
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 19 April 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1162014
This article is part of the Research Topic Hormones and Biostimulants in Plants: Physiological and Molecular Insights on Plant Stress Responses View all 9 articles
 Chen Feng1†
Chen Feng1† Hongtao Gao2,3†
Hongtao Gao2,3† Yonggang Zhou2,3†
Yonggang Zhou2,3† Yan Jing2,3
Yan Jing2,3 Senquan Li2,3
Senquan Li2,3 Zhao Yan1
Zhao Yan1 Keheng Xu2,3
Keheng Xu2,3 Fangxue Zhou2,3
Fangxue Zhou2,3 Wenping Zhang2,3
Wenping Zhang2,3 Xinquan Yang4
Xinquan Yang4 Muhammad Azhar Hussain2,3*
Muhammad Azhar Hussain2,3* Haiyan Li1,2,3*
Haiyan Li1,2,3*The increasing sodium salts (NaCl, NaHCO3, NaSO4 etc.) in agricultural soil is a serious global concern for sustainable agricultural production and food security. Soybean is an important food crop, and their cultivation is severely challenged by high salt concentration in soils. Classical transgenic and innovative breeding technologies are immediately needed to engineer salt tolerant soybean plants. Additionally, unfolding the molecular switches and the key components of the soybean salt tolerance network are crucial for soybean salt tolerance improvement. Here we review our understandings of the core salt stress response mechanism in soybean. Recent findings described that salt stress sensing, signalling, ionic homeostasis (Na+/K+) and osmotic stress adjustment might be important in regulating the soybean salinity stress response. We also evaluated the importance of antiporters and transporters such as Arabidopsis K+ Transporter 1 (AKT1) potassium channel and the impact of epigenetic modification on soybean salt tolerance. We also review key phytohormones, and osmo-protectants and their role in salt tolerance in soybean. In addition, we discuss the progress of omics technologies for identifying salt stress responsive molecular switches and their targeted engineering for salt tolerance in soybean. This review summarizes recent progress in soybean salt stress functional genomics and way forward for molecular breeding for developing salt-tolerant soybean plant.
Soybean (Glycine max (L.) Merr.) is an important legume crop, and over 333 million tonnes are produced globally (Sugiyama, 2019). Soybean, seeds are enriched with essential amino acids, proteins (40%), lipids and metabolites (isoflavones and saponins), contribute 56% of the world’s edible oil for human consumption (Sedivy et al., 2017; Singh et al., 2017). Despite their great importance, field-grown soybean faces great environmental challenges from germination to final harvest. Among these environmental challenges, worldwide soil salinization and alkalinity are growing issues for economic crops, including soybean (Zhao et al., 2020; Feng et al., 2021a). According to current estimations, approximately 1-billion-hectare about 19.5% of farmland is affected by salt worldwide (Li et al., 2019b). In China, more than 100 million hac are under saline-alkaline stress (Cai et al., 2022). In the future, sustainable food supplies will be a serious task to feed a population of 9 billion by 2050 (Zhou et al., 2015). Therefore, breeding soybean varieties with high production, quality and saline-alkaline stress tolerance to guarantee world food security remains an ongoing task.
Soybean is cultivated in tropical, subtropical and temperate climatic regions. The world top ranked soybean producing countries are enlisted in Table 1. Soil salinity threatened the soybean’s seed germination, growth and developmental phases. A higher salt concentration causes various damage to soybean, such as high osmotic stress, water loss, homeostasis and ion imbalances. Morphologically, salt-stressed soybean plants exhibited leaf chlorosis, necrosis and scorching (Zhang et al., 2019a). Salt stress affected the nitrogen fixation efficiency by decreasing the number and biomass of root nodules. Soybean yield is significantly affected if the soil salinity exceeds 5 dS/m. Salt stress reduced the quality and quantity of free amino acids, protein, sucrose, and starch content in mature soybean seeds (El-Sabagh et al., 2015; Do et al., 2018). Higher transduction of Na+ and Cl− from the root zone caused salt toxicity which decreased upto 40% of soybean yield or complete crop failure (Rasheed et al., 2022). Plants adopted various strategies to minimize the Na+ and Cl− ionic/osmotic stress by minimizing the water loss or sequestering toxic ions to storage vacuoles (Deinlein et al., 2014; van Zelm et al., 2020). When Na+ enters the main root cell through nonselective cation channels (NSCCs), the salt-mediated signalling starts within 5 min to 5h. Salt-mediated signalling waves of Ca+, ROS and 3’,5’-cyclic guanosine monophosphate (cGMP) act as early signalling compounds and make a positive feedback loop for K+ influx (van Zelm et al., 2020). Various kinds of ionic transporters/antiporters like Na+/H+ Exchangers (NHXs), GmNHX1 and GmNHX2 have recently been identified that help to regulate the salt concentration in soybean (Sun et al., 2021a). Although various salt sensors and regulators like GmAKT1 have been reported, additional salt response mechanisms are still required for precise salt-tolerant soybean breeding.
Soybean germplasm is highly diverse and natural variations exist for salt tolerance. Recent omics approaches such as transcriptomics, quantitate traits loci (QTL), and genome-wide association studies (GWAS) help to identify various genes that play a significant role in soybean salt stress tolerance (Zhang et al., 2019a). For example, QTLs studies have identified GmSALT3 and GmSALT18 genes that have major role in soybean salt tolerance (Guo et al., 2021). These genes could be genetically engineered into soybean germplasm to enhance salt tolerance. For instance, CRISPR/Cas9 mediated editing of GmAITR, a negative regulator of salt tolerance, increased the soybean salt stress tolerance (He et al., 2021). Similarly, salt stress affects the epigenetic landscape of gene expression in plants. Recent studies revealed that epigenetic modification, epigenetic memories and AS events are also responsible for genetic variability for salt tolerance in soybean. For example, methylation of GmMYB84 enhanced the soybean salt stress tolerance (Zhang et al., 2020b). Small non-coding RNAs such as miRNA have been identified as critical regulators of salt tolerance through modulation of target gene expression (Islam et al., 2022; Zhang et al., 2022d). For instance, small RNA-Seq analysis of soybean under salt stress treatment identified 17 differentially expressed miRNAs and 31 putative target genes. The miR482bd-5p-HEC1 module was identified as a candidate epigenetic regulatory module in response to salt stress in soybean (Cadavid et al., 2020a). Moreover, miR172c-Glyma01g39520 module enhanced the water stress and salt stress tolerance by modulating the ABA pathway in soybean (Li et al., 2016). Interestingly, a single miRNA can regulate the expression of multiple genes. Thus, genetic engineering of single miRNA could help to improve multiple traits in soybean. Therefore, the investigation and functional validation of miRNA-target gene module in soybean salt stress should be paid more attention. In response to salt stress, proline, melatonin (N-acetyl-5-methoxytryptamine), glycine betaine (GB) and different sugars had osmo-protective roles and largely accumulated in soybean. For instance, soybean cultivars accumulate more proline content to heal the salt stress damage. However, osmo-protective roles played by proline largely dependent on plant growth and development stages, concentration and duration of salt stress (Mansour and Ali, 2017). Melatonin, a pleiotropic signaling molecule, can relieve salinity stress adverse effects by enhancing soybean germination, growth and development, detoxifying ROS, and regulating stress responsive genes (Wei et al., 2015; Imran et al., 2021). Similarly, GB acts as an osmolyte and osmo-protectant to enhance soybean salt tolerance. For example, exogenous application of GB enhances the antioxidant activities of superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT) and peroxidase (POD), to enhance the plants stress tolerance (Hernandez-Leon and Valenzuela-Soto, 2022). Similarly, phytohormones, like methylglyoxal (MG) and reactive oxygen species (ROS) detoxifying ascorbate-glutathione (AsA-GSH) pathway, Brassinosteroide (BR), Gibberellin (GA), Jasmonic acid (JA), and Salicylic acid (SA), which has a cross-talk with master stress regulator ABA, contribute soybean salt stress tolerance (Hossain et al., 2021; Waadt et al., 2022). In future, comprehensive investigations are required to establish the exact role of osmoprotectants, phytohormones and the apprehensive mechanisms by which they enhance soybean salt tolerance.
Therefore, this review covered physiological, molecular and biochemical salt-induced changes from salt perception to downstream signalling at the cellular level in soybean. Subsequently, we highlighted the role of different osmo-protectants and phytohormones in ameliorating the salt effects in soybean. We also explained the natural variation contributing to soybean salt tolerance variability. Finally, we explained the shortcomings and prospects of applying these technologies for developing soybean salt tolerance. Salt tolerance is a complex trait. Therefore, it is imperative to augment different salt stress responsive mechanisms to improve soybean performance under salt adversity.
Salt stress has a broad impact on the morphology and physiology of soybean plants. Salt stress induces ionic stress, particularly (Na+/K+/Cl− imbalance), osmotic stress (dehydration) and secondary stresses, especially oxidative stress (ROS) (Yang and Guo, 2018). Salt ions caused ionic stress, which is toxic to plant cells. Salts ions move from the root zone to shoots and accumulate in leaves. Higher concentrations of Na+ and Cl− ions in leaves imbalanced the cytoplasmic ion levels and metabolic pathways, eventually hindering the photosynthesis process and productivity in soybean (Yang and Guo, 2018; Rasheed et al., 2022; Xu et al., 2022a). Higher salt concentrations in soil or water in the root zone result in hyperosmotic and hyperionic conditions, which reduce the uptake of water or necessary nutrients, thereby causing osmotic stress (van Zelm et al., 2020). Ionic and osmotic stresses lead to secondary stresses such as the accumulation of toxic compounds (ROS) and nutrient imbalances in plant cells under salt stress (Dawood et al., 2022). If not properly managed, generated ROS can be destructive to cellular organs, enzymes, DNA, and lipids. For instance, salt stress disturbs the activity of different enzymes involved in the energy production and conversion process, such as nitrate reductase (NR), glutamate dehydrogenase (GDH) and glutamine synthetase/glutamate synthase (GS/(NADPH) in soybean (Ullah et al., 2019). Phytohormones are critical for plant growth and development and abiotic stress response in plants. Salt stress changed the ABA and GA levels which caused an imbalance in the growth and salt stress response in soybean (Shu et al., 2017; Kataria et al., 2019). Morphologically, salt stress reduced seed germination, seedling emergence, growth, leaf length, plant height, fresh weight, dry biomass, and nodulation of soybean (Shu et al., 2017; Kataria et al., 2019). At the reproduction stage, salt stress affects the pods per plant and the number of seeds per plant in soybean. Salt stress also reduced the quality and quantity of oil and protein contents (Figure 1).
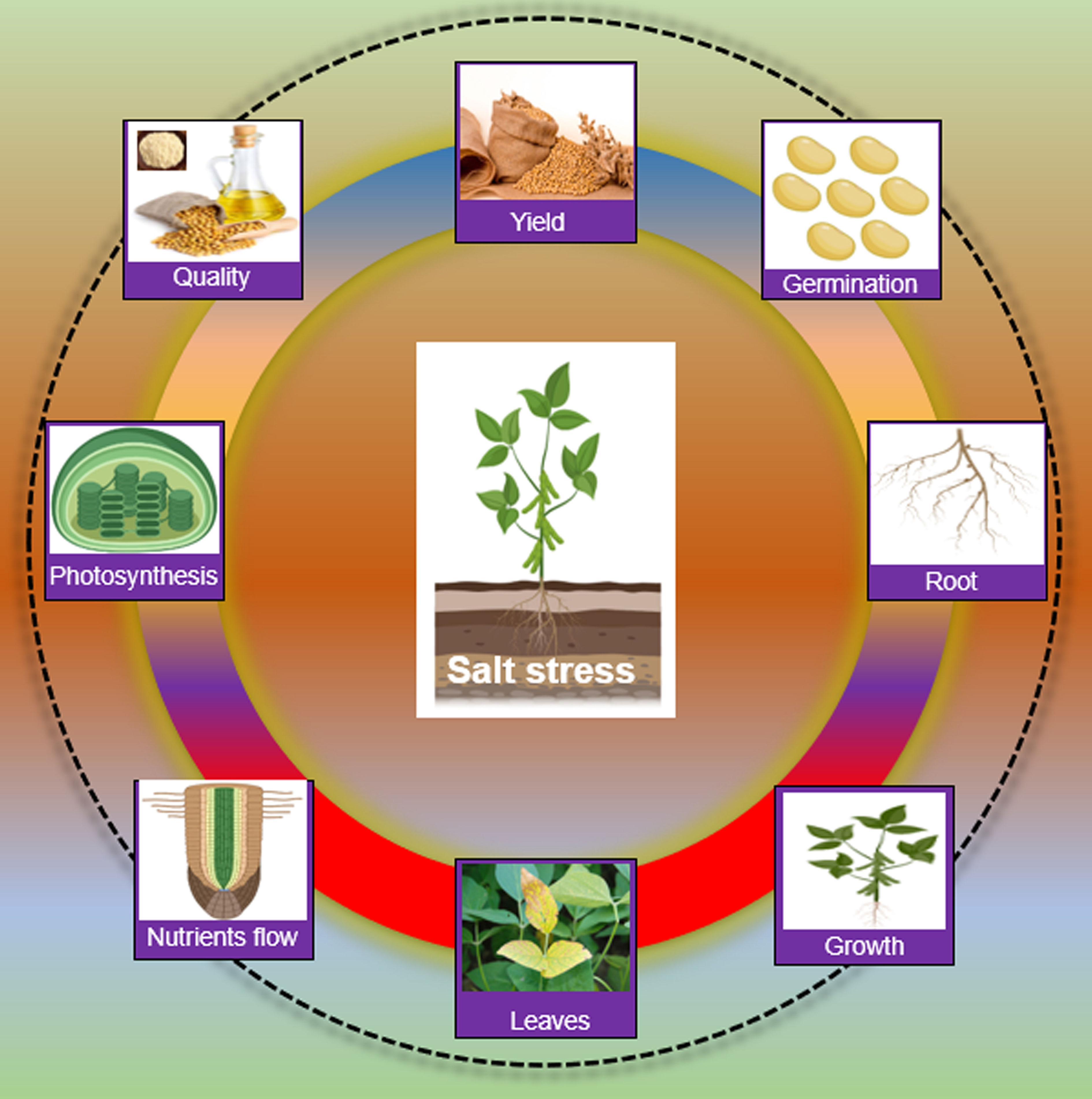
Figure 1 Impact of salt stress on Soybean Salt stress slows down germination, root establishment, seedling growth, causes leaf damage, blockage of nutrient transportation, hinders the photosynthesis process and finally affects the yield and product quality in Soybean.
Photosynthesis is an essential attribute of green plants, which is affected by higher soil salt concentrations. Wild G. soja native to saline soil has halophytic nature. Photosystem I (PSI) and PSII (Fv/Fm) photosynthesis efficiency under 300 mM NaCl treatment were significantly decreased in soybean. However, compared to G. max, G. soja protected the chloroplast ultrastructure and leaf lipid peroxidation by showing rapid recovery after salt stress. Therefore, differences in the photosystems of halophytic soybean could help to understand the salt adaptation mechanisms in cultivated soybean (Yan et al., 2020). Chloroplasts are responsible for photosynthesis related biochemical reactions. Salt stress affects the structures and function of chloroplasts by altering the size, number, lipid and starch accumulation, lamellar organization, and interfering with cross-membrane transduction (Hameed et al., 2021). Salt stress caused an increase in PSI transcripts. PSI is more stress-resistant than PSII. PSI enhanced cyclic electron flow to generate ATP while evading ROS accumulation to impart salt tolerance (Munekage et al., 2004; Johnson, 2011). ATP via cyclic electron flow around PSI helped to prevent the transduction of Na+ in chloroplasts of soybean (He et al., 2015). Therefore, salt toxicity and inhibition of photosynthesis was linked with the hyperaccumulation of Cl− but not that of Na+ in chloroplasts of soybean (Subbarao et al., 2003; Chen and Yu, 2007). Additionally, ROS and chloroplastic metabolites facilitate stress communication signalling between chloroplasts and the nucleus, a process known as ‘retrograde signaling’. Almost 10% to 20% of abiotic stress responsive genes protein are localized to the chloroplasts (Kmiecik et al., 2016). ROS-mediated retrograde signalling depends on factors like generation site, type, dose, timing, and duration in cells. ROS with known retrograde signalling function includes hydrogen peroxide (H2O2), singlet oxygen (1O2), and superoxide anion radical (O2–) (Laloi et al., 2004; Corpas et al., 2017). Lower levels of ROS activate the stress acclimation process, while higher accumulation causes cell death under stress conditions. For example, in response to stress, generated H2O2 is transduced into the nucleus through H2O2-specific aquaporin channels, known as peroxiporins. In the nucleus, H2O2 targets and activates various TFs and stress responsive genes such as DREBs, HSP, WRKY, and cytochrome P450. H2O2 caused activation of either quick response and, afterward induction of many downstream target genes for stress response (Crawford et al., 2018).
Calcium (Ca2+) is a universal secondary messenger at the cellular level. Ca2+ initiates many cellular responses to deal with biotic and abiotic stresses. For instance, salt stress induced early response signalling is mediated by Ca2+ waves, cGMP, and ROS to trigger downstream salt tolerance related genetic pathways and for cell expansion to minimize the stress effects. Recently, Na+ specific, Ca2+ wave influx was identified in plant root cells (Choi et al., 2014). Roots have sodium-specific sensors that regulate root growth direction in saline soil (Galvan-Ampudia et al., 2013). As shown in Figure 2, root plasma membrane-associated NSCCs channels transduce Na+ in plant cells (Demidchik and Tester, 2002; Demidchik and Maathuis, 2007). Salt-induced Ca2+, cGMP and ROS signals regulate NSCCs channel activity. Additionally, plant roots are also inbuilt with extracellular salt sensors such as MONO CATION INDUCED [Ca2+] INCREASES 1 (MOCA1) that sense Na+ ions (Jiang et al., 2019). MOCA1 generates glycosyl inositol phosphorylceramide (GIPC) sphingolipids at the plasma membrane that open unknown channels for Ca2+ influx. The moca1 mutant cannot produce Ca2+ waves in response to Na+, K+, or Li+ ions accumulation and is thus sensitive to salt stress (Kiegle et al., 2000; Donaldson et al., 2004; Miller et al., 2010). Vacuolar Ca2+ is influx by TWO-PORE CHANNEL1 (TPC1) for long-range calcium signals (Figure 2). The tpc1 mutants were found inefficient in speed and long-range Ca2+ wave signals in response to stress (Choi et al., 2014). CBL mediates CIPKs phosphorylation for Ca2+ outfluxes in response to salt stress (Manishankar et al., 2018). Under salt stress, Ca2+ signal-activates SOS pathway for sodium ion homeostasis in cells. Calcium-mediated SOS pathway help to exclude Na+ from cells. For example, Ca2+ mediates SOS3/CBL4 interaction with SOS2/CIPK24. The SOS2-SOS3 complex regulates the phosphorylation of H+/cation antiporter SOS1/NHX7 to release Na+ out of cells (Liu and Zhu, 1998; Halfter et al., 2000; Liu et al., 2000). Similarly, the CBL4-SOSO2 complex squeezes Na+ in the vacuole. The SOS2-LIKE PROTEIN KINASE5 (PKS5) can phosphorylate SOS2. Under salt stress, Ca2+ binds the 14–3–3 proteins and represses the PKS5 activity, thus attenuating the SOS2 (Figure 2). SOS2 caused the phosphorylation of AtANN4 to generate a salt-specific calcium signal (Ma et al., 2019). CPK3-mediated phosphorylation of the vacuolar TWO-PORE K+ CHANNEL 1 (TPK1), or knockout cpk3 and tpk1 mutants, were found salt-sensitive (Latz et al., 2013). A recent study reported that salt-tolerant soybeans strongly activated CBL10-CIPK24-NHX and CBL4-CIPK24-SOS1 complexes to reduce salinity effects (He et al., 2015). Exogenous application of gibberellin biosynthesis inhibitor prohexadione-calcium (Pro-Ca) effectively protects saline-alkali stress damage by regulating seedling phenotype, photosynthetic apparatus, antioxidant defense, and osmoregulation (Feng et al., 2021b).
