- College of Horticulture, Shenyang Agricultural University, Shenyang, China
Introduction: The cuticle wax covering the plant surface is a whitish hydrophobic protective barrier in Chinese cabbage, and the epicuticular wax crystal deficiency normally has higher commodity value for a tender texture and glossy appearance. Herein, two allelic epicuticular wax crystal deficiency mutants, wdm1 and wdm7, were obtained from the EMS mutagenesis population of a Chinese cabbage DH line ‘FT’.
Methods: The cuticle wax morphology was observed by Cryo-scanning electron microscopy (Cryo-SEM) and the composition of wax was determined by GC-MS. The candidate mutant gene was found by MutMap and validated by KASP. The function of candidate gene was verified by allelic variation.
Results: The mutants had fewer wax crystals and lower leaf primary alcohol and ester content. Genetic analysis revealed that the epicuticular wax crystal deficiency phenotype was controlled by a recessive nuclear gene, named Brwdm1. MutMap and KASP analyses indicated that BraA01g004350.3C, encoding an alcohol-forming fatty acyl-CoA reductase, was the candidate gene for Brwdm1. A SNP 2,113,772 (C to T) variation in the 6th exon of Brwdm1 in wdm1 led to the 262nd amino acid substitution from threonine (T) to isoleucine (I), which existed in a rather conserved site among the amino acid sequences from Brwdm1 and its homologs. Meanwhile, the substitution changed the three-dimensional structure of Brwdm1. The SNP 2,114,994 (G to A) in the 10th exon of Brwdm1 in wdm7 resulted in the change of the 434th amino acid from valine (V) to isoleucine (I), which occurred in the STERILE domain. KASP genotyping showed that SNP 2,114,994 was co-segregated with glossy phenotype. Compared with the wild type, the relative expression of Brwdm1 was significantly decreased in the leaves, flowers, buds and siliques of wdm1.
Discussion: These results indicated that Brwdm1 was indispensable for the wax crystals formation and its mutation resulted in glossy appearance in Chinese cabbage.
Introduction
Cuticle wax is a waterproof layer covering the plant’s surface (Lewandowska et al., 2020), which mainly controls non-stomatal water loss. Cuticle wax could be altered by various abiotic or biotic factors (Kosma et al., 2009). Plants grown in drought conditions normally deposit more wax (Kunst and Samuels, 2003). In addition, the waterproof nature of cuticle wax is conducive to removing dust, bacteria, and fungus from plant surfaces (Yang et al., 2018). Leafy vegetables with epicuticular wax crystal deficiency are usually glossy and tender and tend to have higher commercial value. Previous studies have reported glossy mutants in leafy vegetables, such as cabbage, cai-tai, non-heading Chinese cabbage and Chinese cabbage (Ji et al. 2018; Liu et al. 2017b; Liu et al. 2017c; Wang et al. 2019; Wang et al. 2017; Yang et al. 2022a; Yang et al. 2022b; Zhang et al. 2013). Therefore, breeding for the epicuticular wax crystal deficiency products is an approach to improving the commercial quality of leaf vegetables (Ji et al., 2018; Liu et al., 2021a).
The composition of cuticle wax is complex, generally including very-long-chain fatty acids (VLCFAs) and their derivatives (Li et al., 2008). The composition of cuticle wax varies widely across diverse species and environments (Yeats and Rose, 2013). VLCFAs synthesized in epidermal cells are precursors for wax synthesis, which is divided into two stages and carried out in different cellular compartments. In the first stage, in the plastid, the de novo fatty acid synthesis under the action of fatty acid synthase complex (FAS) generates C16 and C18 acyl chains (Kunst and Samuels, 2003; Lewandowska et al., 2020). In the second stage, C16 and C18 fatty acids generate VLCFAs by the act of the fatty acid elongases (FAE) which are composed of four enzymes β-ketoacyl-CoA synthase (KCS) (James et al., 1995), β-ketoacyl-CoA reductase (KCR) (Xu et al., 1997), β-hydroxyacyl-CoA dehydratase (HCD), and enoyl-CoA reductase (ECR) (Yeats and Rose, 2013). There are two pathways of VLCFAs modification. One is the alkane-forming pathway, which produces aldehydes, alkanes, secondary alcohols, and ketones, and the other is the alcohol-forming pathway, which is responsible for the synthesis of primary alcohols and esters (Jetter and Kunst, 2008).
In Arabidopsis, many key enzyme genes involed in the VLCFAs modification were cloned and functionally verified. CER1, CER3, and MAH1 have been identified as key genes in the alkane-forming pathway. The CER1 gene encodes an aldehyde decarboxylase converting fatty acids into alkanes (Bourdenx et al., 2011). Expression of CER1 is induced by osmotic and ABA stress, both of which can rapidly regulate the alkane biosynthesis to accumulate more wax (Bourdenx et al., 2011). Similar results were obtained in wheat (Li et al., 2019), apple (Qi et al., 2018), and rice (Yu et al., 2008). The amino acid sequences between CER1 and CER3 proteins shared a high consistency. Strict co-expression of CER1 and CER3 has been reported to synthesize VLC alkane synthesis in yeast strains (Bernard et al., 2012). MAH1 encodes a midchain alkane hydroxylase and oxidizes VLCFAs to secondary alcohols, which in turn generates ketones (Greer et al., 2007). Whereas, the wax component produced from the alcohol-forming pathway in Arabidopsis leaves only accounts for approximately 20% of the total wax (Jetter and Kunst, 2008). Consequently, enzyme genes encoding relevant enzymes in alcohol synthesis pathways are less than those in alkane synthesis pathways. WSD1 and CER4 have been reported to participate in the alcohol synthesis pathway. WSD1 is a bifunctional enzyme whose main activity is wax ester synthase (WS) activity, acting on the alcohol-forming pathway (Abdullah et al., 2021). CER4 encodes an alcohol-forming fatty acyl-CoA reductase, responsible for primary alcohol and alkyl ester synthesis (Doan et al., 2009). The AtCER4 protein can promote the accumulation of C24:0 and C26:0 primary alcohols, which has been verified in yeast (Saccharomyces cerevisiae). Subcellular localization showed that the AtCER4 protein was localized to the endoplasmic reticulum. The result of qRT-PCR figured that AtCER4 was expressed in flower, stem, leaf, root, and silique (Rowland et al., 2006).
Although many wax biosynthetic genes have been cloned and their functions had been verified in Arabidopsis, few genes have been characterized in Chinese cabbage. In this study, we used a pair of allelic epicuticular wax crystal deficiency mutant wdm1 and wdm7 as materials to clone the mutant gene in Chinese cabbage. We observed the cuticle wax morphology by Cryo-scanning electron microscopy (Cryo-SEM), determined the composition of wax by GC-MS, and analyzed the genetic characteristics of the epicuticular wax crystal deficiency trait. We found the candidate mutant gene by MutMap and validated it by KASP. Finally, we verified its function by allelic variation.
Materials and methods
Plant materials
‘FT’ is a Chinese cabbage DH line and derives from the variety Fukuda 50. The germinated seeds of ‘FT’ were treated with 0.8% EMS (Gao et al., 2022), and the epicuticular wax crystal deficiency mutant was screened in its M2. A total of 8 mutants with stably inherited epicuticular wax crystal deficiency were identified and named as wdm1, wdm2, wdm3, wdm4, wdm5, wdm6, wdm7, and wdm8. Compared with ‘FT’, the epicuticular wax crystal deficiency mutant exhibited less epidermal wax on the whole plant, which was more obvious at the bolting and flowering stages. Other than that, there are no other phenotypical differences between mutant and wild type. Among these mutants, the wdm1 and wdm7 were selected as experimental materials. All plants were grown in a greenhouse at Shenyang Agriculture University. In late August, the seeds were placed on infiltrated filter paper until germination and then were transferred to a 4-degree refrigerator for 15-day vernalization. Subsequently, the vernalized seeds were sowed in the plug and grew in a greenhouse. After 25 days, the seedlings were transplanted into the nutrition bowl in a greenhouse.
Cryo-SEM analysis and GC–MS analysis
Cryo-scanning electron microscopy was used to observe the epidermal wax morphology of ‘FT’ and mutants. Fresh leaves were cut into 1 mm × 3 mm strips and fastened to the sample disk. The sample disk was first loaded into the transmission device, and then immersed in liquid nitrogen for precooling. The transmission device was installed in the preparation chamber, and the sample disk was transferred to the preparation chamber after the vacuum is pumped. Sublimate the water in the sample and spray the sample with gold before observing (Liu et al., 2021a).
Equal-area leaves of ‘FT’ and mutant was cut and applied in the wax analysis. The wax extraction method was as previously reported (Liu et al., 2021a). The composition of the sample was determined by AGILENT 6890-5973N installed with TG-5ms (30 m*250 um*0.25 μm). The GC program utilized was on the strength of previous reports (Wang et al., 2017).
