
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 May 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1165646
This article is part of the Research Topic The Contribution of Molecular Priming to Abiotic Stress Tolerance in Plants View all 6 articles
Oxidative stress causes cellular damage and genomic instability through the accumulation of reactive oxygen species (ROS) in plants, resulting in reduced crop production. Chemical priming, which can enhance plant tolerance to environmental stress using functional chemical compounds, is expected to improve agricultural yield in various plants without genetic engineering. In the present study, we revealed that non-proteogenic amino acid N-acetylglutamic acid (NAG) can alleviate oxidative stress damage in Arabidopsis thaliana (Arabidopsis) and Oryza sativa (rice). Exogenous treatment with NAG prevented chlorophyll reduction induced by oxidative stress. The expression levels of ZAT10 and ZAT12, which are regarded as master transcriptional regulators in response to oxidative stress, increased following NAG treatment. Additionally, Arabidopsis plants treated with NAG showed enhanced levels of histone H4 acetylation at ZAT10 and ZAT12 with the induction of histone acetyltransferases HAC1 and HAC12. The results suggest that NAG could enhance tolerance to oxidative stress through epigenetic modifications and contribute to the improvement of crop production in a wide variety of plants under environmental stress.
As sessile organisms, plants are exposed to various environmental stress factors, such as heat, drought, cold, and salinity, which inhibit growth and reduce crop biomass. In response to environmental stress in plants, oxidative stress, which is caused by the accumulation of reactive oxygen species (ROS) derived from electron energy in the chloroplasts, mitochondria, and peroxisomes, causes cellular damage through membrane oxidation and the collapse of photosynthesis. ROS generation is induced by both biotic and abiotic stresses, such as microbial pathogens (Kim and Hwang, 2014; Yang et al., 2017). Abiotic and biotic stresses disrupt the equilibrium between production and removal of intracellular ROS. This disruption of ROS equilibrium leads the overaccumulation of ROS, which causes oxidative stress. Oxidative stress also threatens the genome stability of plants owing to genotoxic stress, leading to effects such as replication error, cell cycle arrest, and cell death (Baxter et al., 2014). Therefore, plants must respond rapidly and plastically to oxidative stress to maintain their stability at the cellular and genomic levels. In response to plant oxidative stress, low molecular antioxidants, such as flavonoids, anthocyanin, ascorbate and glutathione, are accumulated in tissues, therefore preventing cellular damage caused by ROS (Gechev et al., 2006). Arabidopsis overexpressing flavonol regulator MYB12/PEG1 and anthocyanin regulator MYB75/PAP1 accumulates flavonoids and anthocyanins, such as glycosides of kaempferol, quercetin, and cyanidin, which increases its tolerance to oxidative and drought stress (Nakabayashi et al., 2014). In addition to non-enzymatic ROS scavenging systems, ROS is removed by several antioxidant enzymes in plants. Superoxide anion () generated in chloroplasts is altered to O2 or H2O2 by superoxide dismutases (SODs), and then H2O2 is detoxified by ascorbate peroxidases (APXs) to maintain photosynthesis activity, under stress conditions (Alscher et al., 2002; Shigeoka et al., 2002). Conversely, in mitochondria, the detoxification of H2O2 by glutathione peroxidases is achieved to maintain cellular respiration, suggesting that genetic engineering to improve the expression of such antioxidant enzymes in plants confers tolerance to oxidative stress (Anjum et al., 2016). Overexpression of Populus ascorbate peroxidase (PpAPX) in Nicotiana tabacum alleviated not only oxidative stress but also salinity and drought stress through increases in chlorophyll content and APX activity (Li et al., 2009). Additionally, Ipomoea batatas overexpression of lbAPX results in the acquisition of tolerance against salinity stress, with a reduction in ROS accumulation (Yan et al., 2016).
In addition to genetic engineering, chemical priming, which is a strategy that enhances tolerance to environmental stress based on the regulation of gene expression using functional chemical compounds, is expected to improve crop production in a wide range of plant species (Sako et al., 2021). Exogenous treatment with acetic acid increases the expression levels of jasmonate signalling pathway genes, conferring tolerance to drought stress in several plant species, such as Arabidopsis thaliana (Arabidopsis), Zea mays, Triticum aestivum, Oryza sativa (rice), and Brassica napus (Kim et al., 2017). Arabidopsis and Lactuca sativa plants treated with ethanol exhibit enhanced tolerance to heat stress, which is associated with the stimulation of response to unfolded proteins (Matsui et al., 2022). Heat stress is also modulated following treatment with β-aminobutyric acid (BABA) in Arabidopsis and Brassica rapa, through the induction of heat shock proteins and the improvement of photosynthesis performance and antioxidant systems, respectively (Zimmerli et al., 2008; Quan et al., 2022). Although chemical priming is an effective strategy of alleviating damage caused by environmental stress in plants without genetic engineering, little is known about the chemical compounds that enhance tolerance to oxidative stress in plants.
Studies using Saccharomyces cerevisiae (yeast) have shown that arginine metabolism plays a role in protection against damage caused by oxidative stress. N-acetyltransferase MPR1, which converts proline metabolism intermediate Δ1-pyrroline-5-carboxylate (P5C)/glutamate-γ-semialdehyde (GSA) to N-acetyl-GSA in arginine metabolism, is required for the alleviation of oxidative and heat stress in yeast (Nishimura et al., 2010). MPR1 confers tolerance to these stressors by preventing ROS accumulation associated with arginine synthesis. Furthermore, nitric oxide (NO) converted from arginine by the flavoprotein Tah-18 contributes to stress tolerance in yeast suggesting that the enhancement of arginine metabolism could confer tolerance to oxidative stress in plants (Nishimura et al., 2013). In fact, overexpression Solanum lycopersicum N-acetylglutamate synthase1 (SlNAGS1), which converts glutamic acid (Glu) to N-acetylglutamic acid (NAG) in the first step of arginine metabolism in chlorophyll, alleviates the damage caused by drought and salt stress through the accumulation of intermediates of arginine metabolism in Arabidopsis (Kalamaki et al., 2009). However, little is known about the effects and the underlying molecular mechanisms of arginine metabolism intermediates during chemical priming.
The expression of stress-responsive genes in plants is plastically and rapidly regulated by histone modifications, especially histone acetylation which is closely related to transcriptional activation (Sako et al., 2021). Histone H3 lysine 9 acetylation (H3K9ac) and H4 acetylation (H4ac) increase Arabidopsis T87 cell and tobacco BY-2 cell in response to cold and salinity stress (Sokol et al., 2007; Kurita et al., 2017). Deficiency in HDA9, which is one of the histone deacetylases, results in reduction of heat stress tolerance due to deposition of H3K9ac at heat stress-responsive genes in Arabidopsis (Niu et al., 2022). Additionally, the inhibitor of class I-type histone deacetylase (HDAC) alleviates salinity stress in Arabidopsis by upregulation of Na+/H+ antiporter SOS1, in response to the activation of H4ac (Sako et al., 2016). These findings suggest that the fine-tuning of histone acetylation is important for plant response to various environmental stress.
