
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 March 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1133616
This article is part of the Research Topic Economic Plant Genome and Database Construction and Research View all 10 articles
 Yaolei Mi1
Yaolei Mi1 Ruikun He2
Ruikun He2 Huihua Wan1
Huihua Wan1 Xiangxiao Meng1
Xiangxiao Meng1 Di Liu3
Di Liu3 Wenjun Huang4
Wenjun Huang4 Yanjun Zhang4
Yanjun Zhang4 Zubaida Yousaf5
Zubaida Yousaf5 Hongwen Huang6
Hongwen Huang6 Shilin Chen1,7
Shilin Chen1,7 Ying Wang8*
Ying Wang8* Wei Sun1,7*
Wei Sun1,7*Introduction: Flower color is an ideal trait for studying the molecular basis for phenotypic variations in natural populations of species. Epimedium (Berberidaceae) species exhibit a wide range of flower colors resulting from the varied accumulation of anthocyanins and other pigments in their spur-like petals and petaloid sepals.
Methods: In this work, the anthocyanidins of eight different Epimedium species with different floral pigmentation phenotypes were analyzed using HPLC. Twelve genes involved in anthocyanin biosynthesis were cloned and sequenced, and their expression was quantified.
Results: The expression levels of the catalytic enzyme genes DFR and ANS were significantly decreased in four species showing loss of floral pigmentation. Complementation of EsF3’H and EsDFR in corresponding Arabidopsis mutants together with overexpression of EsF3’5’H in wild type Arabidopsis analysis revealed that these genes were functional at the protein level, based on the accumulation of anthocyanin pigments.
Discussion: These results strongly suggest that transcriptional regulatory changes determine the loss of anthocyanins to be convergent in the floral tissue of Epimedium species.
Accumulation of the secondary metabolite anthocyanin is predominantly responsible for red, blue, and purple pigmentation in angiosperms. Pigmentation is a major determinant of a species’ pollination syndrome, which refers to the selection of particular floral traits caused by the preference of their pollinators (Fenster et al., 2004). Flower color is intricately regulated by the specific combinations of certain pigment metabolites produced, and is subjected to ecological selection and convergent evolution. Therefore, flower color is an ideal trait for examining ecological and evolutionary selection processes. The anthocyanin biosynthetic pathway (ABP) has been well established in many model species, such as Arabidopsis, petunia (Petunia hybrida E. Vilm.), and snapdragon (Antirrhinum majus L.) (Buer et al., 2010; Pollastri & Tattini, 2011). Most of the knowledge of anthocyanin biosynthesis in Arabidopsis has been obtained from the analysis of transparent testa (tt) mutants, which show loss of seed pigmentation (Lepiniec et al., 2006). In the early steps of the pathway, the key enzymes chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H) condense and convert a phenylpropanoid precursor, p-coumaroyl-CoA, along with three molecules of malonyl CoA, to dihydrokaempferol (Lepiniec et al., 2006). Parallel catalyzation by flavonoid-3’-hydroxylase, flavonoid-3’,5’-hydroxylase, dihydroflavonol-4-reductase (DFR), and anthocyanidin synthase (ANS) results in the production of various types of anthocyanidin (Figure 1) (Holton & Cornish, 1995; Boss et al., 1996). The transcriptional regulators controlling flavonoid biosynthetic enzymes have been extensively studied, and include the MYB, the bHLH, and the WD-repeat proteins. Yeast-three-hybrid protein interaction data suggested that a protein complex of the MYB-bHLH-WD40 transcription factors binds the regulatory promoter regions of the flavonoid pathway enzymatic, or structural, genes, to regulate anthocyanin biosynthesis (Gonzalez et al., 2008).
