- 1Fruit Tree Research Institute, Chongqing Academy of Agricultural Sciences, Chongqing, China
- 2Biotechnology Research Institute, Chongqing Academy of Agricultural Sciences, Chongqing, China
Introduction: Blood orange (Citrus sinensis L.) is a valuable source of nutrition because it is enriched in anthocyanins and has high organoleptic properties. Grafting is commonly used in citriculture and has crucial effects on various phenotypes of the blood orange, including its coloration, phenology, and biotic and abiotic resistance. Still, the underlying genetics and regulatory mechanisms are largely unexplored.
Methods: In this study, we investigated the phenotypic, metabolomic, and transcriptomic profiles at eight developmental stages of the lido blood orange cultivar (Citrus sinensis L. Osbeck cv. Lido) grafted onto two rootstocks.
Results and discussion: The Trifoliate orange rootstock provided the best fruit quality and flesh color for Lido blood orange. Comparative metabolomics suggested significant differences in accumulation patterns of metabolites and we identified 295 differentially accumulated metabolites. The major contributors were flavonoids, phenolic acids, lignans and coumarins, and terpenoids. Moreover, transcriptome profiling resulted in the identification of 4179 differentially expressed genes (DEGs), and 54 DEGs were associated with flavonoids and anthocyanins. Weighted gene co-expression network analysis identified major genes associated to 16 anthocyanins. Furthermore, seven transcription factors (C2H2, GANT, MYB-related, AP2/ERF, NAC, bZIP, and MYB) and five genes associated with anthocyanin synthesis pathway (CHS, F3H, UFGT, and ANS) were identified as key modulators of the anthocyanin content in lido blood orange. Overall, our results revealed the impact of rootstock on the global transcriptome and metabolome in relation to fruit quality in lido blood orange. The identified key genes and metabolites can be further utilized for the quality improvement of blood orange varieties.
Introduction
Blood oranges (Citrus sinensis) with distinctive crimson flesh are enriched with anthocyanins (Moufida and Marzouk, 2003; Butelli et al., 2012). The flesh color in blood orange mainly depends on the accumulation of anthocyanins (Habibi et al., 2022). In combination with vitamin C, carotenoids, and fiber, anthocyanins provide a healthy addition to the blood orange dietary properties (Prior and Wu, 2006; De Pascual-Teresa et al., 2010; Paredes-López et al., 2010). Striking color, nutritional properties, and enriched anthocyanins favored blood oranges for health-promoting properties (Kelebek et al., 2008). Anthocyanin-enriched food with high antioxidant activity has distinctive properties against certain diseases such as cancer, heart disease, cholesterol accumulation, and obesity (Hou, 2003; De Pascual-Teresa et al., 2010; Paredes-López et al., 2010; Titta et al., 2010). However, the color and anthocyanin accumulation in blood orange highly depends on the cultivar, environmental attributes, soil, agronomic practices, and rootstock (Rapisarda et al., 2001; Crifò et al., 2011; Siracusa and Ruberto, 2014). Several studies focused on anthocyanin accumulation, composition, and underlying biological pathways in blood oranges have demonstrated potential for quality improvement (Li et al., 2014; Cebadera-Miranda et al., 2019). Aside from anthocyanin enrichment and flesh color, several quality parameters influence consumer choices. These factors include but are not limited to firmness, texture, sugar contents, acids contents, and shelf life (Pallottino et al., 2013; Fabroni et al., 2020).
Citriculture takes advantage of the grafting, and published literature suggests a strong influence of rootstocks on economic traits, such as fruit yield, size, quality, maturation, and postharvest performance (Rodríguez-Gamir et al., 2010; Bowman et al., 2016; Forner-Giner et al., 2020). However, the effect of rootstocks on blood oranges is not well established, with few studies. For instance, Morales et. al., used eight different rootstocks and identified significant differences in metabolite accumulation patterns, specifically phenolics in two orange cultivars (Morales et al., 2021). Another study by Modica et. al., demonstrated differential accumulation patterns of polyphenols, especially anthocyanins, in blood oranges grafted on ten different rootstocks under different environmental conditions (Modica et al., 2022). As fruit quality is highly dependent on genotype and environmental attributes, it is necessary to decipher the underlying mechanism specific to genotype under certain environmental influences.
In recent years, combining omics for integrative analysis of a phenomenon gives more robust outcomes to decipher the underlying genetic mechanisms (Subramanian et al., 2020; Nazir et al., 2021; Raza et al., 2021). A combination of metabolomics and transcriptomics has been widely adapted in horticulture, yielding potential insights for further use in breeding programs for improvement (Su et al., 2019; Tahir Ul Qamar et al., 2020; Zhang and Hao, 2020; Gong et al., 2021). For instance, Gong et. al., utilized an integrative approach combing transcriptomics and metabolomics to understand the flavor of crimson-colored watermelon resulting in the identification of five candidate genes in the glycolytic pathway favoring flavor enhancement (Gong et al., 2021). Although few studies have addressed the quality attributes of red blood oranges in response to different rootstocks, Chinese red blood cultivars have not been characterized for changes in quality parameters due to different rootstocks. Therefore, evaluating existing germplasm and understanding the regulatory mechanisms for utilization in further breeding programs is necessary.
The current study aimed at deciphering the molecular mechanism underlying differential metabolic profiles of the lido blood orange cultivar, which were grafted on four different rootstocks. Lido blood orange fruits at eight different stages were characterized for metabolic and transcriptomic profiles, and conjoint analysis of metabolome and transcriptome further narrowed down the significant variations concerning the fruit quality of lido red blood oranges.
Materials and methods
Plant material and sample collection
Lido blood orange variety grafted on four rootstocks, ‘Trifoliate orange’ (P. trifoliata L. Raf., Pt), ‘Ziyang Xiangcheng’ (C. junos Sieb. ex Tanaka, Cj), C. reticulata Blanco, and ‘Citrange’ (Poncirus trifoliate L. raf. × Citrus sinensis L. Osbeck.) were used in this study. These four rootstocks are hereinafter referred to as Z, X, H, and ZC, respectively. The experimental orchard was established at the regional test base of citrus varieties of Chongqing Academy of Agricultural Sciences, Jiangjin District, Chongqing, China (116° 34 ′ E, 36° 50 ′ N). The experimental region is located in a warm, humid monsoon climate. The region experiences an average annual temperature of 18.4°C, 1001.2 mm of precipitation, 805.5 mm of evaporation, and 206 days without a frost. The citrus orchard in the research region has purple soil type and sandy loam soil texture (6% sand, 89% silt, and 5% clay) with pH 6.43, available potassium 103.83 mg.kg-1, available calcium 0.61 g.kg-1, available magnesium 0.09 g.kg-1, available copper 2.78 mg.kg-1, available iron 191.80 mg.kg-1, available manganese 9.67 mg.kg-1, available zinc 3.47 mg.kg-1, and available boron 1.05 mg.kg-1. The fertilization rate is 1 kg of compound fertilizer plus 0.5 kg of urea plus 1.5 kg of organic fertilizer per plant; before the middle of July in the summer, apply fertilizer for fruit expansion; 1 kg of high potassium compound fertilizer per plant is applied in the ditch; and water-soluble middle and trace elements are supplemented on the leaves.
Fruits were obtained from 4 different rootstocks at eight-time points (harvesting time from October to February). The time points for fruit sampling were T1 (October 12), T2 (November 17), T3 (November 30), T4 (December 15), T5 (December 30), T6 (January 1), T7 (January 29), and T8 (February 15). The tree age was six years at the sampling time, and the planting density was 3 m × 4 m. Three replicates were used for each sample group. Nine blood orange trees with the same growth and similar fruit-bearing capacity were selected as the sample trees from 20 Lido blood oranges with different rootstocks. One sample fruit was collected from the southeast and northwest of each sample tree. The 15 fruits of each three sample trees were mixed into one biological sample of three sets. After the 15 fruits of each biological replicate were transversely cut, we used a scalpel to pick up a small amount of pulp, mix it evenly, and use a juicer to extract juice from the rest. The pulp is used for real-time quantitative analysis of the transcriptome, metabolome, and candidate genes, and the juice was used for the determination of the total solid solids (TSS), vitamin C (VC), acid concentration (A-conc.) and other basic qualities.
The appearance color, soluble solids to acidity ratio, acid concentration, total soluble solids, and vitamin C contents of each replicate fruit were determined. The total soluble solids were measured using a refractometer (Yu et al., 2022) and acid concentration (A-conc.) by acid-base titration by Norminkoda Biotechnology Co., Ltd., Wuhan, China. The Vitamin C was extracted and quantified using the plant ascorbic acid content detection kit (Norminkoda Biotechnology Co., Ltd. Wuhan, China) (Sun et al., 2023).
Metabolomic profiles
Metabolomic profiling was performed by Wuhan Metware Biotechnology Co., Ltd (https://www.metware.cn) following the company’s standard procedures with a series of procedures, including extraction, identification, and quantification of metabolites (Cao et al., 2019; Wang et al., 2019). Cryo-preserved fruit samples (juice and pulp mixture) were weighed and extracted with 1.0 ml of 70% methanol. The methanol extracts were subjected to liquid chromatography mass-spectrometry/M.S. analysis (LC-MS/MS, UPLC, Shim-pack UFLC SHIMADZU CBM30A system; MS, Applied Biosystems 6500 QTRAP). Metware’s metabolite database and public metabolite database were used to identify the metabolites, and quantification was done accordingly. To further access, the differential accumulation patterns of metabolites between different samples were determined using orthogonal partial least squares discriminant analysis. Metabolites with |Log2 Foldchange| ≥ 1 and VIP (variable importance in project) ≥ 1 were defined as DAMs.
Transcriptomic profiles
Total RNA was extracted from the samples (with three biological replicates) with the RNA Extraction kit (TIANGEN, Beijing, China). The RNA quality and concentration were assessed with agarose gel electrophoresis and NanoDrop2000 spectrophotometer. Quality testing, library construction, and sequencing for each sample were done at Metware Biotechnology Co., Ltd (https://www.metware.cn), following the company’s standard procedures. Low-quality data were removed for downstream analysis, and high-quality clean reads were used for transcriptome quantification. The clean reads were localized using Hisat2 to obtain unigenes (Sirén et al., 2014). Reads per kilobase mapping (FPKM) for all genes were used to determine gene expression values, and further screening for differentially expressed genes (DEGs) was performed. The differentially expressed genes (DEGs) were identified by R package DESeq2 for subsequent analysis. The genes featuring FDR < 0.05 and |Log2 Fold change|≥ 1 were considered DEGs. The identified DEGs were further enriched by KEGG analysis. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were applied using Tbtools software (Chen et al., 2020). Heat maps were generated using the OmicStudio tools at https://www.omicstudio.cn/tool.