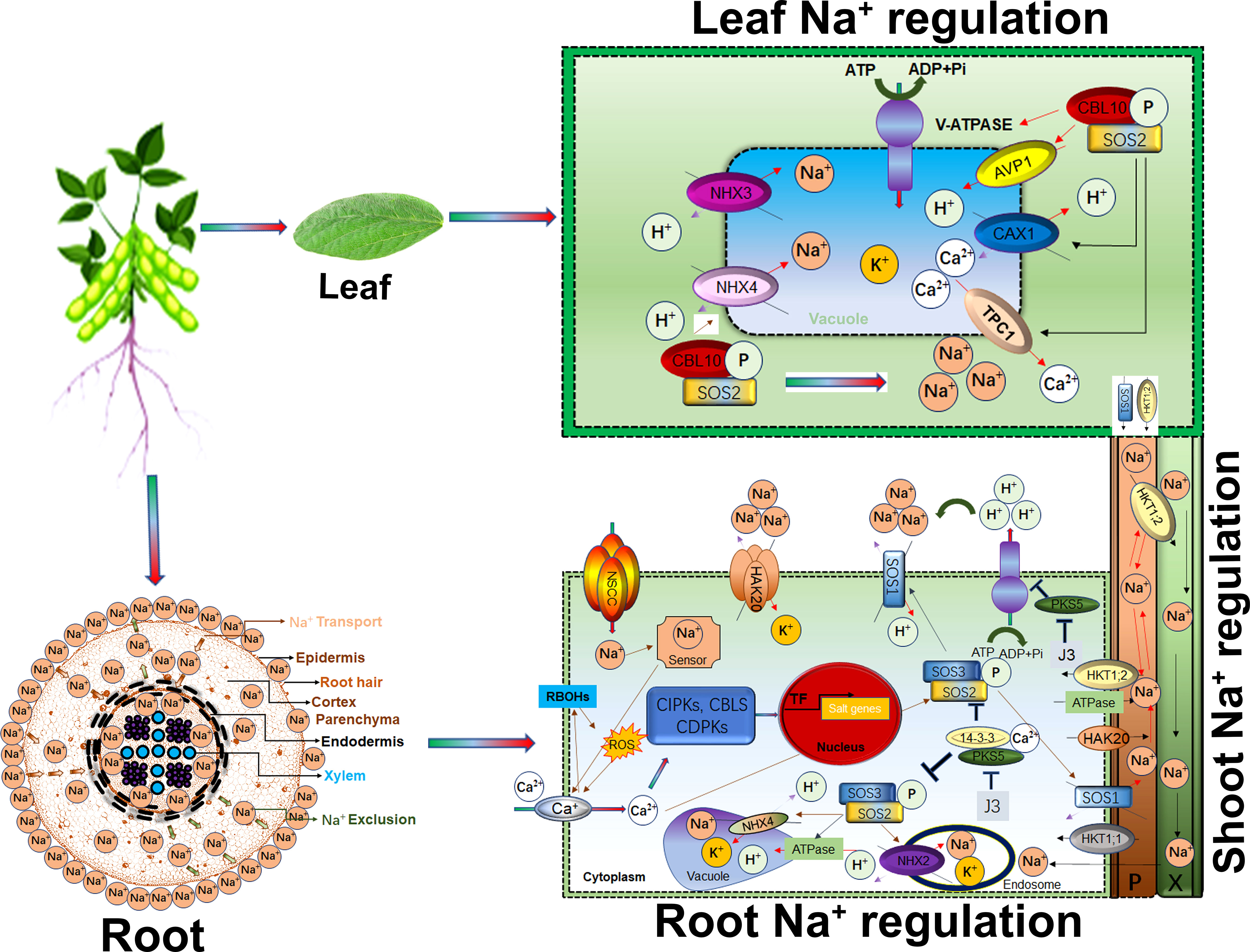
Figure 2 Salt (Na+) regulation in root, shoots transportation and leaves response Higher Na+ concentration around the root epidermis is eventually transported through membrane pores. Apoplastic and symplastic pathways are critical in Na+ ions movement in membrane. Ca2+, RBOHs and ROS cascade induces Ca2+ import upon higher accumulation of Na+ through NSCC within cytoplasm. Ca2+ mediates alteration of CIPKs-CBLs-CDPKs modules transcriptional profile thus enhanced the Na+/H+ and K+/H+ transportation and vacuole storage. SOS2-SOS3 complex activate the SOS1 for Na+ partitioning, exclusion and transportation to leaves. In leaves, CBL10-SOS2 complex trigger the storage or Na+ exclusion to shoots by activation of different transporters. HKT1;2 is involved Na+ recirculation from shoots to roots.
Cytosolic pH homeostasis is indispensable for plant growth and abiotic stress responses (Zhou et al., 2021). Proton pumps-maintained pH homeostasis through cellular ion transport and Na+ sequestration (Li et al., 2022b). P-typeATPases (P-ATPases) are localized in the cellular membrane and improved the Na+ sequestration in the vacuole (Figure 2). Similarly, P-ATPases mediated enhanced Na+/proton antiporter SOS1/NHX7 activity increased the salt and alkaline stresses tolerance through Na+ exclusion from root (van Zelm et al., 2020). The cellular homeostasis between Na+ and K+ is maintained by replacing K+ with Na+, which is mediated by ion channels, transporters, and antiporters during salt stress (Figure 2). The GmNHX1 localized on vacuolar membranes, predominantly transports Na+ to leaves and reduces Na+ absorption in roots. Soybean plants overexpressing or knockout GmNHX1 exhibited higher salt tolerance or susceptibility, respectively. Additionally, GmNHX1 transformed plants showed higher Na+ efflux rate and maintained a higher K+/Na+ ratio after salt treatment than wild-type in addition to induction of osmotic stress-related genes, SKOR, SOS1 and AKT1 to enhance soybean salt tolerance (Sun et al., 2019b). Apart from the vacuolar membranes localized GmNHX1, the other members of the NHX family GmNHX5 localized to Golgi apparatus can transport both K+ and Na+, maintaining a higher Na+/K+ ratio which has demonstrated relevance for salt tolerance in soybean. Mechanistically, GmNHX5 also regulates osmotic stress-related genes, such as GmSOS1, GmSKOR, and GmHKT1 by maintaining a higher Na+/K+ ratio to improve the salt tolerance compared to CRISPR/Cas9 mediated knockout GmNHX5 Soybean (Sun et al., 2021a). The promoter of GmNHXs contained Amiloride binding motifs, an inhibitor of Na+/H+ exchange activity in soybean. Induction of GmNHX2 after 200 mM NaCl stress also indicated its significant role in salt tolerance, which needs to be determined in soybean (Joshi et al., 2021). GmSOS1 localized in the plasma membrane is a Na+ efflux transporter in salt stress. CRISPR-Cas9 mediated gmsos1 mutants that significantly accumulated higher Na+ in the roots resulting in the imbalance of Na+ and K+ efflux under salt stress (Figure 2). GmSOS1 is a Na+/H+ transporter and plays a critical role in soybean salt tolerance by maintaining Na+ homeostasis (Zhang et al., 2022a). Further, the Cation/H+ -exchanger (CHX) had a protective role in salt stress. In soybean a major QTL for salt-tolerance has the causal gene GmCHX1 which is a close paralog of GmCHX20a. Interestingly, overexpression of GmCHX20a led to increased salt sensitivity mainly due to increasing Na+ uptake into the root. However, on the contrary, GmCHX1 overexpression enhanced salt tolerance via Na+ exclusion under salt stress. Thus, the concerted effects of GmCHX20a and GmCHX1 reduced the osmotic and ionic stress in response to elevated salinity in soybean (Jia et al., 2021). However, our current knowledge is limited to transporters that have a role in Na+/K+ homeostasis. In response to salt stress, several cation transporters change their expression; however, their biological role is still elusive in soybean. Recently the development of high-resolution molecular reporters provided deep insights into calcium transport and in vivo function. Similar high-resolution molecular reporters for Na+ and K+ could yield insights into the functions of transporters and the relevance of tissue-specific Na+/K+ regulation and function (van Zelm et al., 2020).
Potassium (K+) is an essential macronutrient involved in plant drought and salt response (Xu et al., 2006; Wang and Wu, 2013). Physiologically and genetically K+ regulation occurs through K+ channels (Shaker K+) and K+ transporters under abiotic stress in plants (Véry and Sentenac, 2003; Véry et al., 2014). In saline soil, plant survival depends on the cellular balance between Na+ and K+ ratio. The intra-cellular K+ level is regulated through high- and low-affinity systems depending upon the external concentration of K+ (Voelker et al., 2010). Although molecular similarity exists between K+ and Na+, they have different roles in cellular processes responding to salt stress. In plants, excessive Na+ accumulation causes K+ deficiency under salt stress (Assaha et al., 2017). Therefore, maintaining a high K+/Na+ ratio is a permissive task for plants to survive under salt-stress conditions. A hyperpolarization-activated voltage-dependent K+ channel known as AKT1 maintains the intracellular Na+/K+ ratio under salt stress (Ardie et al., 2010; Assaha et al., 2017; Ma et al., 2017). AKT1, the first characterized K+ channel in Arabidopsis, is responsible for cellular K+ efflux/influx during salt/osmotic stresses (Figure 3). Salt stress impairs AKT1 channel activity and K+ uptake in the sos1 mutant (Qi and Spalding, 2004). Similarly, SCaBP8/CBL10 inhibits AKT1 activity under salt stress (Ren et al., 2013). Arabidopsis akt1 mutant seedlings were found sensitive to salt stress but could be rescued by exogenous K+ application (Qi and Spalding, 2004). AtKC1 also negatively regulates AKT1-mediated K+ uptake in Arabidopsis (Wang and Wu, 2013). AKT1 mutant showed an imbalance K+/Na+ ratio and salt sensitivity in Zygophyllum xanthoxylum (Ma et al., 2017). However, complementation of Arabidopsis akt1 mutant with SmAKT1 recovered the salt-tolerant phenotype via enhanced K+ uptake (Li et al., 2019b). Overexpression of GmAKT1, OsAKT1 and HvAKT1 improved the root zone K+ uptake and subsequently removed the salt stress effects in soybean, rice and barley, respectively (Ahmad et al., 2016a; Feng et al., 2020b; Feng et al., 2021a; Wang et al., 2021a). Physiologically, intracellular K+ concentration determined the AKT1 phosphorylation state for K+ influx under salt conditions. For instance, Calcineurin B-like protein (CBL) mediated cascade such as CBL1/CBL9 activates CBL-interacting serine/threonine-protein kinase 23 (CIPK23) to phosphorylate AKT1 to boost K+ influx in response to lower cellular K+ in stress conditions (Figure 3) (Xu et al., 2006; Li et al., 2014; Behera et al., 2017). AKT1 forms either homo or hetero-tetrameric channel with pore-forming subunits or K+ channel α-subunit AtKC1, respectively. AtKC1 formed the AKT1-AtKC1 complex to inhibit the K+ transduction activity of AKT1 which is also dependent on the CBL1/9-CIPK23-AKT1 phosphorylation cascade (Duby et al., 2008; Wang et al., 2010; Jeanguenin et al., 2011; Wang et al., 2016; Lu et al., 2022). Additionally, various other K+ channels-related genes were identified in different plants such as AKT5, SPIK, KAT1, AKT2, KC, SKOR GORK, VvK1.2 and FaAKT1 (Anderson et al., 1992; Sentenac et al., 1992; Gambale and Uozumi, 2006; Cuéllar et al., 2013; Garriga et al., 2017). However, these Shaker K+ channel genes have an independent role; for instance, AKT1 and SKOR are expressed in roots and stem and transduce K+ (Lagarde et al., 1996; Johansson et al., 2006). While GORK is expressed in guard cells to regulate stomata and SPIK is mainly expressed in pollen for K+ uptake (Hosy et al., 2003). OsKAT1 acts in concert with K+ channels genes to actively stabilize the cytosolic cation homeostasis, thus protecting cells from salt effects (Obata et al., 2007). These findings suggested that AKT1 is a master regulator of K+ uptake under low K+ concentration and maintains the Na+/K+ homeostasis to ameliorate the salt stress effects (Feng et al., 2021a).
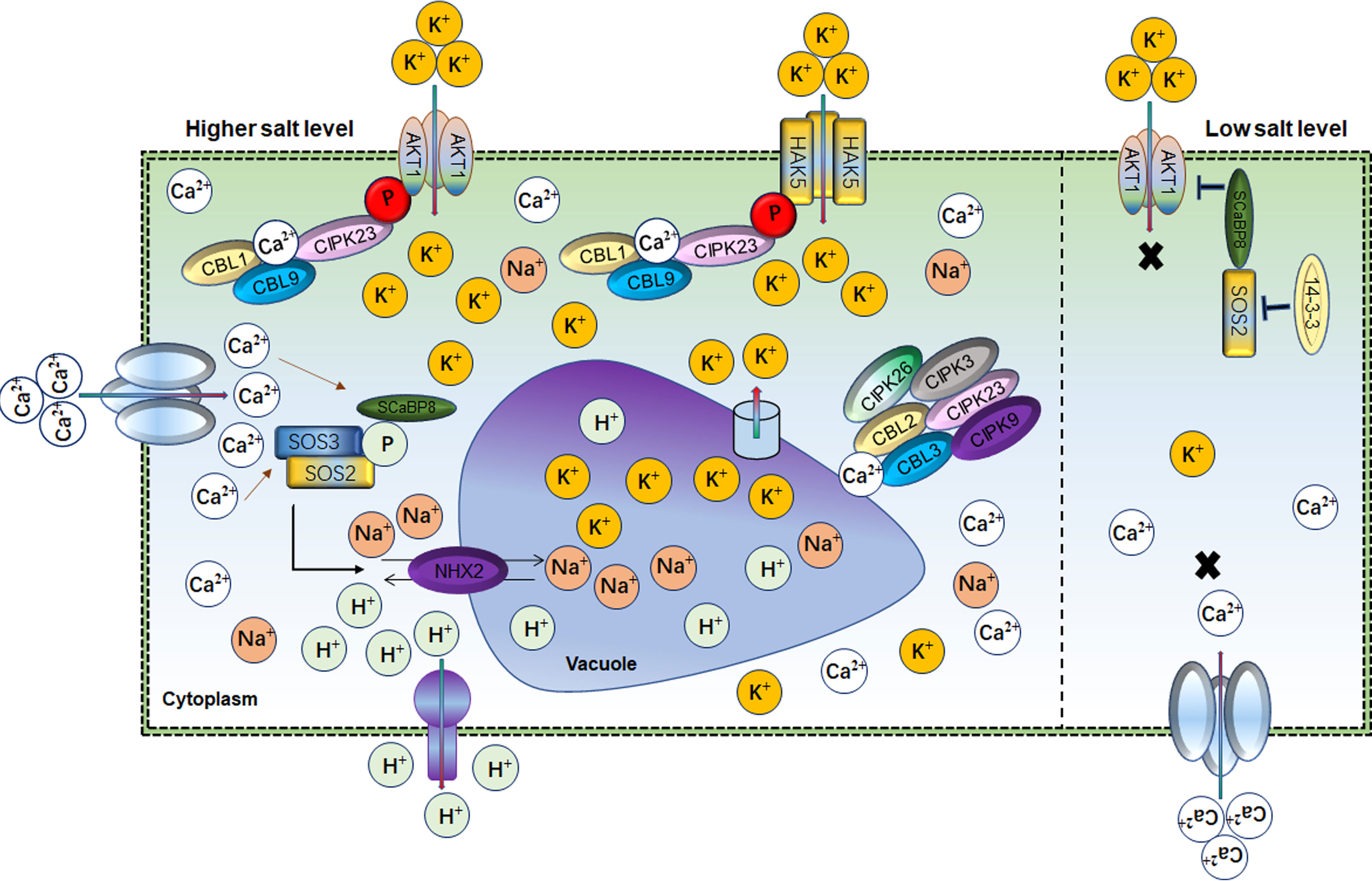
Figure 3 Role of Ca2+ and AKT1 channels in balancing the Na+/K+ homeostasis Cellular ionic homeostasis under lower and higher salt conditions mediated by Ca2+ and AKT1 channels. 14-3-3-SOS2- SCaBP8 module blocked AKT1 activity under low salt levels thus inhibiting the K+ import into the cell. However, under salt stress, Ca2+ activates SOS2 for SCaBP8 phosphorylation to separate from AKT1. SOS2 also had a key role in Na+/H+ vacuolar partitioning. At the same time, the CBL–CIPK cascade activated across the differential gradients of K+, which mediate Ca2+ influx through AKT1 and HAK5 to increase cellular K+. In parallel, vacuolar K+ remobilized through the interaction of CBL2/3 with four CIPKs through K+ channels to maintain the K+/Na+ homeostasis.
Investigating the molecular and genetic basis of soybean saline-alkaline stress tolerance is crucial to expand the cultivation and production in saline-alkaline-affected soil. In contrast to salt (NaCl) stress, alkaline stress is exerted by higher soil carbonate (CO3−2), bicarbonate (NaHCO3) and pH, which has more deteriorating effects on soybean growth (Cao et al., 2017). Soil saline-alkaline stress caused ionic imbalance by inhibiting the absorption of K+, Ca2+, and Mg2+, thus enhancing the Na+ and Cl− uptake under higher root zone pH (Guo et al., 2019). Under saline-alkaline stress, root plasma membrane H+ deficiency reduces the rhizosphere Na+/H+ exchange activity, thereby increasing in vivo Na+ uptake to a hazardous level (Wang et al., 2015a). In planta, saline-alkaline stress induces ionic and osmotic adjustment signals, including Ca2+ signals at the cellular level, to reduce the stress effects (Figure 4). Ca2+ spiking facilitates Na+/H+ antiporter SOS1 phosphorylation by the SOS3-SOS2 complex under sodic stress (El Mahi et al., 2019). Ca2+ responsive SOS1 and plasma membrane H+-ATPase modulate Na+/H+ influx from NaHCO3 to enhance the saline-alkali tolerance (Ni et al., 2020). SOS2 also triggers tonoplast Na+/H+ antiporter NHX1 to squeeze the Na+ into the cell vacuole (Assaha et al., 2017). Furthermore, members of the NHX gene, NHX1-5, including GmSOS1, regulate the compartmentalization of cytoplasmic Na+ and K+ (Fukuda et al., 2011). SOS3-SOS2 complex inhibited the AtHKT1 activity, whereas SOS4 regulates ion channels and transporters to regulate Na+ and K+ homeostasis (Shi and Zhu, 2002; Horie et al., 2006). Similarly, plasma membrane localized GsCHX19.3 regulates the K+ and Na+ under NaHCO3 stress in soybean (Jia et al., 2017a). In soybean, some genetic factors also regulate NaHCO3 rather than pH. For instance, an alkaline stress-induced slow anion channels homolog (SLAH) GsSLAH3 and B transporters GsBOR2 exerted higher tolerance to NaHCO3 and KHCO3 but not to pH (Duan et al., 2018a; Duan et al., 2018b). However, GsBOR2 direct role in K+ regulation under NaHCO3 and KHCO3 stress remained unclear. The Gshdz4-GsNAC019-GsRD29B module conferred alkaline stress tolerance rather than high pH (Cao et al., 2016; Cao et al., 2017). Some transporter selectively transduces ion in soybean under saline-alkaline stress (Figure 4). For example, GsCLC-c2 transports Cl– and NO3−, and GmCHX1 transport Na+, K+, and Cl– (Qu et al., 2021) under saline-alkaline conditions (Wu et al., 2018b; Qu et al., 2021). Under alkaline stress, cellular ROS are regulated by modifying the expression of ROS stress signaling genes in soybean. A NAC transcription factor SALT INDUCED NAC1 (GmSIN1) directly induced the expression of Respiratory burst oxidase homolog B (GmRbohB) to generate ROS to transduce salt stress signals (Li et al., 2019c). Similarly, methionine sulfoxide reductase B (GsMSRB5a) could regulate the expression of ROS signaling genes under saline-alkaline stress (Sun et al., 2016). Phytohormones such as Ascorbic acid acts as ROS scavenger under saline-alkaline stress. L-myoinositol-1-phosphate synthases (GsMIPS) 2 overexpression conferred salt and NaHCO3 stresses tolerance compared to atmips2 mutant (Chen et al., 2015). The overexpression of glutathione S-transferases (GSTs) genes, GsGST13/14/19, enhanced the saline-alkaline stress tolerance (Wang, 2012; Wang et al., 2012; Jia et al., 2016). In response to salt stress, detoxification of Methylglyoxal (MG) and ROS through MG detoxifying glyoxalase and the ROS detoxifying ascorbate-glutathione (AsA-GSH) pathways played protecting effects against salt stress in plants (Hossain et al., 2021). Glutathione (GSH) is a nonenzymatic antioxidant system activated against abiotic stress in plants. The glutathione peroxidases (GPXs) catalyze the oxidation of GSH to produce oxidized glutathione (GSSG), and convert H2O2 to H2O and alcohol to protect ROS-mediated oxidative damage. In soybean, GsGPX10.1 and GsSAMS2 (S-Adenosyl-L-Methionine Synthetases) activity is closely related to GSH content and respond to salt stress, indicating a protecting effect by removal of ROS (Hasanuzzaman et al., 2017; Aleem et al., 2022). These studies suggested that GSH content could be increased by overexpressing SAMS genes and GsGPX10.1, which can help soybean plants to cope with ROS damage. Furthermore, exogenous GSH application minimizes oxidative stress and improves soybean yield-related traits and salt stress tolerance (Akram et al., 2017). Ascorbic acid (AsA) acts as a ROS scavenger in the AsA-GSH cycle. Myo-inositol is one of the main precursors of AsA biosynthesis. MIPSs genes are involved in myo-inositol biosynthesis and respond to NaHCO3 and salt stress treatment in soybean. For instance, compared to the GsMIPS2 T-DNA mutant, GsMIPS2 overexpression plants exhibited higher NaHCO3 and NaCl stress tolerance in Arabidopsis. These results showed that AsA biosynthesis is critical in salt-alkaline stress tolerance in soybean (Cai et al., 2022). Additionally, different RLKs (receptor-like kinases), SnRKs (sucrose non-fermenting1-related protein kinases), MAPKs (mitogen-activated protein kinases) and transcription factors contributed towards soybean saline and alkaline stresses tolerance. For instance, calcium/calmodulin-binding receptor-like kinase (CBRLK) activated by Ca2+/Calmodulins (CaM) complex to form CBRLK-CPI14 complex to confer the alkaline stress tolerance in soybean (Yang et al., 2010; Sun et al., 2014). Co-overexpression of GsSnRK1.1 and GsERF7 significantly improved soybean saline-alkaline stresses tolerance (Feng et al., 2020a). Overexpression of GsTIFY10, GsWRKY20 and GsWRKY15 also improved the saline-alkaline stress tolerance (Zhu et al., 2011). Although, these genes contribute saline-alkaline stress tolerance in soybean. However, “their exact regulatory mechanisms” such as; How they maintain ionic and ROS homeostasis is still elusive. Identifying their impacts on soybean yields in saline-alkaline fields trials is also imperative.
