- 1Centre for Crop Health, Institute for Life Sciences and the Environment, University of Southern Queensland, Toowoomba, QLD, Australia
- 2Centre for Crop & Food Innovation, Murdoch University, Perth, WA, Australia
- 3Department of Agricultural Biotechnology and Molecular Biology, Dr. Rajendra Prasad Central Agricultural University, Pusa, India
- 4School of Agriculture and Environmental Science, Faculty of Health, Engineering and Science, University of Southern Queensland, Toowoomba, QLD, Australia
Pratylenchus thornei is an economically important species of root-lesion nematode adversely affecting chickpea (Cicer arietinum) yields globally. Integration of resistant crops in farming systems is recognised as the most effective and sustainable management strategy for plant-parasitic nematodes. However, breeding for P. thornei resistance in chickpea is limited by the lack of genetic diversity. We deployed a genome-wide association approach to identify genomic regions and candidate genes associated with P. thornei resistance in 285 genetically diverse chickpea accessions. Chickpea accessions were phenotyped for P. thornei resistance in replicated glasshouse experiments performed for two years (2018 and 2020). Whole genome sequencing data comprising 492,849 SNPs were used to implement six multi-locus GWAS models. Fourteen chickpea genotypes were found to be resistant to P. thornei. Of the six multi-locus GWAS methods deployed, FASTmrMLM was found to be the best performing model. In all, 24 significant quantitative trait nucleotides (QTNs) were identified, of which 13 QTNs were associated with lower nematode population density and 11 QTNs with higher nematode population density. These QTNs were distributed across all of the chickpea chromosomes, except chromosome 8. We identified, receptor-linked kinases (RLKs) on chromosomes 1, 4 and 6, GDSL-like Lipase/Acylhydrolase on chromosome 3, Aspartic proteinase-like and Thaumatin-like protein on chromosome 4, AT-hook DNA-binding and HSPRO2 on chromosome 6 as candidate genes for P. thornei resistance in the chickpea reference set. New sources of P. thornei resistant genotypes were identified that can be harnessed into breeding programs and putative candidate P. thornei resistant genes were identified that can be explored further to develop molecular markers and accelerate the incorporation of improved P. thornei resistance into elite chickpea cultivars.
Introduction
Chickpea (Cicer arietinum) is an ancient legume that originated around the time when agriculture began during the Neolithic period in the Fertile Crescent (Abbo et al., 2003) in present day south-east Turkey and adjoining Syria. Domestication activities further led to diversification and adaptation of the crop to suit different environments. Presently, chickpea is an economically important crop, produced in more than 55 countries and ranks third in terms of area under harvest among legumes after soybean and dry beans (FAOSTAT, 2021). It is a climate resilient crop, cultivated worldwide due to its adaptation to arid and semi-arid regions. It is a valuable legume due to its ability to fix atmospheric nitrogen and also to increase biodiversity in cropping systems with cereals when used in rotation to break disease cycles (Mehta, 2014). The root-lesion nematode Pratylenchus thornei is an important pathogen of chickpea worldwide (Zwart et al., 2019). Pratylenchus thornei, using its stylet, a needle like hollow structure, pierces the root cells, enters in, feeds, and releases enzymatic secretions inside the plant cells. It is a migratory endoparasitic nematode and hence can traverse within and outside the root tissue intermittently. Upon infection, Pratylenchus spp. can complete an entire life cycle within the roots. About 4-5 generations of Pratylenchus spp. are completed in a growing season, leading to heavy build-up of the population density, which damages the root tissue and thereby limits the nutrient access to the plant (Fosu-Nyarko and Jones, 2016). As the severity of P. thornei infection increases, plants become prone to secondary infections by soil-borne fungal pathogens including Fusarium oxysporum f. sp. ciceris causing Fusarium wilt (Castillo et al., 1998) and Rhizoctonia bataticola causing dry root rot (Ali, 1997). Economic losses due to P. thornei infestation of up to 30% and 25% have been reported for the world’s largest chickpea producing countries India and Australia, respectively (Ali, 1997; Reen et al., 2014).
Among nematode management practices, breeding for resistance is the most sustainable approach, which is receiving more attention due to increasing climate change and environmental concerns associated with the use of chemical nematicides (Li et al., 2015). The pre-requisite for breeding for disease resistance is the availability of suitable genetic resistance. In the case of chickpea, there are limited sources of P. thornei resistance that have been identified to date, for example; in a study reported by Thompson et al. (2011), out of 182 chickpea accessions from the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), 16 accessions (including 5 wild Cicer spp.) showed resistance or moderate resistance towards P. thornei, with landrace accession ICC11323 showing the lowest population density of P. thornei. Reen et al. (2019) reported accessions of wild Cicer species C. reticulatum and C. echinospermum with improved levels of P. thornei resistance than previously identified by Thompson et al. (2011). Quantitative trait loci for P. thornei resistance have been mapped on Ca7 and Ca4 of C. arietinum in a bi-parental population (Khoo et al., 2021). The limited sources of P. thornei resistance in C. arietinum are partly due to the narrow genetic base of chickpea as a result of the domestication bottleneck (Abbo et al., 2003; Spillane and Gepts, 2000) and genetic drift (Cooper et al., 2000).
In such a scenario, plant genetic resources are an important pool from which novel sources of disease resistance can be identified. The largest collections of chickpea germplasm are held in genebanks at ICRISAT and the International Center for Agricultural Research in the Dry Areas (ICARDA) (Smýkal et al., 2015). In 2001, a core subset consisting of 1,956 chickpea accessions from the total 16,991 accessions in the ICRISAT germplasm collection was developed to capture the genetic diversity of the collection and enhance ultilisation of the germplasm resources for mining traits of interest in chickpea improvement programs (Upadhyaya et al., 2001). Later, a global composite collection of 3,000 chickpea accessions was developed, which consisted of the core subset from ICRISAT and additional accessions selected from the ICARDA genebank based on diversity in phenotypic traits and agroclimatological data linked to geographic origin (Upadhyaya et al., 2006). From the global composite collection, a reference set of 300 accessions, which captured 78% of the genetic diversity in the composite collection, was selected (Upadhyaya et al., 2008). The chickpea reference set provides a practical number of genetically diverse accessions to screen for traits of interest. The predominance of germplasm from Asia in the ICRISAT genebank is reflected in the chickpea reference set, with 66% of the accessions originating from Asia, in particular India and Iran (Upadhyaya et al., 2008). Approximately 90% of accessions in the chickpea reference collection are landraces (Upadhyaya et al., 2008; Varshney et al., 2019). Landraces are primitive varieties with a high capacity to tolerate biotic and abiotic stresses, resulting in high stability and intermediate yield level under low input agricultural systems (Zeven, 1998). Landraces therefore harbor a spectrum of alleles, which if used in breeding programs can help develop agronomically superior cultivars with enhanced stress tolerance and disease resistance.
P. thornei resistance in chickpea is a polygenic trait (Channale et al., 2021) and genome-wide association study (GWAS) serves as an ideal method for genetic dissection of complex traits. GWAS takes advantage of evolutionary recombination events in crops to determine the molecular variations associated with the traits including disease responses (Nordborg and Weigel, 2008; Braulio and Sylvie, 2012). It has been widely used in different crop species to map genomic loci associated with economically important traits (Gangurde et al., 2022). In chickpea, genomic regions associated with Ascochyta blight (Li et al., 2017; Farahani et al., 2022; Raman et al., 2022), Pythium ultimum (Agarwal et al., 2022), nutrient traits (Diapari et al., 2014; Jadhav et al., 2015; Upadhyaya et al., 2016a; Upadhyaya et al., 2016b) and abiotic stress (Thudi et al., 2014) have been mapped using GWAS.
Single nucleotide polymorphism (SNP) markers are popular markers in GWAS due to their genome-wide abundance and flexibility for high- to ultra-high-throughput detection platforms, which enables genome-wide scanning, and fine mapping of target regions (Mammadov et al., 2012). With the advent of low-cost genotyping platforms, GWAS is a preferred method to dissect genetics of resistance in different crop species against different plant parasitic nematodes. To the best of our knowledge P. thornei resistance using GWAS has been reported only in common wheat (Triticum aestivum L.). Kumar et al. (2021) reported nine genes with putative roles in P. thornei resistance in common wheat using pan-Indian wheat germplasm. In addition, Dababat et al. (2016) and Sohail et al. (2022) reported nine and four markers, respectively, linked to P. thornei resistance in spring bread wheat collections held by CIMMYT (International Maize and Wheat Improvement Center). Among other plant parasitic nematode species, marker-trait associations have been identified using GWAS for resistance to root-knot nematode (Meloidogyne javanica) resistance in soybean (Alekcevetch et al., 2021), root-knot nematode (M. graminicola) resistance in Indian wild rice accessions (Hada et al., 2020), and cyst nematode (Heterodera filipjevi) in wheat (Pariyar et al., 2016).
The above-mentioned studies were performed with single-locus and multi-locus models using the Bonferroni correction method. Bonferroni correction is a conservative approach that may result in many important associations being eliminated because they fail to satisfy the stringent criteria of significant tests for complex traits in crops (Wang et al., 2016). Recent multi-locus model methods for GWAS do not require Bonferroni correction and instead use LOD = 3.0 (or P = 0.0002) as a significance cutoff to balance the high power and low false positive rate to detect significant associations (Zhang et al., 2019). Several multi-locus GWAS models have been developed, including multi-locus random-SNP-effect mixed linear model (mrMLM) (Wang et al., 2016), fast multi-locus random-SNP-effect efficient mixed model analysis (FASTmrEMMA) (Wen et al., 2018; Wen et al., 2020), and Iterative modified-sure independence screening expectation-maximization-Bayesian least absolute shrinkage and selection operator (ISIS EM-BLASSO), (Tamba et al., 2017). These multi-locus GWAS models have been shown to have increased statistical power over single locus GWAS methods for the detection of quantitative trait nucleotides (QTNs) of small effect associated with complex traits. For example, candidate genes related to drought stress in cotton (Gossypium hirsutum L.) were identified using a multi-locus method GWAS (Hou et al., 2018). Also, salt tolerant loci were identified in rice (Oryza sativa L.) by using a multi-GWAS approach (Cui et al., 2018).
