- 1Biotechnology Division, CSIR-Institute of Himalayan Bioresource Technology, Palampur, India
- 2Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India
- 3Incubatee at Chief Minister Startup Scheme, Shimla, India
There is a renewed interest in sustainable agriculture wherein novel plant growth-promoting rhizobacteria (PGPR) are being explored for developing efficient biostimulants. The key requirement of a microbe to qualify as a good candidate for developing a biostimulant is its intrinsic plant growth-promoting (PGP) characteristics. Though numerous studies have been conducted to assess the beneficial effects of PGPRs on plant growth under normal and stressed conditions but not much information is available on the characterization of intrinsic traits of PGPR under stress. Here, we focused on understanding how temperature stress impacts the functionality of key stress tolerant and PGP genes of Bacillus sp. IHBT-705 isolated from the rhizosphere of saffron (Crocus sativus). To undertake the study, Bacillus sp. IHBT-705 was grown under varied temperature regimes, their PGP traits were assessed from very low to very high-temperature range and the expression trend of targeted stress tolerant and PGP genes were analyzed. The results illustrated that Bacillus sp. IHBT-705 is a stress-tolerant PGPR as it survived and multiplied in temperatures ranging from 4°C-50°C, tolerated a wide pH range (5-11), withstood high salinity (8%) and osmolarity (10% PEG). The PGP traits varied under different temperature regimes indicating that temperature influences the functionality of PGP genes. This was further ascertained through whole genome sequencing followed by gene expression analyses wherein certain genes like cspB, cspD, hslO, grpE, rimM, trpA, trpC, trpE, fhuC, fhuD, acrB5 were found to be temperature sensitive while, cold tolerant (nhaX and cspC), heat tolerant (htpX) phosphate solubilization (pstB1), siderophore production (fhuB and fhuG), and root colonization (xerC1 and xerC2) were found to be highly versatile as they could express well both under low and high temperatures. Further, the biostimulant potential was checked through a pot study on rice (Oryza sativa), wherein the application of Bacillus sp. IHBT-705 improved the length of shoots, roots, and number of roots over control. Based on the genetic makeup, stress tolerance potential, retention of PGP traits under stress, and growth-promoting potential, Bacillus sp. IHBT-705 could be considered a good candidate for developing biostimulants.
Introduction
PGPRs are now widely recognized for promoting sustainable agriculture (Gouda et al., 2018) for their ability to enhance plant productivity in multiple ways (Raza et al., 2016). PGPRs enable higher nutrient acquisition through the solubilization of phosphate (Ahemad and Khan, 2012), the production of siderophore for better uptake of iron (Beneduzi et al., 2012), the synthesis of plant growth regulators like indole acetic acid, the release of volatile organic compounds, the production of antifungal enzymes like chitinase, glucanase, and ACC-deaminase (1-Aminocyclopropane-1-carboxylate) and inducing resistance against pathogens (Choudhary et al., 2011; García-Fraile et al., 2015). Numerous studies have illustrated the beneficial application of PGPR under stress (Sapre et al., 2018; Ali and Khan, 2021). PGPRs improved yield, and tolerance against salinity stress in wheat (Lastochkina et al., 2017; Ansari et al., 2019), enhanced tolerance to drought in chickpeas (Vurukonda et al., 2016; Khan et al., 2019), and enhanced the growth of the tomato under cold stress (Subramanian et al., 2016). Under heat stress, thermotolerant PGPRs stimulated heat-responsive genes (GmHSP, GmLAX3, and GmAKT2) in soybean (Khan et al., 2020). Despite the multitude of positive traits, the commercial application of PGPR as a biostimulant experiences certain limitations, the most common being the variability of performance under field conditions (Timmusk et al., 2017; Glick, 2018). This is primarily due to the mode of selection process wherein PGP traits of microbes are assessed at a laboratory scale under highly controlled conditions, while field conditions are markedly different (Vasseur-Coronado et al., 2021). In fields, multiple factors interact, hence the PGP potentials of microbes are often lost or masked leading to underperformance (Backer et al., 2018; Mellidou and Karamanoli, 2022). Hence, the selection of the right microbe which does not lose its PGP traits under field conditions and various stresses is crucial (Amaya-Gómez et al., 2020). Unraveling the genetic makeup of PGPR can help in understanding its stress tolerance potential and predict its functionality as a biostimulant. The expression trend of genes will have a direct effect on their PGP potential and field performance (Bruto et al., 2014). Hence in this study, we analyzed the PGP traits of Bacillus sp. IHBT-705 under stressed conditions and carried out gene expression studies of key PGP genes over a wide range of temperatures to understand how temperature impacts their functionality.
Bacillus sp. IHBT-705 was isolated from the rhizosphere of saffron (Crocus sativus) cultivated in Patti village, Kangra District, Himachal Pradesh India since 2018. Among several PGPRs isolated from the region, Bacillus sp. IHBT-705 was found to highly stress tolerant and interestingly its key PGP traits like phosphate and potassium solubilizing potential, siderophore IAA and ACC deaminase, were evident over varied temperature regimes. Hence, the present study focused on assessing the stress tolerance capacity and analyzing the PGP trait of the bacteria, Bacillus sp. IHBT-705. We also wanted to understand the underlining functioning of key PGP genes in response to different temperature regimes i.e., very low (4°C) to very high (50°C). To elucidate the functional characteristics of PGP and stress-related genes, whole genome sequencing of Bacillus sp. IHBT-705 was carried out using a long-read sequencing system, PacBio RS II (Pacific Bioscience, USA). Genome sequencing followed by data mining lead to the identification of several key genes associated with stress tolerance and PGP traits. Further, the expression analyses of key PGP genes were studied in detail under varied temperature regimes. This study provided new insight into understanding the functionality of PGP-related genes of Bacillus sp. IHBT- 705 over a wide range of temperatures. For the first time, the study illustrated that certain PGP genes are highly sensitive to temperature while some genes are versatile and can function well over wide temperature regimes.
Materials and methods
Sampling and physiochemical properties of soil
Soil samples were collected from the rhizosphere of saffron (Crocus sativus) grown at Patti village, Palampur (HP), India (32.0708° North & 76.5415° East) (Supplementary Figure S1) from a depth of 8- 15cm. Five independent soil samples were collected from five different places in the field, wherein each sample consisted of four subsamples pooled together from the four sides of the saffron (Crocus sativus) rhizosphere from the same site. Physiochemical properties of soil i.e pH, electrical conductivity (EC), Distribution of Nitrogen (N), Phosphorus (P), Organic carbon (OC), Potassium (K), Iron (Fe), Manganese (Mn), Zinc (Zn), Calcium (Ca) and Copper (Cu) were analyzed. The pH of the soil and electrical conductivity were measured by immersing the electrodes into the soil: water-1:2.5 (w:v) extraction by using a calibrated pH meter (Thermo Scientific Eutech PC450, USA) and conductivity meter (Thermo Scientific Eutech Cyber Scan CON45, USA). Total N in the soil was determined by the micro-Kjeldahl method after digestion in concentrated sulfuric acid. The total P was determined by Bray’s method (Bray and Kurtz, 1945), whereas the OC was estimated as per the standard dichromate oxidation method (Nelson and Sommers, 1983). Available K, Fe, Mn, Zn, Ca, and Cu was determined by using a flame photometer (model BWB XP, BWB Technologies UK Ltd., UK) after Mehlich -3 extraction (Mehlich, 1984).
Isolation of PGP bacteria
Soil samples were serially diluted (10-8), spread on Tryptone Soya Agar (TSA) media plates (Himedia, India) incubated in an incubator shaker (Innova 12444, New Brunswick Scientific, USA) at 28 ± 0.1°C for 48 hrs for the isolation of PGP bacteria. The morphology of isolated colonies was observed for shape, texture, color, margin, opacity, and elevation. Gram’s reaction was performed as per the method optimized by Vincent (Vincent, 1970). Pure colonies were preserved in glycerol stock (−80°C) till further use.
Stress tolerance potential of Bacillus sp. IHBT-705
To check the temperature tolerance, Bacillus sp. IHBT-705 was grown in Tryptone Soya Broth (TSB) media (Himedia, India) at 180 rpm for 48 hrs in an incubator shaker at ambient temperature (28°C). After 48 hrs, the optical density of bacterial suspension was observed at 600nm and found to be 1 (OD-1) and 100 µl of this bacterial suspension was inoculated in 100 ml of TSB media and incubated at targeted temperatures i.e very low (4°C and15°C), low (20°C), optimum (28°C), high (42°C) and very high (50°C) in an incubator shaker. For pH tolerance, pH (4, 5, 6, 7, 8, 9, 10, 11, and 12) was adjusted in TSB media by 1N NaOH and 1M HCl. Thereafter 100 µl of 48 hrs old Bacillus sp. IHBT-705 suspension (OD-1) was inoculated in 100 ml of selected pH media and incubated at targeted temperatures in an incubator shaker (Getahun et al., 2020). For salinity tolerance, concentrations of sodium chloride (0%, 2%, 4%, 6%, 8%, 10%, 12%, and 15%) were varied in TSB media and 100 µl of 48 hrs old Bacillus sp. IHBT-705 suspension (OD-1) was inoculated in 100 ml of selected NaCl TSB media and incubated at targeted temperatures in an incubator shaker (Ramadoss et al., 2013). For osmotic tolerance, concentrations of polyethylene glycol (PEG-10,000MW) (0%, 5%, 10%, 15%, 20%, 25%, and 30%) were varied in TSB media. Thereafter, 100 µl of 48 hrs old Bacillus sp. IHBT-705 suspension (OD-1) was inoculated in 100 ml of selected PEG TSB media and incubated at targeted temperatures in an incubator shaker (Niu et al., 2018). The optical density of bacterial growth was measured with a microplate reader (Synergy™ LX Multi-Mode BioTek) and incubated in an incubator shaker (Innova 12444, New Brunswick Scientific, USA) at 180 for 48 hrs for all the abiotic stresses mentioned above at targeted all temperatures.
