
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 08 May 2023
Sec. Plant Development and EvoDevo
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1157619
This article is part of the Research TopicMolecular Basis of Stem Cells Underlying Plant VitalityView all 7 articles
Several Kalanchoë species reproduce asexually by forming plantlets in the leaf crenulations. Some species produce plantlets incessantly via somatic embryogenesis and organogenesis, whereas others exclusively develop plantlets after leaf detachment, presumably through organogenesis. SHOOT MERISTEMLESS (STM), which mediates SAM functions, appears to be involved in Kalanchoë plantlet formation, suggesting that meristem genes may be essential for plantlet formation. However, the genetic regulatory network for establishing and maintaining plantlet primordia in Kalanchoë remains elusive. Here, we showed that meristem genes were differentially expressed in the leaf crenulations of K. pinnata during plantlet development after leaf detachment. The regulatory interactions among these meristem genes are largely conserved in K. pinnata crenulations. Moreover, transgenic antisense (AS) plants with lower expression of these key meristem genes formed significantly fewer plantlets with some morphological defects, suggesting that the meristem genes play an important role in plantlet formation and development. Our research revealed that key meristem genetic pathways were co-opted to the leaf margin to facilitate the unique asexual reproduction mechanism in K. pinnata. This also highlights how evolutionary tinkering invents new structures such as epiphyllous buds and plantlets by rewiring pre-existing genetic pathways.
Plants maintain a source of undifferentiated stem cells in the shoot apical meristem (SAM) and root apical meristem (RAM), which differentiate for growth and tissue self-renewal (Sablowski, 2004; Zhu, 2017). Plants have also secondary stem cell niches, such as axillary buds and epiphyllous buds (EBs), which are established post-embryonically in the existing organs, remain dormant and under stress or other external conditions acquire SAM or RAM functions (Dinneny and Benfey, 2008; Ten Hove et al., 2015). Environmental cues, positional cues and phytohormones influence the fate of stem cells (Gaillochet and Lohmann, 2015; Sengupta et al., 2018). The establishment and maintenance of stem cells in secondary niches involve the reacquisition of stem cell identity by a group of cells, followed by morphogenetic processes. Transcriptional regulators, hormones and mobile signals from neighbouring cells can revert differentiation and maintain stem cell identity (Gaillochet and Lohmann, 2015; Galli and Gallavotti, 2016). Previous studies have reported the presence of undifferentiated cells within the parenchyma of the leaves of most succulent plants, which enabled the regeneration of new plants from leaf-cuttings (Gorelick, 2015).
The SAM activity requires a balance between the number of cells recruited for organ formation and the cells to self-sustain (Heidstra and Sabatini, 2014). The main regulatory mechanism in the SAM to preserve stem cells includes the WUSCHEL (WUS) and CLAVATA (CLV) gene negative feedback loop (Schoof et al., 2000; Somssich et al., 2016; Fletcher, 2018). In Arabidopsis thaliana, WUS is required for the maintenance of undifferentiated cells in the SAM (Mayer et al., 1998; Bäurle and Laux, 2005). WUS is expressed in the organising centre (OC) and triggers the expression of CLV3 in the central zone (CZ), which simultaneously restricts WUS to the OC (Bäurle and Laux, 2005). CLV3 binds to CLV1 leucine-rich repeat-receptor-like kinase (LRR-RLK) heterodimers with CLV2 LRRreceptor-like protein (RLP) and its co-receptor CORYNE (CRN) protein to inhibit WUS expression (Fletcher, 2018). It has been shown that CLV3 also binds to CLV2-CRN to inhibit WUS independently from CLV1 (Bleckmann et al., 2009; Guo et al., 2010). This CLV/WUS negative feedback loop determines the area of the WUS domain, which appears to be a well-conserved mechanism within the plant kingdom (Bäurle and Laux, 2005; Somssich et al., 2016). In addition, Arabidopsis HAIRY MERISTEM1 (HAM1) and HAM2 genes are conserved cofactors of WUS/WUSCHEL-LIKE HOMEOBOX (WOX) proteins, which are also essential for SAM maintenance (Zhou et al., 2015). HAM1 and HAM2 are co-expressed with WUS in the CZ, however, it has been shown that WUS can only activate CLV3 in the absence of HAM proteins, avoiding WUS repression in the CZ by CLV3 (Yadav et al., 2011). LRR-RLK BARELY ANY MERISTEM 1 (BAM1) sharing 81% similarity to CLV1 (DeYoung et al., 2006), functions analogously to CLV1, binding directly to CLV3. Double knock-out of CLV1 and BAM1 showed stem cell overproliferation, similar to the phenotypes observed in CLV3 loss-of-function (Shinohara and Matsubayashi, 2015).
The ectopic expression of WUS induced the formation of stem-like cells and somatic embryos in Arabidopsis (Schoof et al., 2000). A SAM-like function can be also acquired by the ectopic expression of WUS in the leaves, triggered by the presence of cytokinins (Gordon et al., 2009). wus mutants showed termination of the meristematic activity after forming a few organs due to the lack of stem cell renewal (Endrizzi et al., 1996). In contrast, clv mutants displayed stem cell over-proliferation and disorganised stem cell arrangement (Brand, 2000; Schoof et al., 2000). It has been shown that the external application of cytokinins significantly downregulates CLV1 expression, resulting in upregulated WUS expression, suggesting that WUS might be activated by cytokinins (Lindsay et al., 2006). However, cytokinins application to clv3 and clv1 loss-of-function mutants in Arabidopsis induced WUS expression, suggesting a CLV-independent mechanism of WUS regulation (Gordon et al., 2009).
The KNOX homeodomain SHOOT MERISTEMLESS (STM) gene is essential for SAM maintenance along with WUS (Clark et al., 1996; Long et al., 1996; Lenhard et al., 2002). STM knocked-out phenotypes completely abolished SAM formation in Arabidopsis (Long et al., 1996; Aida et al., 1999; Scofield et al., 2014), whereas mild STM downregulation perturbed the SAM organisation and compromised its post-embryonic maintenance (Endrizzi et al., 1996; Long et al., 1996). WUS and STM ectopically expressed in the leaves of Arabidopsis triggered a robust subset of meristem functions, including the WUS/CLV pathway, and thus organogenesis (Gallois et al., 2002). Alternatively, expression analyses have reported that CUP SHAPED COTYLEDON1 and 2 (CUC1 and 2) are required for STM expression during Arabidopsis embryogenesis (Aida et al., 1999). Loss of function mutants of STM and CUC genes share similar phenotypes with fused cotyledons (Endrizzi et al., 1996; Takada et al., 2001; Scofield et al., 2018). cuc1 and 2 double mutants were unable to develop an embryonic SAM, as STM was not activated (Aida et al., 1999; Takada et al., 2001). Postembryonically, CUC2 interacts with PINFORMED1 (PIN1) and miR164 to form serrations in the leaves of Arabidopsis (Nikovics et al., 2006; Bilsborough et al., 2011).
The genus Kalanchoë comprises several species that reproduce vegetatively by forming plantlets in the leaf margins. While some species such as K. pinnata and K. prolifera only form plantlets after leaf excision (induced plantlet-forming species), other Kalanchoë species such as K. daigremontiana and K. laetivirens form plantlets incessantly from pedestal-like structures located in the leaf serrations (Garces et al., 2007). While K. pinnata can reproduce sexually, under stress conditions, K. pinnata reproduces through the formation of plantlets from bud-like structures, known as epiphyllous buds (EBs) on leaves crenulations (Sawhney et al., 1994; Kulka, 2008; Laura et al., 2013). K. pinnata EBs on the leaf crenulations stay dormant until leaf detachment or severe damage which triggers a dormancy-release mechanism forming plantlets through organogenesis (Garces et al., 2007). Maintaining cell pluripotency on the leaves appears to be the basis for this mode of vegetative reproduction (Zhu, 2017), capable of forming new plants. It is speculated that K. pinnata EBs are organised during leaf formation (Naylor, 1932) and remain latent until the leaf is detached from the mother plant (Sawhney et al., 1994; Rajsekhar et al., 2016). Plantlet initiation in inducible plantlet-forming Kalanchoë species is triggered by hormonal influence (Sawhney et al., 1994). In K. marnierianum, exogenous cytokinin applications promoted dormancy in the EBs, inhibiting plantlet formation after leaves were detached (Kulka, 2006). Moreover, the auxin transport inhibitor, 2, 3, 5-triiodobenzoic acid (TIBA) restricted the formation of roots in plantlets of K. marnierianum, suggesting that the root formation depends on auxin signals from the plantlet SAM (Kulka, 2008).
Molecular studies in Kalanchoë have confirmed that STM expression in the leaves is only present in the species that form plantlets on the leaf margins (Garces et al., 2007). STM knockout abolished plantlet formation in K. daigremontiana (Garces et al., 2007). This might suggest that STM establishes meristematic competency in the leaf crenulations of Kalanchoë enabling the plantlet formation. Incessant (constitutive) plantlet-forming species such as K. daigremontiana also ectopically expressed embryo genes, FUSCA3 (FUS3) and an unfunctional mutated version of LEAFY COTYLEDON1 (LEC1), in leaves, which allowed the formation of leaf embryos and subsequently bypass dormancy (Garces et al., 2007). The expression of a functional AtLEC1 in K. daigremontiana severely disrupted the process of plantlet formation, inducing dormancy in foliar embryos and the accumulation of seed-specific oils in the leaf margins (Garces et al., 2014). LEC1 and LEC2 in Arabidopsis induce the accumulation of seed-specific oils to prevent the zygotic embryo from desiccation and are a major C source for germinating seedling (Pelletier et al., 2017). During Arabidopsis embryogenesis, FUS3 works as a transcriptional activator facilitating embryo development and avoiding early seedling transition (Wang and Perry, 2013). FUS3 and LEC2 form a complex, which binds to the auxin biosynthesis gene, YUCCA4 (YUC4), inducing lateral root development in Arabidopsis (Tang et al., 2017).
Here we showed that key meristem genes such as K. pinnata WUS (KpWUS) and K. pinnata STM (KpSTM) play important roles in the organisation and maintenance of stem cells during plantlet formation in an induced plantlet-forming species K. pinnata. We also showed that K. pinnata CLV2 (KpCLV2) delimited KpWUS expression in the epiphyllous buds, and K. pinnata CUC2 (KpCUC2) modulated the STM expression and its downregulation affected the indentation depth in the leaves of K. pinnata, disturbing the formation of EBs. This work highlights the acquisition of meristem competency and key meristem pathways as a key factor to enable vegetative reproduction in the leaves of K. pinnata.
Kalanchoë pinnata wild-type (WT) and antisense (AS) transgenic plants were potted into Levington’s F2 compost (Scott’s Miracle-Gro, UK), perlite (Sinclair Horticulture, Ltd, UK), and Vermiculite (Sinclair Horticulture, Ltd, UK) mix in a 6:1:1 ratio. The plants were grown in a Percival Scientific growth chamber AR-60L at 23°C, illuminated with fluorescent lights in a short-day condition photoperiod 8 hours/16 hours. 1 cm2 explants were dissected from young fully-grown whole leaves and used for plant transformation. For scanning electron microscopy (SEM), toluidine blue staining (TB), immunolocalisation and RT-qPCR, tissues from margins of smaller than 0.5 cm leaves were used.
