
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 February 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1135321
This article is part of the Research Topic Exploring the Genetic Potential of Thinopyrum Species in Wheat and Durum Wheat Improvement View all 6 articles
 Xianrui Guo1,2
Xianrui Guo1,2 Yuhong Huang2
Yuhong Huang2 Jing Wang2
Jing Wang2 Shulan Fu2
Shulan Fu2 Chunhui Wang2
Chunhui Wang2 Mian Wang2
Mian Wang2 Chen Zhou2
Chen Zhou2 Xiaojun Hu1
Xiaojun Hu1 Tao Wang3
Tao Wang3 Wuyun Yang4*
Wuyun Yang4* Fangpu Han2*
Fangpu Han2*Wheat stripe rust is a destructive disease in many cool and temperate regions around the world. Exploiting novel sources of resistance can provide wheat cultivars with robust and durable resistance to stripe rust. The wheat–Thinopyrum intermedium addition line TAI-14 was proven to carry a stripe rust resistance gene (named as YrT14) on the alien Th. intermedium chromosome. In order to transfer the resistance gene to wheat, wheat–Th. intermedium translocation lines were created by irradiating the pollen of the line TAI-14. We totally obtained 153 wheat–Th. intermedium translocation lines, among which the long alien segmental translocation line Zhongke 78 and the intercalary translocation line Zhongke 15 not only showed good integrated agronomic traits but also were identified as highly resistant to stripe rust in both seedling and adult plant stages. The alien chromatin in Zhongke 15 was identified as an insertion into the satellite of chromosome 6B, a type of translocation never reported before in chromosome engineering. By screening Simple Sequence Repeat (SSR) and Expressed Sequence Tag (EST) markers as well as the markers developed from RNA-sequencing (RNA-Seq) data, 14 markers were identified specific for the alien chromosome and a physical map was constructed. Both Zhongke 78 and Zhongke 15 could be used as a novel source of stripe rust resistance for wheat breeding, and the linked marker T14K50 can be used for molecular marker–assisted breeding. Finally, based on the karyotype, reaction to stripe rust, and genome resequencing data of different wheat–Th. intermedium translocation lines, the stripe rust resistance gene YrT14 was located to an 88.1 Mb interval from 636.7 to 724.8 Mb on Th. intermedium chromosome 19 corresponding to 7J or 7Js.
As one of the most important food crops, common wheat (Triticum aestivum) is cultivated on approximately 220 million ha around the world and feeds more than one-third of the worlds’ population (http://www.fao.org/faostat). In the pursuit of high yield in breeding, the diversity of crops is becoming narrower and narrower, which, in turn, diminishes their potential to survive in both abiotic and biotic stresses (Allard, 1996; Hoisington et al., 1999; Charmet, 2011). Wheat stripe rust, powdery mildew, and Fusarium head blight are three fungal diseases that seriously threaten wheat production globally (Bennett, 1984; Bai and Shaner, 1994; Kolmer, 2005). It is of great importance to exploit novel resistant sources to provide wheat with robust and durable resistance against these diseases.
As a perennial grass, Thinopyrum intermedium is identified as an allohexaploid. Although chromosome composition is controversial, it is generally regarded as JJJsJsStSt (Chen et al., 1998b). The subgenome St is closely related to the genome of Pseudoroegneria strigose, and the J or JS subgenome is highly homologous with the genome Ee of Th. elongatum and Eb of Th. bessarabicum (Liu and Wang, 1993; Chen, 2005). These results were also supported by the fact that researchers found that one set of chromosomes was labeled as the St type while the rest were all labeled as the non-St type by employing the probe St2-80 to detect the chromosomes in Th. intermedium (Wang et al., 2017). A consensus map with 21 linkage groups containing 10,029 markers was produced by using genotyping-by-sequencing (GBS). Through mapping the sequence tags from Th. intermedium to the diploid barley reference genome, high collinearity and synteny between the three homoeologous Th. intermedium chromosomes and each of the seven barley chromosomes were discovered (Kantarski et al., 2017). Referring to the consensus map and a set of 350,885 75-mer markers, the assembled scaffolds of Th. intermedium were oriented, ordered, and joined together to form chromosomes (http://phytozome.jgi.doe.gov/). This released genomic resource would accelerate the development of markers and gene cloning.
Th. intermedium has been used as a source for wheat improvement for decades. As Th. intermedium can easily be crossed with wheat, numerous useful genes and traits have been transferred into wheat in the form of partial amphiploids, addition lines, substitution lines, and translocation lines. Wheat–Th. intermedium partial amphiploids containing a complete set of wheat and a set of Th. intermedium chromosomes have been reported, such as Zhong1 to Zhong5, TAF46, TE261-1, TE266-1, TE346-1, TAI8355, and 363-1-21 (Chen et al., 1998a; Chen et al., 1999; Chen et al., 2003; Han et al., 2004; Chang et al., 2010; Guo et al., 2015; Cui et al., 2021). Partial amphiploids were usually unstable in cytology and conserved many grass characteristics that were undesirable in breeding exercise. Thus, a great number of addition and substitution lines were produced by crossing the partial amphiploids with wheat. He et al. (1989) developed two sets of wheat–Th. intermedium addition lines TAI-11~17 and TAI-21~27 by crossing the wheat–Th. intermedium partial amphiploid Zhong series lines with common wheat. The resulting progeny lines of TAI-12 and TAI-15 presented longer seeds and spikes than control (He et al., 1989). In addition, another three progeny lines TAI-23, TAI-25, and TAI-27 were identified as resistant to stem rust, leaf rust, and the wheat yellow dwarf virus, respectively (He et al., 1989). New high- molecular-weight subunits were identified in three wheat–Th. intermedium addition lines W210, W211, and W212 that were derived from the hybrids of common wheat Yannong 15 and Th. intermedium H-2 (Wang and Wang, 2016). In addition to wheat–Th. intermedium addition lines, some wheat–Th. intermedium substitution lines were produced, such as the 1St#2 (1D) substitution line AS1677 showing higher kernel protein content and the 4Ai#2(4A) substitution line CI 15092 with eyespot resistance (Li et al., 2005; Li et al., 2016). Based on these intermediate materials, many wheat–Thinopyrum translocation lines derived from centric breakage–fusion, recombination between homoeologous chromosomes, or physical irradiation were reported by different groups. For example, a new compensating wheat–Th. intermedium Robertsonian translocation line CH13-21 with resistance to powdery mildew and stripe rust was derived from a spontaneous centric breakage fusion (Zhan et al., 2015). Relying on ph1b-induced homoeologous recombination, a shortened Th. intermedium segment containing the Wsm3 gene was translocated to chromosome 7B and conferred wheat resistance to the wheat streak mosaic virus (Danilova et al., 2017). In addition, P961341, a wheat–Th. intermedium translocation line with resistance to the yellow dwarf virus was produced by irradiating with γ rays from 60Co (Ohm et al., 2005).
Isozymes Cpxe-Agi2 and Est-Agi3 whose homoeologs in wheat were located on 7AS and 7BS, respectively, were discovered in the addition line TAI-14, which indicated that the alien chromosome in TAI-14 was homoeologous with wheat group-7 chromosomes (Benito et al., 1987; Gao and Hao, 1993; Vladova and Petkolicheva, 1996). In this study, the wheat–Th. intermedium addition line TAI-14 was identified as resistant to stripe rust. Genetics analysis revealed that there was a stripe rust resistance gene (named as YrT14) on the alien Th. intermedium chromosome. In order to transfer the resistance gene to wheat, a series of wheat–Th. intermedium translocation lines were developed, among which the long alien segmental translocation line Zhongke 78 and the intercalary translocation line Zhongke 15 showed good integrated agronomic traits and resistance to stripe rust and could be used as a novel resource for wheat stripe rust breeding. By screening SSR and EST markers as well as the markers developed from RNA-Seq data, we developed specific markers and constructed a physical map for the alien chromosome according to the size of the alien chromatin in different wheat–Th. intermedium translocation lines. Finally, by combining the karyotype, reaction to stripe rust, and genome resequencing data of different wheat–Th. intermedium translocation lines, the stripe rust resistance gene YrT14 was located to an 88.1 Mb interval between 636.5 and 724.8 Mb on Th. intermedium chromosome 19 that was proven to correspond to chromosome 7J or 7Js.
TAI-14 was derived from the wheat–Th. intermedium partial amphiploid Zhong2 (He et al., 1989). It is a disomic addition line, containing a whole set of 42 wheat chromosomes and a pair of chromosomes derived from Th. intermedium (He et al., 1989; Han et al., 1998). We referred to the plant carrying two alien chromosomes as T14-44, the one carrying one alien chromosome as T14-43 and the one without an alien chromosome as T14-42, respectively. The wheat cultivar Jimai 22 was used as the recurrent parent to cross with the translocation line. Wheat cultivars Fielder and Jimai 20 were susceptible to stripe rust and used as parents to generate F2 populations. Two highly stripe rust susceptible wheat varieties Mingxian 169 and Mianyang 11 were used as control in strip rust resistance evaluation. Th. intermedium accession PI 440001, T. urartu accession TMU38, and Aegilops tauschii accession TQ27 were used for the preparation of genomic DNA probes. T. asetivum line Chinese Spring (CS) and Ae. speltoides accession AE739 were used for the preparation of blocking DNA.
At the flowering stage, the spikes of the wheat–Th. intermedium addition line TAI-14 were cut from the plant in the morning, and immediately radiated by γ rays derived from 60Co with a dose of 18 Gy. Then the fresh pollens were used to pollinate the wheat variety Jimai 22 with its stamens removed in advance. The hybrid seeds were harvested at maturity. The translocation lines were identified by utilizing genomic in situ hybridization (GISH). Using Jimai 22 as the recurrent parent, the integrated agronomic traits of the translocation lines were improved by continuous backcrossing.
Seeds were germinated on moist filter paper in a Petri dish at room temperature for 2–3 days. Growing roots were cut from seedlings and treated in nitrous oxide with 15 bars of pressure for approximately 2 h. The roots were subsequently fixed in 90% acetic acid for 5 min and then stored in 70% v/v ethanol at -20°C. Chromosome spread preparation was performed as previously described (Kato et al., 2004).