PGP potential of Bacillus sp. IHBT-705
The initial screening and qualitative estimation of inorganic phosphate (P) solubilization of Bacillus sp. IHBT-705 was done on Pikovskaya’s (PVK) agar plates (Nautiyal, 1999). Qualitative estimation of potassium (K) solubilization assay was done on Aleksandrow agar media (Dhaked et al., 2017). The siderophore production was checked by Chrome Azurol S assay (CAS) (Schwyn and Neilands, 1987). The IAA production ability was evaluated by a colorimetric detection test (Loper, 1986). ACC deaminase assay was done according to the method proposed by Dworkin and Foster (Dworkin and Foster, 1958). All PGPR attributes were done at all targeted temperatures. The qualitative estimation of P, K solubilization and siderophore production was calculated by solubilization index (SI) =colony diameter + halo zone diameter/colony diameter (Paul and Sinha, 2013).
Molecular identification based on 16S rRNA gene sequencing
For molecular identification, the genomic DNA of Bacillus sp. IHBT-705 was extracted by using the GenElute bacterial genomic DNA isolation kit (Sigma-Aldrich, USA) and the quality and quantity of genomic DNA were determined by gel electrophoresis (Bio-Rad, USA) and NanoDrop 2000 (Thermo Scientific, USA) and Qubit version 2.0 fluorometer (Invitrogen, USA) respectively. The 16S rRNA amplification was performed by polymerase chain reaction (PCR) using a genomic DNA template and universal primers 27F (5’AGA-GTT-TGA-TCC-TGG-CTC-AG3’) and 1492R (5’TAC-GGT-TAC-CTT-GTT-ACG-ACT3’) were used to a near-full length, approximately 1500 bp PCR product (Weisburg et al., 1991). For PCR amplification, 25µl standard reaction mixture containing: GoTaq Green Master Mix 1x, primer (forward and reverse primer,1µM each), and DNA template was prepared and cycled at 94°C for 2min for initial denaturation, followed by 35 cycles consisting of denaturation at 94°C for 30sec, annealing at 55°C for 30sec, and elongation at 72°C for 2min; a final 7min elongation step at 72°C. The amplified product was visualized on a 1% agarose gel under UV light for a positive amplification signal, and the PCR product was purified Exosap-IT solution (Applied Biosystem, USA) according to the manufacturer’s instructions. DNA ladder (1Kb, Qiagen, Netherlands) was used for size identification. Big Dye Terminator cycle sequencing kit v3.1 (Applied Biosystems, USA) was used according to the manufacturer’s instructions and its PCR product was purified by Montage sequencing reaction kit (Millipore USA). The purified PCR product was injected into a Genetic analyzer (Applied Biosystems, USA) for Sanger sequencing technology. Sequences obtained were submitted as a query to the BLASTn (https://www.ncbi.nlm.nih.gov/) search algorithm. Strain identification was done at the species level using EzTaxon (https://www.ezbiocloud.net/) and sequences of homologous strains were aligned using CLUSTALW (Larkin et al., 2007), and a phylogenetic tree was constructed with the MEGA version 11 (Tamura et al., 2021) (https://www.megasoftware.net/) using the neighbor-joining method with 1,000 bootstrap replicates (Saitou and Nei, 1987).
Pot experiment
Inoculation of Bacillus sp. IHBT-705 on rice (Oryza sativa)
To carry out the germination assay, seeds of Rice (HPR 2880 B/S) procured from CSKHPKV university Palampur Himachal Pradesh India, were surface sterilized (Nezarat and Gholami, 2009) and the experiment was set up in five replicates with two treatments i.e., treated (Bacillus sp. IHBT-705) and control (without Bacillus sp. IHBT-705). For the treated set, Bacillus sp. IHBT-705 was grown in TSB (Himedia, India) and incubated in an incubator shaker (Innova 12444, New Brunswick Scientific, USA) at 180 rpm for 24 hrs at 28°C, thereafter, centrifuged at 8000 rpm for 10 min. The centrifuged pellet was resuspended in autoclaved water for making OD-1 and used for further experiments. The effect of Bacillus sp. IHBT-705 on the growth of rice was studied by soaking rice seeds in the Bacillus sp. IHBT-705 suspension (OD-1) for 30 min. The seeds of control set were soaked in sterile distilled water. Each pot was filled with 2 kg autoclaved soil and fifty soaked seeds were planted, thereafter10 ml of Bacillus sp. IHBT-705 suspension was added in treated set. Seeds treated with sterile distilled water without bacterial suspension were taken as control. The treatments were arranged in a completely randomized design (CRD) with five replicates and placed in a glass house. After 60 days of sowing, ten plants were randomly uprooted and washed under running water, and root/shoot lengths and numbers of roots were measured.
Complete genome sequencing and annotation of Bacillus sp. IHBT-705
Complete genome sequencing was carried out to understand the genomic complexity of Bacillus sp. IHBT-705. The genomic DNA of Bacillus sp. IHBT-705 was isolated as mentioned above and ~10µg of high-quality intact genomic DNA was used for long reads library preparation using a PacBio SMRTbell template preparation kit with 20Kb insert size (Pacific Biosciences, USA) version 1.0 as per manufacturer’s instructions. Briefly, intact gDNA was fragmented using g-tubes (Covaris, Inc. USA) and the quality of the sheared DNA was checked using gel electrophoresis (Supplementary Figure S2A). Followed by end-repairing, adaptor ligation, and purification of library molecules using Bluepippin size-selection system (Sage Science, USA) called SMRTbell template (Supplementary Figure S2B). The QC of the library was checked with Bioanalyzer DNA 12000 chip (Agilent Technologies, USA) and Qubit Fluorometer v3.0 (Invitrogen, USA) as shown in (Supplementary Figure S2C). Further, genome sequencing of Bacillus sp. IHBT-705 was performed on the PacBio RS II system with polymerase P6-C4 sequencing chemistry. The annotation was performed using Rapid Annotation Using Subsystems Technology (RAST) (Aziz et al., 2008). Denovo assembly of sequenced sub-reads was carried out using HGAP (Hierarchical Genome Assembly Process) (Supplementary Figure S3) protocol version 3.0 in SMRT analysis version 2.2.0 (Pacific Biosciences, USA) and produced a complete circular genome sequence with high coverage.
Whole-genome alignment with closest strain
For comparative genome analysis reference genome sequence of Bacillus altitudinis ASM97268 (Top hit in 16S rRNA Sequencing) was downloaded from the NCBI (Accession number NZ_CP011150.1), and imported into the CLCBio genomics workbench (Qiagen, Netherlands). The complete genome sequence of Bacillus sp. IHBT-705 (CP074101) was used against Bacillus altitudinis (ASM97268) for whole-genome alignment. The alignment was performed using CLC Genomics Workbench (v21.0.3) whole-genome alignment tool with default parameters (minimum initial seed length 15bp and minimum alignment block length (100bp). This tool primarily works by identifying seeds i.e., short stretches of nucleotide sequence that are shared between multiple genomes but not present multiple times in the same genome. These seeds are extended using the HOXD scoring matrix (Chiaromonte et al., 2002) until the local alignment score drops below a fixed threshold.
Genome-based average nucleotide identification with closest strain
The genomic similarity between the Bacillus sp. IHBT-705 and the closest strain B. altitudinis ASM97268 from the NCBI database were calculated using the Orthologous Average Nucleotide Identity algorithm (OrthoANIu) (https://www.ezbiocloud.net).
Expression analysis of stress-tolerant and PGP genes
Genes responsible for stress (cold and heat), pH tolerance, and PGP attributes such as Phosphate solubilization, IAA production, siderophore production, ACC deaminase activity, and root colonization were selected based on the available literature (Table 1) and screened from the whole genome annotation of selected PGP and stress-tolerant bacteria by BLAST to GO (Conesa et al., 2005) for expression studies. Primers for stress-tolerant and PGP genes were designed by using the Primer Express software version 3 (Applied Biosystem USA) as shown in (Supplementary Table S1).
