- 1Hainan Yazhou Bay Seed Laboratory, Sanya, China
- 2State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 3State Key Laboratory of Rice Biology and Breeding, China National Rice Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou, China
As a leading oilseed crop that supplies plant oil and protein for daily human life, increasing yield and improving nutritional quality (high oil or protein) are the top two fundamental goals of soybean breeding. Seed size is one of the most critical factors determining soybean yield. Seed size, oil and protein contents are complex quantitative traits governed by genetic and environmental factors during seed development. The composition and quantity of seed storage reserves directly affect seed size. In general, oil and protein make up almost 60% of the total storage of soybean seed. Therefore, soybean’s seed size, oil, or protein content are highly correlated agronomical traits. Increasing seed size helps increase soybean yield and probably improves seed quality. Similarly, rising oil and protein contents improves the soybean’s nutritional quality and will likely increase soybean yield. Due to the importance of these three seed traits in soybean breeding, extensive studies have been conducted on their underlying quantitative trait locus (QTLs) or genes and the dissection of their molecular regulatory pathways. This review summarized the progress in functional genome controlling soybean seed size, oil and protein contents in recent decades, and presented the challenges and prospects for developing high-yield soybean cultivars with high oil or protein content. In the end, we hope this review will be helpful to the improvement of soybean yield and quality in the future breeding process.
1 Introduction
Oil and protein are essential nutrients for humans and livestock, with almost 70% of cooking oil and half of feed protein coming from plants. Soybean (Glycine max) provides nearly 60% of global oilseed production and accounts for more than 25% of the protein consumption for food and animal feed worldwide, making it a leading commercial crop for vegetable oil and protein production (Wang et al., 2020b). The cultivated soybean was domesticated from wild soybean (Glycine soja) in central China about 5000 years ago and then spread around the world (Carter et al., 2004; Wilson, 2008). As a dominant oilseed and fodder crop, modern cultivated soybean seeds contain approximately 17% oil, 35% protein (including essential and non-essential amino acids), 31% carbohydrates (including soluble and insoluble carbohydrates), 13% moisture, and 4% ash (Liu, 1997) (Figure 1). The oil content of soybean seeds ranges from 8.3 to 27.9%, and protein concentration varies from 34.1 to 56.8% depending on the soybean varieties and cultivation conditions (Wilson, 2004). Soybean oil is generated and stored mainly as fatty acids (FAs), triacylglycerols (TAGs), and tocopherols (Liu et al., 2022). There are five central FAs presented in soybean seeds, including stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), linolenic acid (C18:3), and palmitic acid (C16:0), whose composition directly determined the soybean oil quality. Soybean seed protein consists mainly of storage proteins such as glycinin (11S globulin) and conglycinin (7S globulin) (Liu et al., 2022).
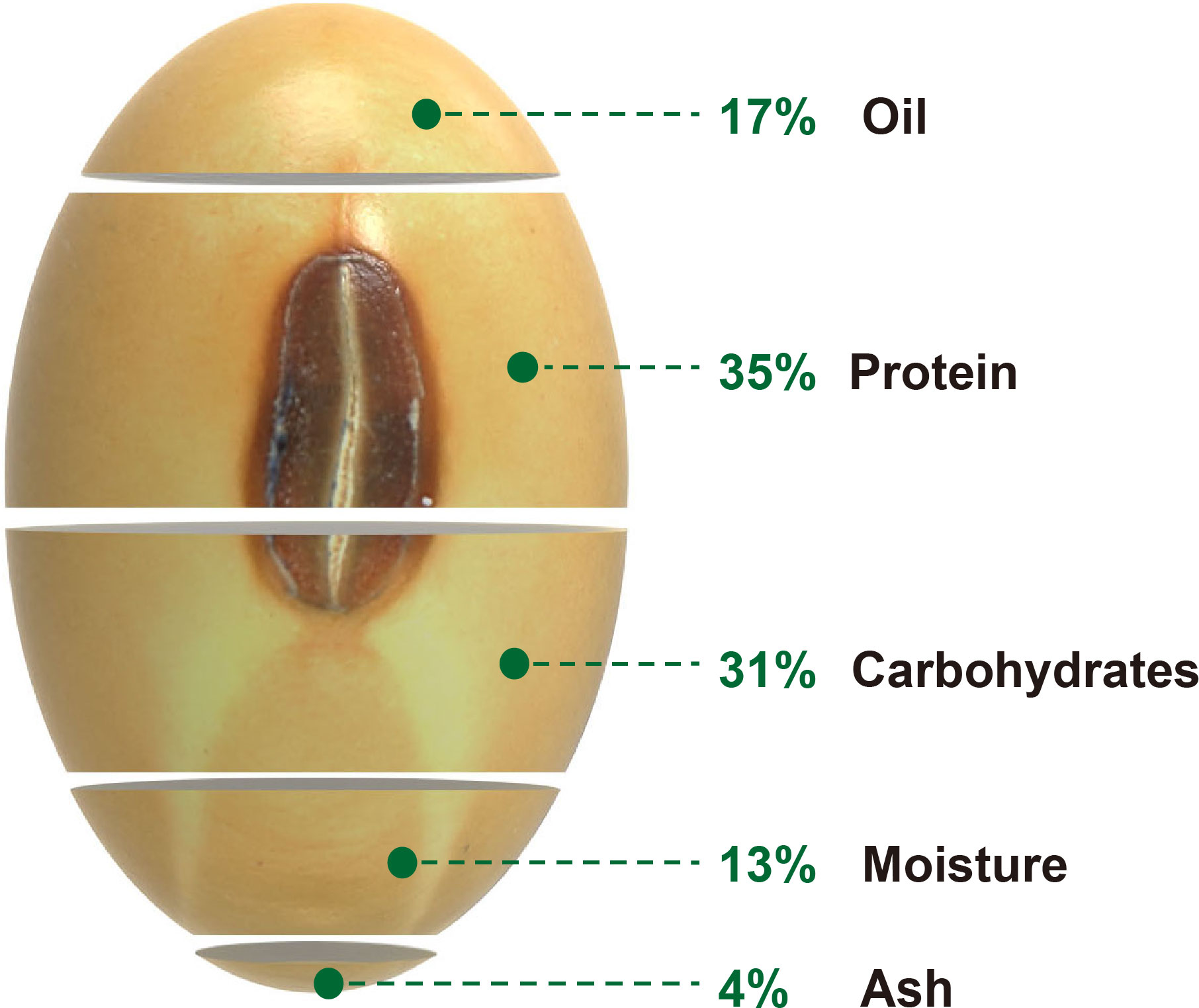
Figure 1 Composition of stored mature soybean seeds. The percentage value indicates the relative weight of the corresponding component in a seed (Liu, 1997).
Recent advances have shown that global crop yields need to be doubled by 2050 to keep up with the growing population and consumption (Godfray et al., 2010; Tilman et al., 2011), which means a 2.4% increase in crop production per year. However, soybean production seriously lags behind the projected demand, growing by an average of only 1.3% per year (Ray et al., 2013). Compared with staple crops, including rice, wheat, and maize, soybean yield is about one-third to one-half as much. Therefore, improving soybean yield is an essential and urgent task for soybean breeding. Increasing seed size is one of the crucial ways to boost soybean yield. Soybean seed size can be described using length (diameter parallel to the hilum), width (diameter from the hilum to the abaxial surface of seed), and thickness (diameter vertical to the hilum), and the composition and content of seed storage reserves directly determine it. Cultivated soybeans generally produce larger seeds with a higher oil level (Wang et al., 2020b). Wild soybeans have smaller seeds with lower oil content than cultivated soybeans. However, the seed protein content is not increased in the large-seed soybean cultivars (Wang et al., 2020b). Therefore, soybean improvement involves parallel increases in seed size, oil accumulation, and a possibly accompanying change in protein level.
For decades, increasing seed size, oil accumulation, and protein content have been the essential objectives of soybean breeding programs. The publication of the soybean reference genome (Williams 82) in 2010 has extensively promoted the development of soybean functional genomics (Zhang et al., 2022). Here, we review the advances in soybean functional genomics on seed size, oil accumulation, and protein content. In addition, we also discuss the challenges and prospects for developing high-yield soybean cultivars with high oil or protein content. As the biochemical synthesis of oils in the seed has been widely studied and well-reviewed (Bates et al., 2013; Xu and Shanklin, 2016; Song et al., 2017; Liu et al., 2022; Yang et al., 2022a), we will not repeat these comments here.