Genetic analysis and allele test
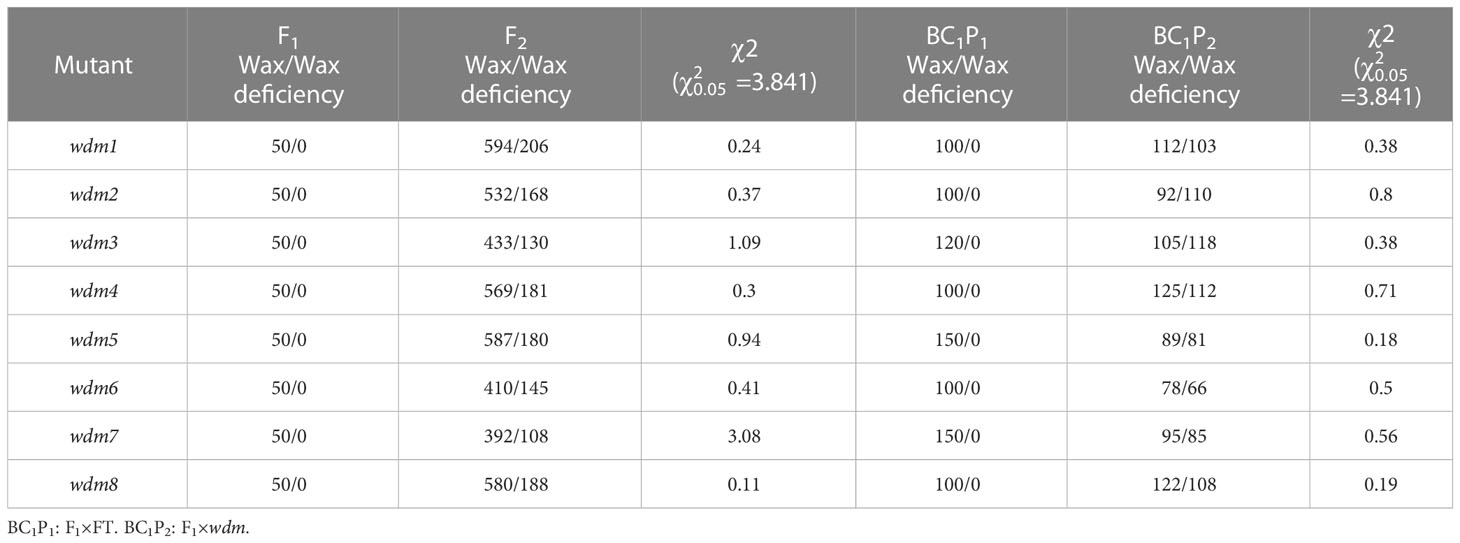
Eight mutants were separately crossed with ‘FT’ to construct F1, F2, and BC1 progenies to carry out the genetic analysis. F2 and BC1 segregation ratios of trait were analyzed by the Chi-square test. Mutant gene function could be verified by allelic mutants (Fu et al., 2020; Zhao et al., 2021). To verify whether there were allelic mutations, we crossed the eight materials with each other. All plants were grown in a greenhouse at Shenyang Agricultural University.
MutMap and data analysis
We employed a modified MutMap method to identify the candidate gene(Gao et al., 2020). Fifty epicuticular wax crystal deficiency mutant plants from F2 were chosen for DNA extraction. Equal amounts of DNA were extracted from each sample and mixed to form an epicuticular wax crystal deficiency mutant pool (HC-pool). DNA from the two parental plants and the HC-pool were re-sequenced by adopting a NovaSeq 6000 System sequencer (Illumina, San Diego, USA). Introducing Burrows-Wheeler Aligner (BWA), sequencing results were aligned to the Brassica reference genome sequence (http://brassicadb.cn/#/Download_genome/Brassica_Genome_data/Brara_Chiifu_V3.0). GATK was used to detect SNPs and INDELs (Liu et al., 2021a). Gene function annotations referred to Brassica Database (BRAD) (Wang et al., 2020).
SNP genotyping by KASP
Kompetitive allele-specific PCR (KASP) was used to detect whether the SNPs identified by MutMap were co-segregated with epicuticular wax crystal deficiency traits and to recognize candidate genes. The DNA samples of 89 F2 glossy individuals from wdm1, 3 F1 individuals, 2 ‘FT’ individuals and 2 wdm1 were prepared for genotyping analysis. The DNA samples of 90 glossy individuals from F2 of wdm7, 2 F1 individuals, 2 ‘FT’ individuals and 2 wdm7 were used for genotyping analysis.
Cloning candidate gene in wdm1 and wdm7
The full length of candidate genes were amplified by full-lengths PCR primers in ‘FT’, wdm1, and wdm7. After the purification of PCR products with a Gel Extraction Kit (CWBIO, Beijing, China), we introduced the purified PCR products into a pGEM-T Easy Vector (Promega, USA). Then, the recombinant vector was transformed into the Top 10 competent cells (CWBIO, Beijing, China). The positive clones were sequenced by GENEWIZ (Tianjin, China).
Bioinformatic analysis of Brwdm1
The conserved domain of Brwdm1, sequenced from the Brassica database, was analyzed online at GSDS2.0 (http://gsds.gao-lab.org/), as well as SMART (https://smart.embl-heidelberg.de/). 50 amino acid sequences of Brwdm1 were put to use in constructing the three-dimensional structure through SWISS-MODEL (https://swissmodel.expasy.org/). The amino acid sequence of Brwdm1 homologs was acquired from Brassica database BLAST. We brought it to DNAMAN V6 (Lynnon BioSoft, Canada) for sequence alignment.
RNA isolation and relative expression analysis
The expression of candidate genes in the root, stem, leaf, flower bud and silique parts were detected by quantitative real-time PCR (qRT-PCR). Using an RNA extraction Kit (TIANGEN, Beijing, China), total RNA was extracted from wdm1 and ‘FT’. FastQuant RT Super Mix (TIANGEN) was used to synthesize first-strand cDNA. Specific primers design and qRT-PCR were performed as previously reported (Liu et al., 2021a).
Results
Characteristics of epicuticular wax crystal deficiency mutants
WT (‘FT’) is a Chinese cabbage (Brassica campestris L. ssp. pekinensis) DH line with a whitish appearance. The epicuticular wax crystal deficiency mutants were obtained in an EMS-mutagenized population of ‘FT’. During the bolting and flowering stage, all the aerial organs, including siliques, buds, stems, and leaves were glaucous, while mutants displayed glossy phenotype in the corresponding organs. Except for the wax trait, there were no obvious differences between mutant and ‘FT’ (Figure 1A).
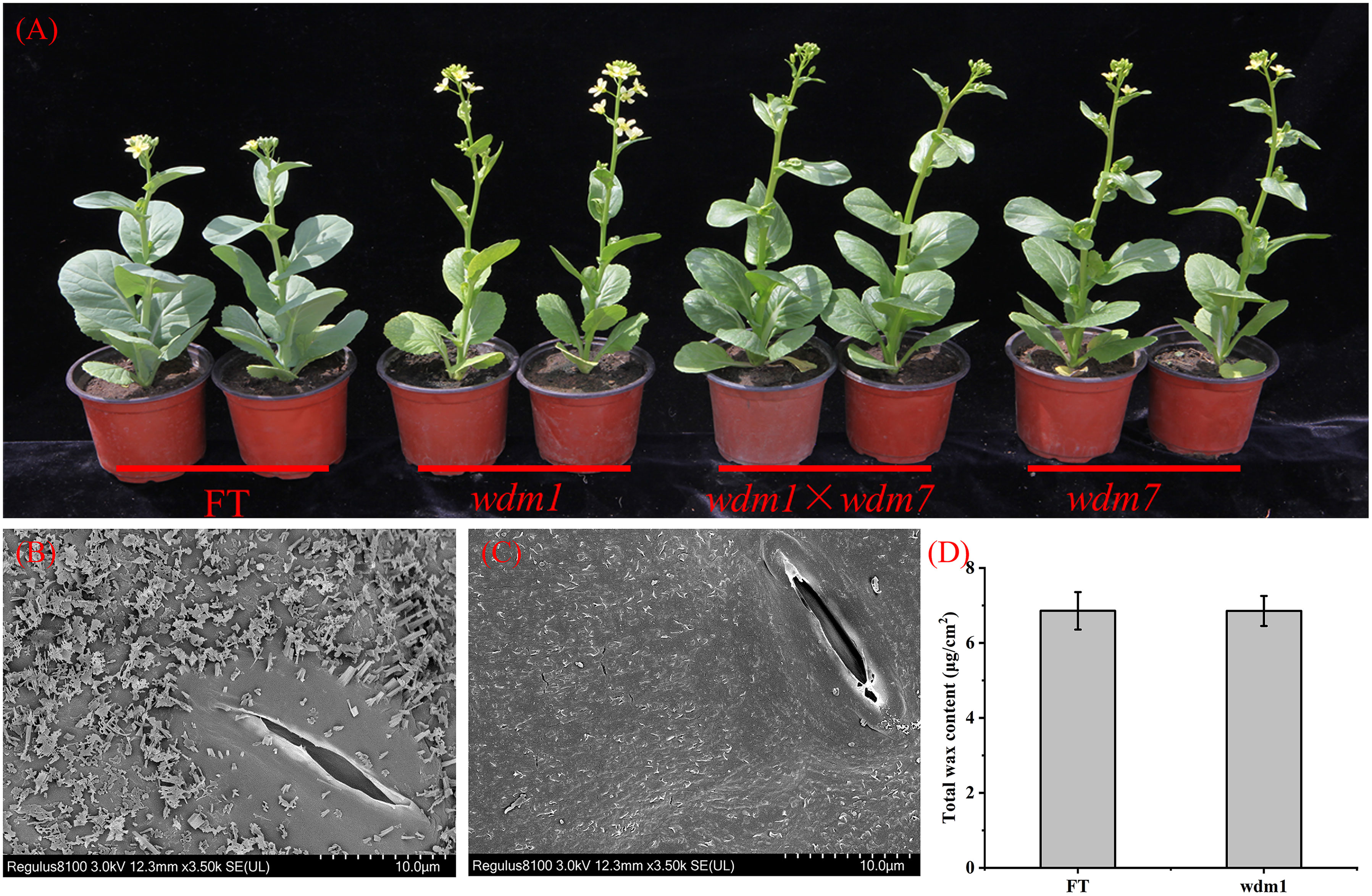
Figure 1 Phenotypic characterization of the ‘FT’ and mutants. (A) All the aerial organs of ‘FT’ were glaucous, while mutants displayed epicuticular wax crystal deficiency. The hybrid progenies from wdm1 and wdm7 appeared epicuticular wax crystal deficiency. SEM images of (B) ‘FT’ leaf and (C) wdm1 leaf. The wdm1 had less epidermal wax. (D) Total cuticle wax coverage and Wax coverage is expressed as μg/cm–2 leaf surface area. Values are means ± SE (n = 3).
