- 1Key Laboratory of Forage Cultivation, Processing and High Efficient Utilization of Ministry of Agriculture and Rural Affairs, Key Laboratory of Grassland Resources of the Ministry of Education, College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot, China
- 2College of Agriculture and Forestry, Hulunbuir University, Hulunbuir, China
- 3Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences, Grassland Research Institute, Hohhot, China
Objective: The objective of this study was to isolate lactic acid bacteria (LAB) from native grasses and naturally fermented silages, determine their identity, and assess their effects on silage quality and bacterial communities of the native grasses of three steppe types fermented for 60 days.
Methods: Among the 58 isolated LAB strains, Limosilactobacillus fermentum (BL1) and Latilactobacillus graminis (BL5) were identified using 16S rRNA sequences. Both strains showed normal growth at 15- 45°C temperature, 3-6.5% NaCl concentration, and pH 4-9. Two isolated LAB strains (labeled L1 and L5) and two commercial additives (Lactiplantibacillus plantarum and Lentilactobacillus buchneri; designated as LP and LB, respectively) were added individually to native grasses of three steppe types (meadow steppe, MS; typical steppe, TS; desert steppe, DS), and measured after 60 d of fermentation. The fresh material (FM) of different steppe types was treated with LAB (1 × 105 colony forming units/g fresh weight) or distilled water (control treatment [CK]).
Results: Compared with CK, the LAB treatment showed favorable effects on all three steppe types, i.e., reduced pH and increased water-soluble carbohydrate content, by modulating the microbiota. The lowest pH was found in the L5 treatment of three steppe types, at the same time, the markedly (p < 0.05) elevated acetic acid (AA) concentration was detected in the L1 and LB treatment. The composition of bacterial community in native grass silage shifted from Pantoea agglomerans and Rosenbergiella nectarea to Lentilactobacillus buchneri at the species level. The abundance of Lentilactobacillus buchneri and Lactiplantibacillus plantarum increased significantly in L1, L5, LP, and LB treatments, respectively, compared with CK (p < 0.05).
Conclusion: In summary, the addition of LAB led to the shifted of microbiota and modified the quality of silage, and L. fermentum and L. graminis improved the performance of native grass silage.
Introduction
Meadow steppe, typical steppe, and desert steppe are three common steppe types in northern temperate Asia, with relatively taste ability and the capacity to meet livestock growth (Hou et al., 2017). Moreover, these steppe types are tolerant to cold, drought, sand, and biotic stresses (Pang et al., 2011; Yuan et al., 2012; Fu et al., 2014), and play a key role in meeting the needs of animal production and, indirectly, protecting human health (Wang et al., 2022). Traditionally, these steppes have been commonly used to make hay or as pasture for grazing ruminants (Zhang et al., 2018). Generally, native grass hay is the traditional method of native grass; however, because the quality and palatability of natural hay is sensitive to environmental factors, it is not feasible to address the seasonal and annual imbalances through hay preparation techniques (Li et al., 2015). Ensiling, a traditional method of preserving animal feed and fresh fodder crops, provides feed for animals all year round, effectively reducing nutritional losses and extending storage time (Wang et al., 2022). Silage was mainly lactic acid bacteria (LAB) in the anaerobic environment using soluble carbohydrates for fermentation, produce lactic acid (LA), leading to low pH, inhibit aerobic bacteria, effective preservation of nutrients and improves feed palatability (Cai et al., 2020).
Our previous study showed that obtaining high-quality silage from different native grass steppes through direct fermentation is difficult (You et al., 2021). The addition of LAB is an effective way to overcome the poor fermentation characteristics of native grasses (Jaipolsaen et al., 2021). Additionally, LAB can inhibit the proliferation of harmful microorganisms and reduce the pH value, improving the quality of silage (Xian et al., 2022). Nevertheless, one of the decisive elements for the successful LAB inoculation of forage silage is the good positive interaction between microorganisms and raw materials (Avila et al., 2014). Previous studies showed that the effect of LAB obtained from stylo, oat, and sweet sorghum is comparable with or even better than that of commercial bacteria (Liu et al., 2012; Sifeeldein et al., 2019; Li et al., 2021). These studies suggest that the forage crop is the best source of suitable isolates. Thus, the selection and application of LAB for making different types of silage is essential, but few LAB have been isolated from different the native grasslands of the Mongolian Plateau. The complex and diverse geographical environment and climatic conditions of the Mongolian Plateau have bred rich and unique LAB resources.
In recent years, the third-generation Pacific Biosciences (PacBio) single-molecule real-time (SMRT) DNA sequencing technology has improved classification accuracy and speed by producing ultra-long reads, directly detecting epi-modification sites and avoiding template amplification (Xu et al., 2018). The SMRT technology can identify organisms to the species level and has been applied to the silages of Italian ryegrass and stylo (Wang et al., 2019; Yan et al., 2019). Consequently, the effects of different LAB on changes in the microbial composition of silage could be evaluated more accurately. Additionally, the effect of LAB obtained from natural sources on the diversity patterns of bacterial communities of native grass silage has been evaluated using the PacBio SMRT method in different steppe types.
In our study, we selected two high-potential strains, Limosilactobacillus fermentum and Latilactobacillus graminis, isolated from native grasses of the Mongolian Plateau. These two strains use multiple carbohydrates, produce more LA and acetic acid (AA) in an anaerobic environment and grow under low pH conditions; however, how these strains change the fermentation quality of different native grass steppes remains unclear. We hypothesized that LAB isolated from their own material might be better for improving the quality of three steppe types native grass silage relative to commercial LAB. In this study, we aimed to 1) determine the effects of different LAB on the fermentation characteristics of three steppe types of native grass silage; 2) evaluate the changes in bacterial communities and their functional properties; and 3) investigate the priority effects and mechanisms of action of L. fermentum and L. graminis on silage bacterial communities.
Materials and methods
LAB strains
A total of 58 LAB strains were isolated from 48 samples of native grass and naturally fermented silage, according to the method of Cai et al. (1998). Two representative strains of LAB(BL1 and BL5) were selected from 58 isolates based on their morphological and physiological tests, growth curve tests, and acid production curve tests. Native grasses in the meadow steppe (MS), typical steppe (TS), and desert steppe (DS) of the Inner Mongolia Plateau in China were harvested at the milk stage in July 2019 and 2020. The following species were predominant in the three steppes: Baical Needlegrass (Stipa baicalensis Roshev.) and Chinese Leymus (Leymus chinensis [Trin.] Tzvel.) in MS; Larch Needlegrass (Stipa grandis P. Smirn.) and Chinese Leymus (L. chinensis [Trin.] Tzvel.) in TS; and Short-flowered Needlegrass (Stipa breviflora Griseb.) and Mongolian Leek (Allium mongolicum Regel.) in DS. Each sample (10 g) was mixed with 90 ml of sterile distilled water. The dilutions were spread on Deman, Rugose, Sharp (MRS) agar (Difco Laboratories, Detroit, MI, USA), and incubated under anaerobic conditions at 35°C for 48 h to isolate LAB. In addition, putative homogeneous and heterogeneous LAB strains were stored in MRS containing 25% glycerol at -80°C by strain purification on MRS agar plates.
Physiological and morphological tests
Gram staining, catalase reaction, colony morphology, and glucose-based gas production tests were performed as described by Kozaki et al. (1992). Temperature, acid and salt tolerance tests of the strains were performed using MRS broth (Cai et al., 1999). Carbohydrate assimilation of the strains was determined using API 50 CH (BioMerieux, Marcy l’Etoile, France) (Zhang et al., 2015).
Identification of LAB strains by 16S rRNA sequence analysis
Genomic DNA of the screened strains was extracted using the Bacterial DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China). The isolated DNA was subjected to polymerase chain reaction (PCR) using primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGCTACCTTGTTACGACT-3’) (Tohno et al., 2012; Pitiwittayakul et al., 2021). Then, 16S rRNA sequences were identified using BLAST analysis in the GenBank database (Ennahar et al., 2003).
Preparation of silage
Native grasses in MS (119°13′E, 44°04′N), TS (116°28′E, 44°05′N), and DS (113°34′42″E, 44°6′N) of the Inner Mongolia Plateau, China, were harvested at the milk stage in July 2021. A sample square (50 m × 100 m) divided into five paired quadrants (100 cm × 100 cm) was selected and used to collect fresh forage samples from the three steppe types. Freshly harvested material was cut into 20-30-mm long pieces using a manual forage chopper (Mode-8, 200; Minghong Business, Shandong, China), placed in polythene bags, and vacuum sealed. Two screened LAB strains (BL1 and BL5; designated as L1 and L5, respectively) and two commercial LAB additives (Lactiplantibacillus plantarum and Lentilactobacillus buchneri; designated as LP and LB, respectively (Shandong Zhongke Jiayi Biological Engineering Co. Ltd., China) were added individually at a concentration of 105 cfu/g of fresh material (FM), with distilled water as a control (CK). The FM and additives were mixed thoroughly, and silage (approximately 250 g) was packed in polyethylene bags (Shenyang Huasheng Plastic Packaging Products Co., Ltd., China). The bags were then sealed with a vacuum sealer to extract the air. Three replicates were made for each treatment, and the silage of different treatments were measured after 60 days of ensiling at room temperature (25°C).
Analyses of chemical composition, microorganism and fermentation parameter
Three replicates were set up for each native grass sample. Dry matter (DM) and content crude protein (CP) were measured following the method of Zhang et al. (2016) and Du et al. (2019). Determination of neutral detergent fiber (NDF) and acid detergent fiber (ADF) content was performed according to the method of Van Soest et al. (1991) by using the Ankom A2000i fiber analyzer (Ankom Technology, Macedon, NY, USA). The water soluble carbohydrates (WSC) content was measured following the method of Chen et al. (2015).
A sample of silage (10 g) was mixed with 90 g deionized water following the description of Yuan et al. (2020), and stored in a refrigerator at 4°C for 24 h. The leachate was filtered through four layers of gauze and filter paper, with measurements of pH, ammonia nitrogen (NH3-N) and organic acids in the leachate. pH was measured using a glass electrode pH meter (Leici pH S-3C, Shanghai, China). The content of LA, AA, propionic acid (PA) and butyric acid (BA) in silages was determined by high performance liquid chromatography (HPLC; model: Waters e2695, Milford, USA) (Cheng et al., 2020). The method of Broderick and Kang (1980) was used to determine NH3-N concentrations. Microbial populations (LAB, yeasts, mold, anerobic bacteria, and coliform bacteria) in the FM were assessed as described in a previous report (Guo et al., 2021a).
DNA extraction, PCR and sequencing
The bacterial community composition of native grass fermented for 60 days was analyzed by 16S rRNA gene sequencing. Based on the investigation of Liu et al. (2019), total DNA was extracted from fresh and silage samples of native grass. The metagenomic DNA extraction and PCR amplification procedures of bacterial 16S ribosomal RNA genes were performed as per Guo et al. (2021).
DNA was amplified with primers 27F (5’-AGRGTTTGATYNTGGCTCAG-3’) and 1492R (5’-TASGGHTACCTTGTTASGACTT-3’). PCR conditions were an initial denaturation at 98°C for 2 min, 30 cycles of denaturation at 98°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 60 s, followed by a final extension at 72°C for 5 min. The PCR products were purified for sequencing and analysis. Each treatment was performed in triplicate. 16S rRNA gene sequence were stored in NCBI with BioProject accession number PRJNA912573.
