- 1Department of Plant and Soil Sciences, Mississippi State University, Mississippi State, MS, United States
- 2Department of Plant and Soil Sciences, Oklahoma State University, Stillwater, OK, United States
- 3United States Department of Agriculture – Agricultural Research Service (USDA ARS) Western Regional Plant Introduction Station, Pullman, WA, United States
Bermudagrass (Cynodon spp.) breeding and cultivar development is hampered by limited information regarding its genetic and phenotypic diversity. To explore diversity in bermudagrass, a total of 206 Cynodon accessions consisting of 193 common bermudagrass (C. dactylon var. dactylon) and 13 African bermudagrass (C. transvaalensis) accessions of worldwide origin were assembled for genetic characterization. Genotyping-by-sequencing (GBS) was employed for genetic marker development. With a minor allele frequency of 0.05 and a minimum call rate of 0.5, a total of 37,496 raw single nucleotide polymorphisms (SNPs) were called de novo and were used in the genetic diversity characterization. Population structure analysis using ADMIXTURE revealed four subpopulations in this germplasm panel, which was consistent with principal component analysis (PCA) and phylogenetic analysis results. The first three principal components explained 15.6%, 10.1%, and 3.8% of the variance in the germplasm panel, respectively. The first subpopulation consisted of C. dactylon accessions from various continents; the second subpopulation was comprised mainly of C. transvaalensis accessions; the third subpopulation contained C. dactylon accessions primarily of African origin; and the fourth subpopulation represented C. dactylon accessions obtained from the Oklahoma State University bermudagrass breeding program. Genetic diversity parameters including Nei’s genetic distance, inbreeding coefficient, and Fst statistic revealed substantial genetic variation in the Cynodon accessions, demonstrating the potential of this germplasm panel for further genetic studies and cultivar development in breeding programs.
Introduction
Cynodon L. C. Rich. is a genus consisting of nine warm-season grass species in the Cynodonteae tribe, Chloridoideae subfamily, and grass family (Poaceae) (Harlan et al., 1970; Wu, 2011). Cynodon is characterized by a globally extensive distribution and harbors rich genetic diversity. Among these species, common bermudagrass (C. dactylon var. dactylon) is the most important species economically and ecologically. Cynodon dactylon var. dactylon is a C4 perennial grass used as a turfgrass, forage, and soil stabilizer (Harlan and De Wet, 1969). It has also been reported to be used in ethno-medicinal and traditional medical practices—it is used in Ayurveda, Unani, Nepalese, and Chinese medical systems and has both antiviral and antimicrobial properties (Shendye and Gurav, 2014). Cynodon dactylon is a weed in agriculture (Horowitz, 1996; Nelson and Burns, 2006), as well as in various maintained turfgrass settings (McElroy and Breeden, 2006; Uddin et al., 2010). Another important species in the Cynodon genus is Cynodon transvaalensis Burtt-Davy, commonly known as African bermudagrass. African bermudagrass itself is not widely used for turfgrass due to high water and fertilizer input requirements and poor performance under extreme temperature conditions (Taliaferro, 1992). African bermudagrass does however contain useful traits (i.e., fine texture, high density, and tolerance to low mowing) for turf cultivar development, and it can readily cross with common bermudagrass to generate interspecific hybrids; thus, it is often used in turfgrass breeding programs.
Ploidy levels and chromosome numbers of the genus Cynodon vary widely. Flow cytometry studies revealed diploidy to hexaploidy levels in Cynodon (Wu et al., 2006; Kang et al., 2008; Gulsen et al., 2009; Jewell et al., 2012; Grossman et al., 2021). Forbes and Burton (1963) confirmed the base chromosome number as 9 in Cynodon species. According to the widely accepted revised taxonomy of Cynodon (Harlan et al., 1970a), diploidy (2n = 2x = 18) and tetraploidy (2n = 4x = 36) are common but hexaploidy (2n = 6x = 54) is rare. Species including C. barberi, C. dactylon var. aridus, C. incompletus var. incompletus, C. plectostachyus, and C. transvaalensis are predominantly diploid (2n = 2x = 18). Species including C. arcuatus, C. dactylon var. dactylon, C. dactylon var. coursii, C. dactylon var. elegans, and C. dactylon var. polevansii are largely tetraploid (2n = 4x = 36). Other species with both diploid and tetraploid cytotypes are C. aethiopicus, C. dactylon var. afghanicus, C. incompletus var. hirsutus, C. nlemfuensis var. nlemfuensis, and C. nlemfuensis var. robustus (Harlan et al., 1970a). A majority of the common bermudagrass germplasm in breeding programs are tetraploids (Mutlu et al., 2014). Although the classification of C. dactylon species as allotetraploid versus autotetraploid is debated (Harlan and De Wet, 1969; Fang et al., 2020; Chaves et al., 2022; Bethel et al., 2006; Harris-Shultz et al., 2010; Guo et al., 2015; Khanal et al., 2017), a recent high-density genetic map developed using genotyping-by-sequencing (GBS) provides a clearer interpretation of the C. dactylon allotetraploid structure (Fang et al., 2020).
Polyploidy creates higher genetic diversity in bermudagrass, and it has been reported that genetic diversity within-population was highest at low latitudes (Zhang et al., 2019). Population diversity, structure, and relationships can be studied using DNA markers, which are increasingly used in basic genomic studies and plant breeding programs. In previous studies, genetic diversity in Cynodon was largely assessed using traditional molecular markers like amplified length fragment polymorphism (AFLP) (Zhang et al., 1999; Wu et al., 2006), simple sequence repeat (SSR) (Ling et al., 2012; Wang et al., 2013), random amplified polymorphic DNA (RAPD) (Al-Humaid and Motawei, 2004), inter-simple sequence repeat (ISSR) (Farsani et al., 2011; Li et al., 2011), and sequence related amplified polymorphism (SRAP) markers (Wang et al., 2009; Huang et al., 2014; Zheng et al., 2017). Molecular markers have been used to study variation among Cynodon species, bermudagrass genotypes, cultivar identification, and off-type confirmation (Caetano-Anollés et al., 1995; Assefa et al., 1999; Zhang et al., 1999; Karaca et al., 2002; Roodt et al., 2002; Wang et al., 2010; Farsani et al., 2011; Dong et al., 2022). The use of genotyping-by-sequencing (GBS) to evaluate off-type grasses in hybrid bermudagrass has also been proposed (Reasor et al., 2018).
Genetic mapping studies are conducted to delineate the framework of chromosomes and provide resources to identify genomic regions that underly traits of interest in bermudagrass. The F1 progeny population derived from C. dactylon ‘T89’ (4x = 36) × C. transvaalensis ‘T574’ (2x = 18) was analyzed to construct genetic maps for each parent (Bethel et al., 2006; Harris-Shultz et al., 2010; Khanal et al., 2017). Using the same population, early results regarding the genetic architecture of canopy height, stolon internode length, length of the longest stolon, and leaf traits (leaf length and leaf blade) were reported (Khanal et al., 2019). Recently, GBS was used in a first-generation, selfed common bermudagrass population, and a high-density genetic map with 3,544 single nucleotide polymorphism (SNP) markers was reported (Fang et al., 2020). More recently, a high-density genetic map for African bermudagrass was created using a GBS approach and QTLs for sod establishment rate were identified (Yu et al., 2021).
Although previous studies provided valuable insights into the genetic diversity, population structure, and genetic architecture of traits in Cynodon, most focused on small germplasm collections and accessions that were location or country specific, with low numbers of markers. Individual bi-parental or selfed populations only sample limited allelic diversity of selected parents. Therefore, a Cynodon germplasm panel encompassing accessions of worldwide origins evaluated with a large volume of molecular markers would provide novel insights into the genomic diversity of this important genus. As such, a Cynodon germplasm panel of 206 accessions consisting of 193 common bermudagrass and 13 African bermudagrass accessions was assembled to study the genetic diversity at the genus level using the GBS approach. The objectives were to obtain high quality SNP markers and to assess the population structure, genetic relatedness, and evolutionary relationship among accessions of the germplasm panel, and to explore their potential for genome-wide association studies.
