
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 May 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1163271
This article is part of the Research Topic Advances in Integrated Disease Management (IDM) For Soil-Borne Plant Pathogens: Innovative Approaches and Underlying Action Mechanism at Molecular Level View all 13 articles
 Liqun Song1,2,3
Liqun Song1,2,3 Xingxing Ping1,2
Xingxing Ping1,2 Zhenchuan Mao1,2
Zhenchuan Mao1,2 Jianlong Zhao1,2
Jianlong Zhao1,2 Yuhong Yang1,2
Yuhong Yang1,2 Yan Li1,2
Yan Li1,2 Bingyan Xie1,2*
Bingyan Xie1,2* Jian Ling1,2*
Jian Ling1,2*Introduction: Root-knot nematodes (RKN) disease is a devastating disease in Cucumis crops production. Existing studies have shown that resistant and susceptible crops are enriched with different rhizosphere microorganisms, and microorganisms enriched in resistant crops can antagonize pathogenic bacteria. However, the characteristics of rhizosphere microbial communities of Cucumis crops after RKN infestation remain largely unknown.
Methods: In this study, we compared the changes in rhizosphere bacterial communities between highly RKN-resistant Cucumis metuliferus (cm3) and highly RKN-susceptible Cucumis sativus (cuc) after RKN infection through a pot experiment.
Results: The results showed that the strongest response of rhizosphere bacterial communities of Cucumis crops to RKN infestation occurred during early growth, as evidenced by changes in species diversity and community composition. However, the more stable structure of the rhizosphere bacterial community in cm3 was reflected in less changes in species diversity and community composition after RKN infestation, forming a more complex and positively co-occurrence network than cuc. Moreover, we observed that both cm3 and cuc recruited bacteria after RKN infestation, but the bacteria enriched in cm3 were more abundant including beneficial bacteria Acidobacteria, Nocardioidaceae and Sphingomonadales. In addition, the cuc was enriched with beneficial bacteria Actinobacteria, Bacilli and Cyanobacteria. We also found that more antagonistic bacteria than cuc were screened in cm3 after RKN infestation and most of them were Pseudomonas (Proteobacteria, Pseudomonadaceae), and Proteobacteria were also enriched in cm3 after RKN infestation. We hypothesized that the cooperation between Pseudomonas and the beneficial bacteria in cm3 could inhibit the infestation of RKN.
Discussion: Thus, our results provide valuable insights into the role of rhizosphere bacterial communities on RKN diseases of Cucumis crops, and further studies are needed to clarify the bacterial communities that suppress RKN in Cucumis crops rhizosphere.
Nematodes were reported to be the second most numerous in the animal kingdom residing in multiple ecosystems, with up to one million species (Bongers and Bongers, 1998; Mitreva et al., 2005). Root-knot nematodes (RKN) were among the top ten plant parasitic nematodes in the world found in almost all vascular plants, causing severe crop losses through direct injury and transmission of pathogenic microorganisms (Chalivendra, 2021). The most devastating RKN species were M. incognita, M. arenaria, M. hapla and M. javanica, causing hundreds of billions of dollars in crop losses each year (Nicol et al., 2011; Singh et al., 2015). Variety of the plant affected their response to RKN parasitism, which usually induced knotted gall tumors in the host plant (Jones et al., 2013). RKN penetrated root cells and induced the production of giant cells near the vascular bundle, from which nematodes to absorb nutrients by mouthparts (Moens et al., 2009).
Cucumber (Cucumis sativus), an economically important crop, in the Cucurbitaceae family is widely grown around the world (Kaur et al., 2023). The RKN was one of the most important diseases in cucumber (Cucumis sativus) growing regions around the world. It was estimated that RKN caused yield losses of 88% in cucumbers under greenhouse cultivation conditions (Expósito et al., 2019). The ubiquitous RKN developed different lifestyles and feeding strategies, which, made them difficult to manage (Topalović and Vestergård, 2021). Chemical nematicides have been widely used to control plant-parasitic nematodes, but the harmful effects of these chemicals on the environment have led to a significant reduction in their frequency of use. Currently, host plant resistance was an effective method for managing plant-parasitic nematodes, but in some cases, high genetic diversity within and between nematodes populations confounded the use of available resistant species and limited their effectiveness (Li et al., 2011). To date, there was no cultivated cucumber cultivars with RKN resistance (Weng, 2010). In contrast, a high resistance to RKN were found in African horn cucumber (Cucumis metuliferus), a relative of African-endemic Cucumis (Walters et al., 2006). The C. metuliferus was a member of the Cucumoraceae, which included several economically important crops such as cucumber, melon, squash and watermelon (Nakata et al., 2005). It was a potential genetic resource for Cucumoraceae crops improvement as it contains genes for resistance to a variety of Cucumoraceae crops pests and diseases (Yagi et al., 2014). And it has been reported to be resistant to RKN, gummy stem blight and Fusarium wilt (Chen et al., 2020). In this case, resistance to RKN in C. metuliferus was associated with reduced nematode penetration, developmental retardation and hypersensitive necrosis (Ye et al., 2017). However, the attempt of interspecific hybridization between C. metuliferus and C. sativus was not successful (Walters et al., 2006).
Biological control was a means of suppressing pests and pathogens by using other organisms that can be natural enemies, such as predators, parasitic organisms and competitors, with the advantages of environmental safety and precise targeting, of which microorganisms were receiving increasing attention (Vega, 2018; Chalivendra, 2021). Microorganisms that colonize plants in numbers that far exceed the number of plant cells were called the plant microbiome, which played an important role in plant growth and development (Mendes et al., 2013). There were several interaction ways between microorganisms and nematodes. Microbial competition for space, nutrients and water reduced nematode activity and reproductive capacity (Pal and McSpadden Gardener, 2006). And microorganisms produced and released certain antibiotics or toxins which may adversely affect the nematode during the infection stage (Lugtenberg and Kamilova, 2009). Many bacterial products can induce systemic resistance in plants to protect the whole plant from pathogenic bacteria and nematodes (Junaid et al., 2013). Some bacteria can also penetrate the cuticle and killed nematodes by the action of enzymes (Tian et al., 2007). In addition, microorganisms contributed to plant development by improving the solubility, uptake and absorption of nutrients, thus helping to increase the tolerance of plant roots to plant-parasitic nematodes (Schouteden et al., 2015). Many antagonistic microorganisms, including Purpureocillium lilacinum, Streptomyces, Pseudomonas, have been tested and widely used to suppress a number of plant pathogens and nematodes (Sarwar et al., 2018; Aiello et al., 2019; Jiao et al., 2019).
The assembly of the microbiome included dynamic changes in species composition and abundance, on the other hand, the steady-state composition of spatially distinct compartments. The rhizosphere, the narrow area around and affected by plant roots, was an important ecological niche in plant microbiome studies and one of the most complex ecosystems on Earth (Hinsinger et al., 2009; Raaijmakers et al., 2009). “Rhizosphere effect”, a phenomenon in which the rhizosphere microbial community differed from the community in the bulk soil, implied that plant roots recruited specific microorganisms including nitrogen-fixing bacteria, biocontrol microorganisms and protozoa, plant-promoting rhizobacteria (PGPR) from the bulk soil to the rhizosphere (Hein et al., 2008; Mendes et al., 2013). The interactions between these rhizosphere microorganisms that were beneficial to plant growth can impact their effects. In addition, the rhizosphere was the area where roots and their exudates affected various biological and ecological processes by interacting with microorganisms (Fang et al., 2013). Microbial interactions, complexity and diversity were of considerable importance to the formation and homeostasis of microbial communities (Cordovez et al., 2019). Also, microbial composition can be influenced by plant genotype, developmental stage (Bulgarelli et al., 2012; Zachow et al., 2014), fertilization management (Saha et al., 2008) and soil type (Schlaeppi et al., 2014).
Infestation by pathogens or RKN can disrupt the original rhizosphere microbial community structure of the crop and alter microbial diversity. A previous study of the Arabidopsis microbiome showed that plants can specifically recruit a set of beneficial microbes that induce resistance and promote growth in response to pathogen infection (Berendsen et al., 2018m). Besides, the Flavobacterium dominated the rhizosphere of resistant tomatoes after Ralstonia solanacearum infestation and tested that Flavobacterium could antagonise Ralstonia solanacearum (Kwak et al., 2018). The bacterial wilt outbreaks modified microbial composition and diversity as well as reduce the abundance of beneficial microorganisms in the soil (Wang et al., 2017). Fusarium wilt-diseased banana had a higher abundance and diversity of fungi or bacteria than disease-free soils, showing a change in the dominant phylum (Zhou et al., 2019). A study showed that tobacco composition of the root microbial community was significantly associated with RKN infection (Cao et al., 2022). In contrast, variations in the rhizosphere microbial community resulting from RKN infestation of Cucumis crops remain unclear.
Previous studies showed that C. metuliferus had the highest number of disease-resistance-associated NBS-LRR genes in Cucurbitaceae by assembling and analyzing the chromosomal level genome of C. metuliferus (Ling et al., 2021), and that root volatiles of C. metuliferus had potential applications against RKN (Xie et al., 2022), which implied the resistance to RKN in Cucumis crops was a complex and integrated mechanism. Therefore, this study compared and analyzed the response of rhizosphere bacterial community to RKN infestation in resistant C. metuliferus and susceptible C. sativus.
