
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 March 2023
Sec. Plant Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1156356
This article is part of the Research Topic Plant and Algal Lipids: Recent Trends and Challenges in a Changing World View all 9 articles
Suberin, a complex polyester deposited in the seed coat outer integument, acts as a hydrophobic barrier to control the movement of water, ions, and gas. However, relatively little is known about the signal transduction involved in suberin layer formation during seed coat development. In this study, the effect of the plant hormone abscisic acid (ABA) on suberin layer formation in seed coats was investigated by characterizing mutations in Arabidopsis related to ABA biosynthesis and signaling. Seed coat permeability to tetrazolium salt was noticeably elevated in aba1-1 and abi1-1 mutants, but not significantly altered in snrk2.2/3/6, abi3-8, abi5-7, and pyr1pyl1pyl2pyl4 quadruple mutants compared with that in the wild-type (WT). ABA1 encodes a zeaxanthin epoxidase that functions in the first step of ABA biosynthesis. aba1-1 and aba1-8 mutant seed coats showed reduced autofluorescence under UV light and increased tetrazolium salt permeability relative to WT levels. ABA1 disruption resulted in decreased total seed coat polyester levels by approximately 3%, with a remarkable reduction in levels of C24:0 ω-hydroxy fatty acids and C24:0 dicarboxylic acids, which are the most abundant aliphatic compounds in seed coat suberin. Consistent with suberin polyester chemical analysis, RT-qPCR analysis showed a significant reduction in transcript levels of KCS17, FAR1, FAR4, FAR5, CYP86A1, CYP86B1, ASFT, GPAT5, LTPG1, LTPG15, ABCG2, ABCG6, ABCG20, ABCG23, MYB9, and MYB107, which are involved in suberin accumulation and regulation in developing aba1-1 and aba1-8 siliques, as compared with WT levels. Together, seed coat suberization is mediated by ABA and partially processed through canonical ABA signaling.
The seed coat surrounds the outermost layer of the seed, and the immature seed coat is comprised of five distinctive cell layers, specifically two layers of outer integument and three layers of the inner integument. Each of the layers has its own disparate developmental paths, but all of them eventually merge into two layers of dead cells (Haughn and Chaudhury, 2005). Through and beyond maturation, the seed coat provides protection against mechanical stress, various pathogens, and herbivores to the embryo and seed itself. It also enhances germination, dormancy, and seed dispersal and transmits signals from the outside to the embryo. These roles played by the seed coat contribute to the proper maturation and preservation of the seed to the next generation (Haughn and Chaudhury, 2005; Radchuk and Borisjuk, 2014). During seed coat development, the outer integument undergoes cell wall thickening (Beeckman et al., 2000), wherein suberin, a lipophilic polyester, is deposited (Molina et al., 2006; Molina et al., 2008). Suberin in the seed coat, shown to be highly concentrated in the chalazal region, contributes to seed dormancy and imparts seed coat impermeability to tetrazolium (TZ) salts (Beisson et al., 2007). It was also recently reported that seed coat suberin forms a physical barrier against heavy metals, such as Cr3+, in the soil (de Silva et al., 2021). Suberin is mainly comprised of ester-linked glycerol, very-long-chain fatty acids (VLCFAs), ω-hydroxy fatty acids, α,ω-dicarboxylic acids, primary alcohols, and ferulate. Unlike root suberin, which is dominated by monomers of C16, C18:1, and C22, seed coat suberin contains monomers of C22 and C24 as its major components (Molina et al., 2006; Li-Beisson et al., 2013).
For the synthesis of suberin aliphatic precursors in the endoplasmic reticulum, 3-ketoacyl-CoA synthase KCS2/DAISY and KCS20 contribute redundantly to the elongation of C20 to C22 VLCFAs, and KCS9 extends the C22 to C24 VLCFAs in Arabidopsis roots and seed coats (Franke et al., 2009; Lee et al., 2009; Kim et al., 2013). Further, potato KCS6 is involved in potato tuber periderm suberization (Serra et al., 2009). Moreover, the levels of C18, C20, and C22 fatty acids are specifically reduced by fatty acyl-CoA reductases FAR5, FAR4, and FAR1 to form respective suberin alcohols (Domergue et al., 2010; Vishwanath et al., 2013). In addition, the conversion of fatty acids to ω-hydroxy fatty acids, mediated by the hydroxylation of ω-carbon, is catalyzed by fatty acid ω-hydroxylase CYP86A1/HORST and CYP86B1/RALPH, where the former has specificity for substrates of ≤ C18 and the latter for substrates of > C20 (Höfer et al., 2008; Compagnon et al., 2009; Molina et al., 2009). ω-Hydroxy fatty acids and α,ω-dicarboxylic acids are then activated to acyl-CoA forms, which are then acylated to generate glycerol-3-phosphate by glycerol-3-phosphate acyltransferase 5 (GPAT5) (Beisson et al., 2007; Li et al., 2007; Yang et al., 2010; Yang et al., 2012). Meanwhile, phenolic monomers, including ferulic acids, are synthesized via the phenylpropanoid pathway, processed in the cytoplasm. Knock-out analysis of BAHD-family genes encoding aliphatic suberin feruloyl transferase/ω-hydroxyacid hydroxycinnamoyl transferase (ASFT/HHT) revealed that ASFT contributes to the incorporation of ferulate into suberin polyester by catalyzing feruloyl-CoA transfer to ω-hydroxy fatty acids and primary alcohols (Gou et al., 2009; Molina et al., 2009). The resulting suberin monomers are secreted through the plasma membrane to be deposited inside the cell wall. ATP binding cassette transporters ABCG1, ABCG2, ABCG6, and ABCG20 contribute to suberin monomer export in roots and/or seed coats (Yadav et al., 2014; Fedi et al., 2017; Shanmugarajah et al., 2019). Arabidopsis lipid transfer protein, AtLTPI-4, is involved in the transport of long-chain (LC) and very-long-chain (VLC) suberin monomers in the Arabidopsis crown gall (Deeken et al., 2016), and glycosylphosphatidylinositol (GPI)-anchored lipid transfer protein 15 (LTPG15) is associated with the export of VLC suberin monomers in seed coats (Lee and Suh, 2018). Upstream of aliphatic and aromatic suberin precursor biosynthesis, seed-specific MYB9 and MYB107 activate the expression of suberization-associated genes (Lashbrooke et al., 2016; Gou et al., 2017). Moreover, MYB transcription factors such as MYB36, MYB39, MYB41, MYB53, MYB92, and MYB93 function as regulators of root endodermal suberization (Kosma et al., 2014; Cohen et al., 2020; To et al., 2020; Wang et al., 2020; Shukla et al., 2021). A NAC family transcription factor, ANAC046 was reported to regulate suberization in Arabidopsis roots (Mahmood et al., 2019).
Abscisic acid (ABA) has long been implicated in the regulation of suberization. This compound is a xanthophyll-derived phytohormone (Nambara and Marion-Poll, 2005) that is actively synthesized during the seed maturation stage and contributes to the deposition of seed storage reserves, improvements in desiccation tolerance, and achieving primary seed dormancy (Kermode, 2005). ABA biosynthesis begins in the plastids with the epoxidation of zeaxanthin to form violaxanthin, which is catalyzed by zeaxanthin epoxidase, encoded by the ABA1 locus of Arabidopsis (Rock and Zeevaart, 1991). Its stimulating role was first observed in suberization of the potato disk and tissue culture upon treatment with exogenous ABA (Soliday et al., 1978). Cottle and Kolattukudy (1982) further observed the increased accumulation of suberin monomers and the induction of enzymes associated with suberization in ABA-treated tissue cultures of potato tubers. Later, the effect of ABA was detected in various suberized tissues, such as the wound-healing sites of tomato (Leide et al., 2012; Tao et al., 2016), potato tuber (Lulai et al., 2008; Boher et al., 2013), and kiwifruit (Han et al., 2017; Han et al., 2018; Han et al., 2019; Wei et al., 2020a; Wei et al., 2020b) and in Agrobacterium-induced tumors in Arabidopsis (Efetova et al., 2007). The formation of Arabidopsis root suberin was determined to be highly stimulated by exogenous ABA treatment, and this response was hindered with the endodermis-specific expression of abi1-1, which suppresses ABA signaling, suggesting the involvement of ABA signaling in the regulation of Arabidopsis root suberization (Barberon et al., 2016).
Despite several preceding studies on the relationship between ABA signaling and suberization, little is known about whether ABA is involved in seed coat suberization. Considering that ABA signaling and seed coat suberin both significantly contribute to proper seed maturation and dormancy, it is reasonable to speculate on the relationship between them. In this study, the seed coat permeability of an ABA biosynthetic mutant, aba1-1 and several ABA signaling mutants was tested through TZ staining. To further investigate the consequences of ABA1 disruption in terms of seed coat suberization, an additional T-DNA insertional mutant allele of aba1 was isolated and named aba1-8. Here, UV autofluorescence observations, TZ staining, and suberin monomer analysis were performed using wild-type, aba1-1, and aba1-8 seeds, which were followed by RT-quantitative PCR analysis. We revealed that the disruption of ABA1 hinders proper seed coat suberization via downregulation of the expression of genes involved in suberin biosynthesis.
All experiments were performed using Arabidopsis thaliana plants. The seed surfaces were sterilized by inverting them in 0.05% Triton X-100 in 75% EtOH for 30 seconds, followed by rinsing them in 100% EtOH 2~3 times. The sterilized seeds were stratified at 4°C for 3 days and grown on 0.6% ½ MS agar medium supplemented with 1% sucrose and then transferred to soil after 7 days if needed. The plants were grown at 23°C, with 50~60% humidity under long-day conditions (16 h light/8 h dark). The T-DNA insertional mutant line, aba1-8 (SAIL_310_A04, At5g67030), was obtained from ABRC (Arabidopsis Biological Resource Center) and selected on ½ MS agar medium containing 10 µg mL-1 phosphinothricin. Homogenous mutants were isolated via PCR screening with the primers listed in Table S1, and knockout of the ABA1 gene was verified through RT-PCR, described below. The rest of the ABA biosynthesis- or signaling-defective mutant [aba1-1, QC3 (pyr1pyl1pyl2pyl4 with Col-0 background), QL3 (pyr1pyl1pyl2pyl4 with Ler background), abi1-1, snrk2.2/3/6+, abi3-8, abi5-7] seeds were obtained from Prof. Soo Young Kim (Chonnam National University).
