- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Sciences, Northwest A&F University, Yangling, China
- 2Hybrid Rapeseed Research Centre of Shaanxi Province, Yangling, China
The Gly-Asp-Ser-Leu (GDSL)-type esterase/lipase proteins (GELP) are one of the most important families of lipolytic enzymes and play prominent roles in seed germination and early seedling establishment through mobilizing the lipids stored in seeds. However, there are no comprehensive studies systematically investigating the GELP gene family in Brassica napus (BnGELP), and their biological significance to these physiological processes are far from understood. In the present study, a total of 240 BnGELP genes were identified in B. napus cultivar “Zhongshuang 11” (ZS11), which is nearly 2.3-fold more GELP genes than in Arabidopsis thaliana. The BnGELP genes clustered into 5 clades based on phylogenetic analysis. Ten BnGELPs were identified through zymogram analysis of esterase activity followed by mass spectrometry, among which five clustered into the clade 5. Gene and protein architecture, gene expression, and cis-element analyses of BnGELP genes in clade 5 suggested that they may play different roles in different tissues and in response to different abiotic stresses. BnGELP99 and BnGELP159 were slightly induced by cold, which may be attributed to two low-temperature responsive cis-acting regulatory elements present in their promoters. An increased activity of esterase isozymes by cold was also observed, which may reflect other cold inducible esterases/lipases in addition to the ten identified BnGELPs. This study provides a systemic view of the BnGELP gene family and offers a strategy for researchers to identify candidate esterase/lipase genes responsible for lipid mobilization during seed germination and early seedling establishment.
Introduction
Rapeseed (Brassica napus L., genome AACC, 2n = 4x = 38) is the third most important oil crop across the world (Ding et al., 2019a), and is an allotetraploid derived from an interspecific hybridization between Brassica rapa (genome AA, 2n = 20) and Brassica oleracea (genome CC, 2n = 18), with subsequent chromosomal doubling (Sun et al., 2017; Chen et al., 2021). Seed oil content (OC), one of the most important economic and nutritional traits for rapeseed, is mainly determined by a dynamic balance between lipid synthesis and breakdown during seed maturation (Ding et al., 2019a; Wang et al., 2021). In rapeseed, lipid biosynthesis is initiated just after embryo formation, and its reduction and decomposition occurs during the late maturation stage (Wang et al., 2021). Oil bodies, or lipid droplets, are the main reservoir of lipids in seeds of B. napus.
While breeding for cultivars with high OC, seed germination and seedling development are also targeted traits due to their impact on successful propagation (Wang et al., 2021). In B. napus, seed germination and early development of the seedling are mainly dependent on the catabolism of stored lipids, which is initiated by the action of esterases/lipases (Clauss et al., 2008; Zhu et al., 2017; Ding et al., 2019a; Wang et al., 2021). GDSL-type esterases/lipases (GELPs) are one type of lipolytic enzyme that mobilizes the stored lipid in oil bodies during germination and early seedling establishment (Akoh et al., 2004; Clauss et al., 2008; Ding et al., 2019a; Ding et al., 2019b; Wang et al., 2021; Shen et al., 2022). GELPs are named after the highly conservative GDSL motif, GDSxxDxG, mainly present close to the N-terminus of the protein (Akoh et al., 2004; Ding et al., 2019b; Shen et al., 2022). GELPs usually harbor five conserved sequence blocks (I-V), which are also important for their classification (Akoh et al., 2004; Ding et al., 2019b; Shen et al., 2022). There are four strictly invariant residues present in each of the four of the conserved blocks, serine (S) in block I, glycine (G) in block II, asparagine (N) in block III and histidine (H) in block V (Akoh et al., 2004; Ding et al., 2019b; Shen et al., 2022). For this reason, GELPs are also called SGNH hydrolases (Akoh et al., 2004; Ding et al., 2019a; Ding et al., 2019b; Shen et al., 2022). GELPs are hydrolytic enzymes that display broad substrate specificity and regio-specificity. Therefore, it is easy to detect the biochemical activity of different GELPs present in a total protein extract from seedlings in a polyacrylamide gel, such as through determining the zymogram of esterase/lipase isozymes (Akoh et al., 2004; Zhu et al., 2017; Ding et al., 2019b; Shen et al., 2022).
GELPs are widely distributed in microbes, animals and plants (Akoh et al., 2004). GELPs have been suggested to play significant roles in controlling nearly all aspects of plant growth and development as well as response to abiotic and biotic stresses (Shen et al., 2022). A crucial first step in transferring the knowledge acquired for model species to non-model species is homology identification. To date, the GELP family has been identified in diverse plant species including many with fully sequenced genomes, such as Arabidopsis thaliana (105 members), Dasypyrum villosum (193), Sedum alfredii (80), Nicotiana tabacum (159), Glycine max (194), Oryza sativa (114), Dendrobium catenatum (52), Carya illinoensis (87), Zea mays (103), Vitis vinifera (83), Brassica rapa (121), and six species in the Rosaceae family (Pyrus bretschneideri, Malus domestica, Prunus mume, Prunus persica, Fragaria vesca and Prunus avium; for a total of 597 GELPs) (Chepyshko et al., 2012; Dong et al., 2016; Lai et al., 2017; Ding et al., 2019b; Su et al., 2020; Lv et al., 2021; Wang et al., 2021; Zhang et al., 2021; Cenci et al., 2022; Jiao et al., 2022; Shen et al., 2022). Some GELP genes have been cloned and functionally characterized in B. napus, such as BnLIP2 (Ling et al., 2006), BnSCE3 (Clauss et al., 2008), BnGDSL1 (Ding et al., 2019a), and BnSFAR1-5 (Karunarathna et al., 2020). However, no comprehensive study aiming to systematically investigate the GDSL gene family has been conducted in B. napus, particularly in cultivar ZS11, for which a high-quality genome assembly has been recently released (Chen et al., 2021). ZS11 is a cultivar with stable yield and its recently published genome assembly is more accurate and complete than the previous genomes (Chalhoub et al., 2014; Bayer et al., 2017; Sun et al., 2017), providing an excellent opportunity for reliable, systematic exploration of all the genes belonging to a specified family.
In the current study, a total of 240 GDSL-like genes were identified from the genome of B. napus cultivar ZS11. A systematic analysis of the BnGDSL gene family, including phylogenetic relationships, chromosomal location, gene synteny, gene and protein structure, including the cis-elements present in the promoters, and expression profiles in different tissues and in response to different abiotic stresses was conducted to gain insights into the evolutionary and functional characteristics of the BnGELP gene family.
Materials and methods
Identification of the GELP gene family in Brassica napus
Protein sequences of the B. napus cultivar (ZS11) were download from the National Genomics Data Center (NGDC) (accession number: PRJCA002883) (Chen et al., 2021), and a local protein database was established using Hidden Markov model (HMM) profiles as seeds. The putative GELP protein family in B. napus (BnGELP) was predicted from ZS11 with the HMM profile corresponding to the Pfam GDSL domain (PF00657) by running the HMMER 3.0 software (with E-value < e–10). The preliminarily searched candidate protein sequences were then submitted to the Pfam website, the NCBI-CDD server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and the SMART (Simple Modular Architecture Research Tool, http://smart.embl-heidelberg.de/) database for GDSL domain verification as previously described (Song et al., 2022).
Analysis of predicted GELP proteins
The full-length protein sequences of the GELPs from B. napus cultivar ZS11 and A. thaliana were aligned by the MUSCLE tool, and a phylogenetic tree was generated using MEGA 11 with the neighbor-joining method (bootstrap replications, n=1000). Visualization of the phylogenetic tree was performed with FigTree v1.4.4 software. The online program EXPASY (https://web.expasy.org/protparam/) was used to calculate the molecular weight, theoretical isoelectric point (pI) and instability index of the BnGELP protein family members. The online software Wolfpsort (https://wolfpsort.hgc.jp/) was used for signal peptide and subcellular localization prediction of the BnGELP proteins. Conserved motifs and protein architecture were predicted by the on-line tool MEME (Multiple Em for Motif Elicitation, https://meme-suite.org/) and visualized by TBtools software (Bai et al., 2016; Chen et al., 2020).
Genome-scale synteny analysis of GELP genes
All 240 BnGLEP and 105 AtGELP protein sequences were aligned using BLASTP with a threshold value of e–10. Duplications of the BnGELP genes were determined using BLAST and MCScanX with default parameters, and divided into tandem and segmental duplications as previously reported (Chen et al., 2020; Song et al., 2022). Similarly, the synteny between the AtGELP and BnGELP genes were also examined using TBtools. Collinearity diagrams were drawn as previously reported (Chen et al., 2020; Song et al., 2022).
Chromosome localization and gene structure analyses
The physical positional information of each gene (GFF file and genome file) in B. napus cultivar ZS11 was downloaded from NGDC (accession number: PRJCA002883). Distribution of the BnGELP genes on the 19 chromosomes of B. napus were then deciphered with Mapchart, as previously described (Chen et al., 2020). The predicted introns and exons of the clade 5 BnGELP genes were extracted from the GFF file, and their intron/exon structures were drawn using Gene Structure View (Advanced) from TB tools as previously described (Song et al., 2022).
