- 1State Key Laboratory of Genetic Engineering and Engineering Research Center of Gene Technology (Ministry of Education), School of Life Sciences, Fudan University, Shanghai, China
- 2Ministry of Education Key Laboratory for Biodiversity Science and Ecological Engineering, Department of Ecology and Evolutionary Biology, School of Life Sciences, Fudan University, Shanghai, China
- 3Institute of Crop Breeding and Cultivation, Shanghai Academy of Agricultural Sciences, Shanghai, China
- 4Ministry of Education, Key Laboratory of Crop Physiology, Ecology and Genetic Breeding College of Agronomy, Jiangxi Agricultural University, Nanchang, China
Plant height and grain size are important agronomic traits affecting rice yield. Various plant hormones participate in the regulation of plant height and grain size in rice. However, how these hormones cooperate to regulate plant height and grain size is poorly understood. In this study, we identified a brassinosteroid-related gene, hfr131, from an introgression line constructed using Oryza longistaminata, that caused brassinosteroid insensitivity and reduced plant height and grain length in rice. Further study showed that hfr131 is a new allele of OsBRI1 with a single-nucleotide polymorphism (G to A) in the coding region, leading to a T988I conversion at a conserved site of the kinase domain. By combining yeast one-hybrid assays, chromatin immunoprecipitation-quantitative PCR and gene expression quantification, we demonstrated that OsARF17, an auxin response factor, could bind to the promoter region of HFR131 and positively regulated HFR131 expression, thereby regulating the plant height and grain length, and influencing brassinosteroid sensitivity. Haplotype analysis showed that the consociation of OsAFR17Hap1/HFR131Hap6 conferred an increase in grain length. Overall, this study identified hfr131 as a new allele of OsBRI1 that regulates plant height and grain length in rice, revealed that brassinosteroid and auxin might coordinate through OsARF17–HFR131 interaction, and provided a potential breeding target for improvement of rice yield.
Introduction
Rice is among the most important food crops worldwide, providing staple food for more than half of the world’s population (Qiao et al., 2021; Huang et al., 2022). However, the global population is growing rapidly and is forecast to attain 9.7 billion by 2050 and 10.4 billion by 2100 (Economic and Social Affairs, 2022). Furthermore, with ongoing economic development, the environment is deteriorating, and the arable land area is decreasing. Therefore, it is particularly important to increase the rice yield per unit land area to keep pace with the projected population growth (McClung, 2014).
Plant height and grain size are two important factors affecting rice yield. An excessively high or low plant height will affect rice yield. A plant that is too tall has poor lodging resistance, whereas if the plant is too short, the plant will produce smaller grains, an increased number of ineffective tillers, and have poor resistance to disease (Liu et al., 2018). Therefore, an appropriate plant height is essential to improve rice yield. This is comparable to the “Green Revolution” wave of semi-dwarf breeding in the 1960s, which doubled food production in much of the world (Hargrove and Cabanilla, 1979; Khush, 1999). The yield per plant of rice is directly affected by three factors: panicle number, number of grains per panicle, and thousand-grain weight. The thousand-grain weight is positively associated with grain size, including grain length, grain width, grain thickness, and degree of grain filling (Xing and Zhang, 2010). Thus, grain size is among the most agronomically important traits in rice breeding.
Plant hormones play crucial roles in the regulation of plant height and grain size in rice, among which brassinosteroids (BRs) are particularly important. Similar in structure to mammalian steroid hormones, BRs are a class of polyhydroxysteroid plant hormones and are involved in multiple biological processes in growth and development (Clouse, 2011). Many BR-related genes have been reported in rice, and most affect both plant height and grain size. The BR synthesis-associated mutants brd2, d11, d2, and brd1 have a reduced plant height and grain size to different degrees (Mori et al., 2002; Hong et al., 2003; Hong et al., 2005; Tanabe et al., 2005). OsBRI1, the rice ortholog of Arabidopsis BRI1, encodes the receptor of BR in rice. Loss-of-function of OsBRI1 results in dwarf culms, reduced grain size, erect leaves, and insensitivity to BR (Yamamuro et al., 2000; Morinaka et al., 2006). OsBAK1 interacts with OsBRI1 to promote BR signal transduction downstream. Overexpression of OsBAK1 increases sensitivity to BR, however, reduces plant height and grain size, which may be caused by repressing GA level (Li et al., 2009; Tong et al., 2014). OsGSK1–OsGSK4, which are homologs of AtBIN2, belong to the GSK3-like kinase family and play an important role in the regulation of BR signaling (Yoo et al., 2006). Loss-of-function of one or several OsGSK genes has revealed that the gsk2, gsk1,2, gsk2,3, and gsk2,4 mutants show increased plant height, whereas the gsk2,3,4 mutant has decreased plant height and that of the gsk1,2,3,4 mutant is not changed significantly. However, the grain length and thousand-grain weight of all types of mutants is significantly increased (Liu et al., 2021). The mutant or knockout lines of the BR-associated transcription factors OsBZR1, OsLIC, and DLT/GS6 all show significant dwarfing; however, the grain size and yield of the mutant or knockout lines of OsBZR1 and OsLIC are significantly decreased, whereas those of the gs6 mutant are significantly increased (Bai et al., 2007; Wang et al., 2008; Tong et al., 2009; Sun et al., 2013; Zhu et al., 2015). Other BR regulators, such as OsBU1 and SG1, are also involved in the regulation of plant height and grain shape (Tanaka et al., 2009; Nakagawa et al., 2011). BR plays an important role in the regulation of multiple phenotypes in rice, such as plant height, grain size, seed germination, etc, involved in complex molecular regulatory networks (Xiao et al., 2020; Niu et al., 2022; Xiong et al., 2022), which is worth of further study. In addition, multiple hormones regulate the plant height and grain size of rice, but how BR coordinates with other hormones requires further study.
In this study, we identified a gene, HFR131, that regulates plant height and grain length in rice. Further study of the hfr131 mutant demonstrated that hfr131 is a new allele of OsBRI1. The replacement of amino acids in the kinase domain of HFR131 caused dwarfing and reduced grain length in the near-isogenic line NIL-hfr131. The auxin response factor OsARF17 was found to regulate plant height and grain length through BR signaling by binding to the promoter of HFR131 and thereby regulating its expression. In addition, the consociation of OsAFR17Hap1/HFR131Hap6 was confirmed to increase the grain length.
Materials and methods
Plant materials and growth conditions
Using wild rice Oryza longistaminata as the donor parent (♀) and indica restorer rice ‘187R’, cultivated by Guangxi Academy of Agricultural Sciences, as the recipient parent (♂), the BC4F1 population was obtained through hybridization and backcrossing with 187R (♂) for four successive generations and the BC4F4 population was obtained by selfing the BC4F1 population for three successive generations. Then we identified the genotype of BC4F4 population with genetic markers and a set of introgression lines were constructed (Figure 1A). Among these lines, by phenotype identification, we identified an introgression line, IL-hfr131, that exhibited significantly reduced plant height, small grain size and erect leaf. The alleles in IL-hfr131 and 187R were therefore designated hfr131 and HFR131, respectively. Subsequently, we backcrossed IL-hfr131 (♀) with its recurrent parent 187R (♂) for four successive generations and selfed for one generation, then identified the genotype of the offspring with genetic markers, obtaining a near-isogenic line, NIL-hfr131, with an introgression fragment on the long arm of chromosome 1 (Supplementary Figure 1). Next, NIL-hfr131 was used to construct a F2 population comprising approximately 4000 plants for fine-mapping and phenotypic analysis (Figure 1A). The plant height (n = 13), leaf angle (n = 10), internode length (n = 13) and panicle length (n = 13) of 187R and NIL-hfr131 were measured. For grain length, grain width and grain weight, each variable was measured on three groups with 50 seeds per group.
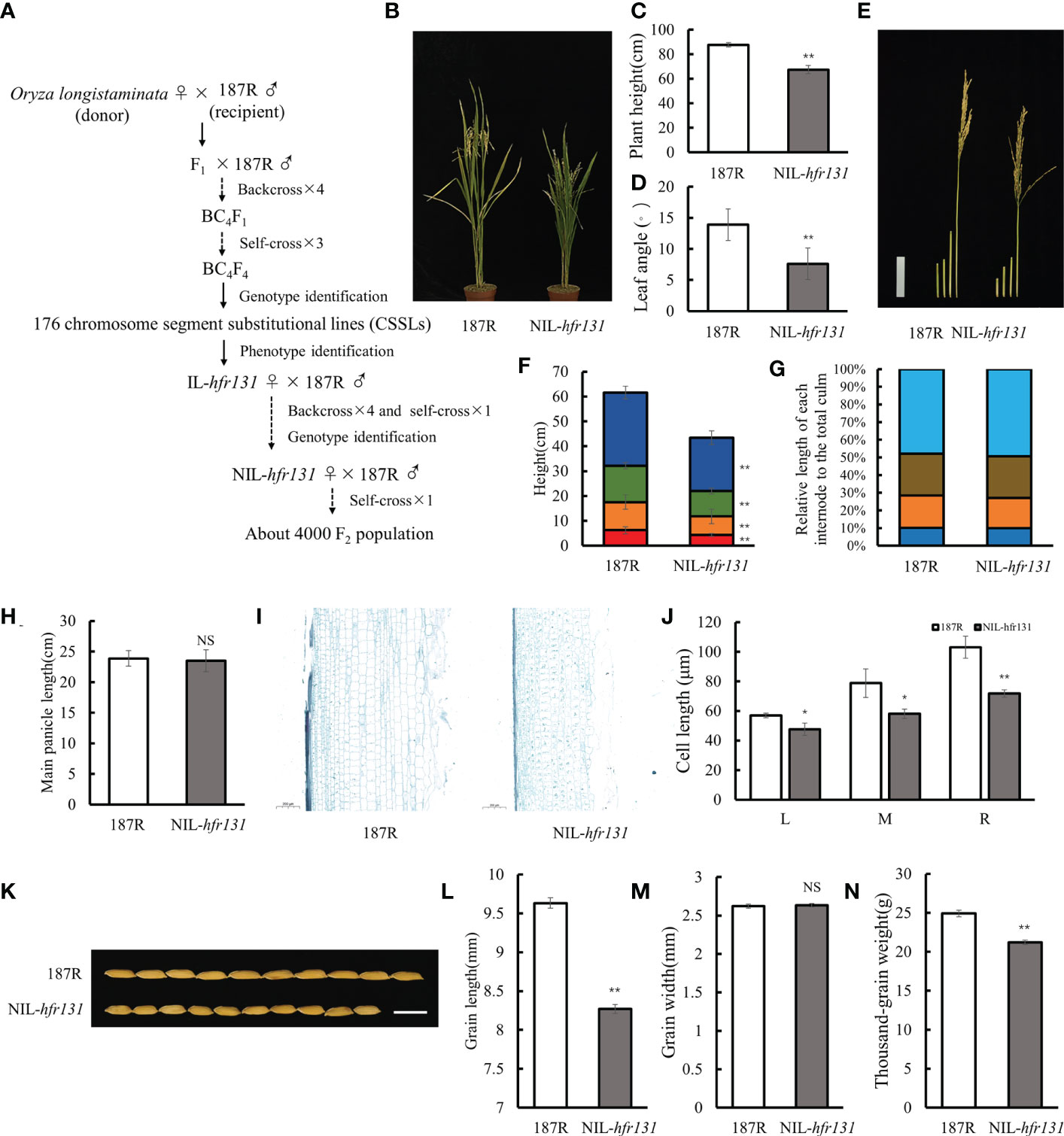
Figure 1 Phenotypes of rice ‘187R’ and NIL-hfr131. (A) Construction of infiltrating lines and F2 population. (B–D) Plant architecture, plant height, and the third leaf angle of 187R and NIL-hfr131. (E–G) Internode morphology (bar = 10 cm), internode length, and internode elongation pattern of 187R and NIL-hfr131. (H) Panicle length of 187R and NIL-hfr131. (I, J) Longitudinal sections of the stem and internode cell length (L, M, and R refer to the left, middle, and right side of the section, respectively) of 187R and NIL-hfr131 (bar = 200 μm). (K–N) Grain morphology (bar = 1 cm), grain length, grain width, and thousand-grain weight of 187R and NIL-hfr131. * P < 0.05, ** P < 0.01, NS non-significant.
