- 1School of Resources and Environmental Sciences, Henan Institute of Science and Technology, Xinxiang, China
- 2School of Horticulture and Landscape Architecture, Henan Institute of Science and Technology, Xinxiang, China
- 3Henan Engineering Research Center of the Development and Utilization of Characteristic Horticultural Plants, Xinxiang, China
Soil salinization, which is the accumulation of salt in soil, can have a negative impact on crop growth and development by creating an osmotic stress that can reduce water uptake and cause ion toxicity. The NHX gene family plays an important role in plant response to salt stress by encoding for Na+/H+ antiporters that help regulate the transport of sodium ions across cellular membranes. In this study, we identified 26 NHX genes in three cultivars of Cucurbita L., including 9 Cucurbita moschata NHXs (CmoNHX1-CmoNHX9), 9 Cucurbita maxima NHXs (CmaNHX1-CmaNHX9) and 8 Cucurbita pepo NHXs (CpNHX1-CpNHX8). The evolutionary tree splits the 21 NHX genes into three subfamilies: the endosome (Endo) subfamily, the plasma membrane (PM) subfamily, and the vacuole (Vac) subfamily. All the NHX genes were irregularly distributed throughout the 21 chromosomes. 26 NHXs were examined for conserved motifs and intron-exon organization. These findings suggested that the genes in the same subfamily may have similar functions while genes in other subfamilies may have functional diversity. The circular phylogenetic tree and collinearity analysis of multi-species revealed that Cucurbita L. had a substantially greater homology relationship than Populus trichocarpa and Arabidopsis thaliana in terms of NHX gene homology. We initially examined the cis-acting elements of the 26 NHXs in order to investigate how they responded to salt stress. We discovered that the CmoNHX1, CmaNHX1, CpNHX1, CmoNHX5, CmaNHX5, and CpNHX5 all had numerous ABRE and G-box cis-acting elements that were important to salt stress. Previous transcriptome data showed that in the mesophyll and veins of leaves, many CmoNHXs and CmaNHXs, such as CmoNHX1, responded significantly to salt stress. In addition, we heterologously expressed in A. thaliana plants in order to further confirm the response of CmoNHX1 to salt stress. The findings demonstrated that during salt stress, A. thaliana that had CmoNHX1 heterologously expression was found to have decreased salt tolerance. This study offers important details that will aid in further elucidating the molecular mechanism of NHX under salt stress.
Introduction
Soil salinization is a serious problem that can have negative impacts on plant growth and development, and it is becoming an increasingly severe global environmental issue. The area of salinized soil is expanding due to a combination of natural environmental changes, such as climate change and changes in water availability, as well as human activities like improper irrigation practices, deforestation, and urbanization. One of the key problems impeding the sustainable growth of agriculture is soil salinization, which has emerged as a severe global environmental issue (Zhu, 2016). The negative effects of salt, which gravely endanger plant growth and development, extend to more than 800 million hectares of land worldwide (Munns and Tester, 2008). The area of salinized soil exhibits a trend of progressive increase under the simultaneous effects of natural environment changes and human activity (Munns and Tester, 2008). China is one of the countries that is particularly affected by soil salinization. According to Qi et al. (2012), China has approximately 36 million hectares of salinized agricultural land, which accounts for around 5% of the country’s total agricultural land. This can have significant implications for food security and agricultural sustainability in China, as crops grown in these areas may experience reduced yields or even failure due to salt stress.
In the process of plant growth, Na+ is a non-essential element. The roots of plants can absorb a lot of the highly mobile Na+ in salinized soil, which can subsequently be transported to the cytoplasm of various tissues and organs to cause salt damage (Rajendran et al., 2010; Roy et al., 2014). Na+ homeostasis in plants is largely maintained by genes from the Na+/H+ antiporter family. Eight NHX genes were discovered in Arabidopsis thaliana, which were separated into the vacuole (Vac) subfamily, the plasma membrane (PM) subfamily, and the endosome nucleolus (Endo) subfamily (Ohnishi et al., 2005; Bassil et al., 2011). All AtNHX genes contain 10-12 transmembrane structures and participate in various biological processes under salt stress (Ohnishi et al., 2005; Bassil et al., 2011). Seven PtNHX genes were identified from Populus tomentosa Carr genome (Tian et al., 2017), and they respond to single or multiple stresses such as drought, heat, cold, salt and ABA. PtNHX7 was involved in salt stress response through calcineurin B-like proteins - calcineurin B-like interacting protein kinase (CBL-CIPK) pathway (Tian et al., 2017). NHX gene—PeSOS1 was found to enhance salt tolerance in Poplus trichocarpa (Meng and Wu, 2018). BvNHX5 may interact with CBL and CIPK to enhance salt tolerance in Beta vulgaris (Wu et al., 2019). A total of 25 NHX genes were identified in Gossypium herbaceum, and GbNHX2 gene was highly expressed under high salt stress. (Akram et al., 2020). These results suggest that NHX genes may play an important role under salt stress in crops.
The Cucurbita genus primarily consists of three crop species: Cucurbita moschata, Cucurbita maxima, and Cucurbita pepo, which are widely planted worldwide due to their excellent disease resistance, established roots, and adaptability (Montero-Pau et al., 2018). The main reason why Cucurbita genus crops, especially Cucurbita moschata, are the main rootstocks of melon crops is that they have salt tolerance and can enhance the stress resistance of melon crops. Na+/H+ antiporter family genes play an important role in maintaining Na+ homeostasis in plants. Since the genomes of three domesticated species of the Cucurbita genus were published (Sun et al., 2017), the identification of the Na+/H+ reverse transporter gene family and their responses to salt stress have not been reported. Herein, this study investigated the evolutionary relationships, gene structures, and regulatory elements of NHX genes in three cultivars of the Cucurbita genus, and examined the transcriptional changes in two of these cultivars, Cucurbita moschata and C. maxima, in response to salt stress. We also identified a differentially expressed gene, CmoNHX1, which was found to respond to salt stress. To confirm the function of this gene, we genetically modified Arabidopsis thaliana, a model plant species, and observed its response to salt stress. These findings have significant implications for developing salt-tolerant cultivars of Cucurbita plants, as well as enhancing our understanding of the mechanisms underlying salt tolerance in these plants. By identifying the specific genes involved in salt tolerance, researchers may be able to develop more effective breeding strategies and genetic engineering techniques to enhance salt tolerance in Cucurbita crops.
Materials and methods
Identification of NHX family genes in Cucurbita
Eight NHX protein sequences of A. thaliana were obtained from the TAIR database (https://www.arabidopsis.org/) (Philippe et al., 2012). On this basis, the local BLAST program was used to search for NHX proteins in the database of three cultivars of Cucurbita genome (C. moschata, C. maxima, C. pepo) (http://cucurbitgenomics.org/). The NHX candidate proteins must meet the requirements: the protein sequence identity of AtNHX protein with other protein more than 70% and e<10−10. Na+_H+_Exchanger domain (PF00999) of NHX candidate proteins from Cucurbita were confirmed by PFAM (http://pfam.xfam.org/) (Finn et al., 2016) and SMART (http://smart.emblheidelberg.de/) (Letunic et al., 2012). Theoretical isoelectric point (pI) and molecular weight (MW) of NHX candidates were predicted by ExPASy software (https://web.expasy.org/compute\upi/) (Bjellqvist et al., 1993). The TMHMM server v. 2. 0 software (http://www.cbs.dtu.dk/services/TMHMM/) (Moller et al., 2001)was used to predict the membrane domain of all proteins.
Phylogenetic relationship of NHX family proteins in several species
To elucidate the phylogenetic relationships of NHX protein families in Cucurbita moschata, C. maxima, C. pepo, P. trichocarpa and A. thaliana, NHX proteins from P. trichocarpa were firstly extracted from previous literature (Tian et al., 2017). Moreover, NHXs protein sequences in C. moschata (Cmo), C. maxima (Cma), C. pepo (Cp), P. trichocarpa (Pt) and A. thaliana (At) were blast by ClustalW program (Larkin et al., 2007). Finally, a circular phylogenetic tree was constructed using MEGA 7.0 software (Kumar et al., 2016) and the Maximum Likelihood method (Guindon and Gascuel, 2003) was applied with bootstrap value set to 1000 replicates.
Conservative domain and gene structure analysis of NHX family members in Cucurbita
To further analyze the conserved domains of NHX family members in Cucurbita, the MEME suite online program (MEME 5.3.3, http://meme-suite.org/tools/meme) (Bailey et al., 2006) was used to analyze and draw the motifs. The operating parameter were set as: the base width was 6-50 aa and the maximum number of motifs was 15.
To clarify the structural characteristics of NHX genes in Cucurbita, the CDS sequences of 26 NHX genes were compared with the corresponding genomic DNA sequences from the corresponding genomic database. Finally, the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) (Hu et al., 2014)was used to map exon-intron structure of NHX genes.
Chromosome location and collinearity analysis of NHXs in Cucurbita
To clarify the distribution of NHX gene on chromosomes in Cucurbita, we first obtained the starting positions of CmoNHX, CmaNHX, and CpNHX genes on chromosomes from the genome databases of three Cucurbita cultivars, and finally the analysis were performed using TBtools (https://github.com/CJ-Chen/TBtools) (Chen et al., 2020).
To further analyze the collinearity of NHX genes among C. moschata (Cmo), C. maxima (Cma) and C. pepo (Cp), and their collinearity with A. thaliana NHX genes, MCScanX software and Circos-0.69 software (Krzywinski et al., 2009) were used to analyze and visualize the collinearity of NHX genes.
Analysis of Cis-acting elements of NHXs in Cucurbita
To identify the cis-acting elements of NHX genes in Cucurbita, the promoter sequences (1500 bp sequence upstream of start codon) of all CmoNHXs, CmaNHXs and CpNHXs were extracted from the genome of Cucurbita (http://cucurbitgenomics.org/). The cis-acting elements of these NHX genes were then predicted using the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). We analyzed the cis-acting elements related to growth and development, hormone and abiotic stress, with emphasis on the cis-acting elements related to salt stress.