Total RNA was extracted from 0.3 cm2 tissues at the leaf notches from 3 biological replicates of K. pinnata wild type at different time points after leaf detachment (0, 4, 24 and 48 hours). 0 hours after leaf detachment was used as a control in addition to tissue samples obtained from the mid-section margin of the leaves 48 hours after leaf detachment, where no plantlet formation may occur. RNA extraction and sequencing analysis were performed as described by (Jácome-Blásquez et al. (2022).
KpWUS, KpCLV2, KpSTM, and KpCUC2 clones were isolated using gene-specific primers based on WUS, CLV2, STM and CUC2 orthologs from K. laxiflora and K. fedtschenkoi sequences (found on Phytozome v12.1, JGI, University of California); Supplementary Table S1. A 455 bp fragment of KpWUS located in exon 1 (Accession OQ674700), a 942 bp fragment of KpCLV2 in exon 2 (Accession OQ621804), a 258 bp fragment of KpCUC2 in exon 1 (Accession OQ621805) and a 333 bp fragment of KpSTM in exon 4 (Accession OQ674701) were cloned to make AS constructs. The sequences were amplified and cloned using Q5® High-Fidelity DNA Polymerase (New England Biolabs, USA). The PCR products were cleaned up (Nucleospin® gel and PCR Clean-Up Kit; Macherey-Nagel, Germany) and then ligated into pGEM®-T Easy (Promega, USA). The fragments were ligated using Golden Gate assembly in AS orientation with the cauliflower mosaic virus (CaMV) 35S Promoter and Terminator for KpWUS and KpCLV2 AS constructs, and CaMV 35S Promoter and the Nopaline Synthase (NOS) Terminator for KpCUC2 and KpSTM AS constructs. The constructs were then inserted into a modified pBI121 vector. Subsequently, the constructs were transformed into Escherichia coli strain DH5α for selection. Correct constructs were then transformed into Agrobacterium tumefaciens strain LBA4404 by electroporation and confirmed with culture PCR.
WT K. pinnata plants were transformed with 35S::KpWUS, 35S::KpCLV2, 35S::KpCUC2 and 35S::KpSTM AS as previously described (Garcěs and Sinha, 2009). Transformed Agrobacterium (LB4404 strain) was grown for 48 hours in Luria Bertani (LB) medium without NaCl supplemented with 50 mg/L Rifampicin, 100 mg/L Kanamycin and 100 mg/L Streptomycin on an orbital shaker, at 30°C and 250 rpm in the dark. OD600 was confirmed until 0.5 o.u. was achieved. The cells were centrifuged at 5500 RFC for 15 minutes and resuspended in 0.5 Murashige and Skoog (MS) medium supplemented with 100 μm of acetosyringone. 1 cm2 leaf fragments previously disinfected with absolute ethanol and commercial bleach (13%) were co-cultured for 2 hours in an orbital shaker in the dark. Inoculated leaf tissues were cultured in the dark in MS medium without the addition of antibiotics for two days, afterwards transferred to SIM (shoot-inducing media) supplemented with 100 mg/L TDZ and 10 mg/L IAA and antibiotics 40 mg/L Kanamycin and 500 mg/L Carbenicillin. After two weeks, these were transferred to SIM with 100 mg/L Kanamycin and 500 mg/L Carbenicillin. We did subculture every 15 days for four months. Once leaves were formed, the explants were transferred to RIM (root-inducing media) containing 0.5 MS media supplemented with 30 g/L sucrose, 7.5 g/L agar, and 5.8 pH adjusted. Roots formed after three weeks, and then the plants were established ex vitro plants into a mix of Levington’s F2 compost (Scott’s Miracle-Gro, UK), perlite (Sinclair Horticulture, Ltd, UK), and Vermiculite (Sinclair Horticulture, Ltd, UK) in a 6:1:1 ratio.
Quick DNA prep for PCR protocol was performed to obtain DNA (Weigel and Glazebrook, 2002). PCR was implemented with Q5® High-Fidelity DNA polymerase and BioTaq™ polymerase (Bioline, UK). The primers used were KpWUS, KpCLV2, KpCUC2, KpSTM and M13 (Supplementary Table S2). Successful incorporation of the insert was also confirmed detecting NTPII forward and reverse primers. PCR settings were the ones recommended in the Q5® protocol, with an annealing temperature of 56°C and an extension time of 30 seconds for 35 cycles.
Leaves were excised from the mother plant, arranged on a dry white paper sheet, and kept in a growth chamber. New plantlet regeneration from the leaf primordia was scored every 3 days up to 21 days for WT and AS lines. New plantlets were scored when they became visible, approximately 0.5 mm in length. A total of 25 leaves from 3 - 5 independent AS lines (several leaves from the same plant) were used for this experiment. Plantlet emergence was scored with Microsoft Excel version 16.16.27, and GraphPad Prism 9.2.0 was used to plot the graphs and perform the ANOVA analysis with Dunnett’s Multiple Comparisons.
Scanning electron microscopy (SEM) was performed on leaf crenulations. Fresh tissue was fixed according to the previous protocol (H. Garces & Sinha, 2009). Samples were examined using a Quanta 650 FEG ESEM with an Energy-Dispersive Spectroscopy (EDS) Bruker XFlash® 6/30 SDD by FEI/ThermoFisher SEM.
EBs of WT K. pinnata and individual KpWUS, KpCLV2, KpSTM and KpCUC2 AS lines were excised and frozen in liquid nitrogen. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, USA) according to a modification (Gehrig et al., 2000; Garces and Sinha, 2009a). RNA was treated with RQ1 DNase (Promega, USA) and cDNA synthesis was achieved at 45°C for 1 hour with the Tetro cDNA Synthesis kit (Bioline, UK). The RT-qPCR reaction was prepared using 100 ng of cDNA in triplicate of three biological replicates in a 20 μL reaction containing 10 μL of SensiFAST™ SYBR Hi ROX kit (Bioline, UK) and 1 mM of each primer. Primer sequences are shown in Supplementary Table S2. The reaction was performed in a StepOnePlus™ Real-Time PCR machine with StepOne™ software v2.3. 18S Ribosomal RNA gene was used as a control gene with an annealing temperature of 60°C. The data analysis was done using the Comparative CT method. Microsoft Excel version 16.16.27 was used for data processing and GraphPad Prism 9.2.0 to plot the graphs and the ANOVA analysis with Dunnett’s Multiple Comparisons using WT as the control.
Tissue samples from K. pinnata epiphyllous buds were fixed and sectioned for immunolocalisation according to the previous protocol (Garces and Sinha, 2009b). Following the sectioning on slides, paraplast wax (Sigma, USA) was removed with a 100% histological clearing agent (Histoclear II) (National diagnostics, USA). Sections were rehydrated in ethanol series with a final blocking incubation in phosphate-buffered saline (PBS) buffer with 1% bovine serum albumin (BSA). The slides were incubated with a specific WUS antibody previously examined in K. daigremontiana (Santa Cruz Biotechnology, catalogue number-sc12587, USA; 1:500 dilution) in 1xPBS with 1% BSA for 2 hours at 4°C. After three washes in 1xPBS, slides were incubated with AP-conjugated Donkey Anti-Goat IgG secondary antibody (Promega, USA; 1:400 dilution). The samples were developed in the dark for 35 minutes for colourimetric detection. The slides were washed in 1xPBS and mounted with DPX Mountant for Histology (Sigma-Aldrich, USA). The slides were observed and photographed in a compound microscope GXML 2800 with an integrated camera GXCAM HI Chrome-SMII (GT Vision, UK).
K. pinnata leaf primordia in WT and AS lines were photographed using a GXCAM-Eclipse (0654) Wi-Fi camera, attached to S8 APO Stereo Microscope (Leica, Germany). To photograph whole leaves of K. pinnata in WT and AS lines, a 12-megapixel Ultrawide: f/2.4, Wide: f/1.6 Telephoto: f/2.2 aperture phone camera was used. Immunolocalisation slides were photographed with a GXCAM HI Chrome-SMII attached to a compound microscope GXML 2800 (GT Vision, UK).
To study the participation of meristem genes in EBs and subsequently plantlet formation in K. pinnata leaves, we initially examined the leaf crenulations during leaf development. A newly emerged leaf of a few millimetres long started forming crenulations in the distal region (Figure 1A) without any visible epiphyllous buds (Figures 1B, D). The crenulations presented no superficial formation of the EBs, however, Toluidine Blue (TB) staining sections revealed numerous small cells resembling stem cells (SC) congregated at the crenulation areas (Figure 1C). The crenulations in small (< 1 cm) younger leaves were fully formed (Figure 1E), nevertheless, there was still no evident EBs formation (Figures 1F, H). The TB-stained sections showed congregated small presumptive SC around the crenulations (Figure 1G). Growing leaves (< 5 cm; Figure 1I) presented deeper crenulations. The crenulation area at this stage exhibited subtle differences in colour in comparison with the mid-sections of the leaf (Figure 1J). TB sections revealed deep crenulations with presumptive SC concentrated particularly in the bottom of the crenulation gorge (Figure 1K). However, the crenulations captured in SEM images showed no clear evidence of EBs presence (Figure 1L). In contrast, fully grown (> 5 cm) leaves (Figure 1M) exhibited EBs when observed in a dissecting microscope. At the leaf margin where plantlets are speculated to form, EB consists of a globular protuberance located in the abaxial side of the leaf crenulation (Figure 1N). TB-stained sections in mature leaves showed an elongated EB structure positioned in the centre of the leaf crenulation composed of small SC (Figure 1O). These crenulations showed a protuberance with an emerging globular-shaped EB when observed in SEM (Figure 1P). Following the leaf detachment, plantlet leaf primordia (L1 and L2) emerged from the EBs after approximately 9 days (Figure 2A). After 15 days following leaf detachment, the EB had leaf 1 (L1) enlarged, and leaf 2 (L2) and roots (R) became visible to the naked eye (Figure 2B). 20 days after leaf detachment, plantlets formed more leaves (L3 and L4) while L1, L2 and roots continued to develop (Figure 2C) and grow. Figure 2D illustrates a 25-day-old plantlet. The plantlets have functional leaves with crenulations and elongated roots; however, these remain attached to the excised senescent mother leaves.
