- Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu, China
Digitaria ciliaris var. chrysoblephara is one of the most competitive and problematic grass weeds in China. Metamifop is an aryloxyphenoxypropionate (APP) herbicide that inhibits the activity of acetyl-CoA carboxylase (ACCase) of sensitive weeds. Following the introduction of metamifop to China in 2010, it has been continuously used in rice paddy fields, thereby substantially increasing selective pressure for resistant D. ciliaris var. chrysoblephara variants. Here, populations of D. ciliaris var. chrysoblephara (JYX-8, JTX-98, and JTX-99) were observed to be highly resistant to metamifop, with resistance index (RI) values of 30.64, 14.38, and 23.19, respectively. Comparison of resistant and sensitive population ACCase gene sequences revealed that a single nucleotide substitution from TGG to TGC resulted in an amino acid substitution from tryptophan to cysteine at position 2,027 in the JYX-8 population. No corresponding substitution was observed for JTX-98 and JTX-99 populations. The ACCase cDNA of D. ciliaris var. chrysoblephara was successfully obtained by PCR and RACE methods, representing the first amplification of full length ACCase cDNA from Digitaria spp. Investigation of the relative expressions of ACCase gene revealed the lack of significant differences between sensitive and resistant populations before and after herbicide treatments. ACCase activities in resistant populations were less inhibited than in sensitive populations and recovered to the same or even higher levels compared to untreated plants. Whole-plant bioassays were also conducted to assess resistance to other ACCase inhibitors, acetolactate synthase (ALS) inhibitors, auxin mimic herbicide, and protoporphyrinogen oxidase (PPO) inhibitor. Cross-resistance and some multi-resistance were observed in the metamifop-resistant populations. This study is the first to focus on the herbicide resistance of D. ciliaris var. chrysoblephara. These results provide evidence for a target-site resistance mechanism in metamifop-resistant D. ciliaris var. chrysoblephara, while providing a better understanding of cross- and multi-resistance characteristics of resistant populations that will help in the management of herbicide-resistant D. ciliaris var. chrysoblephara.
1. Introduction
Digitaria spp. are annual gramineae weeds that infest turfgrass, roadsides, wastelands, and crop systems like Zea mays, Glycine max, and Saccharum officinarum, among others (Basak et al., 2019). They are globally widely distributed in tropical, subtropical, and temperate regions, including in the eastern, northern, northwestern, and northeastern areas of China. Labor shortages and agricultural mechanization have led to the popularization of dry direct-seeding rice cultivation due to its high potentials for resource conservation and economic returns (Matloob et al., 2015; Rao et al., 2017). Dry direct-seeding rice cultivation accounts for over 35% of rice cultivation in China and has become one of the primary methods of rice cultivation in the Jiangsu Province. Dry-wet changes of soils in dry direct-seeding rice fields benefit weeds growth. Various weeds are present in fields that have larger occurrence and longer symbiotic times, thereby seriously threatening the yields and quality of rice. Digitaria spp. weeds have consequently become the dominant weeds in dry direct-seeding rice fields of the Jiangsu Province due to their adaptation to changes in local cropping systems and ecological environments (Wang et al., 2019).
Acetyl-CoA carboxylase (ACCase) inhibitors are one of the most widely used and important classes of herbicides used to control grass in rice fields. ACCase inhibitors can be divided into three types based on differences in their active components, including aryloxyphenoxypropionate (APP), cyclohexenone (CHD), and phenylpyrazole (PPZ) types (Fang et al., 2020). Plant ACCase catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, thereby providing substrates for the synthesis of fatty acids and numerous secondary metabolites (Adina-Zada et al., 2012). Two forms of ACCase are typically present in plants, including heteromeric and homomeric forms (Takano et al., 2020). Poaceae plants possess homomeric ACCase, while dicotyledonous plants have homomeric form in the cytoplasm and heteromeric form in the plastids. Consequently, ACCase-inhibiting herbicides exhibit specific activities against gramineae plants that can selectively inhibit homomeric plastidic ACCase only found in monocots, but do not inhibit heteromeric plastic nor homomeric cytosolic forms, allowing dicots to become tolerant to them (Kukorelli et al., 2013; Takano et al., 2020).
Previous studies have shown that the application of ACCase inhibitors leads to the gradual development of resistance after continuous use over 6 to 10 years (Huang et al., 2003), suggesting a relatively high risk of herbicide resistance. Mechanisms of weed resistance can be divided into target-site resistance (TSR) and non-target-site resistance (NTSR). TSR to ACCase inhibitors is primarily associated with changes in amino acids caused by mutations of ACCase gene, leading to changes in proteins that bind to herbicides, and ultimately leading to the resistance of weeds to herbicides. Sixteen amino acid substitutions at seven sites have been identified in the carboxyl-transferase (CT) domain of plastidic ACCase, including: Ile-1781-Leu, Ile-1781-Val, Ile-1781-Thr, Trp-1999-Cys, Trp-1999-Leu, Trp-1999-Ser, Trp-2027-Cys, Trp-2027-Ser, Ile-2041-Asn, Ile-2041-Val, Ile-2041-Thr, Asp-2078-Gly, Asp-2078-Glu, Cys-2088-Arg, and Gly-2096-Ala, and Gly-2096-Ser (Collavo et al., 2011; Kaundun et al., 2013a; Kaundun et al., 2013b; Papapanagiotou et al., 2015; Guo et al., 2017; Li et al., 2017; Wang et al., 2021). The overexpression of ACCase gene was also identified as a TSR mechanism that effectively increases the amount of target protein to mitigate the toxic effects of herbicide on weeds, as has been primarily shown for glyphosate resistance (Kupper et al., 2017; Zhang et al., 2018). However, few studies have evaluated ACCase gene expression involved in herbicide resistance. Notably, Laforest et al. observed that the overexpression of ACCase gene led to the resistance of Digitaria sanguinalis to ACCase inhibitors (Laforest et al., 2017). NTSR mechanisms are more complicated and include reduced penetration and translocation, sequestration, and enhanced herbicide metabolism (Delye et al., 2011a). These mechanisms can confer complex cross-resistance to herbicides via different modes of action (Yu and Powles, 2014). Further, various enzymes are involved in conferring non-target-site resistance to herbicides, including cytochrome P450 monooxygenases and gluthathione-S-transferases (Brazier et al., 2002).
Metamifop is a type of APP herbicide that has been used continuously following its introduction to China. The herbicide features advantages of low toxicity, high efficiency, environmental safety, and excellent miscibility, among other characteristics. It is a common agent used to control most annual gramineae weeds and exhibits good control of Digitaria spp. weeds. Continuous and extensive use of metamifop has led to increased selection pressures on weeds, eventually leading to a serious risk of herbicide resistance. Analogously, Echinochloa spp. and Leptochloa chinensis have developed resistance to metamifop in China and elsewhere (Won et al., 2014). Mutations of ACCase gene corresponding to Ile-1781-Leu, Trp-1999-Ser, and Trp-2027-Cys are the most common TSR mechanisms identified in metamifop-resistant Echinochloa spp. in China. Furthermore, NTSR mechanisms of Echinochloa crus-galli to metamifop have also been identified (Xia et al., 2016). Moreover, it has been reported that Leptochloa chinensis developed resistance to metamifop due to the ACCase gene mutation Trp-2027-Ser (Yuan et al., 2021). Few studies have evaluated metamifop resistance in D. ciliaris. Yu et al. observed that D. ciliaris can be resistant to sethoxydim and cross-resistant to fenoxaprop and fluazifop (Yu et al., 2017). Further, Basak et al. observed that the Ile-1781-Leu mutation of the ACCase gene led to pinoxaden resistance (Basak et al., 2019). However, herbicide-resistant D. ciliaris var. chrysoblephara populations have not yet been observed in China.
In this study, three D. ciliaris var. chrysoblephara populations that are resistant to metamifop were collected from dry direct seeding rice fields in Jiangsu Province, China. The aims of the present study were consequently to: (1) determine the level of resistance to metamifop in putatively resistant D. ciliaris var. chrysoblephara populations; (2) identify the mechanisms responsible for metamifop resistance in D. ciliaris var. chrysoblephara based on ACCase gene sequencing, gene expression, and enzyme activity; and (3) characterize cross- and multi-resistance of the resistant populations to various herbicides with different modes of action.
2. Materials and methods
2.1. Plant materials
Seeds of D. ciliaris var. chrysoblephara suspected of being resistant to metamifop were collected from dry direct-sown rice fields in October 2020 (Table 1). Seeds of plants from each field were pooled to represent a population. JYX-8, JTX-98, and JTX-99 were presumed to be resistant populations and JSS-19 was presumed to be a sensitive population based on single dose assay pre-experiments. D. ciliaris var. chrysoblephara seeds were germinated on filter paper in Petri dishes in June 2021 over a 12 h photoperiod with 2,500 xL lighting and incubation at 30/25°C for several days. Fifteen seedlings were then transplanted to a plastic pot (9.5 cm diameter and 16 cm height) containing loam soil and grown in an incubator under the above-mentioned conditions.