Response analysis of CmoNHXs and CmaNHXs to salt stress
To determine the response of CmoNHXs and CmaNHXs to salt stress, we excavated the transcriptome data (BioProject: PRJNA464060) published in 2018 (Niu et al., 2018) and analyzed the transcription profile of NHXs in the leaf mesophyll and leaf vein of the C. moschata cultivar, “Rifu” and C. maxima cultivar, “Rimu” under salt stress. Sequencing samples were obtained from 100 mM NaCl treated for 24 hours. 0 mM NaCl was used as a control. The expression value was calculated in terms of reads per kilobase of exon model per million mapped reads (RPKM).
Subcellular localization analysis of CmoNHX1 protein
We cloned the CmoNHX1 gene’s nucleotide sequence (Table S4) and inserted it into PRI101-GFP to create the recombinant vector PRI101-GFP-CmoNHX1 in order to study the subcellular localization of the CmoNHX1 protein. The recombinant vector, control vector PRI101-GFP and plasma membrane marker: pm-rbCD3-1008 were transformed into Agrobacterium tumefaciens (GV3101) by freeze-thaw method (Jyothishwaran et al., 2007), respectively. According to the method of Wu et al. (2010), CmoNHX1 was transiently expressed in tobacco. After 40 hours of dark culture, laser confocal microscopy was used to detect cell fluorescence.
Construction, genetic transformation and response to salt stress of CmoNHX1- ectopic expression vector
To construct CmoNHX1 ectopic expression vector, we constructed CmoNHX1 fragment into pTCK303 using recombinant vector PRI101-GFP-CmoNHX1 as template (Table S4), and the recombinant vector was termed as pTCK303-CmoNHX1. Transformation of constructed recombinant vector into GV3101 by freeze-thaw method (Jyothishwaran et al., 2007). In addition, transgenic A. thaliana plants were obtained by floral dip method (Xu et al., 2010). T2 generation transgenic plants were finally obtained through multi-generation selfing and GUS staining.
We used A. thaliana plants that were heterologously expressed in CmoNHX1 from the T2 generation as well as wild type plants to better understand how CmoNHX1 responded to salt stress. In order to irrigate the plants at the seedling stage, a 100 mM NaCl solution was made, and 50 mL of it was poured twice, once every three days, into each hole. Water was used as the control. Every time a symptom appeared, pictures were taken. Following the prior procedure, RNA extraction, reverse transcription, and qRT-PCR were carried out at this time. (Yuan et al., 2019).
Results
Identification of NHX gene family in Cucurbita
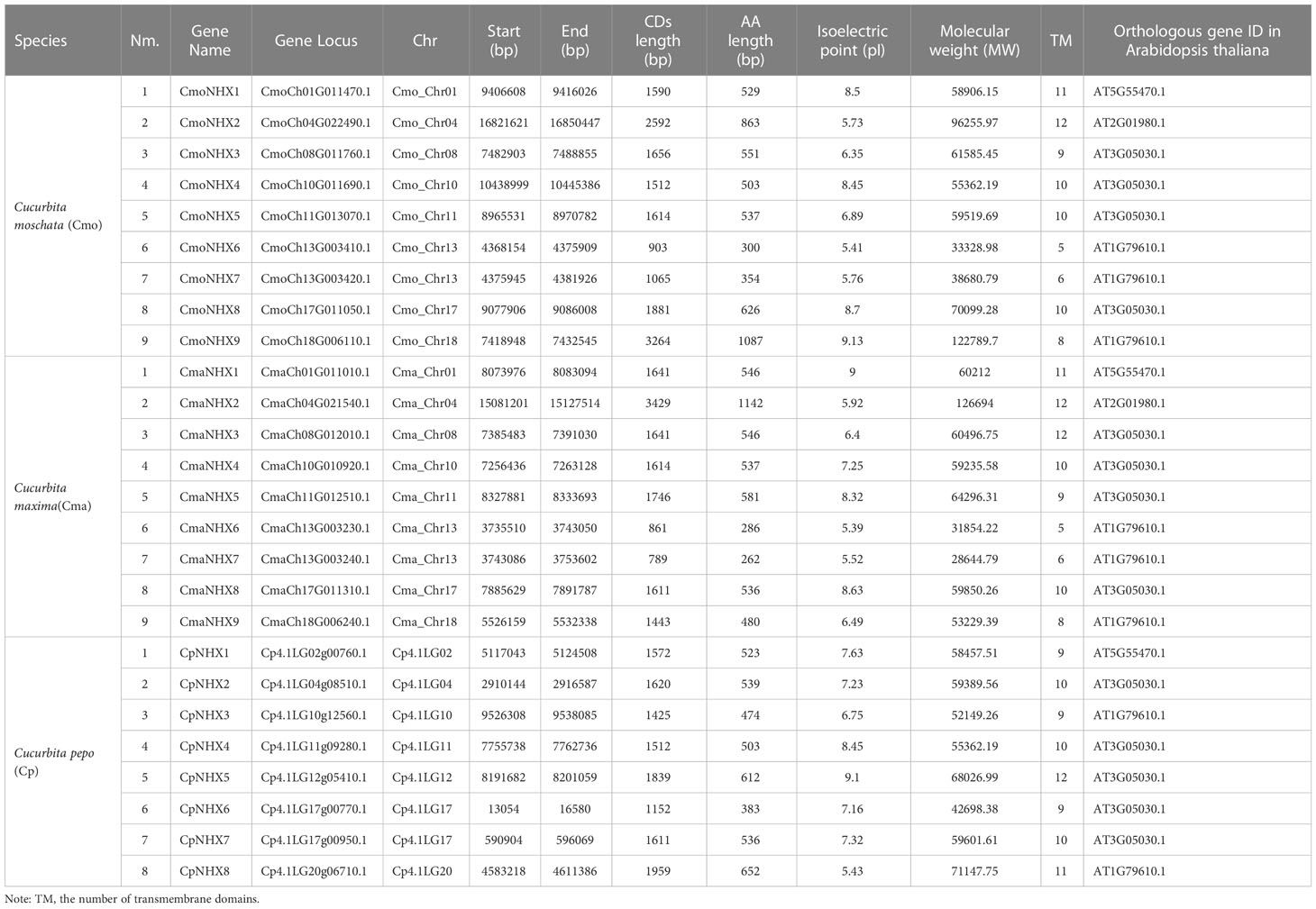
A total of eight AtNHX protein sequences from Arabidopsis thaliana as query sequences were used to identify NHX proteins in three cultivars of the Cucurbita genus, including C. moschata, C. maxima, and C. pepo. Using a series of screening procedures, a total of 26 NHX proteins were discovered, which were named CmoNHX1 through CmoNHX9, CmaNHX1 through CmaNHX9, and CpNHX1 through CpNHX8. The naming convention was based on the name of the corresponding AtNHX genes and their position on the chromosome, from the first to last chromosomes, and from top to bottom. More information about these NHX proteins can be found in Table S1 of the study.
Sequence analysis of 26 NHX genes showed that the length of open reading frame in 9 CmoNHXs ranged from 903 bp (CmoNHX6) to 3264 bp (CmoNHX9), and the corresponding number of amino acids obtained by translation ranged from 300 aa to 1087 aa (Table 1). The pI and MW of CmoNHXs were from 5.41 to 9.13 and from 33.328 98 KDa to 122.789 68 KDa, respectively. Nine CmoNHXs contained 5 to 12 transmembrane domains (Table 1). Table 1 also displays the length of the open reading frame, the length of the amino acid, the size of the pI, and the MW in CmaNHXs and CpNHXs. These findings accurately capture the diversity and conservation of NHXs’ biological and structural features.
Phylogenetic relationships of NHXs in multiple species
The study you mentioned constructed a phylogenetic tree of NHX proteins from Cucurbita moschata, C. maxima, C. pepo, Arabidopsis thaliana, and Populus trichocarpa, to better understand the evolutionary relationships within the plant NHX protein family (Figure 1). Based on the amino acid sequence identity of 92%, the NHX proteins were classified into three subfamilies, namely Endo, Vac, and PM. All the NHX proteins were classified into these subfamilies according to the classification of AtNHX proteins. The phylogenetic analysis revealed that the Endo subfamily contained three CmoNHX proteins, three CmaNHX proteins, two CpNHX proteins, two AtNHX proteins, and one PtNHX protein. The PM subfamily included one CmoNHX protein, one CmaNHX protein, and two AtNHX proteins. The Vac subfamily contained the majority of NHX proteins, including five CmoNHX proteins, five CmaNHX proteins, six CpNHX proteins, four AtNHX proteins, and five PtNHX proteins. The study found that the NHX proteins of Cucurbita, Arabidopsis, and Populus were mostly distributed in the Vac subfamily, followed by the PM subfamily, and finally the Endo subfamily. The PtNHX proteins were mostly found in the Vac subfamily, followed by the Endo subfamily, and lastly the PM subfamily. This information provides insights into the evolutionary history and diversification of NHX proteins in plants. The homology of NHXs among the three domesticated species of Cucurbita is significantly higher than that of A. thaliana and P. trichocarpa, when seen from the perspective of evolutionary lineages (Figure 1).
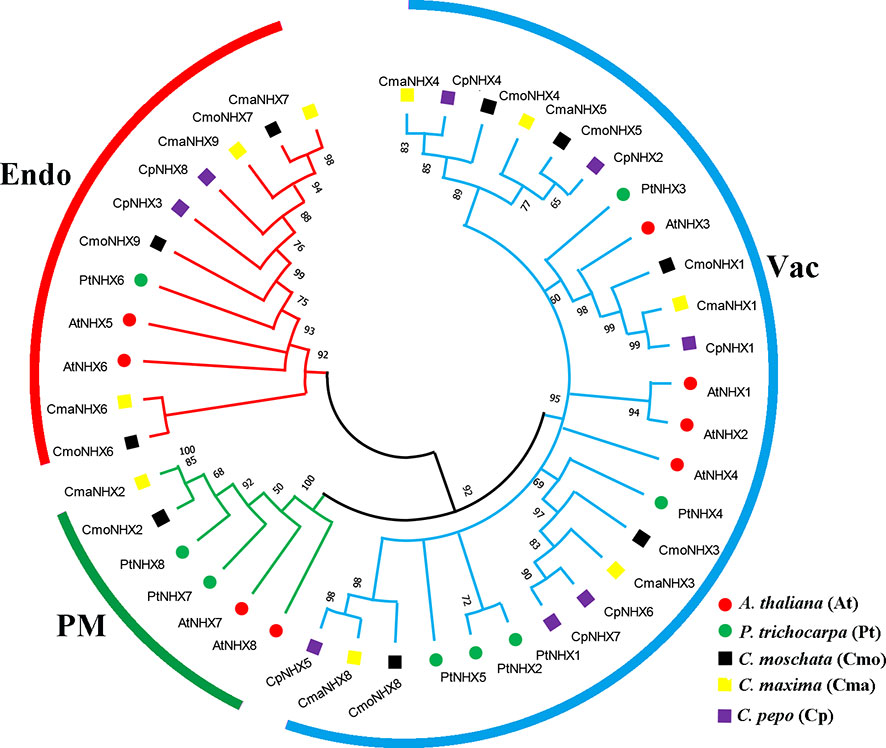
Figure 1 Phylogenetic trees of the NHX gene family in a number of species. Three subfamilies (Endo, PM and Vac) were displayed with evolutionary branches of different colors. The phylogenetic tree was constructed with MEGA 7.0 software using the Maximum Likelihood method with 1000 bootstrap replicates. Cmo, C. moschata; Cma, C. maxima; At, A. thaliana; Pt, P. trichocarpa.