The epicuticular wax crystal deficiency character was controlled by a recessive nuclear gene
‘FT’ was crossed with wdm1 to generate F1. All the F1 plants demonstrated the whitish phenotype consistent with ‘FT’. Among 800 F2 individuals, 594 plants were whitish and 206 plants were epicuticular wax crystal deficiency phenotypes. The segregation ratio was close to 3:1 predicted by the Chi-square test (χ2 = 0.24 < = 3.841). In addition, the segregation ratios of BC1P2 (F1×wdm1) were close to 1:1 (χ2 = 0.38). These results exhibited that the epicuticular wax crystal deficiency character of wdm1 was regulated by a single recessive nuclear gene. The identical approach was employed to analyze the genetic characteristics of the remaining seven mutants, and the results manifested that their epicuticular wax crystal deficiency character was severally controlled by a recessive nuclear gene (Table 1).
wdm1 and wdm7 were allelic epicuticular wax crystal deficiency mutants
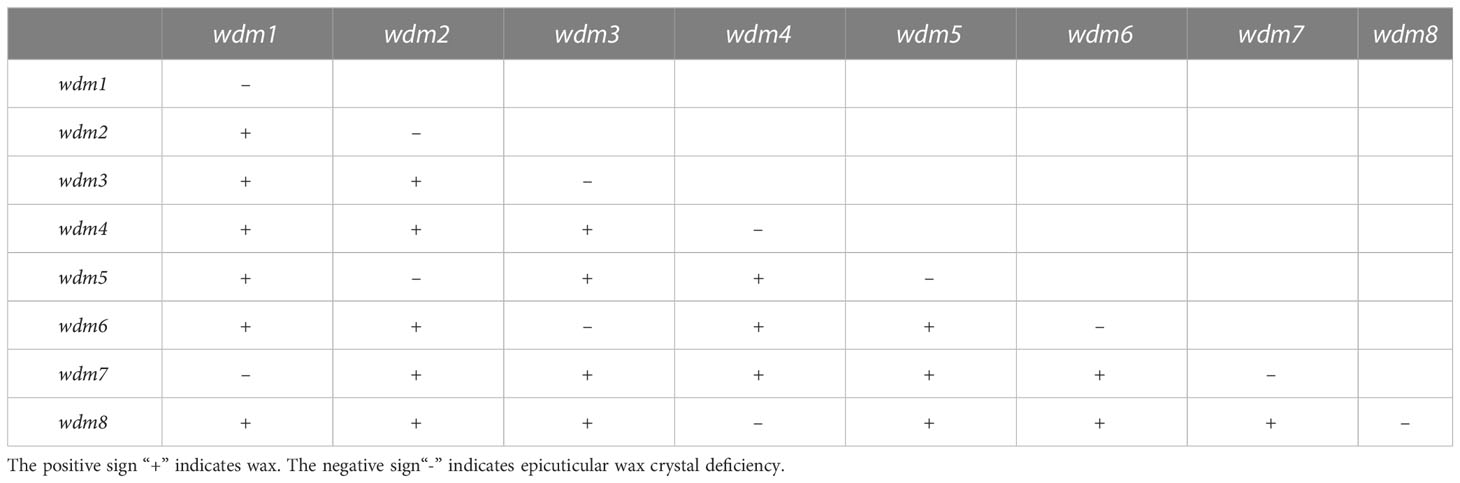
The genetic characterization of these eight mutants was recessive inheritance. After crossing wdm1 and wdm7, we found that all the progenies were epicuticular wax crystal deficiency phenotypes (Figure 1A), which suggested that the epicuticular wax crystal deficiency phenotype of wdm1 and wdm7 was controlled by an identical gene. Similarly, three other groups of allelic mutants: wdm2 and wdm5; wdm3 and wdm6; wdm4 and wdm8, were identified (Table 2).
wdm1 and wdm7 had less epidermal wax crystals
The Cryo-SEM result revealed that flake-and-rod-shaped wax crystals continuously and densely covered the surface of the ‘FT’ leaf, while the filamentous waxy crystals were sparsely distributed on the surface of the wdm1 leaf (Figures 1B, C).
wdm1 decreased in leave primary alcohol and ester
We measured the wax content and composition of ‘FT’ and wdm1 by GC-MS, and surprisingly, the total amount of wax in the leaf epidermis of ‘FT’ and wdm1 was almost the same (Figure 1D). Compared with ‘FT’, the primary alcohol and ester content in wdm1 decreased by 50.5% and 62.4%, respectively, while the secondary alcohol content doubled. In addition, we noticed that the proportion of alkanes in the total wax of wild-type and mutant was large, reaching 77.8% and 78.1%. In particular, C28 alkanes decreased by 37.9% in wdm1, while C26 alkanes increased by 8.5 times (Figure 2).
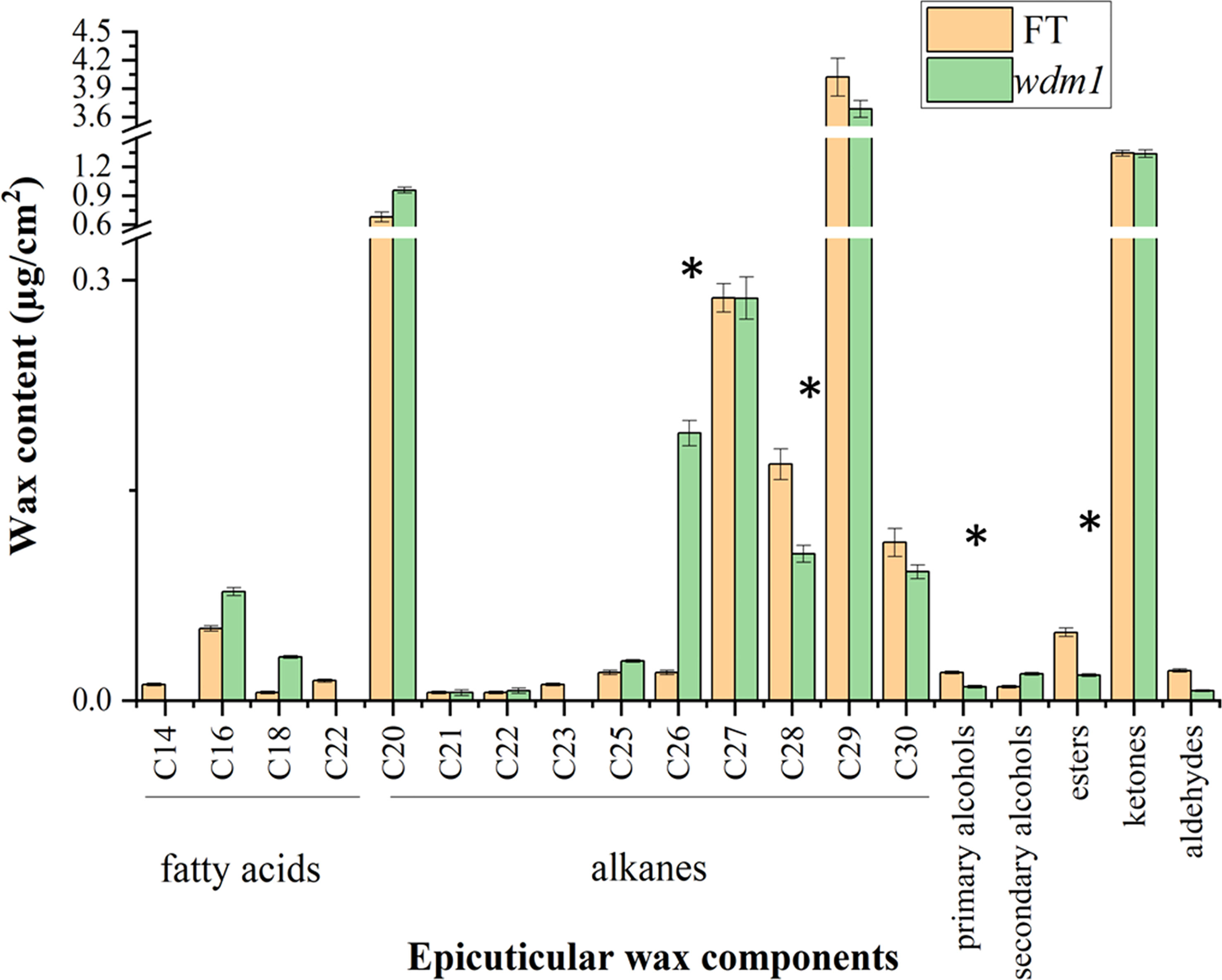
Figure 2 Epicuticle wax components of ‘FT’ and wdm1 leaves. Mutant wdm1 decreased in leave primary alcohol and ester, but elevated secondary alcohol content compared to ‘FT’. Asterisks indicate statistically significant differences determined using a t-test (*P < 0.05).
