- College of Life Sciences, Sichuan Agricultural University, Yaan, China
Pathogen infection is one of the major causes of yield loss in the crop field. The rapid increase of antimicrobial resistance in plant pathogens has urged researchers to develop both new pesticides and management strategies for plant protection. The antimicrobial peptides (AMPs) showed potential on eliminating plant pathogenic fungi and bacteria. Here, we first summarize several overlooked advantages and merits of AMPs, which includes the steep dose-response relations, fast killing ability, broad synergism, slow resistance selection. We then discuss the possible application of AMPs for plant protection with above merits, and highlight how AMPs can be incorporated into a more efficient integrated management system that both increases the crop yield and reduce resistance evolution of pathogens.
Introduction
Since 1950s, antibiotics have been used to control bacterial and fungal pathogens in the crop field. The crisis of antibiotic resistance in agricultural system is looming. Overuse of antibiotics in crop production has dramatically accelerated the evolution of antibiotic resistance of phytopathogen (McManus et al., 2002). The bacterial genera of plant pathogen, which includes Erwinia, Pantoea, Pseudomonas, Xanthomonas, acquired high resistance to streptomycin and oxytetracycline during the past decades (Sundin and Wang, 2018). The increasing resistance of pathogens indirectly plied up overall cost for plant protection. It also raised great concern that resistant bacterial mutants will fail the clinical treatment. For example, several phytopathogenic fungi have evolved high resistance to azole, a class of anti-fungal drug used both in farmland and hospital (Dunne et al., 2017).
In order to suppress the increasing trend of antibiotic resistance in plant protection, researchers have focused on either discovering new antimicrobials or developing new strategies for fighting against plant pathogens (Marcos et al., 2008; Lindsey et al., 2020). In recent years, a large number of natural and synthetic new chemicals are discovered and used in plant protection. Among them, AMPs are good candidates for fighting against phytopathogen (Amso and Hayouka, 2019). Moreover, new ways of drug application with existing antibiotics were also developed based on precisely quantitative methods, which significantly enhanced treatment efficiency and delayed the resistance evolution. For example, streptomycin is re-registered for treating Huanglongbing in the citrus grove. With methods analogues to the pharmacokinetics in pharmacology, recent studies have determined its spatial and temporal variation of drug concentration inside the plant. This helps to identify effective concentration range and duration of effectiveness inside citrus trees (Li et al., 2021). In addition, the patterns of killing of antibiotics are also critical for evaluation of field efficacy of antibiotics. It has been suggested that benchmark dose modeling can be applied to determine the killing effect of pesticides both in laboratory and field in the framework of the dose-response relation (Jensen et al., 2019, Jensen et al., 2022). In fact, this method is akin to pharmacodynamics in pharmacology. Pharmacodynamics of antibiotic provide information on how fast a given drug kills bacteria with respect to the dose. Moreover, dose-response relation also characterizes the ability of resistance selection of the antimicrobials (Yu et al., 2018). Overall, integration of new drug and novel methods of application facilitate the development of integrated pest management, reducing resistance evolution and cutting the total cost of agricultural production.
AMPs have long been proposed as a potential anti-bacteria and anti-fungi reagents in agriculture (Van der Biezen, 2001; Montesinos, 2007; Marcos et al., 2008; Montesinos and Bardají, 2008; Wang et al., 2018; Lobo and Boto, 2022), medicine (Zasloff, 2002; Harris and Coote, 2010; Tam et al., 2015; Hayouka et al., 2017; Annunziato and Costantino, 2020), food industry (Gupta and Srivastava, 2014; Sibel Akalın, 2014; McNulty et al., 2019; Lima et al., 2021; Liu et al., 2021). Here we briefly summarize the general information of AMPs and provide references for readers that are not familiar with AMPs. Most of the AMPs are usually composed of 10 to 50 amino acids with a total net positive charge in the working environments. These amino acids form different secondary structures, namely α-helix, β-sheet, macrocycles and residual-modified structures, which are the important bases of AMPs’ classification (Zasloff, 2002). Also, classification of AMPs in plants is characterized by the disulfide bonds and tertiary structures (Tam et al., 2015). In addition, other standards, such as pathogens that AMPs target, also can be applied to categorize AMPs, i.e. antifungal AMPs and antiviral AMPs (Benfield and Henriques, 2020). AMPs from different origins share similar killing mechanisms when targeting bacterial and fungal pathogens. As summarized by many previous reviews (Zasloff, 2002; Brogden, 2005; Melo et al., 2009; Bocchinfuso et al., 2011; Sibel Akalın, 2014; Benfield and Henriques, 2020; Huan et al., 2020), most of AMPs directly target and rupture the cell membrane to kill pathogens, only a few AMPs enter the cytoplasm to interrupt the cellular physiological process (Rahnamaeian et al., 2015). In general, the membrane-targeting mechanisms of AMPs can be characterized by carpet models, toroidal pore models and barrel-stave models (Epand and Vogel, 1999; Zasloff, 2002; Benfield and Henriques, 2020). Although many previous studies mostly focused on the design, classification, physical structures and chemical properties, as well as killing mechanism of AMPs, which are generally shared by many different AMPs for various usages. We, however, propose that more information and properties of AMPs should be considered especially when using AMPs for plant protection. In this minireview, we attempt to discuss some of the overlooked properties of AMPs including steep dose-response relations, fast killing, slow resistance selection, broad synergism. It should be noted that many of these properties are not discovered in the field of plant protection. For example, the fast killing property of AMPs was revealed by researchers who study biological nano-materials (Fantner et al., 2010), and the slow resistance selection property was discovered by research groups that study ecology and evolution (Perron et al., 2006; Yu et al., 2018; Lazzaro et al., 2020). These properties directly relate to the dosing strategy and effectiveness in different scenario of application (Jensen et al., 2022). We therefor also highlight how these distinctly overlooked properties can be exploited and integrated into the pathogen management protocols (See Figure 1).
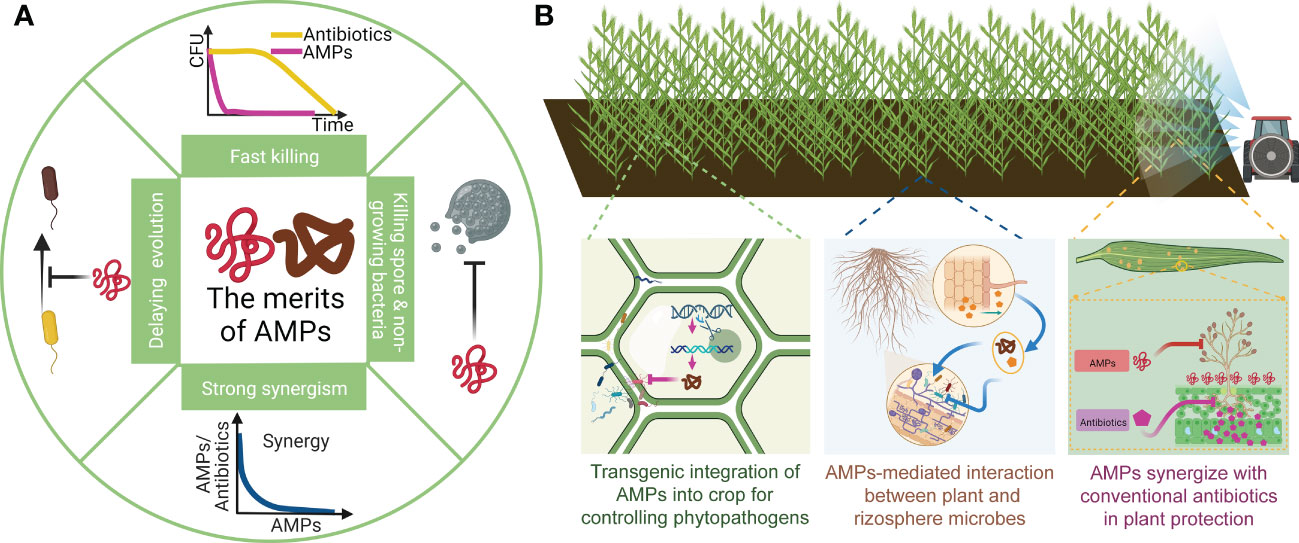
Figure 1 The merits of AMPs and their application in plant protection. (A) An brief illustration of AMPs’ merits. AMPs are able to rapidly kill bacteria within minutes. Due to distinct killing mechanism, they can deactivate non-growing bacterial and fungal spores. In addition, they also broadly synergize with other AMPs and antibiotics. Last, resistance evolution of pathogens to AMPs are particularly slow with high fitness cost. (B) AMPs with above merits can play an important role in integrated pathogen management. An integrated management methods includes endogenously expressing AMPs in plant to directly kill invading pathogens and to regulate symbiosis and root microbiome. Also, spraying AMPs in crop field with conventional antibiotic can form a synergistic effect that more efficiently eliminates pathogens, reduces the use of conventional antibiotics as well as delays pathogen resistance.