2.2. Dose response to metamifop
Dose response experiments were performed when the seedlings reached the four-leaf stage. Presumed resistant and sensitive populations were sprayed using a 3WPSH-500D type bioassay spray tower (Nanjing Institute of Agricultural Mechanization, Ministry of Agriculture and Rural Affairs) using a disc diameter of 50 cm, a nozzle diameter of 0.3 mm, a spray pressure of 0.3 MPa, a droplet diameter of 100 µm, and a sprinkler flow of 90 mL·min−1. Metamifop doses of 0-, 0.03125-, 0.0625-, 0.125-, 0.25-, 0.5-, 1-, and 2-fold the recommended dose (120 g a.i ha−1) were applied to the sensitive population, while 0-, 0.25-, 0.5-, 1-, 2-, 4-, 8-, and 16-fold the recommended dose were applied to the resistant populations. Each pot with 15 seedlings was set up as a replicate. Each treatment comprised four replicates and the experiment was conducted twice.
The above-ground fresh weights of plants were measured 21 days after treatment. Data were pooled since no significant differences between the two repeated experiments were identified based on t-test (p< 0.05) by ANOVA (SPSS v.16.0). The SigmaPlot 12.0 software was used to calculate the GR50 values (herbicide dose causing 50% growth reduction) for different populations of D. ciliaris var. chrysoblephara. A log-logistic model was used to analyze the test data, with the fitting equation as follows:
In the equation, y is the fresh weight percentage of the control, c is the lower limit, d is the upper limit, b is the slope, and x is the herbicide dose (Seefeldt et al., 1995). The resistance index (RI) was obtained based on the GR50 values of the resistant population/GR50 of the sensitive population.
2.3. ACCase CT domain and cDNA sequencing
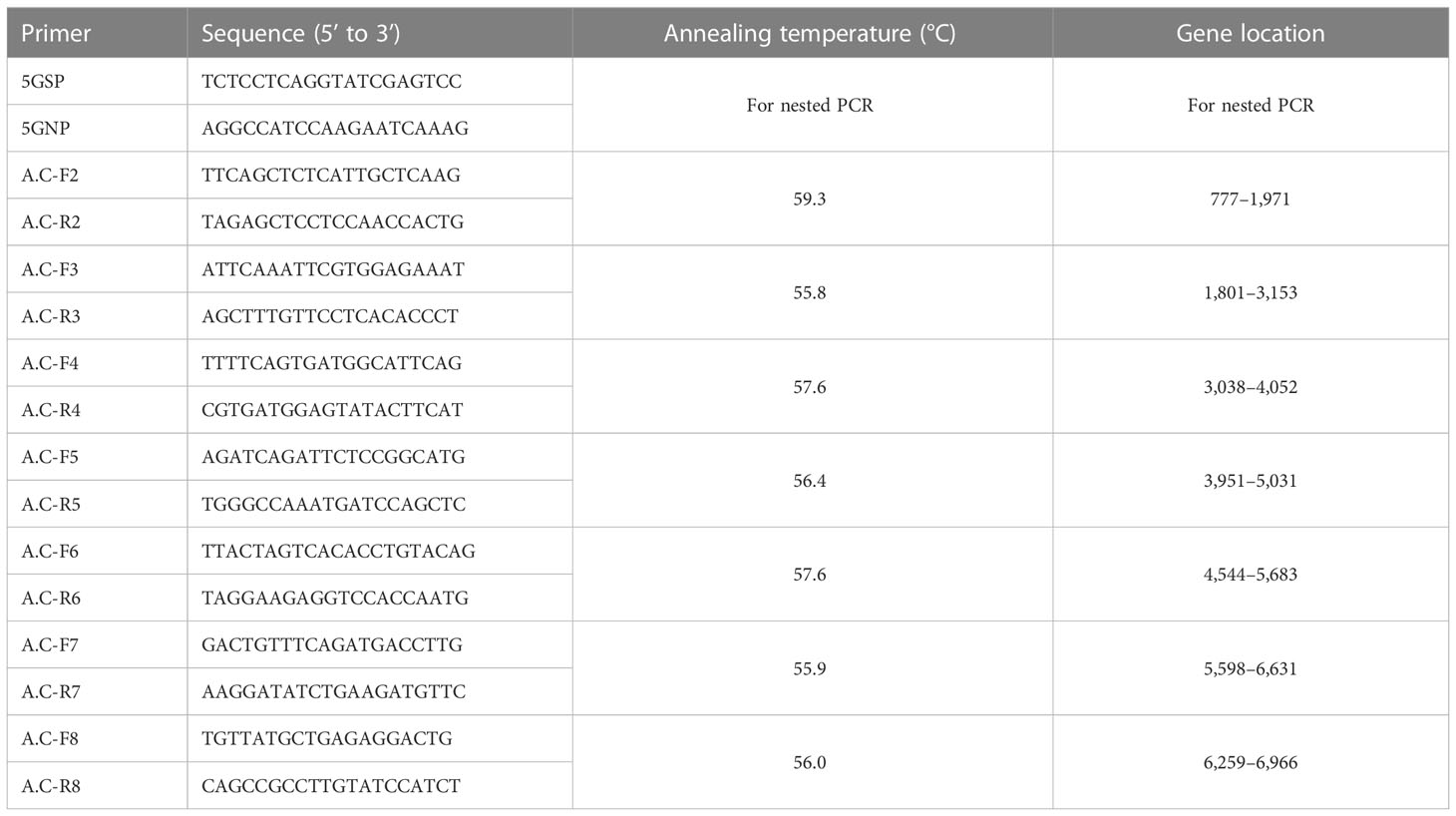
Young leaves from individual plants at the four-leaf stage were subjected to total DNA extraction using a plant genomic DNA extraction kit (Tiangen, China), according to the manufacturer’s instructions. The conserved CT domain of ACCase gene was then amplified using two pairs of the previously designed universal primers ACcp1/ACcp1R and ACcp4/ACcp2R (Delye et al., 2011b). PCR reaction volumes were 50 µL and included 2 µL of DNA, 2 µL of forward and reverse primers (10 µM), 25 µL of 2×PCR long Taq Mix (Vazyme, China), and ddH2O up to 50 µL. Reactions included a pre-denaturation step of 5 min at 95°C, followed by 30 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s. The PCR products were subject to electrophoresis in a 0.75% agarose gel in 1×TAE buffer, and PCR fragments were cloned into the PMD-18T vector (Takara, China) for sequencing. Amplicons from individual plants in each population were sequenced and at least 10 clones from each plant were subjected to sequencing to construct ACCase consensus sequences. Each segment was sequenced in the forward and reverse directions at Invitgen Biotechnology, Ltd. (Shanghai, China) to reduce sequencing errors. The BioEdit sequence alignment editor software was used to align and compare sequence data.
Total RNA was extracted using the RNApre Pure Plant Kit, followed by first strand cDNA generation with the FastQuant RT Kit (Tiangen, China). ACCase cDNA gene sequences of gramineae plants with high homology to D. ciliaris var. chrysoblephara were downloaded from NCBI, including those from Beckmannia syzigachne (GenBank accession number: KF501575), Alopecurus myosuroides (GenBank accession number: AJ310767) and Lolium rigidum (GenBank accession number: AY995232). The sequences were compared with the DNAMAN software. Eight pairs of primers were designed based on homologous sequence comparison of conserved regions (Table 2). In addition, 5’ cDNA terminal rapid amplification (RACE) gene specific primers (Table 2) were designed, and the ACCase cDNA ends were amplified using the HiScript-TS 5’/3’ RACE Kit (Vazyme, China). PCR fragments amplified from resistant populations were cloned into the PMD-18T vector (Takara, China) and amplicons from ten plants of each population were sequenced. For each biological replicate, at least ten clones were sequenced and used to construct ACCase consensus sequences. The BioEdit sequence alignment editor and DNAMAN software were then used to analyze and compare the sequence data.
2.4. ACCase gene expression assay
After growing to the four-leaf stage, plant seedlings from four D. ciliaris var. chrysoblephara populations were sprayed with the recommended dose of metamifop (120 g a.i.ha−1). Seedlings from each population were collected at 0, 12, and 24 h after treatment. Tissues were subjected to RNA extraction and cDNA synthesis, as described above. Primers were designed with the Primer premier 5.0 software package (AC-F: CTGTTGTGGGCAAGGAGGATG; AC-R: TACCAAGCCGAGCAAGATAAG) to amplify 153 bp fragments of target ACCase gene. Actin was used as the internal reference gene and amplified using primers designed using the large crabgrass actin gene (GenBank: KY967696) (Actin-F1: CGGAGAATAGCATGAGGAAGTG; Actin-R1: AGTGGTCGAACAACTGGTATTG). Real-time PCR was conducted with an ABI QuantStudio™ 7 Flex Real-Time PCR System (Thermofisher, USA) using a 2×TSINGKE Master qPCR Mix (SYBR Green I) kit (Tsingke, China). Thermal cycling conditions were used according to the manufacturer’s instructions. Three biological replicates and four technical replicates were used for each sample. The relative expression of ACCase gene were calculated using the 2−ΔΔCT method, while expression from the untreated sensitive population was used as the negative control.
