- Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
Pulsatilla chinensis is an important medicinal herb, its dried radix is used to treat the inflammation since ancient China. Triterpenoid saponins are proved to be the main active compounds of Pulsatilla genus. The triterpenoid saponin contents vary widely in different Pulsatilla species. But no enzyme involved in the triterpenoid saponin biosynthetic pathway was identified in Pulsitilla genus. This seriously limits the explanation of the triterpene content difference of Pulsatilla species. In this article, we obtained two oxidosqualene cyclase (OSC) genes from P. chinensis and P. cernua by touchdown PCR and anchored PCR. These two OSCs converted 2,3-oxidosqualene into different triterpenoids. The OSC from P. cernua is a monofunctional enzyme for β-amyrin synthesis, while the OSC from P. chinensis is a multifunctional enzyme for lupeol and β-amyrin synthesis, and the lupeol is the main product. Then we identified the 260th amino acid residue was the key site for the product difference by gene fusion and site-directed mutant technology. When the 260th amino acid residue was tryptophan (W260) and phenylalanine (F260), the main catalysate was β-amyrin and lupeol, respectively. Then we found that the expression of these two genes was strongly correlated with the lupeol-type and β-amyrin-type triterpenoid contents in P. cernua and P. chinensis. Finally, we found the gene copy number difference of these two genotypes leaded to the triterpenoid diversity in P. cernua and P. chinensis. This study provides useful information for the molecular breeding and quality improvement of P. chinensis and a molecular marker to identify the P. chinensis decoction pieces.
1 Introduction
Pulsatilla chinensis is a species of medicinal and ornamental plant, which belongs to Ranunculaceae, Pulsatilla genus. In China, the dried radix of P. chinensis, which is known as ‘Bai Tou Weng′, is a kind of traditional Chinese medicinal material (TCM), which has been proved could effectively treat dysmenorrhoea, testicular inflammation, and other inflammation in China and other countries from the 19th century up to now (Lydston, 1882; Coley, 1922; Friese et al., 1996; Misra et al., 2021; Niu et al., 2023). The Chinese Pharmacopoeia defines anemoside B4 as the index component of P. chinensis, and the content of anemoside B4 in decoction pieces must be more than 4.6% (dry weight). There are more than 60 species in Pulsatilla genus (Misra et al., 2021). Because of the appearance similarity, the dried radix of P. cernua were usually sold as P. chinensis in the Chinese herbal medicine market, but there were few anemoside B4 in them.
Triterpenoid saponins are the main active compounds of the Pulsatilla genus, which show high biological activity in anticancer (Xu et al., 2012), neuroactive (Yoo et al., 2008), neuroprotective (Liu et al., 2012), antioxidant (Li et al., 2008), antimicrobial (Lee et al., 2001), and cytotoxic agents (Xu et al., 2013). There are abundant triterpenoids, especially lupeol-type and β-amyrin-type triterpenoid saponins in the roots of the Pulsatilla genus (Laska et al., 2019). The lupeol-type triterpenoid saponins abundantly accumulate in the roots of P. chinensis, and the β-amyrin-type triterpenoid saponins abundantly accumulate in the roots of P. cernua. The triterpenoid saponin type difference between P. chinensis and P. cernua make the source and application of decoction pieces more complicated, so clarifying the biosynthesis difference of triterpenoid saponins between P. chinensis and P. cernua is urgent. The main biosynthesis of triterpenoid saponins in plants is clear, and could be divided into four steps (Figure 1). The First step is the synthesis of terpenoid basic units isopentenylpyrophosphate (IPP) and dimethylallypyrophosphate (DMAPP). They were synthesized in two compartmentally separated metabolic pathways, mevalonate (MVA) pathway in the cytoplasm and methylerythritol phosphate (MEP) pathway in the plastid, and these two C5 precursors could interconvert into each other by IPP/DMAPP isomerase (IDI). The second step is that two IPP and one DMAPP units combine to form farnesyl diphosphate (FPP). The third step is that two FPP units synthesize squalene by squalene synthase (SQS), then squalene oxidized into 2,3-oxidesqualene by squalene epoxidase (SE). As the common precursor of triterpenoid and sterol, 2,3-oxidesequalene turned into lupeol, β-amyrin, α-amyrin, and other triterpenoids by oxidosqualene cyclases (OSC) (Krokida et al., 2013; Sun et al., 2013; Wang et al., 2021). The final step is triterpenoids were modified by cytochromes P450 (CYP450) and UDP-dependent glycosyltransferases (UGT) (Seki et al., 2015; Christ et al., 2019; Rahimi et al., 2019). However, no enzyme involved in the triterpenoid saponin biosynthetic pathway was identified in the Pulsitilla genus.
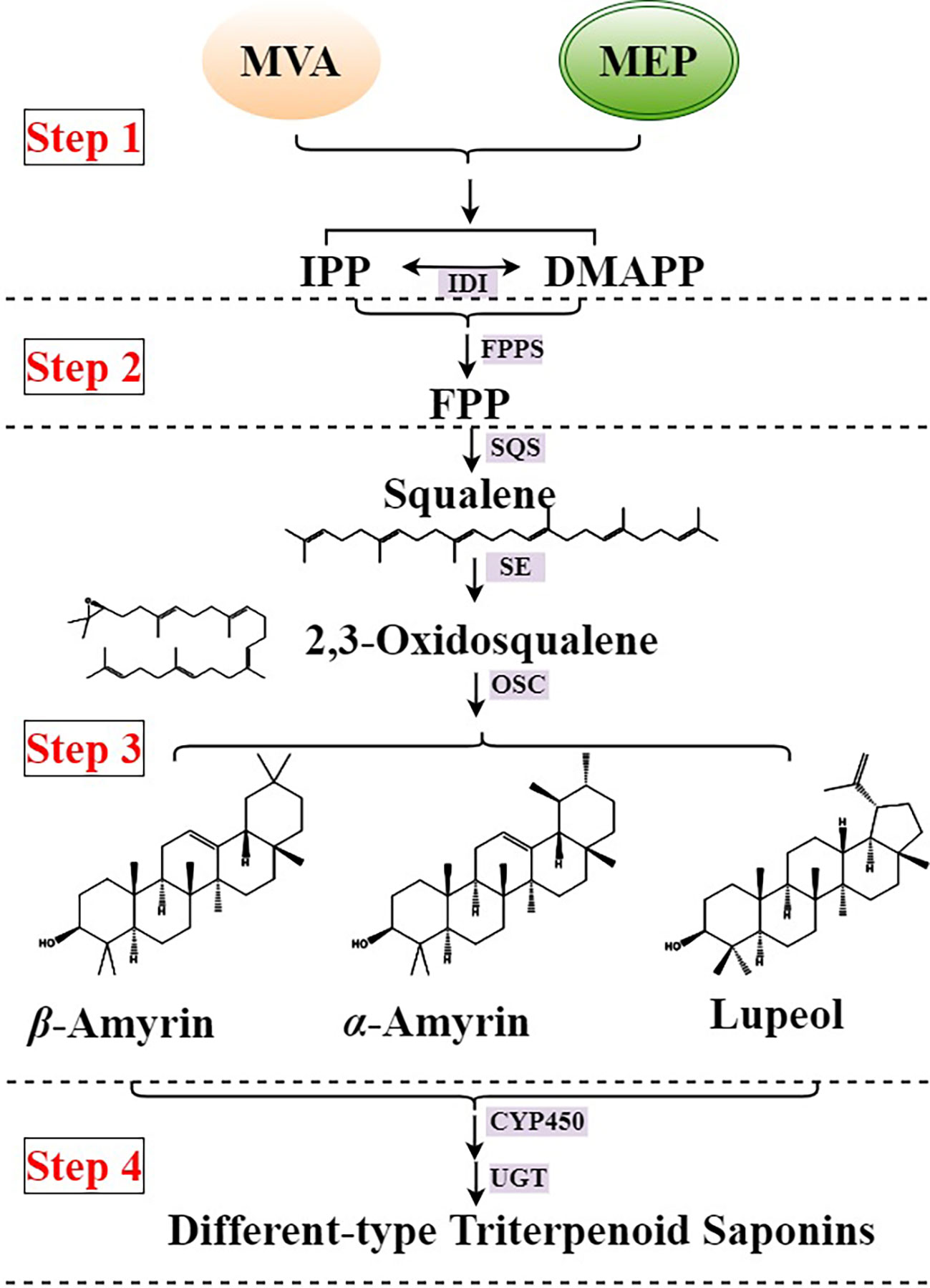
Figure 1 Biosynthesis of triterpenoid saponins in plants. IDI, IPP/DMAPP isomerase; FPPF, farnesyl pyrophosphate synthase; SQS, squalene synthase; SE, squalene epoxidase; OSC, oxidosqualene cyclases; CYP450, cytochromes P450; UGT, UDP-dependent glycosyltransferases.
As the first enzyme caused triterpenoid saponin diversity, OSCs have received much attention since the 19th century. Only one OSC member is responsible for the synthesis of sterol in animals and fungi. While OSC always exists as gene family in plants. For example, there were 13 members in Arabidopsis thaliana, 16 members in Tripterygium wilfordii, and 9 members in Momordica charantia (Husselstein-Muller. et al., 2001; Han et al., 2021; Liu et al., 2021). Until 2022, there were more than 170 OSCs were obtained from 61 plant species (Chen et al., 2021; Wang et al., 2022). The catalysates of plant OSCs include a few monocyclic, bicyclic, tricyclic triterpene alcohols and many tetracyclic and pentacyclic triterpene alcohols (Chen et al., 2021; Wang et al., 2021; Lertphadungkit et al., 2022). In addition, site-directed mutagenesis results of some OSC genes indicate that a single key amino acid mutation is important to its product diversity. As concluded by Chen, et al., key amino acids, which important to the activity of OSC, were divided into 6 groups: Y118, Y410, M(W/L)C(Y/H)CR motif, DCTAE motif, and adjacent residues, C-terminus, and other residues (Chen et al., 2021). These results provide useful information to reveal the triterpenoid saponin biosynthesis difference between P. chinensis and P. cernua.
