- 1Insect Vector Laboratory, Advanced Centre for Plant Virology, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 2Division of Entomology, ICAR-Indian Agricultural Research Institute, New Delhi, India
Bemisia tabaci (Hemiptera: Aleyrodidae) is one of the most important invasive pests worldwide. It infests several vegetables, legumes, fiber, and ornamental crops. Besides causing direct damage by sucking plant sap, B. tabaci is the principal vector of begomoviruses. Chilli leaf curl virus (ChiLCV, Begomovirus) transmitted by B. tabaci is a major constraint in chilli production. B. tabaci genes associated with metabolism, signaling pathways, cellular processes, and organismal systems are highly enriched in response to ChiLCV infection. The previous transcriptome study suggested the association of B. tabaci Toll-like receptor 3 (TLR3) and transducer of erbB2.1 (TOB1) in ChiLCV infection. In the present study, B. tabaci TLR3 and TOB1 were silenced using double-stranded RNA (dsRNA) and the effect on fitness and begomovirus transmission has been reported. Oral delivery of dsRNA at 3 µg/mL reduced the expression of B. tabaci TLR3 and TOB1 by 6.77 and 3.01-fold, respectively. Silencing of TLR3 and TOB1 induced significant mortality in B. tabaci adults compared to untreated control. The ChiLCV copies in B. tabaci significantly reduced post-exposure to TLR3 and TOB1 dsRNAs. The ability of B. tabaci to transmit ChiLCV also declined post-silencing TLR3 and TOB1. This is the first-ever report of silencing B. tabaci TLR3 and TOB1 to induce mortality and impair virus transmission ability in B. tabaci. B. tabaci TLR3 and TOB1 would be novel genetic targets to manage B. tabaci and restrict the spread of begomovirus.
Introduction
Silverleaf whitefly [Bemisia tabaci (Gennadius), Hemiptera: Aleyrodidae] is a key agricultural pest of horticultural and field crops worldwide. Except for Antarctica, B. tabaci has been reported from all the continents. It infests over 900 plant species (Abd-Rabou and Simmons, 2010; Li et al., 2011). Adults and nymphs of B. tabaci cause economic damage to a wide range of vegetables, legumes, fibers, and ornamentals. B. tabaci causes direct damage to plants by sucking sap. Besides, it affects the quality and quantity of the produce by secreting honeydew and transmitting plant viruses (Jones, 2003; Hogenhout et al., 2008; Horowitz et al., 2020). B. tabaci transmits more than 400 plant viruses of the genera Begomovirus (Geminiviridae), Torradovirus (Secoviridae), Carlavirus (Betaflexiviridae), Crinivirus (Colesteroviridae), Ipomovirus (Potyviridae), Polerovirus (Solemoviridae), and Cytorhabdovirus (Rhabdoviridae) (Wang et al., 2020; Ghosh and Ghanim, 2021). B. tabaci is the principal vector of begomoviruses that cause annual losses of around 300 million US$ every year (Varma and Malathi, 2003; Navas-Castillo et al., 2011). The yield losses in vegetable crops caused by begomoviruses range from 20-100% (Polston and Anderson, 1997). B. tabaci transmits begomovirus in a persistent-circulative manner which means the virus particles need to cross the midgut barrier and reach the salivary glands through hemolymph in B. tabaci (Rosen et al., 2015; Czosnek et al., 2017). Several proteins such as heat shock protein 70 (hsp70), cyclophilin B (CypB), and peptidoglycan recognition protein (PGRP) were reported to interact with the coat protein of begomovirus in the midgut of B. tabaci (Götz et al., 2012; Kanakala and Ghanim, 2016; Wang et al., 2016). The current understanding of B. tabaci-begomovirus interactions is largely based on the tomato yellow leaf curl virus (TYLCV). Limited evidence is available on the functions of B. tabaci genes in the transmission of other predominant begomoviruses.
Chilli leaf curl virus (ChiLCV) poses a significant problem in chilli production especially in tropical and sub-tropical countries (Senanayake et al., 2007; Shingote et al., 2022). ChiLCV is a monopartite begomovirus with a circular, single-stranded DNA-A component of 2.7 kb and associated with alphasatellites (~1.4 kb) and/or betasatellites (~1.3 kb). The symptoms of ChiLCV include upward leaf curling, crinkling, puckering, and stunting of the entire plant. Under extreme circumstances, fruit yields have been recorded to drop by up to 100 percent (Thakur et al., 2018). The application of pesticides to curb B. tabaci is the primary control measure in practice (Horowitz et al., 2020). However, chemical insecticides are largely ineffective against B. tabaci due to the quick development of insecticide resistance. In India, an epidemic of B. tabaci-transmitted viral diseases during 2015-16 was mainly due to the failure of chemical pesticides (Verma and Bhattacharya, 2015; Kumar et al., 2020). Chemical insecticides also have the issue of health and environmental hazards.
Implementation of RNA interference (RNAi) would be a novel alternative for the sustainable management of B. tabaci and begomoviruses. RNAi is a eukaryotic gene silencing mechanism that has been employed to impair the development, fecundity, and survival of insect pests by knocking down key genes involved in the processes. However, inadequate information on the gene function of the target pest has restricted the scope of developing an RNAi-based pest management programme. Silencing of B. tabaci hsp70 and fasciclin 2 (fas2) modulates the ChiLCV transmission (Chakraborty and Ghosh, 2022). In our previous study, the differentially expressed genes (DEGs) of B. tabaci in response to ChiLCV have been reported (Nekkanti et al., 2022). DEGs associated with innate immunity such as Toll-like receptor 3 (TLR3), fas2, transducer of erbB2.1 (TOB1), and GMP reductase were highly abundant. Toll receptors induce interferons to confer antiviral resistance in vertebrates (Lester and Li, 2014). TOB1 attenuates IRF3-directed antiviral responses by recruiting HDAC8 in virus-infected macrophages (Yu et al., 2022). However, the role of TLR3 and TOB1 in the virus transmission by B. tabaci or any other arthropods is not known. In the present study, B. tabaci TLR3 and TOB1 were chosen to be silenced through RNAi and the resultant effect on fitness and ChiLCV transmission ability of B. tabaci has been reported. To the best of our knowledge, this is the first evidence of functional validation of B. tabaci TLR3 and TOB1 for ChiLCV transmission.
