
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 March 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1153490
This article is part of the Research Topic Growth Regulation in Horticultural Plants: New Insights in the Omics Era View all 14 articles
Chrysanthemum ×morifolium is well known throughout the world for its diverse and exquisite flower types. However, due to the complicated genetic background of C. ×morifolium, it is difficult to understand the molecular mechanism of its flower development. And it limits the molecular breeding of improving chrysanthemum flower types. C. ×morifolium has the typical radial capitulum, and many researches showed that the members of the MIKCc-type MADS box gene family play a key role in the formation and development of the capitulum. However, it has been difficult to isolate the important MIKCc and investigate their roles in this process due to the lack of genomic information in chrysanthemum. Here, we identified MIKCc-type MADS box genes at whole genome-wide level in C. lavandulifolium, a diploid species closely related to C. ×morifolium, and investigated their roles in capitulum development by gene expression pattern analysis and protein interaction analysis. A total of 40 ClMIKCc were identified and were phylogenetically grouped into 12 clades. Members of all clades showed different enriched expression patterns during capitulum formation. We speculate that the E-class genes in C. lavandulifolium underwent subfunctionalization because they have a significantly expanded, more diverse expression patterns, and specifically tissue expression than AtSEPs. Meanwhile, we detected the C-class expressed in disc floret corolla, which could be the clue to explore the morphological differences between disc and ray floret corolla. In addition, the potential roles of some MIKCcs in complex inflorescence formation were explored by comparing the number and phylogenetic relationship of MIKCc subfamily members in Asteraceae with different capitulum types. Members of the FLC branch in Asteraceae were found to be possibly related to the differentiation and development of the ray floret.
In angiosperms, flowers are the reproductive organs. The yield and quality of crop and ornamental plants are directly affected by the development of their floral organs. In Arabidopsis thaliana, the floral organs have been divided into four rounds according to their location and developmental sequence, which are the sepals, petals, stamens, and pistils from the outside to the inside. Each round of floral organ primordia was under the regulation of a different set of genes to determine its identity (Coen and Meyerowitz, 1991; Rounsley et al., 1995; Causier et al., 2010). Currently, the best explanation for floral organ specification in plants is the ABC(D)E model. This model assumes that the different floral primordia are synergistically regulated by several genes: sepals (A+E), petals (A+B+E), stamens (B+C+E), pistils (C+E) and ovules (D+E). The ABC(D)E model contains the A class gene APETALLA1 (AP1), APETALLA2 (AP2), the B class genes PISTILLATA (PI), APETALLA3 (AP3), the C class gene AGAMOUS (AG), the D class genes SEEDSTICK (STK), SHATTERPROOF1/2 (SHP1/2) and the E class genes SEPALLATA1/2/3/4 (SEP1/2/3/4). They all belong to the MIKCc clade of the MADS box gene family except AP2 (Becker and Theiben, 2003; Theiβen et al., 2016). Other members of the MIKCc clade have also been associated with flowering processes such as flower buds resting (Theiben et al., 2000; Wu et al., 2017; Nishiyama et al., 2021), apical meristematic differentiation (Li et al., 2010), development of inflorescence morphology (Hussin et al., 2021) and flowering time (Lee and Lee, 2010; Wang et al., 2021; Ahmar et al., 2022).
However, the members of the ABC(D)E model differ in their expression patterns in plants with more primitive floral organs or more complex inflorescence structures. In addition, their functions and regulatory mechanisms are still not well characterized (Melzer et al., 2010; Zhang et al., 2017; Zhang L. et al., 2020). The capitulum of the Asteraceae is a complex inflorescence with a highly compressed inflorescence structure. Numerous florets are borne in a spiral pattern on the capitulum in a Fibonacci series with the involucre wrapped around the outside. They can be classified as homogeneous capitulum (discoid or ligulate type) and heterogeneous capitulum (radiate type) depending on the type of florets borne on the receptacle. The capitulum is the most peculiar and important organ of Asteraceae plants, gives them a unique ornamental value. Members of the MIKCc subfamily have been shown to be essential for capitulum development in numerous studies (Elomaa et al., 2018).
Although the capitulum of Asteraceae is known as pseudanthium, the process of developing floral organs and the function of ABC(D)E-class genes in the capitulum differs from that of the single flower (Elomaa et al., 2018; Zhang and Elomaa, 2021). The involucre replaces the first floral organ (sepals) to protect the inner floral organs. This results in the disappearance of the sepals or their transition to crown hairs in the capitulum to aid seed dispersal. The A-class gene AP1 regulates the initiation and development of crown hairs in Taraxacum mongolicum. However, it is also widely expressed throughout the capitulum (Bremer, 1994; Harris, 1995; Wen et al., 2019a; Vijverberg et al., 2021). The structure and morphology of the second floral organ (petal) differ between the two types of florets in the radiate capitulum: Peripheral ray florets mimic the petals of individual flowers to attract pollinators. These petals are ligulate with complete upper and lower epidermis and parenchymatous tissue. The disc florets are responsible for receiving pollen and reproducing, with their petals enclosed in a corolla tube without parenchymatous tissue. B-class genes that determine petal identity are differentially expressed in the corollas of two floret types in C. lavandulifolium and C. ×morifolium, with AP3/PI both highly expressed in the ray floret corollas (Wen et al., 2019a). GDEF2 and GDEF3, the orthologous genes of AP3 in Gerbera hybrida, have a broader domain of expression during the differentiation of the floral organs (Broholm et al., 2010). The third floral organ (stamen) also differs in the two types of florets in the radial capitulum. Disc florets are bisexual and ray florets have only one pistil. The C class gene AG, which determines stamen identity, is conserved in two copies in G. hybrida, C. lavandulifolium, and C. ×morifolium with radial capitulum. However, the expression patterns in these three species are different (Yu et al., 1999; Wen et al., 2019a). All florets on the capitulum have a normally developed fourth floral organ (pistil) and ovule. In Tagetes erecta, TeAGL11-1 shows a partial function of the D-class genes that regulate seed and petal development (Zhang C. et al., 2020). The D-class gene showed more novel function except control the seed development in Asteraceae species. A total of seven SEP-like genes were identified in G. hybrida, indicating a significant expansion of the SEP evolutionary branch. SEPs are highly functionally redundant in A. thaliana, but in G. hybrida they showed specific transcriptional patterns during capitulum formation and floral organ identity determination, suggesting subfunctionalization and neofunctionalization (Zhang et al., 2017). Thus, the relatively conserved expression patterns and functions of ABC(D)E class genes in capitulum and simple plants have resulted in the pseudanthium formation. Meanwhile, their general expanded, differentiated and subfunctionalized in Asteraceae have led to significant differences in the floral organs of capitulum and simple plants. In the meantime, ABC(D)E class genes often regulate floral organ differentiation and development as protein tetramers. Their protein interactions in Asteraceae need to be further investigated.
MIKCc members showed significant neofunctionalization during capitulum and floret development in Asteraceae compared to model plants. ClCAL and ClFUL are specifically highly expressed in C. lavandulifolium at the stage of floret primordia initiation. They may be involved in the differentiation of ray and disc florets (Wen et al., 2022). The B class gene CmPI/CmAP3 is differentially expressed in the ray and disc florets of C. ×morifolium. It leads to the localization of carotenoid accumulation in the petals (Lu et al., 2022). The C-class gene ScAG and the D-class gene ScAGL11 can inhibit anthocyanin synthesis in ray florets of Senecio cruentus, which in turn affects the formation of flower spots (Qi et al., 2022). AGL6 plays the role of the A class gene in basal angiosperms (Kim et al., 2005). However, GhGRCD3, the AGL6 ortholog in G. hybrida, functions as both an A- and E-class gene, affecting crown hair and corolla identity and development, as well as inflorescence meristem maintenance and floral meristem differentiation (Ruokolainen et al., 2010a; Zhang et al., 2017). The key lowering integrator SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) affects ray floret development in G. hybrida. Overexpression of GhSOC1 causes ray floret shortening and discoloration (Ruokolainen et al., 2011). The current research indicates that MIKCc-type MADS box genes have a relatively conserved function during flower development in angiosperms. However, due to the complex structure of the capitulum, ABC(D)E-like genes appear to be commonly expanded, neofunctionalized, and subfunctionalized in Asteraceae. Other MIKCc genes have unique functions in capitulum formation and development. Therefore, the potential new functions of these classical flower development genes and their roles in the process of capitulum formation can be discovered through the mining, identification, and functional studies of MIKCc in Asteraceae.
Recently, with the release of the Asteraceae genome information (Badouin et al., 2017; Reyes-Chin-Wo et al., 2017; Song et al., 2018; Liu et al., 2020; Nakano et al., 2021; van Lieshout et al., 2022; Wen et al., 2022), the identification of gene families based on a genome-wide level has become feasible. Here, C. lavandulifolium (2n=2x=18) was selected as the object of study. It’s a perennial herb and a closely related diploid of the cultivated chrysanthemum (C. ×morifolium), often used as a typical material in chrysanthemum studies. In our previous work, we have conducted a detailed study of the capitulum development process of C. lavandulifolium, and the study of the MIKCc gene family of C. lavandulifolium will help us to further elucidate its formation mechanism (Wen et al., 2019a; Wen et al., 2019b; Wen et al., 2022). In this study, the MIKCc-type MADS box gene family members in the C. lavandulifolium genome been identified. We analyzed their protein physicochemical properties, conserved motifs, and phylogenetic relationships. Furthermore, based on transcriptome data, we screened for highly expressed ClMIKCcs during C. lavandulifolium capitulum and floral organ development. And qRT-PCR, yeast two-hybrid (Y2H) and luciferase complementation assay (LCA) were used to analyze the expression patterns and protein interactions of these ClMIKCcs in nine floral organs of C. lavandulifolium. These results will help to gain insight into the specific functions of the MIKCc-type MADS-box gene in complex inflorescences, and the pattern of the ClMIKCc protein complex. They will also provide potential genetic resources for floral shape improvement in C. ×morifolium. Based on this, we further compared the contraction/expansion of MIKCc subfamily members in Asteraceae, discussed the differentiation characteristics of these members in different capitulum type Asteraceae plants, and provided some new perspectives for resolving MIKCc evolution correlated with capitulum types in Asteraceae.