All rice materials in this study were planted in Shanghai (31°20′26″ N, 121°30′26″ E) in summer and in Hainan (18°18′52″ N, 109°03′05″ E) in winter. Standard field management practices were applied for all lines.
Histological analysis
The second internode of rice stems was sampled and fixed with 50% FAA solution (50 mL anhydrous ethanol, 10 mL 37% formaldehyde, 5 mL glacial acetic acid, 35 mL ddH2O). Histological sections of the samples were prepared by the Wuhan Misp Biotechnology Co., Ltd. (Wuhan, China). The internode cell length of the left, middle, and right side of the section were measured and three layers of cells were measured in each region.
BR treatment
Rice seeds were cultured in 0.5% agar semi-solid medium containing 0 or 1 μM/L 24-epibrassinolide (24-eBL) (Zhang et al., 2015), which is commonly used to test the responsiveness of plants to BR, for 1 week in a greenhouse at 26 °C under shading. The coleoptile length and root length were then measured (n = 10). The change in root length was calculated as a measure of the degree of inhibition of root elongation (n = 10) using the following formula: (root length with 1 μM/L 24-eBL treatment − root length without 24-eBL treatment)/root length without 24-eBL treatment.
Map-based cloning
Total DNA was extracted from leaf samples. The corresponding markers were amplified by PCR, and the PCR products were separated by polyacrylamide gel vertical electrophoresis. The primers used for fine mapping are listed in Supplementary Table 3.
RNA isolation and quantitative real-time PCR
Total RNA from rice samples was extracted using the FastPure® Plant Total RNA Isolation Kit (Vazyme, Nanjing, China). First-strand cDNA was generated by reverse transcription using the PrimeScript™ RT Reagent Kit (Takara, Otsu, Japan). Quantitative real-time PCR (qPCR) was conducted using TB Green Premix Ex Taq II (Tli RNaseH Plus) (Takara) and a CFX96 Real-Time PCR Detection system (Bio-Rad, Hercules, CA, USA). The reactions were conducted with 40 cycles of 95°C for 5 s and 60°C for 30 s. Melting curves were prepared and analyzed. OsActin or OsUBQ5 was used as an internal reference gene. Three biological replicates were performed for each sample. The primers used are listed in Supplementary Table 3.
Dual-luciferase reporter assay
The promoter fragment of HFR131 in the same region of 187R (3500 bp) and NIL-hfr131 (3067 bp) was cloned and inserted into the pGreenII 0800-LUC vector using seamless cloning technology to construct the recombinant vectors proHFR131-LUC and prohfr131-LUC. Seeds of rice ‘9311’ were germinated on wet napkins at 37 °C for 14 days. The stem of the seedlings was cut into thin slices and protoplasts were prepared by hydrolyzing the cell walls with cellulase and macerozyme. The recombinant vectors were transferred into the ‘9311’ protoplasts using 40% polyethylene glycol solution and cultured at 28 °C for 12–16 h. The protoplast lysate was collected and the promoter activity of the HFR131 promoter was tested using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) and a Synergy™ 2 Multi-Mode Reader (BioTek, Winooski, VT, USA). The activation efficiency of luciferase was expressed as the ratio of firefly luciferase (LUC) to Renilla luciferase (REN); the higher the ratio, the stronger the promoter activity. Three technical replicates were performed for each sample. The primers used are listed in Supplementary Table 3.
Transcriptome analysis
Total RNA was extracted from young panicles (n = 3) before heading date of 187R and NIL-hfr131 using the FastPure® Plant Total RNA Isolation Kit (Vazyme). The extracted RNA was used as the template for construction of cDNA libraries, which were sequenced using a MGI MGISEQ-2000 platform by Sangon Biotech (Shanghai, China). The relative gene expression level was calculated and normalized to fragments per kilobase of transcript per million mapped reads. Differentially expressed genes (DEGs) were annotated and classified according to their putative or proven function. The transcriptome datasets were submitted to the GEO database as accession nos. GSE224118.
Construction of transformation lines
Overexpression lines of HFR131 were based on the NIL-hfr131 mutant background. For the coding sequences (CDSs) of OsARF17 and the promoter sequences of HFR131 in ‘ZH11’ and 187R were consistent, respectively, and ZH11 had high seed setting rate, which helped us investigate the phenotype of the grains well, we construct OsARF17 knockout and overexpression lines based on ZH11 background. CRISPR/Cas9 knockout lines of OsARF17 were generated by BIOGLE GeneTech (Jiangsu, China), and the target sequence was 5′- CCAATTATCCTAACTTGCCTCCACAACTTATTTGCCAACTTCACAATGTGACG-3′. All knockout lines of the target sequence were cloned by PCR and sequenced. Regarding the overexpression lines, the CDSs of OsARF17 (LOC_Os06g46410.1) and HFR131 (LOC_Os01g52050.1) were cloned and inserted into the PCHF-FLAG vector to construct the overexpression vectors CaMV35-OsARF17-FLAG and CaMV35S-HFR131-FLAG using a seamless cloning technique. CaMV35S-OsARF17-FLAG was transformed by BIOGLE GeneTech and CaMV35S-HFR131-FLAG was transformed by BioRun (Wuhan, China). All transformed lines were verified by qPCR, and eight biological replicates were performed for OsARF17 overexpression lines (T2 generation) and three technical replicates were performed for HFR131 overexpression lines (T0 generation). The primers used are listed in Supplementary Table 3. The plant height, grain length and grain width of transformation lines were measured. Ten biological replicates were performed for T3 generation of OsARF17 knockout lines and T2 generation of OsARF17 overexpression lines. For the grain length and thousand-grain weight of T0 generation of HFR131 overexpression lines, each variable was measured on three groups with 20 seeds per group.
Yeast one-hybrid assay
Eight 150–300 bp DNA fragments containing an auxin response element (AuxRE) in the 3500 bp promoter region of HFR131 were cloned and inserted into the pAbAi vector. The recombinant vectors were transferred into the yeast strain Y1H Gold, and cultured and screened on SD−Ura solid medium. The positive clones were detected by colony PCR using the primers Y1-test and pAbAi (Supplementary Table 3). The positive colonies were diluted and cultured in SD−Ura solid medium containing aureobasidin A (ABAr) at different concentrations ranging from 0 to 1000 ng/ml. The culture with the optimal concentration of ABAr was selected to eliminate self-activation. The CDS of OsARF17 was cloned by PCR and inserted into the pGADT7 vector to construct the AD-ARF17 recombinant vector, which was transformed into yeast Y1H Gold containing one of the DNA fragments mentioned above. The transformed Y1H Gold cells were cultured on SD−Leu solid medium containing the selected ABAr concentration. The empty pGADT7 vector, KAD, was transformed as a negative control. The Yeastmaker Yeast Transformation System 2 (Clontech) was used following the manufacturer’s instructions. The primers used are listed in Supplementary Table 3.
ChIP-qPCR
Approximately 2.5 g rice seeds were weighed, soaked in water at 37 °C for 24 h, and then sown in nutrient soil 48 h after germination. The seeds were incubated at 26 °C for 14 days and then used for chromatin immunoprecipitation–quantitative real-time PCR (ChIP-qPCR). The experimental procedure followed the method of Wang et al. (Wang et al., 2018). The antibody used in this study was the FLAG antibody and the magnetic beads used were protein A/G magnetic beads. OsActin was used as an internal standard for data normalization. Three technical replicates were performed for each sample. The primers used are listed in Supplementary Table 3.
Haplotype analysis
A haplotype network for HFR131 and OsARF17 based on pairwise differences between two adjacent gene–CDS–haplotypes (gcHaps) was constructed using the pegas R package. The gcHaps data used for building the network were downloaded from the Rice Functional Genomics and Breeding (RFGB) database (https://www.rmbreeding.cn). The 3K Phenotype data were downloaded from RFGB (https://www.rmbreeding.cn/Phenotype). One-way analysis of variance (ANOVA) of the gcHap data was conducted for three agronomic traits (plant height, grain length, and thousand-grain weight), followed by Duncan’s multiple-range test for multiple comparisons using the R package agricolae (v.1.3-5). Two-way ANOVA of the gcHaps data was conducted for analysis of grain length, followed by Duncan’s multiple-range test for multiple comparisons using the R package agricolae (v.1.3-5). The probability level P < 0.05 was considered to be statistically significant. The multiple comparisons and the distribution of gcHaps among the five major populations, Aus, Bas, XI, GJ and Adm, were visualized using the R package ggplot2 (v.3.4.0) as boxplots and stacked barplots. The single-nucleotide polymorphism (SNP) dataset used for calculation of nucleotide diversity (π) and Tajima’s D was downloaded from the Rice SNP-Seek Database (https://snp-seek.irri.org/_download.zul). Nucleotide diversity (π) at each window in the target region was estimated using VCFtools (v.0.1.15) with the “–window-pi” and “–window-pi-step” options, and the average value was used as its p-value. Tajima’s D for the target region was calculated using VCFtools (v.0.1.15) with the “–TajimaD” option.
