Abstract
Leaf angle shapes plant architecture, allowing for optimal light interception to maximize photosynthesis and yield, and therefore is a crucial agronomic trait. Here, we show that the rice (Oryza sativa L.) R2R3-type MYB transcription factor OsMYB7 determines leaf angle in a developmental stage-specific manner. OsMYB7-overexpressing lines produced wide-angled leaves and osmyb7 knockout mutants exhibited erect leaves. This phenotype was restricted to the lamina joints at the late developmental stage. In agreement with these observations, OsMYB7 was preferentially expressed in the lamina joints of post-mature leaves. Since OsMYB7 homologs are transcriptional repressors of lignin biosynthesis, we examined whether OsMYB7 might inhibit thickening of secondary cell walls. Although OsMYB7 repressed lignin biosynthesis, it enhanced thickening of sclerenchyma cell walls by elevating cellulose contents at the lamina joints. Furthermore, we found that OsMYB7 affects endogenous auxin levels in lamina joints, and the adaxial cells of lamina joints in OsMYB7-overexpressing lines and osmyb7 knockout mutants exhibited enhanced and reduced elongation, respectively, compared to the wild type. These results suggest that OsMYB7 promotes leaf inclination partially through decreasing free auxin levels and promoting cell elongation at the adaxial side of lamina joints.
Introduction
Leaf angle, the angle formed between the leaf blade and the culm, is a major agronomic trait in rice (Oryza sativa L.) (Donald, 1968). Indeed, erect leaves improve penetration of sunlight into dense fields, thus enhancing photosynthetic efficiency of the canopy and allowing higher planting density for increased grain production (Wang J. et al., 2020). Plant hormones are the primary factors controlling leaf inclination (Xu et al., 2021). Among them, brassinosteroids (BRs) play a dominant role in determining leaf angle, as most of the mutants with altered leaf angles are associated with malfunctions of BR metabolism or signaling. For example, mutants with elevated BR contents or enhanced BR signaling exhibited increased angles of the lamina joints, whereas those with reduced BR contents or diminished BR signaling exhibited erect leaves (Yamamuro et al., 2000; Mori et al., 2002; Wang et al., 2002; Tanabe et al., 2005; Sakamoto et al., 2006; Jang, 2017). These studies also revealed that BRs accelerate elongation and/or division of the adaxial parenchyma cells at lamina joints, indicating that BRs positively regulate leaf inclination.
Indole-3-acetic acid (IAA), the most abundant naturally occurring auxin, is also a crucial plant hormone that controls leaf angle (Li et al., 2020). In contrast to BRs, IAA negatively regulates leaf inclination, based on the observations that higher IAA contents result in upright leaves while lower IAA causes horizontal leaves (Kim et al., 2013; Yoshikawa et al., 2014). IAA homeostasis is coordinately modulated by its biosynthesis and conjugation. Most IAA is synthesized via two enzymatic steps involving tryptophan aminotransferase (TAA) and flavin monooxygenase (YUCCA) family members. In addition, conjugation of IAA with amino acids, such as aspartate and glutamate, is mainly catalyzed by GRETCHEN HAGEN 3 (GH3) family members and effectively reduces the free IAA concentration (Wang et al., 2018; Hayashi et al., 2021). To date, at least five rice IAA homeostatic genes (OsTAA1, OsGH3-1, OsGH3-2, OsGH3-5, and OsGH3-13) have been identified as involved in regulating leaf angle (Zhang et al., 2009; Du et al., 2012; Zhao et al., 2013; Yoshikawa et al., 2014; Zhang et al., 2015). When OsTAA1 was mutated or OsGH3 genes were overexpressed, the resulting deficiency in IAA promoted the expansion or division of parenchyma cells at the adaxial side of lamina joints, thereby increasing leaf angles. In addition to these IAA homeostasis genes, several auxin signaling genes, such as AUXIN-RESPONSE FACTOR 19 (OsARF19) and LEAF INCLINATION 3 (LC3), affect leaf inclination by directly regulating the expression of OsGH3s (Zhang et al., 2015; Chen et al., 2018).
Besides phytohormones, lignocellulose, a main structural component of the secondary cell wall (SCW), modulates leaf angle through determining the mechanical strength of lamina joints; lignocellulose-deficient mutants exhibited wide leaf angles and mutants with increased lignocellulose contents showed an erect-leaf phenotype (Sun et al., 2020; Wang R. et al., 2020; Huang et al., 2021). For instance, OVATE FAMILY PROTEIN 6 (OsOFP6) knockout rice mutants displayed much wider leaf angles due to the lack of lignocellulose at their lamina joints in a BR-independent manner (Sun et al., 2020). In another case, OsARF6 and OsARF17 were shown to maintain SCW-mediated mechanical strength for supporting leaf weight: the osarf6 osarf17 double mutant exhibited a horizontal leaf angle phenotype due to lignocellulose deficiency at the lamina joints (Huang et al., 2021). Recently, several key transcription factors, which are predicted to downregulate lignin biosynthesis during lamina joint development, were proposed to contribute to leaf inclination: the knockout lines of the corresponding genes, such as Os04g0549700, Os02g0656600, Os03g0182800, and Os07g0674800, had more lignin contents in the sclerenchyma cells of their lamina joints, thereby exhibiting upright leaves (Wang R. et al., 2020).
Several MYB transcription factors regulating biosynthetic programs of SCW components have been identified in plants (Miyamoto et al., 2020). Although the functions of the MYB transcription factors in deposition of lignin, cellulose, and xylan have been extensively studied, additional possible role in phytohormone metabolism or cell growth is not well documented. OsMYB7 encodes a MYB transcriptional repressor with cell wall–associated functions and in this study, we investigated the plants which overexpressed or knockout of OsMYB7. Our findings demonstrate that OsMYB7 increases leaf angle: two independent OsMYB7 overexpressors displayed wide leaf angles, whereas osmyb7-1 knockout mutant showed narrow leaf angles. OsMYB7 inhibited lignin accumulation in the lamina joints. However, further studies revealed that OsMYB7 slightly increases the thickness of the SCW, a primary determinant of mechanical strength, by promoting cellulose deposition. Our results suggest that the alteration in leaf angle may not be primarily due to OsMYB7-mediated modulation of the SCW components, although we cannot exclude the possibility that the leaf angle phenotypes of OsMYB7 overexpressors and osmyb7-1 knockout mutant are due to altered levels of phenylpropanoids. We also found that the contents of free auxin in lamina joints were changed in OsMYB7-overexpressing lines and osmyb7 knockout mutants and that the cell size at the adaxial side of lamina joints was altered in these plants compared to that of WT. Thus, we propose that OsMYB7 increases rice leaf angles partially by decreasing auxin contents and accelerating parenchyma cell elongation at the adaxial side of lamina joints. Collectively, our findings provide a new perspective on the biological functions of cell wall-associated MYB transcription factors and offer a potential biotechnological avenue for breeding new gene-edited rice varieties with erect leaves for high-density cropping.
Materials and methods
Plant materials and growth conditions
The activation-tagged T-DNA insertion line of OsMYB7, designated OsMYB7-OE1 (PFG_4A-03610), was obtained from the Salk Institute Genomics Analysis Laboratory (http://signal.salk.edu/cgi-bin/RiceGE) (Jeon et al., 2000; Jeong et al., 2002). Plants of OsMYB7-OE1, OsMYB7-OE2, osmyb7-1, and the parental japonica cultivar ‘Dongjin’ (referred to as wild type; WT) rice (Oryza sativa) were cultivated under natural long day conditions (around 14 h light per day) in a paddy field (Suwon, Republic of Korea, 37°N latitude). The seedlings were initially grown for one month in a greenhouse after sowing on seed beds, and subsequently transplanted into the paddy field following standard agricultural practices for Korean rice varieties.
Vector construction and rice transformation
To generate the OsMYB7-OE2 transgenic line, the full-length coding sequence of OsMYB7 was amplified by PCR from cDNA obtained from total RNA extracted from lamina joints of WT as template and gene-specific primers (Supplementary Table S4). The PCR product was subcloned into the entry vector pCR™8/GW/TOPO® (Invitrogen, Carlsbad, CA, USA) and then transferred into the gateway-compatible binary destination vector pMDC32 (Curtis and Grossniklaus, 2003) by performing an LR recombination reaction using a Gateway™ LR Clonase™ II Enzyme Mix (Invitrogen). To generate the osmyb7-1 mutant, an OsMYB7-specific 20-nt spacer sequence, TGTCAGGTGGTCTCTGATCG, was designed using the CRISPRdirect software (https://crispr.dbcls.jp/) (Naito et al., 2015) and subcloned into the single guide RNA expression cassette of the pOs-sgRNA entry vector (Miao et al., 2013). The resulting cassette was then transferred into the pH-Ubi-cas9-7 destination vector containing a Cas9 expression cassette (Miao et al., 2013) through LR reaction.
The plasmid constructs were introduced into Agrobacterium tumefaciens strain LBA4404 (Ooms et al., 1982) via the freeze-thaw method (Höfgen and Willmitzer, 1988). Mature seed embryos of WT were used to generate calli, which were then transformed with the Agrobacterium-mediated transformation protocol described by Jeon et al. (2000). The transformed calli were selected on 2N6 medium supplemented with 50 mg L-1 hygromycin (Duchefa Biochemie, Haarlem, the Netherlands), and the regenerated plants were confirmed as transgenic rice.
Transactivation and trans-repression assays
For transactivation assay, we cloned the full-length coding sequence of OsMYB7 into the vector pGBKT7 (BD Biosciences Clontech) between SalI and NotI sites, and that of OsbHLH079 into the same vector between EcoRI and NotI sites. For trans-repression assay, the activation domain of GAL4 transcription factor in the vector pGADT7 (BD Biosciences Clontech) was cloned in frame between NcoI and EcoRI sites in the vector pGBKT7 (BD Biosciences Clontech) to generate the vector rGAL4, as previously described (Mathew et al., 2016). Next, we cloned the full-length coding sequences of OsMYB7 and ONAC026 into the vector rGAL4 between SalI and NotI sites. All the resulting constructs were transformed into the Saccharomyces cerevisiae strain AH109. Subsequently, yeast β-galactosidase liquid assays were performed according to the Yeast Protocols Handbook (BD Biosciences Clontech) using chlorophenol red-β-D-galactopyranoside (CPRG; Roche, Basel, Switzerland) as substrate. Briefly, we incubated yeast extracts in 8 mM CPRG (Roche) for 30 min at 30°C in the dark and measured the absorbance of extracts at a wavelength of 574 nm using a UV/VIS spectrophotometer (PowerWave X, BioTek, Winooski, USA). Primers details are provided in Supplementary Table S4.
Subcellular localization
The full-length coding sequence of OsMYB7 subcloned into the entry vector pCR™8/GW/TOPO® (Invitrogen) was transferred into the Gateway-compatible plant destination vector pEarleyGate 104 (Earley et al., 2006) by the LR reaction. The resulting vector was introduced into onion (Allium cepa) epidermal cells by particle bombardment with a Biolistic PDS-1000/He instrument (Bio-Rad, Hercules, CA, USA). After bombardment, the onion epidermal layers were incubated on Murashige and Skoog (MS) medium (pH 5.7) for 16 h at 22°C in the dark. The nuclei were then stained with 300 nM of 4’,6-diamidino-2-phenylindole (DAPI; Invitrogen) in phosphate-buffered saline (PBS) for 3 min in darkness before observation. Fluorescence emission was analyzed using a Leica TCS SP8 X confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) with the following parameters: excitation at 458 nm and emission at 514 nm for YFP, and excitation at 405 nm and emission at 488 nm for DAPI.
Gene expression analysis (qPCR)
Total RNAs from rice plants were extracted using a MG Total RNA Extraction Kit (MGmed, Seoul, Republic of Korea) according to the manufacturer’s instructions and subjected to reverse transcription with an Oligo(dT)15 Primer (Promega, Madison, WI, USA) and M-MLV Reverse Transcriptase (Promega) to generate first-strand cDNAs. The resulting product mixtures were diluted with distilled water by a factor of four. Quantitative PCR (qPCR) was performed with a GoTaq® qPCR Master Mix (Promega) using a LightCycler® 480 System (Roche) as described in the instruction manual. The reaction mix of a 20-μl final volume for qPCR was prepared by combining the 2 μl of first-strand cDNA mixture, 10 μl of GoTaq® qPCR Master Mix, 0.4 μl of 10 μM forward primer, 0.4 μl of 10 μM reverse primer, and 7.2 μl of nuclease-free water. The gene-specific primers are listed in Supplementary Table S4, and the qPCR conditions were as follows: 95°C for 2 min followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min. Data obtained by qPCR were analyzed using the 2–ΔΔCT method (Livak and Schmittgen, 2001) with GAPDH as a reference for normalization (Jain et al., 2006).
Histochemical detection of GUS enzymatic activity
The lamina joints of OsMYB7-OE1 carrying a β-glucuronidase (GUS) reporter gene driven by the OsMYB7 promoter (Supplementary Figure S3A) were subjected to histochemical GUS staining according to the previously described method (Jefferson et al., 1987) with some modifications. Lamina joints at developmental stages S3, S4, and S5 were sampled and vacuum-infiltrated with a X-Gluc reaction buffer containing 100 mM sodium phosphate buffer (pH 7.0), 10 mM EDTA, 0.1% Triton X-100, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, and 1 mM X-Gluc (Sigma-Aldrich, Saint Louis, MO, USA) for 30 min in darkness, followed by overnight incubation at 37°C under dark conditions. The GUS histochemical staining buffer was then replaced with 70% (v/v) aqueous ethanol to remove chlorophylls. After complete decolorization, images of lamina joints were capture with a digital camera, and transverse hand-cut sections of lamina joints were subsequently photographed using a stereo microscope (SteREO Discovery.V12, Carl Zeiss Microscopy GmbH, Jena, Germany) with dark filter.