To verify the aba1 T-DNA insertion knockout mutant, total RNA was isolated from 9-day-old seedlings using the RNeasy® Plant Mini Kit (Qiagen, 74904) following the manufacturer’s instructions. For RT-qPCR analysis, total RNA was isolated from developing siliques at 4–10 DAF, using the modified version of the methods reported by Oñate-Sánchez and Vicente-Carbajosa (2008). Here, 550 µL extraction buffer (0.2 M LiCl, 200 mM Tris-HCl pH 8.0, 25 mM EDTA, 1% SDS) and 550 µL chloroform were added to approximately 50 mg of ground tissues, vortexed vigorously, and centrifuged for 3 minutes at RT. The supernatant was mixed with 700 µL phenol/chloroform/isoamyl alcohol (IAA; Ambion, AM9720), vortexed, and centrifuged for 3 minutes at RT to remove the remaining carbohydrates. The supernatant was mixed with the same volume of chloroform and centrifuged to additionally remove carbohydrates if needed. Next, the supernatant was mixed with a 1/3 volume of 8 M LiCl and incubated at −20°C for 1 hour to precipitate the nucleic acids. The mixture was centrifuged at 4°C for 30 minutes to retrieve the nucleic acids as pellets. The supernatant was removed completely, the pellets were dissolved in RNase free water and treated with RNase free DNaseI (Qiagen, 79254), and the sample was incubated at 25°C for 1 hour. After incubation, a 1/3 volume of phenol/chloroform/IAA was added to the mixture, vortexed, and centrifuged to remove the enzymes. The supernatant was mixed with a 1/10 volume 3 M NaoAc, pH 5.2, and two volumes of 100% ethanol, inverted, and incubated at −20°C for at least 1 hour. The mixture was centrifuged at 4°C for 20 minutes, and the pellets were washed with 70% ethanol. Washed pellets were air-dried and dissolved in RNase free water. Then, 2 µg of isolated total RNA was used to synthesize complementary DNA using GoScript Reverse Transcriptase (Promega, A5003), following the manufacturer’s instructions. The obtained cDNA was used for RT-PCR and RT-qPCR analyses. PP2AA3 (At1g13320) was used as a reference gene for RT-PCR to normalize RNA expression levels. RT-qPCR was performed using TOPreal™ qPCR 2X Premix (SYBR Green with low ROX; Bio-Rad, RT500M) following the manufacturer’s instructions, and the expression level of each gene was normalized to that of ACTIN7 (At5g09810). The primers were used as listed in Table S1.
For suberin autofluorescence visualization, 100~110 seeds were observed at a time under ultraviolet light (365 nm excitation) and photographed using a digital camera. For the seed coat permeability assay, wild-type and aba1 mutant seeds were immersed in 1% (w/v) TZ red (2, 3, 5-triphenyltetrazolium chloride) solution and incubated at 30°C in the dark. Stained seeds were observed after 8 and 24 hours of incubation and photographed using a digital camera equipped with a micro-lens. To quantify the amount of formazan produced, 50 mg of seeds were immersed in 500 µL of 1% (w/v) TZ solution and incubated at 30°C in the dark for the intended amount of time. After incubation, the seeds were washed in 1 mL of DW and resuspended in 1 mL of 95% ethanol. The seeds were finely ground with mortars and pestles, and the ground seeds were collected in a 2 mL Eppendorf tube. Then, 95% ethanol was added up to 2 mL, and the sample was centrifuged for 3 minutes at 15,000 x g. Next, 1 mL of supernatant was transferred for the measurement of absorbance at 485 nm with a spectrophotometer (Eppendorf BioSpectrometer® fluorescence). For mucilage staining, seeds were soaked in 50 mM EDTA for 2 hours in advance and incubated in 0.01% ruthenium red (w/v) in 10 mM Tris-HCl (pH 8.0) for 1-2 hours. Stained seeds were washed with 10 mM Tris-HCl (pH 8.0) and photographed with a micro-lens-equipped digital camera. For proanthocyanidin staining, seeds were incubated in 1% vanillin in a 6 M HCl solution for 24 hours, washed with DW, and photographed with a micro-lens-equipped digital camera. Three replicates of each experiment were performed for statistical analysis.
Dried seeds were used for seed coat polyester analysis, following the method described by Lee and Suh (2018). Seeds were immersed in 80°C boiling isopropanol (25 mL g-1 of seeds) for 10 minutes and ground thoroughly with a mortar. Ground tissues were delipidated successively using chloroform:methanol (2:1, v/v) and chloroform:methanol (1:2, v/v) with the daily exchange of the solvent and then finally extracted with methanol. Delipidated tissues were dried under nitrogen gas and desiccated for at least 24 hours using a vacuum dessicator until a constant weight was achieved. Delipidated dry residues were depolymerized via the base catalysis method. Internal standards (methyl heptadecanoate and ω-pentadecalactone, 5 µg each) and 2 mL of reaction solvent (3.8 mL methanol, 1.3 mL 28% sodium methoxide in methanol, 0.9 mL methyl acetate per 6 mL of solvent) were added to the dry residues and heated for 2 hours at 60°C. After cooling at RT, 4 mL of dichloromethane, 0.5 mL of glacial acetic acid, and 1 mL of 0.9% NaCl (w/v) in 100 mM Tris-HCl pH 8.0 were added, followed by centrifugation (600 x g, 10 min). The lower organic phase was washed at least twice with 2 mL of 0.9% NaCl (w/v) in 100 mM Tris-HCl pH 8.0. The final extract was dehydrated over anhydrous sodium sulfate and dried under a stream of nitrogen gas. The dried sample was acetylated by dissolving it in 100 µL of pyridine and 100 µL acetic anhydride and heating it for 2 hours at 60°C. After evaporation of the solvent under nitrogen gas, derivatized suberin monomers were dissolved in 50 µL heptane:toluene (1:1, v/v) and analyzed using a GC-flame ionization detector using protocols described by Lee et al. (2009).
To examine whether ABA is an important signaling molecule for the formation of Arabidopsis seed coat suberin, seeds of one ABA biosynthetic mutant and six different ABA signaling mutants were immersed in 1% (w/v) TZ salt for 8 and 24 h, after confirming the mutations in all tested plants except abi3-8 and abi5-7 (Figure S1). gpat5-2, which is defective in seed coat suberin, was used as a positive control (Beisson et al., 2007). Surprisingly, among the six ABA signaling mutants, namely QC3, QL3, abi1-1, snrk2.2/3/6+, abi3-8, and abi5-7, only abi1-1 was slightly stained, whereas the only ABA biosynthetic mutant, aba1-1, showed a conspicuously intense level of staining (Figure 1). This result implied that ABA1 knock-out increases the permeability of the seed coat, likely through a disruption in seed coat suberin (Beisson et al., 2007; Vishwanath et al., 2013; Yadav et al., 2014; Gou et al., 2017). However, the deletion of major ABA signaling factors (PYR1, PYL1, PYL2, PYL4, PP2C, and SnRK2.2, 2.3, 2.6) constituting the canonical ABA signaling pathway (Leung et al., 1994; Meyer et al., 1994; Parcy et al., 1994; Gosti et al., 1999; Finkelstein and Lynch, 2000; Merlot et al., 2001; Fujii and Zhu, 2009; Fujita et al., 2009; Ma et al., 2009; Nakashima et al., 2009; Park et al., 2009) resulted in no or weakly significant alterations in seed coat permeability. The increased permeability of the seed coat induced by ABA1 disruption prompted a further investigation of whether ABA1 is involved in the formation of suberin in seed coats.
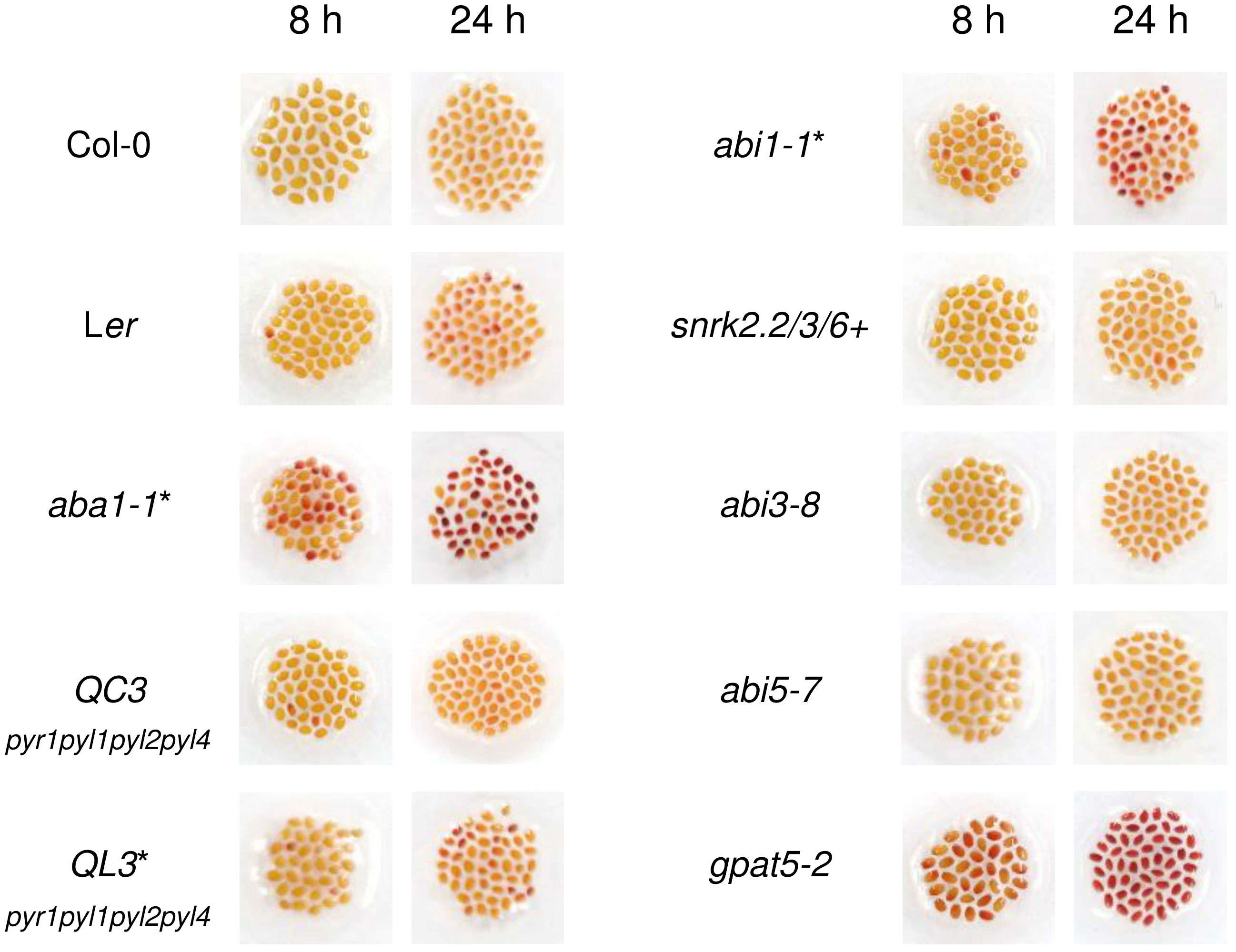
Figure 1 Col-0, Ler, aba1-1, and several abscisic acid (ABA) signaling mutants were immersed in 1% (w/v) tetrazolium salt solution for 8 and 24 hours. After incubation, seeds were photographed with a digital camera. gpat5-2 was used as a positive control. Those with Ler as their background are indicated with asterisks. QC3, pyr1pyl1pyl2pyl4 quadruple mutant with Col-0 background; QL3, pyr1pyl1pyl2pyl4 quadruple mutant with Ler background; abi1-1, PP2C mutant; snrk2.2/3/6+, snrk2.2snrk2.3snrk2.6+ triple mutant.