C. sativus inbred line 9930 (cuc) and C.metuliferus inbred line CM3 (cm3) were provided by Institute of Vegetable and Flowers, Chinese Academy of Agricultural Sciences (IVF-CAAS; Beijing, China). The M. incognita used in this study was obtained from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (IVF-CAAS; Beijing, China). The soil with a multi-year history of nematodes infestation was sampled from a greenhouse in Langfang city (Hebei, China). Then, the plants were maintained in a sterile mixture of soil: vermiculite: perlite (2:2:1, vol/vol/vol) in a glass room of a phytotron in April 2021 with day/night temperatures 28/18°C.
M. incognita was inoculated on the roots of the hollow cabbage. Approximately 45 days after inoculation, the M. incognita egg masses were picked with forceps and disinfected with 0.5% NaClO for 15 seconds. The egg masses were rinsed several times with sterile distilled water so that no NaClO remained on the surface, then placed in a sterile Petri dish with 20 mL of sterile water and incubated in a light-proof incubator at 28°C. The second-stage M. incognita juveniles (J2s) were counted using a stereomicroscope for bioassays.
The cuc and cm3 seeds were soaked in 0.5% NaClO for 2-3 minutes and then rinsed several times with sterile distilled water until no NaClO remained. The surface sterilised seeds were laid flat in a petri dish with sterile filter paper, soaked with a small amount of sterile distilled water (not over the seeds) and incubated in the dark at 28°C in an incubator. The seedling substrate and vermiculite were sterilised twice in an autoclave (121°C, 60 min) and the germinated seeds were sown into the sterilised substrate, which consisted of grass charcoal soil and vermiculite in a 2:1 volume ratio. Then the seedling cavity trays were grown in a greenhouse to obtain soil-grown seedlings for subsequent experiments.
The experiment consisted of five experimental groups, cm3, cuc, cm3 inoculated with nematodes (cm3J), cuc inoculated with nematodes (cucJ) and bulk soil. One sample (rhizosphere soil mix of five randomly selected plants) from each treatment at a time was taken and five replicates were set up. Each plant was inoculated with 500 M. incognita in cm3J and cucJ groups. Approximately 30 days (T1) and 60 days (T2) after inoculation, each plant was uprooted to prepare for rhizosphere samples. The roots without large pieces of soil were placed in a 250 ml sterilized conical flasks with phosphate buffer saline (pbs) buffer submerged. The conical flasks were placed on a shaker (20 min, 25°C, 160 r) to remove the soil still attached to the roots. Afterwards, the conical flasks were left to stand in a refrigerator at 4°C for 24 h. Finally, the lower soil solution was collected as the rhizosphere sample. In total, 40 rhizosphere and 10 bulk soil samples were collected.
Total bacterial genomic DNA from 0.5 g of soil for each sample was extracted using a Fast DNA SPIN extraction kit (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions. The DNA was stored at -20°C until further analysis. The quantity and quality of extracted DNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. The V3-V4 region of bacterial 16S rRNA genes was amplified by PCR using the forward primer 515F (5’-GTGCCAGCMGCCGCGGGTAA-3’) and the reverse primer 907R (5’-CCGTCAATTCMTTTRAGT1T-3’). The primers were incorporated into sample specific 7 bp barcodes for multiplex sequencing. After individual quantification, equal amounts of amplicons were pooled and pair-end 2x300 bp sequencing was performed on the Illumina MiSeq platform using the MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China). The 16S sequence datasets for this study can be found in the NGDC with accession number: PRJCA014808 (https://ngdc.cncb.ac.cn).
A total of 5,262,484 raw reads were obtained from 50 samples, with sequence lengths greater than 400 bp. After splicing the raw reads, double-ended primers were excised, and low-quality reads with a Q scores less than 30 were filtered. Operational taxonomic units (OTU) were clustered using usearch (v10.0) with a 97% similarity cutoff, and chimeric sequences were identified and removed using vsearch (v2.8) based on gold.fa (http://drive5.com/uchime/rdp_gold.fa). Each bacterial 16S rRNA gene was annotated in the greengenes database (gg_16s_13.5) using vsearch (v2.8). The OTU without annotation and annotations for Chloroplast, Mitochondria, Archaea were deleted. Bacterial reads were classified into 4531 OTU after quality filtering. Then, the OTU with relative abundance less than 0.001% were filtered and flattened according to sample_size 52353. We calculated bacterial alpha diversity using the Shannon indices and observed_otus by vsearch (v2.8), and bacterial beta diversity using principal co-ordinates analysis (PCoA) based on the Bray–Curtis dissimilarity matrix by edgeR and vegan packages. Data on the relative abundance differences between groups of bacteria at the phylum level were processed with the dplyr and reshape2 packages. The unique and shared OTU of different groups were plotted by the ggVennDiagram package (Gao et al., 2021). The volcano plot drawn with the edgeR and ImageGP packages showed OTU enrichment differences between groups. Bacterial co-occurrence networks of different groups at different stages visualised correlations with |RHO| > 0.9 and P < 0.05 using the ggCLusterNet package (Wen et al., 2022). Stamp, linear discriminant analysis effect size (LEfSe) analysis and circos plot were performed using the OmicStudio tools at https://www.omicstudio.cn/tool.
Samples from each group were mixed and enriched for bacteria by the following steps. 1) 100 ml of soil solution was added to 150 ml of pre-cooled distilled water, then slowly filtered through a double layer of gauze after mixing well. 2) The supernatant obtained by centrifugation of the filtered solution (700 r, 4°C, 5 min) was collected. 3) The precipitate was resuspended and centrifuged twice more, and the supernatants of the three times were combined. 4). After centrifugation (1000 r, 4°C, 10 min) of all the supernatants, the precipitate was resuspended with 15 ml of 0.8% NaCl. 5) The resuspension was centrifuged (11000 r, 4°C, 30 min) after the slow addition of 10 mL of Nycodenz (0.8 g/mL). 6) After centrifugation, it was divided into Nycodenz-soil mixed particles (lower layer), water (upper layer), and bacteria (middle layer). 7) The bacterial layer, resuspended by the addition of 10 mL of sterilised distilled water, was centrifuged (10,000 r, 4°C, 20 min) to obtain the Nycodenz-free bacteria.
The enriched bacteria were cultured on LB and R2A medium plates, and 20 single colonies of different sizes and morphologies were picked from each plate. Then 600 single colonies were obtained in 3 replicates. The nematicidal activity of the strains was tested in a 24-well plate. The 900 μL of bacterial broth was mixed with 100 μL of nematodes suspension (approximately 100 M. incognita) in individual wells. Additionally, 100 μL of nematodes suspension (approximately 100 M. incognita) mixed with 900 μL of sterilised distilled water and culture medium respectively were added to other wells to serve as untreated controls. The 24-well plates were incubated at 28°C for 24 h. Nematodes were considered dead if the bodies of nematodes were straight and did not move when stimulated with 0.5 M NaOH (Harada and Yoshiga, 2015). The corrected mortality rate was reference Yin (Yin et al., 2021). Bacteria with the corrected mortality rate greater than 85% compared to the corresponding medium were considered as antagonistic bacteria. Genomic DNA of strains were extracted using the TIANamp Bacteria DNA Kit (Tiangen, Beijing China) and the 16S ribosomal DNA was amplified by PCR using 2x Rapid Taq Master Mix (Vazyme, Nan- jing China) and primers 27F and 1492R (Alberoni et al., 2019). The following conditions were used: denaturation at 95°C for 3 min; followed by 34 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and extension at 72°C for 5 min. The product was stored at 4°C until sequencing. The sequence results were analyzed using the NCBI BLAST tool (https://www.ncbi.nlm.nih.gov/).
At the operational taxonomic unit (OTU) level, alpha diversity of microorganisms in the rhizosphere of plants was measured using observed OTU and Shannon index. The Shannon index and the observed OTU index showed that the cucJ group was significantly higher than the cuc group and the bulk soil was significantly higher than the cuc and cm3 groups at T1 (Wilcoxon rank sum test P < 0.05; Figures 1A, B). However, the Shannon index was significantly higher in the bulk group than in the cm3 group (Wilcoxon rank sum test P < 0.05), but the observed OTU index was not significantly different at T2 (Wilcoxon rank sum test P > 0.05; Figures 1C, D).
