- 1Hebei Key Laboratory of Crop Stress Biology, Hebei Normal University of Science and Technology, Qinghuangdao, Hebei, China
- 2Institute of Food Corps, Hubei Academy of Agricultural Sciences, Wuhan, Hubei, China
The MYB transcription factors regulate plant growth, development, and defense responses. However, information about the MYB gene family in Ipomoea species is rare. Herein, we performed a comprehensive genome-wide comparative analysis of this gene family among seven Ipomoea species, sweet potato (I. batatas), I. trifida, I. triloba, I. nil, I. purpurea, I. cairica, and I. aquatic, and identified 296, 430, 411, 291, 226, 281, and 277 MYB genes, respectively. The identified MYB genes were classified into five types: 1R-MYB (MYB-related), 2R-MYB (R2R3-MYB), 3R-MYB (R1R2R3-MYB), 4R-MYB, and 5R-MYB, and the MYB-related or R2R3-MYB type was the most abundant MYB genes in the seven species. The Ipomoea MYB genes were classed into distinct subgroups based on the phylogenetic topology and the classification of the MYB superfamily in Arabidopsis. Analysis of gene structure and protein motifs revealed that members within the same phylogenetic group presented similar exon/intron and motif organization. The identified MYB genes were unevenly mapped on the chromosomes of each Ipomoea species. Duplication analysis indicated that segmental and tandem duplications contribute to expanding the Ipomoea MYB genes. Non-synonymous substitution (Ka) to synonymous substitution (Ks) [Ka/Ks] analysis showed that the duplicated Ipomoea MYB genes are mainly under purifying selection. Numerous cis-regulatory elements related to stress responses were detected in the MYB promoters. Six sweet potato transcriptome datasets referring to abiotic and biotic stresses were analyzed, and MYB different expression genes’ (DEGs’) responses to stress treatments were detected. Moreover, 10 sweet potato MYB DEGs were selected for qRT-PCR analysis. The results revealed that four responded to biotic stress (stem nematodes and Ceratocystis fimbriata pathogen infection) and six responded to the biotic stress (cold, drought, and salt). The results may provide new insights into the evolution of MYB genes in the Ipomoea genome and contribute to the future molecular breeding of sweet potatoes.
1 Introduction
The MYB transcription factor (TF) family, as one of the most prominent TF families in the plant, is composed of three conservative functional domains: DNA-binding domain (DBD), transcriptional activation domain (TAD), and incompletely defied negative regulatory region (NRD). Among the three, DBD is the most conservative and is generally known as the MYB domain (Frampton, 2004). The MYB domain consists of 51–52 conserved amino acids and spacing coding sequences, and every 18 amino acids has a regular spacing of tryptophan residues. This amino acid residue expands the MYB domain into a helix (HTH) structure (Frampton, 2004). According to the total number of adjacent MYB repetitions, MYB TF can be divided into four categories, namely, 1R-MYB (MYB-related), 2R-MYB, 3R-MYB, and 4R-MYB, which contain the corresponding number of MYB repeats, respectively (Dubos et al., 2010).
1R-MYB, also called MYB-related, generally but not always include a single or partial MYB iteration (Klempnauer et al., 1982). Based on the highly conserved motif, the MYB-related TFs can be divided into subgroups (Liu et al., 2020b; Arce-Rodríguez et al., 2021; Yan et al., 2022). The MYB-related TFs play essential roles in plant seed germination and root growth (Zhao et al., 2019), leaf senescence and epidermal cell patterning (Lee and Schiefelbein, 1999; Machado et al., 2009; Park et al., 2018), biosynthesis (Sinaga et al., 2021), the regulation of circadian rhythms (Alabadı́ et al., 2001; Mizoguchi et al., 2002; Okada et al., 2009; Kamioka et al., 2016), and stress responses (Dubos et al., 2010; Park et al., 2018; Zhao et al., 2019; Hoseinpour et al., 2021).
The 2R-MYB genes, named R2R3-MYB, were usually the largest MYB gene subfamily in green plants (Martin and Paz-Ares, 1997). They play essential roles in plant lifetime, including (1) primary and secondary metabolites, such as phenylpropanoid metabolism (Liu et al., 2015; Tang et al., 2021), flavonoid biosynthesis (Czemmel et al., 2012; Lin et al., 2021; Zhao et al., 2022b), anthocyanin biosynthesis (Naing and Kim, 2018; Upadhyaya et al., 2021; Wang et al., 2022c), chlorogenic acid biosynthesis (Tang et al., 2021), indolic and aliphatic glucosinolate biosynthesis (Gigolashvili et al., 2009), and apigenin biosynthesis (Wang et al., 2022b); (2) cell fate and identity, for instance, epidermis and root formation (Du et al., 2009); (3) developmental processes, like embryogenesis (Wang et al., 2009), pollen development (Liu et al., 2021), male sterility (Yu et al., 2021), and plant trichome development (Shangguan et al., 2021); and (4) responses to biotic and abiotic stresses, including diseases (Shen et al., 2017; Gu et al., 2020; Hawku et al., 2022), high salinity (Wang et al., 2021a; Du et al., 2022), cold (Dong et al., 2021), heat (Wu et al., 2021b), and drought (Lv et al., 2021; Wu et al., 2021a).
The 3R-MYB TFs in plants recognize mitosis-specific activator (MSA) elements (Ito et al., 1998; Ma et al., 2009) and play an essential role in cell-cycle regulation. The plant 3R-MYB TFs regulate the G2/M transition (Ito et al., 2001). The recognized DNA element (MSA) was in the upstream promoter region of G2/M-phase-specific genes, which is both necessary and sufficient for driving G2/M-phase-specific gene expression (Ito et al., 2001; Kato et al., 2009). The plant 3R-MYB TFs were often classed into three subgroups: A, B, and C (Ito, 2005). The 3R-MYB TFs in the A and B subgroups were reported to be involved in cell-cycle regulation (Ito et al., 2001; Kato et al., 2009; Haga et al., 2011), and the ones in the C group participated in both cell-cycle and abiotic stresses (Dai et al., 2007; Ma et al., 2009). It was also found that 3R-MYB inhibited plant trichome development and inhibited flavonoid biosynthetically (Dubos et al., 2008; Matsui et al., 2008). The 3R-MYB gene, PhMYBx in petunia, downregulated anthocyanin synthesis (Koes et al., 2005; Chezem and Clay, 2016). The 4R-MYB TFs, representing the smallest subfamily of MYB TFs, contain four R1/R2-like repeats. Little is known about their functions in plants. Recently, the 4R-MYB protein SNAPc4 in Arabidopsis was reported as part of the SNAP complex involved in snRNA gene transcription, and AtSNAPc4 is proved to be an essential gene in gametophyte and zygote development (Thiedig et al., 2021).
Since the first plant MYB gene (ZmMYBC1) was cloned and reported to be involved in pigment biosynthesis in Zea maize (Paz-Ares et al., 1987), the characterization and analysis of MYB genes have been conducted in different plant species. In most cases, the 2R-MYB genes were the most abundant, followed by MYB-related genes and 3R-MYB genes, and the 4R-MYB gene group was the smallest (Dubos et al., 2010). However, the number of MYB genes varied widely among species, ranging from less than 100 to more than 500. For instance, 198 MYB genes in Arabidopsis (Chen et al., 2006), 183 in Oryza sativa (Chen et al., 2006), 475 in Brassica rapa ssp. Pekinensis (Saha et al., 2016), 524 in Gossypium hirsutum (Salih et al., 2016), 174 in Morella rubra (Cao et al., 2021), 217 in Solanum tuberosum (Li et al., 2021c), 182 in Casuarina equisetifolia (Wang et al., 2021b), and 54 in Mangifera indica (Zhang et al., 2022) were identified and analyzed. The number of MYB genes greatly varied in the species of the same genus. For example, in the Solanum genus, 127 MYB genes were reported in S. lycopersicum (Li et al., 2016) whereas 217 in S. tuberosum (Li et al., 2021c); in the Musa genus, 305 MYB genes were identified in M. acuminate whereas 251 in M. balbisiana (Tan et al., 2020). To date, information about the comparative analysis of MYB genes in closely related species (the same genus) is still limited.
The genus Ipomoea (family Convolvulaceae) contains 600–700 species (Nimmakayala et al., 2011), many of which are of considerable importance either as medicinal plants or as ornamental plants (Nimmakayala et al., 2011). The sweet potato (I. batatas), serving as the seventh most important crop in the world, is an essential source of calories, proteins, vitamins, and minerals for humanity (Bovell-Benjamin, 2007; Yang et al., 2017). I. trifida and I. triloba, two diploid relatives of sweet potatoes, can be used as model plants to facilitate sweet potato breeding (Wu et al., 2018). I. nil and I. purpurea are ideal plants for researching photoperiodic flowering and flower coloration (Hoshino et al., 2016). I. cairica (L.) is a perennial vine plant that blooms all year round and has been widely introduced to subtropical, subtropical, and temperate regions as ornamental plants for the landscape. It also has the characteristics of medicinal value because it contains a large number of bioactive compounds. In many countries, a decoction of the whole plant is used to treat tuberculosis, cough, asthma, liver cirrhosis, and jaundice (Jiang et al., 2022). Water spinach (I. aquatica) is one of Asia’s most popular green leafy vegetables, with both aquatic and terrestrial characteristics (Hao et al., 2021).
Related information is limited compared to the importance of the MYB gene family and Ipomoea species, particularly for sweet potatoes. For the sweet potato, the researchers mainly focused on researching a few 2R-MYB genes in the past years. IbMYB1 increases the anthocyanin content and the resistance ability of potatoes (Cheng et al., 2013); the IbMYB1a gene induces anthocyanin accumulation in Arabidopsis (Chu et al., 2013); and overexpression of the IbMYB1 gene in an orange-fleshed sweet potato cultivar produces a dual-pigmented transgenic sweet potato with improved antioxidant activity (Park et al., 2015). IbMYB116 was reported to enhance the drought tolerance of Arabidopsis (Zhou et al., 2019). A single amino acid mutant in the EAR motif of IbMYB44.2 could reduce the inhibition of anthocyanin accumulation in the purple-fleshed sweet potato (Li et al., 2021a). IbMYB48 (a sweet potato R2R3-MYB gene) confers enhanced tolerance to salt and drought stresses in transgenic Arabidopsis (Zhao et al., 2022a). IbMYB308 improves salt stress tolerance in transgenic tobacco (Wang et al., 2022a). However, there is no comparative analysis of the MYB gene family of Ipomoea species, and knowledge of MYB genes of this genus is limited.
This study conducted a genome-wide comparative analysis of the MYB gene family in seven Ipomoea species. There were 296, 430, 411, 291, 226, 281, and 277 MYB genes identified from sweet potato (I. batatas), I. trifida, I. triloba, I. nil, I. purpurea, I. cairica, and I. aquatic, respectively. The identified MYB genes were then subjected to phylogenetic analysis, gene structure investigation, chromosome location, syntenic analysis, non-synonymous substitution (Ka) to synonymous substitution (Ks) [Ka/Ks] calculation, cis-regulatory element (CRE) detection, and expression profile analysis. Then, 10 sweet potato MYB different expression genes (DEGs) were selected for qRT-PCR analysis. The results showed that four responded to biotic stress (stem nematodes and Ceratocystis fimbriata pathogen infection) and six responded to abiotic stress (cold, drought, and salt). These results are likely to give a new view on the evolution of the MYB gene in Ipomoea species. They are conducive to the development of molecular breeding in sweet potatoes in the future.
2 Materials and methods
2.1 Data resources
The whole genome sequences and annotated files of the seven Ipomoea species were obtained from the following open-access databases: sweet potato from the I. batatas Genome Browser (http://public-genomes-ngs.molgen.mpg.de/SweetPotato/), I. trifida and I. triloba from GenBank BioProject (accessions numbers PRJNA428214 and PRJNA428241), I. nil from the I. nil web (http://viewer.shigen.info/asagao/index.php), I. purpurea from the CoGe platform (https://genomevolution.org/coge/GenomeInfo.pl?gid=58735), I. cairica from AGIS (ftp://ftp.agis.org.cn/~fanwei/Ipomoea_cairica_genome_v1), and I. aquatic from BIGD (PRJCA002216). The MYB protein sequences of Arabidopsis were downloaded from the TAIR database (https://www.arabidopsis.org) (Chen et al., 2006).
2.2 Identification of MYB genes in Ipomoea species
To identify the MYB genes in Ipomoea species, two strategies were adopted. First, the protein sequences of the seven Ipomoea species were searched for the MYB domain (Pfam accession number: PF00249) using HMMsearch (ver. 3.1b2) with default parameters. Second, the MYB protein sequences of Arabidopsis were used as queries to a search against the protein database of each Ipomoea species by BLASTP (ver. 2.10.0+) with an E-value cutoff of 1e-10 (Altschul et al., 1990). The protein sequences obtained from the two strategies were merged, and the redundant ones were removed. Then, all of the MYB proteins were subjected to a search against the Pfam database (release 35; adopted Pfam A) using PfamScan.pl (v1.6) with default parameters, search against the Conserved Domain Database (CDD) database (ver. 3.20) employing Reverse Position-Specific BLAST (RPS–BLAST) (ver. 2.10.0+) with an e-value cutoff of 1e-10, and search against the Simple Modular Architecture Research Tool (SMART) database (http://smart.embl-heidelberg.de/) by performing SMART_batch.pl (http://smart.embl-heidelberg.de/help/SMART_batch.pl) with default parameters. The proteins confirmed by all three databases were considered candidate MYB TFs.
2.3 Molecular weight, isoelectric point, and subcellular localization analysis of Ipomoea MYB proteins
The molecular weight (MW) and isoelectric point (pI) for each MYB protein were analyzed by using ExPASy (http://www.expasy.ch/tools/pi_tool.html). The online website WoLF PSORT (https://wolfpsort.hgc.jp/) was employed for predicting the subcellular localization of the Ipomoea proteins.
2.4 Identification of conserved motifs of the Ipomoea MYB genes
The MYB protein sequence motif analysis was performed using the online MEME Suite (https://meme-suite.org/meme/) (Bailey et al., 2009). The maximum detecting number of motifs was designed to identify 20 motifs and the site distribution was set as any, whereas other parameters were set as default.