2 Genetic mapping associated with seed size, oil and protein contents
Seed size, oil and protein contents are complex traits controlled by genetic and environmental factors during seed development and maturation. Given their importance in soybean breeding, researchers have performed extensive linkage analysis to identify quantitative trait loci (QTL) associated with these three seed traits using various bi-parental derived populations, such as F2 population, recombinant inbred lines (RILs), chromosome segment substitution lines (CSSLs), and near-isogenic lines (NILs) (Han et al., 2012; Eskandari et al., 2013a; Eskandari et al., 2013b; Qi et al., 2014; Warrington et al., 2015; Wang et al., 2015a; Yang et al., 2019; Cui et al., 2020; Kumawat and Xu, 2021; Kumar et al., 2022; Luo et al., 2022; Yang et al., 2022b). So far, hundreds of QTLs related to seed size (including seed weight), oil accumulation, and protein content have been documented in the SoyBase Genome Database (http://www.soybase.org). For instance, there are 396 QTLs for seed size and weight (Figure 2; Supplementary Table 1), 333 QTLs for seed oil content (Figure 2; Supplementary Table 2), and 234 QTLs for seed protein content (Figure 2; Supplementary Table 3). Among these QTLs, some of the seed size, oil accumulation, and protein content-related QTLs shared overlapping regions, suggesting the presence of pleiotropic regulatory genes in these QTLs. However, due to the low-resolution and low-density molecular markers and limited population size, most QTLs were mapped in a large chromosome region, making these QTLs less effective in pinpointing the specific gene for crop improvement. At present, only a few genes involved in seed size, oil accumulation, and protein content have been isolated from QTL mappings, such as GmPP2C-1 (Lu et al., 2017), GmB1 (Zhang et al., 2018a), and Glyma.20G85100 (also known as GmSWEET39) (Zhang et al., 2020; Fliege et al., 2022). In addition, two genes related to seed size/weight were identified through mutant-dependent map-based cloning or comparative genome hybridization (CGH) analysis, including GmSSS1 (Zhu et al., 2022) and GmKIX8-1 (Nguyen et al., 2021).
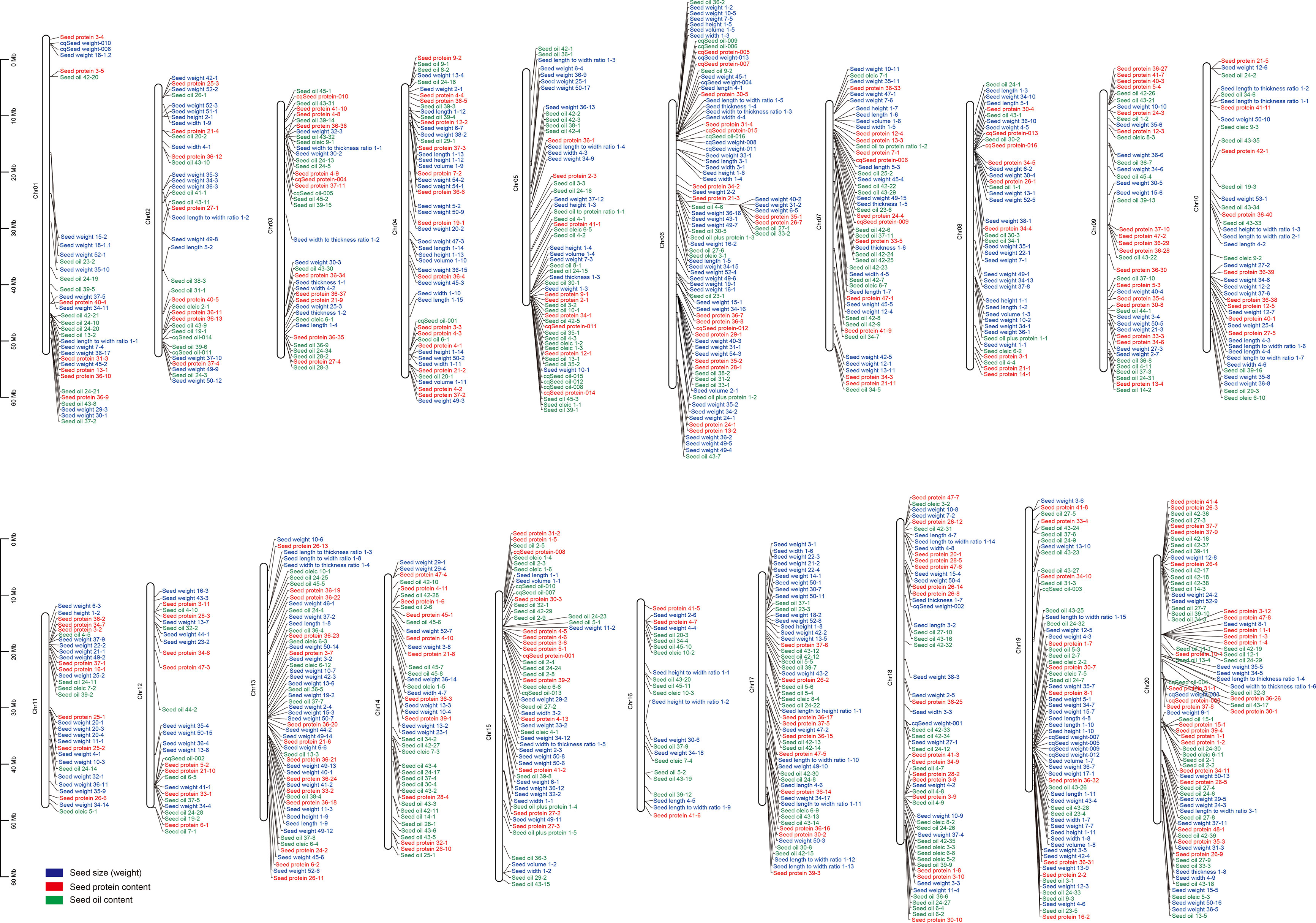
Figure 2 QTLs related to seed size (weight), oil accumulation, and protein content in soybean. These QTLs are derived from the SoyBase database (https://soybase.org/).
With the development of omics, genome-wide association study (GWAS) has become a powerful gene or QTL mapping approach for analyzing complicated agronomic traits in crops. Compared with conventional QTL mapping or linkage analysis, GWAS offers significant advantages: 1) GWAS does not need to build a mapping population. 2) GWAS population includes more natural variation than the bi-parental population. 3) GWAS can achieve higher mapping resolution due to high-density molecular markers and diverse historical recombination events (Wang et al., 2020a; Li et al., 2022b). Over the past decade, dozens of GWAS have been performed to identify QTLs or quantitative trait nucleotides (QTNs) involving seed size, lipid accumulation, and protein level in soybean (Hwang et al., 2014; Zhou et al., 2015; Zhang et al., 2016a; Yan et al., 2017; Zhang et al., 2018b; Lee et al., 2019; Zhao et al., 2019; He et al., 2021; Zhang et al., 2021; Hong et al., 2022). Based on this approach, GmOLEO1 (Zhang et al., 2019b), GmPDAT (Liu et al., 2020a), GmSWEET10a (also known as GmSWEET39) (Miao et al., 2020; Wang et al., 2020b), and GmST05 (Duan et al., 2022) have been identified and confirmed to relate to these seed traits, suggesting this way is more effective. Although GWAS has advantages in genetic mapping, the population structure and individual relationships are likely to produce false positive results in association analysis. Therefore, it is better to integrate linkage mapping and GWAS analysis for dissecting complex traits. Mixed analysis methods have successfully employed and mapped QTLs or QTNs associated with these seed traits in soybean (Cao et al., 2017; Zhang et al., 2019c), and further cloned GmSWEET39 (Zhang et al., 2020), GmGA3ox1 (Hu et al., 2022), GmST1 (Li et al., 2022a), and POWR1 (Goettel et al., 2022).
3 Regulatory genes of seed size
The seeds of higher plants consist of the embryo, endosperm, and seed coat, among which the embryo and endosperm are generated from the fertilized egg cell and central cell, respectively. In contrast, the seed coat is developed from the sporophytic integument. Therefore, seed size is determined by the integrated signals of maternal and zygotic tissues that influence the coordinated growth of the embryo, endosperm, and seed coat (Li et al., 2019). Several signaling pathways that maternal control seed size have been identified in Arabidopsis and rice, such as G-protein signaling, ubiquitin-proteasome signaling, mitogen-activated protein kinase (MAPK) signaling, phytohormone signaling, and some transcriptional regulators. Meanwhile, the HAIKU (IKU) pathway and some phytohormones partially regulate the zygotic tissues’ growth (Li et al., 2019). However, compared with Arabidopsis and rice, the molecular networks regulating seed size in soybean are still lagging behind.