Materials and methods
Plant materials
A total of 206 bermudagrass accessions (193 common bermudagrass (C. dactylon) and 13 African bermudagrass (C. transvaalensis) genotypes) were studied (Table S1), among which 145 accessions were procured from the USDA National Plant Germplasm System (NPGS), 40 accessions were obtained from the Oklahoma State University (OSU) bermudagrass breeding program, and 21 accessions were from the Mississippi State University (MSU) germplasm collection. The ploidy level of accessions ranged from diploid to tetraploid. These accessions are of worldwide origin (covering five continents) representing different geographical locations (29 countries) and genetic diversity (Figure 1).
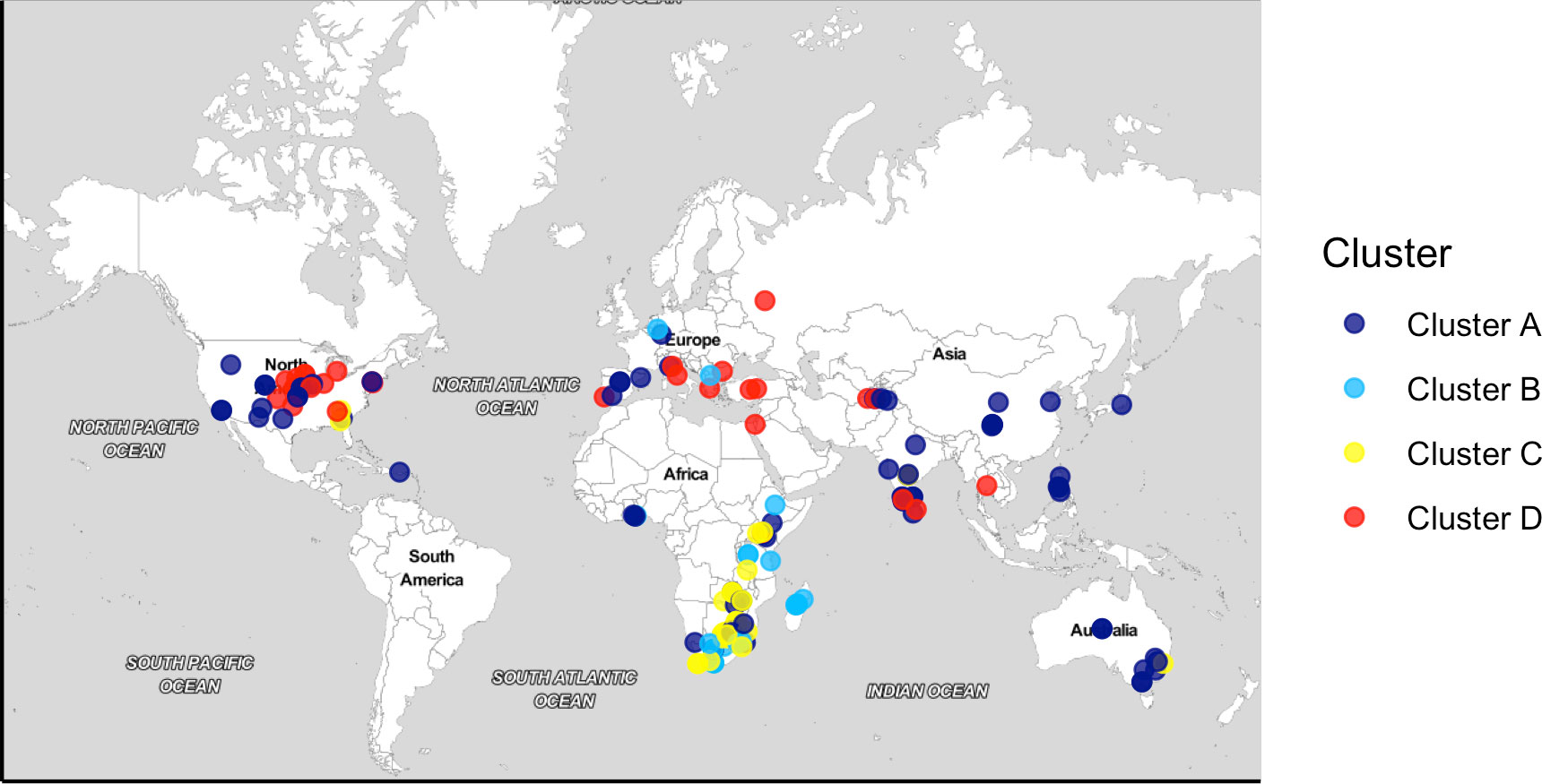
Figure 1 Geographic map of 183 bermudagrass (Cynodon spp.) accessions in the germplasm panel. Accessions were colored according to four subpopulations revealed by ADMIXTURE analysis.
DNA extraction, library construction, and genotyping-by-sequencing
The germplasm was cultured in separate containers under greenhouse conditions at Mississippi State University. Plant leaf material was harvested and freeze-dried. Leaf samples were then shipped to the University of Minnesota Genomic Center (UMGC), where DNA extraction, library preparation, and sequencing were performed. Genomic sequencing libraries were prepared following the GBS protocol with ApeKI enzyme (Elshire et al., 2011). A total of 206 unique barcodes (i.e., corresponding to 206 bermudagrass accessions) were ligated to fragmented DNA sequences. DNA libraries were sequenced with 101 bp single end reads on an Illumina Novaseq SP platform at UMGC.
SNP calling
Raw sequence data were investigated for base quality using FastQC v0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All GBS libraries were sequenced with high-quality reads. The raw GBS data generated by the UMGC did not have inline barcodes, but instead contain variable length padding sequences (0–10 bp) added in front of the enzyme cut site. Therefore, a Perl script (gbstrim.pl, https://bitbucket.org/jgarbe/gbstrim/src/master/) was used to trim padding sequences. The SNPs were then called de novo using the UNEAK pipeline (Lu et al., 2013) of TASSEL 3 standalone. Calling parameters included an error tolerance rate of 0.03, a minor allele frequency of 0.05, and a minimum call rate of 0.5 (i.e., no more than 50% of the bermudagrass accessions in the germplasm panel should have missing data at a given data point).
Population structure and genetic diversity analysis
Data were analyzed using the software ADMIXTURE (Alexander et al., 2009), TASSEL 5.2.77 (Bradbury et al., 2007), adegenet (Jombart et al., 2010; Jombart and Ahmed, 2011), ape (Paradis et al., 2004), pegas (Paradis, 2010), and customized R scripts for plot visualization. To evaluate the hierarchical population structure, the maximum likelihood-based estimation of admixed ancestry with K (subpopulations) ranging from 1 to 10 was conducted in ADMIXTURE v1.3 (Alexander et al., 2009). The optimum number of subpopulations was determined based on a five-fold cross-validation (CV) at K with a minimum CV error. Cynodon accessions were assigned into subpopulations (K1-K4) based on its inferred ancestry from ADMIXTURE. In brief, a Cynodon accession was assigned to a given population if its ancestry fraction from this subpopulation was highest among four subpopulations. Pedigree relationship and ancestry coefficients (Q) from the optimum K were used for visualization of population structure. Principal component analysis (PCA) was also conducted in TASSEL 5.2.77 (Bradbury et al., 2007), and the resulting PC matrix was visualized in R with customized scripts. An identity by state (IBS) matrix was calculated in TASSEL, and a neighbor joining (NJ) tree was visualized in Interactive Tree Of Life (iTOL) v5 (Letunic and Bork, 2021). Based on the results of population structure analyses, fixation index (Fst) between subpopulations were calculated using the adegenet package (Jombart et al., 2010; Jombart and Ahmed, 2011). Observed heterozygosity (Ho), expected heterozygosity (He), number of private alleles, inbreeding coefficients (Fis), Nei’s genetic distance and Fst statistics were calculated using the hierfstat package (Goudet, 2005; Goudet and Jombart, 2015).
