
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 January 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1136616
This article is part of the Research Topic Growth Regulation in Horticultural Plants: New Insights in the Omics Era View all 14 articles
For most fruit and forest species vegetative propagated from elite genotypes, adventitious rooting is essential. The ability to form adventitious roots significantly decreased during the juvenile to adult phase change. Apart from the miR156-SPL pathway, whether there is another regulation mechanism controlling age-dependent adventitious rooting ability remained largely unknown. In the present study, we showed that MdWRKY87 expression level was positively correlation with adventitious rooting ability. In addition, over-expressing of MdWRKY87 in tobacco leads to enhanced adventitious rooting ability, more adventitious root number and accelerated adventitious rooting process. Comparative transcriptome profiling indicated that MdWRKY87 overexpression can activate the expression of adventitious rooting-induced genes, such as WOX11 and AIL. In addition, MdWRKY87 overexpression can inhibit the transcription of adventitious rooting-repressed genes, such as AUX/IAAs and type-B cytokinin RRs. Collectively, here we demonstrated that higher expression level of MdWRKY87 contributes to age-dependent adventitious rooting-competent in juvenile apple rootstock.
The root system is able to adapt its architecture and morphology to the soil environment and physiological requirements of the plant in a flexible manner (Casimiro et al., 2003). Plant root systems typically consist of primary roots, lateral roots, and adventitious roots (Ji et al., 2022). Among these root types, ARs display high phenotypic plasticity in response to a variety of environmental stimuli (Li et al., 2022). Monocotyledons, like rice and maize, produce adventitious roots during normal development. In most tree species, especially poplars, cuttings are the primary means of propagation, and species differ in their ability to form adventitious root significantly. For most fruit and forest species propagated from elite genotypes, adventitious rooting is essential for proliferation. Plant tissue totipotency enables the adventitious root formation from non-root tissues, which has been extensively utilized for vegetative propagation of agricultural and forestry plants (Bellini et al., 2014). Most fruit species, such as apple rootstock, adventitious root formation is not easy to limit good varieties of asexual reproduction. Despite some preliminary studies on the mechanism of adventitious roots formation in difficult-to-root trees, and more in-depth research is needed.
In most tree species, the ability to form adventitious roots significantly decreased during the juvenile to adult phase change. Extensive research has been conducted to overcome problems related to the loss or reduction of the ability of difficult-to-root trees to form adventitious roots (Levy et al., 2014; Xu et al., 2017; Wang et al., 2019; Li et al., 2021). In our previous study, we have showed that the rooting rates of cuttings from juvenile and rejuvenated donor plants were significantly higher than those of cuttings from adult trees in Malus xiaojinensis (Xu et al., 2017). The high expression of miR156 is positively correlated with auxin-induced adventitious roots formation (Levy et al., 2014; Xu et al., 2017). MiR156 functions via its target gene MxSPL26 in regulating adventitious root formation (Xu et al., 2017). Our previous research has indicated that MxSPL26 inhibited MxHB13 expression by directly binding to its promoter (Li et al., 2021). During the adult phase, MxSPL26 interacts with auxin-induced MxTIFY9 and co-represses MxHB13 expression, leading to reduced AR formation (Li et al., 2021). Although several pathways and components of auxin-mediated molecular regulatory networks underlying adventitious root formation in apple have been identified, but the molecular mechanisms need to be investigated further.
The WRKY proteins are a superfamily of transcription factors found exclusively in plant (Rushton et al., 2010). The name of WRKY is derived from the highly conserved amino acid sequence containing WRKYGQK and the zinc finger-like motifs (Cys2-His2 or Cys2-HisCys) (Eulgem et al., 2000; Rushton et al., 2010). Based on both the number of WRKY domains and their zinc-finger motif, WRKYs can be divided into three distinct groups (Eulgem et al., 2000). The WRKYs have been found to function in seed dormancy, embryo and trichome formation, senescence, hormone synthesis, signal transduction, defense responses and abiotic stresses (Johnson et al., 2002; Lagace and Matton, 2004; Zhang et al., 2004; Li et al., 2015; Singh et al., 2017; Hu et al., 2018; Liang et al., 2020; Kang et al., 2021; Zhu et al., 2021). To date, only a small number of research have reported that WRKYs participate in adventitious roots formation. The group IIe WRKY gene of Catalpa Scop, CbNN1 expression increased with increasing adventitious rooting ability (Wang et al., 2019). PuWRKY75, as a transcription activator, controls the low phosphorus driven adventitious root formation through up-regulating PuLRP1 and PuERF003 transcription in Populus ussuriensis (Wang et al., 2022). The functions of WRKYs in adventitious root formation remain to be investigated.
In this study, we identified a group IIe subfamily WRKY gene, MdWRKY87 from apple rootstock. The MdWRKY87 protein was found located in the nucleus and functions as a transcriptional repressor in both yeast and plant cells. Our results also indicated that MdWRKY87 promoted adventitious rooting through regulating root-related gene involved in auxin signaling pathway.
The leafy stem cuttings of M. xiaojinensis (Mx) was used as the materials, because Mx has a high apomictic rate to ensure the juvenile materials stability (Xu et al., 2017). Semi-lignified leafy cuttings (8-10 cm in length) were excised from basal suckers (juvenile phase, Mx-J) and shoots from the canopy of reproductively mature trees (adult phase, Mx-A). The bases of leafy cuttings were immersed 1~2 cm in depth into a 3.0 g L-1 indole butyric acid (IBA, Sigma-Aldrich, St. Louis, MO, USA) solution for 1 min (Xu et al., 2017). Cutting dipped in IBA-free water was used as a control. After plugging the cuttings into 50 cell trays containing fine sand, they were incubated in a solar greenhouse. The rooting ability was evaluated at 35 days after treatment. Three biological replicates, each with at least 50 leafy cuttings, were used for the experiment to manage experiment errors.
Tissue-cultured ‘M9T337’ plantlets were sub-cultured in Murashige and Skoog (MS) medium containing 7.5 g L-1 agar and 30 g L-1 sugar (pH 5.8) with 0.5 mg L-1 IBA and 0.2 mg L-1 6-benzylaminopurine (6-BA) (Cheng et al., 2020). After 30 days, stem cuttings were transferred into 1/2 medium containing rooting 7.5 g L-1 agar and 30 g L-1 sugar (pH 5.8) with 0.5 mg L-1 IBA and 0.1 mg L-1 1-naphthalene acetic acid (NAA) for rooting. The tobacco (Nicotiana tabacum) plants were sub-cultured in MS medium without hormone. The plantlets were grown under a 16 h light/8 h dark photoperiod with day/night temperatures of 25 ± 1°C and 20 ± 1°C.
Paraffin sections of stem bases were prepared as previously described (Xu et al., 2017; Cheng et al., 2020; Jin et al., 2022), with some modifications. The bases of ‘M9T337’ stem cuttings were collected at 6, 9, and 12 days after transplanting on 1/2 MS medium with 0.5 mg L-1 IBA and 0.1 mg L-1 NAA. The bases of tobacco stem cuttings were excised at 2, 4 and 6 days after subculture on hormone-free MS medium. The samples were fixed in FAA solution (70% ethanol: formaldehyde: acetic acid, 95:5:5 [v/v/v]) for 2 days at room temperature, and store at 4 °C. Then samples were dehydrated with a graded ethanal series (50%, 70%, 85%, 95%, and 100%), infiltrated with xylene, and embedded in paraffin. Cross sections with a 10 μm in thickness were cut with a Leica RM2245 (Leica Microsystems, Wetzlar, Germany) rotary microtome, transferred onto glass slides, deparaffined with xylene, and re-hydrated through an ethanol series, and stained with toluidine blue. Slides were observed using an optical microscope DM2500 (Leica Microsystems, Wetzlar, Germany) and photos were obtained using an attached digital camera DFC420 (Leica Microsystems, Wetzlar, Germany).
Total RNA was extracted from approximately 0.5 g of frozen sample using the TIANGEN Plant RNA Kit (TIANGEN biotech CO., LTD, Beijing, China, DP305). For each sample, 1 μg DNase-treated RNA was used to synthesize first-strand cDNA with oligo d(T) or random primer and HiScript® II Q RT SuperMix (Cat. R223-01, Vazyme, China). A LightCycler 480 instrument (Roche, Basel, Switzerland) and ChamQ SYBR Color qPCR Master Mix (Vazyme, Nanjing, China) were used for qRT-PCR. The relative expression levels of genes were normalized to the reference gene EF1α and calculated using the 2 -ΔΔCt method (Livak and Schmittgen, 2001). All reactions were performed with at least three biological replicates. The primers are listed in Supplemental Table S1.