Materials and methods
B. tabaci Asia II 1 population
An isofemale population of B. tabaci Asia II 1 being maintained at the whitefly rearing facility, Advanced Centre for Plant Virology, Indian Agricultural Research Institute (IARI), New Delhi since 2015 was used in the present study. The iso-female line was reared on eggplants, Solanum melongena (var. Navkiran, Mahyco, India) at 28 ± 2°C temperature, 60 ± 10% RH, and 16 hr light - 8 hr dark photoperiod. The identity of the pure culture was confirmed by sequencing of mitochondrial cytochrome oxidase subunit I (mtCOI).
ChiLCV culture
The initial inoculum was taken from a pure culture of ChiLCV maintained at the laboratory by B. tabaci-inoculation. The culture was maintained on chilli plants (var. Priti, Nunhems) in insect-proof conditions. The identity of the virus was further confirmed by sequencing the DNA-A component amplified in PCR using primer pair, Begomo F-Begomo R (Akhter et al., 2009).
Designing and synthesis of dsRNA
In our previous study, the expression of B. tabaci TLR3 and TOB1 was found highly abundant in response to ChiLCV infection (Nekkanti et al., 2022). In the present study, dsRNAs were designed to knock down B. tabaci TLR3 and TOB1. The conserved regions were identified by aligning the sequences of B. tabaci TLR3 and TOB1 available in NCBI. Putative siRNAs in the conserved regions were identified using the siRNA Wizard online tool (https://www.invivogen.com/sirna-wizard, accessed on 12-12-2021). The regions with the maximum number of siRNAs were selected for designing dsRNA. The dsRNA stretch was further investigated for off-target effects with other organisms like humans, mice, birds, ants, and bees. A dsRNA targeting Thrips palmi collagen alpha-1(III) chain-like (TpCOL3A1) and not specific to B. tabaci was used as negative control.
The primer pairs were designed using the NCBI primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast) to amplify the dsRNA stretches. The primers were validated and PCR conditions were optimized in a gradient PCR. The primer pairs used in the study are listed in Table 1. A 25 µL PCR mixture contained 1X PCR buffer (Thermo Fisher Scientific, USA), 0.4 µM of each forward and reverse primer (GCC Biotech, India), 0.26 mM dNTP mix (Thermo Fisher Scientific), 50 ng DNA template of B. tabaci, and 2 U of DreamTaq DNA polymerase (Thermo Fisher Scientific). PCR was performed in a T100 thermocycler (Bio-Rad, USA) with initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 95°C for 40 s, annealing at 55°C for 40 s, extension at 72°C for 40 s, and a final extension at 72°C for 10 min. The amplified PCR products were resolved on 2% agarose gel along with 1 kb plus DNA ladder (Thermo Fisher Scientific) and visualized under a Gel documentation system (MaestroGen Inc, Taiwan).
The amplified PCR products were eluted, ligated in the L4440 expression vector between two T7 promoters, and sequenced for further confirmation. The recombinant plasmids were transformed into RNase III mutant E. coli HT115 cells. The recombinant E. coli HT115 cells were induced with 0.8 M isopropyl-β-D-1-thiogalactopyranoside (IPTG) and cultured overnight at 37°C in a shaking incubator. Total RNA from the induced HT115 cells was extracted using Trizol reagent (Invitrogen, CA, USA) and resuspended in nuclease-free water. The dsRNA was purified by incubating with 1 U of RNase A, DNase and protease-free (Thermo Fisher Scientific) and 1 U of DNase I, RNase-free (Thermo Fisher Scientific) for 1 hr at 37°C in the presence of 500 mM sodium chloride as described by Chakraborty and Ghosh (2022). The enzymes were inactivated by chloroform extraction. The purified dsRNA was quantified in a spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific), and visualized on 2% native agarose gel stained with GoodView (BR Biochem, India).
Oral delivery of dsRNAs to B. tabaci
The purified TLR3 and TOB1 dsRNAs were separately delivered to B. tabaci via the oral feeding method described by Chakraborty and Ghosh (2022). Briefly, around 30 flies were collected in each cylindrical pet bottle (3.5 cm diameter, 16 cm height). The open end of the bottle was sealed with a stretched UV-sterilized Parafilm M. Based on our previous study, purified dsRNA at 3.0 μg/mL was supplemented with the artificial diet comprised of 20% sucrose and 5% yeast extract. The diet with dsRNA was sandwiched between two layers of stretched Parafilm M membrane. A diet without dsRNA and diet with TpCOL3A1 dsRNA were served as control. For ventilation, a hole was made in the wall of the pet bottle and sealed with a muslin cloth. The pet bottles were kept in the upright position in dark at 26 ± 2°C and 60% RH. Three replicates were maintained for each treatment and repeated nine times. Percent mortality data was recorded 48 hr post dsRNA exposure. Tukey’s test was used to differentiate the mean differences across the categories with a 95% confidence interval using XLSTAT 2014.5.03. The surviving B. tabaci from these replicates were utilized to examine the relative expression of TLR3 and TOB1 mRNA and ChiLCV acquisition and transmission efficiency as described below.
