
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 February 2023
Sec. Plant Bioinformatics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1137126
This article is part of the Research Topic Cucurbitaceae: Multi-omics, Functional Analysis, and Molecular Breeding View all 15 articles
 Weirong Luo1,2
Weirong Luo1,2 Zhenxiang Zhao1,2
Zhenxiang Zhao1,2 Hongzhi Chen3
Hongzhi Chen3 Wenhong Ao1,2
Wenhong Ao1,2 Lin Lu1,2
Lin Lu1,2 Junjun Liu1,2
Junjun Liu1,2 Xinzheng Li1,2
Xinzheng Li1,2 Yongdong Sun1,2*
Yongdong Sun1,2*DELLA gene family plays a key role in regulating plant development and responding to stress. Currently, many DELLA family members have been identified in plants, however, information on DELLA genes in pumpkin (Cucurbita moschata) is scarce. In this study, physical and chemical properties, gene structure cis-regulatory elements and expression of CmoDELLA genes were examined in pumpkin. We found that seven CmoDELLA genes were identified in pumpkin, and they were unevenly classified into five chromosomes. CmoDELLA proteins were relatively unstable and their secondary structures were mainly made up α-helix and random coil. All seven CmoDELLA proteins contained typical DELLA domain and GRAS domain, however, motif numbers between CmoDELLA proteins were unevenly distributed, implying the complex evolution and functional diversification of CmoDELLA proteins. Cis-regulatory elements analysis revealed that CmoDELLA genes might play an essential role in regulating plant growth and development, and response to stress in pumpkin. Transcriptome data in the roots, stems, leaves and fruits demonstrated that CmoDELLA2, CmoDELLA3 and CmoDELLA7 were related to the stems development, CmoDELLA1, CmoDELLA4, CmoDELLA5 and CmoDELLA6 were associated with the fruits development. Furthermore, we found that CmoDELLA1 and CmoDELLA5 were up-regulated under NaCl stress. CmoDELLA1, CmoDELLA2, CmoDELLA3, CmoDELLA5, CmoDELLA6 and CmoDELLA7 were remarkably induced under waterlogging stress. While, all of the 7 CmoDELLA genes showed significantly induced expression under cold stress. The expression patterns under abiotic stress suggested that CmoDELLA genes might mediate the stress response of pumpkin to NaCl, waterlogging and cold, however, the functions of different CmoDELLA genes varied under different stress. Overall, our study provides valuable information for further research about the potential functions and regulatory networks of CmoDELLA genes in pumpkin.
DELLA proteins play an essential role in regulating plant development and stress response, as key negative regulators of GA signaling (Zhou et al., 2020). DELLA protein sequences usually contain two conserved domains: DELLA domain and GRAS domain, which are distributed on the N-terminal region and C-terminal region, respectively. DELLA domain is known as the sensing domain of GA signaling for binding to GID1. And GRAS domain contributes mainly to repress GA responses by interacting with transcription factors (TFs), which is crucial for maintaining the functions of DELLA proteins (Xue et al., 2022). DELLA proteins do not contain the typical DNA-binding domain, however, they can interact with many TFs, such as PIFs, BZR1, EXP2, DREB1B, JAZs, and TCPs, to participate in almost all the processes of plant development and stress response (Thines et al., 2007; Navarro et al., 2008; Li et al., 2012; Li et al., 2016; Sechet et al., 2016; Liang et al., 2019). Identification of DELLA proteins firstly began in Arabidopsis, and AtRGL1, AtGAI, AtRGL2, AtRGA and AtRGl3, have been found as the members of the Arabidopsis DELLA family (Chen et al., 2013). DELLA proteins were subsequently identified from pear (Liu H. et al., 2016), strawberry (Li W.J. et al., 2018), cassava (Li X.L. et al., 2018), Camellia sinensis (Han et al., 2020), litchi (Wang et al., 2020), Brassica napus (Sarwar et al., 2021), Chinese cabbage (Guan et al., 2021) and other species.
DELLA genes play vital roles in regulating seed germination, hypocotyl elongation, plant height, flowering, fruit quality and stress response (Xue et al., 2022). Some studies have shown that DELLA genes can control seed germination by regulating the expression of multiple protein kinase genes (Cao et al., 2006). For instance, ABI3 and ABI5 can interact with DELLA proteins to activate SOMNUS and downstream target genes under high temperature, thereby inhibiting seed germination in Arabidopsis (Lim et al., 2013). DELLA proteins interacting with ARF6 and PIFs regulate cell elongation of the hypocotyl, which leading to short hypocotyl in Arabidopsis (Feng et al., 2008; Oh et al., 2014; Liu et al., 2018). Ethylene interacts with DELLA proteins to inhibit root growth and maintain apical hook-like structure of Arabidopsis (Achard et al., 2003). Furthermore, DELLA proteins interact with TCP to influence plant height through regulation of inflorescence apex growth (Davière et al., 2014), and interact with MONOCULM1 to affect tiller number and plant height of rice (Liao et al., 2019). DELLA proteins can also delay the floral transition by interacting with SPLs under long day conditions (Wang et al., 2009; Wu et al., 2009), and repress flowering by inhibiting LFY and SOC1 genes expression under short day conditions (Achard et al., 2007). In addition, Fruit development can be regulated by DELLA proteins. Researchers have concluded that DELLA proteins are implicated in the fruit initiation by interacting with SlARF7/SlIAA9 (Hu et al., 2018), and silencing of DELLA gene results in parthenocarpic fruit in tomato (Martí et al., 2007). According to previous studies, DELLA proteins participate in abiotic stress response and improve the plant survival by regulating reactive oxygen species (ROS) levels during adverse environments (Achard et al., 2008a; Achard et al., 2008b). Study has shown that DELLA protein enhances the salt tolerance of wheat seedling by increasing superoxide dismutase (SOD) activity under salt stress (Wang et al., 2016).
Pumpkin (Cucurbita moschata) is an annual vegetable crop, which has edible, medicinal and ornamental values. Pumpkin is widely cultivated across the globe, and the top producer is China, which produced 7.7 million tonnes of pumpkin (Worldmapper, 2021). Pumpkin is also widely used as a grafting rootstock for other cucurbit vegetable crops, including cucumber, watermelon and melon, which promotes plant growth and strengthens biotic and abiotic stress tolerance (Liu S. S. et al., 2016; Li et al., 2017; Zhang et al., 2019). In recent years, during cultivation, extreme weather and unfavorable environment conditions such as inappropriate temperature, drought stress, waterlogging stress and salt stress, seriously limited the growth and development of cucurbit vegetable crops, leading to the decline of yield and quality. Therefore, it is of significant importance to elucidate the stress response mechanism of pumpkin and screen pumpkin resistant rootstock for strengthening the stress resistance of cucurbit vegetable crops. However, there is no report of DELLA gene family in pumpkin development and stress response.
This main purpose of this study is to identify and characterize DELLA gene family in pumpkin, and to uncover their potential functions under abiotic stress. In this current study, seven CmoDELLA genes were identified in pumpkin. Moreover, the chromosomal localization, protein properties, phylogenetic analysis, gene structure, promoter cis-regulatory elements, protein interaction networks and expression of CmoDELLA genes were investigated. Additionally, physiological changes of pumpkin seedlings under waterlogging stress and cold stress were detected. This is the first report on the genome-wide characterization and expression of CmoDELLA genes in pumpkin, which provides valuable clues on the biological functions of CmoDELLA genes in regulating plant development and stress response for further research.
Pumpkin protein sequences were downloaded from the cucurbit genomics database (http://cucurbitgenomics.org/). Hidden Markov Model (HMM) profile of the DELLA domain (PF12041) was retrieved from the Pfam database (http://pfam.xfam.org). Then the pumpkin protein sequences were searched for this profile using hmmsearch tool of Tbtools software. AtDELLA genes were acquired by searching their gene IDs from TAIR (https://www.arabidopsis.org/), and then they were used as search queries to carry out BLASTp with the E-value of 1e-5 against pumpkin protein sequences. Subsequently, all the candidate protein sequences gained with the above two methods were submitted to CDD (http://ncbi.nlm.nih.gov/cdd) in NCBI to reconfirm the CmoDELLA proteins.
The chromosomal localization information of CmoDELLA family genes was obtained through cucurbit genomics database, and mapped using TBtools v 1.0986961 software (Chen et al., 2020). According to their chromosomal position, CmoDELLA family members were renamed. The physical and chemical properties, such as the coding sequence (CDS) lengths, amino acids number (AA), molecular weight (MW), isoelectric point (pI), grand average of hydropathicity (GRAVY) values and instability index, were predicted through Expasy (https//www.expasy.org/). Additionally, secondary structure prediction was executed via NPS@:SOPMA (https://npsa-prabi.Ibcp.fr/cgi-bin/npsa_automat.sopm.html).
DELLA protein sequences of pumpkin (7), cucumber (4), melon (4), watermelon (4), Arabidopsis (5), soybean (7), Brassica napus (13), rice (1), tomato (2) and maize (3) were retrieved from cucurbit genomics database, TAIR and Ensembl database (http://plants.ensembl.org/index.html), respectively. Based on multiple sequence alignment, phylogenetic tree was created using the neighbor-joining (NJ) method with MEGA 7.0. The conserved domains were analyzed by the NCBI CDD, and the conserved motifs were predicted by MEME (https://meme-suite.org/meme/). Furthermore, the distribution maps of conserved domains and conserved motifs were visualized using TBtools v 1.0986961. Promoter sequences (2 kp before the start codon) of CmoDELLA genes were analyzed through online PlantCare (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to obtain cis-regulatory elements. Interaction networks between CmoDELLA proteins and other proteins were conducted through online STRING (https://string-db.org/), using Arabidopsis as reference species.
The pre-published pumpkin transcriptome sequencing data under four tissues (roots, stems, leaves and fruits) (PRJNA385310) and NaCl stress (PRJNA437579) were obtained from the cucurbit genomics database for exploring the transcriptional profiles. Transcriptional levels were normalized by the reads per kilobase of exon per million reads mapped (RPKM) method. Expression heatmap of CmoDELLA genes was generated with TBtools v1.0986961.