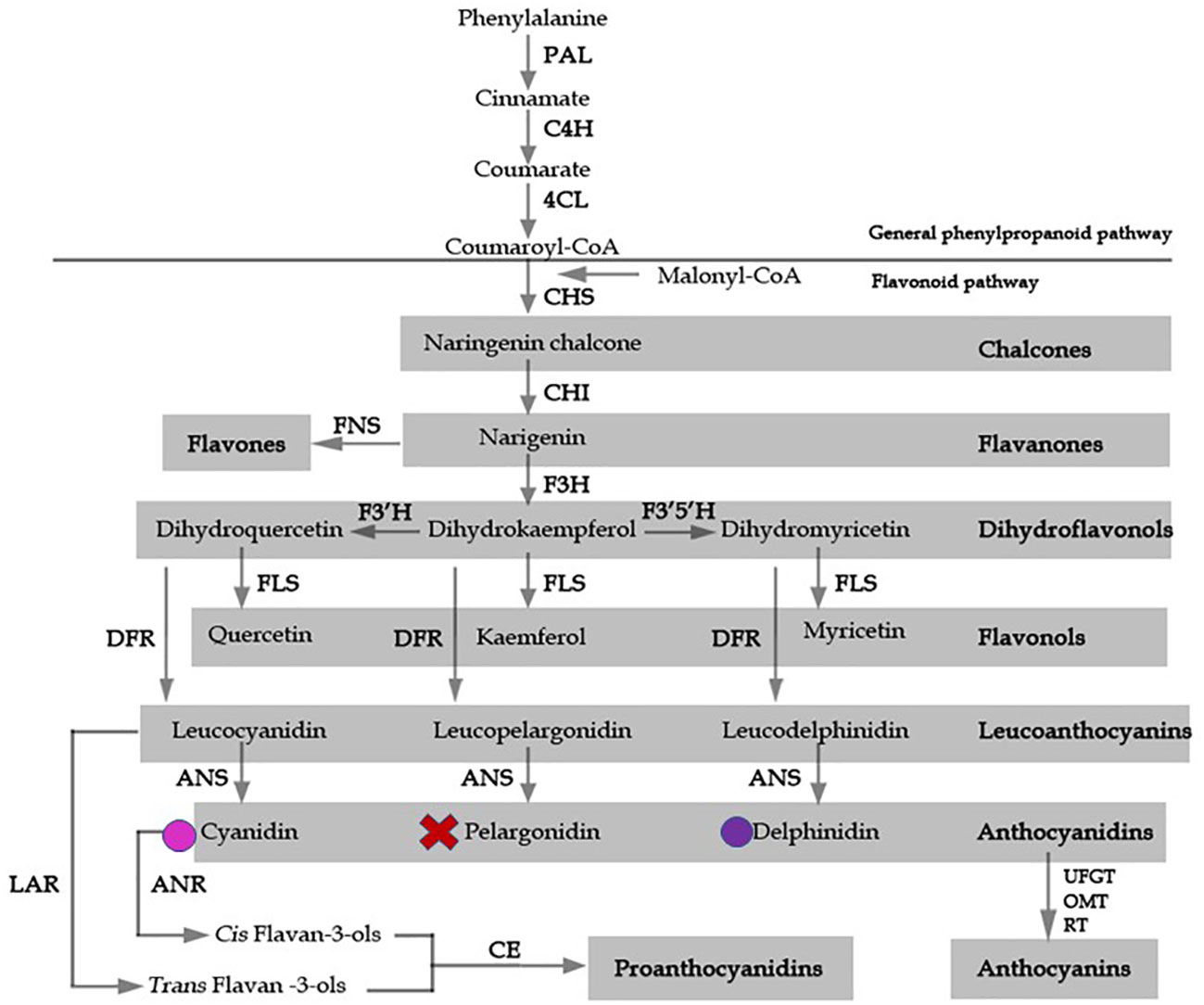
Figure 1 A model for flavonoid biosynthesis in Epimedium flowers based on classic investigation. Pathway enzymes are listed as an abbreviation beside arrows, and include 4CL, 4-coumarate: coenzyme A ligase; ANS, anthocyanidin synthase; C4H, cinnamate-4-hydroxylase; CHS, chalcone synthase, CHI, chalcone isomerase; DFR, dihydroflavonol 4-reductase; F3H, flavavone 3-hydroxylase; F3’H, flavonoid 3’-hydroxylase; F3’5’H, flavonoid 3’5’ hydroxylase; FLS, flavonol synthase; FNS, flavone synthase; LAR, leucoanthocyanidin reductase; UFGT, UDP flavonoid gulcosyl transferase; OMT, O-methyltransferase; PAL, phenylalanine ammonia lyase; RT, rhamnosyl transferase. The products of each enzymatic reaction are listed below the arrows. Colored circles indicate the presence of delphinidin and cyanidin anthocyanidins, X represents absence of pelargonidin.
The evolutionary basis for the loss of anthocyanin pigments in floral tissue has been investigated by characterizing major floral pigmentation loci using controlled cross segregating populations (Schwinn, 2006; Whittall et al., 2006; Hoballah et al., 2007; Streisfeld and Rausher, 2009; Smith and Rausher, 2011). Evidence suggests that flower color transition is affected by the transcriptional regulation of several anthocyanin structural genes expression. For example, altered activity of specific transcriptional factors accounts for altered patterns of pigmentation in white Petunia axillaris and some Antirrhinum species (Schwinn, 2006; Hoballah et al., 2007). Cis-regulatory changes in the F3’H gene promoter cause down-regulation of F3’H transcription and altered flux in the anthocyanin pathway, resulting in increased production of the red pigment, pelargonidin, instead of blue, in Ipomoea horsfalliae Hook. (Des Marais & Rausher, 2010). Although it has been suggested that mutations in structural genes may incur higher deleterious pleiotropy than those in cis-regulatory elements or transcription factors, the possibility that enzyme coding sequence variation is involved in flower color transition cannot be excluded (Streisfeld and Rausher, 2009).
The Epimedium genus (Berberidaceae), known as “Yinyang Huo” by Chinese druggists, is one of the most popular traditional Chinese medicinal herb genera (Sun et al., 2014; Zhang et al., 2021). A monophyletic group of 50 species of Epimedium is found in western and central China (Huang et al., 2013a; Huang et al., 2013b; Huang et al., 2015; Huang et al., 2016). Epimedium species display a vast range of flower colors; from white and yellow to rose, crimson, and violet (Figure 2). These color pigments are distributed in petaloid sepals or petals or both. In this study, we studied the phenotypic variation of color in Epimedium species distributed in the Hubei province of China. The expression of genes involved in the anthocyanin biosynthetic pathway (ABP) was also analyzed for the association with the different flower color polymorphisms. Our study focused on answering two questions: (1) Has anthocyanin pigment loss, or variation, in different species resulted from the same mechanism? (2) Which candidate genes are involved in anthocyanin pigmentation in E. sagittatum?