2.5. ACCase activity assay
After growing to the four-leaf stage, seedlings of four D. ciliaris var. chrysoblephara populations were sprayed with the recommended dose of metamifop (120 g a.i.ha−1). Seedlings were collected from each population at 0, 2, 12, 24, 48, and 72 h after treatment, respectively. ACCase catalyzes the transformation of Acetyl-CoA, NaHCO3, and ATP to generate Malonyl-CoA, ADP, and inorganic phosphorus. The interaction of molybdenum blue and phosphate can generate products with characteristic absorption peaks at 660 nm. Thus, ACCase activity was determined based on inorganic phosphorus levels using the ammonium molybdate method. Specifically, ACCase activity was quantified using an ACCase activity assay kit (Biobox, China) and a microplate spectrophotometer (Agilent BioTek Epoch2, USA). Crude enzyme was prepared by adding 1 mL of extracting solution to 0.1 g of leaf tissue. The enzymatic reactions and phosphate quantification were then conducted based on the manufacturer’s instructions. Absorbance values at 660 nm were determined for experimental reactions, in addition to those for negative controls, blank controls, and to establish a standard curve. Three biological replicates and three technical replicates were used for each sample.
The unit of ACCase activity was calculated based on sample mass, as defined by the amount of 1 μmol of inorganic phosphorus generated for 1 g tissue over 1 h. Specifically, ACCase activity was calculated with the following equation:
In the formula, Vtotal is the total volume of the enzymatic reaction (0.1 mL), Vsample is the volume of added sample (0.01 mL), Vtotal sample is the volume of extracting solution (1 mL), T is the time of enzymatic reaction (0.5 h), and W is the fresh weight of sample (0.1 g).
2.6. Cross- and multi-resistance to other herbicides
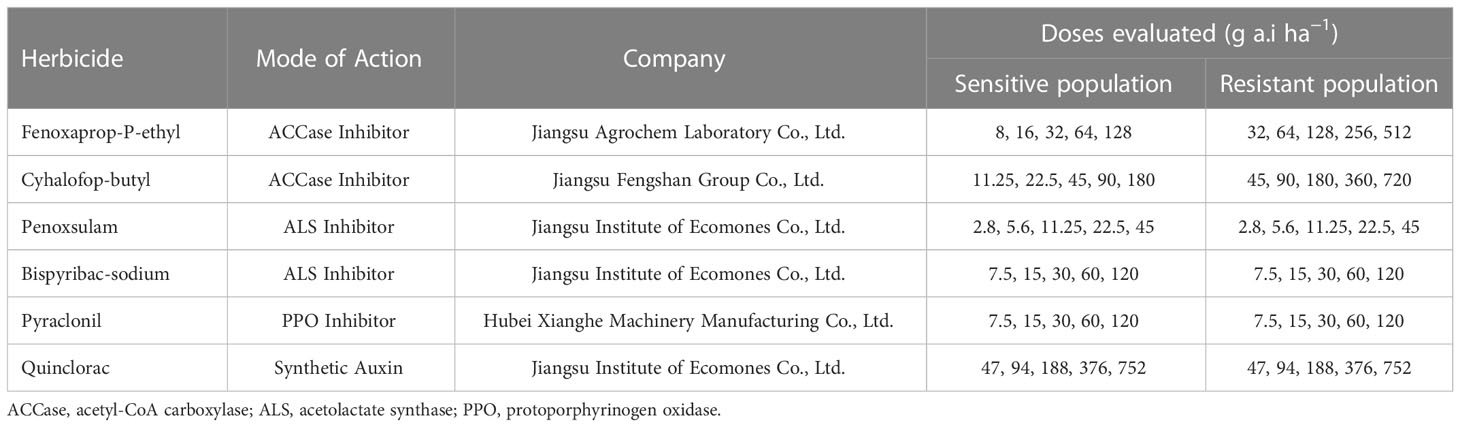
Dose-response experiments were performed to determine whether the D. ciliaris var. chrysoblephara populations exhibited cross- or multi-resistance to other herbicides. Experiments were described as in section 2.2, and the other evaluated herbicides are listed in Table 3.
3. Results
3.1. Dose response to metamifop exposure
Dose response experiments revealed that the JYX-8, JTX-98, and JTX-99 populations were resistant to metamifop (Figure 1; Table 4). Specifically, the GR50 values of these populations were much higher than the recommended dosage of metamifop (120 g a.i.ha−1). Moreover, their RI values were 30.64, 14.38, and 23.19, respectively.
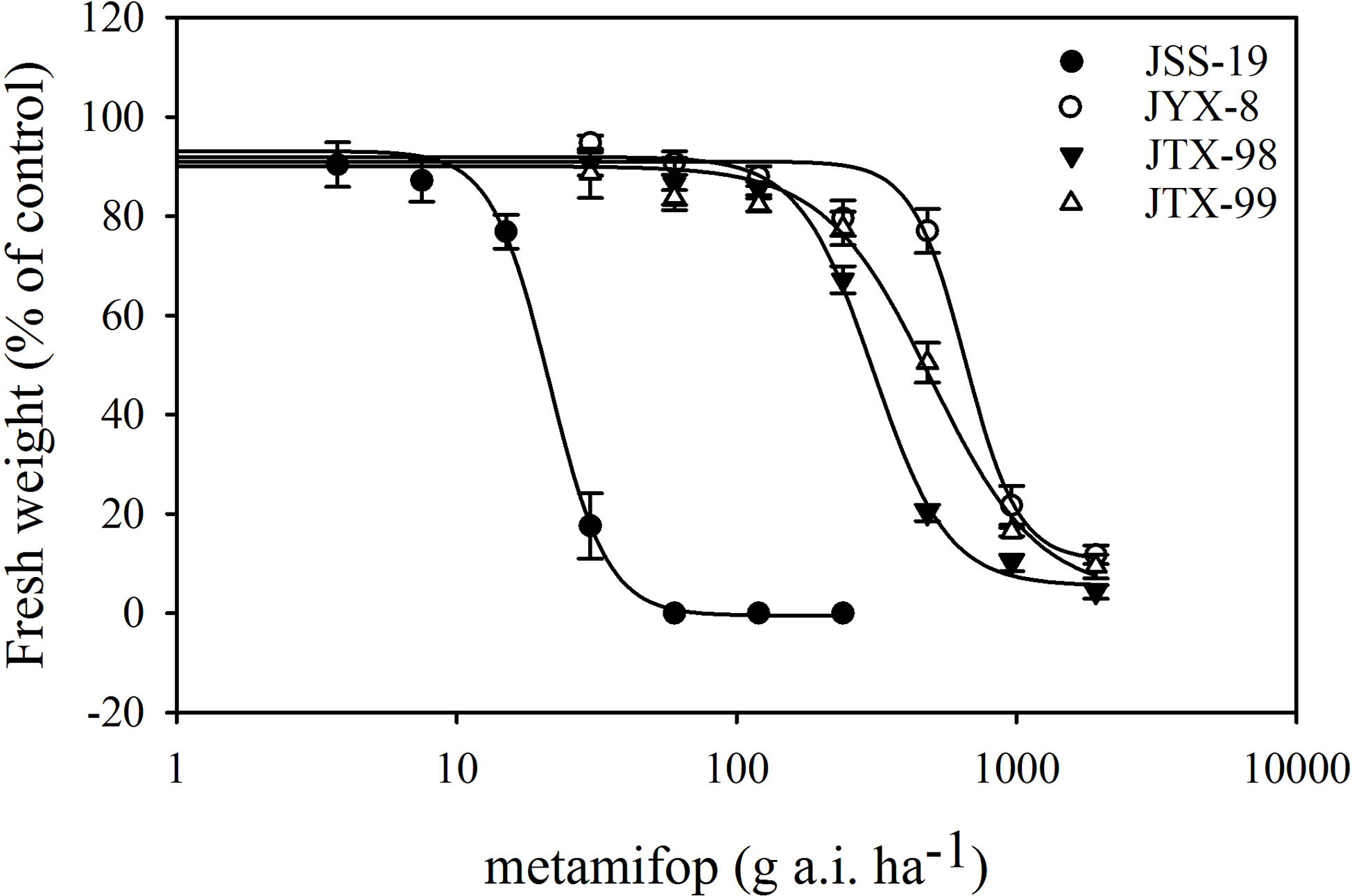
Figure 1 Dose response curves for four populations of D. ciliaris var. chrysoblephara to metamifop generated from dose response experiments. A log-logistic model was used to analyze the test data (the fresh weight percentage of the control). The GR50 values of resistant populations (JYX-8, JTX-98, and JTX-99) were significantly higher than in the sensitive population JSS-19. Vertical bars indicate standard errors.
