- 1School of Life Science, Anhui Agricultural University, Hefei, China
- 2Tobacco Research Institute, Anhui Academy of Agricultural Sciences, Hefei, China
- 3Institute of Plant Protection and Agro-products Safety, Anhui Academy of Agricultural Sciences, Hefei, China
Terpenes are the main class of secondary metabolites produced in response to pest and germ attacks. In maize (Zea mays L.), they are the essential components of the herbivore-induced plant volatile mixture, which functioned as a direct or indirect defense against pest and germ attacks. In this study, 43 maize terpene synthase gene (ZmTPS) family members were systematically identified and analyzed through the whole genomes of maize. Nine genes, including Zm00001d032230, Zm00001d045054, Zm00001d024486, Zm00001d004279, Zm00001d002351, Zm00001d002350, Zm00001d053916, Zm00001d015053, and Zm00001d015054, were isolated for their differential expression pattern in leaves after corn borer (Ostrinia nubilalis) bite. Additionally, six genes (Zm00001d045054, Zm00001d024486, Zm00001d002351, Zm00001d002350, Zm00001d015053, and Zm00001d015054) were significantly upregulated in response to corn borer bite. Among them, Zm00001d045054 was cloned. Heterologous expression and enzyme activity assays revealed that Zm00001d045054 functioned as d-limonene synthase. It was renamed ZmDLS. Further analysis demonstrated that its expression was upregulated in response to corn borer bites and Fusarium graminearum attacks. The mutant of ZmDLS downregulated the expressions of Zm00001d024486, Zm00001d002351, Zm00001d002350, Zm00001d015053, and Zm00001d015054. It was more attractive to corn borer bites and more susceptible to F. graminearum infection. The yeast one-hybrid assay and dual-luciferase assay showed that ZmMYB76 and ZmMYB101 could upregulate the expression of ZmDLS by binding to the promoter region. This study may provide a theoretical basis for the functional analysis and transcriptional regulation of terpene synthase genes in crops.
Introduction
Volatile terpenes are widely exist in plant tissues such as leaves, flowers, and fruits. They play critical roles in pollinator attraction, herbivore predator attraction, and predator repellent. They function as essential substances for plant growth, development, and disease resistance (Dudareva et al., 2005; Yu and Utsumi, 2009; Nagegowda, 2010; Murali-Baskaran et al., 2022).
The biosynthesis of terpenoid occurs via two pathways, namely the mevalonate pathway (MVA) in the cytoplasm and the methylerythritol 4-phosphate pathway (methylerythritol pathway (MEP)) located in the plastid (Cheng et al., 2007). The synthesis of terpenoids consists of three main parts: the synthesis of intermediate isopentenyl pyrophosphate (IPP) and allyl isomer dimethyl allyl pyrophosphate (DMAPP), the formation of precursor substances, including geranylgeranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP), and the generation of the terpenes (Vranova et al., 2013; Bergman et al., 2019). Terpene synthase (TPS) catalyzed the synthesis of the terpenes at the last step by using FPP, GPP, and GGPP as substrates.
Plant terpenoids play an important role in direct and indirect defense against biotic stress. In rice, OsTPS46 catalyzed the synthesis of constitutive limonene and (E)-β-farnesene, which may play a crucial role in protection against Rhopalosiphum padi (Sun et al., 2017). OsTPS19 functions as an (S)-limonene synthase. The products of OsTPS19 (S)-limonene could inhibit the growth of Magnaporthe oryzae spores in vitro, which protected the plants from the infection of rice blast disease (Chen et al., 2018). In maize, TPS10 products (E)-α-bergamotene and (E)-β-farnesene could be consistently induced by herbivory. In addition, overexpression of TPS10 in Arabidopsis generated a volatile signal that attracted Cotesia marginiventris (Köllner et al., 2009). Similarly, the emissions of constitutive volatiles from ZmTPS8 were attractive to C. marginiventris (Fontana et al., 2011). Further analysis revealed that ZmTPS8 is a multiproduct α-copaene synthase. Its product, cubebol, exhibited significant antifungal activity against both Fusarium graminearum and Aspergillus parasiticus in vitro (Saldivar et al., 2023). Otherwise, biosynthesis and emission of (E, E)-α-farnesene triggered by caterpillars and aphids at a high density functioned as a phytochemical signal, attracting their natural enemies, Diadegma semiclausum parasitoids in Arabidopsis (Kroes et al., 2017). In addition, three terpene alcohols farnesol, nerolidol, and plaunotol, are toxic and have antibacterial effects on Staphylococcus aureus (Inoue et al., 2004).
Recently, TPS genes and their families have been identified in the genomes of Arabidopsis thaliana, Solanum lycopersicum, and Oryza sativa (Aubourg et al., 2002; Chen et al., 2011; Falara et al., 2011). In maize, more than a dozen TPS genes have been isolated and analyzed (Shen et al., 2000; Ren et al., 2016; Block et al., 2019; Köllner et al., 2020). In this study, the TPS gene families were systematically identified and analyzed. The genes expressed explicitly in the leaf were tested in response to corn borer bite. One TPS gene named ZmDLS was cloned and functionally analyzed. These results may provide new insight into the vital role of TPS genes in plants’ responses to insect and pathogen attacks.
Materials and methods
Plant material
The maize-inbred line B73 and its zmdls mutant were obtained from the maize Ethyl methanesulfonate (EMS)-induced mutant database (http://www.elabcaas.cn/memd/public/index.html#/). The seedlings were cultivated in a greenhouse at 26°C with 16/8-h light/dark cycle. The leaves were collected at the eighth leaf stage for the following experiments.
Identification of TPS genes from the maize genome
The hidden Markov model (HMM) file was generated using two TPS domains, PF03936 and PF01397, acquired from the Pfam database (http://pfam.xfam.org/). It was then used as a query to blast against the B73 RefGen v4 protein database (https://download.maizegdb.org/Zm-B73-REFERENCE-GRAMENE-4.0/) by HMMER v3.3.2 (http://www.hmmer.org/). The candidate sequences were validated using the SMART and NCBI CDD (http://smart.embl-heidelberg.de/). Finally, the ExPASy (http://web.expasy.org/protparam/) was employed to examine candidate sequences. Also, Wolfpsort (https://wolfpsort.hgc.jp/) was applied to predict the subcellular localization of TPS genes.
The alignment and phylogenetic analysis of TPS proteins
Based on the Cluster W tool, the candidate sequences were aligned with TPS proteins from Oryza sativa and Arabidopsis thaliana (Aubourg et al., 2002; Chen et al., 2011). The neighbor-joining tree was constructed with a 1,000 bootstrap value using MGEA-X.
Analysis of gene structure and recognition of conserved motifs
The TBtools (http://www.tbtools.com/) were employed to analyze the exon-intrinsic structure. The MEME software (http://meme-suite.org/) was used to predict the conservative pattern of ZmTPS as described by Bailey et al. (2009). The number of discovered motifs ranges from 1 to 10. The width of the motifs was set as greater than 6 and less than 200, respectively.
Identification of cis-acting elements in ZmTPS promoter
A 2,000-bp upstream promoter sequence was downloaded for each TPS gene from the maize genome. The online tools PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and New PLACE (https://www.dna.affrc.go.jp/PLACE/?action=newplace) were used to identify the cis-acting regulatory elements.
Chromosomal distribution and genetic interactions of TPS gene
The chromosomal locus information for the TPS gene was obtained from the Maize Genetics and Genomics Database (https://www.maizegdb.org/) (Portwood et al., 2019). The physical locations and genetic interactions were predicted by TBtools software (Chen et al., 2020).
Tissue-specific expression analysis of ZmTPS genes
To analyze the tissue-specific expression pattern of the ZmTPS family genes, raw data of the transcriptome of 23 different tissues (mature leaf, primary root, mature pollen, and silk, etc.) for B73 was downloaded from qTeller of maizeGDB (https://qteller.maizegdb.org/). Fragments per kilobase of transcript per million mapped read (FPKM) values were used to evaluate the relative transcription abundance. The TBtools were employed to draw the heat maps. The gradient color from blue to red indicates the log2 fold change value from FPKM data.
Prokaryotic expression and enzyme activity assays
The constructed pMAL-c2x-Zm00001d045054 recombinant plasmid was introduced into BL21(DE3) expression strains and screened for positive clones. Subsequently, 50 μL of the bacterial solution was aspirated with a pipette under aseptic conditions and added into 5 mL of Luria–Bertani (LB) medium containing ampicillin in a centrifuge tube. After incubating at 37 °C and shaking at 200 r/min overnight, 2 mL of turbid bacterial solution was added into 100 mL of LB medium containing the ampicillin and shaking at 200 r/min for 2–3 h at 37 °C until the bacterial solution reached an OD600 = 0.6–0.8.
Isopropyl β-d-thiogalactoside (IPTG) with a final concentration of 0.5 mM was used to induce the recombinant protein expression. The bacteria solution was induced overnight at 18 °C and then centrifuged at 11,000×g at 4 °C for 10 min. The precipitate was resuspended in a 3-mL buffer solution containing 20 mM Tris-HCl at pH 8.0, 10 mM dithiothreitol (DTT), 5 mM Na2S2O5, and 10 % glycerin to generate a crude enzyme solution after subsequent ultrasonication (five pulses of 20 s at 15 W, 4 °C) (Yahyaa et al., 2015b). The catalytic activity of recombinant Zm00001d045054 was analyzed using geranyl diphosphate (GPP) as a substrate. Enzyme activity was determined in a headspace vial containing 500 µL of crude enzyme, 50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 10 µM MnCl2, and 10 µM GPP in a final total volume of 1 mL (Yahyaa et al., 2015a). The solid-phase microextraction (SPME) with a 100-µm polydimethylsiloxane (PDMS) fiber was then inserted into the headspace bottle. After extraction for 60 min at room temperature, the samples were analyzed using GC-MS to identify volatile terpenes (Li et al., 2021). The components of volatile terpenes were verified by comparing them with the National Institute of Standards and Technology (NIST) 2011 Standard Spectrum Library (https://www.nist.gov/srd/nist-standard-reference-database-1a).