2.5 Sequence alignment and phylogenetic analysis of Ipomoea MYB proteins
The Ipomoea and Arabidopsis MYB protein sequences were aligned using Clustal Omega (Sievers et al., 2011; Sievers and Higgins, 2018). The obtained aligned sequences were submitted to IQ-TREE for phylogenetic analysis using the maximum likelihood approach (Nguyen et al., 2015). The branch support values were calculated by SH-aLRT and UFBoot2 with 1,000 bootstrap replicates (Anisimova et al., 2011).
2.6 Protein motif compositions and gene structures of Ipomoea MYB genes
Based on the results of the online MEME Suite, phylogenetic analysis, and the gff3 files of the genomes, the Ipomoea MYB genes identified above were submitted to TBtools for protein motif compositions and gene structure analysis and picture drawing (Chen et al., 2020).
2.7 Chromosome distribution and duplication pattern analysis of the Ipomoea MYB genes
The MYB genes with chromosomal positions were mapped on the chromosomes of seven Ipomoea species with MapChart (ver. 2.30) (Voorrips, 2002). The potential duplicated MYB genes in the Ipomoea genome were analyzed with MCScanX software (Wang et al., 2012). During this stage, the protein sequences of Ipomoea species were compared against themselves by running the BLASTP (ver. 2.10.0+) program with an E-value of 1e–10 (Altschul et al., 1990). The final output of the duplicated analysis was visualized with the CIRCOS software (ver. 0.66) (Krzywinski et al., 2009).
2.8 Syntenic analysis of MYB genes in the seven Ipomoea genomes
Syntenic block in the genomes of the seven Ipomoea species was analyzed using MCScan software (Python version3) (Tang et al., 2008) with the default parameters (VanBuren et al., 2018; Li et al., 2021b). The gene models were aligned with LAST, and hits were filtered to locate the best 1:1 syntenic blocks (pairs) and were visualized in the dot-plot script using the JCVI package (Tang et al., 2008).
2.9 Ka/Ks analysis of duplicated and syntenic MYB genes
Both duplicated and syntenic MYB gene pairs of the Ipomoea species were selected for the non-synonymous substitution (Ka) to synonymous substitution (Ks) [Ka/Ks] calculation with TBtools (Chen et al., 2020).
2.10 Promoter analysis of Ipomoea MYB genes
The 1.5-kb promoter sequences of the Ipomoea MYB genes were submitted into PLANT CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 21 March 2021) for identification of the putative cis-elements (Magali et al., 2002).
2.11 Expression profile of MYB Ipomoea genes
Six transcriptome bio project datasets referring to abiotic and biotic stresses were selected for expression profile analysis of the sweet potato MYB genes. Three bio project datasets (PRJNA341328 for cold, PRJNA413661 for drought, and PRJNA631585 for salt) referring to abiotic stress and one bio project (PRJNA429283 for root-knot nematode) referring to biotic stress were obtained from the NCBI database. The other two were our in-house transcriptome datasets (unpublished) for sweet potato stem nematodes and C. fimbriata resistance of four sweet potato cultivars or lines (sweet potato stem nematode-susceptible cultivar, “Tengfei,” sweet potato stem nematode-resistant line, “JK20,” C. fimbriata-susceptible cultivar, “Santiandao,” and C. fimbriata-resistant line, “JK274”). In each comparing case, reads were treated as DEGs if |log2FC|>1 and FDR ≤ 5%. Thus, the mean log2FC value for each DEG was calculated. The heat map was constructed to visualize the distribution of the expression level of genes using the fragments per kilobase per million (FPKM) value in MeV software (Howe et al., 2011). The gene FPKM data of I. trifida and I. triloba were downloaded from the sweet potato database (http://sweetpotato.plantbiology.msu.edu/).
2.12 RNA isolation and quantitative qRT-PCR analysis
Xu32 (susceptible cultivar) and JK328 (resistant line) were selected for cold, salt, and drought stress treatments. The cuttings about 25 cm in length from 6-week-olds of them grown in a field were cultured in the Hoagland solution for 3 days to survive: for cold stress treatment, the cuttings were then placed at 28°C (control) and 16°C (cold stress), respectively; for salt stress treatment, the cuttings were cultured in the Hoagland solution with 0 and 86 mM NaCl, respectively; and for drought stress treatments, the cuttings were cultured in Hoagland solution with 0% and 30% PEG 6000. Samples were collected at seven time points (0, 2, 4, 6, 12, 24, and 48 h) after the treatments. The cultivars Tengfei (susceptible cultivar) and JK20 (resistant line) were inoculated with sweet potato stem nematodes (Gao et al., 2011), and the cultivars Santiandao (susceptible cultivar) and JK274 (resistant line) were inoculated with C. fimbriata (Muramoto et al., 2012). Samples were collected at seven time points (0 h, 6 h, 12 h, 1 day, 2 days, 4 days, and 6 days) after the injection. Root samples without injection were used as a control. Then, the total RNA of the samples was isolated using RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) and first-strand cDNA was synthesized by QuantScript Reverse Transcriptase Kit (Tiangen Biotech, Beijing, China). The sweet potato β-actin gene (GenBank AY905538) was used as a control and to normalize the relative quantities of the three individual targeted DEGs based on its consistency across the different time points of each treatment. Three biological replicates were performed at each time point, and the gene expression changes were calculated using the 2–ΔΔCt method for each sample (Schmittgen and Livak, 2008). Using the primers (Table S1) generated with Primer-BLAST software (Ye et al., 2006), qRT-PCR was performed as described previously (Zhai et al., 2016).
3 Results
3.1 Identification of the MYB genes in the seven Ipomoea species
A total of 2,212 MYB genes were identified from the seven Ipomoea species: 296 from sweet potato (I. batatas), 430 from I. trifida, 411 from I. triloba, 291 from I. nil, 226 from I. purpurea, 281 from I. cairica, and 277 from I. aquatic (Tables S2, S3). The number of MYB TFs in the seven investigated Ipomoea species varied. According to the total number of adjacent MYB repetitions, the identified MYB TFs were classed into five types: MYB-related (1R-MYB), R2R3-MYB (2R-MYB), 3R-MYB, 4R-MYB, and 5R-MYB (Tables S2, S3). Among these types, the MYB-related or R2R3-MYB was the most abundant in the seven Ipomoea species. At the same time, the percentages of 3R-MYB, 4R-MYB, and 5R-MYB were comparatively small and no 4R-MYB TFs were detected in I. nil, I. purpurea, I. cairica, and I. aquatic (Tables S2, S3). In the sweet potato (I. batatas), I. nil, I. cairica, and I. aquatic, the number of R2R3-MYB TFs was more than the MYB-related ones, whereas in I. trifida, I. triloba, and I. purpurea, the MYB-related TFs were more dominant than the R2R3-MYB ones, especially in I. purpurea; the proportion of MYB-related TFs (67.26%) was equivalent to 2.17 times of that of R2R3-MYB ones (Tables S2, S3).
3.2 Molecular weight, isoelectric point, and subcellular localization analysis of Ipomoea MYB proteins
The average length of the Ipomoea MYB TFs was 377.43 amino acids, with an average exon number of 4.41(Tables S2, S3). The average of the calculated molecular weight (MW) of the Ipomoea MYB TFs was 41.96 kDa, ranging from 8.66 to 219.26 kDa. In all of the seven Ipomoea species, the average length, the average exon number, and the average MW of the MYB genes were increased with the increase in the number of adjacent MYB repetitions, except for that of the R2R3-MYB genes which was smaller than that of MYB-related genes (Tables S2, S3). The predicted isoelectric point (pI) of the Ipomoea MYB TFs ranged from 4.15 to 11.23, with an average pI of 7.15. Moreover, the average pI of 4R-MYB and 5R-MYB TFs was larger than that of other types of MYB TFs in the seven Ipomoea species (Tables S2, S3). The grand average of hydropathicity of the Ipomoea MYB TFs was -0.70, ranging from -1.53 to 0.18 (Tables S2, S3). Subcellular localization analysis showed that the majority of the Ipomoea MYB TFs (>90%) were predicted to be localized in the nucleus, with a small set of them predicted to localize in other subcellular locations, such as chloroplast, cytosol, and mitochondrial (Tables S4, S5).
3.3 Conserved motif analysis of the Ipomoea MYB proteins
To understand the conserved domains of the Ipomoea MYB proteins, the Motif Elicitation (MEME) analysis was performed on each type of MYB TFs identified above. Twenty conserved motifs were found in the Ipomoea MYB-related TFs, R2R3-MYB TFs, 3R-MYB TFs, and the other (4R- and 5R-MYB) TFs, respectively (Figure S1). As shown in Figure 1, in addition to the number of adjacent MYB repetitions, the MYB domain of each type of MYB TFs varied. At the same time, many conserved amino acids were also detected, especially for the tryptophan residues (W) (Figure 1). In the R2 and R3 domains of the R2R3-MYB TFs, there were three conserved W in R2 and only two conserved W in R3; the first W of R3 was replaced by phenylalanine (F) in this study (Figure 1).
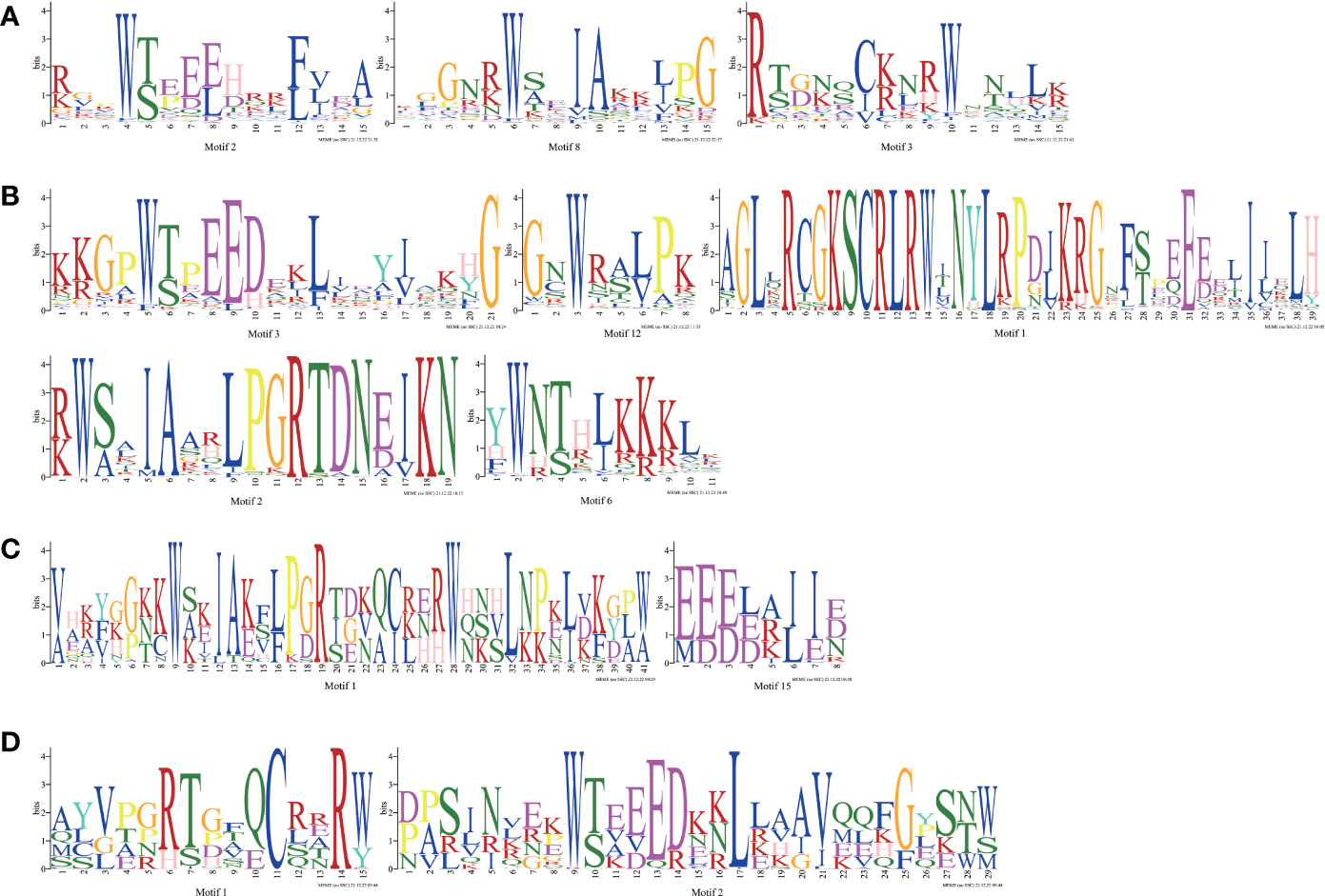
Figure 1 MYB repeats of the proteins in Ipomoea MYB TFs. (A) MYB repeats of Ipomoea MYB-related TFs; (B) MYB repeats of Ipomoea R2R3-MYB TFs; (C) MYB repeats of Ipomoea 3R-MYB TFs; (D) MYB repeats of Ipomoea 4R- and 5R-MYB TFs.
3.4 Phylogenetic analysis of Ipomoea MYB proteins
Phylogenetic trees were constructed with MYB-related TFs, R2R3-MYB TFs, 3R-MYB TFs, and the other (4R- and 5R-MYB) TFs of Ipomoea species and Arabidopsis, respectively. Based on the tree’s topology, the MYB-related TFs of Ipomoea species and Arabidopsis were classed into 27 subgroups, from A1 to A27 (Figure 2). The Arabidopsis MYB-related TFs dispersed in 10 of the 27 subgroups (i.e., A4, A12, A13, A14, A15, A20, A21, A22, A23, and A25) (Figure 2). According to the classification of Arabidopsis MYB-related TFs (Chen et al., 2006), we identified the CCA1-like subfamily (A4 and A13), R-R-type subfamily (A12), I-box-binding-like subfamily (A14 and A25), CPC-like subfamily (A15), TBP-like subfamily (A20, A21and A23), and TRF-like subfamily (A22) (Figure 2). The R2R3-MYB TFs of Ipomoea species and Arabidopsis were divided into 46 subgroups, from B1 to B46 (Figure 3). Based on the previous study of Arabidopsis R2R3-MYB TFs (Dubos et al., 2010), the S1 to S25 subfamilies and potential functions of the genes in each subfamily are indicated in Figure 3; for instance, B11 and B23 were indicated as S11 and S2, respectively, and the gene in the two subgroups might be involved in defense function (Figure 3). The 3R-MYB TFs were classed into 10 subgroups (C1 to C10). All of the 3R-MYB TFs of sweet potato, except for IbMYB275, were clustered together in subgroup C10 (Figure S2). The 3R-MYB TFs belonging to the same species tended to cluster together, for example, subgroups C3, C4, C7, and C10. In comparison, the genes clustered in the subgroups C2, C6, and C8 were always from different species (Figure S2). The results indicated that the members of Ipomoea 3R-MYB TFs might experience different evolutions after the species differentiation: some kept the initial copy number of their ancestors, whereas others increased their copies (Figure S2). The 4R- and 5R-MYB TFs of the seven Ipomoea species were divided into four subgroups, named D1 to D4 (Figure S3).