We evaluated the chickpea reference set (Upadhyaya et al., 2008) for P. thornei resistance in glasshouse experiments in each of two years and identified chickpea lines resistant to P. thornei. Since the chickpea reference set is a global genetic resource that has been phenotyped for different traits, it is of interest to identify accessions that carry multiple disease resistance or other traits of interest. We used reports in the literature to identify accessions in the chickpea reference set with combined resistance to P. thornei and other biotic as well as abiotic stresses. We tested six multi-locus GWAS models to understand the effectiveness of the models and identified robust QTNs associated with P. thornei resistance. Candidate genes for P. thornei resistance were further identified. To the best of our knowledge, this is the first genome-wide association study for P. thornei resistance in chickpea.
Materials and methods
Plant materials
The association panel consisted of a total of 278 chickpea accessions: 276 accessions of the chickpea reference set (Upadhyaya et al., 2006), originating from 32 countries, obtained from the Australian Grains Genebank, Horsham, Victoria, Australia, plus two Australian chickpea varieties (PBA HatTrick and PBA Pistol) (Supplementary Table 1). An additional seven putatively P. thornei resistant C. arietinum accessions (Supplementary Table 2) identified from previous studies were also evaluated (Castillo et al., 2008; Thompson et al., 2011; Rodda et al., 2016), for P. thornei resistance. Well characterized chickpea (9) and wheat (8) lines, covering the range of responses to P. thornei from resistant to very susceptible, were included in the phenotyping experiments as check cultivars. The chickpea check cultivars were Sonali, ILWC 123, ILWC 184, PBA Boundary, Flipper, Yorker, Sona, Kyabra, PBA Seamer. The wheat check cultivars were CP1133872, GS50a, Gauntlet, QT8447, Sunzell, EGA Kidman, Sunguard and Strezlecki. All the accessions were phenotyped twice except for 27 chickpea reference set accessions and three accessions identified as sources of resistance, as they were unavailable at the time of Experiment 1. Unplanted pots inoculated with P. thornei were also included as negative controls.
Glasshouse and plant growth conditions
The association panel and check genotypes were evaluated for P. thornei resistance in inoculated glasshouse pot experiments. Treatments consisted of a single pot (containing one plant of a genotype). Treatments were randomly arranged on glasshouse benches according to a row-column design with three replicates of each treatment. A replicate was equal to one block and each block was allocated to a separate adjacent glasshouse bench. The glasshouse experiment was conducted twice, with Experiment 1 conducted at the Leslie Research Facility, Toowoomba from July to November 2018, and Experiment 2 conducted in a new glasshouse with improved air temperature control at the University of Southern Queensland, Toowoomba from July to November 2020.
All the glasshouse benches were set up with a self-regulating bottom watering system with water flow regulated by a float valve set to a water tension of 20 mm. Capillary matting (Bidim®; Geofabrics Australasia Pty Ltd) was placed over each bench with its edges immersed in the water troughs attached to the bench. Square pots with dimension 70-mm square, 150-mm high, were filled with 330 g (oven-dry equivalent) of a black earth of the Waco clay soil association (Beckmann and Thompson, 1960) pasteurised with aerated steam at 85°C for 30 min (modified from Thompson, 1990). Initially, 80% (252 g) of the total soil was mixed with 30 mL of nutrient solution, to provide 200 mg NO3-N, 25 mg P, 88 mg K, 36 mg Na, 285 mg Ca and 5 mg Zn/kg soil and pots were filled. The pots were placed on the pre-wetted benches and flooded with water to 10-20 mm depth to ensure that capillary action between soil and matting was established. The pots were left overnight to allow the soil to take up water before planting and inoculation. For planting, three seeds of each accession were placed on top of the moist soil and inoculated with 1 mL of commercially available rhizobium culture (Group N, isolate CC1192 [Mesorhizobium ciceri]) prior to inoculation with 3,300 P. thornei (equivalent to 10,000 nematodes/kg oven-dry soil) suspended in 10 mL of water. The remaining 20% (78 g) of soil was placed over the seeds and nematode inoculum as a cap. One week after emergence, plants were thinned to one plant per pot. The extra plants were removed carefully by cutting off the shoot below the crown with a scalpel, leaving the roots in the pot. Air and soil temperatures were maintained between 20 and 25 °C, the optimum temperature for P. thornei reproduction (Thompson et al., 2015), using under-bench heating, shade cloth (as required), evaporative coolers (Experiment 1) and air conditioning (Experiment 2).
Plant harvest, nematode extraction, and enumeration
The plants were grown for 18 weeks. Before harvest, the water supply was turned off to allow the soil to dry down to ~45% moisture content for effective soil sampling and processing for nematode extraction. Plant maturity on the BBCH scale (Lancashire et al., 1991), tiller number, and plant height were recorded at harvest. The plant shoots were cut off at the base just above the soil level, oven dried at 80°C for 48 h and shoot dry biomass was recorded. The pots with soil were packed in polythene bags and stored at 4°C until further processing. The soil and roots from each pot were thoroughly mixed and the roots were cut into approximate 10 mm lengths. The mixture was subsampled with 100 g for determination of gravimetric moisture content and 150 g for nematode extraction. For gravimetric moisture content, the soil mixture was dried at 105°C for 48 h. The nematodes were extracted by the Whitehead tray method (Whitehead and Hemming, 1965). The nematodes were collected on a 20-µm sieve, concentrated in approximately 15 mL water, which was poured into a 30 mL vial and stored at 4°C. Pratylenchus thornei populations extracted from soil and roots were counted under a compound microscope (4x Olympus BX53) using a Peters counting chamber of 1 mL capacity (Chalex Corporation, Portland, Oregon, USA).
Statistical analyses
Due to heterogeneity of variances as assessed by the Shapiro Wilk test for normality, P. thornei counts were transformed by loge (x + 1), where x = number of P. thornei/kg of soil and roots (oven dry equivalent). Pearson’s correlation coefficient was used to determine linear correlation between loge(P. thornei/kg +1) of accessions in Experiment 1 and Experiment 2 in GraphPad Prism (2022). Experiment 1 and Experiment 2 were analyzed together using a linear mixed model (LMM) to obtain best linear unbiased estimates (BLUE) values with the following terms included in the model: genotype as a fixed effect and replication nested in experiment as random effect. Multiple comparisons for REML was applied to obtain Fishers protected least significant differences (l.s.d.) (at 5% significance level). The analyses were performed using Genstat v21 (VSN International, 2020). The range of loge (P. thornei/kg+1) as BLUE values was subdivided into eight equal subranges based on the procedure of Thompson et al. (2020). These subranges were assigned the following ordinal alpha categories, resistant (R), resistant to moderately resistant (R-MR), moderately resistant (MR), moderately resistant to moderately susceptible (MR-MS), moderately susceptible (MS), moderately susceptible to susceptible (MS-S), susceptible (S), susceptible to very susceptible (S-VS) with calibration to this S-VS category based on the wheat cultivar Strzelecki (Thompson et al., 2020). A histogram of frequencies of accessions in these eight classes was produced.
Identification of P. thornei resistant chickpea accessions with resistance to other biotic and/or abiotic stresses
As the chickpea reference collection is a critical resource for studying genetic diversity, these accessions have been phenotyped for various traits by numerous researchers. The chickpea accessions from different phenotyping studies previously reported as R or, MR to the biotic stresses Ascochyta blight, Botrytis grey mould, dry root rot, and Fusarium wilt (Pande et al., 2006), or abiotic stresses (Serraj et al., 2004), or accessions with superior agronomic characteristics (Meena et al., 2010) were compared with the accessions in the current study falling within the categories of R, R-MR, or MR to P. thornei.
Genotyping and population stratification
The SNP dataset for the chickpea reference set germplasm collection was selected for the study as per Varshney et al. (2019). Briefly, the SNP data set for 429 accessions (Varshney et al., 2019) was sorted using the sort tool in the Galaxy Australia web platform at usegalaxy.org.au (Galaxy Community, 2022) to obtain SNP data of the 278 accessions of the association panel. The additional chickpea accessions phenotyped in this study were not included in the 429 accessions genotyped by Varshney et al. (2019) and hence were omitted from the GWAS analysis. The SNP dataset for the 278 chickpea accessions was filtered for missing values (≥20%) and minor-allele frequency (MAF < 5%) using Trait Analysis by aSSociation, Evolution and Linkage (TASSEL) 5.0 (Glaubitz et al., 2014) and the Variant Call Format (VCF) filter tool in Galaxy Australia. Population stratification of the reference set was identified using ADMIXTURE software (version 1.3) (Liu et al., 2020) to measure the proportion of individual ancestry and sub-populations (K). Since the ADMIXTURE algorithm is based on independent loci, pruning of the dataset based on linkage disequilibrium (LD) was carried out using PLINK1.9 (Chang, 2015) command with default parameters. The cross-validation (CV) error was tested for K=1 to 20 for the best value of K. Visual inspection of ADMIXTURE plots generated using the pong tool (Behr et al., 2016) was also used to decide the best K value.