The MdWRKY87 promoter fragment (-2000 bp to -0 bp from the MdWRKY87 ATG start codon) was inserted into pCambia1391 vector generating the proMdWRKY87: GUS construct. Tobacco leaves were transformed with the Agrobacterium tumefaciens strain GV3101 cells harboring a proMdWRKY87: GUS or DR5: GUS construct. Agrobacterium cells were re-suspended in buffer with (10 mM MgCl2, 10 mM MES-KOH, pH 5.6; adding 200 μM acetosyringone immediately prior to use) to an OD600 of 0.8~1.0. After injected with 1 ml needleless syringes, the leaves were treated with 50μM IBA and collected 6 hours later. The leaves were submerged in the GUS staining solution for 24 h at 37°C. After staining, tissues were cleared by immersing in 70% ethanol. All primers used are listed in Supplemental Table S1.
The ORF fragment (stop codon removed) of MdWRKY87 containing SmaI and XbaI sites were inserted into the Super1300-GFP vector to generate the pSuper : MdWRKY87-GFP construct. Subcellular localization was conducted as previously described (Cheng et al., 2021). A. tumefaciens cells (GV3101) expressing pSuper : MdWRKY87-GFP and a Cherry-labelled nuclear marker (NF-YA4-mCherry) was re-suspended using the buffer (10 mM MgCl2, 10 mM MES-KOH, pH 5.6; 200 μM acetosyringone). pSuper : NF-YA4-mCherry was used as a nuclear marker. The tobacco leaves were injected with the re-suspended A. tumefaciens cells using a 1 ml needleless syringe. Three days after infiltration, fluorescence signals of the infiltrated leaves were detected using a laser scanning confocal microscope (Leica TCS SP5 II, Wetzlar, Germany). The primers used for construction are listed in Supplemental Table S1.
The coding fragment of MdWRKY87 were fused to the GAL4-BD in pBD-GAL4 vector. The transcriptional activation analysis was conducted as previously described (Cheng et al., 2021). The transactivation activity was verified by the growth of yeast AH109 harboring full-length of MdWRKY87 on SD/-Trp and SD/-Trp-His plates and was confirmed by a X-α-Gal staining assay. All primers used are listed in Supplemental Table S1.
The MdWRKY87 ORF sequence without stop codon was cloned into the pBD-VP16 vector (Han et al., 2016). The reporter vector contained a GAL4-luciferase (LUC) containing five copies of the GAL4-binding element and a minimal CaMV35S promoter at the 5’ end of the LUC gene (Han et al., 2016).The effector vectors or reporter vectors were introduced into A. tumefaciens strain GV3101. The A. tumefaciens cells was re-suspended to an OD600 of 1.0 using the buffer (10 mM MgCl2, 10 mM MES-KOH, pH 5.6; 200 μM acetosyringone). A. tumefaciens cells harboring effector vector and reporter vector were mixed 1:1, then injected into the tobacco (N. benthamiana) leaves by using a 1 mL needleless syringe. After spraying 1 mM luciferin onto the leaves, luciferase imaging was performed using NEWTON 7.0 (VILBER LOURMAT, Paris, France). An assessment of LUC and REN activities was conducted using the Duo-Lite Luciferase Assay System (DD1205–01, Vazyme, Nanjing, China) and BioStack Ready (BioTek Instruments Inc., Winooski, Vermont, USA). LUC/REN ratio was used to calculate the results. The primers used for construction are listed in Supplemental Table S1.
The MdWRKY87 ORF sequence without stop codon was cloned into pRI101 vector to generate 35S:MdWRKY87-OE construct. The construct was introduced into wild type tobacco (N. tabacum) leaves by A. tumefaciens-mediated transformation as previously described (Zhu et al., 2022). The infected leaves were selected on MS medium containing 100 mg L-1 kanamycin and 300 mg L-1 cefotaxime sodium to generate MdWRKY87-overexpressing (MdWRKY87-OE) transgenic lines. Transgenic plants were propagated by subculture on hormone-free MS medium. The primers used for construction are listed in Supplemental Table S1.
Total RNA was extracted from the stem bases of MdWRKY87-OE and wild type plants using the TIANGEN Plant RNA Kit (TIANGEN biotech CO., LTD, Beijing, China, DP305). A total amount of 3 µg RNA per sample was used in RNA-seq library construction. An Illumina Hiseq (Illumina, CA, USA) system was used for RNA sequencing by Novogene (Novogene, Tianjin, China). A quality assessment was performed on raw data using FastQC. Following Trimmomatic filtering out adapters and unpaired reads, the remaining clean reads were used to calculate the expression of gene by using Kallisto, an RNA-seq quantification program (Bray et al., 2016). The N. tabacum genome was used as the reference genome (Edwards et al., 2017). The count of reads was normalized to Transcripts Per kilobase of exon model per Million mapped reads (TPM). The log2TPM values were subjected to generate the heat map by TBtools software (Chen et al., 2020). The RNA-seq data were deposited in the NCBI Sequence Read Archive (accession number PRJNA917351).
According to our previous research, semi-lignified leafy cuttings from Mx-J and shoots from the canopy of Mx-A were used in this study (Xu et al., 2017; Li et al., 2021). As the previous results (Xu et al., 2017; Li et al., 2021), Mx-J cuttings exhibited a high adventitious rooting ability (Figures 1A–C). After IBA treatment, the rooting percentage of Mx-J cuttings (85.14%) was significantly higher than that of Mx-A cuttings (3.57%, Figure 1B). Neither the cuttings of Mx-A nor Mx-J exhibited the ability of adventitious rooting (Figure 1B). Moreover, the adventitious root number per cutting of Mx-J was significantly more than that of Mx-A (Figure 1C). We previously identified the expression of the WRKY transcription factor family genes in the cutting stems of Mx-A and Mx-J after IBA treatment (Che et al., 2021). The expression of MdWRKY87 gene was significantly induced in the Mx-J cutting treated with IBA (Figure 1C). However, there was no difference in the mRNA levels of MdWRKY87 in the Mx-A cuttings treated with IBA or untreated control (Figure 1C).