To explain the triterpenoid saponin biosynthesis difference between P. chinensis and P. cernua, in this study we obtained two OSC full-length CDS sequences, one from P. chinensis (PchAS) and the other one from P. cernua (PcAS) by touchdown PCR and anchored PCR. There are 27 differential amino acids between them. Then we analyzed the function of these two enzymes. Interestingly, the two OSCs convert 2,3-oxidosqualene into different triterpenoids. PcAS was a monofunctional enzyme for β-amyrin synthesis, while PchAS was a multifunctional enzyme for lupeol and β-amyrin synthesis, and lupeol was the main product. To find the key amino acid for the product diversity, fusion genes of PcAS and PchAS and site-directed mutant of PcAS were expressed in yeast. The final result shows that W260 (tryptophan) is important for β-amyrin synthesis, if this amino acid was mutated to F (phenylalanine), the products changed to lupeol and β-amyrin. Then we found the expressions of two genes were strongly correlated with the lupeol-type and β-amyrin-type triterpenoid content in P. cernua and P. chinensis. Finally, we analyzed the gene copy numbers by qPCR, and the result showed that the gene copy numbers lead to triterpenoid diversity of P. cernua and P. chinensis. This study provides useful information for the quality improvement of P. chinensis and a molecular marker to identify the P. chinensis decoction pieces.
2 Results
2.1 Cloning of putative lupeol and β-amyrin synthase gene from the roots of Pulsatilla chinensis and Pulsatilla cernua
To isolate β-amyrin and lupeol synthase genes, we obtained the full-length sequence of OSCs from P. chinensis and P. cernua by 3 kinds of PCR (Figure 2A). Firstly, we used the degenerate primers (IS1F, IS2R, TAS1F, and TAS1R, Supplementary Table S1) obtained from the previous study (Guhling et al., 2006). Degenerate PCR was performed by using cDNA from roots of P. chinensis and P. cernua as templates. Unfortunately, we didn′t obtain any products. Then we designed codehop primers (NEDG-F 8X, DQDH-R 16X, Supplementary Table S1), which with hybrid structure (5′ consensus and 3′ degenerate). Luckily, we got approximately 890 bp products from both P. chinensis and P. cernua (Supplementary Figure S1A). And this is corresponding to the distance between the codehop primers in the β-amyrin and lupeol synthase genes from the other plants. Then the DNA fragments were subcloned into the zero-background T-vector. 20 independent clones were selected to sequence. When blasted in NCBI database, all the sequences were similar to Aralia elata β-amyrin synthase (Wang et al., 2022), so in this article we named the OSC genes from P. chinensis and P. cernua as PchAS and PcAS, respectively. To obtain the full-length cDNA sequence of PchAS and PcAS, we used 5′- and 3′-RACE technology (because of the high sequence similarity, in this experiment we only used the core fragment of PchAS). We designed two independent primers (5′-GSP-R2-1/R2-2, 3′-GSP-F3-1/F3-2) for each flank and the primer sequences were listed in Supplementary Table S1. The PCR using both of these 5′-GSP primers and 3′-GSP primers could obtain products (Supplementary Figure 1B), and the products were subcloned into the T vector and sequenced. The PchAS cDNA contained an ORF of 2280 bp encoding 759 amino acid residues. At last, we obtained full-length sequences of AS genes from P. chinensis and P. cernua cDNA by Primer F4 and Primer R4 (Supplementary Table S1, Supplementary Figure 1C). The deduced amino acid sequences of PcAS and PchAS showed the highest identity of 86.69% and 85.64% to the Nigella sativa β-amyrin synthase NsbAS1 (Scholz et al., 2009), and both of them have 6 QXXXXXW, 1 MXCYCR, and 1 DCTAE domain, which were the conserved domains for OSC protein sequence. There are 27 different amino acid residues between PcAS and PchAS (Supplementary Figures S2, S3).
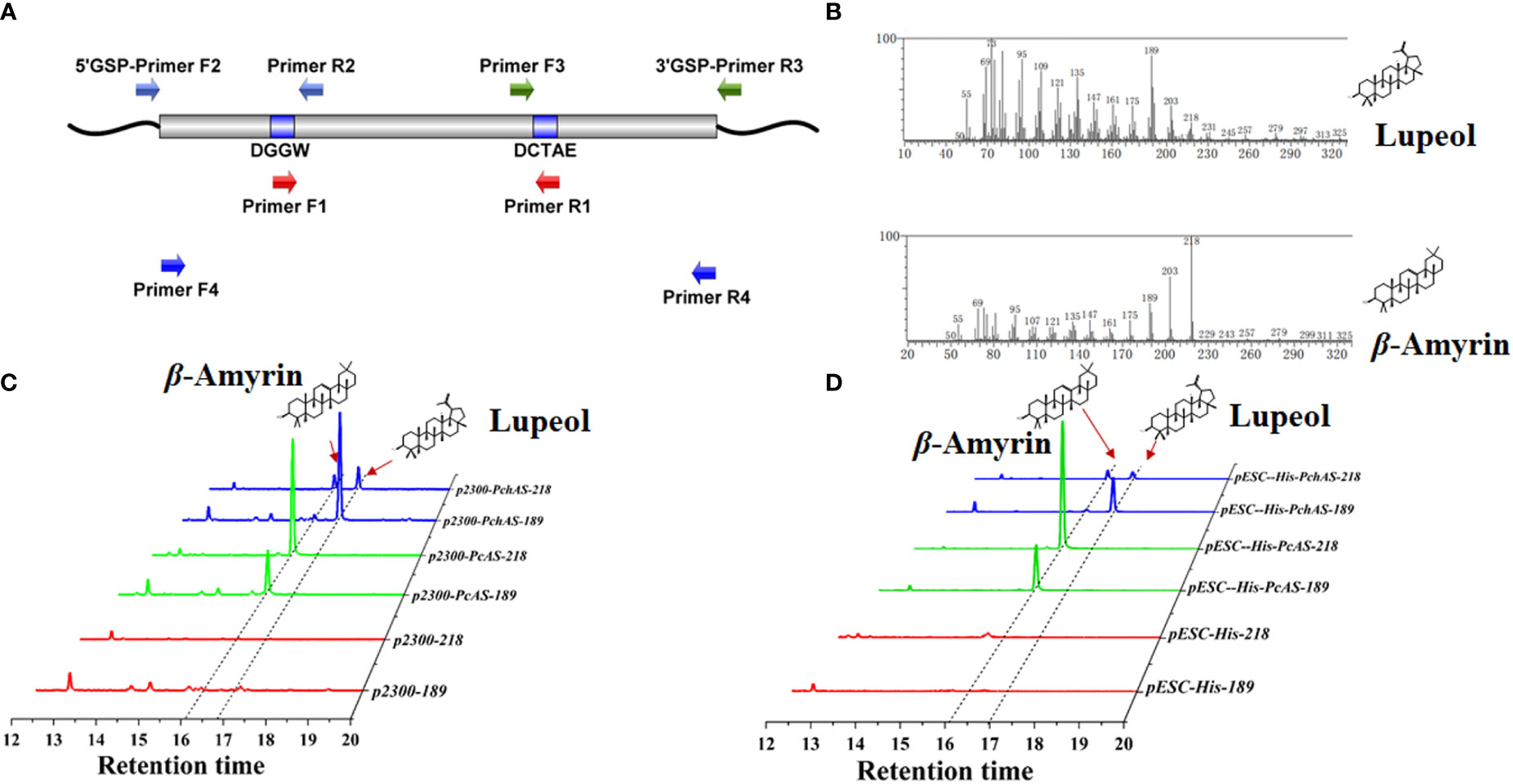
Figure 2 Functional characterization of AS homolog genes from Pulsatilla ceruna and P. chinensis. (A) A simplifed overview of gene cloning of AS from P. ceruna and P. chinensis; (B) Mass spectra of β-amyrin and lupeol detected by GC–MS; (C) GC–MS analysis of extracts from transient transformation Nicotiana benthamiana leaves which expressed AS genes from P. cernua (PcAS) and P. chinensis (PchAS). pCAMBIA2300 empty vector and pSAK277-NbHAMGR co-transformation N. benthamiana leaves were used as control; (D) GC–MS analysis of extracts from engineered yeast which expressed PcAS and PchAS. ERG7-deficient yeast that transformed with pESC-His empty vector was used as control; The 218 and 189 ion counts of β-amyrin and lupeol were shown, respectively.
2.2 Functional analysis of the PcAS and PchAS genes in yeast and Nicotiana benthamiana
In order to elucidate the function of PcAS and PchAS, complete ORFs of them were amplified from root cDNAs using high-fidelity DNA polymerase and gene-specific oligonucleotides and inserted into yeast expression vector pESC-His by homologous recombination technology (AS-pESC-F/R primers for yeast expression vector were listed in Supplementary Table S1). In this vector, PcAS and PchAS genes were driven by the inducible promoter GAL10. Next, we expressed PchAS and PcAS in the lanosterol-deficient yeast strain ATCC40029, which could provide sufficient substrate for PcAS and PchAS. The fragment ion peak of lupeol and β-amyrin standards were shown in Figure 2B and Supplementary Figure S4. As shown in Figure 2D, PcAS produced β-amyrin as the single product (fragment ion peaks and retention time were the same as β-amyrin standard), while PchAS is a multifunctional enzyme, which produced lupeol and β-amyrin (fragment ion peaks and retention time were same as β-amyrin and lupeol standards). The main product of PchAS was lupeol, and β-amyrin was the secondary product (Supplementary Figure S4).