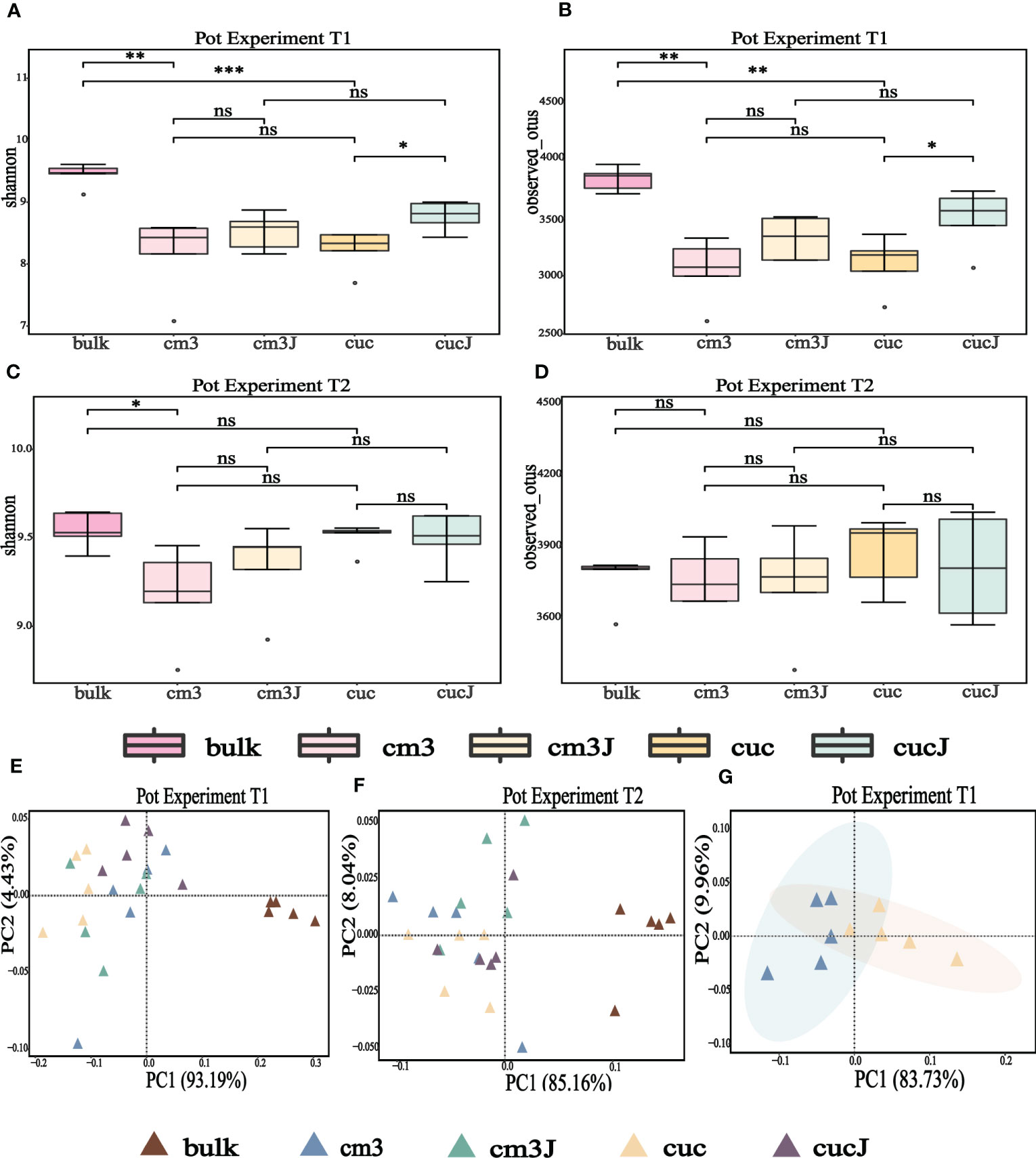
Figure 1 Alpha diversity (shannon indices and observed OTU) of bacteria in the rhizosphere at T1 (A, B) and T2 (C, D). Differences between the cm3, cuc, cm3J, cucJ, and bulk soil were indicated in each figure panel (ns p> 0.05, *p < 0.05, **p < 0.01, and ***p < 0.001). Principal co-ordinates analysis (PCoA) analysis of bacteria at T1 (E, G) and T2 (F) in the rhizosphere of different group on Bray–Curtis distance metrics.
To clarify the differences in the bacterial communities of the different groups, the beta diversity of bacteria was visualized by PCoA based on the Bray-Curtis distance metric. The extremely significant differences in bacterial community composition between bulk soil and plant rhizosphere (PERMANOVA test; P = 0.000050 for T1, P = 0.00010 for T2; Figures 1E, F) suggested that plants influence bacterial communities. Also, PCoA analysis revealed a striking change in the composition of the bacterial communities of the cuc rhizosphere after infestation by RKN (PERMANOVA test; P = 0.0081; Figure 1G).
After RKN infestation, rhizosphere bacterial OTU species varied little, with 93% and 97% of the same OTU for cm3J and cm3 (Figures 2A, B), respectively, and 94% and 98% of the same OTU for cucJ and cuc (Figures 2C, D), respectively, in both periods. In order to gain insight into the taxonomic composition of rhizosphere bacterial community of different treatments, differences in the taxonomic composition of rhizosphere bacteria were compared at the phylum level (Figures 2E, F). The bacterial community at T1 was mainly composed of Proteobacteria, Actinobacteria, Firmicutes, Chloroflexi, Gemmatimonadetes, Bacteroidetes and Acidobacteria. At T1, the abundance of Acidobacteria and Nitrospirae increased in cm3J compared with cm3. Compared with the cuc group, the relative abundance of Actinobacteria, Acidobacteria, TM7, Nitrospirae and Cyanobacteria increased in the cucJ group. The dominant bacterial community at T2 was the same as in T1 period, but no differences in composition ratios were observed. Only the relative abundance of Cyanobacteria increased in cucJ than in cuc. Obviously, the taxonomic ratios of the bacterial community between bulk soil and plant rhizosphere were quite different in each period.

Figure 2 The cm3 and cm3J share the number of OTU species at T1 (A) and T2 (B) and cuc and cucJ share the number of OTU species at T1 (C) and T2 (D). The relative abundance of major bacterial at T1 (phylum level; (E) and T2 (phylum level; (F) taxa present in the rhizosphere of different groups. Circos plot showing the taxonomical relative abundance of the top 10 bacterial microbiome at the class level at T1 (G) and T2 (H). The thickness of each ribbon represents the relative abundance of bacterial assigned to different groups.
Next, the differences in the content of the top 10 bacteria in relative abundance at the class level were compared across the groups (Figures 2G, H). At T1, Gammaproteobacteria abundance appeared to decrease, but Actinobacteria and Bacilli increased to varying degrees in cucJ compared to cuc. Conversely, cm3J was more abundant in Betaproteobacteria but less abundant in Actinobacteria and Bacilli compared to cm3. At T2, Gammaproteobacteria and Betaproteobacteria decreased and Deltaproteobacteria increased in cucJ. Otherwise, Actinobacteria increased and Gammaproteobacteria, Chloroflexi, Betaproteobacteria and Bacilli decreased in cm3J.
The difference in the rhizosphere enrichment of bacterial OTU in cm3J (Figure 3A; Supplementary Figure S1A) and cucJ (Figure 3B; Supplementary Figure S1B) compared with cuc, respectively. The cm3J enriched a quantity of OTU relative to cuc in both periods, while cucJ only enriched a small amount of OTU in T1 period. Next, we compared the top 20 bacteria with differences in abundance in cm3J and cucJ with cuc, respectively. Specifically, the abundance of Armatimonadia, TK10, Dehalococcoidetes, Acidobacteria−5, AT−s54, PRR−12, DA052 and Chloracidobacteria was significantly higher in cm3J than in cuc (t.test, P < 0.05; Figure 3C) at T1. Only the abundance of Actinobacteria was significantly higher in cucJ than in cuc (t.test, P < 0.05; Figure 3D). Cm3J recruited more Chloroflexi and Alphaproteobacteria, while cucJ recruited more Thermomicrobia (Supplementary Figure S1C, D) at T2.