Plant growth, cold treatment and seed oil content measurement
Seeds of B. napus cultivar ZS11 and inbred lines C69, A260, Y196, C145, GH124 and Y172 were placed on petri dishes covered with two layers of filter paper, and grown at 24°C. The whole seedlings were harvested when they were around 5 cm height and stored at -80°C. A set of seedlings of ZS11 were cold treated at 4°C and harvested after 0, 12, 24, 36, 48, 60 and 72 hours.
Mature seeds of B. napus cultivar ZS11 and inbred lines C69, A260, Y196, C145, GH124 and Y172 were harvested. Seed oil content (OC) was determined with near-infrared spectroscopy (ANTARIS II, Thermo Scientific™, USA) as previously described (Wang et al., 2021).
Esterase isozyme electrophoresis and mass spectrometry analysis
Seedlings from ZS11 and the inbred lines were ground into homogenate in an ice-water bath with extraction buffer (100 mM Tris-HCl pH 8.0). The homogenate was then centrifuged at 8000 r/min for 10min. The supernatant was transferred into new tubes. Around 20 μg extracted protein was separated by native polyacrylamide gel electrophoresis (resolving gel 8%, stacking gel 3.6%). After electrophoresis, the gel was first stained with an esterase developing solution (an ethanol solution containing 1% α- naphthyl acetate and 2% β- naphthyl mixed with 2% naphtanil diazo blue B dissolved in 50 mM phosphate buffer at pH 6.4). The gel was destained, fixed with 2% acetic acid after each esterase isozyme band of the zymogram was observed and photographed as previously described (Van Xuan and Jin, 2011; Zhu et al., 2017). Each esterase isozyme band in region I of the zymogram was cut and destained. The protein bands were treated with 100 mM DTT and 60 mM iodoacetamide (IAM) and then digested with trypsin overnight at 37°C. The peptides were collected and desalted using C18 tips. The resulting peptides were analyzed by high-sensitivity LC-MS/MS (QExactive HF-X, ThermoFisher, Waltham, USA) for protein identification using Thermo Scientific Proteome Discoverer 2.4 software as previously described (Wang et al., 2022).
Expression profiles of BnGELP genes determined by transcriptome data
RNA-seq data from 12 tissues (root, stem, leaf, flower, silique, sepal, pistil, stamen, ovule, pericarp, wilting pistil, and blossomy pistil) and under abiotic stresses (dehydration, NaCl, ABA and cold conditions) of ZS11 were downloaded from NGDC (accession numbers PRJNA394926 and CA001775) (Sun et al., 2017; Chen et al., 2021). The FastQC tool was used to check the data quality. Trimmomatic (-0.39) was used to filter the data. All of these RNA-seq data were mapped to the reference genome of ZS11 with HISAT2 2.04 software as previously described (Song et al., 2022). Transcript levels of the BnGELP genes were calculated using transcripts per kilobase of exon model per million mapped reads (TPM) values with the StringTie plug-in in TBtools. A histogram was generated via TBtools software as previously described (Song et al., 2022).
Identification of cis-elements in the promoters of the clade 5 BnGELP genes
To cis-regulatory elements prediction, 2-kb sequences upstream from the start codon of each clade 5 BnGELP gene were extracted from the B. napus cultivar ZS11 genome database. The cis-regulatory elements in the promoters were predicted with the online website PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The obtained results were filtered, categorized, and then visualized using TBtools as previously described (Li et al., 2019; Su et al., 2020).
Cold-reponsive expression of BnGELP genes encoding the proteins identified by mass spectrometry
The expression of the BnGELP genes encoding the proteins from the zymograms and identified by mass spectrometry were determined in response to cold. One-week-old seedlings were moved to a chamber held 4 °C and sampled at 0, 12, 24, 36, 48, 60 and 72 hours. The total RNA was extracted with TRIzol reagent (TaKaRa). The complementary DNA (cDNA) was obtained after reverse transcription with PrimeScript™ RT Master Mix (Perfect Real Time, TaKaRa). Semi-quantitative RT-PCR was performed and normalized to ZS11C02G003910, which is an actin-like gene in B. napus, as previously described (Bai et al., 2016; Li et al., 2018).
Results
Genome-wide identification of BnGELP genes
The HMMER search found 240 putative BnGELP proteins with conserved GDSL-like lipase domains from the genome of B. napus cultivar ZS11, with 119 BnGELP genes encoded in the A subgenome and 121 genes in the C subgenome (Supplementary Table 1). This number of BnGELP proteins is around 2.3-fold more than encoded in A. thaliana. The BnGELPs were named in terms of their location on the chromosomes (Supplementary Table 1). The protein length of the predicted BnGELPs ranged from 158 to 1400 amino acids (aa), with predicted molecular weights (Mw) from 17.16 to 154.33 kDa. The theoretical isoelectric points (pI) ranged from 4.67 to 10.11 (Supplementary Table 1). The isoelectric points of 140 of the BnGELPs (58.33%) were greater than 7. Other characteristics of the BnGELPs, such as gene position, predicted subcellular localization, instability index and accession number, are presented in Supplementary Table 1.
Classification and phylogenetic analyses of BnGELP proteins
To explore the phylogenetic relationships of the GELPs in A. thaliana and B. napus, the protein sequences of 240 BnGELPs and 105 AtGELPs were aligned by the MUSCLE tool, and a phylogenetic tree was generated using MEGA 11 with the neighbor-joining method. As shown in Figure 1, the BnGELP proteins were classified into 5 clades, including 11 in clade 1, 72 in clade 2, 49 in clade 3, 38 in clade 4 and 70 in clade 5 (Supplementary Table 2). The four strictly invariant residues in the blocks of the GDSL motif were present in almost all 240 BnGELP proteins (Figure 2). Clades 2 (72) and 5 (70) were the two largest, and together represented around 60% of all BnGELP family members. In contrast, clade 1 had the fewest members (Supplementary Table 2).
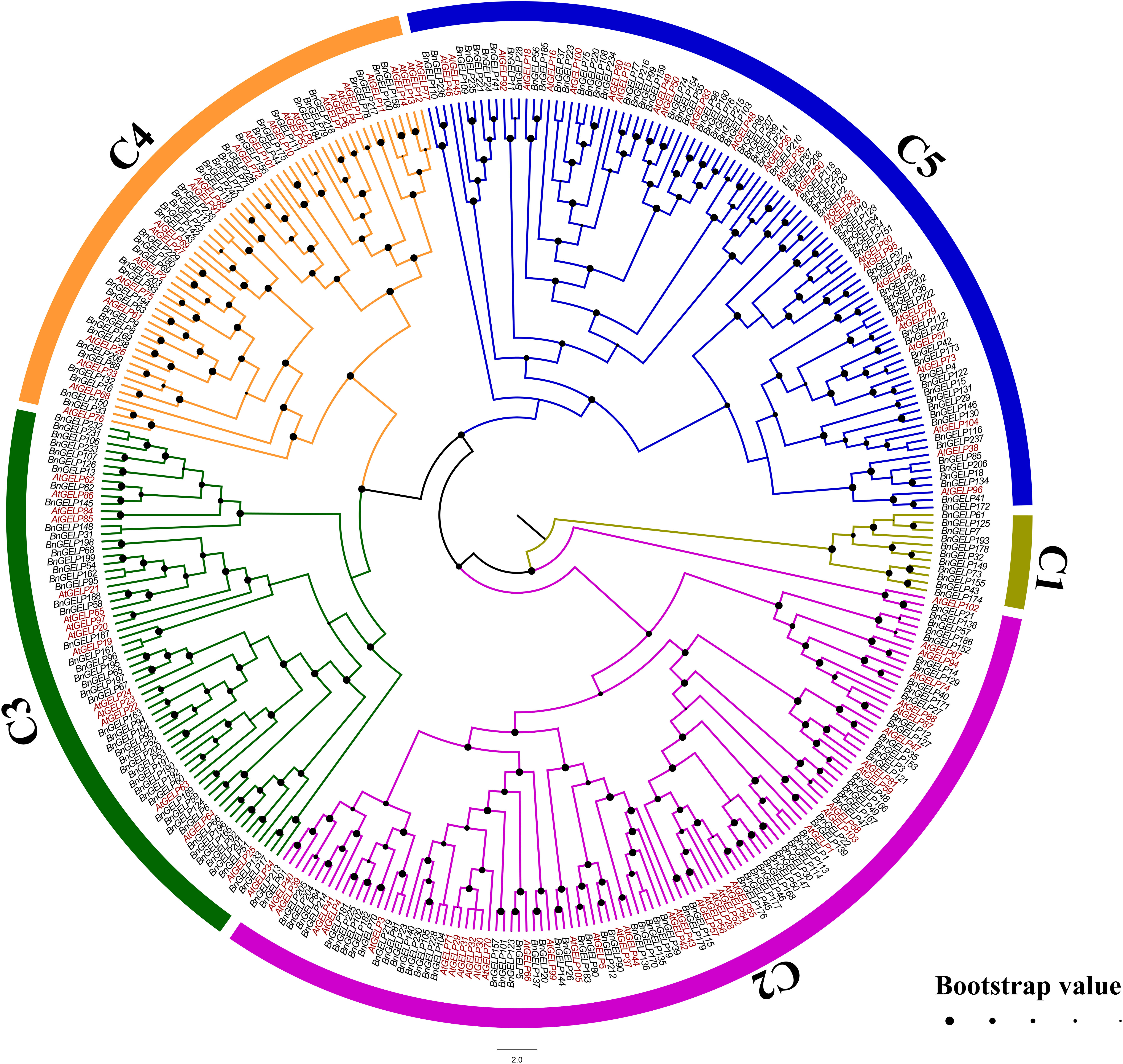
Figure 1 Phylogenetic analysis of GSDL-type Esterases/Lipases from B. napus and A. thaliana. The sequences of 240 BnGELP and 105 AtGELP proteins were aligned by the MUSCLE tool. A phylogenetic tree was generated using MEGA 11 with the neighbor-joining (NJ) method (bootstrap replications, n=1000). The phylogenetic tree was highlighted with FigTree (version 1.4.4). The proteins are clustered into 5 distinct clades designated with a clade number (C1~C5) and labelled with different colors.