Figure 2 Floral phenotypes of accessions of different species within the genus Epimedium. (A–D) are non-pigmentation species (A-); (E–H) are classified as pigmentation species (A+). All photos were taken by W. S. (A) E. sagittatum, (B) E. lishihchenii, (C) E. franchetii, (D) E. wushanense, (E) E. zhushanense, (F) E. epstenii, (G) E. acuminatum, (H) E. leptorrhizum.
Using HPLC, the major pigments from the floral tissues of anthocyanin species (A+) species were found to comprise delphinidin and cyanidin, whereas no detectable anthocyanins were found in the non-anthocyanin species (A-) flowers (Figure 3). To further characterize the mechanism responsible for the non-pigmentation of flowers in A- species, E. sagittatum was used as a model for the enzymatic function.
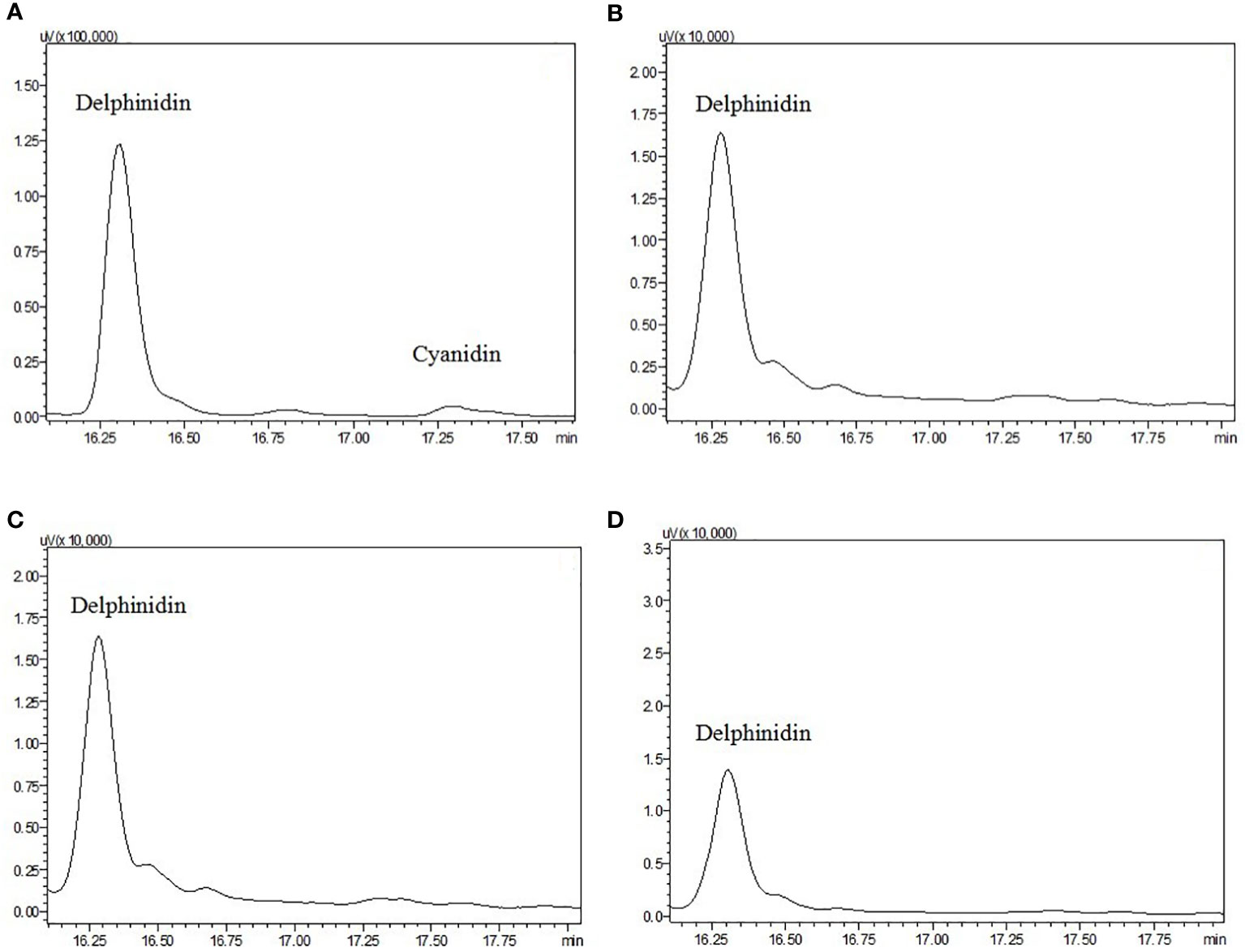
Figure 3 High-performance liquid chromatograms of extracts from petals and sepals of anthocyanidin pigments from (A) E. acuminatum, (B) E. epstenii, (C) E. zhushanense and (D) E. leptorrhizum. Peaks labeled represent the standards of each of the anthocyanin.