With the same method, we constructed the plant expression vectors, in which target genes PcAS or PchAS were driven by the constitutive promoter CaMV35S. In addition, we constructed a plant expression vector that constitutively overexpress NbHMGR gene (pSAK277-NbHMGR) to increase the substrate concentration of PcAS and PchAS. Then, these two genes were co-expressed with NbHMGR in N. benthamiana leaves through Agrobacterium-mediated transient transformation. The results showed that the gene functions in plants were the same as in yeast. That is, PcAS co-expressed with NbHMGR in N. benthamiana leaves produced β-amyrin as the single product. Similarly, PchAS co-expressed with NbHMGR in N. benthamiana leaves produced dominated lupeol and some β-amyrin (Figure 2C and Supplementary Figure S4).
2.3 Identifying the region leading to gene functional diversity by gene fusion
To identify the key regions that lead to the functional diversity between PcAS and PchAS, we separated each of these two genes into 3 fragments, fragment 1: 1-145 amino acid residues, fragment 2: 146-462 amino acid residues, and fragment 3: 462-756 amino acid residues (Figure 3A). Then we fused the different fragments from PcAS and PchAS and obtained six chimera genes by bridge PCR (Figure 3A). As shown in Figure 3B, chimera protein 3 and chimera protein 6 produced lupeol and β-amyrin in yeast (Figure 3B). However, chimera proteins 2, 4, and 5 only produced β-amyrin as the single product in yeast. In other words, when fragment 2 of the chimera protein is from P. chinensis, the chimera protein will show the characteristics of PchAS, which is the multifunctional enzyme, to produce lupeol and β-amyrin. When fragment 2 is from P. cernua, it will show the characteristics of PcAS, which is a monofunctional enzyme, to produce β-amyrin (Figure 3B). Therefore, fragment 2 is the key region for the OSC gene functional diversity between P. chinensis and P. cernua.
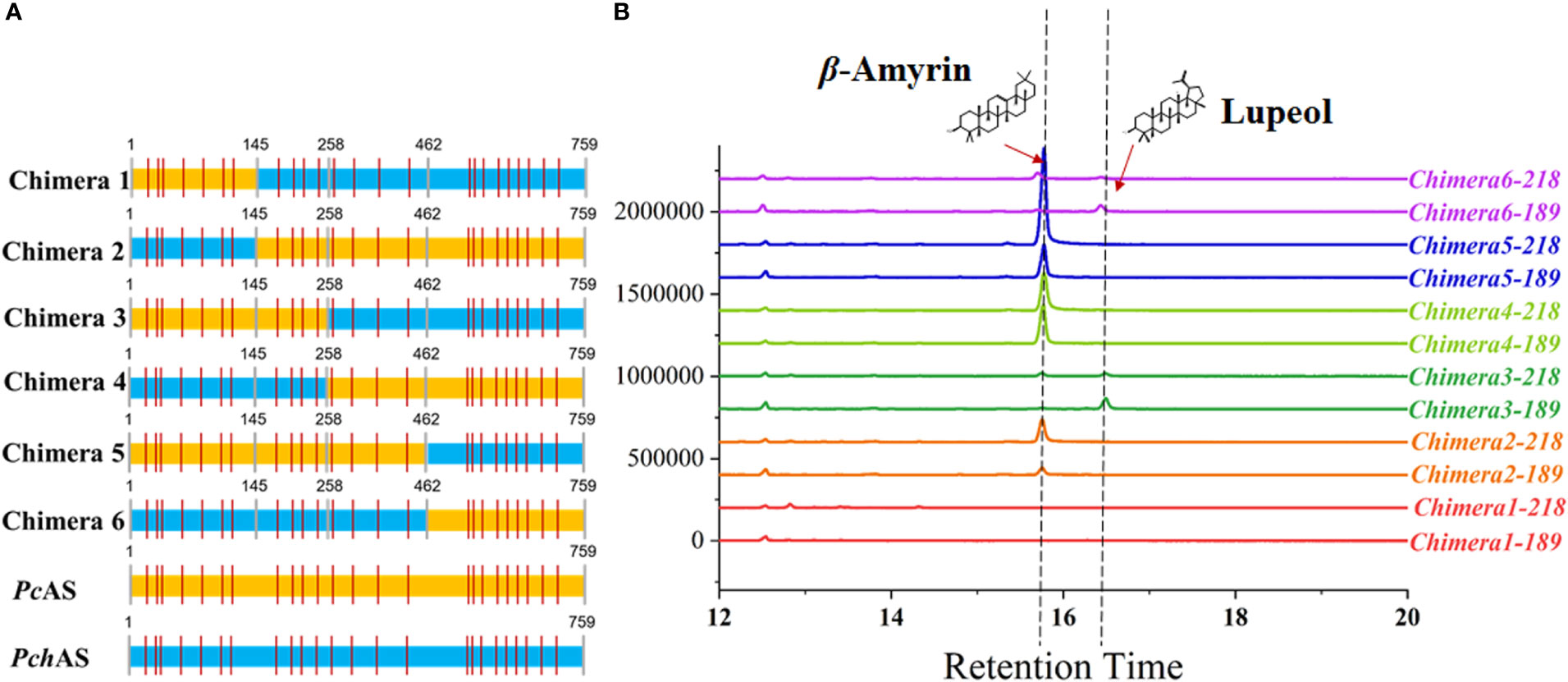
Figure 3 Functional characterization of chimera protein. (A) Diagram of the chimera protein. The colors indicate that fragment source: P. chinensis (blue) and P. cernua (yellow). (B) GC–MS analysis of chimera protein expressed yeast extraction. ERG7-deficient yeast that transformed with pESC-His empty vector was used as control. The 218 and 189 ion counts of β-amyrin and lupeol are shown, respectively. PcAS represent AS gene cloned from P. cernua, PchAS represent AS gene cloned from P. chinensis.
2.4 Identifying the key amino acids of PcAS and PchAS by site-directed mutagenesis
To further identify the key amino acid that led to the product difference between PchAS and PcAS, we directionally mutated the four differential amino acid residues located in fragment 2 of PcAS based on the protein alignment result of PcAS and PchAS (Figure 4A and Supplementary Figure S3). As shown in Figure 4B, mutant proteins (W260F, W260FH296D, W260FI337V, and W260FQ397P) produced one more product, lupeol, compared with the original PcAS. However, mutant proteins (H296D, H296DI337V, H296DQ390P, I337V, I337VQ390P, and Q390P) only produced the single product β-amyrin as PcAS (Supplementary Figures S5A, B). That is to say, either point or multiple mutations of 296, 337, and 397 in fragment 2 couldn′t change the function of PcAS (Figure 4B and Supplementary Figure S5). Only when the 260 tryptophan (W) was mutated to phenylalanine (F), the function of PcAS changed from the single product (β-amyrin) to multiple products (lupeol and β-amyrin). The 260th amino acid of PcAS and PchAS is the site leading to functional difference, which the 781st and 782nd nucleotides of PcAS and PchAS CDS are GG and TT, respectively.
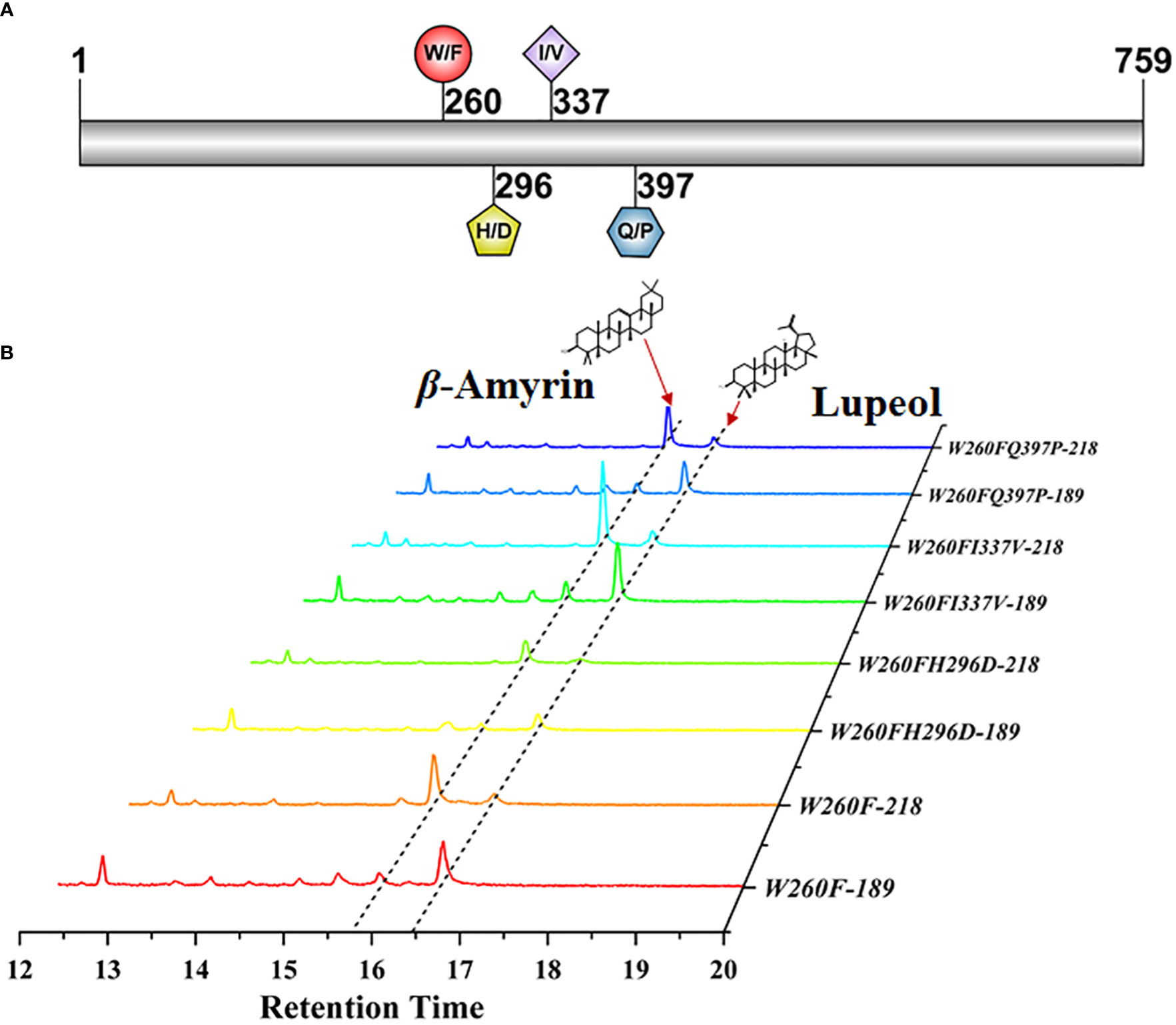
Figure 4 Functional characterization of the directional mutant protein W260F, W260FI337V, W260FH296D, and W260FQ397P. (A) Diagram of the mutant sites of PcAS. (B) GC–MS analysis of mutant protein expressed yeast extraction. The 218 and 189 ion counts of β-amyrin and lupeol are shown, respectively.