Preliminary mapping by MutMap
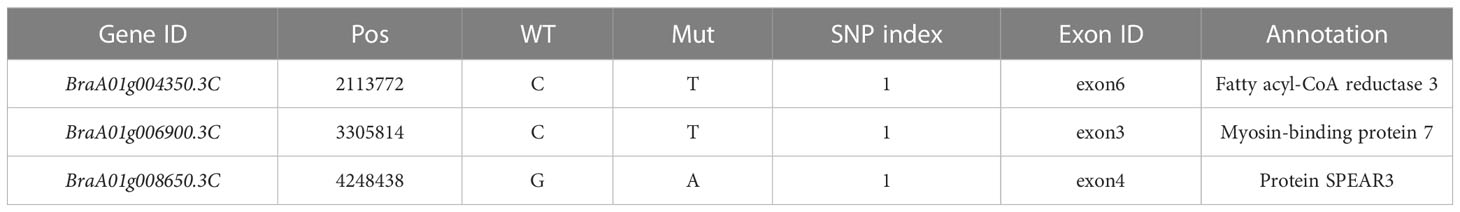
After genome resequencing, we obtained 94,719,968, 54,123,946, and 140,604,782 high-quality reads for wild-type plant ‘FT’, wdm1, and HC pool, respectively. Thereinto, 98%, 96.89%, and 98.97% of high-quality reads from wild-type plant ‘FT’, wdm1 and HC pool were aligned to the Chinese cabbage v. 3.0 reference genome, respectively. Through sequence alignment and mutation analysis software GATK, 1,847,414 SNPs were detected between ‘FT’ and wdm1 pool. Assuming SNP index= 95% as the threshold, an 8.78 Mb region on chromosome A01 was identified as the candidate interval (Figure 3A). Only three SNPs (SNP 2,113,772, SNP 3,305,814, and SNP 4,248,438) were located in exons and caused non-synonymous mutations (Table 3).
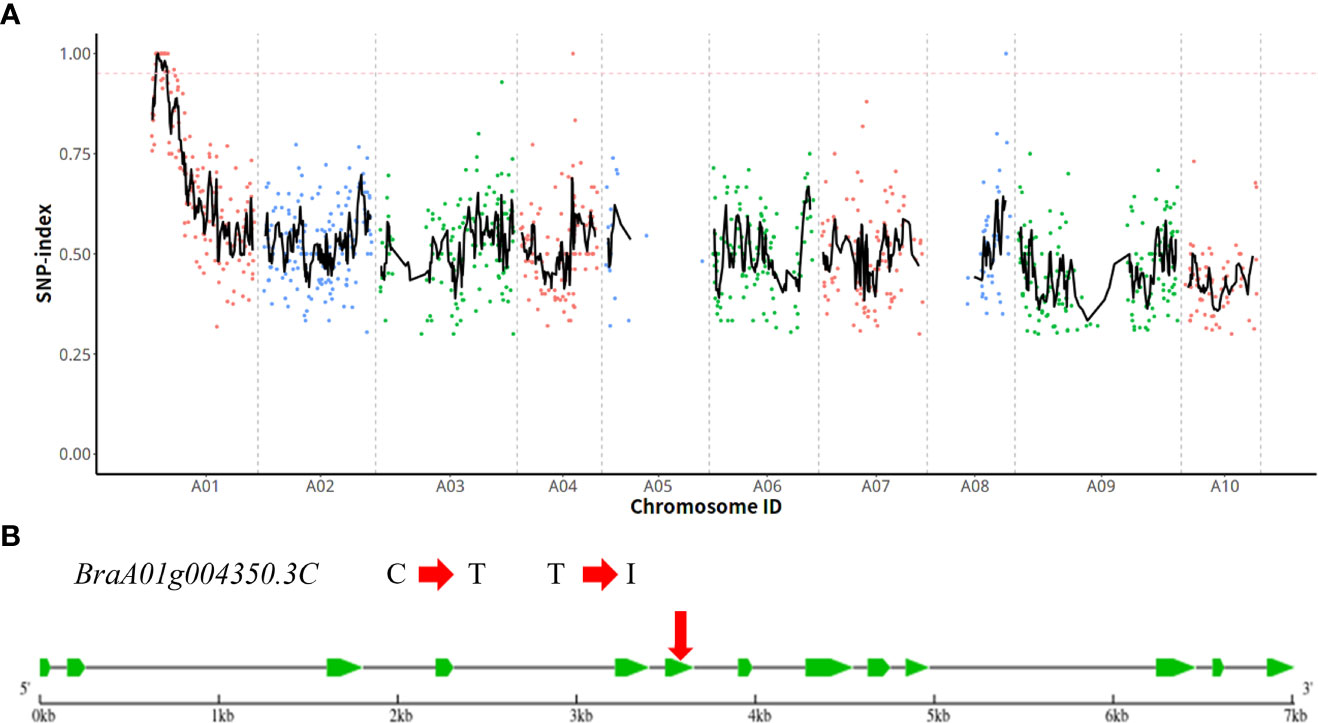
Figure 3 Identification of the candidate gene. (A) MutMap sliding window analysis. The x-axis and y-axis represent the positions of ten chromosomes and the SNP index, respectively. The dotted pink line is the index (0.95) threshold line. (B) Gene structure of BraA01g004350.3C and the location of non-synonymous SNPs in mutant wdm1.
BraA01g004350.3C is the candidate gene
In order to confirm the causal SNP, specific KASP genotyping primers were designed for three SNP mutations. The results depicted that SNP 2,113,772 in wdm1 individuals had a T: T genotype, whereas F1 (‘FT’×wdm1) individuals and ‘FT’ individuals had a C: T and C: C genotype, respectively. The genotypic assay described that SNP 2,113,772 was the T: T genotype in the 87 F2 epicuticular wax crystal deficiency individuals (Figure 4A). By contrast, SNP 3,305,814 were explored as C: T or C: C genotypes and SNP 4,248,438 were detected as G: A genotypes in the 87 F2 epicuticular wax crystal deficiency individuals (Figures 4B, C). Therefore, we considered that the SNP 2,113,772 of BraA01g004350.3C co-segregated with the epicuticular wax crystal deficiency mutant phenotype. BraA01g004350.3C is homologous to Arabidopsis AtCER4 (AT4G33790) and encodes an alcohol-forming fatty acyl-CoA reductase (from the Brassica database). Consequently, we hypothesized that BraA01g004350.3C (Brwdm1) was the candidate causal gene.
