- 1Center of Legume Plant Genetics and System Biology, College of Agronomy, Fujian Agriculture and Forestry University (FAFU), Fuzhou, Fujian, China
- 2College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
- 3Crops Research Institute, Fujian Academy of Agricultural Science, Fuzhou, Fujian, China
- 4College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
Cultivated peanut (Arachis hypogaea) is a leading protein and oil-providing crop and food source in many countries. At the same time, it is affected by a number of biotic and abiotic stresses. O-methyltransferases (OMTs) play important roles in secondary metabolism, biotic and abiotic stress tolerance. However, the OMT genes have not been comprehensively analyzed in peanut. In this study, we performed a genome-wide investigation of A. hypogaea OMT genes (AhOMTs). Gene structure, motifs distribution, phylogenetic history, genome collinearity and duplication of AhOMTs were studied in detail. Promoter cis-elements, protein-protein interactions, and micro-RNAs targeting AhOMTs were also predicted. We also comprehensively studied their expression in different tissues and under different stresses. We identified 116 OMT genes in the genome of cultivated peanut. Phylogenetically, AhOMTs were divided into three groups. Tandem and segmental duplication events played a role in the evolution of AhOMTs, and purifying selection pressure drove the duplication process. AhOMT promoters were enriched in several key cis-elements involved in growth and development, hormones, light, and defense-related activities. Micro-RNAs from 12 different families targeted 35 AhOMTs. GO enrichment analysis indicated that AhOMTs are highly enriched in transferase and catalytic activities, cellular metabolic and biosynthesis processes. Transcriptome datasets revealed that AhOMTs possessed varying expression levels in different tissues and under hormones, water, and temperature stress. Expression profiling based on qRT-PCR results also supported the transcriptome results. This study provides the theoretical basis for further work on the biological roles of AhOMT genes for developmental and stress responses.
Introduction
In Arabidopsis thaliana, O-methyltransferases (OMTs) are heterogeneous enzymes involved in the flavonoid and lignin production pathways (Guo et al., 2001). There are three classes of plant methyltransferases: C- methyltransferases, N- methyltransferases, and O-methyltransferases (Roje, 2006). In plants, OMTs assist the transfer of the methyl group of S-adenosyl-L-methionine (SAM) to the hydroxyl group of numerous organic chemical compounds, ultimately synthesizing the methyl ether variants of these substances (Struck et al., 2012). Based on the molecular weight and bivalent ion dependence, OMTs are divided into Caffeoyl-CoA OMT (CCoAOMT) and Caffeic acid OMT (COMT). COMTs are the main representative of type I, and CCoAOMTs are of type II (Davin and Lewis, 1992). Depending upon the resemblance in sequence and protein motifs, OMT genes are further classified into two separate categories: PL-OMT I and PL-OMT II (CCoAOMT and COMT, respectively) (Joshi and Chiang, 1998). COMT-type proteins bind to a variety of substrates, including caffeoyl CoA ester, caffeic acid, chalcones, myoinositol, scoulerine, 5-hydroxyferuloylester, and 5-hydroxyferulic acid (Ye et al., 1994; Roje, 2006). CCoAOMT-type enzymes use a pair of substrates, caffeoyl CoA and 5-hydroxyferuloyl CoA, to function (Davin and Lewis, 1992). COMT and CCoAOMT both mediate the lignin biosynthesis process. The CCoAOMT enzyme catalyzes an early step in the pathway by converting caffeoyl CoA to feruloyl CoA (Dudareva and Pichersky, 2008), despite the fact that sinapyl alcohol, a key component of S-type lignin, is mostly biosynthesized by COMT proteins at the end of the biosynthetic pathway (Ye et al., 1994; Buer et al., 2010).
Lignin is the second most prevalent biopolymer on the planet and is an essential element of cell walls in certain higher plants (Ralph et al., 2004). It offers mechanical strength to plants and assists water movement throughout whole plant tissues (Liu et al., 2018), and also an excellent barricade for pathogens, fungi, and insects (Peng et al., 2014), so it helps to improve plant response toward environmental calamities (Moura et al., 2010). To understand their significance, OMT genes have been extensively studied in various plants, such as Arabidopsis and rice (Hamberger et al., 2007), citrus and sorghum (Liu et al., 2016b; Rakoczy et al., 2018), switchgrass and dove tree (LIU et al., 2016a), tea plant (Lin et al., 2021) etc. Concerning wheat, Nguyen and his team analyzed the expression profiles of lignin biosynthesis-related genes, including a number of CCoAOMTs, to determine the likely mechanisms behind their expression patterns. They discovered that lignin content was directly linked with lodging resistance, tolerance to various biotic and abiotic stresses, and quality of feedstock biomass (Nguyen et al., 2016). TaCCoAOMT1 regulates lignin biosynthesis (Ma and Luo, 2015); previously, this gene has been reported as a key stem cell growth regulator (Bi et al., 2011). Due to their significant roles in secondary metabolism, intensive work has been done on OMT genes throughout the years (Bout and Vermerris, 2003; Goujon et al., 2003; Kota et al., 2004; Li et al., 2006; Lin et al., 2006; Yoshihara et al., 2008; Ma, 2009; Zhou et al., 2009). A detailed evaluation of the OMT genes in peanut has yet to be performed, despite the fact that the genes’ well-established role offers a good foundation for our research.
Therefore, OMT genes were studied at a genome-wide scale in A. hypogaea and its wild progenitors. One hundred and sixteen OMT genes were found in the cultivated peanut genome. Further, we looked into the evolutionary connections of these AhOMT genes, their conserved domains and motifs, gene structure, and genomic position. We likewise investigated the AhOMT promoters; similarly, expression in different organs under various stress conditions was investigated as well. This study will provide a base for further research on individual genes in peanut and will aid in exploring the biological roles of the OMT genes.
Materials and methods
Identification and characterization of OMT genes in A. hypogaea
OMT genes in the genome of A. hypogaea were comprehensively searched. The protein sequences of AtOMTs were acquired from the TAIR database (https://www.arabidopsis.org/) (Lamesch et al., 2012) and soybean OMTs from Legume Information System (https://legumeinfo.org/) (Gonzales et al., 2005). A. ipaensis and A. duranensis OMT sequences were obtained from the PeanutBase database (https://www.peanutbase.org/home) (Bertioli et al., 2016). The sequences of whole-genome proteins of A. hypogaea were obtained from the Peanut Genome Resource database (PGR) (http://peanutgr.fafu.edu.cn/) (Zhuang et al., 2019). The protein sequences of OMTs from A. duranensis, A. ipaensis, A. thaliana, and G. max were used to search the AhOMTs by BLASTP search with TBtools software (Chen et al., 2020). Further, the HMM search method was also used to search the OMT proteins from A. hypogaea genome. The Pfam database was searched to obtain the HMM files for the OMT family (PF08100 and PF00891) (http://pfam.xfam.org/). The identified proteins were scanned at NCBI and Pfam databases to verify the OMT domain. ProtParam tool (http://web.expasy.org/protparam/) determined the physicochemical characteristics of AhOMTs (Gasteiger et al., 2005). The subcellular localizations of AhOMT proteins in different cell organelles were predicted by the CELLO version v2.5 (http://cello.life.nctu.edu.tw/) (Yu et al., 2006). General Feature Format (GFF3) files were used to view the exon-intron distribution pattern of AhOMTs through TBtools software. Conserved motifs of AhOMT proteins were determined by the MEME database (https://meme-db.org/motifs/) (Bailey et al., 2015).
Phylogenetic and gene duplication analysis of AhOMTs
A phylogenetic tree comprising A. ipaensis, A. duranensis, G. max, A. hypogaea, and A. thaliana proteins was constructed to investigate their phylogenetic connections. Protein sequences were subjected to multiple sequence alignment by MUSCLE method with the help of MEGAX software (https://megasoftware.net/home) (Kumar et al., 2018). A neighbor-joining tree was generated through 1,000 bootstraps with the poisson model. MCScanX was run to identify the duplicated genes. The KaKs Calculator 2.0 program with the MYN approach was used to determine the rates of synonymous and nonsynonymous substitution (Wang et al., 2010). T = ks/2r was used to compute the divergence time with the neutral substitution coefficient r=8.12×10-9 (Bertioli et al., 2016).
Analysis of AhOMT promoters and miRNAs prediction
Promoter sequences up to 2 kb were used to find different binding cites and cis-elements through the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Coding sequences of AhOMTs were used to identify putative miRNAs targeting the AhOMT genes through the psRNATarget database (https://www.zhaolab.org/psRNATarget/home) (Dai et al., 2018).
Genome collinearity and orthologous gene clusters
Comparative synteny was analyzed to examine evolutionary genome conservations between three peanut species and Arabidopsis. The genome and GFF3 files of all these species were subjected to McScanX in TBtools software, and the resulting files were used for multiple synteny analysis. The orthologous OMT proteins were identified in A. hypogea, A. duranensis, A. ipaensis, and A. thaliana through OthroVenn2 (https://orthovenn2.bioinfotoolkits.net/home) (Xu et al., 2019). Protein sequences of Arabidopsis, soybean, and three peanut species were used to identify orthologous genes. The peanut species were assessed individually with each other and with Arabidopsis and soybean to identify orthologous gene clusters.
Functional annotation and prediction of protein-protein interactions
For functional annotation prediction (GO and KEGG), AhOMT proteins were scanned at the EggNOG database (http://eggnog-mapper.embl.de/) (Huerta-Cepas et al., 2019). Enrichment analyses were executed in TBtools software from predicted GO and KEGG annotations.
Protein-protein interactions were predicted based on studied AtOMTs. STRING 11.5 tool (https://www.string-db.org/cgi/) (Szklarczyk et al., 2019) was used to construct the interaction network between peanut and Arabidopsis OMTs. The top 10 interactions were predicted with a medium threshold level (0.4). MCL clustering with inflation parameter 10 was used, and dotted lines were used between cluster edges.
Expression profiling of AhOMT Gsenes
Transcriptome expression data were accessed to view the expression levels of AhOMTs in various organs, phytohormones, water, and temperature treatments. Transcriptome expression data for different tissues (leaf, stem, stem tip, fluorescence, root, root and stem, root tip, root nodule, gynophore/peg, pericarp, testa, cotyledons, and embryo), hormones (ABA, SA, brassinolide, paclobutrazol, ethephon, and ddH2O as control), water (drought and normal irrigation) and temperature treatments (low temperature and room temperature) were accessed from the PGR database. The log2 normalization Fragments per kilobase million (FPKM) of AhOMTs were used to construct the heatmaps.
Stress treatments and qRT-PCR analysis
Seedlings of peanut cultivar Minhua 6 (M-6) were grown in the greenhouse for stress treatments. Four-leaf old M-6 plants were subjected to abscisic acid stress (ABA 10 μg/mL) and low temperature (4°C). Samples were collected before treatment (0h, CK) for both ABA and low temperature and 3, 6, 9, and 12 hours after treatment. RNA was extracted by the CTAB method with some modifications (Sharif et al., 2022). cDNA was synthesized by Evo M-MLV RT Kit (Accurate Biotechnology, Hunan, Co., Ltd. China) following the manufacturer’s protocol. qRT-PCR was performed following our previous study (Sharif et al., 2022), while peanut Actin gene was used as the internal control. Data were analyzed by the 2-ΔΔCT method (Livak and Schmittgen, 2001). Expression levels at different time points were subjected to analysis of variance (ANOVA) and LSD test at α=0.05. Primers used for qRT-PCR are given in Supplementary Table 1.
Results
Identification and characterization of OMT genes in A. hypogaea
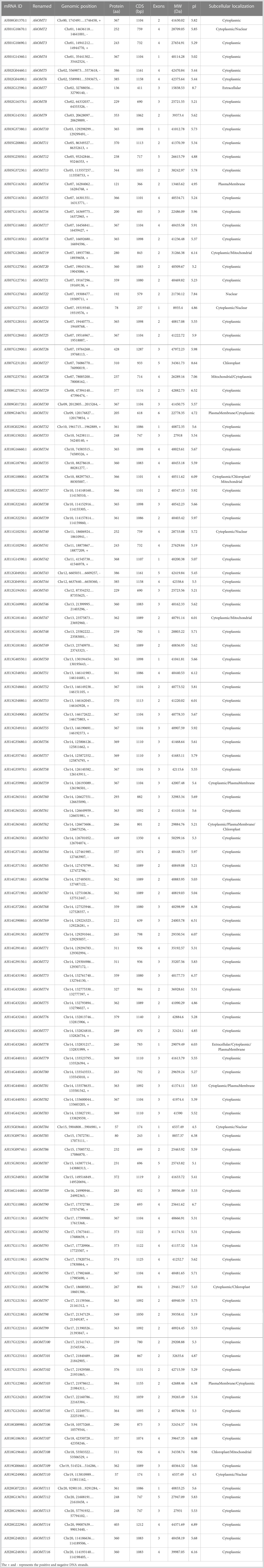
BLASTP and HMM searches were performed to find out the AhOMT family genes. Twenty-four genes were found in Arabidopsis, 55 in G. max, 58 in A. duranensis, and 68 in A. ipaensis through a comprehensive search in their respective genome databases. BLASTP search using these proteins and HMM search identified 116 OMT genes in the A. hypogaea genome. Table 1 shows the details of all 116 AhOMT genes. Briefly, AhOMT genes varied in size, ranging from 57aa (AhOMT84 and AhOMT110) to 449aa (AhOMT63). The same genes possessed the shortest and longest CDS lengths: (AhOMT84, AhOMT110) with 174bp and AhOMT63 with 1350bp. The physicochemical properties of these genes also varied accordingly. The molecular weights were from 6.537 kDa (AhOMT84 and AhOMT110) to 502.99 kDa (AhOMT63), and theoretical isoelectric points varied from 4.5 (AhOMT84, AhOMT110) to 9.06 (AhOMT108). The differences in isoelectric point (pI) and molecular weights (MW) are attributable to post-translational modifications and a high concentration of basic amino acids.
The subcellular localization prediction of AhOMT proteins showed a diverse kind of localization. The main organelle where all OMTs were localized was the cytoplasm, while some AhOMTs were also localized in more than one cell compartment, including the nucleus, mitochondria, chloroplast, plasma membrane, and extracellular spaces. The physicochemical properties of AhOMTs are given in detail in Table 1. Similar patterns of genomic and physicochemical properties were found in the AdOMTs and AiOMTs. The shortest of AdOMTs was AdOMT25 and AdOMT41, with a protein and CDS length of 104 aa and 312 bp, respectively. While the longest AdOMT was AdOMT57, with a protein and CDS length of 1760 aa and 5280 bp, respectively. The other physiochemical properties also varied, as the molecular weight ranged from 11.78 kDa for AdOMT41 to 194.78 kDa for AdOMT57. The theoretical isoelectric points varied from 4.86 for AdOMT43 to 8.51 for AdOMT46. The protein, CDS lengths, and physiochemical properties of AdOMTs are given in Supplementary Table 2. OMTs of A. ipaensis also possessed similar protein, CDS lengths and other properties. Proteins varied from 68 aa (AiOMT43) to 707 aa (AiOMT63), while CDS lengths from 204 bp (AiOMT43) to 2121 bp (AiOMT63). The expected molecular weight for AiOMTs ranged from 7.83 kDa (AiOMT43) to 78.87 kDa (AiOMT63), while the pI varied from 4.56 (AiOMT19) to 9.08 (AiOMT57). Most AiOMTs were located in the cytoplasm, while others were located in mitochondria, endoplasmic reticulum, and nucleus. Supplementary Table 3 shows detailed information about AiOMTs.
