
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 12 April 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1158965
 Syed Sarfaraz Hussain1,2†
Syed Sarfaraz Hussain1,2† Manzar Abbas3†
Manzar Abbas3† Sammar Abbas4†
Sammar Abbas4† Mingke Wei2
Mingke Wei2 Ahmed H. El-Sappah3,5
Ahmed H. El-Sappah3,5 Yuhan Sun1
Yuhan Sun1 Yun Li1*
Yun Li1* Arthur J. Ragauskas6,7,8*
Arthur J. Ragauskas6,7,8* Quanzi Li2*
Quanzi Li2*Alternative splicing (AS) in plants plays a key role in regulating the expression of numerous transcripts from a single gene in a regulatory pathway. Variable concentrations of growth regulatory hormones and external stimuli trigger alternative splicing to switch among different growth stages and adapt to environmental stresses. In the AS phenomenon, a spliceosome causes differential transcriptional modifications in messenger RNA (mRNAs), resulting in partial or complete retention of one or more introns as compared to fully spliced mRNA. Differentially expressed proteins translated from intron-retaining messenger RNA (mRNAir) perform vital functions in the feedback mechanism. At the post-transcriptional level, AS causes the remodeling of transcription factors (TFs) by the addition or deletion of binding domains to activate and/or repress transcription. In this study, we have summarized the specific role of AS in the regulation of gene expression through repression and activation of the transcriptional regulatory network under external stimuli and switch among developmental stages.
Recent studies showed that alternative splicing (AS) is highly pervasive in plants (Reddy et al., 2013), and introns of more than 60% genes undergo this process (Syed et al., 2012). AS plays a key role in the regulation of gene regulatory networks to help plants adapt to environmental fluctuations (Syed et al., 2012). Protein expression in a specific quantity and at a specific time is crucial for the growth and development of eukaryotic organisms. Protein synthesis is regulated by multiple layers of regulation like cell signaling, assembly of spliceosome and transcription factors, transcription, export of mature mRNA to the cytoplasm, and translation (Thatcher et al., 2016). AS plays a fundamental role in the expression of multiple numbers of mRNA and, subsequently, proteins from fewer genes. In recent studies, it has been shown that AS in multicellular organisms causes significant changes in protein isoforms (Stamm et al., 2005). AS of intron-retaining genes contributes to increasing the coding potential and gene expression regulation through multiple mechanisms such as nonsense-mediated decay (NMD) and microRNA-mediated gene regulation (Reddy et al., 2013; El-Sappah et al., 2021).
AS alters the sequences of functional domains of a protein which shows a different function in a gene regulatory network (Severing et al., 2012). In response to severe external stimuli such as heat, salinity, and shade, plants perform extensive alterations in genetic expression and transcriptome diversity via AS to adapt to harsh conditions for their survival (Ding et al., 2020). Kirby (2016) observed that an intense pulsed light treatment of Arabidopsis thaliana in the dark stimulates extensive AS to obscure their metabolism. In another study, Tai et al. proved that AS affects lipid biosynthesis in Picea mariana by retention of one intron out of four, which resulted in termination in ORF at a premature termination codon (Tai et al., 2007).
Coordination between alternative splicing and transcription is an important phenomenon in the regulation of gene expression (Godoy Herz and Kornblihtt, 2019). A fungal pathogen Trichoderma affects the roots of tomatoes which stimulates cytosine methylation, resulting in the activation of AS-regulated differential gene expression (De Palma et al., 2019). Beyond the important role of AS in the increased diversity of proteome by producing novel transcript combinations, presumably, many transcript isoforms have a negative influence over protein structure and function, which suggests its role in the post-transcriptional regulation of gene expression (Campbell et al., 2006). Wang et al. discovered that alternative splicing causes an increased level of snRNA transcripts in pollens which suggests its unique role in the regulation of snRNA biosynthesis in plants (Wang et al., 2020). In this review article, we have summarized recent advances in determining the specific role of AS in the transcriptional regulation of gene expression in plants under the influence of different environmental conditions.
The initiation of transcription and post-transcriptional modifications are subject to genetic and epigenetic factors. Epigenetic factors cause modifications such as chromatin remodeling, DNA methylation, and RNA-associated gene silencing to switch on/off genetic regulatory elements. Transcription factors (TFs) are comprised of cis and trans elements (Sperling, 2007) which are recognized by spliceosomes to perform post-transcriptional modifications. Spliceosome is a complex structure that consists of a group of five (U1, U2, U4, U5, and U6) small nuclear ribonucleoproteins (snRNPs) and several auxiliary proteins such as HnRNP proteins, SR proteins, mnRNP proteins, exon junction complex (EJC), and non-snRNP proteins responsible for the splicing of transcripts (Wahl et al., 2009). The assembly of spliceosome and excision of introns consists of the following steps: (i) the recognition of the splicing site by snRNPs (U1, U2, U4, U5, and U6) and other auxiliary factors (Staley and Woolford, 2009), (ii) the assembly of the spliceosome, (iii) the 5´ splice site cleavage and lariat formation, and finally (iv) the 3´ splice site cleavage and exon ligation (Figure 1) (Jurica and Moore, 2003). After ligation of exons, all components of the spliceosome are disassembled for the assembly of the next spliceosome complex and the splicing of Pre-mRNA.