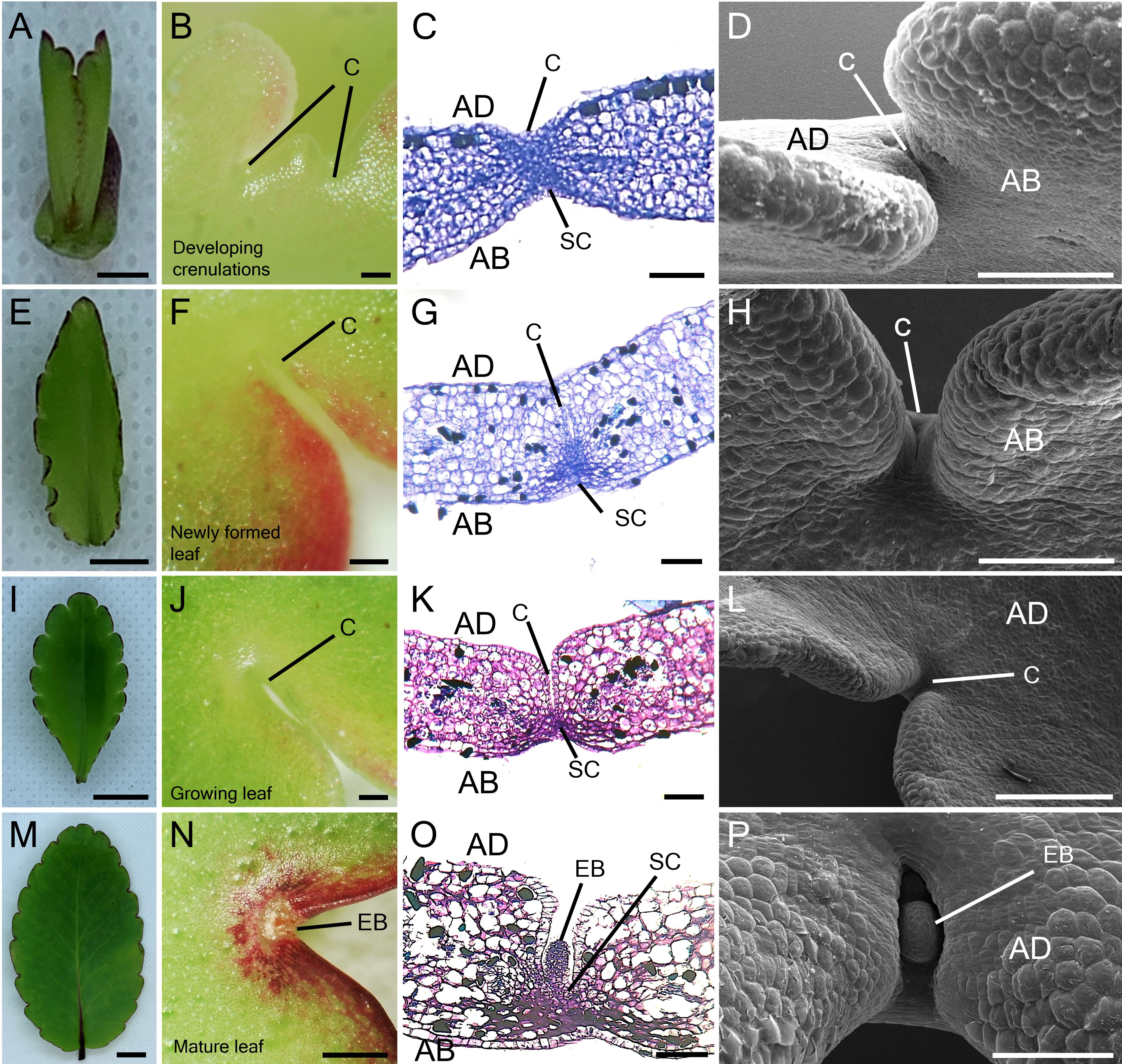
Figure 1 Leaf growth and epiphyllous bud formation in K. pinnata. Morphological appearance of the leaf growth (A, E, I, M). Microscopic imaging of the adaxial side of developing leaf crenulations (B, F, J) and abaxial side (N). TB staining of developing leaf crenulations with small cells resembling stem cells in the crenulation area depicts the presence of stem cells which later develop into EBs (C, G, K, O). SEM imaging of developing crenulations with no superficial epiphyllous bud in the crenulation area (D, H, L) until the leaf is fully developed (P). Scale bar: 1 mm (A, N); 0.5 mm (B, F, J); 0.1 mm (C, G, K); 500 µm (L) and 150 µm (D, H, P). AB; abaxial side, AD; adaxial side, C; Crenulation, SC; Stem cells, EB; Epiphyllous bud.
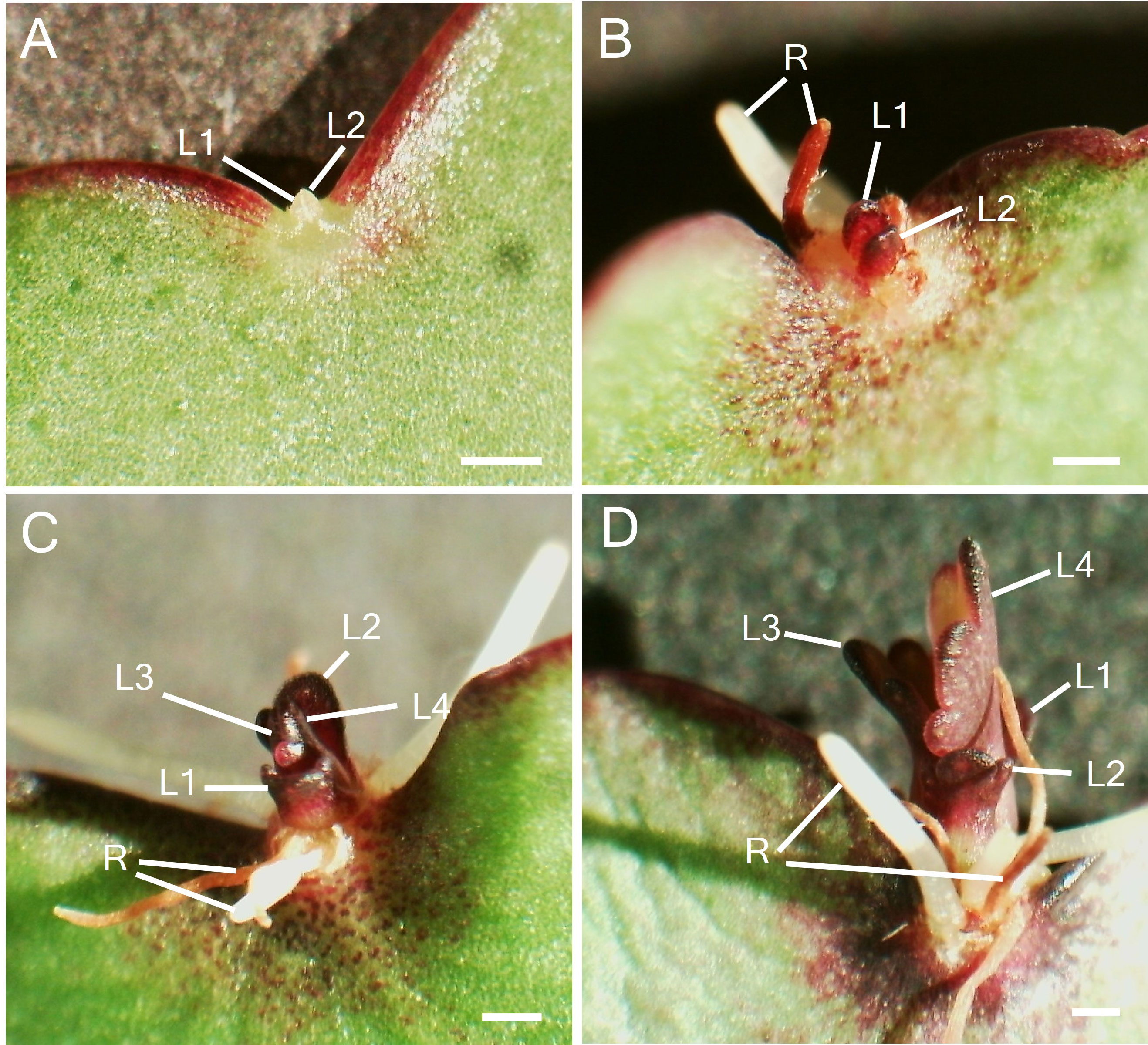
Figure 2 Plantlet formation after leaf detachment. (A). Leaf EB after 9 days of leaf detachment with the emergence of leaves (L1 and L2). (B) EB after 15 days of leaf detachment with enlarged L1 and L2 and root development (C) Emergence of second pair of leaves (L3 and L4) and root growth over time. (D) Growth and development of leaves and roots. Scale bar: 1 mm. L1; Leaf 1, L2; Leaf 2, L3; Leaf 3, L4; Leaf 4, R; root.
To reveal meristem genes involved in plantlet formation, data taken from a previous RNA sequencing analysis was evaluated allowing the detection of early changes in gene expression following the leaf detachment. RNA was extracted from EBs of leaves at 0, 4, 24 and 48 hours, after leaf detachment, and from mid-sections of the detached leaves after 48 hours, with presumably no plantlet formation activity (negative control) (Jácome-Blásquez et al., 2022). Several key meristem genes and closely-related genes were differentially expressed in the EBs after leaf detachment. However, differential expression patterns of these genes varied across time points. From this data, we monitored the expression of meristem genes known to be key in the formation and maintenance of the SAM in Arabidopsis. KpWUS expression was upregulated significantly 24 hours after leaf detachment (p=0.033), followed by downregulation after 48 hours. KpWUS expression was not detected at the mid-sections of the leaves (Figure 3A). WOX genes were also expressed in the EBs. KpWOX4 (p=0.007), KpWOX11 (p=0.033), and KpWOX13 (p=0.013) showed similar expression patterns, being significantly upregulated 48 hours after the leaf detachment. KpWOX4 was undetectable at the mid-sections of the leaves, whereas KpWOX11 and KpWOX13 were slightly expressed (Figures 3B-D). KpHAM3 gene was present in the EBs and the mid-sections of leaves, however, its expression was higher in the EBs (p=0.002), and significantly downregulated 24 hours after leaf detachment (p=0.002; Figure 3E). KpCLV1 and KpCLV2 transcripts were detected in the EBs and downregulated following leaf detachment after 4 and 24 hours (p=0.001; Figure 3F) and 4 hours (Figure 3G), respectively. KpCLAVATA3/ENDOSPERM SURROUNDING REGION-RELATED13 (KpCLE13) was found upregulated in the EBs 48 hours after leaf detachment (p=0.028; Figure 3H). KpCLE16 expression was found first upregulated 4 hours after leaf detachment (p=0.002), then downregulated after 24 and 48 hours (Figure 3I). KpCLE45 was significantly overexpressed in the EBs of attached leaves when compared to the mid-section of the leaves (p=0.032), however, its expression did not vary between the different time points after leaf detachment (Figure 3J). KpBAM1 was upregulated in the EBs when compared to the mid-sections of the leaves. Its expression was downregulated 4 h after the leaf excision (p=0.044; Figure 3K). The KpSTM gene was absent at the mid-section of the leaves but present in the EBs, however, its expression continued unchanging across the different time points after leaf detachment (Figure 3L). KpCUC2 and KpCUC3 transcripts were also present in the EBs. KpCUC2 was completely absent in the mid-sections of the leaves and highly upregulated (p=0.014) after 48 hours following the leaf detachment (Figure 3M). KpCUC3 was upregulated 4 hours (p=0.009) after leaf detachment, following downregulation after 24 hours (Figure 3N). The NAC domain transcription factor ortholog, KpFEZ was detected upregulated in the EBs of attached leaves in comparison to the mid-section (p=0.016). KpFEZ showed a gradual downregulation at 4 hours (p=0.035), 24 hours (p=0.009) and 48 hours (p=0.003) after leaf detachment (Figure 3O).