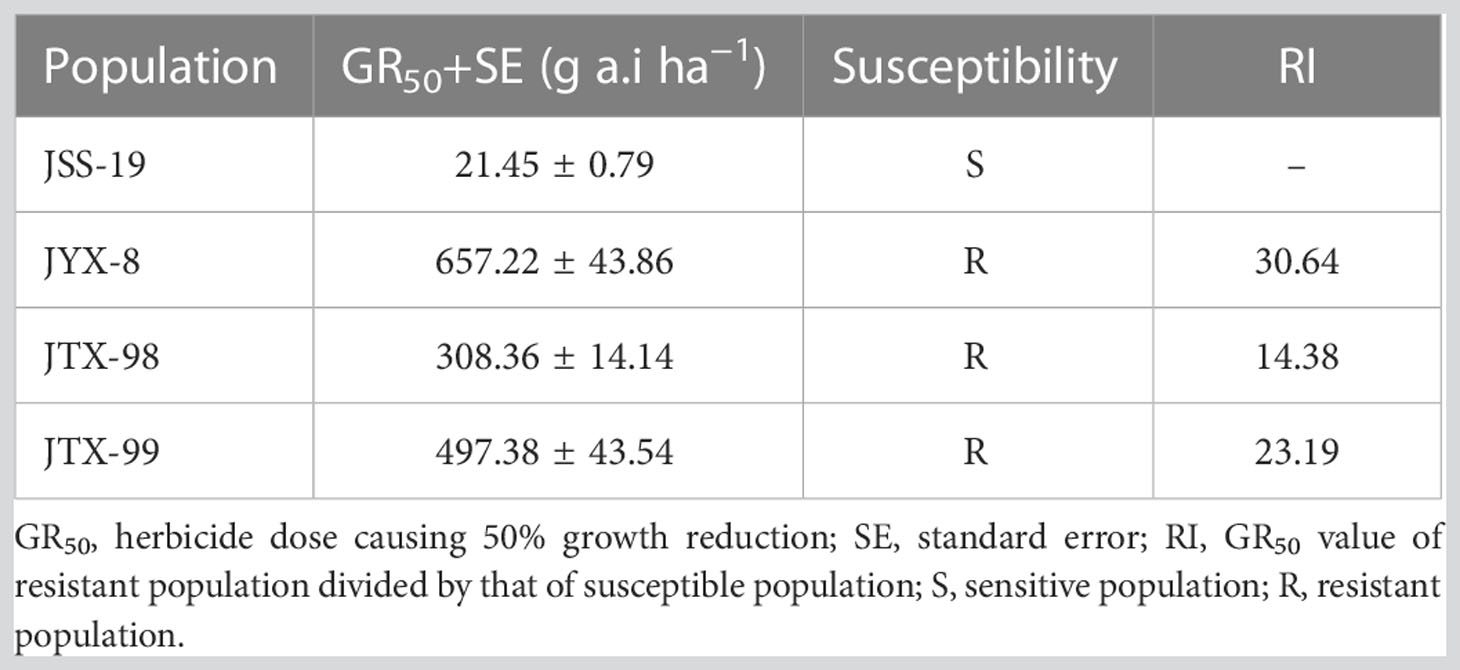
Table 4 GR50 and RI values for four populations of D. ciliaris var. chrysoblephara exposed to metamifop in this study.
3.2. ACCase gene sequencing
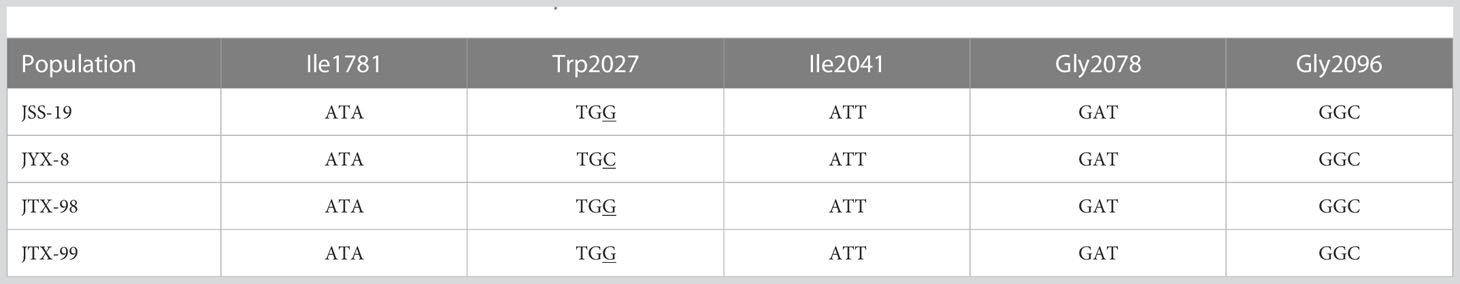
The ACCase CT domain gene fragment amplified by the ACcp1/ACcp1R primers was 551 bp in length and contained a previously reported variable Ile/Leu codon at site 1,781. The gene fragment amplified by the primers ACcp4/ACcp2R was 406 bp in length and contained four previously known variable codons (Trp/Cys, Ile/Asn, Asp/Gly, and Gly/Ala) located at sites 2,027, 2,041, 2,078, and 2,096, respectively. After sequence alignment of gene fragments from the resistant and sensitive populations (Table 5), a single nucleotide substitution was observed from TGG to TGC that resulted in an amino acid substitution at position 2,027 from tryptophan in the susceptible population to cysteine in the resistant population JYX-8. No corresponding substitutions were observed for JTX-98 and JTX-99 ACCase gene.
Using the extracted total RNA as template, eight primer pairs were designed using homologous sequence comparison to amplify ACCase cDNA sequences via PCR and RACE. The sizes of the target fragments amplified by the eight primer pairs were 1,265 bp, 1,304 bp, 1,448 bp, 1,136 bp, 1,144 bp, 1,232 bp, 1,130 bp, and 1,124 bp (Figure 2). The complete ACCase cDNA fragment was 7,277 bp in length and the ORF length was 6,966 bp. BLASTx analysis of the complete sequence against the NCBI database (Table 6) indicated the presence of amino acid sequence homology of 95.17%, 95.04%, 95%, and 94.10% between the ACCase encoded by the gramineae weeds Panicum hallii, Setaria viridis, Setaria italica, and Echinochloa crus-galli, respectively, suggesting that the amplified sequence was indeed an ACCase gene.
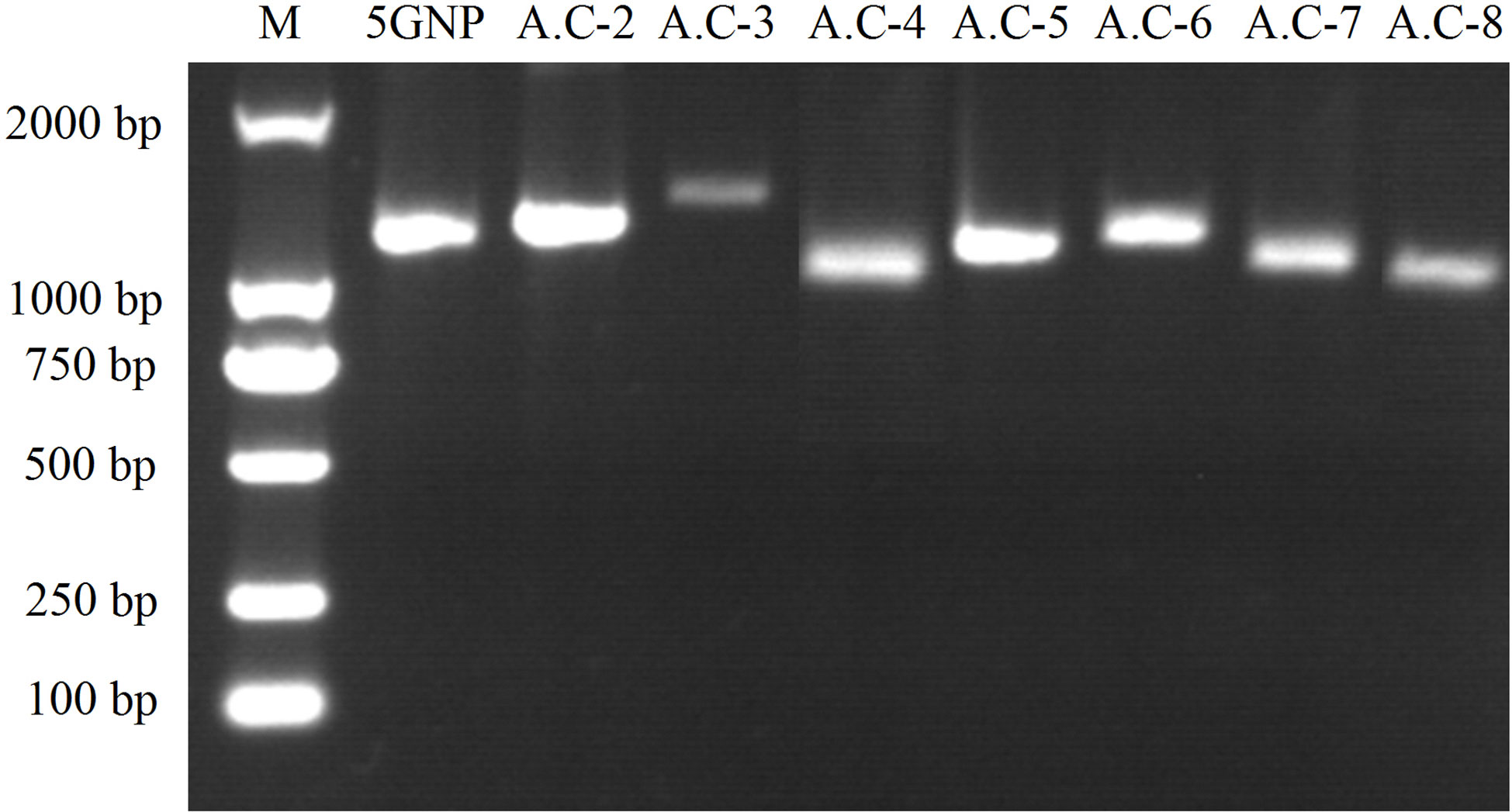
Figure 2 PCR amplification results for ACCase cDNA sequences. Eight primer pairs were designed based on homologous sequence comparison to amplify ACCase cDNA sequences via PCR and RACE. The sizes of the target fragments were 1,265, 1,304, 1,448, 1,136, 1,144, 1,232, 1,130, and 1,124 bp. The abbreviations above the image represent corresponding primers and the numbers on the left-hand side of the image indicate the size of DNA markers.