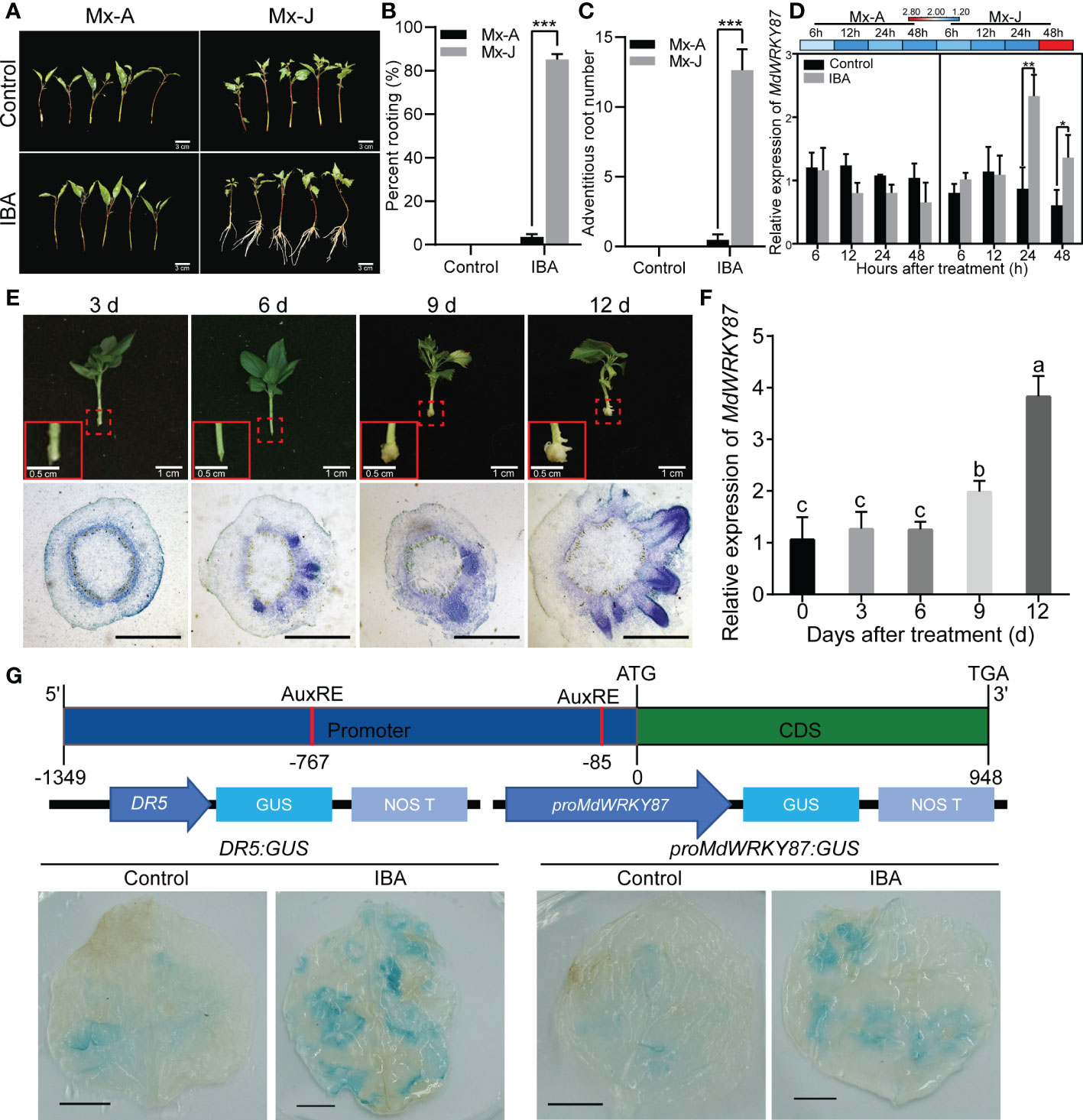
Figure 1 The expression of MdWRKY87 is positively correlated with auxin-induced adventitious root formation. (A) Indole butyric acid (IBA) treatment induced adventitious root formation of leafy cutting from juvenile (Mx-J) not adult (Mx-A) phase in M. xiaojinensis. Scale Bar = 3 cm. (B) Adventitious rooting percentage of leafy cutting from Mx-A and Mx-J phase of M. xiaojinensis after IBA treatment. (C) Number of adventitious roots. For B and C, the mean values ± SD are shown for three biological replicates. Student’s t-test, *** P < 0.001. (D) Relative expression of MdWRKY87 gene in leafy cutting of Mx-A and Mx-J after IBA treatment. The mean values ± SD are shown for three biological replicates. Stem bark samples of 0.5 to 1.0 cm basal sections of 20 Mx-J or Mx-A cuttings treated with IBA-free solution or IBA were pooled together as one biological replicates. Student’s t-test, ** P < 0.01. (E) The process of adventitious root formation of tissue culture plantlets growing on 1/2 MS medium containing IBA. Top, representative pictures of adventitious root formation of plantlets. The red dotted boxes indicate portion of stem base magnified in red solid line boxes (Scale bar = 0.5 cm). Scale bar = 1 cm. Bottom, transverse sections of stem base during the adventitious root formation. Scale bar = 1 mm. (F) Relative expression of MdWRKY87 gene in stem base of tissue culture plantlets. Different letters indicate statistically significant differences (P < 0.05) at by Duncan’s test. (G) GUS activity of N. benthamiana leaves transiently transformed with proMdWRKY87:GUS or DR5:GUS after IBA treatment. Top, schematic representation of MdWRKY87. Medium, schematic representation of the proMdWRKY87:GUS and DR5:GUS constructs. Bottom, GUS activity analysis. Scale bar = 1 cm. *p < 0.05.
To further define the relationship between MdWRKY87 expression levels and adventitious rooting formation, we next examined the expression pattern of MdWRKY87 during adventitious rooting formation of tissue culture plantlets in apple. According to the paraffin sections of stem bases of apple plantlets, we found that primordia with dome-shaped adventitious structures were clearly visible at 6~9 days after transplanting (Figure 1E) as our previous results (Cheng et al., 2020). After 12 days of transplantation, adventitious roots began to appear. As expected, the gene expression levels of MdWRKY87 increased significantly with the emergence of adventitious root from stem base (Figure 1F).
To further validate MdWRKY87 expression in response to IBA treatment, Agrobacterium tumefaciens cells (GV3101) harboring the proMdWRKY87:GUS or DR5:GUS construct (auxin-responsive reporter) were transiently transformed into tobacco leaves. After IBA treatment, the levels of GUS proteins obviously increased in both of leaves transformed with proMdWRKY87:GUS and DR5:GUS construct (Figure 1G), suggesting that the promoter activity of MdWRKY87 responds to auxin. These results indicate high MdWRKY87 expression correlates positively with adventitious rooting and may regulate auxin-mediated adventitious root development.
Multiple sequence alignments of WRKY22 homologs from apple indicated that MdWRKY87 harbored a conserved WRKYGQ domain and a C2H2 (C-X5-C-X23-H-X1-H) zinc-finger motif at its C terminus (Figure 2A) and belonged to WRKY group IIe (Eulgem et al., 2000). To confirm whether MdWRKY87 functions as a transcription factor, we expressed MdWRKY87 fused to green fluorescence protein in tobacco leaves and observed that the fusion protein localized to the nucleus (Figure 2B). A transactivation assay in yeast indicated that the MdWRKY87 protein has transcriptional activation activity (Figure 2C). We also performed a dual-luciferase transactivation assay in tobacco leaves. The results showed that the luciferase activity of co-expression of the reporter with pBD- WRKY87 -VP16 was significantly lower than that in the pBD-VP16 control (Figures 2D, E). These results suggested that MdWRKY87 is indeed a transcription repressor of WRKY group IIe subfamily.
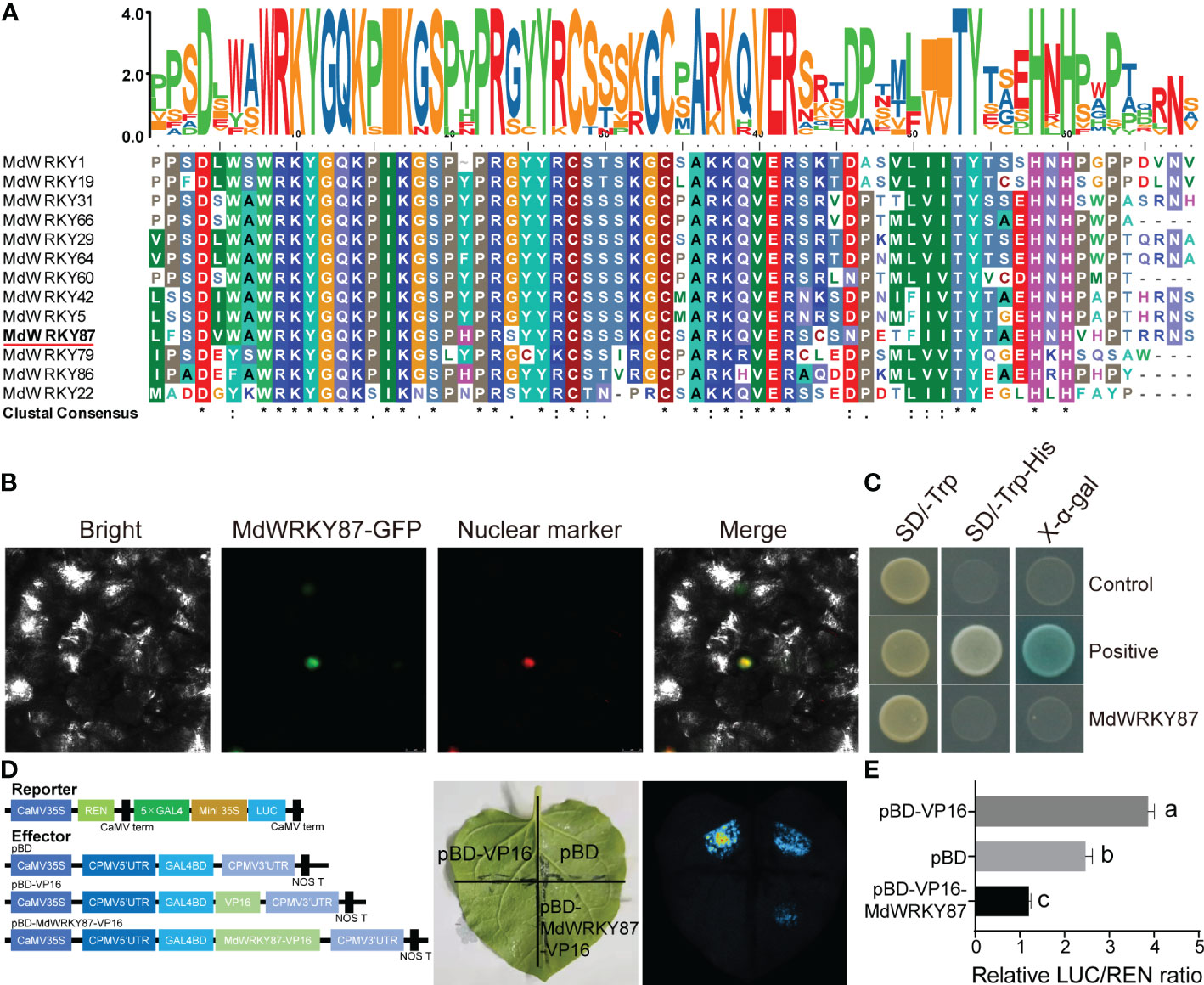
Figure 2 MdWRKY87 is a transcription repressor of WRKY family. (A) Sequence comparison of WRKY domains from WRKY IIe proteins, shaded by conserved amino acids. The accession numbers of all genes used are listed in the Methods section. (B) Subcellular localization of MdWRKY87 in N. benthamiana leaves. Agrobacterium carrying the pSuper : MdWRKY87-GFP and pSuper : NF-YA4-mCherry as a nuclear marker genes was co-infiltrated into N. benthamiana leaves. Images were captured 2 d following agroinfiltration. (C) Transcriptional activity analysis of MdWRKY87 protein in yeast. The transactivation activity was verified by the growth of yeast AH109 harboring MdWRKY87 ORF on SD/-Trp and SD/-Trp-His plates and was confirmed by a X-α-Gal staining assay. (D) Transcriptional activity analysis of MdWRKY87 in N. benthamiana leaves. Reporter and effector constructs were co-infiltrated into N. benthamiana leaves. Left, schematic representation of the effector and reporter constructs. Right, live imaging of N. benthamiana leaves expressing reporter and effector constructs. (E) The dual-luciferase activity assay. The values were determined by calculating the ratio of LUC activity to REN activity (LUC/REN). Different letters indicate statistically significant differences (P < 0.05) at by Duncan’s test.