2.5 The analysis of total triterpenoid content in dried radix of Pulsatilla chinensis and Pulsatilla cernua
To investigate the types and contents of triterpenoid saponins in P. chinensis and P. cernua, we extracted triterpenoids from the roots of P. chinensis and P. cernua by two-phase hydrolysis method and absolutely quantified the content of triterpenoid by HPLC-UV. The results showed that the content of betulinic acid was lowest in triterpenoid acid, which ranged from 0.33 mg/g to 6.57 mg/g. Betulinic acid content in P. ceruna was higher than in P. chinensis (Figures 5A, B). Oleanolic acid content ranged from 0.46 mg/g to 40.22 mg/g, and the content of oleanolic acid in P. ceruna was higher than in P. chinensis (Figures 5A, B). It was higher than betulinic acid but lower than hederagenin and 23-hydroxybetulinic acid in every Pulsatilla material used in this study. The content of hederagenin in P. ceruna was higher than in P. chinensis, which ranged from 22.24 mg/g to 82.26 mg/g. The content of hederagenin was the highest triterpenoid in P. ceruna, while was lower than 23-hydroxybetulinic acid in P. chinensis. 23-Hydroxybetulinic acid was the dominant component in P. chinensis. The content of 23-hydroxybetulinic acid ranged from 3.52 mg/g to 202.47 mg/g. Total triterpenoids in P. chinensis (ranged from 170.35 mg/g to 226.05 mg) were higher than those in P. cernua (ranged from 98.07 mg/g to 111.11 mg/g) (Figures 5A, B). In addition, the contents of lupeol-type triterpenoids in P. chinensis were significantly higher than those of β-amyrin-type, while the contents of β-amyrin-type triterpenoids were significantly higher than the contents of lupeol-type triterpenoids in P. cernua (Figures 5A, B).
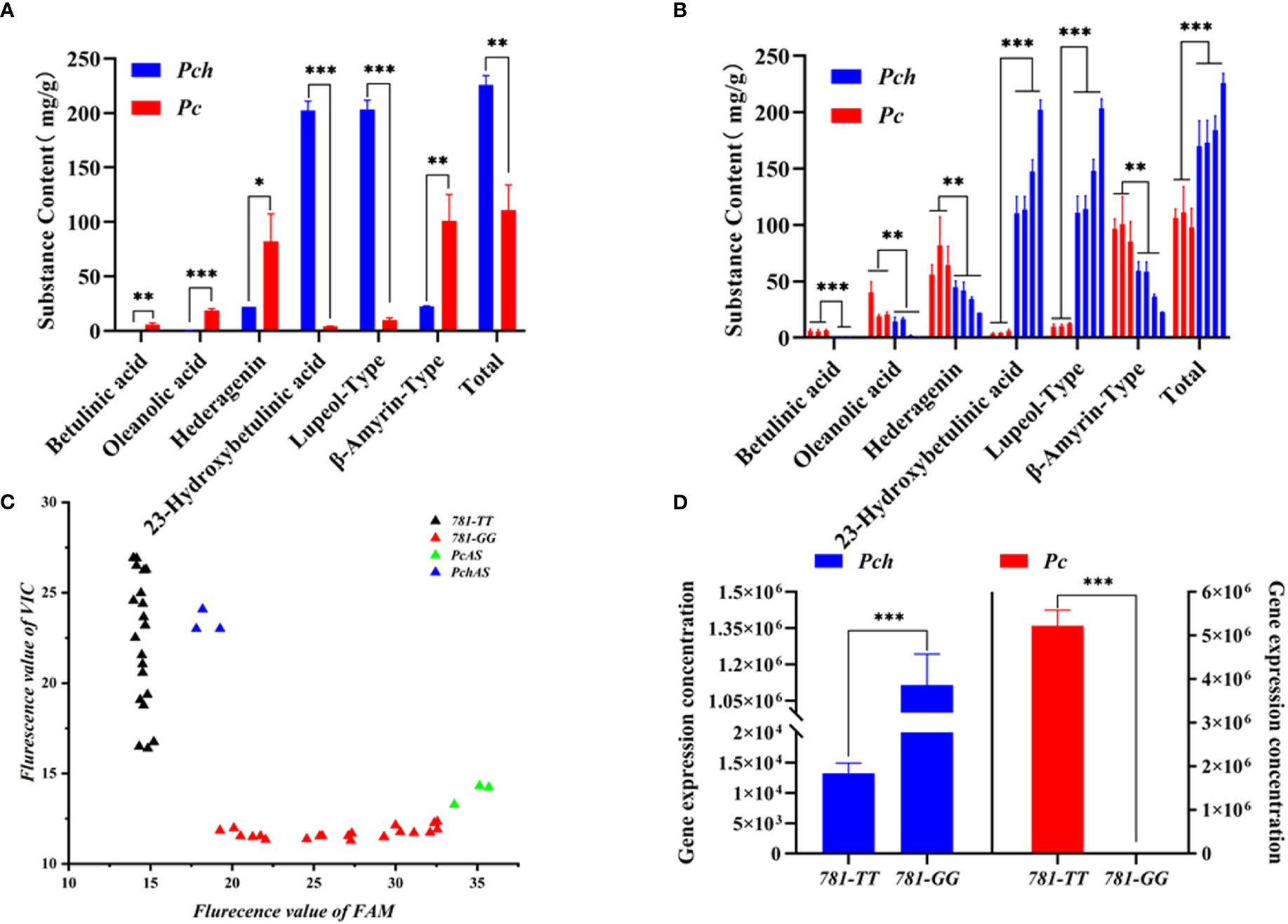
Figure 5 Triterpene acid content, PcAS and PchAS type gene expression in Pulsatilla ceruna and P. chinensis. (A) Different type triterpene acid contents in P. ceruna and P. chinensis freeze-drying roots, respectively; (B) Different type triterpene acid contents in P. ceruna and P. chinensis decoction pieces, respectively; (C) The FAM and VIC fluorescent value of qPCR which P. ceruna, P. chinensis, plasmid pET32a-PcAS and plasmid pET32a-PchAS as template; (D) Absolute gene expression of PcAS and PchAS type in P. ceruna and P. chinensis, 781-TT and 781-GG represent PchAS and PcAS type gene. Student’s t-test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
2.6 Absolute quantification of PcAS and PchAS genes expression in roots of Pulsatilla chinensis and Pulsatilla cernua
Due to the functional differences of the AS genes cloned from P. chinensis and P. cernua, we suspected that the difference of triterpenoid saponin types between P. chinensis and P. cernua were caused by the expression levels of the two gene types. Therefore, we analyzed the expression levels of AS gene type in P. chinensis and P. cernua. Since the site leading to functional difference is the 781st and 782nd nucleotides of PcAS and PchAS (GG/TT), so we used the TaqMan probe method to analyze the gene expression in different species. We designed a pair of specific primers (AS-Tq-F/R), and two probes for 781-GG-type and 781-TT-type. The probes added the MGB marker at 3′ ends to improve the specificity and add FAM fluorescent signal for 781-GG-type and VIC fluorescent signal for 781-TT-type. We first collected these two fluorescent terminal signals of P. chinensis and P. cernua by terminal detection method. As shown in Figure 5C, these two different genotypes were expressed in both P. chinensis and P. cernua. Subsequently, the standard curves of these two genotypes were constructed respectively to absolutely quantitate their expression in P. chinensis and P. cernua (Supplementary Figure S6). As shown in Figure 5D, the expression of 781-TT-type was under the quantitative line and 781-GG-type gene expression was 5.11 × 106 in P. cernua, while the expression of 781-TT-type gene expression level in P. chinensis (1.12 × 106) was about 100 times higher than the 781-GG type (1.32 × 104).
2.7 Gene copy number analysis of 781-TT-type and 781-GG-type in Pulsatilla chinensis, Pulsatilla cernua and Pulsatilla turczaninovii
To explain the 781-TT-type and 781-GG-type gene expression difference in the Pulsatilla genus, we analyzed relative gene copy numbers of the 781-GG-type and 781-TT-type gene in P. chinensis, P. cernua, and P. turczaninovii. We first obtained and sequenced the full-length AS gene sequences from the DNA of these three Pulsatilla species by primer F4 and primer R4 listed in Supplementary Table S1. As shown in Supplementary Figure S7, the AS gene contains 12 introns and 13 extrons. Then we randomly choose four single clones from each species to sequence. The results indicated that there were three clones as 781-TT-type and one clone as 781-GG-type in P. chinensis, while all four clones were 781-GG-type in P. cernua and P. turczaninovii, respectively (Supplementary Figure S8). Finally, we analyzed these two genotypes by qPCR. By the method used to analyze the gene expression levels, we used the plasmids as the standard to quantitate the gene copy numbers. The primers were designed based on gene sequences of PcAS and PchAS (AS-TAQ-F/R). The probes used in this experiment were the 781-GG probe and the 781-TT probe. Then, we collected terminal signals of FAM and VIC fluorescent of P. chinensis, P. cernua and P. turczaninovii, respectively. As the results shown in Figure 6, three different P. chinensis materials have strong fluorescent signals of VIC and weak fluorescent signals of FAM, while the fluorescent signals of VIC and FAM in P. cernua and P. turczaninovii were opposite with in P. chinensis. This result suggests that in these three Pulsatilla species, the gene copy numbers of 781-GG-type and 781-TT-type are different. In P. chinensis, the 781-TT-type gene copy numbers were higher than the 781-GG-type. However, in P. cernua and P. turczaninovii, the 781-TT-type gene copy numbers were lower than the 781-GG-type.