Results
The near-isogenic line NIL-hfr131 exhibited reduced plant height and grain length
Compared with cultivated rice, wild rice has a larger gene pool, which allows adaptation to different environments and is a valuable resource for gene functional research in rice. In this study, we identified an introgression line, IL-hfr131, with significantly reduced plant height and grain size using wild rice Oryza longistaminata as the donor parent (♀) and indica restorer rice 187R as the recipient parent (♂) (Figure 1A), and further constructed a near-isogenic line, NIL-hfr131, containing the gene hfr131 (Supplementary Figure 1). Subsequently, a F2 population containing approximately 4000 plants was constructed using NIL-hfr131 (♀) and 187R (♂) (Figure 1A).
Phenotypic investigation showed that the ratio of dwarf and small-grained plants to high and big-grained plants in the F2 population was close to 1:3, indicating that hfr131 was a recessive gene. Compared with 187R, NIL-hfr131 showed a dwarf plant height accompanied by erect leaves (Figures 1B–D). To determine the direct cause of the plant height reduction, we measured the length of the first four internodes (counting the internode directly connected to the panicle as the first internode) and the panicle length of 187R and NIL-hfr131. The results showed that each internode of NIL-hfr131 was significantly shortened to equal proportions (Figures 1E–G). However, there was no significant difference in panicle length between 187R and NIL-hfr131 (Figure 1H), indicating that the reduced plant height of NIL-hfr131 was caused by the equally proportional shortening of the internodes. The length of the internode is directly related to the growth and development of the internode cells. To further clarify the mechanism of internode shortening of NIL-hfr131, longitudinal sections of the internodes of NIL-hfr131 and 187R were prepared. The results showed that the stem cell length of NIL-hfr131 was significantly shortened both externally and internally (Figures 1I, J), indicating that the reduced internode length of NIL-hfr131 was mainly caused by shortening of the cell length in the stem. Although the panicle length of NIL-hfr131 was not changed significantly, the grain size was significantly decreased. Compared with 187R, the grain length and thousand-grain weight of NIL-hfr131 were significantly decreased, but there was no significant difference in grain width (Figures 1K–N). Taken together, these results showed that the reduced plant height and grain size of NIL-hfr131 was caused by shortening of the internodes and grain length, respectively.
NIL-hfr131 is insensitive to brassinosteroid
Given that NIL-hfr131 plants were reduced in height and had erect leaves, which is similar to the mutant phenotype of genes associated with the BR synthesis and signaling pathways (Hong et al., 2005; Nakamura et al., 2006; Tong et al., 2009), we speculated that HFR131 may be involved in the BR synthesis or signaling pathways. To test this hypothesis, we conducted a BR treatment assay. The results showed that the coleoptile length of 187R was significantly increased after 24-eBL treatment, whereas NIL-hfr131 showed no significant change (Figures 2A, B). Furthermore, after 24-eBL treatment, the root length of 187R was extremely reduced (Figure 2A). Although the root of NIL-hfr131 also showed significant shortening, the inhibitory effect of 24-eBL on NIL-hfr131 was weaker than on 187R (Figures 2A–C). In conclusion, NIL-hfr131 was insensitive to BR, suggesting that HFR131 was involved in the BR signaling pathway rather than the BR synthesis pathway.
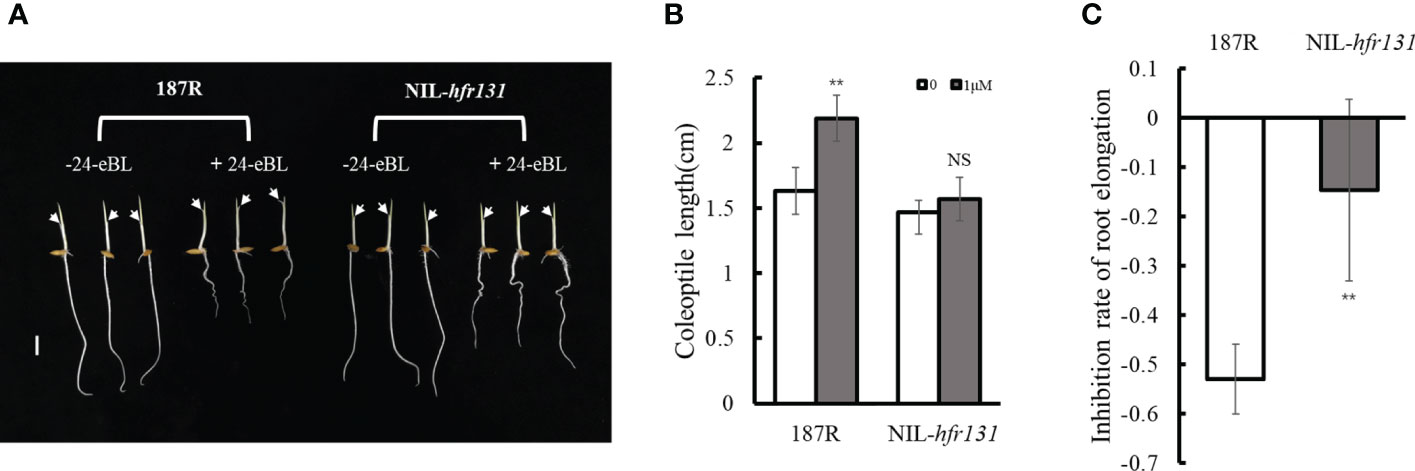
Figure 2 Responses of rice ‘187R’ and NIL-hfr131 to 24-epibrassinolide (24-eBL). (A) Effect of 24-eBL on coleoptile and root elongation in seedlings. The arrows indicate the top of the coleoptile. Bar = 1 cm. (B) Coleoptile length of 187R and NIL-hfr131 seedlings in the presence or absence of 1 μM/L 24-eBL. (C) Inhibition of root elongation of 187R and NIL-hfr131 in response to 1 μM/L 24-eBL. ** P < 0.01, NS non-significant.
hfr131 is a new allele of OsBRI1
To map the location of the HFR131 locus, we constructed two pools comprising 36 dwarf small-grained plants (DS; recessive plants) and 36 tall large-grained plants (HB; dominant plants), selected from the F2 population, and analyzed the two pools using simple sequence repeat (SSR) markers distributed on chromosome 1. The markers RM1297 and RM5931 showed polymorphism between the pools, and exhibited markedly different banding patterns between recessive individuals and dominant individuals (Supplementary Figure 2), indicating the two markers were closely linked to HFR131. Combining these two markers and neighboring SSR markers for further analysis, we preliminarily narrowed the effective locus to the interval between markers RM5931 and RM1152 (Figure 3A).
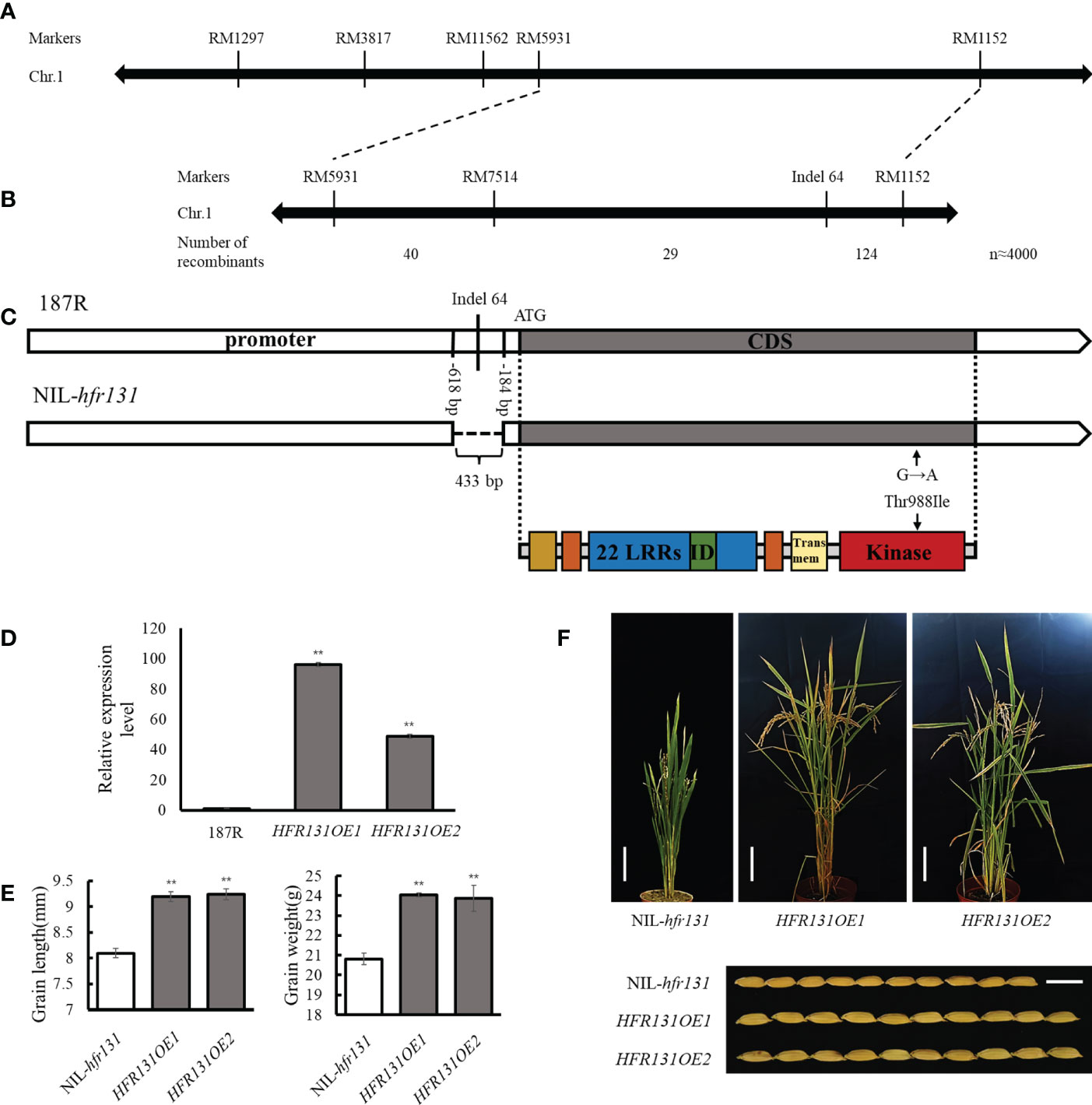
Figure 3 Identification of HFR131. (A) Linkage map of HFR131. (B) Fine mapping of HFR131 using almost 4000 F2 plants. The number of recombinants between adjacent markers is indicated under the linkage map. (C) Gene structure of OsBRI1 in rice ‘187R’ and NIL-hfr131, and the corresponding protein structure redrawn from Nakamura et al. (Nakamura et al., 2006). Positions of the 433 bp deletion are presented by base number before the ATG. (D) Transgene expression level of OsBRI1/HFR131. OsActin was used as an internal reference. (E) Grain length and thousand-grain weight of NIL-hfr131 and HFR131 overexpression lines (T0 generation). (F) Plant architecture (Bar = 10 cm) and grain morphology (Bar = 1 cm) of NIL-hfr131 and HFR131 overexpression lines (T0 generation). The plant height of NIL-hfr131, HFR131OE1 and HFR131OE2 is 50.9 cm, 68 cm and 66 cm, respectively. ** P < 0.01.