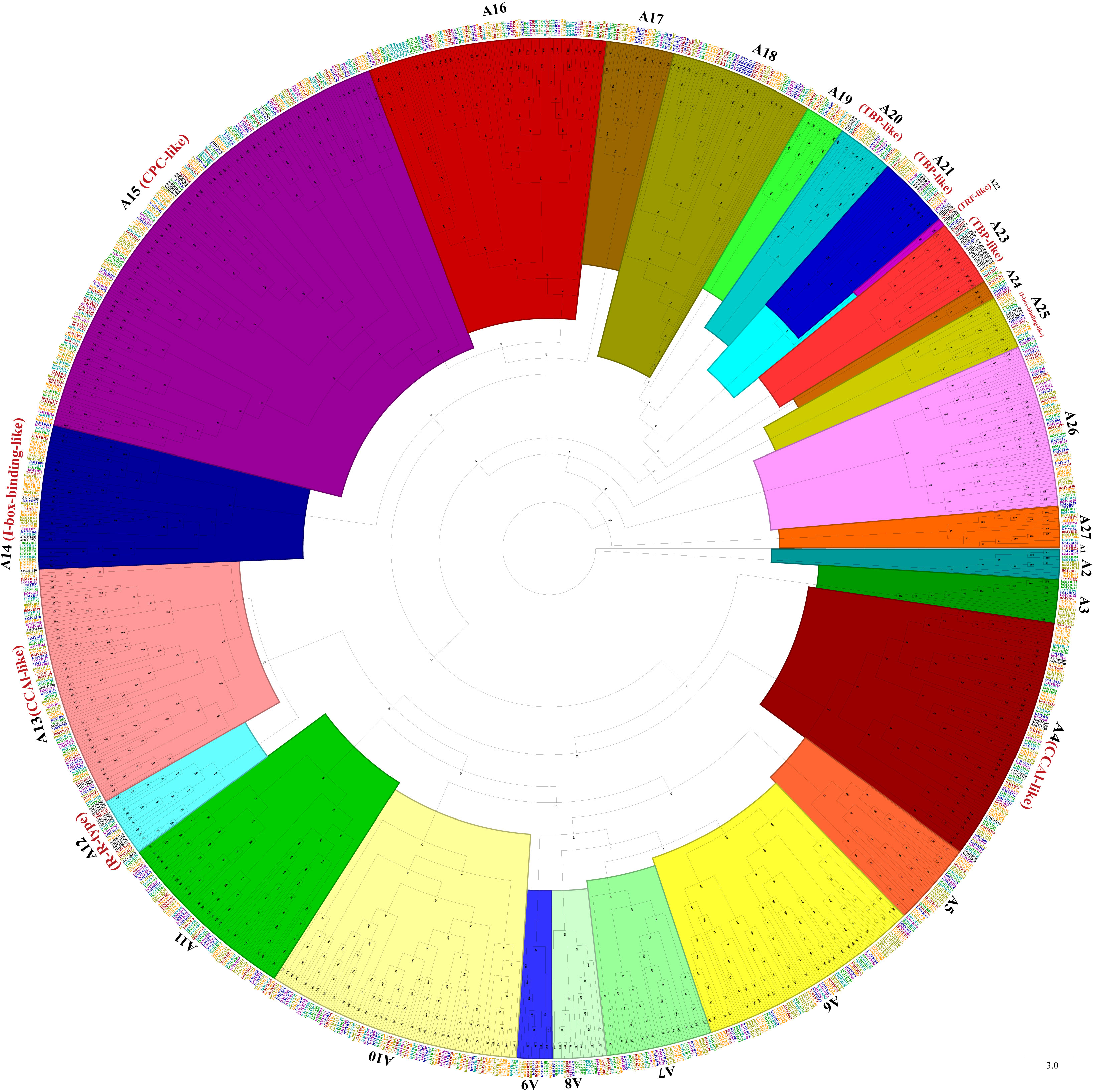
Figure 2 The phylogenetic tree of Ipomoea species and Arabidopsis MYB-related proteins. The proteins were grouped into 27 subgroups, each given a number (e.g., A1 to A27). The “CCA1-like,” “CPC-like,” “I-box-binding-like,” “R-R-type,” “TBP-like,” and “TRF-like” subfamilies were indicated with red name, respectively (Chen et al., 2006).
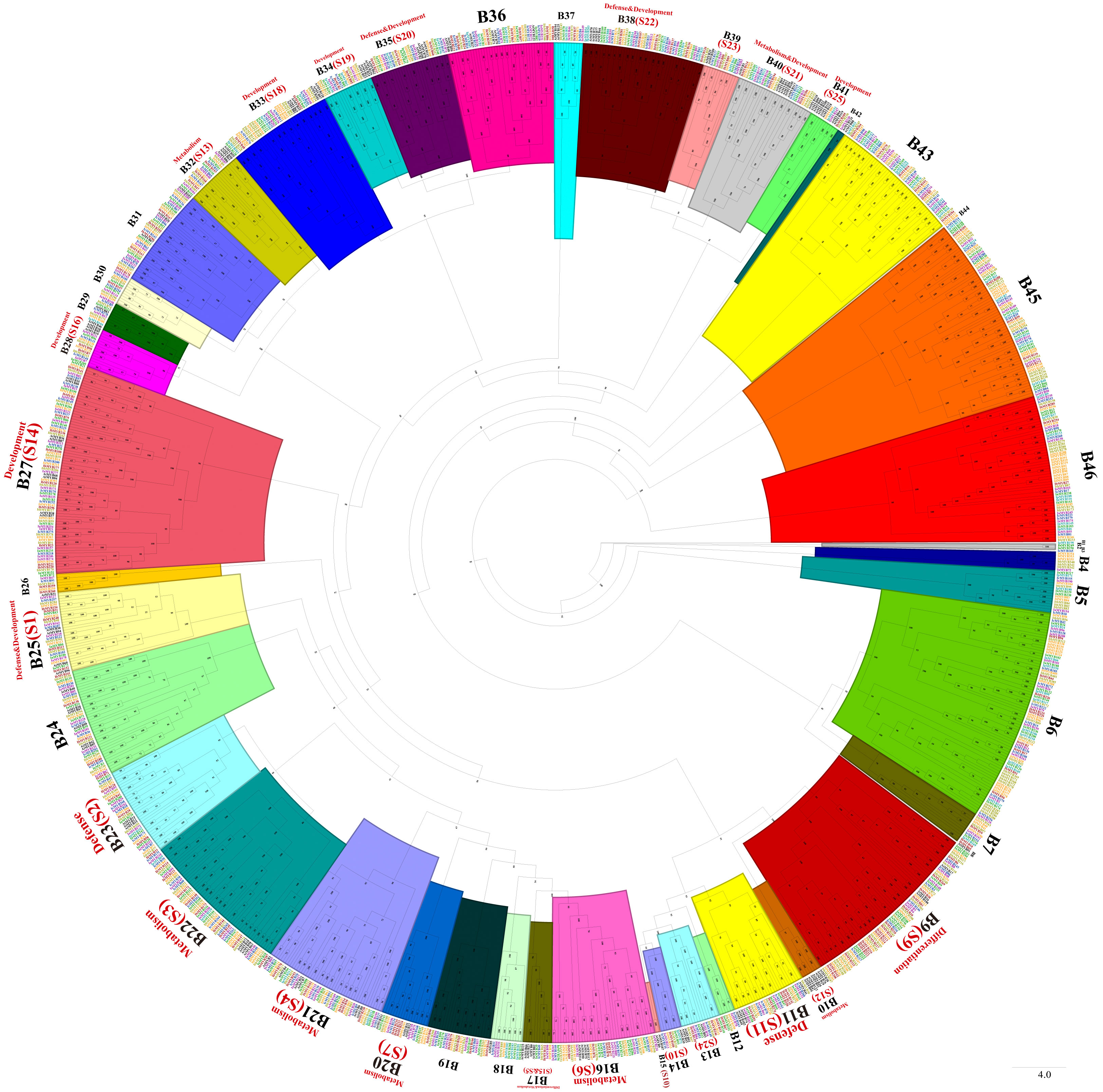
Figure 3 The phylogenetic tree of Ipomoea species and Arabidopsis R2R3-MYB proteins. The proteins were grouped into 46 subgroups, and each group has given a number (e.g., B1 to B46). S1–S25 indicated the Arabidopsis subfamily identified previously and were written in red words, as well as the prospective functions of the Arabidopsis MYB TFs in each subfamily (Dubos et al., 2010).
3.5 Gene structures and motif composition of MYB genes in Ipomoea species
As shown in Figure S4, the gene’s structural diversity might also be a reflection of the evolutional divergence among homologous MYB genes. In other words, the Ipomoea MYB genes clustered in the same subgroups shared a roughly similar exon/intron structure, exon number, and gene length (Figure S4). The MYB-related genes harbored more exons and longer sequence lengths than R2R3-MYB genes (Figure S4, Table S3). The distributions of motifs of MYB proteins in the same subgroup were also roughly similar, suggesting that the motif distributions might be related to functions (Figure S4). The motifs that consist of the core MYB domain were the most conservative: for the MYB-related genes, motif 2 was the most conserved, followed by motif 3, motif 1, and motif 8; for the R2R3-MYB genes, motif 3 was the most conserved, followed by motif 2, motif 6, and motif 12; 19 of the 23 3R-MYB genes contained motif 1 and motif 15; and all of the 4R- and 5R-MYB genes contained motif 1 and motif 2 (Figure S4, Table S6).
3.6 Chromosomal location and duplication analysis of the Ipomoea MYB genes
To understand the genomic distribution of the Ipomoea MYB genes, chromosomal location analysis was performed. As shown in Figure 4, the Ipomoea MYB genes were unevenly distributed across the chromosomes of the Ipomoea species (Figure 4). Duplication analysis showed that 106, 105, and 20 pairs of tandemly duplicated MYB genes were found in I. trifida, I. triloba, and I. nil, respectively. In contrast, no duplicated MYB genes were detected in sweet potato (I. batatas), I. purpurea, I. cairica, and I. aquatic (Figure 4, Table S7). Meanwhile, a total of six, one, one, four, three, three, and seven pairs of segmentally duplicated genes were found in sweet potato (I. batatas), I. trifida, I. triloba, I. nil, I. purpurea, I. cairica, and I. aquatic, respectively (Figure 4, Table S7). These results suggested that both segmental and tandem duplications have played essential roles in MYB gene expansion in I. trifida, I. triloba, and I. nil, and tandem duplications were predominant, whereas in sweet potato (I. batatas), I. purpurea, I. cairica, and I. aquatic, segmental duplications were predominant.

Figure 4 Distribution of MYB genes across the chromosomes of the seven Ipomoea species. (A) Distribution in sweet potato (I. batatas). (B) Distribution in (I.) trifida. (C) Distribution in (I.) triloba. (D) Distribution in (I.) nil. (E) Distribution in (I.) purpurea. (F) Distribution in (I.) cairica. (G) Distribution in (I.) aquatic. The red color indicates the tandemly duplicated MYB genes on the chromosomal positions; the green line connected green square frame indicated segmentally duplicated genes.
3.7 Syntenic analysis of MYB genes in the genomes of Ipomoea species
To determine the evolutionary mechanism of Ipomoea MYB genes, comparative synteny maps of the seven Ipomoea species were constructed (Figure 5). A total of 1,703 Ipomoea MYB genes that formed 4,050 ortholog pairs were detected in the seven Ipomoea species (Tables S8, S9). Of these genes, R2R3-MYB genes were the most (890), followed by MYB-relative (782), 3R-MYB (24), and 5R-MYB (7) genes; when referring to subfamilies, the A15 members were the most (115), followed by the A10 (77), A13 (71), A16 (67), B27 (66), A4 (66), B6 (61), and other subfamily (<60) members (Tables S8, S9). I. trifida and I. triloba harbored the most ortholog MYB gene pairs (248 pairs), followed by I. triloba and I. cairica (240 pairs), I. nil and I. cairica (223 pairs), and I. trifida, and I. cairica (221 pairs) (Table S8). The sweet potato showed more than 200 ortholog MYB gene pairs with I. triloba (214 pairs), I. cairica (214 pairs), and I. trifida (208 pairs) (Table S8). The ortholog MYB genes were distributed in all of the subfamilies except for A1, A22, and D1 (Table S8). In most cases (3,636 of 4,150, 87.61%), two of the ortholog MYB genes were from the same subfamily (Table S10). The A13 subfamily held the most MYB gene pairs (166 pairs), followed by A10 (161 pairs), A4 (161 pairs), A16 (156 pairs), B6 (145 pairs), and B45 (134 pairs) (Table S10). A total of 621 MYB genes (90 from sweet potato, 86 from I. trifida, 90 from I. triloba, 91 from I. nil, 87 from I. purpurea, 89 from I. cairica, and 88 from I. aquatic) can be traced to form ortholog gene pairs between any two of the seven Ipomea species (Figure 5, Table S11). Of the 621 MYB genes, the A4 subfamily members were the most abundant (40), followed by the A16 (36), B6 (36), A15 (33), A10 (30), B45 (29), B43 (24), A13 (23), and other (<20) subfamily members (Table S11).