AMPs protect plant from infections both inside and outside
The immune repository of higher plant contains a group of antimicrobial peptides. Most of the plant AMPs are cystine-rich peptides and cross-linked by disulfide bonds (Tam et al., 2015; Sathoff and Samac, 2019). These plant AMPs, in general, are not structurally different from those from animals. Based on differences in disulfide bonds and tertiary structures, these peptides can be classified into several families, which includes thionins, defensins, hevein-like peptides, knottin-like peptides, lipid transfer proteins and snakins (Tam et al., 2015). Plant antimicrobial peptides widely expressed in variety of plant organs at different developmental stages (Silverstein et al., 2007; Tesfaye et al., 2013), which can be up-regulated upon pathogen infection. AMPs isolated from immune-activated plant showed robust killing effect towards various fungi and bacteria (Segura et al., 1999; Kovalskaya and Hammond, 2009). Overexpression of several defensin-like peptides in plants confer stronger antifungal and antibacterial effect. Moreover, plant defensin peptides both in model and crop plants have shown enhanced and long lasting disease resistance.
Heterologous expression of AMPs in plant using transgenic technologies is one of the efficient and powerful practices to improve plant’s resistance to phytopathogens. The full-length of cDNA of target AMPs was fused to the carrier plasmid, such as pMON22659 and pSAI4 and then transformed in to target plant using Agrobacterium. With this method, researcher integrated alfalfa antifungal peptide (alfAFP) defensin into potato and achieved robust resistance to fungal pathogens Phytophthora cactorum and Fusarium solani, the bacterium Erwinia carotovora both in greenhouse and field (Gao et al., 2000), where the transgenic plant had sixfold lower fungal load. Expression of a variety of plant and animal peptides in plant also confers both bacterial and fungal resistance in different families of plant, including potato (Gao et al., 2000; Osusky et al., 2000), tomato (Chan et al., 2005), rice (Kanzaki et al., 2002), tobacco (Chakrabarti et al., 2003; Swathi Anuradha et al., 2008; Khademi et al., 2020; Zhou et al., 2021), banana (Chakrabarti et al., 2003; Ghag et al., 2012), cotton (Gaspar et al., 2014), and many more other plants in earlier reviews (Keymanesh et al., 2009; Iqbal et al., 2019). This method can potentially reduce the infection and transmission of plant pathogens, and also decrease the incidence of pathogen infection of co-planted non-transgenic plants. Expression of heterologous AMPs in plants requires extra energy and resources and often trade-off with other traits of the plant, such as the yield vegetative growth (Van der Biezen, 2001; Jin et al., 2005). Therefore, it is still awaiting to evaluate that how the environmental factors, such as drought, temperature and many other biotic factors, affect the overall and long-term effect regarding disease resistance in transgenic plants. In addition, evolutionary theory predicts that enhanced host immunity selects more virulent pathogens (Antia et al., 1994; Cressler et al., 2014). Pathogens can evolve, if not direct resistance to host immune effectors, higher virulence and pathogenicity to reduce the expression of immune functions, and eventually break down the disease resistance (Sacristán and García Arenal, 2008; Fleming-Davies et al., 2018). Such breakdown of disease resistance has occurred in the cultivars that integrated R genes (Dodds et al., 2006; Peressotti et al., 2010). It remains elusive that if the transgenic plant with enhanced AMPs expression select virulent pathogens strains that compromises the durable resistance.
In-vitro application of AMPs can be also an effective way to control plant disease. Due to the costly production of AMPs at the moment, large scale field studies and applications of AMPs are rare (Table 1). The synthetic antimicrobial peptide BP15 showed great potential in controlling brown spot disease of pear, showing a disease reduction of about 42%-60% in the serial trials (Puig et al., 2015). Hayouka et al., 2017 synthesized 20-amino-acid-length peptides chain mixture with only two kinds of amino acids with relatively low cost. Those random peptides mixtures are able to rapidly eradicate pathogenic bacteria in plants (Topman et al., 2018; Amso and Hayouka, 2019). Foliar spray of these peptides significantly reduces disease incidence and index, which is almost as effective as commercial copper-based bactericide. In addition, these AMP mixtures showed no toxic effect on beneficial insects and mammalian cells. These studies show that synthesized AMPs have great potential in plant protection. Another possible way is to apply those widely used AMPs to control plant pathogens. For example, the widely-used food preservative, ϵ-poly-L-lysine (ϵ-PL), can successfully control grey mould on tomato in the field (Sun et al., 2017; Sun et al., 2018). Application of peptaibol trichogin with very low concentration (100 µM/129.3 g/ha) in vineyard has significantly reduced the incidence and severity of grapevine downy mildew caused by Plasmopara viticola (Bolzonello et al., 2023). Moreover, the peptide has no phytotoxicity on the plant.
AMPs rapidly kill pathogens in concentrations above the threshold
The fast-killing property of AMPs is vital for its application and often overlooked. Boman firstly recorded that the fast elimination of bacterial pathogens in immune-activated Drosophila (Boman et al., 1972). They found that the “vaccinated” flies with frozen bacteria can kill more than 99.9% of bacteria cells within 7 min after the second infection, then reduce the bacterial load for several orders of magnitudes in next few minutes. Boman and colleagues later proved that the fast killing is mainly achieved by AMPs that expressed by the insect’s immune system (Hultmark et al., 1980). Similar fast-killing property was constantly observed in later isolated AMPs, such as Magainins, the peptide isolated from frog skin (Zasloff, 1987). High-speed atomic force microscope allows one to observe the killing process of AMPs in real time (Fantner et al., 2010). In sufficient high concentrations, AMP can kill bacteria within 30 seconds. Similar fast killing rate was also observed using time-lapse fluorescence microscopy (Barns and Weisshaar, 2013). In contrast, antibiotics take much longer to kill bacteria. For example, bactericidal beta-lactam antibiotics take nearly one hour to kill a single bacterium (Yao et al., 2012). The killing time of many other antibiotics in bulk bacterial culture can range from 12 hours to several days (Ferro et al., 2015; Yang et al., 2018; Vaddady et al., 2019). It’s thus evident that the killing rate of AMPs is order-of-magnitude faster than that of antibiotics.
In addition, AMPs show striking inoculum effect (Nannini et al., 2013; Lenhard and Bulman, 2019; Loffredo et al., 2021). In other words, the killing effects of AMPs depends both on the bacterial density and AMP concentration, i.e. a minimal number of AMPs molecules is required to kill a bacterium (Huang, 2006; Melo et al., 2009). According to quantitative estimation, it requires approximate 107 peptides per cell for PMAP-23 to kill bacteria in bulk culture (Roversi et al., 2014). Moreover, AMP molecules can be unevenly absorbed by bacterial population, which results delayed population growth (Snoussi et al., 2018). Dead bacteria can even bind AMPs molecules and cause the same effect (Wu and Tan, 2019). This indicates that fast depletion of AMPs molecules by some bacterial cells results decreased free AMPs molecules in solution, and protect other bacteria from killing.
The fast-killing and inoculum effect altogether shapes the steep dose-response curve (Yu et al., 2016). It is notable that the concentration range from no effect to full killing is roughly 10 times in various AMPs (Steiner et al., 1981; Zasloff, 1987; Thevissen et al., 1999; Yu et al., 2016; Savini et al., 2017; Savini et al., 2020). This implies that we need to quantitatively evaluate the effective concentration when AMPs are applied in agricultural practice (Mercer et al., 2020; Jensen et al., 2022). Very recently, researchers have borrowed the methods in pharmacokinetics to quantify the spatial and temporal dynamics of antibiotic concentrations used to treat Huanglongbing in citrus (Li et al., 2019; Li et al., 2021). Moreover, the benchmark dose modelling method has also been proposed to quantify the working concentration of pesticides in the field (Jensen et al., 2022).These studies helped design proper dosing strategies in plant protection, which is also applicable for using AMPs in plant protection.
AMPs slowly select mutants with large fitness cost
Bacteria evolve resistance to all antimicrobial agents, but the evolution in AMPs is particularly slow. Experimental evolution revealed that the resistance evolution in AMPs is averagely 10 times slower than in antibiotics (Dobson et al., 2013; Spohn et al., 2019). This can be explained by the AMP’s steep threshold-like dose-response curve (Melo et al., 2009; Yu et al., 2018). When the concentration is higher than the “threshold”, the exposed bacterial can be immediately eliminated (Fantner et al., 2010). If the concentration is lower than the “threshold”, AMPs neither impose physiological stress on bacteria cells nor arrest their growth (Rodríguez-Rojas et al., 2014; Snoussi et al., 2018). It implies that the concentration range that select resistant mutants is rather narrow (Yu et al., 2018).