As critical regulatory components of gene expression, several transcriptional factors (TFs) involved in seed size have been identified in soybean (Figure 3; Table 1). BIG SEEDS1 (BS1) belongs to a group II member of the TIFY TF family. It plays a vital role in controlling the size of seeds, pods, and leaves via a regulatory module that targets cell proliferation in the model legume of Medicago truncatula (Ge et al., 2016). Down-regulation of BS1 orthologous genes (GmBS1 and GmBS2) in soybean resulted in increased seed size and amino acid content. SLB1 encodes an F-box protein that forms part of the SKP1/Cullin/F-box E3 ubiquitin ligase complex. Biochemical and genetic analyses showed that SLB1 interacts with BS1 to control lateral branching and organ growth by regulating BS1 protein stability in Medicago truncatula. In addition, overexpression of SLB1 resulted in increased leaf and seed size in both Medicago truncatula and soybean, suggesting the functional conservation of SLB1 (Yin et al., 2020). Plant WRKY TFs are involved in many biological processes, such as embryogenesis and seed development (Luo et al., 2005). The WRKY15a was differentially expressed during pod development between cultivated and wild soybeans. Four haplotypes (H1-H4) were present in WRKY15a, which varied in the CT-core microsatellite locus at the 5’-untranslated region (5’-UTR) of WRKY15a. The H1 haplotype with six CT-repeats was the only allele in cultivated soybeans, whereas the H3 haplotype with five CT-repeats was the primary allele in wild soybeans. The seed weight with haplotype H1 was heavier than that of wild soybeans harboring haplotypes H2, H3, and H4, and the seed weight was positively correlated with WRKY15a expression, indicating a positive effect of WRKY15a on seed size (Gu et al., 2017). Dt2, encoding a MADS-box TF, plays an essential role in controlling multiple agronomic traits, such as flowering time, stem growth habit, and plant height (Ping et al., 2014; Zhang et al., 2019a). A recent report has shown that Dt2 also determines shoot branching and seed size (Liang et al., 2022). Dt2 knockout lines performed multiple yield-related trait changes, such as the increased seed length and width, heavier seed weight, and higher grain weight per plant, thereby resulting in obviously improved yield per plot. In contrast, the Dt2 overexpression lines exhibited decreased seed length and width.
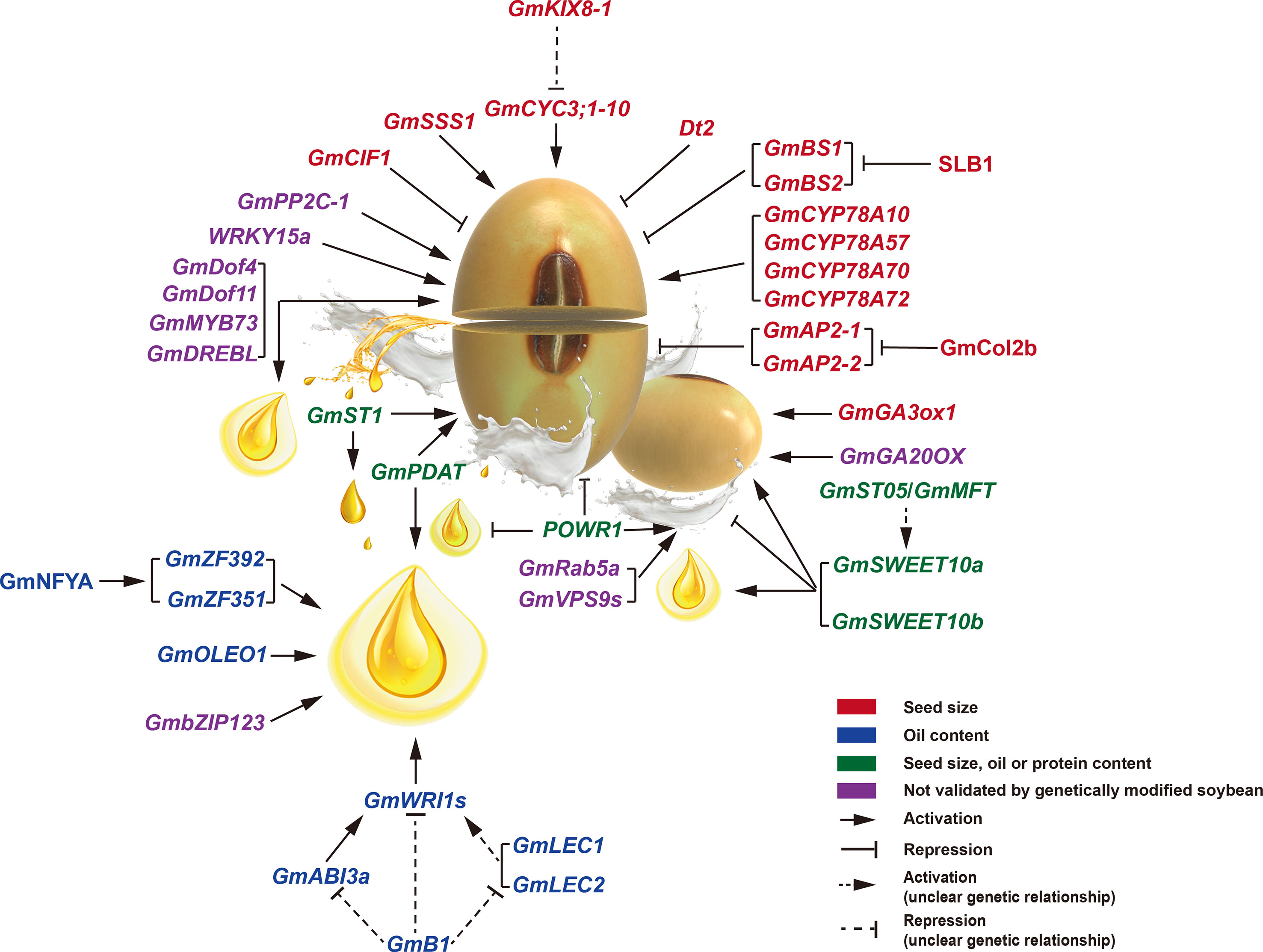
Figure 3 Genetic regulatory network of seed size (weight), oil accumulation, and protein content in soybean. The genes or proteins involving seed size (weight) and oil content are shown in red and blue fonts, respectively. The pleiotropic regulators for seed size (weight), oil accumulation, or protein content are indicated in green fonts. The regulatory genes, whose function has been validated only in Arabidopsis but not soybean, are shown in purple fonts.
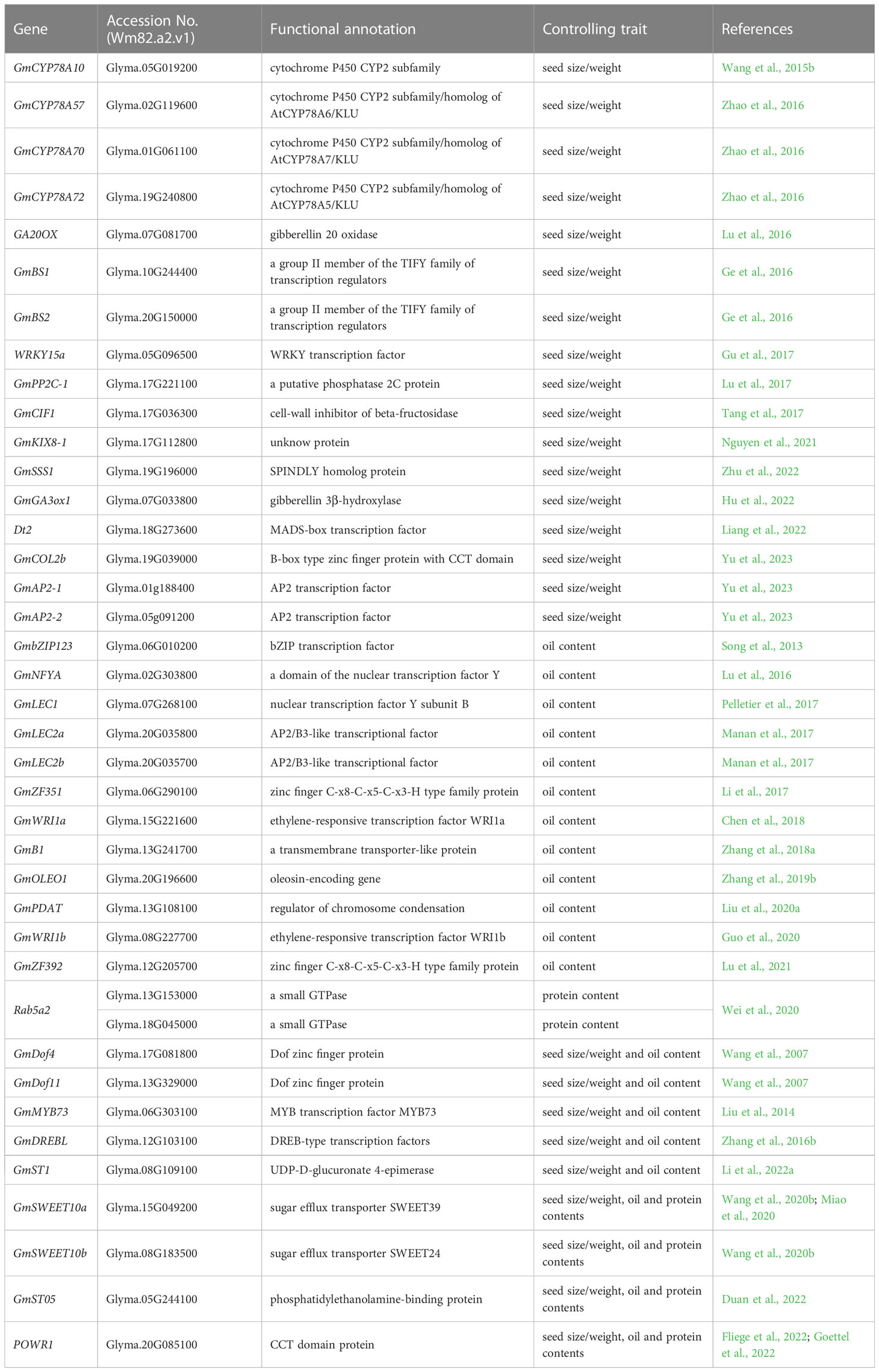
Table 1 Representative genes related to seed size, oil accumulation, and protein content in soybean.