NGS sequencing was performed by Biomarker Technologies (Beijing, China) on a Pacbio_SMRT platform (Pacbio Sequel II, CA, USA). Coverage of α-diversity indicators Chao1 and Good was calculated using QIIME v1.9.1 (Segata et al., 2011). Principal coordinate analysis (PCoA) was performed using the R program (version 3.2.5) on the basis of β-diversity unweighted or weighted unifrac distances. Operational taxonomic units (OTUs) were classified using Ribosome Database Project (RDP) Classifier (version 2.2) against the SILVA (Release 128) 16S rRNA database with a minimum confidence cut off of 0.7, and were then denominated at the phylum, genus, and species levels. Mothur (version v.1.30) software was used to evaluate the α-diversity indices (ACE, Chao 1, Simpson, and Shannon) of the samples. LEfSe (Linear Discriminant Analysis (LDA) effect size) was able to find biomarkers that are statistically different between groups. It was performed using a free online platform (https://international.biocloud.net). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States II (PICRUSt2) software was used to predict microbial functions from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Statistical analysis
The effect of steppe types and ensiling time on silage quality was evaluated by two-way ANOVA and Duncan’s multiple range test. All statistical procedures were performed using SAS 9.3 software (SAS Institute, Inc., Cary, NC, USA). Effects were considered significant when p < 0.05. Figures for microbiota data were performed using the BMK Cloud platform online and GraphPad prism 8.
Results
LAB strain characteristics
Two isolated LAB strains were Gram-positive and catalase negative (Table 1). The strain BL5 was a homo-fermenter, while the strain BL1 was a hetero-fermenter. Additionally, BL1 showed optimal growth at 15-50°C and pH 3.5-9.0 but weak growth at 10°C and pH 3, whereas strain BL5 showed normal growth at 10-45°C and pH 4.0-9.0 but weak growth at 5°C and pH 3.5. The growth of strains BL1 and BL5 was normal at NaCl concentrations of 3.0% and 6.5%, respectively.
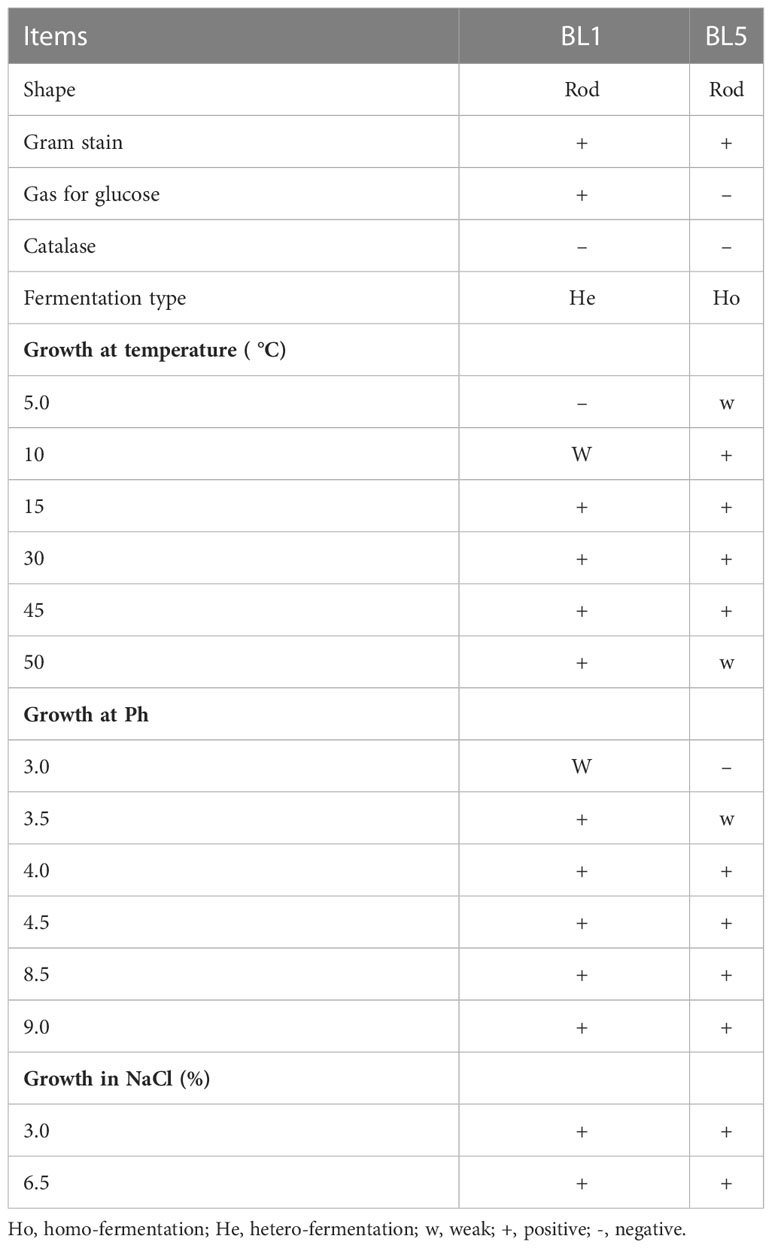
Table 1 he selection of isolated lactic acid bacteria on the base of the morphological and physiological tests.
Both strains exhibited different fermentation patterns, based on the results of gas detection on glucose. The two strains differ in the products of fermentation of carbohydrates such as D-Xylose, D-Mannitol, N-Acetyl Glucosamine, Amygdalin, Esculin, Salicin, Maltose, Lactose, Melibiose, Sucrose, Raffinose, β-Gentiobiose, and Gluconate (Table 2).
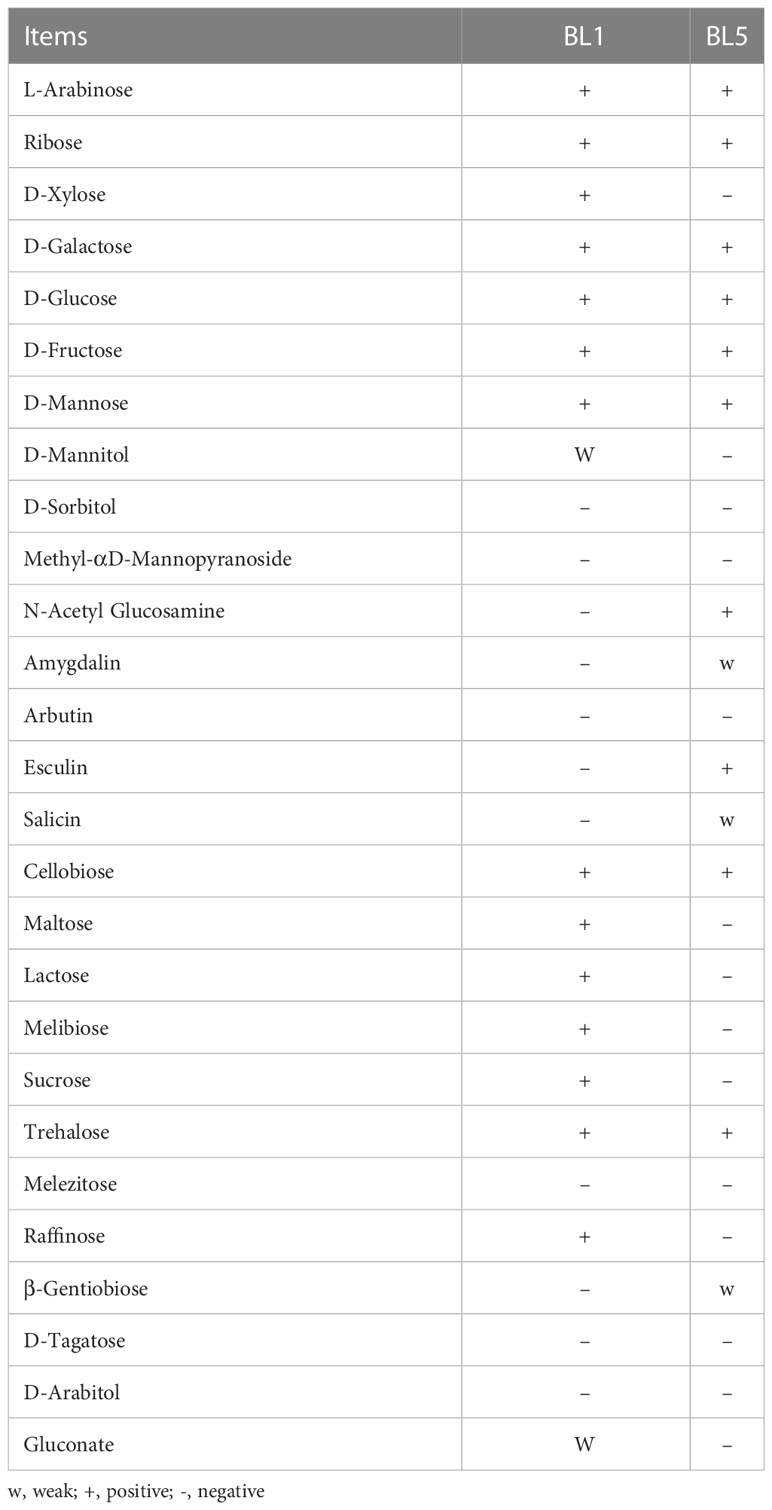
Table 2 The characteristics of isolated lactic acid bacteria on the base of carbohydrate fermentation.
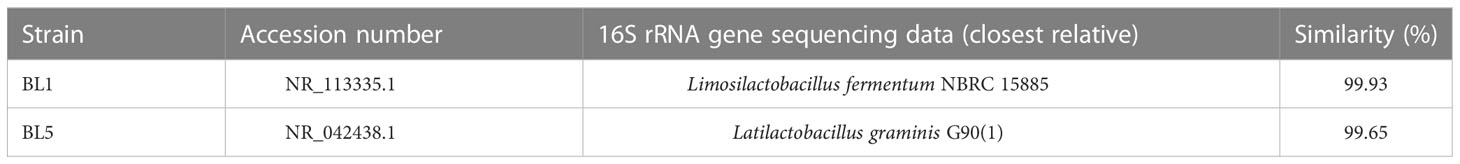
The results of 16S rRNA sequencing were analyzed by BLAST in the GenBank database (Table 3). Strain BL1 showed high similarity to Limosilactobacillus fermentum (99.93%), and strain BL5 expressed high similarity to Latilactobacillus graminis (99.65%). The nucleotide sequences of strains BL1 and BL5 were registered in GenBank under accession numbers OP984707 and OP984708, respectively.
Characteristics of pre-ensiled native grass
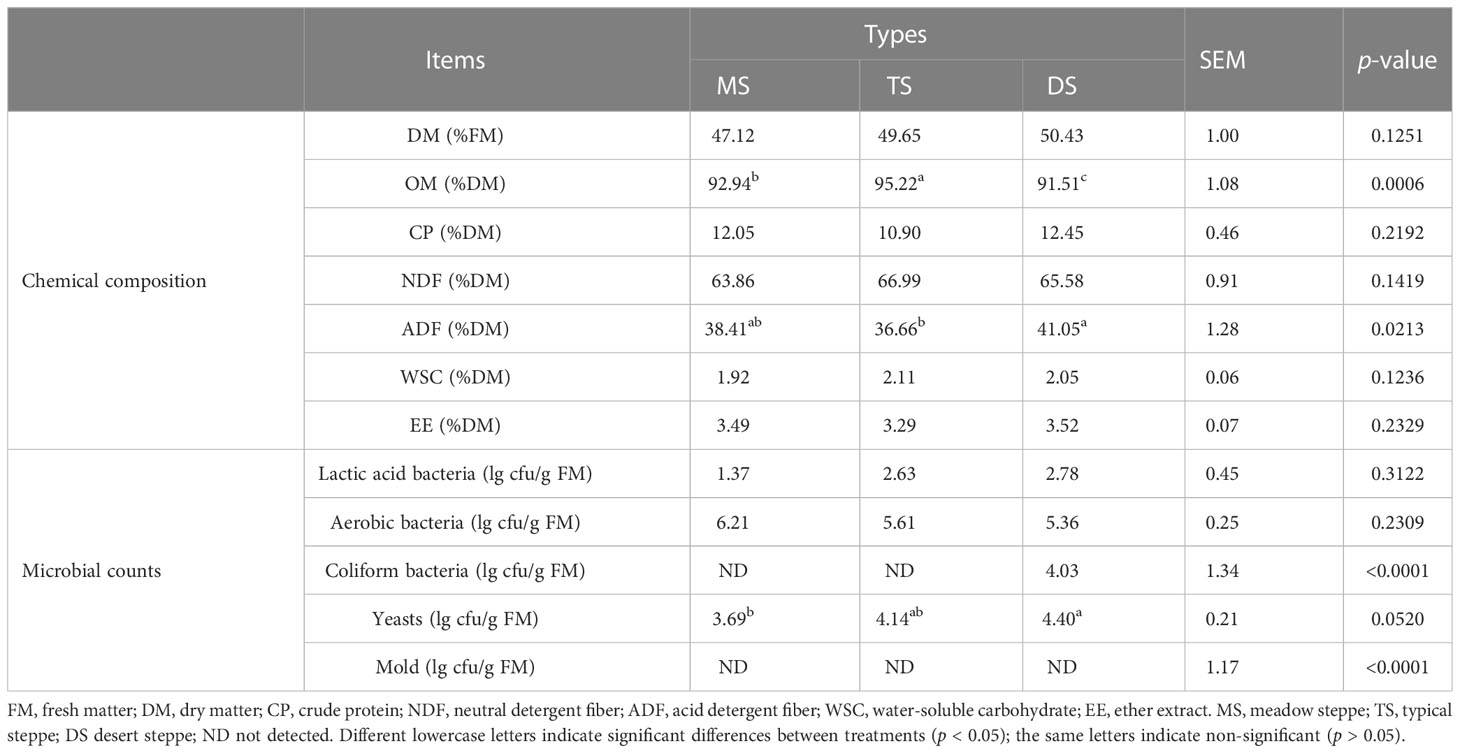
The dry matter (DM) content of the three steppe types varied from 47.12–50.43% and CP content was 12.05% (MS), 10.90% (TS), and 12.45% (DS), respectively (Table 4). Relatively low WSC content (1.92-2.11%) and low LAB number (1.37-2.78 log cfu/g FM) were observed. No coliform bacteria were detected in the raw materials of MS and TS, and no mold was detected in the three steppe types.