Estimating expression of B. tabaci TLR3 and TOB1
The relative expression of B. tabaci TLR3 and TOB1 was estimated 48 hr post dsRNA feeding considering the β-actin gene as endogenous control. The primer pairs, AG301F-AG302R and AG568F-AG569R for TOB1 and TLR3, respectively were used in the RT-qPCR assay (Table 1). Around 30 surviving B. tabaci in three replicates post TLR3 and TOB1 dsRNA exposure were used for total RNA isolation using Trizol reagent. The RNA was quantified in a spectrophotometer (NanoDrop 2000) and used for complementary DNA (cDNA) synthesis using the FIREScript RT cDNA synthesis kit (Solis BioDyne, Estonia). The 20 μL reaction mixture comprised of 1.0 μg RNA template, 5 μM oligo dT primers, 500 μM dNTP mix, 2 μL of 1 X reaction buffer, 10 U FIREScript RT, and 1 U RiboGrip RNase inhibitor. cDNA synthesis was carried out in a T100 thermocycler with reverse transcription at 42°C for 60 min, and enzyme inactivation at 85°C for 5 min. The relative RT-qPCR assay was performed in an Insta Q48M real-time PCR (Himedia, India). A 20 µL reaction mixture contained 1X GoTaq qPCR master mix (Promega, USA), 300 nM CXR passive reference dye, 0.25 µM of each forward and reverse primer, and 2 µL of template cDNA. Thermal cycling was performed at initial denaturation at 95°C for 5 min, 35 cycles of 95°C for 40 s, 55°C for 40 s, and 72°C for 40 s. After each reaction, a dissociation or melting curve was performed to evaluate the specificity of the amplicons. Three biological and two technical replicates were used in the RT-qPCR. The relative expression of mRNA in dsRNA-fed B. tabaci was measured in comparison to untreated control following the method (Livak and Schmittgen, 2001). Microsoft Excel version 2016 was used to perform statistical analysis and prepare graphs. Expression of B. tabaci TLR3 and TOB1 in TpCOL3A1 dsRNA-fed flies was considered as the negative control.
Quantification of virus titer in B. tabaci
A portion of surviving flies post-dsRNA exposure was used to quantify the virus titer in B. tabaci. The flies were allowed to acquire ChiLCV by feeding on a ChiLCV-infected chilli plant (var. Preeti) for 24 hr. The ChiLCV copies acquired by dsRNA-exposed and nonexposed B. tabaci were estimated by absolute quantification in qPCR. DNA was isolated from the batch of 30 adult flies in three replicates using a CTAB extraction buffer as described by Roy et al. (2021) and quantified in a spectrophotometer. qPCR was performed in Insta Q48M real-time PCR with ChiLCV-specific primer pair, AG149F-AG150R (Table 1) (Roy et al., 2021). This was followed by a melting curve analysis to check the specificity of the reaction. Each treatment had three biological and two technical replicates. A standard curve of ChiLCV using primer pair AG149F-AG150R generated in our previous study (Chakraborty and Ghosh, 2022) was used to quantify the ChiLCV copies. The mean CT values obtained in qPCR were fitted into the standard curve and the resulting concentration was used for the calculation of virus copy number in Microsoft Excel 2016 using the following formula.
where N = number of viral copies, x = amount of amplicon in ng, and n = length of linearized plasmid DNA. The mean differences in virus copies were assessed for statistical significance by Tukey’s test at a confidence interval of 95% using XLSTAT 2014.5.03.
Transmission of ChiLCV by TLR3 and TOB1 dsRNA-treated B. tabaci
To check the transmission efficacy of dsRNA-treated B. tabaci, a portion of B. tabaci exposed to ChiLCV for 24 hr was released onto the healthy chilli plants (var. Preeti) at the 3-4 leaf stage. They were allowed for 24 hr of inoculation feeding and eliminated manually. Ten plants in three replicates were used and four adult females per plant were released. The plants were maintained under insect-proof conditions and monitored for symptom development. B. tabaci, not exposed to dsRNA, were used as control. The ChiLCV infection in the inoculated plants was confirmed by ChiLCV-specific PCR at 21 days post-inoculation (dpi).
Results
B. tabaci population and ChiLCV culture
The identity of the B. tabaci population was confirmed by the nucleotide sequence of mtCOI gene. PCR with primer pair C1-J-2195 and L2-N-3014 amplified ~600 bp product as visualized on 1% agarose gel. The sequence analysis with BLASTn showed 100% homology to B. tabaci Asia II 1. The sequence submitted to GenBank can be retrieved by Accession No. OP223446.
PCR amplified a 2.7 kb product of full-length DNA-A segment from ChiLCV-infected plants. The sequence of DNA-A showed 100% homology to ChiLCV isolates upon BLASTn analysis. The sequence can be retrieved by Accession No. OM513903.
Synthesis of dsRNA targeting B. tabaci TLR3 and TOB1
Based on the multiple alignments of B. tabaci TLR3 and TOB1 sequences available in NCBI, the conserved 340 nt and 167 nt stretches of TLR3 (~5.6 kb) and TOB1 (~2.28 kb), respectively were chosen for dsRNA designing. The dsRNA sequences were unique to B. tabaci and no off-target hits were detected with Homo sapiens (taxid: 9605), Formicidae (taxid: 36668), mice (taxid: 10088), honeybees (taxid: 7460), and Aves (taxid: 8782) in blastn analysis.
PCR with primer pairs AG568F-AG569R and AG301F-AG302R produced amplicons of 340 bp and 167 bp for B. tabaci TLR3 and TOB1, respectively (Supplementary Figure 1). The nucleotide sequences of the amplified products showed 100% homology with already available B. tabaci TLR3 and TOB1 sequences. The sequences can be retrieved by Accession No. OP784422 and OP219521. The dsRNA purified from total RNA using DNase I and RNase A produced single specific bands of ~340 bp and ~167 bp, respectively on 2% agarose gel (Figure 1). The concentration of TLR3 dsRNA was 970.0 ng/µL, whereas it was 779.9 ng/µL in the case of TOB1 dsRNA.

Figure 1 Delivery of dsRNA targeting B. tabaci TOB1 and TLR3. Artificial feeding setup for B. tabaci adults. The artificial diet was supplemented with 3.0 μg/mL of dsRNA and sterile water (control). Red arrows show the B. tabaci adults feeding on the artificial diet.