Moreover, resistance to AMPs often suffers significant fitness cost in bacteria. Resistance to AMPs in bacteria is generally caused by the loss of lipids or modification of lipid A on the membrane (Pränting et al., 2008; Andersson et al., 2016; Gao et al., 2016), which substantially impedes nutrition uptake and attenuate bacterial growth. For example, Salmonella typhimurium strains that resistant human peptide LL-37 suffers 25% of growth reduction compare to the wildtype control (Lofton et al., 2013). Colistin-resistant Acinetobacter baumannii strains have fitness cost ranging from 10% to as high as 60% (Mu et al., 2016). High fitness cost makes resistant strains less competitive in low or no antibiotic condition. Bacteria harboring mcr-1 gene can be completely removed within 20 days during thermophilic composting in environment (Gao et al., 2019). Large scale epidemiological survey showed that the frequency of bacteria carrying colistin-resistant genes immediately decreased after banning of colistin in feeding industry as well as in clinics (Shen et al., 2020; Wang et al., 2020; Shen et al., 2021).
AMPs synergize with other antimicrobials and plant immune system
Higher plants deploy a combination of antimicrobial peptides to combat the pathogen invasion. The cocktail of AMPs can be synergistic at different organizational level. AMPs not only synergize with themselves, but also interact with plant host physiological process to eliminate pathogen infection. Here, we categorize the synergism of AMPs into molecular synergism and functional synergism.
Molecular synergism shows that AMPs directly cooperating with other antimicrobials in killing pathogens. Plant AMPs 2S albumins synergize with thionins in inhibiting fungal growth and achieved 2- to 73-fold of increased killing effect compared to single AMP (Terras et al., 1993). Similar synergistic effects among AMPs from different organisms were further observed in eliminating bacteria, fungi and parasites (Westerhoff et al., 1995; McCafferty et al., 1999; Yan and Hancock, 2001; Marxer et al., 2016; Yu et al., 2016). In addition, AMPs also synergize with other antibiotics and fungicides (Arikan et al., 2002; Gonzalez et al., 2002; Mariz-Ponte et al., 2021). Such synergistic combination effect among AMPs can substantially reduce the total amount of molecules that required to kill the pathogen. This implies that application of AMPs in plant protection can reduce the use of agricultural antibiotics and fungicides, as well as the total cost of crop production. Moreover, the combination effect of AMPs can also delay drug resistance evolution and prolong the duration of agricultural chemicals (Maron et al., 2022).
Moreover, synergistic effect of killing can be also achieved by targeting different development stages of pathogens. Most of the agricultural antibiotics and fungicides only act on pathogens on the growing stage, not on the non-growing stage, i.e. the dominant spores. Previous researches showed that AMPs killed non-germinating fungal spores (Levitz et al., 1986; Rioux et al., 2000; Velivelli et al., 2020), thus can substantially reduce the transmission of pathogens. Moreover, spore elimination is particularly important in greenhouse agriculture. Air spora is the main source of plant disease in greenhouse. High density of spore also threatens the health of workers in greenhouse (Ercilla-Montserrat et al., 2017; Madsen et al., 2021). AMPs can be used as a sporicidal to inhibit transmission of plant disease and to protect the health of greenhouse workers. Thus, AMPs can potentially synergize with other antimicrobials, by eliminating the air spora, to reduce the disease prevalence and health risks of agricultural workers.
Functional synergism of AMPs in targeting pathogen is the result of complex interaction between AMPs and pathogen’s physiology. The positively charged AMPs with membrane permeability are able to not only attach on pathogens’ membrane but also target intracellular components of pathogens, which drastically accelerate the killing process. An insect antimicrobial peptide, abaecin, can interact with chaperones in bacteria, which can substantially dampen the bacterial stress response. It enhanced the killing effect of hymenoptaecin for four-fold in terms of inhibition rate, a strong synergistic effect in that combination (Rahnamaeian et al., 2015). In addition, in-vitro assay shows that plant AMPs modulate cellular redox stress in fungi by inducing the accumulation of ROS in vegetative cells and ultimately trigger apoptosis (Van der Weerden et al., 2008; Mello et al., 2011). Moreover, AMPs also activate the MAPK signaling cascade in fungal pathogens to control its growth (Ramamoorthy et al., 2007). Such synergism in fighting against pathogens can also be achieved by AMPs in interacting with plant signal pathways. For example, the cysteine-rich protein designated defensin, SPD1, in sweet potato both regulates the redox status of ascorbate and inhibit fungal and bacterial pathogens (Huang et al., 2008). Plant AMPs also interact with many other regulating pathways to enhance the plant defense and achieve synergism in controlling pathogens (Bolouri Moghaddam et al., 2016).
AMPs directly interacting with plant immune system can be defined as another functional synergism. However, experimental evidence is rare. Previous studies showed that endogenously expressed AMPs can bind with some transcription factors and regulate many signaling pathways (Damon et al., 2012). The consequence of these interactions is not revealed yet. Exogenous expression of a designed AMPs in plant will not elicit the immune response (Badosa et al., 2017; Montesinos et al., 2021), and the plant immunity can only be elicited when that AMPs conjugated with plant immune eliciting peptides, such as flg-15 and flg-22 (Montesinos et al., 2021). In a recent study, a stable antimicrobial peptides from Microcitrus australiasica, the Australian finger lime, was developed to treat Huanglongbing in citrus (Huang C et al., 2021). This peptide, also named as MaSAMP, not only directly target and kill the main pathogens Liberibacter crescens of citrus tree, but also induce plant’s innate immunity to prevent and inhibit infections. Both foliar spray and pneumatic truck injection of MaSAMP to HLB-infected citrus trees significantly reduced disease severity and bacterial load. Moreover, the authors found that MaSAMP application can induce the expression of a set of defense genes, such as pathogenesis-related proteins and the enzyme of SA biosynthesis and phenyl propanoid pathways. This peptide can broadly activate systemic defense responses in tobacco, tomato and citrus trees. This study demonstrated that AMPs can serve bifunctionally as both pathogen eliminators and stimulants of plant immune responses. Such synergistic functions can better protect plant from infections and shows great potential in plant protection.
AMPs regulate symbionts of plant
The nodule-specific cysteine-rich (NCR) AMPs also play an important role in mediating plant-microbe interaction inside the legume plants. These antimicrobial peptides can both maintain and eliminate the viability of nitrogen-fixing bacteroids. At the initiation stage of root nodule, lytic AMPs kill most of the phytopathogenic fungi and bacteria, and maintain the symbiotic bacteria inside root. NCR211 expressed in the nodule interzone II-III promotes differentiation of bacteroids and the formation of nodules. Synthetic NCR211 was further proven inhibiting aggregation of free-living S. meliloti (Kim et al., 2015). In legume plants, nitrogen-fixing symbiotic bacterioids with medium population size can significantly promote the growth of host plant by fixing nitrogen for the host plants. It, however, can also retard host’s growth when the load of bacteroids is high (Sachs et al., 2018). Inside the roots of host plants, a variety of nodule-specific cysteine-rich peptides are secreted to regulate the population of nitrogen-fixing rhizobia (Van de Velde et al., 2010; Wang et al., 2010). Knockout of host’s nodule-specific cysteine-rich peptide demonstrated that these peptides may serve as the extractors of the host plant to harvest nitrogen nutrients synthesized by the bacteroids (Wang et al., 2017; Yang et al., 2017). The direct killing effect of nodule-specific cysteine-rich peptides was measured in vitro as well (Van de Velde et al., 2010). Moreover, these peptides can enter bacterial cytosol and bind with intracellular molecules to slow down the growth of these bacteroids. The balance between plant host and symbionts may be the result of the complex interaction between AMPs and other host peptides (Yang et al., 2017).
All together, these evidence indicates that host plant may harness host peptides to finely tune the symbiosis at different temporal and spatial scale. It is also intriguing that whether these antimicrobial host peptides can be genetically incorporated into non-legume plants to develop symbiotic associations (Pankievicz et al., 2019). In addition, it remains elusive that if these peptides can be secreted by roots and released into soil for mediating broader host-microbe interaction.
Challenges and perspective
Crop yield loss is mainly caused by plant pathogen and pests. The total loss can reaches up to 40% of crop yield at the global level according to a recent survey (Savary et al., 2019), which is apparently far beyond the compensation of any advanced breeding technologies that contributed the yield gain around only 1-3% per year (Tester and Langridge, 2010). This indicates that the most efficient way to promote crop yield is to prevent it from the loss caused by pests and pathogens. Due to the diversity of pathogens at various levels, integrated management strategies are urgently needed to protect the crop. In this short review, we propose that AMPs with key merits can be harnessed as an important part in the integrated strategy.