RT-qPCR based verification of expression of candidate genes
Total RNA was reverse-transcribed in a 20 μL reaction mixture using the HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme). The 20 μL reactions were performed with 10 μL of ChamQTM SYBR® qPCR Master Mix (High ROX Premixed), 1.0 μL 10 mM forward and reverse primers, 7.0 μL of ddH2O and 2.0 μL 5 times diluted cDNA template. The Citrus Actin and EF1 α were used as internal reference genes. The ABI Prism 7500 Fast system was used to perform RT-qPCR. The primers used for RT-qPCR are listed in Supplementary Table 8. The relative expression of genes was calculated by the 2-ΔΔCT method (Schmittgen and Livak, 2008).
Identification of hub genes using WGCNA analyses
Weighted Gene Correlation Network Analysis (WGCNA) (Fuller et al., 2007) was used to identify hub genes. WGCNA analysis was performed with WGCNA-shiny in TBtools (Toolbox for Biologists, v1.082). Cluster analysis was performed on the samples according to the expression levels of DEGs. The correlation between the characteristic gene of the module and the trait was calculated, including the correlation between the gene and the characteristic expression in the module (module membership, MM) and the correlation between each gene and the target trait (gene significance, GS). We used the passing threshold GS.abs >0.5 and MM.abs >0.8 to identify the hub gene of each module. Transcription factor (TF) annotations were searched for the hub genes using PlantTFDB (Jin et al., 2016). Cytoscape (Cytoscape Consortium, USA) software was used to visualize the gene interaction network.
Results
Effect of grafting on physiological and color parameters in lido blood orange
Physiological parameters including total soluble solids (TSS), vitamin C (VC), acid concentration (A-conc.), soluble solids to acidity ratio (SS), and color parameters L (brightness), a (red/green value), and b (Blue/yellow value) from fruit samples were estimated in lido blood orange grafted on four rootstocks Z (Trifoliate orange), ZC (Citrange), H (C. reticulata Blanco), and X (Ziyang Xiangcheng). The data was collected at 8 different time points from October 2021 to February 2022 (T1 to T8) and subjected to analysis of variance. All physiological parameters were significantly different (P < 0.005) between the four rootstocks, implying that the rootstocks altered the nutritional composition of the fruits (Tables S1, S2 and Figure S1). Similarly, we observed a significant effect of the sampling time on the physiological parameters of the fruits. Concerning the flesh color parameters, only L values differed significantly (P < 0.05) between the rootstocks (Tables S1, S2 and Figure S1). However, the sampling time significantly affected all color parameters, indicating that the fruit color changed considerably over the sampling period.
Collectively, fruits samples from Z and X were the most contrasted in terms of color and physiological parameters (Figure S3), therefore, they were selected for downstream analyses.
Differential metabolic profile of lido blood orange grafted to two rootstocks (Z and X)
Metabolites, alongside morpho-physiological attributes such as texture, color, size, and shelf life, are crucial determinants of fruit quality. A comprehensive understanding of these factors could provide valuable insights towards improving the nutritional value and marketability of fruits. Therefore, exploring the roles of metabolites and other traits in fruit quality is a crucial step toward promoting human health and enhancing economic benefits in the fruit industry. Metabolome profiling of fruit samples from rootstocks Z and X (Figure 1A) at eight different time points were analyzed. The analysis identified 966 metabolites belonging to subclasses of alkaloids, amino acids and derivatives, flavonoids, lignans and coumarins, lipids, nucleotides and derivatives, organic acids, phenolic acids, terpenoids, and others (Figure 1B and Table S3). Flavonoids were the most abundant metabolites (35.37%), followed by lipids and phenolic compounds. Comparative metabolomics showed significant differences in the accumulation pattern of flavonoids, phenolic acids, lignans and coumarin, and terpenoids in lido blood orange with the two rootstocks (Figure 2). A total of 295 differentially accumulated metabolites (DAMs) were identified, with most of them being up-accumulated in Z (Figure 1C and Table S3).
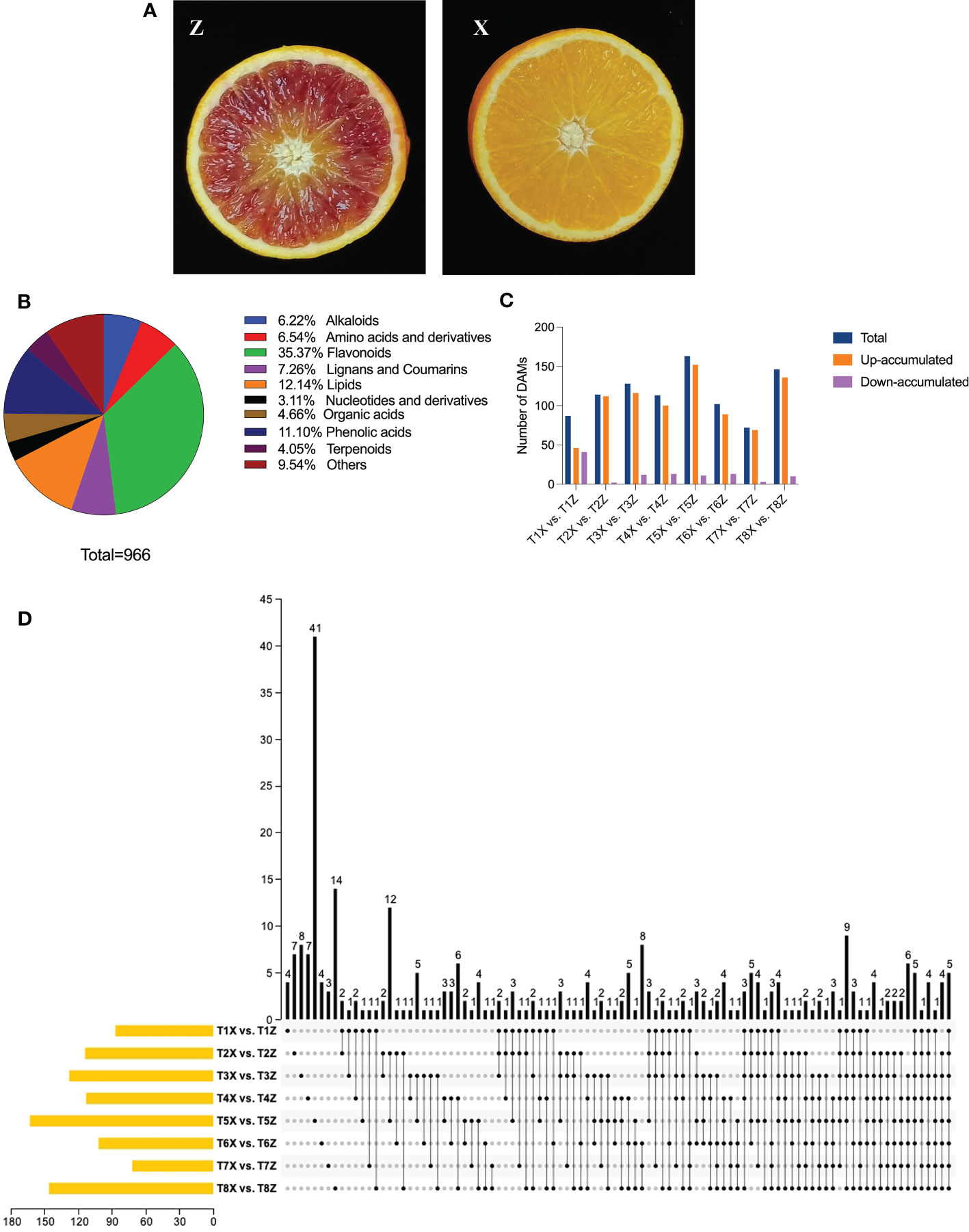
Figure 1 Overview of metabolic profile of lido blood orange on two rootstocks (X and Z) at eight developmental stages (1-8) (A) Pictorial description of lido blood orange fruit with rootstocks Z (Trifoliate orange) and X (Ziyang Xiangcheng) (B) Identification of metabolites and share of each subclass (C) Overview of differentially expressed metabolites between samples at 8 time points (T1-T8) in Lido blood orange with rootstock Z and X (D) Upset plot depicting conserved DAMs between different comparisons.
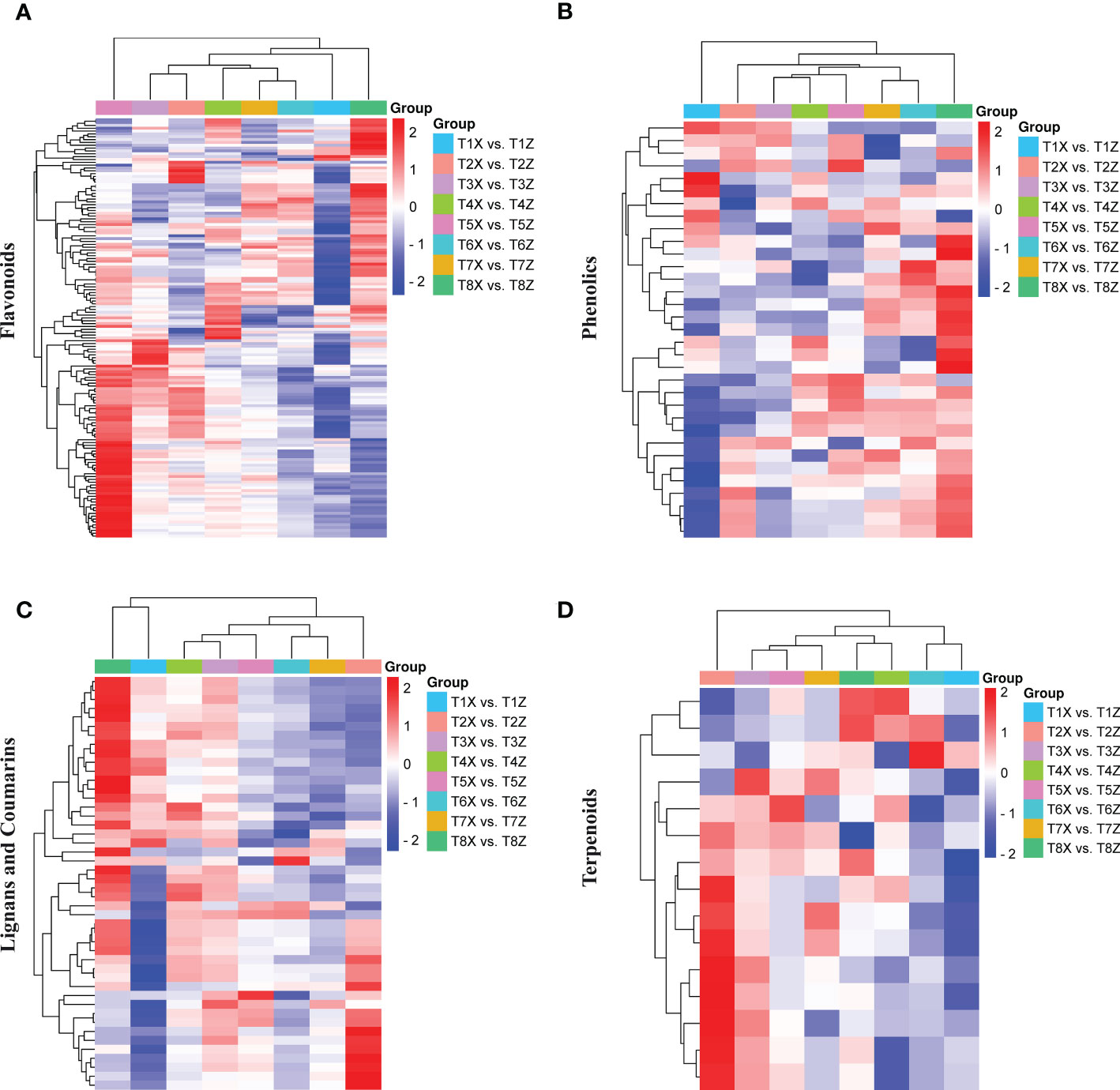
Figure 2 Differential landscape of metabolites in lido blood orange with two rootstocks Z and X (A) Metabolic profile of 149 differentially accumulated flavonoids (B) Metabolic profile of 34 differentially accumulated phenolic acids (C) Metabolic profile of 47 differentially accumulated lignans and coumarins (D) Metabolic profile of 17 differentially accumulated terpenoids.