Genome-wide association and candidate gene analysis
For GWAS, six multi-locus models, namely, mrMLM (Wang et al., 2016), FASTmrMLM, (Tamba and Zhang, 2018), FASTmrEMMA (Wen et al., 2018; Wen et al., 2020), pKWmEB (Ren et al., 2018), pLARmEB (Zhang et al., 2017), ISIS EM-BLASSO (Tamba et al., 2017) were implemented using the mrMLM R package (Zhang et al., 2020) with all the default parameters. The threshold of LOD score for significant QTN was set at 3.0 for all the six methods. The QTNs were considered reliable if they were identified by two or more models. The flanking regions of these QTNs were analysed with the open source computational platform JBrowse2 (Diesh et al., 2022) using track Cicer arietinum CDCFrontier.gnm3.ann1.NPD7.gene_models_main.gff3 (https://cicer.legumeinfo.org). Candidate genes were defined as annotated genes within 315 kb either side of a QTN, based on the LD decay of the chickpea reference set reported by Varshney et al. (2019). The candidate genes were verified for their function based on the literature reporting roles associated with plant-pathogen interactions.
Results
Phenotypic evaluation of Pratylenchus thornei resistance
Pearson’s correlation coefficient indicated that Experiment 1 and Experiment 2, analysed for loge (P. thornei/kg+1) of soil, were moderately correlated (r = 0.3, P < 0.0001, n=264) (Supplementary Figure 1). In order to obtain the most representative phenotypic values, LMM analysis of the combined data sets was used to obtain BLUEs (Supplementary Tables 3, 4). Pratylenchus thornei population densities were normally distributed (Shapiro Wilk test = 0.997; P = 0.789) (Figure 1). Approximately, 5% of the total accessions belonged to the R category (Supplementary Table 5), 9% of the total accessions belonged to R-MR category and 18% of accessions to the MR category. The lowest percentage of accessions (2%) belonged to S-VS category. The well characterized wild relative C. echinospermum ILWC 123, used as a resistant check cultivar had P. thornei population density of 2,660 P. thornei/kg of soil. The only two other wild relatives tested in the study, ICC 20190 and ICC 20183, belonged to R and R-MR categories with P. thornei population densities of 2,012 P. thornei/kg of soil and 6,039 P. thornei/kg of soil, respectively. Chickpea breeding line D05253>F3TMWR2AB001 was in the R-MR category with P. thornei population density of 5,340 P. thornei/kg of soil, and D05293>F3TMWR2AB002 was in MR category with 8,955 P. thornei/kg of soil. MR check cultivar PBA HatTrick had high population density with 16,498 P. thornei/kg of soil and MS-S check cultivar Kyabra produced high P. thornei density of 35,846/kg of soil. Eight of the resistant accessions produced significantly lower P. thornei population densities than the Australian commercial cultivar PBA HatTrick. The wheat check cultivars used in the study to confirm the P. thornei multiplication performed as expected.
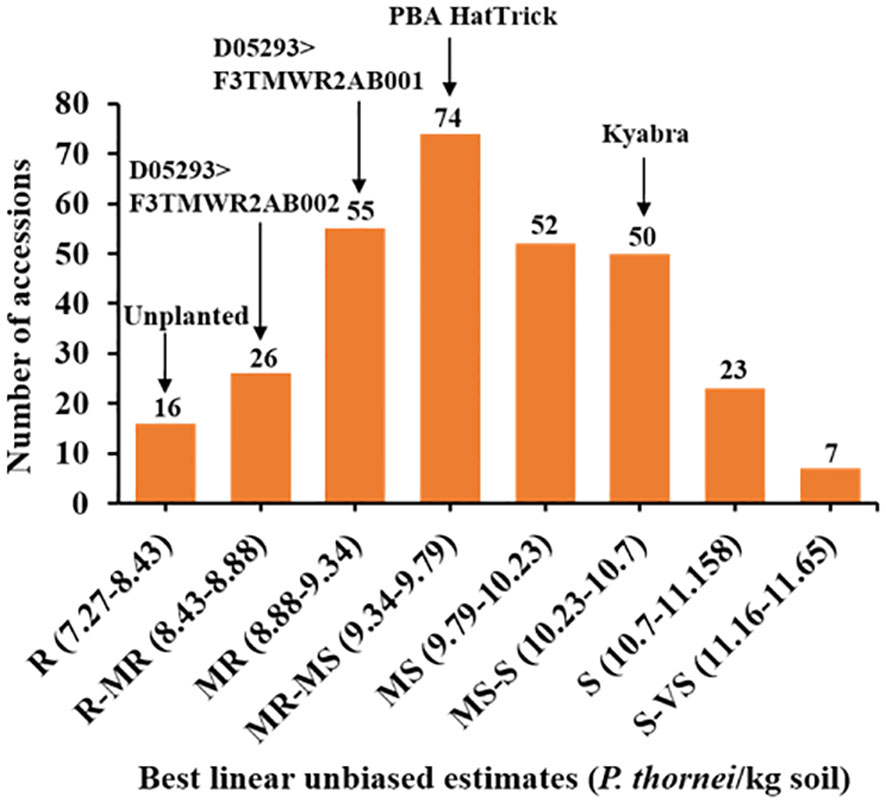
Figure 1 Frequency distribution histogram of the chickpea reference collection and additional resistance sources (total 303 accessions) for P. thornei population densities. Data shown is the best linear unbiased estimate (BLUE) values for combined analysis of Experiment 1 and Experiment 2. Resistant (R), resistant to moderately resistant (R-MR), moderately resistant (MR), moderately resistant to moderately susceptible (MR-MS), moderately susceptible (MS), moderately susceptible to susceptible (MS-S), susceptible (S), susceptible to very susceptible (S-VS).
Identification of P. thornei resistant chickpea accessions with resistance to other biotic and/or abiotic stresses
The chickpea accessions in the present study in R and R-MR ratings for resistance to P. thornei were compared with reports in the literature where the chickpea reference set has been phenotyped for other biotic and abiotic traits (Table 1). ICC 15697 was resistant to P. thornei, moderately resistant to Botrytis grey mould and tolerant to salinity. ICC 8950, ICC 5135, ICC 6816, ICC 95, ICC 14831 were resistant to P. thornei and moderately resistant to Fusarium wilt. ICC 8058 was resistant to P. thornei and Fusarium wilt ICC 12328 was moderately resistant to P. thornei and moderately resistant to Botrytis grey mould, Ascochyta blight, and dry root rot and with superior agronomic traits of grain yield and seed size.
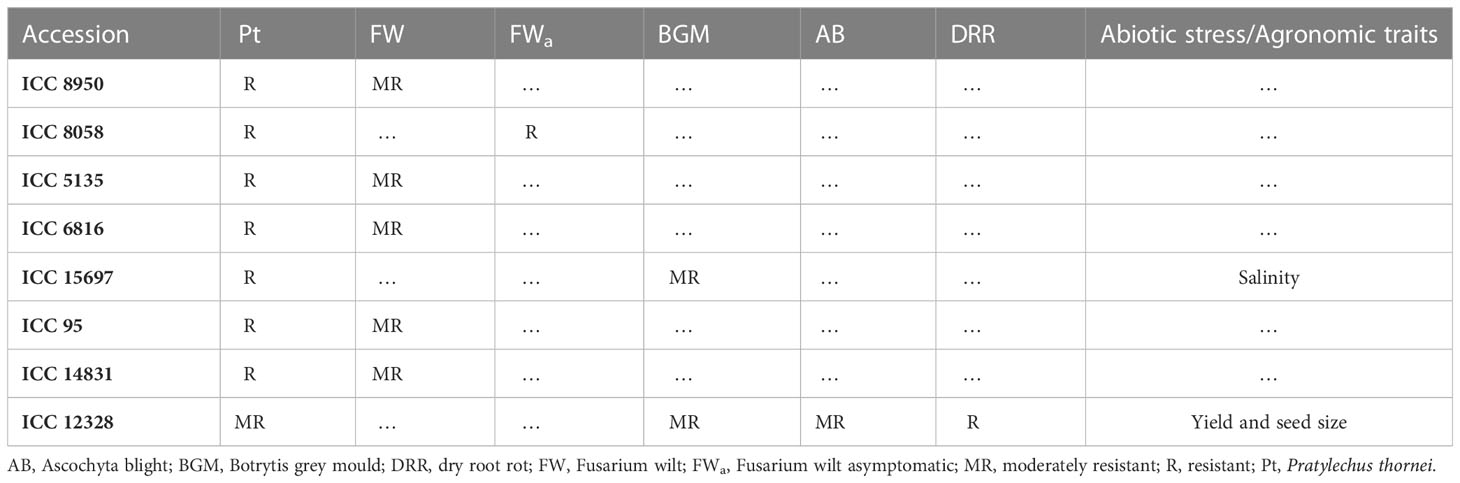
Table 1 The chickpea accessions in resistant (R) and moderately resistant (MR) categories to Pratylenchus thornei of present study collated with reports of multiple disease resistance or tolerance to abiotic stress and/or with superior agronomic traits from the literature.
Genotyping and population stratification
Pruning LD-wise the chickpea association panel of 278 accessions resulted in 112,998 SNPs that were used to assess population stratification. ADMIXTURE is based on maximum likelihood to estimate allele frequencies in each group or sub-population and assign each individual ancestry to one or more of these group or sub-populations. The ADMIXTURE run depicted continuous decline after K=3 (Supplementary Figure 2). The best K value (K=3) was confirmed by visual inspection of the plot generated (Figure 2). The sub-population 1 comprised 107 accessions with most accessions in the group originating from India. The sub-population 2 comprised 120 accessions with the majority of the accessions originating from Iran, whereas sub-population 3 comprised the least number of accessions (n=51) with most accessions originating from Turkey.
