- 1College of Plant Protection, Shenyang Agricultural University, Shenyang, Liaoning, China
- 2Institute of Industrial Crops, Heilongjiang Academy of Agricultural Sciences, Harbin, China
Wheat stem rust is caused by Puccinia graminis f. sp. tritici. This major disease has been effectively controlled via resistance genes since the 1970s. The appearance and spread of new races of P. graminis f. sp. tritici (eg., Ug99, TKTTF, and TTRTF) have renewed the interest in identifying the resistance gene and breeding cultivars resistant to wheat stem rust. In this study, gene postulation, pedigree analysis, and molecular detection were used to determine the presence of stem rust resistance genes in 65 commercial wheat cultivars from Hebei Province. In addition, two predominant races 21C3CTHTM and 34MRGQM were used to evaluate the resistance of these cultivars at the adult-plant stage in 2021–2022. The results revealed that 6 Sr genes (namely, Sr5, Sr17, Sr24, Sr31, Sr32, Sr38, and SrTmp), either singly or in combination, were identified in 46 wheat cultivars. Overall, 37 wheat cultivars contained Sr31. Sr5 and Sr17 were present in 3 and 3 cultivars, respectively. Gao 5218 strong gluten, Jie 13-Ji 7369, and Kenong 1006 contained Sr24, Sr32, and Sr38, respectively. No wheat cultivar contained Sr25 and Sr26. In total, 50 (76.9%) wheat cultivars were resistant to all tested races of P. graminis f. sp. tritici in field test in 2021–2022. This study is important for breeding wheat cultivars with resistance to stem rust.
Introduction
Wheat stem rust, caused by Puccinia graminis Pers. f. sp. tritici Eriks. and E. Henn, is an ancient disease. During the 4th century BCE, the Romans considered April 25 every year as a festival to worship the rust god Robigus. They believed that this would prevent epidemics of cereal rusts by offering red cattle, foxes, and dogs to Robigus (Moscou and van Esse, 2017). The earliest record of the occurrence of wheat stem rust in China can be found in the agricultural book Ma Shou Nong Yan of Qing Dynasty in 1836, in which the disease is termed “jaundice” (Li and Zeng, 2002). This is a common wheat disease globally. Using the ecological niche-model of stem trust developed by Pardey et al. (2013), it was estimated that 66% (1.4 million km2) of the world’s wheat-cultivating region is climatically suitable for the development of the disease. Without durable stem-rust resistance, the average total global loss in wheat production is expected to be 306 million metric tons (MT) out of a total production of 23 billion MT (i.e., a loss of USD 54.7 billion when valued at the average wheat prices in 2010 in the United States) with a 90% chance (the 10th percentile) of losing at least 275 MT during 1918–1960 (Pardey et al., 2013). According to records, the widespread cultivation of a new spring wheat cultivar Marquis in Canada since 1912 led to the worst outbreak of wheat stem rust in the United States and Canada in 1916 (Peterson, 2018). In North Dakota, South Dakota, and Minnesota, the output yield loss in 1916 was 12.7, 10.3, and 9.4 bushels per acre, respectively (Peterson, 2018). On a larger scale, the loss from wheat stem rust was estimated at 200 million bushels in the United States (approximately 40% of crops in the United States) and 100 million bushels in Canada (Campbell and Long, 2001). Historically, several wheat stem rust epidemics have caused devastating losses in wheat yield in China (Wu and Huang, 1987; Lin et al., 2021). According to incomplete statistics, nine epidemics occurred in northeast spring wheat region from 1923 to 1964, the yield loss was 740 and 560 million kg in 1923 and 1948, respectively. In 1956 and 1958, the epidemic in the Jianghuai wheat area caused a wheat yield loss of 1 billion kg (Wu and Huang, 1987).
After the 1950s, wheat stem rust was effectively controlled via the combined effects of wide use of host resistance and eradicating common barberry (Berberis vulgaris; the alternate host of Pgt) in the United States and Europe (Rouse et al., 2012; Peterson, 2018). In the following five decades, except the major epidemic in Ethiopia in 1993 and 1994, wheat production in other regions in the world did not significantly suffer from wheat stem rust. The epidemic in Ethiopia caused significant yield loss of a popular wheat cultivar “Enkoy” (Shank, 1994; Singh et al., 2011). However, a new race of P. graminis f. sp. tritici (Isolate Pgt-Ug99) with virulence to Sr31, a widely present stem-rust resistance gene in wheat in the world, was first detected in Uganda in 1998 and designated as Ug99 (Pretorius et al., 2000). Based on the North American nomenclature system for naming P. graminis f. sp. tritici races, it was initially named TTKS and later redesignated as TTKSK (Wanyera et al., 2006; Jin et al., 2008). The Ug99 group of races (except for races TTKSF and TTKSP) is not only virulent to Sr31 but also has the characteristics of rapid variation in the virulence (Singh et al., 2011). What is more worrying is that new pathogenic races are emerging with combined virulence to Sr31 and some other resistance genes used in globally widely grown commercial wheat cultivars. For example, races TTKST, PTKST, TTHST, and TTKTT have combined virulence to Sr31, Sr24, and Sr38 (Fetch et al., 2016; Nazari et al., 2021; Patpour et al., 2016), and race TTTSK has combined virulence to Sr31, Sr36, and Sr38 (Pretorius et al., 2010). To date, 15 known variants of the Ug99 lineage have been identified, which are currently present in 14 countries. TKTTF, a new race of P. graminis f. sp. tritici, caused a severe wheat stem rust epidemic in 2013–2014 in the “Digalu” wheat cultivation area (approximately 20,000–40,000 ha) in southern Ethiopia, causing 100% yield loss in most wheat cultivation areas (Olivera et al., 2015). A new race TTRTF attacked thousands of hectares of durum wheat in 2016 in Sicily, Italy, causing an unusual epidemic of wheat stem rust in Europe since the 1950s (Bhattacharya, 2017). In recent years, other severe epidemic and outbreaks of wheat stem rust have been reported in Central Asia and the Caucasus region (Olivera Firpo et al., 2017; Olivera et al., 2022; Shamanin et al., 2019), and North Africa (Rust Tracker, 2021). These outbreaks of wheat stem rust indicate that the disease has now returned.