In this current study, pumpkin variety “Hantailang” was used to explore the expression characterization of CmoDELLA genes under waterlogging stress and cold stress. “Hantailang” was cultivated and developed in a climate chamber with growth conditions (16 h light/8 h dark, 25°C daytime/16°C night). The pumpkin seedlings of two-leaf stage were exposed to abiotic stress. Waterlogging stress were conducted with water 2 cm above the soil surface. For cold stress, the seedlings were maintained at 15°C daytime/5°C night. The leaves were collected at day 10 after treatment and stored in -80 °C fridge for qRT-PCR analysis and physiological indicators measurement.
Total RNA of pumpkin leaves was extracted using TaKaRa MiniBEST Plant RNA Extraction Kit (Code No. 9769, TaKaRa, Dalian, China) with the instructions provided in the kit. First chain cDNA was synthesized using PrimeScript™ RT Master Mix (Perfect Real Time) Reagent Kit (Code No. RR036A, Takara, Dalian, China). Subsequently, qRT-PCR experiment was operated by TB Green® Premix Ex Taq™ II (Code No. RR820A, TaKaRa, Dalian, China) to display the expression patterns under waterlogging stress and cold stress. The primer sequences of CmoDELLA genes and reference gene (CmoActin) were listed in Table S1. All the experiments were carried out followed by the instructions. Each sample was replicated three times and the relative expression levels of CmoDELLA genes were analyzed with the 2–ΔΔCt method.
The contents of proline and Malondialdehyde (MDA), catalase (CAT) activity in the leaves of pumpkin were measured using proline (Code No. BC0290, Solarbio, Beijing, China), MDA (Code No. BC0025, Solarbio, Beijing, China) and CAT (Code No. BC0205, Solarbio, Beijing, China) assay kit respectively, following the manufacturer’s protocol. SOD activity was determined with SOD assay kit (Code No. G0104F, Geruisi, Suzhou, China).
A total of 7 CmoDELLA genes were obtained through genome-wide search, and they were successively renamed as CmoDELLA1~7 respectively, according to their chromosomal localization. Seven CmoDELLA genes were located on Chr1, Chr4, Chr11, Chr14 and Chr15, respectively. Among them, there were two CmoDELLA genes on the Chr4 (CmoDELLA2 and CmoDELLA3) and Chr15 (CmoDELLA6 and CmoDELLA7), while only one CmoDELLA gene on the Chr1, Chr11 and Chr14 (Figure 1). The protein properties of CmoDELLA gene family were collected in Table S2. The CDS lengths of CmoDELLA genes varied from 1605 bp to 1854 bp. AA number of CmoDELLA proteins ranged from 534 aa to 617 aa, the MW was between 58316.97 Da and 67383.21 Da, the pI range was 4.70 to 5.52, the GRAVY changed from -0.312 to -0.092. Seven CmoDELLA proteins were defined as unstable proteins with their instability index varying from 42.56 to 53.57. The secondary structures of seven CmoDELLA proteins were all made up α-helix, random coil, extended strand and β-sheets, in which α-helix accounted for the largest proportion (45.52%~49.17%), followed by random coil, extended strand and β-sheets, indicating that the structures of CmoDELLA proteins were the mixed type. Function annotation showed that CmoDELLA1 and CmoDELLA5 were related to the regulation of transcription, whereas CmoDELLA2, CmoDELLA3, CmoDELLA4, CmoDELLA6 and CmoDELLA7 were involved in the regulation of transcription, plant hormone signal transduction pathway, growth and development, and stress response, indicating their pivotal roles in regulating pumpkin development and stress response.
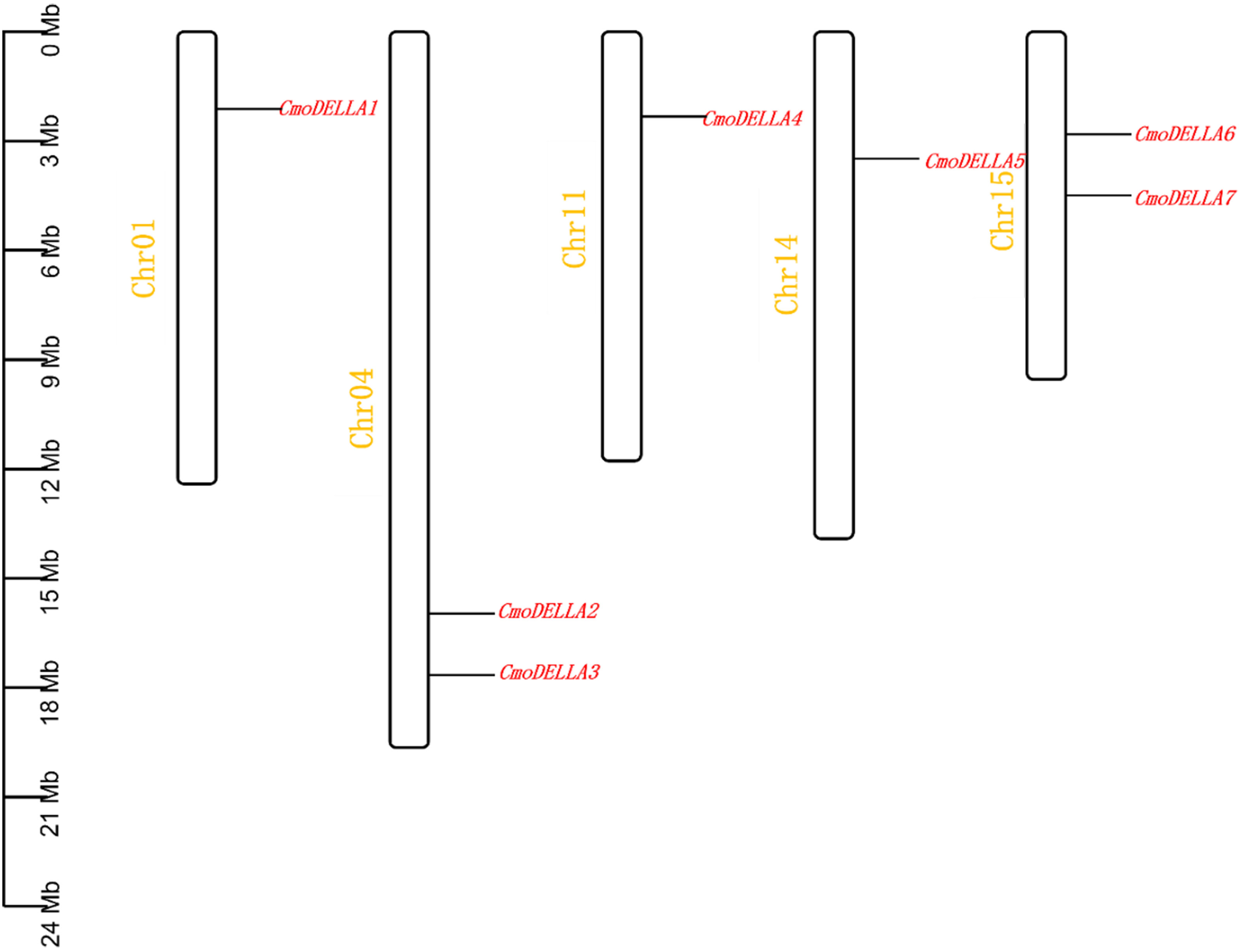
Figure 1 Chromosomal localization of CmoDELLA genes. The names of seven CmoDELLA genes are shown to the right of each chromosome. Gene position can be measured using the scale on the left of the figure in megabases (Mb).
To evaluate the evolutionary relationship of DELLA proteins, phylogenetic tree was created by aligning 50 DELLA protein sequences from pumpkin, cucumber, melon, watermelon, Arabidopsis, soybean, Brassica napus, rice, tomato and maize (Figure 2). According to the phylogenetic relationship listed in the tree, the 50 DELLA proteins could be divided into 4 major classes: Class I, Class II, Class III and Class IV, which contained 8, 11, 15, and 16 members, respectively. As shown in the phylogenetic tree, DELLA proteins from pumpkin exhibited the relatively closer evolutionary relationship with those from watermeolon, melon, cucumber. In addition, CmoDELLA3 and CmoDELLA6 were classified into the Class II, CmoDELLA2 and CmoDELLA7 were clustered into Class III, CmoDELLA1, CmoDELLA4 and CmoDELLA5 belonged to Class IV. Multiple sequence alignment was performed among CmoDELLA proteins (Figure 3). The homology between CmoDELLA2 and CmoDELLA7 was the highest at 94.72%, followed by that between CmoDELLA3 and CmoDELLA6 at 89.37%. The homology of other CmoDELLA proteins was more than 64.66%, indicating the high conservation and complex evolution of CmoDELLA proteins. To better understand the structural differences of CmoDELLA proteins, the conserved domains and conserved motifs were detected. The results showed that CmoDELLA proteins were all composed of N-terminal DELLA domain and C-terminal GRAS domain (Figures 3, 4A), but the conserved motifs between CmoDELLA proteins were unevenly distributed (Figure 4B, Table 1). The number of motifs of different CmoDELLA proteins varied from 11 to 17. Class II memebers contained 17 motifs, Class III memebers included 16 to 17 motifs, whereas Class IV memebers held 11 to 14 motifs. A total of 20 motifs were found in CmoDELLA proteins, and 10 motifs (including motif1, motif2, motif3, motif5, motif6, motif7, motif8, motif9, motif10 and motif14) were highly conserved in all CmoDELLA proteins. Motif4 was identified in all CmoDELLA proteins except CmoDELLA4, and motif13 was present in all CmoDELLA proteins except CmoDELLA5. Moreover, motif15, motif16 and motif17 were detected in CmoDELLA2 and CmoDELLA7. Motif18, motif19 and motif20 were observed in CmoDELLA3 and CmoDELLA6. The different number of motifs between CmoDELLA proteins indicated their functional diversification in pumpkin.