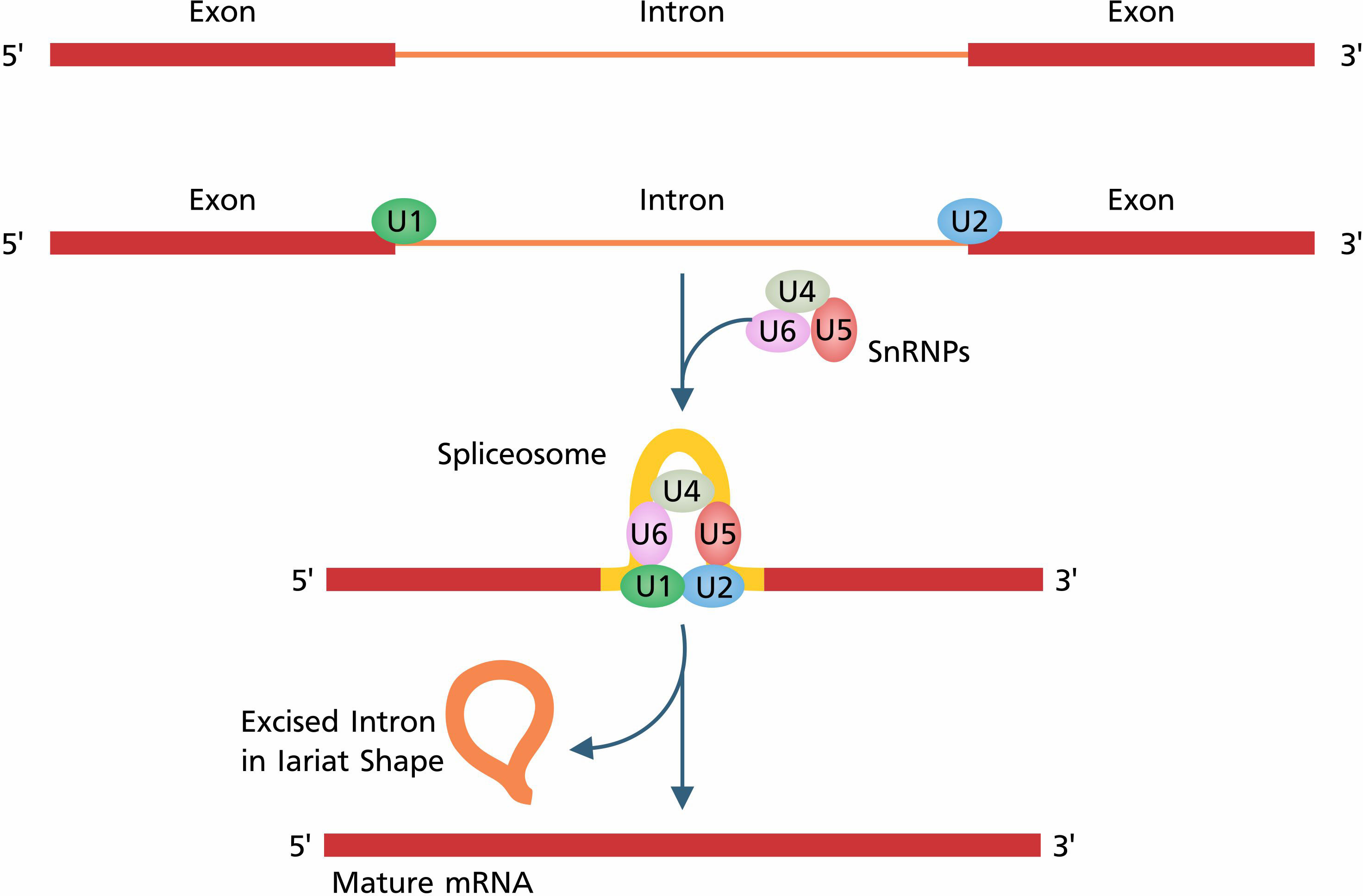
Figure 1 The basic process of constitutive splicing starts with the recognition of splice sites by the spliceosome. U1 and U2 are attached to the 5´ splice site and 3´ splice site, respectively. Splice site recognition is followed by the complete assembly of the spliceosome. After complete assembly of the spliceosome, splicing occurs in two main steps: first, 5´ splice site cleavage and lariat formation, and then, 3´ splice site cleavage and exon ligation.
The precise selection and inclusion of specific exons in mature mRNA are predominantly controlled by cis and trans components which determine protein types or proteoforms to perform specific metabolic processes. Cis and trans factors are comprised of pre-mRNA sequences and protein regulators, respectively. Notably, a clear distinction between splicing factors and elements of constitutive and alternative splicing is under discovery. It is hypothesized that the selection of exon might be influenced by the relative functional strength of splicing sites (Matlin et al., 2005). On the basis of position and function, cis-regulatory elements are divided into the following four categories: exonic splicing silencers (ESSs), exonic splicing enhancers (ESEs), intronic splicing silencers (ISSs), and intronic splicing enhancers (ISEs). Recent studies revealed that despite retaining splicing sites, pseudo-exons do not go under splicing and ESSs/ESEs play their role in determining authentic and pseudo-exons (Zhang and Chasin, 2004).
The serine and arginine-rich (SR) proteins play a pivotal role in the functioning of both constitutive and regulated splicing (Tacke and Manley, 1999). SR proteins are also involved in post-splicing activities such as mRNA translation, nonsense-mediated mRNA decay, and the export of mRNA to an extranuclear matrix (Long and Caceres, 2009). In pre-mRNA splicing, recognition and recruitment of U1AF (U1 auxiliary factor) splicing factor at 5’ and U2AF splicing factor at 3’ splicing sites is of primary importance, which is also regulated by SR protein via protein–protein interaction (Wu and Maniatis, 1993). SR proteins have two functional domains; an N-terminus RNA binding domain which is composed of multiple RNA recognition motifs (RRMs) and an arginine-serine rich C-terminus domain. RRMs initiate sequence-specific RNA binding, and RS domain performs splicing activity (Shen and Green, 2004). ISSS and ESSs are binding sequences of HETEROGENEOUS NUCLEAR RIBONUCLEO PROTEINS (hnRNPs) that bind with RNA by RRM-type and KH-type binding domains (Krecic and Swanson, 1999; Smith and Valcárcel, 2000). The competition between the binding of hnRNPA1 and one of the essential SR protein splicing factors SF2/ASF determines the shift in site selection (Figure 2). SF2/ASF regulates binding of U1 whereas hnRNPA1 inhibits binding of U1 snRNP binding on 5´ splicing site (Eperon et al., 2000). ESSs change the splicing site recognition by inhibiting the use of intron-proximal 5´ and 3´ splicing sites. This alteration of splicing site selection leads to the regulation of intron retention in mature mRNA under AS (Wang et al., 2006).
