
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 March 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1142757
This article is part of the Research TopicDiscovery and Exploration of Genes Related to Important Agronomic Traits in Fruit TreesView all 17 articles
Fruit color is one of the most important traits of jujube (Ziziphus jujuba Mill.). However, the differences in the pigments of different varieties of Jujube are not well studied. In addition, the genes responsible for fruit color and their underlying molecular mechanisms remain unclear. In this study, two jujube varieties, namely “Fengmiguan” (FMG) and “Tailihong” (TLH), were considered. The metabolites from jujube fruits were investigated using ultra-high-performance liquid chromatography/tandem mass spectrometry. Transcriptome was used to screen anthocyanin regulatory genes. The gene function was confirmed by overexpression and transient expression experiments. The gene expression was analyzed by quantitative reverse transcription polymerase chain reaction analyses and subcellular localization. Yeast-two-hybrid and bimolecular fluorescence complementation were used to screen and identify the interacting protein. These cultivars differed in color owing to their respective anthocyanin accumulation patterns. Three and seven types of anthocyanins were found in FMG and TLH, respectively, which played a key role in the process of fruit coloration. ZjFAS2 positively regulates anthocyanin accumulation. The expression profile of ZjFAS2 exhibited its different expression trends in different tissues and varieties. Subcellular localization experiments showed that ZjFAS2 was localized to the nucleus and membrane. A total of 36 interacting proteins were identified, and the possibility of ZjFAS2 interacting with ZjSHV3 to regulate jujube fruit coloration was studied. Herein, we investigated the role of anthocyanins in the different coloring patterns of the jujube fruits and provided a foundation for elucidating the molecular mechanism underlying jujube fruit coloration.
The combination of various pigments leads to the appearance of color in fruits. This appearance is determined by three major types of plant pigments, namely chlorophyll, carotenoids, and flavonoids (Miller et al., 2011). Anthocyanin is a class of flavonoids that are widely distributed in plant seeds as natural water-soluble pigments. These pigments are important secondary metabolites in fruits and have been identified in various fruits such as strawberries, grapes, apples, citruses, peaches, and pomegranates (Huang et al., 2018; Luo et al., 2018; Wang et al., 2018; Gao et al., 2020; Li et al., 2020; Narjesi et al., 2023).
Anthocyanin biosynthesis has been well characterized in plants (Zhang et al., 2014; Tohge et al., 2017). The anthocyanin synthesis pathway is generally divided into five stages. The first stage is the phenylpropanoid pathway, where under the action of acid lyase (PAL), cinnamic acid 4-hydroxylase (C4H), and 4-coumaroyl CoA ligase (4CL), phenylalanine undergoes a series of reactions to form 4-coumaroyl CoA. Furthermore, dihydroflavonol is synthesized from 4-coumaroyl CoA by a series of enzymes such as chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H)/flavanone 3′-hydroxylase (F3′H)/flavanone 3′-5′-hydroxylase (F3′5′H). Subsequently, anthocyanidins are converted from dihydroflavonol by dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS). These anthocyanidins are ultimately modified by UDP-glucose flavonoid 3-glucosyltransferase (UFGT), rhamnosyltransferase (RT), and 3′-O-methyltransferase (OMT). Lastly, these anthocyanins are transported from the cytosol into the vacuoles. The anthocyanin biosynthesis is mainly regulated by three types of transcription factors, namely R2R3-MYB, bHLH, and WD40 repeat protein (Xu et al., 2015). Members of these protein families interact to form a complex (MBW complex containing MYB, bHLH, and WDR) that regulates anthocyanin biosynthesis by binding to the promoter of anthocyanin synthase (Zhang et al., 2014; Gu et al., 2019).
Studies on anthocyanin biosynthesis regulators focus more on R2R3-MYB and bHLH than WD40 repeat protein. Two types of WD40 proteins involved in anthocyanin biosynthesis regulation have been reported. The first type constitutes TTG1 and its homologous genes, such as AN11, PAC1, PFWD, and WDR1/2 (de Vetten et al., 1997; Carey et al., 2004; Hichri et al., 2010; Matus et al., 2010; Yamazaki and Saito, 2011). In Arabidopsis, TTG1 protein interacts with R2R3-MYB (PAP1/2, GL1, MYB5, MYB75, or MYB113/114) and bHLH (GL3, EGL3, or TT8) to form MBW complex, which activates the anthocyanin biosynthesis (Borevitz et al., 2001; Gonzalez et al., 2008; Gonzalez et al., 2009; Qi et al., 2011; Zhou et al., 2012). The second type constitutes COP1 and its homologous genes. COP1 is a key regulator that regulates light-induced anthocyanin accumulation (Li et al., 2012; An et al., 2017; Henry-Kirk et al., 2018).
Chinese jujube (Ziziphus jujuba Mill.) belongs to the family Rhamnaceae and is an economically and ecologically important fruit tree in Asia (Qu and Wang, 1993). Archaeological evidence suggests that the Chinese jujube has been cultivated for more than 3,000 years and has originated in China (Qu and Wang, 1993; Liu and Wang, 2009; Li et al., 2013). Chen et al reported that jujube fruit has high nutritional value and can be used as medicine and food (Chen et al., 2017). A variety of biologically active ingredients have been identified in jujube fruits, such as polysaccharides, flavonoids, phenolic acids, terpenoids, alkaloids, and vitamin C (Pawlowska et al., 2009; Gao et al., 2013; Liu et al., 2014; Chen et al., 2017; Shi et al., 2018; Zhang et al., 2020; Wen et al., 2022; Xue et al., 2022).
Jujube is an ideal natural pigment resource. Studies have shown that these pigments are flavonoids and are mainly found in the peel (Zhang et al., 2010; Choi et al., 2011; Gao et al., 2013; Kou et al., 2015). So far, 40 flavanols, 37 flavonols, 15 anthocyanins, 12 proanthocyanidins, nine dihydroflavones, eight flavanols, seven flavonoid carbonosides, five dihydroflavonols, three isoflavones, and two chalcones have been identified in different jujube fruits (Cheng et al., 2000; Bai et al., 2010; Choi et al., 2011; Choi et al., 2012; Gao et al., 2012; Collado-González et al., 2013; Du et al., 2013; Zozio et al., 2014; Chen et al., 2015; Wojdyło et al., 2016; Shi et al., 2018; Feng et al., 2020; Shi et al., 2020; Zhang et al., 2020; Xue et al., 2022). The jujube fruits studied in several studies were collected from a single variety. Therefore, there is a lack of systematic research on the jujube pigments among different varieties.
ZjANS and ZjUFGT play a crucial role in the accumulation of anthocyanin during the fruit ripening process, which is activated by ZjMYB5, ZjTT8, and ZjTTG1 (Shi et al., 2020; Zhang et al., 2020). ZjDFR is involved in the regulation of postharvest fruit coloration (Kou et al., 2019). The functions of these genes have already been identified in other species. In this study, we identified a novel anthocyanin regulatory gene, ZjFAS2, by analyzing the physiological and biochemical characteristics and performing metabolome and transcriptome profiling, transient expression, and genetic transformation experiments. Moreover, we investigated the expression profile and subcellular localization of ZjFAS2 and identified the proteins that interact with ZjFAS2. The results of this study shed light on the mechanism underlying jujube fruit coloration and provided theoretical support for the genetic improvement of jujube fruit quality.
A total of 28 cultivars were obtained from the jujube germplasm resource of the Luoyang Normal University in Luoyang, Henan, China (Supplementary Table 1). Fruit samples of “Fengmiguan” (FMG) and “Tailihong” (TLH) cultivars were harvested at seven developmental stages on days 20, 35, 50, 65, 80, 90, and 100 after flowering and were named FMG_S1, FMG_S2, FMG_S3, FMG_S4, FMG_S5, FMG_S6, FMG_S7, TLH_S1, TLH_S2, TLH_S3, TLH_S4, TLH_S5, TLH_S6, and TLH_S7, respectively. The new sheet, secondary lateral branch, flower bud, and the seed of FMG and TLH were collected. Fruit samples of the remaining 26 cultivars were harvested at stage S6. Furthermore, the white and red sides of fruit samples were collected and named S6-W and S6-R, respectively. All the fruit samples were brought back to the laboratory in liquid nitrogen containers and stored at −80 °C until further analysis.
The color of the peel of jujube fruits was measured using a colorimeter (SR-64, Shenzhen 3nh Technology Co., Ltd., Shenzhen, China), which provided information according to the Commission Internationale de system in terms of L* (brightness or lightness; 0 = black, 100 = white), a* (−a* = greenness, +a* = redness), b* (−b* = blueness, +b* = yellowness), and hue angle degree (h°) measurements. At least 10 fruits were sampled for FMG and TLH for all the stages.
Total chlorophyll and carotenoid contents were measured as described by Lichtenthaler and Wellburn (1983). Briefly, 100 mg of the sample was ground using 12 mL of 96% ethanol (v/v). The mixture was incubated under dark conditions for 3 h and then centrifuged at 10,000 rpm for 5 min. The absorbance was determined at 663 nm, 646 nm, and 470 nm using an ultraviolet–visible (UV–Vis) spectrophotometer. The total anthocyanin content was measured following the method described by Ai et al. (2016). Briefly, approximately 200 mg of the sample was ground with liquid nitrogen. The anthocyanins were extracted with 8 mL methanol–HCl (99:1 v/v). The absorbance was determined at 530 nm and 657 nm using a UV–Vis spectrophotometer. Cyanidin was used as a reference standard, and the results were expressed as cyanidin equivalents in mg/kg extract. Each sample was analyzed in triplicate. At least five fruits of each cultivar from all the stages were used.
The samples named FMG_S1, FMG_S4, FMG_S5, FMG_S6, FMG_S7, TLH_S1, TLH_S4, TLH_S5, TLH_S6, and TLH_S7 were analyzed using ultrahigh-performance liquid chromatography-electrospray ionization-tandem mass spectrometry (UPLC-ESI-MS/MS). For each sample, three biological replicates were independently analyzed. The primary and secondary spectral data of the metabolites were detected by mass spectrometry. The identification and structural analyses of these metabolites were performed using the MWDB database (Metware Biotechnology Co., Ltd. Wuhan, China) and public databases, namely MassBank (http://www.massbank.jp/), KNAPSAcK (http://kanaya.naist.jp/KNApSAcK/), HMDB (http://www.hmdb.ca/), MoToDB (http://www.ab.wur.nl/moto/), and ChemBank (http://chembank.med.harvard.edu/compounds), PubChem (https://pubchemblog.ncbi.nlm.nih.gov/), NIST Chemistry Webbook (http://webbook.nist.gov/), and METLIN (http://metlin.scripps.edu/index.php). Metabolomics data was processed using Analyst (Version 1.6.3, Sciex, Framingham, MA, USA).
The total RNA extracted from the samples named FMG_S1, FMG_S7, TLH_S1, TLH_S5, and TLH_S7 was collected for transcriptome analysis. For each sample, three biological replicates were independently analyzed. The RNA sequencing (RNA-Seq) was performed by Biomarker Technologies Co., Ltd. (Beijing, China). The transcriptome data were deposited in the NCBI (National Center for Biotechnology Information) Sequence Read Archive database (Accession number: SRR23330575-SRR23330589).
