsif, F., Afridi, M. Z., Shah, F., Wei, F., et al. (2017). Nitrogen nutrition in cotton and control strategies for greenhouse gas emissions: A review. Environ. Sci. pollut. Res. Int. 24 (30), 23471–23487. doi: 10.1007/s11356-017-0131-y
Kiba, T., Feria-Bourrellier, A. B., Lafouge, F., Lezhneva, L., Boutet-Mercey, S., Orsel, M., et al. (2012). The arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell. 24 (1), 245–258. doi: 10.1105/tpc.111.092221
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Lejay, L., Tillard, P., Lepetit, M., Olive, F.d., Filleur, S., Daniel-Vedele, F., et al. (1999). Molecular and functional regulation of two NO3- uptake systems by n- and c-status of arabidopsis plants. Plant J. 18 (5), 509–519. doi: 10.1046/j.1365-313x.1999.00480.x
Lezhneva, L., Kiba, T., Feria-Bourrellier, A. B., Lafouge, F., Boutet-Mercey, S., Zoufan, P., et al. (2014). The arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80 (2), 230–241. doi: 10.1111/tpj.12626
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590 (7847), 600–605. doi: 10.1038/s41586-020-03091-w
Liu, Q., Wu, K., Song, W., Zhong, N., Wu, Y., Fu, X. (2022). Improving crop nitrogen use efficiency toward sustainable green revolution. Annu. Rev. Plant Biol. 73, 523–551. doi: 10.1146/annurev-arplant-070121-015752
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Shah, A. N., Javed, T., Singhal, R. K., Shabbir, R., Wang, D., Hussain, S., et al. (2022). Nitrogen use efficiency in cotton: Challenges and opportunities against environmental constraints. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.970339
Shi, X., Cui, F., Han, X., He, Y., Zhao, L., Zhang, N., et al. (2022). Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar kenong 9204. Mol. Plant 15 (9), 1440–1456. doi: 10.1016/j.molp.2022.07.008
Taochy, C., Gaillard, I., Ipotesi, E., Oomen, R., Leonhardt, N., Zimmermann, S., et al. (2015). The arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 83 (3), 466–479. doi: 10.1111/tpj.12901
Vidal, E. A., Alvarez, J. M., Araus, V., Riveras, E., Brooks, M. D., Krouk, G., et al. (2020). Nitrate in 2020: Thirty years from transport to signaling networks. Plant Cell. 32 (7), 2094–2119. doi: 10.1105/tpc.19.00748
Wang, Y. Y., Cheng, Y. H., Chen, K. E., Tsay, Y. F. (2018). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wang, P., Pu, Y., Abid, M. A., Kang, L., Ye, Y., Zhang, M., et al. (2022). A rapid and efficient method for isolation and transformation of cotton callus protoplast. Int. J. Mol. Sci. 23 (15), 8368. doi: 10.3390/ijms23158368
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wen, B., Xiao, W., Mu, Q., Li, D., Chen, X., Wu, H., et al. (2020). How does nitrate regulate plant senescence? Plant Physiol. Biochem. 157, 60–69. doi: 10.1016/j.plaphy.2020.08.041
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Xia, X., Fan, X., Wei, J., Feng, H., Qu, H., Xie, D., et al. (2015). Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 66 (1), 317–331. doi: 10.1093/jxb/eru425
Xia, X., Wei, Q., Xiao, C., Ye, Y., Li, Z., Marivingt-Mounir, C., et al. (2022). Genomic survey of NPF and NRT2 transporter gene families in five inbred maize lines and their responses to pathogens infection. Genomics 115 (2), 110555. doi: 10.1016/j.ygeno.2022.110555
Zhang, X., Davidson, E. A., Mauzerall, D. L., Searchinger, T. D., Dumas, P., Shen, Y. (2015). Managing nitrogen for sustainable development. Nature 528 (7580), 51–59. doi: 10.1038/nature15743
Zhang, Y., Tateishi-Karimata, H., Endoh, T., Jin, Q., Li, K., Fan, X., et al. (2022). High-temperature adaptation of an OsNRT2.3 allele is thermoregulated by small RNAs. Sci. Adv. 8 (47), eadc9785. doi: 10.1126/sciadv.adc9785
Zhang, G. B., Yi, H. Y., Gong, J. M. (2014). The arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell. 26 (10), 3984–3998. doi: 10.1105/tpc.114.129296
Zhao, Z., Li, M., Xu, W., Liu, J. H., Li, C. (2022). Genome-wide identification of NRT gene family and expression analysis of nitrate transporters in response to salt stress in poncirus trifoliata. Genes 13 (7), 1115. doi: 10.3390/genes13071115
Keywords: nitrate transporters, bio-information analysis, expression model, stress, cotton
Citation: Pu Y, Wang P, Abbas M, Khan MA, Xu J, Yang Y, Zhou T, Zheng K, Chen Q and Sun G (2023) Genome-wide identification and analyses of cotton high-affinity nitrate transporter 2 family genes and their responses to stress. Front. Plant Sci. 14:1170048. doi: 10.3389/fpls.2023.1170048
Received: 20 February 2023; Accepted: 22 March 2023;
Published: 05 April 2023.
Edited by:
Zhenyu Gao, China National Rice Research Institute (CAAS), ChinaReviewed by:
Xueying Guan, Zhejiang University, ChinaXiaorong Fan, Nanjing Agricultural University, China
Copyright © 2023 Pu, Wang, Abbas, Khan, Xu, Yang, Zhou, Zheng, Chen and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quanjia Chen, Y2hxamlhQDEyNi5jb20=; Guoqing Sun, c3VuZ3VvcWluZzAyQGNhYXMuY24=
†These authors have contributed equally to this work
 Yuanchun Pu1,2†
Yuanchun Pu1,2† Peilin Wang
Peilin Wang Mubashir Abbas
Mubashir Abbas Kai Zheng
Kai Zheng Quanjia Chen
Quanjia Chen Guoqing Sun
Guoqing Sun