To characterize the role of MdWRKY87 in adventitious rooting, we generated overexpression lines of MdWRKY87 (MdWRKY87-OE) in tobacco plants by Agrobacterium-mediated transformation (Supplemental Figure S1). We tested the effects of MdWRKY87 overexpression on adventitious root formation in MdWRKY87-OE lines #9, #5 and #6 (Figures 3A, B). During the adventitious rooting process, there were obvious morphological differences between MdWRKY87-OE transgenic plants and non-transformed wild type (WT) (Figure 3A). The adventitious rooting rate, root number and root length per stem were significantly higher in MdWRKY87-OE lines than that in WT plants (Figures 3C–E and Supplemental Movie S1). These results supported the notion that high MdWRKY87 expression correlates positively with adventitious rooting.
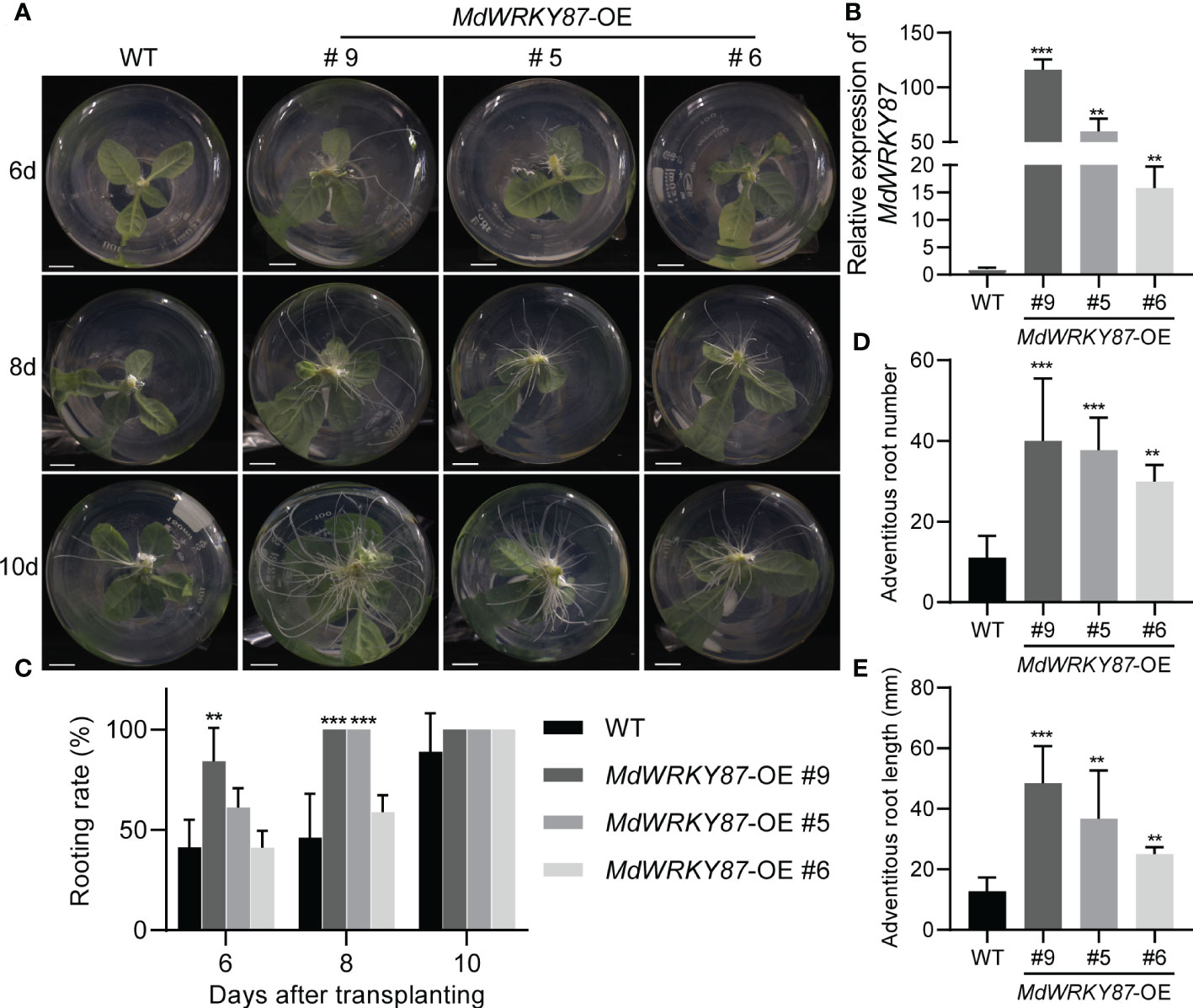
Figure 3 MdWRKY87 accelerates adventitious root formation in transgenic tobacco (N. tabacum) plants. (A) Phenotypes of adventitious roots in wild-type (WT) and independent transgenic lines (MdWRKY87-OE9, MdWRKY87-OE5, and MdWRKY87-OE6) in the absence of auxin at 6, 8 and 10 days after transplanting into hormone-free MS medium. (B) qRT-PCR analysis of MdWRKY87 expression in WT and MdWRKY87-OE lines. (C) Adventitious rooting percentage of wild-type (WT) and independent transgenic lines at 6, 8 and 10 days after transplanting into hormone-free MS medium. (D, E) Number (D) and length (E) of adventitious roots at 10 days after transplanting into hormone-free MS medium. The mean values ± SD are shown for three biological replicates. Asterisks indicate significant differences between WT and each transgenic lines by Student’s t-test (**P< 0.01; ***P< 0.001).
To check whether MdWRKY87 affects the initiation of adventitious root primordia, we conducted the cross sections of the stems of WT and MdWRKY87-OE transgenics lines during adventitious rooting. The initiation of adventitious root primordia was accelerated in MdWRKY87-OE transgenics lines from 2 days after subculture on MS (Figure 4). Moreover, the adventitious root primordium in MdWRKY87-OE transgenics lines were well-developed compared with the WT plantlets at 4 days after subculture, suggesting that high MdWRKY87 expression accelerates the initiation and development of adventitious root primordia.