China is the world’s largest wheat producer and consumer, with wheat production playing a crucial role in national food security (Cao et al., 2019). Wheat is one of the most important crops in China, and flour food is very popular in northern China. Hebei Province, north China, is one of the major wheat-producing regions in the country and plays a significant role in the epidemiology and spread of wheat stem rust in China. According to the data of the National Bureau of Statistics (http://www.stats.gov.cn/), the annual wheat cultivation area in Hebei Province is approximately 2200 thousand ha, nearly one tenth of China’s wheat cultivation area. Historically, wheat stem rust occurred in this region in some years in past (Wu et al., 1963; Wu et al., 1966; Yao et al., 1997). Breeding disease-resistant wheat cultivars is the most economic, effective, and environmentally friendly strategy to control wheat stem rust (Hatta et al., 2021; Luo et al., 2022). Since the emergence of Ug99, we have increased our nationwide research on the variation in the virulence of P. graminis f. sp. tritici, identification of resistance genes, and resistance evaluation of cultivars to mitigate the threat posed by the Ug99 race group to Chinese wheat production (Cao et al., 2019; Li et al., 2016a; Xu et al., 2017; Li et al., 2021; Lin et al., 2021; Luo et al., 2022). However, in Hebei province, stem rust resistance hasn’t been a breeding goal. Therefore, the response of these cultivars to wheat stem rust in the field and the disease-resistance genes present in them are largely unknown. Therefore, in this study, we assessed the on-field infection response of wheat cultivars in Hebei province to two predominant races of P. graminis f. sp. tritici (21C3CTHTM and 34MRGQM) present in China. In addition, gene postulation, molecular testing, and pedigree analysis were performed to identify the genes resistant to wheat stem rust in these wheat cultivars.
Materials and methods
Plant materials and P. graminis f. sp. tritici races
A total of 65 wheat cultivars (Table 1) that were widely grown in Hebei province and 27 monogenic lines were included in this study (Jin et al., 2008). The 65 cultivars were provided by Dr. Hongfei Yan from College of Plant Protection, Hebei Agricultural University, Technological Innovation Center for Biological Control of Crop Diseases and Insect Pests of Hebei Province. The additional 27 monogenic lines include the 19 lines of the North American differential set (Jin et al., 2008) and eight additional lines (SwSr22T.B., Eagle, 73,214,3-1/9*LMPG, Mq(2)5XG2919, RL6082, RL6088, TAF2, and Fed/SrTt3) (Olivera Firpo et al., 2017; Olivera et al., 2022) with known Sr genes (Table 2) were preserved and propagated by the Institute of Plant Immunology of Shenyang Agricultural University Laboratory.
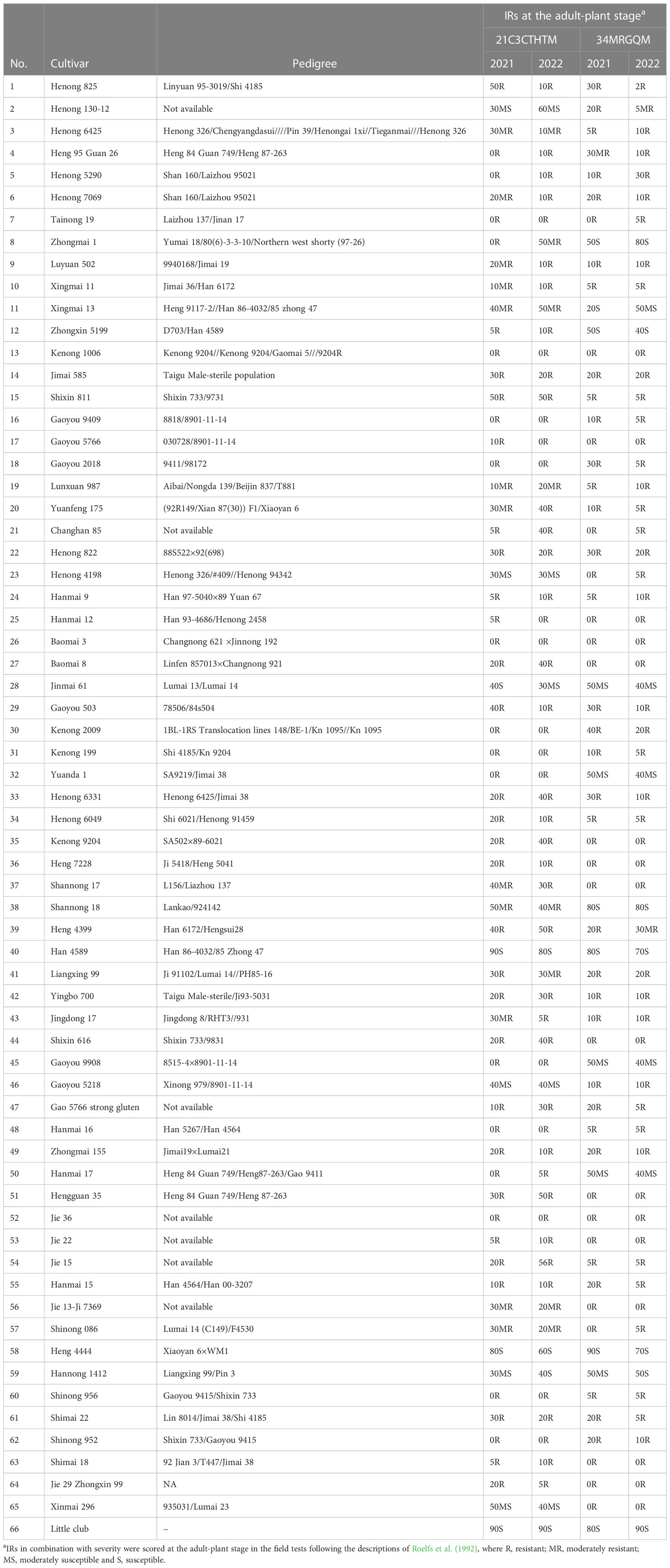
Table 1 Pedigree of tested cultivars, and stem rust infection response (IR) at the adult-plant stage.