In the present study, we revealed that the non-proteogenic amino acid N-acetylglutamic acid (NAG), an intermediate in arginine metabolism in plants, can be used for chemical priming against oxidative stress. In Arabidopsis, exogenous treatment with NAG conferred tolerance to oxidative stress through the upregulation of oxidative stress-responsive genes, with an increase in histone acetylation levels. Additionally, rice treated with NAG exhibited tolerance to oxidative stress, based on the elevated expression levels of antioxidant enzymes, indicating that NAG could be used as a priming compound to enhance tolerance to oxidative stress in plants.
Arabidopsis plants used in the present study were Col-0 accessions. Sterilized Arabidopsis seeds were incubated in D.W. at 4°C for 24 h. After incubation, seeds were sown in liquid medium containing 1/2 Murashige and Skoog (MS) medium and 1% (w/v) sucrose, and then placed in an incubator set at 22°C, with a 16-h light/8-h dark photoperiod. Rice plants used in the present study were Nipponbare cultivar. Sterilized rice seeds were incubated in liquid medium containing 1/2 MS in an incubator set at 30°C, with a 16-h light/8-h dark photoperiod.
Chlorophyll content was measured as previously described (Yamaguchi et al., 2021). To measure Arabidopsis chlorophyll contents, seven-day-old seedlings were treated with 0–0.4 mM N-acetylglutamic acid (NAG, Tokyo Chemical Industry Co., Ltd.), under 5 μM methyl viologen (MV, Sigma-Aldrich), for 48 h. Five seedlings were placed in 1 mL N, N′- dimethylformamide (DMF) in 1.5-mL tubes, and then incubated at 4°C for 24 h. After incubation, the absorbance of the extraction liquid was measured at 646.8 nm and 663.8 nm on an NP80 spectrophotometer (Implen, Munich, Germany). Total chlorophyll contents were calculated using the following formula: Chl a + b (μM) = 19.43 A646.8 + 8.05 A663.8. To measure the chlorophyll content of rice, three-day-old plants were treated with NAG (0.5 mM) for 24 h and then incubated with 2.5 mM MV for 48 h. Three second foliage leaves were placed in 1 mL DMF. After incubation at 4°C for 24 h, the extraction liquid was used as a sample for the measurement of chlorophyll content. Each chlorophyll content (μM/mgFW) was determined relative to the chlorophyll content of plants with 0 mM NAG and 0 μM MV.
DAB staining was carried out as previously described with minor modification (Yamaguchi et al., 2021). Seven-day-old Arabidopsis seedlings were treated with 0.4 mM NAG and 5 µM MV for 24 h. After incubation, seedlings were stained with peroxidase stain DAB Kit (Nacalai) for 2 h in dark with gentle shaking. Stained seedlings were transferred to bleaching solution (60% ethanol, 20% acetic acid and 20% glycerol) and boiled for 15 min. Samples mounted with 50% glycerol were observed with SZX16 (EVIDENT) equipped with DP23 digital camera (EVIDENT). The experiment was conducted using three biological replicates.
Total RNA was isolated from whole seedlings of Arabidopsis and rice using an RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). RNA was extracted according to the manufacturer’s instructions. Genomic DNA was extracted using an RNase-free DNase set (QIAGEN).
For gene expression analysis, cDNA was synthesized from 1 µg total RNA with a Verso cDNA Synthesis Kit (Themo Fisher Scientific, Waltham, MA, USA). RT-qPCR analysis was performed using TB Green Premix EX Taq II (TaKaRa Bio Inc., Shiga, Japan), gene-specific primers, and a Light Cycler 480 (Roche, Basel, Switzerland). PP2AA3 and OsACTIN1 (gene annotation: Os03t0718100) were used as internal controls in Arabidopsis and rice, respectively. Normalized expression levels were calculated using 2^(-delta delta Ct) method. The primers used for the RT-qPCR analyses are listed in Supplementary Table S1.
Chromatin immunoprecipitation (ChIP) was performed as previously described, with minor modifications (Yamaguchi et al., 2014). Seven-day-old seedlings were treated with 0.4 mM NAG for 2 h and fixed with 1% formaldehyde for 15 min. After neutralization of the formaldehyde with glycine for 5 min, 1 g of the tissue was ground in liquid nitrogen using a mortar and pestle. Chromatin was isolated from the nuclear extract using nuclear extraction buffer. Fragmentation of chromatins were performed with Bioruptor® UCD-250 (Cosmo Bio., Ltd., Tokyo, Japan) using power mode H and an on/off cycle of 30 s/60 s, for a total duration 12 min, on ice. Antibodies were added to the fraction of fragmented chromatins after preclearing, and then the samples were rotated overnight at 4°C. Five microliters of anti-H4ac antibody (Merck Millipore: 06-866) and 2 µL of anti-H3K4me3 (Abcam: ab8580) were used in immunoprecipitation.
qPCR analysis was performed using TB Green Premix EX Taq II (TaKaRa), gene-specific primers, and a Light Cycler 480 (Roche). TA3 was used as the negative control. The primers used for the qPCR analysis are listed in Supplementary Table S1. ChIP experiments were repeated three times with three technical replicates. Statistical analysis was performed using Student’s t-test.
To confirm whether NAG has a capacity to alleviate oxidative stress damage in plants, we observed the responses of Arabidopsis to oxidative stress following treatment with NAG. Arabidopsis plants treated with MV, which produces reactive oxygen species (ROS) in chloroplasts, and is used to induce oxidative stress in plants, showed chlorosis of the cotyledons with a reduction in chlorophyll content (Figures 1A, B). In contrast, MV-induced chlorosis and reduction in chlorophyll content were suppressed in plants treated with NAG, in a dose-dependent manner (Figures 1A, B). DAB staining, which could detect H2O2 as a brown signal in tissues, also showed that the frequency of MV-induced H2O2 in seedlings treated with NAG was lower than that of seedlings without NAG (Figure 1C). These results suggest that NAG alleviated oxidative stress by preventing chlorosis and the accumulation of ROS in Arabidopsis.