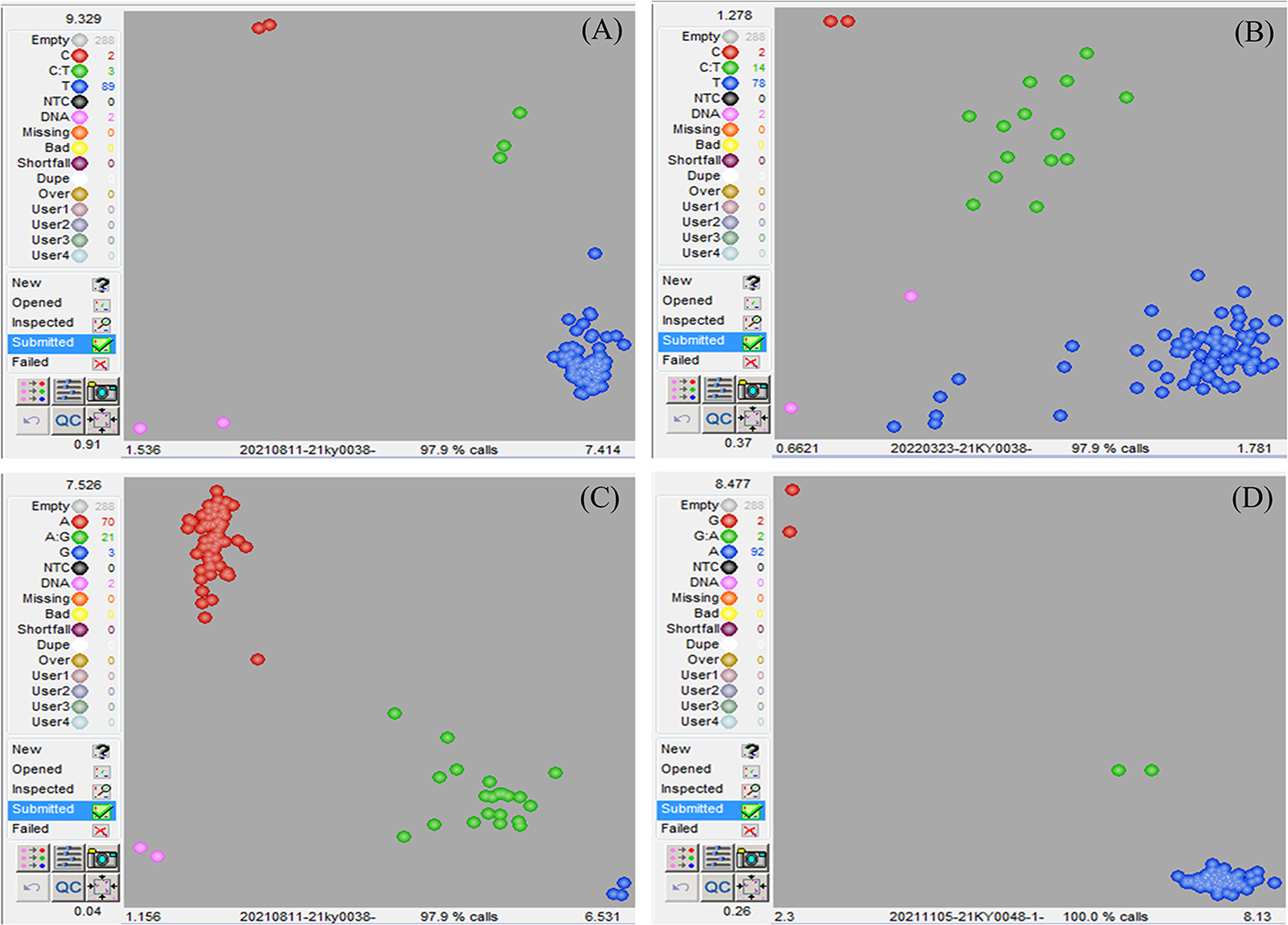
Figure 4 Identification of the candidate gene. 87 epicuticular wax crystal deficiency individuals from F2 of wdm1, 3 F1 individuals, 2 ‘FT’ individuals and 2 wdm1 was used for (A) SNP 2,113,772, (B) SNP 3,305,814, and (C) SNP 4,248,438 genotyping analysis. (A, B) The blue dots, red dots and green dots severally represented T:T genotype, C: C and C: T genotype. (C) The blue dots, red dots and green dots separately stood for G: G genotype, A:A and G: A genotype. The DNA samples of 90 glossy individuals from F2 of wdm7, 2 F1 individuals, 2 ‘FT’ individuals and 2 wdm7 was used for (D) SNP 2,114,994 genotyping analysis.
Mutation of Brwdm1 leads to the epicuticular wax crystal deficiency
Learn from the Brassica database, Brwdm1 was 7015 bp in length and consisted of 13 exons. We cloned the full length of Brwdm1 from ‘FT’, wdm1, and wdm7 (Figure S1). Only the SNP 2,113,772 (C to T) was found for Brwdm1 in wdm1 causing the amino acid change from threonine (T) into isoleucine (I), which was consistent with the MutMap sequencing report (Figure 3B; Figure 4B). Correspondingly, we discovered that there was a SNP (G to A), named SNP 2,114,994, in the 10th exon of Brwdm1 in wdm7, which led to the change of amino acid from valine (V) into isoleucine (I) (Figure 5A). The genotyping results of SNP 2,114,994 showed that wdm7 had a A:A genotype, whereas F1 and ‘FT’ had a G:A and G:G genotype, respectively. And SNP 2,114,994 was the A:A genotype in the 90 F2 glossy individuals (Figure 4D). This verified that the mutation of the gene Brwdm1 lead to the glossy phenotype.
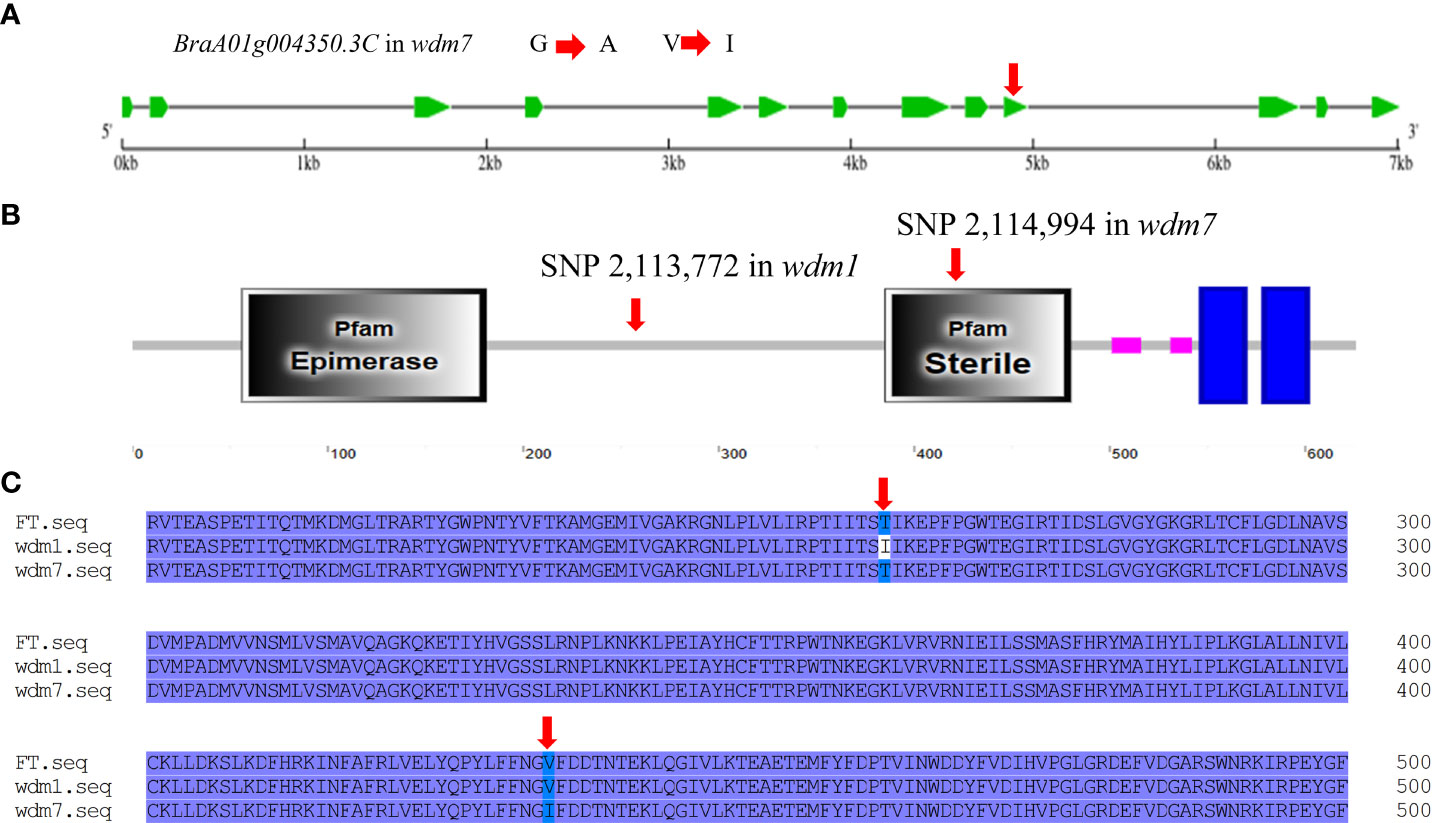
Figure 5 The mutantion of Brwdm1 in wdm7. (A) Gene structure of Brwdm1 and the location of non-synonymous SNP 2,114,994 in wdm7. (B) The conserved domain of Brwdm1 as well as the location of SNP 2,113,772 and SNP 2,114,994. Grey boxes were conserved domains. (C) Partial alignment of protein sequences of Brwdm1 of ‘FT,’ wdm1 and wdm7. wdm1 (the 262nd: T to I). wdm7 (the 434th: V to I).
Structural and phylogenetic analyses of Brwdm1
Brwdm1 protein contains an epimerase domain (56-181), a sterile domain (385-480), and two transmembrane helix regions (547-569 and 579-601). This sterile domain represents the C-terminal domain of fatty acyl CoA reductases, which are involved in cuticle wax biosynthesis. The amino acid transformations in wdm7 (the 434th: V to I) occurred in the sterile domain, while those of wdm1 (the 262nd: T to I) were not (Figures 5B, C). We found several homologs of Brwdm1 from NCBI by BLAST. The amino acid sequences were aligned to understand the relationship between Brwdm1 and these homologs. The results demonstrated that the 262nd amino acid was conservative (Figure 6A). Nucleotide substitutions at both 262nd and 434th issued changes in the three-dimensional structure of the Brwdm1 protein (Figures 6B–E). Therefore, we inferred that the protein conformational changes caused by amino acid substitutions made for the loss of Brwdm1 function, which in turn led to the epicuticular wax crystal deficiency phenotype.