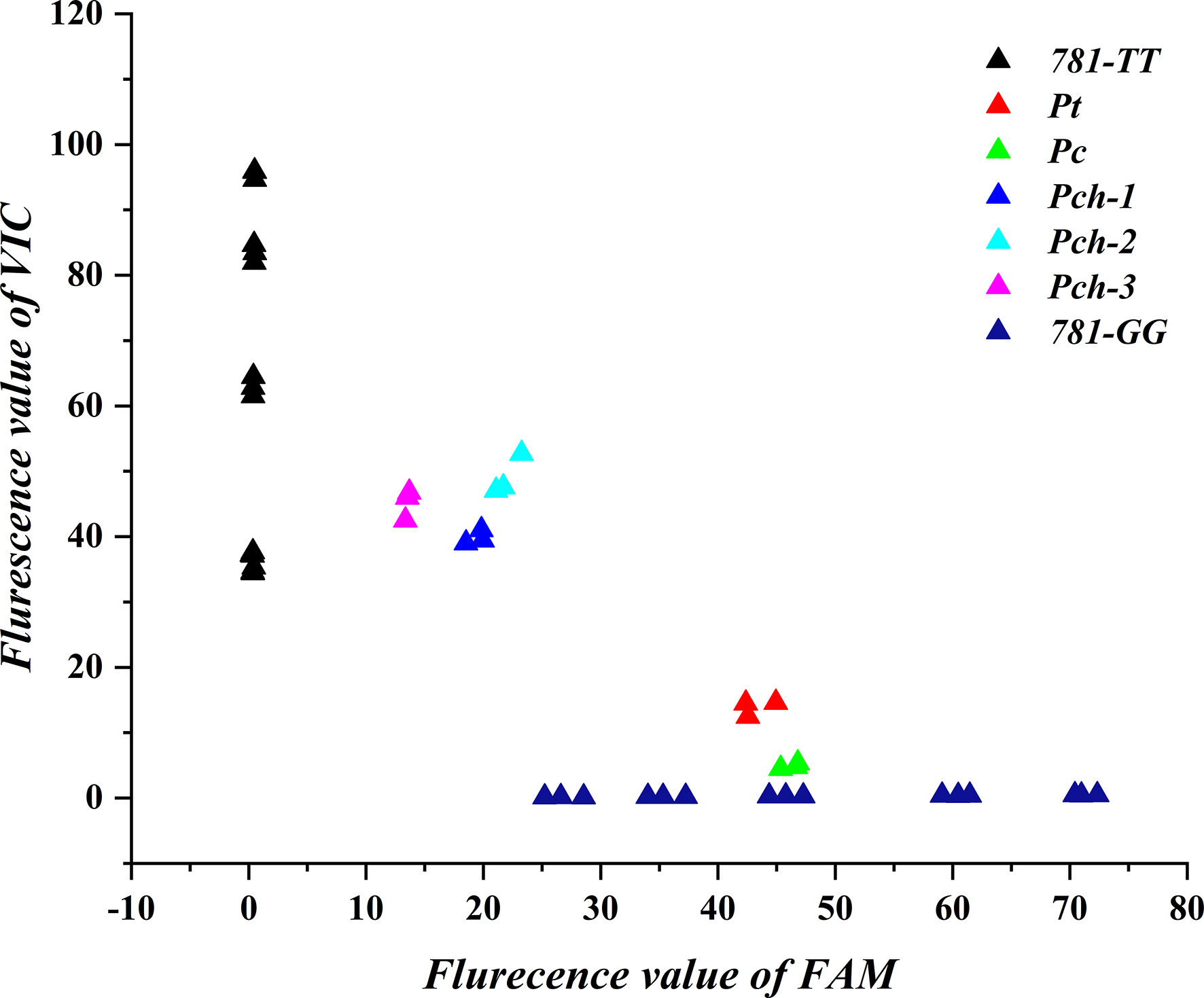
Figure 6 Gene copy number analysis of 781-TT and 781-GG in Pulsatilla ceruna, P. chinensis and P. turczaninovii. Pc, Pch, and Pt represent P. ceruna, P. chinensis and P. turczaninovii, respectively. 781-TT and 781-GG represent PchAS and PcAS type gene standard, respectively.
3 Discussion
The research on the Pulsatilla genus mainly focused on the separation of compounds and the pharmacological activity analysis of the main active components. While up to May 2022, more than 100 triterpenoid saponins have been isolated from the Pulsatilla genus (Wang et al., 2020; Zha et al., 2020; Xu et al., 2022). These isolated compounds are mainly lupeol-type and oleanolic acid-type, and only one compound belongs to ursolic acid-type (Ding et al., 2010). However, the biosynthetic pathway of all separated compounds in Pulsatilla genus has not been analyzed until now, and there was little omics data to support us to reveal it. Therefore, we cloned two 2,3-oxidosqualene cyclase genes from P. chinensis and its closely related species P. cerua through homologous cloning. These two genes are similar to Negro grass β-amyrin synthase gene that has been functionally verified. According to the separate compounds and triterpenoid saponin contents in the Pulsatilla genus (Jin et al., 2018), we speculated that besides the β-amyrin syntheses, there should exist the lupeol synthases in the Pulsatilla genus, especially in P. chinensis. Because the content of anemoside B4, which is a kind of lupeol-type triterpene saponin, is rich in the roots of P. chinensis (Jiang et al., 2021). In order to verify the functions of PcAS and PchAS, we expressed these genes in 2,3-oxisqualene high-yield yeast strain ATCC4021900 and leaves of N. benthamiana. The results shown in Figures 2C, D indicate PcAS is a single functional enzyme that only produces β-amyrin, while PchAS is a multifunctional enzyme, and its main product is lupeol.
At present, about 170 OSC genes from plants have been functionally analyzed (Chen et al., 2021; Wang et al., 2021). At the same time, the researchers also carried out targeted mutation to find the key active sites of OSC genes (Souza-Moreira et al., 2016; Ito et al., 2017; Aiba et al., 2018; Suzuki et al., 2019). The key active sites affecting the products of the family members were mainly located in the N-terminal, MXCYC conservative region, and C-terminal (Guo et al., 2022). For example, in 1999, Kushiro et, al. found that about 20 residues from Cys260 to Trp340 determined the activity of Panax ginseng β-amyrin synthase and Arabidopsis thaliana lupeol synthase. In addition, F474 of Euphorbia tirucalli β-amyrin synthase has a key role in affording the correct folding of the substrate (Ito et al., 2014). While the AS gene from P. chinensis and P. cernua has 27 different amino acid residues in 759 amino acids (Supplementary Figure S3), and these 27 different amino acids are distributed in the whole gene region. Therefore, to determine the main functional diversity region, we fused these two AS gene sequences obtained from P. chinensis and P. cernua, as shown in Figure 3A. Functional analysis of the chimera proteins showed that 145-456 amino acids were important to these two AS functions.
There contains four different amino acid residues in fragment 145-456 between P. chinensis and P. cernua. To further identify the key site leading to its functional difference, we proceed with directional mutation on these different amino acid residues. The results showed that if the 260-position amino acid W of PcAS, which located in the MXCYCR domain, was mutant to F, its catalysate changed from the single product to multiple products. The combination mutation of this site with any other three sites maintained the change (Figure 4). However, when this site did not change, any single mutation or combination mutation at the other three sites will not change the catalysate type (Supplementary Figure S5). Therefore, the 260-position amino acid is considered as the main active site affecting PcAS and PchAS function difference. This result is consistent with previous studies. In 2000, to identify the amino acid residues responsible for β-amyrin (PNY) and lupeol (OEW) synthase product specificity, site-directed mutagenesis on these two synthases was carried out. In this experiment, the 259-position amino acid W of β-amyrin synthase was mutated to Leu, and the 259-position amino acid L of lupeol synthase was mutated to Trp. The catalysate analysis revealed that W259 of β-amyrin synthase and L259 of lupeol synthase were the key amino acid residues (Dang and Prestwich, 2000). Our results provide that besides L, when another key amino acid F exists in the domain of MXCYXR of AS, the catalysate is lupeol.
As the main active component of P. chinensis, anemoside B4 is a kind of lupeol-type triterpenoid saponin. However, the content of anemoside B4 in other species of the Pulsatilla genus cannot reach the stipulation of the Chinese Pharmacopoeia (4.6% dry weight). We speculate that this phenomenon may be due to the insufficient supply of lupeol in P. cernua. However, lupeol is mostly oxidized to form betulinic acid and 23-hydroxybetulinic acid, and β-amyrin is oxidized to form oleanolic acid and hederagenin in plants. Therefore, we used the two-phase extract method to assess extracted saponins and aglycones after acidolysis of P. chinensis and P. cernua. Various aglycones were separated and their contents were accurately quantified (Figures 5A, B). The results showed that lupeol-type triterpenoid saponins were the main products in P. chinensis, and their aglycones were mainly oxidized at 23-position, while oleanane-type triterpenoid saponins were main products in P. cernua.