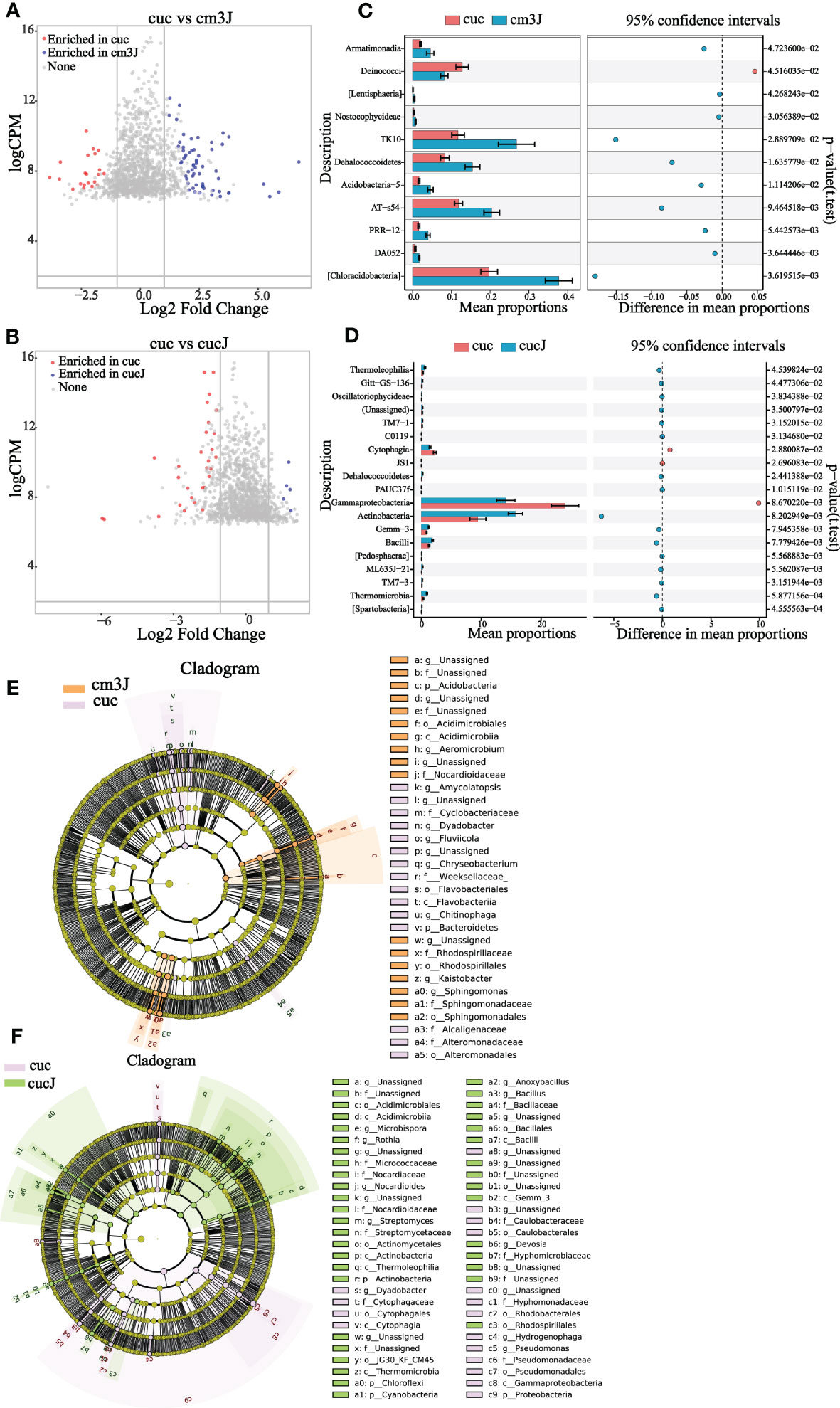
Figure 3 Cm3J (A) and cucJ (B) enriched OTU species compared to cuc at T1. STAMP analysis demonstrates differential enrichment of bacteria (class level) in the cm3J (C) and cucJ (D) at T1. Cladogram showing the bacteria phylogenetic structure of cm3J (E) and cucJ (F) with cuc respectively at T1.
Compositional differences were assessed by calculating the linear discriminant analysis effect size (LEfSe) scores at different levels to find out the changes in the relative abundance of bacteria in resistant and susceptible plants after infection with RKN, respectively (Supplementary Figure S2). Under the condition of LDA threshold ≥ 3.0, cm3J was enriched Acidobacteria, Nocardioidaceae, Rhodospirillales, Sphingomonadales, Acidimicrobiia compared with cuc (Figure 3E) at T1. And cucJ was enriched with a considerable amount of bacteria compared to cuc including Bacilli, Cyanobacteria, Chloroflexi, Actinobacteria, Rhodospirillales, Hyphomicrobiaceae and Gemm_3 (Figure 3F). Thus, it can be found that cucJ enriched more bacterial after inoculation with RKN. Cm3J was mainly enriched with Chloroflexi, Actinobacteria, Stenotrophomonas and Alphaproteobacteria (Supplementary Figure S1E) at T2. Furthermore, there was only little difference between cucJ and cuc compared to rhizosphere bacteria. CucJ was mainly enriched with Thermomicrobia and Microbacteriaceae (Supplementary Figure S1F).
In exploring the interactions of rhizosphere bacterial communities of plants with different treatments at different growth stages, co-occurrence networks were constructed to demonstrate the differences in rhizosphere bacterial interactions. Firstly, it was straightforward to see that the bacterial network of the bulk soil (Figure 4E) was obviously different from the other groups. The taxonomic composition of the network showed no apparent differences among the cm3 (Figure 4A), cuc (Figure 4B), cm3J (Figure 4C) and cucJ (Figure 4D) groups, with most nodes belonging to Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Gemmatimonadetes, Chloroflexi, and Acidobacteria, respectively. The network differences among the groups at T1 were greater than those at T2, so specific analyses were performed for the bacterial networks of the groups at T1.
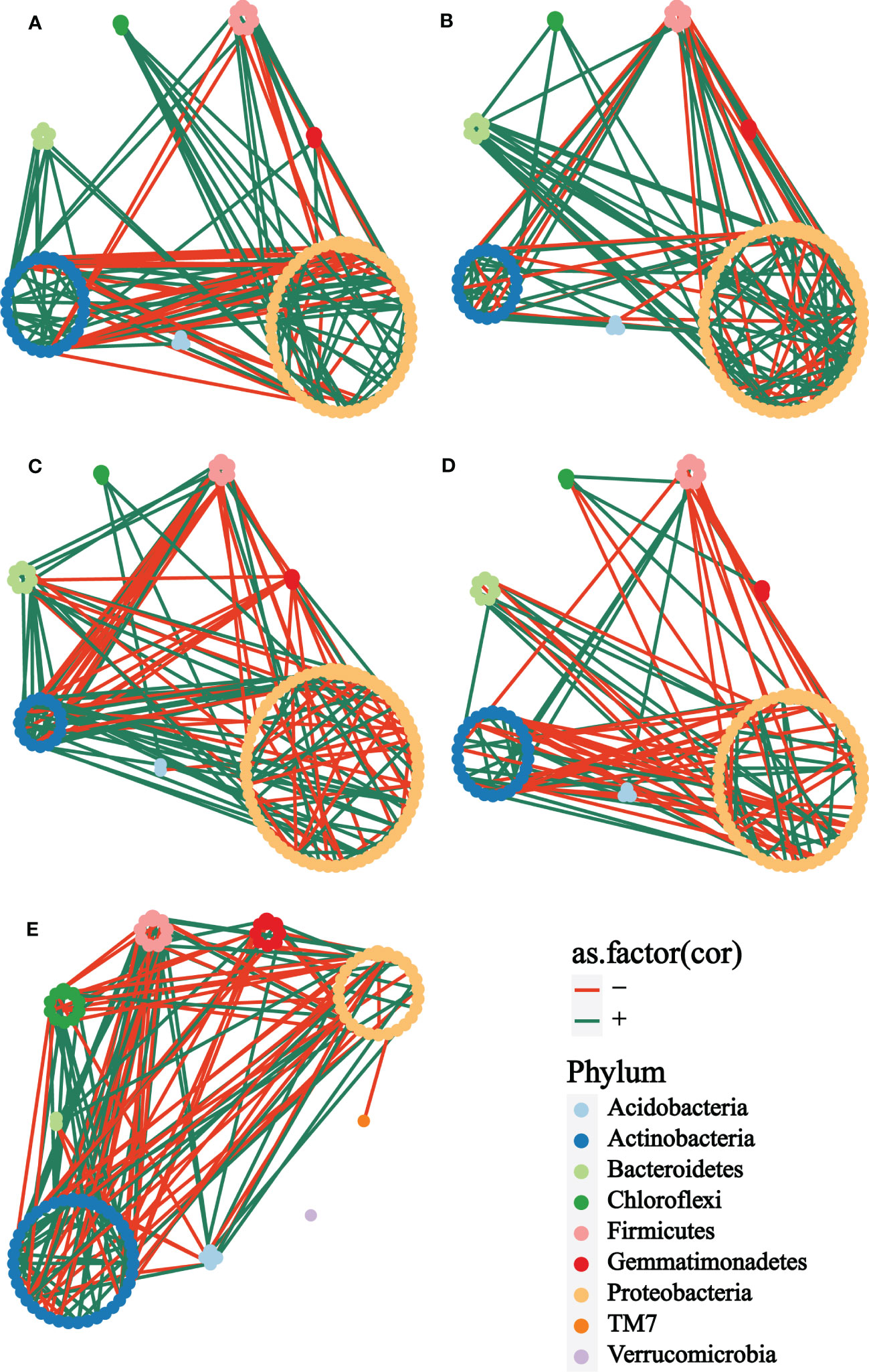
Figure 4 Co-occurrence network analysis of the rhizosphere microbial communities of different groups at T1. (A) cm3, (B) cuc, (C) cm3J, (D) cucJ, (E) bulk. The networks were colored based on the taxonomy taxa of bacteria at the phylum level. Edge s indicated correlations, which were divided into positive (green) or negative (red) correlations.
From cm3J to cucJ, the network complexity gradually decreased. There were 148 edges in the cm3J network, while 115 edges in the cucJ. The average degree and relative.modularity of cm3J were 3.6098 and 1.3185, edge density and diameter were 0.0446 and 1.9494 respectively, while the average.degree and relative.modularity of cucJ were 2.5556 and 0.6200, edge density and diameter were 0.0287 and 2.8704, respectively. More importantly, the positive correlation accounted for 74.32% and the negative correlation accounted for 25.68% in cm3J, while the positive and negative correlations in cucJ were 60.00% and 40.00%, respectively (Supplementary Table S1). Hub nodes were nodes that were more connected with other nodes in the network, and hub microorganisms were those that were closely connected with other microorganisms. The top five OTU with hub_score in the cm3J belonged to Firmicutes, Actinobacteria and Proteobacteria, respectively. Similarly, they belonged to Actinobacteria and Firmicutes respectively in the cucJ (Supplementary Figure S3).