Some genes that encode various enzymes have also been shown to affect soybean seed size (Figure 3; Table 1). A phosphatase 2C-1 (GmPP2C-1) gene from wild soybean helps to increase seed weight or size by improving integument cell size and activating a subset of seed trait-related genes (Lu et al., 2017). In addition, GmPP2C-1 facilitates the accumulation of dephosphorylated GmBZR1 protein, which act as the key transcription factor in BR signaling. Furthermore, overexpression of GmBZR1 can improve seed size or weight in transgenic Arabidopsis. Cell wall invertase (CWI) plays a vital role in sugar signaling and metabolism, affecting the source–sink interaction and seed development (Tang et al., 2017). GmCIF1 encodes a cell wall invertase inhibitor, and suppression of GmCIF1 gene expression exhibited increased CWI activities and larger seed size while with more accumulations of protein, hexoses, and starch in soybean seeds. GmSSS1 encodes a putative O-GlcNAc transferase in soybean. Knockout GmSSS1 resulted in tiny seeds, whereas overexpressing GmSSS1 produced large seeds (Zhu et al., 2022). Modulating GmSSS1 could positively affect cell division and expansion in transgenic plants. GmGA3ox1, a gibberellin (GA) 3β-hydroxylase in soybean, is the critical enzyme in the GA biosynthesis pathway. Knockout of GmGA3ox1 resulted in reduced GA biosynthesis while enhanced photosynthesis (Hu et al., 2022). GmGA3ox1 knockout plants displayed decreased seed weight and length, but improved seed production by increasing branch, pod, and seed numbers. In contrast, overexpression of GmGA3ox1 increased seed weight and length in transgenic soybeans. Similarly, overexpression of GA20OX, encoding a gibberellin 20 oxidase in a rate-limiting step of GA biosynthesis, enhanced the seed size/weight of transgenic Arabidopsis plants (Lu et al., 2016).
Besides the above genes, some homologous soybean genes known to regulate seed size in Arabidopsis have also been shown to control soybean seed size (Figure 3; Table 1). For example, several P450/CYP78A family members are suggested for controlling seed size in Arabidopsis (Wang et al., 2008; Fang et al., 2012). The P450/CYP78A orthologs in soybean, such as GmCYP78A10, GmCYP78A57, GmCYP78A70, and GmCYP78A72, exhibited conserved function to improve seed size or weight (Wang et al., 2015b; Zhao et al., 2016; Du et al., 2017), but the underlying mechanism how they function remains largely elusive. A PPD/KIX/TPL repressor complex consisting of PPD2, KIX8/9, and TPL proteins was shown to affect organ size by modulating meristem proliferation in Arabidopsis (Baekelandt et al., 2018). GmKIX8-1, a soybean AtKIX8 ortholog, is also involved in controlling cell proliferation and organ size. Due to increased CYCLIN D3;1-10 expression and cell proliferation, the GmKIX8-1 loss-of-function mutants displayed an apparent increase in the size of leaves and seeds (Nguyen et al., 2021). Very recently, in both Arabidopsis and soybean, a crucial regulatory cascade involving CO (the central regulator of the photoperiodic pathway) and AP2 (specification of floral meristem identity) was demonstrated to mediate the photoperiod-regulated seed size in a maternal-dependent manner (Yu et al., 2023). GmCOL2b (a soybean CO homolog) positively promoted seed size under short days by directly inhibiting the expression of GmAP2-1 and GmAP2-2.
4 Regulatory genes of seed oil
Seed storage reserves, including oil, protein, and starch, are filled during seed development and maturation. Understanding the storage substance loading into the seeds thus is crucial to improving crop yield and nutritional quality. In the past decades, extensive efforts have been made toward the dissection of molecular pathways for accumulating seed storage reserves, particularly in Arabidopsis. TFs, such as LEC1, LEC2, ABI3, FUS3, and WRI1, and other activators or repressors for storage reserves accumulation during seed development, have been identified in plants (Yang et al., 2022a). However, more details and mechanisms have yet to be clarified, especially for essential crops such as soybean (Figure 3; Table 1).
LEC1 is an atypical TF subunit (NF-YB) that interacts with NF-YA and NF-YC subunits to form an NF-Y TF complex. It is central to controlling seed development, such as embryo morphogenesis, endosperm development, and storage reserve accumulation (Jo et al., 2019). In Arabidopsis, the lec1 null mutants displayed striking defects in embryos and severely restricted protein and lipid accumulation in seeds (Meinke et al., 1994; West et al., 1994). Furthermore, over-expression of LEC1 induced the activation of genes related to the accumulation of storage proteins and lipids, resulting in increased contents of lipids and FAs in the transgenic Arabidopsis (Kagaya et al., 2005). In soybean, GmLEC1 (GmLEC1a or GmLEC1b) transcriptionally regulates the genes involved in distinct cellular processes during seed development and activates seed FAs biosynthesis (Pelletier et al., 2017; Zhang et al., 2017). Further research revealed that GmLEC1 acts in combination with TFs such as GmAREB3, GmbZIP67, and GmABI3 to regulate soybean seed development (Jo et al., 2020).
LEC1 interacts physically with LEC2, a B3 DNA binding domain TF, which has a crucial regulatory role in seed development and in controlling seed protein and oil levels in Arabidopsis (Santos-Mendoza et al., 2008; Angeles-Núñez and Tiessen, 2011; Kim et al., 2015; Jo et al., 2019). The loss-of-function lec2 mutant seeds showed a 30% and 15% decline in oil and protein, respectively, but accumulated more starch and sucrose than wild-type seeds (Angeles-Núñez and Tiessen, 2011). In contrast, in both transgenic Arabidopsis and tobacco plants, AtLEC2 inducible expression increased storage oil accumulation, such as TAGs and FAs (Mendoza et al., 2005; Andrianov et al., 2010; Kim et al., 2015). In soybean, GmLEC2 regulates a subset of genes involving the metabolism of seed storage reserves (Manan et al., 2017). Compared with the control seeds, the TAGs and long-chain FAs contents of GmLEC2a over-expression transgenic Arabidopsis seeds increased by 34% and 4%, respectively.
In the transcriptional network of seed oil accumulation in Arabidopsis, LEC1 and LEC2 synergistically promote WRI1 expression, an AP2 TF gene responsible for the transcriptional regulation of oil biosynthesis, and this regulatory mechanism is conserved in other plant species, for instance, soybean and maize (Baud et al., 2007; Mu et al., 2008; Shen et al., 2010; Manan et al., 2017; Pelletier et al., 2017; Yang et al., 2022a). Its two soybean orthologs, GmWRI1a and GmWRI1b, play a central role in seed oil accumulation. Over-expression of GmWRI1a or GmWRI1b significantly increased total oil and FAs contents and changed FAs composition in the seed, whereas GmWRI1 knockdown hairy roots interfered with lipid biosynthesis (Chen et al., 2018; Chen et al., 2020; Guo et al., 2020; Wang et al., 2022).
GmZF392, a seed-specific tandem CCCH zinc finger (TZF) protein, promotes seed oil accumulation by targeting a bipartite cis-element with TA- and TG-rich sequences in the promoter regions, thereby activating downstream gene expression involving in the lipid biosynthesis (Lu et al., 2021). GmZF392 interacts physically with GmZF351, another activator of lipid accumulation, to additive/synergistic increase the expression of downstream lipid biosynthesis genes (Li et al., 2017; Lu et al., 2021). And both GmZF392 and GmZF351 are positively regulated by GmNFYA, a TF correlated with oil content (Lu et al., 2016; Lu et al., 2021). In addition, GmZF392 and GmZF351 are also direct targets of GmLEC1 (Pelletier et al., 2017). More importantly, GmZF392 and GmZF351 were selected by domestication from wild soybeans to cultivated soybeans.
In addition to the above TFs forming the regulatory module, some functional genes were also involved in regulating seed oil content in soybean (Figure 3; Table 1). Overexpression of a bZIP TF gene (GmbZIP123) enhances lipid accumulation in transgenic Arabidopsis seeds through modulating sugar transport (Song et al., 2013). GmB1, encoding a transporter-like transmembrane protein for the biosynthesis of the bloom in pod endocarp, not only controls seed coat bloom in wild soybeans but also affects oil content in cultivated soybeans (Zhang et al., 2018a). GmOLEO1, a strong artificial-selected oleosin protein-encoding gene, conduces to the improvement in seed oil content during soybean domestication by affecting TAGs metabolism (Zhang et al., 2019b).