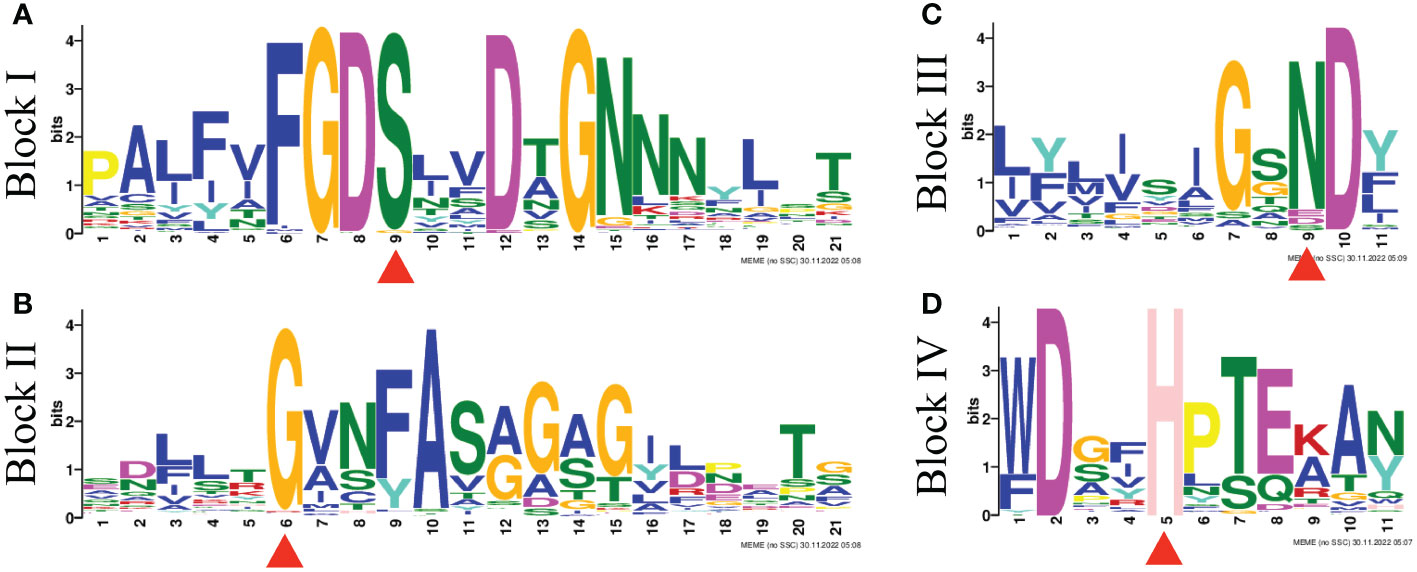
Figure 2 GDSL motif logos of four conserved blocks found in BnGELP proteins. The motif logogram was determined using MEME. The conserved residues Ser, Gly, Asn, and His are in blocks I (A), II (B), III (C) and V (D) all BnGELP proteins and are indicated with red triangles.
Chromosomal distribution of BnGELP genes
The 240 BnGELP genes were mapped onto the 19 chromosomes of B. napus (Supplementary Figure 1). There were 119 BnGELP genes anchored on chromosomes A01 to A10 and 121 BnGELP genes on chromosomes C01 to C09. The BnGELP genes were unevenly scattered on each chromosome at different distribution densities. The chromosomes C03 (21 members), C06 (20 members) and C02 (19 members) had a relatively higher density of BnGELP genes, while chromosomes C08 (4 members), A10 (5 members) and C01 (6 members) had fewer BnGELP genes (Supplementary Figure 1 and Supplementary Table 1).
Gene duplication and collinearity survey of the BnGELP genes
To ascertain the evolution of the BnGELP genes, we first determined the synteny between B. napus and A. thaliana at the whole genome level. A total of 138 collinear gene pairs were observed between the two genomes (Figure 3 and Supplementary Table 3). Most (36 out of 73) AtGELP genes had multiple syntenic genes in B. napus. For instance, both AtGELP22 (At1g54000) and AtGELP23 (At3G14210) had 6 collinear BnGELP genes. However, there were 37 AtGELP genes that had only one syntenic BnGELP gene (Supplementary Table 4).
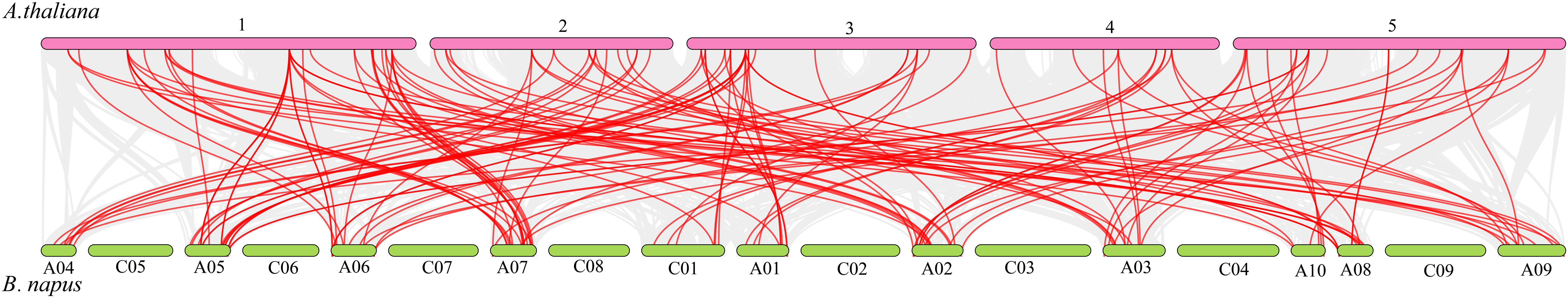
Figure 3 Synteny evaluation of AtGELP and BnGELP genes. The gray lines indicate all collinear blocks, and the red lines highlight the orthologous relationships between AtGELPs and BnGELPs.
To gain an insight into the synteny, we determined the putative tandem and segmental duplication events of the BnGELP family members using BLAST and MCScanX. A total of 2 tandem duplication events and 280 segmental duplications were observed in the B. napus genome (Supplementary Figure 2 and Supplementary Table 3). Only 9 segmental duplication events were observed within the same chromosome, and three of them were observed on chromosome C06. In total, 271 segmental duplications were detected across the chromosomes (Supplementary Table 3). Further analysis revealed that there were 47 duplication events within the A subgenome, 48 events within the C subgenome, and 187 events between the A and C subgenomes (Supplementary Table 3). In terms of the U triangle, allopolyploidization may have played a role in the expansion of the BnGELPs gene family in B. napus.
Several BnGELP proteins are related to esterase isozyme activity during early seedling development
The level of oil content in B. napus is generally correlated with the dynamic balance between lipid biosynthesis and metabolism (Wang et al., 2021). Esterases or lipases are involved in hydrolyzing the stored triacylglycerols and converting them into hexoses and sucrose for the establishment of seedlings (Ding et al., 2019a; Wang et al., 2021). To explore the putative interrelationships between oil content and esterase isozyme activity, we electrophoresed the enzyme extracts from the seeds of different B. napus cultivars and inbred lines with distinct oil content profiles (Figure 4). There were several differences in region I of the zymogram among the different lines. The banding patterns of two high oil content (OC) inbred lines, Y172 (approximately 47% OC) and GH124 (approximately 51% OC), were different than the zymogram of cultivar ZS11 (approximately 49% OC). In contrast to ZS11, GH124 had weak third and fourth bands in region I. For Y172, the fourth band in region I was weak, while the first and second bands were stronger than in ZS11 and GH124 (Figure 4B). Moreover, three inbred lines with low oil content, C69 (approximately 35% OC), A260 (approximately 38% OC) and C145 (approximately 30% OC), also had different banding patterns in the zymogram compared to cultivar ZS11. Compared with ZS11, the third and fourth bands in region I were weak in C69, A260 and C145. However, the first and second bands in C69, A260 and C145 were stronger than in ZS11. Surprisingly, the fifth band in region I was not observed in C145 (Figure 4B).