Results
SNP calling
A total of 600,380,494 reads were generated, with a mean of 2.9 million reads per sample. After trimming padding sequences, a total of 537,127,057 reads were retained. The UNEAK pipeline identified a total of 536,993,374 reads that contained an ApeKI enzyme cut site remnant and a barcode sequence. After variant calling, a total of 37,496 raw SNPs were obtained with minor allele frequency of 0.05 and minimum call rate of 0.5. Across these SNPs, 23,324 (62.20%) have transition substitutions, whereas 14,172 (37.79%) have transversions—a transition to transversions ratio (Ts/Tv) of 1.64:1. The frequency of transitions and transversions observed is given in Table S2.
Across 37,496 SNPs, missing data rate ranged from 0 to 50%, with an average missing data rate of 32.68%. The average minor allele frequency across all SNPs was 0.13. Among 206 Cynodon accessions, missing data rate ranged from 14% to 40%. Although missing data pose a problem, high number of SNPs is desirable in genetic diversity analyses and could partially offset the negative effect of missing data.
Genetic diversity and population structure
One of the major objectives of this study was to assess the relationship among 206 accessions and determine the overall population structure. A total of 37,496 SNPs were analyzed in ADMIXTURE and PCA. ADMIXTURE revealed four bermudagrass subpopulations (K=4) based on maximum likelihood clustering model and five-fold cross-validations, which were designated as subpopulations 1–4 (Figure 2; Figure S1). In brief, subpopulation 1 (84 accessions; 40.7% of the germplasm panel) was comprised of C. dactylon accessions collected from seventeen different countries ranging across Africa, Asia, North America, Australia, and Europe (Table S1). Two C. transvaalensis accessions (PI 647879 and PI 286584) were also assigned to subpopulation 1, suggesting admixture or possibly misidentification of these two accessions. Subpopulation 2 consisted of 36 (17.4%) accessions—22 of African origin, 12 from the United States, and two from Europe. In this cluster, 9 out of 13 C. transvaalensis accessions clustered with 27 C. dactylon accessions. The third cluster (subpopulation 3) contained 54 (26.2%) accessions—52 of which were C. dactylon and two were C. transvaalensis (PI 289922 and PI 647878). Again, hybridization or misidentification should be considered for these two C. transvaalensis. A majority of the accessions in subpopulation 3 were from five African countries, with two from the U.S., one from India, and one from Australia. The fourth cluster (subpopulation 4) contained the remaining 32 (15.5%) accessions of solely C. dactylon. In subpopulation 4, 17 accessions collected in the United States by the Oklahoma State University turfgrass breeding program grouped with 14 accessions collected in neighboring European and Asian countries maintained at the USDA NPGS (Griffin, GA, USA).
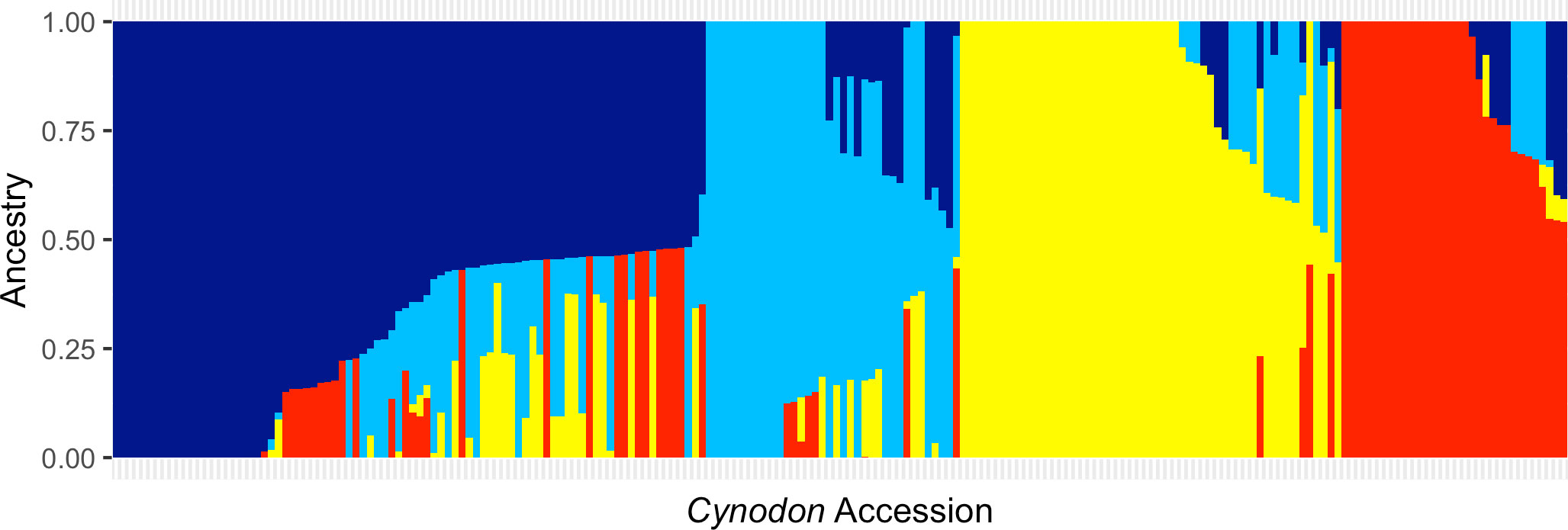
Figure 2 Bar plots from ADMIXTURE analysis. A germplasm panel of 206 Cynodon accessions were clustered into four subpopulations (K=4) based on 37,496 genome-wide SNP markers. Dark blue represents ancestral population 1, light blue represents ancestral population 2, yellow represents ancestral population 3, and red represents ancestral population 3.
Similar patterns were found in PCA and phylogenetic analyses (Figures 3, 4), corroborating K=4 in this Cynodon germplasm panel. Principal component (PC) analysis indicated that PC1, PC2, and PC3 explained 15.6%, 10.1%, and 3.8% of the genetic variation in the panel, respectively (Figure 3). Although there is admixture observed in the PCA analysis (Figure 3), the four clusters can still be distinctly observed. The relatedness and evolutionary relationship of Cynodon accessions are shown in Figure 4. The four groups in the phylogenetic tree were named as A–D (note that some inconsistencies were present but that these generally correspond to 1–4 subpopulations from ADMIXTURE, in corresponding alphanumerical sequence). Detailed comparison between ADMIXTURE results and phylogenetic analysis indicated that groupings of accessions were largely consistent, except for a few differences and for subpopulation 2 (Figure 4). In brief, all the accessions of subpopulation 1 from ADMIXTURE also grouped together in the phylogenetic group A, together with the ten C. dactylon accessions from the Mississippi State University turfgrass breeding program, which were assigned to subpopulation 2 in ADMIXTURE. The group B in the phylogenetic tree corresponded to ADMIXTURE subpopulation 2 but contained three accessions from subpopulation 3. The group C of phylogenetic tree was found to be consistent with ADMIXTURE subpopulation 3, along with three accessions from subpopulation 2. The phylogenetic group D corresponds to ADMIXTURE subpopulation 4 but also has four accessions from subpopulation 3 and seven accessions from subpopulation 2.