Oligo-pSc119.2-1 combined with Oligo-pTa535-1 was used to distinguish the whole set of 42 wheat chromosomes (Tang et al., 2014). Oligo-pSc119.2-1 (10 ng/µl) and Oligo-pTa535-1 (10 ng/µl) were 5’ end-labeled with 6-carboxyfluorescein (6-FAM) and 6-carboxytetramethylrhodamine (6-Tamra) (InvitrogenTM, Shanghai, China), respectively. Genomic DNA was isolated from the leaves of Th. intermedium accession PI 440001, T. urartu accession TMU38, Ae. speltoides accession AE739, Ae. tauschii accession TQ27, and CS using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). The green or red probes with a concentration of 100 ng/µl were prepared according to the nick translation method (Kato et al., 2011). The genomic DNA of Th. intermedium, T. urartu and the plasmid of St2-80 reported by Wang et al. (2017) were labeled with Alexa Fluor-488-5-2'-deoxyuridine 5'-triphosphate (dUTP) (InvitrogenTM, Shanghai, China). The genomic DNA of A. tauschii and the centromeric retrotransposon of wheat (CRW) clone 6C6 was labeled with Texas-red-5-dCTP (InvitrogenTM, Shanghai, China). The genomic DNA of CS and A. speltoides in a concentration of 3,000 ng/µl was used for blocking in multicolor-GISH (mc-GISH). For each slide, FISH was performed in 10 µl reaction volumes, in which 0.2 µl Oligo-pSc119.2-1, 0.2 µl Oligo-pTa535-1, and 0.3 µl 6C6, 0.5 µl St2-80 were used and the 2x SSC, 1x TE buffer was used to adjust the volume. For Th. Intermedium chromatin detection, 10 µl reaction volumes for each slide contain 0.5 µl labeled genomic DNA of PI 440001 and 2.5 µl genomic DNA of CS. For the mc-GISH on wheat, the 10 µl reaction volumes for each slide contain the 2 µl labeled genomic DNA of TMU38, 2 µl genomic DNA of AE739, and 1 µl labeled genomic DNA of TQ27. All chromosomes were counterstained with 4, 6-diamidino-2-phenylindole (DAPI) (Vectashield, Vector Laboratories, Burlingame, CA, USA). Chromosomes on microscope slides were examined using a BX61 fluorescence microscope (Olympus, Tokyo, Japan) equipped with a U-CMAD3 camera (Olympus, Tokyo, Japan) and appropriate filter sets. The signal capture and picture processing were performed using MetaMorph software (Molecular Devices, LLC., San Jose, CA, USA). The final image adjustment was done in Adobe Photoshop CS5 (Adobe Systems Incorporated, San Jose, CA, USA).
In order to get specific markers for the alien chromosome, we screened 197 wheat group-7-specific microsatellite markers reported by Somers et al. (2004) and 88 pairs of sequence-tagged sites-polymerase chain reaction (STS-PCR) primers on wheat group-7 chromosomes (Supplementary Table S1). At the two-leaf stage, 27 plants of T14-44 and 25 plants of T14-42 were collected and separately pooled for RNA isolation using a TRIzol reagent (InvitrogenTM, Shanghai, China), followed by the treatment with DNase I (InvitrogenTM, Shanghai, China). The samples were sequenced using the Illumina Hiseq2500 platform (Berry Genomics, Beijing, China) to generate 125 bp pair-end reads. The de novo assembly of clean reads was performed by using the software Trinity 2.1.1 (Haas et al., 2013). The expression level was calculated by mapping reads to the assembled transcripts employing Trinity scripts, RSEM, and edgeR (Haas et al., 2013). The TransDecoder software package (https://sourceforge.net/projects/transdecoder/) was used to predict the coding region for these transcripts. The transcripts were annotated in the Swiss-Prot database using Blastx. The transcripts expressed in T14-44 but not in T14-42 were extracted. Then, the transcripts annotated as Nucleotide Binding Site–Leucine Rich Repeat (NBS-LRR) protein and protein kinases were used to design primers using the software Primer 5.0 (PREMIER Biosoft, San Francisco, CA, USA).
The conditions of the polymerase chain reaction (PCR) were as follows: initial denaturation at 94°C for 4 min, followed by 35 cycles of 30 s at 94°C, 30 s for annealing at 55°C–60°C, 1 min for extension at 72°C, and a final extension at 72°C for 10 min. Amplified PCR products were separated on 8% non-denaturing polyacrylamide gels stained with silver at 200 V for 1 h and 1.5% agarose gels stained with ethidium bromide at 150 V for approximately 25 min. The D2000 Plus DNA Ladder (GenStar, Beijing, China) and the 100 bp DNA Ladder (TianGen Biotech Co, Beijing, China) were used for the DNA marker in non-denaturing polyacrylamide gel and agarose gel electrophoresis, respectively.
The genome of wheat–Th. intermedium translocation lines Zhongke 15, Zhongke 353, Zhongke 215, Zhongke 16, and Zhongke 55 was resequenced. Genomic DNA for resequencing was isolated by using the CTAB method. A combined reference genome consisted of CS reference genome v1.0 (Consortium (IWGSC) et al., 2018) and three genomic sequences of Chr19-21 extracted from Th. intermedium reference genome v2.1 (http://phytozome.jgi.doe.gov/) were used. Raw reads were first processed with Trimmomatic version 0.36 (with parameters: SLIDINGWINDOW:5:20 LEADING:5 TRAILING:5 MINLEN:25) (Bolger et al., 2014). Cleaned reads were aligned to the combined reference genome with Burrows-Wheeler-Alignment (BWA) mem version 0.7.13-r1126 (Li and Durbin, 2009). Alignments were compressed, sorted, filtered (remove mapping quality less than 20), and indexed by using SAMtools version 1.9 (https://sourceforge.net/projects/samtools/files/samtools/). Duplicate alignments were removed with Picard version 2.25.6 (https://broadinstitute.github.io/picard/). The means of read depth within per 100 kb window were calculated using Mosdepth version 0.2.6 with default parameters (Pedersen and Quinlan, 2018). The chromosome translocation points were identified in Integrative Genomics Viewer (IGV) version 2.12.0 (https://software.broadinstitute.org/software/igv/).
The wheat–Th. intermedium translocation lines Zhongke 78 and Zhongke 15 and the recurrent parent Jimai 22 were planted in rows with a length of 3 m and a spacing of 10 cm in a field in Beijing (E116°42′, N40°10′). Plant height was taken as an average of 10 randomly chosen tillers from five plants. For each line, the tillering number of 10 plants was counted. Ten spikes randomly selected from each line were used to calculate the spike length and kernel number per spike. The thousand kernel weight was measured three times by counting 500 kernels.
The lines T14-44 and T14-42; the wheat–Th. intermedium translocation lines including Zhongke 15, Zhongke 353, Zhongke 215, Zhongke 16, Zhongke 55, and Zhongke 78; and the F2 individuals derived from Zhongke 15/Jimai 20 and Zhongke 78/Fielder were used for stripe rust resistance evaluation in a greenhouse. The wheat variety Mingxian 169 was used as the susceptible control. All plants were planted in the 32-cell trays in a greenhouse set at 16°C with a 16-h photoperiod. Fresh spores of three stripe rust races CYR32, CYR33, and CYR34 were independently suspended in distilled water. When the second leaf expanded, the seedlings were sprayed with the spore suspension. After inoculation, the seedlings were kept moist for 24 h at 12°C–13°C. Then, the plants were moved to a greenhouse set as 16°C with a 16-h photoperiod for approximately 14 days. Infection types were recorded based on a 0–4 scale (He et al., 2009). For the evaluation of adult plant resistance, the adult plant of Zhongke 78 and Zhongke 15 was evaluated in natural conditions in the field in Chengdu, Sichuan Province. The susceptible cultivar Mianyang 11 was used as control.
To characterize the karyotype of the wheat–Th. intermedium addition line TAI-14, we performed GISH and FISH analyses on the mitotic metaphase chromosomes. In addition to 42 chromosomes from wheat stained blue, a pair of chromosomes stained green was also detected in TAI-14, which indicated their origin of Th. intermedium (Figure 1A). Because of its smaller size and no detection of the CRW signal on it, the alien chromosome was classified as a telocentric chromosome (Figure 1A). In spite of having no CRW signals on the alien chromosome, functional centromeres were revealed by the presence of Centromere-specific histone H3 (CENH3) (Guo et al., 2016). However, we observed that the loss of the alien chromosome occurs to a certain extent in the self-crossing offspring, gradually transforming TAI-14 from T14-44 to T14-42. Using the isolates of races CYR32, CYR33, and CYR34, the evaluation on stripe rust resistance was performed on the seedlings of both T14-44 and T14-42. The addition line T14-44 showed resistance to all three races with an obvious hypersensitive response on the seedling leaves (Figure 1B). Conversely, the line T14-42 without an alien chromosome was susceptible to all three races and the vast numbers of fungal spores spread on the seedling leaves (Figure 1B). These results suggested that one genetic locus designated as YrT14 on the alien chromosome was responsible for the stripe rust resistance. Further analysis discovered that the FISH marker St2-80 labeled the terminal region of the telocentric chromosomes in T14-44 (Supplementary Figure 1), which suggested that the alien chromosomes belong to the J or Js subgenome rather than the St subgenome of Th. intermedium. Therefore, we found that one gene (YrT14) from the J or Js subgenome of Th. intermedium conferred wheat with stripe rust resistance.
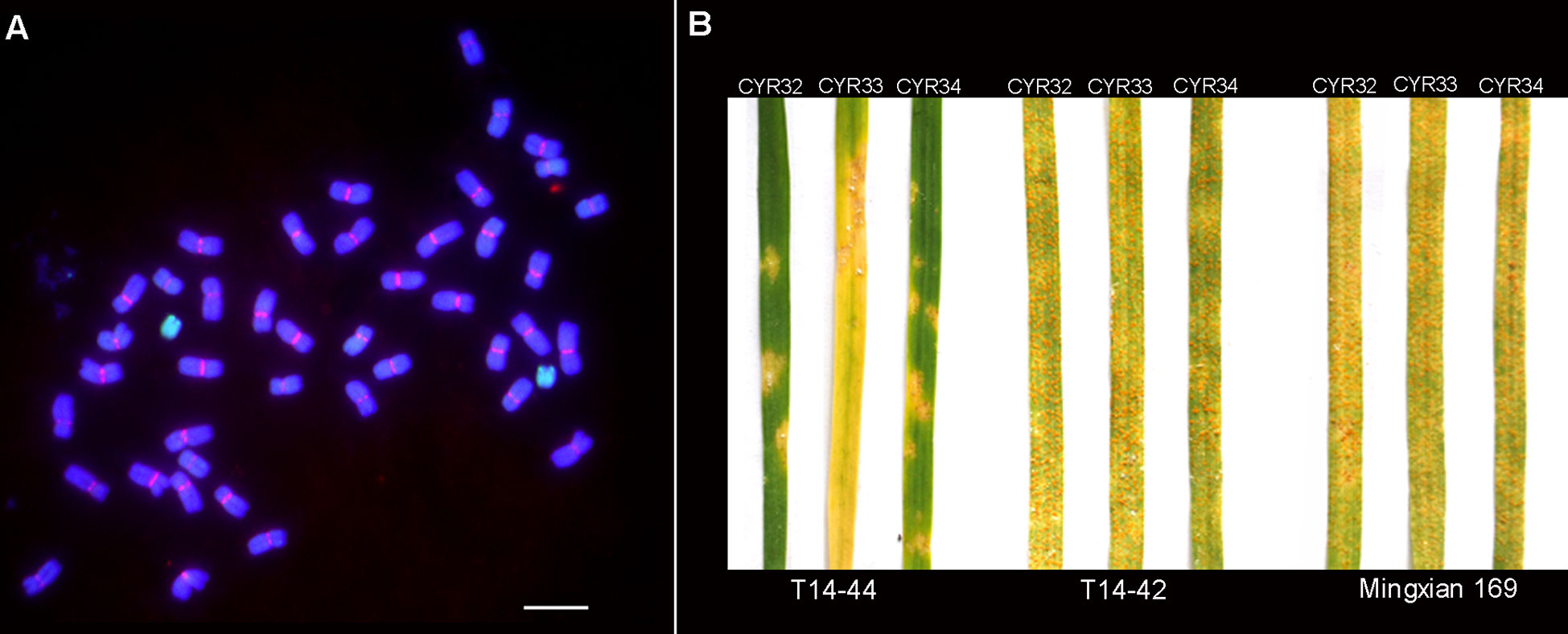
Figure 1 TAI-14 karyotype and its stripe rust resistance. (A) T14-44 karyotype identified by genomic in situ hybridization (GISH) and fluorescence in situ hybridization (FISH). The genomic DNA of Th. intermedium (green) and the wheat centromeric retrotransposon 6C6 (red) were used as a probe. The genomic DNA of Chinese Spring (CS) was used as a blocker. Chromosomes were stained with 4, 6-diamidino-2-phenylindole (DAPI; blue). Scale bar = 10 μm. (B) Stripe rust evaluation on T14-44 and T14-42. Stripe rust tests were performed using three rust races CYR32, CYR33, and CYR34, respectively. Common wheat cultivar Mingxian 169 was used as the susceptible control.