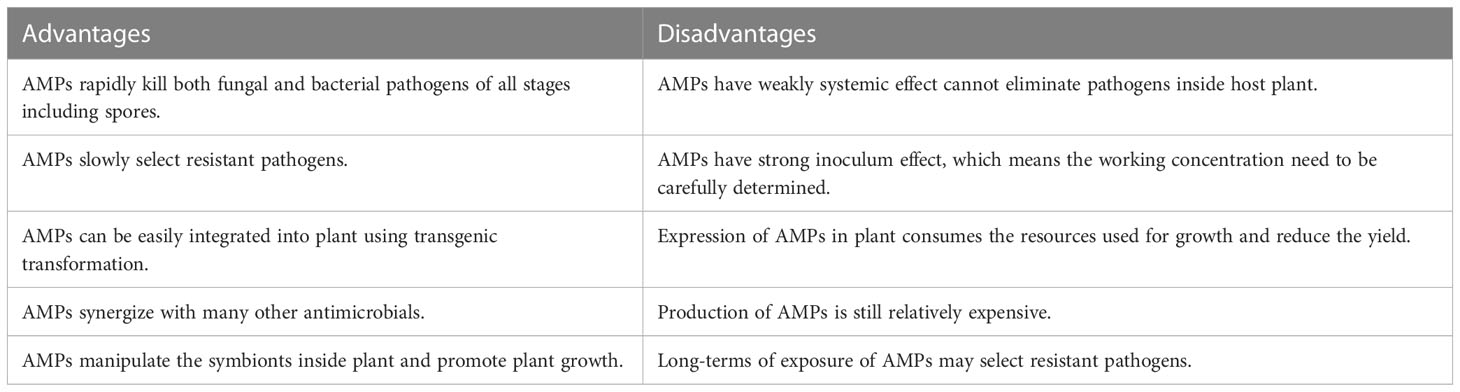
In design of plant protection strategy, no single component or method can accomplish all of the tasks. We argue that an optimal and cost-effective method that makes full use of AMPs’ merits can form a better solution for plant pathogen management (Table 2). With the fast-killing property, AMPs nearly take no time to kill the pathogens. This makes AMPs quickly fulfilling their functions before inactivated by environmental factors. Although the large molecular weight of AMPs may weaken their systemic effects for eliminating infection inside plants, One can combine AMPs with conventional antimicrobials in order to achieve systematic synergistic effect in control plant pathogens. In this case, AMPs eradicate the planktonic bacterial pathogens and fungal spores on the surface of plants in order to stop the spreed of plant disease in the field. The conventional antimicrobials with stronger systemic effects kill pathogen that grows inside plants. In addition, heterologous express of AMPs using transgenic plants is also an option for fighting against phytopathogens. Moreover, the sharp dose-response relation of AMPs suggests that field application with low concentrations may completely fail to kill plant pathogen. It is important that the concentration of AMPs sprayed on plant should be higher than the working critical threshold.
Antibiotic resistance of plant pathogen is always a concern for agricultural production as well as human health. The direct evidence of transfer of resistance pathogen from farm to clinics is rare (Chang et al., 2015). Both experiments and theories proved that AMPs slowly select resistant pathogens with high fitness cost. This indicates that resistant strains can be quickly out-competed by its sensitive counterpart. Seasonal application of AMPs should be less of a concern for the resistance evolution. We also argue that combined application of AMPs with conventional antibiotics not only adds up the overall effect controlling, but also slow down pathogen’s resistance evolution towards conventional antibiotics (Lazzaro et al., 2020)
Large scale application of AMPs requires highly cost-effective production. Recently technological advancement allows one to cheaply produce AMPs either through solid-phase synthesis (Hayouka et al., 2017; Topman et al., 2018), prokaryotic expression (Zhao et al., 2015; Schreiber et al., 2017) and eukaryotic expression (Holaskova et al., 2015; Holásková et al., 2018). Besides, some AMPs, such as Polylysine, that used in food and livestock industry are also ready for plant protection. Although many lab and greenhouse experiments have proven that AMPs are effective against various phytopathogens, one still anticipates large scale field tests in various environmental conditions.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was financially supported by the National Natural Science Foundation of China under Grant No. 32001242 to GY.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amso, Z., Hayouka, Z. (2019). Antimicrobial random peptide cocktails: a new approach to fight pathogenic bacteria. Chem. Commun. 55, 2007–2014. doi: 10.1039/C8CC09961H
Andersson, D. I., Hughes, D., Kubicek-Sutherland, J. Z. (2016). Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Update 26, 43–57. doi: 10.1016/j.drup.2016.04.002
Annunziato, G., Costantino, G. (2020). Antimicrobial peptides (amps): a patent review, (2015–2020). Expert Opin. Ther. Patents 30, 931–947. doi: 10.1080/13543776.2020.1851679
Antia, R., Levin, B. R., May, R. M. (1994). Within-host population dynamics and the evolution and maintenance of microparasite virulence. Am. Nat. 144, 457–472. doi: 10.1086/285686
Arikan, S., Lozano-Chiu, M., Paetznick, V., Rex, J. H. (2002). In vitro Synergy of caspofungin and amphotericin b against aspergillus and fusarium spp. Antimicrob. Agents Chemother. 46, 245–247. doi: 10.1128/AAC.46.1.245-247.2002
Badosa, E., Ferré, R., Francés, J., Bardají, E., Feliu, L., Planas, M. (2009). Sporicidal activity of synthetic antifungal undecapeptides and control of penicillium rot of apples. Appl. Environ. Microbiol. 75, 5563–5569. doi: 10.1128/AEM.00711-09
Badosa, E., Ferre, R., Planas, M., Feliu, L., Besalú, E., Cabrefiga, J., et al. (2007). A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 28, 2276–2285. doi: 10.1016/j.peptides.2007.09.010
Badosa, E., Montesinos, L., Camó, C., Ruz, L., Cabrefiga, J., Francés, J., et al. (2017). Control of fire blight infections with synthetic peptides that elicit plant defense responses. J. Plant Pathol. 99, 65–73. doi: 10.4454/jpp.v99i0.3915
Barns, K. J., Weisshaar, J. C. (2013). Real-time attack of LL-37 on single Bacillus subtilis cells. Biochim. Biophys. Acta Biomembr. 1828, 1511–1520. doi: 10.1016/j.bbamem.2013.02.011
Benfield, A. H., Henriques, S. T. (2020). Mode-of-action of antimicrobial peptides: membrane disruption vs. intracellular mechanisms. Front. Med. Technol. 2. doi: 10.3389/fmedt.2020.610997
Bocchinfuso, G., Bobone, S., Mazzuca, C., Palleschi, A., Stella, L. (2011). Fluorescence spectroscopy and molecular dynamics simulations in studies on the mechanism of membrane destabilization by antimicrobial peptides. Cell. Mol. Life Sci. 68, 2281–2301. doi: 10.1007/s00018-011-0719-1
Bolouri Moghaddam, M. R., Vilcinskas, A., Rahnamaeian, M. (2016). Cooperative interaction of antimicrobial peptides with the interrelated immune pathways in plants. Mol. Plant Pathol. 17, 464–471. doi: 10.1111/mpp.12299
Bolzonello, A., Morbiato, L., Tundo, S., Sella, L., Baccelli, I., Echeverrigaray, S., et al. (2023). Peptide analogs of atrichoderma peptaibol effectively control downy mildew in the vineyard. Plant Dis. doi: 10.1094/PDIS-09-22-2064-RE
Boman, H. G., Nilsson, I., Rasmuson, B. (1972). Inducible antibacterial defence system in drosophila. Nature 237, 232–235. doi: 10.1038/237232a0
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. doi: 10.1038/nrmicro1098
Bundó, M., Montesinos, L., Izquierdo, E., Campo, S., Mieulet, D., Guiderdoni, E., et al. (2014). Production of cecropin a antimicrobial peptide in rice seed endosperm. BMC Plant Biol. 14, 102. doi: 10.1186/1471-2229-14-102
Cavallarin, L., Andreu, D., San Segundo, B. (1998). Cecropin a–derived peptides are potent inhibitors of fungal plant pathogens. Mol. Plant Microbe Interact. 11, 218–227. doi: 10.1094/MPMI.1998.11.3.218
Chakrabarti, A., Ganapathi, T. R., Mukherjee, P. K., Bapat, V. A. (2003). Msi-99, a magainin analogue, imparts enhanced disease resistance in transgenic tobacco and banana. Planta 216, 587–596. doi: 10.1007/s00425-002-0918-y
Chan, Y., Prasad, V., Sanjaya, Chen, K. H., Liu, P. C., Chan, M., et al. (2005). Transgenic tomato plants expressing an arabidopsis thionin (thi2.1) driven by fruit-inactive promoter battle against phytopathogenic attack. Planta 221, 386–393. doi: 10.1007/s00425-004-1459-3
Chang, Q., Wang, W., Regev Yochay, G., Lipsitch, M., Hanage, W. P. (2015). Antibiotics in agriculture and the risk to human health: how worried should we be? Evol. Appl. 8, 240–247. doi: 10.1111/eva.12185
Colombo, M., Masiero, S., Rosa, S., Caporali, E., Toffolatti, S. L., Mizzotti, S., et al. (2020). Nopv1: a synthetic antimicrobial peptide aptamer targeting the causal agents of grapevine downy mildew and potato late blight. Sci. Rep. 10, 17574. doi: 10.1038/s41598-020-73027-x
Cressler, C. E., Nelson, W. A., Day, T., McCauley, E. (2014). Disentangling the interaction among host resources, the immune system and pathogens. Ecol. Lett. 17, 284–293. doi: 10.1111/ele.12229