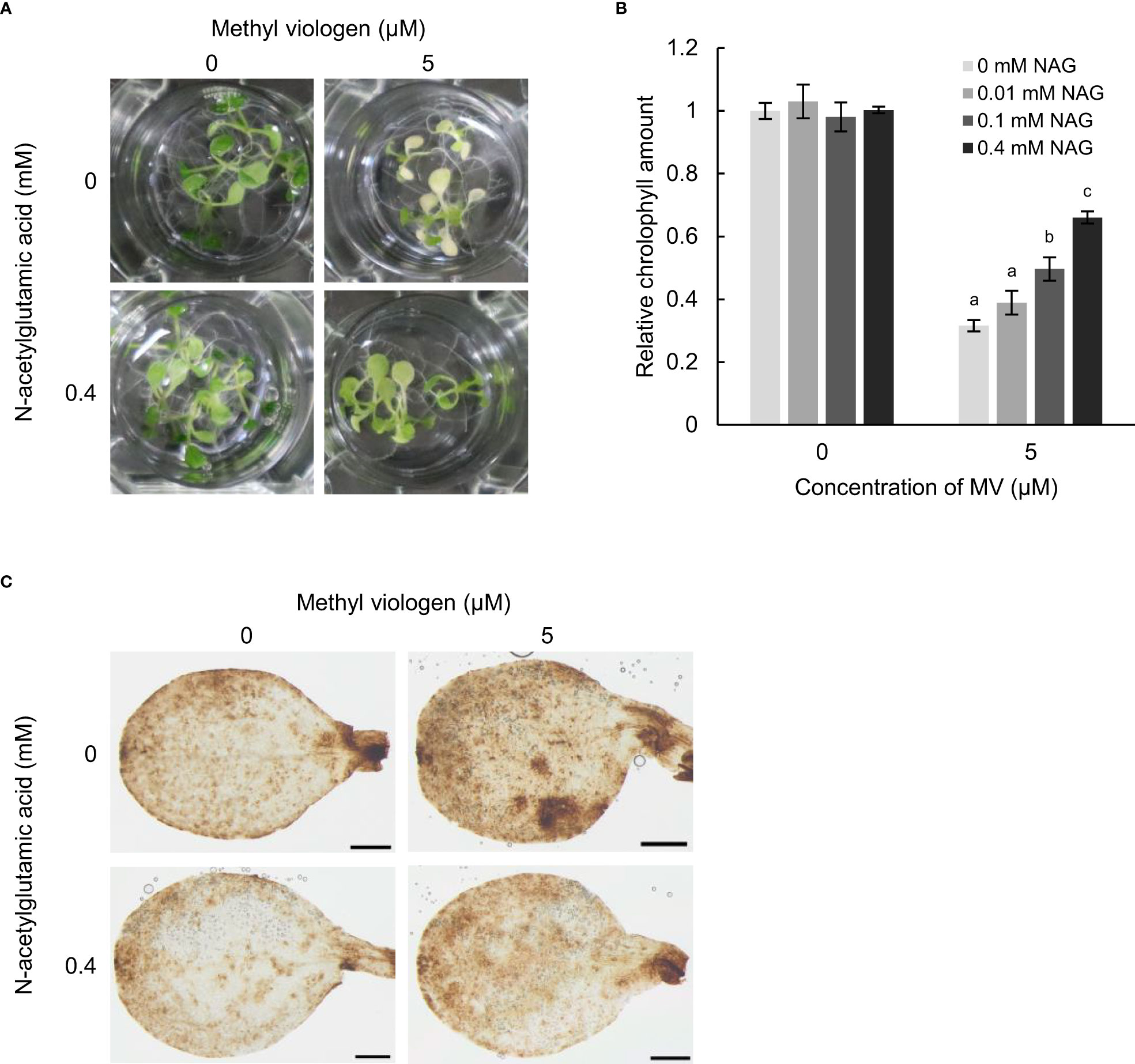
Figure 1 N-acetylglutamic acid enhances oxidative stress tolerance in Arabidopsis thaliana. (A) Seedlings treated with 0, 0.01, 0.1, and 0.4 mM N-acetylglutamic acid (NAG) under 5 μM methyl viologen (MV) for 48 h. (B) Chlorophyll contents in seedlings treated with 0, 0.01, 0.1, and 0.4 mM NAG under 5 μM MV for 48 h. Error bars indicate standard error. n = 15. P < 0.05 (Tukey test). (C) DAB staining for the detection of H2O2 in cotyledons of seedlings treated with or without 0.4 mM NAG and 5 μM MV for 24 h. Three biological replicates were conducted. Scale bar: 0.5 mm.
Several amino acids alleviate environmental stress in plants by activating the expression of stress-response genes. Therefore, we investigated whether NAG activates the expression of genes involved in response to oxidative stress in Arabidopsis. The expression levels of ZAT10 and ZAT12, which are zinc-finger type transcription factors regarded as master regulators in response to oxidative stress in plants, increased with NAG treatment without MV (Figure 2). Furthermore, we observed that exogenous treatment of NAG increased the expression of alternative oxidase 1a (AOX1a) and l-ascorbate peroxidase1 and 2 (APX1 and APX2), which scavenge ROS and modulate cellular damage in response to oxidative stress (Figure 2). Induction of the genes above was observed in plants treated with NAG under control conditions, which was unrelated to MV treatment. According to the results, NAG increased the expression levels of oxidative stress-response genes, independent of oxidative stress damage.
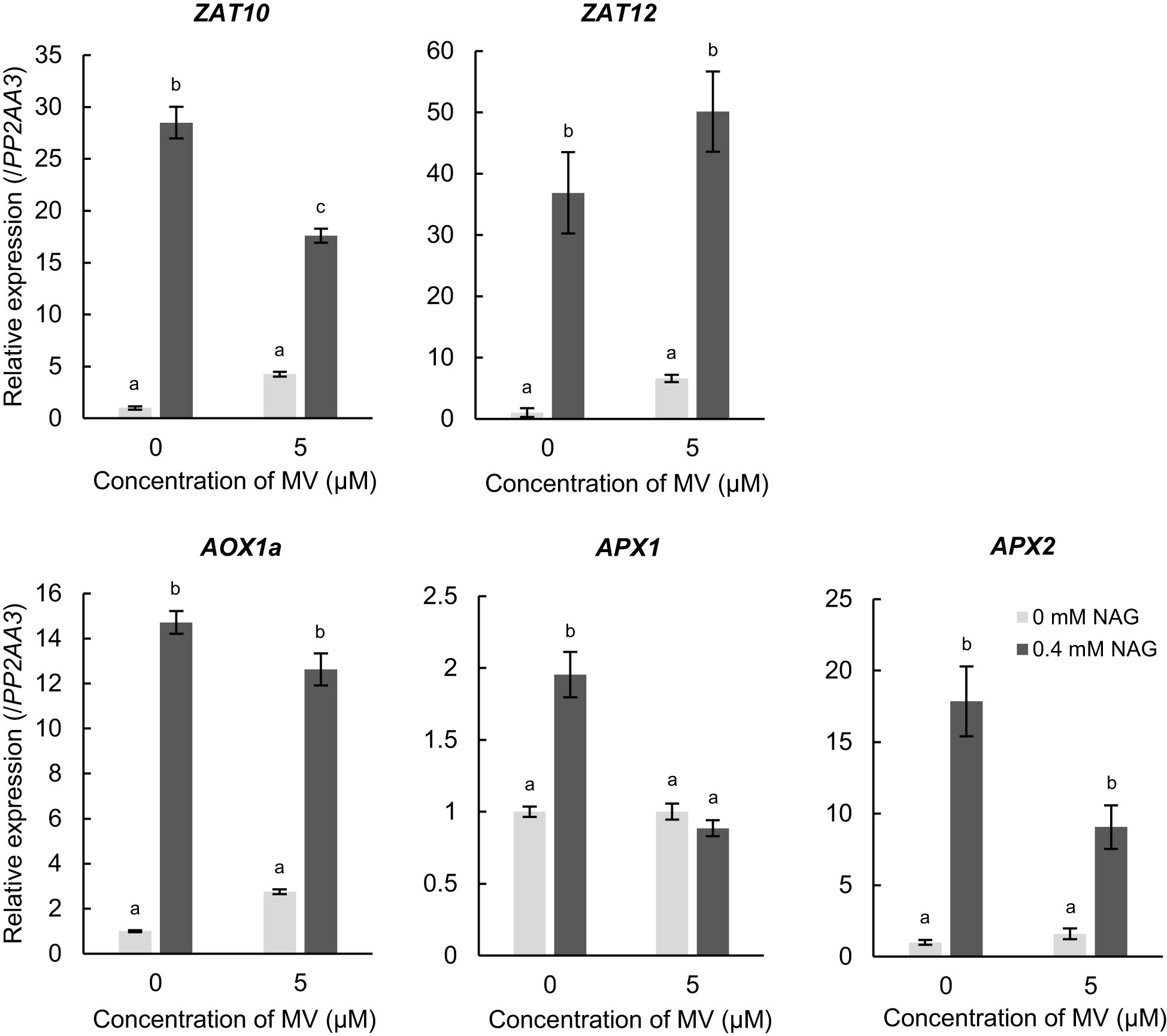
Figure 2 Expression levels of genes involved in responses to oxidative stress increased following treatment with N-acetylglutamic acid (NAG). Expression levels of ZAT10, ZAT12, AOX1a, APX1 and APX2 in seedlings treated with or without 0.4 mM NAG and 5 μM methyl viologen for 2 h. Error bars indicate the standard error. n = 3. P < 0.05 (Tukey test).