Five DAMs, namely isomartynoside, prunetin (5,4’-Dihydroxy-7-methoxy isoflavone), deacetylnomilin, isomexoticin glucoside, and deacetylnomilinic acid, belonging to different subclasses showed a conserved up-accumulation pattern at all eight time points (Figure 1D and Table S3).
Among 341 flavonoids, 149 depicted differential accumulation patterns in at least one comparison (T1-T8) (Figure 2A). The significant up-accumulation of most flavonoid compounds in lido blood orange with rootstock Z is highly suggestive that flavonoid accumulation is directly associated with red color formation. Moreover, among the flavonoids, we identified 15 anthocyanins, 1 flavanol, and 1 flavanone (Figure S4). The relative quantification of anthocyanins showed an increase with fruit maturity and a higher accumulation in Z compared to X (Figure S4).
Among 107 phenolic acids, we only identified 34 differentially accumulated phenolic acids in at least one comparison (Figure 2B).
We further characterized subclasses of lignans and coumarins and terpenoids. Almost half of these compounds were differentially accumulated in at least one comparison (Figures 2C, D). Overall, the metabolome analysis demonstrated the significant influence of rootstock on the fruit quality in lido blood orange.
Differential transcriptomic profile of lido blood orange grafted to two rootstocks (Z and X)
Transcriptome data of X and Z samples from the 8 different time points were analyzed. Expression profiles of 25,701 genes were quantified, generating a total of 457.44 Gb of data. After filtering, 2,799,045,806 (91.79%) clean reads were kept out of 3,049,443,238 raw reads obtained (Table S4). The quality check was performed to confer the reliability and reproducibility of the data. Q20 and Q30 were estimated to be over 97% and 92%, respectively (Table S5). Moreover, GC contents ranged from 43.27 to 44.68%. All the transcriptome data were subjected to principal component analysis to identify the corresponding variation. PC1 and PC2 covered 20.08% and 11.66% of the variation, respectively, and replicates from each group were clustered, showing the good quality of the transcriptome data (Figure 3A).
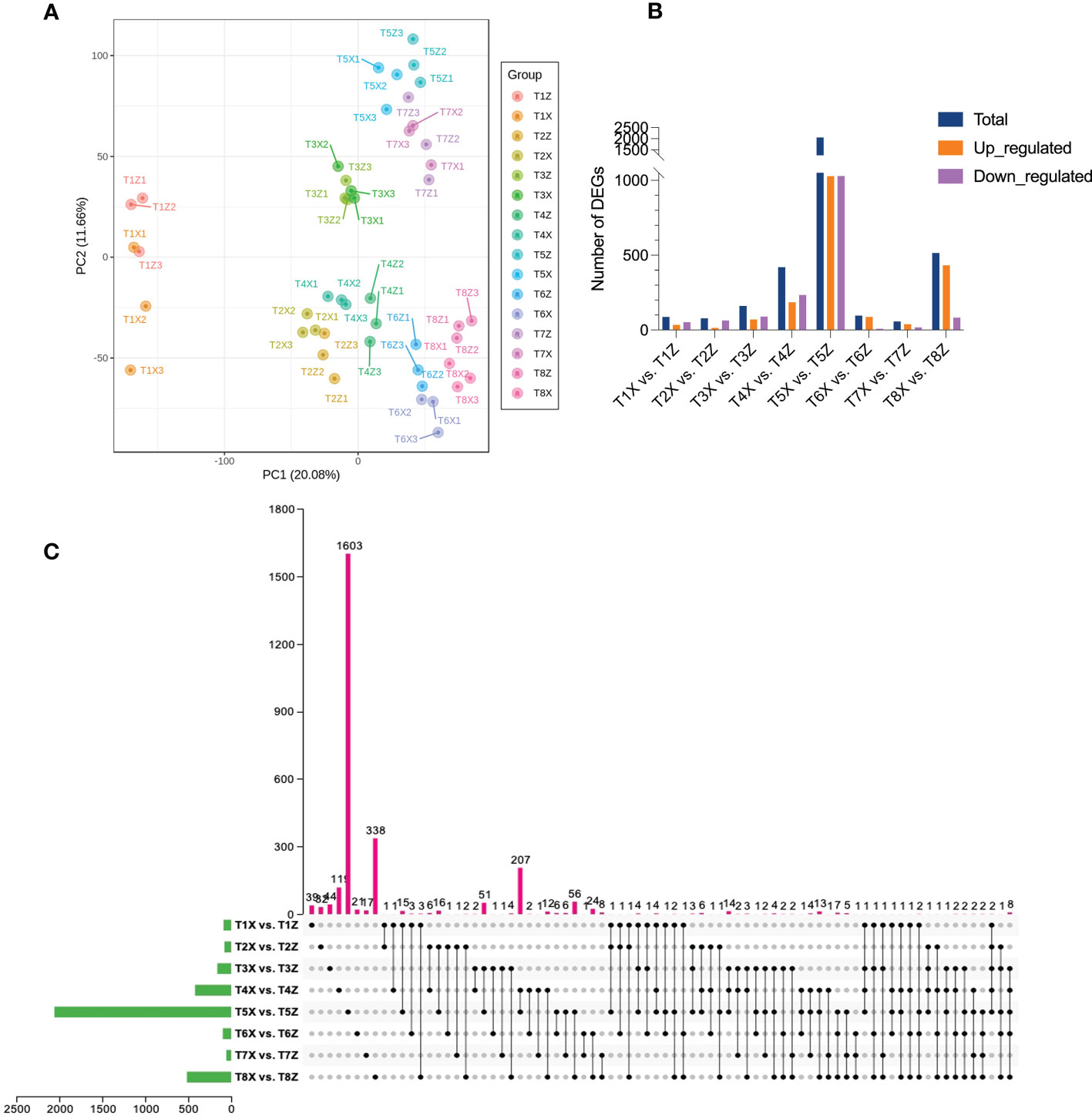
Figure 3 Overview of the transcriptomic profile of lido blood orange on two rootstocks (X and Z) at eight developmental stages (T1-T8) (A) Principal component analysis based on FPKM values of fruit samples of lido blood orange with rootstock Z and X at eight time-points (B) Overview of differentially expressed genes between samples at different stages (C) Upset plot depicting conserved DEGs between different comparisons.
Pairwise comparisons yielded a total of 4179 differentially expressed genes (DEGs) (Figures 3B, C). The expression profiles of all DEGs are presented in Figure S5 and Table S6. Metabolic process and the response to stimulus are the most enriched gene ontology (GO) terms. Next we will investigate changes in specific pathways related to flavonoids, color formation and VC contents.
Insight into the genes regulating flavonoids, anthocyanin and VC in lido blood orange grafted to X and Z
Since flavonoids were the major metabolites differentially accumulated between X and Z fruit samples, we further characterized the 45 DEGs involved in flavonoids biosynthesis. Most the DEGs related to flavonoid biosynthesis were upregulated in Z compared to X (Figure 4). These results suggest that the rootstock Z increased the expression levels of key flavonoids biosynthesis genes leading to high accumulation of flavonoids in fruits.
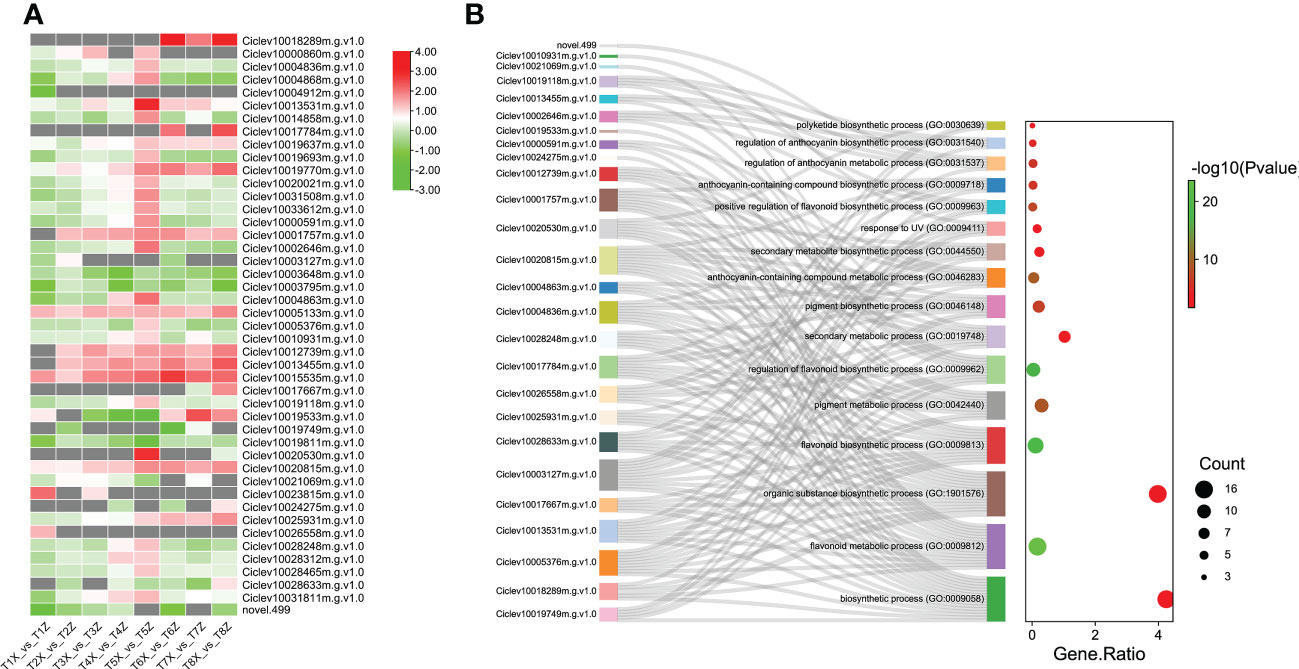
Figure 4 Identification and characterization of DEGs associated with Flavonoid biosynthesis (A) Expression profile of 47 DEGs associated with flavonoid biosynthesis and pigmentation at 8 time points (T1-T8) (B) GO term enrichment for DEGs associated with flavonoid biosynthesis and pigmentation. The y-axis indicates the GO pathways and associated genes.