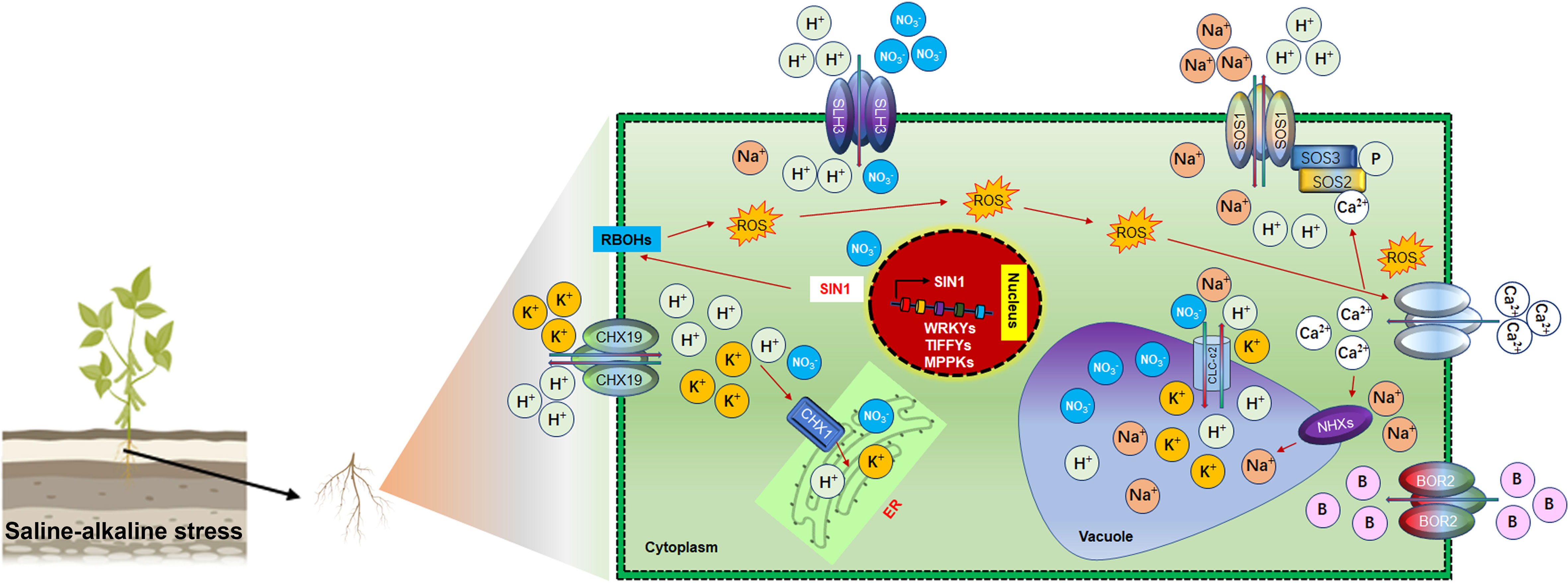
Figure 4 Ionic homeostasis regulation under saline-alkaline stress in Soybean Under saline-alkaline stress, different antiporters and cation exchangers are activated for ionic homeostasis. These transporters transduce ions by choice. For instance, SLH3 is responsible for H+ and NO3-, SOS1 regulate the bidirectional movement of Na+ and H+ ions, CHX19.3 transduce bidirectional movement of K+ and H+ into the endoplasmic reticulum (ER) for packaging and subsequently dispose of through budding process form cell. Activation of RBOH by the nuclear SIN1 gene caused the production of ROS. Stimulating the production of ROS under saline-alkaline stress leads to the activation of Ca2+ channels to mediate the transduction of ions to the vacuole for storage. Interestingly, most of these regulatory pathways are involved in ionic homeostasis rather than pH regulation under saline-alkaline stress in Soybean.
Under salt stress, plants accumulate various organic osmolytes, including proline, glycine betaine, polyamines and sugar alcohols. These osmolytes maintain the intracellular osmotic potential to minimize the harmful effects of salinity stress (Figure 5). Therefore, osmolytes mediated interactive regulatory mechanism along with metabolites, antioxidants and phytohormones will be crucial to understanding soybean salinity tolerance mechanism. Many soybean salt stress-related studies have reported the activation of the proline synthesize gene, Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1) and subsequently, higher accumulation of proline. Proline has an osmotic adjustment capability and plays a protectant role in response to salt stress (Deinlein et al., 2014). Plant metabolism helps proline accumulation, which correlates with plant salt stress response and tolerance (Ahmad et al., 2016a; Nahar et al., 2016). However, recent studies suggested that proline can scavenge only hydroxyl radical but cannot scavenge superoxide, singlet oxygen, nitric oxide, nitrogen dioxide, and peroxynitrite, which puts proline role as a specific ROS scavenger under salt stress conditions (Nahar et al., 2016; Signorelli, 2016; Ahmad et al., 2016b). Furthermore, proline effects are dependent on concentrations, duration and developmental stage of plants under salt stress conditions (Mansour and Ali, 2017). In soybean, overexpression of GmMYB46 induced the expression of salt stress response genes (P5CS1, POD, SOD, and NCED3), thus enhancing the proline and antioxidants accumulation for salt stress tolerance (Liu et al., 2021c). Sugar transport and distribution improve growth and development and stimulates plant salt stress response. Salt-tolerant varieties adjusted their cytoplasmic compatible solutes pool to deal with high salinity levels. Trehalose serves as cells’ metabolic resources and structural components and exhibits hormone-like regulating properties (Sarkar and Sadhukhan, 2022). Sugar synthesis genes, including trehalose-phosphate synthase (TPS), are involved in trehalose production and salt induced osmotic stress adjustment. Under salt stress, trehalose act as a signalling molecule, stabilizes dehydrated enzymes, proteins, and lipid membranes, and activates stress responsive genes for better osmotic stress adaptation in soybean (Cadavid et al., 2020b). Similarly, salt stress induces the sucrose synthesis genes and aromatic amino acids (precursor for phenolic compounds) production. Sucrose helps to balance the osmotic adjustment, and phenolic compounds are potent ROS scavengers in soybean (Noor et al., 2022). Exogenous glycine betaine (GB) application had the otoprotective role and act as a ROS scavenger. GB is an important osmolyte that improves Na+ homeostasis to enhance salt stress tolerance. GB has an excellent role in improving photosynthetic efficiency and antioxidant enzyme activity to mitigate salt-induced growth inhibition. Moreover, GB maintained the Na+/K+ ratio mainly by plummeting the buildup of Na+ in plants (Zhu et al., 2022). GB reduces the membrane peroxidation and improves the photosynthesis and yield under stress conditions in soybean (Khalid et al., 2022). GB treatment increased Na+ efflux in leaf protoplasts and reduced the cellular distribution of Na+. Furthermore, GB improved the vacuolar activity of NHX and V type H+-ATPases genes to improve salt tolerance (Zhu et al., 2022). A previous study reported that GB biosynthesis in plant tissues is highly energy intensive (Bai et al., 2022). Therefore, exogenous spraying of GB is more economic to enhancing soybean salt tolerance. Melatonin has many functions, including omo-protectant, seed priming agent, phytohormone regulator (auxin levels), ion homeostasis, and secondary messengers under abiotic stress. Exogenous application of melatonin reduces the salt effects by increasing the RWC, antioxidant activity, photosynthetic efficiency, cell division, carbohydrates, fatty acids and ascorbate content in soybean (Wei et al., 2015). Recently, a comparative study of GB and melatonin reported that GB is more efficient in relieving the salt effects in Dalbergia odorifera (Cisse et al., 2021). Such kind of studies are lacking in soybean. Therefore, a comparative study that includes a combined effects of both GB and melatonin and the right dose of exogenous spraying will be more beneficial to impart salt tolerance in soybean.
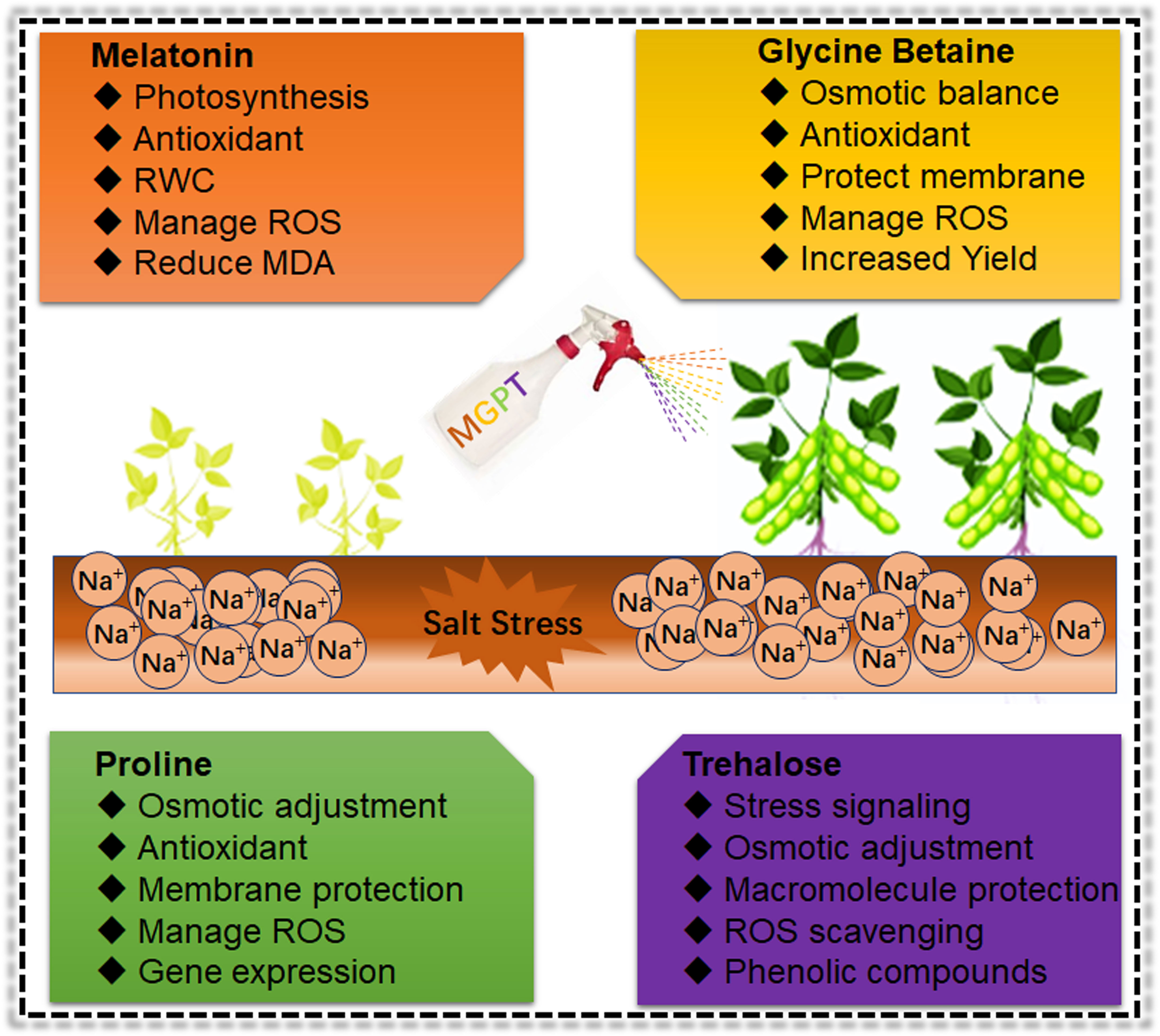
Figure 5 Role of osmoprotectants in ameliorating the Soybean salt stress effects Exogenous application of different biochemical increased the growth and development mainly through enhancement of antioxidant activity, ROS scavenging and protection of cellular structure and membranes under salt stress conditions in Soybean. However, these are short-term approaches to mitigate the salt impacts on Soybean. Therefore, it is necessary to use more advanced biotechnological and breeding approaches to develop salt resilient Soybean germplasm.
Different studies have explained that each plant hormone plays multiple biological roles depending on growth stages, tissues, and environmental conditions (Ku et al., 2018; Yu et al., 2020; Zaid et al., 2021). Here, we summarize how different hormones regulate or ameliorate the salt effects and help to optimize growth, adaptation and final production in soybean (Figure 6). BR is involved in plant adaptation to abiotic stresses. BRI1-EMS suppressor 1 (BES1)/brassinazole-resistant 1 (BZR1) regulates plant signalling pathways in soybean. In soybean, BES1/BZR1 homolog 1 (GmBEH1) interacts with GmBIN2, a putative BR-insensitive 2 (BIN2) to regulate the BR signalling (Yan et al., 2018). Soybean seed priming with BR (24-epibrassinolide, EBL) and nitrogen (N) nutrient solution improved salt stress tolerance. EBL and N triggered the accumulation of different osmolytes such as sugars, proline, and glycine betaine resulting in better protection by preserving tissue water content (Soliman et al., 2020). Therefore, the BR pathway has a cross-talk with osmolytes accumulators in soybean under salt stress. Further, some key enzymes involved in BR synthesis also affect plant salt adaption. For instance, CYP85A1-OE plants exhibited salt tolerance whereas det2-1, SlDWARF, BRI1 or BSK5 knockout plants were found salt-sensitive; however, exogenous BR application rescued the salt-sensitive phenotype of det2-1 and SlDWARF (Cheon et al., 2010; Li et al., 2012; Zhu et al., 2016; Duan et al., 2017). Br also has a cross-talk with ABA; ABA inhibits BR signalling to regulate salt tolerance (Zhang et al., 2009). For example, BRI1 or BSK5 mutants were found sensitive to ABA and BR antagonized the ABA effects for modulating the salt tolerance (Zeng et al., 2010; Li et al., 2012). However, how these pathways respond to soybean salt tolerance is still elusive. Similarly, molecular evidence suggested that a close loop interaction mechanism between SA, GA and BR was activated in response to salt stress, thus conferring plant salt tolerance (Divi et al., 2010; Wang et al., 2019c). These studies suggested a direct or indirect involvement of BR signalling components in salt stress signals through modulation at transcriptional and post-translational levels. Finally, BRs have been proven an essential regulator of plant salt tolerance.
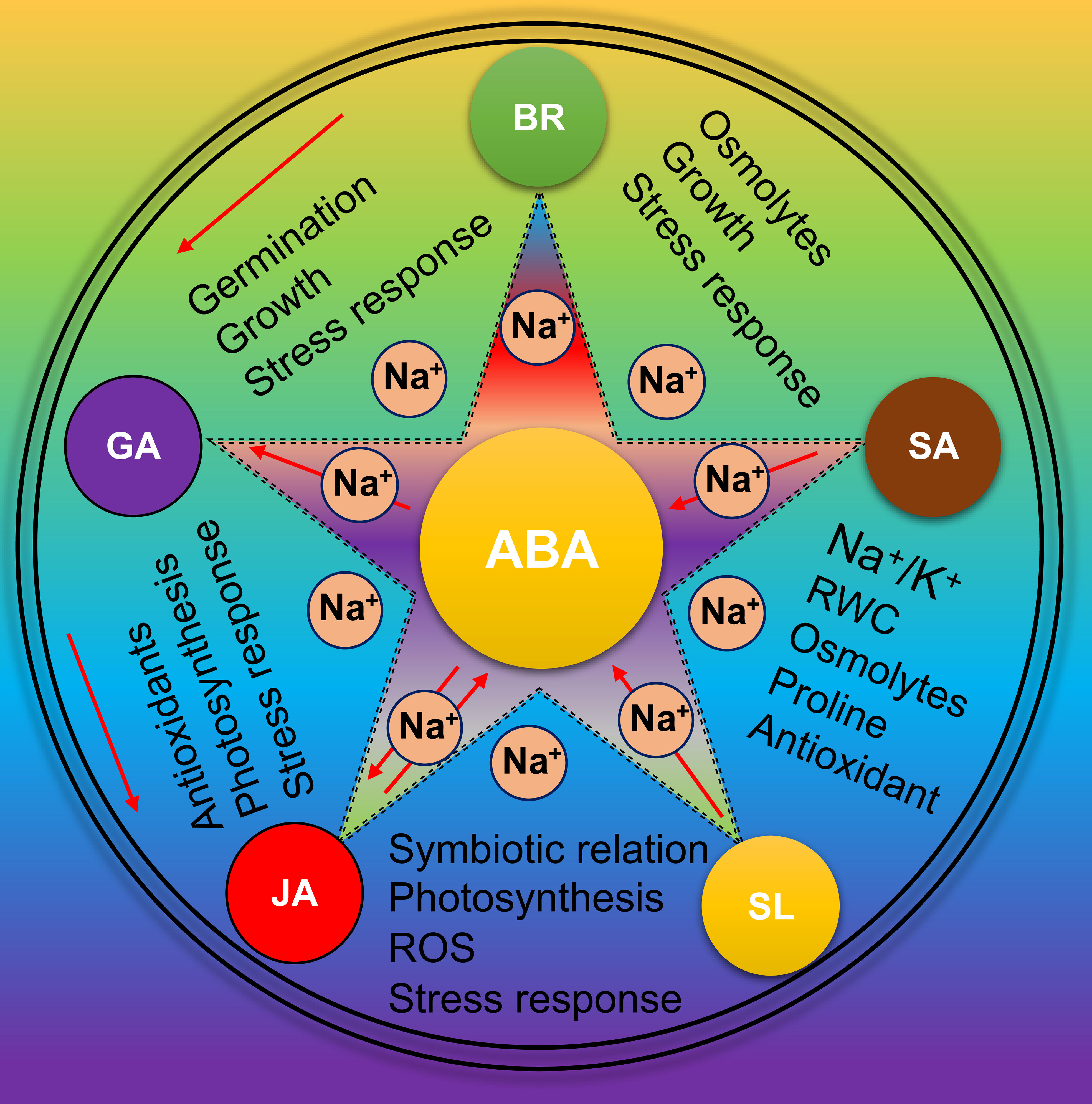
Figure 6 Role of phytohormones in Soybean salt tolerance Under salt stress, various phytohormones naturally activate in Soybean. These phytohormones work in a highly complex interactive network and has cross-talk with each other especially with ABA to reduce the impact of salt stress.
In response to salt stress, activation of JA signaling caused primary root growth inhibition (Zhao et al., 2014). However, jaz3-1 and jasmonates insensitive3 (jai3) mutant increased growth and root cell growth under salt stress (Valenzuela et al., 2016). These results depicted the involvement of the JA pathway in plant salt response. Exogenous JA treatment reduced the salt toxicity by ameliorating the ROS or ion homeostasis and interacting with different phytohormones (Farhangi-Abriz and Ghassemi-Golezani, 2018). Soybean seeds priming with JA, foliar application and/or their combination improved the various physiological traits such as water potential, water use efficiency, osmotic potential, RWC, and photosynthetic rate under salinity stress. JA treatments also improved ABA and GA phytohormones and stress-responsive genes expression of FeSOD, POD, CAT, and APX family under salt stress in soybean. Therefore, JA treatment could efficiently protect seedlings, alleviate salt stress damage, and improve soybean growth against excessive soil salinity (Sheteiwy et al., 2021). Interestingly, JA has a cross-talk with ABA. JA mutant plant demonstrated less accumulation of ABA, while the JAZ1 gene was inducible by ABA treatment to promote salt tolerance (Brossa et al., 2011). Strigolactone (SL) roles in fungi-plant symbiotic interaction, growth and salt tolerance have been well established (Zhang et al., 2015a). For instance, exogenous GR24 enhanced plant growth by increasing the photosynthetic capacity and removing the salt stress effects (Ma et al., 2017). Similarly, exogenous 0.5 μM SL treatment could improve the alkaline and salt tolerance of soybean seedlings by managing the MDA and H2O2 levels and improving the antioxidant system and phenylpropanoid biosynthetic pathway. Furthermore, SL treatment promotes Na+ transport from the roots to the leaves of soybean seedlings to enhance salt tolerance (Chen et al., 2022). Salt treatment induced the SL biosynthesis-related carotenoid cleavage dioxygenases (CCD7 and CCD8) and MAX2 via an ABA-dependent way and upregulating the SL signaling pathway (An et al., 2016; Wang et al., 2019b). SL treatment also improved the salt tolerance of ABA mutants. These results also indicate a crosstalk between SL and ABA for salt tolerance execution in plants. However, exact mechanism of SL and ABA crosstalk yet need to be explored in soybean.
Salt stress reduces the bioactive GA levels and slows seed germination and plant growth (Zhu et al., 2015). Reduction of growth seems to be an adapted mechanism in response to lower GA to prepare the plant for salt tolerance. For example, Arabidopsis and rice GA metabolism-related genes such as GA2ox7, GA2ox5, OsDSK2a and MYB91 enhanced plant salt tolerance by hampering plant growth (Magome et al., 2008; Shan et al., 2014; Zhu et al., 2015; Wang et al., 2020a). Similarly, in soybean, salt stress delays seed germination by negatively regulating gibberellin (GA) while enhancing the ABA biogenesis, which causes an imbalance in the GA/ABA ratio. Fluridone (FLUN), is an inhibitor of ABA biogenesis that promote soybean seed germination under salt stress by altering the GA1/ABA, GA3/ABA, and GA4/ABA ratios (Shu et al., 2017). The cotton gene GhPLATZ-OE in Arabidopsis speeds up germination by suppressing the ABI4 (Zhang et al., 2018). The ABI4 gene mediated the transcription of GA2ox7 (GA catabolic) and the ABA synthesis gene NCED6 (Shu et al., 2016). Higher GA is required for seed germination but lowered GA levels for salt stress tolerance. Therefore, it is direly necessary to identify a feedback loop mechanism to switch on or off the GA levels at the germination and later growth stages.