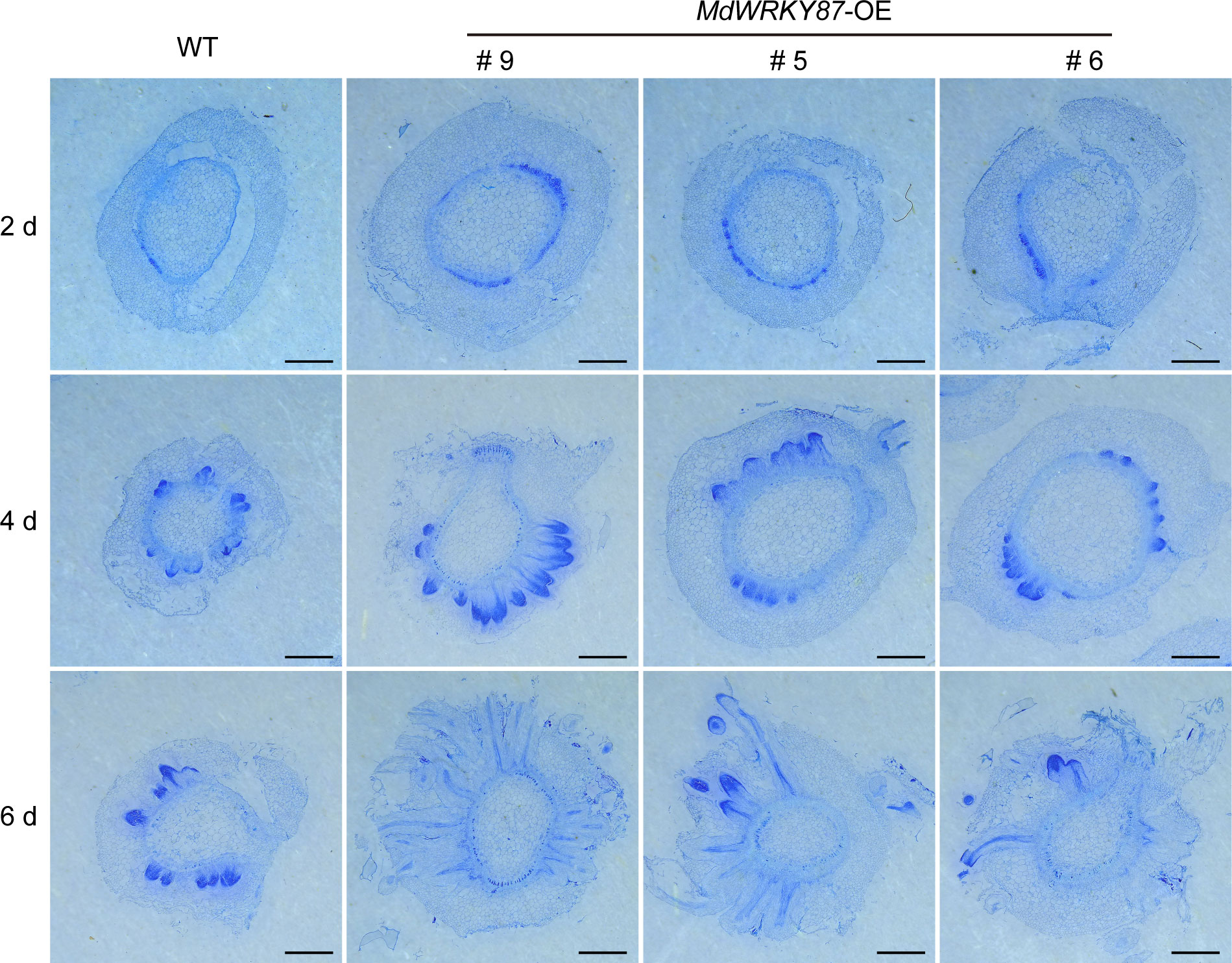
Figure 4 MdWRKY87 accelerates the formation of adventitious root primordium in transgenic tobacco (N. tabacum) plants. Cross-sections of stem bases at 2, 4 and 6 days after transplanting during adventitious root formation in WT and MdWRKY87-OE lines. Sections were stained by toluidine blue. Scale bar = 1 mm.
To determine whether WRKY87 regulates adventitious root formation through modulating auxin polar transport, we examined adventitious rooting capacity in wild-type, MdWRKY87-OE tobacco plants stem cuttings grown on MS medium supplemented with 1-N-naphthylphthalamic acid (NPA). Adventitious rooting was almost absolutely inhibited in both wild-type and MdWRKY87-OE tobacco plants treated with 20µM NPA (Figure 5A). To check whether NPA affects the initiation of adventitious root primordia, we conducted the cross sections of the stems of WT and MdWRKY87-OE lines during adventitious rooting. The initiation of adventitious root primordia was both inhibited in wild-type and MdWRKY87-OE transgenic lines under NPA treatment (Figure 5B). Dense aerial roots developed on the stems of transgenic and wild-type plants under NPA treatment (Figure 5C).
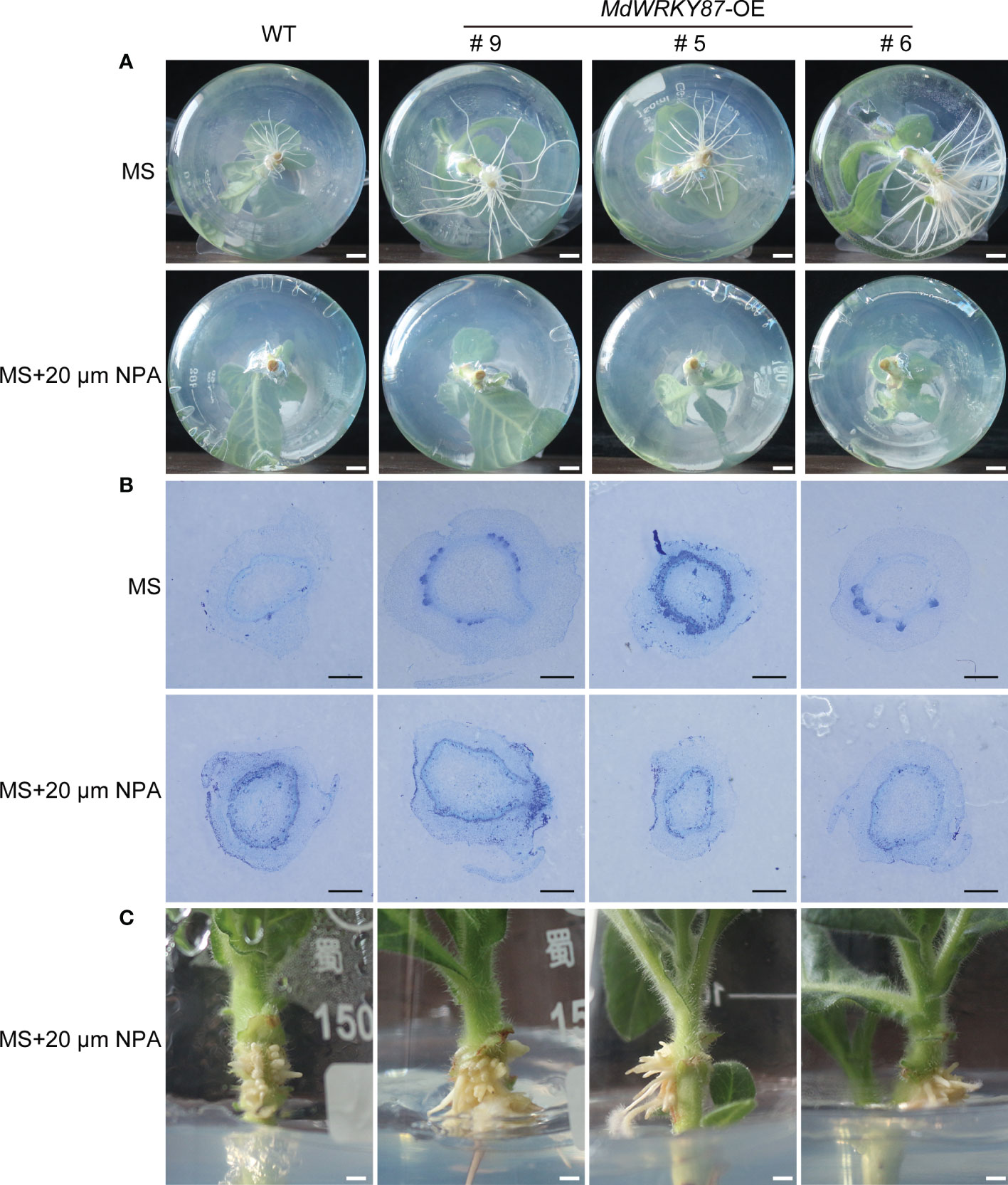
Figure 5 Effects of NPA on adventitious root formation in transgenic tobacco and wild-type plants. (A) Phenotypes of 10-day-old plants (WT, and MdWRKY87-OE lines) grown on MS medium with 20 µM NPA. (B) Cross-sections of stem bases at 5 days after transplanting during adventitious root formation in WT and MdWRKY87-OE lines. Sections were stained by toluidine blue. Scale bars = 500 µm. (C) Stem air root of wild type and MdWRKY87-OE lines at 15 days after transplant to MS medium with 20 µM NPA.
To understand how MdWRKY87 regulates adventitious root development, we conducted a comparative transcriptome analysis of the stem of WT and MdWRKY87-OE tobacco plants. A total 6690 differentially expressed genes (DEGs) in tobacco were identified, including 2000 downregulated and 4690 upregulated genes (Supplemental Figure S2). These DEGs were then subjected to Gene Ontology (GO) functional classification. Within the three GO categories identified, a greatest number of DEGs was significantly enriched in GO categories ‘biological process’ (Supplemental Figure S3). Within the ‘biological process’ GO categories, the top 4 GO terms were “regulation of cellular process”, “RNA biosynthetic process”, “nucleic acid-templated transcription” and “transcription, DNA-templated”, respectively (Figure 6A). To further dig out the key genes regulated by MdWRKY87, we analyzed these four GO categories. Venn diagram analysis showed that 272 overlapping DEGs were identified in these four GO categories (Figure 6B). Most of these overlapping genes were annotated as transcription factor (TF) genes and transcriptional regulator (TR) genes (Figure 6C). Among these overlapping DEGs, there were 242 TF genes, 17 TR genes, and 13 other genes (Figure 6C). Within these TF genes, AP-ERFBP, NAC, and HB type TF genes account for a large proportion (Figure 6C, Supplemental Figure S4). Most of these TR genes are of the types Orphans and AUX/IAA (Figure 6C, Supplemental Figure S4).