Identification of ZmMYBs associated with the control of ZmDLS expression
The raw data of transcriptomes of 23 different tissues for B73 were downloaded from qTeller of maizeGDB (https://qteller.maizegdb.org/). The FPKM values were used to evaluate the relative transcription abundance and construct the gene co-expression network for ZmDLS and the 157 ZmMYBs that have been reported (Du et al., 2012). The cor function in R language (https://www.r-project.org/) was used to calculate the p- and r-values of these gene pairs. The ZmMYBs with values p < 0.01 and r > 0.6 were selected for further analysis.
Yeast one hybrid assay
The ZmDLS promoter cis-acting elements were predicted using the online tools PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and New PLACE (https://www.dna.affrc.go.jp/PLACE/?action=newplace) (Lescot et al., 2002).
The ZmDLS promoter fragment and the full-length ZmMYBs were cloned into pAbAi and pGADT7 vectors, and then the pAbAi-ZmDLS vector was linearized and integrated into the Y1HGold yeast genome. The plasmid pGADT7-ZmMYBs were transformed into decoy strains, screened for positive clones, and coated in SD/-Leu solid medium containing 100 ng·mL−1 Aureobasidin A (AbA) (Supplementary Figure S5) (Li et al., 2017). Three biological replicates were created in the experiment.
Subcellular localization of ZmDLS and ZmMYBs
The full-length CDS of ZmDLS was cloned into the pCAMBIA1305 vector, forming a fusion construct pCAMBIA1305-ZmDLS-GFP. Then, it was transformed into Agrobacterium tumefaciens strain GV3101 and subsequently inoculated in the leaves of 4-week-old Nicotiana tabacum as described by Wang et al. (2021). The empty vector was used as a negative control. In detail, the leaves of Nicotiana tabacum were cut into about 1.0-mm-long strips and then soaked in the enzymatic digestion solution (Supplementary Table S6). To digest the cell walls, the incubation was conducted at room temperature by shaking at 40 rpm/min for 5–7 h. After filtering with a 100-mesh cell strainer, the tube containing the solution was centrifuged at 11,000×g at 4 °C for 10 min. The precipitate was washed with a W5 solution. Finally, the protoplasts were re-suspended in MMg solution (100 μL) and observed under laser confocal microscopy.
Similarly, fusion constructs pCAMBIA1305-ZmMYB-GFP were transformed into Agrobacterium tumefaciens strain GV3101. Then, it was co-cultured with 1 cm2 fresh onions epidermis at 22 °C for 20 h in the dark in the co-culture solid medium (½ MS, 1 % sucrose, 0.03 % casein, 0.28 % proline, 10 μmol/L 2,4-d, 2 μmol/L 6-BA, 200 μmol/L As, and 0.8 % agar) (Thakur et al., 2021). The epidermis was observed and documented under laser confocal microscopy.
Dual-luciferase assay
Approximately 1,200 bp of the ZmDLS promoter were cloned into the pGreenII-0800-LUC vector to create the pZmDLSpro : LUC reporter construct. The full-length cDNAs of ZmMYB76 and ZmMYB101 were cloned into the pGreenII 62-SK vector. The sequences of all primers are provided in Supplementary Table S5. The constructs were introduced into Agrobacterium GV3101 and then co-transformed into the leaves of Nicotiana tabacum. One-half part of the leaf was injected with pGreenII 0800-ZmDLSpro-LUC plus pGreenII 62-SK-ZmMYB infestation solution, and the other part was injected with pGreenII 0800-ZmDLSpro-LUC bacterial solution as the control. The enzymatic activities of firefly and Renilla luciferases were measured with a multifunctional enzyme labeler (Thermo Fisher, Carlsbad, California, USA) using the Dual‐Luciferase Assay Kit (Beyotime, Shanghai, China). The luciferase activity was calculated as described by Yeasn’s instructions.
RNA isolation and qRT-PCR analysis
The maize inbred line B73 and zmdls mutant leaves were collected at the eight-leaf stage. Total RNA was extracted with the RNA easy Plant Kit (Best Technology, Beijing, China) and then reverse transcribed to cDNA using MonScript™ RTIII Super Mix with dsDNase (Monad, Suzhou, Jiangsu, China). The qRT-PCR reaction system contained 50 ng of cDNA, 0.2 μM of forward primer, 0.2 μM of reverse primer, and 1× Super Specificity qPCR Mix with a final volume of 20 μL. qRT-PCR analysis was conducted on a Real-time PCR detection system (Bio-Rad, Hercules, California, USA) using SYBR® Green qPCR mix (MonScript, Monad, Suzhou, Jiangsu, China) following Li’s method (Li Z et al., 2020). The one-way ANOVA was used to estimate the significant differences between control and treatment based on the Tukey method.
Insect feeding experiment
Four leaf discs with a diameter of 2 cm from the zmdls mutant and the wild type were placed on Petri dishes. In total, 20 samples of second-instar larvae of corn borer were transferred to the center of the dish in a chamber at 27 °C, relative humidity 60 %, and light duration 16/8-h light/dark cycle. According to Cao’s method, the percentage of surface area was calculated at 3, 6, 9, and 12 h (Cao et al., 2022). Otherwise, 30 third-instar larvae were fed with leaves from the zmdls mutant and wild type. After 4 days of feeding, each larva was weighed.
In vitro antibacterial assay of d-limonene
The antibacterial activity of d-limonene was determined using the mycelial growth rate method (Li J. et al., 2022). The F. graminearum was placed on PDA mediums containing 0, 100, 200, and 400 μg·L−1 d-limonene, respectively. After incubating at 28 °C for 7 days, the mycelium in control was covered all over the petri dish, and the diameter of the colony was measured to calculate the inhibition rate as described by Zhang et al. (2021). To detect the infection of F. graminearum to the leaves of maize, the F. graminearum was cultured with a synthetic low-nutrient medium (SNA, 7.35 mM KH2PO4, 6.71 mM KCl, 9.89 mM KNO3, 1.11 mM Glucose, 0.58 mM Sucrose, 2.03 mM MgSO4.7H2O, 3.17 mM Agar) and incubated at 28 °C for 5–7 days until the concentration accounted for 106 spores·mL−1. According to Cao’s method, the leaves were inoculated with spore suspension (Cao et al., 2022). Every treatment group contained four leaves and was incubated at 28 °C with 60 % humidity. The infected area of the leaves was calculated and documented with a camera at 3-6 d of incubation.
Results
Identification of the TPS gene family in maize
To systematically identify TPS genes in maize, a hidden Markov model (HMM) profile of the conserved C-terminal (PF03936) and N-terminal (PF01397) domains in TPS proteins was used as a root file to blast against the maize genome database (B73 RefGen v4 protein database). Significant hits with an e-value of < 10−5 were retrieved as candidate TPS proteins. The SMART and NCBI CDD online tools were used to verify the TPS genes. As shown in Figure 1, 43 sequences were identified and annotated as ZmTPS genes directly represented by the gene IDs (Supplementary Table S1). Further analysis revealed that the amino acid sequences of ZmTPSs ranged from 121 to 855 aa. The molecular weights were between 14.27 and 97.57 kDa. The isoelectric point changed from 4.78 to 7.47. The grand average of hydropathicity varied from 0.059 to −0.403, which showed that most of the ZmTPSs are hydrophilic proteins (Supplementary Table S3). Based on phylogenetic analysis, 43 ZmTPS genes were unevenly distributed in five TPS subfamilies. Among them, 25 ZmTPS genes were classified in the TPS-a subfamily, 2 ZmTPS genes belonged to the TPS-b subfamily, 5 ZmTPS genes were grouped into the TPS-c subfamily, 7 ZmTPS genes were categorized into the TPS-e/f subfamily, and 4 ZmTPS genes were classified into the TPS-g subfamily (Figure 1). The members from the same subfamily contained the same conserved motifs and a similar exon–intron structure (Supplementary Figure S1). Chromosomal localization and microsynteny analysis showed that most ZmTPS genes were located on chromosomes 10, 2, 1, and 4 (Supplementary Figure S2). Co-linearity analysis elucidated the relationship between the 43 ZmTPSs. As shown in Supplementary Figure S3, a total of two collinear gene pairs were identified in the maize genome. Among them, Zm00001d037092 on chromosome 6 is co-linear with Zm00001d045054 on chromosome 9. Also, Zm00001d053916 on chromosome 4 is co-linear with Zm00001d015054 on chromosome 5. Otherwise, the identification of cis-elements, including the abscisic acid responsiveness element (ABRE), MeJA-responsive motifs TGACG, gibberellin-responsive motifs, and the stress-related component of the promoter region of ZmTPS genes, predicted the possible role of ZmTPS genes in plant growth, development, and stress response (Supplementary Figure S4).