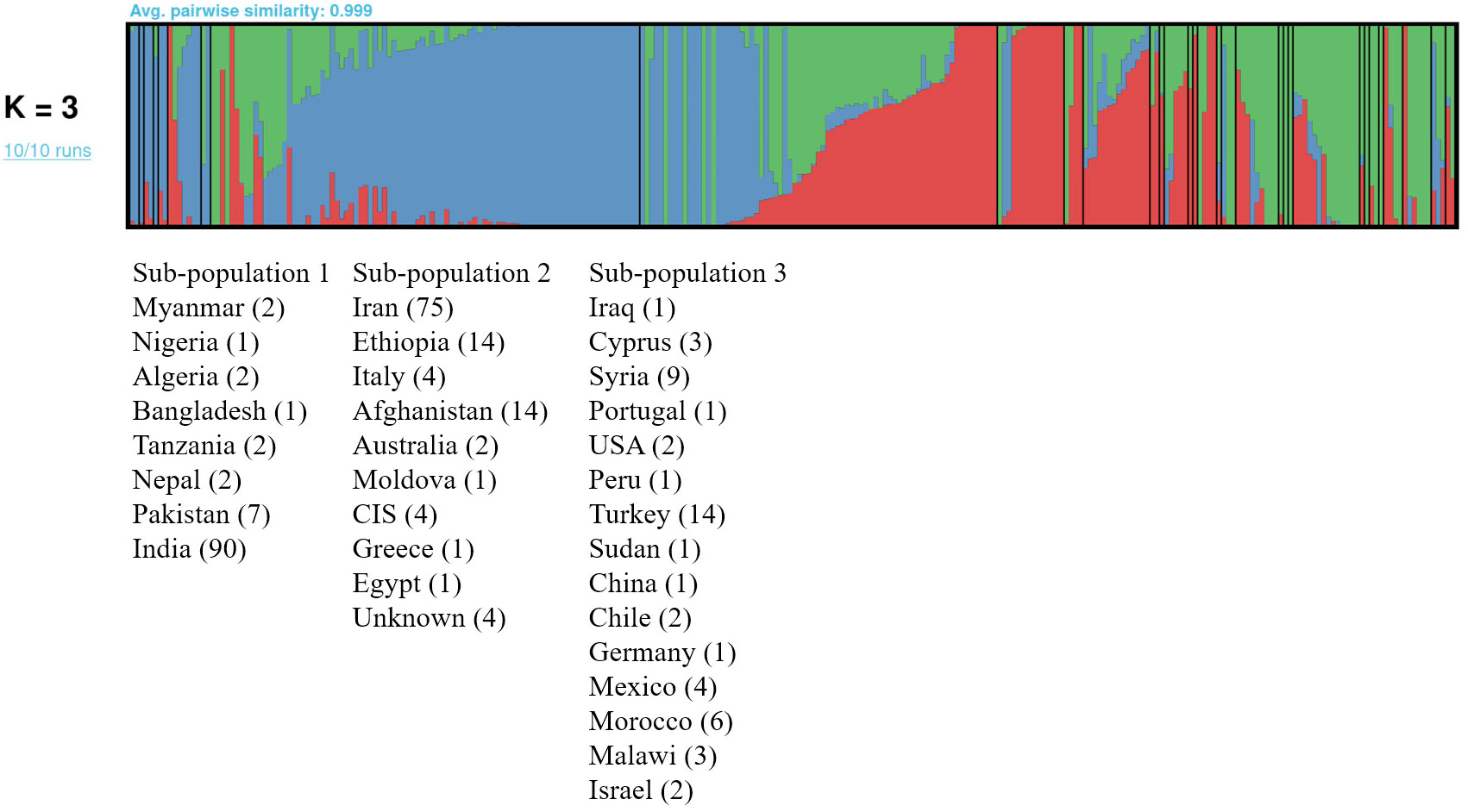
Figure 2 ADMIXTURE analysis of the chickpea association panel of 278 accessions using 112,998 SNPs and the pong tool formed 3 sub-populations: sub-population 1 (Blue), sub-population 2 (Red) and sub-population 3 (Green). The accessions in sub-populations were labelled as per country of origin, with the number of accessions in parentheses. The black vertical lines separate the countries within each sub-population. All the accessions in the respective sub-populations shared more than 50% ancestry except for accessions from Iraq, which had similar ancestry in all sub-populations and that of CIS and Iraq with ancestry between 40 to 50% for respective sub-populations.
Genome-wide association and candidate gene analysis
The SNP data of the chickpea association panel after filtration for missing values (≥20%) and minor-allele frequency (MAF < 5%) resulted in 492,849 SNPs for further analysis. The number of SNPs per chromosome were 74,764 (Ca1), 47,430 (Ca2), 39,254 (Ca3), 14,7829 (Ca4), 42,616 (Ca5), 65,589 (Ca6), 55,798 (Ca7), and 18,569 (Ca8) (Supplementary Figure 3). GWAS using six different models in the mrMLM package resulted in detection of 24 QTNs (Table 2 and Figure 3) distributed on all of the chickpea chromosomes, except for chromosome 8. Ten QTNs were detected by three to five of the six multi-locus GWAS models, with an additional 14 QTNs detected by two models. Among the six multi-locus GWAS models FASTmrMLM detected the highest number of QTNs (19), followed by pKWMEB (16) and ISIS EM-BLASSO (12). The other multi-locus GWAS models detected five or fewer QTNs each. The FASTmrMLM model outperformed other models, co-detecting 14 of the 16 QTNs detected by pKWMEB, and 10 of the 12 QTNs detected by ISIS EM-BLASSO. Mining the results of several multi-locus GWAS has been recommended to improve the detection power and robustness of GWAS (Zhang et al., 2019), particularly for QTNs with small allelic effects (Wang et al., 2016).
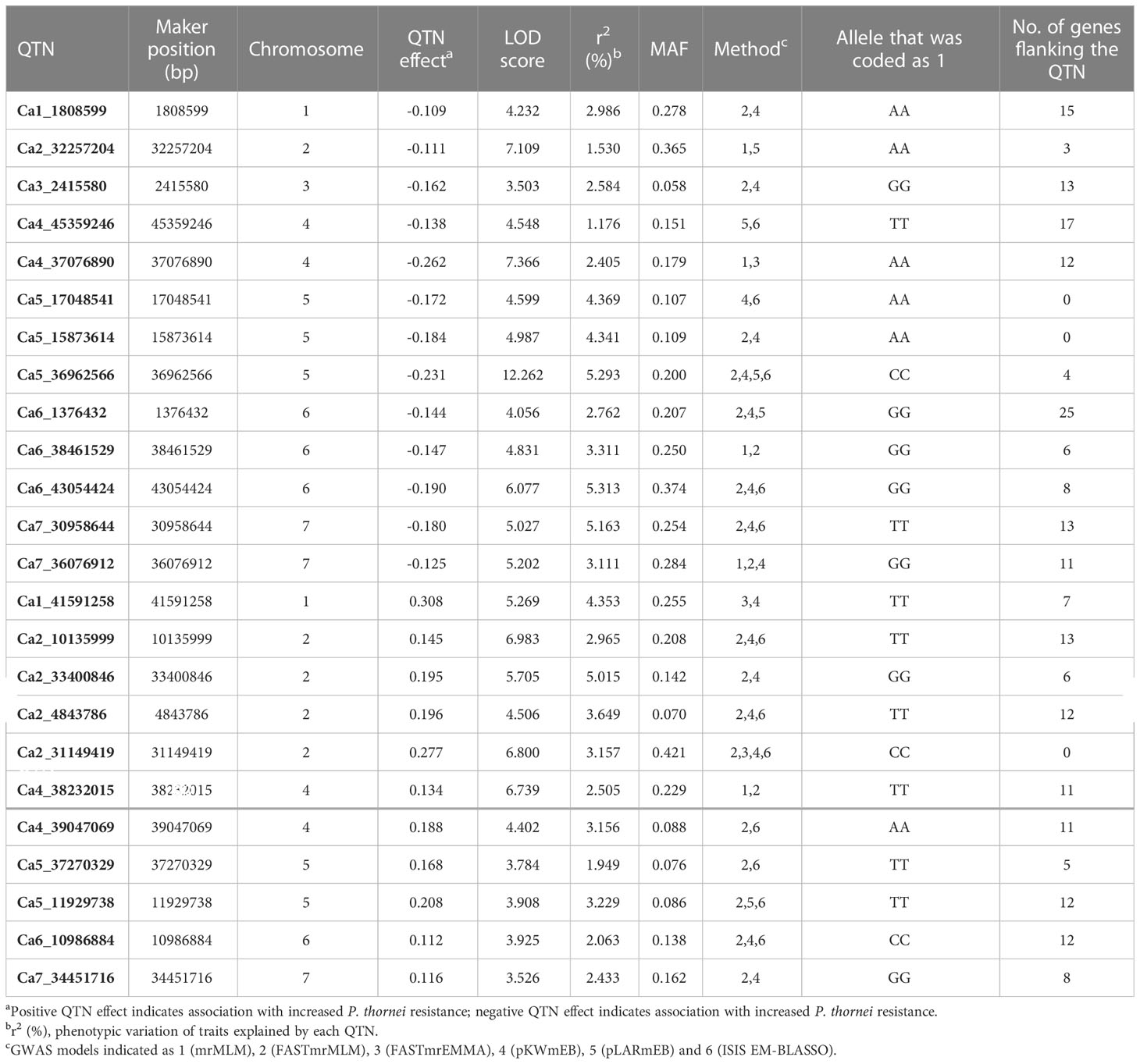
Table 2 The significant quantitative trait nucleotides (QTNs) for Pratylenchus thornei resistance or susceptibility in chickpea detected by two or more multi-locus GWAS methods.