Phloroglucinol-HCl staining and calcofluor white staining
The lamina joints of rice plants were collected at 30 DAH and vacuum-infiltrated immediately in FAA solution (70% [v/v] aqueous ethanol, glacial acetic acid, and formalin in a ratio of 90:5:5 v/v/v) for 30 min, followed by overnight incubation at room temperature with gentle rotation. Hand-cut cross sections were prepared from the fixed lamina joints and washed twice with 70% (v/v) aqueous ethanol for 30 min each with gentle inversion. For histochemical observation of lignin, phloroglucinol-HCl staining was conducted according to a standard protocol (Halpin et al., 1998) with some modifications. Briefly, the sections were immersed in 1% (w/v) solution of phloroglucinol (Sigma-Aldrich) in 70% (v/v) aqueous ethanol for 5 min and then treated with 18% (v/v) aqueous HCl for 5 min before being observed with a stereo microscope (SteREO Discovery.V12, Carl Zeiss Microscopy GmbH). To visualize deposited cellulose, calcofluor white staining was carried out as described previously (Hageage and Harrington, 1984) with a little modification. Briefly, each section was stained with one drop of calcofluor white stain (Sigma-Aldrich) and one drop of 10% (w/v) aqueous potassium hydroxide for 2 min and examined using an inverted phase contrast fluorescence microscope (Axio Observer Z1, Carl Zeiss Microscopy GmbH) under DAPI filter.
Preparation of destarched alcohol-insoluble cell wall residues
The isolation of dsAIRs for the quantification of lignins and celluloses was performed, following the procedure described previously (Zhang et al., 2019) with some modifications. Around 1 g of lamina joints from rice plants at 30 DAH was ground in liquid nitrogen and lyophilized using a freeze dryer (Bondiro, ilShin® Lab Co. Ltd., Yang-Ju, Republic of Korea) for 48 h. Approximately 150 mg of each freeze-dried sample was weighed into a 5-mL snap-cap centrifuge tube and dissolved sequentially with 3 ml of 70% (v/v) aqueous ethanol for two times, 3 ml of chloroform/methanol (1:1 v/v) for one time, and 3 ml of acetone for one time, followed by centrifugation at 1,500 g for 10 min each. The resultant pellet was transferred to a 2 ml screw cap microcentrifuge tube and dried in an oven at 35°C until completely dry. The dried sample was then resuspended in 1.5 ml of 0.1 M sodiumacetate buffer (pH 5.0) and incubated in a heat block for 20 min at 80°C. After cooling on ice, the following agents were added: 35 μl of 0.01% (w/v) sodiumazide in 0.1 M sodiumacetate buffer (pH 5.0), 35 μl of 50 μg/ml amylase (α-Amylase from Bacillus sp., Sigma-Aldrich) in distilled water, and 17 μl of pullulanase (Pullulanase microbial, Sigma-Aldrich). Following overnight incubation in a shaking incubator at 37°C with gentle rpm, the suspension was heated for 10 min using a heat block at 100°C to terminate digestion. The solubilized starch in supernatant was then discarded after centrifugation at 1,500 g for 10 min. The resultant pellet was washed with 1.5 ml of distilled water for three times and 0.5 ml of acetone for two times, followed by centrifugation at 1,500 g for 10 min each. The remaining solvent was totally evaporated in an oven at 35°C. The dried material, dsAIR, was then subjected to the acetyl bromide assay for lignin or anthrone assay for cellulose as described below.
Lignin quantification
To measure the lignin content in dsAIR, an acetyl bromide assay was conducted as previously described (Moreira-Vilar et al., 2014) with a little modification. The 20 mg of dsAIR was weighed into a 5 ml screw cap centrifuge tube and dissolved with 500 μl of 25% (v/v) acetyl bromide (Sigma-Aldrich) in glacial acetic acid for 1 h using an oven at 70°C. After cooling on ice, the following reagents were added successively to the suspension: 0.9 ml of 2 M aqueous NaOH, 0.1 ml of 5 M aqueous hydroxylamine-HCl, and 3.5 ml of glacial acetic acid. The absorbance of each supernatant containing solubilized lignin was measured at a wavelength of 280 nm (Su et al., 2005) using a UV/VIS spectrophotometer (PowerWave X, BioTek) after centrifugation at 1,400 g for 10 min. The content of lignin was subsequently calculated using a standard curve generated with alkali lignin (Sigma-Aldrich).
Cellulose quantification
To quantify cellulose, an anthrone assay was performed after the isolation of crystalline cellulose from dsAIR, following a previously described protocol (Updegraff, 1969) with slight modifications. The 2-3 mg of dsAIR was weighed into a 2 ml screw cap microcentrifuge tube and treated with 250 μl of 2 M trifluoroacetic acid (Sigma-Aldrich) at 121°C for 2 h using a heat block. After cooling on ice, the supernatant was replaced with 1 ml of isopropanol after centrifugation at 11,000 g for 10 min. The sample was then centrifuged at 11,000 g for 10 min, and the supernatant was discarded. The remaining solvent was exhaustively removed using an oven at 35°C. The 1 ml of Updegraff reagent (glacial acetic acid: nitric acid: DW, 8:1:2 v/v/v) was added to the TFA pellet, and the suspension was heated in a heat block at 100°C for 1 h, followed by cooling on ice. The pellet, crystalline cellulose, was obtained after centrifugation at 11,000 g for 15 min and washed once with 1.5 ml of distilled water and three times with 1.5 ml of acetone, followed by centrifugation at 11,000 g for 10 min each. The resultant sample was incubated in an oven at 35°C for 16 h and then completely hydrolyzed into glucose by being treated with 175 μl of 72% (w/w) aqueous sulfuric acid (Alfa Aesar, Ward Hill, MA, USA) at room temperature for 90 min. After the dilution with 825 μl of distilled water, the suspension was centrifuged at 11,000 g for 5 min. The glucose content in resultant supernatant was analyzed using the colorimetric anthrone assay. The 10 μl of supernatant was mixed with 90 μl of distilled water and 200 μl of anthrone reagent in a 96-well polystyrene microtiter plate, followed by incubation in an oven at 80°C for 30 min. The standards prepared with D-(+)-glucose (Sigma-Aldrich) were also subjected to the anthrone assay on the same plate. Absorbance of each sample at 625 nm was read using a UV/VIS spectrophotometer (PowerWave X, BioTek), and the contents of cellulose were calculated based on a standard curve.
Transmission electron microscopy
Sample preparation for TEM was conducted essentially following the conventional method (Inada et al., 1998) but with the help of microwave irradiation using a microwave tissue processor (hereafter termed microwave) (PELCO BioWave® Pro+, Ted Pella, Redding, CA, USA) as previously described (Mowery and Bauchan, 2018) with some modifications. The collars from rice plants were harvested and vacuum-infiltrated in modified Karnovsky’s fixative (2% [w/v] glutaraldehyde and 2% [w/v] paraformaldehyde in 50 mM sodium cacodylate buffer, pH 7.2) for 30 min, followed by overnight incubation at 4°C. After being rinsed three times with 50 mM sodium cacodylate buffer (pH 7.2) at 4°C for 5 min each, the samples were post-fixed by 1% (w/v) osmium tetroxide in 50 mM sodium cacodylate buffer, pH 7.2, using the microwave (2 min On, 2 min Off, 2 min On, 2 min Off, 4 min On, 2 min Off, and 4 min On; 100 W; Tmax = 30°C). Post-fixed specimens were then briefly washed three times with distilled water at room temperature and subjected to microwave-assisted en bloc staining with 0.5% (w/v) uranyl acetate (1 min On, 1 min Off, and 1 min On; 100 W; Tmax = 30°C). The dehydration of samples was performed in the microwave with increasing concentrations of ethanol as follows: single changes in 30%, 50%, 70%, 80%, and 90% ethanol (all v/v) followed by three changes in 100% ethanol (40 s On; 150 W; Tmax = 35°C each). The dehydrated specimens were subsequently incubated twice in propylene oxide under microwave irradiation (40 s On; 150 W; Tmax = 35°C each) and gradually infiltrated with a graded series of increasing concentrations of Spurr’s resin in propylene oxide (once with 30%, 50%, and 80% Spurr’s resin each, followed by two times with 100% Spurr’s resin; all v/v) using the microwave (15 min On; 200 W; Tmax = 40°C each). Next, samples were embedded in pure Spurr’s resin and finally polymerized in an oven at 70°C for 24 h. The specimens in the resulting resin blocks were thinly sectioned (70 nm) using an ultramicrotome (EM UC7, Leica Microsystems) with a Diatome diamond knife, mounted on Formvar-coated copper grids (EMS, Hatfield, PA, USA), and post-stained with 2% (w/v) uranyl acetate for 10 min and Reynolds’ lead citrate for 10 min at room temperature before being observed with a transmission electron microscope (Talos L120C, FEI, Czech Republic) operated at 120 kV.
LC-ESI-MS/MS analysis
The contents of IAA, IAA-Asp, and IAA-Glu were measured following previously described procedures (Matsuda et al., 2005) with some modifications. Approximately 2 g of lamina joints from rice plants was homogenized with a mortar and pestle in liquid nitrogen and lyophilized using a freeze dryer (Bondiro, ilShin® Lab Co. Ltd.) in darkness for 72 h. Around 500 mg of freeze-dried sample was weighed into a 10-mL light safe centrifuge tube and dissolved in 10 mL of 80% (v/v) aqueous acetone containing 2.5 mM diethyl dithiocarbamate using an ultrasonic bath (Powersonic 420, Hwashin Tech Co. Ltd., Gwangju, Republic of Korea) at 0°C for 2 h, followed by gentle rotation at 4°C for 4 h. After centrifugation at 3,500 g for 15 min at 4°C, the supernatant was transferred to a 50-mL light safe centrifuge tube. The extraction procedure was repeated once with the same sample, and the resulting supernatant was combined with the first supernatant in the 50-mL light safe centrifuge tube. As an internal standard, 0.2 nmol of [phenyl-13C6]-indole-3-acetic acid (Cambridge Isotope Laboratories, Andover, MA, USA) was added at the first step of extraction. The combined extract was then passed through s 0.2-μm PTFE membrane filter, and the solvent was completely evaporated in a vacuum. The residue was subsequently resuspended in 500 μL of 10% (v/v) aqueous methanol containing 1% (v/v) acetic acid and subjected to LC-ESI-MS/MS analysis: a Dionex UltiMate 3000 system (Thermo Fisher Scientific, CA, USA) coupled to a triple quadrupole mass spectrometer (TSQ Altis™, Thermo Fisher Scientific) was used for the analysis. A 10-μl aliquot of the sample was injected onto a Cadenza CD-C18 column (75 × 2.0 mm, 3 μm; Imtakt, Kyoto, Japan) and eluted using a mixture of methanol: 0.1% aqueous acetic acid (gradient from 10:90 to 90:10 in 13 min) with a flow rate of 200 μl/min at 30°C. The triple quadrupole mass spectrometer was equipped with an electrospray interface and operated using the multiple reaction monitoring (MRM) in positive ion mode, and the transitions from precursor to product ions for each compound were monitored as follows: [phenyl-13C6]-IAA, m/z of 182.1 to 136.0; IAA, m/z of 176.1 to 130.0; IAA-Asp, m/z of 291.1 to 130.0; IAA-Glu, m/z of 305.1 to 130.0. The levels of IAA, IAA-Asp, and IAA-Glu in each sample were calculated using corresponding calibration curves generated from each external standard. The standard for IAA was purchased from Sigma-Aldrich, and those for IAA-Asp and IAA-Glu were bought from Biosynth Carbosynth® (Compton, Berkshire, UK).
Scanning electron microscopy
Sample preparation for SEM was performed according to a method described previously (Bozzola and Kuo, 2007) with slight modifications. Approximately 1-cm lamina joint segments, containing leaf blade, collar, and leaf sheath, were hand-sectioned longitudinally and immersed overnight in modified Karnovsky’s solution (2% paraformaldehyde and 2% glutaraldehyde in 50 mM sodium cacodylate buffer, pH 7.2) at 4°C. The samples were then washed in three changes of 50 mM sodium cacodylate buffer, pH 7.2, at 4°C for 10 min each, post-fixed with 1% osmium tetroxide in 50 mM sodium cacodylate buffer (pH 7.2) at 4°C for 2 h, briefly rinsed three times in distilled water at room temperature, and dehydrated using a graded series of ethanol consisting of 30% (v/v), 50%, 70%, 80%, 90%, 100%, 100%, and 100% ethanol at 4°C for 10 min each. After the final change of 100% ethanol, the specimens were completely dried with the help of liquid CO2 using a critical point dryer (EM CPD300, Leica Microsystems), mounted on aluminum stubs using conductive carbon adhesive tapes, platinum-coated with a sputter coater (EM ACE200, Leica Microsystems), and finally examined by a field emission scanning electron microscope (Sigma, Carl Zeiss Microscopy GmbH) with an accelerating voltage of 5 kV and a working distance of 7.5 mm.
Accession numbers
Sequence data from this article can be found in the Rice Genome Annotation Project (http://rice.uga.edu/analyses_search_locus.shtml), GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and EMBL’s European Bioinformatics Institute (EMBL-EBI; https://www.ebi.ac.uk/) databases under accession numbers listed in Supplementary Table S3.