Phylogenetic relations of AhOMT genes
The phylogenetic tree containing A. ipaensis, A. duranensis, G. max, A. thaliana, and A. hypogaea OMTs divided them into three main groups (Figure 1). OMTs of all five species were dispersed in all clades of the phylogenetic tree, indicating that the OMTs genes diverged before the divergence of ancestral species. The phylogenetic results revealed that Group I comprised 14 OMT members (two GmOMTs, one AtOMT, four AiOMTs, six AhOMTs, and one AdOMT). Group II comprises 146 OMT members (20 GmOMTs, 21 AtOMT, 31 AiOMTs, 50 AhOMTs, and 24 AdOMTs). Group III contains 160 OMTs members (32 from G. max, two from A. thaliana, 31 from A. ipaensis, 62 from A. hypogaea, and 33 from A. duranensis). In summary, it can be hypothesized from the phylogenetic groupings that OMTs from different species with falling in a similar clade will probably perform similar functions. The greater number of OMTs in cultivated peanut than in its diploid progenitors and other model plants represent a high evolutionary rate in A. hypogaea.
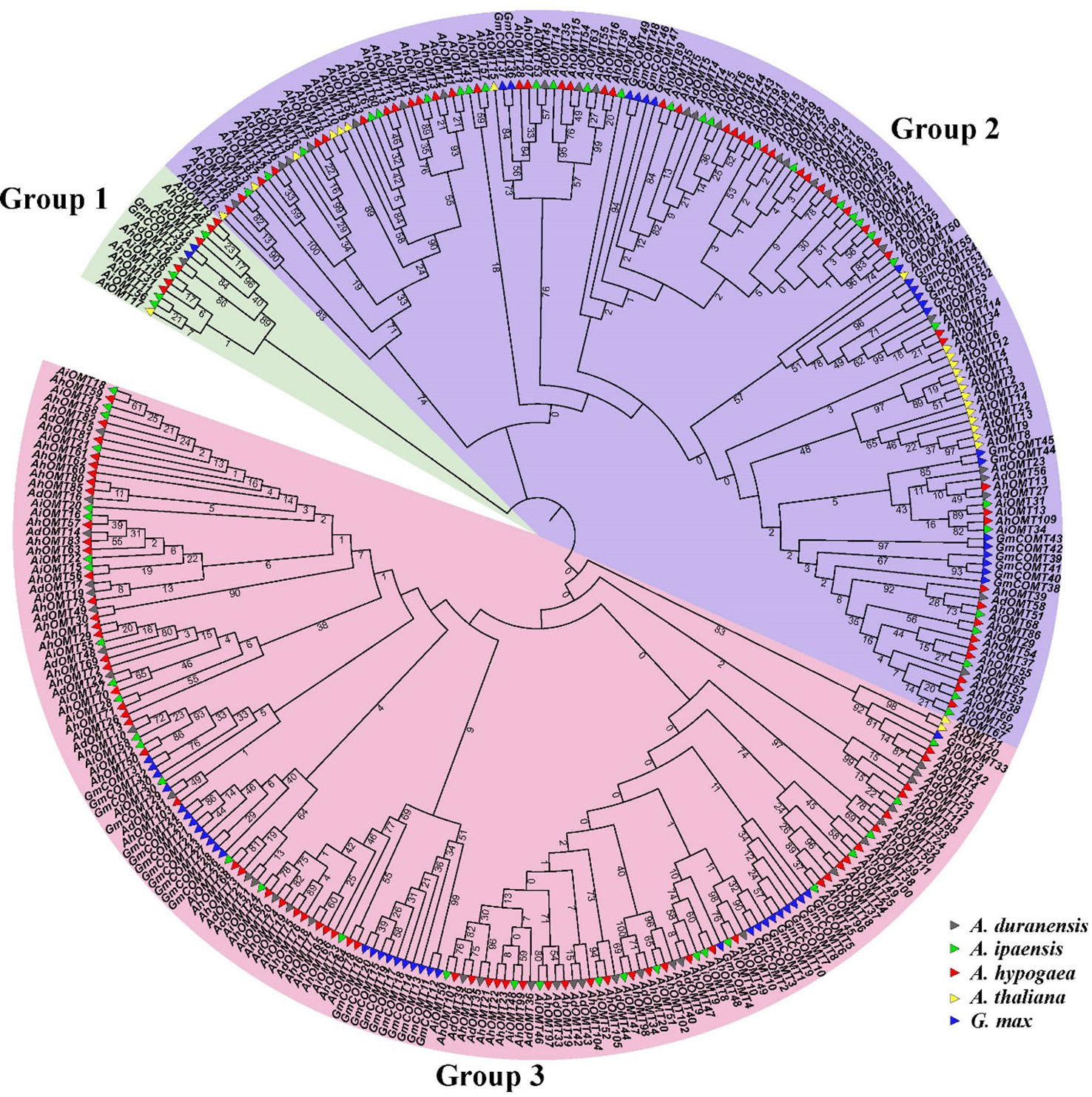
Figure 1 Phylogenetic relationships of O-Methyltransferase genes of A. hypogea, A. duranensis, A. ipaensis, A. thaliana, and G. max. The phylogenetic tree classified all OMTs into three groups, OMTs of all species are present in all clades. Group one is the smallest group as compared to other groups.
Chromosomal locations and gene duplication
Chromosomal location results revealed that all 116 AhOMT genes were dispersed on 18 chromosomes. Chromosomes Chr04 and Chr06 did not possess any OMT gene, while one gene was present on the unassembled genome region (Chr00). Chromosomes Chr00, Chr08, and Chr16 possessed one OMT each, while Chr07 possessed the highest genes in the A subgenome (15 genes) and in the B subgenome on Chr14 (28 genes) and Chr17 (16 genes), and all other chromosomes possessed varying numbers of OMT genes (Figure 2). Chromosomes Chr03, Chr09, and Chr19 had two genes each. Chr01, Chr05, Chr11, Chr12, and Chr18 possessed three genes each, Chr02 possessed four, and Chr15 possessed five AhOMTs. Chr20 is next with six genes, Chr10 with eight genes, and Chr13 with ten genes (Figure 2). The A. duranensis genome possessed 58 OMTs (AdOMTs) unevenly distributed on all ten chromosomes. Only chromosome A09 possessed a single OMT; all other chromosomes contained multiple copies of AdOMTs ranging from 2-19. Chromosome A08 possessed two AdOMTs, while the highest number was present on chromosome A07, which had 19 AdOMTs (Supplementary Figure 1). The genome of A. ipaensis contained 68 copies of OMT genes (AiOMTs) ranging from 2-16 genes. Chromosome B06 had the least number of AiOMTs (two), while chromosomes B04 and B07 possessed the highest number of AiOMTs (16 genes each) (Supplementary Figure 2).
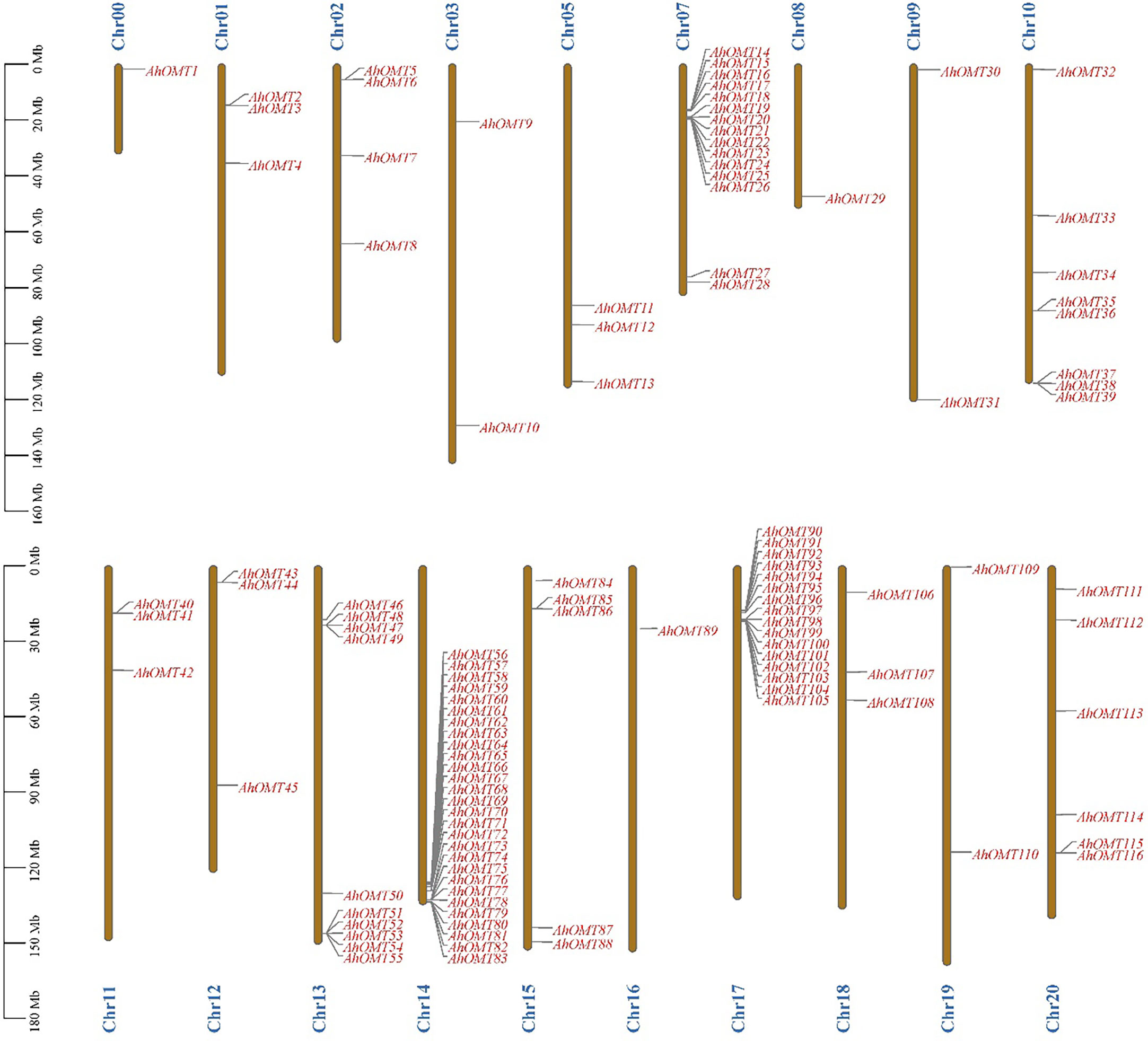
Figure 2 Chromosomal distribution of AhOMTs genes. The highest number of AhOMTs on A-subgenome were present on Chr07 (15 genes) and in B-subgenome on Chr14 (28 genes) and Chr17 (16 genes), and all other chromosomes possessed varying numbers of OMT genes.
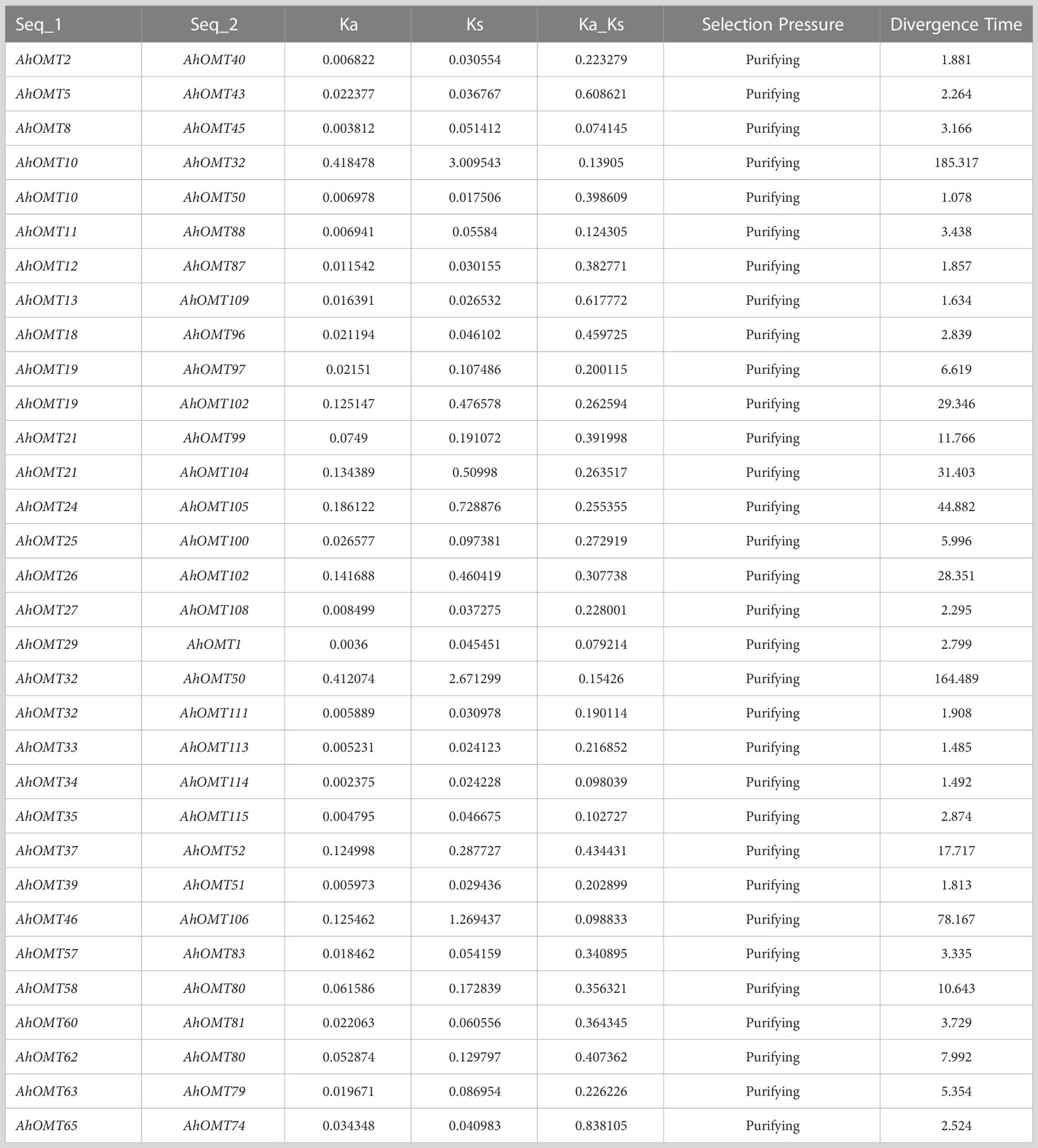
Gene duplication analysis revealed 32 duplicated pairs of AhOMTs. To estimate the molecular evolution rate, the synonymous (Ks) and nonsynonymous (Ka) substitutions were computed for duplicated genes. Positive selection pressure was assumed when Ka/Ks>1, purifying selection when Ka/Ks<1, and neutral selection when Ka/Ks=1 (Yang and Bielawski, 2000). Results showed that mainly purifying selection drove the genome duplication. Furthermore, the duplicated gene pair divergence timeframe was estimated as t=ks/2r. The expected divergence time varied from 1.078 million years ago (mya) for AhOMT10:AhOMT50 to 185.317 MYA for AhOMT10:AhOMT32 (Table 2). Most genes were segmentally duplicated, but some were tandemly duplicated (Figure 3).
