
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 26 April 2023
Sec. Plant Genetics, Epigenetics and Chromosome Biology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1162828
This article is part of the Research TopicPhysiology and Biochemistry of Grain Yield Potential of Rice Concerning Panicle Architectural DesignView all 5 articles
Panicle development is crucial to increase the grain yield of rice (Oryza sativa). The molecular mechanisms of the control of panicle development in rice remain unclear. In this study, we identified a mutant with abnormal panicles, termed branch one seed 1-1 (bos1-1). The bos1-1 mutant showed pleiotropic defects in panicle development, such as the abortion of lateral spikelets and the decreased number of primary panicle branches and secondary panicle branches. A combined map-based cloning and MutMap approach was used to clone BOS1 gene. The bos1-1 mutation was located in chromosome 1. A T-to-A mutation in BOS1 was identified, which changed the codon from TAC to AAC, resulting in the amino acid change from tyrosine to asparagine. BOS1 gene encoded a grass-specific basic helix–loop–helix transcription factor, which is a novel allele of the previously cloned LAX PANICLE 1 (LAX1) gene. Spatial and temporal expression profile analyses showed that BOS1 was expressed in young panicles and was induced by phytohormones. BOS1 protein was mainly localized in the nucleus. The expression of panicle development-related genes, such as OsPIN2, OsPIN3, APO1, and FZP, was changed by bos1-1 mutation, suggesting that the genes may be the direct or indirect targets of BOS1 to regulate panicle development. The analysis of BOS1 genomic variation, haplotype, and haplotype network showed that BOS1 gene had several genomic variations and haplotypes. These results laid the foundation for us to further dissect the functions of BOS1.
Rice (Oryza sativa) is a major food crop feeding more than half of the world’s population (Yoshida and Nagato, 2011; Zeng et al., 2017). With the increase in population and the decrease in arable land, improving rice productivity has become the main goal of breeding (Tilman et al., 2011; Zeng et al., 2017). Rice yield is a complex agronomic trait, which is mainly determined by three traits, such as panicle number, grain number per panicle, and grain weight (Xing and Zhang, 2010). These suggest that panicle development is very important to improve rice yield.
The shoot architecture of plants is dependent on the functions of shoot apical meristem (SAM) and axillary meristems (AMs) (Mcsteen and Leyser, 2005; Wang et al., 2018). In rice, SAM produces the main shoot, while AMs produce new shoot branches called tillers. The main culm and branches determine the biomass and the number of inflorescences and panicles in rice. Inflorescences in rice belong to the raceme class, with long branches called panicles. After the transition from the vegetative to reproductive phase in rice, SAM is changed into an inflorescence meristem (IM; rachis meristem). IM develops into the primary inflorescence axis (rachis) and initiates the primary branch meristem (PBM). PBM generates secondary branch meristem (SBM). At the top of the primary branches (PBs) and secondary branches (SBs), PBMs and SBMs are transformed into spikelet meristem (SM), resulting in terminal spikelets and flowers (Komatsu M.et al., 2003; Itoh et al., 2005; Yoshida et al., 2012). The determination of meristem fate greatly affects the size, morphology of rice panicles, and rice yield.
The transition from SAM to IM is controlled by heading date-related genes. The rice floral transition is promoted by florigens, such as Heading date 3a (Hd3a) and Rice Flowering Locus T 1 (RFT1) (Tsuji et al., 2013; Zhou S. et al., 2021; Zhao et al., 2022). In the SAM, Hd3a and RFT1 are physically associated with bZIP transcription factor OsFD1 and 14-3-3, a member of the Gf14 family, forming florigen activation complexes (FACs). FACs trigger the expression of floral identity genes, such as OsMADS14, OsMADS15, OsMADS18, and OsMADS34, to accelerate floral transition (Tamaki et al., 2007; Komiya et al., 2008; Taoka et al., 2011; Kobayashi et al., 2012). OsMADS34, also called Panicle Phytomer 2 (PAP2), is a member of the grass-specific SEPALLATA (SEP) clade of MADS-box proteins. OsMADS14, OsMADS15, and OsMADS18 are the members of the APETALA 1 (AP1)/FRUITFULL (FUL) family (Zahn et al., 2005; Gao et al., 2010).
Axillary meristems generate panicle branches and florets in rice. Plant hormones, such as auxin and cytokinin (CK), and transcription factors are involved in axillary meristem initiation and outgrowth to control panicle branching in rice (Zhang and Yuan, 2014). Tryptophan Deficient Dwarf 1 (TDD1), an anthranilate synthase beta-subunit, catalyzes the first step of the tryptophan (Trp) biosynthesis and is involved in auxin biosynthesis in rice. The tdd1 mutant shows pleiotropic phenotypes, such as deficiency in Trp and indole-3-acetic acid (IAA), dwarfing, narrow leaves, short roots, and abnormal flowers (Sazuka et al., 2009). Grain number 1a (Gn1a) encodes cytokinin oxidase/dehydrogenase (OsCKX2), which has the enzyme activity to degrade the cytokinin. Downregulated expression of OsCKX2 causes cytokinin accumulation in IMs and increases the number of reproductive organs, resulting in enhanced grain yield (Ashikari et al., 2005). Drought and salt tolerance (DST), a rice zinc-finger transcription factor, directly upregulated the expression of OsCKX2 in the reproductive meristem of rice (Zhang and Yuan, 2014). DST-dependent expression of OsCKX2 regulates CK accumulation in the SAM to control the number of reproductive organs, such as panicle branching and grain number (Li et al., 2013).
Rice Aberrant Spikelet and Panicle 1 (ASP1), a transcriptional corepressor, plays important roles in auxin-related panicle development. The asp1 mutant exhibits abnormal spikelets, which reduced the number of branch and spikelet phenotypes (Yoshida et al., 2012). A plant-specific transcription factor containing a conserved SBP-box DNA-binding domain Squamosa Promoter Binding Protein-Like 14 (OsSPL14), which is also known as Ideal Plant Architecture 1 (IPA1) and Wealthy Farmer’s Panicle (WFP), is involved in panicle branching (Yamasaki et al., 2004; Jiao et al., 2010; Miura et al., 2010). Increasing the OsSPL14 expression in the reproductive stage promotes panicle branching and higher grain yield in rice (Miura et al., 2010). LAX PANICLE 1 (LAX1) is a grass-specific basic helix–loop–helix (bHLH) transcription factor involved in rice reproduction through the auxin-related pathway. LAX1 interacts with LAX2, a plant-specific nuclear protein, to regulate the formation of axillary meristems during reproduction. The lax1, lax2, lax1/lax2 single or double mutants dramatically reduce the number of inflorescence branches and no initiation of lateral spikelet (Komatsu et al., 2001; Komatsu K. et al., 2003; Oikawa and Kyozuka, 2009; Tabuchi et al., 2011). The MADS-box containing transcription factors is also associated with inflorescence and panicle branching in rice. The osmads34 mutants develop abnormal inflorescences and panicles in rice, such as an increased number of primary branches and a decreased number of secondary branches (Zahn et al., 2005; Gao et al., 2010). Suppression of the OsMADS14, OsMADS15, and OsMADS18 genes by RNA interference causes a slight delay in reproductive transition. Further depletion of PAP2 function from these triple knockdown plants inhibited the transition of the meristem to the IM (Kobayashi et al., 2012). OsMADS50 and OsMADS56 (homologous of Suppressor of Overexpression of CONSTANS 1 (SOC1) in Arabidopsis) and OsMADS22, OsMADS47, and OsMADS55 (homologous of Short Vegetative Phase (SVP) or AGAMOUS-Like 24 (AGL24) in Arabidopsis) are involved in regulating of inflorescence branching by repressing the expression of Reduced Culm Number 4 (RCN4), a rice Terminal Flower 1 (TFL1) homolog (Zhu et al., 2022). Knockdown expression of OsMADS50, OsMADS56, OsMADS22, OsMADS47, and OsMADS55 in osmads34 mutant results in an increase in inflorescence branching and the number of secondary and tertiary branches (Liu et al., 2013).
The transition from IM to SM is important for panicle architecture establishment in rice. Aberrant Panicle Organization 1 (APO1), an F-box protein, controls spikelet numbers by inhibiting the transition from IM to SM. The apo1 mutant exhibits decreased number of PBs and spikelets, shortened main axis, and abnormal floral organs (Ikeda et al., 2005; Ikeda et al., 2007). APO1 interacts with APO2 to regulate the development of rice panicles (Abe et al., 2005; Ikeda et al., 2007). The apo2 inflorescence is similar to the apo1 inflorescence, as both exhibit reduced numbers of spikelets and abnormal floral organs (Ikeda et al., 2005; Ikeda-Kawakatsu et al., 2012). TAWAWA 1 (TAW1) belongs to a nuclear protein in Arabidopsis Light-dependent Short Hypocotyls 1 (LSH1) and Oryza Long Sterile Lemma 1 (G1) (ALOG) family. TAW1 regulates rice panicle development by suppressing the transition from IM to SM. TAW1 induces the expression of members of the SVP subfamily of MADS-box genes, including OsMADS22, OsMADS47, and OsMADS55. The dominant mutant, tawawa1-D, shows prolonged branch formation and increased numbers of spikelets because the activity of the IM is extended, and spikelet specification is delayed. In contrast, reductions in TAW1 activity cause IM abortion and spikelet formation, resulting in the formation of small inflorescences (Yoshida et al., 2013).