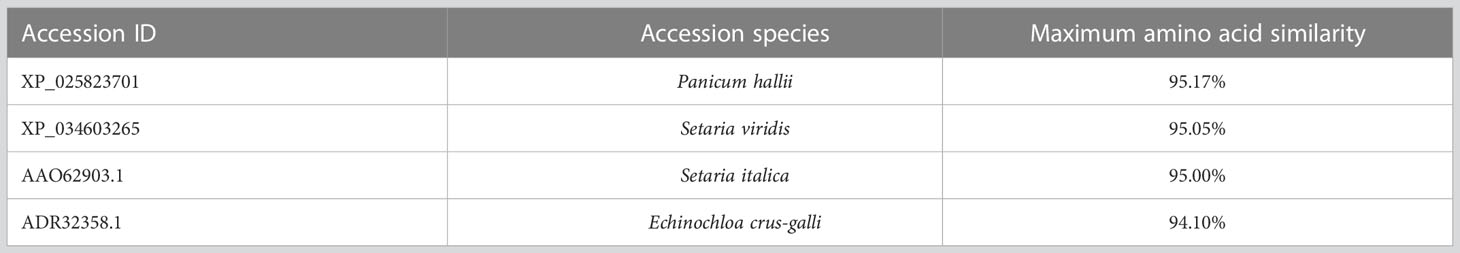
Table 6 BLAST results for D. ciliaris var. chrysoblephara ACCase proteins against the NCBI database.
3.3. Expression of ACCase gene
The expression levels of ACCase gene in resistant and sensitive populations were quantified by RT-qPCR. Relative ACCase gene expression levels for each population are indicated as fold-level values compared to control assays in the sensitive JSS-19 population in the absence of metamifop (Figure 3). Expression levels were not significantly different between control and metamifop treatments, both in the sensitive and resistant populations. Thus, the resistance of the JYX-8, JTX-98, and JTX-99 populations to metamifop was not related to ACCase gene expression.
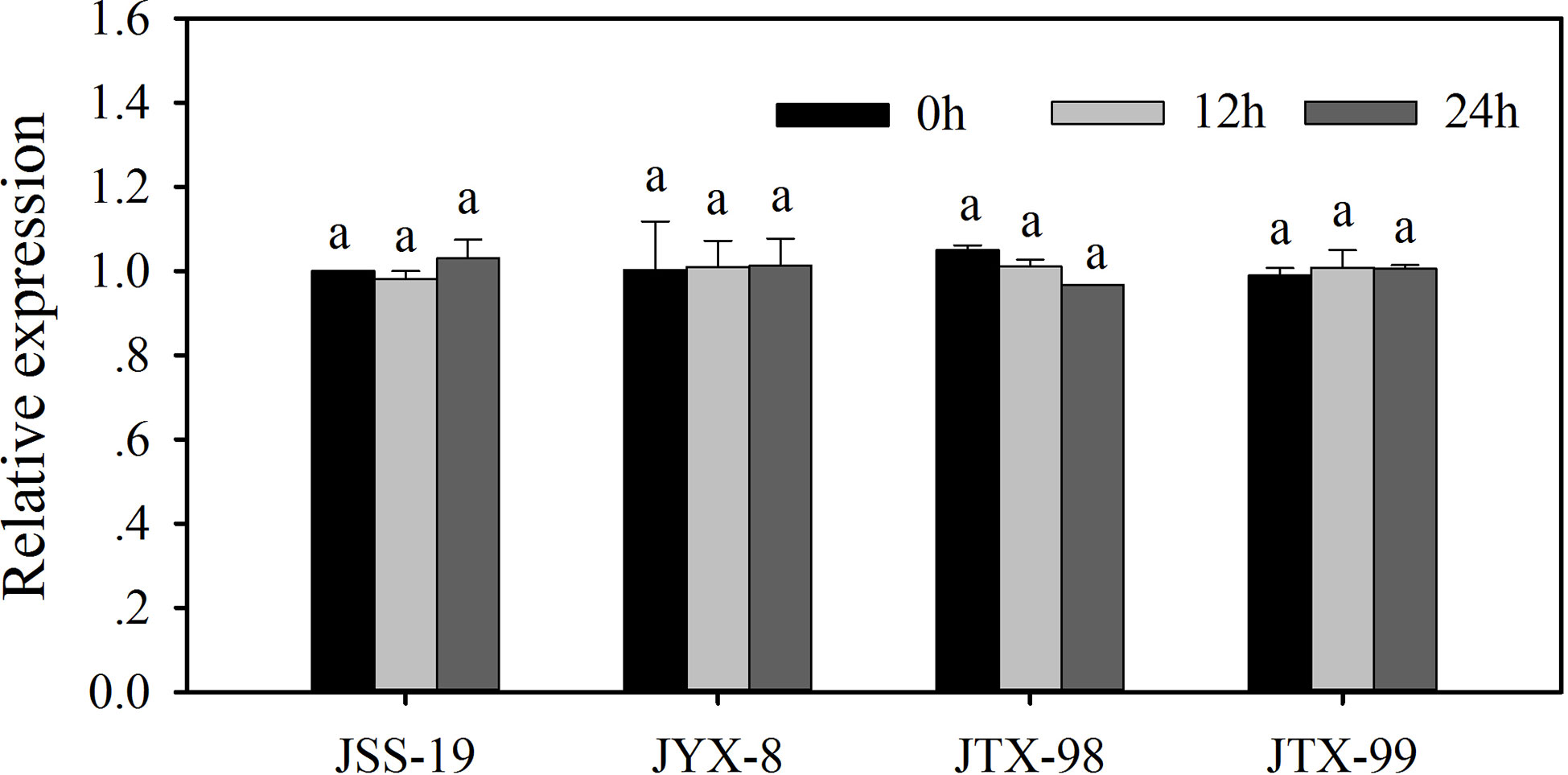
Figure 3 ACCase gene expression in four populations of D. ciliaris var. chrysoblephara after metamifop treatment. Gene expression levels were quantified by RT-qPCR and were not significantly different between control and metamifop treatments, both in the sensitive and resistant populations. Vertical bars indicate standard errors. The same lowercase letters indicate the lack of statistically significant differences based on Tukey’s tests.
3.4. ACCase activity
The influence of metamifop to ACCase activity within four D. ciliaris var. chrysoblephara populations was determined using the ammonium molybdate spectrophotometric method. ACCase activities were not clearly different between 0 and 2 h, and then significantly decreased at 12 and 24 h, followed by a final gradual recovery at 48 and 72 h (Figure 4). The ACCase activity trends were similar between sensitive and resistant populations, although statistically significant differences remained. After treatment for 2 h, their activities were inhibited to 85.85%, 108.43%, 94.69%, and 89.93% of the control levels for the JSS-19, JYX-8, JTX-98, and JTX-99 populations, respectively. The lowest activities were observed at 24 h, with activities inhibited to 64.39%, 77.35%, 80.54%, and 71.84% levels of the controls, respectively. ACCase activity in the sensitive population JSS-19 was most inhibited across the above period. After treatment for 72 h, their activities were recovered to 80.04%, 98.87%, 115.24%, and 97.32% of control levels, respectively. ACCase activities returned to the same or even higher levels than in the control for the resistant populations, but only recovered to 80.04% of that of the control in the sensitive population. These results suggest that ACCase activities in resistant populations were less inhibited than in sensitive populations and recovered to the same or even higher levels compared to untreated plants.