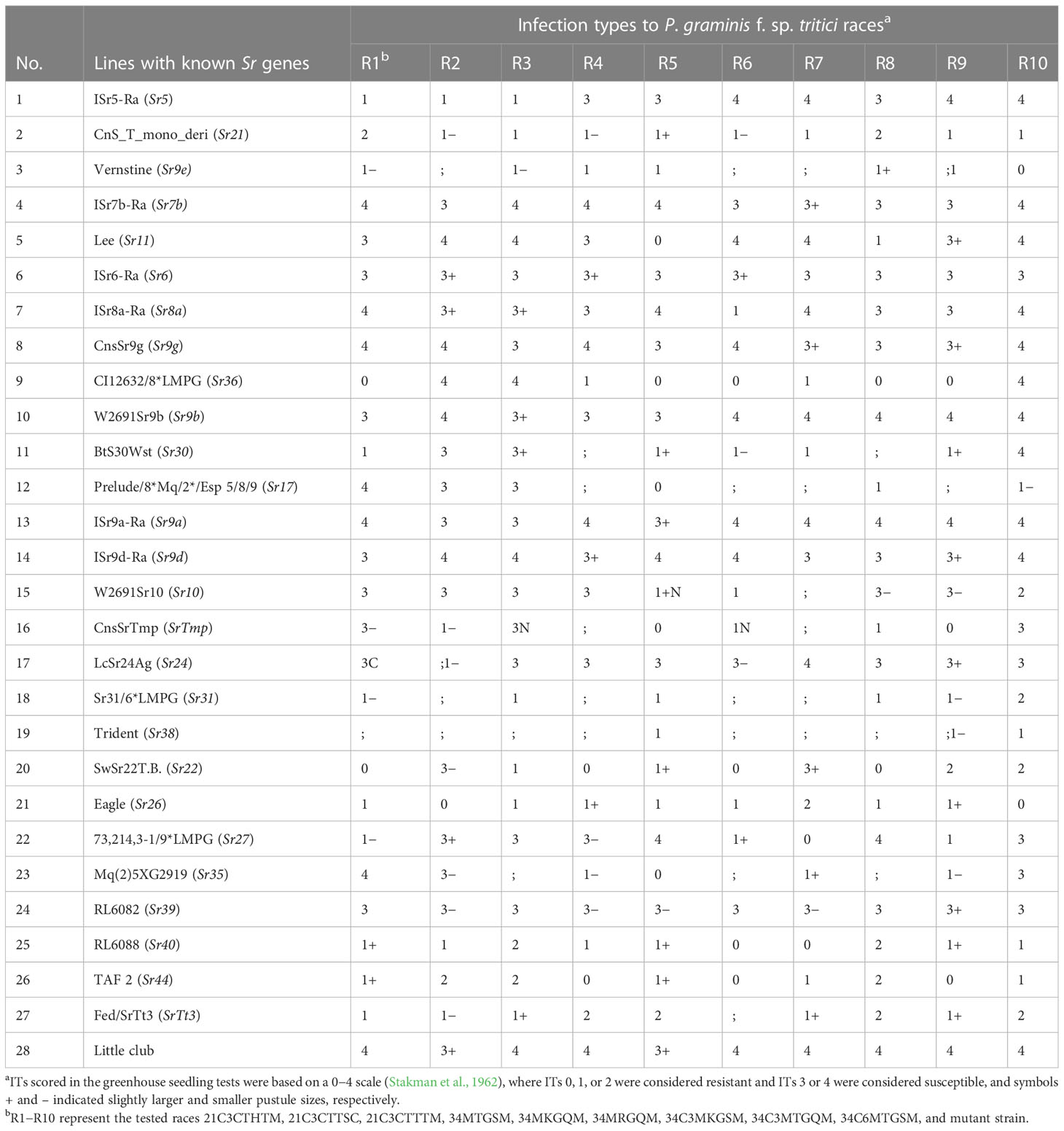
Table 2 The infection types of 34 monogenic lines of wheat with known Sr genes inoculated with 10 P. graminis f. sp. tritici.
The races of P. graminis f. sp. tritici used in this study were 21C3CTHTM, 21C3CTTTM, 21C3CTTSC, 34MKGQM, 34MTGSM, 34MRGQM, 34C3MTGQM, 34C3MKGSM, 34C6MTGSM, and a mutant strain 34MTSRM obtained via artificial irradiation of ultraviolet light. These P. graminis f. sp. tritici races were named as per the North American nomenclature system (Jin et al., 2008; Roelfs and Martens, 1988) and a new Chinese compound differential system for P. graminis f. sp. tritici (Cao et al., 2019).
Field tests
In the third week of March 2021 and 2022, all wheat cultivars were planted in the experimental site of Plant Protection College of Shenyang Agricultural University. The cultivation method was to plant one row of each cultivar (length 1 m and row spacing 25 cm) and one row of Little club (susceptibility check cultivar) for every 10 rows. 21C3CTHTM and 34MRGQM, the races of P. graminis f. sp. tritici dominant in recent two decades in China, were used for the field test. Inoculation was conducted at the jointing stage according to the method by Wu et al. (2020). Maximum severity and infection response (IR) were scored approximately 25 days after inoculation using a modified Cobb scale as described by Roelfs et al. (1992) when Little club exhibited disease severity of 90%–100%. IRs included I for immune, R for resistant, MR for moderately resistant, MS for moderately susceptible, and S for susceptible.
Seedling testing
The 10 races of P. graminis f. sp. tritici were used to inoculate the wheat cultivars, 27 monogenic lines with known Sr genes, and Little club (cultivar for susceptibility check). In total, 8–10 seeds of each genotype were planted in porcelain pots with a diameter of 12 cm in the greenhouse and inoculated with each Pgt race. At the two-leaf stage (one leaf and one sprout), the seedlings were inoculated with each race. Briefly, the leaves were sprayed with 0.05% Tween-20 aqueous solution, dusted with urediniospores mixed with talcum at a ratio of 1:20 (vol/vol), and sprayed again with 0.05% Tween-20 aqueous solution to produce a moisturizing film. The inoculated plants were kept moist in dark at 18–20°C for 16 h and further cultured in a glass greenhouse at 20 ± 1°C. After 2 weeks of inoculation, the infection types (ITs) were recorded based on the 0–4 scale described by Stakman et al. (1962). The ITs 0,;, 1−, 1, 1+, 2, and 2+ were classified as low ITs (resistant reaction), and 3−, 3, 3+, and 4 were classified as high ITs (susceptible reaction). According to the ITs of the cultivar evaluated and those of the wheat lines with known Sr gene, gene postulation was conducted as per the principle proposed by Dubin et al. (1989).
Molecular marker analysis
Genomic DNA of 65 wheat cultivars was extracted using cetyl trimethyl ammonium bromide (CTAB) (Xu et al., 2018). All cultivars were evaluated with molecular markers closely linked to six known genes Sr31, Sr38, Sr32, Sr24, Sr25, and Sr26. Polymerase chain reaction (PCR)-specific primers were synthesized by Shanghai Biotech Co., Ltd, China (Supplementary Table 1). PCR amplification was conducted in 25-µL reaction mixture, which included 1 µL of 10 µmol L−1 of each primer, 0.5 µL of 10 mmol L−1 deoxyribonucleoside triphosphates, 2.5 µL of 10× buffer (Mg2+), 0.2 µL of 5 U µL−1 taq polymerase, and 2 µL of 40 ng µL−1 DNA. The volume was made up to 25 µL using deionized water. The annealing temperature, allele size, and primer sequences for PCR are given in Supplementary Table 1.
Results
Field evaluation
The IRs of 65 wheat cultivars were tested with two predominant races 21C3CTHTM and 34MRGQM during 2021 and 2022 (Table 1). The IRs of all wheat cultivars could be classified into three groups immune, resistance to moderately resistance, and moderately susceptible to susceptible (Table 3). 56 (86.2%) and 55 (84.6%) wheat cultivars were resistant to the races 21C3CTHTM and 34MRGQM, respectively, in the field tests in 2021 and 2022. Overall, 50 (76.9%) wheat cultivars were resistant to all tested races.