Results
OsMYB7 is a nucleus-localized R2R3-type MYB transcriptional repressor
OsMYB7 is an R2R3-type MYB transcription factor in the MYB4/7/32 clade (Miyamoto et al., 2020) (Figure 1A). The MYB transcription factors in this clade, such as OsMYB108 from Oryza sativa (Miyamoto et al., 2019), ZmMYB42, ZmMYB31 from Zea mays (Sonbol et al., 2009; Fornalé et al., 2010), as well as AtMYB4, AtMYB7, and AtMYB32 from Arabidopsis thaliana (Jin et al., 2000; Preston et al., 2004; Fornalé et al., 2014), have been demonstrated to act as negative regulators of lignin biosynthesis. Multiple sequence alignment revealed that the OsMYB7 homologs contain a conserved LxLxL-type of ERF-associated Amphiphilic Repression (EAR) motif, a transcriptional repression motif (Kagale and Rozwadowski, 2011), along with a typical MYB DNA-binding domain composed of two R2 and R3 repeat motifs (Katiyar et al., 2012) (Supplementary Figure S1). In addition, we established that OsMYB7 exhibits strong trans-repression activity with no transactivating property in yeast (Saccharomyces cerevisiae) (Figures 1B, C): OsbHLH079 (Seo et al., 2020), a bHLH transcription factor with transactivation activity, was used as the positive control for transactivation assay, and ONAC026 (Mathew et al., 2016), a NAC transcription factor with trans-repression activity, was used as the positive control for trans-repression assay (see Supplementary Table S3). The results suggest that OsMYB7 acts as a transcriptional repressor in lignin biosynthesis.
Figure 1
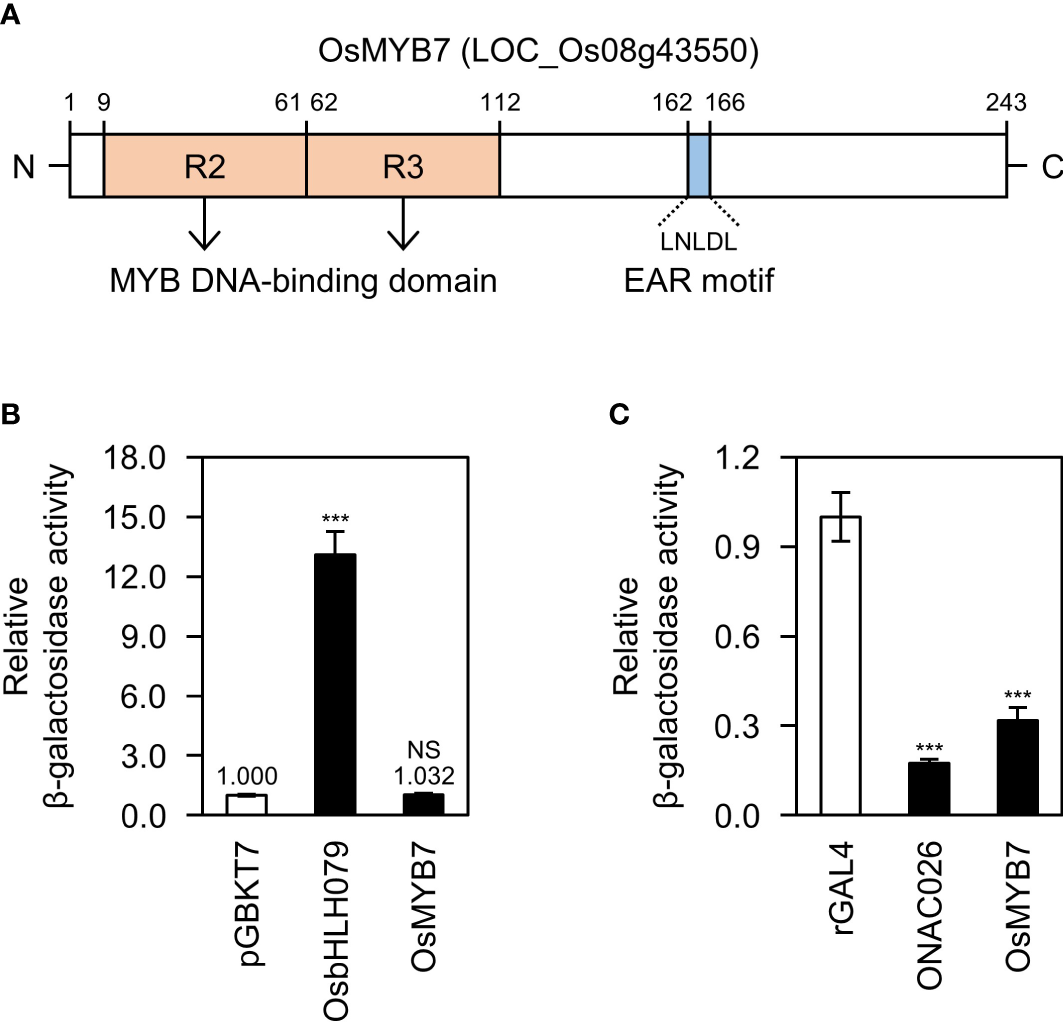
Biochemical characterization of OsMYB7. (A) Schematic diagram of the OsMYB7 protein. The two pale orange boxes indicate the highly conserved R2 and R3 MYB repeats. OsMYB7 also harbors an LxLxL-type EAR transcriptional repression motif, shown by the blue box. EAR, ethylene-responsive element binding factor-associated amphiphilic repression. (B) Transactivation activity assay of OsMYB7 in yeast. The full-length coding sequence of OsMYB7 cloned in the vector pGBKT7 was subjected to a quantitative β-galactosidase liquid assay: OsbHLH079 was used as a positive control, and pGBKT7 was used as a negative control. The β-galactosidase activity of each sample was normalized relative to that of the negative control, which was set to 1. Data are presented as means ± standard deviation (SD) from four independent yeast colonies. Significant differences between means were determined using two-tailed Student’s t-test (***P < 0.001). This experiment was performed twice with similar results. NS, not significant. (C) Trans-repression activity assay of OsMYB7 in yeast. The full-length coding sequence of OsMYB7 cloned in the vector rGAL4 was subjected to a quantitative β-galactosidase liquid assay: ONAC026 was used as a positive control, and rGAL4 was used as a negative control. The β-galactosidase activity of each sample was normalized to that of the negative control, which was set to 1. Data are presented as means ± SD of four independent yeast colonies. Asterisks indicate significant differences as determined by two-tailed Student’s t-test (***P < 0.001). This experiment was conducted twice yielding similar results. NS, not significant.
OsMYB7 and its homologs are predicted to localize to the nucleus in silico (Supplementary Table S1). To clarify the subcellular location of OsMYB7 in vivo, we transiently expressed a construct encoding OsMYB7 fused to yellow fluorescent protein (YFP-OsMYB7) in onion epidermal cells. We detected YFP signals exclusively in the nucleus, confirming that OsMYB7 is a nucleus-localized protein (Supplementary Figure S2).
OsMYB7 promotes leaf inclination at late stages of lamina joint development
To elucidate the physiological function of OsMYB7 in plant development, we obtained two different OsMYB7 overexpressing lines (OsMYB7-OE1 and OsMYB7-OE2) and an osmyb7 knockout mutant (osmyb7-1) (Supplementary Figure S3). The OsMYB7-OE1 line carries an activation-tagged T-DNA insertion at 80 nucleotides upstream of the start codon of OsMYB7, while the OsMYB7-OE2 line contains a 35S::OsMYB7 transgene (Supplementary Figures S3A–C). RT-qPCR analysis confirmed high levels of OsMYB7 transcripts in both overexpressing lines compared to the WT (Supplementary Figure S3D). The osmyb7-1 mutant, generated using CRISPR/Cas9-mediated targeted mutagenesis, harbors a homozygous cytosine insertion in the second exon of OsMYB7, resulting in a truncated protein of 178 amino acids with a disrupted EAR motif and a portion of the DNA binding domain (Supplementary Figures S3E–G).
We observed altered leaf angle phenotypes in these lines under natural long-day conditions in a paddy field. At the booting stage (105 days after sowing [DAS]), we detected significant changes in leaf angles in the 4th and 5th leaves: OsMYB7 overexpressors showed wide leaf angles, whereas the osmyb7-1 mutant had narrow leaf angles compared to the parental japonica rice cultivar ‘Donglin’ (hereafter termed wild type; WT) (Figure 2A). The differences in leaf angles of the 1st, 2nd, and 3rd leaves, also began to emerge at around 10 days after heading (DAH) and became more pronounced at the grain ripening stage (30 DAH) (Figure 2B): all leaves of OsMYB7 overexpressors took on a more horizontal orientation, while those of the osmyb7-1 mutant were in a more vertical orientation compared to WT. Consistent with the field observations, only the 4th and 5th leaf angles showed differences at the booting stage among the examined plants (Figures 2C, E), although the leaf angles of all leaves in OsMYB7 overexpressors and the osmyb7-1 mutant were markedly altered at the ripening stage (Figures 2D, F).
Figure 2
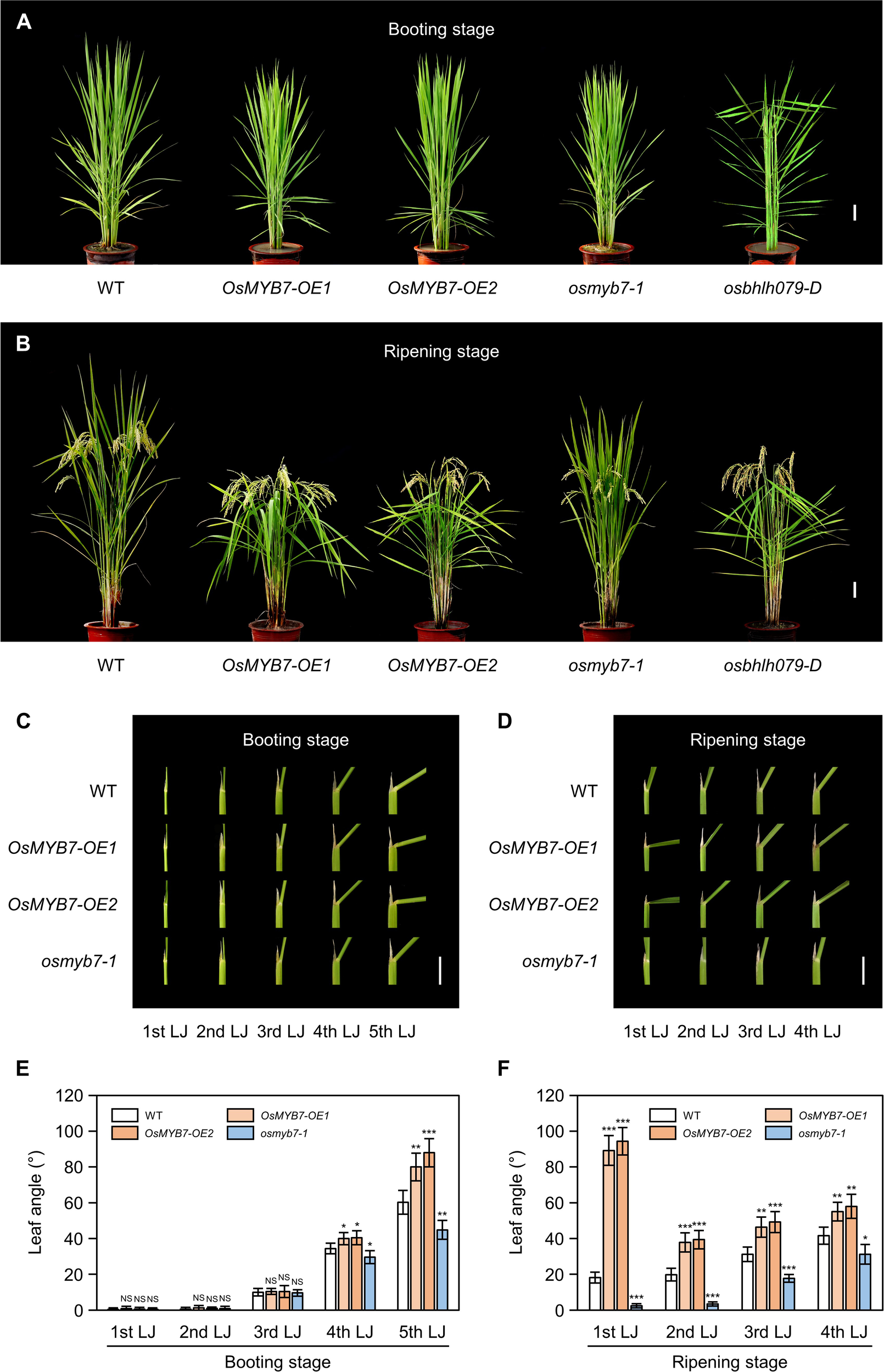
OsMYB7 determines leaf angle at the late stage of lamina joint development. (A, B) Phenotypes of WT, OsMYB7-OE1, OsMYB7-OE2, osmyb7-1, and osbhlh079-D plants at the booting stage (105 DAS) (A) and at the ripening stage (30 DAH) (B) grown under natural long day conditions in a paddy field. The osbhlh079-D plant represents a typical mutant with increased leaf angles due to enhanced BR signaling. Scale: 10 cm. (C, D) Lamina joints of WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants. Samples in (C, D) were obtained from the plants shown in (A,B), respectively. Scale: 1 cm. (E, F) Leaf angle in WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants at the booting stage (105 DAS) (E) and at the ripening stage (30 DAH) (F) grown in a paddy field. Data are presented as means ± SD from five independent plants (5 leaf angles per plant). Asterisks indicate significant differences compared to WT as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). DAH, days after heading; DAS, days after sowing; NS, not significant; nth LJ, lamina joint of nth leaf.