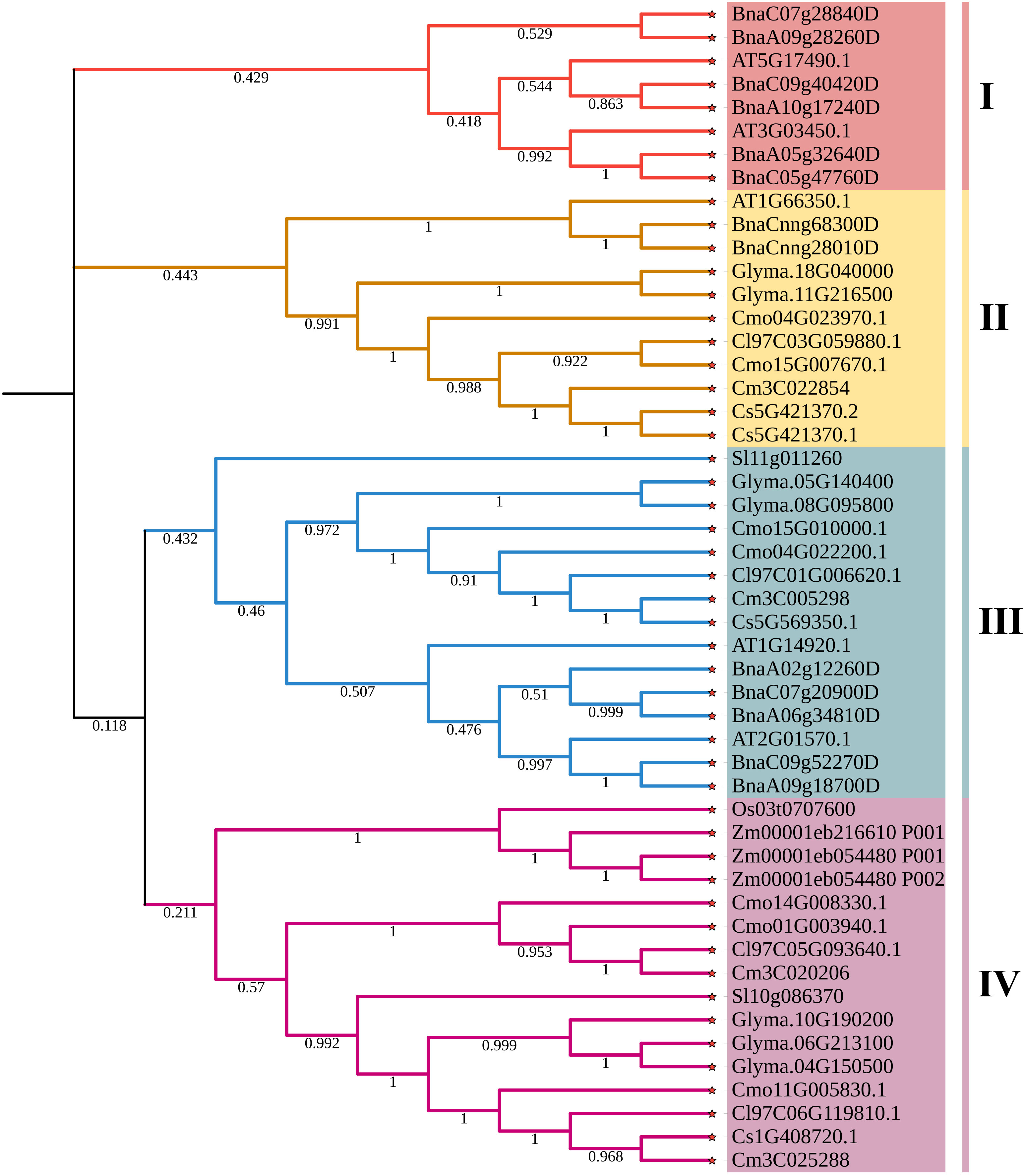
Figure 2 The phylogenetic tree of DELLA proteins from pumpkin, cucumber, melon, watermelon, Arabidopsis, soybean, Brassica napus, rice, tomato and maize. Cmo: pumpkin, Cs: cucumber, Cm: melon, Cl: watermelon, AT: Arabidopsis, Glyma.: soybean, Bna: Brassica napus, Os: rice, Sl: tomato, Zm: maize. The 4 major classes are represented by the different colors.
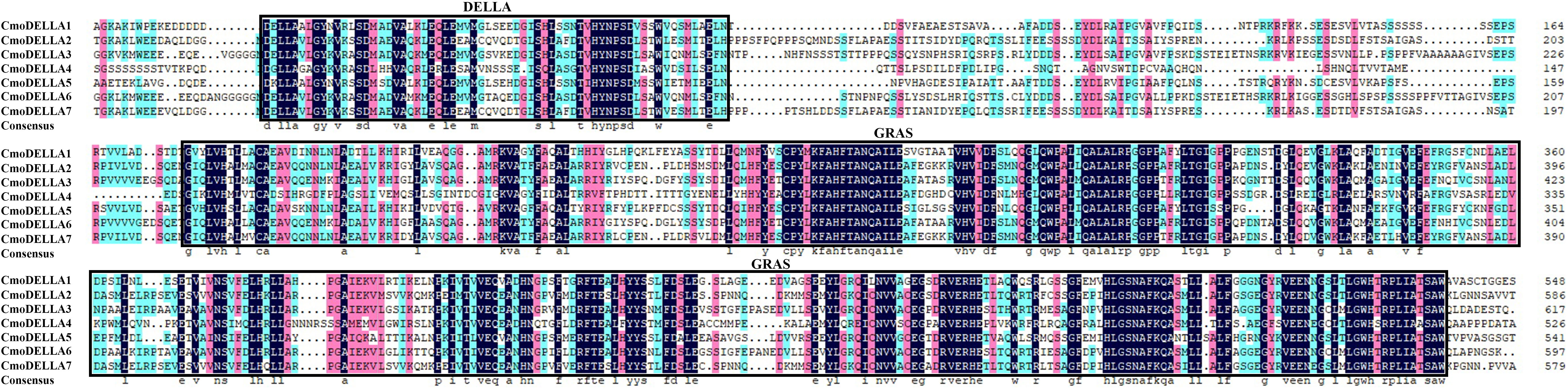
Figure 3 Amino acid sequence alignment of CmoDELLA proteins. The numbers on the right side of the sequence indicate the position of amino acid residues and the colors represent similarities in the protein sequences. The identical amino acid residues are shaded in black background, and the similar amino acid residues are shaded in the red and blue background.
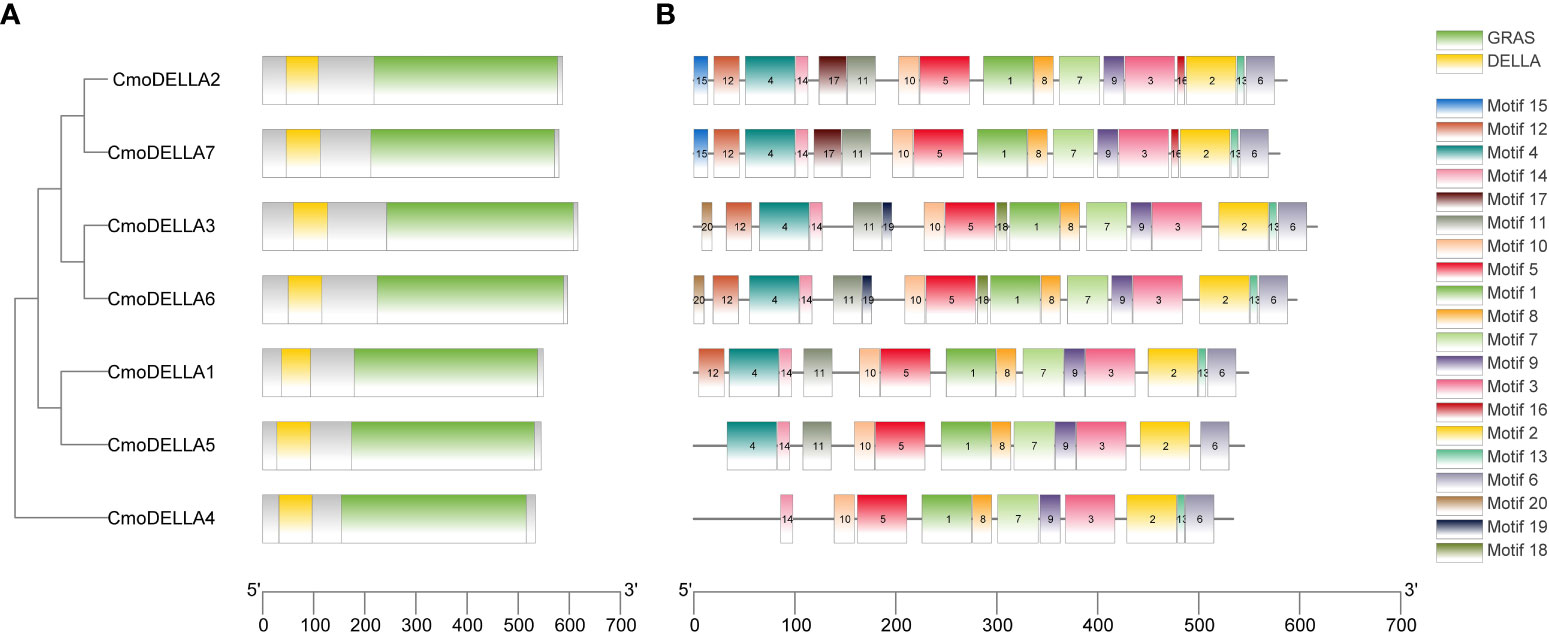
Figure 4 The conserved domains and conserved motifs of CmoDELLA proteins. The lengths of the conserved domains and conserved motifs are shown proportionally. (A) The conserved domains of CmoDELLA proteins. DELLA domain and GRAS domain are represented by yellow and green box, respectively. (B) The conserved motifs of CmoDELLA proteins. The different conserved motifs are indicated by different colored boxes.