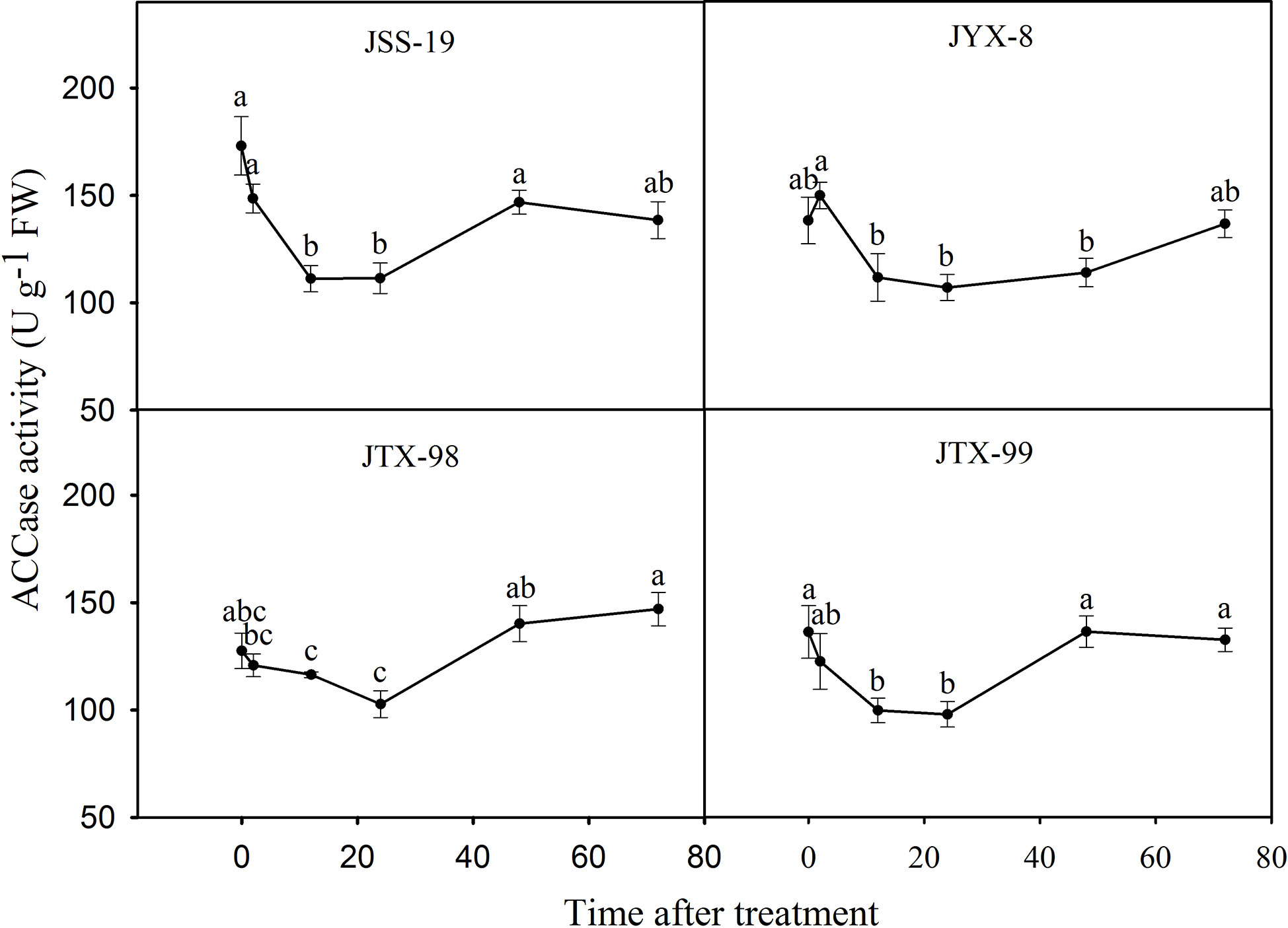
Figure 4 ACCase activities in four populations of D. ciliaris var. chrysoblephara after metamifop treatment. ACCase activities were determined using the ammonium molybdate spectrophotometric method. ACCase activities in resistant populations were less inhibited than in sensitive populations and also recovered to the same or even higher levels compared to untreated plants. Vertical bars indicate standard errors. Different lowercase letters indicate statistically significant differences (P < 0.05) based on Tukey’s tests.
3.5. Cross- and multi-resistance to other herbicides
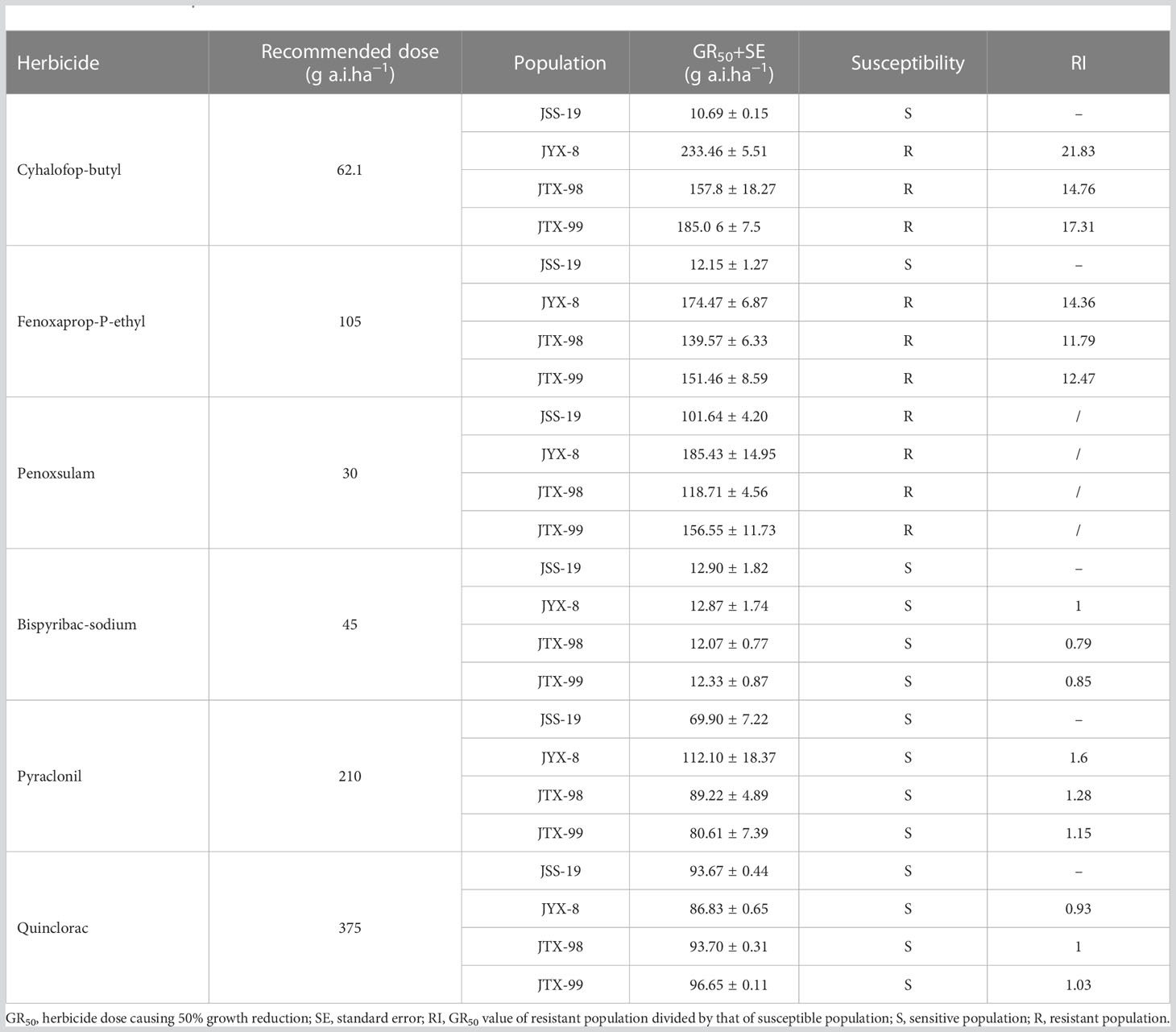
Three metamifop-resistant populations of D. ciliaris var. chrysoblephara were cross-resistant to other ACCase-inhibiting herbicides (Table 7). Specifically, the JYX-8, JTX-98, and JTX-99 populations exhibited high resistance to cyhalofop-butyl, with GR50 values of 233.46, 157.8, and 185.06 g a.i.ha−1 and RI values of 21.83, 14.76, and 17.31, respectively. The populations also exhibited resistance to fenoxaprop-P-ethyl, with GR50 values of 174.47, 139.57, and 151.46 g a.i.ha−1 and RI values of 14.36, 11.79, and 12.47, respectively. Thus, the three resistant populations exhibited similar cross-resistance patterns to ACCase inhibitors.
The multi-resistance characteristics of metamifop-resistant D. ciliaris var. chrysoblephara plants were also evaluated by whole-plant bioassays. The GR50 values of the four populations to the ALS inhibitor penoxsulam (101.64-185.43 g a.i.ha−1) were significantly higher than the recommended field dosage (30 g a.i.ha−1), suggesting the existence of multi-resistance to penoxsulam. The GR50 values of the resistant populations to the ALS-inhibiting herbicide bispyribac-sodium, the auxin mimic herbicide quinclorac, and the PPO-inhibiting herbicide pyraclonil were significantly lower than their recommended field dosages. In addition, the RI values were all< 2, suggesting the absence of multi-resistance to these herbicides.
4. Discussion
Jiangsu is one of the most important rice production areas of China. Metamifop has been used to control gramineae weeds in Jiangsu paddy fields for over 10 years. Consequently, metamifop use carries a high risk for herbicide resistance development. Consistently, we observed that continuous high-intensity use of metamifop did not mitigate the rapid spread of D. ciliaris var. chrysoblephara in paddy fields. We hypothesized that this lack of effect may be due to resistance of D. ciliaris var. chrysoblephara to metamifop. A high level of resistance to ACCase-inhibiting herbicides has been previously reported in various weed species including in wild oat (Avena fatua L.), Japanese foxtail (Alopecurus japonicus), and Amazon sprangletop [Leptochloa panicoides (J. Presl) Hitchc.] (Seefeldt et al., 1996; Xu et al., 2013; Tehranchian et al., 2016). D. ciliaris has only been previously observed to be resistant to ACCase herbicides via collection of plants from sod production fields in Georgia (Yu et al., 2017; Basak et al., 2019). In this study, D. ciliaris var. chrysoblephara populations collected from dry direct-seeding rice fields in the Jiangsu province were shown to exhibit high resistance to metamifop. ACCase inhibitors remain the primary herbicides used to control D. ciliaris var. chrysoblephara. However, the continued use of the same herbicides will inevitably lead to increased selection pressure, leading to rapid development of resistant populations and inestimable harm to rice production efforts.