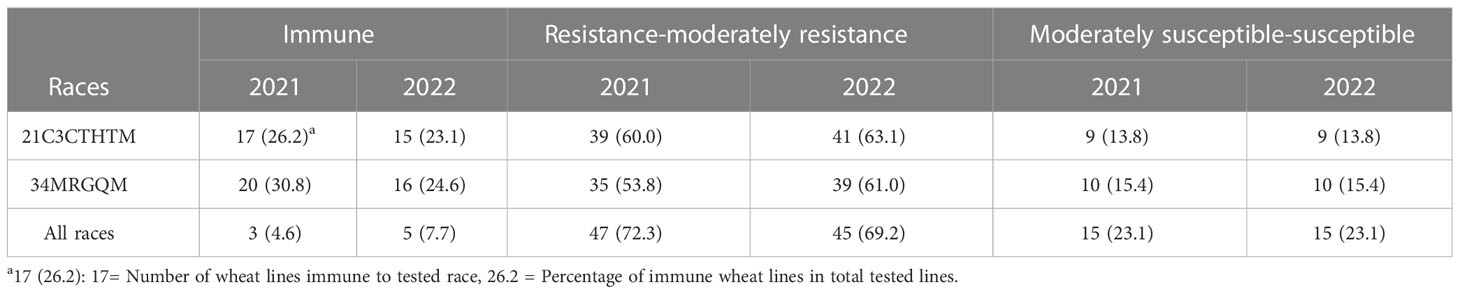
Table 3 Resistance and susceptibility of 65 wheat cultivars to 2 races of P. graminis f. sp. tritici at the at adult-plant stage in 2021 and 2022.
Sr genes in the wheat cultivars based on gene postulation and molecular marker analysis
The monogenic lines with known Sr genes (Sr6, Sr7b, Sr9a, Sr9b, Sr9d, Sr9g, and Sr39) and Little club exhibited high ITs to all Pgt races. In contrast, 9 Sr genes (Sr9e, Sr21, Sr31, Sr38, Sr26, Sr40, Sr44, and SrTt3) exhibited low ITs to all races. Therefore, these 18 Sr genes could not be deduced by gene postulation. However, 13 monogenic wheat lines (each containing Sr5, Sr11, Sr8a, Sr36, Sr30, Sr17, Sr10, SrTmp, Sr24, Sr22, Sr27, Sr32, and Sr35) exhibited high and low ITs for each of the 10 tested Pgt races; therefore, these genes could be used for gene postulation (Table 2). Molecular detection and gene postulation identified 6 Sr genes (Sr5, Sr17, Sr24, Sr31, Sr32, Sr38, and SrTmp), either singly or in combination, in 46 wheat cultivars.
The molecular marker analysis revealed that 37 wheat cultivars carried Sr31 (Table 4; Figure 1). All the lines that tested positive for the Sr31 marker were also resistant to all the evaluated races. None of these races are virulent to Sr31 gene, this again confirmed that Sr31 is present in these cultivars. Three wheat cultivars (Zhongmai 1, Xingmai 13, and Hanmai 17) were postulated to be carrying Sr5. Zhongmai 1 and Xingmai 13 contained only Sr5; these cultivars were resistant to the three races avirulent to Sr5 and were susceptible to 7 races virulent to Sr5 (34MTGSM, 34MKGQM, 34MRGQM, 34C3MKGSM, 34C3MTGQM, 34C6MTGSM, and mutant strain). Hanmai 17 was susceptible to races 34MTGSM and 34MKGQM but resistant to three Sr5-avirulent races (21C3CTHTM, 21C3CTTSC, and 21C3CTTTM) and some other races, indicating that this cultivar contains Sr5 and other unknown Sr gene(s). Three wheat cultivars, namely, Henong 4198, Gao 5218 strong gluten, and Jie 22, might contain Sr17 because they were resistant to all Sr17-avirulent races (34MTGSM, 34MKGQM, 34MRGQM, 34C3MKGSM, 34C3MTGQM, 34C6MTGSM, and mutant strain) and susceptible to three Sr17-virulent races. Although cultivar Heng 4444 exhibited a resistant/susceptible profile that indicates the presence of Sr17 at the seedling stage, in field evaluations against Sr17-avirulent race 34MRGQM exhibited a S response, indicating that Sr17 may not be present in this cultivar. Moreover, Gao 5218 strong gluten was confirmed to contain Sr24 using a specific molecular marker. Two wheat cultivars, namely, Jinmai 61 and Shannong 18, displayed low ITs to three races (21C3CTHTM, 21C3CTTTM, and mutant strain) and high ITs to the remaining 7 races (34C6MTGSM, 34MTGSM, 34MKGQM, 34MRGQM, 34C3MKGSM, 34C3MTGQM, and 21C3CTTSC). This was consistent with the ITs of SrTmp, indicating that they might contain this gene. However, Jinmai 61 exhibited 30MS to 40S susceptible response to race 21C3CTHTM in field tests, therefore, indicating that this cultivar does not carry SrTmp. Two wheat cultivars Jie 13-Ji 7369 and Kenong 1006 contained Sr32 and Sr38, respectively, as revealed by molecular marker analysis. Four wheat cultivars Lunxuan 987, Kenong 199, Heng 4399, and Shimai 22 exhibited resistance to all the races at the seedling stage and an MR to R infection response at the adult-plant stage, this indicated that these cultivars might contain one or more Sr genes (Sr9e, Sr21, Sr31, Sr38, Sr26, Sr40, Sr44, and SrTt3) that had low ITs to all races, as well as unknown effective resistance genes.
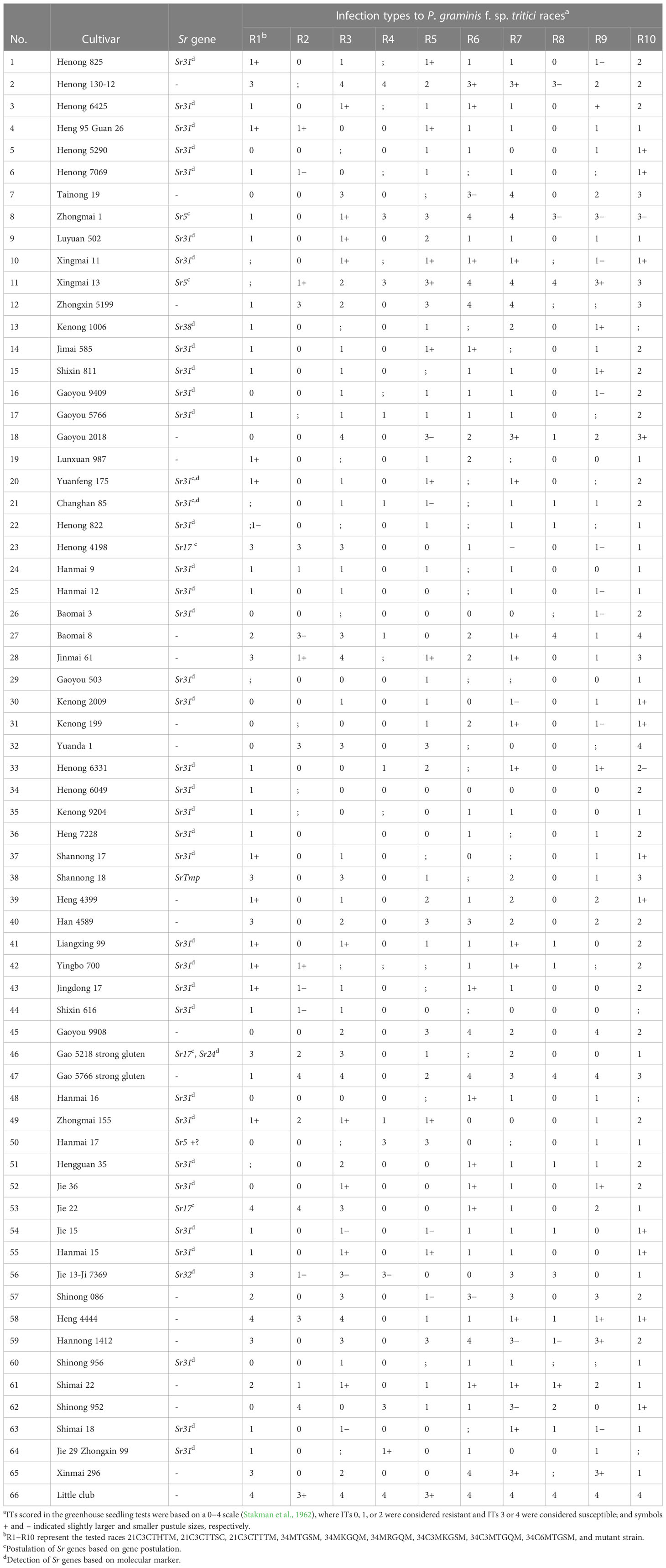
Table 4 Seedling infection types and presence or absence of stem rust resistance (Sr) genes in 65 wheat cultivars based on gene postulation using 10 P. graminis f. sp. tritici and molecular markers.