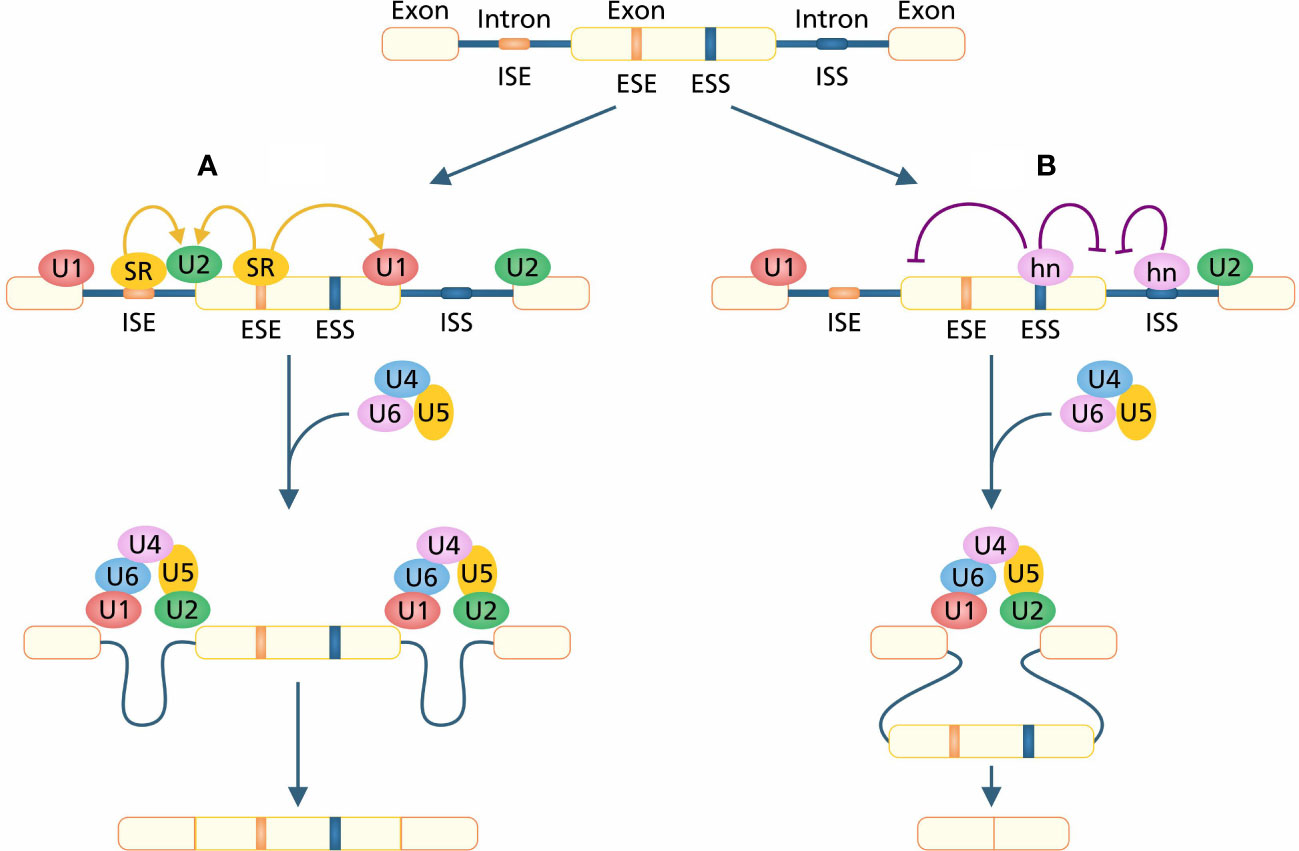
Figure 2 (A) The constitutive splicing in which SR proteins are attached on enhancer sites and SR proteins provide sites for the attachment of U1 and U2 on 5´ and 3´ splice sites, respectively. After complete assembly of the spliceosome, introns are excised out and exons ligated to generate mature mRNA. (B) Silencers play a role in changing the splice site selection. hnRNPs are attached to silencers, inhibiting the attachment of SR proteins which causes a shift in site selection. A shift in site selection leads to the skipping of exons and the generation of exon-missed mRNA.
The Gene regulatory network (GRN) under TFs plays a pivotal role in numerous cellular functions. Certain models of transcriptional regulation have been proposed including context-based binding of multiple transcriptional factors (TFs) and post-transcriptional modifications (PTMs) (Schacht et al., 2014). AS affects the regulation of transcription via the inclusion and exclusion of binding domains, a modification in the stability of transcripts, and protein activity (Garcia et al., 2004). TFs harbor intrinsically disordered proteins (IDPs) and precursor RNAs which are subjected to AS. The coeffects of AS, IDPs, and PTMs cause modifications in proteins involved in cell signaling and cell fate, and, consequently, interrupt the gene regulatory network (Figure 3) (Niklas et al., 2015). In Medicago sativa, the heat shock transcription factor (MsHSF1) is comprised of three introns and four exons which are transcribed into five different isoforms under AS (He et al., 2007). Only MsHSF1b binds with the target gene promoter because all the rest carry premature termination codons and are low-abundance. Isoform MsHSF1b is comprised of the 1st and 4th exons while the other four transcripts retained intervening sequences along with the 1st and 4th exons, which lead to nonsense-mediated mRNA decay (He et al., 2007). Similarly, PtrSND1-A2(WND1B) is a secondary wall thickening associated with TF which is transcribed into two isoforms PtrSND1-A2 (PtrWND1B-s) and PtrSND1-A2ir (PtrWND1B-I) and both are antagonistic in their function. The overexpression of PtrWND1B-s resulted in a thick cell wall, while the overexpression of PtrWND1B-I resulted in a thin cell wall (Zhao et al., 2014). So, AS plays a key role in the transcriptional regulatory network via repression or activation of TFs.
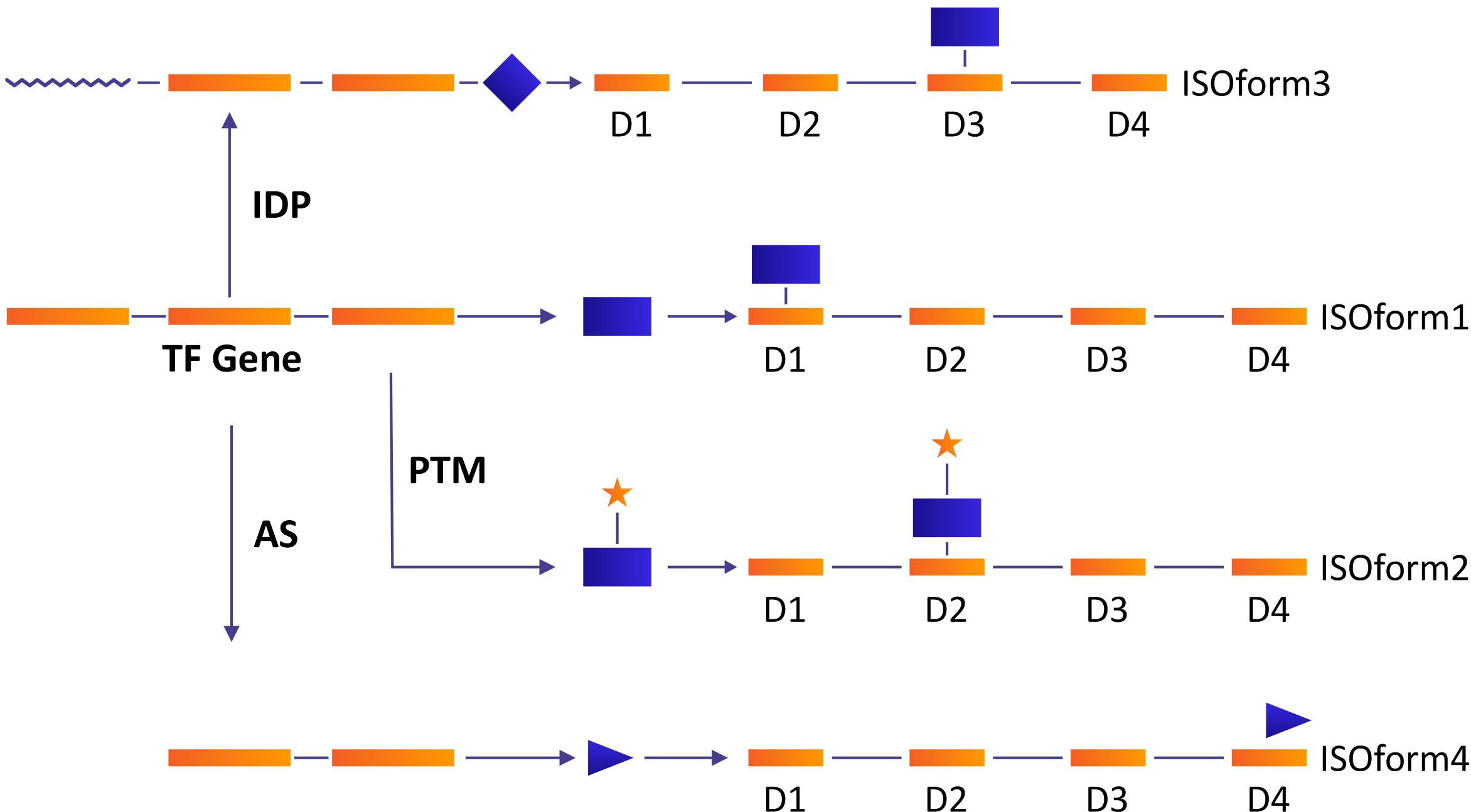
Figure 3 A model of the role of IDP, PTM, and AS in the modulation of gene regulatory network which resulted in modification of protein synthesis. Under normal conditions, transcription factor proteins bind to the promoter of the target gene with extremely high affinity (Isoform 1). The variant as a result of IDPs, PTMs, and AS may change its binding ability and bind to different domains, resulting in a variety of isoforms. A variation in TF protein by PTM (e.g., phosphorylation) may result in the binding ability and production of isoform 2. Isoform 3 may be produced as a result of AS (exclusion of one exon and retention of the intron), and IDP may result in the synthesis of isoform 4.