Analysis of conserved domain of NHX proteins in Cucurbita
Twenty-six Cucurbita NHX proteins are further grouped into three subfamilies (Endo, PM, and Vac) based on the evolutionary tree (Figure 2A). All 26 NHX proteins were found to possess the conserved Na+_H+_Exchanger domain (PF00999) after conducting a conserved domain analysis (Figure 2B). Furthermore, we looked at the motifs of 26 NHX proteins and found a total of 15 motifs (Figure 2C; Figure S2). Motif 11 was present in all NHX proteins. Furthermore, motif 7 and 12 were exclusive to the Vac subfamily, and motif 13 was present only in the PM and Endo subfamilies (Figure 2C). Motifs 5 and 2 are almost always present with motif 11. Overall, PM proteins look very similar to the Vac proteins with some domains missing.
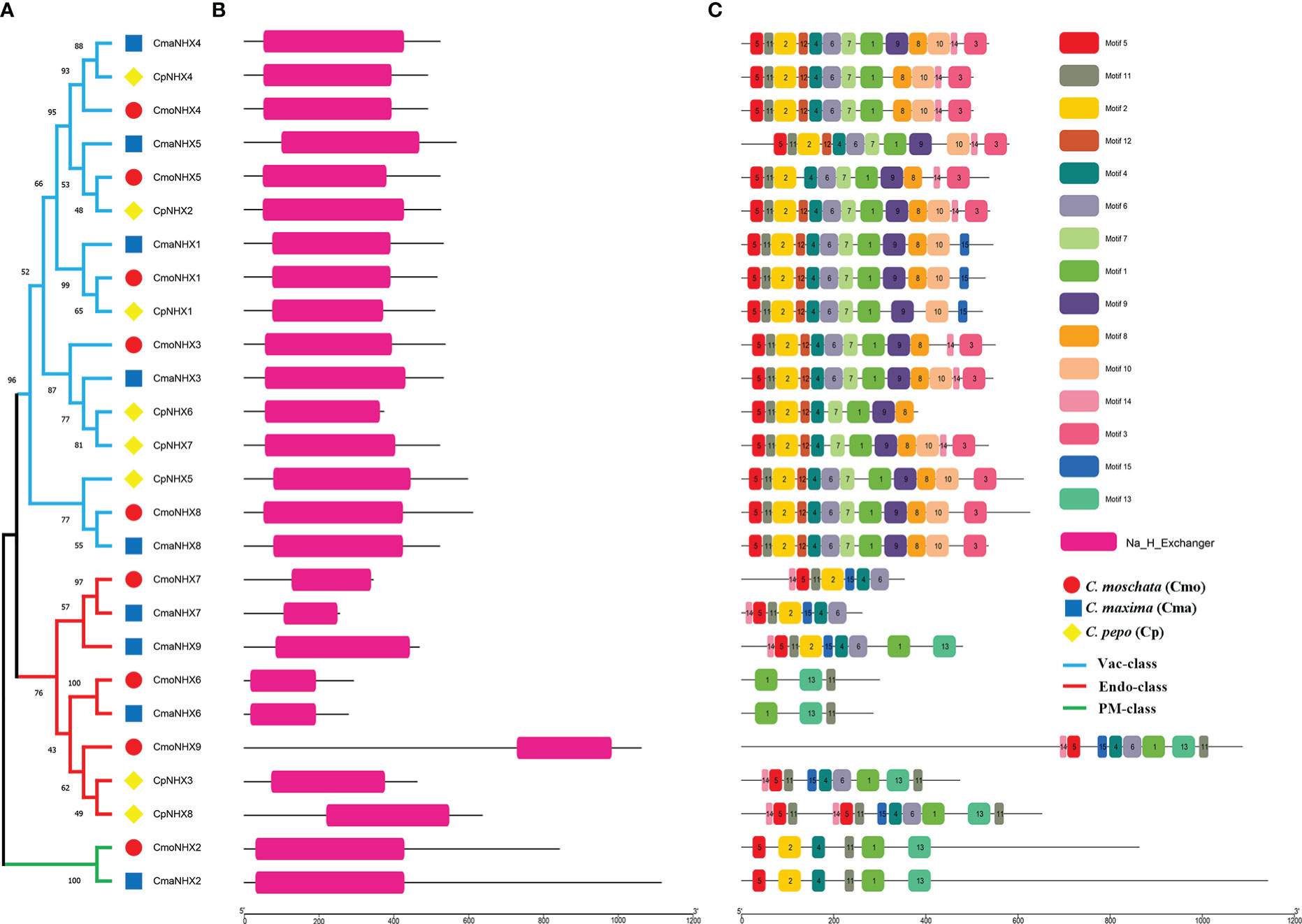
Figure 2 Structural analysis of 26 Cucurbita genera NHX proteins. (A), Phylogenetic tree of NHX protein in 26 NHX proteins in Cucurbita genus. (B), Conserved domains of 26 NHX proteins in Cucurbita genus. (C), Motif analysis of 26 NHX proteins in Cucurbita genus.
Intron-exon structure analysis of 26 Cucurbita NHX genes
A gene’s biological function is significantly influenced by the intron-exon distribution pattern. All NHX genes in the Vac subfamily had 12-16 exons, according to an examination of the intron-exon structure. Exon counts for all NHX genes in the Endo subfamily varied greatly (10-24 exons). There were many exons in the NHX genes of the PM subfamily (20-23). Similar exon numbers and intron lengths can be seen in genes belonging to the same branch, such as CmaNHX4 and CpNHX4, CmaNHX1 and CmoNHX1 (Figure S1). However, certain homologous genes clearly display distinct intron-exon structural variations. As an illustration, CmoNHX2 had 20 exons, but CmaNHX2 had 23 exons and lengthier introns (Figure S1).
The location distribution of 26 NHX genes on chromosomes
The distribution of 26 NHX genes on the chromosomes revealed that 9 CmoNHXs are found on chromosomes Cmo01, Cmo04, Cmo08, Cmo10, Cmo11, Cmo13, Cmo17, and Cmo18 (Figure 3A). The distribution pattern of CmaNHXs and CmoNHX genes is comparable (Figure 3B). On the corresponding chromosomes Cmo13 and Cma13, there are two NHX genes, respectively (Figures 3A, B). The CpNHX genes are primarily found on the chromosomes Cp02, Cp04, Cp10, Cp11, Cp12, Cp17, and Cp20 (Figure 3C), where two genes are located on chromosome Cp17. Differences between the distribution on Cp vs Cmo/Cma reflect larger differences in chromosome synteny.
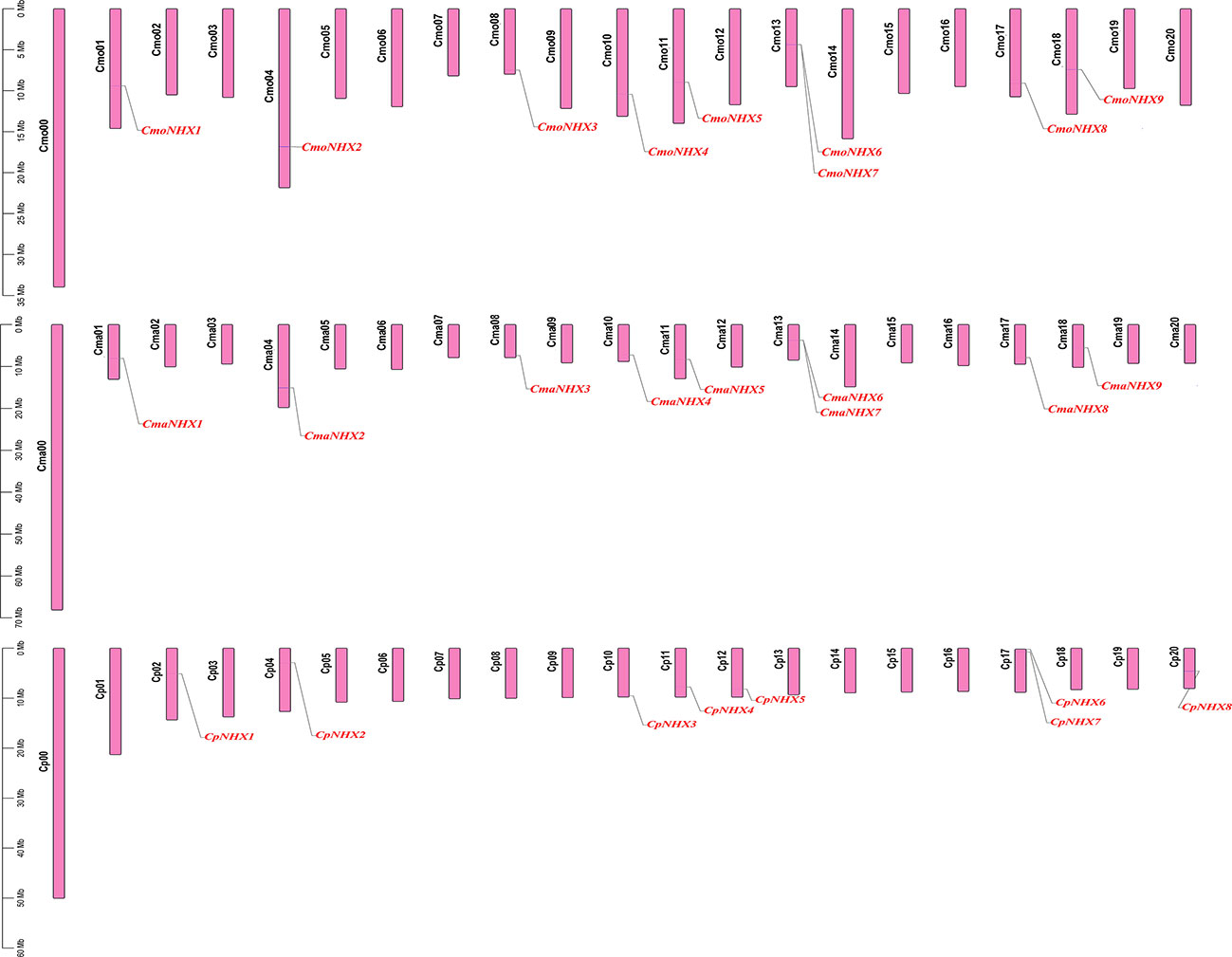
Figure 3 Chromosome distribution map of 26 NHX genes in Cucurbita genus. Black font represents chromosome name, red font represents gene name.