Damon, C., Dmitrieva, J., Muhovski, Y., Francis, F., Lins, L., Ledoux, L., et al. (2012). Interaction network of antimicrobial peptides of Arabidopsis thaliana, based on high-throughput yeast two-hybrid screening. Plant Physiol. Biochem. 58, 245–252. doi: 10.1016/j.plaphy.2012.07.007
Dobson, A. J., Purves, J., Kamysz, W., Rolff, J. (2013). Comparing selection on S. aureus between antimicrobial peptides and common antibiotics. PloS One 8, e76521. doi: 10.1371/journal.pone.0076521
Dodds, P. N., Lawrence, G. J., Catanzariti, A., Teh, T., Wang, C. A., Ayliffe, M. A., et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 8888–8893. doi: 10.1073/pnas.0602577103
Dunne, K., Hagen, F., Pomeroy, N., Meis, J. F., Rogers, T. R. (2017). Intercountry transfer of triazole-resistant Aspergillus fumigatus on plant bulbs. Clin. Infect. Dis. 65, 147–149. doi: 10.1093/cid/cix257
Epand, R. M., Vogel, H. J. (1999). Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta Biomembr. 1462, 11–28. doi: 10.1016/S0005-2736(99)00198-4
Ercilla-Montserrat, M., Izquierdo, R., Belmonte, J., Montero, J. I., Muñoz, P., Linares, C. D., et al. (2017). Building-integrated agriculture: a first assessment of aerobiological air quality in rooftop greenhouses (i-rtgs). Sci. Total Environ. 598, 109–120. doi: 10.1016/j.scitotenv.2017.04.099
Fantner, G. E., Barbero, R. J., Gray, D. S., Belcher, A. M. (2010). Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 5, 280–285. doi: 10.1038/nnano.2010.29
Ferro, B. E., van Ingen, J., Wattenberg, M., van Soolingen, D., Mouton, J. W. (2015). Time-kill kinetics of antibiotics active against rapidly growing mycobacteria. J. Antimicrob. Chemother. 70, 811–817. doi: 10.1093/jac/dku431
Fleming-Davies, A. E., Williams, P. D., Dhondt, A. A., Dobson, A. P., Hochachka, W. M., Leon, A. E., et al. (2018). Incomplete host immunity favors the evolution of virulence in an emergent pathogen. Science 359, 1030–1033. doi: 10.1126/science.aao2140
Gao, A., Hakimi, S. M., Mittanck, C. A., Wu, Y., Woerner, B. M., Stark, D. M., et al. (2000). Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18, 1307–1310. doi: 10.1038/82436
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and mechanism for the mcr-1 colistin resistance. PloS Pathog. 12, e1005957. doi: 10.1371/journal.ppat.1005957
Gao, Y., Lu, C., Shen, D., Liu, J., Ma, Z., Yang, B., et al. (2019). Elimination of the risks of colistin resistance gene (mcr-1) in livestock manure during composting. Environ. Int. 126, 61–68. doi: 10.1016/j.envint.2019.02.015
Gaspar, Y. M., McKenna, J. A., McGinness, B. S., Hinch, J., Poon, S., Connelly, A. A., et al. (2014). Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin nad1. J. Exp. Bot. 65, 1541–1550. doi: 10.1093/jxb/eru021
Ghag, S. B., Shekhawat, U. K. S., Ganapathi, T. R. (2012). Petunia floral defensins with unique prodomains as novel candidates for development of fusarium wilt resistance in transgenic banana plants. PloS One 7, e39557. doi: 10.1371/journal.pone.0039557
Gonzalez, C. F., Provin, E. M., Zhu, L., Ebbole, D. J. (2002). Independent and synergistic activity of synthetic peptides against thiabendazole-resistantfusarium sambucinum. Phytopathology 92, 917–924. doi: 10.1094/PHYTO.2002.92.8.917
Güell, I., Cabrefiga, J., Badosa, E., Ferre, R., Talleda, M., Bardají, E., et al. (2011). Improvement of the efficacy of linear undecapeptides against plant-pathogenic bacteria by incorporation of d-amino acids. Appl. Environ. Microbiol. 77, 2667–2675. doi: 10.1128/AEM.02759-10
Gupta, R., Srivastava, S. (2014). Antifungal effect of antimicrobial peptides (amps lr/14) derived from Lactobacillus plantarum strain lr/14 and their applications in prevention of grain spoilage. Food Microbiol. 42, 1–7. doi: 10.1016/j.fm.2014.02.005
Harris, M. R., Coote, P. J. (2010). Combination of caspofungin or anidulafungin with antimicrobial peptides results in potent synergistic killing of Candida albicans and Candida glabrata in vitro. Int. J. Antimicrob. Agents 35, 347–356. doi: 10.1016/j.ijantimicag.2009.11.021
Hayouka, Z., Bella, A., Stern, T., Ray, S., Jiang, H., Grovenor, C. M., et al. (2017). Binary encoding of random peptide sequences for selective and differential antimicrobial mechanisms. Angew. Chem. Int. Ed. 56, 8099–8103. doi: 10.1002/anie.201702313
Holaskova, E., Galuszka, P., Frebort, I., Oz, M. T. (2015). Antimicrobial peptide production and plant-based expression systems for medical and agricultural biotechnology. Biotechnol. Adv. 33, 1005–1023. doi: 10.1016/j.biotechadv.2015.03.007
Holásková, E., Galuszka, P., Mičúchová, A., Šebela, M., Öz, M. T., Frébort, I., et al. (2018). Molecular farming in barley: development of a novel production platform to produce human antimicrobial peptide LL-37. Biotechnol. J. 13, 1700628. doi: 10.1002/biot.201700628
Huan, Y., Kong, Q., Mou, H., Yi, H. (2020). Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.582779
Huang, H. W. (2006). Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim. Biophys. Acta Biomembr. 1758, 1292–1302. doi: 10.1016/j.bbamem.2006.02.001
Huang, C., Araujo, K., Sánchez, J. N., Kund, G., Trumble, J., Roper, S., et al. (2021). A stable antimicrobial peptide with dual functions of treating and preventing citrus huanglongbing. Proc. Natl. Acad. Sci. U.S.A. 118, e2019628118. doi: 10.1073/pnas.2019628118
Huang, Y., Gao, L., Lin, M., Yu, T. (2021). Recombinant expression of antimicrobial peptides in Pichia pastoris: a strategy to inhibit the Penicillium expansum in pears. Postharvest Biol. Technol. 171, 111298. doi: 10.1016/j.postharvbio.2020.111298
Huang, G., Lai, H., Chang, Y., Sheu, M., Lu, T., Huang, S., et al. (2008). Antimicrobial, dehydroascorbate reductase, and monodehydroascorbate reductase activities of defensin from sweet potato Ipomoea batatas (L.) lam. ‘tainong 57’] storage roots. J. Agric. Food Chem. 56, 2989–2995. doi: 10.1021/jf072994j
Hultmark, D., Steiner, H., Rasmuson, T., Boman, H. G. (1980). Insect immunity. purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora Cecropia. Eur. J. Biochem. 106, 7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x
Iqbal, A., Khan, R. S., Shehryar, K., Imran, A., Ali, F., Attia, S., et al. (2019). Antimicrobial peptides as effective tools for enhanced disease resistance in plants. Plant Cell Tissue Organ Cult. 139, 1–15. doi: 10.1007/s11240-019-01668-6
Jensen, S. M., Kluxen, F. M., Ritz, C. (2019). A review of recent advances in benchmark dose methodology. Risk Anal. 39, 2295–2315. doi: 10.1111/risa.13324
Jensen, S. M., Kluxen, F. M., Ritz, C. (2022). Benchmark dose modelling in regulatory ecotoxicology, a potential tool in pest management. Pest Manage. Sci. 78, 1772–1779. doi: 10.1002/ps.6759
Jin, Z., Hong, J. K., Yang, K. A., Koo, J. C., Choi, Y. J., Chung, W. S., et al. (2005). Over-expression of Chinese cabbage calreticulin 1, brcrt1, enhances shoot and root regeneration, but retards plant growth in transgenic tobacco. Transgenic Res. 14, 619–626. doi: 10.1007/s11248-005-5694-6
Jung, Y., Lee, S., Moon, Y., Kang, K. (2012). Enhanced resistance to bacterial and fungal pathogens by overexpression of a human cathelicidin antimicrobial peptide (hCAP18/LL-37) in chinese cabbage. Plant Biotechnol. Rep. 6, 39–46. doi: 10.1007/s11816-011-0193-0
Kanzaki, H., Nirasawa, S., Saitoh, H., Ito, M., Nishihara, M., Terauchi, R., et al. (2002). Overexpression of the wasabi defensin gene confers enhanced resistance to blast fungus (Magnaporthe grisea) in transgenic rice. Theor. Appl. Genet. 105, 809–814. doi: 10.1007/s00122-001-0817-9