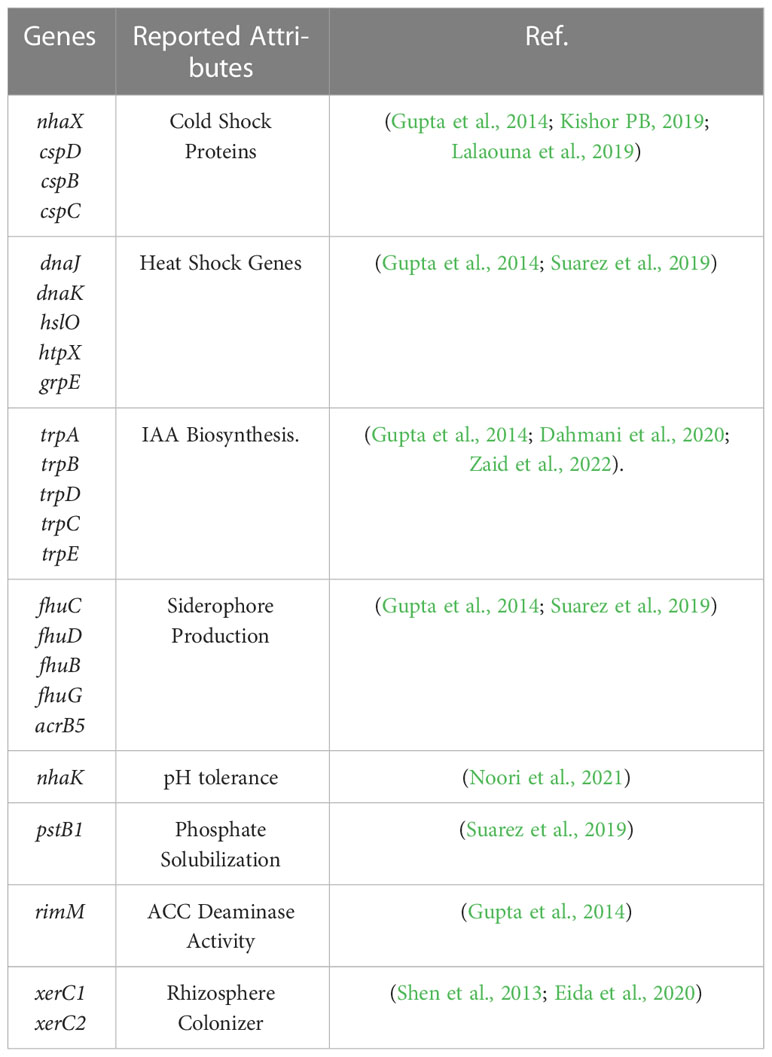
Table 1 List of stress-tolerant and PGP genes selected from the whole genome of Bacillus sp. IHBT-705 for expression studies.
RNA extraction, cDNA synthesis, and qRT-PCR analyses
Bacillus sp. IHBT-705 was grown in TSB media (Himedia India) at all targeted temperatures at 180 rpm for 24 hrs in an incubator shaker and after 24 hrs total RNA was isolated separately from Bacillus sp. IHBT-705 suspension (OD-1) by using RNeasy Mini Kit (Qiagen, Netherlands). The quality of RNA was checked by using RNA nanochip on a bioanalyzer (Agilent Technologies, USA). Further, the RNA quality and quantity were verified by agarose gel electrophoresis (Bio-Rad, USA) followed by ND-1000 NanoDrop (Thermo Scientific, USA) respectively. Total RNA was reverse transcribed into cDNA using a verso cDNA synthesis kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. Briefly, the reaction was prepared in 20µl volume with random hexamer primer conducted at 42˚C for 60min and 95˚C for 2min for heat denaturation of enzymes. Prepared cDNA was confirmed by PCR amplification using 16S primer pair followed by agarose gel electrophoresis (Bio-Rad, USA) (Cárdenas Espinosa et al., 2021). Real-time qRT-PCR was performed with Syber Green PCR Master Mix (Thermo Fisher Scientific, USA) and conducted on Step One Plus 2.0 Real-Time PCR System (Applied Biosystems, USA). The reaction mixture included 2μl of diluted cDNA (1:10), 0.5μl each primer, and SYBR green 5μl. The final reaction volume of 10μl was adjusted with nuclease-free water. The following PCR condition was used: 95˚C for 7min, 40 cycles of 95˚C for 15sec, and 60˚C for 30sec. The melting temperature-determining dissociation step was run at 95˚C for 15sec and 60˚C for 1min and 95˚C for 15sec at the end of the amplification. All the reactions were carried out in triplicate for each cDNA sample (Jian et al., 2020).
Statistical analysis
Relative gene expression calculations of stress-tolerant and PGP genes were calculated according to the comparative critical threshold (2ΔΔCT) method (Livak and Schmittgen, 2001). The expression of two housekeeping genes (16S, secA) was analyzed for stress-tolerant and PGP gene primers. At all targeted temperatures, the expression of the 16S gene was highly stable, and similar results were obtained by using it as a normalization gene at 28˚C (optimum temperature for gene expression) and comparison of Ct-values further revealed that expression of 16S reference gene was stably expressed with constant Ct values (Supplementary Figure S4A, B). Reference gene secA was not stably expressed in the targeted temperatures. Hence, only the 16S reference gene was considered as the internal reference. All experiments were performed in triplicates for gene expression and in five replicates for pot experiments. All data (Significance in fold-change and Pot experiment) was analyzed by using one-way ANOVA followed by Duncan multiple range test (DMRT) (p-value = 0.05) (https://ccari.icar.gov.in/wasp/index.php).
Results
Physicochemical properties of soil
We wanted to understand the nature of soil being the habitat of Bacillus sp. IHBT-705, hence the soil characteristics (pH, EC, available N2, P, OC, K, Fe Mn, Zn, Ca, and Cu) were analyzed (Table 2).
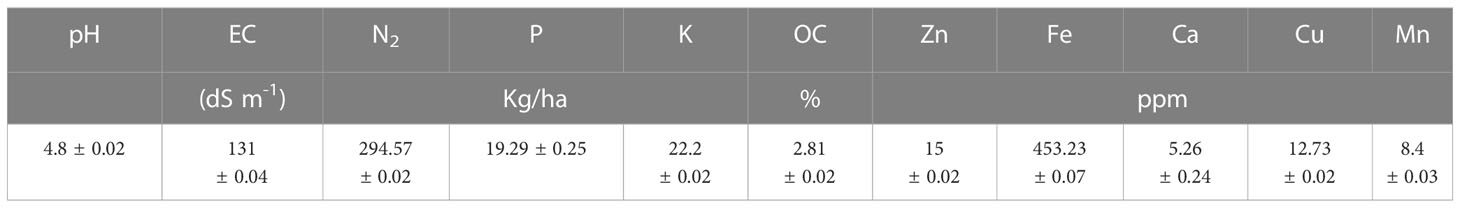
Table 2 Physiochemical properties of rhizosphere soil. Values represented are mean ± standard error of three replicates.
Morphological characterization of Bacillus sp. IHBT-705
Colonies of Bacillus sp. IHBT-705 were off-white, translucent, elevated, and round with a regular margin. Gram staining illustrated it to be rod-shaped gram-positive bacteria.
Stress tolerance potential of Bacillus sp. IHBT-705
Bacillus sp. IHBT-705 exhibited growth on temperatures ranging from 4°C- 50°C on TSA as well as in TSB medium. However, 28°C was found to be the optimum temperature for growth and multiplication (Figure 1A). We wanted to understand the nature of Bacillus sp. IHBT-705 particularly under a stressed environment, so we subjected it to different stresses like temperature, pH, salinity, and osmoticum. Bacillus sp. IHBT-705 exhibited thermotolerance on temperatures ranging from 4°C- 50°C (Figure 1A) and could multiply at pH ranging from 5 to 11 at all the targeted temperatures indicating that it can tolerate and propagate both in acidic as well as the alkaline environment (Figure 1B). The salinity (NaCl) tolerance level of Bacillus sp. IHBT-705 was up to 6% at all the targeted temperatures and increased up to 8% at 28°C and 42°C (Figure 1C). Bacillus sp. IHBT-705 exhibited relatively slow growth in medium supplemented with PEG-10,000MW (10% onwards) from 15°C-42°C and almost no growth at 50°C (Figure 1D). All these stress assessment results indicate Bacillus sp. IHBT-705 to be a stress-tolerant PGP.