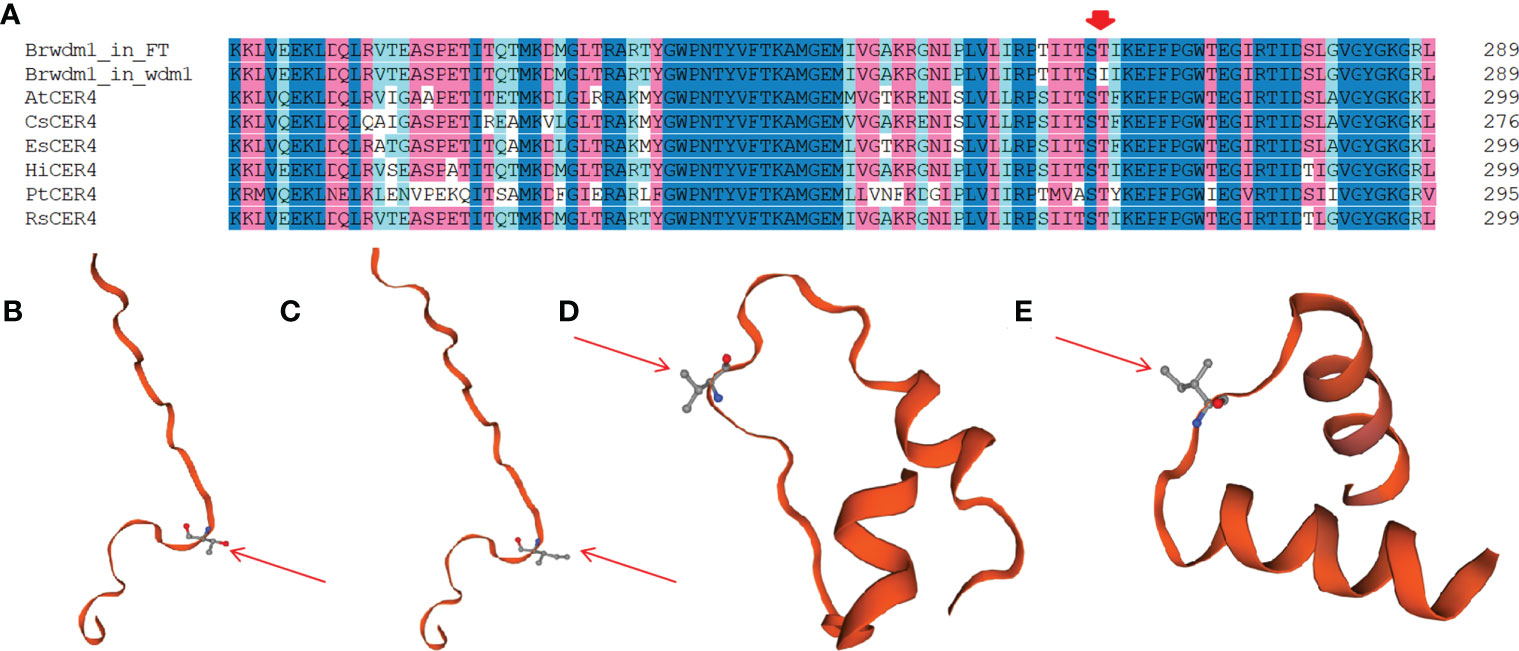
Figure 6 Gene sequence alignment and predicted three-dimensional structure of Brwdm1 protein. (A) Partial aligning to the amino acid sequences of Brwdm1 and these homologues. The CER4 protein accession numbers are as follows: Arabidopsis thaliana, AtCER4 (NP_567936.5); Camelina sativa, CsCER4 (AIE57506.1); Populus tomentosa, PtCER4 (AEV53412.1); Raphanus sativus, RsCER4 (XP_018478019.1); Eutrema salsugineum, EsCER4 (XP_006412311.1) and Hirschfeldia incana, HiCER4 (KAJ0252035.1). Partial three-dimensional structure of BrCER4 protein in ‘FT’ (B) and wdm1 (C) Red arrows: amino acid residue 262 (Thr) of ‘FT’ (B) and (Ile) of wdm1 (C). Partial three-dimensional structure of Brwdm1 protein in ‘FT’ (D) and wdm7 (E) Red arrows: amino acid residue 434 (Val) of ‘FT’ (D) and (Ile) of wdm7 (E).
Relative expression analysis of candidate genes
The relative expression of Brwdm1 of the various organs from ‘FT’ and wdm1 was determined by qRT-PCR. The results showed that the expression levels of Brwdm1 in the leaves of FT and wdm1 was the highest, respectively. Compared with the wild type, the relative expression of Brwdm1 in the leaves, flowers, buds and siliques of wdm1 was significantly decreased (Figure 7).
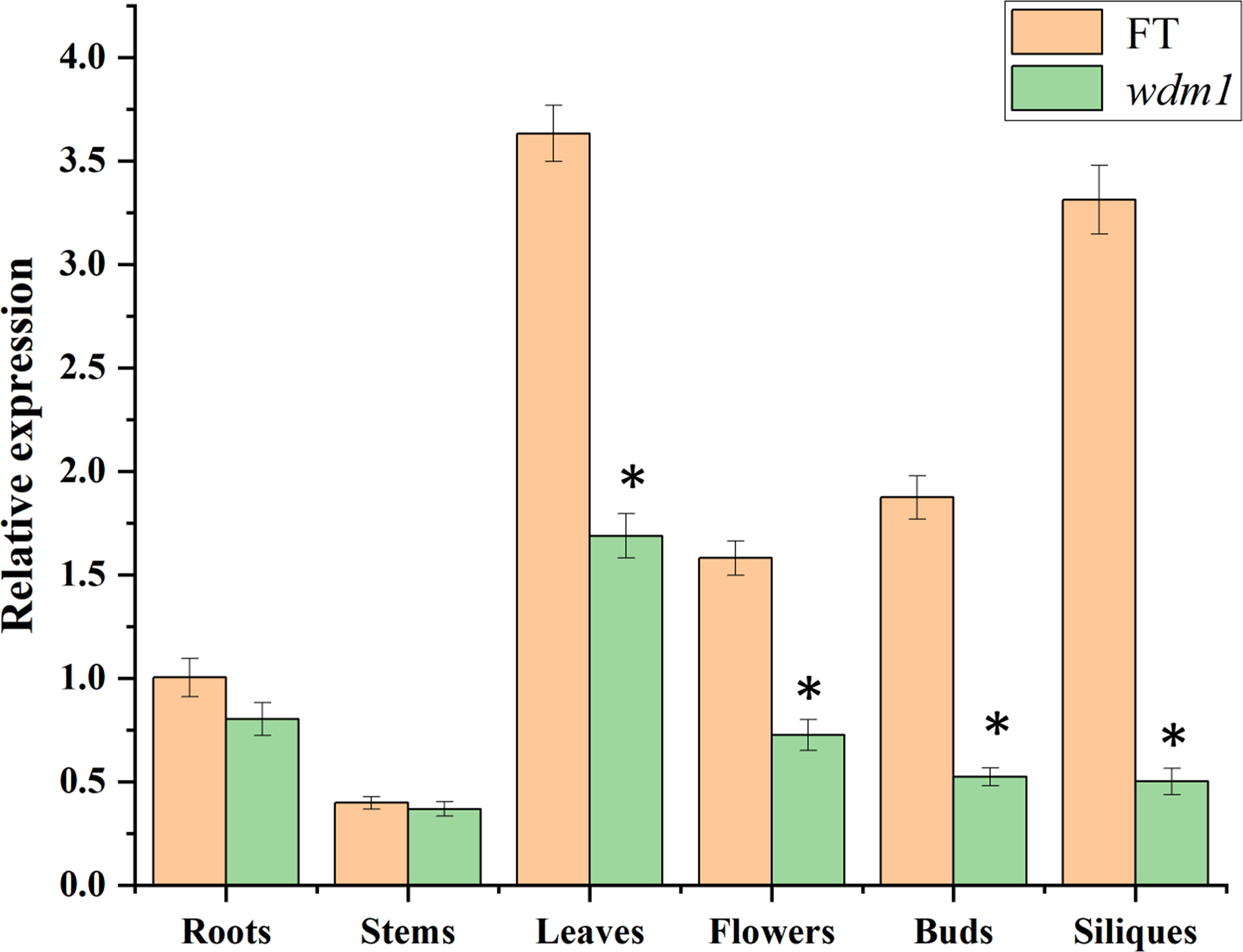
Figure 7 Relative expression levels of Brwdm1 in different organs of ‘FT’ and wdm1. Asterisks indicate statistically significant differences determined using a t-test (*P < 0.05).
Discussion
Plant mutants acquired by artificial mutagenesis possess widely and randomly mutated traits, which are ideal materials for germplasm innovation. As an excellent germplasm resource and its potential breeding value for improved quality, epicuticular wax crystal deficiency mutants have been reported in a variety of plants, such as navel orange, cabbage, cucumber, tomato, and apple (Liu et al., 2012; Wang et al., 2015; Liu et al., 2017c; Zhang et al., 2019; Xiong et al., 2020). The mutants in our experiment were derived from an EMS mutagenesis population. Since the wild-type ‘FT’ was a DH line, our mutants were excellent material for investigating gene functions. In this experiment, eight epicuticular wax crystal deficiency mutants were identified, and their epicuticular wax crystal deficiency traits followed recessive inheritance. For a class of mutants with similar traits and genetic characteristics, the allelism test could be employed to determine whether they were controlled by an identical mutant gene. Among the eight mutants, we recognized four groups of allelic mutants by the allelism test, and one group of them (wdm1 and wdm7) was applied as experimental material to clone the epicuticular wax crystal deficiency mutant gene. Mutmap and KASP tests proved that BraA01g004350.3C (Brwdm1) was the target gene.