Chemical characteristics of native grass silage
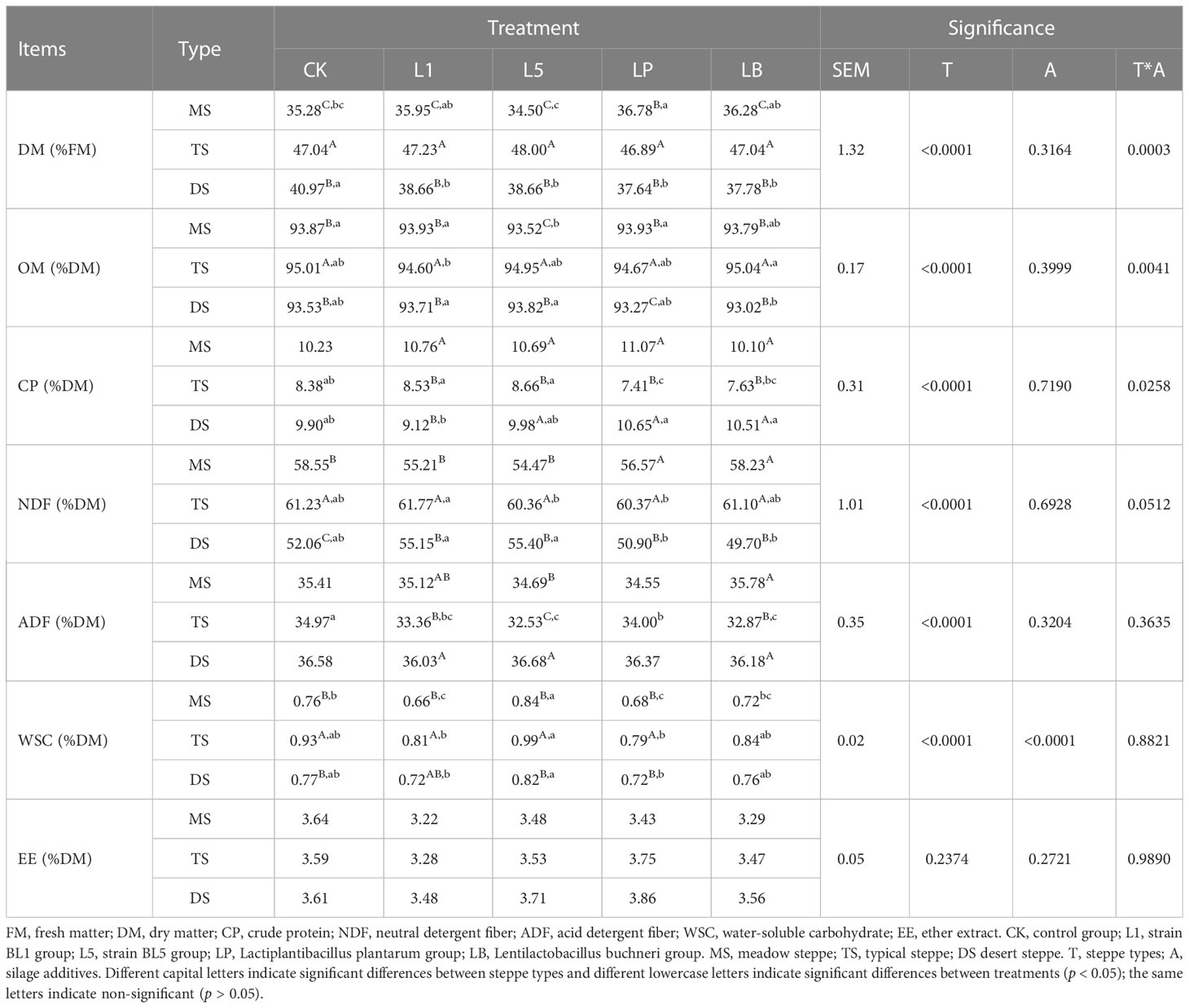
The silage of TS showed higher DM and organic matter (OM) contents than those of MS and DS in all treatments (p < 0.05), and TS silage showed higher NDF and WSC contents than MS and DS in CK and L5 treatment (Table 5). The EE contents showed no significant difference among the three steppe types (p > 0.05). The influence of LAB inoculants on the OM, CP, NDF and WSC of native grass was significant, except effect of LAB inoculants on CP and NDF in MS.
Fermentation quality of native grass silage
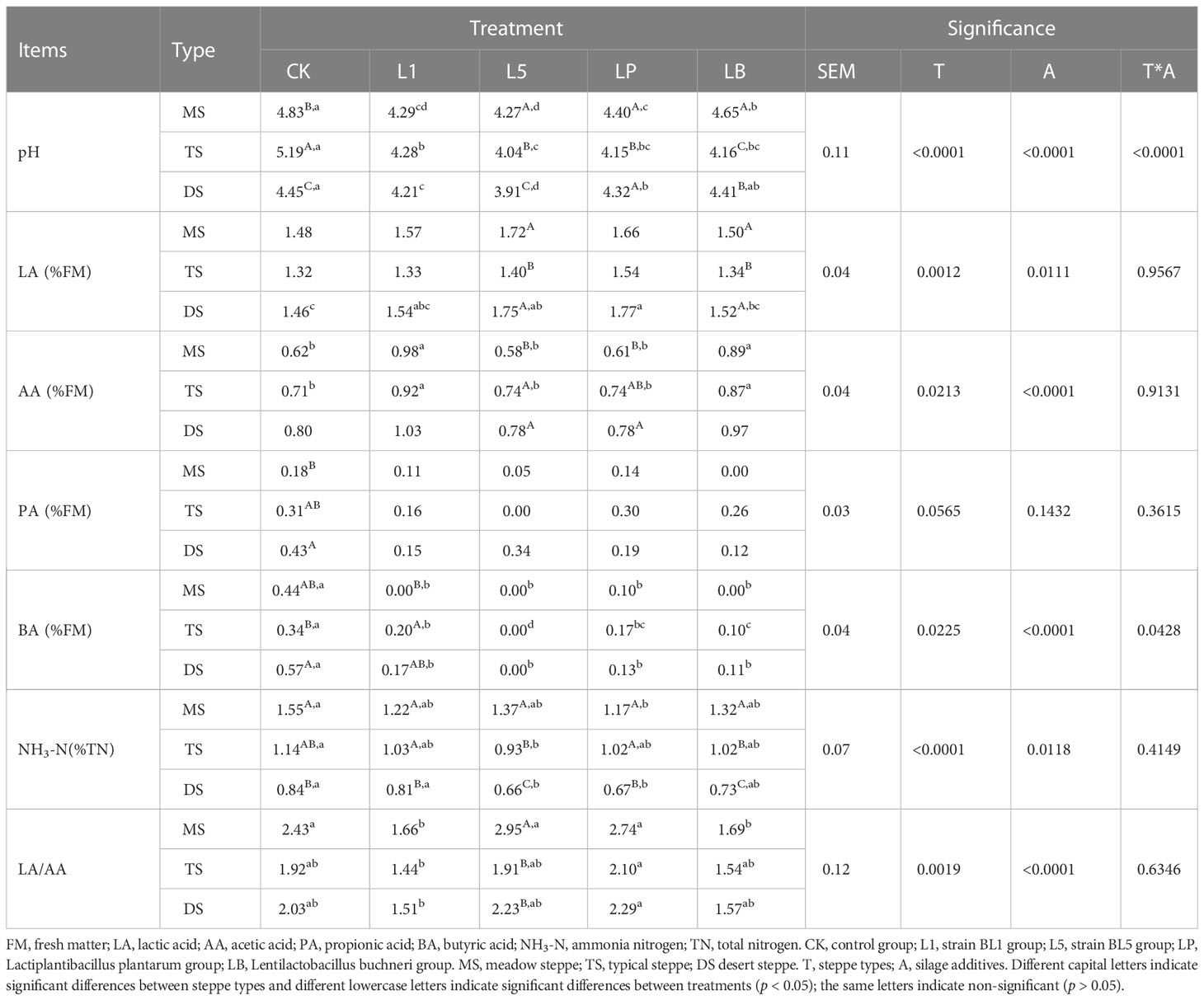
Steppe types had significant effects on the pH (except L1) and NH3-N content of silage (p < 0.05) (Table 6). DS silage showed lower NH3-N content than MS and TS silages (p < 0.05). MS silage showed higher LA/AA than TS and DS in L5 treatment. LAB inoculants had significant effects on pH, PA, BA, NH3-N and LA/AA of silage (p < 0.05). Silage containing LAB inoculant showed significantly lower pH and BA content than CK (p < 0.05). L1 and LB treatments of the MS and TS showed higher AA content than other treatments (p < 0.05).