Phenotype and gene expression analyses revealed that NAG can modulate oxidative stress damage in plants. However, the molecular mechanism controlling the expression of oxidative stress-response genes in NAG remains unknown. In Arabidopsis, exogenous treatment of acetic acid confers drought tolerance accompanied with the activation of some transcription factors including ZAT10 through histone H4 acetylation (Kim et al., 2017). Therefore, we examined whether histone acetylation increased at ZAT10 and ZAT12 following NAG treatment. Chromatin immunoprecipitation – quantitative PCR (ChIP-qPCR) showed the levels of histone H4ac at the 5’ region of ZAT10, and ZAT12 was increased in plants treated with NAG (Figure 3A). In contrast, histone H3 lysine 4 trimethylation (H3K4me3), whose modification levels are correlated with the activation of gene expression, was enhanced only at ZAT12 and not at ZAT10, following NAG treatment (Figure 3B). The results suggest that NAG activates the expression of ZAT10 and ZAT12 by regulating histone acetylation.
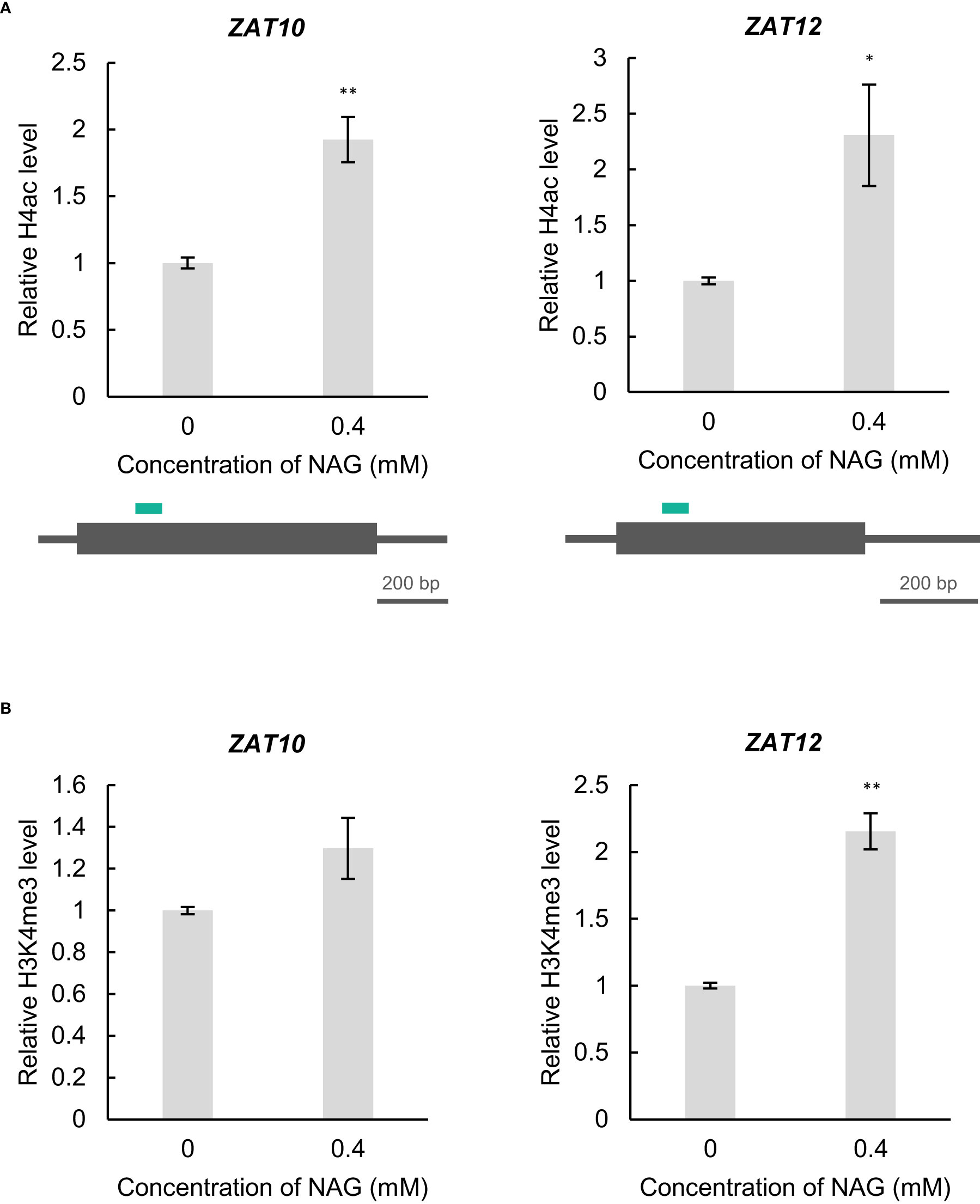
Figure 3 N-acetylglutamic acid (NAG) increases histone acetylation levels at ZAT10 and ZAT12. (A) Top: Levels of histone H4 acetylation (H4ac) at ZAT10 and ZAT12 of seedlings treated with 0.4 mM NAG for 2 h. Error bars indicate standard error. n = 3. **P <0.01, *P < 0.05 (student’s t-test). Bottom: Diagrams of ChIP-qPCR analysis. Green bars indicate the regions amplified by qPCR. (B) Levels of histone H3 lysine 4 trimethylation (H3K4me3) at ZAT10 and ZAT12 in seedlings treated with 0.4 mM NAG for 2 h. Error bars indicate standard error. n = 3. **P < 0.01 (student’s t-test).