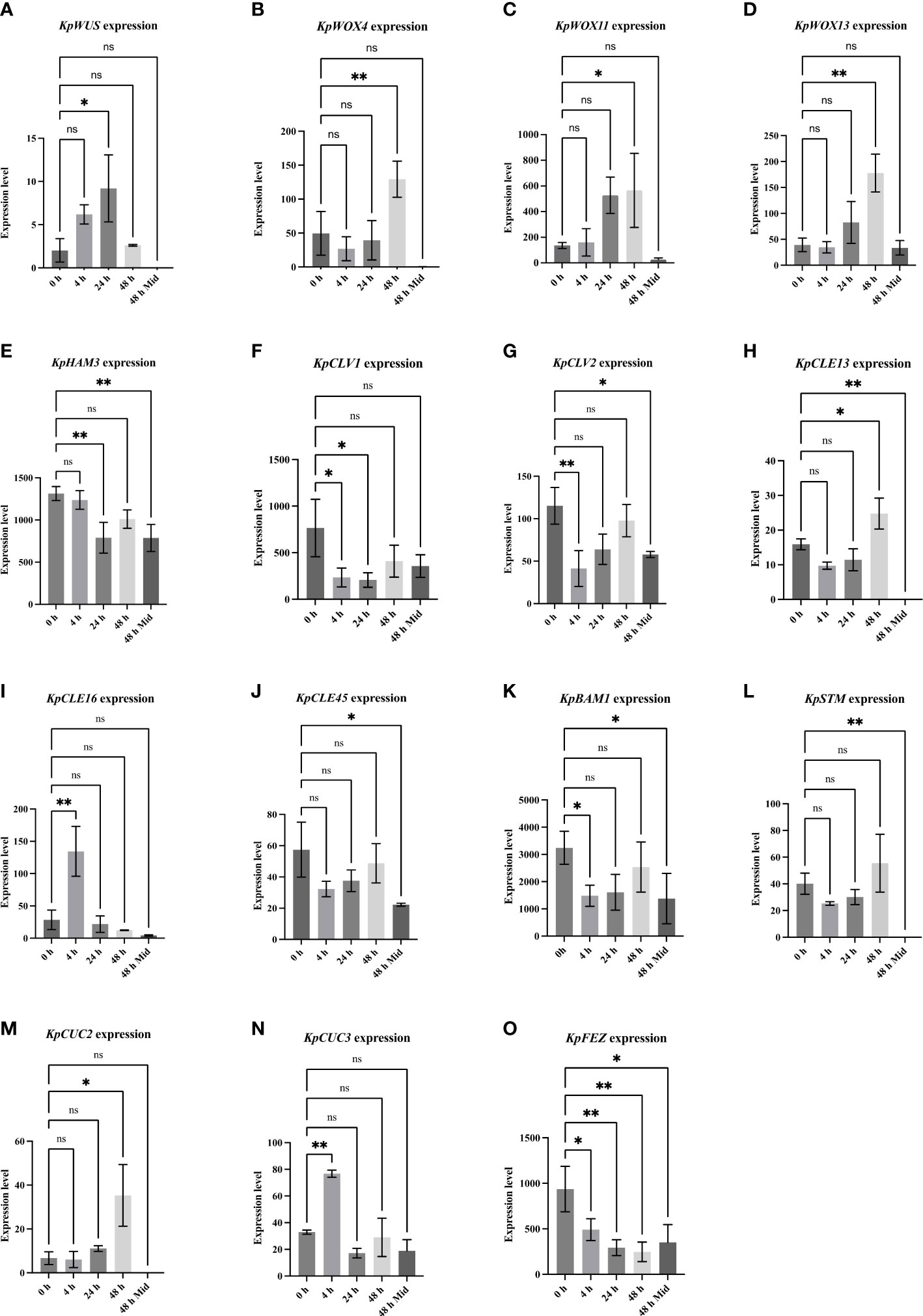
Figure 3 Differential expression of meristem genes at different time points (0, 4, 24, and 48 hours in leaf crenulations and 48 hours in mid-sections of the leaves) after leaf detachment. One-Way ANOVA with Dunnett’s Multiple comparisons, ns (non-significant); * (P-value <0.01); ** (P value <0.001). Errors bars represent the 95% confidence interval. The data was taken from (Jácome-Blásquez et al., 2022).
To elucidate the role of meristem genes in plantlet formation, AS lines were generated by expressing AS fragments of KpWUS, KpCLV2, KpCUC2 and KpSTM under the cauliflower mosaic virus 35S promoter. These genes were chosen as they are key players in the STM pathway which is involved in K. daigremontiana plantlet formation. The AS lines were confirmed by PCR amplifying the transgenes and the NPTII gene (Supplementary Figure S1). We confirmed multiple independent lines: three for KpWUS AS, eight for KpCLV2 AS, five for KpCUC2 AS and five for KpSTM AS (Supplementary Figure S1). K. pinnata WT grows vertically with a decussate leaf arrangement (Figure 4A). Figure 4B shows newly formed leaves emerging from the SAM in WT, and a new leaf pair emerged while the older pair was still growing. The mature leaves of K. pinnata WT have approximately 18 crenulations distributed on the margins (Figure 4C, asterisks). Figure 4D shows a leaf 20 days after detachment with plantlets emerged from the EBs. SEM images show the crenulation with a normal EB development in WT (Figure 4E); a globular-shaped EB was visible in the middle of the leaf crenulation. KpWUS, KdCLV2 and KpCUC2 AS lines had shorter internodes when compared to WT, resulting in shorter plants (Figures 4F, K, P). These AS plants also showed delayed leaf emergence; the youngest developing leaves were barely visible when the previous pair of leaves were already mature (Figures 4G, L, Q, arrow). KpSTM AS transgenic plants were slightly shorter than WT (Figure 4U), but the development of newly emerged leaves was comparable to WT (Figure 4V). This short internode phenotype in AS lines was also confirmed by measuring internode length (Supplementary Figure S1K). The leaves of KpWUS, KpCUC2 and KpSTM AS lines grew slightly wider and shorter in comparison to those in WT (Figures 4H, R, W). The leaves in KpSTM AS lines were bilaterally asymmetrical (Figure 4W). The overall shape of leaves in KpCLV2 AS lines was similar to WT (Figure 4M). AS leaves had fewer leaf crenulations per leaf, except KpSTM AS lines, which had a similar number of leaf crenulations to those in WT (Supplementary Figure S1L). KpWUS AS lines showed severe defects in plantlet formation; detached leaves of KpWUS AS lines after 20 days showed no plantlet formation (Figure 4I). The crenulations in severe phenotypes of KpWUS AS lines showed a cavity without a trace of EBs (Figure 4J). After leaf detachment, leaves of KpCLV2, KpCUC2 and KpSTM AS lines showed fewer plantlets than observed in WT leaves. Some plantlets emerged but ceased development and died (aborted plantlets, labelled with AP; Figures 4N, S, X). SEM images of the leaf crenulations in KpCLV2 AS lines showed that EB was being formed, however, the structure seemed abnormal (Figure 4O); placed in a more exposed position between shallower leaf lobes. Stronger phenotypes in KpCUC2 AS lines only developed roots without leaf initiation from the EBs (Figure 4S) or showed no obvious EB developed (Figure 4T). SEM images of the crenulation area in KpSTM AS lines further showed phenotypic shallowly positioned EBs (Figure 4Y).
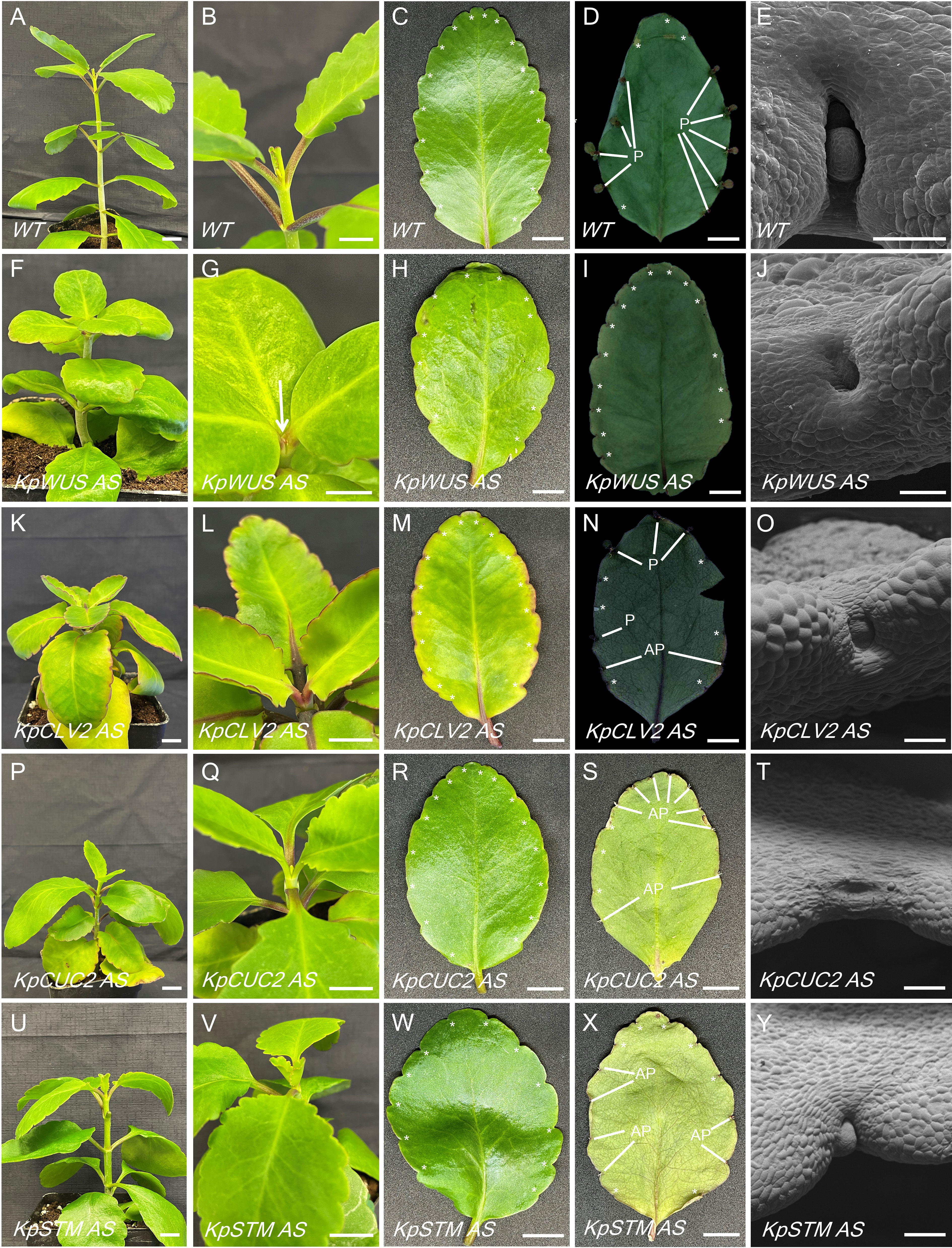
Figure 4 Phenotypic comparison between WT and KpWUS, KpCLV2, KpCUC2 and KpSTM antisense (AS) lines. Full plant images (A, F, K, P, U); newly emerged leaves in the shoot (B, G, L, Q, V); individual leaves imaging at 0 hours (C, H, M, R, W) and 20 days (D, I, N, S, X) following leaf detachment, scale bars: 1 cm. SEM imaging of the leaf crenulation area (E, J, O, T, Y) scale bar: 250 µm. P; plantlet, AP; aborted plantlet, *; leaf crenulation.