It is worth noting that the phenotypes in OsMYB7 overexpressors and the osmyb7-1 mutant were somewhat distinct from those reported in other rice mutants with altered leaf inclinations, such as osbhlh079-D (Seo et al., 2020), OsWRKY53-OE (Tian et al., 2017), and lc2-1 (Zhao et al., 2010), in that the phenotype was restricted to the lamina joints at the late developmental stage. For example, osbhlh079-D, a typical mutant with enhanced BR signaling, showed enlarged leaf angles throughout its life cycle, including at the booting stage (Figures 2A, B). Collectively, these observations indicate that OsMYB7 plays a role in enlarging leaf angles in a developmental stage-specific manner.
OsMYB7 is preferentially expressed in lamina joints at S5 developmental stage
The spatiotemporal pattern of gene expression holds fundamental information for understanding the physiological functions of the encoded protein. Therefore, we determined the tissue specificity of the expression of OsMYB7. Interestingly, OsMYB7 exhibited a gradual increase in expression from the flag lamina joints to the 5th lamina joints (Figure 3A), leading us to hypothesize that OsMYB7 transcript levels in the lamina joints are developmentally regulated. To test this hypothesis, we monitored changes in OsMYB7 expression at the flag- and 2nd-leaf lamina joints across developmental age. Even though the development of lamina joints is divided into six successive stages from initiation (S1) to senescence (S6) based on their morphological features (Zhou et al., 2017) (Supplementary Table S2), we focused on the S3, S4, and S5 stages for simplicity: the lamina joint is encircled by the prior leaf sheath at the S3 stage, and is exposed to the air at the S4 stage, before bending at the S5 stage. Reverse-transcription quantitative PCR (RT-qPCR) analysis revealed that OsMYB7 expression levels gradually increase throughout development, with the sharpest rise at the onset of the S5 stage (Figures 3B, C). Furthermore, subsequent histochemical analysis of GUS (ß-glucuronidase) activity derived from GUS expression driven by the OsMYB7 promoter revealed that the collar at S5 stage exhibited intense GUS staining, while that at S3 stage showed faint GUS expression (Figures 3D–I). These results support that the transcription levels of OsMYB7 in the lamina joints are tightly controlled during development and preferentially upregulated at the S5 stage of lamina joints, in agreement with the phenotypic observations (Figure 2).
Figure 3
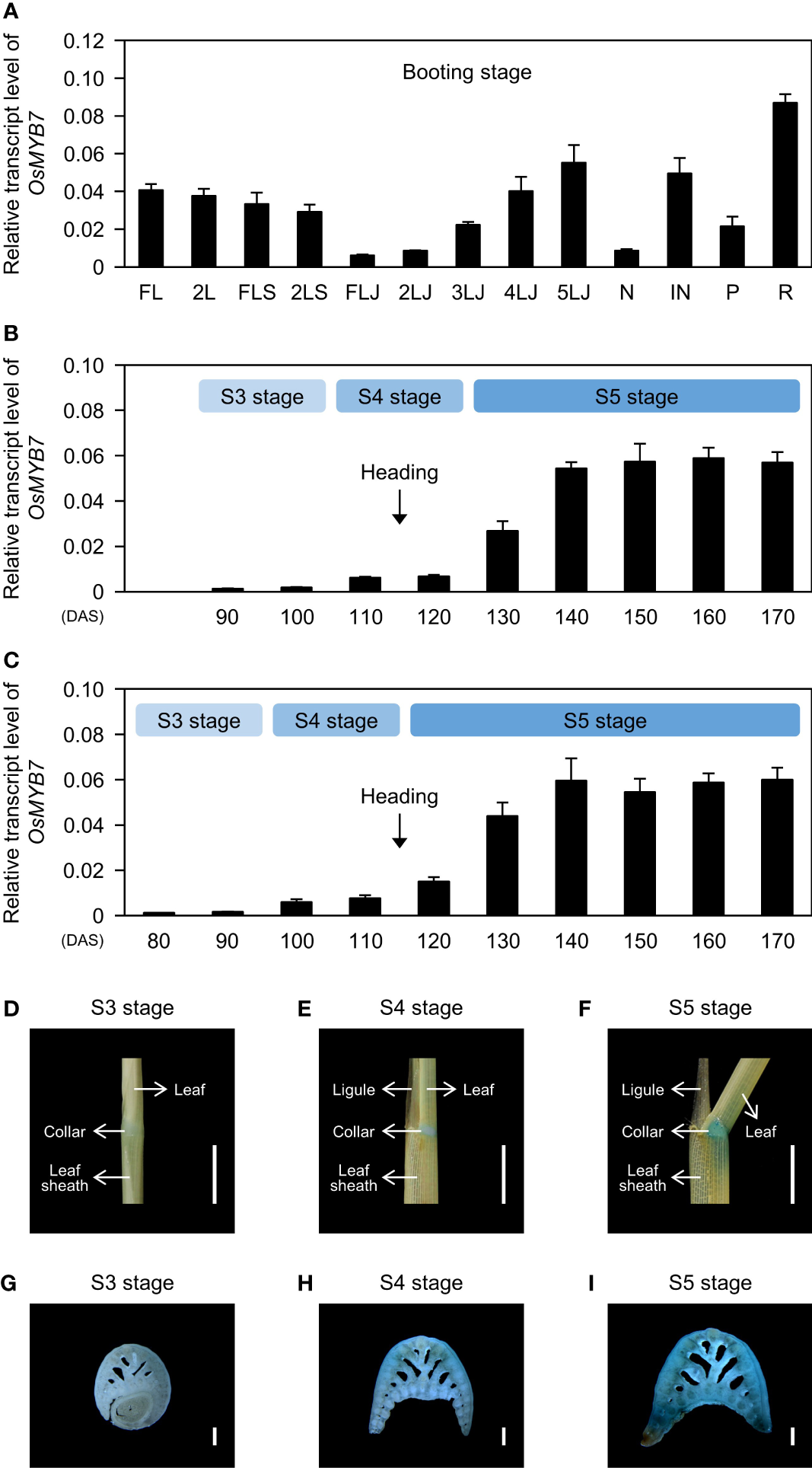
Expression profiles of OsMYB7. (A) Spatial expression patterns of OsMYB7 across tissues. Each tissue designated in (A) was collected from WT at the booting stage grown under natural day-night conditions in a paddy field. The transcript levels of OsMYB7 determined by RT-qPCR analysis were normalized to those of GAPDH and presented as means ± SD from four biological replicates. FL, flag leaf; 2L, 2nd leaf; FLS, flag leaf sheath; 2LS, 2nd leaf sheath; FLJ, lamina joint of flag leaf; nLJ, lamina joint of nth leaf; N, node; IN, internode; P, panicle; R, root. (B, C) Temporal expression patterns of OsMYB7 in the flag leaf lamina joint (B) and the 2nd leaf lamina joint (C) across developmental stages. Each lamina joint was collected every 10 days between S3 and S5 developmental stages from WT grown under natural paddy field conditions. The expression levels of OsMYB7 determined by RT-qPCR analysis were normalized to those of GAPDH and presented as means ± SD from four biological samples (around 30 lamina joints per sample). Each developmental stage is indicated in the light blue boxes. The average time of heading in WT is shown by black arrows. DAS, days after sowing. (D-I) Histochemical analysis of GUS activity derived from the OsMYB7 promoter. GUS activity in the 2nd leaf lamina joint at S3 (D), S4 (E), and S5 (F) stage, and the corresponding transverse section of the 2nd leaf lamina joint at S3 (G), S4 (H), and S5 (I) stages. Samples were collected from OsMYB7-OE1, which harbors a T-DNA with a promoterless β-glucuronidase (GUS) reporter at 80 bp upstream of the ATG initiation site in OsMYB7, grown under natural long-day conditions. A schematic diagram illustrating the position of the T-DNA insertion in OsMYB7-OE1 is shown in Supplementary Figure S3A. Scales: 5 mm (D-F) and 500 μm (G–I). Similar results were repeated in at least three independent samples.
OsMYB7 acts as a pathway-wide transcriptional repressor of lignin biosynthesis
Lignin modulates leaf angle by determining the mechanical strength of lamina joints (Sun et al., 2020; Wang R. et al., 2020). Thus, we speculated that OsMYB7 might dictate leaf angle by controlling lignin biosynthesis. To test whether OsMYB7 function is closely associated with lignin, we examined the transcript levels of 16 lignin biosynthetic genes (see Supplementary Table S3). We determined that transcript abundances of these lignin biosynthetic genes were significantly altered in OsMYB7 overexpressors and the osmyb7-1 mutant in opposite directions: they were downregulated in OsMYB7 overexpressors and upregulated in the osmyb7-1 mutant (Figure 4A).
Figure 4
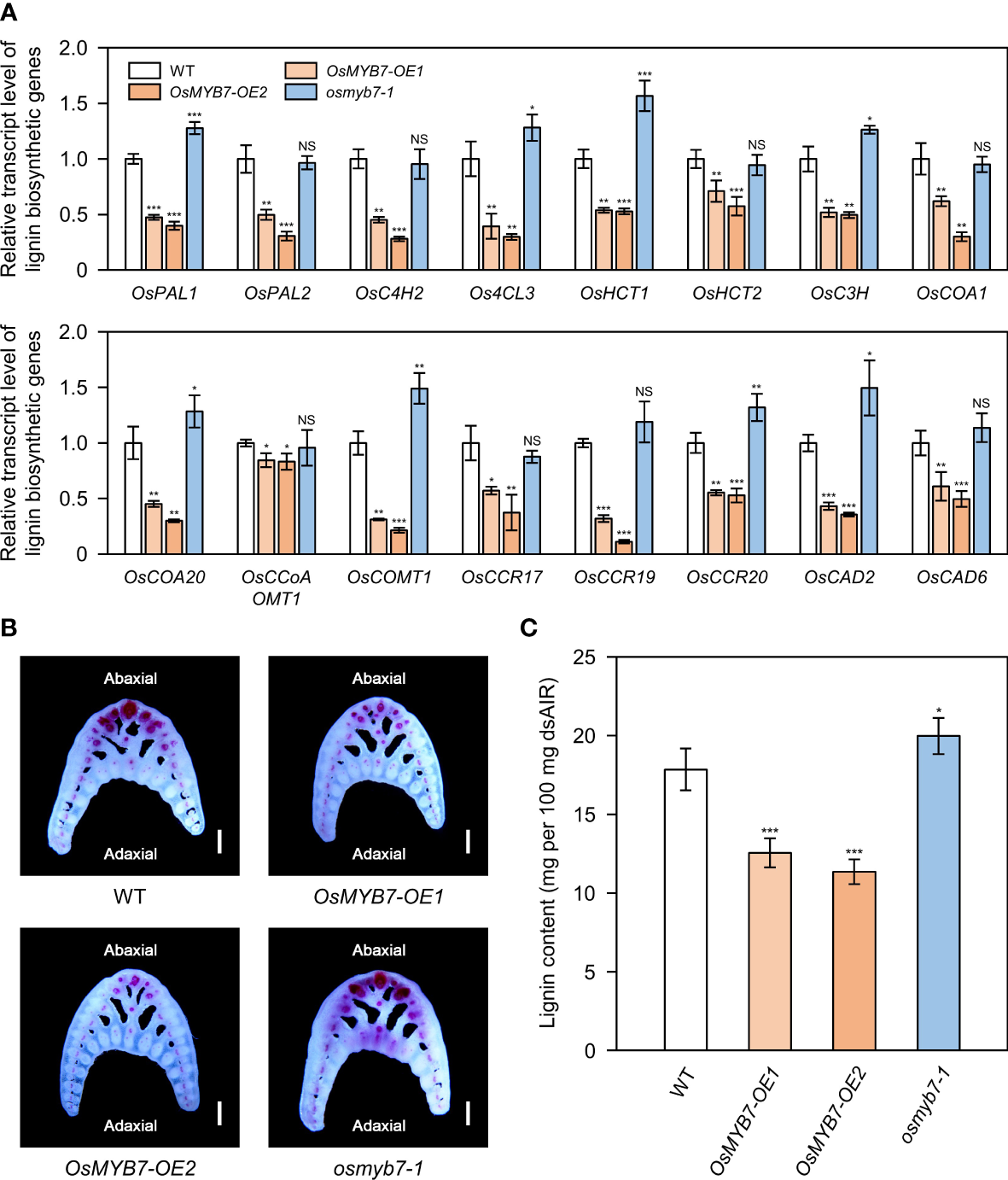
OsMYB7 negatively regulates lignin biosynthesis at the lamina joint. (A) Relative transcript levels of lignin biosynthetic genes in WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants. Samples were harvested from the flag leaf lamina joints at the S5 stage from plants grown under natural long-day conditions in a paddy field. The expression levels of each gene determined by RT-qPCR analysis were normalized to those of GAPDH and represented relative to WT, which was set to 1. Data are presented as means ± SD from four biological samples (around 30 lamina joints per sample). Asterisks indicate significant differences as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). These experiments were conducted twice with similar results. NS, not significant. (B) Histochemical staining of the lamina joint for lignin. Flag leaf lamina joints of WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants at the ripening stage (30 DAH) grown in a natural paddy field were collected, and the corresponding transverse hand sections were subjected to phloroglucinol-HCl staining, which visualizes lignin as a pink to red color. Scale: 500 μm. Similar results were obtained in at least three independent samples. (C) Determination of lignin contents in lamina joints of WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants. Lamina joints of flag leaves at the S5 developmental stage were collected from plants grown under natural long-day conditions and subjected to acetyl bromide assay, which quantifies total lignin contents. Data are presented as means ± SD from six independent samples (around 50 lamina joints per sample); asterisks indicated significant differences as determined by two-tailed Student’s t-test (*P < 0.05 and ***P < 0.001). These experiments were repeated twice, yielding similar results. dsAIR, destarched alcohol-insoluble cell wall residue.