Dimerization modulates the activities of TFs. Homodimers increase the DNA-binding specificity and affinity of TFs, while heterodimers diversify the affinities and specificities of TFs (Kosugi and Ohashi, 2002). Protein-protein interaction and signal transduction are two important functions of dimerization that create functional diversity in various biological and physical processes by different combinations of proteins having distinct regulatory functions (Klemm et al., 1998). AS is also involved in the generation of splicing variants having variable dimerization properties. A biochemically distinct mechanism of negative regulation is the formation of heterodimers by competitive inhibition of small proteins (siPEP) generated by AS (Seo et al., 2011a).
Programmed cell death and secondary cell wall (SCW) formation are two responsible phenomena for wood formation. Wood, which is a primary source of pulping, energy, and wood products, is composed of a secondary cell wall having major components like lignin, cellulose, and hemicelluloses. NAC-MYB is a major complex of transcriptional factors which is involved in the biosynthesis of the secondary cell wall (Nakano et al., 2015). In Populus 6031 and Eucalyptus 2987, mRNAs were transcribed by AS, which shows the key role of AS in wood formation (Xu et al., 2014). AS-mediated NAC (for NAM, ATAF1/2, and CUC2) transcription factors regulate the biosynthesis of SCW in Populus. PtrSND1-A2 is one of the four family members of Secondary Wall-Associated NAC Domain 1 (SND1) TFs which are involved in the activation of genes associated with cell wall biosynthesis. PtrSND1-A2ir is a splice variant retaining a second intron that lacks in binding domain but can still dimerize. (Li et al., 2012). Reportedly, PtrSND1-A2ir (Table 1) represses all members of SND1 and VND6 including Vascular-Related NAC-Domain 6 C1 (PtrVND6-C1) through heterodimerization except PtrSND1-A2 (Lin et al., 2017). Similarly, PtrVND6-C1ir, a member of the PtrVND6-C1 family suppresses all members of the PtrSND1 family including PtrSND1-A2 but has no effect on its cognate PtrVND6-C1. This reciprocal regulation within the gene family suggests a high control of alternative splicing in the regulation of wood formation.
Among environmental stresses, cold is a major stress which affects the growth and development of plants. Plants have developed various strategies and mechanisms to cope with cold stress (Holzhaider et al., 2011). Transcription factors basic-helix-loop-helix (bHLH) play a key role in growth, development, and abiotic stress. In the Eucalyptus hybrid (E. nitens × E. globulus), 44 bHLH transcripts were transcribed under the effect of AS and two unique isoforms during cold acclimation to counter cold stress (Apablaza et al., 2021). Late Elongated Hypocotyl (LHY) and Circadian Clock-Associated 1 (CCA1) coexpress to regulate the transcription of C-Repeat Binding Factor (CBF) to enhance cold tolerance (Dong et al., 2011). Two splice variants transcribed from CCA1 are full-size CCA1α and truncated CCA1β. CCA1α lacks N-terminus DNA binding domain but retains a protein-protein interaction domain for dimer formation. The splice variant CCA1β regulates the transcription of CCA1 and also makes non-functional CCA1α-CCA1β and LHY-CCA1β heterodimers to suppress CCA1α and LHY. The cold stress inhibits AS and restores transcription to full-size CCA1α to enhance cold tolerance (Park et al., 2012).
Plants adjust their flowering time in response to numerous environmental and developmental signals under the effect of AS. Among 12688 differentially expressed genes (DEGs) in an early flowering mutant of trifoliate orange (Poncirus trifoliata), 16343 AS events were identified among which 3´ splice sites in AS were dominant (Ai et al., 2012). In Arabidopsis, CONSTANS (CO) TFs were involved in the regulation of the expression of floral integrator flowering locus T (FT). CO is transcribed into fully spliced COα and intron retaining COβir under the effect of AS. COβir isoform lacked C-terminus DNA binding domain which resulted in premature termination. The E3 group of ubiquitin ligases regulates the stability of CO, while osmotically responsive genes 1 (HOS1) and constitutive photomorphogenic 1 (COP1) degrade CO early in the morning and at night, respectively. Whereas, flavin-binding Kelch repeat F-box1 (FKF1) keeps a stable expression of CO in the late afternoon during long days. COβir makes heterodimer with COα to inhibit its regular binding with the promoter of CO during the daytime to let COP1 and HOS1 suppress CO. In the afternoon during long days, a relative accumulation of both isoforms increases and FKF1 stabilizes the regulation of COα to overcome Coβ-modulated inhibition (Figure 4A) (Gil et al., 2017). This result shows that AS ensures the sustainability of diurnal accumulation dynamics in the photoperiodic flowering of Arabidopsis.