A total of 16 species, including A. thaliana as a reference species, and 15 Asteraceae with three different capitulum types were examined in our study to determine whether the number and classification of MIKCc genes correlated with capitulum type. Among these Asteraceae, radiate type capitulum with both ray florets and disc florets such as Artemisia annua, C. lavandulifolium, C. makinoi, C. nankingense, C. seticuspe, and Helianthus annuus, Stevia rebaudiana, and Smallanthus sonchifolius. Capitulum of the discoid type with disc florets only, such as Arctium lappa, Cynara cardunculus, Carthamus tinctorius, and Mikania micrantha. Capitulum of the ligulate type with only ray florets such as Cichorium endivia, Taraxacum kok-saghyz, and Lactuca sativa.
The reference sequences of A. thaliana MIKCc-type transcription factors were available from the Plant Transcription Factor Database (http://planttfdb.gao-lab; Tian et al., 2020). The genome data of A. annua (PRJNA280557), Arctium lappa (PRJNA764011), C. cardunculus (PRJNA453787), C. lavandulifolium (PRJNA681093), C. endivia (PRJNA797903), C. tinctorius (PRJNA313459), M. micrantha (PRJNA528368), S. rebaudiana (PRJNA436363), and S. sonchifolius (PRJNA798108) were downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/). The genome data of H. annuu (PRJNA345532), L. sativa (PRJNA173551), were downloaded from the EnsemblPlants (http://plants.ensembl.org/index.html). The genome data of T._kok-saghyz (PRJCA005187) was downloaded from Genome Warehouse (https://ngdc.cncb.ac.cn/gwh/). The genome data of C. nankingense was downloaded from Chrysanthemum Genome Database http://www.amwayabrc.com/zh-cn/index.html). C. makinoi genome data was downloaded at https://www.chrysanthemumgenome.wur.nl/. And C. seticuspe (Gojo-0 v1) genome data was downloaded from the Plant Garden database (https://plantgarden.jp/en/list/t1111766/genome/t1111766.G002).
AtMIKCc was used as a reference sequence to search against the genomic protein sequences using TBtools v1.098765 with an E-value cut-off of 1e-5 (Chen et al., 2020) to confirm the candidate MIKCc-type MADS-box genes in the above species. Each predicted sequence was then checked against NCBI and the redundant sequences were removed.
The transcriptome data of C. lavandulifolium (SRR14723013-SRR14723033) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). The six stages represent the six key stages of C. lavandulifolium capitulum development: During the vegetative stage, there are no primordia on the conical-shaped shoot apical meristem (SAM) (stage 1, S1). During the short-day condition, SAMs began to change from conical shape to hemispherical shape (stage 2, S2). At the early stage of floret primordia, only the first-round disc floret primordia differentiated at the margin, which is a key stage for the differentiation of disc and ray florets (stage 5, S5). At the middle stage of floret primordia differentiation, ray floret primordia initiated between the involucre and the outermost disc floret primordia (stage 6, S6). At the middle stage of corolla primordia differentiation, ray floret corollas began to initiate, while disc florets differentiated to produce stamen primordia and pistil primordia (stage 9, S9). At the final stage of corolla primordia differentiation, the corolla formation of the disc florets and the ray florets was completed, and they maintained a closed state (Stage10, S10) (Wen et al., 2019b).
The chromosomal location of ClMIKCc was obtained from genome annotation files (Wen et al., 2022), and TBtools v1.098765 (Chen et al., 2020) was used to map their physical location on C. lavandulifolium chromosomes and display the co-linearity.
The maximum likelihood (ML) method and 5000 bootstrap replicates were used to construct the phylogenetic tree of MIKCc. TBtools v1.098765 (Chen et al., 2020) was used. In this part, sequence alignments were performed with MUSCLE (Edgar, 2004), and trimAI (–automated1 option, Capella-Gutiérrez et al., 2009) helped us trim poorly aligned sequences and reserve reliable comparison results. The best-fit model (JTT+R7) chosen according to BIC and used to construct the phylogenetic tree in IQtree v2 (Minh et al., 2020). The results were further graphically edited using iTOL (https://itol.embl.de/).
The Conserved Domains Search database (https://www.ncbi.nlm.nih.gov/cdd/?term=) was used to predict the conserved domains of MIKCc. The predicted conserved domains were visualized using TBtools v1.098765 (Chen et al., 2020).
The MEME (https://meme-suite.org/meme/tools/meme) was used to identify the conserved motifs in AtMIKCc and ClMIKCc with the following optimal parameters: the width of the motifs was from 6 to 50 amino acids and the maximum number of motifs was 10. TBtools v1.098765 (Chen et al., 2020) was used to visualize the identified motifs.
The molecular weight and theoretical pI of AtMIKCc and ClMIKCc were analyzed using the Expasy ProtParam online software (https://web.expasy.org/protparam/), and the subcellular localization was predicted using the WOLF PSORT online tool (https://wolfpsort.hgc.jp/).
The genome-sequenced C. lavandulifolium G1 line was cultured at Beijing Forestry University for gene expression analysis (Wen et al., 2022). These materials were planted in 10 × 9 cm plastic pots with peat: vermiculite = 1:1. The temperature was 22 ± 1 °C and the light intensity was 4000 lx. The nutritive growth was carried out under long daylight (16 h light/8 h dark). After they grew 14 leaves, they were transferred to short daylight (12 h light/12 h dark) for reproductive growth (Fu et al., 2014). All floral organs (involucre, receptacles, corolla, pistils, ovules, stamens) were sampled at flowering, and four developmental stages of reproductive buds were collected based on Wen’s literature (Wen et al., 2019b). The morphology of the sampled material is shown in Figure 1. All the material was immediately frozen in liquid nitrogen and stored at -80°C for the extraction of total RNA.
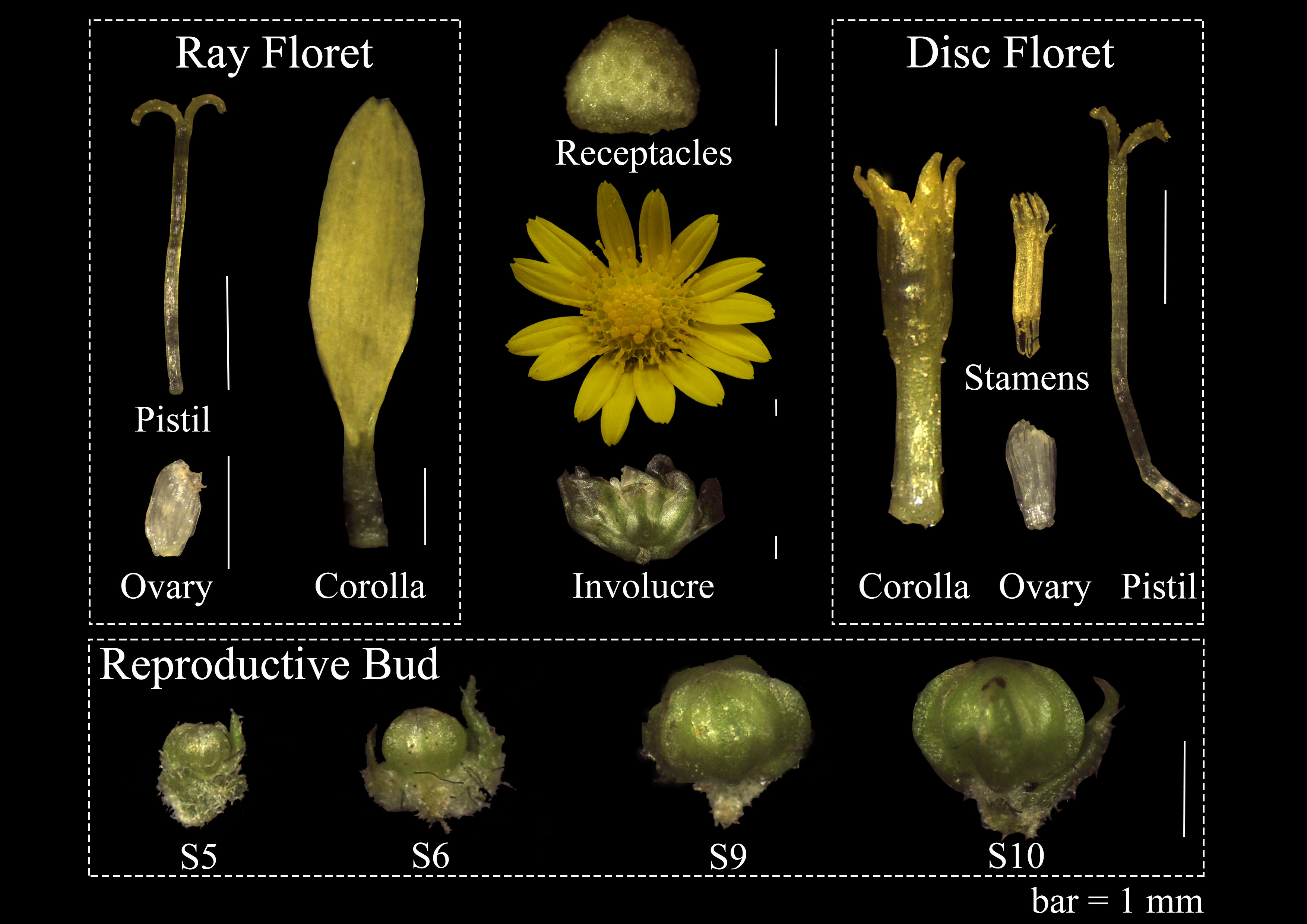
Figure 1 Floral organs on capitulum and four development stages reproductive bud in C. lavandulifolium for qRT-PCR. bar=1mm.
Total RNA was extracted from the collected plant materials using Plant RNA Rapid Extraction Kit (HUAYUEYANG Biotechnology, Beijing, China) and treated with RNase-free DNaseI to digest DNA. Gene expression analysis was performed by Real-time quantitative reverse transcription-PCR (qRT-PCR), which was performed using a Mini Opticon Real-time PCR System (Bio-Rad Laboratories Inc., Hercules, CA, USA) based on the SYBR Premix Ex Taq (Takara Bio Inc., Shiga, Japan). Three biological replicates were used to confirm the reliability of the results. ClSAND was used as an internal control gene for qRT-PCR (Qi et al., 2016). The primers for qRT-PCR were shown in Supplementary Table 2. The ΔΔCt method was used to analyze qRT-PCR data.