AP2 and MADS-box transcription factors are involved in the control of the transition from SM to floral meristems (FMs) in rice. The spikelet is the basic unit of rice panicles. Each spikelet forms one to several small flowers or florets (Yoshida and Nagato, 2011). Rice Indeterminate Spikelet 1 (OsIDS1) is a member of the IDS1 subgroup of the AP2 transcription factors that control inflorescence branching, the initiation of spikelet meristems, and floral meristems (Lee and An, 2012). Rice Supernumerary Bract (SNB) is also a member of the IDS1 subgroup in the AP2 family. Mutations of SNB delay the transition from SM to FM and cause the development of multiple rudimentary glumes in an alternative phyllotaxy as well as aberrant floral shape (Lee et al., 2007). The snb osids1 double mutant generates fewer inflorescence branches and spikelets and delays the transition to FMs (Lee and An, 2012). Frizzy Panicle (FZP)/BRANCHED FLORETLESS 1 (BFL1) is the rice ethylene-response factor (ERF) subfamily of AP2 transcription factors. FZP determines the transition from SM to FM by suppressing the formation of axillary meristems within the spikelet meristem and triggering subsequent floral meristem identity specification. In fzp mutant, the formation of florets is replaced by a sequential round of branching (Komatsu M. et al., 2003; Zhu et al., 2003). Multi-Floret Spikelet 1 (MFS1) is also a factor in the grass-specific ERF subfamily of AP2 transcriptional factors. MFS1 plays important roles in regulating the transformation of spikelet meristems to floral meristems by promoting the expression of SNB, OsIDS1, and G1/Elongated Empty Glume (ELE). The mfs1 mutant shows a delayed transition of spikelet meristems to floral meristems, resulting in an extra hull-like organ and an elongated rachilla (Yoshida et al., 2009; Ren et al., 2013). TONGARI-BOUSHI 1 (TOB1), a protein in the plant-specific YABBY transcription factor family, plays an important role in regulating lateral organ development and spikelet meristem maintenance in rice (Tanaka et al., 2012).
In addition, Rice Circadian Clock Associated 1 (OsCCA1) positively regulates the expression of Teosinte Branched 1 (OsTB1), also known as Fine Culm 1 (FC1), Dwarf 14 (D14), and IPA1/OsSPL14 to repress tiller-bud outgrowth. OsCCA1 also regulates IPA1 expression to mediate panicle and grain development (Wang et al., 2020). Rice Apical Spikelet Abortion (OsASA) encodes a boric acid channel protein that showed the highest expression in inflorescence, peduncle, and anther. The boron distribution maintained by OsASA is required for normal panicle development in a process that involves modulating reactive oxygen species (ROS) homeostasis and salicylic acid (SA) biosynthesis (Zhou D. et al., 2021).
Rice panicle development is a complex process. Although many genes related to panicle development have been cloned, its potential molecular mechanism and genetic regulation network are still unclear. Here, we obtained a mutant with abnormal panicles, named branch one seed 1-1 (bos1-1). The bos1-1 mutant showed pleiotropic panicle defects in panicle development, such as a reduced number of primary and secondary branches and abortion of lateral spikelets, causing a decrease in grain yield. We isolated BOS1 gene through a positional cloning approach, which encoded a bHLH transcription factor. BOS1 protein was mainly localized in the nucleus. BOS1 is expressed in young panicles but is expressed at a lower level in other detected tissues and organs and can be induced by phytohormones. BOS1 controlled panicle development by regulating the expression of OsPIN-FORMED 2 (PIN2), OsPIN3, APO1, and FZP. Genomic variation, haplotype, and haplotype network analyses showed that BOS1 had several genomic variations and haplotypes. These results laid the foundation for further exploring the functions of BOS1 in panicle development in rice.
A japonica rice variety named Shen Nong 9816 (SN9816), bos1-1 mutant in SN9816 background, and a Japanese indica rice variety named Habataki were used as materials in the study. SN9816 variety is widely cultivated in northern China and was used as wild type (WT). The bos1-1 mutant was selected and obtained in the M2 segregation population mutated by ethyl methanesulfonate (EMS) (Zhu et al., 2021). Habataki was used as the male parent to generate the F2 segregation mapping population. WT, bos1-1 mutant, Habataki, F2 population used for genetic analysis, and F2 population used for positional cloning were planted in the experimental station at Shenyang Agricultural University, China (longitude, 123.38°E; latitude, 41.8°N). Rice seeds were first soaked and germinated. Then, the germinated seeds were sown on the seedbed to nurse seedlings. When the seedlings grew to the four-leaf stage, they were transplanted to the paddy field.
To investigate the agronomic traits, WT and bos1-1 mutant were transplanted in three plots in the paddy field. Each plot contains 20 plants. When the rice materials were in the mature stage, five plants of WT and bos1-1 mutant from each plot were selected and sampled for the agronomic trait analysis.
To study the inheritance of bos1-1 mutation, we crossed bos1-1 mutant to WT to generate the first filial generation (F1). Then, the F1 individuals were self-pollinated, and the second filial generation (F2) segregation population was produced. The number of individuals with bos1-1 phenotype and the total number of plants in the F2 population were investigated to calculate the segregate ratio and perform the χ2 test.
To find the locus with bos1-1 mutation, we utilized a positional or map-based cloning strategy. The F2 segregation mapping population was generated by crossing bos1-1 mutant with Habataki. The bulk segregant analysis (BSA) strategy was applied for crude mapping (Yan et al., 2022). The DNA of the F1 hybrid (WT × Habataki) and the DNA pool made from 50 plants with bos1-1 mutant phenotype were used as templates for PCR with molecular markers evenly distributed on the 12 chromosomes of rice. After crude mapping, fine mapping was conducted by developing markers by using polymorphisms between WT and Habataki and enlarging the F2 mapping population. Then, the open reading frames (ORFs) in the region bos1-1 mutation located were amplificated and sequenced to identify bos1-1 mutation.
To accelerate the mapping processes, the MutMap strategy was also used in the study. Thirty plants with bos1-1 phenotype in the F2 population from bos1-1 × WT and WT were used for MutMap as described in the previous study (Abe et al., 2012).
For complementation tests, the CDS of BOS1 fused with a green fluorescent protein (GFP) and the 1,795-bp promoter upstream of ATG were cloned into the pCambia 1300 binary vector with XbaI/BamHI and PstI/SpeI enzymes, respectively. Then, the BOS1p:BOS1c-GFP plasmid was transformed into a bos1-1 mutant through the agrobacterium (Agrobacterium tumefaciens)-mediated transformation of rice mature embryos (Li et al., 2015; Zhu et al., 2021). In the second and third filial transgenic generations (T2 and T3), the phenotypes of the complementary transgenic lines harboring BOS1p:BOS1c-GFP construction were observed. Primers used in the plasmid construction are listed in Supplementary Table S1.
Gene expression was analyzed by the method described before (Wang et al., 2012; Cui et al., 2017). In brief, total RNA was extracted from 100 mg of SAM, 2-mm panicle, and 3–5-mm panicle using Takara RNAiso Plus (Takara Bio Inc., Otsu, Japan; Cat. No. 9108). Total RNA measuring 20 μg was treated with RNase-free DNase I to remove the DNA contaminated (Promega, Madison, WI, USA; Cat. No. M6101). After treatment, 3 μg of RNA was used for first-strand cDNA synthesis with RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA; Cat. No. K1621). OsACT1 (Os03g0718100) was used as a control. Three biological replicates are performed. The formula, 2−delta Ct was used to calculate the relative expression level of genes. Primers in the list were used for the assay (Supplementary Table S2).
Microarray data in Rice Expression Profile Database (RiceXPro, http://ricexpro.dna.affrc.go.jp/) were assayed for the expression of BOS1. The expression of BOS1 in tissues and organs throughout the entire growth period in the field was selected. The expression of BOS1 in tissues and organs, such as roots, stems, leaf blades, leaf sheath, and inflorescence, were selected for the assay. The expression of BOS1 in vegetative, reproductive, and ripening stages at 12:00 was also analyzed; three biological replicates were performed. Four-leaf-old seedlings of rice were treated with 10 μM of IAA, 10 μM of gibberellin 3 (GA3), 1 μM of CK, 1 μM of brassinosteroid (BR), 50 μM of abscisic acid (ABA), and 100 μM of jasmonic acid (JA) for 0, 1, 3, 6, and 12 h; two biological replicates were performed. The shoot of the treated seedlings was used for microarray analysis. The signal intensity detected during hybridization was used for drawing heatmap with GraphPad Prism7 software.
The BOS1 CDS fused with GFP driven by 35S promoter was cloned into pCambia 1300, generating 35S:BOS1-GFP construct. The plasmid harboring 35S:BOS1-GFP cassette was transiently expressed in Nicotiana benthamiana by infiltration according to the method reported before (Wang et al., 2012). In brief, the constructed 35S:BOS1-GFP binary vector was transformed into Agrobacterium strain GV3101 and infiltrates N. benthamiana healthy leaves of 4-week-old seedlings. The pCAMBIA1300-P19 plasmid was co-infiltrated to inhibit RNA interference. The infiltrated N. benthamiana seedlings were grown in a greenhouse with 16-h light/8-h dark at 26°C. After 3 days, the subcellular localization of BOS1 was observed by laser scanning confocal microscopy.