Salicylic acid (SA) is an essential phenolic compound that improves plant growth and defense systems under salt stress by accumulating osmolytes and an enhanced antioxidant system (Filgueiras et al., 2019; Dawood et al., 2022). SA also facilitates nitrogen fixation and microbial symbiotic relationships to enhance photosynthesis and productivity under salt-stress conditions (Ahanger et al., 2019). SA acts in a dosage manner and snc1, sid2, and npr1-5 mutant plants were salt-sensitive (Xu et al., 2010; Jayakannan et al., 2015). Exogenous application of SA induced the proline accumulation through activation of P5CS and involved MYB genes in maintaining higher ABA levels to protect the salt stress damage (Zheng et al., 2018). Interestingly SA reduces lipid peroxidation and interacts with endogenous GAs and ABA to regulate the Na+/K+ balance to enhance salt tolerance (Liu et al., 2022b). It was reported that an exogenous supply of SA improves salt tolerance, growth and yield in soybean. SA enhanced the RWC, osmolytes, protein accumulation, enzymatic and non-enzymatic antioxidants (APX) and ionic homeostasis to enhance salt tolerance in soybean (Nigam et al., 2022). SA also regulated cellular Na+/K+ homeostasis through SA-mediated ionic channels to enhance salt tolerance and interactions with other plant hormones like Cytokinin and GA (Liu et al., 2019c).
During domestication, artificial selection caused the loss of important stress-responsive genetic loci in cultivated soybean. A significant variation exists among cultivated and wild soybeans for the salt response. The allelic diversity and beneficial alleles could be incorporated into domesticated soybeans to improve salt tolerance. Therefore, identifying natural alleles controlling salt tolerance has great potential to improve the soybean yield through genetic improvement. Recently, Glyma03g32900 (GmCHX1/GmSALT3) was identified as a novel shoot sodium ion transporter and associated with salt tolerance (Guan et al., 2014; Qi et al., 2014). GsCLC‐c2 squeezed excessive Cl− into the vacuoles of root cells to improve salt tolerance (Wei et al., 2016). A recent genome-wide study identified 34 salt‐tolerant wild soybeans. A 7‐bp insertion/deletion (InDel) in the promoter of GsERD15B (early responsive to dehydration 15B) regulates the salt tolerance differences in soybean germplasm. GsERD15B contained a PAM2 domain for interaction with poly(A)‐binding (PAB) proteins. The 7‐bp deletion caused up‐regulation of GsERD15B, two GmPAB genes, and various stress‐related genes, including GmABI1, GmABI2, GmCAT4, GmbZIP1, GmP5CS, GmPIP1:6, GmSOS1 and GmMYB84 in response to salt stress. Natural variation in GsERD15B promoter and overexpression of GsERD15B increased the ABA‐signalling, proline content, catalase peroxidase, dehydration response and cation transport to enhance salt tolerance in soybean (Jin et al., 2021b). Natural variations were also identified for dehydration-responsive element-binding (DREB) family transcription factor genes in cultivated soybean. Recently, GWAS identified 103 DREB genes by combining with RNA-sequencing and population genetics of wild, landrace, and cultivated soybean accessions. DREB3a underwent artificial selection, but the DREB3b allele (DREB3b39Del) contributed to salt tolerance in soybean (Hou et al., 2022). Natural variation in gene promoter region also contributes towards salt tolerance in soybean. For instance, four promoter haplotypes of a class B heat shock factor HSFB2b were identified, which repressed the GmNAC2 to improve flavonoid accumulation by activating the flavonoid biosynthesis pathway genes to enhance salt tolerance. Promoter haplotype II was selected during early domestication than haplotype III and had a low distribution frequency (Bian et al., 2020b). Similarly, SNP-based GWAS helped to locate the Glyma03g32900 gene on Chr 3 based on three significant SNP markers, Salt-20, Salt14056 and Salt11655, which were highly correlated with salt-related traits. Additionally, a new minor locus on Chr 8 was also predicted for salt tolerance in soybean (Do et al., 2019b). These results suggested that natural variations exist in highly diverse soybean germplasm, which could be exploited for salt tolerance.
Marker-assisted breeding (MAB) is an efficient approach for developing salt-tolerant soybeans. MAB helps to capture the natural variations in chromosomal regions, termed as QTLs. Major QTLs that contributed to salt tolerance are important for breeding. MAB uses DNA markers associated with particular QTLs that help identify genotypes with the desired alleles (Abbas et al., 2022; Zhang et al., 2015b). Recently, MAB helped to identify a major QTL qST-8 related to salt tolerance which is closely linked with the marker Sat_162 and detected on Chr 8 using a population of 184 recombinant inbred lines (RILs). Similarly, GWAS identified several SNPs in the same genetic region on Chr 8, which were significantly associated with salt tolerance. A candidate gene Glyma.08g102000 (GmCDF1) belong to the cation diffusion facilitator (CDF) family was identified in this region. GmCDF1 affects the K+/Na+ ratio and negatively regulates salt tolerance by regulating the expression of two ion homeostasis-associated genes, GmNHX1 and GmSOS1, in transgenic hairy roots (Zhang et al., 2019a). Similarly, another study identifies major salt-related loci on Chr 3 and Chr 13 using a mapping population of 132 F2 families derived from Williams 82 (salt sensitive) and Fiskeby III (salt tolerant). On Chr 3, three genes were identified based on polymorphic markers, Salt-20, Salt14056, and Salt11655, and significantly associated with salt tolerant phenotype (Do et al., 2018). Similarly, using a RIL population derived from salt-tolerant (Jidou 12) and the salt-sensitive (Ji NF 58) identified a major salt-tolerant QTL, flanked by SSR markers GMABAB and Barcsoyssr_03_1421 on Chr 3, might be useful in marker-assisted selection for soybean salt tolerance (Shi et al., 2018b). A mapping population descendent from a cross between a salt-sensitive Cheongja 3, and a salt-tolerant landrace, IT162669, was analyzed for QTLs conferring salt tolerance. Two novel major loci, qST6, on Chr 6, and qST10, on Chr 10, and eight candidate genes involved in phosphoenolpyruvate carboxylase and an ethylene response factor that control ion toxicity and physiology in response to salt were identified (Cho et al., 2021). A F2:3 mapping populations, was used for QTL mapping, derived from NY36-87 and two salt-sensitive Zhonghuang39 and Peking soybean cultivars. A salt tolerance locus on Chr 3 and Chr 18 that harbored GmSALT3 and GmSALT18 genes co-segregated with the salt tolerance locus was identified using simple sequence repeat SSR markers and bulked segregant analysis (BSA) mapping strategy. This study provides genetic material and novel loci for salt-tolerant soybean breeding (Guo et al., 2021).
Transcriptome analysis is a powerful approach to exploring genome-wide gene expression reprogramming in response to salt stress. The expression and function of genes controlling physiology and metabolism must be modified for plants to endure and recover from salt stress. Recently, under salt stress, RNA-seq analysis identified 15,997 and 15,494 DEGs in the leaves and roots of soybean. The DEGs enrichment analysis divided molecular pathways into different adaptation-related processes under salt stress. The leaf enriched DEGs modules were related to amino acid, protein turnover, carbon fixation, photosynthetic process, phytohormone, and primary nitrogen metabolism. In contrast, root-enriched DEG modules were starch and sucrose metabolism, phenylpropanoid biosynthesis, cell wall and cell membrane composition reactive oxygen species (ROS) scavenging (Liu et al., 2019a). Similarly, another transcriptome analysis revealed 1,235 DEGs under salt stress. Out of a total of 116 TFs, 17 belonged to MYB families. Functional analysis revealed that GmMYB46 localized in the nucleus, up-regulated by salt and mannitol, and enhanced the salt tolerance by ectopic-overexpressing of GmMYB46 in Arabidopsis. Although, GmMYB46 upgraded the expression of salt stress response genes (P5CS1, SOD, POD, and NCED3), but actual mechanism is still vague in soybean and Arabidopsis (Liu et al., 2021c). Transcriptomic analysis of the salt-tolerant ‘Qi Huang No.34’ (QH34) and the salt-sensitive cultivar ‘Dong Nong No.50’ (DN50) identified 17,477 genes responsive to salt stress. Among these, 6644 were DEGs between the two soybean cultivars. These DEGs were enriched in different stress-responsive pathways like phenylpropanoid biosynthesis, phytohormone signalling, the mitogen-activated protein kinase pathway, ribosome metabolism and oxidoreduction, which may play crucial roles in response to salt stress (Hu et al., 2022). The transcript expression profile of a drought-sensitive (C08) and a tolerant (Conquista) after 4 h of salt stress identified 1400 DEGs. Among them, 647 were induced, and 753 were repressed. These DEGs were enriched in ABA, BR signalling pathways, ethylene biosynthesis, DNA repair, and the plastid translation process (Cadavid et al., 2020b). Transcriptomic analysis of RA-452 x Osage mapping population, along with two F4:6 lines with salt tolerant and sensitive lines a total of 2374, 998, 1746, and 630 DEGs in a time series experiment, at 0, 6, 12, and 24 h, respectively. This study identified 13 common DEGs; six were up and seven DEGs were downregulated in the salt-tolerant line. This study reported key potential genes involved in the salt-tolerance such as Glyma.02G228100, Glyma.03G226000, Glyma.03G031000, Glyma.03G031400, Glyma.04G180300, Glyma.04G180400, Glyma.05g204600, Glyma.08G189600, Glyma.13G042200, and Glyma.17G173200 in the soybean salt-tolerant line (Zeng et al., 2019).
Metabolites are low-molecular-weight compounds produced in plants. These metabolites proved to be defensive compounds against biotic and abiotic stresses in plants. During their evolution, plants synthesize new metabolites, and about 200,000 specialized metabolites are produced by plants (Sugiyama, 2019). Salt stress leads to osmotic stress and changes the metabolomic state in soybean. Recently a proteomics study identified salt stress-responsive phosphoproteins in soybean. Upon salt treatment, out of a total of 4698 phosphopeptides, 412 were significantly up-regulated. GmMYB173 protein was differentially phosphorylated at serine 59 upon salt treatment and acted as a substrate for the casein kinase-II. Phosphorylation of MYB binding sites in the promoter of GmCHS5 (flavonoid synthase gene) facilitated GmMYB173 binding. Metabolomics analysis of GmCHS5 catalyzed chalcone and flavonoids revealed that 24 flavonoids of 6745 metabolites were significantly up-regulated upon salt treatment. Overexpression of GmMYB173S59D and GmCHS5 resulted in the accumulation of cyaniding-3-arabinoside chloride belonging to dihydroxy B-ring flavonoid (an antioxidative agent) that enhanced the soybean salt tolerance. Therefore, salt stress-mediated phosphorylation of GmMYB173 increased its binding affinity to the GmCHS5 promoter. Thus, the GmMYB173-GmCHS5 module contributed to the accumulation of dihydroxy B-ring flavonoids and enhanced soybean salt tolerance (Pi et al., 2018). Metabolic profiling of soybean plants revealed that the TCA cycle and amino acid metabolism contributed to the salt response. Soybean varieties with higher nitrogen absorption, assimilation rate, faster TCA cycle activity and more amino acid accumulation for better salt stress response. Rhizospheric secreted soil microbes rapidly degrade metabolites to mediate biological communication. Isoflavones and strigolactones metabolites help to develop a symbiotic relationship between rhizobia and arbuscular mycorrhizal fungi and soybean root (Massalha et al., 2017; Mishra et al., 2022). These symbiotic relationships also help to improve the salt tolerance in soybean.
In planta, genes functional validation is necessary to identify the role of a particular gene in salt tolerance in soybean. Due to the highly complex genome structure of soybean, in the last two decades, soybean genetic transformation progress remained slow and inefficient. Therefore, functional gene validation was primarily performed in Arabidopsis or transient expression in soybean hairy root (Table 2). Several soybean genetic transformation protocols were developed with variable success rates, such as immature cotyledons, hypocotyls, shoot meristems, embryos, half-seed explants and cotyledonary nodes (Zhang et al., 2022b). Soybean Agrobacterium-mediated cotyledonary node (CN) transformation has an average transformation efficiency of 3.8%–8.7% (Li et al., 2017). Recently, some studies reported soybean average transformation efficiency up to 18.7% but still lower than rice and maize (Pareddy et al., 2020; Zhang et al., 2022b). Many factors combinedly affect the soybean transformation efficiency, including varieties, Agrobacterium medium concentration, hormone levels, explant co-cultivation time and explant regeneration capability (Li et al., 2017). Compared with conventional breeding, transgenic approaches have many advantages, including short duration, targeted genome modification and traits improvements (Figure 7). For instance, if transgenic approaches are coupled with speed breeding, the advanced elite germplasm creation process will be more robust and readily available for large-scale cultivation (Hussain et al., 2023).
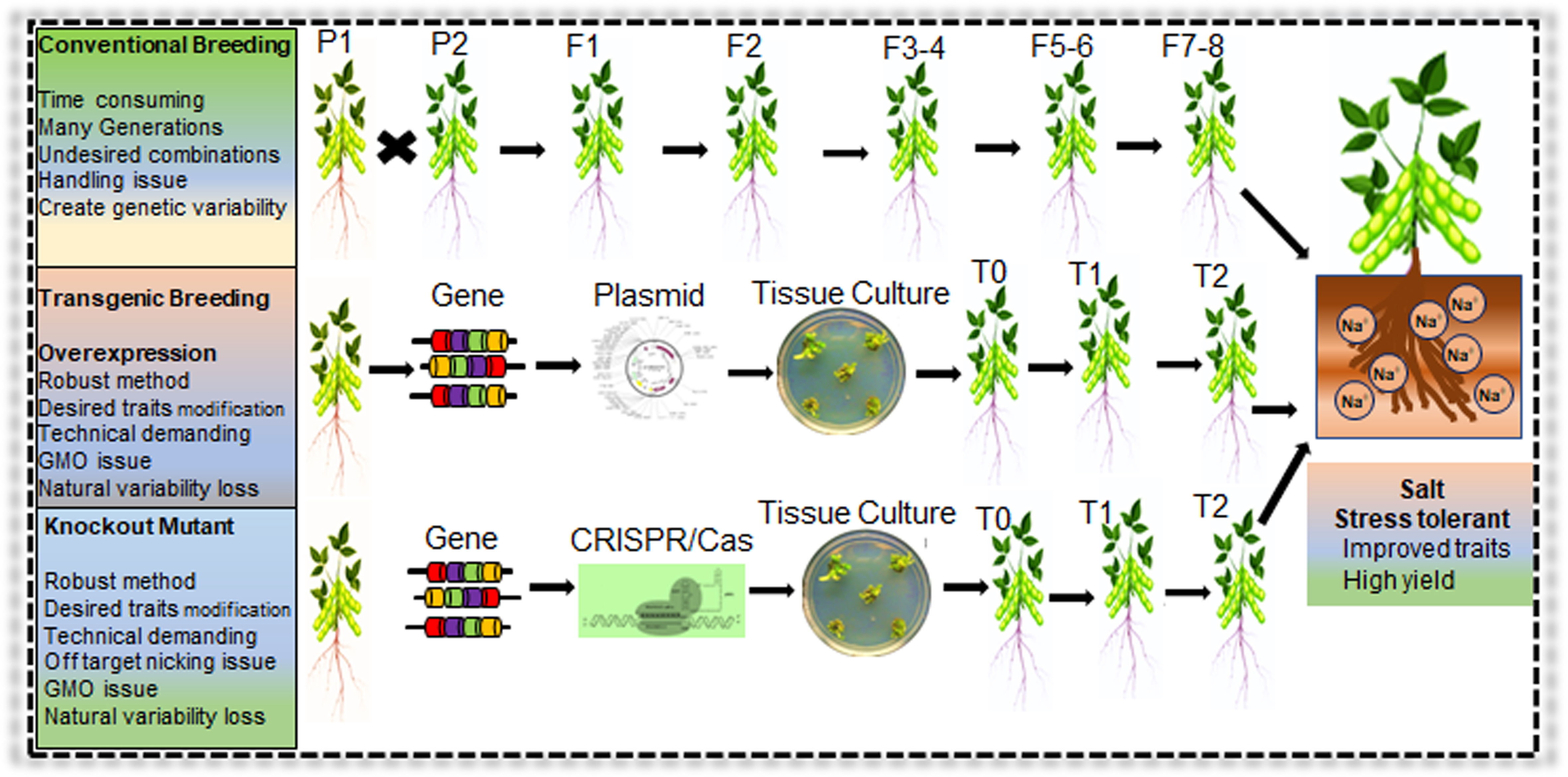
Figure 7 Conventional and modern genetic approaches for development of salt stress tolerant Soybean. Application of conventional breeding, transgenic breeding, and genome editing (CRISPR/Cas) system for salt tolerant Soybean germplasm development. Modern biotechnology approaches are robust that can generate stress responsive germplasm in a short duration compared to conventional breeding methods.