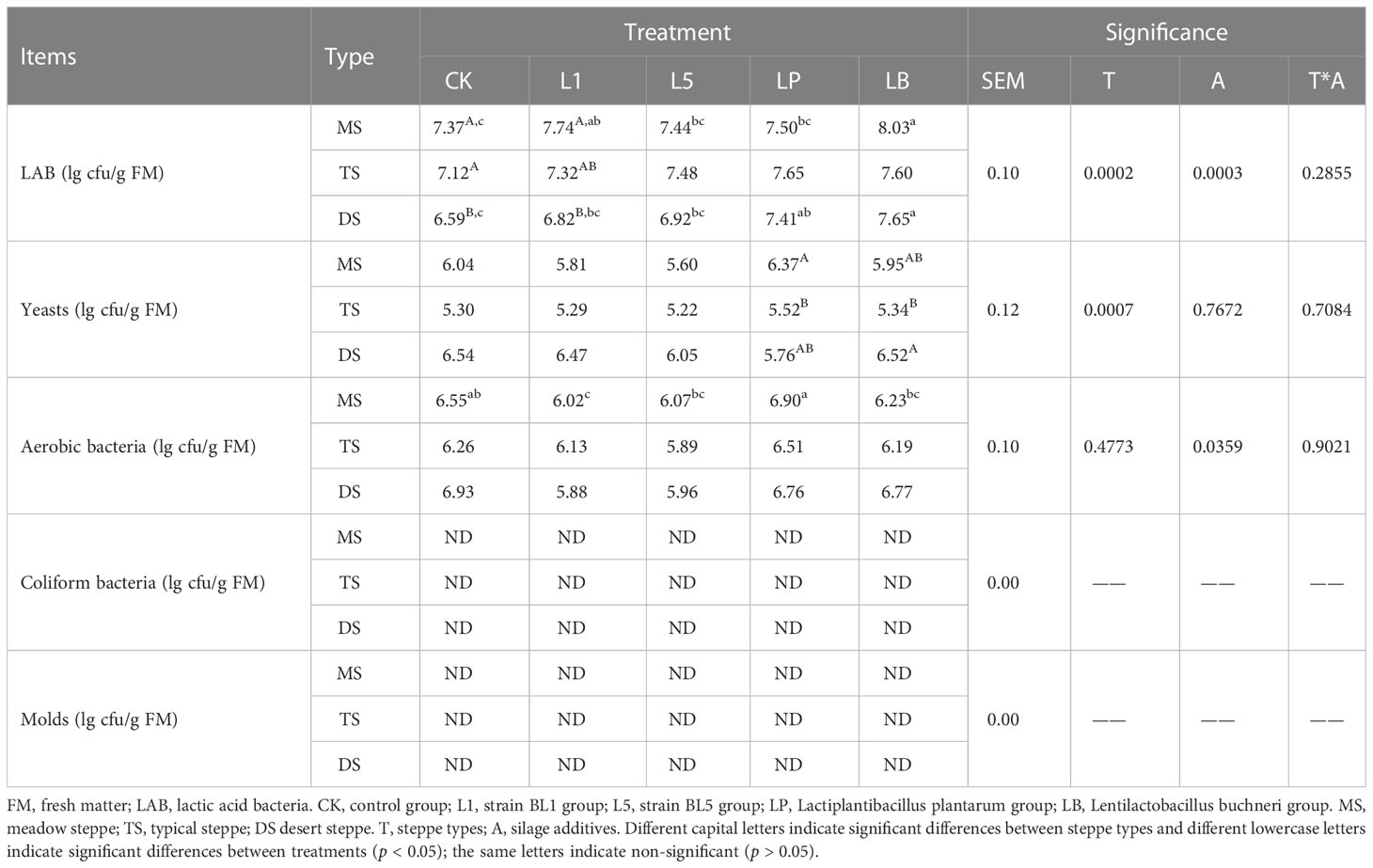
Microbial population of native grass silage
Steppe types significantly influenced LAB counts in CK and L1 treatment (p < 0.05), and significantly influenced yeast counts in LP and LB treatment (p < 0.05), but had no significant effect on aerobic bacteria counts (p > 0.05) (Table 7). Silages of MS and TS showed higher LAB counts than DS in CK treatment. LAB inoculants significantly influenced the number of LAB in MS and DS (p < 0.05), and significantly influenced aerobic bacteria counts in MS (p < 0.05) but had no significant effect on yeast counts (p > 0.05). Additive treatments showed higher LAB counts than CK in MS and DS, and LB silages in these two steppe native grass had higher LAB counts compared with other treatments (p < 0.05). The LP treatment of MS silage had higher aerobic bacteria counts than the other treatments (p < 0.05). Mold and coliform bacteria were not found in the different treatments.
Bacterial community of native grass silage
The 16S rDNA sequences were subjected to high-throughput sequencing to calculate and assess bacterial diversity in the silage of different steppe types (Table 8). The coverage depth for all treatments was above 96%. The ACE index was significantly higher for L1 than FM and LP in silage from MS (p < 0.05), however, the ACE and Chao1 index was significantly higher for L5 than L1 and LB in silage from TS (p < 0.05). The Simpson index of L1 was significantly lower than FM, CK, L5 and LP (p < 0.05) and the Shannon index was significantly lower than FM, CK and L5 (p < 0.05) in silage from MS. The Simpson and Shannon indices were significantly higher (p < 0.05) in TS silage for CK and L5 than for the other treatments. The Simpson and Shannon indices were significantly higher (p < 0.05) but Coverage was significantly lower (p < 0.05) for FM than for the other treatments in the DS silage. In addition, the Simpson and Shannon indices differed between treatments for the three steppe types of natural forage, including FM, with the exception of LB (p < 0.05).
To determine whether bacterial community structure differed between the raw material and silage of different steppe types, principal coordinate analysis (PCoA) was performed based on Unifrac (unweighted) distances (Figure 1). The bacterial communities of raw materials from different steppe types were distinct. Distance was also observed between the bacterial communities of CK and LAB treatments.
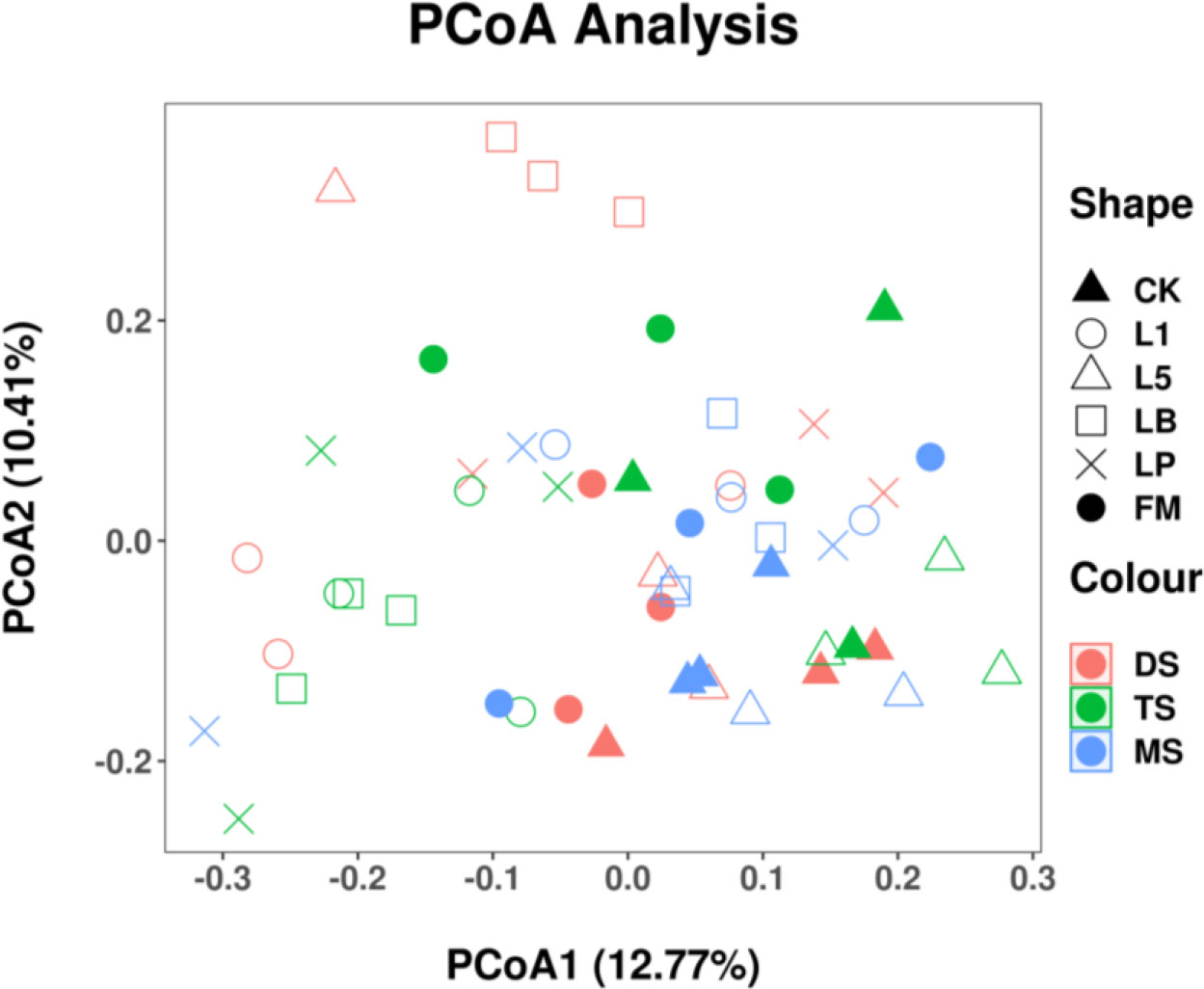
Figure 1 Principal coordinate analysis (PCoA) of the bacterial community of fresh material and native grass on 60 days of ensiling. FM, fresh material; CK, control group; L1, strain BL1 group; L5, strain BL5 group; LP, Lactiplantibacillus plantarum group; LB, Lentilactobacillus buchneri group. MS, meadow steppe; TS, typical steppe; DS desert steppe.
The predominant phylum in the three native grass steppe samples was Proteobacteria (>90%) (Figure 2A). However, their community composition was influenced by ensiling. By the end of the fermentation process, Firmicutes prevailed in all silage samples of the three steppe types, accounting for more than 87% in all groups (except the TSCK treatment). To further reveal phylum-level differences in the bacterial communities of native grass silage based on steppe types and LAB inoculants, one-way analysis of variance (ANOVA) was conducted on the community composition of native grasses before and after ensiling (Figure 2B). Significant differences were observed in the abundance of Acidobacteriota (p < 0.05), and highly significant differences were detected in the abundance of Firmicutes, Proteobacteria, and Actinobacteriota (p < 0.01), which were attached to fresh and silage samples of native grass in different treatments.
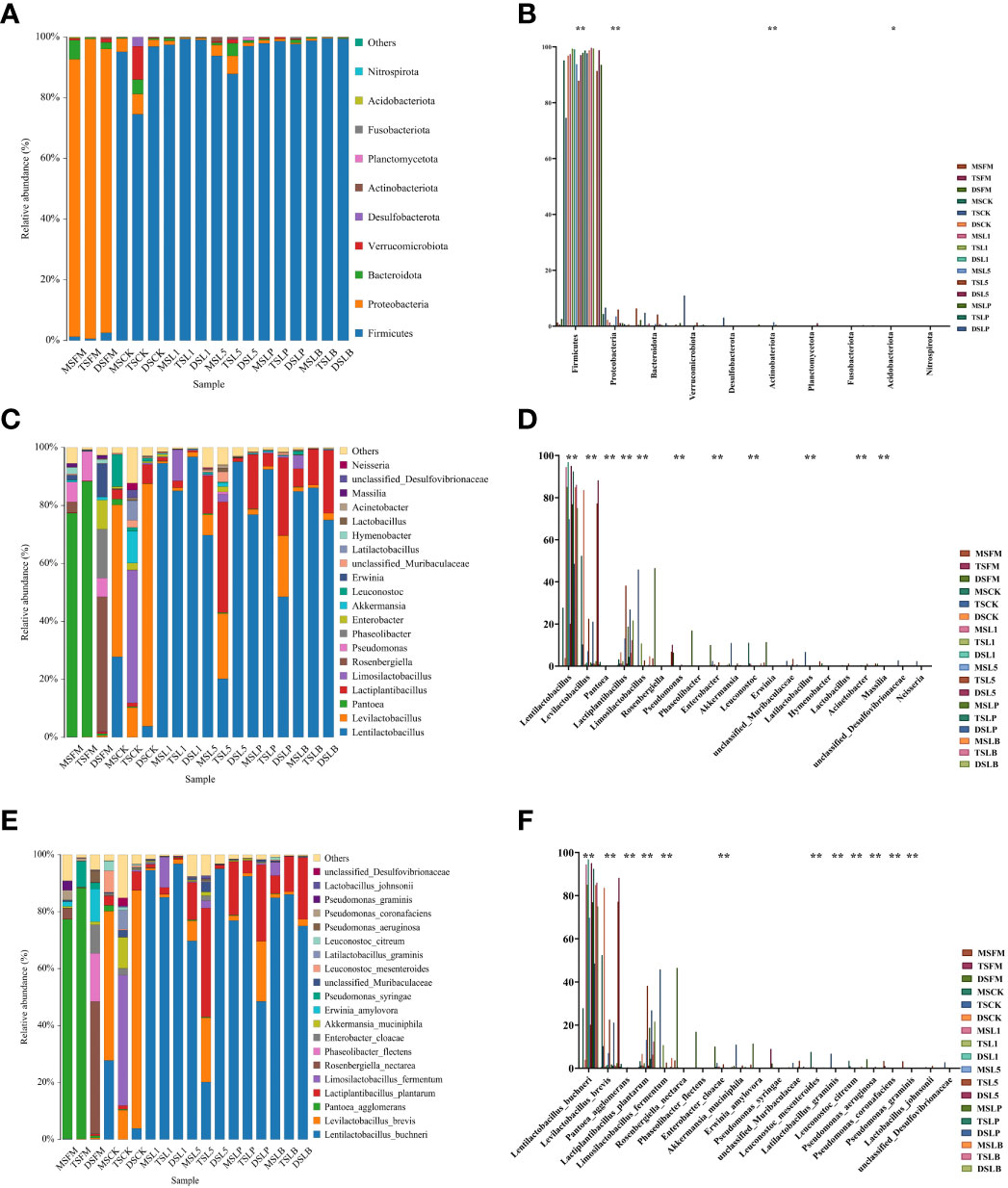
Figure 2 The bacterial community of fresh material and native grass on 60 days of ensiling of different steppe types. (A) The bacterial community was shown at the phylum level. (B) The extended error bar plot displaying the significant differences among groups at the phylum level. (C) The bacterial community was shown at the genus level. (D) The extended error bar plot displaying significant differences among groups at the genus level. (E) The bacterial community was shown at the genus level. (F) The extended error bar plot displaying significant differences among groups at the genus level. *Shows that the significant difference was at p < 0.05 level. **Shows that the significant difference was at p < 0.01 level. FM, fresh material; CK, control group; L1, strain BL1 group; L5, strain BL5 group; LP, Lactiplantibacillus plantarum group; LB, Lentilactobacillus buchneri group. MS, meadow steppe; TS, typical steppe; DS desert steppe.