Currently, limitations still existed in the implementation of the transgenic technology in Chinese cabbage, while allelic mutant analysis could replace the transgenic technology to verify the gene functions. Allelic mutants have been applied in some crops for gene function verification, such as in lettuce (Huo et al., 2016), sorghum (Jiao et al., 2017), and cucumber (Zhang et al., 2018). Through BSR-Seq and whole-genome re-sequencing, gene BrDVR was identified as a candidate gene contributing to whole-plant pale green in Chinese cabbage mutant pem1. In pem2, which was an allelic mutant of pem1, a SNP was identified in BrDVR (Zhao et al., 2021). Herein, we confirmed Brwdm1 was the candidate for epicuticular wax crystal deficiency in wdm1 and found another SNP occurred in Brwdm1 from its allelic mutant wdm7.
Brwdm1 encodes an alcohol-forming fatty acyl-CoA reductase (FAR), involved in cuticle wax biosynthesis. The specific expression of apple CER4 homolog in the fruit epidermis resulted in the abundance of C30, C28 and C26 primary alcohols in the fruit wax of cv Florina (Albert et al., 2012). In navel orange, the reduced expression of CsCER4-LIKE1 and CsCER4-LIKE3 led to a decrease in primary alcohol content in the waxy fruit epidermis (Liu et al., 2015). The CsCER4 gene was cloned in cucumber, and its expression level was significantly diverse between lines 3413 (glossy type) and 3401 (waxy type) (Wang et al., 2018). The substrates of alcohol-forming pathway and the alkane-forming pathway are both very-long-chain fatty acids (VLCFAs). In Arabidopsis cer4 mutants, the content of primary alcohols and wax esters was significantly decreased, and the content of aldehydes, alkanes, secondary alcohols, and ketones was slightly elevated (Rowland et al., 2006). In Arabidopsis cer1 mutants, the content of aldehydes, alkanes, secondary alcohol and ketones, as well as primary alcohols and esters was significantly decreased (Bourdenx et al., 2011). There is a mutual interaction between the two forming pathways. Therefore, mutations in BrCER4 also affected alkane content in Chinese cabbage. Wax crystals and wax films are two forms of wax accumulation in the epidermis. Wax crystals give the plant a glaucous appearance and wax films give the plant a glossy appearance. In our experiment, all the aerial organs of wdm1 and wdm7 were glossy and less wax crystals were observed on the surface of the wdm1 leaf than ‘FT’ under Cryo-scanning electron microscope. The total wax content in our experiment includes not only wax crystals content, but also wax films content. These results were similar to those reported in Arabidopsis (Rowland et al., 2006), indicating that CER4 plays a conserved role across multiple species. Nevertheless, it remains to be explored whether the acyl reduction product is fundamental for the formation of wax crystals.
The homologous genes of AtCER4 have been cloned in Brassica napus, cabbage, and purple cai-tai, and they were reported to be involved in the wax biosynthesis pathway. In Brassica napus, a 157-bp DNA deletion in BnA1.CER4 and a 1905-bp DNA insertion in BnC1.CER4 were confirmed to generate wax deficiency in eceriferum mutant Nilla Glossy, which was controlled by these two recessive alleles (Liu et al., 2021b). In cabbage, a SNP in Bol013612 altered the splice site, giving rise to a six-nucleotide insertion into the cDNA. And this mutation interfered with gene function, which brought about a glossy appearance in mutant LD10G (Liu et al., 2017a). In purple cai-ta, a 39-bp deletion of Bra011470 was responsible for the glossy phenotype (Wang et al., 2019). By aligning Brwdm1 with its homologous proteins, we discovered that the mutant site of Brwdm1 in wdm1 was conserved, and the nucleotide substitution at 262nd changed the three-dimensional structure.
The epicuticular wax crystal deficiency mutants identified in this experiment had the glossy appearance, tender taste, and similar growth potential to the wild-type ‘FT’, and provided an excellent gene resource for commercial quality breeding in Chinese cabbage. Through back-crossing breeding, the glossy and tender characteristics could be transferred into other excellent lines to cultivate high-quality varieties.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/accession: PRJNA941501.
Author contributions
HF and GS designed the experiments. GS performed the experiments and wrote the manuscript. CL and JR assisted in the screening of mutants. HF and BF revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Natural Science Foundation of China (Grant No. 31972405) and the Scientific research funding project of Liaoning Provincial (LJKZ0633).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1161181/full#supplementary-material
Supplementary Figure 1 | Cloning candidate gene BraA01g004350.3C in ‘FT’ and mutants. The full length of 7015 bp was cloned in four segments, which were 1769bp (F: 5’ GGCTTTTACCGATCACCATT 3’; R: 5’ ACGTCCCCATTGACAACAGT 3’), 1802bp (F: 5’ GGGAAGGACTTGTACAGGGTA 3’; R: 5’ GAACCTGGTGAGTCCCATATC 3’), 1655bp (F: 5’ CTCCTGAAACCATCACTCAAA 3’; 5’ AAACAAAGGTTACACAAATGACAT 3’), and 2210bp (F: 5’ AAGACAGAAGCCGAAACCGA 3’; 5’ TTCGGAGAACCTTAGCCCAT 3’), respectively. When designing the primers, we set overlaps between adjacent segments to ensure that the segments can be properly spliced after sequencing. The first 1769bp segment contains part of the promoter sequence, and the last 2210bp segment contains part of the downstream sequence of the gene. After sequencing, the four segments can be spliced together and contain a full length of BraA01g004350.3C (7015 bp).
References
Abdullah, H. M., Rodriguez, J., Salacup, J. M., Castaneda, I. S., Schnell, D. J., Pareek, A., et al. (2021). Increased cuticle waxes by overexpression of WSD1 improves osmotic stress tolerance in Arabidopsis thaliana and Camelina sativa. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22105173
Albert, Z., Ivanics, B., Molnár, A., Miskó, A., Tóth, M., Papp, I. (2012). Candidate genes of cuticle formation show characteristic expression in the fruit skin of apple. Plant Growth Regul. 70, 71–78. doi: 10.1007/s10725-012-9779-y
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J. D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. doi: 10.1105/tpc.112.099796
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Leger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45. doi: 10.1104/pp.111.172320
Doan, T. T., Carlsson, A. S., Hamberg, M., Bulow, L., Stymne, S., Olsson, P. (2009). Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 166, 787–796. doi: 10.1016/j.jplph.2008.10.003
Fu, W., Huang, S., Gao, Y., Zhang, M., Qu, G., Wang, N., et al. (2020). Role of BrSDG8 on bolting in Chinese cabbage (Brassica rapa). Theor. Appl. Genet. 133, 2937–2948. doi: 10.1007/s00122-020-03647-4
Gao, Y., Huang, S., Qu, G., Fu, W., Zhang, M., Liu, Z., et al. (2020). The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Res. 7, 178. doi: 10.1038/s41438-020-00399-6
Gao, Y., Qu, G., Huang, S., Liu, Z., Zhang, M., Fu, W., et al. (2022). Comparison between germinated seed and isolated microspore EMS mutagenesis in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 8. doi: 10.3390/horticulturae8030232
Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L., Kunst, L., et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of. Arabidopsis. Plant Physiol. 145, 653–667. doi: 10.1104/pp.107.107300
Huo, H., Henry, I. M., Coppoolse, E. R., Verhoef-Post, M., Schut, J. W., de Rooij, H., et al. (2016). Rapid identification of lettuce seed germination mutants by bulked segregant analysis and whole genome sequencing. Plant J. 88, 345–360. doi: 10.1111/tpj.13267
James, D. W., Jr, Lim, E., Keller, J., Plooy, I., Ralston, E., Donner, H. K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATIONl (FAEl) gene with the Maize transposon activator. Plant Cell 7, 309–319. doi: 10.1105/tpc.7.3.309
Jetter, R., Kunst, L. (2008). Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. Plant J. 54, 670–683. doi: 10.1111/j.1365-313X.2008.03467.x