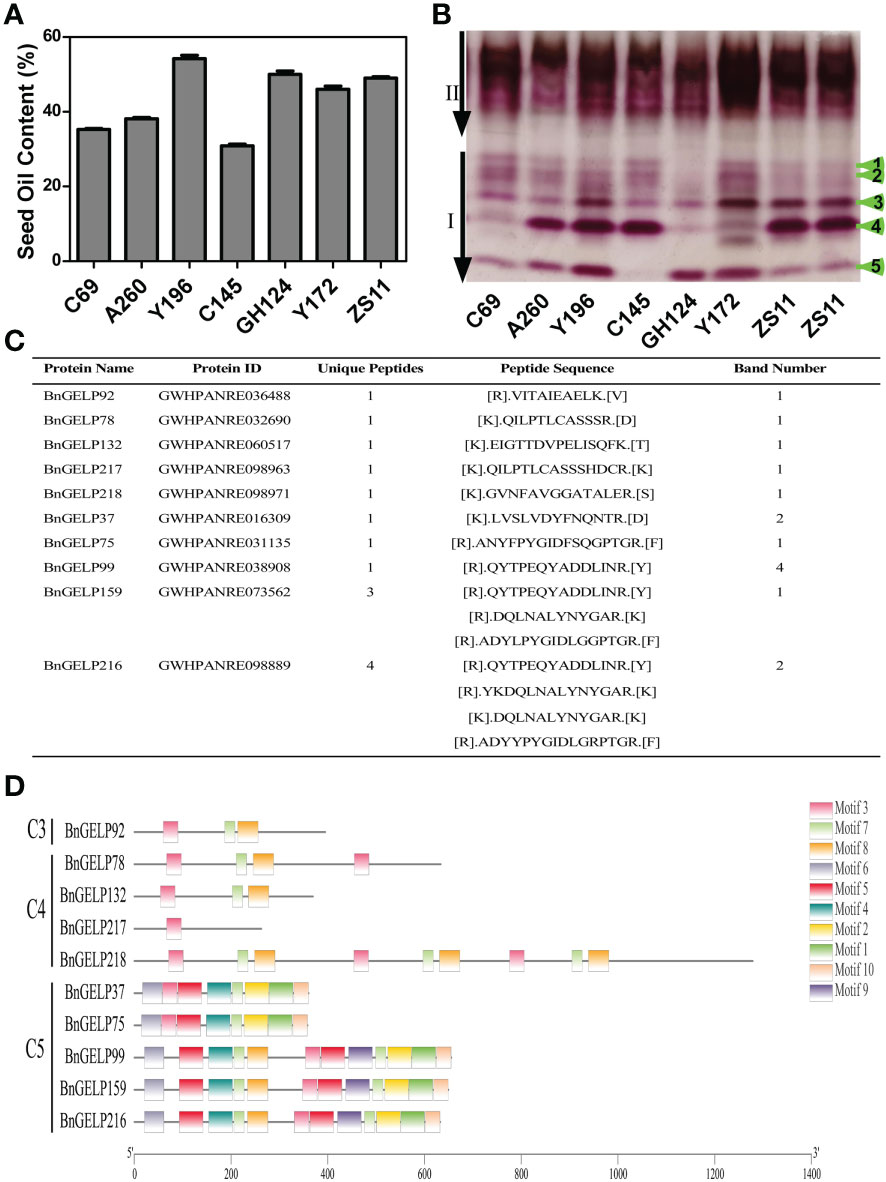
Figure 4 A zymogram of esterase isozymes in different B. napus materials. The seeds were from cultivar ZS11and inbred lines C69, A260, Y196, C145, GH124 and Y172. (A) Seed oil content of the B. napus materials. (B) The zymogram of esterase isozymes in different B. napus materials. (C) Mass spectrometric identification of different bands in region I of the zymogram in (B). Selected unique proteins and the corresponding sequences are indicated. (D) Conserved motifs of the identified BnGELP proteins from region I of the zymogram. Boxes of different colors represent different motifs.
To examine the possible esterases or lipases related to the observed differences in the zymogram, the five indicated bands were cut from the polyacrylamide gel for identification of the proteins via mass spectrometry. Since there are many esterases or lipases associated with lipid metabolism during germination, we performed Gene Ontology (GO) enrichment analysis of the complete portfolio of the identified proteins with respect to esterases or lipases (Supplementary Table 5). One clade 3 BnGELP protein (BnGELP92), four clade 4 BnGELP proteins (BnGELP78, 132, 217 and 218), and five clade 5 BnGELP proteins (BnGELP37, 75, 99, 159 and 216) were identified by at least one unique peptide (Figures 4C, D and Supplementary Tables 5, 6). The identified BnGELP proteins falls into three major bands in region I of the zymogram: six were identified from the first band (BnGELP37, 75, 78, 159, 217 and 218), two from the second band (BnGELP37 and 216), and one from the fourth band (BnGELP99, Figure 4C and Supplementary Table 5).
Gene and protein structure analyses of BnGELPs in the clade 5
Since five of the ten identified BnGELP proteins in region I of the zymogram were from clade 5 (Figure 4 and Supplementary Figure 3), we chose to systemically analyze the 70 proteins in this clade. We first analyzed the chromosomal localization of the clade 5 BnGELP genes. Among the 70- BnGELP genes of clade 5, 36 were encoded on the A subgenome chromosomes and 34 on the C subgenome chromosomes (Supplementary Figure 1). We also determined the overall exon/intron profile of the BnGELP genes within clade 5 (Figure 5). All BnGELP genes were interrupted by at least one intron. BnGELP146 contained the most introns (15 introns), while BnGELP221 had the fewest (one intron). More than 58% of the BnGELP genes (41 genes) in this clade were interrupted by four introns and composed of five exons in their coding regions. Paralogous genes in the phylogenetic tree often had similar gene structureS (Figure 5). All four members of the BnGELP159 subclade (BnGELP77, 216, 99 and 159), two members of the BnGELP37 subclade (BnGELP37 and 223) and four members of the BnGELP75 subclade (BnGELP75, 108, 220 and 234) showed highly similar exon/intron arrangements (Figure 5). We also examined the protein architecture of the five identified BnGELP proteins in clade 5 (BnGELP37, 75, 99, 159 and 216). Consistent with the gene structure, almost all of the homologous proteins in the same subclade displayed similar arrangement of protein motifs (Supplementary Figures 4, 5 and Supplementary Table 7).
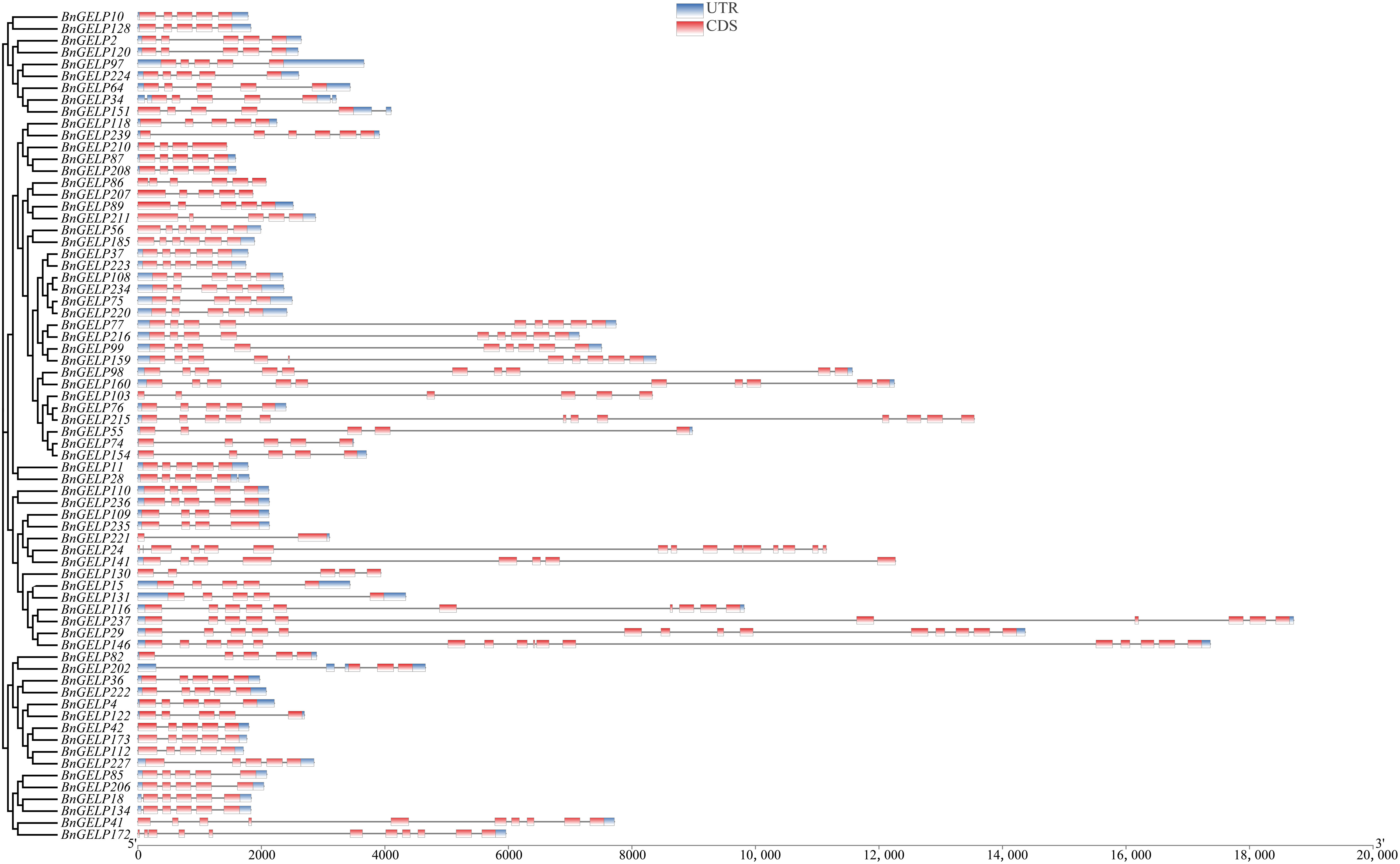
Figure 5 Schematic exon/intron structures of the clade 5 BnGELP genes. The red boxes represent exons and black lines represent introns. The UTR regions are indicated in blue boxes. The length of each CDS can be estimated by the scale at the bottom.