To investigate the molecular mechanisms underlying histone acetylation of oxidative stress-response genes following NAG treatment, we investigated the expression levels of several histone acetyltransferases (HATs). Among the HATs, the expression levels of ARABIDOPSIS HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1 and 12 (HAC1 and HAC12) were specifically increased in plants treated with NAG when compared with that in the control plants (Figure 4). Therefore, NAG facilitated HAC1 and HAC12 to activate the oxidative stress-response genes through histone acetylation.
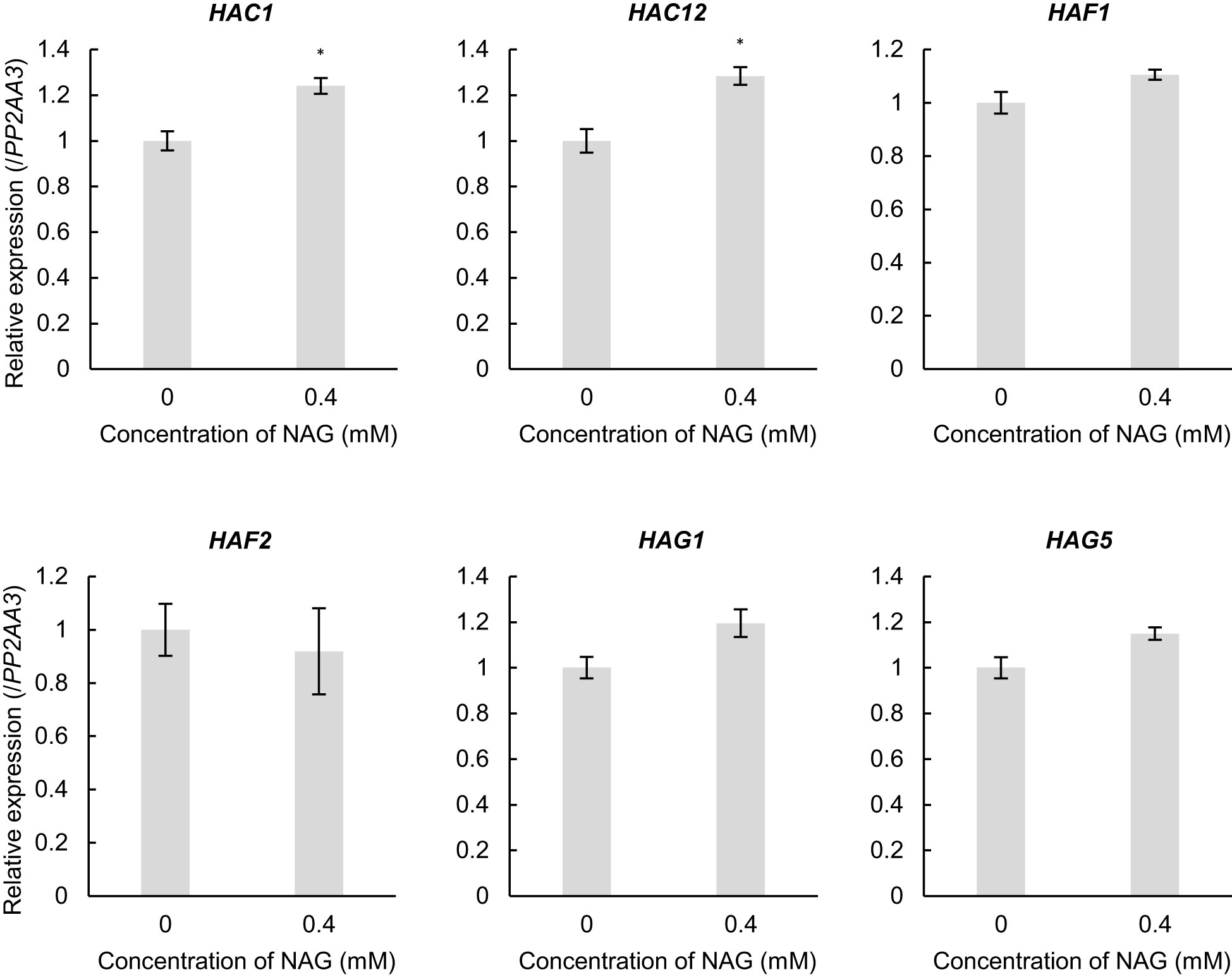
Figure 4 Induction of histone acetyltransferases with the treatment of N-acetylglutamic acid (NAG). Expression levels of HAC1, HAC12, HAF1, HAF2, HAG1 and HAG5 in seedlings treated with or without 0.4 mM NAG for 2 h. Error bars indicate the standard error. n = 3. *P < 0.05 (Student’s t-test).
To examine whether NAG alleviates oxidative stress in plants other than Arabidopsis, we observed response to oxidative stress in the monocotyledonous plant rice. MV-induced chlorosis and reduced chlorophyll contents in the leaves of rice and Arabidopsis (Figures 5A, B). Following to MV, rice plants treated with NAG showed higher amounts of chlorophyll in the leaves than plants not treated with NAG (Figures 5A, B). Additionally, the expression levels of OsAOX1a and OsAOX1b increased in rice plants treated with NAG, which was similar to the results in Arabidopsis (Figure 5C). The results suggest that NAG can alleviate damage caused by oxidative stress by activating oxidative stress-response genes in rice.