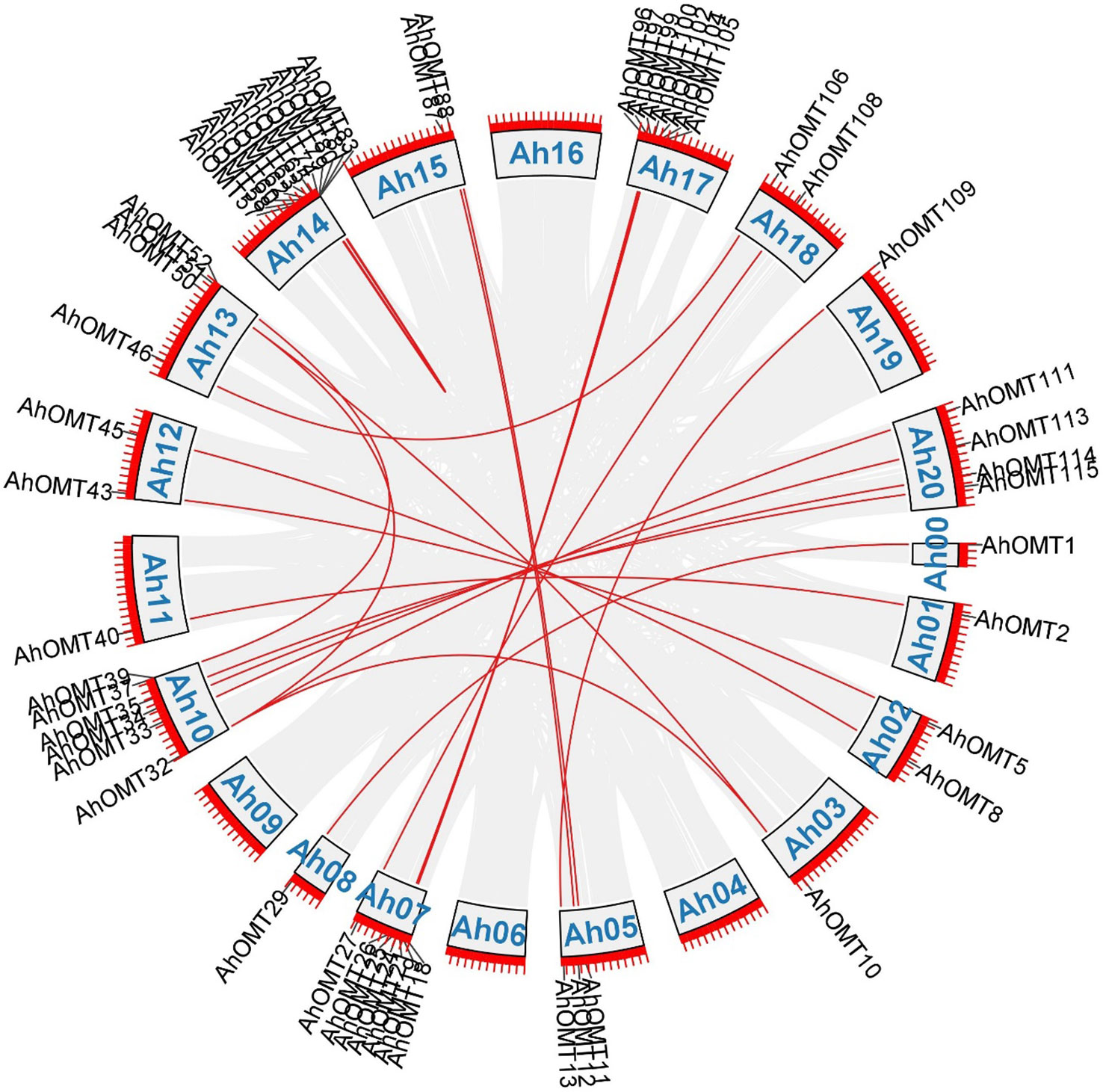
Figure 3 Duplicated gene pairs among AhOMTs. Out of 116 genes, 42 are duplicated genes. Red lines show the duplicated AhOMT pairs, while the gray lines in the background show the syntenic blocks (duplicated pairs) among different chromosomes.
Gene structure and motifs analysis
To better understand the gene structure of AhOMTs, we viewed their exon-intron distribution patterns. According to the findings, the introns in AhOMT genes varied from 0 to 5, and exons from 1 to 6. Many AhOMT genes were composed of a single intron and two exons. Forty-two out of 116 AhOMTs possessed two exons. Three and four exons were also common, as 30 genes possessed three exons while 25 genes had four exons. Thirteen genes were composed of a single exon, and only AhOMT31 comprised six exons (Figure 4). EME server identified conserved motifs inside the full-length protein sequences of AhOMT genes in order to determine structural diversification and functional assessment. Ten conserved motifs were predicted in AhOMT genes (Figure 4). Conserved motifs varied in length as motif 1 was the most extended motif with 39 amino acids, while 4th-6th and 8th-10th motifs were the shortest with 21 amino acid residues (Supplementary Table 4). In a nutshell, conserved motif, phylogenetic, and gene structure analysis indicated that AhOMT proteins comprise extremely well-sustained members of amino acids that remain inside a group. Proteins with similar motifs and structures can therefore be functionally related. The motif distribution patterns and gene structure of OMTs of wild progenitors were as per A. hypogaea OMTs. Information on motifs and structure of AdOMTs are given in Supplementary Figure 3, and on AiOMTs is given in Supplementary Figure 4.
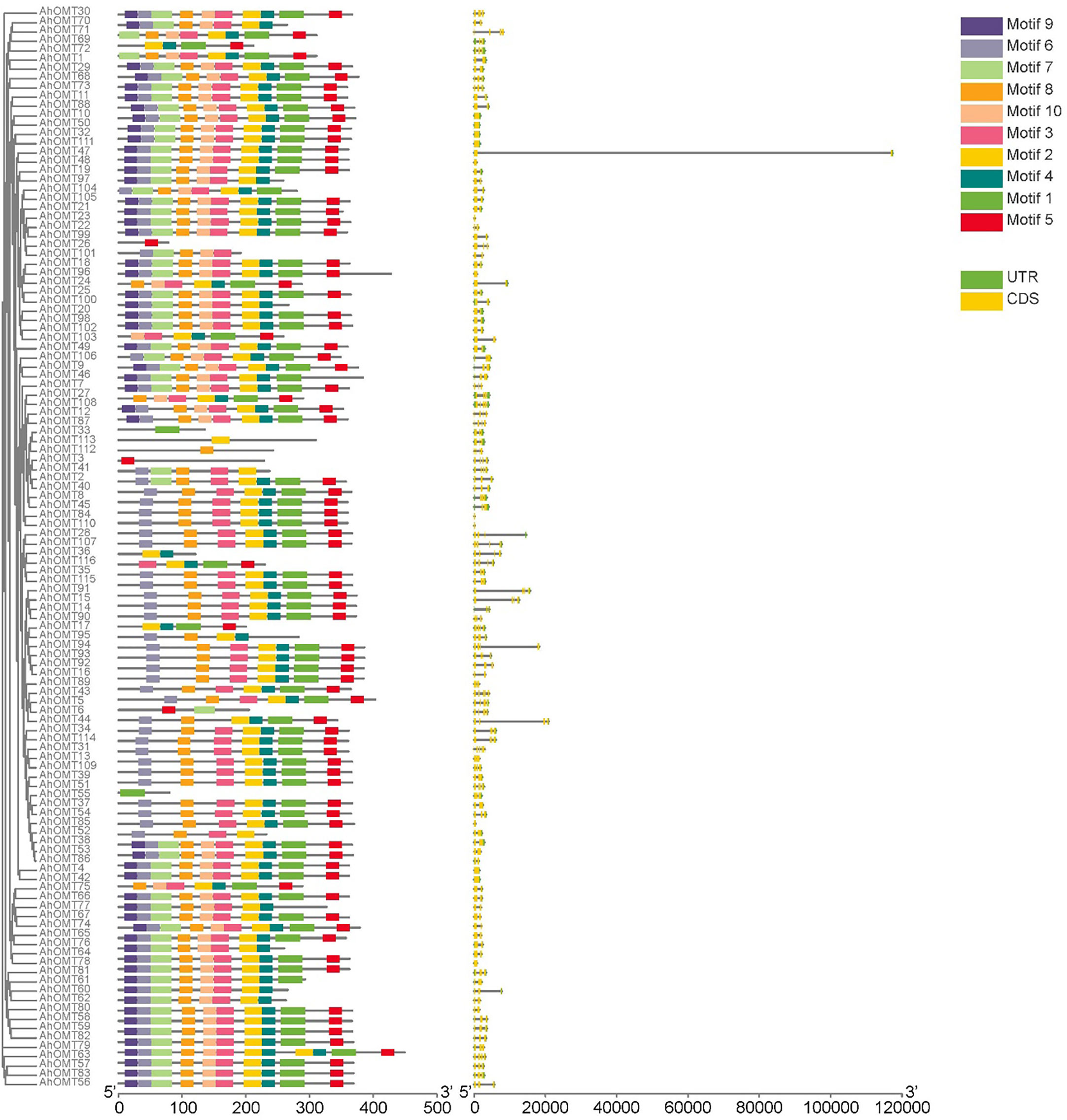
Figure 4 Conserved motifs distribution patterns and gene structure (exons-introns distribution) of AhOMTs genes. Commonly shared motifs among genes tend to cluster in the same groups, referring to their similar functions.
Promoter analysis of AhOMTs genes
The cis-elements of any genes’ promoter are responsible for controlling its expression and functions. We examined cis-acting regions in the AhOMT promoters to know their functional and regulatory roles. Predicted cis-elements showed that aside from the CAAT- and TATA-Box (core promoter elements), a large number of other key elements were also present (Figure 5). We classified these cis-regulatory elements into four groups according to their functions: development and growth-related, hormones-responsive, light-responsive, and stress-related elements. All 116 AhOMTs were enriched with hormones- and light-responsive elements, 108 genes were enriched with growth and development-related elements, and 94 genes were enriched with stress-responsive elements (Figure 6).
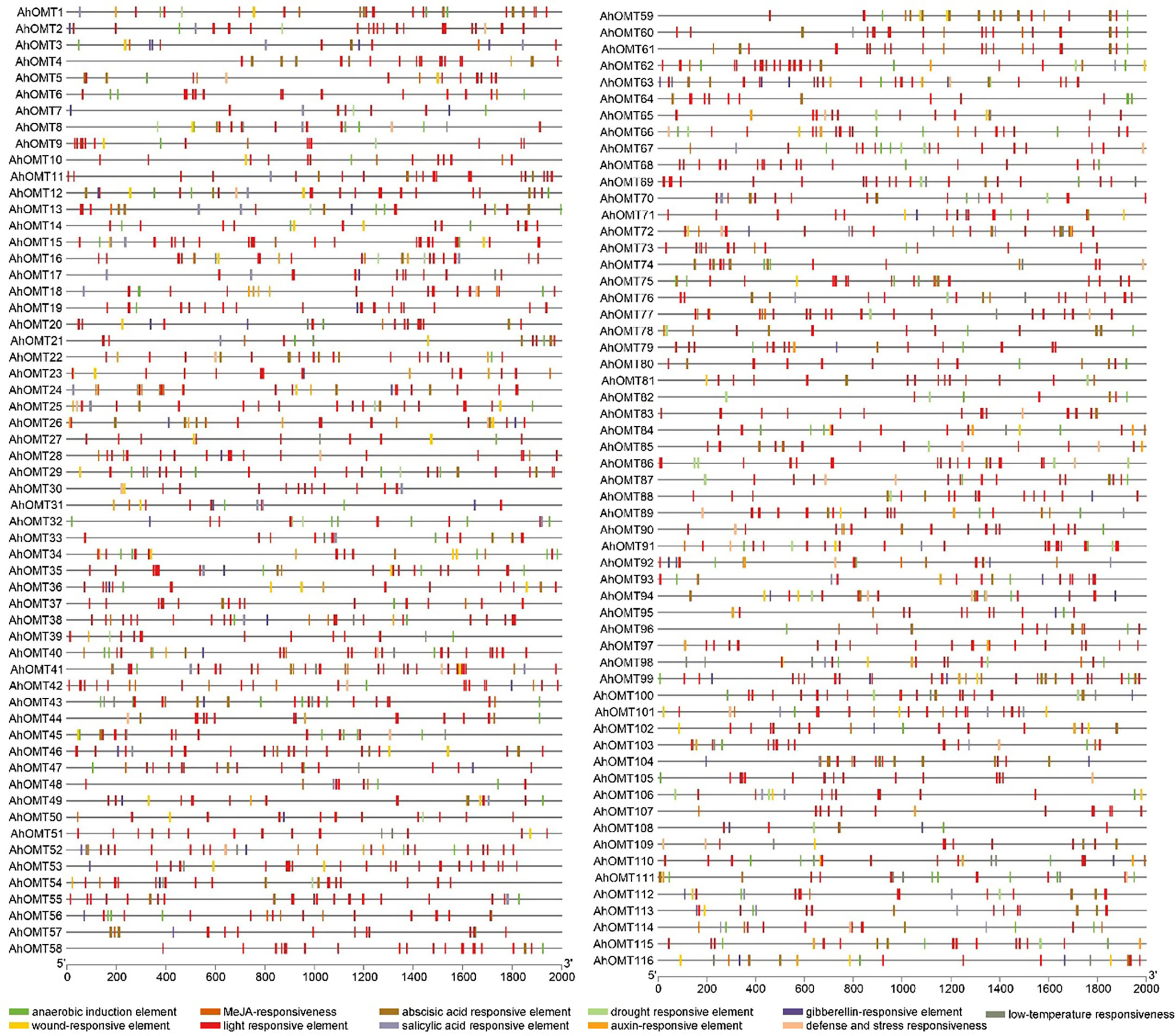
Figure 5 Cis-regulatory elements of AhOMT promoters. Cis-elements analysis revealed important elements responsive to light, hormones, growth and development, and stress responsiveness.