Differential expression analyses of two groups were performed using the DESeq R package (1.10.1). For identifying differentially expressed genes (DEGs), fold change (FC) ≥2 and false discovery rate (FDR) <0.01 were used as the screening criteria. FC represented the ratio of gene expression values in two groups. The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the FDR.
Total RNA was extracted using a plant RNA extraction kit (Cat. No. 9769, TakaRa Biotechnology Inc., China). The quality of RNA was determined using an Agilent Bioanalyser 2100 (Agilent Technologies, Inc., USA), and the concentration was measured using an ND-2000 spectrophotometer (Nanodrop Technologies, Inc., USA). After the treatment of the RNA sample with a gDNA Eraser, the first-strand cDNA was synthesized using the PrimeScript™ RT reagent Kit (Cat. No. RR047A, TakaRa Biotechnology Inc., China). qRT-PCR was performed with gene-specific primers in a real-time PCR reaction system (25 µL) according to the manufacturer’s protocol (Cat. No. RR820A, TakaRa Biotechnology Inc., China). The amplification was monitored on a CFX96 real-time PCR detection system (BIO-RAD). The measurements were obtained using the relative quantification method (Livak and Schmittgen, 2001). Jujube ACTIN (LOC107413530) was used as an internal control. All analyses were based on performed using three biological samples and three replicates of each sample. The primers are listed in Supplementary Table 2.
To construct the overexpression construct of ZjFAS2, the ZjFAS2 cDNA from TLH was digested with XbaI and XhoI and then inserted into the pART-CAM vector. The appropriate constructs were confirmed by sequencing and introduced into the host cells Agrobacterium tumefaciens GV3101. The primers are listed in Supplementary Table 3.
For transgenic validation, the tobacco variety “SR1” was used as the receptor for transformation by Agrobacterium. Tobacco transformation was performed using a previously described method (Horsch et al., 1985; Zong et al., 2019). The transient expression assay in jujube fruits was performed on the basis of a previously described method with slight modifications (Hawkins et al., 2016). The Agrobacterium strain containing the 35S:ZjFAS2 construct was infiltrated on the jujube fruits between stages S4 and S5 for phenotype investigation. Agrobacterium cultures carrying the empty vector served as the negative control. Patches of red color appeared 3–5 days after injection.
The ZjFAS2 cDNA was inserted downstream of the cauliflower mosaic virus (CaMV) 35S promoter through XhoI and EcoRI restriction enzyme sites in the frame with a green fluorescent protein (GFP) in the part-CAM-EGFP vector. The ZjFAS2-GFP fusion construct and nucleus/membrane protein marker (AtH2B-RFP/AtPIP2A-RFP) were cotransformed into tobacco leaves via agrobacterium-mediated transformation. The primers are listed in Supplementary Table 3.
The yeast two-hybrid cDNA library was constructed using the Make Your Own “Mate & Plate™” Library System (Cat. No. 630490, TakaRa Biotechnology Inc., USA). To construct the bait vector for ZjFAS2, the ZjFAS2 cDNA from TLH was digested with EcoRI and BamHI and then inserted into the pGBKT7 vector. The co-transformation of yeast strain Y2HGold was performed after self-activation and toxicity detection to screen for ZjFAS2 interacting proteins. The pGADT7-jujube cDNA and pGBKT7-FAS2 were co-transformed into yeast strain Y2HGold. The yeast cells were cultured in the SD-Trp-Leu-His-Ade medium at 30 °C under dark conditions for three days. The positive colonies were selected and sequenced. The primers are listed in Supplementary Table 3.
The cDNA of ZjFAS2 and ZjSHV3 from TLH was digested with XbaI and KpnI and then inserted into the pCAMBIA1300S-YC and pCAMBIA1300S-YN vectors to produce the cYFP-protein and nYFP-protein constructs, respectively. The BiFC assays were performed using the method described by Guo et al. (2020). The primers are listed in Supplementary Table 3.
To investigate the process of the color change of the jujube fruits, we divided the fruit developmental stages into seven stages (Figure 1A). Harvested between 20 and 100 days after flowering, the fruit weight, length, and width of FMG ranged between 0.58–10.56 g, 13.76–29.37 mm, and 9.16–26.85 mm, respectively, and the fruit weight, length, and width of TLH ranged between 0.56–12.59 g, 16.37–38.05 mm, and 8.00–24.68 mm, respectively (Supplementary Table 4). Maximum fruit size and weight were recorded at stage S7. The color parameters of FMG and TLH ranged between 38.87–69.18 and 29.85–55.64 for L*, –11.10–20.68 and 5.94–19.22 for a*, 15.87–39.85 and 1.76–27.06 for b*, and 36.37–113.05 and 13.54–77.47 for h°. The h° of FMG gradually decreased with fruit development, and the maximum and minimum values were recorded at stages S1 and S7, respectively. However, the h° of TLH showed different patterns, peaking at stage S5 and then decreasing gradually (Supplementary Table 5). These results indicated that the fruit color of FMG and TLH differed considerably during the early stages of fruit development.
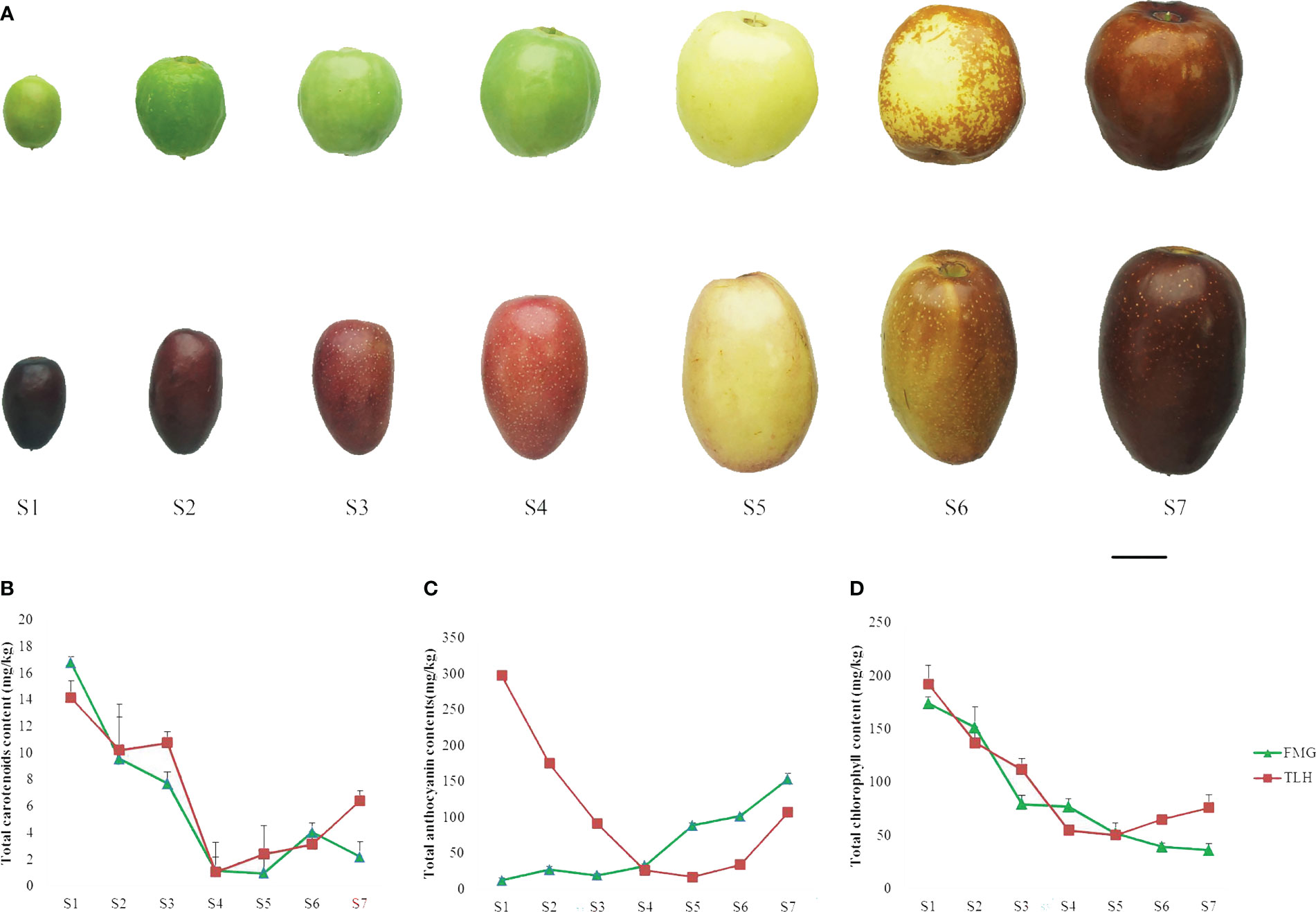
Figure 1 Phenotypic characterization of the jujube cultivars FMG and TLH. (A) Photographs of the seven stages of growth. Bars = 1 cm. (B) Total carotenoid content. (C) Total anthocyanin content. (D) Total chlorophyll content. Error bars indicate standard deviations (SDs) from three biological replications.
A rapid colorimetric assay was performed to evaluate the contents of anthocyanin, chlorophyll, and carotenoid. A small number of carotenoids was detected in the peels (Figure 1B), whereas a large number of anthocyanins and chlorophyll was detected in the peels (Figures 1C, D). The total anthocyanin content in FMG and TLH varied between 143.50–313.75 and 136.60–345.05 mg/kg, respectively. The content of anthocyanins in FMG gradually increased with fruit development, and the highest concentration was recorded at stage S6. However, anthocyanin content in TLH decreased to the lowest level at stage S5 and then increased gradually. The chlorophyll contents of FMG and TLH ranged between 36.25–174.22 and 50.53–192.67 mg/kg, respectively, which decreased rapidly during the ripening process. The correlation analysis showed that only the total anthocyanin content was significantly negatively correlated with h° in both FMG and TLH (Supplementary Table 6).
Taken together, our results indicated that the fruit color of FMG and TLH was mainly determined by anthocyanin content despite the differences in fruit coloration patterns.
To further identify the type of anthocyanins, the metabolites were investigated by UPLC-ESI-MS/MS. A total of 784 putative metabolites were identified from the peels of FMG and TLH (Figures 2A, B; Supplementary Table 7).