To fine map HFR131, we used RM5931 and RM1152 to screen 926 recessive plants selected from the F2 population of almost 4000 plants, and 193 recombinants were identified. The SSR marker RM7514, located between RM5931 and RM1152, was used to analyze the 193 recombinants. The HFR131 locus was located in the region between RM7514 and RM1152. In addition, Indel 64 was designed based on sequence differences between 187R and NIL-hfr131, and was found to co-segregate with NIL-hfr131 (Figure 3B), indicating that HFR131 was located at or adjacent to the site of Indel 64. From a BLAST search of the National Center for Biotechnology Information databases (https://www.ncbi.nlm.nih.gov/), we found that Indel 64 was located in the promoter region of OsBRI1, implying that OsBRI1 may be the candidate gene for HFR131.
We then sequenced the promoter and coding regions of OsBRI1 of 187R and NIL-hfr131. Two main mutant sites were found. One mutation was a 433 bp deletion in the promoter region, and the other was a SNP (G to A transition) in the coding region, converting threonine to isoleucine at position 988 in the kinase domain of OsBRI1 (Figure 3C). These results indicated that HFR131 may be OsBRI1.
To investigate whether OsBRI1 correspond to the phenotypic change in NIL-hfr131, we overexpressed the wild-type OsBRI1, cloned from 187R, in NIL-hfr131 (Figure 3D). The results showed that, compared with NIL-hfr131, the plant height, leaf angle, grain length, and thousand-grain weight of the overexpression lines (HFR131OE1 and HFR131OE2) were significantly increased (Figures 3E, F), indicating that HFR131 is OsBRI1 and that hfr131 is a new allele of OsBRI1.
Amino acid non-synonymous substitution in the HFR131 kinase domain resulted in the reduction of plant height and grain length
Given that there were two mutation sites in hfr131, to determine which of the sites (the SNP in the coding region or the 433 bp deletion in the promoter region) was responsible for the height reduction and short grain length of NIL-hfr131, we first detected the expression levels of HFR131 in the stem and leaf of 187R and NIL-hfr131 by qPCR. Deletion of large fragments in the promoter region often leads to a significant reduction in the gene transcript level, resulting in abnormal gene function. Interestingly, compared with 187R, expression levels of HFR131 in the stem and leaf of NIL-hfr131 were not decreased, but in both were up-regulated significantly (Figure 4A). To further verify whether the 433 bp deletion in the HFR131 promoter enhanced the promoter activity, we conducted a dual-luciferase reporter assay. We constructed two recombinant vectors, proHFR131-LUC and prohfr131-LUC, and transferred them into rice protoplasts. The empty vector was transferred as a negative control. The results showed that the luciferase activation efficiency of the prohfr131-LUC construct was significantly higher than that of the proHFR131-LUC construct (Figure 4B). This result suggested that the 433 bp deletion in the HFR131 promoter region did enhance the promoter activity.
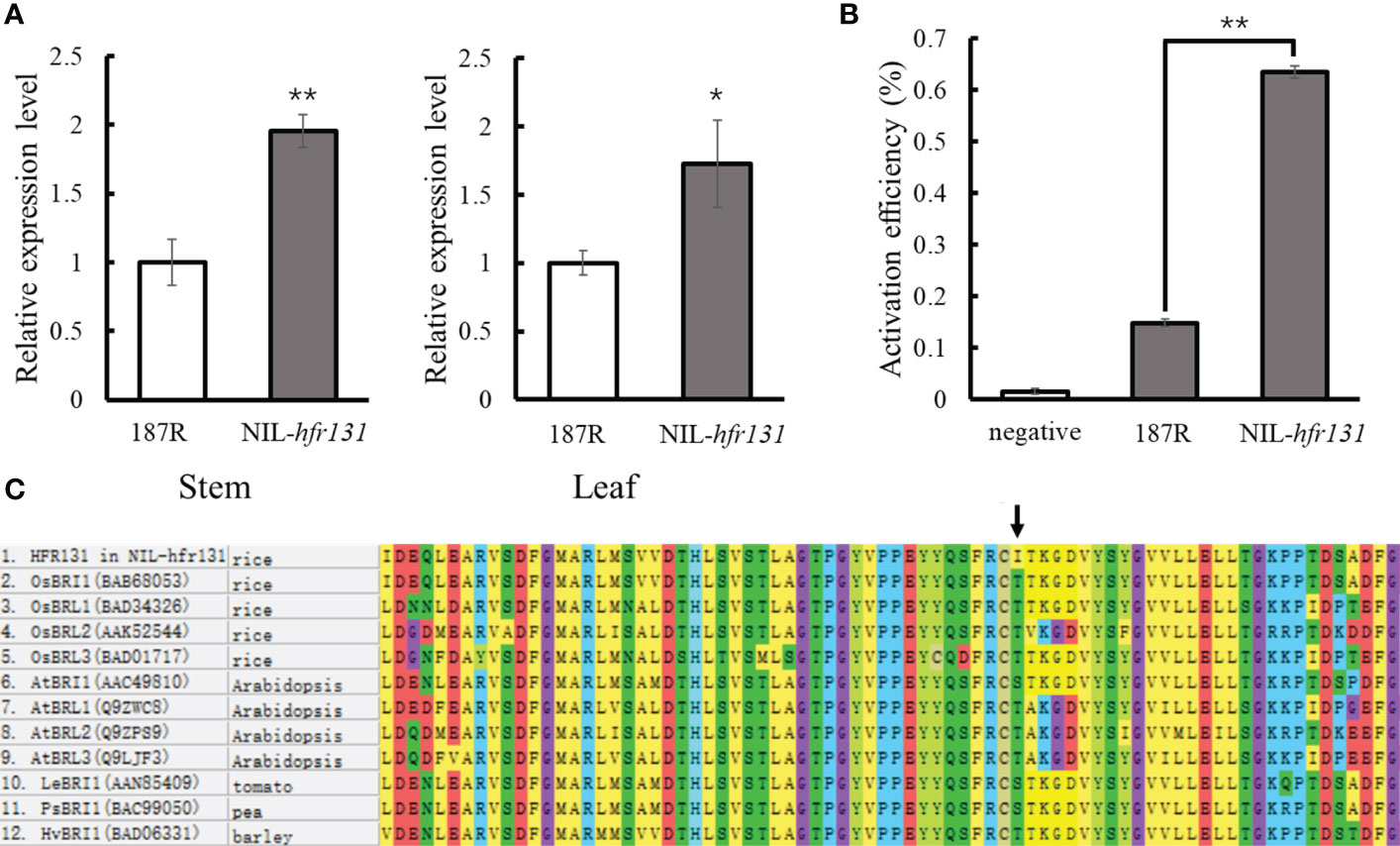
Figure 4 Analysis of the effective mutation site. (A) Expression level of HFR131 in the stem and leaf of 187R and NIL-hfr131. (B) Detection of promoter activity. Activation efficiency was calculated as LUC/REN. (C) Alignment of a portion of the kinase domain of Arabidopsis thaliana, rice, tomato, pea, and barley BRI family members. The arrow indicates the mutation site. * P < 0.05, ** P < 0.01.
To determine which mutation site was the direct factor causing the phenotypic effects of NIL-hfr131, we considered the following three points. First, hfr131 is a recessive gene. Generally, an increase in the expression of a recessive gene does not cause phenotypic effects. Second, we compared the amino acid sequence of HFR131 in NIL-hfr131 with homologous proteins of Arabidopsis, rice, tomato, pea, and barley, and found that the threonine at position 988 of HFR131 is highly conserved (Figure 4C). The mutation site is located in the kinase domain of HFR131, a domain critical to BR signal transduction. However, the amino acid at position 988 of HFR131 was changed to isoleucine in NIL-hfr131, a residue with a quite different structure and physical properties to threonine, implying that this site was the target site causing the phenotypic change of NIL-hfr131. Third, the overexpression of HFR131 in NIL-hfr131 resulted in a significant increase in plant height and grain length (Figures 3E, F), indicating that the increased expression level caused by the deletion of 433 dp in the promoter region of HFR131 was not the direct cause of the decrease in plant height and grain length of NIL-hfr131. In conclusion, the amino acid non-synonymous substitution in the kinase domain of HFR131 was the effective site that led to the decrease in plant height and grain length.
To determine how hfr131 affects plant height and grain length, we selected young panicles before heading date of NIL-hfr131 and 187R for transcriptome analysis and 2444 differentially expressed genes (DEGs) were detected. Among the DEGs, we found that the expression levels of D2, D11, and BRD1, which are associated with BR synthesis, were significantly increased (Supplementary Table 1), which was consistent with the hypothesis of Yamamuro et al. (2000) that plants could compensate for the defects caused by decreased BR sensitivity by increasing BR synthesis. We also observed that genes associated with plant height and grain size showed a significant change in expression level. The positive regulators of grain length GS2 (Hu et al., 2015) and OsLG3 (Yu et al., 2017) were significantly down-regulated. The negative regulators of plant height and grain length OML4 (Lyu et al., 2020) and OsREM4.1 (Gui et al., 2016) were significantly up-regulated. The expression level of OsAP2-39 (Yaish et al., 2010) and OsCYP96B4 (Ramamoorthy et al., 2011), elevating whose expression level would result in dwarf plant height, was significantly upregulated, whereas the expression of LHD2 (Xiong et al., 2006), a positive regulator of plant height, was significantly decreased (Supplementary Table 1). These genes may be the response genes downstream of HFR131 involved in regulating plant height and grain length.