105 antagonistic bacteria were yielded screening of bacteria resistant to M. incognita using R2A and LB medium and classified in 10 genus (Figure 5A), of which 44.76% Pseudomonas, 15.24% Priestia, 16.19% Stenotrophomonas and 10.48% Glutamicibacter (Supplementary Table S2). More antagonistic bacteria were obtained in the cm3J than in the cucJ at T1, and both were less abundant than in the bulk soil. However, the cm3J obtained fewer species of antagonistic bacteria than the cucJ and the bulk soil (genus level). Similar screening results to T1 (Figure 5B) were showed at T2 (Figure 5C). Others, more antagonistic bacteria were obtained in cm3J and cucJ at T1 than T2. Notably, the antagonistic bacteria in the cm3J group were Pseudomonas, Glutamicibacter, and Priestia. The Arthrobacter, Bacillus, Glutamicibacter, Priestia, Pseudomonas and Enterobacter were identified in the cucJ group.
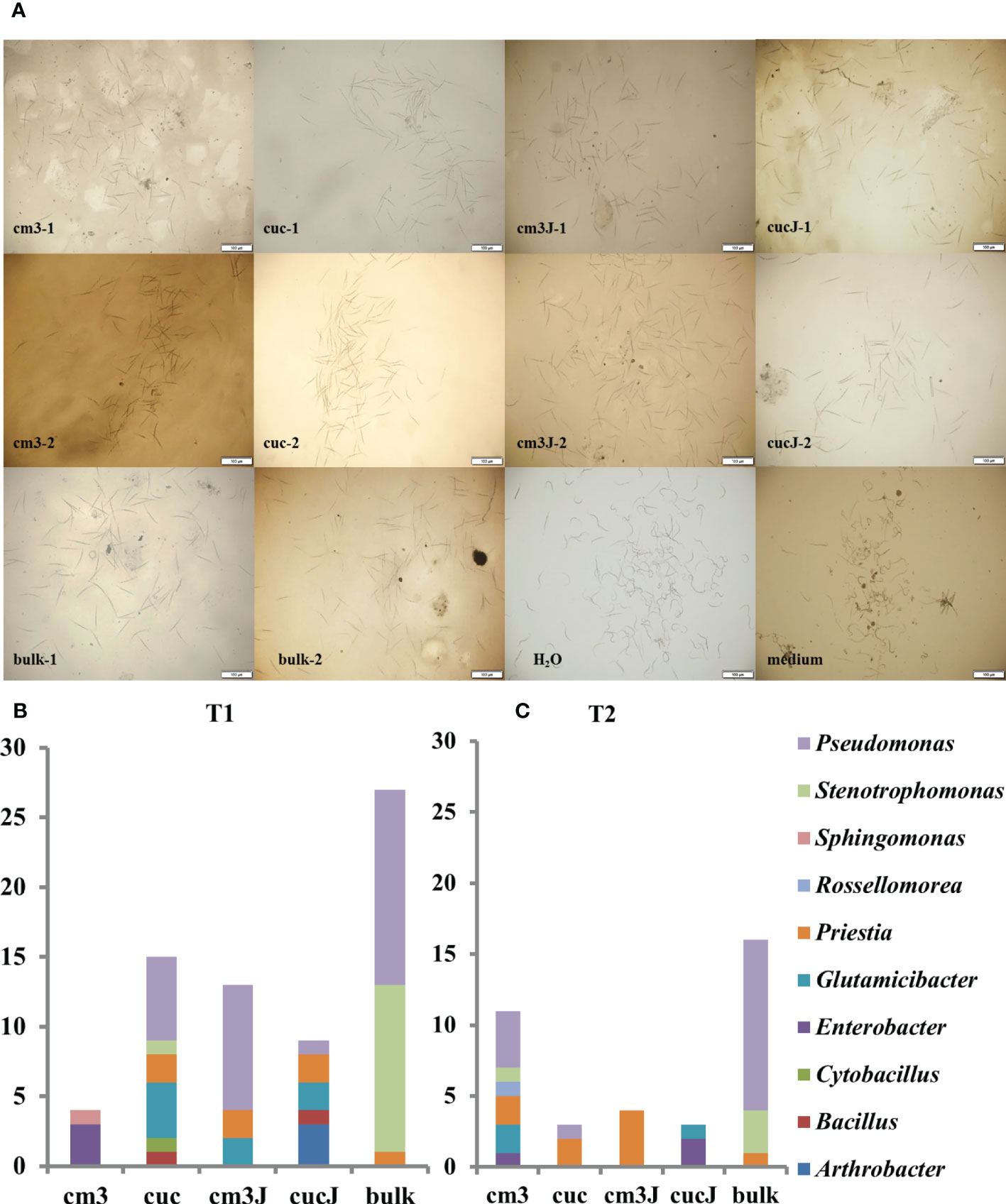
Figure 5 Effectiveness of antagonistic bacteria in killing nematodes (A). Statistics of antagonistic bacteria genus in different groups at T1 (B) and T2 (C). -1: T1; -2: T2.
Changes in soil microbial communities were the most important biological factors influencing the occurrence of soil diseases (Kloeppe et al., 1999; Larkin, 2008). In previous studies, the occurrence of RKN disease was found to be closely related to the interactions between soil microbial communities (Echeverrigaray et al., 2010). Plants infected with RKN responded with resistance by releasing many compounds into the rhizosphere (Čepulytė et al., 2018; Oota et al., 2020; Rutter et al., 2022). Nutrients and metabolites were released through root cells fed by nematodes through the symplast, thereby altering the composition of root exudates (Tian et al., 2015), which in turn may affect the microbial community. Similarly in this study, RKN invaded the plant roots and the rhizosphere bacterial species diversity and community composition changed. Moreover, changes in the rhizosphere bacterial community were more pronounced in the early stages of plant-root knot nematodes interaction, which was consistent with the findings of other studies (Poll et al., 2007). In the early stages of RKN infestation, the species diversity of rhizosphere bacteria increased in cucJ and cm3J, and the composition of the cucJ rhizosphere bacterial community was apparently altered. In addition, there were obvious differences in the rhizosphere bacterial co-occurrence network. These changes may be the result of a combination of attacks by RKN on rhizosphere microbial structures and the plant response in the early stages.
In the study of rhizosphere bacterial community structure, it was found that the rhizosphere network was substantially more complex than the bulk soil, indicating that rhizosphere had greater potential for interaction and ecological niche sharing (Shi et al., 2016). Although the species diversity and community composition of both cm3J and cucJ rhizosphere bacterial communities changed in the face of RKN invasion, the cm3J community structure was more stable with only small changes and no obvious changes in species diversity and community composition, as well as fewer bacterial enrichment species. The homeostasis of soil microorganisms as a dynamic component of the plant-soil system was extremely important for the control of soil-borne diseases. Cooperative and competitive interactions between microbial species and the modularity of the network influence the stability of the community (Faust and Raes, 2012; Coyte et al., 2015). Studies on the rhizosphere microbial community showed that the bacterial community structure of resistant genotypes was more stable than that of sensitive genotypes (Fu et al., 2019; Dong et al., 2021). Likewise, the bacterial community structure of cm3J was more stable than that of cucJ in this study.
The edges connecting two nodes representing different units in a microbial network indicate close associations between the abundance of these units in the whole sample, often interpreted as biological interactions. A great number of microbial interactions have also been confirmed experimentally in the Arabidopsis root community (Durán et al., 2018). In this study, the interactions and positive correlations between rhizosphere bacteria in cm3J were stronger, forming a more complex co-occurrence network. This indicated that the ecological interactions of cm3J rhizosphere dominant bacteria were more positive (Zhang et al., 2018; Lian et al., 2019). Elsewhere, the network complexity (Wagg et al., 2019) and hub taxa in supporting ecosystem function were important (Toju et al., 2018). In contrast, the rhizosphere bacterial community of cucJ was more susceptible to disturbance. When infested by RKN, the cuc rhizosphere bacterial network transmitted the environmental disturbance to the whole network with a short time, in turn destabilizing the network structure. Network inference can provide insights into microbial community composition, but theoretical studies of the effects of some network properties on ecosystem stability still require experimental evidence (Faust, 2021).