5 Regulatory genes of seed protein
Compared with seed size and oil content, only a few genes controlling seed protein or amino acid content have been functionally identified (Figure 3; Table 1) (Krishnan and Jez, 2018). The small GTPase GmRab5a and its guanine exchange factors GmVPS9s are shown to function in the storage protein post-Golgi trafficking in soybean (Wei et al., 2020). Transient over-expression of the dominant negative variant of GmRab5a, or RNAi of either GmRab5a or GmVPS9s, obviously reduced the transport of the cargo marker, which used to reflect storage protein trafficking to protein storage vacuoles in soybean cotyledon cells. In addition, several genes, including POWR1, GmSWEET10a, GmSWEET10b, and GmST05, pleiotropically regulate seed protein, oil content, and seed size (Wang et al., 2020b; Duan et al., 2022; Goettel et al., 2022), which are detailed discussion in the next section.
6 Pleiotropic regulatory genes of seed size, oil and protein contents
Seed size, oil accumulation, and protein content in soybean are highly correlated agronomical traits. However, the selection and underlying molecular basis of these seed-correlated traits during soybean domestication are poorly understood, which is one of the obstacles to soybean yield and quality improvement. So far, several pleiotropic regulatory genes controlling seed size, oil accumulation, and protein content have been cloned and functionally identified in soybean (Figure 3; Table 1).
For instance, the ectopic expression of GmDof4, GmDof11, GmMYB73, and GmDREBL enhanced both seed size/weight and oil accumulation in transgenic Arabidopsis seeds (Wang et al., 2007; Liu et al., 2014; Zhang et al., 2016b). GmPDAT, a phospholipid diacylglycerol acyltransferase encoding gene, was expressed higher in large-seed and high-oil soybean accessions than in small-seed and low-oil accessions. Over-expression of GmPDAT improved seed size and oil level, whereas GmPDAT RNAi plants had reduced seed size and oil accumulation (Liu et al., 2020a). GmST1 encodes a UDP-D-glucuronate 4-epimerase that positively regulates seed size and oil content by modulating pectin biosynthesis and glycolysis pathways, and underwent selection during soybean domestication (Li et al., 2022a).
The sugar transporter SWEET family members play critical roles in seed development (Chen et al., 2015; Wang et al., 2019). A pair of SWEET paralogs in soybean, GmSWEET10a and GmSWEET10b, underwent the stepwise selection that synchronously changed seed size, oil accumulation, and protein level during soybean domestication, by regulating sugar sorting from seed coat to embryo (Zhang et al., 2020; Wang et al., 2020b). Compared with wild-type plants, GmSWEET10a or GmSWEET10b over-expression soybeans displayed significantly increased seed size and higher oil accumulation but decreased protein level, while their knockout plants had reduced seed size and oil content but increased protein level (Wang et al., 2020b). Very recently, a phosphatidylethanolamine-binding protein (PEBP) family member, GmST05 (also known as GmMFT), has been shown to positively regulate seed size and altered oil and protein levels, likely by affecting GmSWEET10a transcription (Li et al., 2014; Duan et al., 2022). In addition, a CCT-domain gene, POWR1, is domesticated and pleiotropically regulates seed quality and yield in soybean, possibly by regulating lipid metabolism and nutrient transport (Goettel et al., 2022). A transposable element (TE) insertion in the CCT-domain of POWR1 resulted in increased seed weight and oil content but decreased protein content. In contrast, over-expression of POWR1 exhibited improved protein content and declined seed weight and oil accumulation in transgenic plants.
7 Challenges and perspectives
Seed size, oil and protein contents are complex quantitative traits governed by multiple genes. Although linkage mapping and GWAS analysis have identified numerous QTLs controlling seed size, oil accumulation, and protein content in soybean, only a few genes have been isolated and functionally validated. One fundamental reason for this phenomenon is that these researchers usually use only one or two approaches, making it hard to pinpoint the target underlying these seed traits. The other key obstacle is the lack of a fast and efficient soybean genetic transformation system for different soybean genotypes, such as Agrobacterium-mediated cotyledonary node soybean transformation, which has been widely used in recently years. The slow and inefficient genetic transformation system makes it more challenging to identify and verify the function of soybean genes (Zhang et al., 2022). That’s why, in some studies, especially those prior to 2015, functional validation was done in Arabidopsis instead of soybean.
With the rapid progress of omics research and the reduction of testing cost, more and more soybean omics data were produced, such as the re-sequencing genome, transcriptome, metabolome, proteome, epigenome, pan-genome, and 3D genome (Ohyanagi et al., 2012; Lin et al., 2014; Shen et al., 2014; Zhou et al., 2015; Liu et al., 2016; Fang et al., 2017; Shen et al., 2018a; Shen et al., 2018b; Liu et al., 2020b; Silva et al., 2021; Ni et al., 2023). These released omics resources will extensively promote the research of soybean functional genomics. Currently, like GWAS, TWAS (transcriptome-wide association study), EWAS (epigenome-wide association study), and PWAS (proteome-wide association study), as well as multi-omics data association studies, such as eGWAS (gene expression-based genome-wide association study) and mGWAS (metabolome-based genome-wide association study) have been successfully developed and applied (Shen et al., 2022). Integration of multiple omics approaches will provide more clues and help narrow the target range underlying these seed traits. However, utilizing these vast omics data that exist in various forms is a considerable challenge. Thus, mathematical methods, like meta-analysis, are expected to address such trouble. Moreover, artificial intelligence (AI) technology or machine learning approach can make mining big data more efficient, for instance, omics data processing, protein structure construction, and pan-omics data integration (Baek et al., 2021; Jumper et al., 2021; Reel et al., 2021).
CRISPR/Cas-based genome editing technology that enables precise modification of genomes to obtain predictable and desired traits has been successfully applied to gene function research and crop germplasm creation. Compared with other crops, such as rice, the soybean genome-editing process is primarily in its infancy; however, successful stories have demonstrated the feasibility of gene editing in soybean (Cai et al., 2018; Bai et al., 2020; Wang et al., 2020b; Nguyen et al., 2021; Bai et al., 2022; Duan et al., 2022; Hu et al., 2022; Liang et al., 2022; Li et al., 2022a). In the future, the improved soybean transformation and more applications of single - or multi-gene ‘base editing’ will greatly facilitate functional research in soybean, ultimately allowing us to decode these complex seed traits and identify critical genes underlying seed size, oil and protein contents.
The ultimate goal of soybean breeding is to cultivate high-yield and high-quality soybean. So far, crop breeding has developed from artificial selection (stage 1.0) and hybrid breeding (stage 2.0) to molecular breeding (stage 3.0). However, to solve the crisis of food shortage caused by the growing population, intelligent breeding (stage 4.0) that can quickly aggregate excellent alleles through precise design is coming (Shen et al., 2022). In previous breeding stages, breeders usually have to stack desirable traits into a single line to create a super variety, which is a huge task. In breeding stage 4.0, optimal and precise design to rapidly pyramid multiple elite alleles with desirable seed traits will facilitate yield, oil, and protein content improvement in soybean.