The genomic DNA variation, haplotype, and haplotype network of BOS1 were analyzed by using RiceVarMap2 (http://ricevarmap.ncpgr.cn/) website. In the haplotype network analysis of BOS1, we selected classification 3, including Indica I, Indica II, Indica III, Indica Intermediate, Aus, Temperate Japonica, Tropical Japonica, Japonica Intermediate, and Intermediate (Intermediate group includes Aromatic and other accessions) for analysis (Huang et al., 2012; Zhao et al., 2015; Zhao et al., 2021).
Two-tailed unpaired t-test with Welch’s correction was used for statistical analysis. Asterisks indicate significant differences between treatments and controls (**p < 0.01, *p < 0.05). The ns represents no significant differences (p > 0.05) between treatments and controls.
The gene accessions are listed in Supplementary Table S3.
A rice panicle consists of a main axis, PBs, SBs, and spikelets. The primary branches are arranged in a spiral phyllotaxy, and spikelets are produced on both the primary and secondary branches (Yoshida et al., 2012). In this study, we identified a mutant with defects in panicle development termed bos1-1 mutant by using EMS mutagenesis. The mutation of BOS1 conferred abnormal pleiotropic panicle phenotypes (Figure 1A). The main defects in bos1-1 mutant were that PBs and SBs only had one terminal spikelet and no lateral spikelets (Figure 1A). Compared with WT, the number of PBs and SBs were significantly decreased by bos1-1 mutation, especially the number of SBs (Figures 1B, C). The panicle length was increased (Figure 1D). Grain sizes, such as grain length, width, and thickness, also increased by bos1-1 mutation (Figures 1E–I). To analyze the effects of bos1-1 mutation on rice yield, seed setting rate, grain number per plant, and 1,000-grain weight were investigated. Compared with WT, the seed setting rate and grain number per plant of bos1-1 mutant were significantly reduced, while the 1,000-grain weight was significantly increased (Figures 1J–L). Finally, compared with WT, the yield per plant of the bos1-1 mutant decreased significantly (Figure 1M); these results indicate that BOS1 was a positive regulator of rice yield.
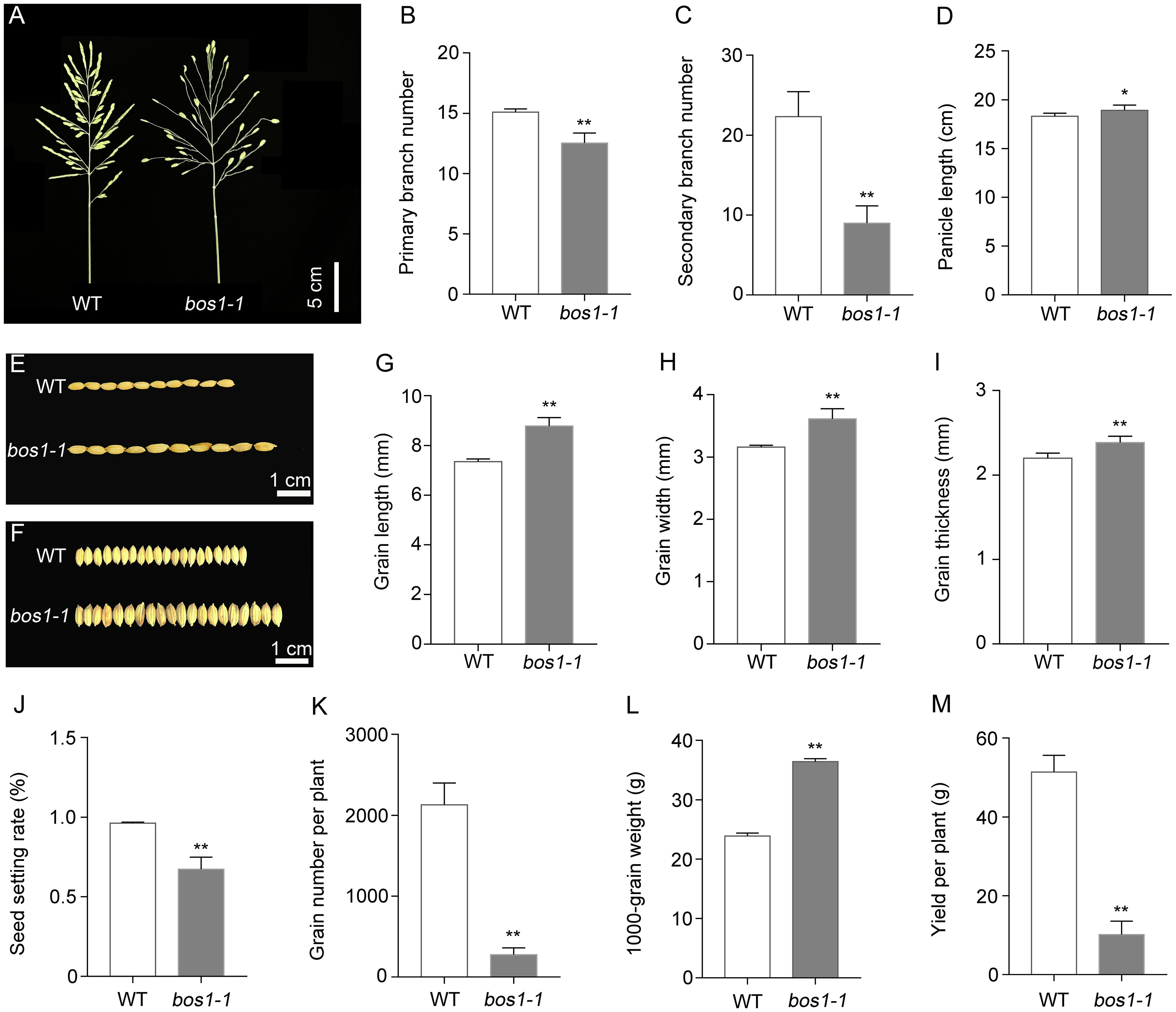
Figure 1 The bos1-1 mutation confers abnormal panicle phenotype in rice. (A) Photos of WT and bos1-1 panicles. Scale bar is 5 cm. (B) Primary branch number. (C) Secondary branch number. (D) Panicle length. (E–I) Grain features include grain length (E, G), grain width (F, H), and grain thickness (I). Scale bar is 1 cm. (J) Seed setting rate. (K) Grain number per plant. (L) 1,000-grain weight. (M) Yield per plant. Data in panels (B–D, G–M) are means ± s.d. The s.d. represents standard deviation. A two-tailed unpaired t-test with Welch’s correction is used for statistical analysis. Asterisks indicate significant differences between WT and bos1-1 mutant (*p < 0.05, **p < 0.01; Student’s t-test). WT, wild type.
Panicle number per plant is an important factor in rice yield composition, which is determined by the number of effective tillers. Compared with WT, the number of effective tillers of the bos1-1 mutant was not changed (Figure 2A). To determine whether bos1-1 affects biomass production, the dry weight and fresh weight of different organs above ground were measured. Compared with WT, the dry and fresh weights of stems, leaves, and sheaths of bos1-1 mutant increased significantly, while the dry and fresh weights of panicles decreased significantly (Figure 2B). Therefore, the economic coefficient of bos1-1 was lower than that of WT (Figure 2C). We also investigated the effects of bos1-1 mutation on plant height and internode length. Compared with WT, the plant height of bos1-1 mutant was significantly reduced (Figures 2D, E), the length of the first elongated internode of bos1-1 mutant increased significantly, and the length of the second, third, fourth, fifth, and sixth elongated internodes decreased significantly (Figure 2F). These results showed that the mutation of BOS1 significantly reduced the economic yield and shortened plant height by reducing internode length in rice.