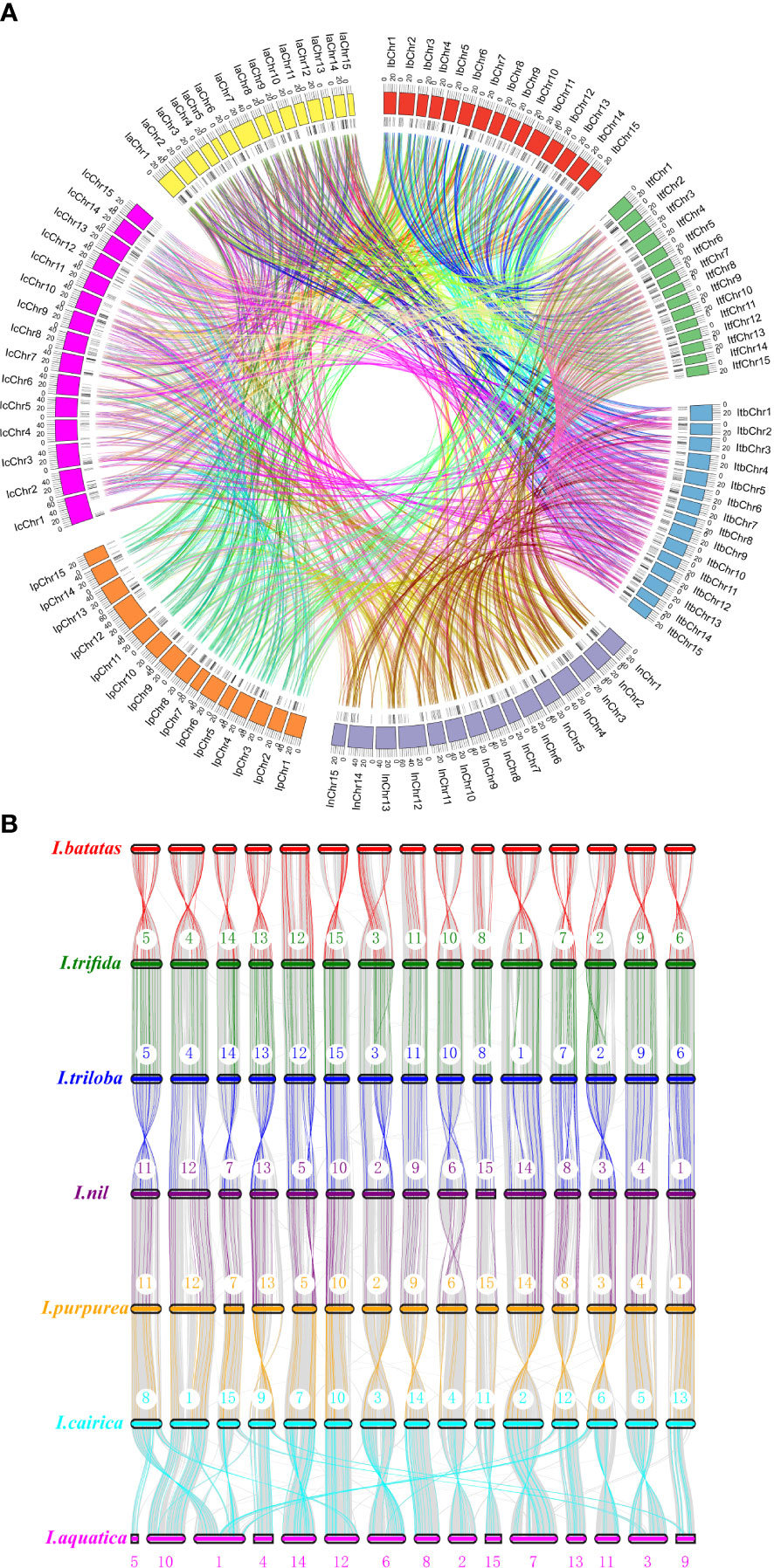
Figure 5 Syntenic analyses of MYB genes in the genomes of Ipomoea species. (A) Chromosomal distribution in the seven Ipomoea species. The outer circle represents the haploid chromosomes of sweet potato (I. batatas) (red), (I.) trifida (green), (I.) triloba (cornflower blue), (I.) nil (medium purple), (I.) purpurea (orange), (I.) cairica (magenta), and (I.) aquatic (yellow), respectively. The second circle (black) represents the matches of MYB genes with the genome of the Ipomoea species. Colorful lines show the collinear MYB gene pairs in the whole genome of the Ipomoea species. (B) Schematic representation of syntenic genes among sweet potato (I. batatas), (I) trifida, (I) triloba, (I) nil, (I) purpurea, (I) cairica, and (I) aquatic. The chromosomes of the seven Ipomoea species were reordered. Gray lines in the background indicate the collinear blocks within Ipomoea genomes, with MYB gene pairs highlighted in chromatic color. The black lines indicated synteny gene pairs between the seven Ipomea species.
3.8 Ka/Ks analysis of duplicated and syntenic MYB genes
To detect whether some MYB genes are under positive selection, Ka/Ks analysis was performed on duplicated and syntenic MYB genes within or between the seven Ipomoea species. Within the seven Ipomoea species, all of the segmentally duplicated genes (23 pairs) harbored a Ka/Ks ratio <1, suggesting that these genes underwent purifying (negative) evolutionary selection. Fifty-one pairs of tandemly duplicated genes’ Ka/Ks <1 underwent purifying (negative) evolutionary selection, and 21 pairs of tandemly duplicated genes possessed Ka/Ks ratio >1 and were subjected to positive selection (Table S12). Between the seven Ipomoea species, 99.5% (4,116 pairs) of the syntenic MYB genes possessed Ka/Ks ratio <1 and the other ones (0.5%, 22 pairs) possessed Ka/Ks ratio >1 (Table S12). These results suggest that the majority of duplicated and syntenic MYB genes were subjected to purifying selection inside duplicated genomic elements during speciation. In contrast, a smaller number of such genes were subjected to positive selection.
3.9 Cis-regulatory elements (CREs) in putative promoter regions of the Ipomoea MYB genes
A 1.5-kb upstream promoter region of Ipomoea MYB genes was scanned for CRE analysis. A lot of CREs related to plant growth and development (light response, cell cycle, endosperm expression, meristem expression, root-specific and seed-specific regulation, and zein metabolism regulation) and stress response (ABA response, anaerobic induction, defense and stress responsiveness, drought stress, flavonoid biosynthetic genes regulation, gibberellin responsiveness, SA responsiveness, low-temperature responsiveness, and MeJA response) were detected (Figure S5, Table S13). Of the CREs related to plant growth and development, the Box 4 motif was the most abundant, followed by the G-Box motif and GT1-motif, and all of them contributed to light response. Of the CREs related to stress response, ABRE motif (ABA response) was the most abundant, followed by ARE (anaerobic induction), TGACG-motif (MeJA response), MBS (drought stress), TCA-element (SA responsive), LTR (low-temperature responsiveness), TGA-element (anaerobic induction), and TC-rich repeats (defense and stress responsiveness) (Figure S5, Table S13).
The entire CREs shown in the head of Table S13 appeared in both MYB-related and R2R3-MYB promoter regions, with Box 4 being the most abundant, followed by G-Box, ABRE, and ARE (Table S14). However, some CREs (3-AF1-binding site, AAAC-motif, chs-Unit 1 m1, GATT-motif, motif I, RY-element, TGA-box, and RY-element were detected in 3R-MYB promoter regions and the AAAC-motif, AT1-motif, ATC-motif, ATCT-motif, CAG-motif, chs-CMA2a, chs-Unit 1 m1, GA-motif, Gap-box, GATT-motif, GTGGC-motif, LS7, MRE, MSA-like, motif I, RY-element, TGA-box, TC-rich repeats, and MBSI) were absent in 3R-, 4R-, and 5R-MYB promoter regions (Table S14). The CGTCA-motif, ABRE, ARE, LTR, and TGACG-motif were the most conserved in the Ipomoea MYB-related promoter regions, and they could be detected in all of the subfamilies of this type of genes (Table S14). Box 4 was the most conserved in the Ipomoea R2R3-MYB promoter regions, followed by TCT-motif, CGTCA-motif, ARE, and TGACG-motif, and they were absent in no more than three of the subfamilies of this type of genes (Table S14). Box 4 and ARE were the most conserved in the subfamilies of 3R-MYB, and chs-CMA1a, G-Box, and ABRE were the most conserved in the subfamilies of 4R- and 5R-MYB (Table S14). However, some CREs showed subfamily specificity; for example, the GATT motif was only detected in the A13, A14, A15, A18, B20, B22, and B37 subfamilies (Table S14). In each subfamily of the MYB promoter regions, the distribution of the CREs was not conservative as the gene-coding sequences; the distribution even showed gene specificity (Figure S6). The same distribution of the CREs occasionally appeared in tandem-duplicated genes; for example, ItbMYB210, ItbMYB211, and ItbMYB212 were tandem-duplicated genes detected in I. triloba, and they shared the same CRE distribution. However, the MYB genes in the same family may show similarity in the number and type of CREs; for instance, ItfMYB68, IbMYB39, and ItfMYB69 in the A4 subfamily, and ItbMYB45, IbMYB141, and ItfMYB42 in the B27 subfamily (Table S13; Figure S6).
3.10 Expression profiles of MYB genes in sweet potato
To explore MYB genes related to stress response, six transcriptome datasets referring to abiotic stresses (cold, drought, and salt) and biotic stresses (sweet potato stem nematodes, Ceratocystis fimbriata, and root-knot nematodes) were analyzed. A total of 101 MYB differentially expressed genes (DEGs) were detected in the transcriptome dataset of chilling injury at low-temperature storage, which was further classed into nine subclasses (1–1 to 1–9) based on their expression (Figure 6A). Compared with the control temperature (CT), the MYB genes in subclass 1-2 were upregulated at 13°C and downregulated at 4°C; the ones in subclass 1-3 were unchanged at 13°C and downregulated at 4°C; the ones in subclasses 1-7 and 1-8 were all unchanged at 13°C and upregulated at 4°C (Figure 6A). A total of 180 MYB DEGs were found in the drought stress transcriptome analysis, and they were classed into 11 subclasses (2-1 to 2-11) (Figure 6B). With the extension of drought time, the MYB genes in subclass 2-5 mainly maintained high expression in drought sense cultivar “Kokei 14,” whereas they were comparably downregulated in both “KT 1” and “I. triloba” (drought-tolerant types) (Figure 6B). A total of 102 MYB DEGs were found in the salt stress transcriptome analysis, and they were classed into seven subclasses (3-1 to 3-7) (Figure 6C). Compared with the control, the MYB genes in subclass 3-1 were upregulated in both “Xu 32” (salt sense cultivar) and “Xu 22” (salt-tolerant cultivar), whereas the MYB genes in subclass 3-4 were downregulated in the two investigated cultivars; compared with the salt-sensitive cultivar “Xu 32,” the MYB genes in subclass 3-5 were kept downregulated under control and salt treatment of “Xu 22”; conversely, the MYB genes in subclass 3-7 were mainly kept upregulated under control and salt treatment on “Xu 22” (Figure 6C).
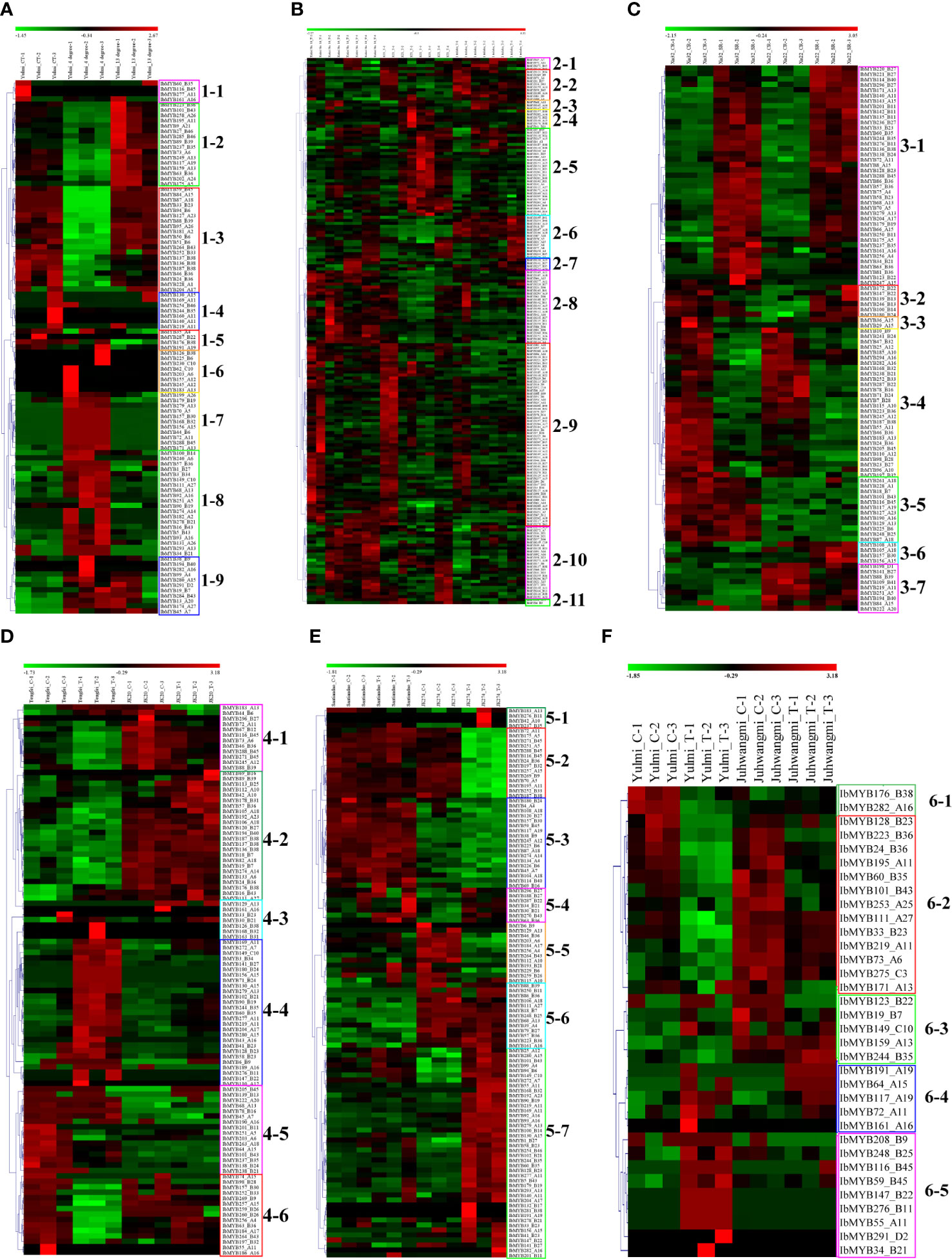
Figure 6 Heatmap of the expression profiles of sweet potato MYB differentially expressed genes (DEGs) in response to abiotic and biotic stresses. (A) MYB DEGs in “Yulmi” under control (CT), 4°C, and 13°C. (B) MYB DEGs in “Kokei 14,” “KT 1,” and “I. triloba” during different times of drought treatments. (C) MYB DEGs in “Xu 32” and “Xu 22” under control and salt treatments. CR, control; SR, treatment. (D) MYB DEGs in “Tengfei” and “JK20” under control and sweet potato stem nematode inoculation. C, control; T, treatment. (E) MYB DEGs in “Santiandao” and “JK274” under control and Ceratocystis fimbriata inoculation. C, control; T, treatment. (F) MYB DEGs in “Yulmi” and “Juhwangmi” under control and root-knot nematode inoculation. C, control; T, treatment. The letters after the MYB gene name indicated its phylogenetic subgroup.