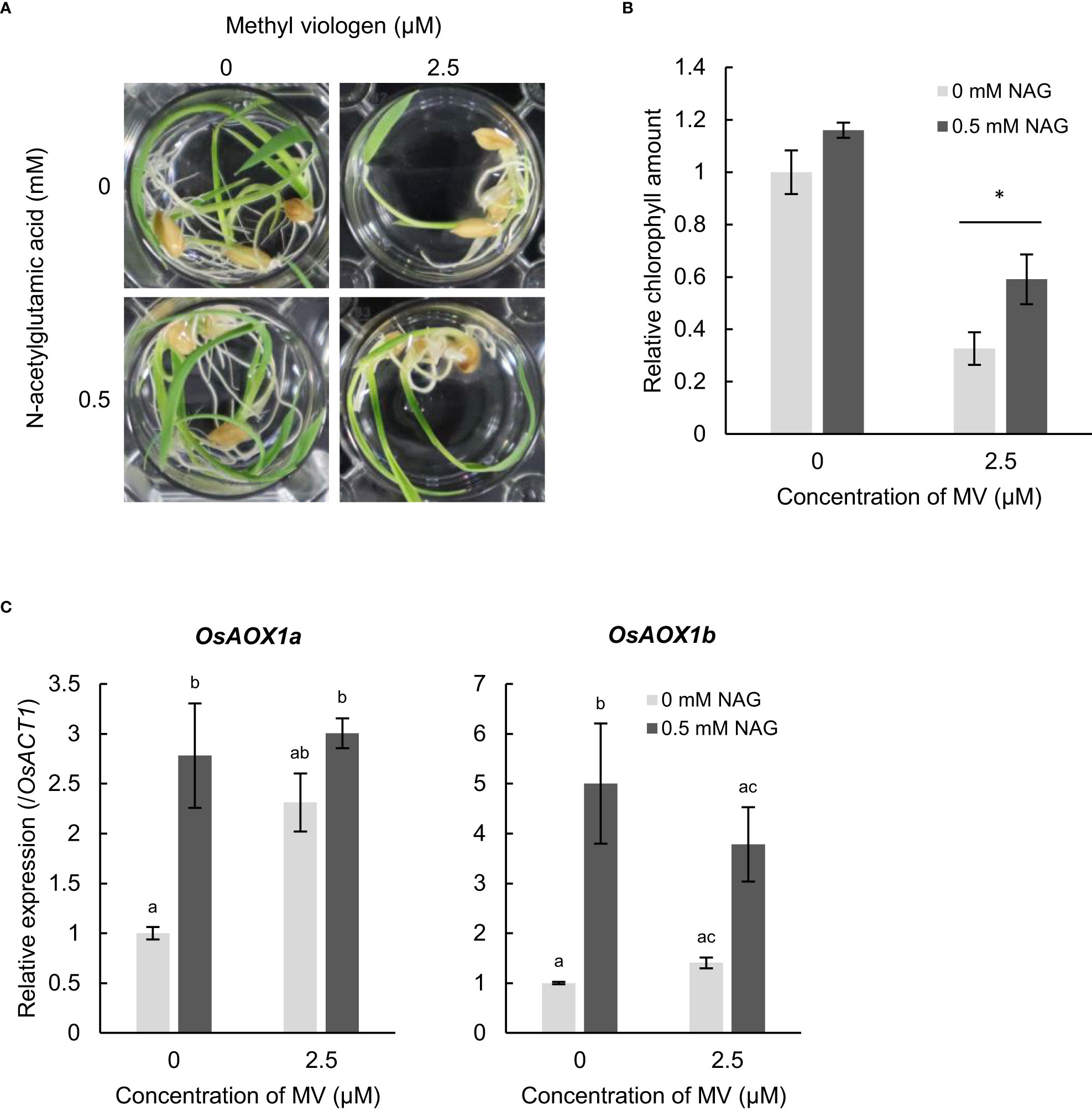
Figure 5 N-acetylglutamic acid (NAG) alleviates oxidative stress damage in Oryza sativa. (A) Seedlings treated with or without 0.5 mM NAG under 2.5 μM methyl viologen (MV) for 48 h. (B) Chlorophyll amounts in seedlings treated with or without 0.5 mM NAG under 2.5 μM MV for 48 h. Error bars indicate standard error. n = 15. *P < 0.05 (Tukey test). (C) Expression levels of OsAOX1a and OsAOX1b in seedlings treated with or without 0.4 mM NAG and 5 μM MV for 2 h. Error bars indicate the standard error. n = 3). P < 0.05 (Tukey test).
In this study, we demonstrated that NAG alleviates oxidative stress by regulating the expression of oxidative stress-response genes in Arabidopsis and rice. NAG is converted from Glu by N-acetylglutamate synthase (NAGS) in the first step of the arginine pathway in plants. SlNAGS1 enhances tolerance to drought and salt stress in Arabidopsis, which is associated with the accumulation of ornithine, a precursor of arginine (Kalamaki et al., 2009). Overexpression of Z. mays N-acetylglutamate kinase (ZmNAGK), which phosphorylates NAG to proceed to arginine metabolism, in Nicotiana tabacum, resulted in greater resistance to drought stress than in wild-type plants with the accumulation of arginine, leading to the hypothesis that exogenous treatment with NAG modulates several environmental stress factors, including oxidative stress, with the activation of arginine metabolism as well as gene regulation activity involving stress-response genes (Liu et al., 2018). In response to heat stress, NO synthesised during arginine metabolism transduces the heat stress signal from the shoot apex to the roots to enhance systemic resistance by upregulating heat stress-response genes in Arabidopsis (He et al., 2022). Therefore, the role of NAG as an activator of systemic resistance via arginine metabolism in response to oxidative stress needs to be examined.
NAG is an intermediate converted from Glu during arginine metabolism in plants, suggesting that Glu-related compounds modulate the damage caused by environmental stress in plants. Exogenous treatment with Glu enhances systemic resistance in the immune system by upregulating defence-related genes, resulting in resistance to pathogen infection in Arabidopsis and rice (Kadotani et al., 2016; Goto et al., 2020). Additionally, deficiency in GLUTAMATE DECARBOXYLASEs (GADs), which synthesise GABA from Glu, results in a reduction in heat stress and high-light stress tolerance in Arabidopsis (Balfagón et al., 2022). Therefore, the application of Glu-related compounds, including NAG, may confer tolerance to several environmental stress factors in plants.
Exogenous treatment of NAG increased expression levels of oxidative stress-response genes in Arabidopsis (Figure 2). Overexpression of ZAT10 or ZAT12 in Arabidopsis induces the upregulation of genes involved in responses to some environmental stress factors, such as heat, oxidative, osmotic, and salinity stress, resulting in tolerance to the stress factors. Deficient mutant of ZAT12 could not enhance the expression levels of APX1 in response to oxidative stress, and APX1 expression was activated with NAG treatment as well as ZAT12 (Figure 2) (Rizhsky et al., 2004). Thus, transcriptional cascade from ZAT12 to APX1 may function in plants treated with NAG in response to oxidative stress. ZAT10 directly regulates the transcriptional activity of genes related to the cadmium (Cd) uptake and detoxification to confer Cd tolerance under higher Cd conditions in Arabidopsis (Dang et al., 2022). Additionally, ZAT12 prevents the overaccumulation of iron (Fe) in Arabidopsis shoots by negatively controlling the transcription involved in Fe uptake (Le et al., 2016). Therefore, NAG may alleviate environmental stress, excluding oxidative stress, through the regulation of ZAT10 or ZAT12 expression.