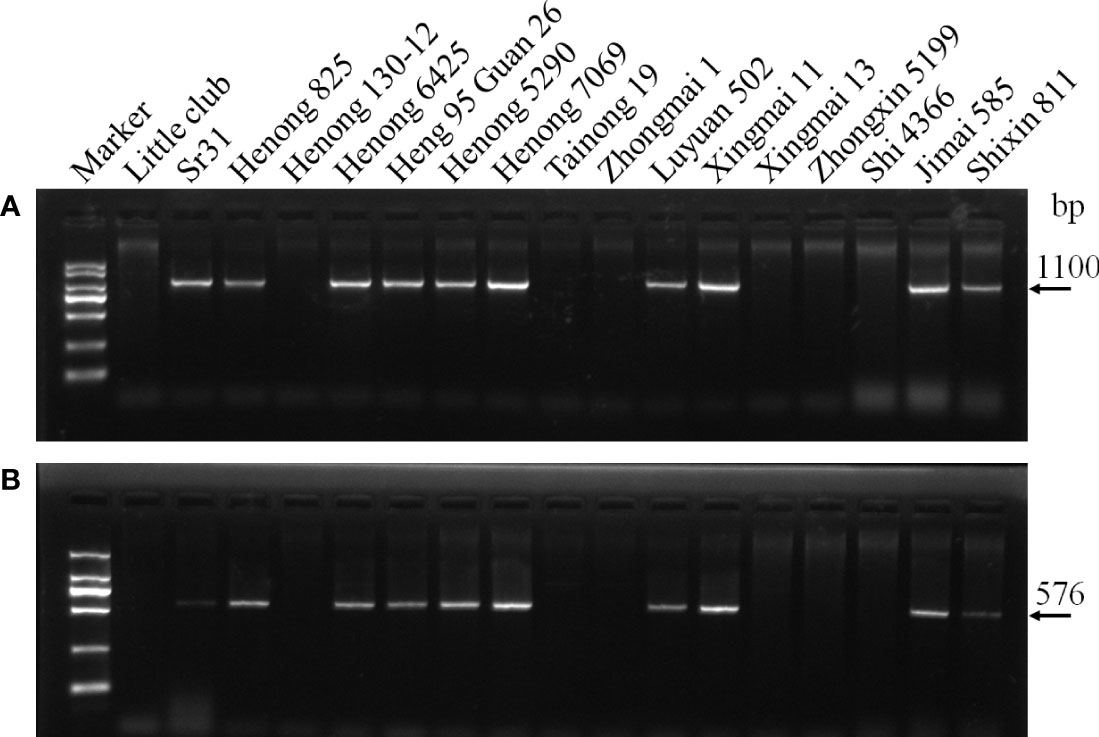
Figure 1 Amplification result for parts of wheat cultivars with markers SCSS30.2576 and Iag95. (A) Iag95, (B) SCSS30.2576.
Discussion
In the past two decades, with the emergence of new races of P. graminis f. sp. tritici, such as Ug99, TKTTF, and TTRTF, wheat stem rust is posing a serious threat to wheat production worldwide (Pretorius et al., 2000; Olivera et al., 2015; Singh et al., 2015; Bhattacharya, 2017; Wu et al., 2020). To mitigate this threat, intensive global efforts are underway to carefully monitor the evolution and migration of races, identify the pathotypes of the races, screen disease-resistant germplasms, and breed wheat cultivars with for durable disease resistance (Singh et al., 2011; Moscou and van Esse, 2017; Cao et al., 2019). Therefore, in this study, 65 wheat cultivars in Hebei Province, which played a bridge role in the epidemiology of wheat stem rust in China, were evaluated for the resistance to wheat stem rust in the field. Gene postulation and molecular marker analysis were conducted to identify the Sr genes present in these cultivars. Overall, 37 wheat cultivars (accounting for 56.9% of the tested cultivars) carrying Sr31 were identified, and these wheat cultivars were resistant to all races in the 2-year field test. Our results are consistent with many previous studies, proving that most Chinese wheat cultivars may contain Sr31 (Li et al., 2016a). This is mainly due to 1BL/1RS translocation (e.g., in wheat lines such as Alondra S, Aftab LeEr, Avorora, Kavkaz, Lovrin 10, Lovrin 13, and Neuzucht) carrying the linkage gene Sr31-Lr26-Yr9-Pm8, which has been widely used in disease-resistant breeding programs in China since the 1960s. Although Ug99 is virulent to Sr31, no race virulent to this gene has been found in China (Li et al., 2018). Overall, 37 wheat cultivars with Sr31 as revealed molecular detection exhibited low ITs to 10 races tested at the seedling stage and to 21C3CTHTM and 34MRGQM in the field test. Further pedigree analysis confirmed that the presence of Sr31 in Henong 825 and Jingdong 17 can be derived from Lovrin 10. 1BL/1RS translation was observed in the pedigrees of Henong 5290, Henong 7069, and Kenong 2009 wheat cultivars. Sr31 in Henong 6049, Shimai 18, and Shimai 22 might be from Avorara or Lovrin 10 because these Sr31 donors were present in the pedigree of these three wheat cultivars. Lovlin 13 and Lumai 14 containing Sr31 were found in the pedigree of Luyuan 205, Zhongmai 155, Liangxing 99, Shannong 17, and Yingbo 700 wheat cultivars, indicating that the presence of Sr31 in these cultivars may be inherited from Lovlin 13 or Lumai 14. Through molecular detection and gene postulation, Gebrewahid et al. (2020) confirmed that Henong 6049, Shimai 18, and Shixin 616 contain Lr26. Because Lr26 and Sr31 are closely linked, this result supported our observations.