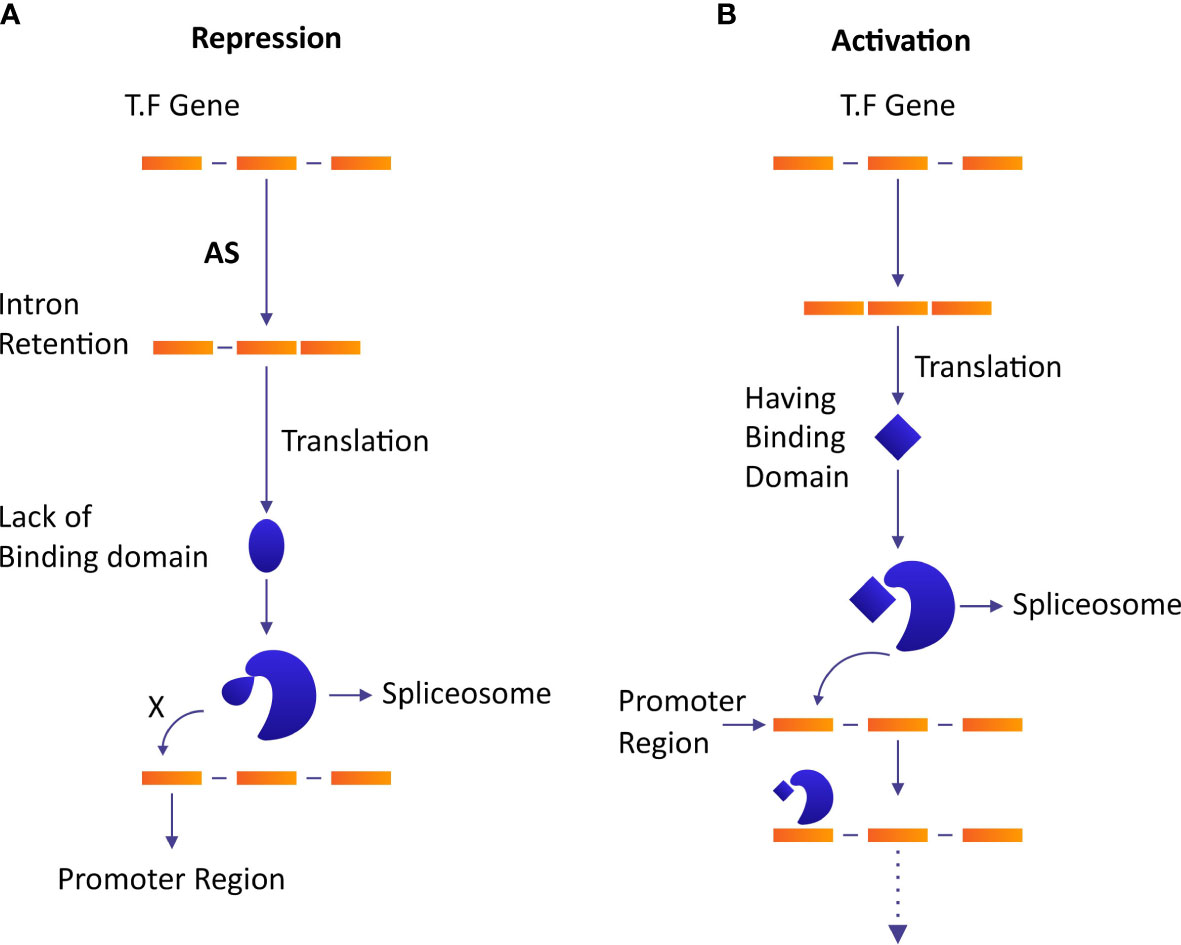
Figure 4 The role of AS in the repression and activation of the transcriptional regulatory network. Homodimers increase the DNA-binding specificity and affinity of TFs, while heterodimers diversify the affinities and specificities of TFs. (A) AS causes a modification of the structure of TF due to intron retention and changes its binding ability which results in an inhibition of the binding of spliceosome complex and represses the regulation of gene expression. (B) The activation of GTN in absence of AS.
Worldwide, the important ornamental white flower plant Chrysanthemum sometimes produces red color fluorites due to the accumulation of anthocyanin color pigment (Li et al., 2021). Flavonoid biosynthetic genes are regulated by special ternary transcriptional factor complexes of MYB-Basic, helix-loop-helix, and WD40 (MBW) (Xu et al., 2015). It has been revealed that mutation in the promotor region of dihydroflavonol-4-reductase (DFR) and alternative splicing of DFR leads to alteration in the accumulation of anthocyanin pigment in the solanum group (Wang et al., 2022). In the chrysanthemum transcriptional factor, cmbHLH2 generates two transcripts: fully transcribed cmbHLH2full and truncated short-length cmbHLH2short. Results showed that short-length alternatively spliced transcript did not have the ability to combine at the promoter region of the cmDFR gene, resulting in the failure in activation of the MBW complex and leading to an inactivation in the biosynthesis of the anthocyanin pigment. These results showed that alternative splicing plays the role of a repressor in the pigmentation of anthocyanin (Lim et al., 2021).
Plant starch is a non-structural carbohydrate that is a major source of carbohydrates during the absence of photosynthesis (Streb and Zeeman, 2012). In Arabidopsis, starch metabolism is regulated by INDERMINATE DOMAIN 14 (IDD14) TF (Seo et al., 2011b). The full-length IDD14α transcript binds with the promoter region of the Qua-Quine Starch (QQS) gene to regulate starch metabolism by generating competitive inhibitors. Under cold stress, the first intron retaining IDD14βir is transcribed because of AS, which lacks a DNA-binding domain and makes a heterodimer with IDD14α. The formation of IDD14α-IDD14βir heterodimer restricts the transcription of the QQS gene and results in starch accumulation. This negative regulation of starch metabolism is a cold stress adaptation strategy to store starch under dark conditions in order to maintain a certain amount of starch to cope with cold stress during day time (Seo et al., 2011b).
A very complex gene regulatory network comprised of hundreds of regulatory factors is involved in the regulation of leaf senescence in plants (Woo et al., 2019). Leaf senescence is a very important stage of development for plant fitness that is regulated by senescence-associated NAC family transcription factors (Sen-NAC TFs) (Wang et al., 2021). AS of Sen-NAC TFs provides an additional layer of post-transcriptional regulation, which maintains the mechanism of leaf senescence. PtRD26, a member of Sen-NAC TFs, is spliced into an intron-retaining variant PrRD26ir in Populus tomentosa. PtRD26ir negatively regulated the functions of senescence-related TFs by repressing their binding ability, which resulted in the delay in leaf senescence (Wang et al., 2021).
In plants, abiotic stress factors cause severe biochemical changes which affect pathways involved in the biosynthesis of proteins. In western poplar, 164 out of 15087 AS-mediated abiotic stress-specific isoforms were identified (Filichkin et al., 2018). Heat shock proteins (Hsp70 & Hsp83) play a pivotal role in heat and heavy metals stress tolerance (El Sappah et al., 2021). Endoplasmic reticulum chaperone OsBiP1 is an unfolded protein response marker that contributes to protein folding (Foresti et al., 2003). In rice, OsHSFA2d regulates the expression of OsBiP1 to regulate signaling pathways of unfolded protein and heat shock response which is spliced into two main variants OsHSFA2dI and OsHSFA2dII by retaining 94 and 270 bp introns, respectively. The OsHSFA2dI variant only activates the transcription of OsBip1 under abiotic stress (Cheng et al., 2015). In Arabidopsis, heat shock factor A2 (HsfA2) is a key regulator of heat stress. Under moderate heat stress (37°C), a 31 nt long intron retaining transcript HsfA2-IIir is transcribed under the effect of AS (Table 2). Under severe heat stress (42°C to 45°C), 1a intron retaining HsfA2-IIIir is expressed which activates the transcription of a small truncated isoform S-HsfA2 responsible for the self-regulation of the transcription of HsfA2 (Liu et al., 2013).