We also confirmed that histone levels H4ac were increased by NAG treatment (Figure 3A). Several synthetic and natural compounds can modulate the epigenetic modification of stress-response genes, resulting in stress tolerance in plants. Treatment with Ky-2, an inhibitor of HDAC, alleviates salinity stress through the activation of the Na+/H+ antiporter SOS1 based on the upregulation of histone H4ac in Arabidopsis (Sako et al., 2016). The induction of JA signalling associated with the hyperacetylation of histones H3 and H4 via the HDAC inhibitor suberoylanilde hydroxamic acid (SAHA) confers salinity stress tolerance in Manihot esculenta. Acetic acid activates jasmonate signalling associated with an increase in histone H4ac levels, resulting in drought stress tolerance in Arabidopsis, rice, Z. mays, T. aestivum, and B. napus (Kim et al., 2017). Radioisotope experiments using acetic acid labelled with 14C show the possibility that acetic acid is used as a substrate for histone H4ac in Arabidopsis. Interestingly, acetic acid is also used as a substrate for acetylation of tRNA catalysed by acetyltransferase tmcAL in Bacillus subtilis, suggesting that NAG might function as a substrate of acetyl group for acetylation at oxidative stress-responsive genes, including histone acetyltransferases (Taniguchi et al., 2018).
Although several studies have explored chemical compounds that alleviate the damage caused by environmental stresses based on histone acetylation, little is known about the effects of chemical compounds involved in epigenetic modification other than histone acetylation. Studies using specific Arabidopsis mutants have demonstrated that histone methylation contributes to stress tolerance. For example, deficiency in LDL1, which is a histone demethylase of histone H3 Lys 4 di-methylation (H3K4me2), causes the accumulation of DNA damage due to genotoxic stress in Arabidopsis (Hirakawa et al., 2019). Additionally, the removal of histone H3 Lys 27 tri-methylation (H3K27me3) on heat stress-response genes by the histone demethylase JUMONJIs (JMJ26, JMJ29, ERF6, REF6) is required for priming against heat stress, indicating that manipulation of histone methylation using chemical compounds could enhance stress tolerance (Yamaguchi et al., 2021).
Although exogenous treatment with NAG specifically increases the expression levels of the histone acetyltransferases HAC1 and HAC12, which are involved in the regulation of flowering time and sugar response, the molecular mechanisms underlying the upregulation of the genes by NAG remains unknown (Figure 4) (Han et al., 2007; Chen et al., 2019). Recently, it has been revealed that defence responses attributed to volatile organic compounds (VOCs) emitted from plants in response to herbivory could be enhanced based on chromatin remodelling (Onosato et al., 2022). Among VOCs, caryophyllene, a sesquiterpene, increases the expression levels of defence-related genes in tobacco BY-2 cells through binding with TOPLESS (TPL)-like protein, which functions as a transcriptional co-repressor by interacting with the transcription factor (Nagashima et al., 2019). The findings of the present study indicate that NAG could activate the expression of oxidative stress-response genes, including histone acetyltransferases, based on chromatin remodelling activities modulated by chromatin modifiers such as TPL-like proteins.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
TH designed the study. TH and ST conducted the experiments. TH analysed the data. TH and KO wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the Kirin Holdings Company, Ltd.
We would like to thank Fuhito Yamazaki (Kirin Holdings Company, Ltd.) for technical assistance with the ChIP experiment and Dr. Noboru Onishi for discussions on this research.
TH, ST, and KO were employed by the Kirin Holdings Company, Ltd.
This study received funding from Kirin Holdings Company Ltd. The funder had the following involvement with the study: the decision to publish.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1165646/full#supplementary-material
Supplementary file S1 | List of primers used in this study
Alscher, R. G., Erturk, N., Heath, L. S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. doi: 10.1093/jexbot/53.372.1331
Anjum, N. A., Sharma, P., Gill, S. S., Hasanuzzaman, M., Khan, E. A., Kachhap, K., et al. (2016). Catalase and ascorbate peroxidase-representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. pollut. Res. Int. 23, 19002–19029. doi: 10.1007/s11356-016-7309-6
Balfagón, D., Gómez-Cadenas, A., Rambla, J. L., Granell, A., de Ollas, C., Bassham, D. C., et al. (2022). γ-aminobutyric acid plays a key role in plant acclimation to a combination of high light and heat stress. Plant Physiol. 188, 2026–2038. doi: 10.1093/plphys/kiac010
Baxter, A., Mittler, R., Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Chen, Q., Xu, X., Xu, D., Zhang, H., Zhang, C., Li, G. (2019). WRKY18 and WRKY53 coordinate with histone ACETYLTRANSFERASE1 to regulate rapid responses to sugar. Plant Physiol. 180, 2212–2226. doi: 10.1104/pp.19.00511
Dang, F., Li, Y., Wang, Y., Lin, J., Du, S., Liao, X. (2022). ZAT10 plays dual roles in cadmium uptake and detoxification in arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.994100
Gechev, T. S., Van Breusegem, F., Stone, J. M., Denev, I., Laloi, C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28, 1091–1101. doi: 10.1002/bies.20493
Goto, Y., Maki, N., Ichihashi, Y., Kitazawa, D., Igarashi, D., Kadota, Y., et al. (2020). Exogenous treatment with glutamate induces immune responses in arabidopsis. Mol. Plant Microbe Interact. 33, 474–487. doi: 10.1094/MPMI-09-19-0262-R
Han, S. K., Song, J. D., Noh, Y. S., Noh, B. (2007). Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J. 49, 103–114. doi: 10.1111/j.1365-313X.2006.02939.x
He, N. Y., Chen, L. S., Sun, A. Z., Zhao, Y., Yin, S. N., Guo, F. Q. (2022). A nitric oxide burst at the shoot apex triggers a heat-responsive pathway in arabidopsis. Nat. Plants 8, 434–450. doi: 10.1038/s41477-022-01135-9
Hirakawa, T., Kuwata, K., Gallego, M. E., White, C. I., Nomoto, M., Tada, Y., et al. (2019). LSD1-LIKE1-mediated H3K4me2 demethylation is required for homologous recombination repair. Plant Physiol. 181, 499–509. doi: 10.1104/pp.19.00530