Ji, J. L., Cao, W. X., Dong, X., Liu, Z. Z., Fang, Z. Y., Zhuang, M., et al. (2018). A 252-bp insertion in BoCER1 is responsible for the glossy phenotype in cabbage (Brassica oleracea L. var. capitata). Mol. Breed. 38. doi: 10.1007/s11032-018-0888-9
Jiao, Y., Burow, G., Gladman, N., Acosta-Martinez, V., Chen, J., Burke, J., et al. (2017). Efficient identification of causal mutations through sequencing of bulked F2 from two allelic bloomless mutants of Sorghum bicolor. Front. Plant Sci. 8, 2267. doi: 10.3389/fpls.2017.02267
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lu, S., Joubes, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of. Arabidopsis. Plant Physiol. 151, 1918–1929. doi: 10.1104/pp.109.141911
Kunst, L., Samuels, A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42(1): 51–80. doi: 10.1016/S0163-7827(02)00045-0
Lewandowska, M., Keyl, A., Feussner, I. (2020). Wax biosynthesis in response to danger: its regulation upon abiotic and biotic stress. New Phytol. 227, 698–713. doi: 10.1111/nph.16571
Li, T., Sun, Y., Liu, T., Wu, H., An, P., Shui, Z., et al. (2019). TaCER1-1A is involved in cuticular wax alkane biosynthesis in hexaploid wheat and responds to plant abiotic stresses. Plant Cell Environ. 42, 3077–3091. doi: 10.1111/pce.13614
Li, F., Wu, X., Lam, P., Bird, D., Zheng, H., Samuels, L., et al. (2008). Identification of the wax ester synthase/acyl-coenzyme a: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in. Arabidopsis. Plant Physiol. 148, 97–107. doi: 10.1104/pp.108.123471
Liu, Z., Fang, Z., Zhuang, M., Zhang, Y., Lv, H., Liu, Y., et al. (2017c). Fine-mapping and analysis of Cgl1, a gene conferring glossy trait in cabbage (Brassica oleracea L. var. capitata). Front. Plant Sci. 8, 239. doi: 10.3389/fpls.2017.00239
Liu, C. H., Song, G. X., Wang, N., Huang, S. N., Gao, Y., Fu, W., et al. (2021a). A single SNP in Brcer1 results in wax deficiency in Chinese cabbage (Brassica campestris L. ssp. pekinensis). Scientia Hortic. 282, 110019. doi: 10.1016/j.scienta.2021.110019
Liu, D., Tang, J., Liu, Z., Dong, X., Zhuang, M., Zhang, Y., et al. (2017a). Cgl2 plays an essential role in cuticular wax biosynthesis in cabbage (Brassica oleracea L. var. capitata). BMC Plant Biol. 17, 223. doi: 10.1186/s12870-017-1162-8
Liu, D. M., Tang, J., Liu, Z. Z., Dong, X., Zhuang, M., Zhang, Y. Y., et al. (2017b). Fine mapping of BoGL1, a gene controlling the glossy green trait in cabbage (Brassica oleracea L. var. capitata). Mol. Breed. 37, 69. doi: 10.1007/s11032-017-0674-0
Liu, D., Yang, L., Zheng, Q., Wang, Y., Wang, M., Zhuang, X., et al. (2015). Analysis of cuticular wax constituents and genes that contribute to the formation of ‘glossy newhall’, a spontaneous bud mutant from the wild-type ‘Newhall’ navel orange. Plant Mol. Biol. 88, 573–590. doi: 10.1007/s11103-015-0343-9
Liu, D. C., Zeng, Q., Ji, Q. X., Liu, C. F., Liu, S. B., Liu, Y. (2012). A comparison of the ultrastructure and composition of fruits’ cuticular wax from the wild-type ‘Newhall’ navel orange (Citrus sinensis [L.] osbeck cv. Newhall) and its glossy mutant. Plant Cell Rep. 31, 2239–2246. doi: 10.1007/s00299-012-1333-x
Liu, J., Zhu, L., Wang, B., Wang, H., Khan, I., Zhang, S., et al. (2021b). BnA1.CER4 and BnC1.CER4 are redundantly involved in branched primary alcohols in the cuticle wax of. Brassica napus. Theor. Appl. Genet. 134, 3051–3067. doi: 10.1007/s00122-021-03879-y
Qi, C.-H., Zhao, X.-Y., Jiang, H., Zheng, P.-F., Liu, H.-T., Li, Y.-Y., et al. (2018). Isolation and functional identification of an apple MdCER1 gene. Plant Cell Tissue Organ Culture (PCTOC) 136, 1–13. doi: 10.1007/s11240-018-1504-8
Rowland, O., Zheng, H., Hepworth, S. R., Lam, P., Jetter, R., Kunst, L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme a reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142, 866–877. doi: 10.1104/pp.106.086785
Wang, C. J., Li, H. L., Li, Y. X., Meng, Q. F., Xie, F., Xu, Y. J., et al. (2019). Genetic characterization and fine mapping BrCER4 in involved cuticular wax formation in purple cai-tai (Brassica rapa L. var. purpurea). Mol. Breed. 39. doi: 10.1007/s11032-018-0919-6
Wang, C. J., Li, Y. X., Xie, F., Kuang, H. H., Wan, Z. J. (2017). Cloning of the Brcer1 gene involved in cuticular wax production in a glossy mutant of non-heading Chinese cabbage (Brassica rapa L. var. communis). Mol. Breed. 37. doi: 10.1007/s11032-017-0745-2
Wang, W., Wang, S., Li, M., Hou, L. (2018). Cloning and expression analysis of Cucumis sativus L. CER4 involved in cuticular wax biosynthesis in cucumber. Biotechnol. Biotechnol. Equip. 32, 1113–1118. doi: 10.1080/13102818.2018.1499444
Wang, N., Zhang, Y., Huang, S., Liu, Z., Li, C., Feng, H. (2020). Defect in Brnym1, a magnesium-dechelatase protein, causes a stay-green phenotype in an EMS-mutagenized Chinese cabbage (Brassica campestris L. ssp. pekinensis) line. Hortic. Res. 7, 8. doi: 10.1038/s41438-019-0223-6
Wang, W., Zhang, Y., Xu, C., Ren, J., Liu, X., Black, K., et al. (2015). Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 87, 219–233. doi: 10.1007/s11103-014-0271-0
Xiong, C., Xie, Q., Yang, Q., Sun, P., Gao, S., Li, H., et al. (2020). WOOLLY, interacting with MYB transcription factor MYB31, regulates cuticular wax biosynthesis by modulating CER6 expression in tomato. Plant J. 103, 323–337. doi: 10.1111/tpj.14733
Xu, X., Dietrich, C. R., Delledonne, M., Xia, Y., Wen, T. J., Robertson, D. S., et al. (1997). Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a beta-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol. 115 (2), 501–510. doi: 10.1104/pp.115.2.501
Yang, C. K., Huang, B. H., Ho, S. W., Huang, M. Y., Wang, J. C., Gao, J., et al. (2018). Molecular genetic and biochemical evidence for adaptive evolution of leaf abaxial epicuticular wax crystals in the genus lithocarpus (Fagaceae). BMC Plant Biol. 18, 196. doi: 10.1186/s12870-018-1420-4
Yang, S., Liu, H., Wei, X., Zhao, Y., Wang, Z., Su, H., et al. (2022a). BrWAX2 plays an essential role in cuticular wax biosynthesis in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor. Appl. Genet. 135 (2), 693–707. doi: 10.1007/s00122-021-03993-x
Yang, S., Tang, H., Wei, X., Zhao, Y., Wang, Z., Su, H., et al. (2022b). BrWAX3, encoding a beta-ketoacyl-CoA synthase, plays an essential role in cuticular wax biosynthesis in Chinese cabbage. Int. J. Mol. Sci. 23. doi: 10.21203/rs.3.rs-1914778/v1
Yeats, T. H., Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Yu, D., Ranathunge, K., Huang, H., Pei, Z., Franke, R., Schreiber, L., et al. (2008). Wax crystal-sparse Leaf1 encodes a beta-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228, 675–685. doi: 10.1007/s00425-008-0770-9
Zhang, C., Chen, F., Zhao, Z., Hu, L., Liu, H., Cheng, Z., et al. (2018). Mutations in CsPID encoding a Ser/Thr protein kinase are responsible for round leaf shape in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 131, 1379–1389. doi: 10.1007/s00122-018-3084-8
Zhang, X., Wang, Q., Zou, C., Liu, Z., Wang, Y., Feng, H. (2013). Genetic analysis and preliminary mapping of wax gene on stem in Chinese cabbage. Mol. Breed. 32, 867–874. doi: 10.1007/s11032-013-9914-0
Zhang, Y. L., Zhang, C. L., Wang, G. L., Wang, Y. X., Qi, C. H., You, C. X., et al. (2019). Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 249, 1627–1643. doi: 10.1007/s00425-019-03115-4
Keywords: Chinese cabbage, EMS mutagenesis, epicuticular wax crystals deficiency, allelic mutants, primary alcohols
Citation: Song G, Liu C, Fang B, Ren J and Feng H (2023) Identification of an epicuticular wax crystal deficiency gene Brwdm1 in Chinese cabbage (Brassica campestris L. ssp. pekinensis). Front. Plant Sci. 14:1161181. doi: 10.3389/fpls.2023.1161181
Received: 08 February 2023; Accepted: 28 April 2023;
Published: 30 May 2023.
Edited by:
Ryo Fujimoto, Kobe University, JapanReviewed by:
Branka Salopek Sondi, Rudjer Boskovic Institute, CroatiaYuxiang Yuan, Henan Academy of Agricultural Sciences, China
Copyright © 2023 Song, Liu, Fang, Ren and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Ren, MjAxOTUwMDAyM0BzeWF1LmVkdS5jbg==; Hui Feng, ZmVuZ2h1aWFhYUBzeWF1LmVkdS5jbg==
 Gengxing Song
Gengxing Song Hui Feng
Hui Feng