Expression patterns of the clade 5 BnGELP genes across different tissues
The expression profiles of all 70 BnGELP genes in clade 5 were determined in 12 different tissues (stem, sepal, pistil, stamen, ovule, pericarp, blossomy pistil, wilting pistil, root, flower, leaf and silique; Supplementary Figure 6), using publicly available RNA-seq data from ZS11 plants (Song et al., 2022). All genes were detected in at least one tissue (Figure 6 and Supplementary Table 8). The gene BnGELP55 was specifically expressed in root, whereas 15 genes were detected in all of the analyzed tissues (Figure 6 and Supplementary Table 8). Forty-nine genes were detected in leaf. The expression of the five identified BnGELP genes in clade 5 (BnGELP37, 75, 99, 159 and 216) was diverse. BnGELP159 displayed higher expression levels in sepal and blossomy pistil, while its close paralogs, BnGELP99 and 216, showed higher levels in pistil and pericarp, respectively (Figure 6 and Supplementary Table 8). BnGELP37 displayed higher expression levels in sepal, while BnGELP75 showed higher levels in pistil and silique (Figure 6 and Supplementary Table 8). These expression patterns suggested that the five BnGELP proteins from region I of the zymogram in the clade 5 may have distinct roles in different tissues.
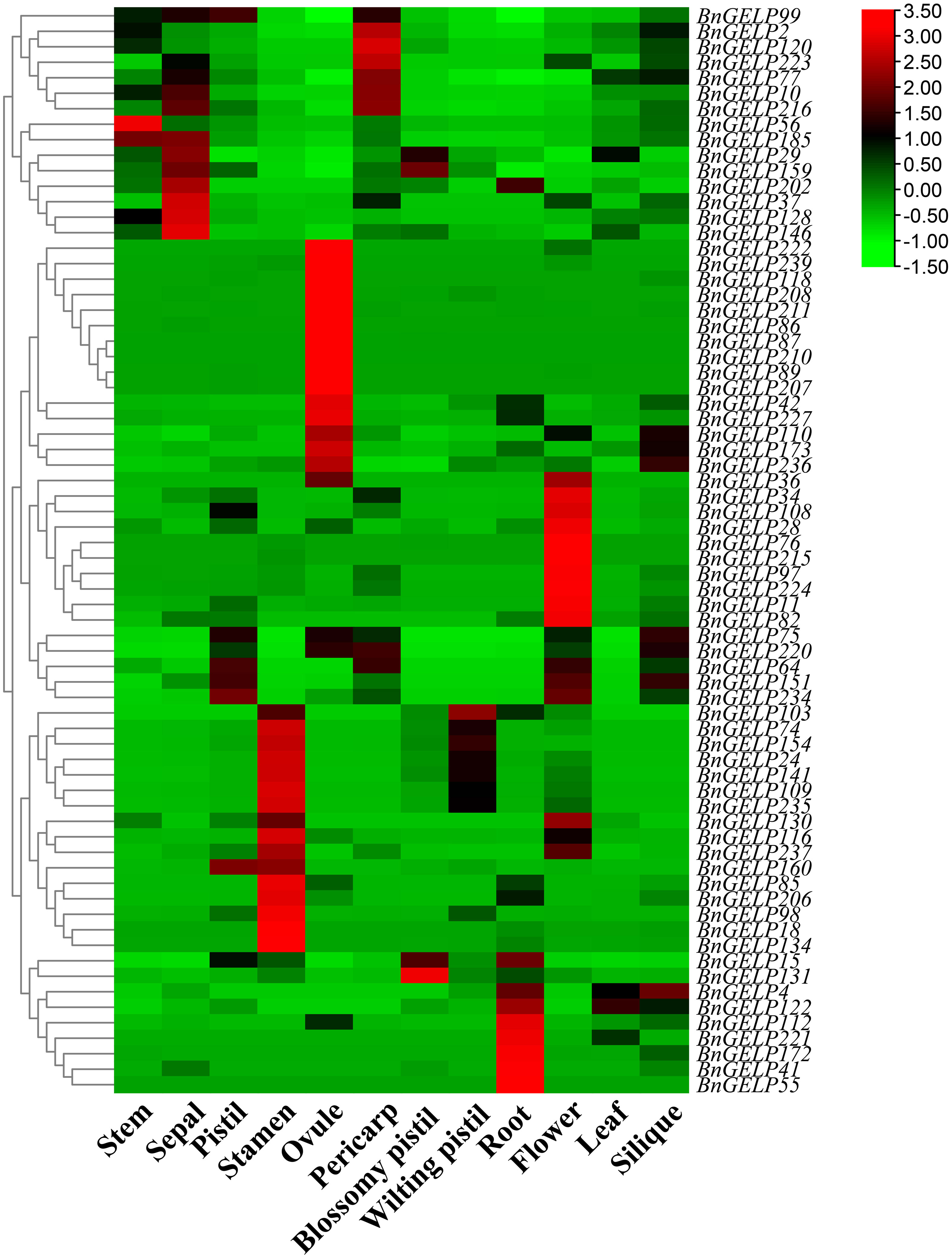
Figure 6 Expression profiles of the clade 5 BnGELP genes in different tissues. Transcript levels for the 70 BnGELP genes were obtained from public transcriptome data obtained from different tissues of B. napus cultivar ZS11. Each relative expression level is equal to the mean values and transformed log2 values for normalization. The color scale represents relative expression levels from low (green colored) to high (red colored).
Expression profiles of the clade 5 BnGELP genes in response to abiotic stresses
To further examine whether the expression levels of the BnGELP genes in the clade 5 were affected by abiotic stresses, we studied the expression profiles of all BnGELP genes under different abiotic stresses (dehydration, NaCl, ABA and cold conditions, Supplementary Figure 7) using the published RNA-seq data (Song et al., 2022). The transcript levels of most of the clade 5 BnGELP genes (60 out of 70) were induced in response to at least one analyzed treatment, while 11 genes were repressed by at least one analyzed treatment (Figure 7 and Supplementary Table 9). Eight genes were not detected in response to any of the analyzed treatments. Moreover, some genes were significantly induced by certain treatments. For instance, BnGELP208 was induced by all the analyzed treatments, and BnGELP29 and BnGELP146 were induced by cold. In contrast, the levels of BnGELP216 and BnGELP77 were decreased with all analyzed treatments (Figure 7 and Supplementary Table 9). The expression levels of the five identified BnGELP genes in clade 5 (BnGELP37, 75, 99, 159 and 216) in response to abiotic stress were diverse. Expression of BnGELP99 and 159 was induced by NaCl, ABA and cold, while the expression of their close paralog, BnGELP216, was significantly repressed in all analyzed treatments (more than 2-fold, Figure 7 and Supplementary Table 9). Expression of BnGELP37 was slightly induced by dehydration but significantly repressed by ABA and cold. However, the expression of its close paralog, BnGELP75, was only slightly induced by NaCl (no more than 2-fold, Figure 7 and Supplementary Table 9). These expression patterns suggested that the five identified BnGELP proteins in the clade 5 may function distinctly in response to abiotic stresses.