Recently, transcriptome analysis identified GmLecRlk (Glyma.07G005700) a candidate salt-responsive gene in soybean. Overexpression of GmLecRlk in hairy root positively affected salt tolerance mainly through antioxidant activity and enhanced ROS scavenging ability in soybean. Furthermore, GmLecRlk improves the salt tolerance by upregulating GmDREB2, GmERF3, and GmbHLH30 and downregulating GmLAMP1, GmGH3.6 and GmPUB8 of soybean (Zhang et al., 2022c). Zinc-finger (VOZ) transcription factors showed differential expression under dehydration, SA and salt stress conditions. Overexpression of GmVOZ1G in soybean hairy roots positively affected drought and salt tolerance by improving antioxidants and maintaining higher RWC. While RNA interference (RNAi) knockout GmVOZ1G soybean plants were sensitive to salt and drought stresses (Li et al., 2020). Modulation of antioxidant genes has positive effects on salt tolerance in soybean. Overexpression of the wild soybean peroxidase GsPRX9 gene improved the morphological traits such as fresh root weight, primary root length, POD, SOD and glutathione enzymes activities, subsequently improving salt tolerance (Jin et al., 2019). Calcineurin B-like protein-interacting protein kinases (CIPKs) are involved in plant adaptation to abiotic stresses. Overexpression or knockout of GmCIPK21 in soybean hairy roots led to increased or decreased salt tolerance, respectively. Further, GmCIPK21 physically interacted with GmCBL4 to scavenge salt-induced ROS and improved ABA response to improve salt tolerance (Li et al., 2022a). Similarly, another CIPK gene (GmPKS4) is upregulated under alkali, salt-alkali, drought, or ABA and localized in the nucleus and cytoplasm in soybean. Overexpressing GmPKS4 in Arabidopsis and soybean hairy roots enhances osmolyte accumulation, ROS scavenging, and salt stress-related gene regulation (Ketehouli et al., 2021). Overexpression of SALT INDUCED NAC1 (GmSIN1) belonging to the NAM/ATAF1/2/CUC2 (NAC) transcription factors promoted root growth, increased yield and salt tolerance in soybean. RNAi soybean for GmSIN1 had the opposite effect. Furthermore, a positive feed-forward system enables the rapid salt signaling through GmSIN1, 9-cis-epoxycarotenoid dioxygenase coding genes (GmNCED3s) involved in ABA synthesis and Respiratory burst oxidase homolog B (GmRbohBs) associated with ROS generation which enables the rapid homeostasis of ABA and ROS signalling for salt tolerance (Li et al., 2019c). Under salt stress, transgenic soybean plants overexpressing GmNAC085 displayed better germination rates due to higher activities of antioxidant enzymes and activation of key stress-responsive proline and dehydrins genes (Hoang et al., 2021). Overexpression of GmMYB118 in soybean hairy roots improved drought and salt tolerance. While CRISPR-mediated knockout plants showed reduced drought and salt tolerance. GmMYB118 improved drought and salt tolerance by regulating osmotic and oxidizing substances and promoting the expression of stress-associated genes (Du et al., 2018). Similarly, overexpression of GmbZIP2 improved the plant resistance to drought and salt stresses through enhanced expression of the stress-responsive genes, including GmDHN15, GmGST1, GmMYB48, GmWD40, and GmLEA in soybean (Yang et al., 2020). Further, some TFs are negative regulators of soybean salt tolerance. For instance, overexpression of GmbZIP15 caused hypersensitivity to salt stress in soybean mainly through repressing effects on stress-responsive genes involved in both ABA-dependent and ABA-independent pathways. GmbZIP15 negatively regulates GmWRKY12 and GmABF1 and positively regulates GmSAHH1 expression in response to abiotic stress (Zhang et al., 2020a). Overexpression of GmTLP8 (Tubby-like proteins 8) improved salt and drought stresses tolerance, whereas GmTLP8-RNAi knockout promoted stress sensitivity in soybean (Xu et al., 2022b). Under salt stress, overexpression of Domain of unknown function 4228-70 (DUF4228) in soybean reduced the leaf curling, wilting MDA, H2O2, and O2- and increased the RWC, proline, and chlorophyll contents to enhance the salt tolerance (Leng et al., 2021). Phospholipase C (PLC) hydrolysis the phospholipids and is involved in plant stresses. soybean overexpressing the GmPI-PLC7 exhibited drought and salt tolerance, while the GmPI-PLC7-RNAi lines exhibited sensitive phenotypes (Chen et al., 2021). Soybean C2 domain-containing proteins GmC2-58, GmC2-88, and GmC2-148 responded to salt, drought, and ABA treatments. Transgenic soybean plants of GmC2-148 maintained higher proline content, lower H2O2, O2- and MDA and increased expression of stress-responsive genes, which enhanced drought and salt tolerance in soybean (Sun et al., 2021b). Calcium-dependent protein kinases (CDPKs) responded to a variety of abiotic stresses. Overexpression of GmCDPK3 improved drought and salt tolerance compared with GmCDPK3-RNAi by reducing the cell membrane damage, MDA and accumulation of higher proline (Pro) and chlorophyll contents in soybean (Wang et al., 2019a). Calmodulin-binding protein 60 (CBP60) is located in the cytomembrane and induced by drought and salt stresses. Soybean hairy roots overexpressing the GmCBP60A-1 increased the proline content, lower electrolyte leakage and MDA, which likely enhanced the drought and salt tolerance, compared to GmCBP60A-1-RNAi soybean (Yu et al., 2021). A novel R2R3-type MYB (GmMYB81) differentially accumulated during embryo development, drought, salt and cold in soybean. GmMYB81 interacts with the abiotic stress regulator GmSGF14l and cooperatively affects soybean abiotic stress tolerance and could be a candidate gene for salt stress tolerance development at seed germination (Bian et al., 2020a). Overexpression of GmTIFY10e and GmTIFY10g genes improved salt tolerance by increasing the PRO, POD, and CAT activity and decreasing the MDA content, contrasting to the GmTIFY10e-GmTIFY10g-RNAi plants which exhibited sensitivity to salt stress in soybean. Further analysis showed that GmTIFY10e and GmTIFY10g regulate the transcript levels of genes related to the ABA signal pathway, such as GmSnRK2, GmMYC2, GmCAT1, GmPP2C and GmPOD (Liu et al., 2022a). GmTGA17, expression was induced by both salt and drought stresses, is a nuclear-localized protein. Overexpression of GmTGA17 in Arabidopsis and soybean enhanced drought and salt stress tolerance. However, soybean GmTGA17-RNAi plants exhibited sensitivity to drought and salt stress. GmTGA17-OE improved the physiological parameters such as chlorophyll and proline contents and decreased the MDA content in soybean compared to RNAi plants (Li et al., 2019a). Similarly, A20/AN1 zinc finger domain containing stress-associated proteins (SAPs) GmSAP16 gene was identified as a novel regulator of water deficit stress, salt, and abscisic acid (ABA) stresses. The overexpression of GmSAP16 enhanced the drought and salt tolerance by modifying the proline and chlorophyll and lowering the MDA contents compared to GmSAP16-RNAi soybean seedlings. GmSAP16-OE and GmSAP16-RNAi also modified the expression of stress-related genes, including GmDREB1B;1, GmRD22, GmDREB2, GmNCED3, GmNHX1, and GmSOS1 (Zhang et al., 2019b). Fatty acid desaturases (FADs) cause the desaturation of fatty acids by introducing double bonds and modulates membrane fluidity in response to abiotic stresses. Similarly, the overexpression of the Omega-3 Fatty Acid Desaturase in soybean showed higher accumulation of JA levels, chlorophyll content, RWC, proline content and higher expression of GmWRKY54 as compared to GmFAD3-silenced plants and exhibited drought and salt tolerance (Singh et al., 2022). Similarly, overexpression of GmNAC109, a homologous of AtAF1 in Arabidopsis, enhanced the drought and salt tolerance. GmNAC109-OE activated the expression of stress marker genes such as DREB1A, DREB2A, AREB1, AREB2, RD29A, and COR15A in transgenic Arabidopsis. ABA-responsive genes ABI1 (ABA INSENSITIVE 1) and ABI5 were also induced, which caused hypersensitivity to ABA in transgenic Arabidopsis. However, GmNAC109 could not induce the ABA-biosynthetic gene NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3) expression and endogenous ABA content. However, GmNAC109-OE significantly increased the expression of AIR3 (AUXIN-INDUCED IN ROOT CULTURES 3) and repressed the ARF2 expression and lateral root formation in transgenic Arabidopsis lines (Yang et al., 2019). Soybean elongation factor 1α (EF1α) includes three structural domains: one GTP-binding domain and two oligonucleotide-binding domains that respond to drought and salt stress. GmEF4-OE soybean showed drought and salt tolerance through improved physiological traits and higher accumulation of proline and lower H2O2, O2-, and MDA (Gao et al., 2019). In soybean GmGRAS37 was significantly induced by drought, salt, ABA and BR treatment. GmGRAS37-OE soybean plants exhibited improved resistance to drought and salt stresses via enhanced expression of drought and salt responsive genes (Wang et al., 2020b). The Catharanthus roseus RLK1-like (CrRLK1L) protein kinase GmCrRLK1L20 responds to drought and salt stress. GmCrRLK1L20-OE improved drought and salt tolerance by activating stress responsive genes GmDREB-like, GmMYB84, GmGST15, GmWRKY40, GmNAC29 and GmbZIP78 (Wang et al., 2021b). Protein phosphatase 2A (PP2A) regulates plants’ intracellular and extracellular ROS signals. Functional analysis demonstrated that GmPP2A-B-71-OE could improve drought and salt tolerance by activating stress responsive genes and ROS elimination in soybeans (Xiong et al., 2021). The DREBs gene belongs to the AP2 family. Overexpression of GmDREB6 into DT84 soybean cultivar enhanced the expression level of the GmP5CS gene and proline accumulation. Subsequently, transgenic soybean plants exhibited higher survival under salt stress treatment (Nguyen et al., 2019). Overexpression of GmNAC06 (Glyma.06g21020.1) caused the accumulation of proline and glycine betaine and alleviating the ROS under salt stress. GmNAC06-OE regulates the Na+/K+ ratios by activating the GmUBC2 and GmHKT1 transporter to maintain ionic homeostasis and improve soybean salt tolerance (Li et al., 2021a). The GsCLC-c2 gene and its promoter are activated in response to salt stress. In Arabidopsis or wild soybean plants overexpressing GsCLC-c2, the salt-induced growth reduction was markedly ameliorated through improved physiological traits compared to GsCLC-c2-RNAi wild soybean. GsCLC-c2 regulated the anionic homeostasis in salt-stressed transgenic Arabidopsis and soybean, thus conferring enhanced salt tolerance (Liu et al., 2021b). GmTGA13 induced the expression of the stress-responsive genes and absorption of K+ and Ca2+, thus regulating the ion homeostasis in the cell (Ke et al., 2022). The MYB-TF involves plant development, secondary metabolism, and abiotic stress responses. GmMYB68-OE showed salt and alkali tolerance via osmotic adjustment and photosynthetic rates and improved yield compared to the GmMYB68-RNAi and WT (He et al., 2020b). The F-box family regulates salt responses and has a developmental role in soybean. At least 12 salt-responding F-box genes were identified in soybean (Jia et al., 2017b). EARLY FLOWERING 3 (ELF3) gene suppressed salt stress response pathways to enhance salt tolerance. In soybean, the J allele is an ortholog of AtELF3 Arabidopsis. Knockout mutants of J-alleles greatly prolong maturity and increase the soybean yield; however, reduced the salt tolerance. Overexpression of J-allele increased salt tolerance by positively regulating the expression of downstream salt stress response genes, including GmNAC, GmSIN1 and GmWRKY12, GmWRKY27, GmWRKY54 in soybean (Cheng et al., 2020). GmWRKY12-OE enhanced drought and salt tolerance via increased proline (Pro) content and decreased MDA of transgenic soybean (Shi et al., 2018a). Ankyrin repeat (ANK) proteins had multiple roles in plant growth and environmental stresses. Soybean GmANK114 belongs to the RING finger (RF) domain-containing ANK-RF subfamily. GmANK114-OE improved the germination rate in Arabidopsis and soybean survival under drought and salt treatments. GmANK114-OE activated the transcription of abiotic stress-related genes WRKY13, NAC11, DREB2, MYB84, and bZIP44, under drought and salt stresses in soybean (Zhao et al., 2020). Stable soybean transgenic plants with ectopically expressing Arabidopsis AtABF3 showed substantial salt stress tolerance by protecting the ion leakage rate and maintaining higher chlorophyll contents (Kim et al., 2018). Ectopic expression of AtARA6 in Shen Nong 9 (SN9) soybean regulated the SNARE complexes in the vesicle transport pathway, which may enhance salt tolerance directly. In transgenic soybeans, MYC2, WRKY6, and WRKY86 were downregulated while four inositol oxygenase genes were induced after salt treatment in transgenic soybeans (Hong et al., 2022). Ectopic over-expression of GmbZIP19 in Arabidopsis increased the sensitivity to salt and drought by destroying the stomata and affecting the stress-related gene expression. GmbZIP19 is a negative regulator of salt and drought stress tolerance (He et al., 2020a). Expression of Cytosolic Glucose-6-phosphate dehydrogenase (G6PD or G6PDH) and GmG6PD7 were induced by NaCl treatment in soybean. Overexpression of GmG6PD7 in Arabidopsis increased the seed germination rate, primary root length and markedly decreased the ROS levels in the transgenic plants. GmG6PD7 affected the glutathione, NADPH and ABA levels and activates ROS scavengers to increase salinity tolerance in Arabidopsis. Complementation assay showed that GmG6PD7 could rescue the seed and root phenotype of Arabidopsis cytosolic G6PD mutant (Atg6pd5 and Atg6pd6) under salt stress (Jin et al., 2021a). GsMYB15 from wild soybean is a typical R2R3-MYB TF that contains multiple stress-related cis-elements and is located in the nucleus. Arabidopsis plants, overexpressing GsMYB15, showed salt tolerance and enhanced resistance to H. armigera larvae. GsMYB15 regulates transgenic plants’ defense and salt stress-related genes (Shen et al., 2018). Furthermore, the Ectopic expression of GmNHX1 improved the morphology and generated more rosette leaves in Arabidopsis under salt stress conditions compared to the wild-type. GmNHX1 increased the Na+ transportation to leaves and caused the reduction of Na+ absorption in roots. Therefore, GmNHX1 maintained a higher K+/Na+ ratio under salt stress conditions (Sun et al., 2019b).
Alternate splicing events could generate multiple mature mRNAs from one gene due to exon skipping, intron retention and AS of 5′ or 3′ sites (Ding et al., 2014; Laloum et al., 2018). The AS events could change genes’ protein structure, metabolic functions and cellular locations. Interestingly, AS variants could compete with normal gene transcript, thus enhancing protein function interference, transcriptome, proteome diversity and stress tolerance (Wani et al., 2020). A recent study has found that abiotic stress-responsive genes are more prone to AS (Yang et al., 2022). Salt stress promotes the alternative gene splicing, 5′ and 3′ splice-site selection introduces the premature termination codons (PTCs) in 49% of all intron-containing genes, thus generating multiple transcripts from the same allele (Ding et al., 2014). For instance, in soybean, salt stress produced the ACYL-COA-BINDING PROTEIN (ACBP) variants through AS, cannot interact with lipoxygenases (LOXs). Overexpression of ACBP AS-variants differentially regulates the expression of LOXs activity, lipid peroxidation and salt tolerance in soybean and Arabidopsis (Lung et al., 2022). Under salt stress, phytohormone ABA accumulates in the plant’s roots and is transported to leaves to regulate stomata or to avoid water loss. ABA also controls plant growth and improves salt stress tolerance through AS events of different transcription factors, protein kinases, and splicing factors. ABA-induced the AS mainly by increasing the number of unconventional splicing sites or by recruiting different splicing factors. ABA inhibits the PP2C phosphatases and activates SnRK2 protein kinases to enhance the ABA signalling. The AS of the HAB1 gene (a negative regulator of SnRK2) is regulated by ABA through intron retention and thus protects the inhibition of SnRK2 protein kinases to respond to salt stress in an ABA-dependent manner (Wang et al., 2015b; Laloum et al., 2018). This evidence indicates that AS modulates salt stress responses by regulating the ABA-dependent and ABA-independent pathways, identifying the novel regulatory layer in plant salt stress tolerance. However, mRNA splicing is an essential cellular process; thus, it will be interesting to identify the regulatory components that transduce the signal to pre-mRNA splicing under salt stress and how they could be helpful for salt tolerance germplasm development. Recent studies have reported that in response to salt stress, AS of a serine/arginine‐rich splicing factor (SR) 45 produced two SR45.1 and SR45.2 variants. SR45.1 was found to be responsive to salt stress and rescued the salt-sensitive phenotype (Figure 8). Further, SR45a-CBP20-CBP80 (cap‐binding protein 20) cascade mediates the splicing of salt stress‐responsive genes by involving ubiquitination and sumoylation process (Kong et al., 2014; Albaqami et al., 2019; Li et al., 2021b). Thus, these studies show that the SR45‐CBP20-CBP80 pathway is a critical regulator in pre‐mRNA splicing in response to salt stress (Wang et al., 2023).
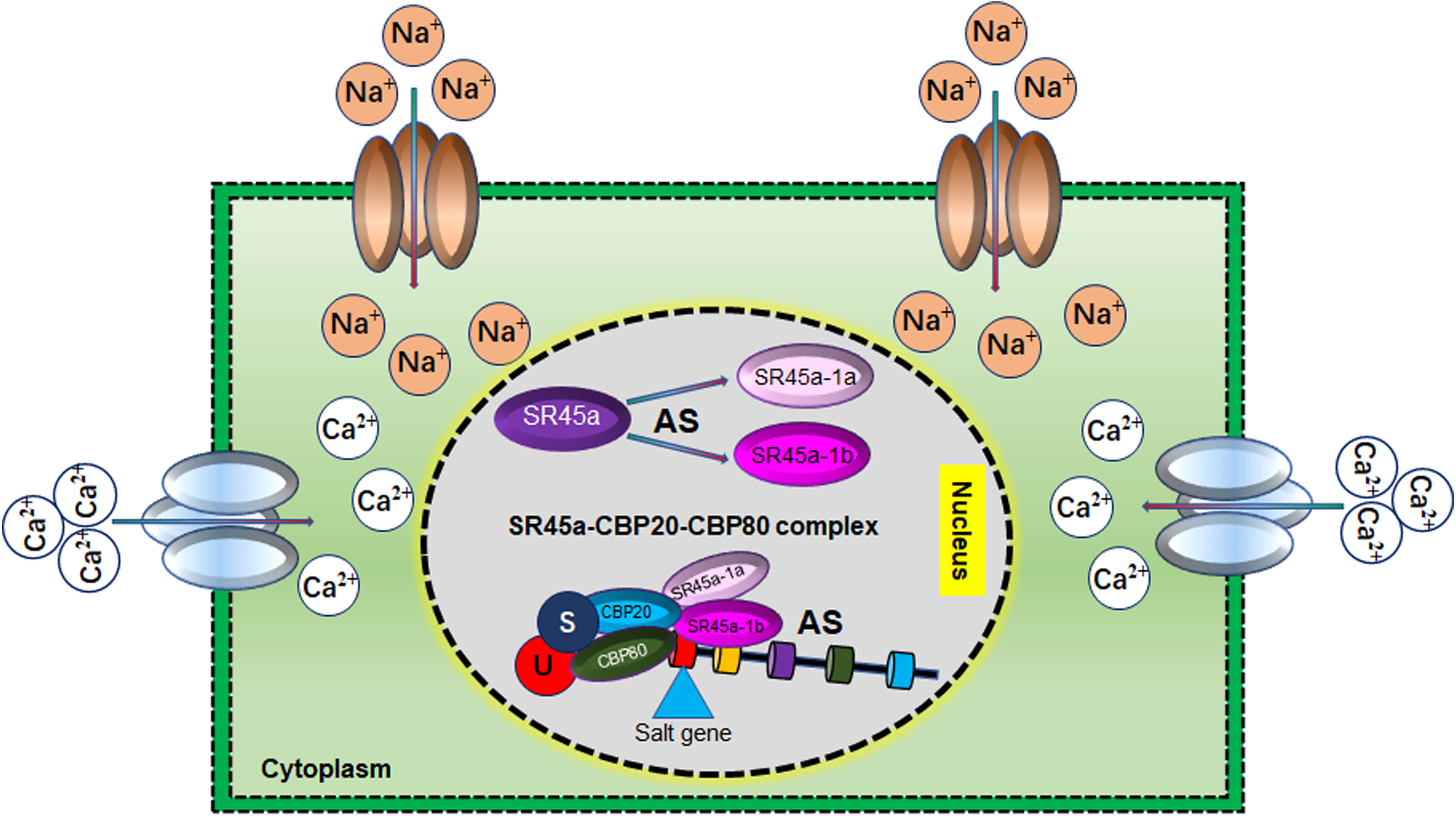
Figure 8 Role of alternative splicing (AS) in Salt tolerance Under salt stress conditions, AS of SR45a generates two variants SR45a-1a and SR45a-1b. These AS variants interact with CBP20 and CBP80 to form a regulatory cascade that regulates the alternative splicing of salt stress‐responsive genes. SR45a-1a and SR45a-1b regulate the salt tolerance in isoform-dependent manner.
Although plants lack a nervous system but are intelligent; plants remember, choose, and make decisions to optimize their resources and enhance their fitness in response to abiotic stresses. Plants inherit this information to the next generation(s) through epigenetic modifications to adapt more efficiently to climate change (Gallusci et al., 2022). Plant memory is solely based on cellular, molecular, and biochemical networks such as metabolic, genetic, and epigenetic memories. Several epigenetic players, such as proteins or RNA, determine the plant epigenetic responses to stresses through histone modifications, DNA methylation, phosphorylation, ubiquitination and chromatin structure, thereby regulating gene expression (Figure 9). For instance, nuclear factor Y subunit GmNFYA mediated histone acetylation improved the salt tolerance by inducing the salt-responsive genes. GmNFYA interacts with GmFVE. GmFVE functions with histone deacetylase GmHDA13 for transcriptional repression by reducing H3K9 acetylation at target loci. GmNFYA also competes with GmHDA13 for interaction with GmFVE. This competition led to the activation and maintenance of histone acetylation for enhanced expression of salt-responsive genes and conferred salt tolerance in soybean (Lu et al., 2021). Similarly, DNA methylation played a critical role in responses to abiotic stress. Salt stress-mediated expression of GmMYB84 relies on DNA methylation (Figure 9). GmMYB84-OE improved the plants traits such as germination rate, osmolytes and antioxidant enzyme activity and K+ levels under salinity stress. EMSA analysis revealed that GmMYB84 physically binds to the cis-regulatory sequences of GmAKT1. Thus, DNA methylation modulates GmMYB84 expression and thereby enhances the salinity stress tolerance of soybean (Zhang et al., 2020b). The ubiquitin/proteasome pathway also played a critical role in soybean development and salt tolerance. For example, over-expression of SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE (SSPP) severely suppressed normal plant growth but improved plant salt tolerance through ROS scavenging. The N-terminal 1-14 residues of ACS7 negatively regulate SSPP protein accumulation through the ubiquitin/proteasome pathway under normal growth conditions. However, ACS7-mediated SSPP protein degradation was repressed by salt and leaf senescence signals through the ubiquitin/proteasome pathway, which providing an effective strategy for salt-tolerant soybean breeding (You et al., 2022). Similarly, histone marks modifications are important for epigenetic memory under salinity stress in soybean. Primed soybean seedlings exhibited a different transcriptomic landscape than non-primed soybean seedlings, indicating some kind of epigenetic memory. Priming induced alterations in histone marks, histone 3 lysine 4 dimethylation (H3K4me2), H3K4me3, and histone 3 lysine 9 acetylation (H3K9ac) coordinately altered the salt stress response through modification ion homeostasis, cell wall and defense-related transcriptional network. Further chemical inhibitors could alter the histone acetylation status and elicit priming-like transcriptional responses in non-primed seedlings. These findings confirmed the importance of histone marks in posttranscriptional regulation and developing the priming response to salt stress in soybean (Yung et al., 2022). Salt stress changed the expression of genes and epigenetic repressive mark, histone H3 lysine 27 trimethylation (H3K27me3) deposition in soybean (Figure 9). Whole-genome ChIP-seq study has identified that the inactivation of genes under salt stress strongly correlates with histone methylation, promoter or coding regions H3K27me3 methylation. Dynamic chromatin regulation through histone modifiers acts as on-and-off switches to respond to salt stress in soybean (Sun et al., 2019a). These epigenetic events control the expression of stress-responsive genes, all of which could regulate transgenerational salt stress response memory in soybean.