We also conducted histochemical staining of lignin from lamina joint cross-sections. Consistent with previous reports (Wang R. et al., 2020; Huang et al., 2021), lignin deposition was mainly observed in sclerenchyma cells and vascular bundles on the abaxial side of lamina joints (Figure 4B). We detected weaker lignin signals in the abaxial sclerenchyma cells of OsMYB7-OE1 and OsMYB7-OE2 lines compared to the WT, while the sclerenchyma cells on the adaxial side were not lignin-rich in both WT and OsMYB7 overexpressors (Figure 4B). In contrast, the functional deficiency of OsMYB7 resulted in noticeable ectopic lignification in the adaxial sclerenchyma cells, although no significant difference in lignin deposition was observed on the abaxial tissues between WT and osmyb7-1, possibly because the lignin on the abaxial side was fully saturated in the WT (Figure 4B). In support of these observations, lignin contents in the lamina joints were 30~36% lower in OsMYB7 overexpressors and 12% higher in the osmyb7-1 mutant, compared to those in WT (Figure 4C). These results thus confirm that OsMYB7 has a negative role in lignin biosynthesis at the lamina joint.
OsMYB7 activates a biosynthetic program for cellulose at lamina joints
Cellulose, together with lignin, controls leaf inclination by affecting the mechanical properties of sclerenchymatous tissues in the lamina joints (Ning et al., 2011; Sun et al., 2020; Huang et al., 2021). In addition, some rice MYB transcriptional activators (OsMYB46, OsMYB58/63, and OsMYB103) of the lignin biosynthetic pathway trigger cellulose biosynthesis (Zhong et al., 2011; Hirano et al., 2013; Noda et al., 2015), leading us to speculate that OsMYB7 may coordinately repress the biosynthesis of SCW components. To assess whether OsMYB7 is involved in cellulose accumulation, we examined the expression levels of 14 genes from the CELLULOSE SYNTHASE/CELLULOSE SYNTHASE-LIKE (OsCESA/CSL) superfamily (see Supplementary Table S3), all of which are expressed abundantly in the lamina joints at the S5 stage (Zhou et al., 2017). Among them, 11 genes showed elevated expression in OsMYB7 overexpressors, and at least 8 genes exhibited decreased expression in the osmyb7-1 mutant, compared to WT (Figure 5A). Moreover, cellulose staining on the transverse sections of lamina joints revealed that OsMYB7 overexpressors and the osmyb7-1 mutant tend to show stronger and weaker signals, respectively, than WT (Figure 5B). We confirmed these observations by anthrone assays, which quantify the contents of total crystalline cellulose; cellulose was overproduced and underproduced in the lamina joints of OsMYB7 overexpressors and the osmyb7-1 mutant, respectively (Figure 5C). Taken together, OsMYB7 promotes cellulose deposition at the lamina joints at the S5 stage.
Figure 5
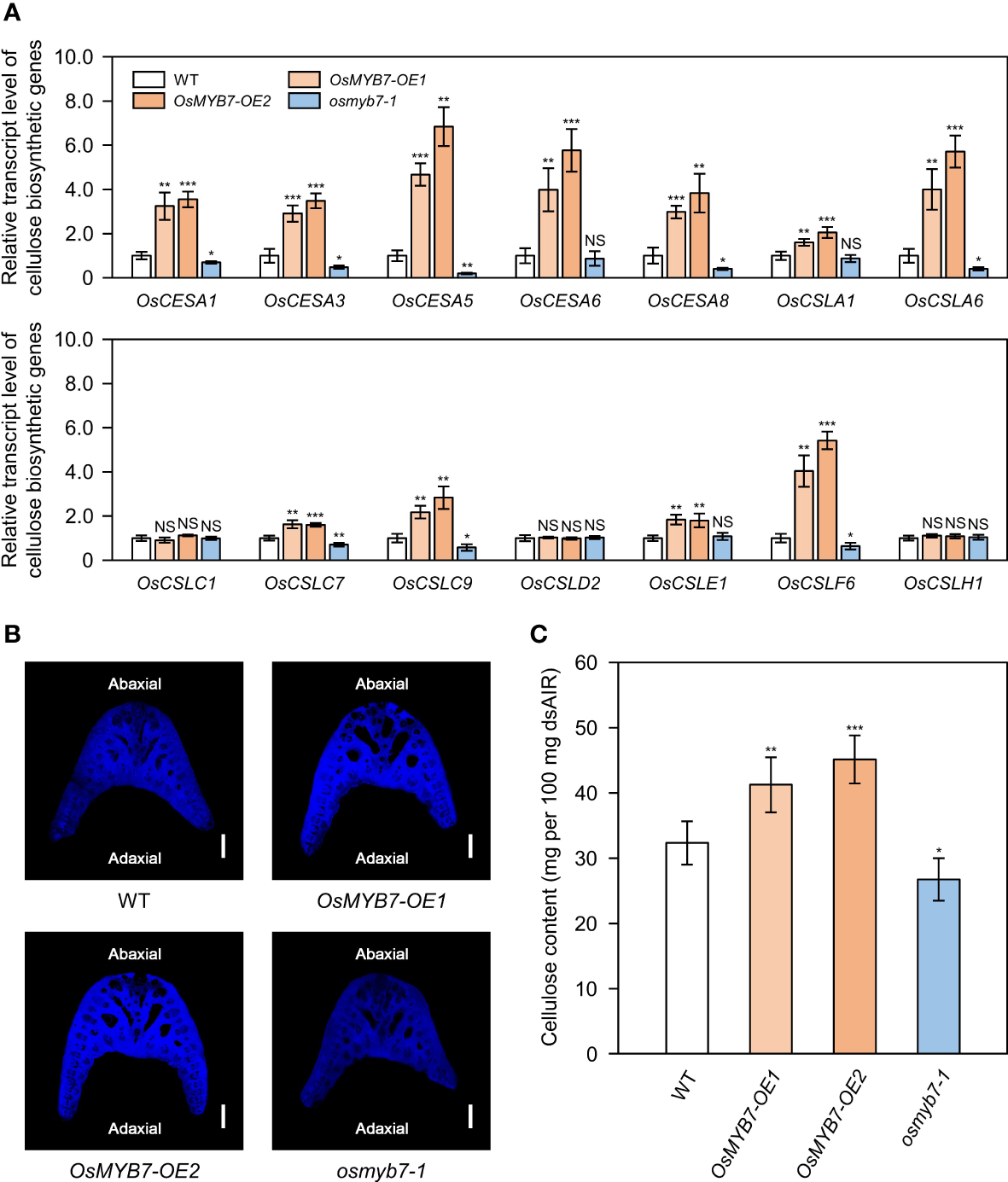
OsMYB7 is a positive regulator of cellulose biosynthesis at the lamina joint. (A) Relative transcript levels of cellulose biosynthetic genes in WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants. Samples obtained in Figure 4A were subjected to RT-qPCR analysis. The expression levels of each gene were normalized to those of GAPDH and presented relative to the WT, which was set to 1. Data are presented as means ± SD. Asterisks indicate significant differences as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). These experiments were conducted twice yielding similar results. NS, not significant. (B) Histochemical staining of the lamina joint for cellulose. Lamina joints of flag leaves at the S5 stage were collected from WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants grown in natural paddy field conditions, and the corresponding transverse sections were subjected to calcofluor white staining, which visualizes cellulose as bright blue color under UV excitation. Scale: 500 μm. Similar results were obtained in at least three independent samples. (C) Determination of cellulose contents in lamina joints of WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants. Flag leaf lamina joints from plants at the ripening stage (30 DAH) grown under natural day-night conditions in a paddy field were harvested and subjected to anthrone assay, which quantifies total cellulose contents colorimetrically. Data are presented as means ± SD from six independent samples (around 50 lamina joints per sample). Asterisks indicate significant differences as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). These experiments were repeated twice with similar results. dsAIR, destarched alcohol-insoluble cell wall residue.
SCW thickening is accelerated by OsMYB7
The thickness of the SCW is determined by the amount of deposited lignin and cellulose and affects leaf angle as a primary decisive factor of mechanical strength (Sun et al., 2020; Huang et al., 2021). However, it was difficult to simply assume that OsMYB7 inhibits thickening of SCW by reducing lignin accumulation, because it promoted deposition of cellulose (Figures 4, 5). To pinpoint the difference in SCW thickness, we examined sclerenchyma cells in the lamina joints using a transmission electron microscope. Interestingly, compared to WT, OsMYB7 overexpressors had moderately thicker SCWs, whereas the osmyb7-1 mutant showed a slight reduction in SCW thickness (Supplementary Figure S4). These results suggest that the inhibition of lignin deposition by OsMYB7 may not be the main cause of alteration in leaf angle. However, we cannot exclude the possibility that other phenylpropanoids, in addition to lignin, may also influence the leaf angle phenotype.
OsMYB7-mediated regulation of leaf angle is independent of BRs
Leaf angle has been widely considered to be determined by the balance between two opposing forces: pushing and supporting forces. The pushing force is primarily derived from preferential cell division/elongation at the adaxial side of lamina joints. The supporting force is based on abundant sclerenchymatous cells with thick SCW in the lamina joints (Wang R. et al., 2020; Xu et al., 2021). Therefore, we focused on the pushing force, which is mainly established under phytohormonal control (Xu et al., 2021). To elucidate whether OsMYB7-mediated leaf angle formation is orchestrated by phytohormones, we examined the transcript levels of representative BR biosynthetic and signaling genes (see Supplementary Table S3), because BRs play a dominant role in adaxial cell division/elongation at the lamina joints (Li et al., 2020). The expression levels of BR-related genes in OsMYB7 overexpressors and the osmyb7-1 mutant were almost identical to those of WT (Supplementary Figure S5). Furthermore, a BR-induced lamina joint inclination assay (Wada et al., 1984) confirmed that the sensitivity of OsMYB7 overexpressors and the osmyb7-1 mutant are not altered by BR treatments, compared to the WT (Supplementary Figure S6). Taken together, these findings suggest that OsMYB7 is involved in the regulation of leaf angles in a BR-independent manner.
Free auxin levels at lamina joints were altered in OsMYB7 overexpressors and osmyb7-1 mutant
Besides BRs, other phytohormones, including auxin, cytokinin, ethylene, gibberellin, and jasmonate, comprehensively participate in controlling leaf inclination (Li et al., 2020; Xu et al., 2021). Accordingly, we investigated whether other phytohormones might affect the expression of OsMYB7 by RT-qPCR. Although OsMYB7 expression was slightly induced in response to 24-epibrassinolide (BL; the active form of BR), it was strikingly upregulated 3.24 and 5.00 times after three and six hours of treatment with auxin (indole-3-acetic acid, IAA), respectively, compared to that in the mock samples (Figure 6A). In support of these results, we identified ten putative auxin-responsive elements (AuxREs), including two canonical AuxREs (Mironova et al., 2014; Lanctot et al., 2020), in the promoter region of OsMYB7 (−2,000 bp to −1 bp from the start codon) (Supplementary Figure S7), suggesting that OsMYB7 is associated with auxin-dependent lamina inclination.