Pairwise yeast two-hybrid analyses were performed to verify the interactions between ClMIKCc. Gene-specific primers (Supplementary Table 3) were used to amplify protein-coding regions for all ClMIKCc tested. The PCR products were then fused into pGADT7 and pGBKT7 vectors. Sequencing was performed to verify the fusion. The constructs were then transformed into the Saccharomyces cerevisiae strain Y2H Gold and sequenced. Only the ClPI and ClSEPd constructs were found to have the ability to self-activate the expression of the HIS3 reporter genes. 10 mmol/L 3-amino-1, 2, 4-triazole (3-AT) was used to eliminate self-activation. For further interaction studies, recombinant vectors were used. Three technological replicates and three different concentrations were used in the protein-protein interaction experiments.
The luciferase complementation assay (LCA) was used to verify the positive results obtained in the Y2H assay (Chen et al., 2008). Target genes were ligated separately to pCAMBIA1300-cLUC and pCAMBIA1300-nLUC using the homologous recombination method. All primer sequence used to construct the vector is shown in Supplementary Table 4. Stop codon removal was required for all genes ligated to pCAMBIA1300-nLUC. The resulting recombinant plasmid was transformed into Agrobacterium tumefaciens (GV3101) using the freeze-thaw method.
Empty pCAMBIA1300-cLUC and pCAMBIA1300-nLUC carriers were used as controls. Subsequently, the Agrobacterium suspension (OD600 = 1.0) was injected into the abaxial leaf surface of three-week-old Nicotiana benthamiana leaves. After 72 h (24 h dark and 48 h light)of incubation in the greenhouse, D-Luciferin potassium salt was applied to the abaxial side of these leaves and then placed in the dark for 5 min. The fluorescence signal was observed using LB983 NightOwl II (Berthold Technologies, Bad Wildbad, Germany).
The 42 possible MIKCc proteins were retrieved from the C. lavandulifolium genome using two different BLAST methods. The retrieved MIKCc proteins were further screened based on the results of multiple sequence comparisons and the Conserved Domain of the NCBI website. The final 40 ClMIKCc proteins were identified. ClMIKCc encodes a polypeptide of 183 to 296 amino acids ranging in molecular weight from 20.73 to 34 kD. The theoretical pI range was 5.07 to 9.58. Subcellular localization predictions showed that 32 of the 40 ClMIKCc proteins were localized in the nucleus, which is typical of transcription factors (Supplementary Table 1).
Based on the genome annotation information of C. lavandulifolium, 38 of the 40 ClMIKCc were localized to all nine chromosomes. Two ClMIKCc were not associated with chromosomes (Figure 2). These ClMIKCc were not evenly distributed among the chromosomes. Chromosomes 1, 2, and 9 contained the fewest members, with two members each, and chromosome 5 contained the most, with eight members. The co-linearity analysis shows that only two MIKCc undergone a tandem repeat (Figure 2).
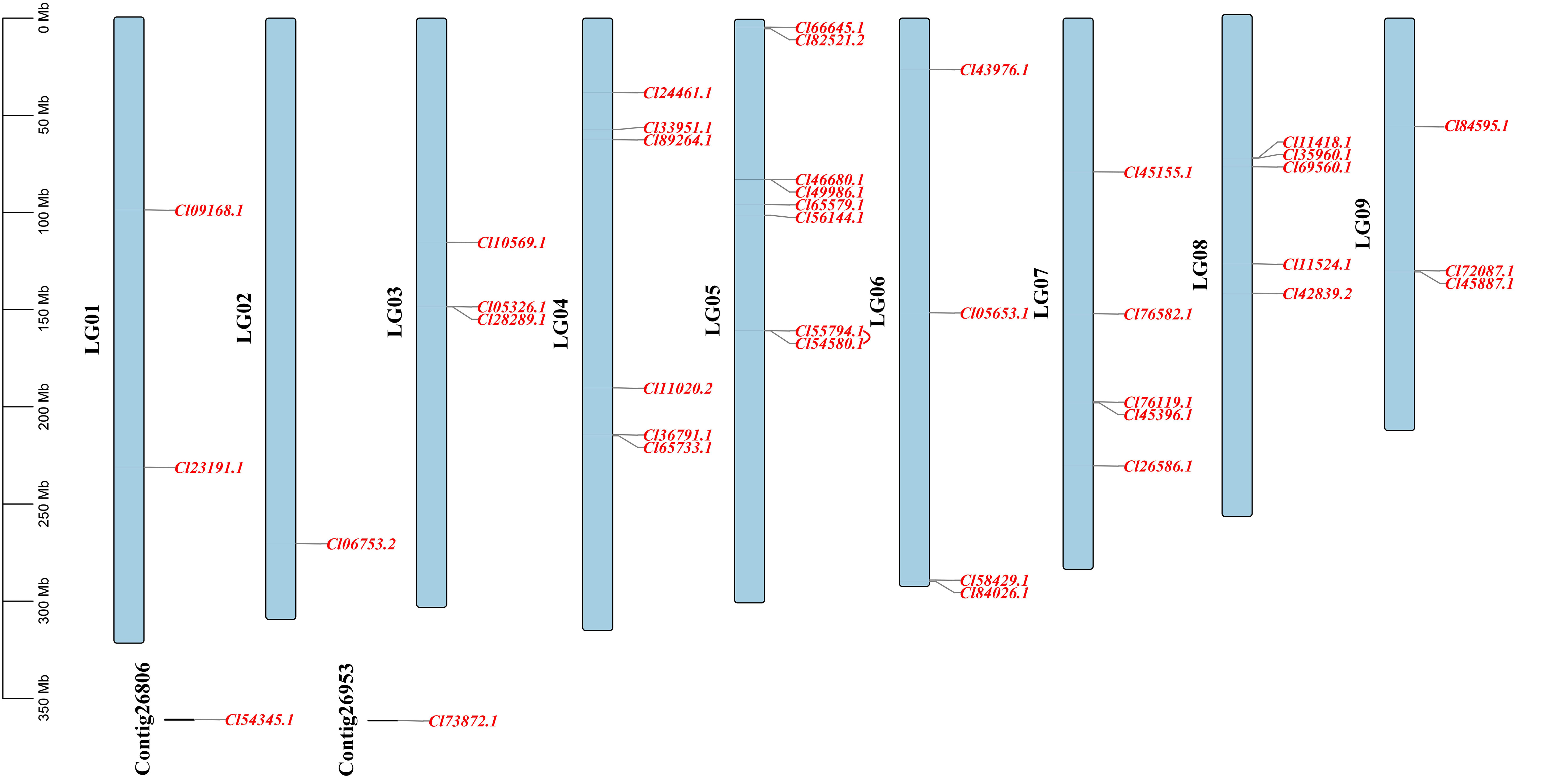
Figure 2 Localization and co-linearity of ClMIKCc on nine C. lavandulifolium chromosomes. The red line shown the co-linearity relationship between genes.
To elucidate the phylogenetic relationships of ClMIKCc, an ML phylogenetic tree was constructed using 40 ClMIKCc and 39 AtMIKCc (Figure 3A). ClMIKCc were distributed in 12 clades (GMM13, AP3/PI, AGL15, SVP, ANR1, FLC, SOC1, AGL12, AG/SHP/STK, AP1/FUL, AGL6, SEP), ClMIKCc underwent contraction in FLC and SOC1 clade and expansion in SVP, FUL and SEP clade.
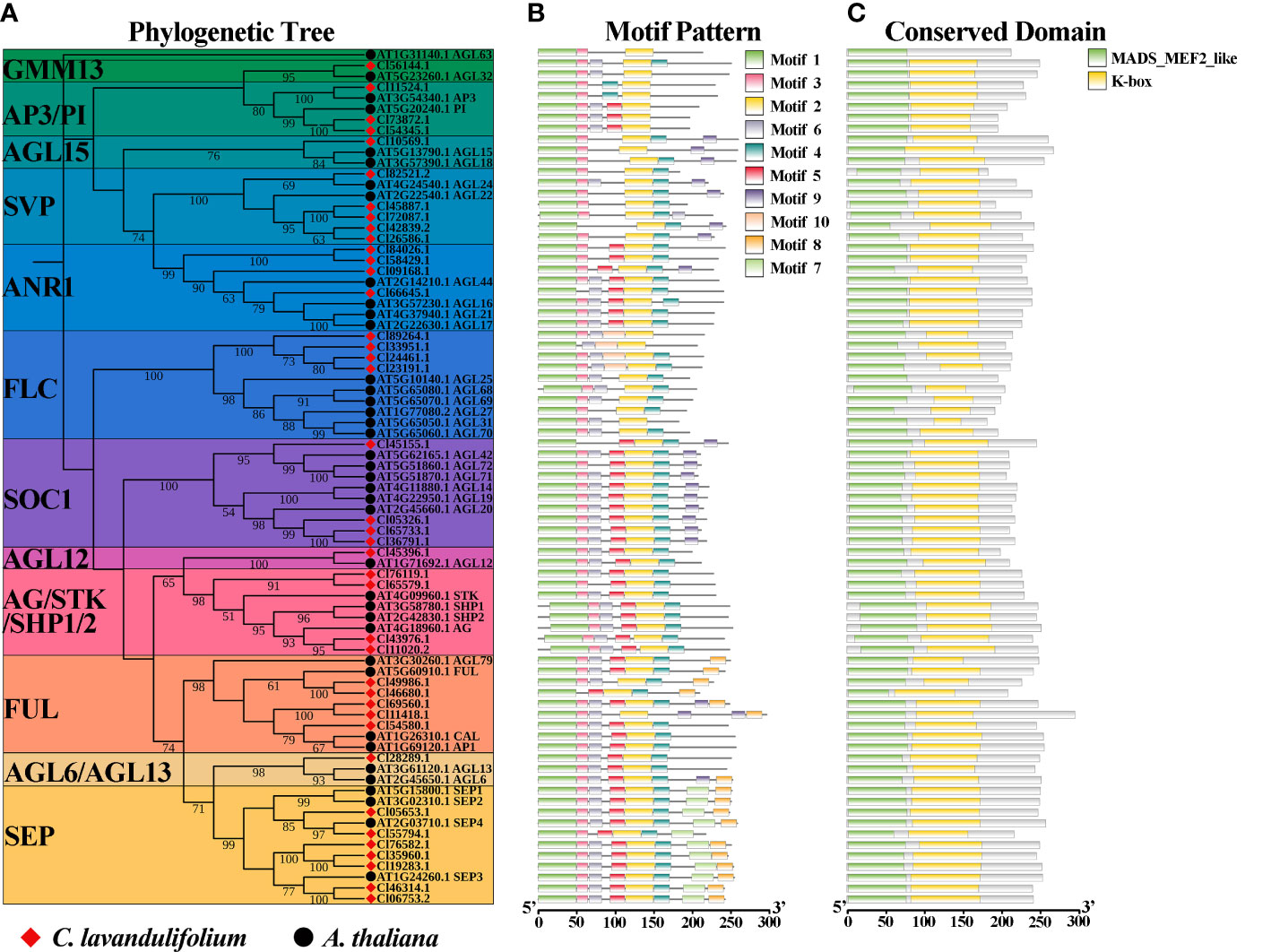
Figure 3 Analysis of phylogenetic, conserved motifs, and conserved structural domains of ClMIKCc and AtMIKCc. (A) ML phylogenetic tree of ClMIKCc and AtMIKCc. The number above branches represented bootstrap value and the values below 50 would not be shown; (B) Conserved motifs in ClMIKCc and AtMIKCc, 10 predicted motifs are indicated by different colored squares; (C) Conserved structural domains in ClMIKCc and AtMIKCc, 2 predicted structural domains are indicated by squares of different colors.