Because the difference in the type of aglycone between P. chinensis and P. cernua is caused by 2,3-oxidoqualene cyclase, we believe that the difference in the contents of triterpenoid saponins between P. chinensis and P. cernua is caused by the different expression of the 781-TT and 781-GG genotypes. Therefore, we absolutely quantitated the expression of these two genotypes in P. chinensis and P. cernua. As shown in Figures 5C, D, these two genotypes were expressed in both P. chinensis and P. cernua, while the 781-TT type was mainly expressed in P. chinensis and the 781-GG type was mainly expressed in P. cernua. This result is consistent with the different type triterpenoid saponin content of P. chinensis and P. cernua. In other words, the expressions of these two genotypes influence the biosynthesis and accumulation of different type triterpenoid saponins in P. chinensis and P. cernua.
In order to further find out whether the expression differences of the 781-GG and 781-TT genotypes in P. chinensis and P. cernua were caused by the difference of the gene copy number, we analyzed the 781-GG and 781-TT genotype copy number in the genome of P. chinensis, P. cernua and P. turczaninovii (anemoside B4 content is under 4.6%) (Song et al., 2021) by the qPCR relative quantitative method. The results showed that 781-GG type copy number was more than the 781-TT type in P. cernua, and the 781-TT type copy number was more than the 781-GG type in P. chinensis.
To sum up, we obtained two OSC genes from P. chinensis and P. cernua through homologous cloning and verified their functions in yeast and N. benthamiana. Then we found out the key amino acid which leads to the function difference of PcAS and PchAS by gene fusion and site-directed mutagenesis. In addition, we clarified the reason for the triterpenoid type difference in P. chinensis and P. cernua by combined analysis of different triterpenoid content and gene expression of 781-TT and 781-GG type AS. At last, we explained that different gene expression of the 781-TT and 781-GG type AS in P. chinensis and P. cernua was caused by the difference of gene copy numbers. This study provided basic information for the molecular breeding of P. chinensis. In addition, our study provides a usable molecular mark to identify the P. chinensis decoction pieces.
4 Materials and methods
4.1 Plant materials
Pulsatilla chinensis and P. cernua plants used for gene expression and metabolite component analysis were obtained from Beijing and Liaoning, China, then planted at the Institute of Botany, Chinese Academy of Science for three months. P.turczaninovii seeds were obtained from the germplasm bank of wild species which were used for DNA extraction. After the seeds germinated in the petri dish with five water-soaking filter papers at 25°C, the seedlings were planted in a 20 cm circular pot with turfy soil, and cultured in the 16 h light/8 h night greenhouse at 23°C.
4.2 RNA and cDNA preparation
Plant total RNA was extracted from the roots by RNAprep pure plant kit (DP441, TIANGEN). 1 µg total RNA was used for first strand cDNA synthesis with HiScript® III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (R312, Vazyme). In addition, in order to obtain the OSC core sequence of P. chinensis and P. cernua, we first used the primers reported in previous studies (Guhling et al., 2006), and the sequences were listed in Supplementary Table S1. Undesirably, these primers were not suitable for OSCs of P. chinensis and P. cernua. Therefore, we designed primers based on highly conserved regions of OSC protein sequences deposited in GenBank used Condehop, which were degenerate at the 3′ core region, and non-degenerate at the 5′ consensus clamp region. The primer sequences were listed in Supplementary Table S1. The degenerate PCR primers F1&R1 and PrimeSTAR® high-fidelity PCR enzyme (R045A, Takara) were used to obtain the core sequence of OSCs by touchdown PCR: 3 min 94°C, (10 s 98°C, 15 s 55°C, 30 s 72°C) × 10 cycles, (10 s 98°C, 5 s 55°C, 30 s 72°C) × 25 cycles, 5 min final extension at 72°C. The resulting PCR products were separated by 1% agarose gel electrophoresis and extracted using the V-ELUTE Gel Mini Purification Kit (ZPV202, ZOMANBIO). Then the purified products were recombined into the pLB vector (VT206, TIANGEN) and transformed into Escherichia coli TOP10 competent cells. The positive clones were sequenced by Sangon Biotech.
5′ and 3′ flanking fragment sequences of OSC were obtained by HiScript-TS 5′/3′ RACE Kit (RA101, Vazyme). Firstly, 5′ RACE-Ready cDNA and 3′ RACE-Ready cDNA were synthesized. Then the 5′ RACE-Ready cDNAs, 10 × Universal Primer Mix (UPM), 5′ GSP Primer R2 were used to extend the 5′ flanking fragment of OSC, and the 3′ RACE-Ready cDNAs, 10 × Universal Primer Mix (UPM), 3′ GSP Primer R2 for 3′ flanking fragment. 5′ GSP-Primer R2 and 3′ GSP-Primer R2 were synthesized according to the obtained OSC core sequence (Supplementary Table S1). All RCR programs used in this stage were referenced in the kit instruction manual. The PCR products were purified, recombined, and finally transformed into E. coli TOP10 competent cells. The positive clones were sequenced by Sangon Biotech.
4.3 OSC full-length CDS sequence cloning and plasmids construction
Based on the OSC gene sequence obtained by RACE PCR from P. chinensis, Primers F4 and Primers R4 were used to obtain OSC CDS full-length sequence. These primers which 5′ flanking sequences add 15 bp homologous sequences of destination vectors (pCAMBIA2300 for gene expression in plant, pET32a for gene expression analysis, and pESC-His for gene expression in yeast) were used to obtain the products with homologous sequences of vectors. Then the purified PCR products were homologously recombined into the linearized vectors by Exnase II (C112, Vazyme). Then the recombined vectors were transformed into E. coli TOP10 competent cells. The positive clones were sequenced by Sangon Biotech. In order to increase the substance of OSC, we constructed a vector (pSAK277-NbHMGRa) which can overexpress N. benthamiana 3-hydroxy-3-methylglutaryl reductase gene (NbHMGRa) in plants (Atsumi et al., 2018).
4.4 Functional characterization of OSCs in yeast
Plasmid pESC-His-PcAS or pESC-His-PchAS were transformed to an ERG7-deficient yeast strain (ATCC4021900) by LiAc/SS-DNA/PEG method (Gietz and Woods, 2002). For the expression of the galactose-inducible constructs, the positive colony was inoculated in 5 mL SD medium lacking uracil and supplemented with 2% glucose, and incubated overnight at 30°C and 220 rpm. Then 300 µL overnight cultures were inoculated into 30 mL of fresh SD medium lacking uracil and containing 2% glucose and grown at 30°C and 220 rpm in a 250 mL flask. After incubated for 24 h, the yeast cells were harvested by centrifugation (5 min, 5000 rpm), and washed twice with sterile water. At last, the cells were suspended at 30 mL SC-His medium with 2% galactose in a new 250 mL flask and incubated at 30°C and 220 rpm for 48 h. Then, the cells were harvested and refluxed with 10 mL 20% KOH/50% EtOH at 90°C for 1 h, then extracted three times with 10 mL of hexane. All hexane solutions were combined and evaporated under N2. Finally, the extractions were resuspended with 1 mL ethyl acetate, and stored at -20°C.
4.5 Functional characterization of OSCs in Nicotiana benthamiana
The plasmids pSAK277-NbHMGR, pCAMBIA2300, pCAMBIA2300-PcAS, and pCAMBIA2300-PchAS were individually transformed into Agrobacterium tumefaciens (strain GV3101) by the freeze-thaw method (Weigel and Glazebrook, 2006). The positive clones were incubated at 28°C, 200 rpm overnight in Luria-Bertani medium containing 50 mg/L kanamycin, 50 mg/L gentamicin, and 50 mg/L rifampicin. A. tumefaciens cells were washed with sterile water twice and resuspended with infiltration buffer (100 µM acetosyringone, 10 mM MgCl2 and 10 mM MES, pH 5.6). Adjust the OD600 of the suspension to 0.6 and staticly incubated at 28°C for 3 h. The suspensions of A. tumefaciens which harbor the plasmid pCAMBIA2300, pCAMBIA2300-PcAS, or pCAMBIA2300-PchAS were mixed with the pSAK277-NbHMGR in equal proportion and infiltrated into N. benthamiana leaves (three leaves per plant). Six days after agro-infiltration, the infiltrated leaves were harvested, frozen in liquid nitrogen immediately, and dried with the freeze-drying technology. 100 mg dried leaf powder was resuspended with 10 mL 20% KOH/50% EtOH solution and incubated at 90°C for 1h, then extracted with 10 mL hexane three times. All hexane solutions were evaporated with N2 and dissolved in 1 mL ethyl acetate. The extracts were stored at -20°C.
4.6 GC-MS analysis of metabolite compounds
Approximately 200 µL engineered yeast or N. benthamiana leaf extracts were evaporated with N2, then dissolved in pyridine and derivatized with N-methyl-N-(trimethylsilyl) trifluoroacetamide at 80°C for 30 min. The derivatized extracts were evaporated with N2 and dissolved in 200 µL hexane for gas chromatography and mass spectrometry (GC/MS) analysis. The GC-MS analysis was carried out as the previous study (Sandeep et al., 2019) with modified. For the GC-MS analysis, an aliquot (2 µL) was injected into the Shimadzu QP2010 gas chromatograph equipped with a WM-5MS capillary column (30 m × 250 µm, film thickness 0.25 µm) in unsplit mode. The injection temperature was 250°C, the carrier gas was helium with a flow rate of 1 mL/min the GC oven temperature was programmed from 180 to 300°C at 20°C min−1 and remained at 15 min. The ion trap temperature was 250°C. The electron energy was 70 eV. Spectra were recorded in the range of 50–750 m/z. All prominent peaks in the GC chromatogram were identified by comparison with a library database and the retention time of authentic standards.