In order to transfer the alien resistance gene YrT14 to common wheat, wheat–Th. intermedium translocation lines were produced by irradiating the pollen of TAI-14 at the flowering stage. The fresh pollens were then pollinated to the recurrent parent Jimai 22. By performing GISH on 8,100 F1 seeds, we obtained 153 wheat–Th. intermedium translocation lines at a frequency of 1.89% (Table 1). According to the size and position of the alien chromatin, the translocation lines were classified into three types (Supplementary Figure 2). If the ratio of the length of the alien fragment in translocation lines to the length of the telocentric chromosome ranged from 0 to 0.5, the translocation lines were classified as short alien segmental translocation lines, such as lines Zhongke 55, Zhongke 61, and Zhongke 316 (Figures 2A–C). If the ratio ranged from 0.5 to 1, they were classified as long alien segmental translocation lines, such as lines Zhongke 843, Zhongke 70, and Zhongke 108 (Figures 2D–F). If one part of the alien chromosome inserted into one wheat chromosome, the translocation lines were classified as intercalary translocation lines, such as lines Zhongke 21, Zhognke 102, and Zhongke 127 (Figures 2G–I). In detail, we obtained 66 short alien segmental translocation lines and 70 long alien segmental translocation lines, which accounted for 43.14% and 45.75%, respectively (Table 1). However, we only screened 17 (11.11%) intercalary translocation lines and the proportion was obviously lower than the other two types (Table 1).
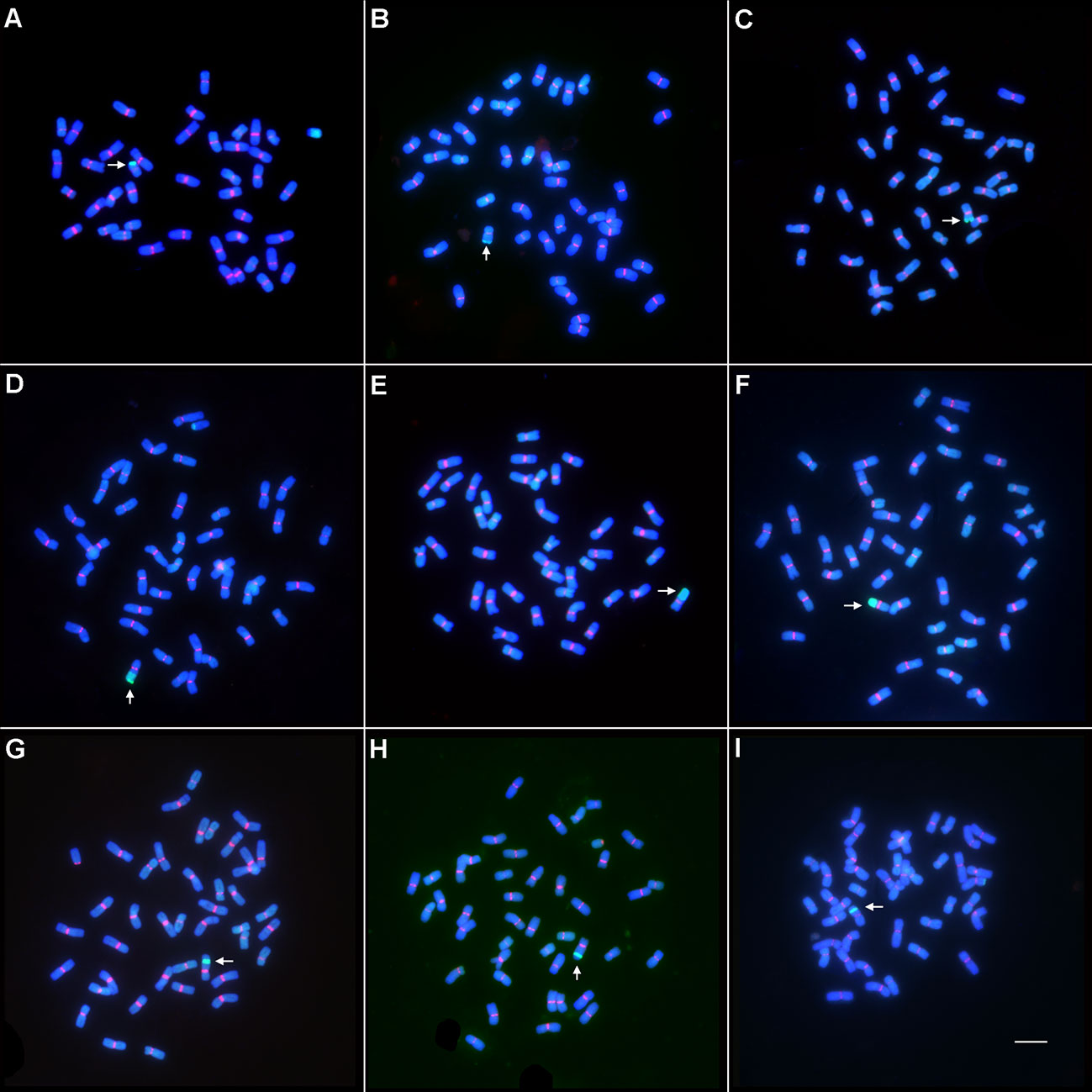
Figure 2 Wheat–Th. intermedium translocation line identified by GISH and FISH. The genomic DNA of Th. intermedium (green) was used as probe for GISH, and the wheat centromeric retrotransposon 6C6 (red) was used as probe for FISH. The genomic DNA of CS was used as a blocker. Chromosomes were stained with DAPI. Arrows indicated the translocated chromosomes. Scale bar = 10 μm. (A–C) The short alien segmental translocation lines identified by GISH and FISH. (A) Zhongke 55; (B) Zhongke 61; (C) Zhongke 316. (D–F) The long alien segmental translocation lines identified by GISH and FISH. (D) Zhongke 843; (E) Zhongke 70; (F) Zhongke 108. (G–I) The intercalary translocation lines identified by GISH and FISH. (G) Zhongke 21; (H) Zhongke 102; (I) Zhongke 127.
To improve the integrated agronomic traits, all wheat–Th. intermedium translocation lines were backcrossed with Jimai 22 and homozygous translocation lines were selected among the offspring. Two homozygous translocation lines Zhongke 78 and Zhongke 15 with good agronomic traits were cytologically analyzed in detail (Figure 3). Zhongke 78 and Zhongke 15 were identified as a long segmental translocation line and an intercalary translocation line, respectively (Figures 3A, D). Combining multicolor GISH, the translocation in Zhongke 78 occurred on chromosome 6A and the alien chromatin nearly replaced the long arm of 6A (Figures 3A–C). For the line Zhongke 15, the inserted alien chromatin was identified on the chromosome 6B by using the probe Oligo-pSc119.2-1 (Figures 3D, E). In addition, the secondary constriction was observed near the inserted position proximal to the centromere (Figure 3D). Therefore, the alien chromatin was found inserted into the satellite of chromosome 6B in the line Zhongke 15.