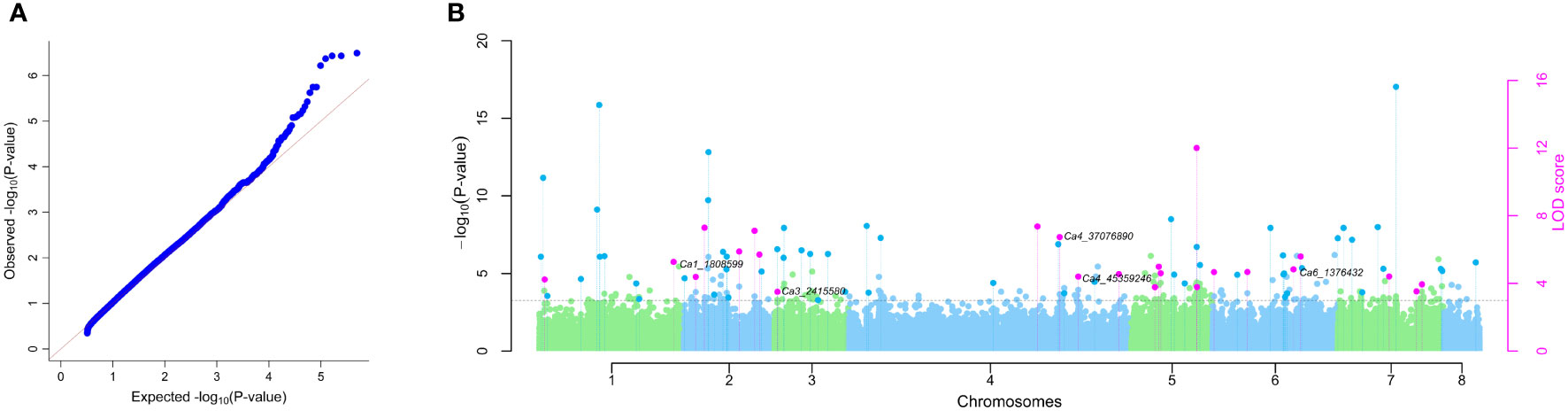
Figure 3 Multi-locus GWAS. (A) Quantile-quantile plot for comparison between expected and observed –log10 (P-values). (B) Manhattan plot for significant QTNs associated with P. thornei resistance in chickpea using mrMLM, FASTmrMLM, FASTmrEMMA, pKWmEB, pLARmEB and ISIS EM-BLASSO models. The -log10 (P-value) and the LOD score of SNP across the genome (y-axes) were plotted against their respective position on each chromosome (x-axis). The pink dots above dotted vertical lines represent all the QTNs commonly identified by multiple approaches and the dark blue dots above dotted vertical lines indicate QTNs identified by a single GWAS model. Candidate gene analysis revealed QTNs Ca1_1808599, Ca4_37076890 and Ca6_1376432 were flanked by Receptor-linked kinases (RLKs). QTN Ca3_2415580 was flanked by GDSL-like Lipase/Acylhydrolase. QTN Ca4_45359246 was flanked by Aspartic proteinase like and Thaumatin-like protein and QTN Ca6_1376432 was flanked by AT-hook DNA-binding and HSPRO2.
The effect size relates to an increase or a decrease in value of the trait under study, meaning a positive effect relates to higher nematode population density and a negative effect relates to lower nematode population density. Thirteen QTNs were associated with lower nematode population density, indicating increased resistance, and 11 QTNs were associated with higher nematode population density, indicating increased susceptibility.
Analysis of single experiment data identified 23 QTNs (6 associated with decreased P. thornei population densities and 17 associated with increased P. thornei population densities) for Experiment 1 (Supplementary Table 6). One of these QTNs (Ca2_31149419) was detected as a significant QTN using BLUEs from combined Experiments 1 and 2 (Supplementary Table 7). Analysis of Experiment 2 data revealed 17 QTNs (10 associated with decreased P. thornei population densities and 7 associated with increased P. thornei population densities). Eight of the QTN detected in the Experiment 2 dataset were in common with analysis of the combined dataset (Supplementary Table 7).
Of the 24 significant QTN positions identified by analysis of the combined dataset, 20 QTNs were flanked by annotated genes (Supplementary Table 8). These candidate genes were analysed for their putative role in P. thornei resistance based on reported functions in biotic and abiotic stress responses in the literature. A total of six candidate genes were identified with previously reported roles in plant-pathogen interactions. Receptor-linked kinases (RLKs), a common multigene family, were located on chromosomes 1, 4 and 6 flanking QTNs Ca1_1808599, Ca4_37076890 and Ca6_1376432, respectively; GDSL-like Lipase/Acylhydrolase was identified on chromosome 3 flanking QTN Ca3_2415580; Aspartic proteinase like and Thaumatin-like protein were identified on chromosome 4 flanking QTN Ca4_45359246; and AT-hook DNA-binding and HSPRO2 were identified on chromosome 6 flanking QTN Ca6_1376432 (Figure 3).
Discussion
A wide variation of resistance and susceptibility to P. thornei was observed among the accessions of the chickpea reference collection. Fourteen accessions were classified as resistant to P. thornei, of which 11 accessions were landraces, two were breeding lines and one a wild relative (C. echinospermum). While wild relatives of chickpea (C. echinospermum and C. reticulatum) have been identified with improved levels of P. thornei resistance over current commercial chickpea cultivars grown in Australia (Thompson et al., 2011; Reen et al., 2019), sources of P. thornei resistance in the C. arietinum cultigen are favoured by plant breeders over the need to create interspecific crosses, as fewer backcrosses are required to introgress resistance while recovering an elite background. Thus, these landrace accessions with P. thornei resistance identified in this study constitute a valuable resource for increasing resistance to P. thornei in chickpea cultivars in Australia and globally.
With limited reports on the mechanisms of resistance against migratory endoparasitic nematodes (Mathew and Opperman, 2020), all annotated genes flanking the QTN were further investigated, with those identified with a reported role in biotic or abiotic stress considered candidate genes for P. thornei. However, all genes in the flanking regions of the significant QTN should be considered for further investigation through differential expression and functional analysis.
Among genes flanking the QTNs, Ca1_1808599, Ca4_37076890 and Ca6_1376432 on chromosomes 1, 4 and 6, respectively, the receptor-linked kinases (RLKs) subtype - lectin RLKs (LecRLKs), was found to be the common multi-gene family. The importance of receptor-like kinases in resistance to P. thornei have been reported in a fine mapping study of P. thornei resistance loci in wheat (Rahman et al., 2020) and overexpression of receptor-like kinase upon P. thornei infection was reported in resistant chickpea genotypes (Channale et al., 2021). LecRLKs act as sensors and mediators of signals from the plant cell surface to intracellular compartments, playing crucial roles in plant development and responses to abiotic and biotic stresses (Sun et al., 2020). In a study by Cheng et al. (2013), OslecRK (Oryzae sativa lectin receptor-like kinase) was found to be a pleiotropic gene involved not only in mounting immune response against biotic stress but also in the germination of seeds.
For Ca3_2415580, a GDSL-like Lipase/Acylhydrolase superfamily protein gene was located in the upstream region of the QTN. GDSL-type esterase/lipase protein is a hydrolytic enzyme that possesses broad substrate specificity to compounds such as thioesters, aryl esters, phospholipids and amino acids (Akoh et al., 2004). The GDSL family is involved in a myriad of roles including plant growth and development, secondary metabolism, plant immunity, and biotic and abiotic stresses (Su et al., 2020). GDSL lipase was demonstrated to have constitutive expression in resistant cultivars of sweet potato (Ipomoea batatas) upon infection with root-knot nematode Meloidogyne incognita. Enhanced expression of GLIP1 (GDSL LIPASE-LIKE 1) increased resistance of Arabidopsis thaliana to Alternaria brassicicola, Erwinia carotovora and Pseudomonas syringae (Kwon et al., 2009).
The genes flanking QTN Ca4_45359246 on chromosome 4 included aspartic proteinase-like, its isoform, and thaumatin-like proteins. Aspartic proteinases are a widely distributed class of proteinases in plants involved in an array of biological processes including plant immunity (Figueiredo et al., 2021). They are also involved in regulating the degradation of proteins induced during stress such as pathogenesis-related proteins like thaumatin to prevent their overaccumulation. They are also known to hydrolyze proteins secreted by invasive pathogens (Schaller and Ryan, 1996). Thaumatin-like protein is a multi-gene family also involved in various functions in host-pathogen interaction like biotic stress, abiotic stress tolerance, and cell signalling (Liu et al., 2010). The role of thaumatin-like protein in P. thornei resistance can also be corroborated from the fact that thaumatin-like protein was overexpressed in a resistant chickpea cultivar on exposure to P. thornei (Channale et al., 2021).
The genes flanking QTN Ca6_1376432 involved in pathogen defence and plant development included Predicted AT-hook DNA-binding and nematode resistance protein-like HSPRO2-like genes. The AT-hook gene family is highly conserved across all land plants being involved in plant growth and development, response to stress and hormonal stimulus (Zhao et al., 2014; Wang et al., 2021). In one of the studies reported by Kim et al. (2007), the AT-hook DNA-binding protein-encoding gene CaATL1 was specifically induced upon pathogen inoculation. Transgenic tomato plants overexpressing CaATL1 showed enhanced resistance against Pseudomonas syringae and Phytophthora capsici (Kim et al., 2007). The HSPRO2-like gene was demonstrated to be a positive regulator of basal resistance, which functions downstream of the salicylic acid pathway in Arabidopsis against P. syringae (Murray et al., 2007). Also, three putative resistance genes were identified encoding nematode resistance HSPRO2-like proteins in response to Pseudomonas syringae pv syringae in Medicago sativa (Nemchinov et al., 2017).
Among candidate genes flanking QTN Ca3_15978889, detected by multiple GWAS methods in Experiment 2, was Bowman-Birk trypsin inhibitor. Bowman-Birk trypsin inhibitor is one of types of serine proteinase inhibitors (PIs) rich in legume seeds (Srinivasan et al., 2005) exhibiting strong inhibitory activity against trypsin, chymotrypsin or both. It plays a major role in plant defence due to its anti-digestive properties (Murdock and Shade, 2002; Ferry et al., 2004; Zavala et al., 2004). In a study reported by Rashed et al., 2008, transcripts with Bowman-Birk trypsin inhibitor were overexpressed in soybean in response to soybean cyst nematode, Heterodera glycines.