To study the role of meristem genes in plantlet formation, we scored plantlet emergence in detached leaves of K. pinnata WT and the AS lines. All AS lines showed fewer plantlets compared to WT (Figure 5A). We calculated the plantlet formation percentage by dividing the number of crenulations with plantlets by the total number of crenulations. To obtain the plantlet formation rate we arcsine-transformed the percentages to statistically analyse them. The plantlet formation percentage after 21 days of leaf detachment was 55.2% in WT, 8% in KpWUS AS lines, 32.5% in KpCLV2 AS lines, 14.4% in KpCUC2 AS lines and 23.5% in KpSTM AS lines. The plantlets became visible in the crenulations 9 days after leaf detachment in WT and the AS lines. Detached leaves of KpWUS, KpCUC2 and KpSTM AS lines formed significantly fewer plantlets than WT between 9 days to 21 days of leaves detachment. Notably, KpCLV2 AS lines showed fewer plantlet phenotypes after 12 days. All AS lines showed a significantly decreased number of plantlets 21 days after detachment, compared to WT: KpWUS AS lines (p=<0.0001), KpCLV2 AS lines (p=0.0018), KpCUC2 (p=0.0001) AS lines and KpSTM AS lines (p=0.0002) (Figure 5B), suggesting that these four meristem genes play a prime role in plantlet formation in the leaves.
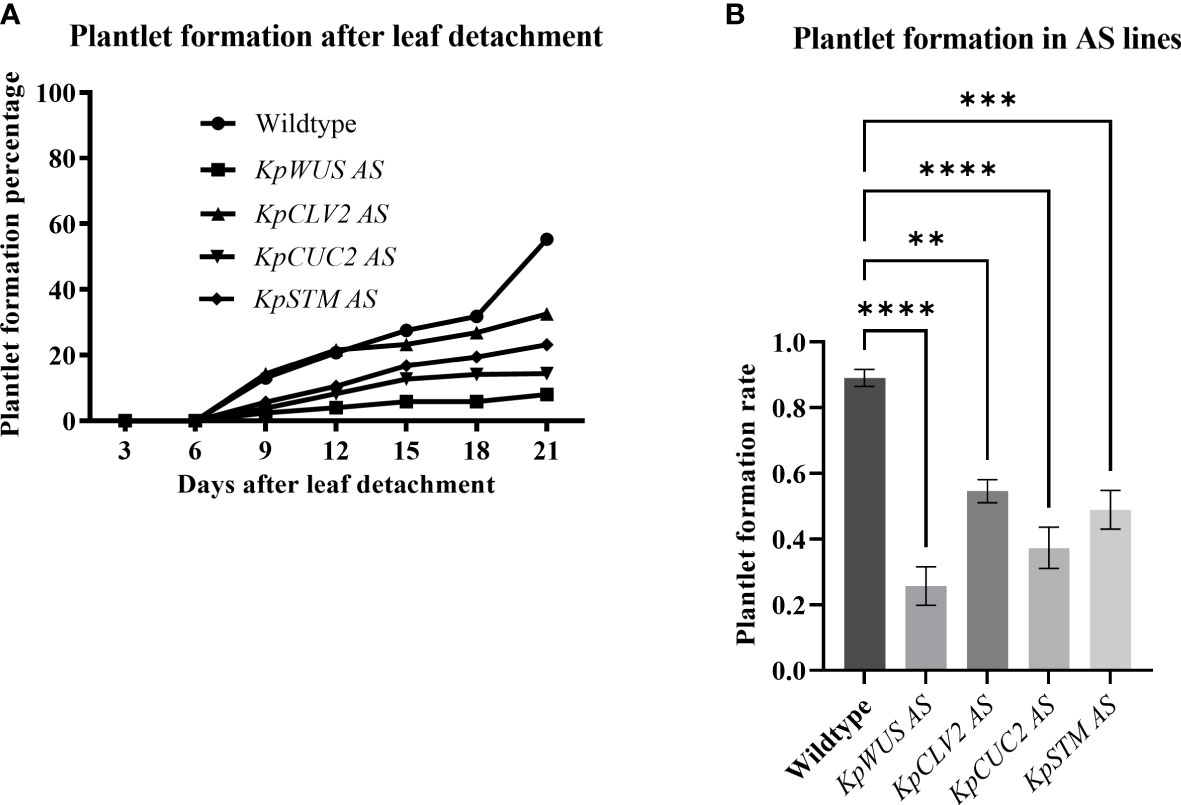
Figure 5 Plantlet formation in antisense (AS) lines. (A) Percentage of plantlet formation with time (up to 21 days) after leaf detachment. (B) Plantlet formation rate in AS lines after 21 days of leaf detachment. The plantlet formation was scored in 25 leaves of 5 different independent lines. The error bars show SE. The percentages were calculated by dividing the number of crenulations with plantlets by the number of crenulations and then multiplied by 100. The rates were obtained by arcsine-transforming the percentages. One-Way ANOVA with Dunnett’s Multiple Comparison, ns (non-significant); * (P-value <0.01); ** (P-value <0.001); *** (P-value <0.0002) and **** (P-value <0.0001).
K. pinnata WT mature leaves presented evident EBs in the middle of the leaf crenulations (Figure 6A). Figure 6B shows developing leaf 1 (L1), L2 and roots 15 days after leaf detachment. Subsequently, L3 and L4 form when L1 and L2 are still growing (Figure 6C), however, L3 and L4 seemed to develop faster (Figure 6D). The leaves of KpWUS, KpCUC2 and KpSTM AS lines showed barely noticeable EBs in mature crenulations when compared to WT leaves (Figure 6E, M, Q). Following 15 days of leaf detachment, KpWUS AS leaves did not show any visible plantlet formation (Figure 6F). Figure 6G shows an EB of the KpWUS AS lines 20 days after the detachment, with only a small root emergence. Other phenotypes of KpWUS AS lines showed small underdeveloped L1 and L2 25 days after leaf detachment (Figure 6H). The EBs in KpCLV2 AS plants were noticeable in mature leaves (Figure 6I). L1 and L2 became visible 15 days after leaf detachment, however, no roots emerged and L2 were larger than L1 (Figure 6J). Figure 6K shows KpCLV2 AS plantlets 25 days after leaf detachment with developed roots but small asymmetric L1 and L2. Other phenotypes of KpCLV2 AS plantlets showed several root development but underdeveloped L1 and L2 (Figure 6L). 15 days after leaf detachment, KpCUC2 AS lines presented abundant root development and underdeveloped and phenotypic L1 and L2 (Figure 6N). Other phenotypes of KpCUC2 AS plants also showed abundant root development and slower development of leaf primordia, L1 and L2 remained undeveloped (Figure 6O). After 25 days of leaf detachment, some phenotypes of KpCUC2 AS lines showed abundant root development, underdeveloped L1 and L2, fused L3 and L4 at the base, with no clear delimitation with newly emerged L5 and L6 (Figure 6P). After 15 days of leaf detachment, strong phenotypes of KpSTM AS lines developed EBs with small asymmetric L1 and L2 (Figure 7R). Other AS phenotypes formed numerous roots and phenotypic L1, L2, L3 and L4 emerged symmetrically (Figure 6S). Other AS phenotypes of KpSTM resembled the KpCUC2 AS ones, without visible divisions between the developing leaf primordia (Figure 6T).

Figure 6 Plantlet phenotypes of KpWUS, KpCLV2, KpCUC2 and KpSTM antisense (AS) lines. Leaf crenulations in attached leaves (A, E, I, M, Q), after 9 days of leaf detachment (B, F, J, N, R). Plantlet formation after 20 days (C, G, K, O, S) and 25 days (D, H, L, P, T) of leaf detachment. WT (A-D), KpWUS AS lines (E-H), KpCLV2 AS lines (I - L), KpCUC2 AS lines (M-P) and KpSTM AS lines (Q-T). (C, D) are the same as Figures 2C, D. Scale bar: 1 mm; L1; leaf 1, L2; leaf 2, L3; leaf 3, L4; leaf 4, L5; leaf 5, L6; leaf 6, R; root, EB; epiphyllous bud.
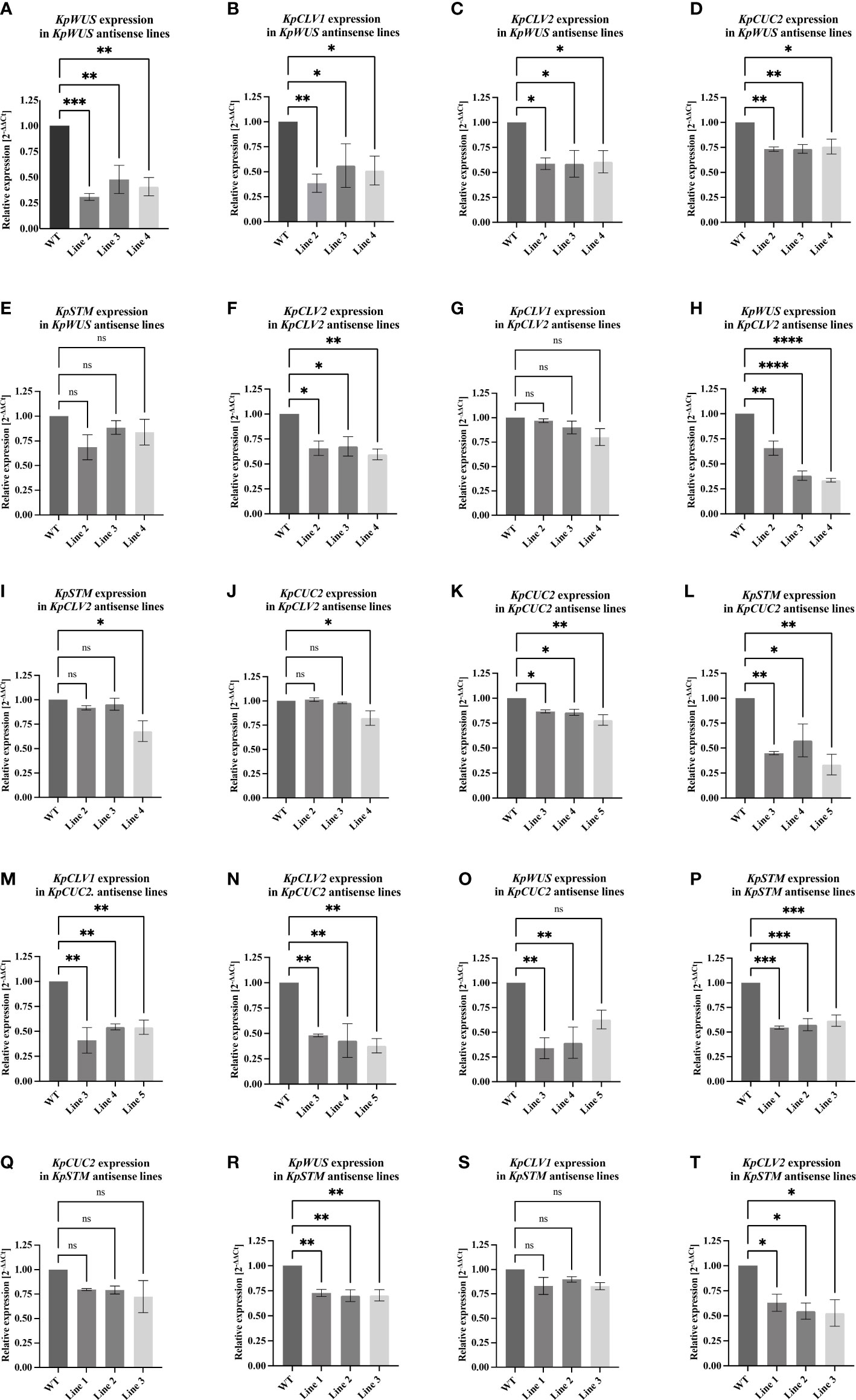
Figure 7 RT-qPCR analysis of KpWUS, KpCLV2, KpCUC2 and KpSTM antisense (AS) lines. Expression of these genes in KpWUS AS lines 2, 3 and 4 (A-E), KpCLV2 AS lines 2, 3 and 4 (F-J), KpCUC2 AS lines 3, 4 and 5 (K-O) and KpSTM AS lines 1, 2 and 3 (P–T). One-Way ANOVA with Dunnett’s Multiple Comparison, ns (non-significant); * (P value <0.01); ** (P value <0.001); *** (P value <0.0002) and **** (P value <0.0001) Errors bars represent the 95% confidence interval.