Collinearity analysis of 26 NHX genes in Cucurbita
MCScanX software was used to examine the collinearity and further investigate the evolutionary relationship between the NHX gene families in C. moschata, C. maxima, C. pepo, and A. thaliana. The findings revealed that, between CmoNHXs and CmaNHXs, CpNHXs, AtNHXs, 13, 11, and 5 syntenic gene pairs were identified, respectively (Figure 4; Table S2). In addition, between CmaNHXs and CpNHXs, AtNHXs, a total of 10, 4 syntenic gene pairs were discovered, respectively (Figure 4; Table S2). Two syntenic gene pairs between CpNHXs and AtNHXs were found (Figure 4, Table S2). We discovered that the homology of C. moschata, C. maxima, and C. pepo was substantially higher than that of A. thaliana based on the collinearity analysis of NHX genes in four species.
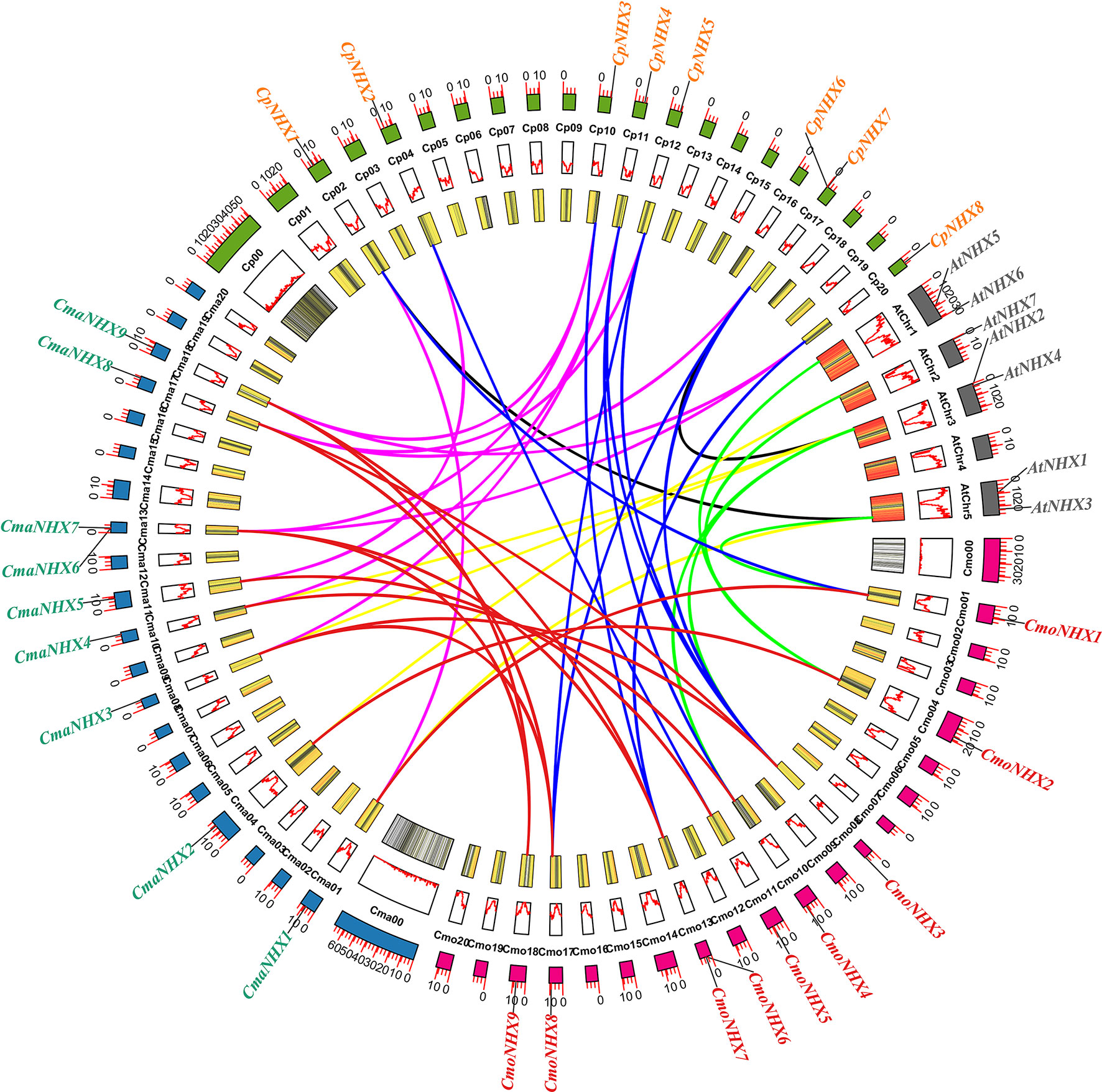
Figure 4 Synteny analysis of the NHX genes among C. moschata, C. maxima, C. pepo and A. thaliana. Cmo, C. moschata; Cma, C. maxima; Cp, C. pepo; At, A. thaliana; The red lines indicated the syntenic NHX gene pairs between C. moschata and C. maxima; The blue lines indicated the syntenic NHX gene pairs between C. moschata and C. pepo; The pink lines indicated the syntenic NHX gene pairs between C. maxima and C. pepo; The green lines indicated the syntenic NHX gene pairs between C. moschata and A. thaliana; The yellow lines indicated the syntenic NHX gene pairs between C. maxima and A. thaliana; The black lines indicated the syntenic NHX gene pairs between C. pepo and A. thaliana. All the data for the various species was extracted from Cucurbit genomics database.
Analysis of Cis-acting elements of NHX gene promoters in Cucurbita
This study examined the CmoNHX, CmaNHX, and CpNHX promoter sequences and discovered 755 cis-acting elements in the promoter regions of 26 NHX genes. They included CmoNHXs, CmaNHXs, and CpNHXs, which each had 252 cis-acting elements, 249 cis-acting elements, and 254 cis-acting elements. The majority of these cis-acting elements were connected to hormone response, abiotic stress, and growth and development factors (Figure 5; Table S3). We focused on examining the cis-acting elements associated with salt stress because of the significance of the NHX gene under salt stress. Although ABRE, TGA-element, TGACG/CGTCA-motif are related to hormone response, G-box and GT1-motif are related to light response, MBS is related to drought stress response (Figure 5; Table S3), but related studies have found that these cis-acting elements are all involved in the response to salt stress (Yamniuk and Vogel, 2004; Saeediazar et al., 2014). In the present study, we discovered that the CmoNHX1, CmaNHX1, and CpNHX1 promoters contain the most ABRE, G-box, and TGACG/CGTCA elements (Figure 5; Table S3). On the basis of the aforementioned analysis, we hypothesized that the genes CmoNHX1, CmaNHX1, CpNHX1, CmoNHX5, CmaNHX5, and CpNHX5 may be crucial in salt stress.
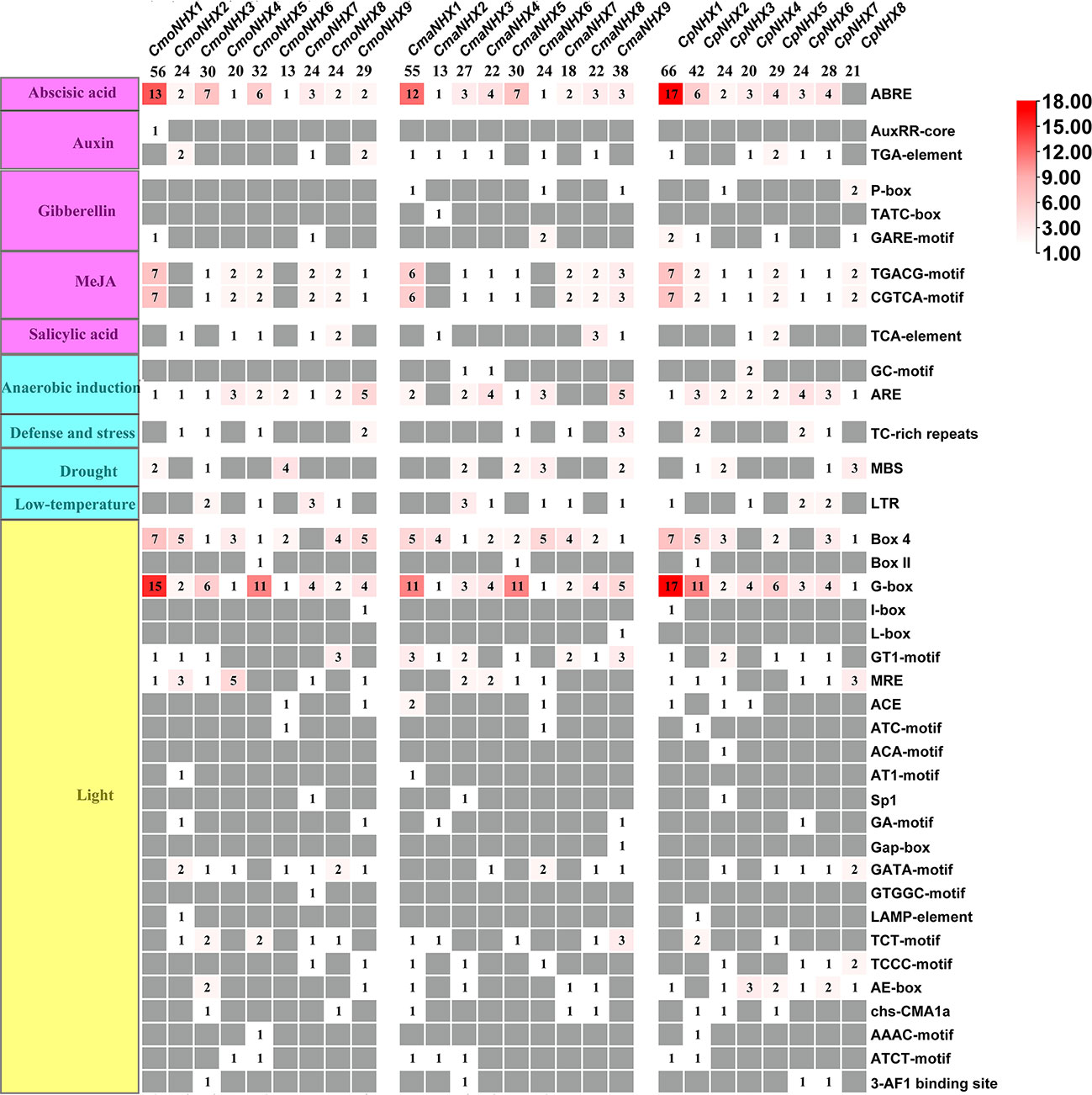
Figure 5 Analysis of promoter cis-acting elements of 26 NHX genes in Cucurbita genus. The leftmost side represents the functional distribution of cis-acting elements; the rightmost side represents the name of cis-acting element.