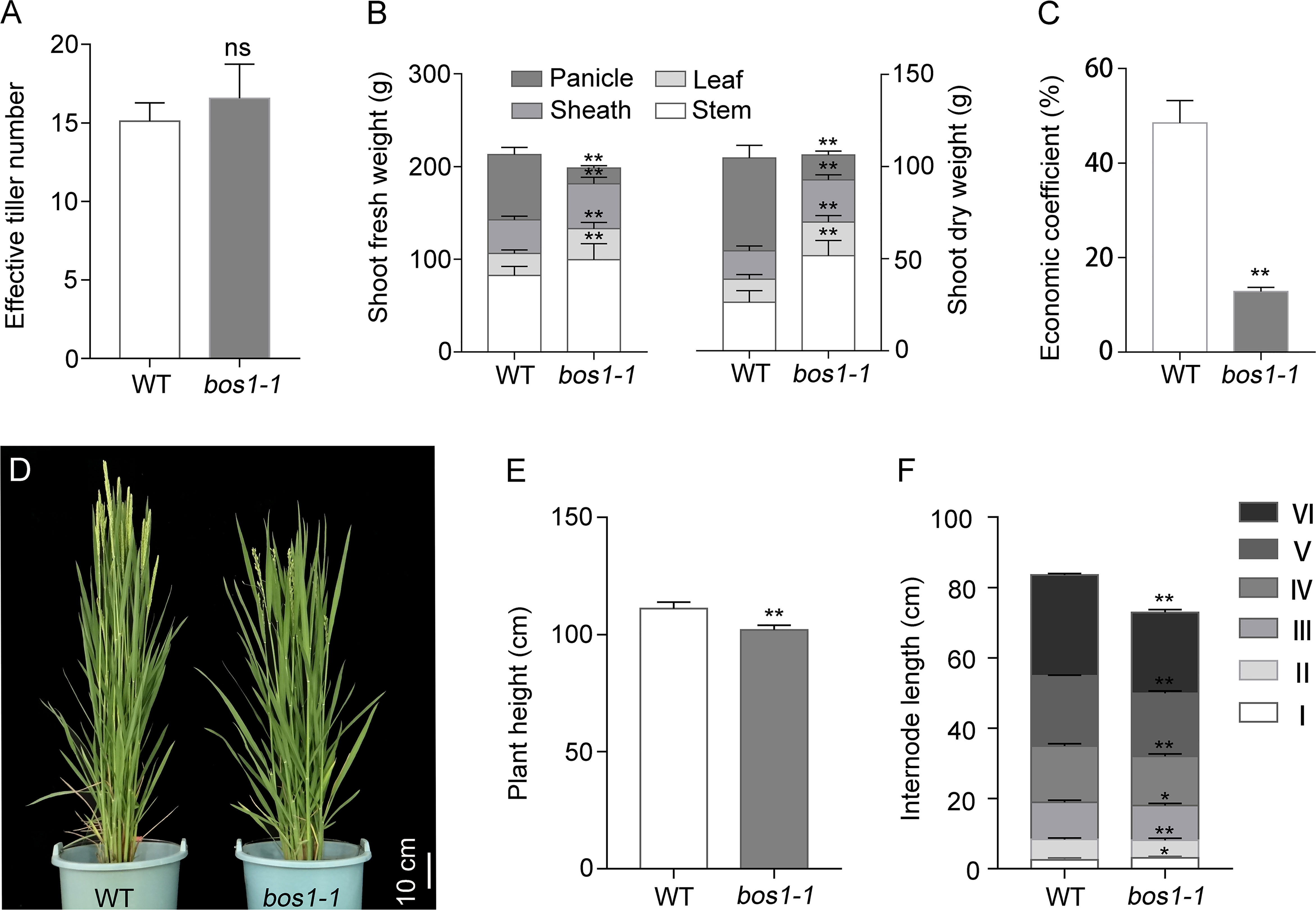
Figure 2 Biomass production in WT and bos1-1 mutant. (A) Effective tiller number. (B) Fresh and dry weight/plant. (C) Economic coefficient. (D–F) Agronomic traits, including photos of rice plant height (D), plant height (E), and internode length (F). Scale bar is 10 cm. Data in panels (A–C, E, F) are means ± s.d. The s.d. represents standard deviation. A two-tailed unpaired t-test with Welch’s correction is used for statistical analysis. Asterisks indicate significant differences between WT and bos1-1 mutant (*p < 0.05, **p < 0.01; Student’s t-test). ns indicates no significant difference. WT, wild type.
To isolate the locus with bos1-1 mutation, it is necessary to determine whether the mutation is dominant or recessive and controlled by single or multiple genes. To carry out genetic analysis, we crossed bos1-1 mutant to WT and generated the first filial (F1) hybrid. The phenotype of all the F1 individuals was consistent with that of WT. After self-pollination of the F1, we obtained an F2 segregating population. In the F2 population, 830 and 270 progenies exhibited WT and bos1-1 mutant phenotypes, respectively. The ratio of individuals with WT phenotype to those with bos1-1 phenotype is 3.07: 1, χ2 = 0.1212 < χ2 (P0.05, 1 = 3.84), suggesting that bos1-1 is a single recessive mutation (Table 1).
To isolate bos1-1 mutation, we applied the positional cloning method. We crossed bos1-1 mutant to an indica rice variety Habataki and produced F1. After the self-pollination of F1, the F2 segregation population was created. BSA was used to map bos1-1 mutation crudely. In the F2 population, 45 individuals with the bos1-1 mutant phenotype were selected to create a DNA pool. Both the F1 (a hybrid of WT × Habataki) DNA and the DNA pool were used as a template to PCR with the polymorphic markers evenly distributed in the 12 chromosomes of rice. The bos1-1 mutation was primarily located between B1-19 and B1-21 molecular markers on chromosome 1, with a physical distance of 4.35 Mb. Subsequently, 270 individual plants with bos1-1 mutant phenotypes were used for fine mapping. The bos1-1 mutation was located between GL1-1B (Chr. 01: 35,424,703) and GL1-3 (Chr. 01: 35,981,090) molecular markers, with a physical distance of 556 kb (Figure 3A).
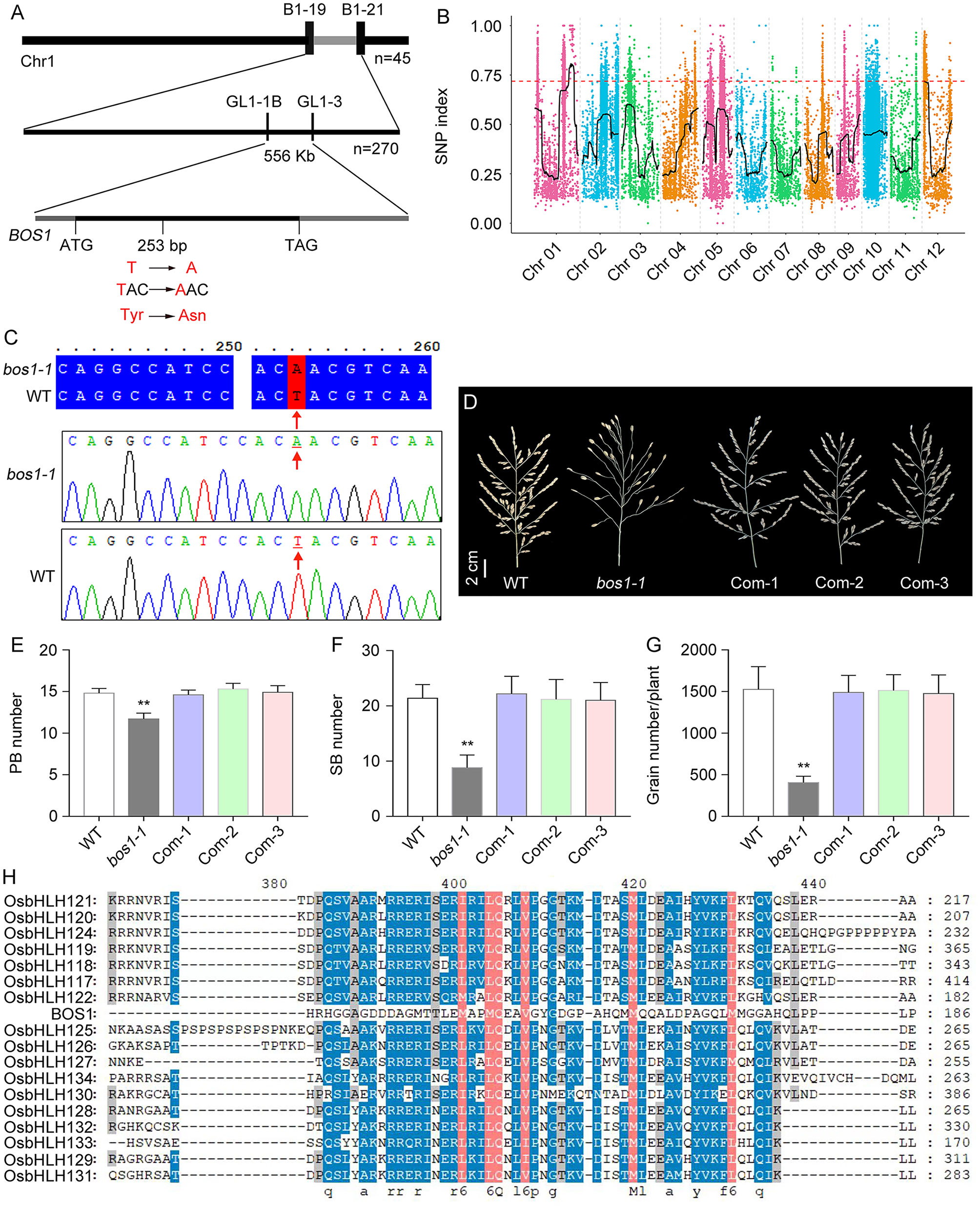
Figure 3 Cloning of BOS1 gene through positional cloning and MutMap strategies. (A) Positional cloning of BOS1 gene. Upper and middle panels represent crude and fine mapping results of bos1-1 mutation, respectively. The numbers of plants with bos1-1 mutant phenotypes used and the molecular markers are shown. Lower panel shows the gene structure of BOS1. Black line represents exons. Gray lines represent the 5′ untranslated region (5′UTR) and 3′UTR. (B) SNP index obtained by MutMap through high-throughput sequencing. (C) Sequence alignment and the trace file showing the mutation identified by sequencing in WT and bos1-1. (D) Photos of panicles in WT, bos1-1, and three complementary transgenic lines. Scale bar is 1 cm. (E, F) The number of PBs (E), SBs (F), and grains (G). (H) Sequence alignment of BOS1 protein and other 17 closely homologous bHLH proteins. SNP, single-nucleotide polymorphism; WT, wild type; PBs, primary branch; SBs, secondary branches. Data in E-G are means ± s.d.. The s.d. represents standard deviation. A two-tailed unpaired t-test with Welch’s correction is used for statistical analysis. Asterisks indicate significant differences (**P < 0.01; Student’s t-test).