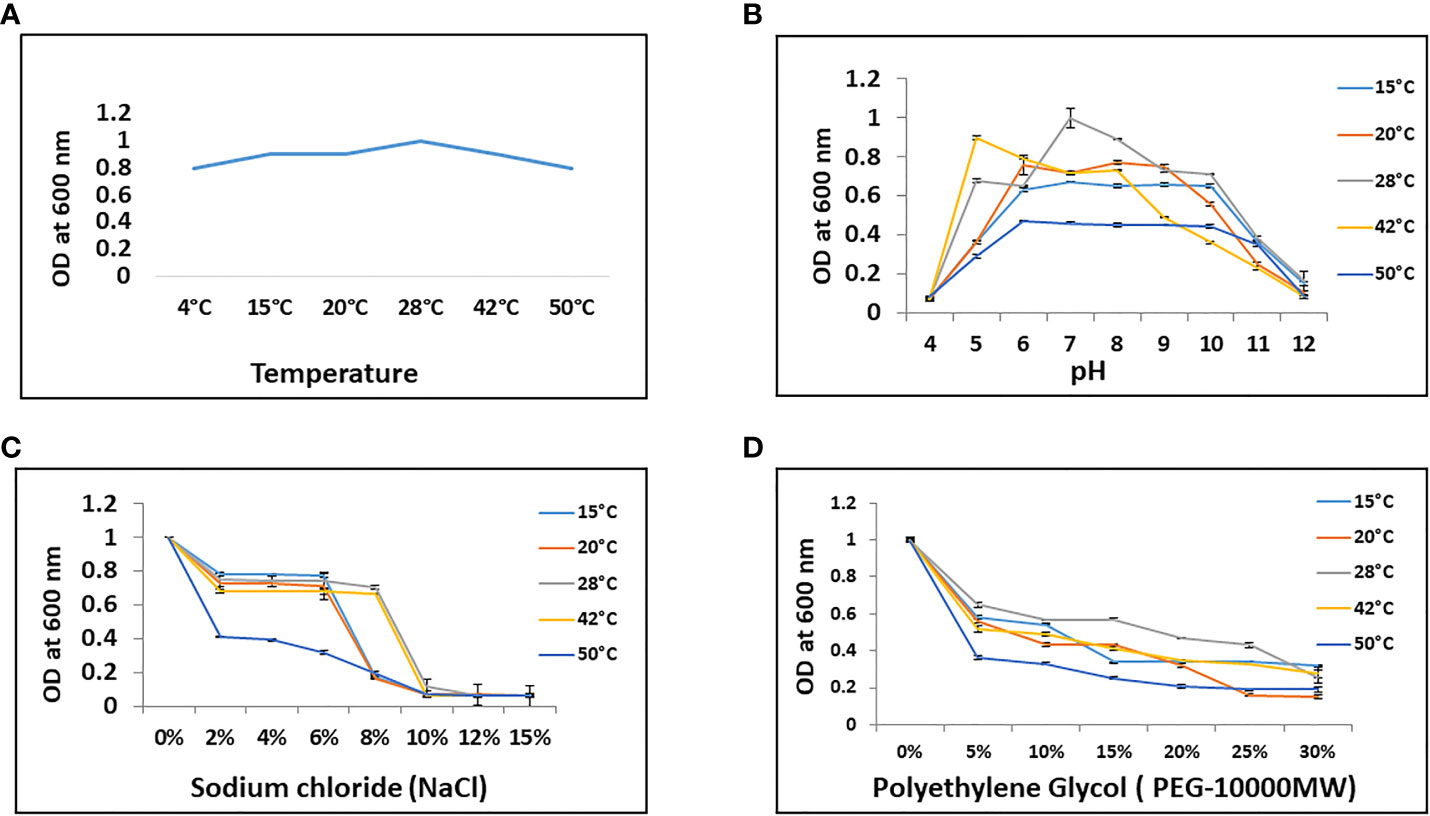
Figure 1 Stress tolerance potential of Bacillus sp. IHBT-705. (A) Thermotolerance potential. (B) pH tolerance. (C) Salt (NaCl) tolerance. (D) Osmotic (PEG 10,000MW) tolerance. All the experiments were done on targeted temperatures and Error bars represent the standard error of three replicates.
PGP potential of Bacillus sp. IHBT-705
Though Bacillus sp. IHBT-705 was found to be a stress-tolerant PGPR, but it was essential to check if it retained its growth-promoting attributes under stress or not so that it can serve as an effective biostimulant. Hence, we checked the key PGP traits under all the targeted temperatures. The P solubilization and siderophore production of Bacillus sp. IHBT-705 was confirmed by a clear zone around the colonies on PKV and blue CAS agar media at all targeted temperatures however, the potential varied. K solubilization by Bacillus sp. IHBT-705 was confirmed by a clear zone around the colonies on Aleksandrow medium at all targeted temperatures except 4°C, IAA production by the pink color formation in the salkowaski reagent at all targeted temperatures except 4°C and ACC deaminase activity was confirmed by the growth of Bacillus sp. IHBT-705 on DF salt minimal medium at all targeted temperatures except 4°C (Table 3).
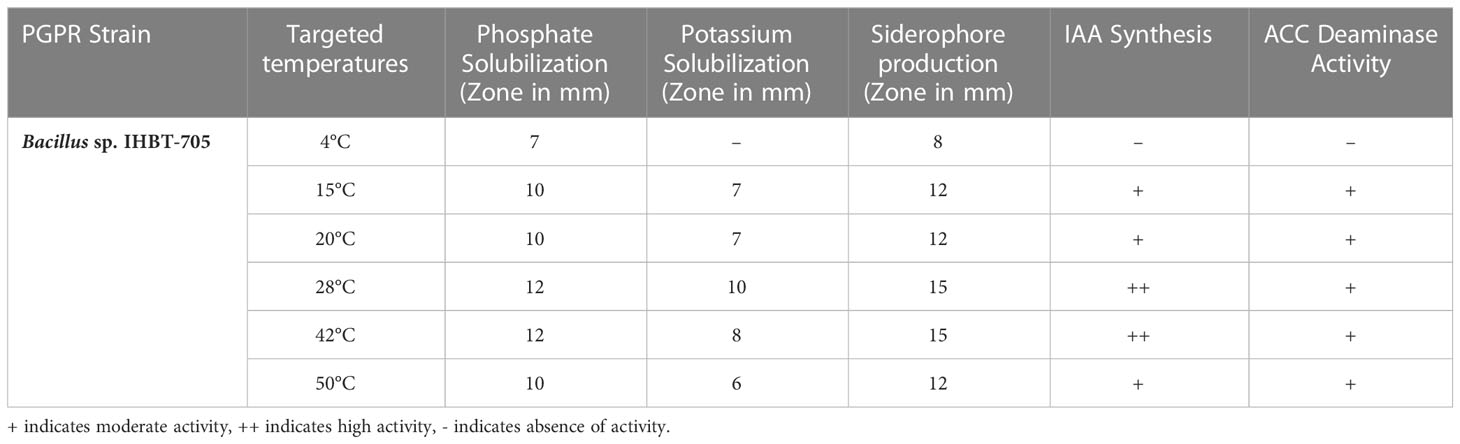
Table 3 PGP Potential of Bacillus sp. IHBT-705. Values represented are the mean of three replicates.
Molecular identification based on 16S rRNA gene sequencing
Phylogenetic analysis of Bacillus sp. IHBT-705 based on 16S rRNA sequence showed a close evolutionary relationship of 99.46% similarity with Bacillus altitudinis (ASJC01000029) as per the closest neighbor-joining method (Figure 2). The16S rRNA nucleotide sequence of Bacillus sp. IHBT-705 has been submitted to the NCBI gen bank under accession number MW959130 and its pure culture was deposited under the accession number MTCC25416 at the Microbial Culture Collection (MCC) at IMTECH, Chandigarh, India.
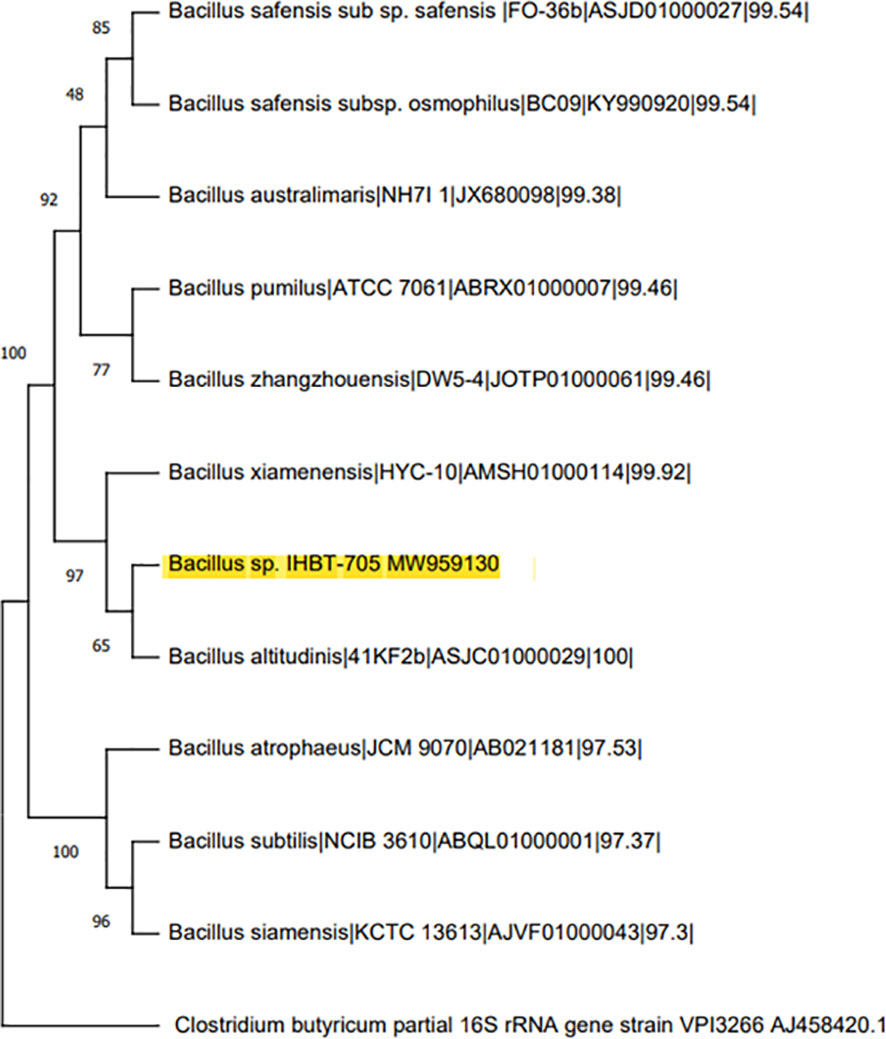
Figure 2 Phylogenetic relationships of 16S sequence of Bacillus sp. IHBT-705 and its homologs to the other species within the genus Bacillus and Clostridium butyricum as an outgroup.
Effect of Bacillus sp. IHBT-705 on rice
Through in vitro assay the Bacillus sp. IHBT-705 was found to exhibit promising plant growth potential this was further investigated through a pot study. Bacillus sp. IHBT-705 inoculation positively affected the growth of rice seedlings (Figure 3A). The treatment of rice with Bacillus sp. IHBT-705 significantly increased the length of shoots (42 cm ±0.1) as compared to the control treatment (28 cm ±0.3). Similarly, the length of roots was higher in Bacillus sp. IHBT-705 treated plants (13.08cm ±0.20) as compared to the control treatment of (8.48 ± 0.40; Figure 3B). Importantly, the numbers of total roots were higher in treated plants (85.58 ± 0.91) as compared to the control treatment (65.2 ± 1.78).