In addition, hormone-related genes were differentially expressed; for example, the gibberellin (GA)-related gene OsGA2ox10 (Lo et al., 2008) was significantly up-regulated and the auxin-related genes OsPIN1d, OsPIN8, and OsAUX2 (Wang et al., 2009; Yu et al., 2015) were significantly down-regulated (Supplementary Table 1), indicating that HFR131 regulated plant height and grain length by coordinating with other hormones.
OsARF17 is involved in the regulation of plant height and grain length by the BR signaling pathway through regulating the expression of HFR131
OsARF17 plays an important role in the regulation of rice architecture, such as leaf angle and tiller angle (Chen et al., 2018; Li et al., 2020; Huang et al., 2021), suggesting that OsARF17 may be an important target gene in rice breeding. However, whether OsARF17 is also involved in the regulation of plant height and grain size, two important agronomic traits, has not been reported. It has been reported that OsARF11 and OsARF19 can regulate the expression of OsBRI1 (Sakamoto et al., 2013; Zhang et al., 2015), so we speculated that OsARF17 may regulate plant height and grain size through HFR131.
We constructed two OsARF17 knockout (KO1 and KO2) and overexpression lines (OE1 and OE2), respectively (Figures 5A, B). Phenotypic observation showed that OsARF17 knockout lines were significantly reduced in plant height compared with ZH11, but OsARF17 overexpression lines did not show the opposite phenotype (Figures 5C, D), which may be related to the interaction of OsARF17 with other proteins in the ARF family, such as OsARF19, whose overexpression lines showed a reduced plant height (Zhang et al., 2015). Whatever, this result showed that OsARF17 was necessary for the maintenance of plant height in rice. Compared with ZH11, the OsARF17 knockout lines had significantly shorter grains and decreased thousand-grain weight, whereas OsARF17 overexpression lines had significantly longer grains and increased thousand-grain weight (Figures 5E–G). These results suggested that OsARF17 affected both plant height and grain length, which was similar to HFR131, indicating that OsARF17 may regulate plant height and grain length through HFR131.
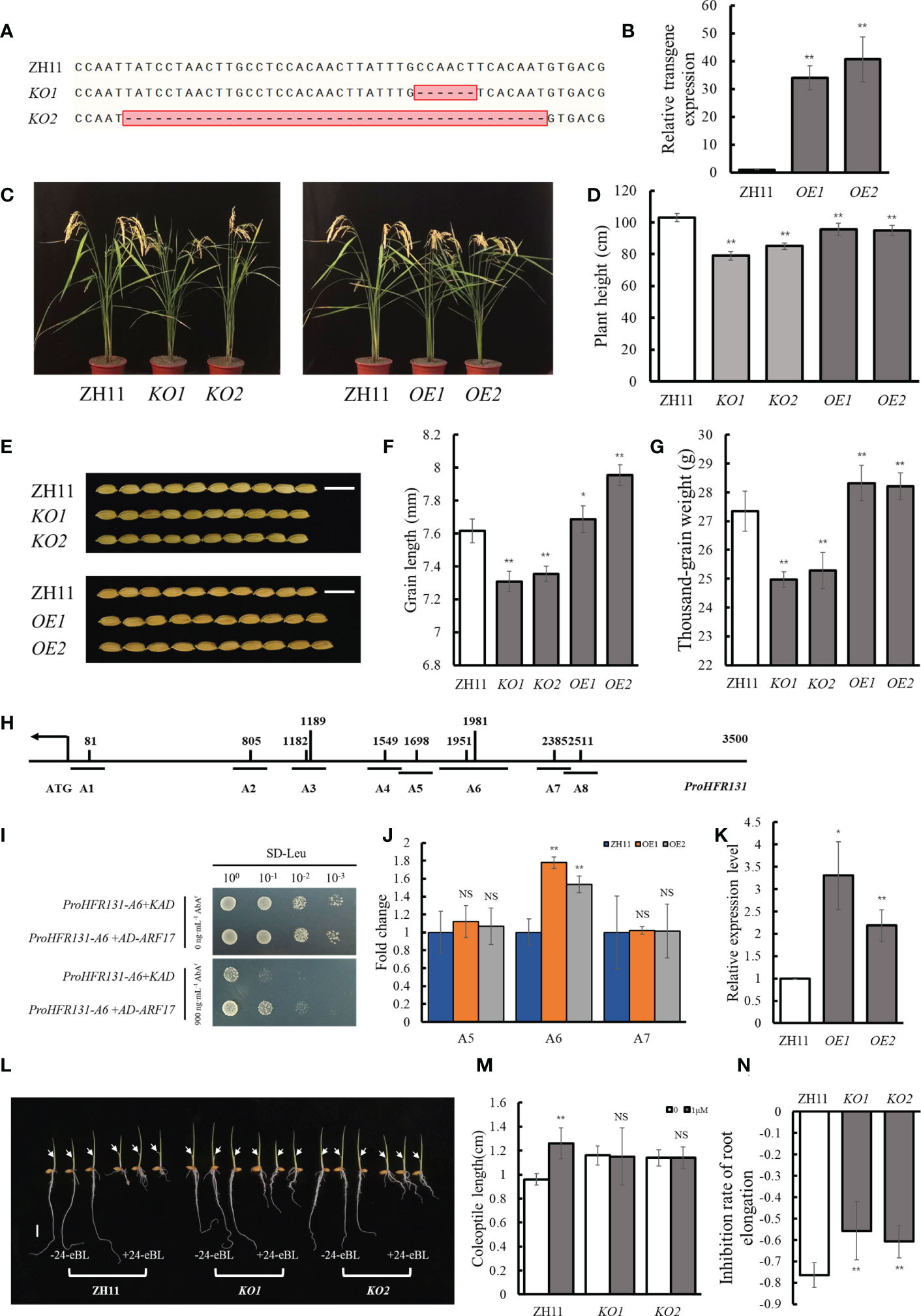
Figure 5 Relationship between OsARF17 and HFR131. (A) Target site in the OsARF17 sequence of rice ‘ZH11’ in the OsARF17 knockout lines. (B) Transgene expression level of OsARF17. OsActin was used as an internal reference. (C, D) Plant architecture and plant height of OsARF17 knockout lines and overexpression lines. (E) Grain morphology of OsARF17 knockout lines and overexpression lines. Bar = 1 cm. (F, G) Grain length and thousand-grain weight of OsARF17 knockout lines and overexpression lines. (H) Positions of 10 auxin response elements in the HFR131 promoter. (I) Yeast one-hybrid assays to detect interaction between OsARF17 and the promoter of HFR131. (J) ChIP-qPCR analysis to detect interaction between OsARF17 and the promoter of HFR131. Fold change is relative to ZH11. OsActin was used as an internal reference. A5, A6, and A7 corresponding to the fragments indicated in Figure 5A. (K) Relative expression level of HFR131 in OsARF17 overexpression lines. OsUBQ5 was used as an internal reference. (L) Effect of 24-eBL on coleoptile and root elongation in seedlings. The arrows indicate the top of the coleoptile. Bar = 1 cm. (M) Coleoptile length of ZH11 and OsARF17 knockout-line seedlings in the presence or absence of 1 μM/L 24- epibrassinolide (24-eBL). (N) Inhibition of root elongation in ZH11 and OsARF17 knockout lines under 1 μM/L 24-eBL treatment. * P < 0.05, ** P < 0.01, NS non-significant.
To verify whether OsARF17 can regulate HFR131, we searched for TGTCT(A/C)C auxin response elements (AuxREs), which are bound by auxin response factors (ARFs) and confer auxin responsiveness (Wang et al., 2007; Shen et al., 2010), within 3500 bp region of HFR131 promoter and 10 AuxREs were found (Figure 5H). We divided the 10 AuxREs into eight fragments with lengths of 150–300 bp (A1–A8) (Figure 5H) and verified by yeast one-hybrid assay and ChIP-qPCR. The yeast one-hybrid assay showed that yeast co-transformed with ProHFR131-A6 and AD-ARF17 grew strongly in the selective medium, whereas yeast co-transformed with ProHFR131-A6 and KAD grew weakly (Figure 5I). The ChIP-qPCR analysis showed that the A6 fragment of the HFR131 promoter was significantly enriched in the two overexpressed lines (Figure 5J). These results indicated that OsARF17 could bind to the A6 fragment of the HFR131 promoter. A qPCR analysis revealed that the expression level of HFR131 was significantly increased in OsARF17 overexpression lines compared with that of the control ZH11 (Figure 5K). These results showed that OsARF17 could bind to the promoter of HFR131 and positively regulated the expression of HFR131.
To further verify whether OsARF17 was involved in the regulation of HFR131, we conducted a BR treatment assay. The results showed that, after treatment with 1 μM/L 24-eBL, root elongation was inhibited more slightly in OsARF17 knockout lines than in the control ZH11. The length of the coleoptile of ZH11 was significantly increased, whereas no significant change was observed in the OsARF17 knockout lines (Figures 5L–N), indicating that plant sensitivity to BR was decreased after OsARF17 knockout. These results indicated that OsARF17 was involved in BR response.
Based on these results, we concluded that OsARF17 could regulate plant height and grain length through regulating the expression of HFR131, that OsARF17 may participate in BR response through HFR131, and that OsARF17–HFR131 interaction provides a bridge for communication between auxin and BR.
Evolutionary correlation analysis of HFR131 and OsARF17
To further understand the relationship between HFR131 and OsARF17 in regulating plant height and grain length, we downloaded the gcHap data for HFR131 and OsARF17 from the RFGB and conducted a haplotype analysis. Eight major Haps (≥30 rice accessions) (Zhang et al., 2021) were resolved based on 20 SNPs distributed among the CDS region and the 3′ untranslated region of HFR131, and a haplotype network was constructed (Figures 6A, B). Xian/Indica (XI) accessions predominantly harbored Hap1, Hap4, Hap7, and Hap8, whereas Geng/Japonica (GJ) accessions predominantly carried Hap2, Hap3, and Hap6, indicating that there was a strong XI–GJ differentiation of HFR131 Haps (Figures 6B, C). Nucleotide diversity (π) among different populations revealed that GJ harbored higher diversity than XI, not only in the gene region of HFR131, but also in the ~500 kb upstream/downstream regions of HFR131 (Figure 6D). Furthermore, HFR131 has not evolved neutrally; the gene was subject to balancing selection in GJ, and directional selection or population expansion in XI (Figure 6E), which indicated that HFR131 may have contributed to XI domestication.