After that, we found a close positive correlation between Bacteroidetes and Proteobacteria in the cm3J network. Among the soil microbiota, the Bacteroidetes tended to be a dominant phylum due to their ability to secrete a variety of carbohydrate-active enzymes (CAZymes) that targeted highly variable glycans in the soil (Larsbrink and McKee, 2020). Bacteroidetes were abundant pathogen suppressor members of the plant microbiome and contributed to rhizosphere phosphorus mobilization (Lidbury et al., 2021). Proteobacteria were also abundant, as typically observed in soil libraries (Janssen, 2006). And multiple positive interactions of Pseudomonadales and Rhizobiales with other bacteria, such as Sphingomonadales. The probiotic Pseudomonas (Gammaproteobacteria, Pseudomonadales) was versatile in terms of plant hosts, soil habitat and improving plant stress response (Kim and Anderson, 2018). The most known effects of Pseudomonas were the protection of plants from fungal diseases and the improvement of plant yield, as well as the recent discovery of interesting aspects regarding insecticidal activity (Ruffner et al., 2013). Rhizobia were a group of soil-borne bacteria that had the ability to fix atmospheric nitrogen for plant growth and promoted root growth (Masson-Boivin et al., 2009; Poupin et al., 2016; Garrido-Oter et al., 2018). Sphingomonas was the main group of rhizosphere and endophytic bacteria with multifaceted functions ranging from remediation of environmental pollution to production of highly beneficial phytohormones involved in rhizosphere remediation of organic matter (Zhang et al., 2013; Feng et al., 2019; Asaf et al., 2020). Additionally, Sphingomonadaceae, Brucellaceae, and Bartonellaceae were also closely associated with other rhizosphere microorganisms. Brucellaceae and Bartonellaceae were associated with nematodes carriage (Bowman, 2011). In summary, the more positive correlation between probiotics and the close interaction with RKN-associated bacteria may be a reason for the resistance of cm3 to RKN infestation.
There was little change in bacterial species when RKN interacted with plant rhizosphere microorganisms; what changed was the abundance of some bacteria. In the early stage, the change in the abundance of bacteria in cucJ was an increase in Actinobacteria and Bacilli, a decrease in Bacteroidetes, Gammaproteobacteria and Betaproteobacteria. Actinobacteria played a role in soil nitrogen fixation, improving nutrient availability, and promoting the production of plant growth regulators (Bhatti et al., 2017). The Bacilli offered a number of advantages for their application in agricultural biotechnology, and some products based on Bacilli, especially Bacillus, have been marketed as microbial pesticides, fungicides or fertilizers (Perez-Garcia et al., 2011). Conversely, Actinobacteria and Bacilli decreased and Betaproteobacteria increased in cm3J. Cm3J was mainly enriched in Gammaproteobacteria and Betaproteobacteria. Differently, cucJ mainly recruited Actinobacteria, Saprospirae, Bacilli and Thermomicrobia.
In addition, Thiobacterales, iii1_15 and Kaistobacter may play a role in cm3J. In cucJ, Thermoleophilia, Streptomyces, Nocardioidaceae, Nocardioides, Micrococcaceae, Acidimicrobiales, Devosia, Gemm_3, Bacillaceae, Cyanobacteria and G30_KF_CM45 may play important roles. Streptomyces was the most abundant and important genus of actinomycetes. And Streptomyces had a beneficial symbiotic relationship with plants, promoting the nutrition and health of the latter (Pang et al., 2022). Nocardioidaceae can degrade a wide range of organic compounds, including aromatic and polyaromatic pollutants and toxic chemicals (Tóth and Borsodi, 2014). Plant-cyanobacterial interactions, as a beneficial symbiotic relationship, have long been demonstrated in rice growing areas. In addition, cyanobacteria may produce or secrete large amounts of biologically active compounds that have the ability to promote plant growth or may make plants more resistant to abiotic or biotic stresses (Bahareh et al., 2021). Most of the antagonistic bacteria screened were Pseudomonas, along with Priestia, Stenotrophomonas, and Glutamicibacter. A large number of Pseudomonas were screened in cm3J. Stenotrophomonas produced similar antibiotics and shared some enzymatic activities, which may make them attractive candidates for biological control of plant diseases and nematodes (Hayward et al., 2010).
We observed that RKN invasion caused recruitment and alteration of probiotic bacteria in both cm3J and cucJ. Cm3J was mainly enriched in the Proteobacteria and cuc was mainly enriched in the Actinobacteria and Bacilli. Moreover, the most abundant bacteria screened for antagonistic bacteria was Pseudomonas (Proteobacteria, Pseudomonadaceae). It was possible that Pseudomonas and other bacteria of the Proteobacteria played a part in RKN infestation of cm3J.
Root exudates of two crops may cause rhizosphere microbial differences. Root exudates include sugars, amino acids, organic acids, fatty acids, and secondary metabolites, which are important ways for plants to communicate with microorganisms and have a major influence on the composition of the rhizosphere microbiome (Bulgarelli et al., 2013; Sasse et al., 2018). Moreover, root exudates recruit microorganisms from the soil to the rhizosphere, where primary metabolites are mainly responsible for attraction and secondary metabolites are mainly responsible for screening the recruited microorganisms (Zhalnina et al., 2018). Furthermore, root exudates were strongly affected by plant species, developmental stage, root physiology, environment, soil type and stress type (Sasse et al., 2018). Beyond that, the assembly of microbial communities in plant roots also depends on microbial interactions (Bai et al., 2022). A study constructed a highly simplified maize SynCom, and single strain exclusion experiments on SynCom showed that no bacterial strains except Enterobacter cloacae caused SynCom collapse, indicating that Enterobacter cloacae was a key member in the community assembly process (Niu et al., 2017). In the case of cm3J and cucJ after infection with RKN in this study, it was possible that different root exudates and microbial interactions contributed to the differences in rhizosphere microbial communities.
In general, the interaction between RKN and rhizosphere bacteria was stronger in the early growth period, which provides a reference for the period of biocontrol. Secondly, a stable and well-connected rhizosphere bacterial community was positive for suppressing RKN infestation. Finally, Pseudomonas and other bacteria in the Proteobacteria in Cucumis crops showed clear changes in response to RKN invasion, which pointed the way for further research on biocontrol agents.
The 16S sequence datasets for this study can be found in the NGDC with accession number: PRJCA014808 (https://ngdc.cncb.ac.cn).
BX and JL designed the experiment. ZM, YL, YY and JZ directed the experiment and the writing. XP and LS performed the experiment and processed the data. LS analysed the data and wrote the manuscript. JL revised the manuscript. All authors contributed to the article and approved the submitted version.
This research is funded by Inner Mongolia Science and Technology Plan Project(2020GG0110), Central Public-interest Scientific Institution Basal Research Fund (No.IVF-BRF2022013), Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China (CARS-25), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2017-IVF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1163271/full#supplementary-material
Aiello, D., Restuccia, C., Stefani, E., Vitale, A., Cirvilleri, G. (2019). Postharvest biocontrol ability of Pseudomonas synxantha against monilinia fructicola and monilinia fructigena on stone fruit. Postharvest Biol. Technol. 149, 83–89. doi: 10.1016/j.postharvbio.2018.11.020
Alberoni, D., Gaggìa, F., Baffoni, L., Modesto, M. M., Biavati, B., Di Gioia, D. (2019). Bifidobacterium xylocopae sp. nov. and bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract. Syst. Appl. Microbiol. 42 (2), 205–216. doi: 10.1016/j.syapm.2018.11.005
Asaf, S., Numan, M., Khan, A. L., Al-Harrasi, A. (2020). Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 40 (2), 138–152. doi: 10.1080/07388551.2019.1709793
Bahareh, N., Bouaicha, N., Metcalf, J. S., Porzani, S. J., Konur, O. (2021). Plant-cyanobacteria interactions: beneficial and harmful effects of cyanobacterial bioactive compounds on soil-plant systems and subsequent risk to animal and human health. Phytochemistry 192, 112959. doi: 10.1016/j.phytochem.2021.112959
Bai, B., Liu, W., Qiu, X., Zhang, J., Zhang, J., Bai, Y. (2022). The root microbiome: community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 64 (2), 230–243. doi: 10.1111/jipb.13226
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., de Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. Isme J. 12 (6), 1496–1507. doi: 10.1038/s41396-018-0093-1
Bhatti, A. A., Haq, S., Bhat, R. A. (2017). Actinomycetes benefaction role in soil and plant health. Microbial Pathogenesis 111, 458–467. doi: 10.1016/j.micpath.2017.09.036