Effect of TLR3 and TOB1 dsRNAs on target mRNA expression
The feeding of TLR3 and TOB1 dsRNAs significantly reduced the target gene expression in B. tabaci adults. In RT-qPCR analysis, the log value of B. tabaci TLR3 expression was 11.55 under normal conditions. Exposure to TLR3 dsRNA significantly declined the TLR3 mRNA level by 6.77-fold compared to the untreated control at 48 hr. The downregulation of target gene expression was consistent in all the biological replicates. Similarly, TOB1 dsRNA significantly down-regulated the TOB1 mRNA level by 3.01-fold (Figure 2). The log2−ΔΔCT value of B. tabaci TOB1 expression was 3.19 under normal conditions. The reduction in the target mRNA expression level of B. tabaci was significantly higher in TLR3 dsRNA exposure than TOB1 dsRNA at 48 hr after oral delivery. The expression of B. tabaci TLR3 and TOB1 post TpCOL3A1 dsRNA exposure was statistically at par with untreated control. There was no significant regulation of the endogenous control gene, β-actin between dsRNA-exposed and non-exposed B. tabaci populations which indicated the specificity of the TLR3 and TOB1 dsRNAs on the target mRNAs. The melting curve analysis in RT-qPCR showed that the primer pairs for TLR3, TOB1 and β-actin did not produce any secondary peaks that indicated the specificity of the reactions.
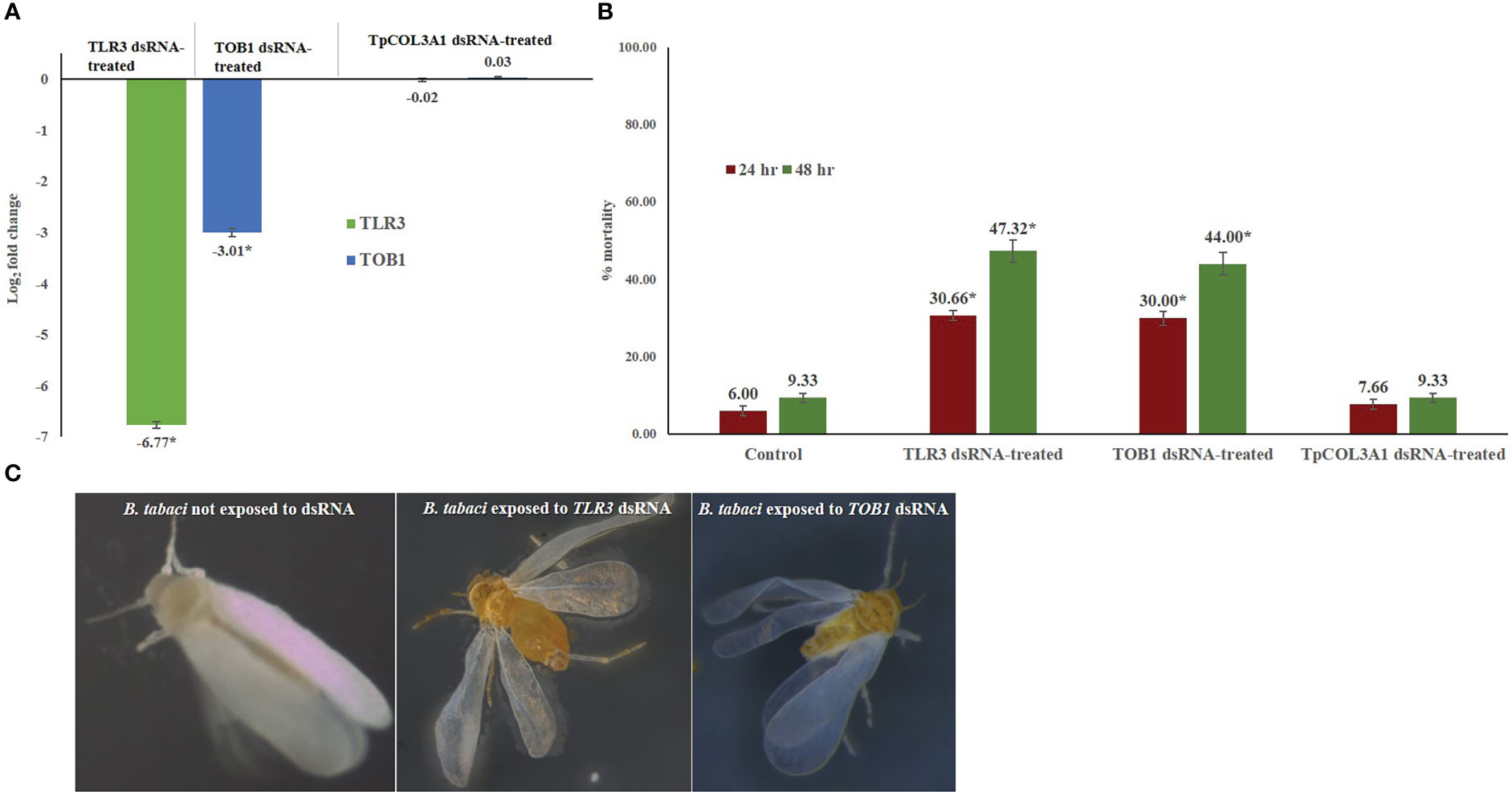
Figure 2 Effect of silencing TOB1 and TLR3 on survival of B. tabaci. (A) Normalized relative expression of B. tabaci TOB1 and TLR3 mRNA post 48 hr of dsRNA feeding. (B) Percent mortality of adult B. tabaci post TLR3 and TOB1 dsRNA feeding at 3.0 µg/mL. The mean denoted by an asterisk (*) indicates a significant difference (p< 0.0001). The error bars are the standard error of the mean (SEM). (C) B. tabaci adults fed on the diet mixed with TLR3 and TOB1 dsRNA and without dsRNA. No morphological deformities were observed in TOB1 and TLR3 dsRNA-fed B. tabaci.
Effect of silencing B. tabaci TLR3 and TOB1 on B. tabaci fitness
Feeding on TLR3 and TOB1 dsRNAs significantly altered the fitness of B. tabaci under controlled conditions (Figure 2). TLR3 and TOB1 dsRNA feeding at a concentration of 3.0 μg/mL induced mortality in B. tabaci adults. A mortality of 30.66% was recorded in TLR3 dsRNA-fed B. tabaci 24 hr post-feeding, whereas it was 29.99% when fed on TOB1 dsRNA. The mortality further increased with an increase in the exposure period. Up to 47.32% mortality was recorded 48 hr post-feeding on TLR3 dsRNA. In the case of TOB1 dsRNA, the mortality increased up to 43.99% compared to B. tabaci fed on a diet without dsRNA (9.33%). However, no morphological deformities were recorded in the TLR3 or TOB1 dsRNA-fed B. tabaci when observed under a microscope (Figure 2). Mortality induced by TLR3 and TOB1 dsRNAs was found to be significant at p<0.0001 at a confidence limit of 95%. There was no significant mortality of B. tabaci post TpCOL3A1 dsRNA exposure compared to untreated control.