Figure 6
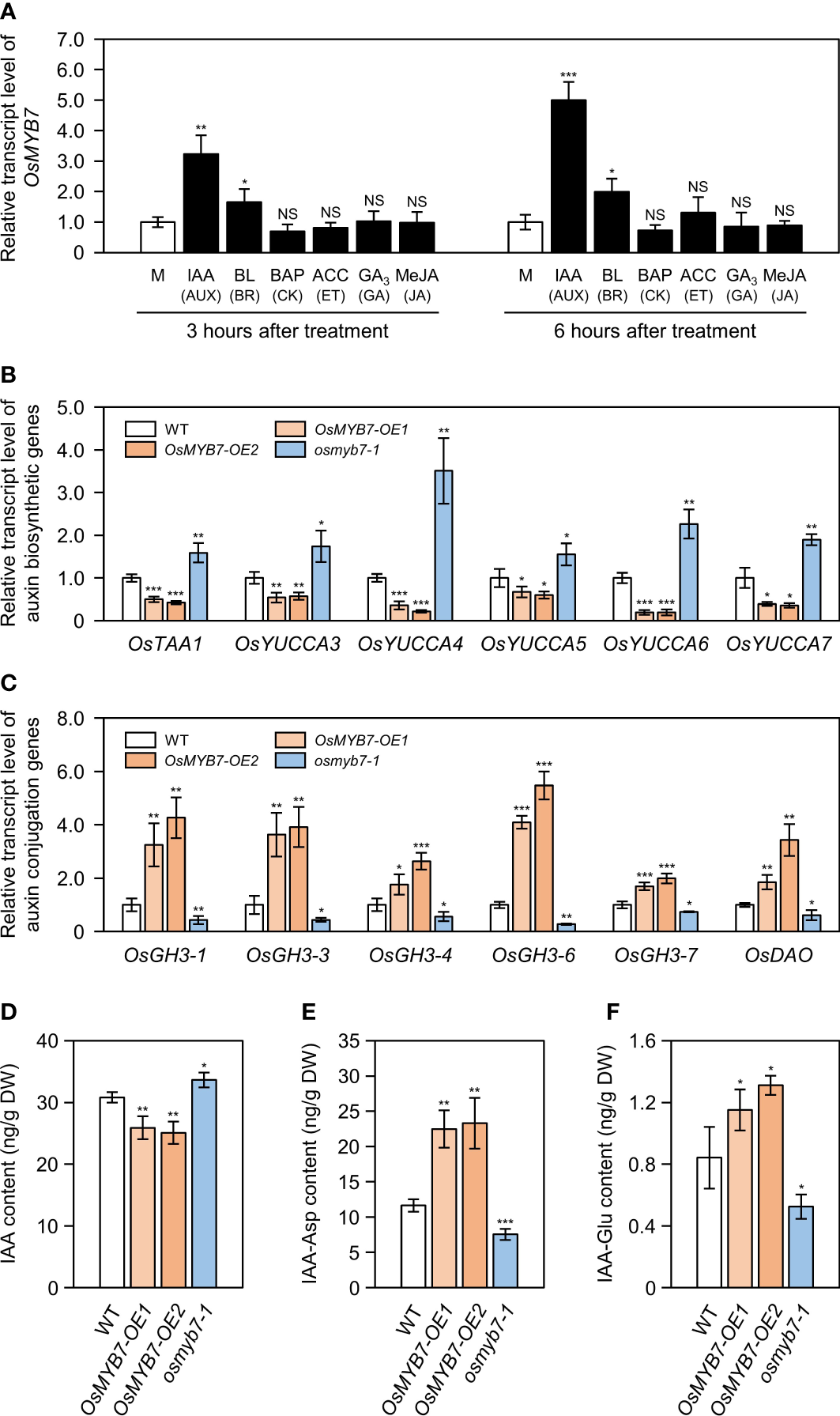
Alteration in free auxin levels at lamina joints may be one cause of different leaf angle phenotypes among WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1. (A) Relative OsMYB7 expression levels in response to various phytohormones determining leaf angle. WT seedlings were grown on half-strength solid MS medium for ten days under constant light conditions at 28°C, followed by incubation in half-strength liquid MS medium containing with 100 μM IAA, 10 μM BL, 10 μM BAP, 10 mM ACC, 100 μM GA3, or 100 μM MeJA. WT seedlings incubated in half-strength liquid MS medium without phytohormone were used as a mock. Total RNA was extracted from shoots collected after 3 or 6 h of treatment, and used for RT-qPCR analysis. The transcript levels of OsMYB7 were normalized to those of GAPDH, with the expression level in the phytohormone-treated group shown relative to that in the mock-treated group, which was set to 1. Data are presented as means ± SD from four biological samples (around 3 shoots per sample). Asterisks indicate significant differences compared to the mock-treated sample, as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). This experiment was performed independently twice and similar results were obtained. AUX, auxin; BR, brassinosteroid; CK, cytokinin; ET, ethylene; GA, gibberellin; JA, jasmonic acid; M, mock; NS, not significant. (B, C) Relative expression of auxin biosynthetic (B) and conjugation (C) genes at the lamina joint. The cDNA samples in Figure 4A were subjected to RT-qPCR analysis. GAPDH was used as an internal control and the relative expression of each gene is shown relative to that in WT (set to 1). Data are presented as means ± SD of four biological replicates. Asterisks indicate significant differences as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). These experiments were conducted twice yielding similar results. (D–F) Contents for free IAA and IAA-amino acid conjugates (IAA-Asp and IAA-Glu). Flag leaf lamina joints at the S5 developmental stage in WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants were collected and subjected to LC-ESI-MS/MS analysis for quantification of free IAA (D), IAA-Asp (E), and IAA-Glu (F). Data are presented as means ± SD from four biological replicates (around 10 lamina joints per plant and around 15 plants per replicates). Asterisks indicate significant differences as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). DW, dry weight; IAA, indole-3-acetic acid; IAA-Asp, IAA-aspartate; IAA-Glu, IAA-glutamate.
To evaluate whether the leaf angle phenotypes of OsMYB7 overexpressors and the osmyb7-1 mutant were associated with endogenous IAA contents, we measured the expression levels of auxin biosynthetic genes and auxin conjugation genes in the lamina joints (see Supplementary Table S3). We determined that auxin anabolic genes (OsTAA1, OsYUCCA3, OsYUCCA4, OsYUCCA5, OsYUCCA6, and OsYUCCA7) are severely downregulated in OsMYB7 overexpressors and upregulated in the osmyb7-1 mutant (Figure 6B). By contrast, auxin conjugation genes (OsGH3-1, OsGH3-3, OsGH3-4, OsGH3-6, OsGH3-7, and OsDAO) were more highly expressed in OsMYB7 overexpressors and less so in the osmyb7-1 mutant compared to the WT (Figure 6C). Consistent with the transcript levels of auxin biosynthetic genes, endogenous free IAA levels in the lamina joints were approximately 1.2-fold lower in OsMYB7 overexpressors and 1.09-fold higher in the osmyb7-1 mutant than the WT (Figure 6D), whereas IAA conjugates, IAA-Asp and IAA-Glu, were more concentrated in the lamina joints of OsMYB7 overexpressors and less concentrated in those of the osmyb7-1 mutant (Figures 6E, F). All of these findings suggest that OsMYB7 may promote rice lamina inclination by inhibiting the accumulation of free auxin in lamina joints.
OsMYB7 affects cell elongation at the adaxial side of the lamina joint
Auxin deficiency boosts parenchymal cell division and/or elongation at the adaxial side of lamina joints, resulting in enlarged leaf angles (Zhao et al., 2013; Zhang et al., 2015). To unravel the cellular mechanism of OsMYB7 in leaf inclination, we investigated the effect of OsMYB7 on the expression of cell division- and cell elongation-associated genes at the lamina joints. The transcript levels of cell division-related genes, such as cyclin and CDK genes (see Supplementary Table S3), were not altered in OsMYB7 overexpressors or the osmyb7-1 mutant (Supplementary Figure S8). By contrast, we observed significant changes in expression of cell elongation-related genes, such as EXPANSIN and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE (XTH) genes (see Supplementary Table S3), in OsMYB7 overexpressors and the osmyb7-1 mutant; these genes were upregulated in OsMYB7 overexpressors and downregulated in the osmyb7-1 mutant (Figure 7). This observation indicated that OsMYB7 may play a positive role in the promotion of lamina inclination by stimulating adaxial cell elongation rather than cell division.
Figure 7
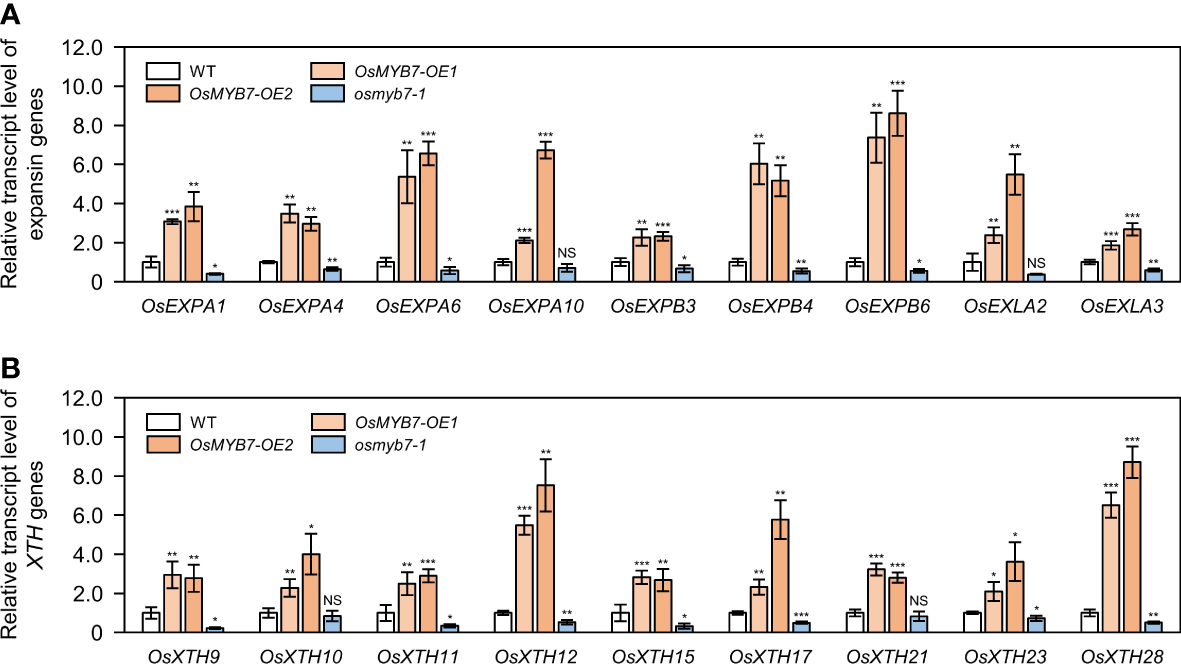
Expression analysis of cell elongation-related genes at the lamina joint. (A, B) Relative transcript levels of expansin (A) and XTH(B) genes in WT, OsMYB7-OE1, OsMYB7-OE2, and osmyb7-1 plants. Total RNA samples obtained in Figure 4A were used for RT-qPCR analysis. The expression level of each examined gene was normalized to that of GAPDH, and shown relative to WT levels, which were set to 1. Data are presented as means ± SD (n = 4); asterisks indicate significant differences as determined by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). These experiments were carried out twice with independent biological replicates, and similar results were obtained. NS, not significant; XTH, xyloglucan endotransglucosylase/hydrolase.
For further verification, we examined longitudinal sections of lamina joints using a scanning electron microscope. In agreement with the gene expression profiles, the adaxial cells in OsMYB7 overexpressors were elongated both longitudinally and transversely to a greater extent than the WT, while those in the osmyb7-1 mutant failed to expand (Figure 8). This result demonstrates that OsMYB7-mediated leaf angle formation at least partially depends on parenchymal cell size at the adaxial side of lamina joints.
Figure 8
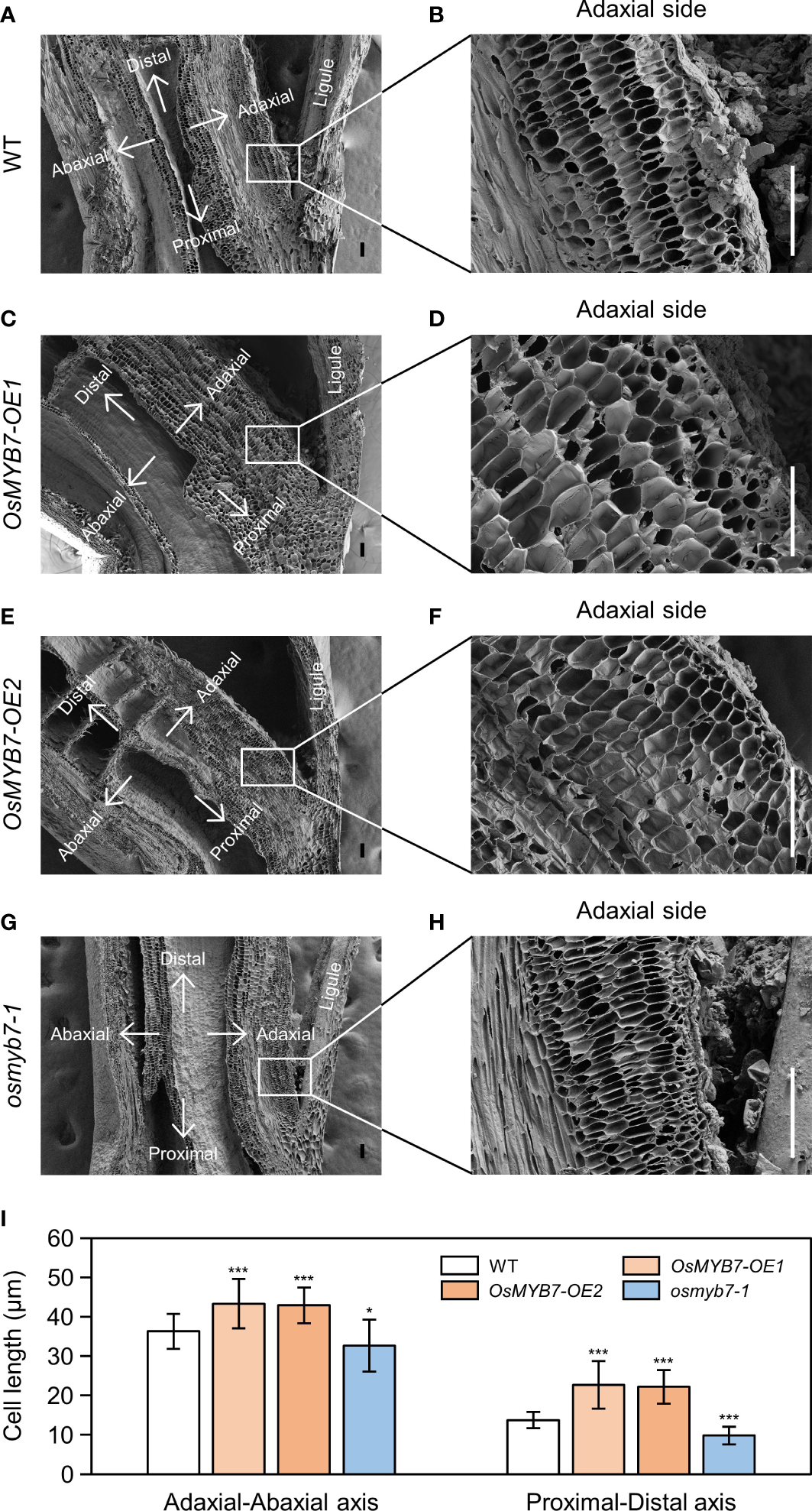
Scanning electron microscopy examination of the lamina joint. (A–H) Longitudinal sections of flag leaf lamina joints at the S5 developmental stage in WT (A), OsMYB7-OE1(C), OsMYB7-OE2(E), and osmyb7-1(G) plants grown under natural long-day conditions in a paddy field. Close-up views of the adaxial region denoted by rectangles in (A, C, E, G) are shown in (B, D, F, H), respectively. Scale: 100 μm. (I) Adaxial cell length in (B, D, F, H). Length of the adaxial cells was measured along the adaxial-abaxial axis and proximal-distal axis. Data are presented as means ± SD from 30 cells. Asterisks indicate significant differences compared to WT, as determined by two-tailed Student’s t-test (*P < 0.05 and ***P < 0.001). Similar results were obtained from at least three independent samples.