Response of NHX genes in Cucurbita to NaCl stress
The different expression patterns of NHX genes in C. moschata and C. maxima suggest that they might have different mechanisms to improve salt tolerance under salt stress. After NaCl treatment, the transcription levels of CmoNHX2, CmoNHX5, CmoNHX6, and CmoNHX7 genes increased greatly in the mesophyll and vein of C. moschata, while the transcription level of CmoNHX1 genes fell dramatically (Figure 6). For instance, after NaCl treatment, the transcription levels of CmoNHX1 in the mesophyll and veins were considerably decreased by 55.44% and 69.04%, respectively, in comparison to the control treatment (Figure 6). In mesophyll, there was no apparent change between the NaCl treatment and the control treatment in the transcript levels of any CmaNHX genes. The transcription levels of CmaNHX6, CmaNHX7, and CmaNHX8 under salt stress were significantly higher than those under control treatment, and the transcription level of CmaNHX1 under salt stress was 51.26% of that under control treatment, despite the fact that CmaNHX2, CmaNHX3, CmaNHX4, CmaNHX5, and CmaNHX9 did not change significantly under salt stress in the veins (Figure 7). These findings imply that NHXs might improve salt tolerance in two cultivars via various expression patterns. Overall, the transcriptional changes of NHX genes in response to salt stress provide insights into the molecular mechanisms of salt tolerance in Cucurbita cultivars.
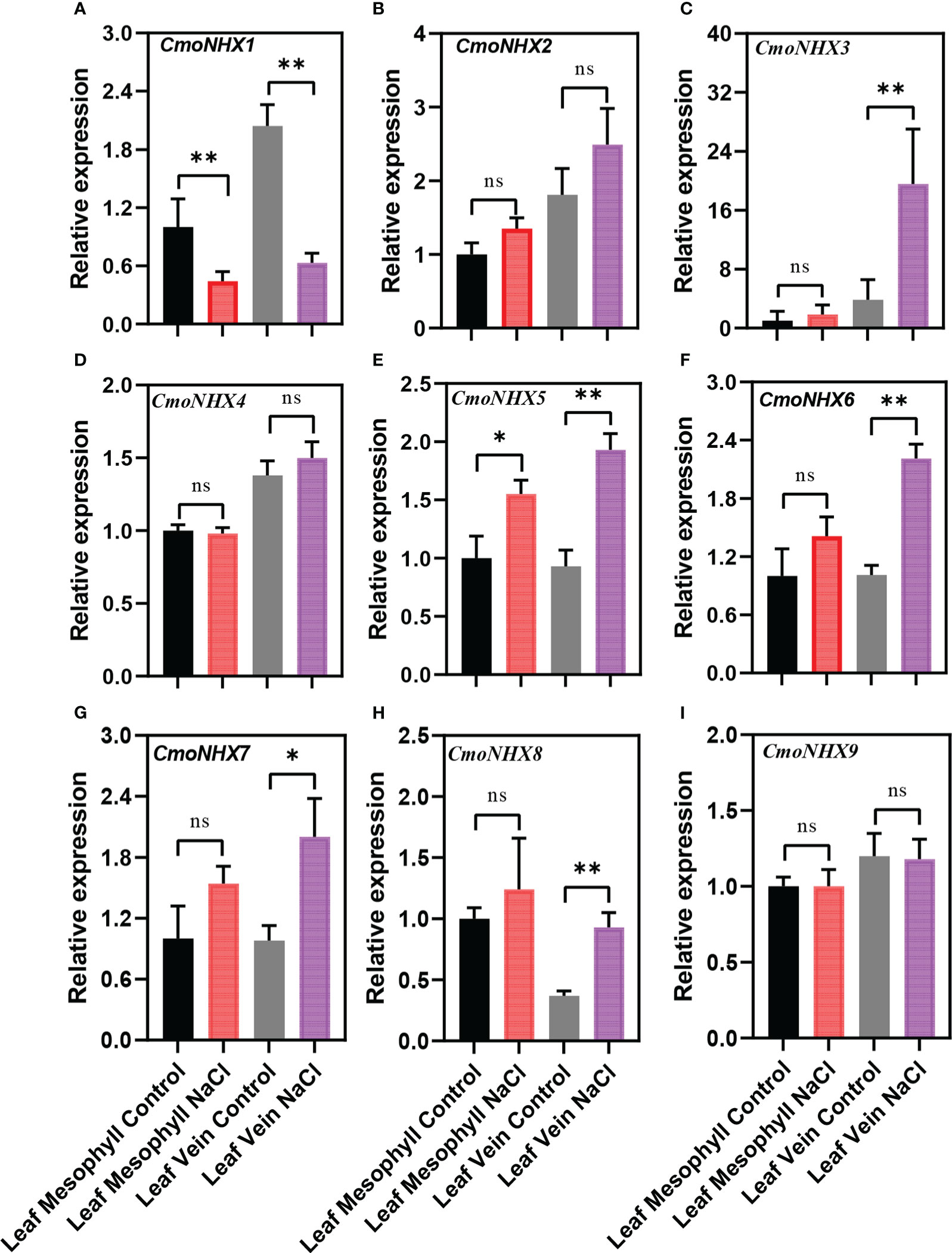
Figure 6 Relative expression of NHX genes from the leaf mesophyll and leaf veins of C. moschata (“Rifu”) in response to treatment with 100 mM NaCl for 24 h. Each data represents the average from three samples. The error bars represent the SDs. * indicates significance at p < 0.05, ∗∗ indicates significance at p < 0.01, NS indicates no significance.
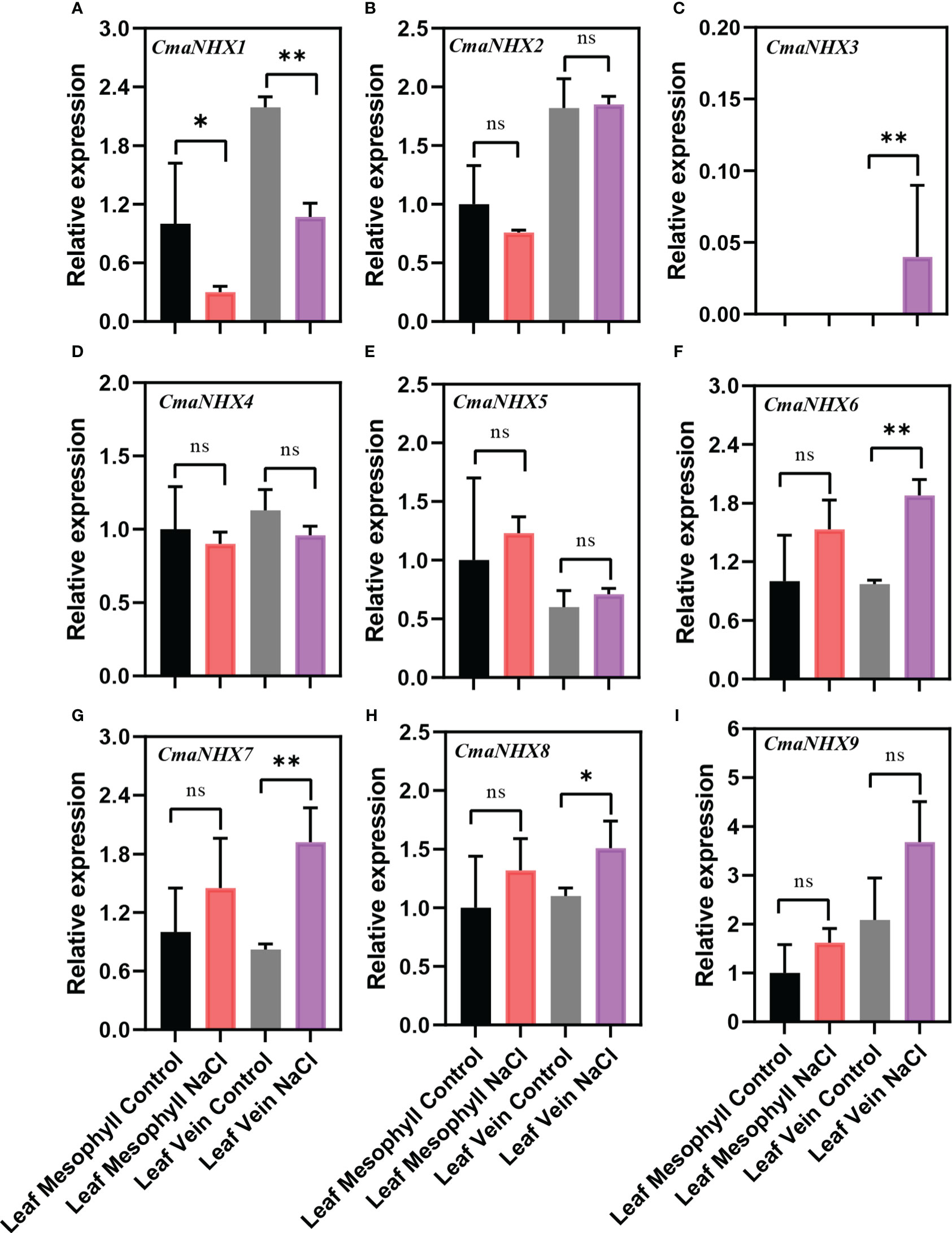
Figure 7 Relative expression of NHX genes from the leaf mesophyll and leaf veins of C. maxima (‘Rimu’) in response to treatment with 100 mM NaCl for 24 h. Each data represents the average from three samples. The error bars represent the SDs. ∗ indicates significance at p < 0.05, ∗∗ indicates significance at p < 0.01, NS indicates no significance.