The genomic organization of aba1-1 and aba1-8 are described in Figure 2A. aba1-1 is a substitution mutant, in which the 2139th G is substituted with an A, resulting in the early termination of translation (Koornneef et al., 1982; Barrero et al., 2005). To carry out our research on ABA1, we attempted to obtain an additional aba1 mutant allele. A T-DNA insertional mutant, SAIL_A03_04, was obtained from ABRC, and T-DNA genotyping was performed to verify that a homozygous mutant line had been obtained. PCR analysis with ABA1_RT_F2 and LB1 primers resulted in a PCR band with the aba1 mutant, but not with the Col-0 wild-type, whereas PCR with a pair of gene-specific primers (ABA1_RT_F2/ABA1_RT_R2) resulted in a PCR band with Col-0 but not with the aba1 mutant, which confirmed the homozygous T-DNA insertion in the aba1 mutant (Figure 2B). Total RNA was isolated from 9-day-old seedlings of the selected homozygotes and analyzed by performing RT-PCR on ABA1, which verified that it was an aba1 knock-out mutant (Figure 2C). RNA quality and quantity were normalized based on the PP2AA3 gene (At1g13320). The selected aba1 knock-out mutant was named aba1-8, and it was found to harbor its T-DNA insertion in the second exon, as shown in Figure 2A.
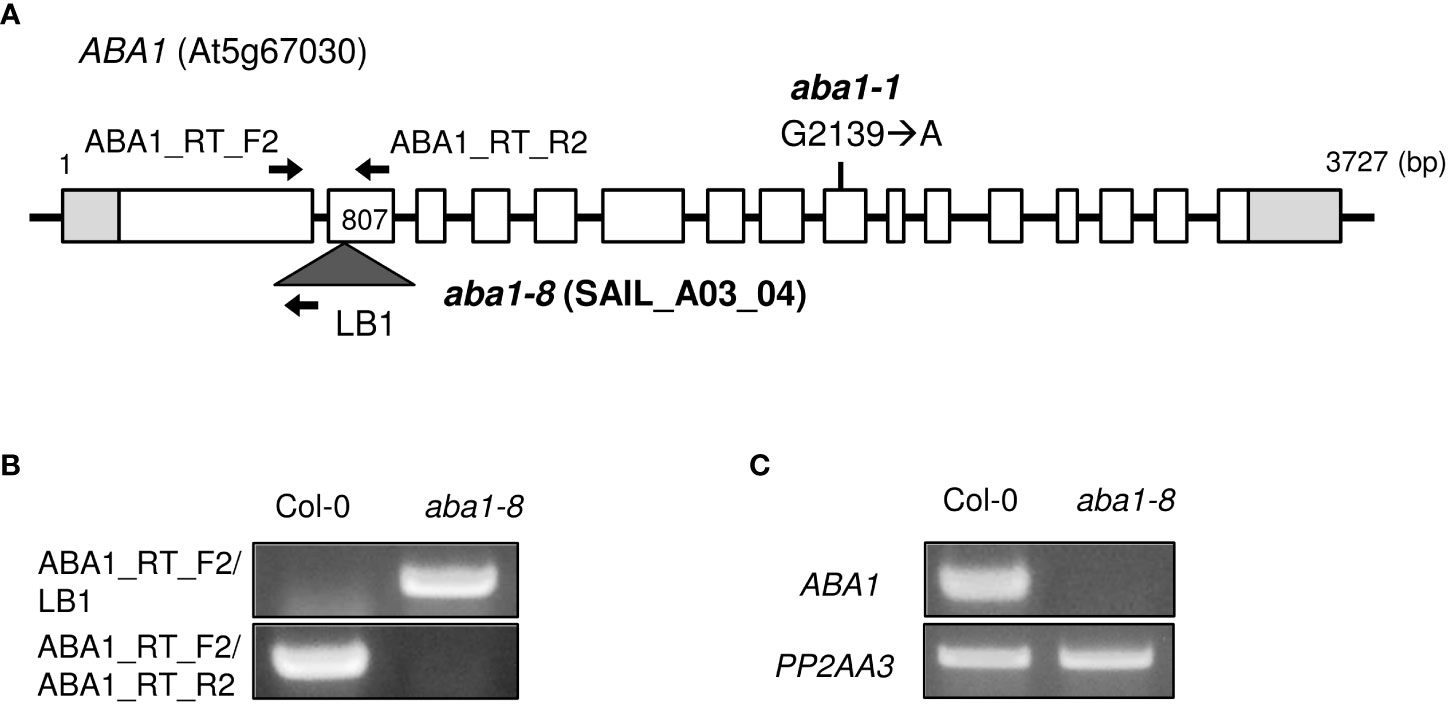
Figure 2 Isolation of T-DNA insertional knock-out mutant aba1-8. (A) Genomic organization of ABA1 gene with a point mutation (G2139 → A) in aba1-1 (Koornneef et al., 1982) and an insertional T-DNA in aba1-8. The white boxes indicate the exons, light gray boxes indicate the untranslated region, and the dark gray triangle indicates the inserted T-DNA. Arrows indicate the binding sites of the primers used for T-DNA mutant isolation and confirmation of the knockout mutation. (B) T-DNA genotyping of Col-0 and aba1-8. ABA1_RT_F2 and ABA1_RT_R2 are ABA1-specific primers and LB1 is a T-DNA-specific primer. (C) RT-PCR analyses of Col-0 and aba1-8. PP2AA3 (At1g13320) was used for normalization.
The morphological and developmental phenotypes of Arabidopsis aba1-1 and aba1-8 were then examined. Arabidopsis wild-types (Landsberg erecta [Ler] and Columbia-0 [Col-0]) and aba1 mutants were photographed at 3 and 6 weeks after germination. Both aba1-1 and aba1-8 plants showed the significant retardation of growth and reductions in sizes (Figures 3A, B). Critically, the aba1 mutants showed abnormal siliques, since silique development was restrained before its actual appearance or in the midst of its elongation (Figure 3B). Then, homozygous aba1-1 and aba1-8 mature seeds were obtained through the application of a plastic canopy on top of the plants to assist in fertilization and silique development, by reducing water loss. To investigate if seed development was hindered, developing siliques at 8 to 10 DAF (days after flowering) were opened, and developing seeds were observed under a microscope. The development of some seeds of aba1-1 and aba1-8 was found to be aborted or the developing seeds were decreased in size (Figure 3C). After harvesting the mature seeds, we observed that some seeds of aba1-1 and aba1-8 had an abnormal shape and relatively small or large sizes (Figure 3D).