The relative abundance of bacterial communities in the pre-ensiled native grass was very low (Figure 2C). The bacterial composition of FM at the genus level differed among the three steppe types. The main epiphytic bacteria associated with the FM of different steppe types were Pantoea in MS and TS, and Rosenbergiella and Phaseolibacter in DS, although the abundance of these genera decreased after ensiling. After 60 days of fermentation, Levilactobacillus was the most common genus in the CK treatments of MS and DS, and Limosilactobacillus was the main genus in the CK treatment of TS. Compared to CK, with the exception of TSL5, all treatments had Levilactobacillus as the dominant genus for all three native grass steppe types, and TSL5 was dominated by the Lactiplantibacillus. The abundance of Lentilactobacillus, Levilactobacillus, Pantoea, Lactiplantibacillus, Limosilactobacillus, Pseudomonas, Enterobacter, Leuconostoc, Latilactobacillus, Acinetobacter, and Massilia showed highly significant differences (p < 0.01) (Figure 2D).
The main epiphytic bacteria associated with the FM of MS and TS were Pantoea agglomerans, and those associated with the FM of DS were Rosenbergiella nectarea and Phaseolibacter flectens, although the abundance of these species decreased after ensiling (Figure 2E). After 60 days of fermentation, Levilactobacillus brevis was the most common species in the CK treatment of MS and DS, while L. fermentum was the main species in the CK treatment of TS. Compared with CK, all LAB silage treatments showed L. brevis, L. fermentum, and L. plantarum as the main species (>80% abundance). The abundance of L. buchneri, L. brevis, Pantoea agglomerans, L. plantarum, L. fermentum, Enterobacter cloacae, Leuconostoc mesenteroides, L. graminis, Leuconostoc citreum, Pseudomonas aeruginosa, Pseudomonas coronafaciens, and Pseudomonas graminis showed highly significant differences (p < 0.01) (Figure 2F).
The LEfSe method was used to evaluate differences in microbial communities of the three steppe types and to explore specific bacterial species in each group (LDA score > 4.0). Steppe type and LAB inoculant had a tremendous influence on the resulting bacterial community (Figure 3). In MS samples, 34 bacteria were significantly enriched, and Lactobacillaceae showed the highest LDA score (5.68) (Figure 3A). In TS samples, 39 bacteria were significantly enriched, and Lactobacillaceae showed the highest LDA score (5.68) (Figure 3B). In DS samples, 38 bacteria were significantly enriched, and Lactobacillaceae showed the highest LDA score (5.64) (Figure 3C).
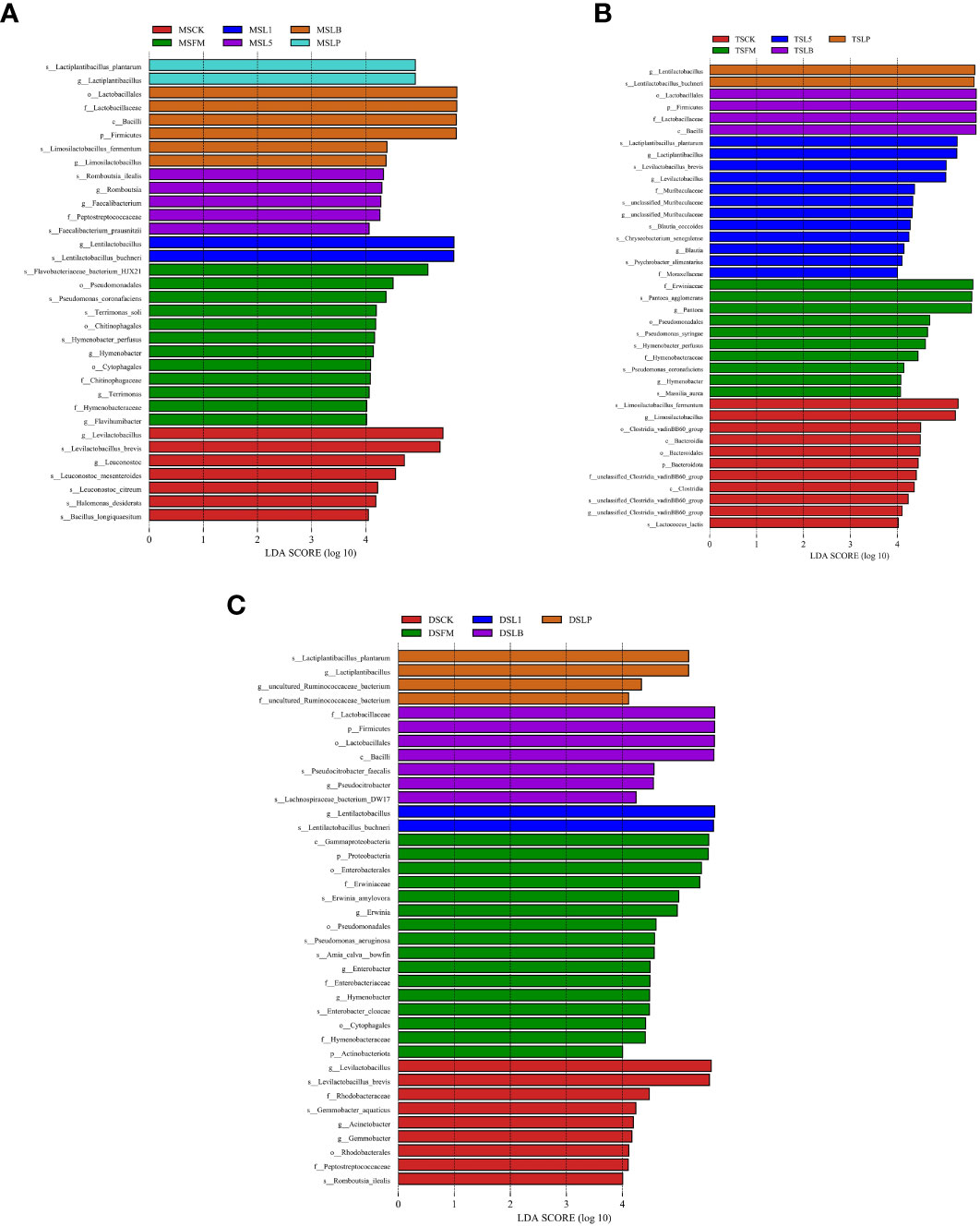
Figure 3 The linear discrimination analysis (LDA) coupled on the bacterial community of fresh material and native grass on 60 days of ensiling, with effect size (LEfSe) analysis. The significant difference in species was estimated by an LDA score greater at default score = 4.0. The length of the histogram shows the LDA score of differences in these groups. FM, fresh material; CK, control group; L1, strain BL1 group; L5, strain BL5 group; LP, Lactiplantibacillus plantarum group; LB, Lentilactobacillus buchneri group. MS, meadow steppe; TS, typical steppe; DS desert steppe. (A) LEfSe analysis between different treatment groups in meadow steppe (MS). (B) LEfSe analysis between different treatment groups in typical steppe (TS). (C) LEfSe analysis between different treatment groups in desert steppe (DS).
PICRUSt predicts the bacterial metabolic function in accordance with the KEGG pathway. In total, six different metabolic pathways were identified in the raw materials and silages of the three native grass steppe types (Figure 4A). Among them, metabolism metabolic abundance accounted for the largest proportion (>75% abundance), followed by environmental information processing and genetic information processing. The top 20 metabolic functions in level 2 had 14 metabolic pathways assigned to metabolism, 3 metabolic pathways assigned to genetic information processing (Figure 4B). There were 2 and 1 metabolic pathways assigned to environmental information processing and cellular processing, respectively. The amino sugar and nucleotide sugar metabolism, pyruvate metabolism, and glycolysis/gluconeogenesis pathways were significantly enriched in the three steppe types after ensiling (level 3), while pentose phosphate pathway and starch and sucrose metabolism were enriched in MS and DS treatments (Figure 4C). In terms of carbohydrate metabolism, the abundance of amino sugar and nucleotide metabolism and pentose phosphate pathway, glycolysis/glycoprotein production, and starch and sucrose metabolism were higher in the CK treatment than in the LAB additive treatments from MS and DS, while the abundance of amino sugar and nucleotide sugar metabolism, glycolysis/gluconeogenesis, and starch and sucrose metabolism was higher in the L5 treatment than in other treatments in TS. In terms of global and overview maps, the abundance of metabolic pathways, biosynthesis of secondary metabolites, biosynthesis of antibiotics, biosynthesis of amino acids, and carbon metabolism was enriched after ensiling. Notably, the abundance of microbial metabolism in diverse environments and carbon metabolism was significantly reduced after ensiling.