In the study, we also used the MutMap strategy, a method based on whole-genome resequencing of pooled DNA from a segregating population of plants with mutant phenotypes, to accelerate gene cloning (Abe et al., 2012). Thirty plants with bos1-1 mutant phenotypes in F2 genetic analysis population (bos1-1 × WT) were chosen to isolate DNA and made a pool to conduct high-throughput sequencing together with WT using an Illumina GAIIx sequencer.
Approximately 54.56 and 84.89 million raw reads, 54.56 and 84.84 million clean reads with an average length of 150 bp, 109.05 and 169.68 million total reads, and 16.34 and 25.42 Giga clean bases with a Q30 > 93% and 41% GC content were obtained from WT and the pool of bos1-1 mutant, respectively. Approximately 99% and 96% of the total reads were mapped and properly mapped to the rice reference genome (IRGSP-1.0, https://rapdb.dna.affrc.go.jp/download/irgsp1.html) (Supplementary Table S4). The average depth (coverage) of the high-throughput sequencing was 41 and 65 for WT and the pool of bos1-1 mutant, respectively. The cover ratio of the high-throughput sequencing was 92% to 97% for WT and the pool of bos1-1 mutant (Supplementary Table S5).
Single-nucleotide polymorphisms (SNPs) and insertion–deletions (indels) between WT and the pool of bos1-1 mutants were analyzed and annotated (Supplementary Figure S1). The association of the SNPs with the bos1-1 mutation was performed by using the SNP index, which was the ratio between the number of reads of a mutant SNP and the total number of reads corresponding to the SNP (Abe et al., 2012). One region was linked with the bos1-1 mutation located in chromosome 1 (Chr. 1) with 4.83 Mb (Figure 3B, Supplementary Table S6).
Approximately 280 differential SNPs were located in Chr. 1. The Chr. 1 region was co-located with the locus mapped by the positional cloning method, suggesting that bos1-1 mutation was in Chr. 1 between GL1-1B and GL1-3 markers. In the co-located region, 30 differential SNPs were identified by the MutMap method in the pool with the bos1-1 mutant phenotype, among which three SNPs led to non-synonymous mutations in three genes, including Os01g0828300, Os01g0831000, and Os01g0836600. Then, the coding regions of the three candidate genes were amplified and sequenced. A T-to-A mutation at 253 bp downstream of ATG was identified, which changed the codon from TAC to AAC, resulting in the change of the 85th amino acid from tyrosine (Tyr) to asparagine (Asn) (Figures 3A, C).
To confirm that the mutation in BOS1 (Os01g0831000) locus was bos1-1, a plasmid containing the BOS1 CDS fused to GFP driven by the native promoter of BOS1 genes (BOS1P:BOS1C-GFP) was constructed. The resulting plasmid was transformed into bos1-1 mutant to obtain the complementary transgenic lines. The bos1-1 mutant phenotypes, such as the number of PBs, SBs, and grains, were rescued in the complementary transgenic lines, indicating that the mutation in BOS1 is responsible for the bos1-1 mutant phenotype (Figures 3D–G). BOS1 encodes a known protein, LAX1, which is a grass-specific basic bHLH domain-containing transcription factor (Komatsu et al., 2001; Komatsu K. et al., 2003; Oikawa and Kyozuka, 2009). A point missense mutation in the bHLH domain disrupted the functions of BOS1.
The rice genome contains 167 bHLH domain-containing proteins, which can be divided into 22 (A to V) subfamilies (Li et al., 2006). bHLH proteins in A and B subfamilies including BOS1 (OsbHLH123) were selected to conduct protein sequence alignment. The results showed that the bHLH proteins were highly conserved, while BOS1 showed lower conservation with other members, even in the bHLH domain (Figure 3H).
To determine the subcellular localization of BOS1, a binary plasmid containing the full-length CDS of BOS1 fused to GFP driven by Cauliflower Mosaic Virus (CaMV) 35S constitutive promoter, 35S:BOS1-GFP, was created. The constructed 35S:BOS1-GFP binary plasmid was transferred into an agrobacterium (A. tumefaciens) strain GV3101 and infiltrated healthy N. benthamiana seedling leaves for transient expression. The results showed that BOS1 was mainly localized in the nucleus, indicating that BOS1 is a nuclear localization protein (Figure 4A).
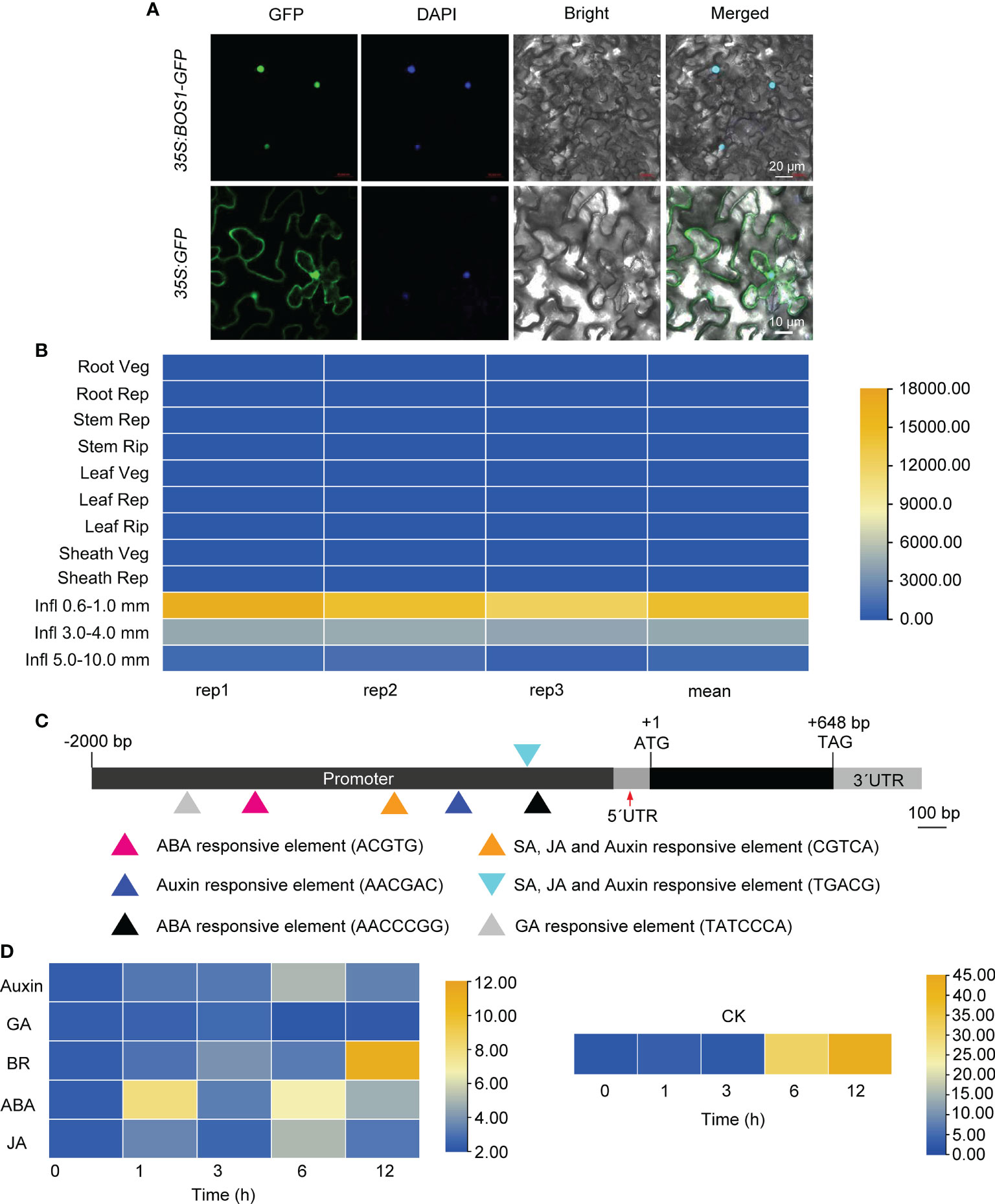
Figure 4 Subcellular localization of BOS1 protein and analysis of BOS1 gene expression profiles. (A) Representative microscopic images showing the subcellular localization of green fluorescent protein (GFP) and BOS1-GFP fusion protein transiently expressed in Nicotiana benthamiana leaves driven by 35S promoter. DAPI was used as a nuclear marker. Scale bars are 20 and 10 μm as labeled. (B) Spatial–temporal of BOS1 gene expression of various tissues/organs throughout entire growth in the field. Three biological replications were performed. (C) Hormone response elements in the promoter region of BOS1. Triangles in different colors represent the elements identified. (D) Heat map of BOS1 gene expression pattern under auxin, GA, CK, BR, ABA, and JA treatment. Two biological replications were performed. CK, cytokinin; BR, brassinosteroid; ABA, abscisic acid; JA, jasmonic acid.
Previous studies have shown that the BOS1 gene is expressed at the boundary between SAM and the new meristem region, indicating that BOS1 gene is a major regulator of axillary meristem formation (Komatsu et al., 2003; Matin and Kang, 2011). To analyze the expression of BOS1 gene, we used the microarray data from the RiceXpro database. The expression of BOS1 in inflorescence was higher than that in other tissues and organs, which corresponds to the severe inflorescence phenotype of the bos1-1 mutant (Figure 4B). To understand whether BOS1 is induced by hormones, we also analyzed cis-acting elements in the BOS1 promoter. The cis-acting elements’ response to hormones was identified (Figure 4C). Then, the expression of BOS1 gene under different hormone treatments, including IAA, GA3, CK, BR, ABA, and JA, was examined. The results showed that the expression of BOS1 was induced by hormones, especially by auxin, CK, BR, ABA, and JA (Figure 4D).