Using the MEME website, the conserved motifs of ClMIKCc and AtMIKCc were predicted. 10 conserved motifs were identified and labeled as motifs 1-10 (Supplementary Figure 1). For ClMIKCc and AtMIKCc, the motifs were relatively consistent and were located in the same clade (Figure 3B). There are 4-8 conserved motifs in each ClMIKCc, and all ClMIKCc and AtMIKCc proteins have Motif 1 and Motif 2. Combined with the predicted results of conserved structural domains, Motif 1 and Motif 2 are the core motifs of the MADS domain and K-box, respectively. Motif 7 is a specific conserved motif in At- and ClSEPs. Motif 10 is present only in ClFLCs (Figure 3C, Supplementary Figure 1).
Based on the public transcriptome dataset of C. lavandulifolium capitulum developmental stages (Wen et al., 2022), the expression pattern of ClMIKCc during C. lavandulifolium capitulum development was analyzed. A total of 26 ClMIKCc are expressed in leaves and six developmental stages (Figure 4A). These ClMIKCc could be mainly classified into four classes according to their expression patterns. Among them, class I, class II, and class IV ClMIKCc were specifically expressed in the capitulum (Supplementary Figure 2). Class I included the B-, C-class genes and ClSEPd/e/f, which were abundantly expressed in the late stage of capitulum development (floral organ differentiation period). These genes are mainly involved in the process of floral organ differentiation in C. lavandulifolium florets. Class II includes ClAP1a, ClSEPa/b/c/f, ClSOC1a, ClSVPa and ClFLCa, which showed high expression levels after the floral transition. They may be involved in both inflorescence primordium development and floral organ differentiation. Class III includes ClFLCb, ClFULa, and ClSVPb/c, which are mainly expressed in the leaf, the apical meristem before and after the floral transition, and the early stage of capitulum development. They may be involved in the floral transition process in C. lavandulifolium. Class IV contains ClANR1a, ClSOC1b/c/d and ClFULb, which are mainly expressed in the middle stage of capitulum development (S2-S6). They may regulate the floral transition and the formation of the inflorescence structure.
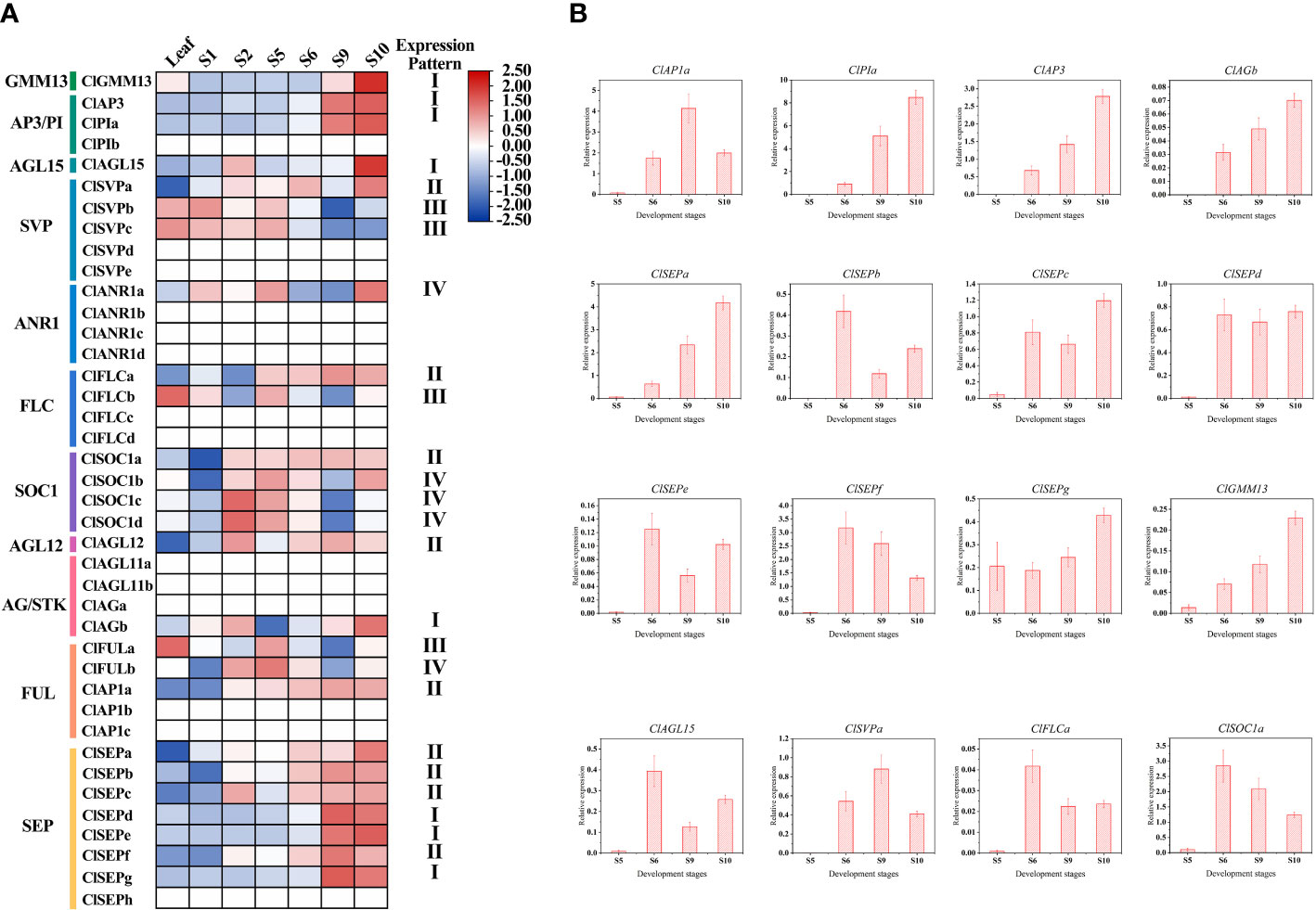
Figure 4 Expression pattern of ClMIKCc during the capitulum development stage. (A) Expression heat map constructed based on transcriptome data during the capitulum development stage of C. lavandulifolium. (B) qRT-PCR analysis of the expression level of class I and class II ClMIKCc in four development stages. Error bars represent the standard errors of three biological replicates.
To further elucidate the role of ClMIKCc in the development of the C. lavandulifolium capitulum, we examined the expression of class I and class II ClMIKCc in the capitulum of C. lavandulifolium at four critical periods after the initiation of the florets primordium by qRT-PCR. As shown in Figure 4B, almost all of these ClMIKCc were first expressed after the initiation of the floret primordium (S5). The expression increased with increasing floret number and floral organ differentiation. Interestingly, with the flowering process of C. lavandulifolium, the expression of ClSVPa and ClFLCa, two genes usually considered as flowering repressors, were also up-regulated. During the development of the C. lavandulifolium inflorescence, they may play a different role than their paralogous genes.
In order to investigate these genes’ roles in the development of different floral organs, 21 ClMIKCc was determined by qRT-PCR in nine floral organs. Class I and II of ClMIKCc were accompanied by the floral organ development, highly expression during the S9 and S10. Those ClMIKCc may be involved in the process of floral organ identity determination and development. The results (Figure 5) showed that the A-class gene ClAP1a was widely expressed in the nine floral organs. There are three B-class genes in C. lavandulifolium, ClAP3, ClPIa, and ClPIb, among which ClAP3 and ClPIa had a typical B-class gene expression pattern that expressed mainly in the stamens and corollas, also a weak expression was detected in the ovules. However, ClPIb was detected weakly expressed in all floral organs. A total of two C-class genes were identified in the C. lavandulifolium genome, ClAGa was not only expressed in the pistil, ovule, and stamen but also partially overlapping expressed with ClAP1a in the corolla of disc floret. ClAGb showed a low expression level and a broad expression domain which is like ClPIb. This expression patterns difference of paralogous genes was also present in two ClAGL11-like genes, ClAGL11a specifically expressed in the ovules, while ClAGL11b expressed in all floral organs but with a lower level. The E-class genes in the SEP branch showed diverse expression patterns. ClSEPa was mainly expressed in ovules and stamens. ClSEPc expressed in all floral organs except corollas and pistils of disc floret. ClSEPd and ClSEPe mainly expressed in ovules and ray floret corollas. ClSEPb, ClSEPf, and ClSEPg expressed in all floral organs.The result also showed that ClAGL12, ClAGL15, and ClFLCa weakly expressed in all floral organs. ClGMM13 and ClSOC1a expressed in pistils, carpels, stamens, involucre, and receptacles. ClSVPa specifically expressed in involucre and receptacles (Figure 5).