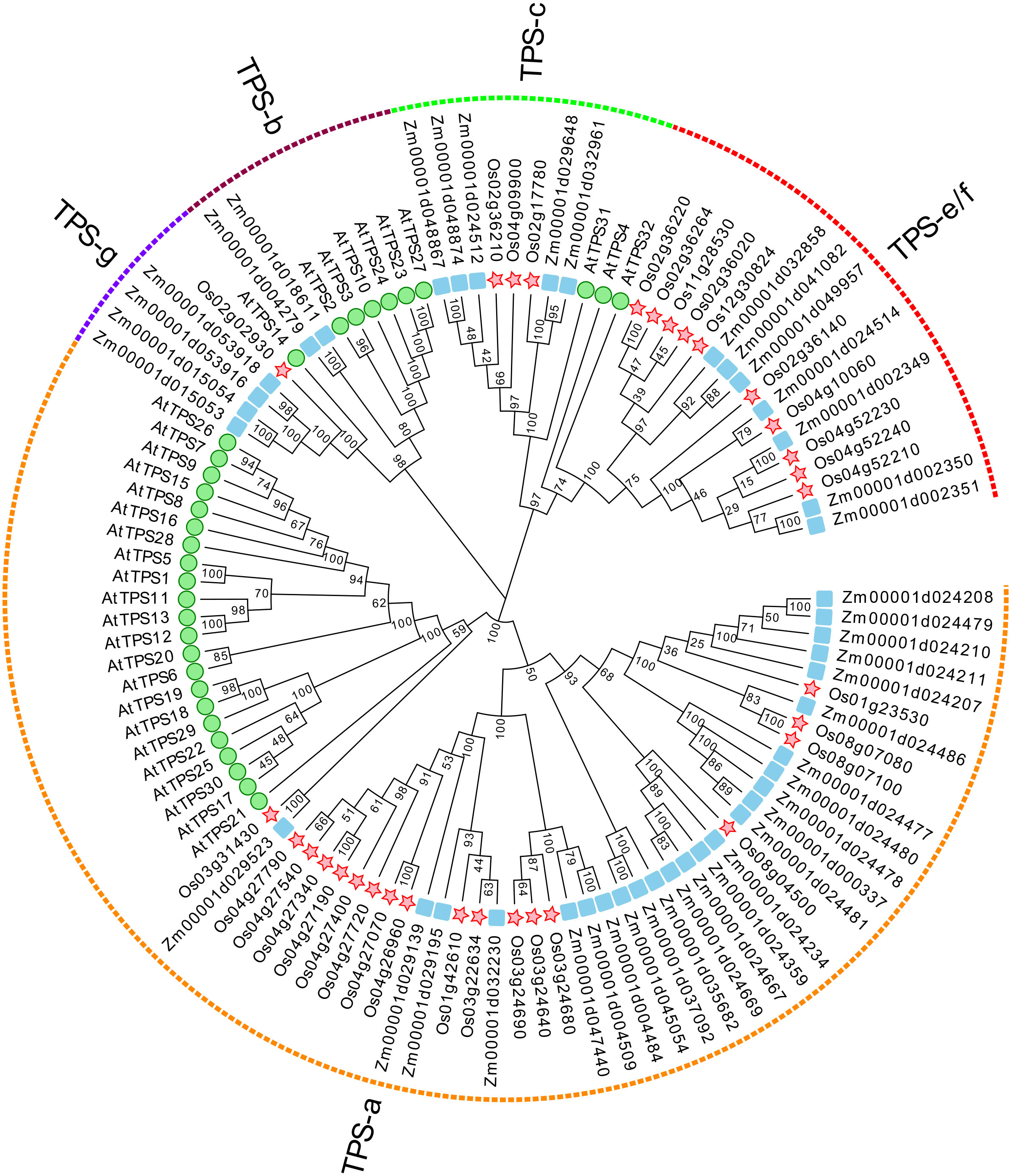
Figure 1 The phylogenetic analysis of ZmTPS proteins. The TPS proteins in Z. mays, A. thaliana, and O. sativa were used. The green circle represents the TPS genes of A. thaliana, the red pentagram represents the TPS genes of O. sativa, and the blue square represents the TPS genes of Z. mays. The neighbor-joining tree was drawn using the MEGA-X. The TPS gene family was divided into five subfamilies, designated TPS a–TPS f.
Identification of tissue-specific expressed ZmTPS genes in maize leaf
As shown in Figure 2, the expression patterns of ZmTPS genes in 23 tissues of maize were analyzed. The results exhibited a distinct tissue-specific expression pattern for 43 ZmTPS genes. Among them, Zm00001d041082, Zm00001d024211, Zm00001d024208, Zm00001d024210, Zm00001d032858, Zm00001d024207, Zm00001d029648, Zm00001d024477, and Zm00001d024478 displayed the highest expression level in the germination of kernels. Zm00001d024481, Zm00001d000337, Zm00001d024479, and Zm00001d024480 presented high expression levels in mature pollen. Zm00001d029139, Zm00001d035682, and Zm00001d004509 showed high expression levels in silk. Zm00001d024359 and Zm00001d029195 displayed the highest expression level in root-cortex. Zm00001d002351, Zm00001d024486, Zm00001d002350, Zm00001d045054, Zm00001d015054, Zm00001d053916, Zm00001d032230, Zm00001d004279, and Zm00001d015053 were found only highly expressed in mature leaf. Zm00001d024512, Zm00001d032961, Zm00001d002349, Zm00001d024669, Zm00001d037092, and Zm00001d047440 expressed explicitly in other tissues.
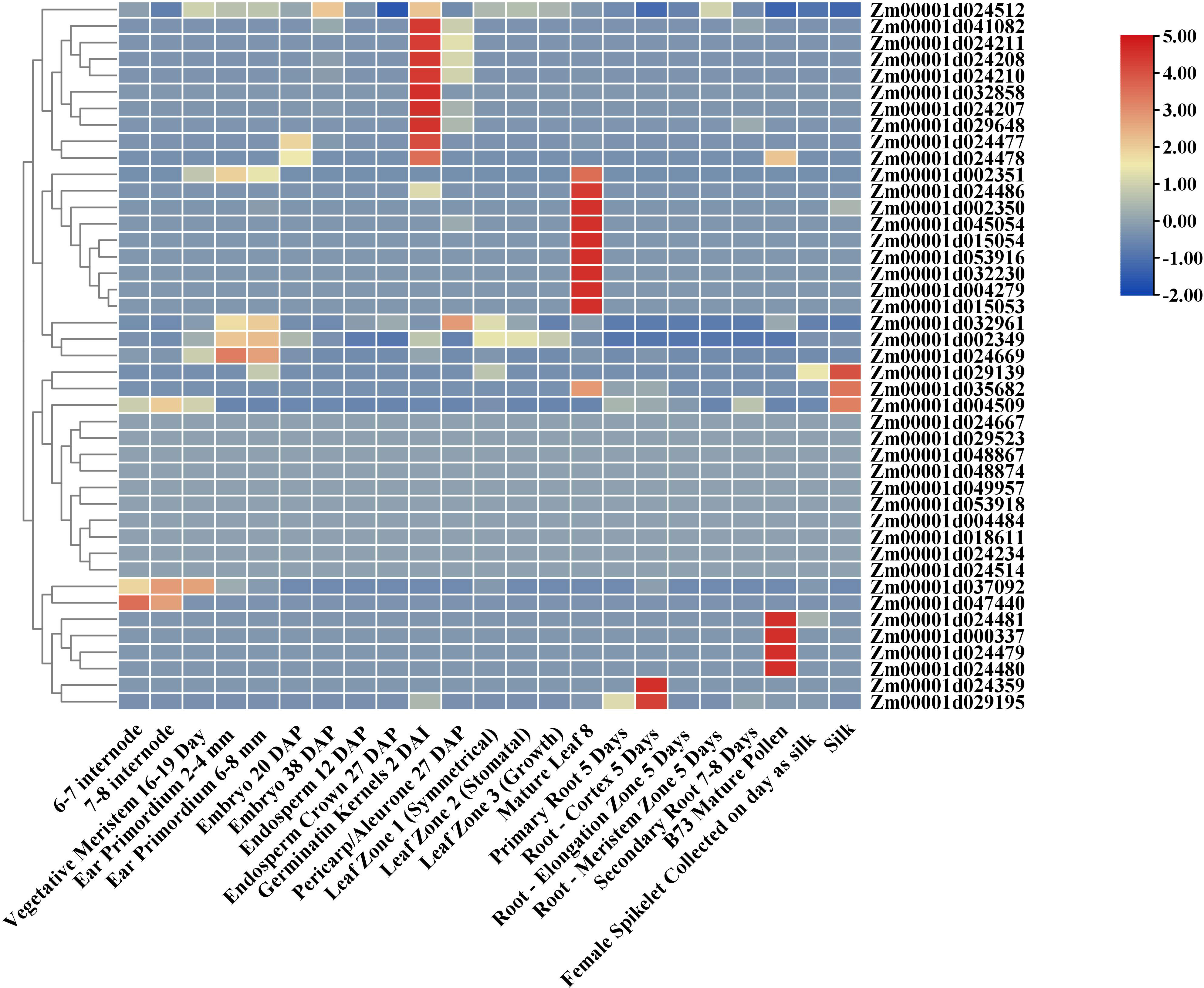
Figure 2 Expression profiles of 43 ZmTPSs in different tissues of maize. The raw data of the transcriptome of 23 different tissues for B73 were downloaded from qTeller of maizeGDB. FPKM values were used to evaluate the relative transcription abundance. The gradient color from blue to red indicates the log2 fold change value from FPKM data.