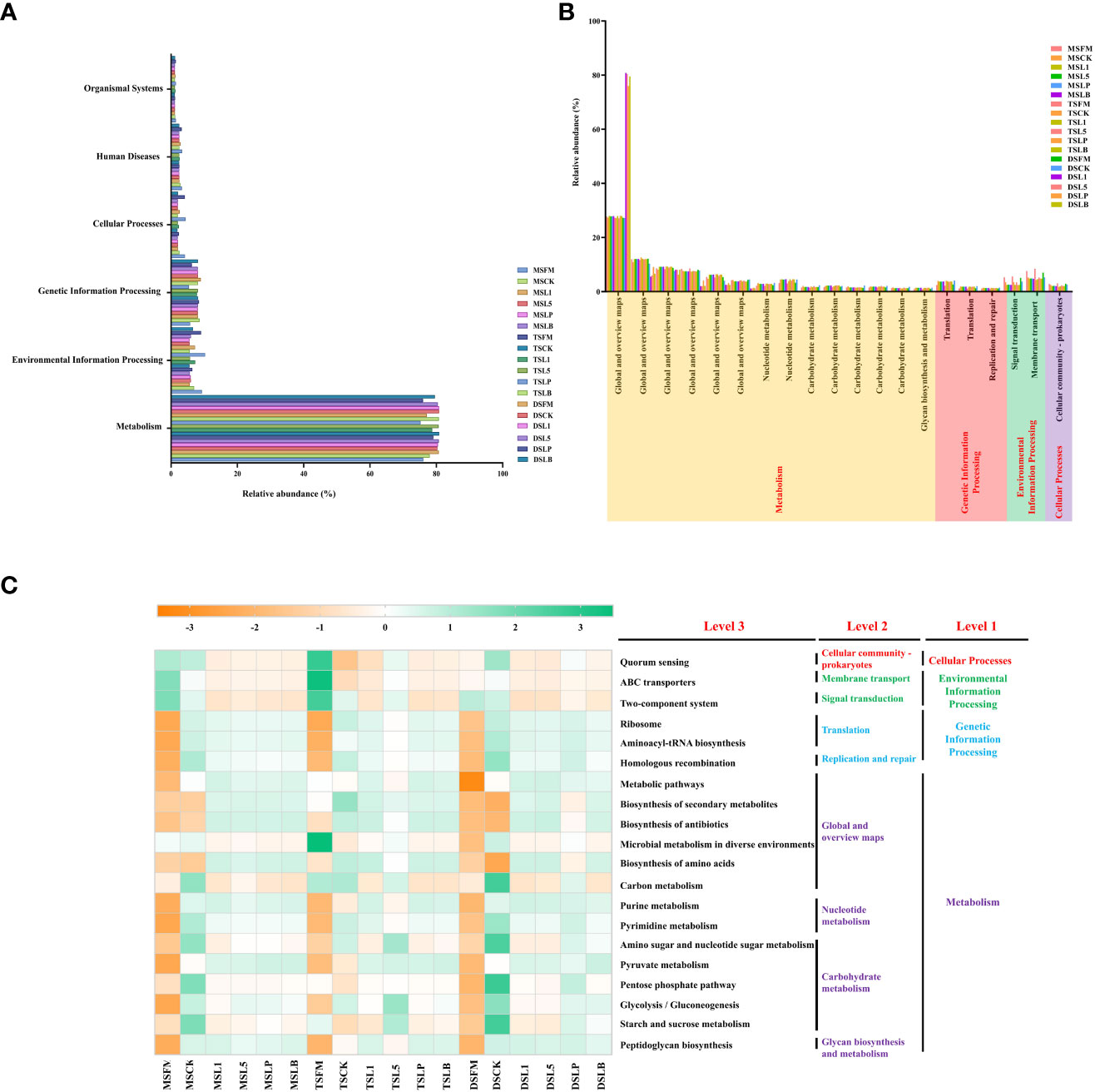
Figure 4 Functional predictions for silage microbiota with significantly different KEGG pathways (p <0.05) among the eighteen groups of silage. KEGG pathways at Level 1 (A), Level 2 (B), and Level 3 (C) are represented. FM, fresh material; CK, control group; L1, strain BL1 group; L5, strain BL5 group; LP, Lactiplantibacillus plantarum group; LB, Lentilactobacillus buchneri group. MS, meadow steppe; TS, typical steppe; DS desert steppe.
Discussion
LAB have been found and identified in forage silage, fermented food, and dairy products. In addition, LAB have been shown to be the dominant bacteria in various forage and silage samples (Burns et al., 2018). Nonetheless, it can be a challenge to identify the differences among dichotomous species based on morphological, physiological, and biochemical tests (Wang et al., 2019a). Therefore, this study combined with actual silage application to further evaluate and screen the selected LAB. The 16S rDNA sequence analysis has widely used to identify organisms at the genus and species levels (Wang et al., 2019b).
Chen et al. (2017) and Chu-Ky et al. (2014) concluded that L. fermentum is highly tolerant to acidity and can grow in low pH environments, which is consistent with the results of this study. Both strains differed not only in their ability to grow in an acidic environment but also in the temperature of the growth environment, which indicates that different LAB differ in their adaptability to the environment in which they grow (Du et al., 2022). In addition, L. fermentum was able to generate more fermentation products compared with strain BL5.
In this study, the DM and WSC contents of native grass was comparatively high, equivalent to that in a dry environment, which could be explained by the different plant species was observed in the three steppes and could support effective acid production for bacteria (Nkosi et al., 2016). A wide range of bacteria attached to the plant surface and different silage materials cultivate different microorganisms, in terms of both composition and population (Khota et al., 2016). Previously published study indicated that the ratio of beneficial to undesirable microorganism was estimated at 1:10 (Lin et al., 1992). In this study, the counts of undesirable microorganism, such as mold and coliform, were lower than the detectable levels in all steppe types, excepted the DS. These results could be included by the compositions of the steppes (Du et al., 2022). In the current study, the numbers of LAB on plants were lower than 5.0 log10 cfu/g FM, which is in accordance with the previous reports (Pang et al., 2012; Du et al., 2022). Therefore, it was necessary to isolate and selecte LAB additives for these steppes.
In the present study, the fermentation characteristics of the three steppe types changed after ensiling, probably because of the complex plant community structure of the native grasslands. Notably, the DM and CP content among these treatments was similar. This higher LA content in these treatments limited the microorganism that could breakdown the DM and CP contents might be the main reason (Muck, 1988; Muck, 2010). In addition, the lack of sufficient substrates for native grass of different steppe types to inhibit protease activity is another reason for this result (Sousa et al., 2019). The NDF and ADF contents showed no significant difference between CK and the four additive treatments, because the majority of LAB additives did not have the ability to produce cellulase and hemi-cellulase to degrade forage cell walls (Xu et al., 2020).
The organic acid (mainly LA and AA) and NH3-N contents, and pH are important indicators for well-preserved forage silage (Hashemzadeh-Cigari et al., 2014). In the high DM silage such as straw forage and native grass, the water activity of LAB is insufficient, so the silage fermentation is reduced due to insufficient metabolic water required for the growth of LAB (Borreani et al., 2018). Four LAB strains produced a large amount of LA and a certain amount of AA to decrease the pH and inhibit the activity of proteases in plants and microbes and reduce proteolysis using a series of substrates (Dong et al., 2022; Lu et al., 2022). Similarly, Si et al. (2019) proved that the homo- and hetero- fermentative LAB inoculants could decrease the NH3-N content and pH during ensiling.
Sequencing of bacteria in fresh native grass and silage samples was carried out by amplicon sequencing. The coverage values for all samples were approximately 0.96. This indicated that the sequencing depth was quite extensive, which justified the probing of characteristics of the bacterial microbial community (Keshri et al., 2019). It is generally believed that these specific operational taxonomic units (OTUs) might be responsible for the differences in silage quality (Yang et al., 2019). In a recent study, a significant reduction in bacterial diversity and species richness was observed as the complex microbial community of FM was eventually replaced by LAB, which dominated the entire fermentation process (Cui et al., 2022). The higher Simpson and Shannon indices for MSL5 and TSL5 and the lower levels exhibited by DSL1 may be due to the higher number and relative abundance of bacteria dominating the fermentation in MSL5 and TSL5, as indicated by the bacterial community composition.
The PCoA plots revealed a clear separation of bacteria in the raw materials and silages of different steppe types, indicating that the bacterial community is altered by the silage technology. The PCoA of silages inoculated with LAB were also separated from the control, which demonstrated a significant effect of the additive on the bacterial community. These results were in agreement with the previous study that reported by Zeng et al. (2020). Therefore, based on the results of α-diversity and β-diversity, the LAB inoculations could influence the microbial diversity and community structure of native grass silage.
As shown, the epiphytic microorganisms could regulates the rate and extent of fermentation. Conversely, the microbial community is formed by the fermentation products. Zhao et al. (2017) concluded that proteobacteria showed the highest relative abundance in the bacterial community of pasture silage feedstock, which is consistent with the results of this study. At the phylum level, the relative abundance of Firmicutes in silage with or without additives increased significantly to become the most important phylum compared to raw materials, while the relative abundance of Proteobacteria decreased significantly, indicating that anaerobic conditions and exogenous additives effectively promote effective acid production by beneficial bacteria in silage and create an acidic environment that inhibits the reproductive activity of spoilage microorganisms (McGarvey et al., 2013).
At the genus level, the dominant epiphytes in fresh native grass were Pantoea and Rosenbergiella, which were similar to the dominant epiphytes in fresh alfalfa (Ogunade et al., 2018) but different from the dominant epiphytes in fresh E. nutans (Ding et al., 2020). It has been suggested that altitude, plant composition and climate can influence the distribution of LAB (Leong et al., 2014). After fermentation process, the portions of Pantoea and Rosenbergiella decreased significantly and were replaced by Lentilactobacillus, Levilactobacillus, Lactiplantibacillus, and Limosilactobacillus in different LAB inoculant treatments, which is in agreement with the results of You et al. (2022). The high sensitivity of Pantoea, Rosenbergiella, and Lactobacillus to the changes in pH may be the main reasons (Li et al., 2022). Furthermore, the abundance of Lentilactobacillus and Lactiplantibacillus increased in four LAB inoculant treatments compared to the CK, indicating that the inoculated strains were not competed out by the microbial populations attached to the plants. This was probably because the inoculated strains dominated and controlled the microbial community during the fermentation process (Wang et al., 2006).
There were significant species-level differences in epiphytic bacteria among the FMs of different steppe types. The dominant species in the raw material of DS was Rosenbergiella nectarea, and the dominant species in both TS and MS was Pantoea agglomerans. This result was different from the results of previous studies (Bao et al., 2016; Guan et al., 2018; Li et al., 2021). The dominant species in the CK treatment of the three steppe types were Levilactobacillus brevis and L. fermentum, which confirmed that harmful epiphytic bacteria in FM were rapidly inhibited under anaerobic environment and low pH stress. There were also some differences in silage containing different LAB species. L. buchneri was the dominant species in BL1 and BL5 treatments. Notably, L. fermentum and L. graminis were not detected among the L1- and L5-attached isolates, mainly because both bacterial species were at the early stages of fermentation and were not present in sufficient numbers to be detected at the late stages of silage fermentation (Beck et al., 1988; Meenakshi Malhotra, 2015). L. plantarum and L. buchneri were the dominant species in LP and LB treatments. This is mainly because, as the fermentation process continues, the temperature of silage sample increases, which usually leads to the transformation of LAB from the homofermentative to the heterofermentative type.
Functional KEGG profiles of bacterial communities were inferred using 16S rRNA gene sequences. The carbohydrate metabolic pathway was enriched after ensiling, which may be because of the fermentation of available carbohydrates into energy and short-chain fatty acids during ensiling (Zhao et al., 2022). The pyruvate metabolism, pentose phosphate pathway, and glycolysis/gluconeogenesis metabolic pathway dominated carbohydrate metabolism in native grass silage treatment of the three steppe types. Du et al. (2022) reported metabolic pathways and their influence on the taste (sweet and sour) and palatability of native grass silage, which is similar to the findings of this study. The enrichment of pyruvate metabolism showed no difference among the silages containing LAB additives. By contrast, the enrichment of pentose phosphate pathway, glycolysis/gluconeogenesis, starch and sucrose metabolism, and amino sugar and nucleotide sugar metabolism increased in the DSLP and TSL5 treatments compared with the other treatments, possibly because of the higher abundance of L. plantarum and Levilactobacillus brevis in these two treatments (Su et al., 2021). Thus, there were showed different abundance in the level 3 of metabolism of carbohydrate metabolism (Table 5). Additionally, the enrichment of starch and sucrose metabolism in the five silage treatments compared with the FM showed different advantages, indicating that the epiphytic LAB utilized the WSC to different degrees in the silage. The enrichment of amino acid metabolism decreased in the TSL5 and DSLP treatments compared with the other treatments. This may be because of two reasons: 1) high CP content (most of the protein is not broken down into NH3-N during fermentation), and 2) higher abundance of L. plantarum, which can promptly decrease pH during the early stages of fermentation and then inhibit amino acid metabolism (Flythe and Russell, 2004).
In summary, bacterial communities of fresh samples of different steppe types of native grasses were found to be dominated by Pantoea agglomerans. The bacterial communities of different steppe types of native grasses with and without the addition of LAB changed significantly after fermentation. The fermentation characteristics of silages supplemented with self-screening strains (L. fermentum and L. graminis) and commercial LAB additives differed considerably due to their different fermentation pathways. The addition of L. fermentum and L. graminis increased the relative abundance of L. buchneri and L. plantarum, reduced the pH and rapidly inhibited undesirable microorganisms. Thus, L. fermentum and L. graminis showed potential to improve the fermentation of native grasses from different steppe types, providing the feasibility of new strains to improve the fermentation of native grass silage by regulating the epiphytic flora in silage to promote the fermentation of LA and AA, improving the fermentation characteristics and metabolic capacity of the microbiota.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA912573.