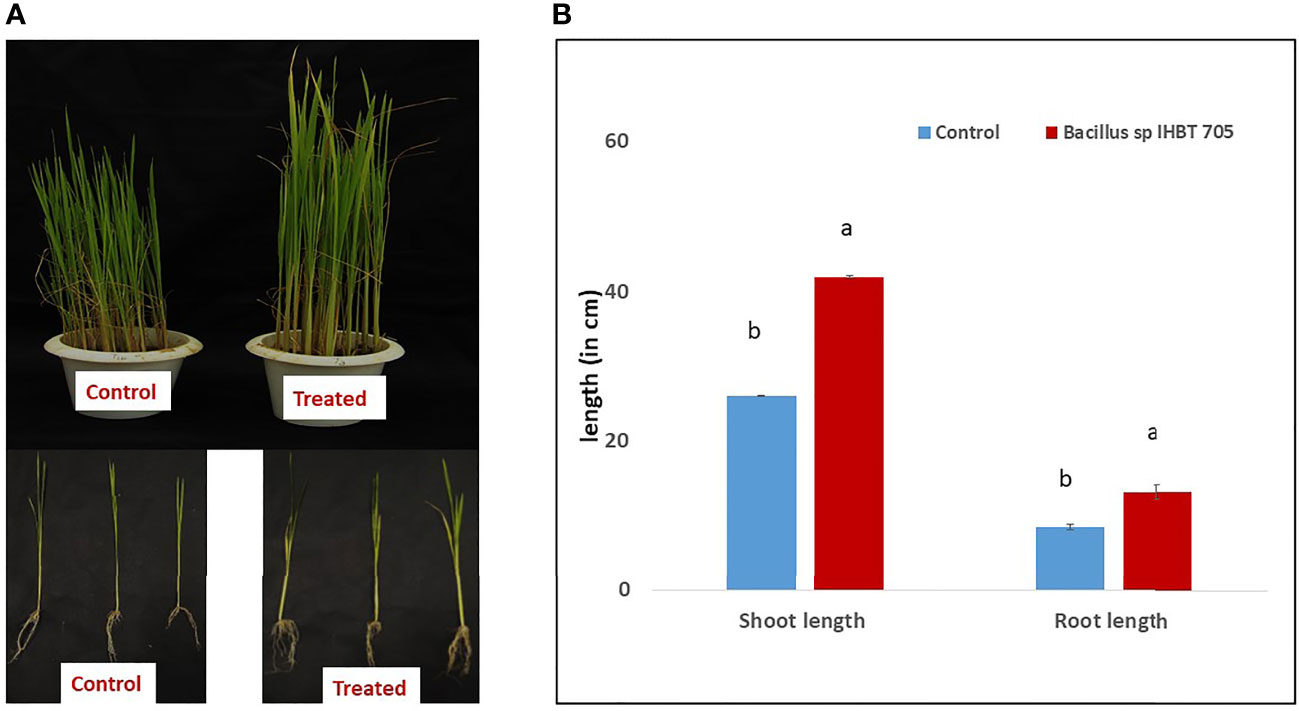
Figure 3 Effect of Bacillus sp. IHBT-705 on Rice (A) growth of rice seedlings under-treated and control conditions (B) shoot and root length. Different letters indicate significant differences among the treated and control treatments at p = 0.05. Data (mean ± SE) are averages of five replicates (n=5).
Complete genome sequencing of Bacillus sp. IHBT-705
The complete genome and plasmid sequences of Bacillus sp. IHBT-705 genome sequences were deposited at DDBJ/EMBL/NCBI GenBank under the accession numbers CP074101 under BioProject PRJNA725988 and BioSample SAMN18917214. Obtained a total of 312,490,430 bases and 39,800 reads (N50 Read length 10,827 bases and mean read length 7,851 bases). Out of which 36,038 reads (91%) were mapped and generated a complete assembly of 3,773,439 bases with 67.91X coverage and 41.3% GC content. Rapid Annotations using the Subsystems Technology (RAST) server predicted 4,015 genes for protein-coding (CDSs), 106 genes for RNAs, and 327 RAST subsystem categories were functionally assigned through the predicted genes as shown (Figure 4A). Out of 1682 features, 319 were found in amino acids and derivatives, 238 in carbohydrates and 206 in protein metabolism. The comparison of the Bacillus sp. IHBT-705 genome sequence with Bacillus altitudinis showed a close alignment using the whole-genome alignment tool as CLCBio Genomics Workbench (v21.0.3) with default parameters (minimum initial seed length 15bp and minimum alignment block length 100bp). The alignment was found to be matched at 5403 blocks (Figure 4B).

Figure 4 Genome annotation of Bacillus sp. IHBT-705. (A) Genome annotation of Bacillus sp. IHBT-705 using RAST (Rapid Annotations using Subsystems Technology). (B) Whole genome alignment of Bacillus sp. IHBT-705 (CP074101.1) with closest species reference genome B. altitudinis (NZ_CP011150.1).
Genome-based average nucleotide identification of Bacillus sp. IHBT-705 with closest strain B. altitudinis ASM97268
The draft genome length of B. altitudinis ASM 97268 was ~3.74 Mbp, while the genome size of Bacillus sp. IHBT-705 was ~3.77 Mbp. Close to 98.57% of total orthologous clusters were shared among two genomes.
Expression analysis of stress-tolerant and PGP genes of Bacillus sp. IHBT-705 under selected temperatures
Having observed that Bacillus sp. IHBT-705 retains its major PGP traits under temperature stress, we wanted to understand the functionality of genes responsible for these traits. Not much information is available on the response of key PGP genes under varying temperature regimes, hence we carried out expression analysis of targeted genes to observe their modulation with the rise and fall of temperature. The observation of the study can pave the way to predict the efficiency of Bacillus sp. IHBT-705 as a biostimulant for plants growing in areas exposed to low and high temperatures. A total of twenty-four genes, governing stress (cold and heat), pH tolerance, and PGP attributes (IAA production, siderophore production, ACC deaminase activity, phosphate solubilization, and root colonization) were targeted based on the information available through literature (Table 1) for expression analysis studies.
Four genes nhaX, cspB, cspC, and cspD were targeted to unravel the cold tolerance capability of Bacillus sp. IHBT-705. Out of four cold tolerant genes, cspC was found to be versatile as it upregulated at 15°C, 20°C, and 42°C. Gene nhaX gene upregulated at 20°C and 42°C. Gene cspB and cspD both upregulated only at 20°C. Gene cspB downregulated at 4°C,15°C, and 42°C while gene cspD downregulated at 4°C and 42°C. All these four cold tolerant genes downregulated at 50°C (Figure 5A). Out of the five heat-tolerant genes (dnaJ, dnaK, hslO, htpX, and grpE), gene dnaJ did not upregulat at any targeted temperature while it stably expressed at 20°C and downregulated at all the other targeted temperatures. Both genes dnaK, and hslO upregulated at 20°C, stably expressed at 42°C and downregulated at 4°C, 15°C, and 50°C. While gene htpX upregulated at 15°C, 20°C, and 42°C, stably expressed at 4°C, and 50°C. Gene grpE upregulated only at 20°C and downregulated at all the other temperatures (Figure 5B). Out of five IAA synthesis genes trpA, trpB, trpD, trpC, and trpE, gene trpA upregulated at 42°C, stably expressed at 15°C and 20°C and downregulated at 4°C, and 50°C. Gene trpB stably expressed at 4°C, 15°C, and 42°C and downregulated at 20°C and 50°C. Gene trpD stably expressed at 20°C and 42°C and downregulated at 4°C, 15°C, and 50°C. Gene trpC, and trpE both upregulated at 20°C. Gene trpC expressed stably at 4°C, 15°C, 42°C and downregulated at 50°C. Gene trpE downregulated at all the targeted temperature except 20°C (Figure 5C). Genes fhuB, fhuC, fhuD, fhuG, and acrB5 targeted for the siderophore production potential and out of these five genes, Genes fhuC and fhuD upregulated at 42°C, fhuC expressed stably at 20°C and fhuD at 4°C, both these genes downregulated on remaining other targeted temperatures. Gene fhuB upregulated at 4°C, 20°C 42°C and 50°C and downregulated at 15°C, while fhuG upregulated at 20°C and 42°C and stably expressed at 4°C and 15°C and downregulated at 50°C. Gene acrB5 upregulated at 20°C and stably expressed at 4°C, 15°C, 42°C, and 50°C indicating retention of siderophore-producing activity at different temperature ranges (Figure 5D). Gene nhaK was targeted for pH tolerance and it stably expressed only at 20°C and downregulated at all the other targeted temperatures. Gene rimM gene targeted for ACC deaminase activity and it upregulated at only 20°C, stably expressed at 42°C and downregulated at all the other targeted temperatures. Gene pstB1 targeted for the phosphate solubilizing potential and it upregulated at 15°C, 20°C, and 42°C and downregulated at 4°C and 50°C. Both the genes for root colonizations xerC1 and xerC2 upregulated at 20°C and 42°C, xerC1 expressed stably and xerC2 were downregulated at 4°C, 15°C and 50°C (Figure 5E).