To determine whether changes in gene expression might be involved in the non-pigmentation phenotype of A- species, the transcript levels of putative anthocyanin biosynthetic enzymes were examined in petal tissue (Figure 4; Supplementary Figure 1A). Expression of CHS1 not CHS2 and CHS3 was found to be significantly lower in E. wushanese (A-) than in other species. Similarly, down-regulation of CHS2 was observed in E. frachetii (A-), suggesting that loss of anthocyanin may result from low levels of expression of different copies of CHS in E. wushanese and E. franchetii. For CHI and F3H, we found no significant correlation between expression level and the loss of anthocyanins in spur tissues of all A- species. Among the structural genes, ANS was the only ABP locus where all A- species had significantly lower expression levels than that of A+ species. This suggests that the lack of pigmentation production in all A- species could be caused primarily by lower ANS expression. The expression level of DFR was significantly lower in A- species than in A+ species, except for E. lishihchenii. It has been reported that substrate competition between FLS and DFR creates a metabolic flux of the flavonoid biosynthetic pathway in Arabidopsis. In this study, low expression of DFR in the A- species E. franchetii and E. wushanese was correlated with increased accumulation of FLS expression. On the other hand, up-regulation of DFR was positively correlated with FLS expression in E. lishihchenii but E. sagittatum showed no correlation with DFR. In summary, these results suggest that the loss of anthocyanin in E. frachetii, E. wushanese and E. sagittatum may be primarily related to alterations at the ANS locus, affecting gene expression.
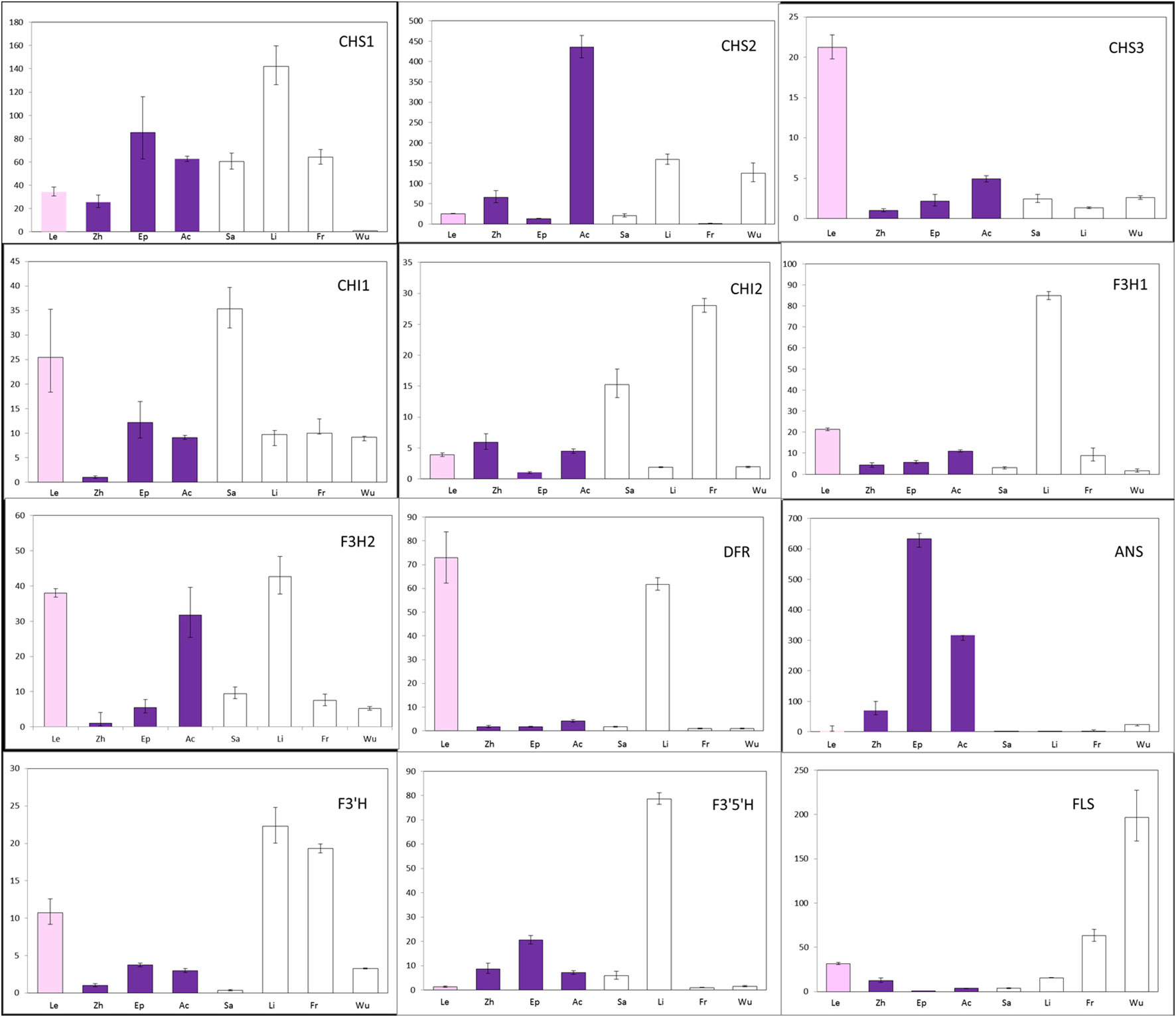
Figure 4 Quantitative expression pattern of ABP structural genes from petal tissue of eight Epimedium species. Colored and empty bars represent A+ and A- species, respectively. Le, Zh, Ep, Ac, Sa, Li, Fr and Wu represent E. leptorrhizum, E. zhushanense, E. epstenii, E.acuminatum, E. sagittatum, E. lishihchenii, E. franchetii and E. wushanense. Data presented here are the mean values of three replicates with error bars indicating SE.