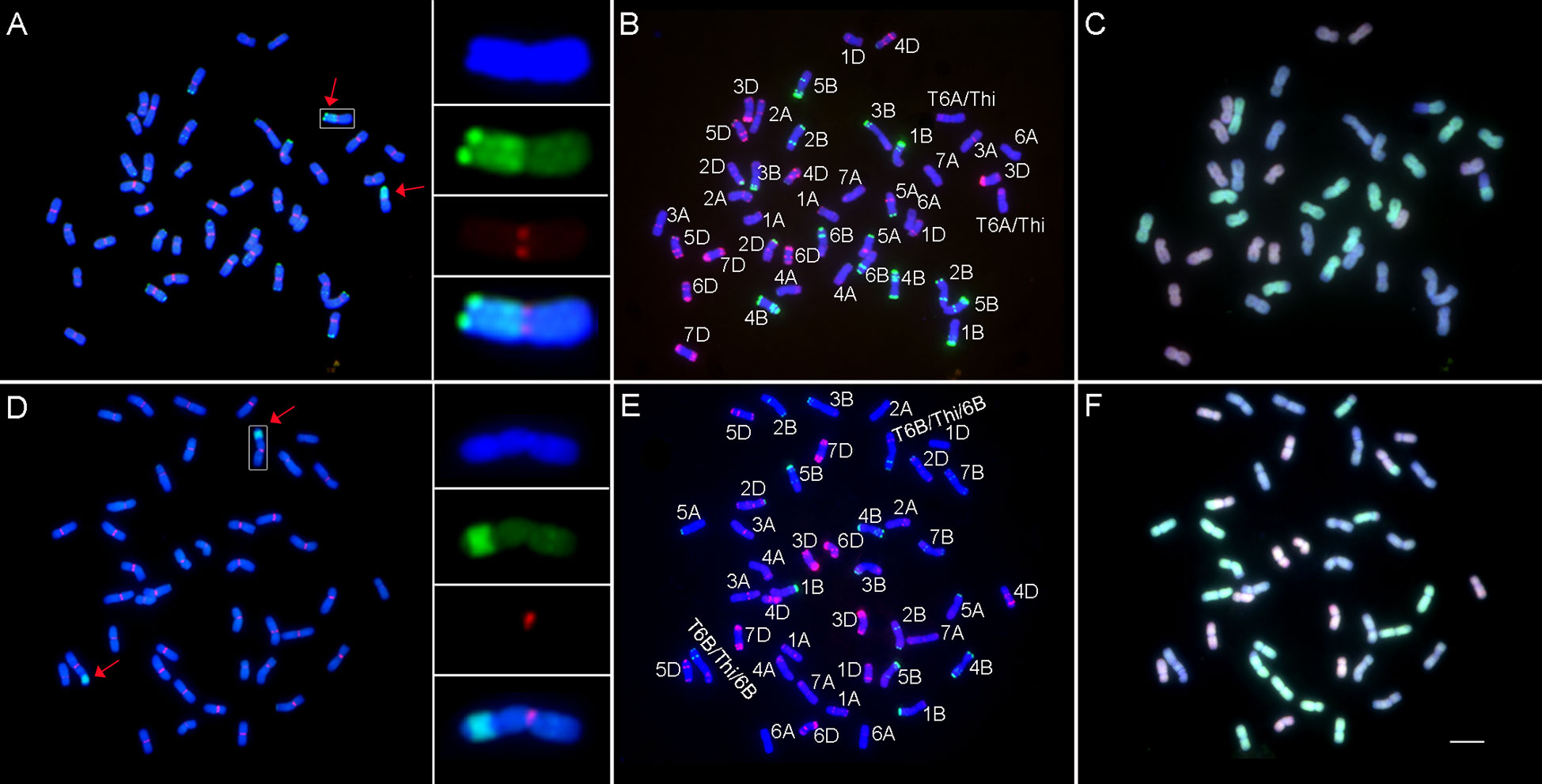
Figure 3 Cytological analysis on lines Zhongke 78 and Zhongke 15. (A–C) Cytological characterization of Zhongke 78. (D–F) Cytological characterization of Zhongke 15. GISH and FISH patterns of Zhongke 78 (A) and Zhongke 15 (D). The genomic DNA of Th. intermedium (green) was used as probe for GISH and the wheat centromeric retrotransposon 6C6 (red) was used as probe for FISH. The genomic DNA of CS was used as a blocker. FISH patterns of Zhongke 78 (B) and Zhongke 15 (E) identified by Oligo-pTa535-1 (red) and Oligo-pSc119.2-1 (green). GISH patterns of Zhongke 78 (C) and Zhongke 15 (F) identified by the genomic DNA of Aegilops tauschii (DD) (red) and the genomic DNA of Triticum urartu (AA) (green). The genomic DNA of Ae. speltoides (SS) was used as a blocker. All chromosomes were counterstained with DAPI. Arrows indicated the translocated chromosomes. Scale bar = 10 μm.
Although both lines contained 42 chromosomes, the subgenomic components of Zhongke 78 were different from that of Zhongke 15 (Figures 3C, F). Upon further analysis, it was discovered that the number of all three subgenome chromosomes in Zhongke 15 was 14 (Figure 3F), whereas Zhongke 78 was found comprised of 16 A subgenome chromosomes, 12 B subgenome chromosomes, and 14 D subgenome chromosomes (Figure 3C). In detail, a pair of chromosomes 7B was absent and one more pair of intact chromosomes 6A was identified in Zhongke 78 (Figure 3C).
Both Zhongke 78 and Zhongke 15 were identified with many tillers, compact plant architecture, and filled seeds in morphology (Figures 4A–C, Supplementary Figure 3A). In detail, both two lines were higher in plant height and longer in spike length than their recurrent parent Jimai 22 (Table 2). Conversely, the kernel number per spike of both two lines were less than that of Jimai 22, with 26.2 of Zhongke 78 and 27.2 of Zhongke 15. In addition, we observed that the line Zhongke 15 exhibited more tillers but lower kernel weight than Jimai 22 while Zhongke 78 showed less tillers but a slightly higher kernel weight than Jimai 22 (Table 2). By inoculating the equally mixed isolates of races CYR32 and CYR34, both two lines were identified as resistant to stripe rust at the seedling stage in the greenhouse (Figure 4D). Furthermore, the adult plant stage resistance was also observed on both lines in the field of Chengdu, Sichuan Province (Figure 4E, Supplementary Figures 3A, B). In order to exclude the effect of the recurrent parent Jimai 22 on the stripe rust resistance, two susceptible cultivars, Fielder and Jimai 20, were employed to produce F2 segregation populations. We performed stripe rust resistance on 218 F2 progenies derived from the cross of Zhongke 78 and Fielder and 225 progenies derived from the cross of Zhongke 15 and Jimai 20 using the isolate of race CYR34. We identified 186 resistant progenies and 32 susceptible progenies from the segregation population derived from Zhongke 78 and Fielder. Similarly, among the F2 progenies derived from Zhongke 15 and Jimai 20, 208 progenies were identified as resistant and 17 were identified as susceptible (Table 3, Supplementary Figure 4). To facilitate the association analysis between stripe rust resistance and alien chromatin, the specific marker T14K50 was used to track the alien chromatins that derived from Zhongke 78 and Zhongke 15 (Figure 5). We found that the marker T14K50 can be only detected in the resistant progenies but not in the susceptible ones for both F2 segregation populations (Table 3, Figure 5). In other words, all F2 progenies carrying the translocated chromosome derived from Zhongke 78 and Zongke 15 were resistant to stripe rust and the translocated chromosome cannot be detected in all susceptible ones. Therefore, the stripe rust resistance was cosegregated with the alien chromatins in Zhongke 78 and Zhongke 15, which confirmed that the stripe rust resistance originated from Th. intermedium.
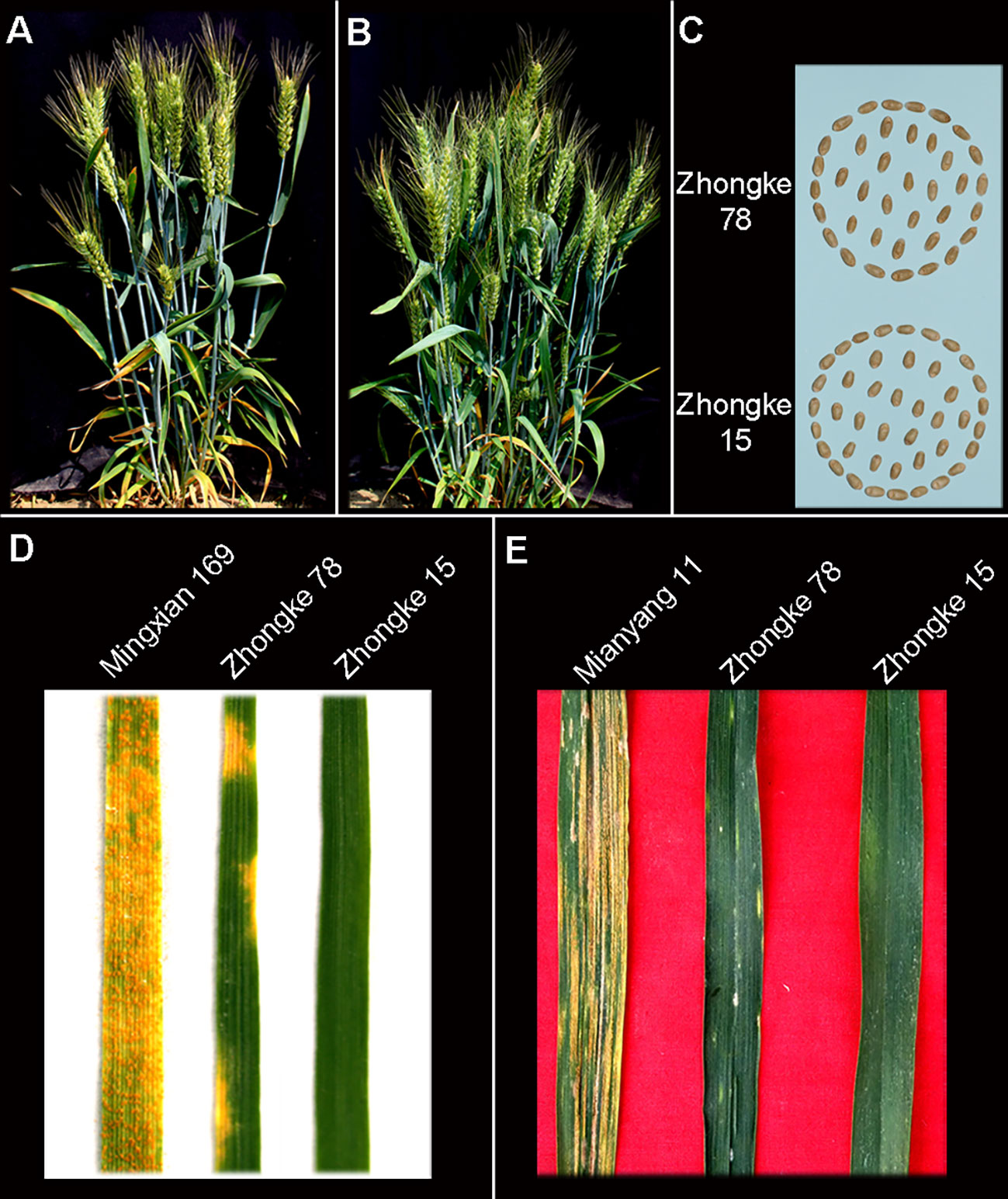
Figure 4 The integrated agronomic traits and stripe rust resistance of homozygous translocation lines Zhongke 78 and Zhongke 15. Phenotype of Zhongke 78 (A) and Zhongke 15 (B). (C) Seeds of Zhongke 78 and Zhongke 15 harvested in the field. (D) Stripe rust resistance evaluation of Zhongke 78 and Zhongke 15 at the seedling stage in a greenhouse by mixed races CYR32 and CYR34. (E) Stripe rust resistance evaluation of Zhongke 78 and Zhongke 15 at the adult plant in the field of Chengdu, Sichuan province.