Kadotani, N., Akagi, A., Takatsuji, H., Miwa, T., Igarashi, D. (2016). Exogenous proteinogenic amino acids induce systemic resistance in rice. BMC Plant Biol. 16, 60. doi: 10.1186/s12870-016-0748-x
Kalamaki, M. S., Alexandrou, D., Lazari, D., Merkouropoulos, G., Fotopoulos, V., Pateraki, I., et al. (2009). Over-expression of a tomato n-acetyl-L-glutamate synthase gene (SlNAGS1) in arabidopsis thaliana results in high ornithine levels and increased tolerance in salt and drought stresses. J. Exp. Bot. 60, 1859–1871. doi: 10.1093/jxb/erp072
Kim, D. S., Hwang, B. K. (2014). An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 65, 2295–2306. doi: 10.1093/jxb/eru109
Kim, J. M., To, T. K., Matsui, A., Tanoi, K., Kobayashi, N. I., Matsuda, F., et al. (2017). Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 3, 17097. doi: 10.1038/nplants.2017.97
Kurita, K., Sakamoto, T., Yagi, N., Sakamoto, Y., Ito, A., Nishino, N., et al. (2017). Live imaging of H3K9 acetylation in plant cells. Sci. Rep. 7, 45894. doi: 10.1038/srep45894
Le, C. T. T., Brumbarova, T., Ivanov, R., Stoof, C., Weber, E., Mohrbacher, J., et al. (2016). ZINC FINGER OF arabidopsis THALIANA12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY-INDUCED transcription factor (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol. 170, 540–557. doi: 10.1104/pp.15.01589
Li, Y.-J., Hai, R.-L., Du, X.-H., Jiang, X.-N., Lu, H. (2009). Over-expression of a populus peroxisomal ascorbate peroxidase (PpAPX) gene in tobacco plants enhances stress tolerance. Plant Breed. 128, 404–410. doi: 10.1111/j.1439-0523.2008.01593.x
Liu, W., Xiang, Y., Zhang, X., Han, G., Sun, X., Sheng, Y., et al. (2018). Over-expression of a maize n-acetylglutamate kinase gene (ZmNAGK) improves drought tolerance in tobacco. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01902
Matsui, A., Todaka, D., Tanaka, M., Mizunashi, K., Takahashi, S., Sunaoshi, Y., et al. (2022). Ethanol induces heat tolerance in plants by stimulating unfolded protein response. Plant Mol. Biol. 110, 131–145. doi: 10.1007/s11103-022-01291-8
Nagashima, A., Higaki, T., Koeduka, T., Ishigami, K., Hosokawa, S., Watanabe, H., et al. (2019). Transcriptional regulators involved in responses to volatile organic compounds in plants. J. Biol. Chem. 294, 2256–2266. doi: 10.1074/jbc.RA118.005843
Nakabayashi, R., Yonekura-Sakakibara, K., Urano, K., Suzuki, M., Yamada, Y., Nishizawa, T., et al. (2014). Enhancement of oxidative and drought tolerance in arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379. doi: 10.1111/tpj.12388
Nishimura, A., Kawahara, N., Takagi, H. (2013). The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem. Biophys. Res. Commun. 430, 137–143. doi: 10.1016/j.bbrc.2012.11.023
Nishimura, A., Kotani, T., Sasano, Y., Takagi, H. (2010). An antioxidative mechanism mediated by the yeast n-acetyltransferase Mpr1: oxidative stress-induced arginine synthesis and its physiological role. FEMS Yeast Res. 10, 687–698. doi: 10.1111/j.1567-1364.2010.00650.x
Niu, Y., Bai, J., Liu, X., Zhang, H., Bao, J., Zhao, W., et al. (2022). Histone deacetylase 9 transduces heat signal in plant cells. Proc. Natl. Acad. Sci. U. S. A. 119, e2206846119. doi: 10.1073/pnas.2206846119
Onosato, H., Fujimoto, G., Higami, T., Sakamoto, T., Yamada, A., Suzuki, T., et al. (2022). Sustained defense response via volatile signaling and its epigenetic transcriptional regulation. Plant Physiol. 189, 922–933. doi: 10.1093/plphys/kiac077
Quan, J., Zheng, W., Wu, M., Shen, Z., Tan, J., Li, Z., et al. (2022). Glycine betaine and β-aminobutyric acid mitigate the detrimental effects of heat stress on Chinese cabbage (Brassica rapa l. ssp. pekinensis) seedlings with improved photosynthetic performance and antioxidant system. Plants (Basel) 11, 1213. doi: 10.3390/plants11091213
Rizhsky, L., Davletova, S., Liang, H., Mittler, R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in arabidopsis. J. Biol. Chem. 279, 11736–11743. doi: 10.1074/jbc.M313350200
Sako, K., Kim, J. M., Matsui, A., Nakamura, K., Tanaka, M., Kobayashi, M., et al. (2016). Ky-2, a histone deacetylase inhibitor, enhances high-salinity stress tolerance in arabidopsis thaliana. Plant Cell Physiol. 57, 776–783. doi: 10.1093/pcp/pcv199
Sako, K., Nguyen, H. M., Seki, M. (2021). Advances in chemical priming to enhance abiotic stress tolerance in plants. Plant Cell Physiol. 61, 1995–2003. doi: 10.1093/pcp/pcaa119
Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. doi: 10.1093/jexbot/53.372.1305
Sokol, A., Kwiatkowska, A., Jerzmanowski, A., Prymakowska-Bosak, M. (2007). Up-regulation of stress-inducible genes in tobacco and arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227, 245–254. doi: 10.1007/s00425-007-0612-1
Taniguchi, T., Miyauchi, K., Sakaguchi, Y., Yamashita, S., Soma, A., Tomita, K., et al. (2018). Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat. Chem. Biol. 14, 1010–1020. doi: 10.1038/s41589-018-0119-z
Yamaguchi, N., Matsubara, S., Yoshimizu, K., Seki, M., Hamada, K., Kamitani, M., et al. (2021). H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in arabidopsis. Nat. Commun. 12, 3480. doi: 10.1038/s41467-021-23766-w
Yamaguchi, N., Winter, C. M., Wu, M. F., Kwon, C. S., William, D. A., Wagner, D. (2014). PROTOCOLS: chromatin immunoprecipitation from arabidopsis tissues. Arabidopsis Book 12, e0170. doi: 10.1199/tab.0170
Yan, H., Li, Q., Park, S. C., Wang, X., Liu, Y. J., Zhang, Y. G., et al. (2016). Overexpression of CuZnSOD and APX enhance salt stress tolerance in sweet potato. Plant Physiol. Biochem. 109, 20–27. doi: 10.1016/j.plaphy.2016.09.003
Yang, C., Li, W., Cao, J., Meng, F., Yu, Y., Huang, J., et al. (2017). Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. doi: 10.1111/tpj.13388
Keywords: N-acetylglutamic acid, chemical priming, oxidative stress, epigenetic regulation, histone acetylation
Citation: Hirakawa T, Tanno S and Ohara K (2023) N-acetylglutamic acid alleviates oxidative stress based on histone acetylation in plants. Front. Plant Sci. 14:1165646. doi: 10.3389/fpls.2023.1165646
Received: 14 February 2023; Accepted: 19 April 2023;
Published: 08 May 2023.
Edited by:
Ece Turhan, Eskişehir Osmangazi University, TürkiyeCopyright © 2023 Hirakawa, Tanno and Ohara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeshi Hirakawa, VGFrZXNoaV9IaXJha2F3YUBraXJpbi5jby5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.