Response of CmoNHX1-heterologously expressed Arabidopsis plants to salt stress
Eight positive seedlings in the T1 generation were grown from CmoNHX1-heterologously expressed A. thaliana plants in order to investigate the function of CmoNHX1. On the leaves of T2 plants, we carried out hygromycin resistance gene identification (Figure 8A) and GUS staining validation (Figure 8B), after which we arbitrarily selected the OE-1 and OE-3 lines for additional investigation.
The transgenic A. thaliana plants and wild-type were both given NaCl treatments in order to study how the CmoNHX1-heterologously expressed A. thaliana plants responded to salt stress. Under water treatment, the phenotype revealed no discernible difference between transgenic plants and wild-type plants. However, when exposed to salt stress, the leaves of A. thaliana plants heterologously expressing CmoNHX1 clearly yellowed and wilted in contrast to wild-type plants (Figure 8C). No significant difference between the transgenic lines OE1 and OE3 (Figure 8E). CmoNHX1 expression under salt stress was considerably lower than it was after water treatment, according to qRT-PCR analyses (Figure 8D). These findings imply that CmoNHX1 decreases the salt tolerance of plants under salt stress.
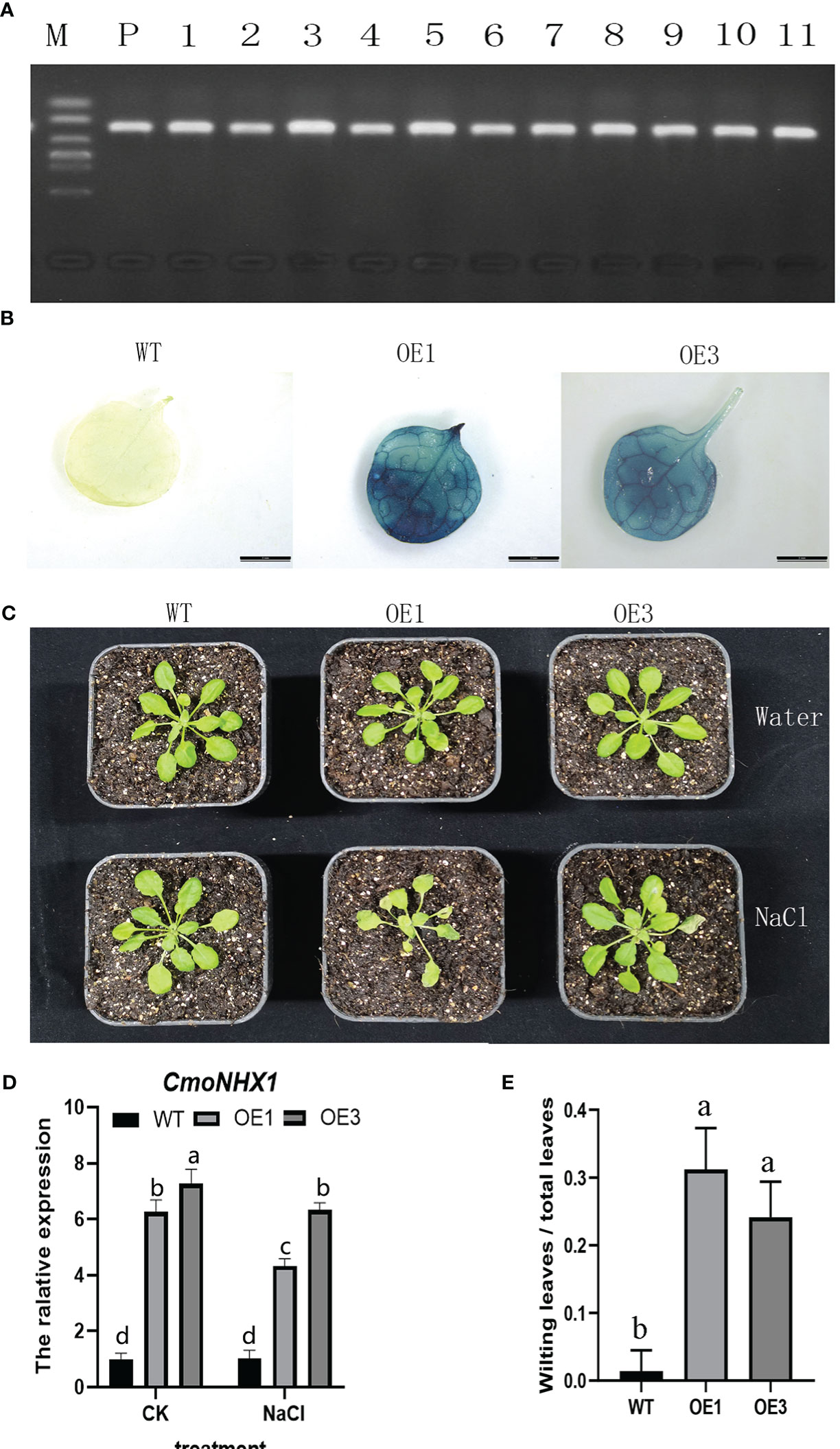
Figure 8 Positive identification and phenotype analysis of CmoNHX1-heterologously expressed A. thaliana plants. (A), Identification of hygromycin resistance genes; (B), GUS staining verification; (C), Analysis of plant phenotype characteristics; (D), Relative expression quantity of CmoNHX1 in transgenic and wild-type plants; (E), The ratio of wilting leaves to total leaves under salt stress. For the wild type, OE1 and OE3 plants, 8 plants were selected respectively for statistics. The data were represented as the means of three replicates, and error bars represented the standard deviations of means. Different letters above the bars indicated significant differences (p < 0.05) between plants.
Subcellular localization of CmoNHX1 protein
The discovery of the CmoNHX1 gene’s temporary expression in tobacco demonstrated that GFP, a fusion protein created with CmoNHX1, emits light on the cytoplasmic membrane and overlaps with the red fluorescent protein of the membrane marker: pm-rbCD3-1008 (Figure 9). This suggests that CmoNHX1 is located on the cytoplasmic membrane.
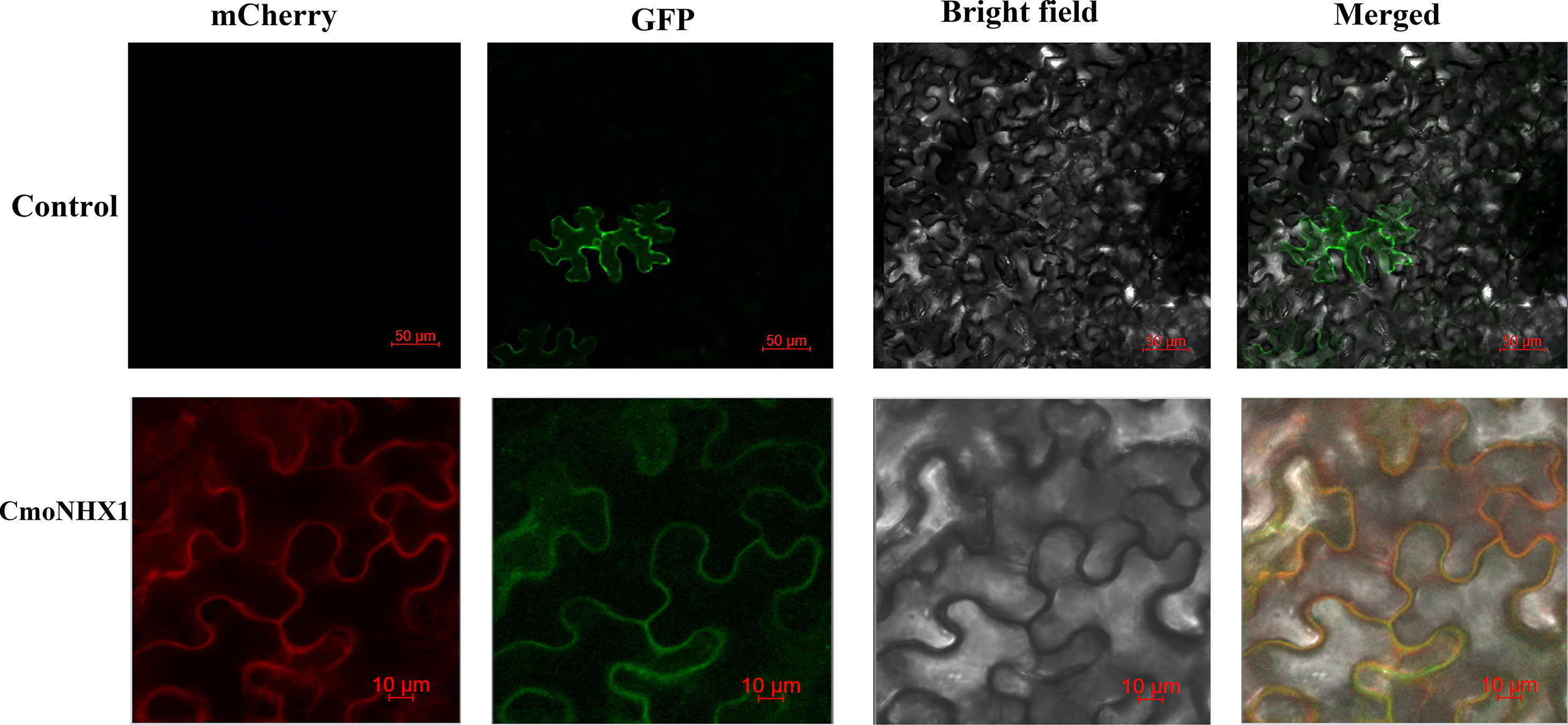
Figure 9 Subcellular localization of CmoNHX1. Transient expression of PRI101-GFP-CmoNHX1 in tobacco leaf cells. The PRI101-GFP construct was used as the control. Control, PRI101-GFP; CmoNHX1, PRI101-GFP-CmoNHX1; mCherry, membrane marker: pm-rbCD3-1008; GFP, green fluorescence of CmoNHX1; Bright field, bright field images of tobacco leaf cells; Merged, overlay of bright field, green fluorescence and red fluorescence images; Bars 50/10 µm.