Sr5, located on chromosome 6D, is present in a standard differential Reliance of P. graminis f. sp. tritici (Sears et al., 1957; Stakman et al., 1962). This gene is widely distributed in global wheat cultivars such as Cheyenne, Chisholm, Justin, Winoka, Arthur, Fortuna, Centurk, Kitt, Thatcher, and Agatha in North America; Catcher, Bindawarra, Aroona, and Avocet in Canada; and Panis, Primus, and Orlando in Europe (McIntosh et al., 1995). Sr5 is the second most commonly used wheat stem rust resistance gene in China after Sr31 and confers valuable resistance against the 21C3 race group of P. graminis f. sp. tritici (Cao et al., 2019). This is mainly because wheat cultivars Minn 2761, Dongfanghong 2, Kefang 1, Dongxie 3, Alondra’S’, Orofen, and their derivatives containing Sr5 have been grown for a long time and continually used in breeding programs (Wu et al., 2020). In this study, Zhongmai 1, Xingmai 13, and Hanmai 17 wheat cultivars were postulated to contain Sr5. Pedigree analysis revealed that the parent Yumai 18 of Zhongmai 1 contained rust-resistant germplasm St2422/464 from Italy (Yang et al., 2009). Therefore, it is speculated that Sr5 contained in Zhongmai 1 may originate from St2422/464.
Sr17, originating from Triticum turgidum var. dicoccum cv. Yaroslav emmer and located on 7BL of the wheat chromosome, is very common in Chinese wheat cultivars and provides resistance to all Sr5-virulent races and some races of the Sr5-avirulence group in China (Cao et al., 2019). Moreover, Sr17 is present in many wheat cultivars from the United States, Canada, India, and Mexico, particularly in those Lr14a and Pm5 as all three genes are linked (Roelfs and Mcvey, 1979). Sr17 is temperature sensitive. It is more effective against P. graminis f. sp. tritici at low temperature. When the temperature is higher than 25°C, the resistance becomes ineffective. In this study, Henong 4198, Gao 5218 strong gluten, and Jie 22 were postulated to contain this gene, and Gao 5218 strong gluten was confirmed to contain Sr24 using a specific molecular marker. Sr24, derived from Thinopyrum ponticum, is completely associated with Lr24. This gene is widely used in global wheat disease resistance breeding programs. Virulent races to Sr24 has been reported in South Africa, Kenya, Mozambique, India, and other countries; however, in North America, it is rarely reported (Jin et al., 2008; Xu et al., 2018). In China, the monogenic wheat line LcSr24Ag (carring Sr24) was first introduced from Cereal Disease Laboratory in University of Minnesota in 1990. When the virulence frequency test was conducted in the same year, 54 of the 89 tested P. graminis f. sp. tritici isolateswere virulent to the Sr24 (Yao et al., 1993). Since then, many P. graminis f. sp. tritici isolates with virulence to Sr24 have been detected, and the virulence frequency is approximately 50% (Wu and Huang, 1987; Cao et al., 2019). Considering the resistance of this gene to new races TKTTF and TTRTF, which caused epidemics in Ethiopia and Italy in 2014 and 2016, respectively (Olivera et al., 2019), we conducted molecular detection using 371 dominant cultivated wheat cultivars (including 65 in this study) in northern China using the molecular marker Sr24 # 12; only 1 wheat cultivar Gao 5218 strong gluten (in this study) may contain this gene. This is consistent with the study by Zhang (2015); screening using molecular markers linked to Lr24 revealed that none of the 406 wheat cultivars included in their study carried this gene (Zhang, 2015). These results revealed that only few wheat cultivars in China contain this gene.
SrTmp, originated from Triumph, was located on chromosome arm 6DS in a region similar to SrCad and Sr42 (Hiebert et al., 2016). This gene was present in commercial cultivars of hard red winter wheat to some extent since 1874 when a wheat cultivar Turkey was introduced (Roelfs and Mcvey, 1979). This cultivar is particularly widely used in the United States, Pakistan, Ethiopia, Nebraska, and other countries (Sajid et al., 2009; Cao et al., 2019). This gene not only has good resistance to many races of P. graminis f. sp. tritici in China but also has resistance to Ug99 and some Ug99 variants (Cao et al., 2019; Li et al., 2016b). In present study, Shannong 18 was postulated to carry SrTmp. Pedigree tracing analysis revealed that Shannong 18 is derived from Yannong 74. Youbaomai, Yannong 74’s parent, contains leaf-rust resistance genes Lr1 and Lr35 and stem-rust resistance gene SrTmp. Therefore, it is speculated that SrTmp contained in these two cultivars comes from Youbaomai (Zhuang, 2003).
Genes Sr25, Sr26, and Sr32 confer resistance to all P. graminis f. sp. tritici races belonging to the Ug99 lineage. Considering the disastrous threat posed by the Ug99 race group on world food security, Nobel Peace Prize laureate Norman Borlaug called for a coordinated global campaign to monitor the populations of Ug99 races and screen wheat germplasms for resistance to Ug99. This can help to control wheat stem rust epidemics and mitigate the potential impact on food security (Singh et al., 2011; Wu et al., 2020; Lin et al., 2021). In view of this, we used molecular markers closely linked to Sr25, Sr26, and Sr32 to screen these genes to identify possible resistant germplasm to Ug99. To date, of the 891 wheat cultivars tested by us, only 1, 2, and 2 wheat cultivars contained Sr25, Sr26, and Sr32, respectively (including 1 in this study). Therefore, very few wheat cultivars from China contain these three genes. Sr38, originated from T. ventricosum, is completely linked with leaf-rust resistance gene Lr37 and stripe rust gene Yr17. Because of the combined resistance to all three rust diseases, Sr38 was widely used in disease-resistance wheat breeding in China (Wu et al., 2020). However, surprisingly, only one of the 65 main wheat cultivars tested in this study possibly contained this gene. Although Sr38 has become ineffective for Ug99 and its variants, no race of P. graminis f. sp. tritici with virulence to this gene has been detected in China.
In summary, our study demonstrated that the wheat cultivars in Hebei Province have very limited genes (most abundant being Sr31) that are resistance to stem rust. These cultivars currently exhibit good resistance to wheat stem rust in Hebei Province. However, if the virulence of P. graminis f. sp. tritici changes in future, particularly once Ug99 race group spreads to China, it will lead to serious epidemics and huge wheat yield losses. Therefore, more disease-resistance genes should be used in disease-resistance breeding to maintain the durable resistance of wheat cultivars.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
TL conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft. HS, XN, RW and ZW performed the experiments, authored or reviewed drafts of the paper. SC, FG and YZ prepared figures and/or tables. XW and YX analyzed the data, prepared figures and/or tables. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Natural Science Foundation of Education Department of Liaoning Province (No. LJKZ0648).