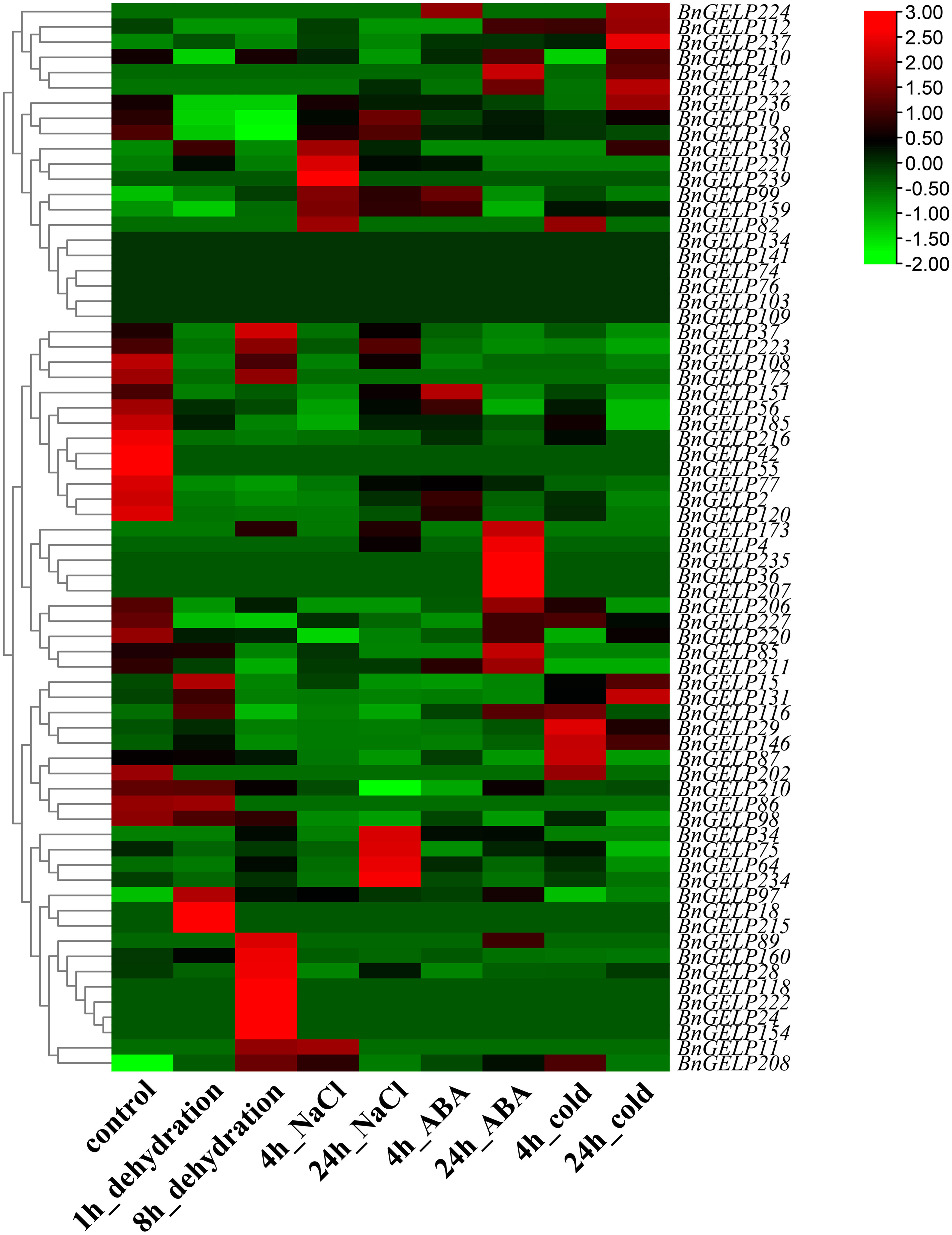
Figure 7 Expression profiles of the clade 5 BnGELP genes under different stresses. Transcript levels for the 70 BnGELP genes were obtained from public transcriptome data obtained from B. napus cultivar ZS11 under different abiotic stresses. Each relative expression level is equal to the mean values and transformed log2 values for normalization. The color scale represents relative expression levels from low (green colored) to high (red colored).
Analysis of cis-acting elements in promoters of the clade 5 BnGELP genes
The cis-acting elements in a promoter can provide clues about the expression profile of the corresponding gene (Shen et al., 2022). As displayed in Figure 8, a total of 12 cis-acting elements were identified in promoters of the 70 BnGELP genes in clade 5. There were cis-acting elements for anaerobic induction in 65 genes and for abscisic acid responsive in 53 genes (Figure 8 and Supplementary Table 10). Forty-eight of the genes had cis-acting elements related to methyl jasmonate (MeJA), while 42 had elements related to low-temperature responses (Figure 8 and Supplementary Table 10). The MYB binding site was present in 45 genes, with 38 that recognized by transcription factors involved in drought response and 7 for those related to flavonoid biosynthesis. Three other hormone-responsive cis-acting regulatory elements were detected in the promoter regions of the clade 5 BnGELP genes, with 41 genes carrying auxin responsive elements, 27 having gibberellin responsive elements, and 31 carrying salicylic acid responsive elements. It is worth noting that 9 out of these 12 cis-acting elements were found in the promoters of the five BnGELP genes in clade 5 (BnGELP37, 75, 99, 159 and 216; Figure 8 and Supplementary Table 10). These included cis-acting elements involved in the responses to abscisic acid, anaerobic induction and low temperatures, suggesting that they may be active during low oxygen or temperature conditions and under the control of plant growth regulators, namely abscisic acid, auxin and gibberellin.
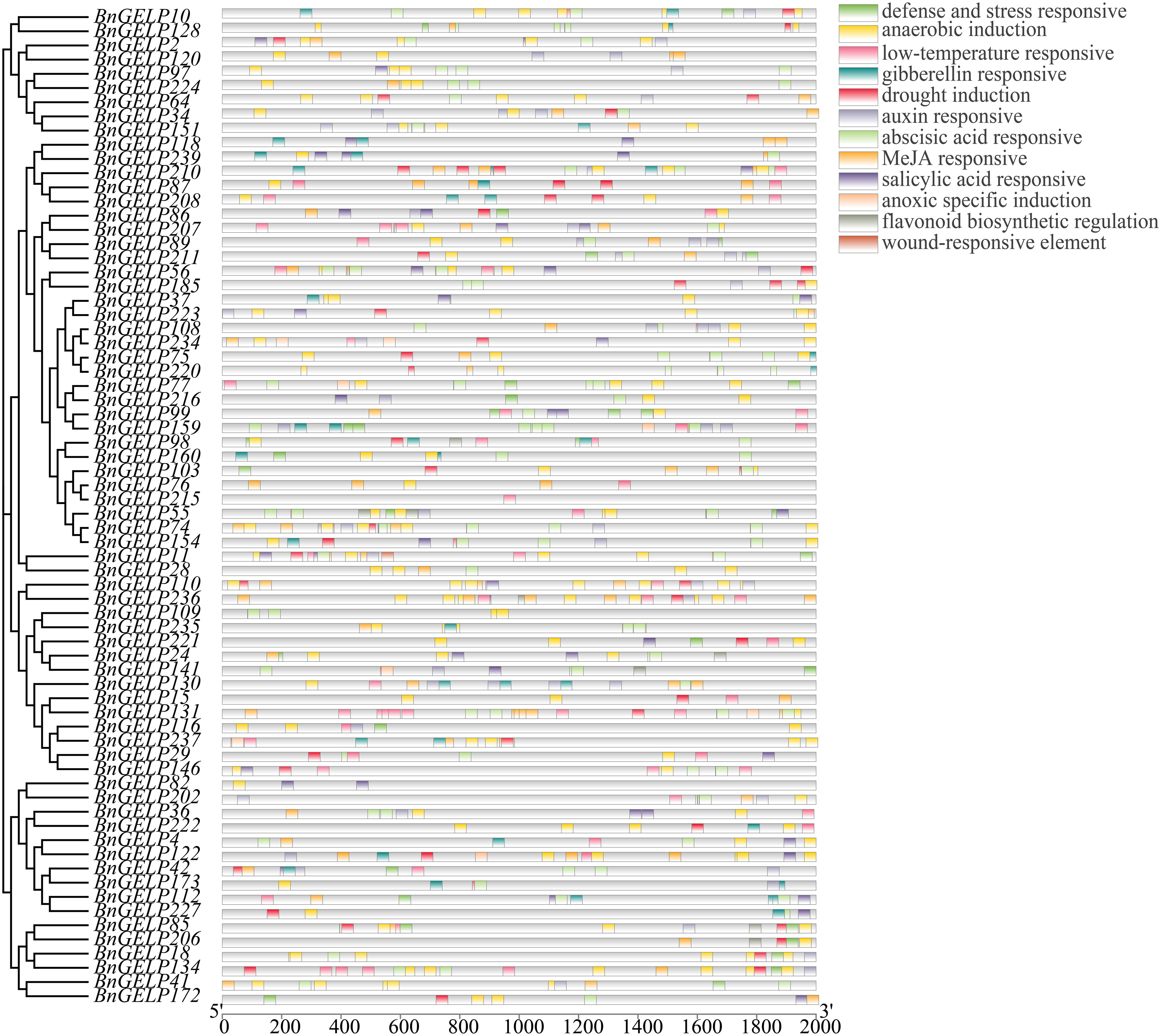
Figure 8 Predicted cis-elements in the promoter regions of the clade 5 BnGELP genes. The name of each regulatory element is shown on the right panel, with different colored boxes representing different cis-elements. The relative location of each element on each promoter can be estimated by the scale at the bottom.
Expression profiles of the BnGELPs identified by mass spectrometry in response to cold
As shown in Figure 8, there were two putative low-temperature responsive elements detected in the promoters of BnGELP99 and 159. To ascertain whether cold affects the expression of the BnGELP genes identified by mass spectrometry, ZS11 was treated with cold, and the expression of them was investigated by qRT-PCR. Inducible expression (more than 2-fold) of BnGELP159 was observed to occur 12 hr following cold treatment, showing a decrease to approximately 1.5-fold at 24 hr, which persisted through 72 hr (Figure 9). Inducible expression (more than 1.5-fold) of BnGELP99 was observed to occur 36 hr following cold treatment (Figure 9). The other two identified BnGELPs, BnGELP92 and 132, do not harbor any low-temperature responsive elements in their promoters (Supplementary Figure 8) and neither were induced by cold (no more than 1.5-fold) during the analyzed treatment time (Figure 9 and Supplementary Figure 8).