Likewise, the DEGs associated with anthocyanin synthesis were further characterized for their potential role in higher accumulation patterns of anthocyanins (blood color formation) in Z. The 17 DEGs showed a differential regulation patterns in Z and X at the 8 time points (Figure 5 and Table S7). PAL, 4CH, 4CL, CHS, CHI, F3H, F3’5’H, DFR, UFGT, and ANS were highly expressed in Z compared to X. Only FLS was slightly higher expressed in X compared to Z. It is well known that FLS competes with DFR to produce flavonols, hence the high FLS expression in X could explain the low anthocyanin content. Overall, the high expression pattern of the major anthocyanin biosynthesis structural genes substantiates the up-accumulation of anthocyanins in Z.
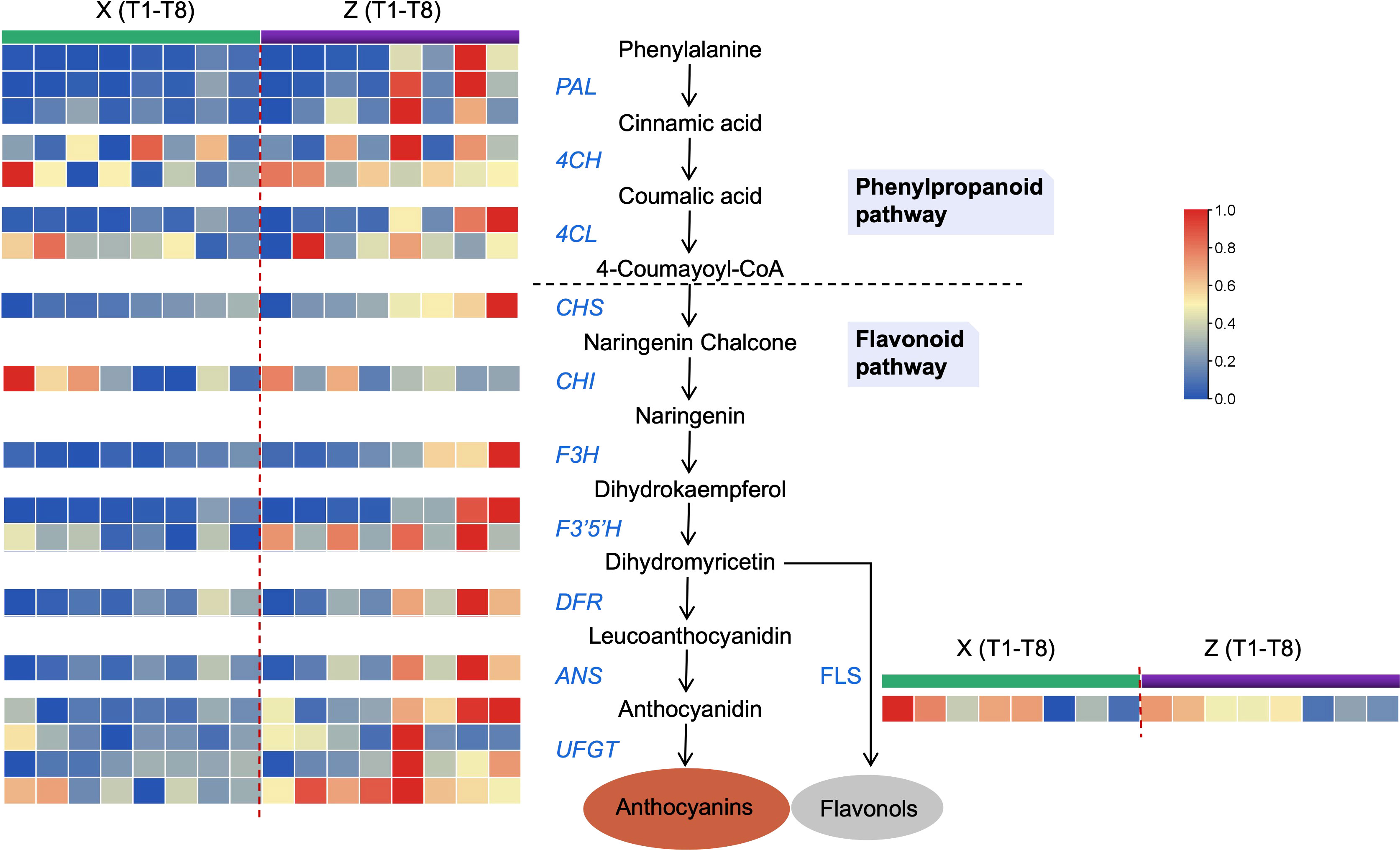
Figure 5 Transcript profiles for genes in the anthocyanin biosynthetic pathway. PAL, phenylalanine ammonia-lyase; 4CH, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3’5’H, flavonoid 3’,5’ hydroxylase; DFR, dihydroflavonol 4-reductase; UFGT, UDP-glucose: flavonoid-3-O-glucosyltransferase; ANS, anthocyanidin synthase; FLS, flavonol synthase. Green and purple represent samples from Z and X, where left to right is the time point from T1 to T8.
Furthermore, we screened the 12 DEGs associated with VC biosynthesis based on GO terms. Majority of these DEGs were up-regulated in Z compared to X (Figure 6 and Table S6), which could explain the higher VC content in lido blood orange grafted on rootstock Z as compared to X.
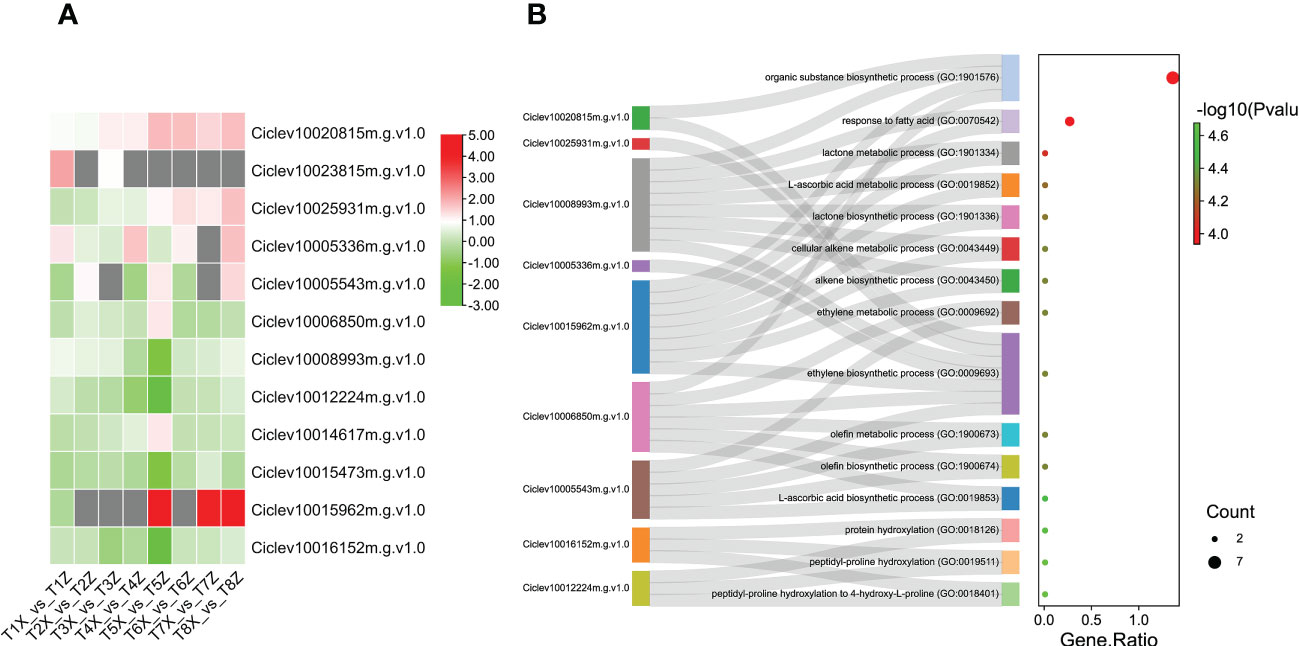
Figure 6 Identification and characterization of DEGs associated with Vitamin C biosynthesis (A) Expression profile of 12 DEGs associated with VC biosynthesis at 8 time points (T1-T8) (B) GO term enrichment for DEGs associated with vitamin C The y-axis indicates the GO pathways and associated genes.
Gene regulatory mechanisms of anthocyanin accumulation
In order to identify regulatory genes involved in anthocyanin accumulation, we performed a weighted gene co-expression network analysis (WGCNA) by constructing a relationship network between DEGs and anthocyanin accumulation. Topology analysis showed that when the threshold β = 13, the scale-free topology fitting index (R2) was close to 90%, indicating that the network was close to a scale-free network (Figure S6). Moreover, 13 modules were identified for further screening of hub genes. Module ME-purple showed a significantly higher correlation with anthocyanin accumulation patterns, and therefore we selected the ME-purple module for further assessment (Figure 7 and Table S8). Enrichment analysis of the ME-purple module suggested significant enrichment of pathways, such as metabolic pathways, phenylpropanoid biosynthesis, flavonoid biosynthesis, and anthocyanin biosynthesis (Figure 7B). A total of 55 genes were identified in this module with seven transcription factors (C2H2, GANT, MYB-related, AP2/ERF, NAC, bZIP, and MYB) and five structural genes associated to the anthocyanin biosynthesis pathway (CHS, F3H, UFGT, and ANS). The degree of association of the above-mentioned transcription factors and hub genes has been presented as a network (Figure 7C). The network analysis revealed that NAC exhibited the highest connectivity with other genes within the module, suggesting that NAC may play a more prominent role in modulating the anthocyanin content.