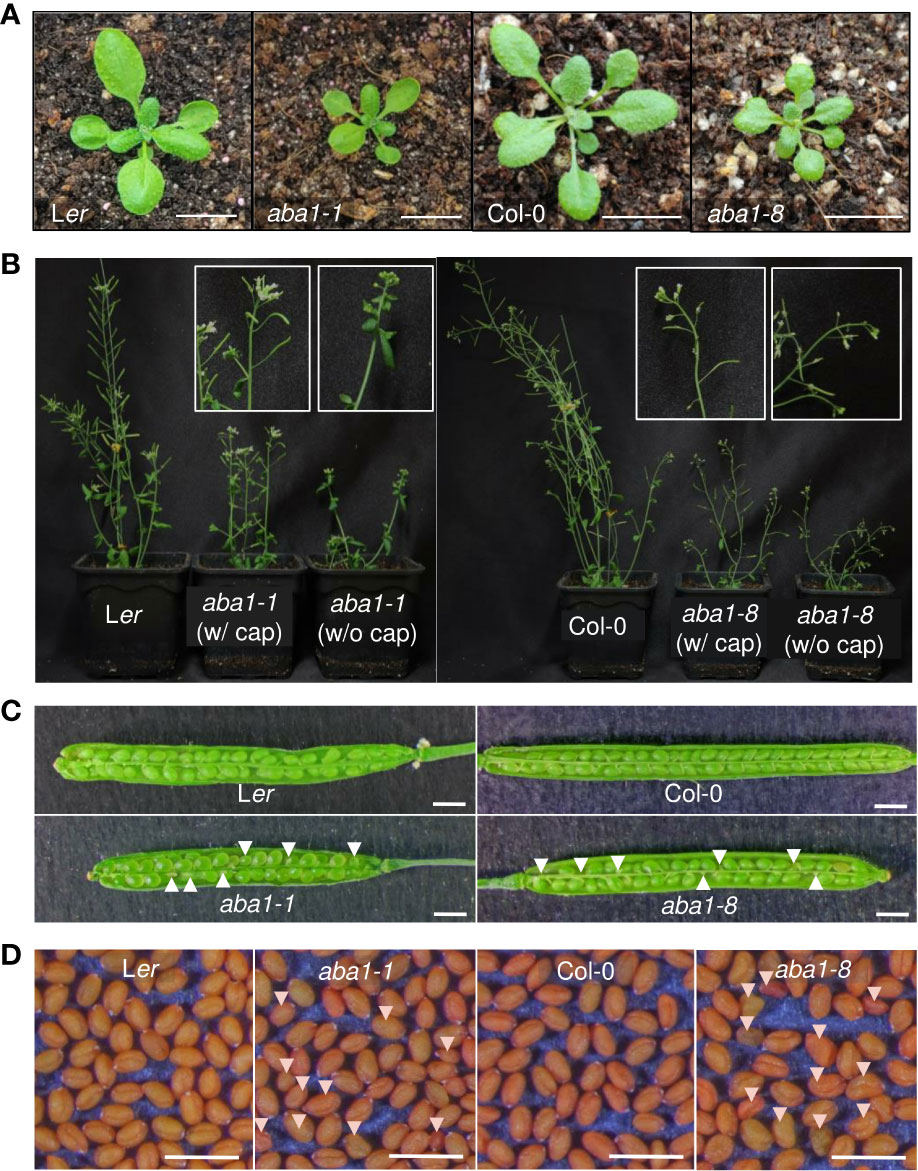
Figure 3 Phenotypes of aba1-1 and aba1-8 compared to their corresponding wild-types. (A) Three-week-old rosette leaves of Ler, aba1-1, Col-0, and aba1-8 (from the left). Bar = 1 cm. (B) Six-week-old Ler, aba1-1, Col-0, and aba1-8 (from the left). Since the mutants had difficulty producing siliques on their own, they were grown with transparent canopies made of plastic wrap to assist in fertilization. White boxes in the figure show a 10× magnified shoot of aba1-1 and aba1-8 grown with and without the plastic canopy (cap), as a representative. (C) Opened developing siliques 8–10 DAF (days after flowering) of Ler, aba1-1, Col-0, and aba1-8. White arrows indicate developing seeds with aborted or shrunken phenotypes. Bar = 1 mm. (D) Mature seeds of Ler, aba1-1, Col-0, and aba1-8. White arrows indicate seeds with an aberrant morphology. Bar = 1 mm.
Because phenolic compounds, such as ferulates, are the components of suberin, suberin is detectable based on its autofluorescence under ultraviolet light (Lee and Suh, 2018). Approximately 100 seeds of the wild-types, aba1-1, and aba1-8 were observed under UV light with a fluorescence microscope. The seeds were sorted into three groups (I, strong; II, weak; and III, none or almost none) based on their autofluorescence signal intensity under UV light. The aba1 mutants had a significantly lower abundance of suberin polyesters accumulated in their seed coats. Moreover, aba1-1 had approximately 18% more seeds that showed no or almost no UV autofluorescence signals (group III) compared to wild-type numbers, and aba1-8 had approximately 58% more seeds belonging to group III compared to numbers in the wild-type (Figure 4A). A TZ permeability assay was performed with both aba1 allele-mutant and wild-type seeds, and the seeds were observed after 8 and 24 h from the onset of staining. TZ-tested seeds were divided into three different groups, namely unstained (Un), weakly stained (StI), and intensely stained (StII) according to the staining intensity. Arabidopsis aba1-1 and aba1-8 had approximately 28% and 54% more intensely stained seeds (group StII) than their corresponding wild-types at 24 h of staining, respectively (Figures 4B, C). This was further confirmed by measuring the amount of formazan produced via the reduction of TZ salt that permeated into the embryos. aba1-1 seeds produced approximately 1.5- and 2-fold more formazan than Ler seeds at 8 and 24 h, respectively. And, aba1-8 seeds produced approximately 4- and 5.6-fold more formazan than Col-0 seeds at 8 and 24 h, respectively (Figure 4D).
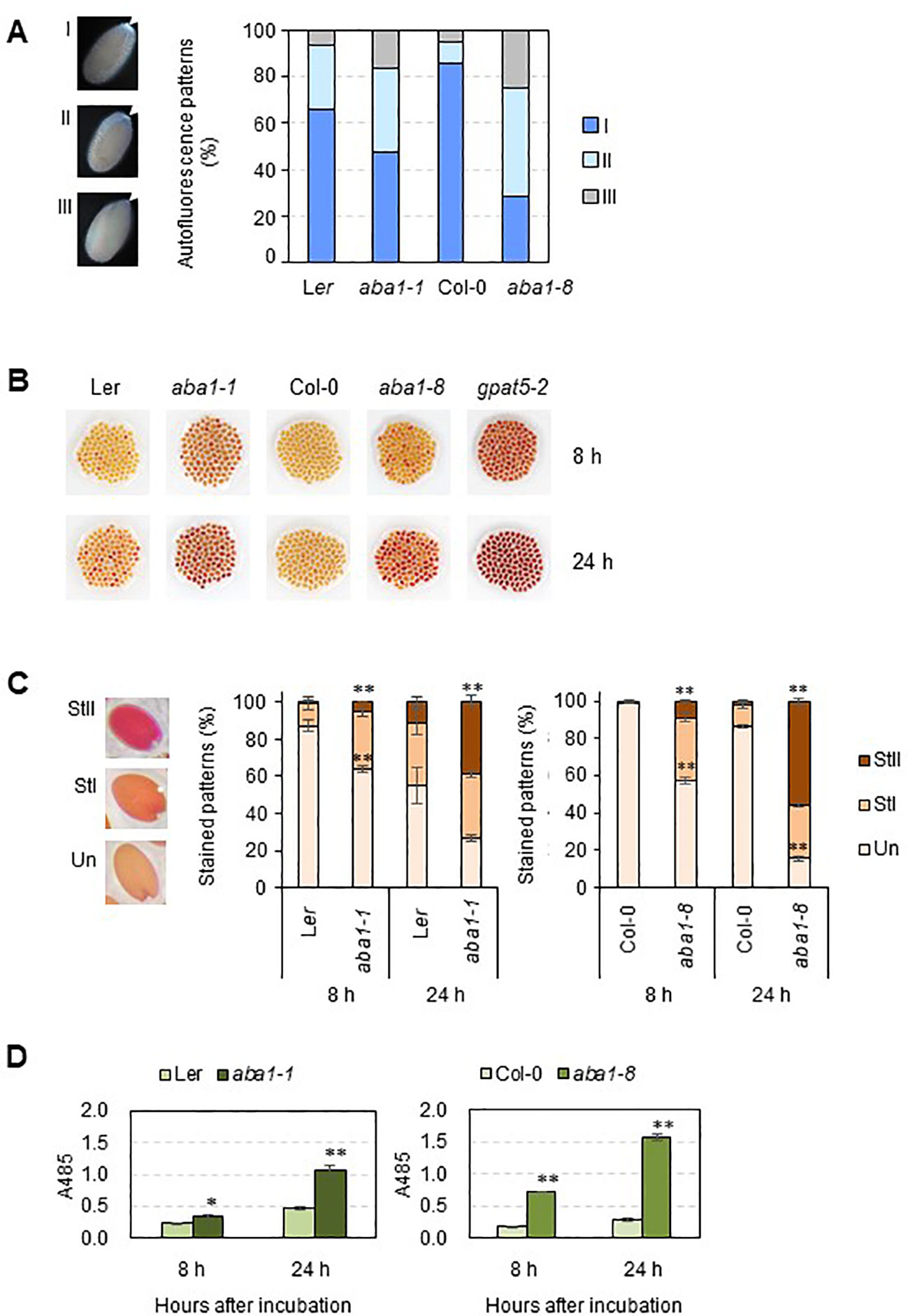
Figure 4 UV autofluorescence observations (A) and seedcoat permeability assay (B–D) on wild-types (Ler and Col-0), aba1-1, and aba1-8. (A) Autofluorescence under UV was observed with a fluorescence microscope. Arrows indicate autofluorescence in the chalazal region of the seed coat. Seeds were divided into three groups (I, strong; II, weak; III, none or almost none) based on their autofluorescence intensity, and one seed from each group is shown as a representative. Percentages of seeds belonging to each group, I, II, and III, were determined and are shown on the right. Values were obtained from 100–110 total seeds. (B) WT and aba1 mutants were immersed in 1% (w/v) tetrazolium salt solution for 8 and 24 h. After the incubation, seeds were photographed with a digital camera. (C) Stained seeds were divided into three groups based on the degree of staining (Un, unstained; StI, weakly stained; StII, strongly stained). Percentages of the seeds belonging to each group were determined. Each value is an average of three replicates with 100–105 seeds per replicate. Error bars indicate ± SE of the average. Asterisks indicate the significance of statistical differences compared to the WT, determined using a Student’s t-test. (*p < 0.05, **p < 0.01). (D) Amount of formazan produced in response to the penetration of tetrazolium salt into the embryos, as measured based on the absorbance at 485 nm at time points of 8 and 24 hours after incubation. Asterisks indicate the significance of statistical differences compared to the WT, determined using a Student’s t-test. (*p < 0.05, **p < 0.01).
To examine whether suberin monomer amounts and composition were altered in aba1 seed coats, suberin monomer analysis was performed. Suberin polyesters were extracted from the seed coats of wild-types (Col-0 and Ler) and aba1 mutants and hydrolyzed, and then, suberin monomers were analyzed through gas chromatography with a flame ionization detector (GC-FID). Interestingly, the levels of C24:0 ω-hydroxy fatty acids and C24:0 α, ω-dicarboxylic acids, which account for the majority of seed coat suberin monomers, were decreased by approximately 16% and 10% in aba1-1, respectively, compared with levels in Ler and by approximately 12% and 10% in aba1-8, respectively, relative to levels in Col-0. The levels of C20:0 and C22:0 α,ω-alkane diols were also decreased by approximately 22% and 15% in aba1-1, respectively, relative to those in Ler and by approximately 15% and 6%, respectively, in aba1-8 compared to those in Col-0. The amount of ferulate decreased by approximately 11% in only aba1-8 relative to Col-0 (Figure 5A). Total suberin monomer amounts were decreased by approximately 3% in both aba1-1 and aba1-8 mutants compared to those in their corresponding wild-types (Figure 5B). Amounts of suberin monomers were also analyzed by totaling the levels of LC (C16-18) and VLC (C20-24) monomers. Amounts of VLC monomers were decreased by approximately 9% and 10% in aba1-1 and aba1-8 seed coats, respectively, compared to those in their wild-types, whereas amounts of LC monomers were increased by approximately 4% and 12% in aba1-1 and aba1-8 seed coats, respectively, compared to those in their corresponding wild-types (Figure 5C).