Keymanesh, K., Soltani, S., Sardari, S. (2009). Application of antimicrobial peptides in agriculture and food industry. World J. Microbiol. Biotechnol. 25, 933–944. doi: 10.1007/s11274-009-9984-7
Khademi, M., Varasteh-Shams, M., Nazarian-Firouzabadi, F., Ismaili, A. (2020). New recombinant antimicrobial peptides confer resistance to fungal pathogens in tobacco plants. Front. Plant Sci. 11, 1236. doi: 10.3389/fpls.2020.01236
Kim, M., Chen, Y., Xi, J., Waters, C., Chen, R., Wang, D.. (2015). An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 15238–15243. doi: 10.1073/pnas.1500123112
Koch, A., Khalifa, W., Langen, G., Vilcinskas, A., Kogel, K., Imani, J., et al. (2012). The antimicrobial peptide thanatin reduces fungal infections in Arabidopsis. J. Phytopathol. 160, 606–610. doi: 10.1111/j.1439-0434.2012.01946.x
Kovalskaya, N., Hammond, R. W. (2009). Expression and functional characterization of the plant antimicrobial snakin-1 and defensin recombinant proteins. Protein Expr. Purif. 63, 12–17. doi: 10.1016/j.pep.2008.08.013
Lazzaro, B. P., Zasloff, M., Rolff, J. (2020). Antimicrobial peptides: application informed by evolution. Science 368, eaau5480. doi: 10.1126/science.aau5480
Lenhard, J. R., Bulman, Z. P. (2019). Inoculum effect of β-lactam antibiotics. J. Antimicrob. Chemother. 74, 2825–2843. doi: 10.1093/jac/dkz226
Levitz, S. M., Selsted, M. E., Ganz, T., Lehrer, R. I., Diamond, R. D. (1986). In vitro Killing of spores and hyphae of Aspergillus fumigatus and Rhizopus oryzae by rabbit neutrophil cationic peptides and bronchoalveolar macrophages. J. Infect. Dis. 154, 483–489. doi: 10.1093/infdis/154.3.483
Li, J., Kolbasov, V. G., Lee, D., Pang, Z., Huang, Y., Collins, N., et al. (2021). Residue dynamics of streptomycin in citrus delivered by foliar spray and trunk injection and effect on ‘Candidatus liberibacter asiaticus’ titer. Phytopathology 111, 1095–1103. doi: 10.1094/PHYTO-09-20-0427-R
Li, J., Pang, Z., Duan, S., Lee, D., Kolbasov, V. G., Wang., N., et al. (2019). The in planta effective concentration of oxytetracycline against ‘Candidatus liberibacter asiaticus’ for suppression of citrus huanglongbing. Phytopathology 109, 2046–2054. doi: 10.1094/PHYTO-06-19-0198-R
Lima, P. G., Freitas, C. D. T., Oliveira, J. T. A., Neto, N. A. S., Amaral, J. L., Silva, A. F. B., et al. (2021). Synthetic antimicrobial peptides control Penicillium digitatum infection in orange fruits. Food Res. Int. 147, 110582. doi: 10.1016/j.foodres.2021.110582
Lindsey, A. P. J., Murugan, S., Renitta, R. E. (2020). Microbial disease management in agriculture: current status and future prospects. Biocatal. Agric. Biotechnol. 23, 101468. doi: 10.1016/j.bcab.2019.101468
Lipsky, A., Joshi, J. R., Carmi, N., Yedidia, I. (2016). Expression levels of antimicrobial peptide tachyplesin I in transgenic ornithogalum lines affect the resistance to Pectobacterium infection. J. Biotechnol. 238, 22–29. doi: 10.1016/j.jbiotec.2016.09.008
Liu, Y., Sameen, D. E., Ahmed, S., Dai, J., Qin, W. (2021). Antimicrobial peptides and their application in food packaging. Trends Food Sci. Technol. 112, 471–483. doi: 10.1016/j.tifs.2021.04.019
Lobo, F., Boto, A. (2022). Host-defense peptides as new generation phytosanitaries: low toxicity and low induction of antimicrobial resistance. Agronomy 12, 1614. doi: 10.3390/agronomy12071614
Loffredo, M. R., Savini, F., Bobone, S., Casciaro, B., Franzyk, H., Mangoni, M. L., et al. (2021). Inoculum effect of antimicrobial peptides. Proc. Natl. Acad. Sci. U.S.A. 118, e2014364118. doi: 10.1073/pnas.2014364118
Lofton, H., Pränting, M., Thulin, E., Andersson, D. I. (2013). Mechanisms and fitness costs of resistance to antimicrobial peptides LL-37, CNY100HL and wheat germ histones. PloS One 8, e68875. doi: 10.1371/journal.pone.0068875
Madsen, A. M., White, J. K., Markouch, A., Kadhim, S., de Jonge, N., Thilsing, T., et al. (2021). A cohort study of cucumber greenhouse workers’ exposure to microorganisms as measured using NGS and MALDI-TOF MS and biomarkers of systemic inflammation. Environ. Res. 192, 110325. doi: 10.1016/j.envres.2020.110325
Marcos, J. F., Muñoz, A., Pérez-Payá, E., Misra, S., López-García, B. (2008). Identification and rational design of novel antimicrobial peptides for plant protection. Annu. Rev. Phytopathol. 46, 273–301. doi: 10.1146/annurev.phyto.121307.094843
Mariz-Ponte, N., Regalado, L., Gimranov, E., Tassi, N., Moura, L., Gomes, P., et al. (2021). A synergic potential of antimicrobial peptides against Pseudomonas syringae pv. actinidiae. Molecules 26, 1461. doi: 10.3390/molecules26051461
Maron, B., Rolff, J., Friedman, J., Hayouka, Z. (2022). Antimicrobial peptide combination can hinder resistance evolution. Microbiol. Spectr. 10, e97322. doi: 10.1128/spectrum.00973-22
Marxer, M., Vollenweider, V., Schmid-Hempel, P. (2016). Insect antimicrobial peptides act synergistically to inhibit a trypanosome parasite. Philos. Trans. R. Soc Lond. B Biol. Sci. 371, 20150302. doi: 10.1098/rstb.2015.0302
McCafferty, D. G., Cudic, P., Yu, M. K., Behenna, D. C., Kruger, R. (1999). Synergy and duality in peptide antibiotic mechanisms. Curr. Opin. Chem. Biol. 3, 672–680. doi: 10.1016/s1367-5931(99)00025-3
McManus, P. S., Stockwell, V. O., Sundin, G. W., Jones, A. L. (2002). Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40, 443–465. doi: 10.1146/annurev.phyto.40.120301.093927
McNulty, M. J., Gleba, Y., Tusé, D., Hahn Löbmann, S., Giritch, A., Nandi, S., et al. (2019). Techno-economic analysis of a plant-based platform for manufacturing antimicrobial proteins for food safety. Biotechnol. Prog. 36, e2896. doi: 10.1002/btpr.2896
Mello, E. O., Ribeiro, S. F. F., Carvalho, A. O., Santos, I. S., Da Cunha, M., Santa-Catarina, C., et al. (2011). Antifungal activity of pvd1 defensin involves plasma membrane permeabilization, inhibition of medium acidification, and induction of ROS in fungi cells. Curr. Microbiol. 62, 1209–1217. doi: 10.1007/s00284-010-9847-3
Melo, M. N., Ferre, R., Castanho, M. A. R. B. (2009). Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 7, 245–250. doi: 10.1038/nrmicro2095
Mercer, D. K., Torres, M., Duay, S. S., Lovie, E., Simpson, L., et al. (2020). Antimicrobial susceptibility testing of antimicrobial peptides to better predict efficacy. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00326
Monroc, S., Badosa, E., Feliu, L., Planas, M., Montesinos, E., Bardají, E., et al. (2006). De novo Designed cyclic cationic peptides as inhibitors of plant pathogenic bacteria. Peptides 27, 2567–2574. doi: 10.1016/j.peptides.2006.04.019
Montesinos, E. (2007). Antimicrobial peptides and plant disease control. FEMS Microbiol. Lett. 270, 1–11. doi: 10.1111/j.1574-6968.2007.00683.x
Montesinos, E., Bardají, E. (2008). Synthetic antimicrobial peptides as agricultural pesticides for plant-disease control. Chem. Biodivers. 5, 1225–1237. doi: 10.1002/cbdv.200890111
Montesinos, L., Bundo, M., Badosa, E., San, S. B., Coca, M., Monte, E., et al. (2017). Production of bp178, a derivative of the synthetic antibacterial peptide bp100, in the rice seed endosperm. BMC Plant Biol. 17, 63. doi: 10.1186/s12870-017-1011-9
Montesinos, L., Bundo, M., Izquierdo, E., Campo, S., Badosa, E., Rossignol, M., et al. (2016). Production of biologically active cecropin a peptide in rice seed oil bodies. PloS One 11, e146919. doi: 10.1371/journal.pone.0146919