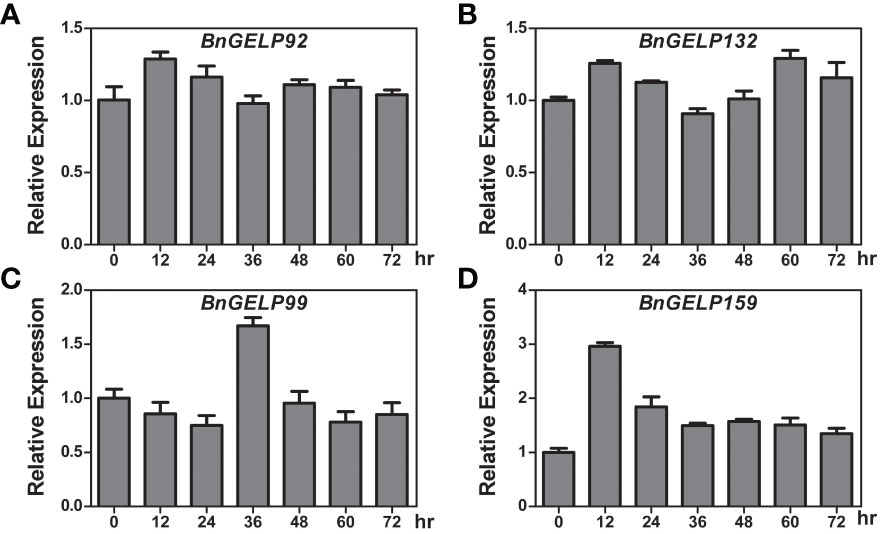
Figure 9 Expression of the BnGELP genes identified by mass spectrometry in response to cold. (A–D) qRT-PCR analysis of the expression of BnGELP92, 132, 99 and 159 in response to cold treatment. Seedlings of B. napus cultivar ZS11 were placed at 4 °C and sampled at 0, 12, 24, 36, 48, 60 and 72 hours. The relative expression levels of BnGELP92, 132, 99 and 159 were normalized to the expression of BnACTIN. The primers are list in Supplementary Table 11.
Because the expression of BnGELP99 and 159 were affected by cold temperature and because of their relationship with the esterase isozyme activity in region I of the zymogram (Figure 9 and Figure 4), we next explored whether cold could impact the esterase activity in ZS11. Cold treatment increased esterase activity in both regions I and II of the zymogram, including the first and second bands in region I (Supplementary Figures 9, 10). These two bands may be related to the identified BnGELP genes in clade 5.
Mobilization of stored lipid and their conversion into soluble sugars (hexoses and sucrose) are crucial for the establishment of seedlings. To explore the putative interrelationships between soluble sugar content and esterase isozyme activity, accumulation of soluble sugars in different B. napus lines and in response to cold was determined. As expected, the soluble sugar contents are higher than control condition after 36 hr following cold treatment (Supplementary Figure 10). Together these results suggested that the cold-inducible expression of the identified BnGELP genes in clade 5 may be correlated with increasing activities of esterase isozymes by cold temperatures.
Discussion
The hydrolysis of the lipid stored in oil bodies by esterase/lipase plays a vital role during seed germination and early post-germinative growth of rapeseed (Ding et al., 2019a; Ding et al., 2019b; Zhan et al., 2022). One class of these esterases/lipases is the GELPs, which have been shown to be involved in the hydrolysis of stored lipids in the initial stage of seed germination in rice (Dolui and Vijayaraj, 2020). GELPs have broad substrate specificity and have a high number of family members within a genome. Multiomics technologies, including genomics, transcriptomics, proteomics and metabolomics, provide opportunities to systematically explore all the genes belonging to a specified gene family within a species (Zhan et al., 2022). Numerous GELP family members have been identified in different plant species (Chepyshko et al., 2012; Dong et al., 2016; Lai et al., 2017; Su et al., 2020; Lv et al., 2021; Wang et al., 2021; Zhang et al., 2021; Cenci et al., 2022; Jiao et al., 2022; Song et al., 2022; Zhan et al., 2022). Recently, a high-quality genome assembly and numerous transcriptome data sets have been released for B. napus cultivar ZS11 (Chen et al., 2021; Liu et al., 2021), making identification and expression analysis of genes such as the BnGELPs more feasible.
In the present study, 240 GDSL-like genes were identified from B. napus cultivar ZS11. The BnGELPs were divided into 5 clades based on phylogenetic tree (C1 to C5, Figure 1 and Supplementary Table 2), which is not consistent with previous reports (Volokita et al., 2011; Lai et al., 2017; Karunarathna et al., 2020; Lv et al., 2021; Cenci et al., 2022). The number of BnGELPs was greater than the number identified in other plant species (Volokita et al., 2011; Chepyshko et al., 2012; Dong et al., 2016; Lai et al., 2017; Su et al., 2020; Lv et al., 2021; Zhang et al., 2021; Cenci et al., 2022; Jiao et al., 2022; Song et al., 2022; Zhan et al., 2022). The BnGELP proteins were not equally distributed into the 5 different phylogenetic clades (Figure 1), with the two largest clades (C2 and C5) representing around 60% of the total BnGELP members (Figure 1 and Supplementary Table 2), suggesting they may be highly differentiated in the B. napus genome. Moreover, the diversity of the molecular weight and isoelectric points of the BnGELP proteins may reflect their functional diversity, which may be attributed to differences in amino acid sequences or domains besides the conserved GDSL domains (Figure 2 and Supplementary Figures 4, 5).
GELPs have a wide range of substrate specificities and are capable of mobilizing multiple lipids, making it easy to detect their biochemical activity using a gel essay called a zymogram (Akoh et al., 2004; Zhu et al., 2017; Ding et al., 2019b; Shen et al., 2022). Ten BnGELPs were detected in region I of the zymogram of ZS11 seedling extracts (Figure 4 and Supplementary Figure 3), providing important clues about the role of the BnGELPs in the regulation of seed germination via hydrolysis of the stored lipids. Further motif, gene expression profiles and cis-elements analyses of the BnGELPs in the clade 5 suggested that members of this clade may play different roles in different tissues and in response to different abiotic stresses. Numerous esterases/lipases have been reported to be involved in the hydrolysis of stored lipids in the initial stage of seed germination, such as serine hydrolases (SHs), GDSL-type esterases/lipases, proteases, Ser carboxypeptidases, ABHD protein, and pectin acetylesterase (Dolui and Vijayaraj, 2020). Interestingly, we also detected other esterases/lipases besides GDSL-type proteins by mass spectrometric analysis (Supplementary Table 5), implying that there should be more esterases/lipases involved in the hydrolysis of stored lipids in the initial stage of seed germination.
A total of 12 cis-acting elements were identified in promoters of the BnGELP genes in clade 5 (Figure 8 and Supplementary Table 10), suggesting that their expression may be induced by low oxygen levels, low temperatures, and abscisic acid, auxin and gibberellin. It is worth noting that most of the clade 5 BnGELP genes (65 out of 70) harbored elements related to anaerobic induction. Any flooding stress during the sowing season may deprive the plant roots of oxygen, which can lead to an adjustment in the expression of genes related to seed germination and early seedling establishment, i.e., genes controlling lipid metabolism, glycolysis, energy metabolism, signal transduction, and photosynthesis. However, no detailed reports have linked the regulation of anaerobic induction and seed germination with respect to fatty acid metabolism. Moreover, a slight cold-inducible expression of BnGELP99 and 159 was observed (Figure 9), which may be attributed to two cis-acting regulatory elements predicted to be involved in low-temperature responsiveness present in their promoters. An increase in the activity of esterase isozymes by cold was also observed (Supplementary Figure 10), which may corelate with other cold-inducible esterase/lipase genes besides our identified BnGELPs. Nevertheless, these hypotheses need to be further verified by conducting further genetic and biochemical studies.
Conclusion
In the current study, a total of 240 GDSL-like genes were identified from B. napus cultivar ZS11. A systematic analysis of the BnGDSL gene family, including phylogenetic relationships, chromosomal location, gene synteny, protein and gene architectures, expression profiles in different tissues and in response to different abiotic stress, as well as cis-elements in the promoters, was conducted to gain insights into the evolutionary and functional characteristics of the clade 5 BnGELPs. Moreover, BnGELPs in clade 5 were linked to the esterase activity in an isozyme zymogram during early seedling establishment via mass spectrometry. In addition, expression of BnGELP99 and 159 may be correlated with the increase in activity of esterase isozymes in response to cold.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
CX and YZ conceived the project and designed the experiment plans. YD, LX, JX, TJ, XT and YW conducted the experiments. CX and YZ analyzed the data and wrote the article. SH, WH and XZ reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the State Basic Research Project from the Department of Science and Technology of Shaanxi Province (2021JM-086), by the State Key Research and Development Project from the Department of Science and Technology of Shaanxi Province (2023-YBNY-023 & 2018NY-097), by the Crop Breeding Key Project of “Two Chains” Fusion of Shaanxi Province (2021LLRH-07), by the Key Research and Development Program of Yangling Seed Industry Innovation Center (Ylzy-yc-2021-02) and by the Science and Innovation Project to Undergraduates by College of Life Sciences, Northwest A&F University.