To study the function of key meristem genes and to reveal the genetic means modulating induced plantlet formation in K. pinnata, we measured the expression of these genes in the AS lines. RT-qPCR of the crenulate leaf region with EBs of AS KpWUS appeared to be significantly downregulated in all KpWUS AS lines (Figure 7A), confirming that KpWUS was successfully downregulated in these AS lines. In the KpWUS AS lines, KpCLV1, KpCLV2 and KpCUC2 were also downregulated (Figures 7B-D). However, KpSTM expression was similar between the KpWUS AS lines and the WT (Figure 7E). KpCLV2 was significantly downregulated in KpCLV2 AS plants (Figure 7F). While KpCLV1 expression was not affected by KpCLV2 downregulation (Figure 7G), KpWUS, KpSTM and KpCUC2 were downregulated in KpCLV2 AS lines (Figures 7H-J). KpCUC2 AS lines showed downregulation in KpCUC2, KpSTM, KpCLV1 and KpCLV2 (Figures 7K-N). KpWUS was only downregulated in lines 3 and 4 in comparison to WT (Figure 7O). KpSTM, KpWUS and KpCLV2 were downregulated in KpSTM AS plants (Figures 7P, R, T), however, KpCUC2 and KpCLV1 expressions were similar to the expression in WT (Figures 7Q, S).
To investigate the location of KpWUS expression in the EBs, we performed immunolocalisation using a WUS antibody on the leaf crenulations of WT, KpWUS and KpCLV2 AS plants. KpWUS expression was first detected in the main vasculature of WT leaves when crenulations were being formed in the leaf lobes (Figure 8A). In growing leaves (< 1 cm long), KpWUS was already detected in a group of small cells in the middle of the leaf crenulation (Figure 8B). Mature leaves showed KpWUS expression localised to the centre of the developing EB and mildly expressed in the vasculature of leaf lobes, presumably in the parenchyma cells (Figure 8C). In fully grown leaves 24 hours after leaf detachment, KpWUS was found to cover a broader expression area including the EBs and the adjacent region (Figure 8D). KpWUS signal was not detected in the crenulations of KpWUS AS lines24 hours after leaf detachment, but the faint expression in the vasculature of the leaf lobes (Figure 8E). In the leaves of KpCLV2 AS lines, 24 hours after leaf detachment, the KpWUS domain appeared to be broad and un-organised around the leaf crenulation, while no obvious EB structure was observed (Figure 8F).
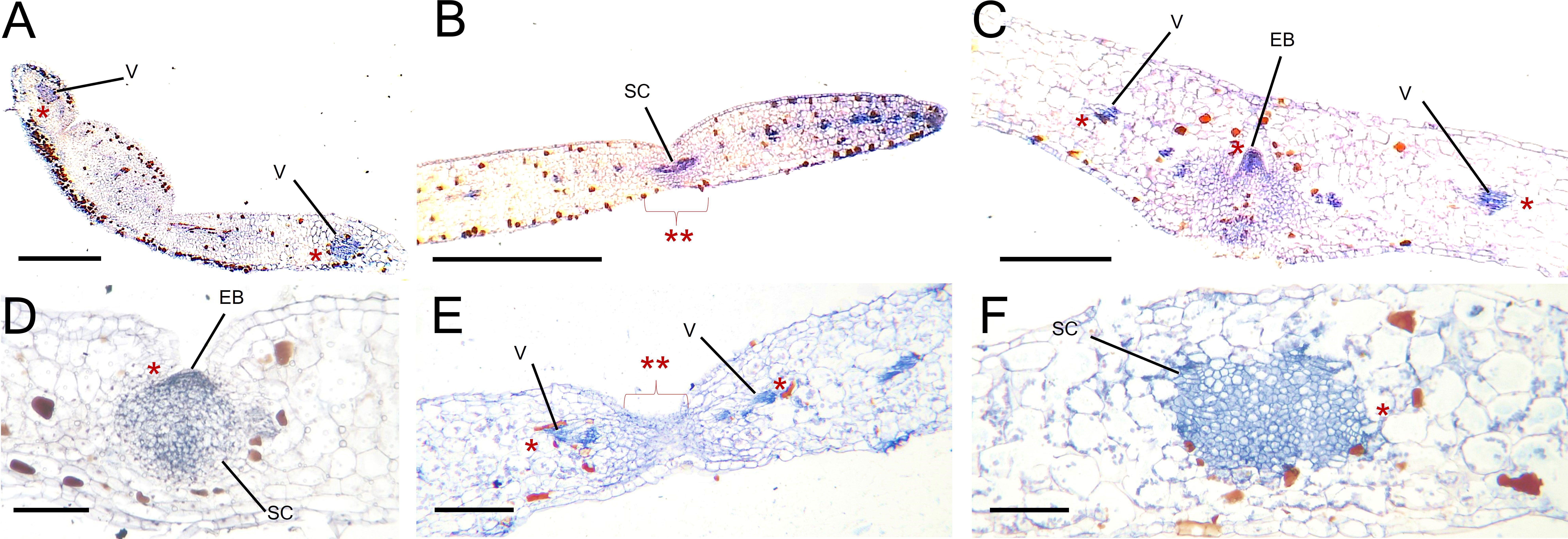
Figure 8 KpWUS signal in epiphyllous buds (EB) of WT, KpWUS and KpCLV2 antisense (AS) lines. KpWUS expression in WT; (A) newly emerged leaves, (B) young leaves <5 cm, (C) fully developed EBs, (D) EBs 24 hours after leaf detachment, (E) in KpWUS AS lines and (F) in KpCLV2 AS lines. The KpWUS signal were detected in the vasculature (V) of the leaves and groups of stem cells (SC) in the middle of the crenulations (marked with *). Note no KpWUS signal in the SC in KpWUS AS lines (marked with ** with braket) in (E) Scale bars: 0.5 mm (A), 1 mm (B-F).
The Kalanchoë genus evolved to reproduce by forming plantlets on the leaf margins (Allorge-Boiteau, 1996; Garces et al., 2007; Garcěs et al., 2009). Whereas basal Kalanchoë species have an obligate sexual reproduction system, more derived inducible plantlet-forming species such as K. pinnata can switch from sexual to asexual reproduction by producing plantlets in the leaf crenulations. Previous work reported that the K. pinnata EBs stay dormant in the leaves until certain conditions such as leaf detachment and stress induce plantlet formation (Naylor, 1932; Sawhney et al., 1994). Our results confirmed that the EBs in K. pinnata are pre-existent in the centre of the leaf crenulations before leaf detachment and stay latent until certain cues trigger plantlet development. It is believed that the disruption of hormone supply to the leaves triggered plantlet formation in K. pinnata (Sawhney et al., 1994). Earlier studies performed in K. pinnata demonstrated that the plant hormone thidiazuron (TDZ) inhibited plantlet development, whereas ethrel (ETH) promoted plantlet formation (Jaiswal and Sawhney, 2006). It was also revealed that cytokinins zeatin, kinetin, and benzyl aminopurine (BAP) severely disrupted plantlet formation in K. marnierianum, an inducible plantlet-forming species (Kulka, 2006).
Molecular studies revealed the expression of STM orthologs in the plantlet primordia of Kalanchoë plantlet forming species, suggesting their role in plantlet formation in various Kalanchoë species (Garces et al., 2007). STM in conjunction with several other genes has proven to be essential for the maintenance of stem cells in the SAM and floral meristem in Arabidopsis (Endrizzi et al., 1996). This suggests that the meristematic activity, presumably modulated by STM during the process of plantlet formation might be also coordinated by other meristem genes. Consistently, our RNA sequencing analysis in the leaf crenulations of K. pinnata revealed several meristem genes being expressed in the plantlet primordia. KpWUS, KpWOX11 and WOX13 being upregulated after leaf detachment, suggesting increasing meristematic activity following the leaf detachment. WOX genes have been shown to have essential roles during Arabidopsis embryo patterning, organogenesis and stem cell initiation and maintenance (Wu et al., 2007; Ueda et al., 2011; Dolzblasz et al., 2016). Overexpressing lines of PeWOX11 in Populus euramericana (Pe) showed an increased number of adventitious root formations (Xu et al., 2015), and PnWOX11 was found upregulated during root formation in P. nigra (Baesso et al., 2020). This suggests that KpWOX11 might be required for root development in the EBs of K. pinnata after leaf detachment. WOX13 in Physcomitrella patens was required for stem cell initiation at the wound margins of detached leaves (Sakakibara et al., 2014). KpWOX13 might be involved in stem cell establishment and maintenance in the leaf crenulations of K. pinnata.
KpCLV1 and KpCLV2 were upregulated in the EBs in comparison with the mid-section of the leaf, however, a significant downregulation was observed 4 hours after leaf detachment for both genes. In Arabidopsis, WUS activates CLV3, which restricts the WUS domain to the OC (Brand, 2000) by binding to the CLV1 ectodomain and the CLV2/CORYNE (CRN) receptor protein complex. Evidence suggests that the CLV1/CLV2 receptor complex interacts with CLV3 to inhibit WUS expression (Müller et al., 2008). The downregulation of KpCLV1 and 2 in the EBs of K. pinnata following the leaf detachment might lead to the upregulation of KpWUS and other KpWOX genes, needed to recruit stem cells for plantlet development.