Author contributions
QL and MZ designed and supervised the study. ZD, QL, and HW drafted the manuscript. XH participated in the production of the article pictures. ZD and QL responded to review comments. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Hainan Yazhou Bay Seed Laboratory Project (B21HJ0002), the National Natural Science Foundation of China (32101755, 32272107), and the Zhejiang Provincial Natural Science Foundation (LY22C130005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1160418/full#supplementary-material
References
Andrianov, V., Borisjuk, N., Pogrebnyak, N., Brinker, A., Dixon, J., Spitsin, S., et al. (2010). Tobacco as a production platform for biofuel: Overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8 (3), 277–287. doi: 10.1111/j.1467-7652.2009.00458.x
Angeles-Núñez, J. G., Tiessen, A. (2011). Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J. Plant Physiol. 168 (16), 1891–1900. doi: 10.1016/j.jplph.2011.05.003
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373 (6557), 871–876. doi: 10.1126/science.abj8754
Baekelandt, A., Pauwels, L., Wang, Z., Li, N., De Milde, L., Natran, A., et al. (2018). Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol. 178 (1), 217–232. doi: 10.1104/pp.18.00327
Bai, M., Yuan, J., Kuang, H., Gong, P., Li, S., Zhang, Z., et al. (2020). Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soybean. Plant Biotechnol. J. 18 (3), 721–731. doi: 10.1111/pbi.13239
Bai, M., Yuan, C., Kuang, H., Sun, Q., Hu, X., Cui, L., et al. (2022). Combination of two multiplex genome-edited soybean varieties enables customization of protein functional properties. Mol. Plant 15 (7), 1081–1083. doi: 10.1016/j.molp.2022.05.011
Bates, P. D., Stymne, S., Ohlrogge, J. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16 (3), 358–364. doi: 10.1016/j.pbi.2013.02.015
Baud, S., Mendoza, M. S., To, A., Harscoët, E., Lepiniec, L., Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON 2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50 (5), 825–838. doi: 10.1111/j.1365-313X.2007.03092.x
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16 (1), 176–185. doi: 10.1111/pbi.12758
Cao, Y., Li, S., Wang, Z., Chang, F., Kong, J., Gai, J., et al. (2017). Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01222
Carter, T. E., Jr., Nelson, R. L., Sneller, C. H., Cui, Z. (2004). Genetic diversity in soybean. Soybeans: Improvement production uses 16, 303–416. doi: 10.2134/agronmonogr16.3ed.c8
Chen, L., Lin, I., Qu, X., Sosso, D., McFarlane, H. E., Londoño, A., et al. (2015). A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27 (3), 607–619. doi: 10.1105/tpc.114.134585
Chen, B., Zhang, G., Li, P., Yang, J., Guo, L., Benning, C., et al. (2020). Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol. J. 18 (1), 155–171. doi: 10.1111/pbi.13183
Chen, L., Zheng, Y., Dong, Z., Meng, F., Sun, X., Fan, X., et al. (2018). Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation. Mol. Genet. Genomics 293 (2), 401–415. doi: 10.1007/s00438-017-1393-2
Cui, B., Chen, L., Yang, Y., Liao, H. (2020). Genetic analysis and map-based delimitation of a major locus qSS3 for seed size in soybean. Plant Breed. 139 (6), 1145–1157. doi: 10.1111/pbr.12853
Du, J., Wang, S., He, C., Zhou, B., Ruan, Y. L., Shou, H. (2017). Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 68 (8), 1955–1972. doi: 10.1093/jxb/erw460
Duan, Z., Zhang, M., Zhang, Z., Liang, S., Fan, L., Yang, X., et al. (2022). Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol. J. 20 (9), 1807–1818. doi: 10.1111/pbi.13865
Eskandari, M., Cober, E. R., Rajcan, I. (2013a). Genetic control of soybean seed oil: I. QTL and genes associated with seed oil concentration in RIL populations derived from crossing moderately high-oil parents. Theor. Appl. Genet. 126 (2), 483–495. doi: 10.1007/s00122-012-1995-3
Eskandari, M., Cober, E. R., Rajcan, I. (2013b). Genetic control of soybean seed oil: II. QTL and genes that increase oil concentration without decreasing protein or with increased seed yield. Theor. Appl. Genet. 126 (6), 1677–1687. doi: 10.1007/s00122-013-2083-z
Fang, C., Ma, Y., Wu, S., Liu, Z., Wang, Z., Yang, R., et al. (2017). Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 18 (1), 161. doi: 10.1186/s13059-017-1289-9
Fang, W., Wang, Z., Cui, R., Li, J., Li, Y. (2012). Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 70 (6), 929–939. doi: 10.1111/j.1365-313X.2012.04907.x
Fliege, C. E., Ward, R. A., Vogel, P., Nguyen, H., Quach, T., Guo, M., et al. (2022). Fine mapping and cloning of the major seed protein quantitative trait loci on soybean chromosome 20. Plant J. 110 (1), 114–128. doi: 10.1111/tpj.15658
Ge, L., Yu, J., Wang, H., Luth, D., Bai, G., Wang, K., et al. (2016). Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. U. S. A. 113 (44), 12414–12419. doi: 10.1073/pnas.1611763113
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327 (5967), 812–818. doi: 10.1126/science.1185383
Goettel, W., Zhang, H., Li, Y., Qiao, Z., Jiang, H., Hou, D., et al. (2022). POWR1 is a domestication gene pleiotropically regulating seed quality and yield in soybean. Nat. Commun. 13 (1), 3051. doi: 10.1038/s41467-022-30314-7
Gu, Y., Li, W., Jiang, H., Wang, Y., Gao, H., Liu, M., et al. (2017). Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. J. Exp. Bot. 68 (11), 2717–2729. doi: 10.1093/jxb/erx147
Guo, W., Chen, L., Chen, H., Yang, H., You, Q., Bao, A., et al. (2020). Overexpression of GmWRI1b in soybean stably improves plant architecture and associated yield parameters, and increases total seed oil production under field conditions. Plant Biotechnol. J. 18 (8), 1639–1641. doi: 10.1111/pbi.13324
Han, Y., Li, D., Zhu, D., Li, H., Li, X., Teng, W., et al. (2012). QTL analysis of soybean seed weight across multi-genetic backgrounds and environments. Theor. Appl. Genet. 125 (4), 671–683. doi: 10.1007/s00122-012-1859-x
He, Q., Xiang, S., Yang, H., Wang, W., Shu, Y., Li, Z., et al. (2021). A genome-wide association study of seed size, protein content, and oil content using a natural population of sichuan and chongqing soybean. Euphytica 217(11), 198. doi: 10.1007/s10681-021-02931-8
Hong, H., Najafabadi, M. Y., Torkamaneh, D., Rajcan, I. (2022). Identification of quantitative trait loci associated with seed quality traits between Canadian and Ukrainian mega-environments using genome-wide association study. Theor. Appl. Genet. 135 (7), 2515–2530. doi: 10.1007/s00122-022-04134-8
Hu, D., Li, X., Yang, Z., Liu, S., Hao, D., Chao, M., et al. (2022). Downregulation of a gibberellin 3β-hydroxylase enhances photosynthesis and increases seed yield in soybean. New Phytol. 235 (2), 502–517. doi: 10.1111/nph.18153
Hwang, E. Y., Song, Q., Jia, G., Specht, J. E., Hyten, D. L., Costa, J., et al. (2014). A genome-wide association study of seed protein and oil content in soybean. BMC Genomics 15, 1. doi: 10.1186/1471-2164-15-1
Jo, L., Pelletier, J. M., Harada, J. J. (2019). Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 61 (5), 564–580. doi: 10.1111/jipb.12806
Jo, L., Pelletier, J. M., Hsu, S. W., Baden, R., Goldberg, R. B., Harada, J. J. (2020). Combinatorial interactions of the LEC1 transcription factor specify diverse developmental programs during soybean seed development. Proc. Natl. Acad. Sci. U. S. A. 117 (2), 1223–1232. doi: 10.1073/pnas.1918441117
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi: 10.1038/s41586-021-03819-2
Kagaya, Y., Toyoshima, R., Okuda, R., Usui, H., Yamamoto, A., Hattori, T. (2005). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46 (3), 399–406. doi: 10.1093/pcp/pci048
Kim, H. U., Lee, K. R., Jung, S. J., Shin, H. A., Go, Y. S., Suh, M. C., et al. (2015). Senescence-inducible LEC2 enhances triacylglycerol accumulation in leaves without negatively affecting plant growth. Plant Biotechnol. J. 13 (9), 1346–1359. doi: 10.1111/pbi.12354
Krishnan, H. B., Jez, J. M. (2018). Review: The promise and limits for enhancing sulfur-containing amino acid content of soybean seed. Plant Sci. 272, 14–21. doi: 10.1016/j.plantsci.2018.03.030
Kumar, R., Saini, M., Taku, M., Debbarma, P., Mahto, R. K., Ramlal, A., et al. (2022). Identification of quantitative trait loci (QTLs) and candidate genes for seed shape and 100-seed weight in soybean [Glycine max (L.) merr.]. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1074245
Kumawat, G., Xu, D. (2021). A major and stable quantitative trait locus qSS2 for seed size and shape traits in a soybean RIL population. Front. Genet. 12. doi: 10.3389/fgene.2021.646102
Lee, S., Van, K., Sung, M., Nelson, R., LaMantia, J., McHale, L. K., et al. (2019). Genome-wide association study of seed protein, oil and amino acid contents in soybean from maturity groups I to IV. Theor. Appl. Genet. 132(6), 1639–1659. doi: 10.1007/s00122-019-03304-5
Li, Q., Fan, C., Zhang, X., Wang, X., Wu, F., Hu, R., et al. (2014). Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PloS One 9 (6), e99642. doi: 10.1371/journal.pone.0099642
Li, Q., Lu, X., Song, Q., Chen, H., Wei, W., Tao, J., et al. (2017). Selection for a zinc-finger protein contributes to seed oil increase during soybean domestication. Plant Physiol. 173 (4), 2208–2224. doi: 10.1104/pp.16.01610
Li, Q., Lu, X., Wang, C., Shen, L., Dai, L., He, J., et al. (2022b). Genome-wide association study and transcriptome analysis reveal new QTL and candidate genes for nitrogen-deficiency tolerance in rice. Crop J. 10 (4), 942–951. doi: 10.1016/j.cj.2021.12.006
Li, N., Xu, R., Li, Y. (2019). Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. doi: 10.1146/annurev-arplant-050718-095851
Li, J., Zhang, Y., Ma, R., Huang, W., Hou, J., Fang, C., et al. (2022a). Identification of ST1 reveals a selection involving hitchhiking of seed morphology and oil content during soybean domestication. Plant Biotechnol. J. 20 (6), 1110–1121. doi: 10.1111/pbi.13791