Discussion
Lignin and cellulose depositions at the lamina joints appear to be regulated under a compensatory balance mechanism
Several cell wall-associated MYB transcription factors regulate lignin accumulation in rice, most of which function as pathway-wide transcriptional activators of lignin biosynthesis (Miyamoto et al., 2020). The rice MYB activators OsMYB46, OsMYB58/63, OsMYB61a, and OsMYB103L have also have been shown to promote cellulose biosynthesis (Zhong et al., 2011; Yang et al., 2014; Noda et al., 2015; Zhao et al., 2019). In Arabidopsis, OsMYB46 overexpression induced ectopic deposition of secondary cell wall components (lignin, cellulose, and xylan) in the walls of the epidermis (Zhong et al., 2011). A knockdown mutation in OsMYB103L decreased lignin and cellulose accumulation, which resulted in brittle leaves (Hirano et al., 2013; Yang et al., 2014). Moreover, there is some evidence that these types of MYB transcription factors directly induce the expression of cellulose synthase genes (Noda et al., 2015). We thus postulated that OsMYB7 might be involved in suppressing the biosynthesis of both lignin and cellulose.
In this study, we demonstrated that OsMYB7 activated the cellulose biosynthetic program, although it inhibited lignin deposition (Figures 4, 5). These findings raised the question as to whether OsMYB7 directly triggered the transcription of cellulose biosynthetic genes: importantly, OsMYB7 did not possess transactivation activity for the direct activation of downstream genes (Figure 1B). In the last few decades, lignin and cellulose deposition has been suggested to be regulated in a compensatory fashion in some plant species, such as rice, maize, and aspen (Populus tremuloides). For example, mutants lacking cellulose, including brittle culm 7 (bc7(t)) in rice and brittle stalk-2 (bk2) or bk-5 in maize, exhibited a significant increase in their lignin contents, and transgenic aspens with lower transcript levels for 4-COUMARATE : COA LIGASE (Pt4CL1), one of the lignin biosynthetic genes, showed a concomitant increase in cellulose (Hu et al., 1999; Sindhu et al., 2007; Wei et al., 2011; Li et al., 2022). Furthermore, in the case of transgenic rice plants overexpressing Arabidopsis SHINE (SHN), the compensatory increase in cellulose was shown to offset any defects in mechanical strength due to lack of lignin (Ambavaram et al., 2011). Conclusively, we suggest that the promotion of cellulose deposition by OsMYB7 may result from compensation for lignin deficiency.
OsMYB7 promotes leaf inclination possibly through accelerating a decline in free auxin levels at the late stage of lamina joint development
MYB proteins comprise one of the largest transcription factor superfamilies in plants and play crucial roles in multiple biological functions, including plant development, phytohormonal signaling, secondary metabolism, and response to environmental stress (Jin and Martin, 1999; Dubos et al., 2010; Liu et al., 2015). Among the 155 putative/known MYB transcription factors in rice (Katiyar et al., 2012), a contribution to leaf angle has only been attributed to one MYB member, GIBBERELLIN-INDUCIBLE MYB-LIKE 2 (OsGAMYBL2), which is responsible for integrating BRs and gibberellin signaling pathways at lamina joints to maintain leaf erectness (Gao et al., 2018). In this study, we identified a new MYB transcription factor, OsMYB7, as influencing leaf inclination through modulating auxin homeostasis (Figures 2, 6). Interestingly, OsMYB7 presented the specific characteristic that its phenotypic effects are restricted to lamina joints at the late developmental stage, coinciding with the findings that expression of OsMYB7 in the lamina joints is drastically upregulated at the S5 stage (Figures 2, 3). Recently, the concentration of active auxin, IAA, was revealed to gradually decline over lamina joint development (Zhou et al., 2017), although the underlying mechanism is largely unknown. Since auxin stimulates cell division and suppresses cell elongation when accumulating at high levels (Chen et al., 2001), it has been widely considered that auxin is responsible for rapid increase in cell number at the developing lamina joints (Zhou et al., 2017). However, auxin induces cell elongation when present at low levels (Chen et al., 2001), and reduced auxin contents in lamina joints at the S5 developmental stage give rise to excessive cell elongation at the adaxial side of lamina joints, promoting lamina joint bending (Zhao et al., 2013; Zhou et al., 2017). Combined with our data, the preferential expression of OsMYB7 in the lamina joints at the late developmental stage may partially contribute to the incremental decrease in auxin contents over lamina joint development, thereby accelerating adaxial cell elongation and leaf inclination at the S5 stage.
Perturbations of auxin homeostasis in OsMYB7 overexpressors and the osmyb7-1 mutant might result from changes in lignin quantity
Auxin acts upstream of the lignin biosynthetic pathway (Qu et al., 2021). Overexpression of auxin biosynthetic genes, such as YUCCA8 and YUCCA9, led to strong lignification of aerial tissues in Arabidopsis (Hentrich et al., 2013). Although the effect of lignin pathway perturbations on auxin homeostasis has been less studied, there is some evidence that altering lignin levels can affect the expression of auxin-related genes. For instance, loss-of-function mutations of lignin biosynthetic genes, such as CINNAMATE 4-HYDROXYLASE (C4H), 4CL1, CAFFEOYL-COA O-METHYLTRANSFERASE 1 (CCoAOMT1), and CINNAMOYL-COA REDUCTASE 1 (CCR1), in Arabidopsis caused a drop in the transcript levels of many auxin-responsive genes (Vanholme et al., 2012). Rogers et al. (2005) revealed that the auxin biosynthetic gene CYTOCHROME P450 79B2 (CYP79B2) was upregulated in all investigated ectopic lignification mutants, including de-etiolated 3 (det3), pom-pom 1 (pom1), and ectopic lignification 1 (eli1), suggesting feedback regulation between lignin and auxin biosynthesis in Arabidopsis. Based on our results, we propose that the alterations in lignin deposition at the lamina joints of both OsMYB7 overexpressors and the osmyb7-1 mutant may contribute to changes in auxin levels. Similarly, low-lignin transgenic tobacco (Nicotiana tabacum) overexpressing CsMYB4a, one of the MYB4/7/32 subgroup members in Tea plant (Camellia sinensis), displayed a series of auxin-deficient phenotypes (Ma et al., 2021, Preprint). Thus, our results provide a new insight into the relationship between lignin and auxin metabolism for the determination of rice leaf angle.
Endogenous auxin may inhibit adaxial cell elongation at lamina joints via negatively regulating the transcription of Expansin and XTH genes
Deficiency in endogenous auxin stimulates the longitudinal elongation of adaxial cells at lamina joints, thereby accelerating leaf inclination (Zhao et al., 2013). However, the underlying mechanism is not well-understood. On the contrary, exogenous auxin treatment promotes expression of cell elongation-related genes, such as Expansin and XTH genes, which results in lamina joint bending (Yokoyama and Nishitani, 2001; Esmon et al., 2006; Nakamura et al., 2009); Expansins induce a pH-dependent relaxation of cell wall non-enzymatically, thus enabling cell elongation (Marowa et al., 2016), and the restructuring of wall-bound xyloglucan by XTHs allows cell expansion (Nishitani et al., 2006). Considering the opposite roles of endogenous and exogenous auxin in the determination of leaf angle (Nakamura et al., 2009; Zhao et al., 2013), we speculate that endogenous auxin at the lamina joints downregulates the transcription of these cell elongation-related genes. Indeed, we demonstrated that the expression of Expansin and XTH genes at the lamina joints is transcriptionally upregulated in OsMYB7 overexpressors and downregulated in the osmyb7-1 mutant (Figure 7). Moreover, OsMYB7 functions to accelerate cell elongation at the adaxial side of lamina joints (Figure 8), in agreement with the expression profiles. These data provide further information on the functional roles of endogenous auxin in controlling the adaxial cell elongation of lamina joints.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
N-CP conceived and supervised the project; S-HK performed the research with the help of JY, HK, and S-JL; S-HK, TK, and KK analyzed the data; S-HK and N-CP wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2022R1A2C1091553 to N-CP) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2022R1A6A3A01086789 to S-HK).
Acknowledgments
We thank Gynheung An (Kyung Hee University, South Korea) for the gift of OsMYB7-OE1 and Li-Jia Qu (Peking University, China) for providing the pOs-sgRNA and pH-Ubi-cas9-7 vectors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1167202/full#supplementary-material
References
1
AmbavaramM. M. R.KrishnanA.TrijatmikoK. R.PereiraA. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol.155, 916–931. doi: 10.1104/pp.110.168641
2
BozzolaJ. J.KuoJ. (2007). “Conventional specimen preparation techniques for scanning electron microscopy of biological specimens,” in KuoJ. Eds. Electron microscopy (Totowa, New Jersey: Humana Press), 449–466. doi: 10.1007/978-1-59745-294-6_22
3
ChenJ. G.UllahH.YoungJ. C.SussmanM. R.JonesA. M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev.15, 902–911. doi: 10.1101/gad.866201
4
ChenS. H.ZhouL. J.XuP.XueH. W. (2018). SPOC domain-containing protein leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PloS Genet.14, e1007829. doi: 10.1371/journal.pgen.1007829
5
CurtisM. D.GrossniklausU. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol.133, 462–469. doi: 10.1104/pp.103.027979
6
DonaldC. M. (1968). The breeding of crop ideotypes. Euphytica17, 385–403. doi: 10.1007/BF00056241
7
DuH.WuN.FuJ.WangS.LiX.XiaoJ.et al. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot.63, 6467–6480. doi: 10.1093/jxb/ers300
8
DubosC.StrackeR.GrotewoldE.WeisshaarB.MartinC.LepiniecL. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci.15, 573–581. doi: 10.1016/j.tplants.2010.06.005
9
EarleyK. W.HaagJ. R.PontesO.OpperK.JuehneT.SongK.et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J.45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
10
EsmonC. A.TinsleyA. G.LjungK.SandbergG.HearneL. B.LiscumE. (2006). A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc. Natl. Acad. Sci. U.S.A.103, 236–241. doi: 10.1073/pnas.0507127103
11
FornaléS.LopezE.Salazar-HenaoJ. E.Fernández-NohalesP.RigauJ.Caparros-RuizD. (2014). AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol.55, 507–516. doi: 10.1093/pcp/pct187
12
FornaléS.ShiX.ChaiC.EncinaA.IrarS.CapelladesM.et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J.64, 633–644. doi: 10.1111/j.1365-313X.2010.04363.x
13
GaoJ.ChenH.YangH.HeY.TianZ.LiJ. (2018). A brassinosteroid responsive miRNA-target module regulates gibberellin biosynthesis and plant development. New Phytol.220, 488–501. doi: 10.1111/nph.15331
14
HageageG. J.HarringtonB. J. (1984). Use of calcofluor white in clinical mycology. Lab. Med.15, 109–112. doi: 10.1093/labmed/15.2.109
15
HalpinC.HoltK.ChojeckiJ.OliverD.ChabbertB.MontiesB.et al. (1998). Brown-midrib maize (bm1) - a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J.14, 545–553. doi: 10.1046/j.1365-313X.1998.00153.x
16
HayashiK. I.AraiK.AoiY.TanakaY.HiraH.GuoR.et al. (2021). The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun.12, 6752. doi: 10.1038/s41467-021-27020-1
17
HentrichM.Sánchez-ParraB.Pérez AlonsoM. M.Carrasco LobaV.CarrilloL.Vicente-CarbajosaJ.et al. (2013). YUCCA8 and YUCCA9 overexpression reveals a link between auxin signaling and lignification through the induction of ethylene biosynthesis. Plant Signal. Behav.8, e26363. doi: 10.4161/psb.26363
18
HiranoK.KondoM.AyaK.MiyaoA.SatoY.AntonioB. A.et al. (2013). Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol.54, 1791–1802. doi: 10.1093/pcp/pct122
19
HöfgenR.WillmitzerL. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res.16, 9877. doi: 10.1093/nar/16.20.9877
20
HuW. J.HardingS. A.LungJ.PopkoJ. L.RalphJ.StokkeD. D.et al. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol.17, 808–812. doi: 10.1038/11758
21
HuangG.HuH.van de MeeneA.ZhangJ.DongL.ZhengS.et al. (2021). AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell33, 3120–3133. doi: 10.1093/plcell/koab175
22
InadaN.SakaiA.KuroiwaH.KuroiwaT. (1998). Three-dimensional analysis of the senescence program in rice (Oryza sativa l.) coleoptiles. Planta206, 585–597. doi: 10.1007/s004250050307