Author contributions
JB: Conceptualization, Investigation, Visualization, Methodology, Formal analysis, Writing -original draft. GG: Conceptualization, Investigation, Formal analysis. ZW: Supervision, Writing-review and editing. YX: Supervision, Writing-review and editing. MZ: Conceptualization, Investigation, Formal analysis. LS: Conceptualization, Investigation, Formal analysis. YW: Conceptualization, Investigation, Formal analysis. JZ: Conceptualization, Investigation, Formal analysis. YJ: Conceptualization, Methodology, Validation, Investigation, Writing-Review and editing, Funding acquisition. SD: Conceptualization, Methodology, Validation, Investigation, Writing-Review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Laboratory of Forage Cultivation and the Processing and Highly Efficient Utilization of the Ministry of Agriculture, the Key Laboratory of Grassland Resources of the Ministry of Education, and funded by the Program for Technology Project of Inner Mongolia (2020GG0032), the National Dairy Technology Innovation Center (2021-National Dairy Innovation Center-1), China, Education Department of Inner Mongolia Autonomous Region (NJZY21238) Excavation and mechanism of action of low temperature tolerant lactic acid bacteria in natural forage silage in Hulunbuir, and China Agriculture Research System of MOF and MARA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Avila, C. L., Carvalho, B. F., Pinto, J. C., Duarte, W. F., Schwan, R. F. (2014). The use of Lactobacillus species as starter cultures for enhancing the quality of sugar cane silage. J. Dairy Sci. 97 (2), 940–951. doi: 10.3168/jds.2013-6987
Bao, W., Mi, Z., Xu, H., Zheng, Y., Kwok, L. Y., Zhang, H., et al. (2016). Assessing quality of medicago sativa silage by monitoring bacterial composition with single molecule, real-time sequencing technology and various physiological parameters. Sci. Rep. 6, 28358. doi: 10.1038/srep28358
Beck, R., Weiss, N., Winter, J. (1988). Lactobacillus graminis sp. nov., a new species of facultatively heterofermentative Lactobacilli surviving at low pH in grass silage. Syst. Appl. Microbiol. 10 (3), 279–283. doi: 10.1016/s0723-2020(88)80013-4
Borreani, G., Tabacco, E., Schmidt, R. J., Holmes, B. J., Muck, R. E. (2018). Silage review: factors affecting dry matter and quality losses in silages. J. Dairy Sci. 101 (5), 3952–3979. doi: 10.3168/jds.2017-13837
Broderick, G., Kang, J. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63 (1), 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Burns, P., Borgo, M. F., Binetti, A., Puntillo, M., Bergamini, C., Paez, R., et al. (2018). Isolation, characterization and performance of autochthonous spray dried lactic acid bacteria in maize micro and bucket-silos. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02861
Cai, Y., Benno, Y., Ogawa, M., Ohmomo, S., Kumai, S., Nakase, T. J. A., et al. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. AEM 64 (8), 2982–2987. doi: 10.1128/AEM.64.8.2982-2987.1998
Cai, Y., Du, Z., Yamasaki, S., Nguluve, D., Tinga, B., Macome, F., et al. (2020). Influence of microbial additive on microbial populations, ensiling characteristics, and spoilage loss of delayed sealing silage of Napier grass. Asian-Australas J. Anim. Sci. 33 (7), 1103–1112. doi: 10.5713/ajas.19.0471
Cai, Y., Kumai, S., Ogawa, M., Benno, Y., Nakase, T. J. A. (1999). Characterization and identification of pediococcus species isolated from forage crops and their application for silage preparation. Microbiol. eAEM 65 (7), 2901–2906. doi: 10.1128/AEM.65.7.2901-2906.1999
Chen, L., Guo, G., Yu, C., Zhang, J., Shimojo, M., Shao, T. (2015). The effects of replacement of whole-plant corn with oat and common vetch on the fermentation quality, chemical composition and aerobic stability of total mixed ration silage in Tibet. Anim. Sci. J. 86 (1), 69–76. doi: 10.1111/asj.12245
Chen, X., Zhao, X., Wang, H., Yang, Z., Li, J., Suo, H. (2017). Prevent effects of Lactobacillus fermentum HY01 on dextran sulfate sodium-induced colitis in mice. Nutrients 9 (6), 545. doi: 10.3390/nu9060545
Cheng, Q., Li, P., Xiao, B., Yang, F., Li, D., Ge, G., et al. (2020). Effects of LAB inoculant and cellulase on the fermentation quality and chemical composition of forage soybean silage prepared with corn stover. Grassl Sci. 67 (1), 83–90. doi: 10.1111/grs.12289
Chu-Ky, S., Bui, T.-K., Nguyen, T.-L., Ho, P.-H. (2014). Acid adaptation to improve viability and X-prolyl dipeptidyl aminopeptidase activity of the probiotic bacterium Lactobacillus fermentum HA6 exposed to simulated gastrointestinal tract conditions. Int. J. Food Sci. Tech 49 (2), 565–570. doi: 10.1111/ijfs.12338
Cui, X., Yang, Y., Zhang, M., Jiao, F., Gan, T., Lin, Z., et al. (2022). Optimized ensiling conditions and microbial community in mulberry leaves silage with inoculants. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.813363
Ding, Z., Bai, J., Xu, D., Li, F., Zhang, Y., Guo, X. (2020). Microbial community dynamics and natural fermentation profiles of ensiled alpine grass elymus nutans prepared from different regions of the qinghai-Tibetan plateau. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00855
Dong, J., Li, S., Chen, X., Sun, Z., Sun, Y., Zhen, Y., et al. (2022). Effects of Lactiplantibacillus plantarum inoculation on the quality and bacterial community of whole-crop corn silage at different harvest stages. Chem. Biol. Technol. Ag 9 (1), 1–16. doi: 10.1186/s40538-022-00326-y
Du, Z., Sun, L., Lin, Y., Yang, F., Cai, Y. (2022). Using PacBio SMRT sequencing technology and metabolomics to explore the microbiota-metabolome interaction related to silage fermentation of woody plant. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.857431
Du, S., You, S. H., Bao, J., Gegentu, Jia, Y. S., Cai, Y. M. (2019). Evaluation of the growth performance and meat quality of Mongolian lamb fed grass, hay or pellets of inner Mongolian native grass. Small Ruminant Res. 181, 34–38. doi: 10.1016/j.smallrumres.2019.10.008
Du, S., You, S., Jiang, X., Li, Y., Jia, Y. (2022). Dynamics of the fermentation quality and microbiotsa in Ephedra sinica treated native grass silage. J. Appl. Microbiol. 133 (6), 3465–3475. doi: 10.1111/jam.15779
Ennahar, S., Cai, Y., Fujita, Y. (2003). Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 69 (1), 444–451. doi: 10.1128/AEM.69.1.444-451.2003
Flythe, M. D., Russell, J. B. (2004). The effect of pH and a bacteriocin (bovicin HC5) on clostridium sporogenes MD1, a bacterium that has the ability to degrade amino acids in ensiled plant materials. FEMS Microbiol. Ecol. 47 (2), 215–222. doi: 10.1016/S0168-6496(03)00259-9
Fu, J., Sun, Y., Chu, X., Xu, Y., Hu, T. (2014). Exogenous 5-aminolevulenic acid promotes seed germination in elymus nutans against oxidative damage induced by cold stress. PloS One 9 (9), e107152. doi: 10.1371/journal.pone.0107152
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in southwest China. Bioresour Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Guo, L., Wang, X., Lin, Y., Yang, X., Ni, K., Yang, F. (2021b). Microorganisms that are critical for the fermentation quality of paper mulberry silage. Food Energy Secur 10 (4), e304. doi: 10.1002/fes3.304
Guo, X., Zheng, P., Zou, X., Chen, X., Zhang, Q. (2021a). Influence of pyroligneous acid on fermentation parameters, CO2 production and bacterial communities of rice straw and stylo silage. Front. Microbiol. 12, 701434. doi: 10.3389/fmicb.2021.701434
Hashemzadeh-Cigari, F., Khorvash, M., Ghorbani, G. R., Ghasemi, E., Taghizadeh, A., Kargar, S., et al. (2014). Interactive effects of molasses by homofermentative and heterofermentative inoculants on fermentation quality, nitrogen fractionation, nutritive value and aerobic stability of wilted alfalfa (Medicago sativa l) silage. J. Anim. Physiol. Anim. Nutr. (Berl) 98 (2), 290–299. doi: 10.1111/jpn.12079
Hou, M., Gentu, G., Liu, T., Jia, Y., Cai, Y. (2017). Silage preparation and fermentation quality of natural grasses treated with lactic acid bacteria and cellulase in meadow steppe and typical steppe. Asian-Australas J. Anim. Sci. 30 (6), 788–796. doi: 10.5713/ajas.16.0578
Jaipolsaen, N., Sangsritavong, S., Uengwetwanit, T., Angthong, P., Plengvidhya, V., Rungrassamee, W., et al. (2021). Comparison of the effects of microbial inoculants on fermentation quality and microbiota in Napier grass (Pennisetum purpureum) and corn (Zea mays l.) silage. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.784535
Keshri, J., Chen, Y., Pinto, R., Kroupitski, Y., Weinberg, Z. G., Sela Saldinger, S. (2019). Bacterial dynamics of wheat silage. Front. Microbiol. 10, 1532. doi: 10.3389/fmicb.2019.01532
Khota, W., Pholsen, S., Higgs, D., Cai, Y. (2016). Natural lactic acid bacteria population of tropical grasses and their fermentation factor analysis of silage prepared with cellulase and inoculant. J. Dairy Sci. 99 (12), 9768–9781. doi: 10.3168/jds.2016-11180
Kozaki, M., Uchimura, T., Okada, S. J. A. (1992). Experimental manual of lactic acid bacteria (Tokyo, Japan: Asakurasyoten), 34–37.