To learn more about the potential functions of CmoDELLA genes involved in different biological process, the promoter sequences of CmoDELLA genes were analyzed to identify cis-regulatory elements. The identified cis-regulatory elements were mainly related to light, plant hormone, stress, and plant growth and development (Figure 5). Among them, light response elements were the most abundant, such as G-Box, TCCC-motif, TCT-motif, Box II, I-box and AE-box. The plant hormone response elements were widely present in the promoter region, including abscisic acid response element (ABRE), TGACG-motif and CGTCA-motif for methyl jasmonic acid response element (MeJA), P-box and GARE-motif for gibberellin response element (GARE), TCA-element for salicylic acid response element (SARE) and AuxRR-core for auxin response element. Stress response elements containing MBS (involved in drought induction), LTR (involved in low temperature response), ARE (involved in anaerobic induction), STRE (involved in heat induction), WUN-motif (involved in wound response), and TC-rich repeats (involved in defense and stress response) were also identified. Simultaneously, we found some growth and development elements, for example, CAT-box associated with meristem, O2-site involved in the regulation of zein metabolism and circadian involved in circadian rhythm regulation.
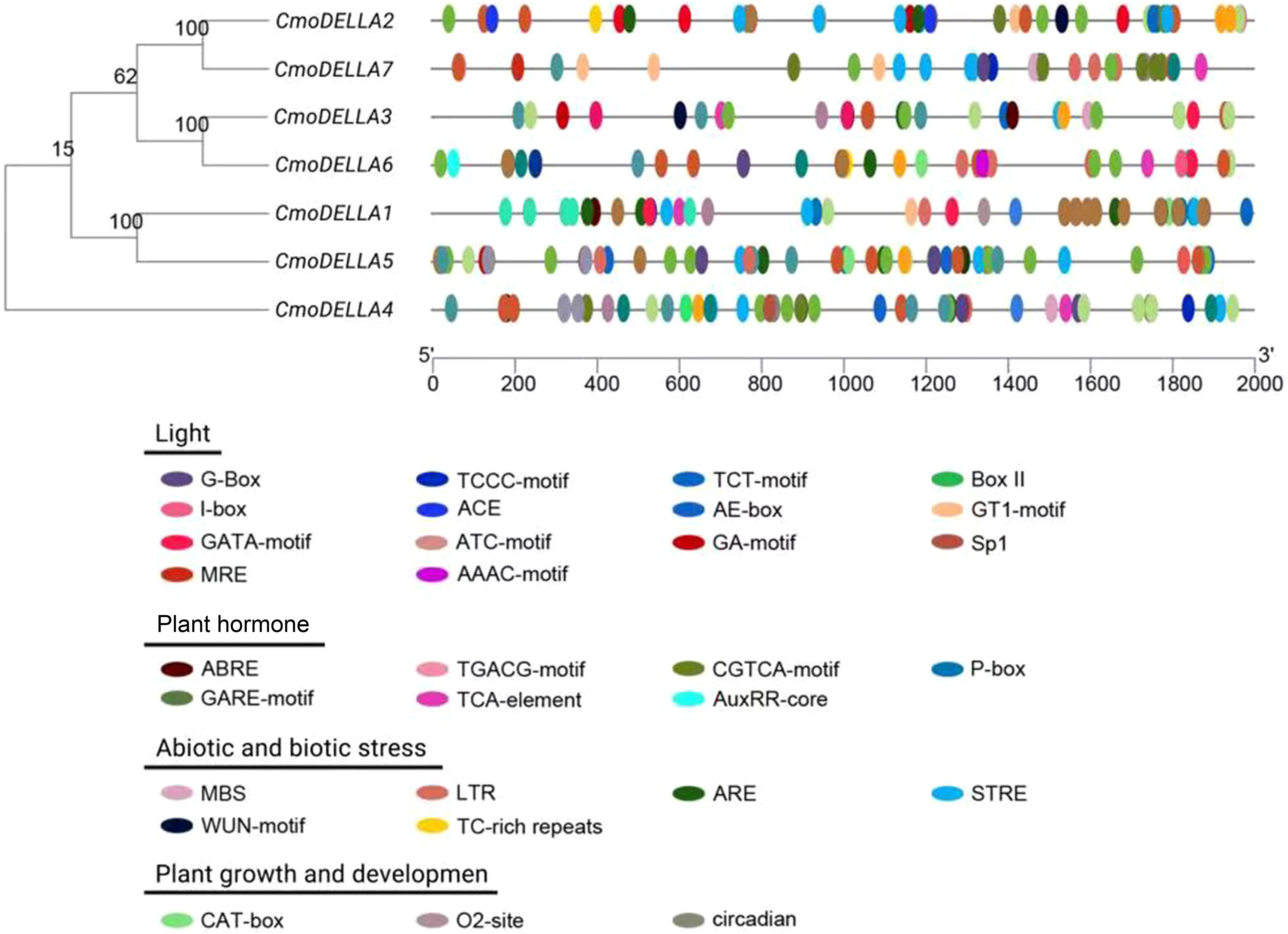
Figure 5 Cis-regulatory elements in the promoter regions of CmoDELLA genes. These cis-regulatory elements are related to light response, plant hormone response, stress response, and plant growth and development. The different colored circles represent the different types and positions of cis-regulatory elements in each CmoDELLA gene.
To better understand the regulatory mechanism of CmoDELLA proteins, protein interaction networks were constructed. It could be seen from Figure 6 that seven CmoDELLA proteins, such as RGL1 (CmoDELLA3), GAI (CmoDELLA2, CmoDELLA4, CmoDELLA6 and CmoDELLA7), RGL2 (CmoDELLA1 and CmoDELLA5), all interacted with GID1A, GID1B, GID1C, SLY1 and PIF3 using Arabidopsis as reference species.
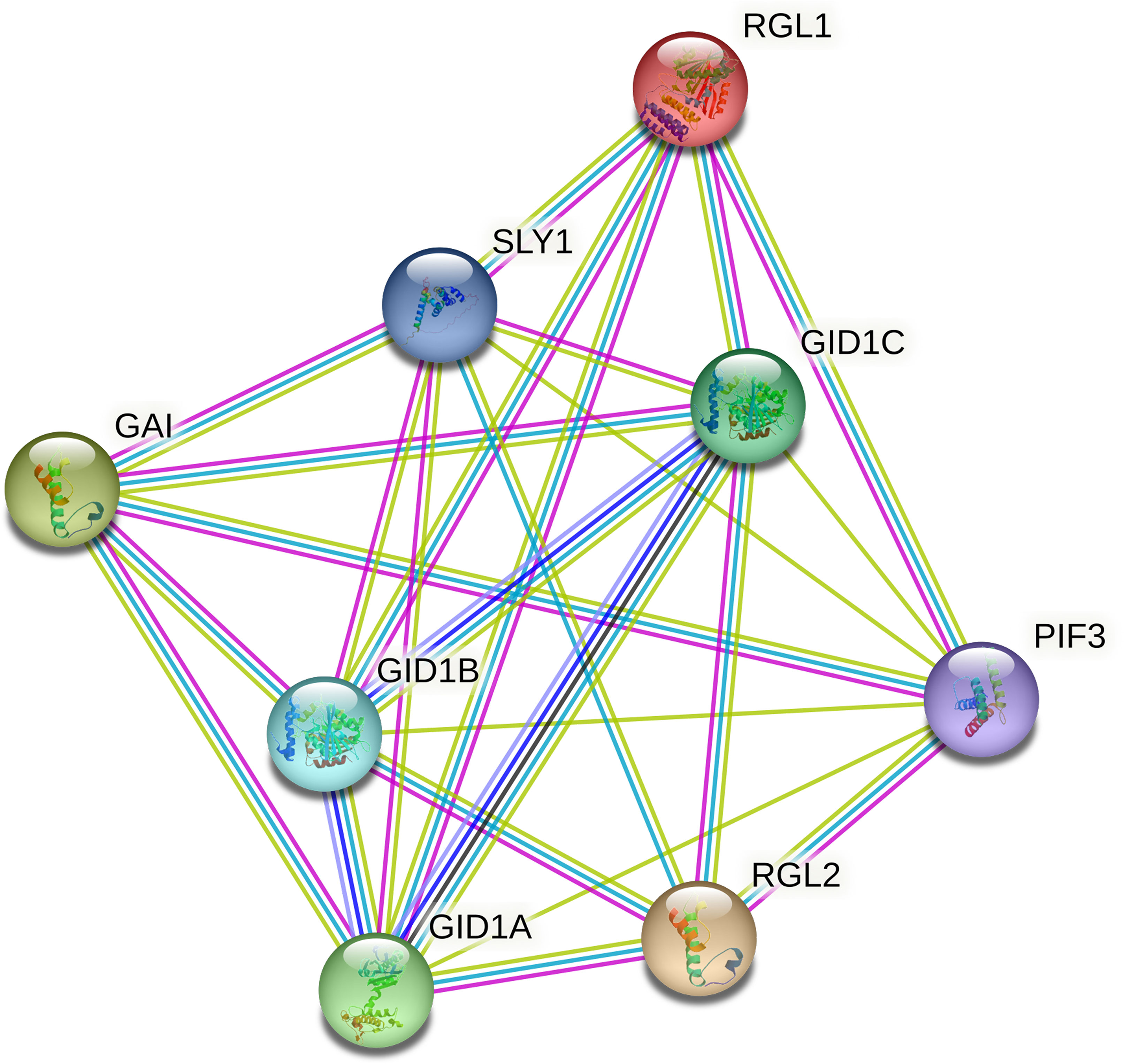
Figure 6 Interaction networks of CmoDELLA proteins. The network nodes represent proteins and the edges indicate protein-protein associations.