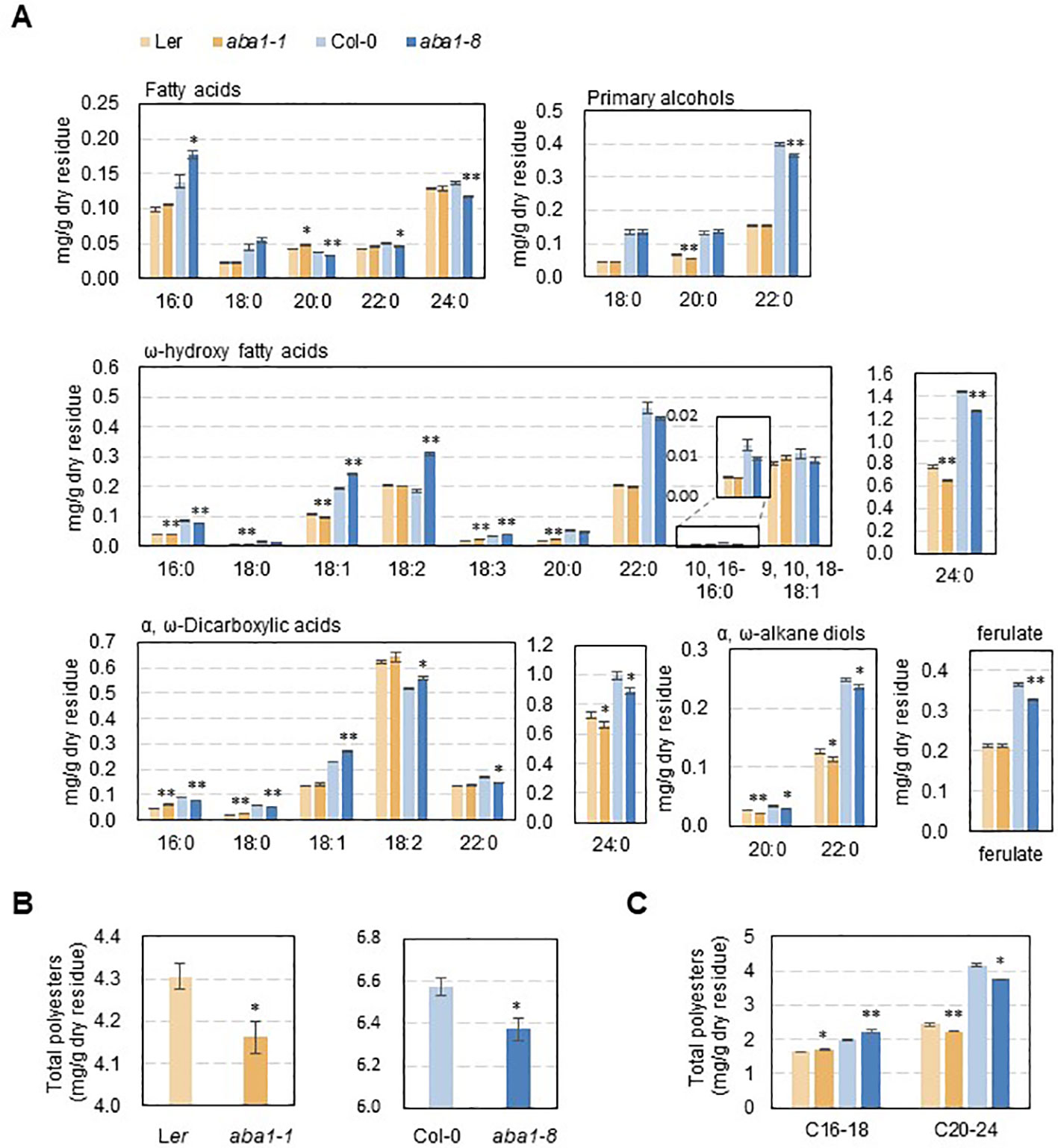
Figure 5 Suberin analysis in wild-type (Ler and Col-0), aba1-1, and aba1-8 seeds. Seed coat polyester monomers (A) and total polyester quantity (B, C) of Ler, aba1-1, Col-0, and aba1-8. The seed dry residue was obtained after grinding and gradual delipidation with chloroform and methanol. Suberin monomers were released from the dry residues by depolymerization through a base catalysis method and analyzed via GC-FID. Error bars indicate ± SE of the average of three to five independent experiments. Asterisks indicate the statistical differences compared to the wild-type, in which *p < 0.05, **p < 0.01.
To investigate if changes in the amounts and composition of suberin monomers were related to the transcript levels of genes involved in suberin formation, RT-qPCR was carried out. Because aba1 seeds were very sensitive to atmospheric conditions (approximately 30 to 40% RH) during the dissection of developing seeds and silique walls, total RNA was isolated from developing siliques at 4–10 DAF, converted to cDNA, and used for the RT-qPCR analysis. KCS1, KCS2, KCS17, KCS20, FAR1, FAR4, FAR5, CYP86A1, CYP86B1, GPAT5, ASFT, LTPG1, LTPG15, ABCG1, ABCG2, ABCG6, ABCG20, ABCG22, ABCG23, MYB9, and MYB107 genes were selected for RT-qPCR analysis based on seed coat-specific or seed coat-preferential expression, based on previous reports or the eFP browser (http://bar.utoronto.ca) (Figure S2) (Beisson et al., 2007; Compagnon et al., 2009; Lee et al., 2009; Molina et al., 2009; Lashbrooke et al., 2016; Gou et al., 2017; Lee and Suh, 2018). Among four β-ketoacyl CoA synthases (KCSs), a dramatic decrease in the transcript levels of KCS17 was observed in aba1-1 and aba1-8, compared with levels in each wild-type. The expression levels of three genes encoding fatty acyl-CoA reductases, FAR1, FAR4, and FAR5, and two fatty acid ω-hydroxylases, CYP86A1 and CYP86B1, were all dramatically decreased in the mutants compared to those in the wild-types. Two types of acyltransferases, aliphatic suberin feruloyl transferase (ASFT) and glycerol-3-phosphate acyltransferase 5 (GPAT5), also showed decreased transcript levels in aba1-1 and aba1-8 compared to wild-type levels, whereas the change in the expression level of ASFT in aba1-1 was not as dramatic as that in aba1-8. In terms of the expression of genes involved in the transport of suberin monomers, expression levels of two LTPGs and six ABCGs, LTPG1, LTPG15, ABCG2, ABCG6, ABCG20, and ABCG23 were significantly downregulated in aba1-1 and aba1-8 compared to levels in their corresponding wild-types. The transcript levels encoding the MYB transcription factors MYB9 and MYB107, which are known to regulate suberin biosynthetic genes mainly in seed coats, were decreased significantly in aba1-1 and aba1-8 relative to levels in their corresponding wild-types (Figure 6).
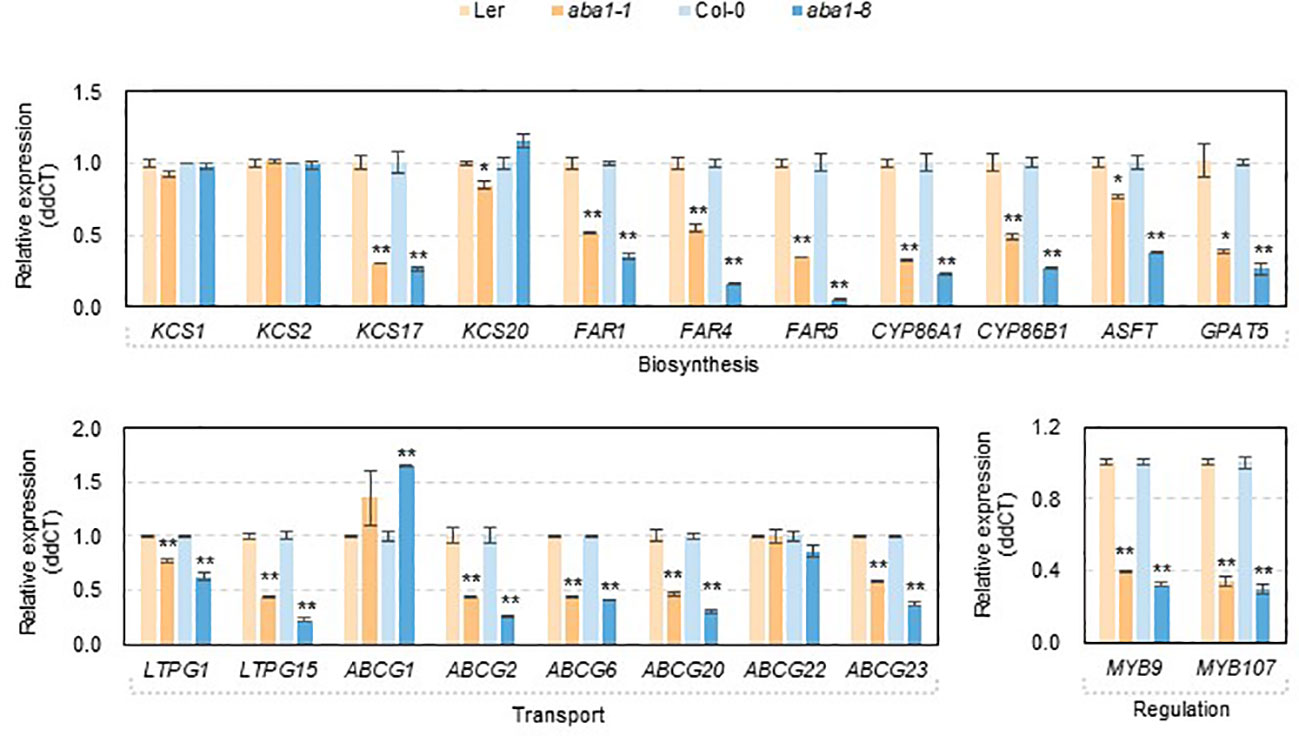
Figure 6 Expression of genes involved in suberin formation in developing siliques of wild-types (Ler and Col-0), aba1-1, and aba1-8. The expression of genes involved in suberin formation in developing siliques (4–10 days after flowering; DAF) in wild-types (Ler and Col-0) and aba1 mutants was analyzed by performing quantitative PCR. The transcription levels were normalized based on the expression level of ACTIN7 (At5g09810), and the raw data were analyzed using the ddCT method. The error bars indicate ± SE of three replicates. A Student’s t-test was used to determine the significance, which is denoted with asterisks (*p<0.05, **p<0.01).