The change of target enzyme activity caused by mutation or overexpression of target gene is the direct cause of weed resistance to herbicide. Herbicide efficacy is determined by the affinity of herbicides to enzymes. Physicochemical interactions are main factors that determine the affinity. For ACCase-inhibiting herbicides, their affinity and efficacy are determined by the interactions between the herbicides and the amino acids at specific positions of the polypeptide chain on ACCase CT domain. Thus, a single nucleotide mutation in the ACCase gene can result in amino acid substitutions imparting resistance to herbicides (Takano et al., 2020). Several amino acid substitutions of ACCase CT domain of grassy weeds have been reported to be involved in resistance to ACCase-inhibiting herbicides, including the Trp-2027-Cys substitution (Deng et al., 2020). Following the first report of the Trp-2027-Cys substitution in Alopecurus myosuroides, it has been observed in many weeds resistant to ACCase-inhibiting herbicides like Avena sterilis, Lolium rigidum, Beckmannia syzigachne, and A. japonicus (Delye et al., 2005; Liu et al., 2007; Yu et al., 2007; Xu et al., 2013; Pan et al., 2015). In this study, the Trp-2027-Cys substitution, resulting from a TGG to TGC nucleotide change, was observed in JYX-8 ACCase. Thus, the interaction between herbicide and target enzyme was influenced by target site mutation of ACCase gene and lead to the resistance of JYX-8 to metamifop. However, similar mutations were not observed for JTX-98 and JTX-99 populations. Overexpression of ACCase gene can confer resistance in large crabgrass to ACCase-inhibiting herbicides (Laforest et al., 2017). In this study, ACCase gene expression levels were not significantly different between control and metamifop treatments, both in the sensitive and resistant populations. ACCase activities were less inhibited in the resistant populations than in the sensitive population. Thus, ACCase from the resistant populations was much less sensitive to metamifop than the sensitive population (Xu et al., 2013). ACCase gene sequencing, gene expression, and enzyme activity investigations suggested that target-site resistance was the most likely mechanism conferring resistance in JYX-8, although the existence of NTSR mechanisms cannot be ruled out. A NTSR mechanism to metamifop, such as enhanced metabolism, may be present in JTX-98 and JTX-99, rather than a directed TSR. The unknown resistance mechanisms require further exploration.
The Trp-2027-Cys substitution can lead to cross-resistance to APP herbicides (Li et al., 2014), consistent with the results of this study. The JYX-8 population carrying the Trp-2027-Cys substitution was resistant to the APP herbicides cyhalofop-butyl and fenoxaprop-P-ethyl, with RI values of 21.83 and 14.36, respectively. In addition, no target site mutations were observed for the JTX-98 and JTX-99 populations, but they also exhibited cross-resistance to the APP herbicides cyhalofop-butyl and fenoxaprop-P-ethyl (RI values > 11.79) via unknown resistance mechanisms. The multi-resistance of the JYX-8, JTX-98, and JTX-99 populations to the ALS inhibitor penoxsulam was observed, but to the ALS inhibitor bispyribac-sodium, the auxin mimic quinclorac, and the PPO inhibitor pyraclonil was not observed. Bispyribac-sodium, quinclorac, and pyraclonil exerted good control on these populations. Thus, these herbicides with different modes of action could still be used to control metamifop-resistant D. ciliaris var. chrysoblephara.
Nevertheless, many studies have shown that different resistance mechanisms (i.e., TSR and/or NTSR mechanisms) may be present in a single weed population that is resistant to a variety of herbicides (Beckie et al., 2012). Multiple target-site resistance to ACCase and ALS inhibitors has been observed in black-grass (Bailly et al., 2012). Further, glyphosate-resistant rigid ryegrass exhibited multiple resistance to ACCase- and ALS- inhibiting herbicides. Specifically, ALS resistance in the population was due to an insensitive target enzyme, while ACCase resistance was due to a non-target-site mechanism (Neve et al., 2004). Consequently, control methods should be carefully evaluated for weed populations with different resistance mechanisms. No evidence of non-target-site resistance has been shown for Digitaria spp. weeds. However, the resistant populations JTX-98 and JTX-99 investigated in this study likely exhibit NTSR mechanisms, warranting additional attention to control methods.
In conclusion, three populations of D. ciliaris var. chrysoblephara collected from dry direct-sown rice fields of the Jiangsu province have evolved resistance to metamifop, and the target-site mutation Trp-2027-Cys is responsible for resistance in the JYX-8 population. To our knowledge, this study represents the first evidence for a molecular mechanism of D. ciliaris var. chrysoblephara resistance to metamifop. In addition, cross- and multi-resistance patterns to other herbicides were also investigated in this study. Non-target-site resistance mechanisms cannot be presently ruled out for the resistance of D. ciliaris var. chrysoblephara to metamifop. Consequently, additional studies are needed to comprehensively understand the mechanism underlying the resistance traits evaluated here.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
HW and YL conceived and designed the experiments; JC and YT performed the lab work, data acquisition, and data analysis; JC and YT wrote the manuscript; TG assisted in performing the experiment; ZZ and GL contributed to the discussion and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31801773 and 31801755) and the Jiangsu Agricultural Science and Technology Innovation Fund [CX(22)3021].
Acknowledgments
We thank LetPub (www.letpub.com) for linguistic assistance and pre-submission expert review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adina-Zada, A., Zeczycki, T. N., Attwood, P. V. (2012). Regulation of the structure and activity of pyruvate carboxylase by acetyl CoA. Arch. Biochem. Biophysics 519, 118–130. doi: 10.1016/j.abb.2011.11.015
Bailly, G., Dale, R., Archer, S., Wright, D., Kaundun, S. (2012). Role of residual herbicides for the management of multiple herbicide resistance to ACCase and ALS inhibitors in a black-grass population. Crop Prot. 34, 96–103. doi: 10.1016/j.cropro.2011.11.017
Basak, S., Mcelroy, J. S., Brown, A. M., Gon Alves, C. G., Patel, J. D., Mccullough, P. E. (2019). Plastidic ACCase ile-1781-Leu is present in pinoxaden-resistant southern crabgrass (Digitaria ciliaris). Weed Sci. 68, 41–50. doi: 10.1017/wsc.2019.56
Beckie, H. J., Warwick, S. I., Sauder, C. A. (2012). Basis for herbicide resistance in Canadian populations of wild oat (Avena fatua). Weed Sci. 60, 10–18. doi: 10.1614/WS-D-11-00110.1
Brazier, M., Cole, D. J., Edwards, R. (2002). O-Glucosyltransferase activities toward phenolic natural products and xenobiotics in wheat and herbicide-resistant and herbicide-susceptible black-grass (Alopecurus myosuroides). Phytochemistry 59, 149–156. doi: 10.1016/S0031-9422(01)00458-7
Collavo, A., Panozzo, S., Lucchesi, G., Scarabel, L., Sattin, M. (2011). Characterisation and management of phalaris paradoxa resistant to ACCase-inhibitors. Crop Prot. 30, 293–299. doi: 10.1016/j.cropro.2010.11.011
Delye, C., Gardin, J., Boucansaud, K., Chauvel, B., Petit, C. (2011a). Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Res. 51, 433–437. doi: 10.1111/j.1365-3180.2011.00864.x
Delye, C., Pernin, F., Michel, S. (2011b). ‘Universal’ PCR assays detecting mutations in acetyl-coenzyme a carboxylase or acetolactate synthase that endow herbicide resistance in grass weeds. Weed Res. 51, 353–362. doi: 10.1111/j.1365-3180.2011.00852.x
Delye, C., Zhang, X. Q., Michel, S., MatJicek, A., Powles, S. B. (2005). Molecular bases for sensitivity to acetyl-coenzyme a carboxylase inhibitors in black-grass. Plant Physiol. 137, 794–806. doi: 10.1104/pp.104.046144