Identification of ZmDLS in response to the maize borer’s bite in the leaf
To study the role of the ZmTPS genes in the leaf, the expression of nine genes, including Zm00001d032230, Zm00001d045054, Zm00001d024486, Zm00001d004279, Zm00001d002351, Zm00001d002350, Zm00001d053916, Zm00001d015053, and Zm00001d015054 were determined after the bite by the maize borer (O.nabilalis). The results showed that the expressions of six genes, including Zm00001d045054, Zm00001d024486, Zm00001d002351, Zm00001d002350, Zm00001d015053, and Zm00001d015054 were significantly upregulated (Figure 3A). The expression of gene Zm00001d032230 was downregulated. To further uncover the function of ZmTPS genes in response to the maize borer’s bite, the Zm00001d045054 was cloned. Heterologous expression and enzyme activity assays of the Zm00001d045054 in Escherichia coli showed that only one terpenoid, d-limonene, was identified in the enzymatic reaction products. The gene specifically catalyzed the synthesis of d-limonene using GPP. So, the gene Zm00001d045054 was named ZmDLS according to the systematic nomenclature (Figure 3B).
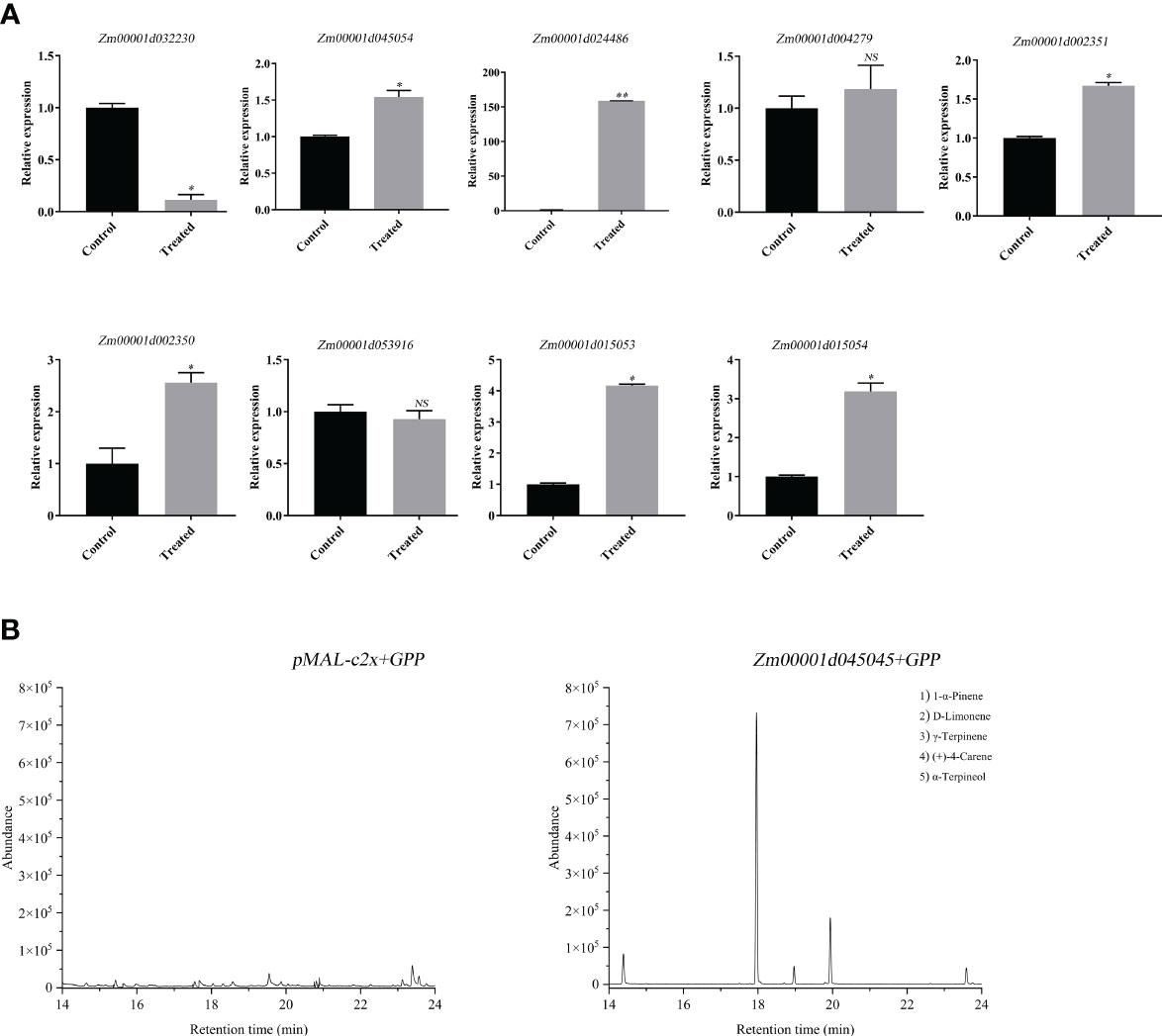
Figure 3 Functional analysis of ZmTPS genes in response to the maize borer’s bite. (A) Expression of ZmTPS genes after maize borer’s bite of the maize inbred line B73. Bars represent the mean values ( ± SD) of three biological replicates. Significance level (*p < 0.05, **p < 0.01); NS means not significant (p > 0.05). (B) Headspace solid-phase microextraction combined with gas chromatography–mass spectrometry (HS-SPME-GC-MS) analysis of products generated in vitro by recombinant ZmDLS protein. GC-MS peaks of volatile compounds were detected using extracts of pMAL-c2x cells (left). GC-MS peaks of volatile compounds were detected using extracts of pMAL-c2x-Zm00001d045054 cells with the addition of GPP (right).
Acquisition and identification of zmdls mutants
To verify the molecular function of the ZmDLS gene, a B73 EMS-mutagenized mutant zmdls was obtained through the maize mutant website (www.elabcaas.cn/memd/). The mutation site was confirmed by sequencing the fragment obtained using PCR amplification. The results showed that the zmdls mutant has a mutation from G to A at 6 bp in the second exon region, which caused premature translation termination at 135 aa and the deletion of the DDXXD structural domain (Figure 4A).
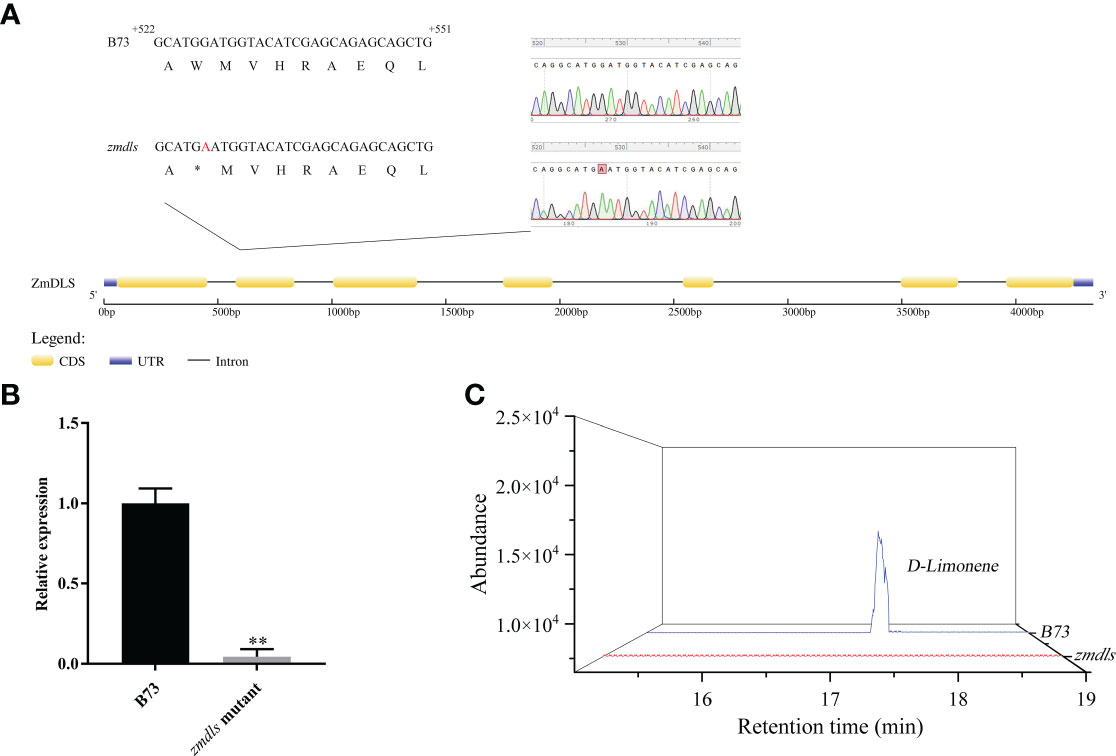
Figure 4 Acquisition and identification of zmdls mutants. (A) Sequence diagram of ZmDLS gene in the maize inbred line B73 and the zmdls mutant. (B) Expression of ZmDLS in the maize inbred line B73 and zmdls mutant. Bars represent the mean values ( ± SD) of three biological replicates. **p < 0.01, significant difference. (C) GC-MS analysis of d-limonene in maize inbred lines B73 and zmdls mutants.
Further experiments determined the content of d-limonene and the expression level of ZmDLS in the zmdls mutant. As shown in Figures 4B, C, the content of d-limonene was lowered, and the expression level of ZmDLS was significantly decreased in the zmdls mutant if compared with that in wildtype B73, which proved that the zmdls mutant was reliable and could be used for subsequent experiments.
Functional analysis of ZmDLS gene
As is shown in Figure 5A and Supplementary Figure S6, the zmdls mutant was more alluring to larvae than the wild type when it was used to feed corn borer (Ostrinia nubilalis). The gene expression analysis revealed that the expression of ZmDLS was significantly downregulated in the mutant (Figures 5B, C). Otherwise, the expressions of Zm00001d002351 and Zm00001d002350 were downregulated too. The expression levels of Zm00001d024486, Zm00001d053916, Zm00001d015053, and Zm00001d015054 were reduced after the bite if compared with that in the wild type (Supplementary Figure S7). The mutant of ZmDLS affected the other ZmTPS genes’ expressions in the leaf.