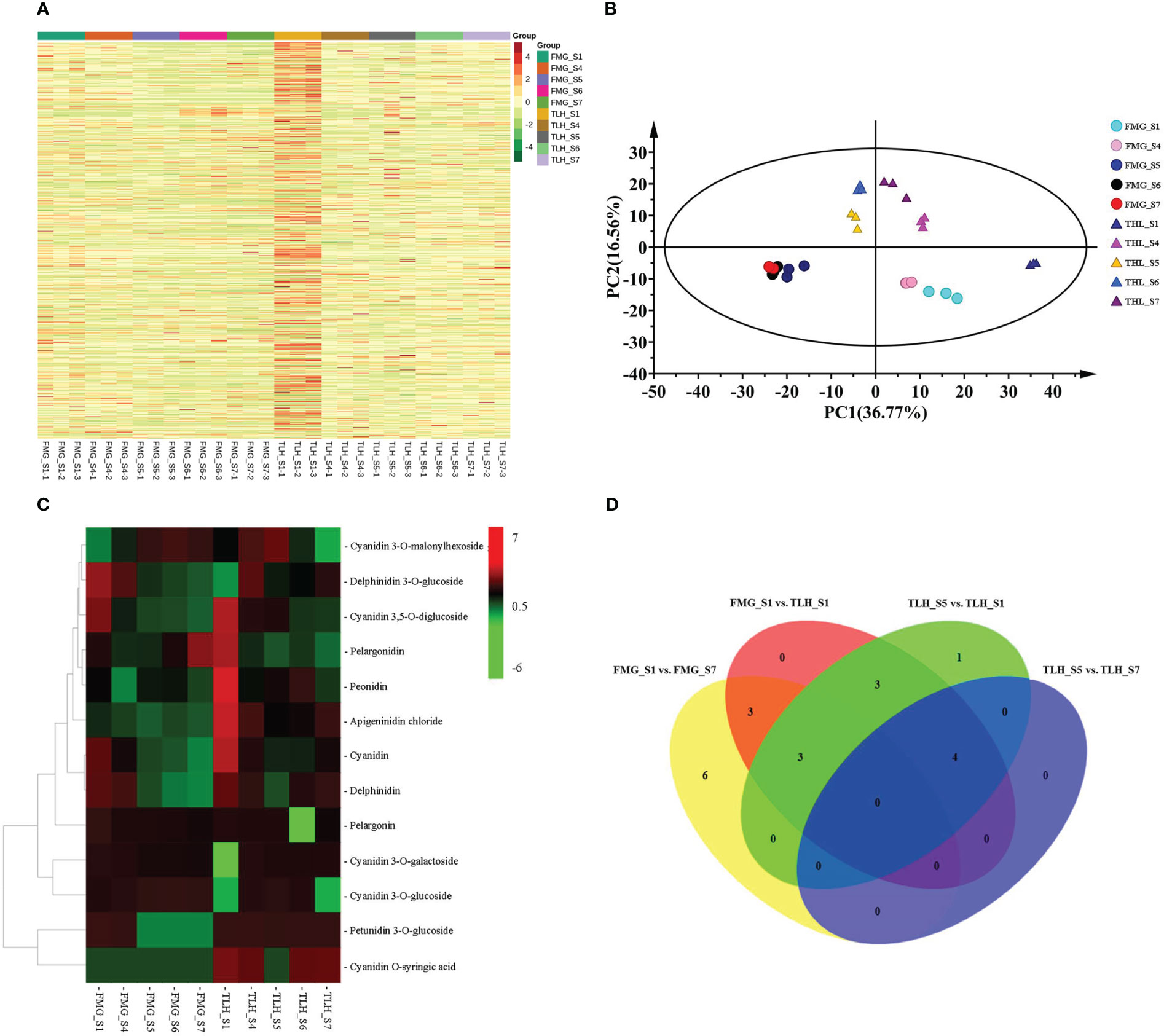
Figure 2 Qualitative and quantitative analysis of the metabolomics data of the peels of FMG and TLH. (A) Heatmap of the quantified identified metabolites. (B) PCA score plot of the metabolites in the FMG and TLH. (C) Heatmap of the quantification of the identified anthocyanins. (D) Venn diagram showing the number of metabolites in FMG_S1, FMG_S7, TLH_S1, TLH_S5, and TLH_S7.
Of note, 12 anthocyanins were identified from the peel extracts of FMG (Figure 2C; Supplementary Table 7). The contents of three of these anthocyanins, namely cyanidin 3-O-glucoside, cyanidin 3-O-malonylhexoside, and pelargonidin, increased with the ripening process and were positively correlated with the total anthocyanin content (Supplementary Table 8). Conversely, nine anthocyanins, namely apigeninidin chloride, cyanidin, cyanidin 3,5-O-diglucoside, cyanidin 3-O-galactoside, delphinidin, delphinidin 3-O-glucoside, pelargonidin, peonidin, peonidin, and petunidin 3-O-glucoside, decreased or remained stable with fruit development and were negatively correlated with the total anthocyanin content (Supplementary Table 8).
We identified 13 anthocyanins from the peel extracts of TLH (Figure 2C; Supplementary Table 7). Nine of these anthocyanins, namely apigeninidin chloride, cyanidin, cyanidin 3,5-O-diglucoside, cyanidin O-syringic acid, delphinidin, pelargonidin, pelargonin, peonidin, and petunidin 3-O-glucoside, were positively correlated with the total anthocyanin content (Supplementary Table 8). Conversely, four anthocyanins, namely cyanidin 3-O-galactoside, cyanidin 3-O-glucoside, cyanidin 3-O-malonylhexoside, and delphinidin 3-O-glucoside, were negatively correlated with the total anthocyanin content (Supplementary Table 8). Multiple comparative analyses showed that apigeninidin chloride, cyanidin, cyanidin 3,5-O-diglucoside, cyanidin O-syringic acid, delphinidin, pelargonidin, and peonidin had a relatively higher content in TLH_S1, whereas apigeninidin chloride, cyanidin O-syringic acid, and delphinidin had a relatively higher content in TLH_S7 (Figure 2D). Together, these results indicated that cyanidin, cyanidin 3,5-O-diglucoside, pelargonidin, and peonidin play a crucial role during the early stages of the fruit coloration process, whereas apigeninidin chloride, cyanidin O-syringic acid, and delphinidin are involved in the complete fruit coloration process.
To evaluate anthocyanin biosynthesis at the transcriptional level, five cDNA libraries were prepared from the peels of FMG and TLH, which were subjected to RNA-Seq analysis (Supplementary Table 9). To verify the accuracy and reproducibility of the anthocyanin biosynthesis-related genes analysis results, 11 anthocyanin structural genes, one R2R3-MYB gene, five bHLH genes, and one WD40 gene were randomly selected and analyzed by qRT-PCR analysis (Supplementary Figure 1). Pearson’s correlation coefficients further indicated that the digital transcript abundance of most genes was significantly correlated with the qRT-PCR results (Supplementary Table 2). These results confirmed the reliability of the RNA-Seq data.
Differential expression of anthocyanin structural genes was performed. In FMG, 16 differentially expressed anthocyanin structural genes were detected in FMG_S1 and FMG_S7. Among these genes, the transcriptional levels of 14 genes decreased with fruit ripening, namely two C4H genes, three CHS genes, one CHI gene, one F3H gene, one F3’5’H gene, two DFR genes, three ANS genes, and one UFGT gene. The expression of one CHI gene and one UFGT gene was upregulated in FMG_S7 (Figure 3A; Supplementary Table 10). In TLH, 17 anthocyanin structural genes showed upregulated expression in TLH_S1, namely two C4H genes, three CHS genes, one CHI gene, one F3H gene, one F3’H gene, one F3’5’H gene, two DFR genes, five ANS genes, and one UFGT gene (Figure 3A; Supplementary Table 11). These 17 genes also showed upregulated expression in TLH_S1 compared with the expression in FMG_S1 (Figure 3A; Supplementary Table 12). During fruit development, one C4H gene, one CHI gene, one F3’H gene, and one UFGT gene were downregulated, and only one UFGT gene was upregulated in TLH_S7 (Figure 3A; Supplementary Table 13). Comparing the gene expression in FMG_S7 and TLH_S7 indicated that one C4H gene, one F3H gene, one DFR gene, and three ANS genes were upregulated, whereas only one F3’H gene was downregulated in TLH_S7 (Figure 3A; Supplementary Table 14).
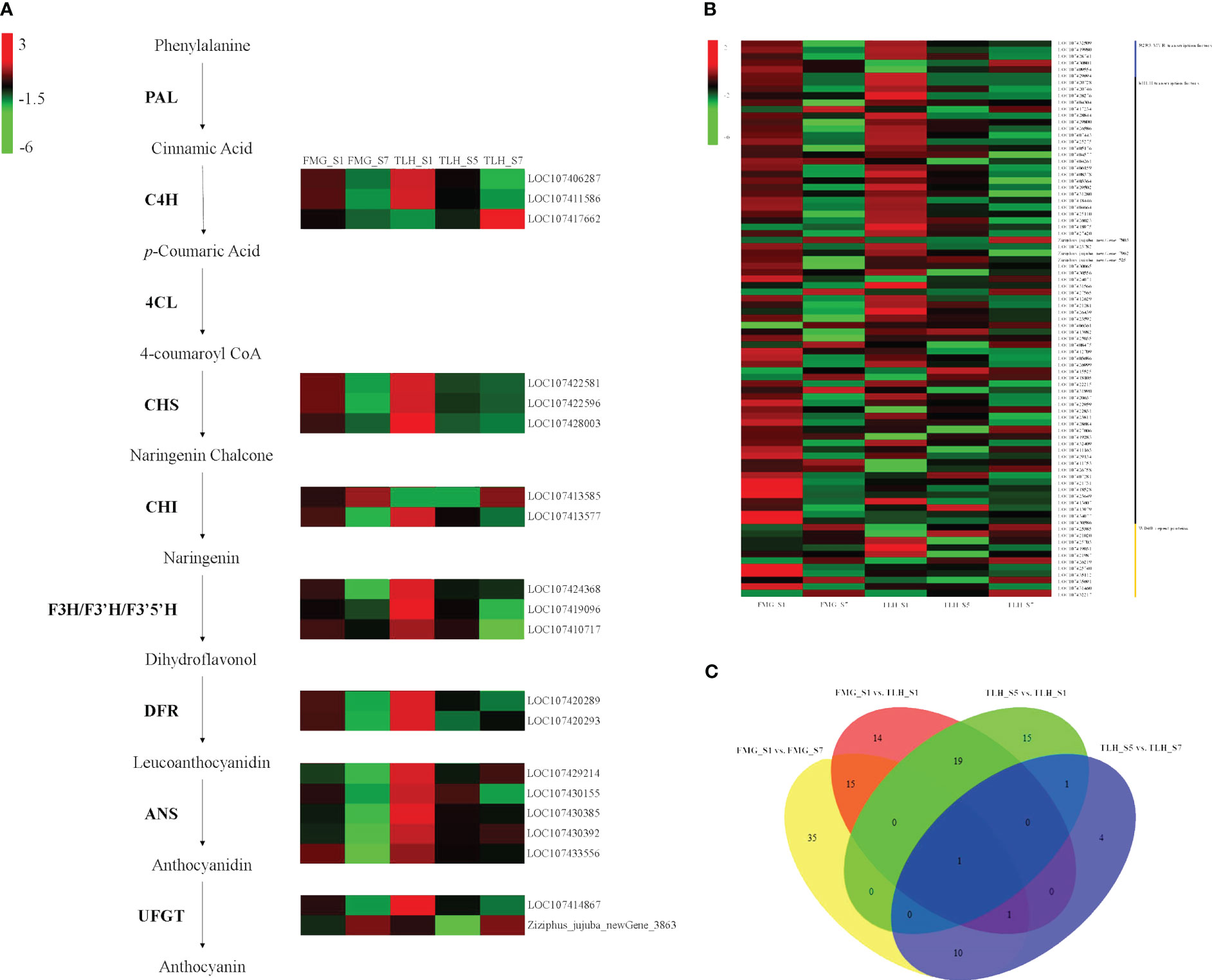
Figure 3 Transcriptome analyses of the peels of FMG and TLH. (A) Heatmap of differentially expressed anthocyanin structural genes. (B) Heatmap of differentially expressed R2R3-MYB, bHLH, and WD40 genes. (C) Venn diagram showing the number of differentially expressed R2R3-MYB, bHLH, and WD40 genes in FMG_S1, FMG_S7, TLH_S1, TLH_S5, and TLH_S7.