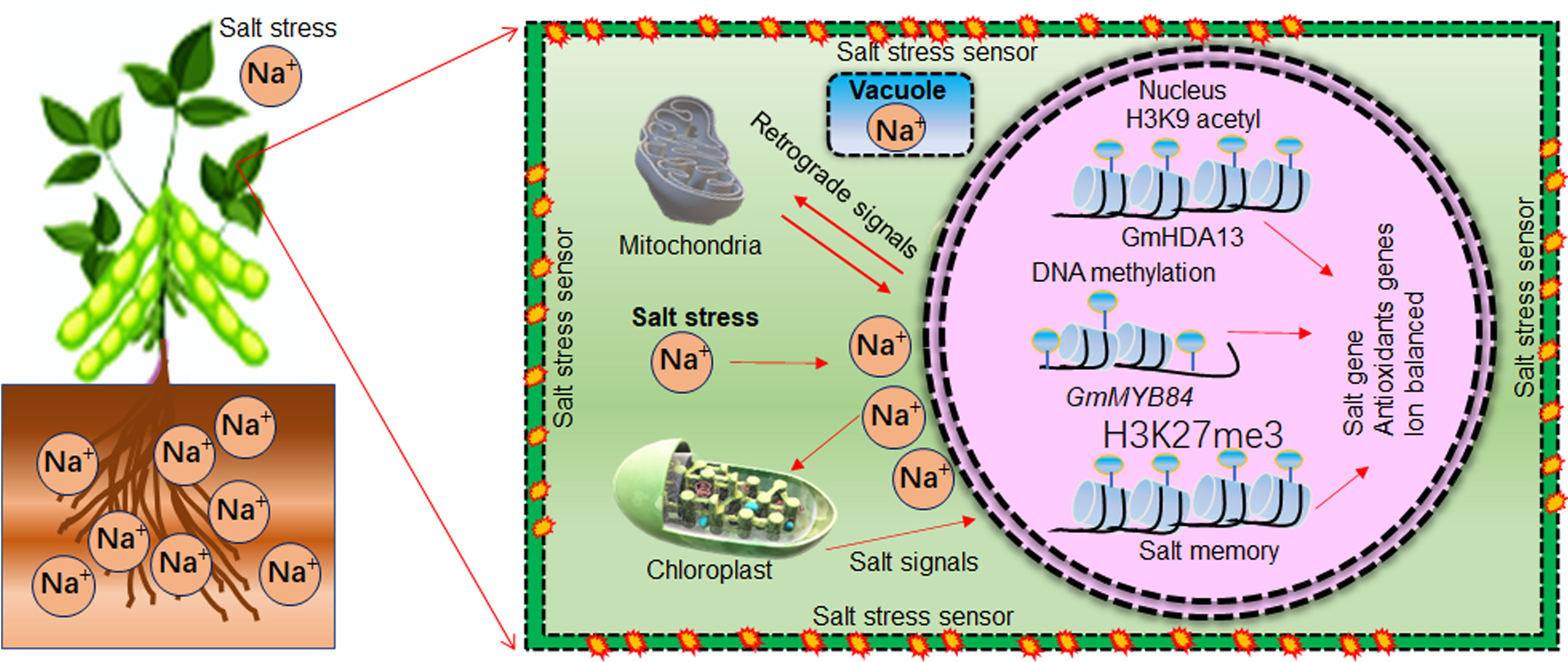
Figure 9 Role of epigenetics and retrograde signaling in Soybean salt tolerance Epigenetic modification alters the transcriptional landscape of salt stress responsive genes through methylation and acetylation. These changes are also a source of epigenetic memory that help to survive the plants under repertoire stresses. Retrograde signaling between different organs like chloroplast and nucleus helps salt homeostasis and activation of different stress-responsive pathways to reduce the salt effects in Soybean.
microRNA genes are involved in various stress responses in soybean. Gma-miR169c-nuclear factor Y-A (NF-YA) modules expressed in different tissues and respond to polyethylene glycol (PEG), high salt, cold stress and abscisic acid (ABA) in soybean. Gma-miR169c exerts a negative regulatory role by repressing its target genes and modifying the various physiological response and stress-responsive genes AtCOR15A, AtRD29A, AtRD22, and AtGSTU25 in Arabidopsis (Yu et al., 2019). miRNA expression was also epigenetically regulated through the process of Histone deacetylation that caused miRNA or target gene repression and silencing. Recently, a small RNA seq library identified 17 miRNAs and 31 putative target genes under salt stress in soybean. The potential targets of miR482bd-5p were HEC1 and BAK1, which exhibited opposite expression patterns. Histone deacetylation analysis of miR482bd-5p showed epigenetic regulation and target gene HEC1 was up-regulated under SAHA-salt treatment in soybean (Cadavid et al., 2020a). Table 3 shows that the miRNA-target genes modules in salt stress response were functionally validated in soybean or Arabidopsis (Sun et al., 2015; Li et al., 2016; Pan et al., 2016; Sahito et al., 2017; Yu et al., 2023).
CRISPR/Cas based genome editing is an efficient approach to improve agronomic traits, including stress tolerance in crops without fitness costs (Zhang et al., 2021). Precise gene editing has become an indispensable routine tool for soybean functional genomics studies (Figure 7). CRISPR/Cas systems have been extensively utilized for traits-genes relation validation in soybean, such as photosynthesis, yield, grain quality, biotic and abiotic stress tolerance (Bao et al., 2019; Do et al., 2019a). For instance, ABA -induced transcription repressors (AITRs) are conserved in angiosperms, regulate ABA signaling, and are localized in the nucleus. ABA treatment increased the expression of GmAITR, and CRISPR/Cas9 edited Cas9-free gmaitr36 double and gmaitr23456 quintuple mutants showed ABA sensitivity and exhibited salt tolerance at seed germination and seedling stage in lab and field experiments (Wang et al., 2021a). In soybean, CRISPR/Cas mediated knockdown of GmSK2-8 induced salt sensitivity during nodule formation. Whereas 2-bp deletion of Gmnsp1b-1 caused frameshift mutation and mutants plants reduced the number of produced nodules compared with WT (He et al., 2021). An efficient CRISPR/Cas9 vector system for multiplex genome editing can target multiple sites in one or multiple genes needed for editing (Ma et al., 2015). CRISPR/Cas9-based edited GmMYB118 mutants accumulated less proline and chlorophyll contents and showed compromised salt tolerance in soybean (Du et al., 2018). Similarly, Recently, the GmNHX5 function was validated through CRISPR/Cas9 mediated mutant in soybean. The edited plants exhibited reduced salt tolerance compared to GmNHX5 overexpression soybean plants. Under salt stress treatment, GmNHX5-OE plants exhibited higher expression of salt responsive genes GmSOS1 and GmSKOR, and maintained a higher Na+/K+ ratio thus improving the soybean salt stress tolerance (Sun et al., 2021a). However, GmNHX5-GmSOS1-GmSKOR modules need further confirmation by genetic analysis. These findings provided a potential candidate gene for developing salt-tolerant soybean germplasm. Likewise, CRISPR/Cas9 mediated editing of GsSOS1 and GsNSCC was performed and the response of mutant plants was validated against salt stress. It was observed that GsSOS1 mutant plants exhibited a changed Na+/K+ ratio compared to GsNSCC mutants, which did not show any significant differences in Na+/K+ ratio under salt stress treatment (Niu et al., 2020). CRISPR-Cas9 mediated edited plants for GmNAC06 exhibited induced proline accumulation to minimize ROS adverse effects and maintained the higher Na+/K+ ratios for ions homeostasis, thus improving the salt stress tolerance (Li et al., 2021a). Although CRISPR/Cas is efficiently utilized, polyploidy and inefficiency of transformation protocol in soybean is a major hurdle in functional genomic validation through CRISPR/Cas. For CRISPR/Cas mediated genome editing, transient hairy root and stable methods have been developed, but their genome editing efficiency largely varies in different soybean cultivars (Liu et al., 2019b). Compared to rice prime base editing by CRISPR/Cas, which allows for base substitutions still not reported in soybean. Consequently, different kinds of CRISPR/Cas systems will provide help for the identification and functional characterization of genes but also provide the opportunity for crop improvement (Chen et al., 2019).
Soybean production is severally affected in salt and alkaline stress soil. The synergetic effects of salt and alkali stress are more deleterious and have become a prime concern for soybean production. Soybean makes a series of modifications to adapt salt stress conditions, for instance, activation of membrane transporters for Na+/K+ homeostasis, Ca2+ influx, antioxidant defense systems, osmotic adjustment, and hormonal regulation to minimize or removal of lethal ion. Active coordination between different cellular organelles through retrograde signalling also proved helpful in coping with salt stress conditions. Developing new salt tolerant soybean varieties with stable and higher yield across unfavorable environments could provide a sustainable supply. Vast genetic diversity and high-quality soybean genome availability help identify salt stress responsive genetic modules/switches, molecular networks and QTLs. These excellent genetic modules/switches could be exploited through transgenic or breeding approaches to develop salt resilient soybean to improve growth and yield under adversity. This review not only comprehensively enriched our understanding of the effect of salt stress on soybean but also provided genetic mechanisms that play role in salt stress tolerance in soybean. Although the investigation of salt stress responses and molecular mechanisms are well defined in cereals, especially in rice and maize plants, it still has a long way to go in soybean crops. For instance, the regulation of identified molecular switches for gene expression and signalling cascade that may regulate Na+/K+ transporters and precise coordination among them remain to be elucidated. However, our current knowledge is limited to transporters that have a role in Na+/K+ homeostasis. In response to salt stress, several cation transporters change their expression; however, their biological role is still elusive in soybean. It is also unclear that salt transporters or antiporters explained here could contribute to plant agronomic performance as well. The molecular pathways regulated by GmAKT1, GmCHXs and CIPKs their cross talk with phytohormone will further help to understand the salt stress response mechanisms in soybean. Domestication process led to the loss of important salt stress-responsive genetic loci in cultivated soybean. Although a significant variation exists among cultivated and wild soybeans for the salt response. The allelic diversity and beneficial alleles of wild soybeans could be incorporated into domesticated soybeans to improve salt tolerance. The complexity of soybean genome and lacking of efficient genetic transformation methods make soybean stress tolerance understandings further complicated. Ultimately, optimizing genetic transformation protocols for in-depth study of downstream and upstream genetic and molecular networks with advanced molecular tools will help elucidate and understand soybean salt tolerance mechanisms. Epigenetic modifications of salt responsive genes through alternative splicing also contribute to the complexity of understanding stress tolerant mechanisms. Therefore, each splicing variant needs to be verified in soybean through genetic engineering. Applying the principles of speed breeding with other efficient crop breeding approaches, like genetic engineering, genotyping by sequencing and genomic selection, could accelerate the soybean improvement for salt resilience. In the future, further improvement and integration of several omics’ approaches, such as small RNA-omics, transcriptomic, proteomic, metabolomics, ion omics and genetic engineering, could warrant to advance soybean varieties with high salt tolerance under saline conditions.
CF, MH and HG conceived the idea and wrote the manuscript. MH, HG, CF and YJ prepared the figures. SL, ZY, KX, WZ, YZ, FZ and YJ helped in the literature search and assistance. MH, XY, WZ and HL supervised the work and edited the manuscript. XY, YZ and HL revised the manuscript and manage the funding resources. All authors contributed to the article and approved the submitted version.
This work was financially funded by the National Natural Science Foundation of China (32171937, 32201716), the Hainan Yazhou Bay Seed Lab (B21HJ0901, B21Y10905, B21Y10906), the Hainan Province Science and Technology Special Fund (ZDYF2022XDNY142), the Scientific Research Foundation of Hainan University Program (Y3AZ20024), and the Hainan Provincial Natural Science Foundation of China (321QN182) and Science and Technology Projects in Guangzhou (202102010387).
CF is thankful to the College of Life Sciences, Jilin Agricultural University Changchun 130118, China for PhD study. I would also like to gratitude all the members of the Soybean Genetics and Breeding Research Group of Sanya Nanfan Research Institute of Hainan University, Sanya Yazhou Bay Area, Hainan province, China for their support throughout the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahanger, M. A., Aziz, U., Alsahli, A. A., Alyemeni, M. N., Ahmad, P. (2019). Influence of exogenous salicylic acid and nitric oxide on growth, photosynthesis, and ascorbate-glutathione cycle in salt stressed vigna angularis. Biomolecules 10, 1–16.
Ahmad, P., Abdel Latef, A. A., Hashem, A., Abd Allah, E. F., Gucel, S., Tran, L. S. (2016b). Nitric oxide mitigates salt stress by regulating levels of osmolytes and antioxidant enzymes in chickpea. Front. Plant Sci. 7, 347.
Ahmad, I., Mian, A., Maathuis, F. J. (2016a). Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J. Exp. Bot. 67, 2689–2698.
Akram, S., Siddiqui, M. N., Hussain, B. M. N., Al Bari, M. A., Mostofa, M. G., Hossain, M. A., et al. (2017). Exogenous glutathione modulates salinity tolerance of soybean [Glycine max (L.) Merrill] at reproductive stage. J. Plant Growth Regul. 36, 877–888.
Albaqami, M., Laluk, K., Reddy, A. S. N. (2019). The arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 100, 379–390.
Aleem, M., Aleem, S., Sharif, I., Wu, Z., Aleem, M., Tahir, A., et al. (2022). Characterization of SOD and GPX gene families in the soybeans in response to drought and salinity stresses. Antioxidants (Basel) 11, 1–23.
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2016). Apple f-box protein MdMAX2 regulates plant photomorphogenesis and stress response. Front. Plant Sci. 7, 1685.
Anderson, J. A., Huprikar, S. S., Kochian, L. V., Lucas, W. J., Gaber, R. F. (1992). Functional expression of a probable arabidopsis thaliana potassium channel in saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 89, 3736–3740.
Assaha, D. V. M., Ueda, A., Saneoka, H., Al-Yahyai, R., Yaish, M. W. (2017). The role of na(+) and k(+) transporters in salt stress adaptation in glycophytes. Front. Physiol. 8, 509.
Ardie, S. W., Liu, S., Takano, T. (2010). Expression of the AKT1-type K(+) channel gene from Puccinellia tenuiflora, PutAKT1, enhances salt tolerance in Arabidopsis. Plant Cell Rep. 29, 865–874.
Bai, M., Zeng, W., Chen, F., Ji, X., Zhuang, Z., Jin, B., et al. (2022). Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize. Biotechnol. Lett. 44, 367–386.
Bao, A., Burritt, D. J., Chen, H., Zhou, X., Cao, D., Tran, L. P. (2019). The CRISPR/Cas9 system and its applications in crop genome editing. Crit. Rev. Biotechnol. 39, 321–336.
Behera, S., Long, Y., Schmitz-Thom, I., Wang, X. P., Zhang, C., Li, H., et al. (2017). Two spatially and temporally distinct Ca(2+) signals convey arabidopsis thaliana responses to k(+) deficiency. New Phytol. 213, 739–750.
Bian, S., Jin, D., Sun, G., Shan, B., Zhou, H., Wang, J., et al. (2020a). Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene 753, 144803.
Bian, X. H., Li, W., Niu, C. F., Wei, W., Hu, Y., Han, J. Q., et al. (2020b). A class b heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 225, 268–283.
Brossa, R., López-Carbonell, M., Jubany-Marí, T., Alegre, L. (2011). Interplay between abscisic acid and jasmonic acid and its role in water-oxidative stress in wild-type, ABA-deficient, JA-deficient, and ascorbate-deficient arabidopsis plants. J. Plant Growth Regul. 30, 322–333.
Cadavid, I. C., da Fonseca, G. C., Margis, R. (2020a). HDAC inhibitor affects soybean miRNA482bd expression under salt and osmotic stress. J. Plant Physiol. 253, 153261.
Cadavid, I. C., Guzman, F., de Oliveira-Busatto, L., de Almeida, R. M. C., Margis, R. (2020b). Transcriptional analyses of two soybean cultivars under salt stress. Mol. Biol. Rep. 47, 2871–2888.
Cai, X., Jia, B., Sun, M., Sun, X. (2022). Insights into the regulation of wild soybean tolerance to salt-alkaline stress. Front. Plant Sci. 13, 1–19.
Cao, L., Yu, Y., Ding, X., Zhu, D., Yang, F., Liu, B., et al. (2017). The glycine soja NAC transcription factor GsNAC019 mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Mol. Biol. 95, 253–268.
Cao, L., Yu, Y., DuanMu, H., Chen, C., Duan, X., Zhu, P., et al. (2016). A novel glycine soja homeodomain-leucine zipper (HD-zip) I gene, Gshdz4, positively regulates bicarbonate tolerance and responds to osmotic stress in arabidopsis. BMC Plant Biol. 16, 184.
Chen, Z. F., Ru, J. N., Sun, G. Z., Du, Y., Chen, J., Zhou, Y. B., et al. (2021). Genomic-wide analysis of the PLC family and detection of GmPI-PLC7 responses to drought and salt stresses in soybean. Front. Plant Sci. 12, 631470.
Chen, C., Sun, X., Liu, A., DuanMu, H. Z., Yu, Y., Xiao, J. L., et al. (2015). Cloning and functional analysis ofGlycine sojaBicarbonate stress responsive GeneGsMIPS2. Acta Agronomica Sin. 41, 1343.
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697.
Chen, C., Xu, L., Zhang, X., Wang, H., Nisa, Z. U., Jin, X., et al. (2022). Exogenous strigolactones enhance tolerance in soybean seedlings in response to alkaline stress. Physiol. Plant 174, e13784.
Chen, X. Q., Yu, B. J. (2007). Ionic effects of na+ and cl- on photosynthesis in glycine max seedlings under isoosmotic salt stress. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 33, 294–300.
Cheng, Q., Gan, Z., Wang, Y., Lu, S., Hou, Z., Li, H., et al. (2020). The soybean gene J contributes to salt stress tolerance by up-regulating salt-responsive genes. Front. Plant Sci. 11, 272.
Cheon, J., Park, S. Y., Schulz, B., Choe, S. (2010). Arabidopsis brassinosteroid biosynthetic mutant dwarf7-1 exhibits slower rates of cell division and shoot induction. BMC Plant Biol. 10, 270.
Cho, K. H., Kim, M. Y., Kwon, H., Yang, X., Lee, S. H. (2021). Novel QTL identification and candidate gene analysis for enhancing salt tolerance in soybean (Glycine max (L.) merr.). Plant Sci. 313, 111085.
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111, 6497–6502.
Cisse, E. H. M., Miao, L. F., Yang, F., Huang, J. F., Li, D. D., Zhang, J. (2021). Gly betaine surpasses melatonin to improve salt tolerance in dalbergia odorifera. Front. Plant Sci. 12, 588847.
Corpas, F. J., Barroso, J. B., Palma, J. M., Rodriguez-Ruiz, M. (2017). Plant peroxisomes: A nitro-oxidative cocktail. Redox Biol. 11, 535–542.
Crawford, T., Lehotai, N., Strand, Å. (2018). The role of retrograde signals during plant stress responses. J. Exp. Bot. 69, 2783–2795.
Cuéllar, T., Azeem, F., Andrianteranagna, M., Pascaud, F., Verdeil, J. L., Sentenac, H., et al. (2013). Potassium transport in developing fleshy fruits: the grapevine inward k(+) channel VvK1.2 is activated by CIPK-CBL complexes and induced in ripening berry flesh cells. Plant J. 73, 1006–1018.
Dawood, M. F. A., Zaid, A., Latef, A. A. H. A. (2022). Salicylic acid spraying-induced resilience strategies against the damaging impacts of drought and/or salinity stress in two varieties of vicia faba l. Seedlings. J. Plant Growth Regul. 41, 1919–1942.
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379.
Demidchik, V., Maathuis, F. J. M. (2007). Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol. 175, 387–404.
Demidchik, V., Tester, M. (2002). Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from arabidopsis roots. Plant Physiol. 128, 379–387.
Ding, F., Cui, P., Wang, Z., Zhang, S., Ali, S., Xiong, L. (2014). Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in arabidopsis. BMC Genomics 15, 431.
Divi, U. K., Rahman, T., Krishna, P. (2010). Brassinosteroid-mediated stress tolerance in arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10, 151.
Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T. N., Stacey, G., Gillman, J. D., et al. (2019a). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 19, 311.
Do, T. D., Vuong, T. D., Dunn, D., Clubb, M., Valliyodan, B., Patil, G., et al. (2019b). Identification of new loci for salt tolerance in soybean by high-resolution genome-wide association mapping. BMC Genomics 20, 318.
Do, T. D., Vuong, T. D., Dunn, D., Smothers, S., Patil, G., Yungbluth, D. C., et al. (2018). Mapping and confirmation of loci for salt tolerance in a novel soybean germplasm. Theor. Appl. Genet. 131, 513–524.
Donaldson, L., Ludidi, N., Knight, M. R., Gehring, C., Denby, K. (2004). Salt and osmotic stress cause rapid increases in arabidopsis thaliana cGMP levels. FEBS Lett. 569, 317–320.
Du, Y. T., Zhao, M. J., Wang, C. T., Gao, Y., Wang, Y. X., Liu, Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18, 320.
Duan, F., Ding, J., Lee, D., Lu, X., Feng, Y., Song, W. (2017). Overexpression of SoCYP85A1, a spinach cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Front. Plant Sci. 8, 1909.
Duan, X., Yu, Y., Duanmu, H., Chen, C., Sun, X., Cao, L., et al. (2018a). GsSLAH3, a glycine soja slow type anion channel homolog, positively modulates plant bicarbonate stress tolerance. Physiol. Plant 164, 145–162.
Duan, X., Yu, Y., Zhang, Y., Chen, C., Duanmu, H., Cao, L., et al. (2018b). A potential efflux boron transporter gene GsBOR2, positively regulates arabidopsis bicarbonate tolerance. Plant Sci. 274, 284–292.
Duby, G., Hosy, E., Fizames, C., Alcon, C., Costa, A., Sentenac, H., et al. (2008). AtKC1, a conditionally targeted shaker-type subunit, regulates the activity of plant k+ channels. Plant J. 53, 115–123.
El Mahi, H., Pérez-Hormaeche, J., De Luca, A., Villalta, I., Espartero, J., Gámez-Arjona, F., et al. (2019). A critical role of sodium flux via the plasma membrane Na(+)/H(+) exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 180, 1046–1065.
El-Sabagh, S. S., Ueda, A., Saneoka H and C, B. (2015). Evaluation of salinity stress effects on seed yield and quality of three soybean cultivars. Azarian J. Agric. 2, 138–141.
Farhangi-Abriz, S., Ghassemi-Golezani, K. (2018). How can salicylic acid and jasmonic acid mitigate salt toxicity in soybean plants? Ecotoxicol Environ. Saf. 147, 1010–1016.