A total of 110 MYB DEGs were found in the dataset of sweet potato stem nematode inoculation transcriptome analysis, and they were further classed into six subclasses (4-1 to 4-6) (Figure 6D). Compared with the sense cultivar “Tengfei,” the MYB genes in subclass 4-2 were mainly kept upregulated under control and sweet potato stem nematode inoculation of “JK20” (sweet potato line); compared to the control, the MYB genes in subclass 4-4 were upregulated in sweet potato stem nematode inoculation of “Tengfei,” whereas they remained downregulated in the same treatment of “JK20” (Figure 6D). There were 101 MYB DEGs found in the dataset of Ceratocystis fimbriata inoculation transcriptome analysis, and they were classed into seven subclasses (5-1 to 5-7) (Figure 6E). Compared with the control, the MYB genes in subclass 5-2 were downregulated in the Ceratocystis fimbriata-resistant line “JK274” after inoculation, whereas they kept a high expression in the sense cultivar “Santiandao”; the MYB genes in subclass 5-7 were upregulated in the Ceratocystis fimbriata-resistant line “JK274” after inoculation, whereas they kept a low expression in the sensitive cultivar “Santiandao”; the MYB genes in subclass 5-3 kept a high expression in “Santiandao” and a low expression in “JK274”; and the MYB genes in subclass 5-6 kept a low expression in “Santiandao” and a high expression in “JK274” (Figure 6E). There were 34 MYB DEGs found in the dataset of root-knot nematode inoculation transcriptome analysis, and they were classed into five subclasses (6-1 to 6-5) (Figure 6F).
During the drought and salt treatments of sweet potato, the DEGs belonging to the A15, B27, and B36 phylogenetic subfamily were the most, whereas in cold stress, the DEGs belonging to the A11, A13, B38, B43, B6, and B36 subfamilies were the most (Figures 6 ,7). During the biotic stress treatments, the DEGs belonging to the A11, A13, A16, and B23 subfamilies were abundant (Figures 6, 7). Both the abiotic and biotic stress treatments detected DEGs in the B11, B23, B25, B35, and B38 subfamilies, the members of which were considered to have the function of defense (Figures 3, 6, 7). To compare the transcriptional changes of various MYB subfamilies between species, the I. trifida and I. triloba MYB genes that responded to abiotic stresses were analyzed. During the cold, drought, and salt treatments, 365, 359, and 366 I. trifida MYB DEGs and 370, 369, and 368 I. triloba MYB DEGs were found, respectively (Tables S15, S16). When comparing the results with that of sweet potato, we discovered that the MYB DEGs of the three Ipomoea species dispersed in 68 phylogenetic subfamilies (Figure 7). Of these subfamilies, 41, 45, and 38 were common in the three Ipomoea species and contained DEG response to cold, drought, and salt treatments, respectively (Figure 7, Table S17). Further analysis revealed that the MYB DEG response to abiotic stresses generally belonged to MYB-related and R2R3-MYB gene types, involving 28 phylogenetic subfamilies, such as A4, A5, A13, A15, B23, B35, and B38 (Figure 7, Table S18).
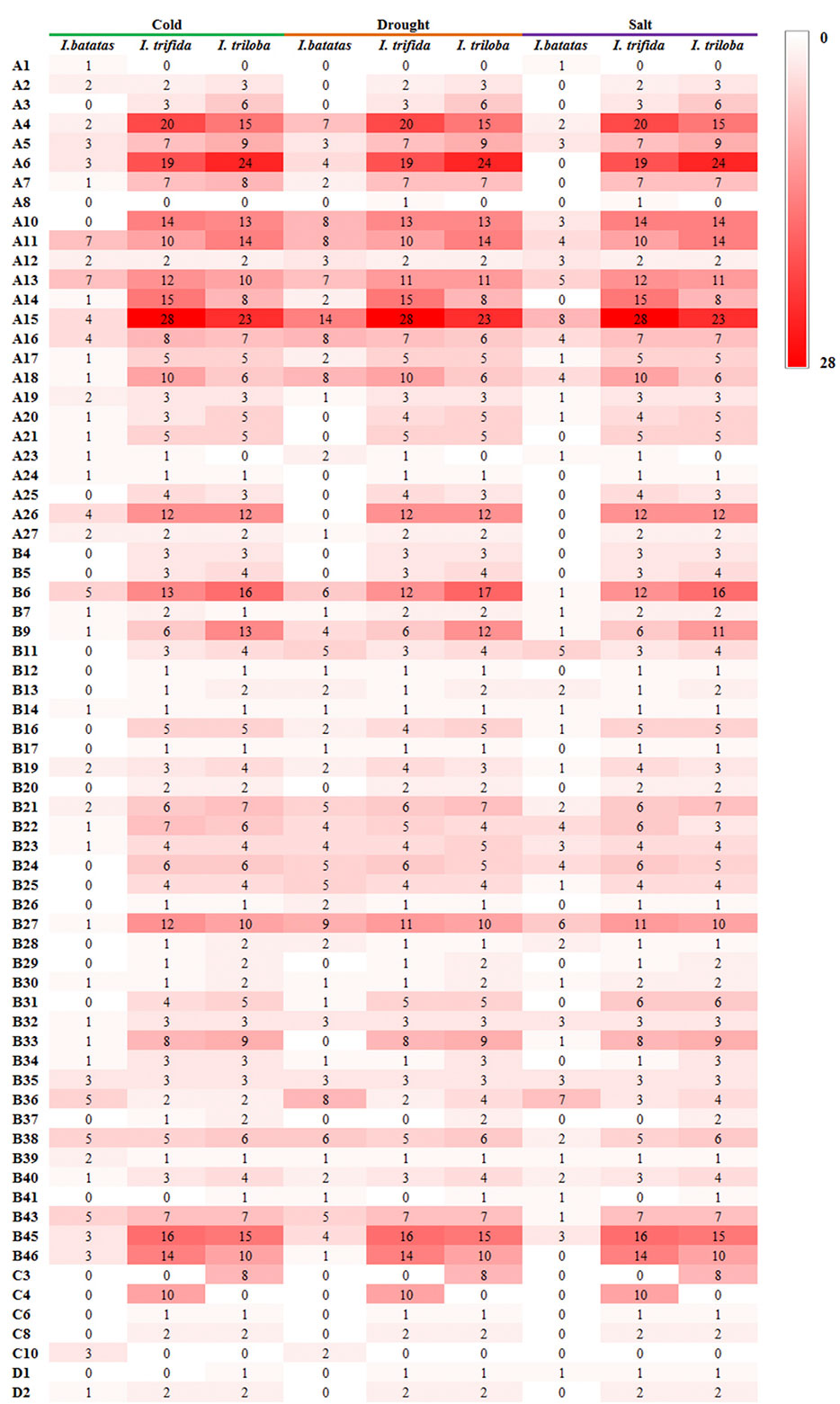
Figure 7 Comparison of subfamily-based gene numbers under abiotic stresses in the sweet potato, I. trifida, and I. triloba.
3.11 Expression analysis of sweet potato MYB genes by quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Based on transcriptome results, IbMYB68, IbMYB237, IbMYB39, IbMYB128, IbMYB127, IbMYB201, IbMYB82, IbMYB187, IbMYB4, and IbMYB18 were selected for further analysis using qRT-PCR. Under cold treatments, compared with the control condition (0 h), the transcripts of IbMYB68 in “JK328” were upregulated and peaked at 12 h after treatments, whereas there was no obvious change in “Xu 32”; the transcripts of IbMYB237 in both “Xu 32” and “JK328” were upregulated and peaked at 6 and 24 h after treatments, respectively (Figure 7). Under drought treatments, compared with the control condition (0 h), the transcripts of IbMYB39 and IbMYB128 in “Xu 32” were upregulated and peaked at 12 and 4 h after treatments, respectively, whereas their transcripts showed no obvious change in “JK328” (Figure 7). Under salt treatments, compared with the control condition (0 h), the transcripts of IbMYB127 in “Xu 32” were upregulated and peaked at 12 h after treatments, whereas the transcripts of IbMYB127 in “JK328” were downregulated and decreased to valley at 12 h after treatments; the transcripts of IbMYB201 in both “Xu 32” and “JK328” were upregulated and peaked at 6 and 12 h after treatments, respectively (Figure 7). Under sweet potato stem nematode infection treatments, compared with the control condition (0 h), the transcripts of IbMYB82 in both “Tengfei” and “JK20” were upregulated and decreased to the valley at 2 days and 12 h after treatments, respectively; the transcripts of IbMYB187 in “Tengfei” were downregulated and peaked at 1 day after treatments, whereas the transcripts of IbMYB187 in “JK20” were upregulated and peaked at 12 h after treatments (Figure 7). Under Ceratocystis fimbriata inoculation treatments, compared with the control condition (0 h), the transcripts of IbMYB4 and IbMYB18 in “JK274” were upregulated and peaked at 2 days and 1 day hours after treatments, respectively, whereas their transcripts showed no obvious changing in “JK274” (Figure 7).
4 Discussion
The MYB transcription factor gene family is one of plants’ most prominent transcription factor families. It is involved in various essential life processes, including plant growth (Zhao et al., 2019), development (Liu et al., 2021; Yu et al., 2021), biosynthesis (Sinaga et al., 2021), and defense responses (Dubos et al., 2010; Lv et al., 2021; Hawku et al., 2022). Therefore, this gene family attracted more and more attention from various plant researchers and has been extensively investigated in many plant species, such as Arabidopsis (Chen et al., 2006), rice (Chen et al., 2006), soybean (Du et al., 2012b), maize (Du et al., 2012a), and potato (Li et al., 2021c). The Ipomoea contains 600–700 species, many of which are of considerable importance as medicinal or ornamental plants (Nimmakayala et al., 2011). The sweet potato is a highly heterozygous hexaploid (2n = 6x = 90) with a large, complex genome approximately 2–3 Gb in size (Ozias-Akins and Jarret, 1994). It is the world’s seventh most important food crop by production quantity (https://www.fao.org/faostat/, accessed 2022 November). I. trifida and I. triloba are two diploid species closely related to the hexaploid sweet potato, with an estimated genome size of 526.4 and 495.9 Mb, respectively (Wu et al., 2018). Due to their smaller genome size and ploidy, they were generally used as model plants to facilitate sweet potato breeding. I. nil is an annual climbing herb-producing blue flower capable of self-pollination (Hoshino et al., 2016). With an estimated genome size of 750 Mb, it has been a model plant for studying photoperiodic flowering and flower coloration. I. purpurea, with an estimated haploid genome size of 814 Mb, is a common agricultural weed exhibiting varying levels of herbicide resistance (Gupta et al., 2021). I. cairica is a perennial creeper introduced as a garden ornamental, and it has an estimated haploid genome size of 730 Mb (Jiang et al., 2022). I. aquatica, with an estimated genome size of 550.03 Mb, is usually called water spinach (Hao et al., 2021). It is one of the most popular green leafy vegetables, with aquatic and terrestrial characteristics (Hao et al., 2021). However, little is known about the MYB gene family in Ipomoea species.
In the present study, a total of 2,212 MYB genes were identified from the seven Ipomoea species, and the method used to identify the Ipomoea MYB genes was reliable and has been reported by previous studies (Zhou et al., 2020; Li et al., 2021c; Wang et al., 2022). Significant variations in MYB family members were observed in our analyzed Ipomoea species. There were 296, 430, 411, 291, 226, 281, and 277 MYB genes identified from sweet potato (I. batatas), I. trifida, I. triloba, I. nil, I. purpurea, I. cairica, and I. aquatic, respectively (Tables S1, S2). The number of MYB TFs in the seven investigated Ipomoea species differed, suggesting that MYBs in different Ipomoea plants have expanded to different degrees (Cao et al., 2021). By comparing the genome size and the number of identified MYB genes in this study, it is found that the number of MYB genes in each Ipomoea species was not highly correlated with the genome size (Yang et al., 2019; Liu et al., 2020a). Moreover, it might also correlate with the quality of the assembled Ipomoea genomes, and the MYB gene number will change along with the improvement of Ipomoea genome quality (Yang et al., 2019; Liu et al., 2020a). In various plants, the number of MYB genes is different; for instance, 198 MYBs were identified in Arabidopsis (Chen et al., 2006), 253 in Hedychium coronarium (Abbas et al., 2021), 235 in Capsicum spp. (Arce-Rodríguez et al., 2021), 174 in M. rubra (Cao et al., 2021), and 155 in Petunia axillaris (Chen et al., 2021). A similar phenomenon has also been reported in other closely related species, for instance, S. lycopersicum (127) and S. tuberosum (217) (Li et al., 2016; Li et al., 2021c), M. acuminate (305), and M. balbisiana (251) (Tan et al., 2020). The identified Ipomoea MYB TFs were classed into five types: MYB-related (1R-MYB), R2R3-MYB (2R-MYB), 3R-MYB, 4R-MYB, and 5R-MYB. Among these types, the MYB-related or R2R3-MYB was the largest in Ipomoea species, and they contain the highest quantity of genes within the MYB family. The R2R3-MYB was usually the largest type in green plants (Martin and Paz-Ares, 1997; Abbas et al., 2021; Arce-Rodríguez et al., 2021; Wang et al., 2021b; Li et al., 2021c), whereas the MYB-related type was also reported as the largest subfamily in some species (Mmadi et al., 2017; Pu et al., 2020).