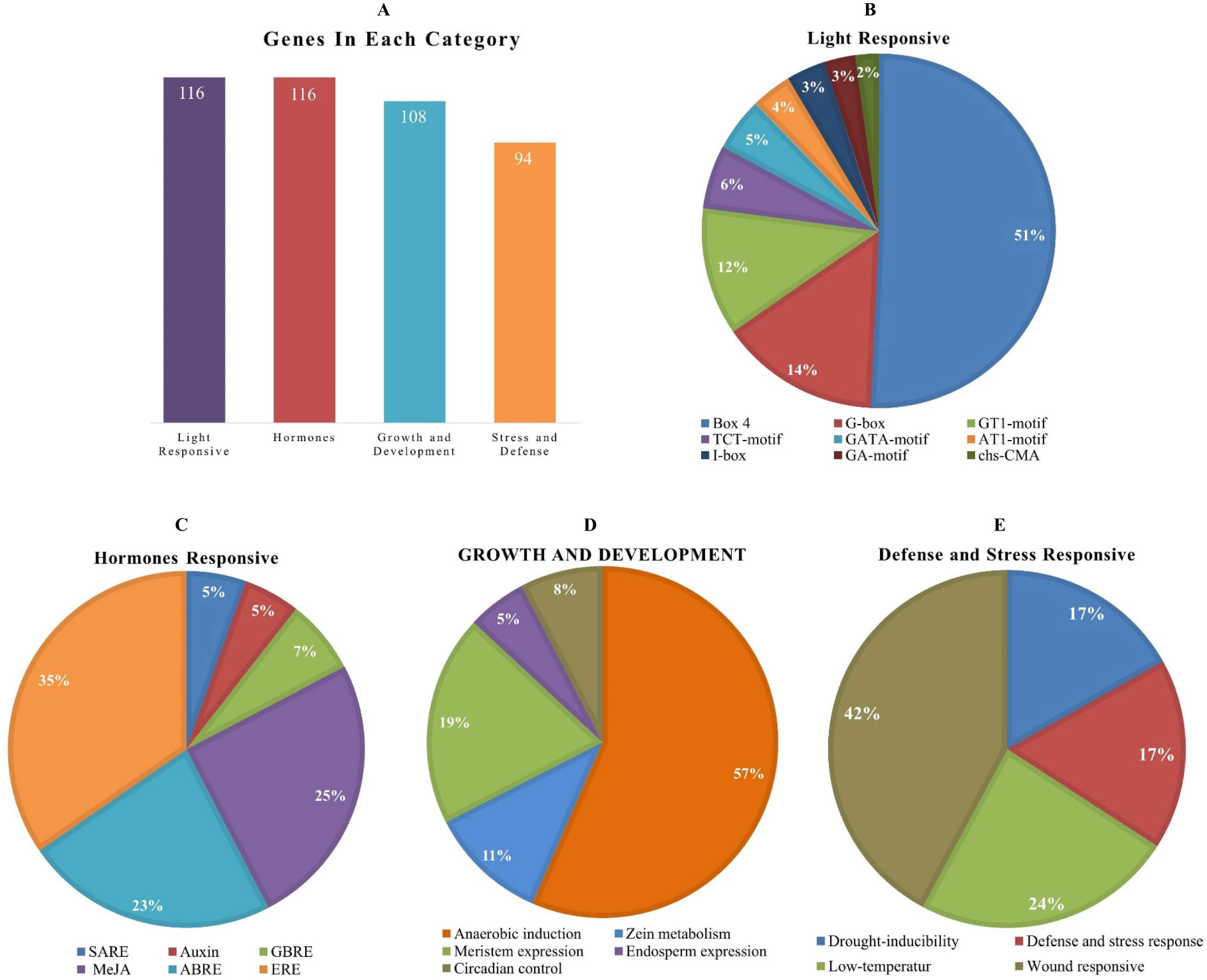
Figure 6 Cis-regulatory elements of AhOMT promoters. (A) The number of genes in different categories of elements. (B) Composition of light-responsive, (C) hormone-responsive, (D) growth and development responsive, and (E) stress-responsive factors.
Elements responsive to light mainly include TCT-motif, GATA-motif, G-box, Box-4, GT1-motif, GA-motif, chs-CMA element, I-box, and AT-1 motif. Other light-responsive elements include 3-AF1 binding site, ATC-motif, AE-box, MRE element, Box II, CAG-motif, CGTCA-motif ATCT-motif, ACE element, Gap-box, TCCC-motif, GTGGC-motif, LAMP-element, LS7 element, and Sp1 element were also present. Hormones responsive class includes ABA-responsive (ABRE), auxin-responsive (AuxRE, AuxRR-core, CGTCA-motif, TGA-box), gibberellins responsive (GARE motif, P- and TATC-box), MeJA-responsive (CGTCA-motif, TGACG-motif), SA-responsive (SARE, TCA-element), and ethylene-responsive (ERE) elements. The growth and development category contained anaerobic induction responsive (ARE), meristem expression responsive (CAT-box), endosperm expression related (GCN4-motif, AACA-motif), circadian control (CAAAGATATC), and zein metabolism-related (O2-site) elements. The stress-responsive class further includes defense and stress response (TC-rich repeats), drought-responsive (MBS), low-temperature responsive (LTR), and wound-related (WUN-motif) elements (Figure 6).
Prediction of miRNAs and synteny analysis
Numerous studies in the last few years have revealed that micro-RNAs regulate the expression of genes under developmental processes and stress responses (Chen et al., 2019; Wani et al., 2020; Raza et al., 2021a). For this reason, we predicted miRNAs targeting AhOMT genes sequentially to get more understanding of miRNA-mediated post-transcriptional regulations of AhOMT genes. Micro RNAs from 12 different families targeted 35 AhOMTs. Supplementary Table 5 contains the complete information on all miRNAs. Two members of the miR156 family targeted AhOMT34, AhOMT37, AhOMT38, AhOMT52-AhOMT55, AhOMT87, and AhOMT114. miR16o-3p was found to target four OMTs. Some of the miRNAs targeting the AhOMTs with their target sites are shown in Figure 7. More research for their expression levels and the genes they target is needed to establish their biological involvement in the peanut genome.
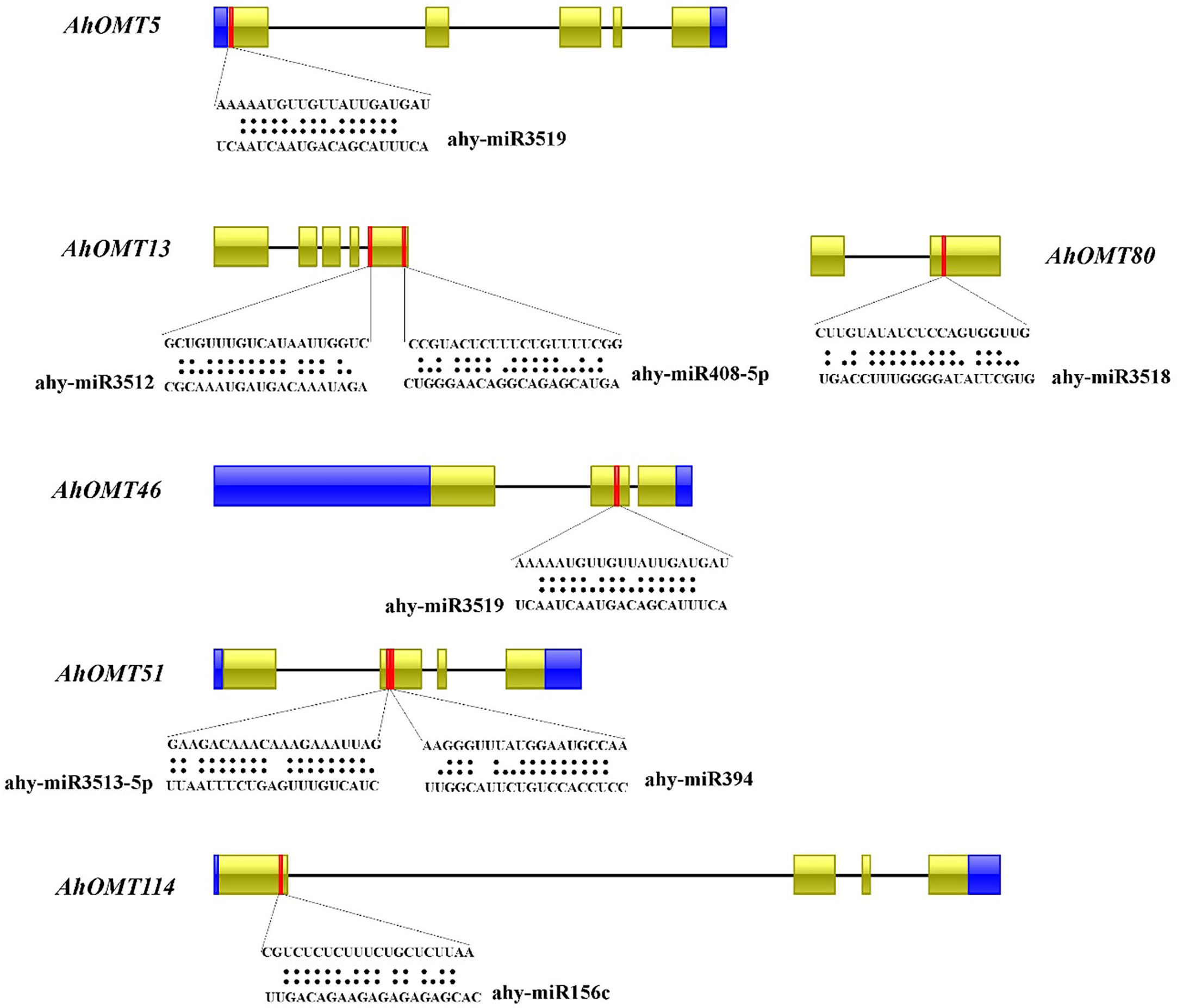
Figure 7 Predicted miRNAs targeting AhOMTs. Schematic representation of some miRNAs targeting AhOMTs, and their target sites.
Comparative synteny analysis among A. hypogaea, diploid peanut species, and A. thaliana represented remarkable evolutionary, duplication, expression, and functional relationships. AhOMTs mainly showed significant syntenic relationships with its wild progenitors and Arabidopsis; however, the syntenic relationships of A. hypogaea were closer to wild peanut species than Arabidopsis. A total of 56 syntenic relationships of A. hypogaea were found in the genome of A. duranensis and 60 in A. ipaensis. In contrast, only four syntenic relationships were found among AhOMTs and AtOMTs. The synteny analysis showed that A. hypogaea is closer to its wild progenitors than Arabidopsis. The syntenic relations of these species are shown in Figure 8.
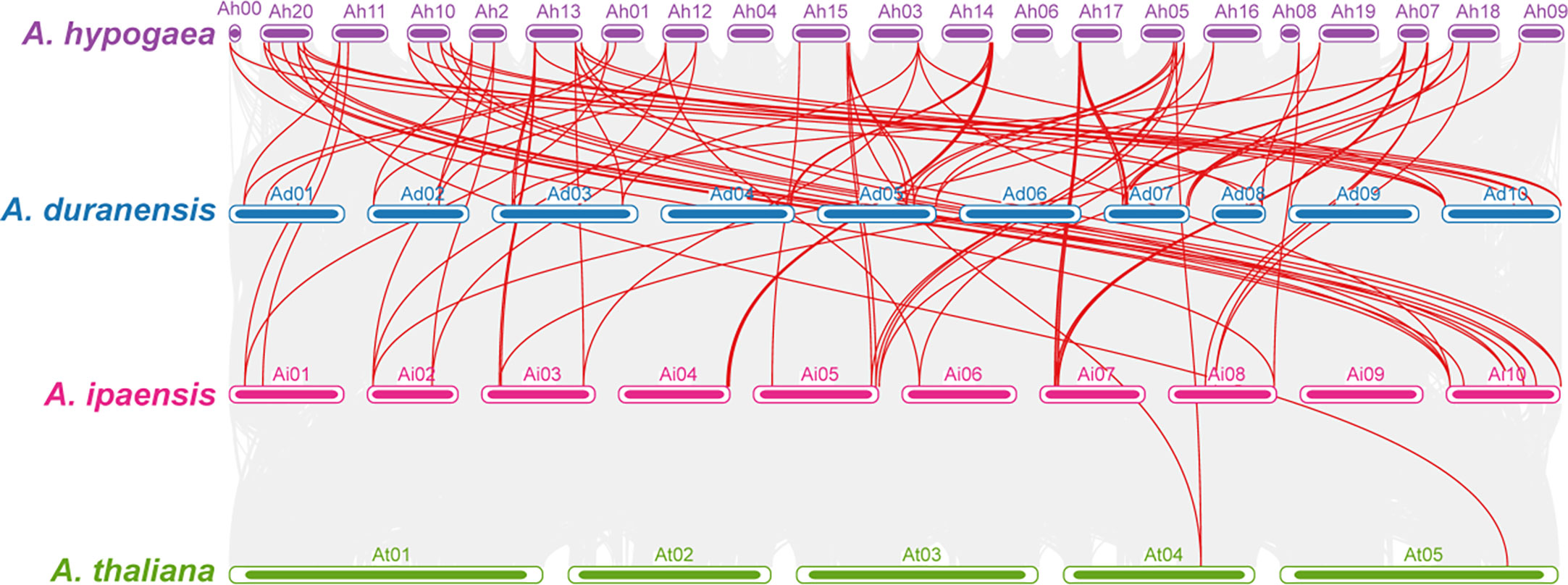
Figure 8 Synteny analysis among A. hypogea, A. duranensis, A. ipaensis, and A. thaliana. Synteny analysis showed key evolutionary relationships of OMTs in diploid and tetraploid peanut species. AhOMTs possessed highly conserved syntenic relationships with other peanut species as compared to Arabidopsis.
Identification of orthologous gene clusters
Identifying orthologous gene clusters is important to assess the polyploidization events during a gene family’s evolution. A relative assessment was developed to identify orthologous gene clusters shared by A. hypogea, A. duranensis, A. ipaensis, G. max, and A. thaliana. The detected gene clusters and their respective overlapping regions are presented in greater detail in Figure 9. A. hypogea recorded maximum clusters, followed by A. ipaensis, A. duranensis, G. max, and A. thaliana. Results showed that three gene clusters are shared among all these species, while 18 gene clusters are solely composed of OMTs found in peanut diploid and tetraploid species, which indicates that polyploidization has evolved new peanut-specific orthologous OMT clusters. We also identified orthologous gene clusters among three peanut species (Supplementary Figure 5). Comparatively, 100, 89, 94, 36, and 21 orthologous OMTs were found in A. hypogea, A. duranensis, A. ipaensis, G. max, and A. thaliana, respectively. Thirty in-paralogs were identified in A. hypogea, and only two were found in A. ipaensis. A duranensis did not show any in-paralogous gene. Surprisingly 32, 14, and 20 singletons were also found in A. hypogea, A. duranensis, and A. ipaensis, respectively (Supplementary Table 6). Results demonstrated that identified orthologous genes decrease with increased phylogenetic distances.