Feng, X., Feng, P., Yu, H., Yu, X., Sun, Q., Liu, S., et al. (2020a). GsSnRK1 interplays with transcription factor GsERF7 from wild soybean to regulate soybean stress resistance. Plant Cell Environ. 43, 1192–1211.
Feng, C., He, C., Wang, Y., Xu, H., Xu, K., Zhao, Y., et al. (2021a). Genome-wide identification of soybean shaker k(+) channel gene family and functional characterization of GmAKT1 in transgenic arabidopsis thaliana under salt and drought stress. J. Plant Physiol. 266, 153529.
Feng, X., Liu, W., Cao, F., Wang, Y., Zhang, G., Chen, Z. H., et al. (2020b). Overexpression of HvAKT1 improves drought tolerance in barley by regulating root ion homeostasis and ROS and NO signaling. J. Exp. Bot. 71, 6587–6600.
Feng, N., Yu, M., Li, Y., Jin, D., Zheng, D. (2021b). Prohexadione-calcium alleviates saline-alkali stress in soybean seedlings by improving the photosynthesis and up-regulating antioxidant defense. Ecotoxicol Environ. Saf. 220, 112369.
Filgueiras, C. C., Martins, A. D., Pereira, R. V., Willett, D. S. (2019). The ecology of salicylic acid signaling: Primary, secondary and tertiary effects with applications in agriculture. Int. J. Mol. Sci. 20, 1–19.
Fukuda, A., Nakamura, A., Hara, N., Toki, S., Tanaka, Y. (2011). Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233, 175–188.
Gallusci, P., Agius, D. R., Moschou, P. N., Dobránszki, J., Kaiserli, E., Martinelli, F. (2022). Deep inside the epigenetic memories of stressed plants. Trends Plant Sci. 28, 142–153.
Galvan-Ampudia, C. S., Julkowska, M. M., Darwish, E., Gandullo, J., Korver, R. A., Brunoud, G., et al. (2013). Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23, 2044–2050.
Gambale, F., Uozumi, N. (2006). Properties of shaker-type potassium channels in higher plants. J. Membr Biol. 210, 1–19.
Gao, Y., Ma, J., Zheng, J. C., Chen, J., Chen, M., Zhou, Y. B., et al. (2019). The elongation factor GmEF4 is involved in the response to drought and salt tolerance in soybean. Int. J. Mol. Sci. 20, 1–16.
Garriga, M., Raddatz, N., Véry, A. A., Sentenac, H., Rubio-Meléndez, M. E., González, W., et al. (2017). Cloning and functional characterization of HKT1 and AKT1 genes of fragaria spp.-relationship to plant response to salt stress. J. Plant Physiol. 210, 9–17.
Guan, R., Qu, Y., Guo, Y., Yu, L., Liu, Y., Jiang, J., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80, 937–950.
Guo, H., Hu, Z., Zhang, H., Min, W., Hou, Z. (2019). Comparative effects of salt and alkali stress on antioxidant system in cotton (Gossypium hirsutum l.) leaves. Open Chem. 17, 1352–1360.
Guo, X., Jiang, J., Liu, Y., Yu, L., Chang, R., Guan, R., et al. (2021). Identification of a novel salt tolerance-related locus in wild soybean (Glycine soja sieb. Zucc.). Front. Plant Sci. 12, 791175.
Halfter, U., Ishitani, M., Zhu, J. K. (2000). The arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U.S.A. 97, 3735–3740.
Hameed, A., Ahmed, M. Z., Hussain, T., Aziz, I., Ahmad, N., Gul, B., et al. (2021). Effects of salinity stress on chloroplast structure and function. Cells 10, 1–22.
Hasanuzzaman, M., Nahar, K., Anee, T. I., Fujita, M. (2017). Glutathione in plants: biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 23, 249–268.
He, Q., Cai, H., Bai, M., Zhang, M., Chen, F., Huang, Y., et al. (2020a). A soybean bZIP transcription factor GmbZIP19 confers multiple biotic and abiotic stress responses in plant. Int. J. Mol. Sci. 21, 1–19.
He, Y., Dong, Y., Yang, X., Guo, D., Qian, X., Yan, F., et al. (2020b). Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max l.). Genome 63, 13–26.
He, Y., Fu, J., Yu, C., Wang, X., Jiang, Q., Hong, J., et al. (2015). Increasing cyclic electron flow is related to na+ sequestration into vacuoles for salt tolerance in soybean. J. Exp. Bot. 66, 6877–6889.
He, C., Gao, H., Wang, H., Guo, Y., He, M., Peng, Y., et al. (2021). GSK3-mediated stress signaling inhibits legume-rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol. Plant 14, 488–502.
Hernandez-Leon, S. G., Valenzuela-Soto, E. M. (2022). Glycine betaine is a phytohormone-like plant growth and development regulator under stress conditions. J. Plant Growth Regulation. doi: 10.1007/s00344-022-10855-3
Hoang, X. L. T., Chuong, N. N., Hoa, T. T. K., Doan, H., Van, P. H. P., Trang, L. D. M., et al. (2021). The drought-mediated soybean GmNAC085 functions as a positive regulator of plant response to salinity. Int. J. Mol. Sci. 22, 1–16.
Hong, Z., Li, Y., Zhao, Y., Yang, M., Zhang, X., Teng, Y., et al. (2022). Heterologous expression of arabidopsis AtARA6 in soybean enhances salt tolerance. Front. Genet. 13, 849357.
Horie, T., Horie, R., Chan, W. Y., Leung, H. Y., Schroeder, J. I. (2006). Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol. 47, 622–633.
Hossain, M. A., Hoque, T. S., Zaid, A., Wani, S. H., Mostofa, M. G., Henry, R. (2021). “Targeting the ascorbate-glutathione pathway and the glyoxalase pathway for genetic engineering of abiotic stress-tolerance in rice,” in Molecular Breeding for Rice Abiotic Stress Tolerance and Nutritional Quality, eds Hossain, M. A., Hassan, L., Iftekharuddaula, K. M., Kumar, A., Henry, R. (Wiley-Blackwell), 398–427. doi: 10.1002/9781119633174.ch21
Hou, Z., Li, Y., Cheng, Y., Li, W., Li, T., Du, H., et al. (2022). Genome-wide analysis of DREB genes identifies a novel salt tolerance gene in wild soybean (Glycine soja). Front. Plant Sci. 13, 821647.
Hu, J., Zhuang, Y., Li, X., Li, X., Sun, C., Ding, Z., et al. (2022). Time-series transcriptome comparison reveals the gene regulation network under salt stress in soybean (Glycine max) roots. BMC Plant Biol. 22, 157.
Hussain, K., Nisa, R. T., Zaid, A., Mushtaq, M. (2023). “The utilization of speed breeding and genome editing to achieve zero hunger,” in Sustainable agriculture in the era of the OMICs revolution. Eds. Prakash, C. S., Fiaz, S., Nadeem, M. A., Baloch, F. S., Qayyum, A. (Cham: Springer International Publishing), 1–15.
Imran, M., Aaqil Khan, M., Shahzad, R., Bilal, S., Khan, M., Yun, B. W., et al. (2021). Melatonin ameliorates thermotolerance in soybean seedling through balancing redox homeostasis and modulating antioxidant defense, phytohormones and polyamines biosynthesis. Molecules 26, 1–21.
Islam, W., Waheed, A., Naveed, H., Zeng, F. (2022). MicroRNAs mediated plant responses to salt stress. Cells 11, 1–21.
Jayakannan, M., Bose, J., Babourina, O., Shabala, S., Massart, A., Poschenrieder, C., et al. (2015). The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in arabidopsis. J. Exp. Bot. 66, 1865–1875.
Jeanguenin, L., Alcon, C., Duby, G., Boeglin, M., Chérel, I., Gaillard, I., et al. (2011). AtKC1 is a general modulator of arabidopsis inward shaker channel activity. Plant J. 67, 570–582.
Jia, Q., Li, M. W., Zheng, C., Xu, Y., Sun, S., Li, Z., et al. (2021). The soybean plasma membrane-localized cation/H(+) exchanger GmCHX20a plays a negative role under salt stress. Physiol. Plant 171, 714–727.
Jia, B., Sun, M., DuanMu, H., Ding, X., Liu, B., Zhu, Y., et al. (2017a). GsCHX19.3, a member of cation/H(+) exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Sci. Rep. 7, 9423.
Jia, B., Sun, M., Sun, X., Li, R., Wang, Z., Wu, J., et al. (2016). Overexpression of GsGSTU13 and SCMRP in medicago sativa confers increased salt-alkaline tolerance and methionine content. Physiol. Plant 156, 176–189.
Jia, Q., Xiao, Z. X., Wong, F. L., Sun, S., Liang, K. J., Lam, H. M. (2017b). Genome-wide analyses of the soybean f-box gene family in response to salt stress. Int. J. Mol. Sci. 18, 1–17.
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca(2+) influx. Nature 572, 341–346.
Jin, J., Li, K., Qin, J., Yan, L., Wang, S., Zhang, G., et al. (2021a). The response mechanism to salt stress in arabidopsis transgenic lines over-expressing of GmG6PD. Plant Physiol. Biochem. 162, 74–85.
Jin, T., Sun, Y., Shan, Z., He, J., Wang, N., Gai, J., et al. (2021b). Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 19, 1155–1169.
Jin, T., Sun, Y., Zhao, R., Shan, Z., Gai, J., Li, Y. (2019). Overexpression of peroxidase gene GsPRX9 confers salt tolerance in soybean. Int. J. Mol. Sci. 20, 1–17.
Johansson, I., Wulfetange, K., Porée, F., Michard, E., Gajdanowicz, P., Lacombe, B., et al. (2006). External k+ modulates the activity of the arabidopsis potassium channel SKOR via an unusual mechanism. Plant J. 46, 269–281.
Johnson, G. N. (2011). Physiology of PSI cyclic electron transport in higher plants. Biochim. Biophys. Acta 1807, 384–389.
Joshi, S., Kaur, K., Khare, T., Srivastava, A. K., Suprasanna, P., Kumar, V. (2021). Genome-wide identification, characterization and transcriptional profiling of NHX-type (Na(+)/H(+)) antiporters under salinity stress in soybean. 3 Biotech. 11, 16.
Kataria, S., Baghel, L., Jain, M., Guruprasad, K. N. (2019). Magnetopriming regulates antioxidant defense system in soybean against salt stress. Biocatalysis Agric. Biotechnol. 18, 101090.
Ke, D., He, Y., Fan, L., Niu, R., Cheng, L., Wang, L., et al. (2022). The soybean TGA transcription factor GmTGA13 plays important roles in the response to salinity stress. Plant Biol. (Stuttg) 24, 313–322.
Ketehouli, T., Zhou, Y. G., Dai, S. Y., Carther, K. F. I., Sun, D. Q., Li, Y., et al. (2021). A soybean calcineurin b-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J. Plant Physiol. 256, 153331.
Khalid, M., Rehman, H. M., Ahmed, N., Nawaz, S., Saleem, F., Ahmad, S., et al. (2022). Using exogenous melatonin, glutathione, proline, and glycine betaine treatments to combat abiotic stresses in crops. Int. J. Mol. Sci. 23, 1–36.
Kiegle, E., Moore, C. A., Haseloff, J., Tester, M. A., Knight, M. R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the arabidopsis root. Plant J. 23, 267–278.
Kim, H. J., Cho, H. S., Pak, J. H., Kwon, T., Lee, J. H., Kim, D. H., et al. (2018). Confirmation of drought tolerance of ectopically expressed AtABF3 gene in soybean. Mol. Cells 41, 413–422.
Kmiecik, P., Leonardelli, M., Teige, M. (2016). Novel connections in plant organellar signalling link different stress responses and signalling pathways. J. Exp. Bot. 67, 3793–3807.
Kong, X., Ma, L., Yang, L., Chen, Q., Xiang, N., Yang, Y., et al. (2014). Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins arabidopsis CBP20 and CBP80 modulate the salt stress response. J. Proteome Res. 13, 2495–2510.
Ku, Y. S., Sintaha, M., Cheung, M. Y., Lam, H. M. (2018). Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 19, 1–35.
Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., et al. (1996). Tissue-specific expression of arabidopsis AKT1 gene is consistent with a role in k+ nutrition. Plant J. 9, 195–203.
Laloi, C., Apel, K., Danon, A. (2004). Reactive oxygen signalling: the latest news. Curr. Opin. Plant Biol. 7, 323–328.
Laloum, T., Martín, G., Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23, 140–150.
Latz, A., Mehlmer, N., Zapf, S., Mueller, T. D., Wurzinger, B., Pfister, B., et al. (2013). Salt stress triggers phosphorylation of the arabidopsis vacuolar k+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 6, 1274–1289.
Leng, Z. X., Liu, Y., Chen, Z. Y., Guo, J., Chen, J., Zhou, Y. B., et al. (2021). Genome-wide analysis of the DUF4228 family in soybean and functional identification of GmDUF4228 -70 in response to drought and salt stresses. Front. Plant Sci. 12, 628299.
Li, M., Chen, R., Jiang, Q., Sun, X., Zhang, H., Hu, Z. (2021a). GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 105, 333–345.
Li, S., Cong, Y., Liu, Y., Wang, T., Shuai, Q., Chen, N., et al. (2017). Optimization of agrobacterium-mediated transformation in soybean. Front. Plant Sci. 8, 246.
Li, Y., Guo, Q., Liu, P., Huang, J., Zhang, S., Yang, G., et al. (2021b). Dual roles of the serine/arginine-rich splicing factor SR45a in promoting and interacting with nuclear cap-binding complex to modulate the salt-stress response in arabidopsis. New Phytol. 230, 641–655.
Li, J., Guo, Y., Yang, Y. (2022b). The molecular mechanism of plasma membrane h(+)-ATPases in plant responses to abiotic stress. J. Genet. Genomics 49, 715–725.
Li, B., Liu, Y., Cui, X. Y., Fu, J. D., Zhou, Y. B., Zheng, W. J., et al. (2019a). Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front. Plant Sci. 10, 549.
Li, J., Long, Y., Qi, G. N., Li, J., Xu, Z. J., Wu, W. H., et al. (2014). The os-AKT1 channel is critical for k+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26, 3387–3402.
Li, S., Wang, N., Ji, D., Zhang, W., Wang, Y., Yu, Y., et al. (2019c). A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 31, 2107–2130.
Li, H., Wang, X. H., Li, Q., Xu, P., Liu, Z. N., Xu, M., et al. (2022a). GmCIPK21, a CBL-interacting protein kinase confers salt tolerance in soybean (Glycine max. L). Plant Physiol. Biochem. 184, 47–55.
Li, W., Wang, T., Zhang, Y., Li, Y. (2016). Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic arabidopsis thaliana. J. Exp. Bot. 67, 175–194.
Li, Z. Y., Xu, Z. S., He, G. Y., Yang, G. X., Chen, M., Li, L. C., et al. (2012). A mutation in arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 426, 522–527.
Li, H., Zhao, Q., Huang, H. (2019b). Current states and challenges of salt-affected soil remediation by cyanobacteria. Sci. Total Environ. 669, 258–272.
Li, B., Zheng, J. C., Wang, T. T., Min, D. H., Wei, W. L., Chen, J., et al. (2020). Expression analyses of soybean VOZ transcription factors and the role of GmVOZ1G in drought and salt stress tolerance. Int. J. Mol. Sci. 21, 1–17.
Liu, J., Gunapati, S., Mihelich, N. T., Stec, A. O., Michno, J. M., Stupar, R. M. (2019b). Genome editing in soybean with CRISPR/Cas9. Methods Mol. Biol. 1917, 217–234.
Liu, J., Ishitani, M., Halfter, U., Kim, C. S., Zhu, J. K. (2000). The arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734.
Liu, J., Li, L., Yuan, F., Chen, M. (2019c). Exogenous salicylic acid improves the germination of limonium bicolor seeds under salt stress. Plant Signal Behav. 14, e1644595.
Liu, X., Liu, F., Zhang, L., Cheng, C., Wei, P., Yu, B. (2021b). GsCLC-c2 from wild soybean confers chloride/salt tolerance to transgenic arabidopsis and soybean composite plants by regulating anion homeostasis. Physiol. Plant 172, 1867–1879.
Liu, Z., Ma, C., Hou, L., Wu, X., Wang, D., Zhang, L., et al. (2022b). Exogenous SA affects rice seed germination under salt stress by regulating Na(+)/K(+) balance and endogenous GAs and ABA homeostasis. Int. J. Mol. Sci. 23, 1–18.
Liu, A., Xiao, Z., Li, M. W., Wong, F. L., Yung, W. S., Ku, Y. S., et al. (2019a). Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 42, 98–114.
Liu, X., Yang, X., Zhang, B. (2021c). Transcriptome analysis and functional identification of GmMYB46 in soybean seedlings under salt stress. PeerJ 9, e12492.
Liu, Y. L., Zheng, L., Jin, L. G., Liu, Y. X., Kong, Y. N., Wang, Y. X., et al. (2022a). Genome-wide analysis of the soybean TIFY family and identification of GmTIFY10e and GmTIFY10g response to salt stress. Front. Plant Sci. 13, 845314.
Liu, J., Zhu, J. K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945.
Lu, L., Wei, W., Tao, J. J., Lu, X., Bian, X. H., Hu, Y., et al. (2021). Nuclear factor y subunit GmNFYA competes with GmHDA13 for interaction with GmFVE to positively regulate salt tolerance in soybean. Plant Biotechnol. J. 19, 2362–2379.
Lu, Y., Yu, M., Jia, Y., Yang, F., Zhang, Y., Xu, X., et al. (2022). Structural basis for the activity regulation of a potassium channel AKT1 from arabidopsis. Nat. Commun. 13, 5682.
Lung, S. C., Lai, S. H., Wang, H., Zhang, X., Liu, A., Guo, Z. H., et al. (2022). Oxylipin signaling in salt-stressed soybean is modulated by ligand-dependent interaction of class II acyl-CoA-binding proteins with lipoxygenase. Plant Cell 34, 1117–1143.
Ma, N., Hu, C., Wan, L., Hu, Q., Xiong, J., Zhang, C. (2017). Strigolactones improve plant growth, photosynthesis, and alleviate oxidative stress under salinity in rapeseed (Brassica napus l.) by regulating gene expression. Front. Plant Sci. 8, 1671.
Ma, L., Ye, J., Yang, Y., Lin, H., Yue, L., Luo, J., et al. (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 48, 697–709.e695.
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284.
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., Oda, K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in arabidopsis. Plant J. 56, 613–626.
Manishankar, P., Wang, N., Köster, P., Alatar, A. A., Kudla, J. (2018). Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 69, 4215–4226.
Mansour, M. M. F., Ali, E. F. (2017). Evaluation of proline functions in saline conditions. Phytochemistry 140, 52–68.
Massalha, H., Korenblum, E., Tholl, D., Aharoni, A. (2017). Small molecules below-ground: the role of specialized metabolites in the rhizosphere. Plant J. 90, 788–807.
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467.
Mishra, A. K., Sudalaimuthuasari, N., Hazzouri, K. M., Saeed, E. E., Shah, I., Amiri, K. M. A. (2022). Tapping into plant-microbiome interactions through the lens of multi-omics techniques. Cells 11, 1–20.
Munekage, Y., Hashimoto, M., Miyake, C., Tomizawa, K., Endo, T., Tasaka, M., et al. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582.
Nahar, K., Hasanuzzaman, M., Fujita, M. (2016). “Roles of osmolytes in plant adaptation to drought and salinity,” in Osmolytes and plants acclimation to changing environment: Emerging omics technologies. Eds. Iqbal, N., Nazar, R., Khan, N. A. (New Delhi: Springer India), 37–68.
Nguyen, Q. H., Vu, L. T. K., Nguyen, L. T. N., Pham, N. T. T., Nguyen, Y. T. H., Le, S. V., et al. (2019). Overexpression of the GmDREB6 gene enhances proline accumulation and salt tolerance in genetically modified soybean plants. Sci. Rep. 9, 19663.
Ni, L., Wang, S., Shen, T., Wang, Q., Chen, C., Xia, J., et al. (2020). Calcium/calmodulin-dependent protein kinase OsDMI3 positively regulates saline-alkaline tolerance in rice roots. Plant Signal Behav. 15, 1813999.
Nigam, B., Dubey, R. S., Rathore, D. (2022). Protective role of exogenously supplied salicylic acid and PGPB (Stenotrophomonas sp.) on spinach and soybean cultivars grown under salt stress. Scientia Hortic. 293, 110654.
Niu, F., Jiang, Q., Cheng, R., Sun, X., Hu, Z., Wang, L., et al. (2020). CRISPR/Cas9-mediated targeted mutagenesis of wild soybean (Glycine soja) hairy roots altered the transcription profile of the mutant. J. Agric. Sci. 12, 14–33.
Noor, J., Ullah, A., Saleem, M. H., Tariq, A., Ullah, S., Waheed, A., et al. (2022). Effect of jasmonic acid foliar spray on the morpho-physiological mechanism of salt stress tolerance in two soybean varieties (Glycine max l.). Plants (Basel) 11, 1–14.
Obata, T., Kitamoto, H. K., Nakamura, A., Fukuda, A., Tanaka, Y. (2007). Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiol. 144, 1978–1985.
Pan, W. J., Tao, J. J., Cheng, T., Bian, X. H., Wei, W., Zhang, W. K., et al. (2016). Soybean miR172a improves salt tolerance and can function as a long-distance signal. Mol. Plant 9, 1337–1340.
Pareddy, D., Chennareddy, S., Anthony, G., Sardesai, N., Mall, T., Minnicks, T., et al. (2020). Improved soybean transformation for efficient and high throughput transgenic production. Transgenic Res. 29, 267–281.