Liang, Q., Chen, L., Yang, X., Yang, H., Liu, S., Kou, K., et al. (2022). Natural variation of Dt2 determines branching in soybean. Nat. Commun. 13 (1), 6429. doi: 10.1038/s41467-022-34153-4
Lin, H., Rao, J., Shi, J., Hu, C., Cheng, F., Wilson, Z. A., et al. (2014). Seed metabolomic study reveals significant metabolite variations and correlations among different soybean cultivars. J. Integr. Plant Biol. 56 (9), 826–836. doi: 10.1111/jipb.12228
Liu, K. (1997). “Chemistry and nutritional value of soybean components,” in Soybean: Chemistry, technology and utilization. (Boston, MA: Springer), 25–113. doi: 10.1007/978-1-4615-1763-4_2
Liu, A., Cheng, S., Yung, W., Li, M., Lam, H. (2022). Genetic regulations of the oil and protein contents in soybean seeds and strategies for improvement. Adv. Bot. Res. 102, 259–293. doi: 10.1016/bs.abr.2022.03.002
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020b). Pan-genome of wild and cultivated soybeans. Cell 182 (1), 162–176.e113. doi: 10.1016/j.cell.2020.05.023
Liu, T., Fang, C., Ma, Y., Shen, Y., Li, C., Li, Q., et al. (2016). Global investigation of the co-evolution of MIRNA genes and micro RNA targets during soybean domestication. Plant J. 85 (3), 396–409. doi: 10.1111/tpj.13113
Liu, Y., Li, Q., Lu, X., Song, Q., Lam, S. M., Zhang, W., et al. (2014). Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol. 14(1), 73. doi: 10.1186/1471-2229-14-73
Liu, J., Zhang, Y., Han, X., Zuo, J., Zhang, Z., Shang, H., et al. (2020a). An evolutionary population structure model reveals pleiotropic effects of GmPDAT for traits related to seed size and oil content in soybean. J. Exp. Bot. 71 (22), 6988–7002. doi: 10.1093/jxb/eraa426
Lu, X., Li, Q., Xiong, Q., Li, W., Bi, Y., Lai, Y., et al. (2016). The transcriptomic signature of developing soybean seeds reveals the genetic basis of seed trait adaptation during domestication. Plant J. 86 (6), 530–544. doi: 10.1111/tpj.13181
Lu, L., Wei, W., Li, Q. T., Bian, X. H., Lu, X., Hu, Y., et al. (2021). A transcriptional regulatory module controls lipid accumulation in soybean. New Phytol. 231 (2), 661–678. doi: 10.1111/nph.17401
Lu, X., Xiong, Q., Cheng, T., Li, Q. T., Liu, X. L., Bi, Y. D., et al. (2017). A PP2C-1 allele underlying a quantitative trait locus enhances soybean 100-seed weight. Mol. Plant 10 (5), 670–684. doi: 10.1016/j.molp.2017.03.006
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 102 (48), 17531–17536. doi: 10.1073/pnas.0508418102
Luo, S., Jia, J., Liu, R., Wei, R., Guo, Z., Cai, Z., et al. (2022). Identification of major QTLs for soybean seed size and seed weight traits using a RIL population in different environments. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1094112
Manan, S., Ahmad, M. Z., Zhang, G., Chen, B., Haq, B. U., Yang, J., et al. (2017). Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01604
Meinke, D. W., Franzmann, L. H., Nickle, T. C., Yeung, E. C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 (8), 1049–1064. doi: 10.1105/tpc.6.8.1049
Mendoza, M. S., Dubreucq, B., Miquel, M., Caboche, M., Lepiniec, L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579 (21), 4666–4670. doi: 10.1016/j.febslet.2005.07.037
Miao, L., Yang, S., Zhang, K., He, J., Wu, C., Ren, Y., et al. (2020). Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol. 225 (4), 1651–1666. doi: 10.1111/nph.16250
Mu, J., Tan, H., Zheng, Q., Fu, F., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148 (2), 1042–1054. doi: 10.1104/pp.108.126342
Nguyen, C. X., Paddock, K. J., Zhang, Z., Stacey, M. G. (2021). GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 229 (2), 920–934. doi: 10.1111/nph.16928
Ni, L., Liu, Y., Ma, X., Liu, T., Yang, X., Wang, Z., et al. (2023). Pan-3D genome analysis reveals structural and functional differentiation of soybean genomes. Genome Biol. 24 (1), 12. doi: 10.1186/s13059-023-02854-8
Ohyanagi, H., Sakata, K., Komatsu, S. (2012). Soybean proteome database 2012: update on the comprehensive data repository for soybean proteomics. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00110
Pelletier, J. M., Kwong, R. W., Park, S., Le, B. H., Baden, R., Cagliari, A., et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. U. S. A. 114 (32), E6710–e6719. doi: 10.1073/pnas.1707957114
Ping, J., Liu, Y., Sun, L., Zhao, M., Li, Y., She, M., et al. (2014). Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26 (7), 2831–2842. doi: 10.1105/tpc.114.126938
Qi, Z., Hou, M., Han, X., Lu, C., Jiang, H., Xin, D., et al. (2014). Identification of quantitative trait loci (QTLs) for seed protein concentration in soybean and analysis for additive effects and epistatic effects of QTLs under multiple environments. Plant Breed. 133 (4), 499–507. doi: 10.1111/pbr.12179
Ray, D. K., Mueller, N. D., West, P. C., Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PloS One 8 (6), e66428. doi: 10.1371/journal.pone.0066428
Reel, P. S., Reel, S., Pearson, E., Trucco, E., Jefferson, E. (2021). Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 49, 107739. doi: 10.1016/j.biotechadv.2021.107739
Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., Lepiniec, L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54 (4), 608–620. doi: 10.1111/j.1365-313X.2008.03461.x
Shen, B., Allen, W. B., Zheng, P., Li, C., Glassman, K., Ranch, J., et al. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153 (3), 980–987. doi: 10.1104/pp.110.157537
Shen, Y., Liu, J., Geng, H., Zhang, J., Liu, Y., Zhang, H., et al. (2018a). De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 61(8), 871–884. doi: 10.1007/s11427-018-9360-0
Shen, Y., Zhang, J., Liu, Y., Liu, S., Liu, Z., Duan, Z., et al. (2018b). DNA Methylation footprints during soybean domestication and improvement. Genome Biol. 19 (1), 128. doi: 10.1186/s13059-018-1516-z
Shen, Y., Zhou, G., Liang, C., Tian, Z. (2022). Omics-based interdisciplinarity is accelerating plant breeding. Curr. Opin. Plant Biol. 66, 102167. doi: 10.1016/j.pbi.2021.102167
Shen, Y., Zhou, Z., Wang, Z., Li, W., Fang, C., Wu, M., et al. (2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26 (3), 996–1008. doi: 10.1105/tpc.114.122739
Silva, E., Belinato, J. R., Porto, C., Nunes, E., Guimaraes, F., Meyer, M. C., et al. (2021). Soybean metabolomics based in mass spectrometry: decoding the plant's signaling and defense responses under biotic stress. J. Agric. Food Chem. 69 (26), 7257–7267. doi: 10.1021/acs.jafc.0c07758
Song, Q., Li, Q., Liu, Y., Zhang, F., Ma, B., Zhang, W., et al. (2013). Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J. Exp. Bot. 64 (14), 4329–4341. doi: 10.1093/jxb/ert238
Song, Y., Wang, X., Rose, R. J. (2017). Oil body biogenesis and biotechnology in legume seeds. Plant Cell Rep. 36 (10), 1519–1532. doi: 10.1007/s00299-017-2201-5
Tang, X., Su, T., Han, M., Wei, L., Wang, W., Yu, Z., et al. (2017). Suppression of extracellular invertase inhibitor gene expression improves seed weight in soybean (Glycine max). J. Exp. Bot. 68 (3), 469–482. doi: 10.1093/jxb/erw425
Tilman, D., Balzer, C., Hill, J., Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U. S. A. 108 (50), 20260–20264. doi: 10.1073/pnas.1116437108
Wang, J., Chen, P., Wang, D., Shannon, G., Zeng, A., Orazaly, M., et al. (2015a). Identification and mapping of stable QTL for protein content in soybean seeds. Mol. Breed. 35 (3), 92. doi: 10.1007/s11032-015-0285-6
Wang, X., Li, Y., Zhang, H., Sun, G., Zhang, W., Qiu, L. (2015b). Evolution and association analysis of GmCYP78A10 gene with seed size/weight and pod number in soybean. Mol. Biol. Rep. 42 (2), 489–496. doi: 10.1007/s11033-014-3792-3
Wang, S., Liu, S., Wang, J., Yokosho, K., Zhou, B., Yu, Y., et al. (2020b). Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 7 (11), 1776–1786. doi: 10.1093/nsr/nwaa110
Wang, J., Schwab, R., Czech, B., Mica, E., Weigel, D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20 (5), 1231–1243. doi: 10.1105/tpc.108.058180
Wang, Q., Tang, J., Han, B., Huang, X. (2020a). Advances in genome-wide association studies of complex traits in rice. Theor. Appl. Genet. 133(5), 1415–1425. doi: 10.1007/s00122-019-03473-3
Wang, Z., Wang, Y., Shang, P., Yang, C., Yang, M., Huang, J., et al. (2022). Overexpression of soybean GmWRI1a stably increases the seed oil content in soybean. Int. J. Mol. Sci. 23 (9), 5084. doi: 10.3390/ijms23095084
Wang, S., Yokosho, K., Guo, R., Whelan, J., Ruan, Y., Ma, J., et al. (2019). The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 180 (4), 2133–2141. doi: 10.1104/pp.19.00641
Wang, H., Zhang, B., Hao, Y., Huang, J., Tian, A., Liao, Y., et al. (2007). The soybean dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 52 (4), 716–729. doi: 10.1111/j.1365-313X.2007.03268.x
Warrington, C. V., Abdel-Haleem, H., Hyten, D. L., Cregan, P. B., Orf, J. H., Killam, A. S., et al. (2015). QTL for seed protein and amino acids in the benning × danbaekkong soybean population. Theor. Appl. Genet. 128 (5), 839–850. doi: 10.1007/s00122-015-2474-4
Wei, Z., Pan, T., Zhao, Y., Su, B., Ren, Y., Qiu, L. (2020). The small GTPase Rab5a and its guanine nucleotide exchange factors are involved in post-golgi trafficking of storage proteins in developing soybean cotyledon. J. Exp. Bot. 71 (3), 808–822. doi: 10.1093/jxb/erz454
West, M., Yee, K. M., Danao, J., Zimmerman, J. L., Fischer, R. L., Goldberg, R. B., et al. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 (12), 1731–1745. doi: 10.1105/tpc.6.12.1731
Wilson, R. F. (2004). “Seed composition,” in Soybeans: Improvement, Production, and Uses, 3rd Edn, eds H. R. Boerma and J. E. Specht (Madison, WI: American Society of Agronomy), 621–677.