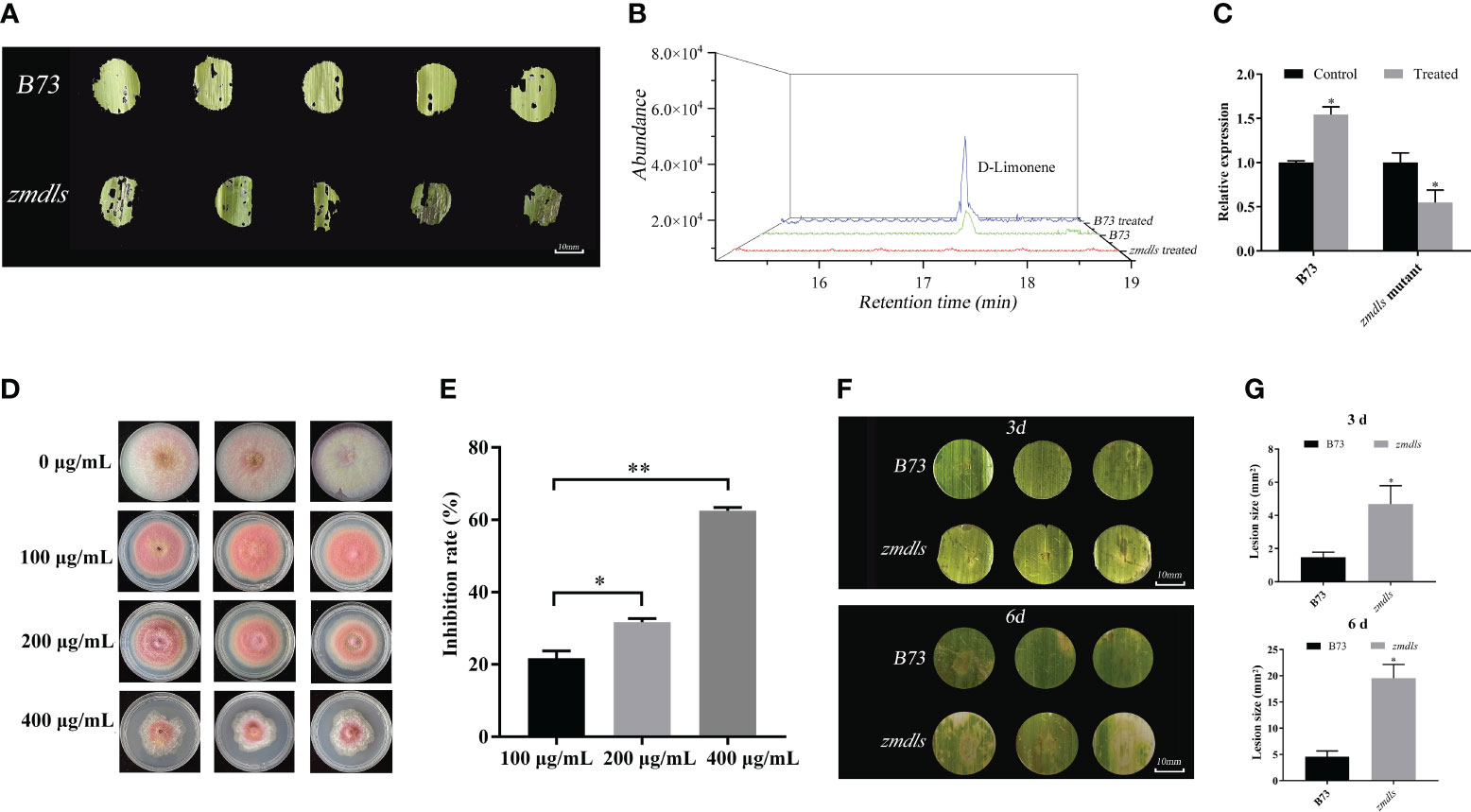
Figure 5 Functional analysis of ZmDLS in response to insect O. nubilalis and fungi F. graminearum attack. (A) The changes of a leaf of wild type and zmdls mutants after maize borer’s bite; each group contains at least three leaves. (B) GC-MS analysis of d-limonene in B73 and zmdls mutants after maize borer’s bite. (C) Expression of ZmDLS after maize borer’s bite in the maize inbred line B73 and zmdls mutants. Bars represent the mean values (± SD) of three biological replicates. *p < 0.05, significant difference. (D) In vitro antibacterial assay of d-limonene. Growth of F. graminearum in PDA medium at final concentrations of 0, 100, 200, and 400 μg/mL d-limonene. (E) In vitro antibacterial assay of d-limonene. There are three replicates in each group. *p < 0.05 and **p < 0.01, significant difference. (F) Wild type and zmdls mutants were inoculated with F. graminearum in vitro. Photographs were taken and the lesion area was measured at 3 and 6 days after inoculation. (G) In vitro inoculation of wild type and zmdls mutants with F. graminearum for 3 or 6 days for disease spot area. Each group contains at least three leaves. *p < 0.05, significant difference.
In addition, the ZmDLS product d-limonene was used to analyze its effects on the fungus F. graminearum. As is shown in Figure 5D, 100 μg·mL−1 of d-limonene inhibited the growth of F. graminearum. The higher concentration of d-limonene had a better inhibitory effect on F. graminearum (Figure 5E). In vitro culture of F. graminearum with the leaves of zmdls mutants showed that the mutant developed more significant necrotic lesions than wild-type leaves (Figures 5F, G).
Analysis of cis-acting elements for ZmDLS promoter
To identify the ZmDLS promoter, the 2,000-bp upstream region of ZmDLS was analyzed. The result showed that the promoter region was rich in cis-acting elements. For example, TGA motif, GARE motif, and ABRE belonged to phytohormone regulatory elements. The WRE3 motif was related to a damage induction element. TCA is a low-temperature regulatory element. The MBS was correlated with the drought response element. GCN4_motif and O2 site were grouped into growth and developmental regulatory elements. Further analysis of the ZmDLS promoter revealed that it mainly contains six typical plant transcription factor families, including MYB, MYC, WRKY, AP2/ERF, bZIP, and Dof (Figure 6A). However, the most enriched element is composed of several MYB binding sites, which possibly conferred the regulation role of MYB transcription factors on ZmDLS.
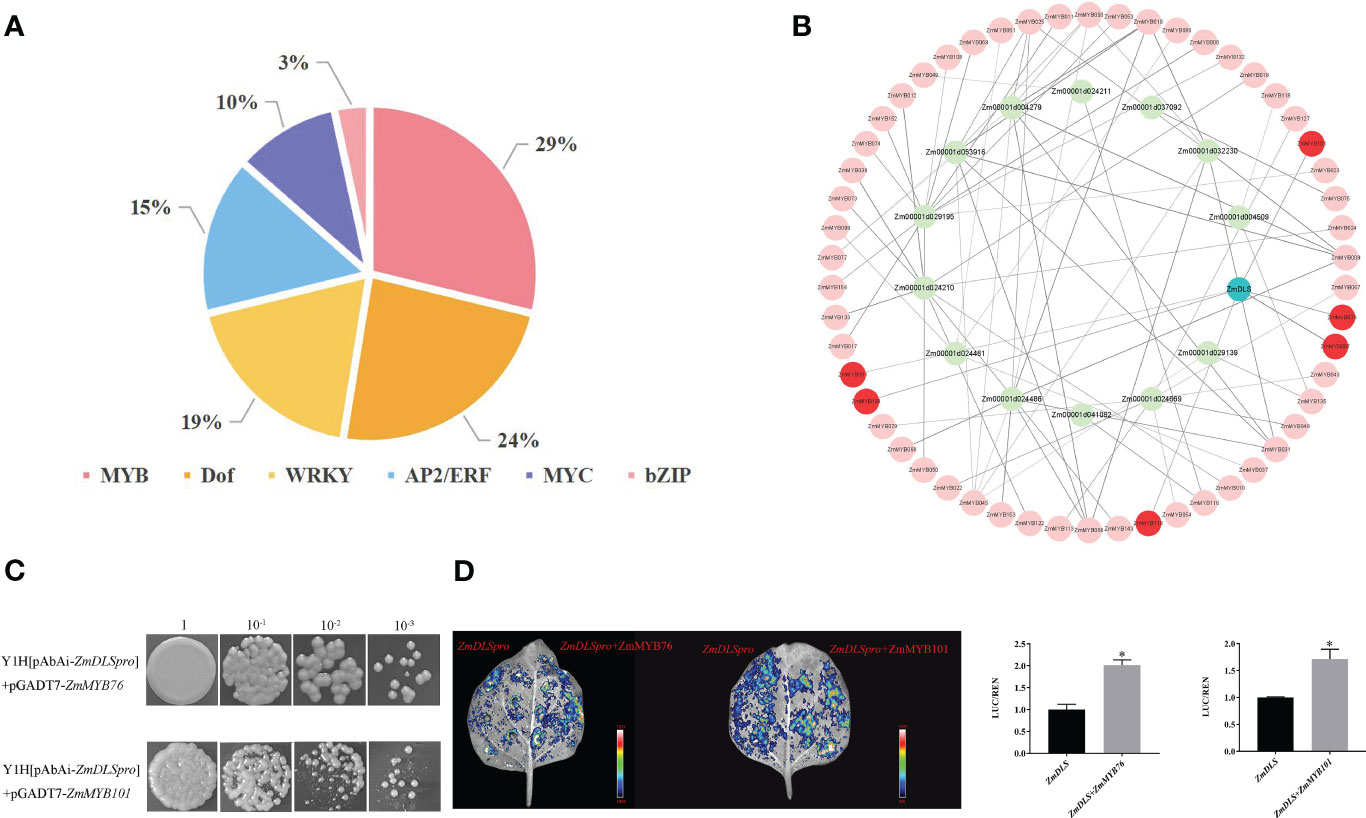
Figure 6 Interaction of ZmMYBs with ZmDLS. (A) ZmDLS promoter cis-acting site. Analysis of the 2,000 bp ZmDLS promoter and counting of the number of six transcription factor families contained in the ZmDLS promoter. (B) Co-expression network diagram of terpenoid synthesis-related genes and ZmMYB transcription factors. (C) Yeast single hybridization analysis of two transcription factors and ZmDLS promoters. Interactions were determined in the presence of AbA (AbA 100 ng/mL) on SD medium lacking leucine. (D) Co-expression of ZmMYB76 and ZmMYB101 with LUC driven by the ZmDLS promoters in the leaves of N. tabacum. An empty vector of pGreenII 62-SK was used as a control. Error bars, mean ± SD of three biological replicates. *p < 0.05, significant difference.