To further analyze the loss of anthocyanin in sepals (Figure 5; Supplementary Figure 1B), gene expression was analyzed across five Epimedium species using the same primers. Expression of DFR was lowest in the three A- species and was correlated with ANS expression, suggesting the expression of DFR and ANS could be regulated by a common transcription factor. CHS1 transcripts were not detected in the sepals of E. wushanese, which also had the lowest CHS1 expression in petals. These observations suggest that negative regulation of the DFR and ANS genes together was also correlated with the lowest CHS1 in sepals and petals in E. wushanese.
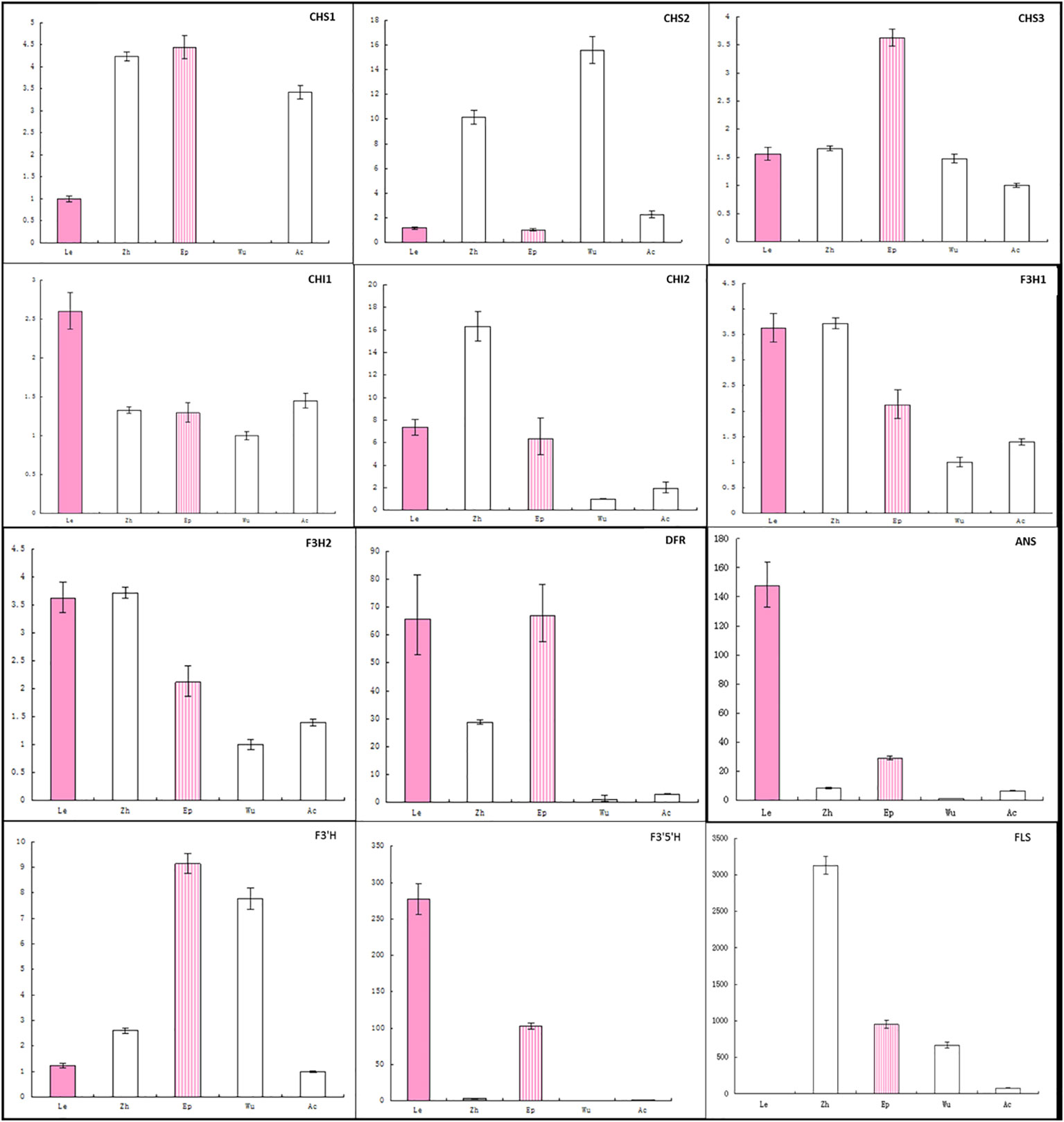
Figure 5 Analysis of expression profiles of anthocyanin genes in petaloid sepals of five Epimedium species using real-time PCR. The cDNA templates are listed as follows: Le, E. leptorrhizum; Zh, E. zhushanense; Ep, E. epstenii; Wu, E. wushanense and Ac, E. acuminatum. Data presented here are the mean values of three replicates with error bars indicating SE.