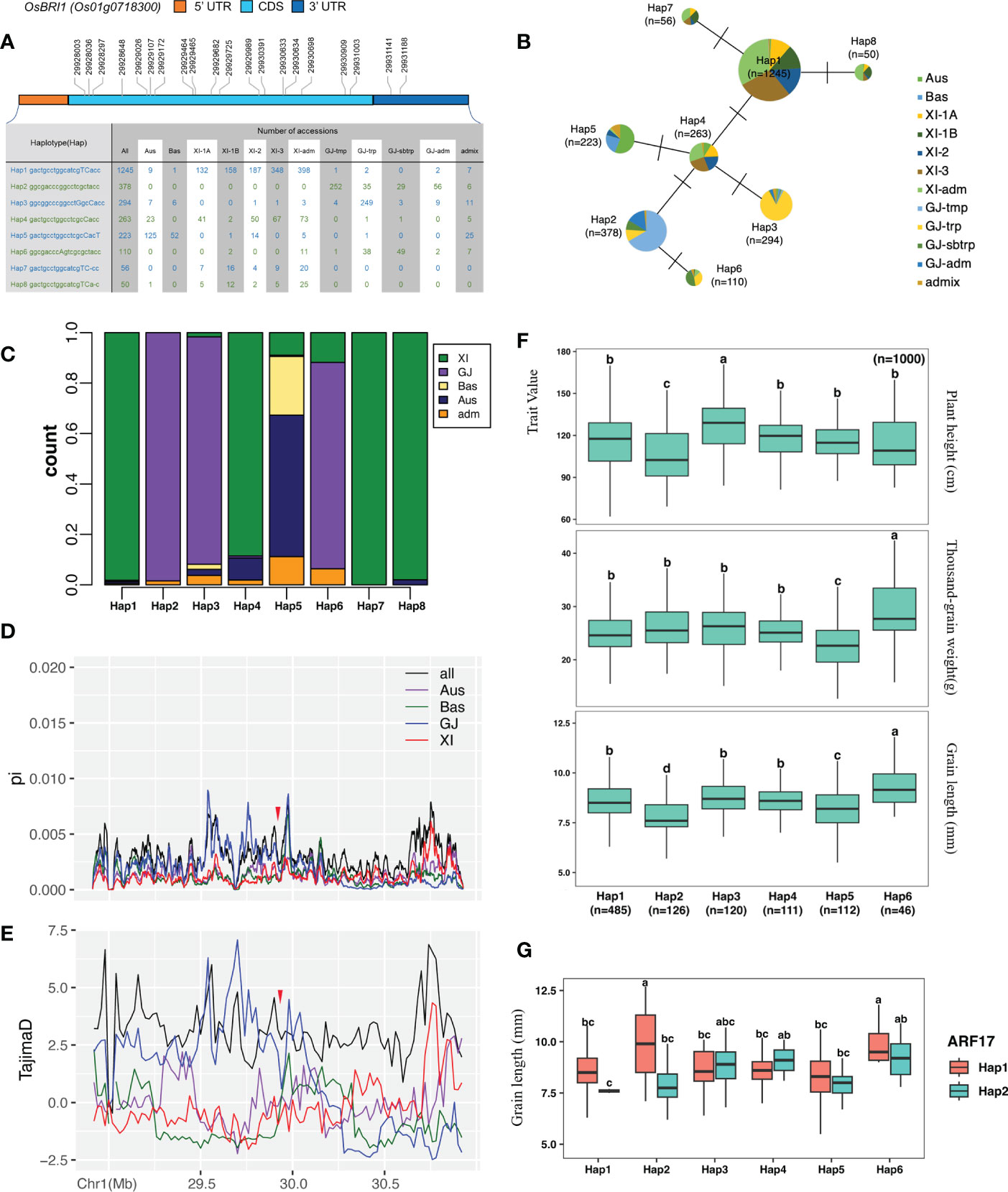
Figure 6 Haplotype analysis of HFR131 and OsARF17. (A) Haplotypes of HFR131/OsBRI1. An uppercase letter in the haplotype sequence indicates a nonsynonymous mutation compared with Hap2. (B) Evolutionary network of HFR131 haplotypes. (C) Frequency of HFR131 haplotypes among subgroups of rice accessions. (D, E) Nucleotide diversity (π) and Tajima’s D for ∼1 Mb genomic region flanking HFR131. The red arrow indicates the position of HFR131. (F) Distribution of plant height, grain length, and thousand-grain weight among rice accessions with different HFR131 haplotypes. (G) Grain length of rice accessions with different haplotypes of HFR131 and OsARF17 Aus, Bas, XI, and GJ refer to different subgroups of rice and Adm (admix) is a fully mixed group. XI-1A, XI-1B, XI-2, XI-3, and XI-adm belong to the XI group. GJ-tmp, GJ-trp, GJ-sbtrp, and GJ-adm belong to the GJ group. Different letters on the boxplots indicate statistically significant differences at P < 0.05 based on Duncan’s multiple range test.
We then investigated the potential associations of major Haps with plant height, grain length, and thousand-grain weight by one-way ANOVA followed by Duncan’s multiple-range tests. Hap3 was associated with the highest average plant height (127.64 ± 18.21 cm), whereas Hap2 was associated with the shortest average plant height (106.18 ± 21.3 cm). Both grain length and thousand-grain weight showed low and high values in Hap5 and Hap6, respectively, inferring that the thousand-grain weight of Hap5 and Hap6 was mainly determined by grain length. Hap7 and Hap8 were not included in the haplotype–trait analysis because the samples with phenotype data for the selected traits were too small (nine and four, respectively) (Figure 6F).
As a regulator of HFR131, we chose two major haplotypes (Hap1 and Hap2) of OsARF17 (≥600 rice accessions) to further investigate its influence on HFR131. OsARF17Hap1 and OsARF17Hap2 were present in 2680 rice accessions and also showed an unequal distribution between XI and GJ (Supplementary Figure 3A). OsARF17Hap1 was present mostly in XI, whereas OsARF17Hap2 was predominantly in GJ, especially in GJ-tmp (GJ from East Asian temperate) and GJ-trp (GJ from Southeast Asian tropical) accessions (Supplementary Figures 3A, B). We performed two-way ANOVA followed by Duncan’s multiple-range test to determine the consociation between HFR131 and OsARF17, and we observed that a significant consociation only existed when considering grain length. Both HFR131Hap2/OsARF17Hap1 and HFR131Hap6/OsARF17Hap1 showed an “a” level in multiple comparisons with long grain length (Figure 6G). However, the HFR131Hap2/OsARF17Hap1 group consisted of only two samples and the grain length of these two samples was divergent (12.7 mm and 7.1 mm, respectively). The grain length of HFR131Hap6 and OsARF17Hap1 was 9.391 mm and 8.567 mm, respectively; however, the grain length of HFR131Hap6/OsARF17Hap1 increased to 9.9 mm (Figure 6G, Supplementary Figure 3C), revealing that the consociation of OsAFR17Hap1/HFR131Hap6 conferred a longer grain length, and thus improved rice yield.
Discussion
The mutation of conserved amino acid in the kinase domain defects the function of OsBRI1
In Arabidopsis thaliana, BRI1 is a receptor of BR, containing a leucine-rich repeat (LRR) domain, a transmembrane domain, and a kinase domain, of which the kinase domain is crucial for the transduction of BR signals (Li and Chory, 1997). BR binds to the extracellular LRR domain of BRI1, and induces association and transphosphorylation between the kinase domains of BRI1 and the co-receptor kinase BAK1. BRI1 activates BAK1 by phosphorylating the activation loop of BAK1, and BAK1 phosphorylates the juxtamembrane and kinase domains of BRI1 to enhance BRI1 signaling in turn. The BR signal is then transmitted downstream (Wang et al., 2012). It has been reported that OsBRI1 and OsBAK1 have a similar structure and function to BRI1 and BAK1, respectively (Yamamuro et al., 2000; Song et al., 2008; Li et al., 2009). In rice, 11 OsBRI1-associated mutants (d61-1 to d61-10 and Fn189) have been reported, among which the Fn189, d61-1, d61-4, and d61-10 mutants harbor mutations in the kinase domain of OsBRI1 (Yamamuro et al., 2000; Nakamura et al., 2006; Zhao et al., 2013). In the present study, we identified a new allele of OsBRI1, hfr131, in which a new mutation site, T988I, in the kinase domain of OsBRI1 was detected, causing significant reductions in plant height and grain length. We compared the protein sequences of the BR receptor and its homologous genes in Arabidopsis thaliana, rice, tomato, pea, and barley, and determined that the mutation site is located in a highly conserved region (Figure 4C), indicating that the site is critical for the function of the OsBRI1 kinase domain and therefore for the function of the gene. Serine/threonine and tyrosine residues are the common phosphorylation sites in BRI1 and BAK1 (Wang et al., 2012). However, T988 in the kinase domain of OsBRI1 was changed to I, an amino acid that cannot be phosphorylated, in NIL-hfr131, indicating that this change may affect the phosphorylation of OsBAK1 to OsBRI1, thus hindering transduction of the BR signal.