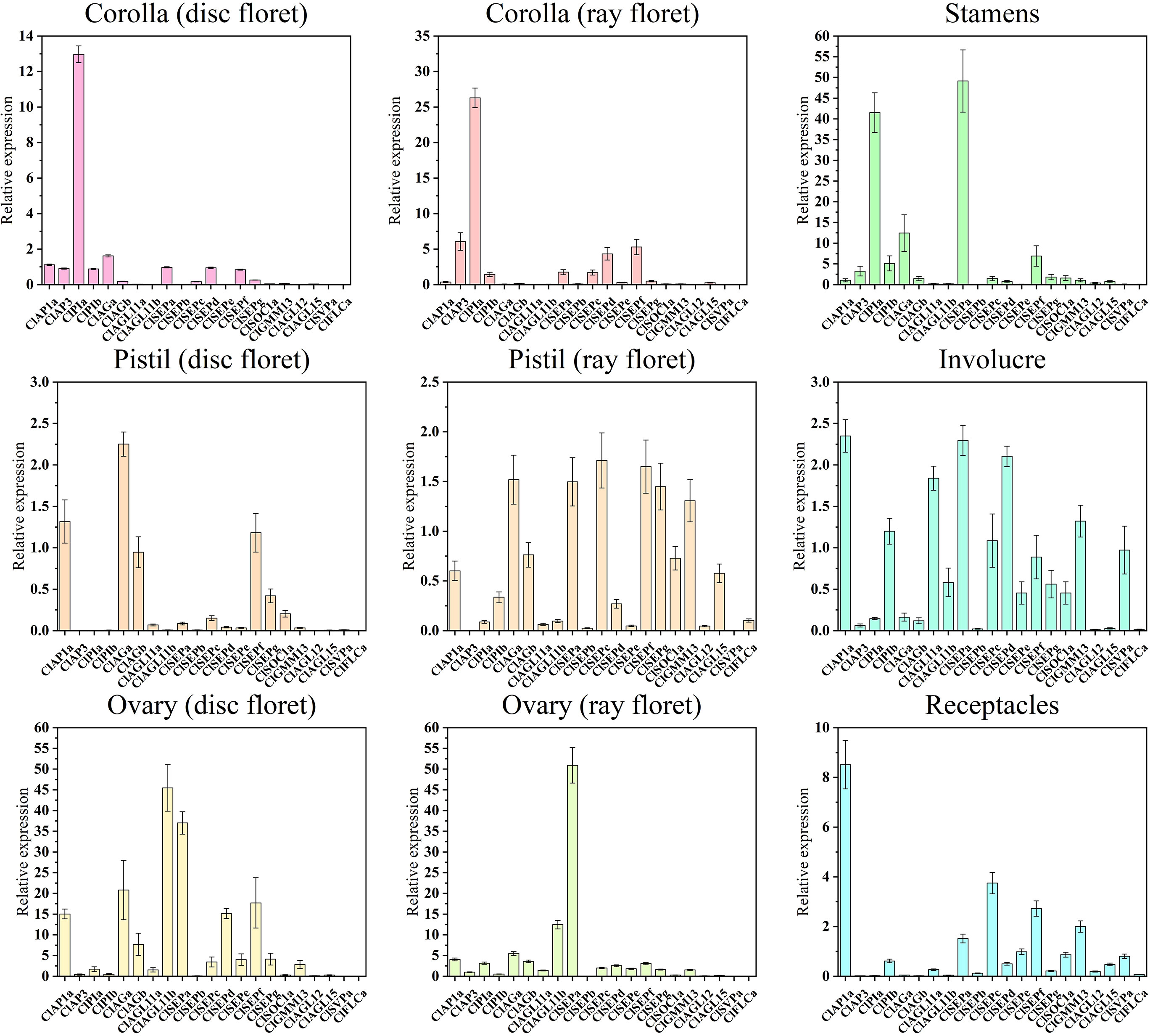
Figure 5 Quantitative RT-PCR analysis of the expression level of 21 ClMIKCc in nine floral organs. Error bars represent the standard errors of three biological replicates.
The expression pattern analysis of 26 ClMIKCc in nine floral organs showed that they had similar patterns in the corresponding floral organs between the two floret types (Figure 5). The expression patterns of ClMIKCc in the floral organs of C. lavandulifolium were generally consistent with the ABC(D)E model in model plants (Figure 5). The B-and E-class genes were predominant in the corolla of both floret types. The stamen development was regulated by B-, C-, and E-class genes. And the pistils and ovules development were regulated by B-, C-, and E-class genes as well as AGL11-like. However, differences in the expression level of same genes or the tissue-specific expression patterns of homologous genes in two floret types may be the cause of their morphology distinction.
Since members of the MIKCc gene subfamily usually form homologous or heterologous protein complexes to function, we performed the yeast two-hybrid assay and LCA to analyze the protein interaction between the four most highly expressed ClMIKCc in each floral organ. The positive results obtained for all Y2H systems are shown in the Supplementary Figure 3. Since the Y2H system may give false positive results, we verified the positive results of the heterologous interactions by the LCA in tobacco (Supplementary Figure 4). These results showed that these four ClMIKCc were able to form protein complexes through different modes of interaction (Supplementary Figure 3-4). Although the A-class gene ClAP1a was widely expressed in the capitulum, it could only interact with ClAGa, ClAGb, and ClAGL11b from the AG clades, as well as the homolog of Bsister (Bs), ClGMM13 (Supplementary Figure 3). Among the B class genes, ClAP3 can only interact with ClPIa. However, ClPIa can extensively form protein dimers with C and E class genes. Similar to B class genes, among two copies of C class gene ClAG, ClAGa was able to form protein dimers with more ClMIKCc. ClAGb was only able to interact with A and C class genes. ClAGL11-like is highly expressed in ovules and can interact with E-class genes. Therefore, it may function as a D-class gene in C. lavandulifolium. ClSEPs are highly expressed in all floral organs, and they also interact to a large extent with members of other clades. Thus, they appear to be key members of the multi-protein complex. ClGMM13 from the Bs evolutionary branch can interact with A- and E-class genes. Its function may also depend on the formation of the protein complex (Figure 6).
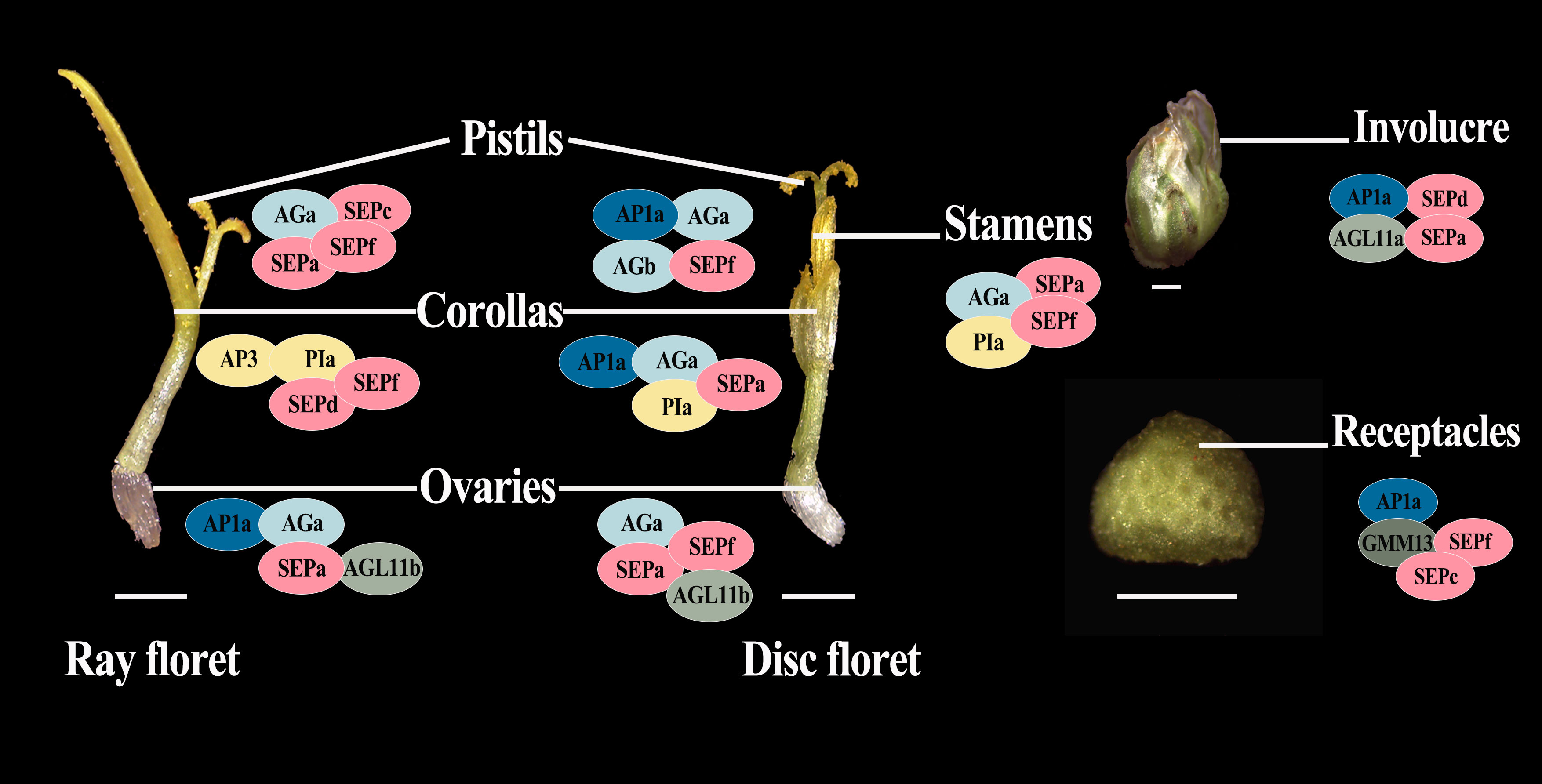
Figure 6 Suppositional interaction model of the four most highly expressed ClMIKCc in the nine floral organs of C. lavandulifolium. Different colors are used to distinguish the members of the different clades. Intersecting ellipses indicate that the protein pair has interactions. All positive results are supported by the Y2H and LCA results. bar=1 mm.