Differential expression of R2R3-MYB, bHLH, and WD40 genes was also detected. We detected 62 differentially expressed genes between FMG_S1 and FMG_S7. Furthermore, 47 genes were downregulated in FMG_S7, namely 11 R2R3-MYB genes, 33 bHLH genes, and three WD40 genes; 15 genes were upregulated in FMG_S7, namely nine bHLH genes and six WD40 genes (Figure 3B; Supplementary Table 10); 36 differentially expressed genes were detected between TLH_S1 and TLH_S5; and 27 genes, namely five R2R3-MYB genes, 20 bHLH genes, and two WD40 genes, were upregulated. However, only six bHLH genes and three WD40 genes were downregulated in TLH_S1 (Figure 3B; Supplementary Table 11). Conversely, only 17 differentially expressed genes were detected between TLH_S5 and TLH_S7. Four bHLH genes and five WD40 genes were upregulated, whereas two R2R3-MYB genes and six bHLH genes were downregulated in TLH_S7 (Figure 3B; Supplementary Table 12). In contrast to the expression of the anthocyanin structural genes, the DEGs related to anthocyanin regulation did not show a clear trend between FMG and TLH during the same fruit developmental stages. Of note, 27 genes, namely six R2R3-MYB genes, 19 bHLH genes, and two WD40 genes, were upregulated, and 23 genes, namely six R2R3-MYB genes, 14 bHLH genes, and three WD40 genes, were downregulated in TLH_S1 compared with FMG_S1 (Figure 3B; Supplementary Table 13). Only six genes were differentially expressed between FMG_S7 and TLH_S7. Among these genes, one R2R3-MYB gene and two bHLH genes were upregulated, and three bHLH genes were downregulated in TLH_S7 (Figure 3B, Supplementary Table 14).
Multiple comparative analyses showed that only one WD40 gene, LOC107425703, showed differential expression among FMG_S1, FMG_S7, TLH_S1, TLH_S5, and TLH_S7 (Figure 3C; Supplementary Tables 10-13). LOC107425703 encodes a WD40 repeat protein homologous to the Arabidopsis FASCIATA2 (FAS2) gene, which was hereafter designated as ZjFAS2. Pearson’s correlation coefficients further indicated that ZjFAS2 expression was significantly positively correlated with the total anthocyanin content (r = 0.906, P <0.05; Figure 3C; Supplementary Figure 1). Together, these results strongly indicated that ZjFAS2 are candidate genes affecting anthocyanin accumulation.
We cloned the full-length complementary DNA (cDNA) of ZjFAS2 from TLH_S1, which is 1,362 bp in length. The predicted ZjFAS2 encodes 453 amino acids and contains seven conserved WD40 repeat domains (Figure 4A). We collected 52 ZjFAS2 homologs from the NCBI (http://www.ncbi.nlm.nih.gov/ which encoded proteins varying from 347 to 593 amino acids in length. The FAS2 homologs can be divided into three major groups on the phylogenetic tree and the similarity of their protein sequences ranged from 63.5 to 75.9% (Figure 4B).
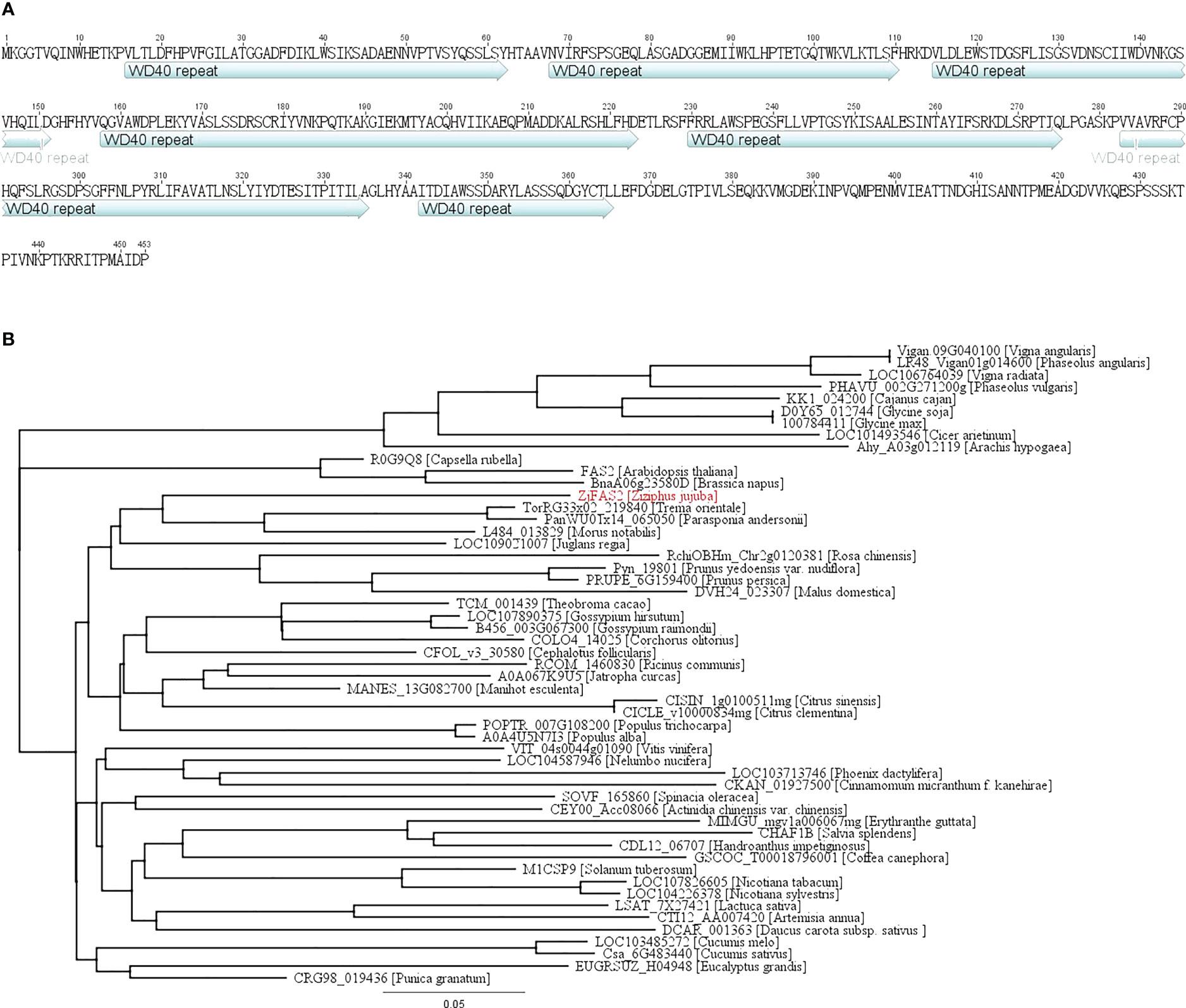
Figure 4 Protein sequence and phylogenetic analysis of ZjFAS2. (A) ZjFAS2 protein sequence. (B) Phylogenetic tree of the plant FAS2 protein. The lengths of the branches refer to the amino acid variation rates.
To validate the genetic function of ZjFAS2, we cloned the cDNA of ZjFAS2 into the pART-CAM vector and introduced the construct into jujube and tobacco via agrobacterium-mediated transformation. ZjFAS2 overexpression significantly increased anthocyanin accumulation in jujube fruits (Figures 5A–C). Furthermore, ZjFAS2 expression was significantly positively correlated with the total anthocyanin content (r = 0.845, P <0.05). In the tobacco transgenic plants, we found that the petals of transgene-positive plants carrying ZjFAS2 had significantly higher total anthocyanin content than the petals of transgene-negative plants (Figures 5D–F). These results provided evidence that ZjFAS2 plays a major role in anthocyanin accumulation.
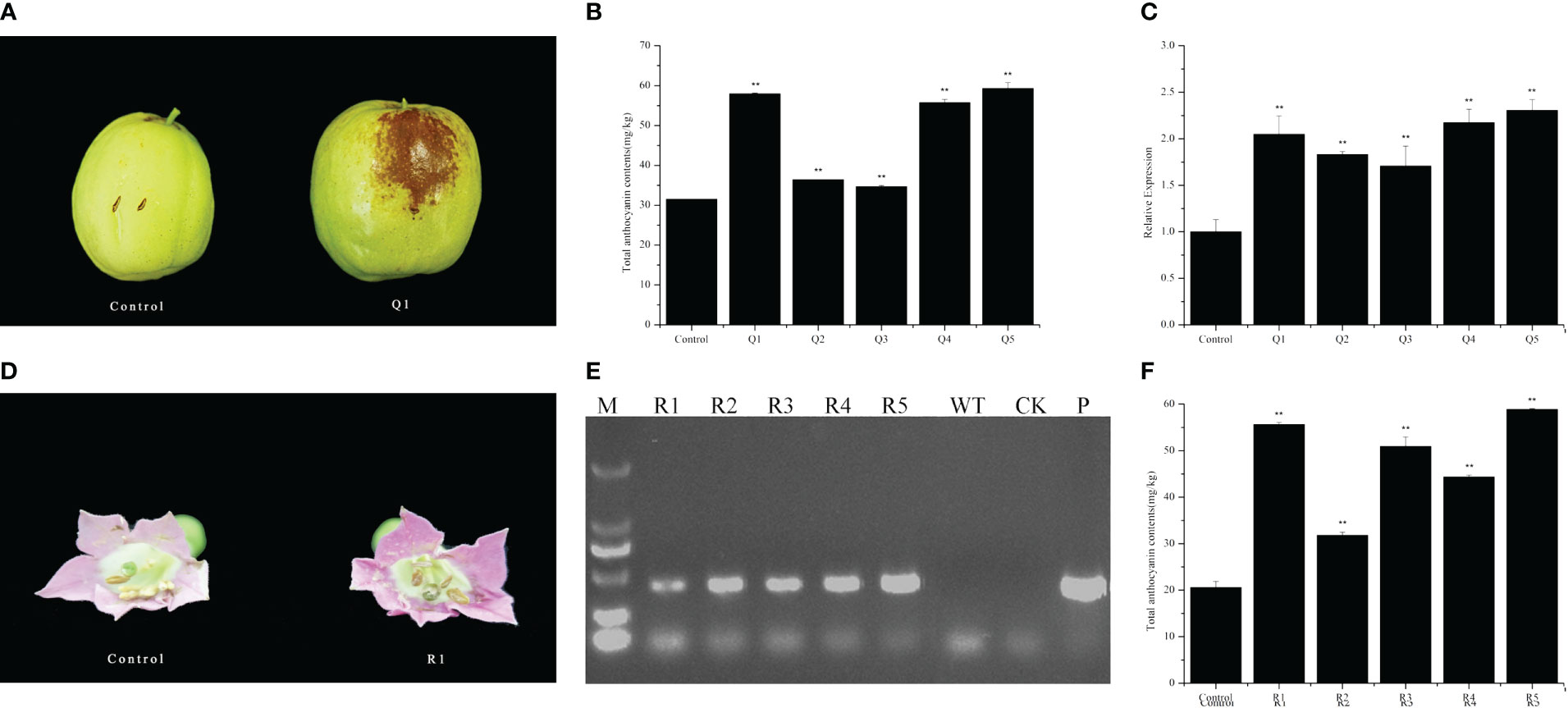
Figure 5 Functional analysis of ZjFAS2. (A) Fruit phenotypes of the FMG-null (Control) and ZjFAS2-transient expression fruit (Q1). (B) Comparison of total anthocyanin content between FMG-null (Control) and ZjFAS2-transient expression fruits (Q1-Q5). (C) Expression of ZjFAS2 in the FMG-null (Control) and ZjFAS2-transient expression fruits (Q1-Q5) by qRT-PCR. (D) Flower phenotypes of the SR1-null (Control) and ZjFAS2-transgenic lines (R1). (E) The examination of ZjFAS2-overexpressed lines (R1-R5), SR1 as negative control (WT), H2O as blank control (CK), and 35S::ZjFAS2 pART-CAM vector (P) by PCR. (F) Expression of ZjFAS2 in the SR1-null (Control) and ZjFAS2-transgenic lines (R1-R5) by qRT-PCR. ** indicates significantly different at P < 0.01. Error bars indicate standard deviations (SDs) from three biological replications.