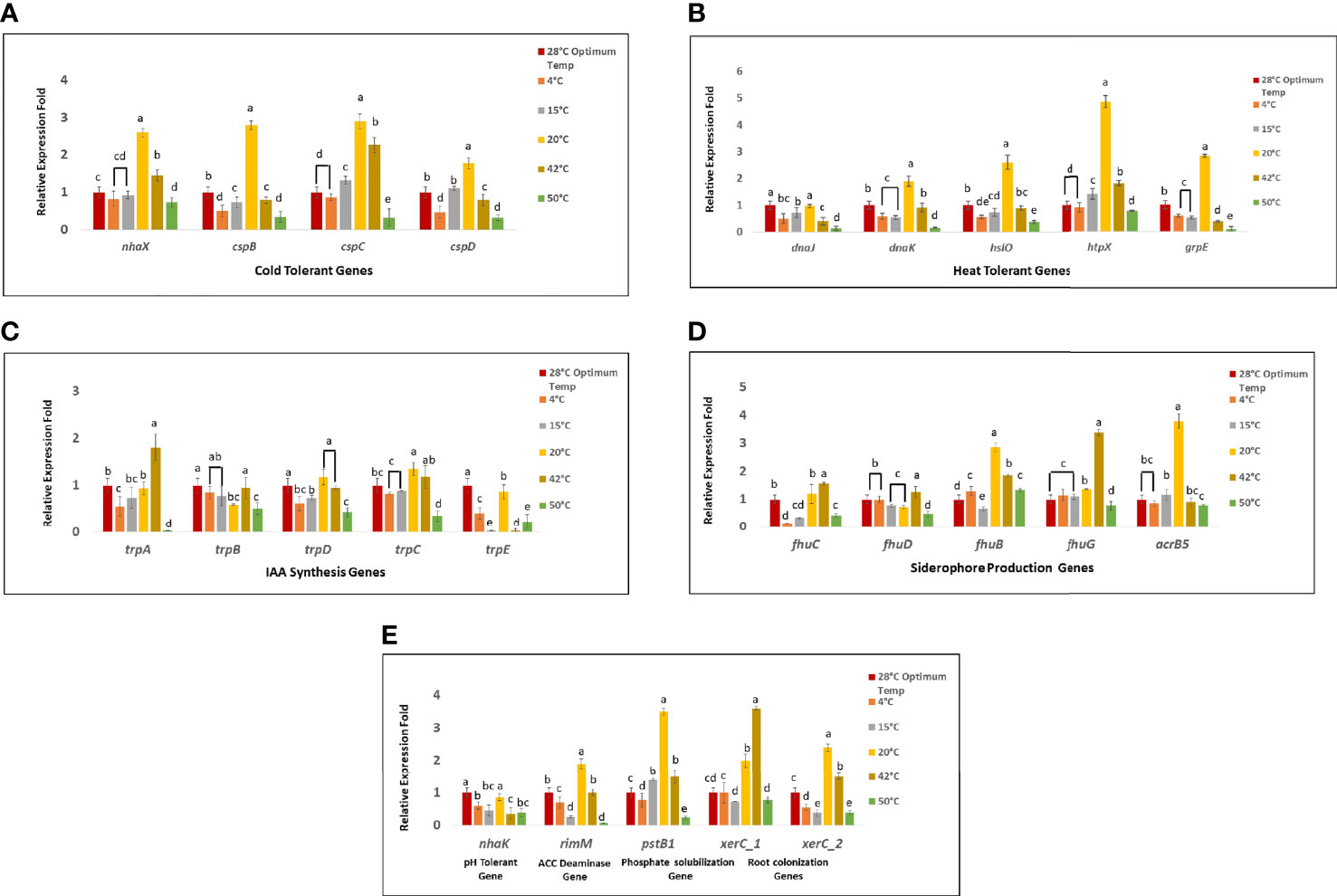
Figure 5 The relative expression of stress-related and PGP genes (A) Cold tolerant genes (B) Heat tolerant genes (C) Genes involved in IAA biosynthesis (D) Genes related to Siderophore-production (E) pH tolerant gene, ACC deaminase gene, phosphate solubilization and genes for root colonization. The results were represented as the mean of three replicates (n=3). Different letters indicate significant differences in gene expression among all targeted temperatures (P = 0.05).
Discussion
Several species of Bacillus are well known for their plant growth-promoting attributes and during the present study plant growth-promoting potentials of Bacillus sp. IHBT-705 was unraveled through in vitro analyses, pot study, and gene expression studies. The bacteria exhibited thermo-tolerance, could multiply in the acidic and alkaline range and could tolerate salt and high PEG concentration indicating that it is a good stress-tolerant PGPR.
Upon whole genome analysis, several key genes like cspB, cspC, cspD, nhaX, and htpX were identified. These genes are known to confer cold stress tolerance in microbes (Kishor PB, 2019; Lalaouna et al., 2019; Suarez et al., 2019), hence it is likely that these genes could have conferred stress tolerance capability to Bacillus sp. IHBT-705. This gene expression study revealed that nhaX upregulated at low as well as high temperatures indicating that it could be responsible for imparting heat and cold tolerance and makes the bacterial strain tolerant to both cold and heat stress. Cold shock proteins (csp) family are known for their stress tolerance potential and found to be constitutively expressed during stress conditions in bacteria and even transgenic plants overexpressing bacterial csp exhibited tolerance to cold, salt, and drought stresses (Gupta et al., 2014; Kishor PB, 2019). In our study gene cspC overexpressed at high as well as low temperature suggesting that it may also impart heat and cold tolerance. Hence, presence of cspC gene makes Bacillus sp. IHBT-705 a valuable microbe from stress tolerance perspective. However, cspB and cspD exhibited a narrow range of functionality as both these genes upregulated only at low temperature.
Literature suggests that genes dnaJ, dnaK, HslO, htpX, and grpE are responsible for heat tolerance (Gupta et al., 2014; Suarez et al., 2019). However, we observed that genes dnaK grpE, hslO upregulated only at low temperature but not at high temperature, hence these genes were found to exhibit a narrow range of functionality. The gene htpX upregulated at low as well as high temperatures indicating its broad range of functionality and could have imparted stress tolerance ability, hence this gene appears to be crucial in conferring thermo-tolerance to Bacillus sp. IHBT-705. As the bacteria could tolerate up to 50°C, but none of the targeted gene upregulated at very high temperature further investigations by targeting other heat-tolerant genes are required to understand its extreme temperature tolerance capability.
The colorimetric tests illustrated that Bacillus sp. IHBT-705 efficiently produces IAA both at low as well as at high temperatures. Earlier studies indicate that trp genes (trpA, trpB, trpD, trpC, trpE) are involved in multiple biological processes including IAA biosynthesis (Gupta et al., 2014; Dahmani et al., 2020; Zaid et al., 2022). Interestingly, the present gene expression study revealed that specific temperature triggers the upregulation of specific sets of IAA-producing genes. Gene trpA was upregulated at high while trpC and trpE were upregulated at low temperature. The expression of trpB largely remained unaltered at both low and high temperatures, hence it could be considered a versatile gene with the ability to function over a broad temperature range with optimum efficacy. The role of microbial IAA in shaping root architecture ultimately benefitting plant productivity has been experimentally proven (Kazan, 2013; Sukumar et al., 2013; Grover et al., 2021), hence IAA is considered an important plant growth-promoting attribute, and in this respect, the complementary functioning of IAA-producing genes at both low and high temperature possibly enable retention of IAA producing capability of Bacillus sp. IHBT-705 under temperature stress which is a beneficial attribute.
Siderophore-mediated iron uptake in plants is a major benefit imparted by PGPRs (Braud et al., 2006; Mandal and Kotasthane, 2014). Siderophores are grouped into three principal categories based on their functional groups which are hydroxamates, catecholate, and carboxylates (Suarez et al., 2019; Kramer et al., 2020). Earlier studies revealed that the iron (III)-hydroxamate ABC transporter cluster fhuCDB and the ferric hydroxamate uptake gene fhuA are responsible for the transport of ferrichrome and other Fe3+ hydroxamate compounds (Gupta et al., 2014; Paul and Lade, 2014), hence in our study fhuCDBG and acrB5 genes associated with the production of siderophore were targeted for expression analysis studies. Among these targeted genes, fhuB was found to be highly versatile as it upregulated from very low to very high temperatures. Similarly, fhuG upregulated from low to high temperature while acrB5 upregulated at very low temperature, hence functionality of gene fhuB is a positive attribute of Bacillus sp. IHBT-705. Compared to IAA-producing genes, siderophore-related genes exhibited better expression at both low and high temperatures. The colorimetric study also illustrated that siderophore producing potential of Bacillus sp. IHBT-705 was active from very low to high temperature which is highly desirable trait from a biostimulant perspective.
The gene nhaK is known for imparting tolerance to pH (Noori et al., 2021), in this study gene nhaK was downregulated at all targeted temperatures thereby indicating a narrow range of functionality with respect to temperature. PGPRs produce extracellular polymeric substances and synthesize 1-aminocyclo-propane-1-carboxylate (ACC) deaminase enzyme or its homologue D-cysteine desulfhydrase, these enzymes lower ethylene accumulation in stressed plants (Sashidhar and Podile, 2010; Wagh et al., 2014) and enable plants to become stress tolerant (Paul and Lade, 2014). Gene rimM responsible for ACC deaminase activity (Gupta et al., 2014), Bacillus sp. IHBT-705 is an ACC- deaminase-producing strain consisting gene rimM and thus can act as a stress buster also. The expression analysis of rimM exhibited upregulation at low as well as a high temperature suggesting that Bacillus sp. IHBT-705 can function efficiently under temperature stress.