Effect of TLR3 and TOB1 silencing on ChiLCV acquisition and transmission by B. tabaci
Silencing of B. tabaci TLR3 and TOB1 significantly decreased the ChiLCV titer within B. tabaci. The mean ChiLCV copy number was 2.84 x 107 in B. tabaci fed on the diet without dsRNA (Figure 3). Exposure to TLR3 dsRNA at 3 μg/mL induced 45.58 -fold reduction (6.23 x 105 copies) in the mean ChiLCV copy. The decrease in ChiLCV copy in TOB1 dsRNA-treated B. tabaci was comparatively lower than in TLR3 dsRNA treatment. The ChiLCV titer in B. tabaci was reduced by 10.75-fold (2.64 x 106 copies) post TOB1 dsRNA exposure. There was no significant change in ChiLCV titer post TpCOL3A1 dsRNA exposure compared to untreated control.
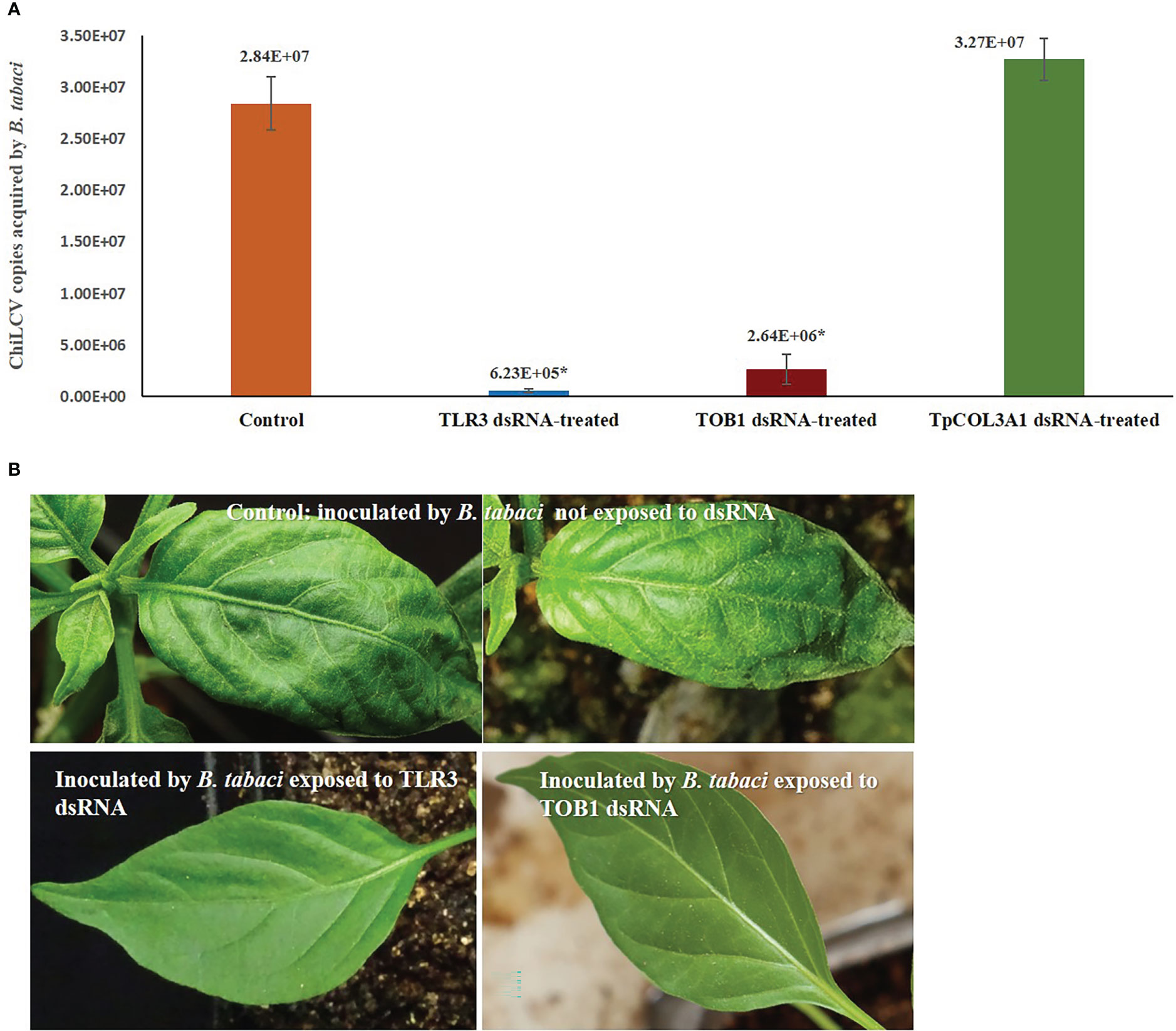
Figure 3 Effect of silencing TLR3 and TOB1 on ChiLCV acquisition and transmission by B. tabaci. (A) Mean ChiLCV copy numbers acquired by TLR3 and TOB1 dsRNA-fed B. tabaci. The mean denoted by an asterisk (*) indicates a significant difference (p< 0.0001). The error bars are the standard error of the mean (SEM). (B) Leaf curl symptoms on chilli plants inoculated by B. tabaci not exposed to dsRNA and exposed to TLR3 and TOB1 dsRNAs.