Acknowledgments
We thank Dr. H.F. Yan at the College of Plant Protection, Hebei Agricultural University, Technological Innovation Center for Biological Control of Crop Diseases and Insect Pests of Hebei Province for providing the 65 wheat cultivars.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1156936/full#supplementary-material
References
Bhattacharya, S. (2017). Deadly new wheat disease threatens europe’s crops. Nature 542, 145–146. doi: 10.1038/nature.2017.21424
Campbell, C. L., Long, D. L. (2001). “The campaign to eradicate the common barberry in the united states,” in Stem rust of wheat: From ancient enemy to modern foe. Ed. Peterson, P. D. (St. Paul, MN: APS Press), 16–50.
Cao, Y. Y., Si, B. B., Zhu, G. Q., Xu, X. F., Li, W. H., Chen, S., et al. (2019). Race and virulence of asexual and sexual populations of Puccinia graminis f. sp. tritici in China from 2009 to 2015. Eur. J. Plant Pathol. 153, 545–555. doi: 10.1007/s10658-018-1581-x
Dubin, H. J., Johnson, R., Stubbs, R. W. (1989). Postulated genes for resistance to stripe rust in selected CIMMYT and related wheats. Plant Dis. 73, 472–475.
Fetch, T., Zegeye, T., Park, R. F., Hodson, D., Wanyera, R. (2016). Detection of wheat stem rust races TTHSK and PTKTK in the Ug99 race group in Kenya in 2014. Plant Dis. 7, 1495. doi: 10.1094/PDIS-11-15-1356-PDN
Gebrewahid, T. W., Zhang, P. P., Yan, Z. J., Li, Z. F., Liu, D. Q. (2020). Identification of leaf rust resistance genes in bread wheat cultivars from Ethiopia. Plant Dis. 109, 2354–2361. doi: 10.1094/PDIS-12-19-2606-RE
Hatta, M. A. M., Arora, S., Ghosh, S., Matny, O., Smedley, M. A., Yu, G., et al. (2021). The wheat Sr22, Sr33, Sr35 and Sr45 genes confer resistance against stem rust in barley. Plant Biotechnol. J. 19 (2), 273–284. doi: 10.1111/pbi.13460
Hiebert, C. W., Kassa, M. T., McCartney, C. A., You, F. M., Rouse, M. N., Fobert, P., et al. (2016). Genetics and mapping of seedling resistance to Ug99 stem rust in winter wheat cultivar triumph 64 and differentiation of SrTmp, SrCad, and Sr42. Theor. Appl. Genet. 129 (11), 2171–2177. doi: 10.1007/s00122-016-2765-4
Jin, Y., Szabo, L. J., Pretorius, Z. A., Singh, R. P., Ward, R., Fetch, T. J. (2008). Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 92, 923–926. doi: 10.1094/PDIS-92-6-0923
Li, T. Y., Cao, Y. Y., Wu, X. X., Xu, X. F., Wang, W. L. (2016a). Seedling resistance to stem rust and molecular marker analysis of resistance genes in wheat cultivars of yunnan, China. PloS One 11 (10), e0165640. doi: 10.1371/journal.pone.0165640
Li, H. G., Hua, L., Rouse, M. N., Li, T. Y., Pang, S. Y., Bai, S. S., et al. (2021). Mapping and characterization of a wheat stem rust resistance gene in durum wheat “Kronos”. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.751398
Li, T. Y., Ma, Y. C., Wu, X. X., Chen, S., Xu, X. F., Wang, H., et al. (2018). Race and virulence characterization of Puccinia graminis f. sp. tritici in China. PloS One 13 (5), e0197579. doi: 10.1371/journal.pone.0197579
Li, T. Y., Wu, X. X., Xu, X. F., Wang, W. L., Cao, Y. Y. (2016b). Postulation of seedling stem rust resistance genes of yunnan wheat cultivars in china. Plant prot. Sci. 52, 242–249. doi: 10.17221/137/2015-PPS
Lin, Q. J., Gao, Y., Wu, X. X., Ni, X. Y., Chen, R. Z., Xuan, Y. H., et al. (2021). Evaluation of resistance to wheat stem rust and identification of resistance genes in wheat lines from heilongjiang province. PeerJ 9, e10580. doi: 10.7717/peerj.10580
Luo, J., Rouse, M. N., Hua, L., Li, H., Li, B., Li, T. Y., et al. (2022). Identification and characterization of Sr22b, a new allele of the wheat stem rust resistance gene Sr22 effective against the Ug99 race group. Plant Biotechnol. J. 20 (3), 554–563. doi: 10.1111/pbi.13737
McIntosh, R. A., Wellings, C. R., Park, R. F. (1995). Wheat rusts: An atlas of resistance genes (Australia: CSIRO). doi: 10.1071/9780643101463
Moscou, M. J., van Esse, E. H. (2017). The quest for durable resistance. Science 358, 1541–1542. doi: 10.1126/science.aar4797
Nazari, K., Al-Maaroof, E., Kurtulus, E., Kavaz, H., Hodson, D., Ozseven, I. (2021). First report of Ug99 race TTKTT of wheat stem rust (Puccinia graminis f. sp. tritici) in Iraq. Plant Dis. 9, 2719. doi: 10.1094/PDIS-02-21-0404-PDN
Olivera, P., Newcomb, M., Szabo, L. J., Rouse, M., Johnson, J., Gale, S., et al. (2015). Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013-14. Phytopathology 105, 917–928. doi: 10.1094/PHYTO-11-14-0302-FI
Olivera, P. D., Sikharulidze, Z., Dumbadze, R., Szabo, L. J., Newcomb, M., Natsarishvili, K., et al. (2019). Presence of a sexual population of Puccinia graminis f. sp. tritici in georgia provides a hotspot for genotypic and phenotypic diversity. Phytopathology 109, 2152–2160.
Olivera, P. D., Szabo, L. J., Kokhmetova, A., Morgounov, A., Luster, D. G., Jin, Y. (2022). Puccinia graminis f. sp. tritici population causing recent wheat stem rust epidemics in Kazakhstan is highly diverse and includes novel virulence pathotypes. Phytopathology 112 (11), 2403–2415. doi: 10.1094/PHYTO-08-21-0320-R
Olivera Firpo, P. D., Newcomb, M., Flath, K., Sommerfeldt-Impe, N., Szabo, L. J., Carter, M., et al. (2017). Characterization of Puccinia graminis f. sp. tritici isolates derived from an unusual wheat stem rust outbreak in Germany in 2013. Plant Pathol. 66, 1258–1266. doi: 10.1111/ppa.12674
Pardey, P. G., Beddow, J. M., Kriticos, D. J., Hurley, T. M., Park, R. F., Duveiller, E., et al. (2013). Right-sizing stem-rust research. Science 340, 147–148. doi: 10.1126/science.122970
Patpour, M., Hovmøller, M. S., Justesen, A. F., Newcomb, M., Olivera, P., Jin, Y., et al. (2016). Emergence of virulence to SrTmp in the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Africa. Plant Dis. 2, 522. doi: 10.1094/PDIS-06-15-0668-PDN
Peterson, P. D. (2018). The barberry eradication program in minnesota for stem rust control: A case study. Annu. Rev. Phytopathol. 56, 203–223. doi: 10.1146/annurev-phyto-080417-050133
Pretorius, Z. A., Bender, C. M., Visser, B., Terefe, T. (2010). First report of a Puccinia graminis f. sp. tritici race virulent to the Sr24 and Sr31 wheat stem rust resistance genes in south Africa. Plant Dis. 94, 784. doi: 10.1094/PDIS-94-6-0784C
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 84, 203. doi: 10.1094/PDIS.2000.84.2.203B
Roelfs, A. P., Mcvey, D. V. (1979). Low infection types produced by Puccinia graminis f. sp. tritici and wheat line with designated genes for resistance. Phytopathology 69, 722–730. doi: 10.1094/phyto-69-722
Roelfs, A. P., Martens, J. W. (1988). An international system of nomenclature for Puccinia graminis f. sp. tritici. Phytopathology 78, 526–533.