Discussion
The presence of NHX genes in various plant species indicates their important role in plant adaptation to stress conditions. The NHX gene family plays a vital role in salt tolerance and pH homeostasis by regulating ion transport in plants (Apse and Blumwald, 2007). Since the discovery of eight NHX genes in A. thaliana, researchers have found NHX gene families in other species, such as P. trichocarpa (Meng and Wu, 2018), Zea mays (Zhang et al., 2018), B. vulgaris (Wu et al., 2019), Anemone vitifolia Buch (Akram et al., 2020) and Triticum aestivum (Sharma et al., 2020). In this study, we discovered a total of 26 NHX genes from three cultivars of the Cucurbita genus. These genes’ physical characteristics, such as their number of amino acids, isoelectric point pI, MW, and transmembrane structure, were quite different from one another, indicating that these cultivars’ genetic diversity occurred during evolution (Table 1). The phylogenetic tree divided 26 NHX proteins into three subfamilies: Vac, Endo and PM. The Vac subfamily had the largest number of NHX proteins (Figure 1), which was similar to the results reported in A. thaliana (Philippe et al., 2012), B. vulgaris (Wu et al., 2019) and P. trichocarpa (Meng and Wu, 2018). This study’s findings showed that some Cucurbita NHX genes’ transcription levels changed under salt stress conditions, suggesting that these genes might be involved in the plants’ response to salt stress. The discovery of NHX genes in various plant species and their diverse physical characteristics highlights the importance of understanding the evolutionary history and genetic diversity of this gene family to elucidate its biological functions fully. Overall, this study provides valuable insights into the diversity and evolution of the NHX gene family in the Cucurbita genus, which could have important implications for developing salt-tolerant crops in the future.
In addition, motif analysis found that all NHX proteins contained Motif 11. Motif 7 and 12 only exist in the Vac subfamily. Motif 13 was also found only in PM and Endo subfamilies (Figure 2C). These results were consistent with those of PtNHX (Tian et al., 2017) and SbNHX (Kumari et al., 2018), indicating that the NHX family genes in Cucurbita were relatively conserved during evolution. Among the 26 NHX genes in Cucurbita, the number of exons from PM subfamily was 20 to 23, which was much larger than that in Vac subfamily, which was consistent with the report in P. trichocarpa (Tian et al., 2017) and Glycine max (Chen et al., 2015). This indicates that the genes of different subfamilies in the same species have structural diversity, so functional diversity may exist. The circular evolutionary tree divides CmoNHXs into three subgroups, with CmoNHX1 located in vacuole (Vac) subfamily but not the plasma membrane (PM) subfamily. However, the transient expression of CmoNHX1 protein in tobacco indicates that CmoNHX1 is located in the cell membrane. These two results are inconsistent, and we speculate that there may be temporally or spatially differentially expressed CmoNHX1.
The primary driving factor behind plant genome evolution is the duplication of single genes, chromosomes, or entire genomes (Paterson et al., 2012). Since NHX genes were duplicated and lost during evolution, plants with the same ancestor have different numbers of NHX genes today. The genes of C. moschata and C. maxima essentially mapped to the same chromosome position (Figure 4), indicating that they may have shared an ancestor (Qanmber et al., 2019), and their evolutionary relationship is closer than that of C. pepo. Nine each and eight NHX genes were found in this study from three Cucurbita cultivars, respectively. Collinearity analysis is a tool that can be used to identify the evolutionary relationship between different species or cultivars based on shared gene sequences or synteny. In this case, the collinearity study suggests that C. moschata and C. maxima are more closely related to each other than they are to C. pepo, and that the number of shared syntenic genes between them decreases as the evolutionary distance between them increases. This information can help researchers to better understand the genetic basis of traits that are shared between different cultivars, as well as the genetic mechanisms that underlie the differences in those traits that have arisen during evolution. The findings of the collinearity study revealed that C. moschata and C. maxima shared 13 pairs of syntenic genes, whereas C. moschata, C. maxima, and C. pepo shared 11 and 10 pairs of syntenic genes, respectively (Figure 5). Before going through two independent phases of diversification, C. pepo originally underwent multiple domestication events in Mexico and the US. (Kates et al., 2017). According to available data, the C. pepo developed at around 30 ± 4 Mya (Schaefer et al., 2009). However, between 26 Mya and 3 Mya, C. moschata and C. maxima diverged from one another (Sun et al., 2017). This suggests that C. pepo, C. moschata, and C. maxima evolved at different rates and that the loss of gene segments or the translocation of chromosomes happened during the evolution process.
The main switch in gene transcription regulation, the cis-acting element, regulates a number of biological processes, including hormone response, abiotic stress response, and developmental process (Ding et al., 2018). Different cis-acting elements play specific functions in gene expression in plants. For instance, according to Ulmasov et al. (1997), auxin induction is primarily mediated by the AuxRE, DR5, AuxRR-core, and TGA-element motifs, while abscisic acid induction is frequently mediated by the ABRE motif (Shu et al., 2019; Wang and Huang, 2019). Inducing drought typically involves MBS and LTR (Dunn et al., 1998; Sazegari et al., 2015). In order to induce light, G-box, AT-rich, GT1-motif, and I-box elements are primarily used (Gilmartin et al., 1992). Furthermore, it was discovered in similar studies that the GT1-motif and TGACG-motif were identified as salt stress response elements (Büyük et al., 2016), and that the ABRE, G-box, MBS, and TGA-element had regulatory effects in salt stress situations (Saeediazar et al., 2014). The number of ABRE in CmoNHX1 was 13. CmoNHX1 contains 15 G-boxes and has seven TGACG/CGTCA elements. Besides, to determine the response of CmoNHXs to salt stress, we excavated the transcriptome data (BioProject: PRJNA464060) published in 2018 (Niu et al., 2018) and analyzed the transcription profile of NHXs in the leaf mesophyll and leaf vein of the C. moschata cultivar, “Rifu” under salt stress. Results revealed that after NaCl treatment, the transcription levels of CmoNHX1 in the mesophyll and veins were considerably decreased by 55.44% and 69.04%, respectively, in comparison to the control treatment (Figure 6). Based on the above analysis, CmoNHX1 may be important players in salt stress, according to research on the NHX family’s potential involvement in a variety of biological processes. We looked at the phenotypes of wild-type and CmoNHX1-heterologously expressed A. thaliana plants in this study and discovered that there was no discernible difference between transgenic and wild-type plants under water treatment. However, when exposed to salt stress, the leaves of CmoNHX1-heterologously expressed A. thaliana plants clearly yellowed and wilted in contrast to wild-type plants (Figure 8C), indicating that CmoNHX1 decreased plants’ ability to withstand salt. Additionally, qRT-PCR results demonstrated that CmoNHX1 expression under salt stress was much lower than it was during water treatment (Figure 8D). Therefore, CmoNHX1 mutants could be created using gene editing technologies in the future, improving the salt tolerance of C. moschata plants.
Under high salt treatment (200 and 300 mM NaCl), the transcription levels of BvNHX3, BvNHX4, and BvNHX5 in barley leaves were substantially greater than those in roots. Additionally, researchers discovered that BvNHX1 and BvNHX3 isolated Na+ in vacuoles to lessen the harm that salt stress caused to plants (Wu et al., 2019). In this study, heterologous expression of NHX1 in A. thaliana reduced plant tolerance to salt stress, a result consistent with the previous cis-acting element analysis and transcriptome data analysis, which demonstrated that NHX1 negatively regulates salt tolerance in plants. The above result was contrary to the expression of BnaNHX1 in cotton (Fu et al., 2020) and BvNHX1 in barley (Wu et al., 2019). Based on the above results, we speculated that the mechanism of NHX1 gene response to salt stress is different in different plant species.
Conclusions
Overall, this study provides important insights into the NHX gene family in Cucurbita species and their response to salt stress. The identification of 26 NHX genes in three cultivars of the Cucurbita genus and the characterization of their physical and structural features, evolutionary relationships, and response to salt stress provide a comprehensive understanding of the NHX gene family in these species. The discovery of the importance of CmoNHX1 in salt tolerance and the negative impact of its overexpression on salt tolerance also has practical implications for breeding salt-tolerant Cucurbita varieties. The findings of this study can serve as a foundation for further functional validation and exploration of the molecular mechanisms underlying the role of NHX genes in plant salt tolerance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
CS: Conceived and designed the research, basic bioinformatics analysis, revised the article. JY: Conceived and designed the research, basic bioinformatics analysis, wrote the original draft of the manuscript. XL: Vector construction, genetic transformation, positive identification of transgenic plants, qRT-PCR analysis of related Genes. RC: Vector construction, genetic transformation, positive identification of transgenic plants, qRT-PCR analysis of related genes. DL: Vector construction, genetic transformation, positive identification of transgenic plants. FW: Provides tools for bioinformatics analysis. XL: Provides methods for identification of transgenic plants. XZL: Revised the article. All authors contributed to the article and approved the submitted version.