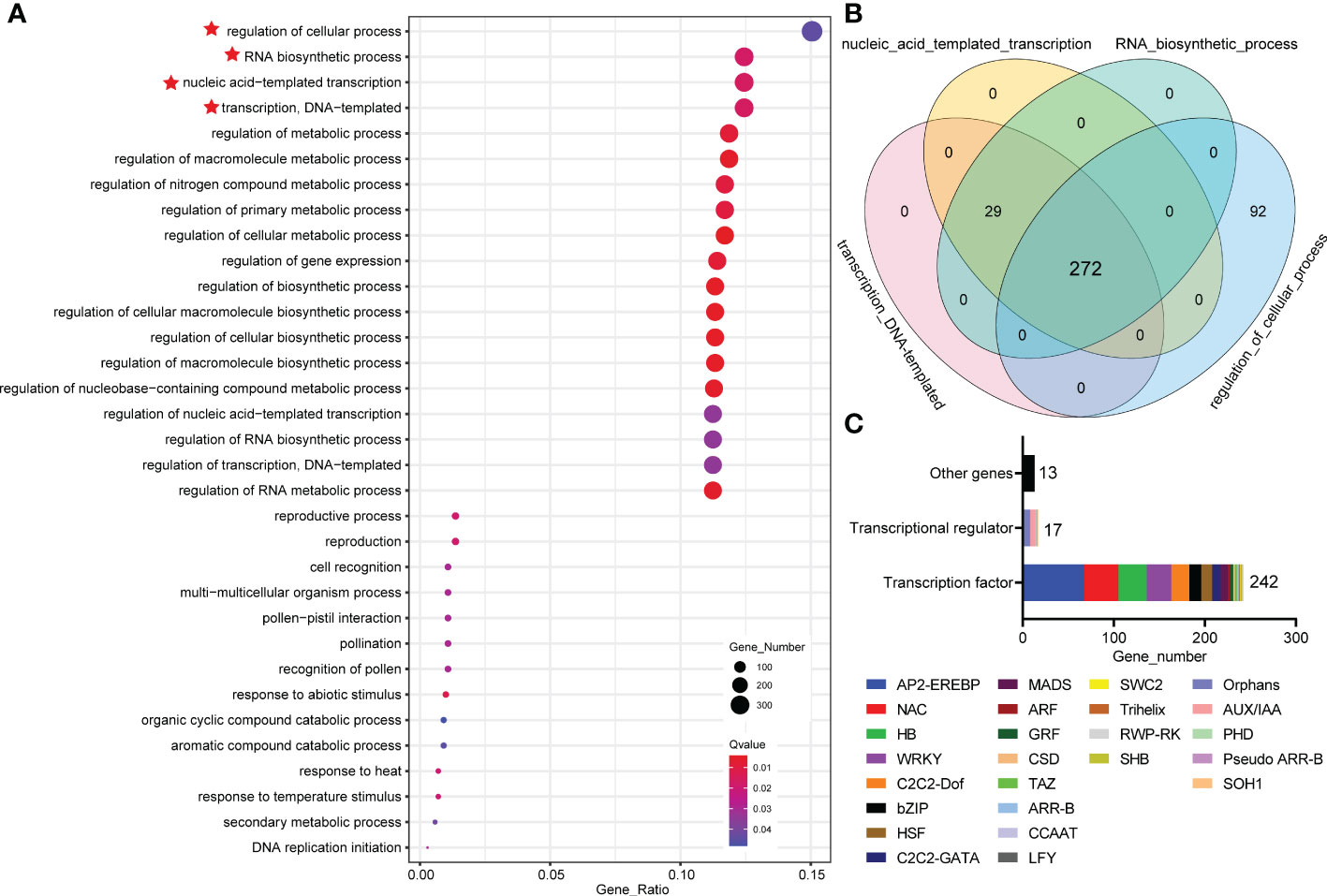
Figure 6 Comparative transcriptome analysis of the stem of WT and MdWRKY87-OE tobacco plants. (A) Functional categorization of DEGs based on the biological process of Gene Ontology (GO). The symbol red star represent the top 4 GO terms. (B) Venn diagram of DEGs in top 4 GO terms based on the biological process of GO. (C) The analysis of overlapping DEGs involved in “transcription, DNA-templated”, “nucleic acid-templated transcription”, “RNA biosynthetic process” and “regulation of cellular process”.
Further analysis indicated these TF and TR genes mainly enriched in auxin and cytokinin signaling pathway. It is well known auxin and cytokinin appear to play antagonistic roles in the adventitious rooting process. Among these TF and TR genes family, previous study has demonstrated that AUX/IAAs, AINTEGUMENTA (ANT), AINTEGUMENTA LIKE1 (AIL), WUSCHEL-RELATED HOMEOBOX (WOX), and type-B cytokinin Response Regulator (RR) family genes involved in adventitious rooting process (Ramírez-Carvajal et al., 2009; Rigal et al., 2012; Lakehal et al., 2019; Geng et al., 2023). In genes up-regulated by MdWRKY87, WUSCHEL-RELATED HOMEOBOX11 (WOX11), and AINTEGUMENTA LIKE1 (AIL) are positive regulators of adventitious root formation. For genes down-regulated by MdWRKY87, AUX/IAAs and type-B Response Regulator genes are negative regulator of adventitious root formation. Hence, negative regulation of AUX/IAAs and type-B Response Regulator genes and positive regulation of WOX11 and AIL by MdWRKY87 contributes to the enhanced adventitious rooting ability in transgenic tobacco plants. The expression level of these genes was confirmed by quantitative real-time (qRT)-PCR, thus supporting the RNA-seq results (Figure 7).
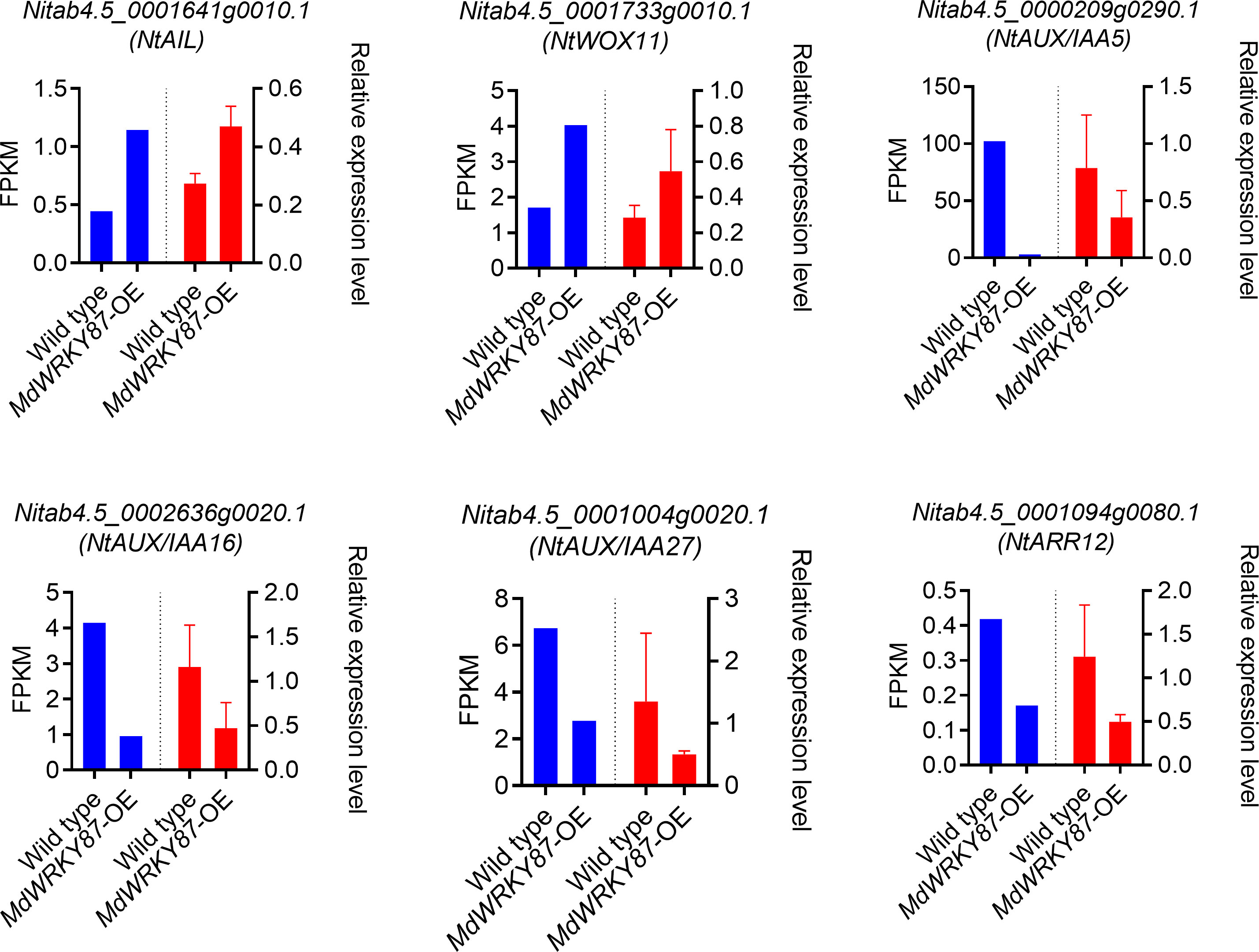
Figure 7 qRT-PCR validation of DEGs in RNA-seq data. Six genes were selected from RNA-seq differentially expressed genes (DEGs) to be validated by qRT-PCR. The data were reported as the means ± SE of three biological replicates. A comparison between the gene expression ratios obtained from RNA-seq data and qRT-PCR.