In the Kalanchoë genus, STM orthologs were found expressed only in the leaf crenulations of plantlet-forming species (Garces et al., 2007). In K. pinnata, KpSTM was found slightly upregulated 48 hours after leaf detachment. In Arabidopsis, STM positively regulates the expression of CUC1, 2 and 3 by binding to their promoter (Spinelli et al., 2011). KpCUC2 andKpCUC3 were significantly upregulated after leaf detachment. These genes in Arabidopsis act redundantly to establish and maintain boundaries around developing organs (Laufs et al., 2004). CUC2 is also responsible for the presence of leaf serrations in Arabidopsis (Kamiuchi et al., 2014). KpCUC2 and KpCUC3 might be playing crucial roles in the formation of epiphyllous buds and the process of plantlet formation via STM-CUC gene pathways.
Our results showed that K. pinnata AS lines expressing AS fragments of meristem genes (KpWUS, KpCLV2, KpCUC2 and KpSTM) resulted in no or defective EBs in the leaf crenulations, suggesting that meristem genes are involved in the EBs formation and maintenance. All the AS lines formed significantly fewer plantlets after leaf detachment when compared to WT. Plantlet formation was strongly disrupted in KpWUS, KpCUC2 and KpSTM AS lines. Conversely, plantlet formation was less severely affected in the KpCLV2 AS lines. Evidence suggests that two parallel CLV3 signaling transduction mechanisms restrict WUS expression in Arabidopsis, either through CLV1 and 2 heterodimers or via independent interaction with CLV1 or 2. Only clv1-2 double mutants resulted in severe phenotypes comparable to the ones observed in CLV3 single mutants (Jeong et al., 1999; Bleckmann et al., 2009). Consistently, our KpCLV2 AS lines formed fewer plantlets, however, the plantlet formation was not as strongly disrupted as when the expression of other meristem genes was downregulated.
Signals to initiate plantlet formation do not seem to be affected in the AS plants, as the plantlet formation in AS lines also starts on day 9 following the leaf detachment. This suggests that the reduced plantlet formation was mainly due to the defect in the formation of EBs during early leaf development before the detachment, and not due to defects in signaling to trigger plantlet initiation. In support of this, all AS lines showed no or defective EBs in most crenulations (Figures 6J, O, T, Y).
Plantlets formed in the AS lines also showed abnormal morphologies. KpWUS AS plantlets terminated meristem activity after forming the first leaf pair and small roots, resembling wus mutants in Arabidopsis (Laux et al., 1996). This suggests that KpWUS is important not only for the EB formation but also for the SAM development of plantlets. KpCUC2 AS plantlets also showed predominant root development and the fusion of plantlet leaf pairs, which reflects the Arabidopsis cuc2 mutant phenotypes. CUC2 in Arabidopsis delimits the boundaries in the developing organs, and its mutation often generates a fusion of lateral organs (Laufs et al., 2004). Auxin may be involved in plantlet root development in some Kalanchoë species (Kulka, 2008), and in Arabidopsis, CUC2 and the auxin accumulation are mutually exclusive (Bilsborough et al., 2011). Therefore, the root overproliferation phenotypes might be due to an increase of auxin in KpCUC2 AS plants. KpSTM AS plantlets also exhibited root over-proliferation and fused emerging leaves, similar to those of KpCUC2 AS lines. It has been shown that STM and CUC genes positively regulate each other in Arabidopsis leaf primordia (Spinelli et al., 2011). Similarly, KpCUC2 might also regulate the expression of KpSTM in the EBs of K. pinnata. Thus, KpSTM AS plants might mimic KpCUC2 AS phenotypes.
The alteration of expression levels of meristem genes in AS backgrounds revealed insight into regulatory relationships among the genes. KpWUS AS lines showed downregulation in KpCLV1, KpCLV2 and KpCUC2, suggesting that KpWUS positively regulated the expression of these genes in the EBs of K. pinnata. Notably, as only the indented regions of the leaf with EBs were harvested for this experiment, it is unlikely that the results could be attributed to the indirect consequences of the decrease of EBs in AS plants. In Arabidopsis, the SAM is maintained by the feedback loop between CLV3 and WUS. A decrease in WUS expression in Arabidopsis leads to fewer stem cells, and thus less CLV3 production (Schoof et al., 2000; Somssich et al., 2016). In Arabidopsis, CLV loss-of-function leads to increased meristematic activity, however, our results indicated that the expression of KpWUS was downregulated in the KpCLV2 attenuated background, suggesting that KpCLV2 influence the expression of KpWUS in the EBs in K. pinnata, in a different way as in the SAM of Arabidopsis.
Conversely, KpSTM was not largely affected by KpWUS downregulation; KpSTM expression levels showed a trend of decrease but were statistically insignificant. However, KpWUS was significantly downregulated in KpSTM downregulated background. This suggests that KpSTM showed a strong reciprocal regulation to establish the meristematic competency in the EBs of K. pinnata, promoting KpWUS and subsequently, KpCLV1 and 2. Moreover, KpCUC2 AS lines showed downregulation in KpWUS, KpSTM, KpCLV1 and KpCLV2. This suggests that KpCUC2 positively modulates the expression of KpWUS, KpCLV1 and 2 via KpSTM in the EBs of K. pinnata. This suggests the role of KpCUC2 in the establishment of meristem competency in the leaves of K. pinnata. Furthermore, CUC2 also controls the leaf margin development in Arabidopsis, and together with the microRNA (miRNA), MiR164A determines the crenulation depth (Nikovics et al., 2006). The leaves of KpCUC2 AS lines showed shallower leaf crenulations. As the presence of crenulations is a prerequisite for plantlet primordia in Kalanchoë, KpCUC2 is also likely to contribute to plantlet development via the formation of crenulations in K. pinnata. Notably, Kalanchoë species such as K. marmorata and K. eriophylla that are unable to produce plantlets possess round leaves without leaf crenulations.
Our results showed that key meristem genes were expressed in the leaf crenulations, giving rise to EBs. Furthermore, immunolocalisation revealed the presence of KpWUS in the crenulations of newly forming leaves and a stronger signal at the centre of the EB in the crenulations of mature K. pinnata WT leaves. This suggests that meristem activity started early when leaves were being formed from the SAM. In addition, KpCLV2 AS plants showed a broad and disorganised KpWUS domain, resulting in a defective EB arrangement. This evidenced the role of KpCLV2 in restricting the KpWUS domain in the EB. The CLV/WUS feedback system prevents the over-proliferation of stem cells preserving the SAM size (Yadav et al., 2011). This revealed that the bud maintenance is modulated by the WUS-CLV complex. Our results also showed that the KpWUS signal increased in the EBs 24 hours following the detachment of the leaves, consistent with our RNA-sequencing analysis. This suggests that KpWUS is also involved during plantlet initiation, increasing meristem activity as a response to leaf.
It appears that K. pinnata recruited meristem pathways to facilitate the novel reproductive strategy such as the formation of EBs in the leaf crenulations. Our results demonstrated the ectopic expression of meristem genes in the leaf crenulations of K. pinnata assists in plantlet formation as a reproductive process. Downregulation in key meristem genes severely disturbs plantlet formation and defective plantlets. Furthermore, Kalanchoë inducible plantlet-forming species represent the first step towards the loss of sexual reproduction within the genus. The importance of K. pinnata as a transitional form from sexual to asexual reproduction instigates the acquisition of meristematic competency to achieve the production of plantlets on the leaves.
The data presented in the study are deposited in the GenBank repository (https://www.ncbi.nlm.nih.gov/genbank/), the accession numbers are: KpWUS Accession OQ674700, KpCLV2 Accession OQ621804, KpCUC2 Accession OQ621805, KpSTM Accession OQ674701.
Conceptualization, MK. Methodology, MK and FJ-B. Experiments, FJ-B. Software, FJ-B. Validation, MK and FJ-B. Formal analysis, FJ-B. Writing—original draft preparation, FJ-B. Writing—review and editing, MK and FJ-B. Supervision, MK. All authors contributed to the article and approved the submitted version.
This work was supported by Biotechnology and Biological Sciences Research Council research grant (BBSRC no. BB/I012982/1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1157619/full#supplementary-material
Supplementary Figure 1 | Genotypes of antisense (AS) lines (A–H). (I) showing stem cells (SC) in the meristem of K. daigremontiana and negative control without the primary antibody (J). Scale bars: 0.5 mm (I) and 1 mm (J). (K) Internode length and (L) the number of crenulations in wildtype and AS lines. One-Way ANOVA with Dunnett’s Multiple Comparison, ns (non-significant); * (P value <0.01); ** (P value <0.001); *** (P value <0.0002) and **** (P value <0.0001) Errors bars represent the 95% confidence interval.
Supplementary Table 1 | Primers for gene cloning and vector assembly.
Supplementary Table 2 | List of primers used for genotyping and RT-qPCR.
Aida, M., Ishida, T., Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570. doi: 10.1242/dev.126.8.1563
Allorge-Boiteau, L. (1996). Madagascar Centre de speciation et d’origine du genre kalanchoe (Crassulaceae). Biogeogr. Madagascar 8, 137–145.