To explore the molecular mechanisms whereby BOS1 controls panicle development in rice, the expression of 19 genes associated with inflorescence development was examined by reverse transcription–polymerase chain reaction (RT-PCR). The results show that the expression of 15 genes in the bos1-1 mutant was similar to that in WT; however, the expression of OsPIN2, OsPIN3, APO1, and FZP genes was changed by bos1-1 mutation (Figure 5A). The alternative splicing of OsPIN2 and APO1 was altered in bos1-1 mutant, leading to increasing pre-mRNA and reducing mature transcripts of them (Figures 5B–E). The expression of OsPIN3 gene was upregulated in bos1-1 mutant (Figure 5F); however, the expression of FZP gene was downregulated in bos1-1 mutant (Figure 5G). The results of their expression patterns showed that higher expression of OsPIN3 and FZP was also in the panicle. The expression of OsPIN2 in the root was higher than in other organs (Supplementary Figure S2). APO1 was also expressed in SAM and young panicles as reported previously (Ikeda et al., 2005). These results indicated that the expression pattern of BOS1 was similar to that of OsPIN3, APO1, and FZP.

Figure 5 Expression of genes related to panicle development in WT and bos1-1 mutant. (A) The expression of genes related to panicle development in WT and bos1-1 mutant by using semi-RT-PCR. Around the shoot apex (ASA), 1–3-mm panicles, and 3–5-mm panicles were sampled for the assay. The pink arrow indicates that the alternative splicing of FZP and APO1 genes has changed. (B–E) The mature transcripts of genes OsPIN2 (B), APO1 (C), FZP (D), and OsPIN3 (E) detected by quantitative RT-PCR. (F, G) The intron retention transcripts of genes OsPIN2I2 (F) and APO1I1 (G) detected by quantitative RT-PCR. Three biological replicates were performed. OsACT1 (Os03g0718100) was employed as an endogenous control. Data in panels B–G are means ± s.d. The s.d. represents standard deviation. A two-tailed unpaired t-test with Welch’s correction is used for statistical analysis. Asterisks indicate significant differences between WT and bos1-1 mutant (*p < 0.05, **p < 0.01; Student’s t-test). ns indicates no significant difference. Primers used in the assay are listed in Supplementary Table S2.
To study genomic variations (GVs) and haplotypes, genomic sequences of BOS1 gene were analyzed through the RiceVarMap2 (ricevarmap.ncpgr.cn) website. In BOS1 locus, there were eight genomic variations caused by SNP. Five of the genomic variations were distributed in the exon of BOS1 gene, four of which were missense mutations (Figures 6A, B).
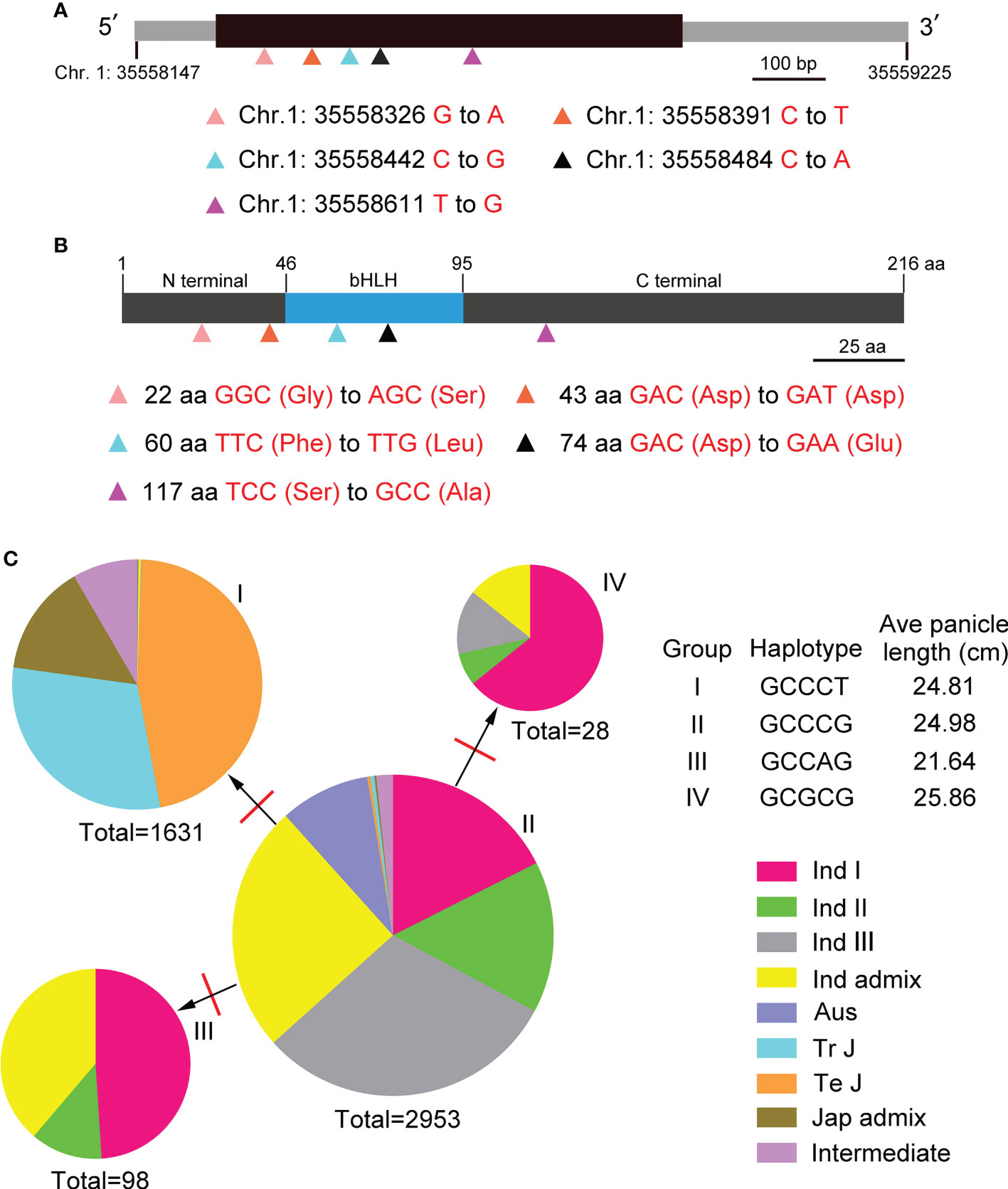
Figure 6 Genetic variation and haplotype network analyses of BOS1. (A) The gene structure of BOS1 and location of the five genomic variations. Black box represents exon, and gray boxes represent the 5′ untranslated region (5′UTR) and 3′UTR. Scale bar is 100 bp. (B) The protein structure of BOS1 and location of the five genomic variations. Gray boxes represent the N-terminal and C-terminal, and blue boxes represent the bHLH domain. Scale bar is 25 aa. (C) A haplotype network of the five genomic variations. The red lines represent the number of mutations between two haplotypes. WT, wild type.
The haplotypes of BOS1 in 4,726 rice landraces, including Indica I, Indica II, Indica III, Indica Intermediate, Aus, Temperate Japonica, Tropical Japonica, Japonica Intermediate, and Intermediate ecotypes, were also analyzed. Four haplotypes were obtained, and a haplotype network was built (Figure 6C, Supplementary Table S4). The proportion of haplotype II in the total number of rice ectypes was the largest, which contained all types of rice, indicating that haplotype II was the main haplotype of BOS1. One mutation changed haplotype II to haplotype I. Haplotype I had the japonica rice. One mutation changed haplotype II to haplotype III and IV. Haplotypes III and IV only had indica rice ecotypes. In addition, we also found that Aus, Temperature Japonica, and Tropical Japonica only contained two haplotypes, while Indica I and Indica II contained four haplotypes, indicating that BOS1 has evolved into high diversity in different varieties (Figure 6C, Supplementary Table S7). We analyzed the average panicle length under different haplotypes and found that the average panicle length of haplotype IV was the longest, the average panicle length of haplotype III was the shortest, and the panicle length of haplotype I and II was similar.
Rice generates raceme panicles and normally comprises primary and secondary branches, which bear single-flowered spikelets (Kyozuka et al., 2014; Zhu et al., 2022). Rice panicle development plays critical roles in yield formation, which is defined by IMs, PBMs, SBMs, SMs, and FMs produced by SAMs and AMs (Itoh et al., 2005; Kyozuka et al., 2014). To further understand the mechanisms of panicle development, a mutant with abnormal panicles, named bos1-1, was obtained using EMS mutagenesis in this study. BOS1 gene was cloned. The regulatory roles of BOS1 on rice panicle development were explored.
We isolated BOS1 gene by applying the positional cloning method and MutMap strategy (Figure 3). BOS1 is a small gene, and the CDS of BOS1 gene was only 648 bp without intron encoding a bHLH domain-containing transcription factor (Figure 3). A single amino acid substitution, Tyr to Asn, in the bHLH domain altered BOS1 functions. The bos1-1 mutation conferred abnormal panicle phenotypes, including abortion of lateral spikelets, increased panicle length and grain size, reduced number of primary branches and secondary branches, and decreased grain number per panicle and seed setting rate (Figures 1, 2). Genomic DNA of BOS1 was able to recover the defects in bos1-1 mutant, suggesting mutation in BOS1 locus is responsible for bos1-1 mutant phenotype. In rice, BOS1 was previously cloned and known as LAX1 (Komatsu et al., 2001; Komatsu K. et al., 2003; Oikawa and Kyozuka, 2009). BOS1 encodes a bHLH domain-containing transcription factor in the B subfamily (Li et al., 2006). However, compared with other members in the A and B subfamily, the sequence similarity of BOS1 was lower, indicating BOS1 may function independently with other members in the B subfamily (Figure 3H). Mutations of BOS1/LAX1 exhibited more severe phenotypes, suggesting that BOS1 plays important roles in panicle development.