For Asteraceae MIKCc phylogenetic analysis, ML trees were constructed using MIKCc proteins from 16 species. These MIKCcs were grouped into 12 clades, and the number of proteins varied greatly among these branches. The SVP, SOC1, SEP, FLC, and AP1/FUL clades had more members (Figure 7A). Furthermore, the number of MIKCc per species in each branch was counted (Figure 7B). The results showed that the number of MIKCc was lowest in C. cardunculus, which has a discoid type capitulum, with 23 members. While the highest number of proteins was found in L. sativa, which had a ligulate type capitulum, with 85 members. SEP clade significant expansion in radial capitulum species, the number generally more than discoid and ligulate type capitulum species. This may be related to the more complex inflorescence structure of the radial capitulum. FLC clade is abundantly retained in Asteraceae with ray floret, whether they were need vernalization.
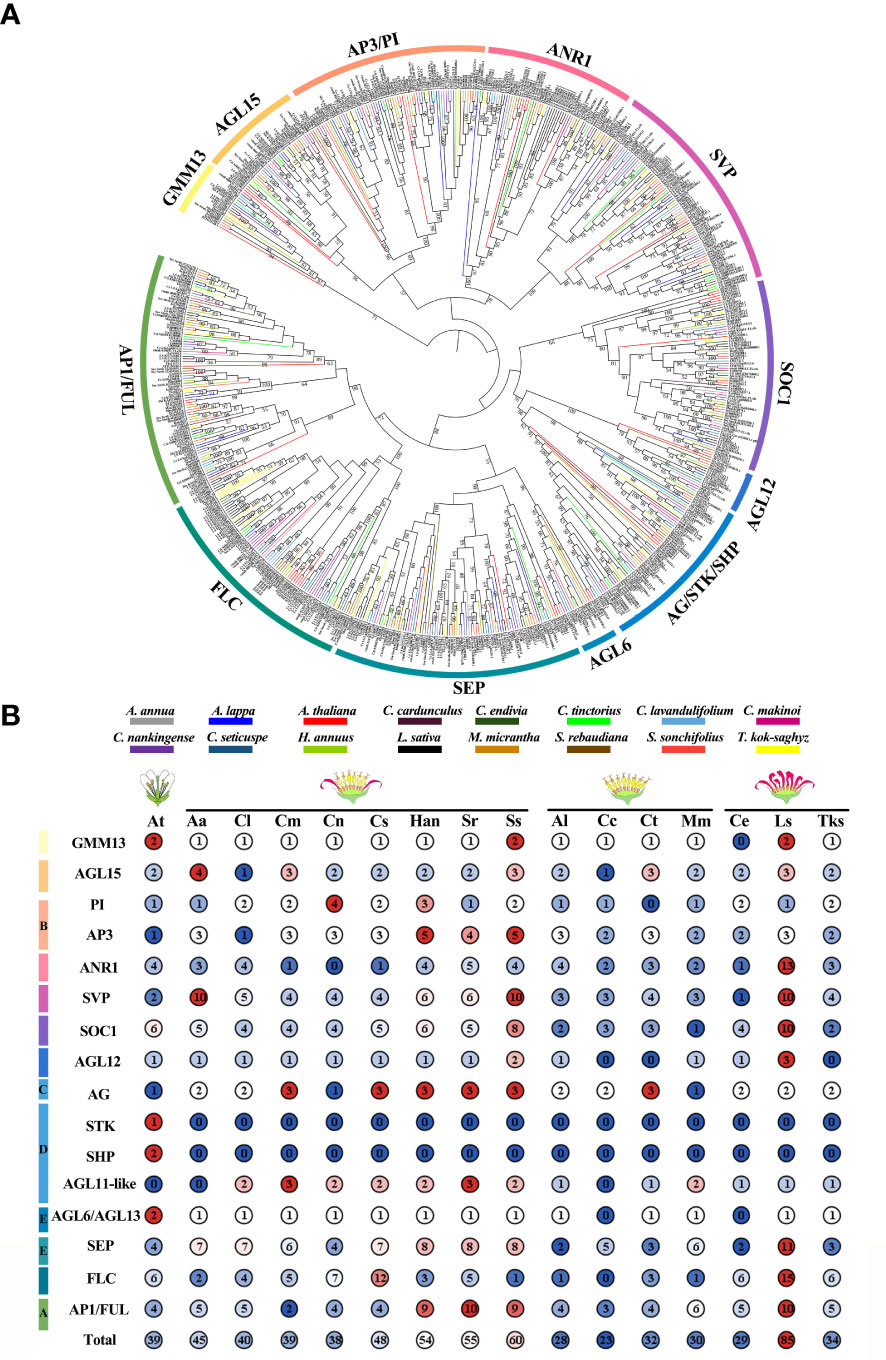
Figure 7 Phylogenetic analysis and statistic of MIKCc in Arabidopsis and fifteen Asteraceae. (A) The ML Phylogenetic tree of MIKCc in 15 Asteraceae. The bootstrap replicates were 5000. The number above branches represented bootstrap value and the values below 50 would not be shown. We used different colors to distinguish species and clades. (B) The member counts of each clade of MIKCc in 16 species. Their respective inflorescence types are indicated on the top. At, A. thaliana; Aa, A. annua; Cl, C. lavandulifolium; Cm, C. makinoi; Cn, C. nankingense; Cs, C. seticuspe; Han, H. annuus; Sr, S. rebaudiana; Ss, S. sonchifolius; Al, A. lappa; Cc, C. cardunculus; Ct, C. tinctorius; Mm, M. micrantha; Ce, C. endivia; Ls, L. sativa; Tks, T._kok-saghyz.
A total of 40 MIKCc-type MADS box genes were identified from the whole genome sequence of C. lavandulifolium. Chromosome localization results showed that ClMIKCcs were unevenly distributed on nine chromosomes of C. lavandulifolium (Figure 2). Based on a phylogenetic tree constructed from ClMIKCc and AtMIKCc, the ClMIKCcs were divided into 11 clades (Figure 3A). The number of A-class genes AP1, B-class genes PI\AP3, C-class genes AG, and flower-forming integrator SOC1 genes in C. lavandulifolium genome was consistent with previous results based on the C. lavandulifolium transcriptome and homologous clones (Wang et al., 2014; Wen et al., 2019a). In addition, we identified many E-class genes in the C. lavandulifolium genome. These SEP clade members showed diverse expression patterns and extensive protein interactions during capitulum and floral organ development (Figures 4–6), which may play a critical role in regulating complex inflorescence formation.
Although C. lavandulifolium does not require vernalization to complete floral transition (Fu et al., 2014), the flowering repressor FLOWERING LOCUS C (FLC), the key integrator of the vernalization pathway, retains four members in the C. lavandulifolium genome. In contrast, other nonvernalizing plants such as Oryza sativa, Cucumis sativus, Zea mays, Sorghum bicolor, and Gossypium hirsutum have experienced a complete loss of members of the FLC members (Arora et al., 2007; Zhao et al., 2011; Hu and Liu, 2012; Ren et al., 2017). Notably, all ClFLCs have a unique motif 10 (Figure 3B, Supplementary Figure 1). This motif is not found in other ClMIKCcs and all AtMIKCcs. These results suggest that the ClFLCs may have a specific function in the flowering process of C. lavandulifolium.
In A. thaliana, members of the MIKCc subfamily not only regulate floral organ identity and development, but also play important roles in several biological processes, such as flowering time, nitrogen utilization, and seed development (Gan et al., 2005; Mizzotti et al., 2014; Theiβen et al., 2016). ClMIKCc has a typical MADS box domain and K box, with similar conserved motifs to the A. thaliana reference sequence (Figure 3), and its function may also be conserved.
However, unlike A. thaliana, the process of capitulum development in C. lavandulifolium involves five stages, which are floral induction, floral transition, involucre differentiation, floral primordia differentiation, and corolla primordia differentiation (Wen et al., 2019b). We found that ClAP1, ClAP3, ClPIa/b, ClAGb, ClSEPs, ClSOC1a, SHORT VEGETATIVE PHASE (ClSVPa), and ClFLCa maintained a high level of expression after the floral primordia (Figure 4B). In A. thaliana, AP1 is an important floral meristem maintenance gene to maintain the limited growth of floral meristem tissues (Pelaz et al., 2001; Litt, 2007; Han et al., 2014), in addition to functioning as an A-class gene to determine the identity of sepals and petals. In G. hybrid, the A- and E-class genes work together to determine the fate of the inflorescence meristem tissue (Uimari et al., 2004; Ruokolainen et al., 2010b). Usually, SOC1 considered an activator in the flowering pathway, and SVP and FLC were repressors (Blümel et al., 2015). In A. thaliana, AGL24, SVP and AP1 function together to promote and maintain the floral meristem (Gregis et al., 2008). SOC1. In contrast, FLC were not found to be associated with inflorescence meristem or floral meristem formation. In view of the fact that there are specific motifs in ClFLCs (Figure 2B) and ClFLCa expression of which is upregulated with C. lavandulifolium flowering (Figure 4B), we hypothesize that part of FLCs are involved in the formation of the inflorescence meristem and floral meristem.
In this study, we found some important genes regulating the developmental process of C. lavandulifolium capitulum, including classical A-, B-, C- and E-class genes. Novel expression patterns were also found for some key genes in the photoperiodic pathway. However, their function needs to be further investigated.