In vivo binding of ZmDLS promoter to ZmMYBs
As shown in Figure 6B, 157 R2R3-MYB genes (named ZmMYB1-157) downloaded from the maize genome database were used to conduct co-expression network analysis with ZmTPSs. Six genes, including ZmMYB97, ZmMYB76, ZmMYB101, ZmMYB104, ZmMYB119, and ZmMYB91, were selected for their high correlation coefficients with ZmDLS (Supplementary Table S7). They were positively correlated with ZmDLS. Gene expression analysis showed that the expression of ZmMYB76 and ZmMYB101 was significantly upregulated after the bite by the maize borer, which was consistent with the expression of ZmDLS (Figure 3A; Supplementary Figure S8). The yeast one-hybrid (Y1H) assay showed that the ZmDLS promoter could be directly bound by ZmMYB76 and ZmMYB101 (Figure 6C). Also, further transient expression assays by agroinfiltration of the leaves of N. tabacum confirmed that the co-expression of LUC driven by ZmMYB76 or ZmMYB101 with ZmDLS promoters significantly increased the LUC/REN ratio, which indicated that ZmMYB76 and ZmMYB101 could upregulate the expression of ZmDLS (Figure 6D).
Subcellular localization of ZmDLS and its regulatory genes in heterologous plants
To investigate the function of ZmDLS, ZmMYB76, and ZmMYB101 in maize, ZmDLS-GFP and ZmMYB-GFP fusion protein were transiently expressed in the leaves of N. tabacum and observed by laser confocal microscopy using a 35S-GFP vector as a control (Figure 7A). The green fluorescent fused ZmDLS-GFP gene was specifically distributed in chloroplasts. The result speculated that the gene might be involved in the MEP pathway and closely related to monoterpene synthesis. Nevertheless, ZmMYB-GFP only displayed a fluorescence signal in the nucleus, indicating that ZmMYB76 and ZmMYB101 are nuclear localization proteins (Figure 7B).
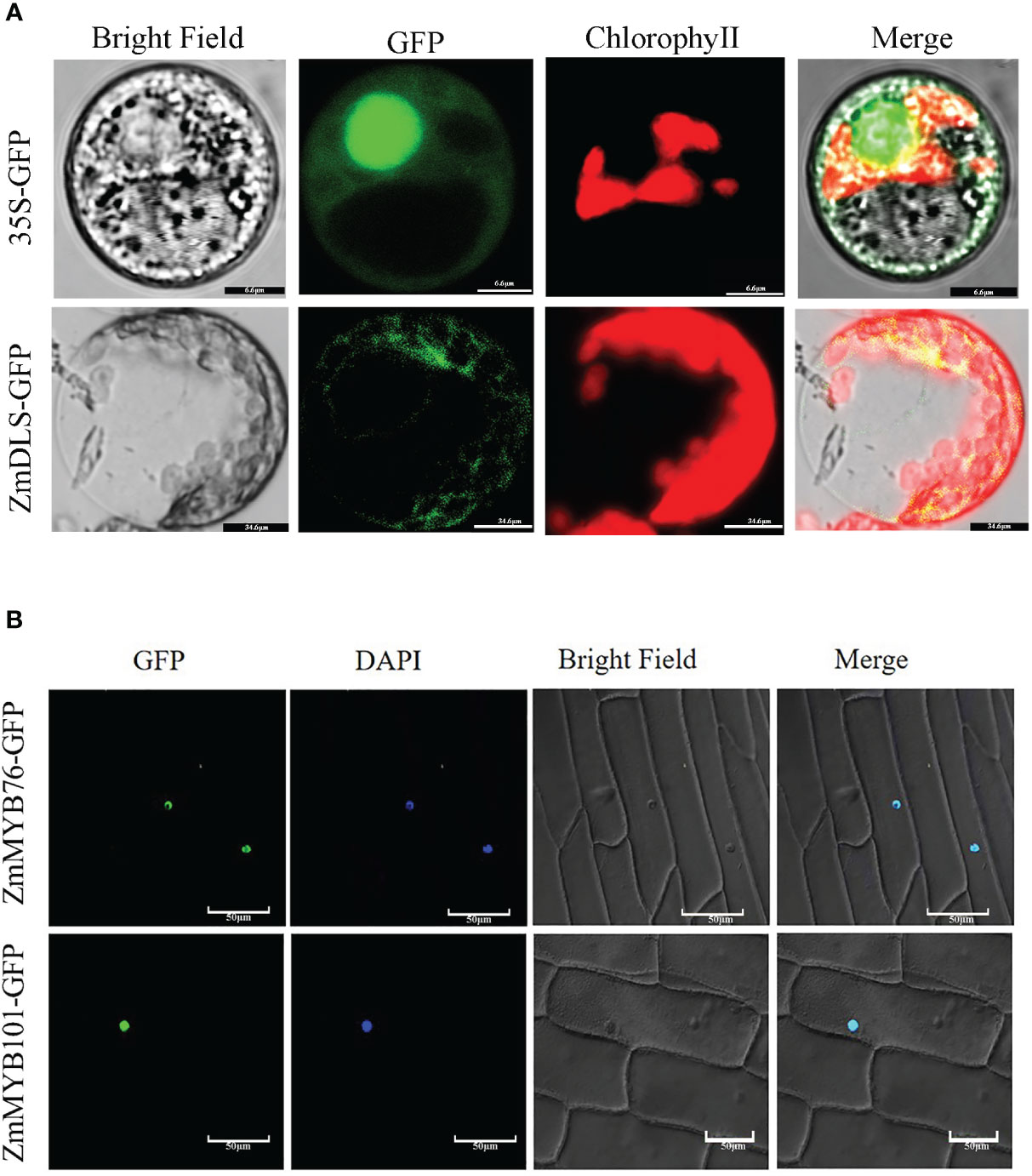
Figure 7 Subcellular localization of ZmDLS and ZmMYBs. (A) Subcellular localization of ZmDLS in the protoplast of Nicotiana tabacum. GFP is used as a control. (B) Subcellular localization of the ZmMYB-GFP fusion protein.
Discussion
Maize is an important food crop and industrial raw material around the world. However, maize is infested with pests and germs, which will cause a large-scale reduction in yields. Terpenoids play a vital role in protecting plants from herbivores and microbial pathogens (Mafu et al., 2018; Block et al., 2019). Many maize terpene synthase genes have been studied previously, but a systematic analysis of the maize TPS gene family has been rarely reported (Ding et al., 2020). In this study, 43 ZmTPS genes were identified and systematically analyzed. These genes were unevenly distributed into TPS-a, TPS-b, TPS-c, TPS-e/f, and TPS-g subfamilies.
d-Limonene (C10H16) is a monocyclic monoterpene commonly found in citrus plants such as lemons, grapes, and oranges (Anandakumar et al., 2021). It was proven to be the chemical composition of the odor with the activities of antioxidants, antibacterials, and anti-insects (Geraci et al., 2017; Yu et al., 2017; Sanei-Dehkordi et al., 2022). DMNT, (E)-β-farnesene, and linalool could attract parasitic wasps or natural enemies, indirectly defending against pests. S-Linalool and (E)-β-caryophyllene have a tendency avoidance effect on brown rice planthopper Nilaparvata lugens (Xiao et al., 2012). In the experiment, ZmDLS catalyzes the synthesis of d-limonene. Further analysis demonstrated that d-limonene inhibited the growth of F. graminearum. The results were in accord with the previous research.
Many terpene synthase (TPS) genes have been identified to study the role of their product in protecting plants from pest and germ attacks. For example, overexpressing GhTPS1 in cotton plants is less attractive to Helicoverpa armigera, Apolygus lucorum, and Aphis gossipii (Zhang et al., 2020). The decreased terpene content in tomato mutant leaves knocked out by SlJIG was more susceptible to H. armigera. The jig-1 and jig-16 mutant leaves produced more necrotic disease in vitro with Botrytis cinerea spores than wild-type leaves (Cao et al., 2022). Overexpression of StTPS18 in potatoes made the plant more susceptible to the bacterial pathogen P. syringae. The tolerance to the Ralstonia solanacearum was enhanced (Dwivedi et al., 2022). Similarly, overexpression of OsTPS19 in rice plants increased resistance to M. oryzae, whereas those with OsTPS19 RNAi had increased susceptibility to this disease (Chen et al., 2018). In the experiment, the expression level of nine TPS genes Zm00001d032230, Zm00001d045054, Zm00001d024486, Zm00001d004279, Zm00001d002351, Zm00001d002350, Zm00001d053916, Zm00001d015053, and Zm00001d015054 was determined after maize borer’s bite of the leaf. Six genes, including ZmDLS, were significantly upregulated in response to corn borer bite. The mutant zmdls downregulated the expressions of Zm00001d024486, Zm00001d002351, Zm00001d002350, Zm00001d015053, and Zm00001d015054. It was more attractive to corn borer bite and susceptible to F. graminearum infection.