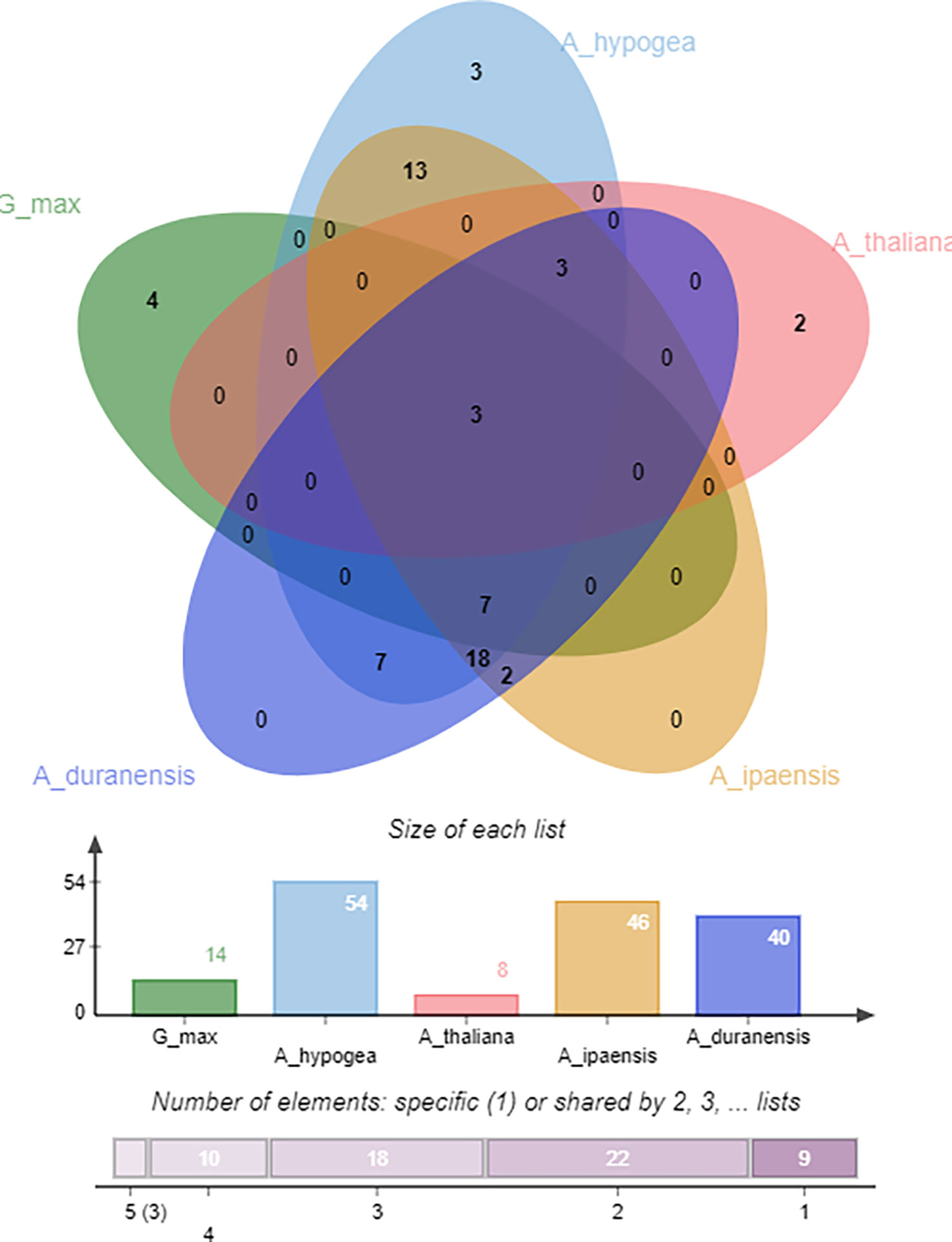
Figure 9 Orthologous genes’ clusters among A. hypogea, A. duranensis, A. ipaensis, A. thaliana, and G. max. A. hypogea recorded maximum clustered. Eighteen gene clusters are solely composed of OMTs found in peanut diploid and tetraploid species, indicating that polyploidization has evolved new peanut-specific orthologous OMT gene clusters.
Prediction of protein-protein interaction network
The Functions of AhOMTs could be speculated based on well-studied Arabidopsis OMTs. Using the STRING database, we performed the interaction network analysis of cultivated peanut OMT proteins relative to orthologues in Arabidopsis to understand their functions. Protein interaction network prediction showed that AhOMT116 has functions related to C4H that regulate carbon flux to essential pigments for pollination or UV protection. AhOMT7 and AhOMT111 may function as Cinnamoyl-CoA reductase 1 (IRX4) involved in lignin biosynthesis at the latter stages. AhOMT87 has CCOAMT-like functions, a putative caffeoyl-CoA O-methyltransferase of Arabidopsis that helps in the biosynthesis of feruloylated polysaccharides. AhOMT77 has 4CL1-related functions (4-coumarate-CoA ligase 1), involved in the later phase of the general phenylpropanoid pathway. AhOMT31 may function as SNC1, a putative disease-resistance protein of the TIR-NB-LRR-type. The interaction network of AhOMTs with well-studied Arabidopsis proteins is given in Figure 10. Some OMTs did not show interactions with reported Arabidopsis proteins, and there is a possibility that these proteins have some other functions yet to be reported.
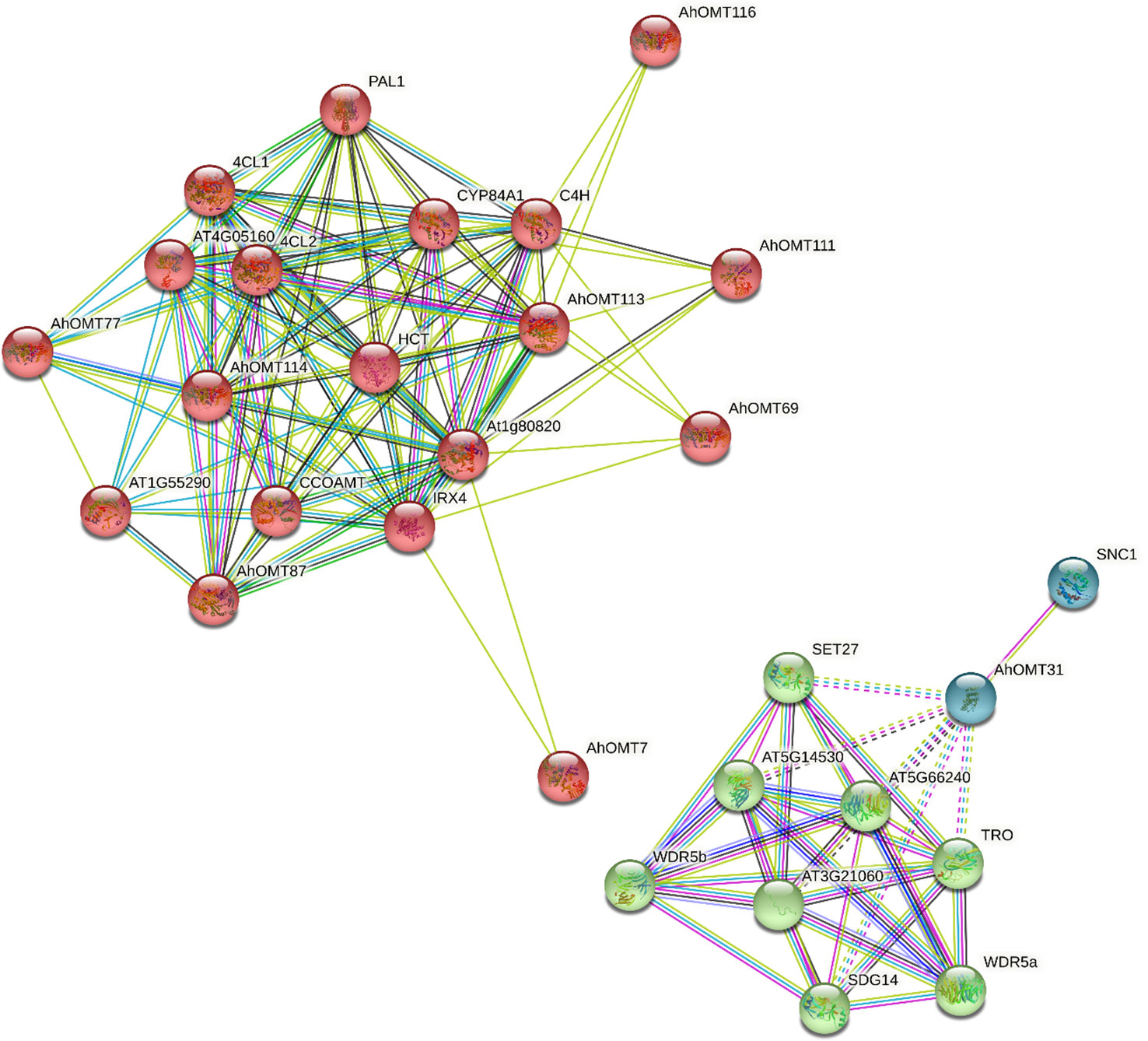
Figure 10 The predicted protein–protein interaction network of AhOMTs using STRING database. Putative protein functions of AhOMTs are predicted on well-studied OMT orthologues in Arabidopsis.
Functional annotation analysis of AhOMTs
GO annotation analysis of AhOMTs was performed to view their possible roles in biological processes (BP), molecular functions (MF), and cellular components (CC). GO enrichment results provided highly enriched terms related to BP, MF, and CC (Figure 11). AhOMTs were mainly involved in MF and BP categories. AhOMTs were highly enriched in transferase activity (GO:0016740), catalytic activities (GO:0003824), methyltransferase activity (GO:0008168, GO:0008171, GO:0042409), and S-adenosylmethionine-dependent methyltransferase activity (GO:0008757) in MF category. In the BP category, AhOMTs were highly enriched in methylation (GO:0032259), biosynthetic process (GO:0009058, GO:0044249), cellular metabolic processes (GO:0044237, GO:0008152), and aromatic compound metabolism (GO:0006725). The KEGG enrichment analysis showed that AhOMTs are mainly involved in metabolic processes, including 01058 acridone alkaloid biosynthesis, 00943 isoflavonoid biosynthesis, B 09110 secondary metabolites biosynthesis, 00380 tryptophan metabolism, 00941 flavonoid production, and amino acid B 09105 metabolism (Figure 11). Collectively, it is evident from functional annotation analysis that AhOMTs play key roles in several cellular, biological, and molecular functions.
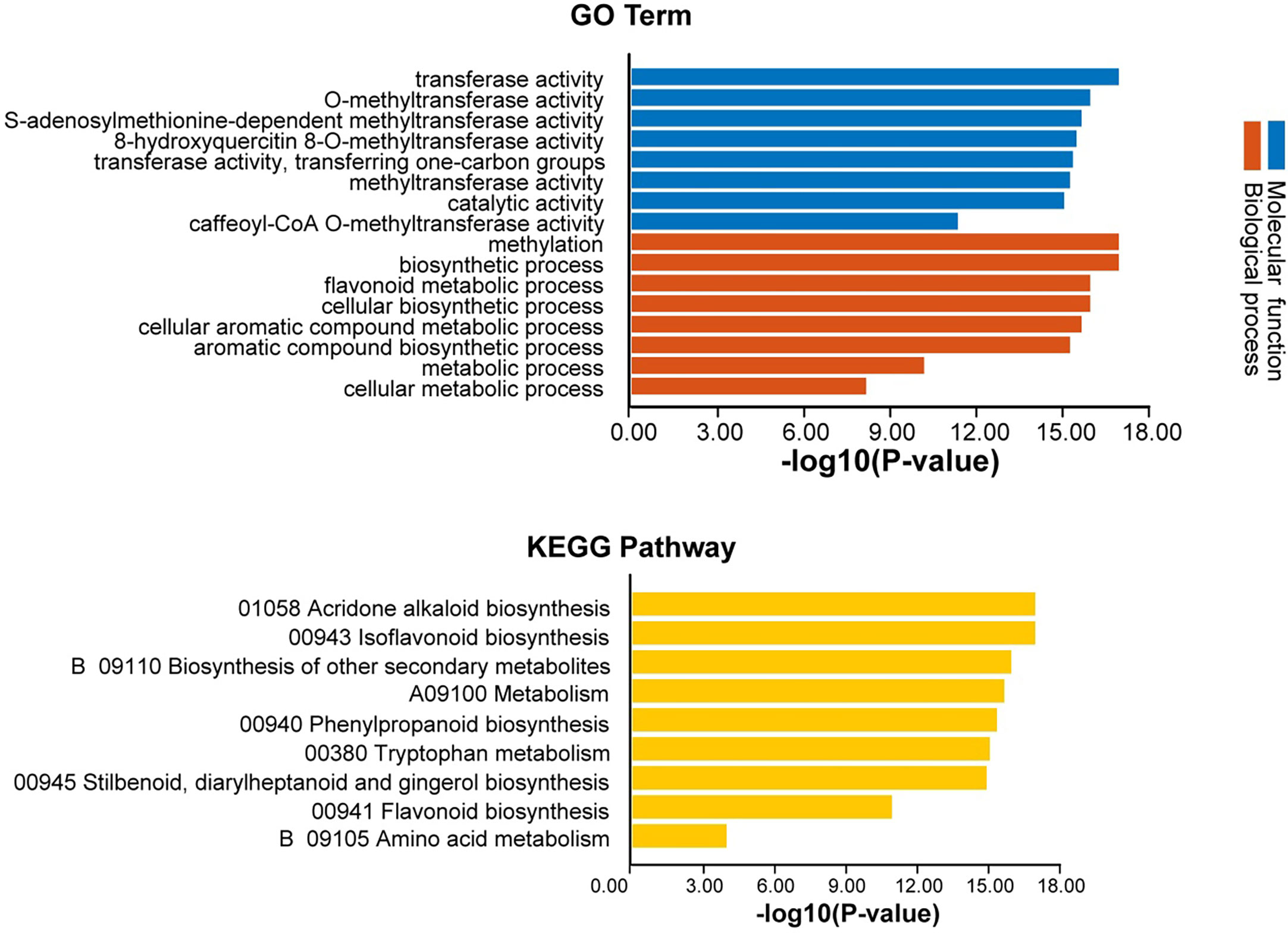
Figure 11 Functional annotation (GO, KEGG) analysis of AhOMTs. The gene ontology analysis showed that AhOMTs are involved in molecular function (MF), and biological processes (BP). The KEGG enrichment analysis showed that AhOMTs are mainly involved in metabolic processes.
Expression profiling of AhOMTs in different organs
AhOMT genes’ expression levels in different organs/tissues, containing leaf, stem, flower, root, root nodule, peg, pericarp, testa, cotyledon, embryo, etc., was determined using the peanut RNA-seq datasets. According to the expression profiling results, there was a noticeable variance in the expression of various tissues. Transcriptome expression results showed that AhOMT32-AhOMT35, AhOMT45, AhOMT71, AhOMT106, AhOMT113, AhOMT114, and AhOMT116 genes showed relatively higher levels of transcriptional abundance in the leaf, stem, flower, root, root nodule, peg, pericarp, testa, cotyledon, and embryo. These genes can be suitable candidates for improving peanut growth and yield. AhOMT9 and AhOMT46 specifically showed high expression in root nodules (Figure 12). It can be speculated that these two genes are good targets to improve nitrogen fixation that can provide good crops by effectively fixing the soil nitrogen. FPKM values of transcriptome expression of AhOMTs are given in Supplementary Table 6.