Roelfs, A. P., Singh, R. P., Saari, E. E. (1992). Rust diseases of wheat: concepts and methods of disease management. (Mexico, D.F.: CIMMYT)
Rouse, M. N., Nava, I. C., Chao, S., Anderson, J. A., Jin, Y. (2012). Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum l.). Theor. Appl. Genet. 125, 877–885. doi: 10.1007/s00122-012-1879-6
Rust Tracker (2021) A global wheat rust monitoring system, pathotype tracker – where is Ug99? Available at: https://rusttracker.cimmyt.org/?page_id=22.
Sajid, A., Syed, J. A., Hidayatur, R., Muhammad, S. S., Muhammad, I., Muhammad, S. (2009). Variability in wheat yield under yellow rust pressure in Pakistan. Turk. J. Agric. For. 33, 537–546. doi: 10.3906/tar-0812-27
Shamanin, V., Shepelev, S., Pozherukova, V., Gultyaeva, E., Kolomiets, T., Pakholkova, E., Morgounov, A. (2009). Primary hexaploid synthetics: Novel sources of wheat disease resistance. Crop Prot. 121, 7–10. doi: 10.1016/j.cropro.2019.03.003
Sears, E. R., Loegering, W. Q., Rodenhiser, H. A. (1957). Identification of chromosomes carrying genes for stem rust resistance in four varieties of wheat. Agron. J. 49(4), 208–212. doi: 10.2134/agronj1957.00021962004900040012x
Shank, R. (1994). Wheat stem rust and drought effects on bale agricultural production and future prospects. report on February 17-28 assessment (Addis Ababa, Ethiopia: United Nations Emergencies Unit for Ethiopia).
Singh, R. P., Hodson, D. P., Huerta-Espino, J., Jin, Y., Bhavani, S., Njau, P., et al. (2011). The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 49, 465–481. doi: 10.1146/annurev-phyto-072910-095423
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: Continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884. doi: 10.1094/PHYTO-01-15-0030-FI
Stakman, E. C., Steward, D. M., Loegering, W. Q. (1962). Identification of physiologic races of Puccinia graminis var. tritici. U.S. Dep. Agric. Agric. Res. Serv., E–617. doi: 10.1080/07060668109501400
Wanyera, R., Kinyua, M. G., Jin, Y., Singh, R. (2006). The spread of stem rust caused by Puccinia graminis f. sp. tritici, with virulence on Sr31 in wheat in Eastern Africa. Plant Dis. 90, 113. doi: 10.1094/PD-90-0113A
Wu, Y. S., Huang, Z. T. (1987). Twenty year’s racial identification and fluctuation analysis of Puccinia graminis var. tritici in China. J. Shenyang Agric. Univ. 18, 105–108.
Wu, Y. S., Huang, Z. T., Wei, S. X., Reng, Y. Z., Xue, L. X. (1966). An analysis of the physiological races of wheat stem rust in 1964 in china. Acta polym. Sin. 5 (1), 59–61.
Wu, Y. S., Huang, Z. T., Wei, S. X., Zeng, G. R., Xue, L. X. (1963). An analysis of the physiological races of wheat stem rust in 1963. Acta Polym. Sin. 4 (3), 294–296.
Wu, X. X., Lin, Q. J., Ni, X. Y., Sun, Q., Chen, R. Z., Xu, X. F., et al. (2020). Characterization of wheat monogenic lines with known Sr genes and wheat lines with resistance to the Ug99 race group for resistance to prevalent races of Puccinia graminis f. sp. tritici in China. Plant Dis. 104 (7), 1939–1943. doi: 10.1094/PDIS-12-19-2736-RE
Xu, X. F., Li, D. D., Liu, Y., Gao, Y., Wang, Z. Y., Ma, Y. C., et al. (2017). Evaluation and identification of stem rust resistance genes Sr2, Sr24, Sr25, Sr26, Sr31 and Sr38 in wheat lines from gansu province in China. PeerJ 5, e4146. doi: 10.7717/peerj.4146
Xu, X. F., Yuan, D. P., Li, D. D., Gao, Y., Wang, Z. Y., Liu, Y., et al. (2018). Identification of stem rust resistance genes in wheat cultivars in China using molecular markers. PeerJ 6, e4882. doi: 10.7717/peerj.4882
Yang, C. L., Hou, J. H., Song, Z. J., Li, X. L., Li, G. Y. (2009). Pedigree analysis of main wheat cultivars and utilization of core gemplasms in henan province. Shandong Agric. Sci. 1, 27–31.
Yao, P., Cao, Y. Y., Liu, W. Z., Wu, Y. S. (1997). Race population trend of Puccinia graminis f. sp. tritici in 1990-1994 in China. Acta Polym. Sin. 24 (4), 297–302.
Yao, P., Cao, Y. Y., Wu, Y. S. (1993). Race identification of Puccinia graminis f. sp. tritici in China in 1990. J. Plant Prot. Res. 20 (1), 65–70.
Zhang, Y. Q. (2015). Identification of wheat leaf rust resistance gene in 460 Chinese wheat cultivars and QTL mapping for adult-plant resistance gene to leaf rust in Italian wheat cultivar libellula (Hebei, China: Hebei Agric. Univ). Master’s thesis.
Keywords: Puccinia graminis f. sp. tritici, wheat cultivars, resistance gene, wheat stem rust, Sr genes
Citation: Sun H, Wang Z, Wang R, Chen S, Ni X, Gao F, Zhang Y, Xu Y, Wu X and Li T (2023) Identification of wheat stem rust resistance genes in wheat cultivars from Hebei province, China. Front. Plant Sci. 14:1156936. doi: 10.3389/fpls.2023.1156936
Received: 02 February 2023; Accepted: 13 March 2023;
Published: 30 March 2023.
Edited by:
Peng Zhang, The University of Sydney, AustraliaReviewed by:
Jianping Zhang, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaPablo Daniel Olivera, University of Minnesota Twin Cities, United States
Copyright © 2023 Sun, Wang, Wang, Chen, Ni, Gao, Zhang, Xu, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianya Li, bGl0aWFueWExMUBzeWF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Huiyan Sun1†
Huiyan Sun1† Tianya Li
Tianya Li