Previous studies demonstrated that many kinds of transcription factors, including WRKY, AP2/ERF, and MYB, directly participated in the terpenes biosynthesis by regulating the expression of TPS genes (Xu et al., 2004; Yu et al., 2012; Sun et al., 2020; Lv et al., 2022). They bind to the TPS gene’s promoter region and positively or negatively regulate terpene biosynthesis (Vom Endt et al., 2002; Cao et al., 2020). For example, CitTPS16 catalyzes the synthesis of geraniol in sweet orange. The transcription factor CitERF71 can activate the expression of CitTPS16 and enhance the accumulation of geraniol synthesis (Li et al., 2017). The transcription factor AaMYC2 can bind to the G-box of CYP71AV1 and DBR2 promoters and regulate artemisinin synthesis in Artemisia annua (Shen et al., 2016). Otherwise, two transcription factors, FhMYB21L1 and FhMYB21L2, can positively regulate FhTPS1 expression. At the same time, FhMYC2 can interact with FhMYB21Ls to form a blocker to inhibit the binding of FhMYB21Ls to the FhTPS1 promoter, which negatively regulates the expression of FhTPS1 (Yang et al., 2020). Previous studies demonstrated that MYB transcription factors regulate terpenoid biosynthesis in plants (Bedon et al., 2010; Shin et al., 2016; Li P. et al., 2020; Li H. et al., 2022; Lv et al., 2022). Two MYB transcription factor genes, FhMYB21L1 and FhMYB21L2, were synchronously expressed with FhTPS1. Overexpression of FhMYB21L1 and FhMYB21L2 could significantly upregulate the expression of FhTPS1 in the Freesia hybrid (Yang et al., 2020). In Hedychium coronarium flowers, the R2R3-MYB transcription factor HcMYB2 could upregulate the expression of the linalool synthase gene HcTPS5 and enhance the emission of volatile compounds (Ke et al., 2021). In the experiment, two MYB transcription factors, ZmMYB76 and ZmMYB101, were verified to play an essential role in the upregulation of the expression of ZmDLS by binding to the region of the promoters, which directly increased the accumulation of d-limonene.
Conclusion
In this study, the TPS gene family was systematically identified and analyzed through maize genomes. A monoterpene synthase gene, ZmDLS, was identified in response to a corn borer bite. ZmDLS was localized in the chloroplast and responsible for the biosynthesis of d-limonene, the chemical component of the odor, with the activities of antioxidant, antibacterial, and anti-insect. In addition, ZmMYB76 and ZmMYB101 could positively regulate the expression of ZmDLS. Otherwise, the experiment demonstrated that ZmDLS participated in the response against the insect Ostrinia nubilalis and the fungus F. graminearum. These findings could supplement a theoretical foundation for the functional study and transcriptional control of TPS genes in crops.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
TL and HF designed the experiment. YW and JZ conducted the experiment. JL, FK and LX helped with phenotypic analysis after the corn borer bite. DX, JXL, HY, and LZ were responsible for cultivating seedlings. All authors have read and approved the final manuscript.
Funding
This work was supported by the Key Research and Development Plan of Anhui Province (202104a06020019), the Anhui University research project (YJS20210230), the Key Research and Development Plan of Anhui Province (202204c06020060), the Young Talent Program of the Anhui Academy of Agricultural Sciences, and the Anhui University Student Innovation and Entrepreneurship Training Program (X202210364150).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1162826/full#supplementary-material
Supplementary Table 8 | Summary of qRT-PCR on data
References
Anandakumar, P., Kamaraj, S., Vanitha, M. K. (2021). D-limonene: a multifunctional compound with potent therapeutic effects. J. Food Biochem. 45 (1), e13566. doi: 10.1111/jfbc.13566
Aubourg, S., Lecharny, A., Bohlmann, J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267 (6), 730–745. doi: 10.1007/s00438-002-0709-y
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 02–08. doi: 10.1093/nar/gkp335
Bedon, F., Bomal, C., Caron, S., Levasseur, C., Boyle, B., Mansfield, S. D., et al. (2010). Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot. 61 (14), 3847–3864. doi: 10.1093/jxb/erq196
Bergman, M. E., Davis, B., Phillips, M. A. (2019). Medically useful plant terpenoids: biosynthesis, occurrence, and mechanism of action. Molecules 24 (21), 3961. doi: 10.3390/molecules24213961
Block, A. K., Vaughan, M. M., Schmelz, E. A., Christensen, S. A. (2019). Biosynthesis and function of terpenoid defense compounds in maize (Zea mays). Planta 249 (1), 21–30. doi: 10.1007/s00425-018-2999-2
Cao, Y., Li, K., Li, Y., Zhao, X., Wang, L. (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biol. (Basel). 9 (3), 61. doi: 10.3390/biology9030061
Cao, Y., Liu, L., Ma, K., Wang, W., Lv, H., Gao, M., et al. (2022). The jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus. J. Integr. Plant Biol. 64 (5), 1102–1115. doi: 10.1111/jipb.13248
Chen, X., Chen, H., Yuan, J. S., Kollner, T. G., Chen, Y., Guo, Y., et al. (2018). The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 16 (10), 1778–1787. doi: 10.1111/pbi.12914
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Tholl, D., Bohlmann, J., Pichersky, E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66 (1), 212–229. doi: 10.1111/j.1365-313X.2011.04520.x
Cheng, A. X., Lou, Y. G., Mao, Y. B., Lu, S., Wang, L. J., Chen, X. Y. (2007). Plant terpenoids: biosynthesis and ecological functions. J. Integr. Plant Biol. 49 (002), 179–186. doi: 10.1111/j.1672-9072.2006.00395.x
Ding, Y., Weckwerth, P. R., Poretsky, E., Murphy, K. M., Sims, J., Saldivar, E., et al. (2020). Genetic elucidation of interconnected antibiotic pathways mediating maize innate immunity. Nat. Plants. 6 (11), 1375–1388. doi: 10.1038/s41477-020-00787-9
Du, H., Feng, B. R., Yang, S. S., Huang, Y. B., Tang, Y. X. (2012). The R2R3-MYB transcription factor gene family in maize. PloS One 7 (6), e37463. doi: 10.1371/journal.pone.0037463
Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., et al. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. PNAS 102 (3), 933–938. doi: 10.1073/pnas.0407360102
Dwivedi, V., Kumar, S. R., Shilpashree, H. B., Krishna, R., Rao, S., Shasany, A. K., et al. (2022). An inducible potato (E, E)-farnesol synthase confers tolerance against bacterial pathogens in potato and tobacco. Plant J. 111 (5), 1308–1323. doi: 10.1111/tpj.15890
Falara, V., Akhtar, T. A., Nguyen, T. T., Spyropoulou, E. A., Bleeker, P. M., Schauvinhold, I., et al. (2011). The tomato terpene synthase gene family. Plant Physiol. 157 (2), 770–789. doi: 10.1104/pp.111.179648
Fontana, A., Held, M., Fantaye, C. A., Turlings, T. C., Degenhardt, J., Gershenzon, J. (2011). Attractiveness of constitutive and herbivore-induced sesquiterpene blends of maize to the parasitic wasp Cotesia marginiventris (Cresson). J. Chem. Ecol. 37 (6), 582–591. doi: 10.1007/s10886-011-9967-7
Geraci, A., Di Stefano, V., Di Martino, E., Schillaci, D., Schicchi, R. (2017). Essential oil components of orange peels and antimicrobial activity. Nat. Prod. Res. 31 (6), 653–659. doi: 10.1080/14786419.2016.1219860
Inoue, Y., Shiraishi, A., Hada, T., Hirose, K., Hamashima, H., Shimada, J. (2004). The antibacterial effects of terpene alcohols on staphylococcus aureus and their mode of action. FEMS Microbiol. Lett. 237 (2), 325–331. doi: 10.1016/j.femsle.2004.06.049
Ke, Y., Abbas, F., Zhou, Y., Yu, R., Fan, Y. (2021). Auxin-responsive R2R3-MYB transcription factors HcMYB1 and HcMYB2 activate volatile biosynthesis in Hedychium coronarium flowers. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.710826
Köllner, T. G., Degenhardt, J., Gershenzon, J. (2020). The product specificities of maize terpene synthases TPS4 and TPS10 are determined both by active site amino acids and residues adjacent to the active site. Plants (Basel). 9 (5), 552. doi: 10.3390/plants9050552
Köllner, T. G., Gershenzon, J., Degenhardt, J. (2009). Molecular and biochemical evolution of maize terpene synthase 10, an enzyme of indirect defense. Phytochemistry 70 (9), 1139–1145. doi: 10.1016/j.phytochem.2009.06.011
Kroes, A., Weldegergis, B. T., Cappai, F., Dicke, M., van Loon, J. J. A. (2017). Terpenoid biosynthesis in arabidopsis attacked by caterpillars and aphids: effects of aphid density on the attraction of a caterpillar parasitoid. Oecologia 185 (4), 699–712. doi: 10.1007/s00442-017-3985-2