Because the disruption of ABA1 caused the defective formation of seed coat suberin, we attempted to observe changes in mucilage and proanthocyanidin deposition in aba1 seed coats. Seeds were stained with 0.01% (w/v) ruthenium red and 1% (w/v) vanillin to visualize mucilage and proanthocyanidin, respectively. The lack of ABA caused a decrease in the thickness of mucilage (Figure S3A), but no noticeable changes in the accumulation of proanthocyanidins, based on mutants of both alleles of aba1 in seeds, relative to those observed in the wild-types (Figure S3B).
ABA is a major phytohormone that regulates seed development. Through an intricate signaling network, it is involved in de-greening and obtaining desiccation tolerance in seeds, as well as in the accumulation of seed storage products, which leads to the maturation and dormancy of the seeds (Ali et al., 2022). In this process, the hydrophobic seed coat polyester is known to play a critical role. Through its osmotic sealing of the seed, seed coat polyesters maintain the impermeability of the seed coat to exclude oxygen, water, heavy metals, and nutrients (Beisson et al., 2007; Fedi et al., 2017; de Silva et al., 2021), which protects the seed itself and maintains seed dormancy. A defective seed coat suberin layer causes increased sensitivity to germination inhibiting factors such as NaCl, paclobutrazol, and ABA, and reduced dormancy at low temperatures (Fedi et al., 2017). Given the relevance of this deep connection between the role of ABA in seed development and the contribution of seed coat polyesters to achieving this role, it could be inevitable that ABA regulates the biosynthesis of seed coat polyesters. In this study, we revealed that ABA is involved in seed coat suberization based on the following evidence: (i) decreased UV autofluorescence signals in the chalazal regions of aba1-1 and aba1-8 seeds, (ii) increased permeability of aba1 seed coats to TZ salts, (iii) significantly decreased levels of suberin monomers, especially C24 α,ω-dicarboxylic acids and C24 ω-hydroxy fatty acids, in aba1 seed coats relative to those in the wild-type, and (iv) decreased expression levels of several genes involved in suberin accumulation and its regulation.
Even though the aba1 mutants showed remarkably increased permeability to TZ salts (Figures 1, 4B–D), the actual amount of suberin monomers was not decreased to the extent that was expected (Figure 5). It was suggested that a decreased amount of suberin monomers is not always correlated with increased permeability of the suberin layers. Rather, the aliphatic monomer arrangement in the suberin microstructure could contribute to the permeability of suberin (Ranathunge and Schreiber, 2011). Although the actual amount of total suberin monomers decreased only by approximately 3% in aba1 mutants, this decrease was mainly caused by the noticeable decrease in the amounts of C24 α,ω-dicarboxylic acids and ω-hydroxy fatty acids. The alterations in the suberin aliphatic and aromatic composition might have resulted in changes to the suberin microstructure, which led to increased permeability. Similar results were observed in a study by Lee and Suh (2018), where only approximately 8% of fatty acids (C20–24), primary alcohols (C20–22), ω-hydroxy fatty acids (C22–24) and α,ω-alkanediols (C20–22) were decreased, and no significant differences in total polyester amounts were observed in ltpg15 seed coats. These slight changes in polyester composition were followed by increased permeability of the ltpg15 seed coat.
The expression of several KCS isoforms was analyzed, as shown in Figure 6. Among these, KCS17 showed a conspicuous decrease in expression in aba1-1 and aba1-8 compared to levels in their corresponding wild-types, whereas other KCSs did not show any decrease (Figure 6). KCS17 was shown to be expressed predominantly in the seed coat (https://bar.utoronto.ca), suggesting that it can be involved in the synthesis of VLCFAs that are required for the formation of seed coat suberins. Downregulated expression of KCS17 is consistent with the decreased amount of total VLC monomers (C20 to C24) along with the increased amount of total long-chain monomers (C16 and C18) in the seed coats of aba1-1 and aba1-8 (Figure 5C). FAR1, FAR4, and FAR5 are involved in the synthesis of C22, C20, and C18 primary alcohols, respectively, which comprise root waxes and suberin polyesters of the root and seed coat in Arabidopsis (Domergue et al., 2010; Vishwanath et al., 2013). FARs showed severely decreased expression in developing siliques of aba1-1 and aba1-8 (Figure 6). This reduced expression is consistent with decreased levels of C20 and C22 primary alcohols in the seed coat polyesters of aba1-1 and aba1-8, respectively. CYP86B1 catalyzes the ω-hydroxylation of VLC fatty acyl-CoAs to ω-hydroxy acids of C22 or C24. This was reflected by the decreased amounts of C24 ω-hydroxy acids in aba1-1 and aba1-8 compared to those in their respective wild-types. Decreased levels of VLC monomers were also observed in suberin layers of ltpg15 seed coats (Lee and Suh, 2018), which aligned with the decreased expression of LTPG15 shown in Figure 6.
Data from the eFP browser database (https://bar.utoronto.ca) showed that the expression of MYB107, MYB9, FAR4, FAR5, CYP86B1, GPAT5, ASFT, LTPG15, ABCG6, ABCG20, and ABCG23 was induced in Arabidopsis seedlings after 3 h of 10 µM ABA treatment (Figure S4). Exogenous ABA treatment was also found to induce expression of the fluorescent protein-encoding gene mCITRINE-SYP122 in Arabidopsis roots under the control of the GPAT5 promoter (Barberon et al., 2016; Shukla et al., 2021; Leal et al., 2022). In kiwifruit, the expression of AchnMYB107, which is a homologue of AtMYB107, is increased in response to exogenous ABA treatment, followed by the induction of AchnCYP86A1 and AchnFAR expression, which are speculated to encode a fatty acid ω-hydroxylase and fatty acyl reductase, respectively, responsible for wound suberization in kiwifruit (Wei et al., 2020a; Wei et al., 2020b). The previous reports, along with the data shown in Figure 6 showing that the expression of several suberization-related genes was decreased in an ABA biosynthetic mutant, suggest that ABA regulates suberization via the direct or indirect induction of several genes involved in suberization-related genes.
Decreased expression of suberization-related genes in aba1 mutants can be considered based on transcriptional regulation via transcription factors. MYB9 and MYB107 were reported to be transcription factors that positively regulate the expression of suberin biosynthetic genes in Arabidopsis seed coats (Lashbrooke et al., 2016; Gou et al., 2017). MYB107 was found to be predominantly expressed in developing Arabidopsis siliques, and its disruption severely increased seed coat permeability to TZ salts. Most suberin monomer components were decreased in myb107 seed coats, and in particular, an approximate 50% decrease in the levels of C24 α,ω-dicarboxylic acids and C24 ω-hydroxy fatty acids were most prominent. The levels of FAR1, FAR4, FAR5, CYP86A1, CYP86B1, GPAT5, ASFT, and LTPG15 were significantly downregulated, but no alterations were observed in the levels of LTPG1 and ABCG2, 6, 20, and 23 transcripts in developing myb107 seeds. In addition, the disruption of Arabidopsis MYB9 causes decreased expression of CYP86A1 in developing seeds (Lashbrooke et al., 2016; Gou et al., 2017). In this study, the expression of MYB9 and MYB107, together with KCS17, FAR1, FAR4, FAR5, CYP86A1, CYP86B1, GPAT5, ASFT, and LTPG15, was severely downregulated in developing aba1-1 and aba1-8 siliques (Figure 6). Taken together, ABA-induced MYB9 and MYB107 are upstream factors involved in the expression of FAR1, FAR4, FAR5, CYP86A1, CYP86B1, GPAT5, ASFT, and LTPG15 in Arabidopsis seed coats. However, the transcriptional factors required for the ABA-dependent expression of ABCG2, 6, 20, and 23 remain to be further investigated.
Interestingly, ABA is likely to regulate the synthesis of seed coat suberins via a seed coat-specific signaling pathway, which has not yet been revealed. The knock-out of ABA1 resulted in defective suberin layers (Figure 1, 4, 5), but the disruption of ABA signaling factors, specifically SnRK2, ABI3, ABI5, and PYR/PYL/RCAR, resulted in no remarkable alterations in seed coat permeability to TZ salt (Figure 1). When ABA-deficient aba3 and ABA-insensitive mutants 112458, abi1-1, snrk2.236, and abf234 were tested for leaf cuticle permeability to toluidine blue O, abf234, an ABA signaling triple mutant with mutations in ABA-responsive element binding factors 2, 3 and 4, was not affected in terms of leaf cuticle permeability, unlike that observed in the other ABA-deficient or ABA-insensitive mutants (Cui et al., 2016). This result suggests that the canonical ABA signaling pathway that responds to SnRK2 kinases is involved in cuticle formation but that there are different downstream ABA-responsive factors involved in the process (Cui et al., 2016). Likewise, another pathway involved in ABA signaling functions in the seed coats, related to ABA in this case, could be suggested. abi1-1, which also displayed increased seed coat permeability (Figure 1), is a dominant negative mutant that has a defective PP2C, continuously sequestering SnRK2 regardless of the presence of ABA. Hence, the canonical ABA signaling factor ABI1 is still considered to be involved in ABA signaling in seed coats. In addition, the ABA receptors PYL3, 6, 8, and 9 could be candidates that transmit the signals involved in seed coat suberization, as they are highly and predominantly expressed in the seed coat or seed coat chalazal region (Figure S5).