In order to investigate the evolutionary relationships of Ipomoea MYB genes, a systematic analysis was performed on MYB-related TFs, R2R3-MYB TFs, 3R-MYB TFs, and the other (4R- and 5R-MYB) TFs of Ipomoea species and Arabidopsis, and four trees of them were constructed, respectively. Based on the topology, the trees were further divided into several subgroups (Figures 2, 3, Figures S2, S3). Supposed that genes belonging to the same branch may experience a standard evolutionary process and possess a conserved function (Li et al., 2021c), this study compared the MYB genes of Ipomoea species and Arabidopsis. Subfamilies containing the MYB members from the Ipomoea species, Arabidopsis, and some species-specific subfamilies were detected. Similar results have been reported by phylogenetic analysis of other species (Wang et al., 2021b; Li et al., 2021c). The phylogenetic trees of MYB protein constructed in this study would help to predict the function of Ipomoea MYB genes with Arabidopsis MYB genes as a reference (Chen et al., 2006; Dubos et al., 2010).
The Ipomoea MYB genes identified in this study were different in amino acid sequence length, molecular weight, and isoelectric point, reflecting the complexity and functional diversity of the Ipomoea MYB genes to some degrees centigrade (Tables S2-5). The structure analysis of the Ipomoea MYB genes was consistent with phylogenetic analysis. Most genes in the same subgroup exhibited similar exon–intron structures (Figure S4). The Ipomoea MYB proteins within the same subgroup showed similar motif compositions, whereas the ones in different subgroups were of high variance (Figure S4). This is consistent with previous reports of other species, such as G. hirsutum (Salih et al., 2016), Prunus salicina (Liu et al., 2020a), and Helianthus annuus (Li et al., 2020). It was also found that the motifs of the core MYB domain were the most conservative (Figure 1, Figure S4). The Ipomoea MYB genes with a close evolutionary relationship might have similar functions (Wang et al., 2021b). Conserved motifs within the same TF family, especially the ones in the same subfamilies, may play essential roles in protein-specific functions (Chen et al., 2021).
The chromosomal location showed that the Ipomoea MYB genes were unevenly distributed across the chromosomes of the Ipomoea species (Figure 4). The uneven distributions of MYB genes have also been detected in chromosomes of other species, such as Jatropha curcas (Zhou et al., 2015), M. rubra (Cao et al., 2021), and C. equisetifolia (Wang et al., 2021b). The gene duplication events, segmental and tandem duplication, played essential roles in gene family expansion and distribution of genes in plants (Cannon et al., 2004; Kong et al., 2007; Jiang et al., 2013). In the present study, segmental and tandem duplications contributed to the MYB gene expansion in I. trifida, I. triloba, and I. nil; tandem duplications were predominant, whereas in sweet potato (I. batatas), I. purpurea, I. cairica, and I. aquatic, segmental duplications were predominant (Figure 4, Table S7). The predominant types of duplications might be species-specific performance, for instance, in H. coronarium (Abbas et al., 2021), Prunus salicina (Liu et al., 2020a), and Helianthus annuus (Li et al., 2020), segmental duplications were dominant, whereas in watermelon (Qing et al., 2018) and palm (Zhou et al., 2020), tandem duplications were predominant.
To further explore the evolutionary relationships of Ipomoea MYB genes, the syntenic maps of the seven Ipomoea species were constructed and compared. There were 1,703 Ipomoea MYB genes that formed 4,050 ortholog pairs detected in the seven Ipomoea species (Tables S8, S9). The results showed that many MYB genes from Ipomoea species presented high levels of collinearity and suggested that these MYB genes probably come from a common ancestor (Liu et al., 2020a; Abbas et al., 2021). It was found that some MYB genes contained more than one counterpart in the Ipomoea species, suggesting that the MYB gene family might undergo different amplification in Ipomoea genomes (Liu et al., 2020a). The orthologous events occurred in all phylogenetic subfamilies except for the A1, A22, and D1 (Table S8). The A1 and A22 subfamily contained only one Ipomoea MYB gene (IbMYB228 and IbMYB290) (Figure 2); the D1 subfamily consisted of three MYB members (IbMYB198, ItfMYB414, and ItbMYB404) (Figure S3). In most cases, the two MYB genes of orthologous pairs were generally from the same subfamily. The A13 subfamily held the most orthologous members, followed by A10, A4, A16, B6, and B45. A total of 621 Ipomoea MYB genes could form orthologous gene pairs between any two of the seven Ipomea species (Figure 5, Table S11).
Ka/Ks analysis of duplicated and syntenic MYB genes within or between the seven Ipomoea species showed that most duplicated and syntenic MYB genes were subjected to purifying selection inside duplicated genomic elements during speciation. In contrast, fewer such genes were subjected to positive selection (Table S12). These results indicated that Ipomoea MYB genes underwent a robust purifying selection during evolution with slight variation after duplication (Zhou et al., 2020). The phenomenon of the high proportion of purifying selection and low proportion of positive selection has been reported in previous MYB gene family studies, such as G. hirsutum (Salih et al., 2016), Elaeis guineensis (Zhou et al., 2020), and H. coronarium (Abbas et al., 2021).
The CREs in promoters of genes are usually involved in transcription initiation and regulation (Ho and Geisler, 2019; Mulat and Sinha, 2021). The present study scanned a 1.5-kb upstream promoter region of Ipomoea MYB genes for cis-regulatory elements analysis. Various CREs related to plant growth, development, and stress response were detected (Figure S5, Table S13). CREs related to stress response, such as ABRE, ARE, TGACG-motif, MBS, TCA-element, LTR, TGA-element, and TC-rich repeats, were abundant in the promoters of the Ipomoea MYB gene. Similar results have been reported by other species, such as Medicago sativa (Zhou et al., 2019), H. coronarium (Abbas et al., 2021), and Petunia axillaris (Chen et al., 2021). It was also found that some CREs were detected in MYB-related and R2R3-MYB promoter regions, whereas they were absent in 3R-, 4R-, and 5R-MYB promoter regions, suggesting that different transcription initiation or regulation might occur between them (Ho and Geisler, 2019; Mulat and Sinha, 2021). In the subfamilies of MYB-related and R2R3-MYB genes, the CGTCA-motif, ABRE, ARE, and TGACG-motif were the most conserved. Moreover, some subfamily-specific CREs were also detected (Table S14). The distribution of the CREs was not conservative as the gene-coding sequences in each subfamily, and the non-conservative distribution of CREs has been reported by other TF gene families, such as NAC (Diao et al., 2018), WRKY (Zheng et al., 2021), and HSP20 (Cui et al., 2021). In this study, the exact distribution of the CREs occasionally appeared in tandemly duplicated genes. However, the MYB genes in the same family may show similarities in the number and type of CREs (Table S13; Figure S6).
Focused on the stress resistance of sweet potatoes, six transcriptome datasets referring to abiotic and biotic stresses were analyzed, and DEGs in each transcriptome dataset were detected. For comparative analysis, the I. trifida and I. triloba MYB genes that responded to abiotic stresses were also detected and analyzed in this study (Figure 7). The results showed that the Ipomoea MYB DEGs’ response to abiotic stresses usually belonged to MYB-related and R2R3-MYB types and was referred to various phylogenetic subfamilies (Figure 7, Table S18). In these subfamilies, some were considered as the defense function subfamilies with Arabidopsis MYB members as the reference (Chen et al., 2006), such as B11, B23, B25, B35, and B38 (Figure 3). During the abiotic stress treatments in the sweet potato, I. trifida and I. triloba, 28 phylogenetic subfamilies contained the MYB members that responded in all the abiotic stress treatments of the three Ipomoea species (Figure 7, Table S18). Based on the results of transcriptome datasets analysis, IbMYB68, IbMYB237, IbMYB39, IbMYB128, IbMYB127, IbMYB201, IbMYB82, IbMYB187, IbMYB4, and IbMYB18 were selected for further analyzing using qRT-PCR (Figure 8). The results of qRT-PCR of the selected IbMYB genes were consistent with the transcriptome analysis. IbMYB68 and IbMYB237 responded to cold treatments, IbMYB39 and IbMYB128 responded to drought treatments, IbMYB127 and IbMYB201 responded to salt treatments, IbMYB82 and IbMYB187 responded to sweet potato stem nematode infection, and IbMYB4 and IbMYB18 responded to Ceratocystis fimbriata infection. These results indicated that the IbMYB genes were essential in biotic and abiotic stress responses.
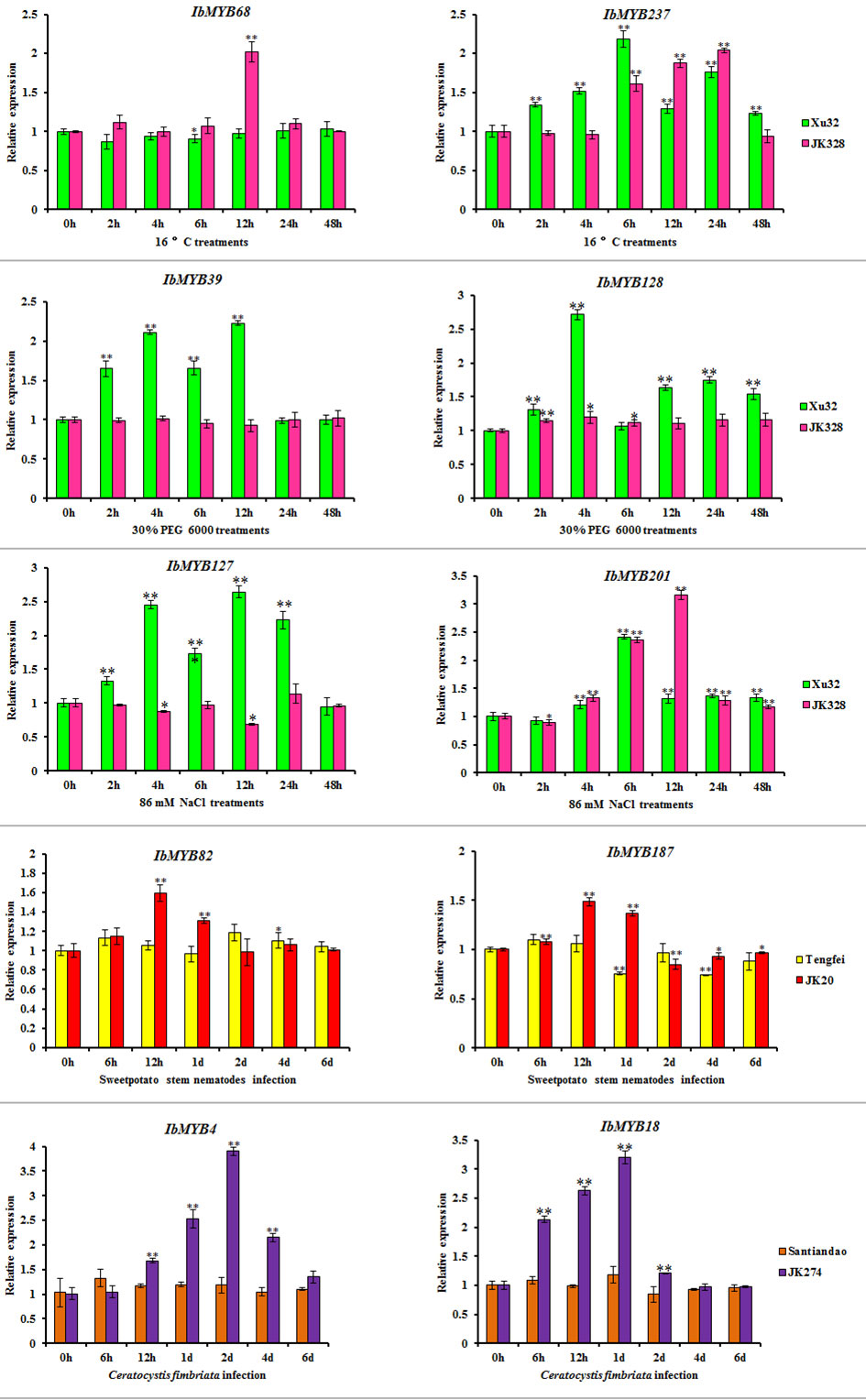
Figure 8 Expression analyses of IbMYB genes in sweet potato cultivars or lines. Denoted the significance of expression levels compared with control were ∗<0.05, ∗∗<0.01. h, hours; d, day(s).
5 Conclusions
This study is the first to systematically analyze the MYB gene family in Ipomoea species. There were 296, 430, 411, 291, 226, 281, and 277 MYB genes were identified from sweet potato (I. batatas), I. trifida, I. triloba, I. nil, I. purpurea, I. cairica, and I. aquatic, respectively. The identified MYB genes were classified into five types and were subjected to molecular weight (MW), isoelectric point (pI), and subcellular localization analysis. Based on the topology, phylogenetic analysis classed the Ipomoea MYB genes into several subgroups using the Arabidopsis MYB genes as a reference. Gene structure, motif organization, duplicated and syntenic analyses, and cis-regulatory element investigation were further performed. Focus on sweet potato, six transcriptome datasets referring to different abiotic and biotic stresses were analyzed and DEGs related to stress responses were detected. Based on the expression profiles of IbMYBs, 10 of the detected DEGs were selected for qRT-PCR analysis under five abiotic and biotic stress treatments. The results were consistent with the transcriptome analysis. This study may help to study the evolution of MYB genes in the Ipomoea species genomes and provide helpful information for the resistance breeding of sweet potatoes.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
Design of the study: ZS and LW; identification of MYB genes in the seven Ipomoea species: ZS and LW; molecular weight (MW) analysis: ZS, LW, and ZJ; isoelectric point (pI) analysis: ZS, LW, and MZ; subcellular localization analysis: ZS, ZJ, and YQ; motif analysis: ZS and YQ; gene structure analysis: ZS, LW, and ZJ; phylogenetic analysis: ZS, LW, and ZJ; chromosome location: ZS, KZ, and ZJ; duplication pattern analysis: ZS, LW, MZ, and ZJ; syntenic analysis: ZS, LW, MZ, and KZ; Ka/Ks analysis: ZS, KZ, MZ, and ZJ; expression profile analysis: ZS, KZ, MZ, and YQ; qRT-PCR analysis: ZS, LW, KZ, and YQ; manuscript preparation: ZS, LW, ZJ, MZ, KZ, and YQ. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Department of Science and Technology of Hebei Province (grant number: 19226335D) and Science Research Foundation of Hebei Normal University of Science and Technology (grant number: 2021JK01).