B. tabaci that were not exposed to TLR3 and TOB1 dsRNAs successfully transmitted ChiLCV to the inoculated plants. 93.33% of the inoculated plants tested positive in PCR with ChiLCV-specific primers. The infected plants showed characteristic ChiLCV symptoms like vein clearing, curling, and twisting of leaves, reduction of leaf size, puckering, reduction in inter-nodal length, thickening of leaves, swelling of veins, and overcrowding of leaves (Figure 3). Chilli plants inoculated by B. tabaci exposed to TLR3 and TOB1 dsRNAs showed no symptoms up to 21 dpi. No amplification specific to ChiLCV was recorded in PCR for the plants inoculated by dsRNA-fed B. tabaci.
Discussion
Several transcripts of B. tabaci are differentially regulated upon ChiLCV infection, yet the functional roles of these genes in virus transmission remain unknown. In ChiLCV-infected B. tabaci, TLR3 and TOB1 were highly abundant in adult B. tabaci which might facilitate the invasion and multiplication of the virus in B. tabaci (Nekkanti et al., 2022). Toll-like receptors are actively involved in innate immune responses against viruses (Perales-Linares and Navas-Martín, 2013; Lester and Li, 2014; He et al., 2021) and TLR3 is involved in protective antiviral responses (Ozato et al., 2002; Fernandes-Santos and Azeredo, 2022). Perales-Linares and Navas-Martín (2013) reported that TLR3 participates in both defense and offense in host immunity to viruses. TOB1 attenuates IRF3-directed antiviral responses by recruiting HDAC8 to specifically suppress IFN-β expression in virus-infected macrophages. TOB1 deficiency enhanced antiviral response and suppressed viral replication in vivo (Yu et al., 2022). The present study aimed to understand the functional role of B. tabaci TLR3 and TOB1 in ChiLCV infection.
dsRNAs targeting B. tabaci TLR3 and TOB1 were orally administered to B. tabaci adults in the present study. The expression of TLR3 and TOB1 mRNA was down-regulated by 6.67- and 3.01-folds, respectively 48 hr post-dsRNA feeding. dsRNA targeting T. palmi COL3A1 was taken as a negative control in B. tabaci. There was no significant regulation in the expression of TL3 and TOB1 post TpCOL3A1 dsRNA exposure. Significant downregulation of target genes by oral delivery of dsRNA was previously reported for B. tabaci ribosomal protein L9 (RPL9), vacuolar-type ATPase subunit A (V-ATPase A), cytochrome P450 family 3 subfamily A polypeptide 1 (Cyp315a1), Cyp18a1, ecdysone receptor gene (EcR5), ecdysone inducible gene (E75), hsp70, and fas2 (Upadhyay et al., 2011; Luan et al., 2013; Vyas et al., 2017; Chakraborty and Ghosh, 2022). Feeding on transgenic plants expressing dsRNA showed a down-regulation of around 90% in B. tabaci aquaporin (AQP) mRNA level 24 hr post-feeding (Raza et al., 2016). The difference in the level of silencing might be due to the variation in concentration of dsRNA, exposure period, target mRNA copies, delivery method, and the host’s defense mechanism (Ramkumar et al., 2021).
Silencing of B. tabaci TLR3 and TOB1 also significantly altered the survival of B. tabaci adults. Silencing of B. tabaci TLR3 induced a mean mortality of 30.66% 24 hr post-dsRNA exposure which increased to 47.32% at 48 hr. Similarly, the silencing of B. tabaci TOB1 caused up to 43.99% mortality compared to B. tabaci (9.33%) fed on a diet without dsRNA and diet with TpCOL3A1 dsRNA. B. tabaci TLR3 and TOB1 are involved in a network of molecular and biological processes. Loss of TLR3 and TOB1 functions due to depletion of mRNA might hamper the critical physiological processes in B. tabaci, leading to mortality. Silencing of actin ortholog, ADP/ATP translocase, α-tubulin, ribosomal protein L9 (RPL9), and V-ATPase A subunit also caused 27-97% mortality in B. tabaci (Upadhyay et al., 2011). Significant mortality was reported in B. tabaci upon silencing AQP, calcitonin (CAL), SWItch/sucrose non-fermentable (SNF7), inhibitor of apoptosis (IAP), hsp20, hsp40, knottin-1 (k-1), CypB, hsp70, and fas2 (Kaur et al., 2020; Chakraborty and Ghosh, 2022). Morphological deformities like twisting wings were reported post-silencing B. tabaci hsp70 (Kanakala et al., 2019). However, no such morphological abnormalities were recorded post-silencing B. tabaci TLR3 and TOB1 in the present study. Probably, TLR3 and TOB1 are not involved in any morphogenesis of B. tabaci or the exposure was too short to induce any morphological deformities.
Further, silencing of B. tabaci TLR3 and TOB1 reduced the ability of B. tabaci to acquire and transmit ChiLCV. The mean ChiLCV copies acquired by B. tabaci were decreased by 45.58 and 10.75- folds post-silencing TLR3 and TOB1 compared to B. tabaci fed on the diet without dsRNA and diet with TpCOL3A1 dsRNA. There were no symptoms in chilli plants up to 21 dpi when inoculated by B. tabaci fed on a diet mixed with TLR3 or TOB1 dsRNA. In contrast to the expectation, the results indicated that TLR3 is not involved in viral defense in B. tabaci as silencing of TLR3 decreases ChiLCV titer and transmission ability. Whereas, a reduction in ChiLCV titer post-silencing TOB1 supports its negative regulatory role in viral defense. Moreover, both target genes are essential for normal physiological functions in B. tabaci. Virus transmission might also be affected due to the poor physiological fitness of B. tabaci resulting from the depletion of TLR3 and TOB1 mRNAs. The results indicated that B. tabaci TLR3 and TOB1 would be novel targets to induce mortality and reduce ChiLCV transmission by B. tabaci. Complete inhibition of the ChiLCV transmission ability of B. tabaci was earlier demonstrated by the spray-on application of naked hsp70 dsRNA under controlled conditions (Chakraborty and Ghosh, 2022). A similar strategy to restrict begomovirus spread by B. tabaci was used by Wang et al. (2016) by silencing B. tabaci defensin-like gene (Btdef) which led to a lower accumulation of tomato yellow leaf curl China virus in B. tabaci. However, the silencing of fas2 increased the ChiLCV acquisition by B. tabaci (Chakraborty and Ghosh, 2022).