Pi, E., Zhu, C., Fan, W., Huang, Y., Qu, L., Li, Y., et al. (2018). Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell Proteomics 17, 1209–1224.
Qi, X., Li, M. W., Xie, M., Liu, X., Ni, M., Shao, G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5, 4340.
Qi, Z., Spalding, E. P. (2004). Protection of plasma membrane k+ transport by the salt overly sensitive1 na+-h+ antiporter during salinity stress. Plant Physiol. 136, 2548–2555.
Qu, Y., Guan, R., Bose, J., Henderson, S. W., Wege, S., Qiu, L., et al. (2021). Soybean CHX-type ion transport protein GmSALT3 confers leaf na(+) exclusion via a root derived mechanism, and cl(-) exclusion via a shoot derived process. Plant Cell Environ. 44, 856–869.
Rasheed, A., Raza, A., Jie, H., Mahmood, A., Ma, Y., Zhao, L., et al. (2022). Molecular tools and their applications in developing salt-tolerant soybean (Glycine max l.) cultivars. Bioengineering (Basel) 9, 1–22.
Ren, X. L., Qi, G. N., Feng, H. Q., Zhao, S., Zhao, S. S., Wang, Y., et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74, 258–266.
Sahito, Z. A., Wang, L., Sun, Z., Yan, Q., Zhang, X., Jiang, Q., et al. (2017). The miR172c-NNC1 module modulates root plastic development in response to salt in soybean. BMC Plant Biol. 17, 229.
Sarkar, A. K., Sadhukhan, S. (2022). Imperative role of trehalose metabolism and trehalose-6-phosphate signaling on salt stress responses in plants. Physiol. Plant 174, e13647.
Sedivy, E. J., Wu, F., Hanzawa, Y. (2017). Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 214, 539–553.
Sentenac, H., Bonneaud, N., Minet, M., Lacroute, F., Salmon, J. M., Gaymard, F., et al. (1992). Cloning and expression in yeast of a plant potassium ion transport system. Science 256, 663–665.
Shan, C., Mei, Z., Duan, J., Chen, H., Feng, H., Cai, W. (2014). OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PloS One 9, e87110.
Shen, X. J., Wang, Y. Y., Zhang, Y. X., Guo, W., Jiao, Y. Q., Zhou, X. A. (2018). Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and helicoverpa armigera in transgenic arabidopsis. Int. J. Mol. Sci. 19, 1–19.
Sheteiwy, M. S., Shao, H., Qi, W., Daly, P., Sharma, A., Shaghaleh, H., et al. (2021). Seed priming and foliar application with jasmonic acid enhance salinity stress tolerance of soybean (Glycine max l.) seedlings. J. Sci. Food Agric. 101, 2027–2041.
Shi, W. Y., Du, Y. T., Ma, J., Min, D. H., Jin, L. G., Chen, J., et al. (2018a). The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 19, 1–20.
Shi, X., Yan, L., Yang, C., Yan, W., Moseley, D. O., Wang, T., et al. (2018b). Identification of a major quantitative trait locus underlying salt tolerance in ‘Jidou 12’ soybean cultivar. BMC Res. Notes 11, 95.
Shi, H., Zhu, J. K. (2002). SOS4, a pyridoxal kinase gene, is required for root hair development in arabidopsis. Plant Physiol. 129, 585–593.
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, P., et al. (2016). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361.
Shu, K., Qi, Y., Chen, F., Meng, Y., Luo, X., Shuai, H., et al. (2017). Salt stress represses soybean seed germination by negatively regulating GA biosynthesis while positively mediating ABA biosynthesis. Front. Plant Sci. 8, 1372.
Signorelli, S. (2016). The fermentation analogy: A point of view for understanding the intriguing role of proline accumulation in stressed plants. Front. Plant Sci. 7, 1339.
Singh, A. K., Raina, S. K., Kumar, M., Aher, L., Ratnaparkhe, M. B., Rane, J., et al. (2022). Modulation of GmFAD3 expression alters abiotic stress responses in soybean. Plant Mol. Biol. 110, 199–218.
Singh, B., Singh, J. P., Singh, N., Kaur, A. (2017). Saponins in pulses and their health promoting activities: A review. Food Chem. 233, 540–549.
Soliman, M., Elkelish, A., Souad, T., Alhaithloul, H., Farooq, M. (2020). Brassinosteroid seed priming with nitrogen supplementation improves salt tolerance in soybean. Physiol. Mol. Biol. Plants 26, 501–511.
Subbarao, G. V., Ito, O., Berry, W. L., Wheeler, R. M. (2003). Sodium–a functional plant nutrient. Crit. Rev. Plant Sci. 22, 391–416.
Sugiyama, A. (2019). The soybean rhizosphere: Metabolites, microbes, and beyond-a review. J. Adv. Res. 19, 67–73.
Sun, T. J., Fan, L., Yang, J., Cao, R. Z., Yang, C. Y., Zhang, J., et al. (2019b). A glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher na(+) efflux rate and K(+)/Na(+) ratio in arabidopsis. BMC Plant Biol. 19, 469.
Sun, T., Ma, N., Wang, C., Fan, H., Wang, M., Zhang, J., et al. (2021a). A golgi-localized Sodium/Hydrogen exchanger positively regulates salt tolerance by maintaining higher K(+)/Na(+) ratio in soybean. Front. Plant Sci. 12, 638340.
Sun, L., Song, G., Guo, W., Wang, W., Zhao, H., Gao, T., et al. (2019a). Dynamic changes in genome-wide Histone3 Lysine27 trimethylation and gene expression of soybean roots in response to salt stress. Front. Plant Sci. 10, 1031.
Sun, X., Sun, M., Jia, B., Qin, Z., Yang, K., Chen, C., et al. (2016). A glycine soja methionine sulfoxide reductase B5a interacts with the Ca(2+)/CAM-binding kinase GsCBRLK and activates ROS signaling under carbonate alkaline stress. Plant J. 86, 514–529.
Sun, Z., Wang, Y., Mou, F., Tian, Y., Chen, L., Zhang, S., et al. (2015). Genome-wide small RNA analysis of soybean reveals auxin-responsive microRNAs that are differentially expressed in response to salt stress in root apex. Front. Plant Sci. 6, 1273.
Sun, X., Yang, S., Sun, M., Wang, S., Ding, X., Zhu, D., et al. (2014). A novel glycine soja cysteine proteinase inhibitor GsCPI14, interacting with the calcium/calmodulin-binding receptor-like kinase GsCBRLK, regulated plant tolerance to alkali stress. Plant Mol. Biol. 85, 33–48.
Sun, Y., Zhao, J. Y., Li, Y. T., Zhang, P. G., Wang, S. P., Guo, J., et al. (2021b). Genome-wide analysis of the C2 domain family in soybean and identification of a putative abiotic stress response gene GmC2-148. Front. Plant Sci. 12, 620544.
Ullah, A., Li, M., Noor, J., Tariq, A., Liu, Y., Shi, L. (2019). Effects of salinity on photosynthetic traits, ion homeostasis and nitrogen metabolism in wild and cultivated soybean. PeerJ 7, e8191.
Valenzuela, C. E., Acevedo-Acevedo, O., Miranda, G. S., Vergara-Barros, P., Holuigue, L., Figueroa, C. R., et al. (2016). Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in arabidopsis primary root. J. Exp. Bot. 67, 4209–4220.
van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433.
Véry, A. A., Nieves-Cordones, M., Daly, M., Khan, I., Fizames, C., Sentenac, H. (2014). Molecular biology of k+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171, 748–769.
Véry, A. A., Sentenac, H. (2003). Molecular mechanisms and regulation of k+ transport in higher plants. Annu. Rev. Plant Biol. 54, 575–603.
Voelker, C., Gomez-Porras, J. L., Becker, D., Hamamoto, S., Uozumi, N., Gambale, F., et al. (2010). Roles of tandem-pore K+ channels in plants - a puzzle still to be solved. Plant Biol (Stuttg) 12(1), 56–63.
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694.
Wang, Z. (2012). Isolation of GsGST19 from glycine soja and analysis of saline-alkaline tolerance for transgenic medicago sativa. Acta Agronomica Sin. 38, 971–979.
Wang, X. P., Chen, L. M., Liu, W. X., Shen, L. K., Wang, F. L., Zhou, Y., et al. (2016). AtKC1 and CIPK23 synergistically modulate AKT1-mediated low-potassium stress responses in arabidopsis. Plant Physiol. 170, 2264–2277.
Wang, X., Chen, X., Wang, Q., Chen, M., Liu, X., Gao, D., et al. (2019c). MdBZR1 and MdBZR1-2like transcription factors improves salt tolerance by regulating gibberellin biosynthesis in apple. Front. Plant Sci. 10, 1473.
Wang, Y., He, L., Li, H. D., Xu, J., Wu, W. H. (2010). Potassium channel alpha-subunit AtKC1 negatively regulates AKT1-mediated k(+) uptake in arabidopsis roots under low-k(+) stress. Cell Res. 20, 826–837.
Wang, Z., Ji, H., Yuan, B., Wang, S., Su, C., Yao, B., et al. (2015b). ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 6, 8138.
Wang, D., Liu, Y. X., Yu, Q., Zhao, S. P., Zhao, J. Y., Ru, J. N., et al. (2019a). Functional analysis of the soybean GmCDPK3 gene responding to drought and salt stresses. Int. J. Mol. Sci. 20, 1–22.
Wang, Q., Ni, J., Shah, F., Liu, W., Wang, D., Yao, Y., et al. (2019b). Overexpression of the stress-inducible SsMAX2 promotes drought and salt resistance via the regulation of redox homeostasis in arabidopsis. Int. J. Mol. Sci. 20, 1–19.
Wang, J., Qin, H., Zhou, S., Wei, P., Zhang, H., Zhou, Y., et al. (2020a). The ubiquitin-binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell 32, 414–428.
Wang, Z. Y., Song, F. B., Cai, H., Zhu, Y. M., Bai, X., Ji, W., et al. (2012). Over-expressing GsGST14 from glycine soja enhances alkaline tolerance of transgenic medicago sativa. Biol. Plantarum 56, 516–520.
Wang, Y., Wu, W. H. (2013). Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 64, 451–476.
Wang, L., Wu, X., Liu, Y., Qiu, Q. S. (2015a). AtNHX5 and AtNHX6 control cellular k+ and pH homeostasis in arabidopsis: Three conserved acidic residues are essential for k+ transport. PloS One 10, e0144716.
Wang, L., Xu, F., Yu, F. (2023). Two environmental signal-driven RNA metabolic processes: Alternative splicing and translation. Plant Cell Environ. 46, 1–15.
Wang, T. T., Yu, T. F., Fu, J. D., Su, H. G., Chen, J., Zhou, Y. B., et al. (2020b). Genome-wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance. Front. Plant Sci. 11, 604690.
Wang, Z. Q., Yu, T. F., Sun, G. Z., Zheng, J. C., Chen, J., Zhou, Y. B., et al. (2021b). Genome-wide analysis of the catharanthus roseus RLK1-like in soybean and GmCrRLK1L20 responds to drought and salt stresses. Front. Plant Sci. 12, 614909.
Wang, X., Zhao, J., Fang, Q., Chang, X., Sun, M., Li, W., et al. (2021a). GmAKT1 is involved in k(+) uptake and Na(+)/K(+) homeostasis in arabidopsis and soybean plants. Plant Sci. 304, 110736.
Wani, S. H., Kumar, V., Khare, T., Guddimalli, R., Parveda, M., Solymosi, K., et al. (2020). Engineering salinity tolerance in plants: progress and prospects. Planta 251, 76.
Wei, W., Li, Q. T., Chu, Y. N., Reiter, R. J., Yu, X. M., Zhu, D. H., et al. (2015). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707.
Wei, P., Wang, L., Liu, A., Yu, B., Lam, H. M. (2016). GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front. Plant Sci. 7, 1082.
Wu, S., Zhu, P., Jia, B., Yang, J., Shen, Y., Cai, X., et al. (2018b). A glycine soja group S2 bZIP transcription factor GsbZIP67 conferred bicarbonate alkaline tolerance in medicago sativa. BMC Plant Biol. 18, 234.
Xiong, Y., Fan, X. H., Wang, Q., Yin, Z. G., Sheng, X. W., Chen, J., et al. (2021). Genomic analysis of soybean PP2A-b ‘‘ family and its effects on drought and salt tolerance. Front. Plant Sci. 12, 784038.
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin b-like proteins, regulates k+ transporter AKT1 in arabidopsis. Cell 125, 1347–1360.
Xu, H. R., Liu, Y., Yu, T. F., Hou, Z. H., Zheng, J. C., Chen, J., et al. (2022b). Comprehensive profiling of tubby-like proteins in soybean and roles of the GmTLP8 gene in abiotic stress responses. Front. Plant Sci. 13, 844545.
Xu, C., Shan, J., Liu, T., Wang, Q., Ji, Y., Zhang, Y., et al. (2022a). CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 191, 2427–2446.
Xu, Q., Zhou, B., Ma, C., Xu, X., Xu, J., Jiang, Y., et al. (2010). Salicylic acid-altering arabidopsis mutants response to NO(2) exposure. Bull. Environ. Contam Toxicol. 84, 106–111.
Yan, K., He, W., Bian, L., Zhang, Z., Tang, X., An, M., et al. (2020). Salt adaptability in a halophytic soybean (Glycine soja) involves photosystems coordination. BMC Plant Biol. 20, 155.
Yan, Q., Wang, L., Li, X. (2018). GmBEHL1, a BES1/BZR1 family protein, negatively regulates soybean nodulation. Sci. Rep. 8, 7614.
Yang, Y., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539.
Yang, L., Ji, W., Zhu, Y., Gao, P., Li, Y., Cai, H., et al. (2010). GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 61, 2519–2533.
Yang, X., Jia, Z., Pu, Q., Tian, Y., Zhu, F., Liu, Y. (2022). ABA mediates plant development and abiotic stress via alternative splicing. Int. J. Mol. Sci. 23, 1–14.
Yang, X., Kim, M. Y., Ha, J., Lee, S. H. (2019). Overexpression of the soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic arabidopsis plants. Front. Plant Sci. 10, 1036.
Yang, Y., Yu, T. F., Ma, J., Chen, J., Zhou, Y. B., Chen, M., et al. (2020). The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int. J. Mol. Sci. 21, 1–19.
You, X., Wang, D., Mei, Y., Bi, J., Liu, S., Xu, W. (2022). N(7) -SSPP fusion gene improves salt stress tolerance in transgenic arabidopsis and soybean through ROS scavenging. Plant Cell Environ. 45, 2794–2809.
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130.
Yu, Q., Liu, Y. L., Sun, G. Z., Liu, Y. X., Chen, J., Zhou, Y. B., et al. (2021). Genome-wide analysis of the soybean calmodulin-binding protein 60 family and identification of GmCBP60A-1 responses to drought and salt stresses. Int J Mol Sci 22, 1–20.
Yu, Y., Ni, Z., Wang, Y., Wan, H., Hu, Z., Jiang, Q., et al. (2019). Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic arabidopsis thaliana. Plant Sci. 285, 68–78.
Yu, Y., Tang, J., Liu, C., Ni, Z. (2023). MicroRNA4359b positively regulates the soybean response to salt stress by targeting the f-box protein GmFBX193. Environ. Exp. Bot. 206, 105177.
Yung, W. S., Wang, Q., Huang, M., Wong, F. L., Liu, A., Ng, M. S., et al. (2022). Priming-induced alterations in histone modifications modulate transcriptional responses in soybean under salt stress. Plant J. 109, 1575–1590.
Zaid, A., Mushtag, M., Wani, S. H. (2021). “Interactions of phytohormones with abiotic stress factors under changing climate,” in Frontiers in Plant-Soil Interaction, eds Aftab, T., Hakeem, K. R. (Amsterdam: Academic Press, Elsevier, Amsterdam: Netherland), 221–236. doi: 10.1016/b978-0-323-90943-3.00010-9
Zeng, A., Chen, P., Korth, K. L., Ping, J., Thomas, J., Wu, C., et al. (2019). RNA Sequencing analysis of salt tolerance in soybean (Glycine max). Genomics 111, 629–635.
Zeng, H., Tang, Q., Hua, X. (2010). Arabidopsis brassinosteroid mutants det2-1 and bin2-1 display altered salt tolerance. J. Plant Growth Regul. 29, 44–52.
Zhang, S., Cai, Z., Wang, X. (2009). The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 4543–4548.
Zhang, M., Cao, J., Zhang, T., Xu, T., Yang, L., Li, X., et al. (2022a). A putative plasma membrane Na(+)/H(+) antiporter GmSOS1 is critical for salt stress tolerance in glycine max. Front. Plant Sci. 13, 870695.
Zhang, Y., Fang, Q., Zheng, J., Li, Z., Li, Y., Feng, Y., et al. (2022c). GmLecRlk, a lectin receptor-like protein kinase, contributes to salt stress tolerance by regulating salt-responsive genes in soybean. Int. J. Mol. Sci. 23, 1–13.
Zhang, W., Liao, X., Cui, Y., Ma, W., Zhang, X., Du, H., et al. (2019a). A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PloS Genet. 15, e1007798.
Zhang, M., Liu, Y., Cai, H., Guo, M., Chai, M., She, Z., et al. (2020a). The bZIP transcription factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean. Int. J. Mol. Sci. 21, 1–19.
Zhang, Y. H., Liu, M. F., He, J. B., Wang, Y. F., Xing, G. N., Li, Y., et al. (2015b). Marker-assisted breeding for transgressive seed protein content in soybean [Glycine max (L.) merr]. Theor. Appl. Genet. 128, 1061–1072.
Zhang, M., Liu, S., Wang, Z., Yuan, Y., Zhang, Z., Liang, Q., et al. (2022b). Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 20, 256–282.
Zhang, Y., Ruyter-Spira, C., Bouwmeester, H. J. (2015a). Engineering the plant rhizosphere. Curr. Opin. Biotechnol. 32, 136–142.
Zhang, W., Wang, N., Yang, J., Guo, H., Liu, Z., Zheng, X., et al. (2020b). The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 291, 110326.
Zhang, S., Yang, R., Huo, Y., Liu, S., Yang, G., Huang, J., et al. (2018). Expression of cotton PLATZ1 in transgenic arabidopsis reduces sensitivity to osmotic and salt stress for germination and seedling establishment associated with modification of the abscisic acid, gibberellin, and ethylene signalling pathways. BMC Plant Biol. 18, 218.
Zhang, D., Zhang, Z., Unver, T., Zhang, B. (2021). CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 29, 207–221.
Zhang, X. Z., Zheng, W. J., Cao, X. Y., Cui, X. Y., Zhao, S. P., Yu, T. F., et al. (2019b). Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in arabidopsis and soybean. Front. Plant Sci. 10, 1453.
Zhang, Y., Zhou, Y., Zhu, W., Liu, J., Cheng, F. (2022d). Non-coding RNAs fine-tune the balance between plant growth and abiotic stress tolerance. Front. Plant Sci. 13, 965745.
Zhao, Y., Dong, W., Zhang, N., Ai, X., Wang, M., Huang, Z., et al. (2014). A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 164, 1068–1076.
Zhao, J. Y., Lu, Z. W., Sun, Y., Fang, Z. W., Chen, J., Zhou, Y. B., et al. (2020). The ankyrin-repeat gene GmANK114 confers drought and salt tolerance in arabidopsis and soybean. Front. Plant Sci. 11, 584167.
Zheng, J., Ma, X., Zhang, X., Hu, Q., Qian, R. (2018). Salicylic acid promotes plant growth and salt-related gene expression in(Caryophyllaceae) grown under different salt stress conditions. Physiol. Mol. Biol. Plants 24, 231–238.
Zhou, J. Y., Hao, D. L., Yang, G. Z. (2021). Regulation of cytosolic pH: The contributions of plant plasma membrane h(+)-ATPases and multiple transporters. Int. J. Mol. Sci. 22, 1–17.
Zhou, Z., Jiang, Y., Wang, Z., Gou, Z., Lyu, J., Li, W., et al. (2015). Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414.
Zhu, D., Bai, X., Chen, C., Chen, Q., Cai, H., Li, Y., et al. (2011). GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol. Biol. 77, 285–297.
Zhu, N., Cheng, S., Liu, X., Du, H., Dai, M., Zhou, D. X., et al. (2015). The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 236, 146–156.
Zhu, T., Deng, X., Zhou, X., Zhu, L., Zou, L., Li, P., et al. (2016). Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 6, 35392.
Keywords: salt stress, Na +/K + homeostasis, phytohormones, epigenetics, soybean
Citation: Feng C, Gao H, Zhou Y, Jing Y, Li S, Yan Z, Xu K, Zhou F, Zhang W, Yang X, Hussain MA and Li H (2023) Unfolding molecular switches for salt stress resilience in soybean: recent advances and prospects for salt-tolerant smart plant production. Front. Plant Sci. 14:1162014. doi: 10.3389/fpls.2023.1162014
Received: 09 February 2023; Accepted: 31 March 2023;
Published: 19 April 2023.
Edited by:
Md Atikur Rahman, Rural Development Administration, Republic of KoreaReviewed by:
Abbu Zaid, Govt. Degree College Doda, IndiaCopyright © 2023 Feng, Gao, Zhou, Jing, Li, Yan, Xu, Zhou, Zhang, Yang, Hussain and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Azhar Hussain, YXpoYXJtYXJvdEBxcS5jb20=; Haiyan Li, aHlsaUBoYWluYW51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.