Qualitative assay phosphate solubilization potential of Bacillus sp. IHBT-705 was observed from very low to very high temperature which were further illustrated through gene expression study. The literature (Hudek et al., 2016; Suarez et al., 2019) suggests that pstB1 plays a functional role in phosphate uptake. In our study gene pstB1 upregulated from very low to high temperature, indicating that this gene is versatile and remains active both under low and high temperatures. This attribute is significant from a biostimulant perspective.
The ability to colonize the rhizosphere is an important attribute of PGPRs and genes xerC1 and xerC2 have been found to code site-specific recombinase which facilitates efficient colonization (Shen et al., 2013; Eida et al., 2020), hence these two genes were targeted. The expression analysis revealed overexpression of xerC1 and xerC2 at low and high temperature indicating that Bacillus sp. IHBT-705 could be a good colonizer both low and high temperatures. Apart from 28°C which is the optimum temperature for the growth of Bacillus sp. IHBT-705, among the targeted genes, the maximum number of genes were upregulated at 20°C and 42°C indicating its PGP potential at both low and high-temperature (Figure 6). Our study suggests that owing to its high-stress tolerance capability, Bacillus sp. IHBT-705 could be used as a potential biostimulant for crops growing in cooler mountainous regions as well as in warmer plains.
Conclusion
Recent years have witnessed an upsurge in microbial-based biostimulant. Several studies on PGP traits illustrate that the intrinsic characteristics of microbes are critical to their biostimulant potential. In this regard, physiological, molecular, and gene expression study of Bacillus sp. IHBT-705 revealed that it could survive both under very low to very high temperatures, low and high pH, and withstand salinity and osmolarity, suggesting that it is a highly stress-tolerant PGPR. Whole genome sequencing revealed the presence of several genes responsible for imparting stress tolerance and multiple genes for a single PGP trait. Furthermore, expression analysis of key PGP genes revealed that temperature is a critical factor governing the upregulation/downregulation of specific PGP genes hence temperature strongly regulates the functionality of PGP traits. As a PGPR, Bacillus sp. IHBT-705 has unique genetic makeup with multiple genes for a specific PGP trait wherein the downregulation of one gene is compensated by the upregulation of complementary genes, making Bacillus sp. IHBT-705 a good candidate for developing biostimulant(s) for agriculture in hilly terrain, warmer plains, and land affected by sodic soils. The genes nhaX, cspC, htpX, fhuB, fhuG, pstB1, xerC1, and xerC2 were found to be versatile genes owing to their expression both under low and high temperatures while cspB, cspD, hslO, grpE, rimM, trpA, trpC, trpE, fhuC, fhuD, acrB5 were found to be temperature sensitive. The presence of stress-tolerant genes may serve as marker genes for selecting other PGPRs as stress tolerant PGPRs for developing biostimulants (s) suitable for agriculture in different terrains.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
NA performed the experimentation, data analysis, and drafted the manuscript. MS contributed to experimentation and data analysis. RV helped in experimentation. PK edited and proofread the manuscript. AP conceptualized, guided, and structured the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors thank the Director, CSIR Institute of Himalayan Bioresource Technology, Palampur (HP) India, for providing R&D facilities, Dr. Amita Bhattacharya for her constant encouragement, Dr. Kunal Singh, Dr. Dharam Singh and Dr. Rakshak Kumar for extending the necessary laboratory facilities to carry out the work. The authors also thank Mr. Ramdeen Prasad for supporting soil analysis. This research article bears Institutional publication No. 4927 under the project number MLP165.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1141538/full#supplementary-material
Supplementary Figure 1 | Map of soil sampling site for isolation of PGPR.
Supplementary Figure 2 | Library validation of gDNA for whole genome sequencing of Bacillus sp. IHBT-705. (A) Gel profile of isolated high-quality gDNA of Bacillus sp. IHBT-705 with 1Kb ladder. (B) Bioanalyzer profile of Bacillus sp. IHBT-705 SMRTbell templates for sequencing with ladder. (C) Bioanalyzer profile of prepared library size (~9.8 Kb) of Bacillus sp. IHBT-705.
Supplementary Figure 3 | Genome assembly features of PacBio RS II data using HGAP v3.0. (A) Polymerase read length distribution of post-filter reads. (B) Polymerase read quality distribution of post-filter reads. (C) Subread filtering of the whole genome of Bacillus sp. IHBT-705 sequencing reads. (D) Coverage across a reference of single contig assembly (with an average coverage of 69X).
Supplementary Figure 4 | Expressions analysis of targeted stress tolerant and PGP genes by qRT-PCR. (A) Amplification plots (B) Melt curves of targeted genes.
References
Ahemad, M., Khan, M. S. (2012). Evaluation of plant-growth-promoting activities of rhizobacterium pseudomonas putida under herbicide stress. Ann. Microbiol. 62, 1531–1540. doi: 10.1007/s13213-011-0407-2
Ali, S., Khan, N. (2021). Delineation of mechanistic approaches employed by plant growth promoting microorganisms for improving drought stress tolerance in plants. Microbiol. Res. 249, 126771. doi: 10.1016/j.micres.2021.126771
Amaya-Gómez, C. V., Porcel, M., Mesa-Garriga, L., Gómez-Álvarez, M. I. (2020). A framework for the selection of plant growth-promoting rhizobacteria based on bacterial competence mechanisms. Appl. Environ. Microbiol. 86, e00760-20. doi: 10.1128/AEM.00760-20
Ansari, F. A., Ahmad, I., Pichtel, J. (2019). Growth stimulation and alleviation of salinity stress to wheat by the biofilm forming bacillus pumilus strain FAB10. Appl. Soil Ecol. 143, 45–54. doi: 10.1016/j.apsoil.2019.05.023
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01473
Beneduzi, A., Ambrosini, A., Passaglia, L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/s1415-47572012000600020
Braud, A., Jézéquel, K., Léger, M.-A., Lebeau, T. (2006). Siderophore production by using free and immobilized cells of two pseudomonads cultivated in a medium enriched with fe and/or toxic metals (Cr, Hg, Pb). Biotechnol. Bioeng. 94, 1080–1088. doi: 10.1002/bit.20937
Bray, R. H., Kurtz, L. T. (1945). Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 59, 39–46. doi: 10.1097/00010694-194501000-00006
Bruto, M., Prigent-Combaret, C., Muller, D., Moënne-Loccoz, Y. (2014). Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related proteobacteria. Sci. Rep. 4, 6261. doi: 10.1038/srep06261
Cárdenas Espinosa, M. J., Schmidgall, T., Wagner, G., Kappelmeyer, U., Schreiber, S., Heipieper, H. J., et al. (2021). An optimized method for RNA extraction from the polyurethane oligomer degrading strain pseudomonas capeferrum TDA1 growing on aromatic substrates such as phenol and 2,4-diaminotoluene. PloS One 16, e0260002. doi: 10.1371/journal.pone.0260002
Chiaromonte, F., Yap, V. B., Miller, W. (2002). Scoring pairwise genomic sequence alignments. Pac. Symp. Biocomput., 115–126. doi: 10.1142/9789812799623_0012
Choudhary, D. K., Sharma, K. P., Gaur, R. K. (2011). Biotechnological perspectives of microbes in agro-ecosystems. Biotechnol. Lett. 33, 1905–1910. doi: 10.1007/s10529-011-0662-0
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., Robles, M. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dahmani, M. A., Desrut, A., Moumen, B., Verdon, J., Mermouri, L., Kacem, M., et al. (2020). Unearthing the plant growth-promoting traits of bacillus megaterium RmBm31, an endophytic bacterium isolated from root nodules of retama monosperma. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00124
Dhaked, B. S., Triveni, S., Reddy, R. S., Padmaja, G. (2017). Isolation and screening of potassium and zinc solubilizing bacteria from different rhizosphere soil. Int. J. Curr. Microbiol. App. Sci. 6, 1271–1281. doi: 10.20546/ijcmas.2017.608.154
Dworkin, M., Foster, J. W. (1958). Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 75, 592–603. doi: 10.1128/jb.75.5.592-603.1958
Eida, A. A., Bougouffa, S., L’Haridon, F., Alam, I., Weisskopf, L., Bajic, V. B., et al. (2020). Genome insights of the plant-growth promoting bacterium cronobacter muytjensii JZ38 with volatile-mediated antagonistic activity against phytophthora infestans. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00369
García-Fraile, P., Menéndez, E., Rivas, R. (2015). Role of bacterial biofertilizers in agriculture and forestry. AIMSBOA 2, 183–205. doi: 10.3934/bioeng.2015.3.183
Getahun, A., Muleta, D., Assefa, F., Kiros, S. (2020). Plant growth-promoting rhizobacteria isolated from degraded habitat enhance drought tolerance of acacia (Acacia abyssinica hochst. ex benth.) seedlings. Int. J. Microbiol. 2020, 8897998. doi: 10.1155/2020/8897998