The present study is the first to demonstrate the involvement of B. tabaci TLR3 and TOB1 in the survival of B. tabaci and ChiLCV transmission by RNAi. It enriches our understanding of the gene functions of B. tabaci in begomovirus transmission. The outcome of the study would enable an in-depth study on the functional genomics of B. tabaci and apprise the B. tabaci-begomovirus relationships.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
AG conceived and designed the research and wrote and edited the final manuscript. PT, SJ, and MK conducted the experiments, recorded the experimental data, and wrote the draft manuscript. AG and SJ reviewed the data. All authors read and approved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The fellowship of PT was supported by the Indian Council of Agricultural Research, New Delhi.
Acknowledgments
The authors are thankful to Vinay Kalia, MS Saharan, and Debjani Dey (IARI) for their advisories. The support received from IARI, New Delhi is thankfully acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1136262/full#supplementary-material
References
Abd-Rabou, S., Simmons, A. M. (2010). Survey of reproductive host plants of Bemisia tabaci (Hemiptera: Aleyrodidae) in Egypt, including new host records. Entomol. News 121, 456–465. doi: 10.3157/021.121.0507
Akhter, A., Qazi, J., Saeed, M., Mansoor, S. (2009). A severe leaf curl disease on chilies in Pakistan is associated with multiple begomovirus components. Plant Dis. 93, 962. doi: 10.1094/PDIS-93-9-0962B
Chakraborty, P., Ghosh, A. (2022). Topical spray of dsRNA induces mortality and inhibits chilli leaf curl virus transmission by bemisia tabaci Asia II 1. Cells 11, 833. doi: 10.3390/cells11050833
Czosnek, H., Hariton-Shalev, A., Sobol, I., Gorovits, R., Ghanim, M. (2017). The incredible journey of begomoviruses in their whitefly vector. Viruses 9, 273. doi: 10.3390/v9100273
Fernandes-Santos, C., Azeredo, E. L. (2022). Innate immune response to dengue virus: Toll-like receptors and antiviral response. Viruses 14, 992. doi: 10.3390/v14050992
Ghosh, S., Ghanim, M. (2021). Factors determining transmission of persistent viruses by bemisia tabaci and emergence of new virus-vector relationships. Viruses 13, 1808. doi: 10.3390/v13091808
Götz, M., Popovski, S., Kollenberg, M., Gorovits, R., Brown, J. K., Cicero, J. M., et al. (2012). Implication of bemisia tabaci heat shock protein 70 in begomovirus-whitefly interactions. J. Virol. 86, 13241–13252. doi: 10.1128/JVI.00880-12
He, Y. J., Lu, G., Qi, Y. H., Zhang, Y., Zhang, X. D., Huang, H. J., et al. (2021). Activation of toll immune pathway in an insect vector induced by a plant virus. Front. Immunol. 11, 613957. doi: 10.3389/fimmu.2020.613957
Hogenhout, S. A., Ammar, e.-D., Whitfield, A. E., Redinbaugh, M. G. (2008). Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359. doi: 10.1146/annurev.phyto.022508.092135
Horowitz, A. R., Ghanim, M., Roditakis, E., Nauen, R., Ishaaya, I. (2020). Insecticide resistance and its management in Bemisia tabaci species. J. Pest Sci. 93, 893–910. doi: 10.1007/s10340-020-01210-0
Jones, D. R. (2003). Plant viruses transmitted by whiteflies. Eur. J. Plant Pathol. 109, 195–219. doi: 10.1023/A:1022846630513
Kanakala, S., Ghanim, M. (2016). Implication of the whitefly Bemisia tabaci cyclophilin b protein in the transmission of tomato yellow leaf curl virus. Front. Plant Sci. 7, 1702. doi: 10.3389/fpls.2016.01702
Kanakala, S., Kontsedalov, S., Lebedev, G., Ghanim, M. (2019). Plant-mediated silencing of the whitefly bemisia tabaci cyclophilin b and heat shock protein 70 impairs insect development and virus transmission. Front. Physiol. 10, 557. doi: 10.3389/fphys.2019.00557
Kaur, R., Gupta, M., Singh, S., Joshi, N., Sharma, A. (2020). Enhancing RNAi efficiency to decipher the functional response of potential genes in Bemisia tabaci AsiaII-1 (Gennadius) through dsRNA feeding assays. Front. Physiol. 11, 123. doi: 10.3389/fphys.2020.00123
Kumar, V., Kular, J. S., Kumar, R., Sidhu, S. S., Chhuneja, P. K. (2020). Integrated whitefly [Bemisia tabaci (Gennadius)] management in bt-cotton in north India: An agroecosystem-wide community-based approach. Curr. Sci. 119 (4), 618–624. doi: 10.18520/cs/v119/i4/618-624
Lester, S. N., Li, K. (2014). Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 426, 1246–1264. doi: 10.1016/j.jmb.2013.11.024
Li, S. J., Xue, X., Ahmed, M. Z., Ren, S. X., Du, Y. Z., Wu, J. H., et al. (2011). Host plants and natural enemies of Bemisia tabaci (Hemiptera: Aleyrodidae) in China. Insect Sci. 18, 101–112. doi: 10.1111/j.1744-7917.2010.01395.x
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, J. B., Ghanim, M., Liu, S. S., Czosnek, H. (2013). Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 43, 740–746. doi: 10.1016/j.ibmb.2013.05.012
Navas-Castillo, J., Fiallo-Olivé, E., Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Nekkanti, A., Chakraborty, P., Ghosh, A., Iquebal, M. A., Jaiswal, S., Baranwal, V. K. (2022). Transcriptomic changes of Bemisia tabaci Asia II 1 induced by chilli leaf curl virus trigger infection and circulation in its vector. Front. Microbiol. 13, 423–466. doi: 10.3389/fmicb.2022.890807