Deng, W., Yang, Q., Chen, Y., Yang, M., Xia, Z., Zhu, J., et al. (2020). Cyhalofop-butyl and glyphosate multiple-herbicide resistance evolved in an eleusine indica population collected in Chinese direct-seeding rice. J. Agric. Food Chem. 68, 2623–2630. doi: 10.1021/acs.jafc.9b07342
Fang, J., He, Z., Liu, T., Li, J., Dong, L. (2020). A novel mutation asp-2078-Glu in ACCase confers resistance to ACCase herbicides in barnyardgrass (Echinochloa crus-galli). Pesticide Biochem. Physiol. 168, 104634. doi: 10.1016/j.pestbp.2020.104634
Guo, W., Zhang, L., Wang, H., Li, Q., Liu, W., Wang, J. (2017). A rare ile-2041-Thr mutation in the ACCase gene confers resistance to ACCase-inhibiting herbicides in shortawn foxtail (Alopecurus aequalis). Weed Sci. 65, 239–246. doi: 10.1017/wsc.2016.32
Huang, S., Wang, Q., Dong, L., Lou, Y. (2003). Acetyl-CoA carboxylase-inhibiting herbicides and resistance of weeds. Weed Sci., 2–6. doi: 10.19588/j.issn.1003-935x.2003.02.001
Kaundun, S. S., Bailly, G. C., Dale, R. P., Hutchings, S.-J., Mcindoe, E. (2013a). A novel W1999S mutation and non-target site resistance impact on acetyl-CoA carboxylase inhibiting herbicides to varying degrees in a UK lolium multiflorum population. PLoS One 8, e58012. doi: 10.1371/journal.pone.0058012
Kaundun, S. S., Hutchings, S.-J., Dale, R. P., Mcindoe, E. (2013b). Role of a novel I1781T mutation and other mechanisms in conferring resistance to acetyl-CoA carboxylase inhibiting herbicides in a black-grass population. PLoS One 8, e69568. doi: 10.1371/journal.pone.0069568
Kukorelli, G., Reisinger, P., Pinke, G. (2013). ACCase inhibitor herbicides – selectivity, weed resistance and fitness cost: a review. Int. J. Pest Manage. 59, 165–173. doi: 10.1080/09670874.2013.821212
Kupper, A., Borgato, E. A., Patterson, E. L., Netto, A. G., Nicolai, M., De Carvalho, S. J., et al. (2017). Multiple resistance to glyphosate and acetolactate synthase inhibitors in palmer amaranth (Amaranthus palmeri) identified in Brazil. Weed Sci. 65, 317–326. doi: 10.1017/wsc.2017.1
Laforest, M., Soufiane, B., Simard, M. J., Obeid, K., Page, E., Nurse, R. E. (2017). Acetyl-CoA carboxylase overexpression in herbicide-resistant large crabgrass (Digitaria sanguinalis). Pest Manage. Sci. 73, 2227–2235. doi: 10.1002/ps.4675
Li, L., Du, L., Liu, W., Yuan, G., Wang, J. (2014). Target-site mechanism of ACCase-inhibitors resistance in American sloughgrass (Beckmannia syzigachne steud.) from China. Pesticide Biochem. Physiol. 110, 57–62. doi: 10.1016/j.pestbp.2014.03.001
Li, L., Luo, X., Wang, J. (2017). Resistance level and target-site mechanism to fenoxaprop-p-ethyl in Beckmannia syzigachne (Steud.) fernald populations from China. Chilean J. Agric. Res. 77, 150–154. doi: 10.4067/S0718-58392017000200150
Liu, W., Harrison, D. K., Chalupska, D., Gornicki, P., O'donnell, C. C., Adkins, S. W., et al. (2007). Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc. Natl. Acad. Sci. 104, 3627–3632. doi: 10.1073/pnas.0611572104
Matloob, A., Khaliq, A., Chauhan, B. S. (2015). Weeds of direct-seeded rice in Asia: Problems and opportunities. Adv. Agron. 130, 291–336. doi: 10.1016/bs.agron.2014.10.003
Neve, P., Sadler, J., Powles, S. B. (2004). Multiple herbicide resistance in a glyphosate-resistant rigid ryegrass (Lolium rigidum) population. Weed Sci. 52, 920–928. doi: 10.1614/WS-04-007R1
Pan, L., Li, J., Xia, W., Zhang, D., Dong, L. (2015). An effective method, composed of LAMP and dCAPS, to detect different mutations in fenoxaprop-p-ethyl-resistant American sloughgrass (Beckmannia syzigachne steud.) populations. Pesticide Biochem. Physiol. 117, 1–8. doi: 10.1016/j.pestbp.2014.10.008
Papapanagiotou, A. P., Paresidou, M. I., Kaloumenos, N. S., Eleftherohorinos, I. G. (2015). ACCase mutations in avena sterilis populations and their impact on plant fitness. Pesticide Biochem. Physiol. 123, 40–48. doi: 10.1016/j.pestbp.2015.01.017
Rao, A. N., Brainard, D. C., Kumar, V., Ladha, J. K., Johnson, D. E. (2017). Preventive weed management in direct-seeded rice: targeting the weed seedbank. Adv. Agron. 144, 45–142. doi: 10.1016/bs.agron.2017.02.002
Seefeldt, S. S., Fuerst, E. P., Gealy, D. R., Shukla, A., Irzyk, G. P., Devine, M. D. (1996). Mechanisms of resistance to diclofop of two wild oat (Avena fatua) biotypes from the Willamette valley of Oregon. Weed Sci. 44, 776–781. doi: 10.1017/S0043174500094704
Seefeldt, S. S., Jensen, J. E., Fuerst, E. P. (1995). Log-logistic analysis of herbicide dose-response relationships. Weed Technol. 9, 218–227. doi: 10.1017/S0890037X00023253
Takano, H. K., Ovejero, R. F. L., Belchior, G. G., Maymone, G. P. L., Dayan, F. E. (2020). ACCase-inhibiting herbicides: mechanism of action, resistance evolution and stewardship. Scientia Agricola 78. doi: 10.1590/1678-992x-2019-0102
Tehranchian, P., Norsworthy, J. K., Korres, N. E., Mcelroy, S., Chen, S., Scott, R. C. (2016). Resistance to aryloxyphenoxypropionate herbicides in Amazon sprangletop: Confirmation, control, and molecular basis of resistance. Pesticide Biochem. Physiol. 133, 79–84. doi: 10.1016/j.pestbp.2016.02.010
Wang, J., Peng, Y., Chen, W., Yu, Q., Bai, L., Pan, L. (2021). The ile-2041-Val mutation in the ACCase gene confers resistance to clodinafop-propargyl in American sloughgrass (Beckmannia syzigachne steud). Pest Manage. Sci. 77, 2425–2432. doi: 10.1002/ps.6271
Wang, H., Xu, P., Sun, Y., Lou, Y., Xu, S. (2019). Status of damage and control strategy for paddy field weeds in jiangsu province. J. Weed Sci. 37, 1–5. doi: 10.19588/j.issn.1003-935X.2019.04.001
Won, O. J., Lee, J. J., Eom, M. Y., Suh, S. J., Park, S. H., Hwang, K. S., et al. (2014). Identification of herbicide-resistant barnyardgrass (Echinochloa crus-galli var. crus-galli) biotypes in Korea. Weed Turfgrass Sci. 3, 110–113. doi: 10.5660/WTS.2014.3.2.110
Xia, X., Tang, W., He, S., Kang, J., Ma, H., Li, J. (2016). Mechanism of metamifop inhibition of the carboxyltransferase domain of acetyl-coenzyme a carboxylase in Echinochloa crus-galli. Sci. Rep. 6, 1–10. doi: 10.1038/srep34066
Xu, H., Zhu, X., Wang, H., Li, J., Dong, L. (2013). Mechanism of resistance to fenoxaprop in Japanese foxtail (Alopecurus japonicus) from China. Pesticide Biochem. Physiol. 107, 25–31. doi: 10.1016/j.pestbp.2013.04.008
Yu, Q., Collavo, A., Zheng, M.-Q., Owen, M., Sattin, M., Powles, S. B. (2007). Diversity of acetyl-coenzyme a carboxylase mutations in resistant lolium populations: evaluation using clethodim. Plant Physiol. 145, 547–558. doi: 10.1104/pp.107.105262
Yu, J., Mccullough, P. E., Czarnota, M. A. (2017). First report of acetyl-CoA carboxylase–resistant southern crabgrass (Digitaria ciliaris) in the united states. Weed Technol. 31, 252–259. doi: 10.1017/wet.2016.34
Yu, Q., Powles, S. (2014). Metabolism-based herbicide resistance and cross-resistance in crop weeds: a threat to herbicide sustainability and global crop production. Plant Physiol. 166, 1106–1118. doi: 10.1104/pp.114.242750
Yuan, G., Tian, Z., Li, T., Qian, Z., Guo, W., Shen, G. (2021). Cross-resistance pattern to ACCase-inhibiting herbicides in a rare trp-2027-Ser mutation Chinese sprangletop (Leptochloa chinensis) population. Chilean J. Agric. Res. 81, 62–69. doi: 10.4067/S0718-58392021000100062
Keywords: Digitaria ciliaris var. chrysoblephara, metamifop, weed resistance, target-site resistance, cross- and multi-resistance
Citation: Cao J, Tao Y, Zhang Z, Gu T, Li G, Lou Y and Wang H (2023) Mechanism of metamifop resistance in Digitaria ciliaris var. chrysoblephara from Jiangsu, China. Front. Plant Sci. 14:1133798. doi: 10.3389/fpls.2023.1133798
Received: 29 December 2022; Accepted: 30 January 2023;
Published: 21 February 2023.
Edited by:
Zhenggang Li, Guangdong Academy of Agricultural Sciences, ChinaReviewed by:
Chun Zhang, Guangdong Academy of Agricultural Sciences, ChinaWeitang Liu, Shandong Agricultural University, China
Copyright © 2023 Cao, Tao, Zhang, Gu, Li, Lou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongchun Wang, SG9uZ2NodW4wMjNAMTI2LmNvbQ==
 Jingjing Cao
Jingjing Cao Yuan Tao
Yuan Tao Hongchun Wang
Hongchun Wang