The plant height of rice mainly depends on the elongation of internodes. Dwarf mutants can be categorized into six types according to the elongation pattern of the upper four or five internodes, namely N-type (wild type), dn-type, dm-type, d6-type, nl-type, and sh-type (Yamamuro et al., 2000). Mutants defective in BR synthesis or signaling are often of the dm-type or d6-type. The d2, d11, and d61-1 mutants exhibit the dm-type internode elongation pattern (Supplementary Table 2), showing a specific shortening of the second internode. The brd2, d61-2, and Fn189 mutants show the d6-type internode elongation pattern (Supplementary Table 2), in which all the internodes are extremely shortened except for the first internode. However, brd1 failed to elongate any internodes (Supplementary Table 2). The T988I conversion in the kinase domain of OsBRI1/HFR131 caused reduction in plant height with equally proportional shortening of all internodes (Figure 1G), which represented the dn-type internode elongation pattern and had not been reported previously in BR synthesis- or OsBRI1-defective mutants (Supplementary Table 2).
hfr131 confers the dn-type internode elongation pattern by coordinating different hormones
The formation of different internode elongation patterns may be associated with differences in the main regulatory hormones at different internodes. GA is considered to play an important role in the elongation of the first internode in rice. The first internode of the GA synthesis-defective d35 mutant is shortened to a significantly greater degree than the other internodes (Itoh et al., 2004). HOX12 and EUI influence elongation of the first internode by regulating the contents of active GAs (Luo et al., 2006; Zhu et al., 2006; Gao et al., 2016). All phenotypic effects of BR result from BR synthesis and perception. BR-synthesis and OsBRI1-related mutants will assist in providing an improved understanding of the role of BR in rice internode regulation. All BR-synthesis and OsBRI1-related mutants reported to date show severe shortening or non-elongation of the second internode, with dm- or d6-type elongation patterns (Supplementary Table 2), indicating that elongation of the second internode may be mainly determined by BR. For the lower internodes, auxin may be the primary functional hormone. Loss-of-function of BR2, which functions in indole-3-acetic acid export (Knöller et al., 2010), disrupts polar auxin transport in the maize stalk, resulting in a reduced plant height with compact lower-stalk internodes (Multani et al., 2003). These findings confirmed that the distribution of auxin plays a crucial role in regulating the lower internodes. In the present study, NIL-hfr131, harboring a new allele of OsBRI1, hfr131, showed a dn-type internode elongation pattern rather than the d6- or dm-type, indicating that this allele can coordinate different hormones. It was further confirmed by transcriptome analysis that OsGA2ox10, which participates in the GA catabolic pathways (Lo et al., 2008), was significantly up-regulated and that OsPIN1d, OsPIN8, and OsAUX2, which are auxin transporters (Wang et al., 2009; Yu et al., 2015), were significantly down-regulated (Supplementary Table 1), implying that there may be coordination between different hormones.
We noticed that the location of HFR131/OsBRI1 was closed to the famous “Green Revolution” gene SD1 (Monna et al., 2002; Sasaki et al., 2002; Su et al., 2021), both of which were located on the long arm of chromosome 1, and the distance of them was nearly 8500 kb (Supplementary Figure 1). Either SD1-mutant or NIL-hfr131 showed a semi-dwarf plant height, but the expression pattern of them were different. SD1 was widely expressed in different organs especially in leaf blades (Monna et al., 2002; Sasaki et al., 2002; Su et al., 2021), however, OsBRI1 was strongly expressed in shoot apex but hardly in leaf blades (Yamamuro et al., 2000), indicating that they regulated plant height through different pathway, also implying that hfr131 may be another useful dwarfing gene resource after sd1.
OsARF17-HFR131 links brassinosteroid and auxin signaling during regulation of plant height and grain length
Auxin and BR coordinate to regulate many aspects of plant growth and development, such as root and bud development (Mazzoni-Putman et al., 2021), and several studies have shown that there is crosstalk between BR and auxin; for example, exogenous auxin improves the sensitivity of rice to BR (Fujioka et al., 1998), and BR affects the transport of auxin (Symons et al., 2007). With extension of the research scope, ARF family proteins were found to be important factors communicating between the BR and auxin signaling pathways. In Arabidopsis thaliana, ARF2, ARF7, and ARF19 are involved in BR and auxin communication through different mechanisms (Vert et al., 2008; Sun et al., 2010; Cho et al., 2014). In rice, OsARF11 and OsARF19 can bind to the OsBRI1 promoter and promote the expression of OsBRI1, thus enhancing the rice response to BR and regulating leaf angle (Sakamoto et al., 2013; Zhang et al., 2015). However, how the BR and auxin communication affects plant height and grain size in rice is poorly understood. In the present study, we confirmed that OsARF17 may be involved in the response to BR through HFR131. OsARF17 can bind to the promoter region of HFR131 and positively regulate the expression of HFR131 (Figures 5I–K). OsARF17 knockout lines showed decreased sensitivity to BR (Figures 5L–N). OsARF17 and HFR131 defective and overexpression lines showed a similar phenotype. Both OsARF17 knockout lines and NIL-hfr131 showed a reduced plant height and grain length, whereas both overexpression lines of OsARF17 and HFR131 exhibited an increased grain length, indicating that BR and auxin regulate plant height and grain length synergistically through OsARF17–HFR131 interaction. In addition, HFR131Hap6/OsARF17Hap1 in combination conferred a longer grain length than both HFR131Hap6 and OsARF17Hap1 individually. This may be the result of efficient coordination of the BR and auxin signaling pathways between the two haplotypes, and highlights the importance of breaking through the limitation of traditional single-gene breeding to increase rice yield.
In summary, this study identified a new allele of OsBRI1, hfr131, which caused a significant decrease in plant height and grain length of rice. It was further found that OsARF17 could regulate the plant height and grain length of rice, and affect the sensitivity of rice to BR, by regulating the expression of HFR131. In addition, the OsAFR17Hap1/HFR131Hap6 consociation was confirmed to contribute to longer grain length. These findings are of great importance for improvement of rice yield by molecular breeding in the future.
Data availability statement
The data presented in the study are deposited in the GEO repository, accession number GSE224118.
Author contributions
XL, YW, DL, and LC contributed to conception and design of the study. DL, LC, ML, FM, and PY performed the experiments. DL, LC, XZ, JH, FN, SH, JC, XY, and JY analyzed and interpreted the data. DL, LC, and ML wrote the manuscript and XL and YW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by National Natural Science Foundation of China (32172043, 31971918 and 31900393), Shanghai Rising-Star Program (20QB1402100), Natural Science Foundation of Shanghai (19ZR1436600) and Innovation Program of Shanghai Municipal Education Commission (2023ZKZD05).
Acknowledgments
We thank Jinying Gou (Fudan University) for providing pAbAi vector and Yan Yan and Yue Hu (Fudan University) for the guidance of yeast one-hybrid assay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1152196/full#supplementary-material
References
Bai, M.-Y., Zhang, L.-Y., Gampala, S. S., Zhu, S.-W., Song, W.-Y., Chong, K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. 104 (34), 13839–13844. doi: 10.1073/pnas.0706386104
Chen, S.-H., Zhou, L.-J., Xu, P., Xue, H.-W. (2018). SPOC domain-containing protein leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PloS Genet. 14 (11), e1007829. doi: 10.1371/journal
Cho, H., Ryu, H., Rho, S., Hill, K., Smith, S., Audenaert, D., et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 16 (1), 66–76. doi: 10.1038/ncb2893
Economic and Social Affairs (2022). World population prospects 2022: Summary of results (NY, USA: United Nations New York).
Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S. (1998). Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49 (7), 1841–1848. doi: 10.1016/S0031-9422(98)00412-9
Gao, S., Fang, J., Xu, F., Wang, W., Chu, C. (2016). Rice HOX12 regulates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 28 (3), 680–695. doi: 10.1105/tpc.15.01021
Gui, J., Zheng, S., Liu, C., Shen, J., Li, J., Li, L. (2016). OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 38 (2), 201–213. doi: 10.1016/j.devcel.2016.06.011
Hargrove, T. R., Cabanilla, V. L. (1979). The impact of semidwarf varieties on Asian rice-breeding programs. BioScience 29 (12), 731–735. doi: 10.2307/1307667
Hong, Z., Ueguchi-Tanaka, M., Fujioka, S., Takatsuto, S., Yoshida, S., Hasegawa, Y., et al. (2005). The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17 (8), 2243–2254. doi: 10.1105/tpc.105.030973
Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15 (12), 2900–2910. doi: 10.1105/tpc.014712
Hu, J., Wang, Y., Fang, Y., Zeng, L., Xu, J., Yu, H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8 (10), 1455–1465. doi: 10.1016/j.molp.2015.07.002
Huang, Y., Dong, H., Mou, C., Wang, P., Hao, Q., Zhang, M., et al. (2022). Ribonuclease h-like gene SMALL GRAIN2 regulates grain size in rice through brassinosteroid signaling pathway. J. Integr. Plant Biol. 64 (10), 1883–1900. doi: 10.1111/jipb.13333
Huang, G., Hu, H., van de Meene, A., Zhang, J., Dong, L., Zheng, S., et al. (2021). AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 33 (9), 3120–3133. doi: 10.1093/plcell/koab175
Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T., Kitano, H., et al. (2004). A rice semi-dwarf gene, Tan-ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54 (4), 533–547. doi: 10.1023/b:plan.0000038261.21060.47
Khush, G. S. (1999). Green revolution: preparing for the 21st century. Genome 42 (4), 646–655. doi: 10.1139/g99-044
Knöller, A. S., Blakeslee, J. J., Richards, E. L., Peer, W. A., Murphy, A. S. (2010). Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J. Exp. Bot. 61 (13), 3689–3696. doi: 10.1093/jxb/erq180