Similarly, candidate genes flanking two QTNs on chromosome 7, Ca7_15462252 and Ca7_4192522 identified by combined GWAS analysis and in Experiment 2 included salicylic acid carboxyl methyltransferase and glutathione S-transferase family protein. Salicylic acid carboxyl methyltransferase is synthesized in plants from salicylic acid and plays important roles in defence against microbial and insect pests (Liu et al., 2010). Salicylic acid dependent immune priming of defense genes was reported in chickpea resistant genotype upon interaction with P. thornei (Channale et al., 2021). Glutathione S transferases are multifunctional enzyme and are highly induced in stress conditions and are also known to be receptors of salicylic acid (Gullner et al., 2018).
The chickpea reference set is a valuable collection capturing the diversity in landrace chickpea germplasm that has been phenotyped by different research groups in glasshouse and field trials for various biotic stresses, including Ascochyta blight, Fusarium wilt, Botrytis grey mould, dry root rot, abiotic stresses including heat, drought, and salinity, and for agronomic traits like yield and seed weight. Accessions previously reported as resistant (ICC 6816, ICC 95, ICC 14831) or asymptomatic (ICC 8058) to Fusarium wilt (Pande et al., 2006) were identified in our study to be resistant to P. thornei. Accessions with dual resistance will be promising sources for chickpea breeding programs to produce superior commercial cultivars for regions where F. oxysporum and P. thornei are known to co-infect chickpea e.g., Southern Spain (Castillo et al., 1998). Similarly, ICC 15697 was resistant to P. thornei in this study and has been reported to be moderately resistant to Botrytis grey mould (Pande et al., 2006) and tolerant to salinity (100mM) (Serraj et al., 2004). Along with multiple disease resistance, it is important that grain yield is also maintained, yet, building a disease-resistant and high-yielding crop is often a major challenge to breeders and scientists (Wang and Dong, 2021). Notably, we identified the high yielding accession ICC 12328 (Meena et al., 2010) as moderately resistant to P. thornei, and this accession was also reported as moderately resistant to Ascochyta blight, dry root rot, and Botrytis grey mould (Pande et al., 2006). Pratylenchus thornei reproduction was reported to be significantly increased in chickpea cultivars susceptible to F. oxysporum f. sp. ciceris (Castillo et al., 1998). Thus, the P. thornei-resistant landraces identified in this study will not only be invaluable for breeding resistant modern cultivars to protect the chickpea crop itself and for integrated management of P. thornei in cropping systems with cereal host crops, but can also be of value in breeding multiple-disease resistant chickpea cultivars.
Conclusion
Six multi-locus GWAS models were used to detect reliable QTNs associated with P. thornei resistance in chickpea landraces. A total of 24 QTNs were co-detected by at least two different multi-locus GWAS models. Candidate gene analysis identified receptor-linked kinases (RLKs) on chromosome 1, 4 and 6, GDSL-like Lipase/Acylhydrolase on chromosome 3, Aspartic proteinase-like and Thaumatin-like protein on chromosome 4, AT-hook DNA-binding and HSPRO2 on chromosome 6 as candidate genes for P. thornei resistance. Following verification of these candidate genes using differential gene expression studies, cost-effective breeder-friendly markers can be developed to aid marker assisted selection. Such molecular tools will facilitate efficient introgression of P. thornei resistance into elite chickpea cultivars and provide important insights into the mechanisms of P. thornei resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
RZ, MT, and JT conceived the experiments. SC conducted the experiments. SC, RZ and MT contributed to data analysis and interpretation. JT and RV reviewed and edited the manuscript. SC drafted the manuscript with input from the other co-authors. All authors critically revised and approved the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Australian Grains Genebank and The International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Telangana, India for supplying the chickpea reference set. We acknowledge the Grains Research and Development Corporation (GRDC), New South Wales (NSW) Department of Primary Industries (DPI) and Queensland Department of Agriculture and Fisheries, Australia, as the co-owners of the chickpea breeding line D05253>F3TMWR2AB001, D05293>F3TMWR2AB002, D05222>F3TMWR2AB001 and acknowledge the breeding by Mr. Ted Knights, NSW DPI and thank Dr. Kristy Hobson for providing seed for this study. We also thank Dr. Kevin Moore, NSW DPI and Dr Tim Sutton, South Australian Research and Development Institute for providing seed. SC was supported by a postgraduate research scholarship from University of Southern Queensland. This work is supported by Galaxy Australia, a service provided by the Australian Biocommons and its partners. The service receives NCRIS funding through Bioplatforms Australia and the Australian Research Data Commons (https://doi.org/10.47486/PL105), as well as The University of Melbourne and Queensland Government RICF funding.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1139574/full#supplementary-material
References
Abbo, S., Berger, J., Turner, N. C. (2003). Evolution of cultivated chickpea: Four bottlenecks limit diversity and constrain adaptation. Funct. Plant Biol. 30 (10), 1081–1087. doi: 10.1071/FP03084
Agarwal, C., Chen, W., Varshney, R. K., Vandemark, G. (2022). Linkage QTL mapping and genome-wide association study on resistance in chickpea to Pythium Ultimum. Front. Genet. 13. doi: 10.3389/fgene.2022.945787
Akoh, C. C., Lee, G.-C., Liaw, Y.-C., Huang, T.-H., Shaw, J.-F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43 (6), 534–552. doi: 10.1016/j.plipres.2004.09.002
Alekcevetch, J. C., de Lima Passianotto, A. L., Ferreira, E. G. C., Dos Santos, A. B., da Silva, D. C. G., Dias, W. P., et al. (2021). Genome-wide association study for resistance to the Meloidogyne javanica causing root-knot nematode in soybean. Theor. Appl. Genet. 134 (3), 777–792. doi: 10.1007/s00122-020-03723-9
Ali, S.S. (1997). Status of nematode problem and research in India. Diagnosis of key nematode pests of chickpea and pigeonpea and their management : Proceedings of a Regional Training Course, 25–30 Nov 1996, ICRISAT, Patancheru, India. (Patancheru, Andhra Pradesh, India: International Crops Research Institute for the Semi-Arid Tropics) 74–82.
Beckmann, G. G., Thompson, C. H. (1960). “Soils and land use in the kurrawa area, darling downs, Queensland,” in Report. division of soils (Melbourne: CSIRO).
Behr, A. A., Liu, K. Z., Liu-Fang, G., Nakka, P., Ramachandran, S. (2016). Pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32 (18), 2817–2823. doi: 10.1093/bioinformatics/btw327
Braulio, J. S.-C., Sylvie, C. (2012). “Association mapping in plant genomes,” in Genetic diversity in plants. Ed. Mahmut, Ç. (Rijeka: IntechOpen), Ch. 3.
Castillo, P., Navas-Cortés, J. A., Landa, B. B., Jiménez-Díaz, R. M., Vovlas, N. (2008). Plant-parasitic nematodes attacking chickpea and their in planta interactions with rhizobia and phytopathogenic fungi. Plant Dis. 92 (6), 840–853. doi: 10.1094/PDIS-92-6-0840
Castillo, P., Vovlas, N., Jiménez-Díaz, R. M. (1998). Pathogenicity and histopathology of Pratylenchus thornei populations on selected chickpea genotypes. Plant Pathol. 47 (3), 370–376. doi: 10.1046/j.1365-3059.1998.00240.x
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., Lee, J. J. (2015). Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 4 (1), s13742–015. doi: 10.1186/s13742-015-0047-8
Channale, S., Kalavikatte, D., Thompson, J. P., Kudapa, H., Bajaj, P., Varshney, R. K., et al. (2021). Transcriptome analysis reveals key genes associated with root-lesion nematode Pratylenchus thornei resistance in chickpea. Sci. Rep. 11 (1), 1–11. doi: 10.1038/s41598-021-96906-3
Cheng, X., Wu, Y., Guo, J., Du, B., Chen, R., Zhu, L., et al. (2013). A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 76 (4), 687–698. doi: 10.1111/tpj.12328
Cooper, H. D., Spillane, C., Hodgkin, T. (2000). “Broadening the genetic base of crops: An overview,” in Broadening the genetic base of crop production. Eds. Cooper, H. D., Spillane, C., Hodgkin, T. (Wallingford: CAB International) 1–23.
Cui, Y., Zhang, F., Zhou, Y. (2018). The application of multi-locus GWAS for the detection of salt-tolerance loci in rice. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01464
Dababat, A. A., Ferney, G. H., Erginbas-Orakci, G., Dreisigacker, S., Imren, M., Toktay, H., et al. (2016). Association analysis of resistance to cereal cyst nematodes (Heterodera avenae) and root lesion nematodes (Pratylenchus neglectus and P. thornei) in CIMMYT advanced spring wheat lines for semi-arid conditions. Breed Sci. 66 (5), 692–702. doi: 10.1270/jsbbs.15158
Diapari, M., Sindhu, A., Bett, K., Deokar, A., Warkentin, T. D., Tar’an, B. (2014). Genetic diversity and association mapping of iron and zinc concentrations in chickpea (Cicer arietinum L.). Genome 57 (8), 459–468. doi: 10.1139/gen-2014-0108
Diesh, C., Stevens, G. J., Xie, P., Martinez, T. D. J., Hershberg, E. A., Leung, A., et al. (2022). JBrowse 2: A modular genome browser with views of synteny and structural variation. BioRxiv. doi: 10.1101/2022.07.28.501447
FAOSTAT (2021). FAOSTAT (Food and Agriculture Organisation of United Nations). Available at: https://www.fao.org/faostat/en/#data/QCL.