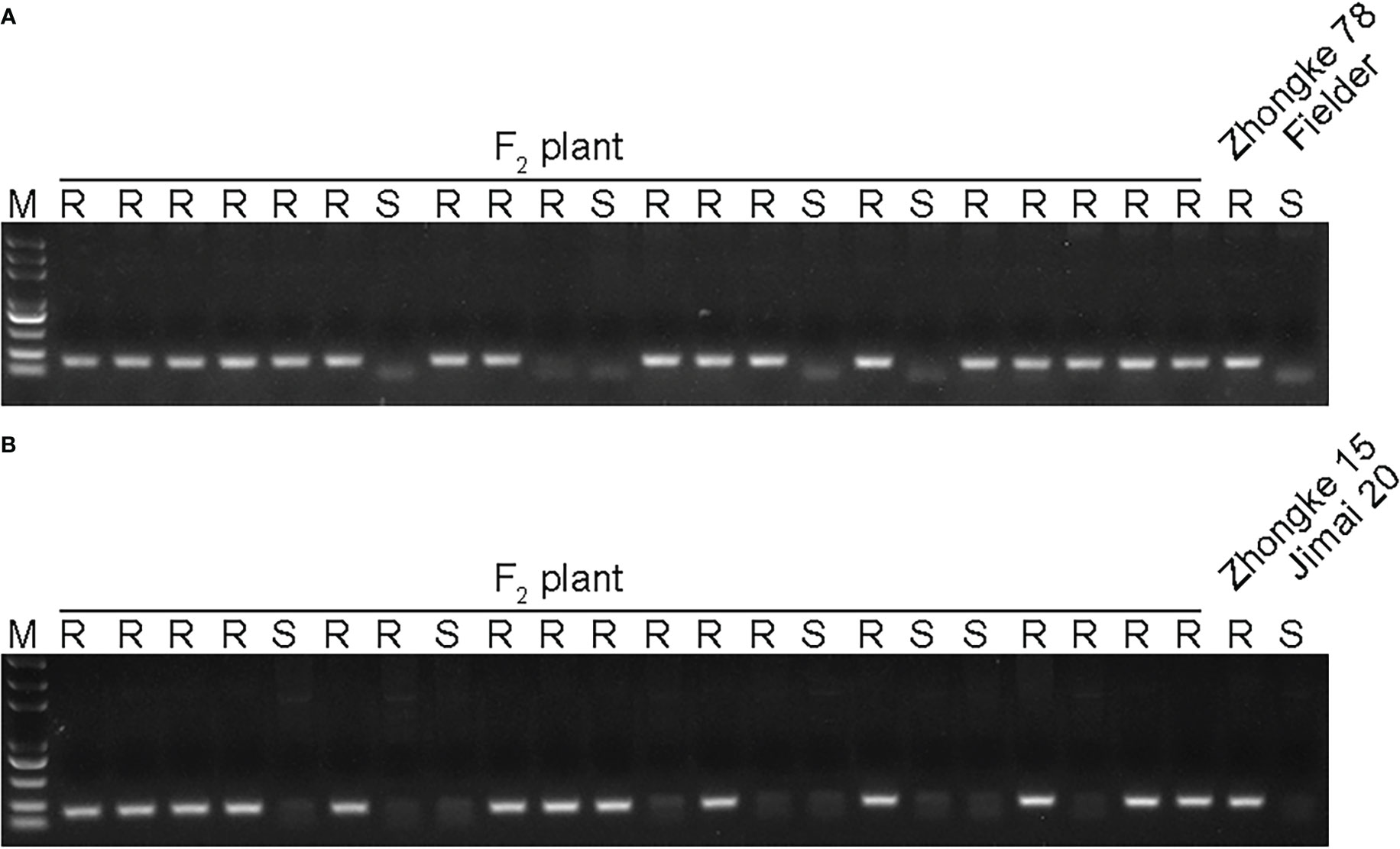
Figure 5 Detecting the alien chromatins among F2 plants derived from Zhongke 78/Field and Zhongke 15/Jimai 20 by using the specific marker T14K50. (A) Detecting the alien chromatin among F2 plants derived from the cross of Zhongke 78 and Fielder. (B) Detecting the alien chromatin among F2 plants derived from the cross of Zhongke 15 and Jimai 20. M, DNA marker. R, resistance to stripe rust. S, susceptible to stripe rust.
To develop specific markers for the alien chromosome to facilitate gene mapping, 197 wheat SSR and 88 EST markers from wheat group 7 were firstly screened. However, only three specific markers for the alien chromosome were obtained, including one SSR marker Xgwm333 and two EST markers BE404744 and BG262960 (Table 4, Figure 6A). In order to obtain more markers for the alien chromosome, we secondly annotated 2,632 de novo transcripts that only expressed in T14-44 but not in T14-42. Among them, 39 transcripts annotated as NBS-LRR protein and 94 transcripts annotated as protein kinase were used to design primers. At last, three markers from the NBS-LRR transcripts and eight markers from the protein kinase transcripts were developed (Table 4, Figure 6A). Totally, we developed 14 specific markers for the alien telocentric chromosome in TAI-14. In addition, we checked the distribution of all markers on seven translocation lines with different translocated patterns by polyacrylamide gel electrophoresis (Figure 6A). Finally, the alien chromosome was divided into four parts and a physical map was constructed with each part anchoring one-to-six markers (Figure 6B). To map the resistance gene YrT14, three wheat–Th. Intermedium lines that were susceptible to stripe rust were selected to exclude the intervals without YrT14 (Figure 7A). The stripe rust–susceptible line Zhongke 16 was proven to carry a large alien chromatin close to the centromere and including regions III and IV of the physical map revealed by molecular markers (Figures 6, 7A). Unlike Zhongke 16, the stripe rust–susceptible line Zhongke 55 was proven to carry a small alien chromatin close to the telomere and only including the regions I of the physical map (Figures 6, 7A). Therefore, based on these results, we roughly mapped the resistance gene YrT14 to the region II of the physical map (Figure 6B).
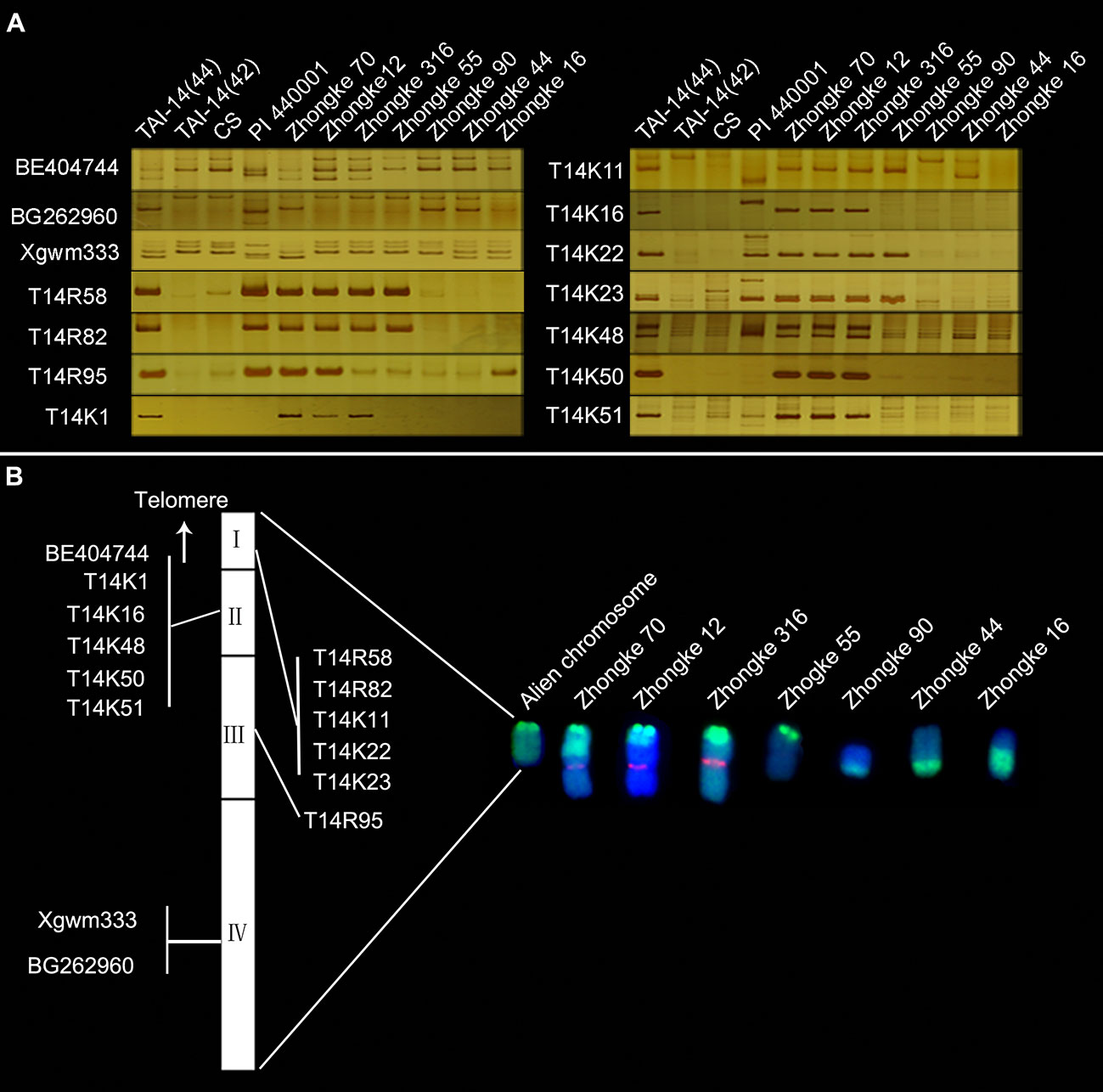
Figure 6 Specific markers and a physical map for the alien chromosome. (A) Specific markers for the alien chromosome and their distribution on seven translocation lines with different patterns. Common wheat CS and the Th. intermedium accession PI 440001 were used as control lines for exploiting specific markers for the alien chromosome in TAI-14. (B) Physical map construction for the alien chromosome.