Our previous study indicated that ZjFAS2 expression and the total anthocyanin content in purple leaves are significantly higher than those in green leaves (Li et al., 2021). To further evaluate the expression profile of ZjFAS2, qRT-PCR and pigment analysis were performed in various organs of TLH and FMG (Figures 6A, B). The total anthocyanin content in the new sheet, secondary lateral branch, flower bud, and seed of TLH was higher than that in the respective tissues of FMG. Conversely, the total anthocyanin content in the sarcocarp of TLH was lower than that in the sarcocarp of FMG. Pearson’s correlation coefficients further indicated that ZjFAS2 expression was significantly positively correlated with the total anthocyanin content (r = 0.848, P <0.01). To examine the expression profile of ZjFAS2 in further detail, the fruits from the semi-red period of 26 cultivars were used in the analysis. Compared with the S6-W, the total anthocyanin content showed a higher concentration in the S6-R of 11 varieties, a lower concentration in the S6-R of 13 varieties, and a similar concentration in the S6-R of two varieties (Figure 6C). However, ZjFAS2 expression showed different patterns. Compared with the ZjFAS2 in S6-W, ZjFAS2 showed upregulated expression in the S6-R of 18 varieties, lower expression in the S6-R of three varieties, and similar expression in the S6-R of five varieties (Figure 6D).
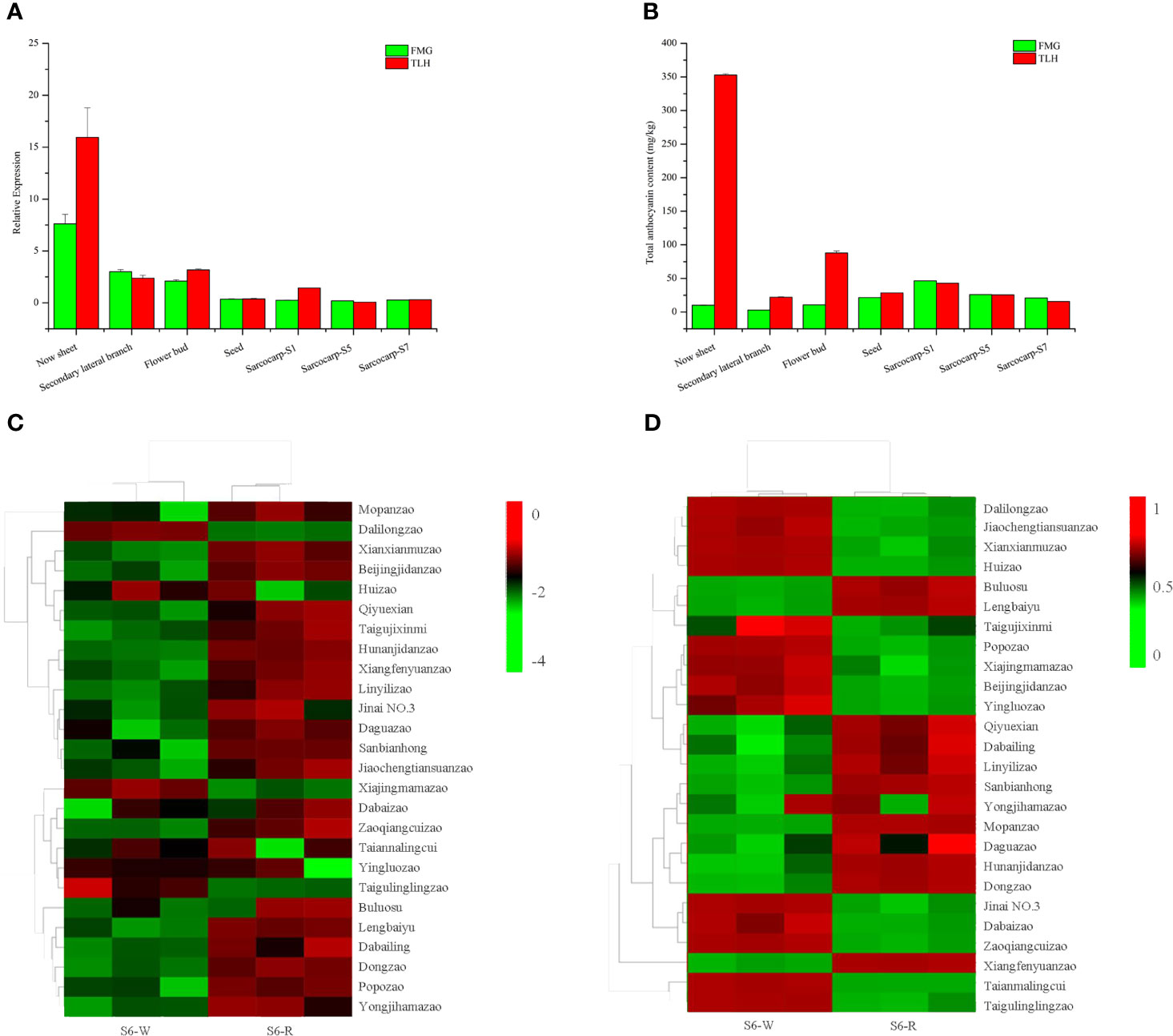
Figure 6 Expression patterns of ZjFAS2. (A) qRT-PCR analysis of ZjFAS2 in Secondary lateral branch, flower bud and seed of FMG and TLH. (B) Total anthocyanin content n Secondary lateral branch, flower bud and seed of FMG and TLH. (C) Heatmap of ZjFAS2 expression level in S6-W and S6-R from 26 varieties. (D) Heatmap of total anthocyanin content in S6-W and S6-R from 26 varieties. Error bars indicate standard deviations (SDs) from three biological replications.
To detect the subcellular localization of ZjFAS2, we constructed a GFP-ZjFAS2 fusion, whose expression was driven by the CaMV 35S promoter. The transient expression experiment in tobacco epidermal cells showed that the GFP-ZjFAS2 fusion protein was colocalized with the markers in both the nucleus and membrane (Figure 7).
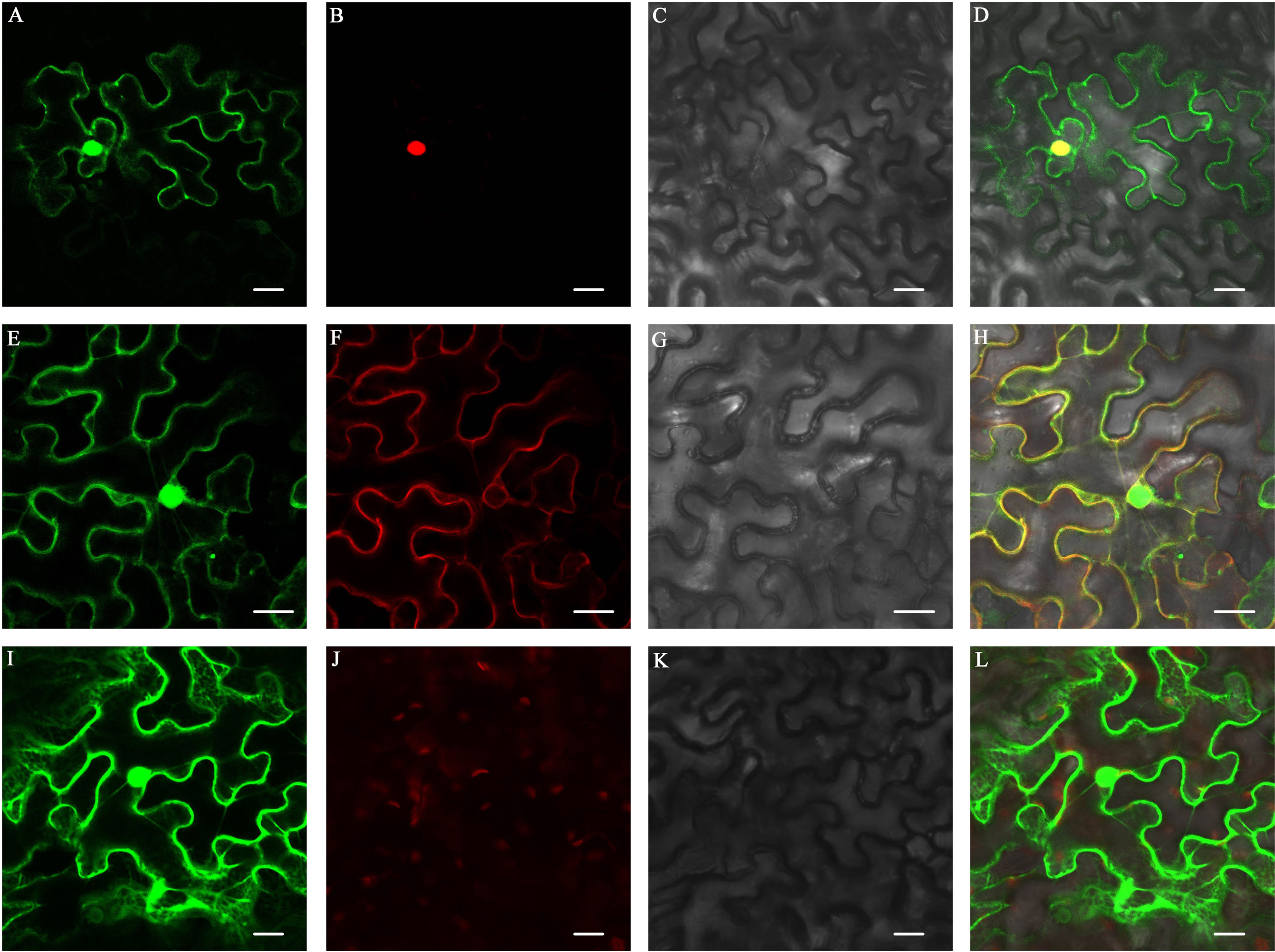
Figure 7 Subcellular localization of ZjFAS2. Colocalization of ZjFAS2-GFP (A) and AtH2B-RFP (B) in the tobacco leaves. The bright-field image (C) and merged image (D) are also shown. Colocalization of ZjFAS2-GFP (E) and AtPIP2A-RFP (F) in the tobacco leaves. The bright-field image (G) and merged image (H) are also shown. (I) The green fluorescent signal of null-GFP in the tobacco leaves. (J) The red fluorescent signal in the same tobacco leaves as (I). (K) The same tobacco leaves as (I) under bright field. (L) The merged image of (I), (J) and (K). Bars = 20 μm.