Montesinos, L., Gascón, B., Ruz, L., Badosa, E., Planas, M., Feliu, L., et al. (2021). A bifunctional synthetic peptide with antimicrobial and plant elicitation properties that protect tomato plants from bacterial and fungal infections. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.756357
Mu, X., Wang, N., Li, X., Shi, K., Zhou, Z., Yu, Y., et al. (2016). The effect of colistin resistance-associated mutations on the fitness of Acinetobacter baumannii. Front. Microbiol. 7, 1715. doi: 10.3389/fmicb.2016.01715
Nadal, A., Montero, M., Company, N., Badosa, E., Messeguer, J., Montesinos, L., et al. (2012). Constitutive expression of transgenes encoding derivatives of the synthetic antimicrobial peptide bp100: impact on rice host plant fitness. BMC Plant Biol. 12, 159. doi: 10.1186/1471-2229-12-159
Nannini, E. C., Singh, K. V., Arias, C. A., Murray, B. E. (2013). In vivo Effects of cefazolin, daptomycin, and nafcillin in experimental endocarditis with a methicillin-susceptible Staphylococcus aureus strain showing an inoculum effect against cefazolin. Antimicrob. Agents Chemother. 57, 4276–4281. doi: 10.1128/AAC.00856-13
Osusky, M., Zhou, G., Osuska, L., Hancock, R. E., Kay, W. W., Misra, S., et al. (2000). Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat. Biotechnol. 18, 1162–1166. doi: 10.1038/81145
Pankievicz, V. C. S., Irving, T. B., Maia, L. G. S., Ané, J. (2019). Are we there yet? the long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol. 17, 99. doi: 10.1186/s12915-019-0710-0
Peressotti, E., Wiedemann-Merdinoglu, S., Delmotte, F., Bellin, D., Di Gaspero, G., Testolin, R., et al. (2010). Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 10, 147. doi: 10.1186/1471-2229-10-147
Perron, G. G., Zasloff, M., Bell, G. (2006). Experimental evolution of resistance to an antimicrobial peptide. Proc. R. Soc B 273, 251–256. doi: 10.1098/rspb.2005.3301
Portieles, R., Ayra, C., Gonzalez, E., Gallo, A., Rodriguez, R., Chacón, O., et al. (2010). Nmdef02, a novel antimicrobial gene isolated from Nicotiana megalosiphon confers high-level pathogen resistance under greenhouse and field conditions. Plant Biotechnol. J. 8, 678–690. doi: 10.1111/j.1467-7652.2010.00501.x
Pränting, M., Negrea, A., Rhen, M., Andersson, D. I. (2008). Mechanism and fitness costs of PR-39 resistance in Salmonella enterica serovar typhimurium LT2. Antimicrob. Agents Chemother. 52, 2734–2741. doi: 10.1128/AAC.00205-08
Puig, M., Moragrega, C., Ruz, L., Montesinos, E., Llorente, I. (2014). Postinfection activity of synthetic antimicrobial peptides Againststemphylium Vesicarium in pear. Phytopathology 104, 1192–1200. doi: 10.1094/PHYTO-02-14-0036-R
Puig, M., Moragrega, C., Ruz, L., Montesinos, E., Llorente, I. (2015). Controlling brown spot of pear by a synthetic antimicrobial peptide under field conditions. Plant Dis. 99, 1816–1822. doi: 10.1094/PDIS-03-15-0250-RE
Rahnamaeian, M., Cytryńska, M., Zdybicka-Barabas, A., Dobslaff, K., Wiesner, J., Twyman, R. M., et al. (2015). Insect antimicrobial peptides show potentiating functional interactions against gram-negative bacteria. Proc. R. Soc B. 282, 20150293. doi: 10.1098/rspb.2015.0293
Ramamoorthy, V., Zhao, X., Snyder, A. K., Xu, J., Shah, D. M. (2007). Two mitogen-activated protein kinase signalling cascades mediate basal resistance to antifungal plant defensins in Fusarium graminearum. Cell Microbiol. 9, 1491–1506. doi: 10.1111/j.1462-5822.2006.00887.x
Rioux, D., Jacobi, V., Simard, M., Hamelin, R. C. (2000). Structural changes of spores of tree fungal pathogens after treatment with the designed antimicrobial peptide d2a21. Can. J. Bot. 78, 462–471. doi: 10.1139/b00-007
Rodríguez-Rojas, A., Makarova, O., Rolff, J. (2014). Antimicrobials, stress and mutagenesis. PloS Pathog. 10, e1004445. doi: 10.1371/journal.ppat.1004445
Roversi, D., Luca, V., Aureli, S., Park, Y., Mangoni, M. L., Stella, L., et al. (2014). How many antimicrobial peptide molecules kill a bacterium? the case of PMAP-23. ACS Chem. Biol. 9, 2003–2007. doi: 10.1021/cb500426r
Rustagi, A., Kumar, D., Shekhar, S., Yusuf, M. A., Misra, S., Sarin, B. N., et al. (2014). Transgenic Brassica juncea plants expressing msra1, a synthetic cationic antimicrobial peptide, exhibit resistance to fungal phytopathogens. Mol. Biotechnol. 56, 535–545. doi: 10.1007/s12033-013-9727-8
Sachs, J. L., Quides, K. W., Wendlandt, C. E. (2018). Legumes versus rhizobia: a model for ongoing conflict in symbiosis. New Phytol. 219, 1199–1206. doi: 10.1111/nph.15222
Sacristán, S., García Arenal, F. (2008). The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. doi: 10.1111/j.1364-3703.2007.00460.x
Sathoff, A. E., Samac, D. A. (2019). Antibacterial activity of plant defensins. Mol. Plant-Microbe Interact. 32, 507–514. doi: 10.1094/MPMI-08-18-0229-CR
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A., et al. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Savini, F., Loffredo, M. R., Troiano, C., Bobone, S., Malanovic, N., Eichmann, T. O., et al. (2020). Binding of an antimicrobial peptide to bacterial cells: interaction with different species, strains and cellular components. Biochim. Biophys. Acta Biomembr. 1862, 183291. doi: 10.1016/j.bbamem.2020.183291
Savini, F., Luca, V., Bocedi, A., Massoud, R., Park, Y., Mangoni, M. L., et al. (2017). Cell-density dependence of host-defense peptide activity and selectivity in the presence of host cells. ACS Chem. Biol. 12, 52–56. doi: 10.1021/acschembio.6b00910
Schreiber, C., Müller, H., Birrenbach, O., Klein, M., Heerd, D., Weidner, T., et al. (2017). A high-throughput expression screening platform to optimize the production of antimicrobial peptides. Microb. Cell. Fact. 16, 29. doi: 10.1186/s12934-017-0637-5
Segura, A., Moreno, M., Madueño, F., Molina, A., García-Olmedo, F. (1999). Snakin-1, a peptide from potato that is active against plant pathogens. Mol. Plant-Microbe Interact. 12, 16–23. doi: 10.1094/MPMI.1999.12.1.16
Shen, C., Zhong, L., Yang, Y., Doi, Y., Paterson, D. L., Stoesser, N., et al. (2020). Dynamics of MCR-1 prevalence and MCR-1-positive Escherichia coli after the cessation of colistin use as a feed additive for animals in china: a prospective cross-sectional and whole genome sequencing-based molecular epidemiological study. Lancet Microbe 1, e34–e43. doi: 10.1016/S2666-5247(20)30005-7
Shen, C., Zhong, L., Zhong, Z., Doi, Y., Shen, J., Wang, Y., et al. (2021). Prevalence of MCR-1 in colonized inpatients, China 2011–2019. Emerg. Infect. Dis. 27, 2502–2504. doi: 10.3201/eid2709.203642
Shi, W., Li, C., Li, M., Zong, X., Han, D., Chen, Y., et al. (2016). Antimicrobial peptide melittin against Xanthomonas oryzae pv. Oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 100, 5059–5067. doi: 10.1007/s00253-016-7400-4
Shu, C., Cui, K., Li, Q., Cao, J., Jiang, W. (2021). Epsilon-poly-l-lysine (ϵ-pl) exhibits multifaceted antifungal mechanisms of action that control postharvest alternaria rot. Int. J. Food Microbiol. 348, 109224. doi: 10.1016/j.ijfoodmicro.2021.109224
Sibel Akalın, A. (2014). Dairy-derived antimicrobial peptides: action mechanisms, pharmaceutical uses and production proposals. Trends Food Sci. Technol. 36, 79–95. doi: 10.1016/j.tifs.2014.01.002
Silverstein, K. A. T., Moskal, W. A., Wu, H. C., Underwood, B. A., Graham, M. A., Town, C. D., et al. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51, 262–280. doi: 10.1111/j.1365-313X.2007.03136.x
Snoussi, M., Talledo, J. P., Del Rosario, N., Mohammadi, S., Ha, B., Košmrlj, A., et al. (2018). Heterogeneous absorption of antimicrobial peptide LL37 in Escherichia coli cells enhances population survivability. Elife 7, e38174. doi: 10.7554/eLife.38174