To uncover the expression profiles of CmoDELLA genes, the transcriptional levels of CmoDELLA genes in the roots, stems, leaves and fruits were evaluated (Figure 7). RNA-seq data revealed that seven CmoDELLA genes were expressed in all analyzed tissues but exhibited different abundance levels. For example, CmoDELLA2, CmoDELLA3, CmoDELLA4, CmoDELLA6 and CmoDELLA7 had higher transcriptional profiles, whereas CmoDELLA1 and CmoDELLA5 demonstrated lower transcriptional levels in the four tissues. Moreover, CmoDELLA2, CmoDELLA3 and CmoDELLA7 were highly expressed in the stems, while CmoDELLA1, CmoDELLA4, CmoDELLA5 and CmoDELLA6 were mainly expressed in the fruits. Differential expression of CmoDELLA genes in different tissues indicated that the tissue specificity and functional divergence of CmoDELLA genes in the growth and development of pumpkin.
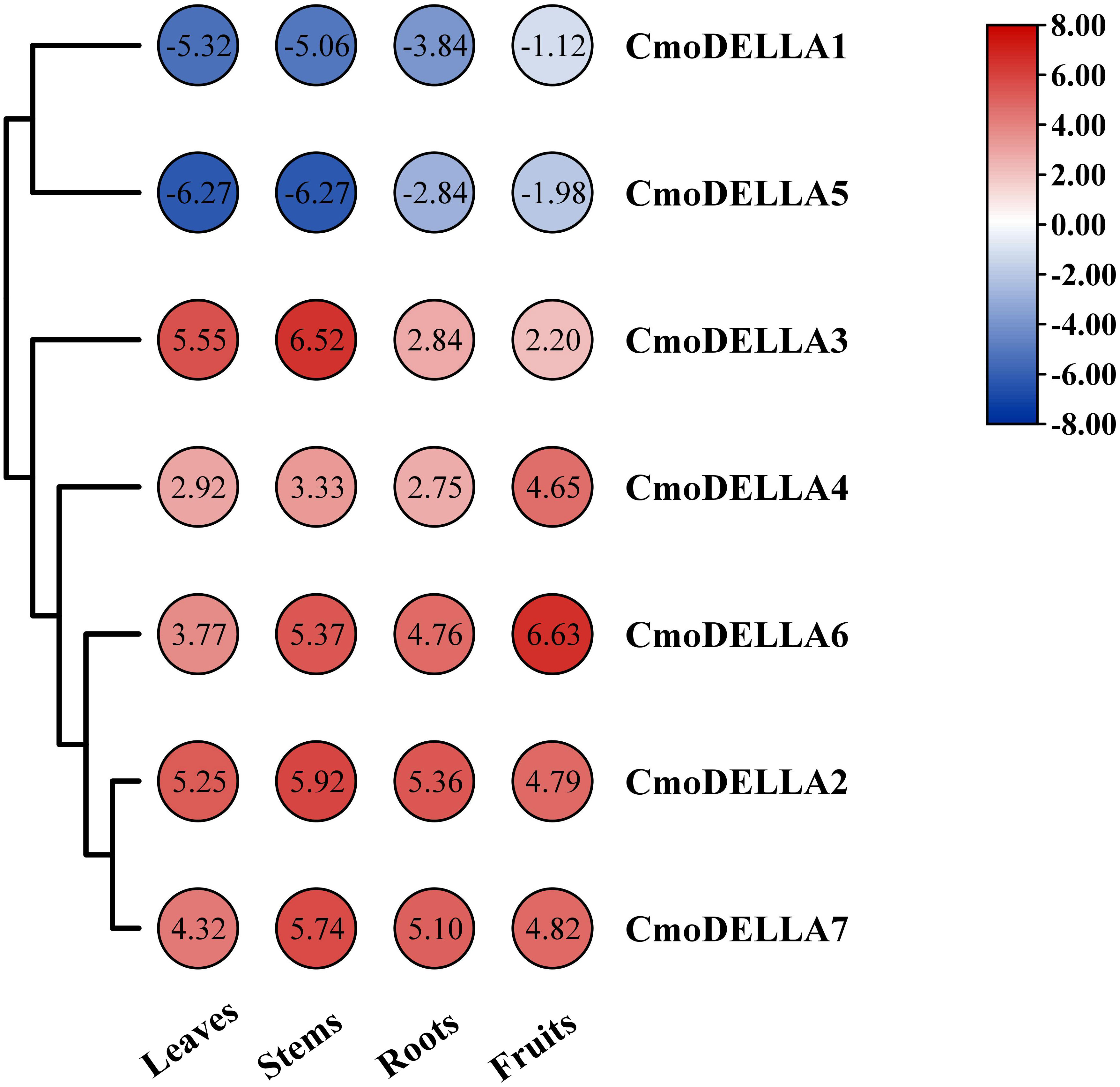
Figure 7 Transcriptional levels of CmoDELLA genes in the different tissues (leaves, stems, roots and fruits). The transcriptional levels are normalized by the reads per kilobase of exon per million reads mapped (RPKM) method. The red color represents the higher expression, while the blue color indicates the lower expression.
CmoDELLA genes not only participated in plant development, but were also involved in various abiotic stress. Promoter cis-regulatory elements analysis also showed that CmoDELLA genes contained some stress response elements. To further explore and gain more insights into possible functions under abiotic stresses, such as salt, waterlogging and cold temperature, transcriptional profiles of CmoDELLA genes under NaCl stress were analyzed based on the pre-published RNA-seq data. CmoDELLA genes showed significant differences in response to NaCl stress (Figure 8). Transcriptional levels of CmoDELLA2, CmoDELLA3, CmoDELLA4, CmoDELLA6 and CmoDELLA7 were down-regulated after 75 mmol/L NaCl stress for 24 h in contrast with the normal condition. In contrast, the expression levels of CmoDELLA1 and CmoDELLA5 showed an increased trend after NaCl stress. In addition, the expression levels of CmoDELLA genes were investigated by qRT-PCR for further understanding the functions in response to waterlogging stress and cold stress. As shown in Figure 9, seven CmoDELLA genes were all up-regulated under waterlogging stress and cold stress after 10 days, compared to the normal condition. Under waterlogging stress, the expression levels of CmoDELLA genes were significantly higher than that without waterlogging treatment, except for CmoDELLA4. For example, CmoDELLA1~7 showed 1.62-, 1.51-, 1.45-, 1.18-, 1.30-, 1.44-, 1.41-fold higher expression levels, respectively, under waterlogging stress. Under cold stress, CmoDELLA1~7 exhited 9.25-, 8.56-, 7.22-, 5.81-, 6.38-, 6.95-, 5.02-fold remarkably higher expression, respectively, compared to the control. The above results suggested that CmoDELLA genes might be implicated in response to NaCl stress, waterlogging stress and cold stress in pumpkin, however, the functions of different CmoDELLA genes varied under different stress.
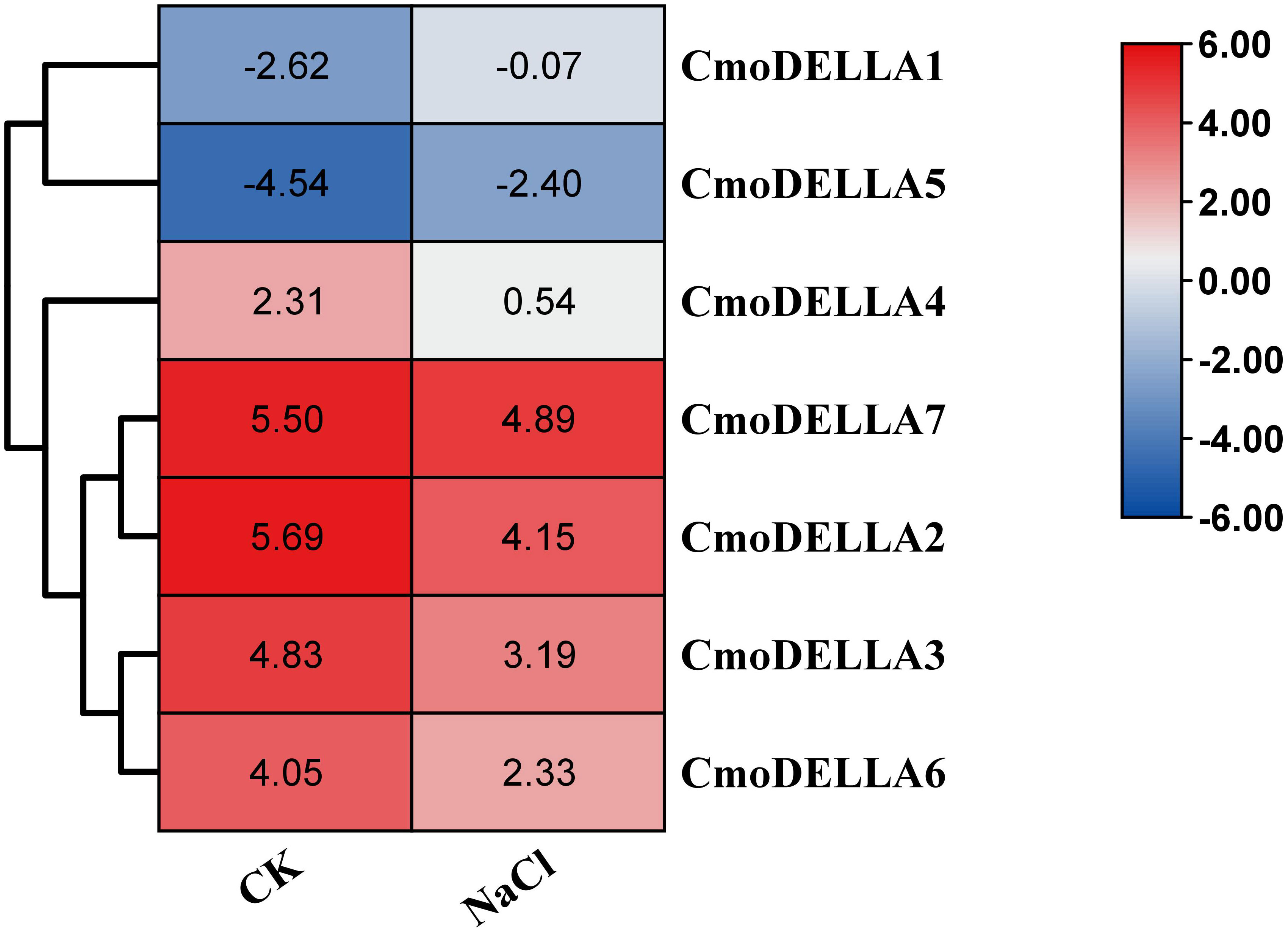
Figure 8 Transcriptional profiles of CmoDELLA genes under NaCl stress after 24 h. The transcriptional levels are normalized by RPKM method. The red color represents the higher expression, while the blue color indicates the lower expression.