No study has reported that FAS2 is involved in the regulation of anthocyanin accumulation. To elucidate the regulatory mechanism underlying the role of ZjFAS2 on jujube fruit coloration, 36 interacting proteins were identified by the yeast two-hybrid library screens (Supplementary Figure 2; Supplementary Table 15). Notably, one of the ZjFAS2 interactors, the lycerophosphodiester phosphodiesterase GDPDL3-like (LOC107434323) is highly homologous with Arabidopsis SHAVEN3 (AtSHV3). The shv3 mutant of Arabidopsis thaliana can increase anthocyanin accumulation (Hayashi et al., 2008). Moreover, we determined the interaction of ZjFAS2 and ZjSHV3 in vivo by the BiFC assay (Figure 8). Therefore, we believe that ZjFAS2 may interact with ZjSHV3 to regulate jujube fruit coloration.
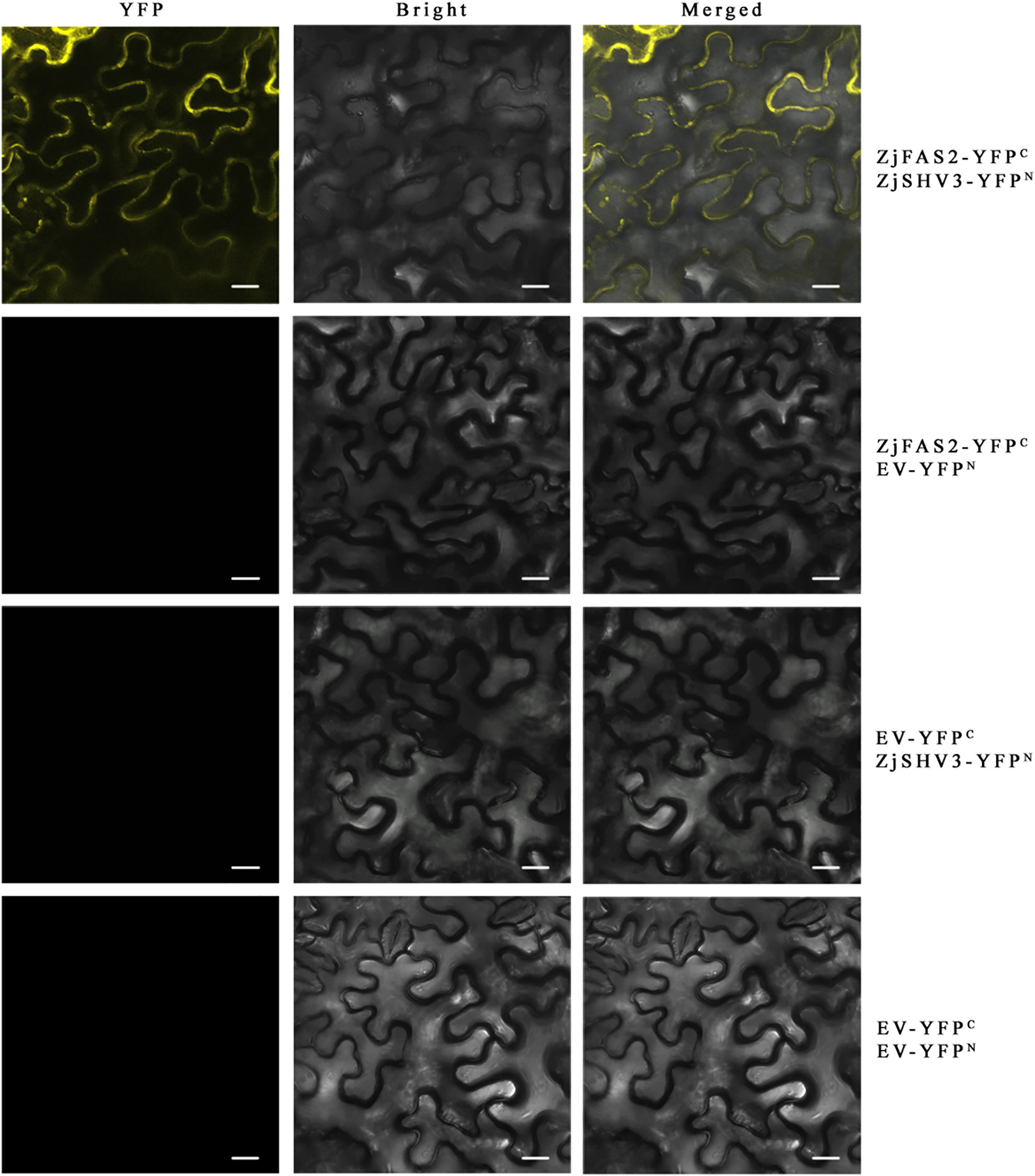
Figure 8 BiFC assay verifies the interaction between ZjFAS2 and ZjSHV3 in the tobacco leaves. Bars = 20 μm.
The fruit color of FMG was mainly determined by chlorophyll during the early stages of fruit development and was mainly determined by anthocyanins during the late stages of fruit development on the basis of the color phenotype and pigment (Figures 1C, D). Compared with FMG, “Junzao” exhibited a similar fruit color change process and chlorophyll change patterns but different anthocyanin change patterns (Shi et al., 2018; Shi et al., 2019; Figures 1B–D). Lutein is considered to determine the fruit color of “Junzao” during the full maturity stage (Shi et al., 2018). Our results indicated that the anthocyanin content differs significantly among the varieties during the semi-red period, which is consistent with the results of a previous study (Zhang et al., 2020; Figure 6C). Moreover, in contrast to the fruit color of FMG and “Junzao,” the fruit color of TLH is determined by anthocyanin content during both the early and late stages of fruit development and was determined by both chlorophyll and anthocyanin contents during the middle of fruit development (Shi et al., 2018; Shi et al., 2019; Figures 1C, D). Our results showed that the fruit color of different varieties is determined by different pigments in jujube, which is consistent with the results of previous studies.
Multiple anthocyanin structural genes were upregulated in TLH_S1, which resulted in significantly higher total anthocyanin content in TLH than in FMG (Supplementary Table 12). Furthermore, FMG and TLH exhibit different fruit coloring patterns. However, most of the transcriptional levels of anthocyanin biosynthetic genes showed the same expression trends with ripening (Figure 3A). In both FMG and TLH, most of the anthocyanin structural genes, namely C4H, CHS, CHI, F3H, F3’H, F3’5’H, DFR, ANS, and UFGT, showed higher transcriptional levels during the immature stage but were downregulated during the ripening period, and only one CHI gene and one UFGT gene were upregulated during the ripe stage, which is consistent with a previous study (Shi et al., 2020). A previous study also showed similar results wherein the anthocyanin structural genes were highly active during the white period (stage S5) but were gradually silenced over the ripening period (stage S7), and only three UFGT genes were gradually activated during the ripening period in “Dongzao” (Zhang et al., 2020). Therefore, the expression pattern of anthocyanin structural genes was not limited to the late fruit development period but was rather involved in the complete fruit development period among the different varieties. This indicates that the increased expression of early biosynthetic genes could promote anthocyanidin accumulation in early fruit development. Furthermore, these anthocyanidins are gradually modified to anthocyanins by UFGT during the fruit ripening process.
In Arabidopsis, FAS2 maintains the morphologies of stem, leaf, and flower and the organization of shoot and root apical meristems (Reinholz, 1966; Ottoline Leyser and Furner, 1992; Kaya et al., 2001). Furthermore, FASCIATA 1 (FAS1), FAS2, and MULTICOPY SUPPRESOR OF IRA1 (MSI1) form the chromatin assembly factor-1 (CAF-1) complex, which is required for DNA replication and nucleotide excision repair (Endo et al., 2006; Exner et al., 2006; Schönrock et al., 2006; Mozgova et al., 2010; Picart-Picolo et al., 2020). Through yeast two-hybrid screening, the interaction of FAS1 and FAS2 was verified in jujube (Supplementary Table 15). Notably, to the best of our knowledge, the role of FAS2 in the regulation of anthocyanin accumulation was reported for the first time. Surprisingly, unlike the reported WD40 proteins, we did not find that FAS2 forms a complex with MYB and bHLH in the nucleus to regulate anthocyanin accumulation. However, the results of subcellular localization and BiFC assays supported that ZjFAS2 may interact with ZjSHV3 to regulate jujube fruit coloration on the cell membrane (Figures 7, 8). Therefore, our results revealed a novel gene function of FAS2 and a new model of anthocyanin regulation.
Fruit color is modulated by environmental and biological factors (An et al., 2018; Henry-Kirk et al., 2018; Behzadi et al., 2021; Mahmoudian et al., 2021). Carbohydrate is an important factor affecting anthocyanin accumulation and sucrose has the most significant regulatory effect (Gu et al., 2019). Among different jujube varieties, sucrose, glucose, and fructose contents show different trends, but the total carbohydrate content gradually increases during fruit maturation (Guo et al., 2015; Song et al., 2019; Feng et al., 2020). In this study, 23 carbohydrates were identified from the peel extracts of FMG and TLH (Supplementary Table 7). Among them, five carbohydrates, namely Ribulose-5-phosphate, D(+)-Melezitose O-rhamnoside, Maltotetraose, Trehalose 6-phosphate, and D(+)-Melezitose, were positively correlated with the total anthocyanin content in FMG. Only one carbohydrate, D(−)-Threose, was positively correlated with the total anthocyanin content in TLH. Furthermore, sucrose exhibited high concentrations during the color change period in both FMG and TLH. The carbohydrates may play a crucial role in the jujube fruit coloration process.
The SHV3 is involved in cellulose biosynthesis, hypocotyl elongation, and anthocyanin accumulation (Hayashi et al., 2008; Yeats and Somerville, 2016a; Yeats et al., 2016b). The deficiency of SHV3 and its paralogs (SVL1)increases sucrose accumulation via the plasma membrane sucrose-proton symporter SUC1 (Yeats et al., 2016b). Conversely, sucrose acts as a signaling molecule that promotes SUC1 phosphorylation and protein activity (Niittyla et al., 2007; Yeats and Somerville, 2016a; Yeats et al., 2016b). Furthermore, SUC1 and an R2R3-MYB transcription factor, MYB75/PAP1, are required for sucrose-induced anthocyanin accumulation (Holton and Cornish, 1995; Teng et al., 2005; Sivitz et al., 2008; Lasin, 2019). In plants with suc1 mutant, anthocyanin structural genes that are downstream of MYB75 were downregulated, which resulted in decreased anthocyanin accumulation (Sivitz et al., 2008). To summarize, we propose a working model of the molecular mechanism of anthocyanin accumulation regulated by FAS2 (Figure 9). When the carbohydrate content reaches a certain threshold, the FAS2 increases the sucrose accumulation via the plasma membrane sucrose-proton symporter SUC1 by inhibiting SHV3 protein activity, resulting in sucrose-induced anthocyanin accumulation.