An interesting work by Ling et al., 2018 investigated a heat shock tolerance strategy of Arabidopsis named heat-shock memory. IR alternative splicing plays a key role in the tolerance of heat in Arabidopsis, but a dramatic change by the plant was observed in response to a second exposure of the plant to non-lethal heat shock. Plants exposed to non-lethal heat shock retained a high level of IR and do not respond to the second heat shock which is favorable for the normal growth and development of the plant. This study also emphasizes the important role of AS in the heat tolerance of plants.
Trace metal elements’ stress has serious effects and consequences on plant growth and animal health upon ingestion. Metal tolerance proteins (MTPs) play an important role in heavy metal stress resistance and metal accumulation (El-Sappah et al., 2021; Abbas et al., 2022). AS regulates gene expression via intron-specific promoters by increasing the diversity of MTPs transcripts which results in a significant halt in the accumulation of trace metal elements in plant tissues. In bean (Phaseolus vulgaris), metal-responsive elements binding the transcription factor PvMTF-1 regulate the transcription of abiotic stress-responsive gene PvSR2 (Yang et al., 2015). In this case, an intronic promoter transcribes PvMTF-1 to complement metal-responsive elements by hybridization at the upstream region of the PvSR2 promoter to regulate its expression (Figure 4B) (Yang et al., 2015).
It has been noticed that SR proteins play an important role in the splicing of mRNA and regulation of alternative splicing (Long and Caceres, 2009). Zhao et al. (2021) studied 24 SR genes of populus trichocarpa and found that these 24 PtSR genes could undergo alternative splicing and generate 45 transcripts under abiotic and hormone stresses. Among these 24 PtSR genes, the overexpression of PtSCL30 negatively regulates the cold and salt stress-responsive genes by making changes in the AS of these genes. In Arabidopsis, 95 transcripts of SR genes were transcribed from 15 genes under the effect of AS (Palusa et al., 2007). Heat stress stimulates AS of several SR genes while hormone stress stimulates AS of only three SR genes (Wang et al., 2021). Protein sequencing analysis revealed that truncated functional domains with distinct functions in different polypeptide isoforms were produced under the effect of AS (Palusa et al., 2007). SR1 gene in Arabidopsis is transcribed into five different isoforms out of which only one is fully spliced. SR1, SR1B, and SR1C were transcribed by targeting alternative 3´ splice sites, and the remaining two SR1D and SR1E were transcribed by targeting 5´ splice sites of intron 9. The higher ratio of SR1B/SR1 under high temperatures revealed the crucial role of SR1B in adaptation to heat stress (Lazar and Goodman, 2000).
AS emerges as a source of diversity in gene expression to cope with biotic and abiotic stresses. Under AS, numerous transcripts are spliced from a single transcript due to (i) choices of alternative site selection by the spliceosome, (ii) intron retention, (iii) selection of alternate exons, and (iv) premature stop codons. In this study, we summarized the interaction of different abiotic stress factors with AS and invited further studies to investigate the signaling pathways involved. Over time, the global environment is adversely changing and is affecting the weather pattern. This change in the climate is having a bad effect on plant growth and all other forms of life. These insights will be helpful in better understanding the adaptation of plants to environmental stresses and the evolution of multicellular organisms. Furthermore, it will be helpful to improve the withholding capacity of plants under different environmental stresses. We also summarized the specific roles of AS involved in the transcriptional regulatory network. Notably, TFs and SR proteins are involved in the constitutive and alternative splicing of different transcripts. Further deep insights into AS of SR proteins in different plant species and the effects of their isoforms on the splicing of mRNAs will open new avenues in understanding the diversity of gene expression. Deep insights into AS involved in cell wall biosynthesis will provide a precise benchmark for the genetic modification of ligno-cellulosic contents in wood.
The increase in the number of genes as a result of alternative splicing makes it more important to understand its mechanism and function in regulation. Numerous examples of AS responding to various stresses like heat, cold, hormones, and heavy metals have been observed in plants, which plays an important role in the adaptation by producing diverse forms of proteins. AS also diversifies protein production by the suppression and activation of the transcriptional regulation of genes and the splicing of SR genes. There is a need to explore events and functions of AS in plants, which can be helpful for the development of desired growth characteristics and adaptation to different stresses in plants.
Conceptualization: SH, MA, and QL; Writing-original draft and drawing figures: SH, MA, SA, YS, YL, and AE-S; Editing and proof reading: AR, YL, and QL; Writing the final manuscript: SH and MA. All authors reviewed and approved the final submission.
This work was supported by the National Natural Science Foundation of China (32071787 and 32171769) and the Scientific Research Project of Yibin University (XJ2020007601).
We are very grateful to the kind administration of Beijing Forestry University, Chinese Academy of Forestry, and Yibin University, China for providing us with such a prestigious and well-equipped platform for research and development. We are also grateful to the kind administration of the National Natural Science Foundation of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbas, M., Li, Y., Elbaiomy, R. G., Yan, K., Ragauskas, A. J., Yadav, V., et al. (2022). Genome-wide analysis and expression profiling of SlHsp70 gene family in solanum lycopersicum revealed higher expression of SlHsp70-11 in roots under Cd(2+) stress. Front. Biosci. (Landmark Ed) 27 (6), 186. doi: 10.31083/j.fbl2706186
Ai, X. Y., Lin, G., Sun, L. M., Hu, C. G., Guo, W. W., Deng, X. X., et al. (2012). A global view of gene activity at the flowering transition phase in precocious trifoliate orange and its wild-type [Poncirus trifoliata (L.) raf.] by transcriptome and proteome analysis. Gene 510 (1), 47–58. doi: 10.1016/j.gene.2012.07.090
Apablaza, H., Solís, M., Conejera, D., Fonseca, A., Cid, J., Tarifeño-Saldivia, E., et al. (2021). bHLH transcription factors undergo alternative splicing during cold acclimation in a eucalyptus hybrid. Plant Mol. Biol. Rep 40, 310–326. doi: 10.1007/s11105-021-01313-7