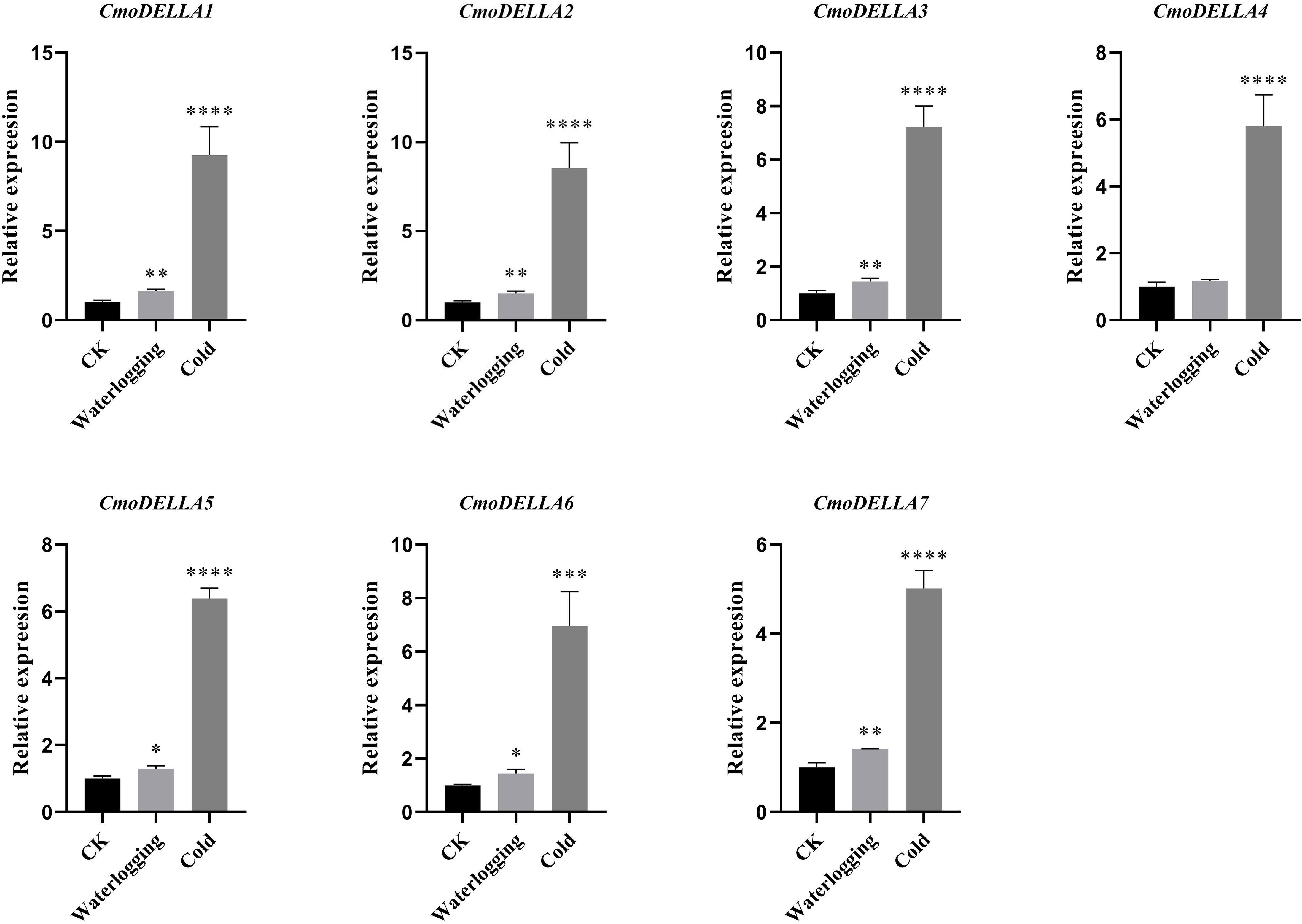
Figure 9 Expression levels of CmoDELLA genes in response to waterlogging stress and cold stress after 10 days. The relative expression levels of CmoDELLA genes are normalized with respect to the reference gene (CmoActin). The X-axis represents the waterlogging stress and cold stress. The Y-axis represents the relative expression levels. The values are denoted as the means ± SDs. The significant difference is represented by asterisks at *P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001.
In this study, we measured the changes of proline, MDA, SOD and CAT in pumpkin leaves under waterlogging stress and cold stress after 10 days (Figure 10). Under waterlogging stress and cold stress, the proline contents were remarkably lower, compared to the control. The MDA contents were 1.44- and 1.62-fold of that in the control, respectively, but did not differ significantly between the control and waterlogging stress. The CAT activities were significantly decreased by 18.16% and 35.43% under waterlogging stress and cold stress, respectively, compared with that in the control. While, SOD activity was lower under waterlogging stress than that in the control, but significantly higher under cold stress than that in the control.
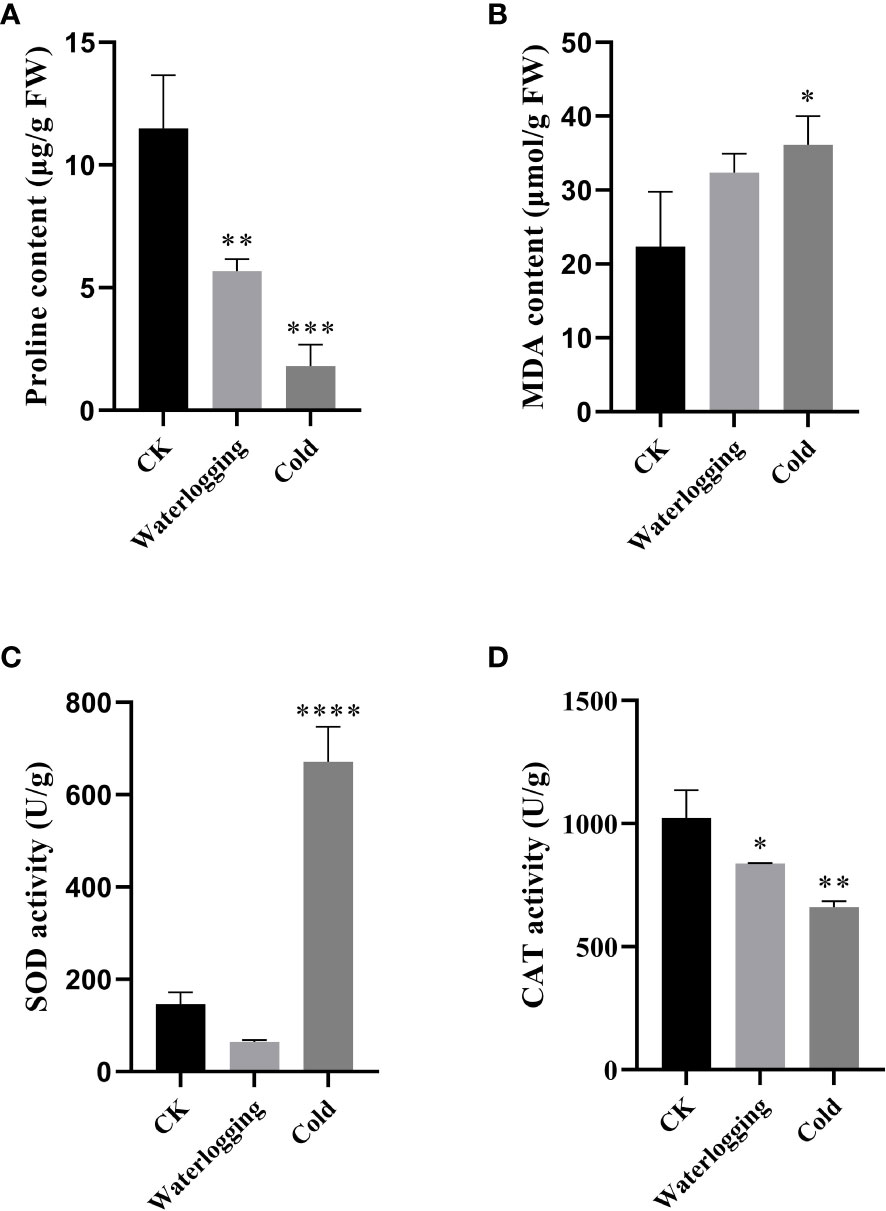
Figure 10 Physiological changes under waterlogging stress and cold stress after 10 days in pumpkin. (A); proline content. (B); MDA content. (C); SOD activity. (D); CAT activity. The values are denoted as the means ± SDs. The significant difference is represented by asterisks at *P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001.
DELLA gene family plays key roles in regulating plant development and stress response. Up to now, DELLA genes have been extensively identified and characterized in plants, but the research of DELLA genes in pumpkin has rarely been reported. In this present study, seven CmoDELLA genes were obtained in pumpkin by genome-wide analysis, which were located on five chromosomes respectively. CmoDELLA proteins were all unstable proteins, and the secondary structures of them were mainly made up α-helix and random coil. The phylogenetic tree displayed that 50 DELLA protein sequences from pumpkin, cucumber, melon, watermelon, Arabidopsis, soybean, Brassica napus, rice, tomato and maize were divided into four subfamilies, and the DELLA proteins of pumpkin shared the closer evolutionary relationship with those of watermeolon, melon, cucumber, which may be due to pumpkin and watermeolon, melon, cucumber belonging to cucurbita family. N-terminal DELLA domain and C-terminal GRAS domain were the typical conserved domains of DELLA family members in various plants (Sarwar et al., 2021). In this study, it was found that seven CmoDELLA proteins held the highly conserved DELLA domain and GRAS domain, shared 10 conserved motifs, however, motif numbers between CmoDELLA proteins were unevenly distributed, indicating the complex evolution and functional diversification of CmoDELLA proteins. These findings were consistent with the study has been found in BnaDELLA proteins (Sarwar et al., 2021).