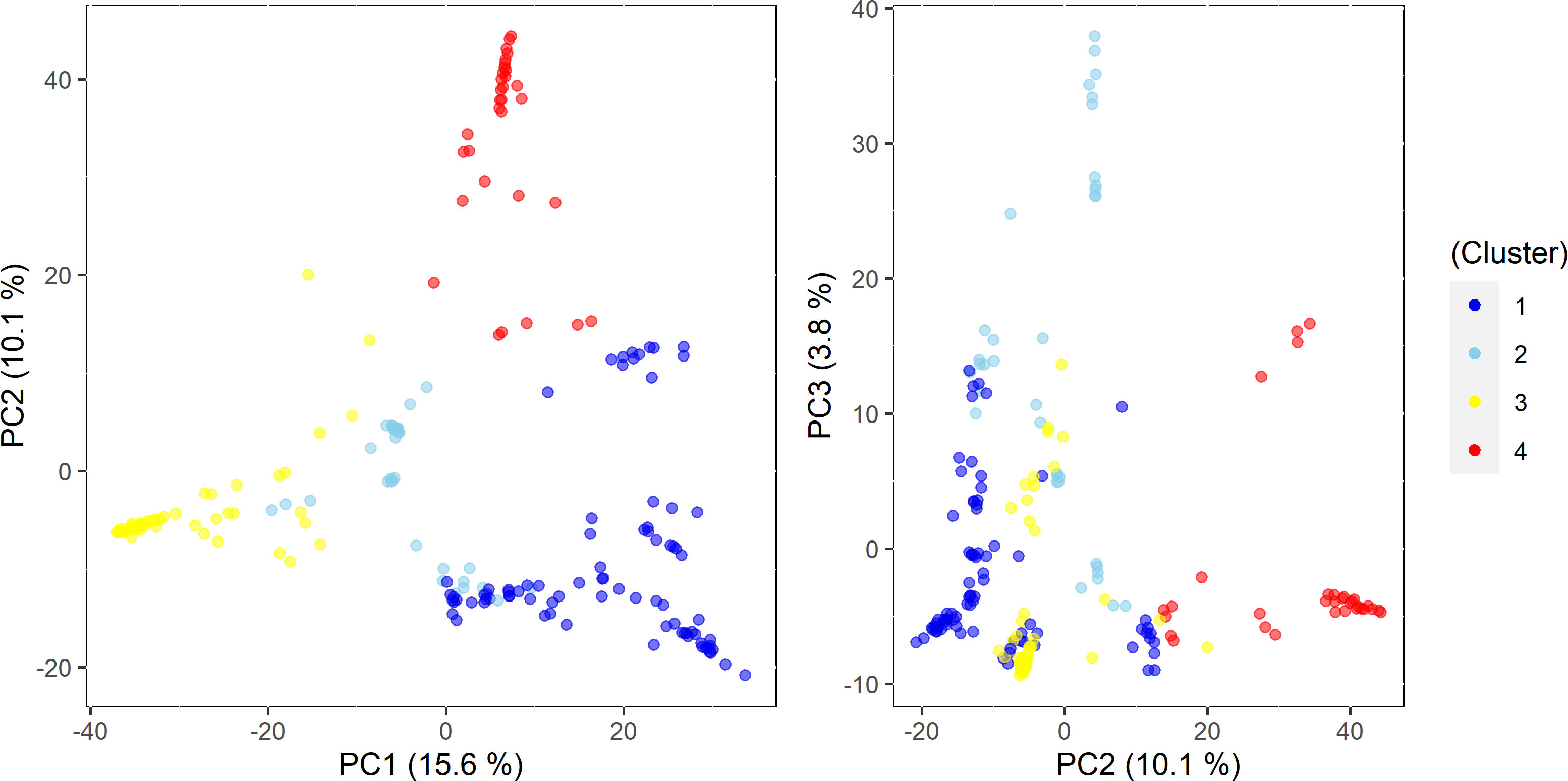
Figure 3 Principal Component (PC) plot extracted from genome-wide SNP markers for this panel and first three principal components explained 15.6%, 10.1%, and 3.8% of the variance in the germplasm panel.
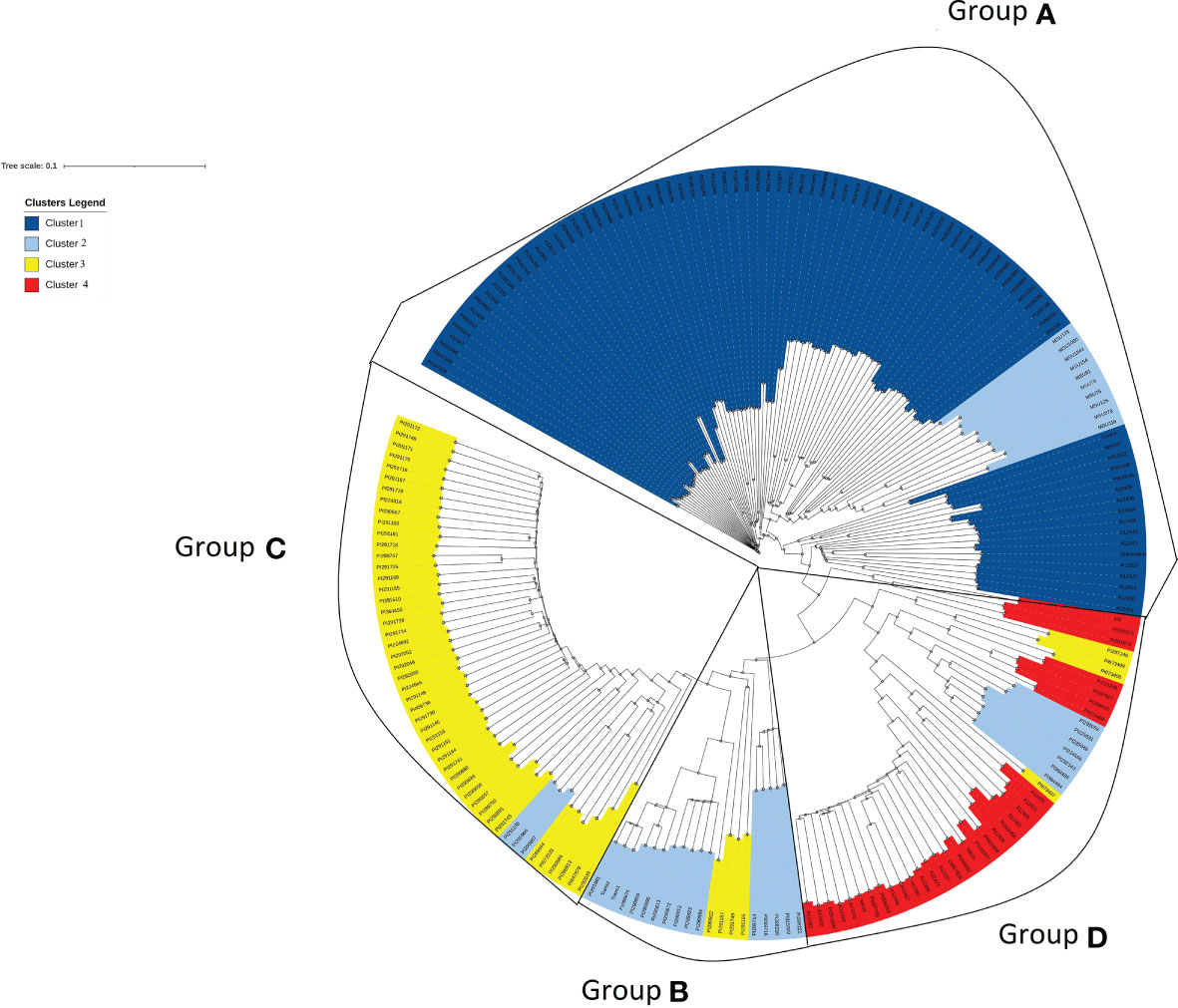
Figure 4 Phylogenetic tree exhibiting relatedness and evolutionary relationship of accessions in the Cynodon germplasm panel. Accessions were colored according to four subpopulations (1–4) revealed by ADMIXTURE analysis. Four distinct groups (A–D) were marked according to phylogenetic clusters and detailed comparison between ADMIXTURE results and phylogenetic analysis indicated that groupings of accessions were largely consistent. Each branch end represents a Cynodon accession.