Although, it is well known that juvenile or rejuvenated phase leafy cuttings are much easier to root than the adult ones in perennial woody plants, the underlying molecular mechanism that mediates these differences is largely unknown.
For rooting recalcitrant woody plants, juvenility is necessary for efficient adventitious rooting. In general, rooting rates in adult plants are usually lower than in juvenile plants (Xu et al., 2017). Recent studies have provided a paradigm for the molecular basis of age-dependent adventitious rooting ability (Sun and Zhu, 2021). According to previous reports, we can understand the molecular basis from at least two independent signaling pathways: (i) via the accumulation of EIN3 protein in adult plants, which directly suppresses expression of WUSCHEL RELATED HOMEOBOX (WOX) genes to inhibit rooting (Ma et al., 2020); (ii) the miR156-SPLs pathway, which modulates root regeneration by crosstalk with auxin signaling pathway (Xu et al., 2016; Xu et al., 2017; Ye et al., 2020a; Ye et al., 2020b; Li et al., 2021). Whether there is other regulation mechanism controlling age-dependent adventitious rooting ability? Here we demonstrated that higher expression level of MdWRKY87 contributing to adventitious rooting-competent in juvenile apple rootstock independent of miR156/SPL pathway. As revealed by qRT-PCR, the expression level of MdWRKY87 was positively correlated with adventitious rooting ability (Figures 1D, F). Over-expressing of MdWRKY87 in tobacco leads to enhanced adventitious rooting ability, more adventitious root number and accelerated adventitious rooting process (Figures 3, 4 and Supplemental Movie S1). In addition, SPL family genes was not found in the transcriptome data of MdWRKY87-OE transgenic plants. It has been widely reported that WRKY transcription factors participate in the regulation of plant growth and development, abiotic stress responses, and disease response. However, the function of WRKY involved in adventitious rooting remains largely unknown. Here, we found WRKY87 transcription factors play essential role in adventitious root formation. In agreement to this result, PuWRKY75 was identified to control the low phosphorus driven adventitious root formation in Populus ussuriensis (Pu) (Wang et al., 2022). In addition, it was interesting that PuWRKY75 act as a transcriptional enhancer, but MdWRKY87 act as a transcriptional inhibitor (Figures 2C–E). There will be more WRKY genes, which be identified involving in adventitious rooting process in the future research.
It was well known that auxin play a dominant role in regulation of adventitious root formation (Lakehal et al., 2019). To further identify how MdWRKY87 regulate adventitious root formation, we analyzed the interaction between MdWRKY87 and auxin. MdWRKY87 has significantly enhanced expression levels in response to exogenous IBA treatment (Figures 1D, F). In addition, pro MdWRKY87: GUS transiently transformed tobacco leaves exhibited induction of GUS activity after spraying with IBA and the promoter region of MdWRKY87 has auxin response element (Figure 1G). These suggest that MdWRKY87 acts downstream of auxin to regulate adventitious root formation. However, the adventitious rooting ability was inhibited in MdWRKY87-OE transgenic tobacco lines upon treatment with the polar auxin transport inhibitor NPA (Figure 5). Taken together, we conclude MdWRKY87 act not only downstream of auxin, but also feedback regulation during adventitious rooting process. Consistent with this, previous data also demonstrate that WRKY71/EXB1 play pivotal roles in shoot branching by regulating auxin pathways (Guo et al., 2015). However, specific mechanism underlying the crosstalk between MdWRKY87 and auxin signaling needs to be further demonstrated.
MdWRKY87 was known to control adventitious root formation (Figure 3), but the regulatory role of MdWRKY87 was unknown. Comparative transcriptome profiling between the wild type and MdWRKY87-OE transgenic plants was conducted in this study (Figure 6). A set of 272 overlapping DEGs were identified through GO enrichment analysis, implying their potential importance for MdWRKY87-dependent adventitious rooting formation (Figure 6B). Among these DEGs, we found MdWRKY87 overexpression can activate the expression of adventitious rooting-induced genes, such as WOX11 and AIL, and the counterparts of WOX11 and AIL promote adventitious root formation in A.thaliana and poplar (Rigal et al., 2012; Hu and Xu, 2016; Geng et al., 2023). In addition, MdWRKY87 overexpression can inhibit the transcription of adventitious rooting-repressed genes, such as AUX/IAAs and RRs, and the counterparts of AUX/IAAs and type-B RRs inhibit adventitious root formation in Arabidopsis, apple, and poplar (Ramírez-Carvajal et al., 2009; Lakehal et al., 2019; Zhao et al., 2020). Similarly, it was demonstrated that PuWRKY75 interacted with PuMYB40 and directly co-regulate PuLRP1 and PuERF003 to promote adventitious root formation in P. ussuriensis (Wang et al., 2022).
In conclusion, a relatively high expression level of MdWKRY87 contribute to improving adventitious rooting ability. Based on this potential mechanisms, artificial methods for adventitious rooting ability improving will be created via manipulating MdWRKY87 gene expression, especially for rooting recalcitrant woody perennial species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XX, and ZL conceived and designed the experiment. QT and MX conducted the experiment and data analysis. DW, CW, XW and QC contributed to the data analysis. QT and MX wrote the manuscript. DW, CW, XW and QC drafted the discussion and revised the manuscript. All authors contributed to the article and approved the final version.
This research was funded by National Natural Science Foundation of China (grant number 32172523, and 31801824); the Breeding Plan of Shandong Provincial Qingchuang Research Team, China (2019); The Fruit innovation team of Shandong modern agricultural industry technology system (SDAIT-06-05); Shandong Provincial Cooperative Promotion Plan of Major Agricultural Technologies (SDNYXTTG-2022-08).
Author XW was employed by Weihai Yingjuval Nursery Limited Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1136616/full#supplementary-material
Supplemental Movie S1 | Time-lapse video showing the adventitious rooting of MdWRKY87-OE transgenic tobacco plants.