Farahani, S., Maleki, M., Ford, R., Mehrabi, R., Kanouni, H., Kema, G. H., et al. (2022). Genome-wide association mapping for isolate-specific resistance to ascochyta rabiei in chickpea (Cicer arietinum L.). Physiol. Mol. Plant Pathol. 121, 101883. doi: 10.1016/j.pmpp.2022.101883
Ferry, N., Edwards, M. G., Gatehouse, J. A., Gatehouse, A. M. (2004). Plant-insect interactions: Molecular approaches to insect resistance. Curr. Opin. Biotechnol. 15 (2), 155–161. doi: 10.1016/j.copbio.2004.01.008
Figueiredo, L., Santos, R. B., Figueiredo, A. (2021). Defense and offense strategies: The role of aspartic proteases in plant–pathogen interactions. Biology 10 (2), 75. doi: 10.3390/biology10020075
Fosu-Nyarko, J., Jones, M. G. K. (2016). Advances in understanding the molecular mechanisms of root lesion nematode host interactions. Annu. Rev. Phytopathol. 54, 253–278. doi: 10.1146/annurev-phyto-080615-100257
Galaxy Community (2022). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50 (W1), W345–W351. doi: 10.1093/nar/gkac247
Gangurde, S. S., Xavier, A., Naik, Y. D., Jha, U. C., Rangari, S. K., Kumar, R., et al. (2022). Two decades of association mapping: Insights on disease resistance in major crops. Front. Plant Sci. 4863. doi: 10.3389/fpls.2022.1064059
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., et al. (2014). TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PloS One 9 (2), e90346. doi: 10.1371/journal.pone.0090346
Gullner, G., Komives, T., Király, L., Schröder, P. (2018). Glutathione s-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01836
Hada, A., Dutta, T. K., Singh, N., Singh, B., Rai, V., Singh, N. K., et al. (2020). A genome-wide association study in Indian wild rice accessions for resistance to the root-knot nematode Meloidogyne graminicola. PloS One 15 (9), e0239085. doi: 10.1371/journal.pone.0239085
Hou, S., Zhu, G., Li, Y., Li, W., Fu, J., Niu, E., et al. (2018). Genome-wide association studies reveal genetic variation and candidate genes of drought stress related traits in cotton (Gossypium hirsutum L.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01276
Jadhav, A., Rayate, S., Mhase, L., Thudi, M., Chitikineni, A., Harer, P., et al. (2015). Marker-trait association study for protein content in chickpea (Cicer arietinum L.). J. Genet. 94 (2), 279–286. doi: 10.1007/s12041-015-0529-6
Khoo, K. H., Sheedy, J. G., Taylor, J. D., Croser, J. S., Hayes, J. E., Sutton, T., et al. (2021). A QTL on the Ca7 chromosome of chickpea affects resistance to the root-lesion nematode Pratylenchus thornei. Mol. Breed. 41 (12), 1–13. doi: 10.1007/s11032-021-01271-8
Kim, S. Y., Kim, Y. C., Seong, E. S., Lee, Y. H., Park, J. M., Choi, D. (2007). The chili pepper CaATL1: An AT-hook motif-containing transcription factor implicated in defence responses against pathogens. Mol. Plant Pathol. 8 (6), 761–771. doi: 10.1111/j.1364-3703.2007.00427.x
Kumar, D., Sharma, S., Sharma, R., Pundir, S., Singh, V. K., Chaturvedi, D., et al. (2021). Genome-wide association study in hexaploid wheat identifies novel genomic regions associated with resistance to root lesion nematode (Pratylenchus thornei). Sci. Rep. 11 (1), 1–14. doi: 10.1038/s41598-021-80996-0
Kwon, S. J., Jin, H. C., Lee, S., Nam, M. H., Chung, J. H., Kwon, S. I., et al. (2009). GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 58 (2), 235–245. doi: 10.1111/j.1365-313X.2008.03772.x
Lancashire, P. D., Bleiholder, H., Boom, T. V. D., Langelüddeke, P., Stauss, R., Weber, E., et al. (1991). A uniform decimal code for growth stages of crops and weeds. Ann. Appl. Biol. 119 (3), 561–601. doi: 10.1111/j.1744-7348.1991.tb04895.x
Li, H., Rodda, M., Gnanasambandam, A., Aftab, M., Redden, R., Hobson, K., et al. (2015). Breeding for biotic stress resistance in chickpea: Progress and prospects. Euphytica 204 (2), 257–288. doi: 10.1007/s10681-015-1462-8
Li, Y., Ruperao, P., Batley, J., Edwards, D., Davidson, J., Hobson, K., et al. (2017). Genome analysis identified novel candidate genes for ascochyta blight resistance in chickpea using whole genome re-sequencing data. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00359
Liu, C.-C., Shringarpure, S., Lange, K., Novembre, J. (2020). Exploring population structure with admixture models and principal component analysis. Methods Mol. Biol. 2090, 67–86. doi: 10.1007/978-1-0716-0199-0_4
Liu, P. P., Yang, Y., Pichersky, E., Klessig, D. F. (2010). Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in arabidopsis. Mol. Plant Microbe Interact. 23 (1), 82–90. doi: 10.1094/mpmi-23-1-0082
Mammadov, J., Aggarwal, R., Buyyarapu, R., Kumpatla, S. (2012). SNP markers and their impact on plant breeding. Int. J. Plant Genomics 2012, 728398. doi: 10.1155/2012/728398
Mathew, R., Opperman, C. H. (2020). Current insights into migratory endoparasitism: Deciphering the biology, parasitism mechanisms, and management strategies of key migratory endoparasitic phytonematodes. Plants 9 (6), 671. doi: 10.3390/plants9060671
Meena, H., Kumar, J., Upadhyaya, H., Bharadwaj, C., Chauhan, S., Verma, A., et al. (2010). Chickpea mini core germplasm collection as rich sources of diversity for crop improvement. J. SAT Agric. Res. 8, 1–5.
Mehta, Y. R. (2014). “Wheat and wheat production constraints,” in Wheat diseases and their management (Cham: Springer).
Murdock, L. L., Shade, R. E. (2002). Lectins and protease inhibitors as plant defenses against insects. J. Agric. Food Chem. 50 (22), 6605–6611. doi: 10.1021/jf020192c
Murray, S. L., Ingle, R. A., Petersen, L. N., Denby, K. J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant-Microbe Interact. 20 (11), 1431–1438. doi: 10.1094/MPMI-20-11-1431
Nemchinov, L. G., Shao, J., Lee, M. N., Postnikova, O. A., Samac, D. A. (2017). Resistant and susceptible responses in alfalfa (Medicago sativa) to bacterial stem blight caused by Pseudomonas syringae pv. syringae. PloS One 12 (12), e0189781. doi: 10.1371/journal.pone.0189781
Nordborg, M., Weigel, D. (2008). Next-generation genetics in plants. Nature 456 (7223), 720–723. doi: 10.1038/nature07629
Pande, S., Kishore, G. K., Upadhyaya, H., Rao, J. N. (2006). Identification of sources of multiple disease resistance in mini-core collection of chickpea. Plant Dis. 90 (9), 1214–1218. doi: 10.1094/PD-90-1214
Pariyar, S. R., Dababat, A. A., Sannemann, W., Erginbas-Orakci, G., Elashry, A., Siddique, S., et al. (2016). Genome-wide association study in wheat identifies resistance to the cereal cyst nematode Heterodera filipjevi. Phytopathology 106 (10), 1128–1138. doi: 10.1094/phyto-02-16-0054-fi
Rahman, M. S., Linsell, K. J., Taylor, J. D., Hayden, M. J., Collins, N. C., Oldach, K. H. (2020). Fine mapping of root lesion nematode (Pratylenchus thornei) resistance loci on chromosomes 6D and 2B of wheat. Theor. Appl. Genet. 133 (2), 635–652. doi: 10.1007/s00122-019-03495-x
Raman, R., Warren, A., Krysinska-Kaczmarek, M., Rohan, M., Sharma, N., Dron, N., et al. (2022). Genome-wide association analyses track genomic regions for resistance to Ascochyta rabiei in Australian chickpea breeding germplasm. Front. Plant Sci. 1314. doi: 10.3389/fpls.2022.877266
Rashed, N. A., Macdonald, M. H., Matthews, B. F. (2008). Protease inhibitor expression in soybean roots exhibiting susceptible and resistant interactions with soybean cyst nematode. J. Nematol. 40 (2), 138–146.
Reen, R. A., Mumford, M. H., Thompson, J. P. (2019). Novel sources of resistance to root-lesion nematode (Pratylenchus thornei) in a new collection of wild cicer species (C. reticulatum and C. echinospermum) to improve resistance in cultivated chickpea (C. arietinum). Phytopathology 109 (7), 1270–1279. doi: 10.1094/PHYTO-02-19-0047-R
Reen, R. A., Thompson, J. P., Clewett, T. G., Sheedy, J. G., Bell, K. L. (2014). Yield response in chickpea cultivars and wheat following crop rotations affecting population densities of Pratylenchus thornei and arbuscular mycorrhizal fungi. Crop Pasture Sci. 65 (5), 428–441. doi: 10.1071/CP13441
Ren, W.-L., Wen, Y.-J., Dunwell, J. M., Zhang, Y.-M. (2018). pKWmEB: Integration of kruskal-Wallis test with empirical bayes under polygenic background control for multi-locus genome-wide association study. Heredity 120, 208–218. doi: 10.1038/s41437-017-0007-4
Rodda, M., Hobson, K., Forknall, C., Daniel, R., Fanning, J., Pounsett, D., et al. (2016). Highly heritable resistance to root-lesion nematode (Pratylenchus thornei) in Australian chickpea germplasm observed using an optimised glasshouse method and multi-environment trial analysis. Australas. Plant Pathol. 45 (3), 309–319. doi: 10.1007/s13313-016-0409-4
Schaller, A., Ryan, C. A. (1996). Molecular cloning of a tomato leaf cDNA encoding an aspartic protease, a systemic wound response protein. Plant Mol. Biol. 31 (5), 1073–1077. doi: 10.1007/BF00040725
Serraj, R., Krishnamurthy, L., Upadhyaya, H. (2004). Screening chickpea mini-core germplasm for tolerence to soil salinity. Int. Chickpea Pigeonpea Newslett. 11, 29–32.