Acknowledgments
We are grateful to Drs Wang Qinhu and Pei Guoliang (Northwest A&F University) for help with bioinformatics analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1139972/full#supplementary-material
References
Akoh, C. C., Lee, G. C., Liaw, Y. C., Huang, T. H., Shaw, J. F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43 (6), 534–552. doi: 10.1016/j.plipres.2004.09.002
Bai, Y., Zhu, W., Hu, X., Sun, C., Li, Y., Wang, D., et al. (2016). Genome-wide analysis of the bZIP gene family identifies two ABI5-like bZIP transcription factors, BrABI5a and BrABI5b, as positive modulators of ABA signalling in Chinese cabbage. PloS One 11 (7), e0158966. doi: 10.1371/journal.pone.0158966
Bayer, P. E., Hurgobin, B., Golicz, A. A., Chan, C. K., Yuan, Y., Lee, H., et al. (2017). Assembly and comparison of two closely related Brassica napus genomes. Plant Biotechnol. J. 15 (12), 1602–1610. doi: 10.1111/pbi.12742
Cenci, A., Concepcion-Hernandez, M., Guignon, V., Angenon, G., Rouard, M. (2022). Genome-wide classification and phylogenetic analyses of the GDSL-type esterase/lipase (GELP) family in flowering plants. Int. J. Mol. Sci. 23 (20), 12114. doi: 10.3390/ijms232012114
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345 (6199), 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Tong, C., Zhang, X., Song, A., Hu, M., Dong, W., et al. (2021). A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 19 (3), 615–630. doi: 10.1111/pbi.13493
Chepyshko, H., Lai, C. P., Huang, L. M., Liu, J. H., Shaw, J. F. (2012). Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa l. japonica) genome: new insights from bioinformatics analysis. BMC Genomics 13, 309. doi: 10.1186/1471-2164-13-309
Clauss, K., Baumert, A., Nimtz, M., Milkowski, C., Strack, D. (2008). Role of a GDSL lipase-like protein as sinapine esterase in brassicaceae. Plant J. 53 (5), 802–813. doi: 10.1111/j.1365-313X.2007.03374.x
Ding, L. N., Guo, X. J., Li, M., Fu, Z. L., Yan, S. Z., Zhu, K. M., et al. (2019a). Improving seed germination and oil contents by regulating the GDSL transcriptional level in Brassica napus. Plant Cell Rep. 38 (2), 243–253. doi: 10.1007/s00299-018-2365-7
Ding, L. N., Li, M., Wang, W. J., Cao, J., Wang, Z., Zhu, K. M., et al. (2019b). Advances in plant GDSL lipases: from sequences to functional mechanisms. Acta Physiologiae Plantarum 41, 151. doi: 10.1007/s11738-019-2944-4
Dolui, A. K., Vijayaraj, P. (2020). Functional omics identifies serine hydrolases that mobilize storage lipids during rice seed germination. Plant Physiol. 184 (2), 693–708. doi: 10.1104/pp.20.00268
Dong, X., Yi, H., Han, C. T., Nou, I. S., Hur, Y. (2016). GDSL esterase/lipase genes in Brassica rapa l.: genome-wide identification and expression analysis. Mol. Genet. Genomics 291 (2), 531–542. doi: 10.1007/s00438-015-1123-6
Jiao, Y., Zhang, J., Pan, C. (2022). Genome-wide analysis of the GDSL genes in pecan (Carya illinoensis k. koch): phylogeny, structure, promoter cis-elements, co-expression networks, and response to salt stresses. Genes (Basel) 13 (7), 1103. doi: 10.3390/genes13071103
Karunarathna, N. L., Wang, H., Harloff, H. J., Jiang, L., Jung, C. (2020). Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 18 (11), 2251–2266. doi: 10.1111/pbi.13381
Lai, C. P., Huang, L. M., Chen, L. O., Chan, M. T., Shaw, J. F. (2017). Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol. Biol. 95 (1-2), 181–197. doi: 10.1007/s11103-017-0648-y
Li, H., Han, X., Qiu, W., Xu, D., Wang, Y., Yu, M., et al. (2019). Identification and expression analysis of the GDSL esterase/lipase family genes, and the characterization of SaGLIP8 in Sedum alfredii hance under cadmium stress. PeerJ 7, e6741. doi: 10.7717/peerj.6741
Li, G., Hu, X., Hou, L., Cao, L., Wang, Q., Wang, D., et al. (2018). Molecular identification of BrHAB2a, one of the two AtHAB2-like proteins in Brassica rapa, is an important component of ABA signaling. Biochem. Biophys. Res. Commun. 503 (2), 495–500. doi: 10.1016/j.bbrc.2018.04.185
Ling, H., Zhao, J., Zuo, K., Qiu, C., Yao, H., Qin, J., et al. (2006). Isolation and expression analysis of a GDSL-like lipase gene from Brassica napus l. J. Biochem. Mol. Biol. 39 (3), 297–303. doi: 10.5483/bmbrep.2006.39.3.297
Liu, D., Yu, L., Wei, L., Yu, P., Wang, J., Zhao, H., et al. (2021). BnTIR: an online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 19 (10), 1895–1897. doi: 10.1111/pbi.13665
Lv, J., Dai, C. B., Wang, W. F., Sun, Y. H. (2021). Genome-wide identification of the tobacco GDSL family and apical meristem-specific expression conferred by the GDSL promoter. BMC Plant Biol. 21 (1), 501. doi: 10.1186/s12870-021-03278-x
Shen, G., Sun, W., Chen, Z., Shi, L., Hong, J., Shi, J. (2022). Plant GDSL esterases/lipases: evolutionary, physiological and molecular functions in plant development. Plants (Basel) 11 (4), 468. doi: 10.3390/plants11040468
Song, M., Linghu, B., Huang, S., Li, F., An, R., Xie, C., et al. (2022). Genome-wide survey of leucine-rich repeat receptor-like protein kinase genes and CRISPR/Cas9-targeted mutagenesis BnBRI1 in Brassica napus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.865132
Su, H. G., Zhang, X. H., Wang, T. T., Wei, W. L., Wang, Y. X., Chen, J., et al. (2020). Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in soybean. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00726
Sun, F., Fan, G., Hu, Q., Zhou, Y., Guan, M., Tong, C., et al. (2017). The high-quality genome of Brassica napus cultivar 'ZS11' reveals the introgression history in semi-winter morphotype. Plant J. 92 (3), 452–468. doi: 10.1111/tpj.13669
Van Xuan, D., Jin, I. (2011). Relationship between esterase isozymes and some agronomic traits in F2 populations derived from the crossing of milyang 23 and ashahi. J. Crop Sci. Biotechnol. 14, 11–15. doi: 10.1007/s12892-010-0001-3
Volokita, M., Rosilio-Brami, T., Rivkin, N., Zik, M. (2011). Combining comparative sequence and genomic data to ascertain phylogenetic relationships and explore the evolution of the large GDSL-lipase family in land plants. Mol. Biol. Evol. 28 (1), 551–565. doi: 10.1093/molbev/msq226
Wang, N., Tang, C., Fan, X., He, M., Gan, P., Zhang, S., et al. (2022). Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185 (16), 2961–2974.e2919. doi: 10.1016/j.cell.2022.06.027
Wang, H., Wang, Q., Pak, H., Yan, T., Chen, M., Chen, X., et al. (2021). Genome-wide association study reveals a patatin-like lipase relating to the reduction of seed oil content in Brassica napus. BMC Plant Biol. 21 (1), 6. doi: 10.1186/s12870-020-02774-w
Zhan, X., Qian, Y., Mao, B. (2022). Identification of two GDSL-type esterase/lipase genes related to tissue-specific lipolysis in Dendrobium catenatum by multi-omics analysis. Life (Basel) 12 (10), 1563. doi: 10.3390/life12101563
Zhang, H., Zhang, X., Zhao, J., Sun, L., Wang, H., Zhu, Y., et al. (2021). Genome-wide identification of GDSL-type esterase/lipase gene family in Dasypyrum villosum l. reveals that DvGELP53 is related to BSMV infection. Int. J. Mol. Sci. 22 (22), 12317. doi: 10.3390/ijms222212317
Keywords: Gly-Asp-Ser-Leu (GDSL)-type esterases/lipases, GELP, lipid mobilization, Brassica napus, seed germination
Citation: Ding Y, Xing L, Xu J, Jiang T, Tang X, Wang Y, Huang S, Hao W, Zhou X, Zhang Y and Xie CG (2023) Genome-wide exploration of the GDSL-type esterase/lipase gene family in rapeseed reveals several BnGELP proteins active during early seedling development. Front. Plant Sci. 14:1139972. doi: 10.3389/fpls.2023.1139972
Received: 08 January 2023; Accepted: 27 February 2023;
Published: 15 March 2023.
Edited by:
Helin Tan, Nanjing Agricultural University, ChinaReviewed by:
Chuchuan Fan, Huazhong Agricultural University, ChinaCunmin Qu, Southwest University, China
Copyright © 2023 Ding, Xing, Xu, Jiang, Tang, Wang, Huang, Hao, Zhou, Zhang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfeng Zhang, emhhbmd5ZmNsQDE2My5jb20=; Chang Gen Xie, Y2hhbmdlbi54aWVAbndzdWFmLmVkdS5jbg==
†These authors have contributed equally to this work
 Yahui Ding1†
Yahui Ding1† Liwen Xing
Liwen Xing Shuhua Huang
Shuhua Huang Yanfeng Zhang
Yanfeng Zhang Chang Gen Xie
Chang Gen Xie