Fixation index (Fst) evaluates genetic differences between subgroups within a population. In plants, Fst values greater than 0.15 are considered to significantly differ while values below 0.05 are considered insignificant (Hartl et al., 1997; Frankham et al., 2002). Fixation index (Fst) value between subpopulations 3 and 4 is 0.4895 (Table 1). Subpopulation 3 mainly consisted of accessions of African origin (tropical regions), while subpopulation 4 contained accessions primarily from the OSU breeding program; thus, most of the OSU accessions in this study were collected from the US Midwest (temperate regions). These high genetic differences observed between subpopulations 3 and 4 are intriguing, which may suggest that the OSU Cynodon accessions could have been enriched for local selective advantages, such as cold tolerance genes, that shaped the genetic difference between these germplasm materials and other Cynodon accessions. Pairwise Fst comparison aligned with Nei’s genetic distance (Table 1).
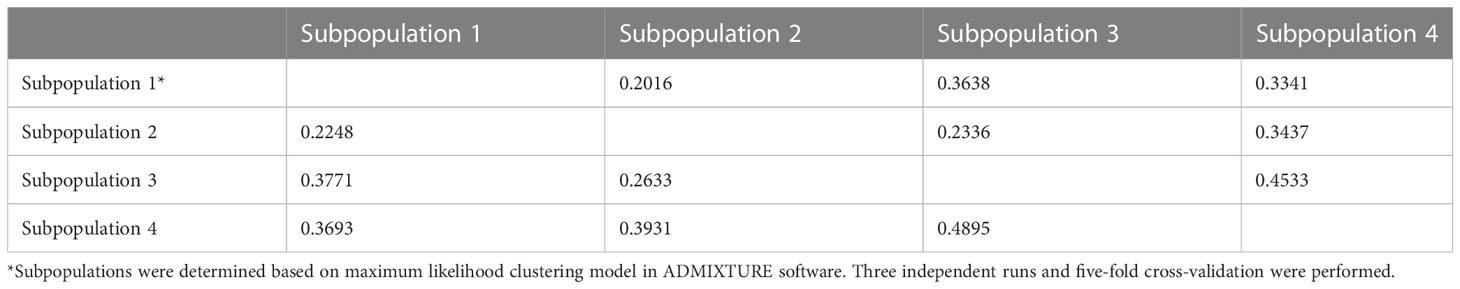
Table 1 Pairwise fixation index (Fst, lower diagonal) and Nei’s genetic distance (upper diagonal) among four subpopulations in the bermudagrass germplasm panel.
Plant breeding practices rely on genetic variation in order to develop new cultivars with different genetic combinations including improved traits. Molecular markers provide reliable tools to evaluate genetic variation in germplasm collections. Various parameters of genetic diversity in this germplasm collection are shown in Table 2. The highest number of segregating sites were found in subpopulation 1 (n = 35343), followed by subpopulation 3 (n = 28240), 2 (n = 25907) and 4 (n = 21782). Rich genetic diversity exists in this germplasm panel (Table 2). Observed heterozygosity (Ho) in four subpopulations (1-4) were 0.12, 0.08, 0.09, and 0.07, respectively. Expected heterozygosity (He) were 0.22, 0.22, 0.17, and 0.19 in subpopulations 1-4, respectively. Subpopulation 1 contained the highest number of private alleles (30,4230), followed by 21,701 in subpopulation 3, 1,185 in subpopulation 2, and 981 in subpopulation 4 (Table 2). Inbreeding coefficients ranged from 0.45 to 0.53 across these four subpopulations.
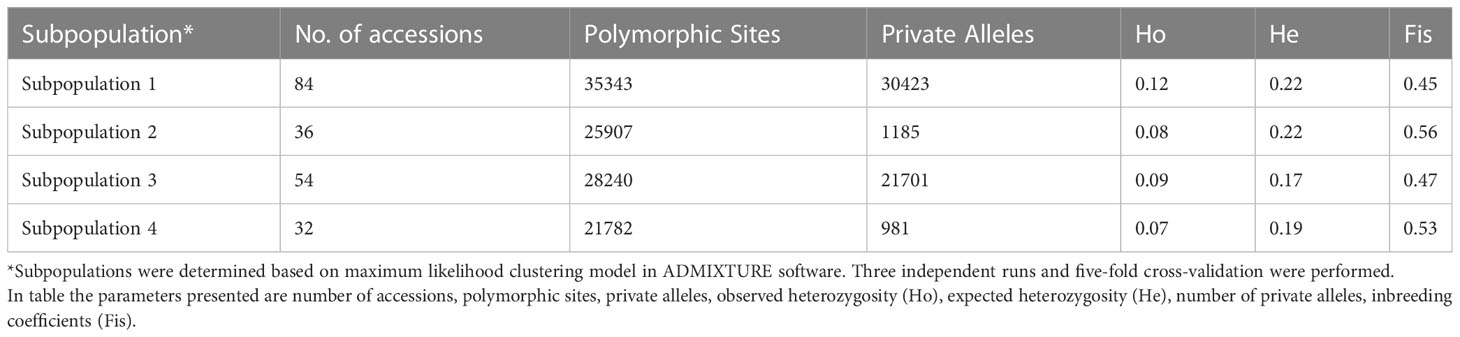
Table 2 Population structure in the bermudagrass (Cynodon spp.) germplasm panel and genetic diversity index of the four subpopulations.
Discussion
Despite recent advancements in high throughput sequencing technologies, genomic resources remain limited in bermudagrass and many other specialty crops. Most previous studies characterizing genetic variation and diversity in Cynodon used traditional markers such as AFLP, ISSR, SRAP and SSRs (Wu et al., 2006; Farsani et al., 2011; Ling et al., 2012; Wang et al., 2013; Zheng et al., 2017). Albeit effective and reproducible, data collection from traditional molecular markers is time-consuming and labor-intensive. In this study, we used genotyping-by-sequencing (GBS) technology and obtained a total of 37,496 high-quality SNP markers to evaluate the genomic diversity in 206 bermudagrass accessions from five continents and 29 countries of the world. GBS has identified genome-wide molecular markers at low cost, and this technology is advantageous for breeders of species like bermudagrass in which limited genomic information is available (Kim et al., 2016; Fang et al., 2020). Although missing data were high in part of the SNPs and/or taxa, high number of SNPs is desirable for genetic diversity assessment and could likely benefit data analyses by surveying more genomic regions.
Previous bermudagrass characterization studies were limited due to the small size or limited geographic coverage of populations studied with small numbers of genetic loci sampled, leading to a limited scope in understanding the genetic structure and variations in Cynodon. For example, Wu et al. (2004) surveyed the genetic diversity of 28 accessions from 11 countries while Jewell et al. (2012) studied 690 accessions, all collected from Australia. The present study utilized a large worldwide germplasm collection, presumably overcoming limitations of previous studies. Four genetic groups were identified in this Cynodon germplasm panel (Figures 2, 3). Similar results were obtained from ADMIXTURE analysis, PCA, and phylogenetic analysis, which corroborates the validity of the genetic structure in this study. Subpopulation 1 consisted of accessions from different geographic locations (Figure 2). Such grouping of genetic materials from different locations was likely due to the exchange of breeding materials between programs and common use of materials from the USDA NPGS, which were originally contributed by multiple Cynodon research groups. Therefore, many of the studied Cynodon accessions may share common ancestry at various levels.