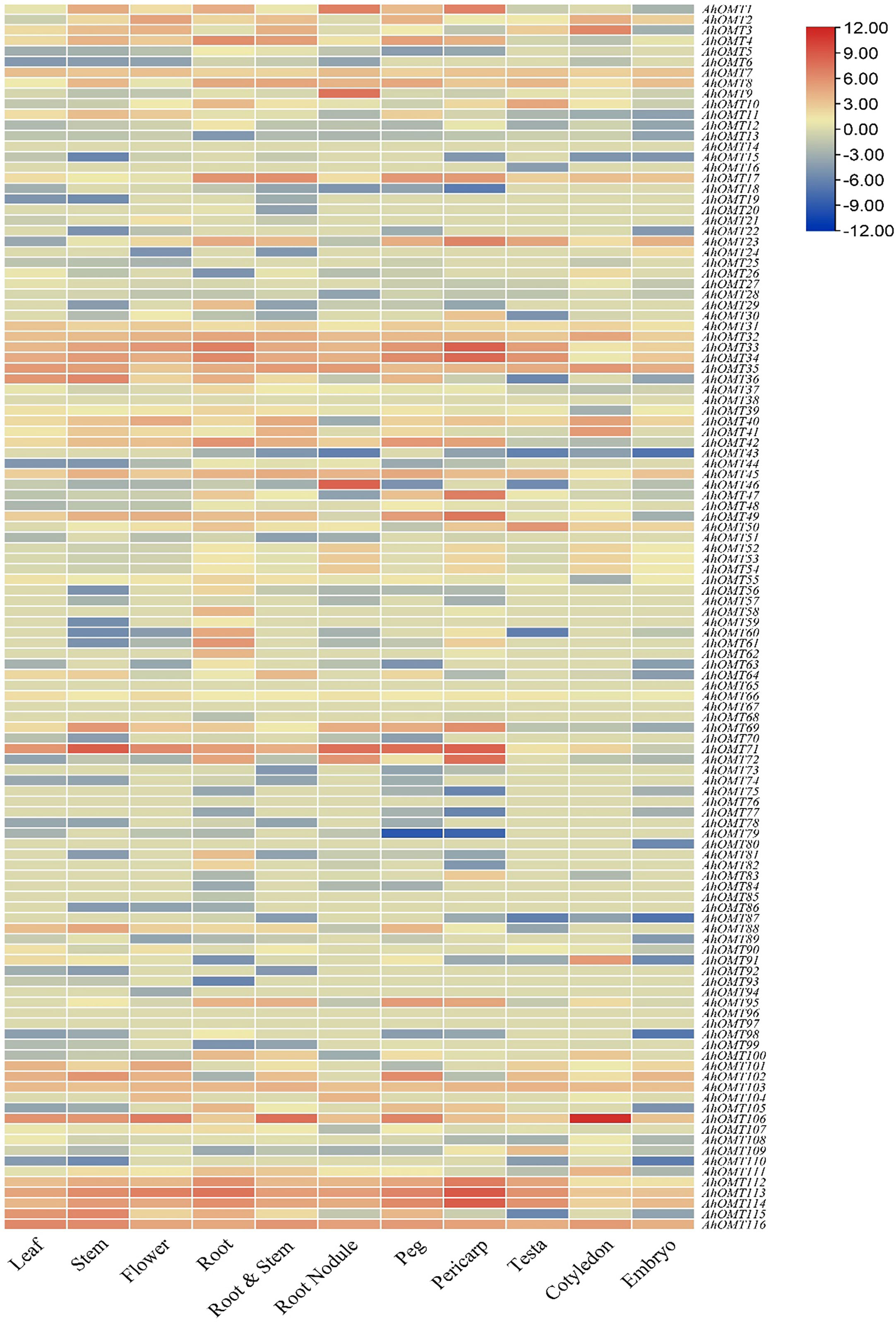
Figure 12 Transcriptome expression of AhOMTs in different tissues. AhOMTs possessed varying expression matrix in different tissues. All genes possessed varying levels of expression in different tissues.
Expression profiling of AhOMTs under hormones, drought, and temperature stress
Transcriptome data provided the expression patterns of 116 AhOMTs for different phytohormones (ABA, SA, Brassinolide, Paclobutrazol, and Ethephon) treatment, water stress (drought and regular irrigation), temperature stress (4°C and 28°C). Under temperature stress, the AhOMT106 gene was highly active, while AhOMT35, AhOMT71, and AhOMT113 were also expressed in most cases, but AhOMT35 did not show expression under drought stress. Almost 16 genes showed expression in response to ABA and SA, 14 genes responded to brassinolide, and 12 genes were responsive to ethephone. Thirteen genes were expressed under decreased temperature, and almost 11 genes were responsive to drought stress (Figure 13). Many genes were non-responsive to the hormones, water and temperature treatments.
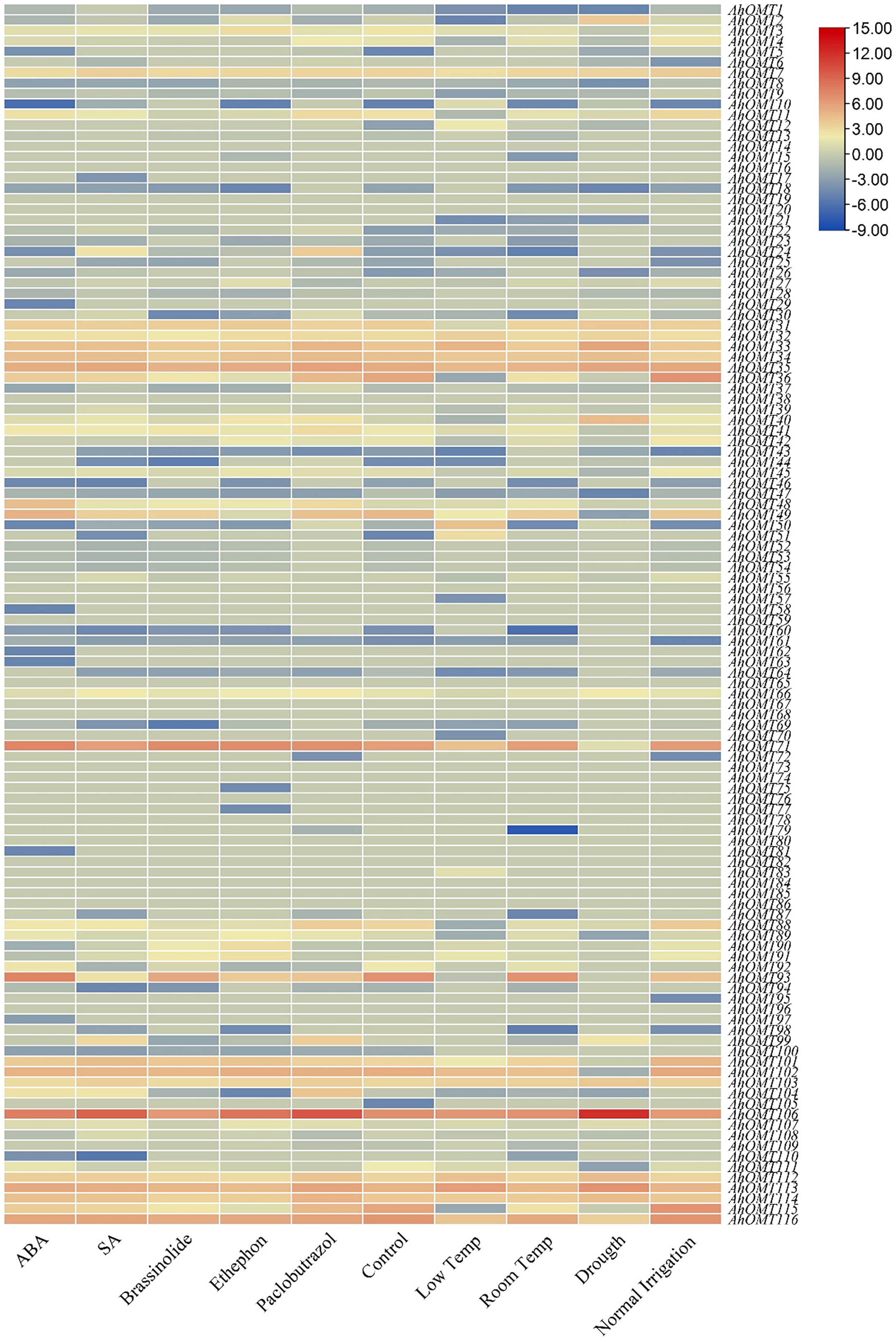
Figure 13 Transcriptome expression of AhOMTs under different hormones and stress conditions. Under the stress conditions, the AhOMT106 gene was most active, while AhOMT35, AhOMT71, AhOMT113 were also expressed in most of cases.
Quantitative expression profiling under ABA and low-temperature treatment
For real-time expression profiling by qRT-PCR, 12 AhOMT genes were randomly selected. These genes included AhOMT-7, AhOMT-18, AhOMT-33, AhOMT-34, AhOMT-35, AhOMT-46, AhMT-61, AhOMT-71, AhOMT-93, AhOMT-106, AhOMT-113, and AhOMT-116. These genes were selected based on their response to hormones, water and temperature stress, while genes with higher and lower expression were considered. Under ABA treatment, the expression of all selected genes corresponds to their transcriptome expression. For instance, AhOMT-7, AhOMT-33, AhOMT-34, AhOMT-35, AhOMT-71, AhOMT-93, AhOMT-106, AhOMT-113, and AhOMT-116 were upregulated under ABA stress, while AhOMT-18, AhOMT-46, and AhOMT-61 were downregulated (Figure 14). Under low temperature, a similar expression was found as of ABA treatment. Although there were some deviations in transcriptome expression and qRT-PCR expression, overall, the expression pattern of all selected genes is in accordance with transcriptome expression (Figure 15). The results of qRT-PCR represent the reliability of transcriptome datasets.
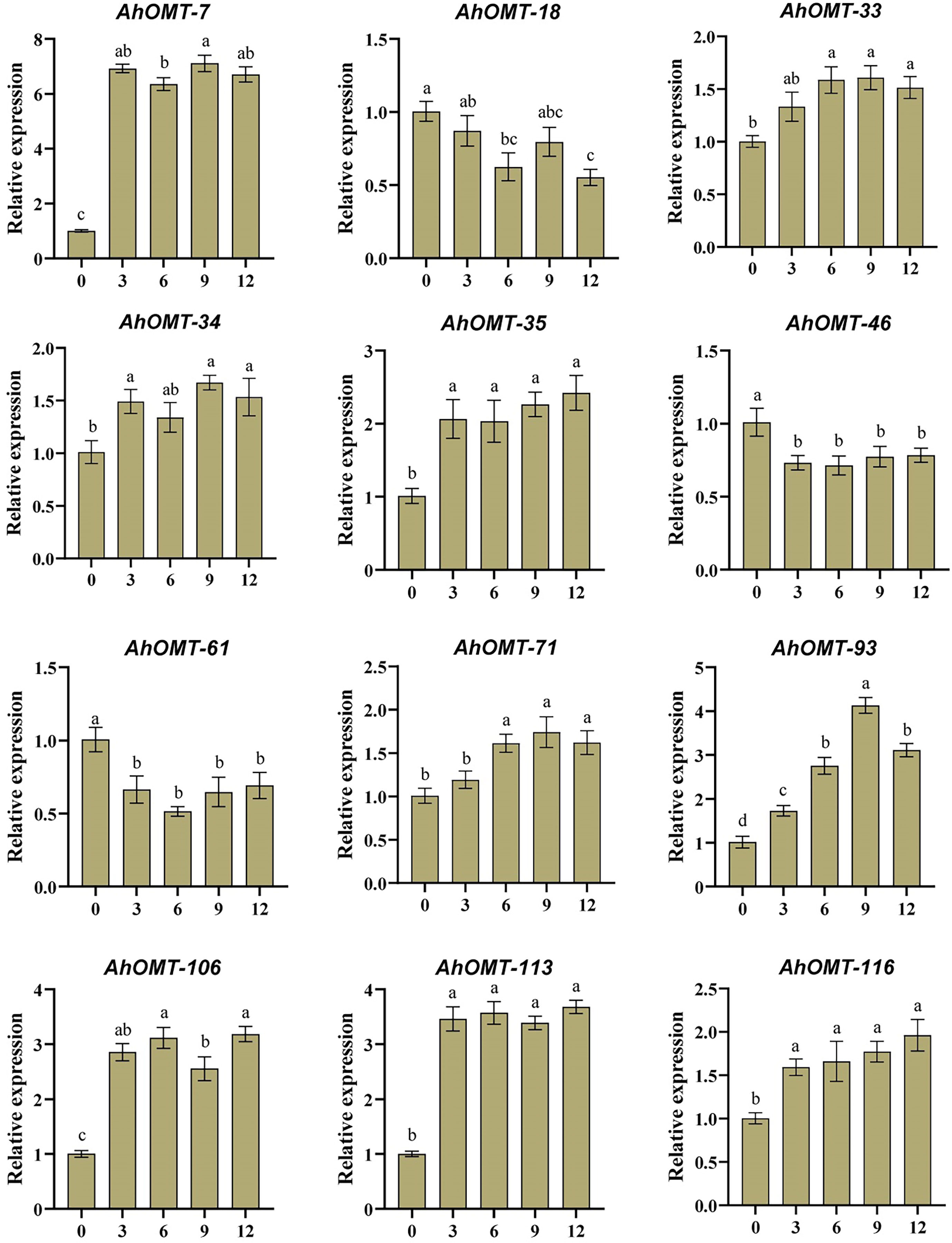
Figure 14 Expression profiling of AhOMT genes in response to ABA treatment. Mainly AhOMT genes recorded increased expression under ABA stress, while some genes were down-regulated. a, b, and c represents the significance levels among expression at different time points.