Acknowledgments
We would like to thank all the editors and the reviewers for their efforts and their valuable comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1155018/full#supplementary-material
Supplementary Figure 1 | Sequence logos for the 20 conserved motifs of the MYB-related, R2R3-MYB, 3R-MYB, and 4R- and 5R-MYB proteins in the seven Ipomoea species, respectively.
Supplementary Figure 2 | The phylogenetic tree of Ipomoea species and Arabidopsis 3R-MYB proteins. The proteins were grouped into 10 subgroups and each group has given a number (C1 to C10).
Supplementary Figure 3 | The phylogenetic tree of Ipomoea species and Arabidopsis 4R- and 5R-MYB proteins.
Supplementary Figure 4 | Motif and gene structure analyses of the MYB genes in the seven Ipomoea species. (A) Motif and gene structure analyses of the MYB-related genes in the seven Ipomoea species. (B) Motif and gene structure analyses of the R2R3-MYB genes in the seven Ipomoea species. (C) Motif and gene structure analyses of the 3R-MYB genes in the seven Ipomoea species. (D) Motif and gene structure analyses of the 4R- and 5R-MYB genes in the seven Ipomoea species.
Supplementary Figure 5 | The summary heatmap of cis-elements involved in plant growth and development and stress responses in Ipomoea MYB promoters.
Supplementary Figure 6 | Cis-elements in the promoter region of MYB genes in Ipomoea species. (A) Cis-elements in MYB-related genes promoter; (B) Cis-elements in R2R3-MYB genes promoter; (C) Cis-elements in 3R-MYB genes promoter; (D) Cis-elements in 4R- and 5R-MYB genes promoter.
References
Abbas, F., Ke, Y., Zhou, Y., Yu, Y., Waseem, M., Ashraf, U., et al. (2021). Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 12, 623742. doi: 10.3389/fpls.2021.623742
Alabadı́, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P., Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the arabidopsis circadian clock. Science 293 (5531), 880–883. doi: 10.1126/science.106132
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anisimova, M., Gil, M., Dufayard, J. F., Dessimoz, C., Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60 (5), 685–699. doi: 10.1093/sysbio/syr041
Arce-Rodríguez, M. L., Martínez, O., Ochoa-Alejo, N. (2021). Genome-wide identification and analysis of the MYB transcription factor gene family in chili pepper (Capsicum spp.). Int. J. Mol. Sci. 22 (5), 2229. doi: 10.3390/ijms22052229
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (Web Server issue), W202–W208. doi: 10.1093/nar/gkp335
Bovell-Benjamin, A. C. (2007). Sweet potato: a review of its past, present, and future role in human nutrition. Adv. Food Nutr. Res. 52, 1–59. doi: 10.1016/S1043-4526(06)52001-7
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Sci. 12, 1244. doi: 10.3389/fpls.2021.691384
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., He, W., Guo, X., Pan, J. (2021). Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia. Int. J. Mol. Sci. 22 (9), 4838. doi: 10.3390/ijms22094838
Chen, Y., Yang, X., He, K., Liu, M., Li, I., Gao, Z., et al (2006) The MYB transcription factor superfamily of arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family Plant Mol. Biol. 60 (1), 107–124.
Cheng, Y.-J., Kim, M.-D., Deng, X.-P., Kwak, S.-S., Chen, W. (2013). Enhanced salt stress tolerance in transgenic potato plants expressing IbMYB1, a sweet potato transcription factor. J. Microbiol. Biotechnol. 23 (12), 1737–1746. doi: 10.4014/jmb.1307.07024
Chezem, W. R., Clay, N. K. (2016). Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131, 26–43. doi: 10.1016/j.phytochem.2016.08.006
Chu, H., Jeong, J. C., Kim, W. J., Chung, D. M., Jeon, H. K., Ahn, Y. O., et al. (2013). Expression of the sweetpotato R2R3-type IbMYB1a gene induces anthocyanin accumulation in arabidopsis. Physiologia Plantarum 148 (2), 189–199. doi: 10.1111/j.1399-3054.2012.01706.x
Cui, F., Taier, G., Wang, X., Wang, K. (2021). Genome-wide analysis of the HSP20 gene family and expression patterns of HSP20 genes in response to abiotic stresses in Cynodon transvaalensis. Front. Genet. 12, 732812. doi: 10.3389/fgene.2021.732812
Czemmel, S., Heppel, S. C., Bogs, J. (2012). R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249 (2), 109–118. doi: 10.1007/s00709-012-0380-z
Dai, X., Xu, Y., Ma, Q., Xu, W., Wang, T., Xue, Y., et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic arabidopsis. Plant Physiol. 143 (4), 1739–1751. doi: 10.1104/pp.106.094532
Diao, W., Snyder, J. C., Wang, S., Liu, J., Pan, B., Guo, G., et al. (2018). Genome-wide analyses of the NAC transcription factor gene family in pepper (Capsicum annuum l.): chromosome location, phylogeny, structure, expression patterns, cis-elements in the promoter, and interaction network. Int. J. Mol. Sci. 19 (4), 1028. doi: 10.3390/ijms19041028
Dong, J., Cao, L., Zhang, X., Zhang, W., Yang, T., Zhang, J., et al. (2021). An R2R3-MYB transcription factor RmMYB108 responds to chilling stress of rosa multiflora and conferred cold tolerance of arabidopsis. Front. Plant Sci. 12, 696919. doi: 10.3389/fpls.2021.696919
Du, H., Feng, B.-R., Yang, S.-S., Huang, Y.-B., Tang, Y.-X. (2012a). The R2R3-MYB transcription factor gene family in maize. PloS One 7 (6), e37463. doi: 10.1371/journal.pone.0037463
Du, B., Liu, H., Dong, K., Wang, Y., Zhang, Y. (2022). Over-expression of an R2R3 MYB gene, MdMYB108L, enhances tolerance to salt stress in transgenic plants. Int. J. Mol. Sci. 23 (16), 9428. doi: 10.3390/ijms23169428
Du, H., Yang, S. S., Liang, Z., Feng, B. R., Liu, L., Huang, Y. B., et al. (2012b). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12 (1), 1–22. doi: 10.1186/1471-2229-12-106
Du, H., Zhang, L., Liu, L., Tang, X.-F., Yang, W.-J., Wu, Y.-M., et al. (2009). Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 74 (1), 1–11. doi: 10.1186/1471-2229-12-106
Dubos, C., Le Gourrierec, J., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55 (6), 940–953. doi: 10.1111/j.1365-313X.2008.03564.x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi: 10.1016/j.tplants.2010.06.005
Frampton, J. (2004). Myb transcription factors: Their role in growth, differentiation and disease (Netherlands: Springer).
Gao, S., Yu, B., Yuan, L., Zhai, H., He, S.-z., Liu, Q.-c. (2011). ). production of transgenic sweetpotato plants resistant to stem nematodes using oryzacystatin-I gene. Scientia Hortic. 128 (4), 408–414. doi: 10.1016/j.scienta.2011.02.015
Gigolashvili, T., Berger, B., Flügge, U.-I. (2009). Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem. Rev. 8 (1), 3–13. doi: 10.1007/s11101-008-9112-6
Gu, K.-D., Zhang, Q.-Y., Yu, J.-Q., Wang, J.-H., Zhang, F.-J., Wang, C.-K., et al. (2020). R2R3-MYB transcription factor MdMYB73 confers increased resistance to the fungal pathogen botryosphaeria dothidea in apples via the salicylic acid pathway. J. Agric. Food Chem. 69 (1), 447–458. doi: 10.21203/rs.3.rs-107830/v1
Gupta, S., Harkess, A., Soble, A., Van Etten, M., Leebens-Mack, J., Baucom, R. (2021). Inter-chromosomal linkage disequilibrium and linked fitness cost loci influence the evolution of nontarget site herbicide resistance in an agricultural weed. bioRxiv 2021.04.04.438381. doi: 10.1101/2021.04.04.438381
Haga, N., Kobayashi, K., Suzuki, T., Maeo, K., Kubo, M., Ohtani, M., et al. (2011). Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in arabidopsis. Plant Physiol. 157 (2), 706–717. doi: 10.1104/pp.111.180836
Hao, Y., Bao, W., Li, G., Gagoshidze, Z., Shu, H., Yang, Z., et al. (2021). The chromosome-based genome provides insights into the evolution in water spinach. Scientia Hortic. 289, 110501. doi: 10.1016/j.scienta.2021.110501
Hawku, M. D., He, F., Bai, X., Islam, M. A., Huang, X., Kang, Z., et al. (2022). A R2R3 MYB transcription factor, TaMYB391, is positively involved in wheat resistance to Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 23 (22), 14070. doi: 10.3390/ijms232214070
Ho, C.-L., Geisler, M. (2019). Genome-wide computational identification of biologically significant cis-regulatory elements and associated transcription factors from rice. Plants 8 (11), 441. doi: 10.3390/plants8110441
Hoseinpour, F., Darvishzadeh, R., Abdollahi Mandoulakani, B., Habibi, N., Dardan, E., Ranjbar, A. (2021). Evaluation of relative changes in the expression level of SOS2, MYB-related and HD-ZIP genes in oil seed sunflower lines under salinity stress. J. Crop Breed. 13 (39), 152–165.
Hoshino, A., Jayakumar, V., Nitasaka, E., Toyoda, A., Noguchi, H., Itoh, T., et al. (2016). Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 7 (1), 1–10. doi: 10.1038/ncomms13295
Howe, E. A., Sinha, R., Schlauch, D., Quackenbush, J. (2011). RNA-Seq analysis in MeV. Bioinformatics 27 (22), 3209–3210. doi: 10.1093/bioinformatics/btr490
Ito, M. (2005). Conservation and diversification of three-repeat myb transcription factors in plants. J. Plant Res. 118 (1), 61–69. doi: 10.1007/s10265-005-0192-8
Ito, M., Araki, S., Matsunaga, S., Itoh, T., Nishihama, R., Machida, Y., et al. (2001). G2/M-phase–specific transcription during the plant cell cycle is mediated by c-Myb–like transcription factors. Plant Cell 13 (8), 1891–1905.
Ito, M., Iwase, M., Kodama, H., Lavisse, P., Komamine, A., Nishihama, R., et al. (1998). A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase–specific transcription. Plant Cell 10 (3), 331–341.