In this germplasm panel, we included 13 African bermudagrass (C. transvaalensis) accessions, many of which clustered in subpopulation 2 with C. dactylon of African origin (Figure 2). As C. dactylon plants are predominantly tetraploids and C. transvaalensis are diploids, mixture of C. dactylon and C. transvaalensis in this subpopulation may shed light on the polyploidization origin of C. dactylon. This finding is novel. We were surprised to find the common bermudagrass and African bermudagrass accessions were grouped together in the same cluster instead of forming two separate groupings relative to other common bermudagrass accessions. It is well known that common bermudagrass can readily cross with African bermudagrass. Their hybrids, however, are sterile triploids, which cannot backcross with common bermudagrass nor African bermudagrass, indicating a strong gene flow barrier between the two species, or that the two species have independent evolutionary pathways. One reasonable speculation is that ancestor(s) of modern diploid African bermudagrass may have substantially contributed to the formation of tetraploid common bermudagrass. When whole genome sequences of the two species are available, comparative analysis may validate this hypothesis. In Miscanthus (Panicoideae subfamily), two major species, M. sinensis (largely diploid) and M. sacchariflorus (largely tetraploid) are used for biomass breeding. Genetic analyses in these two Miscanthus species revealed unidirectional gradient introgression from diploid M. sinensis to tetraploid M. sacchariflorus (Clark et al., 2019). This finding also supports the current mainstream hypothesis that Africa is the center of origin of Cynodon (Burton, 1948; Cui et al., 2021). In subpopulation 3, three C. transvaalensis accessions (PI 251108, PI 291964, and PI 290887) grouped with 46 C. dactylon (Figure 2). In addition to the possibility of close relatedness between these three C. transvaalensis accessions and the C. dactylon in this group, other reasons may include potential mislabeling of species and contamination of materials. Inevitably, further detailed investigation of accessions is needed.
In subpopulation 4 (Figure 2), most accessions from the Midwest and northeastern regions in the US grouped with accessions from European countries and cold/temperate areas in Asia. Bermudagrass is not native to the US (Taliaferro, 1992). This result suggests that the cold hardy, naturalized germplasm collected in OK, KS, NE, IL, MI, MO, and NJ are genetically similar to cold hardy germplasm in Europe and Asia. The genotype A12193 used in this study was collected on the campus of Michigan State University, East Lansing, MI. Bermudagrass on the campus of Michigan State University was a single clone, which was introduced by W. J. Beal in the 1880s. This bermudagrass is amongst the earliest bermudagrass introduced to the region (Gilstrap, 2002; Gilstrap, 2012). The current study indicates that this bermudagrass accession most likely came from Europe or Asia, instead of Africa as originally speculated (Gilstrap, 2002).
Principal component analysis (PCA), ADMIXTURE and phylogenetic analysis all support that most of African accessions formed a unique cluster with the remaining accessions exhibiting admixture (Figures 2–4). The distinct grouping of African accessions indicates that unique genetic variations exist in this germplasm and that they may have important implications for Cynodon breeding. Due to the non-admixture of African accessions with others, they may not have been widely used in breeding programs to date. Genetic diversity within breeding programs decreases due to selection, small population size, genetic drift, and other factors (Reif et al., 2005; Fu, 2015). These accessions can be used in breeding programs to increase genetic variation. Furthermore, most of the African accessions within this PCA cluster are from four nearby countries (South Africa, Zambia, Zimbabwe, Mozambique) in southern Africa, and one accession is from Kenya (East Africa) (Table S1). It is reasonable to hypothesize that other African countries could potentially contribute unique and diverse germplasm as well.
Although Cynodon species are predominantly outcrossing, breeding programs have mainly relied on small subset. Observed heterozygosity ranged from 0.07 to 0.12, which were considerably smaller than expected heterozygosity which ranged from 0.17 to 0.22 (Table 2). This was corroborated by relatively high inbreeding coefficients (0.45-0.56, Table 2). These results further supported the grouping of four subpopulations and were likely due to extensive hybridizations of certain accessions as parental lines.
Results shed new light on the introduction history and origin of unknown accessions of bermudagrass. For example, in subpopulation 2, most of the African accessions clustered together except for 12 accessions from the US and two from Europe. This supports the hypothesis that bermudagrass was introduced to colonial America from Africa (Nelson and Burns, 2006). There is a possibility that many Cynodon accessions in North America or other places are still genetically similar to the ones from their African origin. The geographic origin of some accessions was unknown, especially the MSU accessions. Inferences from the ADMIXTURE analysis and phylogenetic tree could shed light on the origin of these accessions (Figures 2, 4). Twelve of the MSU accessions were clustered in subpopulation 2 with African bermudagrass, indicating the origin might be in Africa. Most of the MSU accessions resulted from hybridizations made during the past 30 years, in the breeding program and are full- or half-siblings, and thus they clustered closely together. The subpopulation 2 from ADMIXTURE is spread all over the phylogenetic tree in different groups suggests that this subpopulation is related to other subpopulations or may contain admixture from other subpopulations.
The 37,496 high-quality SNPs used in the current study allow us to survey more genomic regions of the Cynodon genome and enhances our understanding of this underexplored genus. Moreover, SNP data across the genome in this study assist in the clarification of Cynodon accessions with conflicting information and can help in clarifying the misidentification of accessions. As an example, PI 290812, PI 290813, and PI 290872 accessions are currently named as C. nlemfuensis by Grossman et al. (2021) while the USDA database lists them as C. transvaalensis Burtt Davy. In this study, these three accessions were grouped closely with C. transvaalensis (Figure 4), supporting the USDA data curation.
Conclusions
This study revealed extensive genetic diversity in the two Cynodon species important to breeding turfgrass cultivars by exploiting a bermudagrass germplasm panel of worldwide origin. To the best of our knowledge, this study reports the highest number of genome-wide SNP markers in a bermudagrass germplasm study. Strong and distinct genetic structure, as revealed by multiple analyses, indicates rich genetic variation for further improvement and development of new bermudagrass cultivars to reach a new adaptive peak. The grouping of African bermudagrass with common bermudagrass populations originating in Africa is a new finding and suggests significance in evolution and adaptation in the Africa continent. This study reveals that naturalized bermudagrass in the Midwest and Northeast of the US were genetically similar and likely introductions from Europe or Asia. The separate groupings of common bermudagrass accessions provides information to breeders and geneticists aiding in parental line selection for developing breeding populations. This study provides a valuable guide for allele mining of desirable genes for the traits of interest (e.g., abiotic and biotic stress resistance), which will improve bermudagrass breeding and serve as foundation for genetic mapping of important traits.
Data availability statement
The SNP datasets generated and analyzed in this study are available in the Figshare repository: https://figshare.com/articles/dataset/Genomic_Diversity_Bermudagrass/21941333. R scripts used in data analysis and visualization are available on Github: https://github.com/SinghLovepreet2363/Bermudagrass_GenomicDiversity.
Author contributions
LS and HD conceptualized the study, analyzed the data, drafted the manuscript. YW contributed to the germplasm collection, assisted in interpreting the results and edited the manuscript. MW, JM, BS, BB edited and reviewed the manuscript. HD obtained the funding for the project. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by USDA HATCH fund (MIS-213140) and Mississippi Department of Agriculture and Commerce (Award # G00006342).
Acknowledgments
We thank USDA-ARS Plant Genetic Resources Conservation Unit: Griffin, GA for germplasm and providing us information regarding the accessions. We also thank Mr. Wayne Philley, Mr. Geoffrey Lalk and Mr. Jason Ruffin for maintaining greenhouse and field trials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1155721/full#supplementary-material
References
Alexander, D. H., Novembre, J., Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Al-Humaid, A., Motawei, M. I. (2004). Molecular characterization of some turfgrass cultivars using randomly amplified polymorphic DNA (RAPD) markers. J. Food Agric. Environ. 2, 376–380.