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, N., Dong, Y., Lv, M., Qian, L., Sun, X., Liu, L., et al. (2021). Combined analysis of volatile terpenoid metabolism and transcriptome reveals transcription factors related to terpene synthase in two cultivars of Dendrobium officinale flowers. Front. Genet. 12. doi: 10.3389/fgene.2021.661296
Li, J., Shao, Y., Yang, Y., Xu, C., Jing, Z., Li, H., et al. (2022). The chromatin modifier protein FfJMHY plays an important role in regulating the rate of mycelial growth and stipe elongation in Flammulina filiformis. J. Fungi. (Basel). 8 (5), 477. doi: 10.3390/jof8050477
Li, Z., Tang, J., Srivastava, R., Bassham, D. C., Howell, S. H. (2020). The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell. 32 (11), 3559–3575. doi: 10.1105/tpc.20.00260
Li, P., Xia, E., Fu, J., Xu, Y., Zhao, X., Tong, W., et al. (2022). Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants (Camellia sinensis). Plant J. 110 (4), 1144–1165. doi: 10.1111/tpj.15729
Li, X., Xu, Y., Shen, S., Yin, X., Klee, H., Zhang, B., et al. (2017). Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 68 (17), 4929–4938. doi: 10.1093/jxb/erx316
Li, H. Y., Yue, Y. Z., Ding, W. J., Chen, G. W., Li, L., Li, Y. L., et al. (2020). Genome-wide identification, classification, and expression profiling reveals R2R3-MYB transcription factors related to monoterpenoid biosynthesis in Osmanthus fragrans. Genes (Basel). 11 (4), 353. doi: 10.3390/genes11040353
Lv, M., Sun, X., Li, D., Wei, G., Liu, L., Chen, F., et al. (2022). Terpenoid biosynthesis in Dendrobium officinale: identification of (E)-β-caryophyllene synthase and the regulatory MYB genes. Ind. Crops Prod. 182, 114875. doi: 10.1016/j.indcrop.2022.114875
Mafu, S., Ding, Y., Murphy, K. M., Yaacoobi, O., Addison, J. B., Wang, Q., et al. (2018). Discovery, biosynthesis and stress-related accumulation of dolabradiene-derived defenses in maize. Plant Physiol. 176 (4), 2677–2690. doi: 10.1104/pp.17.01351
Murali-Baskaran, R. K., Mooventhan, P., Das, D., Dixit, A., Sharma, K. C., Senthil-Nathan, S., et al. (2022). The future of plant volatile organic compounds (pVOCs) research: advances and applications for sustainable agriculture. Environ. Exp. Bot. 200, 104912. doi: 10.1016/j.envexpbot.2022.104912
Nagegowda, D. A. (2010). Plant volatile terpenoid metabolism: biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 584 (14), 2965–2973. doi: 10.1016/j.febslet.2010.05.045
Portwood, J. L., 2nd, Woodhouse, M. R., Cannon, E. K., Gardiner, J. M., Harper, L. C., Schaeffer, M. L., et al. (2019). MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Res. 47 (D1), D1146–D1154. doi: 10.1093/nar/gky1046
Ren, F., Mao, H., Liang, J., Liu, J., Shu, K., Wang, Q. (2016). Functional characterization of ZmTPS7 reveals a maize τ-cadinol synthase involved in stress response. Planta 244 (5), 1065–1074. doi: 10.1007/s00425-016-2570-y
Saldivar, E. V., Ding, Y., Poretsky, E., Bird, S., Block, A. K., Huffaker, A., et al. (2023). Maize terpene synthase 8 (ZmTPS8) contributes to a complex blend of fungal-elicited antibiotics. Plants (Basel). 12 (5), 1111. doi: 10.3390/plants12051111
Sanei-Dehkordi, A., Moemenbellah-Fard, M. D., Saffari, M., Zarenezhad, E., Osanloo, M. (2022). Nanoliposomes containing limonene and limonene-rich essential oils as novel larvicides against malaria and filariasis mosquito vectors. BMC Complement. Med. Ther. 22 (1), 140. doi: 10.1186/s12906-022-03624-y
Shen, Q., Lu, X., Yan, T., Fu, X., Lv, Z., Zhang, F., et al. (2016). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210 (4), 1269–1281. doi: 10.1111/nph.13874
Shen, B., Zheng, Z., Dooner, H. K. (2000). A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: characterization of wild-type and mutant alleles. PNAS 97 (26), 14807–14812. doi: 10.1073/pnas.240284097
Shin, D. H., Choi, M. G., Kang, C. S., Park, C. S., Choi, S. B., Park, Y. I. (2016). A wheat R2R3-MYB protein PURPLE PLANT1 (TaPL1) functions as a positive regulator of anthocyanin biosynthesis. Biochem. Biophys. Res. Commun. 469 (3), 686–691. doi: 10.1016/j.bbrc.2015.12.001
Sun, Y., Huang, X. Z., Ning, Y. S., Jing, W. X., Bruce, T. J. A., Qi, F. J., et al. (2017). TPS46, a rice terpene synthase conferring natural resistance to bird cherry-oat Aphid, Rhopalosiphum padi (Linnaeus). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00110
Sun, P. W., Xu, Y. H., Yu, C. C., Lv, F. F., Tang, X. L., Gao, Z. H., et al. (2020). WRKY44 represses expression of the wound-induced sesquiterpene biosynthetic gene ASS1 in Aquilaria sinensis. J. Exp. Bot. 71 (3), 1128–1138. doi: 10.1093/jxb/erz469
Thakur, T., Gandass, N., Mittal, K., Jamwal, P., Muthamilarasan, M., Salvi, P. (2021). A rapid, efficient, and low-cost BiFC protocol and its application in studying in vivo interaction of seed-specific transcription factors, RISBZ and RPBF. Funct. Integr. Genomics 21 (5-6), 593–603. doi: 10.1007/s10142-021-00801-z
Vom Endt, D., Kijne, J. W., Memelink, J. (2002). Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry 61 (2), 107–114. doi: 10.1016/s0031-9422(02)00185-1
Vranova, E., Coman, D., Gruissem, W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. doi: 10.1146/annurev-arplant-050312-120116
Wang, H. L., Wang, T., Yang, Q., Yin, W., Xia, X., Guo, H., et al. (2021). Preparation and transfection of Populus tomentosa mesophyll protoplasts. Bio Protoc. 11 (22), e4220. doi: 10.21769/BioProtoc.4220
Xiao, Y., Wang, Q., Erb, M., Turlings, T. C., Ge, L., Hu, L., et al. (2012). Specific herbivore-induced volatiles defend plants and determine insect community composition in the field. Ecol. Lett. 15 (10), 1130–1139. doi: 10.1111/j.1461-0248.2012.01835.x
Xu, Y. H., Wang, J. W., Wang, S., Wang, J. Y., Chen, X. Y. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-a. Plant Physiol. 135 (1), 507–515. doi: 10.1104/pp.104.038612
Yahyaa, M., Matsuba, Y., Brandt, W., Doron-Faigenboim, A., Bar, E., McClain, A., et al. (2015b). Identification, functional characterization, and evolution of terpene synthases from a basal dicot. Plant Physiol. 169 (3), 1683–1697. doi: 10.1104/pp.15.00930
Yahyaa, M., Tholl, D., Cormier, G., Jensen, R., Simon, P. W., Ibdah, M. (2015a). Identification and characterization of terpene synthases potentially involved in the formation of volatile terpenes in carrot (Daucus carota L.) roots. J. Agric. Food Chem. 63, 4870–4878. doi: 10.1021/acs.jafc.5b00546
Yang, Z., Li, Y., Gao, F., Jin, W., Li, S., Kimani, S., et al. (2020). MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 71 (14), 4140–4158. doi: 10.1093/jxb/eraa184
Yu, Z. X., Li, J. X., Yang, C. Q., Hu, W. L., Wang, L. J., Chen, X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5 (2), 353–365. doi: 10.1093/mp/ssr087
Yu, F., Utsumi, R. (2009). Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell Mol. Life Sci. 66 (18), 3043–3052. doi: 10.1007/s00018-009-0066-7
Yu, L., Yan, J., Sun, Z. (2017). D-limonene exhibits anti-inflammatory and antioxidant properties in an ulcerative colitis rat model via regulation of iNOS, COX-2, PGE2 and ERK signaling pathways. Mol. Med. Rep. 15 (4), 2339–2346. doi: 10.3892/mmr.2017.6241
Zhang, L., Lu, G., Huang, X., Guo, H., Su, X., Han, L., et al. (2020). Overexpression of the caryophyllene synthase gene GhTPS1 in cotton negatively affects multiple pests while attracting parasitoids. Pest Manag. Sci. 76 (5), 1722–1730. doi: 10.1002/ps.5695
Keywords: maize, terpene synthase, corn borer bite, Fusarium graminearum attack, defense response, transcription factor
Citation: Wang Y, Zou J, Li J, Kong F, Xu L, Xu D, Li J, Yang H, Zhang L, Li T and Fan H (2023) Identification and functional analysis of ZmDLS associated with the response to biotic stress in maize. Front. Plant Sci. 14:1162826. doi: 10.3389/fpls.2023.1162826
Received: 10 February 2023; Accepted: 26 June 2023;
Published: 20 July 2023.
Edited by:
Wilfried Rozhon, Anhalt University of Applied Sciences, GermanyReviewed by:
Yiwei Zhou, Guangdong Academy of Agricultural Sciences (GDAAS), ChinaRong Chen, Hangzhou Normal University, China
Copyright © 2023 Wang, Zou, Li, Kong, Xu, Xu, Li, Yang, Zhang, Li and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honghong Fan, hhfan0551@126.com; Tingchun Li, litingchun2003@126.com
†These authors have contributed equally to this work
 Yiting Wang1†
Yiting Wang1† Fanna Kong
Fanna Kong Tingchun Li
Tingchun Li Honghong Fan
Honghong Fan