Brummell, D. A., Chen, R. K. Y., Harris, J. C., Zhang, H., Hamiaux, C., Kralicek, A. V., et al. (2011). Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. J. Exp. Bot. 62 (10), 3519–3534. doi: 10.1093/jxb/err043
Campbell, M. A., Haas, B. J., Hamilton, J. P., Mount, S. M., Buell, C. R. (2006). Comprehensive analysis of alternative splicing in rice and comparative analyses with arabidopsis. BMC Genomics 7, 327. doi: 10.1186/1471-2164-7-327
Cheng, Q., Zhou, Y., Liu, Z., Zhang, L., Song, G., Guo, Z., et al. (2015). An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. (Stuttg) 17 (2), 419–429. doi: 10.1111/plb.12267
De Palma, M., Salzano, M., Villano, C., Aversano, R., Lorito, M., Ruocco, M., et al. (2019). Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus trichoderma harzianum Horticul. Res. 6, 5. doi: 10.1038/s41438-018-0079-1
Ding, Y., Wang, Y., Qiu, C., Qian, W., Xie, H., Ding, Z. (2020). Alternative splicing in tea plants was extensively triggered by drought, heat and their combined stresses. PeerJ 8, e8258. doi: 10.7717/peerj.8258
Dong, M. A., Farré, E. M., Thomashow, M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the c-repeat binding factor (CBF) pathway in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 (17), 7241–7246. doi: 10.1073/pnas.1103741108
El Sappah, A. H., Elbaiomy, R. G., Li, J., Yan, K., Wang, Y., Zhao, X., et al. (2021). Genome-wide identification and expression analysis of metal tolerance protein (MTP) gene family in medicago truncatula under a broad range of heavy metals stress. Front. Genet. 12, 713224. doi: 10.3389/fpls.2021.705249
El-Sappah, A. H., Yan, K., Huang, Q., Islam, M. M., Li, Q., Wang, Y., et al. (2021). Comprehensive mechanism of gene silencing and its role in plant growth and development Front. Plant Sci. 12, 705249. doi: 10.3389/fgene.2021.713224
Eperon, I. C., Makarova, O. V., Mayeda, A., Munroe, S. H., Cáceres, J. F., Hayward, D. G., et al. (2000). Selection of alternative 5' splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell Biol. 20 (22), 8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000
Filichkin, S. A., Hamilton, M., Dharmawardhana, P. D., Singh, S. K., Sullivan, C., Ben-Hur, A., et al. (2018). Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00005
Foresti, O., Frigerio, L., Holkeri, H., de Virgilio, M., Vavassori, S., Vitale, A. (2003). A phaseolin domain involved directly in trimer assembly is a determinant for binding by the chaperone BiP. Plant Cell 15 (10), 2464–2475. doi: 10.1105/tpc.013052
Garcia, J., Gerber, S. H., Sugita, S., Südhof, T. C., Rizo, J. (2004). A conformational switch in the piccolo C2A domain regulated by alternative splicing. Nat. Struct. Mol. Biol. 11 (1), 45–53. doi: 10.1038/nsmb707
Gil, K. E., Park, M. J., Lee, H. J., Park, Y. J., Han, S. H., Kwon, Y. J., et al. (2017). Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in arabidopsis photoperiodic flowering. Plant J. 89 (1), 128–140. doi: 10.1111/tpj.13351
Godoy Herz, M. A., Kornblihtt, A. R. (2019). Alternative splicing and transcription elongation in plants. Front. Plant Sci. 10 (309). doi: 10.3389/fpls.2019.00309
He, Z. S., Xie, R., Zou, H. S., Wang, Y. Z., Zhu, J. B., Yu, G. Q. (2007). Structure and alternative splicing of a heat shock transcription factor gene, MsHSF1, in Medicago sativa. Biochem. Biophys. Res. Commun. 364 (4), 1056–1061. doi: 10.1016/j.bbrc.2007.10.131
Holzhaider, J. C., Sibley, M. D., Taylor, A. H., Singh, P. J., Gray, R. D., Hunt, G. R. (2011). The social structure of new caledonian crows. Anim. Behav. 81 (1), 83–92. doi: 10.1016/j.anbehav.2010.09.015
Jurica, M. S., Moore, M. (2003). Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12 (1), 5–14. doi: 10.1016/S1097-2765(03)00270-3
Kirby, J. N. (2016). The role of mindfulness and compassion in enhancing nurturing family environments. Clin. Psychology: Sci. 23 (2), 142. doi: 10.1111/php.12550. Practice.
Klemm, J. D., Schreiber, S. L., Crabtree, G. R. (1998). Dimerization as a regulatory mechanism in signal transduction. Annu. Rev. Immunol. 16, 569–592. doi: 10.1146/annurev.immunol.16.1.569
Kosugi, S., Ohashi, Y. (2002). DNA Binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30 (3), 337–348. doi: 10.1046/j.1365-313x.2002.01294.x
Krecic, A. M., Swanson, M. S. (1999). hnRNP complexes: Composition, structure, and function. Curr. Opin. Cell Biol. 11 (3), 363–371. doi: 10.1016/s0955-0674(99)80051-9
Lazar, G., Goodman, H. M. (2000). The arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 42 (4), 571–581. doi: 10.1023/a:1006394207479
Li, Q., Lin, Y.-C., Sun, Y.-H., Song, J., Chen, H., Zhang, X.-H., et al. (2012). Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in populus trichocarpa. Proc. Natl. Acad. Sci. 109 (36), 14699–14704. doi: 10.1073/pnas.1212977109
Li, Y., Liu, X., Li, F., Xiang, L., Chen, K. (2021). The isolation and identification of anthocyanin-related GSTs in chrysanthemum. Horticulturae 7 (8), 231. doi: 10.3390/horticulturae7080231
Lim, S. H., Kim, D. H., Jung, J. A., Lee, J. Y. (2021). Alternative splicing of the basic helix–loop–helix transcription factor gene CmbHLH2 affects anthocyanin biosynthesis in ray florets of chrysanthemum (Chrysanthemum morifolium). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.669315
Lin, Y. J., Chen, H., Li, Q., Li, W., Wang, J. P., Shi, R., et al. (2017). Reciprocal cross-regulation of VND and SND multigene TF families for wood formation in populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 114 (45), E9722–e9729. doi: 10.1073/pnas.1714422114
Ling, Y., Serrano, N., Gao, G., Atia, M., Mokhtar, M., Woo, Y. H., et al. (2018). Thermopriming triggers splicing memory in arabidopsis. J. Exp. Bot. 69 (10), 2659–2675. doi: 10.1093/jxb/ery062
Liu, J., Sun, N., Liu, M., Liu, J., Du, B., Wang, X., et al. (2013). An autoregulatory loop controlling arabidopsis HsfA2 expression: Role of heat shock-induced alternative splicing. Plant Physiol. 162 (1), 512–521. doi: 10.1104/pp.112.205864
Long, J. C., Caceres, J. F. (2009). The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417 (1), 15–27. doi: 10.1042/bj20081501
Matlin, A. J., Clark, F., Smith, C. W. J. (2005). Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6 (5), 386–398. doi: 10.1038/nrm1645
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00288
Niklas, K. J., Bondos, S. E., Dunker, A. K., Newman, S. A. (2015). Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Front. Cell Dev. Biol. 3 (8). doi: 10.3389/fcell.2015.00008