23
JainM.NijhawanA.TyagiA. K.KhuranaJ. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun.345, 646–651. doi: 10.1016/j.bbrc.2006.04.140
24
JangS. (2017). A novel trimeric complex in plant cells that contributes to the lamina inclination of rice. Plant Signal. Behav.12, e1274482. doi: 10.1080/15592324.2016.1274482
25
JeffersonR. A.KavanaghT. A.BevanM. W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J.6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
26
JeonJ. S.LeeS.JungK. H.JunS. H.JeongD. H.LeeJ.et al. (2000). T-DNA Insertional mutagenesis for functional genomics in rice. Plant J.22, 561–570. doi: 10.1046/j.1365-313x.2000.00767.x
27
JeongD. H.AnS.KangH. G.MoonS.HanJ. J.ParkS.et al. (2002). T-DNA Insertional mutagenesis for activation tagging in rice. Plant Physiol.130, 1636–1644. doi: 10.1104/pp.014357
28
JinH.CominelliE.BaileyP.ParrA.MehrtensF.JonesJ.et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J.19, 6150–6161. doi: 10.1093/emboj/19.22.6150
29
JinH.MartinC. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol.41, 577–585. doi: 10.1023/A:1006319732410
30
KagaleS.RozwadowskiK. (2011). EAR motif-mediated transcriptional repression in plants. an underlying mechanism for epigenetic regulation of gene expression. Epigenetics6, 141–146. doi: 10.4161/epi.6.2.13627
31
KatiyarA.SmitaS.LenkaS. K.RajwanshiR.ChinnusamyV.BansalK. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and arabidopsis. BMC Genom.13, 544. doi: 10.1186/1471-2164-13-544
32
KimJ. I.BaekD.ParkH. C.ChunH. J.OhD. H.LeeM. K.et al. (2013). Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant6, 337–349. doi: 10.1093/mp/sss100
33
LanctotA.Taylor-TeeplesM.OkiE. A.NemhauserJ. L. (2020). Specificity in auxin responses is not explained by the promoter preferences of activator ARFs. Plant Physiol.182, 1533–1536. doi: 10.1104/pp.19.01474
34
LiQ.NieS.LiG.DuJ.RenR.YangX.et al. (2022). Identification and fine mapping of the recessive gene BK-5, which affects cell wall biosynthesis and plant brittleness in maize. Int. J. Mol. Sci.23, 814. doi: 10.3390/ijms23020814
35
LiX.WuP.LuY.GuoS.ZhongZ.ShenR.et al. (2020). Synergistic interaction of phytohormones in determining leaf angle in crops. Int. J. Mol. Sci.21, 5052. doi: 10.3390/ijms21145052
36
LiuJ.OsbournA.MaP. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant8, 689–708. doi: 10.1016/j.molp.2015.03.012
37
LivakK. J.SchmittgenT. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods25, 402–408. doi: 10.1006/meth.2001.1262
38
MaG. L.WuY. L.JiangC. J.ChenY. F.XingD. W.ZhaoY.et al. (2021). CsMYB4a from Camellia sinensis regulates the auxin signaling pathway by interacting with CsIAA4. BioRxiv. doi: 10.1101/2021.10.11.463959
39
MarowaP.DingA.KongY. (2016). Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep.35, 949–965. doi: 10.1007/s00299-016-1948-4
40
MathewI. E.DasS.MahtoA.AgarwalP. (2016). Three rice NAC transcription factors heteromerize and are associated with seed size. Front. Plant Sci.7, 1638. doi: 10.3389/fpls.2016.01638
41
MatsudaF.MiyazawaH.WakasaK.MiyagawaH. (2005). Quantification of indole-3-acetic acid and amino acid conjugates in rice by liquid chromatography-electrospray ionization-tandem mass spectrometry. Biosci. Biotechnol. Biochem.69, 778–783. doi: 10.1271/bbb.69.778
42
MiaoJ.GuoD.ZhangJ.HuangQ.QinG.ZhangX.et al. (2013). Targeted mutagenesis in rice using CRISPR-cas system. Cell Res.23, 1233–1236. doi: 10.1038/cr.2013.123
43
MironovaV. V.OmelyanchukN. A.WiebeD. S.LevitskyV. G. (2014). Computational analysis of auxin responsive elements in the Arabidopsis thaliana l. genome. BMC Genom.15, S4. doi: 10.1186/1471-2164-15-S12-S4
44
MiyamotoT.TakadaR.TobimatsuY.TakedaY.SuzukiS.YamamuraM.et al. (2019). OsMYB108 loss-of-function enriches p-coumaroylated and tricin lignin units in rice cell walls. Plant J.98, 975–987. doi: 10.1111/tpj.14290
45
MiyamotoT.TobimatsuY.UmezawaT. (2020). MYB-mediated regulation of lignin biosynthesis in grasses. Curr. Plant Biol.24, 100174. doi: 10.1016/j.cpb.2020.100174
46
Moreira-VilarF. C.de Cássia Siqueira-SoaresR.Finger-TeixeiraA.de OliveiraD. M.FerroA. P.da RochaG. J.et al. (2014). The acetyl bromide method is faster, simpler and presents best recovery of lignin in different herbaceous tissues than klason and thioglycolic acid methods. PloS One9, e110000. doi: 10.1371/journal.pone.0110000
47
MoriM.NomuraT.OokaH.IshizakaM.YokotaT.SugimotoK.et al. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol.130, 1152–1161. doi: 10.1104/pp.007179
48
MoweryJ.BauchanG. (2018). Optimization of rapid microwave processing of botanical samples for transmission electron microscopy. Microsc. Microanal.24, 1202–1203. doi: 10.1017/S1431927618006499
49
NaitoY.HinoK.BonoH.Ui-TeiK. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics31, 1120–1123. doi: 10.1093/bioinformatics/btu743
50
NakamuraA.FujiokaS.TakatsutoS.TsujimotoM.KitanoH.YoshidaS.et al. (2009). Involvement of c-22-hydroxylated brassinosteroids in auxin-induced lamina joint bending in rice. Plant Cell Physiol.50, 1627–1635. doi: 10.1093/pcp/pcp106
51
NingJ.ZhangB.WangN.ZhouY.XiongL. (2011). Increased leaf angle1, a raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the lamina joint of rice. Plant Cell23, 4334–4347. doi: 10.1105/tpc.111.093419
52
NishitaniK.VissenbergK.VerbelenJ. P.VissenbergK. (2006). “Roles of the XTH protein family in the expanding cell,” in VerbelenJ. P.VissenbergK.The expanding cell (Berlin: Springer, Berlin, Heidelberg), 89–116. doi: 10.1007/7089_2006_072
53
NodaS.KoshibaT.HattoriT.YamaguchiM.SuzukiS.UmezawaT. (2015). The expression of a rice secondary wall-specific cellulose synthase gene, OsCesA7, is directly regulated by a rice transcription factor, OsMYB58/63. Planta242, 589–600. doi: 10.1007/s00425-015-2343-z
54
OomsG.HooykaasP. J. J.Van VeenR. J. M.Van BeelenP.Regensburg-TuïnkT. J. G.SchilperoortR. A. (1982). Octopine Ti-plasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region. Plasmid7, 15–29. doi: 10.1016/0147-619X(82)90023-3
55
PrestonJ.WheelerJ.HeazlewoodJ.LiS. F.ParishR. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J.40, 979–995. doi: 10.1111/j.1365-313X.2004.02280.x
56
QuG.PengD.YuZ.ChenX.ChengX.YangY.et al. (2021). Advances in the role of auxin for transcriptional regulation of lignin biosynthesis. Funct. Plant Biol.48, 743–754. doi: 10.1071/FP20381
57
RogersL. A.DubosC.SurmanC.WillmentJ.CullisI. F.MansfieldS. D.et al. (2005). Comparison of lignin deposition in three ectopic lignification mutants. New Phytol.168, 123–140. doi: 10.1111/j.1469-8137.2005.01496.x
58
SakamotoT.MorinakaY.OhnishiT.SunoharaH.FujiokaS.Ueguchi-TanakaM.et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol.24, 105–109. doi: 10.1038/nbt1173
59
SeoH.KimS. H.LeeB. D.LimJ. H.LeeS. J.AnG.et al. (2020). The rice basic helix-Loop-Helix 79 (OsbHLH079) determines leaf angle and grain shape. Int. J. Mol. Sci.21, 2090. doi: 10.3390/ijms21062090
60
SindhuA.LangewischT.OlekA.MultaniD. S.McCannM. C.VermerrisW.et al. (2007). Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol.145, 1444–1459. doi: 10.1104/pp.107.102582
61
SonbolF. M.FornaléS.CapelladesM.EncinaA.TouriñoS.TorresJ. L.et al. (2009). The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol.70, 283–296. doi: 10.1007/s11103-009-9473-2
62
SuG.AnZ.ZhangW.LiuY. (2005). Light promotes the synthesis of lignin through the production of H2O2 mediated by diamine oxidases in soybean hypocotyls. J. Plant Physiol.162, 1297–1303. doi: 10.1016/j.jplph.2005.04.033
63
SunX.MaY.YangC.LiJ. (2020). Rice OVATE family protein 6 regulates leaf angle by modulating secondary cell wall biosynthesis. Plant Mol. Biol.104, 249–261. doi: 10.1007/s11103-020-01039-2
64
TanabeS.AshikariM.FujiokaS.TakatsutoS.YoshidaS.YanoM.et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell17, 776–790. doi: 10.1105/tpc.104.024950
65
TianX.LiX.ZhouW.RenY.WangZ.LiuZ.et al. (2017). Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol.175, 1337–1349. doi: 10.1104/pp.17.00946
66
UpdegraffD. M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem.32, 420–424. doi: 10.1016/S0003-2697(69)80009-6
67
VanholmeR.StormeV.VanholmeB.SundinL.ChristensenJ. H.GoeminneG.et al. (2012). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell24, 3506–3529. doi: 10.1105/tpc.112.102574
68
WadaK.MarumoS.AbeH.MorishitaT.NakamuraK.UchiyamaM.et al. (1984). A rice lamina inclination test - a micro-quantitative bioassay for brassinosteroids. Agric. Biol. Chem.48, 719–726. doi: 10.1271/bbb1961.48.719
69
WangJ.JingX.QianQ.GuanghengZ. (2020). Development of rice leaves: how histocytes modulate leaf polarity establishment. Rice Sci.27, 468–479. doi: 10.1016/j.rsci.2020.09.004
70
WangR.LiuC.LiQ.ChenZ.SunS.WangX. (2020). Spatiotemporal resolved leaf angle establishment improves rice grain yield via controlling population density. iScience23, 101489. doi: 10.1016/j.isci.2020.101489
71
WangZ. Y.NakanoT.GendronJ.HeJ.ChenM.VafeadosD.et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell2, 505–513. doi: 10.1016/S1534-5807(02)00153-3
72
WangY.ZhangT.WangR.ZhaoY. (2018). Recent advances in auxin research in rice and their implications for crop improvement. J. Exp. Bot.69, 255–263. doi: 10.1093/jxb/erx228
73
WeiC.XieP.ChenY.YuH.SuY.GuM.et al. (2011). Anatomical and chemical characteristics of culm of rice brittle mutant bc7(t). Funct. Plant Biol.38, 227–235. doi: 10.1071/FP10235
74
XuJ.WangJ. J.XueH. W.ZhangG. H. (2021). Leaf direction: lamina joint development and environmental responses. Plant Cell Environ.44, 2441–2454. doi: 10.1111/pce.14065
75
YamamuroC.IharaY.WuX.NoguchiT.FujiokaS.TakatsutoS.et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell12, 1591–1605. doi: 10.1105/tpc.12.9.1591
76
YangC.LiD.LiuX.JiC.HaoL.ZhaoX.et al. (2014). OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa l.). BMC Plant Biol.14, 158. doi: 10.1186/1471-2229-14-158
77
YokoyamaR.NishitaniK. (2001). A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of arabidopsis. Plant Cell Physiol.42, 1025–1033. doi: 10.1093/pcp/pce154
78
YoshikawaT.ItoM.SumikuraT.NakayamaA.NishimuraT.KitanoH.et al. (2014). The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J.78, 927–936. doi: 10.1111/tpj.12517
79
ZhangS. W.LiC. H.CaoJ.ZhangY. C.ZhangS. Q.XiaY. F.et al. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol.151, 1889–1901. doi: 10.1104/pp.109.146803
80
ZhangS.WangS.XuY.YuC.ShenC.QianQ.et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ.38, 638–654. doi: 10.1111/pce.12397
81
ZhangL.ZhangB.ZhouY. (2019). Cell wall compositional analysis of rice culms. Bio-protoc.9, e3398. doi: 10.21769/BioProtoc.3398
82
ZhaoS. Q.HuJ.GuoL. B.QianQ.XueH. W. (2010). Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res.20, 935–947. doi: 10.1038/cr.2010.109
83
ZhaoK.LinF.Romero-GamboaS. P.SahaP.GohH. J.AnG.et al. (2019). Rice genome-scale network integration reveals transcriptional regulators of grass cell wall synthesis. Front. Plant Sci.10, 1275. doi: 10.3389/fpls.2019.01275
84
ZhaoS. Q.XiangJ. J.XueH. W. (2013). Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol. Plant6, 174–187. doi: 10.1093/mp/sss064
85
ZhongR.LeeC.McCarthyR. L.ReevesC. K.JonesE. G.YeZ. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol.52, 1856–1871. doi: 10.1093/pcp/pcr123
86
ZhouL. J.XiaoL. T.XueH. W. (2017). Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol.174, 1728–1746. doi: 10.1104/pp.17.00413
Summary
Keywords
auxin, cellulose, leaf angle, lignin, OsMYB7, rice
Citation
Kim S-H, Yoon J, Kim H, Lee S-J, Kim T, Kang K and Paek N-C (2023) OsMYB7 determines leaf angle at the late developmental stage of lamina joints in rice. Front. Plant Sci. 14:1167202. doi: 10.3389/fpls.2023.1167202
Received
16 February 2023
Accepted
27 March 2023
Published
14 April 2023
Volume
14 - 2023
Edited by
Stephan Pollmann, National Institute of Agricultural and Food Research and Technology, Spain
Reviewed by
Xu Peng, Xishuangbanna Tropical Botanical Garden (CAS), China; Jeongim Kim, University of Florida, United States
Updates
Copyright
© 2023 Kim, Yoon, Kim, Lee, Kim, Kang and Paek.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nam-Chon Paek, ncpaek@snu.ac.kr
†ORCID: Suk-Hwan Kim, orcid.org/0000-0002-8496-256X; Jungwon Yoon, orcid.org/0000-0002-5326-8460; Hanna Kim, orcid.org/0000-0001-8145-4191; Sang-Ji Lee, orcid.org/0000-0001-9112-5236; Taehoon Kim, orcid.org/0000-0002-0555-5852; Kiyoon Kang, orcid.org/0000-0002-1244-1669; Nam-Chon Paek, orcid.org/0000-0002-9827-2287
This article was submitted to Plant Physiology, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.