Promoter cis-regulatory elements determine the specific function of the genes. Analysis of cis-regulatory elements can provide an insight into exploring the expression and regulation mechanism of genes under different tissues and stress environments. Previous studies have shown that DELLA proteins participate in plant hormone signal transduction pathway, including GA, auxin, abscisic acid, ethylene, and jasmonate (Achard et al., 2006), which affects diverse aspects of plant development and response to environmental stress (Xu et al., 2014). Additionally, some researches have reported the importance of ABRE, SARE and MeJA for abiotic stress tolerance via plant hormone signal transduction pathway (Doornbos et al., 2011; Rivas-San Vicente and Plasencia, 2011). In the current study, CmoDELLA genes contained a lot of promoter cis-regulatory elements involved in light, plant hormone, stress, and plant growth and development, such as ABRE, MeJA, GARE, SARE and ARE, suggesting that the CmoDELLA genes may be responsible for plant development and stress response in pumpkin by plant hormone signal pathway. Similar findings have been also reported in apple (Fan et al., 2017), rice (Muhammad et al., 2019), soybean (Liang et al., 2022) and mango (Wang et al., 2022).
Usually, DELLA proteins play essential roles by interacting with other TFs in plants, as key negative regulators of GA signaling (Zhou et al., 2020). GA signal transduction pathway is mainly involved in GID1, DELLA proteins and F-box family proteins (Xue et al., 2022). GID1A, GID1B and GID1C are soluble GA receptors, and SLY1 belongs to F-box family protein. GID1 interacts with the N-terminal domain of DELLA proteins to form GID1-DELLA complex, and then DELLA proteins are bound by SLY1, which leading to degradation of DELLA proteins (McGinnis et al., 2003; Murase et al., 2008). PIF3 is phytochrome-associated protein, and DELLA proteins bind with the promoter of PIF3 to inhibit the PIF-mediated hypocotyl elongation (de Lucas et al., 2008). Protein interaction networks analysis indicated that seven DELLA proteins all interacted with GID1A, GID1B, GID1C, SLY1 and PIF3, suggested their involvement in the development process via GA signal transduction pathway.
Gene functions are usually understood by detecting tissue-specific expression profiles of genes. Here, the transcriptome data of CmoDELLA genes was analyzed in the roots, stems, leaves and fruits. The results showed that seven genes were all expressed in the four tissues. Moreover, CmoDELLA2, CmoDELLA3 and CmoDELLA7 were highly expressed in the stems, while CmoDELLA1, CmoDELLA4, CmoDELLA5 and CmoDELLA6 were mainly expressed in the fruits. In cucumber, four DELLA genes displayed the distinct expression patterns in the different tissues, and CsGAIP exhibited the higher expression levels in the stems (Zhang et al., 2014). In compliance with this, some researches have also been reported that DELLA genes have a predominant roles in regulating plant stem elongation growth in Arabidopsis (King et al., 2001) and Brassica napus (Zhao et al., 2017). Furthermore, we found that CmoDELLA1 showed significantly up-regulated expression in the pollinated fruits than that in the ovaries without pollination in pumpkin (Luo et al., 2021). Recent study showed that DELLA proteins promoted ovule initiation by interacting with the CUC2 TF (Barro-Trastoy et al., 2022). The results presented here suggested that CmoDELLA2, CmoDELLA3 and CmoDELLA7 were related to the stems development, CmoDELLA1, CmoDELLA4, CmoDELLA5 and CmoDELLA6 were associated with the fruits development. Further investigation should be conducted to verify their functions.
Several studies have reported that DELLA genes participate in the stress response process (Huang et al., 2006). For instance, overaccumulation of DELLA proteins enhances the salt stress (Sakuraba et al., 2017) and cold stress tolerance (Yang et al., 2013), which significantly improves plant fitness. Han et al. (2020) analyzed the transcriptome data of DELLA genes in Camellia sinensis under drought stress, cold stress and NaCl stress, and speculated that DELLA genes were involved in the abiotic stress response. Sarwar et al. (2021) found that BnaDELLA genes exhibited different expression abundance under NaCl stress, suggesting the BnaDELLA genes vital roles in susceptibility to NaCl stress. Consistent with those, this current study showed the distinct transcriptional patterns of CmoDELLA genes under NaCl stress. For instance, CmoDELLA1 and CmoDELLA5 showed an increased expression under NaCl stress after 24 h, indicating CmoDELLA1 and CmoDELLA5 might be crucial in response to NaCl stress. Additionally, in our qRT-PCR analysis, CmoDELLA1, CmoDELLA2, CmoDELLA3, CmoDELLA5, CmoDELLA6 and CmoDELLA7 were remarkably induced under waterlogging stress after 10 days. And all of the 7 CmoDELLA genes showed significantly induced expression under cold stress after 10 days. These results suggested the vital roles of CmoDELLA genes in response to waterlogging stress and cold stress. Many previous studies on AtDELLA genes (Zhou et al., 2017; Blanco-Touriñán et al., 2020) and BnaDELLA genes (Sarwar et al., 2021) have provided evidence of their fundamental roles in regulating plant physiology under cold stress. Moreover, the expression profiles of different BnaDELLA genes varied under different stress treatments (Sarwar et al., 2021). Based on the above results, CmoDELLA genes might mediate the stress response of pumpkin to NaCl, waterlogging and cold, however, the functions of different CmoDELLA genes varied under different stress.
Previous report has shown that adverse environment conditions promote DELLA accumulation (Achard et al., 2008b) and then DELLA proteins increase SOD activity to improve salt stress tolerance in wheat (Wang et al., 2016). We here showed that CmoDELLA genes expression and SOD activity were significantly higher under cold stress than those in the control. Further researches are required to better comprehend the regulatory relationships between CmoDELLA genes expression and SOD activity under abiotic stress.
In summary, seven CmoDELLA genes were obtained in pumpkin by genome-wide analysis. Furthermore, the chromosomal localization, protein properties, phylogenetic tree, gene structure, promoter cis-regulatory elements and protein interaction networks of CmoDELLA genes were conducted. Expression profiles of CmoDELLA genes under different tissues and abiotic stress were determined through RNA-seq data and qRT-PCR. Additionally, physiological changes of pumpkin seedlings were measured under waterlogging stress and cold stress. As a whole, these results revealed the vital roles of CmoDELLA genes in regulating plant development and stress response in pumpkin, which would provide valuable clues for further studying the potential functions and regulatory networks of CmoDELLA genes.
Publicly available datasets were analyzed in this study. This data can be found here: NCBI PRJNA385310 and PRJNA437579.
YS conceived and designed the experiments. WL carried out the experiments. WL, ZZ, HC, WA, LL, and JL analyzed the data, prepared figures and tables. WL wrote the manuscript. YS and XL reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the Key Research and Development Program of Henan Province (No. 212102110130), the Major Public Welfare Projects of Henan Province (No. 201300111300) and the Key Science and Technology Program of Xinxiang City (No. GG2019013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1137126/full#supplementary-material
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., Genschik, P. (2008a). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Achard, P., Liao, L. L., Jiang, C. F., Desnos, T., Bartlett, J., Fu, X. D., et al. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172. doi: 10.1104/pp.106.092254
Achard, P., Renou, J. P., Berthomé, R., Harberd, N. P., Genschik, P. (2008b). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18, 656–660. doi: 10.1016/j.cub.2008.04.034
Achard, P., Vriezen, W. H., van der Straeten, D., Harberd, N. P. (2003). Ethylene regulates arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825. doi: 10.1105/tpc.015685
Barro-Trastoy, D., Gomez, M. D., Blanco-Touriñán, N., Tornero, P., Perez-Amador, M. A. (2022). Gibberellins regulate ovule number through a DELLA-CUC2 complex in Arabidopsis. Plant J. 110, 43–57. doi: 10.1111/tpj.15607
Blanco-Touriñán, N., Legris, M., Minguet, E. G., Costigliolo-Rojas, C., Nohales, M. A., Iniesto, E., et al. (2020). COP1 destabilizes DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 13792–13799. doi: 10.1073/pnas.1907969117
Cao, D. N., Cheng, H., Wu, W., Soo, H. M., Peng, J. R. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142, 509–525. doi: 10.1104/pp.106.082289
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J. H., Cheng, T. L., Wang, P. K., Tian, L., Wang, G. P., Luo, Y. M., et al. (2013). Genome-wide bioinformatics analysis of DELLA-family proteins from plants. Plant Omics 6, 201–207.
Davière, J. M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24, 1923–1928. doi: 10.1016/j.cub.2014.07.012
de Lucas, M., Davière, J. M., Rodríguez-Falcón, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Doornbos, R. F., Geraats, B. P., Kuramae, E. E., Van Loon, L. C., Bakker, P. A. (2011). Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of arabidopsis thaliana. Mol. Plant Microbe Interact. 24, 395–407. doi: 10.1094/mpmi-05-10-0115
Fan, S., Zhang, D., Zhang, L. Z., Gao, C., Xin, M. Z., Tahir, M. M., et al. (2017). Comprehensive analysis of GASA family members in the malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics 18, 827. doi: 10.1186/s12864-017-4213-5
Feng, S. H., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J. L., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Guan, H. L., Huang, X. M., Zhu, Y. N., Xie, B. X., Liu, H. C., Song, S. W., et al. (2021). Identification of DELLA genes and key stage for GA sensitivity in bolting and flowering of flowering Chinese cabbage. Int. J. Mol. Sci. 22, 12092. doi: 10.3390/ijms222212092
Han, Y. X., Dai, H. W., Zheng, S. T., Tong, H. R., Yuan, L. Y. (2020). Identification and expression analysis of the DELLA gene family in camellia sinensis(L.) o. ktze. Plant Sci. J. 38, 644–653. doi: 10.11913/PSJ.2095-0836.2020.50644
Hu, J. H., Israeli, A., Ori, N., Sun, T. P. (2018). The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell 30, 1710–1728. doi: 10.1105/tpc.18.00363
Huang, X. Z., Jiang, C. F., Liao, L. L., Fu, X. D. (2006). Progress on molecular foundation of GA biosynthesis pathway and signaling. Chin. Bull. Bot. 5, 499–510.