Assefa, S., Taliaferro, C. M., Anderson, M. P., de los Reyes, B. G., Edwards, R. M. (1999). Diversity among Cynodon accessions and taxa based on DNA amplification fingerprinting. Genome 42, 465–474. doi: 10.1139/g98-152
Bethel, C. M., Sciara, E. B., Estill, J. C., Bowers, J. E., Hanna, W., Paterson, A. H. (2006). A framework linkage map of bermudagrass (Cynodon dactylon x transvaalensis) based on single-dose restriction fragments. Theor. Appl. Genet. 112, 727–737. doi: 10.1007/s00122-005-0177-y
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Caetano-Anollés, G., Callahan, L. M., Williams, P. E., Weaver, K. R., Gresshoff, P. M. (1995). DNA Amplification fingerprinting analysis of bermudagrass (Cynodon): genetic relationships between species and interspecific crosses. Theor. Appl. Genet. 91, 228–235. doi: 10.1007/BF00220882
Chaves, A. L. A., Carvalho, P. H. M., Ferreira, M. T. M., Benites, F. R. G., Techio, V. H. (2022). Genomic constitution, allopolyploidy, and evolutionary proposal for Cynodon rich. based on GISH. Protoplasma 259, 999–1011. doi: 10.1007/s00709-021-01716-z
Clark, L. V., Jin, X., Petersen, K. K., Anzoua, K. G., Bagmet, L., Chebukin, P., et al. (2019). Population structure of miscanthus sacchariflorus reveals two major polyploidization events, tetraploid-mediated unidirectional introgression from diploid m. sinensis, and diversity centred around the yellow Sea. Ann. Bot. 124, 731–748. doi: 10.1093/aob/mcy161
Cui, F., Taier, G., Li, M., Dai, X., Hang, N., Zhang, X., et al. (2021). The genome of the warm-season turfgrass African bermudagrass (Cynodon transvaalensis). Hortic. Res. 8, 93. doi: 10.1038/s41438-021-00519-w
Dong, H., Philley, H. W., Harris-Shultz, K. (2022). Registration of ‘MSB-264’ and ‘MSB-285’ bermudagrasses. J. Plant Regist. 16, 185–197. doi: 10.1002/plr2.20218
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One 6, e19379. doi: 10.1371/journal.pone.0019379
Fang, T., Dong, H., Yu, S., Moss, J. Q., Fontanier, C. H., Martin, D. L., et al. (2020). Sequence-based genetic mapping of Cynodon dactylon pers. reveals new insights into genome evolution in poaceae. Commun. Biol. 3, 358. doi: 10.1038/s42003-020-1086-y
Farsani, T. M., Etemadi, N., Sayed-Tabatabaei, B. E., Talebi, M. (2011). Assessment of genetic diversity of bermudagrass (Cynodon dactylon) using ISSR markers. Int. J. Mol. Sci. 13, 383–392. doi: 10.3390/ijms13010383
Forbes, J. I., Burton, G. W. (1963). Chromosome numbers and meiosis in some Cynodon species and hybrids 1. Crop Sci. 3 (1), 75–79. doi: 10.2135/cropsci1963.0011183X000300010023x
Frankham, R., Ballou, J. D., Briscoe, D. A., McInnes, K. H. (2002). Introduction to conservation genetics (Cambridge: Cambridge University Press). doi: 10.1017/CBO9780511808999
Fu, Y.-B. (2015). Understanding crop genetic diversity under modern plant breeding. Theor. Appl. Genet. 128, 2131–2142. doi: 10.1007/s00122-015-2585-y
Gilstrap, D. (2002). MSU bermudagrass: what are we waiting for? – funding. lawncare, athletic fields, and commercial turf field day (East Lansing, MI: Michigan State University).
Gilstrap, D. (2012). MSU bermudagrass: looks like a clone, acts like a clone. lawncare, athletic fields, and commercial turf field day (East Lansing, MI: Michigan State University).
Goudet, J. (2005). Hierfstat, a package for r to compute and test hierarchical f-statistics. Mol. Ecol. Notes 5 (1), 184–186. doi: 10.1111/j.1471-8286.2004.00828.x
Grossman, A. Y., Andrade, M. H. M. L., Chaves, A. L. A., Mendes Ferreira, M. T., Techio, V. H., Lopez, Y., et al. (2021). Ploidy level and genetic parameters for phenotypic traits in bermudagrass (Cynodon spp.) germplasm. Agronomy 11, 912. doi: 10.3390/agronomy11050912
Gulsen, O., Sever-Mutlu, S., Mutlu, N., Tuna, M., Karaguzel, O., Shearman, R. C., et al. (2009). Polyploidy creates higher diversity among Cynodon accessions as assessed by molecular markers. Theor. Appl. Genet. 118, 1309–1319. doi: 10.1007/s00122-009-0982-9
Guo, Y., Wu, Y., Anderson, J. A., Moss, J. Q., Zhu, L. (2015). Disomic inheritance and segregation distortion of SSR markers in two populations of Cynodon dactylon (L.) pers. var. dactylon. PloS One 10, e0136332. doi: 10.1371/journal.pone.0136332
Harlan, J. R., De Wet, J. M. J. (1969). Sources of variation in Cynodon dactylon (L). pers. Crop Science. 9, 774–778. doi: 10.2135/cropsci1969.0011183X000900060031x
Harlan, J. R., De Wet, J. M., Huffine, W. W., Deakin, J. R. (1970). Oklahoma Agricultural experiment station, bulletin no. 673, January 1970: a guide to the species of cynodon (Gramineae).
Harlan, J. R., De Wet, J. M., Rawal, K. M., Felder, M. R., Richardson, W. L. (1970a). Cytogenetic studies in Cynodon LC Rich.(Gramineae) 1. Crop Sci. 10, 288–291. doi: 10.2135/cropsci1970.0011183X001000030023x
Harris-Shultz, K. R., Schwartz, B. M., Hanna, W. W., Brady, J. A. (2010). Development, linkage mapping, and use of microsatellites in bermudagrass. J. Amer. Soc Hortic. Sci. 135, 511–520. doi: 10.21273/JASHS.135.6.511
Hartl, D. L., Clark, A. G., Clark, A. G. (1997). Principles of population genetics Vol. 116 (Sunderland: Sinauer associates).
Horowitz, M. (1996). Bermudagrass (Cynodon dactylon): a history of the weed and its control in Israel. Phytoparasitica 24, 305–320. doi: 10.1007/BF02981413
Huang, C., Liu, G., Bai, C., Wang, W. (2014). Genetic analysis of 430 Chinese Cynodon dactylon accessions using sequence-related amplified polymorphism markers. Int. J. Mol. Sci. 15, 19134–19146. doi: 10.3390/ijms151019134
Jewell, M. C., Zhou, Y., Loch, D. S., Godwin, I. D., Lambrides, C. J. (2012). Maximizing genetic, morphological, and geographic diversity in a core collection of australian bermudagrass. Crop Sci. 52, 879–889. doi: 10.2135/cropsci2011.09.0497
Jombart, T., Ahmed, I. (2011). Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. doi: 10.1093/bioinformatics/btr521
Jombart, T., Devillard, S., Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11, 94. doi: 10.1186/1471-2156-11-94
Kang, S. Y., Lee, G. J., Lim, K. B., Lee, H. J., Park, I. S., Chung, S. J., et al. (2008). Genetic diversity among Korean bermudagrass (Cynodon spp.) ecotypes characterized by morphological, cytological and molecular approaches. Mol. Cells (Springer Sci. Business Media BV) 25, 163–171.