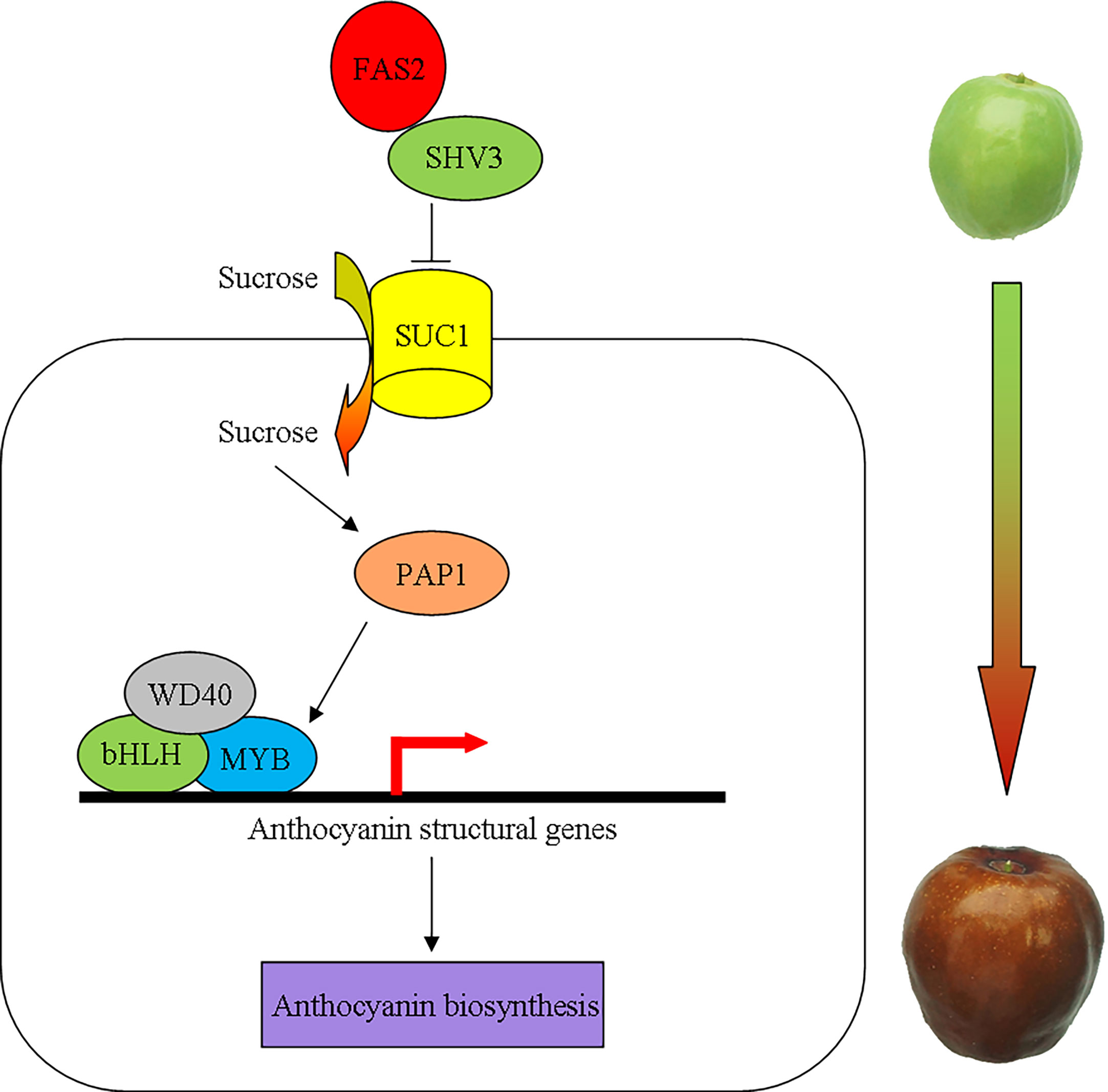
Figure 9 A schematic model for the possible molecular mechanism of anthocyanin accumulation regulated by FAS2. FAS2 enhances the sucrose accumulation via the plasma membrane sucrose-proton symporter SUC1 by inhibiting SHV3 protein activity. The higher sucrose concentration causes an increase in the expression of MYB75 genes. MYB75 interacts with bHLH and WD40 to form the MBW complex positively regulating the expression of anthocyanin structural genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SL designed and supervised the study, and conducted most of the experiments. YS performed fruit color measurement and pigment content analysis. SZ participated in anthocyanin identification and quantification. QZ participated in genetic transformation. LC and YW participated in expression pattern. SL and SZ initiated the project. SL analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by supported by National Natural Science Foundation of China (Grant No. 32101565), and Henan Provincial Science and Technology Research Project (Grant No. 212102110183).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1142757/full#supplementary-material
Ai, T. N., Naing, A. H., Kim, C. K. (2016). Influences of different light sources and light/dark cycles on anthocyanin accumulation and plant growth in petunia. J. Plant Biotechnol. 43, 119–124. doi: 10.5010/JPB.2016.43.1.119
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4, 17023. doi: 10.1038/hortres.2017.23
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., Hao, Y. J. (2018). Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 41, 2678–2692. doi: 10.1111/pce.13393
Bai, H. L., Wang, J., Liu, C. M., Li, L. (2010). Isolation and purification of flavonoids from Ziziphus jujuba by high-speed counter-current chromatography. J. Chin. Chem. Soc. 57, 1071–1076. doi: 10.1002/jccs.201000150
Behzadi, R. P., Roozban, M. R., Karimi, S., Ghahremani, R., Vahdati, K. (2021). Osmolyte accumulation and sodium compartmentation has a key role in salinity tolerance of pistachios rootstocks. Agriculture 11, 708. doi: 10.3390/agriculture11080708
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., Lamb, C. (2001). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 12, 2383–2393. doi: 10.2307/3871236
Carey, C. C., Strahle, J. T., Selinger, D. A., Chandler, V. L. (2004). Mutations in the pale aleurone color1 regulatory gene of the zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell. 16, 450–464. doi: 10.1105/tpc.018796
Chen, J., Chan, P. H., Lam, C. T., Li, Z., Lam, K. Y., Yao, P., et al. (2015). Fruit of Ziziphus jujuba (Jujube) at two stages of maturity: Distinction by metabolic profiling and biological assessment. J. Agric. Food Chem. 63, 739–744. doi: 10.1021/jf5041564
Chen, J., Liu, X., Li, Z., Qi, A., Yao, P., Zhou, Z., et al. (2017). A review of dietary Ziziphus jujuba fruit (Jujube): Developing health food supplements for brain protection. Evidence-Based Complementary Altern. Med. 2017, 1–10. doi: 10.1155/2017/3019568
Cheng, G., Bai, Y., Zhao, Y., Tao, J., Liu, Y., Tu, G., et al. (2000). Flavonoids from Ziziphus jujuba mill var. spinosa. Tetrahedron 56, 8915–8920. doi: 10.1016/S0040-4020(00)00842-5
Choi, S. H., Ahn, J. B., Kim, H. J., Im, N. K., Kozukue, N., Levin, C. E., et al. (2012). Changes in free amino acid, protein, and flavonoid content in jujube (Ziziphus jujube) fruit during eight stages of growth and antioxidative and cancer cell inhibitory effects by extracts. J. Agric. Food Chem. 60, 10245–10255. doi: 10.1021/jf302848u
Choi, S. H., Ahn, J. B., Kozukue, N., Levin, C. E., Friedman, M. (2011). Distribution of free amino acids, flavonoids, total phenolics, and antioxidative activities of jujube (Ziziphus jujuba) fruits and seeds harvested from plants grown in Korea. J. Agric. Food Chem. 59, 6594–6604. doi: 10.1021/jf200371r
Collado-González, J., Cruz, Z. N., Rodríguez, P., Galindo, A., Díaz-Baños, F. G., García de la Torre, J., et al. (2013). Effect of water deficit and domestic storage on the procyanidin profile, size, and aggregation process in pear-jujube (Z. jujuba) fruits. J. Agric. Food Chem. 61, 6187–6197. doi: 10.1021/jf4013532
de Vetten, N., Quattrocchio, F., Mol, J., Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11, 1422–1434. doi: 10.1101/gad.11.11.1422
Du, L. J., Gao, Q. H., Ji, X. L., Ma, Y. J., Xu, F. Y., Wang, M. (2013). Comparison of flavonoids, phenolic acids, and antioxidant activity of explosion-puffed and sun-dried jujubes (Ziziphus jujuba mill.). J. Agric. Food Chem. 61, 11840–11847. doi: 10.1021/jf401744c
Endo, M., Ishikawa, Y., Osakabe, K., Nakayama, S., Kaya, H., Araki, T., et al. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25, 5579–5590. doi: 10.1038/sj.emboj.7601434
Exner, V., Taranto, P., Schönrock, N., Gruissem, W., Hennig, L. (2006). Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133, 4163–4172. doi: 10.1242/dev.02599
Feng, Z., Gao, Z., Jiao, X., Shi, J., Wang, R. (2020). Widely targeted metabolomic analysis of active compounds at different maturity stages of ‘Hupingzao’jujube. J. Food Composition Anal. 88, 103417. doi: 10.1016/j.jfca.2020.103417
Gao, Q., Luo, H., Li, Y., Liu, Z., Kang, C. (2020). Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant Biotechnol. J. 18, 1550–1561. doi: 10.1111/pbi.13317
Gao, Q. H., Wu, C. S., Wang, M. (2013). The jujube (Ziziphus jujuba mill.) fruit: a review of current knowledge of fruit composition and health benefits. J. Agric. Food Chem. 61, 3351–3363. doi: 10.1021/jf4007032
Gao, Q. H., Wu, C. S., Yu, J. G., Wang, M., Ma, Y. J., Li, C. L. (2012). Textural characteristic, antioxidant activity, sugar, organic acid, and phenolic profiles of 10 promising jujube (Ziziphus jujuba mill.) selections. J. Food sci. 77, C1218–C1225. doi: 10.1111/j.1750-3841.2012.02946.x
Gonzalez, A., Mendenhall, J., Huo, Y., Lloyd, A. (2009). TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325, 412–421. doi: 10.1016/j.ydbio.2008.10.005
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Gu, K. D., Wang, C. K., Hu, D. G., Hao, Y. J. (2019). How do anthocyanins paint our horticultural products? Sci. hortic. 249, 257–262. doi: 10.1016/j.scienta.2019.01.034
Guo, S., Duan, J. A., Qian, D., Tang, Y., Wu, D., Su, S., et al. (2015). Content variations of triterpenic acid, nucleoside, nucleobase, and sugar in jujube (Ziziphus jujuba) fruit during ripening. Food Chem. 167, 468–474. doi: 10.1016/j.foodchem.2014.07.013
Guo, T., Lu, Z. Q., Shan, J. X., Ye, W. W., Dong, N. Q., Lin, H. X. (2020). ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell. 32, 2763–2779. doi: 10.1105/tpc.20.00351
Hawkins, C., Caruana, J., Schiksnis, E., Liu, Z. (2016). Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep. 6, 1–11. doi: 10.1038/srep29017
Hayashi, S., Ishii, T., Matsunaga, T., Tominaga, R., Kuromori, T., Wada, T., et al. (2008). The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 49, 1522–1535. doi: 10.1093/pcp/pcn120
Henry-Kirk, R. A., Plunkett, B., Hall, M., McGhie, T., Allan, A. C., Wargent, J. J., et al. (2018). Solar UV light regulates flavonoid metabolism in apple (Malus x domestica). Plant Cell environ. 41, 675–688. doi: 10.1111/pce.13125
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Holton, T. A., Cornish, E. C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 7, 1071. doi: 10.1105/tpc.7.7.1071
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Wallroth, M., Eichholtz, D., Rogers, S. G., et al. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Huang, D., Wang, X., Tang, Z., Yuan, Y., Xu, Y., He, J., et al. (2018). Subfunctionalization of the Ruby2–Ruby1 gene cluster during the domestication of citrus. Nat. plants. 4, 930–941. doi: 10.1038/s41477-018-0287-6
Kaya, H., Shibahara, K. I., Taoka, K. I., Iwabuchi, M., Stillman, B., Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 104, 131–142. doi: 10.1016/S0092-8674(01)00197-0
Kou, X., Chen, Q., Li, X., Li, M., Kan, C., Chen, B., et al. (2015). Quantitative assessment of bioactive compounds and the antioxidant activity of 15 jujube cultivars. Food Chem. 173, 1037–1044. doi: 10.1016/j.foodchem.2014.10.110
Kou, X., He, Y., Li, Y., Chen, X., Feng, Y., Xue, Z. (2019). Effect of abscisic acid (ABA) and chitosan/nano-silica/sodium alginate composite film on the color development and quality of postharvest Chinese winter jujube (Zizyphus jujuba mill. cv. dongzao). Food Chem. 270, 385–394. doi: 10.1016/j.foodchem.2018.06.151
Lasin, P. (2019). AtSUC1 root expression and sucrose response leading to anthocyanin accumulation (Doctoral dissertation, university of Minnesota).