Smýkal, P., Coyne, C. J., Ambrose, M. J., Maxted, N., Schaefer, H., Blair, M. W., et al. (2015). Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 34 (1-3), 43–104. doi: 10.1080/07352689.2014.897904
Sohail, Q., Erginbas-Orakci, G., Ozdemir, F., Jighly, A., Dreisigacker, S., Bektas, H., et al. (2022). Genome-wide association study of root-lesion nematodes Pratylenchus species and crown rot Fusarium culmorum in bread wheat. Life 12 (3), 372. doi: 10.3390/life12030372
Spillane, C., Gepts, P. (2000). “Evolutionary and genetic perspectives on the dynamics of crop genepools,” in Broadening the genetic base of crop production (CABI International), 25–70. doi: 10.1079/9780851994116.0025
Srinivasan, A., Giri, A. P., Harsulkar, A. M., Gatehouse, J. A., Gupta, V. S. (2005). A kunitz trypsin inhibitor from chickpea (Cicer arietinum L.) that exerts anti-metabolic effect on podborer (Helicoverpa armigera) larvae. Plant Mol. Biol. 57 (3) 359–374. doi: 10.1007/s11103-004-7925-2
Su, H.-G., Zhang, X.-H., Wang, T.-T., Wei, W.-L., Wang, Y.-X., Chen, J., et al. (2020). Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in soybean. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00726
Sun, Y., Qiao, Z., Muchero, W., Chen, J.-G. (2020). Lectin receptor-like kinases: The sensor and mediator at the plant cell surface. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.596301
Tamba, C. L., Ni, Y.-L., Zhang, Y.-M. (2017). Iterative sure independence screening EM-Bayesian LASSO algorithm for multi-locus genome-wide association studies. PloS Comput. Biol. 13 (1), e1005357. doi: 10.1371/journal.pcbi.1005357
Tamba, C. L., Zhang, Y. M. (2018). A fast mrMLM algorithm for multi-locus genome-wide association studies. BioRxiv. doi: 10.1101/341784
Thompson, J. (1990). Treatments to eliminate root-lesion nematode (Pratylenchus thornei sher & Allen) from a vertisol. Nematologica 36 (1-4), 123–127. doi: 10.1163/002925990X00095
Thompson, J., Clewett, T., O’Reilly, M. (2015). Temperature response of root-lesion nematode (Pratylenchus thornei) reproduction on wheat cultivars has implications for resistance screening and wheat production. Ann. Appl. Biol. 167 (1), 1–10. doi: 10.1111/aab.12187
Thompson, J. P., Reen, R. A., Clewett, T. G., Sheedy, J. G., Kelly, A. M., Gogel, B. J., et al. (2011). Hybridisation of Australian chickpea cultivars with wild cicer spp. increases resistance to root-lesion nematodes (Pratylenchus thornei and P. neglectus). Australas. Plant Pathol. 40 (6), 601–611. doi: 10.1007/s13313-011-0089-z
Thompson, J. P., Sheedy, J. G., Robinson, N. A. (2020). Resistance of wheat genotypes to root-lesion nematode (Pratylenchus thornei) can be used to predict final nematode population densities, crop greenness, and grain yield in the field. Phytopathology 110 (2), 505–516. doi: 10.1094/PHYTO-06-19-0203-R
Thudi, M., Upadhyaya, H. D., Rathore, A., Gaur, P. M., Krishnamurthy, L., Roorkiwal, M., et al. (2014). Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PloS One 9 (5), e96758. doi: 10.1371/journal.pone.0096758
Upadhyaya, H. D., Bramel, P. J., Singh, S. (2001). Development of a chickpea core subset using geographic distribution and quantitative traits. Crop Sci. 41 (1), 206–210. doi: 10.2135/cropsci2001.411206x
Upadhyaya, H. D., Dwivedi, S. L., Baum, M., Varshney, R. K., Udupa, S. M., Gowda, C. L., et al. (2008). Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol. 8 (1), 1–12. doi: 10.1186/1471-2229-8-106
Upadhyaya, H. D., Dwivedi, S. L., Singh, S., Sahrawat, K. L., Singh, S. K. (2016a). Genetic variation and postflowering drought effects on seed iron and zinc in ICRISAT sorghum mini core collection. Crop Sci. 56 (1), 374–383. doi: 10.2135/cropsci2015.05.0308
Upadhyaya, H., Furman, B., Dwivedi, S., Udupa, S., Gowda, C., Baum, M., et al. (2006). Development of a composite collection for mining germplasm possessing allelic variation for beneficial traits in chickpea. Plant Genet. Resour. 4 (1), 13–19. doi: 10.1079/PGR2005101
Upadhyaya, H. D., Wang, Y.-H., Sastry, D. V., Dwivedi, S. L., Prasad, P. V., Burrell, A. M., et al. (2016b). Association mapping of germinability and seedling vigor in sorghum under controlled low-temperature conditions. Genome 59 (2), 137–145. doi: 10.1139/gen-2015-0122
Varshney, R. K., Thudi, M., Roorkiwal, M., He, W., Upadhyaya, H. D., Yang, W., et al. (2019). Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 51 (5), 857–864. doi: 10.1038/s41588-019-0401-3
VSN International (2020). Genstat for windows. 21st Edition (Hemel Hempstead, UK: VSN International).
Wang, M., Chen, B., Zhou, W., Xie, L., Wang, L., Zhang, Y., et al. (2021). Genome-wide identification and expression analysis of the AT-hook motif nuclear localized gene family in soybean. BMC Genomics 22 (1), 1–26. doi: 10.1186/s12864-021-07687-y
Wang, Y., Dong, S. (2021). A new roadmap for the breeding of disease-resistant and high-yield crops. Stress Biol. 1 (1), 1–3. doi: 10.1007/s44154-021-00023-0
Wang, S.-B., Feng, J.-Y., Ren, W.-L., Huang, B., Zhou, L., Wen, Y.-J., et al. (2016). Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 6 (1), 1–10. doi: 10.1038/srep19444
Wen, Y. J., Zhang, H., Ni, Y. L., Huang, B., Zhang, J., Feng, J. Y., et al. (2018). Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Briefings Bioinf. 19 (4), 700–712. doi: 10.1093/bib/bbw145
Wen, Y., Zhang, Y., Zhang, J., Feng, J., Zhang, Y. (2020). The improved FASTmrEMMA and GCIM algorithms for genome-wide association and linkage studies in large mapping populations. Crop J. 8 (5), 723–732. doi: 10.1016/j.cj.2020.04.008
Whitehead, A., Hemming, J. (1965). A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann. Appl. Biol. 55 (1), 25–38. doi: 10.1111/j.1744-7348.1965.tb07864.x
Zavala, J. A., Patankar, A. G., Gase, K., Hui, D., Baldwin, I. T. (2004). Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol. 134 (3), 1181–1190. doi: 10.1104/pp.103.035634
Zeven, A. C. (1998). Landraces: A review of definitions and classifications. Euphytica 104 (2), 127–139. doi: 10.1023/A:1018683119237
Zhang, J., Feng, J. Y., Ni, Y. L., Wen, Y. J., Niu, Y., Tamba, C. L., et al. (2017). pLARmEB: Integration of least angle regression with empirical bayes for multilocus genome-wide association studies. Heredity 118, 517–524. doi: 10.1038/hdy.2017.8
Zhang, Y.-M., Jia, Z., Dunwell, J. M. (2019). The applications of new multi-locus GWAS methodologies in the genetic dissection of complex traits. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00100
Zhang, Y.-W., Tamba, C. L., Wen, Y.-J., Li, P., Ren, W.-L., Ni, Y.-L., et al. (2020). mrMLM v4. 0.2: An R platform for multi-locus genome-wide association studies. Genomics Proteomics Bioinf. 18 (4), 481–487. doi: 10.1016/j.gpb.2020.06.006
Zhao, J., Favero, D. S., Qiu, J., Roalson, E. H., Neff, M. M. (2014). Insights into the evolution and diversification of the AT-hook motif nuclear localized gene family in land plants. BMC Plant Biol. 14 (1), 1–19. doi: 10.1186/s12870-014-0266-7
Keywords: chickpea, Cicer arietinum, root-lesion nematode, Pratylenchus thornei, GWAS, candidate resistance genes
Citation: Channale S, Thompson JP, Varshney RK, Thudi M and Zwart RS (2023) Multi-locus genome-wide association study of chickpea reference set identifies genetic determinants of Pratylenchus thornei resistance. Front. Plant Sci. 14:1139574. doi: 10.3389/fpls.2023.1139574
Received: 07 January 2023; Accepted: 13 March 2023;
Published: 24 March 2023.
Edited by:
Yuan-Ming Zhang, Huazhong Agricultural University, ChinaReviewed by:
Ya-Wen Zhang, Huazhong Agricultural University, ChinaRajiv Sharma, Scotland’s Rural College, United Kingdom
Yang-Jun Wen, Nanjing Agricultural University, China
Copyright © 2023 Channale, Thompson, Varshney, Thudi and Zwart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca S. Zwart, cmViZWNjYS56d2FydEB1c3EuZWR1LmF1
 Sonal Channale
Sonal Channale John P. Thompson
John P. Thompson Rajeev K. Varshney
Rajeev K. Varshney Mahendar Thudi
Mahendar Thudi Rebecca S. Zwart
Rebecca S. Zwart