Karaca, M., Saha, S., Zipf, A., Jenkins, J. N., Lang, D. J. (2002). Genetic diversity among forage bermudagrass (Cynodon spp.) evidence from chloroplast and nuclear DNA fingerprinting. Crop Sci. 42, 2118–2127. doi: 10.2135/cropsci2002.2118
Khanal, S., Dunne, J. C., Schwartz, B. M., Kim, C., Milla-Lewis, S., Raymer, P. L., et al. (2019). Molecular dissection of quantitative variation in bermudagrass hybrids (Cynodon dactylon x transvaalensis): morphological traits. G3 (Bethesda) 9, 2581–2596. doi: 10.1534/g3.119.400061
Khanal, S., Kim, C., Auckland, S. A., Rainville, L. K., Adhikari, J., Schwartz, B. M., et al. (2017). SSR-enriched genetic linkage maps of bermudagrass (Cynodon dactylon × transvaalensis), and their comparison with allied plant genomes. Theor. Appl. Genet. 130, 819–839. doi: 10.1007/s00122-017-2854-z
Kim, C., Guo, H., Kong, W., Chandnani, R., Shuang, L.-S., Paterson, A. H. (2016). Application of genotyping by sequencing technology to a variety of crop breeding programs. Plant Sci. 242, 14–22. doi: 10.1016/j.plantsci.2015.04.016
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, H., Liu, L., Lou, Y., Hu, T., Fu, J. (2011). Genetic diversity of Chinese natural bermudagrass (Cynodon dactylon) germplasm using ISSR markers. Sci. Hortic. 127, 555–561. doi: 10.1016/j.scienta.2010.12.001
Ling, Y., Zhang, X. Q., Ma, X., Chen, S. Y., Chen, T. T., Liu, W. (2012). Analysis of genetic diversity among wild bermudagrass germplasm from southwest China using SSR markers. Genet. Mol. Res. 11, 4598–4608. doi: 10.4238/2012.October.17.5
Lu, F., Lipka, A. E., Glaubitz, J., Elshire, R., Cherney, J. H., Casler, M. D., et al. (2013). Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PloS Genet. 9, e1003215. doi: 10.1371/journal.pgen.1003215
McElroy, J. S., Breeden, G. K. (2006). Triclopyr safens the use of fluazifop and fenoxaprop on zoysiagrass while maintaining bermudagrass suppression. ats 3. doi: 10.1094/ATS-2006-0502-01-RS
Mutlu, S. S., Mutlu, N. E., Selim, C. E., Hocagil, M. M. (2014). Broadening the genetic base of bermudagrass. Eur. J. Hortic. Sci. 79, 183–194.
Nelson, C. J., Burns, J. C. (2006). Fifty years of grassland science leading to change. Crop Sci. 46, 2204–2217. doi: 10.2135/cropsci2006.04.0278gas
Paradis, E. (2010). Pegas: an r package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Paradis, E., Claude, J., Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in r language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Reasor, E. H., Brosnan, J. T., Staton, M. E., Lane, T., Trigiano, R. N., Wadl, P. A., et al. (2018). Genotypic and phenotypic evaluation of off-type grasses in hybrid bermudagrass [Cynodon dactylon (L.) pers. x c. transvaalensis burtt-Davy] putting greens using genotyping-by-sequencing and morphological characterization. Hereditas 155, 8. doi: 10.1186/s41065-017-0043-3
Reif, J. C., Zhang, P., Dreisigacker, S., Warburton, M. L., van Ginkel, M., Hoisington, D., et al. (2005). Wheat genetic diversity trends during domestication and breeding. Theor. Appl. Genet. 110, 859–864. doi: 10.1007/s00122-004-1881-8
Roodt, R., Spies, J. J., Burger, T. H. (2002). Preliminary DNA fingerprinting of the turf grass Cynodon dactylon (Poaceae: chloridoideae). Bothalia 32, 117–122. doi: 10.4102/abc.v32i1.474
Shendye, N. V., Gurav, S. S. (2014). Cynodon dactylon: a systemic review of pharmacognosy, phytochemistry and pharmacology. Int. J. Pharm. Pharm. Sci. 6, 7–12.
Taliaferro, C. M. (1992). Out of Africa: a new look at "african" bermudagrass. USGA Green Sect. Rec. (USA) 30, 10–12.
Uddin, M., Juraimi, A. S., Ismail, M. R., Brosnan, J. T. (2010). Characterizing weed populations in different turfgrass sites throughout the klang valley of Western peninsular Malaysia. Weed Technol. 24, 173–181. doi: 10.1614/WT-09-046.1
Wang, Z., Liao, L., Yuan, X., Guo, H., Guo, A., Liu, J. (2013). Genetic diversity analysis of Cynodon dactylon (bermudagrass) accessions and cultivars from different countries based on ISSR and SSR markers. Biochem. Syst. Ecol. 46, 108–115. doi: 10.1016/j.bse.2012.09.001
Wang, Z., Wu, Y., Martin, D. L., Gao, H., Samuels, T., Tan, C. (2010). Identification of vegetatively propagated turf bermudagrass cultivars using simple sequence repeat markers. Crop Sci. 50, 2103–2111. doi: 10.2135/cropsci2010.02.0116
Wang, Z., Yuan, X., Zheng, Y., Liu, J. (2009). Molecular identification and genetic analysis for 24 turf-type Cynodon cultivars by sequence-related amplified polymorphism markers. Sci. Hortic. 122, 461–467. doi: 10.1016/j.scienta.2009.05.031
Wu, Y. (2011). “Cynodon,” in Wild crop relatives: genomic and breeding resources. Ed. Kole, C. (Berlin, Heidelberg: Springer Berlin Heidelberg), 53–71. doi: 10.1007/978-3-642-14255-0_4
Wu, Y. Q., Taliaferro, C. M., Bai, G. H., Anderson, M. P. (2004). AFLP analysis of Cynodon dactylon (L.) pers. var. dactylon genetic variation. Genome 47, 689–696. doi: 10.1139/g04-032
Wu, Y. Q., Taliaferro, C. M., Bai, G. H., Martin, D. L., Anderson, J. A., Anderson, M. P., et al. (2006). Genetic analyses of chinese Cynodon accessions by flow cytometry and AFLP markers. Crop Sci. 46, 917–926. doi: 10.2135/cropsci2005.08.0256
Yu, S., Fang, T., Dong, H., Yan, L., Martin, D. L., Moss, J. Q., et al. (2021). Genetic and QTL mapping in African bermudagrass. Plant Genome 14, e20073. doi: 10.1002/tpg2.20073
Zhang, L. H., Ozias-Akins, P., Kochert, G., Kresovich, S., Dean, R., Hanna, W. (1999). Differentiation of bermudagrass (Cynodon spp.) genotypes by AFLP analyses. Theor. Appl. Genet. 98, 895–902. doi: 10.1007/s001220051148
Zhang, J., Wang, M., Guo, Z., Guan, Y., Liu, J., Yan, X., et al. (2019). Genetic diversity and population structure of bermudagrass [Cynodon dactylon (L.) pers.] along latitudinal gradients and the relationship with polyploidy level. Diversity (Basel) 11, 135. doi: 10.3390/d11080135
Keywords: single nucleotide polymorphism (SNP) markers, germplasm, genomic diversity, diploid, tetraploid, ADMIXTURE analysis
Citation: Singh L, Wu Y, McCurdy JD, Stewart BR, Warburton ML, Baldwin BS and Dong H (2023) Genetic diversity and population structure of bermudagrass (Cynodon spp.) revealed by genotyping-by-sequencing. Front. Plant Sci. 14:1155721. doi: 10.3389/fpls.2023.1155721
Received: 31 January 2023; Accepted: 18 May 2023;
Published: 08 June 2023.
Edited by:
Mohan Lal, North East Institute of Science and Technology (CSIR), IndiaReviewed by:
Bochra Amina Bahri, University of Georgia, United StatesRajneesh Paliwal, International Institute of Tropical Agriculture (IITA), Nigeria
Gang Nie, Sichuan Agricultural University, China
Abdurrahim Yılmaz, Abant Izzet Baysal University, Türkiye
Copyright © 2023 Singh, Wu, McCurdy, Stewart, Warburton, Baldwin and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian S. Baldwin, YmJhbGR3aW5AcHNzLm1zc3RhdGUuZWR1; Hongxu Dong, aHhkb25nODlAZ21haWwuY29t
 Lovepreet Singh
Lovepreet Singh Yanqi Wu2
Yanqi Wu2 Brian S. Baldwin
Brian S. Baldwin Hongxu Dong
Hongxu Dong