Previous studies have shown that the BOS1 gene is expressed at the boundary between SAM and the new meristem region, indicating that BOS1 gene is a major regulator of axillary meristem formation (Komatsu M. et al., 2003; Matin and Kang, 2011). In this study, we found that BOS1 was predominantly expressed in young panicles but was expressed at a lower level in other tissues or organs detected, such as root, leaf, sheath, and stem (Figure 4B). The BOS1 expression profile corresponds to the severe panicle phenotypes of bos1-1 mutant. The vegetative branching of bos1-1 and lax1 mutants is normal, but the axillary meristems were severely blocked in the mutants’ reproductive stage, revealing that BOS1 is active in the reproductive stage. The results of subcellular localization showed that BOS1 protein was mainly localized in the nucleus, which is consistent with its potential functions as a transcription factor (Figure 4A). In addition, several hormone response elements, including auxin, GA, ABA, SA, and JA response elements, in the BOS1 promoter were identified, suggesting that BOS1 may be induced by phytohormones (Figure 4C). As expected, the expression of BOS1 was activated by auxin, BR, ABA, SA, and JA, suggesting that BOS1 may control panicle development by regulating hormone biosynthesis or signaling (Figure 4D). More efforts will be put to dissect the principles.
The bHLH domain-containing proteins are one of the largest transcription factor families having 167 members in rice (Li et al., 2006). The bHLH domain has two adjacent regions with distinctive functions: the basic region and the HLH region. The HLH region mediates protein interaction, allowing the formation of homodimers or heterodimers (Pires and Dolan, 2010). The basic region functions as a DNA-binding motif. It has been known that bHLH domain-containing proteins can bind to cis-elements known as E-box, G-box, PIF-binding E (PBE) box, and N-box (Pires and Dolan, 2010). As a potential transcription factor, BOS1 may control rice panicle development by regulating gene expression. In this study, the differential expression of rice panicle development-related genes between WT and bos1-1 was examined. The results showed that APO1 and FZP were differential expressed. FZP determines the transition from SM to FM (Komatsu M. et al., 2003; Zhu et al., 2003). APO1 is a known regulator of the transition from IM to SM (Ikeda et al., 2005; Ikeda et al., 2007; Ikeda-Kawakatsu et al., 2012).
We also found that the expression of OsPIN2 and OsPIN3 is differential between WT and bos1-1 mutant. OsPIN2 gene encodes an auxin efflux carrier that possibly regulates the auxin from the root tip to the root-elongation zone in rice (Inahashi et al., 2018). Overexpression of OsPIN2 leads to a shorter plant height, more tillers, and a large tiller angle (Chen et al., 2012). Expression of OsPIN3 gene is involved in polar auxin transport and induced by osmotic stress in rice. Overexpression of OsPIN3 improves drought tolerance (Zhang et al., 2012). It is also reported that OsPIN1 and OsPIN3 are involved in vegetative axillary meristem specification and outgrowth in rice (Deshpande et al., 2015). Barren Stalk 1 (BA1) is the homolog of BOS1 in maize. BA1 physically interacts with Barren Inflorescence 2 (BIF2) and controls auxin-mediated axillary meristem initiation (Gallavotti et al., 2004; Skirpan et al., 2008). The phenotypes of ba1 are similar to the bif2 mutant (Mcsteen and Hake, 2001; Ritter et al., 2002; Skirpan et al., 2008). The expression of BA1 is induced by auxin. In ba1 mutant, no auxin accumulation in the newly initiated inflorescence meristems is examined (Wu and Mcsteen, 2007; Gallavotti et al., 2008). These results suggest that OsPIN2, OsPIN3, APO1, and FZP may be the direct or indirect targets for BOS1 to control panicle development. More efforts are required to confirm this hypothesis in the future.
BOS1 is a small gene without introns, and the genomic DNA of BOS1 gene is only 1,078 bp. The results of genomic variation analysis in 4,726 rice accessions showed that eight SNPs were found. Five GVs were distributed in exons, four of which were missense mutations, and the remaining one is synonymous mutations (Figures 5A, B). Four haplotypes of the GVs in BOS1 locus were obtained. Haplotype II was the ancestor one because all types of rice had this haplotype and the frequencies were the highest. The haplotype network of BOS1 gene indicated that one mutation changed haplotype II to haplotypes I, III, and IV. BOS1 protein sequence was limitedly conserved with other members, even in the bHLH domain (Li et al., 2006). It was interesting that BOS1 contains a divergent non-synonymous polymorphism (Chr. 1: 35558611 T to G). According to the database of RiceVarMap2, 99.70% of indica varieties contain G, whereas 98.20% of japonica varieties contain T. It is likely that BOS1 might be involved in rice subspecies divergence.
In conclusion, panicle defective mutant bos1-1 was obtained by EMS mutagenesis. Mutations in BOS1 gene caused severe defects in panicle development and other related agronomic traits. BOS1 gene was isolated by combining positional cloning and MutMap methods, which encoded a bHLH domain-containing protein. A single amino acid change disrupted the bHLH domain and conferred the bos1-1 mutant phenotype. BOS1 gene was predominantly expressed in young panicles and was induced by phytohormones. BOS1 protein was localized in the nucleus. The expression of panicle development-related genes, such as OsPIN2, OsPIN3, APO1, and FZP, was changed by bos1-1 mutation, suggesting the genes may be the direct or indirect targets of BOS1 to regulate panicle development. BOS1 gene encountered genomic variations. These results laid the foundation to further dissect the functions of BOS1.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XW and YL designed the study. YL, XZ and YH generated the genetic and mapping population. YL and XZ prepared the samples for MutMap. YL cloned the gene. YL and SL made the construct and generated the transgenic lines. YL observed the phenotype of the mutant and complementary transgenic lines. YL and YY performed expression pattern and subcellular localization experiments. YL and SL aligned the protein sequence. YL examined the expression of panicle development-related genes by quantitative RT-PCR. YL and YH analyzed the GVs, haplotypes, and haplotype networks. XW and YL prepared the figures and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32070642 and 31371222 to Dr. XW).