Glick, B. (2018). Soil microbes and sustainable agriculture. Pedosphere 28, 167–169. doi: 10.1016/S1002-0160(18)60020-7
Gouda, S., Kerry, R. G., Das, G., Paramithiotis, S., Shin, H.-S., Patra, J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 206, 131–140. doi: 10.1016/j.micres.2017.08.016
Grover, M., Bodhankar, S., Sharma, A., Sharma, P., Singh, J., Nain, L. (2021). PGPR mediated alterations in root traits: Way toward sustainable crop production. Front. Sustain. Food Syst. 4. doi: 10.3389/fsufs.2020.618230
Gupta, A., Gopal, M., Thomas, G., Manikandan, V., Gajewski, J., Thomas, G., et al. (2014). Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PloS One 9, e104259. doi: 10.1371/journal.pone.0104259
Hudek, L., Premachandra, D., Webster, W. A. J., Bräu, L. (2016). Role of phosphate transport system component PstB1 in phosphate internalization by nostoc punctiforme. Appl. Environ. Microbiol. 82, 6344–6356. doi: 10.1128/AEM.01336-16
Jian, C., Luukkonen, P., Yki-Järvinen, H., Salonen, A., Korpela, K. (2020). Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PloS One 15, e0227285. doi: 10.1371/journal.pone.0227285
Kazan, K. (2013). Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 1655–1665. doi: 10.1093/aob/mct229
Khan, M. A., Asaf, S., Khan, A. L., Jan, R., Kang, S.-M., Kim, K.-M., et al. (2020). Thermotolerance effect of plant growth-promoting bacillus cereus SA1 on soybean during heat stress. BMC Microbiol. 20, 175. doi: 10.1186/s12866-020-01822-7
Khan, N., Bano, A., Rahman, M. A., Guo, J., Kang, Z., Babar, M. A. (2019). Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum l.) induced by PGPR and PGRs. Sci. Rep. 9, 2097. doi: 10.1038/s41598-019-38702-8
Kishor PB, K. (2019). Bacterial cold shock proteins - the molecular chaperones for multiple stress tolerance. AIBM 12, 57–61. doi: 10.19080/AIBM.2019.12.555837
Kramer, J., Özkaya, Ö., Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Lalaouna, D., Baude, J., Wu, Z., Tomasini, A., Chicher, J., Marzi, S., et al. (2019). RsaC sRNA modulates the oxidative stress response of staphylococcus aureus during manganese starvation. Nucleic Acids Res. 47, 9871–9887. doi: 10.1093/nar/gkz728
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lastochkina, O., Pusenkova, L., Yuldashev, R., Babaev, M., Garipova, S., Blagova, D., et al. (2017). Effects of bacillus subtilis on some physiological and biochemical parameters of triticum aestivum l. (wheat) under salinity. Plant Plant Physiol. Biochem. 121, 80–88. doi: 10.1016/j.plaphy.2017.10.020
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loper, J. (1986). Influence of bacterial sources of Indole3-acetic acid on root elongation of sugar beet. Phytopathology 76, 386–389. doi: 10.1094/Phyto-76-386
Mandal, L., Kotasthane, A. S. (2014). Isolation and assessment of plant growth promoting activity of siderophore producing pseudomonas fluorescens in crops. Intern. Jour. Agricul. Environ. Biotech. 7, 63. doi: 10.5958/j.2230-732X.7.1.009
Mehlich, A. (1984). Mehlich 3 soil test extractant: A modification of mehlich 2 extractant. Commun. Soil Sci. Plant Anal. 15, 1409–1416. doi: 10.1080/00103628409367568
Mellidou, I., Karamanoli, K. (2022). Unlocking PGPR-mediated abiotic stress tolerance: What lies beneath. Front. Sustain. Food Syst. 6. doi: 10.3389/fsufs.2022.832896
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Nelson, D. w., Sommers, L. e. (1983). “Total carbon, organic carbon, and organic matter,” in Methods of soil analysis (John Wiley & Sons, Ltd), 539–579. doi: 10.2134/agronmonogr9.2.2ed.c29
Nezarat, S., Gholami, A. (2009). Plant growth promoting rhizobacteria for improving seed germination, seedling growth and yield of MaizeScreening. Pakistan journal of biological sciences. PJBS 12, 26–32. doi: 10.3923/pjbs.2009.26.32
Niu, X., Song, L., Xiao, Y., Ge, W. (2018). Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02580
Noori, F., Etesami, H., Noori, S., Forouzan, E., Salehi Jouzani, G., Malboobi, M. A. (2021). Whole genome sequence of pantoea agglomerans ANP8, a salinity and drought stress–resistant bacterium isolated from alfalfa (Medicago sativa l.) root nodules. Biotechnol. Rep. 29, e00600. doi: 10.1016/j.btre.2021.e00600
Paul, D., Lade, H. (2014). Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: A review. Agron. Sustain. Dev. 34, 737–752. doi: 10.1007/s13593-014-0233-6
Paul, D., Sinha, S. (2013). Phosphate solubilizing activity of some bacterial strains isolated from jute mill effluent exposed water of river ganga. Indian J. Fundam. Appl. Life Sci. 3, 39–45.
Ramadoss, D., Lakkineni, V. K., Bose, P., Ali, S., Annapurna, K. (2013). Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. Springerplus 2, 6. doi: 10.1186/2193-1801-2-6
Raza, W., Ling, N., Yang, L., Huang, Q., Shen, Q. (2016). Response of tomato wilt pathogen ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain bacillus amyloliquefaciens SQR-9. Sci. Rep. 6, 24856. doi: 10.1038/srep24856
Saitou, N., Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sapre, S., Gontia-Mishra, I., Tiwari, S. (2018). Klebsiella sp. confers enhanced tolerance to salinity and plant growth promotion in oat seedlings (Avena sativa). Microbiol. Res. 206, 25–32. doi: 10.1016/j.micres.2017.09.009
Sashidhar, B., Podile, A. R. (2010). Mineral phosphate solubilization by rhizosphere bacteria and scope for manipulation of the direct oxidation pathway involving glucose dehydrogenase. J. Appl. Microbiol. 109, 1–12. doi: 10.1111/j.1365-2672.2009.04654.x
Schwyn, B., Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shen, X., Hu, H., Peng, H., Wang, W., Zhang, X. (2013). Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in pseudomonas. BMC Genomics 14, 271. doi: 10.1186/1471-2164-14-271
Suarez, C., Ratering, S., Hain, T., Fritzenwanker, M., Goesmann, A., Blom, J., et al. (2019). Complete genome sequence of the plant growth-promoting bacterium hartmannibacter diazotrophicus strain E19T. Int. J. Genomics 2019, 7586430. doi: 10.1155/2019/7586430
Subramanian, P., Kim, K., Krishnamoorthy, R., Mageswari, A., Selvakumar, G., Sa, T. (2016). Cold stress tolerance in psychrotolerant soil bacteria and their conferred chilling resistance in tomato (Solanum lycopersicum mill.) under low temperatures. PloS One 11, e0161592. doi: 10.1371/journal.pone.0161592
Sukumar, P., Legué, V., Vayssières, A., Martin, F., Tuskan, G. A., Kalluri, U. C. (2013). Involvement of auxin pathways in modulating root architecture during beneficial plant–microorganism interactions. Plant. Cell Environ. 36, 909–919. doi: 10.1111/pce.12036
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Timmusk, S., Behers, L., Muthoni, J., Muraya, A., Aronsson, A.-C. (2017). Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00049
Vasseur-Coronado, M., du Boulois, H. D., Pertot, I., Puopolo, G. (2021). Selection of plant growth promoting rhizobacteria sharing suitable features to be commercially developed as biostimulant products. Microbiol. Res. 245, 126672. doi: 10.1016/j.micres.2020.126672
Vincent, J. M. (1970). “A manual for the practical study of the root-nodule bacteria,” in A manual for the practical study of the root-nodule bacteria. IBP Handbk 15 Oxford and Edinburgh: Blackwell Scientific Publications. Available at: https://www.cabdirect.org/cabdirect/abstract/19710700726.
Vurukonda, S. S. K. P., Vardharajula, S., Shrivastava, M., SkZ, A. (2016). Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 184, 13–24. doi: 10.1016/j.micres.2015.12.003
Wagh, J., Shah, S., Bhandari, P., Archana, G., Kumar, G. N. (2014). Heterologous expression of pyrroloquinoline quinone (pqq) gene cluster confers mineral phosphate solubilization ability to herbaspirillum seropedicae Z67. Appl. Microbiol. Biotechnol. 98, 5117. doi: 10.1007/s00253-014-5610-1
Weisburg, W. G., Barns, S. M., Pelletier, D. A., Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Keywords: saffron rhizosphere, stress tolerant PGP potential, long-read sequencing, gene expression, biostimulants for soil and plant health
Citation: Ali N, Swarnkar MK, Veer R, Kaushal P and Pati AM (2023) Temperature-induced modulation of stress-tolerant PGP genes bioprospected from Bacillus sp. IHBT-705 associated with saffron (Crocus sativus) rhizosphere: A natural -treasure trove of microbial biostimulants. Front. Plant Sci. 14:1141538. doi: 10.3389/fpls.2023.1141538
Received: 10 January 2023; Accepted: 14 February 2023;
Published: 27 February 2023.
Edited by:
Manoj Gajanan Kulkarni, University of KwaZulu-Natal, South AfricaReviewed by:
Ni Luh Suriani, Udayana University, IndonesiaSangeeta Paul, Indian Agricultural Research Institute (ICAR), India
Copyright © 2023 Ali, Swarnkar, Veer, Kaushal and Pati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aparna Maitra Pati, YXBhcm5hQGloYnQucmVzLmlu
 Nilofer Ali
Nilofer Ali Mohit Kumar Swarnkar
Mohit Kumar Swarnkar Raj Veer3
Raj Veer3 Priya Kaushal
Priya Kaushal Aparna Maitra Pati
Aparna Maitra Pati