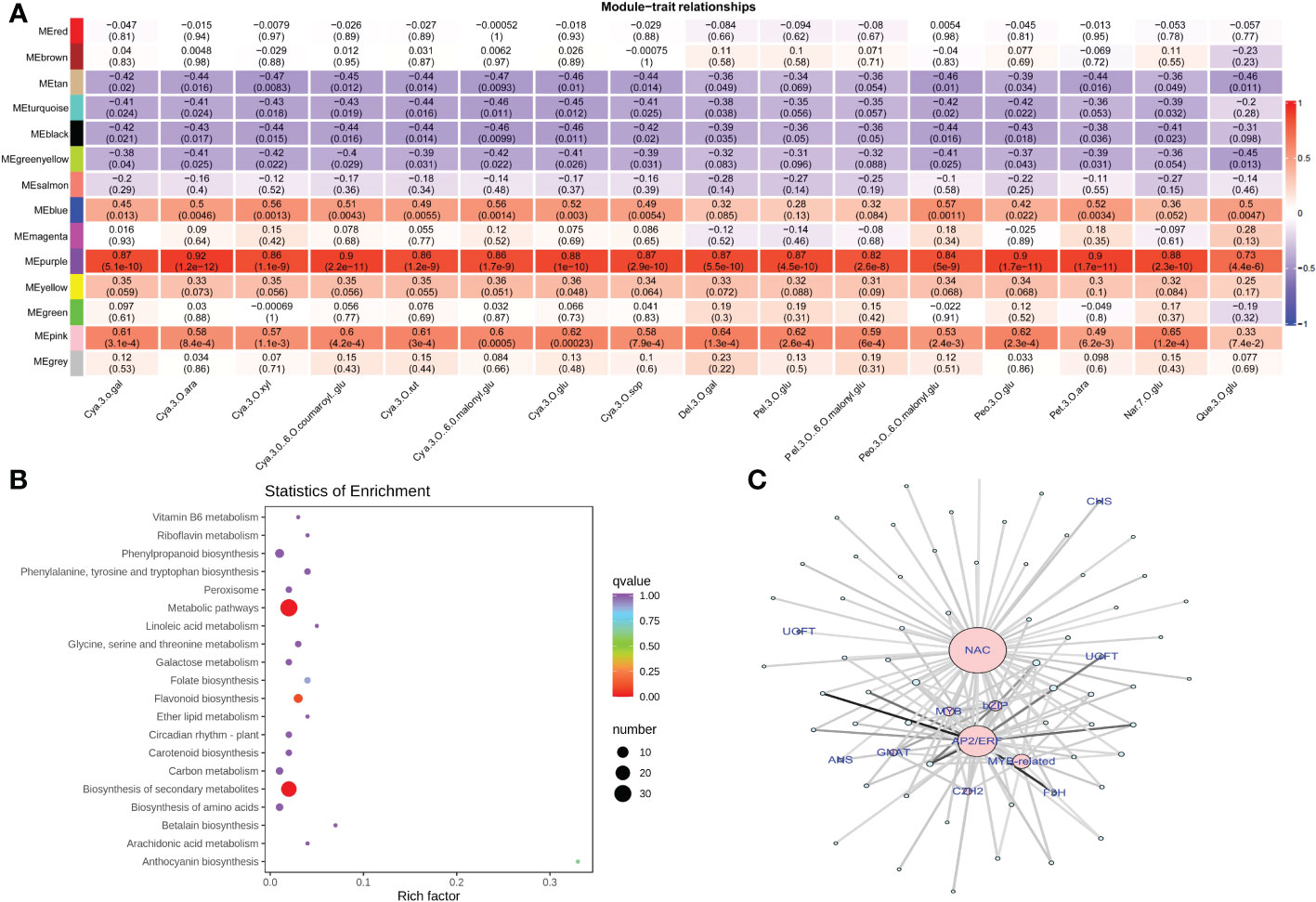
Figure 7 Weighted gene co-expression network analysis for gene mining (A) Module/trait correlations and corresponding p values. (B) Pathway enrichment analysis of genes in ME-purple module (C) The network of the highly connected genes in the ME-purple module. Pink represents transcription factors, and gene names in blue represent the hub genes associated with anthocyanin biosynthesis.
RT-qPCR based verification of expression profile of candidate genes
A total of 17 candidate genes linked to anthocyanin content in blood orange were used for RT-qPCR experiment. The goal was to validate the expression profile obtained from the RNA-seq. The primers for each candidate gene are listed in Table S9. Among the 17 candidate genes, 15 were up-regulated in Z at time points T5-T8 (Figure 8). PAL, C4H, 4CL, CHS, CHI, F3H, F3’5’H, DFR, ANS, WD40, UFGT, GST, Ruby, MYBF1, and NAC were among the 15 up-regulated genes in Z. Moreover, RT-qPCR results correlated well with the RNA-seq data (Pearson correlation R2 = 73%), validating the variation observed in the whole transcriptomic datasets.
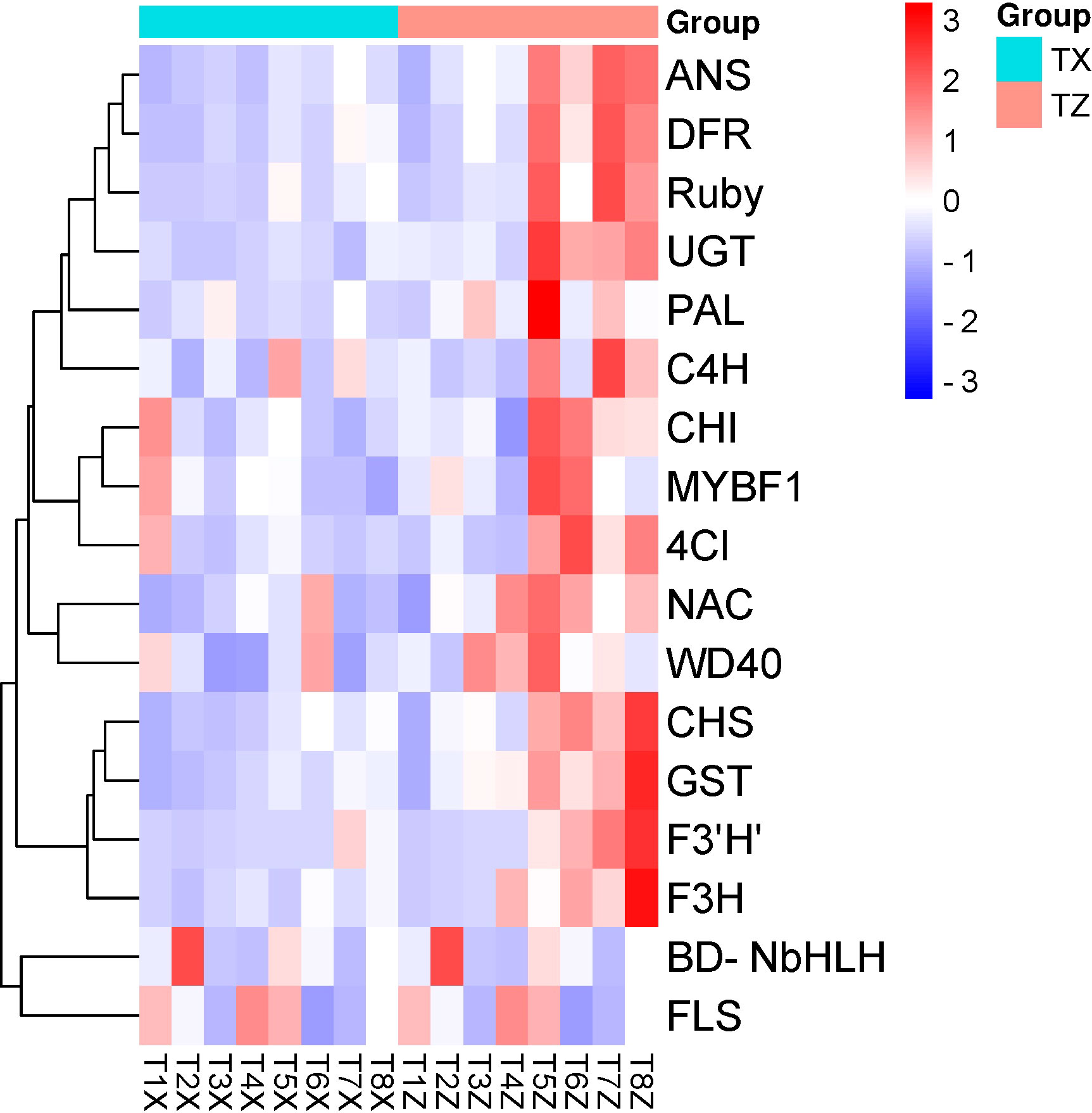
Figure 8 qRT-PCR based expression analysis of selected genes in flavonoid biosynthesis genes at 8 time points (T1-T8) in Lido blood orange with rootstock Z and X. Where PAL: Phenylalanine ammonia-lyase, C4H: Cinnamate 4-hydroxylase, 4CL: 4-Coumarate, CHS: Chalcone synthase, CHI: Chalcone isomerase, F3H: Flavanone 3-hydroxylase, F3’5’H: Flavonoid 3’,5’-hydroxylase, DFR: Dihydroflavonol 4-reductase, ANS: Anthocyanidin synthase, WD40: WD40 repeat-containing protein, UFGT: UDP-glucose: flavonoid 3-O-glucosyltransferase, GST: Glutathione S-transferase, Ruby: Gene involved in cotton fiber development, MYBF1: Myeloblastosis transcription factor 1, NAC: NAM, ATAF, and CUC transcription factor family, FLS: flavonol synthase, and BD-NbHLH: Basic-helix-loop-helix.
Discussion
Blood oranges (Citrus sinensis L.) are characterized by a high accumulation of anthocyanins and polyphenolics, conferring their characteristic red color and a valuable source of available antioxidants (Morales et al., 2021). The yield and quality of citrus depend on the genotype, rootstock, and environmental attributes (Cimen and Yeşiloğl, 2016; Martínez-Cuenca et al., 2016; Song et al., 2020; Morales Alfaro et al., 2021). Inherent rootstock differences such as root distribution, water uptake, and hydraulic conductivity are attributed to the final yield and quality of the fruit (Barry et al., 2004). The TSS content in the blood orange juice fell within the 9.47–11.3°Brix range for ‘Z’ and within the 8.65-10.7°Brix range for ‘X’. These values were at par with the previous study concerning ‘Moro’ and ‘Tarocco Rosso’ blood oranges (Morales et al., 2021) and lower than ‘Tarocco Scirè’, ‘Tarocco Rosso’, ‘Tarocco Ippolito’ and ‘Sanguinello’ (12.57 and 15.30°Brix range) (Continella et al., 2018; Cebadera-Miranda et al., 2019). Similar trends were observed for SS and A-conc. Concerning Vitamin C, the results obtained in this study showed a higher concentration of vitamin C (38.5-77 mg/100 ml) in the fruit compared to previous studies ranging from 34.24-66.9 mg/100g (Arena et al., 2001; Rapisarda et al., 2008; Kafkas et al., 2009; Cebadera-Miranda et al., 2019).
To elucidate the metabolome and transcriptome differences pertaining to rootstocks Z (Trifoliate orange) and X (Ziyang Xiangcheng), we systematically evaluated both genotypes at different harvest time points and identified differential regulation of both metabolome and transcriptome. The analysis of differentially accumulated metabolites suggested higher accumulation patterns in ‘Trifoliate orange’ compared with ‘Ziyang Xiangcheng’, emphasizing that lido blood oranges with ‘Trifoliate orange’ rootstock produced superior quality fruit with higher nutrition values. A total of 87% of the differentially accumulated metabolites were up-accumulated in ‘Trifoliate orange’ blood orange. Previously reports also showed significant differences in metabolite accumulation pertaining to different rootstocks. For instance, Miranda et. al., compared Sanguinello and Tarocco blood oranges and identified Tarocco with the highest flavonoid accumulation (Cebadera-Miranda et al., 2019). Another study by Morales et. al., confirmed rootstock-dependent flavonoid accumulation patterns in blood oranges (Morales et al., 2021).