Ozato, K., Tsujimura, H., Tamura, T. (2002). Toll-like receptor signaling and regulation of cytokine gene expression in the immune system. BioTechniques Suppl. 66-8, 70, 72 passim. doi: 10.2144/Oct0208
Perales-Linares, R. F., Navas-Martín, S. (2013). Toll-like receptor 3 in viral pathogenesis: Friend or foe? Immunology 140, 153–167. doi: 10.1111/imm.12143
Polston, J. E., Anderson, P. K. (1997). The emergence of whitefly-transmitted geminiviruses in tomato in the Western hemisphere. Plant Dis. 81, 1358–1369. doi: 10.1094/PDIS.1997.81.12.1358
Ramkumar, G., Asokan, R., Prasannakumar, N. R., Kariyanna, B., Karthi, S., Alwahibi, M. S., et al. (2021). RNA Interference suppression of v-ATPase b and juvenile hormone binding protein genes through topically applied dsRNA on tomato leaves: Developing biopesticides to control the south American pinworm, Tuta absoluta (Lepidoptera: Gelechiidae). Front. Physiol. 12, 742871. doi: 10.3389/fphys.2021.742871
Raza, A, Malik, H.J., Shafiq, M., Amin, I., Scheffler, J.A., Scheffler, B.E., Mansoor, S. (2016). RNA interference based approach to down regulate osmoregulators of whitefly (Bemisia tabaci): potential technology for the control of whitefly. PLoS ONE 11(4), e0153883. doi: 10.1371/journal.pone.0153883
Rosen, R., Kanakala, S., Kliot, A., Cathrin Pakkianathan, B., Farich, B. A., Santana-Magal, N., et al. (2015). Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 15, 1–8. doi: 10.1016/j.coviro.2015.06.008
Roy, B., Chakraborty, P., Ghosh, A. (2021). How many begomovirus copies are acquired and inoculated by its vector, whitefly (Bemisia tabaci) during feeding? PloS One 16, e0258933. doi: 10.1371/journal.pone.0258933
Senanayake, D. M. J. B., Mandal, B., Lodha, S., Varma, A. (2007). First report of chilli leaf curl virus affecting chilli in India. Plant Pathol. 56, 343–343. doi: 10.1111/j.1365-3059.2007.01513.x
Shingote, P. R., Wasule, D. L., Parma, V. S., Holkar, S. K., Karkute, S. G., Parlawar, N. D., et al. (2022). An overview of chili leaf curl disease: Molecular mechanisms, impact, challenges, and disease management strategies in Indian subcontinent. Front. Microbiol. 29. doi: 10.3389/fmicb.2022.899512
Simon, C., Frati, F., Beckenbach, A.T., Crespi, B.J., Liu, H., Flook, P.K. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am 87, 651–701. doi: 10.1093/AESA/87.6.651
Thakur, H., Jindal, S. K., Sharma, A., Dhaliwal, M. S. (2018). Chilli leaf curl virus disease: A serious threat for chilli cultivation. J. Plant Dis. Prot. 125, 239–249. doi: 10.1007/s41348-018-0146-8
Upadhyay, S. K., Chandrashekar, K., Thakur, N., Verma, P. C., Borgio, J. F., Singh, P. K., et al. (2011). RNA Interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 36, 153–161. doi: 10.1007/s12038-011-9009-1
Varma, A., Malathi, V. G. (2003). Emerging geminivirus problems: A serious threat to crop production. Ann. Appl. Biol. 142, 145–164. doi: 10.1111/j.1744-7348.2003.tb00240.x
Verma, S., Bhattacharya, A. (2015)Whitefly destroys 2/3rd of punjab’s cotton crop, 15 farmers commit suicide. In: Times of India. Available at: http://timesofindia.indiatimes.com/articleshow/49265083.cms?source=contentofinterest&utm_medium=text&utm_campaign=cpps (Accessed 21-02-2023).
Vyas, M., Raza, A., Ali, M. Y., Ashraf, M. A., Mansoor, S., Shahid, A. A., et al. (2017). Knock-down of whitefly gut gene expression and mortality by orally delivered gut gene-specific dsRNAs. PLoSOne 12, e0168921. doi: 10.1371/journal.pone.0168921
Wang, Y., He, Y., Liu, S., Wang, X. (2020). Mechanisms of plant virus transmission by the whitefly Bemisia tabaci. Chin. Sci. Bull. 65, 1463–1475. doi: 10.1360/TB-2019-0854
Wang, Z. Z., Shi, M., Huang, Y. C., Wang, X. W., Stanley, D., Chen, X. X. (2016). A peptidoglycan recognition protein acts in whitefly (Bemisia tabaci) immunity and involves in begomovirus acquisition. Sci. Rep. 6, 37806. doi: 10.1038/srep37806
Keywords: whitefly, RNAi, ChiLCV, Toll-like receptor 3, transducer of erbB2.1, virus-vector relationship
Citation: Thesnim P, Jangra S, Kumar M and Ghosh A (2023) Effect of silencing Bemisia tabaci TLR3 and TOB1 on fitness and begomovirus transmission. Front. Plant Sci. 14:1136262. doi: 10.3389/fpls.2023.1136262
Received: 02 January 2023; Accepted: 22 February 2023;
Published: 14 March 2023.
Edited by:
Muhammad Naeem Sattar, King Faisal University, Saudi ArabiaReviewed by:
Khalid Mehmood, Muhammad Nawaz Shareef University of Agriculture, PakistanLuc Swevers, National Centre of Scientific Research Demokritos, Greece
Walter Leke, Institute for Agricultural Research for Development (IRAD), Cameroon
Tahir Farooq, Guangdong Academy of Agricultural Sciences, China
Copyright © 2023 Thesnim, Jangra, Kumar and Ghosh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amalendu Ghosh, YW1hbDRlbnRvQGdtYWlsLmNvbQ==
†Present address: Sumit Jangra, Tropical Research and Education Centre, University of Florida, Homestead, FL, United States, Manish Kumar, Department of Plant Pathology, College of Agricultural & Environmental Sciences, University of Georgia, Tifton, GA, United States
‡ORCID: Amalendu Ghosh, orcid.org/0000-0001-6634-5771
 Pathukandathil Thesnim
Pathukandathil Thesnim Sumit Jangra
Sumit Jangra Manish Kumar
Manish Kumar Amalendu Ghosh
Amalendu Ghosh