Jiang, S. Y., González, J. M., Ramachandran, S. (2013). Comparative genomic and transcriptomic analysis of tandemly and segmentally duplicated genes in rice. PloS One 8 (5), e63551. doi: 10.1371/journal.pone.0063551
Jiang, F., Wang, S., Wang, H., Wang, A., Xu, D., Liu, H., et al. (2022). A chromosome-level reference genome of a convolvulaceae species Ipomoea cairica. G3 12 (9), jkac187. doi: 10.1093/g3journal/jkac187
Kamioka, M., Takao, S., Suzuki, T., Taki, K., Higashiyama, T., Kinoshita, T., et al. (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the arabidopsis circadian clock. Plant Cell 28 (3), 696–711. doi: 10.1105/tpc.15.00737
Kato, K., Gális, I., Suzuki, S., Araki, S., Demura, T., Criqui, M.-C., et al. (2009). Preferential up-regulation of G2/M phase-specific genes by overexpression of the hyperactive form of NtmybA2 lacking its negative regulation domain in tobacco BY-2 cells. Plant Physiol. 149 (4), 1945–1957. doi: 10.1104/pp.109.135582
Klempnauer, K. H., Gonda, T. J., Bishop, J. M. (1982). Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31 (2), 453–463. doi: 10.1016/0092-8674(82)90138-6
Koes, R., Verweij, W., Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10 (5), 236–242. doi: 10.1016/j.tplants.2005.03.002
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., dePamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50 (5), 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Lee, M. M., Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 (5), 473–483. doi: 10.1016/S0092-8674(00)81536-6
Li, Y., Leveau, A., Zhao, Q., Feng, Q. (2021b). Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat. Commun. 12 (1), 2563. doi: 10.1038/s41467-021-22920-8
Li, Y., Liang, J., Zeng, X., Guo, H., Luo, Y., Kear, P., et al. (2021c). Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Hortic. Plant J. 7 (2), 129–141. doi: 10.1016/j.hpj.2020.12.001
Li, J., Liu, H., Yang, C., Wang, J., Yan, G., Si, P., et al. (2020). Genome-wide identification of MYB genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus l. Ind. Crops Products 143, 111924. doi: 10.1016/j.indcrop.2019.111924
Li, Z., Peng, R., Tian, Y., Han, H., Xu, J., Yao, Q. (2016). Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol. 57 (8), 1657–1677. doi: 10.1093/pcp/pcw091
Li, L.-X., Wei, Z.-Z., Zhou, Z.-L., Zhao, D.-L., Tang, J., Yang, F., et al. (2021a). A single amino acid mutant in the EAR motif of IbMYB44. 2 reduced the inhibition of anthocyanin accumulation in the purple-fleshed sweetpotato. Plant Physiol. Biochem. 167, 410–419. doi: 10.1016/j.plaphy.2021.08.012
Lin, S., Singh, R. K., Moehninsi, M., Navarre, D. A. (2021). R2R3-MYB transcription factors, StmiR858 and sucrose mediate potato flavonol biosynthesis. Horticulture Res. 8 (2021), 25. doi: 10.1038/s41438-021-00463-9
Liu, C., Hao, J., Qiu, M., Pan, J., He, Y. (2020a). Genome-wide identification and expression analysis of the MYB transcription factor in Japanese plum (Prunus salicina). Genomics 112 (6), 4875–4886. doi: 10.1016/j.ygeno.2020.08.018
Liu, J., Osbourn, A., Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8 (5), 689–708. doi: 10.1016/j.molp.2015.03.012
Liu, X., Wu, Z., Feng, J., Yuan, G., He, L., Zhang, D., et al. (2021). A novel R2R3-MYB gene LoMYB33 from lily is specifically expressed in anthers and plays a role in pollen development. Front. Plant Sci. 12, 730007. doi: 10.3389/fpls.2021.730007
Liu, Y., Zeng, Y., Li, Y., Liu, Z., Lin-Wang, K., Espley, R. V., et al. (2020b). Genomic survey and gene expression analysis of the MYB-related transcription factor superfamily in potato (Solanum tuberosum l.). Int. J. Biol. Macromolecules 164, 2450–2464. doi: 10.1016/j.ijbiomac.2020.08.062
Lv, K., Wei, H., Liu, G. (2021). A R2R3-MYB transcription factor gene, BpMYB123, regulates BpLEA14 to improve drought tolerance in Betula platyphylla. Front. Plant Sci. 12, 791390. doi: 10.3389/fpls.2021.791390
Ma, Q., Dai, X., Xu, Y., Guo, J., Liu, Y., Chen, N., et al. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150 (1), 244–256. doi: 10.1104/pp.108.133454
Machado, A., Wu, Y., Yang, Y., Llewellyn, D. J., Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59 (1), 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
Magali, L., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Martin, C., Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13 (2), 67–73. doi: 10.1016/S0168-9525(96)10049-4
Matsui, K., Umemura, Y., Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in arabidopsis. Plant J. 55 (6), 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.-R., et al. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in arabidopsis. Dev. Cell 2 (5), 629–641. doi: 10.1016/S1534-5807(02)00170-3
Mmadi, M. A., Dossa, K., Wang, L., Zhou, R., Wang, Y., Cisse, N., et al. (2017). Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Genes 8 (12), 362. doi: 10.3390/genes8120362
Mulat, M. W., Sinha, V. B. (2021). Distribution and abundance of CREs in the promoters depicts crosstalk by WRKYs in tef [Eragrostis tef (Zucc.) troetter]. Gene Rep. 23, 101043. doi: 10.1016/j.genrep.2021.101043
Muramoto, N., Tanaka, T., Shimamura, T., Mitsukawa, N., Hori, E., Koda, K., et al. (2012). Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant Cell Rep. 31 (6), 987–997. doi: 10.1007/s00299-011-1217-5
Naing, A. H., Kim, C. K. (2018). Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 98 (1), 1–18. doi: 10.1007/s11103-018-0771-4
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Nimmakayala, P., Vajja, G., Reddy, U. K. (2011). “Ipomoea,” in Wild crop relatives: Genomic and breeding resources. Ed. Kole, C. (Berlin, Heidelberg: Springer). doi: 10.1007/978-3-642-21102-7_7
Okada, R., Kondo, S., Satbhai, S. B., Yamaguchi, N., Tsukuda, M., Aoki, S. (2009). Functional characterization of CCA1/LHY homolog genes, PpCCA1a and PpCCA1b, in the moss Physcomitrella patens. Plant J. 60 (3), 551–563. doi: 10.1111/j.1365-313X.2009.03979.x
Ozias-Akins, P., Jarret, R. L. (1994). Nuclear DNA content and ploidy levels in the genus Ipomoea. J. Am. Soc. Hortic. Sci. 119, 110–115. doi: 10.21273/JASHS.119.1.110
Park, S.-C., Kim, Y.-H., Kim, S. H., Jeong, Y. J., Kim, C. Y., Lee, J. S., et al. (2015). Overexpression of the IbMYB1 gene in an orange-fleshed sweet potato cultivar produces a dual-pigmented transgenic sweet potato with improved antioxidant activity. Physiologia Plantarum 153 (4), 525–537. doi: 10.1111/ppl.12281
Park, D.-Y., Shim, Y., Gi, E., Lee, B.-D., An, G., Kang, K., et al. (2018). The MYB-related transcription factor RADIALIS-LIKE3 (OsRL3) functions in ABA-induced leaf senescence and salt sensitivity in rice. Environ. Exp. Bot. 156, 86–95. doi: 10.1016/j.envexpbot.2018.08.033
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P., Saedler, H. (1987). The regulatory c1 locus of zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6 (12), 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Pu, X., Yang, L., Liu, L., Dong, X., Chen, S., Chen, Z., et al. (2020). Genome-wide analysis of the MYB transcription factor superfamily in physcomitrella patens. Int. J. Mol. Sci. 21 (3), 975. doi: 10.3390/ijms21030975
Qing, X., Jie, H., Jianhui, D., Xiaojin, H., Zhang, X. (2018). Genomic survey and expression profiling of the MYB gene family in watermelon. Hortic. Plant J. 4 (1), 1–15. doi: 10.1016/j.hpj.2017.12.001
Saha, G., Park, J.-I., Ahmed, N. U., Kayum, M. A., Kang, K.-K., Nou, I.-S. (2016). Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol. Biochem. 104, 200–215. doi: 10.1016/j.plaphy.2016.03.021
Salih, H., Gong, W., He, S., Sun, G., Sun, J., Du, X. (2016). Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 17 (1), 1–12. doi: 10.1186/s12863-016-0436-8
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3 (6), 1101–1108. doi: 10.1038/nprot.2008.73
Shangguan, X., Yang, Q., Wu, X., Cao, J. (2021). Function analysis of a cotton R2R3 MYB transcription factor GhMYB3 in regulating plant trichome development. Plant Biol. 23 (6), 1118–1127. doi: 10.1111/plb.13299
Shen, X., Guo, X., Guo, X., Zhao, D., Zhao, W., Chen, J., et al. (2017). PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 112, 302–311. doi: 10.1016/j.plaphy.2017.01.015
Sievers, F., Higgins, D. G. (2018). Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 27 (1), 135–145. doi: 10.1002/pro.3290
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Sinaga, D. S., Ho, S.-L., Lu, C.-A., Yu, S.-M., Huang, L.-F. (2021). Knockdown expression of a MYB-related transcription factor gene, OsMYBS2, enhances production of recombinant proteins in rice suspension cells. Plant Methods 17 (1), 1–10. doi: 10.1186/s13007-021-00799-2
Tan, L., Ijaz, U., Salih, H., Cheng, Z., Ni Win Htet, N., Ge, Y., et al. (2020). Genome-wide identification and comparative analysis of MYB transcription factor family in Musa acuminata and Musa balbisiana. Plants 9 (4), 413. doi: 10.3390/plants9040413
Tang, N., Cao, Z., Yang, C., Ran, D., Wu, P., Gao, H., et al. (2021). A R2R3-MYB transcriptional activator LmMYB15 regulates chlorogenic acid biosynthesis and phenylpropanoid metabolism in lonicera macranthoides. Plant Sci. 308, 110924. doi: 10.1016/j.plantsci.2021.110924
Tang, H., Wang, X., Bowers, J. E., Ming, R., Alam, M., Paterson, A. H. (2008). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18 (12), 1944–1954. doi: 10.1101/gr.080978.108
Thiedig, K., Weisshaar, B., Stracke, R. (2021). Functional and evolutionary analysis of the arabidopsi s 4R-MYB protein SNAPc4 as part of the SNAP complex. Plant Physiol. 185 (3), 1002–1020. doi: 10.1093/plphys/kiaa067
Upadhyaya, G., Das, A., Ray, S. (2021). A rice R2R3-MYB (OsC1) transcriptional regulator improves oxidative stress tolerance by modulating anthocyanin biosynthesis. Physiologia Plantarum 173 (4), 2334–2349. doi: 10.1111/ppl.13583
VanBuren, R., Wai, C. M., Keilwagen, J., Pardo, J. (2018). A chromosome-scale assembly of the model desiccation tolerant grass Oropetium thomaeum. Plant Direct 2 (11), e00096. doi: 10.1002/pld3.96
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered 93 (1), 77–78. doi: 10.1093/jhered/93.1.77
Wang, W., Fang, H., Aslam, M., Du, H., Chen, J., Luo, H., et al. (2022). MYB gene family in the diatom Phaeodactylum tricornutum revealing their potential functions in the adaption to nitrogen deficiency and diurnal cycle. J. Phycology 58 (1), 121–132. doi: 10.1111/jpy.13217
Wang, H., Liu, J.-X., Feng, K., Li, T., Duan, A.-Q., Liu, Y.-H., et al. (2022b). AgMYB12, a novel R2R3-MYB transcription factor, regulates apigenin biosynthesis by interacting with the AgFNS gene in celery. Plant Cell Rep. 41 (1), 139–151. doi: 10.1007/s00299-021-02792-4
Wang, X., Niu, Q.-W., Teng, C., Li, C., Mu, J., Chua, N.-H., et al. (2009). Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in arabidopsis. Cell Res. 19 (2), 224–235. doi: 10.1038/cr.2008.276
Wang, S., Shi, M., Zhang, Y., Xie, X., Sun, P., Fang, C., et al. (2021a). FvMYB24, a strawberry R2R3-MYB transcription factor, improved salt stress tolerance in transgenic arabidopsis. Biochem. Biophys. Res. Commun. 569, 93–99. doi: 10.1016/j.bbrc.2021.06.085
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wang, C., Wang, L., Lei, J., Chai, S., Jin, X., Zou, Y., et al. (2022a). IbMYB308, a sweet potato R2R3-MYB gene, improves salt stress tolerance in transgenic tobacco. Genes 13 (8), 1476. doi: 10.3390/genes13081476
Wang, Y., Zhang, Y., Fan, C., Wei, Y., Meng, J., Li, Z., et al. (2021b). Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 21 (1), 1–17. doi: 10.1186/s12870-021-03083-6
Wang, Y., Zhou, L.-J., Wang, Y., Geng, Z., Ding, B., Jiang, J., et al. (2022c). An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of chrysanthemum. Scientia Hortic. 293, 110674. doi: 10.1016/j.scienta.2021.110674
Wu, S., Lau, K. H., Cao, Q., Hamilton, J. P., Sun, H., Zhou, C., et al. (2018). Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 9 (1), 1–12. doi: 10.1038/s41467-018-06983-8
Wu, Y., Li, T., Cheng, Z., Zhao, D., Tao, J. (2021a). R2R3-MYB transcription factor PlMYB108 confers drought tolerance in herbaceous peony (Paeonia lactiflora pall.). Int. J. Mol. Sci. 22 (21), 11884. doi: 10.3390/ijms222111884
Wu, Z., Li, T., Liu, X., Yuan, G., Hou, H., Teng, N. (2021b). A novel R2R3-MYB transcription factor LlMYB305 from lilium longiflorum plays a positive role in thermotolerance via activating heat-protective genes. Environ. Exp. Bot. 184, 104399. doi: 10.1016/j.envexpbot.2021.104399
Yan, X., Ding, W., Wu, X., Wang, L., Yang, X., Yue, Y. (2022). Insights into the MYB-related transcription factors involved in regulating floral aroma synthesis in sweet osmanthus. Front. Plant Sci. 13, 765213–765213. doi: 10.3389/fpls.2022.765213
Yang, K., Li, Y., Wang, S., Xu, X., Sun, H., Zhao, H., et al. (2019). Genome-wide identification and expression analysis of the MYB transcription factor in moso bamboo (Phyllostachys edulis). PeerJ 6, e6242. doi: 10.7717/peerj.6242
Yang, J., Moeinzadeh, M., Kuhl, H., Helmuth, J., Xiao, P., Haas, S., et al. (2017). Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 3 (9), 696–703. doi: 10.1038/s41477-017-0002-z
Ye, J., McGinnis, S., Madden, T. L. (2006). BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34 (Web Server issue), W6–W9. doi: 10.1093/nar/gkl164
Yu, J., Zhao, G., Li, W., Zhang, Y., Wang, P., Fu, A., et al. (2021). A single nucleotide polymorphism in an R2R3 MYB transcription factor gene triggers the male sterility in soybean ms6 (Ames1). Theor. Appl. Genet. 134 (11), 3661–3674. doi: 10.1007/s00122-021-03920-0
Zhai, H., Wang, F., Si, Z., Huo, J., Xing, L., An, Y., et al. (2016). A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 14 (2), 592–602. doi: 10.1111/pbi.12402
Zhang, H., Liu, Z., Luo, R., Sun, Y., Yang, C., Li, X., et al. (2022). Genome-wide characterization, identification and expression profile of MYB transcription factor gene family during abiotic and biotic stresses in mango (Mangifera indica). Plants 11 (22), 3141. doi: 10.3390/plants11223141
Zhao, P., Hou, S., Guo, X., Jia, J., Yang, W., Liu, Z., et al. (2019). A MYB-related transcription factor from sheepgrass, LcMYB2, promotes seed germination and root growth under drought stress. BMC Plant Biol. 19 (1), 1–15. doi: 10.1186/s12870-019-2159-2
Zhao, Y., Zhang, G., Tang, Q., Song, W., Gao, Q., Xiang, G., et al. (2022b). EbMYBP1, a R2R3-MYB transcription factor, promotes flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 13, 946827. doi: 10.3389/fpls.2022.946827
Zhao, H., Zhao, H., Hu, Y., Zhang, S., He, S., Zhang, H., et al. (2022a). Expression of the sweet potato MYB transcription factor IbMYB48 confers salt and drought tolerance in arabidopsis. Genes 13 (10), 1883. doi: 10.3390/genes13101883
Zheng, J., Zhang, Z., Tong, T., Fang, Y., Zhang, X., Niu, C., et al. (2021). Genome-wide identification of WRKY gene family and expression analysis under abiotic stress in barley. Agronomy 11 (3), 521. doi: 10.3390/agronomy11030521
Zhou, C., Chen, Y., Wu, Z., Lu, W., Han, J., Wu, P., et al. (2015). Genome-wide analysis of the MYB gene family in physic nut (Jatropha curcas l.). Gene 572 (1), 63–71. doi: 10.1016/j.gene.2015.06.072
Zhou, L., Yarra, R., Jin, L., Cao, H. (2020). Genome-wide identification and expression analysis of MYB gene family in oil palm (Elaeis guineensis jacq.) under abiotic stress conditions. Environ. Exp. Bot. 180, 104245. doi: 10.1016/j.envexpbot.2020.104245
Keywords: Ipomoea species, MYB gene family, phylogenetic analysis, duplication analysis, gene expression, stresses response, sweet potato
Citation: Si Z, Wang L, Ji Z, Zhao M, Zhang K and Qiao Y (2023) Comparative analysis of the MYB gene family in seven Ipomoea species. Front. Plant Sci. 14:1155018. doi: 10.3389/fpls.2023.1155018
Received: 31 January 2023; Accepted: 06 March 2023;
Published: 20 March 2023.
Edited by:
Meng Kou, Sweet Potato Research Institute (CAAS), ChinaCopyright © 2023 Si, Wang, Ji, Zhao, Zhang and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengzhi Si, c2l6ZW5nemhpQDE2My5jb20=
 Zengzhi Si
Zengzhi Si Lianjun Wang2
Lianjun Wang2