Flavonoids play a crucial role during plant growth and development with various physiological functions such as defense response (Piasecka et al., 2015), post-stress recovery (Havsteen, 2002) and pigmentation (Schiestl and Johnson, 2013). We performed a qualitative and quantitative analysis of secondary metabolites in lido blood orange. Flavonoids were among the most abundant metabolites in this study, and 43% of the flavonoids were identified with differential accumulation patterns. The results are in line with previous reports on metabolite accumulation in blood oranges (Wang et al., 2017). High-performance liquid chromatography has enabled researchers to detect major and minor metabolites of the fruit (Russo et al., 2021). Flavonoids, phenolic acids, and anthocyanins are major characteristic compounds in blood oranges (Lo Piero, 2015). As important subclasses of flavonoids, flavonols, and pro-anthocyanidins are proven to be the main pigment components (Liu et al., 2019), while flavanones are considered major regulators of yellow pigmentations (Peterson et al., 2006; Goulas and Manganaris, 2012). Shi et. al., emphasized that structurally modified anthocyanins and co-pigmented flavonoids are responsible for the purple coloration of tea leaves (Shi et al., 2021). Cyanidin 3-glucoside and cyanidin 3-(6”-malonylglucoside) are considered major anthocyanins in blood oranges, covering 90% of the anthocyanins in blood oranges (Habibi et al., 2022). Various studies have identified other minor anthocyanins in blood oranges, including delphinidin, cyanidin, and peonidin (Lee, 2002). Anthocyanins are also considered a major contributor to the pigmentation of different plant organs (Guo et al., 2009). Reports have emphasized the key role of anthocyanins in tissue color development. For instance, Xue et al. reasoned that red-colored seed-coat formation in peanut is directly associated with anthocyanin accumulation (Xue et al., 2021). Qiu et al. found differential accumulation patterns of anthocyanin in purple fruit and yellow passion fruit (Qiu et al., 2020). Similarly, a study concerning Primula vulgaris outlined a gradual increase in anthocyanins as the color deepened from white, yellow, blue, and pink (Li et al., 2020). Similarly, our results emphasized an increased accumulation of anthocyanins in Z (red flesh color) compared with rootstock X (yellow flesh color). Aside from flavonoids, other subclasses of metabolites, including phenolic acids, lignans, coumarins, and terpenoids, were also characterized for their differential cumulation patterns in lido blood orange. Our results signify the differences in metabolite accumulation pertaining to different rootstocks and emphasize that rootstock plays a crucial role in quality enhancement.
Transcriptome provides a genetic basis for a trait under study, and it has been widely used to decipher the genes underlying a specific trait by utilizing contrasting phenotypes (Gusev et al., 2016). Moreover, weighted gene co-expression network analysis can provide significant insights into the co-regulation of trait-specific DEGs (Ding et al., 2021; He et al., 2021; Li et al., 2021). Combined with the metabolome analysis, chalcone synthase (CHS), flavanone 3-hydroxylase (F3H), UDP glucose: flavonoid-3-O-glucosyltransferase (UFGT), and Anthocyanidin Synthase (ANS) were identified as key structural genes in the flavonoid biosynthesis pathway in Lido blood orange positively regulating the anthocyanin biosynthesis. Wang et. al., observed upregulation of seven co-expressed proteins (PAL, CHS, F3’H, F3’5’H, DFR, ANS, and UFGT) and their corresponding mRNAs (Wang et al., 2017). Interestingly, there were strong positive correlations between the transcription levels of these genes and the protein expression levels. Previously reports are highly suggestive of the active involvement of CHS and UFGT in regulating anthocyanins in different plant tissues (Hu et al., 2022; Mao et al., 2022; Meng et al., 2022; Muhammad et al., 2022; Ohta et al., 2022). Castellarin et. al., while studying red grapes, reported that the expression of Flavonoid 3’,5’-hydroxylase controls the production of red cyanidin-based anthocyanins accumulated as red pigments (Castellarin et al., 2006). Several other reports also suggested the significant role of Flavonoid 3’,5’-hydroxylase in pigmentation (De Vetten et al., 1999; Boase et al., 2010; Ishiguro et al., 2012; Vikhorev et al., 2019). Moreover, several transcription factors (TF), including C2H2, GANT, MYB-related, AP2/ERF, NAC, bZIP, and MYB, were also identified as a regulators of hub genes in this study. Our results suggested that the NAC TF might play a key role in anthocyanin synthesis. Although several studies have demonstrated that the NAC family is involved in the regulation of various biological processes, from plant development to response to stress, a few members have been identified as regulators of anthocyanin biosynthesis (Morishita et al., 2009; Zhou et al., 2015; Mahmood et al., 2016; Jiang et al., 2019; Zhang et al., 2020; Martín-Pizarro et al., 2021). For instance, in peach (Prunus persica), a BLOOD (BL) gene encoding a NAC TF played a key role in determining the blood-flesh trait by activating PpMYB10.1 in the process of peach fruit ripening (Zhou et al., 2015). Similarly, Zhang et al. identified a NAC TF MdNAC42 in red-fleshed apples, which could interact with MdMYB10, an important positive regulator of anthocyanin biosynthesis, to promote anthocyanin accumulation during apple fruit ripening (Zhang et al., 2020). Zou et al. demonstrated that silencing LcNAC002 led to a significant reduction in the expression levels of LcSGR and LcMYB1, inhibiting chlorophyll degradation and anthocyanin accumulation in Litchi chinensis (Zou et al., 2023). Another study by Morishita et. al., demonstrated that LcNAC13 physically interacts with LcR1MYB1 to coregulate anthocyanin biosynthesis-related genes during litchi fruit ripening (Jiang et al., 2019). In strawberry, FaNAC035 could induce anthocyanin accumulation during fruit ripening by promoting ABA accumulation (Martín-Pizarro et al., 2021). Liu et al. reported that MdNAC1 expression is significantly upregulated in response to abscisic acid (ABA) due to an ABRE cis-acting element in its promoter region. Furthermore, the accumulation of anthocyanins in apple co-transformed with MdNAC1 and MdbZIP23 was observed to be enhanced in the presence of ABA (Liu et al., 2023). These findings suggest that MdNAC1 may be involved in the ABA-mediated regulation of anthocyanin biosynthesis in apple. Since the molecular mechanism of NAC TFs in citrus is poorly understood, it is important to investigate whether they can form protein complexes with other transcription factors like MYB, WD40, or bHLH to regulate anthocyanin synthesis.
Citrate accumulation is the main reason for the acidity of citrus fruits. However, the decrease of acidity during the ripening stages is associated with the catabolism of citrate (Cercós et al., 2006). NAD(P) is a key gene family involved in citric acid catabolism, and its upregulation results in reduced citrate accumulation (Guo et al., 2016). The upregulation of NAD(P)-dependent IDH results in increased conversion of isocitrate to α-ketoglutarate, which leads to reduced citrate accumulation in the TCA cycle. This is because isocitrate is a precursor of citrate, and its rapid conversion to α-ketoglutarate reduces the amount of citrate produced in the TCA cycle. Therefore, the upregulation of NAD(P)-dependent IDH can modulate citric acid catabolism and ultimately affect citrate accumulation in the cell. Our results suggested up-regulation of both ATP citrate synthase and NAD(P)-bd_dom domain-containing protein at harvest time-point 8 (T8). Together with the pattern of acid concentration in lido blood orange with different rootstocks, regulation of these genes is highly suggestive of playing a pivotal role in higher acid accumulation in lido blood orange with rootstock Z compared to X.
Conclusion
Together, physiological, metabolic, and transcriptomic characterization revealed that lido blood orange with ‘Trifoliate orange’ rootstock is superior in fruit quality compared with ‘Ziyang Xiangcheng’. Our results also provided key regulators for flavonoids, anthocyanins, and acidity, which can be a valuable source for further utilization in improving blood orange varieties.
Data availability statement
The raw RNA-seq datasets used for this study can be found in the NCBI SRA under the project number: PRJNA898909 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA898909).
Author contributions
Conceptualization, LY, LH, YC, and MW; Formal analysis, YC, Shuang Li, and LG; Funding acquisition, LH; Investigation, HY and HH; Methodology, WW and MW; Resources, YC; Software, HH; Supervision, LY; Validation, YC and LG; Visualization, HY; Writing – original draft, LH and LY; Writing – review and editing, LH and LY. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Chongqing academy of agricultural sciences performance incentive guide project (cqaas2021jxjl01), Chongqing academy of agricultural sciences municipal financial special project (NKY-2022AB005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1169220/full#supplementary-material
Supplementary Figure 1 | Mean comparisons of physiological parameters of Lido blood orange on four rootstocks at eight time points (A) Total soluble solids (TSS), (B) Acid concentration, (C) Vitamin C, and (D) soluble solids to acidity ratio (TSS/Acid concentration).
Supplementary Figure 2 | Mean comparisons of color parameters of Lido blood orange on four rootstocks at eight time points (A) L (brightness), (B) a (red/green value), and (C) b (Blue/yellow value).
Supplementary Figure 3 | Pictorial description of lido blood orange fruit with rootstocks (A) Z (Trifoliate orange) (B) ZC (Citrange), (C) H (C. reticulata Blanco), and (D) X (Ziyang Xiangcheng).
Supplementary Figure 4 | Anthocyanin accumulation pattern in lido blood orange with Z and X rootstocks at eight time points.
Supplementary Figure 5 | Expression profile of identified DEGs in multiple comparisons of lido blood orange with Z and X rootstocks at eight time points.
Supplementary Figure 6 | Weighted gene co-expression network analysis (A) cluster dendrogram (B) Eigengene adjacency heatmap (C) Topology analysis of appropriate weighting coefficient (D) mean connectivity.