4.7 Extraction and determination of total triterpenoid
4.7.1 Extraction method
The diphasic solvent extraction method was used to analyze the contents of total triterpenoid compounds. First, 10 mL MeOH was added to 100 mg dry-root powder of P. chinensis and P. cernua and mixed for approximately 30 s. Ultrasonication was employed to assist and accelerate the extraction of triterpenoids in the ultrasonic bath (100W) at room temperature for 25 min. Then centrifuged at 4500 g for 10 min. The clear supernatant was evaporated to dryness with N2. The residues were resuspended in 1 mL methanol and transferred into a 100 mL flask. 15 mL 20% H2SO4 and 15 mL toluene were added to the flask. The mixture was incubated at 60°C, 150 rpm for 16 h. Then the organic phase was collected and evaporated with N2. The residues were resuspended in 500 µL MeOH and filtered through 0.22 µm filters for HPLC-UV analysis.
4.7.2 HPLC-UV conditions
The chromatographic system consists of an HPLC (high-performance liquid chromatograph) and a UV detector. The stationary phase is a C30 column (250 x 4.6mm, Thermo Fisher Scientific). The column temperature was maintained at 40°C. The separation was performed by means of a linear gradient elution (phase A: water supplemented with 0.05% phosphoric acid, phase B: acetonitrile). The gradient was as follows: 55~60% B in 10 min, 60~80% B in 40 min, 95% B in 45 min, and 95% B for 5 min. The injected volume is 20 µL. All triterpene acids were detected at 210 nm.
4.8 Chimeric protein construction and site-directed mutagenesis
The homolog protein regions of PcAS and PchAS, which contain key sites influence OSC activity based on the previous study (Guo et al., 2022) were separated into three protein segments (Figure 3A). To generate chimeric proteins, the CDS segments of PcAS and PchAS were amplified by PCR using the primer pairs listed in Supplementary Table S1 (AS-1381-F/R, AS-720-F/R, and AS-424-F/R). The segments were assembled and cloned into the vector pESC-His to obtain 6 chimeric proteins (Figure 3A).
After the key region of OSC was identified, the single and double site-directed mutations of PcAS were constructed by the Mut Express®IIFast Mutagenesis Kit (C214, Vazyme). The primer pairs used were listed in Supplementary Table S1 (781-F/R, 892-F/R, 1013-F/R, and 1160-F/R).
4.9 Gene expression analysis
TaqMan-qPCR and absolute quantification were used to analyze the expression of PcAS and PchAS in different species. Firstly, the plasmids pET32a-PcAS and pET32a-PchAS were constructed as standards for these two gene types. And the gradient dilutions of plasmids pET32a-PcAS and pET32a-PchAS were used to build standard curves for these two gene types, respectively. Secondly, the primers were designed by Primer Express 3.0.1. The primers of PcAS (781-GG) were modified with FAM fluorescent labeling and PchAS (781-TT) were modified with VIC fluorescent labeling in their 5′ ends, respectively. MGB was added to their 3′ ends. The primer sequence informations were listed in Supplementary Table S1. Then ChamQ Geno-SNP Probe Master Mix (Q811, Vazyme) and Roche LightCycler 480 were used to analyze the absolute expression of PcAS and PchAS.
4.10 Gene copy number analysis of 781-TT type AS and 781-GG type AS in Pulsatilla ceruna, Pulsatilla chinensis and Pulsatilla turczaninovii
TaqMan-qPCR was used to analyze the gene copy number in P. ceruna, P. chinensis and P. turczaninovii as previous study (Maron et al., 2013) with modified. Firstly, genomic DNA was isolated from leaves of P. ceruna, P. chinensis and P. turczaninovii by FastPure® Plant DNA Isolation Mini Kit (DC104, Vazyme). Then the primers (AS-Tq-F/R) and probes 781-TT/GG with VIC and FAM fluorescent labeling, respectively, were designed by Primer Express 3.0.1 (Supplementary Table S1). Finally, ChamQ Geno-SNP Probe Master Mix (Q811, Vazyme) and Roche LightCycler 480 were used to analyze the fluorescence of VIC and FAM to explain 781-TT and 781-GG type gene copy number. In this experiment, the standards of 781-TT and 781-GG were the same as those used in gene expression.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JJ and AL designed the experiments and supervised the study. XL and JJ performed most of the experiments and wrote the manuscript. YX and JD performed the HPLC-UV analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32202415), the Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences (CI2021A04404), and the Fundamental Research Funds for the Central public welfare research institutes (ZZ14-YQ-029, ZXKT22002).
Acknowledgments
We thank Professor Wei Gao from Capital Medical University for kindly providing the yeast strain.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1144738/full#supplementary-material
Supplementary Figure 1 | Homologous cloning electrophoretogram of AS genes from Pulsatilla ceruna and P. chinensis. (A) The AS gene core fragment cloning from P. ceruna and P. chinensis; (B) 5′ and 3′ flank fragment of P. chinensis AS genes. (C) Full-length AS genes of P. ceruna and P. chinensis.
Supplementary Figure 2 | Phylogenetic analysis of PcAS and PchAS with the OSCs of other species.
Supplementary Figure 3 | Sequence alignment of AS proteins from Nigella sativa (NsAS), Pulsatilla ceruna (PcAS) and Pulsatilla chinensis (PchAS).
Supplementary Figure 4 | GC-MS analysis of β-amyrin and lupeol standard and the contents of PcAS and PchAS catalysate. The 218 and 189 ion counts of β-amyrin and lupeol are shown, respectively.
Supplementary Figure 5 | Functional characterization of mutant protein of I337V, H296D, Q397P. (A) GC–MS analyses of the yeast extraction expressing the mutant proteins contain the I337V with/without other sites; (B) GC–MS analyses of H296D, Q397P and both these two sites mutant proteins expressed yeast extraction. The 218 and 189 ion counts of β-amyrin and lupeol are shown, respectively.
Supplementary Figure 6 | Standard curve of 781-TT (PchAS) and 781-GG (PcAS) gene type.
Supplementary Figure 7 | Sequence alignment of AS gene DNA full-length sequences and CDS full-length sequences.
Supplementary Figure 8 | The AS gene core fragment sequences of AS genes cloned from the DNA of Pulsatilla ceruna, P. chinensis and P. turczaninovii.
Supplementary Table 1 | Primers used in this article.
References
Aiba, Y., Watanabe, T., Terasawa, Y., Nakano, C., Hoshino, T. (2018). Strictly conserved residues in Euphorbia tirucalli amyrin cyclase: Trp612 stabilizes transient cation through cation- interaction and CH- interaction of Tyr736 with Leu734 confers robust local protein architecture. Chembiochem 19, 486–495. doi: 10.1002/cbic.201700572
Atsumi, G., Kagaya, U., Tabayashi, N., Matsumura, T. (2018). Analysis of the mechanisms regulating the expression of isoprenoid biosynthesis genes in hydroponically-grown Nicotiana benthamiana plants using virus-induced gene silencing. Sci. Rep. 8 (1), 14804. doi: 10.1038/s41598-018-32901-5
Chen, K., Zhang, M., Ye, M., Qiao, X. (2021). Site-directed mutagenesis and substrate compatibility to reveal the structure-function relationships of plant oxidosqualene cyclases. Nat. Prod. Rep. 38, 2261–2275. doi: 10.1039/d1np00015b
Christ, B., Xu, C., Xu, M., Li, F. S., Wada, N., Mitchell, A. J., et al. (2019). Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 10 (1), 3206. doi: 10.1038/s41467-019-11286-7
Coley, F. C. (1922). Pulsatilla in dysmenorrhoea. Br. Med. J. 1 (3184), 13–14. doi: 10.1136/bmj.1.3184.13-a
Dang, T. Y., Prestwich, G. D. (2000). Site-directed mutagenesis of squalene-hopene cyclase: altered substrate specificity and product distribution. Chem. Biol. 7, 643–649. doi: 10.1016/s1074-5521(00)00003-x
Ding, X., Chen, Z., Li, X., Li, X., Xu, Q., Yang, S. (2010). Chemical constituents from Pulsatilla chinensis. Chin. Tradi. Herbal Drugs 41, 1952–1954. doi: 10.7501/j.issn.0253-2670
Friese, K. H., Kruse, S., Moeller, H. (1996). Acute otitis media in children. comparison between conventional and homeopathic therapy. HNO 44, 462–466.