In summary, we revealed that ABA transmits downstream signals via a partially preserved canonical ABA signaling pathway, which eventually activates seed coat suberization through positive transcription factors including MYB107 and MYB9. This in turn can upregulate the expression of several suberization-related genes, resulting in seed coat suberin deposition (Figure 7). This study provides a better understanding of the process of seed coat suberization and a basis to investigate an unknown ABA signaling pathway functioning in the seed coat.
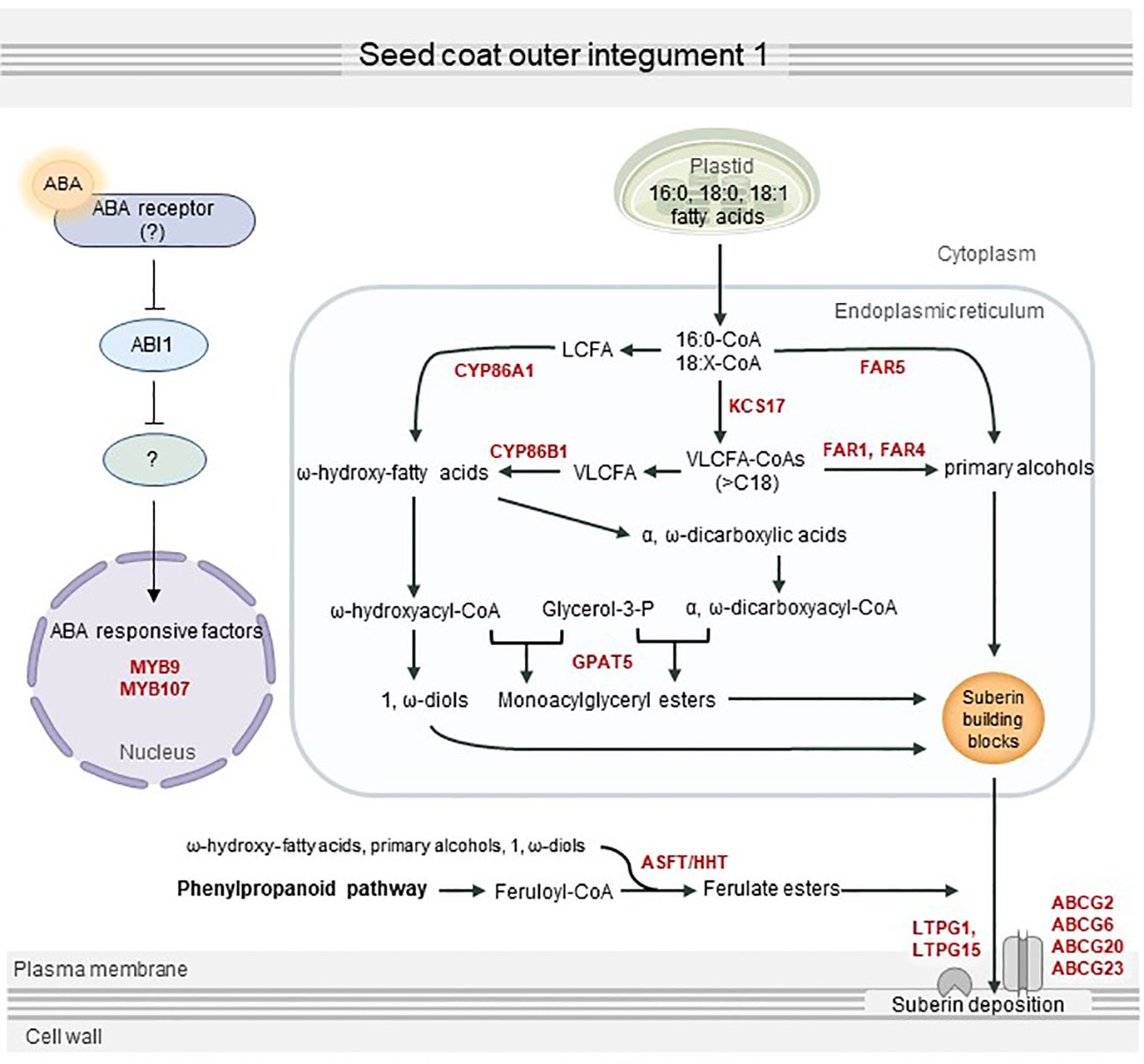
Figure 7 Proposed overview of seed coat suberization regulated by abscisic acid (ABA). ABA transferred to the seed coat is perceived by an unknown ABA receptor, which transmits an ABA signal via the sequestration of ABI1 phosphatase protein 2C. MYB107, MYB9, and other suberization-related transcription factors are directly or indirectly activated by ABA signaling, which in turn upregulates suberin biosynthesis and transport genes, including KCS17, FAR1, FAR4, FAR5, CYP86A1, CYP86B1, GPAT5, ASFT, ABCG2, ABCG6, ABCG20, ABCG23, LTPG1, and LTPG15. This contributes to the proper development of the seed by stimulating suberin deposition in the seed coat.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
MS designed the research. JC and HK performed the experiments and analyzed the data with MS. JC and MS wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Research Foundation (NRF-2021R1A2C1006049 and NRF-2022R1A5A1031361 to MS) of the Republic of Korea.
We greatly appreciate Soo Young Kim (Chonnam National University) for providing Arabidopsis aba1-1, QC3, QL3, abi1-1, snrk2.2/3/6+, abi3-8, and abi5-7 seeds and Ryeo Jin Kim for technical support in suberin analyses.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1156356/full#supplementary-material
Ali, F., Qanmber, G., Li, F., Wang, Z. (2022). Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 35, 199–214. doi: 10.1016/j.jare.2021.03.011
Barberon, M., Vermeer, J. E. M., De Bellis, D., Wang, P., Naseer, S., Andersen, T. G., et al. (2016). Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164, 447–459. doi: 10.1016/j.cell.2015.12.021
Barrero, J. M., Piqueras, P., González-Guzmán, M., Serrano, R., Rodríguez, P. L., Ponce, M. R., et al. (2005). A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J. Exp. Bot. 56, 2071–2083. doi: 10.1093/jxb/eri206
Beeckman, T., De Rycke, R., Viane, R., Inzé, D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113, 139–148. doi: 10.1007/PL00013924
Beisson, F., Li, Y., Bonaventure, G., Pollard, M., Ohlrogge, J. B. (2007). The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of arabidopsis. Plant Cell 19, 351–368. doi: 10.1105/tpc.106.048033
Boher, P., Serra, O., Soler, M., Molinas, M., Figueras, M. (2013). The potato suberin feruloyl transferase FHT which accumulates in the phellogen is induced by wounding and regulated by abscisic and salicylic acids. J. Exp. Bot. 64, 3225–3236. doi: 10.1093/jxb/ert163
Cohen, H., Fedyuk, V., Wang, C., Wu, S., Aharoni, A. (2020). SUBERMAN regulates developmental suberization of the arabidopsis root endodermis. Plant J. 102, 431–447. doi: 10.1111/tpj.14711
Compagnon, V., Diehl, P., Benveniste, I., Meyer, D., Schaller, H., Schreiber, L., et al. (2009). CYP86B1 is required for very long chain ω-hydroxyacid and α,ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 150, 1831–1843. doi: 10.1104/pp.109.141408
Cottle, W., Kolattukudy, P. E. (1982). Abscisic acid stimulation of suberization: Induction of enzymes and deposition of polymeric components and associated waxes in tissue cultures of potato tuber. Plant Physiol. 70, 775–780. doi: 10.1104/pp.70.3.775
Cui, F., Brosché, M., Lehtonen, M. T., Amiryousefi, A., Xu, E., Punkkinen, M., et al. (2016). Dissecting abscisic acid signaling pathways involved in cuticle formation. Mol. Plant 9, 926–938. doi: 10.1016/j.molp.2016.04.001
Deeken, R., Saupe, S., Klinkenberg, J., Riedel, M., Leide, J., Hedrich, R., et al. (2016). The nonspecific lipid transfer protein AtLtpI-4 is involved in suberin formation of Arabidopsis thaliana crown galls. Plant Physiol. 172, 1911–1927. doi: 10.1104/pp.16.01486
de Silva, N. D. G., Boutin, C., Lukina, A. O., Western, T. L., Molina, I., Rowland, O. (2021). Seed coat suberin forms a barrier against chromium (Cr3+) during early seed germination in Arabidopsis thaliana. environ. Exp. Bot. 191, 104632. doi: 10.1016/j.envexpbot.2021.104632
Domergue, F., Vishwanath, S. J., Joubès, J., Ono, J., Lee, J. A., Bourdon, M., et al. (2010). Three arabidopsis fatty acyl-coenzyme a reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 153, 1539–1554. doi: 10.1104/pp.110.158238
Efetova, M., Zeier, J., Riederer, M., Lee, C., Stingl, N., Mueller, M., et al. (2007). A central role of abscisic acid in drought stress protection of agrobacterium-induced tumors on arabidopsis. Plant Physiol. 145, 853–862. doi: 10.1104/pp.107.104851
Fedi, F., O’Neill, C. M., Menard, G., Trick, M., Dechirico, S., Corbineau, F., et al. (2017). Awake1, an ABC-type transporter, reveals an essential role for suberin in the control of seed dormancy. Plant Physiol. 174, 276–283. doi: 10.1104/pp.16.01556
Finkelstein, R. R., Lynch, T. J. (2000). The arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Franke, R., Höfer, R., Briesen, I., Emsermann, M., Efremova, N., Yephremov, A., et al. (2009). The DAISY gene from arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 57, 80–95. doi: 10.1111/j.1365-313X.2008.03674.x
Fujii, H., Zhu, J. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U. S. A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
Gosti, F., Beaudoin, N., Serizet, C., Webb, A. A. R., Vartanian, N., Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11, 1897–1909. doi: 10.1105/tpc.11.10.1897
Gou, M., Hou, G., Yang, H., Zhang, X., Cai, Y., Kai, G., et al. (2017). The MYB107 transcription factor positively regulates suberin biosynthesis. Plant Physiol. 173, 1045–1058. doi: 10.1104/pp.16.01614
Gou, J., Yu, X., Liu, C. (2009). A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 18855–18860. doi: 10.1073/pnas.0905555106
Han, X., Lu, W., Wei, X., Li, L., Mao, L., Zhao, Y. (2018). Proteomics analysis to understand the ABA stimulation of wound suberization in kiwifruit. J. Proteomics 173, 42–51. doi: 10.1016/j.jprot.2017.11.018
Han, X., Mao, L., Lu, W., Wei, X., Ying, T., Luo, Z. (2019). Positive regulation of the transcription of AchnKCS by a bZIP transcription factor in response to ABA-stimulated suberization of kiwifruit. J. Agric. Food Chem. 67, 7390–7398. doi: 10.1021/acs.jafc.9b01609
Han, X., Mao, L., Wei, X., Lu, W. (2017). Stimulatory involvement of abscisic acid in wound suberization of postharvest kiwifruit. Sci. Hortic. 224, 244–250. doi: 10.1016/j.scienta.2017.06.039
Haughn, G., Chaudhury, A. (2005). Genetic analysis of seed coat development in arabidopsis. Trends Plant Sci. 10, 472–477. doi: 10.1016/j.tplants.2005.08.005
Höfer, R., Briesen, I., Beck, M., Pinot, F., Schreiber, L., Franke, R. (2008). The arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 59, 2347–2360. doi: 10.1093/jxb/ern101
Kermode, A. R. (2005). Role of abscisic acid in seed dormancy. J. Plant Growth Regul. 24, 319–344. doi: 10.1007/s00344-005-0110-2
Kim, J., Jung, J. H., Lee, S. B., Go, Y. S., Kim, H. J., Cahoon, R., et al. (2013). Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162, 567–580. doi: 10.1104/pp.112.210450
Koornneef, M., Jorna, M. L., Brinkhorst-van der Swan, D. L. C., Karssen, C. M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 61, 385–393. doi: 10.1007/BF00272861