Bellini, C., Pacurar, D. I., Perrone, I. (2014). Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666. doi: 10.1146/annurev-arplant-050213-035645
Bray, N. L., Pimentel, H., Melsted, P., Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34 (5), 525–527. doi: 10.1038/nbt.3519
Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., et al. (2003). Dissecting arabidopsis lateral root development. Trends Plant Sci. 8 (4), 165–171. doi: 10.1016/S1360-1385(03)00051-7
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, C., Che, Q., Su, S., Liu, Y., Wang, Y., Xu, X. (2020). Genome-wide identification and characterization of Respiratory burst oxidase homolog genes in six Rosaceae species and an analysis of their effects on adventitious rooting in apple. PloS One 15 (9), e0239705. doi: 10.1371/journal.pone.0239705
Cheng, C., Yu, Q., Wang, Y., Wang, H., Dong, Y., Ji, Y., et al. (2021). Ethylene-regulated asymmetric growth of the petal base promotes flower opening in rose (Rosa hybrida). Plant Cell 33 (4), 1229–1251. doi: 10.1093/plcell/koab031
Che, Q., Zhang, Z., Song, W., Li, Q., Xu, X., Wang, Y. (2021). Comparison of WRKY gene family bioinformatics in 'GDDH13' and M. baccata and expression analysis of MdWRKY gene family. Mol. Plant Breed. (China) 19 (5), 1484–1502. doi: 10.13271/j.mpb.019.001484
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18 (1), 448. doi: 10.1186/s12864-017-3791-6
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 (5), 199–206. doi: 10.1016/S1360-1385(00)01600-9
Geng, L., Li, Q., Jiao, L., Xiang, Y., Deng, Q., Zhou, D. X., et al. (2023). WOX11 and CRL1 act synergistically to promote crown root development by maintaining cytokinin homeostasis in rice. New Phytol. 237 (1), 204–216. doi: 10.1111/nph.18522
Guo, D., Zhang, J., Wang, X., Han, X., Wei, B., Wang, J., et al. (2015). The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in arabidopsis. Plant Cell 27 (11), 3112–3127. doi: 10.1105/tpc.15.00829
Han, Y. C., Kuang, J. F., Chen, J. Y., Liu, X. C., Xiao, Y. Y., Fu, C. C., et al. (2016). Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol. 171 (2), 1070–1084. doi: 10.1104/pp.16.00301
Hu, Z., Wang, R., Zheng, M., Liu, X., Meng, F., Wu, H., et al. (2018). TaWRKY51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum l.). Plant J. 96 (2), 372–388. doi: 10.1111/tpj.14038
Hu, X., Xu, L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172 (4), 2363–2373. doi: 10.1104/pp.16.01067
Ji, X. L., Li, H. L., Qiao, Z. W., Zhang, J. C., Sun, W. J., You, C. X., et al. (2022). The BTB protein MdBT2 recruits auxin signaling components to regulate adventitious root formation in apple. Plant Physiol. 189 (2), 1005–1020. doi: 10.1093/plphys/kiac084
Jin, Y., Li, J., Zhu, Q., Du, X., Liu, F., Li, Y., et al. (2022). GhAPC8 regulates leaf blade angle by modulating multiple hormones in cotton (Gossypium hirsutum l.). Int. J. Biol. Macromol 195, 217–228. doi: 10.1016/j.ijbiomac.2021.11.205
Johnson, C. S., Kolevski, B., Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of arabidopsis, encodes a WRKY transcription factor. Plant Cell 14 (6), 1359–1375. doi: 10.1105/tpc.001404
Kang, G., Yan, D., Chen, X., Yang, L., Zeng, R. (2021). HbWRKY82, a novel IIc WRKY transcription factor from Hevea brasiliensis associated with abiotic stress tolerance and leaf senescence in arabidopsis. Physiol. Plant 171 (1), 151–160. doi: 10.1111/ppl.13238
Lagace, M., Matton, D. P. (2004). Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219 (1), 185–189. doi: 10.1007/s00425-004-1253-2
Lakehal, A., Chaabouni, S., Cavel, E., Le Hir, R., Ranjan, A., Raneshan, Z., et al. (2019). A molecular framework for the control of adventitious rooting by TIR1/AFB2-Aux/IAA-dependent auxin signaling in arabidopsis. Mol. Plant 12 (11), 1499–1514. doi: 10.1016/j.molp.2019.09.001
Levy, A., Szwerdszarf, D., Abu-Abied, M., Mordehaev, I., Yaniv, Y., Riov, J., et al. (2014). Profiling microRNAs in Eucalyptus grandis reveals no mutual relationship between alterations in miR156 and miR172 expression and adventitious root induction during development. BMC Genomics 15, 524. doi: 10.1186/1471-2164-15-524
Liang, W., Sun, F., Zhao, Y., Shan, L., Lou, H. (2020). Identification of susceptibility modules and genes for cardiovascular disease in diabetic patients using WGCNA analysis. J. Diabetes Res. 2020, 4178639. doi: 10.1155/2020/4178639
Li, X., Shen, F., Xu, X., Zheng, Q., Wang, Y., Wu, T., et al. (2021). An HD-ZIP transcription factor, MxHB13, integrates auxin-regulated and juvenility-determined control of adventitious rooting in Malus xiaojinensis. Plant J. 107 (6), 1663–1680. doi: 10.1111/tpj.15406
Li, P., Song, A., Gao, C., Jiang, J., Chen, S., Fang, W., et al. (2015). The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol. Biochem. 95, 26–34. doi: 10.1016/j.plaphy.2015.07.002
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li, Q. Q., Zhang, Z., Zhang, C. X., Wang, Y. L., Liu, C. B., Wu, J. C., et al. (2022). Phytochrome-interacting factors orchestrate hypocotyl adventitious root initiation in arabidopsis. Development 149 (10). doi: 10.1242/dev.200362
Ma, Z., Li, W., Wang, H., Yu, D. (2020). WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age-mediated flowering. J. Integr. Plant Biol. 62 (11), 1659–1673. doi: 10.1111/jipb.12946
Ramírez-Carvajal, G. A., Morse, A. M., Dervinis, C., Davis, J. M. (2009). The cytokinin type-b response regulator PtRR13 is a negative regulator of adventitious root development in populus. Plant Physiol. 150 (2), 759–771. doi: 10.1104/pp.109.137505
Rigal, A., Yordanov, Y. S., Perrone, I., Karlberg, A., Tisserant, E., Bellini, C., et al. (2012). The AINTEGUMENTA LIKE1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in poplar. Plant Physiol. 160 (4), 1996–2006. doi: 10.1104/pp.112.204453
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 (5), 247–258. doi: 10.1016/j.tplants.2010.02.006
Singh, A. K., Kumar, S. R., Dwivedi, V., Rai, A., Pal, S., Shasany, A. K., et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215 (3), 1115–1131. doi: 10.1111/nph.14663
Sun, L., Zhu, Z. (2021). The molecular basis of age-modulated plant de novo root regeneration decline in Arabidopsis thaliana. Plant Cell Physiol. 62 (1), 3–7. doi: 10.1093/pcp/pcaa134
Wang, P., Ma, L., Wang, S., Li, L., Wang, Q., Yang, R., et al. (2019). Identification and analysis of a candidate WRKY transcription factor gene affecting adventitious root formation using association mapping in Catalpa scop. DNA Cell Biol. 38 (4), 297–306. doi: 10.1089/dna.2018.4528
Wang, H., Pak, S., Yang, J., Wu, Y., Li, W., Feng, H., et al. (2022). Two high hierarchical regulators, PuMYB40 and PuWRKY75, control the low phosphorus driven adventitious root formation in Populus ussuriensis. Plant Biotechnol. J. 20 (8), 1561–1577. doi: 10.1111/pbi.13833
Xu, M., Hu, T., Zhao, J., Park, M. Y., Earley, K. W., Wu, G., et al. (2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PloS Genet. 12 (8), e1006263. doi: 10.1371/journal.pgen.1006263
Xu, X., Li, X., Hu, X., Wu, T., Wang, Y., Xu, X., et al. (2017). High miR156 expression is required for auxin-induced adventitious root formation via MxSPL26 independent of PINs and ARFs in Malus xiaojinensis. Front. Plant Sci. 8, 1059. doi: 10.3389/fpls.2017.01059
Ye, B. B., Shang, G. D., Pan, Y., Xu, Z. G., Zhou, C. M., Mao, Y. B., et al. (2020a). AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 32 (1), 226–241. doi: 10.1105/tpc.19.00378
Ye, B. B., Zhang, K., Wang, J. W. (2020b). The role of miR156 in rejuvenation in Arabidopsis thaliana. J. Integr. Plant Biol. 62 (5), 550–555. doi: 10.1111/jipb.12855
Zhang, Z. L., Xie, Z., Zou, X., Casaretto, J., Ho, T. H., Shen, Q. J. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134 (4), 1500–1513. doi: 10.1104/pp.103.034967
Zhao, D., Wang, Y., Feng, C., Wei, Y., Peng, X., Guo, X., et al. (2020). Overexpression of MsGH3.5 inhibits shoot and root development through the auxin and cytokinin pathways in apple plants. Plant J. 103 (1), 166–183. doi: 10.1111/tpj.14717
Zhu, H., Jiang, Y., Guo, Y., Huang, J., Zhou, M., Tang, Y., et al. (2021). A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 160, 175–183. doi: 10.1016/j.plaphy.2021.01.014
Keywords: auxin, adventitious root formation, transcriptome, apple, rootstock
Citation: Tian Q, Xu M, Wu D, Wang C, Wang X, Che Q, Li Z and Xu X (2023) Integrated transgene and transcriptome reveal the molecular basis of MdWRKY87 positively regulate adventitious rooting in apple rootstock. Front. Plant Sci. 14:1136616. doi: 10.3389/fpls.2023.1136616
Received: 03 January 2023; Accepted: 17 January 2023;
Published: 26 January 2023.
Edited by:
Lin Xi, University of Hohenheim, GermanyReviewed by:
Bowen Liang, Hebei Agricultural University, ChinaCopyright © 2023 Tian, Xu, Wu, Wang, Wang, Che, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaozhao Xu, MjAxODAxMDA2QHFhdS5lZHUuY24=; Zhengnan Li, bGl6aGVuZ25hbkBpbWF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.