Funding
This work was mainly supported by the Henan Province Science Foundation for Youths. (222300420160), the National Natural Science Foundation of China (Nos. 32102393), the Scientific Research Foundation for High-level Talent (103010620001/015 and 2017034). Funding body has no role in the study design, data collection, analysis and manuscript writing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1136810/full#supplementary-material
References
Akram, U., Song, Y. H., Li, C. Z., Abid, M. A., Askari, M., Myat, A. A., et al. (2020). Genome-wide characterization and expression analysis of NHX gene family under salinity stress in Gossypium barbadense and its comparison with Gossypium hirsutum. Genes. 11 (7), 803. doi: 10.3390/genes11070803
Apse, M. P., Blumwald, E. (2007). Na+ transport in plants. FEBS letters 581 (12), 2247–2254. doi: 10.1016/j.febslet.2007.04.014
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, 369–373. doi: 10.1093/nar/gkl198
Bassil, E., Ohto, M. A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., et al. (2011). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell. 23 (1), 224. doi: 10.1105/tpc.110.079426
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J. C., et al. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 14 (10), 1023–1031. doi: 10.1002/elps.11501401163
Büyük, I., Inal., B., Ilhan, E., Tanriseven, M., Aras, S., Erayman, M. (2016). Genome-wide identification of salinity responsive HSP70s in common bean. Mol. Biol. Rep. 43 (11), 1–16. doi: 10.1007/s11033-016-4057-0
Chen, H. T., Chen, X., Wu, B. Y., Yuan, X. X., Zhang, H. M., Cui, X. Y., et al. (2015). Whole-genome identification and expression analysis of k+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J. Integr. Agr. 14, 1171–1183. doi: 10.1016/S2095-3119(14)60918-7
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Ding, X., Li, J. H., Pan, Y., Zhang, Y., Ni, L., Wang, Y. L., et al. (2018). Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int. J. Mol. Sci. 19 (6), 1583. doi: 10.3390/ijms19061583
Dunn, M. A., White, A. J., Vural, S., Hughes, M. A. (1998). Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare l.). Plant Mol. Biol. 38 (4), 551–564. doi: 10.1023/a:1006098132352
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 (1), 279–285. doi: 10.1093/nar/gkv1344
Fu, X. K., Lu, Z. Y., Wei, H. L., Zhang, J. J., Yang, X., Wu, A. M., et al. (2020). Genome-wide identification and expression analysis of the NHX (sodium/hydrogen antiporter) gene family in cotton. Front. Genet. 11. doi: 10.3389/fgene.2020.00964
Gilmartin, P. M., Memelink, J., Hiratsuka, K., Kay, S. A., Chua, N. H. (1992). Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell. 4 (7), 839–849. doi: 10.1105/tpc.4.7.839
Guindon, S., Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 (5), 696–704. doi: 10.1080/10635150390235520
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Jyothishwaran, G., Makoto., K., Selvaraj, T., Srideshikan, S. H. (2007). A modified freeze-thaw method for the efficient transformation of Agrobacterium tumefaciens. Curr. Sci. India. 93, 770–772. doi: 10.1371/journal.pone.0000936
Kates, H. R., Soltis, P. S., Soltis, D. E. (2017). Evolutionary and domestication history of Cucurbita (pumpkin and squash) species inferred from 44 nuclear loci. Mol. Phylogenet. Evol. 111, 98–109. doi: 10.1016/j.ympev.2017.03.002
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Kumari, P. H., Kumar, S. A., Ramesh, K., Reddy, P. S., Nagaraju, M., Prakash, A. B., et al. (2018). Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHX) homologs in sorghum bicolor. Genes. 9 (5), 236. doi: 10.3390/genes9050236
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics. 23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Doerks, T., Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, 302–305. doi: 10.1093/nar/gkr931
Meng, K., Wu, Y. X. (2018). Footprints of divergent evolution in two Na+/H+ type antiporter gene families (NHX and SOS1) in the genus populus. Tree Physiol. 38 (6), 813–824. doi: 10.1093/treephys/tpx173
Moller, S., Croning, M. D., Apweiler, R. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 17 (7), 646–653. doi: 10.1093/bioinformatics/17.7.646
Montero-Pau, J., Blance, J., Bombarely, A., Ziarsolo, P., Esteras, C., Martí-Gómez, C., et al. (2018). De-novo assembly of zucchini genome reveals a whole genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 16 (6), 1161–1171. doi: 10.1111/pbi.12860
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Niu, M. L., Xie, J. J., Chen, C., Cao, H. H., Sun, J. Y., Kong, Q. S., et al. (2018). An early ABA-induced stomatal closure, na+ sequestration in leaf vein and k+ retention in mesophyll confer salt tissue tolerance in cucurbita species. J Exp. Bot. 69 (20), 4945–4960. doi: 10.1093/jxb/ery251
Ohnishi, M., Fukada-Tanaka, S., Hoshino, A., Takada, J., Inagaki, Y., Iida, S. (2005). Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory. Plant Cell Physiol. 46 (2), 259–267. doi: 10.1093/pcp/pci028
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 492 (7429), 423–427. doi: 10.1038/nature11798
Philippe, L., Berardini, T. Z., Li, D. H., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, 1202–1210. doi: 10.1093/nar/gkr1090
Qanmber, G., Lu, L. L., Liu, Z., Yu, D. Q., Zhou, K. H., Huo, P., et al. (2019). Genome-wide identification of GhAAI genes reveals that GhAAI66 triggers a phase transition to induce early flowering. J. Exp. Bot. 70 (18), 4721–4736. doi: 10.1093/jxb/erz239
Qi, P. P., Ran, S. H., Zhang, K. (2012). Effects of different irrigation methods and crop types on soil salinization of cultivated land in arid areas of northwest China. J. Agro-environment Sci. 31 (4), 6. CNKI:SUN:NHBH.0.2012-04-023.
Rajendran, K., Tester, M., Roy, S. J. (2010). Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 32 (3), 237–249. doi: 10.1111/j.1365-3040.2008.01916.x
Roy, S. J., Negrão, S., Tester, M. (2014). Salt resistant crop plants. J. Curr. Opin. Biotechnol. 26, 115–124. doi: 10.1016/j.copbio.2013.12.004
Saeediazar, S., Zarrini, H. N., Ranjbar, G., Heidari, P. (2014). Identification and study of cis regulatory elements and phylogenetic relationship of TaSRG and other salt response genes. J. Bio. Env. Sci. 5 (1), 1–5.
Sazegari, S., Niazi, A., Ahmadi, F. S. (2015). A study on the regulatory network with promoter analysis for arabidopsis DREB-genes. Bioinformation. 11 (2), 101–106. doi: 10.6026/97320630011101
Schaefer, H., Heibl, C., Renner, S. S. (2009). Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc. Biol. Sci. 276 (1658), 843–851. doi: 10.1098/rspb.2008.1447
Sharma, H., Taneja, M., Upadhyay, S. K. (2020). Identification, characterization and expression profiling of cation-proton antiporter superfamily in Triticum aestivum l. and functional analysis of TaNHX4-b. Genomics. 112 (1), 356–370. doi: 10.1016/j.ygeno.2019.02.015
Shu, W. B., Zhou, H. J., Jiang, C., Zhao, S. T., Wang, L. Q., Li, Q. Z., et al. (2019). The auxin receptor TIR1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/IAA28 in populus. Plant Biotechnol. J. 17 (2), 338–349. doi: 10.1111/pbi.12980
Sun, H. H., Wu, S., Zhang, G. Y., Jiao, C., Guo, S. G., Ren, Y., et al. (2017). Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 10 (10), 1293–1306. doi: 10.1016/j.molp.2017.09.003
Tian, F. ,. X., Chang, E. M., Li, Y., Sun, P., Hu, J. J., Zhang, J. (2017). Expression and integrated network analyses revealed functional divergence of NHX-type Na+/He+ exchanger genes in poplar. Sci. Rep. 7 (1), 2607. doi: 10.1038/s41598-017-02894-8
Ulmasov, T., Murfett, J., Hagen, G., Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 9 (11), 1963–1971. doi: 10.1105/tpc.9.11.1963
Wang, J., Huang, R. F. (2019). Modulation of ethylene and ascorbic acid on reactive oxygen species scavenging in plant salt response. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00319
Wu, Y. J., Jiang, B., Zhang, Y., Li, Y. B., He, L., Wang, Y. C. (2010). Transient expression in tobacco by agrobacterium-mediated transformation. J. Northeast Forestry University. 38 (09), 110–112. doi: 10.13759/j.cnki.dlxb.2010.09.008
Wu, G. Q., Wang, J. L., Li, S. J. (2019). Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris l.) and their regulated expression under salt stress. Genes. 10 (5), 401. doi: 10.3390/genes10050401
Xu, H. M., Zhang, L. J., Liu, C. (2010). Study on the infection of A. thaliana by Agrobacterium tumefaciens dipping in flowers. Northern Horticulture 14)4, 143–146. CNKI:SUN:BFYY.0.2010-14-059.
Yamniuk, A. P., Vogel, H. J. (2004). Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27 (1), 33–57. doi: 10.1385/MB:27:1:33
Yuan, J. P., Liu, T. K., Yu, Z. H., Li, Y., Ren, H. B., Hou, X. L., et al. (2019). Genome-wide analysis of Chinese cabbage IQD gene family and the response of BrIQD5 in drought resistance. Plant Mol. Biol. 99 (6), 603–620. doi: 10.1007/s11103-019-00839-5
Zhang, M., Cao, Y. B., Wang, Z. P., Wang, Z. Q., Shi, J. P., Liang, X. Y., et al. (2018). A retrotransposon in an HKT1 family sodium transporter causes variation of leaf na+ exclusion and salt tolerance in maize. New Phytol. 217 (3), 1161–1176. doi: 10.1111/nph.14882
Keywords: Cucurbita L., Na+/H+ antiporter, evolutionary relationship, NHX1, expression pattern
Citation: Shen C, Yuan J, Li X, Chen R, Li D, Wang F, Liu X and Li X (2023) Genome-wide identification of NHX (Na+/H+ antiporter) gene family in Cucurbita L. and functional analysis of CmoNHX1 under salt stress. Front. Plant Sci. 14:1136810. doi: 10.3389/fpls.2023.1136810
Received: 03 January 2023; Accepted: 28 February 2023;
Published: 14 March 2023.
Edited by:
Sunil Kumar Sahu, Beijing Genomics Institute (BGI), ChinaReviewed by:
Hongxia Zhang, Ludong University, ChinaDixit Sharma, Central University of Himachal Pradesh, India
Copyright © 2023 Shen, Yuan, Li, Chen, Li, Wang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingping Yuan, anB5dWFuNjY2QDE2My5jb20=
 Changwei Shen
Changwei Shen Jingping Yuan
Jingping Yuan Xin Li2,3
Xin Li2,3 Daohan Li
Daohan Li Fei Wang
Fei Wang Xing Liu
Xing Liu