To study the catalytic activity of E. sagittatum ABP gene products, 35S::EsF3’H and 35S::EsDFR genes were individually transferred into their respective Arabidopsis mutants; transparent testa 7 (tt7) lacking flavonoid 3’-hydroxylase, and transparent testa3 (tt3) lacking dihydroflavonol reductase under the control of the cauliflower mosaic virus 35S promoter (Peer et al., 2001). Transgenic and mutant control seedlings were grown under nitrogen stress to determine if the Epimedium genes could rescue the Arabidopsis anthocyanin-null mutant phenotypes. Accumulation of anthocyanins was observed in transgenic seedlings ectopically expressing EsF3’H and EsDFR (Figure 6). However, the tt7 and tt3 mutant controls did not exhibit anthocyanin accumulation in cotyledons. Thus the E. sagittatum genes showed catalytic activity in Arabidopsis. Given the lack of an Arabidopsis mutant for F3’5’H, in order to determine whether EsF3’5’H can function in vivo, we overexpressed 35S::EsF3’5’H in wild-type Arabidopsis. Under normal conditions on 1/2 MS medium, the seedlings overexpressing EsF3’5’H showed comparable anthocyanin production to wild-type controls (Figure 6).

Figure 6 Phenotypes of the Arabidopsis with overexpression of EsF3’H, EsDFR and F3’5’H(A) Image of anthocyanin in tt7 mutant and rescuing line of EsF3’H in tt7 background, (B) Phenotypes of wild-type, and transgenic Arabidopsis seedling with EsF3’5’H, (C) Phenotypes of tt3 mutant, and transgenic Arabidopsis seedling with EsDFR.
The four Epimedium A- species (E. sagittatum, E. lishihchenii, E. franchetii and E. wushanense) investigated in this study appeared to exhibit anthocyanin loss at the phenotypic level via reduced activity of the anthocyanin branch of the flavonoid pathway. In all species, this appears to involve reduced transcriptional activity of pathway genes, similar to studies in Mimulus aurantiacus (Streisfeld & Rausher, 2009). Interestingly, one A- species (E. lishihchenii) expressed all ABP loci except for ANS at a high level.
While the data linking conserved gene regulation changes to anthocyanin level changes are purely correlative, we found no evidence for the role of coding-region mutations in determining different anthocyanin levels. In A- specie E. sagittatum, the F3’H and DFR enzymes were shown to rescue anthocyanin production in their corresponding Arabidopsis mutants, suggestive of adequate catalytic function (Huang et al., 2012). Accumulation of anthocyanin in 35S::EsDFR in this study and 35S::EsMYBA1 transformed Arabidopsis indicated functionality of the EsDFR and EsMYBA1 coding region (Huang et al., 2013a). Thus, we concluded that the loss of flower color in E. sagittatum (A-) was due to a tissue-specific regulatory change affecting EsDFR and EsANS transcription and not coding-region mutations of EsDFR, EsANS, or EsMYBA1. We also suggested that the changes responsible for the loss of pigmentation in E. franchetii and E. wushanense flowers were shared with E. sagittatum, based on similar correlative gene expression patterns and anthocyanin production in leaves. Functional assays of the putative cis-elements and trans-regulators involved in DFR or ANS transcription were required to determine the precise regulatory mechanisms resulting in reduced ANS and DFR gene expression in A- species.
Downregulation of CHS was a major cause of white flowers in natural populations of Aquilegia flavellata and Parrya nudicaulis (Whittall et al., 2006; Dick et al., 2011). Although we found an association between the A- phenotype and downregulation of CHS1 in yellow-flowered E. wushanese, DFR, and ANS were also downregulated, which may also have contributed to the A- phenotype. Therefore, the A- phenotype in four Epimedium species was also proposed to be due to alteration at the regulatory level, rather than functional mutations in ABP enzymes. The loci regulating anthocyanin in Epimedium were currently being fine-mapped and confirmed by transformation assays.
Eight Epimedium species (E. acuminatum, E. franchetii, E. leptorrhizum, E. epstenii, E. sagittatum, E. lishihchenii, E. wushanense, and E. zhushanense) grown in the specialized Epimedium nurseries of Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China (Figure 2). All plants were transplanted from wild populations and growing under the same environmental conditions. Floral tissues including petaloid sepals and spur-like petals were collected in the spring of 2011. The eight species were separated into two groups corresponding to anthocyanin (A+) (E. acuminatum, E. leptorrhizum, E. epstenii and E. zhushanense) and non-anthocyanin (A-) (E. franchetii, E. sagittatum, E. lishihchenii, E. wushanense) based on visual observation of the floral tissues. The samples were weighed, packaged in aluminum foil, flash-frozen in liquid nitrogen, and then stored at -80°C.