Bongers, T., Bongers, M. (1998). Functional diversity of nematodes. Appl. Soil Ecol. 10 (3), 239–251. doi: 10.1016/S0929-1393(98)00123-1
Bowman, D. D. (2011). Introduction to the alpha-proteobacteria: wolbachia and bartonella, rickettsia, brucella, ehrlichia, and anaplasma. Top. Companion Anim. Med. 26 (4), 173–177. doi: 10.1053/j.tcam.2011.09.002
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for arabidopsis root-inhabiting bacterial microbiota. Nature 488 (7409), 91–95. doi: 10.1038/nature11336
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren van Themaat, E., Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Cao, Y., Yang, Z. X., Yang, D. M., Lu, N., Yu, S. Z., Meng, J. Y., et al. (2022). Tobacco root microbial community composition significantly associated with root-knot nematode infections: dynamic changes in microbiota and growth stage. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.807057
Čepulytė, R., Danquah, W. B., Bruening, G., Williamson, V. M. (2018). Potent attractant for root-knot nematodes in exudates from seedling root tips of two host species. Sci. Rep. 8 (1), 10847. doi: 10.1038/s41598-018-29165-4
Chalivendra, S. (2021). Microbial toxins in insect and nematode pest biocontrol. Int. J. Mol. Sci. 22 (14), 7657. doi: 10.3390/ijms22147657
Chen, J. R., Ou, S. L., Nieh, T. I., Lu, C. Y., Ku, H. M. (2020). Molecular dissection of Cucumis metuliferus resistance against Papaya Ringspot Virus by grafting. Plants (Basel) 9 (12), 1666. doi: 10.3390/plants9121666
Cordovez, V., Dini-Andreote, F., Carrión, V. J., Raaijmakers, J. M. (2019). Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 73 (1), 69–88. doi: 10.1146/annurev-micro-090817-062524
Coyte, K. Z., Schluter, J., Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350 (6261), 663–666. doi: 10.1126/science.aad2602
Dong, Z., Guo, Y., Yu, C., Zhixian, Z., Rongli, M., Deng, W., et al. (2021). The dynamics in rhizosphere microbial communities under bacterial wilt resistance by mulberry genotypes. Arch. Microbiol. 203 (3), 1107–1121. doi: 10.1007/s00203-020-02098-1
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote arabidopsis survival. Cell 175 (4), 973–983.e914. doi: 10.1016/j.cell.2018.10.020
Echeverrigaray, S., Zacaria, J., Beltrão, R. (2010). Nematicidal activity of monoterpenoids against the root-knot nematode meloidogyne incognita. Phytopathology 100 (2), 199–203. doi: 10.1094/phyto-100-2-0199
Expósito, A., García, S., Giné, A., Escudero, N., Sorribas, F. J. (2019). Cucumis metuliferus reduces Meloidogyne incognita virulence against the Mi 1.2 resistance gene in a tomato-melon rotation sequence. Pest Manag. Sci. 75 (7), 1902–1910. doi: 10.1002/ps.5297
Fang, S., Liu, D., Tian, Y., Deng, S., Shang, X. (2013). Tree species composition influences enzyme activities and microbial biomass in the rhizosphere: a rhizobox approach. PloS One 8 (4), e61461. doi: 10.1371/journal.pone.0061461
Faust, K., Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10 (8), 538–550. doi: 10.1038/nrmicro2832
Faust, K. (2021). Open challenges for microbial network construction and analysis. Isme J. 15 (11), 3111–3118. doi: 10.1038/s41396-021-01027-4
Feng, Y., Yuan, Q., Yang, Z., Zhang, T., Liu, X., Li, C., et al. (2019). Effect of sphingomonas sp. strain on degradation of polyphenols in redried tobacco leaves. Acta Tabacaria Sin., 25, 19–24. doi: 10.16472/j.chinatobacco.2018.038
Fu, L., Ou, Y., Shen, Z., Wang, B., Li, R., Shen, Q. (2019). Stable microbial community and specific beneficial taxa associated. J. Microbiol. Biotechnol. 29 (10), 1624–1628. doi: 10.4014/jmb.1904.04061
Gao, C. H., Yu, G., Cai, P. (2021). ggVennDiagram: an intuitive, easy-to-Use, and highly customizable r package to generate Venn diagram. Front. Genet. 12. doi: 10.3389/fgene.2021.706907
Garrido-Oter, R., Nakano, R. T., Dombrowski, N., Ma, K. W., AgBiome, T., McHardy, A. C., et al. (2018). Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24 (1), 155–167 e155. doi: 10.1016/j.chom.2018.06.006
Harada, Y., Yoshiga, T. (2015). Distinguishing between inactivated and dead second stage juveniles of Meloidogyne incognita using the NaOH method. Nematological Res. (Japanese J. Nematology) 45 (1), 51–55. doi: 10.3725/jjn.45.51
Hayward, A. C., Fegan, N., Fegan, M., Stirling, G. R. (2010). Stenotrophomonas and Lysobacter: ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J. Appl. Microbiol. 108 (3), 756–770. doi: 10.1111/j.1365-2672.2009.04471.x
Hein, J. W., Wolfe, G. V., Blee, K. A. (2008). Comparison of rhizosphere bacterial communities in arabidopsis thaliana mutants for systemic acquired resistance. Microb. Ecol. 55 (2), 333–343. doi: 10.1007/s00248-007-9279-1
Hinsinger, P., Bengough, A. G., Vetterlein, D., Young, I. M. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321 (1), 117–152. doi: 10.1007/s11104-008-9885-9
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72 (3), 1719–1728. doi: 10.1128/aem.72.3.1719-1728.2006
Jiao, Y., Li, Y., Li, Y., Cao, H., Mao, Z., Ling, J., et al. (2019). Functional genetic analysis of the leucinostatin biosynthesis transcription regulator lcsL in Purpureocillium lilacinum using CRISPR-Cas9 technology. Appl. Microbiol. Biotechnol. 103 (15), 6187–6194. doi: 10.1007/s00253-019-09945-2
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14 (9), 946–961. doi: 10.1111/mpp.12057
Junaid, J., Dar, N. A., Bhat, T., Bhat, A., Bhat, M. (2013). Commercial biocontrol agents and their mechanism of action in the management of plant pathogens. Int. J. Modern Plant&Anim. Sci. 1 (2), 39–57.
Kaur, H., Manchanda, P., Kumar, P., Dhall, R. K., Chhuneja, P., Weng, Y. (2023). Genome-wide identification and characterization of parthenocarpic fruit set-related gene homologs in cucumber (Cucumis sativus l.). Sci. Rep. 13 (1), 2403. doi: 10.1038/s41598-023-29660-3
Kim, Y. C., Anderson, A. J. (2018). Rhizosphere pseudomonads as probiotics improving plant health. Mol. Plant Pathol. 19 (10), 2349–2359. doi: 10.1111/mpp.12693
Kloeppe, J. W., Rodríguez-Kábana, R., Zehnder, A. W., Murphy, J. F., Sikora, E., Fernández, C. (1999). Plant root-bacterial interactions in biological control of soilborne diseases and potential extension to systemic and foliar diseases. Australas. Plant Pathol. 28 (1), 21–26. doi: 10.1071/AP99003
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Larkin, R. P. (2008). Relative effects of biological amendments and crop rotations on soil microbial communities and soilborne diseases of potato. Soil Biol. Biochem. 40 (6), 1341–1351. doi: 10.1016/j.soilbio.2007.03.005
Larsbrink, J., McKee, L. S. (2020). Bacteroidetes bacteria in the soil: glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol. 110, 63–98. doi: 10.1016/bs.aambs.2019.11.001
Li, J., Todd, T. C., Lee, J., Trick, H. N. (2011). Biotechnological application of functional genomics towards plant-parasitic nematode control. Plant Biotechnol. J. 9 (9), 936–944. doi: 10.1111/j.1467-7652.2011.00601.x
Lian, T., Ma, Q., Shi, Q., Cai, Z., Zhang, Y., Cheng, Y., et al. (2019). High aluminum stress drives different rhizosphere soil enzyme activities and bacterial community structure between aluminum-tolerant and aluminum-sensitive soybean genotypes. Plant Soil 440 (1), 409–425. doi: 10.1007/s11104-019-04089-8
Lidbury, I., Borsetto, C., Murphy, A. R. J., Bottrill, A., Jones, A. M. E., Bending, G. D., et al. (2021). Niche-adaptation in plant-associated bacteroidetes favours specialisation in organic phosphorus mineralisation. ISME J. 15 (4), 1040–1055. doi: 10.1038/s41396-020-00829-2
Ling, J., Xie, X., Gu, X., Zhao, J., Ping, X., Li, Y., et al. (2021). High-quality chromosome-level genomes of Cucumis metuliferus and Cucumis melo provide insight into Cucumis genome evolution. Plant J. 107 (1), 136–148. doi: 10.1111/tpj.15279
Lugtenberg, B., Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Masson-Boivin, C., Giraud, E., Perret, X., Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17 (10), 458–466. doi: 10.1016/j.tim.2009.07.004
Mendes, R., Garbeva, P., Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37 (5), 634–663. doi: 10.1111/1574-6976.12028
Mitreva, M., Blaxter, M. L., Bird, D. M., McCarter, J. P. (2005). Comparative genomics of nematodes. Trends Genet. 21 (10), 573–581. doi: 10.1016/j.tig.2005.08.003
Moens, M., Perry, R. N., Starr, J. L. (2009). Meloidogyne species - a diverse group of novel and important plant parasites. Root-knot Nematodes 1-17. doi: 10.1079/9781845934927.0001
Nakata, E., Staub, J. E., López-Sesé, A. I., Katzir, N. (2005). Genetic diversity of Japanese melon cultivars (Cucumis melo l.) as assessed by random amplified polymorphic DNA and simple sequence repeat markers. Genet. Resour. Crop Evol. 52 (4), 405–419. doi: 10.1007/s10722-005-2258-9
Nicol, J. M., Turner, S. J., Coyne, D. L., Nijs, L.d., Hockland, S., Maafi, Z. T. (2011). “Current nematode threats to world agriculture,” in Genomics and molecular genetics of plant-nematode interactions. Eds. Jones, J., Gheysen, G., Fenoll, C. (Dordrecht: Springer Netherlands), 21–43.