The involucre is formed by a cluster of bracts. It is an important basis for the classification of Asteraceae (Carlquist, 1976), based on its shape and the number of whorls. The involucre is located on the outermost whorl of the capitulum. It envelops and protects the capitulum (Yang and Sun, 2009). It is similar to the first whorl of the floral organs (sepals) in individual flowers in morphology, position, and function. We observed a high expression level of A- and E-class genes expressed in the involucre of C. lavandulifolium (Figure 5). This expression pattern is also similar to that of sepals (Theißen et al., 2016). Interestingly, we also found that the D-class gene ClAGL11a is highly expressed in the involucre (Figure 5). It can also interact with ClAP1a and ClSEPa to form a protein complex (Figures 5, 6). In contrast, ectopic expression of STK/AGL11 in A. thaliana leads to the transformation of sepals into carpel-like organs with ovules, indicating the antagonism between D-class genes and sepals (Favaro et al., 2003; Pinyopich et al., 2003). This result suggests that the involucre of C. lavandulifolium is similar to the first whorl of floral organs (sepals) of A. thaliana, but not in terms of the pattern of gene expression. The A-class gene ClAP1a is widely expressed in all floral organs. This result is similar to previous findings in C. lavandulifolium, C. ×morifolium, and G. hybrida (Ruokolainen et al., 2010b; Wen et al., 2019a).
There is a clear difference in corolla morphology between ray and disc florets (Figure 1). B class genes have a higher expression level in the ray floret corolla. Interestingly, we observed an overlap in the expression of A- and C-class genes in the disc floret corolla of C. lavandulifolium (Figure 5). C-class genes are usually considered to have a negative effect on corolla formation (Wollmann et al., 2010). In the classical ABC model, the A- and C-class genes are antagonistic to each other, with the A-class genes repressing the expression of the C-class genes in order to maintain petal/stamen differentiation (Coen and Meyerowitz, 1991). However, in the disc floret corolla, ClAP1a and ClAGa not only have similar expression patterns. They can also form a heterodimer. We suggest that ClAGa may be a candidate to explain such differences, considering the obvious differences in length and structure between ray and disc corollas. Meanwhile, E-class genes are also differentially expressed between the two floral types (Figure 5). Studies on G. hybrid have also shown that E-class genes can directly affect floral organ identity and morphology (Zhang et al., 2017).
Except for the C class genes that were found to be expressed in the corolla of disc florets, the B, C, D, and E class genes in C. lavandulifolium have similar tissue-specific expression to the model plant (Figure 5). We also found that the retention pattern of C. lavandulifolium is conserved for paralogs, with some members of all four gene classes (ClPIb, ClAGb, ClAGL11b, ClSEPc, ClSEPg) showing a consistent expression pattern with lower expression levels and broader expression domains (Figure 5). In Asteraceae (Figure 7), expansions and duplications of ABC(D)E class genes are widespread. Expansions of A-, B-, C-, and E-class genes were also found in G. hybrid. Some members have low expression levels and broad expression domains. Inhibition of these members results in little phenotype alteration (Broholm et al., 2010; Ruokolainen et al., 2010a; Ruokolainen et al., 2010b; Zhang et al., 2017). These results suggest that, in response to the expansion and duplication of ABC(D)E class genes, this strategy of subfunctionalization is conserved in Asteraceae.
The number of MIKCc gene subfamily members showed significant differences among Asteraceae plants with distinct capitulum types (Figure 7B). The largest account of MIKCc was 85 in L. sativa which has the ligulate type capitulum. The smallest number of MIKCc was in the discoid type capitulum species C. cardunculus, at 23. ANR1, SVP, SOC1 FLC, and SEP clades all underwent a significant expansion in L. sativa due to a specific whole genome triplication (WGT) event, with many MADS-box genes distributed within the triplication region (Reyes-Chin-Wo et al., 2017). In H. annuus, which also underwent WGT (Badouin et al., 2017), the number of MIKCcs was also significantly higher than that in other Asteraceae. These results suggest that genome duplication events are the main reason for the expansion of MIKCc gene family in Asteraceae.
In perennial Brassicaceae plants (e.g. Arabis alpine), FLC expression is repressed by low temperatures of winter and reactivated in spring, conferring a seasonal flowering pattern to the plant (Leijten et al., 2018). In this study, the amount of FLC branch members were found to be significantly different among seven Asteraceae tested. Many FLCs retained in Asteraceae species with ray florets, although they did not need to undergo a low-temperature period to complete the flowering transition. Whereas in two discoid type capitulum species, C. cardunculus and M. micrantha, the FLC was significantly contracted or completely lost (Figure 7B). Previous studies have shown that C. lavandulifolium flowering is strictly dependent on short-day induction rather than low temperature (Fu et al., 2014), but four FLC-like (Figure 3) are retained in the genome and these genes contain a specific motif 10 (Figure 3B, Supplementary Figure 1). Transcriptomic data on the capitulum development stage showed that ClFLCa highly expressed at the stage of the florets and the corolla primordium differentiation (Figure 4A), which is the same expression pattern with A- and E-class genes (Figure 4). It might play a novel role in the capitulum. CiMFL in Cichorium intybus, the ortholog of AtFLC, was found to fail to recover the early flowering phenotype of the flc3 mutant of A. thaliana, but instead resulted in abnormal leaf organ morphology. However, the morphological changes in the floral organs were not observed due to a continuous reduction in the CiMFL expression level during flowering in transgenic lines (Locascio et al., 2009). This result also suggests that members of the FLC in Asteraceae have a novel function in controlling organ morphology. Accordingly, we hypothesize that the FLC may have novel functions in the development of capitulum in Asteraceae, especially in the development of ray florets.
In this study, a total of 40 ClMIKCc proteins were identified and classified into 12 branches. Transcriptomic data and qRT-PCR indicated that most ClMIKCc are specifically expressed in the capitulum, and ClFLCa may have novel functions in regulating the differentiation and development of ray floret. C. lavandulifolium has a conservative coping strategy for the redundant MIKCc gene, where members of the same branch have similar expression patterns, but only one member is highly expressed. Y2H and LCA results revealed the interactions of ClMIKCc in different floral organs of C. lavandulifolium. That will help to determine the tetrameric model in C. lavandulifolium floral organs. The results of phylogenetic analysis of MIKCc in Asteraceae indicated that there may be a correlation between the number of SEPs and the complexity of capitulum structure, as the SEP clade was significantly expanded in Asteraceae with radial capitulum. However, FLC was almost lost in Asteraceae with discoid capitulum, which, combined with the expression pattern of ClFLCa in C. lavandulifolium, further indicates the potential role of FLC in ray floret development in Asteraceae.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SD conceived and designed the study. JL performed most of the experiments and data analysis. QZ performed part of gene expression analysis. JL wrote the manuscript, and DK, YP, and XW edited it. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (No. 31530064).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1153490/full#supplementary-material
Ahmar, S., Zhai, Y., Huang, H., Yu, K., Khan, M. F. U., Shahid, M., et al. (2022). Development of mutants with varying flowering times by targeted editing of multiple SVP gene copies in Brassica napus l. Crop J. 10, 67–74. doi: 10.1016/j.cj.2021.03.023
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 8 (1), 1–21. doi: 10.1186/1471-2164-8-242
Badouin, H., Gouzy, J., Grassa, C. J., Murat, F., Staton, S. E., Cottret, L., et al. (2017). The sunflower genome provides insights into oil metabolism, flowering and asterid evolution. Nature 546, 148–152. doi: 10.1038/nature22380
Becker, A., Theiβen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. doi: 10.1016/S1055-7903(03)00207-0
Blümel, M., Dally, N., Jung, C. (2015). Flowering time regulation in crops–what did we learn from arabidopsis? Curr. Opin. Biotech. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Broholm, S. K., Pöllänen, E., Ruokolainen, S., Tähtiharju, S., Kotilainen, M., Albert, V. A., et al. (2010). Functional characterization of b class MADS-box transcription factors in Gerbera hybrida. J. Exp. Bot. 61, 75–85. doi: 10.1093/jxb/erp279
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15), 1972–1973. doi: 10.1093/bioinformatics/btp348
Carlquist, S. (1976). Tribal interrelationships and phylogeny of the asteraceae. Aliso 8, 465–492. doi: 10.5642/aliso.19760804.10
Causier, B., Schwarz-Sommer, Z., Davies, B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73–79. doi: 10.1016/j.semcdb.2009.10.005
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1093/nar/gkh340
Elomaa, P., Zhao, Y., Zhang, T. (2018). Flower heads in asteraceae–recruitment of conserved developmental regulators to control the flower-like inflorescence architecture. Hortic. Res. 5. doi: 10.1038/s41438-018-0056-8
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Fu, J., Wang, L., Wang, Y., Yang, L., Yang, Y., Dai, S. (2014). Photoperiodic control of FT-like gene ClFT initiates flowering in Chrysanthemum lavandulifolium. Plant Physiol. Bioch. 74, 230–238. doi: 10.1016/j.plaphy.2013.11.004
Gan, Y., Filleur, S., Rahman, A., Gotensparre, S., Forde, B. G. (2005). Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222 (4), 730–742. doi: 10.1007/s00425-005-0020-3