Palusa, S. G., Ali, G. S., Reddy, A. S. (2007). Alternative splicing of pre-mRNAs of arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 49 (6), 1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x
Park, M. J., Seo, P. J., Park, C. M. (2012). CCA1 alternative splicing as a way of linking the circadian clock to temperature response in arabidopsis. Plant Signal Behav. 7 (9), 1194–1196. doi: 10.4161/psb.21300
Penfield, S., Josse, E. M., Halliday, K. J. (2010). A role for an alternative splice variant of PIF6 in the control of arabidopsis primary seed dormancy. Plant Mol. Biol. 73 (1-2), 89–95. doi: 10.1007/s11103-009-9571-1
Posé, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503 (7476), 414–417. doi: 10.1038/nature12633
Qin, Z., Wu, J., Geng, S., Feng, N., Chen, F., Kong, X., et al. (2017). Regulation of FT splicing by an endogenous cue in temperate grasses. Nat. Commun. 8, 14320. doi: 10.1038/ncomms14320
Reddy, A. S. N., Marquez, Y., Kalyna, M., Barta, A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25 (10), 3657–3683. doi: 10.1105/tpc.113.117523
Schacht, T., Oswald, M., Eils, R., Eichmüller, S. B., König, R. (2014). Estimating the activity of transcription factors by the effect on their target genes. Bioinformatics 30 (17), i401–i407. doi: 10.1093/bioinformatics/btu446
Seo, P. J., Hong, S. Y., Kim, S. G., Park, C. M. (2011a). Competitive inhibition of transcription factors by small interfering peptides. Trends Plant Sci. 16 (10), 541–549. doi: 10.1016/j.tplants.2011.06.001
Seo, P. J., Kim, M. J., Ryu, J. Y., Jeong, E. Y., Park, C. M. (2011b). Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2 (1), 303. doi: 10.1038/ncomms1303
Severing, E. I., van Dijk, A. D., Morabito, G., Busscher-Lange, J., Immink, R. G., van Ham, R. C. (2012). Predicting the impact of alternative splicing on plant MADS domain protein function. PloS One 7 (1), e30524. doi: 10.1371/journal.pone.0030524
Shen, H., Green, M. R. (2004). A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16 (3), 363–373. doi: 10.1016/j.molcel.2004.10.021
Smith, C. W., Valcárcel, J. (2000). Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25 (8), 381–388. doi: 10.1016/s0968-0004(00)01604-2
Sperling, S. (2007). Transcriptional regulation at a glance. BMC Bioinf. 8 (6), S2. doi: 10.1186/1471-2105-8-S6-S2
Staley, J. P., Woolford (2009). Assembly of ribosomes and spliceosomes: Complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 21 (1), 109–118. doi: 10.1016/j.ceb.2009.01.003
Stamm, S., Ben-Ari, S., Rafalska, I., Tang, Y., Zhang, Z., Toiber, D., et al. (2005). Function of alternative splicing. Gene 344, 1–20. doi: 10.1016/j.gene.2004.10.022
Streb, S., Zeeman, S. C. (2012). Starch metabolism in arabidopsis. Arabidopsis book/American Soc. Plant Biologists 10, 435–445. doi: 10.1199/tab.0160
Syed, N. H., Kalyna, M., Marquez, Y., Barta, A., Brown, J. W. (2012). Alternative splicing in plants–coming of age. Trends Plant Sci. 17 (10), 616–623. doi: 10.1016/j.tplants.2012.06.001
Tacke, R., Manley, J. L. (1999). Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11 (3), 358–362. doi: 10.1016/s0955-0674(99)80050-7
Tai, H. H., Williams, M., Iyengar, A., Yeates, J., Beardmore, T. (2007). Regulation of the beta-hydroxyacyl ACP dehydratase gene of picea mariana by alternative splicing. Plant Cell Rep. 26 (1), 105–113. doi: 10.1007/s00299-006-0213-7
Thatcher, S. R., Danilevskaya, O. N., Meng, X., Beatty, M., Zastrow-Hayes, G., Harris, C., et al. (2016). Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170 (1), 586–599. doi: 10.1104/pp.15.01267
Wahl, M. C., Will, C. L., Lührmann, R. (2009). The spliceosome: Design principles of a dynamic RNP machine. Cell 136 (4), 701–718. doi: 10.1016/j.cell.2009.02.009
Wang, X., Chen, X., Luo, S., Ma, W., Li, N., Zhang, W., et al. (2022). Discovery of a DFR gene that controls anthocyanin accumulation in the spiny Solanum group: Roles of a natural promoter variant and alternative splicing. Plant J. 111 (4), 1096–1109. doi: 10.1111/tpj.15877
Wang, W., Pu, X., Yang, S., Feng, Y., Lin, C., Li, M., et al. (2020). Alternative splicing of DSP1 enhances snRNA accumulation by promoting transcription termination and recycle of the processing complex Proc. Nat. Acad. Sci. 117, 33, 20325–20333. doi: 10.1073/pnas.2002115117
Wang, Z., Xiao, X., Van Nostrand, E., Burge, C. B. (2006). General and specific functions of exonic splicing silencers in splicing control. Mol. Cell 23 (1), 61–70. doi: 10.1016/j.molcel.2006.05.018
Wang, H.-L., Zhang, Y., Wang, T., Yang, Q., Yang, Y., Li, Z., et al. (2021). An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in populus. Plant Cell 33 (5), 1594–1614. doi: 10.1093/plcell/koab046
Woo, H. R., Kim, H. J., Lim, P. O., Nam, H. G. (2019). Leaf senescence: Systems and dynamics aspects Ann. Rev. Plant Biol. 70, 347–376. J.A.R.o.P.B. doi: 10.1146/annurev-arplant-050718-095859
Wu, J. Y., Maniatis, T. (1993). Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75 (6), 1061–1070. doi: 10.1016/0092-8674(93)90316-i
Xu, W., Dubos, C., Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20 (3), 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu, P., Kong, Y., Song, D., Huang, C., Li, X., Li, L. (2014). Conservation and functional influence of alternative splicing in wood formation of populus and eucalyptus. BMC Genomics 15 (1), 780. doi: 10.1186/1471-2164-15-780
Yang, W., Bei, X., Liu, M., Qi, X. (2015). Intronic promoter-mediated feedback loop regulates bean PvSR2 gene expression. Biochem. Biophys. Res. Commun. 463 (4), 1097–1101. doi: 10.1016/j.bbrc.2015.06.064
Zhang, X. H., Chasin, L. A. (2004). Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 18 (11), 1241–1250. doi: 10.1101/gad.1195304
Zhao, Y., Sun, J., Xu, P., Zhang, R., Li, L. (2014). Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 164 (2), 765–776. doi: 10.1104/pp.113.231134
Keywords: alternative splicing, transcriptional regulation, intron retaining, gene regulatory network, spliceosome
Citation: Hussain SS, Abbas M, Abbas S, Wei M, El-Sappah AH, Sun Y, Li Y, Ragauskas AJ and Li Q (2023) Alternative splicing: transcriptional regulatory network in agroforestry. Front. Plant Sci. 14:1158965. doi: 10.3389/fpls.2023.1158965
Received: 04 February 2023; Accepted: 13 March 2023;
Published: 12 April 2023.
Edited by:
Changlin Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Guiyan Yang, Northwest A&F University, ChinaCopyright © 2023 Hussain, Abbas, Abbas, Wei, El-Sappah, Sun, Li, Ragauskas and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Li, eXVubGlAYmpmdS5lZHUuY24=; Arthur J. Ragauskas, YXJhZ2F1c2tAdXRrLmVkdQ==; Quanzi Li, bGlxekBjYWYuYWMuY24=
†These authors have contributed equally to this work and share the first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.