King, K. E., Moritz, T., Harberd, N. P. (2001). Gibberellins are not required for normal stem growth in arabidopsis thaliana in the absence of GAI and RGA. Genetics 159, 767–776. doi: 10.1093/genetics/159.2.767
Li, K. L., Yu, R. B., Fan, L. M., Wei, N., Chen, H. D., Deng, X. W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7, 11868. doi: 10.1038/ncomms11868
Li, L., Shu, S., Xu, Q., An, Y. H., Sun, J., Guo, S. R. (2017). NO accumulation alleviates H2O2-dependent oxidative damage induced by Ca(NO3)2 stress in the leaves of pumpkin-grafted cucumber seedlings. Physiol. Plant 160, 33–45. doi: 10.1111/ppl.12535
Li, Q. F., Wang, C. M., Jiang, L., Li, S., Sun, S. S., He, J. X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal 5, ra72. doi: 10.1126/scisignal.2002908
Li, W. J., Zhang, J. X., Sun, H. Y., Wang, S. M., Chen, K. Q., Liu, Y. X., et al. (2018). FveRGA1, encoding a DELLA protein, negatively regulates runner production in fragaria vesca. Planta 247, 941–951. doi: 10.1007/s00425-017-2839-9
Li, X. L., Liu, W., Li, B., Liu, G. Y., Wei, Y. X., He, C. Z., et al. (2018). Identification and functional analysis of cassava DELLA proteins in plant disease resistance against cassava bacterial blight. Plant Physiol. Biochem. 124, 70–76. doi: 10.1016/j.plaphy.2017.12.022
Liang, N. S., Zhan, Y. G., Yu, L., Wang, Z. Q., Zeng, F. S. (2019). Characteristics and expression analysis of FmTCP15 under abiotic stresses and hormones and interact with DELLA protein in fraxinus mandshurica rupr. Forests 10, 343–343. doi: 10.3390/f10040343
Liang, S., Chen, Q. S., Zhu, Z. K., Li, D. D., Qi, Z. M., Xin, D. W. (2022). Identification and analysis of soybean DELLA gene family. Chin. J. Oil Crop Sci. 44, 996–1005. doi: 10.19802/j.issn.1007-9084.2021224
Liao, Z. G., Yu, H., Duan, J. B., Yuan, K., Yu, C. J., Meng, X. B., et al. (2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 10, 2738. doi: 10.1038/s41467-019-10667-2
Lim, S., Park, J., Lee, N., Jeong, J., Toh, S., Watanabe, A., et al. (2013). ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. doi: 10.1105/tpc.113.118604
Liu, H., Yang, H. Z., Li, L., Wang, F., Wu, S. H., Li, Y. Y. (2016). Cloning and expression analysis of PpGAI genes of DELLA protein related to dormancy from pyrus. Mol. Plant Breed 14, 1995–2002. doi: 10.13271/j.mpb.014.001995
Liu, K., Li, Y. H., Chen, X. N., Li, L. J., Liu, K., Zhao, H. P., et al. (2018). ERF72 interacts with ARF6 and BZR1 to regulate hypocotyl elongation in Arabidopsis. J. Exp. Bot. 69, 3933–3947. doi: 10.1093/jxb/ery220
Liu, S. S., Li, H., Lv, X. Z., Ahammed, G. J., Xia, X. J., Zhou, J., et al. (2016). Grafting cucumber onto luffa improves drought tolerance by increasing ABA biosynthesis and sensitivity. Sci. Rep. 6, 20212. doi: 10.1038/srep20212
Luo, W. R., Li, Y. Y., Sun, Y. D., Lu, L., Zhao, Z. X., Zhou, J. G., et al. (2021). Comparative RNA-seq analysis reveals candidate genes associated with fruit set in pumpkin. Sci. Hortic. 288, 110255. doi: 10.1016/j.scienta.2021.110255
Martí, C., Orzáez, D., Ellul, P., Moreno, V., Carbonell, J., Granell, A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52, 865–876. doi: 10.1111/j.1365-313X.2007.03282.x
McGinnis, K. M., Thomas, S. G., Soule, J. D., Strader, L. C., Zale, J. M., Sun, T. P., et al. (2003). The Arabidopsis SLEEPY1 gene encodes a putative f-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130. doi: 10.1105/tpc.010827
Muhammad, I., Li, W. Q., Jing, X. Q., Zhou, M. R., Shalmani, A., Ali, M., et al. (2019). A systematic in silico prediction of gibberellic acid stimulated GASA family members: a novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice. J. Plant Physiol. 234–235, 117-132. doi: 10.1016/j.jplph.2019.02.005
Murase, K., Hirano, Y., Sun, T. P., Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. doi: 10.1038/nature07519
Navarro, L., Bari, R., Achard, P., Lisón, P., Nemri, A., Harberd, N. P., et al. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18, 650–655. doi: 10.1016/j.cub.2008.03.060
Oh, E., Zhu, J. Y., Bai, M. Y., Arenhart, R. A., Sun, Y., Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the arabidopsis hypocotyl. Elife 3, e03031. doi: 10.7554/eLife.03031
Rivas-San Vicente, M., Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Sakuraba, Y., Bülbül, S., Piao, W. L., Choi, G., Paek, N. C. (2017). Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 92, 1106–1120. doi: 10.1111/tpj.13747
Sarwar, R., Jiang, T., Ding, P., Gao, Y., Tan, X. L., Zhu, K. M. (2021). Genome-wide analysis and functional characterization of the DELLA gene family associated with stress tolerance in b. napus. BMC Plant Biol. 21, 286. doi: 10.1186/s12870-021-03054-x
Sechet, J., Frey, A., Effroy-Cuzzi, D., Berger, A., Perreau, F., Cueff, G., et al. (2016). Xyloglucan metabolism differentially impacts the cell wall characteristics of the endosperm and embryo during Arabidopsis seed germination. Plant Physiol. 170, 1367–1380. doi: 10.1104/pp.15.01312
Thines, B., Katsir, L., Melotto, M., Niu, Y. J., Mandaokar, A., Liu, G. H., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Wang, J. W., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, R. Q., Fan, X. C., Song, M. F., Xiao, Y., Guo, L., Meng, F. H., et al. (2016). A wheat DELLA gain-of-function mutant aibian 1 promotes seedlings salt tolerance. Acta Agronomica Sin. 42, 1721–1726. doi: 10.3724/SP.J.1006.2016.01721
Wang, Y., He, S., Wei, Y. Z., Dong, C., Liu, L. Q., Jue, D. W., et al. (2020). Molecular and functional characterization of two DELLA protein-coding genes in litchi. Gene 738, 144455. doi: 10.1016/j.gene.2020.144455
Wang, Y. Y., Yu, H. P., Zhao, Z. C., Gao, A. P., Zhang, Y., Zhou, K. B. (2022). Cloning and analysis of. DELLA-GAI Gene its promoter mango. Mol. Plant Breed 20, 2526–2533. doi: 10.13271/j.mpb.020.002526
Worldmapper (2021). Available at: http://worldmapper.org/maps/pumpkinproduction/ (Accessed December 2, 2022).
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Xu, H., Liu, Q., Yao, T., Fu, X. D. (2014). Shedding light on integrative GA signaling. Curr. Opin. Plant Biol. 21, 89–95. doi: 10.1016/j.pbi.2014.06.010
Xue, H. D., Gao, X., He, P., Xiao, G. H. (2022). Origin, evolution, and molecular function of DELLA proteins in plants. Crop J. 10, 287–299. doi: 10.1016/j.cj.2021.06.005
Yang, D. L., Dong, W. X., Zhang, Y. Y., He, Z. H. (2013). Gibberellins modulate abiotic stress tolerance in plants. Sci. Sin. 43, 1119–1126. doi: 10.1360/052013-321
Zhang, J., Yang, J. J., Yang, Y., Luo, J., Zheng, X. Y., Wen, C. L., et al. (2019). Transcription factor CsWIN1 regulates pericarp wax biosynthesis in cucumber grafted on pumpkin. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01564
Zhang, Y., Liu, B., Yang, S., An, J. B., Chen, C. H., Zhang, X. L., et al. (2014). A cucumber DELLA homolog CsGAIP may inhibit staminate development through transcriptional repression of b class floral homeotic genes. PloS One 9, e91804. doi: 10.1371/journal.pone.0091804
Zhao, B., Li, H. T., Li, J. J., Wang, B., Cheng, D., Wang, J., et al. (2017). Brassica napus DS-3, encoding a DELLA protein, negatively regulates stem elongation through gibberellin signaling pathway. Theor. Appl. Genet. 130, 727–741. doi: 10.1007/s00122-016-2846-4
Zhou, M. Q., Chen, H., Wei, D. H., Ma, H., Lin, J. (2017). Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 7, 39819. doi: 10.1038/srep39819
Keywords: Cucurbita moschata, DELLA, characterization, gene expression, abiotic stress
Citation: Luo W, Zhao Z, Chen H, Ao W, Lu L, Liu J, Li X and Sun Y (2023) Genome-wide characterization and expression of DELLA genes in Cucurbita moschata reveal their potential roles under development and abiotic stress. Front. Plant Sci. 14:1137126. doi: 10.3389/fpls.2023.1137126
Received: 04 January 2023; Accepted: 13 February 2023;
Published: 23 February 2023.
Edited by:
Qiusheng Kong, Huazhong Agricultural University, ChinaReviewed by:
Yunsong Lai, Sichuan Agricultural University, ChinaCopyright © 2023 Luo, Zhao, Chen, Ao, Lu, Liu, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongdong Sun, eWRzdW4yMDE5QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.