The profiles of anthocyanidins from the samples of A- species and A+ species were analyzed using HPLC. The precursors of anthocyanin pigments were extracted from 100 mg of fresh corolla tissue. For each sample, 20 μL of supernatant was injected into a Shimadzu LC-20 AT liquid chromatograph (Shimadzu Corporation, Japan) and a 250×4.6 mm reverse phase C18 column (Sigma-Aldrich, USA) at a flow rate of 1 ml min-1. The organic solvent was composed of acetonitrile and 0.1% trifluoroacetic acid, and the polar solvent was 0.1% trifluoroacetic acid in HPLC-grade water. The anthocyanin was measured at 550 nm. The chemical compounds cyanidin, delphinidin, malvidin, pelargonidin, peonidin, and petunidin (Poypehenols Laboratories, Norway), were used as anthocyanidin standards.
In total, 12 genes from E. sagittatum involved in the ABP were cloned following RT-PCR amplification using degenerate primers or specific primers based on our previous investigation(Zeng et al., 2010; Huang et al., 2013a; Huang et al., 2013b; Huang et al., 2015). These genes were CHS1, CHS2, CHS3, CHI1, CHI2, F3H1, F3H2, F3’H, F3’5’H, FLS, DFR, ANS. In this study, all pairs of primer from E. sagittatum were transferred to other Epimedium species.
Total RNA was extracted from inner sepals and petals at anthesis, at which time the biosynthesis of anthocyanin is completed. First-strand cDNA was synthesized using PrimeScript RT reagent Kit (Takara, Japan) following the manufacturer’s instructions. In each 20 μL qRT-PCR reaction, 50 ng of cDNA was amplified using SYBR® Premix Ex TaqTM II (Takara, Japan) and 100 mM of primers in an ABI7500 Real-Time PCR machine (ABI, USA) as per the manual. Actin was amplified as the control gene. The samples from three tissues were used and three technical replicates were performed for each sample. Data were analyzed by ABI7500 software. In this study, all pairs of primer (CHS1, CHS2, CHS3, CHI1, CHI2, F3H1, F3H2, F3’H, F3’5’H, FLS, DFR, and ANS) from E. sagittatum were transferred in other Epimedium species. All primers used in this manuscript are listed in the supplementary database.
For functional analyses, the E. sagittatum (A-) genes EsF3’H and EsDFR were overexpressed in their respective Arabidopsis thaliana (ecotype Landsberg) mutants, each lacking anthocyanins at the seedling stage. EsF3’5’H, The coding regions of EsF3’H, EsF3’5’H, and EsDFR were cloned into pMD19-T (Takara, Japan). The SalI and SacI digested fragment of each gene was purified and ligated into the pMV plasmid (derived from pBI121) behind the cauliflower 35S promoter. The plasmids were then transformed into Agrobacterium strain EHA105. Arabidopsis wild-type and mutants (tt3 and tt7) were transformed by the floral dip infiltration method(Zhang et al., 2006). Transformants were selected on 1/2 Murashige and Skoog medium supplemented with 50 μg/mL kanamycin. Resistant seedlings were then transferred into the soil to harvest seeds. T1 seedlings were screened on 1/2 MS medium minus nitrogen for observation of anthocyanin accumulation.
Publicly available datasets were analyzed in this study. This data can be found here: Reference.
WS, HH, and YW conceived and designed the experiments. WS and YM performed the experiments.WS, WH, ZY, XM, and HW analyzed the data. WS wrote the paper. XM and HW revised the paper. YZ provided Figure 2 and collected species. RH and SC supervised this investigation. All authors contributed to the article and approved the submitted version.
The authors are grateful for finical support from the Scientific and Technological Innovation Project of the China Academy of Chinese Medical Sciences (CI2021A04806 and CI2021A04008) and National Key Research and Development Program of China (2022YFC3501703).
We thank Ms. Xiaoping Pan for assistance with qRT-PCR analysis, Dr. Sen Lin and Guoxiang Dong for assistance with HPLC analysis, Ms. Xiaomin Hu for assistance with screening Arabidopsis.
Author RH was employed by company By-Health Institute of Nutrition and health. By-health Co., Ltd.