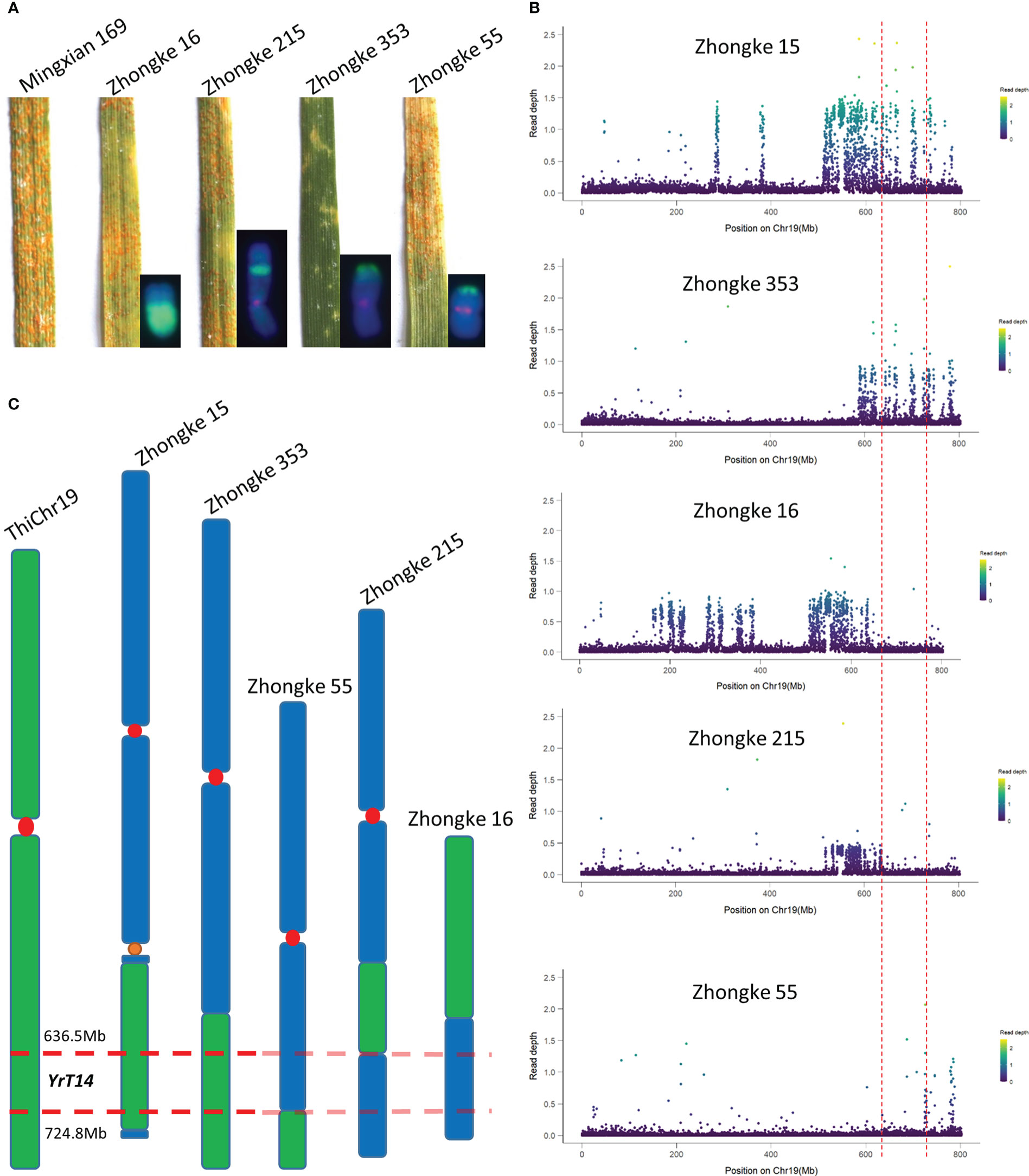
Figure 7 The physical mapping of the stripe rust resistance gene YrT14. (A) Stripe rust resistance evaluation on the wheat–Th. intermedium translocation lines Zhongke 16, Zhongke 215, Zhongke 353 and Zhongke 55. (B) Genome resequencing and the read depth analysis on Zhongke 15, Zhongke 353, Zhongke 16, Zhongke 215, and Zhongke 55. (C) Mapping YrT14 by relying on the overlaps among different wheat–Th. intermedium lines.
To further narrow down the candidate interval of YrT14, five wheat–Th. intermedium translocation lines including the said two stripe rust–susceptible ones as well as Zhongke 215, Zhongke 353, and Zhongke 15 were selected to perform genome resequencing. Compared with the line Zhongke 15, Zhongke 215 was also identified as an intercalary translocation line but susceptible to stripe rust (Figure 7A). Except for Zhongke 15 and Zhongke 78, the translocation line Zhongke 353 was also identified as resistant to stripe rust and confirmed by the association analysis between stripe rust and the alien chromatin (Figure 7A, Table S2). After mapping the reads to the reference genome, we found that the alien chromatin in our developed translocation lines was corresponding to the chromosome 19 (Chr19) of the assembled reference genome of Th. intermedium (Figure 7B). Read depth analysis revealed that the stripe rust–resistant lines Zhongke 15 and Zhongke 353 shared a common interval ranging from 586.9 to 782.0 Mb (Figure 7B). Based on the results of the stripe rust–susceptible line Zhongke 55, we excluded the intervals from 724.8 Mb to the telomere at the end of the alien chromosome (Figure 7B). Similarly, the interval from the centromere to 636.7 Mb was also excluded according to the results of the stripe rust–resistant lines Zhongke 16 and Zhongke 215 (Figure 7B). Therefore, relying on the overlap among the five above-mentioned translocation lines, the stripe rust resistance gene YrT14 was narrowed down to an approximately 88.1 Mb interval from 636.7 to 724.8 Mb on Th. intermedium Chr19 corresponding to chromosome 7J or 7Js (Figure 7C, Supplementary Figure 1).
Exploiting genes from wild relatives is an important way for wheat improvement. As a wild relative species containing many beneficial genes for both biotic and abiotic stresses, Th. intermedium has been widely used in wheat improvement because of its crossability with wheat. It was reported that several stripe rust resistance genes have been mapped on the different St chromosomes of Th. intermedium, including chromosomes 1St, 2St, 3St, and 7St (Hu et al., 2011; Song et al., 2013; Nie et al., 2019; Wang et al., 2022). Except for the St subgenome, the subgenome J or Js has also been proven to carry the resistance genes for stripe rust. A gene for stripe rust resistance in the wheat–Th. intermedium addition line Z4 was located to the short arm or in the proximal region of the long arm of chromosome 7Js (Lang et al., 2018). Two other stripe rust resistance genes Yr50 and YrL693 were reported in wheat–Th. intermedium introgression lines CH223 and L693, respectively. However, both genes were putatively derived from Th. intermedium based on molecular mapping while the Th. intermedium chromatin cannot be detected on wheat chromosomes (Liu et al., 2013; Huang et al., 2014). In this study, the genetic analysis revealed the location of a resistance gene for stripe rust on the alien chromosome in the wheat–Th. intermedium addition line TAI-14. It was demonstrated that the alien chromosome in TAI-14 was homoeologous with wheat group-7 chromosomes (Gao and Hao, 1993). This was confirmed in our study by the fact that three markers from the long arms of wheat group-7 chromosomes, Xgwm333, BE404744, and BG262960, were identified specific for the alien chromosome. We demonstrated that the alien chromosome in TAI-14 did not belong to the St subgenome by using the FISH marker St2-80 reported by Wang et al. (2017). In addition, the genome resequencing data supported that the alien telocentric chromosome was corresponding to the long arm of Chr19. More importantly, the resistance gene YrT14 was located to an interval ranging from 636.5 to 724.8 Mb, which was far away from the short arm and the proximal region of the long arm reported by Lang et al. (2018). Therefore, in this study, we demonstrated that the long arm of chromosome 7J or 7Js from Th. intermedium was also responsible for stripe rust resistance and the gene YrT14 on it was a novel gene that was not reported before.
Translocation lines made it possible to transfer useful genes from related species to wheat. Linkage drag is a key factor affecting the success of the alien gene transfer (Faris et al., 2008). No differences on agronomic traits were observed between T14-44 and T14-42 in the field (Supplementary Figure 5), which indicated that the alien telocentric chromosome did not carry genes that have deleterious effects on yield and quality. However, the loss of the alien chromosome in TAI-14 occurred to a certain extent in the self-crossing offspring, gradually transforming TAI-14 from T14-44 to T14-42 and finally resulting in losing resistance to stripe rust. Therefore, in this study, we chose to transfer the stripe rust-resistance gene YrT14 to common wheat by the mean of the translocation line.
Physical radiation can break both wheat and wild relative chromosomes at a different position in addition lines. It was reported that classic Non-homologous end joining (NHEJ) junctions produced translocation lines during Double strand break (DSB) repair (Gillert et al., 1999). We obtained 153 wheat–Th. intermedium translocation lines by irradiating the pollen of the addition line TAI-14, including 66 short alien segmental translocation lines, 70 long alien segmental translocation lines, and 17 intercalary translocation lines. Up to date, two wheat–Th. intermedium translocation lines Zhongke 15 and Zhongke 78 with good integrated agronomic traits have been selected from the backcrossing offspring. We employed F2 populations to exclude the effect of the recurrent parent Jimai 22 on the resistance to stripe rust and confirmed that the alien chromatins in both Zhongke 15 and Zhongke 78 were cosegregated with the stripe rust resistance.
China is one of the largest stripe rust epidemic regions in the world (Stubbs, 1988; Wan et al., 2004). It was reported that the CYR32 and CYR33 races were accounted the highest frequencies among all tested races from 1997 to 2007 in China because of their parasitic fitness on leading commercial cultivars (Wan et al., 2007; Wang et al., 2009). The appearance of CYR34 has overcome the resistance of many wheat cultivars with Yr26 located on chromosome 1BS (Ma et al., 2001; Wang et al., 2008; Han et al., 2015). It was predicted to develop into a new predominant race and may cause stripe rust epidemics in China (Bai et al., 2018; Wang et al., 2019). Therefore, it is of great importance to exploit new resistant sources that can protect wheat from races CYR32, CYR33, and CYR34. In our study, the gene YrT14 was identified highly resistant to all three races. Therefore, Zhongke 15 and Zhongke 78 should be incorporated into the pool of germplasm as novel valuable resources for wheat stripe rust–resistant breeding.
The usefulness of the translocation lines depended on whether the alien fragment can compensate for the replaced wheat segments as well as the linkage drag (Faris et al., 2008). As the alien chromosome in TAI-14 was homoeologous to the wheat group-7 chromosome, it may compensate the loss of 7B chromosome to some extent in Zhongke 78. Except for compensated translocation lines, the intercalary translocation line with an alien chromatin carrying targeted genes and no deleterious genes inserting into a wheat chromosome and without the loss of any wheat chromatin would be an ideal type. A wheat–Agropyron cristatum intercalary translocation line Pubing 3035 was reported to enhance thousand-grain weight and spike length in two populations (Zhang et al., 2015). In our study, Zhongke 15 was identified as an intercalary translocation line without the loss of any wheat chromosome. More interestingly, we found that a segment of the alien chromatin was inserted into the satellite of chromosome 6B, making the satellite much bigger than before. Several intercalary translocation lines between wheat and wild relatives have been reported (Mukai et al., 1993; Chen et al., 2008; Luan et al., 2010; <xr rid="r8">Chen et al., 2012</xr>). However, this is the first time to our knowledge that an intercalary translocation into the satellite region has been reported.
In China, the 1BL/1RS translocation lines were introduced from Europe and Russia and widely used in wheat breeding because of their stripe rust and powdery mildew resistance. Several important genes were found located on the satellite of chromosome 1RS, such as the resistance genes against powdery mildew (Pm8), stripe rust (Yr9), leaf rust (Lr26), and stem rust (Sr31) (Mago et al., 2005; Sharma et al., 2011). To a certain extent, the newly formed satellite with YrT14 in Zhongke 15 is similar to the satellite of chromosome 1R. All these suggested that the intercalary translocation line Zhongke 15 had great potential for wheat stripe rust resistance breeding.