Li, J., Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 (5), 929–938. doi: 10.1016/S0092-8674(00)80357-8
Li, Y., Li, J., Chen, Z., Wei, Y., Qi, Y., Wu, C. (2020). OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol. J. 18 (10), 2015–2026. doi: 10.1111/pbi.13360
Li, D., Wang, L., Wang, M., Xu, Y.-Y., Luo, W., Liu, Y.-J., et al. (2009). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7 (8), 791–806. doi: 10.1111/j.1467-7652.2009.00444.x
Liu, F., Wang, P., Zhang, X., Li, X., Yan, X., Fu, D., et al. (2018). The genetic and molecular basis of crop height based on a rice model. Planta 247 (1), 1–26. doi: 10.1007/s00425-017-2798-1
Liu, D., Yu, Z., Zhang, G., Yin, W., Li, L., Niu, M., et al. (2021). Diversification of plant agronomic traits by genome editing of brassinosteroid signaling family genes in rice. Plant Physiol. 187 (4), 2563–2576. doi: 10.1093/plphys/kiab394
Lo, S.-F., Yang, S.-Y., Chen, K.-T., Hsing, Y.-I., Zeevaart, J. A. D., Chen, L.-J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20 (10), 2603–2618. doi: 10.1105/tpc.108.060913
Luo, A., Qian, Q., Yin, H., Liu, X., Yin, C., Lan, Y., et al. (2006). EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47 (2), 181–191. doi: 10.1093/pcp/pci233
Lyu, J., Wang, D., Duan, P., Liu, Y., Huang, K., Zeng, D., et al. (2020). Control of grain size and weight by the GSK2-LARGE1/OML4 pathway in rice. Plant Cell 32 (6), 1905–1918. doi: 10.1105/tpc.19.00468
Mazzoni-Putman, S. M., Brumos, J., Zhao, C. S., Alonso, J. M., Stepanova, A. N. (2021). Auxin interactions with other hormones in plant development. Cold Spring Harbor Perspect. Biol. 13 (10), a039990. doi: 10.1101/cshperspect.a039990
McClung, C. R. (2014). Making hunger yield. Science 344 (6185), 699–700. doi: 10.1126/science.1254135
Monna, L., Kitazawa, N., Yoshino, R., Suzuki, J., Masuda, H., Maehara, Y., et al. (2002). Positional cloning of rice semidwarfing gene, sd-1: rice "green revolution gene" encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9 (1), 11–17. doi: 10.1093/dnares/9.1.11
Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T., Sugimoto, K., et al. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130 (3), 1152–1161. doi: 10.1104/pp.007179
Morinaka, Y., Sakamoto, T., Inukai, Y., Agetsuma, M., Kitano, H., Ashikari, M., et al. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141 (3), 924–931. doi: 10.1104/pp.106.077081
Multani, D. S., Briggs, S. P., Chamberlin, M. A., Blakeslee, J. J., Murphy, A. S., Johal, G. S. (2003). Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302 (5642), 81–84. doi: 10.1126/science.1086072
Nakagawa, H., Tanaka, A., Tanabata, T., Ohtake, M., Fujioka, S., Nakamura, H., et al. (2011). SHORT GRAIN1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 158 (3), 1208–1219. doi: 10.1104/pp.111.187567
Nakamura, A., Fujioka, S., Sunohara, H., Kamiya, N., Hong, Z., Inukai, Y., et al. (2006). The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140 (2), 580–590. doi: 10.1104/pp.105.072330
Niu, M., Wang, H., Yin, W., Meng, W., Xiao, Y., Liu, D., et al. (2022). Rice DWARF AND LOW-TILLERING and the homeodomain protein OSH15 interact to regulate internode elongation via orchestrating brassinosteroid signaling and metabolism. Plant Cell 34 (10), 3754–3772. doi: 10.1093/plcell/koac196
Qiao, J., Jiang, H., Lin, Y., Shang, L., Wang, M., Li, D., et al. (2021). A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 14 (10), 1683–1698. doi: 10.1016/j.molp.2021.06.023
Ramamoorthy, R., Jiang, S.-Y., Ramachandran, S. (2011). Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner. PloS One 6 (11), e28069. doi: 10.1371/journal.pone.0028069
Sakamoto, T., Morinaka, Y., Inukai, Y., Kitano, H., Fujioka, S. (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant Cell 73 (4), 676–688. doi: 10.1111/tpj.12071
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). A mutant gibberellin-synthesis gene in rice. Nature 416 (6882), 701–702. doi: 10.1038/416701a
Shen, C., Wang, S., Bai, Y., Wu, Y., Zhang, S., Chen, M., et al. (2010). Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa l.). J. Exp. Bot. 61 (14), 3971–3981. doi: 10.1093/jxb/erq208
Song, D., Li, G., Song, F., Zheng, Z. (2008). Molecular characterization and expression analysis of OsBISERK1, a gene encoding a leucine-rich repeat receptor-like kinase, during disease resistance responses in rice. Mol. Biol. Rep. 35 (2), 275–283. doi: 10.1007/s11033-007-9080-8
Su, S., Hong, J., Chen, X., Zhang, C., Chen, M., Luo, Z., et al. (2021). Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice. Plant Biotechnol. J. 19 (11), 2304–2318. doi: 10.1111/pbi.13661
Sun, Y., Fan, X.-Y., Cao, D.-M., Tang, W., He, K., Zhu, J.-Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19 (5), 765–777. doi: 10.1016/j.devcel.2010.10.010
Sun, L., Li, X., Fu, Y., Zhu, Z., Tan, L., Liu, F., et al. (2013). GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J. Integr. Plant Biol. 55 (10), 938–949. doi: 10.1111/jipb.12062
Symons, G. M., Ross, J. J., Jager, C. E., Reid, J. B. (2007). Brassinosteroid transport. J. Exp. Bot. 59 (1), 17–24. doi: 10.1093/jxb/erm098
Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17 (3), 776–790. doi: 10.1105/tpc.104.024950
Tanaka, A., Nakagawa, H., Tomita, C., Shimatani, Z., Ohtake, M., Nomura, T., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-Loop-Helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151 (2), 669–680. doi: 10.1104/pp.109.140806
Tong, H., Jin, Y., Liu, W., Li, F., Fang, J., Yin, Y., et al. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58 (5), 803–816. doi: 10.1111/j.1365-313X.2009.03825.x
Tong, H., Xiao, Y., Liu, D., Gao, S., Liu, L., Yin, Y., et al. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26 (11), 4376–4393. doi: 10.1105/tpc.114.132092
Vert, G., Walcher, C. L., Chory, J., Nemhauser, J. L. (2008). Integration of auxin and brassinosteroid pathways by auxin response factor 2. Proc. Natl. Acad. Sci. 105 (28), 9829–9834. doi: 10.1073/pnas.0803996105
Wang, Z.-Y., Bai, M.-Y., Oh, E., Zhu, J.-Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46 (1), 701–724. doi: 10.1146/annurev-genet-102209-163450
Wang, J.-R., Hu, H., Wang, G.-H., Li, J., Chen, J.-Y., Wu, P. (2009). Expression of PIN genes in rice (Oryza sativa l.): Tissue specificity and regulation by hormones. Mol. Plant 2 (4), 823–831. doi: 10.1093/mp/ssp023
Wang, Y., Luo, X., Sun, F., Hu, J., Zha, X., Su, W., et al. (2018). Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun. 9 (1), 3516. doi: 10.1038/s41467-018-05829-7
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394 (1-2), 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, L., Xu, Y., Zhang, C., Ma, Q., Joo, S.-H., Kim, S.-K., et al. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PloS One 3 (10), e3521. doi: 10.1371/journal.pone.0003521
Xiao, Y., Zhang, G., Liu, D., Niu, M., Tong, H., Chu, C. (2020). GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant J. 102 (6), 1187–1201. doi: 10.1111/tpj.14692
Xing, Y., Zhang, Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61 (1), 421–442. doi: 10.1146/annurev-arplant-042809-112209
Xiong, G. S., Hu, X. M., Jiao, Y. Q., Yu, Y. C., Chu, C. C., Li, J. Y., et al. (2006). LEAFY HEAD2, which encodes a putative RNA-binding protein, regulates shoot development of rice. Cell Res. 16 (3), 267–276. doi: 10.1038/sj.cr.7310034
Xiong, M., Yu, J., Wang, J., Gao, Q., Huang, L., Chen, C., et al. (2022). Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 189 (1), 402–418. doi: 10.1093/plphys/kiac043
Yaish, M. W., El-kereamy, A., Zhu, T., Beatty, P. H., Good, A. G., Bi, Y.-M., et al. (2010). The APETALA-2-Like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PloS Genet. 6 (9), e1001098. doi: 10.1371/journal.pgen.1001098
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 (9), 1591–1605. doi: 10.1105/tpc.12.9.1591
Yoo, M.-J., Albert, V. A., Soltis, P. S., Soltis, D. E. (2006). Phylogenetic diversification of glycogen synthase kinase 3/SHAGGY-like kinase genes in plants. BMC Plant Biol. 6 (1), 3. doi: 10.1186/1471-2229-6-3
Yu, C., Sun, C., Shen, C., Wang, S., Liu, F., Liu, Y., et al. (2015). The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in cd stress responses in rice (Oryza sativa l.). Plant J. 83 (5), 818–830. doi: 10.1111/tpj.12929
Yu, J., Xiong, H., Zhu, X., Zhang, H., Li, H., Miao, J., et al. (2017). OsLG3 contributing to rice grain length and yield was mined by ho-LAMap. BMC Biol. 15 (1), 28. doi: 10.1186/s12915-017-0365-7
Zhang, F., Wang, C., Li, M., Cui, Y., Shi, Y., Wu, Z., et al. (2021). The landscape of gene-CDS-haplotype diversity in rice: Properties, population organization, footprints of domestication and breeding, and implications for genetic improvement. Mol Plant. 14 (5), 787–804. doi: 10.1016/j.molp.2021.02.003
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38 (4), 638–654. doi: 10.1111/pce.12397
Zhao, J., Wu, C., Yuan, S., Yin, L., Sun, W., Zhao, Q., et al. (2013). Kinase activity of OsBRI1 is essential for brassinosteroids to regulate rice growth and development. Plant Sci. 199, 199–200. doi: 10.1016/j.plantsci.2012.10.011
Zhu, X., Liang, W., Cui, X., Chen, M., Yin, C., Luo, Z., et al. (2015). Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of carbon starved anther, a MYB domain protein. Plant J. 82 (4), 570–581. doi: 10.1111/tpj.12820
Keywords: plant height, grain length, brassinosteroid, auxin, OsARF17, OsBRI1, rice
Citation: Lan D, Cao L, Liu M, Ma F, Yan P, Zhang X, Hu J, Niu F, He S, Cui J, Yuan X, Yang J, Wang Y and Luo X (2023) The identification and characterization of a plant height and grain length related gene hfr131 in rice. Front. Plant Sci. 14:1152196. doi: 10.3389/fpls.2023.1152196
Received: 27 January 2023; Accepted: 13 March 2023;
Published: 24 March 2023.
Edited by:
Jinsong Bao, Zhejiang University, ChinaReviewed by:
Xu Feifei, Zhejiang University, ChinaDivya Balakrishnan, Indian Institute of Rice Research (ICAR), India
Xiaoli Fan, Chengdu Institute of Biology (CAS), China
Copyright © 2023 Lan, Cao, Liu, Ma, Yan, Zhang, Hu, Niu, He, Cui, Yuan, Yang, Wang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojin Luo, bHVveGpAZnVkYW4uZWR1LmNu; Ying Wang, d2FuZ195QGZ1ZGFuLmVkdS5jbg==
†These authors contributed equally to this work and share first authorship
 Dengyong Lan
Dengyong Lan Liming Cao3†
Liming Cao3† Ying Wang
Ying Wang Xiaojin Luo
Xiaojin Luo