Spohn, R., Daruka, L., Lázár, V., Martins, A., Vidovics, F., Grézal, G., et al. (2019). Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 10, 4538. doi: 10.1038/s41467-019-12364-6
Steiner, H., Hultmark, D., Engström, Å., Bennich, H., Boman, H. G. (1981). Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292, 246–248. doi: 10.1038/292246a0
Sun, G., Wang, H., Shi, B., Shangguan, N., Wang, Y., Ma, Q., et al. (2017). Control efficiency and expressions of resistance genes in tomato plants treated with ϵ-poly-l-lysine against botrytis cinerea. Pest. Biochem. Physiol. 143, 191–198. doi: 10.1016/j.pestbp.2017.07.007
Sun, G., Yang, Q., Zhang, A., Guo, J., Liu, X., Wang, W., et al. (2018). Synergistic effect of the combined bio-fungicides ϵ-poly- l -lysine and chitooligosaccharide in controlling grey mould (Botrytis cinerea) in tomatoes. Int. J. Food Microbiol. 276, 46–53. doi: 10.1016/j.ijfoodmicro.2018.04.006
Sundin, G. W., Wang, N. (2018). Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 56, 161–180. doi: 10.1146/annurev-phyto-080417-045946
Swathi Anuradha, T., Divya, K., Jami, S. K., Kirti, P. B. (2008). Transgenic tobacco and peanut plants expressing a mustard defensin show resistance to fungal pathogens. Plant Cell Rep. 27, 1777–1786. doi: 10.1007/s00299-008-0596-8
Tam, J., Wang, S., Wong, K., Tan, W. (2015). Antimicrobial peptides from plants. Pharmaceuticals 8, 711–757. doi: 10.3390/ph8040711
Terras, F., Schoofs, H., Thevissen, K., Osborn, R. W., Vanderleyden, J., Cammue, B., et al. (1993). Synergistic enhancement of the antifungal activity of wheat and barley thionins by radish and oilseed rape 2s albumins and by barley trypsin inhibitors. Plant Physiol. 103, 1311–1319. doi: 10.1104/pp.103.4.1311
Tesfaye, M., Silverstein, K. A., Nallu, S., Wang, L., Botanga, C. J., Gomez, S. K., et al. (2013). Spatio-temporal expression patterns of Arabidopsis thaliana and Medicago truncatula defensin-like genes. PloS One 8, e58992. doi: 10.1371/journal.pone.0058992
Tester, M., Langridge, P. (2010). Breeding technologies to increase crop production in a changing world. Science 327, 818–822. doi: 10.1126/science.1183700
Thevissen, K., Terras, F. R. G., Broekaert, W. F. (1999). Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl. Environ. Microbiol. 65, 5451–5458. doi: 10.1128/AEM.65.12.5451-5458.1999
Topman, S., Tamir-Ariel, D., Bochnic-Tamir, H., Stern Bauer, T., Shafir, S., Burdman, S., et al. (2018). Random peptide mixtures as new crop protection agents. Microb. Biotechnol. 11, 1027–1036. doi: 10.1111/1751-7915.13258
Vaddady, P. K., Trivedi, A., Rathi, C., Madhura, D. B., Liu, J., Lee, R., et al. (2019). Dynamic time-kill curve characterization of spectinamide antibiotics 1445 and 1599 for the treatment of tuberculosis. Eur. J. Pharm. Sci. 127, 233–239. doi: 10.1016/j.ejps.2018.11.006
Van der Biezen, E. A. (2001). Quest for antimicrobial genes to engineer disease-resistant crops. Trends Plant Sci. 6, 89–91. doi: 10.1016/S1360-1385(01)01870-2
Van der Weerden, N. L., Lay, F. T., Anderson, M. A. (2008). The plant defensin, nad1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 283, 14445–14452. doi: 10.1074/jbc.M709867200
Van de Velde, W., Zehirov, G., Szatmari, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. doi: 10.1126/science.1184057
Velivelli, S. L. S., Czymmek, K. J., Li, H., Shaw, J. B., Buchko, G. W., Shah, G. M., et al. (2020). Antifungal symbiotic peptide ncr044 exhibits unique structure and multifaceted mechanisms of action that confer plant protection. Proc. Nat. Acad. Sci. U. S. A. 117, 16043–16054. doi: 10.1073/pnas.2003526117
Vu, T. T., Kim, H., Tran, V. K., Vu, H. D., Hoang, T. X., Han, J. W., et al. (2017). Antibacterial activity of tannins isolated from Sapium baccatum extract and use for control of tomato bacterial wilt. PloS One 12, e181499. doi: 10.1371/journal.pone.0181499
Wang, W., Deng, L., Yao, S., Zeng, K. (2018). Control of green and blue mold and sour rot in citrus fruits by the cationic antimicrobial peptide PAF56. Postharvest Biol. Technol. 136, 132–138. doi: 10.1016/j.postharvbio.2017.10.015
Wang, D., Griffitts, J., Starker, C., Fedorova, E., Limpens, E., Ivanov, S., et al. (2010). A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327, 1126–1129. doi: 10.1126/science.1184096
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in china: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wang, Q., Yang, S., Liu, J., Terecskei, K., Ábrahám, E., Gombár, A., et al. (2017). Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Nat. Acad. Sci. U. S. A. 114, 6854–6859. doi: 10.1073/pnas.1700715114
Westerhoff, H. V., Zasloff, M., Rosner, J. L., Hendler, R. W., de Waal, A., Gomes, A. V., et al. (1995). Functional synergism of magainins pgla and magainin-2 in Escherichia coli, tumor cells and liposomes. Eur. J. Biochem. 228, 257–264. doi: 10.1111/j.1432-1033.1995.00257.x
Wu, F., Tan, C. (2019). Dead bacterial absorption of antimicrobial peptides underlies collective tolerance. J. R. Soc Interface 16, 20180701. doi: 10.1098/rsif.2018.0701
Yan, H., Hancock, R. E. W. (2001). Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob. Agents Chemother. 45, 1558–1560. doi: 10.1128/AAC.45.5.1558-1560.2001
Yang, Z., Choi, H., Weisshaar, J. C. (2018). Melittin-induced permeabilization, re-sealing, and re-permeabilization of e. coli membranes. Biophys. J. 114, 368–379. doi: 10.1016/j.bpj.2017.10.046
Yang, S., Wang, Q., Fedorova, E., Liu, J., Qin, Q., Zheng, Q., et al. (2017). Microsymbiont discrimination mediated by a host-secreted peptide inmedicago truncatula. Proc. Nat. Acad. Sci. U. S. A. 114, 6848–6853. doi: 10.1073/pnas.1700460114
Yao, Z., Kahne, D., Kishony, R. (2012). Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol. Cell 48, 705–712. doi: 10.1016/j.molcel.2012.09.016
Yu, G., Baeder, D. Y., Regoes, R. R., Rolff, J. (2016). Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 60, 1717–1724. doi: 10.1128/AAC.02434-15
Yu, G., Baeder, D. Y., Regoes, R. R., Rolff, J. (2018). Predicting drug resistance evolution: insights from antimicrobial peptides and antibiotics. Proc. R. Soc B 285, 20172687. doi: 10.1098/rspb.2017.2687
Zasloff, M. (1987). Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cdna sequence of a precursor. Proc. Nat. Acad. Sci. U. S. A. 84, 5449–5453. doi: 10.1073/pnas.84.15.5449
Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. doi: 10.1038/415389a
Zhao, C., Dwyer, M. D., Yu, A. L., Wu, Y., Fang, S., Middelberg, A., et al. (2015). A simple and low-cost platform technology for producing pexiganan antimicrobial peptide in E. coli. Biotechnol. Bioeng. 112, 957–964. doi: 10.1002/bit.25505
Keywords: plant protection, antimicrobial peptide (AMP), phytopathogen, rapid killing, resistance evolution, synergism
Citation: Tang R, Tan H, Dai Y, Li L, Huang Y, Yao H, Cai Y and Yu G (2023) Application of antimicrobial peptides in plant protection: making use of the overlooked merits. Front. Plant Sci. 14:1139539. doi: 10.3389/fpls.2023.1139539
Received: 07 January 2023; Accepted: 07 April 2023;
Published: 19 July 2023.
Edited by:
Spyridon Alexandros Petropoulos, University of Thessaly, GreeceReviewed by:
Piyush Baindara, University of Missouri, United StatesCopyright © 2023 Tang, Tan, Dai, Li, Huang, Yao, Cai and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guozhi Yu, Z3VvemhpLnl1QHNpY2F1LmVkdS5jbg==
 Rui Tang
Rui Tang Hui Tan
Hui Tan Yan Dai
Yan Dai Lin’ai Li
Lin’ai Li Yan Huang
Yan Huang Huipeng Yao
Huipeng Yao Guozhi Yu
Guozhi Yu