Kosma, D. K., Murmu, J., Razeq, F. M., Santos, P., Bourgault, R., Molina, I., et al. (2014). AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J. 80, 216–229. doi: 10.1111/tpj.12624
Lashbrooke, J., Cohen, H., Levy-Samocha, D., Tzfadia, O., Panizel, I., Zeisler, V., et al. (2016). MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. Plant Cell 28, 2097–2116. doi: 10.1105/tpc.16.00490
Leal, A. R., Belo, J., Beeckman, T., Barros, P. M., Oliveira, M. M. (2022). The combined effect of heat and osmotic stress on suberization of arabidopsis roots. Cells 11, 2341. doi: 10.3390/cells11152341
Lee, S. B., Jung, S. J., Go, Y. S., Kim, H. U., Kim, J. K., Cho, H. J., et al. (2009). Two arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60, 462–475. doi: 10.1111/j.1365-313X.2009.03973.x
Lee, S. B., Suh, M. C. (2018). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein 15 affects seed coat permeability in arabidopsis. Plant J. 96, 1206–1217. doi: 10.1111/tpj.14101
Leide, J., Hildebrandt, U., Hartung, W., Riederer, M., Vogg, G. (2012). Abscisic acid mediates the formation of a suberized stem scar tissue in tomato fruits. New Phytol. 194, 402–415. doi: 10.1111/j.1469-8137.2011.04047.x
Leung, J., Bouvier-Durand, M., Morris, P., Guerrier, D., Chefdor, F., Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264, 1448–1452. doi: 10.1126/science.7910981
Li, Y., Beisson, F., Koo, A. J. K., Molina, I., Pollard, M., Ohlrogge, J. (2007). Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. U. S. A. 104, 18339–18344. doi: 10.1073/pnas.0706984104
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11, e0161. doi: 10.1199/tab.0161
Lulai, E. C., Suttle, J. C., Pederson, S. M. (2008). Regulatory involvement of abscisic acid in potato tuber wound-healing. J. Exp. Bot. 59, 1175–1186. doi: 10.1093/jxb/ern019
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mahmood, K., Zeisler-Diehl, V., Schreiber, L., Bi, Y., Rothstein, S. J., Ranathunge, K. (2019). Overexpression of ANAC046 promotes suberin biosynthesis in roots of Arabidopsis thaliana. int. J. Mol. Sci. 20, 6117. doi: 10.3390/ijms20246117
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi: 10.1046/j.1365-313x.2001.00965.x
Meyer, K., Leube, M. P., Grill, E. (1994). A protein phosphatase 2C involved in ABA signal transduction in arabidopsis thaliana. Science 264, 1452–1455. doi: 10.1126/science.8197457
Molina, I., Bonaventure, G., Ohlrogge, J., Pollard, M. (2006). The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochem. 67, 2597–2610. doi: 10.1016/j.phytochem.2006.09.011
Molina, I., Li-Beisson, Y., Beisson, F., Ohlrogge, J. B., Pollard, M. (2009). Identification of an arabidopsis feruloyl-coenzyme a transferase required for suberin synthesis. Plant Physiol. 151, 1317–1328. doi: 10.1104/pp.109.144907
Molina, I., Ohlrogge, J. B., Pollard, M. (2008). Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J. 53, 437–449. doi: 10.1111/j.1365-313X.2007.03348.x
Nakashima, K., Fujita, Y., Kanamori, N., Katagiri, T., Umezawa, T., Kidokoro, S., et al. (2009). Three arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083
Nambara, E., Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Oñate-Sánchez, L., Vicente-Carbajosa, J. (2008). DNA-Free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes. 1, 93–93. doi: 10.1186/1756-0500-1-93
Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., Giraudat, J. (1994). Regulation of gene expression programs during arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. doi: 10.1105/tpc.6.11.1567
Park, S., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Radchuk, V., Borisjuk, L. (2014). Physical, metabolic and developmental functions of the seed coat. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00510
Ranathunge, K., Schreiber, L. (2011). Water and solute permeabilities of arabidopsis roots in relation to the amount and composition of aliphatic suberin. J. Exp. Bot. 62, 1961–1974. doi: 10.1093/jxb/erq389
Rock, C. D., Zeevaart, J. A. (1991). The ABA mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 88, 7496–7499. doi: 10.1073/pnas.88.17.7496
Serra, O., Soler, M., Hohn, C., Franke, R., Schreiber, L., Prat, S., et al. (2009). Silencing of StKCS6 in potato periderm leads to reduced chain lengths of suberin and wax compounds and increased peridermal transpiration. J. Exp. Bot. 60, 697–707. doi: 10.1093/jxb/ern314
Shanmugarajah, K., Linka, N., Gräfe, K., Smits, S. H. J., Weber, A. P. M., Zeier, J., et al. (2019). ABCG1 contributes to suberin formation in Arabidopsis thaliana roots. Sci. Rep. 9, 11381. doi: 10.1038/s41598-019-47916-9
Shukla, V., Han, J., Cléard, F., Lefebvre-Legendre, L., Gully, K., Flis, P., et al. (2021). Suberin plasticity to developmental and exogenous cues is regulated by a set of MYB transcription factors. Proc. Natl. Acad. Sci. U. S. A. 118, e2101730118. doi: 10.1073/pnas.2101730118
Soliday, C. L., Dean, B. B., Kolattukudy, P. E. (1978). Suberization: Inhibition by washing and stimulation by abscisic acid in potato disks and tissue culture. Plant Physiol. 61, 170–174. doi: 10.1104/pp.61.2.170
Tao, X., Mao, L., Li, J., Chen, J., Lu, W., Huang, S. (2016). Abscisic acid mediates wound-healing in harvested tomato fruit. Postharvest Biol. Technol. 118, 128–133. doi: 10.1016/j.postharvbio.2016.04.002
To, A., Joubès, J., Thueux, J., Kazaz, S., Lepiniec, L., Baud, S. (2020). AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J. 103, 660–676. doi: 10.1111/tpj.14759
Vishwanath, S. J., Kosma, D. K., Pulsifer, I. P., Scandola, S., Pascal, S., Joubès, J., et al. (2013). Suberin-associated fatty alcohols in arabidopsis: Distributions in roots and contributions to seed coat barrier properties. Plant Physiol. 163, 1118–1132. doi: 10.1104/pp.113.224410
Wang, C., Wang, H., Li, P., Li, H., Xu, C., Cohen, H., et al. (2020). Developmental programs interact with abscisic acid to coordinate root suberization in arabidopsis. Plant J. 104, 241–251. doi: 10.1111/tpj.14920
Wei, X., Lu, W., Mao, L., Han, X., Wei, X., Zhao, X., et al. (2020a). ABF2 and MYB transcription factors regulate feruloyl transferase FHT involved in ABA-mediated wound suberization of kiwifruit. J. Exp. Bot. 71, 305–317. doi: 10.1093/jxb/erz430
Wei, X., Mao, L., Lu, W., Wei, X., Han, X., Guan, W., et al. (2020b). Three transcription activators of ABA signaling positively regulate suberin monomer synthesis by activating cytochrome P450 CYP86A1 in kiwifruit. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01650
Yadav, V., Molina, I., Ranathunge, K., Castillo, I. Q., Rothstein, S. J., Reed, J. W. (2014). ABCG transporters are required for suberin and pollen wall extracellular barriers in arabidopsis. Plant Cell 26, 3569–3588. doi: 10.1105/tpc.114.129049
Yang, W., Pollard, M., Li-Beisson, Y., Beisson, F., Feig, M., Ohlrogge, J. (2010). A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc. Natl. Acad. Sci. U. S. A. 107, 12040–12045. doi: 10.1073/pnas.0914149107
Keywords: ABA signaling, abscisic acid, Arabidopsis, seed coat, suberin
Citation: Choi J, Kim H and Suh MC (2023) Disruption of the ABA1 encoding zeaxanthin epoxidase caused defective suberin layers in Arabidopsis seed coats. Front. Plant Sci. 14:1156356. doi: 10.3389/fpls.2023.1156356
Received: 01 February 2023; Accepted: 01 March 2023;
Published: 15 March 2023.
Edited by:
Eric Marechal, UMR5168 Laboratoire de Physiologie Cellulaire Vegetale (LPCV), FranceReviewed by:
Stephen Beungtae Ryu, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Republic of KoreaCopyright © 2023 Choi, Kim and Suh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mi Chung Suh, bWNzdWhAc29nYW5nLmFjLmty
†Present address: Hyojin Kim, Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.