GISH and FISH are two powerful cytogenetic techniques for identifying alien chromatin on a wheat background. However, they cannot distinguish the accurate size of the alien fragment at the molecular level. In this study, by resequencing five wheat–Th. intermedium translocation lines and relying on their overlap, we mapped the stripe rust resistance gene YrT14 to an approximately 88.1 Mb interval ranging from 636.7 to 724.8 Mb on Th. intermedium Chr19. Although the candidate interval was narrowed down, it was still too large to exploit the gene YrT14. To resolve the issues, on the one hand, we could create a mutant to produce genetic populations to fine-map the resistance gene YrT14. On the other hand, with the insertion of the alien chromatin, the translocated chromosome T6B/Thi/6B in the line Zhongke 15 was obviously larger in size than other chromosomes, which opened up the possibility of using mutants in conjunction with chromosome sorting and sequencing to facilitate the cloning of the candidate gene (Dolezel et al., 2014; Sánchez-Martín et al., 2016). In addition, the read depth analysis in this study revealed great differences between the alien chromatin in TAI-14 and Chr19 in the reference genome, which might be caused by the difference between Th. intermedium accessions and the incomplete reference genome. Therefore, the stripe rust–susceptible mutant and a more complete reference genome of Th. intermedium are the prerequisites for the cloning of YrT14.
In this study, the wheat–Th. intermedium addition line TAI-14 was identified as resistant to three dominant stripe rust races CYR32, CYR33, and CYR34. Furthermore, we demonstrated the presence of a novel resistance gene (YrT14) for stripe rust on the alien chromosome. To transfer the resistance gene to common wheat, γ irradiation was performed on the line TAI-14. A great number of wheat–Th. intermedium translocation lines were developed, including 66 short alien segmental translocation lines, 70 long alien segmental translocation lines, and 17 intercalary translocation lines. Among them, the long alien segmental translocation line Zhongke 78 and the intercalary translocation line Zhongke 15 were identified as resistant to stripe rust and showing good integrated agronomic traits. Cytological analysis revealed that the alien chromatin nearly replaced the long arm of chromosome 6A in Zhongke 78, while a segment of the alien chromatin was inserted into the satellite of chromosome 6B in Zhongke 15. Both lines could be used as novel resources for wheat stripe rust resistance breeding. In addition, by screening SSR and EST markers as well as the markers developed from RNA-Seq data, 14 specific markers were identified for the alien chromosome and a physical map was constructed. The linked marker T14K50 can be used for molecular marker–assisted breeding. Finally, based on the karyotype, reaction to stripe rust, and genome resequencing data of different wheat–Th. intermedium translocation lines, the stripe rust resistance gene YrT14 was located to an 88.1 Mb interval ranging from 636.7 to 724.8 Mb on Th. intermedium chromosome 19.
The wheat–Th. intermedium translocation lines developed in this study were available from the corresponding author to be required for academic study.
The raw sequencing data was deposited in NCBI database under number PRJNA911033. All other data are included in the article.
XG, WY, and FH planned and designed the research. XG, YH, JW, SF, CW, MW, CZ, XH, and TW performed experiments, conducted fieldwork, and performed evaluation on stripe rust resistance. XG and FH wrote the manuscript with contributions from all authors. All authors contributed to the article and approved the submitted version.
This research was funded by the Major Program of National Agricultural Science and Technology of China (NK20220607).
We thank Yang Zhou (Institute of Crop Science, Chinese Academy of Agricultural Sciences) and Xiangqi Zhang (Institute of Genetics and Developmental Biology, Chinese Academic of Sciences) for their good suggestions on this research. We thank Dengcai Liu (Triticeae Research Institute, Sichuan Agricultural University) for help on stripe rust resistance evaluation. We thank Nathan Han (Washington University in St. Louis) for critically reading and editing this manuscript. We would like to thank Dr. Chaolan Fan (Institute of Genetics and Developmental Biology, Chinese Academic of Sciences) for critically reading this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1135321/full#supplementary-material
Allard, R. (1996). “Genetic basis of the evolution of adaptedness in plants,” in Adaptation in plant breeding (Springer), 1–11.
Bai, B. B., Liu, T. G., Liu, B., Gao, L., Chen, W. Q. (2018). High relative parasitic fitness of G22 derivatives is associated with the epidemic potential of wheat stripe rust in China. Plant Dis. 102 (3), 483–487. doi: 10.1094/pdis-04-17-0511-sr
Bai, G., Shaner, G. (1994). Scab of wheat: Prospects for control. Plant Dis. 78 (8), 760–766. doi: 10.1094/PD-78-0760
Benito, C., Figueiras, A. M., González-Jaén, M. T. (1987). Location of genes coding isozyme markers on Aegilops umbellulata chromosomes adds data on homoeology among Triticeae chromosomes. Theor. Appl. Genet. 73 (4), 581–588. doi: 10.1007/BF00289198
Bennett, F. G. (1984). Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol. 33 (3), 279–300. doi: 10.1111/j.1365-3059.1984.tb01324.x
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Chang, Z. J., Zhang, X. J., Yang, Z. J., Zhan, H. X., Li, X., Liu, C., et al. (2010). Characterization of a partial wheat-Thinopyrum intermedium amphiploid and its reaction to fungal diseases of wheat. Hereditas 147 (6), 304–312. doi: 10.1111/j.1601-5223.2010.02156.x
Charmet, G. (2011). Wheat domestication: Lessons for the future. C. R. Biol. 334 (3), 212–220. doi: 10.1016/j.crvi.2010.12.013
Chen, Q. (2005). Detection of alien chromatin introgression from Thinopyrum into wheat using s genomic DNA as a probe-a landmark approach for Thinopyrum genome research. Cytogenetic Genome Res. 109 (1-3), 350–359. doi: 10.1159/000082419
Chen, S., Chen, P., Wang, X. (2008). Inducement of chromosome translocation with small alien segments by irradiating mature female gametes of the whole arm translocation line. Sci. China C. Life Sci. 51 (4), 346–352. doi: 10.1007/s11427-008-0048-2
Chen, Q., Conner, R. L., Ahmad, F., Laroche, A., Fedak, G., Thomas, J. B. (1998a). Molecular characterization of the genome composition of partial amphiploids derived from Triticum aestivum × Thinopyrum ponticum and T. aestivum × Th. intermedium as sources of resistance to wheat streak mosaic virus and its vector, Aceria tosichella. Theor. Appl. Genet. 97 (1), 1–8. doi: 10.1007/s001220050860
Chen, Q., Conner, R. L., Laroche, A., Ji, W. Q., Armstrong, K. C., Fedak, G. (1999). Genomic in situ hybridization analysis of Thinopyrum chromatin in a wheat-Th. intermedium partial amphiploid and six derived chromosome addition lines. Genome 42 (6), 1217–1223. doi: 10.1139/g99-090
Chen, Q., Conner, R. L., Laroche, A., Thomas, J. B. (1998b). Genome analysis of Thinopyrum intermedium and Thinopyrum ponticum using genomic in situ hybridization. Genome 41 (4), 580–586. doi: 10.1139/g98-055
Chen, Q., Conner, R. L., Li, H., Sun, S., Ahmad, F., Laroche, A., et al. (2003). Molecular cytogenetic discrimination and reaction to wheat streak mosaic virus and the wheat curl mite in zhong series of wheat-Thinopyrum intermedium partial amphiploids. Genome 46 (1), 135–145. doi: 10.1139/g02-109
Chen, P., You, C., Hu, Y., Chen, S., Zhou, B., Cao, A., et al. (2012). Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol. Breed. 31 (2), 477–484. doi: 10.1007/s11032-012-9804-x
Consortium (IWGSC) TIWGS, Investigators IR principal, Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191.
Cui, Y., Xing, P., Qi, X., Bao, Y., Wang, H., Wang, R. R., et al. (2021). Characterization of chromosome constitution in three wheat-Thinopyrum intermedium amphiploids revealed frequent rearrangement of alien and wheat chromosomes. BMC Plant Biol. 21 (1), 129. doi: 10.1186/s12870-021-02896-9
Danilova, T. V., Zhang, G., Liu, W., Friebe, B., Gill, B. S. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of a wheat streak mosaic virus and Triticum mosaic virus resistance gene Wsm3 from Thinopyrum intermedium to wheat. Theor. Appl. Genet. 130 (3), 549–556. doi: 10.1007/s00122-016-2834-8
Dolezel, J., Vrana, J., Capal, P., Kubalakova, M., Buresova, V., Simkova, H. (2014). Advances in plant chromosome genomics. Biotechnol. Adv. 32 (1), 122–136. doi: 10.1016/j.biotechadv.2013.12.011
Faris, J. D., Xu, S., Cai, X., Friesen, T. L., Jin, Y. (2008). Molecular and cytogenetic characterization of a durum wheat-Aegilops speltoides chromosome translocation conferring resistance to stem rust. Chromosome Res. 16 (8), 1097–1105. doi: 10.1007/s10577-008-1261-3
Gao, M., Hao, S. (1993). Isozymic studies of the homoeology between Triticum aestivum and Elytrigia intermedia chromosomes in set I wheat-wheatgrass alien addition lines. Acta Genetica Sin. 20 (5), 462–467.
Gillert, E., Leis, T., Repp, R., Reichel, M., HoÈsch, A., Breitenlohner, I., et al. (1999). A DNA damage repair mechanism is involved in the origin of chromosomal translocations t (4; 11) in primary leukemic cells. Oncogene 18 (33), 4663. doi: 10.1038/sj.onc.1202842
Guo, X., Shi, Q., Wang, J., Hou, Y., Wang, Y., Han, F. (2015). Characterization and genome changes of new amphiploids from wheat wide hybridization. J. Genet. Genomics 42 (8), 459–461. doi: 10.1016/j.jgg.2015.06.006
Guo, X., Su, H., Shi, Q., Fu, S., Wang, J., Zhang, X., et al. (2016). De novo centromere formation and centromeric sequence expansion in wheat and its wide hybrids. PloS Genet. 12 (4), e1005997. doi: 10.1371/journal.pgen.1005997
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8 (8), 1494–1512. doi: 10.1038/nprot.2013.084
Han, F., He, M., Hao, S., Ma, Y., Xin, Z. (1998). Variation of the wheatgrass chromosomes in wheat-wheatgrass disomic addition line TAI-14 revealed by fluorescence in situ hybridization. Acta Botanica Sin. 40 (1), 33–36.