Wilson, R. F. (2008). “Soybean: market driven research needs,” in Genetics and Genomics of Soybean, ed. G. Stacey (New York, NY: Springer), 3–15. doi: 10.1007/978-0-387-72299-3_1
Xu, C., Shanklin, J. (2016). Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 67, 179–206. doi: 10.1146/annurev-arplant-043015-111641
Yan, L., Hofmann, N., Li, S., Ferreira, M. E., Song, B., Jiang, G., et al. (2017). Identification of QTL with large effect on seed weight in a selective population of soybean with genome-wide association and fixation index analyses. BMC Genomics 18 (1), 529. doi: 10.1186/s12864-017-3922-0
Yang, Y., Kong, Q., Lim, A. R. Q., Lu, S., Zhao, H., Guo, L., et al. (2022a). Transcriptional regulation of oil biosynthesis in seed plants: current understanding, applications, and perspectives. Plant Commun. 3 (5), 100328. doi: 10.1016/j.xplc.2022.100328
Yang, Y., La, T. C., Gillman, J. D., Lyu, Z., Joshi, T., Usovsky, M., et al. (2022b). Linkage analysis and residual heterozygotes derived near isogenic lines reveals a novel protein quantitative trait loci from a Glycine soja accession. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.938100
Yang, H., Wang, W., He, Q., Xiang, S., Tian, D., Zhao, T., et al. (2019). Identifying a wild allele conferring small seed size, high protein content and low oil content using chromosome segment substitution lines in soybean. Theor. Appl. Genet. 132(10), 2793–2807. doi: 10.1007/s00122-019-03388-z
Yin, P., Ma, Q., Wang, H., Feng, D., Wang, X., Pei, Y., et al. (2020). SMALL LEAF AND BUSHY1 controls organ size and lateral branching by modulating the stability of BIG SEEDS1 in Medicago truncatula. New Phytol. 226 (5), 1399–1412. doi: 10.1111/nph.16449
Yu, B., He, X., Tang, Y., Chen, Z., Zhou, L., Li, X., et al. (2023). Photoperiod controls plant seed size in a CONSTANS-dependent manner. Nat. Plants 9(2), 343–354. doi: 10.1038/s41477-023-01350-y
Zhang, H., Goettel, W., Song, Q., Jiang, H., Hu, Z., Wang, M., et al. (2020). Selection of GmSWEET39 for oil and protein improvement in soybean. PloS Genet. 16 (11), e1009114. doi: 10.1371/journal.pgen.1009114
Zhang, M., Liu, S., Wang, Z., Yuan, Y., Zhang, Z., Liang, Q., et al. (2022). Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 20 (2), 256–282. doi: 10.1111/pbi.13682
Zhang, J., Song, Q., Cregan, P., Jiang, G. (2016a). Genome-wide association study, genomic prediction and marker-assisted selection for seed weight in soybean (Glycine max). Theor. Appl. Genet. 129(1), 117–130. doi: 10.1007/s00122-015-2614-x
Zhang, D., Sun, L., Li, S., Wang, W., Ding, Y., Swarm, S. A., et al. (2018a). Elevation of soybean seed oil content through selection for seed coat shininess. Nat. Plants 4 (1), 30–35. doi: 10.1038/s41477-017-0084-7
Zhang, D., Wang, X., Li, S., Wang, C., Gosney, M. J., Mickelbart, M. V., et al. (2019a). A post-domestication mutation, Dt2, triggers systemic modification of divergent and convergent pathways modulating multiple agronomic traits in soybean. Mol. Plant 12 (10), 1366–1382. doi: 10.1016/j.molp.2019.05.010
Zhang, J., Wang, X., Lu, Y., Bhusal, S. J., Song, Q., Cregan, P. B., et al. (2018b). Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding. Mol. Plant 11 (3), 460–472. doi: 10.1016/j.molp.2017.12.016
Zhang, T., Wu, T., Wang, L., Jiang, B., Zhen, C., Yuan, S., et al. (2019c). A combined linkage and GWAS analysis identifies QTLs linked to soybean seed protein and oil content. Int. J. Mol. Sci. 20 (23), 5915. doi: 10.3390/ijms20235915
Zhang, W., Xu, W., Zhang, H., Liu, X., Cui, X., Li, S., et al. (2021). Comparative selective signature analysis and high-resolution GWAS reveal a new candidate gene controlling seed weight in soybean. Theor. Appl. Genet. 134 (5), 1329–1341. doi: 10.1007/s00122-021-03774-6
Zhang, D., Zhang, H., Hu, Z., Chu, S., Yu, K., Lv, L., et al. (2019b). Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PloS Genet. 15 (7), e1008267. doi: 10.1371/journal.pgen.1008267
Zhang, Y., Zhao, F., Li, Q., Niu, S., Wei, W., Zhang, W., et al. (2016b). Soybean GmDREBL increases lipid content in seeds of transgenic Arabidopsis. Sci. Rep. 6 (1), 34307. doi: 10.1038/srep34307
Zhang, D., Zhao, M., Li, S., Sun, L., Wang, W., Cai, C., et al. (2017). Plasticity and innovation of regulatory mechanisms underlying seed oil content mediated by duplicated genes in the palaeopolyploid soybean. Plant J. 90 (6), 1120–1133. doi: 10.1111/tpj.13533
Zhao, B., Dai, A., Wei, H., Yang, S., Wang, B., Jiang, N., et al. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90 (1-2), 33–47. doi: 10.1007/s11103-015-0392-0
Zhao, X., Dong, H., Chang, H., Zhao, J., Teng, W., Qiu, L., et al. (2019). Genome wide association mapping and candidate gene analysis for hundred seed weight in soybean [Glycine max (L.) Merrill]. BMC Genomics 20(1), 648. doi: 10.1186/s12864-019-6009-2
Zhou, Z., Jiang, Y., Wang, Z., Gou, Z., Lyu, J., Li, W., et al. (2015). Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33 (4), 408–414. doi: 10.1038/nbt.3096
Keywords: soybean, seed size, oil, protein, QTL, functional genome
Citation: Duan Z, Li Q, Wang H, He X and Zhang M (2023) Genetic regulatory networks of soybean seed size, oil and protein contents. Front. Plant Sci. 14:1160418. doi: 10.3389/fpls.2023.1160418
Received: 07 February 2023; Accepted: 24 February 2023;
Published: 07 March 2023.
Edited by:
Xiaobo Wang, Anhui Agricultural University, ChinaReviewed by:
Dan Zhang, Henan Agricultural University, ChinaYingpeng Han, Northeast Agricultural University, China
Copyright © 2023 Duan, Li, Wang, He and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Li, bGlxaW5nMTk4NjEwMkAxNjMuY29t; Min Zhang, emhhbmdtaW5AZ2VuZXRpY3MuYWMuY24=
†These authors have contributed equally to this work
 Zongbiao Duan
Zongbiao Duan Qing Li
Qing Li Hong Wang
Hong Wang Xuemei He
Xuemei He Min Zhang
Min Zhang