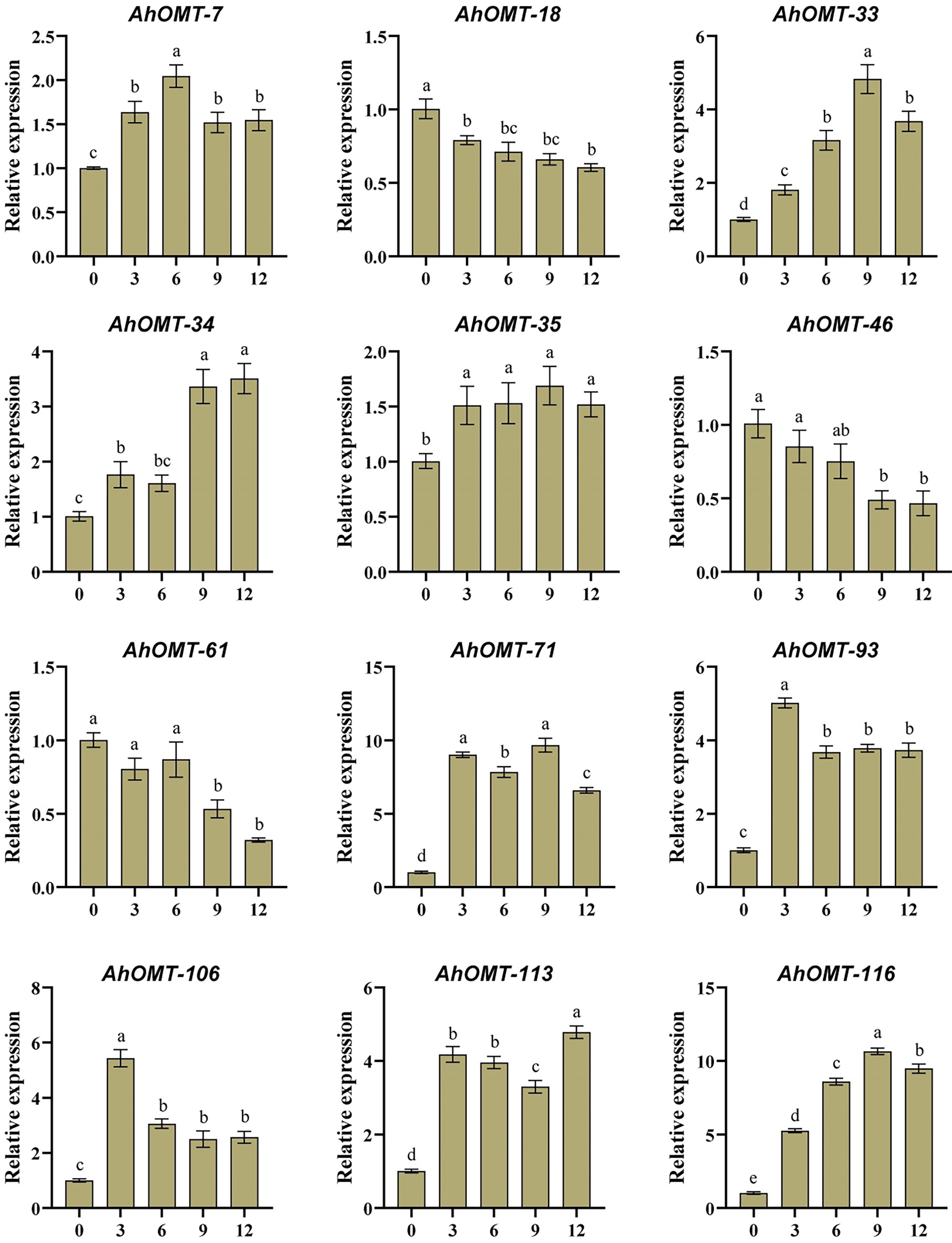
Figure 15 Expression profiling of AhOMT genes in response to cold stress. Mainly AhOMT genes recorded increased expression under cold stress, while some genes were down-regulated. a, b, and c represents the significance levels among expression at different time points.
Discussion
Several plants, including A. thaliana, B. distachyon, B. napus, P. trichocarpa, O. sativa, and others, have been studied at the whole-genome level to determine the presence and possible roles of OMT family genes. Because of their importance for synthesizing S-type lignin, the roles of OMTs have been well established. Lignin is the cell wall’s most important component to cope with environmental and biological stress (Boerjan et al., 2003). Reduced lignin production poses the plant to a lodging state (Hu et al., 2015). Reduced lignin concentration in legumes reduces stalk strength which ultimately reduces diseases and pathogens resistance (Bellaloui, 2012). Genome size, genome duplication, and gene distribution all have a significant influence on genetic diversity. Genetic duplication has been recognized for years as a source of the expression, originality, and variety found in gene families across species (Wang et al., 2012). Additionally, some AhOMT duplications may be crucial to their multiplication as they can bring neofunctionalization and diversity in gene families (Lavin et al., 2005; Chapman et al., 2008).
Some gene families have originated and extended due to tandem or segmental duplications. Gene family’s evolution in this manner is crucial for their diversification (Cannon et al., 2004). The opposite is also true: gene function may have an impact on copy number and genome structure, resulting in widely disparate patterns of segmental or tandem duplication (Cannon et al., 2004). After tandem duplication, genes occur in clusters (Savard et al., 2011). It is important to understand the evolution of gene clusters to provide updated information on evolutionary history. Previously occurrence of tandem duplication was confirmed in pomegranates by gene mapping by Yuan and coworkers. They identified three OMT genes (PgOMT01 to PgOMT03). Relatively large scale duplication of the pomegranate genome resulted in forming the PgOMT tandem duplications (Yuan et al., 2018). To a certain extent, tandem duplication has evolved the PgAOMT family.
Exon numbers and distribution patterns have a key role in the expression of any gene (Kolkman and Stemmer, 2001). In our investigation, most AhOMTs had fewer introns, and members of the same evolutionary group tended to have exon–intron patterns comparable. For instance, the presence of two or more introns in AhOMT genes demonstrates that the OMT gene development may be directly tied to the diversity of gene architectures. A similar set of findings has also been observed for the OMT gene family in Chinese jujube (Song et al., 2017). Several studies have found that genes with lesser introns expressed rapidly as introns can influence expression by delayed transcript synthesis in three different means, by (1) splicing, (2) increasing the length of the growing transcript, or (3) increasing the energy requirement of the transcript of lengthy transcripts (Jeffares et al., 2008). Less number of introns in most AhOMTs than its progenitors indicates a possible quicker response to induction; however, additional research is required to confirm this hypothesis. OMT proteins from five species used in this study were clustered into three phylogenetic groups. Conferring to the phylogenetic tree, three unique groups represents substrate specificity according to their functional traits (Joshi and Chiang, 1998).
To control gene transcription, various proteins must bind to cis-regions of the promoter. GT1-motif (Gao et al., 2004), GATA-motif (Argüello-Astorga and Herrera-Estrella, 1998), I-box (Donald and Cashmore, 1990), and G-box (Giuliano et al., 1988), are cis-regions needed for light-mediated transcription. According to our findings, S-type lignin may be controlled by AhOMT genes, which may interact with light-induced proteins and have circadian patterns in their gene promoters. The circadian rhythm regulates many genes in higher plants, including those involved in photosynthesis and starch mobilization. Hormones highly influence plant growth and development. According to Kim and coworkers, the kenaf OMT gene (Hibiscus cannabinus) is expressed after six hours of SA, ABA, auxin, ethylene, and GA treatment (Kim et al., 2013). Their findings also support our results, as AhOMT genes were generally influenced by hormone treatments. With this, SA-related factors were discovered in the AhOMT promoters, implying their key function in the hormonal regulation of AhOMT. When they studied the OMT gene, they observed that it could be stimulated by H2O2, cold, and salt, which showed that hormonal and abiotic stimuli might affect the OMT genes’ transcription. We also found similar findings for cold stress, as AhOMTs were highly influenced (up- and down-regulated). Another study indicated that Brassica napus OMT family genes were more highly expressed under drought-stressed circumstances than in regular irrigation (Li et al., 2016). The cold and drought have been shown to significantly increase the expression of an OMT gene in Ligusticum chuanxiong (Li et al., 2015). Some OMT promoter sequences included stress-related motifs such as ARE, LTR, and MBS. OMT genes are influenced by salt, and cold stress (Kim et al., 2013) and the presence of stress-responsive elements suggests that OMT genes might play a role in neutralizing the abiotic stresses. Some AhOMT gene promoters were revealed to have heat-responsive and MBS sights that can collectively induce drought tolerance. In addition to these CREs, stress response involves TC-rich repeats, W1-BOx, ARE, and LTR (Zhang et al., 2015). In light of these studies, it could be speculated that abiotic stress may promote AhOMT genes's expression, although more work is required for its confirmation. Micro-RNAs have got wide attention for their developmental and stress-tolerance roles. We identified miRNAs ahy-miR156a, ahy-miR167-3p, ahy-miR3513-5p, ahy-miR3521, ahy-miR156a, ahy-miR160-3p, ahy-miR3508, ahy-miR3513-3p, ahy-miR3518, ahy-miR3519 etc., targeting AhOMTs (Supplementary Table 5). ahy-miR3521 have been reported to target the AhOPT3.2, this gene is also targeted by ahy-miR156a. additionally ahy-miR156a also targets and down-regulates the AhOPT3.3 and AhOPT3.4. ahy-miR167-3p targets AhYSL3.2, AhYSL3.4, and AhYSL3.7. all of these miRNAs downregulates their corresponding genes by mRNA cleavage (Wang et al., 2022). Our miRNAs prediction results also revealed their cleaving activity.
The OMT genes in plants have earlier been shown to be vital genes that regulate the expression of a protein necessary for development and growth (Zhang et al., 2021). Gene expression in different organs and tissues was investigated in this research. AhOMTs demonstrated diverse expressions in time- and space-defined manners. The expression differences in different tissues indicate the functional differences between OMT genes (Zhang et al., 2021). This research also demonstrated that the expression of these genes might be triggered by a certain environment or may be highly unique to a particular organ or developmental stage. Among various abiotic stresses, low temperature and drought stress significantly impair the plant growth and production (Raza et al., 2021b; Raza et al., 2022a; Raza et al., 2022b; Raza et al., 2023) Owing to this, the OMT expression under these stressful environments was investigated. According to our findings, the expression of AhOMT-7, AhOMT-33, AhOMT-34, AhOM-35, AhOMT-71, AhOM-93, AhOMT106, AhOMT-113, and AhOMT116 increased when exposed to low temperatures and hormones treatment. Under drought stress, some AhOMTs were up-regulated, and others were down-regulated. Our findings are in agreement with previous reports such as OMTs were upregulated in response to drought stress in grape barriers (Giordano et al., 2016) and down-regulated in Brassica napus (Li et al., 2016). In terms of the mechanism of this event, further research is needed in this area as well. In the near future, the integration of genomics and genome editing technologies could be coupled to improve the production of orphan crops including peanut (Yaqoob et al., 2023). As a result, evolutionary links, structure, and expression of AhOMT genes were thoroughly investigated in this work, revealing that these genes played a critical role in peanut stress tolerance and offered a theoretical basis for peanut breeding efforts.
Conclusion
This study identified 116 OMT genes in cultivated peanut. Sequentially to get well perceptive of the AhOMT genes, we conducted a wide range of genomic analyses, including evolutionary and genomic characterization, genes structural analysis, cis-acting regions, prediction of miRNAs, and conserved motifs analysis. A combination of gene structure and phylogenetic analysis revealed three main groups of AhOMTs. In addition, these genes’ expression was profiled across different tissues against low temperature, hormones, and drought stress. Furthermore, the AhOMT genes expression demonstrated that AhOMT-7, AhOMT-33, AhOMT-34, AhOM-35, AhOMT-71, AhOM-93, AhOMT106, AhOMT-113, and AhOMT116 played a vital role against low temperature, hormones, and drought treatments. This study establishes the framework for future work into the functional study of AhOMT in peanut breeding programs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA480120.
Author contributions
WZ and HC conceived the idea and designed the study. TC, YS, YZ, QY, and XC analyzed the data and wrote the manuscript. KC, YC, MG, HD, YP, AR, and CZ helped in literature search, revision, and provided technical guidance. WZ, HC, and YZ supervised the work and edited the final version. TC and YS equally contributed to the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation (NSFC) of China (U1705233, 32072103, 32272155, 31701463, and 31601337), the Science and Technology Foundation of Fujian Province of China (2021N5007 and 2017N0006), the Special Fund for Scientific and Technological Innovation of Fujian Agriculture and Forestry University (KFb22010XA and KFb22011XA), and Foreign Cooperation Projects for Fujian Academy of Agricultural Sciences (DWHZ2021-20).
Acknowledgments
Authors are thankful to Center of Legume Plant Genetics and system Biology, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China for providing favorable conditions for this work and technical guidance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1145624/full#supplementary-material
References
Argüello-Astorga, G., Herrera-Estrella, L. (1998). Evolution of light-regulated plant promoters. Annual Review of Plant Physiology and Plant Molecular Biology 49, 525–555. doi: 10.1146/annurev.arplant.49.1.525
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Research 43, W39–W49. doi: 10.1093/nar/gkv416
Bellaloui, N. (2012). Soybean seed phenol, lignin, and isoflavones partitioning as affected by seed node position and genotype differences. Food Nutr. Sci. 3, 447. doi: 10.4236/fns.2012.34064
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of arachis duranensis and arachis ipaensis, the diploid ancestors of cultivated peanut. Nature Genetics 48, 438–446. doi: 10.1038/ng.3517
Bi, C., Chen, F., Jackson, L., Gill, B. S., Li, W. J. (2011). Expression of lignin biosynthetic genes in wheat during development and upon infection by fungal pathogens. Plant Molecular Biology Reporter 29, 149–161. doi: 10.1007/s11105-010-0219-8
Boerjan, W., Ralph, J., Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bout, S., Vermerris, W. (2003). A candidate-gene approach to clone the sorghum brown midrib gene encoding caffeic acid O-methyltransferase. Molecular Genetics and Genomics 269, 205–214. doi: 10.1007/s00438-003-0824-4
Buer, C. S., Imin, N., Djordjevic, M. A. (2010). Flavonoids: new roles for old molecules. Journal of Intigrative Plant Biology 52, 98–111. doi: 10.1111/j.1744-7909.2010.00905.x
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. J. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in arabidopsis thaliana. BMC Plant Biology 4, 1–21. doi: 10.1186/1471-2229-4-10
Chapman, M. A., Leebens-Mack, J. H., Burke, J. M. (2008). Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol. Biol. Evol. 25, 1260–1273. doi: 10.1093/molbev/msn001
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Yang, Q., Chen, K., Zhao, S., Zhang, C., Pan, R., et al. (2019). Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea l.). BMC Genomics 20, 1–17. doi: 10.1186/s12864-019-5770-6
Dai, X., Zhuang, Z., Zhao, P. X. (2018). psRNATarget: A plant small RNA target analysis server, (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Davin, L. B., Lewis, N. G. (1992). “Phenylpropanoid metabolism: Biosynthesis of monolignols, lignans and neolignans, lignins and suberins,” in Phenolic metabolism in plants (New York: Springer), 325–375.