Leong, K. H., Chen, Y. S., Pan, S. F., Chen, J. J., Wu, H. C., Chang, Y. C., et al. (2014). Diversity of lactic acid bacteria associated with fresh coffee cherries in Taiwan. Curr. Microbiol. 68 (4), 440–447. doi: 10.1007/s00284-013-0495-2
Li, X., Chen, F., Wang, X., Sun, L., Guo, L., Xiong, Y., et al. (2021). Impacts of low temperature and ensiling period on the bacterial community of oat silage by SMRT. Microorganisms 9 (2), 274. doi: 10.3390/microorganisms9020274
Li, D., Ni, K., Pang, H., Wang, Y., Cai, Y., Jin, Q. (2015). Identification and antimicrobial activity detection of lactic acid bacteria isolated from corn stover silage. Asian-Australas J. Anim. Sci. 28 (5), 620–631. doi: 10.5713/ajas.14.0439
Li, P., Tang, X., Liao, C., Li, M., Chen, L., Lu, G., et al. (2021). Effects of additives on silage fermentation characteristic and In vitro digestibility of perennial oat at different maturity stages on the qinghai Tibetan. Microorganisms 9 (11), 2403. doi: 10.3390/microorganisms9112403
Li, H., Zeng, T., Du, Z., Dong, X., Xin, Y., Wu, Y., et al. (2022). Assessment on the fermentation quality and bacterial community of mixed silage of faba bean with forage wheat or oat. Front. Microbiol. 13, 875819. doi: 10.3389/fmicb.2022.875819
Lin, C., Bolsen, K., Brent, B., Hart, R., Dickerson, J., Feyerherm, A., et al. (1992). Epiphytic microflora on alfalfa and whole-plant corn. J. Dairy Sci. 75 (9), 2484–2493. doi: 10.3168/jds.S0022-0302(92)78010-2
Liu, Q., Chen, M., Zhang, J., Shi, S., Cai, Y. (2012). Characteristics of isolated lactic acid bacteria and their effectiveness to improve stylo (Stylosanthes guianensis sw.) silage quality at various temperatures. Anim. Sci. J. 83 (2), 128–135. doi: 10.1111/j.1740-0929.2011.00937.x
Liu, B., Huan, H., Gu, H., Xu, N., Shen, Q., Ding, C. (2019). Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresour Technol. 273, 212–219. doi: 10.1016/j.biortech.2018.10.041
Lu, Y., Li, P., Bai, S., Chen, S., Zhao, M., Gou, W., et al. (2022). Effect of phenyllactic acid on silage fermentation and bacterial community of reed canary grass on the qinghai Tibetan plateau. BMC Microbiol. 22 (1), 83. doi: 10.1186/s12866-022-02499-w
McGarvey, J. A., Franco, R. B., Palumbo, J. D., Hnasko, R., Stanker, L., Mitloehner, F. M. (2013). Bacterial population dynamics during the ensiling of Medicago sativa (alfalfa) and subsequent exposure to air. J. Appl. Microbiol. 114 (6), 1661–1670. doi: 10.1111/jam.12179
Meenakshi Malhotra, I. K. (2015). Identification of Lactobacillus fermentum strains with potential against colorectal cancer by characterizing short chain fatty acids production, anti-proliferative activity and survival in an intestinal fluid: In vitro analysis. J. Bioa Biom 7. doi: 10.4172/1948-593x.1000132
Muck, R. J. (1988). Factors influencing silage quality and their implications for management. J. Dairy Sci. 71 (11), 2992–3002. doi: 10.3168/jds.S0022-0302(88)79897-5
Muck, R. E. (2010). Microbiologia da silagem e seu controle com aditivos. Rev. Bras. Zootecnia 39, 183–191. doi: 10.1590/S1516-35982010001300021
Nkosi, B. D., Meeske, R., Langa, T., Motiang, M. D., Modiba, S., Mkhize, N. R., et al. (2016). Effects of ensiling forage soybean (Glycine max (L.) merr.) with or without bacterial inoculants on the fermentation characteristics, aerobic stability and nutrient digestion of the silage by damara rams. Small Ruminant Res. 134, 90–96. doi: 10.1016/j.smallrumres.2015.12.001
Ogunade, I. M., Jiang, Y., Pech Cervantes, A. A., Kim, D. H., Oliveira, A. S., Vyas, D., et al. (2018). Bacterial diversity and composition of alfalfa silage as analyzed by illumina MiSeq sequencing: effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 101 (3), 2048–2059. doi: 10.3168/jds.2017-12876
Pang, H., Qin, G., Tan, Z., Li, Z., Wang, Y., Cai, Y. (2011). Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst. Appl. Microbiol. 34 (3), 235–241. doi: 10.1016/j.syapm.2010.10.003
Pang, H., Tan, Z., Qin, G., Wang, Y., Li, Z., Jin, Q., et al. (2012). Phenotypic and phylogenetic analysis of lactic acid bacteria isolated from forage crops and grasses in the Tibetan plateau. J. Microbiol. 50 (1), 63–71. doi: 10.1007/s12275-012-1284-5
Pitiwittayakul, N., Bureenok, S., Schonewille, J. T. (2021). Selective thermotolerant lactic acid bacteria isolated from fermented juice of epiphytic lactic acid bacteria and their effects on fermentation quality of stylo silages. Front. Microbiol. 12, 673946. doi: 10.3389/fmicb.2021.673946
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), 1–18. doi: 10.1186/gb-2011-12-6-r60
Si, H., Liu, H., Li, Z., Nan, W., Jin, C., Sui, Y., et al. (2019). Effect of Lactobacillus plantarum and Lactobacillus buchneri addition on fermentation, bacterial community and aerobic stability in lucerne silage. Anim. Prod Sci. 59 (8), 1528–1536. doi: 10.1071/an16008
Sifeeldein, A., Wang, S., Li, J., Dong, Z., Chen, L., Kaka, N. A., et al. (2019). Phylogenetic identification of lactic acid bacteria isolates and their effects on the fermentation quality of sweet sorghum (Sorghum bicolor) silage. J. Appl. Microbiol. 126 (3), 718–729. doi: 10.1111/jam.14123
Sousa, D. O., Hansen, H. H., Nussio, L. G., Nadeau, E. (2019). Effects of wilting and ensiling with or without additive on protein quality and fermentation of a lucerne-white clover mixture. Anim. Feed Sci. Tech 258, 114301. doi: 10.1016/j.anifeedsci.2019.114301
Su, W., Jiang, Z., Hao, L., Li, W., Gong, T., Zhang, Y., et al. (2021). Variations of soybean meal and corn mixed substrates in physicochemical characteristics and microbiota during two-stage solid-state fermentation. Front. Microbiol. 12, 688839. doi: 10.3389/fmicb.2021.688839
Tohno, M., Kobayashi, H., Nomura, M., Uegaki, R., Cai, Y. (2012). Identification and characterization of lactic acid bacteria isolated from mixed pasture of timothy and orchardgrass, and its badly preserved silage. Anim. Sci. J. 83 (4), 318–330. doi: 10.1111/j.1740-0929.2011.00955.x
Van Soest, P. V., Robertson, J. B., Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74 (10), 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, X., Haruta, S., Wang, P., Ishii, M., Igarashi, Y., Cui, Z. (2006). Diversity of a stable enrichment culture which is useful for silage inoculant and its succession in alfalfa silage. FEMS Microbiol. Ecol. 57 (1), 106–115. doi: 10.1111/j.1574-6941.2006.00099.x
Wang, Y., He, L., Xing, Y., Zheng, Y., Zhou, W., Pian, R., et al. (2019a). Dynamics of bacterial community and fermentation quality during ensiling of wilted and unwilted moringa oleifera leaf silage with or without lactic acid bacterial inoculants. mSphere 4 (4). doi: 10.1128/mSphere.00341-19
Wang, Y., He, L., Xing, Y., Zhou, W., Pian, R., Yang, F., et al. (2019b). Bacterial diversity and fermentation quality of moringa oleifera leaves silage prepared with lactic acid bacteria inoculants and stored at different temperatures. Bioresour Technol. 284, 349–358. doi: 10.1016/j.biortech.2019.03.139
Wang, C., He, L., Xing, Y., Zhou, W., Yang, F., Chen, X., et al. (2019). Fermentation quality and microbial community of alfalfa and stylo silage mixed with moringa oleifera leaves. Bioresour Technol. 284, 240–247. doi: 10.1016/j.biortech.2019.03.129
Wang, F., Li, Z., Fu, B., Lu, Y., Liu, G., Wang, D., et al. (2022). Short-term grazing exclusion alters soil bacterial Co-occurrence patterns rather than community diversity or composition in temperate grasslands. Front. Microbiol. 13, 824192. doi: 10.3389/fmicb.2022.824192
Wang, S., Li, J., Zhao, J., Dong, Z., Dong, D., Shao, T. (2022). Effect of epiphytic microbiota from napiergrass and Sudan grass on fermentation characteristics and bacterial community in oat silage. J. Appl. Microbiol. 132 (2), 919–932. doi: 10.1111/jam.15293
Xian, Z., Wu, J., Deng, M., Wang, M., Tian, H., Liu, D., et al. (2022). Effects of cellulase and Lactiplantibacillus plantarum on the fermentation parameters, nutrients, and bacterial community in cassia alata silage. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.926065
Xu, D., Ding, W., Ke, W., Li, F., Zhang, P., Guo, X. (2018). Modulation of metabolome and bacterial community in whole crop corn silage by inoculating homofermentative Lactobacillus plantarum and heterofermentative lactobacillus buchneri. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03299
Xu, D., Ding, Z., Wang, M., Bai, J., Ke, W., Zhang, Y., et al. (2020). Characterization of the microbial community, metabolome and biotransformation of phenolic compounds of sainfoin (Onobrychis viciifolia) silage ensiled with or without inoculation of Lactobacillus plantarum. Bioresour Technol. 316, 123910. doi: 10.1016/j.biortech.2020.123910
Yan, Y., Li, X., Guan, H., Huang, L., Ma, X., Peng, Y., et al. (2019). Microbial community and fermentation characteristic of Italian ryegrass silage prepared with corn stover and lactic acid bacteria. Bioresour Technol. 279, 166–173. doi: 10.1016/j.biortech.2019.01.107
Yang, L., Yuan, X., Li, J., Dong, Z., Shao, T. (2019). Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresour Technol. 275, 280–287. doi: 10.1016/j.biortech.2018.12.067
You, S., Du, S., Ge, G., Wan, T., Jia, Y. (2021). Selection of lactic acid bacteria from native grass silage and its effects as inoculant on silage fermentation. Agron. J. 113 (4), 3169–3177. doi: 10.1002/agj2.20720
You, J., Zhang, H., Zhu, H., Xue, Y., Cai, Y., Zhang, G. (2022). Microbial community, fermentation quality, and in vitro degradability of ensiling caragana with lactic acid bacteria and rice bran. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.804429
Yuan, X., Li, J., Dong, Z., Shao, T. (2020). The reconstitution mechanism of napier grass microiota during the ensiling of alfalfa and their contributions to fermentation quality of silage. Bioresour Technol. 297, 122391. doi: 10.1016/j.biortech.2019.122391
Yuan, X., Yu, C., Shimojo, M., Shao, T. (2012). Improvement of fermentation and nutritive quality of straw-grass silage by inclusion of wet hulless-barley distillers’ grains in Tibet. Asian-Australas J. Anim. Sci. 25 (4), 479–485. doi: 10.5713/ajas.2011.11435
Zeng, T., Li, X., Guan, H., Yang, W., Liu, W., Liu, J., et al. (2020). Dynamic microbial diversity and fermentation quality of the mixed silage of corn and soybean grown in strip intercropping system. Bioresour Technol. 313, 123655. doi: 10.1016/j.biortech.2020.123655
Zhang, Q., Wu, B., Nishino, N., Wang, X., Yu, Z. (2016). Fermentation and microbial population dynamics during the ensiling of native grass and subsequent exposure to air. Anim. Sci. J. 87 (3), 389–397. doi: 10.1111/asj.12427
Zhang, Q., Wu, Z., Yu, Z., Na, R. (2018). Effect of lactic acid bacteria inoculants on fermentation characteristics and aerobic stability of three native grasses in Mongolian steppe. Grassl Sci. 64 (3), 199–206. doi: 10.1111/grs.12197
Zhang, Q., Yu, Z., Wang, X. (2015). Isolating and evaluating lactic acid bacteria strains with or without sucrose for effectiveness of silage fermentation. Grassl Sci. 61 (3), 167–176. doi: 10.1111/grs.12097
Zhao, X., Liu, J., Liu, J., Yang, F., Zhu, W., Yuan, X., et al. (2017). Effect of ensiling and silage additives on biogas production and microbial community dynamics during anaerobic digestion of switchgrass. Bioresour Technol. 241, 349–359. doi: 10.1016/j.biortech.2017.03.183
Keywords: native grass, steppe types, lactic acid bacteria, silage quality, bacterial community
Citation: Bao J, Ge G, Wang Z, Xiao Y, Zhao M, Sun L, Wang Y, Zhang J, Jia Y and Du S (2023) Effect of isolated lactic acid bacteria on the quality and bacterial diversity of native grass silage. Front. Plant Sci. 14:1160369. doi: 10.3389/fpls.2023.1160369
Received: 22 February 2023; Accepted: 02 June 2023;
Published: 06 July 2023.
Edited by:
Denise Tieman, University of Florida, United StatesReviewed by:
Everlon Cid Rigobelo, São Paulo State University, BrazilCláudia Braga Pereira Bento, Universidade Federal dos Vales do Jequitinhonha e Mucuri, Brazil
Copyright © 2023 Bao, Ge, Wang, Xiao, Zhao, Sun, Wang, Zhang, Jia and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yushan Jia, anlzX25tQHNpbmEuY29t; Shuai Du, ZHVzaHVhaV9ubUBzaW5hLmNvbQ==
 Jian Bao
Jian Bao Gentu Ge
Gentu Ge Zhijun Wang
Zhijun Wang Yanzi Xiao2
Yanzi Xiao2 Muqier Zhao
Muqier Zhao Lin Sun
Lin Sun Yushan Jia
Yushan Jia Shuai Du
Shuai Du