Baesso, B., Terzaghi, M., Chiatante, D., Scippa, G. S., Montagnoli, A. (2020). WOX genes expression during the formation of new lateral roots from secondary structures in populus nigra (L.) taproot. Sci. Rep. 10, 18890. doi: 10.1038/s41598-020-75150-1
Bäurle, I., Laux, T. (2005). Regulation of WUSCHEL transcription in the stem cell niche of the arabidopsis shoot meristem. Plant Cell 17, 2271–2280. doi: 10.1105/tpc.105.032623
Bilsborough, G. D., Runions, A., Barkoulas, M., Jenkins, H. W., Hasson, A., Galinha, C., et al. (2011). Model for the regulation of arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. 108, 3424–3429. doi: 10.1073/pnas.1015162108
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A. M., Simon, R. (2009). Stem cell signaling in arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176. doi: 10.1104/pp.109.149930
Brand, U. (2000). Dependence of stem cell fate in arabidopsis on a feedback loop regulated by CLV3 activity. Sci. (80-.) 289, 617–619. doi: 10.1126/science.289.5479.617
Clark, S. E., Jacobsen, S. E., Levin, J. Z., Meyerowitz, E. M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in arabidopsis. Development 122, 1567–1575. doi: 10.1242/dev.122.5.1567
DeYoung, B. J., Bickle, K. L., Schrage, K. J., Muskett, P., Patel, K., Clark, S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in arabidopsis. Plant J. 45, 1–16. doi: 10.1111/j.1365-313X.2005.02592.x
Dinneny, J. R., Benfey, P. N. (2008). Plant stem cell niches: standing the test of time. Cell 132, 553–557. doi: 10.1016/j.cell.2008.02.001
Dolzblasz, A., Nardmann, J., Clerici, E., Causier, B., van der Graaff, E., Chen, J., et al. (2016). Stem cell regulation by arabidopsis WOX genes. Mol. Plant 9, 1028–1039. doi: 10.1016/j.molp.2016.04.007
Endrizzi, K., Moussian, B., Haecker, A., Levin, J. Z., Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10, 967–979. doi: 10.1046/j.1365-313X.1996.10060967.x
Fletcher, J. (2018). The CLV-WUS stem cell signaling pathway: a roadmap to crop yield optimization. Plants 7, 87. doi: 10.3390/plants7040087
Gaillochet, C., Lohmann, J. U. (2015). The never-ending story: from pluripotency to plant developmental plasticity. Development 142, 2237–2249. doi: 10.1242/dev.117614
Galli, M., Gallavotti, A. (2016). Expanding the regulatory network for meristem size in plants. Trends Genet. 32, 372–383. doi: 10.1016/j.tig.2016.04.001
Gallois, J.-L., Woodward, C., Reddy, G. V., Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in arabidopsis. Development 129, 3207–3217. doi: 10.1242/dev.129.13.3207
Garces, H. M. P., Champagne, C. E. M., Townsley, B. T., Park, S., Malho, R., Pedroso, M. C., et al. (2007). Evolution of asexual reproduction in leaves of the genus kalanchoe. Proc. Natl. Acad. Sci. U.S.A. 104, 15578–15583. doi: 10.1073/pnas.0704105104
Garces, H. M. P., Koenig, D., Townsley, B. T., Kim, M., Sinha, N. R., Garcês, H. M. P., et al. (2014). Truncation of LEAFY COTYLEDON1 protein is required for asexual reproduction in kalanchoë daigremontiana. Plant Physiol. 165, 196–206. doi: 10.1104/pp.114.237222
Garcěs, H., Sinha, N. (2009). Transformation of the plant kalanchoë daigremontiana using agrobacterium tumefaciens. Cold Spring Harb. Protoc. 4, 4–7. doi: 10.1101/pdb.prot5303
Garces, H., Sinha, N. (2009a). Extraction of RNA from the plant kalanchoe daigremontiana. Cold Spring Harb. Protoc. 2009, pdb.prot5305–pdb.prot5305. doi: 10.1101/pdb.prot5305
Garces, H., Sinha, N. (2009b). Fixing and sectioning tissue from the plant kalanchoe daigremontiana. Cold Spring Harb. Protoc. 2009, pdb.prot5301–pdb.prot5301. doi: 10.1101/pdb.prot5301
Gehrig, H. H., Winter, K., Cushman, J., Borland, A., Taybi, T. (2000). An improved RNA isolation method for succulent plant species rich in polyphenols and polysaccharides. Plant Mol. Biol. Rep. 18, 369–376. doi: 10.1007/BF02825065
Gordon, S. P., Chickarmane, V. S., Ohno, C., Meyerowitz, E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the arabidopsis shoot meristem. Proc. Natl. Acad. Sci. 106, 16529–16534. doi: 10.1073/pnas.0908122106
Gorelick, R. (2015). Why vegetative propagation of leaf cuttings is possible in succulent and semi-succulent plants. Haseltonia 20, 51–57. doi: 10.2985/026.020.0109
Guo, Y., Han, L., Hymes, M., Denver, R., Clark, S. E. (2010). CLAVATA2 forms a distinct CLE-binding receptor complex regulating arabidopsis stem cell specification. Plant J. 63, 889–900. doi: 10.1111/j.1365-313X.2010.04295.x
Heidstra, R., Sabatini, S. (2014). Plant and animal stem cells: similar yet different. Nat. Rev. Mol. Cell Biol. 15, 301–312. doi: 10.1038/nrm3790
Jácome-Blásquez, F., Ooi, J. P., Zeef, L., Kim, M. (2022). Comparative transcriptome analysis of two kalanchoë species during plantlet formation. Plants 11, 1643. doi: 10.3390/plants11131643
Jaiswal, S., Sawhney, S. (2006). Modulation of TDZ-induced morphogenetic responses by anti-auxin TIBA in bud-bearing foliar explants of kalanchoe pinnata. Plant Cell. Tissue Organ Cult. 86, 69–76. doi: 10.1007/s11240-006-9099-x
Jeong, S., Trotochaud, A. E., Clark, S. E. (1999). The arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1933. doi: 10.1105/tpc.11.10.1925
Kamiuchi, Y., Yamamoto, K., Furutani, M., Tasaka, M., Aida, M. (2014). The CUC1 and CUC2 genes promote carpel margin meristem formation during arabidopsis gynoecium development. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00165
Kulka, R. G. (2006). Cytokinins inhibit epiphyllous plantlet development on leaves of bryophyllum (Kalanchoe) marnierianum. J. Exp. Bot. 57, 4089–4098. doi: 10.1093/jxb/erl180
Kulka, R. G. (2008). Hormonal control of root development on epiphyllous plantlets of bryophyllum (Kalanchoe) marnierianum: role of auxin and ethylene. J. Exp. Bot. 59, 2361–2370. doi: 10.1093/jxb/ern106
Laufs, P., Peaucelle, A., Morin, H., Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131(17), 4311-4322. doi: 10.1242/dev.01320
Laura, M., Borghi, C., Regis, C., Cassetti, A., Allavena, A. (2013). Ectopic expression of Kxhkn5 in the viviparous species kalanchoe × houghtonii induces a novel pattern of epiphyll development. Transgenic Res. 22, 59–74. doi: 10.1007/s11248-012-9628-9
Laux, T., Mayer, K. F., Berger, J., Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in arabidopsis. Development 122, 87–96. doi: 10.1242/dev.122.1.87
Lenhard, M., Jürgens, G., Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in arabidopsis shoot meristem regulation. Development 129, 3195–3206. doi: 10.1242/dev.129.13.3195
Lindsay, D. L., Sawhney, V. K., Bonham-Smith, P. C. (2006). Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in arabidopsis. Plant Sci. 170, 1111–1117. doi: 10.1016/j.plantsci.2006.01.015
Long, J. A., Moan, E. I., Medford, J. I., Barton, M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of arabidopsis. Nature 379, 66–69. doi: 10.1038/379066a0
Müller, R., Bleckmann, A., Simon, R. (2008). The receptor kinase CORYNE of arabidopsis transmits the stem cell–limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946. doi: 10.1105/tpc.107.057547
Mayer, K. F. X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the arabidopsis shoot meristem. Cell 95(6), 805-815. doi: 10.1016/S0092-8674(00)81703-1
Naylor, E. (1932). The morphology of regeneration in bryophyllum calycinum. Am. J. Bot. 19, 32–40. doi: 10.1002/j.1537-2197.1932.tb09634.x
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Pelletier, J. M., Kwong, R. W., Park, S., Le, B. H., Baden, R., Cagliari, A., et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. 114, E6710–E6719. doi: 10.1073/pnas.1707957114
Rajsekhar, P. B., Arvind Bharani, R. S., Ramachandran, M., Jini Angel, K., Rajsekhar, S. P. V. (2016). The “wonder plant” kalanchoe pinnata (linn.) pers.: a review. J. Appl. Pharm. Sci. 6, 151–158. doi: 10.7324/JAPS.2016.60326
Sablowski, R. (2004). Plant and animal stem cells: conceptually similar, molecularly distinct? Trends Cell Biol. 14, 605–611. doi: 10.1016/j.tcb.2004.09.011
Sakakibara, K., Reisewitz, P., Aoyama, T., Friedrich, T., Ando, S., Sato, Y., et al. (2014). WOX13 - like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss physcomitrella patens. Development 141, 1660–1670. doi: 10.1242/dev.097444
Sawhney, S., Bansal, A., Sawhney, N. (1994). Epiphyllous bud differentiation in kalanchoe pinnata: a model developmental system. Indian J. Plant Physiol. 37, 229–236.
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jürgens, G., Laux, T. (2000). The stem cell population of arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
Scofield, S., Dewitte, W., Murray, J. A. (2014). STM sustains stem cell function in the arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1. Plant Signal. Behav. 9, e28934. doi: 10.4161/psb.28934
Scofield, S., Murison, A., Jones, A., Fozard, J., Aida, M., Band, L. R., et al. (2018). Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development 145 (9), dev157081. doi: 10.1242/dev.157081
Sengupta, S., Kizhakedathil, M. P. J., Sankar, D. P. (2018). Plant stem cells-regulation and applications: a brief review. Res. J. Pharm. Technol. 11, 1535. doi: 10.5958/0974-360X.2018.00286.X
Shinohara, H., Matsubayashi, Y. (2015). Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 82, 328–336. doi: 10.1111/tpj.12817
Somssich, M., Je, B., Simon, R., Jackson, D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248. doi: 10.1242/dev.133645
Spinelli, S. V., Martin, A. P., Viola, I. L., Gonzalez, D. H., Palatnik, J. F. (2011). A mechanistic link between STM and CUC1 during arabidopsis development. Plant Physiol. 156, 1894–1904. doi: 10.1104/pp.111.177709
Takada, S., Hibara, K., Ishida, T., Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. doi: 10.1242/dev.128.7.1127
Tang, L. P., Zhou, C., Wang, S. S., Yuan, J., Zhang, X. S., Su, Y. H. (2017). FUSCA 3 interacting with LEAFY COTYLEDON 2 controls lateral root formation through regulating YUCCA 4 gene expression in Arabidopsis thaliana. New Phytol. 213, 1740–1754. doi: 10.1111/nph.14313
Ten Hove, C. A., Lu, K.-J., Weijers, D. (2015). Building a plant: cell fate specification in the early arabidopsis embryo. Development 142, 420–430. doi: 10.1242/dev.111500
Ueda, M., Zhang, Z., Laux, T. (2011). Transcriptional activation of arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 20, 264–270. doi: 10.1016/j.devcel.2011.01.009
Wang, F., Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of arabidopsis seed development. Plant Physiol. 161, 1251–1264. doi: 10.1104/pp.112.212282
Weigel, D., Glazebrook, J. (2002). Arabidopsis: a laboratory manual (New York: Cold Spring Harbor Laboratory Press).
Wu, X., Chory, J., Weigel, D. (2007). Combinations of WOX activities regulate tissue proliferation during arabidopsis embryonic development. Dev. Biol. 309, 306–316. doi: 10.1016/j.ydbio.2007.07.019
Xu, M., Xie, W., Huang, M. (2015). Two WUSCHEL-related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant 155, 446–456. doi: 10.1111/ppl.12349
Yadav, R. K., Perales, M., Gruel, J., Girke, T., Jonsson, H., Reddy, G. V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the arabidopsis shoot apex. Genes Dev. 25, 2025–2030. doi: 10.1101/gad.17258511
Zhou, Y., Liu, X., Engstrom, E. M., Nimchuk, Z. L., Pruneda-Paz, J. L., Tarr, P. T., et al. (2015). Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517, 377–380. doi: 10.1038/nature13853
Keywords: asexual and vegetative reproduction, leaf crenulations, plantlet formation, SHOOTMERISTEMLESS (STM), WUSCHEL, stem cells, meristem, Kalanchoë species
Citation: Jácome-Blásquez F and Kim M (2023) Meristem genes are essential for the vegetative reproduction of Kalanchoë pinnata. Front. Plant Sci. 14:1157619. doi: 10.3389/fpls.2023.1157619
Received: 02 February 2023; Accepted: 04 April 2023;
Published: 08 May 2023.
Edited by:
Mary Byrne, The University of Sydney, AustraliaReviewed by:
Chihiro Furumizu, Hiroshima University, JapanCopyright © 2023 Jácome-Blásquez and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minsung Kim, bWluc3VuZy5raW1AbWFuY2hlc3Rlci5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.