We thank the editors and reviewers for their comments on the manuscript. We also thank the National Natural Science Foundation of China (32070642 and 31371222 to Dr. XW) for its support of this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1162828/full#supplementary-material
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Chen, Y., Fan, X., Song, W., Zhang, Y., Xu, G. (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149. doi: 10.1111/j.1467-7652.2011.00637.x
Cui, Z., Tong, A., Huo, Y., Yan, Z., Yang, W., Yang, X., et al. (2017). SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in arabidopsis. BMC Biol. 15, 80. doi: 10.1186/s12915-017-0422-2
Deshpande, G. M., Ramakrishna, K., Chongloi, G. L., Vijayraghavan, U. (2015). Functions for rice RFL in vegetative axillary meristem specification and outgrowth. J. Exp. Bot. 66, 2773–2784. doi: 10.1093/jxb/erv092
Gallavotti, A., Yang, Y., Schmidt, R. J., Jackson, D. (2008). The relationship between auxin transport and maize branching. Plant Physiol. 147, 1913–1923. doi: 10.1104/pp.108.121541
Gallavotti, A., Zhao, Q., Kyozuka, J., Meeley, R. B., Ritter, M. K., Doebley, J. F., et al. (2004). The role of barren stalk1 in the architecture of maize. Nature 432, 630–635. doi: 10.1038/nature03148
Gao, X., Liang, W., Yin, C., Ji, S., Wang, H., Su, X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711
Huang, X., Kurata, N., Wei, X., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Ikeda, K., Ito, M., Nagasawa, N., Kyozuka, J., Nagato, Y. (2007). Rice ABERRANT PANICLE ORGANIZATION 1, encoding an f-box protein, regulates meristem fate. Plant J. 51, 1030–1040. doi: 10.1111/j.1365-313X.2007.03200.x
Ikeda, K., Nagasawa, N., Nagato, Y. (2005). ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 282, 349–360. doi: 10.1016/j.ydbio.2005.03.016
Ikeda-Kawakatsu, K., Maekawa, M., Izawa, T., Itoh, J., Nagato, Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180. doi: 10.1111/j.1365-313X.2011.04781.x
Inahashi, H., Shelley, I. J., Yamauchi, T., Nishiuchi, S., Takahashi-Nosaka, M., Matsunami, M., et al. (2018). OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant 164, 216–225. doi: 10.1111/ppl.12707
Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., et al. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46, 23–47. doi: 10.1093/pcp/pci501
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kobayashi, K., Yasuno, N., Sato, Y., Yoda, M., Yamazaki, R., Kimizu, M., et al. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24, 1848–1859. doi: 10.1105/tpc.112.097105
Komatsu, M., Chujo, A., Nagato, Y., Shimamoto, K., Kyozuka, J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850. doi: 10.1242/dev.00564
Komatsu, M., Maekawa, M., Shimamoto, K., Kyozuka, J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373. doi: 10.1006/dbio.2000.9988
Komatsu, K., Maekawa, M., Ujiie, S., Satake, Y., Furutani, I., Okamoto, H., et al. (2003). LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. U.S.A. 100, 11765–11770. doi: 10.1073/pnas.1932414100
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Kyozuka, J., Tokunaga, H., Yoshida, A. (2014). Control of grass inflorescence form by the fine-tuning of meristem phase change. Curr. Opin. Plant Biol. 17, 110–115. doi: 10.1016/j.pbi.2013.11.010
Lee, D. Y., An, G. (2012). Two AP2 family genes, supernumerary bract (SNB) and osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69, 445–461. doi: 10.1111/j.1365-313X.2011.04804.x
Lee, D. Y., Lee, J., Moon, S., Park, S. Y., An, G. (2007). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49, 64–78. doi: 10.1111/j.1365-313X.2006.02941.x
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Li, D., Xu, H., Sun, X. X., Cui, Z. B., Zhang, Y., Bai, Y. G., et al. (2015). Differential transformation efficiency of japonica rice varieties developed in northern China. Crop Breed. Appl. Biotechnol. 15, 162–168. doi: 10.1590/1984-70332015v15n3a28
Li, S. Y., Zhao, B. R., Yuan, D. Y., Duan, M. J., Qian, Q., Tang, L., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. U. S. A. 110, 3167–3172. doi: 10.1073/pnas.1300359110
Liu, C., Teo, Z. W., Bi, Y., Song, S., Xi, W., Yang, X., et al. (2013). A conserved genetic pathway determines inflorescence architecture in arabidopsis and rice. Dev. Cell 24, 612–622. doi: 10.1016/j.devcel.2013.02.013
Matin, M. N., Kang, S. G. (2011). Genetic and phenotypic analysis of lax1-6, a mutant allele of LAX PANICLE1 in rice. J. Plant Biol. 55, 50–63. doi: 10.1007/s12374-011-9189-0
Mcsteen, P., Hake, S. (2001). Barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128, 2881–2891. doi: 10.1242/dev.128.15.2881
Mcsteen, P., Leyser, O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56, 353–374. doi: 10.1146/annurev.arplant.56.032604.144122
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Oikawa, T., Kyozuka, J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21, 1095–1108. doi: 10.1105/tpc.108.065425
Pires, N., Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Ren, D., Li, Y., Zhao, F., Sang, X., Shi, J., Wang, N., et al. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 162, 872–884. doi: 10.1104/pp.113.216044
Ritter, M. K., Padilla, C. M., Schmidt, R. J. (2002). The maize mutant barren stalk1 is defective in axillary meristem development. Am. J. Bot. 89, 203–210. doi: 10.3732/ajb.89.2.203
Sazuka, T., Kamiya, N., Nishimura, T., Ohmae, K., Sato, Y., Imamura, K., et al. (2009). A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 60, 227–241. doi: 10.1111/j.1365-313X.2009.03952.x
Skirpan, A., Wu, X., Mcsteen, P. (2008). Genetic and physical interaction suggest that BARREN STALK 1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J. 55, 787–797. doi: 10.1111/j.1365-313X.2008.03546.x
Tabuchi, H., Zhang, Y., Hattori, S., Omae, M., Shimizu-Sato, S., Oikawa, T., et al. (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287. doi: 10.1105/tpc.111.088765
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Tanaka, W., Toriba, T., Ohmori, Y., Yoshida, A., Kawai, A., Mayama-Tsuchida, T., et al. (2012). The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 24, 80–95. doi: 10.1105/tpc.111.094797
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Tilman, D., Balzer, C., Hill, J., Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Tsuji, H., Taoka, K., Shimamoto, K. (2013). Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr. Opin. Plant Biol. 16, 228–235. doi: 10.1016/j.pbi.2013.01.005
Wang, F., Han, T., Song, Q., Ye, W., Song, X., Chu, J., et al. (2020). The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell 32, 3124–3138. doi: 10.1105/tpc.20.00289
Wang, B., Smith, S. M., Li, J. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
Wang, X., Wu, F., Xie, Q., Wang, H., Wang, Y., Yue, Y., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in arabidopsis. Plant Cell 24, 3278–3295. doi: 10.1105/tpc.112.100081
Wu, X., Mcsteen, P. (2007). The role of auxin transport during inflorescence development in maize (Zea mays, poaceae). Am. J. Bot. 94, 1745–1755. doi: 10.3732/ajb.94.11.1745
Xing, Y., Zhang, Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. doi: 10.1146/annurev-arplant-042809-112209
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2004). A novel zinc-binding motif revealed by solution structures of DNA-binding domains of arabidopsis SBP-family transcription factors. J. Mol. Biol. 337, 49–63. doi: 10.1016/j.jmb.2004.01.015
Yan, B., Zheng, H., Sang, Y., Wang, Y., Sun, J., Li, F., et al. (2022). A single amino acid substitution in MIL1 leads to activation of programmed cell death and defense responses in rice. Int. J. Mol. Sci. 23 (16), 8853. doi: 10.3390/ijms23168853
Yoshida, H., Nagato, Y. (2011). Flower development in rice. J. Exp. Bot. 62, 4719–4730. doi: 10.1093/jxb/err272
Yoshida, A., Ohmori, Y., Kitano, H., Taguchi-Shiobara, F., Hirano, H. Y. (2012). Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 70, 327–339. doi: 10.1111/j.1365-313X.2011.04872.x
Yoshida, A., Sasao, M., Yasuno, N., Takagi, K., Daimon, Y., Chen, R., et al. (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. U.S.A. 110, 767–772. doi: 10.1073/pnas.1216151110
Yoshida, A., Suzaki, T., Tanaka, W., Hirano, H. Y. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. U.S.A. 106, 20103–20108. doi: 10.1073/pnas.0907896106
Zahn, L. M., Kong, H., Leebens-Mack, J. H., Kim, S., Soltis, P. S., Landherr, L. L., et al. (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223. doi: 10.1534/genetics.104.037770
Zeng, D., Tian, Z., Rao, Y., Dong, G., Yang, Y., Huang, L., et al. (2017). Rational design of high-yield and superior-quality rice. Nat. Plants 3, 17031. doi: 10.1038/nplants.2017.31
Zhang, Q., Li, J., Zhang, W., Yan, S., Wang, R., Zhao, J., et al. (2012). The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 72, 805–816. doi: 10.1111/j.1365-313X.2012.05121.x
Zhang, D., Yuan, Z. (2014). Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65, 553–578. doi: 10.1146/annurev-arplant-050213-040104
Zhao, H., Li, J., Yang, L., Qin, G., Xia, C., Xu, X., et al. (2021). An inferred functional impact map of genetic variants in rice. Mol. Plant 14, 1584–1599. doi: 10.1016/j.molp.2021.06.025
Zhao, H., Yao, W., Ouyang, Y., Yang, W., Wang, G., Lian, X., et al. (2015). RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res. 43, D1018–D1022. doi: 10.1093/nar/gku894
Zhao, C., Zhu, M., Guo, Y., Sun, J., Ma, W., Wang, X. (2022). Genomic survey of PEBP gene family in rice: Identification, phylogenetic analysis, and expression profiles in organs and under abiotic stresses. Plants (Basel) 11 (12), 35736727. doi: 10.3390/plants11121576
Zhou, D., Shen, W., Cui, Y., Liu, Y., Zheng, X., Li, Y., et al. (2021). APICAL SPIKELET ABORTION (ASA) controls apical panicle development in rice by regulating salicylic acid biosynthesis. Front. Plant Sci. 12, 636877. doi: 10.3389/fpls.2021.636877
Zhou, S., Zhu, S., Cui, S., Hou, H., Wu, H., Hao, B., et al. (2021). Transcriptional and post-transcriptional regulation of heading date in rice. New Phytol. 230, 943–956. doi: 10.1111/nph.17158
Zhu, Q. H., Hoque, M. S., Dennis, E. S., Upadhyaya, N. M. (2003). Ds tagging of BRANCHED FLORETLESS 1 (BFL1) that mediates the transition from spikelet to floret meristem in rice (Oryza sativa l). BMC Plant Biol. 3, 6. doi: 10.1186/1471-2229-3-6
Zhu, M., Hu, Y., Tong, A., Yan, B., Lv, Y., Wang, S., et al. (2021). LAZY1 controls tiller angle and shoot gravitropism by regulating the expression of auxin transporters and signaling factors in rice. Plant Cell Physiol. 61, 2111–2125. doi: 10.1093/pcp/pcaa131
Keywords: BOS1, bos1-1, bHLH transcription factor, panicle development, panicle branch, rice
Citation: Lv Y, Zhang X, Hu Y, Liu S, Yin Y and Wang X (2023) BOS1 is a basic helix–loop–helix transcription factor involved in regulating panicle development in rice. Front. Plant Sci. 14:1162828. doi: 10.3389/fpls.2023.1162828
Received: 10 February 2023; Accepted: 03 April 2023;
Published: 26 April 2023.
Edited by:
Chengzhen Liang, Institute of Biotechnology (CAAS), ChinaReviewed by:
Wenchao Yin, Institute of Crop Sciences (CAAS), ChinaCopyright © 2023 Lv, Zhang, Hu, Liu, Yin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxue Wang, d2FuZ3h4QHN5YXUuZWR1LmNu; eGlhb3h1ZXdhbmc2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.