References
Arena, E., Fallico, B., Maccarone, E. (2001). Evaluation of antioxidant capacity of blood orange juices as influenced by constituents, concentration process and storage. Food Chem. 74, 423–427. doi: 10.1016/S0308-8146(01)00125-X
Barry, G. H., Castle, W. S., Davies, F. S. (2004). Rootstocks and plant water relations affect sugar accumulation of citrus fruit via osmotic adjustment. J. Am. Soc. Hortic. Sci. 129, 881–889. doi: 10.21273/JASHS.129.6.0881
Boase, M. R., Lewis, D. H., Davies, K. M., Marshall, G. B., Patel, D., Schwinn, K. E., et al. (2010). Isolation and antisense suppression of flavonoid 3', 5'-hydroxylasemodifies flower pigments and colour in cyclamen. BMC Plant Biol. 10, 1–12. doi: 10.1186/1471-2229-10-107
Bowman, K. D., Mccollum, G., Albrecht, U. (2016). Performance of ‘Valencia’orange (Citrus sinensis [L.] osbeck) on 17 rootstocks in a trial severely affected by huanglongbing. Scientia Hortic. 201, 355–361. doi: 10.1016/j.scienta.2016.01.019
Butelli, E., Licciardello, C., Zhang, Y., Liu, J., Mackay, S., Bailey, P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255. doi: 10.1105/tpc.111.095232
Cao, H., Ji, Y., Li, S., Lu, L., Tian, M., Yang, W., et al. (2019). Extensive metabolic profiles of leaves and stems from the medicinal plant dendrobium officinale kimura et migo. Metabolites 9, 215. doi: 10.3390/metabo9100215
Castellarin, S. D., Di Gaspero, G., Marconi, R., Nonis, A., Peterlunger, E., Paillard, S., et al. (2006). Colour variation in red grapevines (Vitis vinifera l.): genomic organisation, expression of flavonoid 3'-hydroxylase, flavonoid 3', 5'-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genomics 7, 1–17. doi: 10.1186/1471-2164-7-12
Cebadera-Miranda, L., Domínguez, L., Dias, M. I., Barros, L., Ferreira, I. C., Igual, M., et al. (2019). Sanguinello and tarocco (Citrus sinensis [L.] osbeck): bioactive compounds and colour appearance of blood oranges. Food Chem. 270, 395–402. doi: 10.1016/j.foodchem.2018.07.094
Cercós, M., Soler, G., Iglesias, D. J., Gadea, J., Forment, J., Talón, M. (2006). Global analysis of gene expression during development and ripening of citrus fruit flesh. a proposed mechanism for citric acid utilization. Plant Mol. Biol. 62, 513–527. doi: 10.1007/s11103-006-9037-7
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cimen, B., Yeşiloğl, T. (2016). Rootstock breeding for abiotic stress tolerance in citrus. Abiotic biotic Stress plants-recent Adv. Future Perspect 527–563. doi: 10.5772/62047
Continella, A., Pannitteri, C., La Malfa, S., Legua, P., Distefano, G., Nicolosi, E., et al. (2018). Influence of different rootstocks on yield precocity and fruit quality of ‘Tarocco scirè’pigmented sweet orange. Scientia Hortic. 230, 62–67. doi: 10.1016/j.scienta.2017.11.006
Crifò, T., Puglisi, I., Petrone, G., Recupero, G. R., Piero, A. R. L. (2011). Expression analysis in response to low temperature stress in blood oranges: implication of the flavonoid biosynthetic pathway. Gene 476, 1–9. doi: 10.1016/j.gene.2011.02.005
De Pascual-Teresa, S., Moreno, D. A., García-Viguera, C. (2010). Flavanols and anthocyanins in cardiovascular health: a review of current evidence. Int. J. Mol. Sci. 11, 1679–1703. doi: 10.3390/ijms11041679
De Vetten, N., Ter Horst, J., Van Schaik, H.-P., De Boer, A., Mol, J., Koes, R. (1999). A cytochrome b 5 is required for full activity of flavonoid 3′, 5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. 96, 778–783. doi: 10.1073/pnas.96.2.778
Ding, T., Zhang, R., Zhang, H., Zhou, Z., Liu, C., Wu, M., et al. (2021). Identification of gene co-expression networks and key genes regulating flavonoid accumulation in apple (Malus× domestica) fruit skin. Plant Sci. 304, 110747. doi: 10.1016/j.plantsci.2020.110747
Fabroni, S., Amenta, M., Timpanaro, N., Todaro, A., Rapisarda, P. (2020). Change in taste-altering non-volatile components of blood and common orange fruit during cold storage. Food Res. Int. 131, 108916. doi: 10.1016/j.foodres.2019.108916
Forner-Giner, M. A., Continella, A., Grosser, J. W. (2020). “The citrus genome,” in "Citrus rootstock breeding and selection," (Springer Nature Switzerland: Springer), 49–74.
Fuller, T. F., Ghazalpour, A., Aten, J. E., Drake, T. A., Lusis, A. J., Horvath, S. (2007). Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome 18, 463–472. doi: 10.1007/s00335-007-9043-3
Gong, C., Diao, W., Zhu, H., Umer, M. J., Zhao, S., He, N., et al. (2021). Metabolome and transcriptome integration reveals insights into flavor formation of ‘Crimson’watermelon flesh during fruit development. Front. Plant Sci. 12, 629361. doi: 10.3389/fpls.2021.629361
Goulas, V., Manganaris, G. A. (2012). Exploring the phytochemical content and the antioxidant potential of citrus fruits grown in Cyprus. Food Chem. 131, 39–47. doi: 10.1016/j.foodchem.2011.08.007
Guo, J., Han, W., Wang, M. H. (2009). Ultraviolet and environmental stresses involved in the induction and regulation of anthocyanin biosynthesis: a review. Afr. J. OF Biotechnol. 725, 4966–4972.
Guo, L.-X., Shi, C.-Y., Liu, X., Ning, D.-Y., Jing, L.-F., Yang, H., et al. (2016). Citrate accumulation-related gene expression and/or enzyme activity analysis combined with metabolomics provide a novel insight for an orange mutant. Sci. Rep. 6, 1–12. doi: 10.1038/srep29343
Gusev, A., Ko, A., Shi, H., Bhatia, G., Chung, W., Penninx, B. W., et al. (2016). Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252. doi: 10.1038/ng.3506
Habibi, F., García-Pastor, M. E., Puente-Moreno, J., Garrido-Auñón, F., Serrano, M., Valero, D. (2022). Anthocyanin in blood oranges: a review on postharvest approaches for its enhancement and preservation. Crit. Rev. Food Sci. Nutr. 1-13, 1–13. doi: 10.1080/10408398.2022.2098250
Havsteen, B. H. (2002). The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 96, 67–202. doi: 10.1016/S0163-7258(02)00298-X
He, Y., Wang, Z., Ge, H., Liu, Y., Chen, H. (2021). Weighted gene co-expression network analysis identifies genes related to anthocyanin biosynthesis and functional verification of hub gene SmWRKY44. Plant Sci. 309, 110935. doi: 10.1016/j.plantsci.2021.110935
Hou, D.-X. (2003). Potential mechanisms of cancer chemoprevention by anthocyanins. Curr. Mol. Med. 3, 149–159. doi: 10.2174/1566524033361555
Hu, H., Shi, B., Zhu, W., Zheng, B., Zhou, K., Qian, M., et al. (2022). Genome-wide identification, characterization and expression analysis of mango (Mangifera indica l.) chalcone synthase (CHS) genes in response to light. Horticulturae 8, 968. doi: 10.3390/horticulturae8100968
Ishiguro, K., Taniguchi, M., Tanaka, Y. (2012). Functional analysis of antirrhinum kelloggii flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase genes; critical role in flower color and evolution in the genus antirrhinum. J. Plant Res. 125, 451–456. doi: 10.1007/s10265-011-0455-5
Jiang, G., Li, Z., Song, Y., Zhu, H., Lin, S., Huang, R., et al. (2019). LcNAC13 physically interacts with LcR1MYB1 to coregulate anthocyanin biosynthesis-related genes during litchi fruit ripening. Biomolecules 9, 135. doi: 10.3390/biom9040135
Jin, J., Tian, F., Yang, D.-C., Meng, Y.-Q., Kong, L., Luo, J., et al. (2016). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45, 1040–1045. doi: 10.1093/nar/gkw982
Kafkas, E., Ercisli, S., Kemal, K. N., Baydar, K., Yilmaz, H. (2009). Chemical composition of blood orange varieties from Turkey: a comparative study. Pharmacognosy Magazine 5, 329. doi: 10.4103/0973-1296.58155
Kelebek, H., Canbas, A., Selli, S. (2008). Determination of phenolic composition and antioxidant capacity of blood orange juices obtained from cvs. Moro and sanguinello (Citrus sinensis (L.) osbeck) grown in Turkey. Food Chem. 107, 1710–1716. doi: 10.1016/j.foodchem.2007.10.004
Lee, H. S. (2002). Characterization of major anthocyanins and the color of red-fleshed Budd blood orange (Citrus sinensis). J. Agric. Food Chem. 50, 1243–1246. doi: 10.1021/jf011205+
Li, X., Cheng, Y., Wang, M., Cui, S., Guan, J. (2021). Weighted gene coexpression correlation network analysis reveals a potential molecular regulatory mechanism of anthocyanin accumulation under different storage temperatures in ‘Friar’plum. BMC Plant Biol. 21, 1–14. doi: 10.1186/s12870-021-03354-2
Li, Y., Van Den Ende, W., Rolland, F. (2014). Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Mol. Plant 7, 570–572. doi: 10.1093/mp/sst161
Li, L., Ye, J., Li, H., Shi, Q. (2020). Characterization of metabolites and transcripts involved in flower pigmentation in primula vulgaris. Front. Plant Sci. 11, 572517. doi: 10.3389/fpls.2020.572517
Liu, W., Mei, Z., Yu, L., Gu, T., Li, Z., Zou, Q., et al. (2023). The ABA-induced NAC transcription factor MdNAC1 interacts with a bZIP-type transcription factor to promote anthocyanin synthesis in red-fleshed apples. Horticulture Res. doi: 10.1093/hr/uhad049
Liu, M., Zhang, C., Duan, L., Luan, Q., Li, J., Yang, A., et al. (2019). CsMYB60 is a key regulator of flavonols and proanthocyanidans that determine the colour of fruit spines in cucumber. J. Exp. Bot. 70, 69–84. doi: 10.1093/jxb/ery336
Lo Piero, A. R. (2015). The state of the art in biosynthesis of anthocyanins and its regulation in pigmented sweet oranges [(Citrus sinensis) l. osbeck]. J. Agric. Food Chem. 63, 4031–4041. doi: 10.1021/acs.jafc.5b01123
Mahmood, K., Xu, Z., El-Kereamy, A., Casaretto, J. A., Rothstein, S. J. (2016). The arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci., 1548 7. doi: 10.3389/fpls.2016.01548
Mao, W., Han, Y., Chen, Y., Sun, M., Feng, Q., Li, L., et al. (2022). Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of chalcone synthase 1. Plant Cell 34, 1226–1249. doi: 10.1093/plcell/koac006