Donald, R., Cashmore, A. R. (1990). Mutation of either G box or I box sequences profoundly affects expression from the arabidopsis rbcS-1A promoter. The EMBO Journal 9, 1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x
Dudareva, N., Pichersky, E. J. (2008). Metabolic engineering of plant volatiles. Current Opinion in Biotechnology 19, 181–189. doi: 10.1016/j.copbio.2008.02.011
Gao, Y., Li, J., Strickland, E., Hua, S., Zhao, H., Chen, Z., et al. (2004). An arabidopsis promoter microarray and its initial usage in the identification of HY5 binding targets in vitro. Plant Molecular Biology 54, 683–699. doi: 10.1023/B:PLAN.0000040898.86788.59
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., Bairoch, A. J. (2005). Protein identification and analysis tools on the ExPASy server. 571–607. doi: 10.1385/1-59259-890-0:571
Giordano, D., Provenzano, S., Ferrandino, A., Vitali, M., Pagliarani, C., Roman, F., et al. (2016). Characterization of a multifunctional caffeoyl-CoA O-methyltransferase activated in grape berries upon drought stress. Plant Physiol. Biochem. 101, 23–32. doi: 10.1016/j.plaphy.2016.01.015
Giuliano, G., Pichersky, E., Malik, V., Timko, M., Scolnik, P., Cashmore, A. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. PNAS 85, 7089–7093. doi: 10.1073/pnas.85.19.7089
Gonzales, M. D., Archuleta, E., Farmer, A., Gajendran, K., Grant, D., Shoemaker, R., et al. (2005). The legume information system (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res. 33, D660–D665. doi: 10.1093/nar/gki128
Goujon, T., Sibout, R., Pollet, B., Maba, B., Nussaume, L., Bechtold, N., et al. (2003). A new arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Molecular Biology 51, 973–989. doi: 10.1023/A:1023022825098
Guo, D., Chen, F., Inoue, K., Blount, J. W., Dixon, R. A. (2001). Downregulation of caffeic acid 3-o-methyltransferase and caffeoyl CoA 3-o-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and s lignin. The Plant Cell 13, 73–88. doi: 10.1105/tpc.13.1.73
Hamberger, B., Ellis, M., Friedmann, M., de Azevedo Souza, C., Barbazuk, B., Douglas, C. (2007). Genome-wide analyses of phenylpropanoid-related genes in populus trichocarpa, arabidopsis thaliana, and oryza sativa: The populus lignin toolbox and conservation and diversification of angiosperm gene families. Canadian Journal of Botany 85, 1182–1201. doi: 10.1139/B07-098
Hu, D., Liu, X., Wang, C., Yang, H., Li, H., Ruan, R., et al. (2015). Expression analysis of key enzyme genes in lignin synthesis of culm among different lodging resistances of common buckwheat (Fagopyrum esculentum moench.). Sci. Agric. Sin. 48, 1864–1872. doi: 10.3864/j.issn.0578-1752.2015.09.20
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Research 47, D309–D314. doi: 10.1093/nar/gky1085
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends in Genetics 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Joshi, C. P., Chiang, V. L. (1998). Conserved sequence motifs in plant s-adenosyl-L-methionine-dependent methyltransferases. Plant Molecular Biology 37, 663–674. doi: 10.1023/A:1006035210889
Kim, J., Choi, B., Cho, B.-K., Lim, H.-S., Kim, J. B., Natarajan, S., et al. (2013). Molecular cloning, characterization and expression of the caffeic acid O-methyltransferase (COMT) ortholog from kenaf (Hibiscus cannabinus). Plant Omics 6, 246–253.
Kolkman, J. A., Stemmer, W. P. (2001). Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 19, 423–428. doi: 10.1038/88084
Kota, P., Guo, D., Zubieta, C., Noel, J., Dixon, R. A. (2004). O-Methylation of benzaldehyde derivatives by “lignin specific” caffeic acid 3-o-methyltransferase. Phytochemistry 65, 837–846. doi: 10.1016/j.phytochem.2004.01.017
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. J. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35, 1547. doi: 10.1093/molbev/msy096
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Lavin, M., Herendeen, P. S., Wojciechowski, M. F. (2005). Evolutionary rates analysis of leguminosae implicates a rapid diversification of lineages during the tertiary. Systematic Biology 54, 575–594. doi: 10.1080/10635150590947131
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, W., Lu, J., Lu, K., Yuan, J., Huang, J., Du, H., et al. (2016). Cloning and phylogenetic analysis of brassica napus l. caffeic acid O-methyltransferase 1 gene family and its expression pattern under drought stress. PloS One 11, e0165975. doi: 10.1007/s11103-006-0029-4
Li, H. M., Rotter, D., Hartman, T. G., Pak, F. E., Havkin-Frenkel, D., Belanger, F. C. (2006). Evolution of novel O-methyltransferases from the vanilla planifolia caffeic acid O-methyltransferase. Plant Molecular Biology 61, 537–552. doi: 10.1584/jpestics.31.47
Li, J.-J., Zhang, G., Yu, J.-h., Li, Y.-y., Huang, X.-h., Wang, W.-J., et al. (2015). Molecular cloning and characterization of caffeic acid 3-o-methyltransferase from the rhizome of ligusticum chuanxiong. Biotechnology Letters 37, 2295–2302. doi: 10.1007/s10529-015-1917-y
Lin, F., Yamano, G., Hasegawa, M., Anzai, H., Kawasaki, S., Kodama, O. (2006). Cloning and functional analysis of caffeic acid 3-o-methyltransferase from rice (Oryza sativa). Journal of Pesticide Science 31, 47–53. doi: 10.1584/jpestics.31.47
Lin, S.-J., Yang, Y.-Z., Teng, R.-M., Liu, H., Li, H., Zhuang, J. (2021). Identification and expression analysis of caffeoyl-coenzyme a O-methyltransferase family genes related to lignin biosynthesis in tea plant (Camellia sinensis). Protoplasma 258, 115–127. doi: 10.1007/s00709-020-01555-4
LIU, S.-j., HUANG, Y.-h., HE, C.-j., Cheng, F., ZHANG, Y.-w. (2016a). Cloning, bioinformatics and transcriptional analysis of caffeoyl-coenzyme a 3-o-methyltransferase in switchgrass under abiotic stress. Journal of Integrative Agriculture 15, 636–649. doi: 10.1016/S2095-3119(16)61363-1
Liu, X., Luo, Y., Wu, H., Xi, W., Yu, J., Zhang, Q., et al. (2016b). Systematic analysis of O-methyltransferase gene family and identification of potential members involved in the formation of O-methylated flavonoids in citrus. Gene 575, 458–472. doi: 10.1016/j.gene.2015.09.048
Liu, Q., Luo, L., Zheng, L. (2018). Lignins: biosynthesis and biological functions in plants. International Journal of Molecular Sciences 19, 335. doi: 10.3390/ijms19020335
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Q.-H. (2009). The expression of caffeic acid 3-o-methyltransferase in two wheat genotypes differing in lodging resistance. Journal of Experimental Botany 60, 2763–2771. doi: 10.1093/jxb/erp132
Ma, Q.-H., Luo, H.-R. (2015). Biochemical characterization of caffeoyl coenzyme a 3-o-methyltransferase from wheat. Planta 242, 113–122. doi: 10.1007/s00425-015-2295-3
Moura, J. C. M. S., Bonine, C. A. V., de Oliveira Fernandes Viana, J., Dornelas, M. C., Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. Journal of Integrative Plant Biology 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Nguyen, T.-N., Son, S., Jordan, M. C., Levin, D. B., Ayele, B. T. (2016). Lignin biosynthesis in wheat (Triticum aestivum l.): Its response to waterlogging and association with hormonal levels. BMC Plant Biol. 16, 1–16. doi: 10.1186/s12870-016-0717-4
Peng, D., Chen, X., Yin, Y., Lu, K., Yang, W., Tang, Y., et al. (2014). Lodging resistance of winter wheat (Triticum aestivum l.): Lignin accumulation and its related enzymes activities due to the application of paclobutrazol or gibberellin acid. Field Crops Research 157, 1–7. doi: 10.1016/j.fcr.2013.11.015
Rakoczy, M., Femiak, I., Alejska, M., Figlerowicz, M., Podkowinski (2018). Sorghum CCoAOMT and CCoAOMT-like gene evolution, structure, expression and the role of conserved amino acids in protein activity. Mol. Genet. Genomics 293, 1077–1089. doi: 10.1007/s00438-018-1441-6
Ralph, J., Lundquist, K., Brunow, G., Lu, F., Kim, H., Schatz, P. F., et al. (2004). Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochemistry Review 3, 29–60. doi: 10.1023/B:PHYT.0000047809.65444.a4
Raza, A., Charagh, S., Abbas, S., Hassan, M. U., Saeed, F., Haider, S., et al. (2023). Assessment of proline function in higher plants under extreme temperatures. Plant Biol. doi: 10.1111/plb.13510
Raza, A., Charagh, S., García-Caparrós, P., Rahman, M. A., Ogwugwa, V. H., Saeed, F., et al. (2022a). Melatonin-mediated temperature stress tolerance in plants. GM Crops Food 13, 196–217. doi: 10.1080/21645698.2022.2106111
Raza, A., Mubarik, M. S., Sharif, R., Habib, M., Jabeen, W., Zhang, C., et al. (2022b). Developing drought-smart, ready-to-grow future crops. Plant Genome, e20279. doi: 10.1002/tpg2.20279
Raza, A., Su, W., Gao, A., Mehmood, S. S., Hussain, M. A., Nie, W., et al. (2021a). Catalase (CAT) gene family in rapeseed (Brassica napus l.): Genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. Int. J. Mol. Sci. 22, 4281. doi: 10.3390/ijms22084281
Raza, A., Tabassum, J., Kudapa, H., Varshney, R. K. (2021b). Can omics deliver temperature resilient ready-to-grow crops? Crit. Rev. Biotechnol. 41, 1209–1232. doi: 10.1080/07388551.2021.1898332
Roje, S. (2006). S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67, 1686–1698. doi: 10.1016/j.phytochem.2006.04.019
Savard, O. T., Bertrand, D., El-Mabrouk, N. (2011). “Evolution of orthologous tandemly arrayed gene clusters,” in BMC bioinformatics, 12, 1–12. (Galway, Ireland: BioMed Central)
Sharif, Y., Chen, H., Deng, Y., Ali, N., Khan, S., Zhang, C., et al. (2022). Cloning and functional characterization of a pericarp abundant expression promoter (AhGLP17-1P) from peanut (Arachis hypogaea l.). Front. Genet. 12. doi: 10.3389/fgene.2021.821281
Song, S., Zhou, H., Sheng, S., Cao, M., Li, Y., Pang, X. (2017). Genome-wide organization and expression profiling of the SBP-box gene family in Chinese jujube (Ziziphus jujuba mill.). 18, 1734. doi: 10.3390/ijms18081734
Struck, A. W., Thompson, M. L., Wong, L. S., Micklefield, J. (2012). S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemBioChem 13, 2642–2655. doi: 10.1002/cbic.201200556
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Wang, C., Wang, X., Li, J., Guan, J., Tan, Z., Zhang, Z., et al. (2022). Genome-wide identification and transcript analysis reveal potential roles of oligopeptide transporter genes in iron deficiency induced cadmium accumulation in peanut. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.894848
Wang, Y., Wang, X., Paterson, A. H. (2012). Genome and gene duplications and gene expression divergence: a view from plants. Ann. New York Acad. Sci. 1256, 1–14. doi: 10.1111/j.1749-6632.2011.06384.x
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics, Proteomics & Bioinformatics 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wani, S. H., Kumar, V., Khare, T., Tripathi, P., Shah, T., Ramakrishna, C., et al. (2020). miRNA applications for engineering abiotic stress tolerance in plants. Biologia 75, 1063–1081. doi: 10.2478/s11756-019-00397-7
Xu, L., Dong, Z., Fang, L., Luo, Y., Wei, Z., Guo, H., et al. (2019). OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research 47, W52–W58. doi: 10.1093/nar/gkz333
Yang, Z., Bielawski, J. (2000). Statistical methods for detecting molecular adaptation. Trends in Ecology and Evolution 15, 496–503. doi: 10.1016/S0169-5347(00)01994-7
Yaqoob, H., Tariq, A., Bhat, B. A., Bhat, K. A., Nehvi, I. B., Raza, A., et al. (2023). Integrating genomics and genome editing for orphan crop improvement: a bridge between orphan crops and modern agriculture system. GM Crops Food 14, 1–20. doi: 10.1080/21645698.2022.2146952
Ye, Z.-H., Kneusel, R. E., Matern, U., Varner, J. (1994). An alternative methylation pathway in lignin biosynthesis in zinnia. The Plant Cell 6, 1427–1439. doi: 10.1105/tpc.6.10.1427
Yoshihara, N., Fukuchi-Mizutani, M., Okuhara, H., Tanaka, Y., Yabuya, T. (2008). Molecular cloning and characterization of O-methyltransferases from the flower buds of iris hollandica. Journal of Plant Physiology 165, 415–422. doi: 10.1016/j.jplph.2006.12.002
Yu, C. S., Chen, Y. C., Lu, C. H., Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins: Structure, Function, and Bioinformatics 64, 643–651. doi: 10.1002/prot.21018
Yuan, Z., Fang, Y., Zhang, T., Fei, Z., Han, F., Liu, C., et al. (2018). The pomegranate (Punica granatum l.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnology Journal 16, 1363–1374. doi: 10.1111/pbi.12875
Zhang, X., Chen, B., Wang, L., Ali, S., Guo, Y., Liu, J., et al. (2021). Genome-wide identification and characterization of caffeic acid O-methyltransferase gene family in soybean. Plants 10, 2816. doi: 10.3390/plants10122816
Zhang, J., Li, Y., Jia, H.-X., Li, J.-B., Huang, J., Lu, M.-Z., et al. (2015). The heat shock factor gene family in salix suchowensis: a genome-wide survey and expression profiling during development and abiotic stresses. Frontiers in Plant Science 6, 748. doi: 10.3389/fpls.2015.00748
Zhou, J.-M., Seo, Y. W., Ibrahim, R. K. (2009). Biochemical characterization of a putative wheat caffeic acid O-methyltransferase. Plant Physiology and Biochemistry 47, 322–326. doi: 10.1016/j.plaphy.2008.11.011
Keywords: bioinformatics, environmental stress, functional annotation, gene duplication, micro-RNAs, peanut genomics, phylogenetic tree
Citation: Cai T, Sharif Y, Zhuang Y, Yang Q, Chen X, Chen K, Chen Y, Gao M, Dang H, Pan Y, Raza A, Zhang C, Chen H and Zhuang W (2023) In-silico identification and characterization of O-methyltransferase gene family in peanut (Arachis hypogaea L.) reveals their putative roles in development and stress tolerance. Front. Plant Sci. 14:1145624. doi: 10.3389/fpls.2023.1145624
Received: 16 January 2023; Accepted: 10 March 2023;
Published: 31 March 2023.
Edited by:
Yihua Wang, Nanjing Agricultural University, ChinaReviewed by:
Muhammad Aamir Manzoor, Anhui Agricultural University, ChinaGarima Kushwaha, Guardanthealth, Inc., United States
Copyright © 2023 Cai, Sharif, Zhuang, Yang, Chen, Chen, Chen, Gao, Dang, Pan, Raza, Zhang, Chen and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijian Zhuang, d2VpamlhbnpAZmFmdS5lZHUuY24=
†These authors have contributed equally to this work
 Tiecheng Cai
Tiecheng Cai Yasir Sharif
Yasir Sharif Yuhui Zhuang
Yuhui Zhuang Qiang Yang
Qiang Yang Xiangyu Chen
Xiangyu Chen Kun Chen
Kun Chen Yuting Chen1
Yuting Chen1 Hao Dang
Hao Dang Ali Raza
Ali Raza Chong Zhang
Chong Zhang Hua Chen
Hua Chen Weijian Zhuang
Weijian Zhuang