Niu, B., Paulson, J. N., Zheng, X., Kolter, R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U.S.A. 114 (12), E2450–E2459. doi: 10.1073/pnas.1616148114
Oota, M., Tsai, A. Y., Aoki, D., Matsushita, Y., Toyoda, S., Fukushima, K., et al. (2020). Identification of naturally occurring polyamines as root-knot nematode attractants. Mol. Plant 13 (4), 658–665. doi: 10.1016/j.molp.2019.12.010
Pal, K. K., McSpadden Gardener, B. (2006). Biological control of plant pathogens. Plant Health Instructor 2, 1–25. doi: 10.1094/PHI-A-2006-1117-02
Pang, F., Solanki, M. K., Wang, Z. (2022). Streptomyces can be an excellent plant growth manager. World J. Microbiol. Biotechnol. 38 (11), 193. doi: 10.1007/s11274-022-03380-8
Perez-Garcia, A., Romero, D., de Vicente, A. (2011). Plant protection and growth stimulation by microorganisms: biotechnological applications of bacilli in agriculture. Curr. Opin. Biotechnol. 22 (2), 187–193. doi: 10.1016/j.copbio.2010.12.003
Poll, J., Marhan, S., Haase, S., Hallmann, J., Kandeler, E., Ruess, L. (2007). Low amounts of herbivory by root-knot nematodes affect microbial community dynamics and carbon allocation in the rhizosphere. FEMS Microbiol. Ecol. 62 (3), 268–279. doi: 10.1111/j.1574-6941.2007.00383.x
Poupin, M. J., Greve, M., Carmona, V., Pinedo, I. (2016). A complex molecular interplay of auxin and ethylene signaling pathways is involved in arabidopsis growth promotion by burkholderia phytofirmans PsJN. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00492
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321 (1), 341–361. doi: 10.1007/s11104-008-9568-6
Ruffner, B., Pechy-Tarr, M., Ryffel, F., Hoegger, P., Obrist, C., Rindlisbacher, A., et al. (2013). Oral insecticidal activity of plant-associated pseudomonads. Environ. Microbiol. 15 (3), 751–763. doi: 10.1111/j.1462-2920.2012.02884.x
Rutter, W. B., Franco, J., Gleason, C. (2022). Rooting out the mechanisms of root-knot nematode-plant interactions. Annu. Rev. Phytopathol. 60, 43–76. doi: 10.1146/annurev-phyto-021621-120943
Saha, S., Prakash, V., Kundu, S., Kumar, N., Mina, B. L. (2008). Soil enzymatic activity as affected by long term application of farm yard manure and mineral fertilizer under a rainfed soybean–wheat system in n-W himalaya. Eur. J. Soil Biol. 44 (3), 309–315. doi: 10.1016/j.ejsobi.2008.02.004
Sarwar, A., Latif, Z., Zhang, S., Zhu, J., Zechel, D. L., Bechthold, A. (2018). Biological control of potato common scab with rare isatropolone c compound produced by plant growth promoting Streptomyces A1RT. Front. Microbiol. 9, 1126. doi: 10.3389/fmicb.2018.01126
Sasse, J., Martinoia, E., Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23 (1), 25–41. doi: 10.1016/j.tplants.2017.09.003
Schlaeppi, K., Dombrowski, N., Oter, R. G., Ver Loren van Themaat, E., Schulze-Lefert, P. (2014). Quantitative divergence of the bacterial root microbiota in arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. U.S.A. 111 (2), 585–592. doi: 10.1073/pnas.1321597111
Schouteden, N., De Waele, D., Panis, B., Vos, C. M. (2015). Arbuscular mycorrhizal fungi for the biocontrol of plant-parasitic nematodes: a review of the mechanisms involved. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01280
Shi, S., Nuccio, E. E., Shi, Z. J., He, Z., Zhou, J., Firestone, M. K. (2016). The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol. Lett. 19 (8), 926–936. doi: 10.1111/ele.12630
Singh, S., Singh, B., Singh, A. P. (2015). Nematodes: a threat to sustainability of agriculture. Proc. Environ. Sci. 29, 215–216. doi: 10.1016/j.proenv.2015.07.270
Tian, B. Y., Cao, Y., Zhang, K. Q. (2015). Metagenomic insights into communities, functions of endophytes, and their associates with infection by root-knot nematode, meloidogyne incognita, in tomato roots. Sci. Rep. 5, 17087. doi: 10.1038/srep17087
Tian, B., Yang, J., Zhang, K. Q. (2007). Bacteria used in the biological control of plant-parasitic nematodes: populations, mechanisms of action, and future prospects. FEMS Microbiol. Ecol. 61 (2), 197–213. doi: 10.1111/j.1574-6941.2007.00349.x
Toju, H., Tanabe, A. S., Sato, H. (2018). Network hubs in root-associated fungal metacommunities. Microbiome 6 (1), 116. doi: 10.1186/s40168-018-0497-1
Topalović, O., Vestergård, M. (2021). Can microorganisms assist the survival and parasitism of plant-parasitic nematodes? Trends Parasitol. 37 (11), 947–958. doi: 10.1016/j.pt.2021.05.007
Tóth, E. M., Borsodi, A. K. (2014). “The family nocardioidaceae,” in The prokaryotes: actinobacteria. Eds. Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E., Thompson, F. (Berlin, Heidelberg: Springer Berlin Heidelberg), 651–694.
Vega, F. E. (2018). The use of fungal entomopathogens as endophytes in biological control: a review. Mycologia 110 (1), 4–30. doi: 10.1080/00275514.2017.1418578
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., van der Heijden, M. G. A. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10 (1), 4841. doi: 10.1038/s41467-019-12798-y
Walters, A., Wehner, T., Daykin, M. E., Barker, K. R. (2006). Penetration rates of root-knot nematodes into cucumis sativus and c. metuliferus roots and subsequent histological changes. Nematropica 36, 231–242.
Wang, R., Zhang, H., Sun, L., Qi, G., Chen, S., Zhao, X. (2017). Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 7 (1), 343. doi: 10.1038/s41598-017-00472-6
Wen, T., Xie, P., Yang, S., Niu, G., Liu, X., Ding, Z., et al. (2022). ggClusterNet: an r package for microbiome network analysis and modularity-based multiple network layouts. iMeta 1, e32. doi: 10.1002/imt2.32
Weng, Y. (2010). Genetic diversity among Cucumis metuliferus populations revealed by cucumber microsatellites. HortScience 45 (2), 214–219. doi: 10.21273/HORTSCI.45.2.214
Xie, X., Ling, J., Mao, Z., Li, Y., Zhao, J., Yang, Y., et al. (2022). Negative regulation of root-knot nematode parasitic behavior by root-derived volatiles of wild relatives of Cucumis metuliferus CM3. Hortic. Res. 9, uhac051. doi: 10.1093/hr/uhac051
Yagi, K., Siedlecka, E., Pawelkowicz, M., Wojcieszek, M., Przybecki, Z., Tagashira, N., et al. (2014). Karyotype analysis and chromosomal distribution of repetitive DNA sequences of cucumis metuliferus using fluorescence in situ hybridization. Cytogenet. Genome Res. 144 (3), 237–242. doi: 10.1159/000369183
Ye, D. Y., Qi, Y. H., Cao, S. F., Wei, B. Q., Zhang, H. S. (2017). Histopathology combined with transcriptome analyses reveals the mechanism of resistance to meloidogyne incognita in Cucumis metuliferus. J. Plant Physiol. 212, 115–124. doi: 10.1016/j.jplph.2017.02.002
Yin, N., Zhao, J. L., Liu, R., Li, Y., Ling, J., Yang, Y. H., et al. (2021). Biocontrol efficacy of Bacillus cereus strain bc-cm103 against meloidogyne incognita. Plant Dis. 105 (8), 2061–2070. doi: 10.1094/PDIS-03-20-0648-RE
Zachow, C., Müller, H., Tilcher, R., Berg, G. (2014). Differences between the rhizosphere microbiome of beta vulgaris ssp. maritima-ancestor of all beet crops-and modern sugar beets. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00415
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3 (4), 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, X., Lin, L., Zhu, Z., Yang, X., Wang, Y., An, Q. (2013). Colonization and modulation of host growth and metal uptake by endophytic bacteria of sedum alfredii. Int. J. Phytoremediation 15 (1), 51–64. doi: 10.1080/15226514.2012.670315
Zhang, B., Zhang, J., Liu, Y., Shi, P., Wei, G. (2018). Co-Occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem. 118, 178–186. doi: 10.1016/j.soilbio.2017.12.011
Keywords: time dynamic, co-occurrence network, rhizosphere bacteria, Cucumis crops, root-knot nematodes
Citation: Song L, Ping X, Mao Z, Zhao J, Yang Y, Li Y, Xie B and Ling J (2023) Variation and stability of rhizosphere bacterial communities of Cucumis crops in association with root-knot nematodes infestation. Front. Plant Sci. 14:1163271. doi: 10.3389/fpls.2023.1163271
Received: 10 February 2023; Accepted: 03 May 2023;
Published: 30 May 2023.
Edited by:
Musharaf Ahmad, University of Agriculture, Peshawar, PakistanReviewed by:
Mariantonietta Colagiero, National Research Council (CNR), ItalyCopyright © 2023 Song, Ping, Mao, Zhao, Yang, Li, Xie and Ling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Ling, bGluZ2ppYW5AY2Fhcy5jbg==; Bingyan Xie, eGllYmluZ3lhbkBjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.