The reviewer HP declared a shared affiliation with the authors YM,SC, and WS to the handling editor at the time of review China Academy of Chinese Medical Sciences, Beijing, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1133616/full#supplementary-material
Boss, P. K., Davies, C., Robinson, S. P. (1996). Analysis of the expression of anthocyanin pathway genes. Plant Physiol. 111, 1059–1066. doi: 10.1104/pp.111.4.1059
Buer, C. S., Imin, N., Djordjevic, M. A. (2010). Flavonoids: New roles for old molecules. J. Integr. Plant Biol. 52, 98–111. doi: 10.1111/j.1744-7909.2010.00905.x
Des Marais, D. L., Rausher, M. D. (2010). Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evol. (N. Y). 64, 2044–2054. doi: 10.1111/j.1558-5646.2010.00972.x
Dick, C. A., Buenrostro, J., Butler, T., Carlson, M. L., Kliebenstein, D. J., Whittall, J. B. (2011). Arctic Mustard flower color polymorphism controlled by petal-specific downregulation at the threshold of the anthocyanin biosynthetic pathway. PLoS One 6. doi: 10.1371/journal.pone.0018230
Fenster, C. B., Armbruster, W. S., Wilson, P., Dudash, M. R., Thomson, J. D. (2004). Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 35, 375–403. doi: 10.1146/annurev.ecolsys.34.011802.132347
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Hoballah, M. E., Gubitz, T., Stuurman, J., Broger, L., Barone, M., Mandel, T., et al. (2007). Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19, 779–790. doi: 10.1105/tpc.106.048694
Holton, T. A., Cornish, E. C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071. doi: 10.2307/3870058
Huang, W., Khaldun, A. B. M., Lv, H., Du, L., Zhang, C., Wang, Y. (2016). Isolation and functional characterization of a R2R3-MYB regulator of the anthocyanin biosynthetic pathway from Epimedium sagittatum. Plant Cell Rep. 35, 883–894. doi: 10.1007/s00299-015-1929-z
Huang, W., Sun, W., Lv, H., Luo, M., Zeng, S., Pattanaik, S., et al. (2013a). A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway. PLoS One 8. doi: 10.1371/journal.pone.0070778
Huang, W., Sun, W., Lv, H., Xiao, G., Zeng, S., Wang, Y. (2013b). Isolation and molecular characterization of thirteen R2R3-MYB transcription factors from Epimedium sagittatum. Int. J. Mol. Sci. 14, 594–610. doi: 10.3390/ijms14010594
Huang, W., Sun, W., Wang, Y. (2012). Isolation and molecular characterisation of flavonoid 3’-hydroxylase and flavonoid 3’, 5’-hydroxylase genes from a traditional Chinese medicinal plant, Epimedium sagittatum. Gene 497, 125–130. doi: 10.1016/j.gene.2011.11.029
Huang, W., Zeng, S., Xiao, G., Wei, G., Liao, S., Chen, J., et al. (2015). Elucidating the biosynthetic and regulatory mechanisms of flavonoid-derived bioactive components in Epimedium sagittatum. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00689
Lepiniec, L., Debeaujon, I., Routaboul, J.-M., Baudry, A., Pourcel, L., Nesi, N., et al. (2006). Genetics and biochemistry of deed flavonoids. Annu. Rev. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252
Peer, W. A., Brown, D. E., Tague, B. W., Muday, G. K., Taiz, L., Murphy, A. S. (2001). Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 126, 536–548. doi: 10.1104/pp.126.2.536
Pollastri, S., Tattini, M. (2011). Flavonols: Old compounds for old roles. Ann. Bot. 108, 1225–1233. doi: 10.1093/aob/mcr234
Schwinn, K. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18, 831–851. doi: 10.1105/tpc.105.039255
Smith, S. D., Rausher, M. D. (2011). Gene loss and parallel evolution contribute to species difference in flower color. Mol. Biol. Evol. 28, 2799–2810. doi: 10.1093/molbev/msr109
Streisfeld, M. A., Rausher, M. D. (2009). Genetic changes contributing to the parallel evolution of red floral pigmentation among Ipomoea species. New Phytol. 183, 751–763. doi: 10.1111/j.1469-8137.2009.02929.x
Sun, W., Huang, W., Li, Z., Song, C., Liu, D., Liu, Y., et al. (2014). Functional and evolutionary analysis of the AP1/SEP/AGL6 superclade of MADS-box genes in the basal eudicot Epimedium sagittatum. Ann. Bot. 113, 653–668. doi: 10.1093/aob/mct301
Whittall, J. B., Voelckel, C., Kliebenstein, D. J., Hodges, S. A. (2006). Convergence, constraint and the role of gene expression during adaptive radiation: Floral anthocyanins in Aquilegia. Mol. Ecol. 15, 4645–4657. doi: 10.1111/j.1365-294X.2006.03114.x
Zeng, S., Xiao, G., Guo, J., Fei, Z., Xu, Y., Roe, B. A., et al. (2010). Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. et zucc.) maxim. BMC Genomics 11. doi: 10.1186/1471-2164-11-94
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Keywords: gene expression, Epimedium, anthocyanin, spur, sepal
Citation: Mi Y, He R, Wan H, Meng X, Liu D, Huang W, Zhang Y, Yousaf Z, Huang H, Chen S, Wang Y and Sun W (2023) Genetic and molecular analysis of the anthocyanin pigmentation pathway in Epimedium. Front. Plant Sci. 14:1133616. doi: 10.3389/fpls.2023.1133616
Received: 29 December 2022; Accepted: 07 March 2023;
Published: 27 March 2023.
Edited by:
Mark Chapman, University of Southampton, United KingdomReviewed by:
Fangyuan Zhang, Southwest University, ChinaCopyright © 2023 Mi, He, Wan, Meng, Liu, Huang, Zhang, Yousaf, Huang, Chen, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Wang, eWluZ3dhbmdAc2NpYi5hYy5jbg==; Wei Sun, d3N1bkBpY21tLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.