Gietz, R. D., Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. doi: 10.1016/s0076-6879(02)50957-5
Guhling, O., Hobl, B., Yeats, T., Jetter, R. (2006). Cloning and characterization of a lupeol synthase involved in the synthesis of epicuticular wax crystals on stem and hypocotyl surfaces of Ricinus communis. Arch. Biochem. Biophys. 448, 60–72. doi: 10.1016/j.abb.2005.12.013
Guo, C. F., Xiong, X. C., Dong, H., Qi, X. Q. (2022). Genome-wide investigation and transcriptional profiling of the oxidosqualene cyclase (OSC) genes in wheat (Triticum aestivum). J. Syst. Evol. 60, 1378–1392. doi: 10.1111/jse.12738
Han, Y., Yang, Y., Li, Y., Yin, X., Chen, Z., Yang, D., et al. (2021). Genome-wide identification of OSC gene family and potential function in the synthesis of ursane- and oleanane-type triterpene in Momordica charantia. Int. J. Mol. Sci. 23, 196. doi: 10.3390/ijms23010196
Husselstein-Muller., T., Schaller, H., Benveniste, P. (2001). Molecular cloning and expression in yeast of 2,3-oxidosqualenetriterpenoid cyclases from arabidopsis thaliana. Plant Mol. Biol. 45, 75–92. doi: 10.1023/a:1006476123930
Ito, R., Masukawa, Y., Nakada, C., Amari, K., Nakano, C., Hoshino, T. (2014). Beta-amyrin synthase from Euphorbia tirucalli. steric bulk, not the pi-electrons of phe, at position 474 has a key role in affording the correct folding of the substrate to complete the normal polycyclization cascade. Org. Biomol. Chem. 12, 3836–3846. doi: 10.1039/c4ob00064a
Ito, R., Nakada, C., Hoshino, T. (2017). Beta-amyrin synthase from Euphorbia tirucalli l. functional analyses of the highly conserved aromatic residues Phe413, Tyr259 and Trp257 disclose the importance of the appropriate steric bulk, and cation-pi and CH-pi interactions for the efficient catalytic action of the polyolefin cyclization cascade. Org. Biomol. Chem. 15, 177–188. doi: 10.1039/c6ob02539k
Jiang, H. H., Zhang, W. D., Yang, J. X., Xue, G. R., Su, S. S., Li, C., et al. (2021). Miniaturized solid-phase extraction using a mesoporous molecular sieve SBA-15 as sorbent for the determination of triterpenoid saponins from Pulsatilla chinensis by ultrahigh-performance liquid chromatography-charged aerosol detection. J. Pharm. Biomed. Anal. 194, 13810. doi: 10.1016/j.jpba.2020.113810
Jin, M. M., Zhang, W. D., Song, G. S., Xu, Y. M., Du, Y. F., Guo, W., et al. (2018). Discrimination and chemical phylogenetic study of four Pulsatilla herbs using UPLC-ESI-MS/MS combined with hierarchical cluster analysis. J. Chromatogr. Sci. 56, 216–224. doi: 10.1093/chromsci/bmx102
Krokida, A., Delis, C., Geisler, K., Garagounis, C., Tsikou, D., Peña-Rodríguez, L. M., et al. (2013). A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis. New Phytol. 200, 675–690. doi: 10.1111/nph.12414
Laska, G., Sienkiewicz, A., Stocki, M., Zjawiony, J. K., Sharma, V., Bajguz, A., et al. (2019). Phytochemical screening of Pulsatilla species and investigation of their biological activities. Acta Soc Bot. Pol. 88, 3613. doi: 10.5586/asbp.3613
Lee, H. S., Beon, M. S., Kim, M. K. (2001). Selective growth inhibitor toward human intestinal bacteria derived from Pulsatilla cernua root. J. Agric. Food Chem. 49, 4656–4661. doi: 10.1021/jf010609z
Lertphadungkit, P., Qiao, X., Ye, M., Bunsupa, S. (2022). Characterization of oxidosqualene cyclases from Trichosanthes cucumerina l. reveals key amino acids responsible for substrate specificity of isomultiflorenol synthase. Planta 256, 58. doi: 10.1007/s00425-022-03972-6
Li, H. B., Wong, C. C., Cheng, K. W., Chen, F. (2008). Antioxidant properties in vitro and total phenolic contents in methanol extracts from medicinal plants. Lwt-Food Sci. Technol. 41, 385–390. doi: 10.1016/j.lwt.2007.03.011
Liu, J. Y., Guan, Y. L., Zou, L. B., Gong, Y. X., Hua, H. M., Xu, Y. N., et al. (2012). Saponins with neuroprotective effects from the roots of Pulsatilla cernua. Molecules 17, 5520–5531. doi: 10.3390/molecules17055520
Liu, Y., Tu, L., Lu, Y., Xia, M., Gao, W. (2021). Identification and functional characterization of 2,3-oxidosqualene cyclase genes family in Tripterygium wilfordii. Acta Pharmacol. Sin. 56, 3370–3376. doi: 10.16438/j.0513-4870.2021-1628
Maron, L. G., Guimaraes, C. T., Kirst, M., Albert, P. S., Birchler, J. A., Bradbury, P. J., et al. (2013). Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. U.S.A. 110, 5241–5246. doi: 10.1073/pnas.1220766110
Misra, P., Nayak, C., Chattopadhyay, A., Palit, T. K., Gupta, B., Sadhukhan, S., et al. (2021). Individualized homeopathic medicines in chronic rhinosinusitis: Randomized, double-blind, placebo-controlled trial. Homeopathy 110, 13–26. doi: 10.1055/s-0040-1715842
Niu, C., Hu, X. L., Yuan, Z. W., Xiao, Y., Ji, P., Wei, Y. M., et al. (2023). Pulsatilla decoction improves DSS-induced colitis via modulation of fecal-bacteria-related short-chain fatty acids and intestinal barrier integrity. J. Ethnopharmacol. 300, 115741. doi: 10.1016/j.jep.2022.115741
Rahimi, S., Kim, J., Mijakovic, I., Jung, K. H., Choi, G., Kim, S. C., et al. (2019). Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv. 37, 107394. doi: 10.1016/j.biotechadv.20190.04.016
Sandeep, M. R.C., Chanotiya, C. S., Mukhopadhyay, P., Ghosh, S. (2019). Oxidosqualene cyclase and CYP716 enzymes contribute to triterpene structural diversity in the medicinal tree banaba. New Phytol. 222, 408–424. doi: 10.1111/nph.15606
Scholz, M., Lipinski, M., Leupold, M., Luftmann, H., Harig, L., Ofir, R., et al. (2009). Methyl jasmonate induced accumulation of kalopanaxsaponin I in Nigella sativa. Phytochemistry 70, 517–522. doi: 10.1016/j.phytochem.2009.01.018
Seki, H., Tamura, K., Muranaka, T. (2015). P450s and UGTs: Key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol. 56, 1463–1471. doi: 10.1093/pcp/pcv062
Song, Y. M., Yang, Y. Y., Zhang, T. T., Xu, L., Xing, Y. P., Zhao, R., et al. (2021). Determination and fingerprint of anemoside B4 in baitouweng (Pulsatillae radix) by HPLC. J. Tradit. Chin. Med. 39, 127–131. doi: 10.13193/j.issn.1673-7717.2021.02.033
Souza-Moreira, T. M., Alves, T. B., Pinheiro, K. A., Felippe, L. G., De Lima, G. M. A., Watanabe, T. F., et al. (2016). Friedelin synthase from Maytenus ilicifolia: Leucine 482 plays an essential role in the production of the most rearranged pentacyclic triterpene. Sci. Rep. 6, 36858. doi: 10.1038/srep36858
Sun, J., Xu, X., Xue, Z., Snyder, J. H., Qi, X. (2013). Functional analysis of a rice oxidosqualene cyclase through total gene synthesis. Mol. Plant 6, 1726–1729. doi: 10.1093/mp/sst038
Suzuki, A., Aikawa, Y., Ito, R., Hoshino, T. (2019). Oryza sativa parkeol cyclase: Changes in the substrate-folding conformation and the deprotonation sites on mutation at Tyr257: Importance of the hydroxy group and steric bulk. Chembiochem 20, 2862–2875. doi: 10.1002/cbic.201900314
Wang, A. D., Bao, Y., Wang, X., Li, M. C., Ren, X. H., Liu, J. Y., et al. (2020). A new triterpenoid saponin from Pulsatilla cernua predicted by NMR-based mosaic method. Nat. Prod. Rep. 34, 909–914. doi: 10.1080/14786419.2018.1538995
Wang, J., Guo, Y., Yin, X., Wang, X., Qi, X., Xue, Z. Y. (2021). Diverse triterpene skeletons are derived from the expansion and divergent evolution of 2,3-oxidosqualene cyclases in plants. Crit. Rev. Biochem. Mol. Biol. 57, 113–132. doi: 10.1080/10409238.2021.1979458
Wang, Y., Zhang, H., Ri, H. C., An, Z., Wang, X., Zhou, J. N., et al. (2022). Deletion and tandem duplications of biosynthetic genes drive the diversity of triterpenoids in Aralia elata. Nat. Commun. 13, 2224. doi: 10.1038/s41467-022-29908-y
Weigel, D., Glazebrook, J. (2006). Transformation of agrobacterium using the freeze-thaw method. CSH Protoc. 7), pdb.prot4666. doi: 10.1101/pdb.prot4666
Xu, L., Deng, R., Feng, Y., Yang, X., Zhang, W., Liu, P. (2022). Chemical constituents from pulsatillae radix. J. Tradit. Chin. Med. 47, 5550–5555. doi: 10.19540/j.cnki.cjcmm.20220412.202
Xu, Q. M., Shu, Z., He, W. J., Chen, L. Y., Yang, S. L., Yang, G., et al. (2012). Antitumor activity of Pulsatilla chinensis (Bunge) regel saponins in human liver tumor 7402 cells in vitro and in vivo. Phytomedicine 19, 293–300. doi: 10.1016/j.phymed.2011.08.066
Xu, K., Shu, Z., Xu, Q. M., Liu, Y. L., Li, X. R., Wang, Y. L., et al. (2013). Cytotoxic activity of Pulsatilla chinensis saponins and their structure-activity relationship. J. Asian Nat. Prod. Res. 15, 680–686. doi: 10.1080/10286020.2013.790901
Yoo, H. H., Lee, S. K., Lim, S. Y., Kim, Y., Kang, M. J., Kim, E. J., et al. (2008). LC-MS/MS method for determination of hederacolchiside e, a neuroactive saponin from Pulsatilla koreana extract in rat plasma for pharmacokinetic study. J. Pharm. Biomed. Anal. 48, 1425–1429. doi: 10.1016/j.jpba.2008.09.012
Keywords: triterpenoid saponin, Pulsatilla chinensis, Pulsatilla cernua, oxidosqualene cyclase (OSC), gene copy number
Citation: Liu X, Xu Y, Di J, Liu A and Jiang J (2023) The triterpenoid saponin content difference is associated with the two type oxidosqualene cyclase gene copy numbers of Pulsatilla chinensis and Pulsatilla cernua. Front. Plant Sci. 14:1144738. doi: 10.3389/fpls.2023.1144738
Received: 15 January 2023; Accepted: 07 February 2023;
Published: 23 February 2023.
Edited by:
Fei Shen, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Zhichao Wu, National Institutes of Health (NIH), United StatesJie Wu, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences (CAS), China
Hongna Zhang, Hainan University, China
Copyright © 2023 Liu, Xu, Di, Liu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinzhu Jiang, anpqaWFuZ0BpY21tLmFjLmNu; An Liu, YWxpdUBpY21tLmFjLmNu
 Xianju Liu
Xianju Liu Yudi Xu
Yudi Xu An Liu
An Liu Jinzhu Jiang
Jinzhu Jiang