Martínez-Cuenca, M.-R., Primo-Capella, A., Forner-Giner, M. A. (2016). Influence of rootstock on citrus tree growth: effects on photosynthesis and carbohydrate distribution, plant size, yield, fruit quality, and dwarfing genotypes. Plant Growth 16, 107. doi: 10.5772/64825
Martín-Pizarro, C., Vallarino, J. G., Osorio, S., Meco, V., Urrutia, M., Pillet, J., et al. (2021). The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell 33, 1574–1593. doi: 10.1093/plcell/koab070
Meng, Y., Zhang, H., Fan, Y., Yan, L. (2022). Anthocyanins accumulation analysis of correlated genes by metabolome and transcriptome in green and purple peppers (Capsicum annuum). BMC Plant Biol. 22, 1–11. doi: 10.1186/s12870-022-03746-y
Modica, G., Pannitteri, C., Di Guardo, M., La Malfa, S., Gentile, A., Ruberto, G., et al. (2022). Influence of rootstock genotype on individual metabolic responses and antioxidant potential of blood orange cv. tarocco scirè. J. Food Composition Anal. 105, 104246. doi: 10.1016/j.jfca.2021.104246
Morales, J., Bermejo, A., Navarro, P., Forner-Giner, M.Á., Salvador, A. (2021). Rootstock effect on fruit quality, anthocyanins, sugars, hydroxycinnamic acids and flavanones content during the harvest of blood oranges ‘Moro’and ‘Tarocco rosso’grown in Spain. Food Chem. 342, 128305. doi: 10.1016/j.foodchem.2020.128305
Morales Alfaro, J., Bermejo, A., Navarro, P., Quinones, A., Salvador, A. (2021). Effect of rootstock on citrus fruit quality: a review. Food Rev. Int., 1–19. doi: 10.1080/87559129.2021.1978093
Morishita, T., Kojima, Y., Maruta, T., Nishizawa-Yokoi, A., Yabuta, Y., Shigeoka, S. (2009). Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 50, 2210–2222. doi: 10.1093/pcp/pcp159
Moufida, S. D., Marzouk, B. (2003). Biochemical characterization of blood orange, sweet orange, lemon, bergamot and bitter orange. Phytochemistry 62, 1283–1289. doi: 10.1016/S0031-9422(02)00631-3
Muhammad, N., Luo, Z., Yang, M., Li, X., Liu, Z., Liu, M. (2022). The joint role of the late anthocyanin biosynthetic UFGT-encoding genes in the flowers and fruits coloration of horticultural plants. Scientia Hortic. 301, 111110. doi: 10.1016/j.scienta.2022.111110
Nazir, M. F., He, S., Ahmed, H., Sarfraz, Z., Jia, Y., Li, H., et al. (2021). Genomic insight into the divergence and adaptive potential of a forgotten landrace g. áhirsutum l. purpurascens. J. Genet. Genomics 48, 473–484. doi: 10.1016/j.jgg.2021.04.009
Ohta, Y., Atsumi, G., Yoshida, C., Takahashi, S., Shimizu, M., Nishihara, M., et al. (2022). Post-transcriptional gene silencing of the chalcone synthase gene CHS causes corolla lobe-specific whiting of Japanese gentian. Planta 255, 1–12. doi: 10.1007/s00425-021-03815-w
Pallottino, F., Menesatti, P., Lanza, M. C., Strano, M. C., Antonucci, F., Moresi, M. (2013). Assessment of quality-assured tarocco orange fruit sorting rules by combined physicochemical and sensory testing. J. Sci. Food Agric. 93, 1176–1183. doi: 10.1002/jsfa.5871
Paredes-López, O., Cervantes-Ceja, M. L., Vigna-Pérez, M., Hernández-Pérez, T. (2010). Berries: improving human health and healthy aging, and promoting quality life–a review. Plant foods Hum. Nutr. 65, 299–308. doi: 10.1007/s11130-010-0177-1
Peterson, J. J., Dwyer, J. T., Beecher, G. R., Bhagwat, S. A., Gebhardt, S. E., Haytowitz, D. B., et al. (2006). Flavanones in oranges, tangerines (mandarins), tangors, and tangelos: a compilation and review of the data from the analytical literature. J. Food Composition Anal. 19, S66–S73. doi: 10.1016/j.jfca.2005.12.006
Piasecka, A., Jedrzejczak-Rey, N., Bednarek, P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206, 948–964. doi: 10.1111/nph.13325
Prior, R. L., Wu, X. (2006). Anthocyanins: structural characteristics that result in unique metabolic patterns and biological activities. Free Radical Res. 40, 1014–1028. doi: 10.1080/10715760600758522
Qiu, W., Su, W., Cai, Z., Dong, L., Li, C., Xin, M., et al. (2020). Combined analysis of transcriptome and metabolome reveals the potential mechanism of coloration and fruit quality in yellow and purple passiflora edulis sims. J. Agric. Food Chem. 68, 12096–12106. doi: 10.1021/acs.jafc.0c03619
Rapisarda, P., Bellomo, S. E., Intelisano, S. (2001). Storage temperature effects on blood orange fruit quality. J. Agric. Food Chem. 49, 3230–3235. doi: 10.1021/jf010032l
Rapisarda, P., Bianco, M. L., Pannuzzo, P., Timpanaro, N. (2008). Effect of cold storage on vitamin c, phenolics and antioxidant activity of five orange genotypes [Citrus sinensis (L.) osbeck]. Postharvest Biol. Technol. 49, 348–354. doi: 10.1016/j.postharvbio.2008.02.002
Raza, A., Tabassum, J., Kudapa, H., Varshney, R. K. (2021). Can omics deliver temperature resilient ready-to-grow crops? Crit. Rev. Biotechnol. 41, 1209–1232. doi: 10.1080/07388551.2021.1898332
Rodríguez-Gamir, J., Primo-Millo, E., Forner, J. B., Forner-Giner, M. A. (2010). Citrus rootstock responses to water stress. Scientia Hortic. 126, 95–102. doi: 10.1016/j.scienta.2010.06.015
Russo, M., Bonaccorsi, I. L., Arigò, A., Cacciola, F., De Gara, L., Dugo, P., et al. (2021). Blood orange (Citrus sinensis) as a rich source of nutraceuticals: investigation of bioactive compounds in different parts of the fruit by HPLC-PDA/MS. Natural Product Res. 35, 4606–4610. doi: 10.1080/14786419.2019.1696329
Schiestl, F. P., Johnson, S. D. (2013). Pollinator-mediated evolution of floral signals. Trends Ecol. Evol. 28, 307–315. doi: 10.1016/j.tree.2013.01.019
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shi, J., Simal-Gandara, J., Mei, J., Ma, W., Peng, Q., Shi, Y., et al. (2021). Insight into the pigmented anthocyanins and the major potential co-pigmented flavonoids in purple-coloured leaf teas. Food Chem. 363, 130278. doi: 10.1016/j.foodchem.2021.130278
Siracusa, L., Ruberto, G. (2014). “Polyphenols in plants,” in "Plant polyphenol profiles as a tool for traceability and valuable support to biodiversity," (Elsevier), 15–33. do: 10.1016/B978-0-12-397934-6.00002-4
Sirén, J., Välimäki, N., Mäkinen, V. (2014). HISAT2-fast and sensitive alignment against general human population. IEEE/ACM Trans. Comput. Biol. Bioinforma 11, 375–388.
Song, F., Bai, F., Wang, J., Wu, L., Jiang, Y., Pan, Z. (2020). Influence of citrus scion/rootstock genotypes on arbuscular mycorrhizal community composition under controlled environment condition. Plants 9, 901. doi: 10.3390/plants9070901
Su, J., Jiang, J., Zhang, F., Liu, Y., Ding, L., Chen, S., et al. (2019). Current achievements and future prospects in the genetic breeding of chrysanthemum: a review. Horticulture Res. 6. doi: 10.1038/s41438-019-0193-8
Subramanian, I., Verma, S., Kumar, S., Jere, A., Anamika, K. (2020). Multi-omics data integration, interpretation, and its application. Bioinf. Biol. Insights 14, 1177932219899051. doi: 10.1177/1177932219899051
Sun, Y., Zhou, M., Luo, L., Pan, H., Zhang, Q., Yu, C. (2023). Metabolic profiles, bioactive compounds and antioxidant activity of rosehips from xinjiang, China. LWT 114451. doi: 10.1016/j.lwt.2023.114451
Tahir Ul Qamar, M., Faryad, A., Bari, A., Zahid, B., Zhu, X., Chen, L.-L. (2020). “Environment, climate, plant and vegetation growth,” in "Effectiveness of conventional crop improvement strategies vs. omics," (Springer Nature Switzerland: Springer), 253–284.
Titta, L., Trinei, M., Stendardo, M., Berniakovich, I., Petroni, K., Tonelli, C., et al. (2010). Blood orange juice inhibits fat accumulation in mice. Int. J. Obes. 34, 578–588. doi: 10.1038/ijo.2009.266
Vikhorev, A. V., Strygina, K. V., Khlestkina, E. K. (2019). Duplicated flavonoid 3’-hydroxylase and flavonoid 3’, 5’-hydroxylase genes in barley genome. PeerJ 7, e6266. doi: 10.7717/peerj.6266
Wang, J.-H., Liu, J.-J., Chen, K.-L., Li, H.-W., He, J., Guan, B., et al. (2017). Comparative transcriptome and proteome profiling of two citrus sinensis cultivars during fruit development and ripening. BMC Genomics 18, 1–13. doi: 10.1186/s12864-017-4366-2
Wang, Y., Zeng, X., Xu, Q., Mei, X., Yuan, H., Jiabu, D., et al. (2019). Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. AoB Plants 11, plz021. doi: 10.1093/aobpla/plz021
Xue, Q., Zhang, X., Yang, H., Li, H., Lv, Y., Zhang, K., et al. (2021). Transcriptome and metabolome analysis unveil anthocyanin metabolism in pink and red testa of peanut (Arachis hypogaea l.). Int. J. Genomics 2021, 5883901. doi: 10.1155/2021/5883901
Yu, H., Li, H., Wei, R., Cheng, G., Zhou, Y., Liu, J., et al. (2022). Widely targeted metabolomics profiling reveals the effect of powdery mildew on wine grape varieties with different levels of tolerance to the disease. Foods, 2461 11. doi: 10.3390/foods11162461
Zhang, S., Chen, Y., Zhao, L., Li, C., Yu, J., Li, T., et al. (2020). A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 40, 413–423. doi: 10.1093/treephys/tpaa004
Zhang, C., Hao, Y.-J. (2020). Advances in genomic, transcriptomic, and metabolomic analyses of fruit quality in fruit crops. Hortic. Plant J. 6, 361–371. doi: 10.1016/j.hpj.2020.11.001
Zhou, H., Lin-Wang, K., Wang, H., Gu, C., Dare, A. P., Espley, R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. doi: 10.1111/tpj.12792
Keywords: blood orange, flavonoids, pigmentation, gene expression, quality improvement
Citation: Yang L, Chen Y, Wang M, Hou H, Li S, Guan L, Yang H, Wang W and Hong L (2023) Metabolomic and transcriptomic analyses reveal the effects of grafting on blood orange quality. Front. Plant Sci. 14:1169220. doi: 10.3389/fpls.2023.1169220
Received: 19 February 2023; Accepted: 02 May 2023;
Published: 01 June 2023.
Edited by:
Karthikeyan Adhimoolam, Jeju National University, Republic of KoreaCopyright © 2023 Yang, Chen, Wang, Hou, Li, Guan, Yang, Wang and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Hong, bG9xdWF0dmFsdWVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Lei Yang1†
Lei Yang1† Yang Chen
Yang Chen Haijian Yang
Haijian Yang