Han, F., Liu, B., Fedak, G., Liu, Z. (2004). Genomic constitution and variation in five partial amphiploids of wheat-Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor. Appl. Genet. 109 (5), 1070–1076. doi: 10.1007/s00122-004-1720-y
Han, D., Wang, Q., Chen, X., Zeng, Q., Wu, J., Xue, W., et al. (2015). Emerging Yr26-virulent races of Puccinia striiformis f. tritici are threatening wheat production in the sichuan basin, China. Plant Dis. 99 (6), 754–760. doi: 10.1094/pdis-08-14-0865-re
He, R., Chang, Z., Yang, Z., Yuan, Z., Zhan, H., Zhang, X., et al. (2009). Inheritance and mapping of powdery mildew resistance gene Pm43 introgressed from Thinopyrum intermedium into wheat. Theor. Appl. Genet. 118 (6), 1173–1180. doi: 10.1007/s00122-009-0971-z
He, M., Xu, Z., Zou, M., Zhang, H., Chen, D., Piao, Z., et al. (1989). The establishment of two sets of alien addition lines of wheat-wheatgrass. Sci. China (Ser. B.) 32 (6), 695–705.
Hoisington, D., Khairallah, M., Reeves, T., Ribaut, J. M., Skovmand, B., Taba, S., et al. (1999). Plant genetic resources: What can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci. U. S. A. 96 (11), 5937–5943. doi: 10.1073/pnas.96.11.5937
Hu, L., Li, G., Zeng, Z., Chang, Z., Liu, C., Zhou, J., et al. (2011). Molecular cytogenetic identification of a new wheat-Thinopyrum substitution line with stripe rust resistance. Euphytica 177 (2), 169–177. doi: 10.1007/s10681-010-0216-x
Huang, Q., Li, X., Chen, W., Xiang, Z., Zhong, S., Chang, Z., et al. (2014). Genetic mapping of a putative Thinopyrum intermedium-derived stripe rust resistance gene on wheat chromosome 1B. Theor. Appl. Genet. 127 (4), 843–853. doi: 10.1007/s00122-014-2261-7
Kantarski, T., Larson, S., Zhang, X., DeHaan, L., Borevitz, J., Anderson, J., et al. (2017). Development of the first consensus genetic map of intermediate wheatgrass (Thinopyrum intermedium) using genotyping-by-sequencing. Theor. Appl. Genet. 130 (1), 137–150. doi: 10.1007/s00122-016-2799-7
Kato, A., Lamb, J. C., Albert, P. S., Danilova, T., Han, F., Gao, Z., et al. (2011). Chromosome painting for plant biotechnology. Methods Mol. Biol. 701, 67–96. doi: 10.1007/978-1-61737-957-4_4
Kato, A., Lamb, J. C., Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U. S. A. 101 (37), 13554–13559. doi: 10.1073/pnas.0403659101
Kolmer, J. A. (2005). Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 8 (4), 441–449. doi: 10.1016/j.pbi.2005.05.001
Lang, T., La, S., Li, B., Yu, Z., Chen, Q., Li, J., et al. (2018). Precise identification of wheat- Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 61 (3), 177–185. doi: 10.1139/gen-2017-0229
Li, H. J., Arterburn, M., Jones, S. S., Murray, T. D. (2005). Resistance to eyespot of wheat, caused by Tapesia yallundae, derived from Thinopyrum intermedium homoeologous group 4 chromosome. Theor. Appl. Genet. 111 (5), 932–940. doi: 10.1007/s00122-005-0025-0
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25 (14), 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, G., Wang, H., Lang, T., Li, J., La, S., Yang, E., et al. (2016). New molecular markers and cytogenetic probes enable chromosome identification of wheat-Thinopyrum intermedium introgression lines for improving protein and gluten contents. Planta 244 (4), 865–876. doi: 10.1007/s00425-016-2554-y
Liu, J., Chang, Z., Zhang, X., Yang, Z., Li, X., Jia, J., et al. (2013). Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor. Appl. Genet. 126 (1), 265–274. doi: 10.1007/s00122-012-1979-3
Liu, Z., Wang, R. (1993). Genome analysis of Elytrigia caespitosa, Lophopyrum nodosum, Pseudoroegneria geniculata ssp. scythica, and Thinopyrum intermedium (Triticeae: Gramineae). Genome 36 (1), 102–111. doi: 10.1139/g93-014
Luan, Y., Wang, X., Liu, W., Li, C., Zhang, J., Gao, A., et al. (2010). Production and identification of wheat-Agropyron cristatum 6P translocation lines. Planta 232 (2), 501–510. doi: 10.1007/s00425-010-1187-9
Ma, J., Zhou, R., Dong, Y., Wang, L., Wang, X., Jia, J. (2001). Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum l. using microsatellite markers. Euphytica 120 (2), 219–226. doi: 10.1023/A:1017510331721
Mago, R., Miah, H., Lawrence, G. J., Wellings, C. R., Spielmeyer, W., Bariana, H. S., et al. (2005). High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor. Appl. Genet. 112 (1), 41–50. doi: 10.1007/s00122-005-0098-9
Mukai, Y., Friebe, B., Hatchett, J., Yamamoto, M., Gill, B. (1993). Molecular cytogenetic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 102 (2), 88–95. doi: 10.1007/BF00356025
Murray, M. G., Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 (19), 4321–4326. doi: 10.1093/nar/8.19.4321
Nie, L., Yang, Y., Zhang, J., Fu, T. (2019). Disomic chromosome addition from Thinopyrum intermedium to bread wheat appears to confer stripe rust resistance. Euphytica 215 (3), 56. doi: 10.1007/s10681-019-2376-7
Ohm, H., Anderson, J., Sharma, H., Ayala, L. (2005). Registration of yellow dwarf viruses resistant wheat germplasm line P961341. Crop Sci. 45 (2), 805. doi: 10.2135/cropsci2005.0805
Pedersen, B. S., Quinlan, A. R. (2018). Mosdepth: Quick coverage calculation for genomes and exomes. Bioinformatics 34 (5), 867–868. doi: 10.1093/bioinformatics/btx699
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17 (1), 221. doi: 10.1186/s13059-016-1082-1
Sharma, S., Xu, S., Ehdaie, B., Hoops, A., Close, T. J., Lukaszewski, A. J., et al. (2011). Dissection of QTL effects for root traits using a chromosome arm-specific mapping population in bread wheat. Theor. Appl. Genet. 122 (4), 759–769. doi: 10.1007/s00122-010-1484-5
Somers, D. J., Isaac, P., Edwards, K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum l.). Theor. Appl. Genet. 109 (6), 1105–1114. doi: 10.1007/s00122-004-1740-7
Song, X. J., Li, G. R., Zhan, H. X., Liu, C., Yang, Z. J. (2013). Molecular identification of a new wheat-Thinopyrum intermedium ssp. trichophorum addition line for resistance to stripe rust. Cereal Res. Commun. 41 (2), 211–220. doi: 10.1556/CRC.2013.0009
Stubbs, R. (1988). “Pathogenicity analysis of yellow (stripe) rust of wheat and its significance in a global context,” in Breeding strategies for resistance to the rusts of wheat (Mexico: CIMMYT), 23–38.
Tang, Z., Yang, Z., Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55 (3), 313–318. doi: 10.1007/s13353-014-0215-z
Vladova, R., Petkolicheva, K. (1996). Use of isozymes and seed storage proteins as chromosome markers in the study of tetraploid triticales. Cereal Res. Commun. 24 (4), 417–424.
Wan, A., Chen, X., He, Z. (2007). Wheat stripe rust in China. Aust. J. Agric. Res. 58 (6), 605. doi: 10.1071/ar06142
Wan, A., Zhao, Z., Chen, X., He, Z., Jin, S., Jia, Q., et al. (2004). Wheat stripe rust epidemic and virulence of puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 88 (8), 896–904. doi: 10.1094/PDIS.2004.88.8.896
Wang, B., Li, G., Li, Q., Wang, F., Shi, Y., Liu, Q., et al. (2009). Parasitic fitness of the major epidemic strains of puccinia striiformis f. sp. tritici in China. Acta Phytopathol. Sin. 103 (1), 82–87.
Wang, L., Shi, Q., Su, H., Wang, Y., Sha, L., Fan, X., et al. (2017). St2-80: A new FISH marker for St genome and genome analysis in Triticeae. Genome 60 (7), 553–563. doi: 10.1139/gen-2016-0228
Wang, L., Tang, X., Wu, J., Shen, C., Dai, M., Wang, Q., et al. (2019). Stripe rust resistance to a burgeoning puccinia striiformis f. sp. tritici race CYR34 in current Chinese wheat cultivars for breeding and research. Euphytica 215 (4), 68. doi: 10.1007/s10681-019-2383-8
Wang, Y., Wang, H. (2016). Characterization of three novel wheat-Thinopyrum intermedium addition lines with novel storage protein subunits and resistance to both powdery mildew and stripe rust. J. Genet. Genomics 43 (1), 45–48. doi: 10.1016/j.jgg.2015.10.004
Wang, S., Wang, C., Feng, X., Zhao, J., Deng, P., Wang, Y., et al. (2022). Molecular cytogenetics and development of St-chromosome-specific molecular markers of novel stripe rust resistant wheat-Thinopyrum intermedium and wheat-Thinopyrum ponticum substitution lines. BMC Plant Biol. 22 (1), 111. doi: 10.1186/s12870-022-03496-x
Wang, C., Zhang, Y., Han, D., Kang, Z., Li, G., Cao, A., et al. (2008). SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica 159 (3), 359–366. doi: 10.1007/s10681-007-9524-1
Zhan, H., Zhang, X., Li, G., Pan, Z., Hu, J., Li, X., et al. (2015). Molecular characterization of a new wheat-Thinopyrum intermedium translocation line with resistance to powdery mildew and stripe rust. Int. J. Mol. Sci. 16 (1), 2162–2173. doi: 10.3390/ijms16012162
Keywords: wheat, Thinopyrum intermedium, translocation line, stripe rust, gene mapping
Citation: Guo X, Huang Y, Wang J, Fu S, Wang C, Wang M, Zhou C, Hu X, Wang T, Yang W and Han F (2023) Development and cytological characterization of wheat–Thinopyrum intermedium translocation lines with novel stripe rust resistance gene. Front. Plant Sci. 14:1135321. doi: 10.3389/fpls.2023.1135321
Received: 31 December 2022; Accepted: 13 February 2023;
Published: 24 February 2023.
Edited by:
Azahara Carmen Martin, John Innes Centre, United KingdomReviewed by:
Mahmoud Said, Institute of Experimental Botany of the Czech Academy of Sciences, CzechiaCopyright © 2023 Guo, Huang, Wang, Fu, Wang, Wang, Zhou, Hu, Wang, Yang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangpu Han, ZnBoYW5AZ2VuZXRpY3MuYWMuY24=; Wuyun Yang, eWFuZ3d1eXVuQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.