Li, S., Deng, B., Tian, S., Guo, M., Liu, H., Zhao, X. (2021). Metabolic and transcriptomic analyses reveal different metabolite biosynthesis profiles between leaf buds and mature leaves in Ziziphus jujuba mill. Food Chem. 347, 129005. doi: 10.1016/j.foodchem.2021.129005
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. doi: 10.1104/pp.112.199703
Li, D. K., Niu, X. W., Tian, J. B. (2013). The illustrated germplasm resources of Chinese (Beijing: China Agriculture Press).
Li, C., Wu, J., Hu, K. D., Wei, S. W., Sun, H. Y., Hu, L. Y., et al. (2020). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 7, 37. doi: 10.1038/s41438-020-0254-z
Lichtenthaler, H. K., Wellburn, A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 11, 591–592. doi: 10.1042/bst0110591
Liu, M. J., Wang, M. (2009). Chinese Jujube germsplasm resources (Beijing: China Forestry Publishing House Press).
Liu, M. J., Zhao, J., Cai, Q. L., Liu, G. C., Wang, J. R., Zhao, Z. H., et al. (2014). The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 5, 5315. doi: 10.1038/ncomms6315
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, H., Dai, C., Li, Y., Feng, J., Liu, Z., Kang, C. (2018). Reduced anthocyanins in petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. botany. 69, 2595–2608. doi: 10.1093/jxb/ery096
Mahmoudian, M., Rahemi, M., Karimi, S., Yazdani, N., Tajdini, Z., Sarikhani, S., et al. (2021). Role of kaolin on drought tolerance and nut quality of Persian walnut. J. Saudi Soc. Agric. Sci. 20, 409–416. doi: 10.1016/j.jssas.2021.05.002
Matus, J. T., Poupin, M. J., Cañón, P., Bordeu, E., Alcalde, J. A., Arce-Johnson, P. (2010). Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera l.). Plant Mol. Biol. 72, 607–620. doi: 10.1007/s11103-010-9597-4
Miller, R., Owens, S. J., Rørslett, B. (2011). Plants and colour: flowers and pollination. Optics Laser Technol. 43, 282–294. doi: 10.1016/j.optlastec.2008.12.018
Mozgova, I., Mokroš, P., Fajkus, J. (2010). Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell. 22, 2768–2780. doi: 10.1105/tpc.110.076182
Narjesi, V., Fatahi, Moghadam, J., Ghasemi-Soloklui, A. A. (2023). Effects of photo-selective shade net color and shading percentage on reducing sunburn and increasing the quantity and quality of pomegranate fruit. Int. J. Hortic. Sci. Technol. 10 (2), 25–38. doi: 10.22059/ijhst.2022.343648.567
Niittyla, T., Fuglsang, A. T., Palmgren, M. G., Frommer, W. B., Schulze, W. X. (2007). Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics. 6, 1711–1726. doi: 10.1074/mcp.M700164-MCP200
Ottoline Leyser, H. M., Furner, I. J. (1992). Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116, 397–403. doi: 10.1242/dev.116.2.397
Pawlowska, A. M., Camangi, F., Bader, A., Braca, A. (2009). Flavonoids of Zizyphus jujuba l. and Zizyphus spina-christi (L.) willd (Rhamnaceae) fruits. Food Chem. 112, 858–862. doi: 10.1016/j.foodchem.2008.06.053
Picart-Picolo, A., Grob, S., Picault, N., Franek, M., Llauro, C., Halter, T., et al. (2020). Large Tandem duplications affect gene expression, 3D organization, and plant-pathogen response. Genome Res. 30, 1583–1592. doi: 10.1101/gr.261586.120
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 23, 1795–1814. doi: 10.1105/tpc.111.083261
Qu, Z. Z., Wang, Y. H. (1993). China Fruit’s monograph-Chinese jujube volume (Beijing: China Forestry Publishing House Press).
Reinholz, E. (1966). Radiation induced mutants showing changed inflorescence characteristics. Arab. Inf. Serv. 3, 19–20.
Schönrock, N., Exner, V., Probst, A., Gruissem, W., Hennig, L. (2006). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281, 9560–9568. doi: 10.1074/jbc.M513426200
Shi, Q., Du, J., Zhu, D., Li, X., Li, X. (2020). Metabolomic and transcriptomic analyses of anthocyanin biosynthesis mechanisms in the color mutant Ziziphus jujuba cv. tailihong. J. Agric. Food Chem. 68, 15186–15198. doi: 10.1021/acs.jafc.0c05334
Shi, Q., Li, X., Du, J., Li, X. (2019). Anthocyanin synthesis and the expression patterns of bHLH transcription factor family during development of the Chinese jujube fruit (Ziziphus jujuba mill.). Forests 10, 346. doi: 10.3390/f10040346
Shi, Q., Zhang, Z., Su, J., Zhou, J., Li, X. (2018). Comparative analysis of pigments, phenolics, and antioxidant activity of Chinese jujube (Ziziphus jujuba mill.) during fruit development. Molecules 23, 1917. doi: 10.3390/molecules23081917
Sivitz, A. B., Reinders, A., Ward, J. M. (2008). Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 147, 92–100. doi: 10.1104/pp.108.118992
Song, J., Bi, J., Chen, Q., Wu, X., Lyu, Y., Meng, X. (2019). Assessment of sugar content, fatty acids, free amino acids, and volatile profiles in jujube fruits at different ripening stages. Food Chem. 270, 344–352. doi: 10.1016/j.foodchem.2018.07.102
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. doi: 10.1104/pp.105.066688
Tohge, T., de Souza, L. P., Fernie, A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. botany. 68, 4013–4028. doi: 10.1093/jxb/erx177
Wang, N., Qu, C., Jiang, S., Chen, Z., Xu, H., Fang, H., et al. (2018). The proanthocyanidin-specific transcription factor md MYBPA 1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 96, 39–55. doi: 10.1111/tpj.14013
Wen, C., Zhang, Z., Shi, Q., Yue, R., Li, X. (2022). Metabolite and gene expression analysis underlying temporal and spatial accumulation of pentacyclic triterpenoids in jujube. Genes 13, 823. doi: 10.3390/genes13050823
Wojdyło, A., Carbonell-Barrachina, Á.A., Legua, P., Hernández, F. (2016). Phenolic composition, ascorbic acid content, and antioxidant capacity of Spanish jujube (Ziziphus jujube mill.) fruits. Food Chem. 201, 307–314. doi: 10.1016/j.foodchem.2016.01.090
Xu, W., Dubos, C., Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xue, X., Zhao, A., Wang, Y., Ren, H., Li, Y., Li, D., et al. (2022). Metabolomics-based analysis of flavonoid metabolites in Chinese jujube and sour jujube fruits from different harvest periods. J. Food sci. 87, 3752–3765. doi: 10.1111/1750-3841.16290
Yamazaki, M., Saito, K. (2011). Molecular genetic study on the anthocyanin chemotypes of Perilla frutescens var. crispa. Natural Product Commun. 6, 423–427. doi: 10.1002/ejlt.201000308
Yeats, T. H., Somerville, C. R. (2016a). A dual mechanism of cellulose deficiency in shv3svl1. Plant Signaling behavior. 11, e1218108. doi: 10.1080/15592324.2016.1218108
Yeats, T. H., Sorek, H., Wemmer, D. E., Somerville, C. R. (2016b). Cellulose deficiency is enhanced on hyper accumulation of sucrose by a h+-coupled sucrose symporter. Plant Physiol. 171, 110–124. doi: 10.1104/pp.16.00302
Zhang, Y., Butelli, E., Martin, C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. doi: 10.1016/j.foodchem.2019.125903
Zhang, H., Jiang, L., Ye, S., Ye, Y., Ren, F. (2010). Systematic evaluation of antioxidant capacities of the ethanolic extract of different tissues of jujube (Ziziphus jujuba mill.) from China. Food Chem. toxicol. 48, 1461–1465. doi: 10.1016/j.fct.2010.03.011
Zhang, Q., Wang, L., Liu, Z., Zhao, Z., Zhao, J., Wang, Z., et al. (2020). Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba mill. peel coloration. Food Chem. 312, 125903. doi: 10.1016/j.foodchem.2019.125903
Zhou, L. L., Shi, M. Z., Xie, D. Y. (2012). Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta. 236, 825–837. doi: 10.1007/s00425-012-1674-2
Zong, Y., Zhu, X., Liu, Z., Xi, X., Li, G., Cao, D., et al. (2019). Functional MYB transcription factor encoding gene AN2 is associated with anthocyanin biosynthesis in Lycium ruthenicum murray. BMC Plant Biol. 19, 1–9. doi: 10.1186/s12870-019-1752-8
Keywords: jujube (Ziziphus jujuba Mill.), fruit color, anthocyanin, WD40 repeat protein, ZjFAS2
Citation: Li S, Shen Y, Zheng S, Zhu Q, Cai L, Wang Y and Zhao X (2023) ZjFAS2 is involved in the fruit coloration in Ziziphus jujuba Mill. by regulating anthocyanin accumulation. Front. Plant Sci. 14:1142757. doi: 10.3389/fpls.2023.1142757
Received: 12 January 2023; Accepted: 27 February 2023;
Published: 10 March 2023.
Edited by:
Matthew Clark, University of Minnesota Twin Cities, United StatesReviewed by:
Kourosh Vahdati, University of Tehran, IranCopyright © 2023 Li, Shen, Zheng, Zhu, Cai, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shipeng Li, U2hpcGVuZ2xpQGx5bnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.