Gregis, V., Sessa, A., Colombo, L., Kater, M. M. (2008). AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in arabidopsis. Plant J. 56, 891–902. doi: 10.1111/j.1365-313X.2008.03648.x
Han, Y., Zhang, C., Yang, H., Jiao, Y. (2014). Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 6840–6845. doi: 10.1073/pnas.131853211
Harris, E. M. (1995). Inflorescence and floral ontogeny in asteraceae: a synthesis of historical and current concepts. Bot. Rev. 61, 93–278. doi: 10.1007/BF02887192
Hu, L., Liu, S. (2012). Genome-wide analysis of the MADS-box gene family in cucumber. Genome 55, 245–256. doi: 10.1139/g2012-009
Hussin, S. H., Wang, H., Tang, S., Zhi, H., Tang, C., Zhang, W., et al. (2021). SiMADS34, an e-class MADS-box transcription factor, regulates inflorescence architecture and grain yield in Setaria italica. Plant Mol. Biol. 105, 419–434. doi: 10.1007/s11103-020-01097-6
Kim, S., Koh, J., Yoo, M. J., Kong, H., Hu, Y., Ma, H., et al. (2005). Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 43, 724–744. doi: 10.1111/j.1365-313X.2005.02487.x
Lee, J., Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Leijten, W., Koes, R., Roobeek, I., Frugis, G. (2018). Translating flowering time from Arabidopsis thaliana to brassicaceae and asteraceae crop species. Plants 7, 111. doi: 10.3390/plants7040111
Li, Z. M., Zhang, J. Z., Mei, L., Deng, X. X., Hu, C. G., Yao, J. L. (2010). PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata l. raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Mol. Biol. 74, 129–142. doi: 10.1007/s11103-010-9660-1
Litt, A. (2007). An evaluation of a-function: evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 168, 73–91. doi: 10.1086/509662
Liu, B., Yan, J., Li, W., Yin, L., Li, P., Yu, H., et al. (2020). Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 11, 1–13. doi: 10.1038/s41467-019-13926-4
Locascio, A., Lucchin, M., Varotto, S. (2009). Characterization of a MADS FLOWERING LOCUS c-LIKE (MFL) sequence in cichorium intybus: a comparative study of CiMFL and AtFLC reveals homologies and divergences in gene function. New Phytol. 182, 630–643. doi: 10.1111/j.1469-8137.2009.02791.x
Lu, C., Qu, J., Deng, C., Liu, F., Zhang, F., Huang, H., et al. (2022). The transcription factor complex CmAP3-CmPI-CmUIF1 modulates carotenoid metabolism by directly regulating the carotenogenic gene CmCCD4a-2 in chrysanthemum. Hortic. Res. 9. doi: 10.1093/hr/uhac020
Melzer, R., Wang, Y. Q., Theiβen, G. (2010). The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Sem. Cell Dev. Biol. 21, 118–128. doi: 10.1016/j.semcdb.2009.11.015
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Bio. Evol. 37 (5), 1530–1534. doi: 10.1093/molbev/msaa131
Mizzotti, C., Ezquer, I., Paolo, D., Rueda-Romero, P., Guerra, R. F., Battaglia, R., et al. (2014). SEEDSTICK is a master regulator of development and metabolism in the arabidopsis seed coat. PloS Genet. 10 (12), e1004856. doi: 10.1371/journal.pgen.1004856
Nakano, M., Hirakawa, H., Fukai, E., Toyoda, A., Kajitani, R., Minakuchi, Y., et al. (2021). A chromosome-level genome sequence of chrysanthemum seticuspe, a model species for hexaploid cultivated chrysanthemum. Commun. Biol. 4 (1), 1167. doi: 10.1038/s42003-021-02704-y
Nishiyama, S., Matsushita, M. C., Yamane, H., Honda, C., Okada, K., Tamada, Y., et al. (2021). Functional and expressional analyses of apple FLC-like in relation to dormancy progress and flower bud development. Tree Physiol. 41, 562–570. doi: 10.1093/treephys/tpz111
Pelaz, S., Gustafson-Brown, C., Kohalmi, S. E., Crosby, W. L., Yanofsky, M. F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. doi: 10.1046/j.1365-313X.2001.2641042.x
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Qi, F., Liu, Y., Luo, Y., Cui, Y., Lu, C., Li, H., et al. (2022). Functional analysis of the ScAG and ScAGL11 MADS-box transcription factors for anthocyanin biosynthesis and bicolour pattern formation in Senecio cruentus ray florets. Hortic. Res. 9. doi: 10.1093/hr/uhac071
Qi, S., Yang, L., Wen, X., Hong, Y., Song, X., Zhang, M., et al. (2016). Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00287
Ren, Z., Yu, D., Yang, Z., Li, C., Qanmber, G., Li, Y., et al. (2017). Genome-wide identification of the MIKC-type MADS-box gene family in Gossypium hirsutum l. unravels their roles in flowering. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00384
Reyes-Chin-Wo, S., Wang, Z., Yang, X., Kozik, A., Arikit, S., Song, C., et al. (2017). Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 8 (1), 1–11. doi: 10.1038/ncomms14953
Rounsley, S. D., Ditta, G. S., Yanofsky, M. F. (1995). Diverse roles for MADS box genes in arabidopsis development. Plant Cell 7, 1259–1269. doi: 10.1105/tpc.7.8.1259
Ruokolainen, S., Ng, Y. P., Albert, V. A., Elomaa, P., Teeri, T. H. (2010a). Large Scale interaction analysis predicts that the gerbera hybrida floral e function is provided both by general and specialized proteins. BMC Plant Bio. 10, 1–13. doi: 10.1186/1471-2229-10-129
Ruokolainen, S., Ng, Y. P., Albert, V. A., Elomaa, P., Teeri, T. H. (2011). Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann. Bot. 107, 1491–1499. doi: 10.1093/aob/mcr112
Ruokolainen, S., Ng, Y. P., Broholm, S. K., Albert, V. A., Elomaa, P., Teeri, T. H. (2010b). Characterization of SQUAMOSA-like genes in gerbera hybrida, including one involved in reproductive transition. BMC Plant Bio. 10, 1–11. doi: 10.1186/1471-2229-10-128
Song, C., Liu, Y., Song, A., Dong, G., Zhao, H., Sun, W., et al. (2018). The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol. Plant 11, 1482–1491. doi: 10.1016/j.molp.2018.10.003
Theiβen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Münster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Theiβen, G., Melzer, R., Rümpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143 (18), 3259–3271. doi: 10.1242/dev.134080
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Uimari, A., Kotilainen, M., Elomaa, P., Yu, D., Albert, V. A., Teeri, T. H. (2004). Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc. Natl. Acad. Sci. U.S.A. 101, 15817–15822. doi: 10.1073/pnas.040684410
van Lieshout, N., van Kaauwen, M., Kodde, L., Arens, P., Smulders, M. J., Visser, R. G., et al. (2022). De novo whole-genome assembly of Chrysanthemum makinoi, a key wild chrysanthemum. G3 12 (1), jkab358. doi: 10.1093/g3journal/jkab358
Vijverberg, K., Welten, M., Kraaij, M., van Heuven, B. J., Smets, E., Gravendeel, B. (2021). Sepal identity of the pappus and floral organ development in the common dandelion (Taraxacum officinale; asteraceae). Plants 10, 1682. doi: 10.3390/plants10081682
Wang, Y., Huang, H., Ma, Y., Fu, J., Wang, L., Dai, S. (2014). Construction and de novo characterization of a transcriptome of Chrysanthemum lavandulifolium: analysis of gene expression patterns in floral bud emergence. Plant Cell Tissue Organ Cult. 116, 297–309. doi: 10.1007/s11240-013-0404-1
Wang, J., Jiu, S., Xu, Y., Sabir, I. A., Wang, L., Ma, C., et al. (2021). SVP-like gene PavSVP potentially suppressing flowering with PavSEP, PavAP1, and PavJONITLESS in sweet cherries (Prunus avium l.). Plant Physiol. Bioch. 159, 277–284. doi: 10.1016/j.plaphy.2020.12.013
Wen, X., Li, J., Wang, L., Lu, C., Gao, Q., Xu, P., et al. (2022). The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Horti. Res. 9. doi: 10.1093/hr/uhab022
Wen, X., Qi, S., Huang, H., Wu, X., Zhang, B., Fan, G., et al. (2019a). The expression and interactions of ABCE-class and CYC2-like genes in the capitulum development of Chrysanthemum lavandulifolium and C.× morifolium. Plant Growth Regul. 88, 205–214. doi: 10.1007/s10725-019-00491-5
Wen, X., Qi, S., Yang, L., Hong, Y., Dai, S. (2019b). Expression pattern of candidate genes in early capitulum morphogenesis of Chrysanthemum lavandulifolium. Sci. Hortic. 252, 332–341. doi: 10.1016/j.scienta.2019.03.064
Wollmann, H., Mica, E., Todesco, M., Long, J. A., Weigel, D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633–3642. doi: 10.1242/dev.036673
Wu, R., Tomes, S., Karunairetnam, S., Tustin, S. D., Hellens, R. P., Allan, A. C., et al. (2017). SVP-like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00477
Yang, Y., Sun, H. (2009). The bracts of Saussurea velutina (Asteraceae) protect inflorescences from fluctuating weather at high elevations of the hengduan mountains, southwestern China. Arct. Antarct. Alp. Res. 41, 515–521. doi: 10.1657/1938-4246-41.4.515
Yu, D., Kotilainen, M., Pöllänen, E., Mehto, M., Elomaa, P., Helariutta, Y., et al. (1999). Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J. 17, 51–62. doi: 10.1046/j.1365-313X.1999.00351.x
Zhang, L., Chen, F., Zhang, X., Li, Z., Zhao, Y., Lohaus, R., et al. (2020). The water lily genome and the early evolution of flowering plants. Nature 577, 79–84. doi: 10.1038/s41586-019-1852-5
Zhang, T., Elomaa, P. (2021). Don’t be fooled: false flowers in asteraceae. Curr. Opin. Plant Biol. 59, 101972. doi: 10.1016/j.pbi.2020.09.006
Zhang, C., Wei, L., Wang, W., Qi, W., Cao, Z., Li, H., et al. (2020). Identification, characterization and functional analysis of AGAMOUS subfamily genes associated with floral organs and seed development in marigold (Tagetes erecta). BMC Plant Bio. 20, 1–17. doi: 10.1186/s12870-020-02644-5
Zhang, T., Zhao, Y., Juntheikki, I., Mouhu, K., Broholm, S. K., Rijpkema, A. S., et al. (2017). Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol. 216, 939–954. doi: 10.1111/nph.14707
Keywords: chrysanthemum, Chrysanthemum lavandulifolium, capitulum development, MIKCc-type gene family, complex inflorescences
Citation: Li J, Zhang Q, Kong D, Pu Y, Wen X and Dai S (2023) Genome-wide identification of the MIKCc-type MADS-box gene family in Chrysanthemum lavandulifolium reveals their roles in the capitulum development. Front. Plant Sci. 14:1153490. doi: 10.3389/fpls.2023.1153490
Received: 29 January 2023; Accepted: 14 March 2023;
Published: 23 March 2023.
Edited by:
Lin Xi, University of Hohenheim, GermanyReviewed by:
Xu Xiaozhao, China Agricultural University, ChinaCopyright © 2023 Li, Zhang, Kong, Pu, Wen and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silan Dai, c2lsYW5kYWlAc2luYS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.